Abstract
Mycobacterium tuberculosis (Mtb) infection is among the world’s deadliest infectious diseases. Developing effective treatments and biomarkers for tuberculosis requires a deeper understanding of its pathobiology and host responses. Here, we report a comprehensive characterization of circulating short non-coding RNAs (sncRNAs) in plasma samples from Mtb-infected patients. We achieved this by pre-treating plasma RNAs with T4 polynucleotide kinase to convert all RNA ends to those compatible with sncRNA sequencing. We discovered a global and drastic upregulation of plasma sncRNAs in Mtb-infected patients, with tRNA-derived sncRNAs representing the most dramatically elevated class. Most of these tRNA-derived sncRNAs originated from a limited subset of tRNAs, specifically from three tRNA isoacceptors, and exhibited skewed patterns to 5′-derived fragments, such as 5′ halves, 5′ tRNA fragments (tRFs), and internal tRFs (i-tRFs) from the 5′ regions. Further, Mtb-infected patients displayed markedly upregulated and distinct profiles of both rRNA- and mRNA-derived sncRNAs. Some of these sncRNAs, which are abundant and specific to Mtb-infected patients, robustly activated human macrophages via Toll-like receptor 7 and induced cytokine production. This drastic accumulation of circulating, immunostimulatory sncRNAs in the plasma of Mtb-infected patients offers insights into the sncRNA-driven aspects of host immune response against infectious diseases and suggests a pool of potential therapeutic targets and biomarkers.
Keywords: MT: Non-coding RNAs, short non-coding RNA, circulating RNA, tRNA half, tRF, rRF, mRF, TLR7, plasma, tuberculosis, cytokine
Graphical abstract
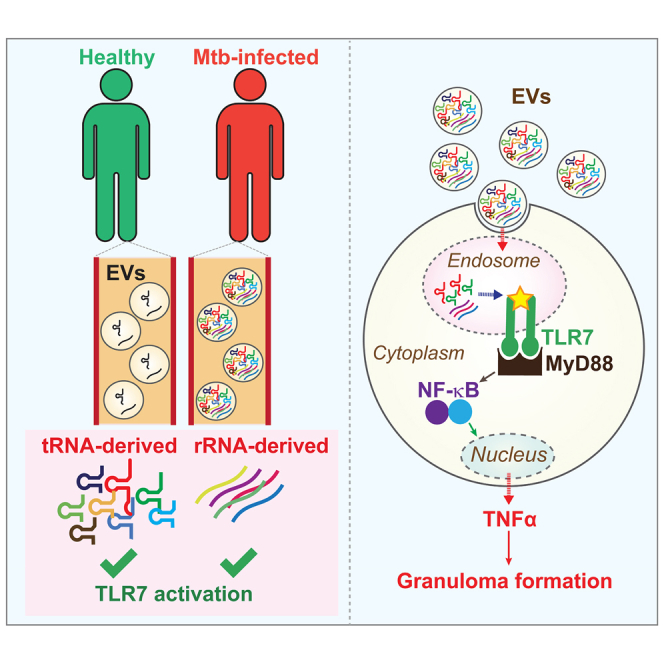
Yohei Kirino and colleagues report a profound accumulation of tRNA- and rRNA-derived small RNAs in the plasma of patients infected with Mycobacterium tuberculosis. The small RNAs include potent TLR7 activators capable of inducing cytokine secretion. These findings unveil a previously unrecognized aspect of tuberculosis pathology and promote future mechanistic studies.
Introduction
Despite treatments and cures, tuberculosis, an infectious disease caused by Mycobacterium tuberculosis (Mtb) infection, remains a formidable global health challenge. It is one of the most lethal infectious diseases, claiming approximately 1.3 million lives annually.1,2,3 The treatment and cure of tuberculosis can be a challenging and drawn-out process, in part due to Mtb’s propensity to enter a non-replicative state resistant to antibiotic treatment.4 Further, the persistence of drug-resistant Mtb strains adds another layer of complexity.5 The intricate interplay between host and pathogen in tuberculosis underscores the need for a better understanding of the body’s immune response to Mtb. Comprehending this dynamic is imperative for the development of novel biomarkers and treatment strategies to combat this significant public health threat.
One of the initial means by which the innate immune system detects and responds to invading microbes involves the pattern recognition receptors (PRRs), which are stimulated by either exogenous pathogen-associated molecular patterns (PAMPs) or endogenous damage-associated molecular patterns (DAMPs).6,7 Toll-like receptors (TLRs), a specific class of PRRs expressed in both immune cells (e.g., macrophages and dendritic cells) and non-immune cells (e.g., epithelial cells and fibroblast cells), respond to an array of ligands.8,9,10 Of the 10 TLRs characterized in humans, TLR7 and -8 reside in endosomes and specifically recognize single-stranded RNAs (ssRNAs) as their ligands.11 Activation of TLR7 and -8 by ssRNA triggers a signaling cascade through MyD88, leading to IRF7- and nuclear factor κB (NF-κB)-mediated transcription and downstream induction of interferon and cytokine production.9,10 One such cytokine, tumor necrosis factor alpha (TNF-α), promotes the formation of granulomas, clusters of cells containing a core of infected macrophages surrounded by other macrophages, which play a critical role in the immune response to Mtb infection.12,13,14,15,16 Notably, activation of TLR7 has been shown to promote the eradication of Mycobacterium bovis by macrophages.17
Typically, the ssRNA ligands identified as activators of TLR7 and -8 have been “foreign” molecules, originating from invading microbes, such as bacterial transfer RNAs (tRNAs).18,19,20,21,22,23 However, multiple reports suggest that “endogenous” ssRNAs from host cells, such as microRNAs (miRNAs), can further act as endosomal TLR ligands. For recognition by TLR7 and -8, ssRNAs must be located inside the endosomes where TLR7 and -8 reside, which can be achieved via extracellular vesicle (EV)-mediated delivery of ssRNAs to endosomes. In fact, various miRNAs are incorporated into EVs, and these EV-packaged extracellular (ex-) miRNAs can reach and activate endosomal TLR7 and -8 in recipient cells.24,25 Such miRNA-mediated activation of endosomal TLRs has been found to promote immune response in the context of infection, as well as in conditions such as neurodegeneration, cancer, myocardial ischemia, and sepsis.24,26,27,28 Given that EVs carry various RNA species besides miRNAs, including mRNAs, tRNAs, and Y-RNAs,29,30 it is plausible that other EV-incorporated host-derived ssRNAs could be delivered to endosomal TLRs and function as their ligands, although this possibility remains largely underexplored.
The unexplored nature of EV-incorporated ex-ssRNAs as endosomal TLR ligands could be partly attributed to the technical limitations of RNA sequencing (RNA-seq). Current standard RNA-seq for short non-coding RNAs (sncRNAs), originally designed for miRNA sequencing, involves the ligation of 5′ and 3′ adaptors (ADs) to the 5′-monophosphate (P) and 3′-hydroxyl (OH) ends of sncRNAs. Consequently, while sncRNAs with the 5′-P/3′-OH ends, such as miRNAs, are efficiently sequenced by this method, sncRNAs with different terminal forms, such as 5′-OH, 3′-P, and 2′,3′-cyclic phosphate (cP), cannot be efficiently captured.31,32 This is especially important for the sequencing analyses of ex-sncRNAs, as many ex-sncRNAs lack 5′-P or 3′-OH.33,34 To overcome this limitation, pre-treatment of RNAs with T4 polynucleotide kinase (T4 PNK), which converts the RNA termini to 5′-P/3′-OH ends (thus rendering them available for 5′-/3′-AD ligation), has been used to capture whole ex-sncRNAs in human plasma samples.33,35,36 In our recent sequencing study of sncRNAs in EVs secreted from human monocyte-derived macrophages (HMDMs), pre-treatment of RNA samples with a mutant T4 PNK, which lacks 3′-dephosphorylation activity, dramatically reduced cDNA yield.34 This suggests that most ex-sncRNAs in HMDM EVs contain 3′-P- or cP, while miRNAs and other 3′-OH-containing RNAs constitute only a minor proportion, further confirming the necessity of T4 PNK treatment to capture the entire ex-sncRNAome.
Our recent studies suggested the involvement of 5′-tRNA half molecules, which contain a cP and thus are not captured by standard RNA-seq, in infection and immune response.34,37 Mycobacterial infection and accompanying immune response lead to upregulation of NF-κB-mediated transcription of the gene angiogenin (ANG), a member of the RNase A superfamily with known role of tRNA cleavage,38 inducing an accumulation of tRNA halves in HMDMs.34 Among these generated tRNA halves, 5′-tRNA halves are selectively and abundantly packaged into EVs of HMDMs and subsequently delivered into the endosomes of recipient HMDMs. Within the endosomes, specific 5′-tRNA halves, such as those from tRNAHisGUG and tRNAValCAC/AAC (5′-halfHisGUG and 5′-halfValCAC/AAC), are capable of activating TLR7.34,37 The strength of this activity was equal to or even greater than that of HIV-derived ssRNA40, a widely used positive-control ssRNA known as a strong activator of endosomal TLRs,11 suggesting that 5′-halfHisGUG and 5′-halfValCAC/AAC can induce cytokine production and immune response.34,37 Our TaqMan RT-qPCR quantification demonstrated that the levels of 5′-tRNA halves were drastically increased by approximately 1,000-fold in patients infected with Mtb,39 suggesting that the upregulation and secretion of 5′-tRNA halves are not limited to HMDMs in vitro but constitute an actual pathological feature of Mtb infection. However, neither the complete ex-sncRNA repertoire nor the potential for ex-sncRNAs to activate endosomal TLRs has been examined in Mtb patients.
Here, we present a comprehensive analysis of ex-sncRNAs in plasma from both healthy individuals and patients infected with Mtb. Previous investigations of ex-sncRNA expression in Mtb patients relied on standard RNA-seq without T4 PNK treatment,40,41,42 thus focusing mainly on miRNAs. By capturing the complete expression profile of plasma sncRNAs through sncRNA sequencing of plasma RNAs pre-treated with T4 PNK, we revealed drastic changes in both the quantities and profiles of ex-sncRNAs, derived most prominently from tRNA, mRNA, and rRNA, in Mtb patients. We further investigated the ability of Mtb-specific ex-sncRNAs to activate TLR7. Our characterization of a fuller breadth of sncRNAs in Mtb patients sheds light on hitherto neglected but highly abundant classes of ex-sncRNAs with potential roles as immune activators in Mtb infection.
Results
The levels of plasma sncRNAs are drastically upregulated in patients infected with Mtb, with tRNA-derived sncRNAs constituting the most dramatically upregulated class
Given that the expression of tRNA halves and other sncRNAs can be affected by sex hormones43 and aging,44 we limited our study to males between 30 and 35 years old to minimize potential impacts of sex and age on sncRNA expression. We obtained plasma samples for sequencing from four healthy individuals (H1–4) and four patients infected with Mtb (M1–4). Using our established multiplex version of TaqMan RT-qPCR, which enables simultaneous quantification of specific tRNA halves and an internal control in limited sample quantities,45 we observed a drastic upregulation of 5′-tRNA halves in plasma from Mtb-infected patients (Figure S1). Specifically, the levels of 5′-tRNA half from tRNAGlyGCC (5′-halfGlyGCC) and 5′-halfHisGUG increased by approximately 500- to 1,000-fold and 4,000- to 8,000-fold, respectively. These results recapitulated our prior findings, which involved examining four healthy and four Mtb plasma samples,37,45 and prompted us to undertake a comprehensive characterization of all circulating sncRNA species in plasma from healthy individuals and Mtb-infected patients.
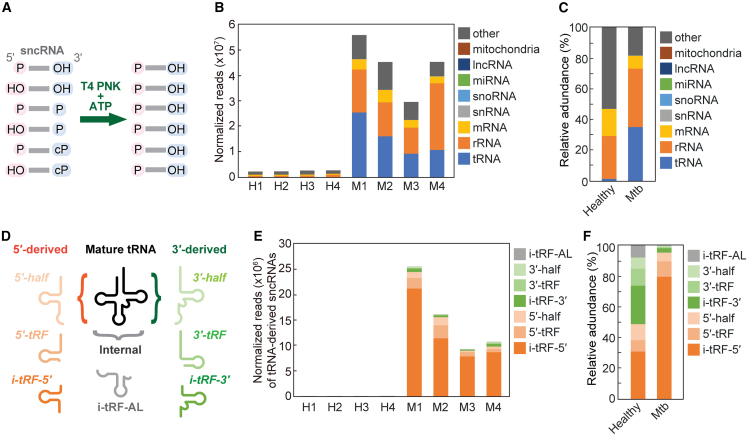
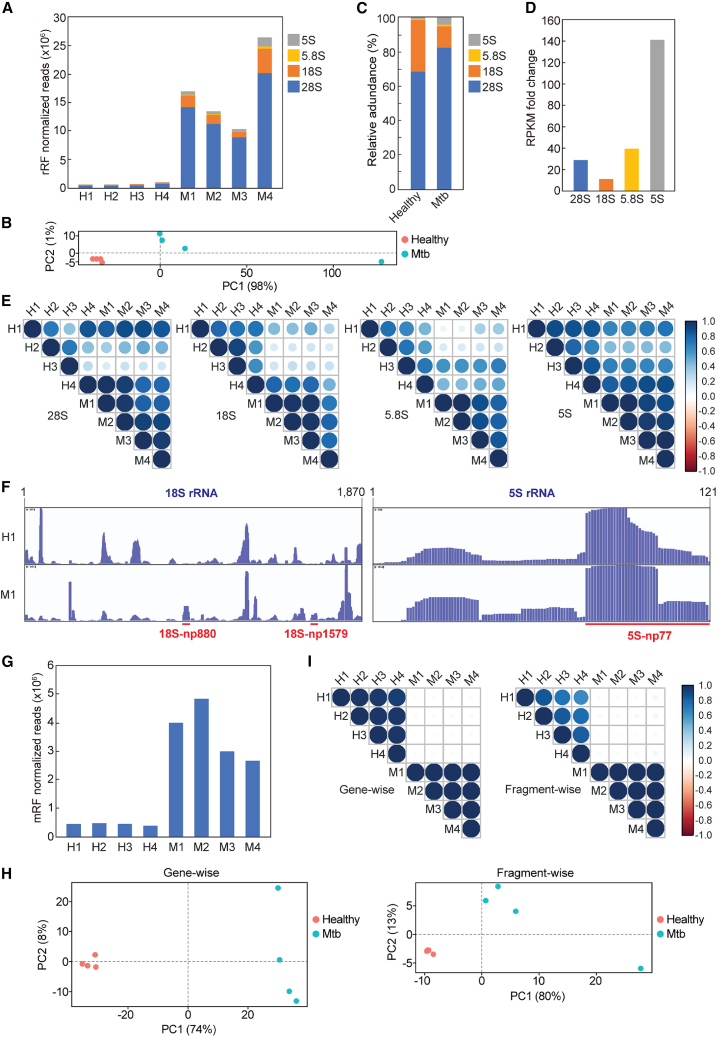
In our plasma sncRNA sequencing, an identical volume of plasma from each sample was mixed with equal amounts of spike-in control RNAs. Subsequently, the plasma sncRNAs were isolated and treated with T4 PNK in the presence of ATP to convert the RNA ends to 5′-P/3′-OH (Figure 1A), which was followed by AD ligation and subsequent RT-PCR for cDNA amplification. When T4 PNK treatment was omitted, cDNA yields significantly decreased, confirming that most plasma sncRNAs lack 5′-P/3′-OH ends, as suggested by previous studies.33,34,35,36 Illumina sequencing and analysis of the obtained reads, normalized based on the read counts of the spike-in control RNAs, revealed drastic upregulation of various sncRNAs in Mtb-infected patients (Figure 1B). Among the various substrate RNA species, tRNAs, rRNAs, and mRNAs served as rich sources of plasma sncRNAs. On average, reads derived from tRNA, rRNA, and mRNA constituted approximately 35%, 38%, and 8% in Mtb patients, respectively (Figure 1C). These ratios significantly differed from those observed in healthy individuals (Figure 1C), suggesting an Mtb-infection-induced alteration in global plasma sncRNA profiles. Within the three main classes (i.e., tRNA-, rRNA-, and mRNA-derived sncRNAs), tRNA-derived reads were particularly enriched in Mtb samples, showing an approximate 24-fold increase as a percentage of occupancy (Figure 1C) and ∼431-fold increase in normalized reads (Figure 1B). Conversely, rRNA- and mRNA-derived reads did not show as significant a difference in the percentage of occupancy (Figure 1C) despite still exhibiting ∼24- and 8-fold increases in normalized reads, respectively (Figure 1B). These results indicate that Mtb-infected patients display a drastic increase in the levels of global ex-sncRNAs in plasma, with a particularly marked enrichment of tRNA-derived sncRNAs.
Figure 1.
Sequencing of plasma sncRNAs from healthy individuals and Mtb-infected patients
(A) Schematic representation of the sncRNA sequencing for T4 PNK-treated plasma RNAs. (B) Abundance of plasma sncRNAs annotated to the indicated RNAs. The read counts of these sncRNAs were normalized using the read counts of the spike-in RNAs. (C) Proportions of plasma sncRNAs annotated to the indicated RNAs. Average values from the four samples are shown. (D) Schematic showing examples of the different classes of tRNA-derived sncRNAs. (E) Abundances of tRNA-derived sncRNAs subclassified into the indicated categories. Read counts, normalized using the spike-in RNAs, are shown. (F) Proportion of tRNA-derived sncRNAs, represented as average values across the four samples.
5′-Derivatives of tRNA-derived sncRNAs are enriched in Mtb-infected patients
The significant enrichment of tRNA-derived sncRNAs in Mtb-infected patients led us to further profile these molecules using a tRNA fragment (tRF) classification method,46,47,48 incorporating modifications from our previous study44 (Figure 1D). tRNA halves are produced by anticodon-loop cleavage, with the 5′ half and 3′ half retaining their respective intact 5′ and 3′ ends. tRFs encompass all other tRNA-derived sncRNAs, excluding tRNA halves. The 5′-tRF and 3′-tRF retain intact mature 5′ and 3′ ends, respectively, while internal tRF (i-tRF) is wholly derived from the internal region of mature tRNAs. We further subclassified i-tRFs into three categories: i-tRF-5′, i-tRF-3′, and i-tRF-AL. i-tRF-5′ and i-tRF-3′ originate from the 5′ and 3′ regions of mature tRNAs, respectively, while i-tRF-AL encompasses the entire anticodon loop. We categorize 5′ half, 5′-tRF, and i-tRF-5′ as 5′ derivatives, and 3′ half, 3′-tRF, and i-tRF-3′ as 3′ derivatives (Figure 1D).
While plasma tRNA-derived sncRNAs in healthy individuals showed roughly equivalent levels of 5′ and 3′ derivatives, interestingly, most (∼91%–97%) in Mtb-infected patients were identified as 5′ derivatives (Figures 1E and 1F). In Mtb samples, i-tRFs-5′ constituted the most abundant subclass within the 5′ derivatives, accounting for ∼71%–84% of tRNA-derived sncRNAs (Figures 1E and 1F). The levels of 5′ halves and 5′-tRFs were also significant at ∼3%–10% and ∼6%–16%, respectively. In contrast, although 3′ derivatives on average comprised 43% of the total tRNA-derived sncRNAs in healthy samples, they were rendered negligible in Mtb-infected samples (Figures 1E and 1F). These results suggest that Mtb infection leads to skewed accumulation of 5′ derivatives in plasma tRNA-derived sncRNAs.
The majority of tRNA-derived sncRNAs originate from a focused subset of tRNAs in Mtb-infected patients
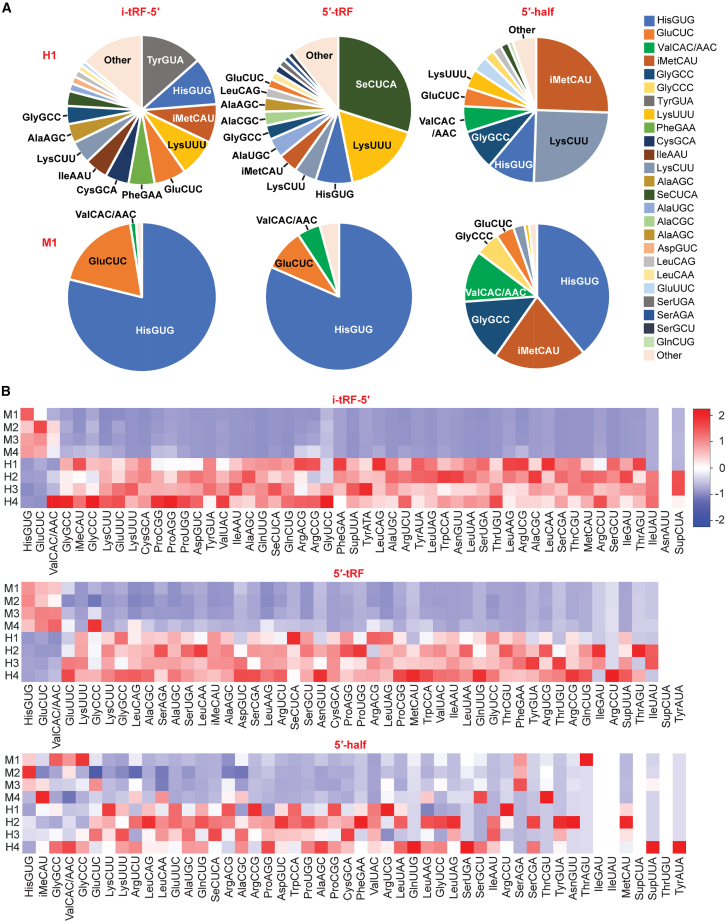
Although humans have more than 40 tRNA isoacceptors, in Mtb-infected patients, i-tRF-5′, the most abundantly accumulated subclass, mainly originated from just three tRNAs: tRNAHisGUG, tRNAGluCUC, and tRNAValCAC/AAC (Figure 2A). In contrast, healthy samples exhibited a more balanced profile, with i-tRFs-5′ arising from a diverse range of tRNAs (Figure 2A). These patterns were consistent across all eight sequenced samples (Figures 2B and S2). We observed a similar reduction in source isoacceptor diversity for 5′-tRFs and 5′ halves in Mtb samples (Figures 2A, 2B, and S2). Across all 5′ derivatives, the same three tRNAs (tRNAHisGUG, tRNAGluCUC, and tRNAValCAC/AAC) served as the primary sources, collectively constituting 95%–99%, 83%–98%, and 21%–81% of i-tRFs-5′, 5′-tRFs, and 5′ halves, respectively (Figures 2A and 2B).
Figure 2.
Analysis of the 5′ derivatives of tRNA-derived sncRNAs
(A) Proportion of reads corresponding to 5′ derivatives, sourced from respective cytoplasmic tRNA species, in both H1 and M1 samples. The abundant 5′ derivatives from all eight samples are shown in Figure S2. (B) Heatmap representing read distribution for 5′ derivatives originating from respective tRNAs. Coloration is based on the column’s Z score. Note that these analyses are based on reads per million (RPM) values, not on normalized abundances.
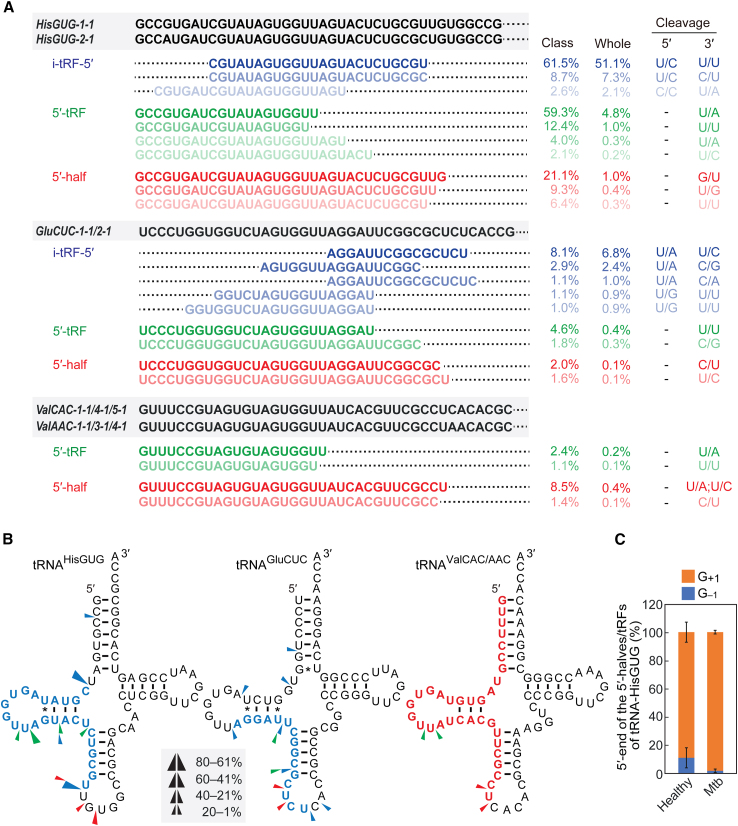
In Mtb samples, the majority of sncRNAs from these three major tRNA species were produced from specific isodecoders (Figure S3) and limited to specific tRFs (Figure 3A). For instance, only three i-tRFHisGUG-5′ species, originating from nucleotide position (np) 9–32 and np 3–23, collectively made up 72.8% and 60.5% of i-tRFs-5′ and whole-tRNA-derived sncRNAs, respectively. Similarly, only two to four specific species dominated the composition of i-tRFs-5′, 5′-tRFs, and 5′ halves derived from all three major tRNAs (Figure 3A). The cleavages leading to the production of these specific molecules primarily occurred between a 5′-pyrimidine (typically uridine) and a 3′-purine (Figures 3A and 3B). Exceptions include G-U cleavage for the production of 5′-halfHisGUG and the cleavages between pyrimidines for generating several i-tRFs-5′. We noted some overlap in the cleavage positions responsible for the production of i-tRFs-5′, 5′-tRFs, and 5′ halves (Figures 3A and 3B), implying that at least some of i-tRFs-5′ and 5′-tRFs are produced from 5′ halves, and some i-tRFs-5′ may arise from 5′-tRFs.
Figure 3.
Abundant species of the 5′ derivatives
(A) Sequences of highly abundant tRNA-derived sncRNAs (comprising >1% of each class) are shown alongside the sequences of their corresponding tRNAs (full tRNA sequences are shown in Figure S3). The percentage values for each class (i.e., i-tRF-5′, 5′-tRF, or 5′ half) as well as the overall proportion within the whole pool of tRNA-derived sncRNAs are indicated. The 5′ and 3′ nucleotides, between which cleavage occurs to generate the 5′ and 3′ ends of each sncRNA, are also displayed. (B) Cleavage sites in the indicated tRNAs were predicted based on the terminal positions of the identified i-tRFs-5′ (blue arrowheads), 5′-tRFs (green arrowheads), and 5′ halves (red arrowheads). Sequences of the most abundant tRNA-derived sncRNA are highlighted in colors: blue for i-tRF-5′ and red for 5′ half. (C) Proportion of the 5′-terminal nucleotide of the 5′ halves and 5′-tRFs of tRNAHisGUG.
tRNAHisGUG is unique in its possession of an extra guanosine at the 5′ end, known as G–1, which is added post-transcriptionally by a specific guanyltransferase, THG1.49 Our previous study demonstrated that human cell lines contain a fraction of mature tRNAHisGUG molecules that lack this G–1. Moreover, a substantial portion of the 5′-halvesHisGUG lack G–1 and instead have G+1 as the 5′-terminal nucleotide in both human cell lines and mouse tissues.34,44,50,51 Consistent with these findings, all abundant circulating 5′-tRFs and 5′ halves of tRNAHisGUG in the plasma of Mtb-infected patients lacked the G–1 nucleotide (Figure 3A). Analyses encompassing all circulating 5′-tRFs and 5′ halves of tRNAHisGUG showed that, in healthy samples, approximately 89% of these sncRNAs were missing the G–1 nucleotide (Figure 3C). This figure rose to over 98% in Mtb samples (Figure 3C). These results indicate a preferential accumulation of G–1-lacking 5′-tRFHisGUG and 5′-halfHisGUG in the plasma, which is even more pronounced in Mtb patients.
5′-tRF and 5′ half of tRNAValCAC/AAC are potent activators of macrophage TLR7
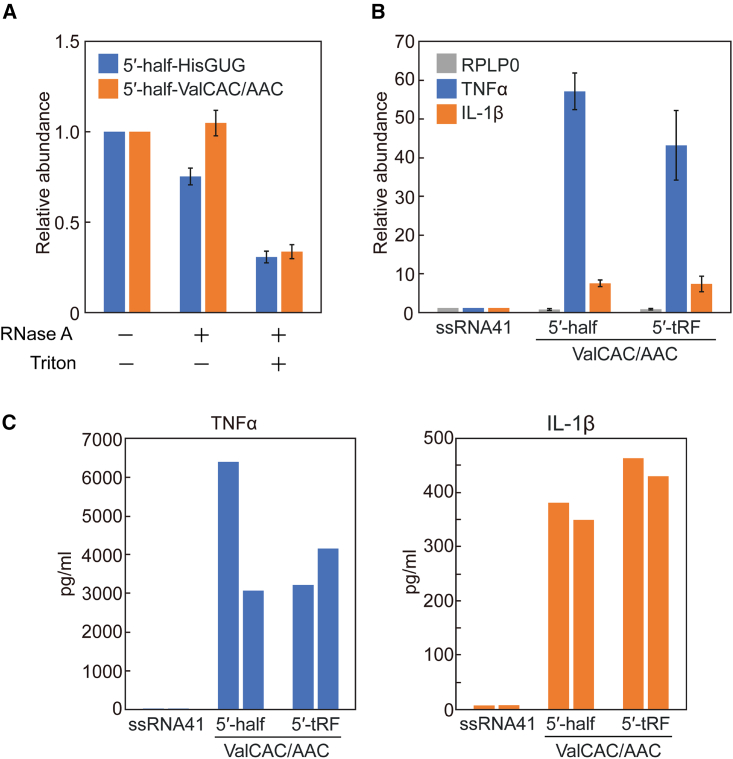
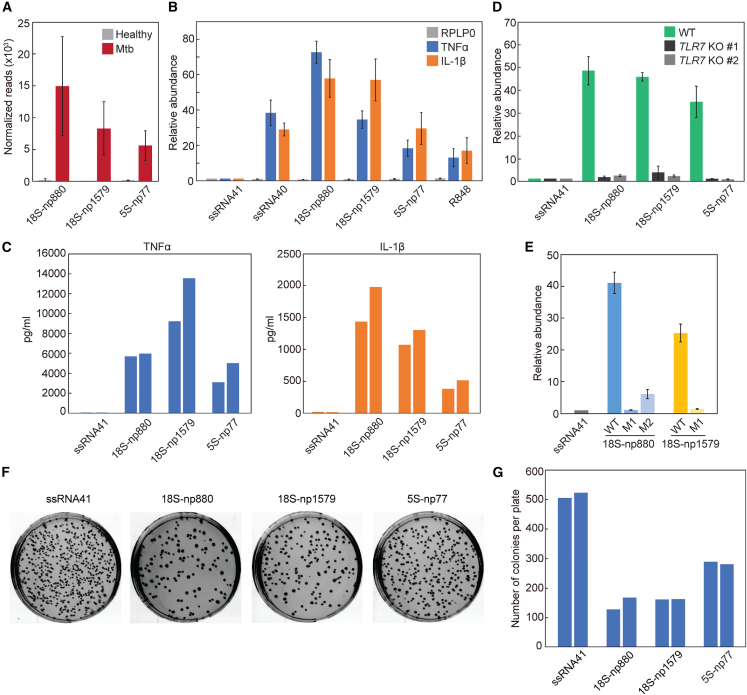
Upon Mtb infection, macrophages engulf invading bacteria and act as important mediators of pathogen recognition and antimicrobial immune response through PRRs, such as surface TLR2 and endosomal TLR7.52,53,54 Our recent studies demonstrated that 5′-tRNA halves are abundantly accumulated in the EVs of HMDMs, and the ex-5′ halves of two out of the three major tRNAs identified in Mtb plasma samples, namely 5′-halfHisGUG and 5′-halfValCAC/AAC, can activate TLR7 when delivered into the endosomes of HMDMs, inducing production of TNF-α and other cytokines.34,37 While Mtb plasma treated only with RNase A and untreated plasma yielded comparable amplification signals in TaqMan RT-qPCR for the detection of both 5′-tRNA halves, the plasma treated with both RNase A and detergent showed a marked decrease in these signals (Figure 4A), indicating that the majority of the detected 5′-tRNA halves are shielded from RNase degradation in the absence of detergent. This protection suggests that the 5′-tRNA halves are most likely localized inside the EVs.
Figure 4.
The activity of tRNAValCAC/AAC-derived sncRNAs in inducing cytokine production in macrophages
(A) Mtb plasma sample was treated with RNase A and/or Triton X-100 and then subjected to TaqMan RT-qPCR for quantification of 5′-tRNA halves. The graph displays average values of relative abundance, with data from untreated sample set as 1. Error bars indicate the SD. (B) After DOTAP-mediated endosomal delivery of the indicated RNAs, cytokine mRNA levels were quantified using RT-qPCR. The graph displays average values of relative abundance, with data for ssRNA41 set as 1. Error bars indicate the SD. (C) After DOTAP-mediated endosomal delivery of the indicated RNAs, culture medium was analyzed by ELISA to measure concentrations of the indicated cytokines. Two independent experiments were performed, the results of which are shown as separate bars.
Both sites of successive uridines at np 19–20 and 32–33 in 5′-halfHisGUG are required for its TLR7 simulation activity,34,37 suggesting that i-tRFHisGUG-5′ and 5′-tRFHisGUG detected in Mtb samples may not robustly stimulate TLR7. In contrast, the 5′-terminal GUUU sequence was shown to be necessary and sufficient for the activity of 5′-halfValCAC/AAC,37 suggesting that 5′-tRFValCAC/AAC retaining the 5′-terminal sequences could activate TLR7. To investigate this, we delivered the most abundantly identified 5′-tRFValCAC/AAC, as well as 5′-halfValCAC/AAC, into the endosomes of HMDMs using a cationic liposome reagent, 1,2-dioleoyloxy-3-trimethylammonium-propane (DOTAP), which mimics exosomes and has been widely employed for endosomal delivery of sncRNAs in previous studies.34,55,56,57 As a negative control, an inactive, 20-nt mutant version of HIV-1-derived ssRNA, termed ssRNA41,11 was utilized. mRNA levels of both examined cytokines, TNF-α and IL-1β, were significantly increased upon endosomal delivery of 5′-tRFValCAC/AAC and 5′-halfValCAC/AAC (Figure 4B). Moreover, dramatically increased levels of these cytokines were secreted into the culture medium following endosomal delivery of these 5′ derivatives of tRNAValCAC/AAC (Figure 4C). These results indicate that 5′-tRFValCAC/AAC, as well as 5′-halfValCAC/AAC, which retains the 5′-GUUU, strongly activates endosomal TLR7 in HMDMs, inducing cytokine production.
Differential profiles of plasma rRNA- and mRNA-derived sncRNAs between healthy individuals and Mtb-infected patients
Circulating sncRNAs can be derived from various types of transcripts, not limited to tRNAs, and both circulating rRNA- and mRNA-derived fragments (rRFs and mRFs) are considered potential biomarkers in various diseases.36,58,59,60 In Mtb-infected patients, rRF levels were dramatically elevated (Figures 1B and 5A). Principal-component analysis (PCA) of rRFs demonstrated marked distinctions of their profile between healthy and Mtb-infected samples (Figure 5B). Although most rRFs originated from 28S and 18S rRNAs in both healthy and Mtb-infected samples (Figure 5C), an increase in 5S rRNA-derived reads was particularly pronounced in the Mtb-infected samples compared to the healthy cohort (Figure 5D). Fragment-wise correlation analysis revealed consistently strong correlations in rRF profiles among Mtb-infected samples, while healthy samples exhibited greater variability and weaker correlation among them (Figure 5E). The weakest correlations were observed between healthy and Mtb samples, suggesting the existence of distinct, Mtb-specific rRF profiles. Indeed, sequence alignments demonstrated the presence of specific rRFs in Mtb-infected patients, such as those derived from 18S rRNA starting from np 880 and np 1579, which are here termed 18S-np880 and 18S-np1579, respectively (Figures 5F and S4).
Figure 5.
Analysis of sequencing reads mapped to rRNAs and mRNAs
(A) Abundance of rRFs originating from the indicated rRNAs. Read counts, normalized to those of spike-in RNAs, are shown. (B) PCA of rRFs. (C) Proportions of rRFs derived from the indicated rRNAs. Average values from the four samples are shown. (D) Fold change of the rRFs (Mtb vs. healthy samples) derived from respective rRNAs. Note that these analyses are based on reads per kilobase of each rRNA, per million (RPKM) values, not on normalized abundances. (E) Pearson correlation analysis among rRFs derived from each rRNA. The color and size of each dot correspond to the correlation coefficient. (F) Read alignments of rRFs mapped to 18S and 5S rRNAs in samples H1 and M1 were visualized using the IGV. The positions of the three rRFs examined in this study are indicated by red lines. Alignments of rRFs across all rRNAs for the eight libraries are shown in Figure S4. (G) The normalized abundance of mRFs. (H) PCA of mRFs using mRNA IDs (gene-wise, left) or unique fragment counts (fragment-wise, right). (I) Pearson correlation analysis of mRFs, both gene-wise and fragment-wise.
mRF levels were also elevated in Mtb-infected patients, although the extent of this increase was far less pronounced than that of tRNA- and rRNA-derived sncRNAs (Figure 5G). Both gene-wise and fragment-wise PCA of mRFs exhibited clear differences between healthy and Mtb-infected samples (Figure 5H). Correlational analysis revealed stark differences in mRF profiles between healthy and Mtb-infected samples (Figure 5I). Gene-wise correlation data suggest that the mRNA sources of mRFs are consistent within both healthy and Mtb-infected samples; however, they diverge when comparing the two groups (Figure 5I). Fragment-wise correlation results supported these results, revealing that plasma-circulating mRF profiles are consistent within healthy or Mtb-infected groups while being different between the two groups.
Infection-specific rRFs are potent activators of macrophage TLR7
Given that tRNA-derived sncRNAs can activate TLR7 (Figures 4B and 4C), we aimed to investigate the potential immune-activating roles of rRFs. Of particular interest were the 18S-np880 and 18S-np1579, originating from the np 880–906 and 1,579–1,618 of 18S rRNA, respectively. These rRFs not only specifically accumulated in Mtb samples (Figures 5F, 6A, and S4) but also feature tandem uridine sequences (Figures S5A and S5B), which are known to be preferable ligands of TLR7.11 We also focused on 5S-np77, an rRF derived from 3′-terminal region (np 77–120) of 5S rRNA (Figures 5F and S4). This rRF was also significantly upregulated upon Mtb infection (Figure 6A) and contains tandem uridine sequences (Figures S5A and S5B). DOTAP-mediated delivery of these selected rRFs into the endosomes of HMDMs caused a dramatic increase in the mRNA levels of TNF-α and interleukin (IL)-1β (Figure 6B). The increases induced by 18S-np880 and 18S-np1579 were significantly higher than those induced by ssRNA40, a widely used HIV-derived ssRNA known to strongly activate endosomal TLRs.11 Furthermore, in our experiments at a specific time point, the activity of all three examined rRFs was stronger than that of R848, a TLR7 and -8 agonist used in cancer and infectious disease therapies.61,62,63,64 These results suggest that these rRFs function as potent immunostimulatory molecules. This was confirmed not only by elevated cytokine mRNA levels but also by increased cytokine secretion following endosomal delivery of these rRFs (Figure 6C). The cytokine induction was significantly reduced when these rRFs were transfected into TLR7 KO HMDMs (Figure 6D), corroborating that these rRFs induce cytokine production via TLR7.
Figure 6.
Characterization of TLR7-activating rRFs
(A) Normalized read counts for the three selected rRFs. (B) After DOTAP-mediated endosomal delivery of the indicated RNAs, cytokine mRNAs were quantified using RT-qPCR. The graph displays average values of relative abundance, with the data for ssRNA41 set as 1. Error bars indicate the SD. (C) After DOTAP-mediated endosomal delivery of the indicated RNAs, the culture medium was analyzed using ELISA to measure the concentrations of the indicated cytokines. Results from two independent experiments are shown as separate bars. (D) The DOTAP experiments were performed using two different TLR7 KO THP-1 cell clones (#1 and #2) followed by quantification of cytokine mRNAs using RT-qPCR. The average values of relative abundance are shown, with data for ssRNA41 set as 1. Error bars indicate the SD. (E) The indicated wild-type and mutant rRFs were subjected to DOTAP experiments, followed by quantification of TNF-α mRNA levels. (F and G) After DOTAP-mediated endosomal delivery of the indicated RNAs, HMDMs were subjected to bacterial infection and invasion assay. Representative images of plates with E. coli colonies (F) and bar graphs of the counted colony numbers (G) are shown. Results from two independent experiments are shown as separate bars.
We next explored sequence determinants of these active rRFs. Consecutive uridine sequences (e.g., UU and UUU) in ligand ssRNAs have been established as crucial for TLR7 binding.65 18S-np880 contains two instances each of UU and UUUU sequences. Replacing all these uridines with adenines (designated as 18S-np880-M1; Figure S5C) resulted in loss of TLR7 activation (Figure 6E), confirming the essential role of these consecutive uridine sequences for the activity. We then generated a mutant, 18S-np880-M2, that retains secondary structure of 18S-np880 and two UUUU sequences but lacks the two UU sequences due to their replacement with CC or CA (Figure S5C). This mutant exhibited TLR7-stimulating activity, albeit significantly diminished compared to the wild-type 18S-np880 (Figure 6E), indicating that, while both UU sequences are important for full activity, the presence of two UUUU sequences alone can still confer partial activity. 18S-np1579 contains three UU sequences, and their replacement with adenines (designated as 18S-np1579-M1; Figure S5C) abolished the TLR7-simulating activity of 18S-np1579 (Figure 6E), further emphasizing the importance of sequential uridines for the activity.
We further probed whether rRF-mediated TLR7 activation possesses functional immune-stimulating and antimicrobial properties by conducting bacterial elimination assays. Following DOTAP-mediated endosomal delivery of the three active rRFs, HMDMs were infected with Escherichia coli. After incubation, the E. coli-infected HMDMs were lysed, and viable E. coli colonies were enumerated on Luria-Bertani (LB) agar plates. As shown in Figures 6F and 6G, the number of E. coli colonies was significantly reduced on plates corresponding to HMDMs transfected with the active rRFs compared to those transfected with the negative control, ssRNA41. These results suggest that rRF-mediated activation of macrophages can effectively boost cellular immunity in response to infection.
Discussion
While circulating sncRNAs are increasingly garnering attention as potential biomarkers and functional molecules in various diseases, a significant portion of these extracellular sncRNAs contain either a cP or 3′-P,34 thereby eluding detection by standard RNA-seq.32 Given that even a single species of cP-containing tRNA half, rRF, or mRF can be much more abundant than total miRNAs in tissues44 and EVs,34 standard RNA-seq data for circulating sncRNAs are considered to be inherently biased, as they exclude the majority of sncRNAs. In this study, building upon previous efforts by other groups and us,33,34,35,36 we employed T4 PNK pre-treatment of plasma RNA prior to sncRNA sequencing. This allowed us to comprehensively capture human plasma sncRNAs and to compare their differential profiles between healthy individuals and patients infected with Mtb. Our research provides the first full sequencing and characterization of circulating sncRNAs in Mtb-infected patients and reveals a drastic and global accumulation of sncRNAs, with tRNA-derived sncRNAs emerging as the most dramatically upregulated class. We do acknowledge certain limitations in the present study, including the small sample size and the absence of detailed clinical information for the samples collected from men testing positive for Mtb immunoreactivity. However, due to the magnitude of difference we observed between healthy and Mtb samples, we are confident that our results reveal genuine features of tuberculosis pathology. Supporting this assertion, DESeq2 analysis showed markedly high fold changes in Mtb patients and tremendously low p values for the sncRNAs highlighted in this study (Table S1).
It is noteworthy that the accumulation of tRNA-derived sncRNAs in Mtb-infected patients is skewed toward 5′ derivatives, in contrast to those in healthy individuals. Although recent studies have provided evidence that certain tRNA halves exist in the form of nicked mature tRNAs,66,67 the scarcity of 3′ derivatives suggest that 5′ and 3′ derivatives of tRNA-derived sncRNAs can be regulated through distinct and independent pathways. The existence of a specific pathway for 5′ derivatives aligns well with multiple studies that have reported a much higher accumulation of 5′-tRNA halves compared to 3′-tRNA halves in various cell systems, including U2OS cells under oxidative stress, ANG-overexpressed HEK293T cells, Epstein-Barr virus-infected B lymphoblastic cells, as well as in diverse mouse tissues and serum.68,69,70,71,72
Our previous study demonstrated that the majority of 5′-halfHisGUG and 5′-halfGluCUC in human plasma were RNase resistant under detergent-free conditions,34 while they were susceptible to degradation by RNase in the presence of detergent. We here observed the same phenomena with 5′-halfHisGUG and 5′-halfValCAC/AAC in Mtb patient plasma, suggesting that they are protected by incorporation into EVs. After the anticodon cleavage of tRNAs in cells and tissues, 5′ halves, but not 3′ halves, may be selectively stabilized by their binding proteins and/or packaged into EVs. These stabilized 5′ halves may subsequently be cleaved to generate other 5′ derivatives such as i-tRFs-5′ and 5′-tRFs. Indeed, selective packaging of 5′ halves into HMDM EVs was observed in our previous study,34 and a greater abundance of 5′ halves compared to 3′ halves in human serum was reported previously.68 Although ANG has been identified as an endoribonuclease that cleaves the anticodon loop of mature tRNAs to produce tRNA halves,38 further research is needed to determine whether ANG or other RNases are responsible for the drastic upregulation of 5′ halves in Mtb-infected patients. The enzymes producing i-tRFs-5′ and 5′-tRFs also remain unidentified, although our sequencing analysis suggests preferential cleavage between pyrimidines and purines. Further investigation is required to elucidate the molecular mechanisms underlying the selective accumulation of 5′ derivatives as well as the enzymes responsible for their production and any interacting proteins that contribute to their stabilization.
Further investigation is also needed to clarify why only a limited number of tRNA-derived sncRNAs, specifically derived from three tRNAs (tRNAHisGUG, tRNAGluCUC, and tRNAValCAC/AAC), are dramatically enriched in Mtb-infected patients. It is noteworthy that the 5′ halves of two out of these three tRNA species, tRNAHisGUG and tRNAValCAC/AAC, have been identified as immunostimulatory molecules that activate macrophage TLR7.34,37 Six other 5′ halves examined, such as tRNAGluCUC, did not stimulate endosomal TLRs,34,37 leading us to speculate that immune-active molecules may be selectively accumulated in Mtb-infected patients through an unknown mechanism. Our study strengthened this assumption by demonstrating that 5′-tRFValCAC/AAC, as well as 5′-halfValCAC/AAC, can serve as potent TLR7 activators. It has been reported that 5′ halvesGluCUC dimerize, rendering them relatively stable,73 which may account for the abundant accumulation of tRNAGluCUC-derived sncRNAs.
In this study, we further demonstrated that the accumulation of circulating rRFs and mRFs is dramatically enhanced in Mtb-infected patients. The correlational analysis results were particularly striking for mRFs, which displayed a highly consistent and infection-specific pattern of production. Although less definitive, a similar trend was observed for rRFs. Of particular interest to us were certain infection-specific rRFs that proved to be potent activators of TLR7, the finding of which aligned with our assumption that immunostimulatory molecules can be selectively accumulated in Mtb-infected patients. Our findings are also in line with a previous study showing that rRNA-derived sncRNAs act as circulating DAMPs, which are released under various conditions, both within and outside of EVs.74 The biogenesis of these rRFs is still not fully understood. While RNase1 cleaves extracellular rRNA,75 this activity seems to have an anti-inflammatory effect.
Previous studies have demonstrated that post-transcriptional modifications of RNAs can impede or alter their immunogenicity.22,23,76,77 However, out of our three focused rRFs, two (18S-np1579 and 5S-np77) are expected to be unmodified. This is based on the comprehensive identification of rRNA modifications,78 which indicated that the regions in 18S and 5S mature rRNAs remain unmodified. The remaining rRF, 18S-np880, is expected to contain pseudouridine (Ψ) at np 897. However, this modification was found only at low levels (23%) in mature 18S rRNA,78 suggesting that the unmodified 18S-np880 is likely the dominant form. Therefore, we believe that our experiments using synthetic unmodified rRFs closely represent endogenous phenomena.
While further studies are needed to determine whether the levels of TLR7-stimulating sncRNAs are sufficient to affect the proinflammatory environment in the blood upon Mtb infection, our previous study has proved that 5′-halfHisGUG is present at a physiologically relevant concentration capable of inducing cytokine production via TLR7 activation.34 Upon TLR7 stimulation, it is plausible that ANG generation could be induced through NF-κB-mediated transcription of the ANG gene.34 Additionally, TLR7 stimulation has been shown to produce reactive oxygen species,54 which may generate oxidative stress that triggers ANG-generated tRNA cleavage to produce stress-induced tRNA halves.38,79 In both potential scenarios, a feedforward mechanism may exist that promotes further production of immunostimulatory molecules upon TLR7 activation, potentially resulting in the observed drastic upregulation of these molecules in Mtb-infected patients.
The induction of TNF-α via TLR7 activation by circulating tRNA- and rRNA-derived sncRNAs highlights a potential physiological role these sncRNAs may have within the context of tuberculosis. TNF-α accumulates in the serum of Mtb patients and has been shown to be a key factor in the formation of granulomas, wherein a core of infected macrophages is surrounded by other macrophages and lymphocytes.12,13,14,15,80 Granuloma formation is a crucial step in the immune response to tuberculosis; its disruption, for example by anti-TNF-α monoclonal antibodies (mAbs), is associated with the resuscitation of dormant Mtb.16 We postulate that highly abundant sncRNAs, capable of activating TLR7, promote the immune response to tuberculosis by enhancing TNF-α production and subsequent granuloma formation. Moreover, TLR7 activation, either through ssRNA or imiquimod, can promote the death of Mtb by autophagy, suggesting a direct mechanism whereby the sncRNA molecules we have identified here may contribute to the elimination of the invading bacteria.53,54 The ability of our rRNA-derived sncRNAs to impede bacterial infection further supports the hypothesis that these molecules might function as part of an RNA-mediated antimicrobial response to Mtb infection.
Although not capable of activating TLR7, tRNAGluCUC-derived fragments, which were highly accumulated in Mtb patients, may also play a role in the immune response to Mtb infection. In the context of respiratory syncytial virus (RSV) infection, the 5′-halfGluCUC binds to mRNA and inhibits the translation of apoER2, which is involved in disruption of viral genome synthesis, thus implicating the 5′ half in immune evasion by RSV.81 Further, evidence suggests that signaling through apoER2 drives the polarization of macrophages to an anti-inflammatory M2 phenotype.82 The inhibitory activity of 5′-halfGluCUC in apoER2 expression is dependent upon the first 18 nt of 5′-halfGluCUC, a sequence found in approximately 10% of the tRNAGluCUC-derived sncRNAs in our Mtb samples.
Our finding that both 5′-halfValCAC/AAC and a shorter derivative, 5′-tRFValCAC/AAC, possess TLR7-activating potential provides insights not only into the pathophysiology of Mtb infection but also into other diseases beyond tuberculosis. For example, a previous study found that the circulating 5′-halfValCAC/AAC detected in the serum of amyotrophic lateral sclerosis (ALS) patients is associated with slow disease progression,83 although the details of this connection are yet to be clarified. Increased levels of 5′-halfValCAC/AAC have been observed in the spinal cord of SODG93A mice, an ALS model, which exhibit a slower disease progression.83 It is possible that the tRNAValCAC/AAC-derived sncRNAs might induce the production of TNF-α, whose protective role has been demonstrated in SODG93A mice84 and that is present at elevated levels in ALS patients.85 Therefore, the circulating sncRNA molecules highlighted in our study may be implicated in the pathophysiology of a diverse array of diseases.
The activity and abundance of the sncRNAs highlighted in our study imply potential therapeutic applications. If these sncRNAs drive pathological inflammation, targeting and inhibiting them may offer a means to mitigate such inflammation. Conversely, if they have beneficial health implications, they could be explored as therapeutic agents. Several small-molecule activators of TLR7 have been studied for cancer treatment.86 One such molecule, R848 (resiquimod), has been shown to reduce tumor burden and extend survival in a pancreatic cancer mouse model.87 We observed that all of our rRFs induced a stronger TNF-α and IL-1β response than R848 in our experiments at a specific time point. Future research should explore the potential of these circulating sncRNAs as non-invasive diagnostic biomarkers. For Mtb infection, markers capable of distinguishing between degrees of active infection or identifying patients on the road to recovery and those with persistent infection would be especially valuable. To achieve this, studies involving larger cohorts with varied clinical parameters are essential. A more profound understanding of the biogenesis, packaging, and stabilization of circulating sncRNAs, coupled with insights into their biological roles, will deepen our appreciation of their potential applications.
Materials and methods
Ethical approval, human plasma samples, and RNA isolation
The Office of Human Research (OHR) of Thomas Jefferson University (TJU) approved our use of human plasma samples without any private information, adhering to all federal, institutional, and ethical guidelines. We obtained the de-identified plasma samples from a biological specimen company, BioIVT. The human plasma samples were sourced from males between the ages of 30 and 35 years for both healthy individuals and patients infected with Mtb (Table S2). RNA isolation from these plasma samples was carried out as described previously.34,45 First, 500-μL aliquots of plasma samples were centrifuged at 16,100 × g for 5 min, after which 400 μL of supernatant were taken and mixed with synthetic spike-in RNAs (Table S3), followed by RNA extraction using TRIzol LS (Invitrogen). Further purification of the extracted RNAs was achieved using the miRNeasy Mini Kit (Qiagen).
Multiplex quantification of 5′-tRNA halves
The levels of 5′-tRNA halves were quantified using a multiplex TaqMan RT-qPCR method that we developed for specific quantification of 5′-tRNA halves using limited starting materials.45 Briefly, 400 μL of plasma were mixed with 1 fmol of spike-in control RNA (5′-GGGAGGCAAGCCCGACGUCGUCCAGAUUGUCCGC-3′) and subjected to RNA extraction as described above. The RNA pellet was resuspended in 8 μL of RNase-free water. Subsequently, 2 μL of this solution were used for T4 PNK treatment and 3′-AD ligation as described previously.45 Multiplex TaqMan RT-qPCR was performed using One Step PrimeScript RT-PCR Kit (Takara Bio) on a StepOne Plus Real-time PCR machine (Applied Biosystems). The Ct value of the Spike-in RNA was used for normalization. The sequences of targeted 5′-tRNA halves, primers, and TaqMan probes are shown in Tables S4 and S5. The information on sncRNA “license plates”88 and names via tDRnamer89 for the targeted sncRNAs in this study are included in Tables S4. To confirm the presence of 5′-tRNA halves in EVs, Mtb plasma sample was incubated with PureLink RNase A (400 ng/μL, Thermo Fisher Scientific) with or without 0.5% Triton X-100 at 37°C for 30 min.
Pre-treatment of plasma RNA with T4 PNK, sncRNA sequencing, and data analysis
Plasma RNA mixed with spike-in RNAs, extracted as described above, was treated with T4 PNK in the presence of 1 mM ATP at 37°C for 40 min, followed by phenol/chloroform/isoamylalcohol extraction and ethanol precipitation. The RNA was then subjected to cDNA amplification using the TruSeq Small RNA kit (Illumina). The amplified cDNAs were gel purified, and their quality and amount were assessed by a Bioanalyzer High Sensitivity DNA chip (Agilent) and Qubit (Thermo Fisher Scientific). The cDNA libraries were sequenced on Illumina NextSeq 500 at the MetaOmics Core Facility of the Sidney Kimmel Cancer Center at TJU.
Bioinformatic analyses were performed as described previously.44,51 In brief, we utilized the cutadapt tool (DOI: https://doi.org/10.14806/ej.17.1.200) to remove the 3′-AD. After selecting 15- to 60-nt reads, we used Bowtie2 (2.3.5) for mapping.90 Mapped read numbers were normalized by those of spike-in RNAs. Data analysis and visualization were carried out using R packages: gplots for heatmap analysis and corrplot for Pearson correlation analysis. Differential expression analysis for rRFs and mRFs was carried out, and PCA plots were prepared using DESeq2.91 rRF sequence alignments were prepared using the Integrative Genomics Viewer (IGV).92
In vitro RNA synthesis
The synthetic RNAs used in this study (Table S4) were synthesized in vitro as described previously.34,50 dsDNA templates were prepared using PrimeSTAR GXL DNA Polymerase (Takara Bio) and the primers shown in Table S5. In vitro transcription reaction with T7 RNA polymerase (New England Biolabs) was carried out at 37°C for 6 h. For 18S 1579-1618 and 5S 77-120, the reaction mixtures were further subjected to three cycles of 90°C for 2 min and 37°C for 30 min to induce ribozyme cleavage. The synthesized RNAs were then gel purified using denaturing PAGE with single-nucleotide resolution.
Cell culture, endosomal delivery of RNA, and quantification of cytokines
THP-1 human acute monocytic leukemia cells (American Type Culture Collection) were cultured in RPMI 1640 medium (Corning) with 10% FBS and differentiated into HMDMs using phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) as described previously.34,93 TLR7 KO THP-1 cell lines, whose TLR7 expression is completely depleted, were previously generated via CRISPR-Cas9.34 Before endosomal delivery of RNAs, the cells were primed with 100 units/mL of interferon gamma (Thermo Fisher Scientific) for 24 h.94 To deliver RNAs to endosomes, we used the cationic liposome DOTAP (Sigma-Aldrich) as previously described.25,34,95 In brief, 230 pmol of synthetic RNAs or R848 (Invitrogen) were mixed with 60 μL of HEPES buffered saline (HBS) and 15 μL of DOTAP reagent and incubated for 15 min. The RNA-DOTAP solution was then added to 1 mL of RPMI 1640 medium with 2% FBS, followed by incubation of the cells for 16 h.
For quantification of cytokine mRNAs by RT-PCR, as in our previous study,34 total RNA was extracted from the cells using TRIsure (Bioline), treated with DNase I (Promega) and subjected to reverse transcription using RevertAid Reverse Transcriptase (Thermo Fisher Scientific) and a reverse primer. The synthesized cDNAs were then subjected to PCR using 2×qPCR Master Mix (Bioland Scientific) and forward and reverse primers, as described in our previous study.34 The abundance of the target mRNA was calculated as a ratio to GAPDH mRNA and further normalized to the control experiment (transfection of ssRNA41). Cytokine concentrations in the culture media of HMDMs were measured by Multiplexing LASER Bead Technology (Eve Technologies).
Bacterial infection and elimination assay
After 16 h of DOTAP transfection of RNAs, HMDMs (1 × 106 cells) were plated on six-well plates and incubated with E. coli (with a multiplicity of infection [MOI] of 10) in RPMI 1640 (without antibiotics) for 1 h at 37°C. HMDMs were then washed with PBS three times and incubated with RPMI 1640 containing a high concentration (3×) of penicillin-streptomycin (Thermo Fisher Scientific) at 37°C for 1 h. Medium was then replaced with RPMI 1640 containing a normal concentration (1×) of penicillin-streptomycin followed by further incubation at 37°C for 24 h. HMDMs were then washed and lysed with 0.5% Triton X-100. Intracellular bacteria were enumerated by plating on LB agar plates.
Data and code availability
The obtained sequence reads are publicly available from the NCBI Sequence Read Archive (BioProject: PRJNA986180).
Acknowledgments
We are grateful to the members of Kirino lab for helpful discussions. This study was supported in part by National Institutes of Health grants (GM106047, HL150560, AI151641, AI168975, and AI171366 to Y.K.) and American Cancer Society Research Scholar Grant (RSG-17-059-01-RMC, to Y.K.).
Author contributions
The project was conceived by Y.K. Experiments were designed by J.G., M.S., and Y.K. and performed by J.G. and T.K. Bioinformatics and data analysis were carried out by J.G. and M.S. The manuscript was written by J.G. and Y.K. with contributions from T.K. and M.S. Funding support was provided by Y.K.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102156.
Supplemental information
References
- 1.Furin J., Cox H., Pai M. Tuberculosis. Lancet. 2019;393:1642–1656. doi: 10.1016/S0140-6736(19)30308-3. [DOI] [PubMed] [Google Scholar]
- 2.Houben R.M.G.J., Dodd P.J. The Global Burden of Latent Tuberculosis Infection: A Re-estimation Using Mathematical Modelling. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behr M.A., Edelstein P.H., Ramakrishnan L. Revisiting the timetable of tuberculosis. BMJ. 2018;362 doi: 10.1136/bmj.k2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connolly L.E., Edelstein P.H., Ramakrishnan L. Why is long-term therapy required to cure tuberculosis? PLoS Med. 2007;4 doi: 10.1371/journal.pmed.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean A.S., Tosas Auguet O., Glaziou P., Zignol M., Ismail N., Kasaeva T., Floyd K. 25 years of surveillance of drug-resistant tuberculosis: achievements, challenges, and way forward. Lancet Infect. Dis. 2022;22:e191–e196. doi: 10.1016/S1473-3099(21)00808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brubaker S.W., Bonham K.S., Zanoni I., Kagan J.C. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G.Y., Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satoh T., Akira S. Toll-Like Receptor Signaling and Its Inducible Proteins. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.MCHD-0040-2016. [DOI] [PubMed] [Google Scholar]
- 11.Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 12.Kindler V., Sappino A.P., Grau G.E., Piguet P.F., Vassalli P. The inducing role of tumor necrosis factor in the development of bactericidal granulomas during BCG infection. Cell. 1989;56:731–740. doi: 10.1016/0092-8674(89)90676-4. [DOI] [PubMed] [Google Scholar]
- 13.Garcia I., Miyazaki Y., Marchal G., Lesslauer W., Vassalli P. High sensitivity of transgenic mice expressing soluble TNFR1 fusion protein to mycobacterial infections: synergistic action of TNF and IFN-gamma in the differentiation of protective granulomas. Eur. J. Immunol. 1997;27:3182–3190. doi: 10.1002/eji.1830271215. [DOI] [PubMed] [Google Scholar]
- 14.Flynn J.L., Goldstein M.M., Chan J., Triebold K.J., Pfeffer K., Lowenstein C.J., Schreiber R., Mak T.W., Bloom B.R. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 15.Ehlers S., Benini J., Kutsch S., Endres R., Rietschel E.T., et al. Pfeffer K. Fatal granuloma necrosis without exacerbated mycobacterial growth in tumor necrosis factor receptor p55 gene-deficient mice intravenously infected with Mycobacterium avium. Infect. Immun. 1999;67:3571–3579. doi: 10.1128/iai.67.7.3571-3579.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapoor N., Pawar S., Sirakova T.D., Deb C., Warren W.L., Kolattukudy P.E. Human granuloma in vitro model, for TB dormancy and resuscitation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgado M.A., Elmaoued R.A., Davis A.S., Kyei G., Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund J.M., Alexopoulou L., Sato A., Karow M., Adams N.C., Gale N.W., Iwasaki A., Flavell R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melchjorsen J., Jensen S.B., Malmgaard L., Rasmussen S.B., Weber F., Bowie A.G., Matikainen S., Paludan S.R. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J. Virol. 2005;79:12944–12951. doi: 10.1128/JVI.79.20.12944-12951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Triantafilou K., Orthopoulos G., Vakakis E., Ahmed M.A.E., Golenbock D.T., Lepper P.M., Triantafilou M. Human cardiac inflammatory responses triggered by Coxsackie B viruses are mainly Toll-like receptor (TLR) 8-dependent. Cell Microbiol. 2005;7:1117–1126. doi: 10.1111/j.1462-5822.2005.00537.x. [DOI] [PubMed] [Google Scholar]
- 21.Gantier M.P., Irving A.T., Kaparakis-Liaskos M., Xu D., Evans V.A., Cameron P.U., Bourne J.A., Ferrero R.L., John M., Behlke M.A., Williams B.R.G. Genetic modulation of TLR8 response following bacterial phagocytosis. Hum. Mutat. 2010;31:1069–1079. doi: 10.1002/humu.21321. [DOI] [PubMed] [Google Scholar]
- 22.Jöckel S., Nees G., Sommer R., Zhao Y., Cherkasov D., Hori H., Ehm G., Schnare M., Nain M., Kaufmann A., Bauer S. The 2'-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 2012;209:235–241. doi: 10.1084/jem.20111075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehrig S., Eberle M.E., Botschen F., Rimbach K., Eberle F., Eigenbrod T., Kaiser S., Holmes W.M., Erdmann V.A., Sprinzl M., et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 2012;209:225–233. doi: 10.1084/jem.20111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann S.M., Krüger C., Park B., Derkow K., Rosenberger K., Baumgart J., Trimbuch T., Eom G., Hinz M., Kaul D., et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 25.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J., et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabbri M., Paone A., Calore F., Galli R., Croce C.M. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 2013;10:169–174. doi: 10.4161/rna.23144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y., Zou L., Yan D., Chen H., Xu G., Jian W., Cui P., Chao W. Extracellular MicroRNAs Induce Potent Innate Immune Responses via TLR7/MyD88-Dependent Mechanisms. J. Immunol. 2017;199:2106–2117. doi: 10.4049/jimmunol.1700730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou L., He J., Gu L., Shahror R.A., Li Y., Cao T., Wang S., Zhu J., Huang H., Chen F., et al. Brain innate immune response via miRNA-TLR7 sensing in polymicrobial sepsis. Brain Behav. Immun. 2022;100:10–24. doi: 10.1016/j.bbi.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shurtleff M.J., Yao J., Qin Y., Nottingham R.M., Temoche-Diaz M.M., Schekman R., Lambowitz A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA. 2017;114:E8987–E8995. doi: 10.1073/pnas.1712108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nolte-'t Hoen E.N.M., Buermans H.P.J., Waasdorp M., Stoorvogel W., Wauben M.H.M., 't Hoen P.A.C. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shigematsu M., Kawamura T., Kirino Y. Generation of 2',3'-Cyclic Phosphate-Containing RNAs as a Hidden Layer of the Transcriptome. Front. Genet. 2018;9:562. doi: 10.3389/fgene.2018.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shigematsu M., Kirino Y. Making invisible RNA visible: discriminative sequencing methods for RNA molecules with specific terminal formations. Biomolecules. 2022;12:611. doi: 10.3390/biom12050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giraldez M.D., Spengler R.M., Etheridge A., Goicochea A.J., Tuck M., Choi S.W., Galas D.J., Tewari M. Phospho-RNA-seq: a modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J. 2019;38 doi: 10.15252/embj.2019101695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pawar K., Shigematsu M., Sharbati S., Kirino Y. Infection-induced 5'-half molecules of tRNAHisGUG activate Toll-like receptor 7. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin Y., Yao J., Wu D.C., Nottingham R.M., Mohr S., Hunicke-Smith S., Lambowitz A.M. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA. 2016;22:111–128. doi: 10.1261/rna.054809.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akat K.M., Lee Y.A., Hurley A., Morozov P., Max K.E., Brown M., Bogardus K., Sopeyin A., Hildner K., Diacovo T.G., et al. Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis. JCI Insight. 2019;5 doi: 10.1172/jci.insight.127317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawar, K, Kawamura, T, and Kirino, Y (submitted). The tRNAVal half: a strong endogenous Toll-like receptor 7 ligand with a 5′-terminal universal sequence signature. [DOI] [PMC free article] [PubMed]
- 38.Yamasaki S., Ivanov P., Hu G.F., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawamura T., Shigematsu M., Kirino Y. In vitro production and multiplex quantification of 2',3'-cyclic phosphate-containing 5'-tRNA half molecules. Methods. 2022;203:335–341. doi: 10.1016/j.ymeth.2021.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Guo J., Fan S., Li Y., Wei L., Yang X., Jiang T., Chen Z., Wang C., Liu J., et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakrabarty S., Kumar A., Raviprasad K., Mallya S., Satyamoorthy K., Chawla K. Host and MTB genome encoded miRNA markers for diagnosis of tuberculosis. Tuberculosis. 2019;116:37–43. doi: 10.1016/j.tube.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Lyu L., Zhang X., Li C., Yang T., Wang J., Pan L., Jia H., Li Z., Sun Q., Yue L., et al. Small RNA Profiles of Serum Exosomes Derived From Individuals With Latent and Active Tuberculosis. Front. Microbiol. 2019;10:1174. doi: 10.3389/fmicb.2019.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honda S., Loher P., Shigematsu M., Palazzo J.P., Suzuki R., Imoto I., Rigoutsos I., Kirino Y. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. USA. 2015;112:E3816–E3825. doi: 10.1073/pnas.1510077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shigematsu M., Morichika K., Kawamura T., Honda S., Kirino Y. Genome-wide identification of short 2',3'-cyclic phosphate-containing RNAs and their regulation in aging. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawamura T., Shigematsu M., Kirino Y. In vitro production and multiplex quantification of 2',3'-cyclic phosphate-containing 5'-tRNA half molecules. Methods. 2022;203:335–341. doi: 10.1016/j.ymeth.2021.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson B., Dutta A. Function and Therapeutic Implications of tRNA Derived Small RNAs. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.888424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magee R., Rigoutsos I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res. 2020;48:9433–9448. doi: 10.1093/nar/gkaa657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackman J.E., Gott J.M., Gray M.W. Doing it in reverse: 3'-to-5' polymerization by the Thg1 superfamily. RNA. 2012;18:886–899. doi: 10.1261/rna.032300.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shigematsu M., Kirino Y. 5'-Terminal nucleotide variations in human cytoplasmic tRNAHisGUG and its 5'-halves. RNA. 2017;23:161–168. doi: 10.1261/rna.058024.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shigematsu M., Kirino Y. Oxidative stress enhances the expression of 2',3'-cyclic phosphate-containing RNAs. RNA Biol. 2020;17:1060–1069. doi: 10.1080/15476286.2020.1766861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thoma-Uszynski S., Stenger S., Takeuchi O., Ochoa M.T., Engele M., Sieling P.A., Barnes P.F., Rollinghoff M., Bolcskei P.L., Wagner M., et al. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291:1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 53.Bao M., Yi Z., Fu Y. Activation of TLR7 Inhibition of Mycobacterium Tuberculosis Survival by Autophagy in RAW 264.7 Macrophages. J. Cell. Biochem. 2017;118:4222–4229. doi: 10.1002/jcb.26072. [DOI] [PubMed] [Google Scholar]
- 54.Lee H.J., Kang S.J., Woo Y., Hahn T.W., Ko H.J., Jung Y.J. TLR7 Stimulation With Imiquimod Induces Selective Autophagy and Controls Mycobacterium tuberculosis Growth in Mouse Macrophages. Front. Microbiol. 2020;11:1684. doi: 10.3389/fmicb.2020.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z., Ohto U., Shibata T., Krayukhina E., Taoka M., Yamauchi Y., Tanji H., Isobe T., Uchiyama S., Miyake K., Shimizu T. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity. 2016;45:737–748. doi: 10.1016/j.immuni.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 56.Tanji H., Ohto U., Shibata T., Taoka M., Yamauchi Y., Isobe T., Miyake K., Shimizu T. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 2015;22:109–115. doi: 10.1038/nsmb.2943. [DOI] [PubMed] [Google Scholar]
- 57.Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 58.Li M., Zeringer E., Barta T., Schageman J., Cheng A., Vlassov A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jia J., Yang S., Huang J., Zheng H., He Y., Wang L. Distinct Extracellular RNA Profiles in Different Plasma Components. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.564780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinn J.F., Patel T., Wong D., Das S., Freedman J.E., Laurent L.C., Carter B.S., Hochberg F., Van Keuren-Jensen K., Huentelman M., et al. Extracellular RNAs: development as biomarkers of human disease. J. Extracell. Vesicles. 2015;4 doi: 10.3402/jev.v4.27495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S.Y., Kim S., Kim J.E., Lee S.N., Shin I.W., Shin H.S., Jin S.M., Noh Y.W., Kang Y.J., Kim Y.S., et al. Lyophilizable and Multifaceted Toll-like Receptor 7/8 Agonist-Loaded Nanoemulsion for the Reprogramming of Tumor Microenvironments and Enhanced Cancer Immunotherapy. ACS Nano. 2019;13:12671–12686. doi: 10.1021/acsnano.9b04207. [DOI] [PubMed] [Google Scholar]
- 62.Dowling D.J., Barman S., Smith A.J., Borriello F., Chaney D., Brightman S.E., Melhem G., Brook B., Menon M., Soni D., et al. Development of a TLR7/8 agonist adjuvant formulation to overcome early life hyporesponsiveness to DTaP vaccination. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-20346-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J., Wu C.C.N., Lee K.J., Chuang T.H., Katakura K., Liu Y.T., Chan M., Tawatao R., Chung M., Shen C., et al. Activation of anti-hepatitis C virus responses via Toll-like receptor 7. Proc. Natl. Acad. Sci. USA. 2006;103:1828–1833. doi: 10.1073/pnas.0510801103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nian H., Geng W.Q., Cui H.L., Bao M.J., Zhang Z.N., Zhang M., Pan Y., Hu Q.H., Shang H. R-848 triggers the expression of TLR7/8 and suppresses HIV replication in monocytes. BMC Infect. Dis. 2012;12:5. doi: 10.1186/1471-2334-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z., Ohto U., Shibata T., Taoka M., Yamauchi Y., Sato R., Shukla N.M., David S.A., Isobe T., Miyake K., Shimizu T. Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands. Cell Rep. 2018;25:3371–3381.e5. doi: 10.1016/j.celrep.2018.11.081. [DOI] [PubMed] [Google Scholar]
- 66.Costa B., Li Calzi M., Castellano M., Blanco V., Cuevasanta E., Litvan I., Ivanov P., Witwer K., Cayota A., Tosar J.P. Nicked tRNAs are stable reservoirs of tRNA halves in cells and biofluids. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2216330120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X., Wolin S.L. Transfer RNA halves are found as nicked tRNAs in cells: evidence that nicked tRNAs regulate expression of an RNA repair operon. RNA. 2023;29:620–629. doi: 10.1261/rna.079575.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dhahbi J.M., Spindler S.R., Atamna H., Yamakawa A., Boffelli D., Mote P., Martin D.I.K. 5' tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su Z., Kuscu C., Malik A., Shibata E., Dutta A. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J. Biol. Chem. 2019;294:16930–16941. doi: 10.1074/jbc.RA119.009272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mo X., Du S., Chen X., Wang Y., Liu X., Zhang C., Zhu C., Ding L., Li Y., Tong Y., et al. Lactate Induces Production of the tRNA(His) Half to Promote B-lymphoblastic Cell Proliferation. Mol. Ther. 2020;28:2442–2457. doi: 10.1016/j.ymthe.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krishna S., Yim D.G., Lakshmanan V., Tirumalai V., Koh J.L., Park J.E., Cheong J.K., Low J.L., Lim M.J., Sze S.K., et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019;20 doi: 10.15252/embr.201947789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen L., Xu W., Liu K., Jiang Z., Han Y., Jin H., Zhang L., Shen W., Jia S., Sun Q., Meng A. 5' Half of specific tRNAs feeds back to promote corresponding tRNA gene transcription in vertebrate embryos. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abh0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tosar J.P., Gámbaro F., Darré L., Pantano S., Westhof E., Cayota A. Dimerization confers increased stability to nucleases in 5' halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. 2018;46:9081–9093. doi: 10.1093/nar/gky495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Preissner K.T., Fischer S. Functions and cellular signaling by ribosomal extracellular RNA (rexRNA): Facts and hypotheses on a non-typical DAMP. Biochim. Biophys. Acta Mol. Cell Res. 2023;1870 doi: 10.1016/j.bbamcr.2022.119408. [DOI] [PubMed] [Google Scholar]
- 75.Preissner K.T., Fischer S., Deindl E. Extracellular RNA as a Versatile DAMP and Alarm Signal That Influences Leukocyte Recruitment in Inflammation and Infection. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.619221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jung S., von Thülen T., Laukemper V., Pigisch S., Hangel D., Wagner H., Kaufmann A., Bauer S. A single naturally occurring 2'-O-methylation converts a TLR7- and TLR8-activating RNA into a TLR8-specific ligand. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 78.Taoka M., Nobe Y., Yamaki Y., Sato K., Ishikawa H., Izumikawa K., Yamauchi Y., Hirota K., Nakayama H., Takahashi N., Isobe T. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018;46:9289–9298. doi: 10.1093/nar/gky811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hartmann A., Kunz M., Köstlin S., Gillitzer R., Toksoy A., Bröcker E.B., Klein C.E. Hypoxia-induced up-regulation of angiogenin in human malignant melanoma. Cancer Res. 1999;59:1578–1583. [PubMed] [Google Scholar]
- 80.Mirzaei A., Mahmoudi H. Evaluation of TNF-alpha cytokine production in patients with tuberculosis compared to healthy people. GMS Hyg. Infect. Control. 2018;13:Doc09. doi: 10.3205/dgkh000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deng J., Ptashkin R.N., Chen Y., Cheng Z., Liu G., Phan T., Deng X., Zhou J., Lee I., Lee Y.S., Bao X. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol. Ther. 2015;23:1622–1629. doi: 10.1038/mt.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baitsch D., Bock H.H., Engel T., Telgmann R., Müller-Tidow C., Varga G., Bot M., Herz J., Robenek H., von Eckardstein A., Nofer J.R. Apolipoprotein E induces antiinflammatory phenotype in macrophages. Arterioscler. Thromb. Vasc. Biol. 2011;31:1160–1168. doi: 10.1161/ATVBAHA.111.222745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hogg M.C., Rayner M., Susdalzew S., Monsefi N., Crivello M., Woods I., Resler A., Blackbourn L., Fabbrizio P., Trolese M.C., et al. 5'ValCAC tRNA fragment generated as part of a protective angiogenin response provides prognostic value in amyotrophic lateral sclerosis. Brain Commun. 2020;2:fcaa138. doi: 10.1093/braincomms/fcaa138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brambilla L., Guidotti G., Martorana F., Iyer A.M., Aronica E., Valori C.F., Rossi D. Disruption of the astrocytic TNFR1-GDNF axis accelerates motor neuron degeneration and disease progression in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2016;25:3080–3095. doi: 10.1093/hmg/ddw161. [DOI] [PubMed] [Google Scholar]
- 85.Tateishi T., Yamasaki R., Tanaka M., Matsushita T., Kikuchi H., Isobe N., Ohyagi Y., Kira J.i. CSF chemokine alterations related to the clinical course of amyotrophic lateral sclerosis. J. Neuroimmunol. 2010;222:76–81. doi: 10.1016/j.jneuroim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 86.Schön M.P., Schön M. TLR7 and TLR8 as targets in cancer therapy. Oncogene. 2008;27:190–199. doi: 10.1038/sj.onc.1210913. [DOI] [PubMed] [Google Scholar]
- 87.Michaelis K.A., Norgard M.A., Zhu X., Levasseur P.R., Sivagnanam S., Liudahl S.M., Burfeind K.G., Olson B., Pelz K.R., Angeles Ramos D.M., et al. The TLR7/8 agonist R848 remodels tumor and host responses to promote survival in pancreatic cancer. Nat. Commun. 2019;10:4682. doi: 10.1038/s41467-019-12657-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pliatsika V., Loher P., Telonis A.G., Rigoutsos I. MINTbase: a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics. 2016;32:2481–2489. doi: 10.1093/bioinformatics/btw194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holmes A.D., Chan P.P., Chen Q., Ivanov P., Drouard L., Polacek N., Kay M.A., Lowe T.M. A standardized ontology for naming tRNA-derived RNAs based on molecular origin. Nat. Methods. 2023;20:627–628. doi: 10.1038/s41592-023-01813-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson J.T., Thorvaldsdottir H., Turner D., Mesirov J.P. igv.js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV) Bioinformatics. 2023;39 doi: 10.1093/bioinformatics/btac830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pawar K., Hanisch C., Palma Vera S.E., Einspanier R., Sharbati S. Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci. Rep. 2016;6 doi: 10.1038/srep19416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gantier M.P., Tong S., Behlke M.A., Xu D., Phipps S., Foster P.S., Williams B.R.G. TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J. Immunol. 2008;180:2117–2124. doi: 10.4049/jimmunol.180.4.2117. [DOI] [PubMed] [Google Scholar]
- 95.Honda K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The obtained sequence reads are publicly available from the NCBI Sequence Read Archive (BioProject: PRJNA986180).