Abstract
The orphan nuclear receptor Nur77 is a critical regulator of the survival and death of tumor cells. The pro-death effect of Nur77 can be regulated by its interaction with Bcl-2, resulting in conversion of Bcl-2 from a survival to killer. As Bcl-2 is overexpressed in various cancers preventing them from apoptosis and promoting their resistance to chemotherapy, targeting the apoptotic pathway of Nur77/Bcl-2 may lead to new cancer therapeutics. Here, we report our identification of XS561 as a novel Nur77 ligand that induces apoptosis of tumor cells by activating the Nur77/Bcl-2 pathway. In vitro and animal studies revealed an apoptotic effect of XS561 in a range of tumor cell lines including MDA-MB-231 triple-negative breast cancer (TNBC) and MCF-7/LCC2 tamoxifen-resistant breast cancer (TAMR) in a Nur77-dependent manner. Mechanistic studies showed XS561 potently induced the translocation of Nur77 from the nucleus to mitochondria, resulting in mitochondria-related apoptosis. Interestingly, XS561-induced accumulation of Nur77 at mitochondria was associated with XS561 induction of Nur77 phase separation and the formation of Nur77/Bcl-2 condensates. Together, our studies identify XS561 as a new activator of the Nur77/Bcl-2 apoptotic pathway and reveal a role of phase separation in mediating the apoptotic effect of Nur77 at mitochondria.
Key words: XS561, Nur77, Bcl-2, Apoptosis, Mitochondria, Phase separation, TNBC, TAMR
Graphical abstract
XS561-induced mitochondrial-related apoptosis depends on Nur77 phase separation and the formation of Nur77/Bcl-2 condensates at mitochondria.
1. Introduction
Nur77 (also named TR3, NGFIB, or NR4A1), is an orphan nuclear receptor because its natural ligand has not yet been found. Nur77 contains three conserved structural domains similar to classical nuclear receptors (NRs), including the N terminus intrinsically disordered region (IDR), the ligand-binding domain (LBD) and the central DNA-binding domain (DBD). Nur77 is critical in controlling the survival and death of tumor cells1,2. Regardless of the expression of the estrogen receptor (ER), Nur77 is overexpressed in breast cancer patients3,4. Ectopic expression of Nur77 increases invasion and metastasis in triple-negative breast cancer (TNBC) cells by activating TGF-/SMAD signaling5,6, whereas ectopic Nur77 expression in ER-positive MCF-7 breast cancer cells promotes their tumor growth in animals7. Interestingly, translocation of Nur77 from the nucleus to the cytoplasm resulted in apoptosis of in breast cancer cells8. Emerging evidence indicates that the subcellular localization of Nur77 has a significant effect in both the survival and death of tumor cells2,9,10. Thus, understanding the mechanism of Nur77 action in breast cancer may provide novel strategies for the development of breast tumor therapies including TNBC.
The nuclear action of Nur77 is often associated with its growth-promoting effect, whereas the death effect of Nur77 involves its epistatic translocation from the nucleus into mitochondria11. In mitochondria, Nur77 can interact to the LOOP of Bcl-2, and induce Bcl-2 conformational change, leading to the exposure of the BH3 domain of Bcl-2, thereby converting Bcl-2 from an anti-apoptotic molecule to a pro-apoptotic protein12. Bcl-2 is one pro-survival member belonging to the Bcl-2 family proteins13, 14, 15, which is overexpressed in various cancers, including ER-positive16 and TNBC breast cancer17, serving to maintain the survival of tumor cells. Overexpression of Bcl-2 also suppresses tamoxifen-induced apoptosis in breast tumor cells, leading to their resistance to tamoxifen therapy18. Abnormally overexpressed Bcl-2 can prevent cancer cells from apoptosis and/or mediate their resistance to chemotherapy and endocrine therapy, making Bcl-2 an attractive target for developing cancer therapeutics19, 20, 21, 22. Interestingly, upon binding of Nur77, Bcl-2 is transformed from a tumor cell survivor to a tumor cell killer23, raising an intriguing opportunity to target the conversion of Bcl-2 phenotype using Nur77 modulators24, 25, 26. This largely depends on our identification of Nur77 modulators that promote the Nur77/Bcl-2 apoptotic pathway, and the understanding of the molecular mechanism by which Nur77 converts Bcl-2 phenotype.
Liquid–liquid phase separation (LLPS) is considered as a new principle of cellar separation of their biomolecules into membrane-less organelles to regulate multiple biological processes with cells27. We recently reported that Nur77 could mediate the autophagy of mitochondria by phase separation with SQSTM1/p62 through the N-terminal IDR of Nur77 and the PB1 of SQSTM1/p62 upon binding to celastrol28,29. Interestingly, deletion of the IDR of Nur77 completely abolished its mitochondrial association and apoptosis induction11. This together with our recent finding that BI1071, which can activate the Nur77/Bcl-2 apoptotic pathway, could promote Nur77 phase separation29,30, suggesting the involvement of Nur77 phase separation in the Nur77/Bcl-2 apoptotic pathway. Nevertheless, whether the LLPS of Nur77 and Bcl-2 function in the Nur77/Bcl-2 apoptotic pathway and how it acts remain to be determined.
Structure-based virtual screening is an effective method to find hit compound of target protein when the three-dimensional structure of the target protein is available31, 32, 33. Although it has been a dozen years since the first report of the crystal structure of the Nur77-LBD (PDB ID: 1YJE)34, structure-based discovery of Nur77 ligands via virtual screening is not always successful. One reason is that, unlike other nuclear receptor such as ER and androgen receptor (AR), no classical ligand-binding pocket (LBP) is found in the Nur77-LBD34. However, recent determination of Nur77-LBD/ligand complex crystal structures found that some grooves on the surface of Nur77 can serve as binding sites for small molecule ligands35,36, providing possibilities for the application of structure-based virtual screening. As a pilot exercise, we have screened 1.5 million compounds from the ChemDiv chemical database, aiming to identify Nur77 ligands that could have good binding affinity to Nur77 to activate the Nur77/Bcl-2 apoptotic pathway. Here, we report our identification and characterization XS561 as a novel ligand of Nur77 via virtual screening. Our studies showed that XS561 bound to Nur77 to regulate Nur77/Bcl-2 phase separation, leading to Nur77 mitochondrial targeting and apoptosis in tumor cells.
2. Materials and methods
2.1. Antibodies and reagents
For Western blotting, antibodies of anti-Ki67 (Abcam plc, ab15580), anti-Hsp60 (Abcam plc, ab46798), anti-β-actin (Cell Signaling Technology, 4970S), anti-cleaved caspase-3 (Cell Signaling Technology, 9661S), anti-Nur77 (Cell Signaling Technology, 3960S), anti-Myc (9E10) (Santa Cruz Biotechnology, Sc-40), anti-PCNA (Santa Cruz Biotechnology, sc-7907), anti-α-Tubulin (Santa Cruz Biotechnology, sc-8035), anti-Bcl-2 (Santa Cruz Biotechnology, sc-783), anti-Bcl-2 antibody (Abmart, T40056F), anti-PARP (Santa Cruz Biotechnology, sc-7150) were purchased and all the above antibodies were used in the dilution ratio of 1:1000. For immunoprecipitation, anti-Flag (Sigma–Aldrich, F1804, dilution: 1:100), anti-Bcl-2 (Santa Cruz Biotechnology, sc-65392, dilution: 1:50) was utilized. For immunofluorescence staining, Fluorescent Probes of Mito-tracker deep red (ThermoFisher Scientific, M22426, dilution: 1:20,000), JC-1 Probe (Thermo Fisher Scientific, T3168, 1:500) and Mito-SOX Red Mitochondrial Superoxide Indicator (Thermo Fisher Scientific, M36008, dilution: 1:3000) were utilized.
2.2. Cell culture and cell lines
MCF-7 breast cancer, MDA-MB-231, BT549 TNBC breast cancer, MCF-7/LCC2 (TAMR subline of MCF-7 cell line), HepG2 liver cancer, HeLa ovarian cancer, and HEK293T cell, mouse embryonic fibroblast (MEF) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) at 37 °C in a humidified mixture of 5%:95% (v/v) CO2 and air. MDA-MB-231, HepG2, HeLa and HEK293T cells came from Chinese Academy of Science Shanghai Cell Bank. MCF-7 and MCF-7/LCC2 were obtained from Wen Liu's Lab in Xiamen University. MEF and Bcl-2−/−MEF, HeLa and Nur77−/−HeLa were stored in our laboratory. Culture of the above cells was at 37 °C in a humidified mixture of 5%:95% (v/v) CO2 and air.
2.3. Generation of Nur77 knockdown cell lines by CRISPR/Cas9 system
The CRISPR/Cas9 system was used to knockout Nur77 in HeLa cells. gRNA target sequences were designed and synthesized for Nur77 (5′-ACCTTCATGGACGGGCTACAC-3′). The gRNA cloning vector Px330 (Addgene, 71707) was used for cloning. To identify cells with Nur77 knockout, HeLa cells were transfected with both a control vector and a gRNA expression vector, and subjected to G418 (0.5 mg/mL) selection. Western blotting of individual colonies was performed using anti-Nur77 antibody.
2.4. Cell genotyping
Total RNAs of MEF-WT, Bcl2−/−MEF, Hela-WT and Nur77−/−HeLa cells were prepared using Trizol Reagent (Takara Biomedical Technology) according to the Manufacturer's Instructions. The cDNAs were obtained by using a Reverse Transcription Kit (Yeasen Biotechnology). Primers used for PCR amplification using DNA Polymerase (Vazyme Biotech) were based on the DNA sequences of Bcl-2 and Nur77 from the PubMed Database:
Nur77 (F: 5′- ATGCCCTGTATCCAAGCCCAA-3′, S: 5′-TCAGAAGGGCAGCGTGTCCAT-3′);
Bcl-2 (F: 5′-ATGGCGCAAGCCGGGAGAACA-3′, S: 5′- TCACTTGTGGCCCAGGTATGC-3′),
and the amplified products were identified by Agarose Gel Electrophoresis (Biosharp).
2.5. Plasmids, siRNAs, transfection, and rescue experiments
Plasmids GFP-Nur77, pcmv-Myc-Nur77, GFP-Nur77-LBD, GFP-Nur77-IDR, GFP-Nur77-ΔIDR, Myc-Nur77-LBD, Myc-Nur77-IDR, Myc-Nur77-ΔIDR, Flag-cmv-Bcl-2, pcmv-Myc-Bcl-2, GFP-Bcl-2 were constructed using standard methods as reported previously29,30. GFP-Nur77-LBD/H372A, pcmv-Myc-Nur77/H372A, GFP-Nur77/H372A, pcmv-Myc-Bcl-2-TSS/EEE (Bcl-2-3E), pcmv-Myc-Bcl-2-TSS/AAA (Bcl-2-3A), pcmv-Myc-Bcl-2-ΔLOOP, pRC-cmv-Bcl-2, pRC-cmv-Bcl-2-3E, pRC-cmv-Bcl-2-3A, GFP-Bcl-2-3E were constructed using the QuikChang Site-Directed Mutagenesis Kit (ThermoFisher Scientific). Nur77 (NM_002135, siRNA ID: SASI_Hs02_00333289, SASI_Hs02_00333290, and SASI_Hs02_00333291), Bcl-2 (NM_000633, siRNA ID: SASI_Hs01_00119086, SASI_Hs01_00119087, and SASI_Hs01_00119088) and control siRNAs from Sigma–Aldrich. Lipo 2000 was used to transfect cells following the instructions from manufacturer.
For rescue experiments, MDA-MB-231 cells were transfected with Nur77 or Bcl-2 siRNA for 24 h, followed by transfection with Nur77 or Bcl-2 (WT and mutants) for 24 h and treatment with XS561 (10 μmol/L) for 6 h.
2.6. Western blotting and Mn(II)-Phosbind SDS-PAGE
Cell lysates were prepared using RAPI medium including 25 mmol/L Tris·HCl (pH 7.6), 0.1% SDS, 1% NP-40, 1% sodium deoxycholate, 150 mmol/L NaCl, and proteinase inhibitor cocktail, separated via SDS-PAGE, and electrotransfer to Nitrocellulose filter (NC) membrane. Membranes were incubated in TBST [0.1% Tween-20, 50 mmol/L Tris–HCl (pH 7.4) and 150 mmol/L NaCl] with 5% skim milk for 1 h, followed with primary antibodies at 4 °C overnight. They were then incubated with corresponding secondary antibodies followed by detection using an ECL system (ThermoFisher Scientific).
The phosphorylation of Bcl-2 was analyzed by Mn (II)-Phosbind SDS-PAGE (APExBIO Technology LLC, F4002), which can bind phosphate ions under neutral pH conditions. In the configuration of sodium dodecyl sulfate-polyacrylamide gels, 20 μmol/L Phos-tag and 40 μmol/L MnCl2 were added. After electrophoresis of cell lysates in phos-tag gel, the phos-tag gel was immersed in transmembrane buffer containing 10 mmol/L EDTA and washed to eliminate manganese ions, then transferred onto NC membrane, and analyzed by Western blotting.
2.7. Co-immunoprecipitation (Co-IP)
HEK293T cells transfected expression plasmids were treated with or without XS561 for 3 h, and lysed with Western/IP buffer (Beyotime Biotechnology). 5% of cell lysate was used as input. The remaining lysates were incubated with appropriate antibodies at 4 °C overnight followed by incubation with Protein A/G-Sepharose beads (Santa Cruz Biotechnology) for 3 h. PBS was used to wash the beads five times before adding loading buffer. Samples were then boiled and analyzed by SDS-PAGE for immunoblotting.
2.8. Preparation of mitochondrial fractions
To prepare nuclear, mitochondrial, and cytoplasmic fractions, and cells were suspended in Sucrose Gradient Buffer D (250 mmol/L sucrose, 2 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L DTT,10 mmol/L Tris–HCl pH 7.5) with 1% protease inhibitor for 15 min. Cells were then homogenized by douncing 20 times slowly and smoothly using a Dounce Homogenizer. The homogenate was spun at 600 × g for 5 min to obtain nuclear pellet. Supernatant was centrifuged at 5000 × g for 10 min to mitochondrial constituent. SDS-PAGE was used to analyze the various fractions.
2.9. Flow cytometry
Flow cytometry was conducted as described23. The total cells treated with or without XS561 were collected with centrifugation at 400 × g.
For apoptosis detection, cells were stained with Annexin V/FITC for 15 min, followed by staining of propidium iodide (PI) for 5 min (Yeasen Biotechnology). After fluorescence compensation, cells were instantly detected through FITC and PC5.5 fluorescence channels. The proportion of early apoptosis (PI–/Annexin V+) and late apoptosis (PI+/Annexin V+) was determined as the percentage of apoptotic cells.
For mitochondrial membrane potential (ΔΨm) assay, cells were stained with 5 μg/mL 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1, ThermoFisher Scientific) for 30 min, and subsequently detected by FITC and PE fluorescence channels.
For mitochondrial reactive oxygen species (Mito-ROS) detection, cells were stained with Mito-SOX (Thermo Fisher Scientific) for 20 min, followed by PE fluorescence channel detection. Data were analyzed using a cytoFLEX Flow Cytometry System (Beckman–Coulter, Miami, FL, USA).
2.10. Immunostaining
Cells incubated on slides were labeled with Mito-Tracker (ThermoFisher Scientific, Deep Red) at 1:20,000 dilution for 30 min before being fixed for 0.5 h in 4% paraformaldehyde solution. Cells were then permeabilized on ice using PBS with 0.1% Triton X-100 for 10 min, blocked by PBS with 1% bovine serum albumin (BSA) for 30 min at room temperature, and then incubated with primary antibody at 4 °C overnight. Following three rounds of PBS washing, cells were incubated with secondary antibodies that bind to FITC or Cy3-binding for 2 h at room temperature. A Leica TCS SP8 system fluorescence microscope was used to gather and examine fluorescence images.
2.11. Animal studies
The right flank of each nude mouse (BALB/c, SPF grade, 18–20 g, 8-week-old, female) was injected with log-phase tumor cells (HepG2 cells or MDA-MB-231, 5 × 106 cells in 100 μL PBS) for the xenograft nude mice study. Mice were then randomly separated to different groups (n = 5–6) and given oral XS561 treatment starting 10 days after transplantation, when the mean tumor volume had increased to about 100 mm3. Tumor size and body weight were measured every 2 days. XS561 was first completely dissolved by using DMSO, followed by dilution with saline containing 5.0% Tween-80 (v/v) to obtain a final concentration of XS561 at 0.05 mg/mL. Saline with 5.0% Tween-80 and DMSO was used as vehicle control.
Prior to immunohistochemistry analysis, tumor xenograft tissues were immediately fixed with 4% paraformaldehyde. All experiments involving animals were carried out with permission by the Xiamen University Animal Care and Use Committee. All mice were treated consistent with the "Principles for the Utilization and Care of Vertebrate Animals”, as well as the "Guide for the Care and Use of Laboratory Animals.".
2.12. Zebrafish embryo toxicity study
Wild-type zebrafish (AB strain) were feeding in a circulation tank with fish water (0.17 mmol/L KCl, 5.0 mmol/L NaCl, 0.16 mmol/L MgSO4 and 0.4 mmol/L CaCl2) at 14/10 h photoperiod in the fish room. Embryos were gained from spawning mature fish set in a spawning box in groups of one female and two males overnight. Healthy embryos were selected and transferred to 24-well plates with 20–22 embryos per well and exposed to different compounds at different concentrations and 0.1% DMSO is set as control. The time points of 24, 48 and 72 hpf were selected for our observation, and the images were collected using a stereomicroscope to record the number of surviving embryos or juveniles and the development of embryos to further evaluate the embryo mortality and hatching rate.
2.13. Virtual screening and molecular docking experiments
A grid-based docking program, Schrodinger's Glide (see www.schrodinger.com), was used for virtual screening and the docking study of XS561 to Nur77. We used the Nur77-LBD crystal structure containing a cytosporone B analog (TMPA) (PPDB ID: 3V3Q) for simulation35. Schrödinger's Maestro was used for our visualization of virtual screening and the results of docking.
2.14. Surface plasmon resonance (SPR)
SPR assay was conducted as described30. Purified Nur77-LBD protein was solubilized in 50 mmol/L NaOAc (pH 5.0) to a 0.05 mg/mL concentration, followed by immobilizing on a CM5 sensor chip via amine coupling by following the instruction from the manufacturer (GE Healthcare) at about 10,000 RU density. The kinetics of binding between XS561 and Nur77-LBD was tested (25 °C) on a BIAcore T200 Instrument (GE Healthcare). Following fitting the kinetic data globally for compound gradient concentrations, the dissociation constant (Kd) between XS561 and protein was calculated. This was done in accordance with the conventional 1:1 interaction model.
2.15. Fluorescence titration analyses
All fluorescence measurements were performed using an Agilent Cary Eclipse fluorescence spectrophotometer. The experiments were conducted utilizing a consistent Nur77-LBD protein concentration of 1 μmol/L, while the concentrations of compounds were systematically varied in the range of 0.01–1 μmol/L for comparison. Fluorescence spectra of Nur77-LBD, both in the absence and presence of additional compounds, were meticulously recorded within the wavelength spectrum of 300–500 nm, with excitation occurring at a wavelength of 280 nm. All gathered fluorescence data underwent thorough analysis via the Origin software and the kinetic profiles were shown in the text.
2.16. Cellular thermal shift assay (CETSA)
MDA-MB-231 cells were cultured in 10 cm plates, treated with DMSO, XS561 (15 μmol/L), celastrol (4 μmol/L), respectively for 1 h, and resuspended with PBS supplemented with protease inhibitor (Roche). Divide each group into eleven portions, one portion without heat treatment, and the remaining portions were heat-treated for 3 min at 52, 53.7, 55.6, 58.2, 60.8, 63.2, 65.8, 68.4, 70.3, and 72 °C, freeze–thawed three times in liquid nitrogen, and then centrifuged at 20,000 × g for 20 min at 4 °C. The supernatants were obtained, added with loading buffer, and analyzed by Western blotting.
2.17. Phosphatase assay
To determine the nature of Bcl-2 modification, lysates from cells treated with XS561 for 6 h were incubated with thermosensitive alkaline phosphatase (TAP) for 0.5 h. Following the incubation period, the reactions were subjected to boiling in SDS sample loading buffer and examined using Western blotting.
2.18. FRAP
For in vivo experiments, the Zeiss LSM 980 microscope with 63 × oil immersion objective was used to perform the FRAP of GFP-Bcl-2-3E and GFP-Bcl-2. Images were taken every 3 s while the bleaching was being done at 80% laser power (488 nm) for 100/120 s. Using the FIJI plug-in FRAP Profiler, the fluorescence intensity of unbleached control spots, bleached spots, and background was quantified. Photobleaching during image acquisition was controlled by determining whether the fluorescence values of the unbleached spots were stable. The values of the bleached points were obtained by subtracting the background intensity. Every data point shows the fluorescence intensity with standard and mean deviation of three bleached (experimental) particles.
2.19. Statistical analysis
Data are shown as mean ± standard deviation (SD) or mean ± standard error of mean (SEM) from at least three technical replicates. Determination of the statistical significance of differences among several groups of methods used the Student's t-test. Differences were regarded to be statistically significant at P < 0.05 (N.S., P > 0.05; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001).
3. Results
3.1. XS561 binds selectively to the LBD of Nur77
We screened a chemical collection of 1.5 million compounds commercially available from ChemDiv (www.chemdiv.com) company for Nur77 binders (Supporting Information Fig. S1A). The human Nur77-LBD crystal structure complexed with TMPA retrieved via the Protein Data Bank (www.rcsb.org) (PDB code: 3V3Q) was used in our virtual screening process35. The binding site B, identified based on the position of the co-crystallized ligand TMPA was selected for docking grid box (Fig. 1B). 1.5 million compounds from ChemDiv database were initially docked using high throughput virtual screening (HTVS) mode because of its high efficiency. HTVS docked compounds at Top 5% were then chosen for the docking of standard precision (SP). The top 3% compounds with best scores from SP docking were imported into Canvas 2.7 for hierarchical clustering analysis using their two-dimensional (2D) fingerprints (Fig. S1A). After comparing with the TMPA binding mode in the crystal structure (PDB code: 3V3Q), 4 clusters of molecules with 10 compounds (Fig. S1B) were selected and purchased for biological evaluation.
Figure 1.
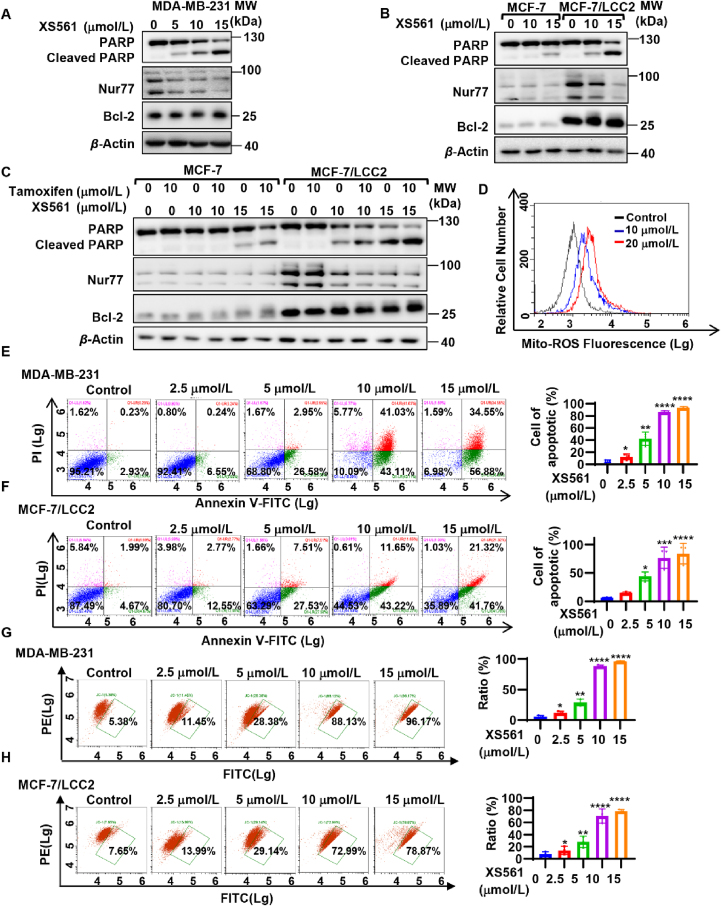
XS561 specifically binds to Nur77 proteins. (A) Chemical structure of XS561. (B) The novel binding site B of Nur77-LBD. (C, D) Molecular modeling of XS561 binding site on Nur77-LBD. (E) SPR analysis of the binding between XS561 and Nur77-LBD. (F) Fluorescence titration analysis of the binding between XS561 and Nur77-LBD. (G, H) XS561 (15 μmol/L) and celastrol (4 μmol/L) influences the thermal stability of Nur77 in MDA-MB-231 cells as assayed by CETSA (n = 3).
Our goal was to identify compounds that bind Nur77 to initiate the Nur77/Bcl-2 apoptotic pathway. Thus, we first evaluated if the selected compounds could activate apoptosis in MDA-MB-231 TNBC cells. Our results demonstrated that only XS561 (Fig. 1A) could effectively induce the PARP cleavage, an indication of apoptosis, at a concentration of 10 μmol/L, while other compounds had no such apoptotic effect (Fig. S1C). We then determined the binding of these compounds to Nur77 using SPR assay, and found that XS561 had the highest binding affinity to Nur77, with a Kd of 8.2 μmol/L (Fig. S1D and Fig. 1E). For comparison, the binding Kd of TMPA and celastrol is 0.38 and 0.26 μmol/L, respectively, which is at same range reported previously28,35, indicating the accuracy of our binding study (Supporting Information Fig. S2A and S2B). XS561 did not show apparent binding to ERα and PPARγ, with the fitted Kd values of 206 and 143 μmol/L, respectively (Fig. S2C and S2D). Thus, XS561 selectively binds Nur77. Fluorescence titration analyses further confirmed the binding of XS561 to Nur77 with a comparable Kd of 2.2 ± 1.6 μmol/L (Fig. 1F). XS561 binding to Nur77 was further investigated by CETSA, with celastrol as a positive control. Our results revealed that treatment of MDA-MB-231 cells with XS561 or celastrol effectively altered the thermal stability of Nur77, demonstrating the ability of XS561 to bind Nur77 protein in cells (Fig. 1G and H).
There are two chiral carbon atoms in XS561 (Fig. 1A and Supporting Information Fig. S3A–S3C), which can result in four different stereoisomers (Supporting Information Fig. S4A). To study which stereoisomer binds to Nur77, we used molecular docking to predict the binding mode of the four stereoisomers. Among the four docking results, the XS561_14R28R was scored the highest with −7.390 kcal/mol (Fig. S4B and S4C). Hydrophobic carbon chain structure on XS561_14R28R displayed a binding mode similar to TMPA. However, the phenol structure bound to a deeper cavity in the protein where TMPA did not occupy. In addition, the polar hydrogens on XS561_14R28R formed H-bond interactions with His372 and Leu373 (Fig. 1C and D). Hydrophobic group of XS561_14R28R made van der Waals contact with His163, Val167, Val169, Arg450, Tyr453, Arg454, Lys456 and Thr379. Thus, we predict that XS561_14R28R is the most important active component of XS561 for binding Nur77.
3.2. XS561 effectively induces apoptosis of TNBC and TAMR breast cancer cells
We systematically evaluated the apoptotic effect of XS561 in various cancer cell lines, including TNBC and TAMR cells. PARP cleavage of MDA-MB-231 TNBC cells could be induced by XS561 in a dose-dependent manner with significant induction of PARP cleavage at 10 μmol/L concentration (Fig. 2A). XS561 was also effective in HepG2 liver cancer and HeLa cervical cancer cells (Supporting Information Fig. S5A and S5B). In ER-positive MCF-7 cells, induction of apparent PARP cleavage by XS561 occurred at 15 μmol/L concentration. Thus, XS561 is effective both in MDA-MB-231 TNBC cells and ER-positive MCF-7 cells. We also evaluated the apoptotic effect of XS561 in tamoxifen-resistant MCF-7/LCC2 cells developed from MCF-7 cells37. To our surprise, XS561 was even more effective in MCF-7/LCC2 cells than in the parental MCF-7 cells, which might be due to elevated expression of Nur77 and Bcl-2 in MCF-7/LCC2 cells than MCF-7 cells (Fig. 2B). We also confirmed the pro-apoptotic effect of XS561 by MTT assay, showing the compound was effective against MCF-7 (IC50: 9.267 ± 0.28 μmol/L), MCF-7/LCC2 (IC50: 3.353 ± 0.65 μmol/L), and TNBC cell lines MDA-MB-231 (IC50: 5.884 ± 0.80 μmol/L) and BT549 (IC50: 3.992 ± 0.65 μmol/L) (Fig. S5C). Furthermore, XS561 showed no apparent effect in the MCF-10A cells, the normal mammary cell line (Fig. S5D). Interestingly, the expression levels of Nur77 and Bcl-2 in MCF-10A cells were significantly lower than those in MCF-7, MCF-7/LCC2, MDA-MB-231, and BT549 cells (Fig. S5E). These data suggest a role of Nur77 and Bcl-2 in mediating the apoptotic effect of XS561. XS561 and tamoxifen exhibited a strong synergistic effect in apoptosis induction in MCF-7/LCC2 cells (Fig. 2C). A similar synergistic effect was also observed when HA14-1, a Bcl-2 inhibitor38, was used (Fig. S5F). Enhanced Bcl-2 expression was responsible for the resistance of MCF-7 breast cancer cells to endocrine therapy20. Our finding that XS561 was more effective in breast cancer cells expressing elevated Bcl-2 suggests that targeting Bcl-2 by XS561 may overcome tamoxifen resistance in breast cancer.
Figure 2.
Apoptotic effect of XS561 in TNBC and TAMR cells. (A, B) Induction of PARP cleavage of TNBC and TAMR cells. MDA-MB-231 (A) or MCF-7 and MCF-7/LCC2 cells (B) are treated with indicated concentration OF XS561 for 6 h and then analyze by Western blotting for PARP cleavage, Nur77 and Bcl-2 expression. (C) Synergistic effect of XS561 and tamoxifen on apoptosis of TAMR cells. The combination effect of tamoxifen and XS561 in MCF-7 and MCF-7/LCC2 cells were indicated via PARP cleavage. (D) XS561 induces production of Mito-ROS. The production of Mito-ROS in MDA-MB-231 cells dealt with XS561 (10, 20 μmol/L) for 6 h were staining with Mito-SOX and then analyzed by flow cytometry. (E, F) Apoptotic effect of XS561 revealed via Annexin V/PI double staining. Left graph, Representative images of MDA-MB-231(E) and MCF-7/LCC2 (F) cells were treated with or without indicated concentration of XS561 for 6 h before conducting Annexin V/PI double staining assay and subsequent analysis flow cytometry. Right graph, Quantitative analysis of the ratio of the apoptotic cells. (G, H) XS561 induces the potential loss of mitochondrial membrane. Left graph, Representative images of MDA-MB-231 (G) and MCF-7/LCC2 (H) cells treated with or without the indicated concentration of XS561 for 6 h were stained with JC-1 and measured via flow cytometry. Right graph, Quantitative analysis of the ratio of cells with mitochondrial potential loss. Data are expressed as mean ± SD, n = 3. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001 vs. Control group.
To further evaluate the pro-apoptotic effect of XS561 in TNBC and TAMR cancer cells, we studied its effect on production of Mito-ROS known to promote cellar apoptosis. Flow cytometry analysis revealed that XS561 dose-dependently induced Mito-ROS generation in MDA-MB-231 cells (Fig. 2D). As shown in the Annexin V/PI apoptosis experiments23, the apoptotic MDA-MB-231 cells increased from 3.16% to 91.43%, whereas apoptotic MCF-7/LCC2 cells increased from 6.66% to 63.08%, when they were treated with 15 μmol/L XS561 for 6 h (Fig. 2E and F). The decline in mitochondrial membrane potential (ΔΨm) of cells is a hallmark in mitochondria-related cell apoptosis. We determined the effect of XS561 on ΔΨm by staining cells with JC-1, which displays red fluorescence when membrane potential is high and shows green fluorescence as the membrane potential is low. Fig. 2 (G and H) shows that XS561 could effectively cause mitochondrial membrane dysfunction in MCF-7/LCC2 and MDA-MB-231 cells, as the increase of green fluorescence correlated positively with the increase of XS561 concentration. These data demonstrate that XS561 induced mitochondria-associated apoptosis in TNBC and TAMR cells.
3.3. XS561 inhibits tumor growth in animals
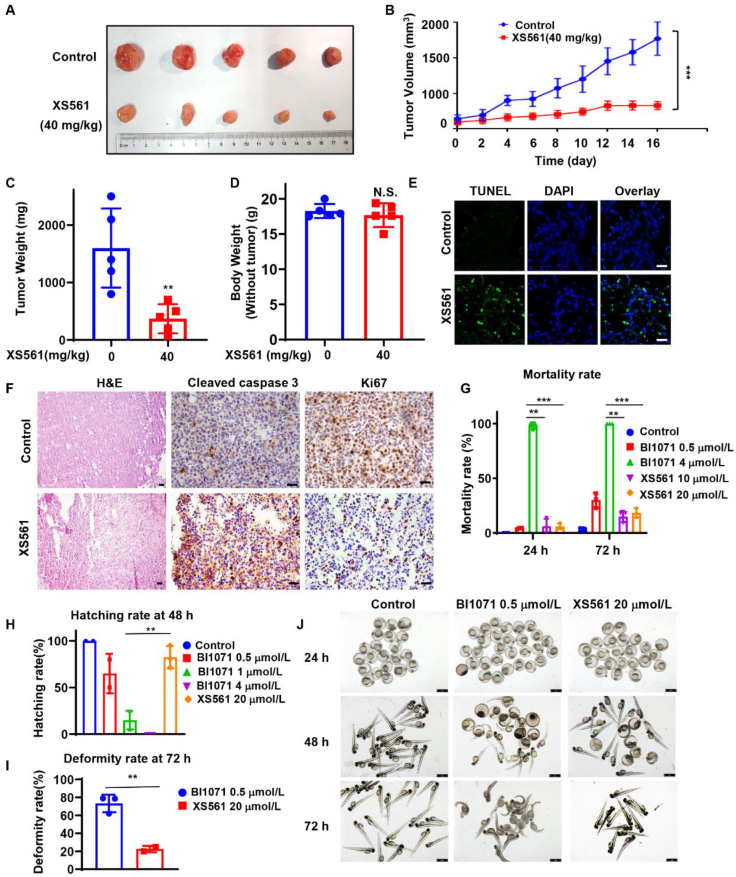
To evaluate the anticancer effect of XS561 in vivo, nude mice possessing MDA-MB-231 tumor xenografts were separated into control and XS561-treated (40 mg/kg) groups. XS561 administration significantly reduced the growth rate of MDA-MB-231 tumor in a time-dependent manner (Fig. 3A and B). The growth of MDA-MB-231 tumor was inhibited by 76.9% after treated with XS561 for 16 days (Fig. 3C). Notably, the XS561 treatment did not show apparent loss of body weight (Fig. 3D). TUNEL assay showed that XS561-treated tumor specimens had a high degree of cell death (Fig. 3E). H&E staining indicated that XS561 could promote nuclear crinkling of tumor cells. Immunostaining revealed that XS561 enhanced the expression of cleaved caspase 3 and reduced the expression of Ki67 in tumors (Fig. 3F). Similar anti-tumor effect of XS561 was observed in mice bearing HepG2 tumor xenografts (Supporting Information Fig. S6A–S6E). These data demonstrated a potent anti-tumor effect of XS561 in animals mainly through its apoptosis induction.
Figure 3.
Anti-tumor activity of XS561 in xenograft tumor model and toxicity study of XS561. (A) Representative photos of MDA-MB-231 tumors from mice administered with XS561 for 16 days. (B) Growth curves of MDA-MB-231 tumor in mice administered with or without XS561. (C) Weight of the MDA-MB-231 tumor in mice treated for 16 days with or without XS561. (D) Weight of mice after receiving either XS561 or a placebo for 16 days. For the mice study, Data are expressed as mean ± SEM, n = 5. N.S., P > 0.05; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001 vs. Control group. (E) Representative images show TUNEL assay for apoptosis of tumor cells after treatment with XS561. (F) For H&E and immunohistochemistry staining of Ki67 and cleaved caspase 3, MDA-MB-231 tumors from mice given either XS561 or a placebo for 16 days were gathered. Tumors were chosen from different groups randomly and representative images were shown. Scale bar, 50 μm. (G) Statistics on the mortality rate at 24 or 72 hpf. Data are expressed as mean ± SEM, n = 3. ∗∗P < 0.01; ∗∗∗P < 0.001 vs. BI1071 4 μmol. (H) Data on the 48 hpf hatching rate. (I) Data on the deformity rate in 72 hpf. Data are expressed as mean ± SD, n = 3. ∗∗P < 0.01 vs. BI1071 1 μmol. (J) Morphology of BI1071 or XS561-treated 24 hpf embryos or 72 hpf zebrafish larvae. Scale bar, 1 mm. Data are expressed as mean ± SD, n = 3. ∗∗P < 0.01 vs. BI1071 0.5 μmol.
3.4. Safety evaluation of XS561 in zebrafish
The potent anti-tumor activity of XS561 described above prompted us to evaluate its safety in zebrafish, a model for rapidly assessing compound toxicity in vivo. For comparison, BI1071, another Nur77 ligand that can induce apoptosis of tumor cells30 was used. We first evaluated the toxic effects of XS0561 with BI1071 on developing zebrafish embryos and larvae, including mortality rate, hatching rate, deformity rate and body morphology. As shown in Fig. 3G, XS561 did not exhibit significant zebrafish embryonic lethality at concentrations up to 20 μmol/L during the 24 h post-fertilization period, while embryos exposed to BI1071 showed nearly 100% mortality at only 4 μmol/L. The hatching rate of zebrafish embryos was recorded at 48 h after fertilization (Fig. 3H). Embryos exposed to 1 μmol/L BI1071 were already difficult to survive, with a hatching rate of less than 20%. The hatching rate of embryos exposed to 0.5 μmol/L BI1071 was similar to that of embryos exposed to 20 μmol/L XS561 (Fig. 3H). We also studied the morphology of zebrafish larvae 72 h after fertilization. Although the mortality rate of zebrafish exposed to 0.5 μmol/L BI1071 was not high, the larvae in this environment already showed obvious deformities, exhibiting phenotypic characteristics of large curved spine and short tail length, and the larval deformity rate was close to 80%. In contrast, larvae exposed to 20 μmol/L XS561 had a deformity rate of less than 20% (Fig. 3I) and showed less deformity in the larvae with deformed characteristics than those in the 0.5 μmol/L BI1071 environment (Fig. 3J). Together, XS561 was less toxic than BI1071, representing a better lead compound with for future optimization.
3.5. XS561 induces Nur77 mitochondrial localization and Nur77 dependent apoptosis
To study the molecular mechanism by which XS561 induces Nur77-Bcl-2 apoptotic pathway, we examined whether XS561 induced Nur77 translocation from the nucleus to mitochondria, a critical step in Nur77-mediated apoptosis11. Cellular fractionation assays demonstrated that upon XS561 treatment Nur77 was distributed throughout both the nucleus and mitochondria, whereas Nur77 was predominantly nuclear without treatment (Fig. 4A). The ability of XS561 to induce Nur77 mitochondrial targeting suggested its activation of Nur77-mediated mitochondrial pathway. To study the role of Nur77, we knocked down Nur77 in MDA-MB-231 by transfection Nur77 siRNA and depleted Nur77 from HeLa cells by CRISPR/Cas9 technology (Supporting Information Fig. S7A). The resulting cells, MDA-MB-231-siNur77 and Nur77−/−HeLa were evaluated for their response to XS561. Our results showed that XS561 induction of PARP cleavage (Fig. 4B, Fig. S7B) and mitochondrial ROS release (Fig. S7C) was largely attenuated in MDA-MB-231-siNur77 cells and Nur77−/−HeLa cells as compared to their respective parental cells. Annexin V/PI staining revealed that the percentage of XS561-induced apoptotic MDA-MB-231 cells dropped from 36.28% to 11.44% upon Nur77 knocking down (Fig. 4C, Fig. S7F). Similarly, the percentage of XS561-induced apoptotic HeLa cells decreased from 37.42% to 7.42% when Nur77 was depleted from HeLa cells (Fig. S7D). JC-1 staining also showed that inhibiting Nur77 expression largely impaired the effect of XS561 on reducing ΔΨm, with loss of ΔΨm from 42.02% to 11.70% in MDA-MB-231 and from 56.64% to 4.30% in HeLa cells (Fig. 4D, Fig. S7E and S7G).
Figure 4.
XS561 binds Nur77 to induce Nur77-targeting mitochondria and Nur77-dependent apoptosis. (A) Transfection of Myc-Nur77 into HEK293T cells was treated with or without XS561 (10 μmol/L) for 2 h. Whole cell lysate (WCL) and mitochondrial fractions (Mito) were prepared to conduct Western blotting analysis. Expression of mitochondrial protein Hsp60 and nuclear protein PARP indicated the quality of the mitochondrial fraction. (B)–(D) MDA-MB-231-siNC and MDA-MB-231-siNur77 cells treated with XS561 (10 μmol/L) for 6 h, then PARP cleavage was analyzed by Western blotting (B), Annexin V/PI (C) and JC-1 staining (D) were detected via flow cytometry. Data represent at least three independent experiments. (E)–(G) MDA-MB-231-siNur77 cells were transfected with Myc-cmv, Myc-Nur77 and Nur77 mutant plasmids and treated with XS561 (10 μmol/L) for 6 h, then PARP cleavage (E) was detected by westering blotting, and Annexin V/PI (F) and JC-1 (G) were detected by flow cytometry.
Analysis of XS561-Nur77 interactions showed XS561 forms H-bond interactions with His372 site of Nur77. Thus, we transfected into MDA-MB-231-siNur77 cells with Nur77 or mutant with His372 replaced with Ala (Nur77-H372A) to determine the requirement of XS561 binding to Nur77. Transfection of Nur77 could rescue the effect of XS561 on inducing PARP cleavage, cell death, and loss of ΔΨm in MDA-MB-231-siNur77 cells. In contrast, the expression of Nur77-H372A failed to recover the effect of XS561. The effect is specific as transfection of Myc-Nur77-C566K, a mutant with Cys566 substituted with Lys, which affects another ligand-binding groove28,29, could confer cells with sensitivity to XS561 (Fig. 4E–G, Fig. S7H and S7I). These data therefore reveal the importance of XS561 binding to Nur77 in its induction of apoptosis.
3.6. The role of Nur77 phase separation in XS561-induced apoptosis
The N-terminal IDR is essential for Nur77 to undergo mitochondrial targeting and phase separation11,29. To study whether Nur77 phase separation plays a role in XS561 induction of Nur77 mitochondrial targeting and apoptosis, we tested whether XS561 could induce Nur77 LLPS, and found that Nur77 formed punctate structures in the nucleus of MCF-7/LCC2 cells, MDA-MB-231 cells, and MCF-7 cells. However, upon XS561 treatment, Nur77 formed puncta both in the nucleus and cytoplasm (Supporting Information Fig. S8A). To determine the role of IDR of Nur77 in mediating XS561-induced Nur77 phase separation, GFP-Nur77, GFP-Nur77-LBD and GFP-Nur77-ΔIDR were transfected (Fig. 5A). As we reported previously, Nur77 puncta were formed in the nucleus, indicating the occurrence of phase separation (Fig. 5B, left). On the contrary, GFP-Nur77-LBD or GFP-Nur77-ΔIDR showed much reduced ability to form punctate structures29 (Fig. 5C and D, left), revealing a role of IDR. We next investigated how the binding of XS561 to Nur77 affected its phase separation. Upon XS561 treatment, GFP-Nur77 and Nur77-LBD formed Nur77 droplets in the cytoplasm (Fig. 5B and C), where they colocalized with mitochondria (Fig. 5G and H). The effect of XS561 was lost when Nur77-H372A or Nur77-LBD-H372A, two mutants with impaired XS561 binding, were analyzed (Fig. 5E, F, H and I). Although XS561 failed to induce nuclear export of Nur77-ΔIDR, a mutant lacking IDR, it still induced the mutant from a diffuse state to a spherical particle structure in the nucleus (Fig. 5D and G). Similar results were obtained when BI1071 was used (Fig. 5B–F). Taken together, these data demonstrate that XS561 binding to Nur77 could promote its phase separation either in the cytoplasm or in the nucleus.
Figure 5.
The ability of Nur77 phase separation is requited for XS561 induced apoptosis. (A) Schematic representation of Nur77 and its mutants. (B)–(F) GFP-Nur77 or mutants form droplets in HeLa cells after exposure to the target compounds (XS561: 10 μmol/L, BI1071: 0.5 μmol/L). Left: Illustrations of GFP-Nur77 and its mutants in transfected droplets. A magnified view of the inset is also shown. Scale bar, 3 μm. Right: Quantification of GFP-Nur77 and its mutants' droplet formation. The results come from at least three different experiments. (G)–(I) Representative images of colocalization of Nur77 and its mutants with mitochondria in XS561-treated cells. Scale bar, 3 μm. Data are representative of at least three independent experiments. (J)–(K) MDA-MB-231-siNur77 cells were transfected with Myc-cmv, Myc-Nur77 and Nur77 mutant plasmids and treated with XS561 (10 μmol/L) for 6 h, then PARP cleavage (J) was detected by westering blotting, and Annexin V/PI were detected by flow cytometry (K).
We next transfected into MDA-MB-231-siNur77 cells with Nur77 and mutants with different ability to respond to XS561 in forming droplets and migrating to mitochondria. Our data showed that transfection of Nur77 and Nur77-LBD, which form droplets and target mitochondria in response to XS561 (Fig. 5A–H, Fig. S8B–S8D), could rescue the effects of XS561 on inducing PARP cleavage, apoptotic cell increase and mitochondrial membrane potential. In contrast, transfection of Nur77-IDR, Nur77-ΔIDR could not rescue the effects of XS561 (Fig. 5J and K, Fig. S8E–S8G). These data reveal the importance of Nur77 mitochondrial targeting and phase separation in XS561-induced apoptosis.
3.7. XS561-induced Nur77-mediated apoptosis is Bcl-2 dependent
Mitochondrial Nur77 induces apoptosis by interacting with Bcl-212. Our observation that MCF-7/LCC2 cells with higher expression of Nur77 and Bcl-2 compared with the parental MCF-7 cells were more sensitive to XS561 (Fig. 2B) suggested that not only Nur77 but also Bcl-2 could mediate the pro-apoptotic effect of XS561. Thus, we evaluated the apoptotic effect of XS561 in mouse embryonic fibroblast (MEF) depleted with Bcl-2 (Bcl-2−/−MEF) (Supporting Information Fig. S9A) and in MDA-MB-231 transfected with Bcl-2 siRNA (MDA-MB-231-siBcl-2). Similar to the impact of Nur77 inhibition, down regulation of Bcl-2 in MEFs or MDA-MB-231 impaired the effect of XS561 on activating PARP cleavage (Fig. 6A and B) and inducing Mito-ROS accumulation (Fig. 6C). Annexin V/PI staining demonstrated that compared to MDA-MB-231-siNC and MEFs, the apoptotic effect of XS561 in MDA-MB-231-siBcl-2 cells and Bcl-2−/−MEFs was significantly reduced from 48.29% to 11.45% and from 68.86% to 0.24%, respectively (Fig. 6D, Fig. S9B). The reduction of ΔΨm by XS561 in MDA-MB-231 and MEFs was also inhibited in MDA-MB-231-siBcl-2 cells (from 79.71% to 28.04%) and Bcl-2−/−MEFs (from 87.94% to 7.08%) (Fig. 6E, Fig. S9C). These results demonstrate that Bcl-2 is also critical in XS561-induced apoptotic effect.
Figure 6.
XS561-induced Nur77-related apoptosis is Bcl-2 and its phosphorylation modification dependent. (A)–(E) Bcl-2 deletion lessens the apoptotic impact of XS561. After being exposed to 10 μmol/L XS561 for 6 h, MEF and Bcl-2−/−MEF cells were tested for PARP cleavage by Western blot (A), and Mito-ROS (C). Also, MDA-MB-231-siNC and MDA-MB-231-siBcl-2 cells treated with XS561 (10 μmol/L) for 6 h, then PARP cleavage were analyzed by Western blotting (B), Annexin V/PI (D) and JC-1 staining (E) were detected via flow cytometry. Data are expressed as mean ± SD, n = 3. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001. (F) WB analysis of TAP effect on the stability of p-Bcl-2 expressed in XS561 treated MCF-7/LCC2 cells. (G) Activation of MAPK and JNK by XS561. MCF-7/LCC2 cells treated with 10 and 15 μmol/L of XS561 for 3 h were analyzed for the activation of p-JNK, p-p38 MAPK and p-ERK MAPK by Western blotting. The levels of β-actin were used for loading control. (H) MCF-7/LCC2 cells were incubated for 6h with XS561 (15 μmol/L), in the presence or absence of JNKI SP600125 (10 μmol/L) or p38I SB203580 (10 μmol/L), and analyzed by Western blotting for PARP cleavage and Bcl-2 phosphorylation. (I) MCF-7/LCC2 cells were transfected with control siRNA or Nur77 siRNA for 24 h, then transfected with Myc, Myc-Nur77, Myc-Nur77-H372A or Myc-Nur77-C566K were treated with XS561 (15 μmol/L) for 6 h, then analyzed by Western blotting for PARP cleavage and Bcl-2 phosphorylation.
Interestingly, we noticed that a shifted modification of Bcl-2 occurred in XS561-treated MCF-7/LCC2 cells (Fig. 2B and C). Since Bcl-2 phosphorylation could convert Bcl-2 to a proapoptotic form39,40, we speculated that Bcl-2 phosphorylation might be modified during XS561 treatment. Considering the small difference in band position between regular Bcl-2 and modified Bcl-2, we used Mn(II)-Phosbind SDS-PAGE for the Western blotting assay, which can better retain phosphorylation-modified proteins to enhance the difference between regular and phosphorylation-modified proteins on SDS-PAGE. The up-shifted Bcl-2 band disappeared when lysates were treated with TAP (Fig. 6F), demonstrating that XS561 induced phosphorylation of Bcl-2. Bcl-2 can be phosphorylated by JNK and MAPK signaling pathway39,40. Thus, we investigated whether XS561 could activate JNK, p38 MAPK, and ERK MAPK in MCF-7/LCC2 cells. Fig. 6G shows that both p-JNK and p-p38 MAPK were activated by XS561, while ERK activation was inhibited by XS561. We next examined whether XS561 activation of JNK and MAPK was required for XS561-induced Bcl-2 phosphorylation and apoptotic effect by using chemical inhibitors of JNK (JNKI, SP6001125) and MAPK (p38I, SB203580). Treatment with JNKI completely blocked XS561 activation of JNK, Bcl-2 phosphorylation and PARP cleavage, whereas treatment with p38I had no effect (Fig. 6H). We also examined whether XS561 induced Bcl-2 phosphorylation by directly binding to Nur77. Nur77-H372A, but not Nur77 or Nur77-C566K, diminished the effect of XS561 on induction of Bcl-2 phosphorylation and PARP cleavage (Fig. 6I). Together, these data demonstrate that XS561 binding to Nur77 resulted in JNK activation and subsequent Bcl-2 phosphorylation.
3.8. Bcl-2 phase separation promotes Nur77-Bcl-2 apoptotic pathway
Phosphorylation of Thr69, Ser70 and Ser84 (TSS) sites in the LOOP of Bcl-2 plays a crucial role in determining the death effect of Bcl-239,40. Since phosphorylation is also important in regulating phase separation41,42, we asked whether Bcl-2 phosphorylation modulated its phase separation. We first used the program of PONDR (VSL2) (http://www.pondr.com/)43 to predict whether Bcl-2 has the ability to phase separate, and found that it displays the highest disorder tendency in its LOOP that is responsible for binding to Nur77 (Fig. 7A). To study the role of phosphorylation in Bcl-2 phase separation, we substituted the TSS sites of Bcl-2 with Alanine (Bcl-2-3A) or Glutamic acid (Bcl-2-3E), and tested their effect on Bcl-2 body formation. Bcl-2 and Bcl-2-3E formed more droplets with addition of XS561 or BI1071, whereas Bcl-2-3A and Bcl-2-ΔLOOP exhibited amorphous condensates (Fig. 7B). To study whether Bcl-2 and Bcl-2-3E underwent LLPS in vivo, we transfected GFP-Bcl-2-3E and GFP-Bcl-2 plasmids in HeLa cells. FRAP analysis revealed that GFP-Bcl-2 (t1/2 = 10.46 s) and GFP-Bcl-2-3E (t1/2 = 6.34 s) redistributed quickly to from unbleached to bleached area (Fig. 7C and D). Thus, Bcl-2 and Bcl-2-3E exhibited liquid-like properties via LLPS. This result together with data showing that deleting LOOP from Bcl-2 (Bcl-2-ΔLOOP) or dephosphorylation of TSS sites (Bcl-2-3A) impaired the effect of XS561 in inducing Bcl-2 lose phase separation reveal a critical role of Bcl-2 phosphorylation in XS561 induction of Bcl-2 phase separation.
Figure 7.
Bcl-2 phase separation promotes Nur77-Bcl-2 apoptotic pathway. (A) The Intrinsic disorder tendency of Bcl-2. Bcl-2 contains four short conserved functional domains BH (1–4), TM, transmembrane domains, and loosely structured LOOP ring regions. (B) Bcl-2 and its mutations formed droplets when HeLa cells were treated with different compounds (XS561: 10 μmol/L, BI1071: 0.5 μmol/L). Left: Illustrations of transfected Bcl-2 and its mutations in droplets. A magnified picture of the inset displays as well. Scale bar, 3 μm. Right: Quantification of Bcl-2 and its mutations' droplet formation. Data represent at least three independent experiments. (C, D) The evolution of the fluorescence intensity of GFP-Bcl-2 (C) and GFP-Bcl-2-3E (D) droplets following photobleaching. Data are presented as mean values ± SEM (n = 3 independent experiments). Scale bar, 1.5 μm. (E)–(G) After being exposed to 10 μmol/L XS561 for 6 h, MDA-MB-231-siBcl-2 cells transfected with Myc-cmv, Myc-Bcl-2, and their mutants were examined for PARP cleavage by Western blotting (E), JC-1 staining (F) and annexin V/PI (G) staining by flow cytometry. Data represent at least three independent experiments.
We also examined whether XS561 induced Bcl-2 phase separation by binding to Nur77. XS561 did not induce the formation of Bcl-2 body and Nur77-Bcl-2 condensates in Nur77−/− HeLa cells (Fig. S9D). However, transfection of Nur77 but not Nur77-H372A into Nur77−/− HeLa cells could rescue the effects of XS561 (Fig. S9E and S9F), suggesting that XS561 promoted the body formation of Bcl-2 by binding to Nur77. In studying the role of Bcl-2 phosphorylation in XS561-induced Nur77/Bcl-2 apoptotic pathway, we found that re-expressing Bcl-2-3E or Bcl-2 in MDA-MB-231-siBcl-2 cells and Bcl-2−/−MEFs could rescue the effects of XS561 on inducing PARP cleavage (Fig. 7E, Fig. S9I), activating apoptosis assessing by Annexin V/PI staining (Fig. 7F, Fig. S9G and S9J), and decreasing ΔΨm (Fig. 7G, Fig. S9H and S9K) in these cells, whereas transfection of Bcl-2-3A or Bcl-2-ΔLOOP could not (Fig. 7E–G, Fig. S9G–S9K). Thus, the phosphorylation of Bcl-2 is a critical event in XS561-induced Nur77/Bcl-2 apoptotic pathway.
3.9. XS561 promotes the formation of Nur77/Bcl-2 condensates
In studying the effect of XS561 in Nur77 interaction with Bcl-2 and the formation of Nur77/Bcl-2 condensates, we found that XS561 could promote Nur77 interaction with Bcl-2 in MCF-7/LCC2 cells revealed by co-IP assay (Fig. 8A). XS561 also promoted the interaction of ectopically-expressed Nur77 with endogenous Bcl-2 in MDA-MB-231 cells and Flag-Bcl-2 in HEK293T cells. For comparison, celastrol, which induces Nur77-dependent mitophagy28,29, did not show any effect on Nur77 interaction with Bcl-2 (Supporting Information Fig. S10A and S10B), demonstrating the specific effect of XS561. Immunofluorescence analysis demonstrated that XS561 but not celastrol induced the co-localization between Nur77 and Bcl-2 (Fig. S10C). By contrast, celastrol but not XS561 induced the formation of Nur77/p62 condensates, a critical event for Nur77-mediated mitophagy28,29. Consistently, celastrol but not XS561 induced Nur77 interaction (Fig. S10D) and colocalization (Fig. S10E) with p62. Thus, XS561 selectively induced Nur77 condensation with Bcl-2 but not p62.
Figure 8.
XS561 promotes the Nur77/Bcl-2 condensates. (A) Co-IP assay was used to study the endogenous interaction of Bcl-2 with Nur77 in MCF-7/LCC2 cells treated with or without XS561. (B)–(E) HeLa cells transfected with GFP-Nur77 or mutants and Flag-Bcl-2 were treated with or without XS561 (10 μmol/L) for 2 h, followed by staining with anti-Bcl-2 antibodies, and then observed by confocal microscopy. Scale bar, 3 μm. (C) HEK293T cells transfected with GFP-Nur77 or mutants and Flag-Bcl-2 were treated with or without XS561 (10 μmol/L) for 2 h, and then examined by co-IP assay by using anti-Flag antibodies. IgG was used as a negative control. (D) Co-IP assay was used to study the interaction of Bcl-2 with Nur77 or Nur77-H372A in HEK293T cells treated with or without XS561. (F) HeLa cells transfected with Bcl-2 or mutants and GFP-Nur77 were treated with or without XS561 (10 μmol/L) for 2 h, followed by staining with anti-Bcl-2 antibodies, and then observed by confocal microscopy. Scale bar, 3 μm. (G) HEK293T cells were transfected with Bcl-2, Bcl-2 mutants, Nur77, and their interaction was examined by co-IP assay.
Confocal microscopy analysis demonstrated that XS561 induced the formation of GFP-Nur77/Bcl-2 condensates at mitochondria (Fig. 8B). However, Nur77-LBD capable of interacting with Bcl-2 (Fig. 8C) formed spherical particle with Bcl-2 at mitochondria, demonstrating the importance of IDR in Nur77 condensation with Bcl-2. Nur77-ΔIDR, which predominantly resided in the nucleus (Fig. 5), and Nur77-IDR, which cannot interact with Bcl-2 (Fig. 8C), failed to form condensates with Bcl-2 at mitochondria (Fig. 8B). Consistent with their inability to bind XS561, Nur77-H372A or Nur77-LBD-H372A were incapable of forming condensates with Bcl-2 (Fig. 8D and E). Thus, the condensation of Nur77 with Bcl-2 at mitochondria requires not only their interaction but also the phase separation of Nur77. To study whether the phase separation of Bcl-2 was required for its condensation with Nur77, the condensation of Bcl-2 and mutants was analyzed. Bcl-2 and Bcl-2-3E could interact with Nur77 and form condensates with Nur77 upon XS561 stimulation, whereas Bcl-2-3A or Bcl-2-ΔLOOP displayed impaired Nur77 interaction and formation of Nur77/Bcl-2 condensates (Fig. 8F and G, Fig. S10F and S10G). Thus, the phase separation of Bcl-2 also contributed to formation of Nur77/Bcl-2 condensates.
4. Discussion
Nur77 translocating from the nucleus to mitochondria and its subsequent interaction with Bcl-2 can convert Bcl-2 from a survival to a killer of tumor cells11,12, representing a promising strategy to develop new cancer therapeutics9,44. Although several small molecules that bind Nur77 to trigger Nur77-dependent apoptosis have been identified, their efficacy and selectivity remain to be improved30,45, 46, 47. Given that Nur77 lacks classical LBP34,48, the search for small molecules that specifically bind to Nur77 to activate its apoptotic pathway remains a great challenge. We report here our identification of XS561 as a selective Nur77 binder with potent effect on inducing apoptosis of tumor cells including TNBC and TAMR cells. Our mechanistic studies revealed a critical role of the phase separation of Nur77 and Bcl-2 in mediating the apoptotic effect of XS561 through the formation of Nur77/Bcl-2 condensates at mitochondria.
Although the classical ligand-binding pocket of Nur77 is fulfilled with bulky side chains of amino acid, preventing it from binding small molecules48, recent development have demonstrated that some grooves on the surface of Nur77 can serve as binding sites for small molecule ligands35,36. In this study, we used Glide to perform docking study of 1.5 million compounds commercially available from ChemDiv. After comparing with the TMPA binding mode in the crystal structure (PDB code: 3V3Q), finally 10 compounds (Fig. S1B) were purchased for biological evaluation. Binding assays confirmed that XS561 could bind Nur77 with a Kd of low micromolar. Molecular docking suggested that XS561 bound to a native groove formed among helices 1, 5, 7 and 8 of Nur77 surface and formed H-bond interactions with His372 and Leu373 (Fig. 1C and D). In line with our previous study, the His372 site is the key residue involving in Nur77-Bcl-2-apoptotic pathway30. Mutagenesis studies confirmed that His372 was the key residue involving in the binding of XS561. Compared to the Nur77-LBD/TMPA complex, XS561 occupied a deeper groove and formed H-bond interactions with His372 and Leu373 that were not observed in the binding of TMPA to Nur7735. The specific binding model of XS561 to Nur77 provides a proper site for Bcl-2 to interact, which promotes their phase separation (Figure 5, Figure 7) and interaction (Fig. 8), and finally induces cell apoptosis in tumor cells (Figure 2, Figure 4, Figure 6).
Apoptosis induction through the Nur77//Bcl-2 pathway has been reported in prostate12, lung cancer49,50, ovarian51 and ER-positive breast cancer23. TNBC has few targeted therapies available, and hence there has been tremendous interest in identifying new suitable targeted therapies for tumor type. As Bcl-2 is overexpressed in TNBC17, Bcl-2 inhibitors or its combination with cytotoxic agents have been extensively investigated for treating TNBC52. The Bcl-2 inhibitor ABT-199 either alone or in combination showed great effectiveness in inhibiting TNBC cells growth in vitro and in animals53. As Nur77 is also expressed in TNBC cells54 (Fig. 2A), targeting Nur77 to convert Bcl-2 phenotype may provide an effective way to induce apoptosis of TNBC. Our data demonstrate that XS561 strongly induced apoptosis of MDA-MB-231 cells in vitro and inhibited the growth of MDA-MB-231 xenograft tumor in animals. Previous studies showed that overexpression of Bcl-2 contributed to the development of resistance of breast cancer to tamoxifen20. Interestingly, we found that Nur77 was also elevated during the development of tamoxifen resistance in MCF-7 cells (Fig. 2B), suggesting the feasibility of using XS561 to overcome tamoxifen resistance. Indeed, our results showed that XS561 was even more effective in inducing apoptosis in MCF-7/LCC2 TAMR cells than in MCF-7 cells (Fig. 2B). XS561 also showed a strong synergistic effect on inducing apoptosis of MCF-7/LCC2 when it was used in combination with tamoxifen (Fig. 2C). Our results therefore identify XS561 as a promising agent to overcome tamoxifen resistance of breast cancer cells with elevated levels of Bcl-2 and Nur77.
Biomolecular condensates mediated through LLPS affect various biological processes during tumorigenesis, including DNA damage repair, transcription, chromatin changes and tumor suppression. These changes in phase separation may lead to abnormal cellular activities such as promoting angiogenesis, unrestricted proliferation, invasion and metastasis, and resistance to death55, 56, 57. For example, ERα is selectively concentrated in an estrogen-dependent manner into a MED1 condensate, through which tumor cells efflux and dilute tamoxifen, leading to drug resistance58. Although phase separation was initially elucidated through different pathways to promote tumor growth, strategies to target phase separation for tumor therapy remain to be developed. Our data demonstrated that XS561 promoted the phase separation of Nur77-IDR by binding to Nur77-LBD. In addition, it enhanced the phase separation of the LOOP of Bcl-2 by inducing Bcl-2 phosphorylation. XS561-induced phase separation of Nur77 and Bcl-2 resulted in their condensation at mitochondria, thereby leading to induction of mitochondria-mediated apoptosis. Notably, the LOOP of Bcl-2 is an important determinant of Bcl-2 function12,40. For example, phosphorylation of Bcl-2 can promote survival by facilitating dissociation with Becline-1 and promote autophagy and thus survival40. Phosphorylation of Bcl-2 can also promote apoptosis by enhancing its interaction with Bax40. However, how the flexible LOOP domain of Bcl-2 regulates its function remains largely undefined. In the present study, we found for the first time that LOOP phosphorylation of Bcl-2 promotes its phase separation and condensation with Nur77 at mitochondria. Thus, our finding that the LOOP domain can undergo phosphorylation-dependent phase separation offers a plausible explanation for regulating its complex biological activities.
5. Conclusions
Our studies identify XS561 as a selective Nur77 binder that effectively induces Nur77/Bcl-2 apoptotic pathway. By targeting the Nur77/Bcl-2 apoptotic pathway, XS561 not only effectively promotes apoptosis of tumor cells including triple-negative breast cancer cells but also overcomes the resistance of breast cancer cells to tamoxifen. With its improved safety profile, XS561 represents a promising drug lead that warrants further investigation. Our mechanistic studies reveal a crucial role of phase separation of Nur77 and Bcl-2 in mediating the formation of Nur77/Bcl-2 condensates at mitochondria, providing a mechanistic insight into Nur77/Bcl-2-mediated apoptosis. Our finding that the flexible LOOP domain of Bcl-2 can undergo phosphorylation-dependent phase separation offers a plausible explanation for complex and diverse function of Bcl-2 and its regulation.
Acknowledgments
This work was partially supported by grants from the The National Natural Science Foundation of China, China (81672749 and 22207093), Xiamen Bureau of Science & Technology, Xiamen city, China (3502Z20193004 and 3502Z20150007, China), Youth Innovation Fund from Xiamen City, Fujian Province, China. (3502Z20206033) and the Fujian Provincial Key Laboratory of Innovative Drug Target Research, Fujian Province, China (FJ-YW-2022KF01, China).
Author contributions
Xiaohui Chen, Meichun Gao and Yongzhen Xia conducted molecular, cellular and animal experiments and analyzed the data; Xin Wang, Zhiping Zeng and Ying Su conducted the compound discovery experiments. Jingbo Qin, Hongying He, Weirong Liu, Xiaowei Zhang and Shuangzhou Peng provided technical assistance. Xiaohui Chen, Ying Su and Xiaokun Zhang conceived and designed the study, Xiaohui Chen, Zhiping Zeng, Ying Su and Xiaokun Zhang wrote the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.11.017.
Contributor Information
Zhiping Zeng, Email: zengzhiping@xmu.edu.cn.
Ying Su, Email: ying.su@nucmito.com.
Xiaokun Zhang, Email: xkzhang@xmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kolluri S.K., Bruey-Sedano N., Cao X., Lin B., Lin F., Han Y.H., et al. Mitogenic effect of orphan receptor TR3 and its regulation by MEKK1 in lung cancer cells. Mol Cell Biol. 2003;23:8651–8667. doi: 10.1128/MCB.23.23.8651-8667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson A.J., Arango D., Mariadason J.M., Heerdt B.G., Augenlicht L.H. TR3/Nur77 in colon cancer cell apoptosis. Cancer Res. 2003;63:5401–5407. [PubMed] [Google Scholar]
- 3.Hedrick E., Lee S.O., Doddapaneni R., Singh M., Safe S. Nuclear receptor 4A1 as a drug target for breast cancer chemotherapy. Endocr Relat Cancer. 2015;22:831–840. doi: 10.1530/ERC-15-0063. [DOI] [PubMed] [Google Scholar]
- 4.Muscat G.E., Eriksson N.A., Byth K., Loi S., Graham D., Jindal S., et al. Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Mol Endocrinol. 2013;27:350–365. doi: 10.1210/me.2012-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F., Drabsch Y., Dekker T.J., de Vinuesa A.G., Li Y., Hawinkels L.J., et al. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-β signalling. Nat Commun. 2014;5:3388. doi: 10.1038/ncomms4388. [DOI] [PubMed] [Google Scholar]
- 6.Hedrick E., Safe S. Transforming growth factor β/NR4A1-inducible breast cancer cell migration and epithelial-to-mesenchymal transition is p38α (mitogen-activated protein kinase 14) dependent. Mol Cell Biol. 2017;37 doi: 10.1128/MCB.00306-17. 00306-00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie L., Jiang F.Q., Zhang X.D., Alitongbieke G., Shi X.L., Meng M.J., et al. Honokiol sensitizes breast cancer cells to TNF-α induction of apoptosis by inhibiting Nur77 expression. Br J Pharmacol. 2016;173:344–356. doi: 10.1111/bph.13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu G., Lu L., Gan J., Zhang D., Liu J., Huang G. Dual roles of orphan nuclear receptor TR3/Nur77/NGFI-B in mediating cell survival and apoptosis. Int Rev Cell Mol Biol. 2014;313:219–258. doi: 10.1016/B978-0-12-800177-6.00007-4. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X.K. Targeting Nur77 translocation. Expert Opin Ther Targets. 2007;11:69–79. doi: 10.1517/14728222.11.1.69. [DOI] [PubMed] [Google Scholar]
- 10.Moll U.M., Marchenko N., Zhang X.K. p53 and Nur77/TR3—transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 11.Li H., Kolluri S.K., Gu J., Dawson M.I., Cao X., Hobbs P.D., et al. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289:1159–1164. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- 12.Lin B., Kolluri S.K., Lin F., Liu W., Han Y.H., Cao X., et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 13.Czabotar P.E., Lessene G., Strasser A., Adams J.M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 14.Chipuk J.E., Moldoveanu T., Llambi F., Parsons M.J., Green D.R. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Youle R.J., Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 16.Dawson S.J., Makretsov N., Blows F.M., Driver K.E., Provenzano E., Le Quesne J., et al. BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer. 2010;103:668–675. doi: 10.1038/sj.bjc.6605736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 18.Kumar R., Mandal M., Lipton A., Harvey H., Thompson C.B. Overexpression of HER2 modulates bcl-2, bcl-XL, and tamoxifen-induced apoptosis in human MCF-7 breast cancer cells. Clin Cancer Res. 1996;2:1215–1219. [PubMed] [Google Scholar]
- 19.Cory S., Huang D.C., Adams J.M. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 20.Raha P., Thomas S., Thurn K.T., Park J., Munster P.N. Combined histone deacetylase inhibition and tamoxifen induces apoptosis in tamoxifen-resistant breast cancer models, by reversing Bcl-2 overexpression. Breast Cancer Res. 2015;17:26. doi: 10.1186/s13058-015-0533-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu D., Li Y., Zheng L., Xiao H., Ouyang L., Wang G., et al. Small molecules targeting protein–protein interactions for cancer therapy. Acta Pharm Sin B. 2023;13:4060–4088. doi: 10.1016/j.apsb.2023.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng J. How to unleash mitochondrial apoptotic blockades to kill cancers?. Acta Pharm Sin B. 2017;7:18–26. doi: 10.1016/j.apsb.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolluri S.K., Zhu X., Zhou X., Lin B., Chen Y., Sun K., et al. A short Nur77-derived peptide converts Bcl-2 from a protector to a killer. Cancer Cell. 2008;14:285–298. doi: 10.1016/j.ccr.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flemming A. New strategies to tip the Bcl-2 balance. Nat Rev Drug Discov. 2008;7:977. [Google Scholar]
- 25.Martz L. Bcl-2 double take. SciBX. 2008;1:938. [Google Scholar]
- 26.Qi B., Hardwick J.M. Bcl-2 turns deadly. Nat Chem Biol. 2008;4:722–723. doi: 10.1038/nchembio1208-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Case L.B., Zhang X., Ditlev J.A., Rosen M.K. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science. 2019;363:1093–1097. doi: 10.1126/science.aau6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu M.J., Luo Q., Alitongbieke G., Chong S.Y., Xu C.T., Xie L., et al. Celastrol-induced Nur77 interaction with TRAF2 alleviates inflammation by promoting mitochondrial ubiquitination and autophagy. Mol Cell. 2017;66:141–153. doi: 10.1016/j.molcel.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng S.Z., Chen X.H., Chen S.J., Zhang J., Wang C.Y., Liu W.R., et al. Phase separation of Nur77 mediates celastrol-induced mitophagy by promoting the liquidity of p62/SQSTM1 condensates. Nat Commun. 2021;12:5989. doi: 10.1038/s41467-021-26295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X.H., Cao X.H., Tu X.H., Alitongbieke G., Xia Z.B., Li X.T., et al. BI1071, a novel Nur77 modulator, induces apoptosis of cancer cells by activating the Nur77–Bcl-2 apoptotic pathway. Mol Cancer Therapeut. 2019;18:886–899. doi: 10.1158/1535-7163.MCT-18-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lionta E., Spyrou G., Vassilatis D.K., Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr Top Med Chem. 2014;14:1923–1938. doi: 10.2174/1568026614666140929124445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., Zhu Y., Chen J., Yang Y., Zhu L., Zhao J., et al. Identification of a novel PAK1 inhibitor to treat pancreatic cancer. Acta Pharm Sin B. 2020;10:603–614. doi: 10.1016/j.apsb.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flaig R., Greschik H., Peluso-Iltis C., Moras D. Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain. J Biol Chem. 2005;280:19250–19258. doi: 10.1074/jbc.M413175200. [DOI] [PubMed] [Google Scholar]
- 35.Zhan Y.Y., Chen Y., Zhang Q., Zhuang J.J., Tian M., Chen H.Z., et al. The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nat Chem Biol. 2012;8:897–904. doi: 10.1038/nchembio.1069. [DOI] [PubMed] [Google Scholar]
- 36.Wang W.J., Wang Y., Chen H.Z., Xing Y.Z., Li F.W., Zhang Q., et al. Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway. Nat Chem Biol. 2014;10:133–140. doi: 10.1038/nchembio.1406. [DOI] [PubMed] [Google Scholar]
- 37.Brunner N., Frandsen T.L., Holst-Hansen C., Bei M., Thompson E.W., Wakeling A.E., et al. MCF7/LCC2: a 4-hydroxytamoxifen resistant human breast cancer variant that retains sensitivity to the steroidal antiestrogen ICI 182,780. Cancer Res. 1993;53:3229–3232. [PubMed] [Google Scholar]
- 38.Wang J.L., Liu D., Zhang Z.J., Shan S., Han X., Srinivasula S.M., et al. Structure-based discovery of an organic compound that binds Bcl-2 protein and induces apoptosis of tumor cells. Proc Natl Acad Sci U S A. 2000;97:7124–7129. doi: 10.1073/pnas.97.13.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blagosklonny M.V. Unwinding the loop of Bcl-2 phosphorylation. Leukemia. 2001;15:869–874. doi: 10.1038/sj.leu.2402134. [DOI] [PubMed] [Google Scholar]
- 40.Wei Y., Sinha S., Levine B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy. 2008;4:949–951. doi: 10.4161/auto.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu H., Yu D., Hansen A.S., Ganguly S., Liu R., Heckert A., et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558:318–323. doi: 10.1038/s41586-018-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emperador-Melero J., Wong M.Y., Wang S.S.H., de Nola G., Nyitrai H., Kirchhausen T., et al. PKC-phosphorylation of Liprin-α3 triggers phase separation and controls presynaptic active zone structure. Nat Commun. 2021;12:3057. doi: 10.1038/s41467-021-23116-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pearce M.C., Satterthwait A.C., Zhang X.K., Kolluri S.K. Cancer therapeutics based on BCL-2 functional conversion. Apoptosis. 2019;24:1–2. doi: 10.1007/s10495-018-1504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu X., Chen X., Zhang D., Gao M., Liang J., Bao G., et al. Optimization of novel oxidative DIMs as Nur77 modulators of the Nur77–Bcl-2 apoptotic pathway. Eur J Med Chem. 2021;211 doi: 10.1016/j.ejmech.2020.113020. [DOI] [PubMed] [Google Scholar]
- 46.Chen X., Tu X., Zhang X., Cao B., Liu W., Zhang J., et al. SAR study of oxidative DIMs analogs targeting the Nur77-mediated apoptotic pathway of cancer cells. Bioorg Chem. 2022;129 doi: 10.1016/j.bioorg.2022.106156. [DOI] [PubMed] [Google Scholar]
- 47.Qin J., Chen X., Liu W., Chen J., Liu W., Xia Y., et al. Discovery of 5-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)-1H-indole-2-carboxamide derivatives as novel anti-cancer agents targeting Nur77. Eur J Med Chem. 2022;244 doi: 10.1016/j.ejmech.2022.114849. [DOI] [PubMed] [Google Scholar]
- 48.Kurakula K., Koenis D.S., van Tiel C.M., de Vries C.J. NR4A nuclear receptors are orphans but not lonesome. Biochim Biophys Acta. 2014;1843:2543–2555. doi: 10.1016/j.bbamcr.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 49.Han Y.H., Cao X., Lin B., Lin F., Kolluri S.K., Stebbins J., et al. Regulation of Nur77 nuclear export by c-Jun N-terminal kinase and Akt. Oncogene. 2006;25:2974–2986. doi: 10.1038/sj.onc.1209358. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y.Q., Zhao W., Xie G.B., Huang M.F., Hu M.J., Jiang X., et al. Induction of Nur77-dependent apoptotic pathway by a coumarin derivative through activation of JNK and p38 MAPK. Carcinogenesis. 2014;35:2660–2669. doi: 10.1093/carcin/bgu186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J., Wang G.H., Duan Y.H., Dai Y., Bao Y.Z., Hu M.J., et al. Modulation of the Nur77–Bcl-2 apoptotic pathway by p38α MAPK. Oncotarget. 2017;8:69731–69745. doi: 10.18632/oncotarget.19227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumari M., Krishnamurthy P.T., Sola P. Targeted drug therapy to overcome chemoresistance in triple negative breast cancer. Curr Cancer Drug Targets. 2020;20:559–572. doi: 10.2174/1568009620666200506110850. [DOI] [PubMed] [Google Scholar]
- 53.Inao T., Iida Y., Moritani T., Okimoto T., Tanino R., Kotani H., et al. Bcl-2 inhibition sensitizes triple-negative human breast cancer cells to doxorubicin. Oncotarget. 2018;9:25545–25556. doi: 10.18632/oncotarget.25370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Q., Dawson M.I., Zheng Y., Hobbs P.D., Agadir A., Jong L., et al. Inhibition of trans-retinoic acid-resistant human breast cancer cell growth by retinoid X receptor-selective retinoids. Mol Cell Biol. 1997;17:6598–6608. doi: 10.1128/mcb.17.11.6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong X., Tang R., Xu J., Wang W., Zhao Y., Yu X., et al. Liquid–liquid phase separation in tumor biology. Signal Transduct Targeted Ther. 2022;7:221. doi: 10.1038/s41392-022-01076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 57.Bradner J.E., Hnisz D., Young R.A. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klein I.A., Boija A., Afeyan L.K., Hawken S.W., Fan M., Dall'Agnese A., et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.