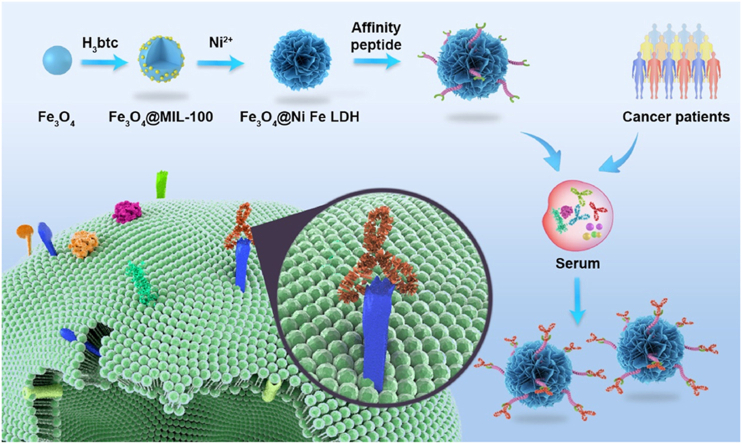
Abstract
Due to low immobilized ligand density, limited binding capacity, and severe interference from serum proteins, developing ideal peptide-based biomaterials for precise recognition and in vivo analysis of biopharmaceuticals remains a huge challenge. In this study, mimotope peptide modified pompon mum-like biomimetic magnetic microparticles (MMPs, 3.8 μm) that mimic the specific functionalities of CD20 on malignant B cells were developed for the first time. Benefit from the numerous ligand binding sites (Ni2+) on the pompon mum-like MMPs, these novel materials achieved ≥10 times higher peptide ligand densities (>2300 mg/g) and antibody binding capacities (1380 mg/g) compared to previous reported biomaterials. Leveraging the high specificity of the mimotope peptide, rituximab can be precisely recognized and enriched from cell culture media or serum samples. We also established an LC‒MS/MS method using the MMPs for tracking rituximab biotransformation in patient serum. Intriguingly, deamidation of Asn55 and Asn33, as well as oxidation of Met81 and Met34 were observed at the key complementarity determining regions of rituximab, which could potentially influence antibody function and require careful monitoring. Overall, these versatile biomimetic MMPs demonstrate superior recognition and enrichment capabilities for target antibodies, offering interesting possibilities for biotransformation analysis of biopharmaceuticals in patient serum.
Key words: Therapeutic monoclonal antibody, Mimotope peptide, Precise recognition, Peptide-based biomaterials, Biotransformation, Patient serum
Graphical abstract
Mimotope peptide modified pompon mum-like biomimetic MMPs that mimic the specific functionalities of CD20 on malignant B cells were developed and applied for precise recognition and biotransformation analysis of rituximab.
1. Introduction
Depending on the 3D structure of peptides, their superior biochemical activity, and diverse functionalities, peptide based biomaterials exhibited fascinating prospects in various application fields, including biomedicine, bioanalysis, bionics, etc.1. For instance, polymer‒peptide conjugates with long circulation time were engineered for tumor targeted delivery to improve therapeutic efficacy2,3 or self-assembled peptide nanomaterials were designed for cancer diagnostics and therapy4,5. In particular, with the rapid development of precision medicine and biopharmaceuticals, peptides as important recognition elements were also used to construct biosensors for clinical diagnosis and therapeutic monitoring6, 7, 8 or fabricate sorbents for the purification and bioanalysis of therapeutic proteins in bioengineering9, 10, 11, 12, 13, 14. Therefore, the exploitation of peptide based biomaterials with desired properties will certainly gain more attention in future.
Recently, short antigenic epitopes or mimotope peptides, which can bind to the antigen-binding fragment (Fab) of the corresponding antibody, have emerged as promising “surrogate antigens” for precise recognition and in vivo analysis of antibodies15, 16, 17. For instance, to rapidly monitor severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) seroconversion, Tedrick et al.18 successfully developed epitope-functionalized gold nanoparticles for selective detection of SARS-CoV-2 IgGs in patient plasma. Some mimotope peptide functionalized affinity membranes or monolithic materials were also designed for the specific enrichment and quantification of different monoclonal antibodies (mAbs), such as trastuzumab, bevacizumab, or infliximab, in complex biological fluids11,19, 20, 21, 22. Although the use of a mimotope peptide can overcome the high manufacturing costs and ligand instability of the native antigen, the low immobilized ligand density and limited antibody binding capacity of the resulting materials will limit their further application to in vivo analysis of antibodies and related products20,23,24.
A few magnetic microparticles (MMPs, larger than 1 μm) with biomimetic porous structures, such as wrinkled flower-like magnetic graphene microspheres, and cabbage-like MMPs, were recently developed, which possess higher magnetic field strength and larger surface area compared to traditional magnetic nanoparticles (MNPs), membrane, or monolithic materials25,26. Therefore, biomimetic MMPs could immobilize affinity ligands with higher density to drive specific binding of more target protein or more precisely recognize low concentrations of the target from complex samples27. For instance, Yang et al. designed wrinkled flower-like magnetic graphene microspheres (6 μm) for the saturation adsorption (317.58 mg/g) of bovine serum albumin (BSA) by strong interactions between the abundant carboxyl groups of dopamine on the surface and the amino groups of BSA25. Liu et al. reported mannatide-zinc-antigen hybrid microparticles (M-Zn-OVA HMPs) with cabbage-like morphology (1.7 μm), which improved the loading capacity of the antigen, and thus resulted in excellent prophylactic and therapeutic efficacy against B16-OVA and E. G7-OVA tumors26. Thanks to their characteristics, biomimetic MMPs could overcome the inherent shortcomings of traditional materials to promote the development of in vivo analytical technology for antibodies. However, to the best of our knowledge, mimotope peptide modified biomimetic MMPs have not been reported to date.
The human cluster of differentiation 20 (CD20), as an important membrane protein and drug target, is overexpressed on malignant B cells (about 5 μm)28,29. Therefore, a series of anti-CD20 antibodies, such as rituximab, obinutuzumab, and mosunetuzumab were developed to treat B-cell related non-Hodgkin's lymphoma, certain types of leukemia and autoimmune diseases. However, some challenges remain for the specific enrichment and in vivo analysis of anti-CD20 antibodies, including the non-specific adsorption of IgGs and other serum proteins, the low concentration of the target antibody, and the low enrichment capacity of traditional materials. In this study, inspired by CD20-rituximab specific interactions, mimotope peptide (HN19: HHHHHHGSGSGSWPRWLEN or HE24: HHHHHHGSGSGSQDKLTQWPKWLE) modified pompon mum-like MMPs (3.8 μm, similar to B-cells) were designed to mimic the specific functionalities of CD20 for the first time. The biomimetic MMPs were fabricated via a metal-organic framework (MOF)-mediated cascade derivation strategy, and then the CD20 mimotope peptide was immobilized by metal chelation interaction between the histidine tag of the peptide and nickel ion on the MMP surface. Afterwards, the specificity, binding capacity and anti-fouling ability of the biomimetic MMPs were systematically evaluated, a comparison between the MMPs and the previously reported materials was made to highlight the advantages of the novel material. Moreover, the application potential of the MMPs to the precise enrichment of rituximab was evaluated by testing a series of spiked cell culture mediums and human serum samples. In particular, the biotransformation analysis of rituximab in non-Hodgkin's lymphoma patient serum samples was monitored by combining selective capture on the novel MMPs, trypsin digestion, and LC‒QTOF-MS.
2. Materials and methods
2.1. Chemicals and reagents
Ferric chloride (FeCl3·6H2O, AR), sodium formate (HCOONa, 98%), monosodium phosphate (NaH2PO4, 99%), disodium hydrogen phosphate (Na2HPO4, 99%), sodium chloride (NaCl, 98%), sodium hydroxide (NaOH, 98%), sodium acetate (CH3COONa, 98%), urea (CH4N2O, ≥99.5%), trimesic acid (H3btc, 98%) were purchased from Aladdin (Shanghai, China). Nickel (II) sulfate hexahydrate (NiSO4·6H2O, ≥98.5%), polyethylene glycol (PEG 6000), ethylene glycol ((CH2OH)2, 98%) were bought from Macklin reagent (Shanghai, China). Mimotope HN19 and mimotope HE24 were synthesized by GenScript (Nanjing, China) with purity over 95%. Rituximab (MabThera®) was purchased from China Roche (Shanghai, China). Human serum albumin (HSA, 95%) was from Equitech-bio (Beijing, China). Myoglobin (MYO, ≥90%) was from yuanye Bio-Technology (Shanghai, China). Human immunoglobulin G (hIgG, >95%) and β-lactoglobulin (β-Lg, >90%) were bought from Sigma‒Aldrich (Shanghai, China). SDS-PAGE gel kit and BCA assay kit were purchased from Beyotime (Shanghai, China). Sequencing grade modified trypsin was obtained from Promega (Beijing, China). RPMI-1640 cell culture mediums were gifted by other groups from Jinan University (Guangzhou, China). Non-Hodgkin's lymphoma patients' serum samples were obtained from The First Affiliated Hospital of Jinan University (Guangzhou, China). Samples were collected under appropriate ethical approval and after written informed consent of patients.
2.2. Synthesis of biomimetic MMPs
The biomimetic MMPs was prepared by solvothermal method in three steps. Firstly, Fe3O4 nanoparticles and Fe3O4@MIL-100(Fe) were synthesized according to the method in the Supporting Information. Subsequently, 50 mg Fe3O4@MIL-100(Fe), 200 mg NiSO4·6H2O, and 120 mg urea were successively added into 40 mL water with stirring at room temperature to form a homogeneous suspension. Then, this suspension was transferred into a sealed Teflon-lined stainless steel autoclave at 120 °C for 12 h. After cooling to room temperature, the resulting orange product was separated using an external magnet and washed several times with deionized water. Finally, the dried pompon mum-like Fe3O4@NiFe LDH MMPs were obtained.
Then, 2.0 mg Fe3O4@NiFe LDH MMPs were suspended in 500 μL deionized water and the HN19 mimotope peptide (500 μL of 1 mg/mL solution) was added. The mixture was rotated and incubated at room temperature for 12 h. The Fe3O4@NiFe LDH@HN19 were separated from the supernatant in a magnetic field and washed thrice with deionized water to remove the unbound peptide. The preparation process of Fe3O4@NiFe LDH@HE24 was the same as that of Fe3O4@NiFe LDH@HN19.
2.3. Specificity and anti-fouling ability of biomimetic MMPs
To investigate the specificity of the biomimetic material for rituximab, 2 mg MMPs were added to a protein mixture containing 100 μL of 4 mg/mL BSA, 100 μL of 1 mg/mL rituximab or hIgG, 100 μL of 1 mg/mL myoglobin and 100 μL of 1 mg/mL β-lactoglobulin, mixed and incubated for 3 h, and then separated in a magnetic field. Then, the washing buffer (20 mmol/L PB, 150 mmol/L NaCl, pH 8.0) was added to the MMPs. After magnetic separation, the elution buffer (10 mmol/L HCOONa, 100 mmol/L NaCl, pH 3.05) was used, and the obtained supernatant was analyzed by SDS-PAGE.
To investigate the non-specific adsorption of impurity protein, FITC-labeled BSA was selected as model protein. 2 mg MMPs were added to 1 mL BSA solution (1 mg/mL), mixed and incubated for 3 h, separated in a magnetic field, and their fluorescence was observed under UV light.
2.4. Binding performances of Fe3O4@NiFe LDH@HN19/HE24 MMPs
For binding performance evaluation, 0.2 mg of MMPs were immersed in 10 mL of 0.1 mg/mL rituximab solution, mixed and incubated (0–6 h). At the specified time intervals, 1 mL of the mixture was removed, magnetically separated, and the remaining antibody concentration was determined. Then, the binding capacity was calculated according to Eq. (1):
| (1) |
where C0 and Ct are the rituximab concentrations initially and at time t, respectively, V is the volume of the rituximab solution, and m is the amount of MMPs.
In the isothermal binding experiment, 0.2 mg of MMPs was added to 10 mL rituximab solutions of different concentrations (ranging from 0.01 to 0.1 mg/mL), mixed and incubated for 3 h. After magnetic separation, the remaining antibody concentration was determined, and the binding capacity was calculated according to Eq. (1).
2.5. Enrichment and analysis of rituximab in complex biological fluids
RPMI-1640 cell culture medium and diluted human serum spiked with rituximab were used to evaluate the application potentials of the biomimetic MMPs. Specifically, rituximab (1 mg/mL, 100 μL) was added to RPMI-1640 cell culture medium or ten-fold diluted human serum, mixed and incubated with different MMPs for 3 h. The mixture solution were separated in a magnetic field, and after the washing and elution steps, the resulting supernatant was subjected to further analysis (including SDS-PAGE, circular dichromatic, and bioactivity analysis).
2.6. Biotransformation analysis of rituximab in non-Hodgkin's lymphoma patient serum samples
For peptide mapping analysis, the eluates containing rituximab captured from clinically diluted five-fold serum samples were first desalted and the solvent was exchanged for water using the Amicon® Ultra-0.5 centrifugal filter unit (10 kDa MWCO, Merck Millipore, Burlington, MA, USA). Subsequently, 10 mol/L urea in 100 mmol/L Tris-HCl buffer pH 8.0 and 10 mmol/L dithiothreitol were added for denaturation and reduction of disulfide bonds, respectively, followed by 20 mmol/L iodoacetamide for alkylation. After changing the solvent to 100 mmol/L Tris-HCl pH 8.0 through ultrafiltration, the proteins were digested with trypsin at 37 °C for 3 h and the digestion was quenched by adding 5% FA. The peptides resulting from enzymatic digestion were separated on an ACQUITY UPLC BEH C18 column (2.1 mm I.D. × 150 mm, 1.7 μm, 130 Å, Waters, Milford, MA, USA) using an ExionLC system coupled to the SCIEX ZenoTOFTM 7600 system (Framingham, MA, USA). The mobile phase A was composed of 0.1% FA in water while the mobile phase B consisted of 0.1% FA in ACN. A 120 min gradient elution at a flow rate of 200 μL/min and a column temperature of 60 °C was carried out for peptide separation. Detailed method parameters were summarized in Supporting Information Table S1. For qualitative and relatively quantitative analysis, SCIEX OS and BioPharma view software were used.
3. Results and discussion
3.1. Preparation of pompon mum-like biomimetic MMPs
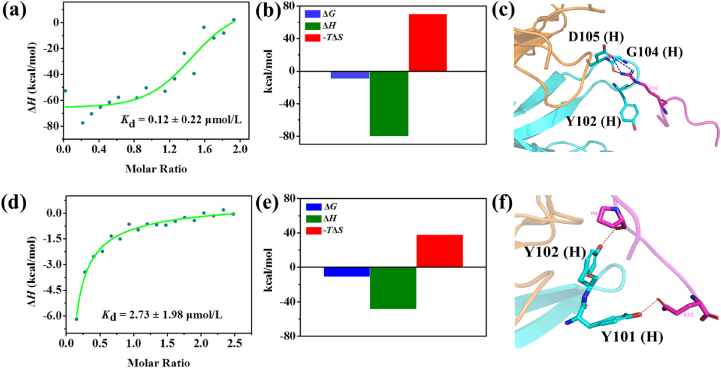
To mimic the functionalities of CD20 on malignant B cells, two mimotope peptides (HN19: HHHHHHGSGSGSWPRWLEN and HE24: HHHHHHGSGSGSQDKLTQWPKWLE) were moderately modified and designed for rituximab recognition according to the previously reported studies17,29. Firstly, the affinities of the two peptides to rituximab were evaluated by isothermal titration calorimetry (ITC) assay. As shown in Fig. 1, the affinity of HN19 (Kd = 0.12 ± 0.22 μmol/L) to rituximab was slightly higher than that of HE24 (Kd = 2.73 ± 1.98 μmol/L). Moreover, as shown in Fig. 1b and e, ΔG < 0 and |ΔH| > |TΔS|, which indicated that the environments of the binding between the mimotope peptide and rituximab were consistent with the enthalpy‒entropy compensation of enthalpy drive on driving process30, and therefore the hydrogen bond and electrostatic interactions could dominate their binding behavior. The binding models for these mimotope peptides with the Fab region of rituximab were then investigated (Fig. 1c and f). For the key recognition sequence (WPRWLEN) of HN19, residue Arg3 was a key site, mainly involved in binding with residues Tyr102, Gly104, and Asp105 in the Fab region of the heavy chain of rituximab through salt bridge and hydrogen bond interactions. For the key recognition sequence (QDKLTQWPKWLE) of HE24, residues Pro8 and Glu12 were involved in binding with residues Tyr102 and Tyr101 in the heavy chain of rituximab (Fab region) through hydrogen bond interactions. Compared to the binding behavior of HE24, HN19 exhibited more interactions (salt bridges) with rituximab Fab, which indicated that the binding affinity of HN19 to rituximab could be higher than that of HE24. The above results showed that these peptides could efficiently mimic the functionalities of CD20 and lead to the development of versatile biomaterials.
Figure 1.
Binding analysis of mimotope peptide with rituximab. (a) ITC analysis of HN19 with rituximab; (b) Energy changes during binding between HN19 and rituximab; (c) Molecular docking of key recognition sequence of HN19 with rituximab Fab; (d) ITC analysis of HE24 with rituximab; (e) Energy changes during binding between HE24 and rituximab; (f) Molecular docking of key recognition sequence of HE24 with rituximab Fab.
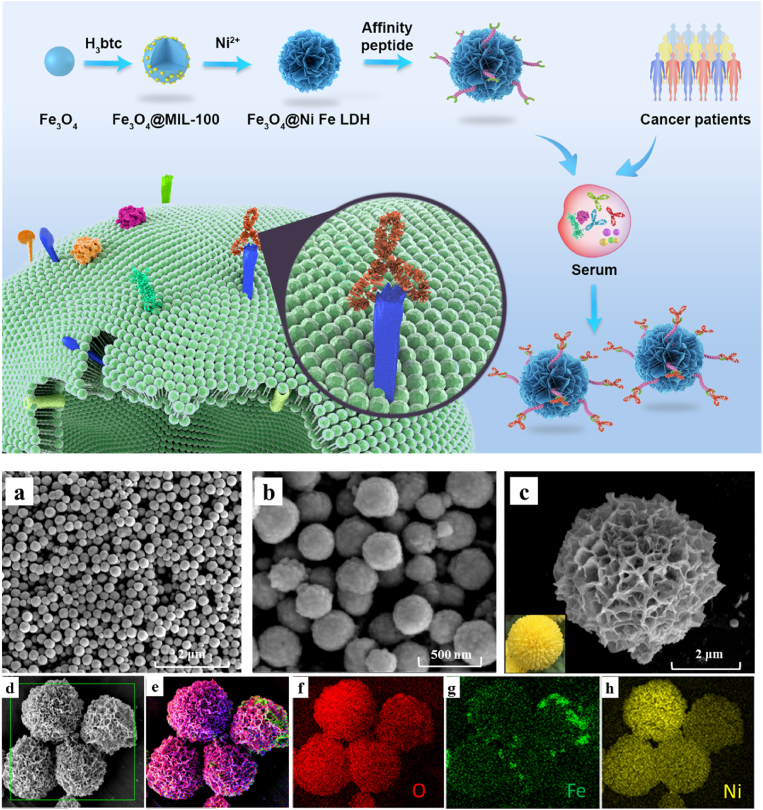
Subsequently, mimotope peptide functionalized MMPs were prepared in four steps. As shown in Fig. 2, the Fe3O4 nanoparticles were first synthesized, and then trimesic acid (H3btc) was added and reacted with Fe3+ ions on the surface of these nanoparticles to produce uniform Fe3O4@MIL-100(Fe) in situ. Then, the Fe ions in the MOF layer of Fe3O4@MIL-100(Fe) were reacted with nickel sulfate hexahydrate (NiSO4·6H2O) to produce pompon mum-like MMPs (Fe3O4@NiFe LDH, μm level, Fig. 2c). The generation of the Ni layer and the formation of microparticles could be attributed to the fact that hydroxide ions in alkaline environment (urea at high temperature) can slowly coprecipitated with nickel ions through high-temperature hydrothermal decomposition, coupled with the continuous connection of individual nanoparticles over time and the aggregation effect of MNPs31. According to the previous report32, the pompon mum-like microparticles possess high porosity and large specific surface area to provide numerous ligand binding sites, which was beneficial for the improvement of the resultant material's functionality. Finally, the HN19 or HE24 modified biomimetic MMPs (Fe3O4@NiFe LDH@peptide) were prepared by metal chelation interaction between the histidine tag of the peptide ligand and Ni2+ on the Fe3O4@NiFe LDH MMPs.
Figure 2.
Characterization of the materials. Scanning electron microscopy (SEM) images of (a) Fe3O4, (b) Fe3O4@MIL-100(Fe) and (c) Fe3O4@NiFe LDH. (d) SEM image of Fe3O4@NiFe LDH and Energy dispersive spectrometer (EDS) images for (e) the superposition of the elements, (f) O element, (g) Fe element, (h) Ni element.
3.2. Characterization of the pompon mum-like biomimetic MMPs
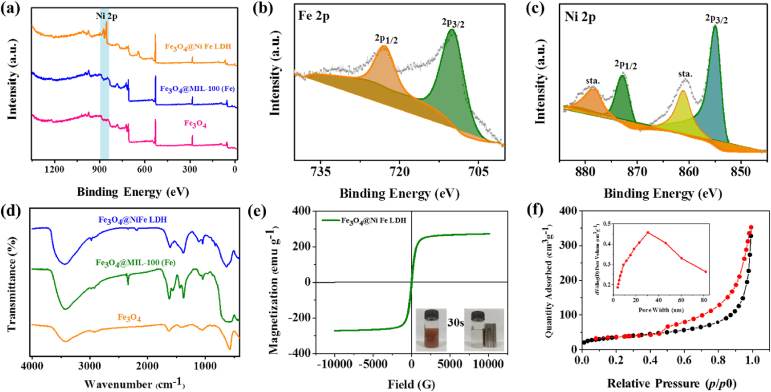
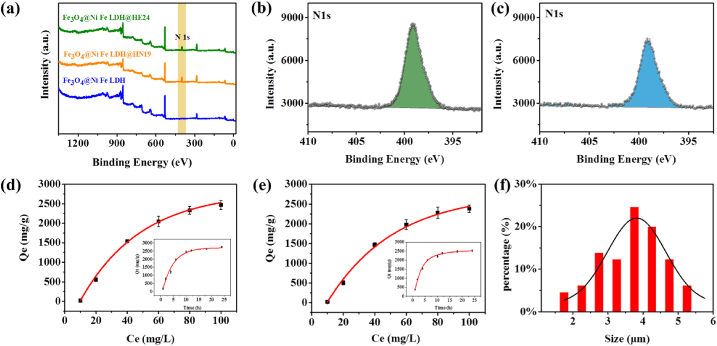
To verify the successful preparation of the biomimetic MMPs, their morphological changes during the synthesis of the material were first monitorized by SEM (Fig. 2a‒c). With the nucleation of Fe3O4, the generation of Fe3O4@MIL-100(Fe), followed by the continuous coprecipitation and aggregation of Ni layer on Fe3O4@MIL-100(Fe), the nanoparticles eventually grew into pompon mum-like Fe3O4@NiFe LDH microparticles. The particle size of the corresponding materials were gradually increased from 100 nm (Fe3O4) to 300 nm (Fe3O4@MIL-100(Fe)), and after aggregation finally grew to 3.8 μm (Fe3O4@NiFe LDH) (Fig. 2c). EDS results further confirmed that the abundant Ni element (yellow part, Fig. 2h) was evenly distributed on the material surface. X-ray photoelectron spectroscopy (XPS) results (Fig. 3a) also showed that the characteristic peaks of Ni at 855.0 and 872.0 eV were observed in the high-resolution XPS spectrum of Fe3O4@NiFe LDH in comparison to that of Fe3O4@MIL-100(Fe). The high-resolution XPS spectrum of Ni 2p exhibited four peaks, corresponding to Ni 2p3/2, Ni 2p1/2 and two shakeup satellites (Fig. 3c). Moreover, the infrared spectrum of Fe3O4@NiFe LDH was similar to that of Fe3O4@MIL-100(Fe), which could be attributed to the coprecipitation of Ni ions onto the matrix material by non-covalent interaction (Fig. 3d). Therefore, these characterizations symmetrically verified the successful preparation of Fe3O4@NiFe LDH. In particular, the abundant Ni element (37.84 wt%, Supporting Information Table S2), super-strong magnetism (270 emu/g, Fig. 3e), and large specific surface area (128.43 m2/g, Fig. 3f) were also observed on Fe3O4@NiFe LDH, which is beneficial for the high density immobilization of the peptide ligand to produce the versatile biomimetic Fe3O4@NiFe LDH@peptide MMPs.
Figure 3.
Characterization of the materials. (a) XPS survey data of Fe3O4, Fe3O4@MIL-100(Fe) and Fe3O4@NiFe LDH. High-resolution XPS spectra of (b) Fe and (c) Ni of Fe3O4@NiFe LDH. (d) Fourier transform infrared (FT-IR) spectra of Fe3O4, Fe3O4@MIL-100(Fe) and Fe3O4@NiFe LDH. (e) Magnetic hysteresis loops of Fe3O4@NiFe LDH. (f) N2 sorption isotherms of Fe3O4@NiFe LDH.
The efficient immobilization of the mimotope peptide on Fe3O4@NiFe LDH was then monitored by XPS and BCA tests. The characteristic N 1s peak was observed in the full-survey XPS spectrum of the biomimetic MMPs (Fig. 4a), which implied that the peptide ligand (HN19 or HE24) was successfully grafted on the material surface. However, the N intensity of Fe3O4@NiFe LDH@HN19 (Fig. 4b) was higher than that of Fe3O4@NiFe LDH@HE24 (Fig. 4c), which could be due to a difference in peptide density on the biomimetic materials. According to the peptide concentration-dependent experiments, the maximum HN19 density on Fe3O4@NiFe LDH@HN19 was 2467 mg/g. This was found to be slightly higher than the HE24 density of 2382 mg/g on Fe3O4@NiFe LDH@HE24 (Fig. 4d and e). It is interesting to note that these ligand densities are dramatically higher than those of previously reported peptide based affinity materials (Table 1). The ultrahigh peptide ligand density could be attributed to the abundant Ni2+ chelation sites, large specific surface area, and high porosity of Fe3O4@NiFe LDH.
Figure 4.
Characterization of materials and evaluation of maximum peptide ligand density. (a) XPS survey data of Fe3O4@NiFe LDH, Fe3O4@NiFe LDH@HN19 and Fe3O4@NiFe LDH@HE24. High-resolution XPS spectra of (b) Ni on Fe3O4@NiFe LDH@HN19 and (c) Ni on Fe3O4@NiFe LDH@HE24. (d) Maximum HN19 density on Fe3O4@NiFe LDH@HN19. (e) Maximum HE24 density on Fe3O4@NiFe LDH@HE24. (f) Size distribution histogram of Fe3O4@NiFe LDH@peptide.
Table 1.
Comparison of binding performances of the previously reported affinity materials.
| Material | Ligand | Ligand density | Target protein | Binding capacity | Ref. |
|---|---|---|---|---|---|
| Sepharose beads | AviPure ligand | 429 nmol/g | IgG | 53 mg/mL | 38 |
| Agarose-OPS adsorbent | 31.93 mg/mL | IgG | 24.2 mg/mL | 23 | |
| Porous membranes containing immobilized peptide | KGSGSGSQLGPYELWELSH (KH19) | 5.1 ± 1.3 mg/mL | Trastuzumab | 1.3 ± 0.3 mg/mL | 37 |
| Peptide immobilized monolith | HHHHHHGSGSGSQLGPYELWELSH (HH24) | 11 mg/mL | Trastuzumab | 16.4 mg/g | 13 |
| Peptide immobilized monolith | Fmoc-HWEGWV | 155 μmol/mL | IgG | 101.8 mg/mL | 39 |
| Fe3O4@SiO2 magnetic nanoparticles | Protein A | 203 mg/g | anti-EGFR mAb | 112.3 mg/g | 24 |
| Peptide immobilized monolith | Histidine-tagged cyclic peptide | 13.8 mg/mL | IgG | 119.3 mg/g | 11 |
| His-MWNTs | l-Histidine | 210 μmol/g | IgG | 267.8 mg/g | 40 |
| Porous membranes containing immobilized peptide | KGSGSGSWPRWLEN (KN14) | / | Rituximab | 16.5 mg/mL | 19 |
| Fe3O4@NiFe LDH@HN19 Fe3O4@NiFe LDH@HE24 |
HN19 HE24 |
2467 mg/g 2382 mg/g |
Rituximab Rituximab |
1375 mg/g 1194 mg/g |
This work |
3.3. Optimization of bioseparation conditions
The separation performances of the biomimetic MMPs in serum were commonly limited by the low concentration of the target protein and the serious interferences from high abundance proteins33,34. Therefore, to ensure the proper functioning of the obtained Fe3O4@NiFe LDH@peptide MMPs, the separation conditions (including the composition and pH of the washing and elution buffers) were systematically optimized. Firstly, the washing step plays a crucial role in bioseparation process for the elimination of interference proteins. Therefore, the influence of the pH and salt concentration of the washing buffer on the adsorption performances of the biomimetic MMPs was evaluated (Supporting Information Fig. S1). A protein mixture containing 1 mg/mL β-lactoglobulin, 1 mg/mL myoglobin, 1 mg/mL rituximab, and 4 mg/mL HSA was chosen as test analyte. At pH 8.0, the electrostatic repulsion interactions between the mimotope peptide HN19 (pI 7.7, from peptide properties calculator, NovoPro, Shanghai, China) and interference proteins (HSA pI: 4.7, β-lactoglobulin pI: 5.2, myoglobin pI: 6.9) could effectively eliminate their non-specific adsorption on the biomimetic MMP surface (Supporting Information Fig. S2a). Moreover, the highest recovery (98.4%) and purity for rituximab (pI: 9.3)35 was obtained at this pH (Figs. S1a and S2a). The salt concentration of the washing buffer could affect the hydrophobic interactions between the mimotope peptide and rituximab, and therefore the influence of NaCl concentration was also investigated (Figs. S1b and S2b). An appropriate salt concentration (150 mmol/L NaCl) could raise the surface tension of the solution and diminish the electrostatic interaction with interference proteins, however a lower salt concentration (50 mmol/L NaCl) or higher salt concentration (250 mmol/L NaCl) would lead to a rituximab loss in the washing fraction and poor enrichment efficiency36. Therefore, a washing buffer of pH 8.0 consisting of 20 mmol/L PB and 150 mmol/L NaCl, was selected for further studies.
Finally, three kinds of elution buffer were investigated (Fig. S1c), including eluent 1 (10 mmol/L sodium formate, 100 mmol/L NaCl, pH 3.0), eluent 2 (25% ACN, 0.1% formic acid), and eluent 3 (10 mmol/L urea) with strong elution ability. Although the highest recovery was achieved with 10 mmol/L urea, the biomimetic MMPs could be destroyed under harsh conditions. Compared to eluents 2 and 3, an acceptable recovery (>88%) and good stability of the material were obtained with eluent 1. Moreover, these moderate conditions can overcome the drawbacks of the harsh elution conditions (2% sodium dodecyl sulfate, 100 mmol/L dithiothreitol) of the traditional peptide based affinity membranes20, so that the bioactivity of the enriched antibody will be well maintained for further studies of antibody functions, including antigen binding affinity, antibody-dependent cellular cytotoxicity, complement-dependent cytotoxicity, etc. Therefore, elution buffer 1 (10 mmol/L sodium formate, 100 mmol/L NaCl, pH 3.0) was selected for further studies.
3.4. Specificity, anti-fouling ability, and antibody binding capacity of the biomimetic MMPs
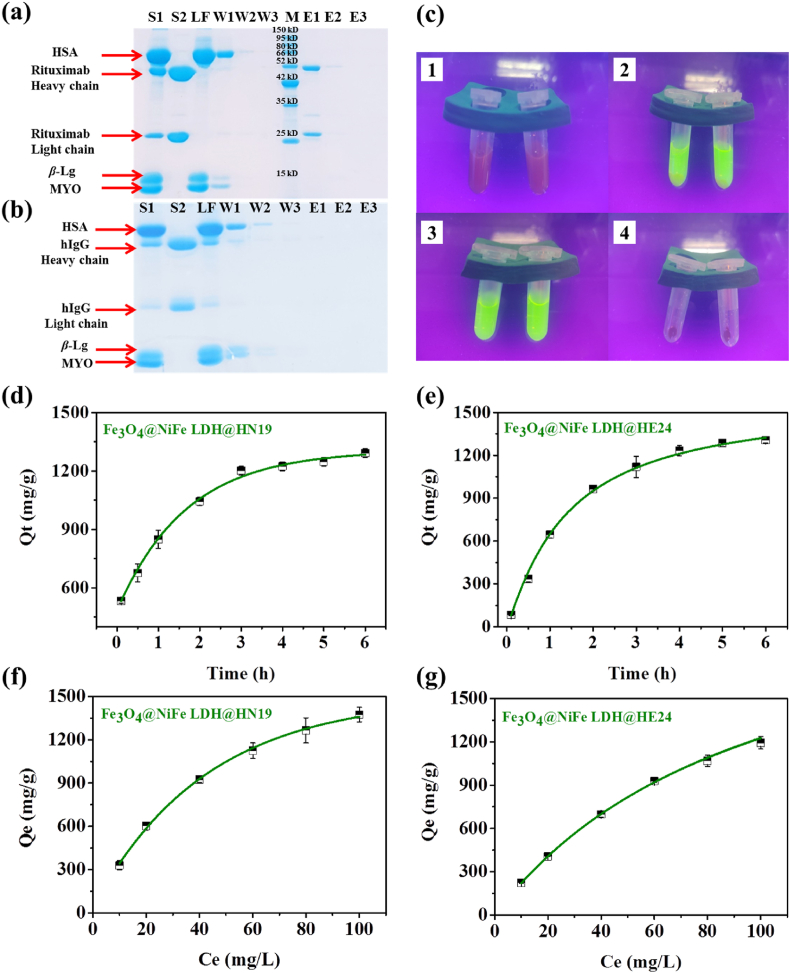
Under the optimized conditions, the selectivity of the biomimetic MMPs was first evaluated using a mixture of proteins (rituximab, HSA, myoglobin, and β-lactoglobulin) as analytes. Fig. 5a shows that only rituximab was detected in the elution fraction and the other proteins were observed in the loading and washing fractions. Moreover, a mixture including hIgG instead of rituximab was used for comparison. As shown in Fig. 5b, no obvious hIgG was detected in the elution fraction. Therefore, the biomimetic MMPs possesses excellent specificity for rituximab. Then, the anti-fouling ability of the biomimetic MMPs was visually evaluated by a FITC-labeled BSA adsorption test. As shown in Fig. 5c-2, a very strong fluorescence was observed after incubation of biomimetic MMPs with FITC-labeled BSA. After magnetic separation, no fluorescence was observed on the biomimetic MMPs (Fig. 5c-4), which confirms their excellent anti-fouling ability.
Figure 5.
(a) Selectivity of Fe3O4@NiFe LDH@HN19 evaluated by SDS-PAGE analysis. S1, feedstock; S2, standard rituximab (1 mg/mL); LF, loading fractions; W1‒W3, washing fractions; M, marker; E1‒E3, elution fractions. (b) Specificity of Fe3O4@NiFe LDH@HN19 evaluated by SDS-PAGE analysis. S1, feedstock; S2, standard hIgG (1 mg/mL); LF, loading fractions; W1‒W3, washing fractions; M, marker; E1‒E3, elution fractions. Stain of protein lane was achieved with Coomassie blue staining solution. (c) Anti-fouling test of Fe3O4@NiFe LDH@HN19 by fluorescence analysis. (1) Fe3O4@NiFe LDH@HN19. (2) Fe3O4@NiFe LDH@HN19 after addition of FITC labeled BSA solution. (3) Supernatant obtained after magnetic separation. (4) Precipitate obtained after magnetic separation. (d) Binding kinetics isotherm of rituximab on Fe3O4@NiFe LDH@HN19. (e) Binding kinetics isotherm of rituximab on Fe3O4@NiFe LDH@HE24. (f) Maximum rituximab binding capacities on Fe3O4@NiFe LDH@HN19. (g) Maximum rituximab binding capacities on Fe3O4@NiFe LDH@HE24.
Considering the excellent specificity of the pompon mum-like biomimetic MMPs, their antibody binding performances, including binding kinetics and equilibrium binding isotherm, were also evaluated. Firstly, the binding kinetics of the biomimetic MMPs for rituximab were assessed in a predetermined time interval, and the biomimetic MMPs (Fe3O4@NiFe LDH@HN19 and Fe3O4@NiFe LDH@HE24) were separately incubated with rituximab and the amount of antibody bound to the MMPs was measured at different times. As illustrated in Fig. 5d and e, Fe3O4@NiFe LDH@HN19 and Fe3O4@NiFe LDH@HE24 showed fast binding rates for rituximab during the first 3 h, then the binding equilibrium was reached after about 4 h. The maximum rituximab binding capacities of these biomimetic MMPs were 1291 mg/g (Fe3O4@NiFe LDH@HN19) and 1306 mg/g (Fe3O4@NiFe LDH@HE24), respectively. Next, the equilibrium adsorption isotherm was applied to evaluate the binding capacities of Fe3O4@NiFe LDH@HN19 and Fe3O4@NiFe LDH@HE24 for rituximab. Similarly, the biomimetic MMPs (Fe3O4@NiFe LDH@HN19 or Fe3O4@NiFe LDH@HE24) were separately incubated with a series of increasing concentrations of rituximab (from 10 to 100 mg/L), and the maximum binding capacity of Fe3O4@NiFe LDH@HN19 was calculated to be 1375 mg/g, and that of Fe3O4@NiFe LDH@HE24 was calculated to be 1194 mg/g (Fig. 5f and g). The higher binding capacity of the former could be due to that the higher affinity and density of HN19 on the material surface.
More importantly, the larger size and abundant apertures of the novel MMPs also resulted in ligand density and antibody binding capacity 10 times higher than those of protein A (203.0 mg/g) immobilized MNPs for capturing monoclonal antibodies (112.3 mg/g)24 and the previously reported fragment crystallizable (Fc)-specific peptide based monoliths for enriching hIgG (119.3 mg/g11 or 101.8 mg/mL39) In particular, the peptide density and binding ability of the biomimetic MMPs were significantly superior than those of the mimotope peptide KH19 (5.1 mg/mL) immobilized affinity membrane towards trastuzumab (1.3 mg/mL)37 and the HH24 (11.0 mg/mL) modified polymeric monolith towards trastuzumab (16.4 mg/g)13, and even the KN14 modified affinity membrane towards rituximab (16.5 mg/mL)19. In a word, Fe3O4@NiFe LDH@peptide MMPs exhibited fascinating performances with higher binding capacity when compared to those of previously reported protein A, Fc-specific peptide, and antigen mimotope peptide functionalized materials as summarized in Table 1.
3.5. Enrichment and analysis of rituximab in complex biological fluids
Inspired by the above results, the actual application potential of the novel MMPs was further investigated by purifying a series of complex biological samples spiked with rituximab and verifying its bioactivity and stability. Firstly, with the increase of culture scale and cell expression level, the upstream production capacity of mAbs was gradually enhanced, so that purification technologies with high efficiency and high loading capacity, such as biomimetic MMPs, were increasingly desired. As shown in Supporting Information Fig. S3, RPMI-1640 cell culture mediums spiked with rituximab (1 mg/mL) were used as test samples and only the heavy and light chains of rituximab were observed in the SDS-PAGE images of all elution fractions after enrichment. Hence, these biomimetic MMPs exhibited a good application prospect for mAb purification in biopharmaceutical manufacturing process.
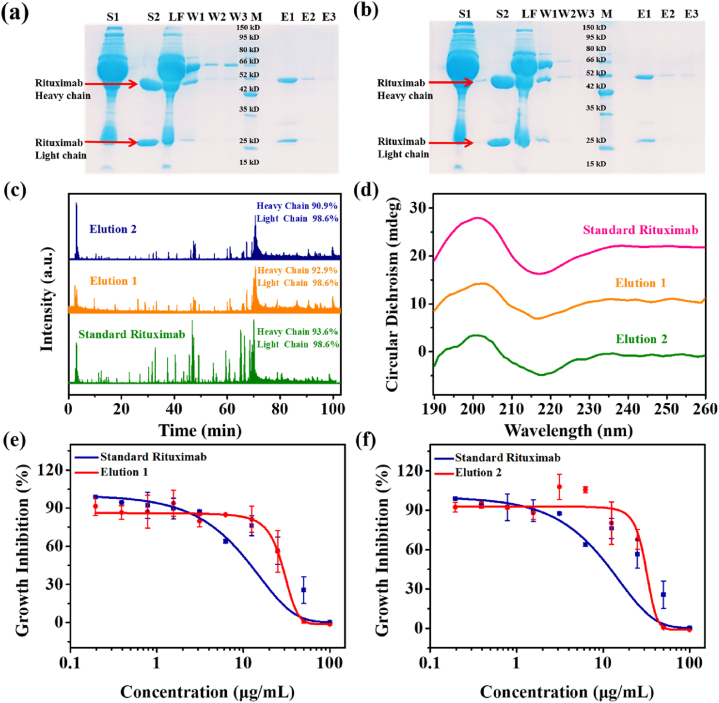
Secondly, some obstacles and challenges, including low in vivo concentrations of antibodies and complex biological matrices containing IgGs, could seriously interfere with antibody quantification and biotransformation analysis in clinical samples41. Therefore, the precise recognition and enrichment ability of the novel MMPs were also evaluated by using 10-fold diluted spiked serum samples. As can be seen in Fig. 6a and b, thanks to the excellent affinity, specificity and enrichment features of the novel material, almost all of interference proteins were eliminated by washing steps and the target antibody was detected with high purity in the elution fractions by SDS-PAGE. In particular, the peptide mapping of the eluted protein was performed by LC‒QTOF-MS after trypsin digestion. Fig. 6c shows that similar peptide sequence coverages were obtained for standard rituximab (98.6% for light chain and 93.6% for heavy chain) and the eluted protein (98.6% for light chains and 90.9%‒92.9% for heavy chains). These results further confirmed the excellent enrichment ability of these biomimetic MMPs. Moreover, the second structure and bioactivity of the eluted rituximab were monitored and compared. As shown in Fig. 6d, the characteristic peaks of β-sheets (200 and 216 nm) were found in CD curves of standard rituximab and the eluted protein. The cytotoxicity assay of the eluted rituximab further indicated that its IC50 values were 23.89 μg/mL (when eluted from Fe3O4@NiFe LDH@HN19) and 28.43 μg/mL (when eluted from Fe3O4@NiFe LDH@HE24), which were close to the IC50 (21.10 μg/mL) of standard rituximab (Fig. 6e and f). Based on the above, these biomimetic MMPs not only have a good application prospect for antibody capture from spiked serum samples, but also the secondary structure and high bioactivity of the eluted rituximab can be well maintained, which is beneficial for further antibody quantification, function study, and even in vivo biotransformation analysis.
Figure 6.
Enrichment process of rituximab in spiked human serum. (a) SDS-PAGE results of rituximab enrichment on Fe3O4@NiFe LDH@HN19 from 10-fold diluted spiked human serum. (b) SDS-PAGE results of rituximab enrichment on Fe3O4@NiFe LDH@HE24 from 10-fold diluted spiked human serum. S1, feedstock; S2, standard rituximab; LF, loading fractions; W1‒W3, washing fractions; M, marker; E1‒E3, elution fractions. Stain of protein lane was achieved with Coomassie blue staining solution. (c) Mass spectra and sequence coverages of standard rituximab and the eluted protein from Fe3O4@NiFe LDH@HN19 and Fe3O4@NiFe LDH@HE24. (d) Circular dichromatic (CD) spectra of standard rituximab and the eluted protein from Fe3O4@NiFe LDH@HN19 and Fe3O4@NiFe LDH@HE24. Bioactivity analysis (cytotoxicity assay) of standard rituximab and the eluted protein from Fe3O4@NiFe LDH@HN19 (e) or from Fe3O4@NiFe LDH@HE24 (f).
3.6. Biotransformation analysis of rituximab in non-Hodgkin's lymphoma patient serum samples
After administration of mAbs to patients, various chemical modifications, such as deamidation, isomerization, oxidation, and glycation, may be formed or accelerated and result in decreased antigen binding affinity, reduced drug activity, and undesirable changes in pharmacokinetics and immunogenicity42,43. Therefore, the in vivo biotransformation analysis of mAbs is of high interest for the development, precise therapeutic use, and risk assessment of biopharmaceuticals. Based on our previously reported study on biotransformation analysis13, a reliable bioanalytical platform was developed by combining rituximab enrichment on biomimetic MMPs, trypsin digestion and peptide mapping by LC‒QTOF-MS. To exclude the potential interference of complex matrices, blank serum and rituximab spiked serum sample were employed as analytes. Some possible peptide modification sites for rituximab were summarized in Supporting Information Table S3. As shown in Supporting Information Fig. S4, no obvious serum protein interference was observed by comparing extracted ion chromatograms (XICs) of blank serum (a) and rituximab-spiked serum (b) sample (all samples were analyzed after treatment with Fe3O4@NiFe LDH@HN19 and trypsin digestion). Moreover, high response intensities were achieved for the potential modified peptides of rituximab, which verify the reliability of the bioanalytical platform.
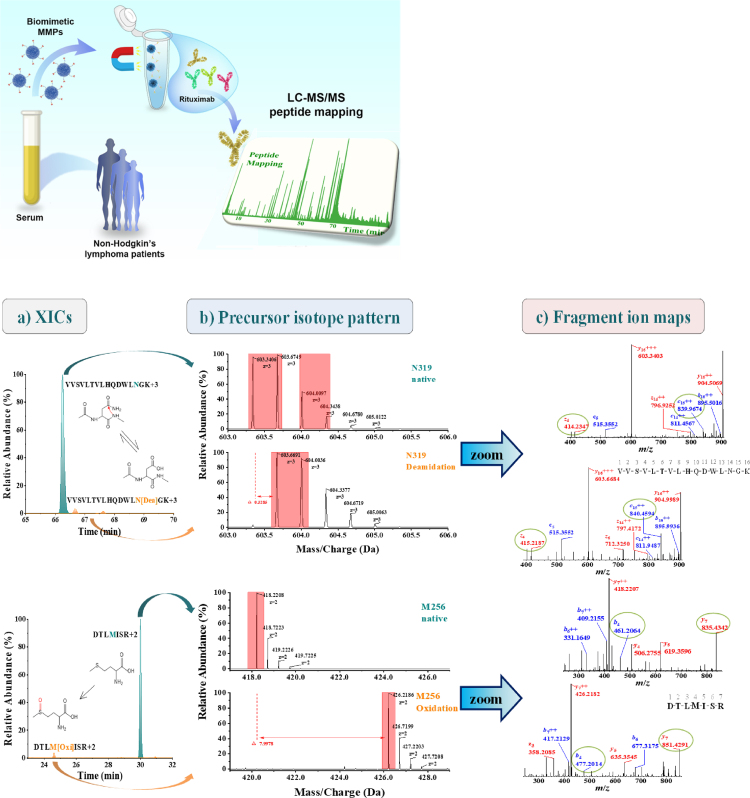
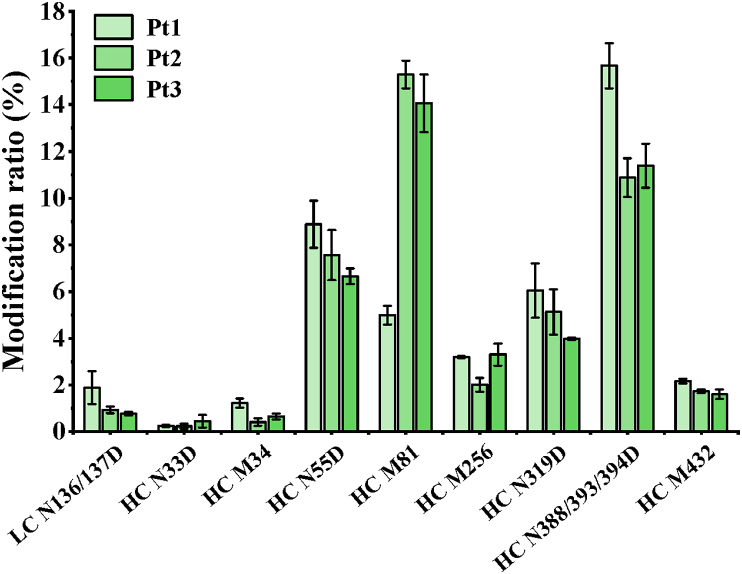
For biotransformation analysis, the two MMPs were then applied to enrich rituximab from 5-fold diluted serum from patients with non-Hodgkin's lymphoma. As shown in Supporting Information Fig. S5, the characteristic heavy and light chains of rituximab were observed in SDS-PAGE images. The obtained protein was then digested by trypsin for peptide mapping. Based on the MS/MS results for the peptide digest, Fig. 7 details the data processing of the detected biotransformation, for which XICs indicate peptide retention times with insets representing the modification mechanism (Fig. 7a). A deamidation of Asn to Asp leads theoretically to 1 Da mass increase, which correlates well with the measured m/z shift of ∼0.33 Da for the isotopic pattern of triply charged ions (Fig. 7b). Moreover, the localization of deamidation at Asn319 was achieved by diagnostic fragment ions (encircled in green in Fig. 7c), such as the appearance of z4+1 in the deamidated derivative. Similarly, a m/z difference of ∼8.00 Da was observed between the native and modified forms of the doubly charged peptide ions containing Met256. This corresponds to 16 Da mass increase for oxidation. Especially, some subtle key modifications at Asn33, Met34, Asn55, and Met81 from complementarity determining regions (CDR) in heavy chains were observed in Supporting Information Figs. S6 and S7. These critical biotransformation should be carefully monitored during quality control, biosimilar development or treatment cycle to guarantee efficacy and safety44, 45, 46, 47, 48.
Figure 7.
Analytical process for non-Hodgkin's lymphoma patient serum and example data of identified and confirmed peptides using ZenoTOFTM 7600 system. (a) XICs of peptides containing N319/D319 and M256/M256 oxidated. (b) Comparison of the isotopic distributions of the triply/doubly charged ions for the native and modified peptides. (c) Representative fragment ions detected in the MS/MS spectrum.
Finally, the modification ratios were calculated by dividing XIC peak areas of the modified peptide by the total XIC peak areas of the unmodified and modified forms to evaluate the extent of biotransformation (Fig. 8). The results showed that rituximab deamidation levels in serum samples were ranging from 0.2% to 15.7% while the range for oxidation was from 0.4% to 15.3%. It is noteworthy that peptide modification degrees varied between patients, which is possibly due to the administration dosage, individual variations, etc. In a word, the bioanalytical platform based on biomimetic MMPs exhibited a great application potential for the in vivo biotransformation analysis of antibodies.
Figure 8.
Biotransformation analysis of rituximab in patient serums using the bioanalytical platform based on biomimetic MMPs.
4. Conclusions
In this study, pompon mum-like biomimetic MMPs (3.8 μm) were developed to mimic the specific functionalities of CD20 on malignant B cells for the precise recognition, enrichment, and biotransformation analysis of rituximab in complex biological fluids. Thanks to the large size and specific surface area, abundant pores and Ni2+ chelation sites, the mimotope peptides were immobilized with ultrahigh density (>2300 mg/g) on the novel material, so that their maximum binding capacities for rituximab were close to 1380 mg/g. These characteristic performances were significantly superior to those of the previously reported peptide functionalized affinity MNPs, membranes, and monolithic materials. In particular, the abundant interference proteins in biological fluids were fast eliminated using the biomimetic MMPs under optimized conditions and the target rituximab could be precisely recognized from serum samples in spite of the high concentration of IgGs. Thanks to these advantages, the biomimetic MMPs not only exhibit excellent enrichment performances for rituximab from spiked cell culture mediums and serums, but also the secondary structure and bioactivity of the enriched antibody can be well maintained. Especially, some key biotransformations, such as the deamidation of Asn55 and Asn33, the oxidation of Met81 and Met34, were successfully monitored in the CDRs of rituximab captured from patient serums using the biomimetic MMPs coupled to trypsin digestion and peptide mapping by LC‒MS/MS. These critical biotransformations need to be carefully followed during quality control, biosimilar development, or treatment cycle to guarantee efficacy and safety. In future, with the development of biopharmaceuticals and precise medicine, different biomimetic MMPs should be designed and applied to in vivo analysis of mAbs and biosimilars, and even antibody‒drug conjugates or bispecific antibodies.
Author contributions
Qiqin Wang, Zhengjin Jiang, Minyi Li designed the research; Jiawen Yang, Aixuan Zhou performed the experiments and carried out data analysis; Qiaoxian He, Jingwei Zhou, and Wentao Wang performed some experiments; Jiawen Yang and Aixuan Zhou wrote the draft manuscript; Jacques Crommen revised the manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82173773, 82273893, 82373829), the Natural Science Foundation of Guangdong Province, China (2021A0505030039, 2021A0505020014), the High-End Foreign Experts Project, China (G2021199005L), and the Science and Technology Program of Guangdong Provincial Medical Products Administration, China (2023TDZ11).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.10.018.
Contributor Information
Zhengjin Jiang, Email: jzjjackson@hotmail.com.
Qiqin Wang, Email: qiqinxtu@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Dhas N., García M.C., Kudarha R., Pandey A., Nikam A.N., Gopalan D., et al. Advancements in cell membrane camouflaged nanoparticles: a bioinspired platform for cancer therapy. J Control Release. 2022;346:71–97. doi: 10.1016/j.jconrel.2022.04.019. [DOI] [PubMed] [Google Scholar]
- 2.Du A.W., Stenzel M.H. Drug carriers for the delivery of therapeutic peptides. Biomacromolecules. 2014;15:1097–1114. doi: 10.1021/bm500169p. [DOI] [PubMed] [Google Scholar]
- 3.Shah A., Malik M.S., Khan G.S., Nosheen E., Iftikhar F.J., Khan F.A., et al. Stimuli-responsive peptide-based biomaterials as drug delivery systems. Chem Eng J. 2018;353:559–583. [Google Scholar]
- 4.Qiao Z.Y., Lin Y.X., Lai W.J., Hou C.Y., Wang Y., Qiao S.L., et al. A general strategy for facile synthesis and in situ screening of self-assembled polymer-peptide nanomaterials. Adv Mater. 2016;28:1859–1867. doi: 10.1002/adma.201504564. [DOI] [PubMed] [Google Scholar]
- 5.Sato K., Hendricks M.P., Palmer L.C., Stupp S.I. Peptide supramolecular materials for therapeutics. Chem Soc Rev. 2018;47:7539–7551. doi: 10.1039/c7cs00735c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M.X., Li L.Y., Zhang L.M., Zhao J.G., Jiang Z.Q., Wang W.Z. Peptide-derived biosensors and their applications in tumor immunology-related detection. Anal Chem. 2022;94:431–441. doi: 10.1021/acs.analchem.1c04461. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J.G., Cao J., Wang W.Z. Peptide-based electrochemical biosensors and their applications in disease detection. J Anal Test. 2022;6:193–203. [Google Scholar]
- 8.Karimzadeh A., Hasanzadeh M., Shadjou N., Guardia Mdl. Peptide based biosensors. TrAC, Trends Anal Chem. 2018;107:1–20. [Google Scholar]
- 9.Zou X.J., Zhang Q.L., Lu H.L., Lin D.Q., Yao S.J. Development of a hybrid biomimetic ligand with high selectivity and mild elution for antibody purification. Chem Eng J. 2019;368:678–686. [Google Scholar]
- 10.Fang Y.M., Lin D.Q., Yao S.J. Review on biomimetic affinity chromatography with short peptide ligands and its application to protein purification. J Chromatogr A. 2018;1571:1–15. doi: 10.1016/j.chroma.2018.07.082. [DOI] [PubMed] [Google Scholar]
- 11.Xu R.R., Lu L., Sun L.J., Liu X., Lei Y.T., Huang S.F., et al. Development of histidine-tagged cyclic peptide functionalized monolithic material for the affinity purification of antibodies in biological matrices. J Chromatogr A. 2021;1635 doi: 10.1016/j.chroma.2020.461707. [DOI] [PubMed] [Google Scholar]
- 12.Lei Y.T., Liu X., Lu L., Liu C.H., Xu R.R., Huang S.F., et al. Rapid preparation of 1-vinylimidazole based non-affinity polymers for the highly-selective purification of antibodies from multiple biological sources. J Chromatogr A. 2020;1632 doi: 10.1016/j.chroma.2020.461607. [DOI] [PubMed] [Google Scholar]
- 13.Lu L., Liu X., Zuo C.Y., Zhou J.W., Zhu C.D., Zhang Z., et al. In vitro/in vivo degradation analysis of trastuzumab by combining specific capture on HER2 mimotope peptide modified material and LC‒QTOF-MS. Anal Chim Acta. 2022;1225 doi: 10.1016/j.aca.2022.340199. [DOI] [PubMed] [Google Scholar]
- 14.Zhu C.D., Han H., Chen Z.W., Shen Y., Zhang Q.X., Bao C., et al. Tetrapeptide-based mimotope affinity monolith for the enrichment and analysis of anti-HER2 antibody and antibody‒drug conjugate. Anal Chim Acta. 2023;1246 doi: 10.1016/j.aca.2023.340892. [DOI] [PubMed] [Google Scholar]
- 15.Lei Y.T., Shen Y., Zuo C.Y., Lu L., Crommen J., Wang Q.Q., et al. Emerging affinity ligands and support materials for the enrichment of monoclonal antibodies. TrAC, Trends Anal Chem. 2022;157 [Google Scholar]
- 16.Li W.N., Ran Y.G., Li M., Zhang K., Qin X., Xue X.C., et al. Mimotope vaccination for epitope-specific induction of anti-VEGF antibodies. BMC Biotechnol. 2013;13:77. doi: 10.1186/1472-6750-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M., Yan Z., Han W., Zhang Y.Q. Mimotope vaccination for epitope-specific induction of anti-CD20 antibodies. Cell Immunol. 2006;239:136–143. doi: 10.1016/j.cellimm.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Perciani C.T., Liu L.Y., Wood L., MacParland S.A. Enhancing immunity with nanomedicine: employing nanoparticles to harness the immune system. ACS Nano. 2021;15:7–20. doi: 10.1021/acsnano.0c08913. [DOI] [PubMed] [Google Scholar]
- 19.Berwanger J.D., Tan H.Y., Jokhadze G., Bruening M.L. Determination of the serum concentrations of the monoclonal antibodies bevacizumab, rituximab, and panitumumab using porous membranes containing immobilized peptide mimotopes. Anal Chem. 2021;93:7562–7570. doi: 10.1021/acs.analchem.0c04903. [DOI] [PubMed] [Google Scholar]
- 20.Liu W.J., Bennett A.L., Ning W.J., Tan H.Y., Berwanger J.D., Zeng X.Q., et al. Monoclonal antibody capture and analysis using porous membranes containing immobilized peptide mimotopes. Anal Chem. 2018;90:12161–12167. doi: 10.1021/acs.analchem.8b03183. [DOI] [PubMed] [Google Scholar]
- 21.Wang X.Y., Xia D.H., Han H., Peng K., Zhu P.J., Crommen J., et al. Biomimetic small peptide functionalized affinity monoliths for monoclonal antibody purification. Anal Chim Acta. 2018;1017:57–65. doi: 10.1016/j.aca.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Huang S.F., Tang R.T., Zhang T.T., Zhao J., Jiang Z.J., Wang Q.Q. Anti-fouling poly adenine coating combined with highly specific CD20 epitope mimetic peptide for rituximab detection in clinical patients' plasma. Biosens Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112678. [DOI] [PubMed] [Google Scholar]
- 23.Bresolin I.T.L., Bueno S.M.A. Evaluation of amino acid O-phosphoserine as ligand for the capture of immunoglubulin G from human serum. Appl Biochem Biotechnol. 2012;167:632–644. doi: 10.1007/s12010-012-9679-7. [DOI] [PubMed] [Google Scholar]
- 24.Hou X.M., Zhao C.J., Tian Y.L., Dou S.L., Zhang X., Zhao J.P. Preparation of functionalized Fe3O4@SiO2 magnetic nanoparticles for monoclonal antibody purification. Chem Res Chin Univ. 2016;32:889–894. [Google Scholar]
- 25.Yang Z.T., Yang K., Cui Y.H., Shah T., Ahmad M., Zhang Q.Y., et al. Synthesis of surface imprinted polymers based on wrinkled flower-like magnetic graphene microspheres with favorable recognition ability for BSA. J Mater Sci Technol. 2021;74:203–215. [Google Scholar]
- 26.Liu K.F., Tao Y.H., Wang L.Y., Lei J.D., Liu J., Liu Q. A novel multifunctional vaccine platform with dendritic cell-targeting and pH-responsive for cancer immunotherapy: antigen-directed biomimetic fabrication of a cabbage-like mannatide-zinc-antigen hybrid microparticles. Chem Eng J. 2021;426 [Google Scholar]
- 27.Eivazzadeh-Keihan R., Bahreinizad H., Amiri Z., Aliabadi H.A.M., Salimi-Bani M., Nakisa A., et al. Functionalized magnetic nanoparticles for the separation and purification of proteins and peptides. TrAC, Trends Anal Chem. 2021;141 [Google Scholar]
- 28.Kumar A., Planchais C., Fronzes R., Mouquet H., Reyes N. Binding mechanisms of therapeutic antibodies to human CD20. Science. 2020;369:793–799. doi: 10.1126/science.abb8008. [DOI] [PubMed] [Google Scholar]
- 29.Perosa F., Favoino E., Caragnano M.A., Dammacco F. Generation of biologically active linear and cyclic peptides has revealed a unique fine specificity of rituximab and its possible cross-reactivity with acid sphingomyelinase-like phosphodiesterase 3b precursor. Blood. 2006;107:1070–1077. doi: 10.1182/blood-2005-04-1769. [DOI] [PubMed] [Google Scholar]
- 30.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 31.Karvelas E.G., Lampropoulos N.K., Benos L.T., Karakasidis T., Sarris I.E. On the magnetic aggregation of Fe3O4 nanoparticles. Comput Methods Progr Biomed. 2021;198 doi: 10.1016/j.cmpb.2020.105778. [DOI] [PubMed] [Google Scholar]
- 32.Kannan P., Maiyalagan T., Lin B.Y., Lei W., Jie C., Guo L.H., et al. Nickel-phosphate pompon flowers nanostructured network enables the sensitive detection of microRNA. Talanta. 2020;209 doi: 10.1016/j.talanta.2019.120511. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L.W., Ma S.J., Chen Y., Wang Y., Ou J.J., Uyama H., et al. Facile fabrication of biomimetic chitosan membrane with honeycomb-like structure for enrichment of glycosylated peptides. Anal Chem. 2019;91:2985–2993. doi: 10.1021/acs.analchem.8b05215. [DOI] [PubMed] [Google Scholar]
- 34.Cui X.L., Mi W., Hu Z.S., Li X.Y., Meng B., Zhao X.Y., et al. Global characterization of modifications to the charge isomers of IgG antibody. J Pharm Anal. 2022;12:156–163. doi: 10.1016/j.jpha.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beck A., Diemer H., Ayoub D., Debaene F., Wagner-Rousset E., Carapito C., et al. Analytical characterization of biosimilar antibodies and Fc-fusion proteins. TrAC, Trends Anal Chem. 2013;48:81–95. [Google Scholar]
- 36.Chen W.Y., Liu Z.C., Lin P.H., Fang C.I., Yamamoto S.C. The hydrophobic interactions of the ion-exchanger resin ligands with proteins at high salt concentrations by adsorption isotherms and isothermal titration calorimetry. Sep Purif Technol. 2007;54:212–219. [Google Scholar]
- 37.Tan H.Y., Yang J.Y., Linnes J.C., Welch C.J., Bruening M.L. Quantitation of trastuzumab and an antibody to SARS-CoV-2 in minutes using affinity membranes in 96-well plates. Anal Chem. 2022;94:884–891. doi: 10.1021/acs.analchem.1c03654. [DOI] [PubMed] [Google Scholar]
- 38.Kangwa M., Yelemane V., Ponnurangam A., Fernandez-Lahore M. An engineered Staphylococcal protein A based ligand: production, characterization and potential application for the capture of immunoglobulin and Fc-fusion proteins. Protein Expr Purif. 2019;155:27–34. doi: 10.1016/j.pep.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Du K.F. Peptide immobilized monolith containing tentacle-type functionalized polymer chains for high-capacity binding of immunoglobulin G. J Chromatogr A. 2014;1374:164–170. doi: 10.1016/j.chroma.2014.11.060. [DOI] [PubMed] [Google Scholar]
- 40.Du Z., Zhang S.L., Zhou C.Y., Liu M., Li G.K. l-Histidine functionalized multi-walled carbon nanotubes for on-line affinity separation and purification of immunoglobulin G in serum. Talanta. 2012;99:40–49. doi: 10.1016/j.talanta.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 41.Yang N., Tang Q., Hu P., Lewis M.J. Use of in vitro systems to model in vivo degradation of therapeutic monoclonal antibodies. Anal Chem. 2018;90:7896–7902. doi: 10.1021/acs.analchem.8b00183. [DOI] [PubMed] [Google Scholar]
- 42.Tran J.C., Tran D., Hilderbrand A., Andersen N., Huang T., Reif K., et al. Automated affinity capture and on-tip digestion to accurately quantitate in vivo deamidation of therapeutic antibodies. Anal Chem. 2016;88:11521–11526. doi: 10.1021/acs.analchem.6b02766. [DOI] [PubMed] [Google Scholar]
- 43.Huang L.H., Lu J.R., Wroblewski V.J., Beals J.M., Riggin R.M. In vivo deamidation characterization of monoclonal antibody by LC/MS/MS. Anal Chem. 2005;77:1432–1439. doi: 10.1021/ac0494174. [DOI] [PubMed] [Google Scholar]
- 44.Strasser L., Oliviero G., Jakes C., Zaborowska I., Floris P., Ribeiro da Silva M., et al. Detection and quantitation of host cell proteins in monoclonal antibody drug products using automated sample preparation and data-independent acquisition LC‒MS/MS. J Pharm Anal. 2021;11:726–731. doi: 10.1016/j.jpha.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kellie J.F., Pannullo K.E., Li Y.H., Fraley K., Mayer A., Sychterz C.J., et al. Antibody subunit LC‒MS analysis for pharmacokinetic and biotransformation determination from in-life studies for complex biotherapeutics. Anal Chem. 2020;92:8268–8277. doi: 10.1021/acs.analchem.0c00520. [DOI] [PubMed] [Google Scholar]
- 46.Mo J.J., Yan Q.R., So C.K., Soden T., Lewis M.J., Hu P. Understanding the impact of methionine oxidation on the biological functions of IgG1 antibodies using hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2016;88:9495–9502. doi: 10.1021/acs.analchem.6b01958. [DOI] [PubMed] [Google Scholar]
- 47.Su Z., Xiao D., Xie F., Liu L.Q., Wang Y.M., Fan S.Y., et al. Antibody–drug conjugates: recent advances in linker chemistry. Acta Pharm Sin B. 2021;11:3889–3907. doi: 10.1016/j.apsb.2021.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song X.W., Li C., Meng Y.F. Mass spectrometry imaging advances and application in pharmaceutical research. Acta Materia Medica. 2022;1:507–533. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.