Abstract
Developing diagnostics and treatments for neurodegenerative diseases (NDs) is challenging due to multifactorial pathogenesis that progresses gradually. Advanced in vitro systems that recapitulate patient-like pathophysiology are emerging as alternatives to conventional animal-based models. In this review, we explore the interconnected pathogenic features of different types of ND, discuss the general strategy to modelling NDs using a microfluidic chip, and introduce the organoid-on-a-chip as the next advanced relevant model. Lastly, we overview how these models are being applied in academic and industrial drug development. The integration of microfluidic chips, stem cells, and biotechnological devices promises to provide valuable insights for biomedical research and developing diagnostic and therapeutic solutions for NDs.
Subject terms: Alzheimer's disease, Huntington's disease, Amyotrophic lateral sclerosis, Parkinson's disease, Neurodegeneration
This review focuses on recent advances in on-chip platforms for patient-like in vitro modeling of the pathology of neurodegenerative diseases, including Alzheimer’s, Parkinson’s, and Huntington’s diseases as well as Amyotrophic lateral sclerosis. The authors advocate for broader usage of these human-relevant models in the academic and pharmaceutical fields.
Introduction
As life expectancy increases with advances in medical care, humanity faces a new crisis related to the rising ageing population1. This leads to a concomitant increase in the incidence of incurable neurodegenerative diseases (NDs)2–4. These diseases generally affect the brain activity in the elderly by diminishing their cognitive and behavioural functions5–8. Tremendous efforts have been made for decades to investigate the underlying mechanisms of these fatal NDs using both in vivo and in vitro systems, despite the complexity of the human brain structure and the limitations of real-time observation. Notably, several pathogenic features, such as specific neuronal loss, gliosis, neuroinflammation, oxidative stress, mitochondrial dysfunction, and early vascular damage tend to overlap in common NDs, including Alzheimer’s disease (AD)9,10, Parkinson’s disease (PD)6,11, amyotrophic lateral sclerosis (ALS)7,12, and Huntington’s disease (HD)13,14. Although the primary initiating factor remains elusive, hypotheses suggest that the accumulation of several dysfunctional proteins is closely related to the onset of each ND15.
Despite advances in our understanding of NDs, the development of disease-modifying treatments (DMT) that target the underlying mechanisms of their pathogenesis remains challenging. Only a few DMTs, such as aducanumab16 and lecanemab17 for AD and riluzole18 and edaravone19 for ALS, have recently been approved. A substantial number of drug trials for NDs that were effective in preclinical studies have yet to demonstrate efficacy in clinical trials, and the limitations of current experimental models are responsible for this failure. Notably, animal models, which have been heavily relied upon for the drug development of NDs, differ fundamentally from humans in terms of their immune systems20 and the ratio, distribution, morphology, and gene expression of their brain cells21. Moreover, current animal models of NDs cannot fully replicate the complex pathogenic lesions of human NDs, and often overlook sporadic cases, which account for most cases22,23. To investigate the pathological processes of NDs, advanced in vitro models that can accurately replicate various physiological conditions, multicell types, and interactive cell-to-cell environments, and provide real-time monitoring modulated by different biochemical and biophysical cues are in high demand.
Over the past decades, stem cell biology and advances in cell culture techniques have rapidly developed a stem cell-based 3D culture platform that generates an “organoid” for disease modelling and drug screening24. Organoids have several advantages over conventional 2D cell cultures, including patient-specific origins and intrinsically complicated 3D compositions that mimic tissues. Several brain organoid subregions have been developed for brain studies, including the cerebral cortex25, hippocampus26, hypothalamus27, forebrain27, midbrain27,28, functional choroid plexus29, and cerebellum30. However, current methods for organoid growth have several limitations that need to be addressed, including insufficient neuronal functionality and cellular microenvironment, lack of vascular and immunological components, low reproductivity and uniformity, and difficulties in long-term maintenance31–33.
Microfluidic organs or organoids-on-chips have provided unique opportunities for researchers to develop an inventive experimental design by allowing the reproduction of critical elements of each organ, such as disease-specific miniaturised environments with high controllability and tunability, biochemical, and biophysical cues, continuous medium flow, and complex interactions between cells relevant to the in vivo system34. Moreover, integrated microfluidic chips are ideal platforms for drug screening and basic life science studies in biomedical research35. Despite the challenges in determining the brain’s critical structural and functional units owing to its complex morphology and physiology, many commercialised and customised microfluidic chips have simplified the complex characteristics of the brain into miniaturised systems22. Existing brain-on-chips mimic diverse levels of brain units, such as the axonal chip36, neuron-glia chip37, blood-brain-on-a-chip38,39, and neurovascular-unit-on-a-chip40. These have been applied in studies on brain tumours41, vascular diseases40, brain injury42, neuroinflammation43, and NDs44. These microfluidic chip designs can be repurposed for desired disease conditions according to the experimental targets, allowing researchers to understand neuropathogenesis at a more profound level.
Recently, the idea of taking advantage of microfluidic chips to overcome the limitations of organoid formation was proposed, with promising results in the development of organoids-on-chips for the pancreas45, liver46, kidney47, stomach48, and brain49–51. This approach provides better in vivo mimicry properties than conventional models, preventing necrosis by controlling organoid expansion with chip size, integration with the vascular bed to provide sufficient nutrients, allowing co-culturing with systemic microglia, applying mechanical and physical cues, delivering a drug through a physiologically relevant barrier, and achieving mature characterisation with continuous fluid flow. However, there is still room for improvement in both culture systems to accelerate their application in drug screening and personalised medicine. In this review, we aim to highlight the potential of integrated microfluidic technologies for advancing research on the neuropathogenesis of NDs (AD, PD, ALS, and HD) to make substantial breakthroughs in disease modelling and drug development.
Current understanding of neurodegenerative diseases
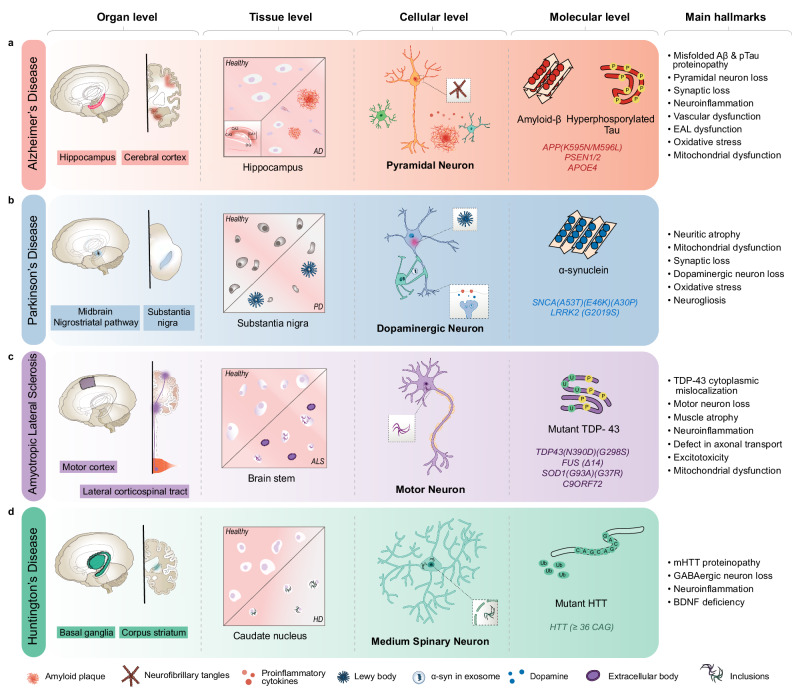
Numerous studies have identified the interconnected pathogenic features that contribute to ND progression over several decades. These include dysfunctional protein-related pathogeneses, gliosis, neuroinflammation, metabolic alterations, oxidative stress, mitochondrial dysfunction, and genetic alterations. However, the primary aetiologies of selective neuronal cell death and synaptic loss in NDs remain to be determined. This has hindered the development of disease-modifying drugs and early diagnostic tools. In this section, we review the current progress in the investigation of AD, PD, HD, and ALS. These diseases share some commonalities and distinctions in their pathogeneses (Fig. 1).
Fig. 1. The similarities and differences in the main hallmarks of neurodegenerative diseases from the organ to molecular levels.
a AD is characterised by the inclusion of misfolded amyloid-β (Aβ) and neurofibrillary tangles in pyramidal neurons, primarily in the hippocampus and cortex regions of the brain. b PD is characterised by Lewy body aggregates composed of misfolded α-synuclein and degeneration of dopaminergic neurons in the substantia nigra region of the brain. c ALS is characterised by including mutant TAR DNA-binding protein 43 (TDP-43) and other proteins, degeneration of motor neurons in the motor cortex and spinal cord, and muscle atrophy with dysfunctional proteins. d HD is characterized by including mutant Huntingtin protein (mHTT) and degeneration of medium spiny neurons in the basal ganglia, and corpus striatum of the brain. AD Alzheimer’s disease, ALS amyotrophic lateral sclerosis, BDNF brain-derived neurotrophic factor, EAL endosomal-autophagic-lysosomal pathway, GABA gamma-aminobutyric acid, HD Huntington’s disease, PSEN presenilin 1, SNCA synuclein alpha.
Alzheimer’s disease
AD has become a major concern worldwide, accounting for 80% of all dementia cases. The prevalence of AD is expected to rapidly increase over the next 30 years52. In AD, symptoms of cognitive and behavioural impairment occur as the first, but memory loss presents much earlier, before clinical diagnosis, especially in the predominant age group (>65 years)53,54. This results from progressive neuronal and synaptic loss mainly in the cortex and hippocampus (Fig. 1a). Unfortunately, when AD is first diagnosed in the clinical setting, several irreversible pathological changes occurring in the brain are already in progress55. One of the main pathological characteristics of AD is the deposition of amyloid plaques and neurofibrillary tangles (NFT) in the brain tissue. On a macroscale, moderate atrophy of the cortex, amygdala, and hippocampus as well as enlargement of the temporal horn of the lateral ventricle have been observed in AD. This ultimately leads to a loss of whole brain volume56. Gene mutations linked to amyloid protein processing pathways, such as APP57, PSEN158, and PSEN259 are responsible for early-onset AD (EOAD). Recent genome-wide association studies (GWAS) have revealed many genetic factors associated with late-onset AD (LOAD), such as BIN1, APH1B, PTK2B, PILRA, CASS4, CCDC6, TSPAN14, NCK2, and SPRED260,61. However, most LOAD cases are attributed to APOE462 and TREM2 R47H variants63,64, which regulate brain inflammation, cholesterol, and glucose metabolism, and microglia function65,66. In this section, we describe the current leading hypotheses of AD, such as amyloid-β (Aβ) and tau protein dysfunction, neuroinflammation, blood-brain barrier (BBB) dysfunction, gut-brain axis, mitochondrial dysfunction, oxidative stress, and metabolism alterations.
Amyloid-β and hyperphosphorylated tau-related pathogenesis
The most prominent histological hallmark of AD is the deposition of misfolded amyloid aggregates or plaques in the extracellular space. The core element of amyloid plaques is the pathological Aβ peptide (~4 kDa), which is produced by altered cleavage of the amyloid precursor protein (APP). APP is a type I transmembrane protein that is typically involved in cell interactions, migration, synapse formation, and neuroprotection. It is mainly produced by neurons, astrocytes, platelets, and vascular endothelial cells67,68. However, in the familial AD brain, the APP cascade is altered due to mutation in protein processing genes (APP, PSEN1, and PSEN2), causing the formation of Aβ peptides67,68. Among Aβ peptides of various lengths, Aβ42 tends to aggregate and form plaques, but Aβ40 is the most abundant. In addition, this abnormal aggregation is induced by the impairment of protein regulation systems, such as the endoplasmic reticulum system, ubiquitin-proteasome system (UPS), and autophagy-lysosomal pathway (ALP)69. Importantly, dysfunction of several regulating proteins, such as LRP170, SNX671, GGA372, and SORLA73 affect APP endocytosis, endosomal trafficking, and Aβ production, which are closely associated with the onset of AD.
Studies have focused on examining differences in the effects of various forms of Aβ on cells, and they reported that intermediate forms of Aβ, such as oligomers and protofibrils, are more neurotoxic than mature fibrils10,74,75. Several in vitro studies have reported that the pathogenicity of oligomers varies depending on their size and structure. These oligomers are known to cause neurotoxicity by binding to neuronal receptors (PrPc76, NMDAR77, β2-AR78, p75NTR79, and α7nACR80), altering cell membranes81, causing mitochondrial dysfunction82, Ca+2 imbalance83, as well as inducing tauopathy84.
AD progression may be linked to Aβ propagation among neurons. In the human AD postmortem models, the positions of Aβ plaques exhibit a regional-specific outer to inner pattern, initially detected at the frontal, and medial parietal cortex, followed by the allocortical region and midbrain. Then, in late clinical phase, Aβ accumulation is found in the brainstem and cerebellum85. Furthermore, positron emission topography (PET) imaging of the AD brain has revealed that Aβ dynamically accumulates in synaptic contact areas, which may be a result of Aβ propagation86. Also, an in vitro study has demonstrated prion-like propagation of Aβ oligomers between neighbouring cells87,88, which is explained by the “seeding-nucleation” theory observed in an in vitro study as a three-step process (seeding, nucleation, and elongation), where Aβ oligomers formed from monomers elongate into fibrils. These fibrils then accelerate the formation of oligomers (nuclei), resulting in increased aggregations89,90. Moreover, current hypotheses have shown three Aβ propagation ways: direct cell contact91, exosomes92–94, and tunnelling nanotubes between cells95–97.
Abnormal intracellular accumulation of tau proteins, NFTs, is another primary pathological marker that is considered to be more closely related to cognitive decline98. Tau, a microtubule-associated protein, is mainly located in the axons and supports axonal transport, maintenance, and generation of microtubules. However, alterations in post-translational modification promote its aberrant aggregation99. Among the several types of post-translational modifications, the relationship between hyperphosphorylated tau (pTau) and AD pathogenesis has been well studied by discovering several phosphorylation sites (Thr175, Ser202, Thr205, Thr212, and Ser422) and its toxic promotion of misfolding and self-aggregation100. Furthermore, other neuroprotective phosphorylated states (Ser214, Ser262, and Ser305) inhibit tau aggregation101,102. However, hyperphosphorylation depends on the starting site, which may accelerate other multisite phosphorylation states103. The leading cause of tau hyperphosphorylation is unknown despite the discovery of several tau-associated kinases and phosphatases, such as GSK-3β, CDK5, ERK2, PKA, and PP2A104–107. Recently, one group found that a 12-amino-acids-long peptide derived from CDK5 disrupts hyperactivated CDK5 (CDK5-p25 complex). This enzyme leads to decreased DNA disruption, pTau levels, and neurodegeneration, and increased behavioural ability in an AD mouse model108.
The spread of toxic tau shows similar characteristics to Aβ, such as prion-like behaviour and propagation between cells via direct contact109, exosomes110, and nanotubules111. In vivo and in vitro studies have shown that tau propagation from the entorhinal cortex (EC) to the hippocampal region is mediated by exosomes released from microglia112. Similarly, one in vitro study has also shown that microglia secreted extracellular vesicles (EVs) containing Aβ propagate through axons, causing synaptic alterations113. Despite the protective effect of microglia exosomes and EVs by promoting clearance of Aβ and tau, studies suggest that these may be a factor in propagation among cell-cell interactions in the AD brain, which makes them an exciting aim for further research.
The relation between Aβ and NFT in AD pathogenesis remains controversial. In the early phase, NFTs were observed in neurons of the more profound parts of the EC and CA1 regions of the hippocampus, followed by other parts of the brain, including the parietal, medial, and lateral occipital cortices, in a reversed pattern of amyloid plaques86. A histological-level study also found that NFT begins to form and spread under the influence of Aβ deposition114. Recent automatic PET imaging analysis has revealed that tau accumulation first appears at the rhinal cortex, independent from Aβ115. Subsequently, tau spreads robustly throughout neocortex when the Aβ level reaches at certain threshold. This finding has been termed ‘Ca-tau-strophe’ and highlights the importance of the tau protein in earlier diagnosis and therapeutic choice115,116. However, the detailed mechanisms underlying tau protein toxicity have not been fully elucidated. However, substantial research findings have proposed an association between Aβ and other proteins (such as Pin1, HSP, Fyn kinase, α-synuclein, and PASCIN1) regarding tau hyperphosphorylation117,118. Aβ accumulation induces tau aggregation and propagation trans-synaptically in vitro studies119. Notably, some studies have suggested that NFT may protect against amyloid plaques117. This emerging evidence has indicated the importance of synergy of Aβ and tau in AD pathogenesis120.
APP-C99, a C-terminal fragment of APP, has recently been suggested as an early AD marker associated with endosomal-autophagic-lysosomal (EAL) malfunction121. C99 accumulation in neurons causes lysosomal and synaptic distortion and cognitive changes by aggregating within EAL vesicle membranes122.
Neuroinflammation and systematic inflammation
Compelling evidence for the association of immune-regulating genes with AD suggests that systemic immune-mediated neurodegeneration is an alternative hypothesis for the pathogenesis of AD. As ageing, circulating proinflammatory cytokines (IL-1β, IL-6, TNFα, and IL-18) induced by infection, chronic diseases, stresses, and cellular senescence may initiate or contribute to the neuropathogenesis of AD123. When these cytokines cross the BBB, they can activate glia and astrocytes, leading to Aβ and tau phosphorylation, oligomerization, neuroinflammation, and neurotransmitter toxicity123. In pathological studies, increased levels of proinflammatory proteins in the cerebrospinal fluid (CSF) have been associated with brain volume loss and thinning of the white matter, corresponding to cognitive dysfunction124.
As Aβ plaques form, the presence of activated microglia and astrocytes also increase125. Therefore, extensive research has focused on the role of astrocytes and microglia in AD progression, including their contribution to Aβ and pTau depositions, neuroinflammation, and Ca+2 imbalance. Microglia exert their neuroprotective activities by clearing misfolded proteins126. However, as AD progresses, microglia transform into a deteriorating state, enhancing pTau-related kinase expression and production of the inflammasome NLRP3 (NOD-, LRR-, and pyrin domain-containing protein 3). Increased production of NLRP3 accelerates the activity of caspase-1 and secretion of proinflammatory cytokines, such as IL-1β and IL-18, and promotes neuronal and glial death via apoptosis127,128. In addition, GWAS identified multiple genes associated with LOAD (CR1, SPI1, MS4As, TREM2, ABCA7, CD33, and INPP5D) that are expressed in microglia, underscoring the important role of microglia in AD pathogenesis129. Specifically, studies have suggested that a mutation of TREM2, lipid sensing transmembrane glycoprotein expressed in microglia, causes the loss of its phagocytosis function130, and diminishes its migration towards Aβ plaques and uptake of Aβ40 and Aβ42131,132. Notably, the observation of APOE as a ligand for TREM2 has revealed new insights into their association with the pathogenesis on Aβ and pTau133,134. Yeh et al. 134 demonstrated that microglia expressing TREM2 variants exhibited reduced internalisation of Aβ and the apolipoprotein or lipoprotein complex. Gratuze et al. 135 recently suggested that microglial activation could worsen tau-associated neurodegeneration in the presence of APOE4 through a TREM2-independent pathway. One group recently developed a successful therapeutic approach based on TREM2 function in microglia activation. They introduced a high-affinity human TREM2 antibody with a monovalent transferrin receptor (TfR) binding site to enable efficient delivery to the brain. This antibody promotes glucose and lipid metabolism in microglia, shifting them to a metabolically responsive state distinct from that triggered by amyloid pathology136.
Reactive astrocytes play a critical role in the progression of AD. It has been suggested that astrocytes become reactive to surrounding plaques, but there are limited data on the exact morphological and functional alteration137,138. A recent PET study revealed a correlation between reactive astrocytes and protein biomarkers (Aβ, and tau) in the CSF of patients with AD, suggesting their contribution to progression139. The expression of APOE in many cell types has gained attention because APOE4 is considered the primary genetic risk factor for sporadic AD140. Astrocytes expressing APOE4 have demonstrated diminished functions, such as autophagy, clearance, and internalisation of Aβ, and support for neurons for their durability and synapse formation141,142. Moreover, a recent report suggests that astrocytes expressing APOE4 immoderately provide cholesterol to neurons by expanding their lipid membrane, increasing APP deposition, and promoting Aβ formation143. Conversely, oligodendrocytes expressing APOE4 exhibit dysfunction in myelination144, coinciding with increased neurotransmitter production and Aβ42 secretions in APOE4 neurons145. Studies have demonstrated an association between APOE4 expression and tauopathy. The deletion of APOE4 leads to the reversal of pathologies, including increased maintenance of the myelin sheath and decreased disease-associated neural subpopulations, and facilitates the severity of tauopathy146.
At the molecular level, using single-cell and single-nucleus RNA sequencing, researchers have recently revealed the specific transcriptional and functional characteristics of astrocytes and microglia, which have been categorised as disease-associated astrocytes (DAA) and microglia (DAM)147,148. The DAA subtype of astrocytes is associated with the early stages of AD and progressively increases over time. In AD mouse models, the level of homoeostatic astrocytes declines, while intermediate and glial fibrillary acidic protein highly expressed DAAs in the hippocampus and cortex region147. DAM showed the same transitioning pattern from a homoeostatic to an activated state. Moreover, one study indicated that phagocytic DAM subtypes can internalise Aβ proteins through a two-step sequential process that involves the downregulation of microglia-regulating genes and upregulation of phagocytic and lipid metabolism pathways148. According to a recent study using 3D in situ RNA sequencing, spatiotemporal variations in glial cells have been observed in the brains of AD mice. This study has revealed a layered position of cells surrounding Aβ plaques, with the first layer being DAM, followed by DAA and oligodendrocytes149.
Blood-brain barrier dysfunction
Research on the role of the BBB in AD is important because of the strong hypothesis of its association to the circulation of misfolded proteins originating from the brain or body system, and other pathogenic features. BBB disruption can be observed during the early stages of AD before regional atrophy occurs, and is emerging as a leading pathogenesis150. Initial defects in brain vessels may initiate various interconnected pathways such as misfolded proteins and neuroinflammation, and directly contribute to neurodegeneration151. Conversely, according to animal studies, pathological Aβ and tau proteins may cause disruption in blood vessels in vulnerable areas of AD during disease progression152,153. BBB disruption begins with changes in permeability, which are associated with a decrease in tight junction proteins in endothelial cells and reduced pericytes151,154. However, Aβ155 and tau accumulation156 disrupt the BBB integrity by altering the level of tight junction proteins. Moreover, microglia-driven neuroinflammation damages the BBB integrity and contribute to disease development157. Therefore, investigating brain vascular pathology is a promising avenue for gaining insight into the initial systemic mechanisms of all NDs.
Glymphatic system dysregulation
The dynamics of CSF and tissue interstitial fluid are supported by the perivascular network known as the glymphatic system158. Bidirectional aquaporin channels (AQP) facilitate continuous fluid flow to assist in circulating proteins and metabolites. The contribution of AQP to AD pathogenesis, associated with the clearance of pathological proteins, has been investigated159,160. In the absence of AQP4, APP mouse had decreased influx of CSF tracer, and accumulation of Aβ and APOE4 proteins in neurons, as well as phagocytic activation of microglia, suggesting the substantial role of AQP4 during disease progression160. Moreover, AQP4 knockout tau-expressing mice demonstrate increased tau aggregation and neurodegeneration, suggesting a highly relevant AQP4-dependent extracellular tau clearance pathway161.
AQP4 is primarily expressed in the endfeet processes of perivascular astrocytes in the BBB and tripartite synapses162,163. It is also found in the epithelial cells of the choroid plexus and the ependymal cells of the ventricular lining in the brain. Human postmortem AD models have shown decreased levels of perivascular AQP4 in the frontal cortex, which is associated with cognitive dysfunction and amyloid and tau pathology164,165. At the cellular level, the effect of mislocalisation of AQP4 channels on Aβ accumulation have been also investigated with deletion of α-syntrophin (SNTA1), important for perivascular localisation166. Several approaches have been proposed to inhibit AQP4 mislocalisation as a treatment for central nervous system (CNS) injury and stroke, with the aim of stabilising the BBB and disrupting the blood-spinal cord barrier disruption167,168. One group tested a calmodulin inhibitor targeting the mislocalisation of AQP4 to the calmodulin-mediated cell surface. This approach has successfully demonstrated reduced oedema and recovery of spinal cord injury. Moreover, the same calmodulin inhibitor decreases the expression of AQP4 proteins and reduces oedema, while increasing glycogen metabolism in the early acute phase in a post-stroke mouse model168. Cerebral oedema is the main hallmark of CNS injury and the main underlying factor for NDs, treating oedema by regulating AQP4 could be the treatment of choice for AD.
Gut-brain axis
The brain and gut communicate via neural, immune, endocrine, and metabolic pathways. Several hypotheses have proposed that the gut microbiome substantially contributes to brain disorders, including AD169. Considerable evidence suggests a connection between changes in gut microbiota and various features of AD, including Aβ, tauopathy, and neuroinflammation, and other pathogenesis-inducing neurodegeneration170,171. The current hypothesis is that gut microbiota indirectly affects BBB homoeostasis by activating peripheral immune cells. Alternatively, it can occur through the direct release of metabolites and hormones that travel through the vagus nerve, leading to neuroinflammation and contributing to proteinopathy171. Notably, a study has reported the antimicrobial effect of Aβ, revealing its protective role in immunity and inflammation172. This antimicrobial function has been consistent in all forms of Aβ peptide from oligomers to fibrils, not only in in vitro cultures, but also in AD mouse brains. Bacterial infection has accelerated the seeding and formation of Aβ deposition, which is in close proximity to the pathogen. These findings suggest that the inflammatory effects of infection and microbial changes may possibly initiate amyloid pathogenesis172.
A recent study demonstrated the impact of the gut microbiota on tauopathy of human APOE isoforms in a tau-transgenic mouse model173. In bacterial-free tauopathy mice, the level of pTau in the hippocampal area decreased, and microglia and astrocytes remained in a homoeostatic state in a sex- and APOE-dependent manner. In particular, when testing the differences in APOE isoforms, only APOE3 expressed in the tau male model showed improvement after treatment with antibiotics, but not the APOE4 isoforms of both sexes. Moreover, short-chain fatty acids in bacterial metabolites play a role in tau-induced neurodegeneration in mice. These findings strongly suggests that the importance of the gut-brain axis on AD pathogenesis, association with Aβ- and tau-induced pathogenesis, and neuroinflammation.
Brain-derived neurotrophic factor deficiency
Brain-derived neurotrophic factor (BDNF) is a critical factor in hippocampal neurogenesis that sustains synapses and neuronal circuits. A deficiency in BDNF is associated with several neuropathogenesis, including Aβ deposition, phosphorylated tau formation, neuroinflammation, and apoptotic cell death174. Moreover, the deficiency in neurogenesis observed in the early stages of AD can be explained by alterations in BDNF signalling; however, the underlying mechanism remains unknown. One study claimed that Aβ monomers maintain the production and release of BDNF by activating IGF receptors175. In addition, when BDNF was restored in a tau mouse model, behavioural changes, neuronal loss, and synaptic loss were reduced, but not hyperphosphorylation176.
Mitochondrial dysfunction and oxidative stress
In the early stages of AD, changes in mitochondrial structure, function, and accumulation have been observed177. Specifically, impairment of the mitophagy process is the major change currently attributed to cholesterol metabolism alterations related to APOE, mitophagy regulation of Aβ, tau protein, and APP-C99 accumulations on mitochondrial-associated membrane177–179. Astrocytes expressing APOE4 exhibit reduced fission and mitophagy180.
The principal characteristic of most NDs is the different vulnerable neurons181. During the progression of AD, neurons of the ECII, pyramidal neurons of the CA1, cholinergic neurons of the basal forebrain, noradrenergic neurons of the locus coeruleus, and rostral neurons are more vulnerable at the onset of the disease, whereas sensory cortical neurons begin to degenerate only at later stages. A recent study identified the molecular basis of vulnerable neuronal subtypes by profiling mRNA and successfully built a data source for modelling AD with specific neuronal circuits182,183. Vulnerability is attributed to oxidative stress, because oxygen-sensitive glutamatergic cells are selectively affected184. When oxidative stress increases, the vulnerable cells tend to experience DNA damage and mitochondrial fragmentation185.
Altered glucose and lipid metabolism
Understanding the complex roles of glucose and lipid metabolism is crucial for understanding neurodegeneration in the brain. According to a recent cohort study, high blood glucose and cholesterol levels increase the risk of AD186. Additionally, as the disease progresses, glucose levels in the brain decrease, whereas cholesterol levels increase187,188. Brain imaging studies have revealed that glucose metabolism is impaired in the hippocampal and cortical areas before the onset of cognitive decline or pathological changes188,189. Reduced glucose uptake is associated with accelerated neurodegeneration and Aβ depositions189–191. A four-month insulin treatment via the intranasal route improved cognitive function in APOE2- and APOE3-male mouse models, but not APOE4-expressed female mice192.
Others suggest that Aβ depositions cause a deficiency of GLUT-1 and GLUT-3 in vulnerable regions193. One group has proposed that BBB dysfunction, which is apparent at the onset of AD, is due to decreased GLUT-1 expression in BBB endothelial cells194. These results suggested a complex association between glucose metabolism and AD pathogenesis.
In the oldest investigation by Alois Alzheimer, lipid accumulation was highlighted as the main pathological feature in AD195. Subsequently, many high-risk genes associated with AD were found to be involved in lipid metabolism, including APOE, TREM2 CLU, and ABCA7. This suggests that lipids are essential for AD progression; however, the complex link between changes in lipid metabolism and AD pathology remains unclear. Cholesterol and sphingomyelin of lipid rafts regulate APP processing in lipid rafts, suggesting their role in the formation of Aβ196,197. During ageing, disruption of the lipid composition of the transmembrane membrane could be one of the earlier pathogeneses of LOAD. Furthermore, studies have indicated the role of glial cells in lipid metabolism, particularly cholesterol143,144. Astrocytes expressing APOE4 increase cholesterol input to neurons, causing an expansion of lipid rafts that ameliorate Aβ formation143. Meanwhile, oligodendrocytes expressing APOE4 show decreased myelinating function due to altered cholesterol metabolism144. Moreover, other studies have suggested a connection between lipid and glucose metabolism in AD owing to reduced glucose uptake caused by high levels of oxidised cholesterol198. These findings demonstrate that the initial pathogenesis of AD, particularly LOAD, may be related to disruption of metabolism in the brain.
Parkinson’s disease
PD currently affects over six million patients worldwide and is the second most prevalent ND164. PD progresses slowly with complex clinical features, affecting not only the motor systems (tremor, bradykinesia, rigidity, and gait disturbance), but also non-motor systems (rapid eye movement sleep behaviour disorder), constipation, hyposmia, and cognitive impairment199,200. These symptoms correlate with neuronal degeneration in certain brain regions, most commonly in the substantia nigra pars compacta (SNpc), where dopamine neurons reside201 (Fig. 1b). Subsequently, patients begin to experience cognitive dysfunction in various extra-nigral regions, including the basal forebrain, thalamus, amygdala, and cortex, which initiates atrophy. Unfortunately, it is often diagnosed in a clinic much later, after the brain has lost approximately 50% of dopaminergic cell death202.
Most cases of PD are sporadic and occur later in life. Less than 10% of the cases related to genetic inheritance occur at an earlier age203. Currently, owing to GWAS, more than 200 genes associated with both early- and late-onset PD have been identified204–207. There were 90 loci related to familial cases, including SNCA, LRRK2, VPS35, PRKN, PINK1, DJ-1 and GBA. Among these genes, point mutations and multiplications discovered in SNCA strongly intensified with the development of PD208. Other genes that govern important biological pathways in dopaminergic neurons, including mitochondrial function and protein degradation (LRRK2, PARK2, PARK7, PINK1, and PRKN), have also been suggested as significant risk factors for PD. Despite notable findings, the early pathogenesis of PD remains unclear because of its complexity. Here, we focus mostly on misfolded α-syn related pathogenesis, and dopaminergic neuron vulnerability, as well as mitochondrial dysfunction occurring in PD.
Accumulation of misfolded α-synuclein
One of the main conspicuous histopathological hallmarks of PD is the abnormal protein deposition called Lewy bodies (LBs) and Lewy neurites, which consist chiefly of aggregated α-syn209 and are found mostly in the extracellular space of SNpc210. A similar inclusion can be found in synucleinopathies, such as dementia with LBs, Parkinson’s disease dementia, and multiple system atrophy211,212. Physiological α-syn (14 kDa) is an intracellular protein, normally expressed by the SNCA gene, and is abundantly found in presynaptic terminal of neurons and erythrocytes. Although the physiological function of α-syn is not fully understood, several studies have suggested that it is associated with the modulation of synaptic functions213 and may have neuroprotective role214. The α-syn structure is generally divided into three regions: amphipathic domain (N terminus, residues 1–60), central non-amyloid component (NAC region, residues 61–95), and carboxyl acidic tails (C terminus, residues 96–140)215. The N terminus of α-syn consists of four 11-amino acid imperfect repeats and consensus sequence, KTKEGV, which is responsible for forming the α-helical structure when binding to the cellular membrane, such as apolipoproteins216. The hydrophobic NAC region is essential for forming β-sheet rich α-syn fibrils, which were first discovered in amyloid plaques and induce the formation of aggregated forms of α-syn217. However, the toxicity of LBs is still controversial, as some suggest that it is merely an accumulated trace of the cytoprotective response against PD progression218. Misfolded α-syn aggregation is considered the culprit of complicated pathogenesis of PD. Misfolded oligomeric α-syn is responsible for triggering neuronal toxicity by interfering with cell-to-cell signal transmission, mitochondrial pathways, and disruption of protein degradation pathways rather than mature fibrils219–222. Moreover, the physiological conformation of α-syn in vivo is an α-helical tetramer (58 kDa), rather than an unfolded α-helical monomer structure, that is resilient against aggregation223,224.
One of the notable theories of aggregation pathways is that misfolded α-syn aggregates may operate similarly to prions and self-propagate by converting normal prion proteins into abnormal ones through a seeding effect87. In the process of prion-like aggregation, α-syn creates a stable seed or nucleus as it combines into insoluble aggregates. This, in turn, accelerates formation of aggregated α-syn by misfolding α-syn and recruiting it into itself. The resulting aggregated α-syn can then be fragmented into small seeds, thereby perpetuating a vicious cycle of aggregation225. Preformed synthetic α-syn aggregates can accelerate the fibrillation of α-syn in buffer conditions226,227. Additionally, administrating preformed α-syn aggregates into mouse brains and cultured neurons facilitates the accumulation of α-syn aggregates in neurons, ultimately leading to the development of PD228–230. α-syn originates in the olfactory bulb or dorsal nucleus of the vagus nerve before propagating to other regions15,231. Notably, some groups have proposed the bottom-up hypothesis of prion-like progression of α-syn pathology, which suggests that the propagation is caused by the transmission through the autonomic nervous system232. This is supported by the earlier gastrointestinal symptoms prior to the onset of motor symptoms in patients with PD. This current research suggests that prion-like behaviour of α-syn may explain the progression of PD.
The factors that contribute to the development of abnormal α-syn aggregates are not yet fully understood. However, familial PD risk genes play a role in triggering aberrant aggregation. Specifically, multiplications (duplication and triplication) of the SNCA gene results in an overexpression of α-syn by disrupting the proteostasis process, including the synthesis, degradation, and clearance of α-syn233,234. In addition, several missense mutations in the SNCA gene, including A53T, A30P, E46K235 and H50Q236, disturb the folding dynamics of α-syn by distorting its intrinsic structure, which can lead to the early onset of PD237,238.
Dysfunction in protein degradation pathways
As LBs accumulate during PD progression, researchers have investigated protein degradation pathways to confirm the link between the impairment of these pathways and the onset of the disease. Under normal physiological conditions, protein dynamics are regulated via protein degradation pathways, such as UPS239 and ALP240. However, when these degradation pathways are disrupted owing to ageing, oxidative stress, inflammation, or other unknown causes, they can lead to the abnormal deposition of misfolded proteins in neurons, causing neuronal toxicity, which is related to various NDs241,242.
In PD, LBs are highly ubiquitinated and the cellular components of the UPS are abundantly identified, suggesting that protein clearance via the UPS is closely related to the pathogenesis of PD243,244. In vitro and in vivo studies have shown that introducing proteasomal inhibitors to disrupt the UPS led to accumulation of misfolded α-syn and neuronal death due to an impaired 26S proteasome pathway245–247. Conversely, overexpression of α-syn in dopaminergic neurons can cause dysfunction in the UPS by disrupting the 26S proteasome. This precedes the onset of behavioural impairment and neurodegeneration248.
Another protein degradation pathway, the ALP pathway, plays an important role in clearing large cellular debris that cannot be removed by the UPS. The ALP system functions through lysosomal acidification via three delivery pathways: macroautophagy, chaperone-mediated autophagy (CMA), and microautophagy. Perturbations of these lysosome-mediated protein degradation pathways are associated with pathogenesis of PD249. The introduction of genetic mutations (ATP13A2 and LAMP2) into the components of lysosomal degradation pathways induces the accumulation of α-syn aggregates250,251. In contrast, overexpression, or modified forms of α-syn could impair the clearance of misfolded proteins by hampering the CMA-mediated protein degradation. This could eventually lead to an increase in the pathogenesis of α-syn252,253.
Degeneration of highly vulnerable dopaminergic neurons
Motor dysfunction in PD is explained by dopamine deficiency caused by the degeneration of dopaminergic neurons in the SNpc. Mainly dopaminergic cells are found to be the most vulnerable to pathogenic protein toxicity, leading to impaired dopaminergic signalling from the basal ganglia to the whole brain area, such as the nigrostriatal pathway. Corresponding to dopamine depletion, several pathological processes that regulate dopamine synthesis occur in the brain254. Compensatory overexpression of postsynaptic dopamine receptors occurs when there is a considerable loss of dopaminergic neurons255. Activated microglia are also observed on-site in dopaminergic neurons, promoting degeneration by releasing reactive oxygen species (ROS)256 and neurotoxic cytokines, and inducing glutamate excitotoxicity257. Furthermore, a deficiency in the dopaminergic circuitry is associated with reduced neurogenesis in the subventricular and subgranular zones6,258. However, molecular evidence for the selective vulnerability of dopaminergic neurons to degeneration is lacking. Recently, researchers profiled the genomics of PD-derived dopaminergic neurons and found that AGTR1 expressed by a specific subgroup in the ventral tier of the pars compacta was highly upregulated in degeneration targets259.
Mitochondrial dysfunction
Mitochondria, which control cell death and energy production, have been linked to the pathogenesis of various NDs260. The chemical toxin 2-methyl-4-phenyl-1,2,3,6-tetrahydrophiridine (MPTP) was first found to cause acute Parkinsonism by disrupting mitochondrial pathways261. Once MPTP enters dopaminergic neurons through the dopamine transporter, it is oxidised by enzymes and converted into 1-methyl-4-phenylpyridinium ions (MPP+). It damages dopaminergic neurons by interfering with mitochondrial metabolism, specifically complex I of the electron transport chain, leading to increased oxidative stress262,263. As this environmental factor causes mitochondrial dysfunction, which is implicated in Parkinsonism, accumulating reports have suggested a substantial correlation between mitochondrial dysfunction and PD264,265.
α-syn plays a central role in pathogenesis of PD by inhibiting mitochondrial pathways226. One study identified α-syn null mice that were resistant to Parkinsonism caused by the administration of MPTP, implying that α-syn is essential for causing toxicity to dopaminergic neurons in conjunction with MPP+ 266,267. Additionally, α-syn aggregates negatively influence mitochondrial function, including mitochondrial fission and fusion, mitochondrial fragmentation, and complex I deficiency268. Exposure of preformed α-syn fibrils induces the recruitment of phosphorylated acetyl-CoA carboxylase 1 (ACC1) at the mitochondrial membranes, causing mitochondrial fragmentation. Oligomeric α-syn specifically compromises the mitochondrial complex I pathway and generates ROS in cybrid cell lines, leading to increased cell death269. Dopaminergic neurons generated from patients with PD carrying the A53T mutation or triplication of the SNCA gene show accumulated α-syn inclusions, as well as impaired cellular morphology and membrane potential270. In addition to impacts of environmental agents and misfolded α-syn on mitochondrial dysfunction in PD, GWAS have discovered that various genetic variants related to mitochondria are implicated in the early onset of PD271,272. Among the genetic factors, autosomal recessive PARK2 and PARK6, which express E3 ubiquitin ligase (Parkin), and PTEN-induced kinase 1 (PINK1), respectively, are strongly associated with mitochondrial dysfunction. Under normal conditions, PINK1 and Parkin are associated with the mitochondrial quality control system, mitophagy, which degrades and recycles damaged mitochondria to maintain mitochondrial homoeostasis273. However, when genetic mutations occur in these genes, the PINK1/Parkin system is unable to prevent oxidative stress and an imbalance in ATP production in mitochondria, eventually leading to the degeneration of dopaminergic neurons274–276.
The impairment of DJ-1, which protects neurons from oxidative stress by acting as a redox sensor and antioxidant, is also associated with the pathogenesis of PD, particularly in familial cases277,278. In PD, due to DJ-1 dysfunction, ROS production increases in the mitochondria, causing damage to neurons. In fact, DJ-1 is localised in the mitochondria and overexpression of DJ-1 relieves oxidative stress, preventing neuronal death279–282. However, when genetic mutations of the DJ-1 gene disrupt its biological pathways, which can lead to neurodegeneration and a monogenic form of Parkinsonism by compromising antioxidant activity277.
Environmental factors and neurotoxicity
Given the increasing impact of environmental pollution, it is crucial to consider the influence of neurotoxic chemicals on the onset and development of age-related NDs. Neurotoxic heavy metals, pesticides, and metal-based nanoparticles contribute to ND pathogenesis283. Chronic exposure to lead, the main component of polluted air, is associated with cognitive dysfunction284. Common neurotoxic mechanisms have been observed in BBB disruption, oxidative stress, mitochondrial dysfunction, and protein aggregation during ND progression285.
Liu et al. 286 recently reported that polystyrene nanoplastic particles induce the α-syn mediated neurodegeneration by invading the brain through BBB. They hypothesised that charged nanoplastics disrupt the lysosome degrading function, and trigger the α-syn protein misfolding, resulting in the intracellular α-syn fibril accumulation in neurons. This study has demonstrated that there is a notable area to investigate concerning the detrimental effects of nanoplastics on the other NDs.
Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis is a chronic, progressive disorder that specifically degenerates upper and lower motor neurons in the nerve skeletal muscles (Fig. 1c). General clinical symptoms begin with motor dysfunction, such as muscle weakness, dysarthria, and dysphagia, and gradually disturb cognitive and behavioural activities, usually leading to immobility due to respiratory failure. Mutations in more than 30 susceptibility genes are associated with familial and sporadic ALS pathogenesis. These corresponding genes were commonly clustered under functions, such as protein homoeostasis, RNA homoeostasis, axonal outgrowth, cellular transport, mitochondrial alteration, oxidative stress, and neuroinflammation287,288. Over 70% of familial ALS cases are due to mutations in the four main genes (SOD1, C9ORF72, TARDB and FUS) associated with RNA metabolism289. Despite the existence of known genetic risk factors, the initial cause and interconnected pathogenetic mechanism of sporadic cases, which account for 90–95% of all cases, have not yet been clearly identified290.
Vulnerable motor neuron degeneration
Neuronal degeneration of ALS often occurs in the corticospinal fibres, partial upper motor cortex, ventral horn of the spinal cord, and precentral gyrus. However, in most cases no significant whole-brain changes were observed291. As the disease progresses, pathological damage in cortical regions mirrors cognitive dysfunction examined in 50–65% of patients292. Histologically, with motor neuron degeneration, astrogliosis appears in the anterior horn of the spinal cord and motor nuclei of the brainstem, and spongiosis and microvacuolation are predominantly observed in the motor cortex. At the lower motor neuron level, the tripartite synapse (neuromuscular junction) consists of muscle fibres and synapse-covering glial cells (Schwann cells), and the axonal terminal of the presynaptic motor neuron is prone to damage, which often progresses to the upper neurons293. The higher vulnerability of these neurons is explained by their fast fatigue and high metabolism185. In particular, long-extruding axons are more sensitive to metabolic impairment. Recent evidence shows that the significance of non-motor (systematic) symptoms progressively increases with the severity of ALS, which may be related to the spreading pathology, similar to other NDs294.
Dysfunctional protein-related pathogenesis
Similar to other representative NDs, the main hallmark of both familial and sporadic ALS is mislocated protein accumulations, including commonly hyperphosphorylated TPD43, and dipeptide repeat (DPR) protein depositions following protein superoxide dismutase (SOD1) deposits found in cytoplasm295. These proteins are found predominantly in motor neurons. These deposition could be explained by the disturbance of the protein degradation mechanism. However, efforts are needed to reveal the consequences of protein aggregation on motor neuron function.
More than 200 mutations in SOD1 are known to cause neurotoxicity in several ways, including protein misfolding, proteasome impairment, excitotoxicity, oxidative stress, endoplasmic reticulum stress, impaired axonal transport, axonopathy, inflammation, altered RNA processing, and mitochondrial dysfunction296. Mutated SOD1 proteins are associated with motor neuron toxicity through the formation of SOD1 aggregations297,298. Furthermore, a recent molecular study on the effects of different mutations in SOD1 indicated that altered conformational variants of SOD1 are more toxic than the full form of SOD1 when associated with clinical phenotypes299. SOD1 aggregates cause selective motor neuronal death300. SOD1 aggregates are found not only in the cytoplasm of motor neurons, but also in astrocytes, microglia, and oligodendrocytes301. Moreover, similar to other NDs, evidence gathered from studies suggests exosome transfer along anatomical pathways302–304 and the potential role of oligodendrocytes in the uptake and prion-like propagation of SOD1 aggregates305. Regardless of this evidence, other components of SOD1-linked ALS need to be studied further because of reported cases of sporadic and familial ALS without SOD1 inclusion in some brain areas306.
Approximately 95% of patients with familial ALS have motor neurons with mutant TDP-43 inclusions in the cytoplasm307,308. Under normal physiological conditions, TDP-43 is localised in the nucleus and regulates gene expression and RNA metabolism. However, hyperphosphorylation, ubiquitination, and generation of C-terminal fragments cause aggregation in the cytoplasm of glia and neurons, leading to mitochondrial dysfunction, ROS production, and nucleoplasmic transport impairment. The double spiral-fold structure of TDP-43 suggests several underlying mechanisms associated with macromolecules, prion-like aggregation, and propagation that need further investigations309. Correlating with an increase in cytoplasmic aggregation, lower levels of nucleus TDP-43 have led to abnormal splicing, especially in UNC13A transcripts, which are crucial for neurotransmitter-meditated communication310–312. However, the pathological functions of UNC13A remain unclear. Notably, TDP-43, which is found in the cytoplasm of myotubes, exerts a protective effect by integrating into muscle formation and regeneration313.
Lastly, hexanucleotide repeat expansion in C9ORF72 is found in 3–7% of sporadic ALS cases (40% of familial ALS) and is a common genetic cause of ALS314. RNA transcripts of these extended regions form five different DPRs and RNA foci deposited in the neurons of the ALS brain exhibit toxicity in several models315–317. DPRs influence transport from the nucleus to the cytoplasm by causing defects in the nuclear membrane and inducing TDP-43 dislocation318. Following this investigation, therapeutic options have been developed. One group recently suggested an antisense oligonucleotide (ASO) that can suppress the expression of repeat groups in transgenic mice, which is considered a primary step towards gene therapy319. Notably, these distinct aggregations may be positive for other phenotypes. DPR aggregates are found along with TDP-43 aggregates in the ALS brain; therefore, it is important to further demonstrate their relationship and differences320.
Neuroinflammation
Imaging and postmortem cell culture studies have shown the critical roles of microglia, astrocytes, other nervous and immune cells, and inflammatory cytokines in ALS pathophysiology321,322. When motor neurons have degenerated, microglia are induced and secrete ROS and proinflammatory cytokines, such as TNF-α, IL-1, and IL-6. However, as the disease progresses, microglial activation becomes detrimental to motor neurons323. Whereas mutated SOD1-expressed astrocytes induced selective toxicity in spinal motor neurons324. Studies have suggested several pathways of astrocyte-induced neurotoxicity such as glutamate toxicity325, lactate impairment326, secretion of inflammatory mediators327,328, and necroptosis329. However, the contribution of mutant proteins to astrocyte dysfunction and motor neuron-specific toxicity remains unknown. Likewise, C9ORF72 and SOD1-expressed oligodendrocytes have also exhibited motor neuron neurotoxicity in soluble and direct ways330.
Mitochondrial dysfunction
Motor neurons are highly energetic; therefore, they tend to be sensitive to age-related metabolic changes and mitochondrial dysfunction. In ALS pathophysiology, motor neurons derived from SOD1, TDP-43, C9ORF72, and FUS show impaired mitochondrial structure, dynamics, and functions that lead to death due to the activation of ROS and intrinsic apoptotic signalling pathways331–333. A study of sporadic ALS patient-derived induced pluripotent stem cells (iPSCs) reported several changes in mitochondrial parameters, including increased ROS, depolarisation of the inner membrane potential, reduced ATP production, impairment of oxidative phosphorylation, and the protein import system334.
Disturbance in RNA metabolism
Many genes linked to ALS contribute to regulation of RNA metabolism, such as SOD1, TDP-43, FUS, and C9ORF72335. As mentioned above, the abnormal expansion of C9ORF72 is highly related to the production of abnormal RNA nuclei loci, which contribute to neurotoxicity. The RNA-binding protein encoded by TDP-43 has a regulatory role in RNA splicing, stability, and transport, especially in the expression of ND-associated (FUS, tau, ATXN2 and progranulin), synaptic, and neurotransmitter-regulating proteins, which suggest an important role in ALS pathogenesis336,337.
Huntington’s disease
HD is a rare disease with an autosomal dominant inherited pattern caused by a mutation in the Huntingtin gene (HTT)338. The progressive loss of striatal and cortical neurons leads to chorea and deterioration in cognitive and behavioural abilities, mostly as a result of the toxic effects of inherited mutant HTT (mHTT)-encoded large HTT proteins. At an early stage, caudate nucleus atrophy occurs and the neurons of the basal ganglia (corpus striatum) and corticostriatal circuit begin to degenerate when patients experience clinical symptoms339 (Fig. 1d). As the disease progresses, the cortical, occipital, and parietal regions become atrophied, corresponding to the clinical characteristics of motor and cognitive alterations339,340. Although the exact triggers of striatal neuron degeneration remain elusive, the majority of studies have focused on earlier pathogenesis features, such as the toxicity of mHTT and impaired DNA regulation of the HTT gene, which inspired several drug candidates, but all have failed at different stages. In addition, other HTT-dependent and HTT-independent pathways have been investigated, including tau dysregulation, mitochondrial dysfunction, excitotoxicity, impaired BDNF, and neuroinflammation. These pathways may have a decisive impact on HD pathogenesis, similar to mHTT.
CAG repeat and mutant Huntingtin protein
HTT, the main cause of HD, HTT gene resides on chromosome 4 and encodes the large Huntingtin protein HTT (348 kDa) with a segment of polyglutamine341. It is normally found in the cytoplasm or nucleus of neurons; however, its specific function has not yet been determined. Some reports have claimed that it may be related to BDNF production, axonal transport, neuroblast migration, and the overall development and communication of the nervous system342,343. However, in its mutated form, the HTT gene is composed of an aberrant number of cytosine, adenosine, and guanine (CAG) repeats ranging from 40 to 250 repeats, which ranges from 6 to 35 in healthy individuals341. Over 1000 CAG repeats have been reported previously344. The instability of CAG repeats was first discovered in the striatal neurons of mice two decades ago and later validated in a human autopsy model345. The number of CAG repeats correlates well with the severity of clinical HD symptoms346. Therefore, longer and more unstable repeats generally result in an earlier onset13. Early-onset HD (Westphal variant) is associated with dystonia, Parkinsonism, and psychiatric symptoms, whereas late-onset HD often correlates with 40–55 CAG repeats, which initially occur with chorea. However, the toxicity of full-length HTT proteins, especially in striatal neurons, is not fully understood, and whether they are the earliest causative factors is still debated.
Current HD pathogenesis has been broadly described as the polyglutamine tract of mHTT generated from CAG expansion and the abnormal post-translational process of HTT, which eventually leads to a cascade of pathological process343,347,348. The deleterious effect of mHTT was first demonstrated in mouse brains, exhibiting intracellular inclusion, striatal shrinkage, and compensatory ventricular enlargement accompanied by progressive motor alteration349. In addition, earlier in vivo studies provided evidence that a decrease in mHTT expression correlates with the recovery of behavioural and pathological alterations350.
However, it remains controversial whether full-length Huntingtin proteins or fragments are toxic to neurons351. When the number of the CAG repeats increases, translated large Huntingtin proteins tend to divide into fragments that aggregate, causing the death of striatal neurons352,353. Evidence has been gathered regarding the pathogenicity of different mHTT protein fragments. The most toxic fragment is the short HTT exon 1 protein generated by either aberrant splicing354 or proteolysis355. This protein aggregates and causes neurotoxicity in the striatal region, resulting in motor and transcriptional dysregulation in mouse models354. This is similar to the toxicity of full-length HTT, but tends to cause more severe neuronal death356.
In contrast, oligomers are more toxic than large inclusions found in cells353,357–359. Current evidence suggests that the N17, polyproline, and polyserine (polyS)-rich domains are crucial for aggregation and neuronal toxicity360–362. In contrast, some reports indicated that monomer mHTT induces apoptotic death, whereas mHTT aggregates exhibit a protective role by withholding neuron necrosis357,363. Additionally, a recent report claimed that HTT aggregates were not linked to degeneration of the cortical and striatal regions364.
Several in vitro studies have demonstrated the ability of mHTT to spread from cell to cell via tunnelling nanotubes365, endocytosis366, and prion-like behaviours to form wild-type HTT aggregate367. Importantly, the malfunction of the proteasome system on the clearance of mHTT has been observed in several in vitro and animal models368,369. Moreover, aggregated polyS and polyleucine-containing proteins exhibit propagation behaviour, which was recently observed in vivo and in vitro studies362.
CAG repeats are not only associated with neurotoxicity at the protein level, but mRNA-containing CAG repeats in the nucleus may also be neurotoxic. Mutant HTT mRNA generates clusters in the nucleus of cortical and striatal neurons in HD mice and in postmortem models370. However, it remains unclear whether the formation of mRNA clusters exerts neurotoxic or preventative effects370.
Tau dysregulation
Recent studies have reported a significant association between tau protein and HD pathogenesis371. Studies have identified tauopathy features, including misfolding, hyperphosphorylated tau aggregates, NFTs, and mHTT oligomers, in the postmortem HD brain. These features correlate with severe cognitive alterations but not with motor dysfunction372. Furthermore, several cellular-level studies have confirmed tau dysregulation in HD models, including altered splicing373 and hyperphosphorylation374,375 of tau proteins induced by mHTT aggregates. Further studies are needed to investigate the advanced protein crosstalk mechanisms between tau and mHTT proteins to understand the pathogenesis of HD.
DNA instability and oxidative stress
Recently, the focus has shifted from polyglutamine tract proteins, the product of HTT gene, to CAG repeats and, most importantly, dysfunction of the DNA repair system, which seems to correlate with much earlier and more severe pathogenesis376. This was shown in a GWAS, where uninterrupted expansion of CAG repeats in DNA, but not polyglutamine, was related to earlier and more severe pathophysiology in patients with HD377. Dysfunction of DNA repair proteins that induce CAG expansion was observed in the highly clustered DNA repair genes MSH3, MLH1, PMS1, PMS2, MLH3, and FAN. Therefore, these genes may be effective targets for the treatment376,377. Small single-stranded RNA, ASOs, restrict MSH3 expression, which lowers the rate of CAG expansion378. In addition, a recent study showed the influence on CAG expansion by inactivating FAN1 with a mutation in an HD model using human iPSCs(hiPSCs)379. This finding shows a promising approach for delaying onset and severity with genetic treatment, since FAN1 is predominantly clustered in the earlier onset of HD.
Notably, the normal HTT protein itself plays a role in transcriptional regulation by binding to support the DNA repair enzymes activated by oxidative stress380. mHTT directly affects the transcriptional processes of cells, mostly in the caudate nucleus of the brain and in some peripheral tissues381. Moreover, there is evidence on the relationship between HTT and several other transcriptional regulators, such as p53, the CREB-binding protein, CBP, and miRNAs. A recent study investigated the miRNA profiles of patients with HD and found that overexpression of hsa-mir-10b-5p inhibited GTPBP10 expression, which may correlate with the gradual loss of mitochondrial dysfunction382. Furthermore, epigenetic and chromatin-modifying factors are suggested to be involved in transcriptional dysregulation. Alterations in transcriptional regulation were observed in an early HD mouse model with downregulated epigenetic regulatory genes383.
Vulnerable medium spiny neurons and excitotoxicity
In patients with HD, neurons and other cells exhibit CAG expansion384. Notably, typically inhibitor GABAergic medium spiny neurons (MSN) of the corticostriatal tract, specifically those in the caudate putamen and globus pallidus, are preferentially degenerated385. The causes of this vulnerability and degeneration still need to be better understood. However, oxidative stress and the toxic effects of mHTT aggregates on synapse formation386,387, and excitotoxicity388, similar to other NDs, may play a role. Morphologically, MSN have many spines on their axons that substantially increase in number and size at an early stage. As the disease progresses, MSN as well as other types of neurons begin to be affected13. Patient-derived MSN neurons displayed age-associated characteristics, such as DNA damage and mitochondrial dysfunction, as well as severe death in the presence of endogenous mHTT aggregates387.
Overproduction of glutamate neurotransmitters generates excitotoxicity, followed by a cascade of alterations in neurons of the corticostriatal pathway388,389. In a proteomic analysis of late-stage HD mouse brain tissue, convincing impairment of proteins corresponding to glutamate signalling, neurotransmitter balance, synaptic transmission, glycolysis, and ATP production was revealed390. Notably, astrocytes display alterations in the glutamate-GABA-glutamine regulatory system, leading to decreased GABA production and increased glutamine release. In addition to MSN, cortical neurons exhibit decreased synaptic activity, increased excitability, and reduced complex dendritic arborisation during disease progression. However, this can be reversed in the second postnatal week and HD symptoms reappear. Treatment with ampakine CX516 stopped reappearing by stabilising glutamate transmission391.
Mitochondrial dysfunction
Various mitochondrial changes occur in neuronal and non-neuronal cells, including changes in mitochondrial structure, ATP production, motility, and biogenesis, which tend to be interdependent on the mHTT protein392–395. The interaction of mHTT with the inner and outer mitochondrial membranes causes abnormalities in Ca+2 homoeostasis and mitochondrial protein dynamics396. Changes in the structure and localisation of mitochondria have been observed in HD models, such as decreased number and fusion, increased fragmentation and fission, and accumulation in the soma with disrupted motility397,398.
Impaired brain-derived neurotrophic factor synthesis and transport
As with other NDs, BDNF impairment is another pathophysiological feature of HD, which correlates with its anatomically vulnerable site. Cortical neurons of the corticostriatal pathway provide sufficient BDNF to the MSN, which has physiologically low BDNF mRNA399. Additionally, altered BDNF formation and transmission in cortical neurons with mHTT aggregates may lead to a reduction in BDNF in MSN400. In addition, evidence suggests that mHTT affects the BDNF transcription factor of neurons401 and astrocytes402.
Neuroinflammation
Whether neuroinflammation is a causative factor or a response to other pathophysiological features in HD remains unclear. Inevitably, owing to their role in extracellular glutamate clearance, glial cells are critical for studying their influence on the highly vulnerable glutamatergic MSN. There is evidence on astrocyte and microglial activation, increased proinflammatory cytokines, and hypothesised systemic inflammation, all of which are associated with disease progression403. Many animal and human models have indicated that astrogliosis and striatal astrocyte dysfunction are considerable components of HD pathogenesis404,405. In an in vivo study, glial cells expressing mHTT induced a more severe disease phenotype and caused hyperexcitability in striatal neurons406. In the HD postmortem model, C3 expressed-A1 astrocytes contributed to the death of neurons and oligodendrocytes while losing some functions, such as synapse formation and function, and phagocytic capacity, which confirmed the neurotoxic function of astrocytes407. Importantly, A1 astrocytes were induced by IL-1A, TNF, and C1q cytokines of reactive microglia and were present in other NDs, such as AD, ALS, PD, and multiple sclerosis, which makes them critical candidates for further research. In other studies, mHTT in astrocytes caused astrogliosis and impaired BDNF release402 and glutamate uptake408, leading to striatal neuronal death when co-cultured with astrocytes409. In a recent study, it was discovered that mHTT aggregates promote proinflammatory cytokines and ROS by activating microglia410.
Neuropathogenesis-on-chips for neurodegenerative diseases
Recently, several alternative culture systems have emerged as substitutes for traditional in vitro models to provide more reliable and representative human systems. These techniques include 3D cell culture, tissue engineering with 3D bioprinting, and microfluidics411,412. Among these, microfluidics stands out for its capability to replicate critical elements of organs with precise control over biochemical and biomechanical aspects, offering a high-throughput, physiologically relevant, and cost-effective solution compared with conventional methods. The representative lung-on-chip replicates the alveolus, which is the key functional unit of lungs413. This two-chamber chip design allows externally controllable mechanical forces to mimic the alveolus movement, and direct monitoring of the interaction between epithelial and endothelial cells separated by semi-permeable membrane413. Moreover, this design has been applied to pulmonary disease414 as well as other organ units with similar mechanical conditions, such as gut-on-a-chip415 and glomerular-on-a-chip416.
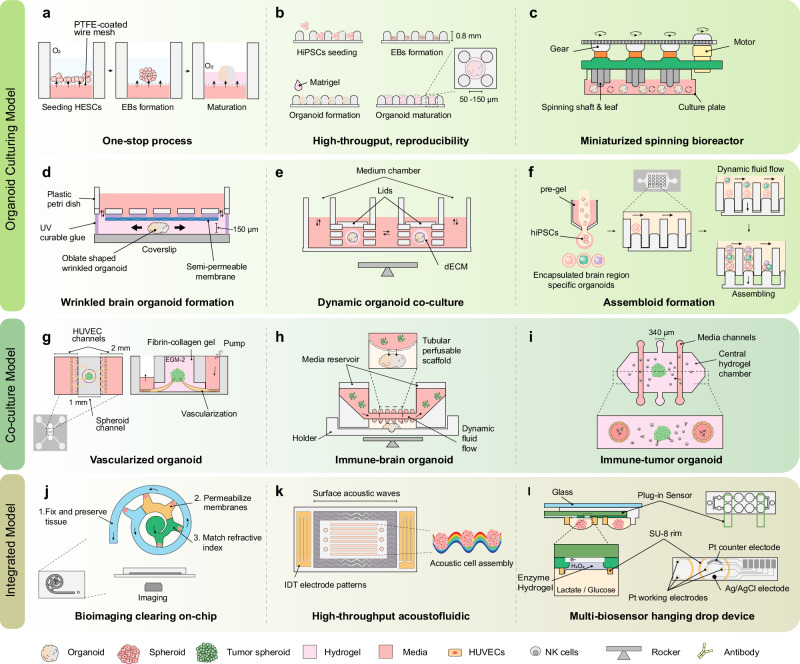
The possibilities of chip-based in vitro modelling are expanding in collaboration with advanced biological and engineering techniques, such as iPSCs, 3D culture, genome editing, hydrogels, 3D bioprinting, integrated biosensors, monitoring and analysis methods, various designs, and fabrication techniques. In this section, we highlight the benefits of using 2D and 3D microfluidic chips to model NDs. First, we provide practical biological and technological guidance for modelling NDs using various designs and introduce existing examples of NDs-on-chips. Furthermore, we present a more reliable and physiologically relevant approach: a 3D culture-on-a-chip (organoid-on-a-chip) design that can be adapted for studying NDs.
Strategy for modelling neurodegenerative diseases on a microfluidic chip
Choosing cell source
Approximately 86 billion neurons are interconnected in a highly ordered manner to form neuronal networks within the human brain417. These networks exhibit distinct activities with support from non-neuronal cells (~9.8 billion) such as astrocytes, microglia, and oligodendrocytes417. Cerebrovascular cells comprise 0.3% of all brain cells and play a crucial role in disease progression. Recent single-cell sequencing data revealed the existence of different subtypes within each cell type, based on their spatial location and pathological conditions at the molecular level, suggesting the complexity of brain cells418. Owing to the limited access to the human brain, there is a demand for the development of methods for source cells with region-specific characteristics. This is particularly important for modelling NDs because they exhibit pathogenesis specific to certain regions.
Owing to the existing animal-based in vivo and in vitro models of NDs, current knowledge was accumulated. These models offer valuable insights into cellular characteristics, disease progression, systemic and age-related changes, and the impact of the environment on the complex pathogenesis of NDs, as described in the preceding section419–422. In particular, mice and rats are widely used because of their closely identical genomes (~85%)423 and the presence of age-dependent behavioural phenotypes that can be compared with humans424. Transgenic mice that overexpressed or knock-in of human genes associated with familial NDs have been mostly developed. AD mouse models are created using transgenic, knock-in, and injection methods, focusing on Aβ pathology (PDAPP, Tg2576, and APP23), tau, and neuroinflammation (JNPL3, rTg4510, PS19, and 3xTg)419. Currently, over 170 mouse models with AD mutations mostly exhibit characteristics of EOAD. Recently, a new sporadic AD mouse model has been generated425. This model includes humanised Aβ by knock-in method without expressing the FAD mutation and exhibits age-dependent impairment of cognition, synaptic, immune response, formation of periodic-acid-schiff granules, as well as transcriptomic changes in energy, metabolism, and neuroplasticity-related genes.
However, the external approach is widely used for NDs, using various chemicals and recombinant pathological proteins. In patients with PD, drugs (reserpine and haloperidol), neurotoxins (6-OHDA, MPTP, and lipopolysaccharide), and agrochemicals (rotetone, paraquat, and manab) induce PD-like characteristics, such as motor deficits, mitochondrial dysfunction, neuroinflammation, and nigrostriatal dopaminergic cell death420. Numerous studies have used exogenous recombinant human and mouse proteins to characterise misfolded protein pathologies both in vitro362 and in vivo426. α-syn preformed fibrils generated from monomeric recombinant α-syn can induce hyperphosphorylation of endogenous α-syn and enhance the spread in vitro427 and in vivo428.
Non-mammalian species, such as Caenorhabditis elegans, Drosophila, and yeasts are simpler and more cost-effective alternative models429. Cytotoxicity, aggregation, and propagation of the α-syn protein has been investigated in these models with overexpressed SNCA regarding PD. Despite the genetic variations in humans, these short-lifespan species are ideal for investigating the effects of ageing on ND pathogenesis429.
Human embryonic stem cells (hESCs) have also been used to model NDs and to provide genetic and molecular information relevant to humans. Human neural progenitor cells (hNPCs) derived from hESCs can differentiate into several subtypes, such as cholinergic, dopaminergic, serotonergic, noradrenergic, and medium spiny striatal neurons, expressing neuron-specific markers430, among which H9 and H1 are dominantly used in research431. However, ethical concerns surrounding hESCs led to the need for alternative solutions. Methods to induce neural differentiation, such as neural induction and direct reprogramming, have been successfully developed since the discovery of iPSCs from somatic cells with transfection factors432,433. Current differentiation protocols utilising genetic or chemical methods have demonstrated promising results in the generation of diverse types of neural cells, including cortical, basal forebrain, dopaminergic, cholinergic, glutamatergic, GABAergic, MSN, and functional motor neurons. Additionally, these protocols can generate other types of brain cells, such as oligodendrocytes, astrocytes, pericytes, vascular endothelial cells, and microglia434,435. HiPSCs have been extensively used in PD studies owing to the availability of current protocols that mainly focus on the induction of dopaminergic neurons. However, for further applications, there is a need to improve and standardise the derivation methods436.
Familial and sporadic patient-derived iPSCs are another candidate cell source. This approach allows the generation of human-relevant pathological phenotypes that cannot be achieved using other animal models. iPSC lines from patients with PD with SNCA, LRRK2 Parkin, CHCCHD2, PARK2, and PINK1 mutations exhibit important phenotypes including mitochondrial dysfunction, oxidative stress, and α-syn accumulation in induced midbrain-like dopaminergic neurons437,438.
High reliance on genetically modified models often overlooks sporadic cases and fails to fully consider other important factors, such as ageing and patient-specific traits. Additionally, there are inevitable limitations due to the differences between human and animal brains at the cellular and tissue levels20,21. While genetically engineered animal-derived cells continue to play a fundamental role in in vitro studies of NDs439, human patient-derived cells show promise as reliable in vitro models for basic scientific research and the development of personalised medicine.
Choosing microfluidic chip design
First chip application on brain research began with a notable compartmentalised in vitro system referred to as the ‘Campenot chamber’440. This device has two fluidically separated chambers and is used to study the effects of nerve growth factors on axonal growth. Subsequently, researchers have converted the complex characteristics of the brain into simple and miniaturised systems by mimicking diverse levels of units of the brain, including axons, neuron-glia cells, the BBB, and the neurovascular-unit22.
Typical microfluidic chips consist of two or more compartments (chambers) for cell co-culture. These compartments were connected by microchannels, porous membranes, and phase guides. This allows for direct or indirect interactions between homogeneous (neural circuits) or heterogeneous (neuron-glia) cell populations which are loaded into fluidically isolated chambers.
The earliest design of microfluidic chips for the CNS involved the separation of a neuronal soma from its neurites using microchannels. This design allowed for directional control of neurite growth36. By incorporating multiple compartments for different neuronal subpopulations, the connectivity and size of the circuits can be regulated, enabling the modelling of neural circuits in a highly ordered manner. Current neuronal chips have been designed with compartments positioned in various geometries441 and different diameters and numbers442,443, microchannels with patterned shape444,445, and controlled fluidic flow446. These features effectively manipulate neurite growth and can create both indirect and direct as well as asymmetric and symmetric neuronal connections. Moreover, valves can be established on the chip to act as a controllable barrier between compartments using external factors, such as air, oil, gel, liquid, temperature, or pressure447–449. Regulated fluid flow can be achieved using extra pump systems and passive hydrostatic pressure, which applies controllable shear stress to the cells. This allows for a gradient of chemicals with varying concentrations throughout the cell compartments, which is useful for disease modelling. In ND research, gradients-on-a-chip used for investigation of pathological protein dynamics and toxicity, synaptic and neurochemical interactions, vesicle transportation, and neurite ingrowth450,451.
Another approach involves the use of a porous membrane-based chip that acts as an interface between two or more compartments. This chip enables indirect interactions mediated by soluble chemicals and direct physical contact. The size, position, and number of the pores were customised according to the research objectives. The thickness of the membrane was adjusted to facilitate the experimental interactions. Based on this design, BBB mimicry has been achieved on chips452,453. It mimics the walls of blood vessels by culturing a monolayer of endothelial cells on a porous membrane, facilitating soluble interactions with brain chambers452. In another example, a porous membrane was placed horizontally between two chambers to culture vascular and brain cells separately453. This design enables more dimensional interactions than a planar culture.
Monitoring and analysing steps
Microfluidic chips offer several advantages over conventional models for monitoring and analysis. These include allowing optical clarity for real-time monitoring under various chemical or physical cues, gas permeability, and continuous flow to balance nutrients or waste for longer viability; fluidic isolation for multi-omics analysis of soluble biomarkers from distinct cell populations; and microengineering to integrate biochemical, electrochemical, and electrophysiological biosensors.
Most microfluidic chips are composed of clear optical materials (polystyrene and polydimethylsiloxane). Additionally, most designs include a pre-determined image acquisition region within their structures, which enables consistent and accurate monitoring. Specific biomarkers can be detected directly from the chip using fluorescence and confocal microscopy by introducing a very small amount of staining dyes and antibodies through the fluidic channel inlet. However, issues of contamination, accumulated cell debris, and air bubbles in tiny channels can pose challenges in maintaining cultures and imaging. These can be prevented by carefully processing the fabrication steps, using trapped air bubble methods, or force-out procedures454.
The integration of various types of biosensors on microfluidic chips enables the continual measurement of the biochemical and electrophysical properties of cell cultures. Various parameters, such as oxygen, pH, and soluble metabolite levels can be analysed using immune- and enzyme-based sensors, allowing the evaluation of cellular survival and metabolism in response to induced pathophysiological conditions454. Moreover, the incorporation of automated electrochemical biosensors in microfluidic chips enables the continuous monitoring and analysis of dynamic cellular activity455.
In the case of the brain-on-a-chip, a transendothelial electrical resistance system, integrating electrodes38,456, and planar electrophysical multielectrode arrays (MEA)457 are often incorporated to measure the integrity and permeability of the BBB and assess neuronal electrical signals, respectively. Off-chip analysis tools, such as mass spectroscopy, liquid chromatography, and biochemical assays, such as ELISA, can be utilised to analyse small amounts of samples isolated from microfluidic chip cultures. Alternatively, whole-cell culture samples can be lysed and collected for further genomic, transcriptomic, and proteomic analyses. In certain cases, rather than opting for the commonly used closed approach, an open-chip design offers various benefits. The open chambers provide easier exposure to additional chemical components, imaging, transfer for analysis, and sample collection. Notably, most open designs tend to lose their dynamic fluid flow, which can limit the controllability and relevance. However, the benefits of using open chips have also been reported in recent studies. Salman et al. 458 developed an open chip for modelling the human brain endothelial microvessel. It enables the unidirectional exchange of media, gas, and nutrients within the collagen gel through a cylindrical hollow lumen. The top opening increases oxygen availability. Furthermore, it simplifies the harvesting of cells and exosomes for subsequent transcriptomic and proteomic analyses and allows for standard and advanced imaging by carefully optimising the dimensions and geometry.
Application in modelling neurodegenerative disease pathogenesis
Based on this research objective, relevant anatomical and physiological units, including the neuromuscular junction (NMJ), corticostriatal pathway, substantia nigra, BBB, glymphatic system, neurovascular unit (NVU), and gut-brain axis, can be replicated using matched chip designs. However, most existing ND studies using microfluidic chips still lack physiological relevance owing to the choice of cell source and designs under 2D and static conditions. These drawbacks limit the wide applicability of microfluidic chips. The limitations of 2D systems include their inability to generate complex interactions, waste removal, and oxygen diffusion. A planar environment reduces morphology, proliferation, differentiation ability, cellular signalling, and gene expression which affects the in vivo relevance of hiPSCs459.
Conversely, the implementation of a 3D extracellular matrix (ECM) gel provides mechanostructural and biochemical cues to cells which have improved relevance in cellular morphology, migration behaviour, cellular signalling, interactions, and substantial gene expressions460. Moreover, it allows time-dependent differentiation and neurodegenerative changes461. The important factor of NDs, ageing, alters properties of the brain ECM460. Aβ induced accumulation and aggregation of pTau has been observed in 3D in vitro condition462,463. This evidence indicates the importance of ECM in modelling NDs.
To characterise a 3D microenvironment that provides not only mechanical, but also biochemical cues, the properties of the ECM, such as pore size, solute retention, attachment motifs, biodegradability, and stiffness, must be considered460. Alterations in these properties can affect the behaviours of various brain cells as well as disease pathogenesis464,465. During AD, owing to altered functional connectivity, vulnerable regions of the brain exhibit altered stiffness, which is correlated with structural changes466. Furthermore, evidence shows that ECM stiffness directly affect the aggregation and propagation of pathogenic molecules, such as Aβ and tau oligomers460.
Various hydrogels have been integrated into microfluidic chips to demonstrate direct interactions while creating a more physiologically relevant brain tissue-like environment467,468. One example of this design utilises a phase guide in the central channel to confine ECM gel-embedded cells from dispersing into side flow channels filled with medium, while still allowing interaction without a barrier461,468. In the next section, we provide an overview of the use of custom and commercial microfluidic chips to model NDs pathogenesis.
Existing examples of neurodegenerative disease-on-chips
Alzheimer’s disease-on-chips
Existing studies using microfluidic chips have investigated substantial AD pathogenesis, including Aβ and tauopathy, mitochondrial dysfunction, and neuroinflammation, providing valuable new insights that were not observed using conventional culture methods (Table 1). In particular, numerous neurons-on-a-chip are used for tau propagation469–473, and Aβ toxicity studies474–476. Neurons-on-a-chip allows the separation of the soma and neurites, facilitating the real-time visualisation of proteinopathy. Some groups have found evidence of tau transfer via exosomes110 and synapses477,478 using 2–3 compartment chips, suggesting prion-like propagation of tau.
Table 1.
Summary of Alzheimer’s disease-on-chips
| Alzheimer’s disease-on-chips | ||||||
|---|---|---|---|---|---|---|
| Neuropathogenesis | Chip model | Ref.Fig. | ||||
| Targeted pathogenesis | Cell source | Modelling approach | Features | Type | Design | |
| Neuroinflammation | hNeu/Ast/MG | FAD(670N/M671L/V717I) Endo Aβ/pTau | 3D/S/C | Circular migration chip | 2CHRs/MCs | 482 Fig. 2b |
| BBB dysfunction | hNeu/Ast/Peri/EC | APP/PSEN1 Endo Aβ | 3D/S/C | BBB-on-a-chip | 5MCs/Bs | 452 Fig. 2c |
| Dysfunction of AQP4 | hAst/HBMEC | Exo Aβ/LPS | 3D/D/C | Glymphatics-on-a-chip | Central CHRs/2MCs | 486 |
| Immune response to Aβ | hNEUT | Exo Aβ mono | 2D/S/C/G | Chemotaxis chip | 3CHRs/3Vs/MCs | 483 |
| Immune response to Aβ | hMG | Exo Aβ mono, oli | 2D/S/O/G | Circular chemotaxis chip | 2CHRs/MCs | 484 |
| Aβ toxicity | pCoNeu | Exo Aβ | 3D/D/C | Neurospheroid chip | 50 wells/ OP | 485 Fig. 2d |
| Aβ toxicity | pCoNeu | Exo Aβ | 2D/D/C/G | Spatial G chip | MCs/OP | 451 Fig. 2a |
| Aβ toxicity | pHiNeu/MG | Exo Aβ oli, fib | 2D/D/O | Circular co-culture chip | 4CHRs/MCs | 479 |
| Aβ toxicity | pCoNeu | Exo Aβ/Glutamate | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 474 |
| Aβ toxicity | pCoNeu | Exo Aβ oli | 2D/S/O | Axon-on-a-chip | 2CHRs/MCs | 475 |
| Aβ toxicity transfer | pCoNeu | Exo Aβ mono | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 574 |
| Aβ effect on tauopathy | pCoNeu/pHiNeu | Exo Aβ/pTau | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 476 |
| Tau transfer | pHiNeu | hTau(P301L) | 2D/S/C | Neuronal circuit chip | 3CHRs/MCs | 478 |
| Tau transfer by exosome | pHiNeu | H GFP-tau exosomes | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 110 |
| Tau transfer, aggregation | pCoNeu | hTau | 2D/S/C | Axon-on-a chip | 3CHRs/MCs | 575 |
| Tau transfer | pCoNeu/PHiNeu | hTau(P301L) | 2D/S/C | Axon-on-a-chip | 3CHRs/MCs | 477 |
| Tau transfer | pCo/HiNeu | hTau | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 469 |
| Tau transfer | pCoNeu | hTau(P301L) | 2D/S/C | Axon-on-a-chip | 3CHRs/MCs | 470 |
| Tau transfer | pCoNeu | hTau | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 471 |
| Tau transfer | pCoNeu | G of OA/pTau | 2D/D/C/G | Axon-on-a- chip | 2CHRs/main MC/24MCs | 473 |
| Tau transfer | hiPSCs Neu | Exo Cy5-Tau mono, oli | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 472 |
AQP4 aquaporin, Ast astrocyte, Aβ amyloid beta, B barrier, BBB blood-brain barrier, C closed, CHR chamber, Cy5 cyanine 5, MC microchannels, D dynamic, EC endothelial cell, endo endogenous, exo exogenous, fib fibril, G gradient, GFP green fluorescent protein, H human, HBMEC human brain microvascular endothelial cell, hiPSC human-induced pluripotent stem cell, LPS lipopolysaccharide, MG microglia, NEUT neutrophil, O open, OA okadaic acid, mono monomer, oli oligomers, OP osmotic pump, pCoNeu primary cortical neuron, Peri pericyte, pHiNeu primary hippocampal neuron, pTau hyperphosphorylation of tau, S static, V valve.
Aβ-related pathology is also being investigated mainly using microfluidic chips consisting of two to four compartments. Substantial evidence of Aβ toxicity has been gathered, including the induction of hyperphosphorylation of tau proteins, degeneration of axons474, and disruption of BDNF retrograde signalling of axons475. Some studies have cultured neurons as well as co-cultured microglia with primary hippocampal neurons in the presence of exogenous Aβ proteins. Ruiz et al. 479 studied the glial clearance of different forms of Aβ, in a 4-compartment circular open microfluidic system. Each open compartment allowed for four parallel experiments and easy seeding co-cultures of different populations, such as microglia and neurons, with various cell densities under gravity-induced dynamic conditions. Aβ oligomers have a higher toxicity to hippocampal neurons compared to that of fibrils. However, only the microglia changes in Aβ exposure; the neuronal aspect is not included. Choi et al. 451 developed a microfluidic chip that can generate dynamic fluid conditions for gradient formation of Aβ aggregates, similar to in vivo studies, by integrating an osmotic pump to induce slow laminar flow (0.25 µL/min), similar to the interstitial flow rate (Fig. 2a). Under dynamic conditions, the cultured primary rat neurons showed a higher level of viability (7.5%). However, when exposed to Aβ oligomers, the neurons lost their synaptic activity and eventually experienced apoptosis. In addition, the fibril form of Aβ was less toxic to neurons than oligomers.
Fig. 2. Examples of microfluidic chips for NDs modelling.
AD - a Gradient chip with interstitial flow. The effect of Aβ oligomers on neurons was observed in gradient concentration condition451. b 3D static neuroinflammation-on-a-chip. Microglia migrated in response to inflammatory cytokines across microchannels to the centre chamber where AD neurons and astrocytes were cultured482. c BBB-on-a-chip. Formation of amyloid plaques, neurofibrillary tangles, Aβ, and increased permeability of brain ECs, were fully recapitulated in a 3D environment452. d Dynamic neurospheroid-on-a-chip. A flow of exogenous Aβ caused by an osmotic pump resulted in axonal degeneration and cell death485. PD- e Navie and GFP-tagged α-syn expressing neuroglioma cells were co-cultured to monitor the spreading of α-syn494. f Gradient chip. It modulated the intracellular α-syn expression of singularly trapped yeasts in system with galactose gradient450. g Dopaminergic neurons-on-a-chip. In a 3D condition, it recapitulated mitochondrial abnormalities and degeneration of dopaminergic neurons with the PD-related mutation461,577. h Substantia nigra and vascular barrier chip. hiPSCs-derived midbrain dopaminergic neurons, primary glia cells and a brain microvascular EC line were separated by a porous membrane to assess BBB on-a-chip dysfunction, progressive neuronal loss, neuroinflammation, and astrogliosis453. ALS- i 3-chamber-chip allowed metabolic interaction between SOD1-mutated astrocytes and cortical neurons through microchannels in a glutamate gradient condition502. j ALS chip. Applying chemotactic and volumetric gradients successfully formed interactions between FUS-mutated motor neurons and mesangioblast-derived myotubes through microchannels503. k An open compartmentalized NMJ device was used to co-culture optogenetic motor neuron and SOD1-mutated astrocytes as a spheroid to assess the muscle denervation pathology of ALS505. l TDP-43 mutated motor neuron spheroid and muscle fibres were co-cultured in a 3D condition between two separate reservoirs, which recapitulate ALS pathologies506. HD- m Impaired BDNF transport due to mHTT expression through cortical axons resulted in striatal neurons degeneration509. n The synaptic connection between cortical axons and striatal dendrites was formed by the different lengths of microchannels and a separate synaptic channel, assessed the early progression pathology507. AD Alzheimer’s disease, ALS amyotrophic lateral sclerosis, BBB blood-brain barrier, EC endothelial cell, HD Huntington’s disease, hiPSC human-induced pluripotent stem cell, NMJ neuromuscular junction, PD Parkinson’s disease.
Several on-chips studies have been conducted using human neural progenitor cells transduced with familial AD-specific mutated genes, such as FAD 670N/M671L/V717, APP, and PSEN1. In AD pathogenesis, astrocytes play an essential role in the progression of AD, with “a double-edged sword” effect on neuroinflammation, owing to their neuroprotective and neurotoxic effects138,480,481. However, the dynamics of astrocytes and pathogenic proteins involved in neuroinflammation are not well understood. Park et al. 482 designed a simple circular chip that has two chambers (central and radial) which allow microglia migration towards to the centre where AD neurons derived chemokines and cytokines (TNFα, IL-1, and IL-6) are located (Fig. 2b). This design created a gradient of Aβ and neuroinflammatory molecules in a 3D environment and showed great potential for further molecular studies, including co-culture with glial cells. Additionally, numerous studies have been conducted on the migration of human neutrophils483 and microglia484 towards exogenous Aβ monomers and oligomers with chemotaxis chips in 2D conditions.
By incorporating various types of cells, a molecular understanding of the neuroinflammatory phenomenon can be obtained using microfluidic chips. Recently, research was focused on well-defined brain features, such as the BBB and the neurovascular unit, in AD neuropathogenesis. Shin et al. 452 developed a BBB-on-a-chip comprising a cell chamber for AD-mutated hNPCs and a tube-shaped endothelial cell chamber separated by a central barrier. As shown in Fig. 2c, the accumulation of Aβ and pTau proteins were observed in the brain channel. Upon removal of the barrier by Matrigel application, the proteins are transferred to endothelial cells, where they are deposited in the cytoplasm. They cause damage to tight junctions, indicating BBB disruption, which is observed in the early stages of AD. These promising results suggest that BBB-on-a-chip can be used as a drug-screening tool in the future.
A microfluidic chip composed of 50 cylindrical wells was developed to culture neurospheroids and integrate them with an osmotic micropump to provide a continuous medium supply similar to the interstitial flow rate485 (Fig. 2d). Medium with exogenous Aβ flows over neurospheroids continuously, causing axonal degeneration, synaptic dysfunction, and neuronal death. The spherical wells of this chip generate more homogeneous spheroids, and continuous flow leads to increased viability, which directly affects cell-cell interactions. This chip demonstrated that integrating microfluidic chips with 3D cultures can create more in vivo-like conditions that are unavailable in other conventional systems.
Unique characteristics of the glymphatic clearance system can be explored with microfluidic chips by allowing dynamic glymphatic flow in the space between astrocytes and endothelial cell co-culture163. Lee et al. 486 recently replicated the gliovascular unit using a microfluidic chip in AD studies. The glymphatics-on-a-chip design allows for the co-culturing of human astrocytes and blood vessels in 3D gel-embedded parallel microchannels. LPS-induced neuroinflammation causes altered fluid drainage and Aβ clearance through matrisome- and cell-dependent fluid transport pathways.
Parkinson’s disease on-chips
The majority of existing PD-on-a-chip studies have focused on recapitulating α-syn related pathogenesis (Table 2). However, only a few have used dopaminergic neurons to increase the relevance of these models. The first on-a-chip PD model was developed using simple single-linear microchannels with a gradient of neurotoxins to model the apoptosis of dopaminergic neurons caused by 6-OHDA. Depending on the concentration, this neurotoxin can induce apoptosis or necrosis487. By generating a back-and-forth continuous flow, the model achieved low and high concentration gradients of 6-OHDA, which resulted in PC12 cell line apoptosis by oxidatively damaging the mitochondria.
Table 2.
Summary of Parkinson’s disease-on-chips
| Parkinson’s disease-on-chips | ||||||
|---|---|---|---|---|---|---|
| Neuropathogenesis | Chip model | Ref Fig | ||||
| Targeted pathogenesis | Cell source | Modelling approach | Features | Type | Design | |
| Inflammation/ BBB disruption | ptPSCsNeu/Peri/Ast | LRRK2-G2019S | 3D/S/C | BBB-on-a-chip | 3CHRs/B | 497 |
| BBB disruption | hiPSCs DANeu/pAst/MG/Peri/HBMECs | Exo h α-syn mono, fib | 3D/D/C | BBB-on-a-chip | 2CHRs/B | 453 Fig. 2h |
| Mitochondrial abnormality/ α-syn toxicity | HNESCs DANeu | LRRK2-G2019S | 3D/S/C | Co-culture chip | CHRs/B | 461 Fig. 2g |
| α-syn transfer, toxicity | H4 cell line | Exo α-syn-GFP | 2D/D/C | Co-culture chip | 2CHRs/1MC/V | 494 Fig. 2e |
| Mitochondrial damage | PC12 cell line | 6-OHDA | 2D/D/C/G | G chip | Linear MCs | 487 |
| α-syn inclusion | S. cerevisiae | GFP-α-syn SNCA(A53T) | 2D/S/C | Automated chip | CHRs/Ts | 496 |
| α-syn inclusion | S. cerevisiae | GFP-α-syn mono,fib | 2D/S/C/G | G generator chip | CHR/Ts | 450 Fig. 2f |
| α-syn transfer | hiPSCs CoNeu | Exo α-syn mono, fib | 2D/S/C | Axon-on-a-chip | 2CHRs/ asymmetric MCs | 495 |
| α-syn transfer | pHiNeu | Exo α-syn fib | 2D/S/C | Axon-on-a-chip | 3CHRs/MCs | 576 |
| α-syn transfer | pHiNeu | Exo α-syn fib | 2D/S/C | Neuronal network chip | 2-6IC/Heart-arrow MCs | 441 |
| α-syn transfer | pCoNeu | Exo α-syn fibrils | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 488 |
| α-syn transfer | pCoNeu | Exo α-syn fib | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 489 |
| Mitochondrial abnormalities | pCoNeu | Rotetone | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 490 |
| Mitochondrial abnormalities | ptPSCsNeu | SNCA(E46K,E57K) α-syn oli | 2D/S/C | Axon-on-a-chip | 2CHRs/MCs | 492 |
6-OHDA 6-hydroxydopamine, Ast astrocyte, B barrier, BBB blood-brain barrier, C closed, CHR chamber, D dynamic, DA Neu dopaminergic neuron, ECM extracellular matrix, exo exogenous, fib fibril, G gradient, GFP green fluorescent protein, hBMEC human brain microvascular endothelial cell, hiPSC human-induced pluripotent stem cell, hNESC human neuroepithelial stem cell, IC interlocking units, MC microchannel, MG microglia, mono monomer, oli oligomer, pCoNeu primary cortical neuron, Peri pericyte, pHiNeu primary hippocampal neuron, ptPSCs patient pluripotent stem cells, S static, T trap, V valve, α-syn alpha synuclein.
The microfluidic chip with two chambers connected through microchannels is the most common method for investigating α-syn inclusion and propagation. This chip design allows for clear observation of the process of α-syn fibril uptake, and its anterograde or retrograde transport through axons to other neurons by exposing neurons to exogenous α-syn488,489. Van Laar et al. 490 used the same microfluidic chip to emulate the dynamics of mitochondrial alterations observed in the earlier stages of PD. In PD, neural degeneration begins in long, less myelinated and distant axons491. This was confirmed by their response to rotenone; mitochondrial biogenesis increased in distal axons, followed by cell bodies and dendrites. Prots et al. 492 also used the same chip to investigate the toxicity of α-syn oligomers in neurons derived from iPSCs from patients with PD with mutant α-syn (E46K and E57K). They observed α-syn oligomerization led to the impairment of anterograde mitochondrial transport through axons, and structural synaptic loss. This result confirmed the role of mutant α-syn oligomers in the early pathogenesis of PD. Additionally, one group cultured preformed primary neurons with exogenous α-syn fibrils for an α-syn transfer study based on evidence that exogenous recombinant α-syn can promote misfolding of physiological α-syn, leading to the formation of LB-like aggregates in neurons493. In this experiment, a three-chamber chip was used to demonstrate the toxic effect of α-syn aggregation and uptake by inhibiting regulator protein of protein trafficking. They also investigated how endogenously released α-syn are toxic to separately cultured neurons in a paracrine way.
An example of a customised chip with micro-valves that allows control fluid dynamics has been developed to demonstrate α-syn uptake and spread494 (Fig. 2e). By co-culturing cells expressing GFP-tagged α-syn and normal cells in two chambers connected through microchannels, the researchers were able to observe α-syn successfully spreading throughout two different populations. Another group reconstructed a human corticocortical neuronal network by culturing hiPSCs on a microfluidic chip495. In this design, neurons in the two-cell chambers interact with each other in a narrow central chamber by projecting their axons through asymmetric microchannels. They indicated that prion-like propagation of physiological α-syn occurs between human neurons.
Two-, and three-, or multichambered chips recapitulate neuron to neuron α-syn spread. A microfluidic chip with two to six interlocked chambers that can be adjusted in several different ways successfully recapitulated the complex neuronal network without barriers and demonstrated phosphorylated α-syn propagation441. A well-known design of microfluidics, the gradient-generator chip, can provide a reliable chemical concentration by creating nine different doses in microchannels450 (Fig. 2f). They induced intracellular α-syn with exogenous α-syn monomer and fibrils inside Saccharomyces cerevisiae which were trapped individually in the chip with different chemical gradient environments. Recently, another group cultured live yeast on a microfluidic chip integrated with automated feedback control, which can regulate the concentration of α-syn aggregation in S. cerevisiae expressing A53T α-syn combined with GAL-inducible promoter with GFP in real-time496. Over a galactose gradient, they determined the threshold of α-syn mutant protein expression that induces the formation of inclusions and their clearance, which is strictly based on autophagy at the single-cell level. This approach can be applied to other quantitative studies of human neuronal cell lines.
As dopaminergic neurons are selectively damaged during the progression of PD, Bolognin et al. 461 studied the effect of the LRRK2-G2019S mutation on dopaminergic neurons (Fig. 2g). In this experiment, they used a “Mimetas” culture chip with a phase guide that allows culturing cells in 3D ECM conditions. The dopaminergic neurons successfully formed structurally relevant networks on the chip. Notably, substantial changes due to the PD mutation, including neurodegeneration and changes in the number and complexity of mitochondria, were only observed after six weeks in the 3D condition. This result demonstrates the importance of mechanical cues in demonstrating physiologically relevant pathogenesis.
Considering the significance of the BBB in PD pathogenesis, one group has successfully demonstrated the BBB dysfunction caused by the toxicity of exogenous human α-syn fibrils453 using a 3D microfluidic chip (Fig. 2h). The design of the chip allows the culturing of brain and vascular cells in tissue-specific ECM cocktails in two separate channels connected through a horizontal semi-permeable membrane. This mimicry of BBB has exhibited formation of intracellular inclusion by internalised α-syn, which leads to a progressive loss of dopaminergic neurons, mitochondrial dysfunction, and neuroinflammation associated with glia cell activation. Brain endothelial cells were cultured in support of pericytes, microglia, and astrocytes, therefore, the BBB was formed with higher maintenance and functionality. Moreover, dopaminergic neurons, the main cells of the substantia nigra, stably secrete dopamine, indicating that functional maturation is not considerable in conventional cultures.
More recently, de Rus Jacquet explored astrocytic activity in both BBB dysfunction and inflammation during PD pathogenesis497. The significance of the chip design is that the ECM gel (collagen) acts as a barrier between the brain vascular channels which allows direct interaction. The phenotypes of vascular changes in vitro which were observed in sporadic PD and human postmortem models were successfully demonstrated and vascular changes depended on LRRK2-G2019S-expressing astrocytes, which exhibit proinflammatory activity. In this model, LRRK2-G2019S-expressing patient-derived astrocytes failed to support BBB integrity and inflammation, particularly angiogenesis, as confirmed by RNA-seq data. Moreover, mutated astrocytes regulated BBB integrity via the MEK1/2 signalling pathway, providing evidence for the importance of astrocytes in PD pathogenesis.
Amyotrophic lateral sclerosis on-chips
Numerous rodent models have been developed to investigate ALS based on newly discovered genetic factors. These models have become indispensable tools for investigating disease mechanisms, although they mostly reflect familial case of ALS498. In vitro modelling, mostly using cells sourced from transgenic mice with familial gene mutations, such as TARDBP, SOD1, and C9ORF72, has also revealed important dysfunctions of cellular processes related to ALS which are also shared with sporadic ALS498. These dysfunctions appeared in protein transport, homoeostasis, RNA transcripts, and mitochondria499. Although such models provide defined mutation-related information, a specific model for sporadic ALS has not been developed421. In ALS, several connected mechanisms lead to motor neuron death, causing diverse phenotypes that are difficult to model. Mutations in RNA-binding proteins, such as TDP-43, FUS, NRPNA1, MATR3, and SETX, alter RNA metabolism500. Therefore, a unified model that reflects a patient’s phenotype is required.
Previous ALS studies have investigated toxic protein aggregation and propagation using simple 2-chamber microfluidic chips (Table 3). Westergard et al. 501 determined that C9ORF72-linked DPRs spread between cortical neurons via exosomes and non-exosomes via axonal anterograde and retrograde transmissions. This study once again proved that the spread of toxic proteins is a common underlying mechanism of progressive NDs.
Table 3.
Summary of the amyotrophic lateral sclerosis-on-chips
| Amyotrophic lateral sclerosis-on-chips | ||||||
|---|---|---|---|---|---|---|
| Neuropathogenesis | Chip model | Ref. Fig. | ||||
| Targeted pathogenesis | Cell source | Modelling approach | Features | Type | Design | |
| NMJ impairment | hiPSCs MN/hMesangioblast | FUS(P525L, R521H) | 3D/S/C/G | NMJ-on-a-chip | 2CHRs/MCs | 503 Fig. 2j |
| NMJ impairment | hiPSCs MN/pMyoblast | C9ORF72 | 2D/S/C | Radial chip | 2CHRs/MCs | 504 |
| NMJ impairment | ESCs MN/Ast/pMyofibres | SOD1(G93A) | 3D/S/O | NMJ-on-a-chip | 3CHRs/MCs/2 anchors | 505 Fig. 2k |
| NMJ impairment | ptMN hiPSCs Myoblasts | TDP-43(G298S) | 3D/S/C | NMJ-on-a-chip | 2CHRs/pillars | 506 Fig. 2l |
| NMJ impairment | pCoNeu/Ast | SOD1(G93A) | 2D/D/C/G | NMJ-on-a- chip | 2CHRs/main MC/MCs | 502 Fig. 2i |
Ast astrocyte, C close, CHR chamber, D dynamic, ESC embryonic stem cell, G gradient, h human, hiPSC human-induced pluripotent stem cell, MC microchannel, MN motor neuron, NMJ neuromuscular junction, O open, P primary, pCoNeu primary cortical neuron, Pt patient, S static.
Similar to other NDs, neuroinflammation is a pathogenic feature of ALS. Accordingly, astrocytes were simulated using microfluidic culture systems. In one of the earliest studies, Kunze et al. 502 co-cultured primary cortical neurons and SOD1-mutated astrocytes without direct contact to demonstrate the effect of astrocytes on neuronal death under a glutamate gradient condition (Fig. 2i). They observed neuronal death and decreased synaptic protein expression in neurons in the presence of astrocytes from patients with ALS, indicating SOD1 toxicity.
Considering that NMJ degeneration is the main hallmark of ALS, the co-culture of myocytes and motor neurons has been mostly recapitulated for ALS studies, taking advantage of the compartmentalised elements of microfluidic chips. Several groups have used different types of culture systems, including monolayer cultures and spheroids derived from genetically modified ESCs and hiPSCs503–506. Radial-shaped two-chamber microfluidic chips demonstrated mitochondrial toxicity, protein synthesis dysfunction, and axonal phosphorylated TDP-43 deposition associated with the activity of C9ORF72-modified hiPSCs neurons504. Another research group successfully established NMJ impairment in ALS by culturing hiPSC-derived FUS-mutated neurons and human primary mesangioblast503 (Fig. 2j). They generated NMJ by creating a volumetric gradient of BDNF, GDNF, and CNTF in microfluidic chambers to induce axonal growth in the muscle cell chambers through a microchannel. They found that FUS neurons exhibited a lower axonal growth rate, leading to fewer NMJ. However, an HDAC6 inhibitor reversed axonal growth in FUS neurons. Machado et al. 505 developed an open microfabricated chip composed of three compartments, which enabled the analysis of interactions between spheroids containing ESC-derived neurons and SOD1 astrocytes as well as primary myofibres (Fig. 2k). They investigated how astrocytes with mutated SOD1 induce neuronal loss, followed by myofibril dysfunction. However, these advanced models lose their physiological relevance when primary rodent myofibres are used, despite the use of human ALS-specific motor neurons. An exceptional example of such a model is co-cultured patient-derived motor neuron spheroids and hiPSC-derived skeletal myoblasts on a custom-made 3D microfluidic chip506 (Fig. 2l). Once skeletal muscle fibres are formed, motor spheroids are introduced into a 3D gel to form NMJ. Moreover, optogenetic technology has been applied to motor neuron spheroids by transfecting them with a light-sensitive ion channel (channelrhodopsin-2), allowing the non-invasive control of motor neuronal activity. The chip design also includes several important features, such as a separate reservoir allowing for the co-culturing of cells. The region for neurite elongation was designed to be longer to achieve a physiologically relevant neurite length. The pillars in these designs provide functions, such as limiting the size of the spheroid, preventing core necrosis, and measuring the strength of muscle contraction by changes in pillar displacement. By generating NMJ, they successfully recapitulated TDP-43 aggregation, neurotoxicity, and muscle atrophy due to synaptic loss.
Huntington’s disease on-chips
Compared with other NDs, there have been relatively few microfluidic chip applications for HD (Table 4). These chips replicate the corticostriatal network, a vulnerable region in HD pathophysiology, with two-406 and three-chambers507,508. To understand the toxic effects of mHTT on the corticostriatal neural circuit, on-a-chip studies commonly use transgenic mouse neurons. One group investigated how mHTT reduced the cortical axonal transport of BDNF and detected a positive effect of the TCP-1 ring complex (TRiC) on decreasing mHTT levels509 (Fig. 2m). Chip contains two separate chambers for cortical and striatal neurons, connected by microchannels. Design allows the observation of striatal neuron degeneration, and decreased synapses. In their three-chamber chip (presynaptic, synaptic, and postsynaptic), Virlogeux et al. 508 established laminin and poly-D-lysine gradients from presynaptic and synaptic chambers. By culturing mHTT-transduced primary neurons, deficiencies in synapses, BDNF, and glutamatergic trafficking, and a decline in mitochondrial transport and vesicle synthesis were observed. Later, this group used the chip in a drug target study by recapitulating the restoration of Huntingtin-dependent BDNF vesicle transport by activating palmitoylation510. They observed improved axonal BDNF trafficking, synapse formation, and viability in the chip channels. Based on an in vivo study of HD knock-in mice that showed improvements in motor deficiency and behavioural changes, the authors claimed that ML348, which inhibits acyl-protein thioesterase 1 (APT1), could be used as a therapy against disease progression. Using the same chip, Migazzi et al. 507 identified distortion in the anterograde axonal transport of HTT-meditated vesicles and neuronal damage due to arginine methylation-defective HTT(Fig. 2n). They identified how PRMT6 expression could restore HTT-mediated axonal trafficking. To model the corticostriatal pathway, they reconstituted two separate cell chambers connected through microchannels of different lengths and the middle “synaptic” chamber.
Table 4.
Summary of Huntington’s disease-on-chips
| Huntington’s disease-on-chips | ||||||
|---|---|---|---|---|---|---|
| Neuropathogenesis | Chip model | Ref. Fig. | ||||
| Targeted pathogenesis | Cell source | Modelling approach | Feature | Type | Design | |
| mHTT toxicity | pCoNeu/StrNeu | mHTT (548-17q r118k) | 2D/S/C | Corticostriatal on-a-chip | 3CHRs/MCs | 507 Fig. 2n |
| Corticostriatal network dysfunction | pCoNeu/StrNeu | mHTT (140 CAG repeat) | 2D/S/C/G | Corticostriatal on-a-chip | 3CHRs/unilateral MCs | 508 |
| Corticostriatal atrophy | pCoNeu/StrNeu | mHTT (97CAG full-length) | 2D/S/C | Corticostriatal on-a-chip | 2CHRs/MCs | 509 Fig. 2m |
C closed, CHR chamber, G gradient, MC microchannel, mHTT mutant Huntingtin protein, pCoNeu primary cortical neuron, S static, StrNeu striatal neuron.
In another study, basal ganglia circuits were composed of the cortex, striatum, globus pallidus internal, substantia nigra reticularis, compacta, and thalamus using a chip with five chambers linked to microchannels511. Notably, the size and location of each regional chamber were designed to mimic a similar ratio of cells and interactions as the 3D anatomical structure of the basal ganglia circuits. They demonstrated the direct neuronal connectivity between five different cell populations using rodent primary neurons. The striatum and its communications with connected regions are important neuronal circuits in HD studies.
Moreover, by integrating an MEA with 256 electrodes, this microfluidic chip analysed the neuronal network functionality in real time. Although the experiment has not yet characterised the disease-specific pathophysiology, it is possible to detect the effect of Huntingtin or α-syn proteins in neural circuit function using this in vivo relevant MEA-integrated regional-neuron network chip. Microfluidic chips can be modified in several ways. One research group recently proposed an electrostimulation microfluidic chip by modifying the chip design for current HD disease modelling. They created holes with electrodes in cell chambers to directly stimulate neurons. This design recapitulated and successfully measured the neuronal activity of the corticostriatal circuit but has not been applied to HD studies yet512.
Microfluidic-based 3D in vitro models
Brain organoids help characterise disease-specific brain regions with distinct cellular makeup, such as the forebrain, midbrain, and hindbrain. Several approaches have been developed to model NDs using brain organoids. To induce Aβ protein accumulation, researchers have treated normal organoids with patient serum513, used small molecule (Aftin-5) for post-modification of the APP protein514, used CRISPR/Cas9 to generate organoids expressing NDs genetic risk factors, or directly created patient-derived organoids515. However, they mostly modelled familial cases516–524 and comparably few sporadic cases515,525.
In the pathophysiological progression of NDs, crosstalk between distinct regions is crucial point to investigate. For such a recapitulation, combining two or more brain region-specific organoids may be useful. By addressing this, Pasca et al. 526 first generated combined organoids termed “assembloid” to better recapitulate the regional interactions, including non-neuronal cells, such as microglia and endothelial cells from different embryonic layers. Following this, brain assembloids have been developed for various regions, including the cortical-striatal, cortical-spinal527, cortical-perictyes528, corticospinal-skeletal muscle529, and disease models, such as Timothy disease530 and AD531,532. Moreover, to overcome the limitation of vascularisation, Kong et al. 531 recently developed a fused cortical vessel assembloid treated with Aβ proteins externally. When exposed to SARS-CoV-2 infection, this assembloid showed an increased number of astrocytes and microglia, indicating how systematic inflammation accelerates neuroinflammation in AD. In another study, instead of regional characterisation, Rickner et al. 532 generated a neuro-astrocyte assembloid (asteroids) composed of neurons cultured with tau oligomers and astrocytes that showed rapid maturation. Assembloid successfully demonstrated several pathological changes, such as tau misfolding, aggregation, and reactive astrogliosis. When treated with an HSP inhibitor, the pathological and neurodegenerative changes declined.
However, the use of organoid models to study NDs remains controversial for several reasons. The current method for developing organoids has only recently achieved anatomical and functional characterisations closer to those of the foetal neonatal human brain after approximately nine to 10 months of culture533,534. Therefore, it is considered more suitable for modelling neurodevelopmental disorders and has been applied to diseases, such as autism spectrum disorder535, epilepsy536, and microencephaly25,537. Nonetheless, encouraging results of organoid maturation have also been observed in terms of cellular composition, anatomical characteristics, such as distinct cortical layers, functional choroid plexus, and brain convolution formation using genetic or mechanical methods538,539.
Recently, various organoid culture platforms have been designed with promising results (Fig. 3). These platforms aim to improve in vivo characterisation, maturation, and uniformity of organoids while reducing the culturing time and labour required. They also have the potential to cooperate with immune and vascular components, bioimaging, and biosensors for effective mimicry and analysis412.
Fig. 3. Representative designs of organoid on-a-chip.
a One-stop microfluidic organoid-culture platform. The four-step organoid maturing process was performed on one platform540. b High-throughput micropillar array. Brain organoids were directly generated from dissociated hiPSCs in a controllable micropillar array with restrained space in the microarray541. c Miniaturised multi-well spinning bioreactor device. The device is cost-effective, high throughput, and has increased maturation and reproducibility542. d With the mechanical stimulation of a microfluidic chip, surface folding appeared in the brain organoid544. e Organoids in a human brain-derived decellularized ECM gel, under the dynamic condition of a microfluidic device, showed higher survival, and uniformity50. f Organoids were encapsulated via microfluidic electrospray, and transferred to multi-layered microfluidic chip to form brain-region-specific assembloid547. g A tumour spheroid was integrated with a perfusable vascular network in a three-line chamber microfluidic chip548,549. h Neuroimmune interaction of cortical organoid and microglia in a 3D-printed organoid culturing device550. i NK cell interaction with tumour organoid in a microfluidic chip552. j Microfluidic device for clearing intact 3D microtissue in 1 d for confocal microscopy553. k Acoustofluidic chip for forming uniform spheroids in a fast and high-throughput manner554. l Hanging drop device with plug-in multi biosensors555. PTFE polytetrafluoroethylene, EB embryonic bodies, ECM extracellular matrix, HESC human embryonic stem cell, hiPSC human-induced pluripotent stem cell, HUVEC human umbilical vein endothelial cell, IDT interdigital transducer pattern, NK natural killer.
Conventional organoid protocols involve several steps that are likely to lead to contamination, damage, sample loss, and dissimilarity. This could be solved with a “one-stop” microfluidic platform that enables all steps of organoid culturing and long-term maintaining process, while monitoring and analysing without any manual additional transferring process540,541. Ao et al. 540 designed a simple device to study the effects of cannabis on prenatal brain organoids (Fig. 3a). This simple “one-stop” chip allows organoids to start from embryonic bodies (EB). With a physical barrier and an air-liquid interface, organoids are prevented from random merging and core necrosis, which leads to better uniformity and maturation. A similar “one-stop” approach has been employed to the micropillar array device, resulting in greater uniformity and reproducibility541 (Fig. 3b). However, organoids eventually exhibit nutrient deficiency in their cores. Furthermore, these models should be transferred to separate analysis tools such as MEA because the design does not include integrated analysis tools. Some research groups have used spinning bioreactors to provide an equal distribution of nutrients to cultured organoids542. To enhance organoid viability, a dynamic medium flow can be achieved by integrating either a rocking platform or a syringe pump49,543. Qian et al. 542. developed a 3D-printed bioreactor that resulted in improved cortical characterisation, such as a human-like outer radial glial layer, compared with static conditions (Fig. 3c). Furthermore, microfluidics can help achieve more human-like characteristics, such as surface wrinkles544 and functional myelinated axons of peripheral neurons545. One group induced mechanical force using a simple chip device to produce wrinkled brain organoids544 (Fig. 3d). Compressed organoids are manipulated to an oblate shape with a surface-wrinkling effect that is not observed in conventional methods. They observed contraction of the cytoskeleton in the core and nuclear expansion in the outer area of the brain organoids.
Another crucial component of 3D culture is the surrounding scaffolds. Supporting biomaterials, such as Matrigel, decellularised ECM (dECM), and other biocompatible hydrogels have been used to accelerate maturation and structural development by providing a brain-like environment50,546. Cho et al. 50 generated organoids with a brain-derived decellularized ECM on a microfluidic-based platform integrated with a rocking platform (Fig. 3e). Compared to the commonly used Matrigel, dECM induces rapid growth and desirable neuron-glia interactions. The microfluidic chip provides an optimal environment for dynamic fluid flow and gas nutrient transfer, thereby accelerating cell viability and growth. Additionally, this platform allows for indirect interactions between organoids which can be applied in co-culture studies of organoids from different regions. There are a few examples of the use of microfluidic chips for assembloid development. However, only recently one group reported a microfluidic-based assembloid-forming device547 (Fig. 3f). They designed a three-layered microfluidic chip (micropillar, microhole, and culture top layer) to generate an assembloid by combining encapsulated organoids. However, before loading onto the chip, EB and different brain regional organoids should be separately generated via a microfluidic encapsulation method. The interaction between the cortical-hippocampal-thalamic regions has been well characterised using assembloid modelling.
Vascularisation and immune interactions are critical for further characterisation of brain organoids. Using vasculogenesis- or angiogenesis-inducing factors, some groups have attempted to vascularise 3D organoids using microfluidic platforms. Nashimoto et al. 548,549 created a blood vessel network merged with tumour spheroids in a fibroblast hydrogel in the middle chamber by culturing human umbilical vein endothelial cells in two side channels (Fig. 3g). The vascular network exhibited high perfusability and viability. In other designs, instead of following the commonly used closed-well approach, Salmon et al. 51 established an open-well organoid chip using 3D printing and successfully generated perfusable neurovascular organoids by simultaneously differentiating iPSCs into vascular and neuronal lineages. One group developed an assembly chip device for the interaction of circulating microglia and brain organoids using a tubular porous scaffold550 (Fig. 3h). Moreover, this chip design was repurposed in different studies to recapitulate organoid vascularisation506,551. Repurposed chips can also be used in brain studies. Tumour spheroid interactions with natural killer cells were recapitulated in chips with vascular-mimicked channels552 (Fig. 3i).
Microfluidic chips can be integrated into bioimaging devices and biosensors for precise monitoring and analysis. However, because of their density and size, 3D cultures have limited imaging properties. One group has proposed on-a-chip clearing techniques for 3D spheroids for better imaging. Sequential clearing steps were performed on the device channels. This approach was 20 times faster than conventional clearing methods553 (Fig. 3j). For precise manipulation of 3D cultures, the surface acoustic wave has been applied to the microfluidic chip, which allows high-throughput production of uniform spheroids and assembling spheroids554 (Fig. 3k). Also, biosensor-integrated microfluidic chips have also been developed for 3D cultures555. This hanging drop chip allowed the monitoring of secreted glucose and lactate levels with electrodes functionalized with oxidase enzymes555 (Fig. 3l).
The outlook of in vitro model in the pharmacological industry
The challenges in neurodegenerative disease drug development
Despite substantial advances in our understanding of disease mechanisms and technological innovations, the development of effective treatments for NDs remains challenging. Recent breakthroughs have been made in the treatment of AD, including the use of aducanumab16, lecanemab17, and donanemab556. However, the effectiveness of the new drugs only appears at certain stages of AD116,556. Moreover, Aβ-against antibodies are controversial in human cognition although it exhibits a considerable decrease of the plaque. While these recent approvals are beneficial in certain groups of patients, there is still much work to be done to expand the effectiveness of these drugs, considering the multifactorial and heterogeneous characteristics of NDs.
In addition, the treatment development process requires a cost-effective and time-saving approach. The AD drug development field has invested approximately $42.5 billion in clinical trials over the past 30 years, which requires a long, dedicated effort, but has yielded few promising results116,557. Several factors have been attributed to drug failure, including intervention at an inappropriate stage of disease progression, insufficient biomarkers, and the use of incorrect research models116. Specifically, preclinical studies conducted on familial genetically engineered mouse models have limitations as they do not fully replicate all the features of NDs, especially sporadic cases. These models exhibit varied clinical and pathological phenotypes that restrict their effectiveness in translation to human physiology558. Moreover, long-term preservation of animals with age-related characteristics can be costly and time-consuming559. Translation to a more physiologically relevant, human-cell-sourced, in vitro model that incorporates most features of certain NDs for studying disease mechanisms and developing drugs has the potential to reduce costs and time for development and produce more representative and reliable test results.
Microfluidic chip in pharmaceutical industry
Over the past two decades, the development of advanced in vitro models has increased in the academic field. However, the pharmaceutical industry has not yet adopted microfluidic chips560,561.
Following the recent FDA policy on the shift towards the use of more human-relevant models in drug testing, both academic and pharmaceutical industries have begun to use organ-on-a-chip and organoid-based models562. Several large pharmaceutical companies (AstraZeneca, Bayer, and Roche) have been using organoid and microfluidic chips to a limited degree to enhance the screening accuracy of potential drug candidates561,563. Moreover, global efforts (USA, Europe, and Japan) have been made to translate academic achievements to the pharmaceutical industry using a standardised and reproducible predictive drug-screening approach564–566. Tissue chip testing centres established by the NIH National Center for Advancing Translational Sciences initiated grantees to validate various types of tissue chip platforms for drug testing566,567. Under these grants, three independent organisations validated microphysiological systems that replicated the kidney, liver, and BBB for drug transport and toxicity studies, with the goal of increasing the reliability, reproducibility, robustness, and throughput of its effective application568.
Microfluidic chip for drug screening at the academic level
Despite ongoing efforts to address reproducibility and scalability in the pharmaceutical industry, drug screening and toxicity studies of potential ND drug components have also been conducted on-a-chip in the academic field. Numerous companies, such as Emulate, TissUse, Mimetas, Nortis, and CN Bio offer various designs of microfluidic chips to researchers for conduct physiologically relevant studies of disease mechanism, drug screening, and toxicity563.
Some research groups have performed drug screening and toxicity studies on previously rejected drug candidates using microfluidic chips. Clinical trials on ASOs to prevent and alleviate the progression of AD, PD, ALS, and HD are currently underway. However, these clinical trials demonstrated toxicity. Nieskens et al. 569 verified ASOs toxicity by testing varying doses on a kidney proximal tubule-on-a-chip using human renal proximal tubule epithelial cells. In addition, one group reported the dose-dependent efficacy of NMJ-blocking drugs for ALS using a commonly used micro-channelled two-compartment design. This chip allows the interaction between motor neurons and muscles only through the axons. Functional ability was revealed by muscle contraction when motor neurons were stimulated using silver electrodes. They tested the several doses of botulinum neurotoxin, Curare, and α-bungarotoxin (α-BTX), and quantified the dose-dependent NMJ dysfunction as dose-response curve570.
Monolayer culture approaches have often been applied to drug discovery and preclinical screening. However, as described in the previous section, 3D cell culture is expected to provide closer in vivo characteristics, increasing the predictability of clinical results. In addition, replacing animal-based drug screening with patient-origin organoids is considered a substantial development in the pharmaceutical field which will increase human reliability and reduce costs and ethical concerns by overcoming the drawbacks of animal models. Recently, several attempts have been made to apply organoids in drug discovery studies in an academic field. Osaki et al. 506 tested the muscle atrophy-reversing effect of bosutinib and rapamycin on spheroids on chips, confirming drug efficacy with caspase-3/7 staining at day 14. Moreover, neuroprotective treatments positively influenced the optogenetic stimulation of muscle contraction. This chip design is beneficial because it evaluates not only the single or combined drug effects on functional 3D co-cultures, but also drug delivery across the EC barrier. This chip design significantly increased throughput by simultaneously screening single and combined drugs. Park et al. 515. introduced a novel organoid-culture drug-screening platform that employed mathematical network analysis. Researchers have investigated the effects of current FDA-approved drug candidates on AD patient-origin cerebral organoids and confirmed the structure, viability, and Aβ and tau depositions. This study demonstrates the potential of patient-origin organoids for drug screening of NDs.
Potentials of advanced microfluidic chip in pharmaceutical industry
Through collaborative global efforts, the integration of microfluidic chips with advanced technologies has the potential to revolutionise the pharmaceutical industry. The organoid-on-a-chip and multiorgan-on-a-chip systems can play crucial roles in various stages of drug discovery, ranging from target identification to preclinical screening (Fig. 4). In addition, we believe that such an advanced model can be useful even in the clinical stage when deciding on the effective DMT for each patient subtype by sourcing patient-derived cells. However, to achieve this, many crucial industrial and technological challenges must be considered, such as reliability, robustness, reproducibility, compatibility with analysis techniques, material choice, scalability, and standardisation. Moreover, there is a great need for defined metrics for validation and benchmarks for modelling each ND.
Fig. 4. The future outlook of NDs-on-a-chip for drug development.
a Patient-derived pluripotent stem cells can provide relevant insights into ND pathophysiology. b 3D culture-on-a-chip combines the in vivo relevant characteristics of patient-derived 3D cultures (neurospheroids, organoids, and assembloids) and the unique engineering possibilities of microfluidic chips that can model interconnected brain regions and multi-organs. c Microfluidic-based, patient-derived in vitro ND models will contribute to drug discovery and preclinical drug development studies by replacing the animal models with the promise of personalised treatment. ND neurodegenerative disease.
An advanced on-a-chip system has the capability to not only validate the effects of drugs on neuropathological features, but also measure functional changes through real-time electrophysiological measurements. Efforts have been made to measure brain waves in real time from brain organoids using invasive and non-invasive electrophysiological biosensors integrated on a high-throughput microfluidic chip534. This integration will provide a clinically relevant assessment of NDs. Moreover, once drug candidates are identified for NDs, their mechanism of action, safety, dose-response relationship, potential side effects, and pharmacokinetics can be tested on even more complex inclusive systems, such as multiorgan-on-a-chip platforms (composed of the brain, kidney, liver, gut, and heart)571. The use of multiorgan-on-a-chip models reduces the dependence on animal models during this stage, while potentially providing more representative, cost-effective, and accurate results, thereby facilitating successful clinical testing. In addition, patient-derived cell sources can facilitate the development of effective treatments for certain patient subsets. Advanced on-a-chip systems could be beneficial for patient stratification testing to identify effective DMT with fewer side effects, especially in challenging multifactorial diseases, such as AD572.
In the future, microfluidic chips will substantially contribute to the translation of brain organoids and assembloids for the development of ND drug discovery and personalised medicine. Notably, in the development of microfluidic chips, there are biological, technical, and commercial challenges for the entire application to the pharmaceutical industry and personalised medicine573. Despite these hurdles, we believe that advanced microfluidic chips and molecular biophysical techniques will advance our understanding of the complexities of ND pathogenesis and challenge our dependence on animal-based tools.
Acknowledgements
This study was supported by the HK Lee Foundation. This study was supported by the Institute for Basic Science of Korea (IBS-R011-D1) and the Advanced Facility Center for Quantum Technology.
Author contributions
C.H. and L.P.L. supervised the study. S.A. wrote this review. H.N. and T.H. made substantial contributions to the discussions on all topics and edited the manuscript. T.S.K. and E.O. wrote and edited the manuscript’s PD and HD sections, respectively. R.S.W. contributed to the discussions on the AD sections and edited the manuscript. K.K.K. reviewed and edited the manuscript.
Peer review
Peer review information
Nature Communications thanks Mootaz Salman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Luke P. Lee, Email: lplee@bwh.harvard.edu
Chaejeong Heo, Email: cjheo@skku.edu.
References
- 1.United Nations, Department of Economic and Social Affairs, Population Division (2019). World Population Prospects 2019: Highlights (ST/ESA/SER.A/423). (United Nations, 2019).
- 2.Nichols E, et al. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: an analysis for the Global Burden of Disease Study 2019. Lancet Public Health. 2022;7:e105–e125. doi: 10.1016/S2468-2667(21)00249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deuschl G, et al. The burden of neurological diseases in Europe: an analysis for the Global Burden of Disease Study 2017. Lancet Public Health. 2020;5:e551–e567. doi: 10.1016/S2468-2667(20)30190-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, et al. Burden of common neurologic diseases in Asian Countries, 1990–2019. An Analysis for the Global Burden of Disease Study 2019. Neurology. 2023;100:e2141–e2154. doi: 10.1212/WNL.0000000000207218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erkkinen, M. G., Kim, M. O. & Geschwind, M. D. Clinical neurology and epidemiology of the major neurodegenerative diseases. Cold Spring Harb. Perspect. Biol.10.1101/cshperspect.a033118 (2018). [DOI] [PMC free article] [PubMed]
- 6.Poewe W, et al. Parkinson disease. Nat. Rev. Dis. Prim. 2017;3:17013. doi: 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- 7.Kiernan MC, et al. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 8.Chaganti SS, McCusker EA, Loy CT. What do we know about late onset Huntington’s disease? J. Huntingt. Dis. 2017;6:95–103. doi: 10.3233/JHD-170247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knopman DS, et al. Alzheimer disease. Nat. Rev. Dis. Prim. 2021;7:33. doi: 10.1038/s41572-021-00269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masters CL, et al. Alzheimer’s disease. Nat. Rev. Dis. Prim. 2015;1:15056. doi: 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- 11.Przedborski S. The two-century journey of Parkinson disease research. Nat. Rev. Neurosci. 2017;18:251–259. doi: 10.1038/nrn.2017.25. [DOI] [PubMed] [Google Scholar]
- 12.Taylor JP, Brown RH, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bates GP, et al. Huntington disease. Nat. Rev. Dis. Prim. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 14.Gan L, Cookson MR, Petrucelli L, La Spada AR. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 2018;21:1300–1309. doi: 10.1038/s41593-018-0237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng C, Trojanowski JQ, Lee VM. Protein transmission in neurodegenerative disease. Nat. Rev. Neurol. 2020;16:199–212. doi: 10.1038/s41582-020-0333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cummings J, et al. Aducanumab produced a clinically meaningful benefit in association with amyloid lowering. Alzheimers Res. Ther. 2021;13:98. doi: 10.1186/s13195-021-00838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Dyck CH, et al. Lecanemab in early Alzheimer’s disease. N. Engl. J. Med. 2022;388:9–21. doi: 10.1056/NEJMoa2212948. [DOI] [PubMed] [Google Scholar]
- 18.Gurney ME, Fleck TJ, Himes CS, Hall ED. Riluzole preserves motor function in a transgenic model of familial amyotrophic lateral sclerosis. Neurology. 1998;50:62–66. doi: 10.1212/WNL.50.1.62. [DOI] [PubMed] [Google Scholar]
- 19.Ito H, et al. Treatment with edaravone, initiated at symptom onset, slows motor decline and decreases SOD1 deposition in ALS mice. Exp. Neurol. 2008;213:448–455. doi: 10.1016/j.expneurol.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 20.Bjornson-Hooper, Z. B. et al. A Comprehensive Atlas of Immunological Differences Between Humans, Mice, and Non-Human Primates. Front. Immunol.10.3389/fimmu.2022.867015 (2022). [DOI] [PMC free article] [PubMed]
- 21.Hodge RD, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bang S, Jeong S, Choi N, Kim HN. Brain-on-a-chip: a history of development and future perspective. Biomicrofluidics. 2019;13:051301. doi: 10.1063/1.5120555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertram L, Tanzi RE. The genetic epidemiology of neurodegenerative disease. J. Clin. Investig. 2005;115:1449–1457. doi: 10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loewa A, Feng JJ, Hedtrich S. Human disease models in drug development. Nat. Rev. Bioeng. 2023;1:545–559. doi: 10.1038/s44222-023-00063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakaguchi H, et al. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian X, et al. Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018;13:565–580. doi: 10.1038/nprot.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jo J, et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellegrini L, et al. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 2020;369:eaaz5626. doi: 10.1126/science.aaz5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10:537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Koo B-K, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofer M, Lutolf MP. Engineering organoids. Nat. Rev. Mater. 2021;6:402–420. doi: 10.1038/s41578-021-00279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JH, Byeun DG, Choi JK. Progress, prospects, and limitations of organoid technology. Organoid. 2022;2:e9. doi: 10.51335/organoid.2022.2.e9. [DOI] [Google Scholar]
- 34.Ingber DE. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022;23:467–491. doi: 10.1038/s41576-022-00466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mou L, et al. Integrated biosensors for monitoring microphysiological systems. Lab Chip. 2022;22:3801–3816. doi: 10.1039/D2LC00262K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor AM, Dieterich DC, Ito HT, Kim SA, Schuman EM. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron. 2010;66:57–68. doi: 10.1016/j.neuron.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, J., Koito, H., Li, J. & Han, A. A multi-compartment CNS neuron-glia Co-culture microfluidic platform. J. Vis. Exp.10.3791/1399 (2009). [DOI] [PMC free article] [PubMed]
- 38.Park T-E, et al. Hypoxia-enhanced blood-brain barrier chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat. Commun. 2019;10:2621. doi: 10.1038/s41467-019-10588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawakita S, et al. Organ-on-a-chip models of the blood–brain barrier: recent advances and future prospects. Small. 2022;18:2201401. doi: 10.1002/smll.202201401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyu Z, et al. A neurovascular-unit-on-a-chip for the evaluation of the restorative potential of stem cell therapies for ischaemic stroke. Nat. Biomed. Eng. 2021;5:847–863. doi: 10.1038/s41551-021-00744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi H-G, et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019;3:509–519. doi: 10.1038/s41551-019-0363-x. [DOI] [PubMed] [Google Scholar]
- 42.Yap, Y. C., Dickson, T. C., King, A. E., Breadmore, M. C. & Guijt, R. M. In Stem Cell Technologies in Neuroscience. Neuromethods (eds Srivastava, A. K., Snyder, E. Y. & Teng, Y. D.) Ch. 10, 145–156 (Springer, New York, 2017).
- 43.Pediaditakis I, et al. A microengineered Brain-Chip to model neuroinflammation in humans. iScience. 2022;25:104813. doi: 10.1016/j.isci.2022.104813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amirifar L, et al. Brain-on-a-chip: recent advances in design and techniques for microfluidic models of the brain in health and disease. Biomaterials. 2022;285:121531. doi: 10.1016/j.biomaterials.2022.121531. [DOI] [PubMed] [Google Scholar]
- 45.Haque MR, et al. Patient-derived pancreatic cancer-on-a-chip recapitulates the tumor microenvironment. Microsyst. Nanoeng. 2022;8:36–36. doi: 10.1038/s41378-022-00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, et al. Modeling Human Nonalcoholic Fatty Liver Disease (NAFLD) with an organoids-on-a-chip system. ACS Biomater. Sci. Eng. 2020;6:5734–5743. doi: 10.1021/acsbiomaterials.0c00682. [DOI] [PubMed] [Google Scholar]
- 47.Homan KA, et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019;16:255–262. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee KK, et al. Human stomach-on-a-chip with luminal flow and peristaltic-like motility. Lab Chip. 2018;18:3079–3085. doi: 10.1039/C8LC00910D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Wang L, Zhu Y, Qin J. Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab Chip. 2018;18:851–860. doi: 10.1039/C7LC01084B. [DOI] [PubMed] [Google Scholar]
- 50.Cho A-N, et al. Microfluidic device with brain extracellular matrix promotes structural and functional maturation of human brain organoids. Nat. Commun. 2021;12:4730. doi: 10.1038/s41467-021-24775-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salmon I, et al. Engineering neurovascular organoids with 3D printed microfluidic chips. Lab Chip. 2022;22:1615–1629. doi: 10.1039/D1LC00535A. [DOI] [PubMed] [Google Scholar]
- 52.2023 Alzheimer’s disease facts and figures. Alzheimers Dement.19, 1598–1695 (2023). [DOI] [PubMed]
- 53.Bature F, Guinn B-A, Pang D, Pappas Y. Signs and symptoms preceding the diagnosis of Alzheimer’s disease: a systematic scoping review of literature from 1937 to 2016. BMJ Open. 2017;7:e015746. doi: 10.1136/bmjopen-2016-015746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rocca WA, Amaducci LA, Schoenberg BS. Epidemiology of clinically diagnosed Alzheimer’s disease. Ann. Neurol. 1986;19:415–424. doi: 10.1002/ana.410190502. [DOI] [PubMed] [Google Scholar]
- 55.Jansen WJ, et al. Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. Jama. 2015;313:1924–1938,. doi: 10.1001/jama.2015.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011;1:a006189. doi: 10.1101/cshperspect.a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang J, et al. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 58.Sherrington R, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 59.Levy-Lahad E, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 60.Schwartzentruber J, et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat. Genet. 2021;53:392–402. doi: 10.1038/s41588-020-00776-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wightman DP, et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 2021;53:1276–1282. doi: 10.1038/s41588-021-00921-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamazaki Y, Zhao N, Caulfield TR, Liu CC, Bu G. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat. Rev. Neurol. 2019;15:501–518. doi: 10.1038/s41582-019-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guerreiro R, et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2012;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giau VV, et al. Genetic analyses of early-onset Alzheimer’s disease using next generation sequencing. Sci. Rep. 2019;9:8368. doi: 10.1038/s41598-019-44848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mendez MF. Early-onset Alzheimer disease. Neurol. Clin. 2017;35:263–281. doi: 10.1016/j.ncl.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Q, Wang Z, Li H, Wiese M, Zheng H. APP physiological and pathophysiological functions: insights from animal models. Cell Res. 2012;22:78–89. doi: 10.1038/cr.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 69.Liu Y, et al. Roles and mechanisms of the protein quality control system in Alzheimer’s disease. Int. J. Mol. Sci. 2021;23:345. doi: 10.3390/ijms23010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou H, et al. Low-density lipoprotein receptor-related protein-1 (LRP1) C4408R mutant promotes amyloid precursor protein (APP) α-cleavage in vitro. Neuromol. Med. 2017;19:300–308. doi: 10.1007/s12017-017-8446-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okada H, et al. Proteomic identification of sorting nexin 6 as a negative regulator of BACE1-mediated APP processing. FASEB J. 2010;24:2783–2794. doi: 10.1096/fj.09-146357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim W, et al. BACE1 elevation engendered by GGA3 deletion increases β-amyloid pathology in association with APP elevation and decreased CHL1 processing in 5XFAD mice. Mol. Neurodegener. 2018;13:6. doi: 10.1186/s13024-018-0239-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andersen OM, et al. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry. 2006;45:2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- 74.Lambert MP, et al. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc. Natl Acad. Sci. USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lacor PN, et al. Synaptic targeting by Alzheimer’s-related amyloid beta oligomers. J. Neurosci. 2004;24:10191–10200. doi: 10.1523/JNEUROSCI.3432-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Younan ND, Chen K-F, Rose R-S, Crowther DC, Viles JH. Prion protein stabilizes amyloid-β (Aβ) oligomers and enhances Aβ neurotoxicity in a Drosophila model of Alzheimer’s disease. J. Biol. Chem. 2018;293:13090–13099. doi: 10.1074/jbc.RA118.003319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liang J, Kulasiri D, Samarasinghe S. Computational investigation of Amyloid-β-induced location- and subunit-specific disturbances of NMDAR at hippocampal dendritic spine in Alzheimer’s disease. PLoS ONE. 2017;12:e0182743. doi: 10.1371/journal.pone.0182743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang D, et al. β2 adrenergic receptor, protein kinase A (PKA) and c-Jun N-terminal kinase (JNK) signaling pathways mediate tau pathology in Alzheimer disease models. J. Biol. Chem. 2013;288:10298–10307. doi: 10.1074/jbc.M112.415141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen L-L, et al. Neurotrophin receptor p75 mediates amyloid β-induced tau pathology. Neurobiol. Dis. 2019;132:104567. doi: 10.1016/j.nbd.2019.104567. [DOI] [PubMed] [Google Scholar]
- 80.Bencherif M, Lippiello PM. Alpha7 neuronal nicotinic receptors: the missing link to understanding Alzheimer’s etiopathology? Med. Hypotheses. 2010;74:281–285. doi: 10.1016/j.mehy.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 81.Bode DC, Baker MD, Viles JH. Ion channel formation by amyloid-β42 oligomers but not amyloid-β40 in cellular membranes. J. Biol. Chem. 2017;292:1404–1413. doi: 10.1074/jbc.M116.762526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H, et al. Parkin overexpression attenuates Aβ-induced mitochondrial dysfunction in HEK293 cells by restoring impaired mitophagy. Life Sci. 2020;244:117322. doi: 10.1016/j.lfs.2020.117322. [DOI] [PubMed] [Google Scholar]
- 83.Sun JL, et al. Co-activation of selective nicotinic acetylcholine receptors is required to reverse beta amyloid–induced Ca2+ hyperexcitation. Neurobiol. Aging. 2019;84:166–177. doi: 10.1016/j.neurobiolaging.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang F, et al. β-amyloid redirects norepinephrine signaling to activate the pathogenic GSK3β/tau cascade. Sci. Transl. Med. 2020;12:eaay6931. doi: 10.1126/scitranslmed.aay6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thal DR, Rüb U, Orantes M, Braak H. Phases of Aβ-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58:1791–1800. doi: 10.1212/WNL.58.12.1791. [DOI] [PubMed] [Google Scholar]
- 86.Pignataro A, Middei S. Trans-Synaptic Spread of Amyloid-β in Alzheimer’s Disease: Paths to β-Amyloidosis. Neural Plast. 2017;2017:5281829–5281829. doi: 10.1155/2017/5281829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501:45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Olsson TT, Klementieva O, Gouras GK. Prion-like seeding and nucleation of intracellular amyloid-β. Neurobiol. Dis. 2018;113:1–10. doi: 10.1016/j.nbd.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 89.Cohen SIA, et al. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl Acad. Sci. USA. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sowade RF, Jahn TR. Seed-induced acceleration of amyloid-β mediated neurotoxicity in vivo. Nat. Commun. 2017;8:512. doi: 10.1038/s41467-017-00579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nath S, et al. Spreading of neurodegenerative pathology via neuron-to-neuron transmission of β-amyloid. J. Neurosci. 2012;32:8767–8777. doi: 10.1523/JNEUROSCI.0615-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sardar Sinha M, et al. Alzheimer’s disease pathology propagation by exosomes containing toxic amyloid-beta oligomers. Acta Neuropathol. 2018;136:41–56. doi: 10.1007/s00401-018-1868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rajendran L, et al. Alzheimer’s disease β-amyloid peptides are released in association with exosomes. Proc. Natl Acad. Sci. USA. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lim CZJ, et al. Subtyping of circulating exosome-bound amyloid β reflects brain plaque deposition. Nat. Commun. 2019;10:1144. doi: 10.1038/s41467-019-09030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y, Cui J, Sun X, Zhang Y. Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ. 2011;18:732–742. doi: 10.1038/cdd.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang K, et al. Intercellular transport of Tau protein and β-amyloid mediated by tunneling nanotubes. Am. J. Transl. Res. 2021;13:12509. [PMC free article] [PubMed] [Google Scholar]
- 97.Valappil, D. K., Mini, N. J., Dilna, A. & Nath, S. Membrane interaction to intercellular spread of pathology in Alzheimer’s disease. Front. Neurosci.10.3389/fnins.2022.936897 (2022). [DOI] [PMC free article] [PubMed]
- 98.Arriagada PV, Growdon JH, Hedley-Whyte ET, Hyman BT. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer’s disease. Neurology. 1992;42:631–639,. doi: 10.1212/WNL.42.3.631. [DOI] [PubMed] [Google Scholar]
- 99.Gong CX, Liu F, Grundke-Iqbal I, Iqbal K. Post-translational modifications of tau protein in Alzheimer’s disease. J. Neural Transm. 2005;112:813–838. doi: 10.1007/s00702-004-0221-0. [DOI] [PubMed] [Google Scholar]
- 100.Alonso ADC, Zaidi T, Grundke-Iqbal I, Iqbal K. Role of abnormally phosphorylated tau in the breakdown of microtubules in Alzheimer disease. Proc. Natl Acad. Sci. USA. 1994;91:5562–5566. doi: 10.1073/pnas.91.12.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schneider A, Biernat J, von Bergen M, Mandelkow E, Mandelkow EM. Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry. 1999;38:3549–3558. doi: 10.1021/bi981874p. [DOI] [PubMed] [Google Scholar]
- 102.Strang KH, et al. Phosphorylation of serine 305 in tau inhibits aggregation. Neurosci. Lett. 2019;692:187–192. doi: 10.1016/j.neulet.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stefanoska K, et al. Alzheimer’s disease: ablating single master site abolishes tau hyperphosphorylation. Sci. Adv. 2022;8:eabl8809. doi: 10.1126/sciadv.abl8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Patrick GN, et al. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402:615–622. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- 105.Mazanetz MP, Fischer PM. Untangling tau hyperphosphorylation in drug design for neurodegenerative diseases. Nat. Rev. Drug Discov. 2007;6:464–479. doi: 10.1038/nrd2111. [DOI] [PubMed] [Google Scholar]
- 106.Goedert M, Jakes R, Qi Z, Wang JH, Cohen P. Protein phosphatase 2A is the major enzyme in brain that dephosphorylates tau protein phosphorylated by proline-directed protein kinases or cyclic AMP-dependent protein kinase. J. Neurochem. 1995;65:2804–2807. doi: 10.1046/j.1471-4159.1995.65062804.x. [DOI] [PubMed] [Google Scholar]
- 107.Liu F, Grundke-Iqbal I, Iqbal K, Gong CX. Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur. J. Neurosci. 2005;22:1942–1950. doi: 10.1111/j.1460-9568.2005.04391.x. [DOI] [PubMed] [Google Scholar]
- 108.Pao P-C, et al. A Cdk5-derived peptide inhibits Cdk5/p25 activity and improves neurodegenerative phenotypes. Proc. Natl Acad. Sci. USA. 2023;120:e2217864120. doi: 10.1073/pnas.2217864120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Clavaguera F, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y, et al. The release and trans-synaptic transmission of Tau via exosomes. Mol. Neurodegener. 2017;12:5. doi: 10.1186/s13024-016-0143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tardivel M, et al. Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological Tau protein assemblies. Acta Neuropathol. Commun. 2016;4:1–14. doi: 10.1186/s40478-016-0386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Asai H, et al. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 2015;18:1584–1593. doi: 10.1038/nn.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gabrielli M, et al. Microglial large extracellular vesicles propagate early synaptic dysfunction in Alzheimer’s disease. Brain. 2022;145:2849–2868. doi: 10.1093/brain/awac083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen G-F, et al. Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017;38:1205–1235. doi: 10.1038/aps.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sanchez JS, et al. The cortical origin and initial spread of medial temporal tauopathy in Alzheimer’s disease assessed with positron emission tomography. Sci. Transl. Med. 2021;13:eabc0655. doi: 10.1126/scitranslmed.abc0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boxer AL, Sperling R. Accelerating Alzheimer’s therapeutic development: the past and future of clinical trials. Cell. 2023;186:4757–4772. doi: 10.1016/j.cell.2023.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ittner LM, Götz J. Amyloid-β and tau — a toxic pas de deux in Alzheimer’s disease. Nat. Rev. Neurosci. 2011;12:67–72. doi: 10.1038/nrn2967. [DOI] [PubMed] [Google Scholar]
- 118.Muralidar S, Ambi SV, Sekaran S, Thirumalai D, Palaniappan B. Role of tau protein in Alzheimer’s disease: the prime pathological player. Int J. Biol. Macromol. 2020;163:1599–1617. doi: 10.1016/j.ijbiomac.2020.07.327. [DOI] [PubMed] [Google Scholar]
- 119.Shin WS, et al. Amyloid β-protein oligomers promote the uptake of tau fibril seeds potentiating intracellular tau aggregation. Alzheimers Res. Ther. 2019;11:86. doi: 10.1186/s13195-019-0541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Busche MA, Hyman BT. Synergy between amyloid-β and tau in Alzheimer’s disease. Nat. Neurosci. 2020;23:1183–1193. doi: 10.1038/s41593-020-0687-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lauritzen I, et al. Intraneuronal aggregation of the β-CTF fragment of APP (C99) induces Aβ-independent lysosomal-autophagic pathology. Acta Neuropathol. 2016;132:257–276. doi: 10.1007/s00401-016-1577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pulina, M. V., Hopkins, M., Haroutunian, V., Greengard, P. & Bustos, V. C99 selectively accumulates in vulnerable neurons in Alzheimer’s disease. Alzheimers Dement.10.1016/j.jalz.2019.09.002 (2019). [DOI] [PubMed]
- 123.Walker KA, Ficek BN, Westbrook R. Understanding the role of systemic inflammation in Alzheimer’s disease. ACS Chem. Neurosci. 2019;10:3340–3342. doi: 10.1021/acschemneuro.9b00333. [DOI] [PubMed] [Google Scholar]
- 124.Taipa R, et al. Proinflammatory and anti-inflammatory cytokines in the CSF of patients with Alzheimer’s disease and their correlation with cognitive decline. Neurobiol. Aging. 2019;76:125–132. doi: 10.1016/j.neurobiolaging.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 125.Serrano-Pozo A, et al. Reactive glia not only associates with plaques but also parallels tangles in Alzheimer’s disease. Am. J. Pathol. 2011;179:1373–1384. doi: 10.1016/j.ajpath.2011.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ries, M. & Sastre, M. Mechanisms of Aβ clearance and degradation by glial cells. Front. Aging Neurosci.10.3389/fnagi.2016.00160 (2016). [DOI] [PMC free article] [PubMed]
- 127.Ising C, et al. NLRP3 inflammasome activation drives tau pathology. Nature. 2019;575:669–673. doi: 10.1038/s41586-019-1769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Venegas C, et al. Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature. 2017;552:355–361. doi: 10.1038/nature25158. [DOI] [PubMed] [Google Scholar]
- 129.Efthymiou AG, Goate AM. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener. 2017;12:43. doi: 10.1186/s13024-017-0184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kleinberger G, et al. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 2014;6:243ra286. doi: 10.1126/scitranslmed.3009093. [DOI] [PubMed] [Google Scholar]
- 131.Jiang T, et al. Upregulation of TREM2 ameliorates neuropathology and rescues spatial cognitive impairment in a transgenic mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2014;39:2949–2962. doi: 10.1038/npp.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ulland TK, Colonna M. TREM2 — a key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol. 2018;14:667–675. doi: 10.1038/s41582-018-0072-1. [DOI] [PubMed] [Google Scholar]
- 133.Bailey CC, DeVaux LB, Farzan M. The triggering receptor expressed on myeloid cells 2 binds apolipoprotein E. J. Biol. Chem. 2015;290:26033–26042. doi: 10.1074/jbc.M115.677286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron. 2016;91:328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 135.Gratuze M, et al. TREM2-independent microgliosis promotes tau-mediated neurodegeneration in the presence of ApoE4. Neuron. 2023;111:202–219.e207. doi: 10.1016/j.neuron.2022.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van Lengerich B, et al. A TREM2-activating antibody with a blood–brain barrier transport vehicle enhances microglial metabolism in Alzheimer’s disease models. Nat. Neurosci. 2023;26:416–429. doi: 10.1038/s41593-022-01240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rodríguez JJ, Olabarria M, Chvatal A, Verkhratsky A. Astroglia in dementia and Alzheimer’s disease. Cell Death Differ. 2009;16:378–385. doi: 10.1038/cdd.2008.172. [DOI] [PubMed] [Google Scholar]
- 138.Ding Z-B, et al. Astrocytes: a double-edged sword in neurodegenerative diseases. Neural Regener. Res. 2021;16:1702–1710,. doi: 10.4103/1673-5374.306064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ferrari-Souza JP, et al. Astrocyte biomarker signatures of amyloid-β and tau pathologies in Alzheimer’s disease. Mol. Psychiatry. 2022;27:4781–4789. doi: 10.1038/s41380-022-01716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mathys H, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Simonovitch S, et al. Impaired Autophagy in APOE4 Astrocytes. J. Alzheimers Dis. 2016;51:915–927. doi: 10.3233/JAD-151101. [DOI] [PubMed] [Google Scholar]
- 142.Zhao J, et al. APOE ε4/ε4 diminishes neurotrophic function of human iPSC-derived astrocytes. Hum. Mol. Genet. 2017;26:2690–2700. doi: 10.1093/hmg/ddx155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee SI, et al. APOE4-carrying human astrocytes oversupply cholesterol to promote neuronal lipid raft expansion and Aβ generation. Stem Cell Rep. 2021;16:2128–2137. doi: 10.1016/j.stemcr.2021.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Blanchard JW, et al. APOE4 impairs myelination via cholesterol dysregulation in oligodendrocytes. Nature. 2022;611:769–779. doi: 10.1038/s41586-022-05439-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lin YT, et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron. 2018;98:1141–1154.e1147. doi: 10.1016/j.neuron.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Koutsodendris N, et al. Neuronal APOE4 removal protects against tau-mediated gliosis, neurodegeneration and myelin deficits. Nat. Aging. 2023;3:275–296. doi: 10.1038/s43587-023-00368-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Habib N, et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 2020;23:701–706. doi: 10.1038/s41593-020-0624-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290.e1217. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 149.Zeng H, et al. Integrative in situ mapping of single-cell transcriptional states and tissue histopathology in a mouse model of Alzheimer’s disease. Nat. Neurosci. 2023;26:430–446. doi: 10.1038/s41593-022-01251-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nation DA, et al. Blood–brain barrier breakdown is an early biomarker of human cognitive dysfunction. Nat. Med. 2019;25:270–276. doi: 10.1038/s41591-018-0297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sweeney MD, Sagare AP, Zlokovic BV. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018;14:133–150. doi: 10.1038/nrneurol.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Montagne A, Zhao Z, Zlokovic BV. Alzheimer’s disease: a matter of blood–brain barrier dysfunction? J. Exp. Med. 2017;214:3151–3169. doi: 10.1084/jem.20171406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bennett RE, et al. Tau induces blood vessel abnormalities and angiogenesis-related gene expression in P301L transgenic mice and human Alzheimer’s disease. Proc. Natl Acad. Sci. USA. 2018;115:E1289–E1298. doi: 10.1073/pnas.1710329115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Armulik A, et al. Pericytes regulate the blood-brain barrier. Nature. 2010;468:557–561. doi: 10.1038/nature09522. [DOI] [PubMed] [Google Scholar]
- 155.Song J, Choi S-M, Whitcomb DJ, Kim BC. Adiponectin controls the apoptosis and the expression of tight junction proteins in brain endothelial cells through AdipoR1 under beta amyloid toxicity. Cell Death Dis. 2017;8:e3102–e3102. doi: 10.1038/cddis.2017.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu C-C, et al. Tau and apolipoprotein E modulate cerebrovascular tight junction integrity independent of cerebral amyloid angiopathy in Alzheimer’s disease. Alzheimers Dement. 2020;16:1372–1383. doi: 10.1002/alz.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang Y, et al. Interleukin-1β induces blood–brain barrier disruption by downregulating sonic hedgehog in astrocytes. PLoS ONE. 2014;9:e110024. doi: 10.1371/journal.pone.0110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Iliff JJ, et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci. Transl. Med. 2012;4:147ra111. doi: 10.1126/scitranslmed.3003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu Z, et al. Deletion of aquaporin-4 in APP/PS1 mice exacerbates brain Aβ accumulation and memory deficits. Mol. Neurodegener. 2015;10:58. doi: 10.1186/s13024-015-0056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Feng W, et al. Microglia prevent beta-amyloid plaque formation in the early stage of an Alzheimer’s disease mouse model with suppression of glymphatic clearance. Alzheimers Res. Ther. 2020;12:125. doi: 10.1186/s13195-020-00688-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ishida, K. et al. Glymphatic system clears extracellular tau and protects from tau aggregation and neurodegeneration. J. Exp. Med. 10.1084/jem.20211275 (2022). [DOI] [PMC free article] [PubMed]
- 162.Iliff J, Simon M. CrossTalk proposal: the glymphatic system supports convective exchange of cerebrospinal fluid and brain interstitial fluid that is mediated by perivascular aquaporin-4. J. Physiol. 2019;597:4417–4419. doi: 10.1113/JP277635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Salman MM, et al. Emerging roles for dynamic aquaporin-4 subcellular relocalization in CNS water homeostasis. Brain. 2021;145:64–75. doi: 10.1093/brain/awab311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zeppenfeld DM, et al. Association of perivascular localization of aquaporin-4 with cognition and Alzheimer disease in aging brains. JAMA Neurol. 2017;74:91–99,. doi: 10.1001/jamaneurol.2016.4370. [DOI] [PubMed] [Google Scholar]
- 165.Simon MJ, et al. Transcriptional network analysis of human astrocytic endfoot genes reveals region-specific associations with dementia status and tau pathology. Sci. Rep. 2018;8:12389. doi: 10.1038/s41598-018-30779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pedersen TJ, Keil SA, Han W, Wang MX, Iliff JJ. The effect of aquaporin-4 mis-localization on Aβ deposition in mice. Neurobiol. Dis. 2023;181:106100. doi: 10.1016/j.nbd.2023.106100. [DOI] [PubMed] [Google Scholar]
- 167.Kitchen P, et al. Targeting aquaporin-4 subcellular localization to treat central nervous system edema. Cell. 2020;181:784–799.e719. doi: 10.1016/j.cell.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sylvain NJ, et al. The effects of trifluoperazine on brain edema, aquaporin-4 expression and metabolic markers during the acute phase of stroke using photothrombotic mouse model. Biochim. Biophys. Acta Biomembr. 2021;1863:183573. doi: 10.1016/j.bbamem.2021.183573. [DOI] [PubMed] [Google Scholar]
- 169.Murray ER, Kemp M, Nguyen TT. The microbiota–gut–brain axis in Alzheimer’s disease: a review of taxonomic alterations and potential avenues for interventions. Arch. Clin. Neuropsychol. 2022;37:595–607. doi: 10.1093/arclin/acac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vogt NM, et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017;7:13537. doi: 10.1038/s41598-017-13601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chandra S, Sisodia SS, Vassar RJ. The gut microbiome in Alzheimer’s disease: what we know and what remains to be explored. Mol. Neurodegener. 2023;18:9. doi: 10.1186/s13024-023-00595-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kumar DKV, et al. Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Sci. Transl. Med. 2016;8:340ra372–340ra372. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Seo D-O, et al. ApoE isoform and microbiota-dependent progression of neurodegeneration in a mouse model of tauopathy. Science. 2023;379:eadd1236. doi: 10.1126/science.add1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gao L, Zhang Y, Sterling K, Song W. Brain-derived neurotrophic factor in Alzheimer’s disease and its pharmaceutical potential. Transl. Neurodegener. 2022;11:4. doi: 10.1186/s40035-022-00279-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zimbone, S. et al. Amyloid beta monomers regulate cyclic adenosine monophosphate response element binding protein functions by activating type-1 insulin-like growth factor receptors in neuronal cells. Aging Cell10.1111/acel.12684 (2018). [DOI] [PMC free article] [PubMed]
- 176.Jiao SS, et al. Brain-derived neurotrophic factor protects against tau-related neurodegeneration of Alzheimer’s disease. Transl. Psychiatry. 2016;6:e907–e907. doi: 10.1038/tp.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mary A, Eysert F, Checler F, Chami M. Mitophagy in Alzheimer’s disease: molecular defects and therapeutic approaches. Mol. Psychiatry. 2023;28:202–216. doi: 10.1038/s41380-022-01631-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Roca-Agujetas V, et al. Cholesterol alters mitophagy by impairing optineurin recruitment and lysosomal clearance in Alzheimer’s disease. Mol. Neurodegener. 2021;16:15. doi: 10.1186/s13024-021-00435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lee S-E, et al. Accumulation of APP-CTF induces mitophagy dysfunction in the iNSCs model of Alzheimer’s disease. Cell Death Discov. 2022;8:1. doi: 10.1038/s41420-021-00796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Schmukler E, et al. Altered mitochondrial dynamics and function in APOE4-expressing astrocytes. Cell Death Dis. 2020;11:578. doi: 10.1038/s41419-020-02776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71:35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 182.Rexach J, Geschwind D. Selective neuronal vulnerability in Alzheimer’s disease: a modern holy grail. Neuron. 2020;107:763–765. doi: 10.1016/j.neuron.2020.08.018. [DOI] [PubMed] [Google Scholar]
- 183.Roussarie J-P, et al. Selective neuronal vulnerability in Alzheimer’s disease: a network-based analysis. Neuron. 2020;107:821–835.e812. doi: 10.1016/j.neuron.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang X, Michaelis EK. Selective neuronal vulnerability to oxidative stress in the brain. Front. Aging Neurosci. 2010;2:12–12. doi: 10.3389/fnagi.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fu H, Hardy J, Duff KE. Selective vulnerability in neurodegenerative diseases. Nat. Neurosci. 2018;21:1350–1358. doi: 10.1038/s41593-018-0221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang X, et al. Midlife lipid and glucose levels are associated with Alzheimer’s disease. Alzheimers Dement. 2023;19:181–193. doi: 10.1002/alz.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lazar AN, et al. Time-of-flight secondary ion mass spectrometry (TOF-SIMS) imaging reveals cholesterol overload in the cerebral cortex of Alzheimer disease patients. Acta Neuropathol. 2013;125:133–144. doi: 10.1007/s00401-012-1041-1. [DOI] [PubMed] [Google Scholar]
- 188.Mosconi L. Brain glucose metabolism in the early and specific diagnosis of Alzheimer’s disease. Eur. J. Nucl. Med. Mol. Imaging. 2005;32:486–510. doi: 10.1007/s00259-005-1762-7. [DOI] [PubMed] [Google Scholar]
- 189.Duran-Aniotz C, Hetz C. Glucose metabolism: a sweet relief of Alzheimer’s disease. Curr. Biol. 2016;26:R806–809. doi: 10.1016/j.cub.2016.07.060. [DOI] [PubMed] [Google Scholar]
- 190.Niccoli T, et al. Increased glucose transport into neurons rescues Aβ toxicity in drosophila. Curr. Biol. 2016;26:2291–2300. doi: 10.1016/j.cub.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Willette AA, et al. Association of insulin resistance with cerebral glucose uptake in late middle–aged adults at risk for Alzheimer disease. JAMA Neurol. 2015;72:1013–1020. doi: 10.1001/jamaneurol.2015.0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Claxton A, et al. Sex and ApoE genotype differences in treatment response to two doses of intranasal insulin in adults with mild cognitive impairment or Alzheimer’s disease. J. Alzheimers Dis. 2013;35:789–797. doi: 10.3233/JAD-122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kyrtata N, Emsley HCA, Sparasci O, Parkes LM, Dickie BR. A systematic review of glucose transport alterations in Alzheimer’s disease. Front Neurosci. 2021;15:626636. doi: 10.3389/fnins.2021.626636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Winkler EA, et al. GLUT1 reductions exacerbate Alzheimer’s disease vasculo-neuronal dysfunction and degeneration. Nat. Neurosci. 2015;18:521–530. doi: 10.1038/nn.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Foley P. Lipids in Alzheimer’s disease: a century-old story. Biochim. Biophys. Acta. 2010;1801:750–753. doi: 10.1016/j.bbalip.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 196.Hicks, D., Nalivaeva, N. & Turner, A. Lipid rafts and Alzheimer’s disease: protein-lipid interactions and perturbation of signaling. Front. Physiol.10.3389/fphys.2012.00189 (2012). [DOI] [PMC free article] [PubMed]
- 197.Feringa, F. M. & van der Kant, R. Cholesterol and Alzheimer’s disease; from risk genes to pathological effects. Front. Aging Neurosci.10.3389/fnagi.2021.690372 (2021). [DOI] [PMC free article] [PubMed]
- 198.Gamba P, et al. A crosstalk between brain cholesterol oxidation and glucose metabolism in Alzheimer’s disease. Front Neurosci. 2019;13:556. doi: 10.3389/fnins.2019.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chaudhuri KR, Healy DG, Schapira AHV. Non-motor symptoms of Parkinson’s disease: diagnosis and management. Lancet Neurol. 2006;5:235–245. doi: 10.1016/S1474-4422(06)70373-8. [DOI] [PubMed] [Google Scholar]
- 200.Schapira AHV, Chaudhuri KR, Jenner P. Non-motor features of Parkinson disease. Nat. Rev. Neurosci. 2017;18:435–450. doi: 10.1038/nrn.2017.62. [DOI] [PubMed] [Google Scholar]
- 201.Damier P, Hirsch EC, Agid Y, Graybiel AM. The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain. 1999;122:1437–1448. doi: 10.1093/brain/122.8.1437. [DOI] [PubMed] [Google Scholar]
- 202.Cheng HC, Ulane CM, Burke RE. Clinical progression in Parkinson disease and the neurobiology of axons. Ann. Neurol. 2010;67:715–725. doi: 10.1002/ana.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wirdefeldt K, Adami H-O, Cole P, Trichopoulos D, Mandel J. Epidemiology and etiology of Parkinson’s disease: a review of the evidence. Eur. J. Epidemiol. 2011;26:1. doi: 10.1007/s10654-011-9581-6. [DOI] [PubMed] [Google Scholar]
- 204.Blauwendraat C, Nalls MA, Singleton AB. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020;19:170–178. doi: 10.1016/S1474-4422(19)30287-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Funayama M, Nishioka K, Li Y, Hattori N. Molecular genetics of Parkinson’s disease: contributions and global trends. J. Hum. Genet. 2023;68:125–130. doi: 10.1038/s10038-022-01058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fung H-C, et al. Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5:911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 207.Simón-Sánchez J, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chartier-Harlin M-C, et al. α-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 209.Pollanen MS, Dickson DW, Bergeron C. Pathology and biology of the Lewy body. J. Neuropathol. Exp. Neurol. 1993;52:183–191. doi: 10.1097/00005072-199305000-00001. [DOI] [PubMed] [Google Scholar]
- 210.Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry. 1988;51:745–752. doi: 10.1136/jnnp.51.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Spillantini MG, et al. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 212.McCann H, Stevens CH, Cartwright H, Halliday GM. α-Synucleinopathy phenotypes. Parkinsonism Relat. Disord. 2014;20:S62–S67. doi: 10.1016/S1353-8020(13)70017-8. [DOI] [PubMed] [Google Scholar]
- 213.Sulzer D, Edwards RH. The physiological role of α-synuclein and its relationship to Parkinson’s Disease. J. Neurochem. 2019;150:475–486. doi: 10.1111/jnc.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chandra S, Gallardo G, Fernández-Chacón R, Schlüter OM, Südhof TC. α-Synuclein cooperates with CSPα in preventing neurodegeneration. Cell. 2005;123:383–396. doi: 10.1016/j.cell.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 215.Bendor, J., Logan, T. & Edwards, R. H. The dunction of α-synuclein. Neuron79, 1044–1066 (2013). [DOI] [PMC free article] [PubMed]
- 216.Bartels T, et al. The N-terminus of the intrinsically disordered protein α-synuclein triggers membrane binding and helix folding. Biophys. J. 2010;99:2116–2124. doi: 10.1016/j.bpj.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 218.Chartier S, Duyckaerts C. Is Lewy pathology in the human nervous system chiefly an indicator of neuronal protection or of toxicity? Cell Tissue Res. 2018;373:149–160. doi: 10.1007/s00441-018-2854-6. [DOI] [PubMed] [Google Scholar]
- 219.Bengoa-Vergniory N, Roberts RF, Wade-Martins R, Alegre-Abarrategui J. Alpha-synuclein oligomers: a new hope. Acta Neuropathol. 2017;134:819–838. doi: 10.1007/s00401-017-1755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Danzer KM, et al. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol. Neurodegener. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ingelsson, M. Alpha-synuclein oligomers—neurotoxic molecules in Parkinson’s disease and other Lewy body disorders. Front. Neurosci.10.3389/fnins.2016.00408 (2016). [DOI] [PMC free article] [PubMed]
- 222.Winner B, et al. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl Acad. Sci. USA. 2011;108:4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang W, et al. A soluble α-synuclein construct forms a dynamic tetramer. Proc. Natl Acad. Sci. USA. 2011;108:17797–17802. doi: 10.1073/pnas.1113260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bartels T, Choi JG, Selkoe DJ. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Narkiewicz J, Giachin G, Legname G. In vitro aggregation assays for the characterization of α-synuclein prion-like properties. Prion. 2014;8:19–32. doi: 10.4161/pri.28125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Yagi H, Kusaka E, Hongo K, Mizobata T, Kawata Y. Amyloid fibril formation of alpha-synuclein is accelerated by preformed amyloid seeds of other proteins: implications for the mechanism of transmissible conformational diseases. J. Biol. Chem. 2005;280:38609–38616. doi: 10.1074/jbc.M508623200. [DOI] [PubMed] [Google Scholar]
- 227.Bousset L, et al. Structural and functional characterization of two alpha-synuclein strains. Nat. Commun. 2013;4:2575. doi: 10.1038/ncomms3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Luk KC, et al. Intracerebral inoculation of pathological α-synuclein initiates a rapidly progressive neurodegenerative α-synucleinopathy in mice. J. Exp. Med. 2012;209:975–986. doi: 10.1084/jem.20112457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mougenot A-L, et al. Prion-like acceleration of a synucleinopathy in a transgenic mouse model. Neurobiol. Aging. 2012;33:2225–2228. doi: 10.1016/j.neurobiolaging.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 230.Tanudjojo B, et al. Phenotypic manifestation of α-synuclein strains derived from Parkinson’s disease and multiple system atrophy in human dopaminergic neurons. Nat. Commun. 2021;12:3817. doi: 10.1038/s41467-021-23682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nat. Rev. Neurol. 2013;9:13–24. doi: 10.1038/nrneurol.2012.242. [DOI] [PubMed] [Google Scholar]
- 232.Visanji NP, Brooks PL, Hazrati L-N, Lang AE. The prion hypothesis in Parkinson’s disease: Braak to the future. Acta Neuropathol. Commun. 2013;1:2. doi: 10.1186/2051-5960-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Polymeropoulos MH, et al. Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science. 1996;274:1197–1199. doi: 10.1126/science.274.5290.1197. [DOI] [PubMed] [Google Scholar]
- 234.Fuchs J, et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68:916–922. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 235.Zarranz JJ, et al. The new mutation, E46K, of α-synuclein causes parkinson and Lewy body dementia. Ann. Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 236.Appel-Cresswell S, et al. Alpha-synuclein p.H50Q, a novel pathogenic mutation for Parkinson’s disease. Mov. Disord. 2013;28:811–813. doi: 10.1002/mds.25421. [DOI] [PubMed] [Google Scholar]
- 237.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 238.Ruggeri FS, et al. The influence of pathogenic mutations in α-synuclein on biophysical and structural characteristics of amyloid fibrils. ACS Nano. 2020;14:5213–5222. doi: 10.1021/acsnano.9b09676. [DOI] [PubMed] [Google Scholar]
- 239.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 240.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 241.Hipp MS, Park S-H, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 242.Hipp MS, Kasturi P, Hartl FU. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019;20:421–435. doi: 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- 243.Kuzuhara S, Mori H, Izumiyama N, Yoshimura M, Ihara Y. Lewy bodies are ubiquitinated. Acta Neuropathol. 1988;75:345–353. doi: 10.1007/BF00687787. [DOI] [PubMed] [Google Scholar]
- 244.Ii K, Ito H, Tanaka K, Hirano A. Immunocytochemical co-localization of the proteasome in ubiquitinated structures in neurodegenerative diseases and the elderly. J. Neuropathol. Exp. Neurol. 1997;56:125–131. doi: 10.1097/00005072-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 245.Bedford L, et al. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J. Neurosci. 2008;28:8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sun F, et al. Proteasome inhibitor MG-132 induces dopaminergic degeneration in cell culture and animal models. NeuroToxicology. 2006;27:807–815. doi: 10.1016/j.neuro.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 247.Schapira AHV, et al. Proteasomal inhibition causes loss of nigral tyrosine hydroxylase neurons. Ann. Neurol. 2006;60:253–255. doi: 10.1002/ana.20934. [DOI] [PubMed] [Google Scholar]
- 248.McKinnon C, et al. Early-onset impairment of the ubiquitin-proteasome system in dopaminergic neurons caused by α-synuclein. Acta Neuropathol. Commun. 2020;8:17. doi: 10.1186/s40478-020-0894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Bellomo G, Paciotti S, Gatticchi L, Parnetti L. The vicious cycle between α-synuclein aggregation and autophagic-lysosomal dysfunction. Mov. Disord. 2020;35:34–44. doi: 10.1002/mds.27895. [DOI] [PubMed] [Google Scholar]
- 250.Usenovic M, Tresse E, Mazzulli JR, Taylor JP, Krainc D. Deficiency of ATP13A2 leads to lysosomal dysfunction, α-synuclein accumulation, and neurotoxicity. J. Neurosci. 2012;32:4240–4246. doi: 10.1523/JNEUROSCI.5575-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Murphy KE, et al. Lysosomal-associated membrane protein 2 isoforms are differentially affected in early Parkinson’s disease. Mov. Disord. 2015;30:1639–1647. doi: 10.1002/mds.26141. [DOI] [PubMed] [Google Scholar]
- 252.Martinez-Vicente M, et al. Dopamine-modified α-synuclein blocks chaperone-mediated autophagy. J. Clin. Investig. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Mazzulli JR, Zunke F, Isacson O, Studer L, Krainc D. α-Synuclein–induced lysosomal dysfunction occurs through disruptions in protein trafficking in human midbrain synucleinopathy models. Proc. Natl Acad. Sci. USA. 2016;113:1931–1936. doi: 10.1073/pnas.1520335113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Cramb KML, Beccano-Kelly D, Cragg SJ, Wade-Martins R. Impaired dopamine release in Parkinson’s disease. Brain. 2023;146:3117–3132. doi: 10.1093/brain/awad064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rinne JO, et al. Increased density of dopamine D2 receptors in the putamen, but not in the caudate nucleus in early Parkinson’s disease: a PET study with [11C]raclopride. J. Neurol. Sci. 1995;132:156–161. doi: 10.1016/0022-510X(95)00137-Q. [DOI] [PubMed] [Google Scholar]
- 256.Block ML, Zecca L, Hong JS. Microglia-mediated neurotoxicity: uncovering the molecular mechanisms. Nat. Rev. Neurosci. 2007;8:57–69. doi: 10.1038/nrn2038. [DOI] [PubMed] [Google Scholar]
- 257.Wang J, Wang F, Mai D, Qu S. Molecular mechanisms of glutamate toxicity in Parkinson’s disease. Front. Neurosci. 2020;14:585584. doi: 10.3389/fnins.2020.585584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Borta A, Höglinger GU. Dopamine and adult neurogenesis. J. Neurochem. 2007;100:587–595. doi: 10.1111/j.1471-4159.2006.04241.x. [DOI] [PubMed] [Google Scholar]
- 259.Kamath T, et al. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat. Neurosci. 2022;25:588–595. doi: 10.1038/s41593-022-01061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 261.Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 262.Langston JW, Irwin I, Langston EB, Forno LS. 1-methyl-4-phenylpyridinium ion (MPP+): identification of a metabolite of MPTP, a toxin selective to the substantia nigra. Neurosci. Lett. 1984;48:87–92. doi: 10.1016/0304-3940(84)90293-3. [DOI] [PubMed] [Google Scholar]
- 263.Watanabe Y, Himeda T, Araki T. Mechanisms of MPTP toxicity and their implications for therapy of Parkinson’s disease. Med. Sci. Monit. 2005;11:RA17–RA23. [PubMed] [Google Scholar]
- 264.Abou-Sleiman PM, Muqit MMK, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat. Rev. Neurosci. 2006;7:207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- 265.Exner N, Lutz AK, Haass C, Winklhofer KF. Mitochondrial dysfunction in Parkinson’s disease: molecular mechanisms and pathophysiological consequences. EMBO J. 2012;31:3038–3062. doi: 10.1038/emboj.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Dauer W, et al. Resistance of α-synuclein null mice to the parkinsonian neurotoxin MPTP. Proc. Natl Acad. Sci. USA. 2002;99:14524–14529. doi: 10.1073/pnas.172514599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Thomas B, et al. Resistance to MPTP-neurotoxicity in α-synuclein knockout mice is complemented by human α-synuclein and associated with increased β-synuclein and Akt activation. PLoS ONE. 2011;6:e16706. doi: 10.1371/journal.pone.0016706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Mullin S, Schapira A. α-synuclein and mitochondrial dysfunction in Parkinson’s disease. Mol. Neurobiol. 2013;47:587–597. doi: 10.1007/s12035-013-8394-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Grassi D, et al. Identification of a highly neurotoxic α-synuclein species inducing mitochondrial damage and mitophagy in Parkinson’s disease. Proc. Natl Acad. Sci. USA. 2018;115:E2634–E2643. doi: 10.1073/pnas.1713849115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zambon F, et al. Cellular α-synuclein pathology is associated with bioenergetic dysfunction in Parkinson’s iPSC-derived dopamine neurons. Hum. Mol. Genet. 2019;28:2001–2013. doi: 10.1093/hmg/ddz038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ryan BJ, Hoek S, Fon EA, Wade-Martins R. Mitochondrial dysfunction and mitophagy in Parkinson’s: from familial to sporadic disease. Trends Biochem. Sci. 2015;40:200–210. doi: 10.1016/j.tibs.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 272.Larsen SB, Hanss Z, Krüger R. The genetic architecture of mitochondrial dysfunction in Parkinson’s disease. Cell Tissue Res. 2018;373:21–37. doi: 10.1007/s00441-017-2768-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Pickrell AliciaM, Youle RichardJ. The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Ge P, Dawson VL, Dawson TM. PINK1 and Parkin mitochondrial quality control: a source of regional vulnerability in Parkinson’s disease. Mol. Neurodegener. 2020;15:20. doi: 10.1186/s13024-020-00367-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 276.Quinn PMJ, Moreira PI, Ambrósio AF, Alves CH. PINK1/PARKIN signalling in neurodegeneration and neuroinflammation. Acta Neuropathol. Commun. 2020;8:189. doi: 10.1186/s40478-020-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Bonifati V, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 278.Dolgacheva LP, Berezhnov AV, Fedotova EI, Zinchenko VP, Abramov AY. Role of DJ-1 in the mechanism of pathogenesis of Parkinson’s disease. J. Bioenerg. Biomembr. 2019;51:175–188. doi: 10.1007/s10863-019-09798-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Guzman JN, et al. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhang L, et al. Mitochondrial localization of the Parkinson’s disease related protein DJ-1: implications for pathogenesis. Hum. Mol. Genet. 2005;14:2063–2073. doi: 10.1093/hmg/ddi211. [DOI] [PubMed] [Google Scholar]
- 281.Taira T, et al. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Canet-Avilés RM, et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl Acad. Sci. USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Nabi M, Tabassum N. Role of environmental toxicants on neurodegenerative disorders. Front. Toxicol. 2022;4:837579. doi: 10.3389/ftox.2022.837579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Ramirez Ortega D, et al. Kynurenine pathway as a new target of cognitive impairment induced by lead toxicity during the lactation. Sci. Rep. 2020;10:3184. doi: 10.1038/s41598-020-60159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Cannon JR, Greenamyre JT. The role of environmental exposures in neurodegeneration and neurodegenerative diseases. Toxicol. Sci. 2011;124:225–250. doi: 10.1093/toxsci/kfr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Liu Z, et al. Anionic nanoplastic contaminants promote Parkinson’s disease–associated α-synuclein aggregation. Sci. Adv. 2023;9:eadi8716. doi: 10.1126/sciadv.adi8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Mejzini R, et al. ALS genetics, mechanisms, and therapeutics: where are we now? Front. Neurosci. 2019;13:1310. doi: 10.3389/fnins.2019.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Suzuki, N., Nishiyama, A., Warita, H. & Aoki, M. Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy. J. Hum. Genet.10.1038/s10038-022-01055-8 (2022). [DOI] [PMC free article] [PubMed]
- 289.Smukowski SN, et al. Progress in amyotrophic lateral sclerosis gene discovery: reflecting on classic approaches and leveraging emerging technologies. Neurol Genet. 2022;8:e669. doi: 10.1212/NXG.0000000000000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Talbott EO, Malek AM, Lacomis D. The epidemiology of amyotrophic lateral sclerosis. Handb. Clin. Neurol. 2016;138:225–238. doi: 10.1016/B978-0-12-802973-2.00013-6. [DOI] [PubMed] [Google Scholar]
- 291.Saberi S, Stauffer JE, Schulte DJ, Ravits J. Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol. Clin. 2015;33:855–876. doi: 10.1016/j.ncl.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Goldstein LH, Abrahams S. Changes in cognition and behaviour in amyotrophic lateral sclerosis: nature of impairment and implications for assessment. Lancet Neurol. 2013;12:368–380. doi: 10.1016/S1474-4422(13)70026-7. [DOI] [PubMed] [Google Scholar]
- 293.Sanes JR, Lichtman JW. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat. Rev. Neurosci. 2001;2:791–805. doi: 10.1038/35097557. [DOI] [PubMed] [Google Scholar]
- 294.Chowdhury A, Mukherjee A, Sinharoy U, Pandit A, Biswas A. Non-motor features of amyotrophic lateral sclerosis: a clinic-based study. Ann. Indian Acad. Neurol. 2021;24:745–753. doi: 10.4103/aian.AIAN_51_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Blokhuis AM, Groen EJN, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125:777–794. doi: 10.1007/s00401-013-1125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Hayashi Y, Homma K, Ichijo H. SOD1 in neurotoxicity and its controversial roles in SOD1 mutation-negative ALS. Adv. Biol. Regul. 2016;60:95–104. doi: 10.1016/j.jbior.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 297.Forsberg K, et al. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS ONE. 2010;5:e11552. doi: 10.1371/journal.pone.0011552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Paré B, et al. Misfolded SOD1 pathology in sporadic amyotrophic lateral sclerosis. Sci. Rep. 2018;8:14223. doi: 10.1038/s41598-018-31773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Berdyński M, et al. SOD1 mutations associated with amyotrophic lateral sclerosis analysis of variant severity. Sci. Rep. 2022;12:103. doi: 10.1038/s41598-021-03891-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Benkler C, et al. Aggregated SOD1 causes selective death of cultured human motor neurons. Sci. Rep. 2018;8:16393. doi: 10.1038/s41598-018-34759-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Forsberg K, Andersen PM, Marklund SL, Brännström T. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:623–634. doi: 10.1007/s00401-011-0805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ayers JI, Fromholt SE, O’Neal VM, Diamond JH, Borchelt DR. Prion-like propagation of mutant SOD1 misfolding and motor neuron disease spread along neuroanatomical pathways. Acta Neuropathol. 2016;131:103–114. doi: 10.1007/s00401-015-1514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Grad LI, et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl Acad. Sci. USA. 2014;111:3620–3625. doi: 10.1073/pnas.1312245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Münch C, O’Brien J, Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc. Natl Acad. Sci. USA. 2011;108:3548–3553. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Thomas EV, Fenton WA, McGrath J, Horwich AL. Transfer of pathogenic and nonpathogenic cytosolic proteins between spinal cord motor neurons in vivo in chimeric mice. Proc. Natl Acad. Sci. USA. 2017;114:E3139–E3148. doi: 10.1073/pnas.1701465114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Da Cruz S, et al. Misfolded SOD1 is not a primary component of sporadic ALS. Acta Neuropathol. 2017;134:97–111. doi: 10.1007/s00401-017-1688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 308.Xu Z, Yang C. TDP-43-the key to understanding amyotrophic lateral sclerosis. Rare Dis. 2014;2:e944443. doi: 10.4161/21675511.2014.944443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Arseni D, et al. Structure of pathological TDP-43 filaments from ALS with FTLD. Nature. 2022;601:139–143. doi: 10.1038/s41586-021-04199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Lipstein N, et al. Munc13-1 is a Ca2+-phospholipid-dependent vesicle priming hub that shapes synaptic short-term plasticity and enables sustained neurotransmission. Neuron. 2021;109:3980–4000.e3987. doi: 10.1016/j.neuron.2021.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Brown A-L, et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature. 2022;603:131–137. doi: 10.1038/s41586-022-04436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Ma XR, et al. TDP-43 represses cryptic exon inclusion in the FTD–ALS gene UNC13A. Nature. 2022;603:124–130. doi: 10.1038/s41586-022-04424-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Vogler TO, et al. TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature. 2018;563:508–513. doi: 10.1038/s41586-018-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Keller BA, et al. Co-aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism. Acta Neuropathol. 2012;124:733–747. doi: 10.1007/s00401-012-1035-z. [DOI] [PubMed] [Google Scholar]
- 315.Choi SY, et al. C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nat. Neurosci. 2019;22:851–862. doi: 10.1038/s41593-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.May S, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128:485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Mizielinska S, et al. C9orf72 repeat expansions cause neurodegeneration in drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Ryan S, Rollinson S, Hobbs E, Pickering-Brown S. C9orf72 dipeptides disrupt the nucleocytoplasmic transport machinery and cause TDP-43 mislocalisation to the cytoplasm. Sci. Rep. 2022;12:4799. doi: 10.1038/s41598-022-08724-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tran H, et al. Suppression of mutant C9orf72 expression by a potent mixed backbone antisense oligonucleotide. Nat. Med. 2022;28:117–124. doi: 10.1038/s41591-021-01557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Liu, J. & Wang, F. Role of neuroinflammation in amyotrophic lateral sclerosis: cellular mechanisms and therapeutic implications. Front. Immunol.10.3389/fimmu.2017.01005 (2017). [DOI] [PMC free article] [PubMed]
- 322.Liu, E., Karpf, L. & Bohl, D. Neuroinflammation in amyotrophic lateral sclerosis and frontotemporal dementia and the interest of induced pluripotent stem cells to study immune cells interactions with neurons. Front. Mol. Neurosci.10.3389/fnmol.2021.767041 (2021). [DOI] [PMC free article] [PubMed]
- 323.Liao B, Zhao W, Beers DR, Henkel JS, Appel SH. Transformation from a neuroprotective to a neurotoxic microglial phenotype in a mouse model of ALS. Exp. Neurol. 2012;237:147–152. doi: 10.1016/j.expneurol.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Nagai M, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Howland DS, et al. Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS) Proc. Natl Acad. Sci. USA. 2002;99:1604–1609. doi: 10.1073/pnas.032539299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Ferraiuolo L, et al. Dysregulation of astrocyte–motoneuron cross-talk in mutant superoxide dismutase 1-related amyotrophic lateral sclerosis. Brain. 2011;134:2627–2641. doi: 10.1093/brain/awr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Hensley K, et al. Primary glia expressing the G93A-SOD1 mutation present a neuroinflammatory phenotype and provide a cellular system for studies of glial inflammation. J. Neuroinflamm. 2006;3:1–9. doi: 10.1186/1742-2094-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Marchetto MC, et al. Non-cell-autonomous effect of human SOD1G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 329.Re DianeB, et al. Necroptosis drives motor neuron death in models of both sporadic and familial ALS. Neuron. 2014;81:1001–1008. doi: 10.1016/j.neuron.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Ferraiuolo L, et al. Oligodendrocytes contribute to motor neuron death in ALS via SOD1-dependent mechanism. Proc. Natl Acad. Sci. USA. 2016;113:E6496–E6505. doi: 10.1073/pnas.1607496113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Nakaya T, Maragkakis M. Amyotrophic lateral sclerosis associated FUS mutation shortens mitochondria and induces neurotoxicity. Sci. Rep. 2018;8:1–15. doi: 10.1038/s41598-018-33964-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Magrané J, Cortez C, Gan W-B, Manfredi G. Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum. Mol. Genet. 2013;23:1413–1424. doi: 10.1093/hmg/ddt528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Lopez-Gonzalez R, et al. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron. 2016;92:383–391. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Singh T, et al. Neuronal mitochondrial dysfunction in sporadic amyotrophic lateral sclerosis is developmentally regulated. Sci. Rep. 2021;11:18916. doi: 10.1038/s41598-021-97928-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Butti, Z. & Patten, S. A. RNA Dysregulation in amyotrophic lateral sclerosis. Front. Genet.10.3389/fgene.2018.00712 (2019). [DOI] [PMC free article] [PubMed]
- 336.Buratti, E. & Baralle, F. E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci.13, 867–878 (2008). [DOI] [PubMed]
- 337.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Ross CA, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol. 2014;10:204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- 339.Johnson EB, et al. Dynamics of cortical degeneration over a decade in Huntington’s disease. Biol. Psychiatry. 2021;89:807–816. doi: 10.1016/j.biopsych.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Rosas HD, et al. Cerebral cortex and the clinical expression of Huntington’s disease: complexity and heterogeneity. Brain. 2008;131:1057–1068. doi: 10.1093/brain/awn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell72, 971–983 (1993). [DOI] [PubMed]
- 342.Nasir J, et al. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 343.Zuccato C, Cattaneo E. Huntington’s disease. Handb. Exp. Pharmacol. 2014;220:357–409. doi: 10.1007/978-3-642-45106-5_14. [DOI] [PubMed] [Google Scholar]
- 344.Kennedy L, et al. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 345.Mangiarini L, et al. Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nat. Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- 346.Rosenblatt A, et al. Age, CAG repeat length, and clinical progression in Huntington’s disease. Mov. Disord. 2012;27:272–276. doi: 10.1002/mds.24024. [DOI] [PubMed] [Google Scholar]
- 347.Aviolat H, et al. Assessing average somatic CAG repeat instability at the protein level. Sci. Rep. 2019;9:19152. doi: 10.1038/s41598-019-55202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Kim M. Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues. Prion. 2013;7:221–228. doi: 10.4161/pri.23807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 350.Harper SQ, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Sathasivam K, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl Acad. Sci. USA. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Benn CL, et al. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington’s disease. Hum. Mol. Genet. 2005;14:3065–3078. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- 353.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/S0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 354.Neueder A, et al. The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington’s disease patients. Sci. Rep. 2017;7:1307. doi: 10.1038/s41598-017-01510-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Hoffner G, Island ML, Djian P. Purification of neuronal inclusions of patients with Huntington’s disease reveals a broad range of N-terminal fragments of expanded huntingtin and insoluble polymers. J. Neurochem. 2005;95:125–136. doi: 10.1111/j.1471-4159.2005.03348.x. [DOI] [PubMed] [Google Scholar]
- 356.Yang H, et al. Truncation of mutant huntingtin in knock-in mice demonstrates exon1 huntingtin is a key pathogenic form. Nat. Commun. 2020;11:2582. doi: 10.1038/s41467-020-16318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 358.Miller J, et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat. Chem. Biol. 2011;7:925–934. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Nucifora LG, et al. Identification of novel potentially toxic oligomers formed in vitro from mammalian-derived expanded huntingtin exon-1 protein. J. Biol. Chem. 2012;287:16017–16028. doi: 10.1074/jbc.M111.252577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.DiGiovanni LF, Mocle AJ, Xia J, Truant R. Huntingtin N17 domain is a reactive oxygen species sensor regulating Huntingtin phosphorylation and localization. Hum. Mol. Genet. 2016;25:3937–3945. doi: 10.1093/hmg/ddw234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Moldovean SN, Chiş V. Molecular dynamics simulations applied to structural and dynamical transitions of the Huntingtin protein: a review. ACS Chem. Neurosci. 2020;11:105–120. doi: 10.1021/acschemneuro.9b00561. [DOI] [PubMed] [Google Scholar]
- 362.Owada R, Mitsui S, Nakamura K. Exogenous polyserine and polyleucine are toxic to recipient cells. Sci. Rep. 2022;12:1685. doi: 10.1038/s41598-022-05720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Ramdzan YM, et al. Huntingtin inclusions trigger cellular quiescence, deactivate apoptosis, and lead to delayed necrosis. Cell Rep. 2017;19:919–927. doi: 10.1016/j.celrep.2017.04.029. [DOI] [PubMed] [Google Scholar]
- 364.Layburn FE, et al. N-terminal mutant huntingtin deposition correlates with CAG repeat length and symptom onset, but not neuronal loss in Huntington’s disease. Neurobiol. Dis. 2022;174:105884. doi: 10.1016/j.nbd.2022.105884. [DOI] [PubMed] [Google Scholar]
- 365.Costanzo M, et al. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J. Cell Sci. 2013;126:3678–3685. doi: 10.1242/jcs.126086. [DOI] [PubMed] [Google Scholar]
- 366.Babcock DT, Ganetzky B. Transcellular spreading of huntingtin aggregates in the drosophila brain. Proc. Natl Acad. Sci. USA. 2015;112:E5427–E5433. doi: 10.1073/pnas.1516217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Pearce MMP, Spartz EJ, Hong W, Luo L, Kopito RR. Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat. Commun. 2015;6:6768. doi: 10.1038/ncomms7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Lin JT, et al. Regulation of feedback between protein kinase A and the proteasome system worsens Huntington’s disease. Mol. Cell. Biol. 2013;33:1073–1084. doi: 10.1128/MCB.01434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 370.Ly S, et al. Mutant huntingtin messenger RNA forms neuronal nuclear clusters in rodent and human brains. Brain Commun. 2022;4:fcac248. doi: 10.1093/braincomms/fcac248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Gratuze M, Cisbani G, Cicchetti F, Planel E. Is Huntington’s disease a tauopathy? Brain. 2016;139:1014–1025. doi: 10.1093/brain/aww021. [DOI] [PubMed] [Google Scholar]
- 372.Vuono R, et al. The role of tau in the pathological process and clinical expression of Huntington’s disease. Brain. 2015;138:1907–1918. doi: 10.1093/brain/awv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Fernández-Nogales M, et al. Huntington’s disease is a four-repeat tauopathy with tau nuclear rods. Nat. Med. 2014;20:881–885. doi: 10.1038/nm.3617. [DOI] [PubMed] [Google Scholar]
- 374.Blum D, et al. Mutant huntingtin alters Tau phosphorylation and subcellular distribution. Hum. Mol. Genet. 2015;24:76–85. doi: 10.1093/hmg/ddu421. [DOI] [PubMed] [Google Scholar]
- 375.Gratuze M, et al. Tau hyperphosphorylation and deregulation of calcineurin in mouse models of Huntington’s disease. Hum. Mol. Genet. 2015;24:86–99. doi: 10.1093/hmg/ddu456. [DOI] [PubMed] [Google Scholar]
- 376.Lahue RS. New developments in Huntington’s disease and other triplet repeat diseases: DNA repair turns to the dark side. Neuronal Signal. 2020;4:Ns20200010. doi: 10.1042/NS20200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Lee J-M, et al. CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell. 2019;178:887–900.e814. doi: 10.1016/j.cell.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Bunting E, et al. I01|Msh3-targeting antisense oligonucleotides halt CAG repeat expansions in Huntington’s disease IPSC-derived neurons. J. Neurol. Neurosurg. Psychiatry. 2022;93:A85–A85. [Google Scholar]
- 379.McAllister B, et al. Exome sequencing of individuals with Huntington’s disease implicates FAN1 nuclease activity in slowing CAG expansion and disease onset. Nat. Neurosci. 2022;25:446–457. doi: 10.1038/s41593-022-01033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Maiuri T, et al. Huntingtin is a scaffolding protein in the ATM oxidative DNA damage response complex. Hum. Mol. Genet. 2016;26:395–406. doi: 10.1093/hmg/ddw395. [DOI] [PubMed] [Google Scholar]
- 381.Hodges A, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 382.Guo S, et al. MicroRNA editing patterns in Huntington’s disease. Sci. Rep. 2022;12:3173. doi: 10.1038/s41598-022-06970-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Hervás-Corpión I, et al. Early alteration of epigenetic-related transcription in Huntington’s disease mouse models. Sci. Rep. 2018;8:9925. doi: 10.1038/s41598-018-28185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Mouro Pinto R, et al. Patterns of CAG repeat instability in the central nervous system and periphery in Huntington’s disease and in spinocerebellar ataxia type 1. Hum. Mol. Genet. 2020;29:2551–2567. doi: 10.1093/hmg/ddaa139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Vonsattel JP, et al. Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 386.Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 387.Victor MB, et al. Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat. Neurosci. 2018;21:341–352. doi: 10.1038/s41593-018-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Cicchetti F, Soulet D, Freeman TB. Neuronal degeneration in striatal transplants and Huntington’s disease: potential mechanisms and clinical implications. Brain. 2011;134:641–652. doi: 10.1093/brain/awq328. [DOI] [PubMed] [Google Scholar]
- 389.Olney JW, Gubareff TD. Glutamate neurotoxicity and Huntington’s chorea. Nature. 1978;271:557–559. doi: 10.1038/271557a0. [DOI] [PubMed] [Google Scholar]
- 390.Skotte NH, et al. Integrative characterization of the R6/2 mouse model of Huntington’s disease reveals dysfunctional astrocyte metabolism. Cell Rep. 2018;23:2211–2224. doi: 10.1016/j.celrep.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 391.Braz BY, et al. Treating early postnatal circuit defect delays Huntington’s disease onset and pathology in mice. Science. 2022;377:eabq5011. doi: 10.1126/science.abq5011. [DOI] [PubMed] [Google Scholar]
- 392.Kim J, et al. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum. Mol. Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Johri A, Chandra A, Flint Beal M. PGC-1α, mitochondrial dysfunction, and Huntington’s disease. Free Radic. Biol. Med. 2013;62:37–46. doi: 10.1016/j.freeradbiomed.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Browne SE, Beal MF. The energetics of Huntington’s disease. Neurochem. Res. 2004;29:531–546. doi: 10.1023/B:NERE.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 395.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 396.Panov AV, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 397.Manczak M, Reddy PH. Mitochondrial division inhibitor 1 protects against mutant huntingtin-induced abnormal mitochondrial dynamics and neuronal damage in Huntington’s disease. Hum. Mol. Genet. 2015;24:7308–7325. doi: 10.1093/hmg/ddv429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Haun F, et al. S-nitrosylation of dynamin-related protein 1 mediates mutant huntingtin-induced mitochondrial fragmentation and neuronal injury in Huntington’s disease. Antioxid. Redox Signal. 2013;19:1173–1184. doi: 10.1089/ars.2012.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Altar CA, et al. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature. 1997;389:856–860. doi: 10.1038/39885. [DOI] [PubMed] [Google Scholar]
- 400.Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 401.Zuccato C, et al. Loss of Huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 402.Hong Y, Zhao T, Li XJ, Li S. Mutant Huntingtin impairs BDNF release from astrocytes by disrupting conversion of Rab3a-GTP into Rab3a-GDP. J. Neurosci. 2016;36:8790–8801. doi: 10.1523/JNEUROSCI.0168-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Rocha, N. P. et al. Microglia activation in basal ganglia is a late event in Huntington disease pathophysiology. Neurol. Neuroimmunol. Neuroinflamm.10.1212/nxi.0000000000000984 (2021). [DOI] [PMC free article] [PubMed]
- 404.Khakh BS, et al. Unravelling and exploiting astrocyte dysfunction in Huntington’s disease. Trends Neurosci. 2017;40:422–437. doi: 10.1016/j.tins.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Lee H-G, Wheeler MA, Quintana FJ. Function and therapeutic value of astrocytes in neurological diseases. Nat. Rev. Drug Discov. 2022;21:339–358. doi: 10.1038/s41573-022-00390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Benraiss A, et al. Human glia can both induce and rescue aspects of disease phenotype in Huntington disease. Nat. Commun. 2016;7:11758. doi: 10.1038/ncomms11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Faideau M, et al. In vivo expression of polyglutamine-expanded huntingtin by mouse striatal astrocytes impairs glutamate transport: a correlation with Huntington’s disease subjects. Hum. Mol. Genet. 2010;19:3053–3067. doi: 10.1093/hmg/ddq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Shin J-Y, et al. Expression of mutant Huntingtin in glial cells contributes to neuronal excitotoxicity. J. Cell Biol. 2005;171:1001–1012. doi: 10.1083/jcb.200508072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.O’Regan GC, et al. Human Huntington’s disease pluripotent stem cell-derived microglia develop normally but are abnormally hyper-reactive and release elevated levels of reactive oxygen species. J. Neuroinflamm. 2021;18:94. doi: 10.1186/s12974-021-02147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Tan H-Y, Cho H, Lee LP. Human mini-brain models. Nat. Biomed. Eng. 2021;5:11–25. doi: 10.1038/s41551-020-00643-3. [DOI] [PubMed] [Google Scholar]
- 412.Fang, G., Chen, Y.-C., Lu, H. & Jin, D. Advances in spheroids and organoids on a chip. Adv. Funct. Mater.33, 2215043 (2023).
- 413.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Huh D, et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174,. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 416.Musah, S. et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed. Eng.10.1038/s41551-017-0069 (2017). [DOI] [PMC free article] [PubMed]
- 417.Azevedo FA, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 2009;513:532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- 418.Piwecka M, Rajewsky N, Rybak-Wolf A. Single-cell and spatial transcriptomics: deciphering brain complexity in health and disease. Nat. Rev. Neurol. 2023;19:346–362. doi: 10.1038/s41582-023-00809-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Yokoyama, M., Kobayashi, H., Tatsumi, L. & Tomita, T. Mouse models of Alzheimer’s disease. Front. Mol. Neurosci.10.3389/fnmol.2022.912995 (2022). [DOI] [PMC free article] [PubMed]
- 420.Dovonou A, et al. Animal models of Parkinson’s disease: bridging the gap between disease hallmarks and research questions. Transl. Neurodegener. 2023;12:36. doi: 10.1186/s40035-023-00368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Fisher EMC, et al. Opinion: more mouse models and more translation needed for ALS. Mol. Neurodegener. 2023;18:30. doi: 10.1186/s13024-023-00619-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Pouladi MA, Morton AJ, Hayden MR. Choosing an animal model for the study of Huntington’s disease. Nat. Rev. Neurosci. 2013;14:708–721. doi: 10.1038/nrn3570. [DOI] [PubMed] [Google Scholar]
- 423.Uhl EW, Warner NJ. Mouse models as predictors of human responses: evolutionary medicine. Curr. Pathobiol. Rep. 2015;3:219–223. doi: 10.1007/s40139-015-0086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Semple BD, Blomgren K, Gimlin K, Ferriero DM, Noble-Haeusslein LJ. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 2013;106-107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Baglietto-Vargas D, et al. Generation of a humanized Aβ expressing mouse demonstrating aspects of Alzheimer’s disease-like pathology. Nat. Commun. 2021;12:2421. doi: 10.1038/s41467-021-22624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Balducci C, et al. Synthetic amyloid-β oligomers impair long-term memory independently of cellular prion protein. Proc. Natl Acad. Sci. USA. 2010;107:2295–2300. doi: 10.1073/pnas.0911829107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Luk KC, et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Chung HK, Ho HA, Pérez-Acuña D, Lee SJ. Modeling α-synuclein propagation with preformed fibril injections. J. Mov. Disord. 2019;12:139–151. doi: 10.14802/jmd.19046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Shadrina, M. & Slominsky, P. Modeling Parkinson’s disease: not only rodents? Front. Aging Neurosci.10.3389/fnagi.2021.695718 (2021). [DOI] [PMC free article] [PubMed]
- 430.Nistor G, et al. Derivation of high purity neuronal progenitors from human embryonic stem cells. PLoS ONE. 2011;6:e20692. doi: 10.1371/journal.pone.0020692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Rivetti di Val Cervo P, Besusso D, Conforti P, Cattaneo E. hiPSCs for predictive modelling of neurodegenerative diseases: dreaming the possible. Nat. Rev. Neurol. 2021;17:381–392. doi: 10.1038/s41582-021-00465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 433.Kang S, et al. Characteristic analyses of a neural differentiation model from iPSC-derived neuron according to morphology, physiology, and global gene expression pattern. Sci. Rep. 2017;7:12233. doi: 10.1038/s41598-017-12452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Penney J, Ralvenius WT, Tsai L-H. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol. Psychiatry. 2020;25:148–167. doi: 10.1038/s41380-019-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Chen SW, et al. Efficient conversion of human induced pluripotent stem cells into microglia by defined transcription factors. Stem Cell Rep. 2021;16:1363–1380. doi: 10.1016/j.stemcr.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Marton RM, Ioannidis JPA. A comprehensive analysis of protocols for deriving dopaminergic neurons from human pluripotent stem cells. Stem Cells Transl. Med. 2019;8:366–374. doi: 10.1002/sctm.18-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Wang Y, et al. Generation of induced pluripotent stem cell line (ZZUi007-A) from a 52-year-old patient with a novel CHCHD2 gene mutation in Parkinson’s disease. Stem Cell Res. 2018;32:87–90. doi: 10.1016/j.scr.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 438.Chung SY, et al. Parkin and PINK1 patient iPSC-derived midbrain dopamine neurons exhibit mitochondrial dysfunction and α-synuclein accumulation. Stem Cell Rep. 2016;7:664–677. doi: 10.1016/j.stemcr.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Jankowsky JL, Zheng H. Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol. Neurodegener. 2017;12:89. doi: 10.1186/s13024-017-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Campenot RB. Local control of neurite development by nerve growth factor. Proc. Natl Acad. Sci. USA. 1977;74:4516. doi: 10.1073/pnas.74.10.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Megarity D, et al. A modular microfluidic platform to enable complex and customisable in vitro models for neuroscience. Lab Chip. 2022;22:1989–2000. doi: 10.1039/D2LC00115B. [DOI] [PubMed] [Google Scholar]
- 442.Peyrin J-M, et al. Axon diodes for the reconstruction of oriented neuronal networks in microfluidic chambers. Lab Chip. 2011;11:3663–3673,. doi: 10.1039/c1lc20014c. [DOI] [PubMed] [Google Scholar]
- 443.Renault R, Durand J-B, Viovy J-L, Villard C. Asymmetric axonal edge guidance: a new paradigm for building oriented neuronal networks. Lab Chip. 2016;16:2188–2191. doi: 10.1039/C6LC00479B. [DOI] [PubMed] [Google Scholar]
- 444.Courte J, et al. Reconstruction of directed neuronal networks in a microfluidic device with asymmetric microchannels. Methods Cell Biol. 2018;148:71–95. doi: 10.1016/bs.mcb.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 445.le Feber, J., Postma, W., de Weerd, E., Weusthof, M. & Rutten, W. L. C. Barbed channels enhance unidirectional connectivity between neuronal networks cultured on multi electrode arrays. Front. Neurosci.10.3389/fnins.2015.00412 (2015). [DOI] [PMC free article] [PubMed]
- 446.Gu L, et al. Microfluidic control of axonal guidance. Sci. Rep. 2014;4:6457. doi: 10.1038/srep06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Kim J, Kang M, Jensen EC, Mathies RA. Lifting gate polydimethylsiloxane microvalves and pumps for microfluidic control. Anal. Chem. 2012;84:2067–2071. doi: 10.1021/ac202934x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 449.Sesen M, Rowlands CJ. Thermally-actuated microfluidic membrane valve for point-of-care applications. Microsyst. Nanoeng. 2021;7:48. doi: 10.1038/s41378-021-00260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Fernandes JTS, et al. Modulation of alpha-synuclein toxicity in yeast using a novel microfluidic-based gradient generator. Lab Chip. 2014;14:3949–3957. doi: 10.1039/C4LC00756E. [DOI] [PubMed] [Google Scholar]
- 451.Choi YJ, et al. Neurotoxic amyloid beta oligomeric assemblies recreated in microfluidic platform with interstitial level of slow flow. Sci. Rep. 2013;3:1921. doi: 10.1038/srep01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Shin Y, et al. Blood–brain barrier dysfunction in a 3D in vitro model of Alzheimer’s disease. Adv. Sci. 2019;6:1900962. doi: 10.1002/advs.201900962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Pediaditakis I, et al. Modeling alpha-synuclein pathology in a human brain-chip to assess blood-brain barrier disruption. Nat. Commun. 2021;12:5907. doi: 10.1038/s41467-021-26066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Leung CM, et al. A guide to the organ-on-a-chip. Nat. Rev. Methods Prim. 2022;2:33. doi: 10.1038/s43586-022-00118-6. [DOI] [Google Scholar]
- 455.Aleman J, Kilic T, Mille LS, Shin SR, Zhang YS. Microfluidic integration of regeneratable electrochemical affinity-based biosensors for continual monitoring of organ-on-a-chip devices. Nat. Protoc. 2021;16:2564–2593. doi: 10.1038/s41596-021-00511-7. [DOI] [PubMed] [Google Scholar]
- 456.Henry OYF, et al. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip. 2017;17:2264–2271. doi: 10.1039/C7LC00155J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Moutaux E, Charlot B, Genoux A, Saudou F, Cazorla M. An integrated microfluidic/microelectrode array for the study of activity-dependent intracellular dynamics in neuronal networks. Lab Chip. 2018;18:3425–3435. doi: 10.1039/C8LC00694F. [DOI] [PubMed] [Google Scholar]
- 458.Salman MM, et al. Design and validation of a human brain endothelial microvessel-on-a-chip open microfluidic model enabling advanced optical imaging. Front. Bioeng. Biotechnol. 2020;8:573775. doi: 10.3389/fbioe.2020.573775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Centeno EGZ, Cimarosti H, Bithell A. 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling. Mol. Neurodegener. 2018;13:27. doi: 10.1186/s13024-018-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Hebisch M, et al. The impact of the cellular environment and aging on modeling Alzheimer’s disease in 3D cell culture models. Adv. Sci. 2023;10:2205037. doi: 10.1002/advs.202205037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Bolognin S, et al. 3D cultures of Parkinson’s disease-specific dopaminergic neurons for high content phenotyping and drug testing. Adv. Sci. 2019;6:1800927. doi: 10.1002/advs.201800927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Choi SH, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Kwak SS, et al. Amyloid-β42/40 ratio drives tau pathology in 3D human neural cell culture models of Alzheimer’s disease. Nat. Commun. 2020;11:1377. doi: 10.1038/s41467-020-15120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Baruffaldi D, Palmara G, Pirri C, Frascella F. 3D cell culture: recent development in materials with tunable stiffness. ACS Appl. Bio Mater. 2021;4:2233–2250. doi: 10.1021/acsabm.0c01472. [DOI] [PubMed] [Google Scholar]
- 465.Segel M, et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature. 2019;573:130–134. doi: 10.1038/s41586-019-1484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Murphy MC, et al. Regional brain stiffness changes across the Alzheimer’s disease spectrum. Neuroimage Clin. 2016;10:283–290. doi: 10.1016/j.nicl.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Sobrino A, et al. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 2016;6:31589. doi: 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Trietsch SJ, et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun. 2017;8:262. doi: 10.1038/s41467-017-00259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Dujardin S, et al. Neuron-to-neuron wild-type Tau protein transfer through a trans-synaptic mechanism: relevance to sporadic tauopathies. Acta Neuropathol. Commun. 2014;2:14. doi: 10.1186/2051-5960-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Takeda S, et al. Neuronal uptake and propagation of a rare phosphorylated high-molecular-weight tau derived from Alzheimer’s disease brain. Nat. Commun. 2015;6:8490. doi: 10.1038/ncomms9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Wu JW, et al. Small misfolded Tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. J. Biol. Chem. 2013;288:1856–1870. doi: 10.1074/jbc.M112.394528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Usenovic M, et al. Internalized tau oligomers cause neurodegeneration by inducing accumulation of pathogenic tau in human neurons derived from induced pluripotent stem cells. J. Neurosci. 2015;35:14234–14250. doi: 10.1523/JNEUROSCI.1523-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Kunze A, Meissner R, Brando S, Renaud P. Co-pathological connected primary neurons in a microfluidic device for alzheimer studies. Biotechnol. Bioeng. 2011;108:2241–2245. doi: 10.1002/bit.23128. [DOI] [PubMed] [Google Scholar]
- 474.Annweiler C, Brugg B, Peyrin J-M, Bartha R, Beauchet O. Combination of memantine and vitamin D prevents axon degeneration induced by amyloid-beta and glutamate. Neurobiol. Aging. 2014;35:331–335. doi: 10.1016/j.neurobiolaging.2013.07.029. [DOI] [PubMed] [Google Scholar]
- 475.Poon WW, et al. β-Amyloid impairs axonal BDNF retrograde trafficking. Neurobiol. Aging. 2011;32:821–833. doi: 10.1016/j.neurobiolaging.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Deleglise B, et al. β-amyloid induces a dying-back process and remote trans-synaptic alterations in a microfluidic-based reconstructed neuronal network. Acta Neuropathol. Commun. 2014;2:145. doi: 10.1186/s40478-014-0145-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Wu JW, et al. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat. Neurosci. 2016;19:1085–1092. doi: 10.1038/nn.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Calafate S, et al. Synaptic contacts enhance cell-to-cell tau pathology propagation. Cell Rep. 2015;11:1176–1183. doi: 10.1016/j.celrep.2015.04.043. [DOI] [PubMed] [Google Scholar]
- 479.Ruiz A, et al. Testing Aβ toxicity on primary CNS cultures using drug-screening microfluidic chips. Lab Chip. 2014;14:2860–2866. doi: 10.1039/C4LC00174E. [DOI] [PubMed] [Google Scholar]
- 480.Maragakis NJ, Rothstein JD. Mechanisms of disease: astrocytes in neurodegenerative disease. Nat. Clin. Pract. Neurol. 2006;2:679–689. doi: 10.1038/ncpneuro0355. [DOI] [PubMed] [Google Scholar]
- 481.Liu, J. et al. Microglia: a double-edged sword in intracerebral hemorrhage from basic mechanisms to clinical research. Front. Immunol.10.3389/fimmu.2021.675660 (2021). [DOI] [PMC free article] [PubMed]
- 482.Park J, et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 2018;21:941–951. doi: 10.1038/s41593-018-0175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Baik SH, et al. Migration of neutrophils targeting amyloid plaques in Alzheimer’s disease mouse model. Neurobiol. Aging. 2014;35:1286–1292. doi: 10.1016/j.neurobiolaging.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Cho H, et al. Microfluidic chemotaxis platform for differentiating the roles of soluble and bound amyloid-β on microglial accumulation. Sci. Rep. 2013;3:1823. doi: 10.1038/srep01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Park J, et al. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer’s disease. Lab Chip. 2015;15:141–150. doi: 10.1039/C4LC00962B. [DOI] [PubMed] [Google Scholar]
- 486.Soden PA, Henderson AR, Lee E. A microfluidic model of AQP4 polarization dynamics and fluid transport in the healthy and inflamed human brain: the first step towards glymphatics-on-a-chip. Adv. Biol. 2022;6:e2200027. doi: 10.1002/adbi.202200027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Seidi A, et al. A microfluidic-based neurotoxin concentration gradient for the generation of an in vitro model of Parkinson’s disease. Biomicrofluidics. 2011;5:022214. doi: 10.1063/1.3580756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Brahic M, Bousset L, Bieri G, Melki R, Gitler AD. Axonal transport and secretion of fibrillar forms of α-synuclein, Aβ42 peptide and HTTExon 1. Acta Neuropathol. 2016;131:539–548. doi: 10.1007/s00401-016-1538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Freundt EC, et al. Neuron-to-neuron transmission of α-synuclein fibrils through axonal transport. Ann. Neurol. 2012;72:517–524. doi: 10.1002/ana.23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Van Laar, V. S. et al. Evidence for compartmentalized axonal mitochondrial biogenesis: mitochondrial DNA replication increases in distal axons as an early response to Parkinson’s disease-relevant stress. J. Neurosci.38, 7505–7515 (2018). [DOI] [PMC free article] [PubMed]
- 491.Braak H, Ghebremedhin E, Rüb U, Bratzke H, Del Tredici K. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004;318:121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- 492.Prots I, et al. α-Synuclein oligomers induce early axonal dysfunction in human iPSC-based models of synucleinopathies. Proc. Natl Acad. Sci. USA. 2018;115:7813–7818. doi: 10.1073/pnas.1713129115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Volpicelli-Daley LA, Luk KC, Lee VM. Addition of exogenous α-synuclein preformed fibrils to primary neuronal cultures to seed recruitment of endogenous α-synuclein to Lewy body and Lewy neurite-like aggregates. Nat. Protoc. 2014;9:2135–2146. doi: 10.1038/nprot.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Fernandes, J. T. S., Chutna, O., Chu, V., Conde, J. P. & Outeiro, T. F. A novel microfluidic cell co-culture platform for the study of the molecular mechanisms of Parkinson’s disease and other synucleinopathies. Front. Neurosci.10.3389/fnins.2016.00511 (2016). [DOI] [PMC free article] [PubMed]
- 495.Gribaudo S, et al. Propagation of α-synuclein strains within human reconstructed neuronal network. Stem Cell Rep. 2019;12:230–244. doi: 10.1016/j.stemcr.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Perrino G, Wilson C, Santorelli M, di Bernardo D. Quantitative characterization of α-synuclein aggregation in living cells through automated microfluidics feedback control. Cell Rep. 2019;27:916–927.e915. doi: 10.1016/j.celrep.2019.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.de Rus Jacquet A, et al. The contribution of inflammatory astrocytes to BBB impairments in a brain-chip model of Parkinson’s disease. Nat. Commun. 2023;14:3651. doi: 10.1038/s41467-023-39038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Todd TW, Petrucelli L. Modelling amyotrophic lateral sclerosis in rodents. Nat. Rev. Neurosci. 2022;23:231–251. doi: 10.1038/s41583-022-00564-x. [DOI] [PubMed] [Google Scholar]
- 499.Cook C, Petrucelli L. Genetic convergence brings clarity to the enigmatic red line in ALS. Neuron. 2019;101:1057–1069. doi: 10.1016/j.neuron.2019.02.032. [DOI] [PubMed] [Google Scholar]
- 500.Hardiman O, et al. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Prim. 2017;3:17071. doi: 10.1038/nrdp.2017.71. [DOI] [PubMed] [Google Scholar]
- 501.Westergard T, et al. Cell-to-cell transmission of dipeptide repeat proteins linked to C9orf72-ALS/FTD. Cell Rep. 2016;17:645–652. doi: 10.1016/j.celrep.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Kunze A, et al. Astrocyte–neuron co-culture on microchips based on the model of SOD mutation to mimic ALS. Integr. Biol. 2013;5:964–975. doi: 10.1039/c3ib40022k. [DOI] [PubMed] [Google Scholar]
- 503.Stoklund Dittlau K, et al. Human motor units in microfluidic devices are impaired by FUS mutations and improved by HDAC6 inhibition. Stem Cell Rep. 2021;16:2213–2227. doi: 10.1016/j.stemcr.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Altman T, et al. Axonal TDP-43 condensates drive neuromuscular junction disruption through inhibition of local synthesis of nuclear encoded mitochondrial proteins. Nat. Commun. 2021;12:6914. doi: 10.1038/s41467-021-27221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Machado CB, et al. In vitro modeling of nerve–muscle connectivity in a compartmentalized tissue culture device. Adv. Biosyst. 2019;3:1800307. doi: 10.1002/adbi.201800307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Osaki T, Uzel SGM, Kamm RD. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci. Adv. 2018;4:eaat5847. doi: 10.1126/sciadv.aat5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Migazzi A, et al. Huntingtin-mediated axonal transport requires arginine methylation by PRMT6. Cell Rep. 2021;35:108980. doi: 10.1016/j.celrep.2021.108980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Virlogeux A, et al. Reconstituting corticostriatal network on-a-chip reveals the contribution of the presynaptic compartment to Huntington’s disease. Cell Rep. 2018;22:110–122. doi: 10.1016/j.celrep.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 509.Zhao X, et al. TRiC subunits enhance BDNF axonal transport and rescue striatal atrophy in Huntington’s disease. Proc. Natl Acad. Sci. USA. 2016;113:E5655–E5664. doi: 10.1073/pnas.1603020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Virlogeux, A. et al. Increasing brain palmitoylation rescues behavior and neuropathology in Huntington disease mice. Sci. Adv.10.1126/sciadv.abb0799 (2021). [DOI] [PMC free article] [PubMed]
- 511.Maisonneuve BGC, et al. Deposition chamber technology as building blocks for a standardized brain-on-chip framework. Microsyst. Nanoeng. 2022;8:86. doi: 10.1038/s41378-022-00406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Han S, Bang S, Kim HN, Choi N, Kim SH. Modulating and monitoring the functionality of corticostriatal circuits using an electrostimulable microfluidic device. Mol. Brain. 2023;16:13. doi: 10.1186/s13041-023-01007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Chen X, et al. Modeling sporadic Alzheimer’s disease in human brain organoids under serum exposure. Adv. Sci. 2021;8:2101462. doi: 10.1002/advs.202101462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Pavoni S, et al. Small-molecule induction of Aβ-42 peptide production in human cerebral organoids to model Alzheimer’s disease associated phenotypes. PLoS ONE. 2018;13:e0209150. doi: 10.1371/journal.pone.0209150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Park JC, et al. A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids. Nat. Commun. 2021;12:280. doi: 10.1038/s41467-020-20440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Raja WK, et al. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS ONE. 2016;11:e0161969. doi: 10.1371/journal.pone.0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Gonzalez C, et al. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry. 2018;23:2363–2374. doi: 10.1038/s41380-018-0229-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Arber C, et al. Familial Alzheimer’s disease mutations in PSEN1 lead to premature human stem cell neurogenesis. Cell Rep. 2021;34:108615. doi: 10.1016/j.celrep.2020.108615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Shimada H, et al. A next-generation iPSC-derived forebrain organoid model of tauopathy with tau fibrils by AAV-mediated gene transfer. Cell Rep. Methods. 2022;2:100289. doi: 10.1016/j.crmeth.2022.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Yin J, VanDongen AM. Enhanced neuronal activity and asynchronous calcium transients revealed in a 3D organoid model of Alzheimer’s disease. ACS Biomater. Sci. Eng. 2021;7:254–264. doi: 10.1021/acsbiomaterials.0c01583. [DOI] [PubMed] [Google Scholar]
- 521.Son MY, et al. Distinctive genomic signature of neural and intestinal organoids from familial Parkinson’s disease patient-derived induced pluripotent stem cells. Neuropathol. Appl. Neurobiol. 2017;43:584–603. doi: 10.1111/nan.12396. [DOI] [PubMed] [Google Scholar]
- 522.Smits LM, et al. Modeling Parkinson’s disease in midbrain-like organoids. npj Parkinsons Dis. 2019;5:5. doi: 10.1038/s41531-019-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Pereira JD, et al. Human sensorimotor organoids derived from healthy and amyotrophic lateral sclerosis stem cells form neuromuscular junctions. Nat. Commun. 2021;12:4744. doi: 10.1038/s41467-021-24776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Szebényi K, et al. Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nat. Neurosci. 2021;24:1542–1554. doi: 10.1038/s41593-021-00923-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Zhao J, et al. APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer’s disease patient iPSC-derived cerebral organoids. Nat. Commun. 2020;11:5540. doi: 10.1038/s41467-020-19264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Marton RM, Pașca SP. Organoid and assembloid technologies for investigating cellular crosstalk in human brain development and disease. Trends Cell Biol. 2020;30:133–143. doi: 10.1016/j.tcb.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 527.Miura Y, et al. Engineering brain assembloids to interrogate human neural circuits. Nat. Protoc. 2022;17:15–35. doi: 10.1038/s41596-021-00632-z. [DOI] [PubMed] [Google Scholar]
- 528.Wang L, et al. A human three-dimensional neural-perivascular ‘assembloid’ promotes astrocytic development and enables modeling of SARS-CoV-2 neuropathology. Nat. Med. 2021;27:1600–1606. doi: 10.1038/s41591-021-01443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Andersen J, et al. Generation of functional human 3D cortico-motor assembloids. Cell. 2020;183:1913–1929.e1926. doi: 10.1016/j.cell.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Birey F, et al. Dissecting the molecular basis of human interneuron migration in forebrain assembloids from Timothy syndrome. Cell Stem Cell. 2022;29:248–264.e247. doi: 10.1016/j.stem.2021.11.011. [DOI] [PubMed] [Google Scholar]
- 531.Kong D, et al. Cortical-blood vessel assembloids exhibit Alzheimer’s disease phenotypes by activating glia after SARS-CoV-2 infection. Cell Death Discov. 2023;9:32. doi: 10.1038/s41420-022-01288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Rickner HD, et al. Single cell transcriptomic profiling of a neuron-astrocyte assembloid tauopathy model. Nat. Commun. 2022;13:6275. doi: 10.1038/s41467-022-34005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Gordon A, et al. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat. Neurosci. 2021;24:331–342. doi: 10.1038/s41593-021-00802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Trujillo CA, et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019;25:558–569.e557. doi: 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Mariani J, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Birey F, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Luo J, Li P. Human pluripotent stem cell-derived brain organoids as in vitro models for studying neural disorders and cancer. Cell Biosci. 2021;11:99. doi: 10.1186/s13578-021-00617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Li Y, et al. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell. 2017;20:385–396.e383. doi: 10.1016/j.stem.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Scott G, Huang Y. Engineering cerebral folding in brain organoids. Neural Regen. Res. 2022;17:2420–2422,. doi: 10.4103/1673-5374.335789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Ao Z, et al. One-stop microfluidic assembly of human brain organoids to model prenatal cannabis exposure. Anal. Chem. 2020;92:4630–4638. doi: 10.1021/acs.analchem.0c00205. [DOI] [PubMed] [Google Scholar]
- 541.Zhu Y, et al. In situ generation of human brain organoids on a micropillar array. Lab Chip. 2017;17:2941–2950. doi: 10.1039/C7LC00682A. [DOI] [PubMed] [Google Scholar]
- 542.Qian X, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Ao Z, et al. Tubular human brain organoids to model microglia-mediated neuroinflammation. Lab Chip. 2021;21:2751–2762. doi: 10.1039/D1LC00030F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Karzbrun E, Kshirsagar A, Cohen SR, Hanna JH, Reiner O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 2018;14:515–522. doi: 10.1038/s41567-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Sharma AD, et al. Engineering a 3D functional human peripheral nerve in vitro using the nerve-on-a-chip platform. Sci. Rep. 2019;9:8921. doi: 10.1038/s41598-019-45407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Sood D, et al. Functional maturation of human neural stem cells in a 3D bioengineered brain model enriched with fetal brain-derived matrix. Sci. Rep. 2019;9:17874. doi: 10.1038/s41598-019-54248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Zhu Y, Zhang X, Sun L, Wang Y, Zhao Y. Engineering human brain assembloids by microfluidics. Adv. Mater. 2023;35:2210083. doi: 10.1002/adma.202210083. [DOI] [PubMed] [Google Scholar]
- 548.Nashimoto Y, et al. Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device. Integr. Biol. 2017;9:506–518. doi: 10.1039/C7IB00024C. [DOI] [PubMed] [Google Scholar]
- 549.Nashimoto Y, et al. Vascularized cancer on a chip: the effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials. 2020;229:119547. doi: 10.1016/j.biomaterials.2019.119547. [DOI] [PubMed] [Google Scholar]
- 550.Ao Z, et al. Understanding immune-driven brain aging by human brain organoid microphysiological analysis platform. Adv. Sci. 2022;9:2200475. doi: 10.1002/advs.202200475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Zhang S, Wan Z, Kamm RD. Vascularized organoids on a chip: strategies for engineering organoids with functional vasculature. Lab Chip. 2021;21:473–488. doi: 10.1039/D0LC01186J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Ayuso JM, et al. Evaluating natural killer cell cytotoxicity against solid tumors using a microfluidic model. Oncoimmunology. 2019;8:1553477. doi: 10.1080/2162402X.2018.1553477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Chen YY, et al. Clarifying intact 3D tissues on a microfluidic chip for high-throughput structural analysis. Proc. Natl Acad. Sci. USA. 2016;113:14915–14920. doi: 10.1073/pnas.1609569114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Chen B, et al. High-throughput acoustofluidic fabrication of tumor spheroids. Lab Chip. 2019;19:1755–1763. doi: 10.1039/C9LC00135B. [DOI] [PubMed] [Google Scholar]
- 555.Misun PM, Rothe J, Schmid YRF, Hierlemann A, Frey O. Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsyst. Nanoeng. 2016;2:16022. doi: 10.1038/micronano.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Sims JR, et al. Donanemab in early symptomatic Alzheimer disease: the TRAILBLAZER-ALZ 2 randomized clinical trial. JAMA. 2023;330:512–527. doi: 10.1001/jama.2023.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Cummings JL, Goldman DP, Simmons-Stern NR, Ponton E. The costs of developing treatments for Alzheimer’s disease: a retrospective exploration. Alzheimers Dement. 2022;18:469–477. doi: 10.1002/alz.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Akhtar A, et al. Preclinical models for Alzheimer’s disease: past, present, and future approaches. ACS Omega. 2022;7:47504–47517. doi: 10.1021/acsomega.2c05609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Watamura N, Sato K, Saido TC. Mouse models of Alzheimer’s disease for preclinical research. Neurochem. Int. 2022;158:105361. doi: 10.1016/j.neuint.2022.105361. [DOI] [PubMed] [Google Scholar]
- 560.Vulto P, Joore J. Adoption of organ-on-chip platforms by the pharmaceutical industry. Nat. Rev. Drug Discov. 2021;20:961–962. doi: 10.1038/s41573-021-00323-0. [DOI] [PubMed] [Google Scholar]
- 561.Marx U, et al. Biology-inspired microphysiological systems to advance patient benefit and animal welfare in drug development. ALTEX. 2020;37:365–394. doi: 10.14573/altex.2001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Wadman M. FDA no longer has to require animal testing for new drugs. Science. 2023;379:127–128. doi: 10.1126/science.adg6276. [DOI] [PubMed] [Google Scholar]
- 563.Mastrangeli M, Millet S, Orchid partners, T. van den Eijnden-van Raaij J. Organ-on-chip in development: towards a roadmap for organs-on-chip. ALTEX. 2019;36:650–668. doi: 10.14573/altex.1908271. [DOI] [PubMed] [Google Scholar]
- 564.Figarol A, et al. Interstitial flow regulates in vitro three-dimensional self-organized brain micro-vessels. Biochem. Biophys. Res. Commun. 2020;533:600–606. doi: 10.1016/j.bbrc.2020.09.061. [DOI] [PubMed] [Google Scholar]
- 565.Mastrangeli M, et al. Building blocks for a European Organ-on-Chip roadmap. ALTEX. 2019;36:481–492. doi: 10.14573/altex.1905221. [DOI] [PubMed] [Google Scholar]
- 566.Rusyn I, et al. Microphysiological systems evaluation: experience of TEX-VAL Tissue Chip Testing Consortium. Toxicol. Sci. 2022;188:143–152. doi: 10.1093/toxsci/kfac061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Schurdak M, et al. Applications of the microphysiology systems database for experimental ADME-Tox and disease models. Lab Chip. 2020;20:1472–1492. doi: 10.1039/C9LC01047E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Sakolish C, et al. Analysis of reproducibility and robustness of a renal proximal tubule microphysiological system OrganoPlate 3-lane 40 for in vitro studies of drug transport and toxicity. Toxicol. Sci. 2023;196:52–70. doi: 10.1093/toxsci/kfad080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Nieskens TTG, et al. Nephrotoxic antisense oligonucleotide SPC5001 induces kidney injury biomarkers in a proximal tubule-on-a-chip. Arch. Toxicol. 2021;95:2123–2136. doi: 10.1007/s00204-021-03062-8. [DOI] [PubMed] [Google Scholar]
- 570.Santhanam N, et al. Stem cell derived phenotypic human neuromuscular junction model for dose response evaluation of therapeutics. Biomaterials. 2018;166:64–78. doi: 10.1016/j.biomaterials.2018.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Tavakol DN, Fleischer S, Vunjak-Novakovic G. Harnessing organs-on-a-chip to model tissue regeneration. Cell Stem Cell. 2021;28:993–1015. doi: 10.1016/j.stem.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Abdelnour C, et al. Perspectives and challenges in patient stratification in Alzheimer’s disease. Alzheimers Res. Ther. 2022;14:112. doi: 10.1186/s13195-022-01055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Low LA, Mummery C, Berridge BR, Austin CP, Tagle DA. Organs-on-chips: into the next decade. Nat. Rev. Drug Discov. 2021;20:345–361. doi: 10.1038/s41573-020-0079-3. [DOI] [PubMed] [Google Scholar]
- 574.Song H-L, et al. β-Amyloid is transmitted via neuronal connections along axonal membranes. Ann. Neurol. 2014;75:88–97. doi: 10.1002/ana.24029. [DOI] [PubMed] [Google Scholar]
- 575.Nobuhara CK, et al. Tau antibody targeting pathological species blocks neuronal uptake and interneuron propagation of tau in vitro. Am. J. Pathol. 2017;187:1399–1412. doi: 10.1016/j.ajpath.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Wang B, et al. 14-3-3 proteins reduce cell-to-cell transfer and propagation of pathogenic α-synuclein. J. Neurosci. 2018;38:8211–8232. doi: 10.1523/JNEUROSCI.1134-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Moreno EL, et al. Differentiation of neuroepithelial stem cells into functional dopaminergic neurons in 3D microfluidic cell culture. Lab Chip. 2015;15:2419–2428. doi: 10.1039/C5LC00180C. [DOI] [PubMed] [Google Scholar]