ABSTRACT
Although genetic manipulation is one of the hallmarks of model organisms, its applicability to non-model species has remained difficult due to our limited understanding of their fundamental biology. For instance, manipulation of a cell line originated from the black-legged tick Ixodes scapularis, an arthropod that serves as a vector for several human pathogens, has yet to be established. Here, we demonstrate the successful genetic modification of the commonly used tick ISE6 line through ectopic expression and clustered regularly interspaced palindromic repeats [(CRISPR)/CRISPR-associated protein 9 (Cas9)] genome editing. We performed ectopic expression using nucleofection and attained CRISPR-Cas9 editing via homology-dependent recombination. Targeting the E3 ubiquitin ligase x-linked inhibitor of apoptosis (xiap) and its substrate p47 led to an alteration in molecular signaling within the immune deficiency network and increased infection of the rickettsial agent Anaplasma phagocytophilum in I. scapularis ISE6 cells. Collectively, our findings complement techniques for the genetic engineering of I. scapularis ticks, which currently limit efficient and scalable molecular genetic screens in vivo.
IMPORTANCE
Genetic engineering in arachnids has lagged compared to insects, largely because of substantial differences in their biology. This study unveils the implementation of ectopic expression and CRISPR-Cas9 gene editing in a tick cell line. We introduced fluorescently tagged proteins in ISE6 cells and edited its genome via homology-dependent recombination. We ablated the expression of xiap and p47, two signaling molecules present in the immune deficiency (IMD) pathway of Ixodes scapularis. Impairment of the tick IMD pathway, an analogous network of the tumor necrosis factor receptor in mammals, led to enhanced infection of the rickettsial agent Anaplasma phagocytophilum. Altogether, our findings provide a critical technical resource to the scientific community to enable a deeper understanding of biological circuits in the black-legged tick I. scapularis.
KEYWORDS: tick-borne diseases, ticks, rickettsia, Lyme disease, Borrelia burgdorferi
OBSERVATION
The black-legged tick Ixodes scapularis is a medically relevant chelicerate that transmits several bacteria, viruses, and protozoa to humans and other animals (1, 2). To date, inefficient methods for genetic manipulation in I. scapularis make this organism mostly intractable, which leaves significant fundamental gaps in the biology of this ectoparasite. As an example, ectopic expression is a robust tool for elucidating gene function and discovering new phenotypes. However, tick cell lines are reportedly refractory to established transfection methods (3). Additionally, the use of clustered regularly interspaced short palindromic repeats [(CRISPR)/CRISPR-associated protein 9 (Cas9)] (4) remains challenging despite the recent strides made in genome sequencing (5, 6) and the documented in vivo application of CRISPR-Cas9 editing to score morphological phenotypes in I. scapularis (7). Currently, RNA interference (RNAi) is a widely accepted technique to study functional genomics in ticks (8), but this approach presents limitations, such as off-target effects and transient or low knockdown efficiency (9). Thus, there is a pressing need for the development of genetic tools to manipulate the biology of I. scapularis and understand interactions between this arthropod and the microbes it encounters. Here, we report the ectopic expression and CRISPR-Cas9 gene editing of the commonly used ISE6 cell line originated from I. scapularis. We indicate the role of the E3 ubiquitin ligase x-linked inhibitor of apoptosis (xiap) and p47 in activating the immune deficiency pathway (10–12). The IMD network is analogous to the tumor necrosis factor receptor pathway in mammals (13, 14) and acts as a primary defense against infection of Gram-negative bacteria in ticks (10–12).
RESULTS
Ectopic expression and XIAP-p47 interactions in a tick cell line
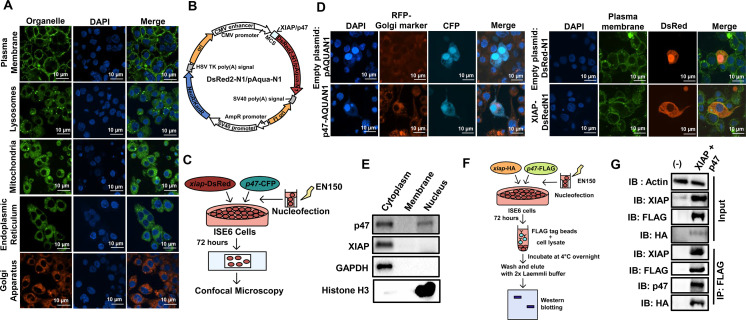
To determine whether fluorescent probes might be used to visualize subcellular structures in I. scapularis, we stained organelles within ISE6 cells through commonly used molecular dyes for the plasma membrane, lysosome, mitochondria, endoplasmic reticulum, and Golgi apparatus (Fig. 1A). We then characterized two previously identified proteins from I. scapularis: XIAP and p47 (10–12). p47 is an enzymatic substrate of the E3 ubiquitin ligase XIAP and activates the tick IMD pathway through Kenny (also known as IKKγ/NEMO) in response to infection (10). The impairment of p47 expression through RNAi in I. scapularis reduces Kenny accumulation, lessens phosphorylation of IKKβ (IRD5), and diminishes cleavage of the nuclear factor-κB molecule Relish in I. scapularis (10).
Fig 1.
Ectopic expression in the ISE6 cell line. (A) Confocal images of ISE6 cells stained with different molecular dyes. (B) Cartoon depicting plasmids used for confocal microscopy. Discosoma red-N1 (DsRed2-N1) contains the fluorescent gene DsRed, whereas p-Aquamarine-N1 (pAqua-N1) carries the fluorescent gene Aqua. (C) Schematic representation of ectopic expression in tick cells. (D) Ectopic expression of Aqua-tagged p47 and DsRed-tagged XIAP in ISE6 cells. ISE6 cells were nucleofected with the plasmid containing p47-Aqua, xiap-DsRed, or the empty vector (DsRed-N1 or pAqua-N1); Golgi (red), plasma membrane (green), and DAPI (blue). (E) Sub-cellular fractionation of ISE6 cells. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and histone H3 were used as cytosolic and nuclear markers, respectively. (F) Schematic representation of pull-down in ISE6 cells. xiap-HA and p47-FLAG constructs were nucleofected in ISE6 cells (3 × 107). Co-transfected cells were harvested after 72 hours, and 10 mg of the lysate was incubated with 50 µL of FLAG beads for 18 hours at 4°C. (G) The complex was immunoprecipitated (IP) using the 3X-FLAG peptide and subjected to immunoblotting (IB). Data represent one of two independent experiments.
We cloned p47 into the pAquamarine-N1 (p47-Aqua) plasmid and xiap in the Discosoma red-N1 (XIAP-DsRed) plasmid (Fig. 1B). We successfully developed a protocol to nucleofect these plasmids into tick cells (Fig. 1C). Confocal microscopy revealed that the recombinant protein p47-Aqua localized in the nucleus and the cytosol, whereas XIAP-DsRed was predominantly detected in the cytosol of ISE6 cells (Fig. 1D). Sub-cellular fractionation of nucleofected cells independently confirmed the location of p47 and XIAP in ISE6 cells (Fig. 1E). McClure et al. (10) demonstrated that XIAP binds and ubiquitinylates p47 in a lysine (K)-63 dependent manner in human embryonic kidney 293T cells. To take advantage of the developed protocol for ectopic expression in tick cells, we next performed co-immunoprecipitation using the XIAP-HA and p47-FLAG vectors (Fig. 1F). We detected molecular interactions between XIAP and p47 via affinity purification in ISE6 cells (Fig. 1G). These data demonstrate the ectopic expression of immune molecules in ISE6 cells and confirm protein-protein interactions in I. scapularis.
CRISPR-Cas9 genome editing in the ISE6 cell line
We performed xiap and p47 genome editing via CRISPR-Cas9 in the ISE6 cell line. We isolated genomic DNA from ISE6 cells and designed PCR primers to amplify 1,000 base pairs (bp) of the target regions. Specifically, two single guide RNAs (sgRNAs) per gene were made for xiap and p47 (Tables S1 and S2). We developed a methodology for CRISPR-Cas9 through homology-directed repair (HDR) (Fig. 2; Fig. S1–S4). Genome manipulation via HDR displays a precise editing mechanism when a template is introduced into cells as a donor for homologous recombination (15). We used a cassette with an antibiotic marker and a reporter gene, along with DNA fragments homologous to the target gene. These homologous regions were positioned on either side of the donor cassette to facilitate recombination (Fig. 2A; Fig. S3A). Due to the low efficiency associated with transfecting tick lines, we employed a ribonucleoprotein (RNP) delivery method to introduce the Cas9 endonuclease protein and sgRNAs in ISE6 cells (Fig. S1). Following nucleofection, cells were split and selected with puromycin (Fig. S2). We chose antibiotic selection over cell sorting because it remains technically unfeasible to culture tick cells from a single clone.
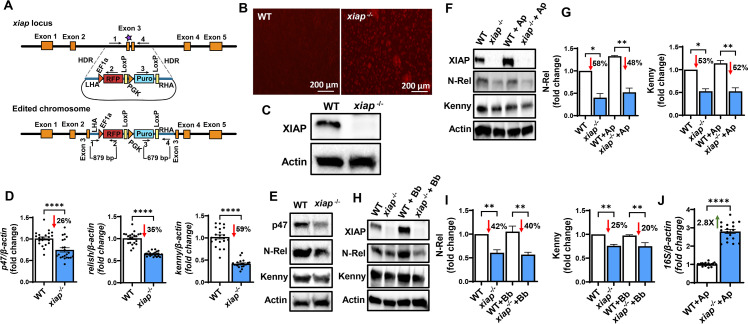
Fig 2.
CRISPR-Cas9 xiap editing in ISE6 cells. (A) Schematic representation of the xiap locus and the donor construct. The orange boxes represent the five exons of xiap with sgRNA binding and the Cas9 cleavage site on exon 3 (purple star). The donor construct contains the red fluorescent protein (RFP) and puromycin cassette (RFP-Puro) along with the DNA fragments of ~600 bp in length, homologous to the xiap locus, flanking the Cas9 cleavage site on the 5′ and 3′ ends for HDR. The arrows with numbers 1–4 represent the primers for the amplification analysis. (B and C) Editing was confirmed via fluorescence microscopy (B) and western blotting (C). (D and E) Functional disruption of xiap impaired the expression of molecules associated with the IMD pathway at both transcriptional (D) and translational levels (E). (F–I) 3 × 106 wild-type (WT) and xiap-/- cells were plated in a 6-well plate and stimulated with Anaplasma phagocytophilum [multiplicity of infection (MOI) 50] or Borrelia burgdorferi (MOI 50) for 15 minutes. Disruption of XIAP signaling impairs Kenny accumulation and Relish cleavage in response to (F and G) A. phagocytophilum infection or (H and I) B. burgdorferi stimulation. For data normalization, Kenny and N-Rel band densities were normalized to actin, and values were divided by the uninfected WT control densitometry. Western blot images are a representative image of at least two independent experiments. (J) WT and xiap-/- cells were infected with A. phagocytophilum (MOI 50). ISE6 cells were harvested after 72 hours of infection. The A. phagocytophilum 16S rRNA transcript was quantified by qRT-PCR, and the expression data were normalized to I. scapularis β-actin. The qRT-PCR data show the combination of three independent experiments. Results are represented as a mean ± SEM. Statistical significance was evaluated by unpaired t test with Welch’s correction (D and J) and one-way ANOVA with post hoc Tukey test (G and I). *P < 0.05; **P < 0.01; and ****P < 0.0001.
For validation of the editing event, a primer set was designed to target both p47- and xiap-edited cells amplifying the left homology arm and the red fluorescent protein gene. A second primer pair was engineered to cover the puromycin cassette and the right homology arm (Fig. 2A; Fig. S3A). PCR amplification detected the donor cassette insertion in the edited p47-/- and xiap-/- ISE6 cells (Fig. S3B and S4A). Knock-in events were orthogonally confirmed through Sanger sequencing (Fig. S3C and S4B) and fluorescence microscopy (Fig. 2B and S3D). Western blot revealed the absence of the wild-type (WT) protein bands for p47 and XIAP in edited ISE6 cells (Fig. S3E and 2C). Disruption of a p47 homolog in the budding yeast Saccharomyces cerevisiae, named Shp1, leads to lethality (16). Similarly, edited p47-/- cells did not survive the puromycin selection procedure, likely due to the involvement of p47 in growth. These findings confirmed the delivery of Cas9 RNPs and independent ablation of both p47 and xiap in ISE6 cells.
Functional disruption of xiap impairs IMD signaling pathway in I. scapularis
We then asked whether cells deficient in xiap (xiap-/-) were impaired for the IMD signaling pathway (Fig. S5) (10–12). Editing of xiap affected the transcription and translation of p47, relish, and kenny (IKKγ/NEMO) and the cleavage of the NF-κB molecule Relish in ISE6 cells (Fig. 2D and E). We determined the effect of xiap editing in ISE6 cells during microbial stimulation with the Lyme disease spirochete Borrelia burgdorferi or the rickettsial agent Anaplasma phagocytophilum. A significant decrease in the accumulation of Kenny and reduced nuclear translocation of N-Rel were observed in xiap-/- cells during microbial stimulation (Fig. 2F through I), indicating that functional disruption of xiap results in the impairment of the IMD pathway in ticks. Importantly, WT and xiap-/- ISE6 cells were infected with A. phagocytophilum for 72 hours, and bacterial burden was assessed by RT-qPCR. We observed an increase in A. phagocytophilum load in xiap-/- ISE6 cells compared to the control treatment (Fig. 2J). These data obtained through CRISPR editing implicated the E3 ubiquitin ligase XIAP as an important molecule for the I. scapularis IMD pathway.
DISCUSSION
Genetic manipulation of ticks lags behind insects and our study expands the genetic toolbox in I. scapularis. We (i) expressed two tick proteins in ISE6 cells, XIAP and p47 (10–12); (ii) confirmed their previous molecular interactions (10); and (iii) successfully detected their subcellular location. We also developed a protocol for CRISPR-Cas9 gene editing in ISE6 cells and provided evidence that disrupting components of the IMD pathway resulted in an increase in A. phagocytophilum burden in I. scapularis. Our approach follows an earlier report in which Kurtti et al. (17) used cationic lipid-based transfection reagents to deliver a red fluorescent protein and a selectable marker, neomycin phosphotransferase, into ISE6 cells.
Recent advancements used embryo injection of CRISPR-Cas9 through the Receptor-Mediated Ovary Transduction of Cargo (ReMOT Control) (18) for direct delivery of the Cas9-RNP complex into I. scapularis (7). Although this technology is a breakthrough for the tick community, there are significant limitations for its applicability in vivo, including low survival and efficiency in addition to the long life cycle of I. scapularis ticks. Our study provides a complementary approach and paves the way for exploring ancillary CRISPR-Cas9 technologies, including CRISPR activation (19) and CRISPR interference (20). The development of genetic tools for tick research offers unique avenues to identify crucial genes related to physiology, immune signaling, and the detection of microbes. Ectopic expression and CRISPR technologies in cells might aid in the identification of antigen targets for the development of tick-based vaccines (21), epitopes associated with the α-gal allergy to red meat (22), and proteins linked to acaracide resistance (23).
ACKNOWLEDGMENTS
We acknowledge members of the Pedra laboratory for providing insightful discussions and manuscript feedback. We thank Jon Skare (Texas A&M University Health Science Center) for providing the B. burgdorferi B31 strain, clone MSK5; Ulrike G. Munderloh (University of Minnesota) for supplying the ISE6 cell line; Joseph Mauban (University of Maryland School of Medicine) for aiding in confocal microscopy; Biopolymer/Genomics core facility for Sanger sequencing, and Francy E. Cabrera Paz (University of Maryland School of Medicine) for providing administrative support.
This work was supported by grants from the National Institutes of Health (NIH) to A.J.O. (F31AI152215), L.R.B. (F31AI167471), H.J.L.-Y. (T32AI162579), J.H.F.P. (R01AI134696, R01AI116523, and P01AI138949), N.P., S.E.M., and J.H.F.P. (R21 AI168592). N.P., S.E.M., and J.H.F.P. were also supported in kind by the Fairbairn Family Lyme Research Initiative. N.P. is an investigator of the Howard Hughes Medical Institute.
Fig. S5 was created with BioRender.com.
The content is solely the responsibility of the authors and does not represent the official views of the NIH, the Department of Health and Human Services or the United States government.
N.S. and J.H.F.P. designed the study. N.S., A.R., A.J.O., L.R.B., H.J.L.-Y., and S.S. performed the experiments. N.S. and J.H.F.P. wrote the manuscript. L.R.B. and H.J.L.-Y. helped in creating schematics. M.B., S.E.M., and N.P. provided intellectual insights and scientific advice. All authors analyzed the data and contributed to the editing of the manuscript. J.H.F.P. supervised the study.
Contributor Information
Joao H. F. Pedra, Email: jpedra@som.umaryland.edu.
Gregory D. Ebel, Colorado State University, Fort Collins, Colorado, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.02479-23.
Supplemental text, figures, and tables.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Eisen L. 2020. Stemming the rising tide of human-biting ticks and tickborne diseases, United States. Emerg Infect Dis 26:641–647. doi: 10.3201/eid2604.191629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eisen RJ, Eisen L. 2018. The blacklegged tick, Ixodes scapularis: an increasing public health concern. Trends Parasitol 34:295–309. doi: 10.1016/j.pt.2017.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O’Neal AJ, Singh N, Mendes MT, Pedra JHF. 2021. The genus Anaplasma: drawing back the curtain on tick-pathogen interactions. Pathog Dis 79:ftab022. doi: 10.1093/femspd/ftab022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang F, Wen Y, Guo X. 2014. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet 23:R40–R46. doi: 10.1093/hmg/ddu125 [DOI] [PubMed] [Google Scholar]
- 5. Miller JR, Koren S, Dilley KA, Harkins DM, Stockwell TB, Shabman RS, Sutton GG. 2018. A draft genome sequence for the Ixodes scapularis cell line, ISE6. F1000Res 7:297. doi: 10.12688/f1000research.13635.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De S, Kingan SB, Kitsou C, Portik DM, Foor SD, Frederick JC, Rana VS, Paulat NS, Ray DA, Wang Y, Glenn TC, Pal U. 2023. A high-quality Ixodes scapularis genome advances tick science. Nat Genet 55:301–311. doi: 10.1038/s41588-022-01275-w [DOI] [PubMed] [Google Scholar]
- 7. Sharma A, Pham MN, Reyes JB, Chana R, Yim WC, Heu CC, Kim D, Chaverra-Rodriguez D, Rasgon JL, Harrell RA, Nuss AB, Gulia-Nuss M. 2022. Cas9- mediated gene-editing in the black-legged tick, Ixodes scapularis, by embryo injection and ReMOT control. iScience 25:103781. doi: 10.1016/j.isci.2022.103781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de la Fuente J, Kocan KM, Almazán C, Blouin EF. 2007. RNA interference for the study and genetic manipulation of ticks. Trends Parasitol 23:427–433. doi: 10.1016/j.pt.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 9. Sledz CA, Williams BRG. 2005. RNA interference in biology and disease. Blood 106:787–794. doi: 10.1182/blood-2004-12-4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McClure Carroll EE, Wang X, Shaw DK, O’Neal AJ, Oliva Chávez AS, Brown LJ, Boradia VM, Hammond HL, Pedra JHF. 2019. p47 licenses activation of the immune deficiency pathway in the tick Ixodes scapularis. Proc Natl Acad Sci U S A 116:205–210. doi: 10.1073/pnas.1808905116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaw DK, Wang X, Brown LJ, Chávez ASO, Reif KE, Smith AA, Scott AJ, McClure EE, Boradia VM, Hammond HL, Sundberg EJ, Snyder GA, Liu L, DePonte K, Villar M, Ueti MW, de la Fuente J, Ernst RK, Pal U, Fikrig E, Pedra JHF. 2017. Infection-derived lipids elicit an immune deficiency circuit in arthropods. Nat Commun 8:14401. doi: 10.1038/ncomms14401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Neal AJ, Singh N, Rolandelli A, Laukaitis HJ, Wang X, Shaw DK, Young BD, Narasimhan S, Dutta S, Snyder GA, Samaddar S, Marnin L, Butler LR, Mendes MT, Cabrera Paz FE, Valencia LM, Sundberg EJ, Fikrig E, Pal U, Weber DJ, Pedra JHF. 2023. Croquemort elicits activation of the immune deficiency pathway in ticks. Proc Natl Acad Sci U S A 120:e2208673120. doi: 10.1073/pnas.2208673120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. 1995. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci U S A 92:9465–9469. doi: 10.1073/pnas.92.21.9465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kleino A, Silverman N. 2014. The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol 42:25–35. doi: 10.1016/j.dci.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang F, Doudna JA. 2017. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529. doi: 10.1146/annurev-biophys-062215-010822 [DOI] [PubMed] [Google Scholar]
- 16. Zhang S, Guha S, Volkert FC. 1995. The Saccharomyces SHP1 gene, which encodes a regulator of phosphoprotein phosphatase 1 with differential effects on glycogen metabolism, meiotic differentiation, and mitotic cell cycle progression. Mol Cell Biol 15:2037–2050. doi: 10.1128/MCB.15.4.2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kurtti TJ, Mattila JT, Herron MJ, Felsheim RF, Baldridge GD, Burkhardt NY, Blazar BR, Hackett PB, Meyer JM, Munderloh UG. 2008. Transgene expression and silencing in a tick cell line: a model system for functional tick genomics. Insect Biochem Mol Biol 38:963–968. doi: 10.1016/j.ibmb.2008.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaverra-Rodriguez D, Macias VM, Hughes GL, Pujhari S, Suzuki Y, Peterson DR, Kim D, McKeand S, Rasgon JL. 2018. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat Commun 9:3008. doi: 10.1038/s41467-018-05425-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159:647–661. doi: 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154:442–451. doi: 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sajid A, Matias J, Arora G, Kurokawa C, DePonte K, Tang X, Lynn G, Wu MJ, Pal U, Strank NO, Pardi N, Narasimhan S, Weissman D, Fikrig E. 2021. mRNA vaccination induces tick resistance and prevents transmission of the Lyme disease agent. Sci Transl Med 13:eabj9827. doi: 10.1126/scitranslmed.abj9827 [DOI] [PubMed] [Google Scholar]
- 22. Commins SP. 2020. Diagnosis & management of α-gal syndrome: lessons from 2,500 patients. Expert Rev Clin Immunol 16:667–677. doi: 10.1080/1744666X.2020.1782745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al-Rofaai A, Bell-Sakyi L. 2020. Tick cell lines in research on tick control. Front Physiol 11:152. doi: 10.3389/fphys.2020.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, figures, and tables.