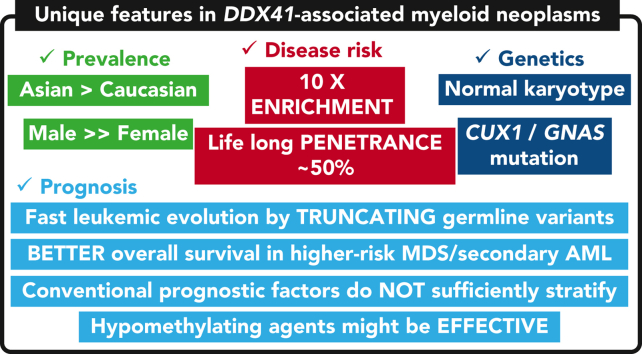
Key Points
-
•
DDX41 germ line mutations explain ∼80% of known germ line predisposition to MNs in adults, and the life-long risk was ∼50%.
-
•
DDX41-mutated MDS patients rapidly progressed to AML, which was however, confined to those having truncating variants.
Visual Abstract
Abstract
Germ line DDX41 variants have been implicated in late-onset myeloid neoplasms (MNs). Despite an increasing number of publications, many important features of DDX41-mutated MNs remain to be elucidated. Here we performed a comprehensive characterization of DDX41-mutated MNs, enrolling a total of 346 patients with DDX41 pathogenic/likely-pathogenic (P/LP) germ line variants and/or somatic mutations from 9082 MN patients, together with 525 first-degree relatives of DDX41-mutated and wild-type (WT) patients. P/LP DDX41 germ line variants explained ∼80% of known germ line predisposition to MNs in adults. These risk variants were 10-fold more enriched in Japanese MN cases (n = 4461) compared with the general population of Japan (n = 20 238). This enrichment of DDX41 risk alleles was much more prominent in male than female (20.7 vs 5.0). P/LP DDX41 variants conferred a large risk of developing MNs, which was negligible until 40 years of age but rapidly increased to 49% by 90 years of age. Patients with myelodysplastic syndromes (MDS) along with a DDX41-mutation rapidly progressed to acute myeloid leukemia (AML), which was however, confined to those having truncating variants. Comutation patterns at diagnosis and at progression to AML were substantially different between DDX41-mutated and WT cases, in which none of the comutations affected clinical outcomes. Even TP53 mutations made no exceptions and their dismal effect, including multihit allelic status, on survival was almost completely mitigated by the presence of DDX41 mutations. Finally, outcomes were not affected by the conventional risk stratifications including the revised/molecular International Prognostic Scoring System. Our findings establish that MDS with DDX41-mutation defines a unique subtype of MNs that is distinct from other MNs.
Makishima and colleagues report on a comprehensive characterization of 346 patients with myeloid neoplasms (MN) in the context of germ line DDX41 mutation and 525 first-degree relatives. Development of MN increased rapidly after age 40, with 49% developing MN by age 90. Computations were different from DDX41 wild-type patients, and when present, did not worsen the prognosis associated with this subset of MN.
Introduction
The 2016 revision to the World Health Organization classification includes myeloid neoplasms (MNs) with germ line predisposition.1 The addition of this section was justified by the growing evidence that a substantial proportion of patients with MNs including myelodysplastic syndromes (MDS), myelodysplastic/myeloproliferative neoplasms (MDS/MPNs), and acute myeloid leukemias (AMLs) have recurrent, inherited pathogenic traits.2, 3, 4, 5 Recognition of such germ line predisposition alleles has clinical relevance because it affects cascade testing among family members, dictates cancer surveillance strategies for individuals achieving long-term remissions, and donor selection for allogeneic hematopoietic stem cell transplantation (HSCT).3,4,6, 7, 8
In the recent years, among these alleles with a predisposition to leukemia, those affecting DDX41 have been of particular interest.9, 10, 11, 12, 13, 14, 15, 16, 17, 18 This is because, unlike mutations in other cancer predisposition genes, those in DDX41 are responsible for late-onset MNs with a peak in the sixth to seventh decades of life and explain a much larger etiologic fraction than other variants. In fact, there have been many follow-up publications after our original report of DDX41-mutant MNs. Nonetheless, the current knowledge about DDX41-mutated MNs is still largely limited.1,19, 20, 21, 22 The major reason for this being the invariably small number of DDX41-mutated cases included in these publications. Thus, several important questions remain to be answered, including the frequency of both germ line and somatic DDX41 variants depending on disease subtype and variant type, their impacts on clinical pictures and outcomes, the relative incidence of those variants compared with that of other known germ line risk variants, and among others, the magnitude of the risk (penetrance) of the development of MNs conferred by DDX41 pathogenic variants. All these are indispensable information for the management of patients with DDX41 variants and their relatives.
Here we reveal the spectrum of common pathogenic DDX41 germ line variants across multiple ethnicities, estimate the age-related risk of MN development ascribed to pathogenic DDX41 germ line variants, and unveil their unique genetic and clinical features, through a large international collaboration accruing 346 DDX41-mutated MN cases.
Materials and methods
Study subjects
In total, 9082 patients with MNs were enrolled from 56 collaborating institutes or projects across 7 countries (MN cohort), in which 50 from 42 families were confirmed to have additional relatives with MNs. Tumor-derived DNA was collected from all cases at 1 or more time points. Diagnostic or treatment–naïve tumor samples were obtained in 7698 cases, of which 2360 had survival data (supplemental Table 1, available on the Blood website) and were used for survival outcome analysis. Germ line DNA was available for 1350 patients selected for the establishment of criteria useful to discriminate either the germ line or somatic nature of DDX41 variants present in the whole MN cohort. Of these, a total of 1039 Japanese patients aged ≥20 years were constitutively enrolled at Kyoto University and 26 collaborative institutes between 2010 and 2021 who were diagnosed with either of AML (including therapy-related and secondary AML [sAML]), MDS, MDS/MPN, or MPN and whose germ line DNA was successfully extracted from buccal smear for predisposition analysis (Table 1; supplemental Figures 1-2). Bone marrow (BM)- or blood-derived tumor DNA was subjected to whole genome and/or targeted sequencing using different driver gene panels, which included DDX41 and an additional 36 genes in common (supplemental Table 2). For population studies as control, we enrolled 20 238 individuals from 3 large Japanese biobanks, including the Tohoku Medical Megabank (TOMMO) (https://www.megabank.tohoku.ac.jp), the Nagahama-cho cohort (http://hi.med.kyoto-u.ac.jp/nagahama.html), and BioBank Japan (BBJ) (https://biobankjp.org/en/index.html), in which frequency of individual having germ line DDX41 variants was available on the basis of whole genome or targeted sequencing.23, 24, 25 For subjects in BBJ, information on sex was estimated by the analysis of SNP-array data.23 As general population control, gnomAD data set (ver.2.1.1) was used for the analysis of the regional distribution and the minor allele frequency of each DDX41 variant (https://gnomad.broadinstitute.org/). All samples were obtained according to the protocols approved by the ethics board of the participating institutions or biobanks. Then, we collected information related to the first-degree relatives of these cases, if available, identifying 525 cases from 169 families (defined as kin cohorts). In this study, we defined the first-degree relatives as parents, siblings, and children, who all share risk alleles with probands at 50% probability. We calculated the penetrance of DDX41 risk alleles in the kin cohorts whereas all other analyses were performed on the MN cohort. All the participating institutions in this study received informed consent under each of their institutional review boards approval, which follows the Declaration of Helsinki.
Table 1.
Patient characteristics
| Variables | DDX41-WT | DDX41-mutated | Total |
|---|---|---|---|
| n (%) | 8736 (96.2%) | 346 (3.8%) | 9082 (100%) |
| Sporadic | 8730 | 302 | 9032 |
| Familial (proband) | 6 (6) | 44 (36) | 50 (42) |
| Age (median, range) | 65, 1-94 | 68, 15-94 | 65, 1-94 |
| Sex (male/female/unknown) | 3666/2324/2746 | 274/72/0 | 3940/2396/2746 |
| OS | |||
| n of informative cases | 2159 | 201 | 2360 |
| Median observation, range | 1.2 y, 0-40.4 | 1.3 y, 0-22.7 | 1.2 y, 0-40.4 |
| Leukemic progression | |||
| n of informative cases | 2128 | 174 | 2302 |
| Median observation, range | 1.6 y, 0.01-32.3 | 1.7 y, 0.03-22.7 | 1.6 y, 0.01-32.3 |
| Diagnosis | |||
| MDS | 4146 | 201 | 4347 |
| AML | 2704 | 136 | 2840 |
| MDS/MPN | 658 | 8 | 666 |
| MPN | 1228 | 1 | 1229 |
| Country | |||
| Japan | 4289 | 172 | 4461 |
| United States | 1767 | 105 | 1872 |
| United Kingdom | 1669 | 36 | 1705 |
| Italy | 240 | 13 | 253 |
| Thailand | 67 | 6 | 73 |
| The Netherlands | 36 | 2 | 38 |
| Germany | 668 | 12 | 680 |
| Status at sampling | |||
| At diagnosis | 6961 | 275 | 7236 |
| Before treatment | 430 | 32 | 462 |
| After treatment | 877 | 39 | 916 |
| Unknown | 468 | 0 | 468 |
| Germ line DNA (%)∗ | |||
| Available | 1003 (20.4%) | 36 (20.9%) | 1039 (23.3%) |
| Not available | 3286 (79.6%) | 136 (79.1%) | 3422 (76.7%) |
| Treatment | |||
| Hypomethylating agents (HMA) | 771 | 108 | 879 |
| Transplantation | 1424 | 94 | 1518 |
| Chemotherapy | 927 | 73 | 1000 |
Number of Japanese cases.
Genetic analysis
Sequencing method
5768 samples were sequenced at Kyoto University by targeted-capture sequencing using either of 5 panels as previously described.26,27 Genomic DNA was enriched for target regions by liquid-phase hybridization using the SureSelect custom kit (Agilent Technologies). The purified library was subjected to high-throughput sequencing analysis with HiSeq 2500, NovaSeq 6000 (Illumina), or DNBSEQ-G400RS (MGI). 1836 samples were sequenced at the Cleveland clinic by multiamplicon deep sequencing (TruSeq) for 61 genes with MiSeq (Illumina) as described in our previous publication.28 1705 samples were sequenced at King’s College London by multiamplicon deep sequencing (QiaSeq; QIAGEN) for 44 genes and NextSeq 550 (Illumina). At the University of Chicago, individuals underwent germ line genetic testing at the Genetic Services Laboratory (Department of Human Genetics, The University of Chicago) to identify 35 individuals with deleterious germ line DDX41 variants. All healthy controls were sequenced in Japan. MN samples were independently gathered from different sites and sequenced at each participating institution.
Detection of genetic variations
Single nucleotide variants, insertions, and deletions were detected using our established pipeline (Genomon pipeline for GRCh37/hg19) as previously reported.26,27,29, 30, 31 For somatic mutation detection, germ line polymorphisms and sequencing artifacts were excluded as previously described,26 followed by visual inspection on the Integrative Genomics Viewer (http://software.broadinstitute.org/software/igv/).
Copy-number analysis
For evaluation of somatic copy-number alterations (CNAs), we performed conventional karyotyping, single nucleotide polymorphism array, or sequencing-based copy-number analysis. For sequencing-based copy-number analysis, we included 1158 to 1428 single nucleotide polymorphisms probes in RNA baits allowing for detection of total copy-number changes (gain or loss) and copy-neutral allelic imbalances (supplemental Figure 3). The pipeline for sequencing-based copy-number analysis (CNACS) is available at https://github.com/papaemmelab/toil_cnacs.26,32 After automated detection of CNAs, manual inspection was performed to exclude erroneous calls.
Adjustment of variant allele frequencies
Variant allele frequencies of somatic mutations were adjusted to reflect the actual clone size based on the copy-number state of the locus of interest. Detailed methods have been described in our previous publication.26
Classification of DDX41 variants
First, we established the criteria to classify DDX41 (MN_016222) variants into germ line and somatic variants in 75 cases as a training set and validated the criteria using an independent set of 35 patients (validation set) (supplemental Figure 2). Using the criteria, 557 nonsynonymous/splice-site/indel DDX41 variants were identified and classified into germ line and somatic variants. We classified the germ line variants into either P/LP or variants with unknown significance based on the guidelines from the American College of Medical Genetics.33.
Statistical analysis
All the statistical analyses were performed using the R statistical platform (https://www.r-project.org/) v.4.0.4. All statistical tests were 2-sided. Benjamini & Hochberg multiple testing correction was applied when appropriate. Box plots for continuous values indicate the median, first and third quartiles (Q1 and Q3), and whiskers extend to the furthest value between Q1 – 1.5 × the interquartile range and Q3 + 1.5 × interquartile range.
Calculation of odds ratio (OR) for individual DDX41 variants
To estimate the risk conferred by DDX41 germ line variants, we calculated ORs and 95% confidence intervals (CIs) for individual variants by comparing their frequencies in Japanese patients of MNs (n = 4461) and those in normal subjects (n = 20 238). As for the normal subjects, we used whole-genome sequencing data of 1008 subjects from the Nagahama-cho cohort (http://hi.med.kyoto-u.ac.jp/nagahama.html), and targeted-sequencing data of 10 850 subjects from BBJ (https://biobankjp.org/en/index.html), and publicly available summary statistics of 8380 subjects from Tohoku Medical Megabank (TOMMO) (https://www.megabank.tohoku.ac.jp). Germ line variants observed in ≥3 subjects with or without MNs were included in the analysis.
Calculation of penetrance
We collected the information of first-degree relatives of patients with MNs with or without DDX41 risk alleles. Cumulative incidence of MNs in first-degree relatives was estimated by Fine-Gray regression, considering death from other causes as a competing risk. Based on the notion that first-degree relatives of carriers inherited the risk allele with a 50% probability, the penetrance can be calculated by the following equation as previously described in kin-cohort retrospective studies.34, 35, 36, 37, 38
In this equation, S indicates the age-dependent function of penetrance. R+ and R− indicate the cumulative incidence of MNs in first-degree relatives of DDX41-mutation carriers and noncarriers, respectively.
Survival analysis
We performed an analysis on overall survival (OS) and the risk of leukemic progression using the R package, Survival (http://cran.r-project.org/web/packages/survival/index.html). Scores of the revised/molecular International Prognostic Scoring System (IPSS-R/M) were calculated according to the previous publications.39,40 OS was estimated by Kaplan-Maier method, and the differences among patient groups were examined by log-rank test or Cox proportional hazard regression. To reduce phenotypic bias between cases with and without available survival data (supplemental Table 1), survival analysis was performed among matching disease phenotypes. To resolve sample size bias between DDX41-wild-type (WT) and patients with DDX41 mutation, we performed confirmatory analyses in WT patients randomly selected with sample numbers adjusted to mutated ones (supplemental Methods). The cumulative incidence of leukemic progression was estimated by Fine-Gray method with nonleukemic death as a competing risk.
Results
DDX41 variants in cases with MNs
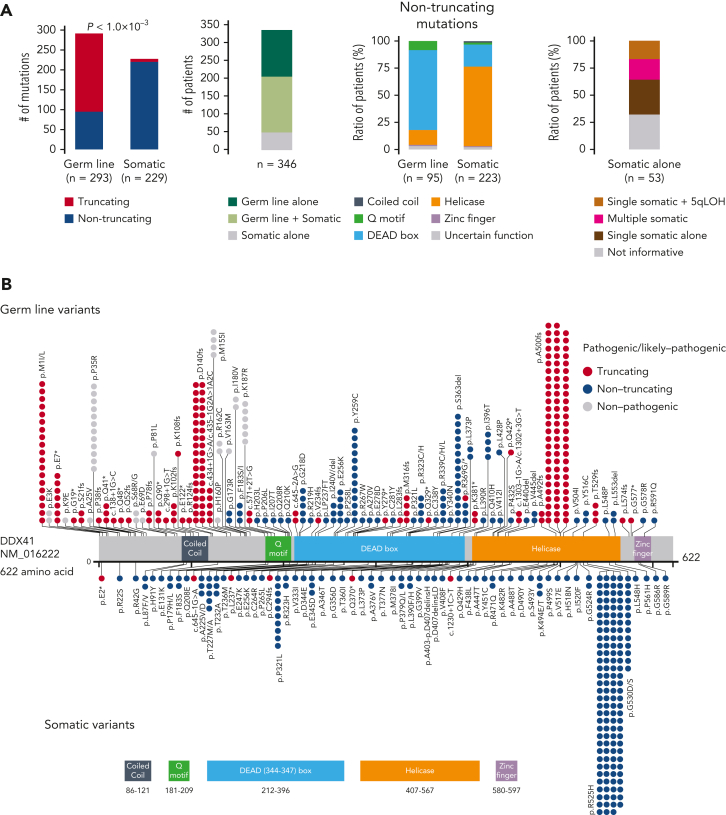
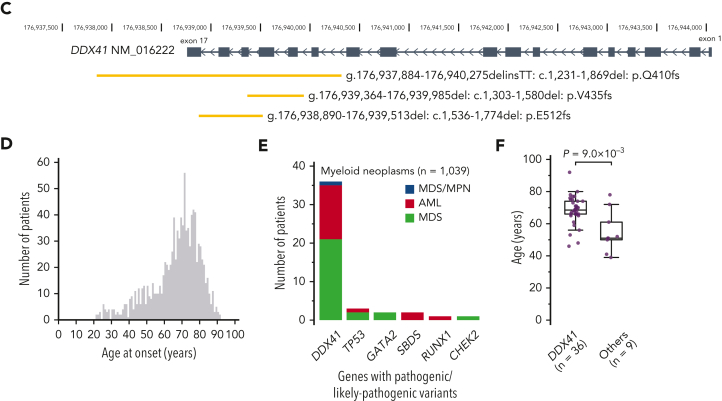
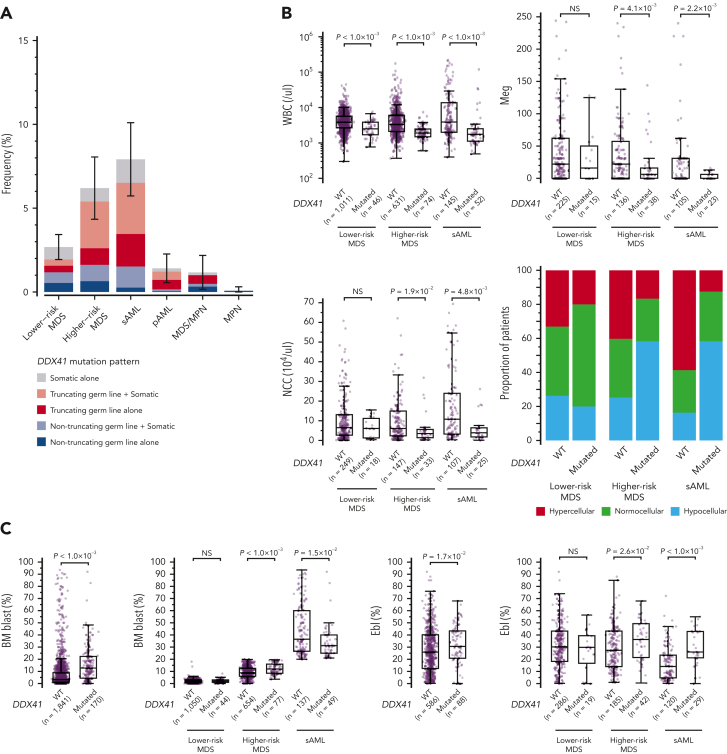
Among a total of 9082 patients with different MNs of divergent ethnicity who enrolled in the study (Table 1), we identified 557 nonsynonymous/splice site variants, including small and large indels, of DDX41 having <1% of minor allele frequency in the publicly available datasets of general populations (“Materials and methods”). Their germ line/somatic origins were determined in 110 cases using germ line DNA, based on which we established the criteria to infer the origin of the remaining variants (supplemental Figure 2). Thus, all variants were classified as either germ line (n = 328) or somatic (n = 229) (Figure 1A-B). According to the guidelines from the American College of Medical Genetics and Genomics,33 we then classified 293 germ line variants in 293 patients as pathogenic (P)/likely pathogenic (LP), whereas the remaining 35 variants were assigned as variants of uncertain significance. Most (96.8%) of the P/LP missense variants were within functional domains, mostly in the DEAD box domain (∼70%), sparing the N-terminal 180 amino acids, where most variants of uncertain significance were located. Of interest, 3 patients had large deletions encompassing multiple exons, which were confirmed to be of germ line origin (Figure 1C). Eight amino acid positions (eg, p.F183 and p.R323), all located in functional domains, were affected by both germ line and somatic variants. As many as 54% of the cases with P/LP germ line DDX41 variants carried a second, somatic DDX41 mutation of which 64% were p.R525H. A minority (n = 53) of cases had somatic mutations alone. Among these, half of the informative cases had a single somatic mutation and the remaining half had either biallelic alterations, (Figure 1A; supplemental Table 3) caused by loss of heterozygosity at chromosome band 5q35.3 (n = 9) where DDX41 is located (supplemental Figure 3) or multiple mutations (n = 10). Most somatic variants (223/229) were nontruncating (Figure 1A), and all truncating somatic variants were heterozygous and not accompanied by an additional germ line truncating alteration, suggesting that homozygous/compound truncating alleles are not compatible with cell viability. Conversely, as many as two-thirds of germ line variants were truncating (Figure 1A). Furthermore, nontruncating germ line hits were enriched in the DEAD box and Q domains, whereas nontruncating somatic mutations were mostly mapped within the helicase domain (Figure 1A-B). Overall, 346 out of 9082 (3.8%) enrolled patients were positive for P/LP DDX41 mutations in either fashion of germ line alone, germ line plus somatic, or somatic alone.
Figure 1.
P/LP germ line variants and somatic mutations in DDX41 found in 346 cases with MNs. (A) Frequency of truncating and nontruncating variants within P/LP germ line variants and somatic mutations (left), cases with P/LP germ line and/or somatic variants of DDX41 (middle left), nontruncating variants involving known functional domains (middle right), and cases with somatic DDX41 mutations alone with mono and biallelic involvement (right). (B) Distribution of 325 germ line variants (top) and 229 somatic mutations (bottom). Truncating (red) and nontruncating (blue) P/LP germ line variants were defined based on the American College of Medical Genetics and Genomics criteria. Gray color indicates those classified as germ line variants with undetermined significance. Amino acid locations of relevant functional domains are indicated at the bottom. (C) Germ line variants with deletions including multiple exons identified in 3 cases. Genomic positions of DDX41 (NM_016222) and affected regions were shown according to the GRCh37/hg19 reference. (D) Distributions of age at disease onset in 1039 patients with MN in whom germ line samples were examined. (E) The number of cases with P/LP alleles of DDX41 and other 22 known leukemia predisposing genes in 1039 Japanese cases with MN. The germ line origin of each variant was confirmed in buccal smear samples. (F) Distributions of age at disease onset in those with P/LP germ line variants in DDX41 and the other genes. Statistical significance was tested by 2-sided Wilcoxon test.
Size of DDX41-variant–associated risk of MN development
We then wanted to evaluate the relative impact of DDX41 compared with other germ line predisposition genes in MNs3. To do so, we investigated P/LP germ line variants in 23 genes implicated in MNs among 1039 Japanese patients with MN aged ≥20 years using targeted or whole-genome sequencing of paired tumor/germ line DNA (Figure 1D and supplemental Figure 4). Overall, we identified 45 P/LP germ line variants based on the American College of Medical Genetics and Genomics criteria, in which the vast majority (80%) were explained by DDX41 (n = 36) with only minor contributions from other variants (Figure 1E), confirming that DDX41 is the most common target of germ line predisposition variants among adult MNs cases. Notably, individuals with germ line DDX41 mutations were diagnosed at older ages (median age, 69 years) compared with those with other deleterious germ line variants (median age, 51 years) (P = 9.0 × 10−3) (Figure 1F).
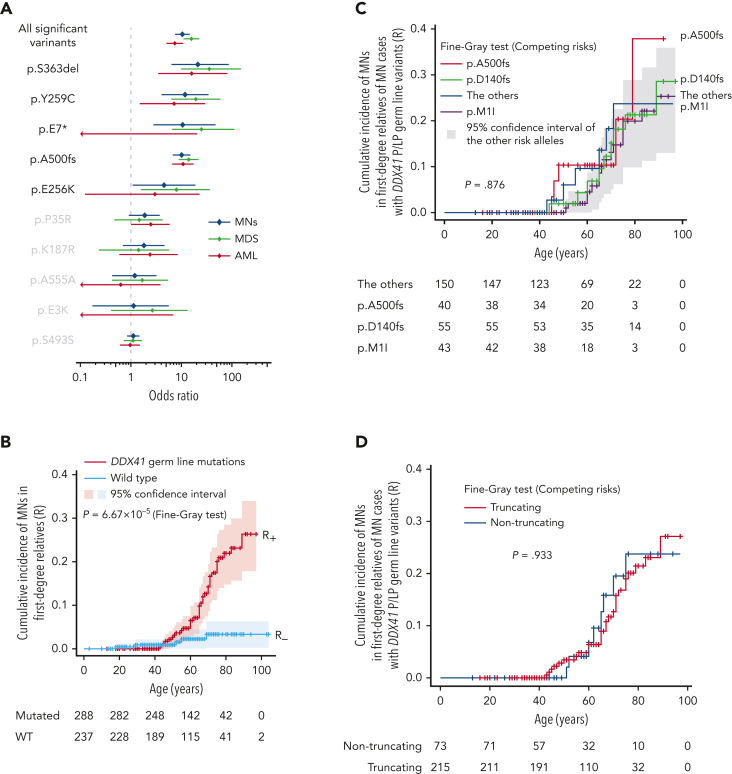
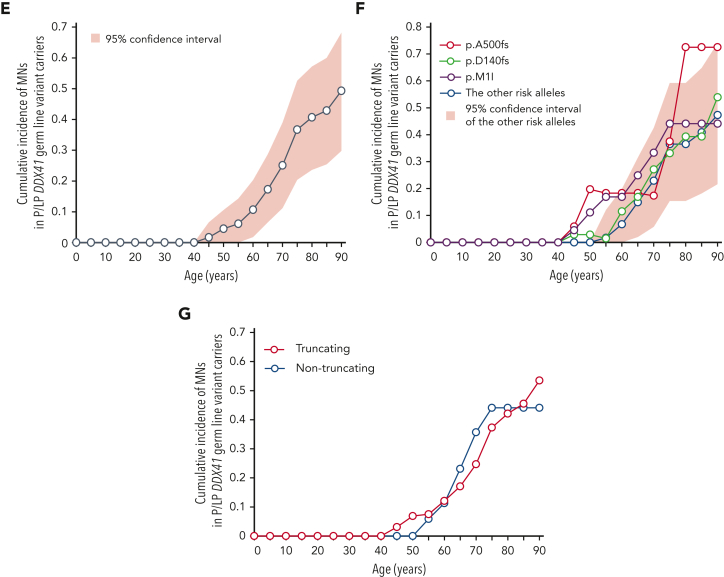
Next, to estimate the size of the risk of developing MNs associated with P/LP DDX41 germ line variants, we calculated the enrichment of different DDX41 germ line variants within 4461 Japanese patients with MN compared with an ethnicity-matched control population (n = 20 238). Among 10 variants found in ≥3 cases in the combined cohort, 5 (encoding p.E256K, p.A500fs, p.Y259C, p.E7∗, and p.S363del) were significantly enriched in MNs with a median OR of 10.6 (4.5-22.8) and therefore likely to be pathogenic, whereas 3 nonsynonymous (encoding p. E3K, p.K187R, and p.P35R) and 2 synonymous (encoding p.S493S and p.A555A) variants showed no significant enrichment (Figure 2A; supplemental Table 4). We also estimated the cumulative incidence (penetrance) of MN development among DDX41 variant carriers by calculating the cumulative incidence of MNs among first-degree relatives of DDX41-mutated MN (R+) and randomly selected DDX41-WT (R−) probands (Figure 2B and supplemental Figure 5).34,38 For 3 major variants, p.D140fs, p.M1I, and p.A500fs, R+ remains negligible under 40 years of age but increases with age to reach 28.6% (95% CI, 11.4-48.5), 23.7% (95% CI, 8.8-42.6), and 37.9% (95% CI, 6.3-71.1) by the age of 90, respectively (Figure 2C). Combining all other P/LP variants, P/LP germ line variants showed a similar R+ profile (Figure 2C). No significant difference in R+ was observed between truncating and nontruncating variants (Figure 2D). Finally, subtracting R− (<3.3%), which was estimated from first-degree relatives of DDX41-WT patients and therefore overestimated actual R−, we projected the expected average penetrance (S = 2R+ − R−) for P/LP DDX41 germ line variants as 10.7% (95% CI, 1.8-20.3) and 49.3% (95% CI, 29.8-68.2) by the age of 60 and 90 years, respectively (Figure 2E). The penetrance in carriers of major variants, including p.D140fs, p.M1I, and p.A500fs, reached as high as 53.9% (95% CI, 15.4-95.8), 44.1% (95% CI, 10.2-84.0), and 72.5% (95% CI, 5.2-141) by the age of 90, respectively (Figure 2F). Combining all other P/LP variants, P/LP germ line variants showed a comparable penetrance of 47.3% (95% CI, 21.6-74.0) (Figure 2F). Notably, no significant difference was observed between truncating and nontruncating variants (Figure 2G).
Figure 2.
DDX41-variant–associated risk of MN development. (A) Enrichment of 10 major germ line DDX41 alleles in 4461 Japanese cases with MNs compared with 20 238 cases from ethnicity-matched controls. ORs are plotted with 95% CIs for each allele. (B-D) Cumulative incidence of MNs calculated by Fine-Gray test of ages at disease onset and death in the first-degree relatives of MN cases with (red) or without (blue) germ line DDX41 mutations are shown in the whole kin cohorts (B), in each P/LP variant (C), and truncating vs nontruncating variants (D). (E-G) Cumulative incidences of MNs in carriers of DDX41 risk alleles estimated by kin-cohort analysis are demonstrated in the whole kin cohorts (E), in each P/LP variant (F), and truncating vs nontruncating variants (G).
Effect of DDX41 variants on hematological phenotype
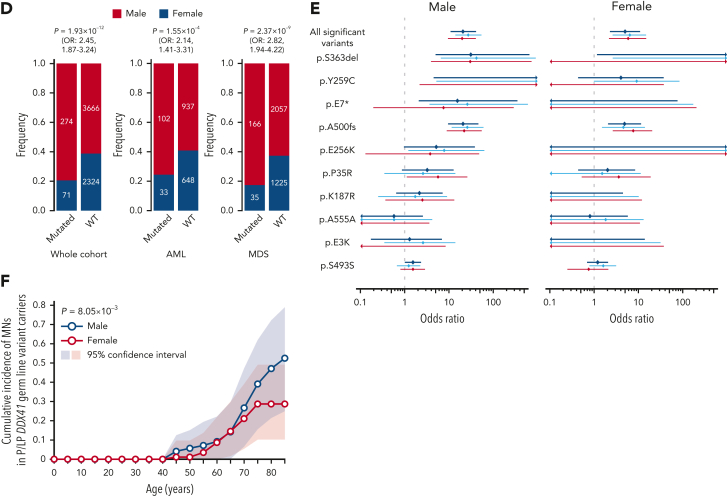
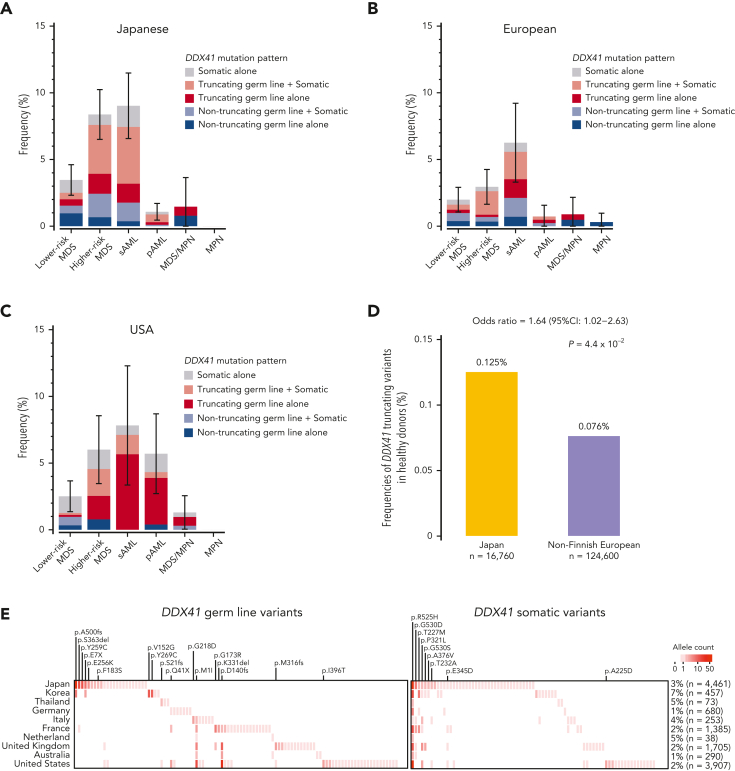
Overall, 3.8% of MN cases had P/LP germ line and/or somatic DDX41 variants (Table 1 and supplemental Table 3). The estimated frequency of DDX41 mutations was highest in sAML and higher-risk MDS (HR-MDS), followed by lower-risk MDS (LR-MDS), primary AML, and MDS/MPN. Mutations were rarely found in MPN cases (Figure 3A). After disease subtypes were stratified, DDX41 mutations were found to be more frequent among Japanese than in Caucasian cases (Figure 4A-C and supplemental Figure 6A), likely explained by a higher prevalence of P/LP DDX41 alleles in the former population (Figure 4D). The distribution of germ line DDX41 variants also showed substantial geographical/ethnic variations. For example, p.A500fs was most prevalent in Japan and Korea but not found in Western countries. The p.D140fs was the major germ line variant in United Kingdom, United States, and France, but was rarely encountered in other European countries and was not detected at all in Asians, whereas p.M1I was more widely observed across multiple Western countries, suggesting an older ancestral history (Figure 4E; supplemental Table 5). No significant geographical variations were observed in somatic DDX41 variants (Figure 4E).
Figure 3.
Demographic features of DDX41-mutated patients. (A) Frequency of DDX41-mutated patients in different subtypes of MNs including LR- and HR-MDS, sAML, pAML, MDS/MPN, and MPN with DDX41-mutation status in terms of mono vs biallelic and truncating vs nontruncating variants. (B-C) Comparison of WBC counts, BM NCC, megakaryocyte (Meg) counts, BM cellularity (B), BM blast and erythroblast (Ebl) counts (C) between DDX41-mutated and unmutated cases. P values are provided using the Wilcoxon rank-sum test. (D) Male and female distributions in disease phenotypes. P values and ORs are provided using the Fisher exact test. (E) Enrichment of 10 major germ line DDX41 alleles in Japanese MN cases compared with control Japanese populations were separately shown for male/female subjects. ORs are plotted with 95% CI for each allele. (F) Cumulative incidence of MNs in male vs female carriers of DDX41 risk alleles estimated by kin-cohort analysis. P value was provided using the Wilcoxon signed-rank test. NCC, nucleated cell counts; pAML, primary AML.
Figure 4.
Geographic distribution of P/LP germ line variants and somatic mutations in DDX41. (A-C) Disease-specific frequencies of DDX41 variants in different geographical areas including Japan (A), Europe (B), and USA (C). (D) Frequency of truncating germ line variants in DDX41 in the Japanese and European general populations. (E) Geographic distribution of major P/LP germ line DDX41 alleles found in 13 249 cases with MNs (left) is compared with that of somatic DDX41 variants (right). The number of distinct DDX41 alleles/variants (horizontal axis) in each country is shown in color gradient. Highly recurrent major alleles/variants are indicated.
The elevated mutation frequency in sAML and HR-MDS was in large part explained by truncating variants (Figure 3A and supplemental Figure 6B). DDX41-mutated cases had lower white blood cell counts than WT counterparts, and except in LR-MDS cases, had reduced BM cellularity, nucleated cell, and megakaryocyte counts (Figure 3B-C). In HR-MDS and sAML, DDX41-mutants had higher erythroblast counts than WT cases (Figure 3C and supplemental Figure 7A). Although BM blasts were slightly increased in DDX41-mutated than unmutated cases among HR-MDS, a lower percentage was registered among sAML cases (Figure 3C), which was more conspicuous in cases with truncating than nontruncating variants (supplemental Figure 7A). Moreover, DDX41-mutated MDS cases were more likely classified into higher IPSS-R risk groups than DDX41-WT cases (supplemental Figure 7B).
Gender distributions in DDX41-mutated MNs
We noticed a strong male predominance in both DDX41-mutated (79.4%) and WT (61.2%) cohorts, which however, was more remarkable among DDX41-mutated cases (P = 1.93 × 10−12; OR, 2.45; 95% CI, 1.87-3.24) (Figure 3D). Remarkably, no difference in gender distribution was found between healthy individuals with and without DDX41 risk alleles (supplemental Table 6). A male dominance was observed across all disease subtypes including LR-MDS (P = 3.53 × 10−3; OR, 2.42; 95% CI, 1.28-4.89), HR-MDS (P = 5.53 × 10−7, OR, 3.15; 95% CI, 1.92-5.43), sAML (P = 4.26 × 10−2; OR, 1.71; 95% CI, 1.01-2.97), and primary AML (P = 2.60 × 10−2; OR, 2.58; 95% CI, 1.07-7.18) (supplemental Figure 8A). Accordingly, looking at the risk of developing MNs in terms of enrichment of P/LP variants in MN vs general population, an increase was observed in males (OR, 20.7; 95% CI, 10.9-41.0) as opposed to females (OR, 5.00; 95% CI, 2.27-10.9), although the enrichment was still highly significant in females (Figure 3E). Similarly, the penetrance associated with DDX41 risk alleles was significantly higher in male than female (P = 8.05 × 10−3), achieving 52.5% vs 28.7% by the age of 85 years, respectively (Figure 3F). Notably, the frequency of each DDX41 risk allele was not different between male and female in the general population (supplemental Table 6).
Unique comutation patterns of DDX41 mutations
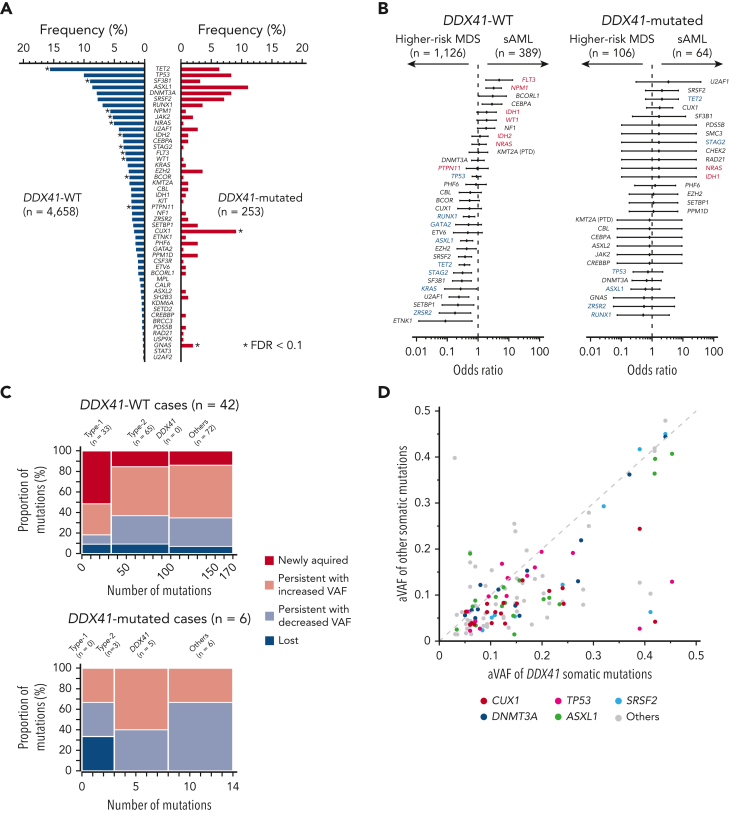
In the current study, patients were enrolled from multiple institutes and hospitals and therefore, they were analyzed for gene mutations using different targeted gene panels. These included DDX41 and additional 36 common genes, which are most frequently mutated in MNs (supplemental Table 2). Overall, 85% of DDX41-mutated cases had 1 or more somatic driver mutations or CNAs implicated in MNs (supplemental Figures 9-10), with somatic DDX41 mutations representing the only detectable acquired driver mutations in 15% of such cases (supplemental Figure 9B). No known alterations in myeloid drivers or CNAs other than the germ line DDX41 mutation were detected instead in the remaining 15%. Including somatic DDX41 mutations, patients with DDX41 mutation had a higher number of somatic mutations than DDX41-WT patients (supplemental Figure 9C). Looking at cytogenetics, the majority of the patients with DDX41 mutation harbored a normal karyotype (66.9% vs 48.2% in WT cases; OR, 2.17; 95% CI, 1.65-2.87; P = 7.08 × 10−9) overall and in each disease phenotype (supplemental Figure 10). Excluding DDX41, the most frequent targets of somatic mutations associated with germ line DDX41 lesions included ASXL1, TP53, CUX1, SRSF2, TET2, and DNMT3A, of which CUX1 mutations were more enriched in cases with biallelic DDX41 configurations (supplemental Figure 9D). Compared with WT cases, DDX41-mutants showed an over-representation of GNAS and CUX1 mutations and under-representation of STAG2, NRAS, NPM1, SF3B1, and TET2 mutations, and the trend was largely similar across different MN subtypes (Figure 5A and supplemental Figure 11). Previously, we reported unique sets of driver mutations implicated in leukemic progression of MDS, designated as “Type-1” and “Type-2,” which were found to be enriched in sAML and HR-MDS compared with HR- and LR-MDS, respectively.28 Intriguingly, none of these mutations was enriched in sAML or HR-MDS in respective comparisons (Figure 5B and supplemental Figure 12). In line with this, in an analysis of longitudinal samples during progression to sAML (n = 48), DDX41-WT patients with MDS were more likely to acquire than lose driver mutations, particularly of the Type-1, whereas none of the DDX41-mutated patients with MDS acquired new driver alterations (Figure 5C and supplemental Figure 13). These observations imply the presence of a distinct mechanism of disease progression in DDX41-mutated MDS. The size of tumor clones as inferred from the largest variant allele frequency of somatic mutations was substantially smaller in patients with biallelic DDX41 mutations or somatic mutations alone, compared with that in those with monoallelic mutations or DDX41-WT cases (supplemental Figure 14). Notably, the size of the somatic DDX41-mutant clones tended to be larger than that of other driver mutations (Figure 5D), suggesting that the acquisition of the second, somatic DDX41 mutation preceded phenomena of clonal evolution.
Figure 5.
Comutation patterns of DDX41 mutated and unmutated cases with MNs. (A) Frequency of co-occurring driver mutations in DDX41-mutated and unmutated cases. Significant difference (q < 0.1) is shown by an asterisk. (B) Comparison of the frequency of driver mutations between HR-MDS and sAML in DDX41-mutated and -unmutated cases in forest plots of ORs and their 95% CI. Type-1 (FLT3, NRAS, WT1, NPM1, IDH1, IDH2, and PTPN11) and Type-2 (GATA2, KRAS, TP53, RUNX1, STAG2, ASXL1, ZRSR2, and TET2) genes are indicated by red and blue characters, respectively. (C) Compositions of Type-1, Type-2, somatic DDX41, and other mutations are shown for each set of mutations that were newly acquired, that persisted with increased or decreased clone size, and that were lost in the second sampling. (D) Diagonal plot comparing variant allele frequencies adjusted by CNAs (aVAF) of somatic DDX41 mutations (the horizontal axis) and co-occurring driver mutations in other genes indicated by colors (the vertical axis).
Clinical effects of DDX41 mutations
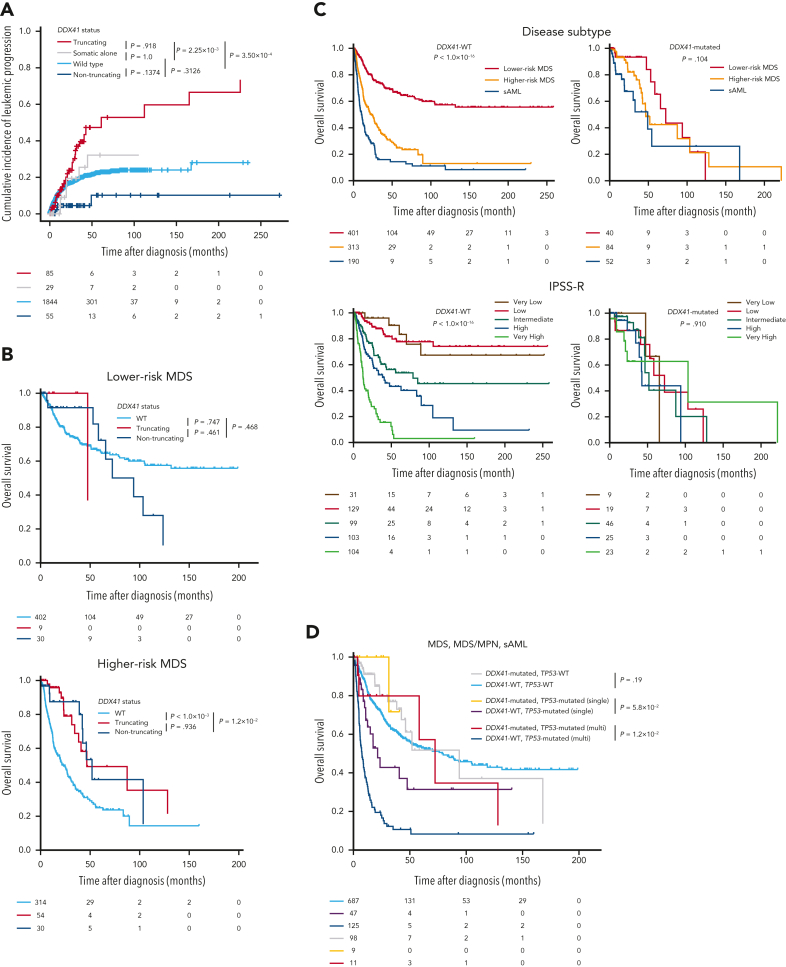
As a whole, DDX41-mutated cases progressed to sAML faster than DDX41-WT (supplemental Figure 15A). However, the effect was confined to P/LP germ line truncating variants. In sharp contrast to such faster AML progression, the leukemic evolution in patients carrying P/LP germ line nontruncating variants or with somatic mutation alone was no faster than that in DDX41-WT cases (Figure 6A). This is in accordance with a significant enrichment of truncating variants in HR-MDS and sAML compared with LR-MDS (supplemental Figure 6B). However, in multivariable analysis including all MDS cases, the effect of truncating variants on leukemia progression was independent of the effects of disease subtype and IPSS-R. By contrast, within DDX41-mutated MDS cases alone, the presence of truncating variants remained the only significant predictor of AML progression (supplemental Table 7). Despite such a large difference in the effect on leukemia progression, OS was similar between P/LP germ line truncating and nontruncating variants (Figure 6B). Analogous results were found when looking at OS in different disease subtypes and IPSS-R/M subgroups with, if ever, a much less conspicuous difference among DDX41-mutated than among DDX41-WT cases (Figure 6C and supplemental Figure 15B-E). Overall, OS of patients with DDX41 mutation was better than that of patients with DDX41-WT HR-MDS/sAML and comparable to that of patients with DDX41-WT LR-MDS (Figure 6C and supplemental Figure 15F), regardless of treatments including hypomethylating agent (HMA), chemotherapy, and HSCT (supplemental Figures 16-19). Although 63.8% of cases in our cohort received both chemotherapy and HMA treatments, such a better prognostic impact of receiving HMA treatments was observed in patients with DDX41-mutation (supplemental Figure 19), even when studying patients underwent HSCT (supplemental Figure 18B). These results suggest that the better prognosis in patients with DDX41-mutation might be associated with effectiveness of HMA treatment. With regard to other prognostic factors, the impact of other well-known predictors of OS in unselected MDS cases was not recapitulated in our DDX41-mutated study cohort. For instance, even multihit TP53 status did not negatively affect OS of DDX41-mutants (Figure 6D and supplemental Figure 20A). Similarly, no other mutation influenced patients’ outcomes (supplemental Figure 20B).
Figure 6.
Clinical impacts of DDX41 germ line and somatic variants. (A) Cumulative incidence of leukemic progression in MNs patients with P/LP germ line truncating/nontruncating DDX41 variants, those with somatic alone, and those without any DDX41 mutation. (B) Kaplan-Meier curves of OS in MDS with and without DDX41 mutations (truncating and nontruncating variants). LR- and HR-MDS were separately shown. (C) Kaplan-Meier curves of OS depending on disease subtype and IPSS-R. Patients with or without any DDX41 mutations were separately shown. (D) Kaplan-Meier curves of OS in MDS, MDS/MPN, and sAML depending on the allelic status of TP53 and the presence of DDX41 mutations. Patients not examined by copy-number analysis, for whom the allelic status of TP53 could not be determined, were excluded from the analysis. P values are provided using Gray test in panel A, and the log-rank test in panels B-D.
Discussion
By taking advantage of a large multinational cohort, our study significantly advances our understanding of DDX41-mutated MNs.
We estimated for the size of the leukemia predisposition ascribed to major DDX41 germ line variants through the analyses of large case-control cohorts and first-degree relatives of DDX41-mutated and unmutated probands, providing instrumental information for the precise management of patients with DDX41-mutation and relatives. Accounting for ∼80% of known leukemia predisposition variants in Japanese cases, P/LP DDX41 variants were 10 times more enriched in MN cases compared with controls and conferred an ∼50% life-time risk of leukemia development. The risk is almost negligible up to the age of 40 but rapidly increases to 25% and 49% by the age of 70 and 90, respectively, when many carriers may die of nonleukemic causes. Together with the significantly older age of onset compared with other leukemia predisposition genes, this further highlights the unique association of DDX41 with late-onset MNs.9, 10, 11, 12, 13, 14, 15,18 Therefore, it is paramount to routinely screen for germ line variants of DDX41, which should be included in molecular profiling panels for elderly patients with MNs, particularly those with higher-risk diseases. It should also be kept in mind that only 44 out of our 292 patients with MN with germ line DDX41 variants had confirmed family history, further emphasizing how the late-onset phenomenon may affect the recognition of this inherited trait.
Furthermore, the large difference in leukemia progression between truncating and nontruncating variants is another major finding. Curiously, despite this large difference in leukemia progression, OS of both variant types did not differ substantially. This indicates that leukemia progression may not necessarily predict a poor OS among patients with DDX41-truncating variants and/or that nonleukemic mortality contributes more to OS than leukemia progression. Moreover, we noted that DDX41-mutant sAML cases have lower blast counts than DDX41-WT sAML cases, suggesting that disease progression of patients with DDX41-truncating variants tends to be stable, keeping relatively low blast counts, even after leukemia progression.
As previously reported,9,12,13,16,22 we noticed a strong male predominance in both DDX41-mutated and WT cohorts, which was more remarkable among DDX41-mutated cases. Accordingly, when considering the enrichment of P/LP variants in MN vs general population as a surrogate for the risk of developing MNs, an increase was observed in males as opposed to females. Similarly, the penetrance associated with DDX41 risk alleles was significantly higher in male than in female subjects. Nevertheless, the frequency of each major DDX41 risk allele did not differ between genders in the general population. Furthermore, no difference in gender distribution was found between healthy individuals with and without DDX41 risk alleles. Taken together, these results suggest that DDX41 P/LP variants have a larger effect on predisposition to MNs in males than in females.
Finally, we elucidated many previously unknown features germane to DDX41-mutated MNs. These tumors show unique hematological phenotypes, including higher degrees of peripheral cytopenia, BM hypocellularity, and erythroid skewing. In addition, they have better OS, which unlike in DDX41-WT cases, does not seem to be substantially affected by IPSS-R/M disease subtype, or even the state of TP53 mutations, although it is important for these findings to be confirmed in independent cohorts in future studies. DDX41-mutated cases have a distinct comutation pattern, compared with DDX41-WT cases, characterized by a higher frequency of CUX1 mutations and lower frequencies of many of other mutations commonly seen in WT cases, regardless of disease subtype. For example, the enrichment of AML–associated Type-1 mutations typically observed in progression from HR-MDS to sAML28 was not recapitulated in DDX41-mutants, suggesting a distinct mechanism of leukemia transformation. In this scenario, AML progression is not associated with mutations in RAS pathway and other signaling molecules (NPM1/IDH1/2), almost a rule in patients with DDX41-WT MDS. Taken together, apart from the impact of heritability, these findings highlight the relevance of correctly recognizing DDX41-mutation status in the management of patients with MN.
Although we acknowledge that the retrospective study design is one of the major caveats of the current study, we believe that the findings presented here are of extreme clinical importance. Our data emphasize the need for recognition of the importance of germ line predisposition to MNs, now feasible thanks to the more widespread adaptation of germ line genetic testing in individuals with MNs and their family members.
Conflict-of-interest disclosure: S.O.: Leadership position/advisory role for: KAN Research Institute, Inc, ChordiaTherapeutics, Inc. Stockholder in: Asahi Genomics Co, Ltd. Grant/Research funding from: KAN Research Institute, Inc, ChordiaTherapeutics, Inc, Sumitomo Dainippon Pharma Co, Ltd, Otsuka Pharmaceutical Co, Ltd, Eisai Co, Ltd. Accepted a researcher from: ChordiaTherapeutics, Inc. The remaining authors declare no competing financial interests.
Acknowledgments
This work was supported by the Japan Agency for Medical Research and Development (AMED) (JP15cm0106056h0005, JP19cm0106501h0004, JP16ck0106073h0003, JP19ck0106250h0003 to S.O.; JP17km0405110h0005, JP19ck0106470h0001, JP21cm0106586h0001 to H.M.; JP19ck0106353h0003 to Y.N.) and the Core Research for Evolutional Science and Technology (CREST) (JP19gm1110011 to S.O.); the Ministry of Education, Culture, Sports, Science and Technology of Japan; the High Performance Computing Infrastructure System Research Project (hp160219, hp170227, hp180198, and hp190158 to S.O. and S.M.) (this research used computational resources of the K computer provided by the RIKEN Advanced Institute for Computational Science through the HPCI System Research project); the Japan Society for the Promotion of Science (JSPS); Scientific Research on Innovative Areas (JP15H05909 to S.O. and S.M.; JP15H05912 to S.M.) and KAKENHI (JP26221308 and JP19H05656 to S.O.; JP16H05338 and JP19H01053 to H.M.; JP15H05707 to S.M.); the Takeda Science Foundation (S.O., H.M., and T.Y.). S.O. is a recipient of the JSPS Core-to-Core Program A: Advanced Research Networks. DNA samples and subjects’ clinical data were provided by Biobank Japan, the Institute of Medical Science, the University of Tokyo. The supercomputing resource was provided by the Human Genome Center, the Institute of Medical Science, the University of Tokyo. This work was supported by a grant from the Edward P. Evans Foundation (to C.G.).
Authorship
Contribution: H.M., Y. Nannya, Y. Momozawa, M.K., M.C., L.M., A.G.K., L.A.G., J.P.M., and S.O. designed the study; S. Korotev, C.G., Y. Momozawa, S.B., P.K., Y.A., M.O., H.I., Y. Miyazaki, T.I., H. Tsurumi, S. Kasahara, C.M.-T., A.T.-K., K.O., T.K., F.M., J.H.J., C.P., P.B., T.H., M.K., M.C., A.G.K., L.A.G., and J.P.M. provided DNA samples and/or clinical data; Y. Nannya, R.S., T.Y., Y. Shiozawa, K.Y., K.C., H. Tanaka and Y.K. performed copy-number analysis; J.T., Y. Momozawa, Y.I.-Y., K.Y., Y. Shiraishi, Y. Nagata, N.K., A.K., Y.O., M.M.N., R.O., T.M., A.Y., M.S., C.P., and P.B. performed sequencing; H.M., Y. Nannya, R.S., Y. Nagata, K.Y., S.M., and S.O. performed bioinformatics analysis; H.M., R.S., Y. Nannya, M.C., L.A.G., J.P.M., and S.O. prepared the manuscript; and all authors participated in discussions and interpretation of the data and results.
Footnotes
The sequencing data from this study are available in the Japanese Genome-phenotype Archive under accession code JGAS000293, the National Center for Biotechnology Information under accession number PRJNA203580, and the European Genome-Phenome Archive under accession number EGAS00001001949.
Data are available on request from the corresponding author, Seishi Ogawa (sogawa-tky@umin.ac.jp).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Supplementary Material
References
- 1.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Cazzola M. Myelodysplastic syndromes. N Engl J Med. 2020;383(14):1358–1374. doi: 10.1056/NEJMra1904794. [DOI] [PubMed] [Google Scholar]
- 3.Godley LA, Shimamura A. Genetic predisposition to hematologic malignancies: management and surveillance. Blood. 2017;130(4):424–432. doi: 10.1182/blood-2017-02-735290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Churpek JE, Pyrtel K, Kanchi KL, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood. 2015;126(22):2484–2490. doi: 10.1182/blood-2015-04-641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogawa S. Genetics of MDS. Blood. 2019;133(10):1049–1059. doi: 10.1182/blood-2018-10-844621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger G, van den Berg E, Sikkema-Raddatz B, et al. Re-emergence of acute myeloid leukemia in donor cells following allogeneic transplantation in a family with a germline DDX41 mutation. Leukemia. 2017;31(2):520–522. doi: 10.1038/leu.2016.310. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi S, Kobayashi A, Osawa Y, et al. Donor cell leukemia arising from preleukemic clones with a novel germline DDX41 mutation after allogenic hematopoietic stem cell transplantation. Leukemia. 2017;31(4):1020–1022. doi: 10.1038/leu.2017.44. [DOI] [PubMed] [Google Scholar]
- 9.Polprasert C, Schulze I, Sekeres MA, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27(5):658–670. doi: 10.1016/j.ccell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewinsohn M, Brown AL, Weinel LM, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127(8):1017–1023. doi: 10.1182/blood-2015-10-676098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardoso SR, Ryan G, Walne AJ, et al. Germline heterozygous DDX41 variants in a subset of familial myelodysplasia and acute myeloid leukemia. Leukemia. 2016;30(10):2083–2086. doi: 10.1038/leu.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebert M, Passet M, Raimbault A, et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood. 2019;134(17):1441–1444. doi: 10.1182/blood.2019000909. [DOI] [PubMed] [Google Scholar]
- 13.Quesada AE, Routbort MJ, DiNardo CD, et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am J Hematol. 2019;94(7):757–766. doi: 10.1002/ajh.25486. [DOI] [PubMed] [Google Scholar]
- 14.Qu S, Li B, Qin T, et al. Molecular and clinical features of myeloid neoplasms with somatic DDX41 mutations. Br J Haematol. 2021;192(6):1006–1010. doi: 10.1111/bjh.16668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alkhateeb HB, Nanaa A, Viswanatha D, et al. Genetic features and clinical outcomes of patients with isolated and comutated DDX41-mutated myeloid neoplasms. Blood Adv. 2022;6(2):528–532. doi: 10.1182/bloodadvances.2021005738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, White T, Xie W, et al. AML with germline DDX41 variants is a clinicopathologically distinct entity with an indolent clinical course and favorable outcome. Leukemia. 2022;36(3):664–674. doi: 10.1038/s41375-021-01404-0. [DOI] [PubMed] [Google Scholar]
- 17.Choi EJ, Cho YU, Hur EH, et al. Unique ethnic features of DDX41 mutations in patients with idiopathic cytopenia of undetermined significance, myelodysplastic syndrome, or acute myeloid leukemia. Haematologica. 2022;107(2):510–518. doi: 10.3324/haematol.2020.270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feurstein S, Trottier AM, Estrada-Merly N, et al. Germ line predisposition variants occur in myelodysplastic syndrome patients of all ages. Blood. 2022;140(24):2533–2548. doi: 10.1182/blood.2022015790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Churpek JE, Smith-Simmer K. In: GeneReviews. Adam MP, Ardinger HH, Pagon RA, et al., editors. University of Washington; 2021. DDX41-associated familial myelodysplastic syndrome and acute myeloid leukemia; pp. 1993–2022. [Google Scholar]
- 20.Voso MT, Gurnari C. Have we reached a molecular era in myelodysplastic syndromes? Hematology Am Soc Hematol Educ Program. 2021;2021(1):418–427. doi: 10.1182/hematology.2021000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duployez N, Largeaud L, Duchmann M, et al. Prognostic impact of DDX41 germline mutations in intensively treated acute myeloid leukemia patients: an ALFA-FILO study. Blood. 2022;140(7):756–768. doi: 10.1182/blood.2021015328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P, Brown S, Williams M, et al. The genetic landscape of germline DDX41 variants predisposing to myeloid neoplasms. Blood. 2022;140(7):716–755. doi: 10.1182/blood.2021015135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saiki R, Momozawa Y, Nannya Y, et al. Combined landscape of single-nucleotide variants and copy number alterations in clonal hematopoiesis. Nat Med. 2021;27(7):1239–1249. doi: 10.1038/s41591-021-01411-9. [DOI] [PubMed] [Google Scholar]
- 24.Tadaka S, Saigusa D, Motoike IN, et al. jMorp: Japanese multi omics reference panel. Nucleic Acids Res. 2018;46(D1):D551–D557. doi: 10.1093/nar/gkx978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagata M, Setoh K, Takahashi M, et al. Association of ALPL variants with serum alkaline phosphatase and bone traits in the general Japanese population: the Nagahama Study. J Hum Genet. 2020;65(3):337–343. doi: 10.1038/s10038-019-0712-3. [DOI] [PubMed] [Google Scholar]
- 26.Yoshizato T, Nannya Y, Atsuta Y, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129(17):2347–2358. doi: 10.1182/blood-2016-12-754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makishima H, Yoshizato T, Yoshida K, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49(2):204–212. doi: 10.1038/ng.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 30.Yoshizato T, Dumitriu B, Hosokawa K, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373(1):35–47. doi: 10.1056/NEJMoa1414799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiraishi Y, Sato Y, Chiba K, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41(7):e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernard E, Nannya Y, Hasserjian RP, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med. 2020;26(10):1549–1556. doi: 10.1038/s41591-020-1008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Struewing JP, Hartge P, Wacholder S, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336(20):1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 35.Whittemore AS, Gong G, Itnyre J. Prevalence and contribution of BRCA1 mutations in breast cancer and ovarian cancer: results from three U.S. population-based case-control studies of ovarian cancer. Am J Hum Genet. 1997;60(3):496–504. [PMC free article] [PubMed] [Google Scholar]
- 36.Levy-Lahad E, Catane R, Eisenberg S, et al. Founder BRCA1 and BRCA2 mutations in Ashkenazi Jews in Israel: frequency and differential penetrance in ovarian cancer and in breast-ovarian cancer families. Am J Hum Genet. 1997;60(5):1059–1067. [PMC free article] [PubMed] [Google Scholar]
- 37.Easton D. Breast cancer genes--what are the real risks? Nat Genet. 1997;16(3):210–211. doi: 10.1038/ng0797-210. [DOI] [PubMed] [Google Scholar]
- 38.Wacholder S, Hartge P, Struewing JP, et al. The kin-cohort study for estimating penetrance. Am J Epidemiol. 1998;148(7):623–630. doi: 10.1093/aje/148.7.623. [DOI] [PubMed] [Google Scholar]
- 39.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernard E, Tuechler H, Greenberg PL, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid. 2022;1(7) doi: 10.1056/EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.