Abstract
The M26 meiotic recombination hot spot in the ade6 gene of Schizosaccharomyces pombe is activated by the heterodimeric M26 binding protein Mts1-Mts2. The individual Mts1 (Atf1, Gad7) and Mts2 (Pcr1) proteins are also transcription factors involved in developmental decisions. We report that the Mts proteins are key effectors of at least two distinct classes of developmental decisions regulated by the mitogen-activated protein (MAP) kinase cascade. The first class (osmoregulation, spore viability, and spore quiescence) requires the Spc1 MAP kinase and the Mts1 protein but does not require the Mts2 protein. The second class (mating, meiosis, and recombination hot spot activation) requires the Spc1 kinase and the Mts1-Mts2 heterodimer. Northern and Western blotting eliminated any significant role for the Spc1 kinase in regulating the expression levels of the Mts proteins. Gel mobility shift experiments indicated that the Mts1-Mts2 heterodimer does not need to be phosphorylated to bind to ade6-M26 DNA in vitro. However, in vivo dimethyl sulfate footprinting demonstrated that protein-DNA interaction within cells is dependent upon the Spc1 MAP kinase, which phosphorylates the Mts1 protein. Thus, the Spc1 kinase helps regulate the effector activities of the Mts1-Mts2 heterodimer in part by modulating its ability to occupy the M26 DNA site in vivo. Meiotic recombination hot spot function is likely the result of DNA conformational changes imparted by binding of the Mts1-Mts2 meiotic transcription factor.
Homologous recombination hot spots are cis-acting DNA sites that increase the frequency of recombination in their vicinity and thereby influence patterns of genomic recombination (22, 44). Mechanisms of recombination initiation involving double-strand DNA breaks (3, 5, 18, 28, 40), regulation of recombination involving competition between nearby hot spots (9, 53, 54), and possible control of the timing and distribution of recombination (20, 51, 52) have all been revealed by study of hot spots. An emerging paradigm is that recombination hot spots are activated by regions of chromatin accessibility, possibly due to the assembly of the transcription machinery in promoter regions (27).
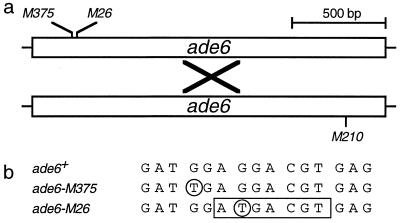
The M26 hot spot in the ade6 gene of the fission yeast Schizosaccharomyces pombe was identified as an auxotrophic mutation that increases ade6 gene conversion to approximately 10-fold the level obtained from crosses harboring other ade6 alleles (14). The hot spot is active only during meiosis (31, 36) and promotes both reciprocal exchange and gene conversion (12, 14, 36). Together, the genetic data indicate that M26 enhances the initiation of recombination at or very near the M26 site (12, 14, 33). A discrete 7-bp nucleotide sequence (5′-ATGACGT-3′) at M26 is essential for hot spot function (Fig. 1) (37). A heterodimeric protein composed of subunits Mts1 and Mts2 (for “M-twenty-six binding protein”) was identified and purified based upon binding to M26 (45). Cloning of the mts1 and mts2 genes, their disruption, and genetic analyses demonstrated that the Mts1-Mts2 heterodimer activates the hot spot (20).
FIG. 1.
ade6-M26 meiotic recombination hot spot. (a) Schematic of the ade6 gene showing the positions of the alleles used. Recombination events (×) generate a wild-type, selectable ade6 gene. Genetic crosses involving M26 (recombination hot spot) generate about 10- to 20-fold more recombinants than crosses involving M375 (basal recombination) (14, 20). (b) The M375 and M26 alleles were created by single G-to-T mutations (circled) that generate translational stops mapping to adjacent codons (31, 41). A specific 7-bp site created by M26 (boxed) is essential for hot spot function (37). The site is bound by the heterodimeric Mts1-Mts2 protein (45), and hot spot function is strictly dependent upon Mts1-Mts2 (20).
An S. pombe gene identical to mts1 has been independently cloned as a gene (atf1) identified by the genome sequencing project (42), as a gene (gad7) required for normal sexual development (17), and as a plasmid multicopy suppressor (atf1) of the partial mating defect in spc1 mutants (39). The mts2 gene was also independently cloned as a weak plasmid multicopy suppressor (pcr1) of a sporulation defect in spo5 mutants (48). The mts1 and mts2 genes encode proteins with basic leucine zipper (bZIP) DNA binding and protein dimerization motifs. The bZIP portions of the predicted polypeptides have about 50% sequence identity with members of the activating transcription factor-cyclic AMP (cAMP) responsive element binding protein (ATF/CREB) family, but the remainder of the proteins have no significant homology to other proteins currently in computer databases.
In addition to their role in M26 hot spot activation (20, 45), the individual Mts1 (Atf1, Gad7) and Mts2 (Pcr1) proteins have roles in sexual development and a number of different stress responses and are required for appropriate transcriptional regulation of a variety of genes (6, 17, 39, 42, 48, 49). This transcriptional regulation requires the mitogen-activated protein (MAP) kinase cascade, and Mts1 is directly phosphorylated by the MAP kinase Spc1 (39, 49). Additional signals converge upon the Mts proteins via the cAMP-dependent protein kinase A pathway and via the Pat1 kinase, which is a repressor of meiotic development (2). Possible input from other signals, the likely differential phosphorylation and the sites at which they occur, and the mechanism by which signals bifurcate at Mts1 and Mts2 remain to be determined. The roles for Mts1 and Mts2 in transcriptional regulation suggest that M26 hot spot activation may be related to transcription regulation. However, steady-state levels of the ade6 transcript do not correlate with hot spot function (13, 20).
While a heterodimer of Mts1 and Mts2 binding to a discrete DNA site at ade6-M26 is proven to activate the recombination hot spot (20, 37, 45), the dimerization partners for the other Mts1-dependent and Mts2-dependent biological functions have not been reported. Because there is a correlation between open chromatin in promoter regions and recombination hot spot activity (27, 44), we explored the possibility that transcriptional regulation and hot spot activation are mechanistically coupled through the use of a common factor, the Mts1-Mts2 heterodimer. We also examined whether hot spot activation, like transcriptional regulation, is dependent upon signals from the meiotic MAP kinase cascade. We report that the Mts1-Mts2 heterodimer is required for mating, meiosis, and meiotic recombination hot spot activities and that the Spc1 MAP kinase regulates these effector functions by modulating DNA binding in vivo. This suggests that hot spot function is a consequence of meiotic chromatin remodeling associated with the transcriptional-regulatory activities of the Mts1-Mts2 heterodimer.
MATERIALS AND METHODS
Strains, culture media, and genetic techniques.
The S. pombe strains used for this study are listed in Table 1. All of the spc1, mts1, and mts2 mutants harbored deletions of the respective genes and were therefore null mutants. Strains were cultured in nitrogen base liquid (NBL) or on nitrogen base agar (NBA) (0.67% Difco yeast nitrogen base without amino acids and with ammonium sulfate, 1% glucose, and 2% agar for solid media); mating and meiosis were conducted on synthetic sporulation agar (SPA) (1% glucose, 0.1% K2HPO4, 10 ng of biotin per ml, 1 μg of pantothenic acid per ml, 10 μg of nicotinic acid per ml, 10 μg of inositol per ml, and 3% agar) (15). NBA, NBL, and SPA were supplemented with the necessary purines, pyrimidines, and amino acids (100 μg/ml) as required. Strain constructions, meiotic crosses, preparation of free ascospores, and analysis of recombination frequencies were done as described previously (14, 20, 31, 36).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source or referencea |
|---|---|---|
| KS1366 | h− spc1::ura4 leu1-32 ura4-D18 | 38 |
| WSP547 | h− ade6-M210 his3-D1 leu1-32 ura4-D18 | 20 |
| WSP550 | h+ ade6-M210 his3-D1 leu1-32 ura4-D18 | 20 |
| WSP571 | h+ ade6-M26 his3-D1 leu1-32 ura4-D18 | 20 |
| WSP578 | h+ ade6-M375 his3-D1 leu1-32 ura4-D18 | 20 |
| WSP598 | h+ his3-D1 leu1-32 ura4-D18 | This studyb |
| WSP599 | h− his3-D1 leu1-32 ura4-D18 | 20 |
| WSP640 | h+ mts2-D1::his3+ his3-D1 leu1-32 ura4-D18 | WSP599 × WSP656 |
| WSP643 | h− ade6-M210 mts1-D15::ura4+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP644 | h+ ade6-M26 mts1-D15::ura4+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP646 | h+ ade6-M375 mts1-D15::ura4+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP649 | h− ade6-M210 mts2-D1::his3+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP650 | h+ ade6-M26 mts2-D1::his3+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP652 | h+ ade6-M375 mts2-D1::his3+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP656 | h+ ade6-M210 mts2-D1::his3+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP671 | h+ ade6-M210 mts1-D15::ura4+ mts2-D1::his3+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP675 | h− ade6-M26 mts1-D15::ura4+ mts2-D1::his3+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP678 | h− ade6-M375 mts1-D15::ura4+ mts2-D1::his3+ his3-D1 leu1-32 ura4-D18 | 20 |
| WSP1269 | h+ mts1-D15::his3+ his3-D1 leu1-32 ura4-D18 | T of WSP598 |
| WSP1037 | h− ade6-M210 spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP550 × KS1366 |
| WSP1040 | h+ ade6-M26 spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP571 × KS1366 |
| WSP1041 | h− ade6-M26 spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP571 × KS1366 |
| WSP1044 | h+ ade6-M375 spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP578 × KS1366 |
| WSP1045 | h− ade6-M375 spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP578 × KS1366 |
| WSP1305 | h− ade6-M210 mts1-D15::his3+ spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP1269 × WSP1037 |
| WSP1306 | h+ ade6-M26 mts1-D15::his3+ spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP1269 × WSP1041 |
| WSP1307 | h+ ade6-M375 mts1-D15::his3+ spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP1269 × WSP1045 |
| WSP1096 | h− ade6-M210 mts2-D1::his3+ spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP640 × WSP1037 |
| WSP1097 | h+ ade6-M26 mts2-D1::his3+ spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP640 × WSP1041 |
| WSP1099 | h+ ade6-M375 mts2-D1::his3+ spc1::ura4+ his3-D1 leu1-32 ura4-D18 | WSP640 × WSP1045 |
Strains were derived from standard genetic crosses or by transformation (T) of the indicated strains.
Complete genealogies available upon request.
Construction of the mts1-D15::his3+ allele.
Targeted replacement was achieved with the same strategy as used to obtain the mts1-D15::ura4+ allele (20) except that selection was for histidine prototrophy instead of for uracil prototrophy. The genomic changes at the mts1 locus are identical except for the inserted selectable marker gene. Targeted replacement was confirmed by a combination of Southern blotting and PCR analyses, as described previously (20).
Determination of osmosensitivity.
Individual colonies were inoculated into 5 ml of appropriately supplemented NBL and grown to a density of 107 cells/ml at 32°C, serial dilutions were made, and the dilutions were plated in parallel on supplemented NBA medium containing various concentrations of NaCl. After incubation for 3 days at 32°C, the plates were examined to determine plating efficiencies.
Analysis of mating and meiosis.
Five-milliliter cultures of heterothallic strains were grown in NBL at 32°C to a density of 107 cells/ml; the cells to be mated were mixed, harvested by centrifugation, washed once with double-distilled H2O, and resuspended at 109 cells/ml in double-distilled H2O, and duplicate aliquots containing 108 cells were plated on SPA. The mating mixtures were incubated at 23°C, and at various time points individual aliquots were collected to monitor the efficiency of mating and sporulation. Mating efficiencies were determined from the frequency of conjugants in the culture, and sporulation efficiencies were determined from the frequency of asci and spores, both determined microscopically with the aid of a hemocytometer. The relative plating efficiency of a known number of spores was used to determine spore viability. Upon counting the spore yields microscopically it was noted that some spores in some mutant backgrounds had germinated prematurely. Because unmated parental cells were excluded from the spore counts and some germinated spores may have grown enough to assume the appearance of unmated cells, it was not possible to determine the precise frequency of premature germination. The reported frequencies are for obviously germinating spores per total visible spores.
Northern blot analyses.
Cells used for mRNA analyses were grown at 32°C with vigorous agitation in supplemented PM synthetic minimal medium (47) and were harvested in early log phase (≤107 cells/ml). In some cases, the cultures were shifted to nitrogen-free PM for 6 h prior to preparation of RNA. Harvesting of cells, disruption by agitation in the presence of glass beads, processing of total cellular RNA, and Northern blotting were done as described previously (23, 24). RNA samples (10 μg) were fractionated on 0.8% agarose–7% formaldehyde gels and transferred to a Hybond-N membrane (Amersham) prior to hybridization. Probes were derived from gel-purified DNA fragments and were labeled by incorporation of [α-32P]dCTP using random hexanucleotide primer extension.
Western blot analysis.
The Mts1 and Mts2 proteins were overexpressed in Escherichia coli and purified by nickel affinity chromatography as previously described (20). The proteins were further purified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, the gels were stained with Coomassie, the bands were excised, and the gel slices were macerated and used directly for immunization. Polyclonal antisera were raised in rabbits by a commercial laboratory (Cocalico Laboratories). Total cellular lysate (10 μg/lane) was fractionated on SDS–10% polyacrylamide gels, transferred to nitrocellulose membranes by semidry electroblotting, probed with primary antisera, amplified with a secondary goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (R14745; Transduction Laboratories), and visualized by chemiluminescence as described previously (16). Each experiment was repeated at least twice with different extracts to confirm the results and gauge potential sample loading errors. Duplicate antisera raised against each of the proteins gave similar results. The antiserum titers used were 1:2,000. Specificity was inferred by lack of signal with preimmune serum, appearance of bands of appropriate sizes with specific antisera, and loss of those bands in extracts from the mts1-D15 and mts2-D1 deletion mutants.
In vivo footprinting.
Our in vivo dimethyl sulfate (DMS) footprinting procedure for S. pombe was developed by modifying those previously reported for budding yeast (7, 35). Cells were grown with vigorous aeration at 32°C in supplemented NBL minimal medium to a density of 107 cells/ml. A 100-ml aliquot of each culture was harvested by centrifugation (1,000 × g, 10 min) and resuspended in 3 ml of NBL at 22°C. Half was stored on ice for subsequent preparation of naked genomic DNA, and the remaining 1.5 ml was treated with 1 μl of DMS (D5279; Sigma) in vivo for 2 min with gentle agitation. The reaction was stopped by addition of 25 ml of ice-cold TEN (10 mM Tris [pH 7.9], 1 mM EDTA, 40 mM NaCl), and then the cells were harvested by centrifugation, resuspended in 1.5 ml of NBL, and stored on ice for subsequent DNA preparation. As a control for the in vivo DMS treatment, highly purified genomic DNA (described below) from untreated cells was treated in vitro with DMS. For each control, a 10-μg aliquot of DNA in a 50-μl reaction volume was treated with 1 μl of DMS for 1 min at 22°C with gentle agitation. The reactions were stopped by the addition of 1 ml of TEN, and the DNAs were extracted with 2 volumes of phenol-chloroform (1:1), precipitated by the addition of 2.5 volumes of ethanol and centrifugation for 15 min, rinsed once with 70% ethanol, and then resuspended in TE (10 mM Tris [pH 7.5], 0.1 mM EDTA).
High-quality genomic DNA was prepared in parallel from DMS-treated and control cells. Cells were washed once with 5 ml of spheroplasting buffer (10 mM Tris [pH 7.5], 10 mM EDTA, 30 mM β-mercaptoethanol, 1 M sorbitol), collected by centrifugation, and resuspended in 5 ml of spheroplasting buffer containing 2 mg of yeast lytic enzyme (152270; ICN) per ml. After 15 min of incubation at 32°C with gentle agitation, the spheroplasts were harvested by centrifugation and resuspended in 0.5 ml of 50 mM Tris (pH 7.9), 20 mM EDTA. Cell lysis was achieved by the addition of 75 μl of 10% SDS and incubation at 65°C for 30 min. Then, 250 μl of 5 M potassium acetate was added, the samples were incubated on ice for 30 min, and the precipitates were removed by centrifugation at full speed in an Eppendorf centrifuge for 15 min at 22°C. The supernatants were collected to clean tubes, an equal volume (∼750 μl) of isopropanol was added to each tube, the samples were incubated at 22°C for 10 min, and nucleic acids were collected by centrifugation at 22°C for 10 min. The pellets were rinsed once with 70% ethanol, air dried briefly, resuspended in 50 μl of TE containing 50 μg of DNase-free RNaseA per ml, and incubated at 37°C for 16 h. Proteinase K (5 μl of a 10-mg/ml stock) and 12.5 μl of 10% SDS were added, the samples were incubated at 65°C for 1 h, 125 μl of 8-ammonium acetate was added, and the samples were incubated on ice for 15 min. The SDS-protein precipitates were removed by centrifugation for 10 min at 4°C, the supernatants were transferred to new tubes, an equal volume of isopropanol was added, the DNAs were precipitated by centrifugation, the pellets were rinsed once with 70% ethanol, and the genomic DNAs were resuspended in 150 μl of TE. Excess salts and ribonucleotides were removed by passage through a Sephadex G-50 column (1-154-560; 5 Prime→3 Prime), and then the DNA concentrations were determined by fluorimetry.
The patterns of DMS reactivity were visualized by using Taq DNA polymerase-mediated radiolabeled primer extension (4). Reactions were in a volume of 50 μl (62.6 mM Tris-HCl [pH 8.8], 16.6 mM NH4SO4, 10 mM β-mercaptoethanol, 6.7 mM MgCl2, 2 mg of bovine serum albumin per ml) and contained 5 μg of genomic DNA, 1 pmol of 5′-end-labeled primer (specific activity, 2 × 103 to 10 × 103 cpm/fmol), and 2 units of AmpliTaq DNA polymerase (United States Biochemical Corp.). Five cycles of primer extension were performed as follows: denaturation at 94°C for 1 min, annealing at 55°C for 2 min, and extension at 75°C for 3 min. After extension, 6 μl of a stop solution (1 mg of proteinase K per ml, 100 mM EDTA, 0.1% SDS) was added to each tube and the tubes were incubated at 65°C for 30 min. E. coli tRNA (5 μg) was added as a carrier, the extension products were precipitated with 2.5 volumes of ethanol and centrifugation, the pellets were rinsed once with 70% ethanol, and then the samples were resuspended in 10 μl of denaturing gel loading buffer (50 mM NaOH, 50 mM NaCl, 0.5 mM EDTA, 10 M urea, 0.075% xylene cyanol, 0.075% bromphenol blue).
As a control for priming specificity, and to locate the relative positions of methylation-induced polymerase stop sites, untreated genomic DNA was sequenced by a thermocycle dideoxy chain termination sequencing protocol (New England Biolabs). Reaction conditions and processing of the extension products were identical to those used to visualize methylation sites.
Primer extension products from DNA methylated in vivo, DNA methylated in vitro, and the genomic DNA sequencing reactions were boiled for 5 min and then fractionated on denaturing polyacrylamide gels (6% polyacrylamide, 10 M urea, 89 mM Tris, 89 mM borate, 1 mM EDTA). Extension products were visualized by autoradiography of dried gels with Kodak XAR-5 film.
RESULTS
Activation of the ade6-M26 meiotic recombination hot spot requires a heterodimer of the Mts1 and Mts2 proteins (20, 45). We therefore used genetic epistasis analyses to determine whether the Mts1-Mts2 heterodimer was the functional moiety for a variety of Spc1 MAP kinase-dependent developmental decisions.
Osmoregulation by Spc1 MAP kinase and Mts1 is genetically distinct from M26 hot spot activation.
The Mts1 and Spc1 proteins are required for the osmotic stress response (20, 39, 49). However, no quantitative comparisons of the various single and double mutants have been reported. To determine the dependence of osmoregulation upon these proteins and to reveal potential genetic interactions, we compared the efficiencies of plating (EOP) of single and double null (deletion) mutants on media containing various concentrations of NaCl (Fig. 2).
FIG. 2.
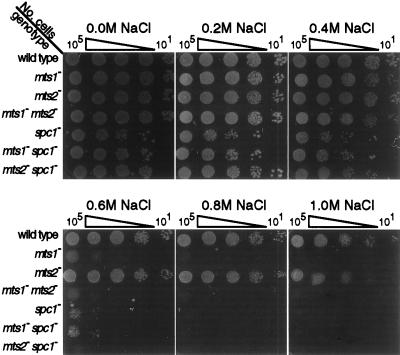
Requirement for mts1 and spc1 in the osmotic stress response. The indicated numbers of cells from healthy log-phase cultures were spotted onto supplemented NBA minimal medium containing the indicated concentrations of NaCl and were incubated for 3 days at 32°C prior to being photographed.
Wild-type cells and mts2 mutant cells were osmotolerant and exhibited high EOP even at high osmolarity (Fig. 2). In contrast, the spc1 and mts1 single mutants were sensitive to osmostress and exhibited similar concentration-dependent EOP curves (Fig. 2). The spc1 mts1 double-mutant EOP results were very similar to those of the spc1 and mts1 single mutants, suggesting that Spc1 and Mts1 function in the same linear pathway. Since the Mts1 protein is directly phosphorylated by the Spc1 MAP kinase in response to osmotic stress (39, 49), we infer that the Mts1 protein has a major role in affecting osmoregulation in response to signals from the MAP kinase cascade. Because the mts2 mutants were osmotolerant (Fig. 2), we conclude that this stress response is not achieved by the Mts1-Mts2 heterodimer. Hot spot activation, which strictly requires a heterodimer of Mts1 and Mts2 binding to the M26 site (20, 45), is genetically distinct from osmoregulation.
Spc1 MAP kinase, Mts1, and Mts2 function together in the same pathway of sexual development.
The fission yeast life cycle is predominantly haploid, and the external signals that trigger sexual development, principally nitrogen starvation, lead to both sexual conjugation and a coupled entry into meiosis. To determine whether the Mts1, Mts2, and Spc1 proteins function in the same or different pathways of sexual development, we determined the efficiencies of mating and meiosis in strains harboring various combinations of deletion mutations. Heterothallic haploid cells were grown to early log phase, were mixed, and were incubated on synthetic sporulation medium (15). The cultures were scored for mating efficiency (the frequency of conjugants), for the efficiency of meiosis (the frequency of ascus and spore formation), and for spore viability.
The mts1, mts2, and spc1 single mutants were all partially sterile and mated with 6 to 16% of the efficiency of wild-type cells (Fig. 3a). Furthermore, while clearly different from wild-type frequencies, the frequencies from each of the single- and double-mutant combinations were statistically indistinguishable. The lack of phenotypic additivity (Fig. 3a), and the proven phosphorylation of Mts1 by Spc1 kinase (39, 49), suggests that the Mts1-Mts2 heterodimer functions directly downstream of Spc1 MAP kinase in a linear pathway of signal transduction required for conjugation.
FIG. 3.
Roles for mts1, mts2, and spc1 in mating, meiosis, and spore viability. (a) Mating efficiencies. Data are average frequencies of conjugants and incipient asci observed from days 1 through 5 of mating between heterothallic strains. (b) Efficiencies of meiosis. Data are total ascus and spore yields after 5 days and are adjusted for the relative mating efficiencies of each cross. (c) Spore viabilities determined from the EOP of known numbers of spores on nonselective medium.
To explore the roles of the proteins in meiosis, we determined what fraction of successful conjugants went on through meiosis to generate asci and spores. The requirements for Spc1, Mts1, and Mts2 in meiosis (Fig. 3b) were very similar to the requirements in conjugation. The single mutants each had a significant defect, ranging from 19 to 41% of wild-type sporulation levels, and there were no significant differences between the meiotic efficiencies of the various single and double mutants. These data suggest that the Mts1-Mts2 heterodimer is also a principal effector of meiotic induction controlled by Spc1 MAP kinase, which directly phosphorylates the Mts1 protein (39, 49). Use of the same pathway for both conjugation and meiotic induction provides a mechanistic basis for the coupling of mating and meiosis in S. pombe.
Spore quiescence and spore viability require Spc1 MAP kinase and Mts1, but not Mts2.
While mating and meiosis were dependent upon the simultaneous presence of Mts1, Mts2, and Spc1, the protein requirements for spore viability (Fig. 3c) were different. Wild-type cells and mts2 mutant cells each produced spores with similar, high viabilities. In contrast, the spc1 and mts1 single mutants and the spc1 mts1 double mutant produced spores with reduced viabilities, and there was no significant difference in the magnitude of their respective defects (Fig. 3c). We infer that Spc1 and Mts1 function in the same linear pathway required for spore viability. Furthermore, because the mts2 mutants had wild-type spore viabilities, we conclude that this viability function does not require a heterodimer of the Mts1 and Mts2 proteins.
Microscopic examination of day 5 mating mixtures revealed that about 30% of the spores in the spc1 mutant, mts1 mutant, and spc1 mts1 double mutant mating mixtures had germinated prematurely (data not shown). In contrast, the wild-type and mts2 mutant spores remained quiescent. We conclude that spore quiescence, which depends upon the spore’s ability to sense poor extracellular nutritional conditions, is regulated (at least in part) by signals from the MAP kinase cascade impinging upon the Mts1 protein. Since the genetic requirements for osmoregulation (Fig. 2), spore viability (Fig. 3c), and spore quiescence (data not shown) are identical, it seems likely that each of these processes depends upon a common pathway requiring Spc1 and Mts1.
In summary, the genetic epistasis experiments (Fig. 2 and 3) revealed that the Mts1 and Mts2 proteins are involved in at least two distinct classes of developmental decisions regulated by the MAP kinase cascade. The first class (osmoregulation, spore viability, and spore quiescence) requires the Spc1 MAP kinase and the Mts1 protein but does not require the Mts2 protein. The second class (mating and meiosis) requires the Spc1 kinase and the Mts1-Mts2 heterodimer. This suggests that the meiotic recombination hot spot activity, which also requires the Mts1-Mts2 heterodimer (20, 45), might be a consequence of the meiotic transcriptional-regulatory activities of the Mts1-Mts2 heterodimer. We therefore tested whether hot spot activation, like the developmental decisions, was dependent upon the Spc1 MAP kinase.
Hot spot activation requires the simultaneous presence of M26, Mts1, Mts2, and Spc1.
To determine whether Spc1 is required for activation of the M26 hot spot, we measured recombination between two sets of ade6 alleles (Fig. 1). Crosses between strains harboring the ade6-M375 and the ade6-M210 alleles were used to reveal basal recombination levels, and crosses between strains harboring the ade6-M26 and the ade6-M210 alleles were used to reveal hot spot recombination levels (14, 20). Wild-type cells (mts1+ mts2+ spc1+) produced a basal recombination frequency of 6.8 × 10−4 and a hot spot-enhanced frequency of 78 × 10−4 (Table 2, line 1). The ratio of these two values, called the “hot spot ratio,” is a measure of the enhancement of recombination conferred by the M26 site. In wild-type meiosis (mts1+ mts2+ spc1+) we observed a hot spot ratio of 11, indicative of a functional M26 hot spot.
TABLE 2.
Requirement for M26, Spc1, Mts1, and Mts2 in hot spot meiotic recombination
| Relevant genotypea
|
Ade+ recombination frequency (104)b
|
Hot spot ratioc | |||
|---|---|---|---|---|---|
| mts1/mts1 | mts2/mts2 | spc1/spc1 | M375 × M210 | M26 × M210 | |
| +/+ | +/+ | +/+ | 6.8 ± 1.5 | 78 ± 9.0 | 11 |
| −/− | +/+ | +/+ | 6.5 ± 1.1 | 5.0 ± 0.6 | 0.8 |
| +/+ | −/− | +/+ | 4.4 ± 0.9 | 4.8 ± 0.9 | 1.1 |
| +/+ | +/+ | −/− | 5.2 ± 1.4 | 3.3 ± 1.0 | 0.6 |
| −/− | −/− | +/+ | 9.6 ± 4.0 | 9.6 ± 4.0 | 1.0 |
| −/− | +/+ | −/− | 5.5 ± 2.6 | 6.7 ± 1.2 | 1.2 |
| +/+ | −/− | −/− | 4.9 ± 3.3 | 4.3 ± 2.1 | 0.9 |
Plus signs indicate mts1+, mts2+, or spc1+; minus signs indicate deleted mts1 (mts1-D15::his3+ or mts1-D15::ura4+) (Table 1) (20), deleted mts2 (mts2-D1::his3+) (20), or deleted spc1 (spc1::ura4+) (38). All strains also have the genotype leu1-32 ura4-D18 his3-D1 (Table 1).
See Fig. 1 for positions of ade6 alleles. Standard genetic crosses (12, 14, 37) were conducted, and spores were plated on supplemented NBA minimal medium containing adenine (100 μg/ml) to determine the total viable spore titer (T) and on NBA medium lacking adenine to determine the ade6+ recombinant titer (R). At least 100 colonies of each type were counted for each cross. The recombination frequency for each experiment is R/T. Data are means ± standard deviations of recombination frequencies from four experiments.
Ratio of recombination frequency from the M26 × M210 (hot spot) cross to that from the M375 × M210 (basal recombination) cross.
Cells that were homozygous mutants for mts1 (Table 2, line 2), homozygous mutants for mts2 (Table 2, line 3), or homozygous mutants for both mts1 and mts2 (Table 2, line 5) produced basal recombination frequencies indistinguishable from that of wild-type cells, indicating that the Mts1 and Mts2 proteins are not required for basal recombination. In contrast, the hot spot recombination frequencies in the mts1 mutants, mts2 mutants, and mts1 mts2 double mutants fell to the frequency of basal recombinants, demonstrating a need for Mts1, Mts2, and M26 in hot spot activation. Because a heterodimer of Mts1 and Mts2 is required for high-affinity binding to the M26 site (45), we conclude that the Mts1-Mts2-M26 complex is an essential component of hot spot function, as previously reported (20).
The basal recombination frequency from cells that were homozygous mutants for spc1 (Table 2, line 6) was similar to that from crosses of wild-type (spc1+) strains (Table 2, line 1). We conclude that the spc1 mutants are recombination proficient and have an intact (wild-type) basal recombination machinery. However, the spc1− deletion mutation abolished hot spot activity (Table 2, line 6), demonstrating that the Spc1 MAP kinase is essential for hot spot activation. To determine whether this requirement is related to the requirement for the Mts proteins, we determined recombination frequencies from crosses of the mts1 spc1 and mts2 spc1 double mutants (Table 2, lines 6 and 7). In each case, hot spot activity was abolished and the recombination frequencies of the double mutants were indistinguishable from those of the single mutants. The lack of phenotypic additivity in the double mutants indicates that the Spc1 kinase acts in a single, linear pathway with the Mts1-Mts2 heterodimer during hot spot activation.
Regulation of the activities of the Mts1 and Mts2 proteins occurs principally by posttranslational mechanisms.
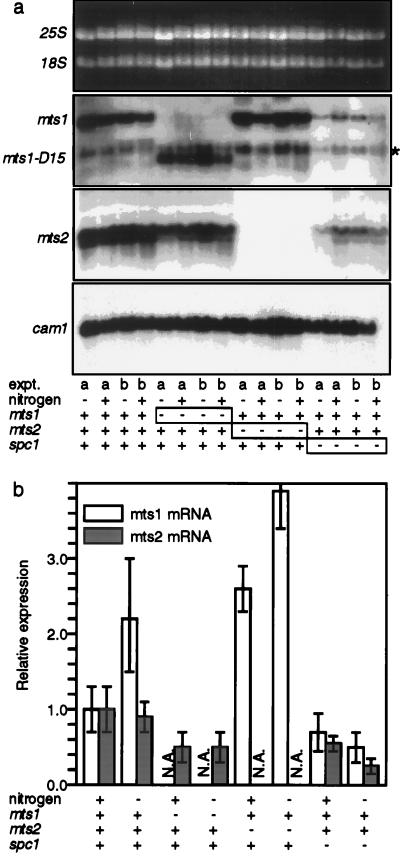
Because the Mts1-Mts2 heterodimer has major roles in Spc1-dependent mating, meiosis, and hot spot activation induced by nitrogen starvation (Fig. 3; Table 2), we investigated whether the Spc1 kinase regulates the expression levels of the Mts1 and Mts2 proteins. We initially determined the mRNA levels in cells grown in the presence of nitrogen or after 6 h of nitrogen starvation. The mts1 gene was induced about twofold by nitrogen starvation, whereas mts2 gene expression levels were unchanged (Fig. 4a). The expression levels of mts1 and mts2 were only two- to fivefold lower in spc1 deletion mutants (Fig. 4a). Similar two- to fourfold effects upon expression were observed in the mts1 and mts2 mutants (Fig. 4a). We conclude that the Spc1 kinase and the Mts1 and Mts2 proteins are not essential for mts1 and mts2 gene expression, although they may have a nominal role in modulating the relative expression levels.
FIG. 4.
Expression of mts1 and mts2 mRNA in wild-type and mutant cells. (a) Parallel experiments were conducted with two different strains (a and b) for each of the indicated genotypes. The cells were either wild type (+) or deletion mutants (−) for the indicated loci. Cells were grown in minimal medium plus nitrogen (+) or for an additional 6 h after a shift to nitrogen-free (−) medium. Panels show fractionated total cellular RNA stained with ethidium bromide and autoradiographs of the RNA probed with mts1, mts2, and the internal loading control cam1. The mts1-D15 strains express a truncated mts1 message. An asterisk marks the position of the cam1 hybridization signal that was inefficiently removed prior to hybridization with the mts1 probe. (b) Relative expression levels. Expression levels were determined by phosphorimage analysis and are normalized to the expression levels in wild-type (mts1+ mts2+ spc1+) cells. Data are means ± standard deviations from duplicate experiments. N.A., not available.
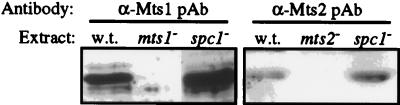
To directly gauge Mts1 and Mts2 protein expression levels, we raised polyclonal antibodies against the proteins and used Western blotting. As shown in Fig. 5, the Mts1 and Mts2 protein levels were not significantly altered by the spc1 deletion mutation. Under a variety of different culture conditions, the steady-state levels of the Mts1 and Mts2 proteins in the mutants were always within a few fold of their expression levels in wild-type cells (data not shown). The identical result of normal Mts1 protein levels in spc1 deletion mutants has recently been reported by another laboratory (6). We conclude that Spc1 does not indirectly regulate the biological functions of the Mts1-Mts2 heterodimer by affecting its expression level. Because the Spc1 kinase is essential for all known Mts1-Mts2-dependent functions (Fig. 2 and 3; Table 2) and Spc1 is known to directly phosphorylate Mts1 (39, 49), we infer that the Spc1 kinase directly regulates Mts1-Mts2 function by posttranslational modification.
FIG. 5.
Expression of Mts1 and Mts2 proteins in wild-type and mutant cells. Equal amounts of total cellular lysate were fractionated on SDS-polyacrylamide gels and subjected to Western blotting with polyclonal anti-Mts1 and anti-Mts2 antisera.
Spc1 MAP kinase is required for ade6-M26 DNA site occupancy by the Mts1-Mts2 protein in vivo.
Phosphorylation of the Mts1-Mts2 heterodimer by the Spc1 MAP kinase could regulate its DNA binding activity, its subcellular localization, or some transactivation function. We have previously shown that Mts1-Mts2 protein expressed in and purified from E. coli is capable of binding to the M26 site (20), suggesting that phosphorylation is not required for protein-DNA interaction in vitro. To confirm this finding, we highly purified Mts1-Mts2 protein from yeast (45), treated it with varying concentrations of each of three different phosphatases, and used a gel mobility shift assay (20, 45, 46) to gauge the DNA binding affinities. In each case the binding was equivalent to that of untreated protein (data not shown), indicating that Mts1-Mts2 heterodimer expressed in and purified from S. pombe does not need to be phosphorylated in order to bind to the M26 site in vitro.
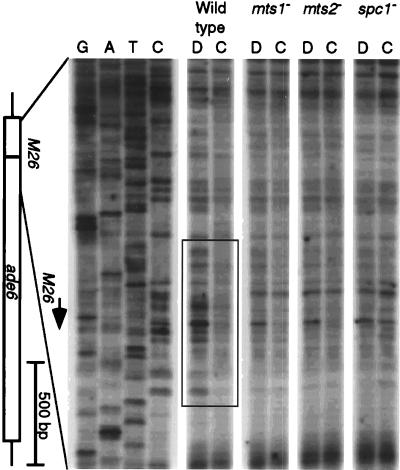
We therefore used DMS footprinting to determine whether the ade6-M26 site was occupied by the Mts1-Mts2 protein in vivo. In wild-type cells, the DNA surrounding the M26 site was partially protected from methylation, relative to the naked DNA control (Fig. 6). The footprint was centered on the M26 site and covered approximately 35 bp, and the degree of protection suggested ≥50% site occupancy. The footprint was dependent upon both the Mts1 and Mts2 proteins, demonstrating that the M26 site was occupied by the Mts1-Mts2 heterodimer. Site occupancy was also dependent upon the presence of the Spc1 MAP kinase. We conclude that the Spc1 kinase regulates effector functions at least in part by modulating the in vivo occupancy of the M26 DNA site by the Mts1-Mts2 protein.
FIG. 6.
In vivo DMS footprinting in wild-type and mutant cells. Cells were in log phase (107 cells/ml) in NBL minimal medium at the time of DMS treatment. Methylation of DNA within living cells (C, chromatin) and of purified DNA (D, DNA) was revealed by thermocycle extension of radiolabeled primer. Thermocycle dideoxynucleotide triphosphate DNA sequencing of untreated, purified genomic DNA was conducted in parallel as a control for priming specificity and to locate the M26 site. The box indicates a DMS footprint of approximately 35 bp, at the M26 site, that is dependent upon the simultaneous presence of the Mts1, Mts2, and Spc1 proteins.
DISCUSSION
M26 is the first meiotic recombination hot spot with essential cis- and trans-acting components defined by mutagenesis (20, 37). Intriguingly, the individual Mts1 and Mts2 proteins are also transcription factors required for a variety of developmental decisions, including one leading to meiotic entry (17, 39, 42). This raised the issue of whether the proteins have multiple, distinct functions or whether recombination hot spot activation is coupled to the transcriptional-regulatory functions of the proteins. To further reveal the mechanisms of recombination hot spot activation, we posed the following three questions. (i) Are the protein requirements for various developmental decisions the same as or different than those for hot spot activation (i.e., Mts1-Mts2 heterodimer)? (ii) Is hot spot activation by the Mts proteins, like the developmental decisions, dependent upon signals from the MAP kinase Spc1? (iii) By what mechanism does Spc1 kinase regulate the biological functions of the Mts proteins?
Mts1 and Mts2 are key effectors of developmental decisions under regulation of the MAP kinase cascade.
The individual Mts proteins are presumptive transcription factors (6, 39, 42, 48, 49). However, the discrete DNA sites involved in Mts1- or Mts2-dependent transcriptional regulation of endogenous genes are not known. Nor have the dimerization partners involved in transcriptional regulation requiring either Mts1 or Mts2 been reported. Our results (Fig. 2 and 3) prove that a heterodimer of Mts1 and Mts2 has no significant role in osmoregulation, spore quiescence, or spore viability. These functions require Mts1, presumably acting as a homodimer or as a heterodimer with some bZIP protein other than Mts2. On the other hand, the Mts1-Mts2 heterodimer is almost certainly the moiety involved in regulating mating and meiosis functions (Fig. 3). Presumably, these developmental decisions are achieved by the Mts1-Mts2 heterodimer binding to M26 sites (or closely related sites) at specific genes to regulate their expression.
The differential use of Mts1 or Mts2 in several disparate functions provides a good example of the economy of nature and can also explain why the proteins are constitutively expressed, but the mutants exhibit phenotypes only during adverse conditions, such as nutritional starvation leading to sexual development. The Mts1 and Mts2 proteins are multifunctional developmental switches that help to activate one of several specific developmental programs in response to intracellular and extracellular conditions. In times of crisis, when the cells may be unable to efficiently synthesize Mts1 and Mts2 de novo, signal transduction impinging upon Mts1, Mts2, or both triggers the appropriate developmental response. A second possible explanation for the constitutive expression is that the Mts1 protein may also function as a transcriptional repressor during periods of normal mitotic growth (6; see also Fig. 6 in reference 42). The failure of spc1 and mts1 mutants to maintain spore quiescence and viability (Fig. 3) is consistent with roles for the Mts1 protein in transcriptional repression, although these phenotypes could conceivably be an indirect consequence of a transcriptional activation defect.
Phosphorylation of Mts1 by Spc1 is implicated in stress responses, sexual development, and M26 recombination hot spot activation (Fig. 2 and 3; Table 2) (6, 39, 49). Our results and the proven phosphorylation of Mts1 by Spc1 (39, 49) suggest strongly that this is a direct interaction. Additional signals from the cAMP-dependent kinase pathway and the Pat1 kinase (a repressor of meiotic entry) also converge upon Mts1 (39, 42, 49) and Mts2 (48). Thus, differential input from various signal transduction pathways may provide additional mechanisms whereby S. pombe can use the same set of factors for the largest number of functions. The biochemical nature of these additional signals and their molecular mechanisms in regulating effector functions remain to be determined.
Spc1 MAP kinase modulates the in vivo DNA binding activity of the Mts1-Mts2 heterodimer to regulate effector functions.
We have shown directly that ade6-M26 site occupancy in vivo is dependent upon Spc1 MAP kinase (Fig. 6), which phosphorylates Mts1 (6, 39, 49). While most reported roles for phosphorylation are in the effector functions of transcription factors, phosphorylation can directly regulate the DNA binding activities of some bZIP proteins. For example, dephosphorylation of ATF-1 and CREB reduces in vitro binding of protein homodimers and heterodimer to the ATF/CREB site, and the DNA binding activity is restored upon phosphorylation of ATF-1 (19, 25). Recently it was found that direct, intramolecular interactions between the DNA binding domain and a portion of the activation domain can render some bZIP proteins inactive (1, 21, 50). Thus, the transactivation domain and the DNA binding domain antagonize each other’s function, which provides a coordinate regulation with several potential benefits (e.g., see references 21 and 50 and references therein). Two models have been suggested. First, binding of coactivator proteins could disrupt the intramolecular interaction between bZIP and the activation domain, thereby making the DNA binding and transactivation domains available. Second, phosphorylation could directly disrupt intramolecular interaction. However, we found that Mts1-Mts2 protein purified from E. coli is capable of DNA binding (20), protein in extracts of yeast cultured under various nutritional conditions (including nitrogen starvation) has roughly equivalent DNA binding activities (20, 45), and treatment of highly purified yeast protein with each of three different phosphatases does not significantly alter DNA binding in vitro (data not shown). Together, the data suggest that differential phosphorylation of the Mts1-Mts2 heterodimer by the Spc1 MAP kinase regulates in vivo site occupancy by allowing sequestered Mts1-Mts2 protein to gain access to the DNA.
Meiotic recombination hot spot activation as a consequence of conformational changes imparted by the Mts1-Mts2 protein and additional meiotic factors.
Our finding that the Mts1-Mts2 heterodimer is a key regulator of mating and meiosis (Fig. 3) suggests that the heterodimer’s activity as a meiotic transcription factor may be responsible for recombination hot spot activation. For example, increasing the transcription of ade6, driven by a heterologous promoter, increases recombination at the locus (13). Perhaps the M26 site functions as a transcriptional enhancer in ade6, and this indirectly promotes recombination. However, this does not seem to be the case. M26 still functions as a hot spot when basal recombination levels are elevated by increased transcription (13). Furthermore, transcription levels of ade6 are similar in the presence or absence of the M26 site (13, 20) and do not change significantly in mts mutants (20), so hotspot activation is not apparently due to increased ade6 transcription.
The connection between the Mts1-Mts2 transcription factor and M26 hot spot activation is probably via conformational changes in meiotic chromatin structure, which are thought to serve as preferential loading sites for meiotic enzymes that initiate recombination by introducing double-strand DNA breaks (3, 18, 27, 29, 52). Meiotically induced M26 site-dependent increases in the accessibility of DNA within chromatin are observed at the ade6-M26 site itself, as well as at a site in the promoter region (26). This chromatin remodeling is dependent upon the Mts1-Mts2 protein (29a). Thus, the Mts1-Mts2 transcription factor remodels local meiotic chromatin structure, probably with the aid of additional meiotic proteins, during hot spot activation.
Our current view is that M26 acts as an enhancer of meiotic recombination. As for transcriptional enhancers, the M26 site functions in a position- and orientation-independent fashion; each of eight M26 sites created by mutagenesis of one or a few base pairs has hot spot activity (11, 14, 31, 41). Furthermore, while basal recombination does not require the Mts1-Mts2 heterodimer (Table 2) (20), hot spot activation does require components of the basal recombination machinery. There are at least 18 genes known to be required for meiotic recombination in S. pombe (8, 10, 32, 34, 43). Mutations of those genes reduce both basal (ade6-M375) and hot spot (ade6-M26) recombination. While the majority of the hyporecombination mutants still exhibit hot spot activity, six of the mutants exhibit no hot spot activity. One of these six genes, rec12, is homologous to SPO11 of Saccharomyces cerevisiae, which encodes a topoisomerase II-like protein thought to catalyze double-strand DNA break formation (3, 18) and to initiate most meiotic recombination (22, 30, 44). We suggest that the Mts1-Mts2 heterodimer increases the local concentration or specific activity of one or more of those six meiotic recombination proteins, including Rec12. This may be achieved by direct protein-protein interactions, or recruitment may be via M26 site-dependent, Mts1-Mts2 protein-dependent, meiotically induced conformational changes in chromatin at the hot spot (26, 29a).
Summary.
The Mts proteins are key effectors of at least two distinct classes of developmental decisions regulated by the MAP kinase cascade. The Spc1 MAP kinase regulates in vivo ade6-M26 DNA site occupancy by the Mts1-Mts2 heterodimer, which is a meiotic transcription factor that activates a meiotic recombination hot spot. This provides a mechanistic basis for the observation that most recombination hot spots are located in promoter regions (52). It also raises the question of whether there are additional connections between the meiotic transcription machinery and that of meiotic recombination or whether hot spot activation is simply a consequence of altered chromatin structure caused by DNA binding and/or transcriptional transactivation.
ACKNOWLEDGMENTS
We thank Charlie Albright, Jeff Flick, Wallace Sharif, and Ron Wisdom for helpful discussions and Steve Lindsey and Calley Hardin for laboratory assistance.
This work was supported by a grant from the National Institutes of Health (GM54671). M.D.K. was supported in part by a training grant from the National Institutes of Health (CA09582), and W.P.W. was a Leukemia Society of America Special Fellow (3021-94) for a portion of this research.
REFERENCES
- 1.Abdel-Hafiz H A, Heasley L E, Kyriakis J M, Avruch J, Kroll D J, Johnson G L, Hoeffler J P. Activating transcription factor-2 DNA-binding activity is stimulated by phosphorylation catalyzed by p42 and p54 microtubule-associated protein kinases. Mol Endocrinol. 1992;6:2079–2089. doi: 10.1210/mend.6.12.1337144. [DOI] [PubMed] [Google Scholar]
- 2.Beach D, Rodgers L, Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- 3.Bergerat A, de Massy B, Gadelle D, Varoutas P-C, Nicolas A, Forterre P. An atypical topoisomerase II from archaea with implication for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 4.Brewer A C, Marsh P J, Patient R K. A simplified method for in vivo footprinting using DMS. Nucleic Acids Res. 1990;18:5574. doi: 10.1093/nar/18.18.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 6.Degols G, Russell P. Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:3356–3363. doi: 10.1128/mcb.17.6.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Densmore L, Payne W E, Fitzgerald-Hayes M. In vivo genomic footprint of a yeast centromere. Mol Cell Biol. 1991;11:154–165. doi: 10.1128/mcb.11.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeVeaux L C, Hoagland N A, Smith G R. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics. 1992;130:251–262. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Q Q, Xu F, White M A, Petes T D. Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics. 1997;145:661–670. doi: 10.1093/genetics/145.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox M E, Smith G R. Control of meiotic recombination in Schizosaccharomyces pombe. Prog Nucleic Acid Res Mol Biol. 1998;61:325–378. doi: 10.1016/s0079-6603(08)60831-4. [DOI] [PubMed] [Google Scholar]
- 11.Fox M E, Virgin J B, Metzger J, Smith G R. Position- and orientation-independent activity of the Schizosaccharomyces pombe meiotic recombination hot spot M26. Proc Natl Acad Sci USA. 1997;94:7446–7451. doi: 10.1073/pnas.94.14.7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm C, Bahler J, Kohli J. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics. 1994;136:41–51. doi: 10.1093/genetics/136.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimm C, Schaer P, Munz P, Kohli J. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1991;11:289–298. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutz H. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics. 1971;69:331–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King R C, editor. Handbook of genetics. Vol. 1. New York, N.Y: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 17.Kanoh J, Watanabe Y, Ohsugi M, Iino Y, Yamamoto M. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells. 1996;1:391–408. doi: 10.1046/j.1365-2443.1996.d01-247.x. [DOI] [PubMed] [Google Scholar]
- 18.Keeney S, Giroux C N, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi M, Shimomura A, Hagiwara M, Kawakami K. Phosphorylation of ATF-1 enhances its DNA binding and transcription of the Na,K-ATPase alpha 1 subunit gene promoter. Nucleic Acids Res. 1997;25:877–882. doi: 10.1093/nar/25.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kon N, Krawchuk M D, Warren B G, Smith G R, Wahls W P. Transcription factor Mts1/Mts2 (Atf1/Per1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in S. pombe. Proc Natl Acad Sci USA. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X Y, Green M R. Intramolecular inhibition of activating transcription factor-2 function by its DNA-binding domain. Genes Dev. 1996;10:517–527. doi: 10.1101/gad.10.5.517. [DOI] [PubMed] [Google Scholar]
- 22.Lichten M, Goldman A S H. Meiotic recombination hotspots. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 23.Lin Y, Larson K L, Dorer R, Smith G R. Meiotically induced rec7 and rec8 genes of Schizosaccharomyces pombe. Genetics. 1992;132:75–85. doi: 10.1093/genetics/132.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y, Smith G R. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics. 1994;136:769–779. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merino A, Buckbinder L, Mermelstein F H, Reinberg D. Phosphorylation of cellular proteins regulates their binding to the cAMP response element. J Biol Chem. 1989;264:21266–21276. [PubMed] [Google Scholar]
- 26.Mizuno K, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- 27.Nicolas A. Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc Natl Acad Sci USA. 1998;95:87–89. doi: 10.1073/pnas.95.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolas A, Treco D, Schultes N P, Szostak J W. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 29.Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Ohta, K., and W. P. Wahls. Unpublished observations.
- 30.Osman F, Subramani S. Double-strand break-induced recombination in eukaryotes. Prog Nucleic Acid Res Mol Biol. 1998;58:263–299. doi: 10.1016/s0079-6603(08)60039-2. [DOI] [PubMed] [Google Scholar]
- 31.Ponticelli A S, Sena E P, Smith G R. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics. 1988;119:491–497. doi: 10.1093/genetics/119.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponticelli A S, Smith G R. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schar P, Kohli J. Preferential strand transfer and hybrid DNA formation at the recombination hotspot ade6-M26 of Schizosaccharomyces pombe. EMBO J. 1994;13:5212–5219. doi: 10.1002/j.1460-2075.1994.tb06852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt H, Kapitza P, Gutz H. Switching genes in Schizosaccharomyces pombe: their influence on cell viability and recombination. Curr Genet. 1987;11:303–308. [Google Scholar]
- 35.Schroeder S C, Wang C K, Weil P A. Identification of the cis-acting DNA sequence elements regulating the transcription of the Saccharomyces cerevisiae gene encoding TBP, the TATA box binding protein. J Biol Chem. 1994;269:28335–28346. [PubMed] [Google Scholar]
- 36.Schuchert P, Kohli J. The ade6-M26 mutation of Schizosaccharomyces pombe increases the frequency of crossing over. Genetics. 1988;119:507–515. doi: 10.1093/genetics/119.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuchert P, Langsford M, Kaslin E, Kohli J. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 1991;10:2157–2163. doi: 10.1002/j.1460-2075.1991.tb07750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- 39.Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2228. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 40.Sun H, Treco D, Schultes N P, Szostak J W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 41.Szankasi P, Heyer W D, Schuchert P, Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination hot spot allele ade6-M26. J Mol Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- 42.Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tavassoli M, Shayeghi M, Nasim A, Watts F Z. Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 1995;23:383–388. doi: 10.1093/nar/23.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahls W P. Meiotic recombination hotspots: shaping the genome and insights into hypervariable minisatellite DNA change. Curr Top Dev Biol. 1998;37:37–75. doi: 10.1016/s0070-2153(08)60171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahls W P, Smith G R. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 1994;8:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- 46.Wahls W P, Swenson G, Moore P D. Two hypervariable minisatellite DNA binding proteins. Nucleic Acids Res. 1991;19:3269–3274. doi: 10.1093/nar/19.12.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe Y, Iino Y, Furuhata K, Shimoda C, Yamamoto M. The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 1988;7:761–767. doi: 10.1002/j.1460-2075.1988.tb02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe Y, Yamamoto M. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol Cell Biol. 1996;16:704–711. doi: 10.1128/mcb.16.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkinson M G, Samuels M, Takeda T, Toone W M, Shieh J-C, Toda T, Millar J B A, Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- 50.Williams S C, Baer M, Dillner A J, Johnson P F. CRP2 (C/EBP beta) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu T C, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu T C, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 53.Yoshino M, Sagai T, Lindahl K F, Toyoda Y, Shiroishi T, Moriwaki K. No dosage effect of recombinational hotspots in the mouse major histocompatibility complex. Immunogenetics. 1994;39:381–389. doi: 10.1007/BF00176154. [DOI] [PubMed] [Google Scholar]
- 54.Zahn-Zabal M, Lehmann E, Kohli J. Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim. Genetics. 1995;140:469–478. doi: 10.1093/genetics/140.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]