Abstract
The DNA sequence motifs pac1 [an A-rich region flanked by poly(C) runs] and pac2 (CGCGGCG near an A-rich region) are conserved near herpesvirus genomic termini and are believed to mediate cleavage of genomes from replicative concatemers. To determine their importance in the cleavage process, we constructed a number of recombinant murine cytomegaloviruses with a second cleavage site inserted at an ectopic location within the viral genome. Cleavage at a wild-type ectopic site occurred as frequently as at the natural cleavage site, whereas mutation of this ectopic site revealed that some of the conserved motifs of pac1 and pac2 were essential for cleavage whereas others were not. Within pac1, the left poly(C) region was very important for cleavage and packaging but the A-rich region was not. Within pac2, the A-rich region and adjacent sequences were essential for cleavage and packaging and the CGCGGCG region contributed to, but was not strictly essential for, efficient cleavage and packaging. A second A-rich region was not important at all. Furthermore, mutations that prevented cleavage also blocked duplication and deletion of the murine cytomegalovirus 30-bp terminal repeat at the ectopic site, suggesting that repeat duplication and deletion are consequences of cleavage. Given that the processes of genome cleavage and packaging appear to be highly conserved among herpesviruses, these findings should be relevant to other members of this family.
Herpesviruses have large (130- to 235-kb) double-stranded linear DNA genomes that circularize shortly after infection (18, 27, 39). Studies of herpes simplex virus type 1 (HSV-1), pseudorabies virus, and human cytomegalovirus (HCMV) suggest that viral DNA synthesis leads to the formation of concatemers of head-to-tail-linked genomes (8, 10, 21, 26, 27, 40, 43, 64). Evidence from HCMV suggests that DNA packaging initiates when a concatemer end associates with an empty capsid. Concatemeric DNA is then translocated into the capsid until a signal is reached and the DNA is cleaved, releasing the concatemer and leaving a unit-length genome within the nucleocapsid (27). Cleavage occurs at sites defined by specific cis-acting signals in the DNA; however, the observation that additional cleavage sites within the genomes of HSV-1 and HCMV and within HSV-1 defective genomes are only rarely utilized by the cleavage machinery suggests that cleavage is also restricted by a head-full packaging constraint (17, 39, 60). In the rare instances that subgenomic-sized fragments are packaged, data from HSV-1 defective genomes suggest that an additional mechanism prevents these capsids from leaving the nucleus (60).
In HSV-1, the cis-acting cleavage signals are located within the terminal repeat or a sequence, that is present as a single copy adjacent to the c sequence at the short-component terminus (ca), as one or more reiterated copies adjacent to the b sequence at the long-arm terminus (anb), and as one or more copies in inverted orientation (relative to the terminal a sequences) at the junction between the long and short components (b′a′mc′) (40). The a sequence is necessary and sufficient to direct cleavage and packaging of HSV-1 genomes (31–33).
Studies using HSV-1 amplicons, which consist of small tandemly repeated subgenomic segments containing an a sequence linked to a viral origin of replication, have defined the role of a sequences in cleavage and packaging (7, 15, 17, 49, 51). The importance of the a sequence in cleavage and packaging was demonstrated by the observation that DNA replication occurred following transfection of HSV-1 origin-containing plasmids into HSV-1-infected cells, but that concatemeric plasmid DNA was not cleaved or packaged unless ba, ca (51), or a sequences alone (15) were included in the plasmid.
Two sequence motifs designated pac1 and pac2 were first identified by their conservation at herpesvirus termini (14). They are found within the HSV-1 a sequence and at the termini of all herpesvirus genomes for which sequence data have been reported. pac1 motifs consist of a 3- to 7-bp A- or T-rich region flanked on each side by 5 to 7 C’s (14, 52); pac2 motifs consist of a 5- to 10-bp A-rich region that is often associated with a nearby CGCGGCG (14). Both pac1 and pac2 are located 30 to 35 bp from the genome termini, generally at opposite ends of the viral genome. Consequently, for most herpesviruses, cleavage to release genomes from concatemers occurs between pac1 and pac2 (14). Comparisons of naturally occurring cleavage sites (13, 19, 28, 44) and deletion mutagenesis of cleavage sites in defective genomes (14, 15, 34, 57, 65) or recombinant viruses (28, 48, 57) have shown that pac1 and pac2 lie within regions of DNA that are coincident with cis-acting cleavage elements. The most detailed of these studies relied on progressive deletions to map essential cleavage elements within the HSV-1 a sequence, but interpretation of these results was compromised by a-sequence duplication and recombination (48). Neither pac1 nor pac2 has been subjected to site-directed mutagenesis to evaluate its contribution to cleavage.
We addressed the roles of pac1 and pac2 in cleavage and packaging during murine cytomegalovirus (MCMV) replication because of the simplicity of both its cleavage site and its genome structure. The MCMV cleavage site consists of pac1 and pac2 flanking either one or two copies of a 30-bp repeat (25) and lacks the multiple direct repeats characteristic of the HSV-1 (32) and HCMV (30, 53) a sequences. Since the MCMV class F genome (39) lacks internal inverted repeats, invertible genome segments, and reiterated cleavage sites (16, (29), recombination between an ectopic cleavage site and the wild-type terminal sequences was anticipated to be minimal. We constructed a recombinant MCMV containing an additional, ectopic cleavage site and found that it was cleaved efficiently. We then constructed additional recombinant MCMVs with ectopic cleavage sites containing mutations within the conserved components of pac1 and pac2 and tested these viruses for cleavage at their ectopic cleavage sites. Here, we report the effects of mutations within the conserved components of pac1 and pac2 on cleavage and packaging.
MATERIALS AND METHODS
Cells and virus culture.
Except where otherwise noted, recombinant MCMVs were propagated in murine NIH 3T3 cells (ATCC CRL1658) as previously described (50).
Virion and infected cell DNA preparation.
Murine NIH 3T3 cells (107) were infected with recombinant viruses at a multiplicity of infection of 0.1 and incubated for 7 days, at which time virion and infected cell DNAs were prepared as previously described for guinea pig cytomegalovirus (GPCMV) (28).
Plasmid construction and mutagenesis.
Plasmid pON432 contains a 5.4-kb HpaI-EcoRI fragment cloned from MCMV DNA (nucleotides 184315 to 189670 in the MCMV genomic sequence [38]) and includes the HindIII site located at nucleotide position 187889 between the HindIII L and J fragments of the MCMV genome (50). Plasmid pMA34 contains an 860-bp HindIII-XhoI fragment from HindIII J (nucleotides 187889 to 188752 [see Fig. 1]) subcloned from pON432 into HindIII-XhoI-digested pMA10, a cloning vector derived by blunt-end ligation of an XhoI-containing linker (P-CCGCTCGAGCGG) into the HincII site of pUC18 (62).
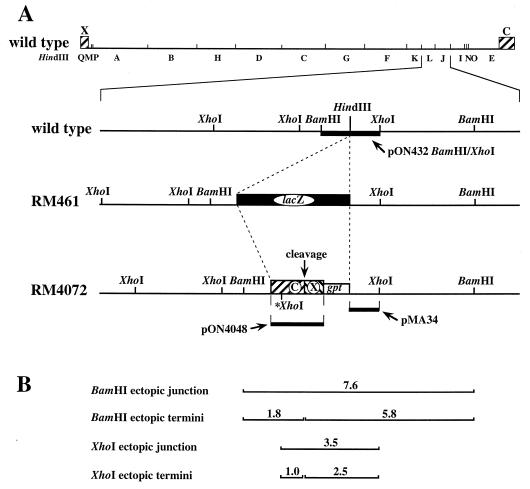
FIG. 1.
Structures of recombinant viruses containing ectopic cleavage sites. Shown is a representation of the wild-type MCMV genome and HindIII restriction map, with X- and C-terminal sequences shown as hatched boxes. The expanded regions show the HindIII site between L and J in wild-type virus, the lacZ insertion (black box) at this HindIII site in recombinant virus RM461, and the ectopic cleavage site inserted at the same location in recombinant virus RM4072. The ectopic site consists of a fusion of C- and X-terminal sequences (hatched boxes labeled X and C) adjacent to a gpt expression cassette (open box). The point at which cleavage is predicted to occur (where C and X sequences are fused) is indicated. The XhoI site marked with an asterisk was engineered into this region (see Materials and Methods). The thick lines indicate the DNA sequences contained in the indicated hybridization probes. (B) BamHI and XhoI fragments generated from recombinant viruses containing ectopic cleavage sites are predicted based on the locations of BamHI and XhoI sites and the predicted point of cleavage. The fragment sizes in kilobases are indicated.
All plasmids used in recombinant virus construction were derived from plasmid pON4072, which has the same structure, within the region of the ectopic cleavage site, as virus RM4072 (Fig. 1A). The ectopic cleavage site introduced into pON4072 was originally derived from plasmid E′ (kindly provided by D. Spector), which contains a fusion of the MCMV C and X termini (named with reference to the EcoRI C and X fragments found at these termini) (24). A 1.9-kb EcoRI-HindIII fragment (CX), consisting of the HindIII Q-terminal fragment (nucleotides 1 to 543) fused to the EcoRI C-terminal fragment (nucleotides 228920 to 230278), was subcloned from plasmid E′ into pUC18 to make pON4047. A KpnI site 1 kb from the point of fusion between C and X sequences (nucleotide position 229283) was converted to an XhoI site (indicated by an asterisk in Fig. 1A) by digestion of pON4047 with KpnI, treatment with T4 DNA polymerase and deoxynucleoside triphosphates (4), and blunt-end ligation to an XhoI linker (P-CCGCTCGAGCGG), to make pON4048. In plasmid pON4051, the modified CX fragment from pON4048 was cloned adjacent to an expression cassette from the Escherichia coli xanthine-guanine phosphoribosyltransferase (gpt) gene in pON1101 (58) by ligation of a HindIII-BsaI fragment from pON4048 (which contains the CX fragment, E. coli origin of replication, and part of the β-lactamase gene) to a HindIII-BsaI fragment from pON1101 (which contains the gpt cassette and the remainder of the β-lactamase gene). Finally, to make plasmid pON4072, an AseI-BamHI fragment from pON4051 containing the gpt/CX region was ligated to HindIII-digested pON432 after making all ends blunt by treatment with T4 DNA polymerase and deoxynucleoside triphosphates (4).
Plasmids described here have the same ectopic site sequences as those of recombinant viruses with the same number (replace pON with RM in designations). The sequences of all mutations as well as the locations of relevant restriction enzyme sites and PCR primers are shown in Fig. 2. To make pON4074, one of the two 30-bp repeats in pON4072 was deleted by digestion with ApaI (which cleaves pON4072 twice, once within each 30-bp repeat) followed by ligation of the ends. Plasmids pON4077 and pMA39 were constructed by digestion of pON4074 with XbaI and ApaI, followed by ligation to 50-bp double-stranded synthetic oligonucleotides engineered with XbaI and ApaI cohesive ends (for pON4077, 5′-CTAGAGGACAAAAATATAGCCCCCCCATGGCGTTCCCCCCCGGGGGGCC; for pMA39, 5′-CTAGAGGACAAAAATATAGCCATGGCAATCAAAATACCCCCCCGGGGGGCC).
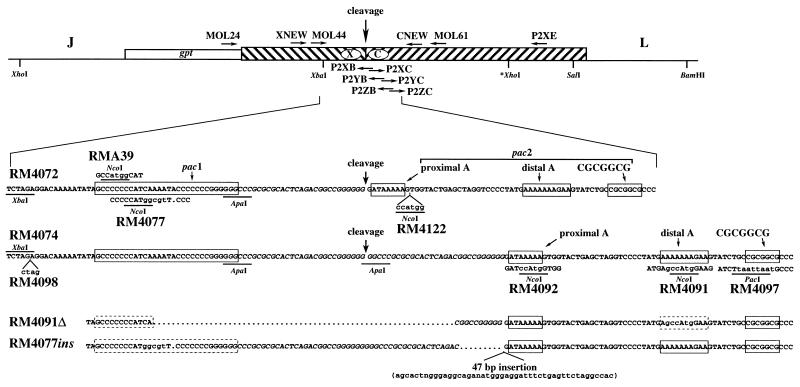
FIG. 2.
Sequences of ectopic cleavage sites and mutations. The ectopic cleavage site with flanking HindIII L and J sequences is shown as in Fig. 1 except that the orientation has been reversed. The predicted point of cleavage, restriction enzyme sites used in plasmid construction, and locations of oligonucleotide primers (horizontal arrows) used for PCR, mutagenesis, and sequencing are indicated. The expanded region shows the wild-type sequences (25) for cleavage sites with one or two copies of the 30-bp repeat (italics) found in plasmids used to make RM4072 and RM4074. The cleavage point predicted for each site is indicated with an arrow, and the predicted components of pac1 and pac2 are boxed. Mutations introduced into the ectopic cleavage sites of the indicated recombinant viruses are shown immediately above (RMA39) or below (RM4077, RM4122, RM4098, RM4092, RM4091, and RM4097) the wild-type sequences. Bases differing from wild type are in lowercase, and deleted bases are indicated by dots. Restriction sites used for mutagenesis or created by mutagenesis are underlined. The sequences of the ectopic cleavage sites of viruses RM4091Δ and RM4077ins which contain spontaneous deletions and an insertion are depicted at the bottom. Boxes enclose predicted components of pac1 and pac2 that have wild-type sequences (solid borders) or contain mutations (dashed borders).
The mutation in plasmid pON4091 was created by cloning two PCR products amplified from pON4048 DNA into the T/A cloning vector pCRII (Invitrogen). The primers contained mutations that introduced NcoI sites such that when the two fragments were cloned adjacent to one another by using NcoI, a complete cleavage site containing an NcoI mutation resulted. Plasmid pON4081 contains the PCR product from primers P2YC (5′-TATGAgccATgGAAGTATCTGCCGCGGCG; mutations shown in lowercase) and P2XE (5′-ACACGAACATCGTTATTACCT), and pON4083 contains the PCR product from primers P2YB (5′-ACTTCcaTggcTCATAGGGGACCTAGCCT) and XNEW (5′-TGCCACGCCCTCGGTGACGTGC). The two PCR-generated sequences were joined at the NcoI site by insertion of the XbaI-NcoI fragment from pON4083 into XbaI-NcoI-digested pON4081 to make pON4087. An XbaI-XhoI fragment containing the combined PCR-generated sequences from pON4087 was then used to replace the analogous XbaI-XhoI fragment in pON4048 to make pON4088; finally, an XbaI-SalI fragment from pON4088 was used to replace the analogous XbaI-SalI fragment of pON4072 to make pON4091.
Plasmid pON4092 was created in a similar way, using different primers. Plasmid pON4085 contains the PCR product from primers P2XC (5′-GGATccAtgGTGGTACTGAGCTAGGTC) and P2XE, and pON4082 contains the PCR product from primers P2XB (5′-CCACcaTggATCCCCCCCGGCCGTCTGA) and XNEW. In pON4089, the two PCR-generated sequences replaced analogous sequences in pON4048 by trimolecular ligation of XhoI-XbaI-digested pON4048 DNA, the NcoI-XbaI fragment of pON4082, and the NcoI-XhoI fragment of pON4085. Finally, an XbaI-SalI fragment from pON4089 was used to replace the analogous XbaI-SalI fragment of pON4072 to make pON4092.
The mutation in plasmid pON4097 was made the same way except that different PCR primers introduced a PacI site instead of an NcoI site. Plasmid pON4086 contains the PCR product from primers P2ZC (5′-TATCTtaattaatGCCCTCGGCGGCAAAAAACTG) and P2XE, and pON4084 contains the PCR product from primers P2ZB (5′-AGGGCattaattaAGATACTTCTTTTTTTCATA) and XNEW. In pON4093, the two PCR-generated sequences were joined at the PacI site by insertion of the XbaI-PacI fragment from pON4084 into XbaI-PacI-digested pON4086. An XbaI-XhoI fragment from pON4093 was then used to replace the analogous XbaI-XhoI fragment in pON4048 to make pON4094; finally, an XbaI-SalI fragment from pON4094 was used to replace the analogous XbaI-SalI fragment of pON4072 to make pON4097.
Plasmid pON4122 was constructed by digestion of pON4092 with ApaI and NcoI followed by ligation to a 43-bp double-stranded synthetic oligonucleotide engineered with ApaI and NcoI cohesive ends (5′GGCCCGCGCGCACTCAGACGGCCGGGGGGGATAAAAAGCCATG). Ectopic cleavage site mutations in all plasmids were confirmed by sequencing.
Virus construction.
Recombinant viruses were constructed by using gpt to select for homologous recombination of plasmid DNAs into the viral genome as described by Vieira et al. (58). For each virus construction, parental plasmid DNA was linearized with SpeI (which cuts all of these plasmids once within MCMV HindIII L sequences 3 kb from the ectopic cleavage site insertion) and electroporated into NIH 3T3 cells as previously described (28). Twenty-four hours after electroporation, cells were infected with MCMV recombinant virus RM461 at a multiplicity of infection of 5. RM461 contains a lacZ insertion at the same HindIII site as the gpt/CX insertion (Fig. 1A), and although this interrupts a 0.85-kb gamma transcript and disrupts expression of reading frames M129 and M131 (38), it does not alter viral growth in cultured cells (50). Therefore, it was anticipated that viruses with gpt/CX insertions at the same site would also grow normally in cultured cells. Three days after infection with RM461, the culture medium was clarified by low-speed centrifugation (800 × g for 5 min) and used to infect a fresh 25-cm2 flask of confluent NIH 3T3 cells. Three hours after infection, the cells were washed once with medium and incubated with 5 ml of medium containing 40 μM mycophenolic acid (Bethesda Research Laboratories) and 278 μM xanthine (Sigma) to select for gpt+ viruses. Three days after infection, the culture medium was clarified by centrifugation and used to infect fresh NIH 3T3 cells under the same conditions. When significant viral cytopathic effect was evident (typically 6 to 10 days after infection), clarified culture supernatants were used to infect fresh NIH 3T3 cells under the same conditions. When significant viral cytopathic effect was evident (typically 6 days after infection), virus clones were isolated by plating serial 10-fold dilutions of clarified culture supernatants into 96-well plates containing NIH 3T3 cells. Seven to ten days later, virus containing wells from plates with the fewest virus positive wells were screened for β-galactosidase expression (β-galactosidase negative viruses are likely to have replaced lacZ in RM461 with gpt/CX from the plasmid). Wells were incubated at 37°C for 1 to 3 h with 100 μM methylumbelliferyl β-d-galactoside (Sigma) in the medium, and fluorescence was observed on a UV transilluminator. Typically more than 50% of the virus-positive wells were β-galactosidase negative (failed to fluoresce) at this stage. β-Galactosidase-negative isolates were screened for the correct sequence at the ectopic site by PCR and sequencing (see below). For each virus constructed, one isolate was subcloned a second time by limiting dilution and sequenced (see below) prior to final assignment of a name.
DNA blot hybridization.
Restriction enzyme-digested virion or infected cell DNAs were electrophoresed on 0.6% SeaKem agarose (FMC) and transferred to nylon Nytran membranes (Schleicher & Schuell) as previously described (27). Hybridization probes, shown in Fig. 1A, were 32P labeled by random hexamer priming and hybridized to membranes as previously described (27).
PCR.
PCR was used to screen virus isolates for insertion of the gpt/CX cassette by amplification between oligonucleotide MOL24 (5′-CACTCCCTGAAGCTC) within the gpt sequence and oligonucleotide CNEW (5′-CCCACTCCACGCCATTCACTTG) or MOL61 (5′-CCGCACACCTCATCCTCAGCA) within the C-terminal sequence (Fig. 2). PCR was performed on infected cell lysates as previously described (28) except that the temperature cycles were as follows: 72°C for 2 min, 95°C for 1 min, and 63°C for 1 min. After amplification, 5-μl aliquots of the PCR products were separated by electrophoresis on 1.5% agarose gels and visualized with ethidium bromide and UV light. NcoI or PacI restriction sites introduced by mutations were detected by NcoI or PacI digestion of 5 μl of PCR product prior to electrophoresis. PCR was used to determine the number of 30-bp repeats in 1 ng of virion or plasmid DNAs, using primers MOL24 and CNEW and PCR conditions described above. PCR products were digested with SmaI, separated on a 6% polyacrylamide gel, and visualized as described above. For mutagenesis, PCR was carried out under the conditions specified above except that template DNA consisted of 500 ng of pON4048 plasmid DNA.
Nucleotide sequence analysis.
Sequencing was performed by the Sanger dideoxy method (41), using an fmol or TaqTrack sequencing kit (Promega) with 5′-end 32P-labeled synthetic oligonucleotide primers according to the manufacturer’s instructions. Ectopic cleavage sites in plasmids and PCR products derived from recombinant viruses were sequenced by using oligonucleotide primers MOL44 (5′-ACAGCGCCCTCTAGACCACA) and XNEW, near the X terminus, and CNEW, near the C terminus (Fig. 2). PCR products from recombinant viruses were extracted once with phenol-chloroform and once with chloroform and then washed three times with TE (10 mM Tris [pH 8.0], 1 mM EDTA), using a Centricon 30 filter (Amicon), prior to sequencing.
RESULTS
Cleavage at an ectopic cleavage site.
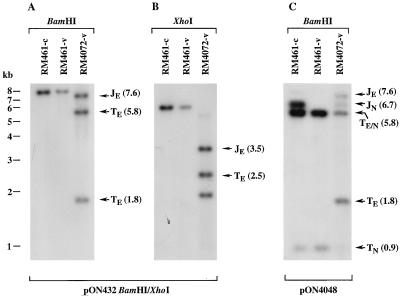
To determine whether a fragment containing a fusion of MCMV termini would be recognized by the cleavage and packaging machinery, we constructed recombinant virus RM4072 (Fig. 1A). Based on the head-full packaging constraints implied by studies with HSV-1 defective genomes (15), we predicted that RM4072 concatemeric replicative intermediates would be cleaved in either of two frames, producing genomes with either natural or ectopic termini. Using DNA blot hybridization, we detected restriction fragments consistent with cleavage at ectopic as well as natural sites. DNAs from RM4072 and the parental virus RM461 were digested with BamHI or XhoI, separated electrophoretically, and hybridized with a 32P-labeled gel-purified BamHI/XhoI fragment probe (Fig. 1A), which contained sequences flanking the ectopic cleavage site. BamHI-digested RM4072 virion DNA contained 7.6-kb ectopic junction and 5.8-, and 1.8-kb ectopic terminal fragments in approximately equimolar amounts (Fig. 3A). XhoI-digested RM4072 virion DNA contained equimolar amounts of a 3.5-kb ectopic junction and a 2.5-kb ectopic terminal fragment (Fig. 3B). A 1.0-kb ectopic terminal fragment was also generated (data not shown) but did not hybridize with this probe (Fig. 1B). The detection of ectopic terminal fragments at levels similar to those for ectopic junction fragments demonstrated that about 50% of RM4072 genomes were formed by cleavage at the ectopic sites, suggesting that ectopic sites were utilized as frequently as natural sites. Thus, the fused MCMV termini represented an authentic cleavage and packaging signal.
FIG. 3.
Cleavage at the ectopic cleavage site of RM4072. Autoradiograms show cell-associated (-c) and virion (-v) DNAs from the parental virus RM461 and from RM4072 digested with BamHI or XhoI and hybridized with 32P-labeled probes following electrophoresis and transfer to nylon. (A and B) Results of hybridization with a BamHI/XhoI fragment from pON432 which detected fragments from ectopic junctions and termini; (C) results of hybridization with pON4048 DNA to detect fragments from both ectopic and natural cleavage sites and termini. The positions of molecular size markers are shown on the left of panel A, and the locations and sizes in kilobases (in parentheses) of BamHI and XhoI ectopic junction (JE), natural junction (JN), ectopic terminal (TE), natural terminal (TN), and comigrating ectopic and natural terminal (TE/N) fragments are indicated. Note that in panel B, an irrelevant 2.0-kb XhoI fragment adjacent to the ectopic cleavage site also hybridizes to the probe.
Whereas the parental virus RM461 cleaved natural sites completely, recombinant RM4072 appeared to cleave at natural sites only 50% of the time. To provide additional evidence that cleavage occurred at ectopic sites within RM4072 concatemeric DNA, BamHI-digested RM4072 was hybridized to a probe consisting of the ectopic cleavage site from pON4048. This probe detected a natural junction fragment of 6.7 kb and natural termini of 5.8 and 0.9 kb in digested RM461-infected cell DNA but only the terminal fragments in virion DNA. In contrast, RM4072 virion DNA contained the natural junction and natural terminal fragments as well as the ectopic junction fragment of 7.6 kb and ectopic terminal fragments of 5.8 and 1.8 kb (Fig. 3C). These results were consistent with our prediction that a head-full restriction would cause cleavage to occur in two frames, each frame leaving alternate cleavage sites uncleaved, and confirmed that ectopic cleavage sites were recognized in recombinant virus RM4072. The similar amounts of ectopic and natural junction fragments indicated that cleavage was equally efficient at natural and ectopic sites (Fig. 3C).
Mutations in the cleavage site.
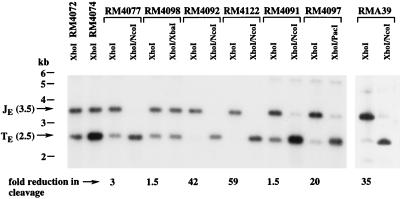
Cleavage occurs at sites with either one or two copies of a 30-bp repeat that are flanked on one side by pac1 and on the other by pac2 (25). Within this region are two A-rich regions which may constitute the A-rich component of pac2, one close to the C terminus (proximal A-rich region) the other more distant (distal A-rich region) (Fig. 2). To determine which sequence elements of pac1 and pac2 were necessary for cleavage and packaging, mutations were introduced into the pac1 A-rich and left poly(C) regions, the pac2 proximal and distal A-rich regions, and the pac2 CGCGGCG motif. In each case, mutations created a new restriction site that significantly altered the natural nucleotide sequence character. A-rich regions and the pac1 left poly(C) region were converted to NcoI sites (CCATGG), and the CGCGGCG motif was converted to an A/T-rich PacI site (TTAATTAA). In addition, a 6-bp NcoI site was inserted adjacent to the pac2 proximal A-rich region, and a 4-bp insertion was made into an XbaI site in a region lacking any conserved sequence elements (Fig. 2). Each recombinant virus was sequenced through the ectopic cleavage site to confirm the predicted sequence and exclude the possibility of spontaneous mutations, and the introduced restriction enzyme sites were used to confirm the presence of the mutations within the virion DNAs. The level of cleavage at the ectopic sites was assessed by hybridization of XhoI-digested virion DNAs with 32P-labeled pMA34 DNA (Fig. 1), a probe from the HindIII J region that was predicted to hybridize to the 3.5-kb ectopic junction and 2.5-kb ectopic terminal fragments (Fig. 1). Mutations that reduced the efficiency of cleavage at ectopic sites would reduce the abundance of the 2.5-kb terminal fragments and increase the abundance of 3.5-kb junction fragments. The ratio of 2.5-kb to 3.5-kb fragment intensities for RM4072 was determined by densitometric quantitation of the 3.5- and 2.5-kb fragments and was then divided by the ratio for each mutant virus to determine the fold reductions in cleavage efficiency for mutant cleavage sites relative to the wild-type cleavage site. Results were confirmed by hybridization of BamHI-digested virion DNAs to the BamHI/XhoI pON432 probe, which detected both ectopic terminal fragments and ectopic junction fragments (not shown).
Mutations in or near the pac2 proximal A-rich region had the most profound effects on cleavage, as evidenced by the virtual absence of 2.5-kb fragments. Densitometric quantitation estimated that the RM4092 mutation, which carried a clustered mutation within the pac2 proximal A-rich region (AAAAA to CCATG), reduced cleavage efficiency 42-fold, and the RM4122 mutation, which carried an insertion mutation adjacent to the proximal A-rich region, reduced efficiency 59-fold (Fig. 4). Mutation of the pac2 CGCGGCG motif to ATTAATTG in virus RM4097 reduced cleavage efficiency 20-fold (Fig. 4). Mutations within the pac1 left poly(C) region (CCCCCCC to CCATGGC) in RMA39 reduced cleavage 35-fold (Fig. 4). Somewhat surprisingly, disruption of the pac1 A-rich region (AAAAA to GGCGGCG) in RM4077 had a modest threefold effect on cleavage (Fig. 4), a level that was consistently observed when progeny from independent infections were analyzed (not shown). In contrast, mutation of the pac2 distal A-rich region (AAAAAA to GCCATG) in RM4091 and insertion of 4 bp into an XbaI site in nonconserved sequences 14 bp to the left of pac1 in RM4098 reduced cleavage efficiency only 1.5-fold (Fig. 4). These findings demonstrated that only the proximal A-rich region was required for cleavage and packaging; the distal region was dispensable. Thus, the proximal region should be considered to be a functional component of pac2. Sequences to the right of the proximal A-rich region were also important for cleavage, and although not strictly required, the CGCGGCG motif contributed significantly to the frequency of cleavage. Within pac1, the left poly(C) region was of considerable importance but the A-rich region was not.
FIG. 4.
Cleavage at mutated ectopic cleavage sites. Autoradiograms of virion DNAs from recombinant viruses digested with the indicated restriction enzymes and hybridized with 32P-labeled pMA34 DNA to detect ectopic junction and terminal fragments following electrophoresis and transfer to nylon. For each virus, the fold reduction in cleavage is estimated from densitometric quantitation of the 2.5- and 3.5-kb fragments in the lane above (for details, see Results). The positions of molecular size markers are shown on the left, and arrows indicate the 3.5-kb XhoI ectopic junction fragments (JE) and 2.5-kb XhoI ectopic terminal (TE) fragments. Analysis of RMA39 was carried out in an experiment separate from the other viruses.
Spontaneous mutations at the ectopic cleavage site.
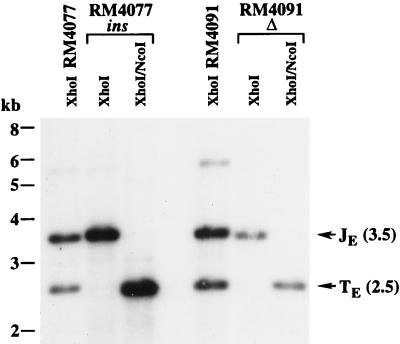
During construction of RM4077 and RM4091, a single isolate of each exhibited a complete failure to cleave ectopic sites. Nucleotide sequencing revealed that these two isolates contained adventitious mutations. One isolate, designated RM4077ins, contained the expected pac1 A-rich region mutation and had a 47-bp insertion disrupting one of the two 30-bp repeats (Fig. 2). The first 13 nucleotides of the 47-bp insertion matched nucleotides 139721 to 139733 of the MCMV genomic sequence, within the M96 reading frame (38), but the remaining 34 bp were apparently not derived from the MCMV genome. The other isolate, designated RM4091Δ, contained the expected mutation in pac2 and had undergone a spontaneous 35-bp deletion removing most of the pac1 A-rich region and 19 bp of the 30-bp repeat (Fig. 2). Cleavage assays confirmed that both RM4077 and RM4091 cleaved efficiently, whereas the viruses with additional mutations failed to cleave at the ectopic site to any detectable level (Fig. 5). The deletion in RM4091Δ may have removed additional sequences required for cleavage, such as the pac1 right poly(C) region or the 30-bp repeat, and the 47-bp insertion in RM4077ins may have inactivated the pac2 A-rich region; however, an alternative explanation is that these mutations altered critical spacing between pac1 and pac2 and thus disrupted their cleavage function.
FIG. 5.
Cleavage at ectopic cleavage sites containing spontaneous mutations. The autoradiogram shows virion DNAs from recombinant viruses digested with the indicated restriction enzymes and hybridized with 32P-labeled pMA34 DNA to detect ectopic junction and terminal fragments following electrophoresis and transfer to nylon. The positions of molecular size markers are shown on the left, and arrows indicate the 3.5-kb XhoI ectopic junction fragments (JE) and 2.5-kb XhoI ectopic terminal (TE) fragments.
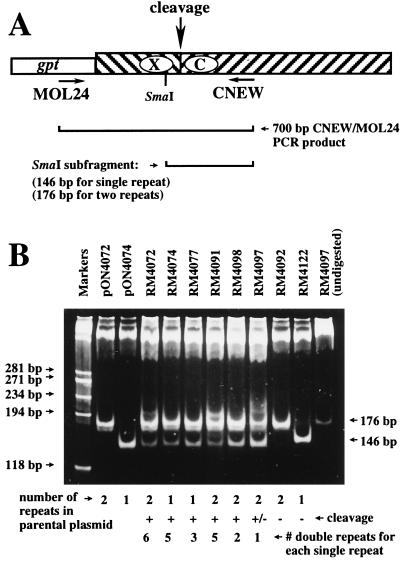
Deletion and duplication of the 30-bp terminal repeat.
The natural cleavage sites of wild-type MCMV contain either single or double 30-bp repeats (25). Virus RM4072 was constructed from a plasmid (pON4072) that contained two copies of the 30-bp repeat element. To determine whether cleavage would occur at an ectopic cleavage site derived from a single 30-bp repeat element, RM4074 was constructed from a plasmid that is identical to pON4072 but contained a single 30-bp repeat. The ectopic cleavage site of virus RM4074 was cleaved as efficiently as the site in RM4072 (Fig. 4). To determine whether RM4072 and RM4074 retained the number of 30-bp repeats present in the plasmids from which they were derived, virion DNAs were PCR amplified by using primers CNEW and MOL24 (Fig. 6A). The resulting 700-bp PCR products were digested with SmaI and separated on a 6% polyacrylamide gel. The PCR product derived from plasmid pON4074 contained a 146-bp SmaI fragment predicted for a single 30-bp repeat, and the PCR product derived from plasmid pON4072 contained a 176-bp SmaI fragment (Fig. 6B). The PCR product derived from either RM4072 or RM4074 DNA contained two SmaI fragments identical in size to those derived from pON4072 and pON4074, indicating that both single and double repeats were present at the ectopic cleavage sites of virus RM4072 (Fig. 6B). Thus, deletion or duplication of the 30-bp repeat generated genomes containing single and double 30-bp repeats at the ectopic cleavage sites of recombinant viruses. Only ectopic sites that were efficiently cleaved exhibited a mixture of single and double 30-bp repeats. Viruses with ectopic cleavage sites that were not cleaved (RM4092 and RM4122) retained the same number of repeats as the plasmids used in their construction (Fig. 6B). The ratio of single- to double-repeat-containing fragments in viruses that were cleaved was estimated by densitometry and ranged from 1:6 for RM4072 to 1:1 for RM4097 (Fig. 6B). The process by which the 30-bp repeat is duplicated appears to be integral to the process of cleavage and packaging.
FIG. 6.
Distribution of 30-bp repeats at ectopic cleavage sites of recombinant viruses. (A) Representation of the ectopic cleavage site as in previous figures, showing the positions of primers MOL24 and CNEW, the predicted PCR product, and SmaI restriction digest products. Ectopic cleavage sites with one 30-bp repeat are predicted to result in 146-bp SmaI fragments, and those with two 30-bp repeats are predicted to result in 176-bp SmaI fragments. (B) Ethidium bromide-stained polyacrylamide gel showing SmaI-digested PCR products amplified from plasmids pON4072 and pON4074 or from virion DNAs of recombinant viruses, using MOL24 and CNEW. The final lane contains the PCR product from RM4097 virion DNA without SmaI digestion. Below the lanes are indicated the number of 30-bp repeats in plasmids pON4072 and pON4074 or in the plasmids used to construct each recombinant virus, whether the ectopic cleavage site is cleaved (+), not cleaved (−), or inefficiently cleaved (±), and the estimated ratio of double- to single-repeat-containing fragments. The sizes of HaeIII fragments of φX174 replicative DNA (markers) are indicated on the left, and arrows on the right indicate the 146- and 176-bp SmaI fragments.
DISCUSSION
Although the signals for herpesvirus genome cleavage and packaging have been localized to regions within the HSV-1 a sequence (14, 48, 57) and are proposed to include the conserved sequence elements pac1 and pac2 (14), definition of HSV-1 cleavage/packaging signals by mutagenesis has been hindered by the structure of the HSV-1 genome, the variable number of a sequences at genome cleavage sites, and the complexity of sequences within the a sequence. In the present study, cleavage/packaging signals were evaluated in the context of a herpesvirus with minimal cleavage site complexity and a simple genome structure which does not undergo inversion. A 1.9-kb fusion of sequences from each end of the MCMV genome was shown to support efficient cleavage and packaging. This finding demonstrated that all of the cis signals necessary for cleavage and packaging are contained within these sequences and that ectopic placement of the cleavage site does not affect the cleavage/packaging process. Mutations targeting the conserved sequence elements pac1 and pac2 demonstrated that these elements are coincident with cis cleavage/packaging elements. These results indicate that this system provides an effective tool for defining herpesvirus cleavage and packaging sequences on the basis of function rather than homology.
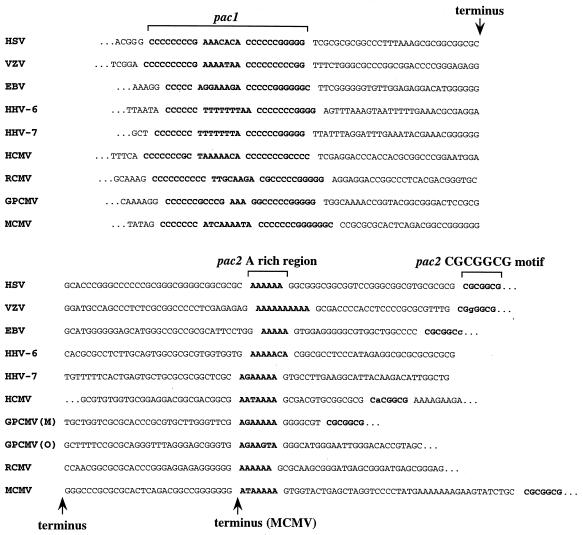
The importance of pac1 in cleavage and packaging was demonstrated by a mutation in the left poly(C) region that virtually eliminated cleavage and packaging at the ectopic site. A mutation in the adjacent A-rich region reduced cleavage and packaging only threefold. This was surprising since A- or T-rich regions are invariably located between poly(C) tracts in herpesvirus pac1 elements (13, 14, 25, 28, 30, 33, 42, 52, 55, 59, 65). These A- or T-rich regions, however, vary significantly in size and composition (Fig. 7), suggesting that A or T richness rather than specific sequences is important for this region. The pac2 A-rich and CGCGGCG components were also found to have important roles in cleavage and packaging. Mutation of the proximal A-rich region virtually eliminated cleavage and packaging but mutation of the distal A-rich region had no significant effect, indicating that the proximal A-rich region should be considered the A-rich component of the MCMV pac2. Mutation of the CGCGGCG motif did not eliminate cleavage and packaging but reduced its frequency 20-fold, indicating that the CGCGGCG motif contributes significantly to the efficiency of cleavage and packaging but is not strictly essential. The observation that the A-rich region is more important than the CGCGGCG motif for cleavage and packaging may suggest that these elements have separate functions. The fact that they are separated by sequences having no role in cleavage or packaging (the distal A-rich region) supports the notion that they are distinct elements. Thus, designation of the A-rich region and CGCGGCG motif as components of pac2 may be artificial.
FIG. 7.
Conserved sequences at herpesvirus termini. Alignments of the terminal sequences of HSV-1 (33), varicella-zoster virus (VZV) (13), Epstein-Barr virus (EBV) (65), HCMV (30), human herpesvirus (HHV-6) (55), human herpesvirus rat (HHV-7) (42), cytomegalovirus (RCMV) (59), GPCMV (28), and MCMV (25). Genomic termini are indicated, with the exception of the HCMV pac2-containing terminus, which has an additional 97 bp between the left end of the sequence and the terminus. The conserved components of pac1 and pac2 are shown in boldface and set off by spaces. Bases within the CGCGGCG motifs of varicella-zoster virus and Epstein-Barr virus that differ from consensus are in lowercase. GPCMV has two pac2-containing termini called M and O. An alternative terminus found in some MCMV genomes is indicated (25).
Our results confirm the premise that conserved sequences at herpesvirus termini (pac1 and pac2) function in cleavage and packaging, which was implied by sequence conservation near the cleavage sites of a variety of herpesviruses (13, 19, 28, 44), and was reinforced by deletion analysis using defective genomes (14, 15, 34, 57, 65) and recombinant viruses (28, 48, 57). In the most detailed of these studies (48), Smiley et al. inserted a-sequence segments with progressive deletions into an ectopic site in the HSV-1 genome and observed a marked decline in cleavage even before the deletions encroached on pac1 or pac2. Larger deletions that removed pac1 or pac2 had no additional effect (48). In our work, both pac1 and pac2 were critical for cleavage and packaging when flanking sequences were left intact. The results of Smiley et al. (48) can be reconciled with our results by a need for proper spacing between pac1 and pac2 since the deletions reported by Smiley et al. (48) may have reduced cleavage efficiency not by removing important sequences but rather by altering the spacing of pac1 relative to pac2. More detailed mutagenesis is needed to accurately define the sequence elements involved in MCMV cleavage and packaging and to determine the importance of their spatial relationships.
Our experiments did not segregate cleavage from packaging, and therefore the sequence elements identified could function in cleavage, packaging, or both. Evidence that pac1 elements and pac2 A-rich regions are located conserved distances from their respective termini (Fig. 7), that spacing between pac1 and pac2 is important (discussed above), and that cleavage is metered a specific distance from pac1 and pac2 (57) suggests that cleavage simultaneously involves pac1 and pac2. After cleavage, cis-acting sequences may be required at concatemer ends for initiation of concatemer packaging, as suggested by evidence that replicative concatemers are packaged directionally (27, 47). Examination of concatemeric DNAs from HSV-1 (26, 43, 64), equine herpesvirus type 1 (47), GPCMV (20), and MCMV (20) revealed that in each case, termini on concatemeric DNA contain pac2. Taken together, these data suggest that proteins bind pac1 and pac2 to initiate assembly of a complex that cleaves the intervening DNA at a precise location, generating a pac1-containing end on the newly formed genome and a pac2-containing end on the concatemer. Subsequently, pac2 may direct binding of the concatemer end to capsid proteins to initiate the next round of packaging.
Cleavage has been proposed to duplicate the HSV-1 a sequence (15, 57), but this has been difficult to prove owing to the variable number of a sequences at HSV-1 termini and junctions. Our observation that cleavage is required for duplication and deletion of the MCMV 30-bp terminal repeat is the first data to link these processes by mutagenesis of the cleavage site. For viruses with wild-type ectopic sites, the 6:1 ratio of double- to single-repeat-containing sites that we observed in a PCR-based assay is higher than the 2:1 ratio estimated by Marks and Spector using DNA blot hybridization to detect natural MCMV cleavage sites (25). This difference may be attributable to differences between the natural and ectopic sites and may suggest that sequences at the natural cleavage site, but outside the region duplicated at the ectopic site, have some impact on repeat duplication. Alternatively, the difference could result from differences in the techniques used to measure the ratios. Alteration of the ratio by mutations is not related to the number of repeats in the plasmids used for construction of the viruses (Fig. 6) and is most pronounced for a mutation in the CGCGGCG motif, for which the ratio is 1:1. This may indicate that the CGCGGCG motif has a role in repeat duplication since inhibition of duplication would result in an increase in the prevalence of single-repeat-containing ectopic sites. How cleavage results in duplication of terminal repeats is not known, but a possible role for cleavage in deletion of terminal repeats is suggested by the observation that fragments containing HSV-1 a sequences, authentically cleaved at each end, accumulate concomitantly with the beginning of cleavage (56). This implies that repeats may be occasionally excised when cleavage occurs on both sides of the repeat.
Failure to cleave half of the ectopic and natural cleavage sites suggests that cleavage occurs at alternate sites and implies that MCMV has a head-full packaging restriction; however, data from defective HSV-1 genomes (60) raise the possibility that occasional cleavages at successive sites (one natural and one ectopic, or vice versa) could generate subgenomic fragments that are packaged but fail to mature from the nucleus. As intracellular DNAs were not examined, the extent to which this may occur is not known; however, the efficient growth of recombinant viruses indicates that cleavage at alternate sites occurs frequently.
The 1.9 kb of terminal sequences that comprise our ectopic site may contain additional cis cleavage/packaging elements that are not coincident with pac1 and pac2. A 6-bp sequence located between pac1 and pac2 is strongly conserved among rodent cytomegaloviruses (i.e., murine, rat, and guinea pig cytomegaloviruses) and is associated with pac2 at the termini of other herpesviruses (28). Its function and importance have not been determined. Other data suggest that sequences outside the pac1-pac2 region can be important for cleavage and packaging. In Epstein-Barr virus, a plasmid DNA packaging assay revealed that sequences within an 84-bp region on the distal side of pac1 (relative to the point of cleavage) are necessary for cleavage and packaging (65). In varicella-zoster virus, a region of the natural cleavage site containing pac1 and pac2 is duplicated at the junction between the long and short arms of the genome, yet this site is cleaved only 5% of the time, suggesting that important sequences present outside the pac1-pac2 region at the natural cleavage site must be lacking from the duplicated site (13).
Finally, the proteins that interact with cis sequence elements to carry out these functions have not been clearly identified. Mutations in herpesvirus-conserved genes represented by HSV-1 open reading frames UL6, UL15, UL25, UL28, UL32, and UL33 block cleavage and packaging (1–3, 5, 6, 23, 36, 37, 45, 46, 54, 61, 63), but it is not known if the proteins encoded by these genes have direct roles in cleavage and packaging or if they bind to cis-acting DNA sequences. HSV-1 infection induces a 21-kDa protein and a 22-kDa protein that interact with the HSV-1 a sequence, but their roles in cleavage or packaging have not been clearly established (12). The ICP1 protein of HSV-1 (UL36) binds to pac2 in a complex that includes an unidentified 140-kDa infected cell protein (11), but a temperature-sensitive mutation in UL36 suggests it is involved in release of DNA from the capsid after infection (9), and a role for ICP1 in cleavage has not been pursued. Kemble and Mocarski failed to identify viral proteins binding to the HCMV cleavage site but found a cellular factor that binds to the pac2 A-rich region (22). These and recent findings of cellular proteins binding to the MCMV pac1 (35) suggest that the DNA binding components of the cleavage/packaging machinery may be derived from the host cell.
We have developed a simple system for defining the DNA sequences required for cleavage and packaging of herpesvirus genomes and have shown that both pac1 and pac2 are cis-acting cleavage/packaging elements. Accurate determination of these cis DNA elements may lead to identification of the proteins which interact with these sequences to carry out cleavage and packaging. Further experiments will be needed to determine their functions and to elucidate the mechanisms by which cleavage and packaging occur. The remarkable conservation of the cleavage/packaging elements pac1 and pac2 indicates that the processes of cleavage and packaging are highly conserved within the Herpesviridae. Therefore, information gained from analysis of the MCMV cleavage site should be applicable to other members of this family.
ACKNOWLEDGMENTS
We are grateful to Maria Kirichenko for valued technical assistance and to Deborah Spector for providing plasmid E′.
M.A.M. was supported in part by Public Health Service grant 5T32AIO7328. This work was supported by Public Health Service grant RO1AI20211 (to E.S.M.).
REFERENCES
- 1.Addison C, Rixon F J, Palfreyman J W, O’Hara M, Preston V G. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology. 1984;138:246–259. doi: 10.1016/0042-6822(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 2.Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 3.Al-Kobaisi M F, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 5.Baines J D, Cunningham C, Nalwanga D, Davison A J. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines J D, Poon A P W, Rovnak J, Roizman B. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnett J W, Epstein D A, Chan H W. Class I defective herpes simplex virus DNA as a molecular cloning vehicle in eucaryotic cells. J Virol. 1983;48:384–395. doi: 10.1128/jvi.48.2.384-395.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bataille D, Epstein A. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology. 1994;48:384–388. doi: 10.1006/viro.1994.1498. [DOI] [PubMed] [Google Scholar]
- 9.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Porat T. Replication of herpesvirus DNA. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Press; 1983. pp. 81–106. [Google Scholar]
- 11.Chou J, Roizman B. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J Virol. 1989;63:1059–1068. doi: 10.1128/jvi.63.3.1059-1068.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalziel R G, Marsden H S. Identification of two herpes simplex virus type 1-induced proteins (21k and 22k) which interact specifically with the a sequence of herpes simplex virus DNA. J Gen Virol. 1984;65:1467–1475. doi: 10.1099/0022-1317-65-9-1467. [DOI] [PubMed] [Google Scholar]
- 13.Davison A J. Structure of the genome termini of varicella-zoster virus. J Gen Virol. 1984;65:1969–1977. doi: 10.1099/0022-1317-65-11-1969. [DOI] [PubMed] [Google Scholar]
- 14.Deiss L P, Chou J, Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986;59:605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deiss L P, Frenkel N. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986;57:933–941. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebling A, Keil G M, Knust E, Koszinowski U H. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J Virol. 1983;47:421–433. doi: 10.1128/jvi.47.3.421-433.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frenkel N. Defective interfering herpesviruses. In: Hahmias A H, Dowdle W R, Schinazy R S, editors. The human herpesviruses—an interdisciplinary perspective. New York, N.Y: Elsevier Science Publishing, Inc.; 1981. pp. 91–120. [Google Scholar]
- 18.Garber D, Beverly S, Coen D. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology. 1993;197:459–462. doi: 10.1006/viro.1993.1612. [DOI] [PubMed] [Google Scholar]
- 19.Hammerschmidt W, Ludwig H, Buhk H. Specificity of cleavage in replicative-form DNA of bovine herpesvirus 1. J Virol. 1988;62:1355–1363. doi: 10.1128/jvi.62.4.1355-1363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hur, J., D. E. Nixon, and M. A. McVoy. Unpublished results.
- 21.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemble G W, Mocarski E S. A host cell protein binds to a highly conserved sequence element (pac-2) within the cytomegalovirus a sequence. J Virol. 1989;63:4715–4728. doi: 10.1128/jvi.63.11.4715-4728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamberti C, Weller S K. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 24.Marks J R, Spector D H. Fusion of the termini of the murine cytomegalovirus genome after infection. J Virol. 1984;52:24–28. doi: 10.1128/jvi.52.1.24-28.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marks J R, Spector D H. Replication of the murine cytomegalovirus genome: structure and role of the termini in generation and cleavage of concatenates. J Virol. 1988;162:98–107. doi: 10.1016/0042-6822(88)90398-4. [DOI] [PubMed] [Google Scholar]
- 26.Martinez R, Sarisky R, Webber P, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McVoy M A, Adler S P. Human cytomegalovirus DNA replicates after early circularization and inversion occurs within the concatemer. J Virol. 1994;68:1040–1061. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McVoy M A, Nixon D E, Adler S P. Cleavage and circularization of guinea pig cytomegalovirus genomes. J Virol. 1997;71:4209–4217. doi: 10.1128/jvi.71.6.4209-4217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercer H A, Marks J R, Spector D H. Molecular cloning and restriction endonuclease mapping of the murine cytomegalovirus genome (Smith strain) Virology. 1983;129:94–106. doi: 10.1016/0042-6822(83)90398-7. [DOI] [PubMed] [Google Scholar]
- 30.Mocarski E S, Liu A C, Spaete R R. Structure and variability of the a sequence in the genome of human cytomegalovirus (Towne strain) J Gen Virol. 1987;68:2223–2230. doi: 10.1099/0022-1317-68-8-2223. [DOI] [PubMed] [Google Scholar]
- 31.Mocarski E S, Post L E, Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980;22:243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- 32.Mocarski E S, Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci USA. 1981;78:7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mocarski E S, Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization, and generation of virion DNA. Cell. 1982;31:89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- 34.Nasseri M, Mocarski E S. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology. 1988;167:25–30. doi: 10.1016/0042-6822(88)90050-5. [DOI] [PubMed] [Google Scholar]
- 35.Nixon, D. E., and M. A. McVoy. 1997. Unpublished data.
- 36.Patel A H, Rixon F J, Cunningham C, Davison A J. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology. 1996;217:111–123. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- 37.Poon A P W, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus type 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawlinson W D, Farrell H E, Barrell B C. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roizman B, Sears A E. The herpesviridae, a brief introduction. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 1–9. [Google Scholar]
- 40.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, editor. Fundamental virology. New York, N.Y: Raven Press; 1996. pp. 1048–1066. [Google Scholar]
- 41.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Secchiero P, Nicholas J, Deng H, Xiaopeng T, VanLoon N, Ruvolo V, Berneman Z N, Reitz M S, Dewhurst S. Identification of human telomeric repeat motifs at the genome termini of human herpesvirus 7: structural analysis and heterogeneity. J Virol. 1995;69:8041–8045. doi: 10.1128/jvi.69.12.8041-8045.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Severini A, Morgan A R, Tovell D R, Tyrrell D L J. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–436. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 44.Shafiqul I C, Buhk H, Ludwig H, Hammerschmidt W. Genomic termini of equine herpesvirus 1. J Virol. 1990;64:873–880. doi: 10.1128/jvi.64.2.873-880.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherman G, Bachenheimer S. DNA processing in temperature sensitive morphogenic mutants of HSV-1. Virology. 1987;158:427–430. doi: 10.1016/0042-6822(87)90214-5. [DOI] [PubMed] [Google Scholar]
- 46.Sherman G, Bachenheimer S. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 47.Slobedman B, Simmons A. Concatemeric intermediates of equine herpesvirus type 1 DNA replication contain frequent inversions of adjacent long segments of the viral genome. Virology. 1997;229:415–420. doi: 10.1006/viro.1997.8447. [DOI] [PubMed] [Google Scholar]
- 48.Smiley J R, Duncan J, Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus 1 KOS DNA. J Virol. 1990;64:5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spaete R R, Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982;30:295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- 50.Stoddart C A, Cardin R D, Boname J M, Manning W C, Abenes G B, Mocarski E S. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J Virol. 1994;68:6243–6253. doi: 10.1128/jvi.68.10.6243-6253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stow N, McMonagle E, Davison A J. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 1983;11:8205–8820. doi: 10.1093/nar/11.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamashiro J C, Filpula D, Friedmann R, Spector D H. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169. J Virol. 1984;52:541–548. doi: 10.1128/jvi.52.2.541-548.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamashiro J C, Spector D H. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J Virol. 1986;59:591–604. doi: 10.1128/jvi.59.3.591-604.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomson B J, Dewhurst S, Gray D. Structure and heterogeneity of the a sequences of human herpesvirus 6 strain variants U1102 and Z29 and identification of human telomeric repeat sequences at the genomic termini. J Virol. 1994;68:3007–3014. doi: 10.1128/jvi.68.5.3007-3014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Umene K. Excision of DNA fragments corresponding to the unit-length a sequence of herpes simplex virus type 1 and terminus variation predominate on one side of the excised fragment. J Virol. 1994;68:4377–4383. doi: 10.1128/jvi.68.7.4377-4383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varmuza S L, Smiley J R. Signals for site-specific cleavage of HSV-1 DNA maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985;41:793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- 58.Vieira J, Farrell H E, Rawlinson W D, Mocarski E S. Genes in the HindIII J fragment of the murine cytomegalovirus genome are dispensable for growth in cultured cells: insertion mutagenesis with the lacZ/gpt cassette. J Virol. 1994;68:4037–4046. doi: 10.1128/jvi.68.8.4837-4846.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vink C, Beuken E, Bruggeman C A. Structure of the rat cytomegalovirus genome termini. J Virol. 1996;70:5221–5229. doi: 10.1128/jvi.70.8.5221-5229.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vlazny D A, Kwong A, Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci USA. 1982;79:1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weller S K, Carmichael E P, Aschman D P, Goldstein D J, Schaffer P A. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology. 1987;161:198–210. doi: 10.1016/0042-6822(87)90186-3. [DOI] [PubMed] [Google Scholar]
- 62.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 63.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann J, Hammerschmidt W. Structure and role of the terminal repeats of Epstein-Barr virus. J Virol. 1995;69:3147–3155. doi: 10.1128/jvi.69.5.3147-3155.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]