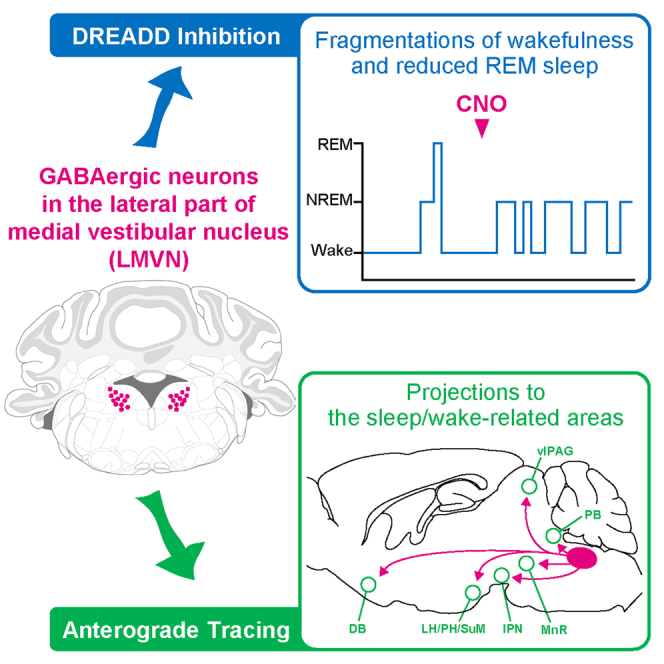
Summary
Body rocking can either induce sleep or arousal. That is, the vestibular sense influences sleep-wake states. Neuronal interactions between sleep-wake systems and vestibular systems, however, remain unclear. In this study, we found that GABAergic neurons in the lateral part of the medial vestibular nucleus (LMVN), a primary vestibular afferent projection site, control sleep-wake states. Specific inhibition of LMVN GABAergic neurons revealed that the firing of LMVN GABAergic neurons underlies stable wakefulness and smooth transitions from non-rapid-eye-movement (NREM) sleep to rapid eye movement (REM) sleep and that LMVN GABAergic neurons do not affect body balance control in freely moving conditions. Selective axonal tracing of LMVN GABAergic neurons indicated that LMVN GABAergic neurons send axons not only to areas involved in vestibular and oculomotor functions but also to areas regulating sleep-wake states. Our findings suggest that LMVN GABAergic neurons stabilize wakefulness and gate the entry into REM sleep through the use of vestibular information.
Subject areas: Behavioral neuroscience, Biochemistry, Biological sciences, Cell biology, Natural sciences, Neuroscience, Systems neuroscienc
Graphical abstract
Highlights
-
•
Discovery of a novel sleep/wake-regulatory population: LMVN GABAergic neurons
-
•
LMVN GABAergic neurons stabilize wakefulness
-
•
LMVN GABAergic neurons gate the entry into REM sleep in the light phase
-
•
LMVN GABAergic neurons project to the sleep/wake-related areas
Behavioral neuroscience; Biochemistry; Biological sciences; Cell biology; Natural sciences; Neuroscience; Systems neuroscienc
Introduction
Sleep has a vital role not only in the rest of the body but also in the individual life,1,2 yet the neural mechanisms governing sleep-wake states remain largely unknown. Many cell groups in the brain have already been experimentally identified as sleep-wake regulatory neurons.3,4,5 Based on physiological and anatomical observations of the defined neurons involved in sleep-wake control, several neural circuit models have also been proposed to explain how the brain organizes sleep and wakefulness,.6,7,8,9 Still, these models do not provide an integrated picture of sleep. This raises the possibility that existing concepts have overlooked different cell groups critical for sleep-wake control. Therefore, searching for and discovering a novel sleep-wake regulatory neural population is necessary to understand sleeping and waking behaviors comprehensively. Sleep-wake states and physical states are reciprocally influencing. Maintenance of posture requires stable wakefulness, and body rocking can either promote sleep or arousal.10,11 These suggest neuronal interactions between the sleep-wake systems and the vestibular systems, but their physiological and anatomical relationships remain largely unresolved. In this study, we focused on the vestibular nucleus (VN) which is a large key brain area for the vestibular sense and might be involved in sleep.12,13,14,15 Targeting GABAergic neurons, which constitute small and diverse subgroups of VN neurons and whose functions are not fully understood,16,17,18 we performed GABAergic neuron-specific exploration of novel sleep-wake-regulatory neurons in the VN with Glutamate decarboxylase 1 (GAD1 or also called GAD67)-Cre mice in which Cre is expressed only in GABAergic neurons19 and an inhibitory Designer Receptor Exclusively Activated by a Designer Drug (DREADD) hM4Di20,21 and Cre recombinase (Cre)-inducible adeno-associated viral (AAV) vectors. We systematically and selectively suppressed firing of GABAergic neurons in and around the VN and assessed the effects on sleep-wake states, revealing that the lateral part of the medial vestibular nucleus (LMVN) contains a novel GABAergic component for sustaining wakefulness.
Results
Targeted suppression of GABAergic neurons in the lateral part of the medial vestibular nucleus
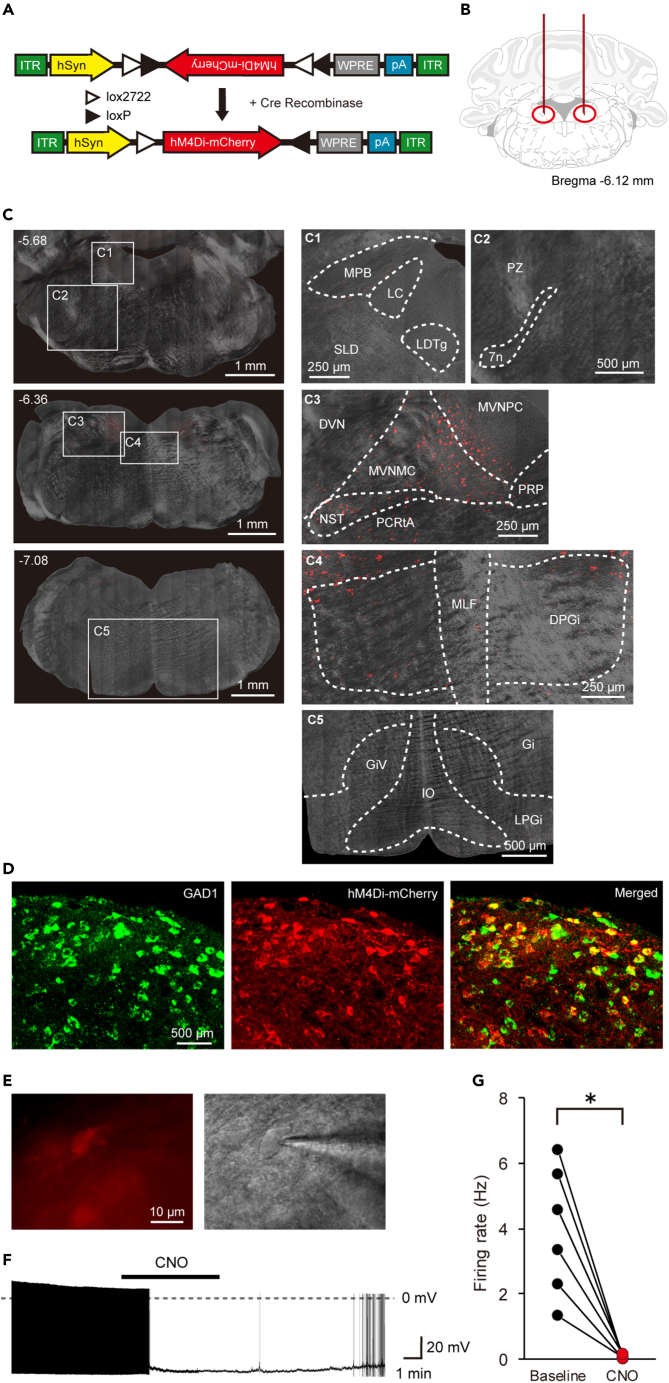
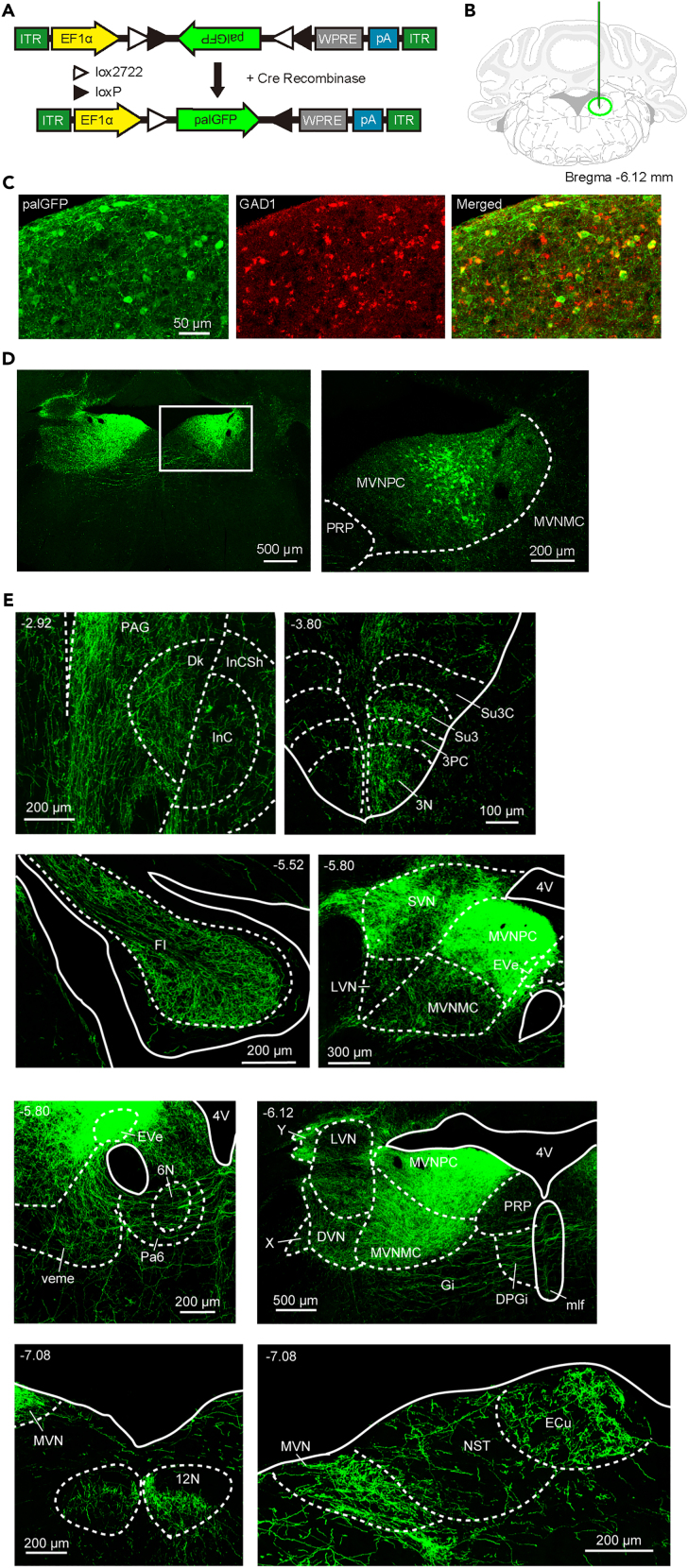
To elucidate how the vestibular system is involved in the sleep-wake system, we locally suppressed neural activity in the areas within or around the medial vestibular nucleus (MVN), a pivotal center for vestibular sensory signal processing located in the medulla, and searched for neuronal populations affecting the sleep-wake state. Here, we report the investigation of GABAergic neurons in the lateral part of the medial vestibular nucleus (LMVN). To achieve the selective inhibition of GABAergic neurons in the LMVN, Cre-dependent AAV vectors encoding hM4Di fused to mCherry were injected bilaterally into the LMVN of GAD1-Cre mice (called GAD1/LMVN/M4 mice here) (Figures 1A and 1B). The expression patterns of hM4Di indicated by mCherry fluorescence were analyzed in the brains of all GAD1/LMVN/M4 mice used for electroencephalogram and electromyogram (EEG/EMG) recordings. For the following sleep analysis (Figures 2, 3, and 4), we accepted only data from the GAD1/LMVN/M4 mice in which hM4Di was expressed bilaterally in the LMVN but not or rarely in the surrounding areas which have been already reported to be involved in the regulation of sleep-wake states: the medial parabrachial nucleus (MPB), locus coeruleus (LC), laterodorsal tegmental nucleus (LDTg), sublaterodorsal tegmental nucleus (SLD), parafacial zone (PZ), dorsal and lateral paragigantocellular reticular nucleus (DPGi and LPGi), gigantocellular reticular nucleus (Gi), and Gi ventral part (GiV) (Figures 1C and S1).7,23,24,25,26 Although AAV vectors could induce retrograde gene transduction via axonal infection in some cases,27 no evident expression of hM4Di was detected in areas distant from the LMVN in GAD1/LMVN/M4 mice (Figures 1C and S1; other areas, data not shown). Histological analysis of the LMVN of GAD1/LMVN/M4 mice confirmed that hM4Di was expressed exclusively in GABAergic neurons (Figure 1D; 93.6% of mCherry-positive cells expressed GAD1 mRNA, 772 of 825 cells from 3 mice). In the LMVN of GAD1/LMVN/M4 mice, hM4Di-expressing neurons exhibited spontaneous action potentials (Figures 1E and 1F). Application of the selective agonist of hM4Di clozapine N-oxide (CNO) almost completely inhibited the spontaneous firing of LMVN neurons expressing hM4Di (Figures 1F and 1G; before 3.93 ± 0.80 Hz; during 0.06 ± 0.03 Hz, n = 6 cells, p = 0.03). These results indicate that the administration of CNO into GAD1/LMVN/M4 mice effectively and specifically inhibits the firing of GABAergic neurons in the LMVN, an unexplored area regarding sleep-wake control.
Figure 1.
Experimental design and confirmation of selective inhibition of LMVN GABAergic neurons
(A) Schematic diagram of Cre-inducible AAV vector expressing hM4Di fused to mCherry under control of human synapsin I promoter. DIO, double-floxed inverted open reading frame; ITR, inverted terminal repeat; hSyn, human synapsin I promoter; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; pA, poly-A tail.
(B) Schematic of bilateral injection of AAV. The outline adapted from the brain atlas of Franklin and Paxinos22 represents a coronal section at the level of the medulla oblongata corresponding to 6.12 mm posterior to bregma. Red vertical lines and circles indicate the injection capillaries and targets, respectively.
(C) Representative images of the coronal brain sections of a GAD1/LMVN/M4 mouse. Confocal fluorescence images (red) and differential interference contrast (DIC) images (gray) are merged. (Left) Wide-field images of the coronal sections. Each value in the upper left corner indicates the anterior-posterior (AP) coordinate from bregma (mm). (Right) Magnified images corresponding to five frames with numbers in left wide-field images (C1-5). Fluorescence images are captured in z stack and projected with maximum intensity. Anatomical abbreviations are listed in Table 1. See also Figure S1.
(D) Representative projected z stack confocal images of fluorescence in situ hybridization (FISH) for Gad1 mRNA (Left, green) and immunohistochemistry (IHC) for hM4Di-mCherry (Middle, red) in the LMVN of a GAD1/LMVN/M4 mouse. Merged image of (Left) and (Middle) displays colocalization (Right).
(E) Fluorescence (Left, red) and Dodt gradient contrast (Right, gray) images of the LMVN of a GAD1/LMVN/M4 mouse in the same field of view. The patch pipette is seen on the right side.
(F) Representative recording from an hM4Di-expressing LMVN neuron of a GAD1/LMVN/M4 mouse in the current-clamp configuration. The solid bar indicates the duration of 5 μM CNO application (5 min). The dotted line denotes 0 mV. Spontaneous action potentials in LMVN neurons expressing hM4Di were inhibited by the application of CNO and recovered after washout.
(G) The mean firing rate of the LMVN neuron expressing hM4Di before (black circles, Baseline) and during (red circles, CNO) the application of CNO. Data obtained from the same cell are connected with a line. ∗p < 0.05, Wilcoxon signed-rank test.
Figure 2.
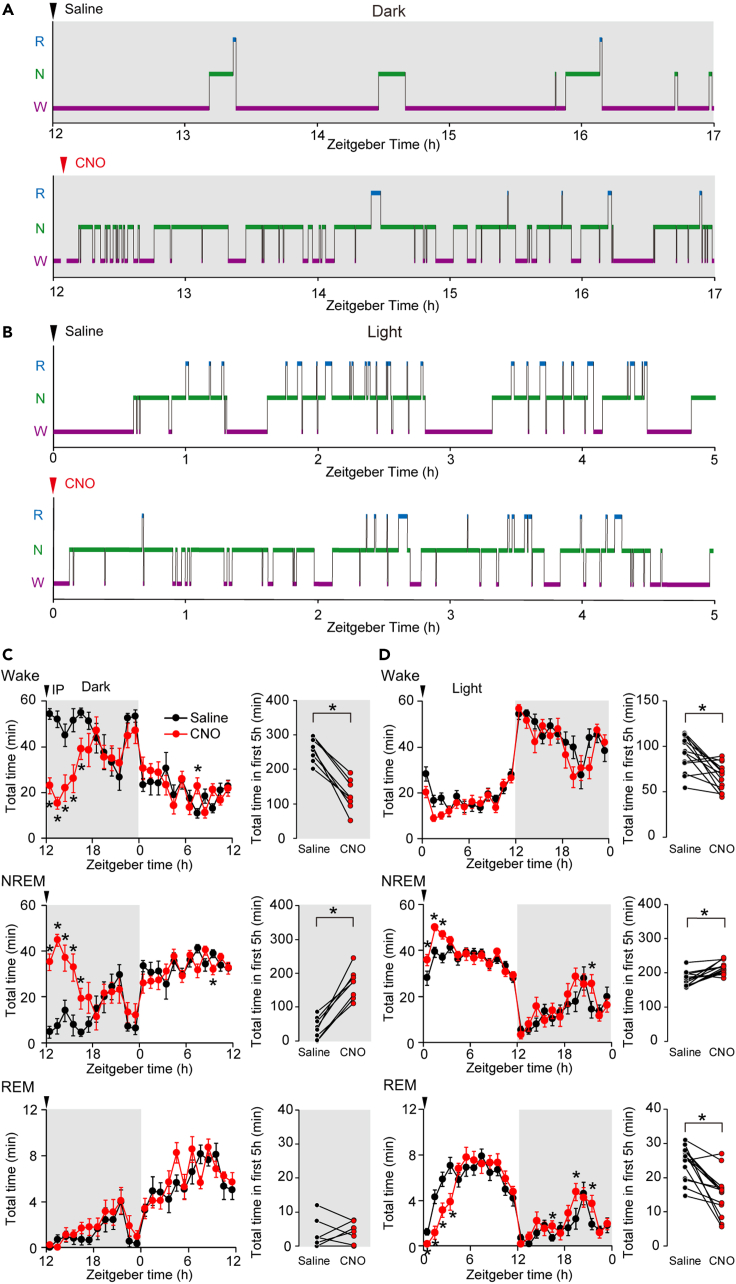
Suppression of LMVN GABAergic neurons decreases the total amounts of wakefulness and REM sleep
(A) The representative hypnograms after the administration of saline (upper) or CNO (lower) into a GAD1/LMVN/M4 mouse at ZT12. Purple, green, and light blue lines indicate the duration of wakefulness (W), NREM sleep (N), and REM sleep (R), respectively. Arrowheads on the upper side denote the timing of administration. Recordings during IP administration were not shown (blank).
(B) Same as (A), but the administration at ZT0.
(C) The plots in the left column show hourly mean amounts of wakefulness (upper), NREM sleep (middle), and REM sleep (lower) in GAD1/LMVN/M4 mice during the first 24 h following the administration of saline (black circles) or CNO (red circles) at ZT12 (n = 8 mice). Arrowheads denote the timing of administration. Error bars indicate ±SEM. The total values for the first 5 h following the administration of saline (black circles) or CNO (red circles) at ZT12 are shown in the right column, in which data obtained from the same mouse are connected with a line. Wake, NREM, and REM represents wakefulness, NREM sleep, and REM sleep, respectively, in this and the following figures.
(D) Same as (C), but administered at ZT0 (n = 14 mice). The hourly amount of wakefulness in GAD1/LMVN/M4 mice was significantly different between saline and CNO groups (F(1,13) = 6.504, p = 0.02), however, there was no statistically significant interaction effect between group (saline and CNO) and time point throughout 24 h following administration (F(23,299) = 1.439, p = 0.09). ∗p < 0.05, two-way repeated measures ANOVA followed by Bonferroni’s multiple comparison test (C left and D left) or Wilcoxon signed-rank test (C right and D right). The light gray background indicates data obtained from experiments during the dark phase in this and the following figures.
Figure 3.
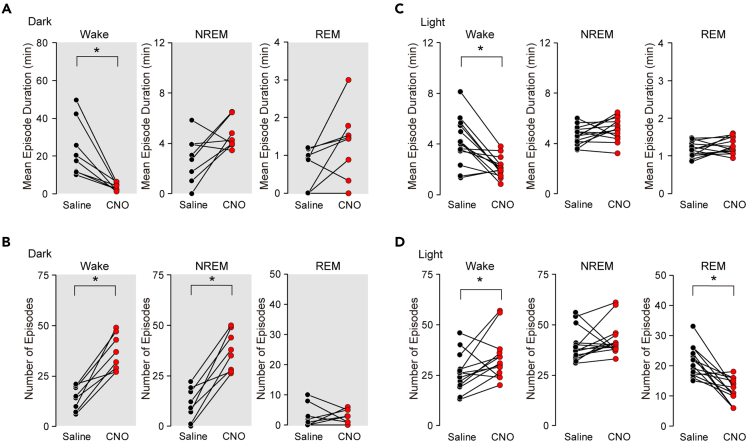
Inhibition of LMVN GABAergic neurons prevents sustained wakefulness
(A) The mean episode duration of wakefulness (left), NREM sleep (middle), and REM sleep (right) in GAD1/LMVN/M4 mice for the first 5 h following the administration of saline (black circles) or CNO (red circles) at ZT12. Data obtained from the same mouse are connected with a line (n = 8 mice).
(B) The number of episodes of wakefulness (left), NREM sleep (middle), and REM sleep (right) in GAD1/LMVN/M4 mice for the first 5 h following the administration of saline (black circles) or CNO (red circles) at ZT12. Data obtained from the same mouse are connected with a line (n = 8 mice).
(C) Same as (A), but administered at ZT0 (n = 11 mice).
(D) Same as (B), but administered at ZT0 (n = 11 mice). ∗p < 0.05, Wilcoxon signed-rank test.
Figure 4.
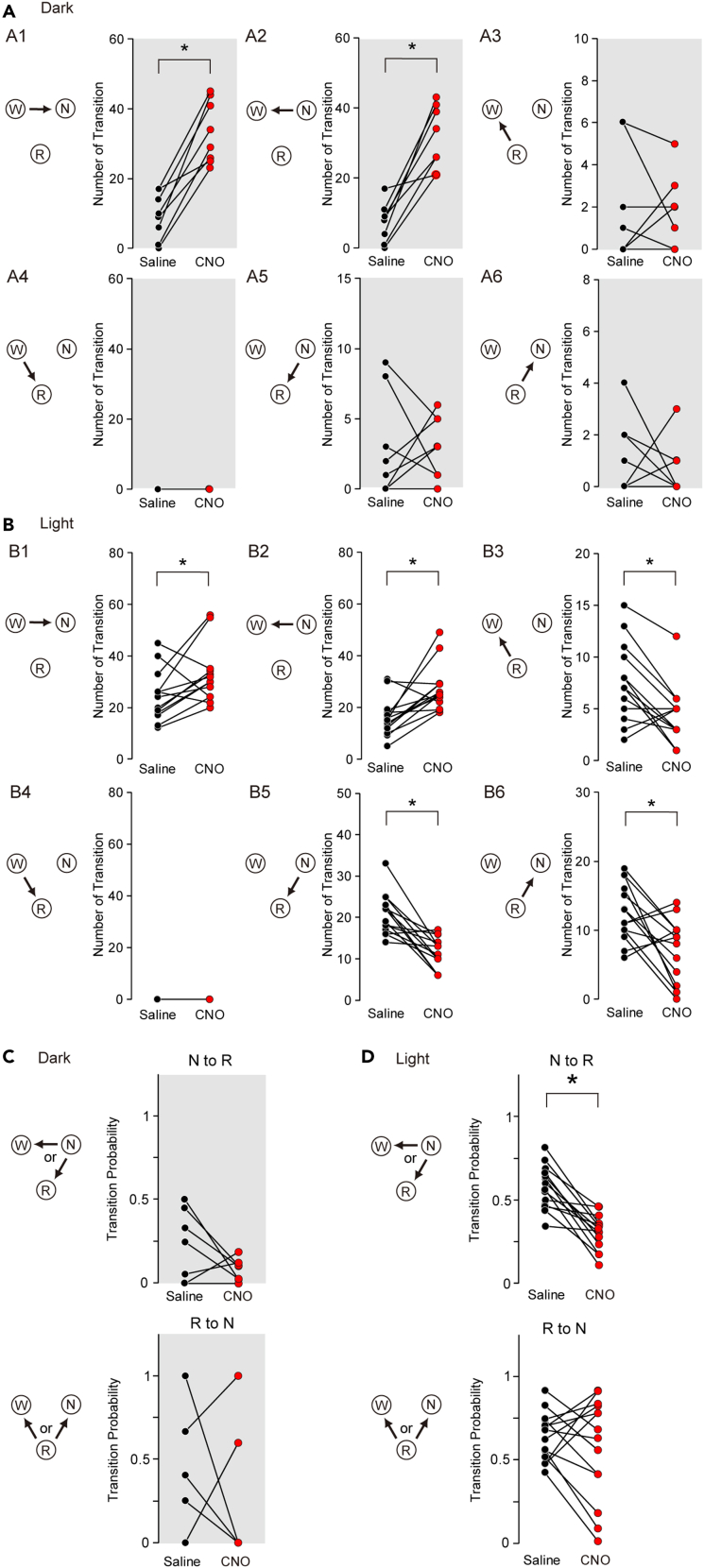
Suppression of LMVN GABAergic neurons promotes wake-to-NREM transinsions but impedes NREM-to-REM transitions in the light phase
(A) The number of transitions between sleep-wake states in GAD1/LMVN/M4 mice for the first 5 h following the administration of saline (black circles) or CNO (red circles) at ZT12. Data obtained from the same mouse are connected with a line (n = 8 mice). W, N, and, R indicate wakefulness, NREM sleep, and REM sleep, respectively. Arrows denote the direction of the transition. A1, W to N; A2, N to W; A3, R to W; A4, W to R; A5, N to R; A6, R to N.
(B) Same as (A) but the administration at ZT0 (n = 14 mice).
(C) Probability of transitions between sleep-wake states in GAD1/LMVN/M4 mice for the first 5 h following the administration of saline (black circles) or CNO (red circles) at ZT12. Transition probability (as an example, N to R) was calculated by dividing the number of the N to R transitions by the total number of transitions from N to W or R. The mice in which N or R was not observed for the first 5 h following the administration of saline or CNO were discarded from the analysis of transition probability (N to R, n = 7 mice; R to N, n = 5 mice).
(D) Same as (C), but for administration at ZT0 (n = 14 mice; no mice were discarded). ∗p < 0.05, Wilcoxon signed-rank test.
Lateral part of the medial vestibular nucleus GABAergic neurons maintain wakefulness during the active phase
To examine if LMVN GABAergic neurons have an intrinsic ability to regulate sleep-wake states, the sleep-wake patterns of GAD1/LMVN/M4 mice were observed for 24 h following the intraperitoneal (IP) administration of CNO or saline. First, GAD1/LMVN/M4 mice were administered CNO at the onset of the dark phase zeitgeber time 12 (ZT12) when mice are most active. Administration of CNO at ZT12 exerted unexpected effects on sleep-wake patterns in GAD1/LMVN/M4 mice: NREM sleep occupied a large part of the 5 h following the administration of CNO (Figure 2A; note that effects of CNO administration on GAD1/LMVN/M4 mice were evaluated in compared to that of saline, unless otherwise stated). The hourly amounts of NREM sleep in GAD1/LMVN/M4 mice were markedly increased at the expense of wakefulness for the first 5-h periods (ZT12–17) following the administration of CNO at ZT12 (Figure 2C; Wake F(1,7) ≥ 13.07, p < 0.01; NREM F(1,7) ≥ 14.66, p < 0.01; n = 8 mice). No significant changes were observed in hourly amounts of wakefulness and NREM sleep over the subsequent hours, but for two time periods (Figure 2C; an increase in Wake for ZT19–20, F(1,7) = 6.45, p = 0.04; a decrease in NREM for ZT21–22, F(1,7) = 7.39, p = 0.03; for all other periods, Wake, F(1,7) ≤ 5.33, p > 0.05; NREM, F(1,7) ≤ 5.44, p > 0.05). Thus, the data acquired during the first 5 h after the administration of CNO or saline were used for a simple comparison between conditions. Also, in the 5-h data, the administration of CNO at ZT12 remarkably increased the total time of NREM sleep at the expense of wakefulness in GAD1/LMVN/M4 mice (Figure 2C; Wake, Δ (= CNO – saline) −132.8 ± 14.4 min, p = 0.01; NREM, Δ +131.0 ± 13.9 min, p = 0.01). A large increase in the total time of NREM sleep in GAD1/LMVN/M4 mice following the administration of CNO at ZT12 was due to an augmentation not in the duration but in the number of NREM sleep episodes (Figures 3A and 3B; the duration, Δ +1.9 ± 0.8 min, p = 0.69; the number, Δ +26.2 ± 3.5, p = 0.01). In addition, the administration of CNO at ZT12 decreased the duration and increased the number of wake episodes in GAD1/LMVN/M4 mice (Figures 3A and 3B; the duration, Δ −19.8 ± 4.9 min, p = 0.01; the number, Δ +22.4 ± 3.4, p = 0.01), reflecting severely fragmented wakefulness due to the frequent appearance of NREM sleep episodes (Figure 2A). These findings indicate that LMVN GABAergic neurons are essential for maintaining wakefulness in the active phase. In addition, in GAD1/LMVN/M4 mice, hM4Di was expressed in a part of the MVN (Figures 1C and S1). Thus, the inhibition of LMVN GABAergic neurons could potentially impact the vestibular function of mice. GAD1/LMVN/M4 mice exhibit no obvious sign of dizziness after the administration of CNO at ZT12 (Videos S1 and S2). Quantifying neck EMG activity during wakefulness, which reflects eye-head gaze shifts,28 also indicated no significant difference in head orientation behavior between the administration of saline and CNO (Figure S4; variance, p = 0.15; integral, p = 0.25, n = 7 mice). These results suggest that the chemogenetic inhibition of LMVN GABAergic neurons has a negligible influence on known vestibular functions at least in freely moving conditions.
The movie was captured using a video camera with a night shot function (29 frames/s, HDR-CX560, Sony). The EEG/EMG electrodes were attached to the mouse.
The mouse was the same as in Video S1.
Also, in the rest phase, lateral part of the medial vestibular nucleus GABAergic neurons sustain wakefulness
Circadian rhythm is crucial in regulating the sleep-wake cycle, which emerged as a distinct sleep-wake pattern during the light and dark phases.29,30,31,32 To investigate whether LMVN GABAergic neurons contribute to sleep-wake control not only in the dark phase but also in the light phase, GAD1/LMVN/M4 mice (n = 14 mice) were next subjected to the administration of CNO at the onset of the light phase ZT0 when mice are falling asleep. During the initial 5 h following the administration of CNO at ZT0, the total amount of wakefulness was decreased, the duration of wake episodes was also reduced, and the number of wake episodes was increased in GAD1/LMVN/M4 mice (Figures 2D, 3C, and 3D; the total time, Δ −25.9 ± 6.3 min, p = 0.004; the episode duration, Δ −2.0 ± 0.3 min, p = 0.004; the number of episodes, Δ +7.93 ± 3.08, p = 0.04), which were similar to that of the administration of CNO at ZT12 (Figures 2C, 3A, and 3B) but the changes were significantly more minor compared with administration at ZT12 (Δ at ZT12 versus Δ at ZT0; the total time, p = 0.0002; the episode duration, p = 0.0002; the number of episodes, p = 0.01; Wilcoxon rank-sum test). In addition, changes in the hourly amounts of wakefulness in GAD1/LMVN/M4 mice did not reach statistical significance over the 24 h following the administration of CNO at ZT0 (see legends to Figure 2D). The hourly amounts of NREM sleep in GAD1/LMVN/M4 mice were significantly increased during the first 3 h following the administration of CNO at ZT0 (ZT0–3, F(1,13) ≥ 5.10, p < 0.04), but not during the subsequent hours (F(1,13) ≤ 3.91, p > 0.07) except for during ZT21–22 (F(1,13) = 4.89, p = 0.046) (Figure 2D). During the first 5 h following the administration of CNO at ZT0, the total amount of NREM sleep in GAD1/LMVN/M4 mice was also increased (Figure 2D; Δ +33.7 ± 7.2 min, p = 0.004), but the change was smaller than that of administration at ZT12 (Δ at ZT12 versus Δ at ZT0, p = 0.0002, Wilcoxon rank-sum test). Unlike administration at ZT12, both the duration and the number of NREM sleep episodes in GAD1/LMVN/M4 mice were not altered following the administration of CNO at ZT0 (Figures 3C and 3D; the duration, Δ +0.5 ± 0.3 min, p = 0.96; the number, Δ +2.93 ± 2.66 p = 0.12), of which, however, no significant changes in the number of NREM sleep episodes could attribute no “net” effect of CNO administration on the transition to NREM sleep (see later in discussion and Figure 4). Administration of CNO at ZT0 produced similar but smaller effects on wakefulness and NREM sleep in GAD1/LMVN/M4 mice as compared to the administration at ZT12, suggesting that the firing of LMVN GABAergic neurons has a role in the maintenance of wakefulness throughout the day and night, but the need in the rest period is less than in the active period.
Lateral part of the medial vestibular nucleus GABAergic neurons control the number, but not the duration, of rapid eye movement sleep episodes in the rest phase
Under natural conditions, REM sleep follows NREM sleep. However, the administration of CNO at ZT12 in GAD1/LMVN/M4 mice caused the frequent appearance of NREM sleep episodes (Figures 2C, 3A, and 3B), but had no significant effect on any of the measures of REM sleep (Figures 2C, 3A, and 3B; the hourly amount, F(1,7) ≤ 4.13, p > 0.08; the total time for the first 5 h, Δ +1.0 ± 1.5 min, p = 0.31; the episode duration, Δ +0.7 ± 0.3 min, p = 0.91; the number of episodes, Δ 0.00 ± 1.56, p = 0.87), suggesting that the appearance and maintenance of REM sleep during the active period is controlled normally even when the states of wakefulness and NREM sleep are highly disrupted by the inhibition of LMVN GABAergic neurons. Unlike administration at ZT12, the hourly amounts of REM sleep in GAD1/LMVN/M4 mice were continuously decreased during the initial 4-h periods following the administration of CNO at ZT0 (ZT0–4 F(1,13) ≥ 5.65, p < 0.02), followed by sporadic increases in REM sleep (ZT16–17, 19–20, and 21–22; F(1,13) ≥ 5.04, p < 0.05; other time points; F(1,13) ≤ 2.35, p > 0.14) (Figure 2D). Reflecting the changes in the hourly amount of REM sleep, the cumulative amount of REM sleep in GAD1/LMVN/M4 mice also decreased after the administration of CNO at ZT12 but recovered to the control level (administration of saline) within 24 h of administration (Figure S2; ZT0–11, and ZT12–13, F(1,13) ≥ 5.65, p < 0.04; ZT19–24/0, F(1,13) ≤ 0.17, p > 0.70), suggesting that the inhibition of LMVN GABAergic neurons in the light period reduces the amount of REM sleep, but does not disturb the following homeostatic recovery responses. During the first 5 h following the administration of CNO at ZT0, GAD1/LMVN/M4 mice showed a large reduction in the total amount of REM sleep (Figure 2D; Δ −9.1 ± 1.9 min, p = 0.001), which was due to a decrease in not the duration but the number of REM sleep episodes (Figures 3C and 3D; the duration, Δ +0.1 ± 0.1 min, p = 0.15; the number, Δ −9.5 ± 1.8, p = 0.001). These findings suggest that the activity of LMVN GABAergic neurons controls the appearance, but not the maintenance, of REM sleep in the light phase.
Lateral part of the medial vestibular nucleus GABAergic neurons gate the entry into rapid eye movement sleep in the rest phase
To clarify the process of fragmented wakefulness and less appearance of REM sleep by the inhibition of LMVN GABAergic neurons, the transitions between sleep and wake states in GAD1/LMVN/M4 mice for the first 5 h following the administration of CNO were calculated. As predicted from fragmented wakefulness (Figures 2 and 3), the transitions of wakefulness to and from NREM sleep in GAD1/LMVN/M4 mice were increased following the administration of CNO at ZT12 (Figure 4A; Δ[W to N] +24.1 ± 3.1, p = 0.01; Δ[N to W] +24.0 ± 37, p = 0.01) and ZT0 (Figure 4B; Δ[W to N] +7.7 ± 3.2, p = 0.04; Δ[N to W] +11.4 ± 2.9, p = 0.004), indicating that LMVN GABAergic neurons stabilize the state of wakefulness via suppression of wake-to-NREM transitions during both the light and dark periods. NREM-to-REM transitions in GAD1/LMVN/M4 mice were decreased by the administration of CNO at ZT0 but not at ZT12 (Figures 4A and 4B; Δ[N to R], ZT12, -0.1 ± 0.9, p = 0.93; ZT0, -9.2 ± 1.8, p = 0.001), raising the possibility that the activity of LMVN GABAergic neurons affect the diurnal variation of appearance of REM sleep (e.g., not only the number but also the probability of the NREM-to-REM transitions is increased during the light phase as compared to during the dark phase in the rodents.33). The circadian alterations in the probability of NREM-to-REM transitions (see legends to Figure 4C) were also observed in GAD1/LMVN/M4 mice during the first 5 h following the administration of saline (Figures 4C and 4D; ZT0, 0.58 ± 0.03, n = 7 mice; ZT12, 0.23 ± 0.08, n = 14 mice; p = 0.002, Wilcoxon rank-sum test), which were significantly decreased following the administration of CNO at ZT0 (Figure 4D; Δ −0.27 ± 0.04, p = 0.001) but not at ZT12 (Figure 4C; Δ −0.14 ± 0.09, p = 0.12). These results suggest that the activity of LMVN GABAergic neurons increases the probability of entry into REM sleep following NREM sleep, especially in the rest phase. As a consequence of less appearance of REM sleep in GAD1/LMVN/M4 mice following the administration of CNO at ZT0 (Figures 3 and 4), the number of REM-to-NREM transitions was decreased (Figures 4A and 4B; Δ[R to N], ZT0, -9.2 ± 1.8, p = 0.001; ZT12, -0.1 ± 1.5, p = 0.93), which resulted in no net change in the number of the transition to NREM sleep (the net change, Δ[W to N] + Δ[R to N], ZT0, 2.1 ± 2.7; ZT12, 24.0 ± 3.5) and thereby no significant change in the number of NREM sleep episodes following the administration of CNO at ZT0 (Figures 3D and 4B). The probability of REM-to-NREM transitions in GAD1/LMVN/M4 mice was not altered following the administration of CNO (Figures 4C and 4D; ZT12, Δ −0.06 ± 0.08, p = 0.36; ZT0, Δ −0.14 ± 0.28, p = 0.69), suggesting that LMVN GABAergic neurons do not influence the decision to enter either wakefulness or NREM sleep after REM sleep episodes.
Some reports have shown that stimulation and lesions of vestibular systems affect oscillatory activity in EEG.34,35,36,37 The delta, theta, beta, and alpha waves were changed, albeit with small differences, following the administration of CNO in GAD1/LMVN/M4 mice during some sleep-wake states (Figure S3), suggesting that the activity of LMVN GABAergic neurons also influences the EEG waves. However, a significant increase in the delta wave activity during NREM sleep, an indication of sleep need,38,39,40 was not observed in GAD1/LMVN/M4 mice during NREM sleep following the administration of CNO (Figure S3), suggesting that sleep need detectable by EEG is not enhanced during the inhibition of LMVN GABAergic neurons.
Lateral part of the medial vestibular nucleus GABAergic neurons send their axons to the sleep-wake systems as well as the vestibular and ocular systems
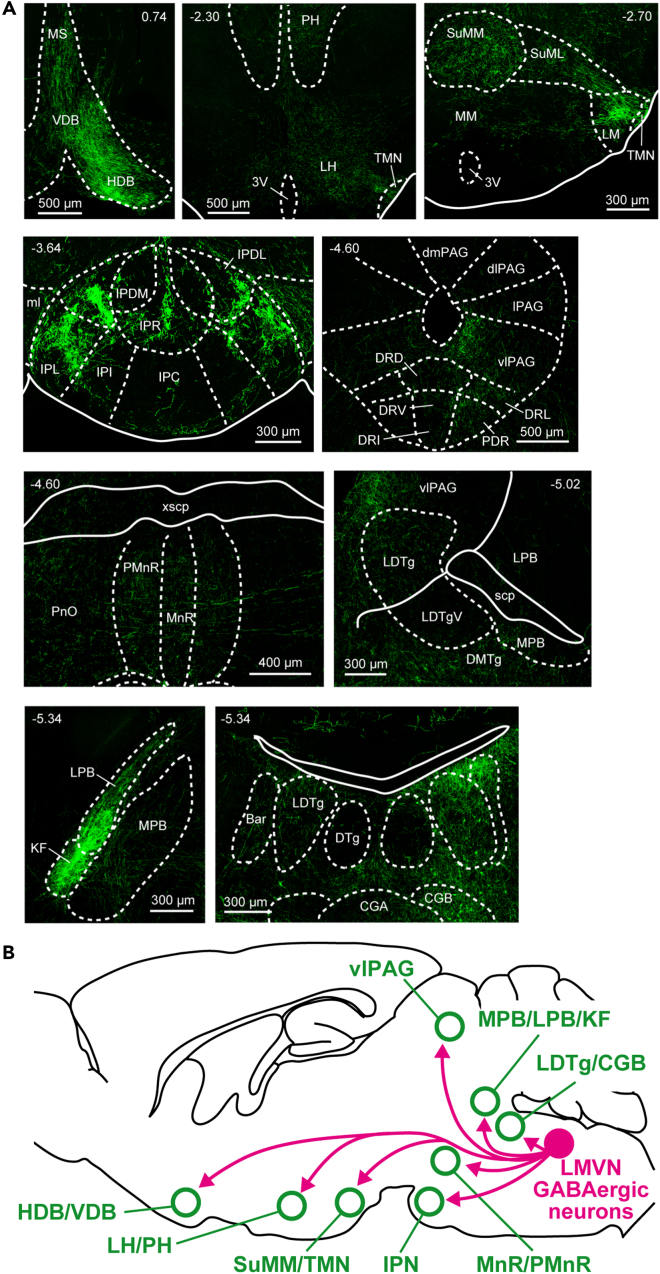
In GAD1/LMVN/M4 mice, most hM4Di-expressing cells lay in the MVN (Figures 1 and S1). MVN GABAergic neurons are components of the vestibular commissural, vestibulo-ocular, vestibulo-olivary, and vestibule-spinal pathways.41,42 A recent report has clarified efferent distribution of MVN GABAergic neurons to the vicinity of the medulla,43 however the long-distance and cell-type specific projection patterns from the LMVN have yet to be elucidated. To understand how LMVN GABAergic neurons organize sleep-wake states, the axonal projections of LMVN GABAergic neurons were traced using Cre-driven AAV vectors encoding GFP fused to the palmitoylation domain of growth-associated protein 43 (palGFP) (Figure 5A). Viral vector-mediated gene transfer of palGFP enables precise visualization of the axonal arbors in the mammalian brain.44,45,46 Cre-inducible AAV vectors encoding palGFP were unilaterally injected into the LMVN of GAD1-Cre mice (Figure 5B), resulting in the selective expression of palGFP in LMVN GABAergic neurons (Figure 5C; 98.7% of palGFP-expressing cells in the LMVN were GAD1 mRNA-positive, n = 1444 cells from 3 mice). GFP-labeled cell bodies were found in the LMVN on the ipsilateral side of the injection. The projections to the contralateral MVN were also observed (Figure 5D), corresponding to the inhibitory commissural pathways between the bilateral MVN.47,48,49,50 Axonal projections of LMVN GABAergic neurons were examined from the forebrain to the medulla (Table 1; n = 3 mice). Axons of LMVN GABAergic neurons were detected in the components of the vestibular and ocular systems: the vestibular nuclei (the medial, lateral, superior, and descending vestibular nucleus (MVN, LVN, SVN, and DVN)), nucleus x and y, nuclei of cranial nerves (the oculomotor (3N), trochlear (4N), abducens (6N), and hypoglossal nucleus (12N)), medial longitudinal fasciculus (MLF) ascending to the interstitial nucleus of Cajal (InC) and nucleus of Darkshevich (Dk), and cerebellar flocculus (Fl) (dark blue in Table 1; Figure 5E), which are consistent with previous reports using other techniques.41,42 Unexpectedly, dense projections from LMVN GABAergic neurons were found in other areas (Figure 6A). The nuclei of the horizontal and vertical limb of the diagonal band (HDB and VDB), and medial septal nucleus (MS), in which LMVN GABAergic axons were found (magenta in Table 1; Figure 6A), are involved in the generation of the EEG theta rhythm but not sleep-wake control,51,52,53,54,55 suggesting that the modulation of the theta component of EEG wave in GAD1/LMVN/M4 mice following CNO administration (Figure S3) is caused by blockage of GABAergic transmission from the LMVN to MS, HDB, and VDB. Axons of LMVN GABAergic were detected in the lateral and posterior hypothalamic areas (LH and PH), mammillary nuclei, interpeduncular nuclei (IPN), ventrolateral PAG (vlPAG), median raphe nucleus (MnR), dorsomedial tegmental area (DMTg), parabrachial area (PB), LDTg, and pons central gray (PCG) (magenta in Table 1; Figure 6A), which are through to regulate sleep-wake states.56,57,58 The anatomical connectivity between the MVN and sleep-wale-related areas is consistent with a histological study in the hamster.59 These findings suggest that LMVN GABAergic neurons could control sleep-wake states through direct connectivity with hypothalamic and brainstem sleep-wake regulatory areas (see discussion).
Figure 5.
Experimental design and confirmation of specific tracing of LMVN GABAergic axons
(A) Schematic diagram of Cre-inducible AAV vector expressing palGFP under control of EF1α promoter. Other abbreviations are defined in Figure 1A.
(B) Schematic of unilateral injection of AAV. The outline is the same as Figure 1B. Green vertical line and circle indicate the injection capillary and target, respectively.
(C) Representative projected z stack confocal images of IHC for palGFP (Left, green) and FISH for Gad1 mRNA (Middle, red) in the LMVN of the GAD1-Cre mouse injected with Cre-inducible AAV vector expressing palGFP. Merged image of (Left) and (Middle) displays colocalization (Right).
(D) Representative images of the coronal brain section of a GAD1-Cre mouse injected unilaterally with Cre-inducible AAV vectors encoding palGFP into the LMVN. (Left) Wide-field images of the coronal sections. The value in the upper left corner indicates the AP coordinates from bregma (mm). AAV vectors were injected into the right side (a white frame). The contralateral projections were observed on the left side. (Right) Magnified images corresponding to a white frame in the left wide-field image.
(E) Confocal fluorescence images of the coronal sections in which a high density of GFP-positive axons was detected. Each value in the upper side indicates the AP coordinates from bregma (mm). Fluorescence images are captured in z stack and projected with maximum intensity. The right side of the image corresponds to the ipsilateral side of the injection. Anatomical abbreviations are listed in Table 1.
Table 1.
Projection areas from LMVN GABAergic neurons
| Brain area | Axon | Brain area | Axon |
|---|---|---|---|
| Cerebral cortex | Ventromedial preoptic nucleus | + | |
| Agranular insular cortex | – | Hypothalamus | |
| Cingulate cortex | – | Accessory olfactory tract | + |
| Dorsal peduncular cortex | – | Anterior hypothalamic area | + |
| Dysgranular insular cortex | – | Anteroventral periventricular nucleus | + |
| Granular insular cortex | – | Arcuate nucleus of the hypothalamus | + |
| Indusium griseum | – | Dorsal hypothalamic area | + |
| Infralimbic cortex | – | Dorsal tuberomammillary nucleus | ++ |
| Neocortex (all four lobes) | – | Dorsomedial hypothalamic nucleus | ++ |
| Prelimbic cortex | – | ♦ Lateral hypothalamic area (LH) | +++ |
| Rhinencephalon | ○ Lateral mammillary nucleus (LM) | +++ | |
| Organum vasculosum | + | Lateral part of the supramammillary nucleus (SuML) | ++ |
| Piriform area | – | Lateral preoptic area | ++ |
| Tenia tecta | – | Medial mammillary nucleus (MM) | + |
| Basal forebrain and septum | ♦ Medial part of the supramammillary nucleus (SuMM) | +++ | |
| Fornix | + | Median eminence | – |
| Lambdoid septal zone | + | Nigrostriatal tract | + |
| Lateral septal nucleus | + | Parasubthalamic nucleus | + |
| Medial forebrain bundle | + | Paraventricular nucleus of the hypothalamus | + |
| Medial septal nucleus (MS) | ++ | Pedunclar part of lateral hypothalamus | ++ |
| Nucleus basalis of Meynert | + | ♦ Posterior hypothalamic area (PH) | +++ |
| ♦ Nucleus of the horizontal limb of the diagonal band (HDB) | +++ | Premammillary nucleus | ++ |
| ♦ Nucleus of the vertical limb of the diagonal band (VDB) | +++ | Retrochiasmatic area | + |
| Septofimbrial nucleus | + | Septohypothalamic nucleus | ++ |
| Substantia innominata (SI) | ++ | Stria terminalis | + |
| Ventral pallidum | + | Subincertal nucleus | ++ |
| Basal ganglia and claustrum | Subparaventricular zone | + | |
| Claustrum | – | Suprachiasmatic nucleus | + |
| Endopiriform claustrum | + | Supraoptic decussations | ++ |
| Globus pallidus | – | Supraoptic nucleus | + |
| Nucleus accumbens | + | ♦ Tuberomammillary nucleus (TMN) | +++ |
| Striatum | – | Ventromedial hypothalamic nucleus | + |
| Subthalamic nucleus | + | Amygdala and bed nucleus of the stria terminalis | + |
| Epithalamus | Basolateral nucleus of the amygdala | – | |
| Lateral habenula | – | Bed nucleus of the stria terminalis | + |
| Medial habenula | – | Cortex-amygdala transition zone | – |
| Stria medullaris | + | Cortical amygdaloid area | + |
| Thalamus | Extended amygdala | ++ | |
| Anterior pretectal nucleus | ++ | Medial amygdaloid nucleus | + |
| Dorsal lateral geniculate nucleus | + | Mentral amygdaloid nucleus | + |
| Intergeniculate leaf | – | Nucleus of the lateral olfactory tract | + |
| Medial lemniscus (mL) | + | Hippocampus | |
| Mediodorsal thalamic nucleus, lateral part | + | CA1 field | – |
| Nucleus of the fields of Forel | + | CA2 field | – |
| Nucleus of the posterior commissure | ++ | CA3 field | – |
| Nucleus reuniens | + | Dentate gyrus | – |
| Paracentral thalamic nucleus | + | Septohippocampal nucleus | + |
| Parafascicular thalamic nucleus | + | Midbrain | |
| Paratenial nucleus | + | Anterior pretectal nucleus | + |
| Posterior intralaminar thalamic nucleus | + | Central nucleus of the inferior colliculus | – |
| Posterior thalamic nuclear group | + | Deep gray layer of the superior colliculus | + |
| Precommissural nucleus | + | Deep white layer of the superior colliculus | + |
| Rhomboid nucleus | + | Dorsal motor nucleus of the vagus nerve | + |
| Submedius thalamic nucleus | + | Dorsal raphe nucleus (DR) | ++ |
| Subparafascicular thalamic nucleus | + | Dorsolateral periaqueductal gray (dlPAG) | + |
| Thalamic reticular nucleus | + | Dorsomedial periaqueductal gray (dmPAG) | + |
| Subthalamus | External nucleus of the inferior colliculus | + | |
| Anteromedial thalamic nucleus | + | Interfascicular nucleus | + |
| Centrolateral thalamic nucleus | + | Interpeduncular nucleus, caudal subnucleus (IPC) | + |
| Centromedial thalamic nucleus | + | Interpeduncular nucleus, dorsolateral subnucleus (IPDL) | – |
| Fields of Forel | + | Interpeduncular nucleus, dorsomedial subnucleus (IPDM) | + |
| Interanteromedial thalamic nucleus | ++ | ♦ Interpeduncular nucleus, intermediate subnucleus (IPI) | +++ |
| Paraventricular thalamic nucleus | ++ | ♦ Interpeduncular nucleus, lateral subnucleus (IPL) | +++ |
| Zona incerta | ++ | Interpeduncular nucleus, rostral subnucleus (IPR) | ++ |
| Preoptic Area | ○ Interstitial nucleus of Cajal (InC) | +++ | |
| Magnocellular preoptic nucleus | + | Lateral periaqueductal gray (lPAG) | ++ |
| Medial preoptic area | + | ♦ Median raphe nucleus (MnR) | +++ |
| Medial preoptic nucleus | + | Mesencephalic reticular formation | ++ |
| Median preoptic nucleus | + | ○ Nucleus of Darkshevich (Dk) | +++ |
| Parastrial nucleus | + | Nucleus of the brachium of the inferior colliculus | + |
| Ventrolateral preoptic nucleus | ++ | ○ Oculomotor nucleus (3N) | +++ |
| ○ Oculomotor nucleus, parvicellular part (3PC) | +++ | ○ Paraabducens nucleus (Pa6) | +++ |
| Parabigeminal nucleus | + | Parvocellular reticular nucleus (PCRt) | ++ |
| ♦ Paramedian raphe nucleus (PMnR) | +++ | Pedunculopontine nucleus (PPT) | ++ |
| Paranigal nucleus | + | Pontine nuclei | + |
| Pararubral nucleus | + | Pontine reticular nucleus (Pn) | + |
| Paratrochlear nucleus | + | Posterodorsal tegmental nucleus | ++ |
| Pre-Edinger-Westphal nucleus | ++ | Principal sensory trigeminal nucleus | ++ |
| Red nucleus | + | Raphe interpositus nucleus | + |
| Retrorubral field | + | Superior cerebellar peduncle | + |
| Retrorubral nucleus | ++ | Superior olive | + |
| Rostral linear nucleus | + | ○ Superior vestibular nucleus (SVN) | ++++ |
| Subpeduncular tegmental nucleus | ++ | Supragenual nucleus | ++ |
| Substantia nigra | + | Supratrigeminal nucleus | ++ |
| Superficial gray layer of the superior colliculus | + | Trigeminal motor nucleus | + |
| Superior colliculus | + | Trigeminal nerve | ++ |
| ○ Supraoculomotor periaqueductal gray (Su3) | +++ | Trigeminal transition zone | + |
| Supraoculomotor cap (Su3C) | ++ | Ventral cochlear nucleus | + |
| Trochlear nucleus (4N) | ++ | Vestibular nerve | + |
| Ventral tegmental area | + | ○ Vestibulomesencephalic tract (veme) | +++ |
| ♦ Ventrolateral periaqueductal gray (vlPAG) | +++ | Medulla | |
| Pons | Ambiguous nucleus | + | |
| A5 cell group | + | Caudoventrolateral reticular nucleus | + |
| A7 cell group | + | Cuneate nucleus | ++ |
| ○ Abducens nucleus (6N) | +++ | ○ Descending vestibular nucleus (DVN) | ++++ |
| Anterior tegmental nucleus | + | Dorsal paragigantocellular nucleus (DPGi) | ++ |
| Barrington’s nucleus | ++ | Dorsal part of the medullary reticular nucleus | + |
| Central gray, alpha part (CGA) | ++ | ○ External cuneate nucleus (ECu) | +++ |
| ♦ Central gray, beta part (CGB) | +++ | Gigantocellular reticular nucleus (Gi) | ++ |
| Central gray, gamma part (CGG) | ++ | Gigantocellular reticular nucleus, alpha part (GiA) | + |
| Dorsal cochlear nucleus | ++ | Gigantocellular reticular nucleus, ventral part (GiV) | + |
| Dorsal lateral parabrachial nucleus, | + | ○ Hypoglossal nucleus (12N) | +++ |
| Dorsal tegmental nucleus (DTg) | + | Inferior olive | ++ |
| Dorsomedial spinal trigeminal nucleus | + | Intermediate reticular nucleus | + |
| ○ Dorsomedial tegmental area (DMTg) | +++ | Intermedius nucleus of the medulla | + |
| External lateral parabrachial nucleus | ++ | Interposed cerebellar nucleus | + |
| Facial nucleus | – | Lateral paragigantocellular nucleus (LPGi) | + |
| Intermediate nucleus of the lateral lemniscus | ++ | Lateral reticular nucleus | ++ |
| ♦ Kolliker-Fuse nucleus (KF) | ++++ | ○ Medial longitudinal fasciculus (MLF) | +++ |
| ♦ Lateral parabrachial nucleus (LPB) | +++ | Nucleus of Roller | + |
| ○ Lateral vestibular nucleus (LVN) | ++++ | Nucleus prepositus (PRP) | ++ |
| ♦ Laterodorsal tegmental nucleus (LDTg) | +++ | Nucleus raphe obscurus | + |
| Locus coeruleus (LC) | ++ | ○ Nucleus x (X) | +++ |
| Longitudinal fasciculus of the pons | + | ○ Nucleus y (Y) | +++ |
| ♦ Medial parabrachial nucleus (MPB) | +++ | Parvicellular part of the lateral cerebellar nucleus | ++ |
| ○ Medial vestibular nucleus magnocellular part (MVNMC) | ++++ | Retroambiguus nucleus | + |
| ○ Medial vestibular nucleus, parvicellular part (MVNPC) | +++ | Rostral ventral respiratory group | + |
| Motor root of the trigeminal nerve | + | Solitary nucleus (NST) | ++ |
| ○ Nucleus of origin of efferents of the vestibular nerve (EVe) | +++ | Spinal trigeminal nucleus | ++ |
| Nucleus of the trapezoid body | + | Cerebellum | |
| Nucleus raphe magnus | + | ○ Flocculus (FI) | +++ |
| Nucleus raphe pontis | + | Lobule I, II, III, IV, and V | + |
| Nucleus reticularis tegmenti pontis | ++ | Medial cerebellar nucleus | + |
The relative densities of GFP-positive axons in individual brain areas (n = 3 mice). ++++, highly dense; +++, dense; ++, moderately dense; +, sparse; -, highly sparse. Of the areas with dense (≥+++) axons of LMVN GABAergic neurons, black diamonds and open circles are shown in sleep-wake-related areas and vestibular-related areas, respectively. See also Figures 5 and 6.
Figure 6.
LMVN GABAergic neurons project to the sleep-wake systems
(A) Confocal fluorescence images of the coronal sections in which a high density of GFP-positive axons was detected. Each value in the upper side indicates the AP coordinates from bregma (mm). Fluorescence images are captured in z stack and projected with maximum intensity. The right side of the image corresponds to the ipsilateral side of the injection. Anatomical abbreviations are listed in Table 1.
(B) A schematic of LMVN GABAergic projections to sleep-wake-regulatory areas.
Discussion
We discovered a novel sleep-wake-regulatory GABAergic population residing in the LMVN. Selective suppression of LMVN GABAergic neurons revealed that they are crucial in maintaining wakefulness and entry into REM sleep during the rest period. Furthermore, specific tracing of axons of LMVN GABAergic neurons disclosed a novel GABAergic circuit from LMVN to brain areas critical for sleep-wake control.
Lateral part of the medial vestibular nucleus GABAergic neuronal population is a novel regulatory component for wakefulness
Cre-dependent AAV vectors encoding hM4Di were randomly injected into the brainstem of GAD1-Cre mice to search for a novel GABAergic population critical for sleep-wake control. In GAD1/LMVN/M4 mice, hM4Di-mCherry was expressed mainly in the LMVN (Figures 1 and S1), which could be due to the non-uniform distribution of GABAergic neurons in the dorsal medulla: GABAergic neurons comprise less than 10% of neurons in the vestibular nuclei, and the largest number is located in the MVN.16,17,18 Acute inhibition of LMVN GABAergic neurons showed that LMVN GABAergic neurons suppress wake-to-NREM transitions, underlying maintenance of wakefulness (Figures 2, 3, and 4). This finding does not conflict essentially with the transection studies (e.g., the animals transected at the pontomedullary junction frequently show a quiescent state60). Still, it is inconsistent with a lesion study showing that the lesion of vestibular nuclei does not alter sleep-wake patterns in cats.61 This inconsistency could be due to a homeostatic compensation commonly observed in sleep-wake control following the ablation of brain areas (e.g., chronic dysfunctions of a wake-promoting center the LC have less effect on sleep-wake states than acute ones) and specific inactivation of LC noradrenergic neurons.62,63,64,65 Activation of MVN neurotensin neurons, presumable GABAergic/glycinergic neurons, increases NREM sleep but decreases REM sleep.13 The different effects and locations from LMVN GABAergic neurons suggest that there are multiple populations of inhibitory neurons with distinct functions for sleep regulation in the MVN.
In general, the stabilization of wakefulness is mediated through lowering homeostatic sleep drive so-called sleep need or enhancing neuropeptide orexin/hypocretin signaling. Suppression of LMVN GABAergic neurons caused the fragmented wakefulness but did not alter the supposed representation of sleep need: the duration of NREM sleep episodes and delta-wave activity in EEG during NREM sleep (Figures 3, 4, and S3). Fragmented wakefulness is also observed in mice lacking neuropeptide orexin signaling.66,67,68,69 However, loss of orexin signaling also exhibits other sleep abnormalities, such as direct wake-to-REM transitions, suggesting that roles of LMVN GABAergic neurons in sleep-wake control might be overlapped with but not entirely equivalent to those of orexin neurons. Taken together, the activity of LMVN GABAergic neurons maintains wakefulness through mechanisms somewhat independent of sleep need and orexin system.
Potential mechanisms underlying maintenance of wakefulness through lateral part of the medial vestibular nucleus GABAergic neurons
To understand the mechanisms underlying LMVN GABAergic maintenance of wakefulness, it is first necessary to clarify the anatomical relationships. VN GABAergic projection to the brainstem has been the subject of many studies, including recent genetic methods,43 but the whole-brain analysis has been lacking. Our whole-brain analysis of LMVN GABAergic projections uncovered unknown anatomical connectivity (Table 1), providing insights into the functional relevance to several possible mechanisms for the stabilization of wakefulness through LMVN GABAergic neurons. One possible mechanism is the direct interference with thalamocortical oscillations,9 which is unfortunately unlikely due to rare projections of LMVN GABAergic neurons found in the cerebral cortex and the thalamus. Another possible mechanism is the suppression of sleep-promoting neurons by LMVN GABAergic neurons, as firing rate of MVN neurons increases during wakefulness and decreases during NREM sleep.70,71 Dense projections from LMVN GABAergic neurons were detected in vestibular and oculomotor-related areas and the connected areas such as the LM, Dk, ECu, DMTg, X, and Y (see Table 1 for non-abbreviated names).72,73,74,75,76 Interestingly, in the hypothalamus and the brainstem, all non-vestibular-ocular-related areas with dense LMVN GABAergic projections were sleep-wake-regulatory areas: the hypothalamus, the LH, PH, SuMM, and TMN; the brainstem, IPN, vlPAG, MnR, PB, KF, LDTg, and PCG (Table 1), of which an effective area for the LMVN GABAergic suppression of NREM sleep might be the vlPAG where NREM sleep-promoting neurons reside.77 The ventrolateral preoptic nucleus (VLPO), a moderately dense projection site of LMVN GABAergic neurons, is well known as a sleep-promoting area,78 thus the VLPO could also be an effective efferent site for the LMVN GABAergic suppression of NREM sleep.
Another possible mechanism for stabilizing wakefulness is the optimization of wake-promoting neuron activity via GABAergic inhibitory tone. For example, the activity of LC noradrenergic (NA) neurons, a regulator of arousal,63,79,80 is adjusted by local GABAergic interneurons in the LC,81 and the excessive activation and the inactivation of LC NA neurons result in behavioral arrests and increased wake-to-NREM sleep transitions, respectively.63 Thus, to stabilize wakefulness, the activity of wake-promoting neurons, such as LC NA neurons, should be optimized to a limited range. Because of LMVN GABAergic moderate projections to the LC and dense projections to several wake-promoting areas such as the LH, PH, SuMM, TMN, and MPB,82,83 LMVN GABAergic neurons could stabilize the state of wakefulness through direct inhibitory modulation or indirect control of wake-promoting neurons via inhibitory local interneurons.
A role of lateral part of the medial vestibular nucleus GABAergic neurons in controlling rapid eye movement sleep
REM sleep was also affected by the inhibition of LMVN GABAergic neurons: the transition probability of NREM-to-REM decreased despite an increase in wake-to-NREM transitions (Figures 3 and 4), suggesting that the activity of LMVN GABAergic neurons opens the gate of the entry into REM sleep. The duration of REM episodes was not changed by the inhibition of LMVN GABAergic neurons (Figure 3), indicating that LMVN GABAergic neurons do not contribute to maintenance of REM sleep. The inhibition of the entry into REM sleep caused by the suppression of LMVN GABAergic neurons was more pronounced in the light phase simply because the probability of the transitions from NREM to REM sleep is increased in rodents during the light phase.33 Electrophysiological and c-Fos immunohistochemical studies have revealed that MVN neurons increase their activity during spontaneous REM sleep, rebound REM sleep, and pharmacologically induced REM-like states.70,71,84,85,86 Dense projections of LMVN GABAergic neurons were found in REM-regulatory areas such as the LH, PH, SuMM, IPN, and vlPAG (Table 1).83,87,88 Taken together, these results suggest that the transitions from NREM to REM sleep is triggered by LMVN GABAergic neurons through direct or indirect control of multiple REM-regulatory neurons.
Physiological and pathological interaction between the sleep-wake system and the vestibular system
Chemogenetic inhibition of LMVN GABAergic neurons, a kind of loss-of-function experiment, disrupted sleep-wake architecture, indicating that sleep-wake state is regulated by LMVN GABAergic neurons under physiological conditions. The MVN is well-known to contain neurons that respond to head rotation and eye movement,89,90,91 whereas behavior of MVN neurons, especially specific neuron types, during sleep-wake cycles is largely unknown. LMVN GABAergic neurons exhibited spontaneous firing at a regular frequency (Figure 1), consistent with previous reports using the MVN slices.92,93,94 Thus, there might not be much difference between LMVN GABAertgic neurons and most MVN neurons in terms of basic firing properties. Spontaneous firing of MVN neurons is modulated by sleep-wake regulators monoamines and acetylcholine,95,96,97,98 which might shape sleep-wake state-dependent firing patterns of most MVN neurons: high-frequency spiking during wakefulness and REM sleep, and low-frequency spiking during NREM sleep.70,71 Therefore, firing of LMVN GABAergic neurons across sleep-wake states may have similar firing characteristics to the majority of MVN neurons.
Suppression of LMVN GABAergic neurons did not cause large changes in walking, posture of sleep, and head movement (Videos S1 and S2, and Figure S4), which is consistent with a recent optogenetic study: activation of GABAergic neurons in the vestibular nuclei does not contribute to postural maintenance.99 Thus, unlike other types of MVN neurons, LMVN GABAergic neurons may not actively affect vestibular functions in freely moving conditions without specific strong or prolonged vestibular stimuli. However, LMVN GABAergic neurons are not unrelated to the vestibular system because the dense anatomical connections with the vestibular-related areas were observed (Figure 5; Table 1). In addition, several LMVN GABAergic projection sites, such as the PB, KF, and DMTg, are involved in both sleep-wake and vestibular functions.100,101,102 The role of the vestibular system is to sense body movement and maintain body balance. Wakefulness is a state of coordination between the active brain state and the tonus of skeletal muscles, especially antigravity muscles. Thus, when the body is in a state of great motion, the brain should be awake to coordinate muscle tonus. In other words, wakefulness is maintained by the mechanism in response to vestibular inputs. Assuming that LMVN GABAergic neurons are responsible for the mechanism, when LMVN GABAergic neurons are suppressed, wakefulness dependent on vestibular inputs becomes fragmented.
Strong vestibular stimulation promotes wakefulness, whereas specific low-frequency vestibular stimulation induces sleep.11,103 Low-frequency stimulation depotentiates synaptic efficacy in MVN neurons,104 implying that slow rocking-induced sleep might be mediated by the reduction of LMVN GABAergic neuron activity. Prolonged vestibular stimulation leads to a pathological condition called sopite syndrome (e.g., drowsiness, apathy, and sleep disturbances).105,106 Synaptic inputs to MVN neurons exhibit opposite long-term changes, potentiation and depression, depending on the input pattern,107 thus dysfunctions of LMVN GABAergic neurons under particular prolonged vestibular stimulation might underlie sopite syndrome. Further study for LMVN GABAergic neurons would help in understanding these physiological or pathological relationships between sleep-wake systems and vestibular systems.
Limitations of the study
This study explores and discovers a novel population, LMVN GABAergic neurons, linking the vestibular systems and the sleep-wake systems, but does not fully demonstrate the vestibular contribution to sleep. LMVN GABAergic neurons project to the vestibulo-ocular and sleep-wake regulatory areas, but this study has not been able to show whether the projections to each area are from the identical, distinct, or overlapping neuronal populations. In addition, experiments that manipulate specific neuron activity do not yet reveal what information LMVN GABAergic neurons encode. Further detailed anatomical and physiological experiments, especially recording of firing of LMVN GABAergic neurons in vivo, would address these limitations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-FITC antibody conjugated to HRP | Jackson ImmunoResearch Labs | Cat# 200-032-037; RRID:AB_2314402 |
| Rabbit anti-RFP antibody | MBL International | Cat# PM005; RRID:AB_591279 |
| Streptavidin-conjugated Alexa Fluor 488 | Thermo Fisher Scientific | Cat# S32354; RRID:AB_2315383 |
| Goat Alexa Fluor 594-conjugated anti-rabbit IgG antibody | Thermo Fisher Scientific | Cat# A-11012; RRID:AB_2534079 |
| Donkey Alexa Fluor 594-conjugated anti-rabbit IgG antibody | Thermo Fisher Scientific | Cat# A-21207; RRID:AB_141637 |
| Streptavidin-conjugated Alexa Fluor 594 | Thermo Fisher Scientific | Cat# S32356 |
| rabbit anti-GFP antibody (for double in situ hybridization-immunohistochemistry staining) | Santa Cruz Biochemistry | Cat# sc-8334; RRID:AB_641123 |
| Alexa Fluor 488-conjugated anti-rabbit IgG antibody | Thermo Fisher Scientific | Cat# A-11008; RRID:AB_143165 |
| Rabbit anti-GFP antibody (for Immunohistochemistry) | Thermo Fisher Scientific | Cat# G10362; RRID:AB_2536526 |
| Bacterial and virus strains | ||
| AAV-hSyn-DIO-hM4Di-mCherry | Krashes et al.21 | N/A |
| AAV-EF1α-DIO-palGFP | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Clozapine N-oxide (CNO) | Enzo Life Science | Cat# BML-NS105 |
| Fluorescein RNA Labeling Mix | Sigma-Aldrich/Roche | Cat# 11685619910 |
| Blocking reagent | Sigma-Aldrich/Roche | Cat# 11096176001 |
| Critical commercial assays | ||
| TSA Plus Biotin Kit | PerkinElmer | NEL749A001KT |
| Experimental models: Cell lines | ||
| HEK293-A cell | ThermoFisher | RRID:CVCL_6910 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Mouse: GAD1-Cre mice (Gad1tm2(cre)Tama) | Higo et al.19 | MGI:4833682 |
| Oligonucleotides | ||
| Forward primer for amplification of GAD1 cDNA 5′-GCACCCGTGTTTGTTCTCAT-3′ |
This paper | N/A |
| Reverse primer for amplification of GAD1 cDNA 5′-AGCTTTGATCTTGGGAGCCA-3′ |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pAAV-hSyn-DIO-hM4Di-mCherry | Krashes et al.21 | RRID:Addgene_44362 |
| Plasmid: pAAV-EF1α-DIO-palGFP | This paper | N/A |
| Plasmid: pCAG-mGFP | Matsuda and Cepko108 | RRID:Addgene_14757 |
| Plasmid: pAAV-EF1a-double floxed-hChR2(H134R)-mCherry-WPRE-HGHpA | a gift from Karl Deisseroth | RRID:Addgene_20297 |
| Plasmid: pHelper (AAV Helper-Free System) | Agilent | Cat# 240071 |
| Plasmid: pAAV-RC (AAV Helper-Free System) | Agilent | Cat# 240071 |
| Software and algorithms | ||
| Fiji | Schindelin et al.109 | RRID:SCR_002285 |
| pCLAMP 11 Software Suite (Clampex and Clampfit) | Molecular Devices | RRID:SCR_011323 |
| SPSS software version 23 | IBM | RRID:SCR_002865 |
| JMP 11 | JMP | RRID:SCR_014242 |
| R | R Project for Statistical Computing | RRID:SCR_001905 |
Resource availability
Lead contact
Further information and requests should be directed to the lead contact, Takeshi Kanda (kanda.takeshi@naramed-u.ac.jp).
Material availability
All materials reported in this paper will be shared by the lead contact upon reasonable request.
Data and code availability
-
•
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the lead contact.
-
•
All code used in this paper will be shared by the lead contact upon reasonable request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
Experimental model and study participant details
Animals
All experimental procedures involving animals were approved by the institutional animal care and use committees of the University of Tsukuba. GAD1-Cre mice19 were used in all experiments. All mice were maintained on a C57BL/6 background and genotyped by PCR of tail DNA using primers for Cre recombinase. Mice were maintained under a strict 12-h light-dark cycle (light on at 9:00 a.m. and off at 9:00 p.m.) in a temperature- (22°C) and humidity-controlled room and fed ad libitum.
Method details
Construction of AAV plasmids
The AAV-ITR plasmid for DREADD pAAV-hSyn-DIO-hM4Di-mCherry was provided by Brian Roth (Krashes et al., 2011). The AAV-ITR plasmid for axonal tracing pAAV-EF1α-DIO-palGFP was constructed by subcloning the coding region of palGFP (mGFP) from pCAG-mGFP (a gift from Connie Cepko; Addgene plasmid #14757)108 into the pAAV-EF1α-DIO cassette made from pAAV-EF1a-double floxed-hChR2(H134R)-mCherry-WPRE-HGHpA (a gift from Karl Deisseroth; Addgene plasmid #20297).
Production of AAV vectors
To package the AAV genome into the AAV serotype 2 capsid, HEK-293A cells were cotransfected with an AAV-ITR plasmid, pHelper (Agilent), and pAAV-RC (Agilent) using calcium phosphate precipitation. Two days later, HEK-293A cells were harvested, centrifuged, and suspended in artificial cerebrospinal fluid (aCSF). The collected cells were lysed by freeze-thaw cycles following the vortexing. To isolate AAV vectors, cell debris was removed by centrifugation, and the supernatant was subjected to DNase/RNase-treatment. The titer of isolated AAV was >1012 genomic copies/ml, which was determined by quantitative PCR.
Microinjection of AAV vectors
AAV vectors were injected into GAD1-Cre mice aged 4 weeks or older. During surgery, mice were anesthetized with isoflurane (induction with 4% and maintenance with 2% through a mask at a rate of 1 mL/min), and the heads were fixed in a stereotaxic apparatus (SR-AM, Narishige), and then the bodies were placed on a heating pad (FHC-HPS-MO, Muromachi Kikai) for maintenance of body temperature at 37°C. After the skin incision along the midline and removal of the soft tissues above the skull, small craniotomies (0.5 mm diameter) were made in the skull over the areas of interest (bregma coordinates; AP from −5.3 to −6.1 mm; ML from ±0.6 to ±1.0 mm unilaterally or bilaterally) and the dura mater was removed. The coordinates were adjusted based on the bregma-lambda distance. The tip of the injection pipette filled with AAV vector solution was first placed on the brain’s surface and advanced vertically to a depth of 3.00 mm–3.93 mm from the surface with a motorized micromanipulator (EMM-3NV, Narishige). AAV vector solution (0.3–0.5 μL) was injected at the rate of >15 min/μL with a microinjector (BJ110, BEX) controlled by a pulse generator (BT200, BEX). After the withdrawal of the pipette, the skin incision was closed with silk sutures, and the mouse was returned to the home cage.
Brain slice preparation and patch-clamp recordings
Acute brain slices containing the LMVN were prepared from 6− to 7−week−old GAD1/LMVN/M4 mice injected with AAV vectors >2 weeks before patch-clamp recordings. The mice were decapitated under isoflurane anesthesia. The brains were rapidly removed into an ice-cold cutting solution that contained (in mM): 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 11 glucose, 210 sucrose, 0.5 CaCl2, 5 MgCl2 (pH 7.4, when bubbled with 95% O2 and 5% CO2). The brains were cut coronally into 200-300 μm-thick slices with a vibratome (VT-1200S, Leica). Before recordings, the slices containing the LMVN were incubated at 37°C for 1 h in artificial cerebrospinal fluid (aCSF) that contained (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 11 glucose, 2 CaCl2, 1 MgCl2 (pH 7.4, when bubbled with 95% O2 and 5% CO2). Neurons in the LMVN were visualized with a 40× water immersion objective (W Plan-Apochromat, 1.0 NA, Zeiss) and Dodt-Gradient-Contrast optics using an upright microscope (Axio Examiner D1, Zeiss) and a charge-coupled device (CCD) camera (AxioCam MRm, Zeiss). The slices were continuously perfused with oxygenated aCSF. Recording patch pipettes were pulled from glass capillaries (1B150F-4, World Precision Instruments) using a micropipette puller (P-97, Sutter Instrument) to give a resistance of 2.9–6.4 MΩ. The patch pipettes were filled with a solution containing (in mM): 135 K-gluconate, 6 NaCl, 2 MgCl2, 0.2 ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 10 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 4 Mg-Adenosine triphosphate (Mg-ATP), 0.4 Na-Guanosine-5′-triphosphate (Na-GTP), and 10 Na2-phosphocreatine (pH adjusted to 7.3 with KOH, the osmolality adjusted to 275–285 mOsm with K-gluconate). All patch-clamp recordings were performed at room temperature (RT) using a computer-controlled amplifier (MultiClamp 700B, Molecular Devices). The signals were digitized with the A/D converter (Digidata 1440A, Molecular Devices) and acquired with Clampex software (Molecular Devices) at a sampling rate of 50 kHz and low-pass filtered at 5 kHz. Whole-cell patch-clamp recordings were made from LMVN neurons expressing hM4Di which were identified by the fluorescence of mCherry. The whole-cell recordings were initially made under voltage-clamp conditions and then switched to current-clamp mode for recording action potentials. The aCSF containing 5 μM CNO was perfused after 3 min stable recordings of action potentials. Data obtained from whole-cell patch recordings were analyzed using Clampfit software (Molecular Devices). The mean firing rates in Baseline (Figure 2D) and CNO (Figure 2D) were calculated from the number of action potentials for 1 min periods from 3.5 min before and 2.5 min after the onset of CNO application (the left edge of the solid bar in Figure 2C). All data in the text are given as the mean ± SEM.
Implantation of EEG/EMG electrodes
EEG and EMG electrodes were implanted in GAD1/LMVN/M4 mice which were injected with AAV vectors >1 week before surgery. The implant surgery was performed in the same way as the AAV injection described above. The custom-made 6-pin connector consisting of four male and two female terminals (3 rows by 2 columns in the AP axis) was used as an EEG/EMG electrode. Two flexible wires (AS633, Cooner Wire Company) were soldered to two female terminals in the middle row of the 6-pin connector prior to the implantation. Four small craniotomies (0.5 mm diameter) were made in the skull over the frontal and parietal lobes in both hemispheres (2 lobes by 2 hemispheres) for mounting the 6-pin connector on the skull. Four male terminals were implanted in the craniotomies, two of which were used for EEG recordings (between ipsilateral frontoparietal electrodes), and the remaining two were not used in this study. The flexible wires were inserted into the neck muscles for EMG recordings. The 6-pin connector was glued with dental cement (3M) to the skull, and the skin was sutured. The mouse was put on a warming plate until fully recovered from anesthesia and transferred to an individual cage.
EEG/EMG recording and data analysis
Following 3–7 days of recovery from the implant surgery of EEG/EMG electrodes, mice were connected to a flexible recording cable assembly and acclimated for >5 days to the EEG/EMG recording conditions, in which a constant white-noise sound was present to avoid disruption in the sleep-wake cycle by a sudden noise. During the acclimation and the experimental period, mice were housed at a constant temperature, humidity, and light-dark cycle (same as home-cage conditions described above) with ad libitum access to food and water gel. After the acclimation, a flexible cable assembly was connected to an analog amplifier (AB-611J, Nihon Kohden). EEG signals were amplified 100000x (20x and 5000x) and band-pass filtered between 0.5 and 100 Hz. EMG signals were amplified 40000x (20x and 2000x) and band-pass filtered between 5 and 300 Hz. EEG/EMG signals were digitized at 250 Hz with a 16 bit analog-to-digital converter (NI PCIe-6320, National Instruments). EEG/EMG were recorded for >2 weeks, and EEG/EMG data for 24 h from the onset of the light or dark phase onset when CNO or saline was injected into the mice (see below) were analyzed. EEG/EMG data were split into epochs of 20 s and scored for sleep-wake states using a MATLAB (MathWorks)-based program according to the following criteria: wakefulness by low-voltage fast EEG and high-amplitude EMG; NREM sleep by high-voltage and low-frequency (1–4 Hz) EEG and low-amplitude EMG; REM sleep by low-voltage EEG with clear theta activity (6–8 Hz) and dramatic suppression of EMG with occasional muscle twitches. The EEG power spectrum was computed by fast-Fourier transform (FFT). The EEG powers in 1-Hz bins were summed in the frequency bands of 1–4 Hz (delta) or 6–8 Hz (theta), 10–15 Hz (alpha), or 10–30 Hz (beta). All data are given as the mean ± SEM.
Quantification of EMG
To evaluate vestibular-related behavior, neck EMG activity was recorded in freely-moving mice. Neck EMG recordings were the same as above. All EMG data was rectified. EMG data was split into 20-s epochs and only epochs of wakefulness were quantified by the variance and the integral. The EMG variance for each epoch was calculated as follows: the variance for the 4 consecutive of the 0.5-s summed values of rectified EMG was calculated (the 2-s variance value), and the median of the consecutive 10 of the 2-s variance values was used as the EMG variance. The EMG integral for each epoch was the mean of the consecutive 5 of the 4-s summed values of rectified EMG.
Intraperitoneal administration
CNO (Enzo Life Science) was dissolved in saline to a concentration of 0.5 mg/mL and intraperitoneally (ip) injected into mice at a dose of 5 mg/kg body weight (10 mL/kg body weight). Mice were subjected to ip injection of saline (10 mL/kg body weight) at least 3 times before EEG/EMG recordings for acclimation to the procedure. CNO or saline was intraperitoneally injected into GAD1/LMVN/M4 mice at ZT0 or ZT12. EEG/EMG recordings were resumed within 3 min after ip administration. The administration interval was >3 days. Effects of CNO administration on sleep-wake states in GAD1/LMVN/M4 mice were evaluated by comparison to that of saline administration into the same mice at the same time point unless otherwise stated.
Synthesis of a probe for in situ hybridization
A fragment of GAD1 cDNA was polymerase chain reaction (PCR)–amplified from a mouse brain cDNA library by the following primers: sense 5′-GCACCCGTGTTTGTTCTCAT-3′ and antisense 5′-AGCTTTGATCTTGGGAGCCA-3′, and cloned into the multi-cloning site of pGEM-T Easy vector (Promega). The plasmid vector containing a fragment of GAD1 cDNA was digested with a restriction enzyme NcoI to linearize at the 5′ end of a cDNA fragment, which was used as a template for PCR with SP6 promoter primer from its 3′ end to synthesize the probe for GAD1 mRNA. The probe was labeled with fluorescein (FITC) (11685619910, Sigma-Aldrich/Roche).
Brain slice preparation for histology
The mice injected with AAV vectors >2 weeks before histology were anesthetized with isoflurane and transcardially perfused with saline, followed by 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS, 0.1 M). The brains were removed from the skull after the perfusion and post-fixed with the same fixative for >12 h at 4°C. After post-fixation, the brains were placed in a 30% sucrose in PBS for >24 h and then stored with optimal cutting temperature compound (Tissue-Tek OTC compound, Sakura Finetek) at −80°C. The brains were sectioned coronally at a thickness of 40 μm for double ISH/IHC staining or 60 μm for single IHC staining with a cryostat (CM3050S, Leica). The sections were washed with phosphate buffer (PB, 0.1 M) three times for 10 min each before the following FISH and IHC procedures.
Double in situ hybridization-immunohistochemistry staining
All steps were performed at RT unless otherwise indicated. The brain sections containing the LMVN of GAD1-Cre/LMVN/M4 mice were used. The sections were re-fixed with 4% PFA for 15 min, rinsed with PBS three times for 10 min each, permeabilized with 0.3% Triton X-100, and then rinsed with PBS twice for 5 min each. For the pretreatment, the sections were digested with 1 μg/mL proteinase K at 37°C for 30 min, neutralized with 0.75% glycine twice for 15 min each, rinsed with PBS twice for 5 min each, acetylated with 0.25% acetic anhydride for 15 min, and then rinsed with PBS twice for 5 min each. The pretreated sections were pre-hybridized for 1–2 h and hybridized overnight at 60°C with a FITC-labeled probe for GAD1 mRNA in hybridization buffer (50% deionized formamide (dFA), 5x saline sodium citrate buffer (SSC), 0.1% N-lauroylsarcosine (NLS), 0.1% sodium dodecyl sulfate (SDS), and 2% blocking reagent (11096176001, Sigma-Aldrich/Roche)). The hybridized sections were rinsed with 2x SSC/50% dFA/0.1% NLS at 50°C twice for 20 min each. After rinsing, the sections were incubated with RNase A buffer (10 mM Tris-HCl [pH 8.0], 1 mM ethylenediaminetetraacetic acid (EDTA), 0.5 M NaCl) for 5 min and treated with RNase A in RNase A buffer for 20 min at 37°C followed by washing in 2x SSC/0.1% NLS at 37°C twice for 20 min each, in 0.2x SSC/0.1% NLS at 37°C twice for 20 min each, and in tris-buffered saline (TBS, 50 mM) for 5 min at RT. Endogenous peroxidase activity was quenched with 0.3% H2O2 in TBS for 20 min. After rinsing with TBS for 5 min, the sections were blocked with 1% blocking reagent for 1 h and sequentially incubated with a mouse anti-FITC antibody conjugated to horseradish peroxidase (HRP) (1:80000; 200-032-037, Jackson ImmunoResearch Labs) and a rabbit anti-RFP antibody (1:1,000; PM005, MBL International) diluted in 1% blocking reagent/0.5% Triton X-100 in TBS at 4°C overnight. Following washing with TNT (TBS, 0.15 M NaCl, and 0.05% Tween 20) three times for 10 min each, the HRP signal was enhanced using the tyramide signal amplification (TSA) biotin system (TSA Plus Biotin Kit, PerkinElmer). After rinsing with TNT, the sections were incubated with a streptavidin-conjugated Alexa Fluor 488 (1:1000; S32354, Thermo Fisher Scientific) and an Alexa Fluor 594-conjugated anti-rabbit IgG antibody (1:1000; A-11012 or A-21207, Thermo Fisher Scientific) diluted in 1% blocking reagent in TBS for 4−5 h. Double ISH/IHC staining of GAD1 mRNA and palGFP was performed in the same way, but using a streptavidin-conjugated Alexa Fluor 594 (1:1000; S32356, Thermo Fisher Scientific) for detection of GAD1 mRNA, a rabbit anti-GFP antibody (1:200; sc-8334, Santa Cruz Biochemistry) as a primary antibody for IHC of palGFP, and an Alexa Fluor 488-conjugated anti-rabbit IgG antibody (1:1000; A-11008, Thermo Fisher Scientific) as the secondary antibody.
Immunohistochemistry for axonal tracing and data analysis
All steps were performed at RT unless otherwise indicated. The sections made from the brains of GAD1-Cre mice unilaterally injected with Cre-inducible AAV vectors encoding palGFP into the LMVN were used. The sections were washed with 0.1 M PB three times for 10 min each, blocked with 3% bovine serum albumins (BSA) in TBS for 1h, and immunostained with a rabbit anti-GFP antibody (G10362; Thermo Fisher Scientific/Molecular Probes) diluted in 0.5% Triton X-100 in 0.5 mM TBS at 4°C overnight. The sections were rinsed with TBS three times for 10 min each and incubated with Alexa Fluor 488-conjugated anti-rabbit IgG secondary antibody (1:1000, A11008, Thermo Fisher Scientific/Molecular) diluted in TBS for >5 h at 4°C.
Confocal imaging and image analysis
After double ISH/IHC or single IHC staining, the sections were rinsed with 0.1 M PB three times for 10 min each and mounted with an anti-photobleaching medium (H-1200, Vector Labs). Imaging was performed using a confocal laser scanning microscope (Axio Imager Z2 and LSM700, Zeiss). In some sections, lambda scan mode was used to remove autofluorescence signals in the confocal images. Image analysis was performed with Fiji (RRID:SCR_002285).109 GFP-positive axon density was estimated based on the appearance, not the fluorescence intensity, of GFP-positive axons in the region of interest. The density was scored on 0–4 scales (4, highly dense; 3, dense; 2, moderately dense; 1, sparse; 0, highly sparse). The symbols in Table 1 indicate the total value of the axon density from 3 mice (++++, >10; +++, >7 ∩ ≤10; ++, >4 ∩ ≤7; +, >0 ∩ ≤4; -, 0). Drawings were adapted from the brain atlas of Franklin and Paxinos.22 Anatomical abbreviations are listed in Table 1.
Quantification and statistical analysis
Statistical analysis
All statistical analyses were carried out using SPSS software version 23 (IBM), JMP 11 (JMP), and R. Wilcoxon signed-rank test was used for paired comparisons (Figures 1G, 2C, 2D, 3, 4, and S4). Wilcoxon rank-sum test was applied for intergroup comparisons (between administration times of drugs). Two-way repeated-measures ANOVA followed by Bonferroni test was used for statistical analysis of hourly sleep data (Figures 2C, 2D, and S2) and EEG power spectrum (Figures S3A and S3B) as follows: if there was a significant interaction between the administrated drugs (CNO and saline) and time (ZT) or frequency (Hz), then the main effect of the administered drug at each time point or frequency was tested. EEG powers in the several frequency bands were analyzed by Student’s paired t-test. The level of significance was set at two-tailed p < 0.05.
Acknowledgments
This work was supported by World Premier International Research Center Initiative (WPI) from JSPS, JSPS KAKENHI (20K06922 and 23K05998 to T. K; 22H04918 to M.Y.), and the Food Science Institute Foundation (to T. K.).
We appreciate the technical assistance and advice of Junya Fukuoka, Tatsuya Korokawa, Miyo Kakizaki, and Mari Hondo (University of Tsukuba). We thank Kenji Sakimura (Niigata University) for providing GAD1-Cre mice.
Author contributions
Conceptualization, Methodology, Formal analysis, and Investigation, D.N. and T.K.; Software, M.S.; Resources, Y.C. and Y.I.; Supervision, M.Y.; Funding Acquisition, T.K. and M.Y.; Discussion and Writing, all authors.
Declaration of interests
The authors declare no competing interests.
Published: February 21, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109289.
Supplemental information
References
- 1.Everson C.A., Bergmann B.M., Rechtschaffen A. Sleep Deprivation in the Rat: III. Total Sleep Deprivation. Sleep. 1989;12:13–21. doi: 10.1093/sleep/12.1.13. [DOI] [PubMed] [Google Scholar]
- 2.Rechtschaffen A., Gilliland M.A., Bergmann B.M., Winter J.B. Physiological correlates of prolonged sleep deprivation in rats. Science. 1983;221:182–184. doi: 10.1126/science.6857280. [DOI] [PubMed] [Google Scholar]
- 3.Eban-Rothschild A., Appelbaum L., de Lecea L. Neuronal Mechanisms for Sleep/Wake Regulation and Modulatory Drive. Neuropsychopharmacol. 2018;43:937–952. doi: 10.1038/npp.2017.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Bon O. Relationships between REM and NREM in the NREM-REM sleep cycle: a review on competing concepts. Sleep Med. 2020;70:6–16. doi: 10.1016/j.sleep.2020.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Saper C.B., Fuller P.M. Wake–sleep circuitry: an overview. Curr. Opin. Neurobiol. 2017;44:186–192. doi: 10.1016/j.conb.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill S., Tononi G. Modeling Sleep and Wakefulness in the Thalamocortical System. J. Neurophysiol. 2005;93:1671–1698. doi: 10.1152/jn.00915.2004. [DOI] [PubMed] [Google Scholar]
- 7.Luppi P.-H., Gervasoni D., Verret L., Goutagny R., Peyron C., Salvert D., Leger L., Fort P. Paradoxical (REM) sleep genesis: The switch from an aminergic–cholinergic to a GABAergic–glutamatergic hypothesis. J. Physiol. Paris. 2006;100:271–283. doi: 10.1016/j.jphysparis.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Saper C.B., Chou T.C., Scammell T.E. The sleep switch: hypothalamic control of sleep and wakefulness. Trends Neurosci. 2001;24:726–731. doi: 10.1016/S0166-2236(00)02002-6. [DOI] [PubMed] [Google Scholar]
- 9.Steriade M., McCormick D.A., Sejnowski T.J. Thalamocortical Oscillations in the Sleeping and Aroused Brain. Science. 1993;262:679–685. doi: 10.1126/science.8235588. [DOI] [PubMed] [Google Scholar]
- 10.Besnard S., Tighilet B., Chabbert C., Hitier M., Toulouse J., Le Gall A., Machado M.-L., Smith P.F. The balance of sleep: Role of the vestibular sensory system. Sleep Med. Rev. 2018;42:220–228. doi: 10.1016/j.smrv.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Kompotis K., Hubbard J., Emmenegger Y., Perrault A., Mühlethaler M., Schwartz S., Bayer L., Franken P. Rocking Promotes Sleep in Mice through Rhythmic Stimulation of the Vestibular System. Curr. Biol. 2019;29:392–401.e4. doi: 10.1016/j.cub.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Bell B.J., Wang A.A., Kim D.W., Xiong J., Blackshaw S., Wu M.N. Characterization of mWake expression in the murine brain. J. Comp. Neurol. 2021;529:1954–1987. doi: 10.1002/cne.25066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashiwagi M., Kanuka M., Tatsuzawa C., Suzuki H., Morita M., Tanaka K., Kawano T., Shin J.W., Suzuki H., Itohara S., et al. Widely Distributed Neurotensinergic Neurons in the Brainstem Regulate NREM Sleep in Mice. Curr. Biol. 2020;30:1002–1010.e4. doi: 10.1016/j.cub.2020.01.047. [DOI] [PubMed] [Google Scholar]
- 14.Black R.D., Rogers L.L. Sensory Neuromodulation. Front. Syst. Neurosci. 2020;14 doi: 10.3389/fnsys.2020.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gervasoni D., Peyron C., Rampon C., Barbagli B., Chouvet G., Urbain N., Fort P., Luppi P.-H. Role and Origin of the GABAergic Innervation of Dorsal Raphe Serotonergic Neurons. J. Neurosci. 2000;20:4217–4225. doi: 10.1523/JNEUROSCI.20-11-04217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumoi K., Saito N., Tanaka C. Immunohistochemical localization of γ-aminobutyric acid- and aspartate-containing neurons in the guinea pig vestibular nuclei. Brain Res. 1987;416:22–33. doi: 10.1016/0006-8993(87)91492-2. [DOI] [PubMed] [Google Scholar]
- 17.Nomura I., Senba E., Kubo T., Shiraishi T., Matsunaga T., Tohyama M., Shiotani Y., Wu J.-Y. Neuropeptides and γ-Aminobutyric acid in the vestibular nuclei of the rat: an immunohistochemical analysis. I. Distribution. Brain Res. 1984;311:109–118. doi: 10.1016/0006-8993(84)91403-3. [DOI] [PubMed] [Google Scholar]
- 18.Walberg F., Ottersen O.P., Rinvik E. GABA, glycine, aspartate, glutamate and taurine in the vestibular nuclei: an immunocytochemical investigation in the cat. Exp. Brain Res. 1990;79:547–563. doi: 10.1007/BF00229324. [DOI] [PubMed] [Google Scholar]
- 19.Higo S., Akashi K., Sakimura K., Tamamaki N. Subtypes of GABAergic neurons project axons in the neocortex. Front. Neuroanat. 2009;3:25. doi: 10.3389/neuro.05.025.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander G.M., Rogan S.C., Abbas A.I., Armbruster B.N., Pei Y., Allen J.A., Nonneman R.J., Hartmann J., Moy S.S., Nicolelis M.A., et al. Remote Control of Neuronal Activity in Transgenic Mice Expressing Evolved G Protein-Coupled Receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krashes M.J., Koda S., Ye C., Rogan S.C., Adams A.C., Cusher D.S., Maratos-Flier E., Roth B.L., Lowell B.B. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franklin K.P., Paxinos G. 2008. The Mouse Brain in Stereotaxic Coordinates. [Google Scholar]
- 23.Anaclet C., Lin J.-S., Vetrivelan R., Krenzer M., Vong L., Fuller P.M., Lu J. Identification and Characterization of a Sleep-Active Cell Group in the Rostral Medullary Brainstem. J. Neurosci. 2012;32:17970–17976. doi: 10.1523/JNEUROSCI.0620-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anaclet C., Ferrari L., Arrigoni E., Bass C.E., Saper C.B., Lu J., Fuller P.M. The GABAergic parafacial zone is a medullary slow wave sleep–promoting center. Nat. Neurosci. 2014;17:1217–1224. doi: 10.1038/nn.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luppi P.-H., Peyron C., Fort P. Not a single but multiple populations of GABAergic neurons control sleep. Sleep Med. Rev. 2017;32:85–94. doi: 10.1016/j.smrv.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Weber F., Chung S., Beier K.T., Xu M., Luo L., Dan Y. Control of REM sleep by ventral medulla GABAergic neurons. Nature. 2015;526:435–438. doi: 10.1038/nature14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothermel M., Brunert D., Zabawa C., Díaz-Quesada M., Wachowiak M. Transgene Expression in Target-Defined Neuron Populations Mediated by Retrograde Infection with Adeno-Associated Viral Vectors. J. Neurosci. 2013;33:15195–15206. doi: 10.1523/JNEUROSCI.1618-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bizzi E., Polit A., Morasso P. Mechanisms underlying achievement of final head position. J. Neurophysiol. 1976;39:435–444. doi: 10.1152/jn.1976.39.2.435. [DOI] [PubMed] [Google Scholar]
- 29.Laposky A.D., Bass J., Kohsaka A., Turek F.W. Sleep and circadian rhythms: key components in the regulation of energy metabolism. FEBS Lett. 2008;582:142–151. doi: 10.1016/j.febslet.2007.06.079. [DOI] [PubMed] [Google Scholar]
- 30.Pace-Schott E.F., Hobson J.A. The Neurobiology of Sleep: Genetics, cellular physiology and subcortical networks. Nat. Rev. Neurosci. 2002;3:591–605. doi: 10.1038/nrn895. [DOI] [PubMed] [Google Scholar]
- 31.Saper C.B., Scammell T.E., Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 32.Wulff K., Gatti S., Wettstein J.G., Foster R.G. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat. Rev. Neurosci. 2010;11:589–599. doi: 10.1038/nrn2868. [DOI] [PubMed] [Google Scholar]
- 33.Benington J.H., Heller H.C. REM-sleep timing is controlled homeostatically by accumulation of REM-sleep propensity in non-REM sleep. Am. J. Physiol. 1994;266:R1992–R2000. doi: 10.1152/ajpregu.1994.266.6.R1992. [DOI] [PubMed] [Google Scholar]
- 34.Bardy C., van den Hurk M., Eames T., Marchand C., Hernandez R.V., Kellogg M., Gorris M., Galet B., Palomares V., Brown J., et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl. Acad. Sci. USA. 2015;112:E2725–E2734. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins W.E., Posner J.B. Electroencephalogram Alpha-activity during Mild Vestibular Stimulation. Nature. 1963;199:933–934. doi: 10.1038/199933a0. [DOI] [PubMed] [Google Scholar]
- 36.Kim D.J., Yogendrakumar V., Chiang J., Ty E., Wang Z.J., McKeown M.J. Noisy Galvanic Vestibular Stimulation Modulates the Amplitude of EEG Synchrony Patterns. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell N.A., Horii A., Smith P.F., Darlington C.L., Bilkey D.K. Lesions of the Vestibular System Disrupt Hippocampal Theta Rhythm in the Rat. J. Neurophysiol. 2006;96:4–14. doi: 10.1152/jn.00953.2005. [DOI] [PubMed] [Google Scholar]
- 38.Borbély A.A. A two process model of sleep regulation. Hum. Neurobiol. 1982;1:195–204. [PubMed] [Google Scholar]
- 39.Franken P., Chollet D., Tafti M. The Homeostatic Regulation of Sleep Need Is under Genetic Control. J. Neurosci. 2001;21:2610–2621. doi: 10.1523/JNEUROSCI.21-08-02610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tobler I., Borbély A.A. Sleep EEG in the rat as a function of prior waking. Electroencephalogr. Clin. Neurophysiol. 1986;64:74–76. doi: 10.1016/0013-4694(86)90044-1. [DOI] [PubMed] [Google Scholar]
- 41.Gliddon C.M., Darlington C.L., Smith P.F. GABAergic systems in the vestibular nucleus and their contribution to vestibular compensation. Prog. Neurobiol. 2005;75:53–81. doi: 10.1016/j.pneurobio.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Holstein G.R. Inhibitory amino acid transmitters in the vestibular nuclei. Neurochemistry of the vestibular system. 2000:143–162. [Google Scholar]
- 43.Shi X.-B., Wang J., Li F.-T., Zhang Y.-B., Qu W.-M., Dai C.-F., Huang Z.-L. Whole-brain monosynaptic outputs and presynaptic inputs of GABAergic neurons in the vestibular nuclei complex of mice. Front. Neurosci. 2022;16 doi: 10.3389/fnins.2022.982596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furuta T., Tomioka R., Taki K., Nakamura K., Tamamaki N., Kaneko T. In Vivo Transduction of Central Neurons Using Recombinant Sindbis Virus: Golgi-like Labeling of Dendrites and Axons with Membrane-targeted Fluorescent Proteins. J. Histochem. Cytochem. 2001;49:1497–1508. doi: 10.1177/002215540104901203. [DOI] [PubMed] [Google Scholar]
- 45.Okada A., Lansford R., Weimann J.M., Fraser S.E., McConnell S.K. Imaging Cells in the Developing Nervous System with Retrovirus Expressing Modified Green Fluorescent Protein. Exp. Neurol. 1999;156:394–406. doi: 10.1006/exnr.1999.7033. [DOI] [PubMed] [Google Scholar]
- 46.Tamamaki N., Nakamura K., Furuta T., Asamoto K., Kaneko T. Neurons in Golgi-stain-like images revealed by GFP-adenovirus infection in vivo. Neurosci. Res. 2000;38:231–236. doi: 10.1016/S0168-0102(00)00176-0. [DOI] [PubMed] [Google Scholar]
- 47.Kasahara M., Mano N., Oshima T., Ozawa S., Shimazu H. Contralateral short latency inhibition of central vestibular neurons in the horizontal canal system. Brain Res. 1968;8:376–378. doi: 10.1016/0006-8993(68)90057-7. [DOI] [PubMed] [Google Scholar]
- 48.Kasahara M., Uchino Y. Selective mode of commissural inhibition induced by semicircular canal afferents on secondary vestibular neurones in the cat. Brain Res. 1971;34:366–369. doi: 10.1016/0006-8993(71)90288-5. [DOI] [PubMed] [Google Scholar]
- 49.Mano N., Oshima T., Shimazu H. Inhibitory commissural fibers interconnecting the bilateral vestibular nuclei. Brain Res. 1968;8:378–382. doi: 10.1016/0006-8993(68)90058-9. [DOI] [PubMed] [Google Scholar]
- 50.Shimazu H., Precht W. Inhibition of central vestibular neurons from the contralateral labyrinth and its mediating pathway. J. Neurophysiol. 1966;29:467–492. doi: 10.1152/jn.1966.29.3.467. [DOI] [PubMed] [Google Scholar]
- 51.Boyce R., Glasgow S.D., Williams S., Adamantidis A. Causal evidence for the role of REM sleep theta rhythm in contextual memory consolidation. Science. 2016;352:812–816. doi: 10.1126/science.aad5252. [DOI] [PubMed] [Google Scholar]
- 52.Buzsáki G. Theta Oscillations in the Hippocampus. Neuron. 2002;33:325–340. doi: 10.1016/S0896-6273(02)00586-X. [DOI] [PubMed] [Google Scholar]
- 53.Gerashchenko D., Salin-Pascual R., Shiromani P.J. Effects of hypocretin–saporin injections into the medial septum on sleep and hippocampal thetaq. Brain Res. 2001;10 doi: 10.1016/s0006-8993(01)02792-5. [DOI] [PubMed] [Google Scholar]
- 54.Petsche H., Stumpf C., Gogolak G. The significance of the rabbit’s septum as a relay station between the midbrain and the hippocampus I. The control of hippocampus arousal activity by the septum cells. Electroencephalogr. Clin. Neurophysiol. 1962;14:202–211. doi: 10.1016/0013-4694(62)90030-5. [DOI] [PubMed] [Google Scholar]
- 55.Vertes R.P., Kocsis B. Brainstem-diencephalo-septohippocampal systems controlling the theta rhythm of the hippocampus. Neuroscience. 1997;81:893–926. doi: 10.1016/s0306-4522(97)00239-x. [DOI] [PubMed] [Google Scholar]
- 56.Funato H., Sato M., Sinton C.M., Gautron L., Williams S.C., Skach A., Elmquist J.K., Skoultchi A.I., Yanagisawa M. Loss of Goosecoid-like and DiGeorge syndrome critical region 14 in interpeduncular nucleus results in altered regulation of rapid eye movement sleep. Proc. Natl. Acad. Sci. USA. 2010;107:18155–18160. doi: 10.1073/pnas.1012764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan P.J., Ma S., Olucha-Bordonau F.E., Gundlach A.L. Nucleus incertus--an emerging modulatory role in arousal, stress and memory. Neurosci. Biobehav. Rev. 2011;35:1326–1341. doi: 10.1016/j.neubiorev.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 58.Saper C.B., Fuller P.M., Pedersen N.P., Lu J., Scammell T.E. Sleep State Switching. Neuron. 2010;68:1023–1042. doi: 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horowitz S.S., Blanchard J., Morin L.P. Medial vestibular connections with the hypocretin (orexin) system. J. Comp. Neurol. 2005;487:127–146. doi: 10.1002/cne.20521. [DOI] [PubMed] [Google Scholar]
- 60.Siegel J.M., Tomaszewski K.S., Nienhuis R. Behavioral states in the chronic medullary and midpontine cat. Electroencephalogr. Clin. Neurophysiol. 1986;63:274–288. doi: 10.1016/0013-4694(86)90095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pompeiano O., Morrison A.R. Vestibular influences during sleep. I. Abolition of the rapid eye movements of desynchronized sleep following vestibular lesions. Arch. Ital. Biol. 1965;103:569–595. [PubMed] [Google Scholar]
- 62.Blanco-Centurion C., Gerashchenko D., Shiromani P.J. Effects of Saporin-Induced Lesions of Three Arousal Populations on Daily Levels of Sleep and Wake. J. Neurosci. 2007;27:14041–14048. doi: 10.1523/JNEUROSCI.3217-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carter M.E., Yizhar O., Chikahisa S., Nguyen H., Adamantidis A., Nishino S., Deisseroth K., de Lecea L. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat. Neurosci. 2010;13:1526–1533. doi: 10.1038/nn.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones B.E., Harper S.T., Halaris A.E. Effects of locus coeruleus lesions upon cerebral monoamine content, sleep-wakefulness states and the response to amphetamine in the cat. Brain Res. 1977;124:473–496. doi: 10.1016/0006-8993(77)90948-9. [DOI] [PubMed] [Google Scholar]
- 65.Lidbrink P. The effect of lesions of ascending noradrenaline pathways on sleep and waking in the rat. Brain Res. 1974;74:19–40. doi: 10.1016/0006-8993(74)90109-7. [DOI] [PubMed] [Google Scholar]
- 66.Chemelli R.M., Willie J.T., Sinton C.M., Elmquist J.K., Scammell T., Lee C., Richardson J.A., Williams S.C., Xiong Y., Kisanuki Y., et al. Narcolepsy in orexin Knockout Mice. Cell. 1999;98:437–451. doi: 10.1016/S0092-8674(00)81973-X. [DOI] [PubMed] [Google Scholar]
- 67.Hara J., Beuckmann C.T., Nambu T., Willie J.T., Chemelli R.M., Sinton C.M., Sugiyama F., Yagami K., Goto K., Yanagisawa M., Sakurai T. Genetic Ablation of Orexin Neurons in Mice Results in Narcolepsy, Hypophagia, and Obesity. Neuron. 2001;30:345–354. doi: 10.1016/S0896-6273(01)00293-8. [DOI] [PubMed] [Google Scholar]
- 68.Mieda M., Hasegawa E., Kisanuki Y.Y., Sinton C.M., Yanagisawa M., Sakurai T. Differential Roles of Orexin Receptor-1 and -2 in the Regulation of Non-REM and REM Sleep. J. Neurosci. 2011;31:6518–6526. doi: 10.1523/JNEUROSCI.6506-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willie J.T., Chemelli R.M., Sinton C.M., Yanagisawa M. To Eat or to Sleep? Orexin in the Regulation of Feeding and Wakefulness. Annu. Rev. Neurosci. 2001;24:429–458. doi: 10.1146/annurev.neuro.24.1.429. [DOI] [PubMed] [Google Scholar]
- 70.Bizzi E., Pompeiano O., Somogyi I. Vestibular Nuclei: Activity of Single Neurons during Natural Sleep and Wakefulness. Science. 1964;145:414–415. doi: 10.1126/science.145.3630.414. [DOI] [PubMed] [Google Scholar]
- 71.Bizzi E., Pompeiano O., Somogyi I. SPONTANEOUS ACTIVITY OF SINGLE VESTIBULAR NEURONS OF UNRESTRAINED CATS DURING SLEEP AND WAKEFULNESS. Arch. Ital. Biol. 1964;102:308–330. [PubMed] [Google Scholar]
- 72.Chubb M.C., Fuchs A.F. Contribution of y group of vestibular nuclei and dentate nucleus of cerebellum to generation of vertical smooth eye movements. J. Neurophysiol. 1982;48:75–99. doi: 10.1152/jn.1982.48.1.75. [DOI] [PubMed] [Google Scholar]
- 73.Brodal A., Pompeiano O. The vestibular nuclei in the cat. J. Anat. 1957;91:438–454.1. [PMC free article] [PubMed] [Google Scholar]
- 74.Krout K.E., Belzer R.E., Loewy A.D. Brainstem projections to midline and intralaminar thalamic nuclei of the rat. J. Comp. Neurol. 2002;448:53–101. doi: 10.1002/cne.10236. [DOI] [PubMed] [Google Scholar]
- 75.Barmack N.H. Central vestibular system: vestibular nuclei and posterior cerebellum. Brain Res. Bull. 2003;60:511–541. doi: 10.1016/S0361-9230(03)00055-8. [DOI] [PubMed] [Google Scholar]
- 76.Brown J.E., Card J.P., Yates B.J. Polysynaptic pathways from the vestibular nuclei to the lateral mammillary nucleus of the rat: substrates for vestibular input to head direction cells. Exp. Brain Res. 2005;161:47–61. doi: 10.1007/s00221-004-2045-4. [DOI] [PubMed] [Google Scholar]
- 77.Zhong P., Zhang Z., Barger Z., Ma C., Liu D., Ding X., Dan Y. Control of Non-REM Sleep by Midbrain Neurotensinergic Neurons. Neuron. 2019;104:795–809.e6. doi: 10.1016/j.neuron.2019.08.026. [DOI] [PubMed] [Google Scholar]
- 78.Sherin J.E., Shiromani P.J., McCarley R.W., Saper C.B. Activation of Ventrolateral Preoptic Neurons During Sleep. Science. 1996;271:216–219. doi: 10.1126/science.271.5246.216. [DOI] [PubMed] [Google Scholar]
- 79.Hayat H., Regev N., Matosevich N., Sales A., Paredes-Rodriguez E., Krom A.J., Bergman L., Li Y., Lavigne M., Kremer E.J., et al. Locus coeruleus norepinephrine activity mediates sensory-evoked awakenings from sleep. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aaz4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kjaerby C., Andersen M., Hauglund N., Untiet V., Dall C., Sigurdsson B., Ding F., Feng J., Li Y., Weikop P., et al. Memory-enhancing properties of sleep depend on the oscillatory amplitude of norepinephrine. Nat. Neurosci. 2022;25:1059–1070. doi: 10.1038/s41593-022-01102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Breton-Provencher V., Sur M. Active control of arousal by a locus coeruleus GABAergic circuit. Nat. Neurosci. 2019;22:218–228. doi: 10.1038/s41593-018-0305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu Q., Wang D.-R., Dong H., Chen L., Lu J., Lazarus M., Cherasse Y., Chen G.-H., Qu W.-M., Huang Z.-L. Medial Parabrachial Nucleus Is Essential in Controlling Wakefulness in Rats. Front. Neurosci. 2021;15 doi: 10.3389/fnins.2021.645877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adamantidis A.R., de Lecea L. Sleep and the hypothalamus. Science. 2023;382:405–412. doi: 10.1126/science.adh8285. [DOI] [PubMed] [Google Scholar]
- 84.Boissard R., Gervasoni D., Schmidt M.H., Barbagli B., Fort P., Luppi P.-H. The rat ponto-medullary network responsible for paradoxical sleep onset and maintenance: a combined microinjection and functional neuroanatomical study: REM sleep brainstem structures in rats. Eur. J. Neurosci. 2002;16:1959–1973. doi: 10.1046/j.1460-9568.2002.02257.x. [DOI] [PubMed] [Google Scholar]
- 85.Verret L., Léger L., Fort P., Luppi P.-H. Cholinergic and noncholinergic brainstem neurons expressing Fos after paradoxical (REM) sleep deprivation and recovery. Eur. J. Neurosci. 2005;21:2488–2504. doi: 10.1111/j.1460-9568.2005.04060.x. [DOI] [PubMed] [Google Scholar]
- 86.Yamuy J., Mancillas J.R., Morales F.R., Chase M.H. C-fos expression in the pons and medulla of the cat during carbachol- induced active sleep. J. Neurosci. 1993;13:2703–2718. doi: 10.1523/JNEUROSCI.13-06-02703.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weber F., Hoang Do J.P., Chung S., Beier K.T., Bikov M., Saffari Doost M., Dan Y. Regulation of REM and Non-REM Sleep by Periaqueductal GABAergic Neurons. Nat. Commun. 2018;9:354. doi: 10.1038/s41467-017-02765-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sapin E., Lapray D., Bérod A., Goutagny R., Léger L., Ravassard P., Clément O., Hanriot L., Fort P., Luppi P.-H. Localization of the Brainstem GABAergic Neurons Controlling Paradoxical (REM) Sleep. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duensing F., Schaefer K.-P. Die Aktivität einzelner Neurone im Bereich der Vestibulariskerne bei Horizontalbeschleunigungen unter besonderer Berücksichtigung des vestibulären Nystagmus. Arch. F. Psychiatr. U. Z. Neur. 1958;198:225–252. doi: 10.1007/BF00941383. [DOI] [PubMed] [Google Scholar]
- 90.Gernandt B. RESPONSE OF MAMMALIAN VESTIBULAR NEURONS TO HORIZONTAL ROTATION AND CALORIC STIMULATION. J. Neurophysiol. 1949;12:173–184. doi: 10.1152/jn.1949.12.3.173. [DOI] [PubMed] [Google Scholar]
- 91.Shimazu H., Precht W. TONIC AND KINETIC RESPONSES OF CAT’S VESTIBULAR NEURONS TO HORIZONTAL ANGULAR ACCELERATION. J. Neurophysiol. 1965;28:991–1013. doi: 10.1152/jn.1965.28.6.991. [DOI] [PubMed] [Google Scholar]
- 92.Bagnall M.W., Stevens R.J., du Lac S. Transgenic Mouse Lines Subdivide Medial Vestibular Nucleus Neurons into Discrete, Neurochemically Distinct Populations. J. Neurosci. 2007;27:2318–2330. doi: 10.1523/JNEUROSCI.4322-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.du Lac S., Lisberger S.G. Membrane and firing properties of avian medial vestibular nucleus neuronsin vitro. J. Comp. Physiol. 1995;176:641–651. doi: 10.1007/BF01021584. [DOI] [PubMed] [Google Scholar]
- 94.Todaka H., Tatsukawa T., Hashikawa T., Yanagawa Y., Shibuki K., Nagao S. Heterotrimeric guanosine triphosphate-binding protein-coupled modulatory actions of motilin on K + channels and postsynaptic γ-aminobutyric acid receptors in mouse medial vestibular nuclear neurons. Eur. J. Neurosci. 2013;37:339–350. doi: 10.1111/ejn.12051. [DOI] [PubMed] [Google Scholar]
- 95.Phelan K.D., Gallagher J.P. Direct muscarinic and nicotinic receptor-mediated excitation of rat medial vestibular nucleus neurons in vitro. Synapse. 1992;10:349–358. doi: 10.1002/syn.890100410. [DOI] [PubMed] [Google Scholar]
- 96.Chen Z.-P., Zhang X.-Y., Peng S.-Y., Yang Z.-Q., Wang Y.-B., Zhang Y.-X., Chen X., Wang J.-J., Zhu J.-N. Histamine H1 Receptor Contributes to Vestibular Compensation. J. Neurosci. 2019;39:420–433. doi: 10.1523/JNEUROSCI.1350-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnston A.R., Murnion B., McQueen D.S., Dutia M.B. Excitation and inhibition of rat medial vestibular nucleus neurones by 5-hydroxytryptamine. Exp. Brain Res. 1993;93:293–298. doi: 10.1007/BF00228397. [DOI] [PubMed] [Google Scholar]
- 98.Licata F., Li Volsi G., Maugeri G., Ciranna L., Santangelo F. Effects of noradrenaline on the firing rate of vestibular neurons. Neuroscience. 1993;53:149–158. doi: 10.1016/0306-4522(93)90293-O. [DOI] [PubMed] [Google Scholar]
- 99.Montardy Q., Wei M., Liu X., Yi T., Zhou Z., Lai J., Zhao B., Besnard S., Tighilet B., Chabbert C., Wang L. Selective optogenetic stimulation of glutamatergic, but not GABAergic, vestibular nuclei neurons induces immediate and reversible postural imbalance in mice. Prog. Neurobiol. 2021;204 doi: 10.1016/j.pneurobio.2021.102085. [DOI] [PubMed] [Google Scholar]
- 100.Valencia Garcia S., Libourel P.-A., Lazarus M., Grassi D., Luppi P.-H., Fort P. Genetic inactivation of glutamate neurons in the rat sublaterodorsal tegmental nucleus recapitulates REM sleep behaviour disorder. Brain. 2017;140:414–428. doi: 10.1093/brain/aww310. [DOI] [PubMed] [Google Scholar]
- 101.Bonis J.M., Neumueller S.E., Krause K.L., Kiner T., Smith A., Marshall B.D., Qian B., Pan L.G., Forster H.V. A role for the Kölliker-Fuse nucleus in cholinergic modulation of breathing at night during wakefulness and NREM sleep. J. Appl. Physiol. 2010;109:159–170. doi: 10.1152/japplphysiol.00933.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qiu M.H., Chen M.C., Fuller P.M., Lu J. Stimulation of the Pontine Parabrachial Nucleus Promotes Wakefulness via Extra-thalamic Forebrain Circuit Nodes. Curr. Biol. 2016;26:2301–2312. doi: 10.1016/j.cub.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ohmura N., Okuma L., Truzzi A., Shinozuka K., Saito A., Yokota S., Bizzego A., Miyazawa E., Shimizu M., Esposito G., Kuroda K.O. A method to soothe and promote sleep in crying infants utilizing the transport response. Curr. Biol. 2022;32:4521–4529.e4. doi: 10.1016/j.cub.2022.08.041. [DOI] [PubMed] [Google Scholar]
- 104.Grassi S., Pettorossi V.E., Zampolini M. Low-Frequency Stimulation Cancels the High-Frequency-Induced Long-Lasting Effects in the Rat Medial Vestibular Nuclei. J. Neurosci. 1996;16:3373–3380. doi: 10.1523/JNEUROSCI.16-10-03373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Graybiel A., Knepton J. Sopite syndrome: a sometimes sole manifestation of motion sickness. Aviat Space Environ. Med. 1976;47:873–882. [PubMed] [Google Scholar]
- 106.Matsangas P., McCauley M.E. Sopite Syndrome: A Revised Definition. Aviat Space Environ. Med. 2014;85:672–673. doi: 10.3357/ASEM.3891.2014. [DOI] [PubMed] [Google Scholar]
- 107.Scarduzio M., Panichi R., Pettorossi V.E., Grassi S. Synaptic Long-Term Potentiation and Depression in the Rat Medial Vestibular Nuclei Depend on Neural Activation of Estrogenic and Androgenic Signals. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Matsuda T., Cepko C.L. Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movie was captured using a video camera with a night shot function (29 frames/s, HDR-CX560, Sony). The EEG/EMG electrodes were attached to the mouse.
The mouse was the same as in Video S1.
Data Availability Statement
-
•
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the lead contact.
-
•
All code used in this paper will be shared by the lead contact upon reasonable request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.