To the Editor:
G protein-coupled receptors (GPCRs) are the largest group of membrane proteins with over 800 members, characteristic of a seven transmembrane domain1. By playing crucial roles in regulation of various physiological processes, GPCRs have been implicated in many diseases including diabetes, obesity, depression and cancer. To initiate different intracellular responses, GPCRs mainly interact with three families of effector proteins upon agonist binding: the heterotrimeric G proteins, G protein-coupled receptor kinases (GRKs) and arrestins1. As for G proteins, there exist four Gα families (Gs, Gi/o, Gq/11 and G12/13) and each of them leads to the generation or depletion of different second messengers1. For example, activation of Gs heterotrimer results in the accumulation of cAMP, while that of Gi causes a decrease in cAMP production1. Likewise, Gq/11 activation elevates inositol triphosphate (IP3) and diacylglycerol (DAG) levels, leading to protein kinase C (PKC) and Ca2+ signaling1. GRKs phosphorylate the activated receptors, and arrestins subsequently bind to phosphorylated receptors and trigger receptor desensitization1.
GPR160 (also annotated as GPCR1 or GPCR150) is a member of class A GPCR family with a length of 338 amino acids. Our previous studies showed that GPR160 mRNA was overexpressed in prostate cancer tissue, and knockdown of the receptor induced apoptosis, cell cycle arrest as well as tumor growth2. Examination of fresh tissue samples from over two hundred patients suggested that GPR160 is a potential biomarker for prostate cancer3. In 2020, Yosten et al.4 identified cocaine- and amphetamine-regulated transcript peptide (CARTp) as an endogenous ligand of GPR160 in mice as it participated in neuropathic pain. Their claim of GPR160 deorphanization was mainly based on the correlation between GPR160 and CARTp demonstrated by ERK phosphorylation and coimmunoprecipitation. However, a recent study disputed the claim because no specific binding of radiolabeled CARTp (125I-CARTp) was observed in human cancer cell lines either endogenously expressing or artificially transfected with GPR1605. This discrepancy led us to explore the relationship between GPR160 and CARTp in terms of clinical significance and signal transduction. Our data indicate that CARTp does not induce GPR160 signaling in human cells and is thus unlikely involved in prostate cancer development.
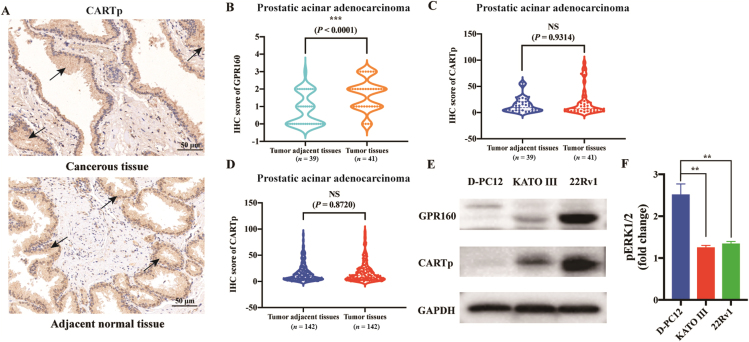
First, we examined the presence of CARTp and GPR160 in human prostate tissue samples by immunohistochemistry (IHC). As shown in Fig. 1A, CARTp was expressed in both cancerous and adjacent normal tissues along with GPR160. But the level of GPR160 was significantly higher in cancerous tissues according to the assessment of 41 tumor and 39 tumor adjacent tissues from patients (P < 0.0001, Fig. 1B), consistent with our previous findings (representative images of GPR160 expression in prostate tissues are exhibited in Supporting Information Fig. S1)3. Further analysis of these samples demonstrated that the expression of CARTp in cancerous vs. adjacent normal tissues was not statistically different (P = 0.9314, Fig. 1C). This observation was also made in a larger sample size involving 142 patients with both tumor and tumor adjacent tissues (P = 0.8720, Fig. 1D). The results suggest that the expression pattern of CARTp in human prostate tissues is distinct from that of GPR160.
Figure 1.
Expression of CARTp in human prostate tissues and signal transduction induced by CARTp in cancer cell lines. (A) Images of CARTp expression in human prostate cancerous (prostatic acinar adenocarcinoma) and adjacent normal tissues detected by immunohistochemistry (IHC), scale bars, 50 μm. Black arrows in the figures indicate stained CARTp. (B, C) 41 samples containing tumor and 39 samples with tumor adjacent tissues from patients were used for IHC analysis of GPR160 (B) and CARTp (C), with unpaired t test. ∗∗∗P < 0.0001. NS, no significance. (D) Samples of 142 patients with both tumor and adjacent tissues were used for immunohistochemistry analysis of CARTp expression, with unpaired t test. NS, no significance. Data (means ± SEM) represent three independent reading. (E) Expression of CARTp and GPR160 in three cancer cell lines (differentiated PC12 cells, 22Rv1 cells and KATO III cells). D-PC12, differentiated PC12 cells. (F) Activation of the ERK1/2 phosphorylation (pERK1/2) induced by 100 μmol/L CARTp in the three cancer cell lines. Data shown are means ± SEM of at least three independent experiments, normalized with respect to baseline signal (i.e., vehicle treatment). D-PC12, differentiated PC12 cells. Statistical analysis was performed by one-way ANOVA followed by Dunnett's multiple comparison test. ∗∗P < 0.01.
We then chose three cancer cell lines that endogenously express GPR160 to examine the CARTp expression and its signaling property. It was shown that CARTp was expressed in both human prostate carcinoma epithelial cells (22Rv1) and gastric carcinoma cells (KATO III), but not differentiated rat pheochromocytoma cells (PC12) (Fig. 1E). Since specific CARTp binding was reported in PC12 cells5 and the peptide could increase ERK1/2 phosphorylation (pERK1/2) levels in this cell line4, we measured the effects of CART(42–89) (one of the active forms of CARTp) on pERK1/2 (Fig. 1F) which confirmed the previous results4. Increased pERK1/2 was consistently observed regardless of the CARTp source (GL Biochem vs. Phoenix Pharmaceuticals, Supporting Information Fig. S2A). However, CART(42–89) did not elicit similar responses in 22Rv1 and KATO III cells measured by the AlphanScreen SureFire pERK1/2 assay kit, and the results were confirmed by western blotting of pERK1/2 (Supporting Information Fig. S2B and S2C). This observation aroused us to investigate the canonical signaling pathways of GPR160.
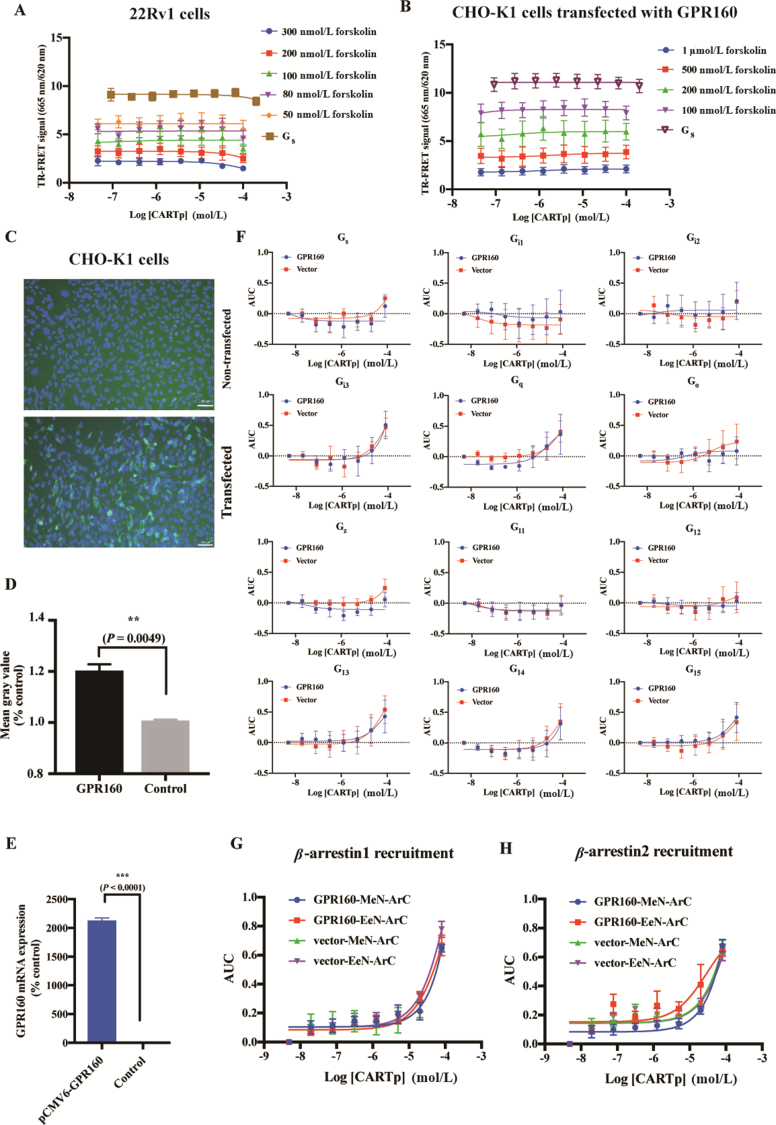
Earlier studies revealed that CARTp signaling involves Gi/o and Gs protein coupling6,7. We next examined if CART(42–89) could influence cAMP in 22Rv1 cells and the results showed that CART(42–89) at concentrations ranging from 100 nmol/L to 100 μmol/L did not affect cAMP levels induced by forskolin (Fig. 2A). Commercial CARTp (Phoenix Pharmaceuticals) was also used in this assay and the response in 22Rv1 cells was in agreement with that synthesized by GL Biochem (Supporting Information Fig. S3). We also performed this experiment in CHO-K1 cells stably transfected with GPR160 and obtained the same conclusion (Fig. 2B), i.e., CARTp failed to activate GPR160 through Gs or Gi/o pathways.
Figure 2.
Signaling profiles of GPR160 induced by CARTp. (A, B) CARTp-induced cAMP signaling in 22Rv1 cells and CHO-K1 cells transfected with human GPR160. Data shown are means ± SEM of three independent experiments. (C–E) Efficiency of GPR160 transfected into CHO-K1 cells using the plasmid (pCMV6-GPR160-Flag) was confirmed by immunofluorescence staining in protein level (green = GPR160-Flag, blue = nuclear staining) (C, D) and RT-qPCR (E) in mRNA level. The scale corresponds to 50 μm. Data shown are means ± SEM of three independent experiments. Statistical analysis was performed by paired t test. ∗∗P < 0.01, ∗∗∗P < 0.0001. (F) Functional analysis of the G protein couplings of GPR160 induced by CARTp. CARTp-induced dissociations of 12 heterotrimeric G proteins were measured using NanoBiT assay. Concentration-response curves were expressed as area-under-the-curve (AUC) across the time–course response curve (0–15 min) for each concentration. Data shown are means ± SEM of three independent experiments. (G, H) CARTp-induced β-arrestin1/2 recruitment. Concentration–response curves were expressed as AUC across the time–course response curve (0–15 min) for each concentration. Data shown are means ± SEM of three independent experiments.
To directly measure GPR160-mediated G protein signaling, we used a NanoBiT-based G protein dissociation assay8. In this system, a large fragment (LgBiT)-fused Gα subunit and a small fragment (SmBiT)-fused Gγ2 subunit, along with an unmodified Gβ1 subunit, were co-expressed in CHO-K1 cells together with the receptor. The efficiency of GPR160 transfection into CHO-K1 cells was determined by immunofluorescence assay (Fig. 2C and D). GPR160 mRNA expression in the same cells was confirmed by RT-qPCR (Fig. 2E). This NanoBiT method was validated using another class A GPCR, cholecystokinin A receptor (CCKAR) (Supporting Information Fig. S4). As shown in Fig. 2F, GPR160 did not elicit meaningful responses among 12 major G protein subtypes upon stimulation of CART(42–89) compared with the control.
Independent of G protein coupling, β-arrestins are involved in GPCRs desensitization and internalization1 and function as scaffold proteins that connect GPCRs to intracellular signaling pathway such as MAPK cascades. Previous studies have suggested that some GPCRs lack functional G protein coupling but exhibit agonist-induced β-arrestins recruitment9. Therefore, we applied a luminescence-based β-arrestin recruitment assay with no fusion tag attached to the C terminus of the receptor to avoid transforming interaction of the receptor with GRKs, arrestins or other proteins10. In this system, the N-terminal NanoLuc fragment is anchored to the membrane (MeN) and the C-terminal NanoLuc fragment (ArC) is fused to β-arrestin1/2. Likewise, the N-terminal NanoLuc fragment attached to the FYVE domain of endofin (EeN) instead of MeN is used to measure β-arrestin1/2 recruitment of internalized receptors in endosomes. In this case, bradykinin type 2 receptor (B2R) was used as a positive control (Supporting Information Fig. S5). Both MeNArC and EeNArC assays showed no response of GPR160 after the addition of different concentrations of CART(42–89), implying that CART(42–89) was unable to stimulate GPR160 to recruit β-arrestin1/2 (Fig. 2G and H).
As mentioned above, the discovery of Yosten et al.4 provoked us to explore signaling pathways of GPR160 in the context of prostate cancer development. Rather than confirming their claim in the murine system, our data are in agreement with another study conducted in human cells5. It appears that CARTp is not an endogenous ligand of GPR160 in the human. It is possible that CARTp might not stimulate GPR160 via classical signaling pathways of GPCRs due to receptor species difference. The human CART(42–89) used in this study is highly similar to the rat CART(55–102)11. CART peptides are evolutionarily conserved in species with high homology between human and rat sequences (95%), while the rodent GPR160 shares only about 65% identity with that of human. Another possibility is that CARTp regulates GPR160 signaling via accessory proteins like the involvement of the melanocortin receptor accessory protein 1 (MRAP1) in the regulation of melanocortin receptor 2 (MC2R)12. Additionally, one CARTp segment [residues 9–28 in CART(42–89)] has been identified as an agonist of GPR68 through activating Gq/11 (calcium mobilization) and Gs (cAMP signaling)13, indicating that CARTp is a possible endogenous ligand of GPR68. Clearly, our work suggests that CARTp is not directly involved in GPR160-related prostate cancer development and the mechanism of which requires further investigations from non-endocrine perspectives, such as effects on the cancer cell mobility3.
Acknowledgments
This work was supported by the National Natural Science Foundation of China 82273961 (Ming-Wei Wang), 82073904 (Ming-Wei Wang), 81872915 (Ming-Wei Wang), 82273985 (Dehua Yang) and 81973373 (Dehua Yang); Shanghai Municipality Science and Technology Development Fund 21JC1401600 (Dehua Yang) and 23XD1400900 (Dehua Yang); National Science & Technology Major Project of China-Key New Drug Creation and Manufacturing Program 2018ZX09735-001 (Ming-Wei Wang) and 2018ZX097110002-002-005 (Dehua Yang); STI2030-Major Project 2021ZD0203400 (Qingtong Zhou); the National Key Basic Research Program of China 2018YFA0507000 (Ming-Wei Wang) and Hainan Provincial Major Science and Technology Project ZDKJ2021028 (Dehua Yang and Qingtong Zhou).
Author contributions
Ming-Wei Wang and Dehua Yang initiated the project, supervised the studies and wrote the manuscript with inputs from all co-authors. Chenyu Ye performed the experiments, drafted figures and participated in manuscript preparation; Qingtong Zhou participated in data analysis and manuscript writing; Shi Lin performed signaling experiments; Wensheng Yang assisted in the immunohistochemistry experiment; Xiaoqing Cai, Yiting Mai and Yanyan Chen took part in method development.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.11.025.
Contributor Information
Dehua Yang, Email: dhyang@simm.ac.cn.
Ming-Wei Wang, Email: mwwang@simm.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Wang J., Hua T., Liu Z.J. Structural features of activated GPCR signaling complexes. Curr Opin Struct Biol. 2020;63:82–89. doi: 10.1016/j.sbi.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Zhou C., Dai X., Chen Y., Shen Y., Lei S., Xiao T., et al. G protein-coupled receptor GPR160 is associated with apoptosis and cell cycle arrest of prostate cancer cells. Oncotarget. 2016;7:12823–12839. doi: 10.18632/oncotarget.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo W., Zhang J., Zhou Y., Zhou C., Yang Y., Cong Z., et al. GPR160 is a potential biomarker associated with prostate cancer. Signal Transduct Targeted Ther. 2021;6:241. doi: 10.1038/s41392-021-00583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yosten G.L., Harada C.M., Haddock C., Giancotti L.A., Kolar G.R., Patel R., et al. GPR160 de-orphanization reveals critical roles in neuropathic pain in rodents. J Clin Invest. 2020;130:2587–2592. doi: 10.1172/JCI133270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freitas-Lima L.C., Pacesova A., Stanurova J., Sacha P., Marek A., Hubalek M., et al. GPR160 is not a receptor of anorexigenic cocaine- and amphetamine-regulated transcript peptide. Eur J Pharmacol. 2023;949:175713. doi: 10.1016/j.ejphar.2023.175713. [DOI] [PubMed] [Google Scholar]
- 6.Yermolaieva O., Chen J., Couceyro P.R., Hoshi T. Cocaine- and amphetamine-regulated transcript peptide modulation of voltage-gated Ca2+ signaling in hippocampal neurons. J Neurosci. 2001;21:7474–7480. doi: 10.1523/JNEUROSCI.21-19-07474.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sathanoori R., Olde B., Erlinge D., Goransson O., Wierup N. Cocaine- and amphetamine-regulated transcript (CART) protects beta cells against glucotoxicity and increases cell proliferation. J Biol Chem. 2013;288:3208–3218. doi: 10.1074/jbc.M112.437145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q., Yang D., Zhuang Y., Croll T.I., Cai X., Dai A., et al. Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor. Nat Chem Biol. 2021;17:1238–1244. doi: 10.1038/s41589-021-00841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey S., Kumari P., Baidya M., Kise R., Cao Y., Dwivedi-Agnihotri H., et al. Intrinsic bias at non-canonical, beta-arrestin-coupled seven transmembrane receptors. Mol Cell. 2021;81:4605–4621. doi: 10.1016/j.molcel.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauge Pedersen M., Pham J., Mancebo H., Inoue A., Asher W.B., Javitch J.A. A novel luminescence-based beta-arrestin recruitment assay for unmodified receptors. J Biol Chem. 2021;296 doi: 10.1016/j.jbc.2021.100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owe-Larsson M., Pawlasek J., Piecha T., Sztokfisz-lgnasiak A., Pater M., Janiuk I.R. The role of cocaine- and amphetamine-regulated transcript (CART) in cancer: a systematic review. Int J Mol Sci. 2023;24:9986. doi: 10.3390/ijms24129986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo P., Feng W., Ma S., Dai A., Wu K., Chen X., et al. Structural basis of signaling regulation of the human melanocortin-2 receptor by MRAP1. Cell Res. 2023;33:46–54. doi: 10.1038/s41422-022-00751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster S.R., Hauser A.S., Vedel L., Strachan R.T., Huang X.P., Gavin A.C., et al. Discovery of human signaling systems: pairing peptides to G protein-coupled receptors. Cell. 2019;179:895–908. doi: 10.1016/j.cell.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.