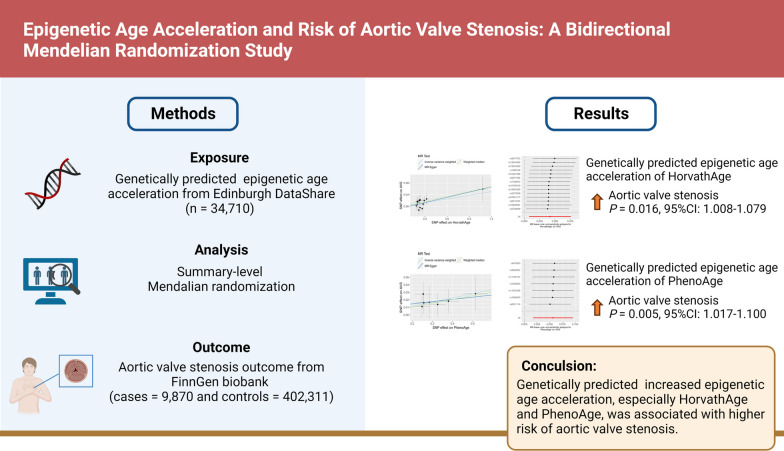
Abstract
Background
Aortic valve stenosis (AVS) is the most prevalent cardiac valve lesion in developed countries, and pathogenesis is closely related to aging. DNA methylation-based epigenetic clock is now recognized as highly accurate predictor of the aging process and associated health outcomes. This study aimed to explore the causal relationship between epigenetic clock and AVS by conducting a bidirectional Mendelian randomization (MR) analysis.
Methods
Summary genome-wide association study statistics of epigenetic clocks (HannumAge, HorvathAge, PhenoAge, and GrimAge) and AVS were obtained and assessed for significant instrumental variables from Edinburgh DataShare (n = 34,710) and FinnGen biobank (cases = 9870 and controls = 402,311). The causal association between epigenetic clock and AVS was evaluated using inverse variance weighted (IVW), weighted median (WM), and MR-Egger methods. Multiple analyses (heterogeneity analysis, pleiotropy analysis, and sensitivity analysis) were performed for quality control assessment.
Results
The MR analysis showed that the epigenetic age acceleration of HorvathAge and PhenoAge was associated with an increased risk of AVS (HorvathAge: OR = 1.043, P = 0.016 by IVW, OR = 1.058, P = 0.018 by WM; PhenoAge: OR = 1.058, P = 0.005 by IVW, OR = 1.053, P = 0.039 by WM). Quality control assessment proved our findings were reliable and robust. However, there was a lack of evidence supporting a causal link from AVS to epigenetic aging.
Conclusion
The present MR analysis unveiled a causal association between epigenetic clocks, especially HorvathAge and PhenoAge, with AVS. Further research is required to elucidate the underlying mechanisms and develop strategies for potential interventions.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-024-01647-5.
Keywords: Aortic valve stenosis, Bidirectional Mendelian randomization, Biological aging, Epigenetic clock, Genome-wide association study
Introduction
Aortic valve stenosis (AVS), the most prevalent valvular heart disease, is characterized by progressive fibro-calcific remodeling and thickening of the aortic valve cusps [1, 2]. It is most often degenerative in pathogenesis, with a prevalence of only 0.02% among subjects aged 18–44 years but 2.8% in patients aged ≥ 75 years [3]. Symptomatic AVS is associated with a dismal prognosis, with a mortality rate of more than 50% at 2 years [4]. However, no pharmacotherapy has been proven to reverse aortic valve calcification effectively [5]. Most patients would eventually require surgical or transcatheter aortic valve repair or replacement [6].
The epigenetic clock is currently the best predictor of biological aging status compared to chronological age and other age-related biomarkers (e.g., telomere length) [7, 8]. Each epigenetic clock reflects biological aging profiles by measuring DNA methylation (DNAm) levels at specific cytosine-phosphate-guanine (CpG) loci. “First generation” epigenetic clocks, like HannumAge and HorvathAge, utilize DNAm levels at CpG loci closely linked to actual age for their calculations [9, 10]. “Second generation” epigenetic clocks, like PhenoAge and GrimAge, exhibit a commendable ability to forecast age-related morbidity and mortality [11, 12]. Epigenetic age acceleration (EAA) is employed to characterize individuals whose estimated physiological age exceeded their actual chronological age, which is strongly related to the development of cardiovascular diseases [13, 14]. Although AVS is associated with senescence [15], no research has been conducted to explore the relationship between the epigenetic aging acceleration and AVS.
Mendelian randomization (MR) leverages genetic variants as instrumental variables (IVs) to support the causal inference without confounding and reverse causation, with random genotype allocation mimicking randomized controlled trials [16]. In this analysis, we performed a bidirectional two-sample MR analysis to investigate the causal association between epigenetic clocks (HannumAge, HorvathAge, PhenoAge, and GrimAge) and the risk of AVS.
Method
Study design
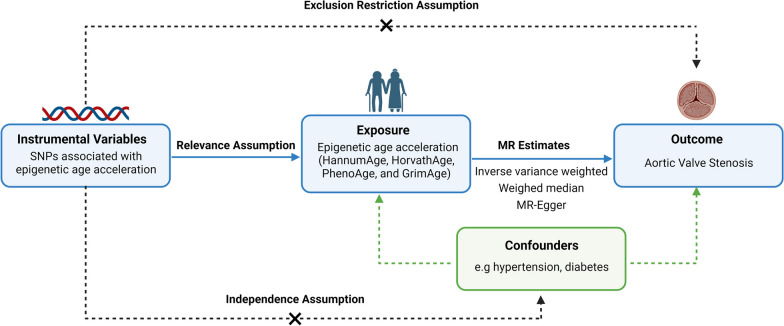
MR analysis should adhere to the following three key assumptions. (1) relevance assumption: IVs must be closely correlated with the exposure phenotype. (2) independence assumption: IVs should be independent of any confounder factors. (3) exclusion-restriction assumption: IVs only influence the outcome through exposure phenotype. An overview of the principle, design, and process of the present MR study is shown in Fig. 1.
Fig. 1.
The flowchart of present study and basic assumptions of MR analysis. The objective of this two-sample bidirectional MR analysis is to investigate the causality between epigenetic age acceleration and AVS. The GWAS meta-analysis utilized in this study is from mixed-sex European cohorts. Abbreviation: AVS: aortic valve stenosis; GWAS: genome-wide association studies; MR: Mendelian randomization; SNPs: single-nucleotide polymorphisms
Data source
The genetic instruments for four epigenetic age measures, namely HannumAge, HorvathAge, PhenoAge, and GrimAge, were obtained from the recent genome-wide association studies (GWAS) meta-analysis based on 28 cohorts of 34,710 European ancestry participants [17]. Summary-level data on AVS were obtained from the GWAS meta-analysis of 412,181 individuals (9870 cases and 402,311 controls) of European descent conducted by FinnGen Project Database. The original data utilized in this study were approved by the ethical committee, and all participants have duly provided their consent forms.
Selection of genetic instrumental variants
Single-nucleotide polymorphisms (SNPs) with genome-wide significant associations with exposure (p < 5 × 10–8) were selected as IVs. Because only four SNPs were selected for GrimAge, the restriction was loosened to a threshold of 5 × 10–6 to identify a suitable number of IVs [18]. Establishing criteria of r2 < 0.001, and kb > 10,000 to eliminate linkage disequilibrium [19]. Then, we deleted IVs surrogating confounders and outcomes to fulfill the second and third MR assumptions by querying the traits proxied of each SNP in the PhenoScannerV2 database. All outlier and palindromic SNPs were removed.
The proportion of phenotypic variations explained by tool variables (R2) and tool strength (F-statistics) were performed to avoid weak shifts in IVs by using the formulas: R2 = [2 × (1 − MAF) × MAF × β2]/(SE2 × N) and F-statistic = [(N − k − 1)/k] × [R2/(1 − R2)], where SE is the standard error, β is the effect size, MAF is the minor allele frequency for each SNP, k presents the number of SNPs, and N presents the sample size. F-statistic ≥ 10 is considered as strong genetic instrument to explain phenotypic variations [20]. Strong genetic instruments were chosen as the IVs of exposure phenotype for MR analysis.
MR analysis
We evaluated the causal associations between epigenetic aging and AVS by using three distinct methods: inverse variance weighted (IVW) with fixed-effects model, MR-Egger, and weighted median (WM) [21]. When statistically significant heterogeneity was present, we used IVW with multiplicative random-effects model for MR analysis [22]. The IVW method combines the Wald estimates of genetically causal associations for each SNP to evaluate the impact of exposure on outcome, which operates under the assumption that all selected SNPs are valid IVs. It can provide the most accurate estimate and is employed as the principal statistical approach to evaluate the causal effect [23]. WM and MR-Egger were applied to complement the MR results. When more than 50% of the selected SNPs are used as IVs, the WM method produces a consistent estimate of the final estimate [24]. The MR-Egger method provides an estimate with adjustment for horizontal pleiotropy if any [25].
Heterogeneity, pleiotropy, and sensitivity assessment
Cochran's Q statistic was performed to assess the heterogeneity among the SNPs for exposure. Q-statistic and I2 (%)-value could quantitatively assess the heterogeneity, which is calculated as I2 = [Q − (K − 1)]/Q (K presents the number of SNPs, Q is Q-statistic) [23]. The horizontal pleiotropy was analyzed by MR-Egger intercept and MR-PRESSO Global test methods. The proximity of the intercept to zero indicates a lower likelihood of horizontal pleiotropy [26]. Employing the leave-one-out sensitivity analysis, we examined whether individual SNPs exerted significant influence on the overall causal estimates by removing each SNP [27].
Statistical analysis
All statistical analyses and result visualizations were implemented by using R software 4.3.1 (R Foundation for Statistical Computing, Vienna, Austria) with the "TwoSampleMR", "LDlinkR", and "forestplot" Packages.
Results
Causal analysis of epigenetic aging on AVS
After calculating F-statistics and querying the proxied traits, we screened significant correlated SNPs as strong IVs of epigenetic aging for MR analyses (HannumAge = 4, R2 = 0.306%, F = 26.587; HorvathAge = 14, R2 = 0.808%, F = 20.189; PhenoAge = 7, R2 = 0.374%, F = 18.632; GrimAge = 3, R2 = 0.102%, F = 11.755) (Additional file 1: Table S1).
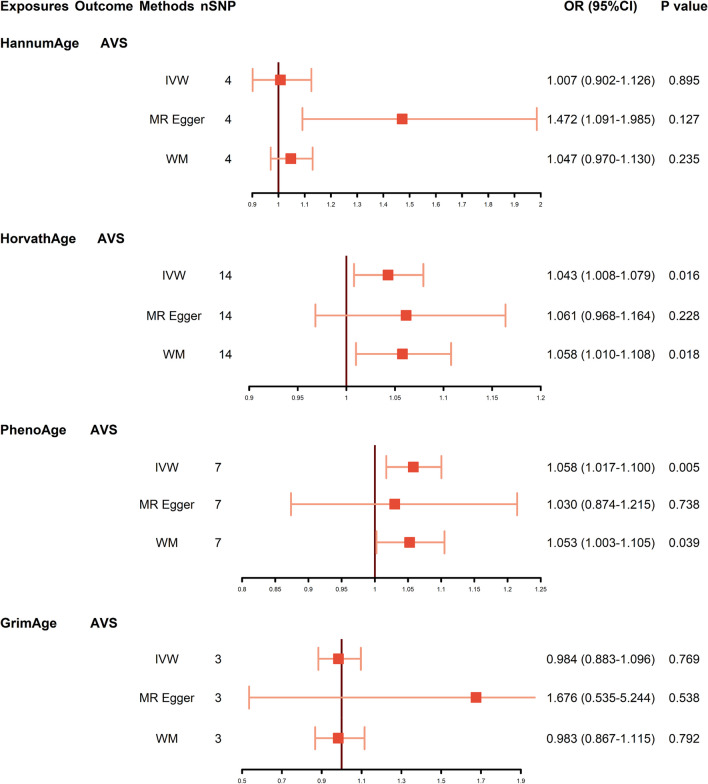
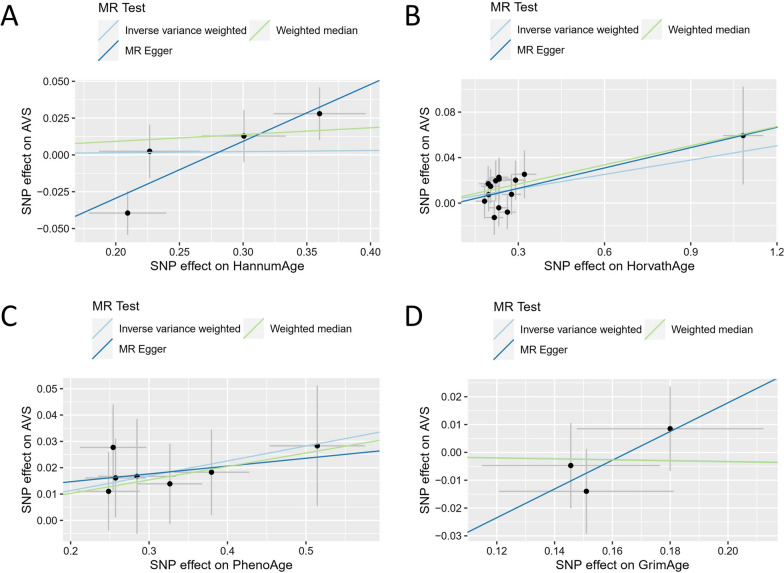
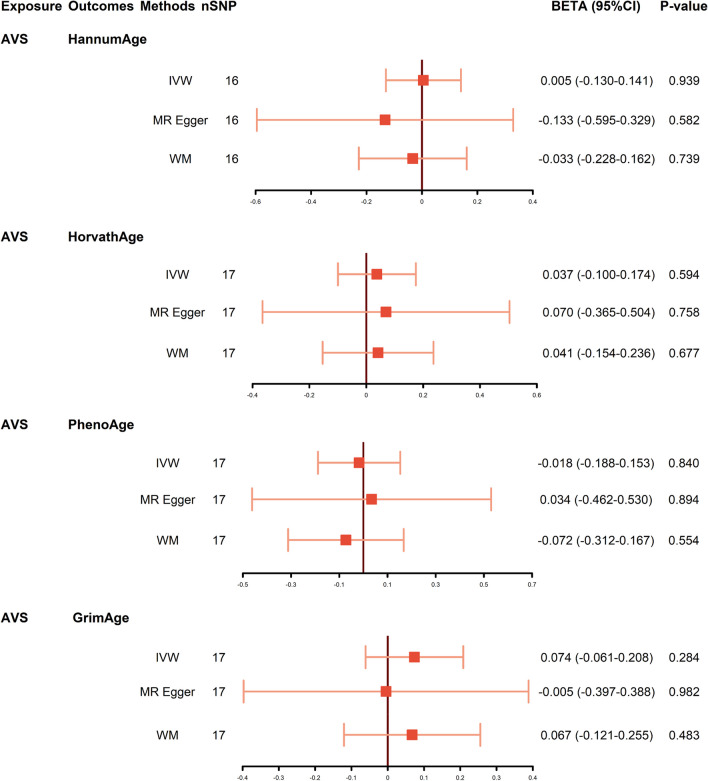
As plotted in Fig. 2, genetically predicted HorvathAge was significantly associated with AVS (95% CI 1.008–1.079, P = 0.016 by IVW; 95% CI 1.010–1.108, P = 0.018 by WM). MR analyses also indicated a causal relationship between PhenoAge and AVS (95% CI 1.017–1.100, P = 0.005 by IVW; 95% CI 1.003–1.105, P = 0.039 by WM). However, we did not observe the causal association between other epigenetic clocks and the odds of AVS (HannumAge: 95% CI 0.902–1.126, P = 0.895; GrimAge: 95% CI 0.883–1.096, P = 0.769). Figure 3 exhibits the scatter plots of the three methods. The trend lines indicated that genetically predicted increased HorvathAge and PhenoAge were related to a higher risk of AVS. The forest plots of individual SNP effect of epigenetic aging on AVS are shown in Additional file 1: Fig. S1.
Fig. 2.
Causal estimates from genetically predicted epigenetic age to AVS. Visualization of the results of three MR analysis methods. Abbreviations: AVS: aortic valve stenosis; CI: confidence interval; IVW: inverse variance weighted; MR: Mendelian randomization; OR: odds ratio; SNP: single-nucleotide polymorphism; WM: weighted median
Fig. 3.
Scatter plots of epigenetic age and AVS. HannumAge (A), HorvathAge (B), PhenoAge (C), and GrimAge (D) as exposure and AVS as outcome. Abbreviations: AVS: aortic valve stenosis; MR: Mendelian randomization; SNP: single-nucleotide polymorphism
Quality control assessment for forward MR analysis
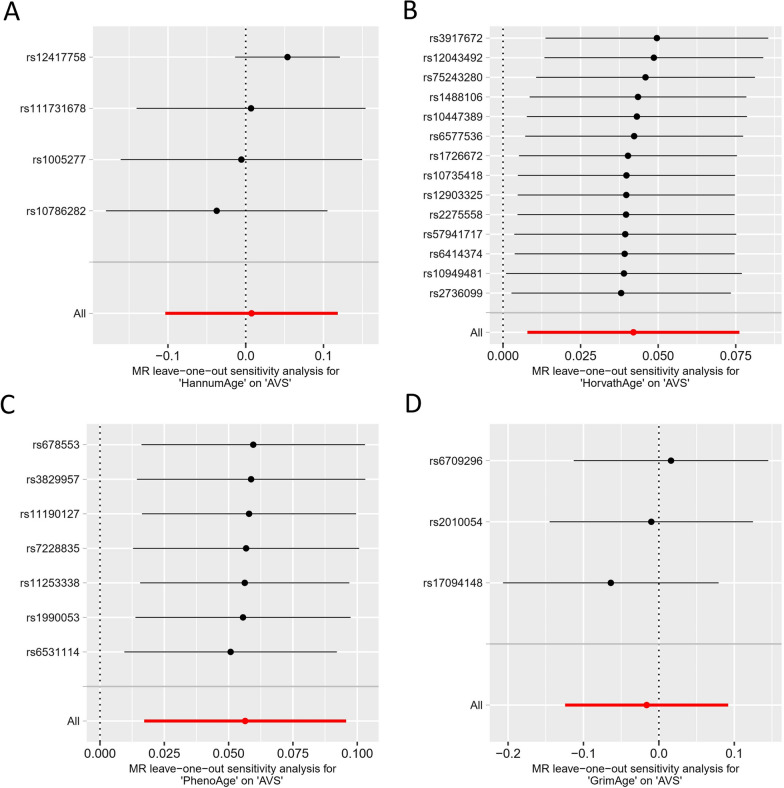
By measuring Cochran’s Q test, heterogeneity was detected in HannumAge (P = 0.018, Q = 10.064, I2 = 70.1%). As a result, we performed IVW with multiplicative random-effects model for MR analysis. No significant heterogeneity between other epigenetic clocks and AVS (HorvathAge: P = 0.897, Q = 7.092, I2 = 0%; PhenoAge: P = 0.991, Q = 0.850, I2 = 0%; GrimAge: P = 0.560, Q = 1.160, I2 = 0%). Table 1 exhibits the results of the MR-Egger intercept and MR-PRESSO Global test, indicating the absence of horizontal pleiotropy among all analyses. After one SNP was removed at a time and the remaining SNPs were analyzed, no significant changes in overall effect estimates were observed (Fig. 4). These findings suggested that our MR results had significant confidence with good robustness and steadiness.
Table 1.
Heterogeneity, pleiotropy, and tool strength of MR analyses
| Exposure | Outcome | Cochran’s Q statistic | Egger intercept | Global test | F-statistic | |||
|---|---|---|---|---|---|---|---|---|
| P value | Q | I2 (%) | P value | Intercept | P value | |||
| GrimAge | AVS | 0.560 | 1.160 | 0.0 | 0.527 | − 0.085 | – | 11.755 |
| HannumAge | AVS | 0.018 | 10.046 | 70.1 | 0.126 | − 0.107 | 0.077 | 26.587 |
| HorvathAge | AVS | 0.897 | 7.092 | 0.0 | 0.693 | − 0.005 | 0.904 | 20.189 |
| PhenoAge | AVS | 0.991 | 0.850 | 0.0 | 0.757 | 0.009 | 0.993 | 18.632 |
| AVS | GrimAge | 0.636 | 13.494 | 0.0 | 0.684 | 0.010 | 0.667 | 20.565 |
| AVS | HannumAge | 0.157 | 20.398 | 26.5 | 0.542 | 0.018 | 0.186 | 20.958 |
| AVS | HorvathAge | 0.339 | 17.739 | 9.8 | 0.879 | − 0.004 | 0.346 | 20.565 |
| AVS | PhenoAge | 0.870 | 9.930 | 0.0 | 0.830 | − 0.007 | 0.887 | 20.565 |
Fig. 4.
Leave-one-out analysis of epigenetic age and AVS. Sensitivity analysis for HannumAge (A), HorvathAge (B), PhenoAge (C), and GrimAge (D) on AVS. Abbreviations: AVS: aortic valve stenosis; MR: Mendelian randomization
Causal analysis of AVS on epigenetic aging
Then, we performed MR analysis with AVS as exposure to explore the possible reverse causality on epigenetic aging. As shown in Fig. 5, genetic predicted AVS was not associated with any epigenetic aging-related traits (HannumAge: P = 0.939, 95% CI − 0.130 to 0.141; HorvathAge: P = 0.594, 95% CI − 0.100 to 0.174; PhenoAge: P = 0.840, 95% CI − 0.188 to 0.153; GrimAge: P = 0.284, 95% CI − 0.061 to 0.208). Neither heterogeneity nor pleiotropy was detected in the reverse directional MR analysis (Table 1). The scatter plots and leave-one-out of the genetic variance are presented in Additional file 1: Figs. S2-3.
Fig. 5.
Causal estimates from genetically predicted AVS to epigenetic age. Visualization of the results of three MR analysis methods. Abbreviations: AVS: aortic valve stenosis; CI: confidence interval; IVW: inverse variance weighted; MR: Mendelian randomization; OR: odds ratio; SNP: single-nucleotide polymorphism; WM: weighted median
Discussion
To the best of our knowledge, this is the first MR analysis to explore the bidirectional causal association between epigenetic clock and AVS. In the present research, we found that increased genetically predicted HorvathAge and PhenoAge were related to a higher risk of AVS. Conversely, the result did not support the causal relationship of AVS on any epigenetic clocks. It is suggested that EAA of HorvathAge and PhenoAge is the risk factor for the prevalence of AVS.
Degenerative lesion is the most common etiology of AVS [28]. Age-related cellular and stress-induced senescence, accompanied by subsequent active processes, constitute crucial elements in the pathomechanism of AVS [29]. Age-related senescent cells release cytokines, chemokines, and matrix metalloproteinases, known as senescence-associated secretory phenotypes. It brings about increased collagen content and leaflet stiffness with extracellular matrix remodeling and structural changes in the valvular tissue [30, 31]. In addition, DNA damage can be triggered by cellular stressors, like excessive mechanical stress, metabolic stress, and oxidative stress, referred to as stress-induced premature cellular senescence [32, 33]. Constant hemodynamic stress-induced endothelial denudation is repaired by circulating endothelially progenitor cells. However, aging-induced reduction in the number of circulating endothelial progenitor cells impedes the clearance of senescent endothelial cells, leading to activate reactive oxygen species, inflammatory responses, and activating the lipid infiltration pathway [34]. Sirtunin1, an NAD (+)-dependent deacetylase, exerts anti-aging effects by controlling mitochondrial biogenesis and oxidative stress [35]. Sophie Carter et.al found that the expression of Sirtunin1 was reduced in explanted valves from AVS patients [36]. Targeted modulation of Sirtunin1 is a potential therapy for AVS [37].
Previous studies have revealed a correlation between DNA methylation and AVS. Based on the Illumina 450 k Beadchip and enzyme-linked immunosorbent assay methods, Nwachukwu et al. identified more than 6,000 differently methylated sites between normal and aortic stenotic tissue. The increased DNA methylation of DNA methyltransferase 3 beta activated the osteogenic pathways in valves [38]. Fayez Hadji et.al reported the dysregulation of DNA methylation in the promoter of H19 by performing multidimensional genomic profiling in human calcific aortic valves. The overexpression of H19 promoted the osteogenic program through impeding NOTCH1 transcription [39]. Takahito Nasu et.al found that the DNA methylation in the region encoding tribbles homolog 1 was lower in the AVS group than in the controls by analyzing epigenome-wide association study of peripheral blood mononuclear cells, which may be the result of hemodynamic overload [40]. In addition, the osteogenic transition of valve interstitial cells was promoted by DNA methylation-mediated downregulation of phospholipid phosphatase 3 [41].
Because there is significant heterogeneity in health aging [42], we need better predictors to understand and measure senescence than chronological age. The epigenetic clock and telomere length are regarded as the most compelling predictors of biological age [8]. Based on the Southern blot hybridization and quantitative polymerase chain reaction, David J Kurz et.al revealed that AVS was associated with shorter telomere length in the elderly [43]. Experiments carried out by Ilona Saraieva also confirmed the result [44]. However, the predictive ability of telomere length is low [8]. Although studies have reported the relationship between the epigenetic clock and cardiovascular diseases, there is no research analyzing the influence of epigenetic aging on the pathogenesis of AVS. A German case-cohort study reported an increased risk of cardiovascular death associated with EAA of HorvathAge [45], contrary to the findings of a study in the Melbourne Collaborative Cohort [46]. What’s more, EAA of PhenoAA, rather than HorvathAA, was related to an increase in the hazard of cardiovascular death in the US Normative Aging Study [47]. These contradictory results may result from the small number of cases and short follow-up time.
In the present research, we explored the causal association between epigenetic clock and AVS by MR analyses. Using the fundamental concept of the random allocation of alleles during zygote formation, MR analysis can establish dependable causal inferences overcoming confounding and reverse causality biases [48]. To mitigate potential bias arising from group stratification, only GWAS data derived from individuals of European descent were utilized in this study. Moreover, our dataset was obtained from the Edinburgh DataShare and the FinnGen Project Database, ensuring no overlap in samples. Quality control assessment demonstrated that our results were reliable and robust. This study expanded the knowledge of risk factors for AVS that EAA of HorvathAge and PhenoAge was associated with higher odds of AVS. Epigenetic clocks may become a surveillance indicator for clinicians and preventive medicine practitioners to assess the risk of developing AVS. Decelerating biological aging has emerged as a novel research focus in the prevention of AVS.
Our study also has inevitable limitations. First, the pooled GWAS data utilized in our study originated from populations of European ancestry, raising concerns about the generalizability of our findings to other ethnic groups. Second, our study could not perform a stratified analysis of the progression and severity of AVS due to the lack of publicly available dataset. Third, the ORs in our findings are low, necessitating cautious interpretation of the results. Finally, since epigenetic aging is essentially related to environmental exposures rather than genetic factors, this highlights the limitations of applying MR in this context.
Conclusion
Our findings suggested the potential causal relationship of accelerated epigenetic clocks, especially HorvathAge and PhenoAge, to the risk of AVS. Slowing down biological aging has emerged as a new research direction in curbing AVS.
Supplementary Information
Additional file 1. Table S1. Summary statistics of the EAA genetic instrumental variables. Table S2. Summary statistics of the AVS genetic instrumental variables in reverse MR analysis. Fig S1. The forest plots of EAA on AVS. Fig S2. The scatter plots of AVS on EAA. Fig S3. The leave-one-out plots of AVS on EAA. Fig S4. The forest plots of AVS on EAA.
Acknowledgements
This investigation utilized data from the Edinburgh DataShare and the FinnGen Project Database. We sincerely appreciate all participants and investigators for their valuable contribution in making statistics accessible and publicly available.
Abbreviations
- AVS
Aortic valve stenosis
- EAA
Epigenetic age acceleration
- GWAS
Genome-wide association studies
- IVW
Inverse variance weighted
- MR
Mendelian randomization
- OR
Odds ratio
- SNPs
Single-nucleotide polymorphisms
- WM
Weighted median
- 95% CI
95% Confidence interval
Author contributions
All authors have read and approved the submission of the manuscript. WC presented research questions. WP and QH participated in the original draft of the manuscript and the literature search. WP, QH, and LZ contributed to the data retrieval, statistical analysis, and visualization of results. JL and XD were involved in the interpretation of the results. WC, TJ, and XQ conducted a thorough critical review and revised the manuscript before its submission.
Funding
This work was supported by the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No.23KJB20019), the Bo-xi Training Program of Natural Science Foundation of China (No. BXQN202211), “Invigorating Health Through Science and Education” youth science and technology project (No. KJXW2022007), and Jiangsu Provincial Health International (Regional) Exchange Support Program.
Availability of data and materials
No original data were generated in the present study. The datasets mentioned in this article are publicly available in the Edinburg DataShare (https://datashare.ed.ac.uk/handle/10283/3645) for GWAS of epigenetic clocks and FinnGen biobank (phenocode: I9_CAVS_OPERATED, https://www.finngen.fi/en/access_results) for GWAS of AVS.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study used the large publicly available GWAS database, which has received approval from their relevant ethical review board and participants.
Competing interest
The authors have declared that there are no commercial or financial conflicts of interest in this work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wanqian Pan, Qi Huang and Le Zhou contributed equally to this work.
Contributor Information
Xiaodong Qian, Email: qianxiaodong@suda.edu.cn.
Tingbo Jiang, Email: jtbsdfyy@163.com.
Weixiang Chen, Email: chenweixiang@suda.edu.cn.
References
- 1.Iung B, et al. A prospective survey of patients with valvular heart disease in Europe: the Euro Heart Survey on Valvular Heart Disease. Eur Heart J. 2003;24(13):1231–1243. doi: 10.1016/S0195-668X(03)00201-X. [DOI] [PubMed] [Google Scholar]
- 2.Goody PR, et al. Aortic valve stenosis: from basic mechanisms to novel therapeutic targets. Arterioscler Thromb Vasc Biol. 2020;40(4):885–900. doi: 10.1161/ATVBAHA.119.313067. [DOI] [PubMed] [Google Scholar]
- 3.Nkomo VT, et al. Burden of valvular heart diseases: a population-based study. Lancet. 2006;368(9540):1005–1011. doi: 10.1016/S0140-6736(06)69208-8. [DOI] [PubMed] [Google Scholar]
- 4.Otto CM, Prendergast B. Aortic-valve stenosis—from patients at risk to severe valve obstruction. N Engl J Med. 2014;371(8):744–756. doi: 10.1056/NEJMra1313875. [DOI] [PubMed] [Google Scholar]
- 5.Bakaeen FG, Rosengart TK, Carabello BA. Aortic stenosis. Ann Intern Med. 2017;166(1):ITC1–ITC16. doi: 10.7326/AITC201701030. [DOI] [PubMed] [Google Scholar]
- 6.Zakkar M, Bryan AJ, Angelini GD. Aortic stenosis: diagnosis and management. BMJ. 2016;355:i5425. doi: 10.1136/bmj.i5425. [DOI] [PubMed] [Google Scholar]
- 7.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 8.Jylhava J, Pedersen NL, Hagg S. Biological age predictors. EBioMedicine. 2017;21:29–36. doi: 10.1016/j.ebiom.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine ME, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10(4):573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu AT, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11(2):303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang R, et al. The association of accelerated epigenetic age with all-cause mortality in cardiac catheterization patients as mediated by vascular and cardiometabolic outcomes. Clin Epigenet. 2022;14(1):165. doi: 10.1186/s13148-022-01380-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ammous F, et al. Epigenetic age acceleration is associated with cardiometabolic risk factors and clinical cardiovascular disease risk scores in African Americans. Clin Epigenet. 2021;13(1):55. doi: 10.1186/s13148-021-01035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cote N, Pibarot P, Clavel MA. Aortic stenosis: what is the role of aging processes? Aging (Albany NY) 2019;11(4):1085–1086. doi: 10.18632/aging.101826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 17.McCartney DL, et al. Genome-wide association studies identify 137 genetic loci for DNA methylation biomarkers of aging. Genome Biol. 2021;22(1):194. doi: 10.1186/s13059-021-02398-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, et al. Effects of iron homeostasis on epigenetic age acceleration: a two-sample Mendelian randomization study. Clin Epigenet. 2023;15(1):159. doi: 10.1186/s13148-023-01575-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, et al. Kidney damage causally affects the brain cortical structure: a Mendelian randomization study. EBioMedicine. 2021;72:103592. doi: 10.1016/j.ebiom.2021.103592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess S, Thompson SG, Collaboration CCG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 21.Burgess S, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. 2019;4:186. doi: 10.12688/wellcomeopenres.15555.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang F, et al. Causality between heart failure and epigenetic age: a bidirectional Mendelian randomization study. ESC Heart Fail. 2023;10(5):2903–2913. doi: 10.1002/ehf2.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35(11):1880–1906. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden J, et al. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greco MF, et al. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34(21):2926–2940. doi: 10.1002/sim.6522. [DOI] [PubMed] [Google Scholar]
- 27.Verbanck M, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ginghina C, et al. Calcific aortic valve disease and aortic atherosclerosis–two faces of the same disease? Rom J Intern Med. 2009;47(4):319–329. [PubMed] [Google Scholar]
- 29.Molnar AA, Pasztor D, Merkely B. Cellular senescence, aging and non-aging processes in calcified aortic valve stenosis: from bench-side to bedside. Cells. 2022;11(21):3389. doi: 10.3390/cells11213389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens EH, et al. Age-related changes in material behavior of porcine mitral and aortic valves and correlation to matrix composition. Tissue Eng Part A. 2010;16(3):867–878. doi: 10.1089/ten.tea.2009.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.VanAuker MD. Age-related changes in hemodynamics affecting valve performance. Am J Geriatr Cardiol. 2006;15(5):277–83; quiz 284-5. [DOI] [PubMed]
- 32.Chen MS, Lee RT, Garbern JC. Senescence mechanisms and targets in the heart. Cardiovasc Res. 2022;118(5):1173–1187. doi: 10.1093/cvr/cvab161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song P, An J, Zou MH. Immune clearance of senescent cells to combat ageing and chronic diseases. Cells. 2020;9(3):671. doi: 10.3390/cells9030671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto Y, et al. Reduced number and function of endothelial progenitor cells in patients with aortic valve stenosis: a novel concept for valvular endothelial cell repair. Eur Heart J. 2009;30(3):346–355. doi: 10.1093/eurheartj/ehn501. [DOI] [PubMed] [Google Scholar]
- 35.Mouchiroud L, et al. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154(2):430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter S, et al. Sirt1 inhibits resistin expression in aortic stenosis. PLoS ONE. 2012;7(4):e35110. doi: 10.1371/journal.pone.0035110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samiei N, et al. Modulatory role of SIRT1 and resistin as therapeutic targets in patients with aortic valve stenosis. Arch Med Res. 2019;50(6):333–341. doi: 10.1016/j.arcmed.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Nwachukwu N, et al. Evidence for altered DNA methylation as a major regulator of gene expression in calcific aortic valve disease (671.15) FASEB J. 2014;28(S1):15. doi: 10.1096/fasebj.28.1_supplement.671.15. [DOI] [Google Scholar]
- 39.Hadji F, et al. Altered DNA methylation of long noncoding RNA H19 in calcific aortic valve disease promotes mineralization by silencing NOTCH1. Circulation. 2016;134(23):1848–1862. doi: 10.1161/CIRCULATIONAHA.116.023116. [DOI] [PubMed] [Google Scholar]
- 40.Nasu T, et al. Epigenome-wide association study identifies a novel DNA methylation in patients with severe aortic valve stenosis. Circ Genom Precis Med. 2020;13(1):e002649. doi: 10.1161/CIRCGEN.119.002649. [DOI] [PubMed] [Google Scholar]
- 41.Mkannez G, et al. DNA methylation of a PLPP3 MIR transposon-based enhancer promotes an osteogenic programme in calcific aortic valve disease. Cardiovasc Res. 2018;114(11):1525–1535. doi: 10.1093/cvr/cvy111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowsky DJ, et al. Heterogeneity in healthy aging. J Gerontol A Biol Sci Med Sci. 2014;69(6):640–649. doi: 10.1093/gerona/glt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurz DJ, et al. Degenerative aortic valve stenosis, but not coronary disease, is associated with shorter telomere length in the elderly. Arterioscler Thromb Vasc Biol. 2006;26(6):e114–e117. doi: 10.1161/atvb.26.6.1303. [DOI] [PubMed] [Google Scholar]
- 44.Saraieva I, et al. Telomere length in valve tissue is shorter in individuals with aortic stenosis and in calcified valve areas. Front Cell Dev Biol. 2021;9:618335. doi: 10.3389/fcell.2021.618335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perna L, et al. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenet. 2016;8:64. doi: 10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dugue PA, et al. Association of DNA methylation-based biological age with health risk factors and overall and cause-specific mortality. Am J Epidemiol. 2018;187(3):529–538. doi: 10.1093/aje/kwx291. [DOI] [PubMed] [Google Scholar]
- 47.Gao X, et al. Comparative validation of an epigenetic mortality risk score with three aging biomarkers for predicting mortality risks among older adult males. Int J Epidemiol. 2019;48(6):1958–1971. doi: 10.1093/ije/dyz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nazarzadeh M, et al. Plasma lipids and risk of aortic valve stenosis: a Mendelian randomization study. Eur Heart J. 2020;41(40):3913–3920. doi: 10.1093/eurheartj/ehaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. Summary statistics of the EAA genetic instrumental variables. Table S2. Summary statistics of the AVS genetic instrumental variables in reverse MR analysis. Fig S1. The forest plots of EAA on AVS. Fig S2. The scatter plots of AVS on EAA. Fig S3. The leave-one-out plots of AVS on EAA. Fig S4. The forest plots of AVS on EAA.
Data Availability Statement
No original data were generated in the present study. The datasets mentioned in this article are publicly available in the Edinburg DataShare (https://datashare.ed.ac.uk/handle/10283/3645) for GWAS of epigenetic clocks and FinnGen biobank (phenocode: I9_CAVS_OPERATED, https://www.finngen.fi/en/access_results) for GWAS of AVS.