Abstract
With our continuous endeavors in seeking potent anti-HIV-1 agents, we reported here the discovery, biological characterization, and druggability evaluation of a class of nonnucleoside reverse transcriptase inhibitors. To fully explore the chemical space of the NNRTI-binding pocket, novel series of dihydrothiopyrano [3,2-d]pyrimidines were developed by employing the structure-based design strategy. Most of the derivatives were endowed with prominent antiviral activities against HIV-1 wild-type and resistant strains at nanomolar levels. Among them, compound 23h featuring the aminopiperidine moiety was identified as the most potent inhibitor, with EC50 values ranging from 3.43 to 21.4 nmol/L. Especially, for the challenging double-mutants F227L + V106A and K103N + Y181C, 23h exhibited 2.3- to 14.5-fold more potent activity than the first-line drugs efavirenz and etravirine. Besides, the resistance profiles of 23h achieved remarkable improvement compared to efavirenz and etravirine. The binding target of 23h was further confirmed to be HIV-1 reverse transcriptase. Molecular modeling studies were also performed to elucidate the biological evaluation results and give guidance for the optimization campaign. Furthermore, no apparent inhibition of the major CYP450 enzymes and hERG channel was observed for 23h. Most importantly, 23h was characterized by good pharmacokinetic properties and excellent safety in vivo. Collectively, 23h holds great promise as a potential candidate for its effective antiviral efficacy and favorable drug-like profiles.
Key words: HIV-1; Reverse transcriptase; Dihydrothiopyrano[3,2-d]pyrimidine; Antiviral agent
Graphical abstract
With our continuous endeavors in seeking potent anti-HIV-1 agents, we report here the discovery of 23h as a potential candidate due to its prominent antiviral activities and favorable druggability profiles.
1. Introduction
Human immunodeficiency virus-1 (HIV-1) is the major pathogen of acquired immune deficiency syndrome (AIDS), which remains a serious epidemic disease threatening global public health1,2. Benefit from the development of antiviral agents for different stages of the HIV-1 replication cycle, the application of combination antiretroviral therapy (cART) has successfully transformed AIDS from a fatal disease to a controllable chronic disease3. Reverse transcriptase (RT) is an attractive HIV-1 therapeutic target due to the validated biochemical mechanism and abundant structural information, which plays a unique role in transcribing ssRNA into dsDNA4,5.
HIV-1 RT inhibitors can be divided into nucleoside RT inhibitors (NRTIs) and nonnucleoside RT inhibitors (NNRTIs) based on distinct binding sites and mechanisms of action6. NRTIs are competitively integrated into the processing DNA to produce chain termination by acting as natural substrate analogues. NNRTIs occupy the induced allosteric site, called the NNRTI-binding pocket (NNIBP), leading to conformational changes in the active site and inhibition of catalytic activity7,8. NNRTIs have become an increasingly essential component of cART for their robust potency, high specificity, and lack of mitochondrial toxicity, and they are available in the first two-drug single-tablet regimen Juluca® (rilpivirine + dolutegravir)9 and the first long-acting injectable formulation Cabenuva® (rilpivirine + cabotegravir)10 for maintenance of virological suppression11.
Diarylpyrimidine (DAPY) derivatives characterized by a central pyrimidine ring and two aromatic wings have attracted widespread attention as the most successful class of HIV-1 NNRTIs12,13. The FDA-approved drugs etravirine (1, ETR) and rilpivirine (2, RPV) possess extraordinarily potent antiviral activities against various resistant strains selected by the earlier NNRTIs and are developed as once-daily oral tablets (Fig. 1)7,12. Another promising drug dapivirine (3, DPV) is later applied as a vaginal microbicide due to its different oral bioavailability and long half-life14. A vaginal ring containing DPV was approved by the European Medicines Agency (EMA) in 2020 as an effective pre-exposure prophylaxis (PrEP) agent for HIV-1 prevention15. Nevertheless, new mutations with decreased susceptibility to ETR or RPV and aggravated cross-resistance continue to emerge in clinical treatment due to the inherent high mutation rate of viral genetic material, for instance, L100I for ETR and Y188L for RPV, especially the most challenging double-mutants F227L + V106A and K103N + Y181C16,17. Moreover, both ETR and RPV suffer from poor water solubility (Sol. < 1 μg/mL) and pharmacokinetic (PK) properties on account of excessive lipophilic rings in the rigid structure, and there are no accurately measured oral bioavailability data18. Consequently, unremitting efforts are still required to focus on extensive structural optimizations of DAPY derivatives to seek novel inhibitors with higher drug–resistance profiles and good pharmacological properties.
Figure 1.
Chemical structures of the approved drugs (ETR, RPV, and DPV).
As is well known, classical structure-based drug design strategies usually focus on the systematic modifications of specific molecular structures to explore the accessible chemical space and enhance the affinity with the target protein19,20. According to the recent advances in crystallographic studies, all DAPY derivatives share a horseshoe-shaped binding conformation and contain four pivotal pharmacophoric features21,22. Therefore, in the present work, extensive molecular elaborations have been conducted based on the “four-point pharmacophore” model by investigating the potential binding sites of tolerant region I, tolerant region II, hydrophobic channel, and hydrogen bonding interaction domain to discover new HIV-1 inhibitors with elevated biological activity and favorable druggability profiles (Fig. 2).
-
(i)
Tolerant region II: The central scaffold of DAPYs is located in the tolerant region II (i.e., entrance channel) composed of amino acid residues Leu100, Glu138, and Val179. During lead optimization, the Fsp3 parameter was introduced as a novel metric to assess drug-like properties by quantifying the carbon saturation and spatial complexity of molecules. It represents the fraction of sp3 hybridized carbon atoms, calculated as the ratio of the number of sp3 hybridized carbon atoms to the total carbon count, which has been proven to be positively correlated with the clinical success rate23,24. However, ETR and RPV have low Fsp3 values because of the introduction of three aromatic rings. Consequently, the saturated dihydrothiopyrano ring with high Fsp3 content and large molecular volume was fused to the central scaffold by cyclizing the pyrimidine ring, in the hope that the newly introduced non-planar structure could reduce the crystal packing of aromatic molecules and fully occupy the binding pocket. Besides, the S atom possesses a comparable atomic radius (rs = 104 pm) and lone pair electron as the Br atom (rBr = 114 pm), which may contribute to the formation of non-polar interaction with Glu138 and electrostatic interaction with Leu100 and Val179.
-
(ii)
Hydrogen bonding interaction domain: The NH linker and the pyrimidine 1′-N atom establish key dual hydrogen bonds with the main chain of Lys101. So the piperazine and aminopiperidine substituted with benzylsulfonamide directly connected to the central scaffold via a carbon−nitrogen bond were introduced to replace the NH linker to determine the necessity of dual hydrogen bonds for the maintenance of activity.
-
(iii)
Hydrophobic channel: The left-wing 2,4,6-trisubstituted phenyl moiety of DAPYs occupies the hydrophobic channel consisting of aromatic residues Tyr181, Phe227, and Trp229. Therefore, three preferred substituents (cyano, cyanovinyl, and methyl) derived from the approved drugs (ETR, RPV, and DPV) were installed on the left-wing to yield effective π−π stacking interactions.
-
(iv)
Tolerant region I: The right-wing p-cyanoaniline fragment of DAPYs points to a broad plastic “groove” surrounded by residues Val106, Leu234, Pro236, and Tyr318, termed as the solvent-exposed tolerant region I (i.e., the Pro236 hairpin loop). Recently, a novel class of catechol diether derivatives was disclosed as potent NNRTIs with robust antiviral activities at picomolar levels and favorable physicochemical properties (Fig. 3), exemplified by compounds 4 (JLJ494, EC50 = 0.055 nmol/L) and 5 (JLJ636, EC50 = 1.9 nmol/L)25,26. Moreover, the crystallographic overlay of DAPYs and catechol diethers indicated that the ethyluracil moiety is oriented towards the tolerant region I. Furthermore, morpholine as a frequently used heterocycle for medicinal chemistry is employed in numerous bioactive molecules, mainly due to its advantageous contribution to the biological and metabolic properties27. Besides, in our previous work, compounds 6 (EC50 = 0.011 μmol/L) and 7 (EC50 = 0.141 μmol/L) with moderate inhibitory potency were discovered by introducing morpholine into the tolerant region I (Fig. 3)28,29. Therefore, uracil and morpholine with a terminal ethylamino group were applied to the right-wing portion as a solvent-friendly fragment to investigate the suitability of tolerant region I. In addition, considering that the flexible piperidine ring could adapt well to the variations of the binding site caused by residue mutations30,31, the p-cyanoaniline was replaced by the privileged piperidine-linked benzyl or phenyl motif to improve the drug resistance profiles.
Figure 2.
Multidimensional optimization of diarylpyrimidine derivatives based on the “four-point pharmacophore” model.
Figure 3.
Chemical structures of the representative NNRTI lead compounds containing uracil (catechol diether derivatives 4 and 5) or morpholine (diarylpyrimidine derivatives 6 and 7).
The energy required to shift from the lowest energy preferred conformation to the pharmacophoric conformation adopted by the ligand to bind to the protein binding site can be compensated by the binding energy released by drug–target interactions32,33. Therefore, the conformational difference between the preferred conformation and the pharmacophoric conformation directly affects the binding affinity of the drug to the target34. Notably, the lowest energy preferred conformation of novel derivatives with distinct scaffolds were found to superimpose well with their pharmacophoric conformation in the RT allosteric binding site, preliminarily validating the rationality of the design idea based on the four-point pharmacophore model (Fig. 4).
Figure 4.
Overlay of the lowest energy preferred conformation (white) and the pharmacophoric conformation (green) of the representative molecules with distinct scaffolds. Predicted binding mode of ligands with the HIV-1 WT RT (PDB code: 6C0N).
Herein, we described the rational design and multidimensional optimization of novel dihydrothiopyrano [3,2-d]pyrimidine derivatives as potent HIV-1 inhibitors. Besides, the preliminary SARs were discussed in detail to guide further work. More importantly, the biological characterization and druggability evaluation of the most promising compound 23h were performed to confirm the feasibility of 23h as a potential drug candidate.
2. Results and discussion
2.1. Chemistry
The newly designed derivatives were prepared via four concise synthetic routes35, 36, 37. As shown in Scheme 1, methyl 2-mercaptoacetate (8) was reacted with methyl 4-chlorobutanoate to afford intermediate 9. Then the cyclization of 9 with NaH generated 10, which was condensed with S-methylthiouronium sulfate to produce 11. Next, 11 was hydrolyzed with HOAc to provide 12, followed by chlorination of the hydroxyl group with POCl3 to give the key intermediate 13. The nucleophilic addition of 13 with different phenols yielded the corresponding intermediates 14a−c. Intermediates 14a or 14b underwent the palladium catalyzed Buchwald−Hartwig reaction with BINAP and Pd2 (dba)3 to provide the final products 15a and 15b. The oxidative products 16a and 16b were prepared by treating 15a with m-CPBA at −78 °C or room temperature.
Scheme 1.
Reagents and conditions: (i) methyl 4-chlorobutanoate, MeONa, KI, MeOH, 70 °C; (ii) NaH, THF, 0 °C to r.t.; (iii) S-methylthiouronium sulfate, KOH, MeOH, r.t.; (iv) HOAc, H2O, 110 °C; (v) POCl3, N,N-dimethylaniline, 110 °C; (vi) K2CO3, DMF, r.t.; (vii) 4-aminobenzonitrile, BINAP, Pd2 (dba)3, Cs2CO3, 1,4-dioxane, 100 °C; (viii) m-CPBA, DCM, −78 °C or r.t.
As depicted in Scheme 2, treatment of 14a with 1-Boc-piperazine or 4-Boc-aminopiperidine in the presence of K2CO3 at 120 °C gave the intermediates 17a or 17b. Removal of the Boc group provided the intermediates 18a or 18b, which then resulted in the target compounds 19a and 19b after nucleophilic substitution with 4-(bromomethyl)benzenesulfonamide. In addition, the intermediates 14a−c were converted into the target compounds 20a−d by reacting with 1-(2-aminoethyl)pyrimidine-2,4(1H,3H)-dione or 2-morpholinoethan-1-amine.
Scheme 2.
Reagents and conditions: (i) K2CO3, DMF, 100 °C; (ii) TFA, DCM, r.t.; (iii) 4-(bromomethyl)benzenesulfonamide, K2CO3, DMF, r.t.; (iv) DIEA, NMP, 120 °C.
As illustrated in Scheme 3, 14a−c were reacted with 4-amino-1-Boc-piperidine in the presence of K2CO3 at 100 °C to provide the intermediates 21a−c, which afforded the key analogues 22a−c after removal of the Boc protecting group. Subsequently, the target compounds 23a−i were obtained from the nucleophilic reaction of 22a−c and various benzyl chlorides/bromides.
Scheme 3.
Reagents and conditions: (i) K2CO3, DMF, 100 °C; (ii) TFA, DCM, r.t.; (iii) K2CO3, DMF, r.t.
As shown in Scheme 4, treatment of 3-fluorobenzonitrile (24) with tert-butyl piperidin-4-ylcarbamate furnished the cyano intermediate 25a and hydrolysis of 25a under NaOH and H2O2 generated the amide intermediate 25b, followed by removing the Boc group to give intermediates 26a and 26b. Finally, 14a was coupled with the above intermediates 26a or 26b via the Buchwald−Hartwig reaction to prepare the target compounds 27a and 27b.
Scheme 4.
Reagents and conditions: (i) tert-butyl piperidin-4-ylcarbamate, K2CO3, DMF, 120 °C; (ii) TFA, DCM, r.t.; (iii) NaOH, H2O2, EtOH, 50 °C; (iv) 3-(4-aminopiperidin-1-yl)benzonitrile or 3-(4-aminopiperidin-1-yl)benzamide, BINAP, Pd2 (dba)3, Cs2CO3, 1,4-dioxane, N2, 100 °C.
2.2. Anti-HIV activity evaluation
All the target compounds were first screened for their anti-HIV activities against the WT (IIIB) and double-mutant K103N + Y181C (RES056) HIV-1 strains. Partially representative compounds with prominent activities were further evaluated for their inhibitory potency against a panel of clinically relevant HIV-1 variants carrying single-mutations L100I, K103N, E138K, Y181C, Y188L, or double-mutations F227L + V106A. Biological evaluation results are presented as EC50 (50% effective concentration of test compound) and CC50 (50% cytotoxic concentration of test compound) values.
As illustrated in Table 1, Table 2, Table 3, most compounds displayed excellent potency against IIIB strain in the single-digit to double-digit nanomolar ranges (EC50 = 1.44−20.0 nmol/L). Among them, compound 16b was the most active IIIB inhibitor with an EC50 of 1.44 nmol/L, being slightly superior to ETR (EC50 = 2.81 nmol/L) and EFV (EC50 = 2.67 nmol/L), and comparable to RPV (EC50 = 1.00 nmol/L). As for the most challenging double-mutant variant RES056, compounds with high potency toward the IIIB strain were also found to be effective at inhibiting the RES056 strain (EC50 = 15.2−445 nmol/L). Five compounds (15a, 15b, 16b, 23h, and 27b) yielded greater potency than EFV (EC50 = 114 nmol/L), with EC50 values of 15.2–85.3 nmol/L. In particular, compound 23h (EC50 = 15.2 nmol/L) was identified as the most powerful inhibitor against RES056 strain, being up to 2.3- and 7.5-fold greater than ETR (EC50 = 35.2 nmol/L) and EFV (EC50 = 114 nmol/L), but slightly weaker than RPV (EC50 = 10.7 nmol/L). Besides, most of the novel compounds had decreased cytotoxicity (CC50 = 3.07−284 μmol/L) compared to ETR (CC50 = 2.20 μmol/L) and RPV (CC50 = 3.98 μmol/L), contributing to the higher SI values and reflecting their sufficient safety profiles at the in vitro cellular level.
Table 1.
Anti-HIV-1 (IIIB and RES056) activity and cytotoxicity of 15a−b and 16a−b in MT-4 cells.
| Compd. | R1 | X | EC50 (nmol/L)a |
CC50 (μmol/L)b | |
|---|---|---|---|---|---|
| IIIB | RES056 | ||||
| 15a | CN | S | 3.38 ± 3.17 | 85.3 ± 54.3 | 4.53 ± 0.72 |
| 15b | CV | S | 3.18 ± 1.21 | 23.5 ± 14.7 | 4.89 ± 0.62 |
| 16a | CN | SO | 7.99 ± 0.95 | 445 ± 95.7 | ≥19.7 |
| 16b | CN | SO2 | 1.44 ± 0.17 | 49.1 ± 14.0 | 18.7 ± 8.01 |
| NVP | – | – | 118 ± 66.9 | 2221 ± 736 | >15.0 |
| EFV | – | – | 2.67 ± 1.73 | 114 ± 55.6 | >6.34 |
| ETR | – | – | 2.81 ± 0.47 | 35.2 ± 19.7 | 2.20 ± 0.015 |
| RPV | – | – | 1.00 ± 0.27 | 10.7 ± 7.96 | 3.98 |

EC50: concentration of compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytopathicity, as determined by the MTT method.
CC50: concentration required to reduce the viability of mock-infected cell cultures by 50%, as determined by the MTT method.
Table 2.
Anti-HIV-1 (IIIB and RES056) activity and cytotoxicity of 19a−b and 20a−d in MT-4 cells.
| Compd. | R1 | Y | EC50 (nmol/L)a |
CC50 (μmol/L)b | |
|---|---|---|---|---|---|
| IIIB | RES056 | ||||
| 19a | CN | N | >3081 | NDc | 19.3 ± 3.04 |
| 19b | CN | CHNH | >16,086 | ND | 3.07 ± 0.30 |
| 20a | CN | – | 233 ± 158 | >248,390 | >284 |
| 20b | CV | – | 20.0 ± 19.8 | 22,659 ± 4857 | 142 ± 31.1 |
| 20c | CH3 | – | 523 ± 466 | >152,494 | 152 ± 28.3 |
| 20d | CN | – | 5690 ± 4760 | ND | 20.5 ± 11.3 |
| NVP | – | – | 118 ± 66.9 | 2221 ± 736 | >15.0 |
| EFV | – | – | 2.67 ± 1.73 | 114 ± 55.6 | >6.34 |
| ETR | – | – | 2.81 ± 0.47 | 35.2 ± 19.7 | 2.20 ± 0.015 |
| RPV | – | – | 1.00 ± 0.27 | 10.7 ± 7.96 | 3.98 |
EC50: concentration of compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytopathicity, as determined by the MTT method.
CC50: concentration required to reduce the viability of mock-infected cell cultures by 50%, as determined by the MTT method.
ND: not determined.
Table 3.
Anti-HIV-1 (IIIB and RES056) activity and cytotoxicity of 23a−i and 27a−b in MT-4 cells.
| Compd. | R1 | R2 | EC50 (nmol/L)a |
CC50 (μmol/L)b | |
|---|---|---|---|---|---|
| IIIB | RES056 | ||||
| 23a | CN | 4-CN | 4.44 ± 1.39 | 263 ± 97.1 | 3.76 ± 0.32 |
| 23b | CN | 3-CN | 6.00 ± 1.03 | 764 ± 86.1 | 3.78 ± 0.63 |
| 23c | CN | 2-CN | 7.10 ± 2.69 | 1158 ± 184 | 3.86 ± 1.14 |
| 23d | CN | 3-SO2NH2 | 4.65 ± 0.65 | 790 ± 134 | 5.61 ± 3.42 |
| 23e | CN | 4-CO2C2H5 | 14.8 ± 13.5 | 4834 ± 517 | 65.3 ± 39.8 |
| 23f | CN | 4-NH2 | 7.00 ± 1.04 | 653 ± 236 | 12.0 ± 6.47 |
| 23g | CN | 4-NHSO2CH3 | 2.37 ± 1.98 | 326 ± 185 | ≥3.87 |
| 23h | CV | 4-SO2NH2 | 3.43 ± 2.00 | 15.2 ± 9.44 | 3.31 ± 1.70 |
| 23i | CH3 | 4-SO2NH2 | 5.43 ± 4.40 | 442 ± 272 | ≥226 |
| 27a | CN | 3-CN | 10.2 ± 7.57 | 836 ± 290 | 8.39 ± 5.22 |
| 27b | CN | 3-CONH2 | 3.76 ± 2.29 | 24.5 ± 8.11 | 5.99 ± 2.95 |
| NVP | – | – | 118 ± 66.9 | 2221 ± 736 | >15.0 |
| EFV | – | – | 2.67 ± 1.73 | 114 ± 55.6 | >6.34 |
| ETR | – | – | 2.81 ± 0.47 | 35.2 ± 19.7 | 2.20 ± 0.015 |
| RPV | – | – | 1.00 ± 0.27 | 10.7 ± 7.96 | 3.98 |
EC50: concentration of compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytopathicity, as determined by the MTT method.
CC50: concentration required to reduce the viability of mock-infected cell cultures by 50%, as determined by the MTT method.
Based on the antiviral evaluation (IIIB and RES056 strains) results, preliminary SAR of these novel derivatives could be delineated: Firstly, with ETR as the lead compound, a non-planar dihydrothiopyrano moiety was introduced into the central ring, with the aim of fully occupying the tolerant region II to improve antiviral activities and physicochemical properties. As depicted in Table 1, compounds 15a−b and 16a−b were able to potently inhibit the HIV-1 IIIB (EC50 = 1.44−7.99 nmol/L) and RES056 strains (EC50 = 23.5−445 nmol/L), while exhibiting reduced cytotoxicity (CC50 ≥ 4.53 μmol/L) compared to ETR and RPV. Analysis of SAR revealed that the nature of substituents at the R1 or X position had a significant influence on the antiviral activity. Pairwise comparison of compounds 15a and 15b indicated that the cyanovinyl group at the R1 position favored the potency over the cyano group, due to enhanced π−stacking interaction with the hydrophobic channel. Sulfonation at the X position resulted in a more potent inhibitor 16b, but the introduction of the sulfoxide group afforded 16a with sharply reduced activity. Thus, the established SAR confirmed that the newly introduced dihydrothiopyrano moiety fits well in the tolerant region II to improve binding affinity.
In continuation of our efforts, we set out to introduce distinct moieties derived from other classes of NNRTIs at the right-wing and linker to replace the p-cyanoaniline fragment, affording derivatives 19a−b and 20a−d based on the molecular hybridization strategy. However, as shown in Table 2, all compounds showed very weak or even loss of activity toward IIIB and RES056 strains. Directly attaching the central scaffold to piperazine and piperidine via a carbon-nitrogen bond to remove the NH linker resulted in the discovery of 19a and 19b with lost potency. Thus, the critical double hydrogen bonds involved in the right imino linker are necessary to maintain the binding force, which also suggests that the inherent conformational flexibility ensures favorable resistance profiles of DAPY NNRTIs. Moreover, to further investigate the tolerant region I, 20a−d were designed by molecular hybridization of DAPYs with catechol diethers and morpholine derivatives. Regrettably, replacement of the p-cyanoaniline moiety of 15a with ethylamino-containing uracil and morpholine yielded 20a and 20d with sharply decreased activities against WT and RES056 strains, probably because the uracil and morpholine were too flexible to fully occupy the tolerant region I. The potency order of 20a−c was as follows: 20b (−CV) > 20a (−CN) > 20c (−CH3), indicating that cyanovinyl and cyano are more favorable than methyl for binding affinity.
In an effort to fully explore the SAR of these dihydrothiopyrano[3,2-d]pyrimidines, we attempted to replace the p-cyanoaniline group with privileged piperidine-linked benzyl or phenyl moieties to improve resistance profiles, considering that the piperidine ring can adapt to the changed binding pockets through flexible conformation. Meanwhile, structurally diverse substituents were introduced to the terminal benzene ring, to reach into the tolerant region I and form additional interactions. As shown in Table 3, most of compounds 23a−i and 27a−b exhibited single-digit nanomolar inhibitory activity against the IIIB strain and maintained moderate potency against the RES056 strain. Further investigation of SAR revealed that the R2 substituents on the phenyl group significantly affected the anti-HIV-1 potency. The comparison between compounds 23a−c indicated that the antiviral activity was dependent on the R2 substitution position of the benzene moiety and that the para-substitution was preferred over the meta- and ortho-substitution. Besides, the compounds with para-substituted phenyl were in the following order of potency: 23a (4-CN) ≈ 23g (4-NHSO2CH3) > 23f (4-NH2) > 23e (4-CO2C2H5), demonstrating that suitably sized substituents containing hydrogen-bond donors and acceptors are more beneficial for inhibitory activity. In agreement with the previously established SAR, the replacement of the methyl at the R1 position with a cyanovinyl led to remarkably improved antiviral activities especially against the RES056 strain, as exemplified by compounds 23i (CH3, EC50(RES056) = 442 nmol/L) and 23h (CV, EC50(RES056) = 15.2 nmol/L). In addition, the contribution of directly linked substituted phenyl on the piperidine moiety to the inhibitory potency was also investigated. Specifically, removal of the methylene linker of compound 23b yielded a slightly less potent compound 27a; while replacing the cyano group of 27a with an amide group significantly elevated the activities (27b, EC50(IIIB) = 3.76 nmol/L, EC50(RES056) = 24.5 nmol/L), but still inferior to those of compound 23h (EC50(IIIB) = 3.43 nmol/L, EC50(RES056) = 15.2 nmol/L).
Based on the preliminary antiviral activities, several potent compounds were further evaluated against various NNRTI-resistant strains. As illustrated in Table 4, all compounds demonstrated excellent potency against HIV-1 variants at nanomolar levels, with EC50 values ranging from 2.30 to 184 nmol/L.
Table 4.
Anti-HIV-1 activity against resistant strains of the representative target compounds.
| Compd. | EC50 (nmol/L)a |
|||||
|---|---|---|---|---|---|---|
| L100I | K103N | Y181C | Y188L | E138K | F227L + V106A | |
| 15a | 16.7 ± 4.51 | 6.41 ± 0.99 | 42.3 ± 4.45 | 20.7 ± 1.88 | 17.4 ± 2.24 | 40.9 ± 0.00 |
| 15b | 7.26 ± 0.51 | 6.42 ± 0.40 | 11.1 ± 3.39 | 23.3 ± 0.48 | 21.7 ± 7.55 | 27.9 ± 9.17 |
| 16b | 21.8 ± 5.65 | 2.30 ± 0.15 | 10.4 ± 1.44 | 12.3 ± 2.27 | 10.9 ± 0.32 | 37.9 ± 21.3 |
| 23f | 55.6 ± 2.68 | 8.69 ± 0.00 | 14.7 ± 1.17 | 26.1 ± 3.53 | 21.6 ± 0.56 | 184 ± 8.33 |
| 23g | 28.0 ± 5.38 | 4.59 ± 0.45 | ≤8.29 | 12.7 ± 2.99 | 8.66 ± 0.54 | 165 ± 40.8 |
| 23h | 4.46 ± 0.36 | 6.13 ± 0.12 | 8.46 ± 1.17 | 16.5 ± 5.18 | 21.4 ± 4.19 | 5.35 ± 0.69 |
| 23i | 39.2 ± 7.66 | 6.56 ± 1.07 | 35.7 ± 5.75 | 43.0 ± 0.26 | 37.6 ± 7.41 | 64.7 ± 13.3 |
| 27b | 13.3 ± 6.79 | 5.46 ± 0.00 | 7.87 ± 1.84 | 7.14 ± 1.50 | 17.1 ± 4.42 | 111 ± 11.4 |
| NVP | 705 ± 451 | 3551 ± 2413 | 3785 ± 2577 | 4416 ± 2469 | 158 ± 112 | 3538 ± 2750 |
| EFV | 57.4 ± 45.9 | 99.2 ± 56.8 | 4.94 ± 1.75 | 93.2 ± 58.9 | 4.67 ± 1.38 | 77.4 ± 55.9 |
| ETR | 9.13 ± 6.10 | 3.38 ± 0.69 | 14.1 ± 5.72 | 15.4 ± 6.07 | 10.7 ± 6.66 | 16.3 ± 6.24 |
| RPV | 1.54 ± 0.00 | 1.31 ± 0.36 | 4.73 ± 0.48 | 79.4 ± 0.77 | 5.75 ± 0.11 | 81.6 ± 21.2 |
EC50: concentration of compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytopathicity, as determined by the MTT method.
In terms of the L100I strain38,39, all selected compounds were more potent than EFV (EC50 = 57.4 nmol/L) with EC50 values of 4.46–55.6 nmol/L, among which 23h (EC50 = 4.46 nmol/L) and 15b (EC50 = 7.26 nmol/L) showed 2.0 times and 1.3 times greater activity than ETR (EC50 = 9.13 nmol/L), although inferior to RPV (EC50 = 1.54 nmol/L). With respect to the K103N strain40,41, all compounds were endowed with excellent potency in the single-digit nanomolar range (EC50 = 2.30−8.69 nmol/L), being more active than EFV (EC50 = 99.2 nmol/L) and comparable to ETR (EC50 = 3.38 nmol/L) and RPV (EC50 = 1.31 nmol/L). In the case of the Y181C strain42,43, all compounds except 15a and 23i were more effective than ETR, while 23h (EC50 = 8.46 nmol/L) demonstrated a 1.7-fold improvement in potency compared to ETR (EC50 = 14.1 nmol/L). As regards the Y188L strain44, all of them inhibited Y188L strain more potently than RPV (EC50 = 79.4 nmol/L) and EFV (EC50 = 93.2 nmol/L), with EC50 values below 43.0 nmol/L, and 23h (EC50 = 16.5 nmol/L) yielded equipotent activity to ETR (EC50 = 15.4 nmol/L). For the E138K strain selected in a high proportion of patients receiving RPV45,46, 23g (EC50 = 8.66 nmol/L) displayed the best potency, which was slightly superior to ETR (EC50 = 10.7 nmol/L) but less potent than RPV (EC50 = 5.75 nmol/L). In terms of the highly resistant double-mutant strain F227L + V106A47,48, most compounds remained very potent inhibition of replication of this strain. In particular, 23h provided optimal efficacy of 5.35 nmol/L, which was up to 15.3- and 14.5-fold better than RPV (EC50 = 81.6 nmol/L) and EFV (EC50 = 77.4 nmol/L), respectively.
Therefore, 23h was identified as the most potent inhibitor against mutant strains. Fig. 5 clearly presented a visual comparison of the anti-HIV-1 potency of 23h versus the reference drugs ETR and EFV, indicating that 23h had higher inhibitory effects on all strains than the reference drugs, especially for the highly resistant double-mutant strains K103N + Y181C and F227L + V106A.
Figure 5.
Comparison of the in vitro antiviral activity of 23h, ETR, and EFV.
Furthermore, the SI and RF values of the selected compounds against the mutant strain are listed in Table 5. To be specific, most compounds had relatively higher SI values of 155–34,451 than ETR (SI = 63−651) and RPV (SI = 49−3045) toward the mutant strains due to their decreased cytotoxicity. Intriguingly, all compounds showed no obvious decline in activity against the K103N strain compared to that against the WT strain, as reflected by an RF value of less than 2.0 (RF = 1.2−2.0). In addition, the RF values of 23h (RF = 1.6 and 4.4) were lower than those of ETR (RF = 5.8 and 12.5), EFV (RF = 29.0 and 42.7), and RPV (RF = 82 and 10.7) for the challenging variants F227L + V106A and K103N + Y181C. More importantly, 23h had a major improvement in drug resistance profiles (RF = 1.3−6.2) compared to ETR (RF = 1.2−12.5), EFV (RF = 1.7−42.7), and RPV (RF = 1.3−82), suggesting that 23h featuring the flexible scaffold is more resilient to clinically relevant NNRTI-resistant mutations.
Table 5.
Selectivity index and resistance fold for HIV-1 variants of the representative target compounds.
| Compd. | SIa (RFb) |
||||||
|---|---|---|---|---|---|---|---|
| L100I | K103N | Y181C | Y188L | E138K | F227L + V106A | RES056 | |
| 15a | 271 (4.9) | 707 (1.9) | 107 (12.5) | 219 (6.1) | 260 (5.1) | 111 (12.1) | 53 (25.2) |
| 15b | 674 (2.3) | 762 (2.0) | 441 (3.5) | 210 (7.3) | 225 (6.8) | 175 (8.8) | 208 (7.4) |
| 16b | 858 (15.1) | 8130 (1.6) | 1798 (7.2) | 1520 (8.5) | 1716 (7.6) | 493 (26.3) | 381 (34.1) |
| 23f | 216 (7.9) | 1381 (1.2) | 816 (2.1) | 460 (3.7) | 556 (3.0) | 65 (26.3) | 18 (93.3) |
| 23g | ≥138 (11.8) | ≥843 (1.9) | ≥467 (≤3.5) | ≥305 (5.4) | ≥447 (3.7) | ≥23 (69.6) | ≥12 (138) |
| 23h | 742 (1.3) | 540 (1.8) | 391 (2.5) | 201 (4.8) | 155 (6.2) | 619 (1.6) | 218 (4.4) |
| 23i | ≥5765 (7.2) | ≥34,451 (1.2) | ≥6331 (6.6) | ≥5256 (7.9) | ≥6011 (6.9) | ≥3493 (11.9) | ≥511 (81.4) |
| 27b | 450 (3.5) | 1097 (1.5) | 761 (2.1) | 839 (1.9) | 350 (4.5) | 54 (29.5) | 244 (6.5) |
| NVP | >21 (6.0) | >4 (30.0) | >4 (32.0) | >3 (37.4) | >95 (1.3) | >4 (30.0) | >7 (18.8) |
| EFV | >110 (21.5) | >64 (37.2) | >1283 (1.9) | >68 (34.9) | >1358 (1.7) | >82 (29.0) | >56 (42.7) |
| ETR | 241 (3.2) | 651 (1.2) | 156 (5.0) | 143 (5.5) | 206 (3.8) | 135 (5.8) | 63 (12.5) |
| RPV | 2575 (1.5) | 3045 (1.3) | 841 (4.7) | 50 (80) | 692 (5.8) | 49 (82) | 371 (10.7) |
SI: selectivity index, the ratio of CC50/EC50.
RF: resistance fold, the ratio of EC50 (mutant strain)/EC50(WT strain).
On the whole, the above biological evaluation results and comprehensive SAR investigations reasonably verified our original design hypotheses: i) the non-planar dihydrothiopyran ring introduced into the central scaffold could accommodated well in the tolerant region II to develop extensive interactions; ii) the cyanovinyl group is the optimal substituent on the left-wing because of the formation of additional π−π stacking interactions; iii) the benzyl-linked aminopiperidine moiety on the right wing proved to be more favorable for improving activity, due to the flexible conformation enables relatively easy adaptation to the residue mutations; and these systematic optimization campaigns led to the identification of the most promising inhibitor 23h with robust antiviral potency and improved resistance profiles against NNRTI-resistant strains.
2.3. HIV-1 RT inhibition assay
To further validate the target of the novel dihydrothiopyrano[3,2-d]pyrimidine derivatives, the representative compounds 15b and 16b featuring p-cyanoaniline and 23h featuring piperidine-linked benzyl sulfonamide were tested for their potency in inhibiting the RT enzyme. As shown in Table 6, all tested compounds demonstrated potent inhibition of RT with IC50 values ranging from 0.027 to 0.529 μmol/L, which were superior or comparable to that of ETR (IC50 = 0.232 μmol/L) and RPV (IC50 = 0.118 μmol/L). In general, the enzymatic assay results were able to suggest that these novel dihydrothiopyrano[3,2-d]pyrimidines showed high binding affinity to RT and belonged to typical HIV-1 NNRTIs.
Table 6.
HIV-1 RT inhibitory activity of 15b, 16b, 23h, NVP, EFV, ETR, and RPV.
| Compd. | IC50 (μmol/L)a | Compd. | IC50 (μmol/L)a |
|---|---|---|---|
| 15b | 0.101 ± 0.012 | EFV | 0.014 ± 0.002 |
| 16b | 0.027 ± 0.011 | ETR | 0.232 ± 0.036 |
| 23h | 0.529 ± 0.142 | RPV | 0.118 ± 0.027 |
| NVP | 0.089 ± 0.015 |
IC50: inhibitory concentration of test compounds required to inhibit biotin deoxyuridine triphosphate (biotin-dUTP) incorporation into HIV-1 RT by 50%.
2.4. Molecular docking analysis
To achieve insight into the theoretical binding modes of novel compounds within RT and elucidate the unique resistance profiles of 23h, molecular docking analysis of 23h and ETR were performed by SYBYL-X 2.0. Four co-crystal structures of WT RT (PDB ID: 6C0N)49, L100I RT (PDB ID: 1S1V)50, F227L + V106A RT (PDB ID: 6DUF)49, and K103N + Y181C RT (PDB ID: 6C0R)49 were chosen as the templates.
As expected, 23h binds to WT, L100I, F227L + V106A, and K103N + Y181C RT in a similar manner. As displayed in Fig. 6, compound 23h adopts a typical U-shaped conformation that resembles other DAPY-typed inhibitors. Several common binding features and protein−ligand interactions were delineated as follows. Firstly, the left-wing of 4-cyanovinyl-2,6-dimethylphenyl moiety as a conjugated system effectively occupies the hydrophobic channel, establishing foremost π−stacking interactions with Tyr181 and Tyr188. Secondly, the central dihydrothiopyrano ring is oriented towards the solvent-exposed tolerant region II, which is stabilized by non-polar interaction with Glu138 and electrostatic interaction with Leu100 and Val179. Thirdly, the pyrimidinyl and the NH linker form the “signature” dual hydrogen-bonds with the amino hydrogen (N⋯H2–O⋯H–N) and carbonyl oxygen (N–H⋯O=C) of the main chain of Lys101, respectively, which are necessary to maintain binding affinity. Fourthly, the benzyl sulfonamide motif stretches into the solvent-exposed tolerant region I, being involved in double hydrogen-bonding interactions with Lys104 (N–H2⋯O=C) and Val106 (S=O2⋯H–N).
Figure 6.
Binding modes and interaction forces of 23h (pink) with WT RT (PDB code: 6C0N).
On the other hand, compound 23h and the lead compound ETR were docked into the mutant RT simultaneously to explain the improved anti-resistance profiles of 23h. Examination of the interaction forces between 23h and L100I RT (Fig. 7a) indicates that the pivotal hydrogen bonds are still retained, despite slight variations in the bond distance and formation condition of the hydrogen bonds. However, one of the key hydrogen bonds between ETR and Lys101 is lost because the disappearance of electrostatic interaction caused by the L100I mutation shifted the pyrimidine ring of ETR away from the main chain of Lys101. As shown in Fig. 7b, the smaller Leu227 and Ala106 further broaden the chemical space of the pocket, allowing 23h to occupy the binding sites through the conformational reorientation of the piperidine ring and the dihydrothiopyrano ring. Furthermore, it is observed that the “water bridge” hydrogen bonds between the piperidine ring and Lys103 (N⋯H2–O⋯H–N) and Pro236 (N⋯H–O–H⋯O=C) and the hydrogen bond between sulfonamide and new Ala106 (S=O2⋯H–N) compensated for the lack of hydrophobic interaction with the highly conserved residue Phe227. By contrast, ETR can only develop one hydrogen bond with Lys101 (N–H2⋯O–H2⋯O=C) through the amino group on the central ring in the presence of a bridging water molecule, whereas the double hydrogen bonds of the pyrimidine nitrogen and the NH linker with Lys101 disappears. With regard to the binding modes in K103N + Y181C RT (Fig. 7c), the nitrogen atom on the pyrimidine ring of ETR generates an intramolecular hydrogen bond (N⋯H2–O⋯H2–N) with its amino group instead of Lys101 via a water molecule, while it has no substantial contribution to the binding affinity. Of particular note are the water bridge hydrogen bonds formed with mutant Asn103 (N⋯H2–O⋯H–N) and Pro236 (N⋯H–O–H⋯O=C) and the dual hydrogen bonds formed with Lys101 (N⋯H2–O⋯H–N; N–H⋯O=C) of 23h continue to be maintained to offset the damage of the K103N mutation. Besides, the left-wing terminal cyano group of 23h develops an additional hydrogen bond with the amino group of Lys223 (C ≡N⋯H2–N), compensating for the affinity loss of the weakened π−π stacking interaction caused by the Y181C substitution.
Figure 7.
(a) Superimposition of 23h (pink) and ETR (blue) with L100I RT (PDB code: 1S1V). (b) Superimposition of 23h (pink) and ETR (blue) with F227L + V106A RT (PDB code: 6DUF). (c) Superimposition of 23h (pink) and ETR (blue) with K103N + Y181C RT (PDB code: 6C0R).
All in all, the molecular docking results clearly revealed the binding modes and interaction forces of 23h and rationalized its potent inhibitory activity against mutant strains compared to ETR, which provides valuable information for further drug design.
2.5. Molecular dynamics simulation
To study the stability state and dynamic behaviors of compound 23h binding to distinct RT, comprehensive molecular dynamics (MD) simulations of 23h in complex with WT RT and various mutant RTs were performed for 200 ns using the software Schrödinger. All the results were visualized by the softwares LigPlot + v.2.2.8 and Schrödinger and the methods were described in the experimental section.
The MD simulation trajectories were clustered using the trajectory clustering program and the most abundant clusters of RT/23h complexes were extracted shown in Fig. 8, still maintaining the horseshoe conformation and interaction profile similar to those in Figure 6, Figure 7. The Root Mean Square Deviation (RMSD) values are commonly used to assess the conformational drift of protein-ligand complexes by measuring the average shift change of atoms in a specific frame relative to the reference frame. As depicted in Fig. 9, the RMSD plots indicated that all the HIV-1 RT-ligand complexes remain stable during the 200 ns simulation process, indicating that the overall structure of the RT proteins did not change significantly due to the binding of the 23h ligand. In addition, the coordinates of 23h fluctuate less than 2.5 Å in each RT/23h complex after 200 ns of MD simulations and consistently bind stably to the HIV-1 RT allosteric site. The Root Mean Square Fluctuation (RMSF) of RT and 23h were further investigated to characterize the local deviation of protein chain and ligand atom positions, respectively. The peaks on the protein RMSF plot indicate the amino acid residues that fluctuate the most during the simulation (Supporting Information Fig. S1), while the ligand RMSF plot shows the ligand fluctuations for each atom of the 23h backbone, corresponding to the two-dimensional structure in the top panel (Supporting Information Fig. S2). In addition, Ramachandran plots showed that all RT residues were located in the most favoured (more than 90%), additional allowed, and generously allowed regions (Fig. 10).
Figure 8.
Binding modes in the most abundant clusters of 23h in complex with WT RT (a, PDB code: 6C0N), L100I RT (b, PDB code: 1S1V), F227L + V106A RT (c, PDB code: 6DUF), and K103N + Y181C RT (d, PDB code: 6C0R).
Figure 9.
RMSD−time profiles of protein-ligand complexes with reference to the first frame for the 23h/WT RT complex (a), 23h/L100I RT complex (b), 23h/F227L + V106A RT complex (c), and 23h/K103N + Y181C RT complex (d) during the 200 ns MD simulations.
Figure 10.
Ramachandran plots of the 23h/WT RT complex (a), 23h/L100I RT complex (b), 23h/F227L + V106A RT complex (c), and 23h/K103N + Y181C RT complex (d) after 200 ns MD simulations.
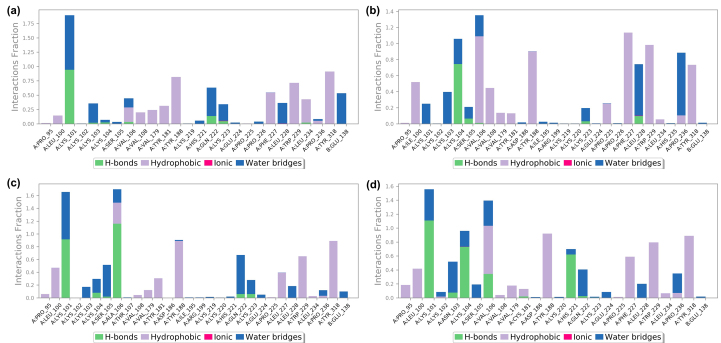
Protein-ligand contacts of 23h with different RTs are illustrated in Figure 11, Figure 12, and Supporting Information Fig. S3, which demonstrate the detailed interactions between the ligand and specific residues, mainly including hydrogen bond, hydrophobic contact, and water bridge. Notably, the interactions fraction plot implies that the specific interaction remains constant over what percent of the 200 ns simulations, while multiple contacts between individual residues and the ligand may yield interaction fraction above 1.0 (Fig. 11). Besides, Fig. 12 visually represents the number of contact forces between 23h and RT in each trajectory frame, as well as the specific residue codes that interact with the ligand, with darker orange reflecting stronger interactions. Consistent with the results in Figure 11, Figure 12, the two-dimensional ligand interaction diagrams demonstrated that 23h establishes signature double hydrogen bonds with the Lys101 backbone of both WT RT and mutant RT and forms extensive hydrophobic contacts with Tyr181, Tyr188, and Trp229, contributing to the critical role of maintaining the protein-ligand binding affinity (Fig. S3).
Figure 11.
Interactions fraction plots of the ligand with specific amino acid residues of HIV-1 WT RT (a), L100I RT (b), F227L + V106A RT (c), and K103N + Y181C RT (d) in MD simulations.
Figure 12.
Number of protein-ligand contacts in each trajectory frame for 23h with HIV-1 WT RT (a), L100I RT (b), F227L + V106A RT (c), and K103N + Y181C RT (d).
Overall, the MD simulation results indicated that the complexes of ligand 23h with both WT RT and mutant RT maintained stable conformations and retained extensive protein-ligand interactions, and Ramachandran plots validated the structural stability of all RT/23h complex, contributing to the high binding affinity between 23h and various RTs.
2.6. CYP450 inhibitory activity assessment
Four main subtypes of cytochrome P450 (CYP) enzymes, CYP2C9, CYP2C19, CYP2D6, and CYP3A4, are widely involved in the metabolism of most drugs in vivo and play a crucial role in detoxification and bioactivation processes51. ETR and RPV have potent CYP inhibitory activity for CYP2C9 and CYP2C19, thus may cause adverse drug–drug interactions in combination with other antiviral agents52,53. Therefore, compound 23h was further evaluated for its CYP enzymatic inhibitory activity to predict the risk of combination administration, with selective enzyme inhibitors as positive controls. As displayed in Table 7, 23h showed no or weak CYP450 inhibitory potency against CYP1A2 (IC50 > 50 μmol/L), CYP2C9 (IC50 = 3.41 μmol/L), CYP2C19 (IC50 = 15.6 μmol/L), and CYP2D6 (IC50 = 36.8 μmol/L), which were much lower than those of ETR (IC50 = 0.277−12.0 μmol/L) and RPV (IC50 = 0.335−9.11 μmol/L). In particular, the inhibitory effects of 23h on CYP2C9 and CYP2C19 were significantly reduced compared to ETR and RPV. Overall, compound 23h showed no apparent inhibition of the major CYP isoforms, with improved CYP inhibition profiles over ETR and RPV.
Table 7.
CYP450 inhibitory activity of compound 23h, approved drugs ETR and RPV, and selective enzyme inhibitors.
| Compd. | IC50 (μmol/L) |
||||
|---|---|---|---|---|---|
| CYP1A2 | CYP2C9 | CYP2C19 | CYP2D6 | CYP3A4 | |
| 23h | >50 | 3.41 | 15.6 | 36.8 | 13.5 |
| ETR | 7.48 | 0.277 | 0.496 | 12.0 | 41.3 |
| RPV | 9.11 | 0.346 | 0.335 | 3.41 | 2.17 |
| α-Naphthoflavone | 0.385 | ||||
| Sulfaphenazole | 0.616 | ||||
| (+)-N-3-Benzylnirvanol | 0.148 | ||||
| Quinidine | 0.142 | ||||
| Ketoconazole | 0.0372 | ||||
2.7. hERG channel inhibition assay
The human ether-à-go-go-related gene (hERG) channel is responsible for the rapid repolarization of the cardiac action potential and plays a crucial role in maintaining normal cardiac function54. Blockade of the hERG channel by non-cardiovascular drugs could induce acquired long QT syndrome and increase the risk of Torsade de Pointes. In the Phase IIb clinical trial NCT00110305 of RPV, dose-dependent prolongation of the QT interval with a delayed onset was observed in healthy subjects receiving daily doses of 75 mg55. Therefore, we further determined the inhibitory activity of compound 23h on the hERG potassium channel to assess the potential cardiac safety risk by using the manual patch-clamp electrophysiology method. As shown in Fig. 13, compound 23h demonstrated an IC50 value of 0.079 μmol/L against the hERG channel, which was weaker than that of the reference drug cisapride (IC50 = 0.012 μmol/L).
Figure 13.
Inhibitory activity of compound 23h against the hERG channel.
2.8. In vivo pharmacokinetics studies
Oral administration remains the most ideal and convenient delivery method for cART12. As shown in Table 8, compound 23h exhibited improved water solubility (sol. = 161 μg/mL, pH = 2.0; sol. = 11.7 μg/mL, pH = 7.0; sol. = 3.04 μg/mL, pH = 7.4), which were higher than those of ETR and RPV at different pH conditions. Therefore, the in vivo pharmacokinetic profiles of the most promising compound 23h and ETR were investigated after iv and po administration (Fig. 14). After iv dose at 2 mg/kg, 23h was characterized by a moderate half-life of 2.09 h, a favorable volume of distribution (V = 49.7 L/kg), and a slow mean clearance rate (CL = 16.6 L/h/kg). After po dose at 20 mg/kg, 23h achieved the maximum plasma concentration (Cmax = 333 μg/L) at 1.50 h, and its area under curve (AUC0−∞) and mean residence time (MRT0−∞) was 1857 μg/L·h and 5.02 h, respectively. Especially, compound 23h (t1/2(iv) = 2.09 h; t1/2(po) = 3.00 h) exhibited a longer half-life than ETR (t1/2(iv) = 1.80 h; t1/2(po) = 1.82 h) after iv and po administration, respectively. Moreover, 23h displayed a favorable oral bioavailability (F) of 15.33%, which was higher than that of ETR (F = 7.17%) and was sufficient for an orally bioavailable candidate (Table 9).
Table 8.
Water solubility of compound 23h, ETR, and RPV.
| Compd. | 23h | ETR | RPV |
|---|---|---|---|
| pH = 7.4 (μg/mL) | 3.04 | <1 | <1 |
| pH = 7.0 (μg/mL) | 11.7 | <1 | 0.02 |
| pH = 2.0 (μg/mL) | 161 | 127 | 103 |
Figure 14.
Drug−time curve of 23h and ETR after iv and po administration.
Table 9.
Pharmacokinetic profiles of compound 23h and ETR.
| Parametera | Unit | 23h (iv)b | 23h (po)c | ETR (iv)b | ETR (po)c |
|---|---|---|---|---|---|
| t1/2 | h | 2.09 ± 0.39 | 3.00 ± 0.74 | 1.80 ± 0.24 | 1.82 ± 0.07 |
| Tmax | h | – | 1.50 ± 0.87 | – | 1.00 ± 0.00 |
| Cmax | μg/L | – | 333 ± 276 | – | 110 ± 34.1 |
| AUC0−t | μg/L·h | 1152 ± 69.7 | 1685 ± 1480 | 576 ± 45 | 395 ± 139 |
| AUC0−∞ | μg/L·h | 1211 ± 78.9 | 1857 ± 1678 | 587 ± 48 | 421 ± 153 |
| V | L/kg | 49.7 ± 9.00 | – | 8.85 ± 1.17 | – |
| CL | L/h/kg | 16.6 ± 1.07 | – | 3.42 ± 0.30 | – |
| MRT0−t | h | 1.56 ± 0.17 | 4.10 ± 0.12 | 0.93 ± 0.23 | 2.55 ± 0.04 |
| MRT0−∞ | h | 2.03 ± 0.41 | 5.02 ± 0.48 | 1.10 ± 0.34 | 3.03 ± 0.14 |
| F | % | 15.33 | 7.17 | ||
PK parameter (mean ± SD, n = 3).
Dosed intravenously at 2 mg/kg.
Dosed orally at 20 mg/kg.
2.9. In vivo safety studies
A suitable drug candidate requires not only excellent antiviral activity and PK profiles, but also favorable in vivo safety properties. Therefore, the single-dose acute toxicity of 23h was investigated to assess its in vivo safety properties. After intragastric administration of 23h at 2000 mg/kg, no death or signs of intoxication were observed in the drug administration mice. Besides, there were no significant body weight loss or abnormal behaviors such as fatigue, cramps, anorexia, and ruffled fur in the treatment group compared with the control group (0 mg/kg). Body weight of treatment and control groups gradually increased in the following 7 days, with no obvious difference in weight gain (Fig. 15). Overall, the in vivo safety studies indicated that 23h was well tolerated at doses up to 2000 mg/kg without acute toxicity.
Figure 15.
The single-dose acute toxicity of 23h after oral administration.
3. Conclusions
Based on the NNRTI “four-point pharmacophore” model, we have designed and synthesized novel series of dihydrothiopyrano[3,2-d]pyrimidines, to fully exploit the potential chemical space and discover potent HIV-1 inhibitors with higher drug–resistance profiles and favorable pharmacological properties. Detailed SAR investigations were derived from the biological evaluation results: (1) the saturated dihydrothiopyran ring introduced into the central scaffold could accommodated well in the tolerant region II to form extensive interactions; (2) the cyanovinyl group is the optimal substituent on the left-wing due to the prolonged conjugation system enhancing the π−stacking interactions; (3) the piperidine-linked benzyl moiety is more favorable for antiviral potency, suggesting that conformational flexibility allows the ligand to adapt for resistant RT mutations; (4) the dual hydrogen bonds formed by the pyrimidinyl nitrogen and the imino linker with Lys101 are essential to maintain binding affinity.
Among these newly developed derivatives, compound 23h was identified as an exceptionally potent inhibitor against WT and resistant HIV-1 strains, with EC50 values of 3.43 nmol/L (IIIB), 4.46 nmol/L (L100I), 6.13 nmol/L (K103N), 8.46 nmol/L (Y181C), 16.5 nmol/L (Y188L), 21.4 nmol/L (E138K), 5.35 nmol/L (F227L + V106A), and 15.2 nmol/L (K103N + Y181C). In the case of the highly resistant double-mutants K103N + Y181C and F227L + V106A, 23h still displayed prominent inhibitory potency of 15.2 and 5.35 nmol/L, being equivalent to RPV (EC50 = 10.7 and 81.6 nmol/L), and more effective than EFV (EC50 = 114 and 77.4 nmol/L) and ETR (EC50 = 35.2 and 16.3 nmol/L). More encouragingly, 23h afforded significant improvement in drug resistance profiles (RF = 1.3−6.2) against various mutant strains compared to EFV (RF = 1.7−42.7), ETR (RF = 1.2−12.5), and RPV (RF = 1.3−82). In the target validation experiment, 23h potently inhibited HIV-1 RT at a low micromolar concentration and thus behaved as a typical NNRTI. Comprehensive molecular docking analysis and MD simulations demonstrated that the molecular mechanism of action of 23h contributes to the elevated binding affinity, providing a reasonable explanation for the improved drug resistance profiles and guiding further structural optimization efforts. In addition, the CYP450 inhibition profiles of 23h were significantly improved over ETR and RPV, with IC50 values in the range of 3.41–50 μmol/L against the main CYP enzymes. Notably, 23h exhibited desirable in vivo pharmacokinetic properties, with a higher oral bioavailability of 15.33% and a longer half-life of 3.00 h compared to ETR (F = 7.17%; t1/2 = 1.82 h). Furthermore, the in vivo safety studies indicated that 23h showed good tolerability at the dose of 2000 mg/kg.
Taken together, the current study validated the design hypothesis of introducing alicyclic rings and hydrophilic groups into the solvent-exposed region to balance the bioactivity and physicochemical properties. Particularly, all in vitro and in vivo data highlight the potential of 23h as an anti-HIV-1 candidate due to its prominent antiviral activities and favorable druggability profiles.
4. Experimental
4.1. Chemistry
Melting point (mp) was determined on the micromelting point apparatus (RY-1G, Tianjin Skylight). 1H NMR and 13C NMR spectra were recorded on the nuclear magnetic resonance spectrometer (Bruker Avance 400 or 600 MHz NMR Spectrometer, Bruker) with DMSO-d6 or CDCl3 as the solvent and tetramethylsilane (TMS) as the internal standard. Mass spectra were obtained on the layer chromatography−mass spectrometry (LC−MS) instrument (TSQ Vantage LC−MS/MS, Thermo Fisher). Thin-layer chromatography (TLC) was conducted on the high-efficiency thin-layer chromatography silica gel plate (Silica Gel 60 GF254, Merck) to monitor the reaction. Silica gel column chromatography was performed on the column packed with 200–300 mesh silica gel. Rotary evaporators (EYELA N-1300D, Tokyo Rikakikai) were used to concentrate the excess solvent. Sample purity was measured on a Shimadzu SPD-20A/20AV HPLC system. All compounds are >95% pure by HPLC analysis.
4.1.1. Synthesis of methyl 4-((2-methoxy-2-oxoethyl)thio)butanoate (9)
To a solution of sodium methoxide (0.15 g, 3.0 mmol) in MeOH (10 mL) was added methyl 2-mercaptoacetate (8, 0.11 g, 1.0 mmol), and the mixture was stirred at room temperature for 30 min. Potassium iodide (1.66 mg, 0.01 mmol) and methyl 4-chlorobutanoate (0.14 g, 1.0 mmol) were added and the solution was stirred at 70 °C for 10 h. After cooling to room temperature, the reaction mixture was filtered and the resultant filtrate was concentrated under vacuum to remove the solvent. Then 10 mL of water was added and the mixture was extracted with EtOAc (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, and concentrated in vacuum to give intermediate 9 as a yellow oil. Yield: 83%. 1H NMR (600 MHz, CDCl3): δ 3.75 (s, 3H, SCH2COOCH3), 3.66 (s, 3H, SCH2CH2CH2COOCH3), 3.21 (s, 2H, SCH2COOCH3), 2.77 (t, J = 7.2 Hz, 2H, SCH2CH2CH2COOCH3), 2.66 (t, J = 7.2 Hz, 2H, SCH2CH2CH2COOCH3), 1.93 (p, J = 7.2 Hz, 2H, SCH2CH2CH2COOCH3). ESI-MS: m/z 207.3 [M + H]+, 224.4 [M + NH4]+, C8H14O4S (206.06).
4.1.2. Synthesis of methyl 3-oxotetrahydro-2H-thiopyran-2-carboxylate (10)
To a solution of sodium hydride (60% in mineral oil, 0.06 g, 1.5 mmol) in THF (10 mL) was slowly added the intermediate 9 (0.21 g, 1.0 mmol) in 2 mL of THF at −10 °C under a nitrogen atmosphere, and the mixture was stirred at room temperature for 2 h. Then the reaction solution was acidified to pH 7 with diluted hydrochloric acid and extracted with EtOAc (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, and concentrated in vacuum to give intermediate 10 as a yellow oil and was used in next step without purification. Yield: 76%. ESI-MS: m/z 175.3 [M + H]+, 192.5 [M + NH4]+, C7H10O3S (174.04).
4.1.3. Synthesis of 2-(methylthio)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-ol (11)
To a solution of potassium hydroxide (0.11 g, 2.0 mmol) in MeOH (10 mL) was added the intermediate 10 (0.17 g, 1.0 mmol) and S-methylthiouronium sulfate (0.29 g, 1.5 mmol), and the mixture was stirred at room temperature for 8 h. Then water (50 mL) and HOAc (1.0 mL) were added to the reaction mixture. The resulting precipitate was collected by filtration, washed with water (3 × 5 mL), and dried to provide intermediate 11 as a white solid. Yield: 80%, mp: 234–236 °C. 1H NMR (400 MHz, DMSO-d6): δ 12.70 (s, 1H, OH), 2.94−2.84 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.61 (t, J = 6.3 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.45 (s, 3H, CH3), 2.08−1.95 (m, 2H, C7-dihydrothiopyranopyrimidine-H). ESI-MS: m/z 215.4 [M + H]+, 237.3 [M + Na]+, C8H10N2OS2 (214.02).
4.1.4. Synthesis of 7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidine-2,4-diol (12)
Intermediate 11 (0.21 g, 1.0 mmol) was suspended in a solution of H2O (10 mL) and HOAc (5.0 mL), and then the mixture was heated to reflux for 24 h. After cooling to room temperature, the resulting precipitate was collected by filtration, washed with water (3 × 5 mL), and dried to afford intermediate 12 as a colorless crystalline solid. Yield: 52%, mp: >300 °C. 1H NMR (400 MHz, DMSO-d6): δ 11.17 (s, 1H, C4–OH), 10.82 (s, 1H, C2–OH), 2.84 (d, J = 6.0 Hz, 2H, C6-dihydrothiopyranopyrimidine-H), 2.43 (t, J = 6.3 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.04−1.91 (m, 2H, C7-dihydrothiopyranopyrimidine-H). ESI-MS: m/z 185.0 [M + H]+, 207.2 [M + Na]+, C7H8N2O2S (184.03).
4.1.5. Synthesis of 2,4-dichloro-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidine (13)
Intermediate 12 (0.18 g, 1.0 mmol) and N,N-dimethylaniline (0.03 g, 0.25 mmol) were dissolved in 10 mL of POCl3 and then the mixture was refluxed at 110 °C for 12 h. After cooling to room temperature, the reaction solution was slowly added to ice and filtered. The crude precipitate was further purified by silica gel column chromatography with EtOAc:PE (1:8) as the eluent to provide intermediate 13 as a white solid. Yield: 69%, mp: 105–107 °C. ESI-MS: m/z 221.3 [M + H]+, C7H6Cl2N2S (219.96).
4.1.6. General procedure for the synthesis of intermediates 14a−c
To a solution of intermediate 13 (0.22 g, 1.0 mmol) and potassium carbonate (0.28 g, 2.0 mmol) in DMF (5 mL), the different material 4-hydroxy-3,5-dimethylbenzonitrile (0.14 g, 1.0 mmol) or (E)-3-(4-hydroxy-3,5-dimethylphenyl)acrylonitrile (0.17 g, 1.0 mmol) or 2,4,6-trimethylphenol (0.14 g, 1.0 mmol) was added and stirred at room temperature for 6 h. Then the reaction mixture was poured into water (20 mL) and extracted with EtOAc (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, and finally purified by silica gel column chromatography with EtOAc:PE (1:6) as the eluent to give the corresponding intermediates 14a−c as a white solid.
4.1.6.1. 4-((2-Chloro-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (14a)
White solid, yield: 67%, mp: 254–256 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.73 (s, 2H, C3,C5-Ph-H), 3.16 (d, J = 5.6 Hz, 2H, C6-dihydrothiopyranopyrimidine-H), 2.92 (t, J = 6.2 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.23−2.13 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.07 (s, 6H, CH3 × 2). ESI-MS: m/z 332.4 [M + H]+, C16H14ClN3OS (331.05).
4.1.6.2. (E)-3-(4-((2-Chloro-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylphenyl)acrylonitrile (14b)
White solid, yield: 86%, mp: 240–242 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.34 (d, J = 16.7 Hz, 1H, ArCH= ), 7.19 (s, 2H, C3,C5-Ph-H), 5.82 (d, J = 16.6 Hz, 1H, =CHCN), 3.17−3.05 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.97 (t, J = 6.3 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.30 (p, J = 6.2 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 2.13 (s, 6H, CH3 × 2). ESI-MS: m/z 358.3 [M + H]+, C18H16ClN3OS (357.07).
4.1.6.3. 2-Chloro-4-(mesityloxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidine (14c)
White solid, yield: 73%, mp: 176–178 °C. ESI-MS: m/z 321.4 [M + H]+, C16H17ClN2OS (320.08).
4.1.7. General procedure for the synthesis of target compounds 15a and 15b
BINAP (0.03 g, 0.05 mmol) and Pd2 (dba)3 (0.05 g, 0.05 mmol) were first suspended in 1,4-dioxane (20 mL) under stirring at room temperature for 15 min. Then the 4-aminobenzonitrile (0.11 g, 0.9 mol) and cesium carbonate (0.97 g, 3.0 mol) were added and the stir was continued for another 10 min. Finally, intermediate 14a (0.33 g, 1.0 mmol) or 14b (0.36 g, 1.0 mmol) was added. The mixture was backfilled with nitrogen fully and heated at 100 °C for 12 h. After cooling to room temperature, the reaction solution was filtered, and the obtained organic layer was purified by silica gel column chromatography with EtOAc:PE (1:4) as the eluent and finally recrystallized from EtOAc−PE to afford the target compounds 15a or 15b as a white solid.
4.1.7.1. 4-((2-((4-Cyanophenyl)amino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (15a)
White solid, yield: 77%, mp: 253–255 °C. 1H NMR (400 MHz, DMSO-d6): δ 9.98 (s, 1H, NH), 7.78 (s, 2H, C3,C5-Ph′′-H), 7.45 (s, 4H, C2,C3,C5,C6-Ph′-H), 3.19−3.05 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.88 (t, J = 6.2 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.26−2.16 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.11 (s, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 163.63, 163.07, 154.68, 154.12, 145.20, 133.10, 133.02, 132.98, 119.92, 119.00, 118.05, 109.09, 104.50, 102.39, 31.74, 26.34, 22.96, 16.12. ESI-MS: m/z 414.5 [M + H]+, 431.5 [M + NH4]+. C23H19N5OS (413.13). HPLC purity: 98.54%.
4.1.7.2. (E)-4-((4-(4-(2-Cyanovinyl)-2,6-dimethylphenoxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)amino)benzonitrile (15b)
White solid, yield: 61%, mp: 278–280 °C. 1H NMR (400 MHz, DMSO-d6): δ 9.95 (s, 1H, NH), 7.68 (d, J = 16.7 Hz, 1H, ArCH = ), 7.54 (s, 2H, C3,C5-Ph′′-H), 7.48 (d, J = 8.6 Hz, 2H, C3,C5-Ph′-H), 7.39 (d, J = 8.8 Hz, 2H, C2,C6-Ph′-H), 6.48 (d, J = 16.7 Hz, 1H, =CHCN), 3.19−3.06 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.87 (t, J = 6.2 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.20 (p, J = 6.0 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 2.08 (s, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 163.47, 163.30, 154.76, 152.47, 150.45, 145.29, 132.97, 131.93, 131.63, 128.64, 119.95, 119.33, 118.06, 104.49, 102.26, 96.93, 31.71, 26.33, 22.98, 16.40. ESI-MS: m/z 440.6 [M + H]+, 457.6 [M + NH4]+, C25H21N5OS (439.15). HPLC purity: 96.28%.
4.1.8. General procedure for the synthesis of target compounds 16a and 16b
A mixture of compound 15a (0.41 g, 1.0 mmol) and 3-chloroperbenzoic acid (0.20 g, 1.2 mmol) in DCM (10 mL) was stirred at −78 °C or room temperature for 6 h. Then saturated sodium bisulfite solution (10 mL) was added and extracted with DCM (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, purified by silica gel column chromatography with EtOAc:PE (1:4) as the eluent, and finally recrystallized from EtOAc−PE to give the target compounds 16a or 16b as a white solid.
4.1.8.1. 4-((2-((4-Cyanophenyl)amino)-5-oxido-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (16a)
White solid, yield: 72%, mp: 247–249 °C. 1H NMR (400 MHz, DMSO-d6): δ 10.52 (s, 1H, NH), 7.80 (d, J = 4.6 Hz, 2H, C3,C5-Ph′′-H), 7.50 (s, 4H, C2,C3,C5,C6-Ph′-H), 3.32−3.19 (m, 2H, C8-dihydrothiopyranopyrimidine-H), 3.09−2.79 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.61−2.51 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.15 (d, J = 14.7 Hz, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 169.09, 167.61, 158.94, 153.63, 144.12, 133.16, 133.08, 119.60, 119.31, 118.95, 111.11, 109.44, 104.19, 60.21, 45.19, 32.08, 16.22, 12.89. ESI-MS: m/z 447.5 [M + NH4]+, 452.3 [M + Na]+, C23H19N5O2S (429.13). HPLC purity: 99.31%.
4.1.8.2. 4-((2-((4-Cyanophenyl)amino)-5,5-dioxido-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (16b)
White solid, yield: 62%, mp: 263–265 °C. 1H NMR (400 MHz, DMSO-d6): δ 10.62 (s, 1H, NH), 7.81 (s, 2H, C3,C5-Ph′′-H), 7.64−7.18 (m, 4H, C2,C3,C5,C6-Ph′-H), 3.69−3.55 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 3.02 (t, J = 6.0 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.34 (d, J = 5.1 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 2.16 (s, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 169.89, 164.70, 158.49, 153.31, 143.82, 133.19, 133.08, 132.92, 119.52, 119.44, 118.92, 112.61, 109.52, 104.48, 52.49, 32.08, 18.98, 16.08. ESI-MS: m/z 463.5 [M + NH4]+, 468.4 [M + Na]+, C23H19N5O3S (445.12). HPLC purity: 95.98%.
4.1.9. General procedure for the synthesis of intermediates 18a and 18b
A mixture of intermediate 14a (0.33 g, 1.0 mmol), tert-butyl piperazine-1-carboxylate (0.23 g, 1.2 mmol), or tert-butyl piperidin-4-ylcarbamate (0.24 g, 1.2 mmol), and potassium carbonate (0.28 g, 2.0 mmol) in 10 mL of DMF was heated at 120 °C for 12 h. After cooling to room temperature, the reaction solution was added to the 50 mL of water. The resulting precipitate was collected by filtration, washed with water (3 × 5 mL), and dried to provide crude products 17a or 17b, which was used directly in the next step without further purification. Subsequently, to a solution of 17a (0.48 g, 1.0 mmol) or 17b (0.50 g, 1.0 mmol) in DCM (10 mL) was added trifluoroacetic acid (1.14 g, 10 mmol) and the mixture was stirred at room temperature for 4 h. Then the reaction solution was alkalized to pH 9 with saturated sodium bicarbonate solution and extracted with DCM (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, and finally purified by silica gel column chromatography with MeOH:DCM (1:10) as the eluent to afford the intermediates 18a or 18b as a white solid.
4.1.9.1. 3,5-Dimethyl-4-((2-(piperazin-1-yl)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)benzonitrile (18a)
White solid, yield: 52%. 1H NMR (400 MHz, DMSO-d6): δ 8.83 (s, 1H, piperazine-NH), 7.69 (s, 2H, C3,C5-Ph-H), 3.53 (t, J = 5.1 Hz, 4H, C2,C6-piperazine-H), 3.12−2.94 (m, 6H, C6-dihydrothiopyranopyrimidine-H, C3,C5-piperazine-H), 2.77 (t, J = 6.3 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.21−2.10 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.08 (s, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 170.78, 163.31, 157.15, 153.73, 132.86, 119.18, 108.83, 101.57, 42.65, 41.11, 32.03, 26.31, 23.18, 21.22, 16.15. C20H23N5OS (381.16).
4.1.9.2. 4-((2-(4-Aminopiperidin-1-yl)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (18b)
White solid, yield: 64%, mp: 234–236 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.87 (s, 2H, NH2), 7.69 (s, 2H, C3,C5-Ph-H), 4.11 (d, J = 13.2 Hz, 2H, piperidine-H), 3.20 (tt, J = 11.0, 3.9 Hz, 1H, piperidine-H), 3.10−3.01 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.80−2.66 (m, 4H, piperidine-H, C8-dihydrothiopyranopyrimidine-H), 2.14 (p, J = 6.1 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 2.08 (s, 6H, CH3 × 2), 1.78 (d, J = 12.0 Hz, 2H, piperidine-H), 1.25 (tt, J = 13.1, 6.6 Hz, 2H, piperidine-H). C21H25N5OS (395.18).
4.1.10. General procedure for the synthesis of target compounds 19a and 19b
Intermediate 18a (0.38 g, 1.0 mmol) or 18b (0.40 g, 1.0 mmol) was dissolved in 10 mL of DMF, followed by the addition of 4-(bromomethyl)benzenesulfonamide (0.28 g, 1.1 mmol) and potassium carbonate (0.17 g, 1.2 mmol). After being stirred at room temperature until completion, the reaction mixture was poured into water (20 mL) and extracted with EtOAc (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, purified by silica gel column chromatography with MeOH:DCM (1:20) as the eluent, and finally recrystallized from EtOAc−PE to give the target compounds 19a or 19b as a white solid.
4.1.10.1. 4-((4-(4-(4-Cyano-2,6-dimethylphenoxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)piperazin-1-yl)methyl)benzenesulfonamide (19a)
White solid, yield: 60%, mp: 141–143 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.77 (d, J = 8.3 Hz, 2H, C3,C5-Ph′-H), 7.66 (s, 2H, C3,C5-Ph′′-H), 7.46 (d, J = 8.3 Hz, 2H, C2,C6-Ph′-H), 7.32 (s, 2H, SO2NH2), 3.50 (s, 2H, N–CH2), 3.36 (s, 4H, C2,C6-piperazine-H), 3.11−2.98 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.73 (dt, J = 178.4, 6.2 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.27 (t, J = 4.5 Hz, 4H, C3,C5-piperazine-H), 2.19−2.09 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.07 (s, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 163.23, 163.02, 157.57, 153.89, 143.26, 142.61, 132.90, 132.73, 129.57, 126.08, 119.10, 108.69, 100.06, 61.70, 52.51, 43.83, 32.05, 26.32, 23.34, 16.18. ESI-MS: m/z 551.2 [M + H]+, C27H30N6O3S2 (550.18). HPLC purity: 96.60%.
4.1.10.2. 4-((1-(4-(4-Cyano-2,6-dimethylphenoxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)piperidin-4-yl)amino)benzenesulfonamide (19b)
White solid, yield: 66%, mp: 227–229 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.74 (d, J = 8.2 Hz, 2H, C3,C5-Ph′-H), 7.68 (s, 2H, C3,C5-Ph′′-H), 7.49 (d, J = 8.2 Hz, 2H, C2,C6-Ph′-H), 7.29 (s, 2H, SO2NH2), 3.97 (d, J = 10.2 Hz, 2H, piperidine-H, NH–CH2), 3.76 (s, 2H, NH–CH2), 3.10−2.98 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.82−2.64 (m, 4H, piperidine-H, C8-dihydrothiopyranopyrimidine-H), 2.13 (m, 4H, piperidine-H, C7-dihydrothiopyranopyrimidine-H), 2.07 (s, 6H, CH3 × 2), 1.71 (d, J = 10.2 Hz, 2H, piperidine-H), 1.05 (q, J = 9.7 Hz, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.07, 157.56, 154.01, 146.12, 142.73, 132.96, 132.70, 128.57, 125.92, 119.13, 108.64, 99.33, 60.22, 53.75, 49.50, 42.58, 32.08, 31.81, 26.33, 23.41, 16.17. ESI-MS: m/z 565.2 [M + H]+, C28H32N6O3S2 (564.20). HPLC purity: 98.13%.
4.1.11. General procedure for the synthesis of target compounds 20a−d
To a solution of intermediate 14a (0.33 g, 1.0 mmol), 14b (0.36 g, 1.0 mmol), or 14c (0.32 g, 1.0 mmol) in 10 mL of NMP was added 1-(2-aminoethyl)pyrimidine-2,4(1H,3H)-dione (0.17 g, 1.1 mmol) and N,N-diisopropylethylamine (0.26 g, 2.0 mmol), and the mixture was stirred at 120 °C for 12 h. After cooling to room temperature, the reaction mixture was poured into water (20 mL) and extracted with EtOAc (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, purified by silica gel column chromatography with EtOAc:PE (1:4) as the eluent, and finally recrystallized from EtOAc−PE to afford the target compounds 20a−c as a white solid. Using a similar synthetic method with the intermediate 14a (0.33 g, 1.0 mmol) and 2-morpholinoethan-1-amine (0.14 g, 1.1 mmol) as starting materials, the target compound 20d was prepared as a white solid.
4.1.11.1. 4-((2-((2-(2,4-Dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethyl)amino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (20a)
White solid, yield: 25%, mp: 203–205 °C. 1H NMR (400 MHz, DMSO-d6): δ 11.21 (d, J = 2.3 Hz, 1H, uracil-NH), 7.74 (s, 2H, C3,C5-Ph-H), 7.29 (s, 1H, C2-uracil-H), 6.93 (s, 1H, NH), 5.41 (s, 1H, C3-uracil-H), 3.69 (s, 2H, NH–CH2), 3.40 (s, 2H, N–CH2), 3.13−3.00 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.74 (t, J = 6.2 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.21−2.15 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.14 (s, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 164.26, 163.16, 158.49, 154.01, 151.33, 146.09, 132.99, 132.84, 119.09, 108.66, 100.59, 99.99, 79.64, 31.76, 26.34, 23.35, 16.16. ESI-MS: m/z 451.5 [M + H]+, 473.3 [M + Na]+, C22H22N6O3S (450.15). HPLC purity: 98.36%.
4.1.11.2. (E)-3-(4-((2-((2-(2,4-Dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethyl)amino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylphenyl)acrylonitrile (20b)
White solid, yield: 37%, mp: 283–285 °C. 1H NMR (400 MHz, DMSO-d6): δ 11.15 (d, J = 7.7 Hz, 1H, uracil-NH), 7.59 (d, J = 16.3 Hz, 1H, ArCH = ), 7.45 (s, 2H, C3,C5-Ph-H), 6.86 (s, 2H, C2-uracil-H, NH), 6.41 (d, J = 16.7 Hz, 1H, =CHCN), 5.31 (d, J = 7.7 Hz, 1H, C3-uracil-H), 3.62 (s, 2H, NH–CH2), 3.17 (s, 2H, N–CH2), 3.05−2.96 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.67 (t, J = 6.3 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.15−2.08 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.05 (s, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 164.26, 163.56, 158.58, 152.37, 151.33, 150.48, 131.63, 131.51, 128.46, 119.40, 96.69, 31.76, 26.35, 23.38, 16.55, 16.44. ESI-MS: m/z 477.4 [M + H]+, 499.5 [M + Na]+, C24H24N6O3S (476.16). HPLC purity: 97.19%.
4.1.11.3. 1-(2-((4-(Mesityloxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)amino)ethyl)pyrimidine-2,4(1H,3H)-dione (20c)
White solid, yield: 37%, mp: 223–225 °C. 1H NMR (400 MHz, DMSO-d6): δ 11.15 (d, J = 7.7 Hz, 1H, uracil-NH), 6.91 (s, 4H, C3,C5-Ph-H, C2-uracil-H, NH), 5.31 (d, J = 7.7 Hz, 1H, C3-uracil-H), 3.61 (s, 2H, NH–CH2), 3.19 (s, 2H, N–CH2), 3.04−2.93 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.66 (t, J = 6.1 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.24 (s, 3H, CH3), 2.09 (dt, J = 12.1, 6.1 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 1.98 (s, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 164.25, 163.91, 158.65, 151.34, 147.88, 134.65, 130.20, 129.39, 100.62, 100.00, 31.73, 26.32, 23.41, 20.82, 16.40. ESI-MS: m/z 440.6 [M + H]+, C22H25N5O3S (439.17). HPLC purity: 98.42%.
4.1.11.4. 3,5-Dimethyl-4-((2-((2-morpholinoethyl)amino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)benzonitrile (20d)
White solid, yield: 68%, mp: 142–144 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.67 (s, 2H, C3,C5-Ph-H), 6.81 (s, 1H, NH), 3.49 (s, 4H, C3,C5-morpholine-H), 3.02 (t, J = 5.5, 4H, NH–CH2, C6-dihydrothiopyranopyrimidine-H), 2.69 (t, J = 6.2 Hz, 2H, C8-dihydrothiopyranopyrimidine-H), 2.51−2.14 (m, 8H, N–CH2, C2,C6-morpholine-H, C7-dihydrothiopyranopyrimidine-H), 2.08 (s, 6H, CH3 × 2). 13C NMR (100 MHz, DMSO-d6): δ 163.43, 163.26, 158.61, 154.11, 133.00, 132.77, 119.06, 108.56, 66.59, 57.69, 53.65, 40.68, 38.06, 31.82, 26.34, 23.45, 16.15. ESI-MS: m/z 426.2 [M + H]+, C22H27N5O2S (425.19). HPLC purity: 99.00%.
4.1.12. General procedure for the synthesis of intermediates 22a−c
A mixture of intermediate 14a (0.33 g, 1.0 mmol), 14b (0.36 g, 1.0 mmol), or 14c (0.32 g, 1.0 mmol), tert-butyl 4-aminopiperidine-1-carboxylate (0.24 g, 1.2 mmol), and potassium carbonate (0.28 g, 2.0 mmol) in 10 mL of DMF was heated at 100 °C for 10 h. After cooling to room temperature, the reaction solution was added to the 50 mL of water. The resulting precipitate was collected by filtration, washed with water (3 × 5 mL), and dried to provide crude products 21a−c, which were used directly in the next step without further purification. Subsequently, to a solution of 21a (0.50 g, 1.0 mmol) or 21b (0.52 g, 1.0 mmol) or 21c (0.48 g, 1.0 mmol) in DCM (10 mL) was added trifluoroacetic acid (1.14 g, 10 mmol) and the mixture was stirred at room temperature for 4 h. Then the reaction solution was alkalized to pH 9 with saturated sodium bicarbonate solution and extracted with DCM (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, and finally purified by silica gel column chromatography with MeOH:DCM (1:10) as the eluent to afford the intermediates 22a−c as a white solid.
4.1.12.1. 3,5-Dimethyl-4-((2-(piperidin-4-ylamino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)benzonitrile (22a)
White solid, yield: 63%, mp: >250 °C. 1H NMR (400 MHz, DMSO-d6): δ 9.00 (s, 1H, piperidine-NH), 7.67 (s, 2H, C3,C5-Ph-H), 7.02 (s, 1H, NH), 3.14 (d, J = 12.5 Hz, 2H, C6-dihydrothiopyranopyrimidine-H), 3.07−2.99 (m, 2H, C8-dihydrothiopyranopyrimidine-H), 2.71 (t, J = 6.3 Hz, 4H, piperidine-H), 2.18−2.09 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.07 (s, 6H, CH3 × 2), 1.81 (s, 2H, piperidine-H), 1.54 (s, 3H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.27, 157.76, 154.08, 132.96, 132.77, 119.08, 108.60, 46.02, 41.99, 31.86, 28.05, 26.31, 23.39, 16.18. ESI-MS: m/z 396.4 [M + H]+, C21H25N5OS (395.18).
4.1.12.2. (E)-3-(3,5-Dimethyl-4-((2-(piperidin-4-ylamino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)phenyl)acrylonitrile (22b)
White solid, yield: 18%. ESI-MS: m/z 422.3 [M + H]+, C23H27N5OS (421.19).
4.1.12.3. 4-(Mesityloxy)-N-(piperidin-4-yl)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-amine (22c)
White solid, yield: 29%, mp: >290 °C. 1H NMR (400 MHz, DMSO-d6): δ 8.91 (s, 1H, piperidine-NH), 6.90 (s, 3H, C3,C5-Ph-H, NH), 3.15 (d, J = 12.6 Hz, 2H, C6-dihydrothiopyranopyrimidine-H), 3.07−2.96 (m, 2H, C8-dihydrothiopyranopyrimidine-H), 2.69 (t, J = 6.3 Hz, 4H, piperidine-H), 2.24 (s, 3H, CH3), 2.12 (p, J = 5.9 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 1.98 (s, 6H, CH3 × 2), 1.82 (s, 2H, piperidine-H), 1.54 (s, 3H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 164.00, 157.90, 147.95, 134.59, 130.15, 129.29, 46.06, 42.15, 31.84, 28.14, 26.30, 23.45, 20.81, 16.44. ESI-MS: m/z 385.5 [M + H]+, C21H28N4OS (384.20).
4.1.13. General procedure for the synthesis of target compounds 23a−i
Intermediate 22a (0.40 g, 1.0 mmol), 22b (0.42 g, 1.0 mmol), or 22c (0.38 g, 1.0 mmol) was dissolved in 10 mL of DMF, followed by the addition of substituted benzyl chloride (or bromide) (1.1 mmol) and potassium carbonate (0.17 g, 1.2 mmol). After being stirred at room temperature until completion, the reaction mixture was poured into water (20 mL) and extracted with EtOAc (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, purified by silica gel column chromatography with MeOH:DCM (1:20) as the eluent, and finally recrystallized from EtOAc−PE to give the target compounds 23a−i as a white solid.
4.1.13.1. 4-((2-((1-(4-Cyanobenzyl)piperidin-4-yl)amino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (23a)
White solid, yield: 81%, mp: 184–186 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.77 (d, J = 8.2 Hz, 2H, C3,C5-Ph′-H), 7.66 (s, 2H, C3,C5-Ph′′-H), 7.47 (d, J = 8.1 Hz, 2H, C2,C6-Ph′-H), 6.89 (br, 1H, NH), 3.48 (s, 2H, N–CH2), 3.12−2.95 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.69 (t, J = 6.3 Hz, 5H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.19−2.02 (m, 8H, CH3 × 2, C7-dihydrothiopyranopyrimidine-H), 1.63 (s, 4H, piperidine-H), 1.32 (s, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.26, 158.00, 154.22, 145.26, 133.01, 132.66, 132.58, 129.87, 119.38, 119.10, 110.08, 108.56, 61.93, 52.59, 48.66, 31.84, 31.66, 26.33, 23.47, 16.16. ESI-MS: m/z 511.6 [M + H]+, C29H30N6OS (510.22). HPLC purity: 95.65%.
4.1.13.2. 4-((2-((1-(3-Cyanobenzyl)piperidin-4-yl)amino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (23b)
White solid, yield: 81%, mp: 164–166 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.72 (d, J = 7.6 Hz, 1H, C4-Ph′-H), 7.68 (s, 1H, C2-Ph′-H), 7.65 (s, 2H, C3,C5-Ph′′-H), 7.62 (d, J = 7.8 Hz, 1H, C6-Ph′-H), 7.53 (t, J = 7.7 Hz, 1H, C5-Ph′-H), 6.87 (s, 1H, NH), 3.45 (s, 2H, N–CH2), 3.09−2.94 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.69 (t, J = 6.3 Hz, 4H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.19−2.09 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.09 (s, 1H, piperidine-H), 2.06 (s, 6H, CH3 × 2), 1.63 (s, 4H, piperidine-H), 1.31 (s, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.26, 157.96, 154.22, 140.90, 134.02, 133.02, 132.68, 132.44, 131.22, 129.89, 119.34, 119.10, 111.65, 108.56, 61.46, 55.38, 52.49, 31.86, 31.64, 31.15, 26.33, 23.47, 21.52, 16.16. ESI-MS: m/z 511.2 [M + H]+, C29H30N6OS (510.22). HPLC purity: 97.91%.
4.1.13.3. 4-((2-((1-(2-Cyanobenzyl)piperidin-4-yl)amino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (23c)
White solid, yield: 78%, mp: 206–208 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.79 (d, J = 7.6 Hz, 1H, C3-Ph′-H), 7.70−7.61 (m, 3H, C3,C5-Ph′′-H, C5-Ph′-H), 7.52 (d, J = 7.7 Hz, 1H, C4-Ph′-H), 7.46 (t, J = 7.6 Hz, 1H, C6-Ph′-H), 6.86 (s, 1H, NH), 3.56 (s, 2H, N–CH2), 3.10−2.94 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.69 (t, J = 6.2 Hz, 4H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.17−2.09 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.09 (s, 1H, piperidine-H), 2.06 (s, 6H, CH3 × 2), 1.59 (s, 4H, piperidine-H), 1.30 (s, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.26, 158.00, 154.22, 142.82, 133.42, 133.41, 133.02, 132.69, 130.48, 128.36, 119.08, 118.11, 112.53, 108.57, 60.36, 60.22, 52.54, 31.88, 31.58, 31.19, 26.33, 23.47, 16.17, 14.56. ESI-MS: m/z 511.3 [M + H]+, C29H30N6OS (510.22). HPLC purity: 97.36%.
4.1.13.4. 3-((4-((4-(4-Cyano-2,6-dimethylphenoxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (23d)
White solid, yield: 74%, mp: 170–172 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.74 (s, 1H, C2-Ph′-H), 7.71 (d, J = 7.4 Hz, 1H, C4-Ph′-H), 7.67 (s, 2H, C3,C5-Ph′′-H), 7.51 (t, J = 7.5 Hz, 1H, C6-Ph′-H), 7.47 (d, J = 7.5 Hz, 1H, C5-Ph′-H), 7.36 (s, 2H, SO2NH2), 6.80 (s, 1H, NH), 3.46 (s, 2H, N–CH2), 3.02 (dd, J = 6.7, 4.1 Hz, 2H, C6-dihydrothiopyranopyrimidine-H), 2.69 (t, J = 6.1 Hz, 4H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.19−2.09 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.08 (s, 1H, piperidine-H), 2.06 (s, 6H, CH3 × 2), 1.66 (s, 4H, piperidine-H), 1.32 (s, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.25, 157.84, 154.22, 144.59, 144.09, 140.31, 133.01, 132.74, 132.47, 129.26, 125.92, 124.68, 119.13, 108.55, 62.05, 52.62, 49.07, 31.82, 31.65, 26.30, 23.45, 16.18. ESI-MS: m/z 565.2 [M + H]+, C28H32N6O3S2 (564.20). HPLC purity: 96.68%.
4.1.13.5. Ethyl 4-((4-((4-(4-cyano-2,6-dimethylphenoxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzoate (23e)
White solid, yield: 57%, mp: 181–183 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.91 (d, J = 8.1 Hz, 2H, C3,C5-Ph′-H), 7.65 (s, 2H, C3,C5-Ph′′-H), 7.41 (d, J = 8.1 Hz, 2H, C2,C6-Ph′-H), 6.87 (br, 1H, NH), 4.30 (q, J = 7.1 Hz, 2H, CO2CH2CH3), 3.46 (s, 2H, N–CH2), 3.09−2.97 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.69 (t, J = 6.2 Hz, 4H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.16−2.09 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.09 (s, 1H, piperidine-H), 2.06 (s, 6H, CH3 × 2), 1.60 (s, 4H, piperidine-H), 1.31 (t, J = 7.1 Hz, 5H, CO2CH2CH3, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 166.11, 163.26, 157.99, 154.24, 144.87, 141.04, 133.02, 132.74, 129.52, 129.29, 128.98, 119.10, 108.56, 62.16, 61.04, 56.50, 52.62, 31.88, 31.65, 31.16, 26.33, 23.47, 16.15, 14.66. ESI-MS: m/z 558.6 [M + H]+, C31H35N5O3S (557.25). HPLC purity: 98.38%.
4.1.13.6. 4-((2-((1-(4-Aminobenzyl)piperidin-4-yl)amino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (23f)
White solid, yield: 64%, mp: 169–171 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.66 (s, 2H, C3,C5-Ph′′-H), 6.87 (d, J = 8.1 Hz, 2H, C2,C6-Ph′-H), 6.49 (d, J = 8.2 Hz, 2H, C3,C5-Ph′-H), 4.94 (s, 2H, NH2), 3.17 (s, 2H, N–CH2), 3.08−2.95 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.68 (t, J = 6.1 Hz, 4H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.12 (d, J = 5.3 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 2.09 (s, 1H, piperidine-H), 2.05 (s, 6H, CH3 × 2), 1.48 (s, 4H, piperidine-H), 1.23 (s, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.24, 158.03, 154.22, 147.95, 133.01, 132.65, 130.16, 125.66, 123.82, 119.11, 114.01, 108.55, 62.55, 52.39, 31.65, 26.31, 23.46, 21.24, 19.03, 16.16, 14.56. ESI-MS: m/z 501.5 [M + H]+, C28H32N6OS (500.24). HPLC purity: 98.40%.
4.1.13.7. N-(4-((4-((4-(4-cyano-2,6-dimethylphenoxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)phenyl)methanesulfonamide (23g)
White solid, yield: 17%, mp: 153–155 °C. 1H NMR (400 MHz, DMSO-d6): δ 9.68 (s, 1H, NHSO2CH3), 7.66 (s, 2H, C3,C5-Ph′′-H), 7.21 (d, J = 8.3 Hz, 2H, C3,C5-Ph′-H), 7.14 (d, J = 8.4 Hz, 2H, C2,C6-Ph′-H), 6.70 (br, 1H, NH), 3.32 (s, 2H, N–CH2), 3.07−2.99 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.96 (s, 3H, NHSO2CH3), 2.69 (t, J = 6.3 Hz, 4H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.11 (t, J = 5.7 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 2.09 (s, 1H, piperidine-H), 2.06 (s, 6H, CH3 × 2), 1.55 (s, 4H, piperidine-H), 1.23 (s, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.25, 137.49, 134.60, 133.01, 132.90, 132.77, 132.70, 130.12, 120.24, 119.13, 108.54, 100.98, 62.08, 52.56, 39.62, 36.25, 31.81, 31.63, 31.17, 26.30, 23.45, 16.16. ESI-MS: m/z 579.2 [M + H]+, C29H34N6O3S2 (578.21). HPLC purity: 98.56%.
4.1.13.8. (E)-4-((4-((4-(4-(2-Cyanovinyl)-2,6-dimethylphenoxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (23h)
White solid, yield: 64%, mp: 237–239 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.77 (d, J = 8.3 Hz, 2H, C3,C5-Ph′-H), 7.59 (d, J = 16.7 Hz, 1H, ArCH = ), 7.43 (s, 4H, C2,C6-Ph′-H, C3,C5-Ph′′-H), 7.30 (s, 2H, SO2NH2), 6.66 (s, 1H, NH), 6.40 (d, J = 16.7 Hz, 1H, =CHCN), 3.43 (s, 2H, N–CH2), 3.08−2.95 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.68 (t, J = 6.2 Hz, 4H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.15−2.09 (m, 2H, C7-dihydrothiopyranopyrimidine-H), 2.06 (s, 1H, piperidine-H), 2.03 (s, 6H, CH3 × 2), 1.61 (s, 4H, piperidine-H), 1.32 (s, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.65, 158.05, 152.59, 150.58, 143.41, 143.14, 131.62, 131.45, 129.43, 128.33, 126.05, 119.42, 96.51, 62.02, 52.67, 49.07, 31.89, 31.68, 26.34, 23.50, 16.46. ESI-MS: m/z 591.2 [M + H]+, C30H34N6O3S2 (590.21). HPLC purity: 98.59%.
4.1.13.9. 4-((4-((4-(Mesityloxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (23i)
White solid, yield: 44%, mp: 218–220 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.77 (d, J = 8.3 Hz, 2H, C3,C5-Ph′-H), 7.44 (d, J = 8.0 Hz, 2H, C2,C6-Ph′-H), 7.30 (s, 2H, SO2NH2), 6.88 (s, 2H, C3,C5-Ph′′-H), 6.60 (s, 1H, NH), 3.45 (s, 2H, N–CH2), 3.08−2.91 (m, 2H, C6-dihydrothiopyranopyrimidine-H), 2.79−2.57 (m, 4H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.23 (s, 3H, CH3), 2.10 (dt, J = 11.8, 5.4 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 1.97 (s, 6H, CH3 × 2), 1.72−1.47 (m, 2H, piperidine-H), 1.46−1.10 (m, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.97, 162.36, 158.15, 148.02, 143.36, 143.13, 134.49, 130.15, 129.42, 129.22, 126.03, 61.99, 52.65, 49.07, 31.86, 31.74, 26.33, 23.54, 20.80, 16.43. ESI-MS: m/z 554.5 [M + H]+, C28H35N5O3S2 (553.22). HPLC purity: 96.82%.
4.1.14. Synthesis of tert-butyl (1-(3-cyanophenyl)piperidin-4-yl)carbamate (25a)
A mixture of 3-fluorobenzonitrile (24, 0.12 g, 1.0 mmol), tert-butyl piperidin-4-ylcarbamate (0.24 g, 1.2 mmol), and potassium carbonate (0.28 g, 2.0 mmol) in 10 mL of DMF was heated at 120 °C for 10 h. After cooling to room temperature, the reaction solution was added to the 50 mL of water. The resulting precipitate was collected by filtration, washed with water (3 × 5 mL), and dried to provide intermediate 25a as a white solid. Yield: 66%. ESI-MS: m/z 302.6 [M + H]+, C17H23N3O2 (301.18).
4.1.15. Synthesis of tert-butyl (1-(3-carbamoylphenyl)piperidin-4-yl)carbamate (25b)
To a solution of intermediate 25a (0.30 g, 1.0 mmol) and sodium hydroxide (0.04 g, 1.0 mmol) in EtOH (10 mL), hydrogen peroxide (5 mL) was dropwise added in and stirred at 50 °C for 2 h. Then the reaction mixture was poured into water (10 mL) and extracted with EtOAc (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, and concentrated in vacuum to give intermediate 25b as a white solid. Yield: 62%, mp: 191–193 °C. ESI-MS: m/z 320.2 [M + H]+, C17H25N3O3 (319.19).
4.1.16. General procedure for the synthesis of intermediates 26a and 26b
To a solution of 25a (0.30 g, 1.0 mmol) or 25b (0.32 g, 1.0 mmol) in DCM (10 mL) was added trifluoroacetic acid (1.14 g, 10 mmol) and the mixture was stirred at room temperature for 4 h. Then the reaction solution was alkalized to pH 9 with saturated sodium bicarbonate solution and extracted with DCM (3 × 10 mL). The combined organic layer was washed with saturated brine (3 × 5 mL), dried over anhydrous Na2SO4, filtered, and concentrated in vacuum to provide intermediates 3-(4-aminopiperidin-1-yl)benzonitrile (26a) or 3-(4-aminopiperidin-1-yl)benzamide (26b) as a viscous oil and was used in next step without purification.
4.1.17. General procedure for the synthesis of target compounds 27a and 27b
BINAP (0.03 g, 0.05 mmol) and Pd2 (dba)3 (0.05 g, 0.05 mmol) were first suspended in 1,4-dioxane (20 mL) under stirring at room temperature for 15 min. Then the intermediate 26a (0.20 g, 1.0 mmol) or 26b (0.22 g, 1.0 mmol) and cesium carbonate (0.97 g, 3.0 mmol) were added and the stir was continued for another 10 min. Finally, intermediate 14a (0.33 g, 1.0 mmol) was added. The mixture was backfilled with nitrogen fully and heated at 100 °C for 12 h. After cooling to room temperature, the reaction solution was filtered, and the obtained organic layer was purified by silica gel column chromatography with MeOH:DCM (1:50) as the eluent and finally recrystallized from EtOAc−PE to afford the target compound 27a or 27b as a white solid.
4.1.17.1. 4-((2-((1-(3-Cyanophenyl)piperidin-4-yl)amino)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)oxy)-3,5-dimethylbenzonitrile (27a)
White solid, yield: 43%, mp: 186–188 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.65 (s, 2H, C3,C5-Ph′′-H), 7.34 (dd, J = 8.5, 7.4, 1H, C5-Ph′-H), 7.26 (s, 1H, C2-Ph′-H), 7.21 (d, J = 8.5 Hz, 1H, C4-Ph′-H), 7.09 (d, J = 7.4 Hz, 1H, C6-Ph′-H), 6.82 (br, 1H, NH), 3.67 (s, 3H, piperidine-H), 3.02 (p, 2H, C6-dihydrothiopyranopyrimidine-H), 2.70 (t, J = 6.0 Hz, 3H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.12 (t, J = 5.7 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 2.08 (s, 1H, piperidine-H), 2.07 (s, 6H, CH3 × 2), 1.69 (s, 2H, piperidine-H), 1.35 (s, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 163.26, 157.91, 154.17, 151.25, 133.01, 132.73, 130.64, 121.53, 120.26, 119.84, 119.12, 118.11, 112.37, 108.58, 56.52, 47.19, 31.85, 31.16, 30.82, 26.31, 23.42, 18.98, 16.18. ESI-MS: m/z 497.2 [M + H]+, C28H28N6OS (496.20). HPLC purity: 97.17%.
4.1.17.2. 3-(4-((4-(4-Cyano-2,6-dimethylphenoxy)-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)benzamide (27b)
White solid, yield: 35%, mp: 232–234 °C. 1H NMR (400 MHz, DMSO-d6): δ 7.91 (s, 1H, C4-Ph′-H), 7.68 (s, 2H, C3,C5-Ph′′-H), 7.38 (s, 1H, C5-Ph′-H), 7.29 (s, 1H, C2-Ph′-H), 7.25 (s, 2H, CONH2), 7.03 (d, J = 4.1 Hz, 1H, C6-Ph′-H), 6.73 (s, 1H, NH), 3.65 (s, 3H, piperidine-H), 3.03 (d, J = 5.4 Hz, 2H, C6-dihydrothiopyranopyrimidine-H), 2.72 (t, J = 6.3 Hz, 3H, C8-dihydrothiopyranopyrimidine-H, piperidine-H), 2.13 (t, J = 5.7 Hz, 2H, C7-dihydrothiopyranopyrimidine-H), 2.09 (s, 1H, piperidine-H), 2.09 (s, 6H, CH3 × 2), 1.72 (s, 2H, piperidine-H), 1.42 (s, 2H, piperidine-H). 13C NMR (100 MHz, DMSO-d6): δ 168.71, 163.27, 157.95, 154.18, 151.18, 135.40, 133.01, 132.75, 129.25, 119.11, 118.66, 117.98, 115.09, 108.58, 56.50, 48.06, 31.88, 31.16, 26.32, 23.46, 19.04, 16.21. ESI-MS: m/z 515.2 [M + H]+, C28H30N6O2S (514.22). HPLC purity: 98.29%.
4.2. In vitro anti-HIV assay
The anti-HIV activity and cytotoxicity of the target compounds were evaluated in MT-4 cells by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method. The MTT method is based on the HIV-induced cytopathogenic effect (CPE) and measures the degree of cell killing on HIV infection. Firstly, stock solutions (10 × final concentration) of test compounds were added in 25 μL volumes to two series of triplicate wells to allow simultaneous evaluation of their effects on mock- and HIV-infected cells. Serial 5-fold dilutions of test compounds were prepared directly in flat-bottomed 96-well microtiter trays by adding 100 μL medium to the 25 μL stock solution and transferring 25 μL of this solution to another well that contained 100 μL medium via a Biomek 3000 robot (Beckman Instruments, Fullerton, CA). Each sample contains untreated control HIV- and mock-infected cell samples. HIV-1 WT strain (IIIB), HIV-1 resistant strains [K103N + Y181C (RES056), L100I, K103N, Y181C, Y188L, E138K, and F227L + V106A] stock (50 μL) at 100−300 CCID50 (50% cell culture infectious dose) or culture medium was added to either the infected or mock-infected wells of the microtiter tray. Mock-infected cells were used to evaluate the effect of test compounds on normal cells to assess their cytotoxicity. Exponentially growing MT-4 cells were centrifuged for 5 min at 1000 rpm and the supernatant was discarded. The MT-4 cells were resuspended at 6 × 105 cells/mL and 50 μL volumes were transferred to the microtiter tray wells. After infecting the MT-4 cells for five days, the viability of mock- and HIV-infected cells was examined spectrophotometrically by the MTT method. The MTT method is based on the reduction of yellow-colored MTT (Acros Organics, Geel, Belgium) by mitochondrial dehydrogenase in metabolically active cells to a blue−purple formazan that can be measured spectrophotometrically. The absorbances were read in an eight-channel computer-controlled photometer (Infinite M1000, Tecan) at two wavelengths (540 nm and 690 nm). All data were calculated using the median absorbance value of three wells. The 50% cytotoxic concentration (CC50) was defined as the concentration of test compounds that reduces the viability of the mock-infected MT-4 cells by 50%. The 50% effective concentration (EC50) was defined as the concentration of test compounds that achieves 50% protection against the cytopathic effect of the virus in infected cells.
4.3. HIV-1 RT inhibition assay
The HIV-1 RT inhibition assay was conducted with an RT assay kit (Roche, Basel, Switzerland) by using the ELISA method. All the reagents for performing the RT reaction came from the kit and the ELSIA procedures for the RT inhibition assay were conducted following the description in the kit protocol. Briefly, the reaction mixture containing HIV-1 RT, reconstituted template and viral nucleotides [digoxigenin (DIG)-dUTP, biotin-dUTP and dTTP] in the incubation buffer with or without inhibitors was incubated for 1 h at 37 °C. The reaction mixture was transferred to a streptavidine-coated microtiter plate (MTP) and incubated for 1 h at 37 °C. The biotin-labeled dNTPs that were incorporated into the cDNA chain in the presence of RT were bound to streptavidin. The unbound dNTPs were removed with washing buffer and anti-DIG-POD working solution was added to the MTPs. The DIG-labeled dNTPs incorporated in cDNA were bound to the anti-DIG-POD antibody after incubation for 1 h at 37 °C. The unbound anti-DIG-PODs were washed out and the peroxide substrate (ABTS) solution was added to the MTPs. A color reaction occurred during cleavage of the substrate catalyzed by POD. The absorbance of the sample was determined using a microtiter plate ELISA reader (Multiskan Sky, Waltham, MA, USA). The inhibition rate of the test compounds was calculated by Eq. (1)
| (1) |
4.4. Cytochrome P450 inhibition assay
Compound 23h was incubated with human liver microsome and NADPH cofactor in the presence of various CYP substrates: phenacetin for CYP1A2, diclofenac for CYP2C9, S-mephenytoin for CYP2C19, dextromethorphan for CYP2D6, and midazolam for CYP3A4. Standard CYP enzyme inhibitors were also tested as positive controls. To begin, a working solution (100 × ) of the test compound 23h and standard inhibitors was prepared. Then, 20 μL of substrate solution was added to the appropriate wells, while 20 μL of potassium phosphate buffer was added to the blank wells. Next, 2 μL of the test compound and positive control working solution was added to their respective wells. For the wells without inhibitors, 2 μL of solvent was added, and for the blank wells, 2 μL of potassium phosphate buffer was added. To prepare the microsome working solution (0.253 mg/mL), 0.114 mL of human liver microsomes was combined with 8.886 mL of potassium phosphate buffer. Next, 158 μL of the HLM working solution was added to all wells of the incubation plate. The plate was pre-warmed for 10 min in a 37 °C water bath. For the NADPH solution (10 mmol/L), 62 mg of NADPH cofactor was added to 6.0 mL of MgCl2 buffer. Then, 20 μL of the NADPH cofactor was added to all incubation wells. The plate was mixed and incubated for 10 min in a 37 °C water bath. At the time point, the reaction should be terminated by adding 400 μL of a cold stop solution (containing 200 ng/mL tolbutamide and 200 ng/mL labetalol in ACN). The samples were then centrifuged at 4000 rpm for 20 min to precipitate the protein. Next, 200 μL of the supernatant was diluted with 100 μL of water and shaken for 10 min before the LC–MS/MS analysis.
4.5. hERG channel inhibition assay
The hERG channel inhibition of compound 23h was determined in HEK293 cells with stably transfected with hERG cDNA. HEK293 cells expressing hERG were plated in F12 dishes for 24 h and maintained at 37 °C under 5% CO2. A micropipette was drawn out from the borosilicate glass to give a tip resistance between 2 and 5 MΩ. For each trial, a single dish of cells was removed from the incubator, washed twice with ECS, and placed on the microscope. HEK293 cells in the exponential growth phase were collected and resuspended in ECS for later use. The whole-cell recordings were conducted with a commercially available patch-clamp amplifier. The HEK293 cells were placed on the microscope stage, and one cell in the recording tank was randomly selected for testing. The perfusion system was fixed on an inverted microscope stage, and the cells were continuously perfused with ECS. Tail currents were evoked once every 15 s by a 1275 ms, −50 mV repolarizing pulse following a 850 ms, +60 mV depolarizing pulse with a hold voltage of −80 mV. The voltage protocol started with a 50 ms depolarization pulse of −50 mV, which served as the baseline for calculating the peak tail current amplitude. Only stable cells with recording parameters exceeding the threshold were used in the experiments. The hERG currents were allowed to stabilize over a 3 min period under the condition that vehicle alone prior to test compound application. The cells were kept in the test solution until the peak tail current was stable (<5% change) for about 3 sweeps. Peak tail amplitudes were then plotted as a function of the sweep number. Before testing the composite application, the average of the three peak tail currents in the steady state was taken as the control current amplitude. Four or five peak tail current measurements at the steady state after test compound application were averaged and used as the remaining current amplitude after inhibition by the test compound. hERG inhibition rate was calculated by Eq. (2):
| (2) |
4.6. In vivo pharmacokinetics assay
All animal treatments were performed strictly in accordance with the institutional guidelines of animal ethical and welfare, after gaining approval from the Medical Ethical Committee of Shandong University. Sprague–Dawley (SD) rats (180−200 g) were purchased from the animal experimental center of Shandong University and were randomly divided into two groups receiving oral administration (20 mg/kg) or intravenous administration (2 mg/kg). Research protocols complied strictly with the institutional guidelines. Compound 23h and ETR were suspended in a mixture solution of PEG-400 and normal saline. Blood samples (200 μL each time) of the intravenous group were collected at 2, 5, 15, 30 min, 1, 1.5, 2, 4, and 8 h after dosing, and blood samples (200 μL each time) of the oral administration group were collected at 5, 15, 30 min, 1, 2, 4, 6, 8, 10, and 12 h after dosing. All the plasma samples were obtained by centrifugation at 8000 rpm for 8 min, and immediately stored at −80 °C until analysis. LC−MS analysis was used to determine the concentration of 23h and ETR in plasma. Briefly, 50 μL of plasma was added to 50 μL of internal standard and 300 μL of methanol in a 5 mL centrifugation tube. All blood samples were centrifuged in an Eppendorf 5415D centrifuge at 3000×g for 10 min and quantified by Agilent 1200 LC/MSD (Agilent, USA). The supernatant layer was collected and a 20 μL aliquot was injected for LC−MS analysis. Standard curves for 23h and ETR in blood were generated by the addition of various concentrations of 23h and ETR together with an internal standard to blank plasma.
4.7. In vivo safety experiment
All animal treatments were performed strictly in accordance with the institutional guidelines of animal ethical and welfare, after gaining approval from the Medical Ethical Committee of Shandong University. Kunming mice (15−19 g) were purchased from the animal experimental center of Shandong University. Research protocols complied strictly with the institutional guidelines. Compound 23h was suspended in a mixture solution of PEG-400 and normal saline at the concentration of 100 mg/mL. All mice had been fasted for 12 h, and then the mice in the test group were administered intragastrically by gavage at a dose of 2000 mg/kg of 23h on the first day. The death, abnormal behaviors, and body weight of these mice were monitored every day during the subsequent 7 days.
Author contributions
Xinyong Liu, Peng Zhan, Dongwei Kang, and Zhao Wang as the supervisors conceived the project and supplied the financial support. Zhao Wang and Heng Zhang conducted the synthesis of compounds and supervised the drug-likeness evaluation experiment. Erik De Clercq and Christophe Pannecouque conducted and supervised the biological activity evaluation assay. Molecular docking and molecular dynamics simulation were performed by Zhen Gao and Zhao Wang. Zhao Wang, Heng Zhang, Zhen Gao, and Zihao Sang conducted the data analysis. Zhao Wang wrote and revised the manuscript.
Conflicts of interest
The authors declare no competing financial interest.
Acknowledgments
We gratefully acknowledge financial support from the National Natural Science Foundation of China (NSFC nos. 81973181 and 81903453), Science Foundation for Outstanding Young Scholars of Shandong Province (ZR2020JQ31, China), Science Foundation for Excellent Young Scholars of Shandong Province (ZR2020YQ61, China), Foreign Cultural and Educational Experts Project (GXL20200015001, China), China Postdoctoral Science Foundation (2022M721948), Shandong Province Natural Science Foundation for Youths (ZR2023QH217, China), Natural Science Foundation of Jiangsu Province (BK20230252, China), Qilu Young Scholars Program of Shandong University, and Taishan Scholar Program at Shandong Province. We thank K. Erven, K. Uyttersprot, and C. Heens for technical assistance with the anti-HIV assays.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.11.023.
Contributor Information
Christophe Pannecouque, Email: christophe.pannecouque@rega.kuleuven.be.
Dongwei Kang, Email: kangdongwei@sdu.edu.cn.
Peng Zhan, Email: zhanpeng1982@sdu.edu.cn.
Xinyong Liu, Email: xinyongl@sdu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.The Lancet H.I.V. UNAIDS strategy aligns HIV priorities with development goals. Lancet HIV. 2021;8:e245. doi: 10.1016/S2352-3018(21)00075-8. [DOI] [PubMed] [Google Scholar]
- 2.Engelman A., Cherepanov P. The structural biology of HIV-1: mechanistic and therapeutic insights. Nat Rev Microbiol. 2012;10:279–290. doi: 10.1038/nrmicro2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauci A.S., Lane H.C. Four decades of HIV/AIDS–much accomplished, much to do. N Engl J Med. 2020;383:1–4. doi: 10.1056/NEJMp1916753. [DOI] [PubMed] [Google Scholar]
- 4.Larsen K.P., Mathiharan Y.K., Kappel K., Coey A.T., Chen D.H., Barrero D., et al. Architecture of an HIV-1 reverse transcriptase initiation complex. Nature. 2018;557:118–122. doi: 10.1038/s41586-018-0055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan P., Pannecouque C., De Clercq E., Liu X. Anti-HIV drug discovery and development: current innovations and future trends. J Med Chem. 2016;59:2849–2878. doi: 10.1021/acs.jmedchem.5b00497. [DOI] [PubMed] [Google Scholar]
- 6.Li G., Wang Y., De Clercq E. Approved HIV reverse transcriptase inhibitors in the past decade. Acta Pharm Sin B. 2022;12:1567–1590. doi: 10.1016/j.apsb.2021.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z., Cherukupalli S., Xie M., Wang W., Jiang X., Jia R., et al. Contemporary medicinal chemistry strategies for the discovery and development of novel HIV-1 non-nucleoside reverse transcriptase inhibitors. J Med Chem. 2022;65:3729–3757. doi: 10.1021/acs.jmedchem.1c01758. [DOI] [PubMed] [Google Scholar]
- 8.Zhuang C., Pannecouque C., De Clercq E., Chen F. Development of non-nucleoside reverse transcriptase inhibitors (NNRTIs): our past twenty years. Acta Pharm Sin B. 2020;10:961–978. doi: 10.1016/j.apsb.2019.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blair H.A. Dolutegravir/Rilpivirine: a review in HIV-1 infection. Drugs. 2018;78:1741–1750. doi: 10.1007/s40265-018-1005-4. [DOI] [PubMed] [Google Scholar]
- 10.Markham A. Cabotegravir plus rilpivirine: first approval. Drugs. 2020;80:915–922. doi: 10.1007/s40265-020-01326-8. [DOI] [PubMed] [Google Scholar]
- 11.Swindells S., Andrade-Villanueva J.F., Richmond G.J., Rizzardini G., Baumgarten A., Masia M., et al. Long-acting cabotegravir and rilpivirine for maintenance of HIV-1 suppression. N Engl J Med. 2020;382:1112–1123. doi: 10.1056/NEJMoa1904398. [DOI] [PubMed] [Google Scholar]
- 12.Ding L., Zhuang C., Chen F. Druggability modification strategies of the diarylpyrimidine-type non-nucleoside reverse transcriptase inhibitors. Med Res Rev. 2021;41:1255–1290. doi: 10.1002/med.21760. [DOI] [PubMed] [Google Scholar]
- 13.Gu S.X., Lu H.H., Liu G.Y., Ju X.L., Zhu Y.Y. Advances in diarylpyrimidines and related analogues as HIV-1 nonnucleoside reverse transcriptase inhibitors. Eur J Med Chem. 2018;158:371–392. doi: 10.1016/j.ejmech.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Seidman D., Weber S., Aaron E. Dapivirine vaginal ring for HIV-1 prevention. N Engl J Med. 2017;376:995. doi: 10.1056/NEJMc1616546. [DOI] [PubMed] [Google Scholar]
- 15.Baeten J.M., Palanee-Phillips T., Mgodi N.M., Mayo A.J., Szydlo D.W., Ramjee G., et al. Safety, uptake, and use of a dapivirine vaginal ring for HIV-1 prevention in African women (HOPE): an open-label, extension study. Lancet HIV. 2021;8:e87–e95. doi: 10.1016/S2352-3018(20)30304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cilento M.E., Kirby K.A., Sarafianos S.G. Avoiding drug resistance in HIV reverse transcriptase. Chem Rev. 2021;121:3271–3296. doi: 10.1021/acs.chemrev.0c00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beyrer C., Pozniak A. HIV drug resistance–an emerging threat to epidemic control. N Engl J Med. 2017;377:1605–1607. doi: 10.1056/NEJMp1710608. [DOI] [PubMed] [Google Scholar]
- 18.Battini L., Bollini M. Challenges and approaches in the discovery of human immunodeficiency virus type-1 non-nucleoside reverse transcriptase inhibitors. Med Res Rev. 2019;39:1235–1273. doi: 10.1002/med.21544. [DOI] [PubMed] [Google Scholar]
- 19.Wu G., Zhao T., Kang D., Zhang J., Song Y., Namasivayam V., et al. Overview of recent strategic advances in medicinal chemistry. J Med Chem. 2019;62:9375–9414. doi: 10.1021/acs.jmedchem.9b00359. [DOI] [PubMed] [Google Scholar]
- 20.Ma Y., Frutos-Beltran E., Kang D., Pannecouque C., De Clercq E., Menendez-Arias L., et al. Medicinal chemistry strategies for discovering antivirals effective against drug-resistant viruses. Chem Soc Rev. 2021;50:4514–4540. doi: 10.1039/d0cs01084g. [DOI] [PubMed] [Google Scholar]
- 21.Lansdon E.B., Brendza K.M., Hung M., Wang R., Mukund S., Jin D., et al. Crystal structures of HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278): implications for drug design. J Med Chem. 2010;53:4295–4299. doi: 10.1021/jm1002233. [DOI] [PubMed] [Google Scholar]
- 22.Das K., Bauman J.D., Clark A.D., Jr., Frenkel Y.V., Lewi P.J., Shatkin A.J., et al. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci U S A. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovering F., Bikker J., Humblet C. Escape from flatland: increasing saturation as an approach to improving clinical success. J Med Chem. 2009;52:6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- 24.Wei W., Cherukupalli S., Jing L., Liu X., Zhan P. Fsp3: a new parameter for drug-likeness. Drug Discov Today. 2020;25:1839–1845. doi: 10.1016/j.drudis.2020.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Frey K.M., Bollini M., Mislak A.C., Cisneros J.A., Gallardo-Macias R., Jorgensen W.L., et al. Crystal structures of HIV-1 reverse transcriptase with picomolar inhibitors reveal key interactions for drug design. J Am Chem Soc. 2012;134:19501–19503. doi: 10.1021/ja3092642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kudalkar S.N., Beloor J., Quijano E., Spasov K.A., Lee W.G., Cisneros J.A., et al. From in silico hit to long-acting late-stage preclinical candidate to combat HIV-1 infection. Proc Natl Acad Sci U S A. 2018;115:E802−E11. doi: 10.1073/pnas.1717932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kourounakis A.P., Xanthopoulos D., Tzara A. Morpholine as a privileged structure: a review on the medicinal chemistry and pharmacological activity of morpholine containing bioactive molecules. Med Res Rev. 2020;40:709–752. doi: 10.1002/med.21634. [DOI] [PubMed] [Google Scholar]
- 28.Huang B., Chen W., Zhao T., Li Z., Jiang X., Ginex T., et al. Exploiting the tolerant region I of the non-nucleoside reverse transcriptase inhibitor (NNRTI) binding pocket: discovery of potent diarylpyrimidine-typed HIV-1 NNRTIs against wild-type and E138K mutant virus with significantly improved water solubility and favorable safety profiles. J Med Chem. 2019;62:2083–2098. doi: 10.1021/acs.jmedchem.8b01729. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Kang D., Feng D., Cherukupalli S., Jiang X., Fu Z., et al. Targeting dual tolerant regions of binding pocket: discovery of novel morpholine-substituted diarylpyrimidines as potent HIV-1 NNRTIs with significantly improved water solubility. Eur J Med Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112811. [DOI] [PubMed] [Google Scholar]
- 30.Kang D., Fang Z., Li Z., Huang B., Zhang H., Lu X., et al. Design, synthesis, and evaluation of thiophene[3,2-d]pyrimidine derivatives as HIV-1 non-nucleoside reverse transcriptase inhibitors with significantly improved drug resistance profiles. J Med Chem. 2016;59:7991–8007. doi: 10.1021/acs.jmedchem.6b00738. [DOI] [PubMed] [Google Scholar]
- 31.Kang D., Fang Z., Huang B., Lu X., Zhang H., Xu H., et al. Structure-based optimization of thiophene[3,2-d]pyrimidine derivatives as potent HIV-1 non-nucleoside reverse transcriptase inhibitors with improved potency against resistance-associated variants. J Med Chem. 2017;60:4424–4443. doi: 10.1021/acs.jmedchem.7b00332. [DOI] [PubMed] [Google Scholar]
- 32.Avgy-David H.H., Senderowitz H. Toward focusing conformational ensembles on bioactive conformations: a molecular mechanics/quantum mechanics study. J Chem Inf Model. 2015;55:2154–2167. doi: 10.1021/acs.jcim.5b00259. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y., Tice C.M., Singh S.B. Conformational control in structure-based drug design. Bioorg Med Chem Lett. 2017;27:2825–2837. doi: 10.1016/j.bmcl.2017.04.079. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn B., Guba W., Hert J., Banner D., Bissantz C., Ceccarelli S., et al. A real-world perspective on molecular design. J Med Chem. 2016;59:4087–4102. doi: 10.1021/acs.jmedchem.5b01875. [DOI] [PubMed] [Google Scholar]
- 35.Goto T., Shiina A., Murata T., Tomii M., Yamazaki T., Yoshida K., et al. Identification of the 5,5-dioxo-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidine derivatives as highly selective PDE4B inhibitors. Bioorg Med Chem Lett. 2014;24:893–899. doi: 10.1016/j.bmcl.2013.12.076. [DOI] [PubMed] [Google Scholar]
- 36.Tang G., Kertesz D.J., Yang M., Lin X., Wang Z., Li W., et al. Exploration of piperidine-4-yl-aminopyrimidines as HIV-1 reverse transcriptase inhibitors. N-Phenyl derivatives with broad potency against resistant mutant viruses. Bioorg Med Chem Lett. 2010;20:6020–6023. doi: 10.1016/j.bmcl.2010.08.068. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z., Zalloum W.A., Wang W., Jiang X., De Clercq E., Pannecouque C., et al. Discovery of novel dihydrothiopyrano[4,3-d]pyrimidine derivatives as potent HIV-1 NNRTIs with significantly reduced hERG inhibitory activity and improved resistance profiles. J Med Chem. 2021;64:13658–13675. doi: 10.1021/acs.jmedchem.1c01015. [DOI] [PubMed] [Google Scholar]
- 38.Melikian G.L., Rhee S.Y., Varghese V., Porter D., White K., Taylor J., et al. Non-nucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance: implications for preclinical evaluation of novel NNRTIs and clinical genotypic resistance testing. J Antimicrob Chemother. 2014;69:12–20. doi: 10.1093/jac/dkt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta S., Fransen S., Paxinos E.E., Stawiski E., Huang W., Petropoulos C.J. Combinations of mutations in the connection domain of human immunodeficiency virus type 1 reverse transcriptase: assessing the impact on nucleoside and nonnucleoside reverse transcriptase inhibitor resistance. Antimicrob Agents Chemother. 2010;54:1973–1980. doi: 10.1128/AAC.00870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhee S.Y., Tzou P.L., Shafer R.W. Temporal trends in HIV-1 mutations used for the surveillance of transmitted drug resistance. Viruses. 2021;13:879. doi: 10.3390/v13050879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClung R.P., Oster A.M., Ocfemia M.C.B., Saduvala N., Heneine W., Johnson J.A., et al. Transmitted drug resistance among human immunodeficiency virus (HIV)-1 diagnoses in the United States, 2014‒2018. Clin Infect Dis. 2022;74:1055–1062. doi: 10.1093/cid/ciab583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vingerhoets J., Tambuyzer L., Azijn H., Hoogstoel A., Nijs S., Peeters M., et al. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled Phase III clinical studies. AIDS. 2010;24:503–514. doi: 10.1097/QAD.0b013e32833677ac. [DOI] [PubMed] [Google Scholar]
- 43.Gupta S., Vingerhoets J., Fransen S., Tambuyzer L., Azijn H., Frantzell A., et al. Connection domain mutations in HIV-1 reverse transcriptase do not impact etravirine susceptibility and virologic responses to etravirine-containing regimens. Antimicrob Agents Chemother. 2011;55:2872–2879. doi: 10.1128/AAC.01695-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuman E.C., Rhee S.Y., Holmes S.P., Shafer R.W. Constrained patterns of covariation and clustering of HIV-1 non-nucleoside reverse transcriptase inhibitor resistance mutations. J Antimicrob Chemother. 2010;65:1477–1485. doi: 10.1093/jac/dkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rimsky L., Vingerhoets J., Van Eygen V., Eron J., Clotet B., Hoogstoel A., et al. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. J Acquir Immune Defic Syndr. 2012;59:39–46. doi: 10.1097/QAI.0b013e31823df4da. [DOI] [PubMed] [Google Scholar]
- 46.Xu H.T., Colby-Germinario S.P., Asahchop E.L., Oliveira M., McCallum M., Schader S.M., et al. Effect of mutations at position E138 in HIV-1 reverse transcriptase and their interactions with the M184I mutation on defining patterns of resistance to nonnucleoside reverse transcriptase inhibitors rilpivirine and etravirine. Antimicrob Agents Chemother. 2013;57:3100–3109. doi: 10.1128/AAC.00348-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith S.J., Pauly G.T., Akram A., Melody K., Ambrose Z., Schneider J.P., et al. Rilpivirine and doravirine have complementary efficacies against NNRTI-resistant HIV-1 mutants. J Acquir Immune Defic Syndr. 2016;72:485–491. doi: 10.1097/QAI.0000000000001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenner B.G., Oliveira M., Ibanescu R.I., Routy J.P., Thomas R. Cell culture selections reveal favourable drug resistance profiles for doravirine and islatravir. J Antimicrob Chemother. 2021;76:2137–2142. doi: 10.1093/jac/dkab126. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y., Kang D., Nguyen L.A., Smithline Z.B., Pannecouque C., Zhan P., et al. Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors. Elife. 2018;7 doi: 10.7554/eLife.36340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren J., Nichols C.E., Chamberlain P.P., Weaver K.L., Short S.A., Stammers D.K. Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors. J Mol Biol. 2004;336:569–578. doi: 10.1016/j.jmb.2003.12.055. [DOI] [PubMed] [Google Scholar]
- 51.Guengerich F.P., Waterman M.R., Egli M. Recent structural insights into cytochrome P450 function. Trends Pharmacol Sci. 2016;37:625–640. doi: 10.1016/j.tips.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Havens J.P., Podany A.T., Scarsi K.K., Fletcher C.V. Clinical pharmacokinetics and pharmacodynamics of etravirine: an updated review. Clin Pharmacokinet. 2020;59:137–154. doi: 10.1007/s40262-019-00830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma M., Saravolatz L.D. Rilpivirine: a new non-nucleoside reverse transcriptase inhibitor. J Antimicrob Chemother. 2013;68:250–256. doi: 10.1093/jac/dks404. [DOI] [PubMed] [Google Scholar]
- 54.Wang W., MacKinnon R. Cryo-EM structure of the open human ether-à-go-go-related K+ channel hERG. Cell. 2017;169:422–430. doi: 10.1016/j.cell.2017.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang C., Lai M.T., Hazuda D. Rational design of doravirine: from bench to patients. ACS Infect Dis. 2020;6:64–73. doi: 10.1021/acsinfecdis.9b00178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.