Abstract
The stable inheritance of the 2μm plasmid in a growing population of Saccharomyces cerevisiae is dependent on two plasmid-encoded proteins (Rep1p and Rep2p), together with the cis-acting locus REP3 (STB). In this study we demonstrate that short carboxy-terminal deletions of Rep1p and Rep2p severely diminish their normal capacity to localize to the yeast nucleus. The nuclear targeting, as well as their functional role in plasmid partitioning, can be restored by the addition of a nuclear localization sequence to the amino or the carboxy terminus of the shortened Rep proteins. Analyses of deletion derivatives of the Rep proteins by using the in vivo dihybrid genetic test in yeast, as well as by glutathione S-transferase fusion trapping assays in vitro demonstrate that the amino-terminal portion of Rep1p (ca. 150 amino acids long) is responsible for its interactions with Rep2p. In a monohybrid in vivo assay, we have identified Rep1p, Rep2p, and a host-encoded protein, Shf1p, as being capable of interacting with the STB locus. The Shf1 protein expressed in Escherichia coli can bind with high specificity to the STB sequence in vitro. In a yeast strain deleted for the SHF1 locus, a 2μm circle-derived plasmid shows relatively poor stability.
The 2μm circle, a relatively small circular plasmid (6,318 bp) present in most common strains of Saccharomyces cerevisiae, has optimized a partitioning system and an amplification system that allow it to be propagated stably in a cell population at a copy number of approximately 60 to 100 per cell (reviewed in reference 2). Genetic analyses suggest that two plasmid-coded proteins, Rep1p and Rep2p, in conjunction with a cis-acting locus STB (also called REP3) contribute to the stability function (16, 17, 19, 23). One plausible mechanism for plasmid stability is that the interaction of Rep1p and Rep2p with the STB element serves to overcome the normal bias in plasmid segregation that tends to favor the mother cell over the daughter cell (22). The evidence for this suspected DNA-protein interaction is quite preliminary and rests almost entirely on the observation that urea-solubilized yeast extracts expressing Rep1p and Rep2p or [cir0] extracts supplemented exogenously with Rep1p and Rep2p can bind STB (14).
The need for plasmid amplification arises only if and when there is a decrease in copy number below the steady-state value. Normally, each plasmid molecule is replicated once, and only once, per cell cycle (35), and the daughter molecules are partitioned efficiently at cytokinesis (27). When there is a drop in copy number, the amplification system overrides the cell cycle restriction of a single round of plasmid replication during one S phase. Plasmid amplification is absolutely dependent on the 2μm circle Flp site-specific recombination system (33). A currently favored model for amplification proposes the recombinational inversion of a bidirectional replication fork and the resultant double-rolling-circle replication mode as the means for obtaining multiple replicas of the plasmid from a single initiation event (9–11, 26). The cessation of amplification would require a second recombination event that can restore the normal direction of fork movement. The time interval between the two successive recombination events would determine the degree of amplification.
How is the amplification system kept silent under normal steady-state growth conditions? And how is it rapidly commissioned into action at short notice? Biochemical answers to these fundamental question are sparse. On the basis primarily of genetic studies, it has been proposed that a regulatory complex containing Rep1p and Rep2p may provide an indirect readout of the copy number and, either at or above a critical concentration, may negatively control amplification by turning down expression of the FLP gene (24, 25, 28, 30). Recently, we have demonstrated self- and cross-interactions between Rep1p and Rep2p by immunoprecipitations of these proteins from mixed extracts of Escherichia coli cells that express them and by baiting assays with hybrid glutathione S-transferase (GST)-Rep proteins (1). Furthermore, these findings were corroborated by in vivo assays in yeast cells. We now present an analysis of the polypeptide regions in Rep1p and Rep2p responsible for their cellular localization as well as for their functional interactions. We also show here that an in vivo assay with the STB locus as a DNA bait fishes out three proteins: the Rep1 and Rep2 proteins, as well as a chromosomally encoded protein (the product of the YIL036W locus in the yeast genome bank). The chromosomal protein can bind to the STB element in vitro as well. Furthermore, its absence in vivo causes high instability of a 2μm circle-derived test plasmid. The potential implications of these findings in the benign parasitism of the 2μm circle plasmid are discussed here.
MATERIALS AND METHODS
Yeast strains.
The following [cir0] and [cir+] yeast strains were used in this study: FVY2-6B (MATα leu2-3,112 ura3-52 his3-d1 [cir0]; Gal+), FVY889 (MATα ade2 leu2-3,112 ura3-52 his5-2 [cir0]; Gal+), FVY93154 (MATa leu2-3,112 ura3-52 trp1-289 ade2 [cir+]; Gal+), CCY666-1A (MATa ade2 his3-Δ200 leu2-3,112 ura3-52 [cir+]; Gal+), CCY666-7B (MATα ade2 his3-Δ200 leu2-3,112 ura3-52 [cir+]; Gal+), and MJY101 (CCY666-7B, shf1Δ::URA3). The cellular localizations of green fluorescent protein (GFP) hybrids were done in strains FVY2 [cir0] and FVY93154 [cir+], respectively. Results were unchanged when FVY889 and CCY666-1A were used as the [cir0] host and the [cir+] host, respectively. In the Results section, only the data from FVY2 [cir0] are presented. The stabilities of the ADE2 containing test plasmids pSTB1 and pSTB2 (see below) were assessed in yeast strains FVY889 and CCY666-1A. The strain EGY48 (his3 trp1 ura3-52 leu2::pLEU2-LexAop6 [cir+]) used for the dihybrid assays was kindly provided by Roger Brent (8), and the strains used for the monohybrid assays were obtained from Clontech Laboratories (Palo Alto, Calif.). The dihybrid test was also performed in a [cir0] strain harboring the requisite markers. The outcomes were identical in the [cir0] and [cir+] strains. The host for the monohybrid assay, supplied by Clontech Laboratories, was a [cir+] strain. This strain was first crossed with a [cir0] partner, and a haploid [cir0] strain with the appropriate markers (ura3, his3, leu2, and trp1) was derived from the cross. The monohybrid analysis was done in this [cir0] derivative.
Plasmids.
The pGFP-Rep plasmids, obtained by joining the 2μm circle REP1 and REP2 coding regions to the 3′ end of the GFP coding region via a short in-frame adapter sequence, have been described previously (1). The cloning manipulations were done in the yeast GFP expression vector pTS408 (an ARS-CEN-URA3 plasmid obtained as a gift from the Botstein laboratory through Clarence Chan). In this plasmid, the GFP gene was controlled by the yeast GAL1-10 promoter. The deletions in the Rep portions of these plasmids were obtained by PCR with primers containing appropriate restriction enzyme sites to facilitate cloning. The nuclear localization signal (NLS) derived from the simian virus 40 (SV40) T antigen was inserted into some of these plasmids in the form of a synthetic DNA fragment encoding the heptapeptide (2HN-PKKKRKV-COOH). The DNA sequence corresponding to this heptapeptide was the same as that in plasmid pGAD424 (from nucleotide positions 452 to 472), which was supplied by Clontech Laboratories.
The plasmids used for assaying stability, pSTB, pSTB1, and pSTB2, were constructed as derivatives of pYES2 (Invitrogen, Carlsbad, Calif.). The pSTB1 and pSTB2 plasmids contained the 2μm circle origin, the STB locus, the yeast LEU2 and ADE2 markers, and one of the two REP loci (REP1 in pSTB1 and REP2 in pSTB2). Neither one of the two REP genes was present in pSTB. The details of their construction have been described earlier (1).
The plasmids pGST-Rep1 and pGST-Rep2 expressing the hybrid proteins used for the GST-baiting assays were described by Ahn et al. (1), who also outlined the features of the plasmids pSRep1 and pSRep2 used to express the Rep proteins containing the S-peptide tag. For the purpose of this study, the Rep coding regions in the pSRep plasmids were replaced by their deletion-harboring counterparts by using PCR-based cloning strategies.
The plasmids for the dihybrid analysis were a gift from the Brent laboratory (8); those for the monohybrid analysis were purchased from Clontech Laboratories.
Fluorescence microscopy of cells.
The protocols for assaying the green fluorescence from GFP-Rep protein fusions and the blue fluorescence from DAPI (4′,6-diamidino-2-phenylindole) complexed with DNA were as detailed by Ahn et al. (1).
Stability assays for plasmids pSTB1 and pSTB2.
Stabilities of the pSTB1 and pSTB2 plasmids in a [cir0] host harboring pGFP-Rep or pGFP-Rep deletion plasmids were assayed as follows. Purified colonies of the plasmid-bearing strains (LEU2 and ADE2 markers on pSTB1 or pSTB2; URA3 on pGFP-Rep) were maintained on SD plates without leucine or uracil-glucose or on SD plates without leucine or uracil-galactose. Single colonies from the glucose and galactose master plates were spread out on yeast extract-peptone-dextrose (YEPD) and YEP-galactose plates, respectively, and grown for 3 days at 30°C. The red and white colonies on each plate were counted. Sectored colonies were grouped with the white colonies if the sector size was smaller than one-fourth the colony size, and with the red colonies if the sector size was larger. The plasmid stability index (SI) was then expressed as the ratio of the white colonies to the sum of the white plus red colonies multiplied by 100. The values of SI given here are for transfer of colonies from selective galactose plates to YEP-galactose plates (see Table 2).
TABLE 2.
Functional competence of deletion derivatives of Rep1p and Rep2p in promoting plasmid stabilitya
| Rep source | Host | Test plasmid | SI |
|---|---|---|---|
| Rep1p | |||
| 2μm circle | [cir+] | pSTB2 | 92 |
| GFP-Rep1p | [cir0] | pSTB2 | 62 |
| ΔN25 | [cir0] | pSTB2 | <10 |
| ΔN50 | [cir0] | pSTB2 | <10 |
| ΔN75 | [cir0] | pSTB2 | <10 |
| ΔN100 | [cir0] | pSTB2 | <10 |
| ΔN125 | [cir0] | pSTB2 | <10 |
| ΔC25 | [cir0] | pSTB2 | <10 |
| ΔC50 | [cir0] | pSTB2 | <10 |
| ΔC75 | [cir0] | pSTB2 | <10 |
| ΔC100 | [cir0] | pSTB2 | <10 |
| ΔC125 | [cir0] | pSTB2 | <10 |
| ΔC150 | [cir0] | pSTB2 | <10 |
| Rep2p | |||
| 2μm circle | [cir+] | pSTB1 | 97 |
| GFP-Rep2p | [cir0] | pSTB1 | 65 |
| ΔN20 | [cir0] | pSTB1 | <10 |
| ΔN120 | [cir0] | pSTB1 | <10 |
| ΔC20 | [cir0] | pSTB1 | <10 |
| ΔC40 | [cir0] | pSTB1 | <10 |
| ΔC60 | [cir0] | pSTB1 | <10 |
| ΔC80 | [cir0] | pSTB1 | <10 |
| ΔC120 | [cir0] | pSTB1 | <10 |
The Rep deletions were tested in the form of their GFP fusion derivatives. It has been shown earlier (1) that a GFP-Rep1p or a GFP-Rep2p fusion can functionally substitute for the corresponding native protein in plasmid maintenance. The plasmid substrates for the stability assay, pSTB1 and pSTB2, contained the 2μm plasmid origin, the STB locus, and the yeast markers ADE2 and LEU2. The pSTB1 plasmid was also a source for Rep1p; similarly, pSTB2 provided Rep2p. The assays were carried out in a [cir+] ade2 leu2 host (Rep1p and Rep2p being provided in their native forms from the endogenous 2μm plasmids) or in an equivalent [cir0] host expressing GFP-Rep fusions. The SI expresses the number of white colonies (indicating retention of the pSTB plasmid-borne ADE2 marker) as a percentage of the total colonies (red and white; red indicating the loss of the ADE2 marker). The values given here are the mean obtained from the transfer of three test colonies from selective galactose plates to nonselective galactose plates. For more details, refer to Materials and Methods.
Stability of the pSTB plasmid in an shf1Δ::URA3 yeast strain.
The test plasmid was pSTB, containing the 2μm plasmid origin, the STB locus, and the yeast ADE2 and LEU2 markers (1). However, it did not contain either REP1 or REP2. Plasmid stability was assayed simultaneously in the parent strain CCY666-7B (wild type for the SHF1 locus) and in its shf1Δ::URA3 derivative (strain MJY101). Individual transformants purified on SD plates lacking leucine and uracil were grown in YEPD liquid medium for approximately 10 generations and plated on YEPD plates, and the colony color was screened after approximately 60 h of growth at 30°C.
Baiting assays with hybrid GST-Rep proteins.
The expression of the GST-Rep hybrid proteins or the S-tagged Rep proteins in E. coli and the preparation of cell supernatants for the baiting assays followed the protocols detailed by Ahn et al. (1). Each baiting mixture contained 1.0 ml of the GST-Rep1p (or GST-Rep2p) plus 1.0 ml of the S-Rep2p (or S-Rep1p) supernatant.
Dihybrid and monohybrid assays in yeast cells.
The dihybrid assays were carried out according to procedures described by Finley and Brent (8). The β-galactosidase activities were assayed according to the method of Guarente (12). The monohybrid assays were done according to the protocols provided by Clontech Laboratories. An approximately 375-bp fragment from the 2μm plasmid spanning the STB locus was amplified by PCR and cloned upstream of the basal promoter of the HIS3 reporter gene. This transcriptional cassette was integrated into the chromosomal HIS3 locus. Titrations with 3-AT (3-amino-1,2,4-triazole) indicated that complete a cessation of growth of the host strain harboring the integration occurred with a 15 mM concentration of the inhibitor. Selection assays with a cDNA-activation domain fusion library from [cir+] yeast were done at an inhibitor concentration of 45 mM. The positive candidates were subjected to two additional rounds of 3-AT selection before they were subcloned into E. coli and characterized.
Western blot analysis of yeast extracts.
Next, 5-ml yeast cultures (optical density at 600 nm of ca. 0.4 to 0.6) were centrifuged, and the pelleted cells were washed twice at 4°C in 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA (pH 8.0), 1 mM phenylmethylsulfonyl fluoride (added from a 200× stock, freshly prepared in isopropanol), and 1× protease inhibitor cocktail from Boehringer Mannheim. The cells were resuspended in 250 μl of the washing buffer and then disrupted by vortexing (six 30-s cycles, with a 30-s intermission on ice between cycles) with acid-washed glass beads (425 to 600 μm in diameter). The supernatants were collected by centrifugation at 4°C in an Eppendorf centrifuge.
Aliquots were fractionated in sodium dodecyl sulfate (SDS)–12% polyacrylamide gels; proteins were then electrophoretically transferred to polyvinylidene difluoride membranes and probed with either a monoclonal antibody against the LexA protein or a polyclonal antiserum against the HA1 (hemagglutinin) epitope tag.
In vitro DNA binding assays.
A 65-bp synthetic DNA, representing one STB repeat element, was obtained by hybridization of two complementary deoxyoligonucleotides and used as the substrate for the in vitro binding reactions. The Shf1 protein tagged with His-6 was expressed in E. coli from the pTrcHis vector (Invitrogen) and was purified by nickel-affinity chromatography. The protein used in the assays was approximately 50% pure. 32P-end-labeled DNA (ca. 0.02 pmol) was incubated with roughly 0.4 pmol of the Shf1 protein in 50 mM HEPES (pH 7.5), 50 mM KCl, 5 mM EDTA, and 10% glycerol (14) in the presence or absence of competitor DNA at 30°C for 30 min. Binding reactions were analyzed by electrophoresis in 12% polyacrylamide gels (acrylamide to bisacrylamide, 29:1) under nondenaturing conditions. The DNA-protein complexes were visualized by autoradiography or phosphorimaging.
RESULTS
Cellular localization and functional competence of deletion derivatives of Rep1 and Rep2 proteins.
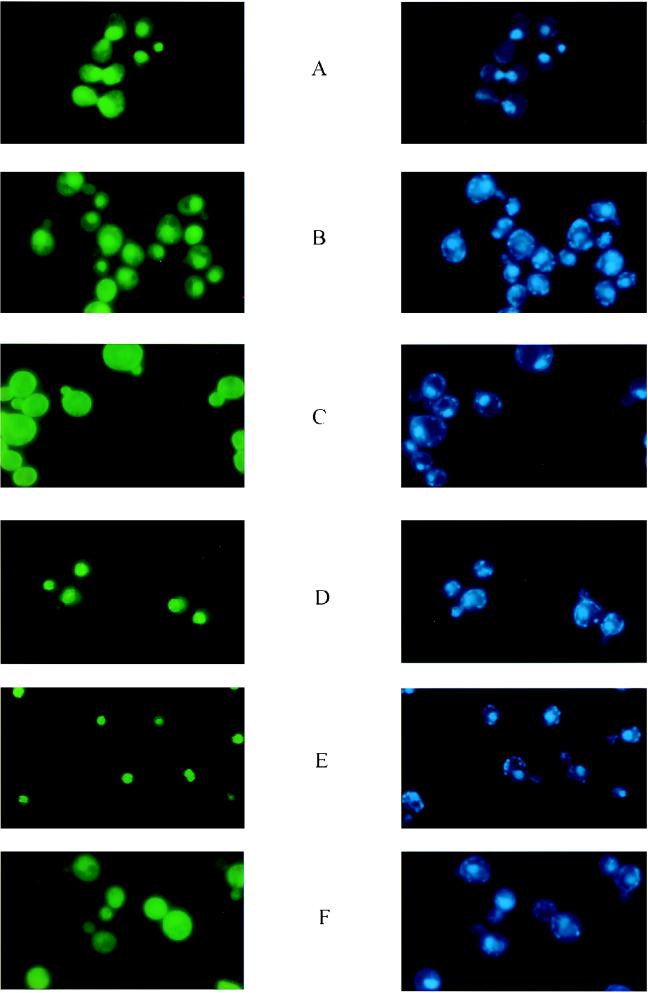
Given that the 2μm plasmid exists primarily in the yeast nucleus (with perhaps only occasional excursions outside of it), it is reasonable to suspect that the logical site of action of the proposed Rep1p-Rep2p complex would be the nucleus. Our recent results that the green fluorescence form of GFP-Rep fusions was primarily localized in the nucleus is consistent with this hypothesis (1). The pattern of fluorescence, though, was distinct for Rep1p and Rep2p. Whereas the GFP-Rep1p fluorescence spilled over weakly into the cytoplasm (Fig. 1A), the GFP-Rep2p fluorescence was much more localized and was confined virtually exclusively to the nucleus in the majority of the cells (Fig. 1D). Relatively large amino-terminal deletions of 125 amino acid residues in Rep1p or 150 amino acids in Rep2p did not affect the compartmentation of the corresponding GFP fusions (Fig. 1B and E, respectively). In sharp contrast, very short carboxy-terminal deletions of 25 amino acids in Rep1p (Fig. 1C) or 20 amino acids in Rep2p (Fig. 1F) abolished the nuclear association of fluorescence from these GFP fusion derivatives. The patterns shown in Fig. 1C and F were indistinguishable from the fluorescence pattern seen with GFP alone. The results obtained with all of the Rep deletions tested are summarized in Table 1.
FIG. 1.
Fluorescence localization from GFP-Rep1p and GFP-Rep2p in [cir+] and [cir0] cells. The cells were prepared for microscopy after 4 h of induction of the GFP-Rep hybrid proteins by transfer to galactose medium as described by Ahn et al. (1). For each pair of panels A through F, the green fluorescence from GFP-Rep proteins is displayed on the left; the blue nuclear fluorescence from the DAPI-DNA is displayed on the right. Panels A and D represent full-length Rep1p and Rep2p, respectively. Panels B and E represent 125- and 150-amino-acid deletions from the amino termini of Rep1p and Rep2p, respectively. These proteins are referred to here as Rep1pΔN125 and Rep2pΔN150. Panels C and F represent deletions of 25 and 20 amino acids from the carboxy termini of Rep1p and Rep2p, respectively (Rep1pΔC20 and Rep2pΔC25). The results presented here are for the longest amino-terminal and shortest carboxy-terminal deletions of each of the two Rep proteins. The fluorescence profiles for the other deletions tested are summarized in Table 1.
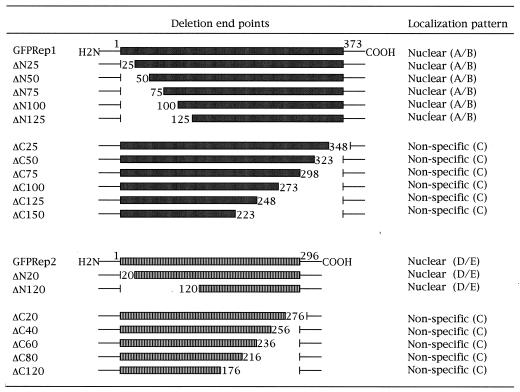
TABLE 1.
Cellular localization of hybrid proteins formed between GFP and Rep deletionsa
The deleted portions of Rep1p or Rep2p are indicated by the blanks in their schematic representations. All of the deletion derivatives were expressed as hybrid proteins fused to the carboxy terminus of GFP. The numbers indicate the extent of the deletions; for example, ΔN125 refers to an amino-terminal deletion that includes the amino acid residue at position 125. The patterns of localization were characterized as A through F, with reference to the profiles shown in Fig. 1.
We have previously shown that GFP-Rep1p and GFP-Rep2p can substitute for the corresponding native proteins in plasmid partitioning with reasonable efficiency. These stability assays were done with pSTB1 and pSTB2, two related plasmids that contain the 2μm circle replication origin, the STB locus, the yeast ADE2 gene, and a second yeast marker (LEU2). Whereas pSTB1 harbored the 2μm circle REP1 gene, pSTB2 harbored the REP2 gene. In these plasmids, expression from the REP1 and REP2 genes was controlled by their native promoters. In a [cir0] ade2 strain, pSTB1 or pSTB2 was lost rapidly under nonselective growth conditions, giving rise to ade2 cells at a very high frequency (easily identified as red colonies or white colonies with large red sectors). In the [cir+] strain, the loss rate was reduced significantly as a result of complementation by the REP1 or REP2 gene function provided by the resident 2μm circle (Table 2).
To test the functional competence of the Rep deletions, we assayed the stabilities of pSTB1 in a [cir0] host that harbored pGFP-Rep2p or its deletion derivatives. Similarly, the stabilities of pSTB2 were assayed in the same host in the presence of pGFP-Rep1p and the deletion constructs obtained from it. The results are summarized in Table 2. Note that, since the GFP plasmids contained a centromere, they were not lost at any appreciable rate even under nonselective growth conditions. A marked drop in plasmid stability was observed with all of the amino-terminal deletions of Rep1p and Rep2p tested. This result suggests that the amino-terminal domains of the Rep proteins are critical for their activity, even though these peptide regions are apparently dispensable in localizing them to the nucleus, the presumed cellular site at which they function. None of the carboxy-terminal deletions of Rep1p or Rep2p assayed were able to sustain the stable propagation of the test plasmids. Either the extreme carboxy-terminal regions of the Rep proteins are directly required for expressing the stability function, or they are required indirectly for steering the Rep proteins to their proper cellular destination.
Restoration of localization and of function in carboxy-terminal Rep deletions by the addition of an NLS sequence.
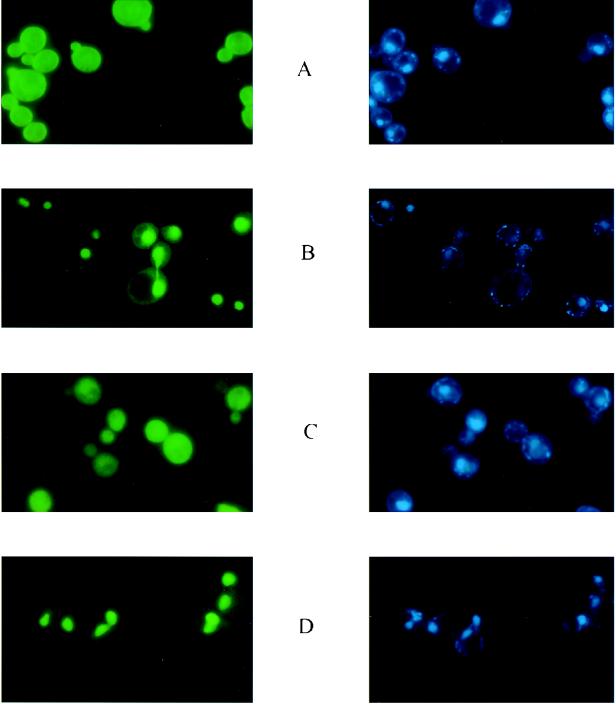
We tested whether the carboxy-terminal deletions of Rep1p and Rep2p can be targeted to the nucleus by the addition of a synthetic heptapeptide derived from the SV40 T antigen that is known to act as an NLS in yeast cells. The results shown in Fig. 2 demonstrate that the two shortest deletions (Rep1ΔC25 and Rep2ΔC20), when provided with the NLS at their carboxy terminus, were efficiently transported to the nucleus. Identical results were obtained when the NLS was placed at the amino terminus of each of these two proteins (data not shown). Furthermore, once they were escorted to their normal cellular locale, both Rep1ΔC25-NLS and Rep2ΔC20-NLS were able to elevate the stability of pSTB2 and pSTB1, respectively, in a [cir0] background (Table 3).
FIG. 2.
Localization of short carboxy-terminal deletions of Rep1p and Rep2p when provided with the NLS from SV40 T antigen. The fluorescence patterns of Rep1pΔC25 and Rep2pΔC20 are shown in panels A and C, respectively, for reference. Panels B and D illustrate the localization of the same deletions when a heptapeptide NLS from an SV40 T antigen was fused to their carboxy termini (B, Rep1pΔC25-NLS; D, Rep1pΔC20-NLS). When the NLS was fused to the amino termini of these deletions, the fluorescence localization was indistinguishable from that shown in panels B and D (data not shown).
TABLE 3.
An exogenous NLS can functionally rescue carboxy-terminal deletions of Rep1p and Rep2p in plasmid maintenancea
| Rep source | Host | Test plasmid | SI | Cellular localization |
|---|---|---|---|---|
| Rep1p | ||||
| pGFP-Rep1p | [cir0] | pSTB2 | 62 | Nuclear |
| Δ25 | [cir0] | pSTB2 | <10 | Nonspecific |
| ΔC25-NLS | [cir0] | pSTB2 | 70 | Nuclear |
| NLS-ΔC25 | [cir0] | pSTB2 | 70 | Nuclear |
| Rep2p | ||||
| pGFP-Rep2p | [cir0] | pSTB1 | 65 | Nuclear |
| ΔC20 | [cir0] | pSTB1 | <10 | Nonspecific |
| ΔC20-NLS | [cir0] | pSTB1 | 72 | Nuclear |
| NLS-ΔC20 | [cir0] | pSTB1 | 72 | Nuclear |
The stability assays were carried out in a [cir0] host as described in Table 2. The NLS prefix indicates fusion of this sequence at the amino terminus, and the NLS suffix indicates fusion to the carboxy terminus. The cellular localization of the deletion protein with and without the NLS is indicated.
Thus, we conclude that the peptide segments comprising the carboxy-terminal extremities of Rep1p and Rep2p are not directly involved in mediating plasmid partitioning. They affect Rep protein function indirectly by their role in targeting these proteins to the proper subcellular location.
Interactions among Rep1p and Rep2p and their deletion derivatives assayed in vivo in yeast cells.
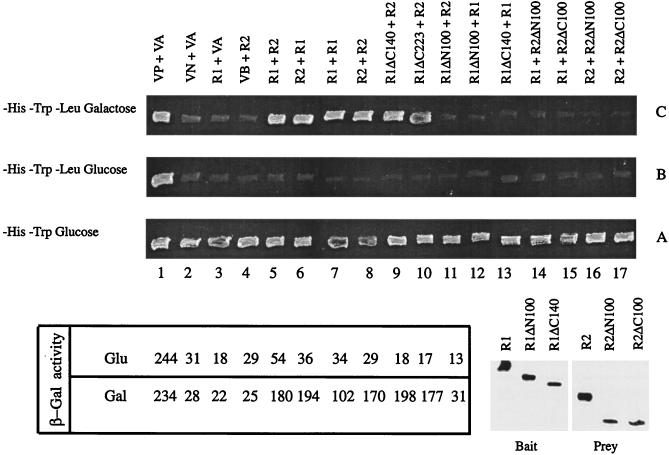
By using the two-hybrid transcriptional activation assay (8), we tested the interactions among pairwise combinations of full-length Rep proteins and a subset of their deletion derivatives. The “bait” protein, a hybrid between LexA and Rep (or a Rep protein containing a deletion), was expressed constitutively from the yeast ADH promoter harbored by an HIS3 plasmid. The prey, a fusion between Rep (or a Rep protein containing a deletion) and a transcriptional activation domain, was expressed from the inducible GAL1 promoter contained within a TRP1 plasmid. The reporter cassette was a chromosomally integrated copy of the yeast LEU2 gene placed under the control of the LexA operator DNA serving as the UAS (upstream activating sequence). The simultaneous presence of the plasmids expressing the bait and the prey was verified for all of the pairwise combinations, as indicated by the growth of the tester strain in SD medium lacking histidine and tryptophan (Fig. 3, row A). The labels above each column in Fig. 3 represent the two proteins under scrutiny (indicated as the bait [LexA fusion] “+” the prey [the activation domain fusion] above each column). A growing patch in row C (LEU2 reporter; SD medium lacking histidine, tryptophan, and leucine-galactose) indicated a positive interaction between a pair of proteins being tested. Note that, whereas the LexA hybrid would be constitutively expressed (from the ADH promoter), the fusion protein containing the transcriptional activator would be induced only in the presence of galactose (from the GAL1 promoter). As glucose would repress expression of the latter, row B should not support growth. Thus, growth in row C and no growth in row B provided the critical criterion for interaction. The positive control used here, a direct fusion of the activation domain to LexA (i.e., column 1), was an exception to this rule. Since this protein was constitutively made from the ADH promoter, the growth of the host strain in rows C and B (column 1) was expected.
FIG. 3.
In vivo baiting assays with Rep1p and Rep2p or their GFP fusion derivatives. Abbreviations: R1, Rep1p; R2, Rep2p; VB, vector containing LexA without Rep fusion; VA, vector containing the transcriptional activation domain without Rep fusion; VP, positive control vector in which the constitutive ADH promoter controlled the expression of the LexA-GAL4 fusion; VN, negative control vector in which LexA fused to a transcriptionally inert protein, Drosophila bicoid, was expressed from the GAL1 promoter. For a given binary protein combination, the protein listed before the plus sign was the bait (fused to LexA), and the protein following the plus sign was the suspected prey (fused to the activation domain). The β-galactosidase units shown in the lower panel were obtained in a variation of the dihybrid assay by using lacZ as the reporter gene. The Western blots (lower right panel) were probed with a monoclonal LexA antibody for the bait proteins and with a polyclonal antiserum to the HA1 epitope for the prey proteins.
The previously established positive interactions between Rep1p and Rep2p (1) were reproduced here for reference (columns 5 and 6). Similarly, by using the same protein as both the bait and the prey, the self-interactions of Rep1p (column 7) and of Rep2p (column 8) were also duplicated here (1). The key results from the cumulative data assembled in Fig. 3 are summarized below. An amino-terminal deletion of 100 amino acids in Rep1p resulted in a loss of self-interaction (column 12), as well as a loss of interaction with Rep2p (column 11). On the other hand, a carboxy-terminal deletion of 140 amino acids in Rep1p resulted in the retention of Rep2p interaction (column 9), but it caused the loss of interaction with full-length Rep1p (column 13). The deletion proteins obtained by removing 100 amino acids from the amino- or the carboxy-terminus of Rep2p failed to interact with either Rep1p (columns 14 and 15) or Rep2p (columns 16 and 17). Finally, a fusion between the amino-terminal 150 residues of Rep1p to LexA (equivalent to a carboxy-terminal deletion of 223 amino acids) was capable of interacting with Rep2p (column 10). These results were consistent with those obtained from a similar assay with a lacZ reporter cassette (the β-galactosidase activity measured for a given protein pair is indicated under the appropriate column in the left lower panel of Fig. 3).
We have verified that the negative results in the interaction assay were not due to a lack of expression of either the bait or the prey (lower right panel of Fig. 3). The bait proteins (fused to LexA) and the prey proteins (containing the HA1 epitope tag) were probed with antibodies to LexA and HA1, respectively. Thus, Rep1ΔN100 and Rep1ΔC140 were expressed as baits, although neither one was able to trap the full-length Rep1p (Fig. 3, row C, columns 12 and 13). Similarly, Rep2ΔN100 and Rep2ΔC100 were expressed as preys, but they were not baited by Rep1p or Rep2p (row C, columns 14 to 17). The full-length Rep1p (fused to LexA) and the full-length Rep2p (fused to the activation domain) were included in the Western blot assays for reference purposes.
We conclude that the 150-amino-acid polypeptide from the amino terminus of Rep1p is sufficient to establish association with Rep2p in vivo in yeast cells. However, this polypeptide alone cannot mediate Rep1p-Rep1p interactions. The interactions for the self-association likely require peptide contributions from both the amino- and the carboxy-terminal regions. In the case of Rep2p, self-association or association with Rep1p is dependent on the integrity of the peptide domains at either end of the protein.
Interactions of the Rep protein deletions assayed in vitro.
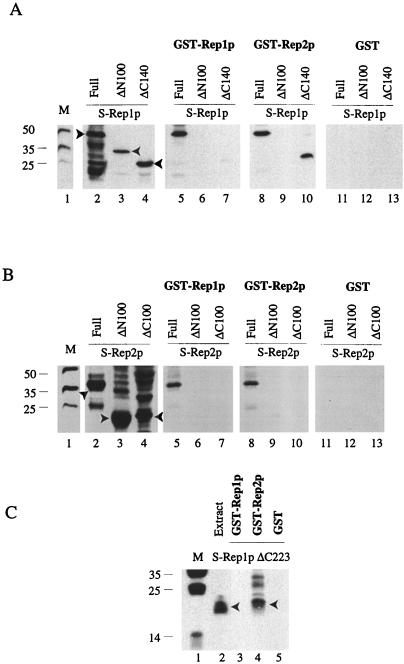
The results from the in vivo assays in yeast cells were verified by a GST-Rep hybrid baiting assay with proteins expressed in E. coli (1). In this analysis, GST-Rep1p or GST-Rep2p was used to trap S-tagged Rep1p, Rep2p, or their deletions from supernatants of the bacterial cell extracts containing them. The results of these baiting assays are displayed in Fig. 4. The electrophoretically fractionated samples were probed with S protein to reveal Rep1p, Rep2p, or their deletions harboring this tag. The presence of each of the S-tagged proteins in the appropriate E. coli extracts used for baiting is shown for reference (Fig. 4A and B, lanes 2, 3, and 4; Fig. 4C, lane 2). When GST-Rep1p was used as the bait, S-Rep1p or S-Rep2p could be pulled down from the respective supernatants (Fig. 4A and B, lanes 5). Similar results were obtained in the reciprocal experiment with GST-Rep2p as the bait (Fig. 4A and B, lanes 8). These results are in conformity with the previous observations of Ahn et al. (1). In the case of the S-tagged deletion proteins, positive interaction was observed between GST-Rep2p and Rep1pΔC140 (Fig. 4A, lane 10). Furthermore, the S-tagged amino-terminal 150-amino-acid peptide from Rep1p (S-Rep1pΔC223) was able to interact with GST-Rep2p (Fig. 4C, lane 4), but not with GST-Rep1p (lane 3). Control assays with GST alone failed to trap any of the S proteins tested (Fig. 4A and B, lanes 11 to 13; Fig. 4C, lane 5).
FIG. 4.
Baiting assays for Rep1p and Rep2p and their deletions performed with GST-Rep1p and GST-Rep2p hybrid proteins. The assays were carried out as described by Ahn et al. (1). After baiting, proteins were fractionated in SDS–12% polyacrylamide gels and probed with the S-protein probe. The lane marked M displays the molecular-weight standards. (A and B) In lanes 2 to 4, proteins extracted by boiling the cells from 200-μl portions of the induced cultures (that were also the starting materials for the assays depicted in lanes 5 to 13) in the SDS sample buffer were run as controls. The S-Rep protein bands of interest in lanes 2 to 4 are indicated by the arrowheads. The data in panel A were obtained with S-tagged Rep1p or its derivatives; the data in panel B were obtained with S-tagged Rep2p or its derivatives. The GST-Rep hybrids used as the baits and the corresponding baited S-Rep proteins (either full length or partially deleted) are indicated above lanes 5 to 10. In the reactions represented in lanes 11 to 13, the baiting was performed with GST and not with the GST-Rep hybrids. (C) Results obtained with Rep1pΔC223 fused to the S-peptide tag. Lane 2 represents the extract from 200 μl of the induced culture expressing Rep1pΔC223; lanes 3 to 5 correspond to baiting assays with GST-Rep1p, GST-Rep2p, and GST, respectively. Detailed protocols of the assays are described in Ahn et al. (1).
Thus, the in vitro assays corroborate the in vivo results that the peptide region responsible for Rep1p-Rep2p interaction maps to the amino terminus of Rep1p. In addition, the in vitro data imply that this interaction is not necessarily dependent upon chromosomally encoded yeast proteins or other plasmid-encoded proteins. Whether the interaction may be modulated by such proteins in a physiologically relevant manner remains to be tested.
Rep1p, Rep2p, and a host-encoded protein interact with the STB locus in vivo.
It has been hypothesized that the STB locus must directly or indirectly interact with the Rep proteins in mediating plasmid stability. The evidence in support of this notion is circumstantial at best. It is clear that the trans-acting Rep proteins and the cis-acting STB locus are functionally tightly interrelated, since mutations in any one of the three loci (REP1, REP2, or STB) lead to the same phenotype, namely, plasmid instability. Preliminary in vitro binding studies (14) have suggested that Rep1p and Rep2p can bind to STB but only in association with a host factor (or factors).
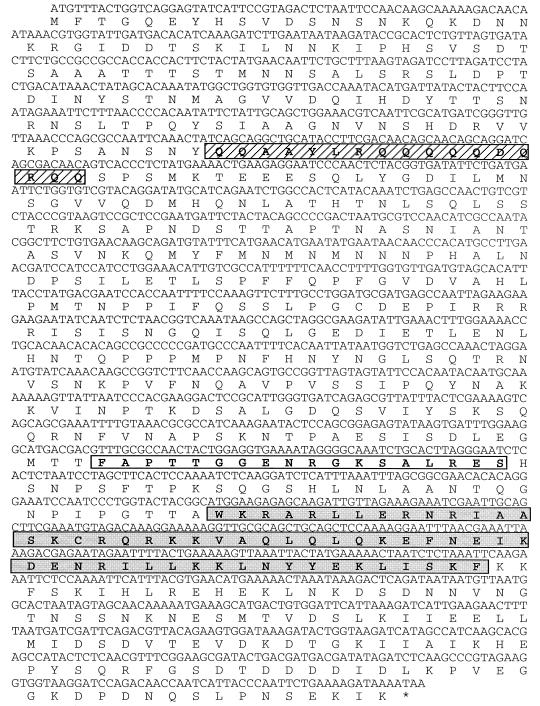
We have therefore searched for STB-binding protein factors in yeast cells by using the monohybrid positive-selection method. The assay was standardized (see Materials and Methods) to establish conditions under which the growth of a yeast colony in the presence of a titrated amount of 3-AT was contingent upon the enhanced transcriptional activation of the HIS3 reporter gene via STB-protein interaction. Under the stringency conditions applied here, only three types of clones were obtained by this procedure. Two of them were the REP1 and REP2 genes, and the third was the as-yet-uncharacterized chromosomal gene YIL036W (designation in the yeast genome bank). The frequencies of REP1, REP2, and YIL036W among the more than 80 positive clones obtained after three rounds of 3-AT selection were approximately 12, 32, and 56%, respectively. The nucleotide sequence of YIL036W, which we designate as SHF1 (STB binding host factor), and the derived amino acid sequence are shown in Fig. 5. Certain interesting structural motifs could be gleaned in the protein (Shf1p) sequence. For example, the peptide region from amino acids 377 to 394 (the open rectangular box in Fig. 5) was suggestive of GTP or ATP binding (as predicted by the UCLA-DOE Structure Prediction Server). Similarly, the peptide stretch from amino acids 421 to 454 (shaded box in Fig. 5; see also Fig. 6) showed strong homology to the cyclic AMP responsive element binding (CREB) motif found in the activating transcription factor (ATF)/CREB family of transcriptional regulators containing the bZIP domain (Fig. 6). Furthermore, there was a significantly high concentration of glutamine residues (65%) in the segment spanning positions 118 through 134 (hatched box in Fig. 5).
FIG. 5.
Chromosomally encoded yeast protein Shf1p that interacts with the STB locus. The monohybrid assay identified the STB-interacting host protein as the product of the YIL036W locus (from the yeast genome bank; now designated as SHF1). The complete nucleotide sequence of the genetically and biochemically uncharacterized SHF1 gene, along with the amino acid sequence of the presumed protein product encoded by it, are shown. The rectangular box (from Phe-377 to Ser-294) indicates a potential GTP or ATP binding sequence (prediction by the UCLA-DOE structure prediction server). The shaded box (from Trp-421 to Phe-454) is highly homologous to the consensus CREB motif present in the ATF/bZIP family of transcriptional regulatory proteins. (For a comparison of this region to homologous regions in a selected set of eukaryotic CREB/bZIP proteins, see Fig. 6.) The abundance of glutamine residues in the hatched box (Gln-118 to Gln-135) is suggestive of the activation patches in a number of transcription factors (6, 7).
FIG. 6.
The CREB motif in the STB-interacting yeast protein Shf1p shows strong homology to the basic and zipper regions of transcriptional regulators from eukaryotes. In the sequence alignment of the Trp-421 to Phe-454 region of Shf1p with similar regions from other eukaryotic ATF/bZIP proteins, amino acid identities or strong conservations are indicated in boldface. The GCN4 and ACR1 proteins are from S. cerevisiae. The MTS proteins are from Schizosaccharomyces pombe, and the ATF series of proteins, CREM and TGA1, are from other organisms. The numbers in parentheses indicate the relevant references.
The fact that the genetic selection employed here yielded Rep1p and Rep2p as two of the three STB-interacting proteins is satisfying. It bolsters our confidence in the assay, and it provides the first strongly suggestive in vivo evidence for this long-speculated DNA-protein interaction. Furthermore, we show below that the Shf1 protein binds directly to STB and that its absence in vivo affects the stability of a 2μm circle-derived plasmid. Note that the in vivo interaction in yeast cells between Shf1p and STB is independent of Rep1p or Rep2p, since the selective procedure was done in the [cir0] strain. By the same reasoning, interaction of Rep1p or Rep2p with STB must also be independent of each other. However, our assays do not reveal whether Shf1p is essential for the interaction of either of the two Rep proteins with STB.
The Shf1 protein binds to the STB locus in vitro.
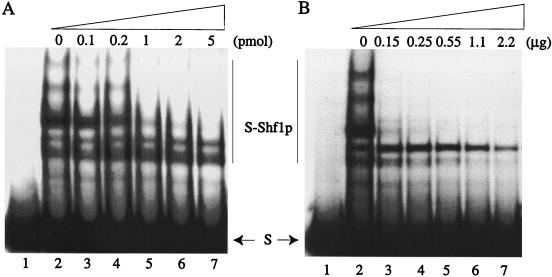
The results from in vitro assays for the binding of Shf1p to the STB locus are shown in Fig. 7. In the 2μm plasmid, the STB sequence is present as a tandem, directly oriented array of 5 to 6 U of a 65-bp consensus element. The binding reactions were done by using a single repeat unit as the substrate. Association between Shf1p and the 65-bp STB element yielded a series of DNA-protein complexes (Fig. 7, lanes 2). This finding suggests that Shf1p may bind to DNA as oligomeric units or that, alternatively, the Shf1p-bound STB elements may associate with each other to produce the hierarchical binding pattern. No specific complexes were observed when binding was done with crude extracts (or extracts fractionated over the nickel column) from IPTG (isopropyl-β-d-thiogalactopyranoside)-induced E. coli cells harboring the expression vector without the SHF1 clone (data not shown). The specificity of Shf1p-STB binding was tested by challenge with non-STB DNA: a 60-bp deoxyoligonucleotide harboring the target site for the 2μm plasmid encoded Flp recombinase (Fig. 7A, lanes 3 to 7) or salmon sperm DNA (Fig. 7B, lanes 3 to 7). At a ca. 5 or 10 M excess of the Flp binding sequence over STB (0.02 pmol per reaction), little or no competition was observed (Fig. 7A, lanes 3 and 4). As the concentration of the competitor was increased, the yield of the higher-order complexes diminished (lanes 5 to 7). However, even at a competitor/STB molar ratio of 250:1, binding was not abolished (lane 7). Comparable results were obtained when salmon sperm DNA was used as the competitor (Fig. 7B). The pattern of competition obtained with 0.15 μg of salmon sperm DNA (equivalent to ca. 5 to 6 pmol of a 60-bp-long DNA) was roughly equivalent to that seen with similar amounts of the Flp DNA target (Fig. 7B, compare lanes 3 and 7). Binding was still evident when the molar ratio of salmon sperm DNA to STB was raised to approximately 4,000:1 (lane 7).
FIG. 7.
Binding of Shf1p in vitro to the STB DNA elements. Approximately 0.02 pmol of the STB DNA (32P-labeled at the 5′ end) was incubated with Shf1p (ca. 0.4 pmol) as described in Materials and Methods. Binding reactions were fractionated by electrophoresis in 12% nondenaturing polyacrylamide gels. For both panels, reactions in lane 1 were controls without the addition of Shf1p; those in lane 2 did not contain competitor DNA. Reactions in lanes 3 to 7 contained the indicated amounts of the competitor DNA: a 60-bp synthetic oligonucleotide in panel A and salmon sperm DNA in panel B. The unbound substrate is marked (S). The bound complexes are denoted by S-Shf1p.
The highly discriminatory (and presumably direct) binding of Shf1p to the STB DNA validates the in vivo assay by which this DNA-protein interaction was inferred initially. We do not know what peptide regions of Shf1p are necessary (and sufficient) for binding to STB. From the sizes of the several independent clones of SHF1 isolated by the 3-AT selection assay in vivo, we surmise that they encode full-length or nearly full-length Shf1p.
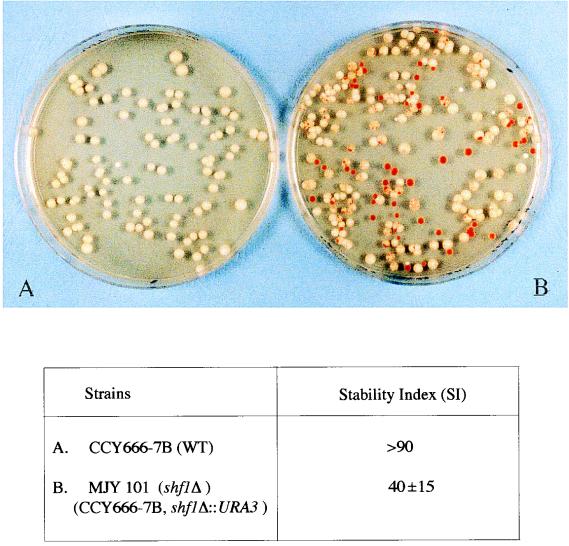
Plasmid stability in a host strain harboring a deletion of the SHF1 locus.
Does the lack of SHF1 function affect the stability of a 2μm circle-derived test plasmid? To address this issue, the maintenance of plasmid pSTB (harboring the 2μm plasmid replication origin and the STB locus) was followed in a [cir+] SHF1 strain (Fig. 8A) and a derivative strain lacking this locus (Fig. 8B) (see Materials and Methods for details). The plasmid loss rate was significantly higher in the shf1 deletion strain, as indicated by the increase in the number of red colonies (indicating the loss of the plasmid-borne ADE2 marker). The results are also expressed as the relative SI (Fig. 8). When the red colonies were transferred to SD plates lacking leucine, they did not grow, thus verifying the simultaneous loss of the ADE2 and LEU2 markers carried on the plasmid pSTB.
FIG. 8.
Maintenance of a 2μm circle-derived plasmid in a host strain deleted for the SHF1 locus. (Top) The stability of the test plasmid pSTB (harboring the ADE2 and LEU2 markers) was assayed as described in Materials and Methods. The host strains (wild type for SHF1 in panel A and shf1Δ in panel B) containing pSTB were grown nonselectively for approximately 10 generations and plated on YEPD plates to assay plasmid loss by colony color. (Bottom) The results from four separate transformants are expressed as the SI as follows: SI = (the number of white colonies/total number of red and white colonies) × 100. A sectored colony was scored as red if the red sector was one-fourth the colony size or larger; otherwise, it was counted as white.
We therefore conclude that the host factor Shf1p plays a role in the stable propagation of the 2μm plasmid in yeast cells. At this time we do not know whether the effect of SHF1 on plasmid stability is direct or indirect, the effect mediated perhaps by its role in controlling the expression and/or activity of the Rep proteins.
DISCUSSION
The 2μm circle plasmid in yeast cells employs a dual strategy for its stable propagation as a benign parasite genome: efficient plasmid partitioning and copy number amplification. Genetic experiments (16, 19, 24, 25, 28) have given rise to a model in which the components of the stability system provide a measure of the plasmid copy number and contribute to the transcriptional regulation of the amplification system (see Fig. 9). Previous experiments (1) have substantiated one central aspect of the model: the assumption that REP1 and REP2 gene products, the two plasmid-coded trans-acting components of the stability system, must physically interact. The analyses of Ahn et al. revealed that the two proteins not only interact in vivo in yeast cells and are targeted to the nucleus but also associate with each other in vitro in the absence of any other yeast proteins. We have now localized the nuclear targeting sequences within Rep1p and Rep2p, functionally replaced them with an exogenous NLS, and mapped (by in vivo and in vitro assays) the peptide regions responsible for Rep protein associations. Furthermore, our studies provide a molecular framework for examining the role of the cis-acting STB locus and of host factors in conferring plasmid stability.
FIG. 9.
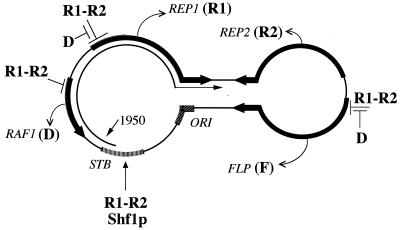
Model for protein-protein and DNA-protein interactions in the high-copy maintenance of the 2μm plasmid. The plasmid is shown in its standard representation, with the parallel lines indicating the 599-bp inverted repeats. The results of early analyses, primarily genetic assays, suggested a potential bipartite Rep1p-Rep2p (R1-R2) regulator that represses transcription from the REP1, FLP, and RAF1 promoters. Direct evidence for the interaction between Rep1p (R1) and Rep2p (R2) in vivo and in vitro was obtained recently (1). The notion that interaction between the cis-acting STB locus and the Rep proteins may be required to promote plasmid partitioning at cell division has been entertained and has received limited experimental support (14). The present study presents evidence for the binding of Rep1p, Rep2p, and a host protein Shf1p to the STB locus. There is some indication that product of the RAF1 gene (D) may antagonize the Rep1p-Rep2p-mediated transcriptional repression. This may be a primary step in triggering copy number amplification of the plasmid. The product of the FLP gene (F) is a site-specific recombinase that is essential for the amplification mechanism (11, 33). The plasmid replication origin is denoted by ORI. The 1,950-nucleotide 2μm circle transcript that originates at STB and spans the REP1 locus is indicated. The diagram, adapted from Ahn et al. (1), summarizes contributions from several laboratories and includes the results of this study (1, 3, 9, 14, 16, 19, 24, 25, 28, 33).
The carboxy-terminal regions of the Rep proteins are essential for their localization.
The cytological assays with GFP and hybrid proteins have revealed that the nuclear compartmentation signals in the Rep proteins are located at the very carboxy-terminal ends of these proteins. Deletions as short as 25 and 20 amino acids from the carboxy-terminal end (of Rep1p and Rep2p, respectively) are sufficient to delocalize them in the cell and to render them nonfunctional in plasmid maintenance. Their capacity for nuclear homing and their activity in plasmid maintenance can be fully restored by an NLS sequence derived from the SV40 T antigen placed either at the amino terminus or at the carboxy terminus. There is some qualitative difference between the patterns of nuclear localization of native Rep1p and the deletion Rep1p harboring the SV40 NLS (compare Fig. 2B to Fig. 1A). This difference, though, is apparently irrelevant to the role of Rep1p in plasmid stability.
Mapping of the Rep1p-Rep2p interaction domains.
The in vivo dihybrid assays and the in vitro GST fusion baiting assays described here have shown the amino-terminal portion of Rep1p consisting of 150 amino acids to be sufficient for interacting with Rep2p. Among the Rep protein deletions in our collection, we have not seen any that have retained the ability for self-interaction. Nor have we identified a deletion of Rep2p that is capable of interacting with the native Rep1p or its deletion variants. It should be clarified that the lack of association between a carboxy-terminal deletion variant of Rep with its full-length version or with the partner Rep (Rep2p in the case of a Rep1p deletion and Rep1p in the case of a Rep2p deletion) cannot be due solely to a lack of the NLS resulting from the deletion. Note that the expression plasmids in the dihybrid system (8) have built-in NLS sequences that are fused in frame to the coding sequences of the proteins being analyzed.
Potential role of host factors in the stable maintenance of the 2μm plasmid.
The isolation of a chromosomally encoded yeast protein Shf1p in an assay designed to select for proteins that bind to the STB locus suggests the potential involvement of a host factor (or host factors) in the persistence of the 2μm plasmid as a benign parasite genome in yeast cells. Note that the same assay has also identified Rep1p and Rep2p as the other two yeast proteins that interact with STB. The Shf1 protein, expressed in E. coli, binds to the STB locus with high specificity, and a host strain lacking SHF1 is compromised in its capacity to maintain a 2μm circle-derived plasmid. The Shf1 protein could well recruit the Rep proteins to the STB locus. Such an activity would agree with the in vitro results (14) that suggest the involvement of a host factor in the binding of the Rep proteins to STB. Whether there is a direct interaction between the Shf1 protein (selected in the monohybrid–3-AT assay) and one or both of the Rep proteins is not known at present.
It should be pointed out that the monohybrid assay with the STB bait has so far failed to select the Raf1 protein (coded for by the open reading frame D of the 2μm plasmid; see Fig. 9) among the several independent, confirmed positive clones. This is somewhat at odds with the results of the in vitro binding assay that suggest rapid, tight, and direct binding of Raf1p to STB (14). We suspect that the outcome of the in vivo assay likely reflects an underrepresentation of the RAF1 locus in the cDNA library used for the monohybrid selection. Preliminary results with a cloned fusion between Raf1p and a transcriptional activation domain in the monohybrid assay suggest a positive interaction between Raf1p and STB (30a).
Based on the functional motifs identified by homology search (Fig. 6) (a sequence element suggestive of nucleotide binding, a glutamine-rich patch, and a characteristic CREB/basic zipper (bZIP) segment), the Shf1 protein qualifies eminently to be a transcriptional regulator. We know that a LexA-Shf1p hybrid protein by itself can promote the transcription of a LEU2 or LacZ reporter in yeast cells when the promoter for either locus is controlled by an upstream LexA operator DNA (30a). In this context, it is relevant to point out that a major 2μm circle transcript originates at STB and is directed towards the REP1 locus (and opposite to the RAF1 locus) (29; see also Fig. 9). We are now testing the hypothesis that the host factor-mediated transcriptional control plays a direct or indirect role in stable plasmid maintenance.
Summary.
As has been pointed out previously, the circular geometry, structural compactness, and functional parsimony of the 2μm plasmid appear to represent an optimized evolutionary solution for the high-copy maintenance of an extrachromosomal selfish DNA element in yeast cells (2). The general picture of the suspected protein-protein and protein-DNA interactions that contribute to this phenomenon, at least some of which have received experimental support, is given in Fig. 9. In a recent study (1) we established by a variety of independent assays that Rep1p and Rep2p can directly interact with each other. We have now extended the potential significance of this finding by demonstrating the interaction of the STB locus with Rep1p, Rep2p, and the product of the chromosomal gene SHF1. The results of the present study therefore offer new insight into the analysis of the molecular contributions made by the host and the plasmid to this benign parasitism.
ACKNOWLEDGMENT
This work was supported by a grant from the Council for Tobacco Research.
ADDENDUM IN PROOF
Scott-Drew and Murray (S. Scott-Drew and J. A. Murray, J. Cell Sci. 111:1779–1789, 1998) have recently demonstrated that Rep1p and Rep2p form a complex. By confocal microscopy they have further demonstrated that the proteins occupy specific sites in the nucleus and that these sites are distributed to both mother and daughter cells during cell division.
REFERENCES
- 1.Ahn Y T, Wu X L, Biswal S, Velmurugan S, Volkert F C, Jayaram M. The 2μm-plasmid-encoded Rep1 and Rep2 proteins interact with each other and colocalize to the Saccharomyces cerevisiae nucleus. J Bacteriol. 1997;179:7497–7506. doi: 10.1128/jb.179.23.7497-7506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broach J R, Volkert F C. Circular DNA plasmids of yeasts: genome dynamics, protein synthesis and energetics. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1991. pp. 297–331. [Google Scholar]
- 3.Cashmore J R, Albury M S, Hadfield C, Meacock P A. Genetic analysis of partitioning functions encoded by the 2μm circle of Saccharomyces cerevisiae. Mol Gen Genet. 1986;203:154–162. [Google Scholar]
- 4.Chen B P, Liang G, Whelan J, Hai T. ATF3 and ATF3ΔZip. Transcriptional repression versus activation by alternatively spliced isoforms. J Biol Chem. 1994;269:15819–15826. [PubMed] [Google Scholar]
- 5.Chevray P M, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courney A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 7.Courney A J, Holtzman D A, Jackson S P, Tjian R. Synergistic activation by glutamine-rich domains of human transcription factor Sp1. Cell. 1988;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 8.Finley R L, Jr, Brent R. Interaction trap cloning with yeast. In: Glover D, Hames B D, editors. DNA cloning-expression systems: a practical approach. Oxford, United Kingdom: Oxford University Press; 1996. pp. 169–203. [Google Scholar]
- 9.Futcher A B, Cox B S. Maintenance of the 2μm circle plasmid in populations of Saccharomyces cerevisiae. J Bacteriol. 1983;154:612–622. doi: 10.1128/jb.154.2.612-622.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Futcher A B. Copy number amplification of the 2μm circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 11.Futcher A B. The 2μm circle plasmid of Saccharomyces cerevisiae. Yeast. 1988;4:27–40. doi: 10.1002/yea.320040104. [DOI] [PubMed] [Google Scholar]
- 12.Guarente L. Yeast promoters and lacZ fusions designed to study the expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 13.Goetz J, Chatton B, Mattei M G, Kedinger C. Structure and expression of the ATFa gene. J Biol Chem. 1996;271:29589–29598. doi: 10.1074/jbc.271.47.29589. [DOI] [PubMed] [Google Scholar]
- 14.Hadfield C, Mount R C, Cashmore A M. Protein binding interactions at the STB locus of the yeast 2μm plasmid. Nucleic Acids Res. 1995;23:995–1002. doi: 10.1093/nar/23.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinnebusch A G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaram M, Sutton A, Broach J R. Properties of REP3: a cis-acting locus required for stable propagation of the Saccharomyces cerevisiae plasmid 2μm circle. Mol Cell Biol. 1985;5:2466–2475. doi: 10.1128/mcb.5.9.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayaram M, Sumida S, Young L-J. Inducible expression of REP1 causes inducible expression of the 2 micron circle stability system. Curr Genet. 1986;11:85–91. doi: 10.1007/BF00378198. [DOI] [PubMed] [Google Scholar]
- 18.Katagiri F, Lam E, Chua N H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi Y. Yeast plasmid requires a cis-acting locus and two plasmid proteins for its stable maintenance. Cell. 1983;35:487–493. doi: 10.1016/0092-8674(83)90182-4. [DOI] [PubMed] [Google Scholar]
- 20.Lee M-R, Chung C S, Liou M L, Wu M, Li W, Hsueh Y P, Lai M Z. Isolation and characterization of nuclear proteins that bind to T cell receptor V-beta decamer motif. J Immunol. 1992;148:1906–1912. [PubMed] [Google Scholar]
- 21.Masquilier D, Foulkes N S, Mattei M G, Sassone-Corsi P. Human CREM gene: evolutionary conservation, chromosomal localization, and inducibility of the transcript. Cell Growth Differ. 1993;4:931–937. [PubMed] [Google Scholar]
- 22.Murray A W, Szostak J W. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- 23.Murray J A H, Cesareni G. Functional analysis of the yeast plasmid partition locus STB. EMBO J. 1986;5:3391–3399. doi: 10.1002/j.1460-2075.1986.tb04655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray J A H, Scarpa M, Rossi N, Cesareni G. Antagonistic controls regulate copy number of the yeast 2μm plasmid. EMBO J. 1987;6:4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds A E, Murray A W, Szostak J W. Roles of the 2μm gene products in stable maintenance of the 2 μm plasmid of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo F D, Scherson I, Broach J R. Direct simulations of yeast 2 micron circle plasmid amplification J. Theor Biol. 1992;155:369–385. doi: 10.1016/s0022-5193(05)80604-6. [DOI] [PubMed] [Google Scholar]
- 27.Sigurdson D C, Garder M E, Livingston D M. Characterization of the transmission during cytoductant formation of the 2 micron DNA plasmid from Saccharomyces. Mol Gen Genet. 1981;183:59–65. doi: 10.1007/BF00270139. [DOI] [PubMed] [Google Scholar]
- 28.Som T, Armstrong K A, Volkert F C, Broach J R. Autoregulation of 2μm circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- 29.Sutton A, Broach J R. Signals for transcription initiation and termination in the Saccharomyces cerevisiae plasmid 2μm circle. Mol Cell Biol. 1985;5:2770–2780. doi: 10.1128/mcb.5.10.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veit B E, Fangman W L. Copy number and partition of the Saccharomyces cerevisiae 2μm plasmid controlled by transcription regulators. Mol Cell Biol. 1988;8:4949–4957. doi: 10.1128/mcb.8.11.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Velmurugan, S., and M. Jayaram. Unpublished data.
- 31.Villarreal X C, Richter J D. Analysis of ATF2 gene expression during early Xenopus laevis development. Gene. 1995;153:225–229. doi: 10.1016/0378-1119(94)00770-s. [DOI] [PubMed] [Google Scholar]
- 32.Vincent A C, Struhl K. ACR1, a yeast ATF/CREB repressor. Mol Cell Biol. 1992;12:5394–5405. doi: 10.1128/mcb.12.12.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volkert F C, Broach J R. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986;46:541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- 34.Wahls W P, Smith G R. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 1994;14:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- 35.Zakian V A, Brewer B J, Fangman W L. Replication of each copy of the yeast 2 micron DNA plasmid occurs during the S phase. Cell. 1979;17:923–934. doi: 10.1016/0092-8674(79)90332-5. [DOI] [PubMed] [Google Scholar]
- 36.Zhu C, Johansen F E, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]