Abstract
The vaccinia virus early transcription factor (VETF) is a DNA binding protein comprised of 70- and 82-kDa subunits encoded by the D6R and A8L genes, respectively. A previous investigation suggested a novel role for the 70-kDa subunit in the morphogenesis of vaccinia virus particles. The principal objectives of the present study were to determine if the 82-kDa subunit of VETF is also required for morphogenesis and, if so, whether the block occurs before or after the incorporation of the genome into the assembling virus particle. To address these and other questions, we constructed and characterized a conditionally lethal recombinant vaccinia virus in which the A8L gene is stringently repressed by the Escherichia coli lac operator system. The amount of 82-kDa protein synthesized could be regulated by the amount of inducer: from undetectable to higher than normal levels. Virus replication, as determined by plaque formation or virus yield upon synchronous infection, was dependent on inducer. Nevertheless, de novo synthesis of the 82-kDa subunit was not required for viral early, intermediate, and late gene expression or DNA replication. Overexpression of the A8L gene alone, produced by high concentrations of inducer, inhibited viral late protein synthesis, whereas overexpression of the D6R gene alone or both VETF genes simultaneously had little inhibitory effect. Laser confocal fluorescence and quantitative electron microscopic analyses revealed that immature and DNA-containing intermediate stage particles accumulated in the absence of inducer, indicating that the A8L protein has a role in morphogenesis of the core and subsequent events.
Vaccinia virus, the prototypic member of the poxvirus family, encodes enzymes and factors needed for replication and expression of its double-stranded DNA genome within the cytoplasm of host cells (25). Viral gene expression is transcriptionally regulated and divided into three successive stages: early, intermediate, and late. The viral DNA-dependent RNA polymerase contains at least eight virus-encoded subunits and is used for transcription of genes belonging to all three classes. The virus-encoded proteins needed for early transcription are present in the infecting virus particles, having been synthesized and assembled along with the viral structural proteins. Thus, de novo synthesis of viral proteins is not required for early transcription, which can occur in the presence of inhibitors of protein synthesis. The translation products of viral early mRNAs include RNA polymerase and factors needed for intermediate transcription, which occurs after DNA replication. Late transcription follows intermediate, because some late transcription factors are encoded by viral intermediate mRNAs.
The factors specific for early transcription have been more thoroughly characterized than those for the other stages. At least two proteins, the RNA polymerase-associated protein of 94 kDa (RAP94 [1, 2, 12]) and the vaccinia virus early transcription factor (VETF [9]), confer early promoter specificity to the RNA polymerase. RAP94, the product of the H4L gene, is thought to interact with VETF, although this has not been directly shown. VETF is a heterodimeric protein with subunits of approximately 70 and 82 kDa encoded by the D6R and A8L genes, respectively (5, 17). VETF binds specifically to early promoters (6, 7, 11) and recruits the RNA polymerase to form the initiation complex (3, 22). Upon addition of ribonucleoside triphosphates, a stable ternary complex is formed (18, 19). The 70-kDa subunit of VETF has an intrinsic ATPase activity (8) that may be involved in promoter clearance (4).
The process by which the components of the early transcription system are specifically incorporated into assembling virions is not yet understood. Insights came from an analysis of a recombinant vaccinia virus with an inducible H4L gene encoding RAP94 (36). In the absence of inducer, noninfectious mature-appearing virus particles lacking RNA polymerase as well as many enzymes involved in mRNA biosynthesis were formed. The finding that VETF is packaged in the absence of RAP94 led to the suggestion that promoter-bound VETF recruits RNA polymerase to the assembly complex, in much the same way as VETF recruits RNA polymerase for transcription. To investigate this possibility, we constructed a conditionally lethal recombinant vaccinia virus with an inducible D6R gene (20). In the presence of inducer, infectious virus particles containing VETF formed. When cells were infected with these particles in the absence of inducer, viral early, intermediate, and late gene expression occurred, demonstrating that only the incoming packaged form of VETF is required for gene expression. In this respect, the phenotype resembled that of the RAP94-inducible mutant. Differences in the phenotypes were apparent, however, when the infected cells were examined by electron microscopy. Immature viral particles accumulated when the D6R gene was repressed, suggesting that the 70-kDa subunit of VETF has a more basic role in morphogenesis than RAP94. The genome of vaccinia virus is thought to enter immature virus particles in the form of an electron-dense nucleoid (24). Because of its specific DNA binding properties, we proposed that VETF, together with other proteins, might serve as a chaperone to escort the genome into assembling particles.
The possibility remained, however, that the role of the 70-kDa polypeptide in morphogenesis was independent of its association with the 82-kDa subunit of VETF. Indeed, there are precedents for vaccinia virus proteins having several independent functions. For example, the cap-specific methyltransferase encoded by the J3R gene exists in monomeric and heterodimeric forms and functions as a cap-specific methyltransferase and a poly(A) polymerase elongation factor (16, 30). It was important, therefore, to determine the phenotype of a mutant vaccinia virus that does not express the 82-kDa subunit of VETF. Here, we describe the construction and characterization of a conditionally lethal recombinant vaccinia virus with an inducible A8R gene and demonstrate the accumulation of immature and DNA-containing intermediate stage particles under nonpermissive conditions.
MATERIALS AND METHODS
Cells and viruses.
Monolayers of BS-C-1 or CV-1 cells were grown in Eagle’s minimum essential medium (EMEM) with 10% fetal bovine serum (FBS). HeLa S3 cells grown in Dulbecco’s minimum essential medium with 10% FBS were used for preparation of virus stocks. Wild-type vaccinia virus (WR strain) and vGW3 (34) were prepared as described previously (20). Inducible mutant viruses were propagated in HeLa S3 monolayers in EMEM supplemented with 2% FBS and 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG).
Construction of recombinant vaccinia virus containing an inducible copy of the A8L gene.
Procedures for the construction of the A8L-inducible virus were similar to those described for a D6R-inducible virus (20). Briefly, the A8L open reading frame was amplified by PCR and inserted into pMITEOlac.20/3 so that it was regulated by a bacteriophage T7 promoter, Escherichia coli lac operator (lacO), and a cDNA copy of the encephalomyocarditis virus leader and flanked by vaccinia virus hemagglutinin (HA) sequences. This plasmid was transfected into cells infected with VT7lacOI (34), which contains a T7 RNA polymerase gene regulated by the vaccinia virus late P11 promoter and a lacO and a vaccinia virus early/late P7.5 promoter-regulated lac repressor (lacI) gene. Several recombinant viruses were plaque purified thrice under agar in the presence of mycophenolic acid. The genotypes of these viruses were verified by PCR. The recombinant virus code named vLJC1 is referred to as vA8ind+end+, signifying the presence of endogenous and inducible copies of the A8L gene.
Deletion of the endogenous copy of the A8L gene was carried out in a manner analogous to that used for the D6R gene (20). The neomycin resistance gene (neo) was obtained by digestion of pVVNEO (13) with SalI and BamHI, subcloned into the pGEM-3 vector to form plasmid pGEM-NEO. DNA on either side of the A8L gene was PCR amplified by using the following primer pairs: (i) GCGCGCGTCGACCTTGGATGATGGATACTTGAGAGTTG and GGGGGGGTCGACATTTATATCGTGGGGTAAAGTGAAAATCTACTACC and (ii) GCGCGCGGATCCCCCAACTCTGGAAGAGCACAAATAAATTAAAC and GGGGGGGGATCCGGAATCGGATACTATATCTTCGGTATCTTGACGCAG. The PCR products were cloned into an intermediate plasmid and subsequently into pGEM-NEO. The resulting plasmid, pA8KO, contains 5′ and 3′ sequences adjacent to the A8L gene at each side of neo with no residual A8L coding sequences. Plasmid pA8KO was mixed with Lipofectamine (Promega) and transfected into vA8ind+end+-infected CV-1 cells; the incubation was continued in the presence of 100 μM IPTG and 2 μg of G418 per ml. After three rounds of plaque purification in the presence of IPTG and G418, deletion of the A8L gene was confirmed by PCR. The recombinant virus was designated vA8ind since it contains a single inducible copy of the A8L gene. Virus stocks were prepared in HeLa cells in the presence of 100 μM IPTG.
Plaque assay.
BS-C-1 monolayers in six-well plates were infected with 10-fold serial dilutions of virus for 1 h and incubated at 37°C for 2 days in EMEM containing 2% FBS and 100 μM IPTG when appropriate. The cells were then stained with crystal violet, and the plaques were counted.
One-step growth of vaccinia viruses.
Monolayers of BS-C-1 cells were infected with 10 PFU of virus per cell for 1 h at 37°C, washed, and incubated at various times with or without IPTG. Cells were harvested, frozen and thawed three times, sonicated, and stored at −80°C. Virus titers were determined by plaque assay.
SDS-PAGE.
BS-C-1 cells in six-well plates were infected with 10 or 20 PFU of recombinant vaccinia viruses per cell at 37°C. Cells were labeled with 100 μCi of [35S]methionine per well for 15 min and lysed in sodium dodecyl sulfate (SDS) electrophoresis buffer for SDS-polyacrylamide gel electrophoresis (PAGE) analysis or in Nonidet P-40 (NP-40) lysis buffer (1.0% NP-40, 150 mM NaCl, 50 mM Tris-HCl [pH 8.0]) at 4°C for 10 min. Insoluble material was removed by sedimentation in a microcentrifuge at 4°C for 5 min, and the supernatant was incubated with antiserum at 4°C for 12 h and then with excess protein A-agarose beads for 2 h. The beads were subsequently washed with NP-40 lysis buffer and 25 mM Tris buffer (pH 8.8). SDS-containing sample buffer was added to the beads and heated at 100°C for 3 min, and the supernatant was applied to an SDS-polyacrylamide gel.
Antibodies.
Rabbit polyclonal antisera to the 70- and 82-kDa subunits of VETF have been described elsewhere (17). Monoclonal antibody (MAb) C3, to the protein encoded by the A27L open reading frame of vaccinia virus (28), was provided by M. Esteban. Fluorescein isothiocyanate (FITC)-labeled rabbit anti-mouse antiserum was purchased from Dako Corporation (Carpinteria, Calif.).
Confocal microscopy.
Viral particles and DNA were colocalized by indirect immunofluorescence and propidium iodide staining. HeLa cells were grown on glass coverslips to approximately 70 to 80% confluency and were infected in the presence or absence of 100 μg of rifampin per ml or 100 μM IPTG. At 18 h after infection, cells were fixed with 3% paraformaldehyde in phosphate-buffered saline for 20 min at room temperature and permeabilized, RNase treated, and processed as previously described (21) except that DNA was stained with 10% propidium iodide and virus particles were visualized with mAb C3 followed by FITC-conjugated rabbit anti-mouse antibody (immunoglobulin G [IgG]). Cells were visualized with a Zeiss Axioplan microscope, and images were captured with a Bio-Rad MRC 1024 laser confocal imaging system.
Electron microscopy.
BS-C-1 monolayers in 60-mm-diameter dishes were infected with 10 PFU of virus per cell in the presence or absence of 100 μM IPTG and fixed at various times in 2% glutaraldehyde (EM Sciences) in 100 mM phosphate buffer (pH 7.4) for 1 h. Cells were then prepared in Epon resin for transmission electron microscopy by osmication, dehydration, and embedding. Thin sections were cut, collected on Formvar-coated copper mesh grids (Polysciences), and stained with 2% uranyl acetate and Reynolds’ lead citrate (26). Samples were viewed with a Philips CM100 electron microscope.
RESULTS
Construction of a mutant vaccinia virus with an inducible A8L gene.
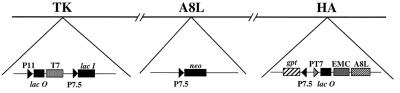
The systems originally used to inducibly regulate expression of vaccinia virus genes had the E. coli lacO placed just downstream of a vaccinia virus promoter (14, 29, 37). Ward et al. (34) developed a more versatile version in which one lacO was placed next to a vaccinia virus promoter that regulates the bacteriophage T7 RNA polymerase gene and a second lacO was placed next to a T7 promoter that regulates a recombinant gene. In addition to greatly enhanced repression in the absence of inducer, due to two lacO-regulated steps, the new system permits overexpression at high inducer concentrations. Following our successful use of the two operator system to regulate the D6R gene, encoding the 70-kDa subunit of VETF (20), we applied a similar strategy to regulate the A8L gene encoding the 82-kDa subunit. Figure 1 shows the important features of the recombinant vaccinia virus, vA8ind, containing an inducible copy of the A8L gene. Of special note are (i) at the thymidine kinase (TK) locus, the vaccinia virus P11 late promoter and lacO regulate the T7 RNA polymerase gene and the constitutively expressed vaccinia virus P7.5 promoter regulates lacI; (ii) the neo gene, regulated by the P7.5 promoter, replaces the endogenous A8L gene; and (iii) at the HA locus, the T7 promoter and lacO regulate the A8L gene and the P7.5 promoter regulates the gpt gene. The neo and gpt genes were used for antibiotic selection. vA8ind was constructed in two steps, starting with a recombinant virus (vlacOI) containing the indicated insertion in the TK gene (Fig. 1). The first step was the construction of the intermediate virus (vA8end+ind+) by insertion of the inducible copy of the A8L gene into the HA locus (Fig. 1). The latter virus was selected with mycophenolic acid, identified by the formation of syncytial plaques due to interruption of the HA gene, and genotyped by PCR. In the second step, the neo gene was inserted into the A8L locus. The recombinant vaccinia virus, vA8ind, was selected in the presence of G418 and IPTG and identified by PCR.
FIG. 1.
Representation of the genome of vA8ind. The TK, A8L, and HA loci are shown. Abbreviations: P11, a vaccinia virus late promoter; lacO, E. coli lac operator; T7, bacteriophage T7 RNA polymerase gene; P7.5, a vaccinia virus early/late promoter; lacI, E. coli lac repressor gene; neo, E. coli neomycin resistance gene; gpt, E. coli gpt gene; PT7, bacteriophage T7 promoter; EMC, cDNA copy of the encephalomyocarditis virus leader. Arrowheads indicate directions of transcription.
Effect of IPTG on virus replication.
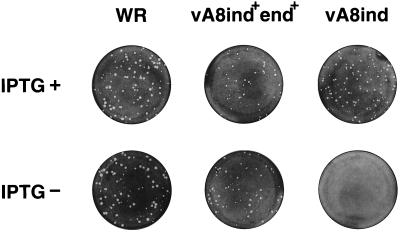
The conditional lethal nature of vA8ind was revealed by plaque assay. IPTG was needed for formation of visible plaques by vA8ind but not for either the standard laboratory strain of vaccinia virus, WR, nor for the intermediate vA8end+ind+, which retains the original A8L gene in addition to the inducible copy (Fig. 2). Microscopic examination of the cell monolayers revealed that vA8ind formed pinpoint foci (not shown) that may be due to secretion of viral growth factors (10) during early gene expression in the absence of IPTG.
FIG. 2.
Effect of IPTG on virus plaque formation. BS-C-1 cell monolayers were infected with the wild-type vaccinia virus (WR), vA8ind+end+, or vA8ind in the presence (+) or absence (−) of 100 μM IPTG. The cells were stained with crystal violet at 48 h after infection.
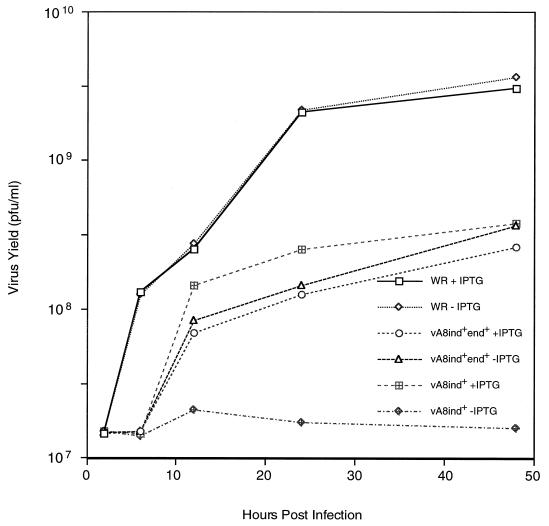
The conditional lethality of vA8ind was further examined by determining one-step virus yields in the presence or absence of 100 μM IPTG (Fig. 3). Without IPTG, little or no increase in titer of vA8ind was detected; with IPTG, there was nearly a 2-log increase. Further experiments indicated that the yield of vA8ind was slightly higher at 20 μM than at 100 μM IPTG (not shown). In contrast, the yield of vA8end+ind+ was unaffected by IPTG and was similar to vA8ind in the presence of inducer. Replication of WR virus was also unaffected by IPTG but was higher than that of the genetically engineered recombinant viruses. For this reason, vA8end+ind+ was the preferred control in some experiments.
FIG. 3.
Effect of IPTG on virus yields. BS-C-1 cells were infected with 10 PFU of wild-type virus (WR), vA8ind+end+, or vA8ind per cell in the presence (+) or absence (−) of 100 μM IPTG. The cells were harvested at the indicated times after infection, and the titers were determined by plaque assay in the presence of 100 μM IPTG.
Inducer-dependent expression of the 82-kDa subunit of VETF.
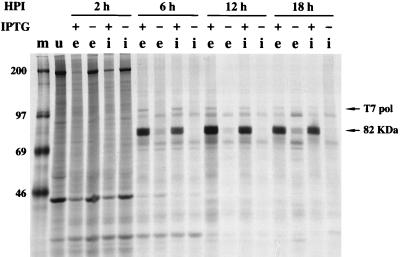
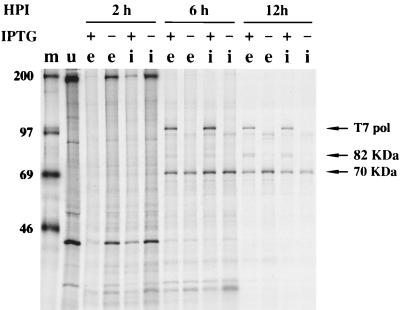
Cells that had been infected with WR, vA8end+ind+, or vA8ind in the presence or absence of 100 μM IPTG were pulse-labeled at intervals with [35S]methionine. Extracts were prepared and incubated with a specific antibody to the A8L protein, and the bound materials were analyzed by SDS-PAGE and autoradiography. A prominent 82-kDa band was synthesized starting at 6 h after infection of cells with vA8ind or vA8end+ind+ in the presence of inducer (Fig. 4). The timing was consistent with the late promoter used for transcription of the T7 RNA polymerase gene. In the absence of inducer, a faint 82-kDa band derived from the endogenous A8L gene was detectable in lysates of cells infected with vA8end+ind+ but not in cells infected with vA8ind (Fig. 4). A similarly faint 82-kDa band was detected when cells were infected with wild-type WR virus (not shown), indicating that the recombinant viruses vA8ind and vA8end+ind+ were overexpressing the protein in the presence of 100 μM IPTG.
FIG. 4.
Inducible expression of the A8L gene. BS-C-1 cells were uninfected (lane u) or infected with vA8ind+end+ (lanes e) or vA8ind (lanes i) in the presence (+) or absence (−) of 100 μM IPTG. The cells were metabolically labeled with [35S]methionine at the indicated times after infection (hours postinfection [HPI]), and lysates were incubated with a polyclonal antibody to the A8L protein and then with protein A beads. The bound proteins were analyzed by SDS-PAGE. The positions of the 82-kDa A8L protein and the bacteriophage T7 RNA polymerase (pol) are indicated. Lane m, marker proteins, with masses indicated in kilodaltons.
Since VETF is a heterodimer of 82- and 70-kDa subunits, their coprecipitation with either an A8L- or a D6R-specific antiserum would be expected. Although the 70-kDa subunit was not clearly resolved in Fig. 4, it was visualized in other experiments using an A8L-specific antiserum (not shown). Using a D6R-specific antiserum, we found that neither under- nor overexpression of the A8L gene grossly affected the synthesis (Fig. 5) or stability (not shown) of the 70-kDa subunit of VETF. When the 82-kDa subunit was overexpressed, it coprecipitated with the 70-kDa subunit when the D6R-specific antiserum was used (Fig. 5). Bands corresponding to T7 RNA polymerase were present in the +IPTG lanes of Fig. 4 and 5, presumably because the recombinant A8L and D6 proteins used to immunize the rabbits had been prepared with a bacteriophage T7 bacterial expression vector, rather than because of coprecipitation with the viral proteins.
FIG. 5.
Expression of the D6R gene is independent of A8L gene expression. The experiment was carried out as described in the legend to Fig. 4 except that rabbit polyclonal antiserum to the D6R protein was used for immunoprecipitation. Arrows indicate the positions of T7 RNA polymerase and the 82- and 70-kDa subunits of VETF. For other details, see the legend to Fig. 4.
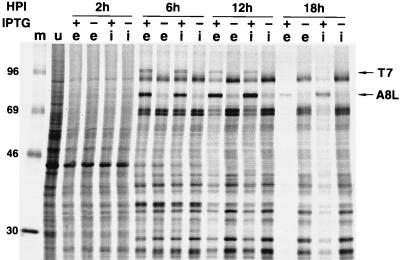
Effect of IPTG on viral protein synthesis.
Since vA8ind was propagated in the presence of IPTG, the infectious virions contain VETF. The expectation was, therefore, that early and late transcription would occur normally in cells infected with vA8ind in the absence of IPTG. The effect of IPTG on the regulation of viral protein synthesis was determined by pulse-labeling cells with [35S]methionine for 15-min intervals at 2, 6, 12, and 18 h after infection with vA8ind or vA8end+ind+. The proteins were resolved by SDS-PAGE and autoradiography (Fig. 6). The results obtained with the control vA8end+ind+ in the absence of IPTG are described first. At 2 h after infection, the labeled protein bands were similar to those of uninfected cells except for decreases in their intensity; as frequently occurs, early viral proteins were not clearly resolved because of the low amounts made and the continued synthesis of host proteins. At 6 h, the prominent pattern produced by the abundant late proteins was established and persisted with little change for the remainder of the infection. In the presence of 100 μM IPTG, bands corresponding to T7 RNA polymerase and the 82-kDa A8L protein were also detected. The intensity of viral bands became progressively weaker at 12 and 18 h in the presence of IPTG. The pattern of labeled viral proteins from cells infected with vA8ind was virtually identical to that of vA8end+ind+ in the presence or absence of IPTG (Fig. 6). Since viral protein synthesis was higher under nonpermissive conditions in the absence of IPTG than under permissive conditions in the presence of IPTG, the defect in virus replication could hardly be caused by a general reduction in viral protein synthesis.
FIG. 6.
Synthesis of viral proteins. Uninfected BS-C-1 cells (lane u) or cells infected with 10 PFU of vA8ind+end+ (lanes e) or vA8ind (lanes i) in the presence (+) or absence (−) of 100 μM IPTG were pulse-labeled for 15 min with [35S]methionine at the indicated times after infection, lysed in SDS sample buffer, and analyzed by SDS-PAGE. Lane m, protein markers, with masses indicated in kilodaltons. The 82-kDa A8L protein and the T7 RNA polymerase (pol) are indicated with arrows.
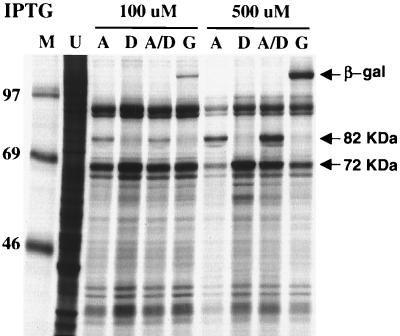
We further investigated the IPTG-induced inhibition of viral protein synthesis caused by the A8L mutant, since this was not apparent with the inducible D6R mutant (20). In our expression system, the amount of target protein produced is proportional to the inducer concentration (34). Infections, in 100 or 500 μM IPTG-containing media, were carried out with 20 PFU of either vA8ind, vD6ind, or lacZ-inducible virus vGW3 per cell or with a mixture of 10 PFU of vA8ind and 10 PFU of vD6ind per cell. The infected cells were labeled with [35S]methionine for 15 min at 12 h after infection, the usual time of maximal vaccinia virus protein synthesis. In cells infected with vA8ind at the higher concentration of IPTG, the 82-kDa band was more intense whereas other viral bands were greatly diminished (Fig. 7, lanes A). The 70-kDa protein encoded by D6R, which migrated slightly ahead of the 69-kDa marker, was not resolved from other viral proteins; nevertheless, comparison of the various lanes indicated that the band including the D6R protein was most intense in lanes D, consistent with its overexpression by the D6R-inducible mutant as previously shown (20). Significantly, other viral proteins were not greatly diminished in cells infected with vD6R at 500 μM IPTG compared to 100 μM IPTG. A strong reduction in labeling of viral proteins was also not seen in cells infected with vGW3, even though β-galactosidase was highly expressed at these IPTG concentrations (Fig. 7, lane G). Therefore, the inhibition of viral protein synthesis was specifically related to overexpression of the A8L gene product. Interestingly, the inhibition of viral protein synthesis did not occur when cells were coinfected with vD6ind and vA8ind even though the 82-kDa protein was overexpressed (Fig. 7, lanes A/D). This was not simply due to coinfection per se, because the inhibition still occurred when cells were coinfected with vA8ind and vGW3 or when the amounts of vD6ind and vA8ind were each doubled for coinfection (data not shown). One explanation for these results is that the inhibitory activity of the 82-kDa subunit was abrogated by complexing with the overexpressed 72-kDa subunit, which itself is not toxic (20).
FIG. 7.
Inhibition of viral late protein synthesis by overexpression of A8L. BS-C-1 cells were infected with 20 PFU of vA8ind (lanes A), vD6ind (lanes D), or vGW3 (lanes G), or with 10 PFU of vA8ind and 10 PFU of vD6ind (lanes A/D), per cell in the presence of 100 or 500 μM IPTG as indicated. After 12 h, the cells were lysed in SDS sample buffer and analyzed by SDS-PAGE. Lane U, lysate of similarly labeled uninfected BS-C-1 cells; lane M, protein markers, with masses indicated in kilodaltons. The β-galactosidase (β-gal) and the 82-kDa and unresolved 70-kDa protein bands are indicated with arrows.
Colocalization of viral DNA and particles.
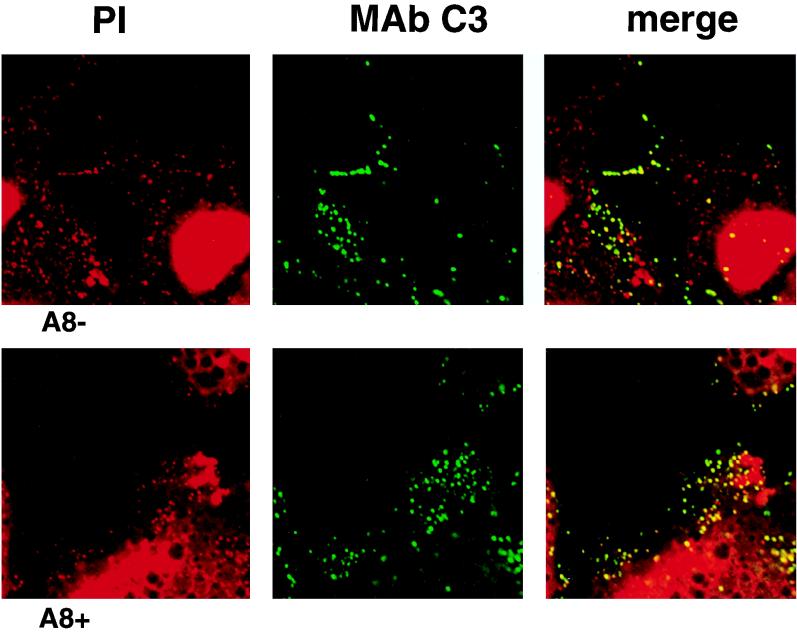
Laser scanning confocal microscopy was used to determine whether DNA-containing viral particles formed under nonpermissive as well as permissive conditions. Virus particles at intermediate and later stages of morphogenesis were labeled by using MAb C3 to the 14-kDa virion surface protein encoded by the A27L gene (31). Cells were infected with vA8ind in the presence or absence of IPTG. Viral and cellular DNA were visualized with propidium iodide, and virus particles were stained with MAb C3 followed by a FITC-conjugated second antibody (IgG). The absence of bleed-through from the green (FITC) channel was verified by using identical settings for infected cells that had not been stained with propidium iodide. Staining of cellular DNA in the nucleus was evident, and viral DNA was clearly visualized in the cytoplasm of infected cells. Two patterns of viral DNA were noted: large perinuclear masses, resembling previous descriptions of Hoechst-stained viral factories, were most clearly demarcated early in infection (not shown), whereas punctate staining particles were only seen late in infection (Fig. 8). Most, but not all, of the punctate DNA colocalized with virus particles detected by the anti-14kDa protein MAb. Correspondingly, nearly all MAb-staining particles appeared to localize with DNA. Similar patterns were obtained in the presence or absence of IPTG, except for an impression that more of the large, discrete DNA factories persisted until later times of infection under the latter conditions. The punctate staining did not occur with either propidium iodide or the MAb when the cells were infected in the presence of rifampin, an inhibitor of an early step in viral morphogenesis, consistent with the identification of these structure as viral particles (data not shown).
FIG. 8.
Confocal microscopic images of cells infected with vA8ind in the absence (A8−) or presence (A8+) of 100 μM IPTG. Infected HeLa cells were stained with either propidium iodide (PI) or MAb C3 followed by FITC-conjugated rabbit anti-mouse IgG. The images obtained with the red (PI), green (MAb C3), and merged channels are shown.
Viral morphogenesis under permissive and nonpermissive conditions.
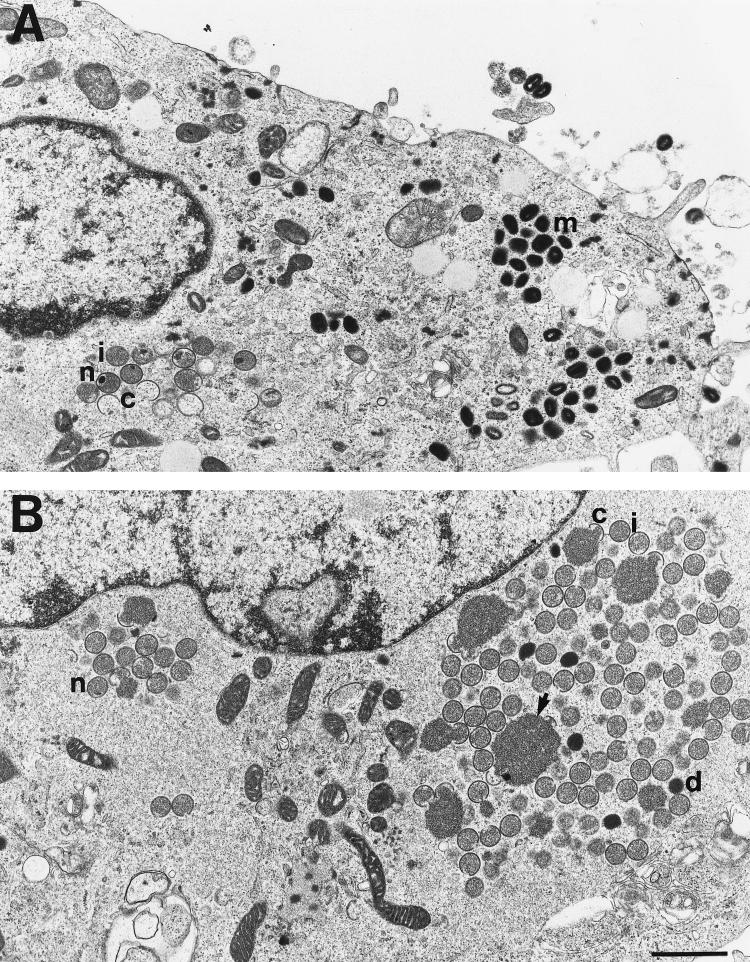
Electron microscopy was used to analyze the viral particles that formed under nonpermissive conditions. High-power images are shown of cells infected with vA8ind in the presence (Fig. 9A) or absence (Fig. 9B) of IPTG. We derived the viral structures into five classes: crescents, spherical immature particles without nucleoids, spherical immature particles with nucleoids, spherical dense particles, and brick-shaped mature particles. In the presence of IPTG, there were large numbers of crescent, immature, and mature forms. In the absence of IPTG, there were mostly crescents, immature forms and dense particles, which lacked the brick shape and internal structure of virions, and few mature particles. The numbers of each type are listed in Table 1. Only a minority of immature particles contained visible nucleoids in the ultrathin sections; however, nucleoids were at least as abundant in the absence of IPTG as in the presence of IPTG. The differences between cells infected in the presence and absence of inducer were noted at 18, 24, and 48 h after infection, indicating that morphogenesis was not merely delayed.
FIG. 9.
Electron microscopic images of vA8ind-infected cells. BS-C-1 cells were infected with vA8ind in the presence (A) or absence (B) of IPTG. Abbreviations: c, crescents; i, immature particles; n, nucleoids; d, dense particles; m, mature particles. The arrow in panel B points to a large granular mass present in areas that contain large numbers of immature particles. The bar represents 1 μm.
TABLE 1.
Quantitation of vA8ind particles resolved by electron microscopy
| IPTG | No. of particles (%)
|
|||||
|---|---|---|---|---|---|---|
| Crescents | Immature | Nucleoids | Dense | Mature | Total | |
| + | 1,274 (36.6) | 914 (26.3) | 39 (1.1) | 90 (2.6) | 1,164 (33.4) | 3,482 (100) |
| − | 1,281 (28.4) | 2,723 (60.4) | 73 (1.6) | 183 (4.0) | 252 (5.6) | 4,511 (100) |
Under abortive conditions, there were also large masses of granular material resembling the internal contents of immature virions. Crescents, partially enclosing some of the granular material, are present at the periphery of the masses (Fig. 9). Such large masses were infrequently encountered in the presence of IPTG at any of the times examined or in wild-type virus infections, but they have been found under a variety of conditions in which virus particle formation is inhibited.
DISCUSSION
The assembly and morphogenesis of infectious poxvirus particles is a complex process requiring the interactions of membranes, numerous proteins, and DNA. One aspect of virion assembly, unique to poxviruses, is the incorporation of a complete stage-specific transcriptional system. We have combined genetic and electron microscopic approaches to gain insights into these processes. Conditionally lethal, inducible mutants with a null phenotype provide an alternative to temperature-sensitive mutants. Such mutants have now been made for the three genes, H4L, D6R, and A8L, that are known to encode proteins that are specific for early transcription. Repression of the H4L gene, encoding RAP94, did not grossly interfere with morphogenesis of virus particles, even though RNA polymerase and numerous other enzymes were not packaged (36). The finding of DNA and VETF in these enzyme-deficient particles suggested that VETF is normally recruited before or independently of RAP94. If the only role of VETF in assembly were to bind RNA polymerase, then repression of either VETF subunit might result in the formation of particles resembling those that formed without RAP94. A previous study showed, however, that repression of D6R interferes with morphogenesis (20). This unexpected finding raised questions that have been addressed in the present study. The first was whether repression of the A8L subunit would give rise to a phenotype similar to that found when D6R was repressed. Presuming that morphogenesis was interrupted, the second question was whether this occurred before or after entry of the DNA genome into viral particles. Given the DNA binding properties of VETF, the former possibility was favored.
To address the first question, we constructed a recombinant vaccinia virus that contains a copy of the A8L gene under lacO control and then deleted the original A8L gene. The resulting virus, vA8ind, was dependent on IPTG for replication. Under nonpermissive conditions, the phenotype of vA8ind was virtually identical to that of the previous vD6ind mutant. Thus, viral protein synthesis proceeded normally in the absence of A8L expression, indicating that the packaged VETF was sufficient for viral early gene expression and that de novo synthesis of the 82-kDa protein was not required for viral intermediate or late gene expression. In the presence of IPTG, the amount of 82-kDa protein was much higher than during wild-type infection, making this virus a possible source of material uncomplexed to the 70-kDa subunit. In a similar manner, vD6ind makes much higher amounts of 70-kDa subunit than occur during infection with wild-type virus (20). Nevertheless, we have not yet tried to purify the individual subunits from soluble extracts.
Masternak and Wittek (23) reported that overexpression of both subunits of VETF, by coinfection of cells with two recombinant viruses, inhibited synthesis of vaccinia virus late proteins. The effects of individually overexpressed subunits were not reported. We observed that overexpression of the 82-kDa protein resulted in inhibition of viral late protein synthesis, whereas overexpression of the 70-kDa protein did not. Moreover, the inhibitory effect caused by overexpression of the 82-kDa protein was prevented by coexpression of 70-kDa subunit. Cross-linking studies have shown that the 82-kDa subunit of VETF can bind DNA in vitro without sequence specificity (11), suggesting a possible general transcriptional inhibitory effect of the overexpressed 82-kDa subunit. In vitro studies with purified subunits would be useful for further investigations of this phenomenon.
When expression of the A8L gene was repressed, the defect in vaccinia virus replication occurred during virion morphogenesis. We wished to know whether the A8L gene was required for incorporating DNA into the assembling virions. Although use of anti-DNA antibodies for immunoelectron microscopy has been described (32), the sensitivity of staining vaccinia virus particles was too low for our purposes. Recently, Vanderplasschen and Smith (33) described the use of laser scanning confocal microscopy to visualize vaccinia virus particles on the cell surface. We used a similar technique to colocalize DNA and virus particles within the cell cytoplasm. Infected cells were permeabilized, digested with RNase, and stained with propidium iodide, a DNA-intercalating agent. Perinuclear viral factories were prominently stained at early times, whereas there was a more dispersed punctate staining pattern at late times. The latter DNA colocalized with particles that bound antibody directed to a virion surface protein. Such DNA-containing particles did not form in the presence of rifampin, an inhibitor of an early step in morphogenesis. However, they did form when A8L gene expression was repressed, indicating that the latter protein was not required for encapsidation of DNA. This colocalization technique may be useful for characterizing other vaccinia virus mutants that have defects in morphogenesis.
Quantitative electron microscopy revealed that there were increased numbers of immature and dense intermediate particles but relatively few apparently mature ones in cells infected with vA8ind in the absence, compared to the presence, of IPTG. The few mature-looking particles that formed in the absence of IPTG could represent either leaky expression of the A8L gene or inefficient VETF-independent maturation, and they may not be infectious. There were similar numbers of immature particles with nucleoids, thought to contain the viral DNA genome (24), in the absence of IPTG as in its presence. The actual number of nucleoids was quite low, however, possibly because they occurred infrequently in the plane of the ultrathin sections. Taking the confocal and electron microscopy results together, we concluded that a block occurs predominantly at an intermediate stage in morphogenesis, after the incorporation of the DNA genome but before the formation of a recognizable core structure. Although we did not quantitate the various types of particles in our previous study of repression of the D6R gene (20), the electron microscopic images were virtually identical to those shown here for vA8ind.
VETF and other DNA binding proteins could have direct roles in morphogenesis of the viral core. An alternative, indirect mechanism for VETF was considered previously in the context of repression of the D6R gene (20). We suggested that de novo synthesis of VETF might be required for transcription of a unique set of late genes that encode proteins required for morphogenesis. Such a late role for VETF could be consistent with the transcriptional reactivation of certain early promoters at late times (15). However, this idea remains speculative, as reactivatable promoters have not been demonstrated in association with genes encoding structural proteins.
To further study the packaging of enzymes and other internal proteins during assembly, we may need to adapt immunoelectron microscopic methods that have been effectively used to monitor the recruitment of viral membrane proteins during morphogenesis (27, 32, 35). Antibodies with high avidities and specificities will be needed, together with procedures to avoid high backgrounds caused by the soluble cytoplasmic proteins.
REFERENCES
- 1.Ahn B-Y, Gershon P D, Moss B. RNA-polymerase associated protein RAP94 confers promoter specificity for initiating transcription of vaccinia virus early stage genes. J Biol Chem. 1994;269:7552–7557. [PubMed] [Google Scholar]
- 2.Ahn B-Y, Moss B. RNA polymerase-associated transcription specificity factor encoded by vaccinia virus. Proc Natl Acad Sci USA. 1992;89:3536–3540. doi: 10.1073/pnas.89.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick C J, Cassetti M C, Harris N, Moss B. Ordered assembly of a functional preinitiation transcription complex, containing vaccinia virus early transcription factor and RNA polymerase, on an immobilized template. J Virol. 1994;68:6052–6056. doi: 10.1128/jvi.68.9.6052-6056.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broyles S S. A role for ATP hydrolysis in vaccinia virus early gene transcription. J Biol Chem. 1991;266:15545–15548. [PubMed] [Google Scholar]
- 5.Broyles S S, Fesler B S. Vaccinia virus gene encoding a component of the viral early transcription factor. J Virol. 1990;64:1523–1529. doi: 10.1128/jvi.64.4.1523-1529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broyles S S, Li J. The small subunit of the vaccinia virus early transcription factor contacts the transcription promoter DNA. J Virol. 1993;67:5677–5680. doi: 10.1128/jvi.67.9.5677-5680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broyles S S, Li J, Moss B. Promoter DNA contacts made by the vaccinia virus early transcription factor. J Biol Chem. 1991;266:15539–15544. [PubMed] [Google Scholar]
- 8.Broyles S S, Moss B. DNA-dependent ATPase activity associated with vaccinia virus early transcription factor. J Biol Chem. 1988;263:10761–10765. [PubMed] [Google Scholar]
- 9.Broyles S S, Yuen L, Shuman S, Moss B. Purification of a factor required for transcription of vaccinia virus early genes. J Biol Chem. 1988;263:10754–10760. [PubMed] [Google Scholar]
- 10.Buller R M L, Chakrabarti S, Moss B, Frederickson T. Cell proliferative response to vaccinia virus is mediated by VGF. Virology. 1988;164:182–192. doi: 10.1016/0042-6822(88)90635-6. [DOI] [PubMed] [Google Scholar]
- 11.Cassetti M C, Moss B. Interaction of the 82-kDa subunit of the vaccinia virus early transcription factor heterodimer with the promoter core sequence directs downstream DNA binding of the 70-kDa subunit. Proc Natl Acad Sci USA. 1996;93:7540–7545. doi: 10.1073/pnas.93.15.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng S, Shuman S. A role for the H4 subunit of vaccinia RNA polymerase in transcription initiation at a viral early promoter. J Biol Chem. 1994;269:14323–14329. [PubMed] [Google Scholar]
- 13.Franke C A, Rice C M, Strauss J H, Hruby D E. Neomycin resistance as a dominant selectable marker for selection and isolation of vaccinia virus recombinants. Mol Cell Biol. 1985;5:1918–1924. doi: 10.1128/mcb.5.8.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst T R, Fernandez M P, Moss B. Transfer of the inducible lac repressor/operator system from Escherichia coli to a vaccinia virus expression vector. Proc Natl Acad Sci USA. 1989;86:2549–2553. doi: 10.1073/pnas.86.8.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcés J, Maternak K, Kunz B, Wittek R. Reactivation of transcription from a vaccinia virus early promoter late in infection. J Virol. 1993;67:5394–5401. doi: 10.1128/jvi.67.9.5394-5401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershon P D, Ahn B Y, Garfield M, Moss B. Poly(A) polymerase and a dissociable polyadenylation stimulatory factor encoded by vaccinia virus. Cell. 1991;66:1269–1278. doi: 10.1016/0092-8674(91)90048-4. [DOI] [PubMed] [Google Scholar]
- 17.Gershon P D, Moss B. Early transcription factor subunits are encoded by vaccinia virus late genes. Proc Natl Acad Sci USA. 1990;87:4401–4405. doi: 10.1073/pnas.87.11.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagler J, Shuman S. Ternary complex formation by vaccinia virus RNA polymerase at an early viral promoter: analysis by native gel electrophoresis. J Virol. 1992;66:2982–2989. doi: 10.1128/jvi.66.5.2982-2989.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagler J, Shuman S. Structural analysis of ternary complexes of vaccinia virus RNA polymerase. Proc Natl Acad Sci USA. 1992;89:10090–10103. doi: 10.1073/pnas.89.21.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X, Carroll L J, Wolffe E J, Moss B. De novo synthesis of the early transcription factor 70-kDa subunit is required for morphogenesis of vaccinia virions. J Virol. 1996;70:7669–7677. doi: 10.1128/jvi.70.11.7669-7677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacs G R, Moss B. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Broyles S S. Recruitment of vaccinia virus RNA polymerase to an early gene promoter by the viral early transcription factor. J Biol Chem. 1993;268:2773–2780. [PubMed] [Google Scholar]
- 23.Masternak K, Wittek R. cis- and trans-acting elements involved in reactivation of vaccinia virus early transcription. J Virol. 1996;70:8737–8746. doi: 10.1128/jvi.70.12.8737-8746.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan C. The insertion of DNA into vaccinia virus. Science. 1976;193:591–592. doi: 10.1126/science.959819. [DOI] [PubMed] [Google Scholar]
- 25.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 26.Reynolds E S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez D, Risco C, Rodríguez J R, Carrascosa J L, Esteban M. Inducible expression of the vaccinia virus A17L gene provides a synchronized system to monitor sorting of viral proteins during morphogenesis. J Virol. 1996;70:7641–7653. doi: 10.1128/jvi.70.11.7641-7653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez J F, Janeczko R, Esteban M. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J Virol. 1985;56:482–488. doi: 10.1128/jvi.56.2.482-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez J F, Smith G L. Inducible gene expression from vaccinia virus. Virology. 1990;177:239–250. doi: 10.1016/0042-6822(90)90477-9. [DOI] [PubMed] [Google Scholar]
- 30.Schnierle B S, Gershon P D, Moss B. Cap-specific mRNA (nucleoside-O2′-)-methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc Natl Acad Sci USA. 1992;89:2897–2901. doi: 10.1073/pnas.89.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sodeik B, Cudmore S, Ericsson M, Esteban M, Niles E G, Griffiths G. Assembly of vaccinia virus: incorporation of p14 and p32 into the membrane of the intracellular mature virus. J Virol. 1995;69:3560–3574. doi: 10.1128/jvi.69.6.3560-3574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van’t Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanderplasschen A, Smith G L. A novel virus binding assay using confocal microscopy: demonstration that intracellular and extracellular vaccinia virions bind to different cellular receptors. J Virol. 1997;71:4032–4041. doi: 10.1128/jvi.71.5.4032-4041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolffe E J, Moore D M, Peters P J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Ahn B-Y, Moss B. Targeting of a multicomponent transcription apparatus into assembling vaccinia virus particles requires RAP94, an RNA polymerase-associated protein. J Virol. 1994;68:1360–1370. doi: 10.1128/jvi.68.3.1360-1370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Moss B. Inducer-dependent conditional-lethal mutant animal viruses. Proc Natl Acad Sci USA. 1991;88:1511–1515. doi: 10.1073/pnas.88.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]