Abstract
To study the regulation of AUUUA-mediated RNA deadenylation and destabilization during Xenopus early development, we microinjected chimeric mRNAs containing Xenopus or mammalian 3′ untranslated region (3′-UTR) sequences into Xenopus oocytes, mature eggs, or fertilized embryos. We found that the AU-rich elements (ARE) of Xenopus c-myc II and the human granulocyte-macrophage colony-stimulating factor gene (GMCSF) both direct deadenylation of chimeric mRNAs in an AUUUA-dependent manner. In the case of the Xenopus c-myc II ARE, mutation of a single AUUUA within an absolutely conserved 11-nucleotide region in c-myc 3′-UTRs prevents ARE-mediated deadenylation. AUUUA-specific deadenylation appears to be developmentally regulated: low deadenylation activity is observed in the oocyte, whereas rapid deadenylation occurs following egg activation or fertilization. Deadenylation results in the accumulation of stable deadenylated RNAs that become degraded only following mid-blastula transition. We conclude that ARE-mediated mRNA deadenylation can be uncoupled from ARE-mediated mRNA decay and that AUUUAs directly signal deadenylation during Xenopus early development.
mRNA destabilization is a major mechanism for the posttranscriptional regulation of gene expression. Many early-response genes, including proto-oncogenes and cytokine and lymphokine genes, are regulated by the instability of their mRNAs. The 3′ untranslated regions (3′-UTR) of these mRNAs contain AU-rich elements (AREs) that function to destabilize the mRNA in mammalian cells (7, 9, 43). These cis-acting destabilizing regions can be quite variable in sequence and length but are characterized by one or more AUUUA motifs intermittently and/or tandemly repeated within an AU-rich region (9, 56). The AUUUA motifs within the ARE are essential for destabilization; deletions or point mutations of the AUUUAs substantially increase the stability of the mRNA, whereas insertion of an AUUUA-rich ARE into the 3′-UTR of a stable mRNA dramatically shortens its half-life (t1/2) (43). Furthermore, increasing the number of tandem copies of the AUUUA motif seems to correlate with greater efficiency of ARE-mediated destabilization (10, 29, 56).
Several studies have suggested that the first step in ARE-mediated decay is mRNA deadenylation, since partially deadenylated products are the only intermediates detected preceding complete mRNA decay for several different ARE-containing mammalian messages (6, 10, 44, 54). These intermediates are short-lived, suggesting that once the mRNA is deadenylated, it is rapidly degraded. In a mammalian in vitro degradation system, an mRNA lacking a poly(A) tail is degraded much more rapidly than the same mRNA containing a poly(A) tail (4, 6). However, since none of these systems uncouple the deadenylation step from decay, it has not been possible to determine whether AREs destabilize mRNAs by promoting the single event of mRNA deadenylation (with resulting obligatory decay of the mRNA) or whether the ARE also recruits an endo- or exonuclease that initiates the degradation of the mRNA.
Many genes that are posttranscriptionally regulated by ARE-mediated decay have been shown to play important roles in cell growth and differentiation. Early development is characterized by cell growth and differentiation and is also a time when gene expression is regulated extensively by posttranscriptional mechanisms (12). Xenopus early development is an attractive system for studying regulatory phenomena because it can be carried out in vitro and occurs rapidly, allowing the sampling of several stages of development in a short period. Previous experiments designed to examine whether ARE-mediated mRNA destabilization occurs in Xenopus laevis used mammalian AREs inserted into the 3′-UTR of chimeric mRNAs; no ARE-mediated decay was observed in the oocyte (27, 28). Moreover, during Xenopus early development, the mammalian ARE was reported not to cause mRNA destabilization but rather to inhibit the translation of the mRNA containing the ARE (27, 33).
In contrast to ARE-mediated mRNA decay, variation in the level of polyadenylation has been documented as an important mechanism for posttranscriptional regulation of gene expression during Xenopus early development (53). In the oocyte nucleus, newly transcribed mRNAs acquire a poly(A) tail directed by the AAUAAA poly(A) signal (52). Once the mRNA travels to the cytoplasm, the length of its poly(A) tail may change. Some mRNAs acquire longer poly(A) tails, whereas others lose their poly(A) tails altogether (5, 17, 24). Egg maturation, which results when oocytes advance to meiosis II, is accompanied by many biochemical changes, including changes in cytoplasmic poly(A) tail length (53). Cytoplasmic lengthening of the poly(A) tail is dependent on a cytoplasmic polyadenylation element (CPE) and the AAUAAA poly(A) signal present in the 3′-UTR of the mRNA (19, 34). Cytoplasmic poly(A) tail shortening is believed to be a nonspecific default process resulting from an imbalance between polyadenylation and deadenylation (20, 48). In most cases, poly(A) tail shortening does not destabilize the mRNA prior to the mid-blastula transition (MBT) (2, 42); however, there are strong correlations between an mRNA’s poly(A) tail length and its translatability during this time (14, 21, 23, 41, 45).
To date, only one specific deadenylation signal has been characterized and shown to act on certain mRNAs during Xenopus early development. The Eg-specific deadenylation element (EDEN) directs the deadenylation of the Eg mRNAs following fertilization. Deadenylation results in the accumulation of stable poly(A)− Eg mRNAs that do not degrade until the mid-blastula stage of development (5, 39).
Our goal here was to ask whether ARE-mediated deadenylation and decay can be documented in Xenopus and, if so, to study the developmental regulation of this process. Since much evidence suggests that deadenylation is the first step of ARE-mediated decay (6, 10, 44, 54), and because poly(A)− transcripts appear to be relatively stable during early development (5, 39), our experiments were designed to detect ARE-mediated effects on mRNA deadenylation as well as stability. We injected chimeric transcripts containing an ARE with or without a poly(A) tail of defined length (100 nucleotides [nt]) into stage VI oocytes, fertilized eggs, or activated mature eggs. Since these transcripts contained neither a CPE nor the AAUAAA polyadenylation signal, AUUUA-mediated deadenylation could be monitored in the absence of polyadenylation. We demonstrate that both a Xenopus and a mammalian AUUUA-containing ARE can direct specific mRNA deadenylation during Xenopus early development. Furthermore, the deadenylation step of ARE-mediated decay is uncoupled from decay of the mRNA body.
MATERIALS AND METHODS
Plasmid construction, structure of RNA substrates, and nomenclature.
All plasmid constructs were derived from the Δ13Tb vector (22). The human β-globin open reading frame was PCR amplified from the pSPkβc vector (46) by using a 5′ primer flanked by an NcoI site (5′hβG) and a 3′ primer flanked by a BglII site followed by an XbaI site (3′hβG). This PCR product was then digested with NcoI and XbaI and inserted into the NcoI/XbaI site of the Δ13Tb vector to replace the cyclin coding region with the coding region of human β-globin. The resulting Δ13Tb#2 construct has a T7 promoter upstream of the Xenopus β-globin 5′-UTR (already present in the vector) followed by the human β-globin coding region. The template for a poly(A) tail of defined length (100 nt) was made by annealing a poly(dA)100 oligonucleotide (5′pA100) to a poly(dT)100 oligonucleotide (3′pA100). The oligonucleotides were flanked by a 5′ XbaI site and a 3′ SacI site to allow insertion of these annealed oligonucleotides into the XbaI/SacI site of Δ13Tb#2; the resulting construct was named Δ13Tb#21-2. All of the constructs used for in vitro transcription were created by inserting the 3′-UTR of choice into the BglII/XbaI site of Δ13Tb#21-2. This insertion places the 3′-UTR between the BglII site following the β-globin stop codon and the XbaI site preceding the poly(A)100 tail. Transcripts initiated at the T7 promoter therefore contain a Xenopus β-globin 5′-UTR (66 nt), a human β-globin coding region (450 nt), a 3′-UTR of choice, and an encoded poly(A)100 tail.
Transcription templates.
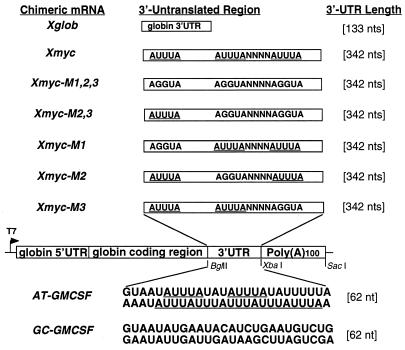
Transcription templates are diagrammed in Fig. 1.
FIG. 1.
Constructs used to make T7 transcripts for microinjection. Plasmids and corresponding chimeric transcripts are named according to the 3′-UTR they contain. All in vitro-transcribed chimeric mRNAs contain the same Xenopus globin 5′-UTR (66 nt) and human β-globin coding region (450 nt); only the 3′-UTR cloned into the BglII/XbaI site of the Δ13Tb#21-2 vector differs as indicated. T7 transcription from an XbaI-cut plasmid yields poly(A)− chimeric transcripts, whereas T7 transcription from a SacI-cut plasmid yields the corresponding poly(A)+ chimeric transcripts with a 100-nt poly(A) tail. All in vitro-transcribed chimeric RNAs were injected in 5′ GpppG capped form.
(i) Xmyc.
The first 342 bp of Xenopus c-myc II (which has been reported to be maternally expressed) (51) 3′-UTR was PCR amplified from genomic Xenopus DNA with a 5′ primer flanked by a BglII site (5′XmycUTR) and a 3′ primer flanked by an XbaI site (3′XmycUTR). The PCR product was cut with BglII/XbaI and cloned into the BglII/XbaI site of ΔTb#21-2.
(ii) Xmyc-M1, Xmyc-M2, Xmyc-M3, Xmyc-M2,3, and Xmyc-M1,2,3.
PCR-based mutagenesis was used to mutate one, two, or all three ATTTA sequences in the 3′-UTR of Xmyc to AGGTA, and these PCR products were flanked by BglII and XbaI sites to be cloned into the BglII/XbaI site of Δ13Tb#21-2. The Xmyc-M1 3′-UTR was PCR amplified by using the 5′ primer 5′Xmyc-M1 and the 3′ primer 3′XmycUTR. The Xmyc-M2 3′-UTR was PCR amplified by using the 5′ primer 5′XmycUTR and the 3′ primer 3′Xmyc-M2, followed by PCR with the 3′ primer 3′XmycUTR. The Xmyc-M3 3′-UTR was PCR amplified by using the 5′ primer 5′XmycUTR and the 3′ primer 3′Xmyc-M3, followed by PCR with the 3′ primer 3′XmycUTR. The Xmyc-M2,3 3′-UTR was PCR amplified by using the 5′ primer 5′XmycUTR and the 3′ primer Xmyc-M2,3, followed by PCR with the 3′ primer 3′XmycUTR. The Xmyc-M1,2,3 3′-UTR was PCR amplified by using the 5′ primer 5′Xmyc-M1 and the 3′ primer Xmyc-M2,3, followed by PCR with the 3′ primer 3′XmycUTR.
(iii) AT-GMCSF.
The AT–granulocyte-macrophage colony-stimulating factor (GMCSF) 3′-UTR sequence was PCR amplified from the pRβGAT vector (43) with a 5′ primer flanked by a BglII site (5′AT-62) and a 3′ primer flanked by an XbaI site (3′AT-62), cut with BglII/XbaI, and cloned into the BglII/XbaI site of Δ13Tb#21-2.
(iv) GC-GMCSF.
The GC-GMCSF 3′-UTR sequence was made by annealing complementary oligonucleotides 5′GC-GMCSF and 3′GC-GMCSF. The annealed fragment was cut with BglII/XbaI and cloned into the BglII/XbaI site of Δ13Tb#21-2.
(v) Xglob.
The Xenopus β-globin 3′-UTR was PCR amplified from the Δ13Tb vector with a 5′ primer flanked by a BglII site (5′XglobUTR) and a 3′ primer flanked by an XbaI site (3′XglobUTR), cut with BglII/XbaI, and inserted into the BglII/XbaI site of Δ13Tb#21-2.
Preparation of RNA substrates, microinjections, and RNA analysis.
Internally labelled and 5′-capped transcripts were synthesized in the presence of GpppG (Pharmacia) with a specific activity of 105 cpm/ng by using [α-32P]UTP (Amersham).
Poly(A)− transcripts were made by digesting the plasmid with XbaI prior to transcription by T7 polymerase. Poly(A)100+ transcripts were made by digesting the plasmid with SacI prior to T7 transcription. Stage VI oocytes were incubated in 1× modified Barth’s saline [80 mM NaCl, 1 mM KCl, 0.8 mM MgSO4(H2O)7, 8 mM NaHCO3, 1 mM HEPES (pH 7.4), 0.7 mM CaCl2] and cytoplasmically injected with 18 nl (20 to 40 pg) of RNA. Eggs, embryos, and activated eggs were obtained by standard procedures (26). Eggs and embryos were incubated in 4% Ficoll in 1/3 MMR (33 mM NaCl, 0.6 mM KCl, 0.3 mM MgCl2, 0.66 mM CaCl2, 1.7 mM Na-HEPES [pH 7.8]) solution prior to cytoplasmic injection with 18 nl (20 to 40 pg) of RNA. Embryos were transferred to 1/3 Marc’s modified Ringer’s solution (MMR) 4 h following fertilization. Oocytes, eggs, or embryos were collected at indicated time points (10 per time point) and frozen at −80°C preceding RNA extraction. RNA was extracted in proteinase K-sodium dodecyl sulfate (SDS) buffer (50 mM NaCl, 50 mM Tris-Cl [pH 7.5], 5 mM EDTA, 0.5% SDS, 0.2 mg of proteinase K/ml) at 50°C followed by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation and was analyzed by 6% denaturing polyacrylamide gel electrophoresis (PAGE).
Poly(A) tail analysis.
Fertilized eggs were injected with 5′-capped and internally labelled ([α32P]ATP) poly(A)+ transcripts (20 to 40 pg). Eggs were collected following injections at 0 and 8 h (for Xmyc) and 0 and 4 h (for AT-GMCSF). RNA was isolated from 20 eggs per time point, and radioactivity was determined by scintillation counting. The equivalent of 5,000 cpm of isolated RNA was subjected to RNase A digestion in a total volume of 10 μl containing 10−4 U of RNase A and 1 μg of tRNA and was incubated for 15 min at 37°C. Uninjected poly(A)+ and poly(A)− transcripts were subjected to the same RNase A treatment for comparison. RNase A digestion products were fractionated by 10% denaturing PAGE, and the resistant poly(A) and several large products (see Fig. 2C and 4C) were quantitated by PhosphorImager analysis to determine the relative levels of the poly(A) tail and the mRNA body.
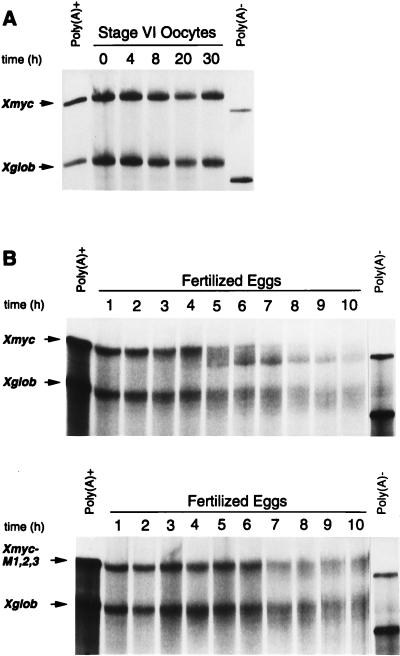
FIG. 2.
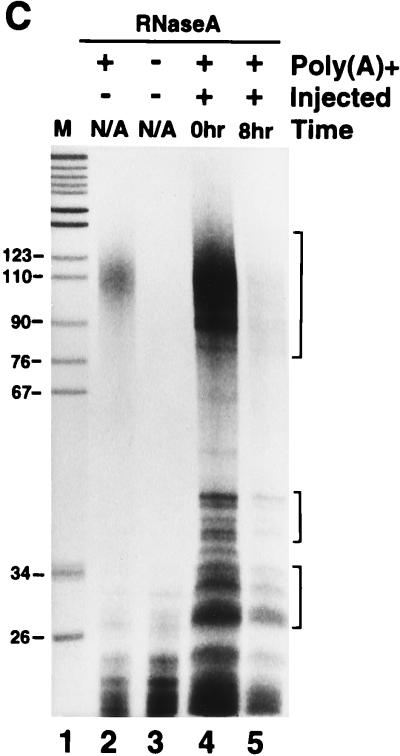
The AU-rich region from the Xenopus c-myc II 3′-UTR directs RNA deadenylation. (A) Poly(A)+ Xmyc (958 nt) and Xglob (749 nt) transcripts were cytoplasmically coinjected into stage VI oocytes. RNA was isolated at the indicated times (in hours) following injection. (B) Poly(A)+ Xmyc and Xglob transcripts (top) or poly(A)+ Xmyc-M1,2,3 and Xglob transcripts (bottom) were coinjected cytoplasmically into fertilized eggs immediately following fertilization. RNA was isolated at the indicated times (in hours) following fertilization. (C) RNase A treatment was used to determine whether the shortening of the Xmyc transcript in fertilized eggs (as seen in panel B, top) was due to poly(A) tail loss. Lane 1, pBR322 MspI DNA marker; lane 2, uninjected [32P]ATP-labelled poly(A)+ Xmyc transcript treated with RNase A; lane 3, uninjected [32P]ATP-labelled poly(A)− Xmyc transcript treated with RNase A; lane 4, 5,000 cpm of RNA isolated from fertilized eggs 0 h after injection with [32P]ATP-labelled poly(A)+ Xmyc transcript treated with RNase A; lane 5, 5,000 cpm of RNA isolated from fertilized eggs 8 h after injection with [32P]ATP-labelled poly(A)+ Xmyc transcript treated with RNase A. Quantitative analysis by PhosphorImager of the distribution of counts per minute in lane 5 relative to lane 4 revealed that incubation for 8 h resulted in a 60 to 70% reduction of the signal for the poly(A) tail relative to common background RNase A digestion products (indicated by brackets). In panels A and B, each lane contains RNA isolated from the equivalent of two injected oocytes or embryos.
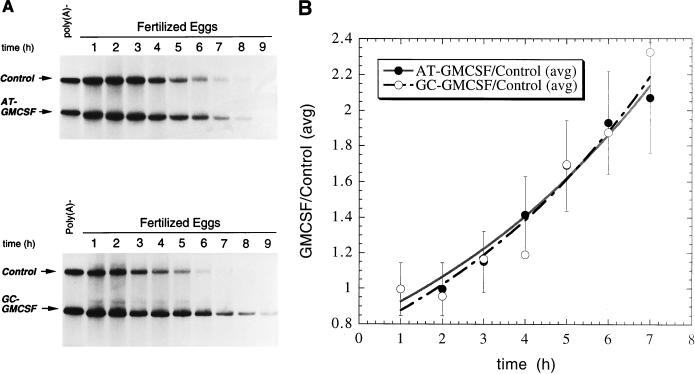
FIG. 4.
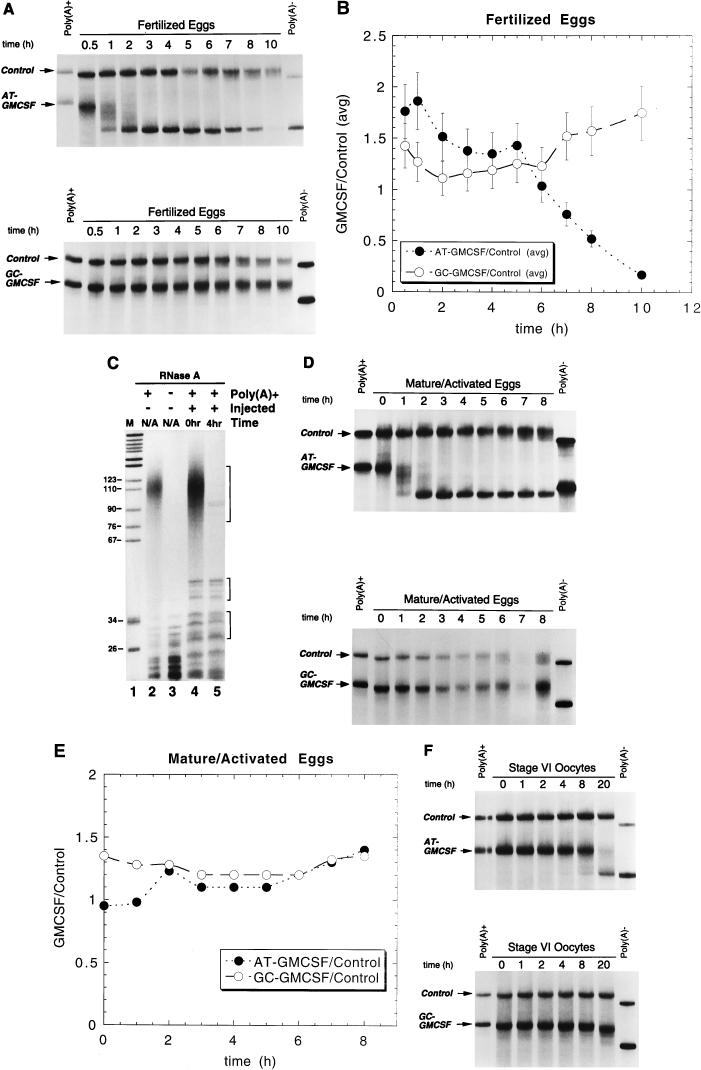
The human GMCSF ARE directs mRNA deadenylation during Xenopus early development. (A) Poly(A)+ AT-GMCSF (top) or GC-GMCSF (bottom) was cytoplasmically coinjected along with a poly(A)+ control mRNA (Xmyc-M2) into eggs immediately following fertilization. RNA was isolated at the indicated times (in hours) following fertilization. (Note: the apparent increase in size of GC-GMCSF is not reproducible.) (B) To determine the relative decay rates for AT-GMCSF and GC-GMCSF in fertilized eggs, the data from panel A and from an equivalent experiment were quantitated by PhosphorImager analysis and averaged (error bars reflect maximum deviations from the averages). The amount of GMCSF transcript remaining divided by the amount of coinjected control remaining for each time point was plotted as a function of time. (C) RNase A treatment was used to determine whether the shortening of the AT-GMCSF transcript in fertilized eggs (as seen in panel A, top) was due to poly(A) tail loss. Lane 1, pBR322 MspI DNA marker; lane 2, uninjected [32P]ATP-labelled poly(A)+ AT-GMCSF mRNA treated with RNase A; lane 3, uninjected [32P]ATP-labelled poly(A)− AT-GMCSF mRNA treated with RNase A; lane 4, RNA (5,000 cpm) isolated from fertilized eggs 0 h after injection with [32P]ATP-labelled poly(A)+ AT-GMCSF mRNA treated with RNase A; lane 5, RNA (5,000 cpm) isolated from fertilized eggs 4 h after injection with [32P]ATP-labelled poly(A)+ AT-GMCSF mRNA treated with RNase A. Quantitative analysis by PhosphorImager of the distribution of counts per minute in lane 5 relative to lane 4 revealed that incubation for 4 h resulted in a >90% reduction of the signal for the poly(A) tail relative to common background RNase A digestion products (indicated by brackets). (D) Poly(A)+ AT-GMCSF (top) or GC-GMCSF (bottom) was cytoplasmically coinjected along with a poly(A)+ control mRNA (Xmyc-M2) into activated mature eggs. RNAs were isolated at the indicated times (in hours) following injection. (E) To determine the relative decay rates for AT-GMCSF and GC-GMCSF in activated mature eggs, the data from panel D were quantitated by PhosphorImager analysis. The amount of GMCSF transcript remaining divided by the amount of coinjected control remaining for each time point was plotted as a function of time. (F) Poly(A)+ AT-GMCSF (top) or poly(A)+ GC-GMCSF (bottom) was cytoplasmically coinjected along with a control mRNA (Xmyc-M2) into stage VI oocytes. RNAs were isolated at the indicated times (in hours) following injection. In panels A, D, and F, each lane contains RNA isolated from the equivalent of two injected oocytes, eggs, or embryos.
Oligonucleotides and primers.
The sequences of the oligonucleotides and primers used in this study are given in parentheses after their designations as follows: 5′hBG (5′-GACACCATGGTGCACCTG-3′), 3′hBG (5′-GCTCTAGAGCAGATCTTAGTGATACTTGTGGGCCAGG-3′), 5′pA100 [5′-GCTCTAGAA(A)96AAGAGCTCCCC-3′], 3′pA100 [5′-GGGGAGCTC(T)99TCTAGAGC-3′], 5′XmycUTR (5′-GAAGATCTTCACAAACTCTTATT-3′), 3′mycUTR (5′-GCTCTAGACAGAGCCAAACTTCAGG-3′), 5′Xmyc-M1 (5′-GAAGATCTTCACAAACTCTTAGGTAACACTTTA-3′), 3′Xmyc-M2 (5′-TAAATGTTTTACCTACAAAGGTTG-3′), 3′Xmyc-M3 (5′-AAGAAAAATACCTGTTTTAAATAC-3′), 3′Xmyc-M2,3 (5′-GTTTATAAGAAAAATACCTGTTTTACCTACAAAAGGTTG-3′), 5′AT-62 (5′-GAAGATCTGATCAGTAATATTTATAT-3′), 3′AT-62 (5′GCTCTAGATGCTTAAATAAATAAA-3′), 5′-GC-GMCSF (5′-GAAGATCTGATCAGTAATATGAATACATCTGAATGTCTGGAATA TTGATTGATAAGCTTAGTCGACCATCTAGAGC-3′), 3′GC-GMCSF (5′GCTCTAGATGGTCGACTAAGCTTATCAATCAATATTCCAGACATTCAGATGTATTCATATTACTGATCAGATCTTC-3′), 5′XglobUTR (5′-GAAGATCTAACCAGCCTCAAG-3′), and 3′XglobUTR (5′-GCTCTAGAGCAGATACGAATGGC-3′).
RESULTS
The AU-rich portion of the Xenopus c-myc II 3′-UTR directs mRNA deadenylation.
The Xenopus c-myc II 3′-UTR was used to test whether a Xenopus AUUUA-rich sequence could direct deadenylation or decay of a chimeric mRNA during Xenopus early development. The c-myc 3′-UTR was chosen because mammalian c-myc mRNA is subject to efficient ARE-mediated decay (6, 10, 25). We focused on the first 342 residues of the Xenopus c-myc II 3′-UTR because this region is AU-rich and contains three AUUUA sequences (the total 3′-UTR is 950 nt). The construct Xmyc, used to generate RNAs for injection into stage VI oocytes and mature or fertilized eggs, is diagrammed in Fig. 1. In vitro transcription from the T7 promoter of the SacI-cut Xmyc vector yields chimeric mRNAs containing a Xenopus globin 5′-UTR, a human globin coding region followed by the first 342 nt of the Xenopus c-myc II 3′-UTR, and a poly(A) tail of 100 nt. The coinjected Xglob mRNA control is identical to Xmyc except that the Xenopus c-myc II 3′-UTR sequences are replaced with the Xenopus globin 3′-UTR, up to but not including the poly(A) signal (Fig. 1). The in vitro-transcribed RNAs contain neither a CPE nor the AAUAAA polyadenylation signal; thus, transcripts should not be subject to poly(A) tail lengthening.
When coinjected into the cytoplasm of stage VI oocytes, both the Xmyc and control Xglob transcript were stable in their polyadenylated forms up to 30 h (Fig. 2A). In oocytes induced to mature in vitro by incubation with progesterone, both transcripts were deadenylated nonspecifically at a similar rate (data not shown). This nonspecific deadenylation activity in mature eggs has been previously described as the default deadenylation of transcripts lacking a CPE or a polyadenylation signal (20, 48).
When the same two polyadenylated transcripts were microinjected into fertilized eggs, Xmyc was shortened to the length of the deadenylated form of the transcript by 7 h (Fig. 2B, top panel). The coinjected control Xglob remained polyadenylated. To confirm that the shortening of the Xmyc transcript was the result of deadenylation, [32P]ATP-labelled polyadenylated transcripts were injected into fertilized eggs; RNA was isolated at 0 and 8 h following injection and subjected to RNase A treatment (Fig. 2C, lanes 4 and 5). RNase A cleaves 3′ of pyrimidine residues and therefore leaves the poly(A)100 tail intact. The shortened Xmyc transcript recovered after 8 h gave an RNase A cleavage pattern similar to that of the Xmyc transcript synthesized without a poly(A) tail (Fig. 2C; compare lanes 3 and 5). Quantitation of poly(A) tail loss was obtained by measuring the counts in the poly(A) digestion product relative to those in the large digestion products from the mRNA body (Fig. 2C). This quantitation demonstrated a 60 to 70% reduction of the poly(A) tail relative to the mRNA body at 0 h versus 8 h following injection into fertilized eggs. Together, these results indicate that the first 342 nt of the Xenopus c-myc II 3′-UTR directs deadenylation of the chimeric transcript.
To determine whether the AUUUA sequences in the subcloned region of the Xenopus c-myc II 3′-UTR are required for deadenylation, all three AUUUA sequences were mutated to AGGUA, creating construct Xmyc-M1,2,3 (Fig. 1). Similar AUUUA→AGGUA mutations in the 3′-UTR of mammalian mRNAs have been shown previously to abolish the destabilizing effect conferred by AREs (43, 56). When microinjected into fertilized eggs, the Xmyc-M1,2,3 transcript was not deadenylated as rapidly as Xmyc, and no prominent deadenylated band appeared (Fig. 2B). Thus, the first 342 nt of the Xenopus c-myc 3′-UTR are capable of directing enhanced deadenylation of an mRNA in an AUUUA-dependent manner in the fertilized Xenopus egg but not in the oocyte.
A highly conserved sequence in the c-myc 3′-UTR is required for deadenylation.
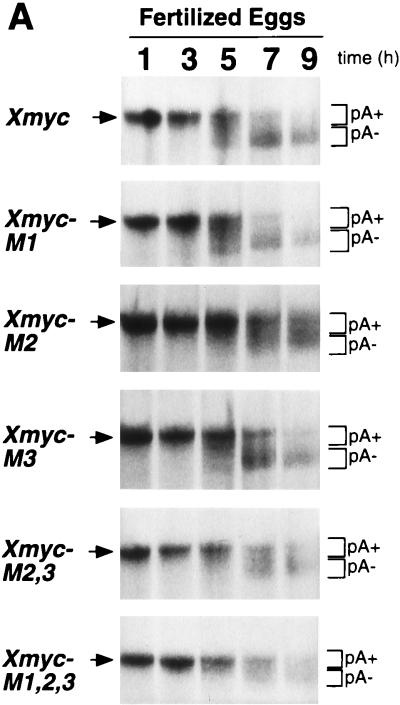
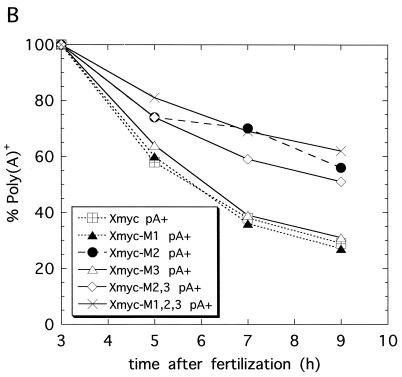
Once we had identified the stage at which AUUUA-specific deadenylation activity appears and had observed that mutation of all three AUUUAs in the Xmyc 3′-UTR to AGGUA prevented this ARE sequence from enhancing mRNA deadenylation, we investigated which of the AUUUA motifs is required for enhanced deadenylation. One or two of the three AUUUAs were mutated to AGGUA (Fig. 1), and the rates of deadenylation of these mutant transcripts were compared with those of Xmyc and Xmyc-M1,2,3 in fertilized eggs (Fig. 3A and B). The data are plotted as percentages of total mRNA remaining polyadenylated versus time in Fig. 3B.
FIG. 3.
A highly conserved sequence in the c-myc 3′-UTR is required for deadenylation. (A) Poly(A)+ (pA+) Xmyc, Xmyc-M1, Xmyc-M2, Xmyc-M3, Xmyc-M2,3, and Xmyc-M1,2,3 mRNAs were each injected cytoplasmically into fertilized eggs immediately following fertilization. RNA was isolated at the times (in hours) indicated, and RNA from two eggs per time point was fractionated on denaturing 6% polyacrylamide gels. (B) Regions indicated by brackets in panel A were quantitated by PhosphorImager analysis and graphed as the percentage of total mRNA transcript remaining polyadenylated {[pA+]/[(pA+) + (pA−)]} versus time to determine the rate of deadenylation for Xmyc and each of the Xmyc mutants. The experiment for which results are shown in panel A was repeated and yielded a similar result. (C) An absolutely conserved 11-nt sequence is present within the 3′-UTR of all known c-myc mRNAs; this conserved sequence includes the AUUUA that is mutated to AGGUA in Xmyc-M2. The locations of the conserved sequences from the start of the 3′-UTR and the total lengths of the 3′-UTR up to but not including the poly(A) signals are as follows: for humans, nt 209 to 219 of 294 nt; for pigs, nt 287 to 297 of 371 nt; for cats, nt 277 to 287 of 362 nt; for mice, nt 231 to 241 of 314 nt; for chickens, nt 283 to 293 of 381 nt; for Xenopus c-myc I, nt 266 to 276 of 950 nt; for Xenopus c-myc II, nt 277 to 287 of 970 nt; and for zebrafish, nt 189 to 199 of 221 nt.
The Xmyc mutants fall into two categories (Fig. 3B). Mutants Xmyc-M2, Xmyc-M2,3, and Xmyc-M1,2,3 all have very slow deadenylation rates, which correspond to the background rate exhibited by the control Xglob transcript (see Fig. 2B). In contrast, mutants Xmyc-M1 and Xmyc-M3 exhibit the same time course of deadenylation as the original Xmyc transcript. The Xmyc-M2 mutant has only a single AUUUA motif mutated to AGGUA; this simple disruption is sufficient to abolish the enhancement of deadenylation conferred by the first 342 nt of the Xmyc II 3′-UTR on the chimeric mRNA. These results suggest that the middle AUUUA of the subcloned portion of Xenopus c-myc II 3′-UTR is instrumental in signaling transcript deadenylation in fertilized eggs. Strikingly, this AUUUA is contained in an 11-nt sequence (Fig. 3C) that is one of three short sequences entirely conserved among the 3′-UTR of all known c-myc homologs (50).
The mammalian GMCSF ARE directs mRNA deadenylation during Xenopus early development.
We next tested whether a mammalian ARE could likewise direct mRNA deadenylation during Xenopus early development. The well-characterized 62-nt mammalian GMCSF ARE contains seven AUUUA sequences and is highly active in signaling ARE-mediated decay in mammalian cells (43). We subcloned this 62-nt sequence to generate the AT-GMCSF vector (Fig. 1). As with the Xmyc vector, in vitro transcription of the SacI-cut AT-GMCSF vector produces chimeric mRNA transcripts with a Xenopus globin 5′-UTR, a human globin coding region, the 62-nt GMCSF ARE 3′-UTR, and a poly(A)100 tail (Fig. 1). In experiments analogous to those presented in Fig. 3, the ability of the mammalian GMCSF ARE sequence to direct deadenylation of the chimeric mRNA was assessed during Xenopus early development (Fig. 4).
We first asked whether AT-GMCSF transcripts are specifically deadenylated in the fertilized egg, the earliest stage at which AUUUA-specific deadenylation of the Xmyc transcript was observed (Fig. 3). The AT-GMCSF RNA was coinjected with the control Xmyc-M2 RNA into fertilized eggs. Figure 4A shows that the AT-GMCSF transcript was deadenylated very rapidly (t1/2 ≈30 min). In contrast, a transcript in which all seven AUUUAs in the GMCSF 3′-UTR had been mutated, GC-GMCSF (Fig. 1), was not deadenylated (Fig. 4A). The rate of deadenylation for the AT-GMCSF transcript was much more rapid (t1/2 ≈30 min) than the rate of degradation (t1/2 ≈5 h) for the Xmyc transcript (Fig. 2B) and is likely to reflect the presence of multiple overlapping AUUUA sequences in its ARE (8, 56).
To determine the effect of deadenylation on transcript stability following fertilization, we quantitated the data from Fig. 4A along with data from an equivalent experiment and graphed the stabilities of the AT-GMCSF and GC-GMCSF transcripts relative to the coinjected control as a function of time in Fig. 4B. After deadenylation, the deadenylated AT-GMCSF transcript is relatively stable for 6 to 7 h following fertilization (t1/2 = 5 h). After this time, which corresponds to the MBT, the deadenylated transcripts decay with a t1/2 of ≈45 min (Fig. 4B). In contrast, the mutant GC-GMCSF and Xmyc-M2 transcripts remain relatively stable and polyadenylated even following MBT, maintaining a consistent decay rate pre- and post-MBT (Fig. 4A and B). Therefore, the AUUUA-rich 3′-UTR appears to destabilize the mRNA at MBT by promoting prior poly(A) tail loss (Fig. 4B). As in the case of Xmyc (Fig. 2C), quantitation of RNase A products demonstrated that the change in size of the AT-GMCSF transcript results from loss of its poly(A) tail (Fig. 4C).
Because activation of mature eggs results in many of the same biochemical changes that occur in fertilized eggs (31, 38), we next tested whether the AT-GMCSF reporter mRNA also becomes deadenylated in activated mature eggs. When microinjected into the cytoplasm of activated mature eggs, the AT-GMCSF transcript was deadenylated with a t1/2 of ≈30 min, leading to the accumulation of a completely deadenylated transcript within 2 h of injection (Fig. 4D, top). The mutant GC-GMCSF transcript was not deadenylated (Fig. 4D, bottom). Quantitative analysis (Fig. 4E) of the data in Fig. 4D demonstrates that AUUUA-directed deadenylation does not alter the transcript stability relative to that of the coinjected Xmyc-M2 control or compared to the relative stability of the mutant GC-GMCSF RNAs, which retain their poly(A) tails. In contrast to the decay that is seen starting at 7 h in the fertilized eggs, all transcripts with or without a poly(A) tail remain stable in activated mature eggs; unfortunately, the experiment could not be carried longer because the eggs begin to die. These data demonstrate that AUUUA-mediated mRNA deadenylation can be uncoupled from the degradation of the rest of the RNA in activated mature eggs. The analogous experiment was done with Xmyc, and it too experienced deadenylation in an AUUUA-dependent manner in the activated mature egg (data not shown).
Finally, we examined the effect of the human GMCSF ARE when the chimeric transcript was cytoplasmically injected into stage VI oocytes. In the oocyte, the AT-GMCSF transcript was deadenylated with a t1/2 of ≈8 h and appeared completely deadenylated after 20 h (Fig. 4F, top). Deadenylation did not have a significant effect on the stability of the AT-GMCSF transcript when it was quantitated relative to the internal polyadenylated control. The mutant GC-GMCSF transcript remained stable and poly(A)+ for up to 20 h following injection (Fig. 4F, bottom). Thus, the rate of deadenylation for the AT-GMCSF transcript in the stage VI oocyte is approximately 20 to 30 times lower than that in activated mature eggs or fertilized eggs.
Since the AT-GMCSF transcript that undergoes deadenylation in Fig. 4F was injected into the cytoplasmic compartment of the stage VI oocyte, it seemed likely that the AUUUA-specific deadenylation activity resides in the cytoplasm. To confirm that deadenylation is a cytoplasmic process, oocytes were enucleated and the resulting cytoplasms were injected with the AT-GMCSF transcript. Deadenylation in the cytoplasm alone occurred at a rate similar to that in the whole oocyte (data not shown). Although this result demonstrates that AUUUA-directed deadenylation activity is present in the oocyte cytoplasm, it does not rule out the possibility that deadenylation also takes place in the nucleus.
Poly(A)− mRNA is not destabilized by an ARE during Xenopus early development.
When polyadenylated AT-GMCSF transcripts were injected into the fertilized egg, they were rapidly deadenylated, but deadenylation appeared to be uncoupled from decay until the embryo reached MBT (Fig. 4B). We therefore asked whether this pattern would be reproduced if poly(A)− AT-GMCSF transcripts were injected into the fertilized egg. Poly(A)− AT-GMCSF and GC-GMCSF transcripts were made by in vitro transcription of the XbaI-cut plasmids (Fig. 1) and coinjected with a poly(A)− control Xmyc-M2 RNA into the fertilized egg (Fig. 5A).
FIG. 5.
Poly(A)− mRNA is not destabilized by an ARE during Xenopus early development. (A) Poly(A)− AT-GMCSF (top) or GC-GMCSF (bottom), along with a poly(A)− control mRNA (Xmyc-M2), was cytoplasmically coinjected into fertilized eggs immediately following fertilization. RNAs were isolated at the indicated times (in hours) following fertilization; each lane contains RNA from two eggs. (B) To determine the relative decay rates for poly(A)− AT-GMCSF and GC-GMCSF in fertilized eggs, the data from panel A and from an equivalent experiment were quantitated by PhosphorImager analysis and averaged (error bars reflect maximum deviations from the averages). The amount of GMCSF transcript remaining divided by the amount of coinjected control Xmyc-M2 remaining was plotted as a function of time.
The relative decay rates for the poly(A)− transcripts in the fertilized egg were determined by quantitative analysis (Fig. 5B) of the data in Fig. 5A and an equivalent experiment. The AT-GMCSF transcript was relatively stable in poly(A)− form until MBT, with a t1/2 of ≈5 h (Fig. 5A). The AT-GMCSF and GC-GMCSF transcripts exhibited similar stabilities in poly(A)− form and were approximately twice as stable as the coinjected Xmyc-M2 RNA in the pre-MBT embryo (Fig. 5B). Together, these data confirm that once the transcript is deadenylated, the wild-type ARE does not further destabilize the mRNA in the pre-MBT embryo.
DISCUSSION
Role of the ARE in Xenopus early development.
We examined the regulation of ARE-mediated mRNA decay during Xenopus early development by injecting reporter transcripts into the Xenopus oocyte, activated mature egg, and fertilized embryo and measuring the effect conferred by the ARE on transcript deadenylation and/or decay. Whereas AREs had previously been assigned a role in controlling translation during Xenopus early development (27), we show here a direct involvement in deadenylation.
We found that after fertilization both the Xenopus c-myc II ARE and the mammalian GMCSF ARE signal the deadenylation of reporter transcripts in an AUUUA-dependent manner. The Xenopus c-myc II AUUUA-containing ARE directs deadenylation in the fertilized or activated mature egg but has no noticeable effect in the stage VI oocyte (Fig. 2). The human GMCSF ARE is a more potent deadenylation signal: it directs mRNA deadenylation in the stage VI oocyte, activated mature egg, and fertilized egg (Fig. 4). The potency of the human GMCSF ARE in directing deadenylation compared to that of the Xmyc ARE is consistent with conclusions drawn from studies in mammalian systems, where more tandem copies of the AUUUA motif correlate with more-efficient ARE-mediated deadenylation and decay (56).
Interestingly, the rate of ARE-mediated deadenylation changes markedly during development. In the fertilized egg and the activated mature egg, the GMCSF ARE directs rapid deadenylation with a t1/2 of ≈20 to 30 min, whereas in the stage VI oocyte the t1/2 is >8 h (≈20 to 30 times slower). We suspect that the c-myc ARE might also direct deadenylation, but very weakly, in the stage VI oocyte; since the t1/2 for deadenylation of transcripts containing this ARE in fertilized eggs is between 4 and 5 h and activity in the oocyte is approximately 20 to 30 times lower, it should take 4 to 5 days for c-myc constructs to be noticeably deadenylated in the oocyte.
The stability of ARE-containing mRNAs also appears to be regulated during Xenopus early development. There is evidence from several organisms that most mRNAs are relatively stable in poly(A)− form prior to the MBT and that they then decay rapidly, while poly(A)+ transcripts are relatively stable following MBT (2, 42). We used the potent ARE from GMCSF to ask whether the deadenylation of transcripts carrying this sequence causes them to become destabilized during early development (Fig. 4B and E). We observed that despite its rapid deadenylation, the GMCSF transcript was not degraded in the stage VI oocyte, activated mature egg, or pre-MBT embryo (Fig. 4). Following MBT, the AT-GMCSF transcript is degraded, presumably because it has been deadenylated. We also tested whether the ARE destabilizes a transcript already in poly(A)− form relative to the same transcript with a mutant ARE (Fig. 5). The wild-type poly(A)− AT-GMCSF transcript was as stable as the mutant poly(A)− GC-GMCSF transcript relative to the coinjected control in pre-MBT cells; post-MBT, all transcripts lacking poly(A) disappear rapidly (Fig. 5). This result demonstrates that the wild-type ARE does not destabilize the poly(A)− form of the transcript pre-MBT. We conclude that AUUUA-rich AREs can function as deadenylation elements uncoupled from transcript decay during early development and suggest that these AREs destabilize mRNAs in post-MBT cells by a mechanism that involves prior loss of the poly(A) tail.
Previous experiments with Xenopus oocytes intriguingly demonstrated that placing a mammalian ARE on a chimeric mRNA resulted in translation inhibition (27). In the fertilized egg, it was found that AREs could inhibit translation and that this effect was independent of whether the transcript was injected in polyadenylated form (33). Our results suggest that the ARE may influence translation of the polyadenylated form of an mRNA at least partially by causing its deadenylation. We have replaced the globin coding region in AT-GMCSF with chloramphenicol acetyltransferase cDNA and have observed that following injection into fertilized eggs, the message is initially translated at the same rate as that for the corresponding GC-GMCSF mutant, but translation then ceases as the AT-GMCSF transcript becomes deadenylated (data not shown). This correlates with previous observations that polyadenylated mRNAs are much more efficiently translated in the oocyte than deadenylated transcripts (15, 21, 23). Conversely, under certain conditions, inhibition of translation can increase the deadenylation rate of an mRNA (35, 37), making it possible that AREs enhance the rate of deadenylation and subsequent mRNA decay by causing inhibition of translation.
Conservation of signals and mechanism of mRNA decay.
We found that the 3′-UTR from the Xenopus c-myc II transcript could direct stage-specific deadenylation during Xenopus early development (Fig. 2). To determine whether the AUUUAs present in the 3′-UTR were important for deadenylation, the AUUUAs were mutated individually and in combination and the effects on deadenylation were measured (Fig. 3A and B). This analysis revealed that the mutation of only one of the AUUUAs disrupted deadenylation directed by the 3′-UTR sequence. This AUUUA resides within an 11-nt sequence (Fig. 3C) previously identified as one of three short sequences (10 to 12 nt long) conserved between Xenopus and mammalian c-myc (50). We propose that this sequence may participate in regulation of the length of the poly(A) tail of endogenous maternally derived c-myc I and c-myc II (51) mRNAs during Xenopus early development; c-myc I mRNA has been reported to be present as a mixed population of polyadenylated and deadenylated forms in the stage VI oocyte and appears completely deadenylated in the mature and the fertilized egg (47). Interestingly, mutation of the conserved AUUUA sequence within the 3′-UTR of mammalian c-myc results in the relocalization of chimeric mRNAs in mammalian fibroblasts (49). Thus, either the localization of the mRNA may have an effect on its deadenylation or deadenylation may cause a change in mRNA localization.
In yeast, deadenylation precedes the degradation of both stable and unstable mRNAs and is often the rate-limiting step of mRNA decay; transcripts that are deadenylated more rapidly tend to decay more rapidly (13). Following deadenylation, the mRNA is decapped and rapidly degraded by a 5′→3′ exonuclease activity (3, 36), although mRNAs are susceptible to a common 3′→5′ degradation machinery as well (1). In mammalian cells, deadenylation is also correlated with mRNA decapping (11). Decapping, in turn, is influenced by the methylation status of the 5′ cap structure (30). Although the experiments on deadenylation and stability in Fig. 2 through 5 were performed by using transcripts initiated with a nonmethylated 5′ cap, we have confirmed that m7GpppG-capped transcripts behave identically (data not shown). Therefore, one explanation for our results is that prior to MBT, some common component of the deadenylation-dependent mRNA decay machinery that is required for the destabilization of ARE-containing transcripts is absent. The missing component is likely to be a protein factor, since inhibition of translation at the blastula stage has been reported to prevent several deadenylated messages from being degraded even after MBT (16). Perhaps a component of the decapping or the 5′→3′ exonuclease activity is absent prior to the mid-blastula stage of Xenopus early development, allowing deadenylated ARE-containing mRNAs to be relatively stable during this time.
How AREs direct deadenylation and decay still remains unclear. As yet, no factors have been identified as causing deadenylation or decay. In contrast, several labs have presented evidence that overexpression of HuR, a human protein with specificity for ARE sequences, can stabilize ARE-containing mRNAs (18, 32, 40). ElrA, the Xenopus homolog of HuR, is present throughout Xenopus early development (55) and could potentially bind and stabilize ARE-containing transcripts during this time. An alternative hypothesis is that ElrA may play a role in directing deadenylation of ARE-containing mRNAs. This hypothesis is supported by the identification of a highly related factor, embryo deadenylation element-binding protein (EDEN-BP), which binds to the EDEN sequence present in the 3′-UTR of the Eg mRNAs and is required for their deadenylation following egg fertilization in Xenopus (39). The EDEN sequence shows little similarity to an ARE; however, the homology between EDEN-BP and ElrA suggests that the two proteins might play similar roles in the deadenylation of ARE-containing mRNAs.
In summary, our results argue that AUUUA-rich ARE-mediated mRNA deadenylation and decay are highly conserved processes. Previously, AUUUA-dependent ARE-mediated deadenylation and decay had been shown to occur only in mammalian tissue culture cells (9, 43). Our findings suggest that the Xenopus oocyte and egg should prove useful as a model system for the study of ARE-mediated deadenylation and for the identification of factors involved in this process and its regulation.
ACKNOWLEDGMENTS
We thank B. DeDecker, D. Enke, C. Fan, M. Frilander, T. McConnell, L. Scharl, and L. Weinstein for many helpful suggestions and careful reading of the manuscript and B. Egan and S. Baserga for plasmids.
This work was supported by grant CA16038 from the National Institutes of Health.
REFERENCES
- 1.Anderson J S J, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires SK12 DEVH box protein and 3′ to 5′ exonuclease of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audic Y, Omilli F, Osborne H B. Postfertilization deadenylation of mRNAs in Xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Mol Cell Biol. 1997;17:209–218. doi: 10.1128/mcb.17.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beelman C, Stevens A, Caponigro G, LaGrandeur T, Hatfield L, Fortner D, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein P, Peltz S, Ross J. The poly(A)–poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvet P, Omilli F, Yannick A-B, Legagneux V, Roghi C, Bassez T, Osborne H B. The deadenylation conferred by the 3′ untranslated region of a developmentally controlled mRNA in Xenopus embryos is switched to polyadenylation by deletion of a short sequence element. Mol Cell Biol. 1994;14:1893–1900. doi: 10.1128/mcb.14.3.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewer G, Ross J. Poly(A) shortening and degradation of the 3′ A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988;8:1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C-Y, Chen T-M, Shyu A-B. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol. 1994;14:416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C-Y, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 10.Chen C-Y, Shyu A-B. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol Cell Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc Natl Acad Sci USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson E. Gene activity in early development. 3rd ed. New York, N.Y: Academic Press; 1986. [Google Scholar]
- 13.Decker C, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 14.Drummond D, Armstrong J, Colman A. The effect of capping and polyadenylation on stability, movement, and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–7394. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond D, Armstrong J, Colman A. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–7394. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duval C, Bouvet P, Omilli F, Roghi C, Dorel C, LeGuellec R, Paris J, Osborne H B. Stability of maternal mRNA in Xenopus embryos: role of transcription and translation. Mol Cell Biol. 1990;10:4123–4129. doi: 10.1128/mcb.10.8.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dworkin M, Dworkin-Rastl E. Changes in RNA titers and polyadenylation during oogenesis and oocyte maturation in Xenopus laevis. Dev Biol. 1985;112:451–457. doi: 10.1016/0012-1606(85)90417-8. [DOI] [PubMed] [Google Scholar]
- 18.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;12:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox C, Sheets M, Wickens M. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- 20.Fox C, Wickens M. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3′ UTR of certain maternal mRNAs. Genes Dev. 1990;4:2287–2298. doi: 10.1101/gad.4.12b.2287. [DOI] [PubMed] [Google Scholar]
- 21.Galili G, Kawata E, Smith L, Larkins B. Role of the 3′-poly(A) sequence in translational regulation of mRNAs in Xenopus laevis oocytes. J Biol Chem. 1988;263:5764–5770. [PubMed] [Google Scholar]
- 22.Gautier J, Solomon J, Booher R, Bazan J F, Kirschner M. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- 23.Huarte J, Stutz A, O’Connell M, Gubler P, Belin D, Darrow A, Strickland S, Vassalli J-D. Transient translational silencing by reversible mRNA deadenylation. Cell. 1992;69:1021–1030. doi: 10.1016/0092-8674(92)90620-r. [DOI] [PubMed] [Google Scholar]
- 24.Hyman L, Wormington M. Translational inactivation of ribosomal protein mRNAs during Xenopus oocyte maturation. Genes Dev. 1988;2:598–605. doi: 10.1101/gad.2.5.598. [DOI] [PubMed] [Google Scholar]
- 25.Jones T, Cole M. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kay B, Peng H B, editors. Methods in cell biology. 36. Xenopus laevis: practical uses in cell and molecular biology. New York, N.Y: Academic Press; 1991. [PubMed] [Google Scholar]
- 27.Kruys V, Marinx O, Shaw G, Deschamps J, Huez G. Translational blockade imposed by cytokine-derived UA-rich sequences. Science. 1989;245:852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- 28.Kruys V, Wathelet M, Poupart P, Contreras R, Fiers W, Content J, Huez G. The 3′ untranslated region of the human interferon-β-mRNA has an inhibitory effect on translation. Proc Natl Acad Sci USA. 1987;84:6030–6034. doi: 10.1073/pnas.84.17.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagnado C, Brown C, Goodall G. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaGrandeur T, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legagneux V, Omilli F, Osborne H B. Substrate-specific regulation of RNA deadenylation in Xenopus embryo and activated egg extracts. RNA. 1995;1:1001–1008. [PMC free article] [PubMed] [Google Scholar]
- 32.Levy N S, Chung S, Furneaux H, Levy A P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 33.Marinx O, Bertrand S, Karsenti E, Huez G, Kruys V. Fertilization of Xenopus eggs imposes a complete translational arrest of mRNAs containing 3′UUAUUUAU elements. FEBS Lett. 1994;345:107–112. doi: 10.1016/0014-5793(94)00413-7. [DOI] [PubMed] [Google Scholar]
- 34.McGrew L, Dworkin-Rastl E, Dworkin M, Richter J. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3:803–915. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- 35.Muckenthaler M, Gunkel N, Stripecke R, Hentze M. Regulated poly(A) tail shortening in somatic cells mediated by cap-proximal translational repressor proteins and ribosome association. RNA. 1997;3:983–995. [PMC free article] [PubMed] [Google Scholar]
- 36.Muhlrad D, Decker C, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 37.Muhlrad D, Decker C, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray A, Kirschner M. Cycline synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- 39.Paillard L, Omilli F, Legagneux V, Bassez T, Manley D, Osborne H B. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng S S-Y, Chen C-Y A, Xu N, Shyu A-B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter J. Translational control in early development. Bioessays. 1991;13:179–183. doi: 10.1002/bies.950130406. [DOI] [PubMed] [Google Scholar]
- 42.Rosenthal E T, Tansey T R, Ruderman J V. Sequence-specific adenylations and deadenylations accompany changes in the translation of maternal messenger RNA after fertilization of Spisula oocytes. J Mol Biol. 1983;166:309–327. doi: 10.1016/s0022-2836(83)80087-4. [DOI] [PubMed] [Google Scholar]
- 43.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 44.Shyu A-B, Belasco J, Greenberg M. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 45.Standart N, Jackson R. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 46.Stolle C, Payne M, Benz E. Equal stabilities of normal β-globin and nontranslatable β-39 thalassemic transcripts in cell-free extracts. Blood. 1987;70:293–300. [PubMed] [Google Scholar]
- 47.Tchang F, Vriz S, Mechali M. Postranscriptional regulation of c-myc RNA during early development of Xenopus laevis. FEBS Lett. 1991;291:177–180. doi: 10.1016/0014-5793(91)81277-f. [DOI] [PubMed] [Google Scholar]
- 48.Varnum S, Wormington M. Deadenylation of maternal mRNAs during Xenopus oocyte maturation does not require specific cis-sequences: a default mechanism for translational control. Genes Dev. 1990;4:2278–2286. doi: 10.1101/gad.4.12b.2278. [DOI] [PubMed] [Google Scholar]
- 49.Veyrune J, Campbell G, Wiseman J, Blanchard J, Hesketh J. A localisation signal in the 3′ untranslated region of c-myc mRNA targets c-myc mRNA and β-globin reporter sequences to the perinuclear cytoplasm and cytoskeletal-bound polysomes. J Cell Sci. 1996;109:1185–1194. doi: 10.1242/jcs.109.6.1185. [DOI] [PubMed] [Google Scholar]
- 50.Vriz S, Mechali M. Analysis of 3′-untranslated regions of seven c-myc genes reveals conserved elements prevalent in post-transcriptionally regulated genes. FEBS Lett. 1989;251:201–206. doi: 10.1016/0014-5793(89)81455-3. [DOI] [PubMed] [Google Scholar]
- 51.Vriz S, Taylor M, Mechali M. Differential expression of two Xenopus c-myc proto-oncogenes during development. EMBO J. 1989;8:4091–4097. doi: 10.1002/j.1460-2075.1989.tb08593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wickens M. How the messenger got its tail: addition of poly(A) in the nuclues. Trends Biochem Sci. 1990;15:277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- 53.Wickens M. In the beginning is the end: regulation of poly(A) addition and removal during early development. Trends Biochem Sci. 1990;15:320–324. doi: 10.1016/0968-0004(90)90022-4. [DOI] [PubMed] [Google Scholar]
- 54.Wilson T, Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 55.Wu L, Good P, Richter J D. The 36-kilodalton embryonic-type cytoplasmic polyadenylation element-binding protein in Xenopus laevis is ElrA, a member of the ELAV family of RNA-binding proteins. Mol Cell Biol. 1997;17:6402–6409. doi: 10.1128/mcb.17.11.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zubiaga A, Belasco J, Greenberg M. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]