Abstract
Translation of the upstream open reading frame (uORF) in the 5′ leader segment of the Neurospora crassa arg-2 mRNA causes reduced initiation at a downstream start codon when arginine is plentiful. Previous examination of this translational attenuation mechanism using a primer-extension inhibition (toeprint) assay in a homologous N. crassa cell-free translation system showed that arginine causes ribosomes to stall at the uORF termination codon. This stalling apparently regulates translation by preventing trailing scanning ribosomes from reaching the downstream start codon. Here we provide evidence that neither the distance between the uORF stop codon and the downstream initiation codon nor the nature of the stop codon used to terminate translation of the uORF-encoded arginine attenuator peptide (AAP) is important for regulation. Furthermore, translation of the AAP coding region regulates synthesis of the firefly luciferase polypeptide when it is fused directly at the N terminus of that polypeptide. In this case, the elongating ribosome stalls in response to Arg soon after it translates the AAP coding region. Regulation by this eukaryotic leader peptide thus appears to be exerted through a novel mechanism of cis-acting translational control.
Short peptide coding regions in the 5′ leaders of prokaryotic mRNAs (leader peptides) and eukaryotic mRNAs (upstream open reading frames [uORFs]) can serve critical regulatory functions. For example, analyses of evolutionarily conserved mechanisms regulating bacterial amino acid biosynthetic gene expression revealed the phenomenon of transcription attenuation, in which the movement of ribosomes over specific leader peptide coding sequences regulates operon expression (14). Studies of fungal genes involved in multiple-pathway control of amino acid biosynthesis, particularly those of Saccharomyces cerevisiae GCN4, have revealed the importance of multiple, cis-acting uORFs in the gene’s mRNA 5′ leader and of modulating the activity of trans-acting translation factors in regulation (10).
The Neurospora crassa arg-2 gene was one of the first amino acid biosynthetic genes to be identified (22). It encodes the small subunit of arginine-specific carbamoyl phosphate synthetase. It is unique among N. crassa Arg biosynthetic genes in that it is negatively regulated by the concentration of Arg in the cell (4). The arg-2 mRNA contains a uORF specifying a 24-residue peptide (20). The uORF encoding this peptide, henceforth called the arg-2 arginine attenuator peptide (AAP) because of its involvement in Arg-specific translational regulation, is evolutionarily conserved; similar AAP sequences are encoded by uORFs in all of the other fungal genes specifying this enzyme that have been characterized so far (1, 21, 27).
Elimination of the uORF initiation codon in the N. crassa arg-2 or the homologous S. cerevisiae CPA1 transcripts shows that uORF translation is crucial for Arg-specific regulation in vivo (9, 19, 27). In N. crassa, the arg-2 uORF reduces translation of ARG2 in vivo by reducing the average number of ribosomes associated with the arg-2 mRNA when Arg is plentiful in the growth medium (18). Arg-specific translational regulation mediated by the arg-2 uORF has been reconstituted in vitro by using a homologous cell-free translation system (25). A primer extension inhibition assay that allowed mapping of ribosomes on RNA at positions corresponding to rate-limiting steps in translation has been developed (26). Through high-resolution mapping of primer extension products, this toeprint assay enabled the localization of ribosomes on RNAs that are engaged in initiation with AUG codons in their P sites and ribosomes engaged in termination with termination codons in their A sites. Ribosome movement that was slowed during elongation by limitation for specific amino acids became stalled with codons for the limiting amino acid in their A sites (26).
Toeprint assays indicate that a high level of Arg causes ribosomes translating the arg-2 uORF to stall with its termination codon in the ribosomal A site (26). This primary event of Arg-regulated stalling of ribosomes is hypothesized to result in Arg-specific negative regulation because the stalled ribosomes block ribosomal scanning from the 5′ end of the mRNA and therefore block trailing ribosomes from reaching the downstream initiation codon. Whether termination is required for Arg-regulated ribosome stalling remains an important question that is not answered by these studies.
Amino acid residues in the arg-2 uORF peptide critical for its function in Arg-specific regulation have been identified. For example, changing Asp-12 of the uORF coding region product to Asn (D12N) eliminates Arg-specific translational control in vivo (9) and in vitro (25, 26). In parallel fashion, mutation of the corresponding Asp residue in the S. cerevisiae CPA1 uORF product eliminates Arg-specific regulation in vivo (5, 27). Other mutations in the N. crassa and S. cerevisiae uORFs that change the predicted amino acid sequence in the evolutionarily conserved region of the uORF peptide also reduce or eliminate Arg-specific translational control (19, 26, 27). When tested in the N. crassa cell-free system, loss of regulation is associated with reduced stalling at the uORF termination codon (26).
In the present study, we investigated the requirements for N. crassa arg-2 uORF function in translational regulation by using the N. crassa cell-free translation system. We first examined the effects of shortening the distance between the uORF termination codon and the downstream initiation codon. Subsequently, we changed the uORF termination codon, which is normally UAA, to UAG and UGA, to determine whether Arg-specific stalling at the termination codon is codon specific. Finally, to test whether negative translational regulation by Arg requires the uORF termination codon and a downstream initiation codon, we fused functional wild-type AAP and nonfunctional mutant AAP peptides directly to the luciferase (LUC) reporter at the LUC N terminus. The results obtained indicate that AAP-mediated, Arg-specific stalling of ribosomes occurs immediately following AAP translation. Translation of the AAP results in a situation in which the movement of ribosomes involved in either termination or elongation is stalled in response to Arg. While stalling requires a specific nascent peptide sequence, it does not appear to require specific RNA sequences distal to the AAP coding region. The results indicate that the nascent peptide encoded by the arg-2 uORF modulates the expression of a downstream gene product by affecting ribosome movement. The regulation of movement of ribosomes involved in either termination or elongation reveals a novel form of eukaryotic translational control.
MATERIALS AND METHODS
Construction of templates for RNA synthesis.
Plasmids were designed to produce capped and polyadenylated synthetic RNA encoding firefly LUC with wild-type or mutant arg-2 sequences in the RNA 5′ leader region (Fig. 1 and Table 1). Mutations were introduced into this region by PCR with mutagenic primers (Table 1) by using procedures described previously (9). PCR products were ligated into the pHLUC+NFS4 vector (Table 1) (25). A plasmid designed to produce capped and polyadenylated synthetic RNA which lacked arg-2 sequences and that encoded sea pansy LUC was constructed by replacing the firefly LUC coding region of pHLUC+NFS4 with the sea pansy LUC coding region of pRL-CMV (Promega).
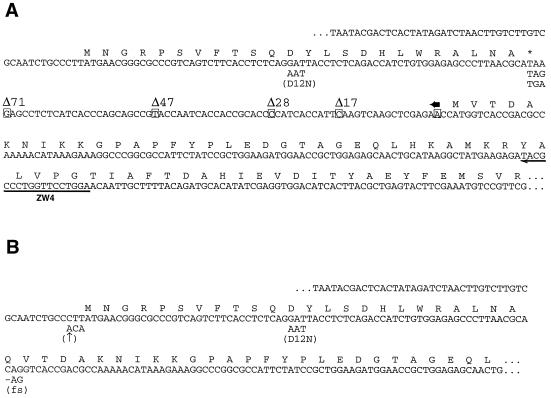
FIG. 1.
The 5′ leader regions of arg-2–LUC genes used in this study (see also Table 1). (A) Sequences of wild-type and mutant templates in which the AAP is encoded by a uORF. The sequence shown begins with the T7 RNA polymerase-binding site and ends within the LUC coding region (26). The amino acid sequences of the arg-2 AAP and the N terminus of LUC are indicated. Point mutations are shown below the wild-type sequence. The endpoints of deletion mutations that shorten the intercistronic region are indicated by the boxed nucleotides. All deletions share the 3′ endpoint, which is indicated by a horizontal arrow above the sequence. The extent of each deletion is indicated (e.g., Δ71 removes the greatest number of nucleotides, leaving 5′-CC-3′ between the uORF termination and LUC initiation codons). The sequence for which the reverse complement was synthesized and used as primer ZW4 for toeprint analysis is indicated by a horizontal arrow below the sequence. (B) Sequences of templates containing wild-type and mutant AAP-LUC fusion genes. The sequence shown begins with the T7 RNA polymerase-binding site and ends within the LUC coding region; the amino acid sequence of the N terminus of the AAP-LUC fusion polypeptide is indicated. Point mutations are shown below the wild-type sequence. The mutation indicated by ↑ improves the initiation context for uORF translation. The fs mutation is a −1 frameshift in which the first nucleotide of the Gln codon bridging the AAP and LUC coding sequences is deleted.
TABLE 1.
Firefly luciferase constructs used in this study
| Construct | 5′ leader structuree | Mutagenic primer (sequence)a | Template | Luciferase activityb |
|---|---|---|---|---|
| pPR101 | Wild type | 2.13 ± 0.14 (4) | ||
| pPS101 | D12N | 0.98 ± 0.03 (3) | ||
| pPR105 | Δ17 | FP6 (CACGC CATGG AATGG TGATG GGGTG CG) | pPR101 | 2.11 ± 0.18 (3) |
| pPR104 | Δ28 | FP5 (CACGC CATGG GGTGC GGTGG TGATT GGT) | pPR101 | 1.97 ± 0.11 (3) |
| pPR103 | Δ47 | FP4 (CACGC CATGG CGGCT GCTGG GTGAT GAG) | pPR101 | 1.83 ± 0.06 (3) |
| pPR102 | Δ71 | FP2 (CACGC CATGG TTATG CGTTA AGGGC TCTCC A) | pPR101 | 2.29 ± 0.11 (3) |
| pSF103 | D12N Δ71 | FP2 | pPS101 | 1.02 ± 0.10 (2) |
| pRF102 | Δ71-TGA | FP15 (CACGC CATGG TCATG CGTTA AGGGC TCTCC A) | pPR101 | 2.24 ± 0.05 (2) |
| pSF101 | D12N Δ71-TGA | FP15 | pPS101 | 0.95 ± 0.01 (2) |
| pRF103 | Δ71-TAG | FP16 (CACGC CATGG CTATG CGTTA AGGGC TCTCC A) | pPR101 | 2.19 ± 0.13 (2) |
| pSF102 | D12N Δ71-TAG | FP16 | pPS101 | 1.02 ± 0.04 (2) |
| pRF107 | AAP-LUC | FP3 (CACGG TGACC TGTGC GTTAA GGGCT CTCCA) | pPR101 | ND |
| pSF104 | D12N AAP-LUC | FP3 | pPS101 | ND |
| pT1011 | ↑AAP-LUC | pT101c | ND | |
| pJC1081 | D12N ↑AAP-LUC | FP3 | pJC108d | ND |
| pPR1071 | fs AAP-LUC | FP17 (CACGG TGACC TTGCG TTAAG GGCTC TCCA) | pPR101 | ND |
The oligonucleotide sequences are shown in the 5′ to 3′ direction. In all cases, mutagenic primers were paired with the forward primer FP1 (5′-CCGCAAGGAATGGTGCAT-3′), which binds to the vector immediately upstream of the sequence shown in Fig. 1, to obtain PCR products from the indicated templates. PCR products were digested with BglII and NcoI (uORF-containing constructs) or BglII and BstEII (AAP-LUC fusion constructs) for ligation into correspondingly digested pHLUC+NFS4 vector (25). The NcoI site is at codon 1 and the BstEII site is at codon 2 of the LUC coding region in this vector.
The ratio of LUC enzyme activity produced after 30 min in reaction mixtures containing 10 μM Arg to that with 500 μM Arg. Values are means ± standard deviations; the numbers of independent experiments analyzed are shown in parentheses. Values for regulation of enzyme synthesis by Arg are given for RNAs containing a uORF specifying the AAP. Analyses of the regulation of AAP-LUC fusion constructs were not done in this way (ND); they were analyzed as described in the text.
The arg-2–LUC plasmid with the wild-type uORF in an improved initiation context (25).
The arg-2–LUC plasmid with the mutant D12N uORF in an improved initiation context (25).
For details, see legend to Fig. 1.
Cell-free translation of RNA and analyses of translation products.
Plasmid DNA templates were purified by equilibrium centrifugation and linearized with Ppu10I. Capped, polyadenylated RNA was synthesized with T7 RNA polymerase from linearized plasmid DNA templates, and the yield of RNA was quantified as described previously (25). The preparation of cell translation extracts from N. crassa and the reaction conditions for in vitro translation were as described previously (25, 26).
Translation was halted by freezing reaction mixtures in liquid nitrogen, and aliquots of the ice-thawed mixtures (5 μl) were used for luciferase assays. Luminometric measurements of enzyme activity were performed for 10 s after a programmed 2-s delay. For measurement of firefly LUC enzyme production only, the luciferase assay system (Promega) was used. For measurement of firefly and sea pansy LUC enzymes produced in the same reaction mixture (dual assay), an aliquot of translation mixture added to 50 μl of luciferase assay reagent II (Promega) and the firefly LUC enzyme was measured. Then, 50 μl of Stop & Glo reagent (Promega) was added manually and the sea pansy enzyme was measured.
For [35S]Met labeling of polypeptides, N. crassa extracts were treated with micrococcal nuclease (25). Quantitation of radiolabeled polypeptides was accomplished by using IPLab Gel and a Molecular Dynamics Phosphorimager to analyze the translation products that were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Primer extension inhibition (toeprint) assays.
The primer extension assays were accomplished as described previously by using primer ZW4 (26); 8 μl of sample instead of 4 μl was loaded onto each gel lane. The gels were dried and exposed to screens of a Molecular Dynamics Phosphorimager for approximately 24 h. All toeprint data shown are representative of multiple experiments.
RESULTS
Effects of reducing the distance between the uORF termination codon and the downstream initiation codon.
Wild-type arg-2 mRNA contains 63 nucleotides (nt) between the uORF stop codon and the ARG2 initiation codon. The arg-2 uORF coding sequence and the intercistronic region were placed upstream of the sequence coding for firefly LUC (another 10 nt of intergenic vector sequences are also present) (Fig. 1). This construct was used to produce capped and polyadenylated synthetic RNA. In vitro translation of this RNA in the presence of a high concentration of Arg (500 μM), in contrast to a low concentration of Arg (10 μM), results in an approximately twofold reduction in LUC polypeptide synthesis (Table 1) (see references 25 and 26). The D12N mutation of the AAP coding sequence (Fig. 1) eliminates the regulatory effect of the AAP (Table 1) (25, 26).
Primer extension inhibition analyses of RNA containing the wild-type arg-2 uORF translated under these conditions have been described previously in detail (26). The longest primer extension products (Fig. 2) represent cDNA extended from the primer to the 5′ end of the synthetic RNA. One set of shorter extension products corresponds to the inhibition of reverse transcription of the RNA template by ribosomes with initiation codons in their P sites (ribosomes at the uORF initiation codon and ribosomes at the LUC initiation codon are indicated in Fig. 2). Other shorter extension products correspond to ribosomes with the uORF termination codon in their A sites (Fig. 2). A comparison of the translation of the arg-2–LUC RNA containing the full intergenic region in reaction mixtures containing a low (10 μM) or high (500 μM) concentration of Arg shows that a high Arg concentration caused a substantial change in the distribution of ribosomes on the RNA (Fig. 2, lanes 1 and 2). These Arg-induced changes did not occur when the D12N mutation was present in the AAP (Fig. 2, lanes 3 and 4). With the wild-type RNA, Arg increased the signal corresponding to ribosomes at the uORF termination codon and caused a cluster of strong toeprints to appear 21 to 30 nt upstream of the termination codon toeprint. This latter cluster of toeprints appears to represent ribosomes whose movement is blocked by ribosomes that are stalled at the termination codon (26). They are likely visible in the toeprint assay because of the dissociation of ribosomes stalled at the termination codon (26).
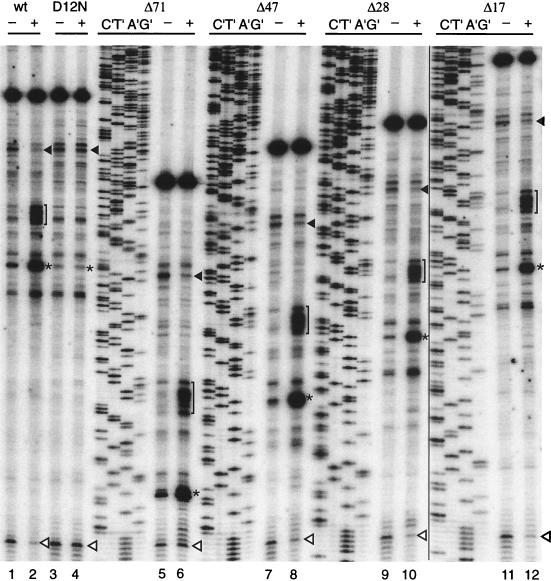
FIG. 2.
Effects of shortening the distance between the uORF termination codon and the downstream LUC initiation codon on Arg-specific regulation. Equal amounts of synthetic RNA transcripts (120 ng) were translated in 20-μl reaction mixtures for 20 min at 25°C. Reaction mixtures contained 10 μM (−) or 500 μM (+) Arg and a 10 μM concentration of each of the other 19 amino acids. The transcripts examined are indicated at the top of the lanes. After 20 min of translation, the translation mixtures were toeprinted with primer ZW4 and analyzed next to dideoxynucleotide sequences of the corresponding DNA template. The nucleotide complementary to the dideoxynucleotide added to each reaction mixture is indicated above the corresponding lane so that the sequence of each template can be directly deduced; the 5′-to-3′ sequence reads from top to bottom. The asterisks indicate the positions of premature transcription termination products corresponding to ribosomes at the uORF termination codon; brackets indicate ribosomes stalled behind those at the termination codon. The closed arrowheads indicate ribosomes at the uORF initiation codon; the open arrowheads indicate ribosomes at the LUC initiation codon. wt, wild-type.
The addition of Arg reduced the toeprint at the LUC initiation codon, consistent with the reduced translation of LUC as determined by a luciferase assay and with the model in which increased stalling of ribosomes at the uORF termination codon decreases ribosomal access to the downstream initiation codon (Fig. 2, lanes 1 and 2). The addition of Arg also reduced the toeprint corresponding to ribosomes at the uORF initiation codon. This decreased signal likely arises as a consequence of the primer extension assay and does not represent reduced binding at this site, for reasons discussed previously (26).
To examine whether the sequence or the distance between the uORF and the downstream start codon were important for regulation, a series of constructs (Fig. 1) were made in which there were progressive deletions of the region between the uORF termination codon and the downstream LUC initiation codon. Translation of RNA containing deletions of 17, 28, 47, or 71 nt of the 73-nt intercistronic region present in the original construct were examined by luciferase and toeprint assays. Progressive deletion of the intercistronic region had negligible effects on Arg-specific regulation as determined by luciferase activity and toeprint analyses. Translation of LUC in all cases was reduced approximately twofold by a high concentration of Arg under standard assay conditions, similar to the regulation observed with the full-length intercistronic region (Table 1). Toeprint analyses showed that the Arg-specific effects on ribosomes translating the uORF were similar in every case (Fig. 2). For all of the mutant mRNAs tested, the addition of Arg increased the toeprint signals corresponding to the uORF termination codon (asterisks), and the appearance of the cluster of toeprints 21 to 30 nt upstream of the toeprint corresponding to the termination codon (brackets), as is observed for the wild-type mRNA. There were few qualitative differences in these signals of each mutant, except for the shortened distance between these signals, as predicted for the size of the deletion.
Consistent with reduced synthesis of LUC, a decrease in the signal corresponding to ribosomes at the LUC initiation codon was observed for most constructs. However, RNA constructs containing the largest deletion, in which only 2 nt separate the uORF termination codon and the LUC initiation codon, did not show a reduced toeprint signal at the LUC initiation codon under high-Arg-concentration conditions (Fig. 2, lanes 5 and 6). The reason for this difference is unknown, but it might arise because ribosomes at the nearby termination codon were contributing to the signal at this position.
Effects of altering the uORF termination codon.
The uORF termination codon, UAA, appears to be the most frequently used in N. crassa (6). To test whether this specific termination codon was important for Arg-specific regulation, plasmids were constructed to produce synthetic RNA in which the wild-type or D12N mutant uORFs were terminated with UAA, UAG, or UGA codons. With the wild-type uORF, all three termination codons showed similar levels of Arg-specific regulation as determined by luciferase activity assays (Table 1, constructs pPR102, pRF102, and pRF103). Comparisons of toeprint assays of each RNA translated in reaction mixtures containing either low or high Arg concentrations indicated that the behaviors of ribosomes at each of these termination codons were indistinguishable (Fig. 3). Regardless of which uORF termination codon was present, the D12N mutant uORF conferred no Arg-specific regulation (Table 1 and Fig. 3). Therefore, while the sequence of the uORF coding region is important for regulation, the type of termination codon used to stop uORF translation did not appear to be important.
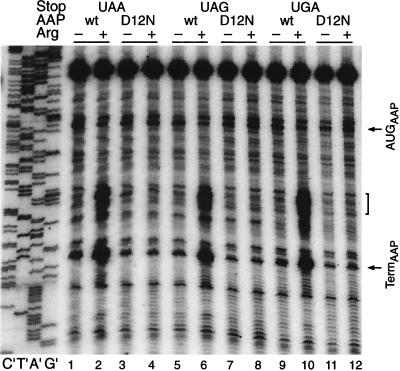
FIG. 3.
Effects of terminating uORF peptide synthesis with each of the three termination codons, UAA (lanes 1 to 4), UAG (lanes 5 to 8), and UGA (lanes 9 to 12), on Arg-specific regulation. Equal amounts of synthetic RNA transcripts (120 ng) were translated in 20-μl reaction mixtures for 20 min at 25°C. Reaction mixtures contained 10 μM (−) or 500 μM (+) Arg and a 10 μM concentration of each of the other 19 amino acids; they were analyzed by toeprinting as described in the legend to Fig. 2. The RNAs are from Δ71 deletion constructs containing either the wild-type (wt) or D12N mutant uORFs terminated with UAA, UAG, or UGA codons (Fig. 1 and Table 1) as indicated at the top of the lanes. Dideoxynucleotide sequencing reactions for the pPR102 template containing the wild-type uORF terminated with UAA are shown on the left (lanes C′, T′, A′, and G′). Arrows indicate the positions of premature transcription termination products corresponding to ribosomes bound at the uORF initiation codon (AUGAAP) and the uORF termination codon (TermAAP); brackets indicate the positions of ribosomes stalled behind those at the termination codon.
Effects of fusing the arg-2 uORF peptide directly to LUC.
The experiments described above indicate that the function of the arg-2 uORF in regulation was retained regardless of the distance between its termination codon and the downstream initiation codon and regardless of which termination codon was used to terminate uORF translation. Therefore, we next investigated whether the termination of uORF peptide synthesis followed by the initiation of new polypeptide synthesis at a downstream start codon was necessary for AAP-mediated translational regulation. Constructs were made in which the normal LUC initiation codon was eliminated and the wild-type AAP coding region was fused directly to the LUC coding region. In such constructs, the AAP initiation codon would be responsible for initiating translation of an AAP-LUC fusion polypeptide.
Three different AAP-LUC fusion constructs containing the wild-type AAP sequence were examined (Fig. 1B). The first contained the AAP initiation codon in its wild-type context, which is relatively inefficient at capturing ribosomes to initiate translation. The second contained the AAP initiation codon in an improved initiation context that is relatively efficient at capturing ribosomes to initiate translation (25, 26). The final construct contained AAP and LUC coding regions deliberately fused out-of-frame with respect to each other, so that, in contrast to the first two constructs, the LUC polypeptide will not be produced by initiation at the AAP initiation codon.
Evidence that these constructs produced fusion polypeptides as predicted was obtained by programming micrococcal nuclease-treated N. crassa translation reaction mixtures with equal amounts of each of the RNAs encoding the fusions and examining [35S]methionine incorporation into translation products (Fig. 4). RNA encoding normal firefly LUC polypeptide produced a major translation product whose migration in SDS-PAGE was consistent with its predicted size of 551 residues (Fig. 4, lane 2). RNA encoding the AAP-LUC fusion polypeptide in its normal initiation context produced a major translation product whose migration in SDS-PAGE was consistent with its predicted size of 574 residues (Fig. 4, lane 3). Improving the AAP-LUC translation initiation context increased the level of the fusion polypeptide (Fig. 4, lane 4). RNA in which the AAP coding region was fused out-of-frame with the LUC coding region did not produce this large fusion polypeptide (Fig. 4, lane 5), as expected. Consistent with these results, measurements of LUC production by enzyme assay showed that, after 30 min of translation, improving the AAP initiation codon increased the level of LUC translation approximately 4-fold and frameshifting reduced the level of LUC translation more than 50-fold (data not shown). These results indicated that, in RNAs encoding AAP-LUC fusion polypeptides, the AAP initiation codon was primarily responsible for initiating translation of active LUC enzyme in vitro.
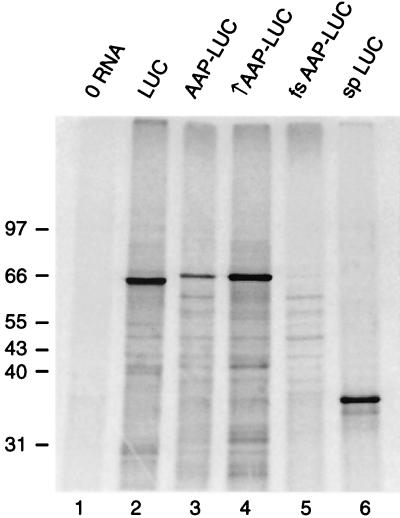
FIG. 4.
Analyses of [35S]methionine-labeled polypeptides produced by translation of synthetic RNA transcripts in N. crassa cell extracts. Micrococcal nuclease-treated N. crassa extracts (20 μl containing 2 μCi of [35S]methionine) were programmed with 120 ng of the indicated RNAs and incubated for 30 min at 25°C. Reactions were stopped by immersing the tube in liquid nitrogen and examined by SDS-PAGE in 10% polyacrylamide gels. Radiolabeled translation products were visualized by phosphorimaging; the positions of molecular mass markers (in kilodaltons) visualized by staining with Coomassie blue are indicated on the left. Lanes: 1, reaction mixture with no added RNA; 2, with RNA encoding firefly LUC; 3, with RNA encoding the AAP-LUC fusion polypeptide in the wild-type initiation context; 4, with RNA encoding the AAP-LUC fusion polypeptide in the improved initiation context; 5, with RNA encoding the AAP coding region frameshifted (fs) with respect to the LUC coding region; 6, with RNA encoding sea pansy (sp) LUC.
We examined the effect of adding 10 or 500 μM Arg to translation reaction mixtures on the synthesis of wild-type AAP-LUC or mutant D12N AAP-LUC fusion polypeptides. Each reaction mixture contained a second capped and polyadenylated RNA specifying sea pansy LUC as an internal control. This RNA lacked arg-2 regulatory sequences. Sea pansy LUC is smaller (311 amino acids) (Fig. 4, lane 6) than the firefly enzyme, and it uses a different substrate to produce light.
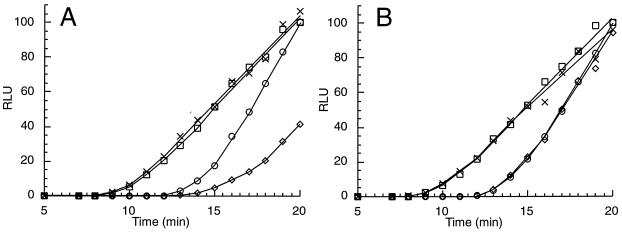
The rates of production of sea pansy LUC were similar in all reaction mixtures (Fig. 5). This indicated that translation of this RNA was unaffected by these levels of Arg. Analyses of the time course of LUC production (Fig. 5) revealed that, in mixtures containing low or high Arg concentrations, the appearance of completed functional sea pansy enzyme synthesis preceded the appearance of firefly enzyme synthesis, consistent with its smaller size. Therefore, excluding protein folding considerations, on the basis of the differences in the sizes of the sea pansy and firefly LUC enzymes and the rates of first appearance of functional enzymes, the elongation rate in the reaction mixtures under these conditions can be estimated to be approximately 1 amino acid per s, comparable to that of other eukaryotic cell-free systems (8). In contrast, translation of LUC from RNA encoding the wild-type AAP fused to LUC but not the D12N mutant AAP fused to LUC was reduced under high concentrations of Arg (Fig. 5). This indicated that the wild-type AAP at the N terminus of a fusion polypeptide functioned to reduce translation and therefore that it functioned in the absence of a termination codon and a downstream initiation codon. In addition, this Arg-responsive AAP activity was cis-acting: while it affected translation of RNA encoding the firefly enzyme, it did not affect translation of RNA encoding sea pansy LUC in the same reaction mixtures.
FIG. 5.
Time courses of translation of RNAs encoding AAP-LUC, D12N AAP-LUC, and sea pansy LUC enzymes in reaction mixtures containing 10 or 500 μM Arg. Translation in reaction mixtures (120 μl) was initiated by using a mixture of RNAs: 150 ng of RNA encoding the wild-type AAP-LUC fusion and 36 ng of RNA encoding sea pansy LUC (A) or 150 ng of RNA encoding the D12N AAP-LUC fusion and 36 ng of RNA encoding sea pansy LUC (B). Reaction mixtures were incubated at 25°C and contained either 10 or 500 μM Arg and a 10 μM concentration of each of the other 19 amino acids. Firefly LUC production (in reaction mixtures containing 10 [○] or 500 [◊] μM Arg) and sea pansy LUC production (in reaction mixtures containing 10 [□] or 500 [×] μM Arg) were determined by using the dual luciferase assay as described in Materials and Methods. RLU, relative light units.
The effect of Arg on the translation of RNA specifying the AAP-LUC fusion was qualitatively different from its effect when the RNA specified the AAP as a uORF product and LUC as a separate, downstream coding region product with its own translation initiation site (Fig. 6). Arg substantially delayed the first appearance of enzymatically active AAP-LUC fusion polypeptide and subsequently lowered the rate of accumulation of LUC (Fig. 6A). In parallel reactions, Arg did not delay the first appearance of enzymatically active LUC when the AAP was present as a uORF product but subsequently lowered the rate of accumulation of LUC (Fig. 6B). This delay in the synthesis of LUC in translation reaction mixtures containing a high instead of a low concentration of Arg and programmed with RNA specifying the AAP at the N terminus of LUC but not RNA specifying the AAP as a separate reading frame product upstream of LUC was highly reproducible, and the length of this delay was extended when translation reaction mixtures were incubated at lower temperatures (data not shown). These differences were also apparent when fusion and uORF constructs containing the AAP in an improved initiation context were compared (data not shown). In these cases, the magnitude of the regulatory effect of Arg increased (data not shown), consistent with the model for regulation in which Arg-stalled ribosomes impede trailing ribosome traffic.
FIG. 6.
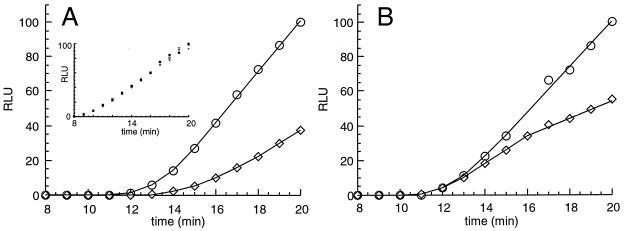
Time courses of translation of RNAs encoding the wild-type AAP as an N-terminal domain and as a uORF product. Reaction conditions were as described in the legend to Fig. 5. (A) Results with 150 ng of RNA encoding the wild-type AAP-LUC fusion and 36 ng of RNA encoding sea pansy LUC; (B) results with 150 ng of RNA encoding the wild-type AAP as a uORF region and LUC as a separate downstream coding region with its own initiation codon and 36 ng of RNA encoding sea pansy LUC. The inset in panel A shows the combined data for translation of the internal control sea pansy LUC RNA in the four reaction mixtures plotted in panels A and B. Firefly LUC production (in reaction mixtures containing 10 [○] or 500 [◊] μM Arg) and sea pansy LUC production were determined by using the dual luciferase assay as described in Materials and Methods. RLU, relative light units.
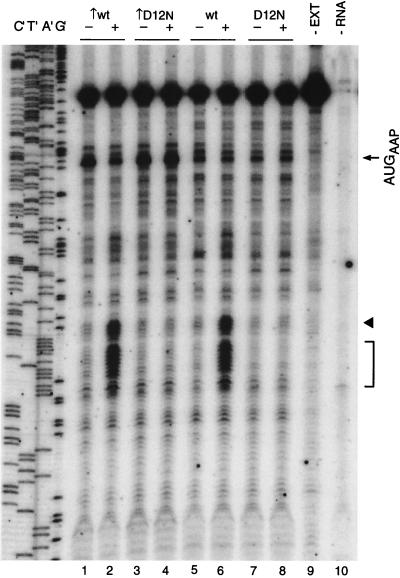
Toeprint analyses were used to examine the effects of Arg on the translation of RNAs specifying the wild-type AAP-LUC and the D12N AAP-LUC polypeptides following initiation at AUG codons in improved and wild-type initiation contexts (Fig. 7). The RNA coding for AAP-LUC in the wild-type initiation context was toeprinted in the absence of extract (Fig. 7, lane 9) as a control. As expected, the primer extension product was predominantly cDNA fully extended to the 5′ end of the RNA template. Also as expected, when toeprint signals from an extract lacking RNA coding for AAP-LUC were examined, near-negligible quantities of primer extension products were observed (Fig. 7, lane 10). Analyses of the wild-type AAP-LUC-specifying RNA translated in translation mixtures containing a low rather than a high Arg concentration (Fig. 7, compare lanes 5 and 6) revealed that a high concentration of Arg substantially increased the intensity of a series of toeprints. One of these corresponded to ribosomes translating the first codon following the AAP coding sequence (as borne out by experiments with the translation inhibitor puromycin as described below). Thus, this stall site in the fusion polypeptide corresponded in position, relative to the AAP coding sequence, to the uORF termination codon. This toeprint site was followed closely by another Arg-induced cluster of toeprints corresponding to ribosomes stalled in the nearby, downstream LUC coding region. The D12N mutation eliminated these Arg-specific effects on toeprints (Fig. 7, compare lanes 7 and 8). These data indicate that AAP-mediated stalling can occur at one or more sites immediately distal to the AAP coding region.
FIG. 7.
Effects of mutations on Arg-specific regulation of AAP-LUC fusion constructs. Equal amounts of synthetic RNA transcripts (120 ng) were translated in reaction mixtures and analyzed by toeprinting as described in the legend to Fig. 2. The transcripts encoded the wild-type (wt) AAP-LUC fusion or the D12N mutant AAP-LUC fusion as indicated in either the wild-type or improved (↑) initiation contexts. Dideoxynucleotide sequencing reactions for the template containing the wild-type AAP-LUC fusion are shown on the left (lanes C′, T′, A′, and G′). The products obtained from primer extension of pure AAP-LUC RNA (18 ng) in the absence of translation reaction mixture (−EXT; lane 9) and from a translation reaction mixture not programmed with RNA (−RNA; lane 10) are shown for comparison. The arrow indicates the position of the premature transcription termination products corresponding to ribosomes bound at the AAP initiation codon (AUGAAP). The arrowhead indicates the position of premature termination products corresponding to ribosomes stalled in the presence of a high level of Arg at the codon immediately following the 24 codons of the AAP. The bracket indicates the position of premature termination products corresponding to ribosomes stalled in the presence of a high level of Arg in the LUC coding region.
Improving the context of the initiation codon for AAP-LUC fusion polypeptide would be expected to increase translation by the more efficient capturing of scanning ribosomes. Consistent with this, we observed increased LUC translation from this RNA as determined by enzyme assay, as described above. Improving the initiation context also increased the toeprint signal corresponding to ribosomes initiating synthesis of the AAP-LUC polypeptide. This effect was observed for RNAs encoding both wild-type and D12N mutant fusions (Fig. 7, lanes 1 to 4). Addition of Arg reduced the toeprint corresponding to ribosomes at the wild-type AAP-LUC initiation codon in either context but not the D12N AAP-LUC initiation codon. This decreased signal in the wild-type cases may be a consequence of the assay procedure (26).
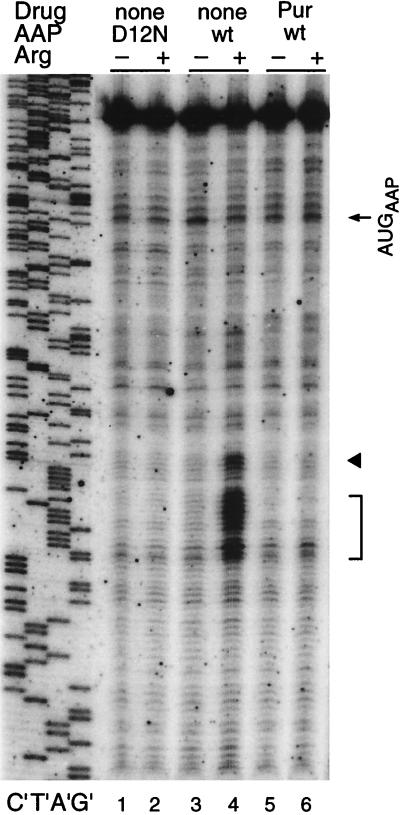
To verify that the Arg-specific toeprints corresponded to ribosomes, we examined the effect of puromycin, which releases ribosomes from mRNA, on these signals (Fig. 8). Translation reaction mixtures containing RNA encoding the wild-type AAP-LUC and either low or high concentrations of Arg were incubated for 20 min. Then, either water (negative control) or puromycin was added and the mixtures were incubated for an additional 5 min. Puromycin caused the loss of Arg-regulated toeprints (Fig. 8, lane 4), indicating that these signals corresponded to ribosomes. Puromycin did not release ribosomes from the AAP-LUC initiation codon as judged by toeprinting; this lack of an effect of puromycin on toeprints corresponding to initiation codons was observed previously (26). The toeprint pattern obtained from RNA coding for wild-type AAP-LUC with puromycin resembled the pattern obtained from RNA coding for D12N AAP-LUC in the absence of drug in that no stalled ribosomes are detected in the region downstream of the AAP region. This indicates that the toeprint assay is detecting ribosomes stalling in response to Arg that is specific to ribosomes which have synthesized the wild-type AAP.
FIG. 8.
Effects of puromycin on Arg-specific regulation of AAP-LUC fusion constructs. Equal amounts of synthetic RNA transcripts (120 ng) were translated in reaction mixtures and analyzed by toeprinting as described in the legend to Fig. 2. The transcripts encoded either the D12N mutant AAP-LUC fusion or the wild-type (wt) AAP-LUC fusion as indicated. Puromycin (Pur) was added where indicated, as described in the text. Dideoxynucleotide sequencing reactions for the template containing the wild-type AAP-LUC fusion are shown on the left (lanes C′, T′, A′, and G′). The arrow indicates the position of the premature transcription termination products corresponding to ribosomes bound at the AAP initiation codon (AUGAAP). The arrowhead indicates the position of premature termination products corresponding to ribosomes stalled in the presence of a high level of Arg at the codon immediately following the 24 codons of the AAP. The bracket indicates the position of premature termination products corresponding to ribosomes stalled in the presence of a high level of Arg in the LUC coding region.
DISCUSSION
The leader region of the N. crassa arg-2 mRNA contains a 24-residue, evolutionarily conserved uORF encoding the AAP. Translation of the AAP coding region appears necessary for Arg-regulated stalling of ribosomes. We observed that translational control through the AAP is cis-acting and does not appear to absolutely require specific downstream translational events to occur. Shortening the distance between the uORF termination codon and the downstream LUC initiation codon did not affect Arg-regulated stalling at the uORF termination codon or the extent of negative regulation conferred on translation initiated at the LUC initiation codon. Changing the uORF termination codon from UAA to UAG or UGA did not affect regulation. Indeed, negative translational regulation was observed when the AAP coding region was fused in-frame directly to the LUC coding region. The data suggest that the nascent AAP acts in cis within the ribosome that has translated it to cause stalling regardless of whether the ribosome is subsequently engaged in termination or elongation.
The regulatory function of the AAP to stall ribosomes was examined in two ways: by examining the kinetics of Arg-specific regulation in the N. crassa cell-free translation system and by mapping the positions of ribosomes on RNA in this system by using a primer extension inhibition (toeprint) assay. When the AAP is encoded by a uORF, the time of initial appearance of functional LUC enzyme (whose synthesis is initiated from a downstream start codon) is the same regardless of whether the reaction mixture contains a low or high concentration of Arg (Fig. 6B). The negative effect of Arg becomes apparent only later, when it reduces the rate of LUC enzyme accumulation. These data are consistent with the proposed model for regulation (26) mediated by the arg-2 uORF in which some of the first scanning ribosomal subunits loaded on the mRNA leak past the codon initiating AAP and initiate instead at the LUC coding region regardless of whether the level of Arg is high or low. In this model, the time at which the first functional LUC polypeptide is translated from such an mRNA pool is not affected by Arg, but Arg reduces LUC polypeptide synthesis subsequently.
In contrast to the situation in which the AAP is encoded by a uORF, for RNA encoding the AAP-LUC fusion polypeptide, ribosomes initiating at the AAP codon are directly responsible for LUC synthesis, as judged by analysis of radiolabeled LUC polypeptide and LUC enzyme activity produced from such RNAs. Thus, all of the ribosomes that initiate LUC synthesis necessarily begin at the AAP start codon. Since each of these ribosomes must translate the AAP, they will be subject to AAP-mediated, Arg-specific ribosome stalling. Therefore, it would be predicted that the time of the first appearance of AAP-LUC fusion polypeptide would be delayed by Arg. This is what is observed (Fig. 6A).
The toeprint data (Fig. 2 and 3) indicate that translation of the wild-type AAP causes ribosomes engaged at termination codons to stall, which has parallels with other uORFs whose peptide coding sequences are important for controlling translation (2, 17, 26). In addition, the AAP also causes ribosomes engaged in elongation to stall (Fig. 7 and 8). The data indicate that stalling can occur in a short region of RNA following the AAP coding region. Previous considerations of how the movement of eukaryotic ribosomes involved in elongation is translationally controlled have focused on the physical structure of the RNA or on the limitation for charged tRNA. Secondary structures in the RNA have been implicated in the blockade of scanning 40S ribosomal subunits (12) and in the slowing of translating 80S ribosomes (23). The presence of pseudoknots or rare codons in the RNA are implicated in ribosome frameshifting (7). It has been hypothesized that rare codons affect ribosome stalling and allow time for the correct folding of nascent domains of a protein containing multiple domains (11, 13, 23).
The relative importance of different cis-acting sequences for N. crassa arg-2 AAP-mediated regulation deduced by using a cell-free translation system from Neurospora correlates with observations on CPA1 regulatory sequence effects on reporter gene expression in vivo in S. cerevisiae (5). In these studies, the activity of the enzyme encoded by the reporter β-galactosidase gene was used to measure regulation; neither the distribution of ribosomes on RNA nor the levels of RNA were measured. Therefore, it was not possible to assess the relative contributions of Arg-specific translational control and Arg-specific effects on CPA1 transcription and CPA1 transcript stability (3). Nevertheless, these observations on regulation in vivo are entirely consistent with observations on regulation in the cell-free translation system. Changing the distance between the CPA1 uORF termination codon and the downstream initiation codon did not alter Arg-specific regulation (5). When the full-length CPA1-encoded AAP was fused directly to the β-galactosidase reporter gene product, Arg-specific regulation was retained (5). These results are also consistent with the in vitro studies on N. crassa arg-2, although, in addition to the caveats listed above, a deliberately frameshifted construct was not tested in vivo to rule out the possibility that another initiation codon than that predicted was used for translation of the functional reporter.
The results presented here indicate that the movement of ribosomes involved in termination or elongation can be regulated by the nascent peptide being produced. The strong conservation of the AAP amino acid sequence among different fungi, with nucleotide differences in the coding region primarily in silent positions, and the demonstrated importance of the evolutionarily conserved peptide sequence for the regulatory function of stalling ribosomes in the Neurospora system indicate a primary role for the nascent peptide. Specific RNA sequences may also contribute to this regulatory process by other means than their capacity to encode polypeptide sequence. No evidence that identifies such sequences has as yet been obtained.
The observation that translation of the AAP can regulate the movement of ribosomes involved in elongation provides evidence for a novel form of eukaryotic translational control. In bacterial systems, there are precedents for the stalling of ribosomes involved in elongation mediated by nascent peptides (17). Nascent peptides can also have other effects on ribosomes involved in elongation: ribosome jumping in the translation of T4 gene 60 requires a 16-residue region of the nascent peptide (15). In eukaryotes, N-terminal nascent peptide domains can interact with the signal recognition particle (24), which halts ribosome movement until docking with membrane of the endoplasmic reticulum. Interestingly, recent evidence shows that the nascent signal polypeptide interacts with the ribosome before it exits the ribosome (16).
Taken together, all of the available data indicate that translation of the AAP is an evolutionarily conserved event of primary importance for regulation. AAP-mediated translational regulation represents an unusual instance in which a nascent peptide appears to regulate the movement of ribosomes.
ACKNOWLEDGMENTS
We thank Geoff Cereghino, Adam Geballe, Uttam L. RajBhandary, Alan Sachs, and Charles Yanofsky for critical reading of the manuscript.
This work was supported by the National Institutes of Health (GM47498).
REFERENCES
- 1.Baek J-M, Kenerley C M. The arg2 gene of Trichoderma virens: cloning and development of a homologous transformation system. Fungal Genet Biol. 1998;23:34–44. doi: 10.1006/fgbi.1997.1025. [DOI] [PubMed] [Google Scholar]
- 2.Cao J, Geballe A P. Ribosomal release without peptidyl tRNA hydrolysis at translation termination in a eukaryotic system. RNA. 1998;4:181–188. [PMC free article] [PubMed] [Google Scholar]
- 3.Crabeel M, LaValle R, Glansdorff N. Arginine-specific repression in Saccharomyces cerevisiae: kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol Cell Biol. 1990;10:1226–1233. doi: 10.1128/mcb.10.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis R H. Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol Rev. 1986;50:280–313. doi: 10.1128/mr.50.3.280-313.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delbecq P, Werner M, Feller A, Filipkowski R K, Messenguy F, Piérard A. A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol Cell Biol. 1994;14:2378–2390. doi: 10.1128/mcb.14.4.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edelmann S E, Staben C. A statistical analysis of sequence features within genes from Neurospora crassa. Exp Mycol. 1994;18:70–81. [Google Scholar]
- 7.Farabaugh P J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Federov A N, Baldwin T O. Protein folding and assembly in a cell-free expression system. Methods Enzymol. 1998;290:1–17. doi: 10.1016/s0076-6879(98)90003-9. [DOI] [PubMed] [Google Scholar]
- 9.Freitag M, Dighde N, Sachs M S. A UV-induced mutation that affects translational regulation of the Neurospora arg-2 gene. Genetics. 1996;142:117–127. doi: 10.1093/genetics/142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch A G. Translational regulation of yeast GCN4: a window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 11.Képès F. The “+70 pause”: hypothesis of a translational control of membrane protein assembly. J Mol Biol. 1996;262:77–86. doi: 10.1006/jmbi.1996.0500. [DOI] [PubMed] [Google Scholar]
- 12.Koloteva N, Muller P P, McCarthy J E. The position dependence of translational regulation via RNA-RNA and RNA-protein interactions in the 5′-untranslated region of eukaryotic mRNA is a function of the thermodynamic competence of 40 S ribosomes in translational initiation. J Biol Chem. 1997;272:16531–16539. doi: 10.1074/jbc.272.26.16531. [DOI] [PubMed] [Google Scholar]
- 13.Komar A A, Jaenicke R. Kinetics of translation of γ B crystallin and its circularly permutated variant in an in vitro cell-free system: possible relations to codon distribution and protein folding. FEBS Lett. 1995;376:195–198. doi: 10.1016/0014-5793(95)01275-0. [DOI] [PubMed] [Google Scholar]
- 14.Landick R, Turnbough C L J, Yanofsky C. Transcription attenuation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Maasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1263–1286. [Google Scholar]
- 15.Larsen B, Peden J, Matsufuji S, Matsufuji T, Brady K, Maldonado R, Wills N M, Fayet O, Atkins J F, Gesteland R F. Upstream stimulators for recoding. Biochem Cell Biol. 1995;73:1123–1129. doi: 10.1139/o95-121. [DOI] [PubMed] [Google Scholar]
- 16.Liao S, Lin J, Do H, Johnson A E. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 17.Lovett P S, Rogers E J. Ribosome regulation by the nascent peptide. Microbiol Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Z, Freitag M, Sachs M S. Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol Cell Biol. 1995;15:5235–5245. doi: 10.1128/mcb.15.10.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Z, Sachs M S. Role of an upstream open reading frame in mediating arginine-specific translational control in Neurospora crassa. J Bacteriol. 1996;178:2172–2177. doi: 10.1128/jb.178.8.2172-2177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orbach M J, Sachs M S, Yanofsky C. The Neurospora crassa arg-2 locus: structure and expression of the gene encoding the small subunit of arginine-specific carbamoyl phosphate synthetase. J Biol Chem. 1990;265:10981–10987. [PubMed] [Google Scholar]
- 21.Shen W-C, Ebbole D J. Cross-pathway and pathway-specific control of amino acid biosynthesis in Magnaporthe grisea. Fungal Genet Biol. 1997;21:40–49. [PubMed] [Google Scholar]
- 22.Srb A M, Horowitz N H. The ornithine cycle in Neurospora and its genetic control. J Biol Chem. 1944;154:129–139. [Google Scholar]
- 23.Thanaraj T A, Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter P, Johnson A E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Sachs M S. Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J Biol Chem. 1997;272:255–261. doi: 10.1074/jbc.272.1.255. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Sachs M S. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werner M, Feller A, Messenguy F, Piérard A. The leader peptide of yeast CPA1 is essential for the translational repression of its expression. Cell. 1987;49:805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]