ABSTRACT
Flavobacterium johnsoniae is a ubiquitous soil and rhizosphere bacterium, but despite its abundance, the factors contributing to its success in communities are poorly understood. Using a model microbial community, The Hitchhikers of the Rhizosphere (THOR), we determined the effects of colonization on the fitness of F. johnsoniae in the community. Insertion sequencing, a massively parallel transposon mutant screen, on sterile sand identified 25 genes likely to be important for surface colonization. We constructed in-frame deletions of candidate genes predicted to be involved in cell membrane biogenesis, motility, signal transduction, and transport of amino acids and lipids. All mutants poorly colonized sand, glass, and polystyrene and produced less biofilm than the wild type, indicating the importance of the targeted genes in surface colonization. Eight of the nine colonization-defective mutants were also unable to form motile biofilms or zorbs, thereby suggesting that the affected genes play a role in group movement and linking stationary and motile biofilm formation genetically. Furthermore, we showed that the deletion of colonization genes in F. johnsoniae affected its behavior and survival in THOR on surfaces, suggesting that the same traits are required for success in a multispecies microbial community. Our results provide insight into the mechanisms of surface colonization by F. johnsoniae and form the basis for further understanding its ecology in the rhizosphere.
IMPORTANCE
Microbial communities direct key environmental processes through multispecies interactions. Understanding these interactions is vital for manipulating microbiomes to promote health in human, environmental, and agricultural systems. However, microbiome complexity can hinder our understanding of the underlying mechanisms in microbial community interactions. As a first step toward unraveling these interactions, we explored the role of surface colonization in microbial community interactions using The Hitchhikers Of the Rhizosphere (THOR), a genetically tractable model community of three bacterial species, Flavobacterium johnsoniae, Pseudomonas koreensis, and Bacillus cereus. We identified F. johnsoniae genes important for surface colonization in solitary conditions and in the THOR community. Understanding the mechanisms that promote the success of bacteria in microbial communities brings us closer to targeted manipulations to achieve outcomes that benefit agriculture, the environment, and human health.
KEYWORDS: Type 9 secretion system, attachment, biofilms, TnSeq, exosortase, zorbs, bacteroidetes
INTRODUCTION
Microorganisms are essential in every ecosystem and often exist in communities where they interact with each other and the environment (1). The soil microbiome drives key biogeochemical cycles, and the rhizosphere microbiome influences plant susceptibility to disease and drought, indicating the critical role of microbial communities in environmental health and agricultural productivity (2, 3). The rhizosphere is the region in soil that is influenced by nutrient-rich plant root secretions or root exudate, which attract microorganisms from the surrounding soil (3). Rhizosphere microorganisms are essential to plant health as they promote plant nutrient uptake, stress tolerance, and disease suppression (4), yet we know relatively little of the genes involved in mediating rhizosphere colonization by many soil bacteria. Hence, understanding microbial colonization in this dynamic environment is an important step toward modifying the soil microbiome for agricultural benefit.
After decades of studying bacterial monocultures, the field of microbiology has begun to explore the significance of multispecies interactions. Advances in high-throughput sequencing technology have enabled the use of metagenomics and metatranscriptomics to determine the taxonomic composition and functional characteristics of microbiomes in soil and rhizospheres (5–9). Nevertheless, the complexity of microbiomes containing thousands of interacting species makes it difficult to determine the functions of genes using classical genetic approaches.
Genetic analysis of a simplified model microbial community enables a mechanistic understanding of community interactions. We previously described The Hitchhikers Of the Rhizosphere (THOR), a three-species model community composed of Pseudomonas koreensis, Bacillus cereus, and Flavobacterium johnsoniae isolated from the rhizospheres of field-grown alfalfa and soybean plants (10). These three species are genetically tractable, making mutant analysis in a community context possible. Additionally, numerous interactions between THOR members have been observed in both field and laboratory settings. For example, P. koreensis and F. johnsoniae were physically associated as biological hitchhikers with B. cereus isolated from soybean roots (10). The three species form more biofilm together than any of the species alone or in pairs (10). B. cereus provides peptidoglycan fragments as a carbon source that enables F. johnsoniae to grow in soybean root exudate (11). F. johnsoniae and P. koreensis induce B. cereus colony expansion (10). F. johnsoniae growth is inhibited by koreenceine, an antibiotic produced by P. koreensis, and koreenceine levels are modulated by the third THOR member, B. cereus (12). Gene expression and metabolite profiles of each member differ when they are in the community vs in pure culture (13, 14). These interactions indicate that THOR members engage in a web of biological interactions.
Flavobacterium spp. are abundant in the rhizosphere and are well studied for their ability to glide on surfaces using cell-surface motility adhesins that are delivered by a unique Type 9 Secretion System (T9SS) (15–18). Some strains of F. johnsoniae are even known to promote plant growth and suppress pathogens in soil, but its behavior in the rhizosphere or on soil particles is poorly understood (19, 20). The elegant work of the McBride lab has provided the framework for genetic analysis in F. johnsoniae (21), enabling us to pinpoint genes important for surface colonization, which emerged as a critical factor in F. johnsoniae survival in the microbial community and biofilm formation on surfaces.
RESULTS
Genetic determinants of surface colonization in F. johnsoniae
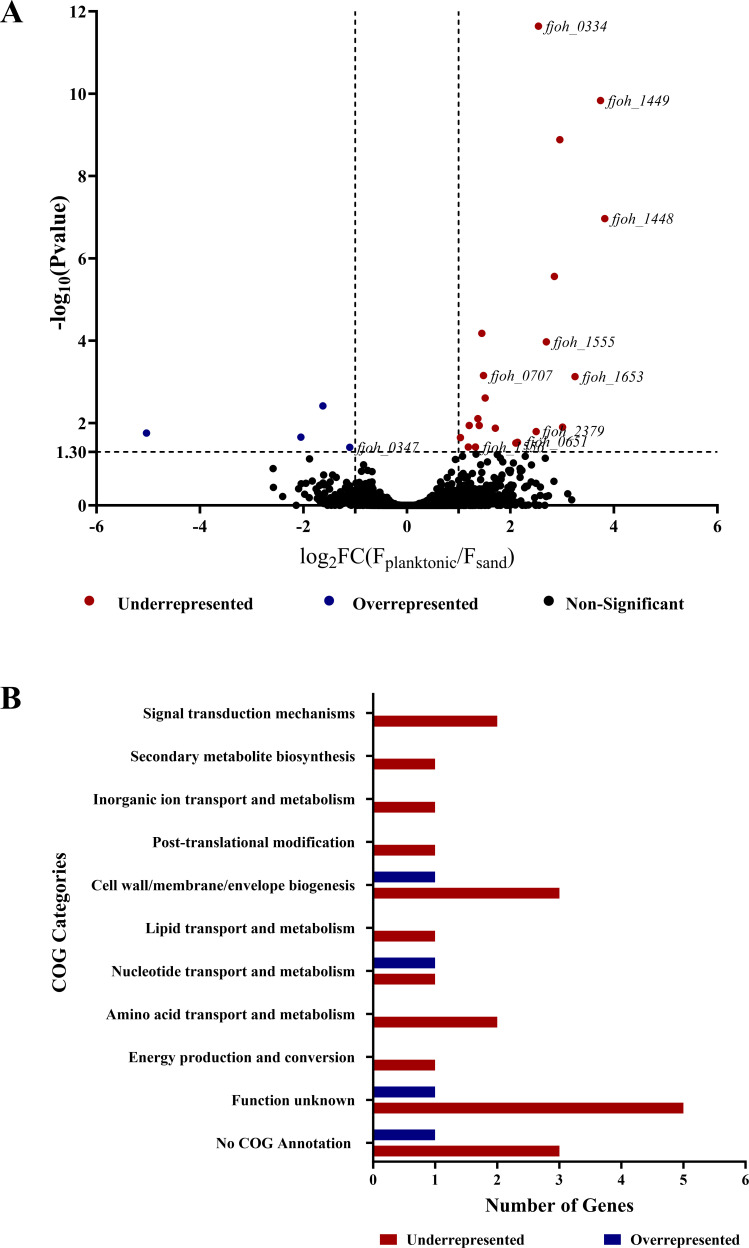
We hypothesized that surface colonization promotes F. johnsoniae success in THOR. Here, we define surface colonization as the ability of bacteria to attach, proliferate, and survive on a substrate. To study surface colonization, we used sand, an abundant, easily manipulated soil particle that Flavobacterium spp. colonize better than other soil particles (22). We leveraged the power of insertion sequencing (INSeq) and conducted a massively parallel screen of F. johnsoniae transposon mutants to identify genes important for sand colonization. We created a library of approximately 75,000 transposon mutants using a modified Bacteroides thetaiotaomicron vector (pSAMFjoh 2) and compared mutant frequencies after growth under planktonic conditions (“input population”) with their frequencies after growth on sand (“output population”) for 48 h. By comparing the abundance of each transposon insertion between the planktonic and sand-colonized populations, we identified 25 candidate genes important for sand colonization. Mutants with transposon insertions in 21 different genes were underrepresented with a minimum fold change (planktonic/sand) of log2 > 1 and adjusted P-value < 0.05, suggesting that deletion of these genes would reduce sand colonization. Mutants with transposon insertions in four genes were overrepresented with a minimum fold change (planktonic/sand) of log2 < −1 with an adjusted P-value < 0.05, suggesting that deletion of these genes would increase colonization (Fig. 1A; Table 1 and Table S1).
Fig 1.
Genes identified by INSeq screen as important for sand colonization. (A) The relative abundance of transposon insertions for each gene was compared between F. johnsoniae grown planktonically and on the sand. The data are presented as a volcano plot with –log10 P-value on the y-axis and log2 fold change (input/output) on the x-axis. The black dots represent mutants whose representation did not differ between conditions (P-value > 0.05), the red represents mutants that are significantly underrepresented on the surface of the sand, and the blue represents mutants that are significantly overrepresented on the surface of the sand (P-value < 0.05 and FC > 1 or FC < −1). The genes affected in mutants whose representation differed between the two conditions were validated by gene deletions; these mutants are labeled by their Gene ID. (B) The genes identified as important in sand colonization are categorized based on Clusters of Orthologous Genes (COG) annotation.
TABLE 1.
Sand colonization genes identified using INSeq screen and validated by mutant analysisa
| Accession ID | Gene | Annotation | COG category | Log2 fold change (input/output) |
|---|---|---|---|---|
| Fjoh_0334 | FJOH_RS01770 | DegT/DnrJ/EryC1/StrS family aminotransferase | E | 2.54 |
| Fjoh_0651 | FJOH_RS03415 | Hypothetical protein | No COG | 2.14 |
| Fjoh_0707 | FJOH_RS03710 porZ | T9SS type A sorting domain-containing protein PorZ | T | 1.48 |
| Fjoh_1448 | xrtF | Exosortase family protein XrtF | S | 3.82 |
| Fjoh_1449 | FJOH_RS07530 | Exosortase F system-associated protein | No COG | 3.74 |
| Fjoh_1555 | porV | T9SS outer membrane channel protein PorV | I | 2.69 |
| Fjoh_1556 | porU | T9SS sortase PorU | S | 1.33 |
| Fjoh_1653 | sprA | Cell surface protein SprA | S | 3.25 |
| Fjoh_2379 | FJOH_RS12370 | LysM peptidoglycan-binding domain-containing protein | M | 2.5 |
List of genes that were validated by constructing in-frame deletions. The complete list of genes identified is provided in Table S1. The COG categories are abbreviated as follows: E, amino acid metabolism and transport; T, signal transduction; S, unknown function; I, lipid transport and metabolism; and M, cell wall/membrane/envelope biogenesis.
Functional annotation revealed that the largest group (40%) of candidate colonization genes had homologs in GenBank annotated as uncharacterized, encoding either hypothetical proteins or proteins of unknown function. A smaller group of genes (16%) encode proteins predicted to be involved in cell wall/membrane/envelope biogenesis, and other genes are predicted to be involved in the transport and biosynthesis of primary and secondary metabolites (Fig. 1B).
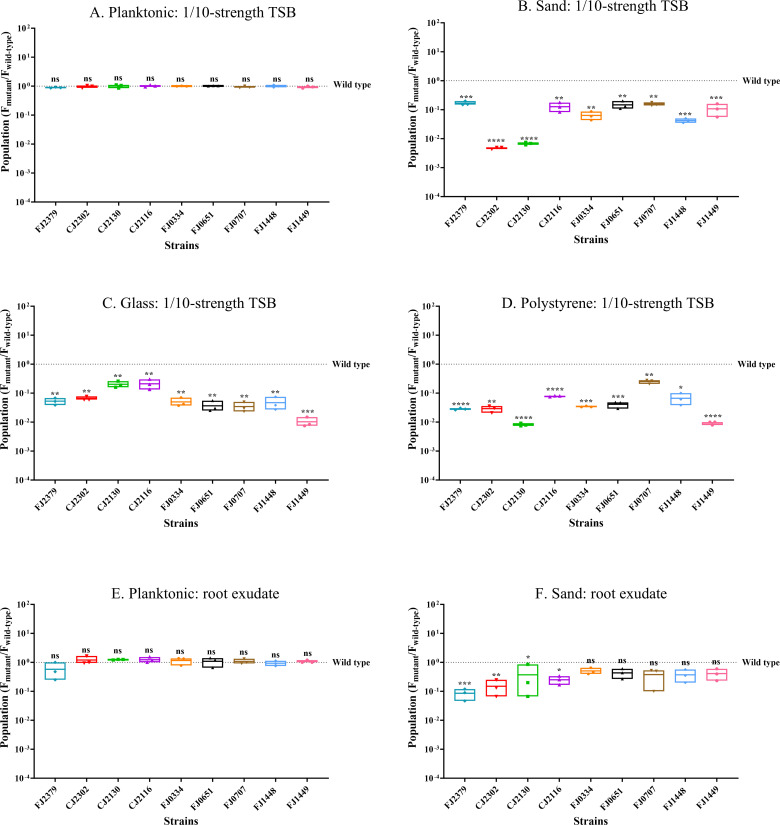
We validated the INSeq screen with in-frame deletions of nine genes that were underrepresented and assessed the mutants’ ability to colonize sand. Planktonic populations of the mutants in tryptic soy broth (TSB) were similar to those of the wild type after 48 h (Fig. 2A), whereas all nine mutants whose genes were underrepresented in the INSeq screen colonized sand poorly, indicating that the affected gene products contribute to sand colonization (Fig. 2B). Complementation of deletions with plasmid-borne copies of the gene of interest restored colonization to wild-type levels (Fig. S1).
Fig 2.
Surface colonization by mutants identified in INSeq analysis. Populations of wild-type F. johnsoniae CJ1827 and deletion mutants are represented as the ratio of mutant to wild type. The initial inoculum was 106 CFU/mL of wild-type F. johnsoniae CJ1827 or mutants (FJ2379, CJ2302, CJ2130, CJ2116, FJ0334, FJ0651, FJ0707, FJ1448, and FJ1449), and populations were determined after 48 h under the following conditions: (A) planktonic: 1/10-strength TSB, (B) sand: 1/10-strength TSB, (C) glass: 1/10-strength TSB, (D) polystyrene: 1/10-strength TSB, (E) planktonic: soybean root exudate, and (F) sand: soybean root exudate. The dotted line indicates wild-type populations, and each data point in the box plots represents one biological replicate. Statistical significance was evaluated using GraphPad Prism. Differences between the mutant and wild type are indicated as ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
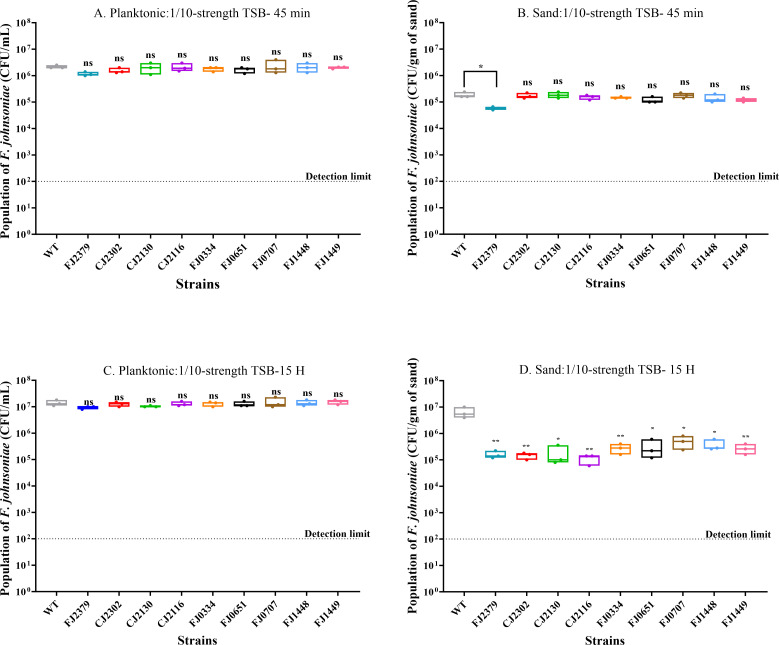
Next, we determined whether the colonization phenotype was caused by a defect in attachment or the inability of the mutants to proliferate after attachment. We first quantified the population of F. johnsoniae CJ1827 (wild type) at several time points to investigate the influence of attachment and growth on colonization (Fig. S2). Based on our observations of the wild type, we measured the effects of gene deletions on attachment by assessing mutant populations during the lag phase (T = 45 min) and the impact on growth by examining mutant populations during the log phase (T = 15 h; Fig. 3). In the planktonic condition, most mutants (FJ2379, CJ2302, CJ2130, CJ2116, FJ0651, FJ0707, FJ1448, and FJ1449) did not exhibit any significant difference in growth rate (Fig. S3) or population size. Mutant FJ0334 had a shorter doubling time than the wild type but reached the stationary phase at a similar time to the wild type, thereby yielding a similar population size in the planktonic phase after 48 h. In contrast to the planktonic condition, eight mutants (CJ2302, CJ2130, CJ2116, FJ0334, FJ0651, FJ0707, FJ1448, and FJ1449) exhibited poor colonization due to growth defect on the sand, whereas only one mutant, FJ2379, displayed deficiencies in initial attachment. This suggests that the observed colonization phenotype is primarily driven by differences in the ability of these mutants to grow and establish themselves on the sand surface rather than their initial attachment capabilities.
Fig 3.
Impact of gene deletions on attachment and growth during sand colonization. Populations of wild type and mutants were determined after 45 min and 15 h under the following conditions: (A) 45 min no solid substrate (planktonic: 1/10-strength TSB), (B) 45 min sand: 1/10-strength TSB, (C) 15 h no solid substrate (planktonic: 1/10-strength TSB), and (D) 15 h sand: 1/10-strength TSB. The dotted line indicates the detection limit (102 CFU/mL), and each data point in box plots represents one biological replicate. Statistical significance was evaluated by comparing the mutants to the wild type with a one-way analysis of variance (ANOVA)followed by Dunnett’s test. Differences between the mutant and wild type are indicated as ns, not significant; *, P < 0.05; **, P < 0.01.
Additionally, since F. johnsoniae is a gliding bacterium, we determined whether any of these mutations affected motility (Fig. S4). We found that only one (CJ2302) of the nine mutants formed nonspreading colonies in PY2 agar, suggesting that not all mutations affecting colonization have an impact on motility.
Sand colonization mutants are altered in the colonization of other substrates
We tested the ability of the sand colonization mutants to colonize other substrates, such as glass, which is chemically similar to sand; polystyrene, which is structurally very different from sand; and sand in soybean root exudate to approximate the rhizosphere (Fig. 2). All nine mutants that colonized sand poorly were also defective in colonizing glass and polystyrene. This suggests that the deleted genes are important for the colonization of surfaces generally. Deletion of genes mostly involved in cell wall/membrane biogenesis and T9SS led to poor sand colonization in root exudate, suggesting their importance in the rhizosphere. The other deletion mutants behaved similarly to the wild type on sand in root exudate.
Biofilm formation correlates with surface colonization ability
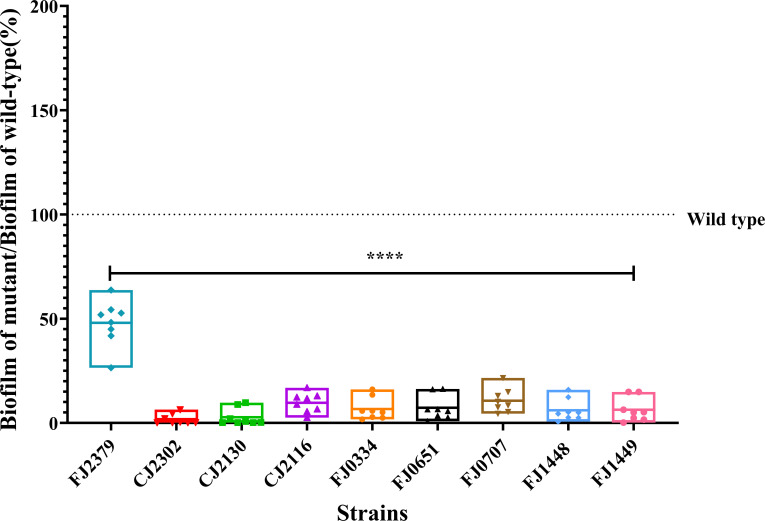
Microorganisms colonize various surfaces in the soil, forming biofilms that enhance fitness by providing protection against predation, desiccation, exposure to antibiotics, and nutrient depletion (23–26). We tested the ability of colonization-defective mutants to produce biofilm using a crystal-violet assay (Fig. 4). All nine mutants that colonized poorly on sand, glass, and polystyrene produced less biofilm compared to the wild type, indicating that surface colonization is important for biofilm formation under these conditions. A linear regression analysis showed an association between biofilm formation and polystyrene colonization with an R2 value of 0.8632 (Fig. S5).
Fig 4.
Colonization-defective mutants form less biofilm. Biofilm was quantified by crystal-violet staining (OD595) 18 h after inoculation. Each point represents one biological replicate comprising the data averaged from eight technical replicates. The data are expressed as a percentage of biofilm produced by the mutant relative to the wild type, where the dotted line indicates the level of wild-type biofilm. The OD595 values were square-root transformed to achieve equal variance, and statistical significance was evaluated by one-way ANOVA followed by Dunnett’s test. Differences between mutants and wild type are indicated as ****, P < 0.0001.
Surface colonization ability correlates with the formation of zorbs
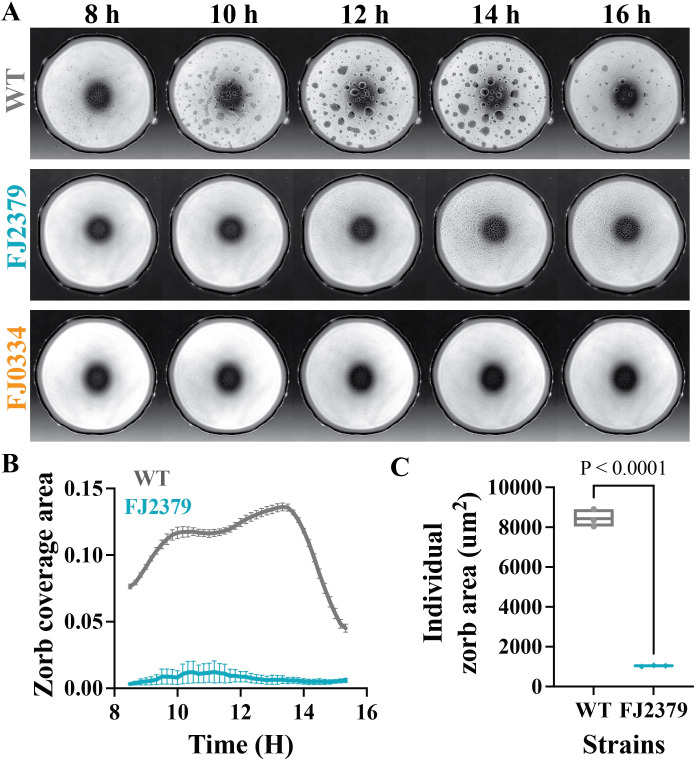
We previously reported that F. johnsoniae forms motile biofilms, designated zorbs, when observed in an under-oil microfluidic device (27). Zorbs move using cells at the base of the structure that are attached to the surface by one pole. Initially, the bacteria largely remain as single cells and then form microcolonies that migrate, merge, grow larger, and eventually disperse into single cells (27). To determine whether defects in static biofilm formation extended to motile biofilms, we monitored zorb formation in the mutants and wild type under oil on a glass substrate with bright-field time-lapse microscopy for 18 h (Fig. 5A; Video S1). In eight of the nine colonization-defective mutants (CJ2302, CJ2130, CJ2116, FJ0334, FJ0651, FJ0707, FJ1448, and FJ1449), zorb formation was completely abolished. Interestingly, mutant FJ2379, which maps in a gene encoding a predicted LysM peptidoglycan-binding domain-containing protein, exhibited less zorbing capacity (the total area covered by zorbs over time; Fig. 5B) and formed smaller zorbs than the wild type (Fig. 5C). These results indicate that genes required for normal surface colonization influence zorb formation and behavior.
Fig 5.
Colonization defects correlate with defects in zorb formation. Zorbing of wild-type and mutant strains was captured by brightfield imaging over 18 h and analyzed for differences in zorbing behavior. (A) Time-lapse images of wild type (CJ1827) and mutants (FJ2379 and FJ0334; representative of all eight mutants that did not form zorbs) between 8 and 16 h. (B) Plots of the total coverage area of zorbs are given as a fraction of the spot area over time. (C) Boxplot of time-averaged individual zorb area. Statistical significance was determined by a one-way ANOVA with multiple comparisons. All plots were generated by quantifying data from independent time-lapse movies of three biological replicates for each strain.
Colonization mutants exhibit altered behavior in a multispecies community
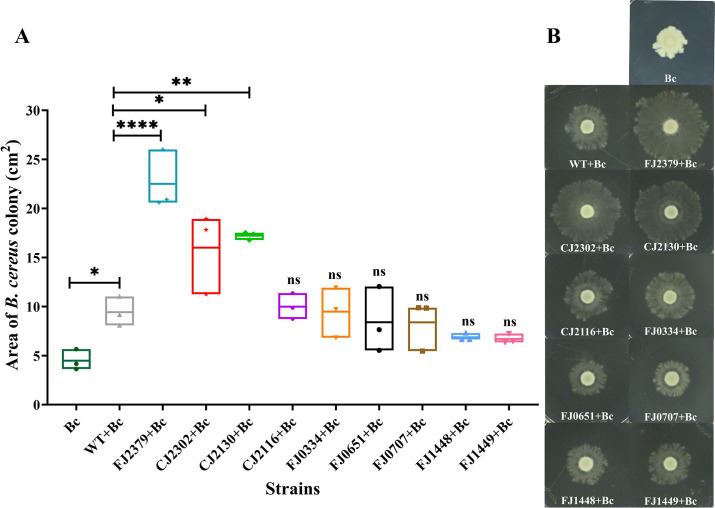
We used THOR to assess the behavior of the surface colonization mutants in a community. THOR members exhibit several community-specific behaviors, including augmented expansion of B. cereus colonies induced by F. johnsoniae (10). Three mutants, CJ2130, CJ2302, and FJ2379, which contain deletions in genes encoding putative outer membrane PorV, SprA, and LysM peptidoglycan-binding domain-containing protein, respectively, induced more expansion of B. cereus colonies than the wild type (Fig. 6). Initially, we hypothesized that expansion could be due to the relative difference in the level of biosurfactant produced by the mutants; however, we did not observe any significant difference in the biosurfactant between the mutants and the wild type (data not shown). Given that PorV and SprA are integral to T9SS-dependent secretion and anchoring proteins to the cell surface, deletion of porV and sprA may have resulted in the loss of secretion of specific cell-surface or outer-membrane proteins. This leads us to suggest that an interaction between the surfaces of the two bacteria mediated by the outer membrane proteins in F. johnsoniae influences the expansion of B. cereus colonies.
Fig 6.
Colonization mutants affect B. cereus colony expansion. Colony expansion of B. cereus in 1/10th-strength tryptic soy agar (TSA) was observed in the presence of wild-type and mutant F. johnsoniae strains. The area of the B. cereus colony is shown as a box plot (A), the experiment was performed three times, and a representative image is shown in panel B. The values (cm2) were square-root transformed for equal variance, and the statistical significance was evaluated with a one-way ANOVA followed by Dunnett’s test. Differences between the mutants and wild type and between the wild type and B. cereus alone are indicated as ns, not significant; *, P < 0.05; **, P < 0.01, or ****, P < 0.0001.
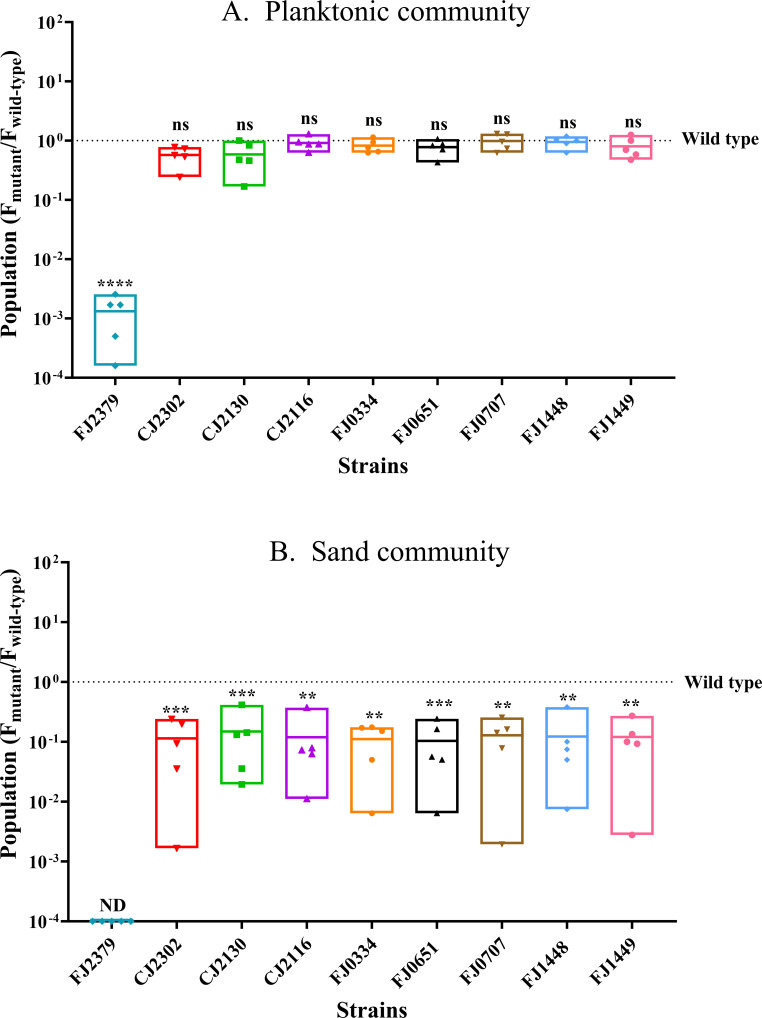
We next assessed whether surface colonization promotes the fitness of F. johnsoniae in the community. In the absence of sand as a substrate (planktonic condition), the fitness of mutants defective in sand colonization was not altered in the full three-member community, as indicated by the mutants and the wild type achieving similar populations in the community. The one exception was FJ2379, whose populations were lower (Fig. 7A) than the wild type in the planktonic community. Consequently, we examined the impact of cell-free supernatants from P. koreensis, B. cereus, and both P. koreensis and B. cereus on the wild type and FJ2379. We found that although FJ2379 did not exhibit any growth defect when cultured in its own or in the culture supernatant of B. cereus, it achieved a significantly lower population when cultured in the culture supernatants of P. koreensis and P. koreensis + B. cereus (data not shown). This suggests that the observed mutant phenotype in the planktonic community is likely mediated by metabolites secreted by P. koreensis, and FJ2379 appears to be relatively sensitive to these metabolites. In contrast, all mutants defective in sand colonization in solitary culture survived poorly in the full community compared to the wild type on sand (Fig. 7B), indicating that the genes required for solitary colonization of surfaces are also required for F. johnsoniae success in a microbial community on a surface.
Fig 7.
Colonization ability affects the fitness of F. johnsoniae in THOR. (A) F. johnsoniae wild type and mutants when grown with P. koreensis and B. cereus planktonically. (B) F. johnsoniae wild type and mutants when grown with P. koreensis and B. cereus on sand. Each data point represents one of five biological replicates, with the dotted line indicating the value for the wild type. The CFU values were log10 transformed for equal variance. The statistical significance was evaluated by comparing the mutants to the wild type with a one-way ANOVA followed by Dunnett’s test. ns = not significant; **, P < 0.01; ***, P < 0.001; or ****, P < 0.0001.
DISCUSSION
In this study, we used INSeq, a genetic screen that couples transposon mutagenesis and high-throughput sequencing to identify genes in F. johnsoniae that are important for sand colonization in solitary culture and in a model community. INSeq (highly similar to transposon sequencing) is a powerful genetic tool to establish relationships between genes and bacterial behavior (28, 29). This tool has been used to simultaneously evaluate the fitness of thousands of discrete mutants and has identified genes crucial for root colonization in several Proteobacteria and genes related to soil colonization in others (30–34). Although INSeq technology was developed in B. thetaiotaomicron, the work presented here is the first INSeq screen on another member of the Bacteroidetes phylum to reveal genes necessary for sand colonization.
Sand is an abundant soil particle, and the presence of F. johnsoniae promotes aggregation of sand particles, thereby reducing soil erosion (35). We identified 25 genes related to sand colonization, most of which are either uncharacterized or encode proteins with unknown functions. Other gene categories found in the screen were annotated as involved in cell wall/membrane/envelope biogenesis and transport and metabolism of primary and secondary metabolites (Fig. 1). Although the majority of these genes have not been explored thoroughly in F. johnsoniae, their predicted functions are similar to genes previously shown to play roles in fitness of other rhizosphere-colonizing bacteria (30, 31).
We validated the INSeq screen with in-frame deletions in nine genes and showed that the mutants’ abilities to colonize various substrates, form biofilms, and survive in a model rhizosphere community were affected by the deletions (Table 2). Based on the predicted functions of the deleted genes, these mutants can be divided into four categories: T9SS, exosortase-system, cell-surface components, and hypothetical proteins.
TABLE 2.
Summary of surface-colonization mutant phenotypes
| Strains | INSeq screen colonization | Sand colonization 1/10-strength TSB | Biofilm | Zorbs | Colony expansion of B. cereus | Community planktonic | Community sand |
|---|---|---|---|---|---|---|---|
| WT | Base level | Base level | Base level | Base level | Base level | Base level | Base level |
| FJ2379 | Underrepresented | Less | Less | Less | More | Less | Less |
| CJ2302 ΔsprA | Underrepresented | Less | Much less | Abolished | More | WT level | Less |
| CJ2130 ΔporV | Underrepresented | Less | Much less | Abolished | More | WT level | Less |
| CJ2116 ΔporU | Underrepresented | Less | Much less | Abolished | WT level | WT level | Less |
| FJ0334 | Underrepresented | Less | Much less | Abolished | WT level | WT level | Less |
| FJ0651 | Underrepresented | Less | Much less | Abolished | WT level | WT level | Less |
| FJ0707 ΔporZ | Underrepresented | Less | Much less | Abolished | WT level | WT level | Less |
| FJ1448 ΔxrtF | Underrepresented | Less | Much less | Abolished | WT level | WT level | Less |
| FJ1449 | Underrepresented | Less | Much less | Abolished | WT level | WT level | Less |
Numerous studies of nongliding bacteria have demonstrated the importance of motility appendages such as flagella and pili in the colonization of surfaces in the rhizosphere (36–39). In contrast, F. johnsoniae glides over surfaces without the aid of flagella or pili, instead using a gliding motility apparatus and adhesins secreted by T9SS (15). Deletion of genes involved in T9SS or gliding motility has been shown previously to affect attachment to glass (sprA, sprE, gldK, gldL, gldM, and porV) and plant roots (gldJ) and biofilm formation (gldA, gldB, gldD, gldF, gldG, gldH, gldI, gldJ, gldN, and porV) (40–45). Our study adds to the existing body of research by demonstrating that deletion of genes associated with T9SS (FJOH RS03710 [porZ ortholog of Porphyromonas gingivalis W83], porV, porU, and sprA) does indeed reduce colonization of various substrates and abolishes biofilm formation. Proteins PorV, PorU, and PorZ are integral components of the T9SS cell surface-exposed attachment complex (PorQUVZ), with PorV playing a pivotal role in shuttling cargo proteins from the SprA translocon to the attachment complex (46–48). Subsequently, these cargo proteins are processed and anchored to the cell surface. Given that the proteins encoded by these genes play a crucial role in the T9SS-mediated secretion of cell-surface and extracellular proteins, deletion of these genes most likely caused loss of secretion of many cell surface proteins or adhesins resulting in the observed defects in surface colonization and biofilm formation.
Several genes from the INSeq screen involved in sand colonization also affected the formation of motile biofilms, or zorbs, produced by F. johnsoniae. Zorbs are self-propelled spherical microcolonies with an extracellular polysaccharide (EPS) core that move using cells at the base of the zorbs (“base cells”) that attach to the surface by one pole of the cell (27). Our data genetically link surface colonization and zorb formation since all mutants were reduced in both phenotypes. The data further connect stationary and motile biofilm formation by showing that zorbing was absent in mutants that did not produce any detectable stationary biofilm. In contrast, mutant FJ0347, which produced more biofilm, formed hypermotile zorbs that merged more than the wild type (Fig. S6). These findings corroborate previous work showing that zorbs are authentic biofilm structures.
We identified two genes (xrtF and FJOH_RS07530) predicted to encode exosortase family proteins. Exosortases are proteases that cleave proteins that contain the carboxy-terminal protein-sorting signal PEP-CTERM and mostly occur in bacterial species with exopolysaccharide biosynthesis gene clusters (49, 50). Although exosortases have been proposed to be associated with biofilm formation based on bioinformatic analysis, our study is the first to provide empirical data indicating that exosortases play a role in surface colonization and biofilm formation. Future research will focus on the substrates for exosortases in F. johnsoniae, which are predicted to be quite dissimilar from PEP-CTERM signal sequences based on genomic analysis.
We additionally identified four colonization genes within a 76.5 kb gene cluster that is predicted to synthesize and export cell-surface polysaccharides (51). This gene cluster contains several transcriptional units, and the deletion of genes from various regions of it had diverse effects on biofilm formation. For instance, deletion of gene FJOH_RS01770, which is part of a transcriptional unit predicted to be involved in the biosynthesis of dTDP-β-L-rhamnose and fucose, abolished biofilm formation, whereas deletion of gene FJOH_RS01835, predicted to be involved in lipopolysaccharide (LPS) biosynthesis, enhanced biofilm formation (Fig. S6A). This difference in phenotype between the two mutants is likely due to FJ0347 having a more hydrophobic cell surface, a characteristic that contributes to adhesion and biofilm formation (Fig. S6B) (27, 52–54).
We predicted that genes that are uniquely involved in interactions between F. johnsoniae and the soil environment would be enriched for those of unknown function since the majority of functional characterization of bacterial genes has been conducted in pure culture in laboratory media. The data support this prediction and demonstrate the power of ecological screens to increase the functional assignments of previously unknown genes. Of five genes (FJOH RS02845, FJOH RS03290, FJOH RS03415, FJOH RS0536, and FJOH RS15790) identified in the INSeq screen that encode hypothetical proteins, we established the role of FJOH RS03415 in sand colonization and biofilm formation.
The study lays the groundwork for understanding F. johnsoniae soil colonization and success in a community. It also suggests several themes to test more broadly among rhizosphere bacteria. First, we provide evidence that the same genes that are required for surface colonization by F. johnsoniae in solitary culture are also important for its survival in the community, a connection not previously addressed with genetic analysis in rhizosphere bacteria. Second, we provide further evidence that cell-surface molecules, such as proteins and polysaccharides, are significant actors in F. johnsoniae surface colonization, which has been established previously in other bacteria. This finding is especially important because of the role of these same molecules in community fitness. Finally, identifying six genes of unknown function in the colonization screen suggests that ecological screens can identify functions for genes that do not play substantial roles in standard laboratory pure culture conditions, which comprise the majority of previous genetic screens in microbiology. Similar work with other species will establish whether these themes are universal principles in microbial ecology.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions
Strains and plasmids used in this study are listed in Table 3, and primers are listed in Table S2. Antibiotics were used at the following concentrations when needed: ampicillin 100 µg mL−1; erythromycin 100 µg mL−1; kanamycin 50 µg mL−1; streptomycin 100 µg mL−1; and tetracycline 20 µg mL−1. Bacteria were propagated on 1/2-strength TSB at 28°C with shaking. For the experimental setup, 106 or 107 cells of each bacterium were inoculated from saturated overnight cultures and grown statically in 1/10-strength TSB at 20°C. Populations of these strains were quantified by dilution plating on Luria–Bertani broth (LB) agar supplemented with gentamicin (10 µg mL−1) for F. johnsoniae, ampicillin (100 µg mL−1) and erythromycin (5 µg mL−1) for P. koreensis, and polymyxin B (5 µg mL−1) for B. cereus. Escherichia coli strains used for cloning were grown in LB at 37°C.
TABLE 3.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Description | Source(s) or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli-S17-1 λpir | Strain used for delivery of mariner transposon | ATCC |
| E. coli-S17-1 | Strain used for cloning/conjugation | ATCC |
| F. johnsoniae UW101 | Wild-type F. johnsoniae ATCC 17061 | Commonly found in soil and freshwater (51) |
| F. johnsoniae CJ1827 | rpsL2; (Smr) parent strain for construction of all deletion mutants | Reference 21 |
| P. koreensis CI12 | Wild-type P. koreensis | Reference 10 |
| B. cereus UW85 | Wild-type B. cereus | Reference 10 |
| FJ0334 | rpsL2 Δfjoh_0334 (Smr) | This study |
| FJ0347 | rpsL2 Δfjoh_0347 (Smr) | This study |
| FJ0651 | rpsL2 Δfjoh_0651 (Smr) | This study |
| FJ0707 ΔporZ |
rpsL2 Δfjoh_0707 (Smr) | This study |
| FJ1448 ΔxrtF |
rpsL2 Δfjoh_1448 (Smr) | This study |
| FJ1449 | rpsL2 Δfjoh_1449 (Smr) | This study |
| CJ2130 ΔporV |
rpsL2 ΔporV (Smr) | Reference 43 |
| CJ2116 ΔporU |
rpsL2 ΔporU (Smr) | Reference 43 |
| CJ2302 ΔsprA |
rpsL2 ΔsprA (Smr) | Reference 40 |
| FJ2379 | rpsL2 Δfjoh_2379 (Smr) | This study |
| Plasmids | ||
| pSAMFjoh_2 | Transposon mutagenesis vector for F. johnsoniae; Apr (Emr) | Reference 27 |
| pRR51 | rpsL-containing suicide vector; Apr (Emr) | Reference 21 |
| pCP23 | E. coli-F. johnsoniae shuttle plasmid; Apr (Tcr) | Reference 55 |
| pCP0334 | 1134 bp fragment spanning fjoh_0334 was amplified with primers Fjoh_0334_CP_UP and Fjoh_0334_CP_DOWN and inserted into BamHI-SphI sites of pCP23; Apr (Tcr) | This study |
| pCP0347 | 729 bp fragment spanning fjoh_0347 was amplified with primers Fjoh_0347_CP_UP and Fjoh_0347_CP_Down and inserted into BamHI-SphI sites of pCP23; Apr (Tcr) | This study |
| pCP0651 | 434 bp fragment spanning fjoh_0651 and its presumed promoter was amplified with primers Fjoh_0651_CP_UP and Fjoh_0651_CP_DOWN and inserted into BamHI-SphI sites of pCP23; Apr (Tcr) | This study |
| pCP0707 | 2283 bp fragment spanning fjoh_0707 was amplified with primers Fjoh_0707_CP_UP and Fjoh_0707_CP_DOWN and inserted into BamHI-SphI sites of pCP23; Apr (Tcr) | This study |
| pCP1448 | 873 bp fragment spanning fjoh_1448 and its presumed promoter was amplified with primers Fjoh_1448_CP_UP and Fjoh_1448_CP_DOWN and inserted into BamHI-SphI sites of pCP23; Apr (Tcr) | This study |
| pCP1449 | 447 bp fragment spanning fjoh_1449 was amplified with primers Fjoh_1449_CP_UP and Fjoh_1449_CP_DOWN and inserted into BamHI-SphI sites of pCP23; Apr (Tcr) | This study |
| pSSK03 | pCP29 carrying porV; Apr (Cfr Emr) | Reference 43 |
| pSSK04 | pCP23 carrying porU; Apr (Tcr) | Reference 43 |
| pSN48 | pCP23 carrying sprA; Apr (Tcr) | Reference 40 |
| pCP2379 | 2,416 bp fragment spanning fjoh_2379 and its presumed promoter was amplified with primers Fjoh_2379_CP_UP and Fjoh_2379_CP_DOWN and inserted into BamHI-SphI sites of pCP23; Apr (Tcr) | This study |
Antibiotic resistances are as follows: streptomycin, Smr; ampicillin, Apr; erythromycin, Emr; tetracycline, Tcr; cefoxitin, Cfr. Antibiotic resistances within parentheses are expressed in F. johnsoniae but not in E. coli, and those without parentheses are expressed in E. coli but not in F. johnsoniae.
Construction of mariner transposon for F. johnsoniae
The F. johnsoniae mariner transposon delivery vector pSAMFjoh_2 was constructed by modifying the resistance cassette and transposase promoter in the B. thetaiotaomicron pSAM-BT42 vector as previously described (27). Briefly, it was modified by replacing the B. thetaiotaomicron transposase promoter with the F. johnsoniae rpoD promoter and adding an F. johnsoniae-compatible erythromycin-resistance (erm) cassette.
Mutant library construction
The transposon mutant library was constructed by conjugating F. johnsoniae UW101 with E. coli-S17-1 λpir containing the MmeI-adapted mariner transposon in pSAM_Fjoh2. Briefly, the donor (E. coli S17-1 λpir) and recipient (F. johnsoniae UW101) strains were grown overnight in LB supplemented with ampicillin (100 µg mL−1) and in casitone yeast extract (CYE) media, respectively. Cultures were aliquoted in a 1:4 donor:recipient ratio, centrifuged, and washed with 1 mL CYE medium. The cells were then resuspended in 100 µL of CYE and spotted on CYE agar. Conjugation took place at 28°C for 12 h. Exconjugants were selected by plating on CYE medium supplemented with erythromycin (100 µg mL−1) and kanamycin (50 µg mL−1) for counterselection against the donor E. coli strain. The plates were incubated at 28°C for 48 h, then ~75,000 clones were pooled into fresh CYE, and aliquots of the mutant library containing 40% glycerol were stored at −80°C until further use.
System for INSeq selection in F. johnsoniae
Overnight cultures of the F. johnsoniae mariner transposon library were made from aliquots of the mutant library in 1/2-strength TSB. Cells were pelleted, washed, and resuspended in 5 mL of 10 mM NaCl. One million cells (OD600 = 0.0008) from the F. johnsoniae mariner transposon library were inoculated in 1 mL of 1/10-strength TSB with and without 0.5 g of sterile sand (50–70 mesh particle size; Sigma-Aldrich) and then incubated without shaking at 20°C for 48 h. The tubes with sand were washed three times with 10 mM NaCl to remove loosely adhered cells and resuspended in 1 mL of 10 mM NaCl. All the tubes were vortexed and sonicated prior to DNA extraction. The QIAGEN DNeasy blood and tissue kit was used to extract DNA, which was used as a template to amplify mariner transposon insertion sites and was ligated with appropriate barcodes as previously described (28). The 125 bp amplicons were adjusted to 1 nM and sequenced using the Illumina HiSeq platform with 2 × 150 bp PE configuration.
INSeq data analysis
Pooled raw sequencing reads were quality processed with fastp (56) using --disable_adapter_trimming and --cut_tail (QC trimming at the 3' end only) parameters and then split into individual fastq files (one for each replicate) by their indexed adapter sequence. Reads were aligned to F. johnsoniae UW101 (accession GCF_000016645.1 ASM1664v1) with bowtie2 (57) using default parameters, and mapped reads were matched to corresponding genes based on F. johnsoniae UW101 coding-sequence annotations (same accession as above). Counts per million were calculated in edgeR (58).
Construction of deletion mutants and complementation plasmids
In-frame deletions in genes FJOH_RS01770, FJOH_RS01835, FJOH_RS03415, porZ, xrtF, FJOH_RS07530, and FJOH_RS12370 were constructed using a gene-deletion strategy that uses dominance of wild-type rpsL on the plasmid pRR51 as a counter-selectable marker over the chromosomal mutant rpsL in F. johnsoniae CJ1827 (21). Briefly, primers were designed with an overlapping sequence of 18 bp to amplify and fuse the regions upstream and downstream of the gene of interest using splicing by overlap extension (SOE) PCR. The 2 kb fusion product was ligated to vector pRR51 and was transformed into E. coli S17 by electroporation. E. coli S17 cells carrying pRR51 with the 2 kb insert were conjugated with F. johnsoniae CJ1827, and exconjugants were selected using CYE supplemented with 100 µg mL−1 erythromycin. Erythromycin-resistant (streptomycin-sensitive) clones were grown overnight in CYE without antibiotics and plated on CYE agar medium supplemented with streptomycin 100 µg mL−1 to select for the loss of plasmid by a second recombination event. Deletion of the target gene in F. johnsoniae CJ1827 was confirmed by PCR amplification using upstream forward and downstream reverse primers and sequencing of the resulting 2 kb product. The wild-type strain used in all mutant comparisons is F. johnsoniae CJ1827 unless otherwise specified.
The plasmids used for complementation were derived from pCP23. The gene of interest with its presumed promoter was amplified using their respective primers. Both the insert and the vector were digested with BamHI-HF and SphI-HF and were ligated into their respective sites in pCP23. The genes that were in a cluster or did not have their own promoter were amplified and cloned into BamHI-HF and SphI-HF sites of pCP23 in the direction of orf1 and were expressed by the vector promoter.
Surface colonization experiments
Surface colonization was assessed using three substrates: sand (0.5 g; 50–70 mesh particle size; Sigma-Aldrich), glass (2 g, 3 mm bead diameter; Fisher Scientific), and polystyrene (24-well plates; CELLTREAT Scientific Products). The wild type (F. johnsoniae CJ1827) and mutants were grown individually for 20 h in 1/2-strength TSB at 28°C; 5 mL of the overnight culture was centrifuged, washed once, and resuspended in 10 mM NaCl. The wild type and mutants (106 CFU mL−1) were inoculated individually in 1/10-strength TSB with and without substrates and grown for 48 h statically at 20°C. The samples were washed three times with 10 mM NaCl to remove loosely adhered cells and were vortexed 30 s, sonicated (Bransonic ultrasonic cleaner 1510R-MT) gently for 2 min with ~50 Hz, and vortexed 30 s to retrieve cells adhered to the substrates. This method was effective in detaching most colonized cells without clumping or lysing them. For polystyrene, each well was scraped 100 times using a sterile toothpick instead of vortexing before and after sonication. The population of cells adhered to the surface was determined by serial dilution and plating on LB containing gentamicin (10 µg mL−1). The soybean root exudate for surface colonization experiments was made as previously described (10). Colonization experiments with planktonic, sand, glass, and polystyrene conditions with 1/10-strength TSB were each performed individually three times with one mutant and wild type at a given time point. We compared each mutant to its wild type by analyzing log10 transformed CFU values using an unpaired two-tailed t-test with Welch’s correction to account for equal variance. Experiments with root exudate were performed three times with all 10 mutants and the wild type. Hence, we log10 transformed the values to achieve equal distribution and performed a one-way ANOVA followed by Dunnett’s test.
Biofilm quantification
F. johnsoniae biofilm formation was quantified using crystal-violet staining. First, 150 µL of a sample containing 107 cells ml−1 of F. johnsoniae strains in 1/10-strength TSB was inoculated into each well in a 96-well plate and incubated without shaking at 20°C for 18 h. The planktonic cells were discarded, and the plate was washed with water. The biofilm was stained with 200 µL of 0.1% crystal-violet solution in 20% methanol for 15 min and washed three times with water. Then 33% acetic acid (200 µL/well) was used to solubilize the crystal violet, and the absorbance was measured at 595 nm.
Zorb microscopy and analysis
An under-oil open microfluidic system (UOMS) device was constructed by chemical vapor-deposition of polydimethylsiloxane (PDMS)-silane onto a glass substrate. The PDMS-silane grafted surface was patterned by O2 plasma treatment to generate patterns of treated and nontreated areas as previously described (59). The device was overlaid with 1.5 mL Fluorinert FC-40 oil, and UOMS spots were seeded with 107 cells (2 µL) of wild type or mutant F. johnsoniae in 1/10-strength TSB. The UOMS device was observed with bright field time-lapse microscopy for 18 h at room temperature as previously described (27).
Time-lapse movies were analyzed using a custom MATLAB code to threshold each frame using Otsu’s segmentation method, then extracting the boundary of each zorb to obtain measures of the zorb area. Zorb speed was quantified using MATLAB code from a previously described tracking method (60) from videos taken at 30-min intervals. Four zorbs were randomly selected and tracked throughout a given time lapse (one in each of the four quadrants to ensure independent zorbs were tracked), with a total of three time-lapse movies analyzed for each strain, resulting in 12 replicates for each strain.
B. cereus motility assay
The expansion of B. cereus colonies in the presence of wild-type or mutant F. johnsoniae was tested using the previously described modified spread-patch method (10). Briefly, B. cereus and F. johnsoniae strains were grown individually for 20 h at 28°C, and 1 mL of culture of each strain was pelleted at 6,000 × g for 6 min and then resuspended in 1 mL of 1× phosphate-buffered saline (PBS). Wild type or mutant F. johnsoniae culture (100 µL) was spread on 1/10-strength TSA plates, which were dried for 2 h at 28°C. B. cereus culture (10 µL) was spotted at the center of each plate and incubated at 28°C. The plates were monitored and imaged on day 5.
Determining fitness of F. johnsoniae in THOR
Fitness of F. johnsoniae in THOR was determined by growing 107 cells mL−1 of F. johnsoniae strains with equal CFU of B. cereus and P. koreensis (1:1:1) statically for 48 h in 1/10-strength TSB with and without sand at 20°C. The samples were processed as described for the surface colonization experiment, and the populations of F. johnsoniae, P. koreensis, and B. cereus were quantified by plating the sample on LB agar supplemented with gentamicin (10 µg mL−1), ampicillin (100 µg mL−1) plus erythromycin (5 µg mL−1), and polymyxin B (5 µg mL−1), respectively.
ACKNOWLEDGMENTS
We thank Mark McBride’s lab for providing strains and plasmids for this work. We also thank the DNA Sequencing Facility at University of Wisconsin-Madison Biotechnology Center for their next-generation sequencing services. We gratefully acknowledge UW-Madison CALS Statistical Consulting for their assistance and thank Dr. Mark McBride and Dr. Edward Ruby for their helpful reviews of a previous version of the manuscript.
This study was supported by the U.S. Army Research Laboratory and the U.S. Army Research Office under contract/grant W911NF1910269, the Vilas Trust, the Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison, the University of Wisconsin Carbone Cancer Center Support Grant NIH P30CA014520, and NIH R01AI59940. A.I.H. was supported by USDA NIFA Grant 2019-2018-08058 (Accession no. 1019190). J.F.N. was supported by an NHGRI training grant from the Genomic Sciences Training Program 5T32HG002760. M.G.C. was supported by USDA NIFA Grant 2020-67012-31772 (Accession no. 1022881). C.L. was supported by NSF EFRI-1136903-EFRI-MKS, NIH R01 CA247479, NIH R01 AI154940, NIH R01 EB010039, NIH R01 CA185251, NIH R01 CA186134, NIH R01 CA181648, NIH R01AI132627, NIH P30CA014520, EPA H-MAP 83573701, and American Cancer Society IRG-15-213-51.
S.M and J.H. designed the research project; S.M performed most of the experiments; A.I.H., J.F.N., M.G.C., C.L., and J.H.S. contributed experimental expertise; S.M., M.G.C., and J.H.S. analyzed data; J.H. and D.J.B. acquired funding; S.M., A.I.H., J.F.N., M.G.C., C.L., and J.H.S. provided methodology; S.M. and J.H. prepared the original draft of the manuscript; S.M., A.I.H., J.F.N., M.G.C., C.L., J.H.S., and D.J.B. reviewed and edited the manuscript.
Footnotes
This article is a direct contribution from Jo Handelsman, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Edward Ruby, Caltech, and Mark McBride, University of Wisconsin-Milwaukee.
Contributor Information
Jo Handelsman, Email: jo.handelsman@wisc.edu.
Kelly T. Hughes, University of Utah, Salt Lake City, Utah, USA
DATA AVAILABILITY
The data are available from the corresponding author. Bacterial strains and mutant libraries are available upon request. The primers used in this study are listed in Table S2. Representative zorb videos can be found in the supplemental material, and the rest will be made available upon request. Insertion sequencing data can be found in the National Center for Biotechnology Information's Sequence Read Archive under BioProject accession PRJNA955258. The files Ama01, Ama02, and Ama03 correspond to three biological replicates in the planktonic condition, and the files Ama04, Ama05, and Ama06 correspond to three biological replicates in the sand condition.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.03428-23.
Sand colonization by deletion mutants complemented with genes of interest.
Sand colonization by F. johnsoniae CJ1827 (wild type) over time.
Growth kinetics of wild type and mutants in 1/10-strength TSB.
Colony spreading of wild type and colonization defective mutants in 1% PY2 agar.
Linear regression analysis of the relationship between polystyrene colonization and biofilm formation.
Characterization of overrepresented mutant FJ0347.
Captions for supplemental figures and movie.
Complete list of sand colonization genes identified using INSeq.
Primers used in this study.
Bacterial zorbing time-lapse videos.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H, Huber DH, Langenheder S, Lennon JT, Martiny JBH, Matulich KL, Schmidt TM, Handelsman J. 2012. Fundamentals of microbial community resistance and resilience. Front Microbiol 3:417. doi: 10.3389/fmicb.2012.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Newman DK, Banfield JF. 2002. Geomicrobiology: how molecular-scale interactions underpin biogeochemical systems. Sci 296:1071–1077. doi: 10.1126/science.1010716 [DOI] [PubMed] [Google Scholar]
- 3. Mendes R, Garbeva P, Raaijmakers JM. 2013. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663. doi: 10.1111/1574-6976.12028 [DOI] [PubMed] [Google Scholar]
- 4. Qu Q, Zhang Z, Peijnenburg WJGM, Liu W, Lu T, Hu B, Chen J, Chen J, Lin Z, Qian H. 2020. Rhizosphere microbiome assembly and its impact on plant growth. J Agric Food Chem 68:5024–5038. doi: 10.1021/acs.jafc.0c00073 [DOI] [PubMed] [Google Scholar]
- 5. Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM. 2014. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J 8:1577–1587. doi: 10.1038/ismej.2014.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akinola SA, Ayangbenro AS, Babalola OO. 2021. The diverse functional genes of maize rhizosphere microbiota assessed using shotgun metagenomics. J Sci Food Agric 101:3193–3201. doi: 10.1002/jsfa.10948 [DOI] [PubMed] [Google Scholar]
- 7. Carvalhais LC, Dennis PG, Tyson GW, Schenk PM. 2013. Rhizosphere metatranscriptomics: challenges and opportunities, p 1137–1144. In Molecular microbial ecology of the rhizosphere. Vol. 2. Wiley Online. [Google Scholar]
- 8. Singh DP, Prabha R, Gupta VK, Verma MK. 2018. Metatranscriptome analysis deciphers multifunctional genes and enzymes linked with the degradation of aromatic compounds and pesticides in the wheat rhizosphere. Front Microbiol 9:1331. doi: 10.3389/fmicb.2018.01331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hayden HL, Savin KW, Wadeson J, Gupta VVSR, Mele PM. 2018. Comparative metatranscriptomics of wheat rhizosphere microbiomes in disease suppressive and non-suppressive soils for rhizoctonia solani AG8. Front Microbiol 9:859. doi: 10.3389/fmicb.2018.00859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lozano GL, Bravo JI, Garavito Diago MF, Park HB, Hurley A, Peterson SB, Stabb EV, Crawford JM, Broderick NA, Handelsman J. 2019. Introducing THOR, a model microbiome for genetic dissection of community behavior. mBio 10:e02846-18. doi: 10.1128/mBio.02846-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peterson SB, Dunn AK, Klimowicz AK, Handelsman J. 2006. Peptidoglycan from Bacillus cereus mediates commensalism with rhizosphere bacteria from the Cytophaga-flavobacterium group. Appl Environ Microbiol 72:5421–5427. doi: 10.1128/AEM.02928-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lozano GL, Park HB, Bravo JI, Armstrong EA, Denu JM, Stabb EV, Broderick NA, Crawford JM, Handelsman J, Elliot MA. 2019. Bacterial analogs of plant tetrahydropyridine alkaloids mediate microbial interactions in a rhizosphere model system. Appl Environ Microbiol 85:e03058-18. doi: 10.1128/AEM.03058-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hurley A, Chevrette MG, Rosario-Meléndez N, Handelsman J, Wright GD. 2022. THOR’s hammer: the antibiotic koreenceine drives gene expression in a model microbial community. mBio 13:e0248621. doi: 10.1128/mbio.02486-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chevrette MG, Thomas CS, Hurley A, Rosario-Meléndez N, Sankaran K, Tu Y, Hall A, Magesh S, Handelsman J. 2022. Microbiome composition modulates secondary metabolism in a multispecies bacterial community. Proc Natl Acad Sci USA 119:e2212930119. doi: 10.1073/pnas.2212930119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McBride MJ, Nakane D. 2015. Flavobacterium gliding motility and the type IX secretion system. Curr Opin Microbiol 28:72–77. doi: 10.1016/j.mib.2015.07.016 [DOI] [PubMed] [Google Scholar]
- 16. Kolton M, Erlacher A, Berg G, Cytryn E. 2016. The Flavobacterium genus in the plant holobiont: ecological, physiological, and applicative insights, p 189–207. In Microbial models: from environmental to industrial sustainability. Springer Singapore. [Google Scholar]
- 17. Johansen JE, Binnerup SJ. 2002. Contribution of Cytophaga-like bacteria to the potential of turnover of carbon, nitrogen, and phosphorus by bacteria in the rhizosphere of barley (Hordeum vulgare L.). Microb Ecol 43:298–306. doi: 10.1007/s00248-002-2006-z [DOI] [PubMed] [Google Scholar]
- 18. Cardinale M, Grube M, Erlacher A, Quehenberger J, Berg G. 2015. Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ Microbiol 17:239–252. doi: 10.1111/1462-2920.12686 [DOI] [PubMed] [Google Scholar]
- 19. Sang MK, Kim KD. 2012. The volatile-producing Flavobacterium johnsoniae strain GSE09 shows biocontrol activity against Phytophthora capsici in pepper. J Appl Microbiol 113:383–398. doi: 10.1111/j.1365-2672.2012.05330.x [DOI] [PubMed] [Google Scholar]
- 20. Soltani A-A, Khavazi K, Asadi-Rahmani H, Omidvari M, Abaszadeh Dahaji P, Mirhoseyni H. 2010. Plant growth promoting characteristics in some Flavobacterium spp. isolated from soils of Iran. J Agric Sci 2:106. doi: 10.5539/jas.v2n4p106 [DOI] [Google Scholar]
- 21. Rhodes RG, Pucker HG, McBride MJ. 2011. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant gliding motility genes remF, remG, remH, and remI. J Bacteriol 193:2418–2428. doi: 10.1128/JB.00117-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hemkemeyer M, Dohrmann AB, Christensen BT, Tebbe CC. 2018. Bacterial preferences for specific soil particle size fractions revealed by community analyses. Front Microbiol 9:149. doi: 10.3389/fmicb.2018.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grinberg M, Orevi T, Kashtan N. 2019. Bacterial surface colonization, preferential attachment and fitness under periodic stress. PLoS Comput Biol 15:e1006815. doi: 10.1371/journal.pcbi.1006815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang W-S, van de Mortel M, Nielsen L, Nino de Guzman G, Li X, Halverson LJ. 2007. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J Bacteriol 189:8290–8299. doi: 10.1128/JB.00727-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santos ALSD, Galdino ACM, Mello TP de, Ramos L de S, Branquinha MH, Bolognese AM, Columbano Neto J, Roudbary M. 2018. What are the advantages of living in a community? a microbial biofilm perspective! Mem Inst Oswaldo Cruz 113:e180212. doi: 10.1590/0074-02760180212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. doi: 10.1038/nrmicro.2016.94 [DOI] [PubMed] [Google Scholar]
- 27. Li C, Hurley A, Hu W, Warrick JW, Lozano GL, Ayuso JM, Pan W, Handelsman J, Beebe DJ. 2021. Social motility of biofilm-like microcolonies in a gliding bacterium. Nat Commun 12:5700. doi: 10.1038/s41467-021-25408-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goodman AL, Wu M, Gordon JI. 2011. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Protoc 6:1969–1980. doi: 10.1038/nprot.2011.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cole BJ, Feltcher ME, Waters RJ, Wetmore KM, Mucyn TS, Ryan EM, Wang G, Ul-Hasan S, McDonald M, Yoshikuni Y, Malmstrom RR, Deutschbauer AM, Dangl JL, Visel A. 2017. Genome-wide identification of bacterial plant colonization genes. PLoS Biol 15:e2002860. doi: 10.1371/journal.pbio.2002860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sivakumar R, Ranjani J, Vishnu US, Jayashree S, Lozano GL, Miles J, Broderick NA, Guan C, Gunasekaran P, Handelsman J, Rajendhran J. 2019. Evaluation of INSeq to identify genes essential for Pseudomonas aeruginosa PGPR2 corn root colonization. G3: Genes, Genomes, Genetics 9:651–661. doi: 10.1534/g3.118.200928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Torres M, Jiquel A, Jeanne E, Naquin D, Dessaux Y, Faure D. 2022. Agrobacterium tumefaciens fitness genes involved in the colonization of plant tumors and roots. New Phytol 233:905–918. doi: 10.1111/nph.17810 [DOI] [PubMed] [Google Scholar]
- 33. Wallner A, Busset N, Lachat J, Guigard L, King E, Rimbault I, Mergaert P, Béna G, Moulin L. 2022. Differential genetic strategies of Burkholderia vietnamiensis and Paraburkholderia kururiensis for root colonization of Oryza sativa subsp.japonica and O. sativa subsp. indica, as revealed by transposon mutagenesis sequencing. Appl Environ Microbiol 88:e0064222. doi: 10.1128/aem.00642-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. do Amaral FP, Tuleski TR, Pankievicz VCS, Melnyk RA, Arkin AP, Griffitts J, Tadra-Sfeir MZ, Maltempi de Souza E, Deutschbauer A, Monteiro RA, Stacey G. 2020. Diverse bacterial genes modulate plant root association by beneficial bacteria. mBio 11:e03078-20. doi: 10.1128/mBio.03078-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Banagan BL, Wertheim BM, Roth MJS, Caslake LF. 2010. Microbial strengthening of loose sand. Lett Appl Microbiol 51:138–142. doi: 10.1111/j.1472-765X.2010.02872.x [DOI] [PubMed] [Google Scholar]
- 36. Capdevila S, Martínez-Granero FM, Sánchez-Contreras M, Rivilla R, Martín M. 2004. Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiol (Reading) 150:3889–3897. doi: 10.1099/mic.0.27362-0 [DOI] [PubMed] [Google Scholar]
- 37. Cooley MB, Miller WG, Mandrell RE. 2003. Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl Environ Microbiol 69:4915–4926. doi: 10.1128/AEM.69.8.4915-4926.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rossi FA, Medeot DB, Liaudat JP, Pistorio M, Jofré E. 2016. In Azospirillum brasilense, mutations in flmA or flmB genes affect polar flagellum assembly, surface polysaccharides, and attachment to maize roots. Microbiol Res 190:55–62. doi: 10.1016/j.micres.2016.05.006 [DOI] [PubMed] [Google Scholar]
- 39. Dörr J, Hurek T, Reinhold-Hurek B. 1998. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol Microbiol 30:7–17. doi: 10.1046/j.1365-2958.1998.01010.x [DOI] [PubMed] [Google Scholar]
- 40. Nelson SS, Glocka PP, Agarwal S, Grimm DP, McBride MJ. 2007. Flavobacterium johnsoniae SprA is a cell surface protein involved in gliding motility. J Bacteriol 189:7145–7150. doi: 10.1128/JB.00892-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rhodes RG, Samarasam MN, Van Groll EJ, McBride MJ. 2011. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion. J Bacteriol 193:5322–5327. doi: 10.1128/JB.05480-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. 2013. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J Bacteriol 195:3201–3212. doi: 10.1128/JB.00333-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kharade SS, McBride MJ. 2015. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J Bacteriol 197:147–158. doi: 10.1128/JB.02085-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kolton M, Frenkel O, Elad Y, Cytryn E. 2014. Potential role of flavobacterial gliding-motility and type IX secretion system complex in root colonization and plant defense. Mol Plant Microbe Interact 27:1005–1013. doi: 10.1094/MPMI-03-14-0067-R [DOI] [PubMed] [Google Scholar]
- 45. Eckroat TJ, Greguske C, Hunnicutt DW. 2021. The type 9 secretion system is required for Flavobacterium johnsoniae biofilm formation. Front Microbiol 12:660887. doi: 10.3389/fmicb.2021.660887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lauber F, Deme JC, Lea SM, Berks BC. 2018. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564:77–82. doi: 10.1038/s41586-018-0693-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Glew MD, Veith PD, Chen D, Gorasia DG, Peng B, Reynolds EC. 2017. PorV is an outer membrane shuttle protein for the type IX secretion system. Sci Rep 7:8790. doi: 10.1038/s41598-017-09412-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gorasia DG, Veith PD, Reynolds EC. 2020. The type IX secretion system: advances in structure, function and organisation. Microorg 8:1173. doi: 10.3390/microorganisms8081173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haft DH, Paulsen IT, Ward N, Selengut JD. 2006. Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol 4:29. doi: 10.1186/1741-7007-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haft DH, Payne SH, Selengut JD. 2012. Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J Bacteriol 194:36–48. doi: 10.1128/JB.06026-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, Staroscik AM, Hoover TR, Cheng YQ, Stein JL. 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol 75:6864–6875. doi: 10.1128/AEM.01495-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krasowska A, Sigler K. 2014. How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol 4:112. doi: 10.3389/fcimb.2014.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Giaouris E, Chapot-Chartier MP, Briandet R. 2009. Surface physicochemical analysis of natural Lactococcus lactis strains reveals the existence of hydrophobic and low charged strains with altered adhesive properties. Int J Food Microbiol 131:2–9. doi: 10.1016/j.ijfoodmicro.2008.09.006 [DOI] [PubMed] [Google Scholar]
- 54. Kochkodan V, Tsarenko S, Potapchenko N, Kosinova V, Goncharuk V. 2008. Adhesion of microorganisms to polymer membranes: a photobactericidal effect of surface treatment with TiO2. Desalin 220:380–385. doi: 10.1016/j.desal.2007.01.042 [DOI] [Google Scholar]
- 55. Agarwal S, Hunnicutt DW, McBride MJ. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci USA 94:12139–12144. doi: 10.1073/pnas.94.22.12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li C, Hite Z, Warrick JW, Li J, Geller SH, Trantow VG, McClean MN, Beebe DJ. 2020. Under oil open-channel microfluidics empowered by exclusive liquid repellency. Sci Adv 6:eaay9919. doi: 10.1126/sciadv.aay9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Crocker JC, Grier DG. 1996. Methods of digital video microscopy for colloidal studies. J Colloid Interface Sci 179:298–310. doi: 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sand colonization by deletion mutants complemented with genes of interest.
Sand colonization by F. johnsoniae CJ1827 (wild type) over time.
Growth kinetics of wild type and mutants in 1/10-strength TSB.
Colony spreading of wild type and colonization defective mutants in 1% PY2 agar.
Linear regression analysis of the relationship between polystyrene colonization and biofilm formation.
Characterization of overrepresented mutant FJ0347.
Captions for supplemental figures and movie.
Complete list of sand colonization genes identified using INSeq.
Primers used in this study.
Bacterial zorbing time-lapse videos.
Data Availability Statement
The data are available from the corresponding author. Bacterial strains and mutant libraries are available upon request. The primers used in this study are listed in Table S2. Representative zorb videos can be found in the supplemental material, and the rest will be made available upon request. Insertion sequencing data can be found in the National Center for Biotechnology Information's Sequence Read Archive under BioProject accession PRJNA955258. The files Ama01, Ama02, and Ama03 correspond to three biological replicates in the planktonic condition, and the files Ama04, Ama05, and Ama06 correspond to three biological replicates in the sand condition.