Abstract
Humanized mouse models, created via transplantation of human hematopoietic tissues into immune-deficient mice, support a number of research applications, including transplantation immunology, virology, and oncology studies. As an alternative to the Bone Marrow Liver Thymus humanized mouse, which uses fetal tissues for generating a chimeric human immune system, the NeoThy humanized mouse employs non-fetal tissue sources. Specifically, the NeoThy model incorporates hematopoietic stem and progenitor cells from umbilical cord blood (UCB) as well as thymus tissue that is typically discarded as medical waste during neonatal cardiac surgeries. Compared to fetal thymus tissue, the abundant quantity of neonatal thymus tissue offers the opportunity to prepare over 1000 NeoThy mice from an individual thymus donor. Here, we describe a protocol for processing of the neonatal tissues (thymus and UCB), creation of NeoThy mice, and all experimental steps from planning to data analysis. This protocol will enable researchers to make effective use of this promising in vivo model of human immune function.
Introduction
Humanized mouse models (also known as human immune system [HIS] mice) enable rigorous in vivo studies of human immune cell function in a tractable mouse host. HIS mice have been utilized in a number of research fields, including transplantation immunology, pluripotent stem cell biology, virology, oncology, and toxicology.1–4 Fundamentally, these mice are created by introducing human hematopoietic tissues into an immune-deficient mouse strain. The Bone Marrow Liver Thymus (BLT) mouse5,6 HIS model incorporates surgical transplantation of human fetal thymus fragments along with injection of fetal liver-derived CD34+ hematopoietic stem and progenitor cells (HSPCs). The thymic tissue, which is implanted under the mouse kidney capsule (with or without the addition of a fragment of fetal liver tissue, depending on the BLT iteration used) and engrafts to form a thymic organoid, provides the developing T cells with human thymic epithelium cell signals needed for positive and negative selection of major histocompatibility complex (MHC)-restricted T cells. The BLT model was therefore an important advance in the history of HIS models, enabling a wide range of scientific insights into HIV pathology7 and other areas of study in multiple fields.8–10
While the BLT model has advanced the understanding of human immune function, biological and ethical concerns have been raised with regard to the use of human fetal tissue in these mice. Research by Mold et al.11 showed that the T cells of the BLT model may be developmentally immature; specifically, they are skewed towards a regulatory phenotype, which may have implications for transplantation immunology and immune-oncology studies where the balance between effector and regulatory function plays critical roles in experimental outcomes. Additionally, the ethical controversy surrounding acquisition of human fetal tissue from pregnancy terminations has limited the availability and utilization of the BLT model in certain states and countries.8 In addition, the scalability for BLT mice is limited, as a single tissue set generally supports the engraftment of 20 to 40 mice on average (Brown, M. E., unpublished observations).
We developed the NeoThy humanized mouse model, which is a BLT-type HIS mouse that incorporates non-fetal humanizing tissues, namely neonatal thymus tissue that has been discarded as medical waste from cardiac surgeries in newborn patients as well as engraftment with umbilical cord blood (UCB) HSPCs.12 In our publication, we conducted several comparative studies between the NeoThy and the BLT models showing general similarities in T cell frequency and phenotype as well as functional capabilities of the respective chimeric human immune cells in transplantation immunology studies. Our group and others are currently conducting additional in-depth comparative studies with the NeoThy, BLT, and related models, with the goal of rigorously documenting strengths and weaknesses of each model (e.g., antigen specific T cell responses) as well as determining which applications are best suited to each type of model. Of particular interest is using these models to define ontogeny-associated differences in T cell development and function.13 Methods for the creation of the BLT model have been previously published,14,15 but our protocol described below will be useful in providing academic researchers with detailed instructions for development and use of the NeoThy model in their laboratories. This includes crucial steps that differentiate the NeoThy from the BLT or other HIS models (e.g., processing neonatal thymus specimens) and that could be problematic if strictly using existing BLT protocols when attempting to make NeoThy mice.
Comparison with other methods
All iterations of humanized mice engrafted with human hematopoietic cells and tissues have two basic components: 1) human tissues (e.g., surgically transplanted thymus tissue and/or HSPCs) that are engrafted into 2) immune-deficient mouse strains that lack functional mouse adaptive immune cells and are therefore amenable to xenotransplantation (e.g., NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ [NSG]). Both the humanizing tissues and the mouse host influence the degree and character of human immune cell engraftment.16 For example, genetic-engineering or exogenous addition of human hematopoietic cytokines to the NSG mouse strain can result in dramatically different human immune cell profiles when using the same donor’s HSPCs (Figure 1). We have successfully used a number of NSG variant (e.g., NSG-SGM3) and NOG variant (e.g., NOG-EXL) strains for NeoThy. Typically, animals are humanized at 6–8 weeks of age and both males and females can be humanized, though, we and others have noted that female mice tend to engraft more robustly at least at earlier timepoints than male mice.17
Figure 1.
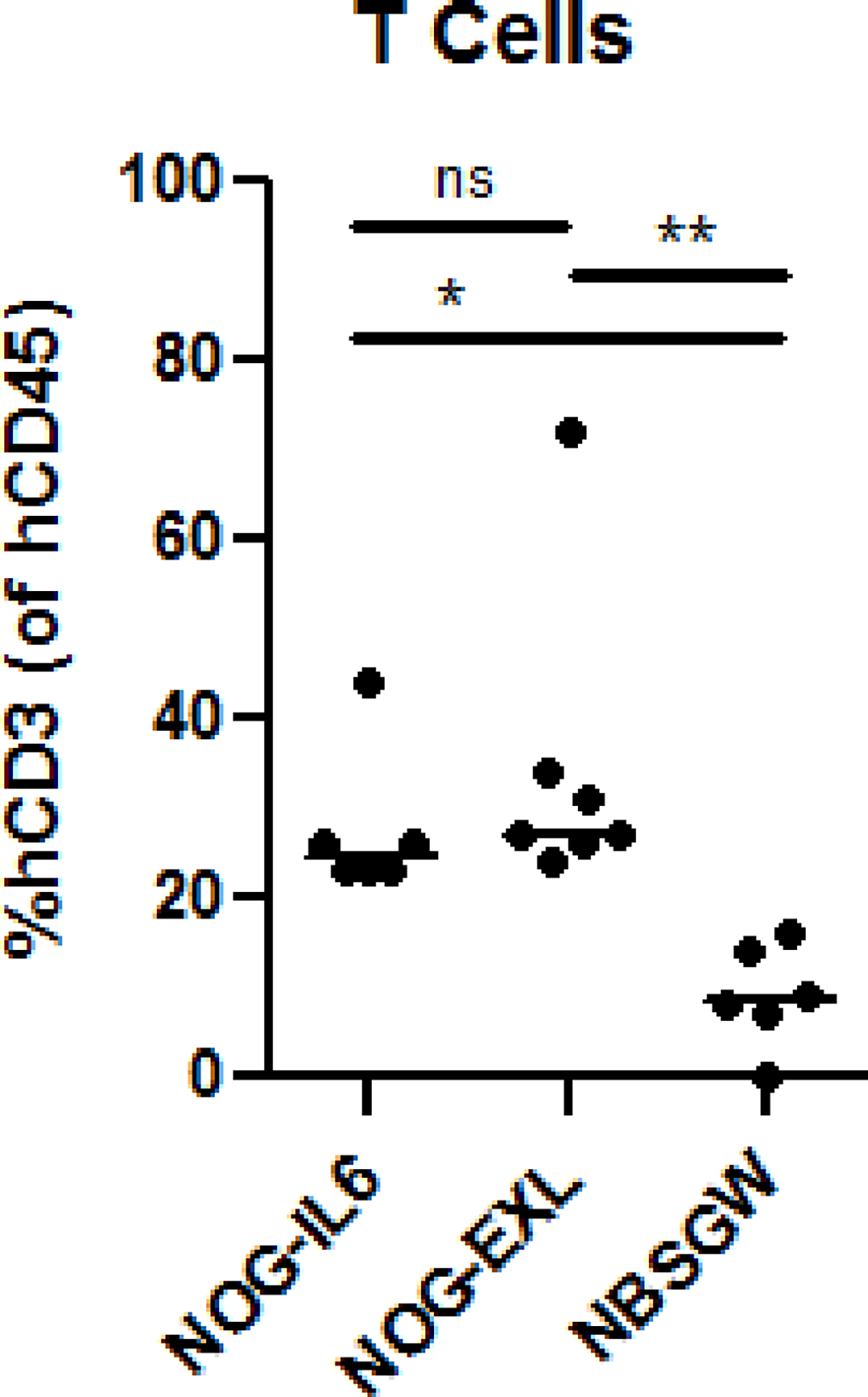
Impact of Immune-Deficient Mouse Host Strain on Human Chimerism. Three strains of immune-deficient mice were humanized with neonatal thymus and HSPCs from the same donor set. NOG-IL6 (n=6), NOG-EXL (n=7), and NBSGW (n=6) were assessed by flow cytometry for human CD45+CD3+ cells at 18–19 weeks post-humanization. One-way ANOVA analysis with Tukey’s multiple comparisons test was performed using GraphPad Prism software. ns=not significant, *=p<0.05, **=p<0.01.
The NeoThy model is an evolution of the BLT mouse, building upon the Kalscheuer et al.18 iteration as well as others before it.19 The NeoThy model utilizes the Kalscheuer approach of using cryopreserved thymus fragments coupled with antibody-mediated depletion of passenger thymocytes, but incorporates neonatal UCB as the HSPC source (rather than from adult bone marrow or fetal liver sources) and thymus tissue from patients up to one year old who are undergoing corrective cardiac surgery.
Importantly, the use of more developmentally-mature neonatal (up to 20 days post-birth) or pediatric (up to 10 years post-birth) thymus tissue20 has beneficial implications with regard to the number of mice that can be generated from one donor. In our Stem Cell Reports publication, we used tissue from seven donors up to one year in age (with a seven-day median age), with a mean thymic weight of 9.3+/−2.9g as compared to a mean fetal thymic weight of 0.6g.12 The relative abundance of tissue in the neonate is not insignificant as there are multiple research applications; this includes induced pluripotent stem cell (iPSC) transplantation studies, where having a large supply of genetically-identical human tissues can enable research that would otherwise be too expensive to reasonably conduct. For example, investigation of the autologous immune response to iPSC-derived cells requires generation of iPSC from human tissue sets.10 Human fetal thymus typically allow for only 20–40 mice per tissue set, so it is time- and cost-prohibitive to create iPSC-derived hematopoietic cell progenitor, explore human leukocyte antigen (HLA) matching and associations with experimental outcomes, and/or perform gene-editing on donor cells and tissues that are so finite in quantity. We anticipate that the ability to generate thousands of NeoThy mice from single HLA-typed and fully characterized thymus donors will enable new areas of research that would otherwise not be possible using low-quantity tissue sets. Importantly, as with the BLT fetal liver, UCB CD34+ cells are a limiting reagent with the NeoThy. We have routinely harvested cord blood samples that allow between 40–100 mice per donor (i.e., 2–5 × 106 CD34+ cells). With HLA matching of multiple UCB donors to a single thymus donor, the NeoThy may offer larger n values for experimental interrogation of thymus-dependent immunological questions (e.g., establishment of self-tolerance).
Applications and limitations
Applications for the NeoThy model, and experimental design of any HIS mouse experiment, should be dictated by the specific research question at hand. The NeoThy model is most useful in those research settings where a full human immune cell repertoire is needed for long-term (>1 month) in vivo studies (e.g., oral gavage drug studies, transplant immunology studies).21 As the human cells in these mice arise in the presence of mouse antigens, the occurrence of graft-vs-host-disease (GVHD) is reduced compared to HIS models made via engraftment of mature immune cells (e.g., hu-PBL mice).22,23 We have not explored alternative mouse strains that may further reduce the incidence of GVHD, such as the C57BL/6 Rag2−/−γc−/−CD47−/− (“TKO”) mouse.24 The use of cryopreserved neonatal thymus fragments, anti-CD2 thymocyte depletion, and non-irradiated NBSGW mouse hosts all help to diminish the occurrence of GVHD in NeoThy mice, though animals should still be monitored for health changes throughout the course of an experiment.25 Additional modifications, such as surgical removal of the murine thymic rudiment prior to humanization26 should be considered in certain instances where interactions between developing human T cells and the anatomically-disorganized murine thymic epithelium may impact downstream experimentation.
The selection of thymus and HSPC donors, including their HLA haplotypes, are important considerations when conducting NeoThy experiments, perhaps more so than with BLT or other HIS mouse studies (when autologous tissue sets are not used). Depending on the frequency of births and neonatal heart surgeries at a particular research center, with institutional review board (IRB) approval it may be feasible to collect matching (autologous) UCB and thymus tissue from the same patient, similar to the autologous liver and thymus tissue typically used in BLT mice. However, in our experience those matched samples may be infrequent due to variation in UCB volumes (i.e., there may not be enough blood volume and HSPCs to humanize suitable numbers of mice). Cadaveric tissue sources may also enable larger-scale autologous studies, but we have only explored living donor tissues to date due to tissue viability concerns. Importantly, even in cases where ample quantities of matched autologous samples can be collected, the abundant quantity of thymus tissue allows for potentially thousands of NeoThy mice to be made from one sample, whereas the HSPCs tend to be the limiting reagent. It is therefore beneficial to use allogeneic cord blood for making the NeoThy model.
We documented in our recent publication that allogeneic UCB could be successfully used, similar to reports using allogeneic liver or bone marrow HSPC sources in BLT mice.18 Given the importance of MHC-restricted antigen presentation and of human MHC molecules on thymic epithelium during T cell selection, we recommend matching cord blood and thymus donors at one HLA class I and one HLA class II allele (Figure 2), even though robust human chimerism is possible in a total mismatch setting (Brown, M. E., unpublished observations).
Figure 2.
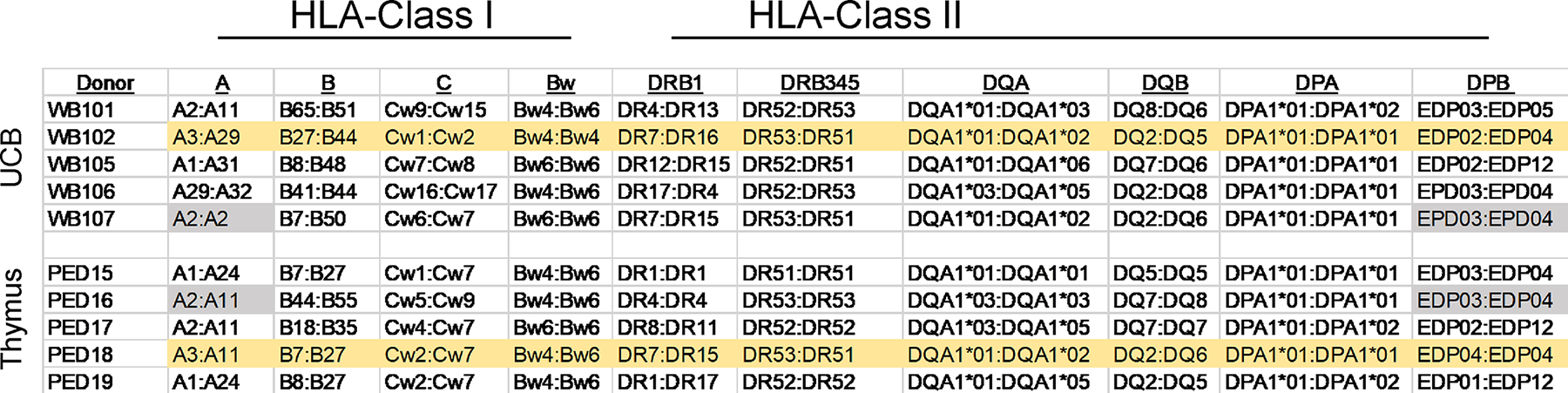
HLA Matching of Allogeneic Thymus and Umbilical Cord Blood HSPCs. Five umbilical cord blood (UCB) donors and five neonatal thymus donors were HLA typed at 11 loci using the LinkSeq Real-Time PCR typing method. When making the NeoThy humanized mouse using allogeneic UCB and thymus, we recommend matching at a minimum of one HLA-Class I allele, and one HLA-Class II allele. For example, UCB donor WB107 is a match to thymus donor PED16 at HLA-A2 and HLA-DPB4 (genes with common alleles shaded in grey), and UCB donor WB102 matches thymus donor PED18 at multiple loci (shaded in yellow). Downstream transplantation studies should consider UCB:Thymus matches together in conjunction with matches/mismatches in HLA type of transplanted tissues.
Given that large, cryopreserved banks of HLA-typed UCB HSPCs and thymic fragments can be compiled and stored for many years, it is therefore possible to not only match thymus and HSPCs, but to select certain HLA types of interest for a particular research application. For example, a specific HLA type (e.g., HLA-B27) can be chosen to investigate contributions of individual haplotypes to HIV pathogenesis outcomes.27 Similarly, for transplant immunology research, care should be given to selecting HLA matched/mismatched combinations that allow for matching/mismatching of cells/tissue that are transplanted into the mice in experiments.4
General limitations of HIS models, including both BLT and NeoThy, are suboptimal B cell maturation, a low degree of antibody class switching, and atypical frequencies of certain other immune cell subsets (e.g., dendritic cells) relative to frequencies seen in healthy adult humans.28 Improving these aspects of HIS models is an active area of research in our lab and in the field.29 Modification of the mouse host strain, such as use of immune-deficient mouse variants that harbor transgenes for human cytokines (e.g., NSG-SGM3,30 NOG-EXL,31 MISTRG32), can augment certain cell populations, as described in Figure 1. If a particular immune cell subset such as Natural Killer (NK) cells is not typically abundant in HIS mice made in a baseline strain (NSG), it is possible that a transgenic strain (NSG-Tg(Hu-IL15) may enhance reconstitution of that cell type,33 enabling functional in vivo experimentation. We are actively developing and evaluating new immune-deficient mouse strains, with the goal of making the NeoThy and other HIS models even more human-like and well-suited for particular research applications.3,34 We are also building upon the lessons learned from the development of the NeoThy to create new xenotransplantation models, such as rhesus macaque primatized mice.35
Development of the protocol
In this protocol, selection of humanizing tissues and mouse strains are critical to effective experimental design. Access to neonatal thymus tissue is likely the key limitation, as not every research institution is affiliated with a children’s hospital that performs neonatal cardiac surgeries on a regular basis. For those institutions where neonatal thymus tissue is available, it is typically possible to obtain this tissue via the institution’s tissue bank. Thymus and other tissues removed during surgery are regarded as medical waste and it is therefore straightforward to obtain the necessary IRB approvals to use the tissue for research applications. For those researchers that do not have local access to a tissue bank and/or a source of thymus tissue, the University of Wisconsin (UW) Humanized Mouse Core (https://www.surgery.wisc.edu/research/uw-humanized-mouse-core-service/) has established a cryobank of thymus tissue. Vials of thymus tissue are available for shipment to researchers seeking to create NeoThy mice at their own institution. While UCB and/or HSPCs are not available from the UW, these cells are available from a number of commercial vendors. Similarly, there are multiple vendors of research animals, from which the strains described below can be obtained. Anti-CD2 antibody is provided via i.v. injection in order to deplete passenger thymocytes within the thymus fragment, as previous described,18 thereby minimizing GVHD and encouraging only de novo T cell production in the animals. The protocol below describes the processing of freshly obtained neonatal thymus and UCB HSPCs, and is most beneficial for those researchers seeking to set up this protocol at their own academic institution.
Overview of the method
This protocol describes the processing of neonatal thymus tissue (Steps 1–12), UCB processing and HSPC separation (Steps 13–47), HLA typing and matching allogenic thymus and UCB tissues (Steps 48–75), humanization surgery (Steps 76–114) and assessment of human immune cell reconstitution (Steps 115–132). HLA typing can be outsourced to a clinical HLA lab, but our method includes a convenient kit-based method that most laboratories can conduct in-house. The overall method described here can be used to create the NeoThy model for most smaller-scale/academic research applications.
Experimental Design
For all experiments involving laboratory animals, we strongly recommend following the ARRIVE Guidelines.36A helpful website is available to assist researchers with important questions of study design, sample size, and other topics: https://arriveguidelines.org/arrive-guidelines. Institutional Animal Care and Use Committee approval is also required in order to insure proper oversight of all aspects of animal care (e.g., appropriate housing) and use (e.g., pain management and allowable blood collection volumes). Additionally, as this protocol utilizes human tissue specimens, all national laws and internal review board guidelines must be followed and informed consent must be obtained from human subjects prior to conducting this research.
Researchers should plan in advance to obtain the appropriate immune-deficient mice used in the experiments. Some mouse providers restrict breeding and it may therefore be necessary to purchase animals on an experiment-by-experiment basis. In the case of in-house breeding, as well as individual animal purchase, it will be necessary to verify that appropriate numbers of mice of a suitable age will be available (and recovered from transportation stress) by the day of surgery. Also, for some experiments, non-humanized control animals will be needed and it will therefore be appropriate to have a number of age-matched controls for the duration of the experiment. Further, for longer-term experiments, vivarium housing space can be an issue (i.e., these animals may require cage rack space for 6 months or longer, limiting the number of new experimental animals that can be housed).
Each base mouse strain is typically available with additional transgenic modifications, as mentioned in “Comparison with other methods,” above. Contact the commercial vendors of the particular strain of interest (see “Biological materials: Mice,” below) for more details regarding transgenic strain availability. The four strains mentioned below engraft with human cells similarly in our hands, and we anticipated that additional strains such as the TKO may engraft similarly but have not yet been tested. For example, when injected with UCB CD34+ HSPCs from the same donor, no statistically significant differences in human immune cell populations are observed (Figure 3). (This point is illustrated without thymus surgery, as we and others have found that overall human engraftment is dictated by the UCB donor and not impacted by the presence or absence of thymus, the latter of which does impact the T cell engraftment, however.) Selection of both males and females in equal numbers will allow for determination of any sex-associated differences in engraftment and experimental outcomes. Female mice, for example, tend to engraft more efficiently than males in certain contexts.37 Specific immune-deficient strain modifications via transgenic expression of human cytokines (e.g., NSG-IL15, NSG-SGM3, NOG-IL6)33 can enable enhanced human chimerism of various immune cell types (e.g., NK cells, myeloid cells). Alternatively, manual dosing of base strain animals with exogenous cytokines38 can achieve similar effects, though this approach can be cost-intensive. Use of any unpublished strain for making the NeoThy should be evaluated in preliminary experiments by the investigator prior to planning larger studies. If a particular strain is not successfully humanized, investigators can confidently use the NSG and NBSGW strains which we have used for a large number of successful experiments.
Figure 3.
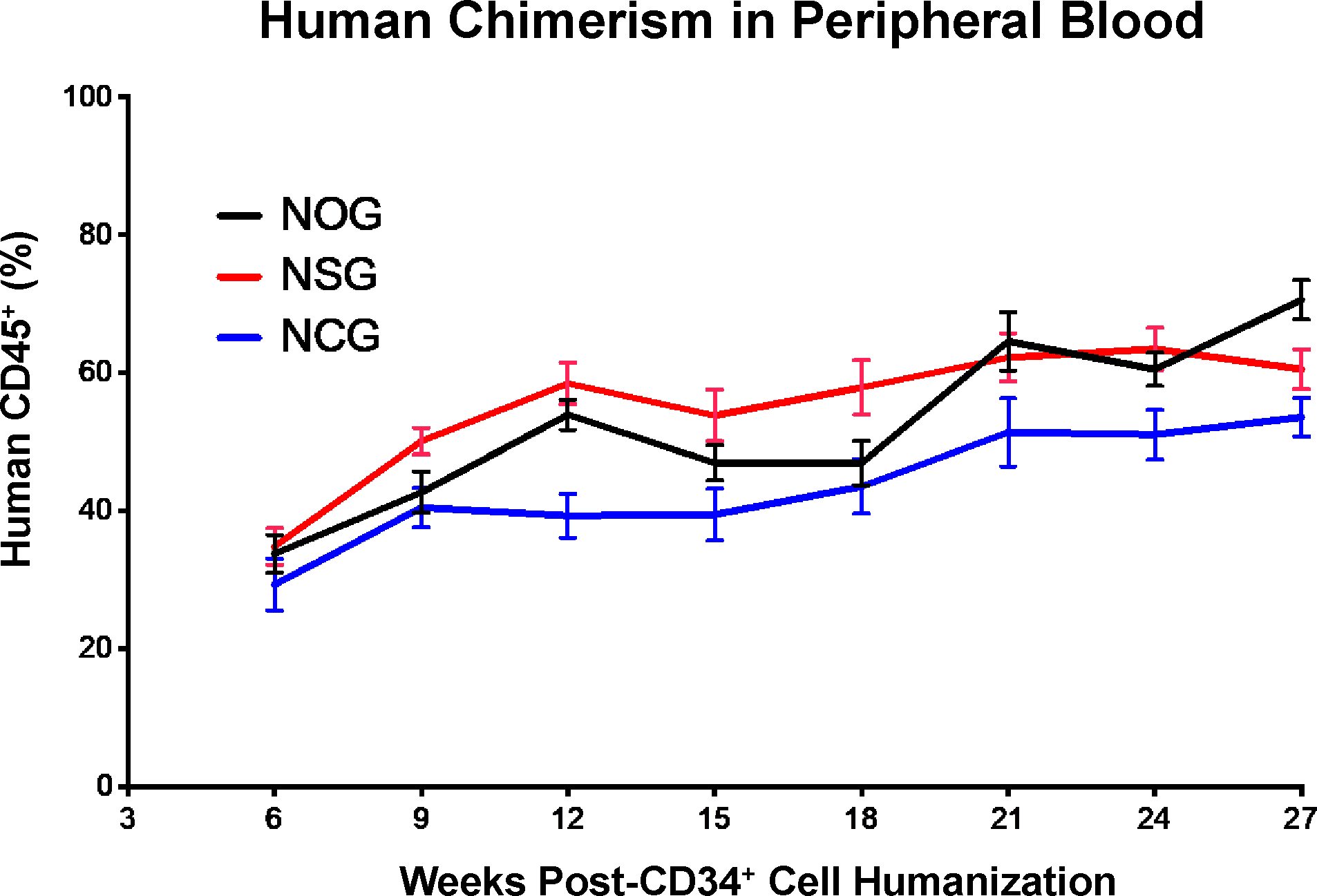
Comparison of Human Immune Cell Chimerism in Three Immune-Deficient Mouse Strains. Female mice from three immune-deficient mice strains (n=10 NOG, n=10 NSG, and n=10 NCG) at 3–4 weeks of age were irradiated with 100cGy, then humanized with 1×105 UCB HSPCs from the same donor. Peripheral blood was assessed by flow cytometry for human CD45+ cells at multiple time points post-humanization (intravenous injection of cells). Mean values are plotted with error bars depicting standard deviation.
Importantly, we also recommend that all humanizing tissues (UCB and thymus) are obtained well in advance of the experiment. HLA considerations (e.g., is a certain HLA-type required and do you have UCB and thymus that match at one HLA class I and one HLA class II allele minimum?) should be decided as early as possible in the experimental design process. A biobank of frozen HLA-typed neonatal thymus tissue is available at the University of Wisconsin-Madison and vials of tissue can be obtained by contacting the corresponding author of this publication. We recommend obtaining thymus tissue, either from the biobank or from local sources, and then using the HLA typing data to guide the purchase of suitable (i.e., HLA-matching) UCB from a commercial vendor and/or obtaining UCB locally and then HLA-typing the samples in-house. In our experience, a number of vendors offer HLA-typed UCB HSPCs for purchase, however, it is often not possible to retroactively HLA-type UCB donors without using precious samples for gDNA isolation.
Lastly, we recommend reading the following experimental notes prior to the start of in-house HLA typing:
Notes for gDNA isolation:
Only isolate gDNA from UCB samples that have more than 2×107 CD34neg cells to ensure sufficient gDNA concentration for HLA typing.
Collect a minimum of 3–4 samples before doing these experiments as it is more efficient to process multiple samples at a time.
Keep RNase Cocktail Enzyme Mix stored at −20°C to preserve activity until ready to use and place back at −20°C immediately after use.
Recommended: keep Proteinase K cool to preserve activity.
Notes for HLA typing:
! CRITICAL Sterility is critical for this protocol. Avoid contamination of the work space by cleaning the area with ethanol. It is recommended to lay down a sheet of aluminum foil cleaned with ethanol within a hood as a sterile work environment.
Store LS Buffer, DNA Polymerase, and 384-well Real-Time PCR trays at −20°C. Keep chilled until immediately before use. Return DNA polymerase to −20°C immediately after use.
Minimize the time between sample addition and the initiation of thermal cycling. If only one instrument is available, set up only one tray.
Transfer any extra sample mix from the tip back into the tube to conserve supply (some repeat pipetters have large dead volumes).
Do not contaminate pipette tips by touching them to the bottom of the Real-Time PCR tray wells. Always dispense sample mixture and No Template Control on the side of the wells.
Make sure that the tray is completely sealed. Any openings in the seal can cause evaporation during amplification.
Use molecular biology grade water.
Materials
Biological materials
Mice.
Immune-deficient mice aged 6–8 weeks should be used for humanization. These “base strains,” a term referring to those animals without additional modifications beyond those which render them immune-deficient, include:
NSG (The Jackson Laboratory, cat. no. 00557)
NBSGW (The Jackson Laboratory, cat. no. 026622)
NOG (Taconic Biosciences, cat. no. NOG)
NCG (Charles River Laboratories, cat. no. 572)
! CAUTION Prior to initiation of experiments, approval from your local institutional animal care and use committee (IACUC) must be obtained. Approval from the UW and University of Massachusetts IACUCs were obtained prior to starting this protocol. ! CRITICAL Most mice base strains require a myeloablative procedure (e.g., irradiation) to allow for engraftment of human cells in the mouse bone marrow.39 In this protocol, for all strains other than the NBSGW, we use irradiation or busulfan dose for myeloablation of certain strains. Busulfan is an option for those labs that do not have access to an irradiator. We have used X-ray and cobalt irradiation, as well as busulfan, and found similar engraftment results with each, as has been previously described.40 It is always recommended that you contact the mouse provider for their recommendations on dosage and/or consult the humanized mouse literature. Ultimately, doses should be experimentally validated and adjusted if needed (e.g., decreasing dose of irradiation if mouse morbidity and/or death is noted at the initial recommended dose).
Thymus and UCB.
! CAUTION National laws and internal review board guidelines must be followed and informed consent must be obtained from human subjects prior to conducting this research. We have followed these guidelines in the work presented here. In cases where matched/autologous UCB and thymus are not available and/or when one UCB donor’s cell stocks are exhausted, it is necessary to use allogeneic UCB sources for creation of NeoThy mice. We choose UCB and neonatal thymus donors that match at least one HLA class I (A, B, or C) allele and at least one HLA class II (DRB or DPB) allele. These particular HLA class I and II genes are the most characterized genes in transplantation immunobiology, and we cannot advise on any benefit of matching other HLA genes (e.g., HLA-DQ). For certain research applications, it may be necessary to do matching at more than one allele from each class. Additionally, cryogenic banking of UCB and thymus fragments allows for selection of certain HLA types that may be of interest for studies associating HLA haplotype to pathogenesis of certain diseases (e.g., HIV). See the HLA typing section below for more details.
Reagents
Anti-CD2 Antibody (Synabs, cat. no. LO-CD2b)
Lymphocyte Separation Medium (Corning, cat. no. 25-072-CV)
Trypan Blue 0.4% Solution (Lonza Bioscience, cat. no. 17-942E)
Lysis Buffer for Whole Blood Staining (BD, cat. no. 555899)
Hanks Balanced Salt Solution without Mg, Ca, or phenol red (Thermo Fisher Scientific, cat. no. 14175-095)
ACK Red Blood Cell Lysis Buffer (Quality Biological, cat. no.118-156-101)
CryoStor CS10 (Stem Cell Technologies, cat. no. 07930)
MACS Bovine Serum Albumin (BSA) Stock Solution (Miltenyi Biotec, cat. no. 130-091-376)
AutoMACS Rinsing Solution (Miltenyi Biotec, order. no. 130-091-222)
FcR Blocking Reagent human (Miltenyi Biotec, order. no. 130-059-901)
CD34 MicroBeads human (Miltenyi Biotec, order. no. 130-097-047)
DMEM/F12 Medium (Gibco, cat. no.11330-032)
- Qiagen QIAamp DNA Blood Mini Kit (Qiagen, cat. no. 51104)
- QIAamp Mini Spin Columns
- 2 mL Collection Tubes
- Buffer AL
- Buffer AW1
- Buffer AW2
Buffer AEQiagen Proteinase K (Qiagen, cat. no. 19131)
RNase Cocktail Enzyme Mix (Thermo Fisher Scientific, cat. no. AM2286)
- LinkSeq™ HLA-ABCDRDQDP+ 384 Kit
- HLA ABCDRDQDP+ 384 Trays
- LinkSeq (LS) Buffer
- DNA Polymerase
- Optical Seal Covers
- Absolute Ethanol, 200 proof, Molecular Biology Grade (Thermo Fisher Scientific, cat. no. T038181000)
- 70% ethanol is achieved via dilution with ultrapure water.
Dulbecco’s Phosphate-Buffered Saline (PBS), 1X without Calcium and Magnesium (Corning, cat. no. 21-031-CV)
Busulfan Injection SD (6mg/ml; Henry Schein, cat. no. 1239588) ! CAUTION Busulfan is a prescription drug. Follow institutional guidelines and practices.
Buprenorphine SR-LAB (0.5mg/ml; Wildlife Pharmaceutical-ZooPharm, LLC,1Z-7400) ! CAUTION Buprenorphine is a controlled substance. Follow institutional guidelines and practices.
Vetbond tissue adhesive (3M, cat. no.1469SB)
Baytril 100 enrofloxacin (Bayer, cat. no. 328RX)
ACD anticoagulant solution (BD Vacutainer, cat. no. 364606)
Isoflurane, USP (Akorn, NDC 59399-106-01)
StemSpan™SFEM II (Stem Cell Technologies, cat. no.09605), aliquoted at 10ml per vial and stored at −20°C.
Human Recombinant Stem Cell Factor (SCF) (Stem Cell Technologies, cat. no.78062.1), aliquoted per manufacturer instructions at 100ng/ul, stored at −20°C.
BD Lysing Buffer (BD, cat. no. 555899)
- Live Dead Fixable Blue (Thermo Fisher Scientific, cat. no. L23105)Flow Cytometry Antibodies (all 5μl per test unless noted)
- APC Mouse Anti-Human CD34 (BD Pharmingen, cat. no. 561209)
- PerCP Rat Anti-Mouse CD45 (mLy5) (BD Pharmingen, cat. no. 557235) 1μl/test
- PerCP Anti-Human CD19 (BD Pharmingen, cat. no. 345778) 5μl/test
- BV605 Mouse Anti-Human CD3 (BD Pharmingen, cat. no. 564712)
- APC-Cy7 Mouse Anti-Human CD45 (Biolegend, cat. no. 304014)
- AmCyan Mouse Anti-Human CD4 (BD Pharmingen, cat. no. 339187)
- APC Mouse Anti-Human CD8 (BD Pharmingen, cat. no. 566853)
- FITC Mouse Anti-Human CD45RA (BD Pharmingen, cat. no. 347723)
- Pacific Blue Mouse Anti-Human CCR7 (Biolegend, cat. no.353209)
Equipment
QuantStuido 5 Real-Time PCR Thermocycler, 384-well (Thermo Fisher Scientific, cat. no. A28140) or similar Real-Time PCR machine, 96 or 384-well
MACS Separator (Miltenyi Biotec, cat. no. 130-090-976)
Microcentrifuge (Eppendorf, cat. no. 5427R)
Microcentrifuge Tubes 1.7mL (VMR, cat. no. 87003-294)
FACS Tube with Cell Strainer Snap Cap (Falcon, cat. no. 352235)
Heparinized Micro-Hematocrit Capillary Tubes (Thermo Fisher Scientific, cat. no. 22362-566)
Bulb for Capillary Tubes (Globe Scientific, cat. no. 51674)
Sterile Cord Blood Collection Unit (Pall Medical, cat. no. 791-08)
Alcohol Swabs (BD, cat. no. 326895)
Sterile Specimen Container (Covidien, cat. no. 2600SA)
1000μl Sterile Barrier Pipet Tips (Dot Scientific, cat. no. UG119LTLR-96RS)
50 mL Conical Centrifuge Tubes (Thermo Fisher Scientific, cat. no. 12-565-270)
Falcon 25 mL Serological Pipette (Thermo Fisher Scientific, cat. no. 13-668-2)
Samco Transfer Pipettes (Thermo Fisher Scientific, cat. no. 204-1S)
15ml Conical Centrifuge Tube (Thermo Fisher Scientific, cat. no. 12-565-268)
MACS LS Columns (Miltenyi Biotec, cat. no. 130-042-401)
Nalegene System 100 Cryogenic Tubes (Thermo Fisher Scientific, cat. no. 5000-1020)
24-well cell culture plate (Thermo Fisher Scientific, cat. no. 142475)
Tissue culture treated dish (Celltreat, cat. no. 229670)
BD PrecisionGlide 23G × 1 inch needles (BD, cat. no. 305145)
(Optional) BD PrecisionGuide 20G × 1 inch needles (BD, cat. no. 305178)
BD 1-mL syringe Luer-Lok tip (BD, cat. no. 309628)
Sterile 1.5-mL conical polypropylene microcentrifuge tubes with attached lid (Thermo Fisher Scientific, cat. no. 05-408-129)
Sterile field (Busse Hospital Disposables, cat. no. 696)
Syringe with 27G needle (BD, cat. no. 305620)
U100 Insulin Syringe 29G (Ultimed Inc., cat. no.09250)
Cotton swabs (Dynarex, cat. no.4302)
5-0 coated VICRYL Suture (Ethicon LLC, cat. no. J385H)
25G needle (BD, cat. no.305125)
5ml Serological pipette (Falcon, cat. no. 357543)
Surgical Blades Stainless (Jorgensen Labs, cat. no. J0581S)
Corning CoolCell FTS30 (Corning, cat. no. 432006)
Sorval Legend X1R Standard Centrifuge (Thermo Fisher Scientific, cat. no. 75004263)
Vortex mixer (Thermo Fisher Scientific, cat. no. 12-812)
Cytoflex Flow Cytometer (Beckman Coulter) or similar unit capable of 8-color acquisition
Water bath (Thermo Fisher Scientific, cat. no. 15-462-20Q) set to 56°C
Cell culture incubator (PHCBI, cat. no. MCO-170AIC)
- Surgical Instruments
- Curved Forceps (Thermo Fisher Scientific, cat. no.16-100-110)
- Scissors (Fine Science Tools, cat. no. 14084-08)
- Straight Forceps (Fine Science Tools, cat. no.11253-20)
- Angled forceps (Fine Science Tools, cat. no.11251-35)
- Iris spatula (Fine Science Tools, cat. no.10094-13)
- Halsey micro needle holder (Fine science tools, cat. no.12500-12)
- Additional instruments may be used according to preferences
Reagent Set Up (Thymus processing):
! CRITICAL Due to the large numbers of thymic fragments per thymus donor, it is critical to have enough cryovials and cryopreservation medium on hand. Plan for a minimum of 2000 fragments, which translates to 100 vials (20 fragments per vial), each in 1ml of cryopreservation medium. Cryovials should be labeled in advance to minimize the amount of time that samples sit in the cryopreservation medium prior to freezing.
Reagent Set Up (HSPC Preparation):
StemSpan SFEM (Liquid aliquots stored at 4°C upon thaw for up to one week, stock bottle aliquoted into 4ml per vial and stored at −20°C), let working liquid aliquot warm at room temperature for 30 minutes, maximum of 2 hours.
DMEM/F12 (store at 4°C), let warm to room temperature for 30 minutes minimum, maximum of 2 hours.
Reagent Set Up (Flow Cytometry):
MACS Buffer
Add 50mL MACS BSA Stock Solution into 1450 mL autoMACS Rinsing Solution. Store at 4°C until ready to use, keeping it on ice in between wash steps/uses during the experiment.
Flow Cytometry Antibody Master Mix Preparation
! CAUTION This step needs to be performed in the dark and quickly as the antibodies used are light and temperature sensitive. The master mix can be made up to 24 hours in advance and stored in the dark at 4°C until ready to use.
! CRITICAL Calculate the volume of master mix using the equation below:
x = number of mice bled (number of samples)
Total volume of master mix = V = (x + 1) * 100μl
Antibody volumes:
PerCP Rat Anti-Mouse CD45 (mLy5) 1μl * (x + 1) = A μl
BV605 Mouse Anti-Human CD3 5μl * (x + 1) = B μl
APC-Cy7 Mouse Anti-Human CD45 5μl * (x + 1) = C μl
AmCyan Mouse Anti-Human CD4 5μl * (x + 1) = D μl
APC Mouse Anti-Human CD8 5μl * (x + 1) = E μl
FITC Mouse Anti-Human CD45RA 5μl * (x + 1) = F μl
Pacific Blue Mouse Anti-Human CCR7 5μl * (x + 1) = G μl
PerCP Anti-Human CD19 5μl * (x + 1) = H μl
Add FACS Buffer to bring total volume to V amount above, accounting for total volume added of A-H.
Vortex the final master mix well and keep it in the dark at 4°C until ready to use.
Reagent setup (HLA Typing):
Remove 1 LS buffer tube and the DNA polymerase vial from −20°C storage and place it in ice to keep it chilled prior to the start of the protocol.
Ensure all gDNA related reagents are at room temperature prior to the start of the procedure.
Equipment Setup (HLA Typing):
Heat the water bath to 56°C prior to starting gDNA processing.
Turn on the Real-Time PCR machine prior to the start of the protocol to minimize the time between sample addition and initiation of thermal cycling. Note: a Real-Time PCR machine is required (i.e., a conventional PCR is not appropriate) for the One Lambda LinkSeq HLA typing method, which relies on melt curve data for typing.
Equipment Setup (Flow Cytometry):
Turn on the CytoFLEX cytometer (or other 8-color cytometer) and perform daily cleaning and manufacturer recommended quality control steps 30 minutes prior to acquiring samples.
Software (Flow Cytometry):
CytoFLEX CytExpert 2.3
Software (HLA Typing):
One Lambda SureTyper v6.1
Protocols (with timing)
! CRITICAL **Sterility is critical in all steps of this protocol—take care to not contaminate any samples, tubes, or reagents*** ! CAUTION National laws and internal review board guidelines must be followed and informed consent must be obtained from human subjects prior to conducting this research. We have followed these guidelines in the work presented here.
Neonatal thymus processing Timing ~4 hours
-
1
Obtain neonatal thymus sample from the operating room. The sample should be placed in a sterile specimen jar and transported on ice to the laboratory. ! CRITICAL Time is of the essence; treat the specimen like an organ transplant and quickly pick it up and transport it to the lab as soon as the surgeons remove it from the patient.
-
2
Using aseptic technique in a Biosafety Level (BSL) 2 biosafety cabinet (BSC), use autoclaved forceps to transfer the thymus from the specimen jar to a sterile petri dish that has been weighed in advance (the weight of the vessel is subtracted to determine the specimen weight).
-
3
Photograph the sample for record keeping, including a ruler for documenting the size of tissue (Figure 4A). Then weigh the specimen by subtracting the weight of the sterile dish from the previous step (Figure 4B).
-
4
Prepare a wet ice-filled container and wrap it with foil to make an ice tray (Figure 4C and 4D). ! CAUTION The foil is important to ensure sterility and also to prevent sample movement as ice melts.
-
5
Disinfect the surface of the ice tray thoroughly with 70% ethanol and place it in the BSC so it can be used to keep the sample cool during dissection.
-
6
After weighing the thymus, put the dish on the ice bed and add 20ml of sterile phosphate-buffered saline (PBS) in the dish.
-
7
Use autoclaved/sterile scissors and forceps to remove the blood vessels (including cauterized tissue), adipose, and the capsule on the surface of the thymus (Figure 4E).
-
8
Use sterilized scissors to cut the thymus first into chunks then into 1 cm × 1 cm pieces/fragments (Figure 4F).
-
9
Continue cutting the thymus into small 1 mm × 1 mm fragments (size used for transplantation) (Figure 4G), moving to the side of the dish for size verification (Figure 4H) and counting. (Optional) Thymocytes released into the buffer can be collected and used for gDNA/HLA typing (see step 48 below) and/or for other experimental uses. ! CAUTION Plan to have enough fragments to fill the amount of vials that will fit into one Corning CoolCell or similar controlled-rate freezing container. For example, if your container holds 24 vials and you want 20 fragments per vial, plan to dissect 20×24=480 fragments before proceeding to next step.
-
10
Transfer all the pieces into sterile cryovials (Figure 4I), taking care to let fragments settle by gravity to the bottom of the tube (Figure 4J). ! CAUTION Thymus fragments can stick to the lid and the sides of the vial, and care needs to be taken during this step to ensure sterility is maintained and/or fragments are not damaged by the screw top. ! CRITICAL Fill all vials needed to fully load your freezing container prior to proceeding to the next step in order to minimize the time that fragments are in the medium prior to freezing.
-
11
Add 1ml sterile CryoStor CS10 freezing medium into each vial. (Figure 4K). ! CAUTION Due to the large number of vials, it is advisable to label tubes in advance.
-
12
Verify all fragments are settled into the freezing medium (Figure 4L) and put all the vials into wet ice until enough samples are prepared to fill a CoolCell controlled rate freezing container. ! CAUTION The samples should not sit for more than 10 minutes prior to storing at-80°C in CoolCell. Store vials at −80°C for 24 hours, then transfer to liquid nitrogen storage.
Figure 4.
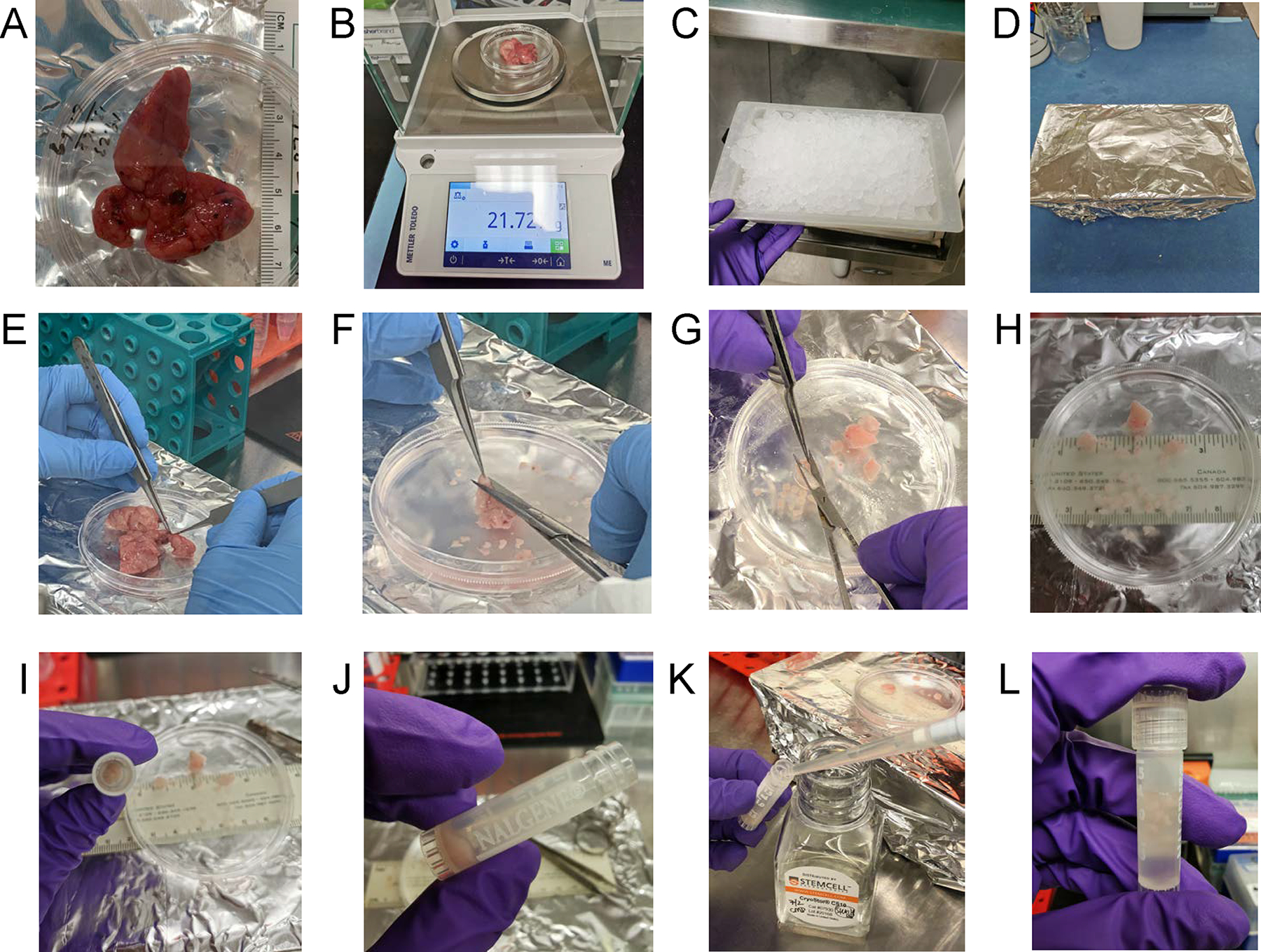
Neonatal Thymus Processing. Using aseptic technique, the thymus tissue is (A) photographed and measured for length and (B) weighed. (C) A wet ice tray is prepared, (D) wrapped in aluminum foil, sprayed with 70% ethanol and placed in the biosafety cabinet. (E) The thymus tissue is placed on ice tray for dissection in cold PBS, whereupon blood vessels, adipose, and cauterized material is removed and then the lobes are cut into large sections. (F) Thymus is dissected into chunks approximately 1–3 cm × 1–3 cm. (G) The chunks are then cut into smaller pieces and moved to the side of the dish for (H) size determination and counting. Fragments are made into 1 mm × 1 mm size for cryopreservation in (I) cryotubes. Typically, 20 fragments per tube are frozen. (J) Fragments are settled by gravity and (K) 1ml of Cryostor CS10 freezing medium is added per vial. (L) The cap is tightened and each vial is inspected to ensure that fragments are submerged in liquid prior to being frozen to −80 degrees Celsius in a Corning CoolCell controlled-rate freezing container.
UCB processing and HSPC separation Timing ~3 hours
! CAUTION This step does not need to be done on the same day as the thymus processing. Both thymus and UCB cells are cryopreserved and can be banked in advance. Thymus and UCB will be HLA typed (see next section), then thawed as needed for humanization in the future. ! CAUTION National laws and internal review board guidelines must be followed and informed consent must be obtained from human subjects prior to conducting this research. We have followed these guidelines in the work presented here.
-
13
Spray the UCB unit bag with 70% ethanol and use an ethanol wipe to sanitize the tubing area to be cut. Cut open the tubing at the top of the bag with a sterilized blade and carefully pour the cord blood sample into 50ml conical tubes. Record the volume of sample.
-
14
Dilute cord blood 1:1 with sterile PBS.
-
15
Add 15ml lymphocyte separation medium (LSM) in each 50ml conical tube.
-
16
Slowly layer 25ml of diluted blood onto the top of the 15ml LSM. Carefully take samples to scale for balancing; do not disturb/mix layers in tubes.
-
17
Balance tubes precisely prior to centrifugation.
-
18
Centrifuge the samples at 400 × g for 30 minutes with low acceleration/brake off. ! CAUTION Having the brake off/low acceleration on the centrifuge is important so as to not disturb the cell layer following centrifugation.
-
19
Using a sterile transfer pipette, collect the buffy coat layer (milky white interface between red lower layer and yellow/clear upper layer) into a new 50ml conical tube.
-
20
Add PBS up to a total volume of 45ml. Cap the tube and gently re-suspend by inverting then centrifuge at 300 × g for 5 minutes using normal acceleration/brake settings. Remove supernatant, taking care not to disturb the cell pellet.
-
21
Repeat PBS Wash step a second time.
-
22
Aspirate supernatant, taking care not to disturb cell pellet, then add 10ml ammonium-chloride-potassium (ACK) lysis buffer to pellet. Gently mix to break up pellet in liquid using a pipette, and then incubate tube at room temperature for 7 minutes.
-
23
Add PBS up to 45ml, then centrifuge at 300 × g for 5 minutes.
-
24
(Optional) Repeat ACK lysis step/wash if there is a significant amount of red blood cells in the pellet (dark red color by eye). Add 5ml ACK lysis buffer to the pellet, gently mix, and leave it at room temperature for 5 minutes.
-
25
Add PBS up to 45ml, and centrifuge at 300 × g for 5 minutes.
-
26
Carefully aspirate the supernatant, and then re-suspend cells (by pipetting up and down a few times; do not vortex) in 20ml PBS. Count the cells via automated cell counter (e.g., Chemometec NucleoCounter NC-200) or hemacytometer/trypan blue exclusion method, then centrifuge at 300 × g for 5 minutes. If greater than 7×107 total cells, proceed to CD34 MACS separation. If less than 7×107, freeze cells in multiple vials of ~2–5×107 cells per vial in CS10 freezing medium for alternative use. ! CAUTION We have found that CD34+ enrichment requires a minimum of 7×107 total cells to achieve >1×106 CD34+ cells for humanization.
-
27
If proceeding to CD34+ (HSPC) positive enrichment, aspirate supernatant after centrifugation, then re-suspend the cell pellet with 108 cells per 300μL of cold, sterile MACS buffer for MACS separation in the next step, or if using a different technology (e.g., Stem Cell Technologies EasySep) follow manufacturer instructions.
-
28
Re-suspend the cell pellet in 300μl of MACS buffer per 1×108 total cells.
-
29
Add 100μl of FcR blocking reagent for up to 1×108 total cells. For >1×108 cells, add by an appropriate factor (2.8×108 = 280μl reagent).
-
30
Add 100μl of CD34 MicroBeads per 1×108 total cells. For >1×108 cells, add by an appropriate factor (2.8×108 = 280μl reagent).
-
31
Mix via gently flicking (do not vortex) and incubate for 30 minutes at at 2–8°C.
-
32
After incubation, transfer the sample to a 15ml conical tube, wash the cells by adding 10ml of cold MACS buffer, and centrifuge at 300 × g for 5 minutes. Aspirate the supernatant completely. Gently break up the pellet by flicking (do not vortex).
-
33
Re-suspend in 500μl MACS buffer per 1 ×108 cells. Flick gently to mix.
-
34
Wipe down the MACS magnet unit with 70% ethanol and place in BSC. Keeping all parts sterile, especially the tip, place MACS LS column (one column needed) in the magnetic field of a suitable MACS separator. Wash the column by rinsing with 3ml MACS buffer, and discard flow-through. If there are multiple samples, carefully label each column on the wide upper portion to discern between samples. Make new, labeled sterile 15ml tubes to collect positive and negative fractions. Place negative fraction tube under the tip of the appropriate column, making sure it does not move when/if bumped. ! CAUTION This step is susceptible to contamination, so take care to prevent touching the tip. It is important to lay out all supplies to allow for ease of movement and insertion of the plunger in Step 38. Position the plunger by removing the paper backing, but leaving the plunger to lie within the sterile clear plastic coating..
-
35
Apply labeled cell suspension (pre-filter if clumps are noticed) onto the prepared LS column. Collect flow-through containing unlabeled “negative fraction” cells into a labeled 15ml conical tube.
-
36
Wash column with 3ml MACS buffer, waiting until drops stop forming at tip, repeating this a total of 3 times. Collect unlabeled “negative fraction” cells that pass through. Note the total volume for cell count calculations. Sample for cell count.
-
37
Being careful to keep things sterile (especially the LS column tip), remove column from the separator and place it on a new 15ml conical tube labeled CD34+. ! CAUTION Keep only the tip inside the opening (i.e., do not place the entire column in the tube even though it fits. As a precaution against contamination, avoid touching it inside the tube).
-
38
Pipette 5ml MACS buffer onto the column. Immediately flush out the magnetically labeled cells by firmly pushing the plunger into the column.
-
39
Collect a small amount of cells from the CD34neg and CD34+ fractions to determine cell number (~300μl). Save cell count samples for CD34 purity test. For the CD34neg population, please see step 47 “gDNA CELL PELLET” before proceeding to step 40.
-
40
Centrifuge the CD34neg and CD34+ flow-through at 300 × g for 5 minutes. Aspirate the supernatant and re-suspend the cell pellet in CS10 freeze medium for freezing.
-
41
Freeze CD34+ cells at 1–1.5 ×106 per vial in CS10 (see thymus fragment freezing step 12 above) if more than 2 ×106 total; otherwise, freeze in one tube.
-
42
Freeze CD34neg cells at 2–2.5 ×107 per vial, using the same method as detailed in steps 41 and 12 above.
-
43
Keep excess cells from both fractions for purity testing in step 46 below. ! CAUTION By using the excess/dregs of the counting and cell recovery tubes, you can minimize using up precious CD34+ for counting and flow cytometry and have more for humanization.
-
44
Transfer to liquid nitrogen storage after 24 hours.
-
45
STERILITY TESTING—Add 2ml of sterile DMEM/F12 cell culture medium to CD34+ tube. Collect medium with residual cells and plate into a 24 well tissue culture plate (add medium alone to a separate well to make sure medium bottle is sterile) then place in a 37°C cell culture incubator. Monitor for 1–5 days to look for any signs of contamination. Make note in lab records that this sample has passed sterility test. If contamination is present, do not use for in vivo experiments (we never use antibiotics in cell cultures, to avoid masking low-level infection). Use for terminal HSC assays instead, noting records accordingly.
-
46
PURITY TESTING—Split residual CD34+ and CD34neg samples from the cell count into two equal aliquots. Stain one CD34+ and one CD34neg sample with APC CD34 for 20 minutes in the cold/dark. The unstained CD34+ and CD34neg samples will be used as controls. After 20 minutes, wash samples with 1ml MACS buffer and then centrifuge at 300 × g for 5 minutes. Pour off the supernatant, break up the pellet, and proceed to run flow. Record the percentage of APC CD34 positive cells. Acceptable purity is >70% CD34+. We have previously determined (unpublished data) that the small number of T cells present at this level are not capable of engrafting an animal.
-
47
Genomic DNA (gDNA) CELL PELLET for HLA typing—Take 2–3 ×107 cells from the CD34neg population to create a cell pellet by centrifuging cells at 300 × g for 5 minutes. Aspirate the supernatant, then label the tube with donor/date/scientist initials. Store at −80°C for HLA typing in the next step. PAUSEPOINT: gDNA isolation and HLA typing can be done at a later date.
HLA typing/matching allogeneic tissues
Procedure:
Genomic DNA (gDNA) Isolation for HLA Typing Timing ~1 hour
-
48
Re-suspend 0.5–2 ×107 cells in 200μl of PBS master mix. For example, assuming 40μl frozen cell/PBS pellet add 160μl master mix (136μl PBS, 20μl Qiagen Proteinase K and 4μl RNase Cocktail Enzyme Mix). Ensure solution is in a sterile 1.7 mL microcentrifuge tube.
-
49
Add 200μl of Buffer AL to samples. Mix by pulse-vortexing for 15 seconds. If the volume of cells and master mix is greater than 200μl, increase the volume of Buffer AL proportionally. ! CRITICAL: Ensure proper mixing until there is no longer a visible cell pellet for best results of lysis of genetic material.
-
50
Incubate at 56°C for 10 minutes. For optimal DNA recovery and size, after about 5 minutes disperse lysed cellular material by forcing through a 23G needle (1mL syringe) until it passes easily through. It will be difficult to draw in the solution. Be careful not to overdraw and/or expel too quickly; this will minimize bubbles. If unable to draw in solution or there are clumps too large for a 23G needle, you can use a 20G needle until the solution passes easily through and then switch to a 23G needle until the solution passes easily through. Then return the solution to 56°C for the final 5 minutes of incubation.
-
51
Briefly centrifuge the 1.7 mL microcentrifuge tube to remove drops from the inside of the lid.
-
52
Add 210μl ethanol (i.e., slightly greater volume of ethanol than Buffer AL) to the sample. Mix by pulse-vortexing for 7–10 times. Rock back and forth to ensure complete mixing; you will be able to see a visible precipitate. After mixing, briefly centrifuge the 1.7 mL microcentrifuge tube to remove drops from the inside of the lid.
-
53
Carefully apply the mixture to the Qiagen Blood mini-spin column (in a 2ml collection tube) without wetting the rim. Incubate for 3 minutes at room temperature. Label the mini-spin column with the sample name. ! CRITICAL: If there are any remaining clumps within the solution, dispose of them and only apply liquid solution into the mini-spin column.
-
54
Centrifuge the mini-spin column at 6000 × g (8000 rpm) for 1 minute. Place the mini-spin column in a clean 2ml collection tube. Discard the tube containing the filtrate.
-
55
Carefully open the mini-spin column and add 500μl of buffer AW1 without wetting the rim. Close the cap and centrifuge at 6000 × g (8000 rpm) for 1 minute. Place the mini-spin column into a clean 2 ml collection tube and discard the collection tube containing the filtrate. Even if the volume of buffer AL and ethanol were increased, the volume of Buffer AW1 should stay the same at 500μl.
-
56
Carefully open the mini-spin column and add 500μl buffer AW2 without wetting the rim. Close the cap and centrifuge at full speed (maximum × g; about 14–16,000 rpm on common centrifuges) for a total of 2 to 3 minutes.
-
57
Place the mini-spin column in a clean, labeled 1.7 mL microcentrifuge tube and discard the 2 mL collection tube containing the filtrate. Label tube 1.7 mL microcentrifuge tube with the sample name.
-
58
Add 75μl buffer AE. Incubate the sample on the column at room temperature for 5 minutes, then centrifuge at 6000 × g for 30 seconds.
-
59
Add a second aliquot of 50μl buffer AE. Incubate the sample on the column at room temperature for 5 minutes, and then centrifuge at 20,000 × g for 30 seconds.
-
60
Proceed to purification analysis and HLA typing or freeze the samples at −20°C.
HLA Typing Timing ~3 hours
Procedure:
-
61
Remove one LinkSeq 384-well Real-Time PCR tray from −20°C storage. Clean the outside of the plate carefully with 70% ethanol and place it within an aseptic BSC working environment. Write down the unique plate identifier, kit lot number, and kit expiration date for later input into SureTyper software. Remove LS buffer and DNA polymerase from −20°C storage and place into ice to remain chilled until use. ! CRITICAL: Keep 384-well Real-Time PCR tray, LS buffer, and DNA polymerase chilled in ice until the beginning of the protocol. Leave LS buffer and DNA polymerase chilled until use.
-
62
After the Real-Time PCR tray has equilibrated to room temperature, centrifuge at 500–2000 × g for 30–60 seconds to ensure primers are at the bottom of all wells. Before centrifuging Real-Time PCR plate, remove LS Buffer from ice to begin thawing at room temperature.
-
63
Begin preparing the sample mix within the LS buffer vial once thawed. Add 92μl of DNA Polymerase to the LS buffer vial and mix to combine. ! CRITICAL: Mix DNA polymerase and LS buffer thoroughly. It is not recommended to vortex this mixture as it will cause an excessive amount of bubbles. Once DNA polymerase has been added to the sample mixture, place the DNA polymerase vial back into −20°C storage to preserve activity.
-
64
Prepare the No Template Control: Transfer 5μl of the LS buffer containing DNA polymerase to a sterile 1.7 mL empty microcentrifuge tube. Add 5μl of molecular biology grade water to the tube and place it in ice to be chilled and set aside until later use.
-
65
To the remaining LS buffer (containing DNA polymerase), add a minimum of 3700ng genomic DNA in 2308μl molecular biology grade water. ! CRITICAL: Ensure thorough mixing of sample mixture before proceeding to the next steps. It is not recommended to vortex this mixture as it will cause an excessive amount of bubbles.
-
66
Pour the thoroughly mixed sample mixture into reagent reservoir. Remove the Real-Time PCR tray plate seal and discard.
-
67
Using an 8-channel pipette, draw up 10μl of sample mixture into each pipette channel. Dispense sample mixture in a downward grid pattern with the first tip at position A1, B1, C1 etc. through P1. Next, align the first pipette tip and dispense into position A2 followed by B2, C2, etc. through P2. Continue this dispensing pattern until last column of the plate, but do not dispense any sample mixture into the No Template Control (NTC) well P24.
-
68
Be careful to pipette the sample to the side of the wells. ! CRITICAL: With every pipette draw, ensure all 8 channels have taken up an equivalent amount of sample mixture. It is common to not get a proper amount of solution in end channel tips. Repeat sample mixture draws until volumes look correct in pipette tips. Do not allow the pipette tips to come into contact with the bottom of the Real-Time PCR tray wells. This will contaminate the primers and invalidate results. You can use the same pipette tips for the entire Real-Time PCR tray as long as the plate is spun down prior to the start of protocol and the pipette tips only touch the upper side of the wells and never the bottom. ! CAUTION Be careful to avoid pipetting the sample mixture into well P24 as that is the No Template Control well. Move quickly when filling the Real-Time PCR tray with sample mixture, but accuracy is most important. This step of the protocol should take less than an hour in total.
-
69
For the last dispense, remove the 8th pipette tip from the multi-channel pipette to prevent dispensing sample mixture into the No Template Control well (well P24).
-
70
Transfer 10μl of No Template Control Mixture to well P24 according to the tray map.
-
71
Gently tap the tray to drive the droplets of sample mixture to the bottom of each well.
-
72
Remove the backing from the new Real-Time PCR optical tray seal. Using a seal applicator, press the seal onto the Real-Time PCR tray. ! CRITICAL: Make sure the PCR tray is completely sealed and wrinkle and bubble-free. Center the Real-Time PCR optical tray seal on all sides to prevent puckering of the tray seal along the edges of the Real-Time PCR tray. Run the seal applicator side to side and up and down several times. Make sure to also run the seal applicator around the border of the outer wells and especially over raised lettering and numbering of the Real-Time PCR tray to avoid puckering of seal while in the Real-Time PCR instrument.
-
73
Centrifuge the sealed Real-Time PCR tray at 500–2000 × g for 30–60 seconds. ! CAUTION Begin next step immediately.
-
74
Set the Real-Time PCR tray into the Real-Time PCR instrument. Begin the amplification process, which should last approximately 75 minutes. ! CRITICAL: Ensure the plate is loaded into the Real-Time PCR instrument in correct orientation according to the lettering and numbering of the Real-Time PCR tray. For the LinkSeq HLA typing kit, a Real-Time PCR is needed because of the melt curve method of typing, so a standard PCR machine is not an acceptable alternative in this protocol.
-
75
After amplification is complete, export raw melt curve data from the Real-Time PCR instrument for input into HLA SureTyper software along with unique plate identification, kit lot number, kit expiration date, and sample name according to the manufacturer’s SureTyper User Manual available online (https://www.thermofisher.com/onelambda/us/en/products.html?articleNumber=STTPGR X). The raw melt curve data should be exported as a text file and contain temperature and derivative data. The SureTyper analysis report for the sample includes typing information for HLA-A, HLA-B, HLA-Bw, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DPA1, HLA-DRB345, HLA-DQB1, and HLA-DPB Epitype. ! CRITICAL: Ambiguous rare test results may be manually selected via allowing SureTyper program to choose the only valid test call that does not require the assignment of a rare allele.
TROUBLESHOOTING
| STEP | PROBLEM | POSSIBLE REASON | POSSIBLE SOLUTION |
|---|---|---|---|
| 50 (gDNA Isolation) | Lysed material is unable to be forced through 23G needle. | Cell pellet has not been lysed small enough during previous steps. | Forced lysed material through 20G needle until the solution passes easily through and then switch to a 23G needle until the solution passes easily through. Move on to next step in protocol. |
| 54 (gDNA Isolation) | Sample mixture has not sufficiently flowed through mini-spin column allowing for collection of filtrate. | Sample mixture may be too thick to flow through the mini-spin column therefore blocking the collection of filtrate into the collection tube. | Centrifuge sample again at 8000 x g for 1.5 minutes. Place the mini-spin column in a clean collection tube. Discard the tube containing the filtrate. Continue to next step in protocol. |
| 60 (gDNA Isolation) | gDNA concentration of sample is too low for HLA Typing. | The cell pellet used for isolation of gDNA contained less than 2x107 CD34neg cells (UCB samples) or thymocytes (Thymus samples). | Redo gDNA isolation using cell pellet containing more than 2x107 CD34neg cells (UCB samples) or thymocytes (Thymus samples). |
| 68 (HLA Typing) | Multi-channel pipette tips come into contact with the bottom of the wells. | Sample mixture was not pipetted to the side of the wells. | Change all pipette tips to avoid contamination of primers and further invalidation of results. |
| 68 (HLA Typing) | Multi-channel pipette tips are drawing up different amounts of sample mixture. | Pipette tips are not filling properly due to reasons such as air bubbles being drawn into certain channels, pipette tips not fitting well, or insufficient pipetting technique. | Repeat sample mixture draws until volumes look correct in pipette tips. If this does not work, change all pipette tips and continue to repeat sample mixture draws until volumes look correct in pipette tips. |
| 72 (HLA Typing) | Evaporation of sample mixture from all or some wells in PCR tray leading to dropout wells in HLA typing analysis. | Plate was not sufficiently sealed and may have contained either air pockets within the center of the tray or puckering of the edge of the optical tray seal. | Repeat HLA typing protocol for sample and ensure PCR tray is sufficiently sealed before placing into PCR machine. Use a seal applicator to press seal over the top of all the wells and along each edge of the PCR tray. Be careful to make sure there is no puckering of the plate seal at each corner of the tray. Ensure there are no air bubbles formed near the raised letters and numbers along the top and left edges of the PCR tray. |
| 75 (HLA Typing) | SureTyper program does not automatically call locus instead flagging it as "ambiguous rare” | More than one genotype can explain the reaction pattern so the program is unable to automatically call the locus. | Ambiguous rare test results may be manually selected via allowing SureTyper program to choose the only valid test call that does not require the assignment of a rare allele. Practice on Supplementary Data 1 data set with Supplementary Data 2 to guide what the finished product should look like. |
NeoThy Surgery
Procedure: Thawing and plating HSPCs one day prior to humanization surgery Timing ~45 minutes ! CAUTION Animal Care and Use Committee approval should be obtained prior to surgery and any procedures (e.g., blood collection) involving mice.
(Optional): Thaw the CD34+ cells on the day of surgery and directly inject the cells. We have found the following overnight plating method to improve engraftment kinetics, but it is possible to get satisfactory engraftment with direct injection. If using direct injection, skip steps 84–92.
-
76
Remove a cryogenic vial of UCB CD34+ HSPCs from the liquid nitrogen storage tank. Record the cell information from the label.
-
77
Place the vial into 37°C water bath. ! CAUTION Do not submerge the cap of the vial in the water as this could contaminate the cells.
-
78
Quickly thaw the cells (1–2 minutes) by gently swirling the vial in the water bath until there is just a small bit of ice left in the vial. ! CAUTION Do not leave in water bath unattended and/or for over 2 minutes as this will result in diminished cell viability.
-
79
Remove vial, spray liberally with 70% ethanol, then wipe down excess liquid. In a BSC, transfer the contents of vial directly to the bottom of a 15ml conical tube using a transfer pipette.
-
80
Collect 8ml DMEM/F12 (with no additives) with a 10ml pipette.
-
81
Add 1ml DMEM/F12 to the empty cell vial.
-
82
Add 7ml DMEM/F12 to the 15ml conical tube. Transfer the medium dropwise into the thawed cells. ! CRITICAL Slow, dropwise addition of thawing medium reduces the osmotic shock to the cells and improves viability. In particular, the 1st and 2nd drops need to be added very slowly (i.e., 1 drop per 2–4 seconds). Gently rock to continually mix the cells as the new medium is added to the tube.
-
83
Centrifuge the cells for 5 minutes at 300 × g.
-
84
While the cells are in the centrifuge, prepare the 24 well cell culture plate by labeling it with the cell information (cells line, thaw date, operator name)
-
85
Bring the pelleted cells back to the BSC, and carefully aspirate the supernatant. Leave ≤ 0.2ml supernatant in the tube.
-
86
Take up 2ml StemSpan SFEM with a 5ml pipet. Add 1mlSFEM to a 2μl aliquot of 100ng/μl SCF.
-
87
Re-suspend the cell pellet very gently by adding another 1ml SFEM and slowing mixing up and down.
-
88
Add the 1ml SFEM+SCF mixture to the cells and pipette up and down 3–4 times.
-
89
After mixing, take out 15μl for cell counting.
-
90
Slowly add all of the ~2ml cell suspension to a 24 well tissue culture plate.
-
91
Using a new pipette, take the remaining 2ml of SFEM and add to a second well in the plate. This will serve as the media-alone control for detection of contamination.
-
92
Place the plate into the 37°C incubator overnight.
Procedure: Surgical Preparation for Up to 20 Mice Timing ~45 minutes
-
93
Prepare anti-CD2 antibody in cell injection buffer: Dilute sterile anti-CD2 antibody to a concentration of 2mg/ml in Hank’s Balanced Salt Solution (HBSS), so that 100ug will be in a volume of 50μl per mouse.
-
94
Collect HSPCs and prepare for injection: Using a sterile transfer pipetter, remove the cells from the incubator and collect by gently swishing up and down, placing cells in 15ml conical tube. Use 2ml of HBSS to wash the cell culture dish to remove residual cells by adding the HBSS to well, swishing it up and down, and adding it back to conical tube with the cells. Note the total volume, mixing by gentle inversion and collect 15μl for cell counting via hemacytometer/trypan blue exclusion method. Aliquot the volume needed for 0.5–1.5 × 105 HSPCs into individual sterile 1.7 ml microcentrifuge tubes, and centrifuge for 5 minutes at 300 × g. (A range of cell numbers are given due to variability in UCB donors as well as number of mice being used in a given experiment. 0.5 × 105 cells minimum are needed for reproducible results, and going over 1.5 × 105 is not advised as the extra cells are not likely to improve engraftment [i.e., will be wasted]. To optimize the number of mice, plan to use 0.5 × 105 cells per mouse). Carefully aspirate the supernatant with a p1000 pipette using filtered barrier tips. Re-suspend the cell pellet in 50μl of sterile HBSS and combine it with 50μl anti-CD2 antibody (100ug) from the previous step. One individual tube of 100μl cells and antibody will be used to inject one mouse. Keep on ice until ready to inject during surgery (see step 112 below).
-
95
Collect and Prepare Thymus Fragments for transplantation: Thaw two vials of 20 thymus fragments in water bath, as in 76–79 above. Prepare 50ml of PBS in a 50ml conical tube. Dump out thymus fragments in freezing media into the PBS, let settle to bottom of tube. Inspect vial to verify no fragments are stuck in vial. If any remain, use a bulb pipette to take some of the PBS and add ~1ml to vial(s), then dump out again. Once all fragments are settled, aspirate out PBS, leaving approximately 10ml at bottom with fragments, being cautious not to aspirate fragments. Add 40ml fresh PBS to wash the fragments again, let settle, then remove down to ~10ml again. Add 10ml more PBS and transfer tube to ice. Transport to surgical suite with other materials. These fragments will be added to sterile dish prior to surgery.
-
96
Weigh each mouse prior to surgery, and record in surgery log.
! CAUTION Do not proceed to the next steps until you are ready to begin surgeries.
-
97
Shave mice: Lay the mouse on its right side while it is under isoflurane anesthesia. Shave the hair of the mouse from the armpit to the hip of the left side using an electric hair clipper (Figure 5A).
-
98Myeloablate Mice (Use one of either of the following options, but not both).
- Via Irradiation. ! CAUTION Ensure that the irradiation vessel is sterile and contained (e.g., within a filter top rat cage) to avoid introduction of pathogens into the mouse colony. For all strains except those with kit mutations (e.g., NBSGW) or other mutations that obviate the need for irradiation, irradiate the mice 4–12 hours prior to surgery with a dose of X-ray irradiation, per vendor recommendations. For NSG mice, we typically use 200cGY. For NOG-EXL or NSG-SGM3 mice, we typically use 55cGY.
- Via Busulfan at 25mg/kg. ! CAUTION Busulfan is a powerful drug, so accurate weights and dosing are critical to avoid accidental deaths. Keep sterile. Prepare and inject as previously described.41
-
99
Inject the Buprenorphine SR (0.6mg/kg) for pain relief subcutaneously via a 27G syringe (Figure 5B). NOTE: this medication can be viscous and difficult to fully remove from tube, so enough volume will be need to allow for retention of material in vial.
Figure 5.
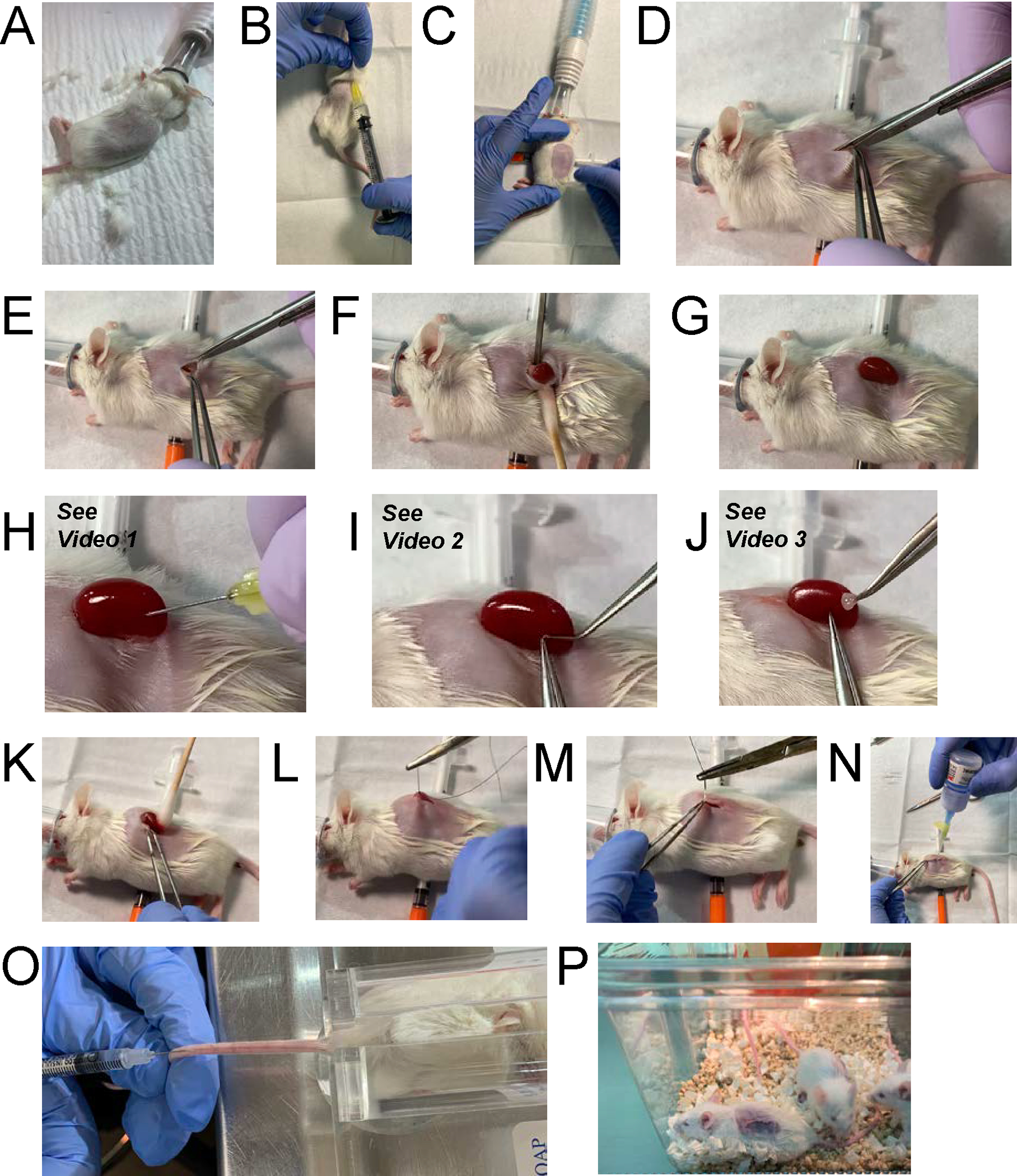
NeoThy Humanization Surgery. (A) Immune-deficient mice are placed under anesthesia and the surgical site is shaved. (B) Mice are subcutaneously administered Buprenorphine SR (0.6mg/kg) for pain-relief. (C) Skin is disinfected with povidone-iodine wipe, followed by 70% ethanol wipe, then allowed to dry. (D) An incision is made in the skin above the left kidney, taking care to leave opening slightly narrower than the length of the kidney (see step G). (E) An incision is made in the abdominal wall. This can be larger than than the opening in the skin. (F) The kidney is exposed using a sterile swab dipped in saline and an iris spatula. (G) The kidney is held in place due to the distal and proximal ends of the kidney resting on the skin due to the small skin incision. (H) A shallow nick is made to cut through the kidney capsule. (I) A tissue pocket is made with a blunt forceps. (J) The thymus tissue is carefully placed under the capsule with a forceps, and slide to the proximal end of the kidney, away from the nick in the capsule. (K) The kidney is gently pushed back into the abdominal cavity using wet swab, as in step F. (L)(M) The abdominal wall then skin are sutured with Vicryl 5–0 absorbable sutures. (N) Vetbond tissue adhesive is added to aid in closing of the skin wound. (O) CD34+ HSPCs and anti-CD2 antibody preparation is i.v. injected into the tail vein. (P) Animals recover under heat lamp.
Procedure: Mouse Surgery for Up to 20 Mice Timing ~15 minutes per mouse = 3 hours
-
100
Completely disinfect the operation table with 70% ethanol. Lay a sterile field on the table. ! CAUTION The surgery is ideally performed in a BSL2 BSC to prevent surgery-acquired infection from the operating suite.
-
101
Place the anesthetized mouse on its right side on the sterile field and keep it under anesthesia during the surgery. (Optional) Use surgical drape to cover the mouse. If used, be sure to monitor the mouse for signs of labored breathing, which may be missed if the head is covered.
-
102
Pour thymus fragments into a sterile dish with lid in the BSC.
-
103
Disinfect the exposed skin twice; first using povidone-iodine wipe, then with 70% ethanol wipe (Figure 5C). Allow to dry briefly.
-
104
Use a forceps to lift the skin under the left rib cage, make a 1cm long slit vertically with scissors through the skin layer first (Figure 5D) and then the abdominal wall layer (Figure 5E). ! CAUTION The abdominal wall layer incision should be smaller than the skin incision and, importantly, be smaller than the length of the kidney. The proximal and distal ends of the kidney will rest on the abdominal wall to keep it in place. This step may take some practice. (Optional) Surgical implantation of the thymic tissue under the kidney capsule may be performed with a surgical trocar to minimize tissue damage and potentially ensure better animal welfare post-surgery.
-
105
Gently squeeze the skin on both sides of the incision with a cotton swab and an iris spatula until the left kidney emerges (Figure 5F) and is retained on the surface of the abdominal wall through the incision (Figure 5G).
-
106
Gently make a 2mm long incision/nick in the capsule of the kidney with a sterile 24G needle (Figure 5H and Video 1).
-
107
Lift the capsule near the incision with forceps in the left hand. With the right hand, insert the tip of a 45° angled/bent forceps under the capsule. Gently separate the capsule from the kidney to make a tissue pocket. Moisten the kidney every 3–5 minutes or as needed to prevent it from getting dry (Figure 5I and Video 2).
-
108
Use the 45° angled/bent forceps to take a 1mm × 1mm thymus fragment and put it between the capsule and the kidney. Push the thymus fragment further under the capsule away from the incision so that it does not slide out (Figure 5J and Video 3). Repeat with second thymus fragment. A surgical trocar could also be used instead of a forceps and potentially less-invasive instrument to promote animal welfare.
-
109
Check if the kidney is bleeding. Gently dab the kidney with a sterile cotton swab until bleeding has subsided. If not bleeding, gently push kidney back into the abdominal cavity of the mouse with a cotton swab (Figure 5K).
-
110
Stitch the incision in the muscle layer with 5–0 coated Vicryl suture (Figure 5L). Repeat with the skin layer (Figure 5M).
-
111
Use Vetbond surgical adhesive (1–3 drops) to cover the incision site, which will help to keep the wound closed in the coming days (Figure 5N).
-
112
Use a 29G syringe to intravenously inject the mixture of 0.5–1.5 × 105 HSPCs and 100ug anti-CD2 antibody prepared in steps 93 and 94 above through the tail vein (Figure 5O). ! CAUTION Give second anti-CD2 injection (only antibody, no cells) at day 7 post-surgery.
-
113
Put the mouse into a clean cage. Keep it warm with a heating light until it wakes up (Figure 5P).
-
114
Give the mouse drinking water supplemented with Baytril 100 enrofloxacin (0.25mg/ml) for 10 days and monitor the wound closely for signs of opening or infection for one week after surgery. ! CAUTION Consult with vet staff if you would like to avoid post-surgical use of antibiotics.
Procedure: Blood Collection for Chimerism Assessment for Up to 20 Mice Timing ~3 minutes per mouse = 1 hour
! CAUTION Mice will start to show evidence of human chimerism starting around week 4 post-humanization. You can assess the kinetics of engraftment by performing the remaining steps of this protocol (blood collection, processing, flow cytometry, and analysis) every 2 weeks, starting at week 4. Alternatively, if the goal is to use the animals for downstream experimentation (e.g., transplantation studies), you can bleed the mice for the first time around week 12. If the goal is to verify engraftment of human T cells in addition to other immune subsets, the first bleed should be at week 16–18. By this timepoint, T cells will have developed in the vast majority of animals. Depending on the downstream application, you may want to set a minimum threshold for humanization (e.g., >20% human CD45+ cells) and for specific human immune cell subsets (e.g., >5% human CD3+ cells). As reported in our Brown et al., 2018 Stem Cell Reports publication,12 initially B cells are the predominant immune cell type in the NeoThy (in the NSG or NBSGW host strain without transgenic cytokines) but T cells gradually increase in frequency starting around week 16.
-
115
Set up the isoflurane anesthesia induction chamber and place the mice to be anesthetized in the chamber. Once the mice are induced with sufficient anesthesia, place it under a nose cone with anesthesia supply. A toe-pinch test can be performed to evaluate their state of consciousness (see Figure 5A).
-
116
Using your thumb and index finger, push the skin near the eye, gently stretching it and causing the eye to slightly protrude. Introduce a capillary tube near the medial canthus of the eye and insert it into the sinus. Slightly twist the tube, allowing blood to fill the tube via capillary action (Figure 6A). ! CAUTION Take care not to damage the eye. Do not push and/or turn the tube with too much force. Facial vein bleeding could be used as a less-traumatic alterantive, however, the blood volumes are less precise that that method and uncontrolled bleeding can be an issue.
-
117
Collect up to two capillary tubes of blood per mouse (140μl). Once collected, slightly apply pressure and close the eyelid in order to prevent further bleeding.
-
118
Place the capillary tubes filled with blood in 200μl of anticoagulant citrate dextrose (ACD) in Eppendorf tubes (Figure 6B).
-
119
(Optional) If serum is needed (e.g., for ELISA), use one of the capillary tubes for serum and the other for flow cytometry. Blood for serum can be collected into an empty sterile tube (i.e., no ACD solution) and allowed to clot for 20 minutes. The tube should then be centrifuged at 1000 × g for 10 minutes. The clear serum supernatant should be removed from the pelleted blood clot. Serum can then be frozen at −80°C for future use.
-
120
Gently flick or shake the tube to mix the blood and ACD. ! CAUTION Clots can form if the ACD is not mixed with the blood shortly after collection. Clots can be manually removed if present after mixing using a sterile pipette tip.
Figure 6.
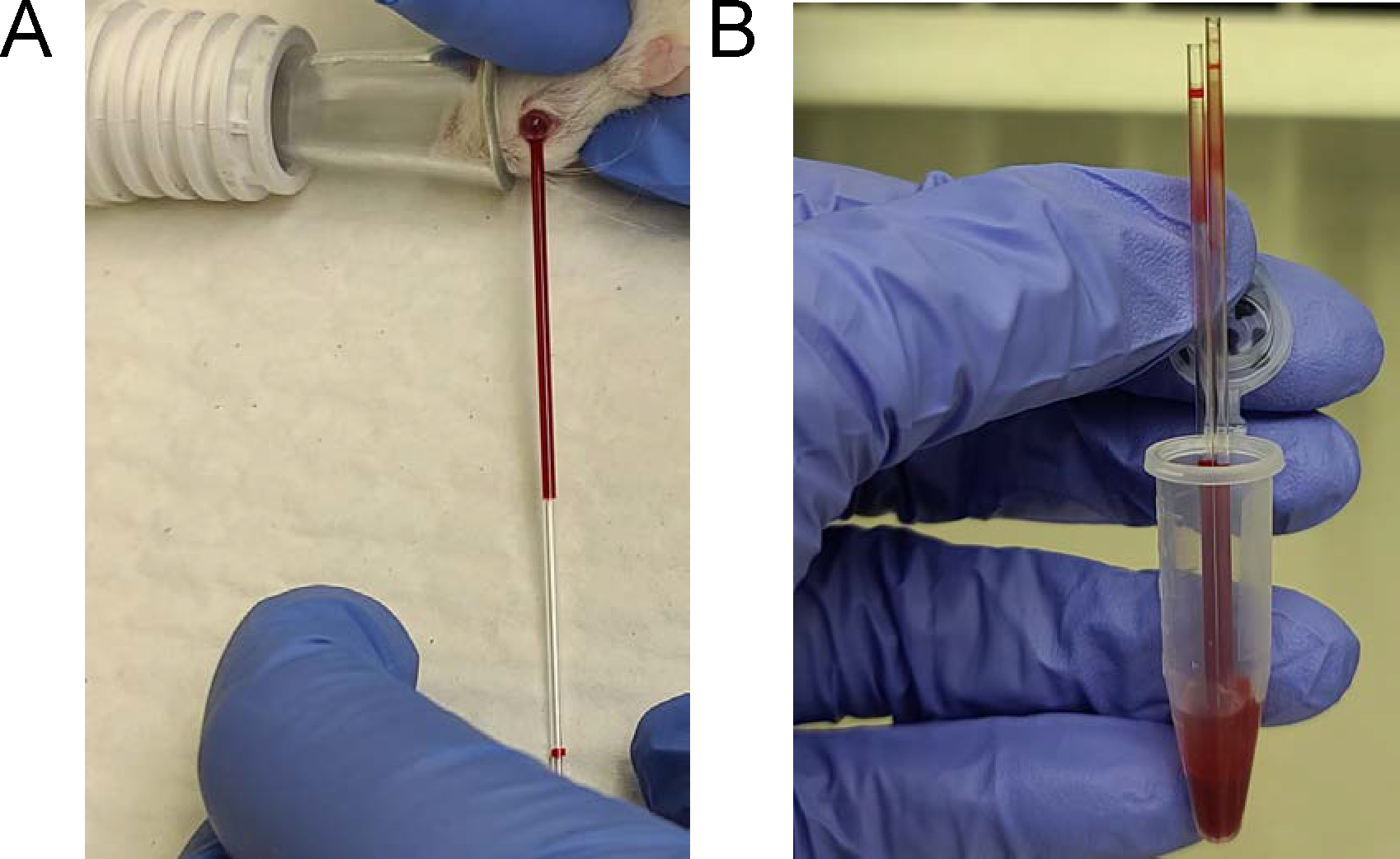
Mouse Retro-orbital Blood Collection. (A) Once mouse has been anesthetized, gently expose the eye. Introduce a glass capillary tube under the eye near the medial canthus of the eye and insert into the sinus. Slightly turn tube to cause slow blood flow. (B) Collect up to two capillary tubes per mouse, flush using bulb, into anticoagulant.
Procedure: Blood Processing for Antibody Staining and Flow Cytometry Acquisition for Up to 20 Mice Timing ~3 minutes per mouse for processing and ~3 minutes for flow cytometry= 2 hours
-
121
After bleeding, add 2ul of FcR blocking reagent per sample, vortex well and incubate for 2 minutes at room temperature in the dark. Then add 100μl antibody master mix directly to each blood sample and then vortex well. Incubate for 15 minutes at room temperature in the dark.
-
122
Add 1ml of BD lysing buffer to each sample. ! CAUTION Add to a maximum of two samples at a time and immediately vortex after adding buffer in order to ensure thorough lysis.
-
123
Incubate samples at 37°C for 3 minutes.
-
124
Centrifuge samples at 300 × g for 5 minutes.
-
125
Pour off the supernatant (leaving ~100μl of sample), vortex to break up the pellet, and then add 1ml of room temperature FACS buffer. Vortex the sample.
-
126
Centrifuge samples at 300 × g for 5 minutes.
-
127
Repeat step 125.
-
128
Repeat steps 122–126 to fully lyse the red blood cells.
-
129
After the second wash, pour off the supernatant (leaving ~100μl of sample) and vortex to break up the pellet. Visually inspect the samples for visible material and, if present, filter with FACS tube with cell strainer snap cap. Proceed to acquire the sample in the flow cytometer. Collect a minimum of 10,000 viable, single cells. Ensure that compensation controls as well as fluorescence minus one (FMO) controls are used to establish gates.
Flow Cytometry Analysis
-
130
First remove doublets via FSC-W vs. FSC-A gating. Singlets are then gated for FSC-A vs. SSC-A, and then viable cells via Live Dead (negative) gating.
-
131
Human CD45+ (hCD45) vs. mouse CD45+ (mCD45/mLy5) cells are plotted to determine human chimerism. To determine human chimerism percentage, we use the equation: %hCD45/(%hCD45+%mCD45). This helps to control for double negative events (e.g., non-leukocyte blood progenitors).
-
132
Immune cell subsets are gated from within the hCD45+ population (Figure 7). Additional immune cell subsets should be determined according to individual experimental needs.
Figure 7.
Flow Cytometry Engraftment Analysis. Gating strategy is shown for determination of common human T cell markers. Viable single human CD45+ cells are subgated into human CD3+ T cell subsets of interest, including CD4+ and CD8+ cells, as well as naïve (CD45RA+CCR7+), effector (CD45RA+CCR7−), central memory (CD45RA−CCR7+), and effector memory (CD45RA−CCR7−) CD8+ T cells.
Timing
Steps 1–12, Neonatal thymus processing: 4 hours
Steps 13–47, UCB processing and HSPC separation: 3 hours
Steps 48–60, Genomic DNA isolation: 1 hour
Steps 61–75, HLA typing: 3 hours (Can be outsourced to a clinical HLA lab)
Steps 76–92, Thawing and plating cells day prior to surgery: 45 minutes
Steps 93–99, Surgical preparation for up to 20 mice: 45 minutes
Steps 100–114, Mouse surgery: 3 hours
Steps 115–118, Blood collection: 1 hour
Steps 119–129, Blood processing for antibody staining and flow cytometry acquisition: 2 hours
Steps 130–132, Flow cytometry analysis: 1+ hours
Anticipated Results
NeoThy mice engraft with human cells with similar kinetics to other humanized mouse models, but may have slightly delayed T cell reconstitution (unpublished data) compared to the BLT.12 This is likely related to the anti-CD2 antibody depleting passenger thymocytes, which can otherwise emigrate from the thymus fragment immediately following the humanization surgery. Traditional BLT models that do not use antibody depletion will have a mixture of passenger thymocyte-derived and de novo T cells, and early emergence of passenger-thymocyte T cells has been documented in a model similar to the NeoThy.42 In the NeoThy produced via the protocol presented here, T cells should be present by 18–20 weeks, if not sooner. Figure 7 shows representative flow cytometry plots illustrating expected values for human vs. mouse CD45+ leukocytes, as well as CD3+ T cell subsets. Engraftment in any humanized mouse is a dynamic process, so it is important to conduct as many blood assessments as needed (typically no more than one per week are advised), according to experimental parameters. ! CRITICAL As the values change with time, it is advisable to assess engraftment as close as possible to downstream experimental procedures. For example, assessing engraftment the day before transplanting tissues into the model for an iPSC immunogenicity study as well as prior to terminal procedures will allow you to verify the degree of humanization at the beginning and end of the experiment, which can then be used in the assessment of the degree and character of immune response to the graft.
Supplementary Material
Supplementary Data 1: HLA Typing Melt Curve Data. This file contains an example of HLA typing data, which can be used as practice for generating a report prior to working on actual samples. It contains an “ambiguous rare” call which can be manually selected by the user, as this may be helpful practice of an issue that can arise with these samples.
Video 1. NeoThy Surgery: Nicking the Kidney Capsule. A shallow nick is made to cut through the kidney capsule.
Video 2. Creating a Tissue Pocket in the Kidney Capsule. A tissue pocket is made by gently grabbing the nicked opening with a fine forceps and using the end of a bent blunt end forceps to create a pocket approximately 1 cm × 1 cm in size. Kidney is moistened with sterile PBS swab to prevent it from drying.
Video 3. Insertion of Neonatal Thymus Fragments Under the Kidney Capsule. By gently grabbing and slightly lifting up the nicked opening with a fine forceps, the bent blunt end forceps is used to grab the edge of the thymus fragment, inserting into the pocket. The rounded back side of the bent blunt forceps is used to push the fragment to the end of the kidney, away from the nick. This procedure is repeated with a second thymus fragment, taking care not to tear the kidney capsule.
Supplementary Data 2: HLA Detailed Report. This file contains an example of the final LinkSeq HLA Typing Report generated from the data in Supplementary Data 1.
Acknowledgments
This study was supported in part by the Wisconsin Alumni Research Foundation, NIH NIAID 75N93021C00004, NIH NHLBI U01HL134764, the UW Carbone Cancer Center Support Grant P30 CA014520 (MEB), CA034196 (LDS), AI132963 (MAB, LDS), OD026440 (MAB, LDS) and NIDDK-supported Human Islet Research Network (HIRN, https://hirnetwork.org) DK104218 (MAB). The authors thank Jennifer Zellner and Dana Maya for manuscript editing assistance, and Kristina Howard for helpful discussion.
Footnotes
Conflict of Interest Disclosure
M.E.B is a consultant for Taconic Biosciences; LDS is a consultant for Dren Bio and Blue Rock Therapeutics; M.A.B is a consultant for The Jackson Laboratory. The other authors declare no conflict of interest.
References
- 1.Shultz LD, Brehm MA, Garcia-Martinez JV & Greiner DL Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol 12, 786–798, doi: 10.1038/nri3311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao LC et al. Creation of PDX-Bearing Humanized Mice to Study Immuno-oncology. Methods Mol Biol 1953, 241–252, doi: 10.1007/978-1-4939-9145-7_15 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Simpson JA & Brown ME Making HIS mice more human-like. J Leukoc Biol 107, 9–10, doi: 10.1002/jlb.5ce1019-262r (2020). [DOI] [PubMed] [Google Scholar]
- 4.Hermsen J & Brown ME Humanized Mouse Models for Evaluation of PSC Immunogenicity. Curr Protoc Stem Cell Biol 54, e113, doi: 10.1002/cpsc.113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan P, Tonomura N, Shimizu A, Wang S & Yang YG Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood 108, 487–492, doi: 10.1182/blood-2005-11-4388 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Melkus MW et al. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med 12, 1316–1322, doi: 10.1038/nm1431 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Honeycutt JB et al. T cells establish and maintain CNS viral infection in HIV-infected humanized mice. The Journal of Clinical Investigation 128, 2862–2876, doi: 10.1172/JCI98968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCune JM & Weissman IL The Ban on US Government Funding Research Using Human Fetal Tissues: How Does This Fit with the NIH Mission to Advance Medical Science for the Benefit of the Citizenry? Stem Cell Reports 13, 777–786, doi: 10.1016/j.stemcr.2019.10.003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deuse T et al. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat Biotechnol 37, 252–258, doi: 10.1038/s41587-019-0016-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao T et al. Humanized Mice Reveal Differential Immunogenicity of Cells Derived from Autologous Induced Pluripotent Stem Cells. Cell Stem Cell 17, 353–359, doi: 10.1016/j.stem.2015.07.021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mold JE et al. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science 330, 1695–1699, doi: 10.1126/science.1196509 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown ME et al. A Humanized Mouse Model Generated Using Surplus Neonatal Tissue. Stem Cell Reports 10, 1175–1183, doi: 10.1016/j.stemcr.2018.02.011 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunis DG et al. Single-Cell Mapping of Progressive Fetal-to-Adult Transition in Human Naive T Cells. Cell Rep 34, 108573, doi: 10.1016/j.celrep.2020.108573 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daharsh L, Zhang J, Ramer-Tait A & Li Q A Double Humanized BLT-mice Model Featuring a Stable Human-Like Gut Microbiome and Human Immune System. J Vis Exp, 10.3791/59773, doi: 10.3791/59773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aryee K-E, Shultz LD & Brehm MA Immunodeficient mouse model for human hematopoietic stem cell engraftment and immune system development. Methods in molecular biology (Clifton, N.J.) 1185, 267–278, doi: 10.1007/978-1-4939-1133-2_18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H et al. A novel humanized mouse model with significant improvement of class-switched, antigen-specific antibody production. Blood 129, 959–969, doi: 10.1182/blood-2016-04-709584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volk V, Schneider A, Spineli LM, Grosshennig A & Stripecke R The gender gap: discrepant human T-cell reconstitution after cord blood stem cell transplantation in humanized female and male mice. Bone Marrow Transplant 51, 596–597, doi: 10.1038/bmt.2015.290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalscheuer H et al. A Model for Personalized in Vivo Analysis of Human Immune Responsiveness. Science Translational Medicine 4, 125ra130, doi: 10.1126/scitranslmed.3003481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCune JM et al. The SCID-hu mouse: murine model for the analysis of human hematolymphoid differentiation and function. Science 241, 1632–1639, doi: 10.1126/science.241.4873.1632 (1988). [DOI] [PubMed] [Google Scholar]
- 20.Varas A et al. Analysis of the Human Neonatal Thymus: Evidence for a Transient Thymic Involution. The Journal of Immunology 164, 6260, doi: 10.4049/jimmunol.164.12.6260 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Sackett SD et al. Genetic Engineering of Immune Evasive Stem Cell-Derived Islets. Transpl Int 35, 10817, doi: 10.3389/ti.2022.10817 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess NJ, Brown ME & Capitini CM GVHD Pathogenesis, Prevention and Treatment: Lessons From Humanized Mouse Transplant Models. Frontiers in Immunology 12, doi: 10.3389/fimmu.2021.723544 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenblatt MB et al. Graft versus host disease in the bone marrow, liver and thymus humanized mouse model. PLoS One 7, e44664, doi: 10.1371/journal.pone.0044664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavender KJ et al. BLT-humanized C57BL/6 Rag2−/−γc−/−CD47−/− mice are resistant to GVHD and develop B- and T-cell immunity to HIV infection. Blood 122, 4013–4020, doi: 10.1182/blood-2013-06-506949 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockridge JL et al. Mice engrafted with human fetal thymic tissue and hematopoietic stem cells develop pathology resembling chronic graft-versus-host disease. Biol Blood Marrow Transplant 19, 1310–1322, doi: 10.1016/j.bbmt.2013.06.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khosravi-Maharlooei M et al. Rapid thymectomy of NSG mice to analyze the role of native and grafted thymi in humanized mice. Eur J Immunol 50, 138–141, doi: 10.1002/eji.201948205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brener J et al. Rapid HIV disease progression following superinfection in an HLA-B*27:05/B*57:01-positive transmission recipient. Retrovirology 15, 7, doi: 10.1186/s12977-018-0390-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stripecke R et al. Innovations, challenges, and minimal information for standardization of humanized mice. EMBO Mol Med 12, e8662, doi: 10.15252/emmm.201708662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jangalwe S, Shultz LD, Mathew A & Brehm MA Improved B cell development in humanized NOD-scid IL2Rγnull mice transgenically expressing human stem cell factor, granulocyte-macrophage colony-stimulating factor and interleukin-3. Immunity, Inflammation and Disease 4, 427–440, doi: 10.1002/iid3.124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryce PJ et al. Humanized mouse model of mast cell-mediated passive cutaneous anaphylaxis and passive systemic anaphylaxis. J Allergy Clin Immunol 138, 769–779, doi: 10.1016/j.jaci.2016.01.049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito R et al. Establishment of a human allergy model using human IL-3/GM-CSF-transgenic NOG mice. J Immunol 191, 2890–2899, doi: 10.4049/jimmunol.1203543 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Rongvaux A et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol 32, 364–372, doi: 10.1038/nbt.2858 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brehm MA et al. Transgenic expression of human IL15 in NOD-<em>scid IL2rg</em><em><sup>null</sup></em> (NSG) mice enhances the development and survival of functional human NK cells. The Journal of Immunology 200, 103.120 (2018). [Google Scholar]
- 34.Vaidya S et al. Enhanced development of functional human innate immune cells in a novel mouse FLT3<sup>null</sup> NSG mouse strain expressing human FLT3L. The Journal of Immunology 204, 223.224 (2020). [Google Scholar]
- 35.Little CJ, Haynes WJ, Huang L, Daffada CM, Wolfe KB, Perrin E, Simpson JA, Kropp Schmidt JA, Hinkle HM, Keding LT, Behrens RT, Evans DT, Kaufman DB, Thomson JA, Golos TG, Brown ME Robust engraftment of fetal non-human primate CD34-positive cells in immune-deficient mice. Journal of Leukocyte Biology In Press (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilkenny C, Browne WJ, Cuthill IC, Emerson M & Altman DG Improving Bioscience Research Reporting: The ARRIVE Guidelines for Reporting Animal Research. PLOS Biology 8, e1000412, doi: 10.1371/journal.pbio.1000412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Notta F, Doulatov S & Dick JE Engraftment of human hematopoietic stem cells is more efficient in female NOD/SCID/IL-2Rgc-null recipients. Blood 115, 3704–3707, doi: 10.1182/blood-2009-10-249326 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Miller JS et al. Expansion and homing of adoptively transferred human natural killer cells in immunodeficient mice varies with product preparation and in vivo cytokine administration: implications for clinical therapy. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation 20, 1252–1257, doi: 10.1016/j.bbmt.2014.05.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McIntosh BE & Brown ME No irradiation required: The future of humanized immune system modeling in murine hosts. Chimerism 6, 40–45, doi: 10.1080/19381956.2016.1162360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayakawa J, Hsieh MM, Uchida N, Phang O & Tisdale JF Busulfan produces efficient human cell engraftment in NOD/LtSz-Scid IL2Rgamma(null) mice. Stem Cells 27, 175–182, doi: 10.1634/stemcells.2008-0583 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montecino-Rodriguez E & Dorshkind K Use of Busulfan to Condition Mice for Bone Marrow Transplantation. STAR Protocols 1, 100159, doi: 10.1016/j.xpro.2020.100159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colas C et al. Generation of functional human T cell development in NOD/SCID/IL2rγ(null) humanized mice without using fetal tissue: Application as a model of HIV infection and persistence. Stem Cell Reports 18, 597–612, doi: 10.1016/j.stemcr.2023.01.003 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1: HLA Typing Melt Curve Data. This file contains an example of HLA typing data, which can be used as practice for generating a report prior to working on actual samples. It contains an “ambiguous rare” call which can be manually selected by the user, as this may be helpful practice of an issue that can arise with these samples.
Video 1. NeoThy Surgery: Nicking the Kidney Capsule. A shallow nick is made to cut through the kidney capsule.
Video 2. Creating a Tissue Pocket in the Kidney Capsule. A tissue pocket is made by gently grabbing the nicked opening with a fine forceps and using the end of a bent blunt end forceps to create a pocket approximately 1 cm × 1 cm in size. Kidney is moistened with sterile PBS swab to prevent it from drying.
Video 3. Insertion of Neonatal Thymus Fragments Under the Kidney Capsule. By gently grabbing and slightly lifting up the nicked opening with a fine forceps, the bent blunt end forceps is used to grab the edge of the thymus fragment, inserting into the pocket. The rounded back side of the bent blunt forceps is used to push the fragment to the end of the kidney, away from the nick. This procedure is repeated with a second thymus fragment, taking care not to tear the kidney capsule.
Supplementary Data 2: HLA Detailed Report. This file contains an example of the final LinkSeq HLA Typing Report generated from the data in Supplementary Data 1.


