Abstract
While the American horseshoe crab, Limulus polyphemus, has robust circadian and circatidal rhythms, virtually nothing is known about the molecular basis of these rhythms in this species or any other chelicerate. In this study, next generation sequencing was used to assemble transcriptomic reads and then putative homologs of known core and accessory circadian genes were identified in these databases. Homologous transcripts were discovered for one circadian clock input gene, five core genes, 22 accessory genes, and two possible output pathways. Alignments and functional domain analyses showed generally high conservation between the putative L. polyphemus clock genes and homologs from Drosophila melanogaster and Daphnia pulex. The presence of both cry1 and cry2 in the L. polyphemus transcriptome would classify its system as an “ancestral”, type 2 clock system. In addition, a novel duplication of CYCLE, and a novel triplication of PERIOD were found. Investigations are currently underway to determine if any of these “circadian” genes also participate in the molecular processes that drive the Limulus circatidal clock.
Keywords: circadian, biological rhythm, clock, transcriptome, horseshoe crab, chelicerate, circatidal, de novo, Illumina, clock genes
1. Introduction.
Endogenously driven biological rhythms have been observed in virtually all organisms in which a concerted effort has been made to look for them (Dunlap, 1999). These rhythmic changes in physiology and behavior are driven by internal biological clocks and help organisms to both anticipate, and synchronize to, predictable rhythmic changes in their environment. These clocks are composed of three essential elements: 1) the input proteins that allow the clock to synchronize to environmental rhythms, such as light:dark cycles; 2) the clock itself, made up of the proteins that allow the clock to keep time; and 3) the output proteins that help to mediate changes in physiology and behavior (Allada and Chung, 2010; Dunlap, 1999). The molecular basis of these three components of the clock have been elucidated in a few model systems such as the fruit fly, Drosophila melanogaster and the mouse, Mus musculus (Zhang and Kay, 2010) and these findings have provided the framework for studies in other organisms. Although the specific molecular mechanisms of the circadian clock vary greatly between distantly related animal models (i.e. M. musculus and D. melanogaster), the general architecture appears to be well preserved across nearly all living organisms (Dunlap, 1999). In plants, fungi, and animals, circadian clocks appear to be composed of interlocking transcription-translation cycles that feedback to control their own transcription.
These circadian clocks involve two sets of proteins: the core proteins (which are part of a negative feedback loop) and the accessory proteins (which modulate the core proteins and are necessary for maintaining the ~24hr periodicity of the core clock). In D. melanogaster the core clock is composed of four proteins: PERIOD (PER), TIMELESS (TIM), CLOCK (CLK), and CYCLE (CYC) (Allada and Chung, 2010). When the per and tim genes are transcribed and translated, they form a heterodimer that acts at the clk promoter to up-regulate CLK expression (Allada and Chung, 2010; Chang and Reppert, 2003). In other insect species Non-photoreceptive(np) CRYPTOCHROME 2 (npCRY2) acts in addition to, or in place of, TIM (Rubin et al., 2006; Yuan et al., 2007). As PER and TIM build up over time, they dimerize and become phosphorylated by accessory genes such as CASEIN KINASE Ie (CKIe). This phosphorylation allows this heterodimer to enter the nucleus and to remove CLK-CYC from the E-box, turning off per and tim (Allada and Chung, 2010). The proteins CLK and CYC (which appear to be constitutively expressed) are, in turn, capable of repressing the activity of per and tim, completing the negative feedback loop.
This core negative feedback loop is also modulated by a host of accessory proteins, such as VRILLE (VRI), CLOCKWORK ORANGE (CWO), and PAR DOMAIN PROTEIN 1e (PDP1e), which form a secondary feedback loop with CLK (Allada and Chung, 2010). Additionally, the proteins SUPERNUMERARY LIMBS (SLIMB) and ARYL HYDROCARBON RECEPTOR NUCLEAR TRANSPORTER (ARNT) also help to modulate the transcription of the core clock components. The protein kinases CASEIN KINASE IIa (CKIIa), and CASEIN KINASE IIß (CKIIß) serve to phosphorylate PER, while TIM is phosphorylated by CKIIa, CKIIß, CASEIN KINASE Ia (CKIa), SHAGGY (SGG), and JETLAG (JET). Along with these kinases, the protein phosphatases PROTEIN PHOSPHATASE 1a (PP1a), PROTEIN PHOSPHATASE 1ß (PP1ß), PROTEIN PHOSPHATASE 2a-MTS (PP2a-MTS), PROTEIN PHOSPHATASE 2a-WBT (PP2a-WBT), PROTEIN PHOSPHATASE 2a-WS (PP2a-TWS) are also crucial for proper clock function (Allada and Chung, 2010).
The input portion of the circadian clock system is provided by the photoreactive(p) protein CRYPTOCHROME 1 (pCRY1). In D. melanogaster, pCRY1 undergoes a conformational change in the presence of blue light (such as in sunlight) and ubiquinates TIM, tagging it for degradation via the proteasome (Emery et al., 1998; Tauber et al., 2004; Yuan et al., 2007). This degradation acts to “reset” the clock, because when TIM is degraded, it can no longer form a heterodimer with PER and the transcriptional regulation of clk is terminated.
The neuropeptides Pigment Dispersing Hormone (PDH) and NEUROPEPTIDE F (NPF) (Lee et al., 2006; Strauss et al., 2011; Taghert and Shafer, 2006) are crucial for the output of the clock in D. melanogaster. Ablation of PDH cells, or mutations of the pdh gene, disrupts the expression of circadian rhythms in this species D. melanogaster (Renn et al., 1999), and these effects are exacerbated by the ablation of NPF containing cells (Hermann et al., 2012). While these peptides are thought to induce numerous physiological and behavioral changes (Lee et al., 2006; Strauss et al., 2011), the mechanism of the control of either NPF or PDH release by the core circadian clock is not known (Depetris-Chauvin et al., 2011).
While much is known about the molecular mechanisms of the circadian clock in D. melanogaster and a few other insect species (Dunlap, 1999; Tomioka and Matsumoto, 2010), much less is known about the circadian clocks of non-insect arthropods, with the exception of Daphnia pulex (Tilden et al., 2011) and Calanus finmarchicus (Christie et al., 2013). One entire sub-phylum of Arthropoda that has been ignored is the chelicerata, which encompasses a range of organisms including spiders, ticks, mites, scorpions, and horseshoe crabs (Giribet and Edgecombe, 2012). The American horseshoe crab, Limulus polyphemus, has long been known to exhibit robust circadian rhythms of lateral eye sensitivity to light and has served as a model species for studies in photophysiology and its circadian control (Barlow, 1983; Barlow et al., 1980). Bob Barlow, his colleagues, and other investigators have shown that more than twenty independent changes occur in the eye of L. polyphemus to achieve an approximately 100,000 fold change in eye sensitivity between night and day (Barlow et al., 1980; Battelle, 2013). Yet, despite decades of study of this system, the molecular mechanisms that drive this clock in L. polyphemus are completely unknown.
Horseshoe crabs also have an additional timing system that serves to synchronize its locomotor activity to the ~ 12.4 hour tidal cycles (Chabot and Watson, 2010; Watson et al., 2008). While circatidal rhythms such as this have been documented in several species, the molecular basis of these clocks is unknown (de la Iglesia and Hsu, 2010; Takekata et al., 2014; Tessmar-Raible et al., 2011). Furthermore, there is some controversy surrounding the nature of the timing system that drives these rhythms. The two primary competing theories that have been put forth to explain the underlying mechanisms that give rise to these 12.4 h rhythms are: 1) The circatidal oscillator theory, which states that two bouts of activity per day are controlled by one, ~ 12.4 h, circatidal clock (which can also be modulated by a circadian clock (Naylor, 1996)) and; 2) the circalunidian theory, which states that each bout of activity is controlled by a separate, ~24.8 hr (the time between successive moonrises), circalunidian oscillator (Palmer and Williams, 1986). This circalunidian model has been well supported by behavioral evidence in L. polyphemus (Chabot et al., 2016; Chabot and Watson, 2010) and our working hypothesis is that each bout of activity is controlled by a modified circadian (Palmer and Williams, 1986) clock that has a slightly longer period than 24 hours. This would be similar to the situation in D. melanogaster, where dawn and dusk bouts of activity are controlled by separate, coordinated, circadian clocks (Stoleru et al., 2004). The goal of this study was to determine if horseshoe crabs possess some of the circadian genes found in other model species. The identification of these genes in L. polyphemus would not only provide the scientific community with a resource to begin studying circadian clock in chelicerates, but may also allow us to begin to understand the molecular basis of circatidal/circalunidian clocks.
2. Methods
2.1. Animals and RNA extraction.
L. polyphemus individuals were collected from Great Bay in Durham NH. Animals were treated in accordance with NIH guide for the care and use of animals (NIH publications No. 8023, revised 1978). The entire central nervous system (brain and ventral nerve cord) was dissected from 4 individuals, immediately snap frozen on dry ice, and stored in 1.5ml Eppendorf tubes at −80°C. Samples were then shipped on dry ice to University of Vermont Cancer Center DNA Analysis Facility (Burlington, VT), where RNA was extracted using Trizol RNA extraction (Invitrogen, Carlsbad, CA) and cleaned using the Qiagen RNAeasy Mini column (Qiagen, Valencia, CA). Quality and concentration of cleaned RNA was determined using Nanodrop spectrophotometer (Thermo Scientific, Madison WI), Qubit Spectrofluorometer (Life Technologies, Carlsbad, CA) and Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA).
2.1. Library construction and sequencing.
A transcriptome library was constructed using Illumina TruSeq RNA Sample Prep LT version 2 (RS-122–2001/2002). 1 ug of each sample was PolyA enriched using magnetic beads and reverse transcribed into cDNA via Superscript II [Invitrogen]. The cDNA was then fragmented, end-repaired, adenylated, and tagged with sequence adaptors for source identification. The cDNA was sequenced using 12pM/flow cell lane with Illumina CBOT and HiSeq1000/1500.
2.2. Transcriptome de novo assembly.
Transcriptome reads were de novo assembled using the CLC genomics workbench proprietary algorithm (CLC version 5.1.2, CLCBio, Boston, MA), as well as SOAP de novo assembly software (Luo et al., 2012) (http://soap.genomics.org.cn/soapdenovo.html, version 1.02) with the default settings.
2.2. Transcriptome mining and gene identification.
The transcriptome was mined with the CLC genomics workbench tBLASTn program (version 5.1.2), using D. melanogaster and other arthropod query sequences. The top hits from the L. polyphemus transcriptome were translated and subjected to a reciprocal BLASTp against the entire UniProtKB protein database to verify sequence homology. A second reciprocal BLASTp was also performed against D. melanogaster and D. pulex circadian proteins exclusively to compare scores to these model organisms.
2.3. Sequence analysis.
Transcripts of interest were translated into protein sequences using CLC genomics workbench (version 5.1.2). These sequences were then aligned to respective orthologs of D. melanogaster and D. pulex, along with an ortholog of a chelicerate species when one was available using CLUSTAL Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/). Functional domains of all proteins of interest in L. polyphemus, D. melanogaster, Daphnia, and chelicerate species when present were identified using SMART Genome program (http://smart.embl-heidelberg.de/ (Letunic et al., 2015; Schultz et al., 1998)). The %Identity/%Similarity of the amino acid sequences these proteins and their respective functional domains were calculated using default settings of the SIAS program (http://imed.med.ucm.es/Tools/sias.html). Sequence alignments from CLUSTAL Omega were pasted into a word document and functional domains were color coded as follows: Light blue:bHLH, Green:PAS, Red:PAC, Grey:Period C, Dark green:Photolyase, Gold:FAD Binding 7, Dark blue:Timeless. Phylogenetic trees were constructed using Mega 5.2 (http://www.megasoftware.net/). Sequences, other than L. polyphemus proteins, were extracted from UniProt database and aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). After alignment a Maximum likelihood tree was generated using default setting and 500 Bootstraps. Tree branches were color coded to specify different clades on the trees: Red:Chelicerata, Blue:Non-chelicerate arthropods, Black:Vertebrates, Violet:Fungi, Green:Nematode. Diamonds were added at gene duplication events.
3. Results.
3.1. Top 5 BLAST hits from UniProtKB
In our analysis of the L. polyphemus (Lp) transcriptome we identified orthologs of all five core clock components (PER, TIM, CLK, CYC, npCRY2), including 3 copies of the per gene [perioda (pera), periodb (perb), and periodc (perc)] and 2 copies of the cyc gene [cycle1 (cyc1) and cycle2 (cyc2)]. Top BLAST hits for all core clock genes are reported in Table 1. A reciprocal BLASTp of all three LpPER paralogs against the UniProt KB Protein database showed closest homology to arthropod PER proteins, including orthologs from three insect species and one chelicerate species (Ixodes scapularis, Black legged tick). BLASTp of LpTIM, LpCLK, LpCYC1, LpCYC2 and LpnpCRY2 showed closest homology to insect orthologs. Top BLAST hits for all core clock components were significant, with e-values ranging from 10^−114 to less than 10^−180, and BLAST scores ranging from 1004 to 2199.
Table 1:
Top 5 BLASTp hits from UniProt.
| Proteins | UniProt KB Protein Database | ||||
|---|---|---|---|---|---|
| Core Proteins | UniProt ID | Species | Protein description | E-value | BLAST score |
| PERIODA (PERA) | |||||
| LpPERA | H7C8F3 | Apteronemobius asahinai | PERIOD, isoform 1 | 1e-121 | 1,058 |
| H7C8F4 | Apteronemobius asahinai | PERIOD, isoform 2 | 2e-121 | 1,054 | |
| B7PDL3 | Ixodes scapularis | Period circadian protein, putative | 4e-118 | 1,013 | |
| Q8MMG2 | Blattella germanica | Circadian clock protein PERIOD | 1e-114 | 1,007 | |
| L0CAI9 | Rhyparobia maderae | Period | 2e-114 | 1,009 | |
| PERIODB (PERB) | |||||
| LpPERB | B7PDL3 | Ixodes scapularis | Period circadian protein, putative | 6e-119 | 1,020 |
| Q8MMG2 | Blattella germanica | Circadian clock protein PERIOD | 8e-118 | 1,031 | |
| A1EA98 | Blattella bisignata | Circadian clock protein period | 2e-115 | 1,008 | |
| H7C8F3 | Apteronemobius asahinai | PERIOD, isoform 1 | 2e-114 | 1,008 | |
| H7C8F4 | Apteronemobius asahinai | PERIOD, isoform 2 | 4e-114 | 1,004 | |
| PERIODC (PERC) | |||||
| LpPERc | H7C8F3 | Apteronemobius asahinai | PERIOD, isoform 1 | 4e-133 | 1,142 |
| H7C8F4 | Apteronemobius asahinai | PERIOD, isoform 2 | 3e-132 | 1,134 | |
| B7PDL3 | Ixodes scapularis | Period circadian protein, putative | 2e-129 | 1,095 | |
| L0CAI9 | Rhyparobia maderae | Period | 9e-129 | 1,115 | |
| A1EA98 | Blattella bisignata | Circadian clock protein period | 5e-124 | 1,071 | |
| TIMELESS (TIM) | |||||
| LpTIM | J9Y3V3 | Clunio marinus | Timeless | 2e-180 | 1,464 |
| G9M9V8 | Thermobia domestica | TIMELESS | 4e-178 | 1,458 | |
| H2D5T9 | Aedes albopictus | Timeless | 6e-165 | 1,370 | |
| E0D5C1 | Gryllus bimaculatus | TIMELESS | 5e-164 | 1,359 | |
| Q05E95 | Aedes aegypti | TIMELESS | 2e-161 | 1,349 | |
| CLOCK (CLK) | |||||
| LpCLK | E2C765 | Harpegnathos saltator | Circadian locomoter output cycles protein kaput | 4e-164 | 1,276 |
| H9KJ84 | Apis mellifera | Uncharacterized protein | 3e-161 | 1,264 | |
| E2AY34 | Camponotus floridanus | Circadian locomoter output cycles protein kaput | 3e-159 | 1,251 | |
| F4WUR0 | Acromyrmex echinatior | Circadian locomoter output cycles protein kaput | 1e-156 | 1,233 | |
| H9HL46 | Atta cephalotes | Uncharacterized protein | 4e-156 | 1,223 | |
| CYCLE 1 (CYC1) | |||||
| LpCYC1 | E0D6T3 | Thermobia domestica | CYCLE | <1e180 | 1,786 |
| A0MH07 | Lutzomyia longipalpis | Cycle | <1e180 | 1,622 | |
| E2A3F0 | Camponotus floridanus | Aryl hydrocarbon receptor nuclear translocator | <1e180 | 1,643 | |
| A9XCF1 | Tribolium castaneum | CYCLE | <1e180 | 1,621 | |
| E2B7K2 | Harpegnathos saltator | Aryl hydrocarbon receptor nuclear translocator | <1e180 | 1,620 | |
| CYCLE 2 (CYC2) | |||||
| LpCYC2 | E0D6T3 | Thermobia domestica | CYCLE | <1e180 | 1,674 |
| E2A3F0 | Camponotus floridanus | Aryl hydrocarbon receptor nuclear translocator | <1e180 | 1,606 | |
| H9K2D4 | Apis mellifera | CYC | <1e180 | 1,562 | |
| A0MH07 | Lutzomyia longipalpis | Cycle | <1e180 | 1,555 | |
| A9XCF1 | Tribolium castaneum | CYCLE | <1e180 | 1,560 | |
| CRYPTOCHROME 2 (npCRY2) | |||||
| LpnpCRY2 | E2BGC2 | Harpegnathos saltator | Cryptochrome-1 | <1e180 | 2,213 |
| L7YAB3 | Solenopsis invicta | Cryptochrome | <1e180 | 2,210 | |
| A8QSC1 | Bombus impatiens | Cryptochrome 2 protein | <1e180 | 2,202 | |
| L0C8K6 | Rhyparobia maderae | Cryptochrome 2 | <1e180 | 2,199 | |
| F4WVC9 | Acromyrmex echinatior | Cryptochrome-1 | <1e180 | 2,199 | |
| Accessory Proteins | |||||
| VRILLE (VRI) | |||||
| LpVRI | E0VHE1 | Pediculus humanus | Transcriptional factor nfil3/e4bp4, putative | 1e-43 | 424 |
| B7PEG8 | Ixodes scapularis | Transcriptional factor nfil3/e4bp4, putative | 9e-43 | 417 | |
| Q699T4 | Antheraea pernyi | Vrille | 6e-42 | 410 | |
| Q1XD36 | Danaus plexippus | Vrille | 1e-41 | 408 | |
| J9K7G7 | Acyrthosiphon pisum | Uncharacterized protein | 4e-40 | 403 | |
| CLOCK WORK ORANGE (CWO) | |||||
| LpCWO | B7PGM4 | Ixodes scapularis | Putative uncharacterized protein | 1e-57 | 528 |
| B4LW47 | Drosophila virilis | GJ23560 | 2e-42 | 430 | |
| Q7QFU3 | Anopheles gambiae | AGAP003844-PA | 2e-41 | 422 | |
| E9GEU4 | Daphnia pulex | Putative uncharacterized protein | 7e-41 | 395 | |
| B4JIN1 | Drosophila grimshawi | GH19128 | 2e-40 | 415 | |
| SUPERNUMERARY LIMBS1 (SLIMB1) | |||||
| LpSLIMB1 | L7M0T0 | Rhipicephalus pulchellus | Uncharacterized protein | <1e180 | 2,159 |
| E9IHL0 | Solenopsis invicta | Putative uncharacterized protein | <1e180 | 2,159 | |
| D2XMQ7 | Saccoglossus kowalevskii | Beta-TCRP E3 ligase | <1e180 | 2,145 | |
| H9HC45 | Atta cephalotes | Uncharacterized protein | <1e180 | 2,142 | |
| D6WA15 | Tribolium castaneum | Supernumerary limbs | <1e180 | 2,138 | |
| SUPERNUMERARY LIMBS2 (SLIMB2) | |||||
| LpSLIMB2 | L7M0T0 | Rhipicephalus pulchellus | Uncharacterized protein | <1e180 | 2,153 |
| E9HMX3 | Daphnia pulex | Putative uncharacterized protein | <1e180 | 2,150 | |
| E9IHL0 | Solenopsis invicta | Putative uncharacterized protein | <1e180 | 2,147 | |
| H3JK44 | Strongylocentrotus purpuratus | Uncharacterized protein | <1e180 | 2,135 | |
| G3I574 | Cricetulus griseus | F-box/WD repeat-containing protein 1A | <1e180 | 2,133 | |
| ARYL HYDROCARBON RECEPTOR NUCLEAR TRANSPORTER1 (ARNT1) | |||||
| LpARNT1 | C7B7E8 | Litopenaeus vannamei | Hypoxia inducible factor 1 beta | <1e180 | 1,799 |
| E0VXW6 | Pediculus humanus | Putative uncharacterized protein | <1e180 | 1,805 | |
| L7MH74 | Rhipicephalus pulchellus | Putative tango | <1e180 | 1,770 | |
| D6W6H5 | Tribolium castaneum | Putative uncharacterized protein | <1e180 | 1,739 | |
| Q1JUI4 | Daphnia magna | Aryl hydrocarbone receptor nuclear translocator | <1e180 | 1,723 | |
| ARYL HYDROCARBON RECEPTOR NUCLEAR TRANSPORTER2 (ARNT2) | |||||
| LpARNT2 | E0VXW6 | Pediculus humanus | Putative uncharacterized protein | <1e180 | 1,900 |
| C7B7E8 | Litopenaeus vannamei | Hypoxia inducible factor 1 beta | <1e180 | 1,858 | |
| Q1JUI4 | Daphnia magna | Aryl hydrocarbone receptor nuclear translocator | <1e180 | 1,850 | |
| Q1JUI5 | Daphnia magna | Aryl hydrocarbone receptor nuclear translocator | <1e180 | 1,838 | |
| E9FQM5 | Daphnia pulex | Putative aryl hydrocarbon receptor nuclear translocator | <1e180 | 1,838 | |
| CASIEN KINASE Iα (CKIα) | |||||
| LpCKIα | Q8JG73 | Danio rerio | Casein kinase 1alpha S | <1e180 | 1,570 |
| Q75WS8 | Carassius auratus | Casein kinase I alpha S | <1e180 | 1,570 | |
| Q8JGT0 | Danio rerio | Casein kinase 1, alpha 1 | <1e180 | 1,568 | |
| Q75WS9 | Carassius auratus | Casein kinase I alpha | <1e180 | 1,568 | |
| B5THN0 | Saccoglossus kowalevskii | Casein kinase 1 protein catalytic subunit | <1e180 | 1,567 | |
| CASIEN KINASE Iε (CKIε) | |||||
| LpXKIε | F6YKD5 | Equus caballus | Uncharacterized protein | <1e180 | 1,422 |
| G1SGL5 | Oryctolagus cuniculus | Uncharacterized protein | <1e180 | 1,418 | |
| G1U4F8 | Oryctolagus cuniculus | Uncharacterized protein | <1e180 | 1,418 | |
| M7AVG9 | Chelonia mydas | Casein kinase I isoform epsilon | <1e180 | 1,416 | |
| H0V5Y3 | Cavia porcellus | Uncharacterized protein | <1e180 | 1,416 | |
| CASIEN KINASE IIα (CKIIα) | |||||
| LpCKIIα | E2AW17 | Camponotus floridanus | Casein kinase II subunit alpha | <1e180 | 1,674 |
| F4WU82 | Acromyrmex echinatior | Casein kinase II subunit alpha | <1e180 | 1,666 | |
| B7PVH2 | Ixodes scapularis | Mitogen-activated protein kinase, putative | <1e180 | 1,671 | |
| E2BH98 | Harpegnathos saltator | Casein kinase II subunit alpha | <1e180 | 1,669 | |
| K7GIG0 | Pelodiscus sinensis) | Uncharacterized protein | <1e180 | 1,665 | |
| CASIEN KINASE IIβ (CKIIβ) | |||||
| LpCKIIβ | Q71U52 | Cyprinus carpio | CK2 beta subunit | 5e-144 | 1,057 |
| Q6DEU1 | Xenopus tropicalis | Casein kinase 2, beta polypeptide | 5e-144 | 1,057 | |
| Q1LXD2 | Danio rerio | Casein kinase 2 beta | 5e-144 | 1,057 | |
| M4AI87 | Xiphophorus maculatus | Uncharacterized protein | 5e-144 | 1,057 | |
| J3SBX9 | Crotalus adamanteus | Casein kinase II subunit beta | 5e-144 | 1,057 | |
| JETLAG (JET) | |||||
| LpJET | K1P9I2 | Crassostrea gigas | F-box only protein 37 | 1e-37 | 354 |
| C3Y7V1 | Branchiostoma floridae | Putative uncharacterized protein | 8e-37 | 356 | |
| C3ZZX8 | Branchiostoma floridae | Putative uncharacterized protein | 6e-35 | 342 | |
| R7VDT7 | Capitella teleta | Uncharacterized protein | 1e-32 | 326 | |
| A7SQL5 | Nematostella vectensis | Predicted protein | 1e-31 | 320 | |
| SHAGGY (SGG) | |||||
| LpSGG | E2C155 | Harpegnathos saltator | Protein kinase shaggy | <1e180 | 1,478 |
| E9H6Q6 | Daphnia pulex | Glycogen synthase kinase 3 beta | <1e180 | 1,475 | |
| E9I8X0 | Solenopsis invicta | Putative uncharacterized protein | <1e180 | 1,469 | |
| G3MII6 | Amblyomma maculatum | Putative uncharacterized protein | <1e180 | 1,453 | |
| F4X3V9 | Acromyrmex echinatior | Protein kinase shaggy | <1e180 | 1,467 | |
| PROTEIN PHOSPHOTASE 1α (PP1α) | |||||
| LpPP1α | K1PXG6 | Crassostrea gigas | Serine/threonine-protein phosphatase | <1e180 | 1,631 |
| A7RVJ0 | Nematostella vectensis | Serine/threonine-protein phosphatase | <1e180 | 1,627 | |
| E9HKA6 | Daphnia pulex | Serine/threonine-protein phosphatase | <1e180 | 1,624 | |
| H2L9G2 | Oryzias latipes | Serine/threonine-protein phosphatase | <1e180 | 1,620 | |
| H9KP59 | Apis mellifera | Serine/threonine-protein phosphatase | <1e180 | 1,618 | |
| PROTEIN PHOSPHOTASE 1β (PP1β) | |||||
| LpPP1β | L7MB61 | Rhipicephalus pulchellus | Serine/threonine-protein phosphatase | <1e180 | 1,470 |
| H2UK23 | Takifugu rubripes | Serine/threonine-protein phosphatase | <1e180 | 1,458 | |
| H3DIM0 | Tetraodon nigroviridis | Serine/threonine-protein phosphatase | <1e180 | 1,457 | |
| H2MDU5 | Oryzias latipes | Serine/threonine-protein phosphatase | <1e180 | 1,457 | |
| G3P7R9 | Gasterosteus aculeatus | Serine/threonine-protein phosphatase | <1e180 | 1,457 | |
| PROTEIN PHOSPHOTASE 2α-mts (PP2α-mts) | |||||
| LpPP2α-mts | L7M975 | Rhipicephalus pulchellus | Serine/threonine-protein phosphatase | <1e180 | 1,336 |
| B7QGY5 | Ixodes scapularis | Serine/threonine-protein phosphatase | <1e180 | 1,335 | |
| E0VKT2 | Pediculus humanus | Serine/threonine-protein phosphatase | <1e180 | 1,333 | |
| R4G8Q4 | Rhodnius prolixus | Putative serine/threonine protein phosphatase | <1e180 | 1,330 | |
| E9IAY8 | Solenopsis invicta | Serine/threonine-protein phosphatase | <1e180 | 1,334 | |
| PROTEIN PHOSPHOTASE 2α-WBT (PP2α-WBT) | |||||
| LpPP2α-WBT | L7M3M4 | Rhipicephalus pulchellus | Putative serine/threonine protein phosphatase… | <1e180 | 2,137 |
| H9J639 | Bombyx mori | Uncharacterized protein | <1e180 | 2,073 | |
| E2AI01 | Camponotus floridanus | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit | <1e180 | 2,045 | |
| H9K2X9 | Apis mellifera | Uncharacterized protein | <1e180 | 2,044 | |
| E0VNG0 | Pediculus humanus | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit | <1e180 | 2,041 | |
| PROTEIN PHOSPHOTASE 2α-TWS (PP2α-TWS) | |||||
| LpPP2α-TWS | H9KM79 | Apis mellifera | Uncharacterized protein | <1e180 | 2,081 |
| I3RJJ3 | Scylla paramamosain | Protein phosphatase 2A regulatory subunit B | <1e180 | 2,074 | |
| G3MLH6 | Amblyomma maculatum | Putative uncharacterized protein | <1e180 | 2,070 | |
| L7M886 | Rhipicephalus pulchellus | Putative serine/threonine protein phosphatase | <1e180 | 2,063 | |
| F4WRB8 | Acromyrmex echinatior | Protein phosphatase PP2A 55 kDa regulatory subunit | <1e180 | 2,055 | |
| Input Pathway Protein | |||||
| CRYPTOCHROME 1 (CRY1)† | |||||
| LpCRY1 | A2A264 | Dianemobius nigrofasciatus | Cryptochrome | 4e-132 | 1,022 |
| K1PRK6 | Crassostrea gigas | Cryptochrome-1 | 3e-120 | 941 | |
| R7V1J3 | Capitella teleta | Uncharacterized protein | 5e-119 | 937 | |
| Q2TJN5 | Danaus plexippus | Cryptochrome | 5e-115 | 908 | |
| J9T2S0 | Mythimna separata | Cryptochrome 1 | 4e-112 | 888 | |
| Output Pathway Proteins | |||||
| NEUROPEPTIDE F RECEPTOR (NPFR)† | |||||
| LpNPFR | T1KR44 | Tetranychus urticae | Uncharacterized protein | 6e-96 | 759 |
| T1IW42 | Strigamia maritima | Uncharacterized protein | 7e-88 | 707 | |
| B0WJ64 | Culex quinquefasciatus | G-protein coupled receptor | 3e78 | 642 | |
| Q16SC4 | Aedes aegypti | AAEL010626-PA | 5e-78 | 636 | |
| T1JTP7 | Tetranychus urticae | Uncharacterized protein | 1e-77 | 642 | |
| NEUROPEPTIDE F (NPF) | |||||
| LpNPF | T1KF83 | Tetranychus urticae | Uncharacterized protein | 23e-12 | 156 |
| A0A087UNM1 | Stegodyphus mimosarum | Uncharacterized protein | 160e-9 | 128 | |
| T1JM02 | Strigamia maritima | Uncharacterized protein | 670e-9 | 125 | |
| F6KM62 | Litopenaeus vannamei | Preproneuropeptide F I | 0.0000013 | 123 | |
| F6KM64 | Melicertus marginatus | Preproneuropeptide F I | 0.0000013 | 123 | |
| PIGMENT DISPERSING HORMONE RECEPTOR (PDHR)† | |||||
| LpPDHR | K7JRN8 | Nasonia vitripennis | Uncharacterized protein | 2.4e-63 | 548 |
| U3U904 | Nilaparvata lugens | Neuropeptide GPCR B2 | 95e-63 | 545 | |
| Q7PQE3 | Anopheles gambiae | AGAP003654-PA | 830e-63 | 537 | |
| A0A0P5AQT3 | Daphnia magna | Class b secretin g-protein coupled receptor | 590e-63 | 533 | |
| A0A0P5CML9 | Daphnia magna | Class b secretin g-protein coupled receptor | 1.4e-60 | 533 | |
indicates partial sequence.
We also identified orthologs of 15 accessory clock components (VRI, CWO, SLIMB, ARNT, CK Iα, CK Iε, CK IIα, CK IrlIβ, JET, SGG, PP1α, PP1β, PP2αMTS, PP2αWBT, PP2αTWS), including duplications of both the LpSLIMB and LpARNT genes, for a total of 17 accessory genes. Only one known accessory clock gene (PDP1) was not found in the L. polyphemus transcriptome. Top BLAST hits for all accessory clock genes are reported in Table 1. A reciprocal BLAST of the L. polyphemus orthologs revealed nearly all clock genes (VRI, CWO, SLIMB1, SLIMB2, ARNT1, ARNT2, CKIα, JET, SGG, PP1α, PP1β, PP2α-MTS, PP2α-MTS, PP2α-TWS) are most closely related to arthropod orthologs. However, CKIα, CKIε, and CKIIβ all show highest levels of homology to vertebrate orthologs. The top 5 BLAST hits of 10 of the17 accessory proteins included at least one known chelicerate ortholog, primarily from the tick family. Top BLAST hits for nearly all accessory clock components were significant, with e-values ranging from 10^−31 to less than 10^−180, and BLAST scores ranging from 320 to 2138 (only LpVRI, LpCWO, and LpJET score below 1000).
We were able to identify partial sequences for the input protein pCRY1, the NPF receptor (NPFR) and the PDH receptor (PDHR), as well as a complete sequence for the output neuropeptide NPF. PDH was not identified in the transcriptome of L. polyphemus. Top BLAST hits for all input and output clock genes are reported in Table 1. The top BLAST hits for LppCRY1 showed that LppCRY1 is most closely related to insect pCRY1 sequences. Despite only being a partial sequence this protein is highly conserved and had an e-value of 10^−132, with a BLAST score of 1022. BLASTp of LpNPF, LpNPFR, and LpPDHR showed closest homology to arthorpod orthologs, including several chelicerates. Top BLAST hits for most output clock components (excluding LpNPF) were significant, with e-values ranging from 10^−60 to 10^−150, and BLAST scores ranging from 533 to 1022. Top BLAST hits for LpNPF showed e-values of 10^−3 and BLAST scores of 84 and 86. The high e-values and low BLAST scores of the LpNPF BLAST are likely because NPF is a short peptide (96 amino acids long).
3.2. Comparisons with D. melanogaster and D. pulex clock genes
Top BLAST hits for all core clock genes against the D. melanogaster and D. pulex databases are reported in Table 2. All putative core clock components identified in this study, with the exception of npCRY2, (PERA, PERB, PERC, TIM, CLK, CYC1, and CYC2,) were found to have the highest homology to the orthologous proteins of D. melanogaster and D. pulex. LpnpCRY2 most closely matched a 6–4 photolyase in D. melanogaster, which is the closest relative to npCRY2 on the cryptochrome gene family tree. This mis-match is the product of the fact that the genus D. melanogaster lost its npCRY2 ortholog during its evolution. However, Apis mellifera retained its npCRY2 ortholog, and the top BLAST hit for LpCRY2 in Apis is npCRY2 (data not shown). Similarly, the closest match to LpnpCRY2 in the D. pulex protein database was “CRY-M”, which is another name for npCRY2. The top BLAST hits for D. melanogaster and D. pulex also showed high levels of homology to our genes of interest, with e-values ranging from 10^−68 to less than 10^−180 and BLAST scores ranging from 653 to 2139.
Table 2:
Top BLASTp hits for Drosophila melanogaster and Daphnia pulex.
| Proteins | Species name | Protein name | UniProt ID | Blast Score | E-value |
|---|---|---|---|---|---|
| CORE PROTEINS | |||||
| PERa | Drosophila melanogaster | Isoform PER-E | P07663 | 735 | 2.0×10–78 |
| Daphnia pulex | PER | E9GW67 | 822 | 1.0×10–90 | |
| PERb | Drosophila melanogaster | Isoform PER-E | P07663 | 653 | 7.0×10–68 |
| Daphnia pulex | PER | E9GW67 | 701 | 4.0×10–75 | |
| PERc | Drosophila melanogaster | Isoform PER-E | P07663 | 765 | 5.0×10–82 |
| Daphnia pulex | PER | E9GW67 | 876 | 1.0×10–97 | |
| TIM | Drosophila melanogaster | Timeless, isoform G | B7Z007 | 1,290 | 3.0×10–152 |
| Daphnia pulex | Putative TIMELESS/TIM-1 protein | E9FZ81 | 956 | 2.0×10–109 | |
| CLK | Drosophila melanogaster | Clk CLOCK jrk PAS1 | O61735 | 1,048 | 1.0×10–125 |
| Daphnia pulex | CLOCK | E9GKD1 | 1,033 | 2.0×10–126 | |
| CYC1 | Drosophila melanogaster | Protein cycle | O61734 | 1,220 | 6.0×10–159 |
| Daphnia pulex | CYCLE | E9FRH8 | 1,500 | <1.0×10–180 | |
| CYC2 | Drosophila melanogaster | Protein cycle | O61734 | 1,227 | 6.0×10–160 |
| Daphnia pulex | CYCLE | E9FRH8 | 1,470 | <1.0×10–180 | |
| npCRY2 | Drosophila melanogaster | phr6-4 | Q8SXK5 | 1,296 | 3.0×10–171 |
| Daphnia pulex | CRY-M | E9GDJ9 | 2,139 | <1.0×10–180 | |
| Accessory Proteins | |||||
| VRI | Drosophila melanogaster | Vri | Q7KTN9 | 306 | 1.0×10–28 |
| Daphnia pulex | Vri† | E9HB85 | 280 | 1.0×10–28 | |
| CWO | Drosophila melanogaster | cwo-RA | B7FNP5 | 312 | 1.0×10–29 |
| Daphnia pulex | Putative uncharacterized protein | E9GEU4 | 395 | 2.0×10–42 | |
| SLIMB1 | Drosophila melanogaster | Slimb | Q9VDE3 | 2,114 | <1.0×10–180 |
| Daphnia pulex | f-box/wd-repeat protein* | E9HMX3 | 2,136 | <1.0×10–180 | |
| SLIMB2 | Drosophila melanogaster | Slimb | Q9VDE3 | 2,107 | <1.0×10–180 |
| Daphnia pulex | f-box/wd-repeat protein* | E9HMX3 | 2,150 | <1.0×10–180 | |
| ARNT1 | Drosophila melanogaster | Aryl hydrocarbon receptor nuclear translocator homolog | O15945 | 1,568 | <1.0×10–180 |
| Daphnia pulex | Putative aryl hydrocarbon receptor nuclear translocator | E9FQM5 | 1,723 | <1.0×10–180 | |
| ARNT2 | Drosophila melanogaster | Aryl hydrocarbon receptor nuclear translocator homolog | O15945 | 1,680 | <1.0×10–180 |
| Daphnia pulex | Putative aryl hydrocarbon receptor nuclear translocator | E9FQM5 | 1,838 | <1.0×10–180 | |
| CKIε | Drosophila melanogaster | Dco dbt | O76324 | 1,282 | 6.0×10–173 |
| Daphnia pulex | Casein kinase i alpha* | E9FS31 | 1,171 | 3.0×10–157 | |
| CKIα | Drosophila melanogaster | Casein kinase I isoform alpha | O76324 | 1,348 | <1.0×10–180 |
| Daphnia pulex | Casien kinase I alpha* | E9HGM4 | 1,526 | <1.0×10–180 | |
| CKIIα | Drosophila melanogaster | Casein kinase II alpha subunit, isoform C | P08181 | 1,590 | <1.0×10–180 |
| Daphnia pulex | Casein kinase ii subunit alpha* | E9GCV0 | 1,659 | <1.0×10–180 | |
| CKIIβ | Drosophila melanogaster | Casein kinase II beta subunit, isoform C | P08182–3 | 1,010 | 6.0×10–137 |
| Daphnia pulex | Casein kinase ii subunit beta* | E9GTE8 | 1,033 | 6.0×10–142 | |
| JET | Drosophila melanogaster | Jetlag, isoform B | Q0E8T8 | 188 | 3.0×10–15 |
| Daphnia pulex | f-box/leucine rich repeat protein* | E9G1Z9 | 155 | 2.0×10–10 | |
| SGG | Drosophila melanogaster | Isoform G of Protein kinase shaggy | P18431 | 1,357 | <1.0×10–180 |
| Daphnia pulex | Glycogen synthase kinase 3 beta | E9H6Q6 | 1,475 | <1.0×10–180 | |
| PP1α | Drosophila melanogaster | Pp1alpha-96A | P48461 | 1,575 | <1.0×10–180 |
| Daphnia pulex | Serine/threonine-protein phosphatase | E9HKA6 | 1,624 | <1.0×10–180 | |
| PP1β | Drosophila melanogaster | Serine/threonine-protein phosphatase beta isofrom | H5V895 | 1,386 | <1.0×10–180 |
| Daphnia pulex | Serine/threonine-protein phosphatase | E9G7U7 | 1,427 | <1.0×10–180 | |
| PP2a-MTS | Drosophila melanogaster | mts PP2A | P23696 | 1,313 | <1.0×10–180 |
| Daphnia pulex | Serine/threonine-protein phosphatase | E9G8K4 | 842 | 5.0×10–110 | |
| PP2-WBT | Drosophila melanogaster | wdb | Q9VB23 | 1,986 | <1.0×10–180 |
| Daphnia pulex | Serine/threonine-protein phosphatase kda regulatory subunit* | E9G2F2 | 1,932 | <1.0×10–180 | |
| PP2-TWS | Drosophila melanogaster | tws aar Pp2A-85F | P36872 | 2,003 | <1.0×10–180 |
| Daphnia pulex | TWS | E9GU66 | 1,974 | <1.0×10–180 | |
| INPUT PATHWAY PROTEINS | |||||
| CRY1† | Drosophila melanogaster | Cryptochrome-1 | O77059 | 754 | 1.0×10–93 |
| Daphnia pulex | CRY-D | E9GSJ7 | 754 | 9.0×10–94 | |
| OUTPUT PATHWAY PROTEINS | |||||
| NPFR† | Drosophila melanogaster | Isoform 6 of Neuropeptide F receptor | Q9VNM1 | 583 | 1.0×10–68 |
| Daphnia pulex | NPFG-protein-coupled receptor | E9GBE9 | 593 | 3.0×10–71 | |
| NPF | Drosophila melanogaster | Neuropeptide F | Q9VET0 | 84 | 5.0×10–3 |
| Daphnia pulex | Putative uncharacterized protein | E9GJI3 | 86 | 2.0×10–3 | |
| PDHR† | Drosophila melanogaster | PDF receptor | Q9W4Y2 | 445 | 1.4 ×10–48 |
| Daphnia pulex | Putative PDF receptor variant 2 | E9FR28 | 532 | 6.3×10–62 | |
indicates partial sequence.
indicates Protein name which originated from D. pulex genome map on Fleabase.
Top BLAST hits for all accessory clock genes against the D. melanogaster and D. pulex databases are reported in Table 2. All putative accessory clock components identified in this study were found to have the highest homology to the predicted orthologous proteins of D. melanogaster and D. pulex. Although on several occasions we found that the closest match to the L. polyphemus genes was an uncharacterized protein (as seen when LpCWO was blasted against the D. pulex database), or the name of the hit simply referred to the gene family (e.g. when the LpSLIMB1 and LpSLIMB2 genes matched the “f-box/wd-repeat protein” in D. pulex. Generally, these problems were only found in some D. pulex orthologs, and so the D. melanogaster (and many of the D. pulex) matches give reliable gene annotations in the BLAST results. With the exception of LpVRI, LpCWO and LpJET, all accessory proteins show significant levels of homology to their respective BLAST hits, with e-values ranging from 10^−110 to less than 10^−180 and BLAST scores ranging from 842 to 2136.
Top BLAST hits for all input and output clock genes against the D. melanogaster and D. pulex databases are reported in Table 2. All putative input and output clock components identified in this study (pCRY1, NPF, NPFR and PDHR) were found to have the highest homology to the predicted orthologous proteins of D. melanogaster and D. pulex. The top BLAST hits for the LppCRY1 showed pCRY1 and CRY-D (D. melanogaster-like cryptochrome also known as pCRY1) as the closest hits in our model systems. Similarly the top hits for all putative output genes (NPF, NPFR, and PDHR) for both D. melanogaster and D. pulex were the expected orthologs for both of these species. With the exception of LpNPF, all output proteins show significant levels of homology to their respective BLAST hits, with e-values ranging from 10^−48 to 10^−71 and BLAST scores ranging from 445 to 583. The high e-values and low BLAST scores of the LpNPF BLAST are likely because NPF is a short peptide (96 amino acids long), additionally the D. pulex NPF ortholog appears to have not yet been characterized as the top BLAST hit showed “Putative uncharacterized protein”.
3.3. Protein and domain alignment
The majority of the core clock components were more homologous to D. pulex core clock proteins than those of D. melanogaster (with the exception of LpCYC1 and LpCYC2 (Table 3; Figure 1 & 2; Supp Figure 1)). The similarity between Limulus clock proteins and those in D. pulex and D. melanogaster were most pronounced when comparing the functional domains of these clock proteins (Table 3). Moreover, the levels of conservation within the functional domains were fairly comparable between D. melanogaster and D. pulex, suggesting that the difference in sequence similarity lies in the regions outside and between domains (Table 3). The two clock protein domains with lowest levels of conservation were the PeriodC domains of all three LpPER paralogs (Table 3; Figure 3 & 4; Supp Figure 2) and the TIMELESS domain of LpTIM (Table 3; Figure 5; Supp Figure 3) (Table 3). LpnpCRY2 showed the highest level of conservation in the core clock proteins (Table 3; Figure 6). The full protein alignment of LpCYC2 to LpCYC1 shows a %Identity and %Similarity of 86% and 89%, respectively (Table 3; Figure 2). The three copies of the period genes also show high levels of homology to one another, with the alignments of the full LpPERA and LpPERB proteins showing a %identity and %similarity of 35% and 48%, LpPERA and LpPERC showing 37% and 49%, and LpPERC and LpPERB showing 65% and 72% (Figure 4). The duplicate per and cyc genes show a greater amount of similarity to their respective paralogs then they do to the respective orthologs of D. melanogaster and D. pulex (per (Figure 3 & 4; Supp Figure 2); cyc (Figure 1 & 2; Supp Figure 1)).
Table 3:
%Identity/%Similarity for L. polyphemus proteins against Drosophila melanogaster and Daphnia pulex.
| Proteins | Drosophila melanogaster | Daphnia pulex | |||
|---|---|---|---|---|---|
| Core proteins | Length | Domains (%Identity/%Similarity) | Total | Domains (%Identity/%Similarity) | Total |
| PERA | 1,001 | PASa(40/53);PASb(44/63);PAC(66/82);PERIODC(N/A) | 26/34 | PASa(40/50);PASb(45/66);PAC(66/84);PERIODC(21/30) | 30/38 |
| PERB | 1,021 | PASa(37/56);PASb(44/55);PAC(75/86);PERIODC(N/A) | 25/31 | PASa(38/57);PASb(49/58);PAC(70/80);PERIODC(20/29) | 32/39 |
| PERC | 1,047 | PASa(38/56);PASb(45/65);PAC(68/82);PERIODC(N/A) | 25/32 | PASa(43/57);PASb(49/61);PAC(73/82);PERIODC(22/31) | 31/39 |
| TIM | 1,100 | TIMELESS(35/45) | 27/38 | TIMELESS(25/40) | 36/45 |
| CLK | 654 | bHLH(67/82);PASa(52/61);PASb(78/82);PAC(75/93) | 32/40 | bHLH(71/84);PASa(55/66);PASb(76/85);PAC(84/91) | 43/50 |
| CYC1 | 623 | bHLH(75/81);PASa(75/87);PASb(53/73);PAC(57/66) | 62/68 | bHLH(78/85);PASa(87/94);PASb(57/66);PAC(57/70) | 31/37 |
| CYC2 | 632 | bHLH(76/81);PASa(76/88);PASb(53/66);PAC(52/66) | 60/66 | bHLH(76/85);PASa(85/93);PASb(59/68);PAC(57/70) | 31/38 |
| npCRY2 | 549 | 6–4 Photolyase(N/A);FAD Binding 7(N/A) | N/A | 6–4 Photolyase(64/76);FAD Binding 7(84/91) | 70/79 |
| Accessory Proteins | |||||
| VRI | 439 | BRLZ(69/80) | 20/28 | BRLZ(68/75) | 42/43 |
| CWO | 536 | bHLH(82/86);ORANGE(32/39) | 16/22 | bHLH(88/95);ORANGE(34/46) | 23/25 |
| SLIMB1 | 533 | b-TrCP D(55/65);FBOX(85/93);WD40(92/92);WD40(84/98); WD40(97/97);WD40(94/94);WD40(97/100);WD40(92/97); WD40(95/95) | 73/78 | b-TrCP D(53/65);FBOX(75/85);WD40(92/95);WD40(84/95); WD40(87/87);WD40(95/97);WD40(95/100);WD40(89/97); WD40(100/100) | 74/80 |
| SLIMB2 | 528 | b-TrCP D(57/68);FBOX(88/93);WD40(92/92);WD40(84/89); WD40(97/97);WD40(95/95);WD40(97/100);WD40(92/97); WD40(92/95) | 74/79 | b-TrCP D(57/68);FBOX(78/85);WD40(92/92);WD40(84/95); WD40(87/87);WD40(95/97);WD40(95/100);WD40(89/97); WD40(97/100) | 76/82 |
| ARNT1 | bHLH(76/81);PASa(74/90);PASb(74/83);PAC(61/75) | 49/57 | bHLH(76/81);PASa(74/88);PASb(75/78);PAC(84/93) | 52/60 | |
| ARNT2 | bHLH(89/91);PASa(76/90);PASb(75/84);PAC(57/70) | 49/57 | bHLH(89/91);PASa(78/88);PASb(74/78);PAC(84/91) | 54/59 | |
| CK Iα | 402 | S TKc(78/84) | 79/84 | S TKc(64/71) | 46/52 |
| CK Iε | S TKc(75/81) | 53/58 | S TKc(73/77) | 64/67 | |
| CK IIα | 329 | S TKc(92/96) | 88/92 | S TKc(94/98) | 86/90 |
| CK IIβ | 198 | CKIIβ(90/93) | 84/86 | CKIIβ(93/95) | 84/86 |
| JET | 234 | LRR(12/35);LRR(15/41);LRR(31/50) | 41/48 | LRR(15/38);LRR(33/48);LRR(31/46) | 14/19 |
| SGG | 364 | S TKc(77/85) | 50/57 | S TKc(83/88) | 69/75 |
| PP1α | 330 | PP2Ac(94/97) | 88/91 | PP2Ac(96/99) | 90/93 |
| PP1β | 274 | PP2Ac(86/88) | 76/79 | PP2Ac(88/89) | 81/82 |
| PP2αMTS | 296 | PP2Ac(79/82) | 80/84 | PP2Ac(54/65) | 51/61 |
| PP2αWBT | 494 | B56(84/91) | 71/78 | B56(81/89) | 74/82 |
| PP2αTWS | 443 | WD40(85/97);WD40(90/93);WD40(93/98);WD40(93/95); WD40(87/90); WD40(93/94);WD40(87/92) | 73/79 | WD40(79/87);WD40(88/100);WD40(93/98);WD40(80/85); WD40(87/92); WD40(87/89);WD40(82/89) | 84/91 |
| Input Pathway Proteins | |||||
| CRY1 | 313† | FAD Binding 7(47/55) | N/A | FAD Binding 7(45/60) | N/A |
| Output Pathway Proteins | |||||
| NPFR | 213† | 7tM GPCR Srsx(53/66) | N/A | 7tM GPCR Srsx(49/61) | N/A |
| NPF | 95 | PAH(36/54) | 23/34 | PAH(41/54) | 29/37 |
| PDHR | 231 † | 7TM-2(40/57) | N/A | 7TM-2(46/63) | N/A |
indicates partial sequence.
Figure 1:

bHLH, PASa, PASb and PAC domains region of putative L. polyphemus CYCLE1 (CYC1) protein. Alignment of L. polyphemus (Lim) CYC1 to Daphnia pulex (Daph) CYC and Drosophila melanogaster (Dros) CYC using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Blue shows bHLH domain, light green shows PAS domains, Red shows PAC domain. For full alignment see Supplemental Figure 4.
Figure 2:

Putative L. polyphemus CYCLE 1 (CYC1) protein aligned against putative CYCLE 2 (CYC2) using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” indicates identical residues, “:” indicates residues with strongly similar properties, “.” indicates residues with weakly similar properties. Light green shows bHLH domain, Blue shows PAS domain, Red shows PAC domain.
Figure 3:

PASa, PASb and PAC domains region of putative L. polyphemus PERIOD (PER) protein. Alignment of L. polyphemus (Lim) PER to Daphnia pulex (Daph) PER, D. melanogaster(Dros) PER, Ixodes scapularis (Ix) PER using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Light green shows PAS domain, Red shows PAC domain. For full alignment see Supplemental Figure 1.
Figure 4:
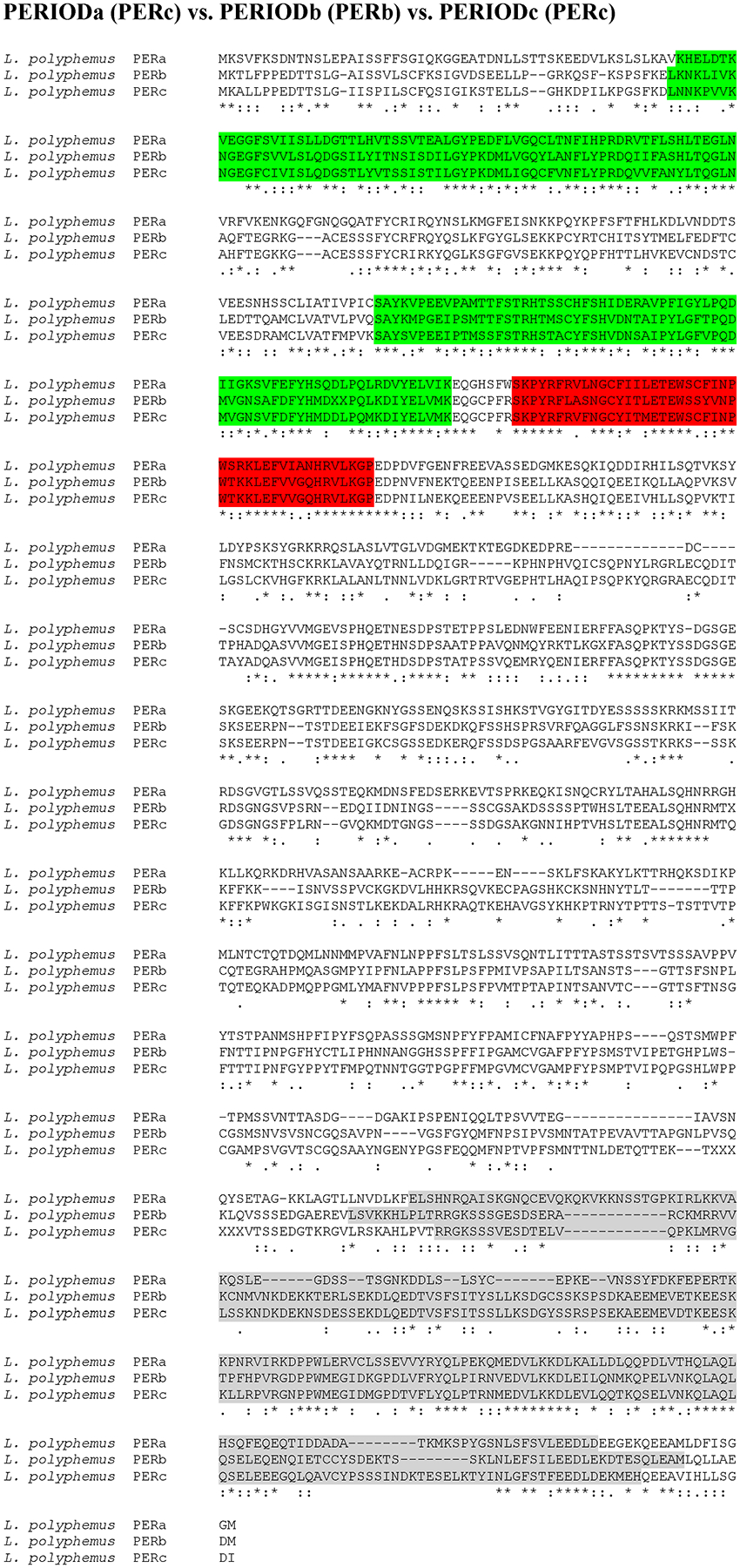
Putative L. polyphemus PERIOD (PER) proteins. Alignment of L. polyphemus (L. polyphemus) PERa to L. polyphemus PERb and PERc using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Green shows bHLH domain, Blue shows PAS domain, Red shows PAC domain, Grey shows Period C domain.
Figure 5:

TIMELESS domain region of putative L. polyphemus TIMELESS (TIM) protein. Alignment of L. polyphemus (Lim) TIM to Daphnia pulex (Daph) TIM and D. melanogaster(Dros) TIM using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” indicates identical residues, “:” indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Dark Blue shows TIMELESS domain. For full alignment see Supplemental Figure 2.
Figure 6:

Putative L. polyphemus CRYPTOCHROME 2 (CRY2) protein. Alignment of L. polyphemus (Lim) CRY2 to Daphnia pulex (Daph) CRY-M and Apis mellifera (Apis) CRY2 using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Dark green shows Photolyase domain, Gold shows FAD Binding 7 domain.
The sequence alignments of the accessory proteins were similar to those of the core proteins. For example, the sequence alignment of the duplicated LpSLIMB paralogs shows an overall %identity/%similarity value of 94/97, and the paralogs of the LpARNT show a value of 85/89. The paralogs of both genes share higher levels of sequence identity to one another than they do to D. melanogaster and D. pulex orthologs. Moreover, the majority of the L. polyphemus orthologs have higher sequence identity to D. pulex then to D. melanogaster, with the higher levels of conservation within the functional domains (Table 3). We found that LpVRI, LpCWO, and LpJET had the lowest levels of conservation in the accessory proteins, but higher levels of conservation in the functional domains of LpVRI and LpCWO (Table 3). Interestingly, we found that the functional domains within the LpJET appear to have less conservation than the non-domain regions (Table 3). Additionally, the kinases and phosphatases tended to be noticeably more conserved than the other accessory proteins, with the most conserved protein (PP1α) having a %Identity/%Similarity of 88/91 for D. melanogaster and 90/93 for D. pulex (Table 3).
The sequence alignments for the input and output proteins showed high levels of homology for both LppCRY1, LpNPFR, and LpPDHR, although the sequence identity for LpNPF was much lower. All of the output proteins had higher levels of homology within the functional domains.
3.4. Phylogenetic trees
All the core clock components for L. polyphemus identified in this study (PERA, PERB, PERC, TIM, CLK, CYC1, CYC2, and npCRY2) clearly fall into the appropriate clades for each gene family (Figure 8, 9, 10 & 11). We also found that whenever the protein sequence for a chelicerate ortholog was available, the L. polyphemus ortholog(s) most closely associated with it. Furthermore, all the chelicerate proteins tended to nest within the clades consisting of arthropod orthologs, but always formed a distinct clade outside the insect and crustacean clades. Within the period gene tree the three paralogs of the L. polyphemus period gene probably originated from two novel duplication events that occurred after the formation of the subphylum chelicerata and after the divergence of Ixodes scapularis (Black legged tick, and a fellow chelicerate) and L. polyphemus (Figure 8). Furthermore, the periodB/periodC duplications probably occurred more recently than the duplication event involving periodA and the common ancestor of periodB and periodC. Similarly, the duplication of the cyc gene also appears to be specific to the subphylum chelicerata (Figure 10). However, without the sequences of any other chelicerate orthologs it is not possible to determine when in the evolution of the chelicerates this duplication occurred. Additionally the phylogenetic tree of the cry gene family showed that LpCRY1 nests within the invertebrate-cry1 clade of the cryptochrome gene family.
Figure 8:
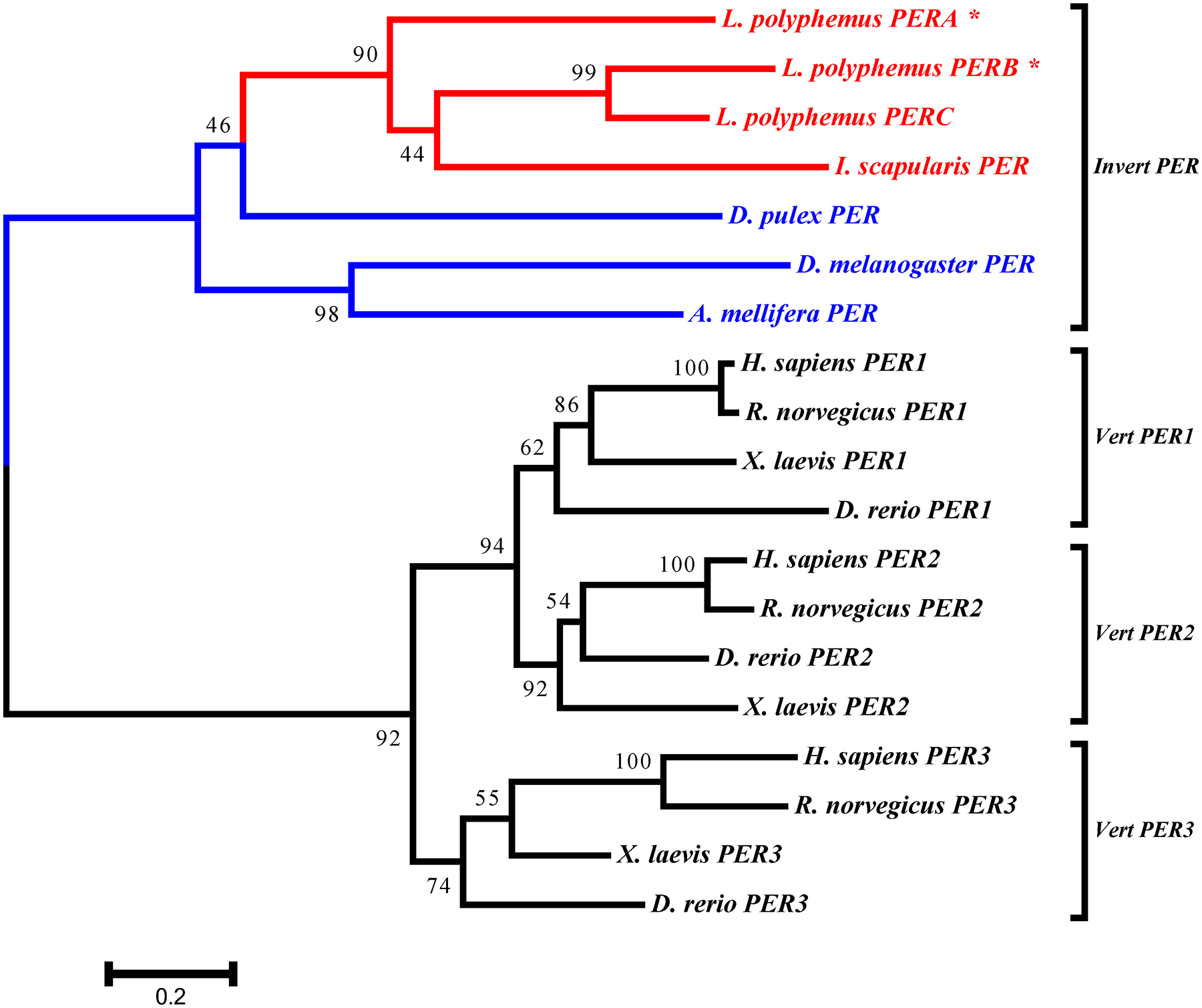
L. polyphemus PERIOD phylograms. L. polyphemus homologs are marked with an “*”, chelicerates shown in red, while all other arthropods are shown in blue, and all vertebrates are labeled in black, the root is a fungi ortholog and is labeled in violet. Major clades are bracketed and labeled. Branch lengths correspond to distance bar on bottom left corner. See Supplemental figure 5 for Cladogram.
Figure 9:
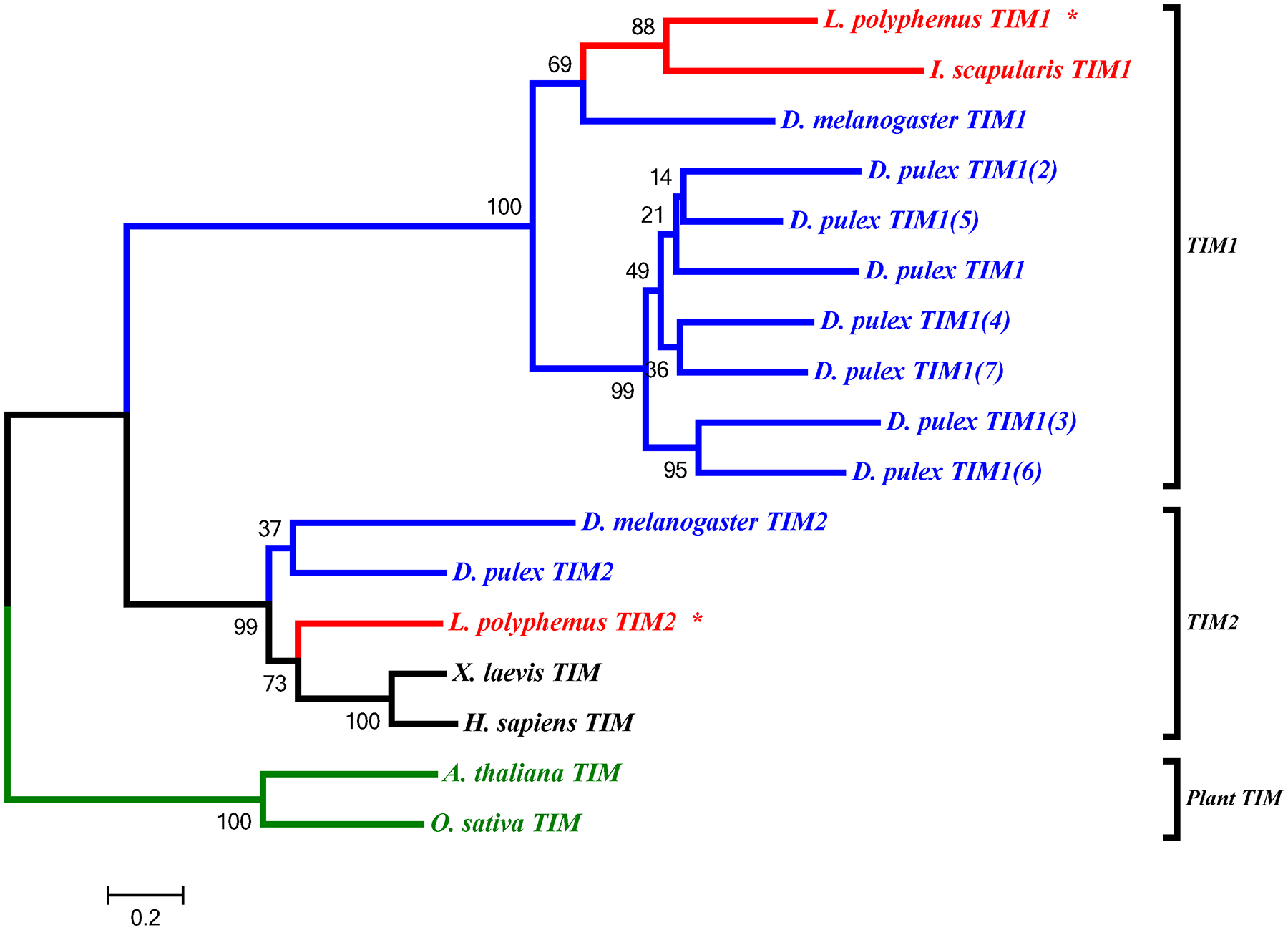
L. polyphemus TIMELESS phylograms. L. polyphemus homologs are marked with an “*”, chelicerates shown in red, while all other arthropods are shown in blue, and all vertebrates are labeled in black, the root is a fungi ortholog and is labeled in violet. Major clades are bracketed and labeled. Branch lengths correspond to distance bar on bottom left corner. See Supplemental figure 6 for Cladogram.
Figure 10:
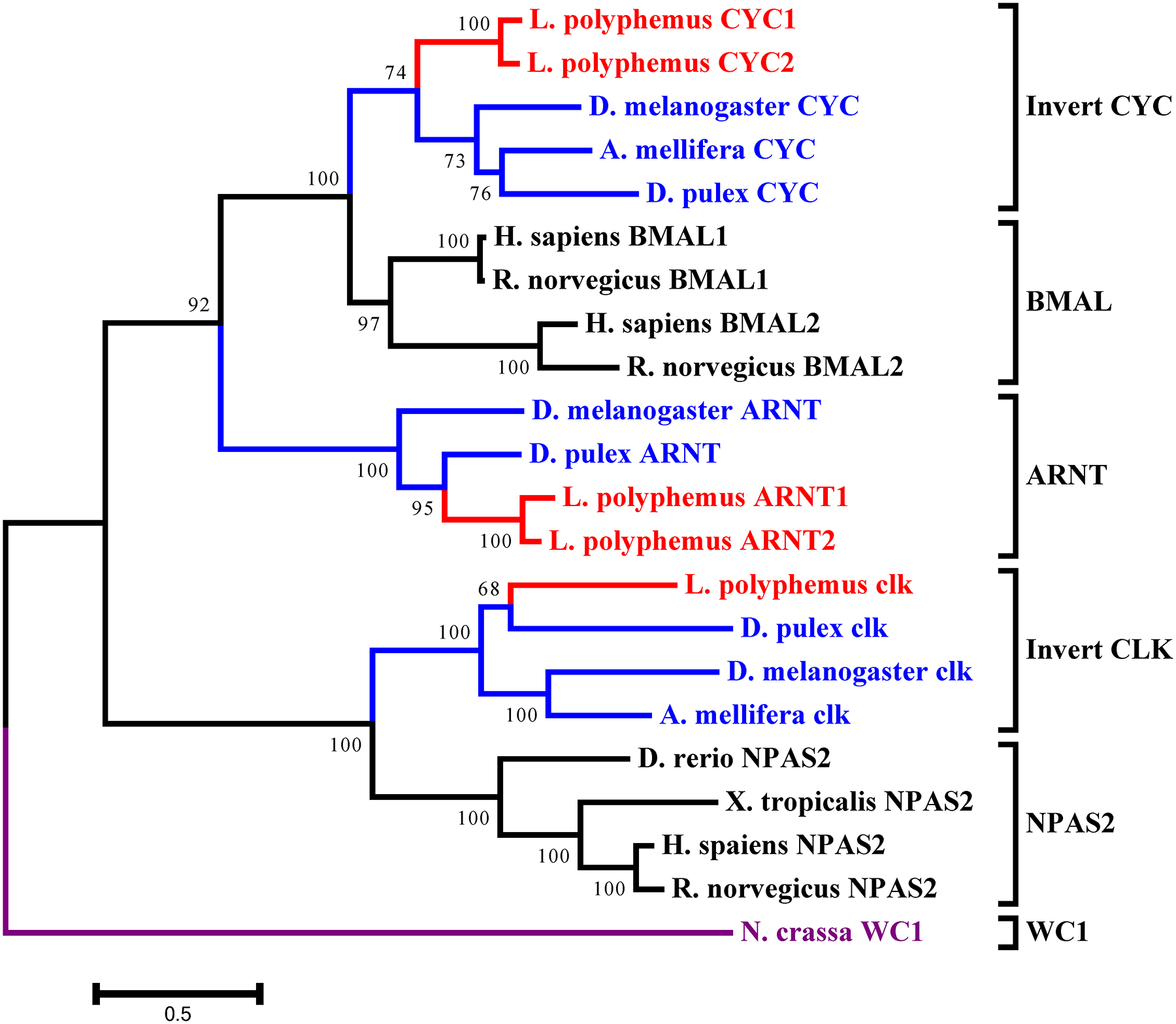
L. polyphemus CLOCK/CYCLE phylograms. L. polyphemus homologs are marked with an “*”, chelicerates shown in red, while all other arthropods are shown in blue, and all vertebrates are labeled in black, the root is a fungi ortholog and is labeled in violet. Major clades are bracketed and labeled. Branch lengths correspond to distance bar on bottom left corner. See Supplemental figure 7 for Cladogram.
Figure 11:
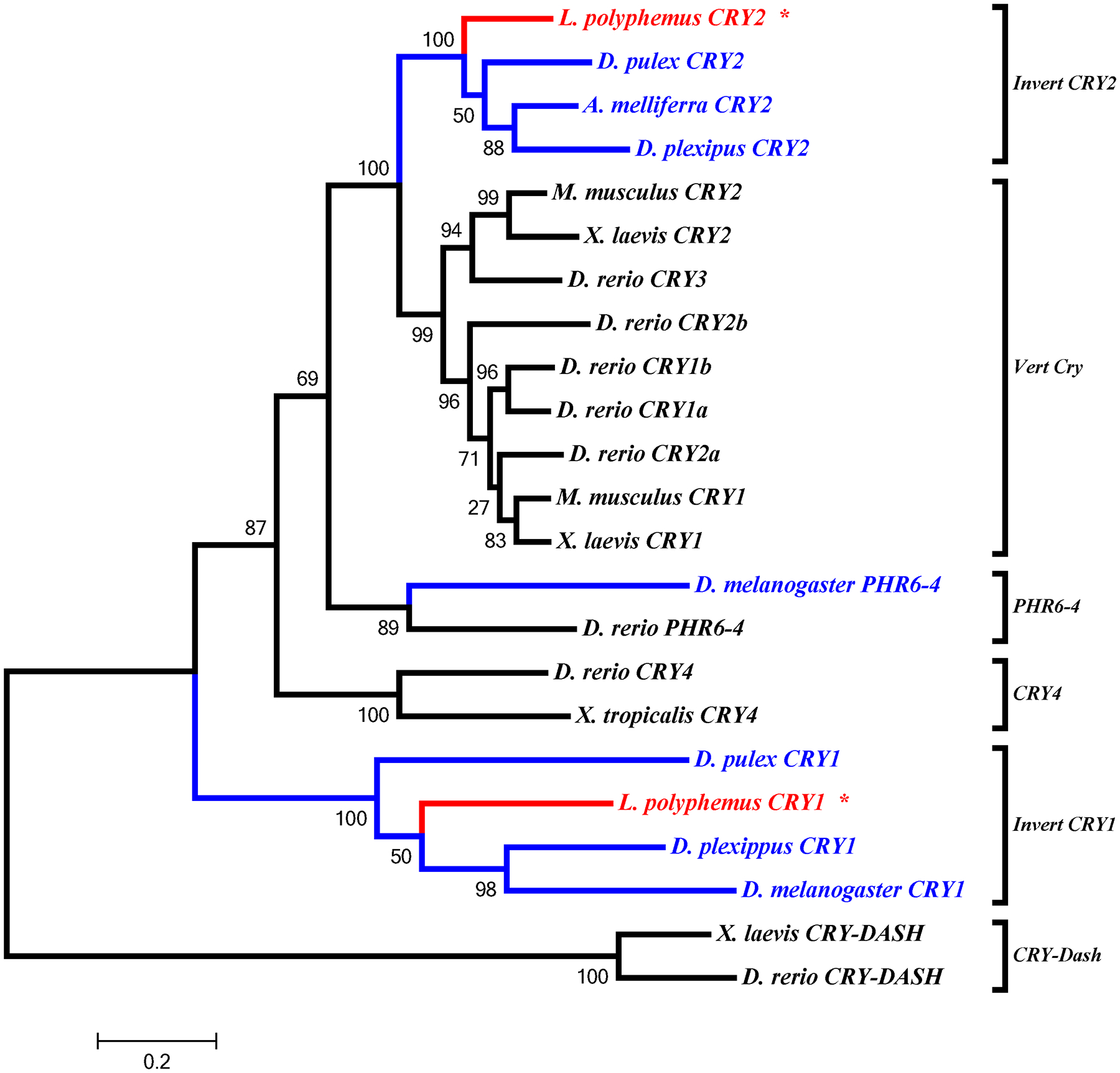
L. polyphemus CRYPTOCHROME phylograms. L. polyphemus homologs are marked with an “*”, chelicerates shown in red, while all other arthropods are shown in blue, and all vertebrates are labeled in black, the root is a fungi ortholog and is labeled in violet. Major clades are bracketed and labeled. Branch lengths correspond to distance bar on bottom left corner. See Supplemental figure 8 for Cladogram.
In the accessory gene group we found two duplicated genes slimb and arnt. Phylogenetic analysis of the LpSLIMB paralogs show that both copies of LpSLIMB nest within the slimb clade (Figure 12). Similarly phylogenetic analysis of the duplicate LpARNT paralogs show that these protein nest within the invertebrate arnt clade (Figure 10). Both of these duplication events appear to have occurred after the divergence of chelicerates, and have not been identified in any other species.
Figure 12:
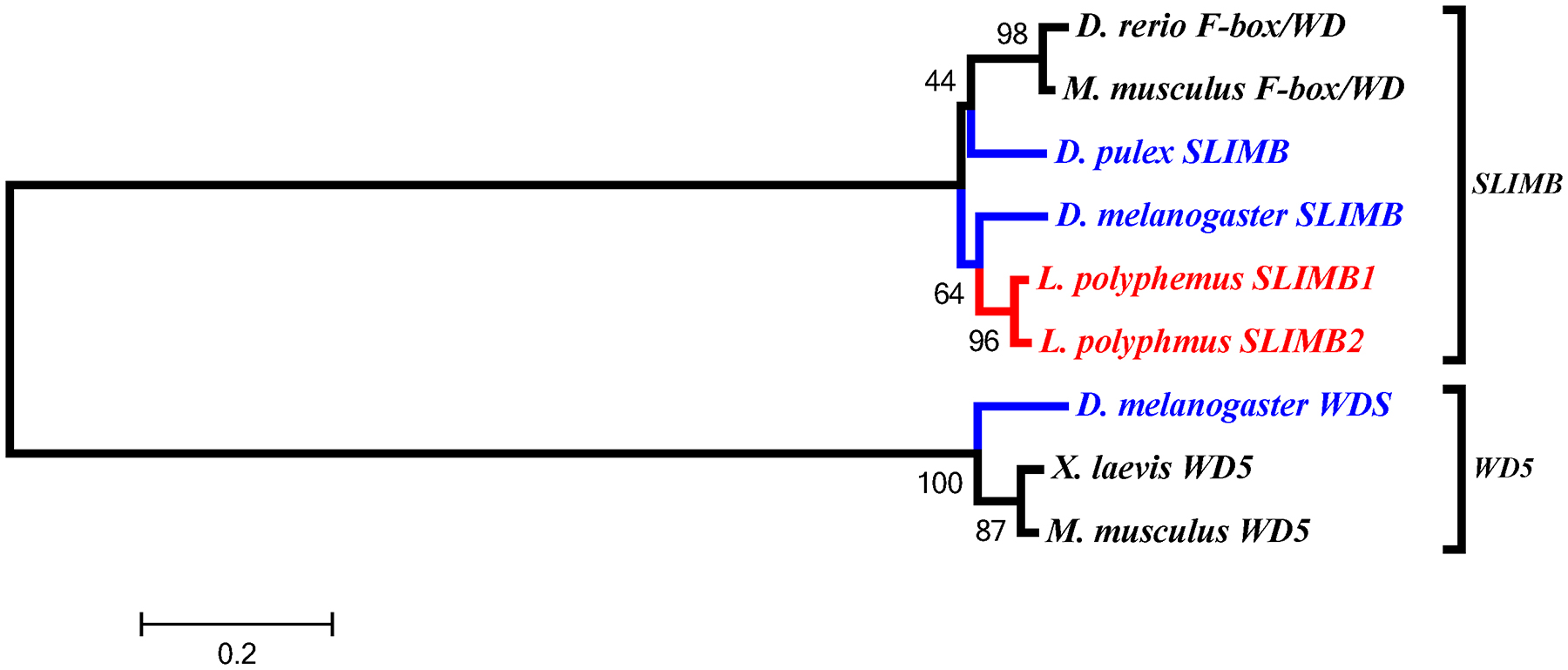
L. polyphemus SLIMB phylograms. L. polyphemus homologs are marked with an “*”, chelicerates shown in red, while all other arthropods are shown in blue, and all vertebrates are labeled in black, the root is a fungi ortholog and is labeled in violet. Major clades are bracketed and labeled. Branch lengths correspond to distance bar on bottom left corner. See Supplemental figure 9 for Cladogram.
4. Discussion.
4.1. Identification of putative circadian proteins in the L. polyphemus de novo transcriptome assembly.
Many arthropods have been shown to exhibit robust physiological and behavioral rhythms (Palmer, 1973). However, the molecular basis of many of these rhythms has been elucidated in a few model species, such as D. melanogaster (Allada and Chung, 2010). While some progress has been made in understanding the architecture of these clocks in non-model insect and crustacean species (Rubin et al., 2006; Tomioka and Matsumoto, 2010; Yuan et al., 2007), little effort has been put forth the investigate the clock mechanisms in chelicerates, including L. polyphemus. Based on the data obtained in this study, it appears that horseshoe crabs possess endogenous clocks that may rely on many of the same molecular components as other, better understood, invertebrate circadian clocks. Moreover, given the fact that horseshoe crabs have been extant for at least 450 million years (Rudkin and Young, 2009), these data might also provide some insight into the evolution of endogenous clocks.
In this study we identified 29 orthologs of circadian clock related proteins in the L. polyphemus transcriptome, corresponding to insect core clock proteins (PER, TIM, CLK, CYC and npCRY2), accessory proteins (VRI, CWO, SLIMBS, ARNT, CKIa, CKIe, CKIIa, CKIIß, JETLAG, SHAGGY, PPT1a, PPT1ß, PPT 2a-MTS, PPT 2a-WBT, and PPT2a-TWS), input proteins (pCRY1), and output proteins (NPF, its receptor (NPFR), and PDHR). Two of the top 5 BLAST hits for LpCYC1, and 1 top hit for LpCYC2 were labeled as “ARNT” proteins. BLASTp of the LpnpCRY2 showed closest homology to proteins labeled pCRY1 and 1 protein labeled as generic “CRY” protein. Despite the mislabeling of these genes, phylogenetic evidence suggests that these BLAST hits are, in fact, orthologs of npCRY2 and CYC, and nest within the appropriate clades of the cryptochrome and bHLH gene families, respectively (data not shown). The mislabeling of the CYC and npCRY2 genes appears to be the result of an inappropriate reference genome during genome annotation (using a mammalian reference instead of an insect). Only two proteins found in insect circadian systems were not identified in the L. polyphemus transcriptome (PDP1e, and PDH). Even though we were unable to identify PDH in the L. polyphemus transcriptome, the presence of its receptor (PDHR) may imply that LpPDH is likely expressed in the L. polyphemus CNS. The absence of LpPDH from the transcriptome is possibly due to the relatively small size of the preprohormone (D. melanogaster - 102 amino acids – Renn et al., 1999; Crayfish - deKleijn et al., 1993). Among the proteins that were identified, the full-length protein sequences were able to be determined for all but LppCRY1 and LpNPFR, and LpPDHR.
The identification of both pCRY1 and npCRY2 orthologs allows us to infer some additional information regarding the architecture of the circadian clock in L. polyphemus, based on what is known about insect clocks. Insect clock systems have been categorized into three types: 1) type 1, which contains cry1 but lacks cry2 (such as in D. melanogaster melanogaster (Hardin, 2005)); 2) type 2, which contains both cry1 and cry2 (such as in the monarch butterfly, Danaus plexipus (Zhu et al., 2005)) and; 3) type 3, which contains cry2 but lacks cry1 (such as in the honeybee, Apis mellifera; (Yuan et al., 2007)). The presence of both cry1 and cry2 in the L. polyphemus transcriptome would classify L. polyphemus as having a type 2 clock system which is considered an “ancestral clock system” (Yuan et al., 2007), which is fitting because L. polyphemus is considered to be a “living fossil” (Rudkin and Young, 2009). Thus, the circadian system of L. polyphemus is most likely organized more like the butterfly and mosquito (Yuan et al., 2007; Zhu et al., 2005), than D. melanogaster (Hardin, 2005) or Apis mellifera (Yuan et al., 2007).
4.2. Conservation and evolution of circadian proteins.
The conservation of the various circadian clock proteins ranged from poor to highly conserved. Nearly all genes showed higher levels of conservation between D. melanogaster and D. pulex than orthologs of either species did to L. polyphemus orthologs. These findings are consistent with the evolutionary relationships of these three subphyla, with insects and crustaceans being more closely related to one another than to chelicerata (Giribet and Edgecombe, 2012). Conservation within functional domains tended to be much higher, in general, indicating that the functions of these proteins within the circadian clock may be well conserved between insects and chelicerata, as well as Crustacea and chelicerata. As such, the proteins which makeup the circadian clock in D. melanogaster may be relatively well conserved across the entire phylum Arthropoda. Interestingly, several domains (PeriodC domain of all three LpPER paralogs, the Timeless domain of LpTIM, and the LRR domains of LpJET) did show surprisingly low levels of conservation. The lack of conservation in these functional domains may confer novel functions of these proteins in the circadian clock of L. polyphemus. For example, the PeriodC domain is a known DNA-binding domain, and the overabundance of mutations in this domain may facilitate new transcriptional targets in the clock cells. Similarly the Timeless and LRR domains are found to be involved in protein-protein interactions, and the high level of mutations in this domain could allow for previously unknown interactions, thus allowing for new clock mechanisms.
Most (72%) L. polyphemus proteins (19 out of the 26, [pCRY1, NPFR, and PDHR were excluded due to incomplete sequences]) showed higher homology to D. pulex orthologs than to D. melanogaster orthologs. Six proteins (CYC1, CYC2, CKIα, CKIIα, JET, and PP2αMTS) show higher levels of sequence identity to D. melanogaster orthologs, and one gene (CKIIβ) showed equal levels of sequence identity to both D. melanogaster and D. pulex. The finding that of 72% of genes share higher sequence identity with D. pulex may be due to the fact that D. pulex and L. polyphemus evolved in similar marine habitats and thus experienced similar environmental pressures. There were six L. polyphemus genes, which show higher homology to D. melanogaster orthologs than to D. pulex. Most of these genes show low levels of conservation in the D. pulex orthologs, which may suggest that these genes may have undergone rapid evolution in the crustacean lineage
Several gene duplications were identified from the L. polyphemus transcriptome. The two duplications of the period gene in L. polyphemus appear to be a novel discovery in invertebrates, and while there have been several duplications of the period gene in vertebrates (Bae et al., 2001), phylogenetic evidence suggests that the duplications this study revealed are unique to the chelicerate lineage, and independent of the vertebrate duplication events. These three paralogs raise an interesting question regarding the possibility of functional divergence following the duplication. Such differences have been observed between two period alleles in D. melanogaster, where each allele has different thermokinetics and varies by latitude (Sawyer et al., 1997). More extreme functional divergences have also been observed following the duplications of the period genes during vertebrate evolution (Bae et al., 2001). The single duplication events of the cycle, arnt, and slimb genes also appear to be unique and previously unknown duplications. These results present intriguing opportunities for potential novel mechanisms and functions within the clock systems of L. polyphemus. Are these different protein isoforms differentially expressed in different cell-types? Could these duplicated genes have been co-opted to create a clock controlling circatidal rhythms in L. polyphemus? Since these duplications do not appear in other chelicerates, such as scorpions and ticks, it is tempting to consider that they might be part of a clock system controlling these rhythms.
The various gene duplications also support the recent discovery of a whole genome duplication event based on genetic mapping of the L. polyphemus genome (Nossa et al., 2014). This genome duplication event appears to have taken place roughly 300 million years ago (Nossa et al., 2014), well after the divergence of Limulidae from the other chelicerate families (~450 million years ago (Rudkin and Young, 2009). This is consistent with our findings, as none of the observed duplications have been found in other species outside the family Limulidae. Given the evidence of at least one genome duplication event, what is even more intriguing than the presence of these duplications is the absence of duplications of the other circadian clock genes. The absence of these duplicate genes following a genome duplication event can have one of two explanations: 1) these genes underwent a loss of function, resulting in the duplicate genes being deleted from the genome or rendered inert, or 2) these genes underwent a functional divergence, resulting in these genes not being expressed in the CNS. In either case we would not expect to find transcripts of these duplicates in our transcriptome assembly.
4.3. Implications and future research.
This study provides the first insight into the molecular mechanisms underlying the biological clocks of L. polyphemus, a member of the subphylum chelicerata. The conservation of these proteins, particularly within functional domains, provides evidence that some or all of these proteins may be involved in the circadian clock, and possibly the circatidal clock, of L. polyphemus. Immunolabelling of putative circadian proteins has been instrumental in localizing circadian clock cells (Siwicki et al., 1988) and their output cells in a variety of insects (Sehadova et al., 2004; Shao et al., 2006) and this study provides the foundation for similar studies in L. polyphemus. This study also provides a resource to aid in the development of qPCR primers and probes for Southern blotting and in situ hybridization. Moreover, the identification of these genes may also allow us to begin to tease apart the mechanisms underlying circatidal/circalunidian clocks (Chabot et al., 2004; Chabot and Watson, 2010), the molecular bases of which are unknown even in model systems (Tessmar-Raible et al., 2011).
Biological clocks act as master regulators of the physiology and behavior of most organisms. The high level of conservation of circadian clock related genes in Limulus indicates that they may play a role in the biological clock system(s) of this species, thus it is likely that the basic architecture of the circadian clock found in insects dates back prior to the divergence of chelicerata on the arthropod phylogenetic tree.
Supplementary Material
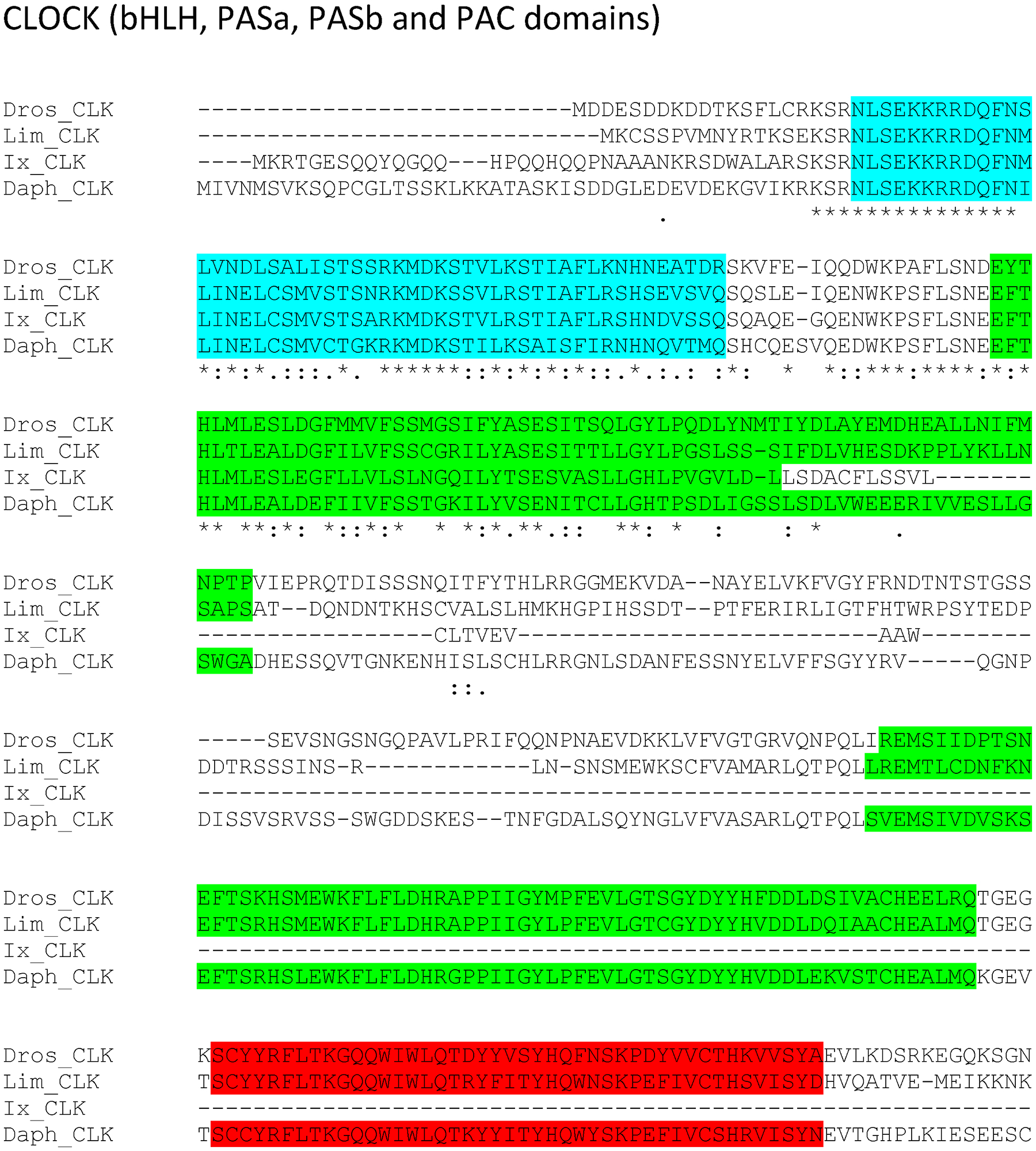
Supplemental Figure 4: Putative L. polyphemus CLOCK (CLK) protein. Alignment of L. polyphemus (Lim) CLK to Daphnia pulex (Daph) CLK, D. melanogaster(Dros) CLK, Ixodes scapularis (Ix) CLK using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Blue shows bHLH domain, Light green shows PAS domain, Red shows PAC domain, Magenta shows Coiled Coil domain.
Supplemental Figure 1: Putative L. polyphemus CYCLE1 (CYC1) protein. Alignment of L. polyphemus (Lim) CYC1 to Daphnia pulex (Daph) CYC and Apis mellifera (Apis) CYC using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Blue shows bHLH domain, light green shows PAS domains, Red shows PAC domain.
Supplemental Figure 2: Putative L. polyphemus PERIOD (PER) protein. Alignment of L. polyphemus (Lim) PER to Daphnia pulex (Daph) PER, D. melanogaster(Dros) PER, Ixodes scapularis (Ix) PER using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Light green shows PAS domain, Red shows PAC domain, Grey shows Period C domain.
Supplemental Figure 3: Putative L. polyphemus TIMELESS (TIM) protein. Alignment of L. polyphemus (Lim) TIM to Daphnia pulex (Daph) TIM and D. melanogaster(Dros) TIM using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” indicates identical residues, “:” indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Dark Blue shows TIMELESS domain.
Figure 7:
bHLH, PASa, PASb and PAC domains region of putative L. polyphemus CLOCK (CLK) protein. Alignment of L. polyphemus (Lim) CLK to Daphnia pulex (Daph) CLK, D. melanogaster(Dros) CLK, Ixodes scapularis (Ix) CLK using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Blue shows bHLH domain, Light green shows PAS domain, Red shows PAC domain. For full alignment see Supplemental Figure 3.
Acknowledgments.
The authors thank Scott Tighe, Ramiro Barrantes, and Tim Hunter of the Vermont Cancer Center for help with the extraction and RNAseq runs and Kelley Thomas and Kazu Okamoto of the Hubbard Center for Genome Studies, UNH for help with the assembly. This work was supported by NSF IOS 0920342 to CCC and WHW and the New Hampshire IDeA Network of Biomedical Research Excellence (NH-INBRE) with an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health: Grant number P20GM103506. The funders had no role in the design, data collection, and analysis, decision to publish, or preparation of the manuscript. The authors state that no competing interests exist.
References.
- Allada R, Chung BY, 2010. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72, 605–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR, 2001. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525–536. [DOI] [PubMed] [Google Scholar]
- Barlow RB, 1983. Circadian rhythms in the Limulus visual system. The Journal of Neuroscience 3, 856–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow RB Jr., Chamberlain SC, Levinson JZ, 1980. Limulus brain modulates the structure and function of the lateral eyes. Science 210, 1037–1039. [DOI] [PubMed] [Google Scholar]
- Battelle BA, 2013. What the clock tells the eye: lessons from an ancient arthropod. Integr Comp Biol 53, 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot CC, C. R-PN, W.H. W, 2016. Circalunidian clocks control tidal rhythms of locomotion in the American horseshoe crab, Limulus polyphemus. Marine and Freshwater Behaviour and Physiology, 75–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot CC, Kent J, Watson WH, 2004. Daily, circadian and tidal rhythms of locomotor activity in the horseshoe crab Limulus polyphemus. Biological Bulliton, 72–75. [Google Scholar]
- Chabot CC, Watson WH, 2010. Circatidal rhythms of locomotion in the American horseshoe crab Limulus polyphemus: Underlying mechanisms and cues that influence them. Curr Zool 56, 499–517. [Google Scholar]
- Chang DC, Reppert SM, 2003. A novel C-terminal domain of drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr Biol 13, 758–762. [DOI] [PubMed] [Google Scholar]
- Christie AE, Fontanilla TM, Nesbit KT, Lenz PH, 2013. Prediction of the protein components of a putative Calanus finmarchicus (Crustacea, Copepoda) circadian signaling system using a de novo assembled transcriptome. Comp Biochem Physiol Part D Genomics Proteomics 8, 165–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Iglesia HO, Hsu YW, 2010. Biological clocks and rhythms in intertidal crustaceans. Front Biosci (Elite Ed) 2, 1394–1404. [DOI] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, Ceriani MF, 2011. Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr Biol 21, 1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, 1999. Molecular bases for circadian clocks. Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M, 1998. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679. [DOI] [PubMed] [Google Scholar]
- Giribet G, Edgecombe GD, 2012. Reevaluating the arthropod tree of life. Annu Rev Entomol 57, 167–186. [DOI] [PubMed] [Google Scholar]
- Hardin PE, 2005. The circadian timekeeping system of Drosophila. Curr Biol 15, R714–722. [DOI] [PubMed] [Google Scholar]
- Hermann C, Yoshii T, Dusik V, Helfrich-Forster C, 2012. Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J Comp Neurol 520, 970–987. [DOI] [PubMed] [Google Scholar]
- Lee G, Bahn JH, Park JH, 2006. Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc Natl Acad Sci U S A 103, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P, 2015. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43, D257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J, 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor E, 1996. Crab clockwork: the case for interactive circatidal and circadian oscillators controlling rhythmic locomotor activity of Carcinus maenas. Chronobiol Int 13, 153–161. [DOI] [PubMed] [Google Scholar]
- Nossa CW, Havlak P, Yue JX, Lv J, Vincent KY, Brockmann HJ, Putnam NH, 2014. Joint assembly and genetic mapping of the Atlantic horseshoe crab genome reveals ancient whole genome duplication. Gigascience 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, 1973. Tidal rhythms: the clock control of the rhythmic physiology of marine organisms. Biological Review 48. [Google Scholar]
- Palmer JD, Williams BG, 1986. Comparative studies of tidal rhythms II. The dual clock control of the locomotor rhythms of two decapod crustaceans. Marine behaviour and physiology, 269–278. [Google Scholar]
- Renn SCP, Park JH, Rosbash M, Hall JC, Taghert PH, 1999. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. [DOI] [PubMed] [Google Scholar]
- Rubin EB, Shemesh Y, Cohen M, Elgavish S, Robertson HM, Bloch G, 2006. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res 16, 1352–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudkin DM, Young GA, 2009. Horseshoe Crabs - An Ancient Ancestry Revealed. Biology and Conservation of Horseshoe Crabs, 25–44. [Google Scholar]
- Sawyer LA, Hennessy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP, 1997. Natural variation in a Drosophila clock gene and temperature compensation. Science 278, 2117–2120. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP, 1998. SMART, a simple modular architecture research tool: Identification of signaling domains. P Natl Acad Sci USA 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehadova H, Markova EP, Sehnal FS, Takeda M, 2004. Distribution of circadian clock-related proteins in the cephalic nervous system of the silkworm, Bombyx mori. J Biol Rhythm 19, 466–482. [DOI] [PubMed] [Google Scholar]
- Shao QM, Sehadova H, Ichihara N, Sehnal F, Takeda M, 2006. Immunoreactivities to three circadian clock proteins in two ground crickets suggest interspecific diversity of the circadian clock structure. J Biol Rhythm 21, 118–131. [DOI] [PubMed] [Google Scholar]
- Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC, 1988. Antibodies to the Period Gene-Product of Drosophila Reveal Diverse Tissue Distribution and Rhythmic Changes in the Visual-System. Neuron 1, 141–150. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M, 2004. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868. [DOI] [PubMed] [Google Scholar]
- Strauss J, Zhang Q, Verleyen P, Huybrechts J, Neupert S, Predel R, Pauwels K, Dircksen H, 2011. Pigment-dispersing hormone in Daphnia interneurons, one type homologous to insect clock neurons displaying circadian rhythmicity. Cell Mol Life Sci 68, 3403–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Shafer OT, 2006. Mechanisms of clock output in the Drosophila circadian pacemaker system. J Biol Rhythm 21, 445–457. [DOI] [PubMed] [Google Scholar]
- Takekata H, Numata H, Shiga S, Goto SG, 2014. Silencing the circadian clock gene Clock using RNAi reveals dissociation of the circatidal clock from the circadian clock in the mangrove cricket. J Insect Physiol 68, 16–22. [DOI] [PubMed] [Google Scholar]
- Tauber E, Last KS, Olive PJW, Kyriacou CP, 2004. Clock gene evolution and functional divergence. J Biol Rhythm 19, 445–458. [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K, Raible F, Arboleda E, 2011. Another place, another timer: Marine species and the rhythms of life. Bioessays 33, 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilden AR, McCoole MD, Harmon SM, Baer KN, Christie AE, 2011. Genomic identification of a putative circadian system in the cladoceran crustacean Daphnia pulex. Comp Biochem Phys D 6, 282–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka K, Matsumoto A, 2010. A comparative view of insect circadian clock systems. Cell Mol Life Sci 67, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson WH, Bedford L, Chabot CC, 2008. Rhythms of locomotion expressed by Limulus polyphemus, the American horseshoe crab: II. Relationship to circadian rhythms of visual sensitivity. Biol Bull-Us 215, 46–56. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Metterville D, Briscoe AD, Reppert SM, 2007. Insect cryptochromes: Gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol 24, 948–955. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Kay SA, 2010. Clocks not winding down: unravelling circadian networks. Nature review, 764–776. [DOI] [PubMed] [Google Scholar]
- Zhu HS, Yuan Q, Briscoe AD, Froy O, Casselman A, Reppert SM, 2005. The two CRYs of the butterfly. Current Biology 15, R953–R954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 4: Putative L. polyphemus CLOCK (CLK) protein. Alignment of L. polyphemus (Lim) CLK to Daphnia pulex (Daph) CLK, D. melanogaster(Dros) CLK, Ixodes scapularis (Ix) CLK using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Blue shows bHLH domain, Light green shows PAS domain, Red shows PAC domain, Magenta shows Coiled Coil domain.
Supplemental Figure 1: Putative L. polyphemus CYCLE1 (CYC1) protein. Alignment of L. polyphemus (Lim) CYC1 to Daphnia pulex (Daph) CYC and Apis mellifera (Apis) CYC using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Blue shows bHLH domain, light green shows PAS domains, Red shows PAC domain.
Supplemental Figure 2: Putative L. polyphemus PERIOD (PER) protein. Alignment of L. polyphemus (Lim) PER to Daphnia pulex (Daph) PER, D. melanogaster(Dros) PER, Ixodes scapularis (Ix) PER using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” Indicates identical residues, “:” Indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Light green shows PAS domain, Red shows PAC domain, Grey shows Period C domain.
Supplemental Figure 3: Putative L. polyphemus TIMELESS (TIM) protein. Alignment of L. polyphemus (Lim) TIM to Daphnia pulex (Daph) TIM and D. melanogaster(Dros) TIM using CLUSTAL Omega. Symbols immediately below sequence indicates functional similarity of residues between protein orthologs. “*” indicates identical residues, “:” indicates residues with strongly similar properties, “.” Indicates residues with weakly similar properties. Dark Blue shows TIMELESS domain.


