Abstract
We developed a polarized hyperspectral microscope to collect four types of Stokes vector data cubes (S0, S1, S2, and S3) of the pathologic slides with head and neck squamous cell carcinoma (HNSCC). Our system consists of an optical light microscope with a movable stage, two polarizers, two liquid crystal variable retarders (LCVRs), and a SnapScan hyperspectral camera. The polarizers and LCVRs work in tandem with the hyperspectral camera to acquire polarized hyperspectral images. Synthetic pseudo-RGB images are generated from the four Stokes vector data cubes based on a transformation function similar to the spectral response of human eye for the visualization of hyperspectral images. Collagen is the most abundant extracellular matrix (ECM) protein in the human body. A major focus of studying the ECM in tumor microenvironment is the role of collagen in both normal and abnormal function. Collagen tends to accumulate in and around tumors during cancer development and growth. In this study, we acquired images from normal regions containing normal cells and collagen fibers and from tumor regions containing cancerous squamous cells and collagen fibers on HNSCC pathologic slides. The preliminary results demonstrated that our customized polarized hyperspectral microscope is able to improve the visualization of collagen on HNSCC pathologic slides under different situations, including thick fibers of normal stroma, thin fibers of normal stroma, fibers of normal muscle cells, fibers accumulated in tumors, fibers accumulated around tumors. Our preliminary results also demonstrated that the customized polarized hyperspectral microscope is capable of extracting the spectral signatures of collagen based on Stokes vector parameters and can have various applications in pathology and oncology.
Keywords: Hyperspectral imaging, polarized hyperspectral imaging, polarized light imaging, Stokes vector, head and neck cancer, collagen
1. INTRODUCTION
Head and neck squamous cell carcinoma (SCC) is originated from the mucosal epithelium in the oral cavity, pharynx and larynx and is a major head and neck malignancy [1]. Computational pathology, also known as digital pathology, is an emerging technology that promises quantitative diagnosis of pathological samples. Traditional computational pathology relies on RGB digitized histology images [2]. Multidimensional optical imaging has grown rapidly in recent years. Rather than measuring only the two-dimensional spatial distribution of light as in the conventional photography, multidimensional optical imaging captures unprecedented information about photons’ spatial coordinates, emittance angles, wavelength, time, and polarization [3].
Hyperspectral imaging (HSI) is an optical imaging method that was originally used in remote sensing, and it has been extended to the applications in several other promising fields including biomedical applications [4]. Hyperspectral imaging acquires the spectra at every pixel in a two-dimensional (2D) image and constructs a three-dimensional (3D) data cube, where rich spatial and spectral information can be obtained simultaneously. Hyperspectral imaging has been implemented on the detection of head and neck cancer [5–17]. Yushkov et al [5] developed an acoustic-optic hyperspectral imaging Polarized Light and Optical Angular Momentum for Biomedical Diagnostics 2023, edited by Jessica C. Ramella-Roman, Hui Ma, Tatiana Novikova, Daniel S. Elson, I. Alex Vitkin, Proc. of SPIE Vol. 12382, 1238204 system with an amplitude mask, which improved the contrast for phase visualization in the stained and unstained histological sections of human thyroid cancer. A pilot study was implemented to test the feasibility of a hyperspectral imaging system for in vivo delineation of preoperative margins of ill-defined basal-cell carcinoma (BCC) on the head and neck region [6]. Our group has investigated several machine learning algorithms for head and neck cancer detection based on hyperspectral imaging, including principal component analysis (PCA) [7], tensor-based computation and modeling [8], the combination of support vector machine (SVM) and minimum spanning forest methods [9, 10], non-negative matrix factorization (NMF) [11], the combination of super pixels, PCA, and SVM [12], as well as convolutional neural networks (CNN) [13, 14, 15, 16, 17].
Polarized light imaging is an effective optical imaging technique to explore the structure and morphology of biological tissues through obtaining their polarization characteristics. It can acquire the 2D spatial polarization information of the tissue, which reflects various physical properties of the tissue, including surface texture, surface roughness, and surface morphology information [18, 19, 20, 21, 22]. The categories of polarized light imaging techniques, namely linear polarization imaging [23, 24, 25], Muller matrix imaging [26, 27], and Stokes vector imaging [28], have been applied on head and neck cancer detection. An orthogonal polarization spectral (OPS) imaging method, which is a type of linear polarization imaging method, was implemented for the evaluation of anti-vascular tumor treatment and oral squamous cell carcinoma on tissue [23, 24]. A multispectral digital microscope (MDM) with an orthogonal polarized reflectance (OPR) imaging mode was developed for in vivo detection of oral neoplasia [25]. A 4×4 Muller matrix imaging and polar decomposition method were applied for diagnosis of oral precancer [27]. Researchers also adopted a 3×3 Muller Matrix imaging method for oral cancer detection [26]. In our previous study, we developed a polarized hyperspectral imaging microscope, which is able to distinguish squamous cell carcinoma from normal tissue on hematoxylin and eosin (H&E) stained slides from larynx based on the spectra of Stokes vector [28].
Polarized hyperspectral imaging (PHSI) is a combination of polarization measurement, hyperspectral analysis, and space imaging technology. It can obtain the polarization, spectral and morphological information of the object simultaneously [29, 30, 31]. We developed a novel dual-modality optical imaging microscope by combining hyperspectral imaging and polarized light imaging. The microscope is capable of acquiring polarization, spectral and spatial information of an object simultaneously, and thus provides more image information for digital pathology compared to RGB digitized histology images.
The tumor microenvironment consists of multiple biochemical, mechanical, and structural signals. One of the major structural components of the tumor microenvironment is the extracellular matrix (ECM). Collagen is the most abundant ECM protein in the human body. A major focus of studying the ECM is the role of collagen in both normal and abnormal function [37]. Collagen tends to accumulate in and around tumors during cancer development and growth [38]. In this paper, we use polarized hyperspectral microscopic imaging to visualize collagens in normal regions containing normal squamous cells and collagen fibers and from the tumor regions containing cancerous squamous cells and collagen fibers on HNSCC pathologic slides.
2. METHODS
2.1. Polarized hyperspectral imaging
The setup of our home-made polarized hyperspectral microscope has been reported by us previously [28]. The system is capable of full Stokes polarized light hyperspectral imaging, which acquires the images of four Stokes vector parameters (S0, S1, S2, and S3) in the wavelength range between 467 nm and 750 nm. The images were collected under 10× magnification with an image size of 1200 × 1200 pixels. The field of view of the imaging system was 656 um × 656 μm. The core components of the imaging system include an optical microscope, two polarizers, two liquid crystal variable retarders (LCVR), and a SnapScan hyperspectral camera. The LCVRs and polarizers are for polarized light imaging. The SnapScan hyperspectral camera is able to acquire data through the translation of the imaging sensor inside of the camera. The polarized light imaging components and hyperspectral imaging components work together in the image acquisition to obtain the Stokes vector parameters in the visible wavelength range. In the polarized hyperspectral imaging dataset obtained by the system, each Stokes vector parameter corresponds to a 3D data cube with two spatial dimensions and one spectral dimension, as shown in Figure 1.
Figure 1.
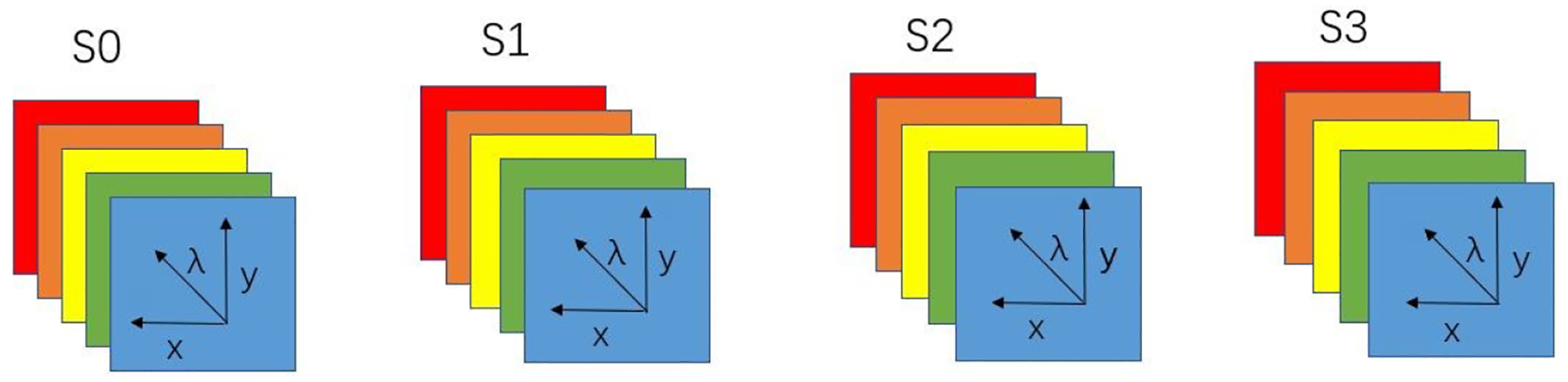
Diagram of full-polarization hyperspectral imaging data cubes. The data cube of each Stokes parameter (S0, S1, S2, and S3) has three dimensions including two spatial dimensions (x, y) and one spectral dimension (λ).
Polarized light imaging is realized by the two polarizers and two LCVRs. Figure 2 demonstrates the schematic of the imaging system with fast axis orientations of the polarizers and LCVRs. Polarizer 1 was set at 45 degrees, and polarizer 2 was set at 0 degrees. LCVR 1 was set at 0 degrees, and LCVR 2 was set at 45 degrees. The system is capable of full Stokes polarimetric imaging, which produces all four components of the Stokes vector. Thus, the system can completely define the polarization properties of transmitted light. The way to calculate the four elements of Stokes vector (S0, S1, S2, and S3) is expressed in the following equation (1):
| (1) |
where Ih represent the light intensity measured with a horizontal linear analyzer, in which the retardations of LCVR 1 and LCVR 2 are both set at 0 rad; Iv represents the light intensity measured with a vertical linear analyzer, in which LCVR 1 is set at 0 rad retardation and LCVR 2 is set at π rad retardation; I45 represents the light intensity measured with a 45 degrees oriented linear analyzer, in which LCVR 1 and LCVR 2 are both set at π/2 rad retardation; Ilc represents the light intensity measured with a left circular analyzer, in which LCVR 1 is set at 0 rad retardation and LCVR 2 is set at π/2 rad retardation. The phase retardation of LCVR is determined by different values of voltage applied on it. In addition, the value of S0 is equal to the value of light intensity.
Figure 2.
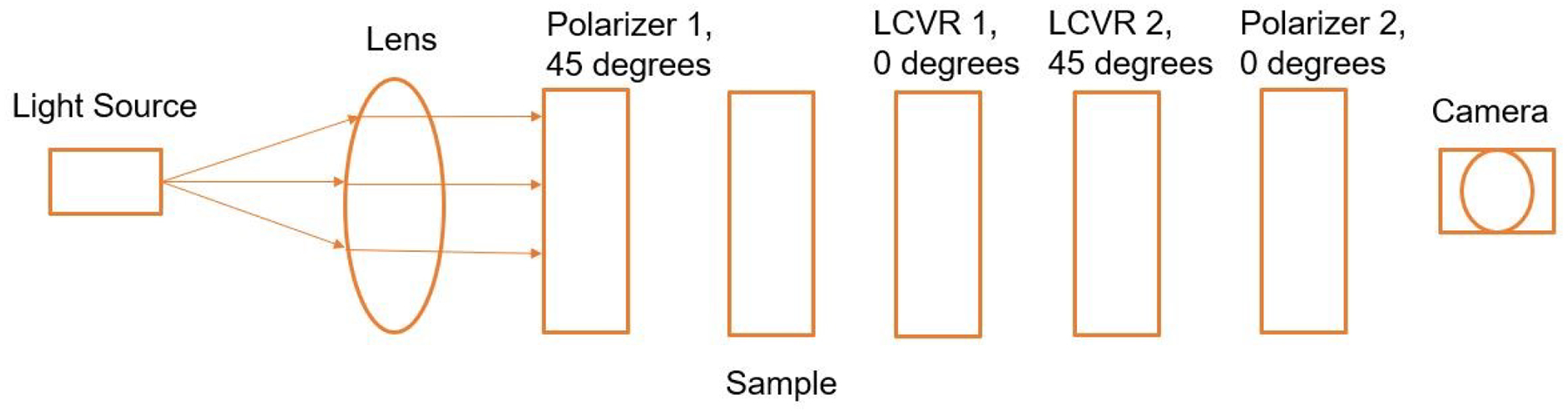
Schematic of the polarized light imaging system. The fast axis orientation of Polarizer 1 was set at 45 degrees, and Polarizer 2 was set at 0 degrees. The fast axis orientation LCVR 1 was set at 0 degrees, and LCVR 2 was set at 45 degrees.
2.2. Sample preparation
Fresh surgical tissue samples were obtained from patients who underwent surgical resection of head and neck cancer, as we described earlier [32]. Of each patient, a sample of the primary tumor, a normal tissue sample, and a sample at the tumor-normal margin were collected. Fresh ex-vivo tissues were formalin fixed, paraffin embedded, sectioned, stained with hematoxylin and eosin, and digitized using whole-slide scanning. Then, a board-certified pathologist specialized in head and neck cancer outlined the cancer margin on the digital slides using Aperio ImageScope (Leica Biosystems Inc, Buffalo Grove, IL, USA). The annotations were used as the histologic reference standard in this study.
2.3. Synthetic RGB images.
To generate synthetic RGB images from Stokes vector data cubes, we adopted an HSI-to-RGB transformation function similar to the spectral response of human eye and modified it for our data to generate the synthetic RGB images [33]. The transformation function is shown in Figure 3. In the transformation process, three different spectral response curves (R,G,B) are multiplied with the data cubes to generated the three images at the three channels (red, green, blue) of synthetic RGB images. We applied this HSI-to-RGB transformation function to all the four Stokes vector parameters (S0, S1, S2, and S3) to generate four sets of PHSI-synthesized RGB images.
Figure 3.
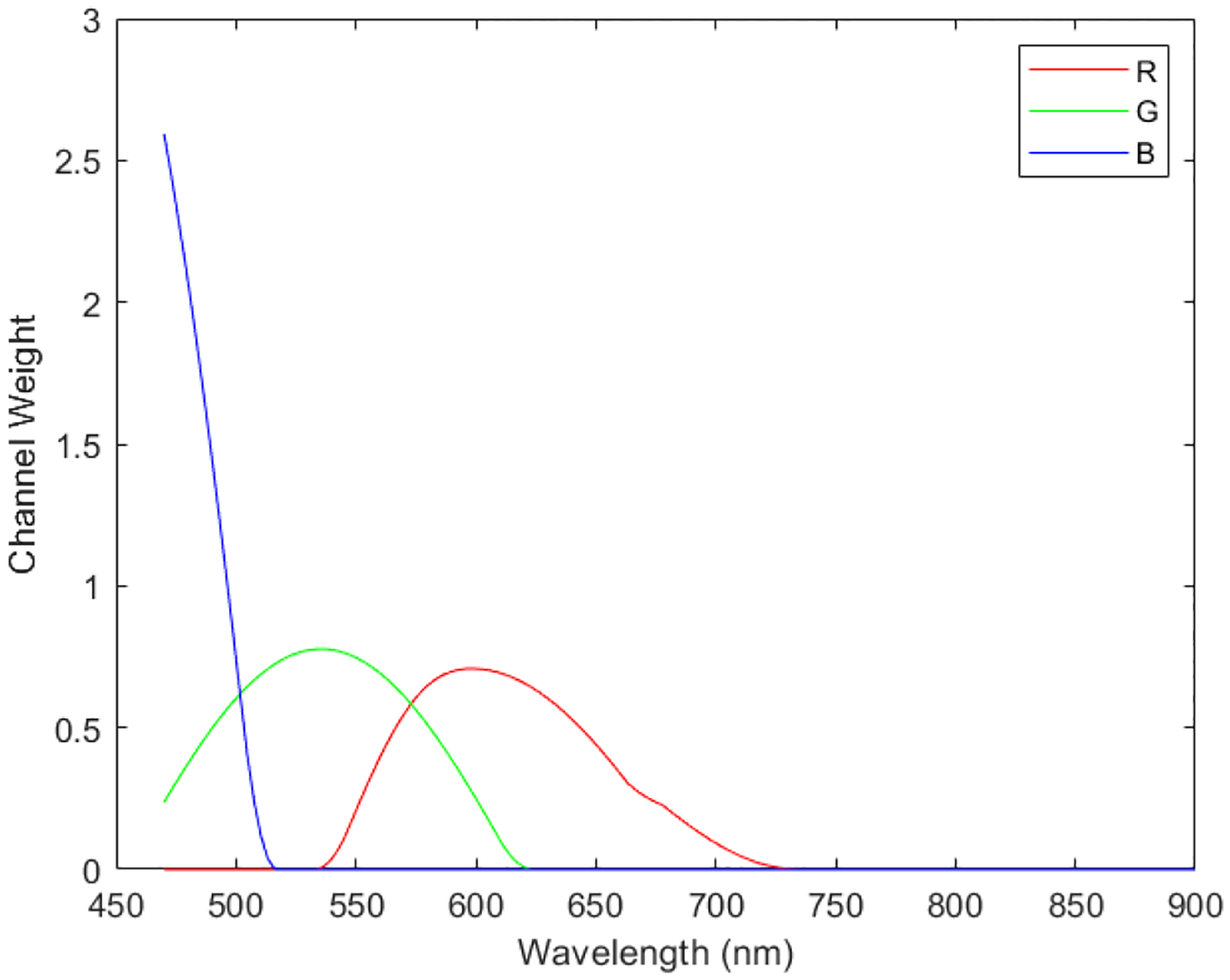
Transformation function to synthesize pseudo-RGB images from the polarized hyperspectral data cubes. In the transformation process, three different spectral response curves (R,G,B) are multiplied with the data cubes to generated the three images at the three channels (red, green, blue) of synthetic RGB images.
3. RESULTS
3.1. PHSI-synthesized RGB images of collagen
Figure 4 demonstrates the PHSI-synthesized synthetic RGB images of the Stokes vector parameters (S0, S1, S2, and S3) of a region containing thick and thin fibers of normal stroma. Figure 5 demonstrates the PHSI-synthesized RGB images of the Stokes vector parameters of a region containing thin fibers of normal stroma and muscle cells. Figure 6 demonstrates the PHSI-synthesized RGB images of the Stokes vector parameters of a region containing fibers accumulated in tumors. Figure 7 demonstrates the PHSI-synthesized RGB images of the Stokes vector parameters of a region containing fibers accumulated around tumors.
Figure 4.
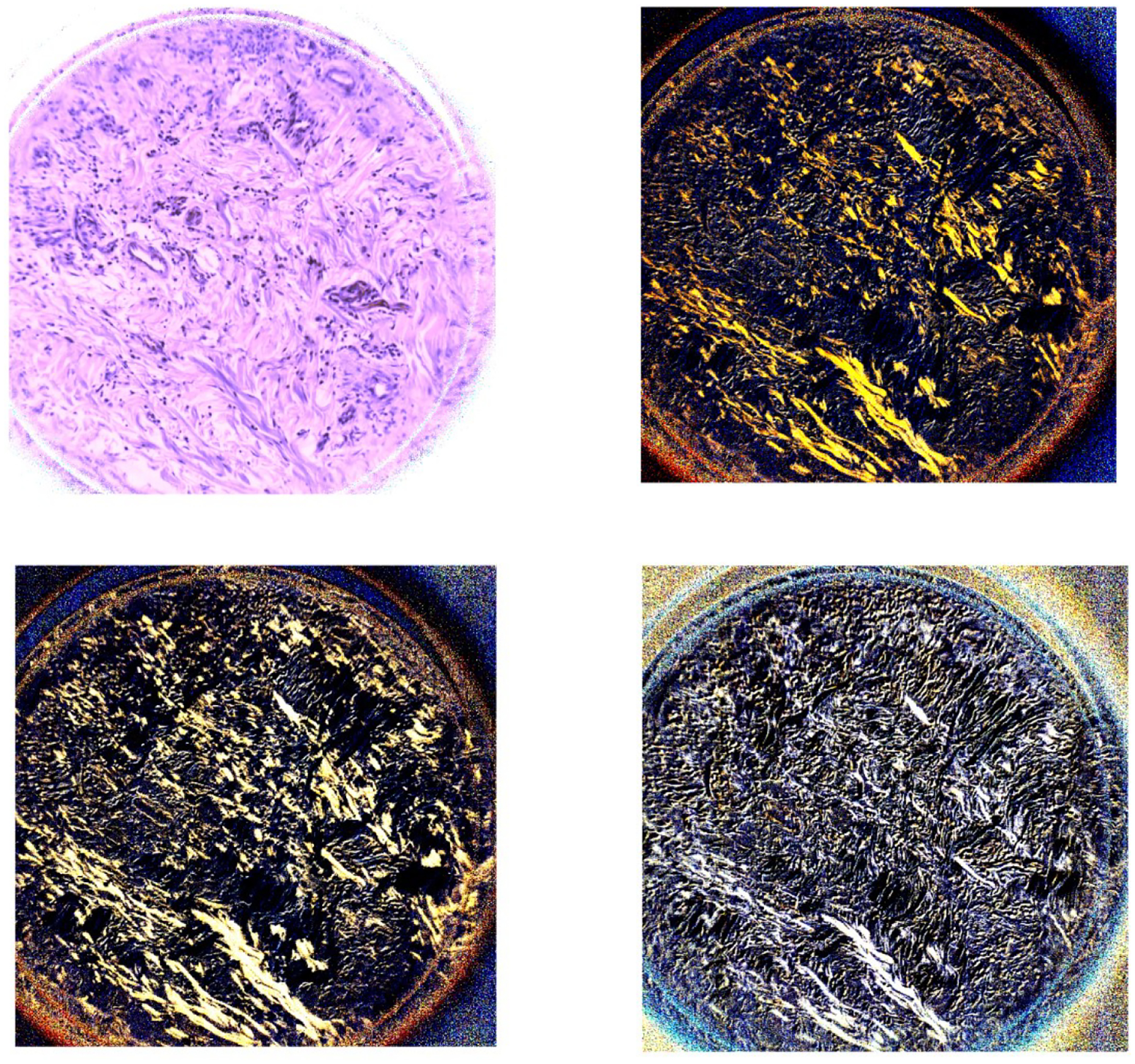
The two images on the first row (left to right) demonstrate the PHSI-synthesized RGB images of S0 and S1 of a region containing thick and thin fibers of normal stroma. The two images on the second row (left to right) demonstrate the PHSI-synthesized RGB images of S2 and S3 of a region containing thick and thin fibers of normal stroma.
Figure 5.
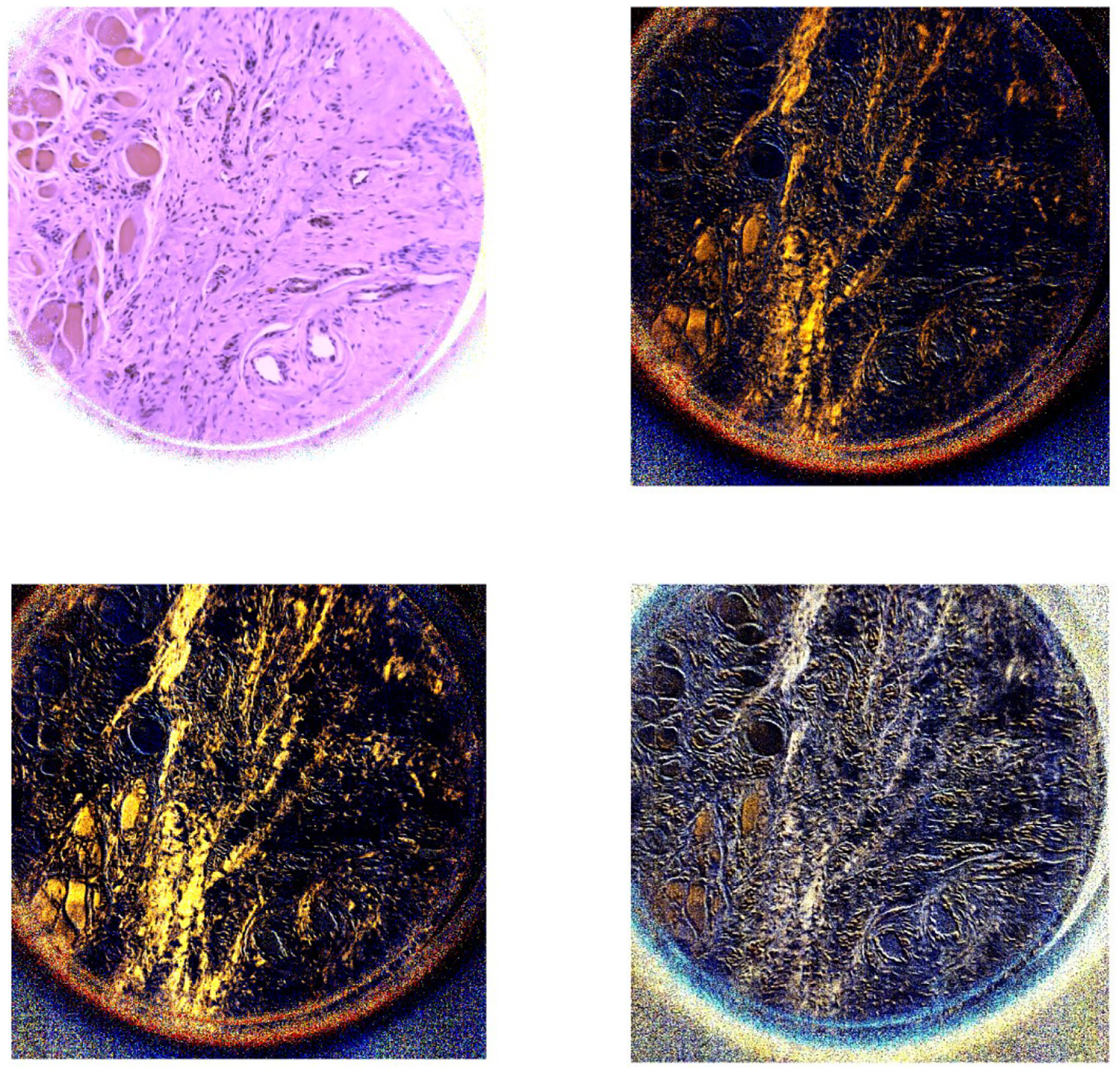
The two images on the first row (left to right) demonstrate the PHSI-synthesized RGB images of S0 and S1 of a region containing thin fibers of normal stroma and muscle cells. The two images on the second row (left to right) demonstrate the PHSI-synthesized RGB images of S2 and S3 of a region containing thin fibers of normal stroma and muscle cells.
Figure 6.
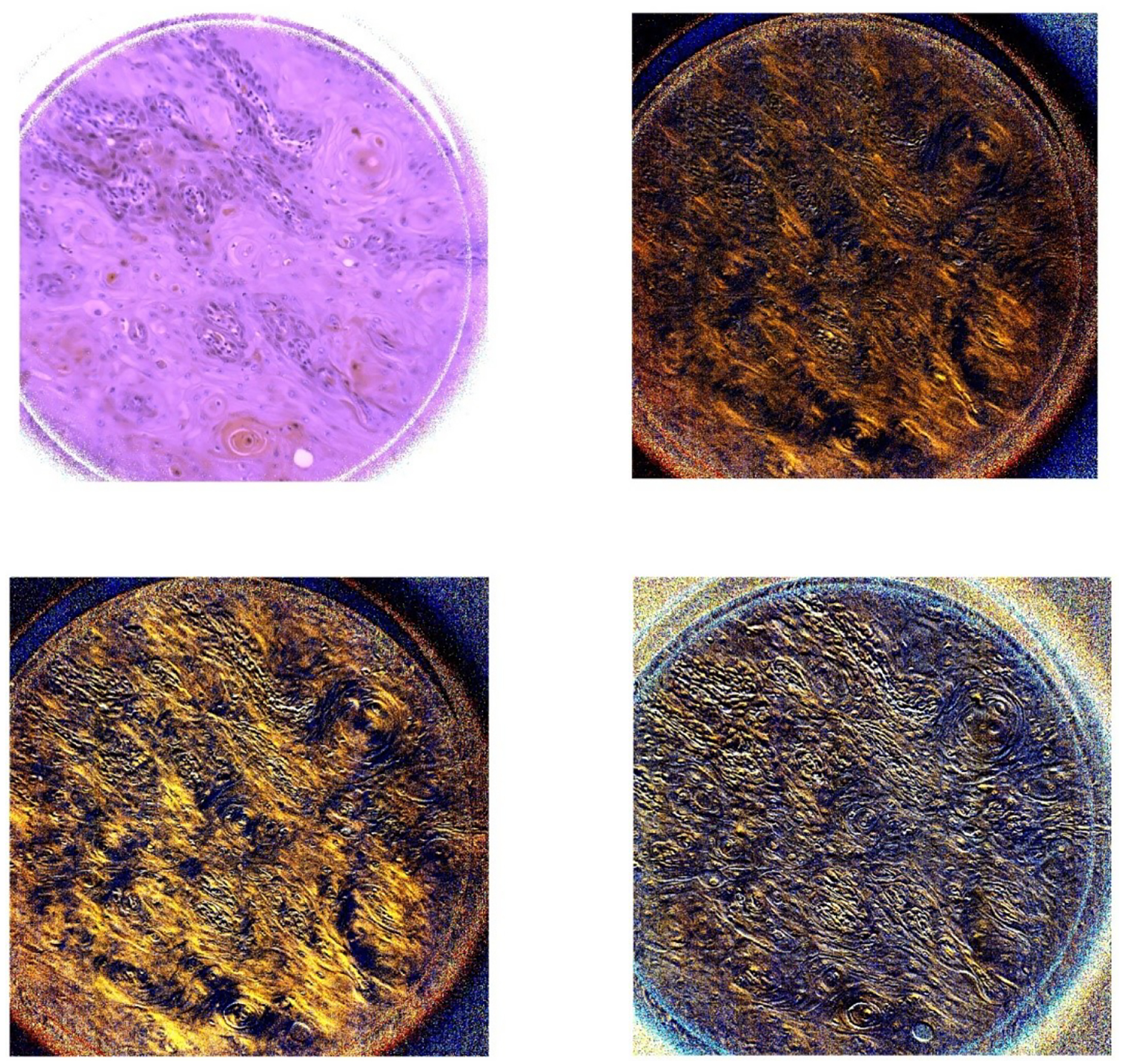
The two images on the first row (left to right) demonstrate the PHSI-synthesized RGB images of S0 and S1 of a region containing fibers accumulated in tumors. The two images on the second row (left to right) demonstrate the PHSI-synthesized RGB images of S2 and S3 of a region containing fibers accumulated in tumors.
Figure 7.
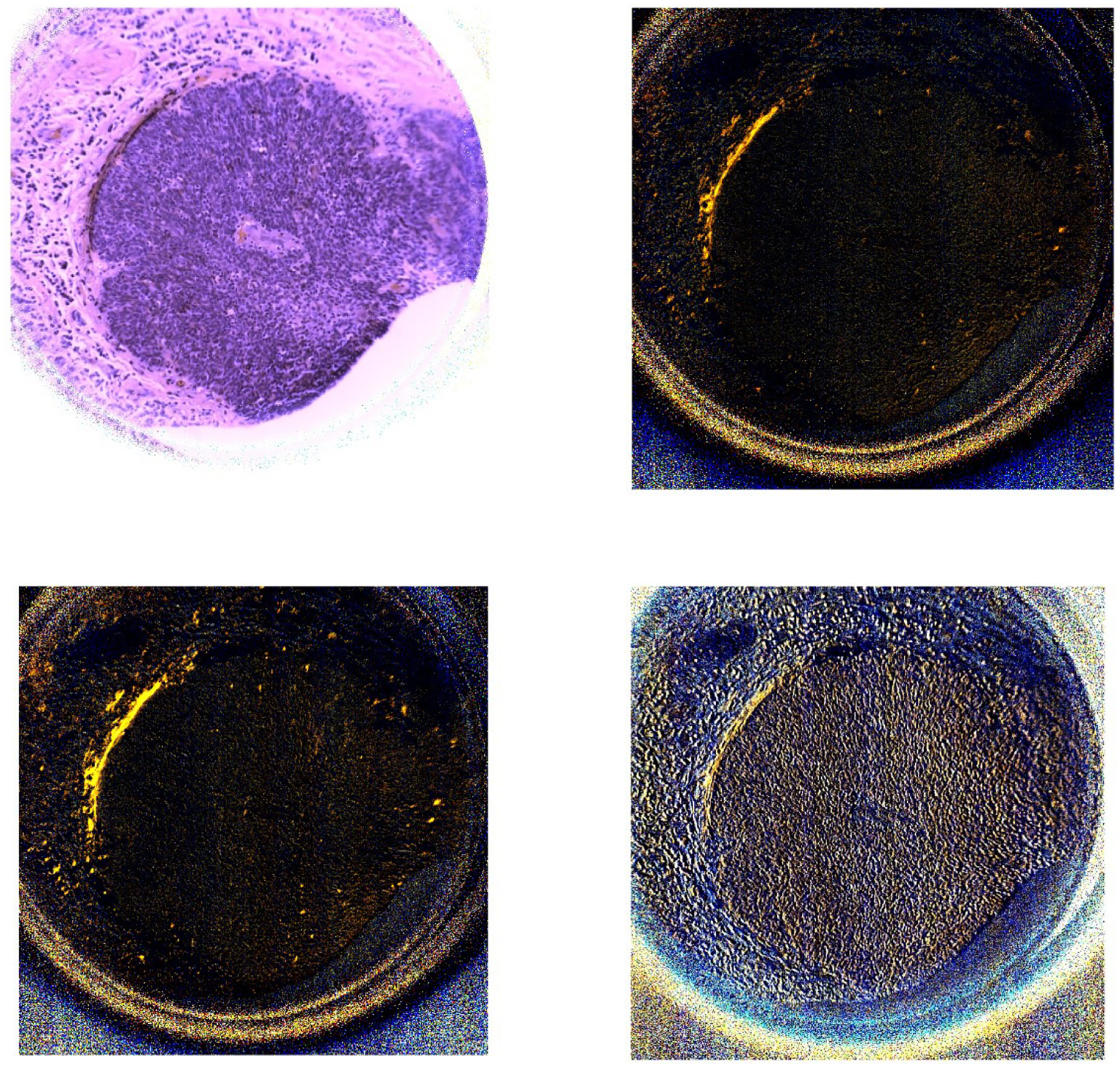
The two images on the first row (left to right) demonstrate the PHSI-synthesized RGB images of S0 and S1 of a region containing fibers accumulated around tumors. The two images on the second row (left to right) demonstrate the PHSI-synthesized RGB images of S2 and S3 of a region containing fibers accumulated around tumors.
The results show that the PHSI-synthesized images of S1, S2, and S3 are able to sensitively detect the fibrillar collagen on pathologic slides, especially on the regions with fibrillar collagen that cannot be seen on S0 (similar to the images of RGB cameras), like the thin fibers of normal stroma (Figure 4 and Figure 5) and the fibers accumulated in tumors (Figure 6). Furthermore, S3 performs the best in enhancing the signals from the regions with collagen while keeping the information from other regions like the regions with small tumor cells (Figure 7).
3.2. PHSI spectra of collagen
With our customized polarized hyperspectral microscope, we are able to extract the spectra based on the mean and standard deviation of Stokes vector parameters of the collagen. Figure 8 demonstrates the PHSI-synthesized RGB images of Stokes vector parameters of a small normal region containing thick fibers and the corresponding four spectra (S0, S1, S2, and S3). Figure 9 shows the PHSI-synthesized RGB images of Stokes vector parameters of a small normal region containing thin fibers and the corresponding four spectra. Figure 10 demonstrates the PHSI-synthesized RGB images of Stokes vector parameters of a small normal region containing fibers on muscle cells and the corresponding four spectra. Figure 11 demonstrates the PHSI-synthesized RGB images of Stokes vector parameters of a small tumor region containing fibers growing within tumor cells and the corresponding four spectra. Figure 12 demonstrates the PHSI-synthesized RGB images of Stokes vector parameters of a small tumor region containing fibers growing around tumor cells and the corresponding four spectra. In these four examples, the four spectra (S0, S1, S2, and S3) show the similar shapes.
Figure 8.
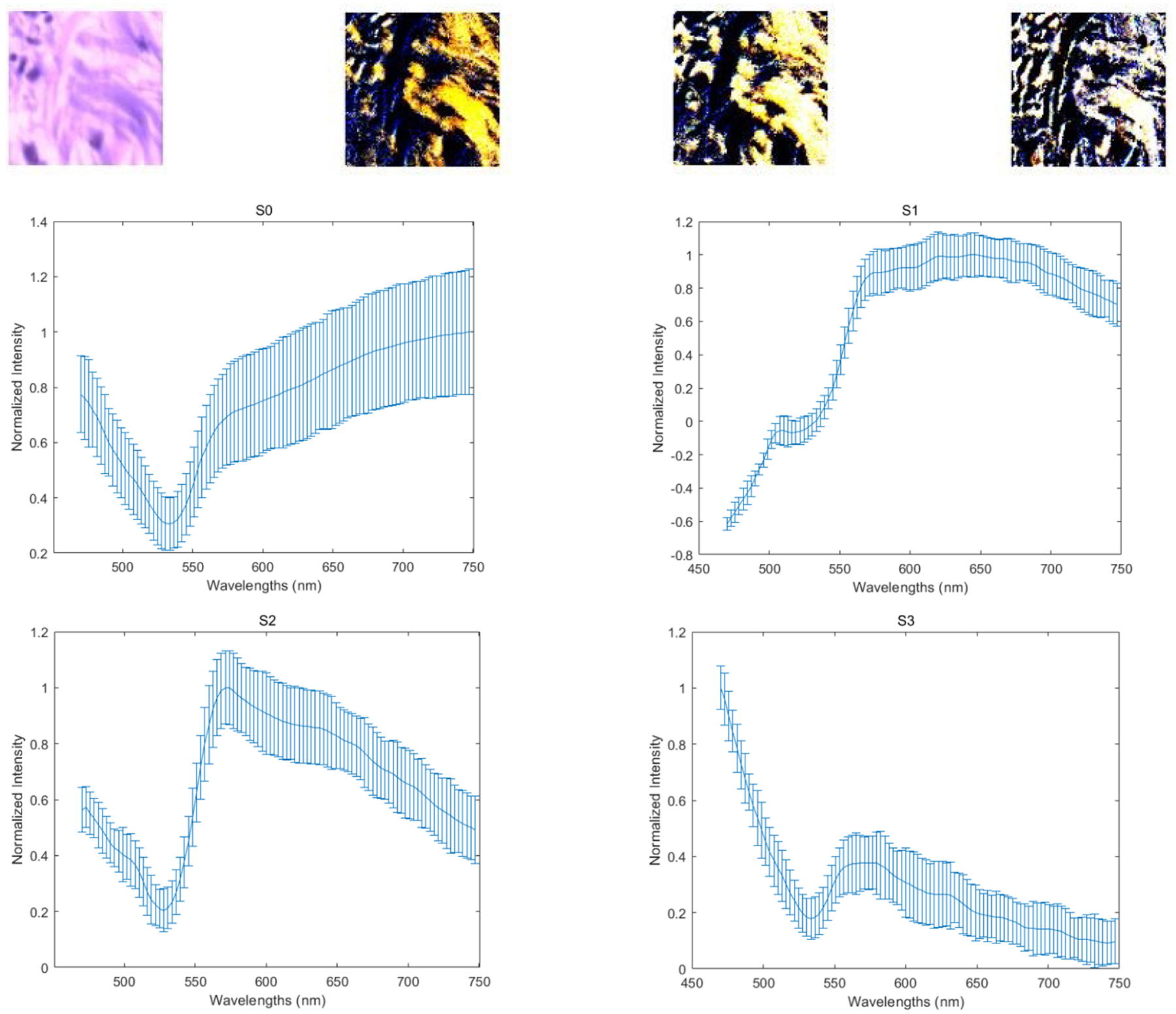
The four images on the first row (left to right) demonstrate the PHSI-synthesized RGB images of S0, S1, S2, and S3 of a small normal region containing thick fibers. The corresponding four spectra (S0, S1, S2, and S3) are located at the second row (S0, S1) and the third row (S2, S3).
Figure 9.
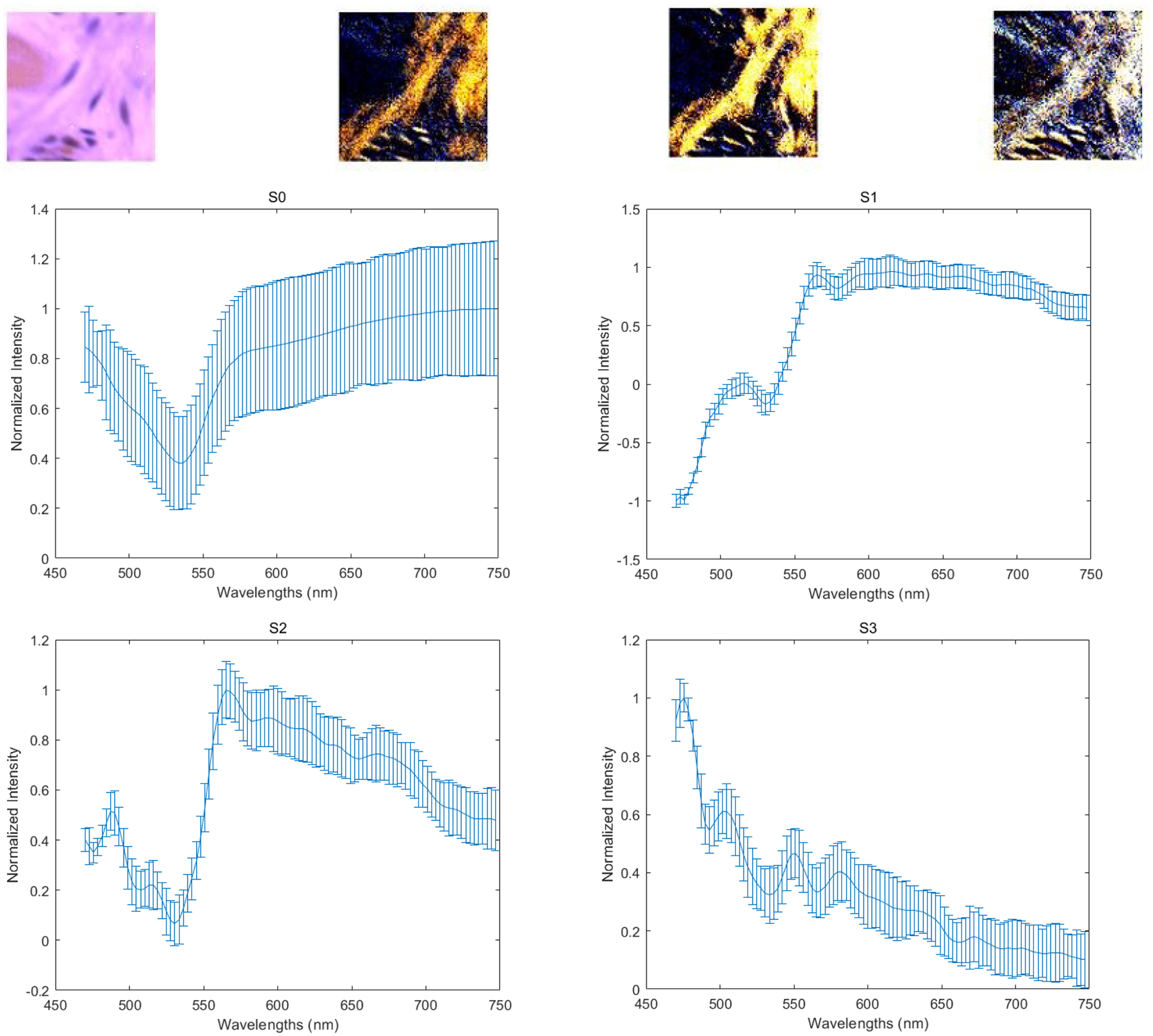
The four images on the first row (left to right) demonstrate the PHSI-synthesized RGB images of S0, S1, S2, and S3 of a small normal region containing thin fibers. The corresponding four spectra (S0, S1, S2, and S3) are located at the second row (S0, S1) and the third row (S2, S3).
Figure 10.
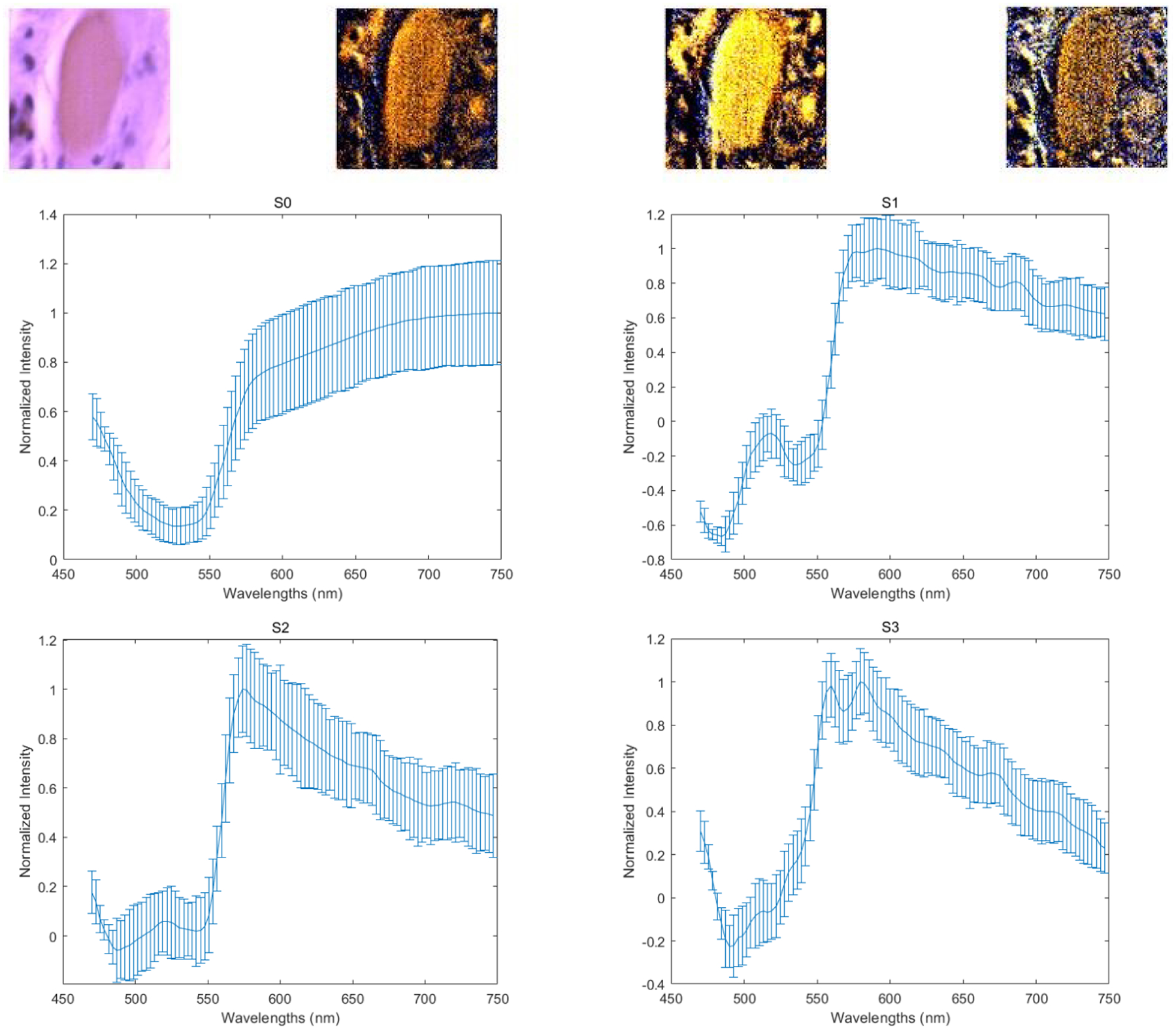
The four images on the first row (left to right) demonstrate the PHSI-synthesized RGB images of S0, S1, S2, and S3 of a small normal region containing fibers on muscle cells. The corresponding four spectra (S0, S1, S2, and S3) are located at the second row (S0, S1) and the third row (S2, S3).
Figure 11.
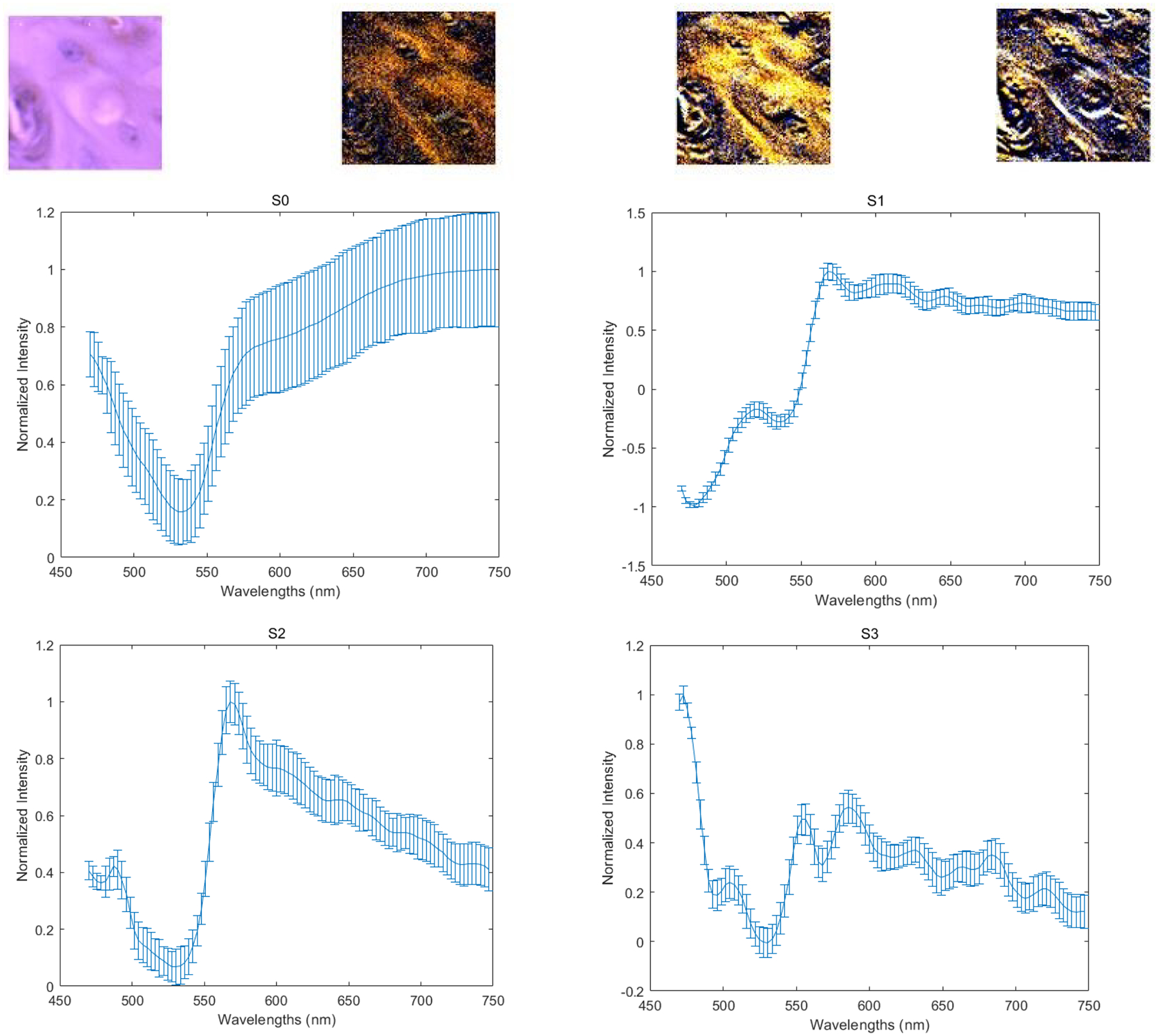
The four images on the first row (left to right) demonstrate the PHSI-synthesized RGB images of S0, S1, S2, and S3 of a small tumor region containing fibers growing within tumor cells. The corresponding four spectra (S0, S1, S2, and S3) are located at the second row (S0, S1) and the third row (S2, S3).
Figure 12.
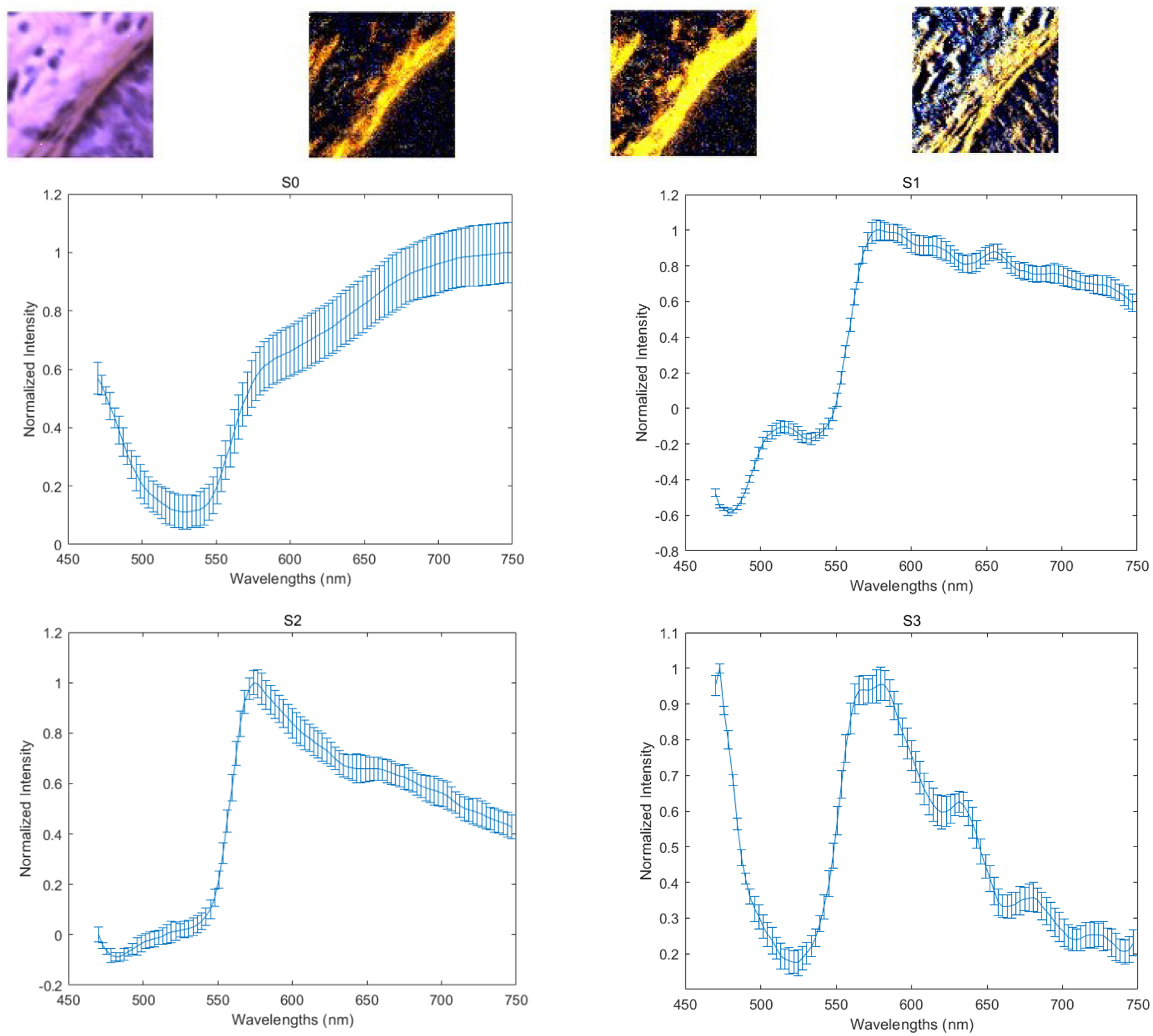
The four images on the first row (left to right) demonstrate the PHSI-synthesized RGB images of S0, S1, S2, and S3 of a small tumor region containing fibers growing around tumor cells. The corresponding four spectra (S0, S1, S2, and S3) are located at the second row (S0, S1) and the third row (S2, S3).
4. DISCUSSION AND CONCLUSION
In this study, we developed a polarized hyperspectral microscopic imaging system for collagen visualization in the normal regions containing normal cells and collagen fibers and in the tumor regions containing cancerous squamous cells and collagen fibers, on HNSCC pathologic slides. Our results demonstrated that our customized polarized hyperspectral microscope is able to improve the visualization of collagen on HNSCC pathologic slides under different situations: thick fibers of normal stroma, thin fibers of normal stroma, fibers on normal muscle cells, fibers accumulated in tumors, fibers accumulated around tumors. To be specific, we find that the PHSI-synthesized images of S1, S2, and S3 are able to sensitively detect the fibrillar collagen on pathologic slides, especially on the regions with fibrillar collagen that cannot be seen on S0 (similar to the images of RGB cameras), like the thin fibers of normal stroma and the fibers accumulated in tumors. Furthermore, S3 performs the best in enhancing the signals from the regions with collagens while keeping the information from other regions like the regions with small tumor cells. Our results also demonstrated that the customized polarized hyperspectral microscope is capable of extracting the spectral signatures of collagen based on Stokes vector parameters (S0, S1, S2, and S3).
To the best of our knowledge, this is the first work to apply polarized hyperspectral imaging to improve the visualization of collagen based on the spatial and spectral information of Stokes vector data cubes. The polarized hyperspectral imaging technique can be further explored to study cancer diagnosis and prognosis based on collagen development in the future.
ACKNOWLEDGMENTS
This research was supported in part by the U.S. National Institutes of Health (NIH) grants (R01CA156775, R01CA204254, R01HL140325, and R21CA231911) and by the Cancer Prevention and Research Institute of Texas (CPRIT) grant RP190588.
REFERENCES
- 1.J. Daniel E et al. , “Head and neck squamous cell carcinoma,” Nature Reviews Disease Primers, 6(1), 1–22 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Samuel O, et al. , “Hyperspectral and multispectral imaging in digital and computational pathology: a systematic review,” Biomedical Optics Express, 11(6), 3195–3233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao Liang, and Wang Lihong V., “A review of snapshot multidimensional optical imaging: measuring photon tags in parallel,” Physics reports, 616, 1–37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu G, and Fei B, “Medical hyperspectral imaging: a review,” Journal of biomedical optics 19(1), 010901 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yushkov KB, and Molchanov VY, “Hyperspectral imaging acousto-optic system with spatial filtering for optical phase visualization,” Journal of biomedical optics 22(6), 066017 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Salmivuori M et al. , “Hyperspectral Imaging System in the Delineation of Ill-defined Basal Cell Carcinomas: A Pilot Study,” Journal of the European Academy of Dermatology and Venereology (2018). [DOI] [PubMed] [Google Scholar]
- 7.Lu G et al. , “Hyperspectral imaging for cancer surgical margin delineation: registration of hyperspectral and histological images.” Medical Imaging 2014: Image-Guided Procedures, Robotic Interventions, and Modeling, International Society for Optics and Photonics, 9036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu G et al. , “Spectral-spatial classification using tensor modeling for cancer detection with hyperspectral imaging,” Medical Imaging 2014: Image Processing, 903413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pike R et al. , “A minimum spanning forest based hyperspectral image classification method for cancerous tissue detection,” Medical Imaging 2014: Image Processing, 90341W (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pike R et al. , “A Minimum Spanning Forest-Based Method for Noninvasive Cancer Detection With Hyperspectral Imaging,” IEEE Trans. Biomed. Engineering 63(3), 653–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu G et al. , “Estimation of tissue optical parameters with hyperspectral imaging and spectral unmixing,” Medical Imaging 2015: Biomedical Applications in Molecular, Structural, and Functional Imaging 94170Q (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung H et al. , “Superpixel-based spectral classification for the detection of head and neck cancer with hyperspectral imaging,” Medical Imaging 2016: Biomedical Applications in Molecular, Structural, and Functional Imaging, 978813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halicek M et al. , “Tumor margin classification of head and neck cancer using hyperspectral imaging and convolutional neural networks,” Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling, International Society for Optics and Photonics, 10576 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halicek M et al. , “Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging,” Journal of biomedical optics, 22(6), 060503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halicek M et al. , “ Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks,” Journal of biomedical optics, 24(3), 060503 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halicek M et al. , “ Hyperspectral imaging of head and neck squamous cell carcinoma for cancer margin detection in surgical specimens from 102 patients using deep learning,” Cancers, 11(9), (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halicek M et al. , “ Tumor detection of the thyroid and salivary glands using hyperspectral imaging and deep learning,” Biomedical Optics Express, 11(3), (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson RR, “Polarized light examination and photography of the skin,” Arch. Dermatol, 127(7), 1000–1005 (1991). [PubMed] [Google Scholar]
- 19.Pierangelo A et al. , “Polarimetric imaging of uterine cervix: a case study,” Optics express 21(12), 14120–14130 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Raković MJ et al. , “Light backscattering polarization patterns from turbid media: theory and experiment,” Applied optics, 38(15), 3399–3408 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Liu B et al. , “Mueller polarimetric imaging for characterizing the collagen microstructures of breast cancer tissues in different genotypes,” Optics Communications, 433, 60–67 (2019). [Google Scholar]
- 22.Chang J et al. , “Division of focal plane polarimeter-based 3× 4 Mueller matrix microscope: a potential tool for quick diagnosis of human carcinoma tissues,” Journal of biomedical optics, 21(5), 056002 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Lindeboom JA, Mathura KR, and Ince C, “Orthogonal polarization spectral (OPS) imaging and topographical characteristics of oral squamous cell carcinoma,” Oral oncology, 42(6), 581–585 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Pahernik S et al. , “Orthogonal polarisation spectral imaging as a new tool for the assessment of antivascular tumour treatment in vivo: a validation study,” British journal of cancer, 86(10), 1622 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roblyer DM et al. , “Multispectral optical imaging device for in vivo detection of oral neoplasia,” Journal of biomedical optics, 13(2), 024019 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manhas S et al. , “Polarized diffuse reflectance measurements on cancerous and noncancerous tissues,” Journal of biophotonics, 2(10), 581–587 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Chung J et al. , “Use of polar decomposition for the diagnosis of oral precancer,” Applied optics, 46(15), 3038–3045 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Zhou X et al. , “Development of a new polarized hyperspectral imaging microscope,” Imaging, Therapeutics, and Advanced Technology in Head and Neck Surgery and Otolaryngology 2020. Vol. 11213. International Society for Optics and Photonics (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong Y et al. , “Quantitatively characterizing the microstructural features of breast ductal carcinoma tissues in different progression stages by Mueller matrix microscope,” Biomedical optics express 8(8), 3643–3655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fartash V et al. , “Polarization-sensitive hyperspectral imaging in vivo: a multimode dermoscope for skin analysis,” Scientific reports, 4(1), 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z et al. , “Polarization-resolved hyperspectral stimulated Raman scattering microscopy for label-free biomolecular imaging of the tooth,” Applied Physics Letters 108(3), 033701 (2016). [Google Scholar]
- 32.Lu G et al. , “Detection of head and neck cancer in surgical specimens using quantitative hyperspectral imaging,” Clinical Cancer Research, 5426–5436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling M, et al. , “Hyperspectral microscopic imaging for automatic detection of head and neck squamous cell carcinoma using histologic image and machine learning,” Medical Imaging 2020: Digital Pathology. Vol. 11320, International Society for Optics and Photonics, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaiming H et al. , “Deep residual learning for image recognition, “ Proceedings of the IEEE conference on computer vision and pattern recognition, 770–778 (2016). [Google Scholar]
- 35.Jeremy W et al. , “Spring research presentation: A theoretical foundation for inductive transfer.” Brigham Young University, College of Physical and Mathematical Sciences, 1(08), (2007). [Google Scholar]
- 36.C. John KC, “The wonderful colors of the hematoxylin–eosin stain in diagnostic surgical pathology,” International journal of surgical pathology 22(1), 12–32 (2014). [DOI] [PubMed] [Google Scholar]
- 37.O. Jonathan N., et al. , “Navigating the collagen jungle: the biomedical potential of fiber organization in cancer,” Bioengineering, 8.2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang C et al. , “Type I collagen deletion in αSMA+ myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer,” Cancer cell, 39.4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


