Abstract
Japanese encephalitis virus (JEV), a mosquito-borne flavivirus, is a zoonotic pathogen that is prevalent in some Southeast Asian countries and causes acute encephalitis in humans. To evaluate the potential application of gene immunization to JEV infection, we characterized the immune responses from mice intramuscularly injected with plasmid DNA encoding JEV glycoproteins, including the precursor membrane (prM) plus envelope (E) proteins and the nonstructural protein NS1. When injected with the plasmid expressing prM plus E, 70% of the immunized mice survived after a lethal JEV challenge, whereas when immunized with the plasmid expressing NS1, 90% of the mice survived after a lethal challenge. As a control, the mice immunized with the DNA vector pcDNA3 showed a low level (40%) of protection, suggesting a nonspecific adjuvant effect of the plasmid DNA. Despite having no detectable neutralizing activity, the NS1 immunization elicited a strong antibody response exhibiting cytolytic activity against JEV-infected cells in a complement-dependent manner. By contrast, immunization with a construct expressing a longer NS1 protein (NS1′), containing an extra 60-amino-acid portion from the N terminus of NS2A, failed to protect mice against a lethal challenge. Biochemical analyses revealed that when individually expressed, NS1 but not NS1′ could be readily secreted as a homodimer in large quantity and could also be efficiently expressed on the cell surface. Interestingly, when NS1 and NS1′ coexisted in cells, the level of NS1 cell surface expression was much lower than that in cells expressing NS1 alone. These data imply that the presence of partial NS2A might have a negative influence on an NS1-based DNA vaccine. The results herein clearly illustrate that immunization with DNA expressing NS1 alone is sufficient to protect mice against a lethal JEV challenge.
Japanese encephalitis virus (JEV), a member of the family Flaviviridae, is transmitted to humans by infected mosquitoes and causes acute encephalitis with fatality rates ranging from 20% to as high as 50% (3, 47). The JEV genome is a single-stranded, positive-sense RNA of approximately 11 kb which contains a single open reading frame encoding a polyprotein. In infected cells, this viral polyprotein is proteolytically cleaved into at least 11 proteins. The virus structural proteins, including the capsid (C), membrane (M; precursor M [prM]), and envelope (E), are encoded by the 5′ one-third of the open reading frame, and the nonstructural (NS) proteins, designated NS1 through NS5, are encoded in the remainder (reviewed in references 5 and 41). NS1 has a predicted molecular mass of 40 kDa; because there are N-linked carbohydrate chains at positions 130 and 207, the actual molecular mass of NS1 detected in JEV-infected cells is approximately 46 kDa. The proteolytic cleavage of E-NS1 ensues as a result of the translocation of NS1 into the lumen of the endoplasmic reticulum ER, and the cleavage of NS1-2A is thought to take place in the lumen of the vesicular compartments (5). An additional NS1-2A-related protein (named NS1′), with a molecular mass of 53 kDa, is often observed in JEV-infected cells (30) and is presumably generated by an unknown protease which recognizes an alternative cleavage site within NS2A (5).
Three of the flavivirus proteins (prM, E, and NS1) are glycosylated and have been reported to be capable of inducing protective immunity (reviewed in reference 34). The E protein, a major structural protein of flavivirus virions, appears to play a dominant role in the generation of neutralizing antibodies and the induction of a protective immune response (2, 20). The prM protein is part of the immature virions, and at the late stages of infection, its proteolytic cleavage to M protein generates mature virions. However, in certain instances this prM cleavage may not be complete, thus allowing the prM protein to be an additional target on virions for neutralizing antibodies (1). In addition to the structural proteins, the flavivirus NS1, which is expressed on the surface of and secreted extracellularly from infected cells (reviewed in reference 41), not only is able to elicit an immune response during the course of flavivirus infections but also confers protection in experimental animals (reviewed in reference 34). This protective phenomenon seems to be dependent on the Fc portion of antibodies, since such NS1-specific antibodies kill infected target cells in a complement-dependent manner (44, 45). One proposed advantage for employing flavivirus NS1 as a subunit vaccine (14) rather than conventional killed JEV vaccines is that NS1 is able to elicit protective immunity in the host without adverse effects involving antibody-dependent enhancement (17).
Effective vaccines to control JEV have been successfully developed by formalin inactivation of JEV cultured in mouse brains. Since the inception of vaccination programs for humans, cases of Japanese encephalitis in many areas such as Japan, Korea, and Taiwan have been greatly reduced (19, 36, 37). However, one of the major problems associated with the use of inactivated JEV vaccines is the lack of long-term immunity (24). To obtain effective protection, multiple boosts of inactivated JEV vaccine are routinely required, making the vaccination program costly; in addition, repeated immunizations with killed vaccines prepared from mouse brains may cause hypersensitivity reactions in vaccinees. Therefore the World Health Organization has recently designated JEV vaccine a high-priority target for further research and development.
DNA-based vaccines have recently been shown to induce protective immune responses against several viral agents, such as human immunodeficiency virus (49, 50), bovine and human herpesviruses (9, 32), hepatitis B virus (10), influenza virus (13, 40, 42, 46), rabies virus (51), hepatitis C virus (25, 28), measles virus (4), St. Louis encephalitis virus (38), and murine cytomegalovirus (15). Endogenous expression of antigen from DNA introduced into host cells leads to the production of structurally and conformationally relevant molecules of the antigen that are able to be presented by major histocompatibility complex classes I and II to activate specific immunity. This cellular process of antigen presentation, in some aspects, resembles the natural course of viral infection. Indeed, in immunized hosts with expressible virus DNA, a broad spectrum of immune responses have been observed, including antibodies, cytotoxic T cells, and helper T cells, as well as protections against challenge with various viruses. The possession of such capacity by the vaccines appears to be particularly crucial for developing DNA vaccines to control viral diseases.
To assess the feasibility of DNA vaccination for JEV infection, in this study we characterized the immunogenicity and the protective capability of plasmid DNA expressing either prM plus E, NS1, or NS1′ with mice as an animal model. Our results revealed that the plasmid expressing NS1, but not its longer derivative NS1′, could induce protective immunity as effectively as could the construct expressing prM plus E. However, our data suggests that the protective mechanisms involved in the NS1 and prME constructs appear to be different. In addition, as a result of comparison of the biochemical properties between recombinant NS1 and NS1′ expressed in cultured cells, we suggest that the ability of JEV glycoproteins to be readily secreted from and effectively expressed on the surface of cells appears to be a pivotal determinant for their ability to act as effective and potent immunogens. The possible role of NS2A in down-regulating the efficacy of an NS1-based gene vaccine is also discussed. The present results not only suggest that DNA immunization can be a potential vaccine approach but also illustrate that gene immunization with JEV NS1 alone is adequate to protect mice against JEV infection.
MATERIALS AND METHODS
Viruses and cell lines.
The Taiwanese local JEV strain NT109 (7), isolated from infected Culex tritaeniorhynchus mosquitoes in 1985, and strain NT113, isolated from infected Culex annulus mosquitoes in 1985, were used for the cloning of JEV genes. A neurovirulent JEV strain, RP-9 (7), was used in the challenge experiments. Virus propagation was carried out with BHK-21 cells in RPMI 1640 medium containing 2% fetal calf serum (GIBCO). Virus titers were determined by a plaque-forming assay on BHK-21 cells.
Construction of plasmids expressing JEV proteins.
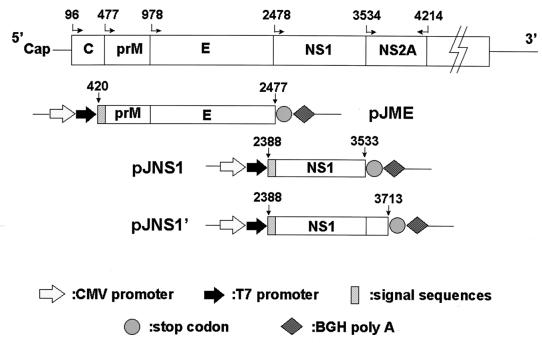
For the expression of JEV prM and E proteins, a cDNA fragment of 2,058 bp was amplified from strain NT113 by reverse transcription-PCR (6) with the primer set 5′-GCGGATCCAGAAGGCTCAATCATGTGGCT-3′ (positive sense) and 5′-GACGCAAGCTTGCTAAGCATGCACATTGGTCG-3′ (negative sense), which hybridize to nucleotides (nt) 420 to 439 and 2461 to 2477, respectively. This fragment comprised the coding regions for both prM and E, as well as the region encoding a 15-amino-acid signal peptide derived from the C terminus of C protein. The cDNA fragment was cloned into a PCR cloning vector, pCRII (Invitrogen), and then the EcoRI insert fragment was excised and subcloned into a eucaryotic expression vector, pcDNA3 (Invitrogen), in which prME is under the control of the enhancer-promoter sequences of the immediately-early gene of human cytomegalovirus. The resulting plasmid construct was named pJME (Fig. 1).
FIG. 1.
Schematic diagram of plasmid constructs expressing various JEV glycoproteins in the mammalian expression vector. The numbers with straight or bent arrows are the nucleotide positions on the JEV genome.
JEV NS1 and NS1′ cDNA fragments were generated by reverse transcription-PCR amplification of the genomic RNA of strain NT109. Two different 3′ primers were used for the NS1 or NS1′ constructions in the first-strand cDNA synthesis step: 5′-GCGGATCCTAAGCATCAACCTGTGA-3′, complementary to nt 3519 to 3533 in the NS1 region, and 5′-CGCTATAGCACCACATACC-3′, complementary to nt 3700 to 3713 in the NS1′ region. The same 5′ primer was used for both constructions during the PCR step: 5′-GCACCATGGGCGTCAACGCA-3′, hybridizing to nt 2388 to 2402. The NS1 cDNA fragment was first cloned to a ddT-tailed, EcoRV-digested pBluescript (Stratagene) generated by a published method (29), and the correct insert was then subcloned to an expression vector pcDNA3 (Invitrogen). The NS1′ cDNA fragment was directly cloned into an expression vector, pCR3 (Invitrogen), by the TA cloning method. The resulting constructs were named pJNS1 and pJNS1′ (Fig. 1); they both encode a so-called signal peptide of 30 amino acids derived from the C terminus of E protein. Plasmid pJNS1′ is almost identical to pJNS1, except that the coding sequence of the former was extended into the NS2A region with 60 amino acids.
RIP.
Briefly, for radioimmunoprecipitation (RIP), BHK-21 cells were infected with virus at a multiplicity of infection (MOI) of 5. At 24 h postinfection, the medium was removed, replaced with warm methionine (Met)- and cystein (Cys)-free RPMI 1640 medium containing 100 μCi of [35S]Pro-mix (Amersham) per ml plus 2% dialyzed fetal bovine serum (GIBCO), and incubated for 2 h at 37°C. The culture fluids were removed, and the cell layers were rinsed with ice-cold phosphate-buffered saline (PBS) and harvested in lysis buffer (1% Nonidet P-40, 150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 1 mM EDTA) containing a cocktail of protease inhibitors (20 μg of phenylmethylsulfonyl fluoride per ml, 2 μg of leupeptin per ml, 2 μg of aprotinin per ml). As positive controls, ascitic fluids containing monoclonal antibody (MAb) specific for the JEV NS1 proteins (6) were used in RIP. The tested serum samples were first incubated with a mixture of protein A-Sepharose and protein G-Sepharose (Pharmacia) at room temperature for 1 h; the 35S-labeled cell lysates were then added and incubated at room temperature for another 1 h. The resulting immune complexes were washed three times with RIPA buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 1% sodium deoxycholate), analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and fluorographed at −70°C.
Western immunoblot analysis.
Cell lysates were mixed with an equal volume of sample buffer without 2-mercaptoethanol, treated by boiling or left untreated, separated by SDS-PAGE, and transferred to a nitrocellulose membrane (Hybond-C Super; Amersham). The nonspecific antibody-binding sites were blocked with 5% skim milk in PBS, and the membranes were reacted with anti-JEV MAb (6). The resulting blot was treated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (Cappel) and developed either with enhanced chemiluminescence reagent (Amersham) or by alkaline phosphatase-conjugated secondary antibody treatment with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP).
Viral proteins expressed by recombinant plasmids.
To verify the viral protein expressed by pJME, the plasmid DNA was transfected into COS-7 cells by the calcium phosphate-DNA coprecipitation method (16). At 44 h postinfection, cytoplasmic extracts were prepared as previously described (8). The samples were analyzed by SDS-PAGE (10% polyacrylamide) under denaturing conditions, transferred to a nitrocellulose membrane, and blotted with MAbs specific for JEV E protein as described above. The specific binding of recombinant E protein to MAbs was revealed by an alkaline phosphatase-conjugated secondary antibody with the enzyme substrates NBT and BCIP. To check for NS1 expression, a transient-expression system involving recombinant vaccinia virus expressing T7 polymerase (vTF7-3) was used. BHK-21 cells were infected with vTF7-3 at MOI 5, and at 1 h postinfection the infected cells were transfected with Lipofectamine (Bethesda Research Laboratories) mixed with either pJNS1 or pJNS1′, both comprising a T7 promoter in front of the genes to be expressed (see Fig. 1A). After overnight incubation, the resulting cells were labeled with [35S]Pro-mix (Amersham), and the cell lysates were isolated and immunoprecipitated as described above. For endoglycosidase F (endo-F) digestion, the immunoprecipitated proteins were boiled in endo-F boiling buffer (50 mM sodium phosphate [pH 7.5], 0.5% SDS, 1% 2-mercaptoethanol) and the resulting proteins were incubated overnight at 37°C with a equal volume of the endo-F incubation buffer (50 mM sodium phosphate [pH 7.5], 2% Nonidet P-40, 0.2% SDS, 1% 2-mercaptoethanol, 25 mM EDTA) with or without endo-F (Boehringer Mannheim).
Indirect immunofluorescence staining of NS1-expressing cells.
For intracellular staining, the cells were fixed in acetone-methanol (1:1) solution for 3 min and then reacted with MAb against JEV NS1 (6). For cell surface staining, the unfixed cells were also reacted with MAb against JEV NS1 at 4°C for 1 h. After being washed with PBS, the cells were further treated with goat anti-mouse fluorescein-conjugated secondary antibody (Cappel), and the resulting cells were examined under a Leitz fluorescent microscope.
Immunization and challenge in mice.
Plasmid DNA was prepared by the Midiprep procedure (Qiagen) as specified by the manufacturer. Groups of 3- to 4-week-old female ICR mice were immunized with 80 μg of recombinant pJME, pJNS1, pJNS1′, or pcDNA3 DNA as a control. DNA in PBS or PBS alone was intramuscularly (i.m.) injected into the thighs of the mice; the mice were subsequently given booster doses by the same method twice at 2-week intervals. At 2 weeks after the final booster dose, each mouse was intraperitoneally (i.p.) challenged with 2 × 107 PFU of a neurovirulent JEV strain RP-9 (about 10 50% lethal doses for 10-week-old ICR mice) in 300 μl of PBS and simultaneously injected intracerebrally (i.c.) with 30 μl of PBS alone into the right hemisphere of the brain (i.p. + i.c. route) (26). To ensure the depth of each i.c. injection, we used 27-gauge one-stop needles (Top Injection Needle, Tokyo, Japan). Mouse mortality was monitored daily for 3 weeks.
Antibody-dependent complement-mediated cytolytic assay.
A modified complement-mediated cytolysis assay was performed to determine the ability of pooled mouse sera to lyse JEV-infected cells. Briefly, target BHK-21 cells grown in 96-well microtiter plates (Corning) were infected with JEV (RP-9) at an MOI of 5 overnight. Pooled sera obtained from the immunized mice were preheated at 56°C for 30 min to inactivate complement, serially diluted, and then incubated with various dilutions of guinea pig complement (Cappel) at 37°C for 60 min before being added to the target cells. After a 4-h incubation, cytolysis was measured by the release of lactate dehydrogenase (LDH), a cytoplasmic enzyme, with a commercial kit (cytotoxicity detection kit; Boehringer Mannheim) as specified by the manufacturer. Briefly, the culture supernatants were clarified by centrifugation, incubated with the reaction mixture (diaphorase/NAD+ and tetrazolium salt INT-sodium lactate) at room temperature for about 30 min, and then read by an enzyme-linked immunosorbent assay reader at 490 nm (microplate reader; Molecular Devices). RPMI 1640 without phenol red (GIBCO) was used throughout this experiment to decrease the background adsorbance. The maximum LDH release was determined from the wells (three wells for each experiment) containing the target cells lysed with 1% Triton X-100; spontaneous LDH release was determined from the wells containing the target cells and medium only. The percent specific lysis was calculated as follows: 100 × (experimental LDH release − spontaneous LDH release)/(maximum LDH release − spontaneous LDH release).
Flow cytometry.
For cell surface staining, the cells were harvested from the culture plates with suspension buffer (PBS containing 0.5 mM EDTA [pH 8.0]) and the cell suspensions were blocked with fluorescence-activated cell sorter (FACS) buffer (1% bovine serum albumin and 0.02% sodium azide in PBS) at 4°C for 30 min. JEV NS1-specific MAb (1:1,000 dilution) (6), a primary antibody, and fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin (1:1,000 dilution) (Cappel), a secondary antibody, were added consecutively to the cell suspensions, which were then incubated at 4°C for 30 min. Cell staining was analyzed with a FACS Calibur (Becton Dickinson) and CELLQuest software.
RESULTS
Construction of plasmids expressing JEV proteins.
By using recombinant vaccinia viruses that specified the synthesis of JEV glycoproteins, the coexpression of prM and E has previously been shown to protect mice from a lethal challenge with JEV (21, 52). In addition to a possible role in eliciting neutralizing antibodies, prM has been implicated in the stabilization of E protein by prevention of its low-pH-induced rearrangement during transport through the acidic compartments of the trans-Golgi network (18). Therefore, to investigate whether the structural glycoproteins of JEV (prM and E) could be used in gene immunization, we constructed a pcDNA3-based plasmid expressing prM plus E under the control of the cytomegalovirus promoter. To obtain proper modifications in the ER-Golgi apparatus complex, a sequence containing a 15-amino-acid signal peptide derived from the C terminus of JEV C protein was added in front of prM plus E, and the resulting construct was named pJME (Fig. 1). To verify the viral protein expression from pJME, the plasmid DNA was transfected into COS-7 cells and cell extracts from transfectants were immunoblotted with a MAb against JEV E protein (6). As shown in Fig. 2A, a single band with a molecular mass of 56 kDa was detected in the pJME-transfected (lane 2) but not in the mock-transfected (lane 1) cell lysate, which correlated with the expected molecular mass of JEV E protein. Moreover, as analyzed by endo-F cleavage, the E protein expressed by pJME appeared to be properly glycosylated, as evidenced by its comigration with the authentic viral E protein (data not shown). The absence of the full-length prME protein in Fig. 2A was probably because the cleavage of prME was both rapid and complete in the ER. These data indicate that pJME could express E glycoprotein indistinguishable from the one expressed by JEV-infected cells. However, it remains unclear why, although abundant E proteins could be found in the lysate of pJME-transfected cells, only small amounts of secreted E proteins were detected in the culture supernatant (data not shown).
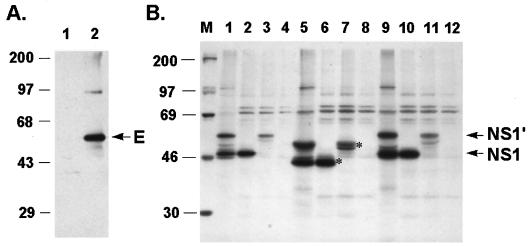
FIG. 2.
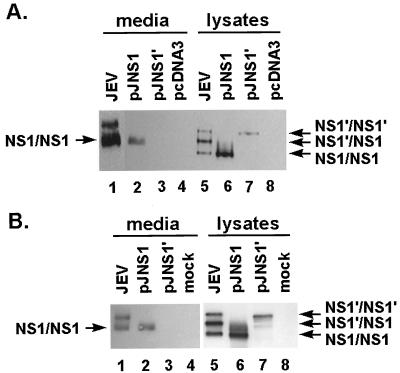
JEV glycoproteins expressed from recombinant plasmids in cell cultures. (A) JEV E protein expressed by pJME. Cell lysates from pJME-transfected (lane 2) or mock-transfected (lane 1) COS-7 were immunoblotted with MAb against JEV E protein as described in Materials and Methods. The position of the E protein is indicated by the arrow. Numbers on the left figure are the molecular mass standards in kilodaltons. (B) JEV NS1 proteins expressed by pJNS1 and pJNS1′ were immunoprecipitated with anti-NS1 MAb and analyzed by SDS-PAGE. Cell lysates were isolated from BHK-21 cells transfected with pJNS1 (lanes 2, 6, and 10), pJNS1′ (lanes 3, 7, and 11), or vector pcDNA3 (lanes 4, 8, and 12); as a control, lysates purified from JEV-infected BHK-21 cells (lanes 1, 5, and 9) were also included. The expression profiles of viral proteins analyzed by RIP are shown in lanes 1 to 4. For endo-F analysis, cell extracts either from JEV-infected cells or from transfected cells were digested with endo-F (lanes 5 to 8), or treated with buffer alone (lane 9 to 12) at 37°C overnight. Lane M contains the molecular weight standards (given in kilodaltons on the left). The positions of the glycosylated NS1 and NS1′ proteins are indicated by the arrows on the right. The asterisks denote the endo-F-sensitive species of NS1 glycoproteins.
To determine whether the JEV nonstructural protein NS1 could be used for DNA immunization, we constructed plasmids expressing either NS1 or NS1′. Similar to pJME, a sequence consisting of a 30-amino-acid signal peptide derived from the C terminus of JEV E was also situated immediately in front of both NS1 and NS1′ constructs, as shown in Fig. 1, and the resulting plasmids were called pJNS1 and pJNS1′, respectively. Plasmid pJNS1′ is identical to pJNS1, except that the coding sequence of the former was extended from the NS1 region into the NS2A region (Fig. 1). The NS1 expression patterns of both constructs were examined by a transient-expression system in BHK-21 cells with recombinant vaccinia virus expressing T7 polymerase to drive the gene tested. From the results shown in Fig. 2B, obtained by immunoprecipitation with the MAb against JEV NS1 (6), protein bands, comigrating with either the authentic NS1 or NS1′ (lane 1) from infected cell lysates, were detected in both the pJNS1-transfected (lane 2) and pJNS1′-transfected (lane 3) cell lysates. No corresponding precipitates could be observed in cells transfected with vector pcDNA3 alone (Fig. 2B, lane 4). pJNS1′ appeared to be able to express only NS1′ but not NS1 (lane 3), implying that NS1′ expressed by this construct was not the precursor of NS1. Alternatively, since the exact C terminus of JEV NS1′ protein is unknown, the cleavage requirements for converting NS1′ to NS1 might not be completely included in this arbitrary pJNS1′ construct.
Both NS1 and NS1′ proteins contain two predicted N-linked glycosylation sites at amino acid positions 130 and 207. We next determined whether recombinant NS1 and NS1′ had undergone proper glycosylation during biosynthesis. After complete digestion by endo-F, the sizes of both NS1 and NS1′, expressed either by JEV infection or by recombinant-plasmid transfection, decreased by approximately 4 to 6 kDa (Fig. 2B, compare lane 1 with lane 5, lane 2 with lane 6, and lane 3 with lane 7). As a negative control, no size alteration could be seen when the samples were treated with just buffer only (lanes 9 to 12). These data confirm the previous finding (12) that NS1 can be targeted to the ER compartment, where it is properly glycosylated in the absence of other viral proteins. Together, these results illustrate that the recombinant NS1 constructs were able to express glycosylated NS1 proteins which were indistinguishable from the ones derived from natural JEV infection.
DNA immunization and JEV challenge in the outbred-mouse model.
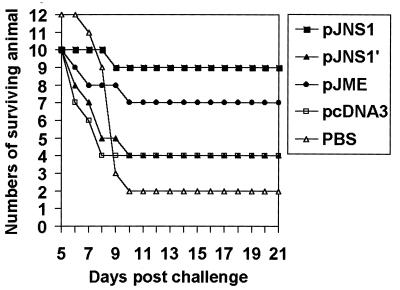
To assess the efficacy of DNA immunization against JEV infection, groups of 3- to 4-week-old female outbred ICR mice were immunized with either recombinant constructs, control vector DNA, or PBS buffer alone. Two weeks after the second immunizing boost, the mice were challenged with a neurovirulent JEV strain, RP-9 (26). The mice were checked daily for survival, and the results are shown in Fig. 3. The survival rates for the different experimental groups at 21 days postchallenge and the results of the statistical analysis are summarized in Table 1. The greatest survival was observed in the group of mice immunized with pJNS1, in which 9 of the 10 tested mice survived the lethal JEV challenge (P < 0.001). The second best survival rate was observed in the pJME-immunized group, whose survival rate was 70% (P < 0.01) (Fig. 3; Table 1). In contrast, pJNS1′, which expressed a longer form of NS1, failed to confer a comparable level of protection on the immunized mice to that conferred by its counterpart pJNS1; instead, pJNS1′, like its parental vector pcDNA3, demonstrated only a 40% survival rate (P < 0.25) (Fig. 3; Table 1). The low level of protective capability observed with pcDNA3, compared to the 17% survival rate of the group of mice which received only PBS buffer, was most probably due to the nonspecific immunostimulatory effect resulting from the bacterial DNA with CpG motifs (23). These results clearly illustrate that immunization with the genes encoding either JEV structural or nonstructural proteins could protect mice from a lethal challenge.
FIG. 3.
Survival of DNA-immunized mice after lethal JEV challenge. Groups of 3- to 4-week-old female ICR mice were immunized with the indicated plasmids or buffer alone and later lethally challenged with JEV as described in Materials and Methods. The JEV-infected mice were monitored daily for survival up to 21 days postchallenge.
TABLE 1.
Survival rate of ICR mice immunized with plasmid DNA expressing JEV glycoproteins after a lethal challengea
| DNA construct | No. of surviving animals/no. of immunized animals (% survival) | Pb |
|---|---|---|
| pJME | 7/10 (70%) | <0.01c |
| pJNS1 | 9/10 (90%) | <0.001d |
| pJNS1′ | 4/10 (40%) | <0.25 |
| pcDNA3 | 4/10 (40%) | <0.25 |
| None (PBS buffer) | 2/12 (17%) |
Groups of 3- to 4-week-old female ICR mice were immunized with plasmid DNA or PBS alone as described in Materials and Methods and monitored daily for 21 days. The survival rate at 21 days p.i. was determined as 100 × (number of survivors)/(number of immunized mice).
P values were obtained by the chi-square test when comparing the survival rate of each group with that of the PBS control group.
P < 0.25 was achieved when comparing the surviving rate of the pJME-immunized group with that of the pcDNA3 group.
P < 0.025 was achieved when comparing the surviving rate of the pJNS1-immunized group with that of the pcDNA3 group.
Characterization of JEV-specific antibody response in immunized mice.
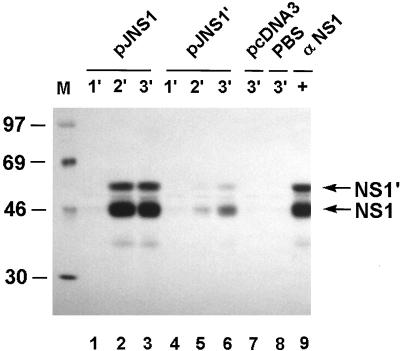
To study the JEV-specific antibody responses elicited by DNA immunization described above, pooled sera from groups of immunized mice were collected and tested by immunoprecipitation with 35S-labeled JEV-infected cell lysates as the antigen. As shown in Fig. 4, in sera from the mice injected with pJNS1, antibodies readily precipitated both NS1 and NS1′ after only one boost (lanes 1 to 3) whereas antisera from the pJNS1′-immunized mice appeared to only weakly precipitate NS1 and NS1′ even after a second boost (lanes 4 to 6). No NS1-specific antibody responses could be detected in the negative controls immunized with either pcDNA3 or PBS buffer only (lanes 7 and 8). The extent of the antibody responses stimulated by pJNS1 and pJNS1′ appeared to correlate well with their protective capabilities, as shown in Table 1 and Fig. 3; that is, pJNS1, rather than pJNS1′, not only elicited a good humoral immunity but also conferred on immunized mice sufficient protection from a subsequent lethal challenge. However, as expected, pJNS1-mediated protection did not appear to be the result of direct neutralization of JEV, since only a basal level of activity (less than a 20-fold serum dilution) could be detected in the sera examined by the 70% plaque reduction neutralization test (data not shown). In pJME-injected mice, we failed to detect any JEV-specific antibody response by immunoprecipitation (data not shown). However, a low titer (40-fold serum dilution) in the plaque reduction neutralization test could be detected (data not shown). Whether this low level of neutralization was responsible for the protection observed in the pJME-immunized group or whether other immune system mechanisms, such as cellular immunity, contributed to this protection remains unclear. The discrepancy between the pJME immunization results and the previously published ones (21, 22, 31) was probably due to different secretion capabilities of the engineered E proteins produced by different recombinant constructs.
FIG. 4.
Seroconversion of mice immunized with pJNS1 or pJNS1′. The antibody responses specific to JEV NS1 in immunized mice were examined by RIP analysis with 35S-labeled, JEV-infected cell lysates. The pooled sera were collected 2 weeks after the primary (lanes 1 and 4), secondary (lanes 2 and 5), and third (lanes 3 and 6 to 8) DNA injections. The serum samples were from mice immunized with pJNS1 (lanes 1 to 3), pJNS1′ (lanes 4 to 6), vector control pcDNA3 (lane 7), or PBS buffer alone (lane 8). A MAb specific for JEV NS1 (αNS1) was used as a positive control (lane 9). The positions of NS1 and NS1′ are indicated by the arrows on the right. Numbers on the left are the molecular mass standards in kilodaltons.
Anti-NS1 antibodies could induce a complement-mediated cytotoxicity in JEV-infected cells.
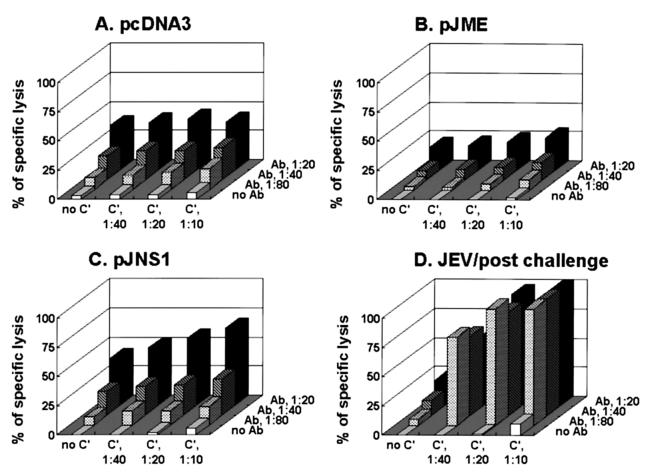
To determine whether NS1-specific antisera can cause cytolysis of JEV-infected cells in the presence of complement, a modified complement-mediated cytotoxicity assay was performed. By measuring the release of a cytoplasmic enzyme, LDH, pooled sera from DNA- or JEV-immunized mice were analyzed for their abilities to lyse JEV-infected BHK-21 cells in the presence of complement. As the results in Fig. 5 indicate, antiserum from pJNS1-immunized mice was able to lyse target JEV-infected cells (Fig. 5C); in contrast, no apparent specific cytolytic could be detected with sera from pJME-immunized (Fig. 5B) and pJNS1′-immunized (data not shown) mice. As controls, sera from JEV-immunized mice displayed a high level of cytolytic activity when complement was included (Fig. 5D) whereas sera from pcDNA3-immunized mice showed no specific activity (Fig. 5A). These results are consistent with previous observations in which flavivirus NS1-induced protective immunity might have resulted from antibody-dependent complement-mediated cytolysis of infected cells (11, 43).
FIG. 5.
The pJNS1-immunized sera exhibited antibody-dependent complement-mediated cytolysis of JEV-infected cells. The sera obtained 2 weeks after the second injections were pooled from ICR mice immunized with pcDNA3 (A), pJME (B), or pJNS1 (C). As a control, sera were also collected from mice that had been infected with JEV (D). In the presence of complement (C′) at different dilutions (none, 1:10, 1:20, or 1:40), the pooled sera were analyzed for their ability to lyse JEV-infected BHK-21 cells at various serum dilutions (no antibody, 1:20, 1:40, or 1:80). The percent specific lysis was determined as described in Materials and Methods.
Biochemical analysis of the differences between JEV NS1 and NS1′ in cultured cells.
In contrast to other flaviviruses, both NS1 and NS1′ are often detected in lysates of JEV-infected cells (30). However, the biological significance of and the biochemical difference between these two NS1 proteins remain uncharacterized. Because JEV NS1′ contains all the genetic information of NS1, it is intriguing to suggest that NS1 and NS1′ from JEV might possess different immune stimulation capabilities. One possible mechanism involved is that NS1 differs from NS1′ in its ability to be secreted from and expressed on JEV-infected cells, a characteristic believed to be important for eliciting an adequate immune response. To test this hypothesis, we further biochemically analyzed the two proteins expressed in cultured cells by a transient- or permanent-expression system. JEV NS1 proteins are secreted from infected cells as dimers which are highly sensitive to heat treatment, as shown by analyzed by Western blotting (12). To assess the amount of secreted NS1 proteins in dimeric form, we prepared the protein samples in the next two experiments without heat treatment before subjecting them to SDS-PAGE. In the JEV-infected cell lysate (Fig. 6A, lane 5), which served as a positive control, three NS1-related bands, corresponding to homodimers of NS1 and NS1′ and a heterodimer between NS1 and NS1′, were detected; however, in the culture supernatant, only two major NS1-related bands could be detected (lane 1), indicating that one of the NS1-related dimers, presumably the NS1′ homodimer, might not be secreted from the cells as efficiently as the other two. When transiently expressed by recombinant vaccinia virus producing T7 polymerase, NS1 derived from pJNS1 construct appeared not only to form dimers intracellularly (lane 6) but also to be secreted as dimers extracellularly (lane 2). The secreted forms of NS1 proteins migrated more slowly than did their counterparts in cell lysates, probably due to the different glycosylation patterns (compare lane 2 with lane 6 and lane 1 with lane 5), as previously described elsewhere (30). In marked contrast, NS1′ expressed by the pJNS1′ construct could form dimers only intracellularly (lane 7) but failed to be secreted in the culture supernatant (lane 3).
FIG. 6.
Intracellular and extracellular protein patterns of JEV NS1 and NS1′ expressed in cell culture. (A) Viral proteins expressed from plasmids by a transient system involving recombinant vaccinia virus vTF7-3. By immunoblotting with MAb against JEV NS1, the culture media (lanes 1 to 4) and their corresponding cell lysates (lanes 5 to 8) were prepared to detect the presence of JEV NS1. Lanes: 1 and 5, samples derived from JEV-infected cells (positive control); 2 and 6, samples from pJNS1-transfected, vTF7-3-infected cells; 3 and 7, samples from pJNS1′-transfected, vTF7-3-infected cells; 4 and 8, samples from pcDNA3-transfected, vTF7-3-infected cells (negative control). Putative homo- and heterodimers of JEV NS1s are marked by arrows. (B) Viral proteins expressed by cell clones containing different plasmids as indicated. JEV NS1s in the culture media (lanes 1 to 4) and in cell lysates (lanes 5 to 8) were analyzed by immunoblotting. Samples to be examined were prepared from cell clones containing pJNS1 (lanes 2 and 6), pJNS1′ (lanes 3 and 7), and no plasmid (lane 4 and 8). Lanes 1 and 5 contain control samples derived from JEV-infected cells.
To exclude any possible effects of vaccinia virus infection, we established permanent cell lines expressing NS1 by transfecting BHK-21 cells with either pJNS1 or pJNS1′. After selection by geneticin (G418 sulfate [GIBCO]) and screening by an indirect immunofluorescence assay (IFA) with MAb against JEV NS1, several cell clones constitutively expressing NS1 or NS1′ were established. A similar experiment to that in Fig. 6A was performed, and representative data obtained from two such cell clones are shown in Fig. 6B. Similar to the result in Fig. 6A, NS1 dimers were readily detected in the cell lysates from both NS1- and NS1′-expressing cell clones (Fig. 6B, lanes 6 and 7). However, only the recombinant NS1 made by the NS1-expressing clone could be released as homodimers extracellularly (lane 2) whereas the secreted form of NS1′ was never detected in the culture medium from NS1′-expressing cells (lane 3). Together, these results clearly indicate that NS1 differs from NS1′ in its ability to be secreted from genetically engineered cells.
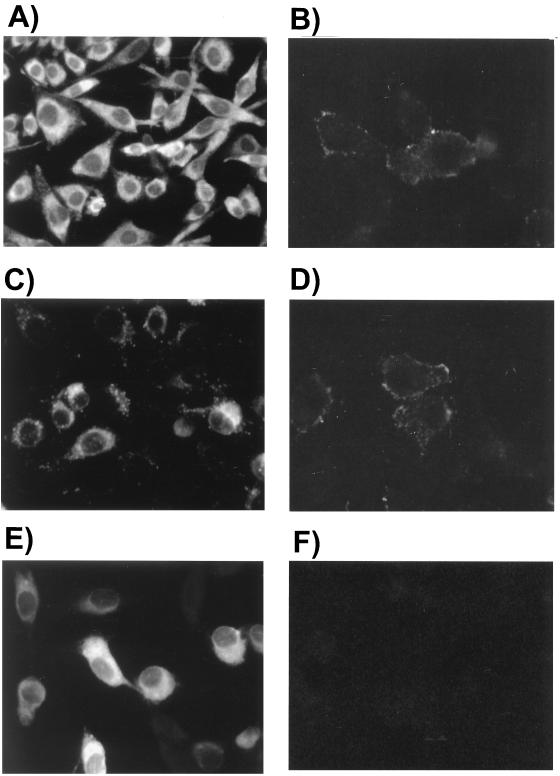
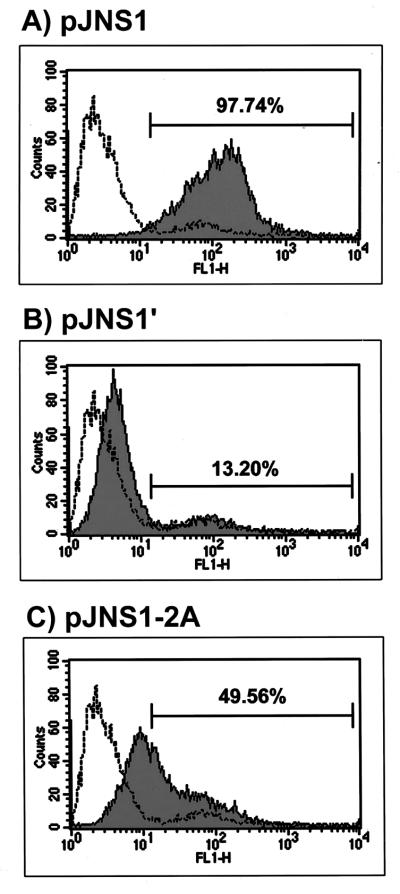
By using IFA with MAb against JEV NS1, the NS1 distribution in NS1- and NS1′-expressing cell clones was also studied. Although cytoplasmic staining of NS1 was observed in both cell clones, the distribution patterns differed; in NS1-expressing cells, some of the recombinant NS1 appeared to aggregate in granules scattered around the cytoplasm (Fig. 7C), while a relatively homogeneous cytoplasmic staining of the engineered NS1′ was observed in NS1′-expressing cells (Fig. 7E). The cytoplasmic pattern of NS1 in JEV-infected cells (Fig. 7A) was also somewhat homogeneous compared to that in NS1-expressing cells (Fig. 7C). The biological significance of this difference remains to be further elucidated. One possible function of NS1 granules seen in the cytoplasm is to act as transport vehicles participating in the secretory process of JEV NS1. Finally, we characterized the surface expression of NS1s on unfixed cells by IFA, and the results demonstrated positive surface staining on the exterior of NS1-expressing cells (Fig. 7D) as well as on JEV-infected cells (Fig. 7B). By contrast, no detectable surface staining could be seen on NS1′-expressing cells (Fig. 7F). These phenomena were further substantiated by FACS analysis, as shown in Fig. 8. The level of NS1 surface expression from cells containing pJNS1 (Fig. 8A) was much higher than that from cells expressing pJNS1′ (Fig. 8B) (97.74 and 13.20%, respectively). Interestingly, only a moderate level (49.56%) of NS1 surface staining could be detected when NS1 and NS1′ coexisted (data not shown) in cells constitutively expressing JEV NS1 and full-length NS2A (Fig. 8C). Taken together, these data strongly suggest that NS1, rather than its counterpart NS1′, could be efficiently expressed on and secreted from pJNS1-transfected cells, and this biochemical discrepancy may contribute to the difference in the efficacy of DNA immunization with pJNS1 and pJNS1′.
FIG. 7.
NS1 protein localization in NS1- or NS1′-expressing cell clones. By using indirect immunofluorescence staining of cells with MAb against JEV NS1, the NS1 expression patterns from cell clones containing pJNS1 (C and D) or pJNS1′ (E and F) were investigated. The NS1 pattern from JEV-infected BHK-21 cells (A and B) was included as a control. The intracellular distribution of NS1 was analyzed from cells fixed with acetone-methanol (1:1) (A, C, and E); on the other hand, cell surface expression of NS1 was analyzed from unfixed cells (B, D, and F) (see Materials and Methods).
FIG. 8.
Flow cytometry of NS1 surface expression from three permanent cell clones. NS1 antigen expressed on the surface of cell clones containing pJNS1 (A), pJNS1′ (B), or pJNS1-2A (C) was analyzed with a FACS Caliber (Becton Dickinson) and CELLQuest software. Cells were stained for surface antigen with a MAb against JEV NS1 followed by fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin (shaded areas). The open area was derived from the staining of parental BHK-21 cells. The numbers above the shaded areas in each panel indicate the percentage of positive staining from tested cells compared to that from parental BHK-21 cells.
DISCUSSION
By passive immunization with MAbs against NS1 and by active immunization with NS1 proteins, flavivirus NS1 has been demonstrated to contain epitopes that are able to stimulate protective immunity (reviewed in references 14, 34, and 48). In the present study, we further demonstrate that DNA immunization with NS1 (as well as with prME) can also confer protection against a lethal JEV challenge in an outbred mouse model. The success of gene vaccination, as shown by our results, clearly indicates the applicability of this approach in the development of a subunit vaccine against flavivirus infection. One obvious advantage of immunizing with DNA over immunizing with purified viral antigens is that JEV glycoproteins expressed on the cell surface from DNA constructs can be correctly presented to the immune system in a native state. Additionally, DNA vaccination can circumvent the detrimental alterations of antigenic integrity during the purification procedure for the preparation of conventional viral antigens. Thus, immunization with genes encoding viral glycoproteins, such as the JEV proteins, can in theory elicit a whole range of immune responses, closely resembling the process of natural virus infection (reviewed in reference 35). It would be of interest to know whether gene immunization with the NS1 gene can also block JEV replication in other viremic amplifying hosts such as birds and pigs.
The present data appear to support the notion that flavivirus NS1 by itself is sufficient to elicit protective immunity, making JEV NS1 a suitable candidate in addition to prME for a subunit vaccine. However, in contrast to the results presented here, several earlier studies in which recombinant viruses were used as an expression vector, including vaccinia virus (21), Sindbis virus (39), and baculovirus (33), have shown that JEV NS1 could induce only low levels of protection in the tested hosts. This discrepancy most probably results from different expression systems being used (DNA immunization versus recombinant viruses) and different coding sequences being included in NS1 constructs. It is possible that JEV NS1 became a relatively weak immunogen when coimmunized with its immunodominant virus vectors, so that the infected host failed to properly recognize and respond to NS1. Alternatively, the possession of extra sequences from NS2A in some cases (21, 39) probably also contributed to the inability of recombinant viruses expressing NS1 to elicit protective immunity. Similar conclusions can be drawn from immunization with pJNS1′ DNA containing about one-quarter of the NS2A sequence constructed in this study, which indeed conferred on the mice only partial protection against a lethal JEV infection (Fig. 3). By contrast, pJNS1 DNA, without any NS2A sequence, was able to provide robust protection against lethal JEV challenge. These data are also consistent with the results of a previous study (11) in which mice immunized with recombinant vaccinia virus expressing dengue virus NS1 alone, but not NS1 plus 15% of NS2A, were fully protected from a subsequent dengue virus challenge.
In JEV-infected cells, both NS1 and NS1′ can be normally generated, yet little is known about their functions in JEV replication and whether NS1′ is the precursor to NS1. Biochemical analyses conducted in this study however, indicate that when these proteins are individually expressed, NS1 differs primarily from its longer version, NS1′, in its ability to be secreted from and expressed on cells (Fig. 6 through 8). In fact, NS1′ is identical to NS1, except that the former contains an extra portion of approximately 60 amino acids from the N terminus of NS2A. JEV NS2A, a small hydrophobic protein, is probably inserted into and spans the lipid bilayer of the ER lumen at least once during biosynthesis. Conceivably, after entry into the lumen of the ER, NS1′ is likely to be retained rather than proceed along the secretory pathway. On the other hand, NS1, which does not seem to contain any recognizable membrane anchorage domains, appears to be proficient in both cell association and secretion (Fig. 6 through 8). This biochemical property of NS1 is particularly relevant because our results clearly demonstrated that NS1 was far more immunogenic than NS1′ with respect to humoral responses (Fig. 4 and 5) and protective immunity (Fig. 3) induced in the immunized hosts. Interestingly, when NS1 and NS1′ coexisted in cells containing pJNS1-2A, the level of cell surface expression of NS1 was significantly lower than that in cells constitutively expressing NS1 alone (Fig. 8). These data seem to imply that the presence of NS1′ might have retarded the normal secretory process for NS1, probably through forming a heterodimer between NS1 and NS1′. It is reasonable to hypothesize that partial NS2A may have a negative impact on NS1-based gene immunization when it is cosynthesized de novo with NS1 in the cell. Exclusion of NS2A would therefore be strategically beneficial for developing NS1 subunit vaccines against JEV infection. However, the motif(s) that dictates the fate of NS1 proteins in cell association or secretion remains to be further elucidated.
With regard to the expression of viral glycoproteins on the cell surface, JEV replication should be able to render infected cells susceptible to antibody-dependent complement-mediated cytolysis and/or antibody-dependent cell-mediated cytotoxicity. Indeed, the fact that immunized sera raised against JEV NS1 caused cytolysis of the JEV-infected cells in a complement-dependent manner (Fig. 5) strongly suggests the involvement of an Fc-mediated cell killing mechanism in the protection of mice from a lethal JEV challenge. Moreover, these data also imply that pJNS1 immunization could stimulate antibody responses with not only the correct epitope specificity but also the appropriate antibody isotype essential for governing the Fc receptor-dependent interactions. Such cytophilic but not neutralizing NS1-specific antibodies could conceivably empower leukocytes involved in the antibody-dependent cell-mediated cytotoxicity reaction important for the elimination of JEV-infected cells. However, the level of specific lysis in pJNS1-immunized sera appeared to be much lower than that in JEV-immunized sera (Fig. 5), indicating that other mechanisms, such as cell-mediated immunity, might also contribute to this protection. Plausibly, NS1 expressed by plasmid DNA from the cells might also be able to elicit specific cytotoxic T-cell responses, as previously suggested by Lobigs et al. (27), whereby cytotoxic T-cell peptide determinants of flavivirus were derived predominantly from viral nonstructural proteins. This may be of interest in JEV vaccination, since flavivirus infections are renowned for inducing MHC class I up-regulation, which leads to down-regulation of virus-specific cytotoxic T-cell memory (reviewed in reference 27). The success of DNA immunization against JEV infection with only portions of viral polyproteins may open avenues for exploring new epitopes that can confer on the immunized host a long-lasting cytotoxic T-cell memory without compromising protection capability.
ACKNOWLEDGMENTS
JEV NT109 and NT113 were kindly provided by the National Institute of Preventive Medicine, Taiwan, Republic of China (ROC).
Y.-L.L. was supported by grant DOH87-TD-1002 from the Department of Health, ROC; grant NSC-87-2314-B-016-090 from the National Science Council (NSC), ROC; and grant (DD01-86IX-CR-501P) from the National Health Research Institute (NHRI) ROC. L.-K.C. was supported by grant NSC 87-2314-B-016-037 M07 from NSC and grant DD01-86IX-CR-501P from NHRI. C.-L.L. was supported by grant NSC 87-2314-B-016-088 from NSC and two grants (DOH87-HR-608 and DD01-86IX-CR-501P) from NHRI.
REFERENCES
- 1.Bray M, Lai C J. Dengue virus premembrane and membrane proteins elicit a protective immune response. Virology. 1991;185:505–508. doi: 10.1016/0042-6822(91)90809-p. [DOI] [PubMed] [Google Scholar]
- 2.Brinton M A. Replication of flaviviruses. In: Schlesinger S, Schlesinger M J, editors. The Togaviridae and Flaviviridae. New York, N.Y: Plenum Press; 1986. pp. 327–374. [Google Scholar]
- 3.Burke D S, Leake C J. Japanese encephalitis. In: Monath T, editor. The arboviruses: epidemiology and ecology. Vol. 3. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 63–92. [Google Scholar]
- 4.Cardoso A I, Blixenkrone-Moller M, Fayolle J, Liu M, Buckland R, Wild T F. Immunization with plasmid DNA encoding for the measles virus hemagglutinin and nucleoprotein leads to humoral and cell-mediated immunity. Virology. 1996;225:293–299. doi: 10.1006/viro.1996.0603. [DOI] [PubMed] [Google Scholar]
- 5.Chambers T J, Hahn C S, Galler R, Rice C M. Flavivirus genome, organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 6.Chen L K, Liao C L, Lin C G, Lai S C, Liu C I, Ma S H, Huang Y Y, Lin Y L. Persistence of Japanese encephalitis virus is associated with abnormal expression of the nonstructural protein NS1 in host cells. Virology. 1996;217:220–229. doi: 10.1006/viro.1996.0109. [DOI] [PubMed] [Google Scholar]
- 7.Chen L K, Lin Y L, Liao C L, Lin C G, Huang Y L, Yeh C T, Lai S C, Jan J T, Chin C. Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology. 1996;223:79–88. doi: 10.1006/viro.1996.0457. [DOI] [PubMed] [Google Scholar]
- 8.Chiang H-Y, Cohen G H, Eisenberg R J. Identification of functional regions of herpes simplex virus glycoprotein gD by using linker-insertion mutagenesis. J Virol. 1994;68:2529–2543. doi: 10.1128/jvi.68.4.2529-2543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox G J, Zamb T J, Babiuk L A. Bovine herpesvirus 1: immune responses in mice and cattle injected with plasmid DNA. J Virol. 1993;67:5664–5667. doi: 10.1128/jvi.67.9.5664-5667.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis H L, Michel M L, Whalen R G. DNA-based immunization induces continuous secretion of hepatitis B surface antigen and high levels of circulating antibody. Hum Mol Genet. 1993;3:1847–1851. doi: 10.1093/hmg/2.11.1847. [DOI] [PubMed] [Google Scholar]
- 11.Falgout B, Bray M, Schlesinger J J, Lai C J. Immunization of mice with recombinant vaccinia virus expressing authentic dengue virus nonstructural protein NS1 protects against lethal dengue virus encephalitis. J Virol. 1990;64:4356–4363. doi: 10.1128/jvi.64.9.4356-4363.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan W F, Mason P W. Membrane association and secretion of the Japanese encephalitis virus NS1 protein from cells expressing NS1 cDNA. Virology. 1990;177:470–476. doi: 10.1016/0042-6822(90)90511-o. [DOI] [PubMed] [Google Scholar]
- 13.Fynan E F, Webster R G, Fuller D H, Haynes J R, Santoro J C, Robinson H L. DNA vaccines: protective immunizations by parenteral, mucosal, and gene-gun inoculations. Proc Natl Acad Sci USA. 1993;90:11478–11482. doi: 10.1073/pnas.90.24.11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson C A, Schlesinger J J, Barrett A D. Prospects for a virus non-structural protein as a subunit vaccine. Vaccine. 1988;6:7–9. doi: 10.1016/0264-410x(88)90004-7. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez Armas J C, Morello C S, Cranmer L D, Spector D H. DNA immunization confers protection against murine cytomegalovirus infection. J Virol. 1996;70:7921–7928. doi: 10.1128/jvi.70.11.7921-7928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorman C M, Moffatt L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halstead S B. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 18.Heinz F X, Stiasny K, Puschner-Auer G, Holzmann H, Allison S L, Mandl C W, Kunz C. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology. 1994;198:109–117. doi: 10.1006/viro.1994.1013. [DOI] [PubMed] [Google Scholar]
- 19.Hoke C H, Nisalak A, Sangawhipa N, Jatanasen S, Laorakapongse T, Innis B L, Kotchasenee S, Gingrich J B, Latendress J, Fukai K, Burke D S. Protection against Japanese encephalitis by inactivated vaccine. N Engl J Med. 1988;319:608–614. doi: 10.1056/NEJM198809083191004. [DOI] [PubMed] [Google Scholar]
- 20.Kimura-Kiroda J, Yasui K. Protection of mice against Japanese encephalitis virus by passive administration with monoclonal antibodies. J Immunol. 1988;141:3606–3610. [PubMed] [Google Scholar]
- 21.Konishi E, Pincus S, Fonseca B A, Shope R E, Paoletti E, Mason P W. Comparison of protective immunity elicited by recombinant vaccinia viruse that synthesize E or NS1 of Japanese encephalitis virus. Virology. 1991;185:401–410. doi: 10.1016/0042-6822(91)90788-d. [DOI] [PubMed] [Google Scholar]
- 22.Konishi E, Pincus S, Paoletti E, Shope R E, Burrage T, Mason P W. Mice immunized with a subviral particle containing the Japanese encephalitis virus prM/M and E proteins are protected from lethal JEV infection. Virology. 1992;188:714–720. doi: 10.1016/0042-6822(92)90526-u. [DOI] [PubMed] [Google Scholar]
- 23.Krieg A M, Yi A K, Matson S, Waldschmidt T J, Bishop G A, Teasdale R, Koretzky G A, Klinman D M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 24.Ku C C, King C C, Lin C Y, Hsu H C, Chen L Y, Yueh Y Y, Chang G J. Homologous and heterologous neutralization antibody responses after immunization with Japanese encephalitis vaccine among Taiwan children. J Med Virol. 1994;44:122–131. doi: 10.1002/jmv.1890440204. [DOI] [PubMed] [Google Scholar]
- 25.Lagging L M, Meyer K, Hoft D, Houghton M, Belshe R B, Ray R. Immune responses to plasmid DNA encoding the hepatitis C virus core protein. J Virol. 1995;69:5859–5863. doi: 10.1128/jvi.69.9.5859-5863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y L, Liao C L, Yeh C T, Chang C H, Huang Y L, Huang Y Y, Jan J T, Chin C, Chen L K. A highly attenuated strain of Japanese encephalitis virus induces a protective immune response in mice. Virus Res. 1996;44:45–56. doi: 10.1016/0168-1702(96)01343-3. [DOI] [PubMed] [Google Scholar]
- 27.Lobigs M, Blanden R V, Mullbacher A. Flavivirus-induced up-regulation of MHC class I antigens; implications for the induction of CD8+ T-cell-mediated autoimmunity. Immunol Rev. 1996;152:5–19. doi: 10.1111/j.1600-065X.1996.tb00908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Major M E, Vitvitski L, Mink M A, Schleef M, Whalen R G, Trepo C, Inchauspe G. DNA-based immunization with chimeric vectors for the induction of immune responses against the hepatitis C virus nucleocapsid. J Virol. 1995;69:5798–5805. doi: 10.1128/jvi.69.9.5798-5805.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason P W. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989;169:354–364. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason P W, Pincus S, Fournier M J, Mason T L, Shope R E, Paoletti E. Japanese encephalitis virus-vaccinia recombinants produce particulate forms of the structural membrane proteins and induce high levels of protection against lethal JEV infection. Virology. 1991;180:294–305. doi: 10.1016/0042-6822(91)90034-9. [DOI] [PubMed] [Google Scholar]
- 32.McClements W L, Armstrong M E, Keys R D, Liu M A. Immunization with DNA vaccines encoding glycoprotein D or glycoprotein B, alone or in combination, induces protective immunity in animal models of herpes simplex virus-2 disease. Proc Natl Acad Sci USA. 1996;93:11414–11420. doi: 10.1073/pnas.93.21.11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCown J, Cochran M, Putnak R, Feighny R, Burrous J, Henchal E, Hoke C. Protection of mice against lethal Japanese encephalitis with a recombinant baculovirus vaccine. Am J Trop Med Hyg. 1990;42:491–499. doi: 10.4269/ajtmh.1990.42.491. [DOI] [PubMed] [Google Scholar]
- 34.Monath T P, Heinz F X. Flaviviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 961–1034. [Google Scholar]
- 35.Murphy B R, Chanock R M. Immunization against virus disease. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 467–497. [Google Scholar]
- 36.National Quarantine Service. Statistics on Japanese encephalitis control in Taiwan areas. Taiwan, Republic of China: National Quarantine Service; 1992. p. 4. [Google Scholar]
- 37.Oya A. Japanese encephalitis vaccine. Acta Pediatr Jpn. 1988;30:175–184. doi: 10.1111/j.1442-200x.1988.tb02516.x. [DOI] [PubMed] [Google Scholar]
- 38.Phillpotts R J, Venugopal K, Brooks T. Immunisation with DNA polynucleotides protects mice against lethal challenge with St. Louis encephalitis virus. Arch Virol. 1996;141:743–749. doi: 10.1007/BF01718332. [DOI] [PubMed] [Google Scholar]
- 39.Pugachev K V, Mason P W, Shope R E, Frey T K. Double-subgenomic Sindbis virus recombinants expressing immunogenic proteins of Japanese encephalitis virus induce significant protection in mice against lethal JEV infection. Virology. 1995;212:587–594. doi: 10.1006/viro.1995.1516. [DOI] [PubMed] [Google Scholar]
- 40.Raz E, Carson D A, Parker S E, Parr T B, Abai A M, Aichinger G, Gromkowski S H, Singh M, Lew D, Yankauckas M A, Baird S M, Rhodes G H. Intradermal gene immunization: the possible role of DNA uptake in the induction of cellular immunity to viruses. Proc Natl Acad Sci USA. 1994;91:9519–9523. doi: 10.1073/pnas.91.20.9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 931–959. [Google Scholar]
- 42.Robinson H L, Hunt L A, Webster R G. Protection against a lethal influenza virus challenge by immunization with a haemagglutinin-expressing plasmid DNA. Vaccine. 1993;11:957–960. doi: 10.1016/0264-410x(93)90385-b. [DOI] [PubMed] [Google Scholar]
- 43.Schlesinger J J, Brandriss M W, Walsh E E. Protection against 17D yellow fever encephalitis in mice by passive transfer of monoclonal antibodies to the nonstructural glycoprotein gp48 and by active immunization with gp48. J Immunol. 1985;135:2805–2809. [PubMed] [Google Scholar]
- 44.Schlesinger J J, Putnak J R, Eckels K H. New approaches to flavivirus vaccine development. Bio/Technology. 1992;20:289–307. doi: 10.1016/b978-0-7506-9265-6.50018-x. [DOI] [PubMed] [Google Scholar]
- 45.Schlesinger J J, Foltzer M, Chapman S. The Fc portion of antibody to yellow fever virus NS1 is a determinant of protection against YF encephalitis in mice. Virology. 1993;192:132–141. doi: 10.1006/viro.1993.1015. [DOI] [PubMed] [Google Scholar]
- 46.Ulmer J B, Donnelly J J, Parker S E, Rhodes G H, Felgner P L, Dwarki V J, Deck R R, DeWitt C M, Friedman A, Hawe L A, Leander K R, Martinez D, Perry H C, Shiver J W, Montgomery D L, Liu M A. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 47.Vaughn D W, Hoke C H., Jr The epidemiology of Japanese encephalitis: prospects for prevention. Epidemiol Rev. 1992;14:197–221. doi: 10.1093/oxfordjournals.epirev.a036087. [DOI] [PubMed] [Google Scholar]
- 48.Venugopal K, Gould E A. Towards a new generation of flavivirus vaccines. Vaccine. 1994;12:966–975. doi: 10.1016/0264-410x(94)90329-8. [DOI] [PubMed] [Google Scholar]
- 49.Wang B, Boyer J, Srikantan V, Coney L, Carrano R, Phan C, Merva M, Agadjanan M, Gilbert L, Ugen K E, Williams W V, Weiner D B. DNA inoculation induces neutralizing immune responses against human immunodeficiency virus type 1 in mice and nonhuman primates. DNA Cell Biol. 1993;12:799–805. doi: 10.1089/dna.1993.12.799. [DOI] [PubMed] [Google Scholar]
- 50.Wang B, Ugen K E, Srikantan V, Agadjanyan M G, Dang K, Refaeli Y, Sato A I, Boyer J, Williams W V, Weiner D B. Gene inoculation generates immune responses against human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1993;90:4156–4160. doi: 10.1073/pnas.90.9.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang Z Q, Spitalnik S, Tran M, Wunner W H, Cheng J, Ertl H C. Vaccination with a plasmid vector carrying the rabies virus glycoprotein gene induces protective immunity against rabies virus. Virology. 1994;199:132–140. doi: 10.1006/viro.1994.1105. [DOI] [PubMed] [Google Scholar]
- 52.Yasuda A, Kimura-Kuroda J, Ogimoto M, Miyamoto M, Sata T, Sato T, Takamura C, Kurata T, Kojima A, Yasui K. Induction of protective immunity in animals vaccinated with recombinant vaccinia viruses that express preM and E glycoproteins of Japanese encephalitis virus. J Virol. 1990;64:2788–2795. doi: 10.1128/jvi.64.6.2788-2795.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]