Abstract
Boundary elements are thought to define the ends of functionally independent domains of genetic activity. An assay for boundary activity based on this concept measures the ability to insulate a bracketed, chromosomally integrated reporter gene from position effects. Despite their presumed importance, the few examples identified to date apparently do not share sequence motifs or DNA binding proteins. The Drosophila protein BEAF binds the scs′ boundary element of the 87A7 hsp70 locus and roughly half of polytene chromosome interband loci. To see if these sites represent a class of boundary elements that have BEAF in common, we have isolated and studied several genomic BEAF binding sites as candidate boundary elements (cBEs). BEAF binds with high affinity to clustered, variably arranged CGATA motifs present in these cBEs. No other sequence homologies were found. Two cBEs were tested and found to confer position-independent expression on a mini-white reporter gene in transgenic flies. Furthermore, point mutations in CGATA motifs that eliminate binding by BEAF also eliminate the ability to confer position-independent expression. Taken together, these findings suggest that clustered CGATA motifs are a hallmark of a BEAF-utilizing class of boundary elements found at many loci. This is the first example of a class of boundary elements that share a sequence motif and a binding protein.
Chromatin appears to be partitioned into chromosomal domains that are operationally defined by bracketing DNA regions called boundary elements or insulators (10; see reference 34 for a review). Boundary elements are presumably necessary to curtail the potentially promiscuous behavior of enhancers, limiting their action to the domain in which they reside. The biological activity of a boundary element is experimentally measured by either position-independent expression or enhancer-blocking assays. If this view of chromosomal organization is correct, boundary elements play a very important functional role. Yet only a few examples have been identified, and each is so far a unique case, as they do not appear to have notable sequence homologies or to have binding activities in common.
The best-characterized boundary elements are the scs and scs′ regions found to bracket the 87A7 hsp70 heat shock puff of Drosophila melanogaster polytene chromosomes (33) and a 340-bp fragment from the gypsy retrotransposon (11). The scs/scs′ and the gypsy-derived elements have a boundary function in both of the assays mentioned above. They confer position-independent expression on a bracketed reporter gene by insulating the transgene from both activating and repressive effects at the site of chromosomal integration, and they block communication between a specific enhancer and promoter when interposed (20, 21, 31). It is important to note that boundary elements do not inactivate promoters or enhancers; they only block communication when interposed (2, 3, 21, 32). For instance, if an enhancer and boundary element are located between two divergently transcribed promoters, the enhancer cannot activate the promoter with the intervening boundary element but can activate the other promoter. Thus, the positional functioning of boundary elements is distinct from the bidirectional repressive effect of silencer elements.
The boundary activity of the gypsy-derived element is known to be mediated by the binding of the zinc finger protein su(Hw) to its reiterated binding sites (31). The su(Hw) protein has been studied in some detail, and regions involved in DNA binding, enhancer blocking, and interactions with mod(mdg4) have been identified (8, 13, 22). Interactions between the mod(mdg4) gene product and the su(Hw) protein are necessary for boundary function (9). In addition to loss of enhancer blocking, it has been suggested that some mod(mdg4) mutations lead to an unmasked activity that represses certain promoters (3).
To address the boundary activity of scs′ at a biochemical level, we previously characterized two cDNAs encoding the related scs′ boundary element-associated factors BEAF-32A and -32B (14, 38). The BEAF activity in Drosophila nuclear extracts appears to be composed predominantly of trimers of one 32A and two 32B subunits. Interactions between BEAF subunits results in cooperative binding to the three CGATA motifs of the high-affinity binding site in scs′ which, in turn, facilitates binding to the lower-affinity binding site located some 200 bp away (14).
Evidence of a role for BEAF in boundary activity derives from an enhancer-blocking assay in Drosophila D1 cells: seven tandem copies of a 48-bp oligonucleotide containing the scs′ high-affinity binding site had enhancer-blocking activity (although less than that obtained by using scs′), while point mutations that eliminated BEAF binding further reduced this activity (38). We immunolocalized BEAF to numerous interbands and puff boundaries on polytene chromosomes, suggesting the existence of a common class of boundary elements in Drosophila and that the band-interband structure of polytene chromosomes could be related to the localization of boundary elements.
In this study, we isolated some of these genomic BEAF binding sites and used transgenic flies to demonstrate that the newly isolated sequences tested represent boundary elements. The only homology found between these candidate boundary elements (cBEs) and scs′ are clusters of CGATA motifs. Despite the varied spacing and orientations of the motifs in the different clusters, BEAF interacts with all of the clusters. We also used transgenic flies to directly establish the functional importance of BEAF binding sites by mutagenesis of CGATA motifs. This strongly indicates that the hundreds of BEAF binding sites in the Drosophila genome represent an abundant class of boundary elements, providing the first example of a class of binding elements that share a sequence motif and a binding protein.
MATERIALS AND METHODS
Plasmids and DNA methods.
Genomic DNA fragments were ligated into EcoRI (cBE28 and -51)- or BamHI (cBE76)-cut pBluescript KS (Stratagene) after five cycles of the enrichment protocol (see below) and selected by alpha complementation. From 101 plasmids, 11 cBEs were obtained, and of these, 4 had sequences related to cBE28. Both strands of the cBEs were dideoxy sequenced, and sequence comparisons and database searches were performed with the GCG Bestfit and Fasta programs. Plasmids for transfections had cBE or control sequences inserted into the BamHI site at −145 of the hsp227 minimal promoter (upstream activating sequence assay) or into the KpnI site at −250 of the hsp27 promoter of a p1200CAT derivative (enhancer-blocking assay; 30). The luciferase-encoding transfection efficiency control plasmid pAcluc has been previously described (38). P-element transposon constructs were made by insertion of the 990-bp PvuII scs fragment (35) into the NsiI site 3′ of the mini-white gene in pCasper4 (27), followed by insertion of different boundary or control elements into the BamHI site 5′ of the mini-white gene. The 215-bp scs′ derivatives M and M* were cloned into a derivative of pSP64 in which an oligonucleotide encoding a BglII site was inserted into the EcoRI site after PCR amplification using primers encoding a BamHI site on one end and a BglII site on the other. M* differs from M only in having the three CGATA motifs of the D site mutated to CTCGA as previously described (14). Dimerization was performed by ligating appropriate BamHI-ScaI and BglII-ScaI fragments of the M or M* plasmid to themselves, resulting in direct repeats. The spacing between the two D sites in the MM construct is the same as that found between the D site and the lower-affinity B site in scs′. The DNA fragment used as a probe in DNase I-hypersensitive site mapping experiments was derived from a plasmid containing the 4 kb of IMPdH genomic sequences, kindly provided by D. Nash (26).
Enrichment protocol for obtaining genomic cBEs.
Genomic DNA was isolated from Kc cells, sonicated to an average size of 0.5 kb, and ligated to a linker prepared from two oligonucleotides, a 25-mer with the sequence 5′GCGGTGACCCGGGAGATCTGAATTC and an 11-mer with the sequence 5′GAATTCAGATC. Affinity-purified, bacterially expressed protein 32B was allowed to bind the DNA in a standard gel shift reaction (38) scaled up by a factor of 2. The protein was immunoprecipitated as previously described by using mouse antibodies raised against the bacterially expressed 32B protein unique amino-terminal domain (14) affinity purified by a filter method (1). Briefly, after a 15-min room temperature binding reaction, 20 μl of protein A-Sepharose beads preincubated with the affinity-purified mouse antibodies was added and the incubation was continued at 4°C for 1 h. The beads were pelleted by 10 s of centrifugation, washed rapidly several times with binding buffer, and digested with proteinase K. The coprecipitated DNA was purified by phenol extraction (yield of about 10 ng) and amplified by ligation-mediated (LM)-PCR (25) with the 25-mer oligonucleotide described above as the primer, and 500 (cycle 1) or 100 (cycles 2, 3, 4, and 5) ng of the resulting amplified DNA was subjected to further cycles of binding, immunoprecipitation, and LM-PCR.
Band shift and DNase I footprinting assays.
Band shift and DNase I footprinting assays were performed as previously described (14). Band shift assays contained either the end-labelled scs′ D subfragment and unlabelled cBE fragments as competitors or end-labelled cBE fragments together with the desired protein (Drosophila nuclear extract or the affinity-purified, bacterially expressed BEAF-32A or -32B protein). Providing end-labelled fragments were of different sizes, up to four probes were mixed in one band shift reaction to limit the total number of assays to be performed and to see their relative affinities (37).
Transfections.
Transient transfections of D1 cells and chloramphenicol acetyltransferase (CAT) and luciferase assays of cell extracts were done essentially as previously described (38), by using 10 μg of CAT plasmids and 1 μg of the transfection efficiency control plasmid pAcluc coprecipitated with calcium phosphate. Briefly, cell extracts were prepared 48 h after adding the DNA, induction with 5 μM ecdysone was done for 24 h, and cells were allowed to recover at 25°C for 12 h after a 2-h heat shock in a 37°C air incubator.
Cells, nuclei, and DNase I treatment.
The Drosophila tissue culture cell lines Kc 161 and D1 were grown as previously described (38). Nuclei were prepared as previously described (24) from exponentially growing Kc cells and used directly for nuclease treatments as already described (19). Nuclei at an A260 of 10 in 850 μl were digested with 0.1-U/ml DNase I or 0.02-U/ml micrococcal nuclease for 0.5 to 6 min at 37°C. For detection of nuclease-hypersensitive sites upstream of the IMPdH gene by the indirect end-labelling method (36), purified DNA samples were digested with PvuII. Genomic DNA digested with PvuII, PvuII plus EcoRI, and PvuII plus XhoI was used for internal size standards. Gel electrophoresis, Southern blotting, and detection of cleavage products were done as previously described (19) by using the purified 742-bp PvuII-XhoI DNA fragment from the IMPdH coding region as a probe. As a control, we successfully mapped the hypersensitive sites in scs (33) by using genomic DNA digested with BamHI and BglII, with additional digestion with NdeI or HpaI for internal size standards. The probe was the 291-bp BamHI-PvuII fragment from the 1.7-kb BamHI-BglII scs fragment (35).
P-element transformation and scoring of reporter gene expression.
Transposon injections into early embryos of Df(1)w67c23(c) flies were done as described by Pirrotta et al. (28), and transformants were selected by rescue of the eye color. Crosses to marked balancer chromosomes were performed to generate stocks and to determine the chromosome of insertion for each line as previously described (35). Levels of mini-white expression of the different transgenic lines were determined from the eye color observed in 2-day-old females of approximately the same size that were heterozygous for the transgene, which allows sensitive detection of differences in eye color (29). To ensure proper analysis of transformant lines, two levels of control were made. First, females of all transformant lines were backcrossed for three generations to the recipient w67c23 flies to allow separation of inserts on the same chromosome by recombination (29). Second, lines that were suspected of containing more than one insert (i.e., any transgenic line with orange or red eyes) were subjected to Southern analyses to see if the phenotype could be due to the additive effects of expression of several P elements (20). All flies were found to have single P-element insertions, except those indicated by asterisks in Fig. 6.
FIG. 6.
cBEs confer position-independent expression on a mini-white reporter gene, and point mutations in a BEAF binding site abolish this activity. Young (∼48-h-old), heterozygous females are shown, each representing one independent transgenic fly line obtained by P-element-mediated transformation with the indicated mini-white constructs. (A) Mini-white gene without bracketing elements. (B) The 990-bp scs PvuII fragment was inserted 3′ of the mini-white gene. (C through G) Derived from B by inserting the following DNA sequences 5′ of the mini-white gene: C, 515-bp scs′ fragment; D, cBE76; E, cBE28; F, scs′-derivative MM dimerized fragment; G, scs′-derivative M*M* dimerized fragment. MM consists of a 227-bp fragment containing the scs′ D (high-affinity) site as a dimer such that the spacing between BEAF binding sites is the same as that found in scs′ for the B (low-affinity) and D sites, and the M*M* sequence differs only in having point mutations in all CGATA motifs to eliminate binding by BEAF.
RESULTS
Isolation of new genomic BEAF binding sites.
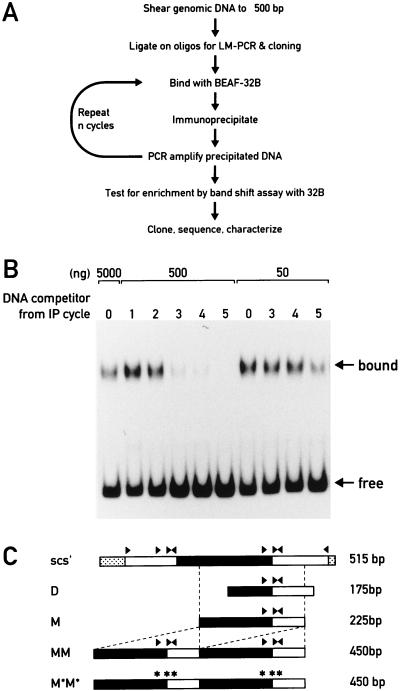
BEAF was previously immunolocalized to numerous interbands and puff boundaries on polytene chromosomes (38). Because BEAF binds to the scs′ boundary element and has been implicated in boundary function, we were interested in characterizing a number of these genomic BEAF binding sites to see if they have boundary activity and to derive a consensus sequence and structure for BEAF binding. Genomic BEAF binding sites were isolated as depicted in Fig. 1A. Briefly, appropriate oligonucleotides were ligated onto sheared genomic DNA to facilitate LM-PCR. Subsequently, specific complexes were formed between the genomic DNA and bacterially expressed BEAF-32B, the 32B protein was immunoprecipitated, and the associated DNA was amplified by LM-PCR. We used 32B since its footprints on scs′ are virtually identical to those of affinity-purified BEAF (14) and we wished to ensure the absence of any contaminating Drosophila DNA binding factors.
FIG. 1.
Isolation of new genomic BEAF binding sites. (A) DNA fractions enriched in BEAF binding sites were prepared from fragmented genomic DNA by an enrichment procedure as outlined schematically in this panel. oligos, oligonucleotides. (B) The relative enrichment for BEAF binding fragments was assayed by competition gel shift analysis. The samples contained a fixed amount of BEAF to titrate about half of the radiolabeled scs′ D subfragment and two levels of competitor DNA (500 and 50 ng) obtained after each enrichment cycle, as indicated. IP, immunoprecipitation. (C) Some features of the scs′ boundary element are indicated. The arrowheads refer to the CGATA motifs (∗ for motifs mutated to CTCGA), the black bar indicates the nuclease-resitant core, and the open bars represent the nuclease-hypersensitive regions. The positions of the D and M subfragments are indicated. The dimerized MM boundary construct and its mutant derivative M*M* are also schematically represented.
The cycle of binding, immunoprecipitation, and amplification was repeated several times, and the relative enrichment for fragments with BEAF binding sites was assessed by a competition band shift experiment (Fig. 1B). Binding reaction mixtures contained the radiolabelled scs′ D subfragment as the probe and a fixed amount of BEAF sufficient to shift about 50% of the DNA probe. Two amounts of amplified DNA obtained after each enrichment cycle were added to the reaction mixtures as competitors. While it took 500 ng of DNA obtained after the third cycle of enrichment to achieve significant dissociation of the probe-BEAF complex, only 50 ng of DNA from the fifth cycle was needed to achieve a similar level of competition (Fig. 1B).
Individual DNA fragments were cloned from the enriched DNA and submitted to gel shift analyses. More than 10% of the 101 clones tested were shifted by 32B (data not shown). Thus, the procedure described above yielded a DNA fraction enriched in cBEs. Here, we focus our attention on three fragments: cBE76, cBE28, and cBE51.
BEAF binds to variably arranged CGATA motifs in the cBEs.
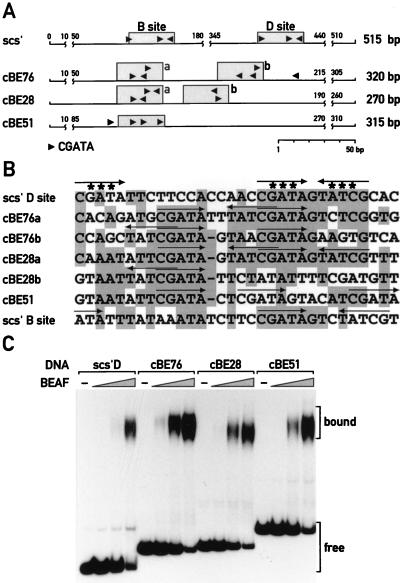
The two BEAF target sequences of the scs′ element are both composed of three CGATA sequences arranged as a palindrome plus a singlet motif. This arrangement is depicted in Fig. 2A for the high- and low-affinity sites (the D and B sites, respectively) present in scs′, where arrowheads represent the CGATA motifs (38). Sequence comparison of scs′ with the new cBEs revealed no homology other than clustered CGATA motifs. Interestingly, the arrangement of these motifs varies (Fig. 2A). Two elements, cBE76 and cBE28, have divergent motifs that overlap at the CG, creating an 8-bp palindrome containing a ClaI site, as indicated by overlapping arrowheads. These extended ClaI sites often have adjacent single CGATA motifs in either orientation. cBE51 is unique in that it does not have an inverted repeat or an extended ClaI site, although it does have three clustered CGATA motifs and a fourth one a short distance away, all in the same orientation. In addition, cBE51 has two ClaI sites that have a 1-bp terminal mismatch with the extended motif. Despite variability in CGATA motif position and orientation between the cBEs and scs′, the sequences can be aligned to indicate a striking level of homology that encompasses as many as four clustered CGATA motifs with one extended ClaI site (Fig. 2B). Note that the low-affinity scs′ B site aligns least well with these other sequences.
FIG. 2.
The selected cBEs share CGATA motifs and contain high-affinity sites for BEAF. (A) The position and arrangement of the CGATA motifs (arrowheads) are shown for the scs′ fragment and the new cBEs cBE76, -28, and -51. The shaded boxes highlight the sequences shown in panel B. (B) The highest sequence homologies found among the various cBEs and scs′ correspond to CGATA clusters. Only the bases shared by at least half of the sequences are shaded. The CGATA motifs are indicated by arrows above the sequences. The positions of the point mutations introduced into the scs′ D site are marked with asterisks. (C) The isolated cBEs contain high-affinity binding sites for BEAF. Affinity-purified BEAF was added in steps increased by a factor of 3. From the relative intensities of the shifted probes, we estimated that BEAF binds the cBEs as well as or better than the high-affinity D site of scs′.
Like bacterially expressed 32B, BEAF purified from Drosophila binds to these cBEs with very high affinity (Fig. 2C). We roughly estimated the affinity for these cBEs to be 2 to 10 times higher than that for the scs′ D subfragment, for which BEAF has a Kd of about 25 pM. The Kd of BEAF for the scs′ B site is about 600 pM. Perhaps this lower affinity is related to the lesser homology with the sequences presented in Fig. 2B.
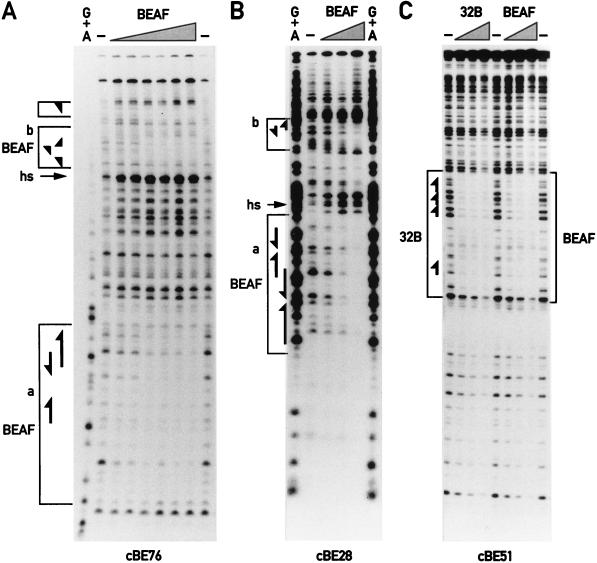
DNase I footprinting experiments confirmed the importance of the CGATA clusters for binding. As for scs′, all CGATA motifs were protected by BEAF from DNase I, while other sequences remained accessible (Fig. 3), and similar footprints were obtained for both BEAF and 32B proteins (Fig. 3C and data not shown). It is of particular interest that the footprints on cBE76 and cBE28 are bipartite, protecting the two CGATA clusters that are separated by about 70 and 50 bp, respectively, but not the sequences in between. As previously observed for the scs′ D site (14), the DNase I footprints of BEAF often have adjacent hypersensitive sites (Fig. 3A and B) that are less prominent or absent when 32B is used (data not shown).
FIG. 3.
Footprint analysis of cBEs: protected regions and hypersensitive sites. Footprint analyses of cBE76 (A), cBE28 (B), and cBE51 (C) were done by using increasing amounts of either affinity-purified BEAF or bacterially expressed 32B protein, as indicated. Induced hypersensitive sites (hs) are marked. Boxed regions indicate the DNase I footprints, and the arrowheads indicate the CGATA motifs.
In conclusion, the different genomic BEAF binding sites consist of several clustered CGATA motifs to which BEAF binds with high affinity despite the varied arrangement of the motifs.
cBEs neither activate transcription nor block upstream enhancers in transient expression assays.
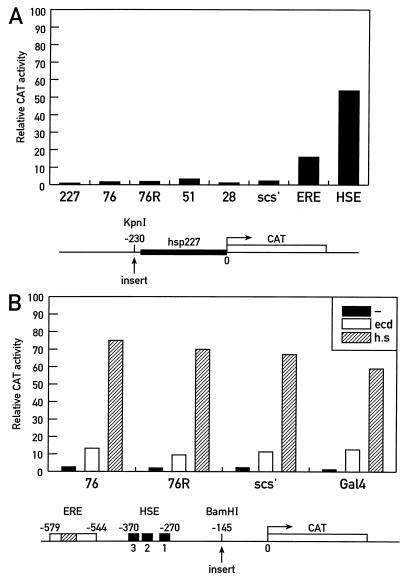
The sequence of cBE76 was present in the Drosophila DNA databases, but those of the other cBEs were not. This 320-bp fragment corresponds to upstream sequences from −350 to −670 of the inosine monophosphate dehydrogenase gene (IMPdH) at the raspberry locus (26; see Fig. 5A). This prompted us to test whether cBEs act as upstream activating sequences. All cBEs were placed 5′ of a 227-bp hsp27 CAT reporter gene which contains no upstream activating elements (30, 38). The full-length 1,200-bp hsp27 promoter, which encompasses an ecdysone response element (ERE), as well as three heat shock elements (HSE; 30), was used as a positive control. Furthermore, all transfections included a plasmid expressing a luciferase gene from an actin 5C promoter to assess the efficiency of transfection. CAT enzyme activity levels were measured after transient transfection of Drosophila D1 cells. In line with previous observations made for the scs′ element (38), the presence of the cBEs did not stimulate CAT significantly above the background. In contrast, high-level expression was observed following activation of the 1,200-bp promoter construct by ecdysone or heat shock (Fig. 4A).
FIG. 5.
Two hypersensitive sites are present in the 5′ region of the IMPdH raspberry gene, one of which overlaps cBE76. Nuclei isolated from Kc cells were treated with DNase I for different lengths of time. The isolated DNA was subjected to indirect end labelling with a PvuII-XhoI fragment from the IMPdH gene as a probe (shown in the map on the right). A major hypersensitive site (hatched box) localizes to the cBE76 fragment (filled box). A second hypersensitive site is located 200 bp closer to the IMPDH gene. Lane EcoRI contained genomic DNA cut with PvuII and EcoRI (which cuts within cBE76) as a size standard.
FIG. 4.
cBEs do not activate transcription, and cBE76 does not block upstream enhancers in transient expression assays. Different CAT reporter constructs were assayed by transient transfection into Drosophila D1 cells. (A) Upstream activating sequence assay. Relative CAT activities obtained when the indicated sequences were inserted upstream of the minimal 227-bp hsp27 CAT reporter gene. Ecdysone or heat shock induction of a 1.2-kb hsp27 promoter (B), which contains the ERE and HSE, served as the positive control. (B) The enhancer-blocking capacities of cBE76 (both orientations), scs′, and a 200-bp fragment containing a Gal4 binding site were tested. These fragments were inserted into the 1,200-bp hsp27 promoter between the promoter and the upstream HSE and ERE as shown. Relative CAT activity obtained following treatment with ecdysone (ecd) or heat shock (h.s.) or without treatment (−) is shown.
We also tested the enhancer-blocking potential of cBE76 by inserting it between the promoter and the upstream ERE and HSE enhancers of the 1,200-bp hsp27 reporter gene (Fig. 4B). A fragment containing Gal4 binding sites was used as a neutral sequence to control for spacing effects. Neither cBE76, in either orientation, nor scs′ affected the CAT activity in extracts prepared from transiently transfected cells after heat shock or ecdysone induction (Fig. 4B). These sequences act as neutral spacers; they do not activate or insulate in transient expression assays. We have previously used the same reporter gene to demonstrate that enhancer blocking by scs′ is only observed following stable genomic integration of the transgene (38).
Boundary elements do not block upstream enhancers in transient expression assays and do not activate transcription. Therefore, these results are consistent with the possibility that the cBEs are boundary elements.
cBE76 contains a nuclease-hypersensitive site.
Regulatory DNA elements often form structures in chromatin that are hypersensitive to nuclease digestion, and the scs and scs′ special chromatin structures were originally identified as major nuclease-hypersensitive sites bracketing the heat shock puff (33). Indeed, it appears that boundary elements are generally associated with hypersensitive sites (4, 6, 18, 34, 35). It was of interest, therefore, to examine cBE76 for nuclease hypersensitivity. Our analysis revealed a major hypersensitive site in the cBE76 region and a second one about 200 bp closer to the transcription start site (Fig. 5A). The sequences in between are notably AT rich. Similar results were obtained with micrococcal nuclease digestions (data not shown). As with the transient expression results, this result is consistent with the possibility that cBE76 is a boundary element.
The two hypersensitive sites upstream of the IMPdH gene are reminiscent of scs and scs′, which contain a pair of sites separated by an AT-rich region. It needs to be stressed, however, that while our data (see below) implicate cBE76 in boundary function, we do not know if sequences encompassing the IMPdH promoter-proximal hypersensitive site could enhance boundary activity or function as a transcription regulation element of this promoter.
cBE76 and cBE28 function as boundary elements in transgenic flies.
We investigated the boundary activity of the cBEs by testing their ability to shield a mini-white reporter gene (27) from chromosomal position effects, as originally done for scs and scs′ by Kellum and Schedl (20). This gene has a minimal promoter which drives low levels of expression, although after chromosomal integration, it is often activated to higher levels through adventitious interactions with endogenous enhancers in the vicinity. Consequently, when the gene is integrated into fly strains that have white eyes, the transformed flies display a range of eye color phenotypes. The relative level of expression is conveniently assessed on the basis of eye color, which changes with increased expression from yellow through orange to dark red (20, 34). When bracketed by boundary elements, the mini-white gene is insulated from these chromosomal position effects so that there is less variability in eye color between independent transgenic lines, and most lines have yellow eyes.
For this assay, we placed cBE76 or cBE28 5′ of a mini-white gene construct that had a 3′ scs element (Fig. 6D and E). As negative controls, we used the mini-white reporter without bracketing elements or with only a 3′ scs element, and as a positive control, we used the mini-white reporter bracketed by the scs′ and scs elements (Fig. 6A, B, and C). At least 10 independent transgenic lines were obtained for each construct by P-element-mediated transformation. We then scored animals from each line for their eye color phenotype. The data are summarized in Table 1, and representative photographs of the different transgenic lines are shown in Fig. 6.
TABLE 1.
Eye phenotypes of transformed lines
 |
No. of lines with eyesa that were:
|
Total no. of lines | ||||
|---|---|---|---|---|---|---|
| Yell | Or | Red | Var | |||
| 2 | 3 | 7 | 12 | |||
| scs | 5 | 7 | 6 | 1 | 19 | |
| scs′ | scs | 8 | 1 | 1 | 10 | |
| 76 | scs | 10 | 2 | 0 | 12 | |
| 28 | scs | 13 | 5 | 1 | 19 | |
| MM | scs | 8 | 1 | 1 | 10 | |
| M*M* | scs | 2 | 1 | 7 | 10 | |
Eye colors are as follows: Yell, light yellow to yellow; Or, light orange to dark orange-brown; Red, light red to wild-type; Var, variegated.
As found previously by others (4, 20), mini-white gene expression levels varied greatly between transgenic lines lacking flanking boundary elements. Among the 12 transgenic lines, 7 had red eyes, 3 had orange eyes, and only 2 displayed yellow eyes. Thus, the expression was strongly skewed toward high levels (red and orange), supposedly due to chromosomal position effects caused by activation by enhancers near the sites of integration. A similar effect was observed for the mini-white reporter flanked only on the 3′ side by scs; eye colors were skewed toward high-level, unbuffered expression (Fig. 6A and B and Table 1). In contrast, as shown originally by Kellum and Schedl (20), bracketing of the mini-white gene by scs′ and scs greatly reduces these position effects so that most transgenic lines have yellow eyes (Fig. 6C and Table 1).
Similar insulated expression was obtained when cBE76 replaced scs′. Among the 12 lines transgenic for cBE76, 10 had yellow eyes, only 2 had (light) orange eyes, and none displayed red eyes (Fig. 6D and Table 1). These data identify cBE76 as a boundary element—hereafter called BE76—that is located upstream of the IMPdH gene.
The 270-bp cBE28 fragment (hereafter called BE28) was also found to have a boundary function. Of 19 lines transgenic for BE28, 13 had yellow eyes, 5 had orange eyes (4 had light orange eyes), and 1 had red eyes (Fig. 6E and Table 1). Southern analyses showed that most lines had a single P-element insertion, but the red-eyed BE28 line had multiple P elements (see Materials and Methods), so in this case, the red-eye phenotype could be due to gene dosage. Thus, both cBEs insulate the mini-white gene from chromosomal position effects, identifying them as new boundary elements.
BEAF binding sites are involved in boundary function.
BEAF binds to the CGATA clusters present in scs′ and the cBEs, so we evaluated the functional importance of BEAF binding sites by mutagenesis of CGATA motifs. A tandem repeat of a 215-bp region of scs′ (fragment M) encompassing the high-affinity D site (Fig. 1C) was constructed such that the spacing between the two binding sites (227 bp) was the same as that between the high (D)- and low (B)-affinity binding sites in scs′ (Fig. 1C). A similar dimer was constructed in which all CGATA motifs were changed to CTCGA; these mutations abolished binding by Drosophila BEAF (14 and data not shown). We refer to the first dimer as MM and the mutated version as M*M*.
These constructs were placed 5′ of the mini-white gene with scs positioned 3′ and tested in flies as described above. We obtained 10 transgenic lines for each construct (Fig. 6F and G and Table 1). The MM construct convincingly buffered mini-white gene expression from position effects; seven lines had yellow eyes, while only two had (light) orange eyes and one had red eyes. Hence, the dimer construct containing the tight binding sites of scs′ serves as an efficient boundary element. In stark contrast, the majority of fly lines containing the M*M* construct had red eyes and only two lines had yellow eyes. Thus, the BEAF binding site is necessary for the boundary element function of this sequence, strongly suggesting that binding by BEAF is necessary.
DISCUSSION
BEAF binds to the scs′ boundary element and immunolocalizes to hundreds of interband regions on Drosophila polytene chromosomes (38). We isolated genomic DNA fragments containing BEAF binding sites in order to assess whether these binding sites possess boundary activity and to identify their consensus sequence motifs. Our demonstration that these sequences protect a reporter gene from chromosomal position effects and that the BEAF binding site is important for this activity provides the first example of a class of boundary elements that have a binding protein in common and argues that the numerous genomic BEAF binding sites represent boundary elements.
Protein binding sites in the cBEs.
Like scs′, the cBEs described here have clustered CGATA motifs. Interestingly, no preferential arrangement of these motifs emerged and perfect inverted repeats, as present in scs′, are not necessary for binding by BEAF. That BEAF primarily recognizes the CGATA motifs was demonstrated by DNase I footprinting experiments. For instance, BE76 and BE28 both have two clusters of motifs separated by 50 to 70 bp and BEAF protects both clusters but not the intervening DNA. We previously reported evidence that BEAF in Drosophila nuclear extracts binds DNA predominantly as trimers composed of one 32A and two 32B subunits, and it is 32B that recognizes the CGATA motif (14). Here we observed that all CGATA motifs present in the cBEs were protected, suggesting that complexes larger than trimers might stabilize binding to low-affinity sites as observed for scs′ (14).
One reason for isolating genomic cBEs was to search for further associated factors. Indeed, an additional binding activity which bound to BE76 and BE28 was detected in Drosophila nuclear extracts. This activity was purified by DNA affinity chromatography and identified by peptide sequencing as transcription factor DREF (15). DREF binds to extended ClaI sites, as found in BE76 and BE28, and is implicated in the regulation of several genes, including some involved in DNA replication (16, 17, 23). The in vitro interaction of DREF with these boundary elements is intriguing. However, neither the transient expression nor the transgenic fly data presented here provided evidence that BE28 or BE76 significantly activated transcription. We also found that binding by BEAF and DREF to these elements was mutually exclusive. It is possible that, depending on the developmental stage or the specific tissue, binding by one excludes any effect of the other and the conditions we assayed did not allow activation by DREF.
Boundary activity of the cBEs and BEAF binding sites.
We have biochemically characterized three of the cBEs, and two were assayed for the ability to confer position-independent expression of the mini-white gene. The ability of a sequence to insulate against chromosomal position effects is inferred by examining the distribution of eye colors obtained from a suitable number of independent transformants. In our case, position-independent expression would result in low-level expression manifested as yellow or light orange eyes in nearly all lines with a particular construct. We have obtained at least 10 lines for each construct and found BE76 and BE28 to be at least as effective as scs′ at buffering mini-white expression (Table 1). The few lines with darker eyes obtained for constructs containing boundary elements presumably result from infrequent integration events near enhancers with particularly strong affinity for the white promoter. It is known that not all enhancer-promoter combinations are blocked in enhancer-blocking assays. For instance, Vazquez and Schedl (35) found that scs′ does not effectively block interactions between the promoter and eye enhancer of the white gene. They went on to show that simple reiteration of a 200-bp scs subfragment up to a tetramer resulted in progressive improvement in enhancer-blocking ability from no blocking to an effectiveness equivalent to that of the original scs element. Although BE28 and BE76 are potent boundary elements despite their small sizes (320 and 270 bp, respectively), it would be of interest to see if reiteration or inclusion of flanking sequences would further improve their potency.
The functional importance of BEAF binding sites was addressed with an artificial construct composed of a dimerized fragment of scs′ containing the high-affinity binding site. This 215-bp region of scs′ was sufficient for boundary activity as a dimer. Mutating the CGATA motifs clearly eliminated boundary activity, demonstrating the importance of these motifs.
Interestingly, the mutations eliminated binding by 32B and BEAF purified from Drosophila but not by bacterially expressed 32A protein. Either 32A does not bind these sequences in vivo despite its ability to do so in vitro, or 32A alone is not sufficient for boundary activity, at least in this sequence context. Immunostaining of polytene chromosomes suggested that the ratio of 32A to 32B varies at different loci (14), raising the possibility that 32A can act without 32B in some sequence contexts. Our transgenic fly data extend the somewhat equivocal results obtained by using cultured cells containing many copies of the integrated transgene per transgenic line (38). In that study, seven tandem copies of a 48-bp oligonucleotide containing the scs′ high-affinity binding site blocked an upstream enhancer, although not as effectively as scs′. Point mutations in the CGATA motifs identical to those used in the present study impaired, but did not completely eliminate, the enhancer-blocking ability of the 7-mer. Perhaps those results were not clearer because the large tandem arrays of transgenes integrated into the chromosomes compromised the assay, or perhaps some aspect of the constructs, such as spacing between BEAF binding sites, was not optimal. The dimerized fragment used here maintained the spacing of the BEAF binding sites of scs′. It will be important to determine the role of spacing between binding sites and to find out whether scs′ sequences, in addition to the BEAF binding sites, are involved. In this regard, neither BE76 nor BE28 has BEAF binding sites separated by 200 bp, as found in scs′. Combined with the BE76 and BE28 data, we propose that the role of BEAF in boundary function relies on a somewhat flexible clustering of CGATA motifs.
The BEAF class of boundary elements.
If the notion of partitioning the genome into functional domains is correct, then one would expect there to be a limited number of classes of boundary elements that bracket the domains. A class would be defined by conserved sequence elements and the proteins involved in establishing boundary activity, although there could be overlap between classes. Yet no notable sequence homology or common binding activity has been found for the few boundary elements described so far, such as the su(Hw) binding sites, scs, scs′, Mcp, and Fab7, although in all cases examined, nuclease-hypersensitive sites localize to these elements (6, 7, 12, 18, 34). This lack of common features, combined with the difficulty of identifying boundary elements despite their proposed importance, has called the functional-domain model into question.
The data presented here identify for the first time a class of boundary elements that have a sequence motif and binding protein in common, i.e., the clustered CGATA sequences to which BEAF binds. Thus, the boundary activity of certain elements is not an isolated phenomenon but appears to occur generally throughout the genome. This is based on the function of the cBEs in transgenic flies, although the physical location of BE76 upstream of a transcription unit is also consistent with its being a domain boundary. BE76 also appears to localize to a band-interband junction in the raspberry locus at 9E3 on polytene chromosomes (5). Significantly, there are roughly 400 copies of BE28 sequences dispersed along the chromosome arms, suggesting that BE28 represents a family of boundary elements within the class that interacts with BEAF (5). We are currently analyzing the structure of the repeat and its genomic distribution in more detail. Perhaps it will be a better model for studying boundary elements and provide a useful tool for gaining insight into the functional organization of chromosomes. It might also help in addressing the notion that the physical organization of polytene chromosomes into bands and interbands may reflect a functional organization into domains.
ACKNOWLEDGMENTS
We are grateful to Vincenzo Pirrotta for sharing expertise and materials to work with flies. We acknowledge the generous advice of Francois Karch, Christophe Tatout, and Martin Müller. We thank Therese Durussel-Jost and I. Hogga for technical expertise and N. Roggli for help with the preparation of the figures.
This work was supported by the Swiss National Fund, the Canton of Geneva, and the Louis Jeantet Foundation.
REFERENCES
- 1.Adachi Y, Laemmli U K. Identification of nuclear prereplication centers poised for DNA synthesis in Xenopus egg extracts: immunolocalization study of replication protein A. J Cell Biol. 1992;119:1–15. doi: 10.1083/jcb.119.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 3.Cai H, Levine M. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 1997;16:1732–1741. doi: 10.1093/emboj/16.7.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung J H, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 5.Cuvier, O., and U. K. Laemmli. Unpublished data.
- 6.Ellis J, Tan-Un K C, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 7.Galloni M, Gyurkovics H, Schedl P, Karch F. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 1993;12:1087–1097. doi: 10.1002/j.1460-2075.1993.tb05750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gdula D A, Corces V G. Characterization of functional domains of the su(Hw) protein that mediate the silencing effect of mod(mdg4) mutations. Genetics. 1997;145:153–161. doi: 10.1093/genetics/145.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerasimova T I, Gdula D A, Gerasimov D V, Simonova O, Corces V G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 10.Gerasimova T I, Corces V G. Boundary and insulator elements in chromosomes. Curr Opin Genet Dev. 1996;6:185–192. doi: 10.1016/s0959-437x(96)80049-9. [DOI] [PubMed] [Google Scholar]
- 11.Geyer P K, Corces V G. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 12.Hagström K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 13.Harrison D A, Gdula D A, Coyne R S, Corces V G. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 1993;7:1966–1978. doi: 10.1101/gad.7.10.1966. [DOI] [PubMed] [Google Scholar]
- 14.Hart C M, Zhao K, Laemmli U K. The scs′ boundary element: characterization of boundary element-associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart, C. M., O. Cuvier, and U. K. Laemmli. Unpublished data.
- 16.Hirose F, Yamaguchi M, Handa H, Inomata Y, Matsukage A. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J Biol Chem. 1993;268:2092–2099. [PubMed] [Google Scholar]
- 17.Hirose F, Yamaguchi M, Kuroda K, Omori A, Hachiya T, Ikeda M, Nishimoto Y, Matsukage A. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J Biol Chem. 1996;271:3930–3937. doi: 10.1074/jbc.271.7.3930. [DOI] [PubMed] [Google Scholar]
- 18.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994;22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Käs E, Laemmli U K. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992;11:705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 21.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Shen B, Rosen C, Dorsett D. The DNA-binding and enhancer-blocking domains of the Drosophila suppressor of Hairy-wing protein. Mol Cell Biol. 1996;16:3381–3392. doi: 10.1128/mcb.16.7.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsukage A, Hirose F, Hayashi Y, Hamada K, Yamaguchi M. The DRE sequence TATCGATA, a putative promoter-activating element for Drosophila melanogaster cell-proliferation-related genes. Gene. 1995;166:233–236. doi: 10.1016/0378-1119(95)00586-2. [DOI] [PubMed] [Google Scholar]
- 24.Mirkovitch J, Mirault M E, Laemmli U K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 25.Mueller P R, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 26.Nash D, Hu S, Leonard N J, Tiong S Y K, Fillips D. The raspberry locus of Drosophila melanogaster includes an inosine monophosphate dehydrogenase like coding sequence. Genome. 1994;37:333–344. doi: 10.1139/g94-046. [DOI] [PubMed] [Google Scholar]
- 27.Pirrotta V. Vectors for P-mediated transformation in Drosophila. In: Rodriguez R L, Denhardt D T, editors. Vectors: a survey of molecular cloning vectors and their uses. London, England: Butterworths; 1983. pp. 437–456. [Google Scholar]
- 28.Pirrotta V, Steller H, Bozzetti M P. Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J. 1985;4:3501–3508. doi: 10.1002/j.1460-2075.1985.tb04109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian S, Pirrotta V. Dosage compensation of the Drosophila white gene requires both the x chromosome environment and multiple intragenic elements. Genetics. 1995;139:733–744. doi: 10.1093/genetics/139.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riddihough G, Pelham H R B. Activation of the Drosophila hsp27 promoter by heat shock and by ecdysones involves independent and remote regulatory sequences. EMBO J. 1986;5:1653–1658. doi: 10.1002/j.1460-2075.1986.tb04408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roseman R R, Pirrotta V, Geyer P K. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott K S, Geyer P K. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein gene. EMBO J. 1995;14:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez J, Farkas G, Gaszner M, Udvardy A, Muller M, Hagstrom K, Gyurkovics H, Sipos L, Gausz J, Galloni M, Hogga I, Karch F, Schedl P. Genetic and molecular analysis of chromatin domains. Cold Spring Harbor Symp Quant Biol. 1993;58:45–53. doi: 10.1101/sqb.1993.058.01.008. [DOI] [PubMed] [Google Scholar]
- 35.Vazquez J, Schedl P. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 37.Xiao H, Perisic O, Lis J T. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- 38.Zhao K, Hart C M, Laemmli U K. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]