Abstract
Simian immunodeficiency virus SIVMne, like human immunodeficiency virus, evolves from a macrophage-tropic, non-syncytium-inducing virus at early times in infection to a T-cell-tropic, syncytium-inducing, cytopathic virus population over the course of progression to AIDS. Because the viruses isolated late in SIVMne infection of macaques include a complex mixture of variants, the viral determinants of such phenotypic changes have not been defined. To identify genetic changes that are important to virus evolution in the host, we constructed chimeric viruses by introducing variant envelope genes representative of proviruses throughout the course of infection and disease into the SIVMne parental clone (SIVMneCL8) that infected the macaque. The chimeric viruses expressed sequences encoding the surface unit of the envelope glycoprotein (Env-SU) of variants cloned between 35 and 170 weeks postinfection. The chimera with Env-SU from 35 weeks postinfection encoded only four changes in V1 compared to SIVMneCL8, whereas the chimeras encoding Env-SU from variants isolated later in infection encoded progressively more mutations both in V1 and elsewhere. Like SIVMneCL8, the chimeras were infectious for CEMx174 cells and macaque peripheral blood mononuclear cells. However, in contrast to SIVMneCL8, the chimeric viruses did not infect macaque macrophages, although each retained the ability to recognize the CCR-5 coreceptor. Thus, these data provide direct evidence that changes which evolve in Env-SU during the course of SIVMne infection do not alter CCR-5 interactions. Viruses encoding Env-SU from the latest times in infection (121 to 170 weeks postinfection), after disease was apparent, were syncytium inducing. However, these viruses were not highly cytopathic, suggesting that additional viral determinants may be required for the rapidly replicating, cytopathic phenotype of the uncloned mixed variant population. Changes in Env-SU did allow the virus to escape serum neutralizing antibodies that recognized the SIVMneCL8 parent. Moreover, the chimera encoding the Env-SU of a virus from 35 weeks postinfection, which differed from SIVMneCL8 only in V1, was not sensitive to neutralization by infected macaque sera, suggesting that V1 may define a portion of the principal neutralizing determinant for SIVMne. Together, these data suggest that SIV variants with changes in the Env-SU may be selected primarily by virtue of their ability to escape neutralizing antibody recognition.
Rapid and continued viral diversity is typical of infections with human and simian immunodeficiency viruses (HIV and SIV). The ability of these lentiviruses to continually evolve in the host may contribute to their ability to persist in an individual despite an active specific immune response against the virus. Unfortunately, persistent lentivirus infections generally lead, with time, to an unremitting disease. Therefore, to design therapeutic approaches that can modulate the course of lentivirus diseases, it is essential not only to characterize the virus variants that evolve during the course of infection but also to understand the basis for their selection in the host.
SIV infection of macaques provides a model system with which to study lentivirus variation as it relates to development of fatal immunodeficiency disease. The value of this model for studies of persistent infection and AIDS pathogenesis is due, in part, to the fact that molecular clones of SIV that cause an immunodeficiency disease very much like HIV-related disease in humans have been identified (22). There are other important parallels between HIV infection in humans and SIV infection in macaques, among them that SIV, like HIV, evolves from a macrophage-tropic (M-tropic) virus at early times in infection to a T-cell-tropic (T-tropic), cytopathic virus mixture over the course of progression to AIDS (36).
In studies of viral diversity in pig-tailed macaques (Macaca nemestrina) that develop AIDS following infection with a clone of the SIVMne isolate, we found that there was strong selection for viruses with particular amino acid changes in the envelope protein (33, 34). Viruses with changes in V1 of the extracellular envelope protein (Env-SU) could be detected prior to onset of disease, and such variants were present in each of five macaques infected with a SIVMne clone (12, 33, 34). Changes in V4 were also occasionally observed, and these changes were found in viruses selected later in infection. A similar pattern of variation is typical of SIVmac and SIVsm infections (8, 23), suggesting that these envelope changes are selected in the context of different infecting viral strains and in different macaque host species.
In studies with a variety of HIV type 1 (HIV-1) variants, the gene coding for the Env-SU was shown to encode the primary viral determinant for tropism and the cytopathic properties of the virus (reviewed in references 27 and 30). In most cases, the determinants mapped to the V3 region of envelope, but in some proviruses, other variable regions, such as V1 or V2, determined the phenotypic properties of the variant (reviewed in references 27 and 30). Interestingly, V3 encodes a principal neutralizing determinant for cell culture-adapted HIV-1 (reviewed in reference 7), although the role of anti-V3 antibodies in the neutralization of viruses within the host remains somewhat unclear. Thus, it is possible that variation in V3 reflects immunological pressures applied against the virus by the host and that V3 changes, although selected for different reasons, in turn affect the cytopathic properties, replication, or tropism of the virus. In SIV, the corresponding V3 region of envelope is conserved (8, 23, 34). Based on the pattern of variation in SIVMne infection, we have previously proposed that the V1 region of SIV envelope may be functionally equivalent to the V3 of HIV-1, or there may be combinations of variable regions in the envelope protein that act in concert to influence the phenotype of either virus (36).
In an attempt to understand lentivirus persistence and its role in pathogenesis, in this study we explored the basis for selection of SIVMne variants with changes in V1 and V4 of the envelope SU. Surprisingly, we found that the Env-SU does not encode the primary determinant for the replication and cytopathic properties of SIVMne, although V1 sequence changes affect syncytium formation and encode a determinant of macrophage tropism. Our studies show that the V1 region of envelope encodes sequences that comprise part of the principal neutralizing determinant for SIVMne, suggesting that humoral immune responses may drive selection for viruses with mutations in V1.
MATERIALS AND METHODS
Construction of chimeric viruses.
Variant envelope genes, which were cloned into M13mp18 following PCR amplification (described in references 34 and 37), were used to construct chimeric viruses with the SIVMneCL8 parent. The NsiI-ClaI fragment of the envelope gene clones, which includes sequences encoding V1 through V5 in the extracellular envelope glycoprotein (Env-SU), was cloned back into SIVMneCL8 in a two-step cloning protocol. First, we generated a KpnI subclone from SIVMneCL8 that spans positions 5239 to 8847 of the parental viral genome, including all of the envelope gene as well as vif-vpx-vpr-tat-rev and portions of nef. This vector was digested with ClaI and then partially digested with NsiI, and a 5.2-kb fragment, which was predicted to have the NsiI cloning site at position 6392 and the ClaI cloning site at position 7546, was gel purified by standard methods. The M13mp18 clones with PCR-generated variant envelope genes were also digested with NsiI and ClaI, and a 1.1-kb fragment encompassing most of the coding sequences for Env-SU, including V1 through V5, was gel purified. This fragment was ligated into the 5.2-kb NsiI-ClaI-digested KpnI subclone of SIVMneCL8. Correct clones were verified by restriction mapping and nucleotide sequence analysis of the V1 sequences. The full-length viral genome was reconstructed by using a unique BstI site 5′ of the envelope coding region (position 5343 in the full-length SIVMneCL8 provirus) and the ClaI site at the end of SU (position 7546). The BstI-ClaI fragment (2.2 kb) was gel purified from the chimeric KpnI subclone(s) and ligated to a similarly digested, gel-isolated 14-kb fragment representing the plasmid encoding the SIVMneCL8 provirus minus its envelope gene. Again, the clones were verified by restriction site analysis and nucleotide sequence analysis of the envelope gene regions spanning V1 through V5.
Transfection of plasmid clones.
Plasmid clones encoding SIVMne chimeras were transfected into CEMx174 cells (a cell line derived from a fusion of a human T-cell line [CEM] and a human B-cell line [721.174]) by using DEAE-dextran. Cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 1 mM glutamine (complete RPMI medium). At approximately 3-day intervals, the cells were counted and diluted in fresh medium to a concentration of 5 × 105/ml. At each of these times, culture supernatant was collected and production of SIV in the supernatant was measured by using an assay specific for SIVp27gag antigen (Immunotech, Westbrook, Maine) as described previously (36). At two time points near the peak level of antigen production, virus supernatants were collected and stored at −70°C. These viral supernatants from transfected CEMx174 cells were used for all subsequent infection studies involving the chimeras and SIVMneCL8. The infectious titer of SIV in the culture supernatants was measured by the sMAGI assay, which previous studies have shown to be a highly sensitive method for determining the tissue culture infectious dose of a variety of SIV variants (10).
Infection of macaque PBMCs.
Peripheral blood mononuclear cells (PBMCs) were isolated from SIV- and simian type D retrovirus-negative M. nemestrina as described previously (36). PBMCs were seeded into 24-well plates at a density of 8 × 105 in 1 ml of complete RPMI medium. Cultures were immediately infected (in duplicate) with virus at a multiplicity of infection (MOI) of 0.01. At 24 h postinfection, cells were washed twice in phosphate-buffered saline and resuspended in 2 ml of complete RPMI medium supplemented with 20 U of human interleukin-2 (Boehringer Mannheim) per ml. At 3- to 4-day intervals, 1 ml of medium was removed and tested for SIV p27gag, and complete RPMI medium plus interleukin-2 was added to a final volume of 2 ml.
Isolation and infection of primary macrophages.
Monocytes/macrophages were isolated from the peripheral blood of a SIV- and simian type D retrovirus-negative M. nemestrina and cultured as described previously (36). The cells were demonstrated to be approximately 90 to 95% monocytes/macrophages by the following criteria: (i) adherence to plastic, (ii) morphology, (iii) nonspecific esterase assay (Sigma Chemical Co.), (iv) reactivity with an anti-human macrophage monoclonal antibody, EBM-11 (anti-CD68; Dako Corp.), and (v) phagocytosis of polystyrene latex beads (Sigma). The monocytes/macrophages (2.5 × 105) were seeded into wells of 48-well plates containing macrophage medium (RPMI 1640 supplemented with 10% human AB serum and 5% fetal bovine serum which were heat inactivated for 30 min, 10% GCT conditioned supernatant, penicillin [100 U/ml], streptomycin [100 μg/ml], and 1 mM glutamine). After 5 days, cultures were infected at an MOI of 0.001 with the different clones of SIV (in duplicate). Two days postinfection, the cells were washed twice with serum-free RPMI 1640 medium and cultured in 1 ml of macrophage medium. Every 3 to 4 days, 0.5 ml of medium was removed and replaced with 0.5 ml of fresh macrophage medium.
Infection of CEMx174 cells.
Infection studies with CEMx174 cells were similar in design to those described previously (36). An equal infectious dose (8 × 103) of each virus was used to infect 8 × 105 CEMx174 cells in a final volume of 1 ml of complete RPMI medium; 1 day later, cells were washed twice in phosphate-buffered saline and resuspended in 2 ml of complete RPMI medium. At 2- to 3-day intervals, cells were counted and diluted to a concentration of 5 × 105/ml with medium and the SIVp27gag levels in the supernatants before dilution were measured. Cell viability was determined by trypan blue exclusion. Syncytia that were at least 5 normal cell diameters across were counted in each culture. Each infection was carried out in duplicate, and infections with each virus were repeated on at least three occasions with similar results.
Infection of MAGI-CCR5 and sMAGI cells.
To test coreceptor specificity in a single-cycle assay that does not require virus spread, we used sMAGI and MAGI cell derivatives (10, 11, 25). sMAGI cells are rhesus macaque CMMT cells that express human CD4 and a HIV–long terminal repeat (LTR)–β-galactosidase (β-gal) reporter gene cassette (10). MAGI cells are human HeLa cells similarly engineered to express human CD4 and the HIV-LTR-β-gal cassette (25). MAGI-CCR5 cells are MAGI cells engineered to express the human CCR-5 β-chemokine receptor (11). In the present study, sMAGI, MAGI, and MAGI-CCR5 cells were cultured and infected as described previously (10, 25). Briefly, 4 × 104 (MAGI or MAGI-CCR5) or 8 × 104 (sMAGI) cells were plated in a 24-well plate a day before infection. Virus was added the next day in the presence of DEAE-dextran at a final concentration 20 μg/ml (MAGI or MAGI-CCR5) or 15 μg/ml (sMAGI) in 150 μl of medium. After 2 (MAGI or MAGI-CCR5) or 3 (sMAGI) days, the cells were stained for β-gal expression as described previously (10, 25).
Neutralization assays.
Neutralization was measured by infecting sMAGI cells with virus in the presence versus absence of serum as described previously (10). Serum samples were obtained from four SIV-infected M. nemestrina macaques; two (macaques 93100 and 92221) were infected with SIVMne, an uncloned mixture from which SIVMneCL8 was derived, and the others (90027 and 92154) were infected with SIVMne E11S, the biological single-cell clone from which the SIVMneCL8 proviral clone was obtained (2, 3). Serum samples were obtained both prior to infection (prebleed) and 5 to 6 months postinoculation. All serum samples were heat inactivated for 30 min at 56°C. Sera was serially diluted two-fold in complete Dulbecco modified Eagle medium (DMEM) (DMEM supplemented with 10% fetal calf serum plus 100 U of penicillin, 100 μg of streptomycin, and 300 μg of glutamine per ml) to a final volume of 25 μl and added to an equal volume of a standard virus stock. For neutralization experiments, 100 to 300 infectious virions harvested from CEMx174 cells transfected with the viral chimeras or SIVMneCL8 were used. The mixture was incubated at 37°C in a 5% CO2 incubator for 45 min. The serum-virus mixture was added, in the presence of 15 μg of DEAE-dextran per ml, to sMAGI cells that had been seeded in a 24-well plate at a density of 8 × 104 cells per well the previous day (10). After 2 h of infection, another 1 ml of complete DMEM was added. The cells were fixed and stained 3 days after infection as described previously (10). The dilution of sera that resulted in 90% neutralization was extrapolated by plotting the serum dilution against the number of infected (β-gal-positive, blue) sMAGI cells in the presence of serum (Vn) over the number in the absence of serum (Vo). At each dilution, duplicate infections were used to determine Vn and Vo, and experiments were repeated on multiple occasions. As a negative control, prebleed sera from each animal was tested against each virus. Although there was some nonspecific activity in prebleed sera at higher dilutions (1:4 and 1:8), it was never above the 90% criterion used to define neutralization titers in this study (data not shown).
RESULTS
Sequences of the envelope gene of the chimeric viruses.
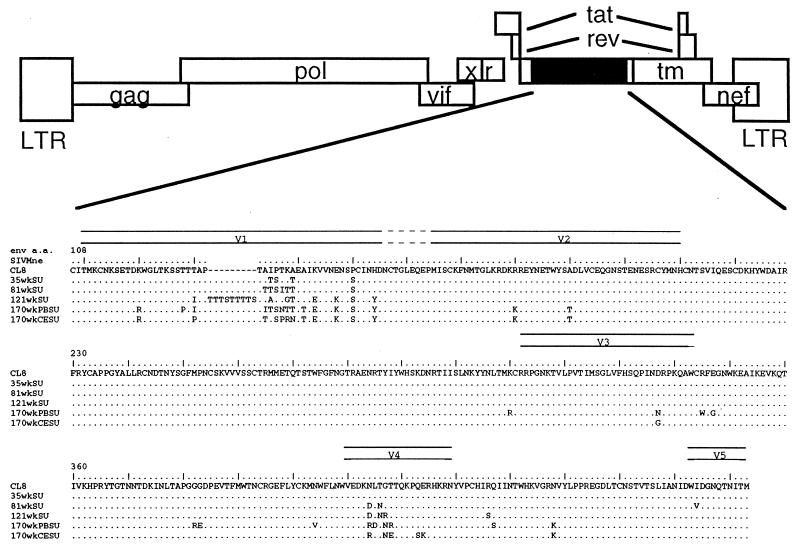
Five chimeric viruses with variant envelope gene sequences isolated from macaque M87004 were constructed (Fig. 1). The particular envelope sequences represented in the chimeras were obtained throughout the course of infection and disease, including viruses present before overt disease, when changes in V1 had just begun to accumulate, through variants present at the terminal stages of disease, when there was more extensive variation in other regions of envelope (33, 34). This macaque developed AIDS at about 1 year postinfection (34). Three clones, from 35, 81, and 121 weeks postinfection, were obtained directly from macaque M87004 PBMCs (34). The two clones from 170 weeks postinfection were isolated from cell cultures expressing a cytopathic, syncytium-inducing virus population that were obtained by briefly coculturing macaque M87004 PBMCs with macaque PBMCs (170wkPBSU virus) or CEMx174 cells (170wkCESU virus) (36, 37). The sequences of V1, V3, and V4 of the variant envelope genes were described previously (34, 37). To identify other changes, the complete sequence of each of the five variant envelope segments expressed in the chimeric genome was determined (Fig. 1B). As reported previously, changes in V1 that encode predicted sites for O-linked and N-linked glycosylation, which are the hallmark of SIVMne envelope variation (33, 34), were common to all of the variants. The envelope gene encoded in the 35wkSU chimera, which was isolated before the onset of AIDS in macaque M87004, contained only four changes, two serines and two threonines in V1. The envelope gene cloned soon after the decline in CD4+ T lymphocytes in macaque M87004 (81 weeks postinfection) had more extensive mutations in V1. The variant from 121 weeks postinfection contained an insertion in V1 that is predicted to encode a nine-amino-acid stretch of serine and threonine residues. Envelope genes cloned from 81 weeks and later times in infection also had acquired mutations encoding potential glycosylation changes in V4. In addition, there were scattered single amino acid changes in the envelope of the two variants from 170 weeks postinfection.
FIG. 1.
Amino acid sequence of the extracellular envelope encoded by the chimeras. (A) The general structure of the chimeric proviruses is depicted schematically. Open boxes denote regions derived from the SIVMneCL8 parent virus, and black boxes denote variants sequences engineered into the chimeras. (B) The predicted amino acid sequence of variant Env-SU in comparison to SIVMneCL8. Only the portion of Env-SU encoded in the chimeras, which includes V1 to V5 as indicated above the sequence, is shown. The number corresponding to the amino acid (a.a.) position in SIVMneCL8 envelope SU is indicated, and the SIVMneCL8 reference sequence is shown in single-letter code. Dashes were introduced to show the position of an insertion in one of the variant envelope clones relative to SIVMneCL8. The sequences of variant envelopes are shown below, and dots are used to indicate identity. The origin of each of the envelope gene subclones used to construct the chimeras has been described previously: 35wkSU, 81wkSU, and 121wkSU represent 35wk:1-1, 81wk:2-3, and 121wk:2-2, respectively (34); 170wkPBSU and 170wkCESU correspond to 170wk B-5 (PB-V1-15) and 170wk D (CE V1-35), respectively (37).
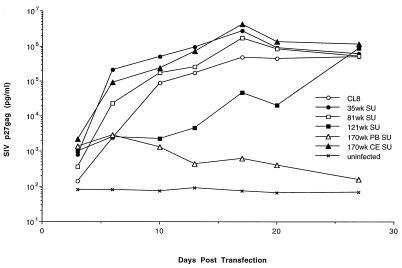
Transfection and expression of chimeric viruses.
To determine if the chimeric viruses were replication competent, we transfected the plasmid DNAs encoding the cloned provirus into CEMx174 cells, which are highly permissive for SIVMne infection (36, 37). Production of infectious virus was monitored at 3- to 4-day intervals over the course of 4 weeks by testing for SIVp27gag in the culture supernatant (Fig. 2).. SIVp27gag antigen could be detected within 4 to 6 days after transfection in each culture, although the amounts of SIVp27gag antigen in the supernatants varied by several orders of magnitude among the viruses tested. By 10 days posttransfection, the 35wkSU, 81wkSU, and 170wkCESU chimeras expressed levels of viral antigen that were similar to that of the parental virus SIVMneCL8. Virus replication was delayed in cells transfected with the 121wkSU chimera, which had an insertion in V1, but they reached the same peak levels as SIVMneCL8 and the other chimeras by 4 weeks posttransfection. The 170wkPBSU chimera expressed very low levels of virus throughout the 4-week period and did not spread in culture. While the defect in the replication of this chimera has not been examined in detail, two charged amino acid residue changes in a highly conserved region of Env-SU could account for this defect (amino acids 382 and 383 [Fig. 1]).
FIG. 2.
Replication following transfection of SIVMne chimeras in CEMx174 cells. CEMx174 cells were transfected with each chimeric virus clone as well as SIVMneCL8. At 3- to 4-day intervals for 4 weeks after transfection, virus replication and spread were monitored by assaying for SIVp27gag in the culture supernatants. To determine the levels of SIVp27gag antigen, dilutions of supernatant were tested in the linear range of the assay. SIVp27gag is plotted as a function of days posttransfection.
The differences in antigen levels were reflected in the amounts of infectious virus that could be detected by sMAGI assay. For example, there were only about 102 infectious particles per ml expressed from cells transfected with the 170wkPBSU chimera, whereas ≥104 to 105 infectious virions were detected at the peak of antigen expression in cells transfected with the other chimeras (Table 1 and data not shown). The envelope protein was expressed and processed normally in these chronically infected cells, except for cells transfected with the 170wkPBSU chimera; in the latter case, the envelope protein could not be detected by immunoprecipitation of radiolabeled cell extracts (data not shown).
TABLE 1.
Infection of MAGI-CCR5 and sMAGI cells
| Virus | Infectious virions/mla
|
||
|---|---|---|---|
| sMAGI | MAGI-CCR5 | MAGI | |
| MneCL8 | 1.2 × 104 | 1.6 × 104 | 0b |
| 35wkSU | 8.7 × 104 | 1.2 × 105 | 0c |
| 81wkSU | 2.2 × 104 | 3.3 × 104 | 0b |
| 170wkCESU | 3.9 × 104 | 3.8 × 104 | 0b |
| MixVar | 9.1 × 105 | 7.6 × 105 | 0c |
| Mac1A11 | 1.0 × 104 | 1.0 × 104 | NTd |
| Mac239 | 2.8 × 104 | 1.4 × 104 | 0b |
| HIV-1SF162 | 0 | 3.2 × 104 | NT |
Average of infections performed in triplicate.
Based on challenge experiments with 2 × 103 sMAGI-infectious virions.
Based on challenge experiments with 2 × 104 sMAGI-infectious virions.
NT, not tested.
Infection and replication in primary macaque lymphocyte and macrophage cultures.
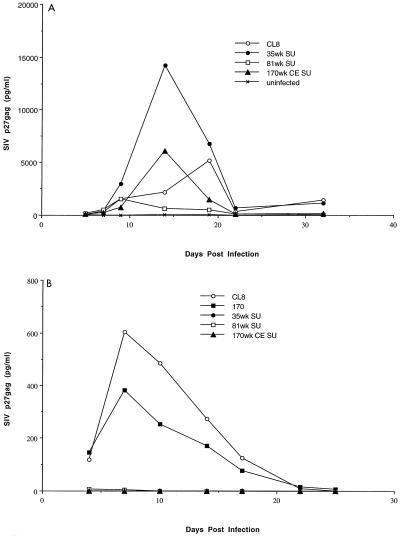
To determine if the envelope of viruses selected over the course of SIVMne infection altered the replication properties of the virus, cultures from primary macaque blood cells were infected with the chimeric viruses. There were low to moderate levels of virus replication, as judged by SIVp27gag expression, in macaque PBMCs infected with the chimeric viruses (Fig. 3A). In general, the level of replication of the chimeras in these cells was similar to that of the parent virus, SIVMneCL8, although two- to threefold differences were observed in some infection experiments. For example, in the experiment shown in Fig. 3A, the peak of antigen production was threefold higher for the 35wkSU chimera and threefold lower for the 81wkSU chimera compared to SIVMneCL8, whereas the peak antigen levels were similar for 170wkCESU and SIVMneCL8. These relatively small differences between viruses were not consistently observed when infections were performed in PBMCs from different macaque donors (data not shown); this donor cell-dependent difference in replication has also been observed with cloned SIVMne proviral variants (24). In light of this finding and the modest differences observed in replication between viruses in cultures from single macaque donor, our studies suggest that there is no consistent replication advantage to the viruses with variant envelope sequences that evolve over time in the host. However, we cannot rule out that there may be animal-specific advantages for replication of one virus over another. Unfortunately, it was not possible to study replication of these viruses in uninfected donor cells from the macaque in which they evolved.
FIG. 3.
Infection of primary macaque cells. M. nemestrina blood-derived cells were infected with virus expressed from CEMx174 cells transfected with the plasmid viral clones. Infection and virus replication were monitored by an assay for SIVp27gag. (A) PBMCs (8 × 105) were infected at an MOI of 0.01; (B) macrophage cultures (2.5 × 105 cells) were infected at an MOI of 0.001. The data represent an average of duplicate infection experiments. In panel B, 170 refers to a full-length molecular clone, SIVMne170, described previously (24).
Our previous studies showed that the virus isolated from macaques at later times in infection was poorly infectious for macaque macrophages compared to the M-tropic parental virus, SIVMneCL8 (36). The tropism and replication of select chimeras were examined in macaque macrophage cultures (Fig. 3B); in these cells, little if any virus replication was detected throughout 25 days in culture. In contrast, virus replication could readily be detected in macrophages infected with SIVMneCL8, as shown previously (36). In addition, a full-length molecular clone isolated from 170 weeks after infection of macaque M87004 (Fig. 3B, 170) (24) replicated in macaque macrophages. This difference between the chimeric viruses and SIVMneCL8 and SIVMne170 was reproducible in macrophage cultures from two different macaques. Interestingly, even the 35wkSU chimera, which encodes only V1 changes, replicated with almost 100-fold-lower efficiency in macaque macrophage cultures compared to the otherwise isogenic parental virus, SIVMneCL8. Because these studies rely on virus spread, it is unclear whether the restriction to macrophage replication is at entry or at later stages in replication. Previous studies with SIVmac239, which encodes an Env-SU with V1 sequences that resemble the variants described here (33), suggest that the restriction to macrophage infection by this virus is at a postentry stage in replication (31).
Taken together, analyses of the replication of the chimeric viruses in primary macaque blood cells suggest that the envelope SU protein of variants that evolve during the course of infection does not, by itself, confer an advantage for virus replication. Moreover, in the context of the parental virus genome, the variant envelope proteins may limit replication in macrophages.
Infection of CEMx174 cells.
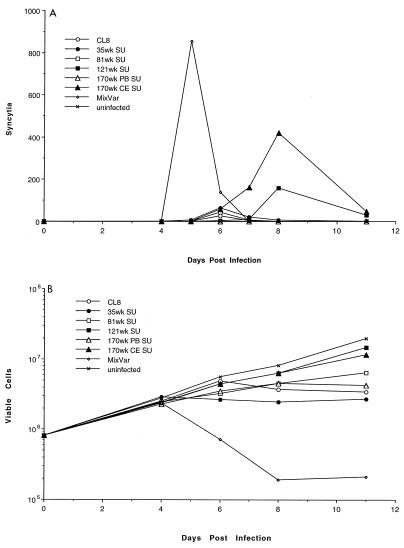
Our previous studies showed that the virus population in SIVMne-infected macaques with AIDS was highly cytopathic and syncytium inducing in CD4+ cell lines, particularly CEMx174 cells, while SIVMneCL8 was not (36). To determine if the changes in envelope were determinants for these cell culture virulence properties, we infected CEMx174 cells with the chimeric viruses and assayed for cell killing and syncytium formation. As a positive control, we used the variant population from macaque M87004 PBMCs at 170 weeks postinfection. This mixed variant population (MixVar) induced syncytia (Fig. 4A) and cytopathic effects (Fig. 4B) in CEMx174 cells by 5 to 6 days postinfection; in contrast, SIVMneCL8 did not induce syncytia, as reported previously (36). Syncytia were also observed in cells infected with chimeric viruses (Fig. 4A). The 35wkSU and 81wkSU chimeras induced a small number of syncytia (peaks of 62 and 44 per well, respectively), whereas the chimeras with envelope SU from proviruses at later stages of infection induced more syncytia (peaks of 156 and 417 per well for 121wkSU and 170wkCESU, respectively). Syncytium induction was delayed in cells infected with the 121wkSU and 170wkCESU viral chimeras, which corresponded to a delay in peak antigen production in these cells compared to cells infected with the variant mixture; in this regard, the kinetics of replication of the chimeras were similar to that of SIVMneCL8, with the exception of SIVMne170wkPBSU, which did not replicate in CEMx174 cells (data not shown).
FIG. 4.
Infections of CEMx174 cells with SIVMneCL8 and the chimeric viruses. Virus harvested from CEMx174 cells transfected with the proviral clones was used to infect 8 × 105 CEMx174 cells at an MOI of 0.01. Infections were performed in duplicate for each virus. In panel A, syncytia are plotted as a function of days postinfection; in panel B, the viable cell counts (log scale) are shown during the 14 days after infection.
Surprisingly, syncytium formation did not correspond with extensive cytopathic effects in these cultures. In cells infected with the variant mixture, there was a dramatic decrease in viable cells starting at 5 to 6 days postinfection, corresponding with the peak of syncytium formation in the cultures. The decrease in the number of viable cells was modest in CEMx174 cells infected with the chimeras or SIVMneCL8 and was independent of whether syncytia were detected in the culture. Similar results have been observed in multiple experiments and in cultures followed for up to 18 days postinfection (data not shown). These data suggest that syncytium formation may not account for the majority of cytopathic effects observed in CEMx174 cells infected with SIVMne late variants. Moreover, these data suggest that while the extracellular envelope glycoprotein may encode determinants for syncytium formation, it is not a major determinant of cytopathic effects in these cells.
Recognition of the CCR-5 coreceptor.
The CCR-5 chemokine receptor protein functions as a coreceptor for M-tropic strains of HIV-1 and for some isolates of SIV, including SIVMne (1, 11, 13, 15, 17–19, 28, 38). Human and macaque CCR-5 receptors function equally well for SIV infection (13). To determine if the changes that evolve in the envelope gene of viruses selected during the course of SIVMneCL8 infection alter coreceptor recognition, we examined human CCR-5 coreceptor specificity in a single-cycle assay that does not require virus spread. For this purpose, we used MAGI and sMAGI cell derivatives (10, 11, 25). sMAGI cells are susceptible to wide variety of SIV variants, including both M-tropic and T-tropic variants (10). Previous studies showed that the viral titers defined by sMAGI cell infection are similar to the tissue culture infectious dose determined by endpoint dilution in CEMx174 cells for both T-tropic and M-tropic SIV strains (10), and so that these cells were used as a positive control. The MAGI cell line is not susceptible to infection by SIVMneCL8, but MAGI cells engineered to express human CCR-5 (MAGI-CCR5 cells) are highly susceptible to SIVMneCL8 infection (11).
Each SIV readily infected the sMAGI cells and MAGI-CCR5 cells, but none of the variants could infect the parental MAGI cells that lacked the exogenous CCR-5 coreceptor (Table 1). This was true for chimeras encoding SU sequences obtained from proviruses throughout the course of infection, through 170 weeks. Similarly, the cytopathic, syncytium-inducing mixed variant population of SIVMne isolated from macaques with AIDS recognized the human CCR-5 coreceptor, as did two clones of SIVmac. The infectivity of all SIV variants in MAGI-CCR5 cells was similar to their infectivity in the highly permissive sMAGI cell line, suggesting that each of the variant envelopes efficiently recognize the CCR-5 chemokine coreceptor. These data suggest that changes in the envelope protein that are selected over time in the host do not alter interactions with the CCR-5 chemokine coreceptor, although experiments with the M. nemestrina receptor gene may be required to more conclusively demonstrate this point.
Lack of recognition of variant envelope proteins by neutralizing antibodies.
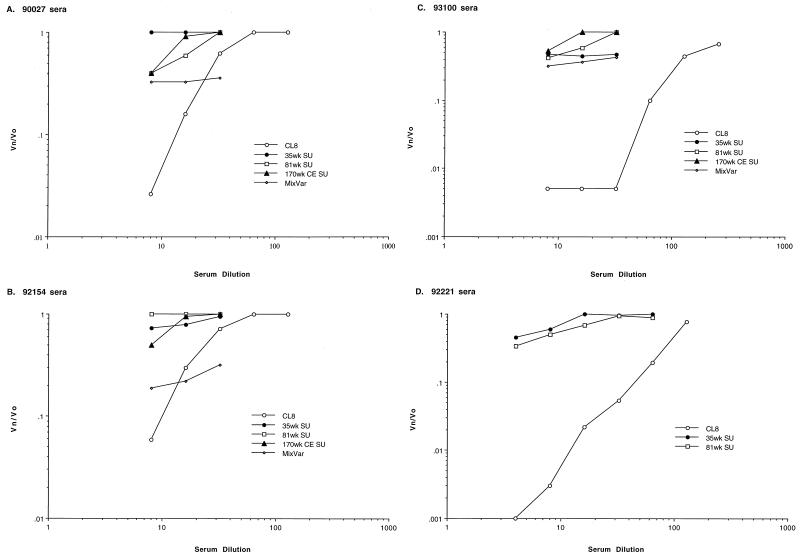
To examine whether the envelope changes encoded by viruses that are selected in the host affect their antibody recognition, neutralization of the chimeras and the parental virus SIVMneCL8 was examined. For this purpose, we analyzed the titer of neutralizing antibodies in sera from SIVMne-infected macaques, using the sMAGI neutralization assay (10). Neutralizing activity could not be detected in the available serum samples from macaque M87004, and we have found that not all SIVMne-infected macaques have detectable neutralizing antibodies in their sera (data not shown). Sera from two macaques (90027 and 92154) infected with the biological clone of SIVMne from which SIVMneCL8 was derived had antibodies that could neutralize SIVMneCL8 but not the chimeric viruses. For example, the 90% neutralizing titer against SIVMneCL8 was 1:12 in macaques 90027 (Fig. 5A) and 92154 (Fig. 5B), but neither serum could neutralize the 35wkSU, 81wkSU, or 170wkCESU chimera. The mixture of late variants isolated from macaque M87004 were also not efficiently neutralized by these sera, although neutralization of this mixed virus population was achieved with a 1:4 dilution of sera from 92154 (Fig. 5B). Because the 35wkSU chimera differs from SIVMneCL8 at only four positions in V1 (Fig. 1), these data show that mutations in V1 alter neutralizing antibody recognition of SIVMne.
FIG. 5.
Neutralizing antibodies titers against SIVMneCL8 and the chimeric viruses in sera from four SIVMne-infected macaques. The number of infectious particles in the presence of sera (Vn) over the number in the absence of sera (Vo) is shown as a function of the serum dilution. The results are an average of duplicate infections in sMAGI cells and are representative of at least two independent experiments. (A) Serum from M. nemestrina 90027, inoculated with SIVMneE11S clone, at 6 months postinfection; (B) serum from M. nemestrina 92154, inoculated with SIVMneE11S, at 6 months postinfection; (C) serum from M. nemestrina M93100, inoculated with uncloned SIVMne, at 5 months postinfection; (D) serum from M. nemestrina 92221, inoculated with uncloned SIVMne, at 5 months postinfection.
These data suggest that animals that have been infected with a SIVMneCL8-like virus may mount a neutralizing antibody response against homologous envelope but not against the heterologous envelope of viruses that evolve later in infection. To determine if neutralizing antibodies that recognize variants with the V1 changes could be detected in monkeys exposed to a complex inoculum, we also examined sera from two macaques (93100 and 92221) inoculated with an uncloned stock of SIVMne. This uncloned SIVMne stock contains a mixture of variants that include viruses with envelope sequences resembling SIVMneCL8, as well as envelope sequences with the V1 serine and threonine changes characteristic of the variants that evolve in SIVMneCL8-infected macaques (38a). The sera from macaque 93100 efficiently neutralized SIVMneCL8 (1:64) but did not neutralize the chimeric 35wkSU, 81wkSU, and 170wkCESU viruses or the variant mixture even at a dilution of 1:8 (Fig. 5C). Similar results were observed with a second macaque (Fig. 5D); SIVMneCL8 was neutralized by macaque 92221 serum, but the chimeric viruses were not. These data suggest that not only do the changes in the variant envelope allow escape from neutralizing responses against the infecting virus, SIVMneCL8, but they also do not appear to elicit a strong humoral response compared to SIVMneCL8 when present in the infecting virus.
DISCUSSION
The evolution and selection of SIV and HIV variants within an infected individual may be influenced by a multitude of interactions that occur between the virus and the host. Genetic diversity is characteristic of HIV and SIV infection, and it undoubtedly reflects a process of selection superimposed upon a high rate of mutation and viral turnover. Variants that emerge over the course of infection are frequently phenotypically and immunologically distinct from the viruses present at earlier times in infection. To begin to understand the host factors that drive evolution of new virus variants in the host, we analyzed the biological properties of viruses encoding envelope SU proteins representative of variants found throughout the course of SIVMne infection and disease. These studies suggest that Env-SU changes do not confer a replication advantage to the selected viruses, but they do allow the virus to escape neutralizing antibody recognition.
The uncloned virus population that emerges in macaques infected with a M-tropic, non-syncytium-inducing clone of SIVMne is T-tropic, cytopathic, syncytium inducing, and poorly infectious for macaque macrophages (36). This phenotypic change in the virus occurs over the course of infection, as AIDS develops, and it is similar to what has previously been observed in HIV-1-infected individuals (36). The uncloned virus mixture is not recognized by macaque sera that can neutralize the SIVMneCL8 parent virus, suggesting that antigenic changes occur in viruses selected during the course of infection. Because these viruses were a mixed population of variants obtained by coculturing PBMCs from the infected animal (37), it was not possible to determine from these experiments which viral gene and/or variant was responsible for the phenotypic and immunological changes observed in the viruses relative to the SIVMneCL8 parental virus.
Our previous studies have shown that the variants that are selected in SIVMne infection of macaques encode characteristic changes in the V1 and V4 regions of envelope (33, 34). To study the biological changes associated with these genetic alterations, a series of chimeric viruses encoding variant envelope sequences in the context of the parental virus were analyzed. We chose variants from one macaque (M87004), infected with SIVMneCL8, because (i) we have extensive data on sequence variation of SIVMneCL8 in this animal over the course of infection and AIDS (33, 34), which allowed us to chose prototype variants from different stages of infection, and (ii) we have shown that the virus populations isolated from the PBMCs of this animal when it had disease were rapidly replicating, cytopathic, and syncytium inducing in T lymphocytes compared to the parental M-tropic, noncytopathic, non-syncytium-inducing SIVMneCL8 virus from which they evolved (36).
The extracellular envelope glycoprotein of viruses that evolve over time in SIVMne infection has acquired mutations that confer the ability to induce syncytium formation. Syncytia were observed in cells infected with viruses encoding envelope proteins from variants present at the later stages of infection and disease in the macaque. In the syncytium phenotype, the chimeric viruses encoding envelope genes from late in infection resembled the uncloned virus isolates from the animal when it had AIDS. However, in contrast to the uncloned isolates, the syncytium-inducing chimeras were not highly cytopathic. These data suggest that the extracellular envelope glycoprotein of SIVMne is not a primary determinant of cytopathic effects. These data also suggest that syncytium formation does not fully account for the cytopathic effects associated with replication of the late variant mixture in CEMx174 cells.
The Env-SU of the late variants does not enhance the replication properties of SIVMne in macaque PBMC cultures. In general, the chimeric viruses replicate with efficiency similar to that of the parental SIVMneCL8 in primary lymphocyte cultures. However, one caveat to the interpretation of these studies is that subtle replication differences cannot be identified in our assays with primary macaque PBMC cultures because of the variability in the relative rates of replication of individual viruses in different macaque donor cells. Surprisingly, the chimeric viruses replicate with lower efficiency than the SIVMneCL8 parent in macaque macrophages. This was true of all chimeras examined, including the 35wkSU chimera, which differed from SIVMneCL8 by only four amino acids, all of which were serines and threonines in V1. This finding may indicate that sequences in V1 affect macrophage tropism of SIVMne, although these experiments do not distinguish whether the replication of these chimeras is restricted at entry or a later stage in infection of macrophages. Interestingly, changes in both a potential N-linked and a potential O-linked glycosylation site in V1 of HIV-1 have also been associated with a change in viral tropism (5, 9). Overall, our data suggest that changes in Env-SU that evolve over the course of SIVMneCL8 infection do not provide a selective advantage for replication of the virus in two major targets cells of the host, lymphocytes and macrophages.
In light of the changes in macrophage tropism of SIVMne variants, we examined the ability of these viruses to recognize the CCR-5 chemokine coreceptor. CCR-5 functions as a coreceptor for infection by M-tropic HIV-1 isolates (1, 15, 17–19, 38), and both the human and macaque CCR-5 proteins can efficiently and interchangeably function as coreceptors for diverse strains of SIV (13, 28). We have shown that human CCR-5 can function as a coreceptor for M-tropic SIVMneCL8 (11). Here we show that human CCR-5 also functions as a coreceptor for SIVMne chimeras that do not replicate in macrophage cultures. While there is a strong correlation between recognition of CCR-5 and macrophage tropism for HIV-1, recognition of CCR-5 itself is not sufficient for macrophage infection (14). Moreover, macrophage infection by dual-tropic HIV-1 may also occur in the absence of functional CCR-5 (35). Our data and the data of Chen et al. (13) suggest that for SIV infection, recognition of CCR-5 by a particular virus variant is also not strictly correlated with the ability of that virus to replicate in primary macrophages. It is possible that these types of studies, using transformed cell lines engineered to express chemokine receptors, do not provide a true picture of the complex interactions that occur between the virus and cellular proteins involved in viral entry into primary cells. Alternatively, the results may suggest that the restriction to replication of the SIVMne chimeras in macrophages is at a postentry stage in replication, as has been reported for SIVmac239 infection in macrophages (31).
Isolates of HIV-1 obtained sequentially during the course of human infections acquire the ability to recognize additional coreceptors but tend to retain the ability to recognize CCR-5 (16, 38). Because the determinant for HIV-1 coreceptor recognition maps to Env-SU (1, 15–19, 39, 40), these data suggest that changes which evolve in SU during the course of HIV-1 infection do not alter CCR-5 interactions. The studies presented here, which include analysis of chimeric viruses encoding SU proteins from different stages of SIVMne infection, directly demonstrate this to be true in the SIV-macaque system. However, our data indirectly suggest that the SIV isolates from late stages of infection do not acquire the ability to recognize the CXCR-4 coreceptor that is recognized by late-stage HIV-1 variants (4, 16, 20) because these SIV isolates do not infect cells which express endogenous CXCR-4 (MAGI cells). Consistent with this, SIVmac also does not use CXCR-4 or any of the other known HIV-1 coreceptors for entry (13). There is strong circumstantial evidence for an additional SIV coreceptor(s) other than CCR-5, at least one of which is expressed in CEMx174 cells (13). Because all of the viruses examined here can replicate in CEMx174 cells, we hypothesize that they would also recognize such a CEMx174 coreceptor. However, in light of the strong parallels in the patterns of phenotypic viral changes observed in HIV-1 infection in humans and SIVMne infection in macaques (36), we suggest that the late SIVMne variants have expanded the repertoire of coreceptors that they can utilize. As new coreceptors for SIV are identified, the panel of viruses described here will provide a powerful tool for defining changes in coreceptor specificity that evolve over time in SIV-infected macaques.
The most striking differences between chimeric viruses encoding envelope variants from infected macaques compared to the parental infecting virus was in the ability to be neutralized by host antibodies. While SIVMneCL8 was neutralized by sera from macaques infected with homologous virus, the chimeric viruses were not. These data illustrate that envelope changes allow the virus to escape humoral immune responses. These studies are consistent with previous studies of SIVmac infections, which showed that variants that emerge during SIVmac239 infection are resistant to neutralization by the host sera (6, 26). Moreover, in macaques infected with uncloned SIVMne, there was detectable neutralizing antibodies to SIVMneCL8 but not to the chimeric viruses, suggesting that envelope changes in viruses that evolve in the host may, in general, make the virus less immunogenic. In support of this hypothesis, a late variant clone isolated from SIVsm infection was inefficiently recognized by a panel of broadly cross-reactive SIV-infected macaque sera, and this virus did not elicit a strong neutralizing antibody response when inoculated into a new host (21).
The differences in neutralizing antibody recognition were mapped to the V1 region; specifically, four predicted serine and threonine changes found in an envelope sequence from 35 weeks postinfection were sufficient for the virus to evade antibody neutralization. Previous studies of SIV have identified several linear and discontinuous B-cell epitopes in SIV envelope (reviewed in reference 7). However, because these studies focused on monoclonal antibodies generated by immunization of mice, it is unclear which of these epitopes may be most immunogenic in macaques. In these analyses, V1 was not identified as an epitope for neutralizing antibody recognition in SIV. The studies presented here suggest that V1 is a principal neutralizing determinant of SIV, although it remains to be seen if V1 defines a linear or discontinuous B-cell epitope or whether changes in V1 are associated with conformational changes that mask an epitope elsewhere. In any case, our data suggest that viruses with changes in V1 may be selected because they can escape the host humoral immune response to the infecting virus.
In the studies presented here, the phenotypic and immunologic changes in viruses that evolve during the course of SIVMne infection have been analyzed in direct comparison with the parental, infecting viral clone. Changes in the V1 region of envelope that occur prior to the onset of AIDS are associated with a restriction for replication of the virus in macrophages and, at the same time, an escape of the virus from neutralizing antibody recognition. Additional envelope changes found in viruses at late-stage AIDS confer the ability to induce syncytium formation, but syncytium induction is not associated with cytopathic effects. The envelope changes in viruses that are selected over time in SIV infection do not, in general, alter the cytopathic properties of the virus, affect replication of the virus in lymphocyte cultures, or alter CCR-5 recognition. These studies may suggest that other viral genes contribute to some of the changes in viral phenotype characteristic of SIV infection in macaques and may explain previous studies suggesting that the pathogenic determinants of SIV infection are complex (29, 32). Together, these data suggest that multiple domains in the SIV genome, which evolve in response to different selective pressures in the host, contribute to evolution of a virus population that is T-tropic, highly cytopathic, and syncytium inducing and that is no longer recognized by the host humoral immune response.
ACKNOWLEDGMENTS
We thank Nancy Haigwood and LaRene Kuller for providing serum samples from infected macaques. Cell lines (CEMx174) and GCT conditioned medium were provided by the AIDS Research and Reference Reagent Program.
This work was supported by the NIH (RO1 AI34251). J.T.K. was supported in part by training grants T32-CA09229 and T32-AI07140 from the NIH.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1a, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Benveniste R E, Arthur L O, Tsai C C, Sowder R, Copeland T D, Henderson L E, Oroszlan S. Isolation of a lentivirus from a macaque with lymphoma: comparison with HTLV-III/LAV and other lentiviruses. J Virol. 1986;60:483–490. doi: 10.1128/jvi.60.2.483-490.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benveniste R E, Hill R W, Eron L J, Csaikl U M, Knott W B, Henderson L E, Sowder R C, Nagashima K, Gonda M A. Characterization of clones of HIV-1 infected HuT 78 cells defective in gag gene processing and of SIV clones producing large amounts of envelope glycoprotein. J Med Primatol. 1990;19:351–366. [PubMed] [Google Scholar]
- 4.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns D P, Collignon C, Desrosiers R C. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J Virol. 1993;67:4104–4113. doi: 10.1128/jvi.67.7.4104-4113.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns D P, Desrosiers R. Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence. Curr Top Microbiol Immunol. 1994;188:185–219. doi: 10.1007/978-3-642-78536-8_11. [DOI] [PubMed] [Google Scholar]
- 8.Burns D P, Desrosiers R C. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 1991;65:1843–1854. doi: 10.1128/jvi.65.4.1843-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrillo A V, Ratner L. Cooperative effects of human immunodeficiency virus type 1 envelope variable loops V1 and V3 in mediating infectivity for T cells. Virology. 1996;6:1310–1316. doi: 10.1128/jvi.70.2.1310-1316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chackerian B, Haigwood N L, Overbaugh J. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology. 1995;213:386–394. doi: 10.1006/viro.1995.0011. [DOI] [PubMed] [Google Scholar]
- 11.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chackerian B, Morton W R, Overbaugh J. Persistence of simian immunodeficiency virus Mne variants upon transmission. J Virol. 1994;68:4080–4085. doi: 10.1128/jvi.68.6.4080-4085.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Zhou P, Ho D D, Landau N T, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;7:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 16.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 18.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 19.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins W R, Montefirori D C. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch V, Johnson P. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 1994;32:183–203. doi: 10.1016/0168-1702(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 23.Johnson P R, Hamm T E, Goldstein S, Kitov S, Hirsch V M. The genetic fate of molecularly cloned simian immunodeficiency virus in experimentally infected macaques. Virology. 1991;185:217–228. doi: 10.1016/0042-6822(91)90769-8. [DOI] [PubMed] [Google Scholar]
- 24.Kimata J T, Overbaugh J. The cytopathicity of a simian immunodeficiency virus Mne variant is determined by mutation in Gag and Env. J Virol. 1997;71:7629–7639. doi: 10.1128/jvi.71.10.7629-7639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinsey N E, Anderson M G, Unangst T J, Joag S V, Narayan O, Zink M C, Clements J E. Antigenic variation of SIV: mutations in V4 alter the neutralization profile. Virology. 1996;221:14–21. doi: 10.1006/viro.1996.0348. [DOI] [PubMed] [Google Scholar]
- 27.Levy J A. Pathogenesis of human immunodeficiency virus infection. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard N, Gerar C, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marthas M L, Ramos R A, Lohman B L, Van Rompay K K, Unger R E, Miller C J, Banapour B, Pedersen N C, Luciw P A. Viral determinants of simian immunodeficiency virus (SIV) virulence in rhesus macaques assessed by using attenuated and pathogenic molecular clones of SIVmac. J Virol. 1993;67:6047–6055. doi: 10.1128/jvi.67.10.6047-6055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miedema F, Meyaard L, Koot M, Klein M R, Roos M L, Groenink M, Fouchier R A M, Van’t-Wout A B, Tersmette M, Sciellekens P T A, Schuitemaker H. Changing virus-host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 31.Mori K, Ringler D J, Desrosiers R C. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by env but is not due to restricted entry. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novembre F J, Johnson P R, Lewis M G, Anderson D C, Klumpp S, McClure H M, Hirsch V M. Multiple viral determinants contribute to pathogenicity of the acutely lethal simian immunodeficiency virus SIVsmmPBj variant. J Virol. 1993;67:2466–2474. doi: 10.1128/jvi.67.5.2466-2474.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overbaugh J, Rudensey L M. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J Virol. 1992;66:5937–5948. doi: 10.1128/jvi.66.10.5937-5948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Hai-Hong G, Jian-Guo D, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudensey L M, Kimata J T, Benveniste R E, Overbaugh J. Progression to AIDS in macaques is associated with changes in the replication, tropism, and cytopathic properties of the simian immunodeficiency virus variant population. Virology. 1995;207:528–542. doi: 10.1006/viro.1995.1113. [DOI] [PubMed] [Google Scholar]
- 37.Rudensey L M, Papenhausen M D, Overbaugh J. Replication and persistence of simian immunodeficiency virus variants after passage in macaque lymphocytes and established human cell lines. J Virol. 1993;67:1727–1733. doi: 10.1128/jvi.67.3.1727-1733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic, and most can use either Lestr or CCR-5 as coreceptors for entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Stallard, G, J. Overbaugh, and S.-L. Hu. Unpublished data.
- 39.Trkola A, Drajic T, Arthos J, Binley L, Olsen W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD-4 dependent, antibody sensitive interactions between HIV-1 and its coreceptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 40.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]