Abstract
We undertook a comprehensive phylogenetic study to establish the genetic relationship among the viruses of the genus Flavivirus and to compare the classification based on molecular phylogeny with the existing serologic method. By using a combination of quantitative definitions (bootstrap support level and the pairwise nucleotide sequence identity), the viruses could be classified into clusters, clades, and species. Our phylogenetic study revealed for the first time that from the putative ancestor two branches, non-vector and vector-borne virus clusters, evolved and from the latter cluster emerged tick-borne and mosquito-borne virus clusters. Provided that the theory of arthropod association being an acquired trait was correct, pairwise nucleotide sequence identity among these three clusters provided supporting data for a possibility that the non-vector cluster evolved first, followed by the separation of tick-borne and mosquito-borne virus clusters in that order. Clades established in our study correlated significantly with existing antigenic complexes. We also resolved many of the past taxonomic problems by establishing phylogenetic relationships of the antigenically unclassified viruses with the well-established viruses and by identifying synonymous viruses.
The genus Flavivirus of the family Flaviviridae comprises over 70 viruses, many of which, such as the dengue (DEN) viruses, Japanese encephalitis (JE) virus, St. Louis encephalitis (SLE) virus, and yellow fever (YF) virus are important human pathogens (22, 31). Dengue and its severe and sometimes fatal forms, dengue hemorrhagic fever and dengue shock syndrome, alone affect nearly 80 million people a year (30). As demonstrated in recent outbreaks of meningitis by West Nile (WN) virus in Algeria and Romania, viruses of this group sometimes cause serious public health concern in unexpected locations (27).
Most of these viruses were serologically classified into eight antigenic complexes, but many viruses, including the prototype of this group, YF virus, could not be affiliated with any complexes (6). Furthermore, many new viruses have been documented since the establishment of the serological classification, but their overall relationship with the other viruses has not been determined. The difficulty encountered with flavivirus classification partly derives from the extensive geographic distribution and the diversity of the arthropod vectors or vertebrates hosts associated with biological transmission of these viruses. Also, it derives from a confusion in virus nomenclature. For example, tick-borne encephalitis virus strains isolated primarily in western parts of Eurasia have been called TBE viruses, but, as clearly pointed out by Calisher (5), no such virus as tick-borne encephalitis virus (or TBE) has ever been registered to an international body dedicated to virus taxonomy. To compound the problem further, an increasing number of viruses have been added as new members of so-called TBE complex without a virus definition provided (16, 18, 29, 42, 52). This practice clearly demonstrates a need for establishing objective criteria for a better classification of those viruses.
Molecular genetic classification of these viruses has been attempted before. In all previous studies, fewer than one-third of the members, primarily mosquito-borne and tick-borne viruses, were used to create phylogenetic trees, which showed evolution of mosquito-borne and tick-borne viruses from the presumed ancestor (3, 11). Since few sequence data were available from other viruses, in particular the viruses without known vectors (hereafter called the non-vector group), those phylogenetic trees provided only partial information.
To establish a comprehensive phylogeny of the genus Flavivirus, we attempted to obtain the genomic sequence of a 1.0-kb segment at the 3′ terminus of the NS5 gene from all viruses whose sequences were not available. We analyzed, together with the other sequence data already published, the genetic relationships among the members of this group. Quantitative criteria based on a combination of the bootstrap support level and the pairwise nucleotide sequence identity were established to define subgeneric taxa. These included cluster, clade, and species. With our new taxonomic definitions, we then compared our genetic classification with the traditional system based on serological data.
MATERIALS AND METHODS
Viruses.
The 58 viruses and one geographic strain of YF virus sequenced in this study and 13 viruses (including cell fusing agent [CFA]) whose NS5 gene sequences had been already available in GenBank are listed in Table 1. The majority of the viruses sequenced were obtained from the World Health Organization (WHO) Collaborating Center for Reference and Research in our institution. For three tick-borne viruses (Kyasanur Forest Disease [KFD]), Russian spring summer encephalitis [RSSE], and Omsk hemorrhagic fever (Omsk HF]), extracted viral RNAs were obtained from the Special Pathogens Branch, Division of Viral and Rickettsial Diseases, Centers for Disease Control and Prevention. Iguape and Kedougou viruses were obtained from the WHO International Reference Center, University of Texas Health Center, Galveston. A total of nine viruses were not used in this study. These included louping ill, Wesselsbron, Spanish sheep tick-borne encephalitis, Greek goat encephalitis, and Turkish sheep tick-borne encephalitis viruses, whose importation and usage is restricted by the U.S. Department of Agriculture. In addition, TBE complex viruses (Absettarov, Kumlinge, Hanzalova, and Hypr), which require higher biosafety facilities, were unavailable in our laboratory.
TABLE 1.
Flaviviruses used in the phylogenetic study
| Virus (strain) | Vector associationa | Distribution | GenBank accession no.b |
|---|---|---|---|
| Alfuy (MRM-3929) | M | Australia | AF013360 |
| Apoi (original) | N (rodent) | Japan | AF013361 |
| Aroa (VenA-1809) | N (sentinel rodent) | Venezuela | AF013362 |
| Bagaza (DakAr B209) | M | Central African Republic | AF013363 |
| Banzi (SAH-336) | M | South Africa and East Africa | L40951c |
| Batu Cave (P70-1459) | N (bat) | Malaysia | AF013369 |
| Bouboui (DakAr B490) | M | Central and West Africa | AF013364 |
| Bukalasa bat (UGBP-111) | N (bat) | Uganda | AF013365 |
| Bussuquara (BeAn 4073) | M | South America | AF013366 |
| Cacipacore (BeAn 327600) | N (bird) | Brazil | AF013367 |
| Carey Island (P70-1215) | N (bat) | Malaysia | AF013368 |
| Cowbone Ridge (W-10986) | N (rodent) | Florida | AF013370 |
| Dakar bat (209) | N (bat) | West and central Africa and Madagascar | AF013371 |
| Dengue type 1 (Singapore S275/90) | M | Tropical and semitropical areas | M87512c |
| Dengue type 2 (Jamaica) | M | Tropical and semitropical areas | M20558c |
| Dengue type 3 (H-87) | M | Tropical and semitropical areas | M93130c |
| Dengue type 4 (814669) | M | Tropical and semitropical areas | M17255c |
| Edge Hill (Aus C-281) | M | Australia | AF013372 |
| Entebbe bat (UgIL-30) | N (bat) | Uganda | AF013373 |
| Gadgets Gully (CSIRO 122) | T | Australia | AF013374 |
| Iguape (SP An71686) | N (sentinel rodent) | Brazil | AF013375 |
| Ilheus (original) | M | Central and South America, Trinidad | AF013376 |
| Israel turkey meningoencephalitis (original) | M | Israel, South Africa | AF013377 |
| Japanese encephalitis (SA-14) | M | Asia and parts of the Pacific | U15763c |
| Jugra (P9-314) | M | Malaysia | AF013378 |
| Jutiapa (JG-128) | N (rodent) | Guatemala | AF013379 |
| Kadam (MP-6640) | T | Uganda, Saudi Arabia | AF013380 |
| Karshi (LEIV-2247) | T | Kazakhstan | AF013381 |
| Kedougou (Dak Aar D1470) | M | Senegal | AF013382 |
| Kokobera (AusMRM 281) | M | Australia | AF013383 |
| Koutango (DakAr D1470) | M, T | Senegal, Central African Republic | AF013384 |
| Kunjin (MRM61C) | M | Australia, Asia | D00246c |
| Kyasanur Forest disease (W371) | T | India | AF013385 |
| Langat (TP21) | T | Southeast Asia, Russia | M86650c |
| Meaban (Brest ART707) | T | France | AF013386 |
| Modoc (M544) | N (rodent) | Western United States | AF013387 |
| Montana myotis leukoencepalitis (40649) | N (bat) | Montana | AF013388 |
| Murray Valley encephalitis (original) | M | Australia, Papua New Guinea | AF013389 |
| Naranjal (25008) | M | Ecuador | AF013390 |
| Negishi (original) | N (human) | Japan and former Soviet Union | AF013391 |
| Ntaya (original) | M | Central and South Africa | AF013392 |
| Omsk hemorrhagic fever (Kubrin) | T | Russia | AF013393 |
| Phnom Penh bat (CAMA-38D) | N (bat) | Cambodia, Malaysia | AF013394 |
| Potiskum (IBAN 10069) | N (rodent) | Nigeria | AF013395 |
| Powassan (LB) | T, M | North America, Russia | L06436c |
| Rio Bravo (M-64) | N (bat) | United States, Mexico | AF013396 |
| Rocio (H-34675) | M | Brazil | AF013397 |
| Royal Farm (EgArt 371) | T | Afghanistan | AF013398 |
| Russian spring summer encephalitis (Sofjin) | T | Russia, Japan | AF013399 |
| Saboya (Dak an D4600) | N (rodent) | Senegal | AF013400 |
| Sal Vieja (38TWM-106) | N (rodent) | Texas | AF013401 |
| San Perlita (71V-1251) | N (rodent) | Texas | AF013402 |
| Saumarez Reef (CSIRO-4) | T | Australia | AF013403 |
| Sepik (MK 7148) | M | Papua New Guinea | AF013404 |
| Sokuluk (LEIV-400K) | N (bat) | Kyrghystan | AF013405 |
| Spondweni (SAAR-94) | M | Africa | AF013406 |
| St. Louis encephalitis (MSI-7) | M | Americas | AF013416 |
| Stratford (AUSC-338) | M | Australia | AF013407 |
| Tick-borne encephalitis—central European subtype (Neudoerfl) | T | Europe | V27495c |
| Tick-borne encephalitis—far eastern subtype (Sofjin) | T | Europe | X07755c |
| Tembusu (MM 1775) | M | Southeast Asia | AF013408 |
| THCArd | M | Thialand | AF013409 |
| Tyuleniy (LEIV-6C) | T | Russia, Oregon | AF013410 |
| Uganda S (original) | M | Africa | AF013411 |
| Usutu (SAAR-1776) | M | Africa | AF013412 |
| West Nile (EG-101) | M, Te | Africa, Asia, Europe | M12294c |
| Yaounde (DakAr Y276) | M | Central African Republic | AF013413 |
| Yellow fever (Asibi) | M, Te | Tropical areas | K02749c |
| Yellow fever (TN-96)f | M | Tropical areas | AF013417 |
| Yokose (Oita 36) | N (bat) | Japan | AF013414 |
| Zika (MR-766) | M | Africa, Asia | AF013415 |
| Cell fusing agent | Unknown | Unknown | M91761c |
M, 1 mosquito; T, 1 tick; N, no known vector (host from which virus was first isolated is in parentheses).
From this study unless otherwise indicated.
From other investigators.
Isolate from mosquitoes in Thailand (20).
Tick is not the principal vector.
Isolate from a Tennessee resident upon return from a travel to the Amazon, Brazil, in 1996.
Reverse transcription (RT)-PCR.
Viral RNA was extracted from 126 μl of 10% suckling mouse brain suspension or cell culture supernatant fluid by using a Qia-HCV kit (Qiagen, Santa Clarita, Calif.) or from 50 μl of bulk mouse brain tissue by using RNeasy (Qiagen). RNA adsorbed on silica membrane was eluted in 50 μl of water.
For cDNA synthesis, 20 μl of viral RNA was mixed with 1 μl each of a forward and a reverse primer (50 μM) as well as 8 μl of water, and the mixture was heated at 92°C for 1 min and then cooled to 45°C. Thirty microliters of enzyme mix (10 μl of 5× reverse transcription buffer [Boehringer Mannheim, Indianapolis, Ind.], 5 μl of deoxynucleoside triphosphate [dNTP] mixture [10 mM each dNTP], 9 U of Rous sarcoma virus reverse transcriptase [RAV-2; Amersham, Cleveland, Ohio], and 4.5 μl of water) was added per tube, and tubes were incubated at 45°C for 45 min.
PCR was performed with a commercial kit (Expand Long Template PCR system; Boehringer Mannheim). Five microliters of cDNA was mixed with 5.0 μl of dNTPs (10 mM each), 1 μl each of forward and reverse primers (50 μM), and 33 μl of water. The reaction mixture was heated to 94°C, and then 50 μl of the enzyme mixture (5 μl of 10× PCR buffer, 0.75 to 1.5 μl of enzymes, 44 μl of water) was added. After heat denaturation at 94°C for 4 min, temperature was shifted to 45°C for 1 min and then to 68°C for 1 min. The thermocycle program was as follows: 3 cycles (94°C for 20 s, 45°C for 1 min, 68°C for 1 min), 10 cycles (94°C for 20 s, 50°C for 30 s, 68°C for 1 min), 16 cycles (94°C for 20 s, 50°C for 30 s, 68°C for 1 min in the first cycle, with an increment of 20 s per cycle thereafter). A final extension was at 68°C for 5 min.
Primers for DNA template amplification.
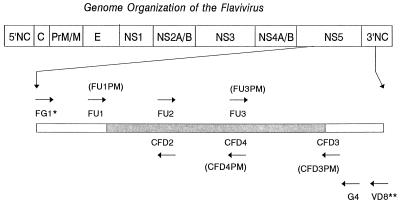
Primers were selected to sequence the genomic regions (nearly 1 kb long) at the 3′ terminus of the NS5 gene delineated between FU1 and cFD3 in Fig. 1. All primers used for DNA template amplification are listed in Table 2, and their relative genomic locations are shown in Fig. 1. For most viruses, a pair of primers (FU1 and cFD3), which had been previously determined (7) produced the desired amplicons. However, for those viruses which did not produce the expected amplicons, templates of various sizes were produced by using the other primers shown in Table 2.
FIG. 1.
Relative genomic positions of primers used for amplification of DNA templates from the NS5 gene of flaviviruses and for sequencing. ∗, From reference 14. ∗∗, The name in parentheses indicates a degenerate primer at the same location corresponding to the primer immediately above. ∗∗∗, From reference 33.
TABLE 2.
Primers for DNA template synthesis and sequencing
| Namea | Sequence (5′→3′) | Genomic positionb |
|---|---|---|
| FU1 (F) | TACAACATGATGGGAAAGAGAGAGAA | 8993 |
| FU1PMc (F) | TACAACATGATGGGVAARAGWGARAA | 8993 |
| cFD3 (R) | AGCATGTCTTCCGTGGTCATCCA | 10077 |
| cFD3PMd (R) | ARCATGTCTTCYGTBGTCATCCA | 10077 |
| FU2 (F) | GCTGATGACACCGCCGGCTGGGACAC | 9233 |
| cFD2 (R) | GTGTCCCAGCCGGCGGTGTCATCAGC | 9258 |
| FU3 (F) | AGCGGAGATGATTGTGTGGT | 9629 |
| FU3PMe (F) | AGYGGAGAYGAYTGYGTNGT | 9629 |
| cFD4 (R) | ACCACACAATCATCTCCGCT | 9648 |
| cFD4PMf (R) | ATNACRCARTCRTCTCCRCT | 9648 |
| FG1g (F) | TCAAGGAACTCCACACATGAGATGTACT | 8270 |
| G4 (R) | CCAGATGTTCTTWGCCCAYTCTGC | 10261i |
| VD8h (R) | GGGTCTCCTCTAACCTCTAG | 10728 |
F, forward primer; R, reverse primer.
Except for primer G4, numbers are genomic positions of YF virus (GenBank accession no. K02749) to which the first bases (5′ end) of primers correspond.
Degenerate primer at the position of FU1.
Degenerate primer at the position of cFD3.
Degenerate primer at the position of FU3.
Degenerate primer at the position of cFD4.
Source, Fulop et al. (14).
Source, Pierre and Deubel (33).
Genomic position of tick-borne encephalitis virus (strain 263) (GenBank accession no. U-27491) to which the first base (5′ end) of the primer corresponds.
Nucleotide sequencing.
Amplicons were purified with a Qiagen PCR purification kit, and aliquots of approximately 60 to 160 ng of the purified DNA templates were used for direct cycle sequencing using an ABI (Foster City, Calif.) Prism DNA sequencing kit for dye terminator cycle sequencing with AmpliTaq-FS enzyme. The sequencing primers including the primers used for DNA template preparation and their corresponding degenerate primers are listed in Tables 2 and 3. Thirty cycles of a thermocycle program (96°C for 15 s, 50°C for 15 s, and 60°C for 4 min) were performed with Gene Amp PCR System 9600 thermocycler (Perkin-Elmer, Norwalk, Conn.). The products were purified in Centri-Sep spin columns (Princeton Separations, Adelphi, N.J.) and directly sequenced with ABI model 377 sequencer. Nucleotide sequences were edited and compilied by using a computer program, DNASIS for Window (version 1.1; Hitachi Software Engineering America, Ltd., South San Francisco, Calif.).
TABLE 3.
Primers used only for sequencing
| Virus | Primera | Sequence (5′→3′) | Genomic positionb |
|---|---|---|---|
| Apoi | F5F230 (F) | GCTCTATGCTGATGACACCGC | 204 |
| Banzi | F8F74 (F) | ATGTGGCTTGGGGCTCGCTTT | 49 |
| F8F322 (F) | CTGAGCACCGACAACTGGCAT | 296 | |
| Cacipacore | F12F808 (F) | TTATGAAGGATGGAAGAAAGC | 779 |
| Jugra | F20F590 (F) | TTGGAGGTGAACGGAGTGAAC | 568 |
| F20F622 (F) | GTAATGGCAGTGAGCGGAGAT | 598 | |
| Kadam | F22F770 (F) | CTTTTGCTCCCACCACTTCCA | 741 |
| Potiskum | F31F353 (F) | TACAAGAACAAGGTGGTCAAA | 337 |
| F31F623 (F) | GCTGTGAGTGGTGATGATTGT | 604 | |
| F31F776 (F) | TCGCACCATTTCCACACGCTG | 757 | |
| Saboya | F34F464 (F) | TTGGGGAATGGGAGGAGAGCA | 704 |
| Uganda S | F44F581 (F) | TGGCTTGAGGTGAATGGCGTA | 565 |
| Sokuluk | F53F63 (F) | AAGAACACAGGGGCTCGGGAC | 410 |
| F53R160 (R) | GACGCCCTCTGCTTCTGCCAT | 487 | |
| F53R498 (R) | ATCCTCGCCTTCCCAATCAGC | 845 | |
| Carey Island | F56F152 (F) | ACTTTGTGGAGGTGGCGTGGA | 123 |
| F56F359 (F) | GGCATACAAAAACAAAGTGGC | 330 |
F, forward primer; R, reverse primer.
The number corresponds to the nucleotide position in the deposited sequence (Table 1) corresponding to the first (5′) base of each primer.
Phylogenetic analysis.
The multiple sequence alignment program Clustal W (version 1.6) (43) was used to obtain an optimal nucleotide or amino acid sequence alignment file. Phylograms for the entire sequence (about 1 kb between primers FU1 and cFD3 [Fig. 1]) were obtained either by MEGA (version 1.01) (25) or PHYLIP (version 3.57c) (12, 13) based on aligned nucleotide or amino acid sequences. MEGA was also applied to analyze a subset of the nucleotide sequence (about 220 bp between primers FU1 and cFD2 [Fig. 1]).
In constructing phylograms with distance methods of MEGA, we determined genetic distance by the proportional distance method (37), Kimura’s two-parameter method (24), and the Tajima-Nei method (40), applying pairwise deletion of gaps and equally weighting both transition and transversion for all three codon positions. A proportional distance matrix was transformed to calculate the pairwise nucleotide sequence identity between all virus pairs. For tree building, various genetic distance matrices were used for the neighbor-joining method (37) which calculated bootstrap confidence intervals of 500 heuristic search replicates and confidence probability of the genetic distance by a standard error test. We also tested a character state tree-building algorithm which consisted of a sequential programs in the PHYLIP package. A strict consensus bootstrap tree was obtained by using the following programs: (i) SEQBOOT to generate 100 reiterated replicas; (ii) DNAPARS or PROTPARS to acquire the most parsimony tree of each reiterated data, (iii) CONSENSE to build a strict consensus bootstrap tree, and (iv) DRAWGRAM to draw the phylogenetic tree.
Virus identification.
Numerous attempts with various primer combinations (Tables 2 and 3) failed to obtain amplicon from Tamana bat virus RNA by RT-PCR. We tried to reidentify the virus by the immunofluorescence technique using 13 polyvalent hyperimmune mouse ascitic fluids prepared against 9 antigenic groups in the family Bunyaviridae and the Tacaribe group of the Arenaviridae, monovalent antiserum against vesicular stomatitis virus, rabies virus, flaviviruses (DEN, Murray Valley encephalitis, YF, WN, Powassan [POW], and Tamana bat viruses), and flavivirus group-reactive monoclonal antibodies 4G2 and 6B6C-1 on virus-infected Vero cells. The infected cells were also embedded in LX-112 Araldite mixture and examined with a model 410 Life Science Phillips electron microscope (Phillips, Eindhoven, The Netherlands) operating at 80 kV as described earlier (9).
RESULTS
PCR primers.
The pair of flavivirus cross-reactive primers (FU1 and cFD3) proved to be highly efficient for generating about 1-kb-long DNA templates near the 3′ terminus of the NS5 gene for most of the viruses by RT-PCR. A small number of viruses that were not amplified or poorly amplified with this pair (Apoi, Karshi, Kokobera, Rio Bravo, Sal Vieja, and Sokuluk viruses) could be sequenced with overlapping DNA templates of various sizes generated with other pairs of primers shown in Table 2.
Molecular classification.
For convenience, throughout the text, the hierarchal levels for molecular systematics of this genus are organized in descending order as follows: cluster, clade, and species. A cluster was designated based on the bootstrap support exceeding 95% and host-vector association. A clade was defined as a group of viruses that share the 69% or higher pairwise nucleotide sequence identity among the members. This 69% quantitative criterion was chosen from the pairwise identity minus 2 standard deviations among four serotypes of DEN virus, because DEN complex viruses are easily separated from other flaviviruses not only by a serologic test but also by analysis of nucleotide sequence data (6, 11). A species was defined as a class of viruses with higher than 84% nucleotide sequence identity among them. The cutoff criterion was derived from two strains of YF viruses, the prototype Asibi and the TN96 isolate, which were separated by 69 years and belonged to two distinct genotypes (7). An extensive envelope gene sequence study on many strains of four DEN, JE, SLE, and YF viruses from all known areas of the world where there viruses are endemic had earlier indicated that YF virus was the most diversified species of all, with a maximum of 14% nucleotide sequence difference among strains. Among 71 viruses of this genus, non-vector, tick-borne, and mosquito-borne clusters contained 14, 15, and 42 viruses, respectively (Table 4). Sixty-eight viruses were further separated into 14 clades, and three viruses, CFA, Apoi, and Kedougou viruses, were not associated with any clade. The following virus pairs had pairwise nucleotide sequence identity of 91, 88, 92, 90, and 95%, respectively, and were determined to be genetic variants of the same virus: Phnom Penh bat and Batu Cave; TBE-central European subtype (TBE-CE) and Negishi; Potiskum and Saboya; THCAr (21) and Tembusu; and Israel turkey meningoencephalitis and Bagaza.
TABLE 4.
Assignment of flaviviruses to clusters and clades
| Virusa | Clade | Antigenic complex |
|---|---|---|
| Non-vector cluster | ||
| CFA | ||
| Apoi | ||
| San Perlita; Jutiapa | I | Modoc |
| Montana myotis leukoencephalitis; Modoc; Cowbone Ridge; Sal Vieja | II | Modoc |
| Bukalasa bat; Dakar bat; Rio Bravo; Carey Island; Phnom Penh bat; Batu Cave | III | Rio Bravo |
| Tick-borne cluster | ||
| Gadgets Gully; Royal Farm; Pow; Karshi; KFD; Langat; Omsk HF; | IV | TBE |
| TBE-far eastern subtype; RSSE; TBE-CE; Negishi | ||
| Kadam; Tyuleniy; Saumarez Reef; Meaban | V | Tyuleniy |
| Mosquito-borne cluster | ||
| Edge Hill; Bouboui; Uganda S; Banzi; Jugra; Saboya; Potiskum | VI | Uganda S |
| Sepik; YF | VII | |
| Sokuluk; Entebbe bat; Yokose | VIII | |
| DEN-1 to -4 | IX | DEN |
| Kedougou | ||
| Zika; Spondweni | X | |
| SLE; Rocio; Ilheus; Tembusu; THCAr; Ntaya; Israel turkey meningoencephalitis; Bagaza | XI | Ntaya |
| Naranjal; Bussuquara; Aroa; Iguape | XII | |
| Kokobera; Stratford | XIII | JE |
| Cacipacore; Yaounde; Koutango; Kunjin; WN; Alfuy; JE; Murray Valley encephalit1is; Usutu | XIV | JE |
The viruses in boldface belong to the corresponding antigenic complex to the right.
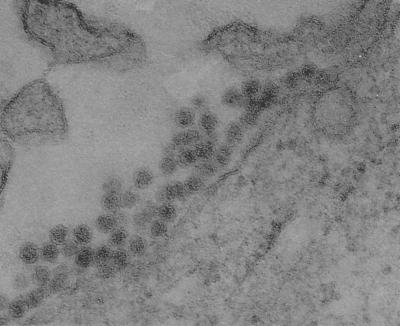
The genome of Tamana bat virus, originally isolated in Trinidad (35), could not be amplified satisfactorily with any combination of the primers used in this study. In immunofluorescence tests, only a weak reactivity was observed with a WN virus monospecific antiserum, and the virus did not react with 20 other polyclonal and 2 flavivirus group-reactive monoclonal antibodies, despite a positive reaction to the homologous polyclonal ascitic fluid against Tamana bat virus (data not shown). However, electron microscopy revealed conclusively that Tamana bat virus-infected Vero cells exhibited numerous virion-like particles that had a morphological characteristics of a typical flavivirus (Fig. 2).
FIG. 2.
Electron micrograph of Vero cells infected with Tamana bat virus (magnification, × 121,125).
Phylograms.
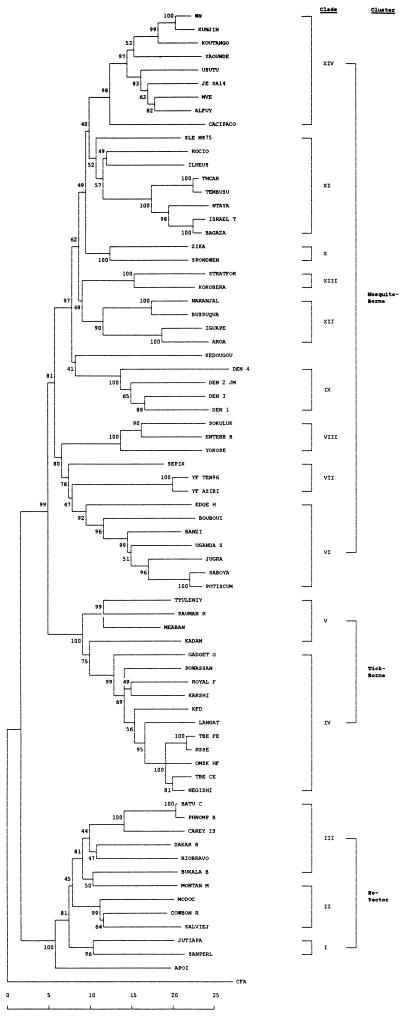
The unrooted neighbor-joining tree based on a proportional distance of 1-kb nucleotide sequence is shown in Fig. 3. The phylogram demonstrates clearly that although the exact host association of CFA virus in nature remains unknown, it is the most distally related flavivirus sequenced so far. CFA virus has pairwise nucleotide sequence identities with viruses of the designated clades (Table 4) as follows: with Apoi virus, 57%; with clade I, 56%; with clade II, 53 to 56%; with clade III, 54 to 57%; with clade IV, 56 to 57%; with clade V, 55 to 57%; with clade VI, 53 to 56%; with clade VII, 55 to 56%; with clade VIII, 54 to 55%; with clade IX, 55%; with clade X, 54 to 55%; with clade XI, 53 to 57%; with clade XII, 54 to 56%; and with clade XIV, 53 to 56%. Furthermore, the phylogram reveals that non-vector and vector-borne clusters emerged first from the putative origin of the genus Flavivirus. The latter further branched off to form tick-borne and mosquito-borne virus clusters. These three clusters are well supported by 99% of bootstrap replicates and 99% confidence probabilities (CPs) of a standard error test.
FIG. 3.
Phylogenetic tree of the genus Flavivirus, using nucleotide sequence. The tree was constructed by the neighbor-joining method of MEGA. Each number at nodes is the percentage of 500 bootstrap replicate support; ∗ indicates confidence probability higher than 90%. Vertical length is arbitrary. Scale is percentage of genetic distance.
The non-vector cluster further branched into three clades with Apoi virus by itself outside any of the three (Fig. 3 and Table 4). San Perlita and Jutiapa viruses in clade I and four viruses in clade II with the exception of Montana myotis leukoencephalitis virus are rodent-associated viruses. The six viruses in clade III are all bat associated. The tick-borne cluster consists of two clades, represented by Gadgets Gully virus and a collection of 10 viruses (clade IV), most of which belong to the so-called TBE complex (10), and Kadam, Tyuleniy, Meaban, and Saumarez Reef viruses (clade V) (Fig. 3). In the aforementioned two clusters, CPs generally parallel bootstrap supports; and even when bootstrap supports are weak, the corresponding CPs are very high, as demonstrated in the clade III (52 versus 99% in Fig. 3).
Nine clades (VI to XIV) comprise the mosquito-borne cluster. Kedougou virus is not associated with any clade and has a range of 60 to 65% nucleotide sequence identities with other mosquito-borne viruses (data not shown). Sepik and YF viruses comprise clade VII; Sokuluk, Entebbe bat, and Yokose viruses comprise clade VIII; Zika and Spondweni viruses comprise clade X; and Naranjal, Busssuquara, Aroa, and Iguape viruses comprise clade XII. None of those viruses have been placed in any antigenic complexes before. Clade VI consists mostly of Uganda S complex viruses and two previously unclassified viruses, Jugra and Saboya viruses. Potiskum virus in this clade is considered a subtype of Uganda S virus by neutralization test (23). The DEN complex, consisting of four serotypes alone, is assigned a separate clade (IX). Former JE complex viruses were separated into clades XI, XIII, and XIV. Clade XI includes four viruses of the Ntaya antigenic complex as well as members of the JE complex, such as SLE, Rocio, and Ilheus viruses. The segregation of the other JE complex viruses, Stratford and Kokobera viruses, into one clade (clade XIII) by themselves agrees well with the previous conclusion that those viruses are distinct from the other JE complex viruses (34). Clade XIV includes Cacipacore virus, Yaounde virus, and the remaining seven JE complex viruses. Bootstrap supports of the clade XI and XII viruses were 53 and 70%, respectively. Nevertheless, the corresponding CPs are both 99%, providing a strong support to our classification.
Among other phylograms created by the combination of distance and tree-constructing methods examined, the tree based on Kimura’s two-parameter method produced a phylogram very similar to those in Fig. 3 and 4. The Tajima-Nei distance method produced a phylogram quite different from those in Fig. 3 and 4 and was judged inappropriate because at the optimal cutoff level for clade (using 65%, rather than 69%, nucleotide sequence identity), Tyuleniy group (clade V) was split to two groups and clade (IV) was further divided, creating a phylogram considerably different from the phylograms obtained by us, as mentioned above, and by other investigators. The phylogram based on nucleotide sequences between FU1 and cFD2 (220 bp) demonstrated a similar tree topology as with 1 kb for most clades (data not shown), despite the low bootstrap supports at some nodes and shift in affiliation of some viruses at the terminal branches.
FIG. 4.
Phylogenetic tree of the genus Flavivirus, using amino acid sequence. The tree was constructed by the neighbor-joining method using MEGA. Each number at nodes is the percentage of 500 bootstrap replicates. Vertical length is arbitrary. Scale is percentage of amino acid distance.
The strict consensus tree, obtained by character-state of the most parsimony algorithm, showed a similar tree topology as by the distance method (data not shown). By this method clade XIII, comprising Stratford and Kokobera viruses, was weakly associated with clade XII. Kedougou virus also was weakly linked to DEN complex viruses (clade IX).
The most notable differences between the amino acid sequence-based tree (Fig. 4) and the nucleotide sequence-based tree are the more distant relation of clade XIII from and the closer relation of clade X to clade XIV in the former tree. A shift of affiliation of Sepik and Montana myotis leukoencephalitis viruses to a different subset of viruses is also observed. Nevertheless, topologies of the trees by both nucleotide and amino acid sequences are essentially identical.
Amino acids and motifs.
In addition to the GDD motif, other highly conserved motifs include YADDTAGWDT, QRGSGQV, DDCVV, TACL, YFHRRDLR, and SAVP (Fig. 5). In other, less conserved motifs, amino acids unique to a particular cluster (or clusters) are found. For a non-vector cluster, they are SF (amino acids [aa] 28 to 29) and G (aa 190) (Fig. 5). For vector-borne clusters, they are A (aa 13) and P (aa 255); for the tick-borne cluster, they are W (aa 57) and C (aa 292).
FIG. 5.
Multiple amino acid sequence alignment of the CFA, Apoi, and Kedougou viruses and one member representing each clade of the genus Flavivirus. Amino acid 1 corresponds to nucleotides 9018 to 9020 of YF virus. A dash indicates missing one amino acid. A dot indicates that the amino acid (or absence of it) in a given amino acid sequence is the same as in the corresponding sequence of CFA virus above of the aligned sequences.
Codon usage.
For analyzing the host association among two clusters (non-vector and vector borne) of viruses, we examined the frequencies of the dinucleotide CpG. When the viruses were classified into two categories, those with ≤9 CG-containing amino acids and those with ≥10 such amino acids, 9 of 14 non-vector group but only 5 of 58 vector-borne viruses belonged to the former category (chi-square test: χ2 = 55.8; P = 0.0000322). When the cutoff number of CG-containing amino acids was changed to 13, the numbers of the viruses in non-vector and vector-borne clusters with ≤13 such amino acids were 11 and 29, respectively (chi-square test: P = 0.053).
DISCUSSION
The phylograms of flaviviruses created in the past were based on the sequences of only about one-third or fewer of the members and thus provided only partial information (3, 11, 15, 28, 48). Nevertheless, the dichotomy between mosquito-borne and tick-borne viruses has been clearly recognized by those investigators and was again confirmed in our study. As shown in our phylograms, the genus Flavivirus presents as a monophyletic tree. Unlike previous studies, however, our study reveals further that from the putative ancestor of the genus Flavivirus, two major branches emerged, non-vector and vector-borne clusters, and that from the latter cluster emerged tick-borne and mosquito-borne clusters. The above topology as well as subsequent branching patterns leading to clades in each cluster were found to be basically identical between the trees based on nucleotide as well as amino acid sequences.
In our study, we constructed trees without selecting CFA virus as an outgroup taxon, as was done before (28). Such an unrooted tree is expected to provide the least biased phylogenetic tree. Irrespective of the difference in requirement of an outgroup in the software used, CFA virus was placed at the root of the tree by MEGA (Fig. 3) and by PHYLIP (data not shown).
The phylogenetic segregation of the viruses into three major clusters was not surprising because of a clear distinction in the size of the sequences between amplimers FU1 and cFD3: members of non-vector cluster all had 1,011 bases, tick-borne viruses had a median length of 1,026 bases, and mosquito-borne viruses had a median length of 1,035 bases. Thus, when all sequences were aligned optimally by introducing gaps to make all lengths equal, 13 aa were missing from all viruses in the non-vector cluster and 8 aa were missing from all viruses of the tick-borne cluster, with all missing amino acids being located at the same sites as the non-vector viruses. On the other hand, the pattern of missing amino acids was more variable in the mosquito-borne cluster. While the majority of the mosquito-borne viruses had 5 aa missing, DEN-1, -2, -3, and -4, Kedougou, Kokobera, and Stratford viruses had an additional missing amino acid. All of these, as well as Zika and Spondweni viruses, had 2 aa missing at the same locations where non-vector and tick-borne viruses similarly were missing amino acids.
Although the flavivirus phylograms produced in the past were primarily based on envelope gene sequences, it has been reported that the topologies based on envelope and NS5 genes showed perfect agreement (28). The envelope gene of flaviviruses is less conserved than the NS5 gene, and this difference is reflected in greater differences in the amino acid sequence. Thus, while the ranges of pairwise amino acid sequence identities in the envelope gene were 72.3 to 80.4% in DEN complex viruses, 81 to 94.6% in JE complex viruses, and 66.3% between Banzi and YF viruses (15), the corresponding ranges for NS5 gene in our study were of 75 to 86%, 83 to 97%, and 72%, respectively, confirming conservative nature of the latter gene.
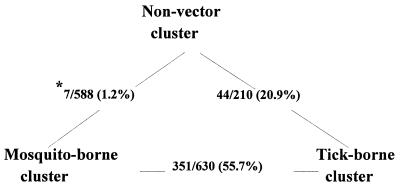
Regarding the evolution of three clusters of viruses, theoretically any cluster could have been ancestral, since unrooted methods were chosen for our phylograms. However, Calisher (5) speculated that “evolutionary pressure may have created a divergence of the virus-vector relationships, perhaps from a common original one.” Blok and Gibbs (3) are of the opinion that the arthropod-mediated transmission of the flaviviruses is an acquired trait, although they also recognized the opposite possibility. One prevailing theory is that tick-borne and mosquito-borne clusters independently evolved from the common ancestor. Marin et al. (28) previously concluded that the Tyuleniy group (Table 4), which branched off early after the vector-borne group split to mosquito-borne and tick-borne clusters, had the traits typical of mosquito-borne viruses, such as the absence of hexapeptide insertion, possession of the common glycosylation site in the envelope gene, and ability to replicate in mosquito cell cultures. We calculated a pairwise nucleotide sequence identity of 63 to 65% between the members of different clusters. A higher percentage of proportional pairwise nucleotide sequence identity could reflect the close genetic and evolutionary relationship between the members of the two clusters. As shown in Fig. 6, the proportion of pairwise nucleotide sequence identities falling in this range was 20.9% between non-vector and tick-borne clusters, whereas it was only 1.2% when the non-vector cluster was compared with the mosquito-borne cluster. On the other hand, when tick-borne and mosquito-borne clusters were compared, as much as 55.7% of the virus pairs had a nucleotide sequence identity in this range. Furthermore, with respect to vector association, while some viruses in the mosquito-borne cluster, such as SLE, WN, and YF viruses, have been sometimes isolated from ticks, the reverse observation has been recorded only in the case of POW virus of the tick-borne cluster. It is noteworthy that none of the members of non-vector cluster replicated in mosquito cell culture (46). Thus, the casual association of mosquito-borne viruses with ticks may be considered a vestigial trait of the past association with ticks before adaptation to mosquitoes. Taken together, the observations provide evidence in support of second possibility that the viruses of this genus evolved from non-vector group to tick-borne and then to mosquito-borne group. Exceptions to the above speculation are Aroa, Entebbe bat, Saboya, and Sokuluk viruses, which are placed in the mosquito-borne cluster in our phylogram despite the absence of arthropod vectors. This may be partly due to the lack of in-depth field investigations to search for arthropod vectors of these viruses. In fact, all of them are known to replicate in a mosquito cell culture (46).
FIG. 6.
Pairwise nucleotide sequence identity relationship among three clusters of the genus Flavivirus. ∗, Number of pairs with 63 to 65% nucleotide sequence identity/total number of pair sequence compared.
It has been recognized that the genomes of higher vertebrates and birds are deficient in the frequency of the dinucleotides CpG, which, in turn, reflect on the biased codon usage containing this dinucleotide. A review on this subject concluded that “there is a partial but not a complete correlation between CG content and evolutionary history of life cycle of different viruses” (39). Among the family Togaviridae, such a deficit was found in many alphaviruses (50). Our analysis similarly confirms a strong relationship between CG deficit and exclusive association with mammalian hosts in the natural transmission cycle when viruses were evaluated in the studied NS5 region.
A cline theory has been proposed to describe the correlation of genetic distance and geographic locations for the so-called TBE complex viruses (10). Assuming that POW virus was more ancestral, the data suggested westward movement of TBE complex viruses to Europe across northern Eurasia. The distribution of the viruses that did not satisfy the geographic movement, such as Negishi and Kyasanur Forest disease viruses, was explained by accidental virus transportation by tick-infested migratory birds. Our results do not support the above cline theory because of geographic distribution of two additional members of the TBE complex (clade IV), Gadgets Gully of Australia and Royal Farm of Afghanistan. Information on geographic distribution of those viruses used for the basis of the above theory was incomplete. For example, indigenous transmission of RSSE virus, which had been previously thought to be confined to eastern parts of Russia, has been confirmed in Japan (41). Furthermore, both Negishi and Langat viruses were reported to have been isolated in the former Soviet Union (36). Regarding the speculations on the origin of Negishi virus, while the role of migratory birds transporting louping ill virus to Far East Asia (47) remains a possibility, it is noted that neutralizing antibody to Negishi virus was detected in mammals there (44). The other speculation that it was actually a reference virus used during identification tests as a result of laboratory contamination or mislabeling (17) is ruled out for the following reason. The published documents reveal that neither TBE-CE nor louping ill virus, most closely related to Negishi virus, was used during virus isolation, passage, and identification phases (1, 19, 32); rather, RSSE and POW viruses were used for identification tests.
Division of a genus to subgeneric levels based on molecular sequence depends on the definition of species. Currently, virus species is defined as “a polythetic class of viruses constituting a replicating lineage and occupying a particular ecological niches” (45), a definition which was adopted by the International Committee on the Taxonomy of Viruses (31). While all classification systems, including serologic technique and nucleotide sequence-based classification, are not without problems (45), combination of those two methods with a minimum amount of discrepancy between them will improve virus classification based on polythetic concept of species definition. As far as quantitative species definition based on nucleotide sequence data is concerned, criteria used for RNA viruses have been variable. For example, bluetongue viruses comprising 24 serotypes are considered one species, while the DEN virus serotypes are considered four distinct species (31). Furthermore, while species are distinguished at a 64.6% nucleotide sequence identity for members of the arenaviruses (4), 67 to 77% identity of the NS5 gene has been adopted for the definition of genotypes within hepatitis C virus (38). The variation of quantitative criteria for various levels of taxa reflects partly the difference in the rate of evolution among different virus groups and partly philosophical difference on the concept of virus species among virologists (45). In our study, the classification into clades using ≥69% pairwise nucleotide sequence identity between viruses as a criterion agreed well with grouping of viruses in the phylogram. Similarly, our definition of >84% pairwise sequence identity as a criterion for species of the members of the genus Flavivirus agreed with the results obtained by neutralization testing. For example, Batu Cave virus, which was shown to be identical to Phnom Penh bat virus according to our definition, had been withdrawn from registration because it was found to be identical to the latter virus by a neutralization test (46). Likewise, THCAr was found to be a subtype of Tembusu virus both by neutralization (23) and sequence analyses.
The application of our criteria should help resolve the confusing taxonomic status of the tick-borne encephalitis viruses primarily isolated in western Eurasia. Although sometimes the Neudoerfl strain is described as the prototype of central European subtype of “TBE virus” (49), neither it nor any virus by the name of tick-borne encephalitis (or TBE) virus has ever been registered (5). In the meanwhile, the number of tick-borne viruses bearing the name TBE virus proliferated. The recently completely sequenced louping ill virus as well as Negishi virus, Spanish sheep tick-borne encephalitis, Turkish sheep tick-borne encephalitis, and Greek goat encephalitis viruses show more than 83% nucleotide sequence identity in the envelope gene region with either the Neudoerfl or the Kumlinge strain (16, 18, 29, 52), and they are serologically indistinguishable (17). The distinction of so-called TBE viruses into two subtypes (far eastern and central or western European) did not help to correct the taxonomic problem because of overlapping geographic distributions (36). Four viruses (Absettarov, Kumlinge, Hanzalova, and Hypr) are registered but considered variants of the same virus by serological classification (5). Thus, it is highly conceivable that when nucleotide sequence data of the unsequenced viruses are made available for comparison, most (if not all) of those tick-borne viruses are determined to be variants of one virus species with its geographic distribution stretching from Far East Asia to the British Isles and from Scandinavia to the countries along the Mediterranean, leaving RSSE as a virus distinct from them. The recently described deer tick virus has a high (>84%) pairwise nucleotide sequence identity in the NS5-3′ untranslated region compared with POW virus (42). For another virus recently described as a new tick-borne virus, Vasilchenko strain, similarly no justification or criteria for classification into a new virus were described (18). Thus, the species status of each of these viruses needs to be carefully reexamined. Then, an appropriate strain must be designated and registered, if not yet done; consequently, all registered synonyms need to be withdrawn from registration. Whatever the outcome of reexamination, when nucleotide sequence identity is high (>80%) compared with a known virus, it is prudent to perform a neutralization test in two directions rather than relying solely on sequence data before one attempts to establish a new virus.
Recently, it was reported that Kunjin virus was a member of WN virus based on short sequence in envelope gene (2). Since those viruses are distinct species according to our classification, we offer our thoughts to identify the possible sources of discrepancy. In the WN virus study above, nucleotide sequence of only one strain of Kunjin virus, which is well known for its close relation to WN virus, was compared with sequences of many strains of WN virus for a phylogenetic study. When only two viruses are compared in a phylogenetic study, it is not surprising that the sole sequence of one virus (Kunjin) is automatically grouped in one of the branches (called lineages in the above study) of the other species, simply because of shared sequence identity. For a more conclusive study, inclusion of more Kunjin virus strains, Asian strains of WN virus, and at least one less related flavivirus is essential, particularly because both viruses are found in Asia. Second, phylograms generated based on very short sequences (<300 bases) are sometimes different from those generated on much longer sequences. For example, while only one genotype was identified for DEN-4 viruses worldwide, using short sequences (8), two genotypes were identified by using the identical criterion (6% divergence), virus strains from the same geographic regions, and a much longer (1.5 kb) sequence of the same viral gene (26). In this study, phylograms based on short sequences were similar to those based on 1-kb sequences, but the bootstrap supports at some nodes were much lower, rendering phylograms unreliable. Thus, a caution was voiced against the use of such short sequences for phylogenetic studies of flaviviruses (51). Regardless, more Asian strains of both viruses are needed to resolve the species status of Kunjin virus, particularly because a Kunjin virus with intermediate characteristics with WN virus was reported (34).
Clades established in our study are not exactly comparable to antigenic complexes in terms of membership (6). For example, Carey Island virus, formerly a member of TBE complex, is now classified as a member of the non-vector cluster, while Saboya virus, formerly a member of the Rio Bravo antigenic complex, now belongs to the mosquito-borne cluster. The discrepancy between molecular and serologic classifications partly reflects the difficulty of achieving a 100% agreement between the two systems based on different principles, given diversity of the viruses involved. Nevertheless, we believe that our molecular classification produces the smallest amount of discrepancy compared with serologic classification and together, the two methods would greatly improve our understanding of the relationship among the members of the genus Flavivirus.
ACKNOWLEDGMENTS
We thank Robert E. Shope, University of Texas, Galveston, for the gift of Iguape and Kedougou viruses; Thomas G. Ksiazek, Special Pathogens Branch, National Center for Infectious Diseases, Centers for Disease Control for Prevention, Atlanta, Ga., for the gift of extracted RNA of KFD, Omsk HF, and RSSE viruses; and Yuki Eshita, Kurume University School of Medicine, Kurume, Japan, for providing documents on Negishi virus.
REFERENCES
- 1.Ando K, Kuratsuka K, Arima S, Hironaka N, Honda Y, Ishii K. Studies on the viruses isolated during epidemic of Japanese B encephalitis in 1948 in Tokyo area. Kitasato Arch Exp Med. 1952;24:49–61. [PubMed] [Google Scholar]
- 2.Berthet, F.-X., H. G. Zeller, M.-T. Drouet, J. Rauzier, J.-P. Digoutte, and V. Deubel. Extensive nucleotide changes and deletions within the envelope glycoprotein gene of Euro-African West Nile viruses. J. Gen. Virol. 78:2293–2297. [DOI] [PubMed]
- 3.Blok J, Gibbs A J. Molecular systematics of the flaviviruses and their relatives. In: Gibbs A, Calisher C H, Garcia-Arenal F, editors. Molecular basis of virus evolution. Cambridge, England: Cambridge University Press; 1995. pp. 270–289. [Google Scholar]
- 4.Bowen M D, Peters C J, Mills J N, Nichol S T. Oliveiros virus: a novel arenavirus from Argentina. Virology. 1996;217:362–366. doi: 10.1006/viro.1996.0124. [DOI] [PubMed] [Google Scholar]
- 5.Calisher C H. Antigenic classification and taxonomy of flaviviruses (family Flaviviridae) emphasizing a universal system for the taxonomy of viruses causing tick-borne encephalitis. Acta Virol. 1988;32:469–478. [PubMed] [Google Scholar]
- 6.Calisher C H, Karabatsos K, Dalrymple J M, Shope R E, Porterfield J S, Westaway E G, Brandt W E. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol. 1989;70:37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- 7.Chang, G.-J. J. Unpublished data.
- 8.Chungue E, Cassar O, Drouet M T, Guzman M G, Laille M, Rosen L, Deubel V. Molecular epidemiology of dengue-1 and dengue-4 viruses. J Gen Virol. 1995;76:1877–1884. doi: 10.1099/0022-1317-76-7-1877. [DOI] [PubMed] [Google Scholar]
- 9.Cropp C B, Prange W C, Monath T P. Le Dantec virus: identification as a rhabodovirus associated with human infection and formation of a new serogroup. J Gen Virol. 1985;66:2749–2754. doi: 10.1099/0022-1317-66-12-2749. [DOI] [PubMed] [Google Scholar]
- 10.de Zanotto P M, Gao G F, Gritsun T, Marin M S, Jiang W R, Venugopal K, Reid H W, Gould E A. An arbovirus cline across the northern hemisphere. Virology. 1995;210:152–159. doi: 10.1006/viro.1995.1326. [DOI] [PubMed] [Google Scholar]
- 11.de Zanotto P M, Gould E A, Gao G F, Harvey P H, Holmes E C. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc Natl Acad Sci USA. 1996;93:548–553. doi: 10.1073/pnas.93.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 13.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 14.Fulop L, Barrett A D T, Phillpotts R, Martin K, Leslie D, Titball R W. Rapid identification of flaviviruses based on conserved NS5 gene sequences. J Virol Methods. 1993;44:179–188. doi: 10.1016/0166-0934(93)90053-t. [DOI] [PubMed] [Google Scholar]
- 15.Fulop L D, Barrett A D T. Nucleotide sequence of NS5 gene of Banzi virus: comparison with other flaviviruses. J Gen Virol. 1995;76:2317–2321. doi: 10.1099/0022-1317-76-9-2317. [DOI] [PubMed] [Google Scholar]
- 16.Gao G F, Hussain M H, Reid H W, Gould E A. Classification of a new member of the TBE flavivirus subgroup by its immunological, pathogenetic and molecular characteristics: identification of subgroup-specific pentapeptides. Virus Res. 1993;30:129–144. doi: 10.1016/0168-1702(93)90002-5. [DOI] [PubMed] [Google Scholar]
- 17.Gould E, Zanotto P M A, Holmes E C. The genetic evolution of flaviviruses. In: Saluzzo J F, Dodet B, editors. Factors in the emergence of arbovirus diseases. Paris, France: Elsevier; 1997. pp. 51–63. [Google Scholar]
- 18.Gritsun T S, Venugopal K, de Zanotto P M, Mikhailov M V, Sall A A, Polkinghorne I, Frolova T V, Pogodina V V. Complete sequence of two tick-borne flaviviruses isolated from Siberia (Vasilchenko strain) and United Kingdom (louping ill): analysis and significance of the 5′ and 3′ UTRs. Virus Res. 1997;49:27–39. doi: 10.1016/s0168-1702(97)01451-2. [DOI] [PubMed] [Google Scholar]
- 19.Honda Y. Comparison of the immunological characters of a new virus “Negishi strain” and Russian Spring Summer encephalitis virus (in Japanese) Kitasato Jikken Igaku. 1951;24:91–93. [PubMed] [Google Scholar]
- 20.Igarashi, A. Unpublished data.
- 21.Igarashi, A. Personal communication.
- 22.Karabatsos N. International catalogue of arboviruses. 3rd ed. San Antonio, Tex: American Society of Tropical Medicine and Hygiene; 1985. [Google Scholar]
- 23.Karabatsos, N. Unpublished data.
- 24.Kimura M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Tamura K, Nei M. Molecular evolutionary genetics analysis version 1.01. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 26.Lanciotti R S, Gubler D J, Trent D W. Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol. 1997;78:2279–2286. doi: 10.1099/0022-1317-78-9-2279. [DOI] [PubMed] [Google Scholar]
- 27.Le Guenno B, Bougermouh A, Azzam T, Bouakaz R. West Nile: a deadly virus? Lancet. 1996;348:1315. doi: 10.1016/s0140-6736(05)65799-6. [DOI] [PubMed] [Google Scholar]
- 28.Marin M S, Mandl C W, Kunz C, Heinz F X. The flavivirus 3′-noncoding region: extensive size heterogeneity independent of evolutionary relationships among strains of tick-borne encephalitis virus. Virology. 1995;213:169–178. doi: 10.1006/viro.1995.1557. [DOI] [PubMed] [Google Scholar]
- 29.Marin M S, McKenzie J, Gao G F, Antoniadis A, Gould E A. The virus causing encephalomyelitis in sheep in Spain: a new member of the tick-borne encephalitis group. Res Vet Sci. 1995;58:11–13. doi: 10.1016/0034-5288(95)90081-0. [DOI] [PubMed] [Google Scholar]
- 30.Monath T P. Yellow fever and dengue-the interactions of virus, vector and host in the re-emergence of epidemic disease. Semin Virol. 1994;5:133–145. [Google Scholar]
- 31.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: classification and nomenclature of virus. New York, N.Y: Spring-Verlag; 1995. pp. 415–421. [Google Scholar]
- 32.Okuno T, Oya A, Ito T. The identification of Negishi virus: a presumably new member of Russian Spring Summer encephalitis virus family isolated in Japan. Jpn J Med Sci Biol. 1961;14:51–59. doi: 10.7883/yoken1952.14.51. [DOI] [PubMed] [Google Scholar]
- 33.Pierre V, Drouet M-T, Deubel V. Identification of mosquito-borne flavivirus sequences using universal primers and reverse transcription/polymerase chain reaction. Res Virol. 1994;145:179–188. doi: 10.1016/s0923-2516(07)80011-2. [DOI] [PubMed] [Google Scholar]
- 34.Poidinger M, Hall R A, MacKenzie J S. Molecular characterization of the Japanese encephalitis serocomplex of the Flavivirus genus. Virology. 1996;218:417–421. doi: 10.1006/viro.1996.0213. [DOI] [PubMed] [Google Scholar]
- 35.Price J L. Isolation of Rio Bravo and a hitherto undescribed agent Tamana Bat virus, from insectivorous bats in Trinidad with serological evidence of infection in bats and man. Am J Trop Med Hyg. 1978;27:153–161. doi: 10.4269/ajtmh.1978.27.153. [DOI] [PubMed] [Google Scholar]
- 36.Rubin S G, Chumakov M P. New data on the antigenic types of tick-borne encephalitis (TBE) virus. Zentralbl Bakteriol Suppl. 1980;9:231–236. [Google Scholar]
- 37.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 38.Simmonds P, Smith D B, McOmish F, Yap P L, Kolberg J, Urdea M S, Holmes E C. Identification of genotypes of hepatitis C virus by sequence comparison in the core, E1, and NS-5 regions. J Gen Virol. 1994;75:1075–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- 39.Strauss E G, Strauss J M, Levine A J. Virus evolution. In: Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Virology. 2nd ed. New York, N.Y: Raven Press; 1990. pp. 167–190. [Google Scholar]
- 40.Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- 41.Takashima I, Morita K, Chiba M, Hayasaka D, Sato T, Takezawa C, Igarashi A, Kariwa H, Yoshimatsu K, Arikawa J, Hashimoto N. A case of tick-borne encephalitis in Japan and isolation of the virus. J Clin Microbiol. 1997;35:1943–1947. doi: 10.1128/jcm.35.8.1943-1947.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Telford S, Armstrong P M, Katavolos P, Foppa I, Olmeda Garcia A S, Wilson M L, Spielman A. A new tick-borne encephalitis-like virus infecting New England deer ticks, Ixodes dammini. Emerging Infect Dis. 1997;3:165–170. doi: 10.3201/eid0302.970209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueda M. A serological survey of cattle and small wild mammals for evidence of tick-borne encephalitis virus infection in Hokkaido, Japan. Jpn J Vet Res. 1980;28:51–52. [Google Scholar]
- 45.Van Regenmortel M H V. Virus species, a much overlooked but essential concept in virus classification. Intervirology. 1990;31:241–254. doi: 10.1159/000150159. [DOI] [PubMed] [Google Scholar]
- 46.Varelas-Wesley I, Calisher C H. Antigenic relationships of flaviviruses with undetermined arthropod-borne status. Am J Trop Med Hyg. 1982;31:1273–1284. doi: 10.4269/ajtmh.1982.31.1273. [DOI] [PubMed] [Google Scholar]
- 47.Venugopal K, Buckley A, Reid H W, Gould E A. Nucleotide sequence of the envelope glycoprotein of Negishi virus shows a close homology to Louping ill virus. Virology. 1992;190:515–521. doi: 10.1016/0042-6822(92)91245-p. [DOI] [PubMed] [Google Scholar]
- 48.Venugopal K, Gritsun T, Lashkevich V A, Gould E A. Analysis of the structural protein gene sequence shows Kyasanur Forest disease virus as a distinct member in the tick-borne encephalitis virus serocomplex. J Gen Virol. 1994;75:227–232. doi: 10.1099/0022-1317-75-1-227. [DOI] [PubMed] [Google Scholar]
- 49.Wallner G, Mandl C W, Ecker M, Holzmann H, Stiasny K, Kunz C K, Heinz F X. Characterization and complete genome sequences of high- and low-virulence variants of tick-borne encephalitis virus. J Gen Virol. 1996;77:1035–1042. doi: 10.1099/0022-1317-77-5-1035. [DOI] [PubMed] [Google Scholar]
- 50.Weaver S C, Hagenbaugh A, Bellew L A, Netesov S V, Volchkov V E, Chang G-J J, Clarke D K, Gousset L, Scott T W, Trent D W, Holland J J. A comparison of the nucleotide sequences of eastern and western equine encephalitis viruses with those of other alphaviruses and related RNA viruses. Virology. 1993;197:375–390. doi: 10.1006/viro.1993.1599. [DOI] [PubMed] [Google Scholar]
- 51.Westaway E G, Blok J. Taxonomy and evolutionary relationships of flaviviruses. In: Gubler D J, Kuno G, editors. Dengue and dengue hemorrhagic fever. Wallingford, England: CAB International; 1997. pp. 147–173. [Google Scholar]
- 52.Whitby J E, Whitby S N, Jennings A D, Stephenson J R, Barrett A D T. Nucleotide sequence of the envelope protein of a Turkish isolate of tick-borne encephalitis (TBE) virus is distinct from other viruses of the TBE virus complex. J Gen Virol. 1993;74:921–924. doi: 10.1099/0022-1317-74-5-921. [DOI] [PubMed] [Google Scholar]