Abstract
Varicella-zoster virus (VZV) causes chicken pox (varicella), becomes latent in dorsal root ganglia, and reactivates decades later to cause shingles (zoster). During latency, the entire VZV genome is present in a circular form, from which genes 21, 29, 62, and 63 are transcribed. Immediate-early (IE) VZV genes 62 and 63 encode regulators of virus gene transcription, and VZV gene 29 encodes a major DNA-binding protein. However, little is known about the function of VZV gene 21 or the control of its transcription. Using primer extensions, we mapped the start of VZV gene 21 transcription in VZV-infected cells to a single site located at −79 nucleotides (nt) with respect to the initiation codon. To identify the VZV gene 21 promoter, the 284-bp region of VZV DNA separating open reading frames (ORFs) 20 and 21 was cloned upstream from the chloramphenicol acetyltransferase gene. In transient-transfection assays, the VZV gene 21 promoter was transactivated in VZV-infected, but not uninfected, cells. Further, the protein encoded by ORF 62 (IE62), but not those encoded by VZV ORFs 4, 10, 61, and 63, transactivates the VZV gene 21 promoter. By use of transient-cotransfection assays in conjunction with 5′ deletions of the VZV gene 21 promoter, a 40-bp segment was shown to be responsible for the transactivation of the VZV gene 21 promoter by IE62. This region was located at −96 to −56 nt with respect to the 5′ start of gene 21 transcription.
Varicella-zoster virus (VZV), a neurotropic alphaherpesvirus, causes childhood chicken pox (varicella), becomes latent in dorsal root ganglia at all levels of the neuraxis, and may reactivate decades later to produce shingles (zoster) (18). The entire 125,884-bp VZV genome has been sequenced, and 71 open reading frames (ORFs) have been identified (14). Transcripts mapping to most of the predicted ORFs have been detected in VZV-infected cells, although fewer than 20 VZV genes have been analyzed in detail (29, 35, 37). The VZV genome is compact: the 71 ORFs are separated by an average of 211 bp, indicating that the promoters are close to the genes they control.
Coordinated control of virus gene expression is a hallmark of herpesvirus lytic replication (21). Because VZV is highly cell associated and does not grow to high titers, experiments involving high multiplicities of infection and single-step virus growth have been difficult. Nevertheless, it appears that like that of the prototype alpha-herpesvirus, herpes simplex virus type 1 (HSV-1), VZV gene transcription during productive infection is highly regulated and follows a complex cascade of events (8). VZV immediate-early (IE) genes are the first to be transcribed, and their promoters are recognized by the existing cellular transcription factors. VZV IE proteins transactivate promoters for the VZV genes involved in virus DNA replication (early proteins). During this time, the input linear herpesvirus DNA circularizes in preparation for replication via a rolling-circle mechanism (3, 25). Following the initiation of virus DNA replication, late viral genes are transcribed and translated. Late gene promoters are not recognized by unmodified cellular transcription factors, and their transactivation by IE proteins is only marginal. However, gene amplification resulting from virus DNA replication overcomes late promoter inefficiency, and late gene transcripts accumulate in infected cells.
During latency, productive VZV replication is blocked, VZV is not seen by electron microscopy in otherwise normal human ganglia, and infectious virus cannot be recovered (17). The VZV genome exists in an episomal form from which at least four virus genes (genes 21, 29, 62, and 63) are transcribed (6, 9, 13). VZV genes 62 and 63 encode IE phosphoproteins which orchestrate virus gene transcription (2, 15, 16, 24, 26, 27, 36), and VZV gene 29 encodes a 130-kDa early DNA-binding protein (28). The function of the VZV gene 21 protein has not been studied, although its HSV-1 homolog, UL37, binds to HSV-1 ICP8, the homolog of the VZV gene 29 protein (39, 40). To begin studying the structure and regulation of VZV gene 21, we used primer extensions to identify the 5′ start of VZV gene 21 transcription and transient-transfection assays to locate the promoter for VZV gene 21 in infected cells.
MATERIALS AND METHODS
Virus and cells.
VZV (Ellen strain) was propagated in a continuous line of African green monkey kidney cells (BSC-1) in Dulbecco’s minimal essential medium supplemented with 10% fetal calf serum. Infected cells were cocultivated with uninfected cells as described previously (19).
RNA extraction and primer extension.
Uninfected and VZV-infected BSC-1 cells were disrupted with guanidine lysis buffer, and total RNA was extracted with acid-phenol (5). Table 1 shows the sequences of the oligonucleotides used for priming cDNA synthesis. T4 polynucleotide kinase and [32P]ATP were used for 5′-end labeling of oligonucleotides; this was followed by electrophoresis on 20% polyacrylamide gels or affinity chromatography (30). For primer extension, 8.9 × 104 cpm of labeled primer was annealed to 5 μg of total RNA at 65°C for 10 min and cDNA was synthesized at 48°C for 60 min with Moloney murine leukemia virus reverse transcriptase (Superscript H−; Gibco-BRL, Gaithersburg, Md.). The extended products along with the DNA sequence (Sequenase; U.S. Biochemicals, Cleveland, Ohio) of the SalI C fragment of VZV DNA were resolved on sequencing gels (11).
TABLE 1.
Oligonucleotide primers
| Primer | Sequence (5′ to 3′)a | 5′ startb | 3′ endb | Use |
|---|---|---|---|---|
| 21pe1 | CTTCTTGTACTTTCAAGTTAC | 30808 | 30828 | Primer extension |
| 21pe2 | GTTTTCTCTGGTGACCATGG | 30853 | 30862 | Primer extension |
| 21pe3 | GGAATTTTCTGGGTAGATCG | 30897 | 30916 | Primer extension |
| 21-mlu | GCGACGCGTAGCTGAGGGGTTAAATTCACA | 30475 | 30497 | 5′-end p21, p21Δ |
| 21-mluA | GCGACGCGTGGTAGGAGGAGCC | 30585 | 30597 | 5′-end p21A |
| 21-mluB | GCGACGCGTGGTTCTTCAACTTACCGTG | 30626 | 30644 | 5′-end p21B |
| 21-mluC | GCGACGCGTGCGTTTTTATTGATGTTAC | 30663 | 30682 | 5′-end p21C |
| 21-mluZ | GCGACGCGTCGGGGTCACGTCCAGCCTGTG | 29979 | 30000 | 5′-end p21Z |
| 21-kpn | GCGGGTACCGGTATATTCTACGCTGACTTAAC | 30758 | 30736 | 3′-end p21, p21A, p21B, p21C |
| 21-kpnA | GCGGGTACCGGCTCCTCCTACC | 30597 | 30585 | 3′-end p21Δ |
| KpnI-MluI-p1 | TACGCGTGGAATTCCGGTACCG | NAc | NA | Modify pCAT3basic |
| KpnI-MluI-p2 | CGCGCGGTACCGGAATTCCACGCGTAGTAC | NA | NA | Modify pCAT3basic |
| 4-HindIII | CAAAATATCTGACAA(GC)TTGCGTGTTTGCAG | 4147 | 4175 | Introduce HindIII site 5′ of ORF 4 |
| 4-SpeI | CAAATTAGTATGTTTTGAC(TAGT)AGCATGAAAAAGG | 2739 | 2771 | Introduce SpeI site 3′ of ORF 4 |
| 10-EcoRI | CTTATTTAAACTAAAGA(A)TT(C)TTACTCTATAAG | 12128 | 12158 | Introduce EcoRI site 5′ of ORF 10 |
| 10-XbaI | CGCGTTAAACGTC(TAG)ATTGGGGTAGAG | 13385 | 13409 | Introduce XbaI site 3′ of ORF 10 |
| 61-HindIII | GAATACAGCCAA(G)CTTGTTACCATGG | 104505 | 104482 | Introduce HindIII site 5′ of ORF 61 |
| 61-SpeI | GAAGTCCTAGTT(AC)T(A)GTTGGGAGGGGG | 103091 | 103067 | Introduce SpeI site 3′ of ORF 61 |
| 62-EcoRI | GGGTACGTCTA(G)AATTCACCCCAG | 109160 | 109138 | Introduce EcoRI site 5′ of ORF 62d |
| 63-HindIII | GGTGCAAAACATGTCC(AAGC)TTGGGGCCGTAGTA | 110592 | 110563 | Introduce HindIII site 5′ of ORF 63 |
| 63-SpeI | TGTATTTATTTATAA(CT)AG(T)ACTACACGCCATGGG | 111435 | 111404 | Introduce SpeI site 3′ of ORF 63 |
| EcoRI-HindIII-p1 | AATTCCTGGA | NA | NA | Link EcoRI site to HindIII site in pCIneo |
| EcoRI-HindIII-p2 | AGCTTCCAGG | NA | NA | Link EcoRI site to HindIII site in pCIneo |
MluI sites are indicated by a single underline. KpnI sites are indicated by a double underline. Parentheses enclose nucleotides added to introduce the desired restriction endonuclease.
Nucleotide position of the oligonucleotide on the VZV genome.
NA, not available.
The 3′ cloning site (XbaI) was supplied by the pAlter-1 vector.
Northern blot analysis.
Total RNA (20 μg) from VZV-infected and control BSC-1 cells was resolved by electrophoresis in 1% agarose gels containing 0.5 mM methylmercury(II) hydroxide (Johnson Matthey, Ward Hill, Mass.), transferred to Zeta-Probe membranes (Bio-Rad, Hercules, Calif.), and probed with either the entire VZV gene 21 ORF, which extends from nucleotide (nt) 30759 to nt 33872 on the VZV genome (14), or a human β-actin cDNA (10, 12). Double-stranded DNA probes were radiolabeled with [32P]dCTP by nick translation (30).
Plasmid construction.
The initiation codons for VZV ORFs 20 and 21 are located at nt 30475 and nt 30759, respectively, and the ORFs are oriented in opposite directions (14). The 284-bp intergenic region separating ORFs 20 and 21 was amplified from pBSalC (11) by PCR with oligonucleotide primers containing either KpnI or MluI restriction endonuclease sites (Table 1). PCR conditions were reported previously (12). The PCR product was resolved by agarose gel electrophoresis, digested with KpnI and MluI, and inserted into a promoterless chloramphenicol acetyltransferase (CAT) reporter plasmid (pCAT3basic; Promega, Madison, Wis.). Before the PCR product was inserted into the reporter plasmid, the multiple cloning site was modified to invert the orientation of the KpnI and MluI sites. Inversion of the KpnI and MluI sites of pCAT3basic was obtained by digesting pCAT3basic with KpnI and MluI and then ligating a double-stranded adapter consisting of annealed oligonucleotides KpnI-MluI-p1 and KpnI-MluI-p2 (Table 1). The fidelity of PCR and cloning was verified by DNA sequencing.
To construct plasmids placing VZV ORFs 4, 10, 61, 62, and 63 under the control of the cytomegalovirus (CMV) IE promoter (pCIneo; Promega), the VZV ORFs, along with 6 to 1,817 bp of flanking sequences, were shuttled into the vector, pAlter-1 (Promega). Following DNA sequencing to determine the orientation of the insert, single-stranded phage DNA and oligonucleotide primers (Table 1) were used to synthesize chimeric double-stranded plasmid DNA. After transformation of Escherichia coli INVαF′ (In Vitrogen, San Diego, Calif.) and selection of tetracycline-sensitive, ampicillin-resistant organisms (antibiotic switch), the plasmid inserts were partially sequenced to confirm the introduction of the desired restriction endonuclease site. The VZV ORFs were then shuttled from pAlter-1 to pCIneo and again the inserts were partially sequenced to confirm the correct construct. Oligonucleotide primers used to introduce the restriction endonuclease sites were designed to leave unchanged the virus DNA sequence around the initiation codon.
DNA transfection and reporter gene assay.
BSC-1 cells (0.7 × 106 cells) were seeded into 90-mm-diameter dishes and grown for 18 to 24 h at 37°C in a humidified CO2 incubator. Supercoiled plasmid DNA, extracted by affinity chromatography (Qiagen, Santa Clarita, Calif.), was diluted to 20 μg in 500 μl of Hanks balanced salt solution. Each transfection reaction mixture consisted of 15 μg of reporter plasmid and 5 μg of β-galactosidase-expressing plasmid DNA (pSV-βGal; Promega) and was precipitated at room temperature for 20 min with the addition of CaCl2 to 0.124 M (20). The DNA-CaPO4 precipitate was added directly to the cell monolayer, and cells were harvested and lysed 48 h after transfection and assayed for total protein and β-galactosidase activity (30). Cell extracts were diluted to yield equal amounts of β-galactosidase activity, and acetylation of [14C]chloramphenicol was determined by ascending thin-layer chromatography (30). Where promoter activity was determined as a response to transactivation by various VZV proteins, equal amounts of cell protein extract were used in CAT assays.
RESULTS
VZV gene 21 transcriptional unit.
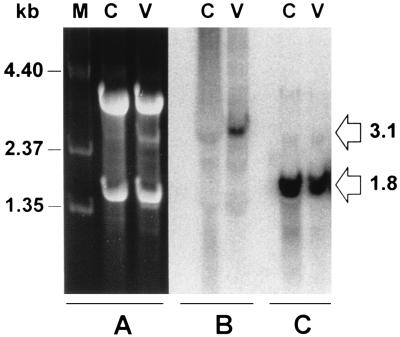
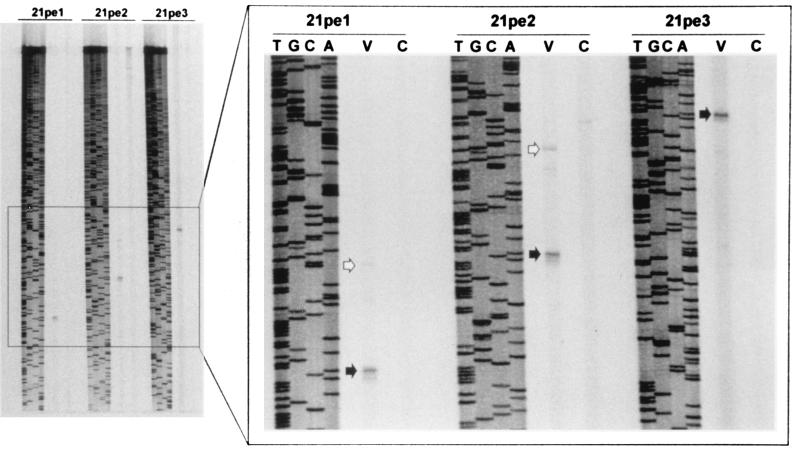
Northern blot analysis of total RNA extracted from VZV-infected cells demonstrated that the VZV gene 21 transcript is a single species of approximately 3.1 kb (Fig. 1). We have previously determined the 3′-terminal structure of VZV gene 21 from both lytically infected cells in tissue culture and from latently infected human trigeminal ganglia (9, 12). The VZV gene 21 ORF is followed by 45 to 52 nt of untranslated RNA containing a typical eukaryotic polyadenylation signal and a poly(A)+ tail. To determine the structure of the VZV gene 21 transcript at the 5′ end, primer extensions were used in which RNA extracted from productively infected cells was reverse transcribed with various 32P-end-labeled oligonucleotides complementary to ORF 21 (Table 1). The sizes of the extended products were compared to those of a DNA sequencing ladder obtained by sequencing the SalI C fragment of VZV DNA with each primer. Figure 2 shows that for each primer used, the reverse transcription product terminated at the identical adenosine (nt 30681), located at −79 with respect to the initiation codon for VZV gene 21. Faint bands were observed at −110 with respect to the initiation codon for VZV gene 21 when the VZV-infected BSC-1 RNA was extended with primer 21pe1 and at −117 when the RNA was extended with primer 21pe2. These products may indicate the presence of minor gene 21 transcripts initiating from a subordinate TATA box; however, since they are not coterminal and not observed when primer 21pe3 is used to extend VZV-infected BSC-1 RNA, these products may be artifacts of the reverse transcription reaction. No product was observed when the primers were used to extend uninfected BSC-1 cell RNA. Identical results were obtained when the experiment was repeated with a different preparation of infected cell RNA. Thus, the VZV gene 21 transcript is a single 3.1-kb poly(A)+ RNA containing a 3,113-nt ORF bounded by untranslated regions of 79 nt at the 5′ end and 45 to 52 nt at the 3′ end.
FIG. 1.
Northern blot analysis of VZV-infected BSC-1 RNA. (A) Total RNA (20 μg) extracted from control BSC-1 cells (lanes C) and VZV-infected BSC-1 cells (lanes V) along with RNA standards (lane M; Gibco-BRL) was resolved in 1% agarose gels containing 0.5 mM methylmercury(II) hydroxide and stained with 0.5 μg of ethidium bromide per ml in 0.5 M ammonium acetate. (B and C) RNA was transferred to a nylon-based membrane and probed for VZV gene 21 transcripts (B) or β-actin transcripts (C). VZV gene 21 transcripts are visible as a discrete 3.1-kb band in VZV-infected cell RNA. Both control and VZV-infected cell RNA contain discrete 1.8-kb β-actin transcripts.
FIG. 2.
Location of the 5′ start of RNA transcription for VZV gene 21. Total RNA (5 μg) from either VZV-infected (lanes V) or uninfected (lanes C) BSC-1 cells was annealed to oligonucleotide primer 21pel, 21pe2, or 21pe3 end labeled with 32P (Table 1). First-strand cDNA was synthesized, and the extended product was resolved by gel electrophoresis. The DNA sequence of the SalI C fragment of VZV DNA primed with the respective oligonucleotides was used to size the cDNA products. The entire gel image as well as an enlargement of the region containing extended products is shown. With all three primers, the cDNA product obtained from VZV-infected cell RNA (closed arrows in lanes V) terminated at the identical adenosine located at nt 30681 on the VZV genome. Minor extended products obtained from VZV-infected cell RNA (open arrows in lanes V) were also observed. No product was observed when uninfected cell RNA was used in the cDNA synthesis reaction (lanes C).
The VZV gene 21 promoter is silent in uninfected cells.
To investigate the VZV gene 21 promoter, the 284-bp DNA segment separating ORF 20 and ORF 21 (Fig. 3) was cloned into the CAT reporter plasmid. Figure 4 shows that in control BSC-1 cells, the VZV gene 21 promoter induces CAT at levels beneath detection under the circumstances used. Further, the function of a VZV-induced protein is required for gene 21 promoter activity.
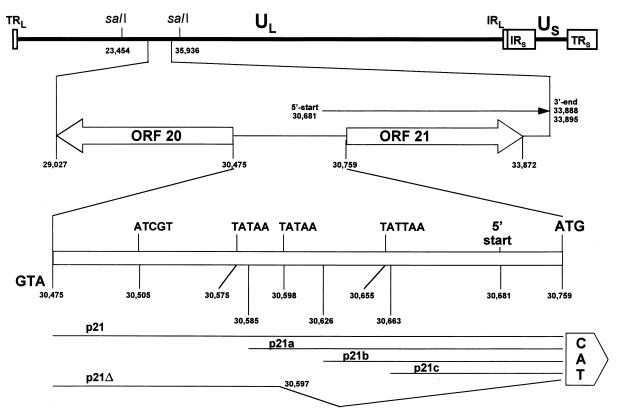
FIG. 3.
Schematic representation of the VZV DNA between ORFs 20 and 21. The VZV genome consists of unique long (UL) and unique short (US) segments of DNA, each bounded by inverted and repeated DNA sequences (TRL/IRL and IRS/TRS). ORFs 20 and 21 are oriented in opposite directions, and both map within the SalI C fragment (positions 23454 to 35936) within the UL. The 284-bp DNA segment separating ORFs 20 and 21 contains one potential IE62 binding site and three TATAA-like boxes. The 5′ start site of gene 21 transcription is located at nt 30681, and the 3′ end of the transcript has been mapped to nt 33888 and nt 33895 (7, 10). The boundary of the CAT reporter constructs used to locate the VZV gene 21 promoter are shown.
FIG. 4.
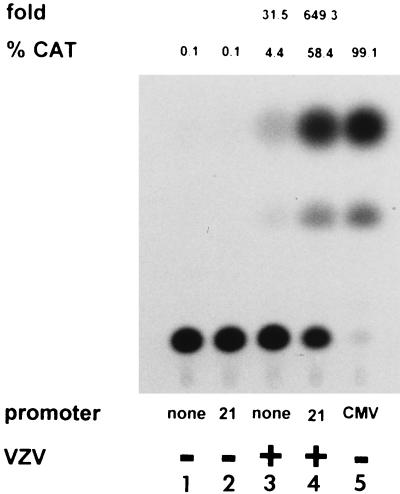
The VZV gene 21 promoter is silent in uninfected cells. The 284-bp VZV DNA segment separating ORFs 20 and 21 was inserted into the CAT reporter plasmid and used to transfect either uninfected cells (lane 2) or VZV-infected cells (lane 4). Controls included the CAT reporter plasmid lacking a promoter transfected into uninfected cells (lane 1) or VZV-infected cells (lane 3) and a CMV IE promoter driving CAT transfected into uninfected cells (lane 5). CAT assays were performed in duplicate, and the average acetylation of chloramphenicol (%CAT) showed that the VZV gene 21 promoter does not function in uninfected cells (−VZV) but is active in VZV-infected cells (+VZV). The amount of promoter activity in infected cells above that in uninfected cells (fold) showed that gene 21 promoter activity was approximately 650-fold higher in VZV-infected cells than in uninfected cells.
The VZV gene 21 promoter is transactivated by the protein encoded by ORF 62 (IE62).
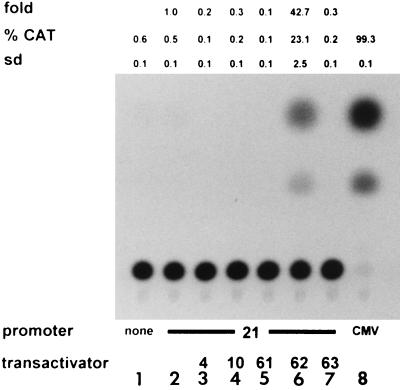
Since the VZV gene 21 promoter is silent in uninfected cells and active during virus infection, one or more VZV-induced proteins must function to transactivate the gene 21 promoter. The most likely candidates are VZV IE proteins or the tegument-associated transactivating protein (encoded by ORF 10). Therefore, we constructed expression plasmids in which VZV IE protein genes corresponding to ORF 4, 61, 62, and 63, along with ORF 10, were placed under the control of the CMV IE3 promoter. Western blot analysis was used to confirm the ability of each construct to express the respective protein in transient-transfection assays (data not shown). Figure 5 shows that the VZV gene 21 promoter is silent in uninfected BSC-1 cells or in BSC-1 cells expressing ORF 4, 10, 61, or 63. However, cotransfection of BSC-1 cells with the gene 21 promoter-CAT construct with the ORF 62 expression plasmid transactivated the VZV gene 21 promoter 42-fold more than cotransfection with the gene 21 promoter alone.
FIG. 5.
The VZV gene 21 promoter is transactivated by IE62. CAT reporter plasmids, either promoterless (lane 1) or containing the 284-bp VZV ORF 20-ORF 21 intergenic region (lanes 2 to 7) or the CMV IE promoter (lane 8), were transfected into cells either alone (lanes 1, 2, and 8) or with plasmids expressing various VZV transactivators (ORFs 4, 10, 61, 62, and 63) (lanes 3 to 7). Duplicate CAT assays indicated that the VZV gene 21 promoter is transactivated by VZV IE62 but not by the proteins encoded by VZV genes 4, 10, 61, and 63. Transactivation of VZV gene 21 promoter by IE62 is ∼42-fold higher than VZV gene 21 promoter activity in the absence of IE62. %CAT, average percent chloramphenicol acetylation; sd, standard deviation.
5′ boundary of the gene 21 promoter.
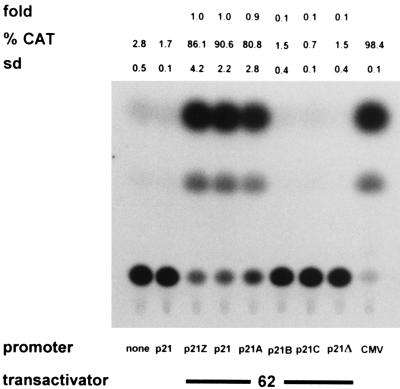
Figure 6 shows the results of transient transfection of BSC-1 cells with various 5′ truncations of the VZV ORF 20-ORF 21 intergenic region inserted into the CAT reporter plasmid. Inspection of the ORF 20-ORF 21 intergenic region indicates the presence of three TATAA-like boxes. The plasmids were constructed to eliminate successively these putative transcriptional regulatory elements. To determine if transcriptional regulatory elements exist upstream of the ORF 20-ORF 21 intergenic region that controls expression of gene 21, a further CAT construct that extended 496 bp into the 5′ end of ORF 20 was made (p21Z-CAT). Since we had determined that IE62 is required for gene 21 promoter activity, all transfections included the IE62 expression vector. The CAT activity of p21Z-CAT was similar to that of p21-CAT, indicating that the VZV gene 21 promoter is contained entirely within the segment of DNA spanning ORFs 20 and 21. Deletion of the 111 bp from position 30475 to position 30585 had little effect on VZV gene 21 promoter activity and reduced CAT activity only from 90.6 to 80.8%. Deletion of the 152 bp from position 30475 to position 30626, however, had a marked effect on VZV gene 21 promoter function and reduced its activity to background levels. Further 5′ truncations of the VZV gene 21 promoter also resulted in background CAT activities. These results indicate that the 5′ boundary of the gene 21 promoter regulatory region of the VZV gene 21 promoter lies between nt 30585 and nt 30626. Since this region contains a TATAA box that could direct the 5′ start of transcription, plasmid p21Δ, which consists of the DNA segment from nt 30475 to nt 30597 inserted into the CAT reporter plasmid, was constructed. In transient-cotransfection assays, p21Δ demonstrated no CAT gene translation products in the presence of IE62 (Fig. 6), indicating a lack of promoter activity.
FIG. 6.
5′ boundary of the gene 21 promoter. Duplicate CAT assays were performed on extracts of uninfected cells that had been transfected with CAT reporter constructs containing either the entire 284-bp DNA segment separating ORF 20 and ORF 21 (p21), a 496-bp extension (p21Z), or various 5′ truncation mutations (p21A, p21B, p21C, and p21Δ) in the presence of the IE62 expression plasmid. The 5′ boundary of the gene 21 promoter was located to a region between nt 30585 (p21A) and nt 30626 (p21B). %CAT, average percent chloramphenicol acetylation; sd, standard deviation.
DISCUSSION
VZV gene 21 consists of a 3,113-nt ORF bounded by a 5′ untranslated region of 79 nt and by a 3′ untranslated region of 45 to 52 nt. During productive infection in tissue culture, VZV gene 21 transcripts appear as a single, discrete 3.1-kb band on denaturing agarose gels. We produced antibodies in rabbits directed against VZV gene 21–glutathione S-transferase fusion proteins and located gene 21 protein predominantly in the cytoplasm as well as in the nucleus of productively infected cells (29a). Although the protein encoded by VZV gene 21 has not been studied, it is homologous (47%) to HSV-1 UL37 (40), a 120-kDa phosphoprotein synthesized after the onset of virus DNA replication (1). In HSV-1-infected cells, UL37 is both cytoplasmic and nuclear and incorporates into the tegument of progeny virions (31, 38). HSV-1 UL37 and ICP8 (the VZV gene 29 product homolog) form a DNA-binding complex (39, 40).
During VZV latency, polyadenylated transcripts mapping to VZV gene 21 are present in human trigeminal ganglia (9, 12). While no animal model of VZV latency and reactivation currently exists, simian varicella virus (SVV) infection in monkeys closely mimics VZV infection in humans (18), and polyadenylated transcripts corresponding to the SVV homolog of VZV gene 21 have been demonstrated in monkey ganglia latently infected with SVV (7).
The consistent detection of VZV gene 21 transcripts in latently infected human ganglia (9, 10, 12) and its SVV homolog in latently infected monkey ganglia (7) suggests that VZV gene 21 is vital to the maintenance of varicella latency or that its transcription is constitutive because cellular transcription factors recognize the promoter. We have identified the VZV gene 21 promoter and have shown that it is silent in uninfected cells, indicating that its activity depends upon a virus-induced protein. We have further identified IE62 as the virus protein capable of transactivating the VZV gene 21 promoter. Along with VZV gene 21 transcripts, polyadenylated transcripts mapping to ORF 29 have been detected in latently infected human ganglia (9, 32). Like the promoter for VZV gene 21, the VZV gene 29 promoter is silent in uninfected cells but is transactivated by VZV IE62 (33, 34). VZV IE62 is a promiscuous transactivator that recognizes numerous VZV, HSV-1, human immunodeficiency virus, and cellular promoters (22, 23, 36). VZV IE62 DNA binding has been located to a nonpalindromic pentamer, ATCGT, and inspection of the VZV gene 21 promoter region shows this potential binding site for region II of IE62 (4, 41, 42). However, deletion of the potential IE62 binding site did not diminish the transactivation of gene 21 promoter by IE62. VZV IE62 transactivation has also been associated with the ubiquitously present cellular transcription factor USF (34). However, the minimal VZV gene 21 promoter domain responsive to IE62 transactivation lacks the consensus USF DNA binding sequence CACGTG. Thus, the promoter for gene 21 may unlock a novel mechanism by which IE62 maintains gene regulation.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants AG 06127 and NS 32623 from the National Institutes of Health.
We thank Paul Kinchington for antisera against proteins encoded by VZV ORFs 4, 10, 61, and 62. We also thank Mary Devlin for editorial review and Cathy Allen for preparation of the manuscript.
REFERENCES
- 1.Albright A G, Jenkins F J. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J Virol. 1993;67:4842–4847. doi: 10.1128/jvi.67.8.4842-4847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudoux L, Defechereux P, Schoonbroodt S, Merville M-P, Rentier B, Piette J. Mutational analysis of varicella-zoster virus major immediate-early protein IE62. Nucleic Acids Res. 1995;23:1341–1349. doi: 10.1093/nar/23.8.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Porat T, Rixon F J. Replication of herpesvirus DNA. IV. Analysis of concatemers. Virology. 1979;94:61–70. doi: 10.1016/0042-6822(79)90438-0. [DOI] [PubMed] [Google Scholar]
- 4.Betz J L, Wydoski S G. Functional interaction of varicella zoster virus gene 62 protein with the DNA sequence bound by herpes simplex virus ICP4 protein. Virology. 1993;195:793–797. doi: 10.1006/viro.1993.1432. [DOI] [PubMed] [Google Scholar]
- 5.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 6.Clarke P, Beer T, Cohrs R, Gilden D H. Configuration of latent varicella zoster virus DNA. J Virol. 1995;69:8151–8154. doi: 10.1128/jvi.69.12.8151-8154.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke P, Matlock W L, Beer T, Gilden D H. A simian varicella virus (SVV) homolog to varicella-zoster virus gene 21 is expressed in monkey ganglia latently infected with SVV. J Virol. 1996;70:5711–5715. doi: 10.1128/jvi.70.8.5711-5715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J J, Straus S E. Varicella-zoster virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippicott-Raven; 1996. pp. 2525–2547. [Google Scholar]
- 9.Cohrs, R., R. Mahalingam, A. N. Dueland, W. Wolf, M. Wellish, and D. H. Gilden. 1992. Restricted transcription of varicella-zoster virus in latently infected human trigeminal and thoracic ganglia. J. Infect. Dis. 166(Suppl. 1):S24–S29. [DOI] [PubMed]
- 10.Cohrs R J, Barbour M, Gilden D H. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J Virol. 1996;70:2789–2796. doi: 10.1128/jvi.70.5.2789-2796.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohrs R J, Barbour M B, Mahalingam R, Wellish M, Gilden D H. Varicella-zoster virus (VZV) transcription during latency in human ganglia: prevalence of VZV gene 21 transcripts in latently infected human ganglia. J Virol. 1995;69:2674–2678. doi: 10.1128/jvi.69.4.2674-2678.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohrs R J, Srock K, Barbour M B, Owens G, Mahalingam R, Devlin M E, Wellish M, Gilden D H. Varicella-zoster virus (VZV) transcription during latency in human ganglia: construction of a cDNA library from latently infected human trigeminal ganglia and detection of a VZV transcript. J Virol. 1994;68:7900–7908. doi: 10.1128/jvi.68.12.7900-7908.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croen K D, Ostrove J M, Dragovic L J, Straus S E. Patterns of gene expression and sites of latency in human nerve ganglia are different for varicella-zoster and herpes simplex viruses. Proc Natl Acad Sci USA. 1988;85:9773–9777. doi: 10.1073/pnas.85.24.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davison A J, Scott A J. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 15.Debrus S, Sadzot-Delvaux C, Nikkels A F, Piette J, Rentier B. Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J Virol. 1995;69:3240–3245. doi: 10.1128/jvi.69.5.3240-3245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felser J M, Kinchington P R, Inchauspe G, Straus S E, Ostrove J M. Cell lines containing varicella-zoster virus open reading frame 62 and expressing the “IE” 175 protein complement ICP4 mutants of herpes simplex virus type 1. J Virol. 1988;62:2076–2082. doi: 10.1128/jvi.62.6.2076-2082.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilden D H, Vafai A. Varicella-zoster. In: Vinken P J, Bruyn G W, Klawans H L, editors. Handbook of clinical neurology. New York, N.Y: Elsevier Science Publishing Co. Inc.; 1989. pp. 229–247. [Google Scholar]
- 18.Gilden D H, Mahalingam R, Dueland A N, Cohrs R. Herpes zoster: pathogenesis and latency. Prog Med Virol. 1992;39:19–75. [PubMed] [Google Scholar]
- 19.Gilden D H, Shtram Y, Friedmann A, Wellish M, Devlin M, Cohen A, Fraser N, Becker Y. Extraction of cell-associated varicella-zoster virus DNA with Triton X-100-NaCl. J Virol Methods. 1982;4:263–275. doi: 10.1016/0166-0934(82)90073-8. [DOI] [PubMed] [Google Scholar]
- 20.Graham R L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 21.Honess R W, Roizman B. Regulation of herpes simplex virus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inchauspe G, Ostrove J M. Differential regulation by varicella-zoster virus (VZV) and herpes simplex virus type-1 trans-activating genes. Virology. 1989;173:710–714. doi: 10.1016/0042-6822(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 23.Inchauspe G, Nagpal S, Ostrove J M. Mapping of two varicella-zoster virus-encoded genes that activate the expression of viral early and late genes. Virology. 1989;173:700–709. doi: 10.1016/0042-6822(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 24.Jackers P, Defechereux P, Baudoux L, Lambert C, Massaer M, Merville-Louis M P, Rentier B, Piette J. Characterization of regulatory functions of the varicella-zoster virus gene 63-encoded protein. J Virol. 1992;66:3899–3903. doi: 10.1128/jvi.66.6.3899-3903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in the nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinchington P R, Bookey D, Turse S E. The transcriptional regulatory proteins encoded by varicella-zoster virus open reading frames (ORFs) 4 and 63, but not ORF 61, are associated with purified virus particles. J Virol. 1995;69:4274–4282. doi: 10.1128/jvi.69.7.4274-4282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinchington P R, Hougland J K, Arvin A M, Ruyechan W T, Hay J. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J Virol. 1992;66:359–366. doi: 10.1128/jvi.66.1.359-366.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinchington P R, Inchauspe G, Subak-Sharpe J H, Robey F, Hay J, Ruyechan W T. Identification and characterization of a varicella-zoster virus DNA-binding protein by using antisera directed against a predicted synthetic oligopeptide. J Virol. 1988;62:802–809. doi: 10.1128/jvi.62.3.802-809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maguire H F, Hyman R W. Polyadenylated, cytoplasmic transcripts of varicella-zoster virus. Intervirology. 1986;26:181–191. doi: 10.1159/000149700. [DOI] [PubMed] [Google Scholar]
- 29a.Mahalingam, R. Personal communication.
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 31.McLauchlan J, Liefkens K, Stow N D. The herpes simplex virus type 1 UL37 gene product is a component of virus particles. J Gen Virol. 1994;75:2047–2052. doi: 10.1099/0022-1317-75-8-2047. [DOI] [PubMed] [Google Scholar]
- 32.Meier J L, Holman R P, Croen K D, Smialek J E, Straus S E. Varicella-zoster virus transcription in human trigeminal ganglia. Virology. 1993;193:193–200. doi: 10.1006/viro.1993.1115. [DOI] [PubMed] [Google Scholar]
- 33.Meier J L, Straus S E. Varicella-zoster virus DNA polymerase and major DNA-binding protein genes have overlapping divergent promoters. J Virol. 1993;67:7573–7581. doi: 10.1128/jvi.67.12.7573-7581.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier J L, Luo X, Sawadogo M, Straus S E. The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol Cell Biol. 1994;14:6896–6906. doi: 10.1128/mcb.14.10.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostrove J M, Reinhold W, Fan C-M, Zorn S, Hay J, Straus S E. Transcription mapping of the varicella-zoster virus genome. J Virol. 1985;56:600–606. doi: 10.1128/jvi.56.2.600-606.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perera L P, Mosca J D, Ruyechan W T, Hay J. Regulation of varicella-zoster virus gene expression in human T lymphocytes. J Virol. 1992;66:5298–5304. doi: 10.1128/jvi.66.9.5298-5304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinhold W C, Straus S E, Ostrove J M. Directionality and further mapping of varicella zoster virus transcripts. Virus Res. 1988;9:249–261. doi: 10.1016/0168-1702(88)90034-2. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz J B, Albright A G, Kinchington P R, Jenkins F J. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology. 1995;206:1055–1065. doi: 10.1006/viro.1995.1028. [DOI] [PubMed] [Google Scholar]
- 39.Shelton L S, Albright A G, Ruyechan W T, Jenkins F J. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J Virol. 1994;68:521–525. doi: 10.1128/jvi.68.1.521-525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shelton L S G, Pensiero M N, Jenkins F J. Identification and characterization of the herpes simplex virus type 1 protein encoded by the UL37 open reading frame. J Virol. 1990;64:6101–6109. doi: 10.1128/jvi.64.12.6101-6109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyler J K, Everett R D. The DNA binding domain of the varicella-zoster virus gene 62 interacts with multiple sequences which are similar to the binding site of the related protein of herpes simplex virus type 1. Nucleic Acids Res. 1993;21:513–522. doi: 10.1093/nar/21.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu C L, Wilcox K W. The conserved DNA-binding domains encoded by the herpes simplex virus type 1 ICP4, pseudorabies virus IE180, and varicella-zoster virus ORF62 genes recognize similar sites in the corresponding promoters. J Virol. 1991;65:1149–1159. doi: 10.1128/jvi.65.3.1149-1159.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]