Abstract
The innermost core of rotavirus is composed of VP2, which forms a protein layer that surrounds the two minor proteins VP1 and VP3, and the genome of 11 segments of double-stranded RNA. This inner core layer surrounded by VP6, the major capsid protein, constitutes double-layered particles that are transcriptionally active. Each gene encoding a structural protein of double-layered particles has been cloned into baculovirus recombinants and expressed in insect cells. Previously, we showed that coexpression of different combinations of the structural proteins of rotavirus double-layered particles results in the formation of virus-like particles (VLPs), and each VLP containing VP1, the presumed RNA-dependent RNA polymerase, possesses replicase activity as assayed in an in vitro template-dependent assay system (C. Q.-Y. Zeng, M. J. Wentz, J. Cohen, M. E. Estes, and R. F. Ramig, J. Virol. 70:2736–2742, 1996). This work reports construction and characterization of VLPs containing a truncated VP2 (VPΔ2, containing amino acids [aa] Met-93 to 880). Expression of VPΔ2 alone resulted in the formation of single-layered Δ2-VLPs. Coexpression of VPΔ2 with VP6 produced double-layered Δ2/6-VLPs. VLPs formed by coexpression of VPΔ2 and VP1 or VP3, or both VP1 and VP3, resulted in the formation of VLPs lacking both VP1 and VP3. The presence of VP6 with VPΔ2 did not result in encapsidation of VP1 and VP3. To determine the domain of VP2 required for binding VP1, far-Western blot analyses using a series of truncated VP2 constructs were performed to test their ability to bind VP1. These analyses showed that (i) full-length VP2 (aa 1 to 880) binds to VP1, (ii) any N-terminal truncation lacking aa 1 to 25 fails to bind VP1, and (iii) a C-terminal 296-aa truncated VP2 construct (aa 1 to 583) maintains the ability to bind VP1. These analyses indicate that the N terminus of rotavirus VP2 is necessary for the encapsidation of VP1 and VP3.
The rotaviruses, members of the family Reoviridae, are triple-layered viruses with a genome of 11 segments of double-stranded RNA (dsRNA) (for a review, see reference 10) and are the most important cause of severe viral gastroenteritis in humans and animals (1, 15). Morphologically and biochemically, the triple-layered virus particles consist of three concentric proteinaceous capsid layers: (i) an outer protein layer of 780 molecules of VP7 from which 60 dimers of the spike protein VP4 project; (ii) a middle protein layer of 260 trimers of VP6; and (iii) an inner protein layer of 120 molecules of VP2 (6, 28). The VP2 inner shell encapsidates the two minor proteins VP1 and VP3 and the 11 segments of genomic dsRNA, and this structure has been called the core of the virion. VP1, VP3, and the 11 segments of genomic dsRNA have been proposed to be organized into a subcore (27). This proposal is supported by the demonstration that much of the genomic dsRNA is organized, forming a dodecahedral structure that is organized around VP1-VP3 complexes located at the fivefold vertices (29).
The VP2 shell of rotavirus plays important roles in the structure and function of the rotavirus core. For example, VP2 interacts with trimers of VP6 that surround the VP2 layer and are perforated by 132 aqueous channels (30, 35), transporting metabolites in and nascent RNA out during transcription. VP2 also binds to viral RNA and may function in the replication and encapsidation of dsRNA (2, 5, 16, 23). The nucleic acid binding domain in VP2 is located between amino acids (aa) 1 and 132 (17), and the bond between Gln-92 and Lys-93 in VP2 is a protease-accessible site (37). Interactions between VP2 or VP1 and nucleic acids appear to be critical for the endogenous enzymatic activity of the core (23, 24). VP1, the minor core protein, is thought to be the viral RNA-dependent RNA polymerase (transcriptase) because (i) temperature-sensitive mutants mapping to gene 1 have an RNA-negative phenotype (13); (ii) VP1 sequences contain the common motif of all RNA polymerases (8, 26); (iii) cross-linking of azido-ATP to VP1 causes a corresponding decrease in the ability of native double-layered particles to synthesize mRNA (33); and (iv) 1/2-VLPs (previously designated VP1/2 particles [38]; VLPs are virus-like particles) expressed from baculovirus recombinants have replicase activity that synthesizes negative-stranded RNA on positive-stranded RNA templates, while VP2 alone lacks this activity (7, 38). Recently, VP1 alone has been reported to lack replicase activity, but VP2 stimulates this activity (24). VP3, the other minor core protein, binds GTP specifically and is thought to be a guanylyltransferase (22, 25). VP1 and VP3 appear to form a complex located on the inner surface of the VP2 layer at the icosahedral fivefold axes (29). Expression of VP2 lacking the N-terminal 92 aa in Spodoptera frugiperda Sf9 cells results in changes of the three-dimensional structure along the inner surface of the 2-VLPs (20).
In this study, N-terminally truncated forms of VP2 expressed from baculovirus recombinants were used to show that interactions with VP2 are necessary for encapsidation of VP1 and VP3 and to map the VP2 domain necessary for these interactions.
MATERIALS AND METHODS
Viruses and cells.
S. frugiperda Sf9 cells were grown and maintained in TNM-FH (Hinks) medium containing 10% fetal bovine serum (FBS) (31). Baculovirus recombinants encoding the following rotavirus proteins were used: pVL941/RF-1 (VP1 of virus strain RF) (8), BacRf2A (VP2 of virus strain RF) (16, 18), BVP2C24 (VP2 Met-93 to aa 880 of virus strain RF) (this work), BVP2D185 (VP2 aa 186 to 880 of virus strain RF) (this work), Bac2AT1777 (VP2 aa 1 to 583 of virus strain RF) (17), Bac2M16.9 (VP2 aa Met-26 to 583 of virus strain RF) (17), Bac2M16.3 (VP2 aa Met-45 to 583 of virus strain RF) (17), pVL1393/SA11-3 (VP3 of virus strain SA11) (22), and pAc461/SA11-6 (VP6 of virus strain SA11) (11, 12).
Construction of baculovirus recombinants encoding VPΔ2 (VP2 aa Met-93 to 880) and VPΔ2′ (VP2 aa 186 to 880).
Two mutants of VP2 with deletions from the N terminus of 92 and 185 aa (designated VPΔ2 and VPΔ2′, respectively) were constructed by using a strategy described previously (17). For VPΔ2, an extra starting codon was added. For VPΔ2′, the starting codon corresponds to Met-186. Briefly, a set of oligonucleotides was designed to amplify the corresponding region of the full-length cDNA encoding VP2 and to generate a BamHI site at the end of the PCR product. These products were cloned into pBS, and part of the recombinant plasmids (pBS2C24Δ and pBSRF2Δ185, corresponding to VPΔ2 and VPΔ2′, respectively) were sequenced to check the junctions. The absence of premature termination mutations in the open reading frame was confirmed by transcription translation using the TnT-T7 coupled system from Promega (Madison, Wis.). Recombinant baculoviruses BVP2C24 and BVP2D185, encoding VPΔ2 and VPΔ2′, respectively, were constructed as previously described (17).
Production and purification of rotavirus single-layered VLPs.
Single-layered VLPs, i.e., Δ2-, Δ2′-, 2-, 1/Δ2-, 1/2-, 1/Δ2/3-, 1/2/3-, Δ2/3-, and 2/3-VLPs, were prepared as previously reported for production of VP2 VLPs (18, 37). For each VLP, 2 × 107 Sf9 cells in a T-75 flask were infected at a multiplicity of infection of 10 PFU per cell for each recombinant and were incubated for 2 h at 27°C for adsorption. The inoculum was removed, and the indicated VLPs were expressed in TNM-FH–10% FBS for 3 to 4 days in the absence or presence of protease inhibitors. Protease inhibitors aprotinin and leupeptin, each at 1 μg/ml, were added daily. The cells were lysed with 2% sodium deoxycholate (DOC) lysis buffer (10 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 2% DOC). The expression levels of VP1 and VP3 in the lysates of VP1/2 versus VP1/Δ2, VP1/2/3 versus VP1/Δ2/3, and VP2/3 versus VPΔ2/3 were examined by testing dilutions of samples, using a dot blot assay for VP1 and [α-32P]GTP binding for VP3. The VLPs were purified and fractionated from the cell lysates by centrifugation for 2 h at 30,000 rpm in a Beckman SW41 rotor through a 5 to 20% sucrose gradient containing 0.2% DOC. Fractions (0.7 ml each) were collected. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining showed that unassembled soluble proteins were located in the top fractions 1 to 4. Although VPΔ2′ (aa 186 to 880) did not form VLPs, all other eight types of single-layered VLPs sedimented as a broad band with a peak at fraction 9, as previously described (37). The yields of the various single-layered VLPs were between 40 and 70 μg per 8 × 107 Sf9 cells. The single-layered VLPs in fraction 9 were kept at 4°C and used within 2 weeks for further experiments.
Production and purification of rotavirus double-layered VLPs.
Double-layered VLPs, i.e., Δ2/6-, 2/6-, 1/Δ2/6-, 1/2/6-, Δ2/3/6-, 2/3/6-, 1/Δ2/3/6-, and 1/2/3/6-VLPs, were prepared essentially as described previously (9, 38). Briefly, Sf9 cells were infected at a multiplicity of infection of 5 PFU per cell for each recombinant. Viruses were adsorbed for 2 h at 27°C. The inoculum was removed by low-speed centrifugation, and VLPs were expressed for 6 days in Grace’s medium–0.5% FBS in the absence or presence of protease inhibitors. The protease inhibitors aprotinin and leupeptin, each at 1 μg/ml, were added daily from days 2 through 5 postinfection. The expression levels of VP1 and VP3 in the lysates of VP1/2/6 versus VP1/Δ2/6, VP1/2/3/6 versus VP1/Δ2/3/6, VP2/3/6 versus VPΔ2/3/6 were examined by testing dilutions of samples 80 h postinfection, using a dot blot assay for VP1 and [α-32P]GTP binding for VP3. The infected cells were harvested 6 days postinfection. The VLPs were released into the medium and purified by pelleting through a 35% sucrose cushion followed by banding by CsCl (refractive index, 1.3610) isopycnic centrifugation. Double-layered VLPs composed of VPΔ2 showed two bands in CsCl gradients, identical to those composed of VP2 (38). The densities were 1.27 g/cm3 for the light particles and 1.30 g/cm3 for the heavy particles. The VLPs in the two bands were pooled, diluted with 10 mM Tris-buffered saline (pH 7.4), and pelleted for 2 h at 30,000 rpm in a Beckman SW41 rotor. The pellets were resuspended in the same Tris-buffered saline and kept at 4°C until used. The yields of the various doubled-layered VLPs were between 200 and 400 μg per 6.5 × 108 Sf9 cells. Double-layered VLPs composed of VPΔ2, like those composed of VP2, were stable for at least 8 months as examined by electron microscopy (EM).
SDS-PAGE.
The protein profiles of all VLPs except those detected by far-Western blotting (see below) were determined by reducing SDS-PAGE using 10% resolving and 4% stacking gels (3, 19). Protein locations were stained with a silver staining kit (Sigma, St. Louis, Mo.) according to the manufacturer’s instructions.
GTP binding assay.
The presence of VP3 in VLPs was determined by a GTP binding assay. The reaction was performed in a 20-μl reaction volume containing ∼2.5 μg of VLP protein, 10 μCi of [α-32P]GTP (Amersham Life Science, Inc., Arlington Heights, Ill.), 2 mM MgCl2, and 10 mM Tris-HCl-buffered saline (pH 7.4) as described previously (22). After a 30-min incubation at room temperature, the reaction was terminated by addition of 5 μl of 5× Laemmli sample buffer. After being boiled for 5 min, the proteins were resolved by SDS-PAGE on a 10% gel and electroblotted onto a nitrocellulose membrane (32). Proteins that bound [α-32P]GTP were visualized by autoradiography.
Production and purification of VP1.
Sf9 cells were infected with baculovirus recombinant pVL941/RF-1 at a multiplicity of infection of 10 PFU per cell and incubated for 2 h at 27°C for adsorption. The inoculum was removed by low-speed centrifugation, and VP1 was expressed for 7 days in serum-free medium SF900 II SFM (Gibco BRL, Grand Island, N.Y.). After 7 days postinfection, the medium containing VP1 was equilibrated by dialysis against 20 mM Tris-HCl (pH 8.1), clarified by high-speed centrifugation, filtered through a 0.22-μm-pore-size filter (Costar Co., Cambridge, Mass.), and then semipurified by fast protein liquid chromatography (FPLC) with a quaternary methylamine anion-exchange column (Water Chromatography Division, Milford, Mass.). Quaternary methylamine-bound proteins were eluted with an NaCl linear gradient, 0 to 1.6 M, and VP1 was eluted with 0.12 M NaCl. VP1-rich fractions were pooled and further purified by passage through an immunoaffinity column containing rabbit immunoglobulin G (IgG) against wild-type baculovirus proteins made in this laboratory. VP1 eluted in the unbound protein portion flowthrough. SDS-PAGE (10% gel) and silver staining were used to analyze the purity of VP1. Analysis of 1 μg of purified VP1 showed a single band with an apparent molecular weight of about 125,000.
Guinea pig antiserum to VP1.
A guinea pig anti-VP1 serum was made by inoculating animals with ∼20 μg of purified VP1 in complete Freund’s adjuvant (Gibco BRL) once, followed by five boosts of ∼20 μg of VP1 each at 3-week intervals in incomplete Freund’s adjuvant (Gibco BRL). The final bleed as primary antibody (1:500 dilution) showed a single VP1 band in VP1 lysates of Sf9 cells infected with baculovirus recombinant pVL941/RF-1 and with 1/2/6-VLPs, as examined by Western blotting. This anti-VP1 serum was used for far-Western blotting to verify the presence of VP1 binding proteins.
Far-Western blotting and VP1 binding test.
A far-Western blotting procedure was established essentially as reported by Lee et al. (21) and Homann et al. (14). Purified VLPs were either denatured with SDS in the presence of β-mercaptoethanol (βME) (Laemmli sample buffer) and boiled for 5 min or denatured with SDS in the absence of βME and kept at 4°C. All the subsequent steps were performed at 4°C. The denatured VLP proteins were resolved on a SDS–10% polyacrylamide gel (19). After electrophoresis, the proteins were blotted to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, Mass.) (32). Proteins on the blots were subsequently renaturated by incubation in standard binding buffer (SBB; 10 mM HEPES [pH 7.4], 10 mM MgCl2, 50 mM EDTA, 1 mM dithiothreitol, 10% [vol/vol] glycerol) for 12 h and then incubated for 6 h in SBB–5% bovine serum albumin (BSA) to saturate and block any free binding sites. For probing, BSA-blocked blots were incubated for 24 h with purified VP1 (10 μg/ml) in SBB, followed by reaction with the guinea pig anti-VP1 serum (1:500 dilution). Reactivity of the anti-VP1 serum was visualized by addition of a goat anti-guinea pig IgG-alkaline phosphatase conjugate (Sigma) and substrate 5-bromo-4-chloro-3-indolylphosphate–p-nitroblue tetrazolium chloride (Amersham Life Science).
EM.
The morphology of all VLPs was examined by EM (37). Collodion-carbon-coated and freshly glow discharged copper grids were used for sample adsorption. Each grid was floated on a drop of sample for 30 min, excess fluid was removed by blotting with filter paper, and the grid was washed for 2 s on a drop of phosphate-buffered saline, floated on a drop of 1% ammonium molybdate for 15 s to stain the VLPs, and finally air dried. All electron micrographs were taken with a Philips CM10 electron microscope operating at 60 kV.
Protein determinations.
Protein concentrations were determined by using the Bio-Rad (Hercules, Calif.) protein assay according to the manufacturer’s instructions. Bovine albumin fraction V (Calbiochem, La Jolla, Calif.) was used as the standard.
RESULTS
The N terminus of VP2 is not required for formation of single- and double-layered VLPs.
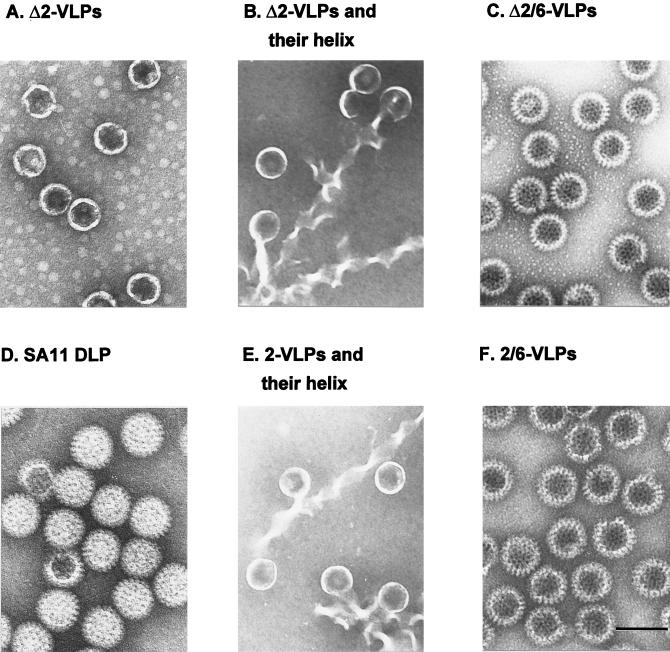
Bovine rotavirus strain RF VPΔ2 containing aa Met-93 to 880 was expressed from baculovirus recombinant BVP2C24 by infection of Sf9 cells. VPΔ2′ containing aa 186 to 880 was expressed from baculovirus recombinant BVP2D185 by using the same expression system. VPΔ2 and VPΔ2′ were expressed at comparable levels. Sucrose gradient fractionation followed by EM examination did not detect any particles in VPΔ2′ (data not shown) but showed that purified VPΔ2 made spherical single-layered core-like particles (Δ2-VLPs) with a smooth surface (Fig. 1A) identical to 2-VLPs (individual particles in Fig. 1E). The apparent diameter of the Δ2-VLPs was 520 ± 20 Å. The purified Δ2-VLPs were morphologically stable for 3 to 4 weeks at 4°C, as reported previously for 2-VLPs (37). Following speed vacuum concentration, Δ2-VLPs revealed a conversion of the structure from individual spherical particles (Fig. 1A) to elongated bristly helix-like structures (Fig. 1B), which has been previously seen to occur in 2-VLPs (Fig. 1E and reference 37). CsCl isopycnic centrifugation banding of Δ2/6-VLPs followed by EM examination showed that purified Δ2/6-VLPs were empty double-layered particles (Fig. 1C) similar to native SA11 double-layered particles (Fig. 1D) and 2/6-VLPs (Fig. 1F). SDS-PAGE followed by silver staining was used to confirm the presence of each structural protein in the purified Δ2-VLPs and Δ2/6-VLPs. Each of the expected proteins was incorporated into the appropriate VLP structure (Fig. 2, lanes 3 and 6).
FIG. 1.
Electron micrographs of the VPΔ2-containing VLPs and their VP2-containing counterparts. Δ2-VLPs were expressed from the recombinant baculovirus BVP2C24 by infection of Sf9 cells and purified on a 5 to 20% sucrose gradient. Δ2/6-VLPs were coexpressed from the recombinant baculoviruses BVP2C24 and pAc461/SA11-6 by infection of Sf9 cells and purified by CsCl isopycnic centrifugation. Shown are the negative-stained structures of the VLPs containing the indicated protein species. (A to C) VPΔ2-containing VLPs; (D) native rotavirus double-layered particles (DLP); (E and F) VP2-containing VLPs as the counterparts of panels A to C. All micrographs are at the same magnification. Bar, 100 nm.
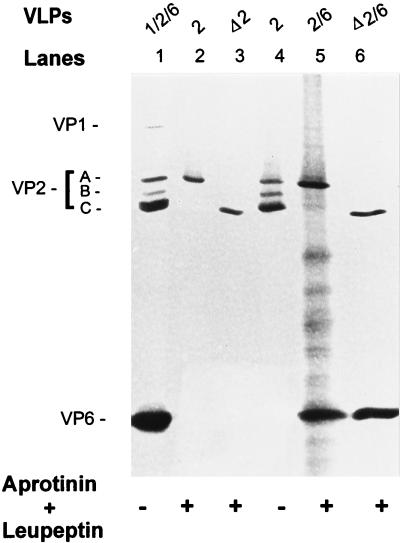
FIG. 2.
Protein content of single- and double-layered VPΔ2-containing VLPs and their VP2-containing counterparts. Shown is a silver-stained SDS–10% polyacrylamide gel with the proteins of purified VLPs resulting from the expression of the indicated genes in the absence or presence of protease inhibitors. Protease inhibitors aprotinin and leupeptin, each at 1 μg/ml, were added daily.
Analysis of the protein profiles of the VLPs containing the VPΔ2 revealed a single band by SDS-PAGE (Fig. 2, lanes 3 and 6) that comigrated with a previously characterized proteolytic cleavage product of VP2 (37). This cleavage product, called band C (Fig. 2, lanes 1 and 4), represents aa 93 to 880 that is derived from the full-length VP2 (band A) (37).
The N terminus of VP2 is required for encapsidation of VP1.
We next examined if VP1 could be encapsidated in a VLP containing the VPΔ2. VLPs obtained from cells infected with recombinants expected to express 1/Δ2-VLPs were structurally composed of only VPΔ2 (Fig. 3, lane 3), while 1/2-VLPs were composed of both VP1 and VP2 (Fig. 3, lane 4). Morphologically both were single-layered types of VLPs appearing to be similar to the particles shown in Fig. 1A, B, and E (data not shown). The VP1 that was not assembled into Δ2-VLPs was found in the top fractions of 5 to 20% sucrose gradients and did not comigrate with Δ2-VLPs (data not shown). Analyses of purified VLPs from cells expressing VP1/Δ2/6 and VP1/2/6 both were morphologically double layered, resembling those seen in Fig. 1C and F (data not shown), but VP1 was present only when the VLPs contained full-length VP2 (Fig. 3, lanes 1 and 2). The expression levels of VP1 in the lysates of VP1/2/6 and VP1/Δ2/6 were comparable at 80 h postinfection. VP1 was still not detected even if a threefold-greater amount of 1/Δ2/6-VLPs and 1/Δ2-VLPs was analyzed (data not shown).
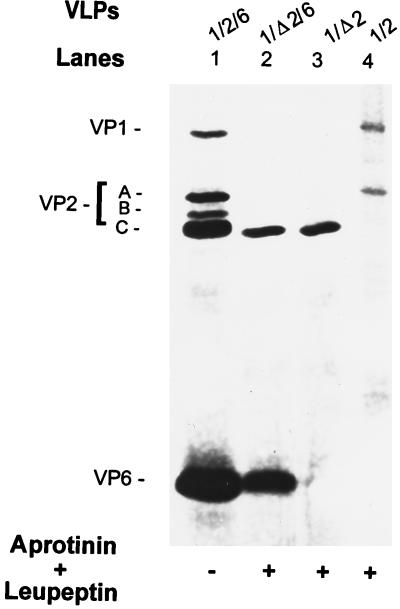
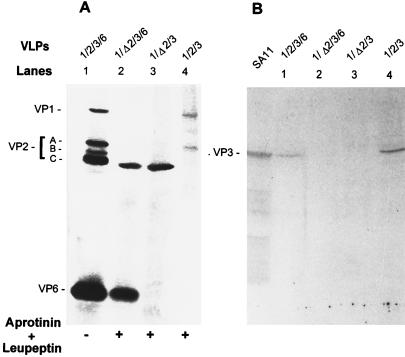
FIG. 3.
Protein content of the coexpressed 1/Δ2-, 1/2-, 1/Δ2/6-, and 1/2/6-VLPs. Shown is a silver-stained SDS–10% polyacrylamide gel with proteins of the purified VLPs resulting from the coexpression of indicated genes in the absence or presence of protease inhibitors. Protease inhibitors aprotinin and leupeptin, each at 1 μg/ml, were added daily. VP1/Δ2 and VP1/2 were coexpressed from the baculovirus recombinant pVL941/RF-1 with recombinant BVP2C24 or BacRF2A, respectively, and purified by centrifugation on a 5 to 20% sucrose gradient. VP1/Δ2/6 and VP1/2/6 were coexpressed from the baculovirus recombinants of pVL941/RF-1, pAc461/SA11-6 with recombinants BVP2C24 and BacRF2A, respectively, and purified by CsCl isopycnic centrifugation. To compare VP1 encapsidation, the protein amount loaded onto each lane was adjusted so that the amount of VPΔ2 was similar to or higher than that of VP2 band A, the VP1 binding protein (Fig. 6C). To create a cleaved VP2 band C for a marker of VPΔ2, 1/2/6-VLPs in lane 1 (also in lanes 1 of Fig. 4 and 5) were produced in the absence of protease inhibitors.
The N terminus of VP2 is required for encapsidation of VP3.
We also examined if VP3 could be encapsidated in a VLP containing VPΔ2. VLPs obtained from cells infected with recombinants that express Δ2/3-VLPs were structurally composed of only VPΔ2 (Fig. 4A and B, lanes 3), while 2/3-VLPs were composed of both VP2 and VP3 (Fig. 4A and B, lanes 4). Morphologically, both types of VLPs were single layered, similar to the particles shown in Fig. 1A, B, and E. Analyses of the VLPs expected to contain VPΔ2/3/6 and VP2/3/6 showed that both were morphologically double layered, similar to those shown in Fig. 1C and F, but VP3 was present only when the VLPs contained full-length VP2 (Fig. 4A and B, lanes 1 and 2). The expression levels of VP3 in the lysates of VP2/3 versus VPΔ2/3 and of VP2/3/6 versus VPΔ2/3/6 were comparable at 80 h postinfection (data not shown). It also appeared that VP3 was encapsidated in parallel to the amount of full-length VP2 in VLPs. Thus, the intensities of labeling of VP3 were comparable (Fig. 4B, lanes 1 and 4) when the amounts of the full-length VP2 in VLPs were comparable (Fig. 4A, lanes 1 and 4).
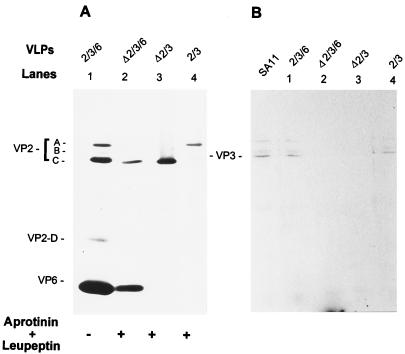
FIG. 4.
Protein content of the coexpressed Δ2/3-, 2/3-, Δ2/3/6-, and 2/3/6-VLPs. (A) Silver-stained SDS–10% polyacrylamide gel with proteins of the purified VLPs resulting from the coexpression of the indicated genes in the absence or presence of protease inhibitors. Protease inhibitors aprotinin and leupeptin, each at 1 μg/ml, were added daily. (B) Autoradiogram of [α-32P]GTP-bound VP3 to visualize the presence of VP3 in the VLPs shown in panel A. VPΔ2/3 and VP2/3 were coexpressed from the baculovirus recombinant pVL1393/SA11-3 with recombinants BVP2C24 and BacRF2A, respectively, and purified on a 5 to 20% sucrose gradient. VPΔ2/3/6 and VP2/3/6 were coexpressed from the baculovirus recombinants pVL1393/SA11-3 and pAc461/SA11-6 with recombinant BVP2C24 or BacRF2A, respectively, and purified by CsCl isopycnic centrifugation. To compare VP3 encapsidation, the protein amount loaded onto each lane was adjusted so that the amount of VPΔ2 was similar to or higher than that of the VP2 band A, the VP1 binding protein (Fig. 6C).
VP1 and VP3 are not, or not efficiently, encapsidated into VLPs lacking the N terminus of VP2.
We further examined if coexpressed VP1 and VP3 could be encapsidated into a VLP containing the VPΔ2. VLPs obtained from cells infected with recombinants expected to express 1/Δ2/3-VLPs were structurally composed of only VPΔ2 (Fig. 5A and B, lanes 3), while 1/2/3-VLPs were composed of VP1, VP2, and VP3 (Fig. 5A and B, lanes 4). Morphologically, both were single-layered types of VLPs resembling the particles shown in Fig. 1A, B, and E. Analyses of the VLPs expected to contain VP1/Δ2/3/6 and VP1/2/3/6 showed that both were morphologically double layered, resembling those seen in Fig. 1C and F, but VP1 and VP3 were present only when the VLPs contained full-length VP2 (Fig. 5A and B, lanes 1 and 2). The expression levels of VP1 and VP3 in the lysates were comparable at 80 h postinfection, and the unassembled VP1 was found in the top fractions of 5 to 20% sucrose gradients of VP1/Δ2/3 (data not shown).
FIG. 5.
Protein content of the coexpressed 1/Δ2/3-, 1/2/3-, 1/Δ2/3/6-, and 1/2/3/6-VLPs. (A) Silver-stained SDS–10% polyacrylamide gel with proteins of purified VLPs resulting from the expression of indicated genes in the absence or presence of protease inhibitors. Protease inhibitors aprotinin and leupeptin, each at 1 μg/ml, were added daily. (B) Autoradiogram of [α-32P]GTP-bound VP3 to visualize the presence of VP3 in the VLPs shown in panel A. VP1/Δ2/3 and VP1/2/3 were coexpressed from the baculovirus recombinants pVL941/RF-1 and pVL1393/SA11-3 with recombinant BVP2C24 or BacRF2A, respectively, and purified by centrifugation on a 5 to 20% sucrose gradient. VPΔ2/3/6 and VP2/3/6 were coexpressed from the baculovirus recombinants pVL941/RF-1, pVL1393/SA11-3, and pAc461/SA11-6 with recombinants BVP2C24 and BacRF2A, respectively, and purified by CsCl isopycnic centrifugation. To compare VP1 and VP3 encapsidation, the protein amount loaded onto each lane was adjusted so that the amount of VPΔ2 was similar to or higher than that of VP2 band A, the VP1 binding protein (Fig. 6C).
Analyses of interactions between VP1 and other proteins in VLPs.
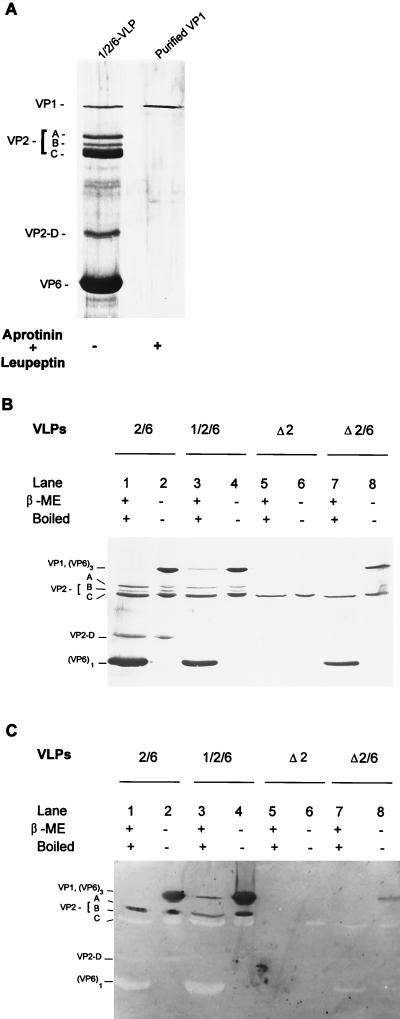
We next investigated the interactions of VP1 with the different proteins in the VLPs by far-Western blotting on PVDF membranes. The blotted proteins, including full-length VP2, VP2 cleavage products, VPΔ2, VP6 monomer, and VP6 trimer, were tested for the ability to bind VP1. A purified preparation of VP1 (Fig. 6A) was used as a probe. To clearly visualize all of the proteins blotted to the PVDF membrane, duplicate samples of those on the PVDF membrane were analyzed by SDS-PAGE (10% gel) followed by silver staining (Fig. 6B). The VP1 binding proteins are shown in Fig. 6C. By comparison with the silver-stained gel (Fig. 6B), only full-length VP2 (VP2-A [Fig. 6C, lanes 1 to 4]) and VP6 trimers (Fig. 6C, lanes 2, 4, and 8) were capable of binding VP1, while the VP2 cleavage products (bands B to D), VPΔ2, and VP6 monomers did not bind VP1 (Fig. 6C). The VP1 visualized in lane 3 in Fig. 6C is thought to be detected by interaction between the anti-VP1 and the VP1 molecules of the 1/2/6-VLPs rather than from the interaction of VP1 as the probe with itself in the VLPs.
FIG. 6.
Interactions between VP1 and other VLP proteins tested by far-Western blotting. (A) Silver-stained SDS–10% polyacrylamide gel of purified VP1 used for far-Western blotting to verify the VP1 binding proteins. VP1 was purified by FPLC followed by passage through an immunoaffinity column of rabbit IgG against wild-type baculovirus proteins. (B) Silver-stained SDS–10% polyacrylamide gel of the proteins in the indicated VLPs. Each purified VLP was (i) mixed with Laemmli buffer containing βME and boiled for 5 min or (ii) mixed with sample buffer without βME, kept at 4°C, and then resolved on an SDS–10% polyacrylamide gel at 4°C. (C) VP1 binding proteins detected on far-Western blots. A duplicate of the gel in panel B was run, and the proteins were transfered to a PVDF membrane for far-Western VP1 binding assay. After renaturation of the proteins blotted to the PVDF membrane and blocking of free binding sites, 10 μg of VP1 per ml was added to probe the proteins capable of binding VP1. The bound VP1 was detected by guinea pig anti-VP1 antibody followed by goat anti-guinea pig IgG-alkaline phosphatase conjugate and enzyme substrates.
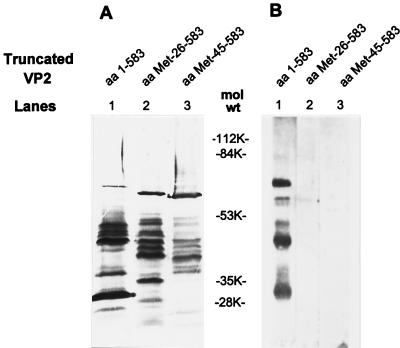
To determine the domain of VP2 required for binding VP1, three truncated constructs of VP2 (aa 1 to 583, aa Met-26 to 583, and aa Met-45 to 583 [truncation constructs P2AT1777, P2M16.9, and P2M16.3 in reference 17) were expressed in Sf9 cells. None of these truncated VP2 construct forms self-assembled into VLPs (17). Cell lysates were Western blotted to nitrocellulose to confirm that the truncated VP2 peptides were expressed. The lysates also were blotted to PVDF membranes for the VP1 binding test. Western blot analysis with monoclonal antibody (MAb) 164AE22, a MAb directed against an epitope on VP2RF between aa 45 and 92, showed that the three truncated forms of VP2RF were expressed well (Fig. 7A). VP1 binding was seen only with VP2 aa 1 to 583 and some of its cleavage products (Fig. 7B, lane 1), while VP2 aa Met-26 to 583 and VP2 aa Met-45 to 583 did not bind VP1 (Fig. 7B, lanes 2 and 3). Six additional truncated constructs of VP2 (aa 1 to 285, aa Met-290 to 880, aa Met-51 to 583, aa Met-61 to 583, aa Met-99 to 583, and aa Met-177 to 583) also were tested for VP1 binding. Only the construct containing aa 1 to 285 showed the ability to bind VP1 (data not shown). These results suggest that (i) a domain within the N terminus of VP2 aa 1 to 25 is essential for binding VP1, (ii) some conformations that form after VP6 trimerization are favorable for VP1 binding, and (iii) VP2 aa 26 to 880 are not sufficient for binding VP1.
FIG. 7.
Mapping the domain of VP2 required for binding VP1 by far-Western blotting. (A) Western blot of three truncated forms of VP2. Lysates of infected cells expressing three N- and C-terminus truncations of VP2 (aa 1 to 583, aa Met-26 to 583, and aa Met-45 to 583) were boiled with Laemmli sample buffer containing βME, resolved on an SDS–10% polyacrylamide gel, and blotted to nitrocellulose. They were probed by MAb 164AE22, which directs a VP2Rf epitope located between aa 45 and 92. (B) VP1 binding of truncated forms of VP2. Three VP2 truncation mutants were mixed with Laemmli buffer containing βME, boiled for 5 min, resolved on an SDS–10% polyacrylamide gel, and blotted to a PVDF membrane. After renaturation of the blots, 10 μg of VP1 per ml was added to probe the peptide fragments capable of binding VP1 as described for Fig. 6C.
DISCUSSION
The native rotavirus VP2 layer, the innermost core shell, serves important roles in the formation of transcriptionally active particles, with VP6 trimers forming the outside shell and VP1, VP3, and the genome of 11 segments of dsRNA being inside the shell. Our goals in this study were (i) to determine the peptide fragment of VP2 necessary for the formation of a single-layered VLP; (ii) to investigate the possible functions of the protease-accessible peptide fragment at the VP2 N terminus; and (iii) to map the domain of VP2 required for the binding and encapsidation of VP1, the RNA-dependent RNA polymerase, and VP3, the guanylyltransferase.
Expression of VP2 from baculovirus recombinants encoding the full-length VP2 by infection of Sf9 cells in the absence of protease inhibitors produces single-layered empty 2-VLPs identical in size and shape to its authentic counterpart (18, 37). This expressed single-layered 2-VLP contains two major cleavage products. One of them represents the peptide fragment of aa 93 to 880. Characterization of various truncated VP2 constructs by Labbé et al. (17) showed that expression of truncated VP2 constructs containing aa 1 to 583, aa 290 to 880, and aa 1 to 290 plus 589 to 880, as well as several other shorter VP2 constructs, did not lead to particle formation in insect cells. Previous work has shown that the rotavirus proteins expressed from baculovirus recombinants in Sf9 cells generally represent their cognate native proteins in both biochemical and functional characteristics (9, 38), and so the inability of those truncated VP2 peptides to form VLPs suggested that these truncations lacked the peptide domain necessary for the formation of single-layered 2-VLPs. To study this possibility, two truncated forms of VP2 that contain aa Met-93 to 880, called VPΔ2, and aa 186 to 880, called VPΔ2′, were created. Expression of VPΔ2′ in Sf9 cells did not result in particle assembly (data not shown). In contrast, expression of VPΔ2 resulted in the formation of single-layered core-like particles, identical to 2-VLPs in yields, appearance, size, and stability, designated Δ2-VLPs. This result suggests that the VP2 N-terminal aa 1 to 92 are not involved in the formation of single-layered 2-VLPs, and aa 93 to 185 are directly or indirectly needed for VP2 self-assembly into a particle. VPΔ2 was capable of forming not only VLPs but also elongated helix structures following concentration as seen previously for 2-VLPs (37). Coexpression of VPΔ2 with VP6 resulted in the formation of double-layered Δ2/6-VLPs identical to 2/6-VLPs in yields, size, shape, and stability. This finding suggests that Δ2-VLPs retain the characteristics necessary for the encapsidation of VP6 trimers and that aa 1 to 92 at the N terminus of VP2 are not involved in the assembly of VP6 trimers into Δ2/6-VLPs.
Coexpression of VPΔ2 plus VP1, VPΔ2 plus VP1 plus VP3, or VPΔ2 plus VP3 resulted in the formation of only single-layered Δ2-VLPs which lacked VP1 and VP3. Coexpression of VPΔ2 plus VP1 plus VP6, VPΔ2 plus VP1 plus VP3 plus VP6, or VPΔ2 plus VP3 plus VP6 produced only double-layered Δ2/6-VLPs which also lacked VP1 and VP3. The expression levels of VP1 and VP3 in 1/Δ2- versus 1/2-VLPs, 1/Δ2/3- versus 1/2/3-VLPs, and Δ2/3- versus 2/3-VLPs, and in 1/Δ2/6- versus 1/2/6-VLPs, 1/Δ2/3/6- versus 1/2/3/6-VLPs, and Δ2/3/6- versus 2/3/6-VLPs, were comparable as determined by dot blot assays for VP1 and by [α-P32]GTP binding for VP3 of dilutions of samples examined at 80 h postinfection. VP1 was detected only in the top fractions of gradients when 1/Δ2- and 1/Δ2/3-VLPs were analyzed by centrifugation through 5 to 20% sucrose gradients, while VP1 or VP3 comigrated with VP2 in the broad band obtained with a peak at fraction 9 when 1/2-, 1/2/3- and 2/3-VLPs were centrifuged in 5 to 20% sucrose gradients (data not shown). It is known that [α-32P]GTP does not label 2-, 1/2-, 2/6-, and 1/2/6-VLPs (38). Therefore, these results clearly suggest that VP2 lacking the N-terminal 92 aa has lost its capability to encapsidate VP1 alone, VP3 alone, or a VP1-VP3 complex. Prasad et al. (29) reported that a flower-shaped VP1-VP3 complex is seen attached to VP2 at the fivefold axes between the 160- and 220-Å radii in both 1/2/3/6-VLPs and native double-layered particles. Recently, Lawton et al. (20) reported a comparison of the inner surface structures between 2-VLPs and Δ2-VLPs; i.e., a loss of mass adjacent to the fivefold axes and a redistribution of mass along the fivefold axes were seen. These results imply that the portion of VP2 at the fivefold axes seen in three-dimensional reconstructions computed from EM images could represent a portion of the N terminus of VP2 extending out from the 2-VLP inner surface to anchor the VP1-VP3 complexes. This could explain why fragments consisting of VP2 aa 93 to 880 are capable of forming stable empty single-layered VLPs.
To map the domain of VP2 required for binding VP1, far-Western blotting, a method to probe protein-protein interactions, was used. Since full-length VP2 (band A) and its three cleavage products (band B [aa 43 to 880], band C [aa 93 to 880, and band D [aa 369 to 880]) are always detected when VP2 is expressed in the absence of protease inhibitors (37, 38) (see below), full-length VP2 and its cleavage products were useful probes for VP1 binding. Far-Western blotting showed that VP1 binds only to full-length VP2 (band A), not to the cleavage bands B, C, and D and VPΔ2. VP1 binding assays with the truncated VP2 consisting of aa 1 to 583, aa Met-26 to 583, and aa Met-45 to 583 showed that only VP2 from aa 1 to 583 is capable of binding VP1. No bands were seen when the BSA-blocked blots were only incubated with the primary or secondary antibody alone before or after incubation with VP1 (data not shown). These facts suggest that (i) a domain at aa 1 to 25 of the VP2 N terminus is required for binding VP1, (ii) aa 584 to 880 at the C terminus of VP2 are not sufficient for VP1 binding, and (iii) the lack of ability of Δ2-VLPs and Δ2/6-VLPs to encapsidate VP1 was not because of an added Met in front of VP2-C. In addition to VP2-A, VP6 trimers, but not VP6 monomers, were capable of interacting with VP1. This finding suggests that (i) a VP1-interacting conformation may exist in VP6 trimers but not in VP6 monomers; (ii) this VP1 binding site on the VP6 trimers may be located close to the VP2 layer, and some portion of VP1 may be associated with VP2 and extend across the VP2 layer into the VP6 layer; and (iii) the preformed VP1-(VP6)3 complexes could not be encapsidated, because Δ2/6-VLPs did not show any evidence of VP1 association. We also probed VP1 from 1/2/6-VLPs with purified VP6 by far-Western blotting. Guinea pig anti-VP6 was used as the primary antibody. The VP6 bound to VP1 and also to the VP2 bands A to C (data not shown). The function, if any, of this interaction between VP1 and VP6 trimers remains unknown. In addition, although it is clear that VP2 aa 1 to 92 are necessary for VP3 encapsidation, we were not able to further map the domain of VP2 required for binding VP3 because the required reagents, e.g., purified VP3, are not available.
In the discussion of a previous report (37), based on its apparent molecular weight in SDS-PAGE, we speculated that VP2-B represented aa 1 to 789 and had a blocked N terminus. This conclusion was reached because the amount VP2-B available was low and difficult to be clearly sequenced. Recently, we obtained additional VP2-B material, blotted it onto an Immobilon-P transfer membrane (Millipore), and sequenced the protein by using an Applied Biosystems 477A protein sequencer in the Protein Sequencing Core Facility of Baylor College of Medicine. An N-terminal sequence was obtained and found to be KKEEVVTDS; this sequence begins at aa 44 of VP2 (16, 36). Recent studies have also detected another cleavage product of VP2 that has a molecular weight of ∼58,000, designated VP2-D. Its N terminus has been sequenced as GINSQAAND beginning at aa 369 of VP2 (4, 16). The significance of these cleavage products remains to be determined.
VP1 is thought to be the RNA-dependent RNA polymerase of rotavirus. It, together with VP3, binds to VP2 at the fivefold axes (29). VP1 also specifically binds to the 3′ end of viral mRNA (24), and the seven nucleotides at the 3′ terminus provide the minimum requirement for replication, at least, of the segment 9 mRNA (34). RNA also binds to the N terminus of VP2 (17), to which VP1 and VP3 also bind (this study). Thus, we know that the polymerase VP1, the guanylyltransferase VP3, and template mRNA all colocalize at the N terminus of VP2 at the fivefold axes, but we do not yet know precisely how they function to bring about RNA transcription, elongation, and exiting from particles.
In summary, we have expressed VPΔ2, an N-terminal 92-aa-deleted VP2, alone and with other species of proteins existing in the single- and double-layered capsids of rotavirus in the baculovirus system, and we have purified and characterized the resulting VLPs. The VP1 and VP3 are not assembled into VLPs with VPΔ2. The N-terminal peptide of VP2 is required for encapsidation of VP1 and VP3.
ACKNOWLEDGMENTS
We gratefully acknowledge the helpful discussions and critical review of the manuscript provided by Sue E. Crawford, J. A. Lawton, and B. V. V. Prasad. We also thank Marie Labbé for preparing the recombinant baculovirus BVP2C24.
This work was supported by Public Health Service grant DK 30144 from the National Institutes of Health and by European Economic Community grant INCO IC18-CT96-0027.
REFERENCES
- 1.Blacklow N R, Greenberg H B. Viral gastroenteritis. N Engl J Med. 1991;325:252–264. doi: 10.1056/NEJM199107253250406. [DOI] [PubMed] [Google Scholar]
- 2.Boyle J F, Holmes K V. RNA-binding proteins of bovine rotavirus. J Virol. 1986;58:561–568. doi: 10.1128/jvi.58.2.561-568.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns J W, Greenberg H B, Shaw R D, Estes M K. Functional and topographical analyses of epitopes on the hemagglutinin (VP4) of the simian rotavirus 11. J Virol. 1988;62:2164–2172. doi: 10.1128/jvi.62.6.2164-2172.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burroughs, M. H., S. E. Crawford, and M. K. Estes. Unpublished data.
- 5.Chen D, Gombold J L, Ramig R F. Intracellular RNA synthesis directed by temperature-sensitive mutant of simian rotavirus SA11. Virology. 1990;178:143–151. doi: 10.1016/0042-6822(90)90387-7. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D., and R. F. Ramig. Unpublished data.
- 7.Chen D, Zeng C Q-Y, Wentz M J, Gorziglia M, Estes M K, Ramig R F. Template-dependent, in vitro replication of rotavirus RNA. J Virol. 1994;68:7030–7039. doi: 10.1128/jvi.68.11.7030-7039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J, Charpilienne A, Chilmonczyk S, Estes M K. Nucleotide sequence of bovine rotavirus gene 1 and expression of the gene product in baculovirus. Virology. 1989;171:131–140. doi: 10.1016/0042-6822(89)90519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford S E, Labbé M, Cohen J, Burroughs M H, Zhou Y J, Estes M K. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5945–5952. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estes M K. Rotavirus and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publisher; 1996. pp. 1625–1655. [Google Scholar]
- 11.Estes M K, Crawford S E, Penaranda M E, Petrie B L, Burns J W, Chan W-K, Ericson B, Smith G E, Summers M D. Synthesis and immunogenicity of the rotavirus major capsid antigen using a baculovirus expression system. J Virol. 1987;61:1488–1494. doi: 10.1128/jvi.61.5.1488-1494.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estes M K, Mason B B, Crawford S, Cohen J. Cloning and nucleotide sequence of the simian rotavirus gene 6 that codes for the major inner capsid protein. Nucleic Acids Res. 1984;12:1875–1887. doi: 10.1093/nar/12.4.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gombold J L, Ramig R F. Assignment of simian rotavirus SA11 temperature-sensitive mutant group A, C, F, and G to genome segments. Virology. 1987;161:463–473. doi: 10.1016/0042-6822(87)90140-1. [DOI] [PubMed] [Google Scholar]
- 14.Homann H E, Willenbrink W, Buchholz C J, Neubert W J. Sendai virus protein-protein interactions studied by a protein-blotting protein-overlay technique: mapping of domains on NP protein required for binding to P protein. J Virol. 1991;65:1304–1309. doi: 10.1128/jvi.65.3.1304-1309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapikian A Z, Flores J, Midthun K, Hoshino Y, Green K Y, Gorziglia M, Nishihawa K, Chanock R M, Potash L, Perez-Schael I. Strategies for the development of a rotavirus vaccine against infantile diarrhea with an update on clinical trials of rotavirus vaccines. Adv Exp Med Biol. 1990;257:67–90. doi: 10.1007/978-1-4684-5712-4_9. [DOI] [PubMed] [Google Scholar]
- 16.Kumar A, Charpilienne A, Cohen J. Nucleotide sequence of the gene encoding for the RNA binding protein (VP2) of RF bovine rotavirus. Nucleic Acids Res. 1989;17:2126. doi: 10.1093/nar/17.5.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labbé M, Baudoux P, Charpilienne A, Poncet D, Cohen J. Identification of the nucleic acid binding domain of the rotavirus VP2 protein. J Gen Virol. 1994;75:3423–3430. doi: 10.1099/0022-1317-75-12-3423. [DOI] [PubMed] [Google Scholar]
- 18.Labbé M, Charpilienne A, Crawford S E, Estes M K, Cohen J. Expression of rotavirus VP2 produces empty core-like particles. J Virol. 1991;65:2945–2952. doi: 10.1128/jvi.65.6.2946-2952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lawton J A, Zeng C Q-Y, Mukherjee S K, Cohen J, Estes M K, Prasad B V V. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee W S, Kao C C, Bryant G O, Liu X, Berk A J. Adenovirus EIA activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991;67:365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- 22.Liu M, Mattion N M, Estes M K. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology. 1992;188:77–84. doi: 10.1016/0042-6822(92)90736-9. [DOI] [PubMed] [Google Scholar]
- 23.Mansell E A, Patton J T. Rotavirus RNA replication: VP2, but not VP6, is necessary for viral replicase activity. J Virol. 1990;64:4988–4996. doi: 10.1128/jvi.64.10.4988-4996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patton J T. Rotavirus VP1 alone specifically binds to the 3′ end of viral RNA, but the interaction is not sufficient to initiate minus-stranded synthesis. J Virol. 1996;70:7940–7947. doi: 10.1128/jvi.70.11.7940-7947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pizarro J L, Sandino A M, Pizarro J M, Fernandez J, Spencer E. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. J Gen Virol. 1991;72:325–332. doi: 10.1099/0022-1317-72-2-325. [DOI] [PubMed] [Google Scholar]
- 26.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prasad B V V, Chiu W. Structure of rotavirus. In: Ramig R F, editor. Rotaviruses. Berlin, Germany: Springer-Verlag; 1994. pp. 9–30. [Google Scholar]
- 28.Prasad B V V, Estes M K. Molecular basis of rotavirus replication: structure-function correlations. In: Chiu W, Burnett R, Garcia R, editors. Structural biology of viruses. Oxford, England: Oxford Press; 1997. pp. 239–268. [Google Scholar]
- 29.Prasad B V V, Rothnagel R, Zeng C Q-Y, Jakara J, Lawton J A, Chiu W, Estes M K. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature (London) 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 30.Prasad B V V, Wang G J, Clerx J P M, Chiu W. Three dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 31.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agricultural Experimental Station bulletin no. 1555. College Station, Tex: Texas A & M University; 1987. [Google Scholar]
- 32.Towbin T, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valenzuela S, Pizzaro J, Sandino A M, Vásquez M, Fernández J, Hernández O, Patton J, Spencer E. Photoaffinity labeling of rotavirus VP1 with 8-azido ATP: identification of the viral RNA polymerase. J Virol. 1991;65:3964–3967. doi: 10.1128/jvi.65.7.3964-3967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wentz M J, Patton J T, Ramig R F. The 3′-terminal consensus sequence of rotavirus mRNA is the minimal promotor of negative-strand RNA synthesis. J Virol. 1996;70:7833–7841. doi: 10.1128/jvi.70.11.7833-7841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeager M, Dryden K A, Olson N H, Greenberg H B, Baker T S. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J Cell Biol. 1990;110:2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng, C. Q.-Y., and M. K. Estes. Unpublished data.
- 37.Zeng C Q-Y, Labbé M, Cohen J, Prasad B V V, Chen D, Ramig R F, Estes M K. Characterization of VP2 particles. Virology. 1994;201:55–65. doi: 10.1006/viro.1994.1265. [DOI] [PubMed] [Google Scholar]
- 38.Zeng C Q-Y, Wentz M J, Cohen J, Estes M K, Ramig R F. Characterization and replicase activity of double-layered and single-layered rotavirus-like particles expressed from baculovirus recombinants. J Virol. 1996;70:2736–2742. doi: 10.1128/jvi.70.5.2736-2742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]