Abstract
A previous analysis of naturally occurring defective interfering (DI) RNA genomes of the prototypic paramyxovirus simian virus 5 (SV5) indicated that 113 bases at the 3′ terminus of the antigenome were sufficient to direct RNA encapsidation and replication. A nucleotide sequence alignment of the antigenomic 3′-terminal 113 bases of members of the Rubulavirus genus of the Paramyxoviridae family identified two regions of sequence identity: bases 1 to 19 at the 3′ terminus (conserved region I [CRI]) and a more distal region consisting of antigenome bases 73 to 90 (CRII) that was contained within the 3′ coding region of the L protein gene. To determine whether these regions of the antigenome were essential for SV5 RNA replication, a reverse genetics system was used to analyze the replication of copyback DI RNA analogs that contained a foreign gene (GL, encoding green fluorescence protein) flanked by 113 5′-terminal bases and various amounts of SV5 3′-terminal antigenomic sequences. Results from a deletion analysis showed that efficient encapsidation and replication of SV5-GL DI RNA analogs occurred when the 90 3′-terminal bases of the SV5 antigenomic RNA were retained, but replication was reduced ∼5- to 14-fold in the case of truncated antigenomes that lacked the 3′-end CRII sequences. A chimeric copyback DI RNA containing the 3′-terminal 98 bases including the CRI and CRII sequences from the human parainfluenza virus type 2 (HPIV2) antigenome in place of the corresponding SV5 sequences was efficiently replicated by SV5 cDNA-derived components. However, replication was reduced ∼20-fold for a truncated SV5-HPIV2 chimeric RNA that lacked the HPIV2 CRII sequences between antigenome bases 72 and 90. Progressive deletions of 6 to 18 bases in the region located between the SV5 antigenomic CRI and CRII segments (3′-end nucleotides 21 to 38) resulted in a ∼25-fold decrease in SV5-GL RNA synthesis. Surprisingly, replication was restored to wild-type levels when these length alterations between CRI and CRII were corrected by replacing the deleted bases with nonviral sequences. Together, these data suggest that a functional SV5 antigenomic promoter requires proper spacing between an essential internal region and the 3′ terminus. A model is presented for the structure of the 3′ end of the SV5 antigenome which proposes that positioning of CRI and CRII along the same face of the helical nucleocapsid is an essential feature of a functional antigenomic promoter.
The paramyxoviruses are a diverse family of nonsegmented negative-sense RNA viruses that includes Sendai virus (SeV), measles virus (MeV), and respiratory syncytial virus (RSV). Simian virus 5 (SV5) is a prototype of the Rubulavirus genus of the Paramyxoviridae family, which includes mumps virus (MuV), SV41, and human parainfluenza virus type 2 (HPIV2). The ∼15-kb paramyxovirus genomic and antigenomic RNAs are tightly bound by the viral nucleocapsid protein (NP) to form nucleocapsid (NC) structures, and it is these ribonucleoprotein complexes that serve as the only templates for viral RNA synthesis. The viral phosphoprotein (P) and the large protein (L) together form the viral RNA-dependent RNA polymerase which is responsible for both transcription to produce mRNAs and replication to produce negative-sense genomes and positive-sense antigenomes (18, 20, 24). For the nonsegmented negative-sense RNA viruses, RNA synthesis initiates from promoters located at the 3′ ends of both the genomic and antigenomic RNAs, but understanding of critical features of the viral template that control the level and particular type of RNA synthesized by the viral polymerase is incomplete.
Two factors that can influence the level of RNA synthesized from the paramyxovirus antigenomic promoter have been described: the total number of nucleotides in the viral RNA and the primary nucleotide sequence at the termini of the viral RNA. The length of a paramyxovirus RNA can be a major factor that determines the level of RNA replication, with genome replication being most efficient when the total number of nucleotides is an even multiple of six (6). The rule of six is thought to reflect critical interactions between the viral polymerase and 3′-terminal promoter sequences, with initiation of RNA replication being most efficient when the last six bases of the 3′-end promoter are bound completely by a single NP (6, 35). The degree to which replication of a particular paramyxovirus genome adheres to the rule of six differs among viruses. For SeV, the rule of six is an apparent strict requisite (6), while RSV shows no particular replicative advantage for genomes having any of the integer lengths tested (41). SV5 is unique in this regard, since the stringency of the rule of six for SV5 defective interfering (DI) RNA replication is intermediate between that found previously for SeV and RSV (26).
A second major factor in paramyxovirus RNA replication is the primary sequence at the 3′ and 5′ termini of the viral RNA, as shown previously by mutagenesis studies (44) and by the structure of paramyxovirus DI RNAs. These subgenomic RNAs contain various segments from the standard nondefective viral genome that are characteristic of an individual DI RNA. However, all naturally occurring DI genomes that are competent for replication retain either the genomic 3′ and 5′ ends in the case of internal deletion of DI RNAs or the genomic 5′ end and its complement for copyback DI RNAs (reviewed in reference 36). The nucleotide sequence in the leader (le) RNA at the 3′ terminus of the genome and in the antitrailer (tr′) RNA at the 3′ terminus of the antigenome have been proposed to be primary determinants of the efficiency of paramyxovirus replication to produce antigenomes and genomes, respectively (7, 44). The levels of RNA synthesized from these two viral promoters are reported to differ significantly (7, 22, 44, 46), but the basis for these differences in promoter activity and important features of the 3′-terminal regions is not completely understood.
The work reported here provides evidence for a third factor that can dramatically influence the level of RNA synthesized from the paramyxovirus antigenomic promoter. We have used a reverse genetics system to analyze features of the 3′-terminal region of the SV5 antigenome that are important for directing RNA replication. The results indicate that a functional SV5 antigenomic promoter requires two discontinuous regions (CRI and CRII) that are located in the 3′-terminal 90 bases of the antigenomic RNA. Most importantly, the relative spacing of these two cis-acting regions is a critical factor in determining the level of SV5 RNA replication. Therefore, the overall number of nucleotides in a viral RNA, the primary nucleotide sequence at the termini, and proper spacing of promoter elements are three features of the viral RNA template that can significantly influence paramyxovirus RNA synthesis. We present a model for the structure of the 3′ end of the SV5 antigenome which proposes that positioning of CRI and CRII along the same face of the helical nucleocapsid is an essential feature of a functional antigenomic promoter.
MATERIALS AND METHODS
Cells and viruses.
Monolayer cultures of A549 cells were grown and infected with virus as previously described (26). Vaccinia virus vTF7.3 (14) was grown and titered in CV1 cells.
Nomenclature.
DNA plasmids encoded antigenomic RNAs flanked on the 3′ and 5′ ends by the hepatitis delta virus (HDV) ribozyme and the T7 RNA polymerase (T7 pol) promoter, respectively (26, 32, 34). Plasmids encoding SV5 DI RNA analogs are named to denote three portions of the antigenome: the 5′ end (proximal to the T7 promoter), the internal segment flanked by complementary termini, and the 3′ end (proximal to HDV ribozyme). Thus, the 113-GL-59 antigenomic RNA contains 113 SV5-specific 5′-terminal nucleotides linked to the Green Lantern (GL; Gibco BRL) open reading frame followed by 59 SV5-specific 3′-terminal nucleotides. Likewise, the DI-SSP antigenomic RNA contains SV5 sequences at the 5′ end (S) and internal position (S), linked to HPIV2-specific 3′ antigenomic RNA sequences (P).
Construction of plasmids encoding the SV5 antigenomes.
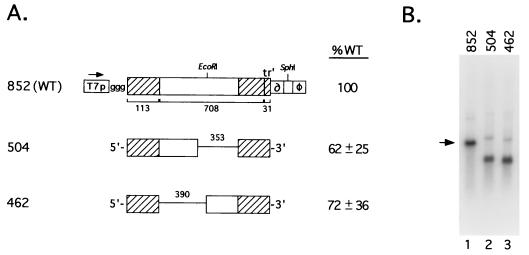
All viral genomes were designed to maintain an overall 6N length. PCR was carried out as described previously (30), using Pwo polymerase (Boehringer Mannheim, Indianapolis, Ind.) along with oligonucleotides listed in Table 1. The sequences of all PCR-derived DNA segments were confirmed by nucleotide sequence analysis (1). The cDNAs encoding copyback genomes DI852 and DI499+5 (DI504) have been described previously (26). To construct deletion mutant DI462, the 5′ antigenomic region of terminal complementarity in pDI852 was amplified in a PCR along with the 462 primer (Table 1) and a primer specific for the T7 promoter sequence in pDI852 (Fig. 1A). The resulting product was digested with EcoRI and cloned into the PvuII and EcoRI sites of pGem3term (a pGem3 plasmid previously modified to contain the T7 terminator sequence downstream from the SP6 promoter [26]. The EcoRI-SphI fragment encoding the 3′ antigenomic end of pDI852 (see Fig. 1 for schematic) was then inserted into the corresponding sites of the subclone to yield pDI462.
TABLE 1.
Oligonucleotide primers used
| Primer | Sequencea | Locationb | Restriction site(s)c |
|---|---|---|---|
| 462 | gcggatccaatattaggcctgaattcgcAATGAAGAAGAAATAGACCGCGGG | 6753–6773 | EcoRI |
| P113 | gcgggatccaggcctgaattcgAATATTAATGAAGAAATAGACCG | 6747–6769 | BamHI, StuI |
| P90 | gcaggcctCGGGATCGATCGATGGCGAGAAAATCT | 6770–6792 | StuI |
| P72 | gcaggcctAATCTAAACATATCAAGAATCAG | 6788–6810 | StuI |
| P59 | gcgggatccaggcctgaattcgCAAGAATCAGAATTAGTTTAAG | 6801–6822 | BamHI, StuI |
| P31 | gcgggatccaggcctgaattcgcgcgGAAGAGGATTAATCTTGGTT | 6829–6848 | StuI |
| HPIV2T7 | gcaggcctggtacctaatacgactcactatagggACCAAGGGGAAAATCAATATG | 6842–6862 | Asp718 |
| HPIV2ribo | gccatgccgacccACCAAGGGGAAAATCAATATG | 6842–6862 | |
| HPIV2del | gctctagatatcTTATGACAACAATGATTA | 6792–6809 | EcoRV |
| GL-TGA | gcgaattccgcggTCACTTGTACAGCTCGTCCATGCCATG | 5002–5027 | EcoRI |
| GL-ATG | gcgaattctctagaaggcctAGCTCACTAGTCGGCGGCCG | 4284–4303 | StuI |
| GL-1 | CCATTCACATCGCCATCCAGT | 4382–4362 | |
| Tr′Bam | gcggatccGAAAAAAGAAGAGGATTAATCT | 6822–6843 | BamHI |
| Tr′del6 | gcggatccAGAAGAGGATTAATCT | 6828–6843 | BamHI |
| Tr′ins6 | gcggatccgatatcGAAAAAAGAAGAGGATTA | 6822–6839 | BamHI |
| Tr′del12 | gcggatccGGATTAATCTTGGTTTTCCCCTTGGTGGGTCGGCATGG | 6834–6859 | BamHI |
| Tr′re12 | gcggatccatttaatctctcGGATTAATCTTGGTTTT | 6834–6850 | BamHI |
| Tr′ins12 | gcggatccgatatcatttaaGAAAAAAGAAGAGGATTAA | 6817–6840 | BamHI |
| Tr′del18 | gcggatccATCTTGGTTTTCCCCTTGGTgggtcggcatgg | 6840–6859 | BamHI |
| Tr′re18 | gcggatccatttaatctctcagtccgaTCTTGGTTTTCCCCTT | 6840–6856 | BamHI |
| Tr′re25 | atctctcagtccgtgaccaaTTTTCCCCTTGGTggg | 6847–6859 |
5′ to 3′; nonviral or non-GL bases are shown as lowercase letters.
Location on antigenomic viral RNA or pGreen Lantern-1 plasmid beginning with 5′ viral or GL-specific nucleotide in primer. Number designations for viral sequences are taken from the published SV5 L (31) and HPIV2 L (19) gene sequences.
Relevant restriction sites used in cloning procedures are listed and underlined in the oligonucleotide sequence.
FIG. 1.
Sequences located internal to the 113-nucleotide terminal complementary regions of DI852 are not required for replication. (A) Structures of SV5 DI genomes DI852, DI504, and DI462. The cDNAs encoding SV5 DI genomes are shown schematically as rectangles, with cross-hatched boxes and lines representing the complementary termini and internal deletions, respectively. The promoter for T7 pol (T7p), the self-cleaving HDV ribozyme (∂), and T7 terminator sequence (ø) are indicated for DI852 only but are included in all cDNAs encoding SV5 DI RNA analogs. Numbers that denote nucleotides composing the 5′ region of terminal complementarity (113 bases), the 3′ end of the L gene (708 bases), the 31-base tr′, and internal deletions are indicated. The horizontal arrow indicates the direction of transcription from the T7 pol promoter to produce a positive-sense DI RNA antigenome containing three nonviral G residues (ggg) at the 5′ end. Replication levels are expressed as percentages (±SD) of that determined for the WT DI852 assayed in parallel. (B) Replication of DI852, DI504, and DI462 genomes. A549 cells were infected with vTF7.3 and then cotransfected with plasmids encoding the L (2.0 μg), P (0.2 μg), and NP (2.0 μg) proteins along with an individual DI antigenome (1.0 μg). Total intracellular RNA was harvested and analyzed by Northern blotting with a 32P-labeled positive-sense riboprobe corresponding to sequences located internal to the 113 bases of terminal complementarity.
Copyback DI RNA analogs containing a foreign gene were constructed by linking DNA fragments from independent PCRs such that a minus-sense copy of the GL open reading frame was inserted between various amounts of SV5-specific 5′ and 3′ termini. For the purposes of nomenclature, the primary T7 transcripts generated from this series of constructs are designated antigenomes and have the SV5 termini in the antigenomic sense flanking the GL segment. To generate cDNAs encoding DI genomes with various amounts of the SV5 termini, we used a three-step cloning procedure that involved the use of primers (P113, P90, P72, P59, and P31 [Table 1]) capable of annealing to opposite strands of both termini of pDI852. However, specific amplification of a single terminus occurs when these primers are used in conjunction with either the SP6 or T7 primer. First, the P113 and P59 primers were used with the T7 primer on the pDI852 template to generate DNA products encoding the antigenomic 113 or 59 SV5 5′-terminal nucleotides. The PCR products resulting from amplification with the T7-P113 and T7-P59 primer combinations were digested with BamHI and cloned into the PvuII and BamHI sites of pGem3term to yield pGem3-113 and pGem3-59. In the second step, the GL open reading frame contained in the pGreen Lantern-1 DNA template (Gibco BRL) was amplified in a PCR with oligonucleotides GL-ATG and GL-TGA. The resulting GL-specific product was cloned into the EcoRI and StuI sites of pGem3-113 and pGem3-59 to give pGem3-113-GL and pGem3-59-GL, respectively. In the third step, which generated the 3′ antigenomic termini of the new constructs, plasmid pDI852 was used as the template in a PCR with primer P113, P90, P72, P59, or P31 and a primer specific for the SP6 promoter, which is located between the SphI site and T7 terminator sequence of pDI852 (Fig. 1A). The StuI-SphI fragments resulting from PCRs using the SP6-P113, -P90, -P72, -P59, and -P31 primer combinations were then ligated into the corresponding sites of pGem3-113-GL to generate the full-length constructs pDI113-GL-113, -90, -72, -59, and -31. Similarly, the StuI-SphI fragment from the P113-SP6 PCR was ligated into the corresponding sites of pGem3-59-GL to produce pDI59-GL-113.
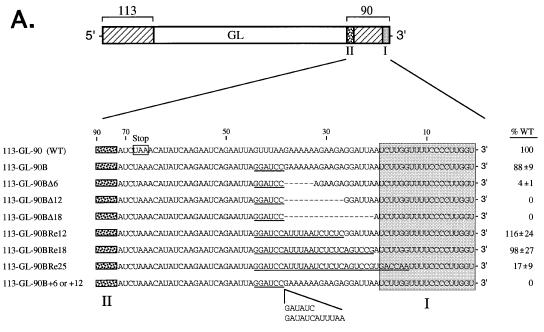
Nucleotide replacements, insertions, or deletions were engineered into SV5 3′ antigenomic segments by using oligonucleotide-directed mutagenesis as previously described (30). Plasmid pDI852 had been previously modified by using the Tr′Bam and SP6 primers to contain a BamHI site (5′-GGATCC) in place of bases 39 to 44 from the 3′ end of the antigenomic RNA. pDI852 containing the BamHI site was used as the template in a PCR with the SP6 and P90 primers. The products were digested with StuI and SphI and cloned into the corresponding sites of pDI113-GL-90 to generate pDI113-GL-90B (see Fig. 7A). pDI113-GL-90B was used in the generation of the length-altered and nucleotide replacement mutants.
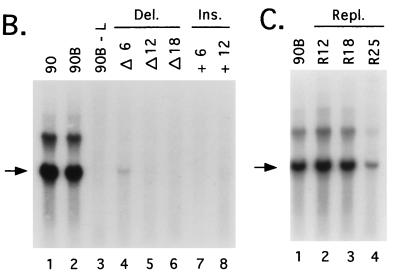
FIG. 7.
Alteration in the spacing between CRII and the 3′ terminus results in SV5 DI RNA analogs that are defective for RNA replication. (A) Sequences of the 3′-terminal regions of SV5 WT and mutant antigenomic RNAs. The structure of the 113-GL-90 DI RNA analog is shown schematically, with the locations of CRI and CRII sequences indicated by shaded and speckled boxes, respectively. The nucleotide sequence of the 3′-terminal 71 bases of the SV5 antigenome is listed below the diagram, with numbers indicating distance from the 3′ terminus. The L protein translational stop codon is boxed. Dashes and underlines in the altered 3′ end sequences denote deletions that created the Δ6, Δ12, and Δ18 antigenomes and nucleotide replacements that created the 113-GL-90B, -BRe12, -BRe18, and -BRe25 antigenomes, respectively. The locations and sequences of 6 or 12 nonviral bases inserted to create the -B+6 and -B+12 antigenomes are shown at the bottom. Relative replication levels are expressed as percentages (±SD) of that determined for the 113-GL-90 (WT) antigenome analyzed in parallel. (B) Replication of SV5-GL DI RNA analogs containing alterations in the length and sequence between CRI and CRII. A549 cells infected with vTF7.3 were transfected with L, P, and NP plasmids along with DNA encoding one of the indicated SV5-GL DI RNA analogs. Total intracellular RNA was harvested and analyzed by Northern blotting with a negative-sense GL-specific riboprobe. Lane 90B-L represents a sample from cells in which the L plasmid was replaced with pGem control DNA. The arrow indicates the position of full-length replication products. Del., deletion mutants; Ins., insertion mutants. (C) Replication of SV5-GL DI RNA analogs containing a replacement of sequences between CRI and CRII. Altered SV5-GL antigenomes containing replacements of bases 14 to 43 were analyzed in the DI replication assay as described above. Repl., replacement mutants.
The pDI113-GL-90BΔ6, -90BΔ12, -90BΔ18, -90BRe12, -90BRe18, -90B+6, or -90B+12 antigenome contained a deletion of 6, 12, or 18 nucleotides (antigenomic bases 33 to 38, 27 to 38, or 21 to 38), a replacement of 12 or 18 nucleotides (antigenomic bases 27 to 38 or 21 to 38), or an insertion of 6 or 12 nucleotides between antigenomic bases 38 and 39, respectively. These altered SV5 antigenomic 3′ segments were prepared by using pDI113-GL-90B as the template in a PCR along with the SP6 primer and primer Tr′del6, Tr′del12, Tr′del18, Tr′re12, Tr′re18, Tr′ins6, or Tr′ins12. The products were digested with BamHI and SphI and cloned into the corresponding sites of pDI113-GL-90B to generate the full-length constructs. The pDI113-GL-90Re25 construct contained a replacement of 25 nucleotides at antigenomic bases 14 to 38. The Tr′re25 primer was used with the SP6 primer in a PCR on template pDI113-GL-90B, and this PCR product was used as a megaprimer (30) in a second PCR for five cycles along with primer GL-1 on template pDI113-GL-90BRe18 (described above). These DNAs were then amplified in a third PCR with the SP6 and GL-1 primers. The resulting products were digested with StuI and SphI and cloned into the corresponding sites of pDI113-GL-90.
To construct chimeric SV5-HPIV2 copyback DI antigenomes, a BamHI fragment encoding the 3′ end of the HPIV2 antigenome (L protein base 5929 to the 3′ terminus [19]) was excised from pBSIISK-HPIV2Lpro (19) and subcloned into the BamHI site of pGem3 to yield pG3-P2L. To construct pDI-PSS, a new T7 pol promoter was fused to the HPIV2 5′ terminus in a PCR using pG3-P2L as the template along with the HPIV2T7 and T7 primers. The PCR product was digested with Asp718 and EcoRV and ligated into the corresponding sites of pDI852 before linking with the SspI to SphI fragment encoding the SV5 3′-terminal antigenomic sequences and HDV ribozyme described previously (26). Plasmid pDI-PSS contained (5′ to 3′) the T7 pol promoter, HPIV2 5′ genomic sequence from positions 1 to 98, a three-base linker, SV5 L gene sequences from bases 6121 to 6859 (31), including the tr′, and the self-cleaving HDV ribozyme. To construct pDI-SSP, pDI852 was used as the template in a PCR along with HPIV2ribo and SP6 primers. The PCR product then served as a megaprimer (30) along with the T7 primer in a PCR using pG3-P2L as the template. The product DNA was digested with EcoRV and SphI and ligated into the corresponding sites of pDI852, before linking with the SspI-to-EcoRV fragment that encodes the SV5 5′-terminal sequences from the wild-type (WT) DI852 genome (26). Plasmid pDI-SSP contained (5′ to 3′) the T7 polymerase promoter, SV5 DI852-specific sequences up to base 6752 of the L protein gene (26), the 98 3′-terminal bases from the HPIV2 antigenomic RNA including the tr′, and the self-cleaving HDV ribozyme. To generate a chimeric DI RNA in which HPIV2-specific CRII had been deleted, pDI-SSP was used as template in a PCR along with the HPIV2del and SP6 primers. The DNA fragment was digested with EcoRV and SphI and reconstructed into pDI852 as described above to yield pDI-SSPt. This plasmid differs from the parental pDI-SSP by containing a three-base linker separating the 3′-terminal 71 bases of the HPIV2 antigenomic RNA from SV5 sequences.
Analysis of in vivo DI RNA replication from cDNA-derived components.
A549 cells in 3.5-cm-diameter dishes were infected (multiplicity of infection of ∼5) for 1 h with vTF7.3 (14) and then cotransfected with DNA encoding the SV5 antigenomes (1.0 μg) along with pGem3-L (2.0 μg), pGem2-P (0.2 μg), and pUC19-NP (2.0 μg) as described previously (26), using a cationic liposome reagent (40). pGem control plasmid was used to normalize for the amount of transfected DNA. Total intracellular RNA (∼20 μg) was harvested at ∼40 to 48 h posttransfection and analyzed by Northern blot analysis as previously described (26).
A positive-sense 32P-labeled riboprobe corresponding to bases 6121 to 6752 of the SV5 L protein gene was generated from plasmid pGEM5-SspI for use in analyzing replication of DI852-derived antigenomes (26). For analyzing replication of DI RNAs carrying the GL sequence, the DNA fragment derived from a PCR with primers GL-ATG and GL-TGA and the pGreen Lantern-1 template described above was digested with EcoRI and StuI and cloned into the corresponding sites of pGem2 (Promega, Madison, Wis.) to generate pGem2-GL. A 219-base riboprobe of antisense polarity (which is of the same polarity as the GL sequence contained in the T7 primary transcripts of the SV5-GL RNAs) was generated from BamHI-linearized pGem2-GL by SP6 RNA polymerase in the presence of [32P]CTP. For detecting primary T7 pol-derived RNA transcripts, a positive-sense 782-base 32P-labeled riboprobe was generated by using T7 pol from the same template that was linearized with EcoRI. Membranes were hybridized with purified riboprobes (∼106 cpm) in ExpressHyb (Clontech, Palo Alto, Calif.) for 1 h at 68°C. After washing (15 min each in 2 × SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate [SDS], 0.2 × SSC–0.1% SDS, and 0.1 × SSC–0.1% SDS), the membrane was exposed at −70°C to radiographic film. Quantitation of RNA replication products was performed with the AMBIS 4000 image acquisition system and software (AMBIS, San Diego, Calif.), with values reported in the figures representing the means from at least three independent experiments ± standard deviations (SD).
For analysis of RNAs from CsCl gradients, duplicate 3.5-cm-diameter dishes of vTF7.3-infected A549 cells were transfected as described above. At 40 h posttransfection, cell lysates were prepared in 0.5 ml of NTE buffer (0.15 M NaCl, 50 mM Tris [pH 7.4], 10 mM EDTA) containing 0.5% Nonidet P-40 and clarified by centrifugation (5 min, 14,000 × g), and 400 μl of the resulting supernatant was layered onto preformed 20 to 40% (wt/wt, in NTE) CsCl gradients. After centrifugation (4 h, 45,000 rpm, 12°C, SW50.1 rotor), samples from the 30% CsCl fraction were collected (10), diluted with NTE, pelleted (6 h, 35,000 rpm, SW41 rotor), and analyzed by Northern blotting as described above.
RESULTS
Sequences located internal to the 113-nucleotide termini of naturally occurring SV5 DI RNAs are not required for replication in vivo.
DI852 is an 852-base copyback DI RNA that was generated during serial undiluted passage of SV5 in tissue culture (26). As shown in Fig. 1A, the DI852 antigenome contains a 113-base 5′ region of terminal complementarity linked to 708 bases from the L protein gene and the 31-base 3′ tr′. DI499 is identical to DI852 with the exception of a 353-base deletion of sequences internal to the 113-base region of terminal complementarity. A five-base insertion into DI499 created a 6N-length antigenome (DI499 + 5 or DI504 [Fig. 1A]) and enhanced replication ∼10-fold (26). This previous result suggested that the 353-base internal deletion in DI499 had not removed a cis-acting sequence that was essential for RNA replication. To determine if the remaining internal sequences in common between DI852 and DI504 are required for SV5 RNA replication, this region was deleted to create pDI462, which encoded an antigenomic RNA with a 390-base deletion (Fig. 1A). DNA segments encoding the full-length DI852 or internally deleted DI499 and DI462 RNAs were inserted between the promoter for T7 pol and the self-cleaving HDV ribozyme (Fig. 1A, T7p and HDV, respectively) such that transcription from the T7 pol promoter would generate a positive-sense antigenomic RNA containing three additional 5′ guanosine (G) residues and an exact 3′ end due to ribozyme self-cleavage (32, 34, 37).
To determine the relative levels of replication for these SV5 DI RNAs, monolayers of A549 cells were first infected with vTF7.3 (14), a recombinant vaccinia virus that expresses the bacteriophage T7 pol. These infected cells were then cotransfected with plasmids encoding the L, P, and NP proteins along with one of the SV5 antigenomic RNAs, each of which was under control of the T7 pol promoter. Northern blot analysis of total intracellular RNA with a positive-sense riboprobe was used to monitor conversion of the positive-sense antigenome synthesized by T7 pol to the negative-sense complementary strand by the SV5 polymerase. As shown in Fig. 1B, RNA from cells cotransfected with the L, P, and NP plasmids and the copyback DI852 antigenome contained a major ∼0.9-kb RNA species that was complementary to the T7 pol-derived RNA transcript, and this RNA was not detected when plasmids encoding L, P, or NP protein were omitted (not shown, but see below). In addition to the ∼0.9-kb DI852 replication product, low levels of a ∼1.8-kb RNA species of unknown origin were detected. The levels of replication for both of the internal deletion mutants were similar to that seen with the WT DI852 (Fig. 1A, 62 and 72% for DI504 and DI462, respectively). These data support the contention that sequences internal to the 3′- and 5′-terminal 113 bases of DI852 are not required for SV5 antigenome replication.
Identification of two regions of sequence identity in the 3′-terminal sequences of the Rubulavirus antigenomic RNA.
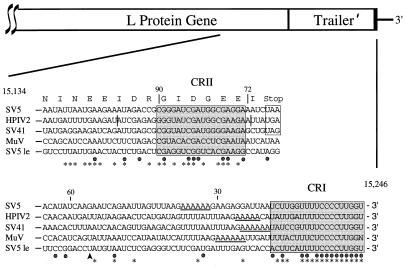
The foregoing results indicated that 113 bases at the 3′ terminus of the SV5 antigenome contain all of the signals necessary for RNA encapsidation and replication. To determine if sequences contained in these 3′-terminal 113 bases were conserved among other members of the Rubulavirus genus of the Paramyxoviridae family, the 3′ ends of the SV5, HPIV2, SV41, and MuV antigenomes were compared. As shown in Fig. 2, the nucleotide sequence alignment identified two regions of sequence identity: bases 1 to 19 at the 3′ terminus of the antigenome (CRI) and a more distal region consisting of antigenome bases 73 to 90 (CRII) that was contained within the 3′ coding region of the L protein gene. Sequence identity in CRII did not solely reflect conservation due to coding of L protein amino acids, since the corresponding 90- to 72-base region from the MuV antigenome is located in the 3′ noncoding segment of the L gene (28). Likewise, when the 3′ ends of the SV5 antigenomic (L-tr′ region) and genomic (le-NP region) RNAs were aligned (Fig. 2, SV5 le sequence), significant sequence identity was found in CRI (16 of 19 bases) and CRII (11 of 18 bases). Thus, CRI and CRII show sequence identity among rubulaviruses and are located at the 3′ termini of both the SV5 genomic and antigenomic RNAs.
FIG. 2.
Nucleotide sequence alignment of the SV5, HPIV2, SV41, and MuV antigenomic 3′ termini. The 3′-terminal 113 bases from the antigenomes of four members of the Rubulavirus genus of paramyxoviruses (SV5, HPIV2, SV41, and MuV) are listed. Amino acids and the translational stop codon (boxed) that are encoded in the 3′ end of the SV5 L gene are designated by one-letter abbreviations above the SV5 sequence, and the site for poly(A) addition is underlined. Solid circles indicate positions of sequence identity between all four viruses. SV5 le is the 3′-proximal 113 nucleotides of the SV5 genomic RNA, with the arrowhead at base 56 denoting the start site for transcription of the NP gene. Asterisks denote positions of sequence identity between the SV5 genomic le-NP and antigenomic L-tr′ regions. CRI (3′ proximal) and CRII (within the SV5 L gene) are indicated by the shaded boxes encompassing positions 1 to 19 and 73 to 90, respectively. Vertical bars in the context of the HPIV2 sequence delineate the region (bases 98 to 72) deleted to generate pDI-SSPt as described in the text. Sequences (references): SV5 (31); HPIV2 (19); SV41 (27); MuV (28).
Efficient replication of copyback DI analogs containing 90 bases from the 3′ terminus of the SV5 antigenome.
To determine if the 113-nucleotide complementary regions of DI852 containing CRI and CRII were capable of directing the encapsidation and replication of a foreign reporter sequence, we constructed a plasmid (pDI113-GL-113) such that a negative-sense copy of the GL open reading frame was flanked on the 5′ and 3′ ends by the 113-nucleotide terminal complementary regions derived from DI852 (Fig. 3A). Plasmid DNA encoding the 113-GL-113 DI RNA (designated an antigenome) was cotransfected along with the L, P, and NP plasmids into vTF7.3-infected A549 cells. Total intracellular RNA was analyzed by Northern blotting with a negative-sense riboprobe specific for the GL sequences contained within the replication product of the SV5-GL DI RNA analog. As shown in Fig. 3B, RNA from cells transfected with this combination of plasmids contained a ∼990-base RNA that hybridized to the negative-sense GL riboprobe (arrow, lane 1), and this RNA species was not detected in control samples from transfected cells in which the L, P, and NP plasmids had been omitted (Fig. 3B, lanes 2 to 4, respectively). As noted previously (26), a faster-migrating GL-specific species was detected in RNA from cells in which the NP plasmid had been omitted from the transfection (lane 4), but the origin of this RNA is not known.
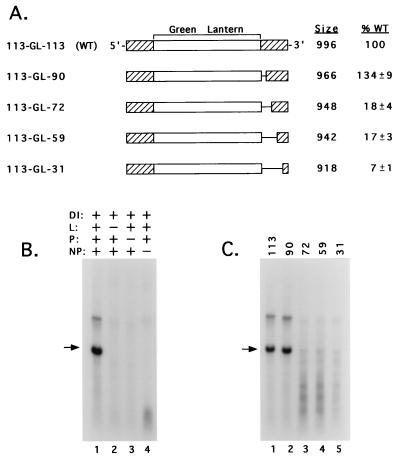
FIG. 3.
Efficient replication of copyback DI analogs having 90 bases from the 3′ terminus of the SV5 antigenome. (A) Structures of the SV5-GL DI RNA analogs. Viral antigenomes are shown schematically as rectangles, with cross-hatched boxes and solid lines representing SV5-specific bases flanking the GL sequences and deletions, respectively. The predicted sizes of the DI RNA analogs and replication level as a percentage (±SD) of that determined for the WT genome analog (113-GL-113) assayed in parallel are shown. (B) Replication of a foreign gene by SV5 cDNA-derived components. vTF7.3-infected A549 cells were cotransfected with plasmids encoding the SV5 L, P, and NP proteins along with plasmids encoding the SV5 DI113-GL-113 genome analog. Total intracellular RNA was harvested and analyzed by Northern blotting using a 32P-labeled negative-sense GL-specific riboprobe of the same polarity as the T7 pol-derived transcript (lane 1). Lanes 2 to 4 represent the replication products generated in cells in which the indicated support plasmids were replaced with pGem control DNA. The arrow indicates the position of full-length replication products. (C) Efficient replication of SV5-GL DI RNA analogs requires viral sequences between antigenomic bases 90 and 72. vTF7.3-infected A549 cells were transfected with plasmids encoding L, P, NP, and the WT SV5-GL DI RNA analog (lane 1) or RNAs that were altered such that they retained 90, 72, 59, or 31 SV5-specific bases at the antigenomic 3′ terminus (lanes 2 to 5, respectively). Replication products were analyzed by Northern blotting as described above.
To determine the minimum continuous 3′-terminal antigenomic sequences that were sufficient for replication of the SV5-GL DI RNA analog, progressive 5′-to-3′ deletions were engineered into pDI113-GL-113 to create antigenomes that contained at their 3′ ends the terminal 90, 72, 59, or 31 bases from the SV5 antigenome (Fig. 3A). When analyzed in the DI replication assay outlined above, an SV5-GL antigenome containing 90 3′-terminal bases replicated to levels that were equivalent to or greater than that of the DI113-GL-113 RNA (Fig. 3C, lane 2; quantitation in Fig. 3A). By contrast, the replication of SV5-GL antigenomes with 72 or 59 bases of viral 3′-terminal sequences was reduced to ∼18% of WT levels (lanes 3 and 4). Further truncation of the 3′ SV5 sequences to the 31-base tr′ region produced an SV5-GL antigenome that was replicated to only ∼7% of WT levels. Together, these data suggest that segments of the SV5 antigenome located between bases 90 and 72 and perhaps 59 and 31 are important for RNA replication. In the case of the SV5-GL antigenomes containing 72 and 59 SV5 3′-terminal bases, distinct RNAs of unknown origin that were smaller than the full-length DI RNA analog were synthesized.
During replication, paramyxovirus RNA synthesis is tightly coupled to encapsidation of the nascent RNA chain by the viral NP protein (reviewed in reference 20). To determine if RNA synthesized from the SV5-GL DI RNA analogs was encapsidated into NC-like structures, vTF7.3-infected A549 cells were transfected with DNA encoding either the DI113-GL-113, -90, or -59 antigenome, along with the L, P, and NP plasmids. Cell lysates were then fractionated by centrifugation on 20 to 40% CsCl gradients. Samples from the 30% CsCl fraction of the gradient were collected and analyzed by Northern blotting with the same GL-specific riboprobe as described above. As shown in Fig. 4, positive-sense GL-specific RNA was recovered from the 30% CsCl fraction of the gradient in the case of the DI113-GL-90 and DI113-GL-113 antigenomes (lanes 1 and 3). GL-specific genome-length RNAs were not detected in the case of DI113-GL-59 (lane 4) or control samples in which the plasmid encoding the SV5 L protein was omitted from the transfected DNA (lane 2). Less-than-genome-length positive-sense GL-specific RNAs were recovered from the DI113-GL-59 CsCl gradient (Fig. 4, lane 4), suggesting that the small RNA species detected previously in samples of total intracellular RNA (Fig. 3C) were encapsidated by NP. Together with the results shown in Fig. 3, these data indicate that 90 nucleotides located at the 3′ terminus of the SV5 antigenomic RNA are capable of directing the encapsidation and synthesis of replication products that have characteristics of bona fide NCs. However, the deletion of a segment containing CRII sequences significantly reduces SV5 RNA replication.
FIG. 4.
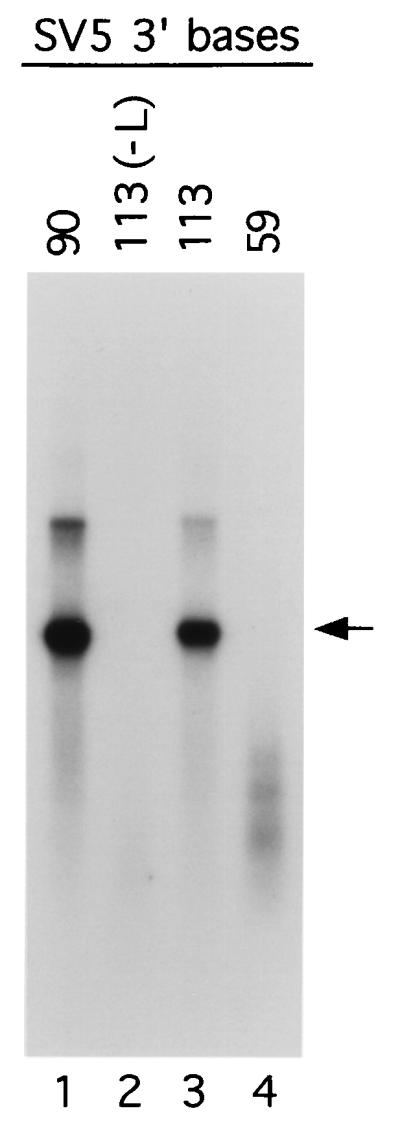
Replication products from the SV5-GL DI RNA analogs have characteristics of viral nucleocapsids. A549 cells infected with vTF7.3 were cotransfected with plasmids encoding the SV5 L, P, and NP plasmids along with the SV5-GL DI RNA analogs containing the indicated lengths of SV5-specific 3′-terminal bases. Cell lysates were prepared and analyzed by CsCl density gradient centrifugation. Samples from the 30% CsCl fraction were collected and analyzed by Northern blotting as described in the legend to Fig. 3. The arrow indicates the position of full-length replication products. Lane 2 represents a sample analyzed in parallel from cells in which the L protein plasmid had been replaced by pGem control plasmid during the transfection.
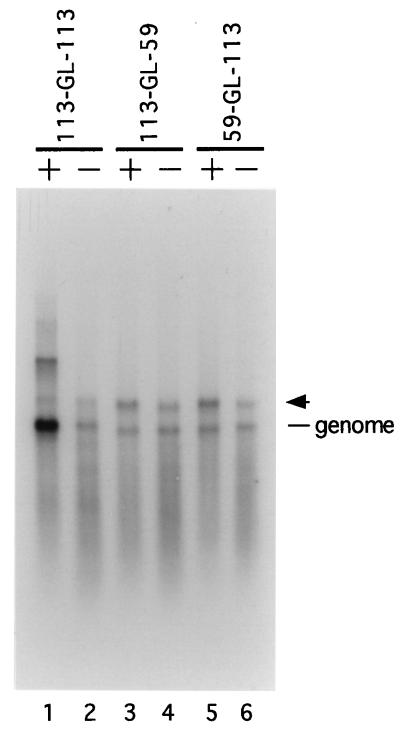
Encapsidation of replication-defective SV5-GL antigenomes that contain truncated terminal regions.
In the foregoing assay for in vivo DI replication, RNA antigenomes synthesized by T7 pol transcription of plasmid DNA must be encapsidated by the viral NP before functioning as a template for replication by the viral polymerase (32, 34). Therefore, a possible explanation for the low replication levels associated with the 3′-terminally truncated SV5-GL antigenomes is that they are defective in NP-mediated encapsidation of the primary T7 pol-derived RNA. To examine this possibility, duplicate dishes of vTF7.3-infected cells were transfected with the P and NP plasmids along with DNA encoding the 113-GL-113, 113-GL-59, or 59-GL-113 antigenome. One set of dishes received the L protein plasmid, while the other received pGem control DNA. In the absence of L protein, it was anticipated that encapsidated antigenomic RNAs would be derived exclusively from T7 pol-derived transcripts, while in the presence of L protein, these encapsidated RNAs would also include replication products generated by the SV5 polymerase. Cell lysates were prepared and analyzed by CsCl gradient centrifugation. Samples from the 30% CsCl fraction of the gradient were analyzed by Northern blotting with a positive-sense riboprobe complementary to the negative-sense GL sequences in the T7 pol-derived RNA.
In the absence of transfected L protein plasmid, the amount of T7 pol-derived RNA recovered from CsCl gradients did not differ significantly between the various SV5-GL antigenomes (Fig. 5, genome RNA species, lanes 2, 4, and 6). A slightly larger GL-specific RNA was also detected in these samples (Fig. 5, arrowhead), and the size of this RNA is consistent with it being a T7 pol-derived antigenome containing uncleaved 3′-terminal HDV ribozyme sequences as described previously for RSV minigenome analogs (16). In the presence of transfected L protein plasmid, the amount of genome-size 113-GL-113 RNA was enhanced, while the level of the corresponding RNA from the 113-GL-59 and 59-GL-113 antigenomes remained relatively constant. The enhanced signal obtained with the replication-competent 113-GL-113 antigenome probably reflects the sum of primary T7 pol-derived RNA synthesis and synthesis of RNA that results from the multiple rounds of genome replication by the SV5 polymerase. Most important, the 59-GL-113 primary RNA transcript contains at the 5′ end the same truncation of SV5-specific terminal bases as is found at the 3′ end of the replication-defective 113-GL-59 antigenome (Fig. 3), and the 59-GL-113 antigenome was encapsidated at levels equivalent to those for the constructs containing the full 113 nucleotides at the 5′ end of the primary transcript. These data indicate that the low replication of SV5-GL antigenomes lacking CRII sequences cannot be accounted for by significant defects in encapsidation of the T7 pol-derived RNA.
FIG. 5.
Encapsidation of replication-defective SV5-GL antigenomes that contain truncated terminal regions. vTF7.3-infected A549 cells were cotransfected with plasmids encoding the SV5 P and NP proteins and the indicated SV5-GL RNA antigenome (113-GL-113, 113-GL-59, or 59-GL-113) in the presence (+) or absence (−) of the L plasmid. Cell lysates were prepared and analyzed by CsCl density gradient centrifugation. Samples recovered from the 30% CsCl fraction of the gradient were analyzed by Northern blotting using a GL-specific 32P-labeled riboprobe complementary to the T7 pol-derived transcript. Genome-length RNA (genome) and a species that corresponds to transcripts with an uncleaved HDV ribozyme (arrowhead) are indicated.
Replication of chimeric DI RNAs containing exchanges between HPIV2 and SV5 antigenomic 3′-terminal sequences.
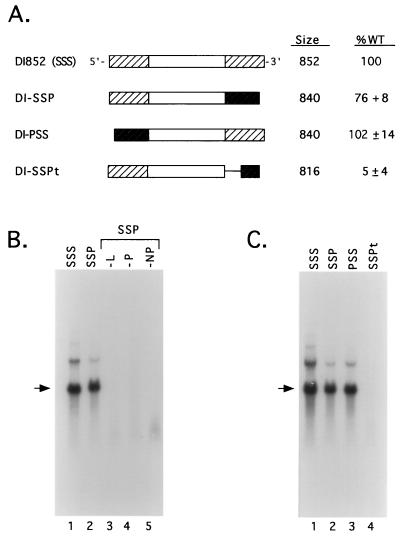
A comparison of SV5 and HPIV2 3′-terminal antigenomic sequences showed a significant degree of sequence identity in CRI (89% over bases 1 to 19) and CRII (83% over bases 73 to 90), whereas the sequence of the intervening region is less well conserved (40% over bases 20 to 72 [Fig. 2]). A plasmid encoding an 840-base chimeric DI RNA was constructed to determine if the HPIV2 antigenomic 3′ end, containing the HPIV2 CRI and CRII sequences, could direct DI RNA replication by the SV5 polymerase. In DI-SSP, 113 3′-terminal bases of the SV5 antigenome were replaced by 98 bases from the corresponding region of the HPIV2 antigenome (Fig. 6A). A549 cells infected with vTF7.3 were cotransfected with the L, P, and NP plasmids along with DNA encoding DI852 (SSS) or DI-SSP. As shown in Fig. 6B, RNA from cells transfected with plasmid encoding DI-SSP contained a major RNA species with an electrophoretic mobility closely matching that of DI852 (lanes 1 and 2). The level of replication for the SV5-HPIV2 chimeric DI RNA was similar to (76% of) that of WT DI852. As with DI852, DI-SSP RNA synthesis was dependent on cotransfection of the SV5 L, P, and NP plasmids (Fig. 6B, lanes 3 to 5, respectively).
FIG. 6.
Replication of chimeric DI RNAs containing exchanges of the SV5 and HPIV2 antigenomic 3′ ends. (A) Schematic representations of chimeric SV5-HPIV2 DI RNAs. Cross-hatched boxes indicate the termini of the DI RNAs, with white and dark-shaded boxes representing SV5- and HPIV2-specific sequences, respectively. The predicted sizes of the DI RNAs are listed, along with the relative replication level expressed as a percentage (±SD) of that determined for DI852 (SSS) analyzed in parallel. (B) Replication of the chimeric DI-SSP RNA requires L, P, and NP proteins. vTF7.3-infected A549 cells were cotransfected with the SV5 L, P, and NP plasmids along with DNA encoding either the DI852 genome (lane 1) or the chimera DI-SSP (lane 2). Total intracellular RNA was harvested and analyzed by Northern blotting using a positive-sense SV5-specific 32P-labeled riboprobe as described in the legend to Fig. 1. Lanes 3 to 5 are samples from cells in which the indicated support plasmids were replaced with pGem control DNA. The arrow indicates the position of genome-length DI852 RNA. (C) Replication of the SV5-HPIV2 chimeric antigenome requires sequences contained within HPIV2 CRII. A549 cells infected with vTF7.3 were transfected with the L, P, and NP plasmids along with DI852 plasmid (SSS; lane 1), DNA encoding the chimera with HPIV2 sequences at the 3′ terminus (SSP; lane 2) or at the 5′ terminus (PSS; lane 3), or a modified DI-SSP RNA in which HPIV2 CRII sequences had been deleted (SSPt; lane 4). Total intracellular RNA was harvested and analyzed by Northern blotting as described above.
As shown in Fig. 6C for the chimeric DI-PSS antigenome, the terminal 98 HPIV2-specific bases were also capable of functioning in replication when positioned at the 5′ end of the viral antigenome (lane 3), and replication levels typically matched that found for the WT DI852 DI RNA (quantitation listed in Fig. 6A). Most importantly, DI RNA replication was reduced ∼20-fold in the case of a truncated SV5-HPIV2 chimera (DI-SSPt [Fig. 6A]) that lacked the HPIV2 CRII sequences between antigenome bases 72 and 90 (Fig. 6C, lane 4). Together, these results indicate that the 3′-terminal 98 bases of the HPIV2 antigenomic RNA can direct encapsidation and replication by the SV5 polymerase components, and RNA replication is dependent on a region of the antigenome that includes HPIV2 CRII sequences. The extent of complementarity between the termini of DI RNAs has been proposed as a factor that can influence the level of replication for vesicular stomatitis virus (VSV) (46). The results presented above suggest that the extent of terminal complementarity is not a major factor in SV5 RNA replication, since the SV5-HPIV2 chimera replicated efficiently despite having continuous terminal complementarity reduced from 113 (in DI852) to 13 bases.
Spacing between CRII and the 3′ terminus of the SV5 antigenome is important for efficient replication of SV5-GL DI RNA analogs.
The SV5 and HPIV2 antigenomic termini have significant sequence identity in CRI and CRII but differ considerably in the intervening region, suggesting that intervening sequences are not critical for replication. To determine the role of sequences between CRI and CRII in SV5 RNA replication, a series of DI RNA analogs was constructed to contain alterations in 3′ antigenomic bases 21 to 44, maintaining an overall 6N-length RNA (26).
A BamHI restriction site was engineered into 3′-terminal bases 39 to 44 of pDI113-GL-90 DNA to facilitate the construction of altered DI analogs. When analyzed in the DI replication assay, the 113-GL-90B DI RNA analog replicated to levels that closely matched that of the WT antigenome (Fig. 7B, lanes 1 and 2), suggesting that the sequence at this position of the 3′-terminal region was not critical for SV5 RNA synthesis. Additional SV5-GL DI RNA analogs were constructed to contain 5′-to-3′ progressive 6-, 12-, and 18-nucleotide deletions of antigenomic RNA bases 38 to 21 (113-GL-90BΔ6, -Δ12, and -Δ18). RNA synthesis from the Δ6 antigenome analog was reduced ∼25-fold compared to the 113-GL-90B RNA (Fig. 7B, lanes 2 and 4; quantitation in Fig. 7A), and replication products from the Δ12 and Δ18 antigenomes were not detected (Fig. 7B, lanes 5 and 6). In the example shown in Fig. 7B, the film was purposely overexposed to illustrate the low level of replication seen in the case of the Δ6 antigenome analog. Likewise, antigenomes containing an insertion of 6 or 12 bases between SV5 antigenomic nucleotides 38 and 39 were defective for replication (Fig. 7B, lanes 7 and 8), and the level of RNA synthesized from these antigenomes was not above that detected in control samples from transfected cells in which the L protein plasmid had been omitted (lane 3).
The reduced replication of the above-described deletion/insertion mutants could result from removal or disruption of an important cis-acting viral sequence or could reflect a change in spacing between the CRI and CRII sequences. To distinguish between these possibilities, nonviral nucleotides were inserted in place of the bases that had been previously deleted in the case of the Δ12 and Δ18 antigenomes, and these replacements restored the overall length of the CRI-CRII intervening region (replacement mutants 113-GL-90BRe12 and -Re18). Length-compensating insertions of 12 (Fig. 7C, lane 2) or 18 (lane 3) nonviral bases restored RNA replication to levels that were equivalent to that of the parental 113-GL-90B antigenome (lane 1). Further extending the mutagenesis to include a replacement of bases 20 to 14 resulted in a ∼6-fold reduction in RNA replication (Fig. 7C, lane 4), consistent with the proposed role for CRI sequences in SV5 RNA synthesis. Together, these data indicate that the dramatic loss in replication of deletion mutants Δ6, Δ12, and Δ18 was due to changes in the spacing between important antigenomic promoter elements and was not due to disruption of a specific sequence requirement per se in this segment.
DISCUSSION
Paramyxovirus DI RNAs have proven to be useful models for the analysis of cis-acting sequences that are necessary for replication of the viral genome. For the paramyxoviruses that have been examined to date, all naturally occurring copyback DI RNAs retain ∼95 bases or more of terminal complementarity (21, 23, 39, 42), suggesting that this length may be at the lower limit of a functional antigenomic replication promoter. The identification of an important cis-acting sequence located between 3′-terminal bases 73 and 90 of the SV5 antigenome provides a possible explanation for the observed ∼90-base minimal extent of terminal complementarity found for paramyxovirus copyback DI RNAs. In support of this possibility, replication of a foreign gene by paramyxovirus cis-acting sequences has been reported for SeV (29), MeV (43), RSV (9), HPIV3 (11, 12), and SV5 (Fig. 3), and in each case, these viral genome analogs retained at least 90 bases from the 3′ terminus of the antigenome.
Sequence alignment identified two conserved regions (CRI and CRII) in the 3′-terminal 90 bases of the Rubulavirus antigenomic RNA, and deletions (CRII) or substitutions (CRI) in these sequences reduced SV5 RNA replication. Several lines of evidence support the proposal that these two important cis-acting elements are separated by sequences that function as a spacer region. First, the sequence of the region located between CRI and CRII is not well conserved between the rubulaviruses SV5 and HPIV2. Nevertheless, a 3′-terminal RNA segment containing the HPIV2 CRII-intervening region-CRI sequence was capable of directing efficient RNA synthesis by the SV5 polymerase components. These results indicate that while the extent of continuous 3′-terminal complementarity in a rubulavirus genome is not a major factor determining the efficiency of RNA replication, the presence of CRI and CRII sequences is essential. Second, the 3′-proximal half of the intervening region located between SV5 CRI and CRII can be replaced by nonviral sequences without compromising RNA replication. Finally, SV5 RNA replication was inhibited by changes of as little as six bases in the length of intervening sequence between CRII and the 3′ terminus, consistent with the proposal that the relative spacing of CRII and CRI is an important feature of the SV5 antigenomic promoter. By contrast, a previous analysis of cis-acting sequences in the le-NP region of the SeV genome showed that six-base insertions at position 47 or 67 of the genomic RNA did not inhibit SeV DI RNA synthesis, but SeV genomes containing 12-base insertions in this region were defective for replication (35). The previous studies and those reported here are not directly comparable since they involved analyses of the genomic (for SeV) and antigenomic (SV5) 3′ termini for these two viruses. Nevertheless, it is possible that the requirement during replication for proper spacing of essential discontinuous cis-acting elements may be a general feature of paramyxovirus promoter function. Further replacement mutagenesis will be required to more precisely map the boundaries of these cis-acting sequences, but these results suggest that the overall length of the segment located 21 to 44 nucleotides from the 3′ end of the antigenome between CRI and CRII, and not the particular sequence per se, may be the most important function of this region during paramyxovirus RNA replication.
The role that SV5 antigenomic CRI and CRII sequences play in RNA replication is not known. During paramyxovirus replication, RNA encapsidation by NP is tightly coupled to RNA synthesis (20, 24), and it is possible that one or both of these regions can function as a nucleation site to initiate NP encapsidation on the nascent RNA. In the case of VSV, previous in vitro work using purified le RNA (3) or synthetic VSV RNAs (25) indicated that signals for encapsidation are located very near the terminus of the viral RNA, but similar studies on NP-mediated encapsidation of paramyxovirus RNAs have not been reported. A deletion of CRII sequences did not compromise the ability of T7 pol-derived SV5-GL antigenomic RNA to be encapsidated in vivo, even when the deletion was engineered into the 5′ region of the antigenome. While this result indicates that CRII sequences are not involved in NP encapsidation of the T7 pol-derived RNA, it is unclear if this also applies to the steps in NP encapsidation of nascent RNA synthesized by the viral polymerase.
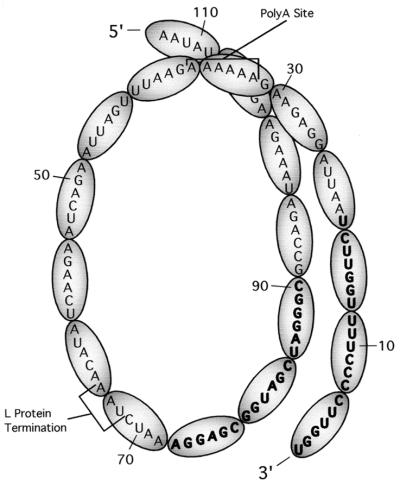
A possible alternative role for SV5 CRI and CRII in RNA replication is suggested by the proposed structure of the SeV NC which was previously derived from electron micrograph images. The SeV NC is thought to exist as a left-handed helix containing an average of 13 NP molecules per turn, and each NP molecule appears to bind six nucleotides (13). Assuming that the SV5 NC structure has these same properties (8, 10), a hypothetical model for the structure of the 3′ terminus of the SV5 antigenome which is consistent with the results of the mutational analysis described here can be proposed. Figure 8 depicts the nucleotide sequence of the 3′-terminal 113 bases of the SV5 antigenome placed in the context of ovals, each of which represents one NP molecule binding to six viral nucleotides, and 13 NP subunits are shown bound to a total of 78 bases (6 times 13) through one turn of a left-handed helix. While speculative, this postulated structure illustrates the positioning of CRI (bases 1 to 19) and CRII (bases 73 to 90) along the same face of the helical NC (Fig. 8). Defects in the replication of antigenomes containing deletions of bases between CRI and CRII can be corrected by compensating insertions of non-SV5 sequences, and these results are consistent with the proposal that RNA synthesis is sensitive to changes in the spatial alignment of CRI and CRII from their optimal position along one face of the helical NC.
FIG. 8.
Hypothetical model for the structure of the 3′-terminal region of the SV5 antigenomic RNA, based on results from the mutational data presented here and assuming that the SV5 NC has properties similar to those proposed previously for the SeV NC by Egelman et al. (13). The nucleotide sequence of the 3′-terminal 113 bases of the SV5 antigenome is listed in the context of ovals that represent NP monomers. The location of the L protein gene translational stop codon (UAA) and poly(A) signal are indicated. The proposed structure aligns SV5 CRI (boldface nucleotides 1 to 19) and CRII (boldface nucleotides 73 to 90) to the same face of the helical NC.
It is possible that the alignment of SV5 antigenomic CRI and CRII sequences creates a site for polymerase binding. Pelet et al. (35) have suggested that the SeV polymerase initiates viral RNA synthesis by interacting with at least two separate regions of the genomic promoter that may be contained within one turn of the helical NC. Genome replication for many RNA viruses, including brome mosaic virus (38), flock house virus (2), bacteriophage Qβ (5), and yeast L-A virus (15), requires discontinuous portions of the viral RNA that are located internal to the 3′ terminus. The influenza virus polymerase has been shown to bind to the 5′-terminal end of the viral RNA (45), and both 5′- and 3′-terminal regions are required for polymerase activity (17). It has been proposed that the internal or 5′-terminal sequences in these viral RNAs may function in the assembly or positioning of the polymerase during the initiation of RNA synthesis, and the alignment of SV5 CRI and CRII along one face of the helical NC may serve a similar function.
The model depicted in Fig. 8 has implications for our understanding of the requirement (6) or the preference (26) for efficient replication of paramyxovirus genomes having an overall 6N length. The rule of six is thought to operate at the level of initiation of RNA synthesis (35), and it is proposed that the 3′ end of the viral RNA may be recognized by the polymerase complex most efficiently when it is precisely assembled with NP and no additional nucleotides protrude from the 3′ end of the NC (6). However, an additional or alternative aspect of the rule of six may be the relative positioning of a particular base within the hexamer of nucleotides bound by an NP molecule as proposed by Pelet et al. (35). As such, non-6N-length alterations distal to the 3′-terminal region of the genomic and antigenomic RNAs may decrease replication by shifting the relative positions of critical bases within the context of an NP monomer. As shown in Fig. 8, a conserved 5′-CGR trinucleotide is found in the 5′-proximal position of each of the SV5 NP-bound hexamers that compose CRII. Preliminary results indicate that substitutions in these trinucleotides result in SV5 RNA analogs that are defective in replication (26a), but it is unclear whether this sequence requirement also involves a position-specific requirement for these nucleotides within an NP monomer.
Sequences analogous to the SV5 CRII cis-acting element may be located in the 3′-terminal region of other paramyxovirus antigenomic RNAs. Previous sequence comparisons of several paramyxoviruses, including SeV, HPIV3, and MeV (4), have identified a conserved sequence located 75 to 95 bases from the end of the RNA (BB box) that is found in the 3′ end of the L protein gene on the antigenome and in the complement of the 5′ end of the NP gene on the genome. However, the nucleotide sequence of the BB box (4) is not closely related to the rubulavirus CRII identified here and, while CRII is clearly essential for RNA synthesis by the SV5 polymerase, the importance of the BB box sequences to RNA replication for these other paramyxoviruses has not been reported.
The results from the analysis of the SV5 antigenomic promoter reported here differ from those reported recently for the prototypic rhabdovirus VSV (22, 33). Using a reverse genetics system, Li and Pattnaik (22) have identified two regions within the 3′-terminal 45 nucleotides of the VSV antigenome that affect copyback DI RNA replication: an essential region I (the 3′-terminal bases 1 to 24) that is sufficient to direct RNA replication and a nonessential region II (bases 25 to 45) that is proposed to play an “enhancer-like” function. By contrast, SV5 CRI and CRII are spaced further apart (within 90 bases) than the corresponding domains in the VSV antigenome (within 45 bases), and a sequence element within CRII as well as proper spacing between CRII and the 3′ terminus are critical factors that influence SV5 RNA replication. It is proposed that the presence of this replication-enhancing sequence in the VSV antigenomic 3′ end may be responsible for a higher level of RNA synthesized from the antigenomic than from the genomic promoter (22). Sequence alignments of the SV5 le and tr′ regions revealed that CRI and CRII are present in the 3′ termini of both the SV5 antigenomic and genomic RNAs. It remains to be determined if differences between the SV5 antigenomic and genomic CRII sequences are a factor that influences the level of RNA synthesized from these two promoter regions.
ACKNOWLEDGMENTS
We thank John Rassa, Si-Yi Chen, Tom Gallagher, and Doug Lyles for helpful comments on the manuscript.
This work was supported by NIH grant AI34329. Oligonucleotide synthesis was performed in the DNA Synthesis Core Laboratory of the Cancer Center of Wake Forest University, supported in part by NIH grant CA-12197.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1995. [Google Scholar]
- 2.Ball L A, Li Y. cis-acting requirements for the replication of flock house virus RNA 2. J Virol. 1993;67:3544–3551. doi: 10.1128/jvi.67.6.3544-3551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg B M, Giorgi C, Kolakofsky D. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell. 1983;32:559–567. doi: 10.1016/0092-8674(83)90475-0. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg B M, Chan J, Udem S A. Function of paramyxovirus 3′ and 5′ end sequences. In theory and practice. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 235–247. [Google Scholar]
- 5.Brown D, Gold L. RNA replication by Qβ replicase: a working model. Proc Natl Acad Sci USA. 1996;93:11558–11562. doi: 10.1073/pnas.93.21.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calain P, Roux L. Functional characterisation of the genomic and antigenomic promoters of Sendai virus. Virology. 1995;212:163–173. doi: 10.1006/viro.1995.1464. [DOI] [PubMed] [Google Scholar]
- 8.Choppin P W, Stoeckenius W. The morphology of SV5 virus. Virology. 1964;23:195–202. doi: 10.1016/0042-6822(64)90282-x. [DOI] [PubMed] [Google Scholar]
- 9.Collins P, Mink M, Stec D. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663–9667. doi: 10.1073/pnas.88.21.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compans R W, Choppin P W. Isolation and properties of the helical nucleocapsid of the parainfluenza virus SV5. Proc Natl Acad Sci USA. 1967;57:949–956. doi: 10.1073/pnas.57.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De B P, Banerjee A K. Rescue of synthetic analogs of genome RNA of human parainfluenza virus type 3. Virology. 1993;196:344–348. doi: 10.1006/viro.1993.1486. [DOI] [PubMed] [Google Scholar]
- 12.Dimmock K, Collins P L. Rescue of synthetic analogs of genomic RNA and replicative-intermediate RNA of human parainfluenza virus type 3. J Virol. 1993;67:2772–2778. doi: 10.1128/jvi.67.5.2772-2778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egelman E, Wu S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;85:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimura R, Wickner R B. Interaction of two cis sites with the RNA replicase of the yeast L-A virus. J Biol Chem. 1992;267:2708–2713. [PubMed] [Google Scholar]
- 16.Grosfeld H, Hill M G, Collins P L. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagen M, Chung T D Y, Butcher J A, Krystal M. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J Virol. 1994;68:1509–1515. doi: 10.1128/jvi.68.3.1509-1515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamaguchi M, Yoshida T, Nishikawa K, Naruse H, Nagai Y. Transcriptive complex of Newcastle disease virus. Virology. 1983;128:105–117. doi: 10.1016/0042-6822(83)90322-7. [DOI] [PubMed] [Google Scholar]
- 19.Kawano M, Okamoto K, Bando H, Kondo K, Tsurudome M, Komada H, Nishio M, Ito Y. Characterizations of the human parinfluenza type 2 virus gene encoding the L protein and the intergenic sequences. Nucleic Acids Res. 1991;19:2739–2746. doi: 10.1093/nar/19.10.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B, Knipe D, Howley P, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1177–1204. [Google Scholar]
- 21.Leppert M, Kort L, Kolakofsky D. Further characterization of Sendai virus DI-RNAs: a model for their generation. Cell. 1977;12:539–552. doi: 10.1016/0092-8674(77)90130-1. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Pattnaik A K. Replication signal in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology. 1997;232:248–259. doi: 10.1006/viro.1997.8571. [DOI] [PubMed] [Google Scholar]
- 23.Mottet G, Roux L. Budding efficiency of Sendai virus nucleocapsids: influence of size and ends of the RNA. Virus Res. 1989;14:175–188. doi: 10.1016/0168-1702(89)90037-3. [DOI] [PubMed] [Google Scholar]
- 24.Moyer S A, Horikami S M. The role of viral and host cell proteins in paramyxovirus transcription and replication. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 249–274. [Google Scholar]
- 25.Moyer S A, Smallwood-Kentro S, Haddad A, Prevec L. Assembly and transcription of synthetic vesicular stomatitis virus nucleocapsids. J Virol. 1991;65:2170–2178. doi: 10.1128/jvi.65.5.2170-2178.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy S K, Parks G D. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the paramyxovirus simian virus 5. Virology. 1997;232:145–157. doi: 10.1006/viro.1997.8530. [DOI] [PubMed] [Google Scholar]
- 26a.Murphy, S. K., and G. D. Parks. Unpublished data.
- 27.Ogawa M, Noriko M, Tsurudome M, Kawano M, Matsumura H, Kusagawa S, Komada H, Nishio M, Ito Y. Nucleotide sequence analysis of the simian virus 41 gene encoding the large (L) protein and construction of a phylogenetic tree for the L proteins of paramyxoviruses. J Gen Virol. 1992;73:2743–2750. doi: 10.1099/0022-1317-73-10-2743. [DOI] [PubMed] [Google Scholar]
- 28.Okazaki K, Tanabayashi K, Takeuchi K, Hishiyama M, Okazaki K, Yamada A. Molecular cloning and sequence analysis of the mumps virus gene encoding the L protein and the trailer sequence. Virology. 1992;188:926–930. doi: 10.1016/0042-6822(92)90555-4. [DOI] [PubMed] [Google Scholar]
- 29.Park K H, Huang T, Correia F F, Krystal M. Rescue of a foreign gene by Sendai virus. Proc Natl Acad Sci USA. 1991;88:5537–5541. doi: 10.1073/pnas.88.13.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parks G D. Mapping of a region of the paramyxovirus L protein required for the formation of a stable complex with the viral phosphoprotein P. J Virol. 1994;68:4862–4872. doi: 10.1128/jvi.68.8.4862-4872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks G D, Ward C D, Lamb R A. Molecular cloning of the NP and L genes of simian virus 5: identification of highly conserved domains in paramyxovirus NP and L proteins. Virus Res. 1992;22:259–279. doi: 10.1016/0168-1702(92)90057-g. [DOI] [PubMed] [Google Scholar]
- 32.Pattnaik A K, Ball L A, Legrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 33.Pattnaik A K, Ball L A, LeGrone A, Wertz G W. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology. 1995;206:760–764. doi: 10.1016/S0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pattnaik A K, Wertz G W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990;64:2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 36.Perrault J. Origin and replication of defective interfering particles. Curr Top Microbiol Immunol. 1981;93:151–207. doi: 10.1007/978-3-642-68123-3_7. [DOI] [PubMed] [Google Scholar]
- 37.Perrotta A T, Been M D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature (London) 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 38.Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc Natl Acad Sci USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Re G. Deletion mutants of paramyxoviruses. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 275–298. [Google Scholar]
- 40.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 41.Samal S, Collins P. RNA replication by a respiratory syncytial virus analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidhu M, Crowley J, Lowenthal A, Karcher D, Menonna J, Cook S, Udem S, Dowling P. Defective measles virus in human subacute sclerosing panencephalitis brain. Virology. 1994;202:631–641. doi: 10.1006/viro.1994.1384. [DOI] [PubMed] [Google Scholar]
- 43.Sidhu M, Chan J, Kaelin K, Spielhofer P, Radecke F, Schneider H, Masurekar M, Dowling P, Billeter M, Udem S. Rescue of synthetic measles virus minireplicons: measles genomic termini direct efficient expression and propagation of a reporter gene. Virology. 1995;208:800–807. doi: 10.1006/viro.1995.1215. [DOI] [PubMed] [Google Scholar]
- 44.Tapparel C, Roux L. The efficiency of Sendai virus genome replication: the importance of the RNA primary sequence independent of terminal complementarity. Virology. 1996;225:163–171. doi: 10.1006/viro.1996.0584. [DOI] [PubMed] [Google Scholar]
- 45.Tiley L S, Hagen M, Matthews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wertz G, Whelan S, LeGrone A, Ball L. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc Natl Acad Sci USA. 1994;91:8587–8591. doi: 10.1073/pnas.91.18.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]