Abstract
The fusion gene CBFB-MYH11 is generated by the chromosome 16 inversion associated with acute myeloid leukemias. This gene encodes a chimeric protein involving the core binding factor β (CBFβ) and the smooth-muscle myosin heavy chain (SMMHC). Mouse model studies suggest that this chimeric protein CBFβ-SMMHC dominantly suppresses the function of CBF, a heterodimeric transcription factor composed of DNA binding subunits (CBFα1 to 3) and a non-DNA binding subunit (CBFβ). This dominant suppression results in the blockage of hematopoiesis in mice and presumably contributes to leukemogenesis. We used transient-transfection assays, in combination with immunofluorescence and green fluorescent protein-tagged proteins, to monitor subcellular localization of CBFβ-SMMHC, CBFβ, and CBFα2 (also known as AML1 or PEBP2αB). When expressed individually, CBFα2 was located in the nuclei of transfected cells, whereas CBFβ was distributed throughout the cell. On the other hand, CBFβ-SMMHC formed filament-like structures that colocalized with actin filaments. Upon cotransfection, CBFα2 was able to drive localization of CBFβ into the nucleus in a dose-dependent manner. In contrast, CBFα2 colocalized with CBFβ-SMMHC along the filaments instead of localizing to the nucleus. Deletion of the CBFα-interacting domain within CBFβ-SMMHC abolished this CBFα2 sequestration, whereas truncation of the C-terminal-end SMMHC domain led to nuclear localization of CBFβ-SMMHC when coexpressed with CBFα2. CBFα2 sequestration by CBFβ-SMMHC was further confirmed in vivo in a knock-in mouse model. These observations suggest that CBFβ-SMMHC plays a dominant negative role by sequestering CBFα2 into cytoskeletal filaments and aggregates, thereby disrupting CBFα2-mediated regulation of gene expression.
The pericentric inversion of chromosome 16 [inv(16)(p13q22)] is a cytogenetic abnormality consistently associated with acute myeloid leukemia (AML) subtype M4Eo (2, 21), a variant of subtype M4 with abnormal eosinophils in the bone marrow and sometimes in the peripheral blood. The inversion results in the reciprocal fusions of two genes: the MYH11 gene (16p13), which encodes the smooth-muscle myosin heavy chain (SMMHC), and the CBFB gene (16q22), which encodes the β subunit of the core binding factor (CBFβ) (24). The chimeric gene CBFB-MYH11 fuses most of the 5′ coding region of CBFB in frame with the 3′ portion of MYH11, resulting in the production of the chimeric protein CBFβ-SMMHC. The reciprocal fusion, MYH11-CBFB, is not believed to be important since its expression is below detectable levels in leukemic cells and it is deleted in some patients with an unbalanced inversion (24, 25, 29).
CBFβ is the heterodimeric partner of CBFα proteins, and together they constitute the core binding factors (CBF). CBF was initially identified as a transcriptional regulator of Moloney murine leukemia virus (50, 51) and polyomavirus (4, 18, 36, 37, 43) in mice, and it was subsequently shown to be an important transcriptional activator of genes involved in mammalian hematopoiesis and bone development (5, 12, 14, 15, 20, 32, 40, 46). Monomeric CBFα proteins bind DNA, albeit weakly (3, 50). Although CBFβ does not make any detectable direct contact with DNA (50), it enhances the DNA binding affinity of the CBFα proteins (4, 36). While CBFβ is expressed from a single gene in the human and mouse, there are three CBFα genes, all of which encode the so-called runt domain (3, 22, 54), which is required for both DNA binding and interaction with CBFβ. One of the three genes, CBFA2, also known as AML1 or PEBP2αB, is located on human chromosome 21 and is involved in several different leukemias as a result of translocations (16, 31, 34, 35, 41). Chromosomal inversions and translocations involving either CBFB or CBFA2 are the most frequent cytogenetic abnormalities in human AMLs (27).
Gene-targeting experiments in mice have demonstrated that the CBFα2 and CBFβ subunits are likely to function together as a complex in vivo. Homozygous disruption of either Cbfa2 (39, 48) or Cbfb (42, 49) in mice produces an identical phenotype: both Cbfa2−/− and Cbfb−/− embryos demonstrate a failure of definitive hematopoiesis in the liver, and in both cases the embryos die at around day 12.5 due to extensive hemorrhages.
The inv(16) chimeric gene CBFB-MYH11 has been shown to exert a dominant negative effect in vivo by a mouse knock-in experiment (8). CBFB-MYH11 was introduced into the mouse genome to replace one copy of the Cbfb gene. The expression of this chimeric gene was controlled by the endogenous Cbfb promoter, thus simulating the condition in leukemic patients. CBFβ-SMMHC was found to dominantly suppress the function of the CBFα2:CBFβ heterodimer, since mouse embryos heterozygous for the knock-in Cbfb-MYH11 gene (CbfbCBFB-MYH11/+) displayed a phenotype similar to that of Cbfb−/− and Cbfa2−/− embryos, i.e., failure of definitive hematopoiesis and midgestation lethality. In vitro, the chimeric protein was shown to retain its ability to interact with CBFα proteins and participate in the formation of protein-DNA complexes (23). Although presence of the chimeric protein reduces CBF DNA-binding activity in cultured Ba/F3 lymphoid and 32D c13 myeloid cells (6), it is not clear how this reduction was achieved. Unlike wild-type CBFβ, CBFβ-SMMHC can potentially form dimers and multimers via the rod-like domain of the myosin chain (23, 25).
Two possible mechanisms could explain the dominant negative effect of the chimeric CBFβ-SMMHC protein. One is that CBFβ-SMMHC, via heterodimerization with CBFα2, can assemble into a ternary complex at the core sites within promoters of target genes and interfere with the regulation of gene expression. The second possibility is that CBFβ-SMMHC, with its capacity to form multimers, can sequester CBFα2 into nonfunctional complexes, thus preventing it from regulating transcription of target genes. Previous studies by our group demonstrated that CBFβ-SMMHC can form rod-like nuclear structures as well as cytoplasmic stress fibers in NIH 3T3 cells stably transfected with a CBFB-MYH11 cDNA construct (53). However, the effect on CBFα2 subcellular localization by CBFβ-SMMHC has not been fully examined. In this study, we used transient-transfection assays in combination with immunofluorescence and green fluorescent protein (GFP) tags to demonstrate that CBFβ-SMMHC does, in fact, sequester CBFα2 in abnormal locations. We also demonstrated that the sequestration requires the abilities of CBFβ-SMMHC to interact with CBFα2 and to multimerize. This observed sequestration can at least partially explain the dominant negative effect of the CBFβ-SMMHC protein on CBF function in leukemogenesis.
MATERIALS AND METHODS
Construction of plasmids.
pEGFP-C2, the vector expressing a modified GFP (EGFP) under the control of the cytomegalovirus (CMV) promoter, was obtained from Clontech. This vector allows for in-frame fusion of target genes at the COOH-terminal end of EGFP. Plasmids pCBFB, pCBFB-MYH11, and pCbfa2, each containing the entire coding region of the respective genes under the control of CMV promoter, have been described previously (23, 26, 45). pCBFB carries the CBFβ187 isoform, pCBFB-MYH11 carries the CBFβ-SMMHC204 isoform (26), and pCbfa2 carries the mouse CBFα2451 isoform. EGFP fusion constructs pEGFP-CBFB, pEGFP-CBFB-MYH11, and pEGFP-Cbfa2 were generated following standard subcloning procedures by using these reported full-length cDNA plasmids.
The deletion construct pCBFB-MYH11ΔN2-11 was generated by PCR-directed mutagenesis. A 436-bp fragment with a 30-bp deletion immediately 3′ to the initiating ATG was amplified with forward primer 5′-ATATGAATTCGGGAAGATGTTCGAGAACGAGGAG-3′ and reverse primer 5′-CAGTAAGCTTACCTCCATTTCCTCCCG-3′ and with pCBFB-MYH11 as the template. This PCR fragment was digested with EcoRI (the recognition site is within the primer) and StuI (the recognition site is within the CBFB-encoding region) and used to replace the EcoRI-StuI fragment of pCBFB-MYH11 to make CBFB-MYH11ΔN2-11, which was subcloned into pBluescript-KS (Stratagene) and pEGFP-C2 by standard methods.
pCBFB-MYH11ΔC390 was generated by digesting pCBFB-MYH11 with SmaI and EcoRI to release the small fragment containing a partial cDNA and ligating with the pCMV vector. pCBFB-MYH11ΔC303 was generated by digesting pCBFB-MYH11 with BglII, filling in the ends with the Klenow subunit of DNA polymerase, and then digesting with EcoRI to release the partial cDNA, which was then ligated to the pCMV vector. pCBFB-MYH11ΔC95 was generated by digesting pCBFB-MYH11 with BsrDI, filling in the ends with the Klenow subunit of DNA polymerase, and digesting with EcoRI to release the partial cDNA, which was then ligated to the pCMV vector.
pEGFP-CBFB-MYH11 was digested with SmaI and BamHI to release the 3′ end of CBFB-MYH11, and then the large fragment was religated to generate pEGFP-CBFB-MYH11ΔC390. pEGFP-CBFB-MYH11ΔC303 was constructed by removing the BglII-BamHI fragment of pEGFP-CBFB-MYH11. pEGFP-CBFB-MYH11ΔC95 was generated by digesting pCBFB-MYH11 with BsrDI, filling in the ends with the Klenow subunit of DNA polymerase, digesting again with EcoRI, and ligating the insert into pEGFP-C2 digested with EcoRI and SmaI.
All the constructs generated were confirmed by restriction digests, and all those involving fusions at their 5′ ends were also confirmed by sequencing across the fusion junctions. In vitro transcription-translation experiments and Western analysis of transiently transfected cells (see below) confirmed the production of proteins of expected sizes.
Antibodies.
α3043, a rabbit polyclonal antibody specific for CBFα2, and β141.2, a mouse monoclonal antibody against CBFβ, have been described previously (5, 49). Texas-red-labeled phalloidin was obtained from Molecular Probes for actin labeling. Fluorescein-labeled affinity purified secondary antibodies against mouse immunoglobulin G (IgG) and rabbit IgG were obtained from Kirkegaard & Perry Laboratories Inc. Texas-red-labeled affinity purified secondary antibodies against mouse IgG and rabbit IgG were obtained from Vector Laboratories. The primary antibodies were diluted in Triton buffer (phosphate-buffered saline [PBS], 0.3% Triton X-100, 2% bovine serum albumin [BSA]) at working dilutions of 1:10, and the secondary antibodies were diluted in the same buffer at the recommended dilutions.
Cell culture and DNA transfection.
NIH 3T3 cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% (vol/vol) heat-inactivated newborn calf serum. DNA transfections were performed with lipofectamine (GIBCO-BRL) according to the manufacturer’s instructions. Fifty thousand cells were plated onto a 15-mm-diameter circular glass coverslip in a 12-well tissue culture dish for 12 h at 37°C before transfection. A maximum of 2.5 μg of DNA was used for each transfection. Twenty percent serum was added to cells at 5 h posttransfection, and the cells were cultured for 12 h at 37°C. The culture medium was replaced with complete medium at this time, and cells were collected for fixation or immunoprecipitation at times ranging from 12 to 48 h after addition of fresh media.
Cell fractionation.
NIH 3T3 cells were plated on a 10-cm-diameter dish at 106 cells per plate and transfected the next day with various expression constructs. Cells were harvested 24 h posttransfection, and cell pellets were resuspended in sucrose/Nonidet P-40 (NP-40) buffer containing 0.32 M sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 0.1 mM EDTA, 10 mM Tris-HCl (pH 8.0), 1 mM dithiothreitol, and 0.5% (vol/vol) NP-40 along with protease inhibitors (13). The lysates were microcentrifuged at 500 × g for 5 min at 4°C, and the supernatant containing the cytoplasm was transferred to a fresh tube. The nuclei were washed with 1 ml of sucrose buffer without NP-40, and then lysed in sodium dodecyl sulfate (SDS) sample buffer by boiling. The protein concentration in each sample was visually quantified on Coomassie-blue-stained SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and equivalent amounts of protein were then separated on an SDS–10% PAGE gel and visualized by Western blotting, as described below. GFP-specific polyclonal antibody from Clontech was used at recommended dilutions to visualize EGFP-CBFα2 and EGFP-CBFβ in the presence and absence of CBFα2. Anti-cytosolic phospholipase A2 (cPLA2) specific monoclonal antibody (Santa Cruz) and anti-retinoblastoma (Rb) specific monoclonal antibody (Pharmingen) were also used at recommended dilutions to probe the blots for ascertaining the purity of the cytoplasmic fraction and nuclear fraction, respectively.
Immunofluorescence.
Coverslips with transfected cells were fixed in two different ways to avoid fixation artifacts. Cells were fixed either in cold (−20°C) methanol for 7 min or in 4% formaldehyde for 30 min. In the latter case, the fixed cells were then permeabilized with 0.3% Triton buffer for 30 min. For experiments involving transfection with GFP fusion constructs, cells were fixed only in formaldehyde followed by permeabilization as described above, since methanol has been shown to inhibit GFP fluorescent activation. Detection of fluorescent signals was performed according to a modification of the procedure described by Cullen (11). Fixed cells were blocked overnight with a solution containing PBS, 10% goat or horse serum (same as that of secondary antibody), 2% milk, and 3% BSA. Transfected cells were incubated with primary antibodies diluted in Tween buffer (PBS, 2% BSA, 0.5% Tween 20) for 2 h at room temperature. Cells were then washed once with PBS for 5 min and twice with Triton buffer (PBS, 2% BSA, 0.3% Triton X-100) for 10 min each. The fluorescence-labeled secondary antibody was diluted in Triton buffer and added to transfected cells for 1 h at room temperature. Cells were then washed once with PBS for 5 min, twice with Triton buffer for 10 min each time, and once with PBS for 10 min before mounting with 4′,6-diamidino-2-phenylindole (DAPI)-antifade on a glass slide for microscopy. Experiments were repeated three or more times.
Immunoprecipitation.
pCBFB, pCBFB-MYH11, and related deletion constructs (pCBFB-MYH11ΔN2-11, pCBFB-MYH11ΔC390, pCBFB-MYH11ΔC303, pCBFB-MYH11ΔC95) were translated in vitro with 35S-labeled methionine by using the Promega reticulocyte lysate translation kit. pCbfa2 was translated in vitro with or without radioactive methionine. Protein A-Sepharose-plus (Pierce) was washed with PBS, then blocked with 2% BSA in PBS. Thirty microliters of the blocked protein A was incubated with antibody α3043 for 2 h at 4°C on a rotator and then washed twice with Triton buffer. The protein A beads were then incubated overnight at 4°C with 35S-labeled CBFβ, CBFβ-SMMHC, and related deletion proteins, in the presence or absence of cold in vitro-translated CBFα2. The Sepharose beads were then collected by centrifugation and washed with Triton buffer four times to remove any residual buffer. The beads were then resuspended and boiled for 5 min in 25 μl of 2× SDS buffer. Ten microliters of this buffer containing the eluted proteins was then analyzed by denaturing PAGE and autoradiography.
Chemical cross-linking.
Proteins were translated in vitro in the presence of 35S-labeled methionine by using the Promega reticulocyte lysate kit. They were then diluted fivefold with PBS and incubated at room temperature for 1 h with glutaraldehyde at a final concentration of 0.0025%. The reactions were stopped by addition of Tris (pH 8) and glycine at final concentrations of 25 and 192 mM, respectively, and analyzed by denaturing PAGE and autoradiography.
Western blot analysis.
Whole-cell extracts in SDS gel loading buffer were prepared from NIH 3T3 cells (106 cells per dish) transfected with the appropriate expression plasmids, separated by SDS–12% PAGE, and transferred to a nitrocellulose membrane. The membrane was blocked with Tris-buffered saline (TBS) buffer (135 mM NaCl, 20 mM Tris [pH 7.5], 5% milk, and 0.1% Tween 20), incubated for 2 h on a shaker at room temperature with an appropriate primary antibody, washed with TBS buffer, and then incubated with a peroxidase-labeled secondary antibody (KPL Inc.) at the recommended dilution in TBS for 1 h at room temperature. Proteins were visualized by developing chemiluminescent signal with reagents from Pierce and exposure to X-ray films.
Analysis of mouse embryonic fibroblasts.
Generation of mice heterozygous for the Cbfa2lz allele will be described elsewhere (33). Briefly, exons 7 and 8 of the Cbfa2 gene were replaced with lacZ-bearing sequences in such a way that a CBFα2-βgal fusion protein was synthesized from the modified Cbfa2 allele. Female mice heterozygous for the Cbfa2lz allele (Cbfa2lz/+) were mated to a male chimeric mouse generated from mouse embryonic stem cells heterozygous for a knock-in inv(16) allele (CbfbCBFB-MYH11/+) (8). Embryos from this cross were isolated at 11.5 days postcoitus (dpc), the yolk sacs and anterior portions (heads) of the embryos were removed, and the rest of each embryo was used to generate mouse embryonic fibroblasts. Embryos were suspended in 5 ml of HEPES-buffered saline and passed through an 18-gauge needle, and the cells were collected by centrifugation, resuspended in 5 ml of Dulbecco’s modified Eagle medium–10% fetal calf serum, and then passed through a 21-gauge needle. Disaggregated cells from individual embryos were plated in one well (35-mm-diameter) of a six-well tissue culture plate and cultured at 37°C. The mouse embryonic fibroblasts were passed one additional time in duplicate plates, and the cells from one plate were processed for β-galactosidase activity following standard protocols. Fibroblasts with both Cbfa2lz/+ and CbfbCBFB-MYH11/+ alleles were further stained with DAPI for nuclear visualization by standard procedures. Inheritance of CbfbCBFB-MYH11 allele was demonstrated by the presence of the CBFβ-SMMHC fusion protein, assayed by Western blot analysis of the mouse embryonic fibroblasts, as described previously (8).
All figures were made with Adobe Photoshop version 4.0 for Macintosh.
RESULTS
Coimmunoprecipitation of CBFβ and of CBFβ-SMMHC and its deletions with CBFα2.
The ability of the CBFβ-SMMHC deletion proteins to interact with CBFα2 was characterized by immunoprecipitation of in vitro-translated proteins. Fig. 1A shows the in vitro-translated 35S-labeled proteins used in the reactions. To assess binding to CBFα2, a polyclonal antibody that recognizes the CBFα2 runt domain (α3043) was first bound to protein A-Sepharose beads. The matrix-bound antibody was then incubated with or without nonradioactive CBFα2 (in vitro-translated from pCbfa2) in conjunction with radioactively labeled CBFβ, CBFβ-SMMHC, CBFβ-SMMHCΔC390, CBFβ-SMMHCΔC303, CBFβ-SMMHCΔC95, and CBFβ-SMMHCΔN2-11 (in vitro-translated from pCBFB, pCBFB-MYH11, pCBFB-MYH11ΔC390, pCBFB-MYH11ΔC303, pCBFB-MYH11ΔC95, and pCBFB-MYH11ΔN2-11, respectively). As seen in Fig. 1B, lanes 2 and 4, CBFβ and full-length CBFβ-SMMHC were immunoprecipitated with CBFα2, as expected. The protein with the N-terminal deletion CBFβ-SMMHCΔN2-11 failed to be immunoprecipitated with CBFα2 (Fig. 1B, lane 12). Those with the C-terminal deletions of CBFβ-SMMHC, which retain the N-terminal end, interacted with CBFα2 and were immunoprecipitated (Fig. 1B, lanes 6, 8, and 10). The 35S-labeled CBFα2 was immunoprecipitated in the presence of the antibody (lane 14) but not in its absence (lane 13), indicating the specificity of the interaction.
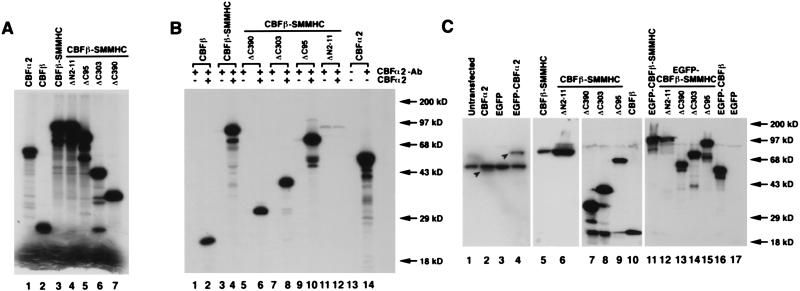
FIG. 1.
Characterization of CBFα2, CBFβ, CBFβ-SMMHC, and the deletion mutants. (A) Generation of protein products by in vitro translation. [35S]methionine-labeled proteins were produced by in vitro translation, separated by gel electrophoresis, and detected by autoradiography. The intense bands at the bottom of the gel are the free [35S]methionine. (B) Interactions between CBFα2 and the deletion mutants. The protein products shown in panel A were immunoprecipitated with polyclonal antibody α3043, in the presence or absence of cold in vitro-translated CBFα2. (C) Expression of cDNA constructs after transient transfection into NIH 3T3 cells. The Western blot with extracts derived from transiently transfected NIH 3T3 cells was probed with α3043 to detect CBFα2 and EGFP-CBFα2 (lanes 1 to 4). The relevant proteins are shown by arrowheads in lanes 2 and 4. Similarly, the antibody β141.2 was used to identify CBFβ, CBFβ-SMMHC, the deletion proteins, and their GFP fusions by Western blotting of cellular extracts (lanes 5 to 17). Protein molecular mass markers are shown on the right sides of panels B and C.
Protein expression from plasmid constructs after transient transfection.
The expression of CBFα2, CBFβ, CBFβ-SMMHC, and the various deletion proteins after transient transfection of NIH 3T3 cells was tested by Western blotting. The polyclonal antibody against CBFα2, α3043, was used to probe whole-cell extracts from untransfected NIH 3T3 cells and from NIH 3T3 cells transfected with pCbfa2, pEGFP-C2 (empty vector), and pEGFP-Cbfa2 (Fig. 1C, lanes 1 to 4). Proteins of expected sizes (49 kDa for CBFα2 and 76 kDa for EGFP-CBFα2) were detected in cells transfected with pCbfa2 and pEGFP-Cbfa2 (Fig. 1C, lanes 2 and 4, respectively). The ∼50 kDa band present in all the samples (Fig. 1C, lanes 1 to 4) most likely results from nonspecific binding of the antibody to an endogenous protein. Cell extracts from NIH 3T3 cells transfected with pCBFB-MYH11 and the deletion constructs with or without EGFP fusion were probed with β141.2, a monoclonal antibody against CBFβ (Fig. 1C, lanes 5 to 17 for plasmids pCBFB-MYH11, pCBFB-MYH11ΔN2-11, pCBFB-MYH11ΔC390, pCBFB-MYH11ΔC303, pCBFB-MYH11ΔC95, pCBFB, pEGFP-CBFB-MYH11, pEGFP-CBFB-MYH11ΔN2-11, pEGFP-CBFB-MYH11ΔC390, pEGFP-CBFB-MYH11ΔC303, pEGFP-CBFB-MYH11ΔC95, pEGFP-CBFB, and pEGFP-C2, respectively). Again, proteins of expected sizes were visible in all the transfected cell samples. Protein bands corresponding to the endogenous CBFβ were detected in certain samples (Fig. 1C, lanes 7 to 9), although the variability suggests that these could be derived from degradation of the higher-molecular-weight proteins.
Subcellular localization of CBFα2, CBFβ, and CBFβ-SMMHC after transient transfection.
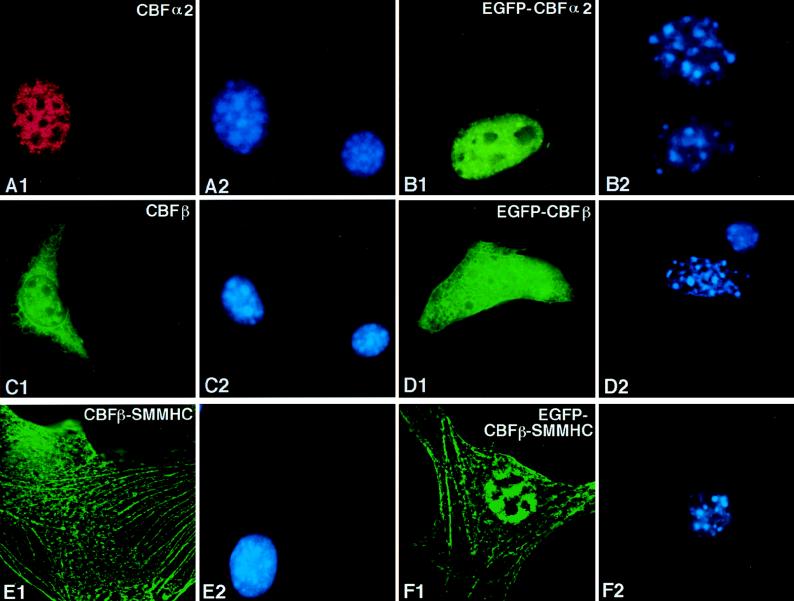
Plasmids containing full-length coding sequences (pCbfa2, pCBFB, and pCBFB-MYH11) were transiently transfected into NIH 3T3 cells to examine the subcellular localization of CBFα2, CBFβ, and CBFβ-SMMHC. Expression and localization of these proteins were detected by primary antibodies α3043 and β141.2, followed by immunofluorescence labeling with fluorescein and Texas-red-conjugated secondary antibodies. The levels of endogenous CBFβ and CBFα2 proteins, though detectable by gel shifts (unpublished data), were below the limits of detection by immunofluorescence. Transient transfection of GFP fusion constructs (pEGFP-Cbfa2, pEGFP-CBFB, and pEGFP-CBFB-MYH11) was used as an independent method of assessing subcellular locations, and the results were in agreement with those of immunofluorescence staining.
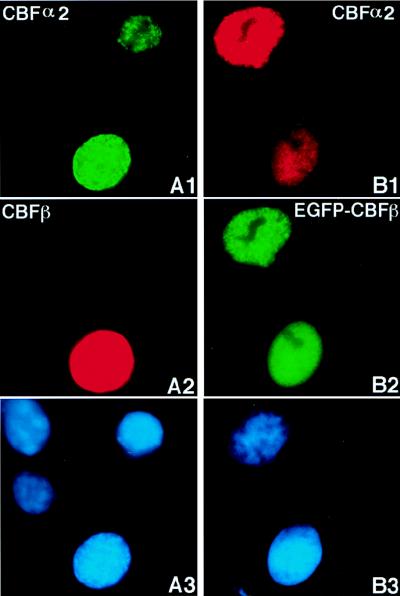
Transiently expressed CBFα2 (from plasmid pCbfa2) was localized completely in the nuclei of transfected NIH 3T3 cells (Fig. 2A1). The fluorescent staining was distributed throughout the nucleus but excluded from nucleoli-like structures (compare Fig. 2A1 and A2). This nuclear localization of CBFα2 was confirmed by pEGFP-Cbfa2 transfection (Fig. 2B1) and was similar to published results for CBFα1 (28). Transiently expressed wild-type CBFβ (from plasmid CBFB) was found evenly distributed in both the cytoplasm and nuclei of transfected cells (Fig. 2C1). This localization pattern was confirmed with EGFP-CBFβ (Fig. 2D1) and was similar to the pattern of EGFP alone (1).
CBFβ-SMMHC (from plasmid pCBFB-MYH11), on the other hand, was localized along stress-fiber-like filaments and aggregates (Fig. 2E1). Observations from pEGFP-CBFB-MYH11 transfection were consistent with the immunofluorescence results (Fig. 2F1). Some of the CBFβ-SMMHC protein was localized in the nuclei in a diffuse pattern in certain transfected cells (Fig. 2F1 and Fig. 4C1), though the majority of the protein showed localization along the filaments and aggregates. The subcellular localization of CBFβ-SMMHC could be classified into three categories based on the level of expression: cells which had a low level of expression showed localization along the filaments (Fig. 2E1 and Fig. 4A1); a moderate level of expression resulted in accumulation of the protein along the filaments, leading to a speckled pattern (Fig. 4B1); and a higher expression level of CBFβ-SMMHC showed aggregation, and the protein seemed to accumulate in higher-order structures (Fig. 4C1).
Full-length CBFα2 can bring CBFβ into the nucleus.
pCBFB and pCbfa2 were cotransfected into NIH 3T3 cells and immunofluorescence labeling was carried out as described above. Two of the four cells shown in Fig. 3A expressed the transiently transfected pCbfa2, as shown by the green signal in Fig. 3A1, while one of these two cells also expressed the transiently transfected pCBFB as shown by the red signal in Fig. 3A2. As shown in Fig. 3A1, CBFα2 was localized completely in the nucleus, similar to the pattern observed when it was expressed alone. Intriguingly, CBFβ also was localized to the nucleus, and it was distributed in a pattern similar to that of CBFα2 (Fig. 3A2). This suggests that full-length CBFα2 is able to bring CBFβ into the nucleus. This nuclear localization of CBFβ by CBFα2 was dose dependent. When equal molar amounts of both plasmid DNAs were used for transfection, as in the cells shown in Fig. 3, approximately 88% of the cells expressing both CBFβ and CBFα2 showed nuclear localization of CBFβ. Increasing the amount of pCbfa2 DNA relative to that of pCBFB increased the proportion of cells in which CBFβ localized to the nucleus. When the amount of pCbfa2 DNA was fivefold molar excess relative to that of pCBFB DNA, in approximately 93% of the cotransfected cells CBFβ and CBFα2 colocalized in the nucleus. Increasing the pCbfa2 DNA amount to ninefold molar excess relative to that of pCBFB DNA resulted in almost 100% of the doubly transfected cells having CBFβ in nuclei along with CBFα2. Cotransfections with pEGFP-CBFB and pCbfa2 gave comparable results (Fig. 3B1, B2, and B3).
FIG. 3.
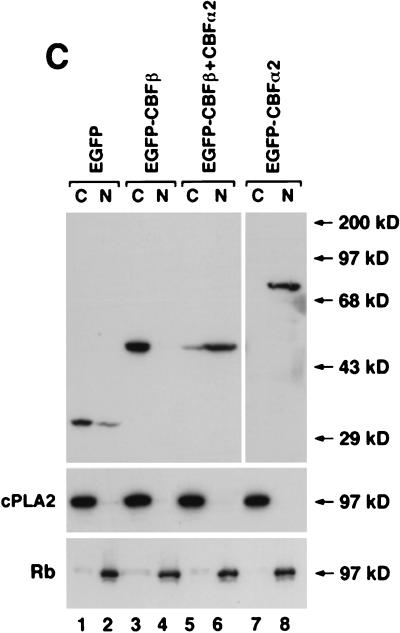
CBFα2 brings CBFβ into the nucleus when coexpressed. Cells were cotransfected with pCbfa2 and pCBFB (A1, A2, and A3) or with pCbfa2 and pEGFP-CBFB (B1, B2, and B3). Panels A1, A2, and A3 represent images of the same field seen through different filters, as do panels B1, B2, and B3. CBFα2 was detected with antibody α3043 and a fluorescein-labeled (A1) or a Texas-red-labeled (B1) secondary antibody. CBFβ localization was detected with β141.2 and a Texas-red-labeled secondary antibody (A2). Panel B2 shows the autofluorescence of EGFP-CBFβ. Cells in panels A3 and B3 were stained by DAPI. (C) Cytoplasmic and nuclear fractionation of the transfected cells is shown. NIH 3T3 cells transfected with pEGFP (lanes 1 and 2), pEGFP-CBFB (lanes 3 and 4), pEGFP-CBFB plus pCbfa2 (lanes 5 and 6), and pEGFP-Cbfa2 alone (lanes 7 and 8) were fractionated into cytoplasmic and nuclear fractions, which were separated on SDS-PAGE gel and probed by Western blotting with a GFP-specific antibody. The quality of cell fractionation was analyzed by probing with antibodies for cPLA2 (cytoplasmic) and Rb (nuclear). C, cytoplasmic fraction; N, nuclear fraction. Molecular mass markers are shown to the right in panel C.
The nuclear transport of CBFβ by CBFα2 was also confirmed by cell fractionation studies. Cytoplasmic and nuclear fractions prepared from NIH 3T3 cells transfected with pEGFP-CBFB and/or pEGFP-Cbfa2 were probed with a GFP-specific antibody on Western blots. As shown in Fig. 3C, transfected EGFP-CBFα2 was predominantly localized in the nuclear fraction (lanes 7 and 8), whereas the transfected EGFP-CBFβ was mainly present in the cytoplasmic fraction (lanes 3 and 4). On the other hand, EGFP-CBFβ was found mostly in the nuclear fraction after cotransfection with pCbfa2 (lanes 5 and 6), consistent with the immunofluorescence data.
CBFβ-SMMHC sequesters CBFα2 along filaments and aggregates.
When CBFα2 and CBFβ-SMMHC were coexpressed in NIH 3T3 cells through transient cotransfection (plasmids pCbfa2 and pCBFB-MYH11) and analyzed by immunofluorescence labeling, CBFβ-SMMHC assembled into the same filamentous or aggregated structures as it did when expressed alone (Fig. 4A1). However, CBFα2 colocalized with CBFβ-SMMHC along the filaments and aggregates (Fig. 4A2). Similar results were obtained when pEGFP-Cbfa2 was cotransfected with pCBFB-MYH11 (Fig. 4B1, B2, and B3) and when pCbfa2 was cotransfected with pEGFP-CBFB-MYH11 (Fig. 4C1, C2, and C3). Occasionally, CBFα2 and CBFβ-SMMHC proteins were found in both the nucleus and cytoplasm (Fig. 4C1 and C2). In more than 95% of the cells that expressed both transfected genes, CBFα2 colocalized with CBFβ-SMMHC along the filaments and aggregates. In cells where the concentration of CBFβ-SMMHC was much lower than that of CBFα2 (based on immunofluorescence intensity), while some CBFα2 colocalized with CBFβ-SMMHC outside the nucleus, the remaining excess of CBFα2 protein localized to the nucleus.
We attempted to confirm the CBFα2 sequestration by CBFβ-SMMHC using cell fractionation analysis, but we found that the CBFβ-SMMHC protein was always present in the nuclear fraction, regardless of whether it was expressed alone or in combination with CBFα2 (1). This result could be explained by the fact that the MHC molecule multimerizes and forms large aggregates at salt concentrations below 200 mM. The low-salt-concentration treatment of cells for the disruption of cytoplasmic membranes (less than 15 mM) (13) would therefore result in the formation of CBFβ-SMMHC aggregates, which would then precipitate with the nuclear fraction in the subsequent centrifugation step. This intrinsic myosin coprecipitation with the nuclear fraction was confirmed, as endogenous nonmuscle MHC (NMMHC) was present mainly in the nuclear fraction in such preparations (1) although NMMHC have been shown to be cytoplasmic proteins associated with actin (19). We also attempted to solubilize myosin by raising the salt concentration during cell lysis, but we were able only to partially solubilize CBFβ-SMMHC, while at the same time nuclear proteins started to leak into the cytoplasmic fraction (1). Therefore, we could not determine with accuracy the percentages of CBFβ-SMMHC and CBFα2 in the cytoplasmic and nuclear compartments using cell fractionation.
Sequestration of CBFα2 results from direct interaction between CBFα2 and CBFβ-SMMHC.
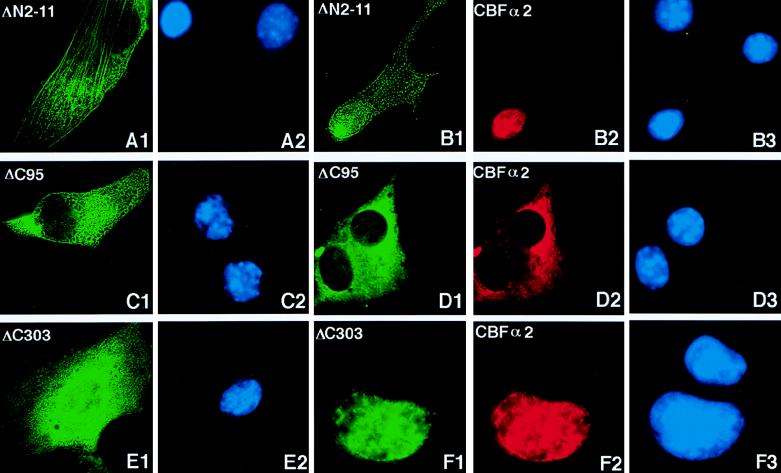
The first 11 amino acids of the CBFβ protein are required for the binding to CBFα2 (44). A CBFβ-SMMHC protein missing this region (CBFβ-SMMHCΔN2-11) failed to interact with CBFα2, as seen by its inability to be coimmunoprecipitated (Fig. 1B). When pCBFB-MYH11ΔN2-11 was transfected into NIH 3T3 cells, the expressed protein localized along the filaments in the cytoplasm, similarly to the full-length protein (Fig. 5A1). However, when coexpressed with CBFα2, the deletion protein did not change the nuclear location of CBFα2. As shown in Fig. 5B, CBFβ-SMMHCΔN2-11 and CBFα2 behaved independently of each other: CBFβ-SMMHCΔN2-11 was distributed mainly in the cytoplasm along filament-like structures, whereas CBFα2 remained exclusively in the nucleus. This observation was confirmed by using EGFP-CBFβ-SMMHCΔN2-11 (1).
FIG. 5.
Immunofluorescence of CBFβ-SMMHC deletion proteins and coexpressed CBFα2. CBFβ-SMMHC deletion proteins and CBFα2 were detected with β141.2 and α3043, respectively, plus appropriate secondary antibodies. (A and B) Immunofluorescence labeling of NIH 3T3 cells transfected with pCBFB-MYH11ΔN2-11, either alone (panels A1 and A2) or with pCbfa2 (panels B1, B2, and B3). Panels A1 and A2 show the same field with the CBFβ-SMMHCΔN2-11 localization in green and DAPI-stained nuclei in blue, respectively. Localization of CBFβ-SMMHCΔN2-11 is again shown in green in panel B1, while cotransfected CBFα2 is shown in red in panel B2. (C1 and C2) Immunofluorescence labeling of cells transfected with only pCBFB-MYH11ΔC95. (D) Cells cotransfected with pCBFB-MYH11ΔC95 (panel D1) and pCbfa2 (panel D2). CBFβ-SMMHCΔC303 was labeled in green when transfected alone (E1) or cotransfected (F1) with CBFα2, which was labeled in red (F2). The panels labeled with the same letter are views of the same fields. (A2, B3, C2, D3, E2, and F3) DAPI staining showing the nuclei of cells in the respective fields.
C-terminal SMMHC domain of CBFβ-SMMHC is required for CBFα2 sequestration.
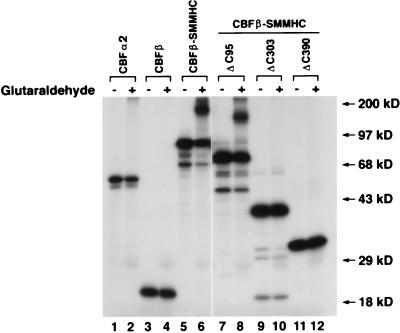
We next determined whether CBFα2 sequestration was dependent on the myosin part of the chimeric protein CBFβ-SMMHC. Constructs with deletions at the C-terminal end of CBFβ-SMMHC (Fig. 1) were used for in vitro cross-linking analysis to test the ability of their protein products to dimerize and multimerize. In vitro-translated proteins were incubated under conditions allowing dimerization in the presence or absence of glutaraldehyde, which covalently cross-links the dimerized proteins. Full-length CBFβ-SMMHC was capable of forming dimers and tetramers as seen by the presence of more-slowly migrating complexes (Fig. 6, lane 6). CBFβ-SMMHCΔC95 was able to dimerize (Fig. 6, lane 8), although with somewhat less efficiency than CBFβ-SMMHC. Removing 175 amino acids from the C-terminal end further decreased the efficiency of dimerization (1). Any further deletion in the C-terminal end abrogated the ability of CBFβ-SMMHC to assemble into dimers, as seen for CBFβ-SMMHCΔC303 (lane 10) and CBFβ-SMMHCΔC390 (lane 12). CBFα2 (lane 2) and CBFβ (lane 4) did not dimerize under these conditions, as expected from previous studies (17, 50).
FIG. 6.
Glutaraldehyde cross-linking of CBFβ, CBFβ-SMMHC, and the deletion proteins. Proteins as labeled at the top of the lanes were translated in vitro in the presence of [35S]methionine. Cross-linking of each protein was accomplished by incubation in the presence (even-numbered lanes) or absence (odd-numbered lanes) of glutaraldehyde. Slow mobility bands were observed for CBFβ-SMMHC and CBFβ-SMMHCΔC95 but not for CBFα2, CBFβ, CBFβ-SMMHCΔC303, and CBFβ-SMMHCΔC390. Protein molecular mass markers are shown on the right.
The subcellular localization of CBFβ-SMMHCΔC95, CBFβ-SMMHCΔC303, and CBFβ-SMMHCΔC390, in the presence or absence of CBFα2, was then determined by immunofluorescence. CBFβ-SMMHCΔC95 (from plasmid pCBFB-MYH11ΔC95), the deletion protein which retains the ability to dimerize, was localized to the cytoplasm in the majority of the cells when transfected alone (Fig. 5C1). Interestingly, CBFβ-SMMHCΔC95 seemed to localize almost entirely to the cytoplasm, with almost no protein evident in nuclei and with no prominent filament-like structures in the cytoplasm. Despite this subtle but consistent difference in subcellular localization compared to full-length CBFβ-SMMHC, CBFβ-SMMHCΔC95 retained the ability to sequester CBFα2 when coexpressed (Fig. 5D1, D2, and D3). On the other hand, deletion proteins CBFβ-SMMHCΔC303 (Fig. 5E1 and E2) and CBFβ-SMMHCΔC390 (1), which failed to form dimers, showed diffuse distribution all over the cells when transfected alone (Fig. 5E1) and were transported into the nucleus when cotransfected with CBFα2 (Fig. 5F1 to F3). These results were also confirmed with EGFP fusion proteins (1). These data suggest that the coiled-coil domain is essential for the subcellular localization of CBFβ-SMMHC and for its ability to sequester CBFα2.
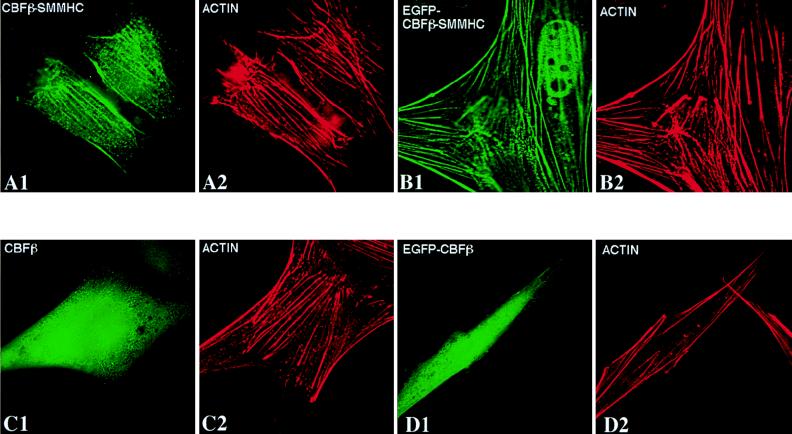
CBFβ-SMMHC colocalizes with actin.
The pattern of localization of the inv(16) fusion protein is similar to that of actin stress fibers. To determine if CBFβ-SMMHC colocalizes with actin, NIH 3T3 cells were transfected with pCBFB-MYH11 or pEGFP-CBFB-MYH11 and subjected to immunofluorescence labeling with β141.2 (for CBFβ-SMMHC) and Texas-red-labeled phalloidin, which specifically interacts with actin. The data in Fig. 7 clearly illustrate that CBFβ-SMMHC and EGFP-CBFβ-SMMHC colocalized with actin filaments (compare Fig. 7A1 with A2 and Fig. 7B1 with B2). Again, some of the EGFP-CBFβ-SMMHC protein can be seen in the nucleus in addition to the filamentous localization in the cytoplasm (Fig. 7B1). NIH 3T3 cells were also transfected with pCBFB or pEGFP-CBFB. In contrast to CBFβ-SMMHC, CBFβ and EGFP-CBFβ were distributed throughout the cell (Fig. 7C1 and D1), the localization showing no correlation with actin localization (Fig. 7C2 and D2).
FIG. 7.
CBFβ-SMMHC colocalizes with actin filaments, while CBFβ does not. NIH 3T3 cells were transfected with pCBFB-MYH11 (A1 and A2), pEGFP-CBFB-MYH11 (B1 and B2), pCBFB (C1 and C2), and pEGFP-CBFB (D1 and D2). Proteins were detected with β141.2 plus a fluorescein isothiocyanate-labeled secondary antibody (panels A1 and C1) and Texas-red-conjugated phalloidin (panels A2, B2, C2, and D2). Panels B1 and D1 show the autofluorescent signal from the GFP fusion proteins.
Colocalization of CBFα2 and CBFβ-SMMHC in vivo.
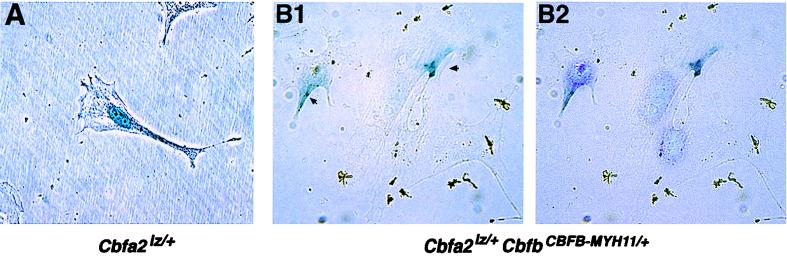
Through homologous recombination in mouse ES cells, the bacterial lacZ gene was fused in frame to the mouse Cbfa2 gene, downstream from the runt domain (33). The expression of the Cbfa2lz allele is under the control of endogenous Cbfa2 promoter, resulting in near-physiological levels of Cbfα2-βgal fusion protein in the cells of Cbfa2lz/+ mice. The chimeric Cbfα2-βgal protein produced retained its ability to interact with CBFβ through the runt domain, while the βgal portion of the protein permitted detection of the protein by the relatively sensitive 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside assay. Matings were set up between Cbfa2lz/+ mice and chimeric mice made with CbfbCBFB-MYH11/+ ES cells, which produce a CBFβ-SMMHC fusion protein (8). Mouse embryonic fibroblasts isolated from 11.5-dpc embryos derived from such matings contained either only the Cbfa2lz allele or both the Cbfa2lz and CbfbCBFB-MYH11/+ alleles. β-Galactosidase activity was detected mainly in the nuclei of mouse embryonic fibroblasts derived from Cbfa2lz/+ embryos (Fig. 8A) (92% of the cells had expression exclusively in the nucleus, and 8% had expression in both the cytoplasm and nucleus), whereas a significant amount of β-galactosidase activity was found in the cytoplasm of fibroblasts containing the CbfbCBFB-MYH11/+ allele (Fig. 8B). Seventy percent of the cells had expression only in the cytoplasm (e.g., the cells shown on the right in Fig. 8B), 29% had it in both the cytoplasm and the nucleus (e.g., the cell shown on the left in Fig. 8B), and 1% had it only in the nucleus. The result demonstrates that cytoplasmic sequestration of CBFα2 by the CBFβ-SMMHC fusion protein also occurs at normal physiological levels of the proteins.
FIG. 8.
In vivo sequestration of Cbfα2 by Cbfβ-SMMHC. Embryonic fibroblasts were isolated from 11.5-dpc mouse embryos, cultured briefly in vitro, and stained for the Cbfα2-βgal protein. (A) A fibroblast from an embryo of Cbfa2lz/+ genotype stained for βgal. (B1) Fibroblasts from an embryo of Cbfa2lz, CbfbCBFB-MYH11/+ genotype stained for βgal. Arrows indicate the βgal staining. (B2) DAPI staining of the same field as shown in panel B1.
DISCUSSION
Genes encoding subunits of the core binding factor CBF are frequently disrupted by chromosome rearrangements in human leukemias. Translocation t(8;21), found in AML subtype M2, generates a fusion protein, AML1-ETO. This protein has been shown to inhibit both AML1/CBFα2- and C/EBPα-dependent transcriptional activation, although the mechanism for this inhibition remains unknown (14, 30, 52). AML1-ETO can also dominantly suppress CBF function, as shown in a mouse knock-in model (38, 55). The AML1-Evi-1 chimeric protein, produced in t(3;21) associated with t-MDS, t-AML, and CML-BC, can suppress transactivation by intact AML1/CBFα2 (56). Again, the mechanism for such suppression is not fully understood. In this study we have addressed the mechanism by which the inv(16) chimeric protein CBFβ-SMMHC plays a dominant negative role. Our results suggest that CBFβ-SMMHC plays a dominant negative role by sequestering CBFα2/AML1 to cytoskeletal structures, thus initiating a process towards leukemia by disrupting transcriptional regulation of genes controlled by CBFα2/AML1.
Upon transient transfection into NIH 3T3 cells, CBFα2 was localized in the nucleus whereas CBFβ was distributed throughout the cell. CBFβ-SMMHC was located predominantly in the cytoplasm, forming filamentous or aggregated structures. We did observe some CBFβ-SMMHC protein in the nuclei of the transfected cells, in addition to the filaments and aggregates in the cytoplasm, but we did not observe the nuclear rod-like structure that we previously found in clonal NIH 3T3 cell lines overexpressing CBFβ-SMMHC (53). This is probably due to the difference in expression level and/or the difference between transient and stable transfections. In fact, the rod-like structures were found only in cell lines expressing high levels of the CBFβ-SMMHC protein and only in a small percentage (10 to 40%) of cells within a cell population (1, 53). On the other hand, the stress fibers formed by CBFβ-SMMHC were evident in most of the cells stably expressing CBFβ-SMMHC, regardless of expression level (1, 53).
In the present study, we demonstrated that CBFβ localized to the nucleus when cotransfected with full-length CBFα2. The likely mechanism for this nuclear localization of CBFβ is that CBFα2 heterodimerizes with CBFβ in the cytoplasm and mediates the transport of the heterodimer into the nucleus. Nuclear localization of CBFα2 can be overridden by CBFβ-SMMHC, which forms homodimers and multimers and interacts with cytoskeletal molecules. The sequestration of CBFα2 by CBFβ-SMMHC is dependent on the heterodimerization domain in CBFβ that mediates interaction with the CBFα subunits. The truncated protein with a deletion of the CBFα2-interacting domain, CBFβ-SMMHCΔN2-11, lost the ability to sequester CBFα2, even though it retained the ability to assemble into cytoskeletal structures (Fig. 5B1 to B3). Sequestration of CBFα2 by CBFβ-SMMHC is also dependent on the presence of the C-terminal SMMHC domain. Deletions of the C-terminal SMMHC domain disrupted the ability of the chimeric protein to dimerize and to interact with other cytoskeletal proteins, most likely other myosin family members such as NMMHC, and compromised its ability to sequester CBFα2. In fact, once the C-terminal SMMHC domain was lost, the truncated CBFβ-SMMHC proteins behaved like wild-type CBFβ in that they were no longer localized along the cytoskeletal structures and localized to the nucleus when coexpressed with CBFα2 (Fig. 5E and F).
Our efforts to confirm these results in the samples from patients with inv(16) yielded equivocal results, most likely because the endogenous levels of the proteins were too low to be identified by immunofluorescence (1). However, CBFα2 sequestration was observed in vivo, in a mouse gene-targeting model. This result not only confirmed the findings obtained with transient transfections in cultured cells which could potentially result from overexpression, but also demonstrated the dominant nature of this sequestration, since both wild-type CBFβ and CBFβ-SMMHC proteins are expressed in the Cbfb-MYH11 knock-in mice (8).
The mechanism for the dominant nature of the CBFα2 sequestration in vivo is still not clear. At least two possibilities exist: one is that CBFβ-SMMHC is more stable than the wild-type CBFβ and therefore has higher concentration in the cells, and the other is that CBFβ-SMMHC binds CBFα2 better than does CBFβ. Results from our previous study (8) showed that CBFβ-SMMHC was expressed at a level similar to that of the wild-type CBFβ in the Cbfb-MYH11 knock-in embryos. CBFβ-SMMHC and CBFβ were found to be expressed at comparable levels in leukemia cells from a number of patients as well (9, 10). However, Western blotting is not very quantitative, and variations up to fivefold have been observed. Therefore, it is possible that the fusion protein is more stable than the wild-type CBFβ. On the other hand, the binding property of CBFβ-SMMHC to CBFα2 has not been studied in detail, since purified full-length CBFβ-SMMHC in large enough quantity for such experiments is not available. Future investigations are needed to find out if either or both of these two factors contribute to the dominant sequestration of CBFα2 by CBFβ-SMMHC.
We observed that transfected CBFβ was localized throughout cells, with some cytoplasmic predominance. In addition, the transfected CBFβ did not colocalize with actin stress fibers (Fig. 7C and D). This differs from a recent study which showed that CBFβ colocalizes with actin by immunofluorescent staining and fractionation (47). The reason for this difference is not clear, but it could be attributed to the differences in the CBFβ isoforms used (CBFβ187 versus CBFβ182), in the cell types used (REF52 cells versus NIH 3T3 cells), and the detergent treatments employed.
A previous study showed that truncated CBFα1/PEBP2αA can bring CBFβ into the nucleus whereas full-length CBFα1/PEBP2αA cannot (28). This leaves open the question as to how CBFβ localizes to the nucleus to enable formation of a nuclear heterodimer with full-length PEBP2αA. One possible explanation for the differences between our data and those reported by Lu et al. (28) is that different CBFα proteins and CBFβ isoforms were used by the two groups. While we used CBFα2, or AML1, the protein product of the gene involved in human leukemias, Lu et al. used PEBP2αA, or CBFα1, which differs from CBFα2 in sequence and has been shown to be important for osteogenesis (12, 20, 32, 40). These two CBFα proteins may have different affinities for CBFβ. In the case of CBFβ, we used the CBFβ187 isoform, while the CBFβ182 isoform was used by Lu et al. (28). These two isoforms have two different C-terminal ends that are the result of alternative splicing, which may be responsible for the differences between the observations made by the two groups. Finally, the concentrations of CBFα1, CBFα2, and CBFβ achieved in the two studies may have differed, which would have influenced the extent of CBFα:CBFβ heterodimerization.
CBFβ-SMMHC-mediated CBFα2 sequestration can at least partially explain the dominant negative suppression of CBF function by CBFβ-SMMHC in hematopoiesis and leukemogenesis. Due to the limited sensitivities of the assays and the nature of transient transfection, we cannot rule out the possibility that free CBFα2 was present in the nuclei of some cells. Therefore, additional mechanisms, such as reduced binding of target DNA sequences by CBFβ-SMMHC:CBFα heterodimers, may also contribute to the dominant negative effect on CBF function of CBFβ-SMMHC. However, it has been postulated that CBFα2 is quantitatively limiting in vivo, since hematopoietic progenitor cells are either reduced in number or have decreased capacity to differentiate in Cbfa2+/− mice (48). Therefore, substantial reduction of functional CBFα2 protein in the nucleus resulting from CBFβ-SMMHC sequestration is probably sufficient to cause defective CBF function, leading to dysregulation of downstream genes and contributing to leukemogenesis.
The functional significance of CBFα2 sequestration by CBFβ-SMMHC is evident in the following two sets of studies. First, it was recently demonstrated that CBFβ-SMMHC can reduce CBF DNA-binding and inhibit the G1-to-S cell cycle transition in cultured myeloid and lymphoid cells (6, 7). These effects were demonstrated to require the ability of CBFβ-SMMHC to interact with CBFα2 and the presence of the C-terminal SMMHC domain, using deletion constructs similar to those described here (7). These effects on CBF DNA-binding and cell cycle could therefore be explained by CBFα2 sequestration. Secondly, CBFα2 sequestration by CBFβ-SMMHC in mouse embryonic cells as described in this study correlates with the ability of CBFβ-SMMHC to block CBF function and disrupt hematopoiesis in mouse embryos. Future efforts will be aimed at confirming whether the sequestration of CBFα2 in vivo does in fact result in pathogenesis. This could potentially be demonstrated by generating knock-in mice expressing chimeric proteins with abrogated ability to dimerize or by substituting for the myosin chain an unrelated cytoskeletal protein. Targeted strategies to overcome or eliminate CBFα2 sequestration will be tested in cell culture and whole-animal models to see if CBFβ-SMMHC pathologic function can be subverted; the results will have clinical implications for leukemia management in the future.
FIG. 2.
Subcellular localization of CBFα2, CBFβ, and CBFβ-SMMHC in transiently transfected cells. NIH 3T3 cells were transfected with pCbfa2 (A1 and A2), pEGFP-Cbfa2 (B1 and B2), pCBFB (C1 and C2), pEGFP-CBFB (D1 and D2), pCBFB-MYH11 (E1 and E2), and pEGFP-CBFB-MYH11 (F1 and F2). Odd-numbered panels show fluorescent signals, while even-numbered panels show DAPI staining of the same fields. Panel A1 shows signal detected with α3043 and a Texas-red-labeled secondary antibody, panels C1 and E1 show signals detected with β141.2 and a fluorescein-labeled secondary antibody, and panels B1, D1, and F1 show the autofluorescent signal of GFP fused to the respective proteins. The antibodies failed to detect the endogenous proteins (compare the untransfected cell in panel A1 with panel A2, as well as that in panel C1 with panel C2).
FIG. 4.
CBFβ-SMMHC sequesters CBFα2 along filaments and aggregates. (A1, A2, and A3) NIH 3T3 cells were cotransfected with pCbfa2 and pCBFB-MYH11. CBFβ-SMMHC localization was detected with antibody β141.2 and a fluorescein-labeled secondary antibody (panel A1), and CBFα2 localization was detected with antibody α3043 and a Texas-red-labeled secondary antibody (panel A2). (B1, B2, and B3) Localization of EGFP-CBFα2 and CBFβ-SMMHC when cotransfected. CBFβ-SMMHC localization is shown by the red signal (β141.2 and a Texas-red-labeled secondary antibody) in panel B1, while EGFP-CBFα2 localization is shown by the green autofluorescent signal in panel B2. (C1, C2, and C3) Cotransfected EGFP-CBFβ-SMMHC (panel C1) and CBFα2 (panel C2), which were detected in a fashion similar to that described for panel B. (A3, B3, and C3) DAPI staining of the respective fields.
ACKNOWLEDGMENTS
We are grateful to Lucio Castilla, Lisa Garrett, Karla Henning, and Trevor Blake for their discussion and assistance throughout the study. We thank Robert Adelstein, James Sellers, and Duane Compton for advice and reagents, David Claxton for sharing unpublished data, Kevin D. Brown for technical assistance, and Francis Collins and Alan Friedman for their critical readings of the manuscript.
N.A.S. is a Scholar and P.P.L. is a Special Fellow of the Leukemia Society of America. N.A.S. is supported by NIH grants RO1 CA58343 and RO1 CA75611.
REFERENCES
- 1.Adya, N., and P. P. Liu. Unpublished data.
- 2.Arthur D C, Bloomfield C D. Association of partial deletion of the long arm of chromosome 16 and bone marrow eosinophilia in acute non-lymphocytic leukemia. Blood. 1983;62:931. . (Letter.) [PubMed] [Google Scholar]
- 3.Bae S-C, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. PEBP2αB/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae S C, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka M, Kagoshima H, Shigesada K, Satake M, Ito Y. Isolation of PEBP2 α B cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–814. [PubMed] [Google Scholar]
- 5.Cameron S, Taylor D S, TePas E C, Speck N A, Mathey-Prevot B. Identification of a critical regulatory site in the human interleukin-3 promoter by in vivo footprinting. Blood. 1994;83:2851–2859. [PubMed] [Google Scholar]
- 6.Cao W, Britos-Bray M, Claxton D F, Kelley C A, Speck N A, Liu P P, Friedman A D. CBF β-SMMHC, expressed in M4Eo AML, reduced CBF DNA-binding and inhibited the G1 to S cell cycle transition at the restriction point in myeloid and lymphoid cells. Oncogene. 1997;15:1315–1327. doi: 10.1038/sj.onc.1201305. [DOI] [PubMed] [Google Scholar]
- 7.Cao W, Liu P P, Friedman A D. Two domains of CBFβ-SMMHC are required for inhibition of CBF DNA-binding and cell cycle progression in hematopoietic cells. Blood. 1997;90:59a. [Google Scholar]
- 8.Castilla L H, Wijmenga C, Wang Q, Stacy T, Speck N A, Eckhaus M, Marin-Padilla M, Collins F S, Wynshaw-Boris A, Liu P P. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 9.Claxton, D. Personal communication.
- 10.Claxton D, Xie Q S, Patel S, Deisseroth A B, Kornblau S. The gene product of CBFB-MYH11. Leukemia. 1996;10:1479–1485. [PubMed] [Google Scholar]
- 11.Cullen B R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 12.Ducy P, Zhang R, Geoffroy V, Ridall A L, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 13.Dyer R B, Herzog N K. Isolation of intact nuclei for nuclear extract preparation from a fragile B-lymphocyte cell line. BioTechniques. 1995;19:192–195. [PubMed] [Google Scholar]
- 14.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer S D. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 15.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR α enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 16.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Crute B E, Sun C, Tang Y Y, Kelley III J J, Lewis A F, Hartman K L, Laue T M, Speck N A, Bushweller J H. Overexpression, purification, and biophysical characterization of the heterodimerization domain of the core-binding factor β subunit. J Biol Chem. 1998;273:2480–2487. doi: 10.1074/jbc.273.4.2480. [DOI] [PubMed] [Google Scholar]
- 18.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y, Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990;64:4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley C A, Sellers J R, Gard D L, Bui D, Adelstein R S, Baines I C. Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities. J Cell Biol. 1996;134:675–687. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R T, Gao Y H, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 21.Le Beau M M, Larson R A, Bitter M A, Vardiman J W, Golomb H M, Rowley J D. Association of an inversion of chromosome 16 with abnormal marrow eosinophils in acute myelomonocytic leukemia. A unique cytogenetic-clinicopathological association. N Engl J Med. 1983;309:630–636. doi: 10.1056/NEJM198309153091103. [DOI] [PubMed] [Google Scholar]
- 22.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 23.Liu P, Seidel N, Bodine D, Speck N, Tarle S, Collins F S. Acute myeloid leukemia with Inv (16) produces a chimeric transcription factor with a myosin heavy chain tail. Cold Spring Harbor Symp Quant Biol. 1994;59:547–553. doi: 10.1101/sqb.1994.059.01.061. [DOI] [PubMed] [Google Scholar]
- 24.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Fusion between transcription factor CBF β/PEBP2 β and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 25.Liu P P, Hajra A, Wijmenga C, Collins F S. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood. 1995;85:2289–2302. [PubMed] [Google Scholar]
- 26.Liu P P, Wijmenga C, Hajra A, Blake T B, Kelley C A, Adelstein R S, Bagg A, Rector J, Cotelingam J, Willman C L, Collins F S. Identification of the chimeric protein product of the CBFB-MYH11 fusion gene in inv(16) leukemia cells. Genes Chromosomes Cancer. 1996;16:77–87. doi: 10.1002/(SICI)1098-2264(199606)16:2<77::AID-GCC1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 27.Look A T. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, Maruyama M, Satake M, Bae S-C, Ogawa E, Kagoshima H, Shigesada K, Ito Y. Subcellular localization of the α and β subunits of the acute myeloid leukemia-linked transcription factor PEBP2/CBF. Mol Cell Biol. 1995;15:1651–1661. doi: 10.1128/mcb.15.3.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marlton P, Claxton D F, Liu P, Estey E H, Beran M, LeBeau M, Testa J R, Collins F S, Rowley J D, Siciliano M J. Molecular characterization of 16p deletions associated with inversion 16 defines the critical fusion for leukemogenesis. Blood. 1995;85:772–779. [PubMed] [Google Scholar]
- 30.Meyers S, Lenny N, Hiebert S W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mundlos S, Otto F, Mundlos C, Mulliken J B, Aylsworth A S, Albright S, Lindhout D, Cole W G, Henn W, Knoll J H, Owen M J, Mertelsmann R, Zabel B U, Olsen B R. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 33.North, T., T. L. Gu, T. Stacy, Q. Wang, L. Howard, E. Dzierzak, M. Binder, M. Marin-Padilla, and N. A. Speck. Cbfa2 is required for the emergence of definitive hematopoietic cells from hemogenic endothelium. Submitted for publication.
- 34.Nucifora G, Begy C R, Erickson P, Drabkin H A, Rowley J D. The 3;21 translocation in myelodysplasia results in a fusion transcript between the AML1 gene and the gene for EAP, a highly conserved protein associated with the Epstein-Barr virus small RNA EBER 1. Proc Natl Acad Sci USA. 1993;90:7784–7788. doi: 10.1073/pnas.90.16.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nucifora G, Begy C R, Kobayashi H, Roulston D, Claxton D, Pedersen-Bjergaard J, Parganas E, Ihle J N, Rowley J D. Consistent intergenic splicing and production of multiple transcripts between AML1 at 21q22 and unrelated genes at 3q26 in (3;21)(q26;q22) translocations. Proc Natl Acad Sci USA. 1994;91:4004–4008. doi: 10.1073/pnas.91.9.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2 β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 α. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 37.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuda T, Cai Z, Yang S, Lenny N, Lyu C J, van Deursen J M, Harada H, Downing J R. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 39.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 40.Otto F, Thornell A P, Crompton T, Denzel A, Gilmour K C, Rosewell I R, Stamp G W, Beddington R S, Mundlos S, Olsen B R, Selby P B, Owen M J. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 41.Romana S P, Mauchauffe M, Le Coniat M, Chumakov I, Le Paslier D, Berger R, Bernard O A. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- 42.Sasaki K, Yagi H, Bronson R T, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor β. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satake M, Inuzuka M, Shigesada K, Oikawa T, Ito Y. Differential expression of subspecies of polyomavirus and murine leukemia virus enhancer core binding protein, PEBP2, in various hematopoietic cells. Jpn J Cancer Res. 1992;83:714–722. doi: 10.1111/j.1349-7006.1992.tb01971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Speck, N. A. Unpublished data.
- 45.Sun W, Graves B J, Speck N A. Transactivation of the Moloney murine leukemia virus and T-cell receptor β-chain enhancers by cbf and ets requires intact binding sites for both proteins. J Virol. 1995;69:4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi A, Satake M, Yamaguchi-Iwai Y, Bae S C, Lu J, Maruyama M, Zhang Y W, Oka H, Arai N, Arai K, Ito Y. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood. 1995;86:607–616. [PubMed] [Google Scholar]
- 47.Tanaka Y, Watanabe T, Chiba N, Niki M, Kuroiwa Y, Nishihira T, Satomi S, Ito Y, Satake M. The protooncogene product, PEBP2β/CBFβ, is mainly located in the cytoplasm and has an affinity with cytoskeletal structures. Oncogene. 1997;15:677–683. doi: 10.1038/sj.onc.1201235. [DOI] [PubMed] [Google Scholar]
- 48.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T L, Huang X, Bushweller J H, Bories J C, Alt F W, Ryan G, Liu P P, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 50.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S W, Speck N A. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol Cell Biol. 1992;12:89–102. doi: 10.1128/mcb.12.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westendorf J J, Yamamoto C M, Lenny N, Downing J R, Selsted M E, Hiebert S W. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-α, inhibits C/EBP-α-dependent transcription, and blocks granulocytic differentiation. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wijmenga C, Gregory P E, Hajra A, Schrock E, Ried T, Eils R, Liu P P, Collins F S. Core binding factor β-smooth muscle myosin heavy chain chimeric protein involved in acute myeloid leukemia forms unusual nuclear rod-like structures in transformed NIH 3T3 cells. Proc Natl Acad Sci USA. 1996;93:1630–1635. doi: 10.1073/pnas.93.4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wijmenga C, Speck N A, Dracopoli N C, Hofker M H, Liu P, Collins F S. Identification of a new murine runt domain-containing gene, Cbfa3, and localization of the human homolog, CBFA3, to chromosome 1p35-pter. Genomics. 1995;26:611–614. doi: 10.1016/0888-7543(95)80185-o. [DOI] [PubMed] [Google Scholar]
- 55.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 56.Zent C S, Mathieu C, Claxton D F, Zhang D E, Tenen D G, Rowley J D, Nucifora G. The chimeric genes AML1/MDS1 and AML1/EAP inhibit AML1B activation at the CSF1R promoter, but only AML1/MDS1 has tumor-promoter properties. Proc Natl Acad Sci USA. 1996;93:1044–1048. doi: 10.1073/pnas.93.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]