ABSTRACT
Dietary patterns and corresponding gut microbiota profiles are associated with various health conditions. A diet rich in polyphenols, primarily plant-based, has been shown to promote the growth of probiotic bacteria in the gastrointestinal tract, subsequently reducing the risk of metabolic disorders in the host. The beneficial effects of these bacteria are largely due to the specific metabolites they produce, such as short-chain fatty acids and membrane proteins. In this study, we employed a metabolomics-guided bioactive metabolite identification platform that included bioactivity testing using in vitro and in vivo assays to discover a bioactive metabolite produced from probiotic bacteria. Through this approach, we identified 5’-methylthioadenosine (MTA) as a probiotic bacterial-derived metabolite with anti-obesity properties. Furthermore, our findings indicate that MTA administration has several regulatory impacts on liver functions, including modulating fatty acid synthesis and glucose metabolism. The present study elucidates the intricate interplay between dietary habits, gut microbiota, and their resultant metabolites.
KEYWORDS: Bifidobacterium, 5’-methylthioadenosine, LC-MS/MS metabolomics, probiotic, prebiotic, postbiotic, polyphenols
Introduction
The global prevalence of obesity has nearly tripled since 1975, primarily because of unhealthy dietary habits.1–4 The growing prevalence of obesity has emerged as a significant global public health concern due to its association with an elevated susceptibility to several chronic diseases.5 Recent research has placed emphasis on the development of microbiome-based interventions aimed at modulating the gut microbiota, as studies have demonstrated its substantial involvement in the pathogenesis of obesity and metabolic illnesses. The predominant microbiome-focused therapy objectives presently encompass dietary therapies, fecal microbiota transplantation, probiotic and prebiotic treatments, as well as microbiome-associated metabolites.6
Diet influences the bidirectional interplay between the microbiome and the host by providing substrates for gut bacteria, and the metabolites provided by gut bacteria can induce a series of physiological and pathological functions. For instance, a western-style diet, which is typically high in fat and animal protein, can increase the abundance of the microbial metabolite trimethylamine (TMA)-producing bacteria in the gut. These bacteria are capable of degrading choline or carnitine into TMA, which is ultimately converted to TMAO, a microbial metabolite linked to an elevated risk of cardiovascular disease and other health conditions.7,8 Besides, protein-rich foods are also high in histidine, which can undergo metabolism by gut bacteria, particularly Escherichia coli, resulting in the production of imidazole propionate.9 Elevated levels of imidazole propionate have been observed in individuals with heart failure and type 2 diabetes.9,10 In contrast, polyphenols are a large group of secondary metabolites commonly present in fruits and vegetables that play a prominent role in human health.11 The two-way interaction between polyphenols and microbiota has been emphasized for its potential to impact host metabolism and lower the risk of cardiometabolic disease.12 Notably, the interaction between gut microbes and polyphenols may play a significant role in the polyphenol-rich fruits’ beneficial health effects since several types of polyphenols have low bioavailability, with 95% of the ingested polyphenols persisting close to the microbiota in the mucus layer.13,14 Dietary polyphenols are metabolized by the gut microbiota to achieve multiple health benefits, including improvement of lipid metabolism, insulin resistance, blood pressure, and systemic inflammation.15,16 In contrast, polyphenols can enhance the growth of beneficial bacteria in the host, which further affects host metabolism.
Other microbiome-based interventions to improve metabolic disorders include beneficial bacteria and their functional metabolites. These bacteria have demonstrated efficacy in mitigating obesity, diabetes, inflammation, allergies, and enhancing cognitive function.17 The functional metabolites produced by these commensal bacteria play pivotal roles in human health. For instance, tryptophan derivatives, including indole and its derivatives, are produced by a variety of intestinal bacteria, including Escherichia and Bacteroides species. These metabolites serve multiple functions, such as modulating gut barrier functions, influencing inflammation and behavior responses in the central nervous system.18
Interestingly, researchers have found that the effects of probiotics are partly induced by specialized metabolites derived from gut microbes, such as short-chain fatty acids and membrane proteins.19,20 Since a growing number of intestinal microbial metabolites have been linked to a variety of diseases, the discovery of bioactive metabolites from microorganisms is a strategy for the development of novel drugs or functional foods for disease therapy.21–24 However, the absence of comprehensive research has hindered the elucidation of the underlying mechanisms and the identification of appropriate pharmaceutical options.
Due to the complexity and dynamics of the gut microbiota, the integration of various therapies targeting the microbiome may yield more favorable results compared to their individual applications. The combination of prebiotics, probiotics, and postbiotics can work synergistically to provide various health benefits to the host. Prebiotics can selectively promote the growth of probiotics and suppress the growth of harmful bacteria, leading to a more balanced gut microbiota community.25 In addition, prebiotics and postbiotics can enhance the survival and growth of probiotics and subsequently promote the attachment and colonization of probiotic bacteria to the intestinal lining.26 Nevertheless, the lack of evidence has limited the investigation of the interaction between prebiotics, probiotics, and postbiotics. Thus, strategies based on a greater understanding of the molecular mechanisms, particularly the mode of action of small molecules derived from microorganisms, are required.23
This study aimed to investigate the bioactive metabolites derived from probiotic strains that were enriched by a polyphenol-rich diet, and to elucidate the interaction between dietary patterns, gut microbiota, and their resultant metabolites. As the discovery of functional metabolites is a complex process,27 we included a series of approaches to gradually evaluating functional metabolites and then confirming their biological activity. To select a specific bacterium that promotes metabolic health, the identification process began with an animal study and analysis of microbial communities. The next step was to use a fractionated extraction method to fully analyze bacterial metabolites. At the same time, in vitro and in vivo bioactivity testing were used to find bioactive metabolites that came from the gut microbiota that was being studied. To gain further insight into how the compound is transformed from dietary components, a stable in vitro isotope-labeled experiment was performed. In addition, alterations in hepatic gene expression were analyzed to elucidate the detailed biological mechanism of the bioactive metabolite. Finally, we identified a phenotype-modulating metabolite produced by probiotic bacteria using a metabolomics-guided bioactive-metabolite identification platform.
Results
Lychee polyphenols attenuated HFD-induced obesity and improved the dysbiosis of the gut microbiota
Lychee (Litchi chinensis Sonn.), a tropical fruit cultivated widely in Taiwan, contains high concentrations of phenolics, proanthocyanins, and flavanols.28,29 In our previous work, the phenolic compounds of lychee were analyzed by the Global Natural Products Social (GNPS), which is a molecular networking method that relies on the concept that molecules with comparable chemical structures exhibit similar MS/MS fragmentation patterns.28 Considering that lychee polyphenols have been shown to have a variety of biological activities, including antimicrobial, anticancer, anti-inflammatory, anti-obesity, and anti-diabetes properties,29,30 we next tested for their ability to modulate the gut in the present study. First, we found that lychee polyphenols alleviated high-fat diet-induced obesity, decreased hyperlipidemia, and ameliorated glucose tolerance and insulin sensitivity (Figure 1a-f and Figure. S1a–e). Administration of lychee polyphenols enriched the diversity and rectified the dysbiosis of microbiota caused by HFD (Figure 1h, i and Figure. S1f). Moreover, lychee polyphenols slightly enriched the abundance of Bifidobacterium spp. and marginally increased Bifidobacterium longum (Figure 1j, k). Since the primary aim of this study was to investigate the specific metabolites produced by probiotics, rather than focusing on the interaction between polyphenols and probiotic bacteria, the polyphenol profile was omitted from this publication.
Figure 1.
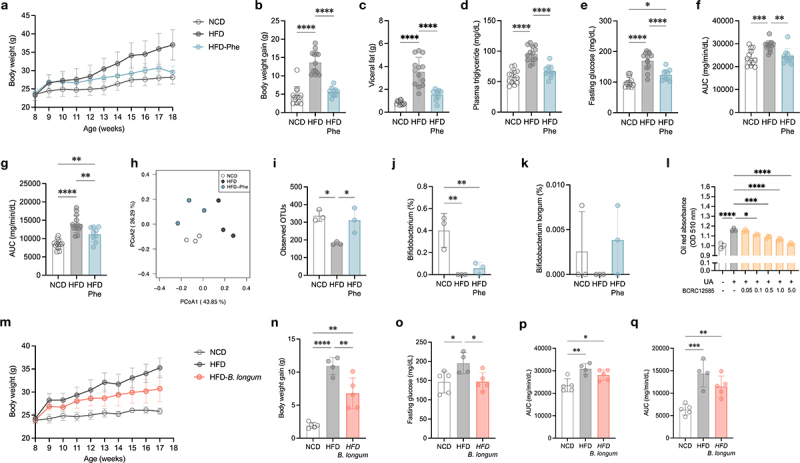
Lychee polyphenols attenuated HFD-induced obesity, improved the dysbiosis of the gut microbiota, and enriched Bifidobacterium longum, which exerted anti-obesity activity.
(a–k) Effect of lychee polyphenols on host metabolism and gut microbiota composition. (a) Body weight changes. (b) Body weight gain during the experimental period. (c) Weight of visceral fat. (d) Plasma triglycerides. (e) Fasting glucose. (f) Area under the curve (AUC) derived from oral glucose tolerance test (OGTT). (g) AUC derived from insulin tolerance test (ITT). (h) Gut microbiota composition as represented by principal-coordinate analysis (PCoA) of Bray-Curtis distances. (i) Observed operational taxonomic unit (OTU). (j) Abundance of Bifidobacterium in feces. (k) Abundance of B. longum in feces. (l) Effect of B. longummetabolites on lipid accumulation in HepG2 cells. (m–q) Effect of B. longum on host metabolism. (m) Body weight changes. (n) Body weight gain during the experimental period. (o) Fasting glucose. (p) AUC derived from OGTT. (q) AUC derived from ITT. Data are means and SD. Statistical analyses were performed by one-way ANOVA with Tukey’s range test (b–g, i–k, and n–q) or two-tailed Student’s t-test (l), respectively (*, P < .05; **, P <.01; ***, P < .001; ****, P < .0001). Treatment groups were compared with uric acid control (l). (a–k) N = 13, 13, 10 mice per group, while number of fecal samples in each group is 3. (m–q) N = 5, 4, 5 mice per group. Abbreviations: AUC, area under the curve; B. longum: Bifidobacterium longum; HFD, high-fat diet; NCD, normal chow diet; OGTT, oral glucose tolerance test; Phe, lychee polyphenols.
Bifidobacterium longum exerted anti-obesity activity
Although Bifidobacterium spp. have been recognized as probiotics that exert an anti-obesogenic effect on the host, the metabolic benefits of their metabolites and the underlying mechanisms remain unclear. Therefore, we explored the regulatory capacity of Bifidobacterium metabolites in host metabolism. To this end, we purchased all commercial Bifidobacterium strains available in Taiwan and colonized them in C57BL/6 germ-free mice (Table S1). By testing the lipid accumulation activity in HepG2 cells, metabolites extracted from Bifidobacterium-treated mouse feces significantly decreased lipid accumulation in a dose-dependent manner (Figure. S2). Metabolites from each strain of Bifidobacterium also exerted anti-obesogenic activity (Figure 1l and Figure. S3). Since we attempted to identify specific chemicals produced by probiotic microorganisms that have therapeutic potential and could be easily scaled up in the industry, Bifidobacterium. longum. subsp. longum BCRC12585 (hereafter referred to as B. longum), a commercial strain of B. longum isolated from humans and easily scaled up, was selected to further explore the bioactive metabolites in a mouse model.
Next, 108 colony-forming units (CFUs) of B. longum were administered to C57BL/6 mice once a day until one week prior to sacrifice. The mice exhibited decreased body weight and weight gain, while visceral and subcutaneous fat were marginally decreased in B. longum-treated mice (Figure 1m, n and Figure. S1g–h). In addition, mice treated with B. longum showed remarkably decreased fasting glucose levels and slightly improved glucose and insulin tolerance (Figure 1o-q and Figure. S1i–j).
Identification of bioactive metabolite produced by B. longum
To further identify the bioactive metabolites produced by B. longum, we applied a fractionation approach followed by liquid chromatography-mass spectrometry (LC-MS) coupled with an in vitro screening strategy (Figure 2a). Bioactivity-guided fractionation is a powerful technique for the separation of natural products with biological activity23. Briefly, 25 fractions derived from B. longum metabolites were prepared using pre-HPLC columns, followed by individual testing of the anti-lipid accumulation activity in HepG2 cells. Interestingly, fractions A–H (Figure. S4), which has a short retention time, strongly inhibited lipid accumulation (Figure 2b and Figure. S5), indicating that the active compounds were mostly small molecules with high polarities. To identify candidate metabolites from each active fraction, further analysis was performed using liquid chromatography coupled with high-resolution mass spectrometry (LC-MS/MS). Fourteen compounds, including nine amino acids (proline, valine, glutamine, glutamic acid (GA), arginine, pyroglutamic acid (PyroGA), methionine, isoleucine, and tryptophan), two peptides (cyclo-(His-Pro), Pyro-glutamyl-valine (Pyro-glu-val)), and adenosine and its two derivatives (3-Adenosine monophosphate and 5’-S-methyl-5’-thioadenosine (MTA)), were identified and further verified using authentic standards (Figure. S6 and Table S2). Among the identified metabolites, seven significantly inhibited lipid accumulation in HepG2 cells (Figure 2c and Figure. S7). Notably, MTA abundance exhibited a negative correlation with most obesity biomarkers in mice (Figure 2d). Collectively, we integrated in vitro, in vivo, and metabolomic data to identify the bioactive metabolites that could be produced by B. longum.
Figure 2.
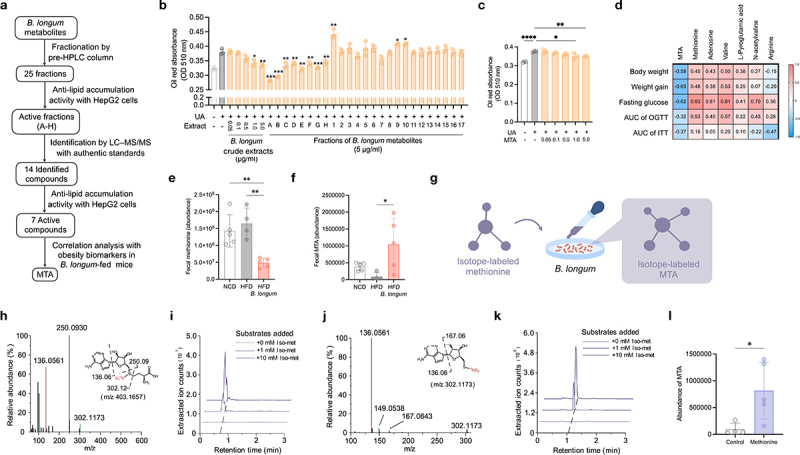
Identification of bioactive metabolite from B. longum.
(a) The workflow to identify bioactive metabolite from B. longum. (b) Effect of crude extract and fractions of B. longum on lipid accumulation in HepG2 cells (n = 3). (c) Effect of 5’-methylthioadenosine (MTA) on lipid accumulation in HepG2 cells (n = 3). (d) Correlation analysis of obesity-related phenotype and the abundance of bioactive metabolites in mice feces (n = 9). The color and number in each column represented the Spearman r. (e–f) The abundance of methionine (e) and MTA (f) in mice feces (n = 4–5). (g–l) B. longum can produce 13C2H3-MTA from 13C2H3-S-adenosyl-L-methionine. (g) The experimental design. B. longum were incubated with isotope-labeled methionine(-13C2H3) in a gradient concentration, and the bacterial metabolites was extracted for detecting isotope-labeled MTA. (h–i) Tandem mass validation (h) and extracted ion chromatogram (i) of the isotope labeled intermediate product S-Adenosyl-L-methionine (m/z 403.1657) produced by B. longum. (j–k) Tandem mass validation (j) and extracted ion chromatogram (k) of the isotope labeled MTA (m/z 302.1173) produced by B. longum. (l) The abundance of MTA from the feces of mice administered with methionine (n = 4). Data are means and SD. Statistical analyses were performed by one-way ANOVA with Tukey’s range test (e, f) or two-tailed Student’s t-test (b, c, l), respectively (*, P < .05; **, P < .01; ***, P < .001; ****, P < .0001). Treatment groups were compared with uric acid control (b, c and l). Abbreviations: AUC, area under the curve; B. longum: Bifidobacterium longum; HFD, high-fat diet; NCD, normal chow diet; OGTT, oral glucose tolerance test.
Interestingly, both MTA and its precursor, methionine, were identified in the active fractions of B. longum and exhibited a lipid-lowering effect in HepG2 cells (Figure 2c and Figure. S7), we investigated whether B. longum can produce MTA from methionine. As the precursor for MTA synthesis, methionine was decreased in the feces of mice treated with B. longum, while MTA was increased (Figure 2e,f). Next, an in vitro isotope-labeled experiment coupled with mass spectrometry was used to determine whether B. longum could convert methionine to MTA. Briefly, B. longum was incubated on a plate coated with 1 mL isotope-labeled methionine (Iso-met, -13C2H3) at 0, 1, and 10 mM (Figure 2g). As a result, 13C2H3-MTA with m/z 302.1173 (M + 4) and its intermediate product 13C2H3-S-Adenosyl-L-methionine with m/z 403.1657 (M + 4) were detected in a dose-dependent manner (Figure 2h-k), suggesting B. longum could synthesize MTA using methionine. Furthermore, the administration of methionine increased the level of MTA in the feces (Figure 2l). In addition, we found that other Bifidobacterium species used in this study, including B. breve, B. adolescentis, B. infantis, B. longum, B. bifidum, and B. animalis, could also synthesize MTA with methionine (Figure. S8). In this study, we discovered that the bioactive metabolite MTA, which has anti-obesogenic potential, can be produced by Bifidobacterium by converting dietary amino acids.
MTA, a methionine-derived metabolite produced by Bifidobacterium longum, ameliorated obesity and metabolic disorders
Next, we investigated the effect of MTA on host metabolism by treating HFD-fed C57BL/6 mice with or without MTA. After administration of MTA, metabolic indicators, including body weight, weight increase over the study period, and visceral and subcutaneous fat mass, were significantly decreased in mice (Figure 3a-e). Similar reductions in glucose tolerance and insulin sensitivity (Figure 3f-i). Moreover, liver weight and H&E staining revealed that MTA alleviated hepatic steatosis in obese mice (Figure 3j,k). These results indicated that MTA may effectively regulate lipid metabolism and insulin sensitivity, thereby reducing obesity and hepatic steatosis.
Figure 3.
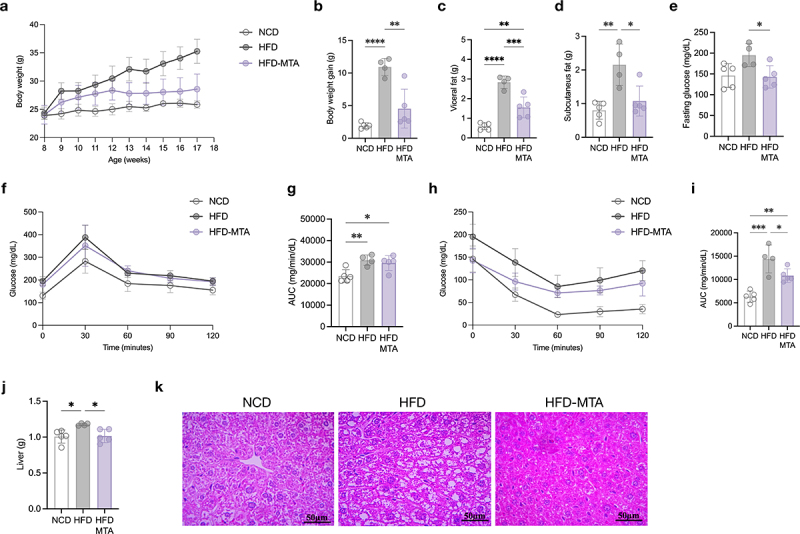
MTA ameliorated obesity and metabolic disorders.
(a) Body weight changes. (b) Body weight gain during the experimental period. (c) Weight of visceral fat. (d) Weight of subcutaneous fat. (e) Fasting glucose. (f) Plasma glucose profile measured during the OGTT. (g) AUC derived from oral glucose tolerance test (OGTT). (h) Plasma glucose profile measured during the ITT. (i) AUC derived from insulin tolerance test ITT. (j) Weight of liver. (k) Representative histological features of H&E-stained liver tissue. Data are means and SD. Statistical analyses were performed by one-way ANOVA with Tukey’s range test (*, P < .05; **, P < .01; ***, P < .001; ****, P < .0001). N = 5, 4, 5 mice per group. Abbreviations: AUC, area under the curve; HFD, high-fat diet; MTA, 5’-methylthioadenosine; NCD, normal chow diet; OGTT, oral glucose tolerance test.
MTA exerts numerous regulatory effects on hepatic energy metabolism
To obtain a more comprehensive understanding of MTA as an anti-obesity molecule, hepatic RNA sequencing was performed on mice treated with both MTA and B. longum to determine its underlying mechanism for preventing obesity. Gene ontology (GO) enrichment analysis of the top 30 differentially expressed molecular functions revealed that MTA considerably regulated the genes involved in fatty acid metabolism (Ltb4r1, Insig2, Elovl3 and Trib3), growth hormone signaling (Enho and Arntl), neurotransmitter signaling (Hcn3 and Adrb2), leptin signaling, and bile acid metabolism (Figure 4a,b). B. longum mainly modulated the genes involved in fatty acid metabolism, bile acid metabolism, and immune response (Figure. S9). Given that the gene expression of bile acid metabolism was regulated in mice treated with MTA (Figure 4a,b), hepatic bile acid analysis was performed to determine whether this regulation contributes to bile acid composition (Figure. S10 and Table S3). Furthermore, mice treated with MTA and B. longum demonstrated a notable trend toward elevated levels of taurine-conjugated primary bile acids in the liver. These included taurocholic acid (TCA), taurodeoxycholic acid (TD-CA), iso-taurocholic acid (iso-TCA), iso-taurodeoxycholic acid (iso-TDCA), tauro-β-muricholic acid (TβMCA), tauroursodeoxycholic acid (TUDCA), taurochenodeoxycholic acid (TCDCA), and taurolithocholic acid (TLCA) as shown in Figure S10. These results revealed that the effect of MTA and B. longum on the prevention of obesity and insulin resistance may be due to a variety of regulatory activities, especially fatty acid metabolism and bile acid metabolism in the liver (Figure 4c).
Figure 4.
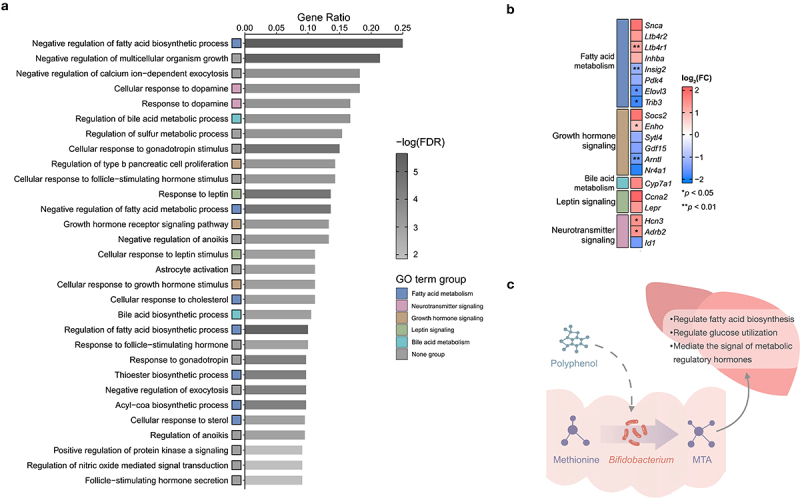
MTA exerts numerous regulatory effects on hepatic energy metabolism.
(a) The top 30 ranked GO terms according to gene ratio. ‘Gene ratio’ is the percentage of total DEGs in the given GO term (only input GO terms of the BP category with at least 10 total genes were included in the calculation). (b) The log2 fold change heat map. Log2 fold change calculate based on the average expression of MTA compared to the control samples (HFD) and red implies increased expression while blue implies decreased expression in MTA samples. Each gene is color-coded based on its major functional category: fatty acid metabolism (blue), growth hormone signaling (brown), bile acid metabolism (apple green), leptin signaling (green), and neurotransmitter signaling (red). (c) Polyphenols may prevent obesity by enriching Bifidobacterium and their amino acid metabolite MTA, which exerts variety of regulation activities of energy metabolism. Two-tailed paired Student t-test p values indicate statistical significance (*p < .05, **p < .01). Abbreviations: BP, biological process; DEG, differential expression gene; FDR, false discovery rate; GO, gene ontology; MTA, 5’-methylthioadenosine.
Discussion
Although previous studies have demonstrated that Bifidobacterium improves metabolic disorders,31,32 the underlying mechanism has been the subject of few in-depth investigations. The currently known mechanisms by which probiotics confer metabolic benefits to the host include modification of the gut microbiota composition, enhancement of the gut epithelial barrier function, competitive adherence to the gut mucosa, modulation of the immune system, reduction of appetite or food intake, and production of antimicrobial substances and beneficial metabolites.31,33–35 Some studies have further explored how probiotic metabolites can regulate host metabolism. For example, Bifidobacterium-derived short-chain fatty acids can improve host nonalcoholic fatty liver disease in a mouse model.20,36 However, the identification of bioactive small molecules is a major challenge in the search for novel microbial metabolites with potential use in drug discovery. Using a bioactivity-guided fractionation platform, we found that polyphenols may prevent obesity by enriching Bifidobacterium and their amino acid metabolite, 5’-methylthioadenosine (MTA).
As a biosynthetic precursor for several key chemicals, MTA plays a role in energy metabolism, including the regulation of the immune system, gene expression, and synthesis of important biomolecules such as DNA and proteins.37,38 The majority of research on MTA suggests that it has potential health benefits, especially anticancer effects.39,40 In contrast, few studies have reported that MTA is associated with a higher body mass index and metabolic disease risk,41–43 yet the role and mechanism of MTA in obesity and metabolic disorders have not been investigated in vivo. Here, we demonstrated that MTA regulated lipid metabolism and insulin sensitivity in mice, as well as reducing obesity and hepatic steatosis. Moreover, we conducted an in-depth analysis of the MTA-regulated genes in the liver to investigate the impact of MTA on hepatic lipid accumulation. Notably, the expression of insulin-induced gene 2 (Insig2), which influences cholesterol metabolism, lipogenesis, and glucose homeostasis in the liver,44–46 decreased in MTA-treated mice. MTA treatment also decreased ELOVL fatty acid elongase 3 (Elovl3), which is responsible for the elongation of very-long-chain fatty acid elongation.47 These findings suggest that suppression of Insig2 and Elovl3 by MTA may contribute to fatty acid remodeling in the liver and aid in body weight regulation. Additionally, the beta-2-adrenergic receptor gene (Adrb2) and leptin receptor gene (Lepr), which promote metabolic signaling associated with energy expenditure via epinephrine and leptin,48,49 respectively, were upregulated in MTA-treated mice. In addition, elevated expression of adropin (encoded by the energy homeostasis-associated gene Enho) in MTA-treated mice may improve glucose metabolism by enhancing glucose utilization.48 Likewise, the overexpression of inhibin beta-A, which is encoded by the Inhba gene, can stimulate mitochondrial energy metabolism and increase energy expenditure. These results suggest that MTA can improve energy metabolism by modulating the signaling of these metabolic hormones and proteins. Collectively, these results revealed that the effect of MTA on the prevention of obesity and insulin resistance may be due to a variety of regulatory activities of energy metabolism, including fatty acid biosynthesis and glucose utilization, which may be regulated by the signaling of hormones and proteins with metabolic regulatory effects.
Our study has limitations. (1) Although MTA has been detected in small amounts in several cell types, such as prokaryotes, yeast, plants, and higher eukaryotes,39,50 the mechanism of MTA synthesis in human tissues has been comprehensively explained. However, there is still a lack of comprehensive knowledge on the specific genes involved in the biosynthesis pathway of MTA in Bifidobacteria. The aforementioned constraint placed a restriction on our future investigation, particularly in investigating the metabolic pathways or bioprocesses of MTA by employing metabolic engineering. (2) The findings of our study indicate that MTA exhibits a beneficial impact on metabolism in a mouse model. However, additional clinical research is required to fully investigate the potential of MTA as a small-molecule medication for the treatment of metabolic diseases. (3) To truly determine the translatability of our findings to humans, subsequent studies in human cohorts and clinical trials would be essential.
In conclusion, we found that lychee polyphenols alleviated high-fat diet-induced metabolic disorders and increased the abundance of Bifidobacterium. We next demonstrated that B. longum, a Bifidobacterium strain, exerts anti-obesogenic activity in both in vitro and in vivo models. To identify a bioactive metabolite produced by B. longum, we applied a fractionation approach followed by LC-MS coupled with an in vitro screening strategy to the identification of bioactive metabolite produced by Bifidobacterium. MTA was identified by in vitro screening and its anti-obesity activity in vivo. Finally, we demonstrated that MTA may prevent obesity and insulin resistance by regulating fatty acid biosynthesis, glucose utilization, and bile acid metabolism. Using multi-omics methods, we present a strategy for identifying bioactive compounds and their biological activities.
Our findings have implications. First, MTA, which is a gut microbiota-derived small molecule, may have potential for use in the development of functional foods and pharmaceuticals to treat a variety of obesity-related diseases with further research and advancement. Second, each of the polyphenols, Bifidobacterium, and MTA showed promise in promoting weight reduction in obese mice. These components could individually serve as prebiotic, probiotic, or postbiotic interventions. This finding may provide a basis for the development of innovative interventions targeting metabolic disorders using microbiome-based approaches.
Materials and methods
Mice (Exp. 1, 2, 4, 9 and 10)
Specific pathogen-free (SPF) (7 weeks old) and germ-free (6 weeks old) C57BL/6JNarl male mice were purchased from the National Laboratory Animal Center in Taiwan. Specific pathogen-free (SPF) mice were maintained at the National Taiwan University Animal Resource Center. Germ-free and gnotobiotic mice were maintained in the gnotobiotic facilities of the National Laboratory Animal Center in Taiwan. All animal experiments were approved by the Institutional Animal Care and Use Committee of National Taiwan University (no. NTU.105EL-00093 for SPF mouse experiments) and the National Laboratory Animal Center (no. NLAC-107-O-006-R2 for germ-free mouse experiments).
C57BL/6JNarl mice anti-obesity model (Exp. 1, 4, 9 and 10)
C57BL/6JNarl 7-week-old mice were randomized and housed simultaneously with a 12-h light/dark cycle. After one-week adaptation period, C57BL/6JNarl mice were fed a chow diet (OpenSource Diets, D12450J, 10% calories from fat) or a high-fat diet (OpenSource Diets, D12492, 60% calories from fat) and treated with phenolic extracts (200 mg/kg/day), methionine (100 mg/kg/day), and 5’-S-methyl-5’-thioadenosine (100 mg/kg/day), respectively. Oral gavage was used to deliver phenolic extracts, Bifidobacterium (11 strains mixed), B. longum, methionine and MTA. The mice in the HFD control group were given sterile PBS through intragastric gavage as a vehicle. All treatments were started at the age of 8 weeks and the last 8–10 weeks.
Extract of lychee polyphenols (Exp. 1)
Lychee fruit (Litchi chinensis Sonn.) was obtained at commercial maturity from the local market in Taipei, Taiwan. After removing the peel and seed manually, the pulp was frozen in liquid nitrogen and dehydrated by a freeze dryer (FM 25EL-85, VirTis, USA). Finally, the pulp was grounded into fine powder. The dried powder (300 g) of Lychee pulp was extracted with 70% methanol aqueous solution (2.4 L) using a ultrasonicator (30 W, 60 kHz) for 30 min. After filtration of the extracts through filter papers, the residual compounds were further extracted twice using the same protocol. Methanol in all filtrates was removed using a rotary evaporator (at 45°C). Polysaccharides in the water filtrates were removed by an Oasis HLB column (20 cc/1 g, Waters, Milford, MA, USA). The remaining compounds were spun down to obtain crude extracts.
Oral glucose tolerance test (OGTT) and insulin tolerance test (ITT) (Exp. 1, 4, 9 and 10)
For OGTT and ITT, mice were fasted for 6 hours before the oral gavage with glucose (2 g/kg body weight) and the injection intraperitoneally with insulin (0.75 U/kg body weight). Blood samples were collected from the tail vein and measured before glucose or insulin administration using an Accu-Check Performa Blood Glucose Meter (Roche Diabetes Care, Inc., USA). Additional blood glucose levels were measured at 30, 60, 90, and 120 minutes.
Histopathological analysis (Exp. 9)
For H&E staining, tissues of mice were fixed with formaldehyde/PBS (4%, v/v) overnight. After dehydration in 70% ethanol, tissues were embedded in paraffin and sectioned at a 5 μm thickness. Hematoxylin and eosin staining was performed according to the standard protocol. For Oil Red O staining, frozen mouse tissues were sectioned at a thickness of 9 μm and stained according to the manufacturer’s protocol (O0625, Sigma Aldrich).
Germ-free mice colonized with Bifidobacterium (Exp. 2)
Germ-free 6-week-old C57BL/6JNarl mice from several litters were randomized housed in three sterilized isolators (6 or 7 mice for each) at 22 ± 1°C with 55–65% relative humidity on a 12-h light/dark cycle. The gnotobiotic mice were orally inoculated with 109 CFUs of Bifidobacterium (11 strains mixed) in 0.2 mL of sterile PBS once a week, and germ-free mice were gavaged with 0.2 mL of sterile PBS as the control group. All the mice were fed a high-fat diet (OpenSource Diets, D12492, 60% calories from fat). he National Laboratory Animal Center Institutional Animal Care and Use Committee in Taiwan approved all procedures. To investigate whether the metabolites of Bifidobacterium could attenuate obesity, the mouse feces (100 mg) were extracted with a 70% methanol aqueous solution (1 mL) containing cholic acid-d4 (2 ppm) as an internal standard. After 30-min of homogenization using an ultrasonicator (30 W, 60 kHz), the extracts were centrifuged at 12,000 rpm for 10 min at 4°C, and the supernatants were collected.
Bacteria cultures (Exp. 2, 7, 8)
Bifidobacterium were incubated in glucose minimal medium (GMM) broth in an anaerobic workstation (Whitley DG250, Don Whitley Scientific Limited, England). For metabolite preparation, bacterial strains were individually grown on agar plates with recommended media in an anaerobic workstation (detailed information is summarized in Table S1).51
Cell cultures (Exp. 2, 3, 5 and 6)
Hepatocellular carcinoma (HepG2) cells were obtained from the Bioresource Collection and Research Center (BCRC), Hsinchu, Taiwan). For all experiments, cells were kept in a humidified 5% CO2 incubator at 37°C (Panasonic, Japan) and maintained in RPMI 1460 medium (Thermo Fisher Scientific) supplemented with 100 U/mL penicillin, 100 mg/mL streptomycin, and 10% fetal bovine serum (FBS). The cells were routinely examined to prevent mycoplasma contamination.
Lipid accumulation assay in HepG2 cells (Exp. 2, 3, 5 and 6)
The lipid accumulation assay was performed according to a previous study with minor modifications (Choi, 2014 #285). Briefly, HepG2 cells at 70% confluence were incubated in RPMI 1640 medium containing 120 μg/mL uric acid for 24 hours, which was used to increase the lipid accumulation. After washing with PBS buffer, the cells were maintained in fresh RPMI 1640 medium containing the test substance (5.0, 1.0, 0.5, 0.1, and 0.05 μg/mL) and 120 μg/mL uric acid for another 24 h. Cells maintained in pure RPMI 1640 were used as controls.
The HepG2 cells after treatment were washed with PBS and fixed with 4% formaldehyde solution for 1 h. After Oil Red O staining (15 min at 60°C), the cells were washed thrice with ddH2O and air-dried. Then, the Oil Red O solution in cells was extracted with 250 μL 100% isopropanol, and 100 μL of it was used for the absorbance measurement at 518 nm with a microplate reader (Synergy H1, BioTek, USA). The percent inhibition of fat accumulation was determined using the following relationship:
Cell viability assay
The sulforhodamine B (SRB) assay was used to evaluate cell viability as previously described with minor modifications.52 The cells were washed three times with ddH2O. After drying, 100 μL SRB (0.04% SRB in 0.1% acetic acid) was added to each well and incubated at room temperature for 1 h. SRB was dissolved for three times with 1% acetic acid before air drying. Bound SRB was dissolved in 150 μL Tris solution (10 mM, PH10.5) for 10 min on a shaker. The absorbance of the Tris solution was recorded at 540 nm wavelength using a plate reader (Synergy H1, BioTek, USA). The cell viability was calculated as follows:
Fraction of the metabolites from gut microbiota (Exp. 5)
B. longum incubated in 140 dishes were collected and extracted with 70% methanol (100 mL) using an ultrasonicator (30 W, 60 kHz) for 30 minutes. After centrifugation at 12,000 rpm for 10 min, the residue was extracted twice, as described above. All supernatants were combined and evaporated using a rotary evaporator under reduced pressure at 60°C to remove methanol and water. The crude extracts were dissolved in methanol at 200 mg/mL for further purification using an HPLC pump (LC-20AD, Shimadzu, Japan) equipped with a manual injector and an ODS C18 analytical column (10 × 250 mm). The elution gradient for separation was as follows: 0% of solvent B, 0 to 5 min; 0%–50% of solvent B, 5 to 20 min; 50%-100% of solvent B from 20–40 min; 100%- 100% of solvent B from 40–50 min; 100%-50% of solvent B from 50–60 min; 50%-0% of solvent B from 70–80 min. A high-resolution Fourier-transform mass spectrometer (Orbitrap Elite, Thermo Scientific) equipped with an ESI source operated in the positive ionization mode was used as the detector. From 0 to 30 min, each fraction was collected every 0.5 min; from 30 to 80 min, each fraction was collected every 2 min. Finally, 26 fractions were obtained after combining the fractions with similar mass spectra. After drying using a rotary evaporator, 26 fractions were used for cell experiments.
Fecal and hepatic metabolite extract (Exp. 2 and Exp. 9)
The mouse feces (100 mg) were extracted with 70% methanol aqueous solution (1 mL) containing cholic acid-d4 (2 ppm) as internal standard. After 30-min of homogenization using an ultrasonicator (30 W, 60 kHz), the extracts were centrifuged at 12,000 rpm for 10 min at 4°C, and the supernatants were collected for LC-MS/MS analysis.
Mouse liver was obtained and washed with dd H2O to remove any residual gallbladder or bile acid. The washed liver tissue was frozen in liquid nitrogen and ground into fine powder. The powder of the liver tissue (100 mg) was extracted with ice-cold methanol (1 mL) containing cholic acid-d4 (2 ppm) as an internal standard. After 30-min of homogenization using an ultrasonicator (30 W, 60 kHz), the extracts were centrifuged at 12,000 rpm for 10 min at 4°C, and the supernatants were collected for LC-MS/MS analysis.
UPLC-MS/MS analysis (Exp. 2 and Exp. 9)
LC-MS/MS analysis was performed using a Dionex U3000 UPLC system coupled with a high-resolution Fourier-transform mass spectrometer (Q Exactive Plus, Thermo Scientific) using an Acquity HSS T3 (2.1 × 100 mm, 1.7 μm) column. The solvent A, 0.1% formic acid in water) and solvent B, 0.1% formic acid in acetonitrile) were used as the mobile phases. The elution gradient for separation was as follows: 20% of solvent B from 0 to 2 min; 20%-52% of solvent B from 2 to 12 min; 52%-65% of solvent B from 12–18 min; 65%-90% of solvent B from 18–22 min; 90%-20% of solvent B from 22–26 min; 20% of solvent B from 26–30 min. The flow rate was set to 0.3 mL/min. Mass spectrometric analysis was performed in the positive ionization mode. The ESI source (Thermo Scientific, Bremen, Germany) was operated at 50°C with a capillary temperature of 260°C, ionization voltage of 3.5 kV and a sheath gas flow rate of 25 L/min. Survey scans were acquired using a Q Exactive Plus mass analyzer operating at 70,000 (FWHM) resolving power. A mass range of 100–1500 m/z and a normalized collision energy of 30 were used for the survey scans. The method was set to analyze the top 10 most intense ions from the data-dependent scans, and dynamic exclusion was enabled for 15 s.
Molecular networking of the metabolites in mouse feces was created using the online workflow at GNPS (http://gnps.ucsd.edu).53 The data were then clustered with a precursor ions tolerance of 0.05 Da and a MS/MS product ions tolerance of 0.5 Da to create consensus spectra. A network with edges having a cosine score above 0.6 was created.
The GNPS spectral libraries were used to identify metabolites in mouse feces and liver. This was done using MS2 spectra and was then confirmed by chemical standards (detailed information is summarized in Table S2–4).
Fecal DNA extraction (Exp. 1)
Since there have been huge numbers of studies showing that the polyphenols from different plant sources could increase the growth or abundance of Bifidobacterium in the gut,54–56 a random selection of three mice in each experimental group was made to analyze the 16S rRNA of fecal samples. Fecal DNA extraction was performed using the QIAamp PowerFecal® DNA kit (QIAGEN, Germany). Briefly, 150 mg of feces from each mouse collected from the rectum was suspended in the provided buffer, homogenized using beads, and the residues were removed by centrifugation. Fecal DNA was collected using DNA-binding columns. After washing with the provided solutions, the DNA was eluted with elution buffer (10 mM Tris-HCl, pH 8.0). A Thermo ScientificTM NanoDropTM 2000/2000c spectrophotometer (Thermo Fisher, USA) was used to measure the quality and concentration of each sample. The ratio of absorbance at 260 nm to 280 nm should fall from 1.6 2.0. The abundance of Bifidobacterium spp. was determined by PCR analysis. Two microliters of fecal DNA was mixed with dNTPs, YEAtaq DNA polymerase (Yeastern Biotech, Taiwan), and genus-specific primer pairs (gene-specific primer sequences are shown in Table S5).57,58 The program was set as follows:95°C preheating, 95°C for 30 s, 50°C for 30 s, 72°C for 1 min, returning to 95°C, repeated 30 times, and 72°C for 15 min for the final extension. The polymerase chain reaction products were then analyzed by 0.8% agarose gel electrophoresis, and the signals of Bifidobacterium signals were detected at approximately 560 bp. The intensity was quantified using ImageJ according to reference59 and the area under the curve (AUC) was determined. The AUC fold-change was calculated by normalizing to the control groups.
16S rRNA gene amplicon sequencing (Exp. 1)
Genomic DNA was extracted from fecal samples using a DNA kit (TIANGEN Biotech Co. Ltd., Beijing, China) and quantified using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, USA). DNA (30–50 ng) was used to generate amplicons using the MetaVx Library Preparation kit. V3, V4, and V5 hypervariable regions of prokaryotic 16S rDNA were selected for generating amplicons and taxonomic analysis. DNA libraries were validated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and quantified a Qubit 2.0 Fluorometer. DNA libraries were multiplexed and loaded on an Illumina MiSeq instrument, according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). Sequencing was performed using paired-end configuration, and image analysis and base calling were conducted using the MiSeq Control Software (MCS) embedded in the MiSeq instrument.
RNA sequencing and analysis (Exp. 9)
Total RNA was extracted using Trizol Reagent (Invitrogen Life Technologies). Sequencing libraries were generated using a TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). The mRNA was fragmentized into 300 bp and then converted to cDNA. The cDNA library was purified using the AMPure XP system (Beckman Coulter,Beverly, CA, USA) to select the preferred cDNA fragments of 300 bp in length. The sequencing library was then sequenced on a HiSeq X platform (Illumina) with a 150-bp paired-end configuration by Personalbio Biotechnology Co., Ltd. (Shanghai, China).
The transcripts were assembled by mapping to Mus musculus (house mouse) genome assembly, GRCm38. The assembled transcripts were then annotated using the NCBI, eggNOG, KEGG, GO, EC, Ensembl, and UniPort databases. All the genes were tested for differential expression levels with DESeq, and the differentially expressed genes were subjected to GO enrichment analysis using the top GO software based on the hypergeometric test. The enrichment networks of significant genes/pathways/GO terms were analyzed using NetworkAnalyst and the R-package igraph.
Quantitative real-time PCR (Exp. 9)
Total RNA was extracted from SPF mouse livers using Trizol Reagent (Invitrogen Life Technologies). cDNA synthesis from 1 μg of total RNA was performed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Japan). TB Green Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa Bio, Japan) was used for qRT-PCR at a final reaction volume of 20 μL in an Applied Biosystems Step One Plus thermocycler (Applied Biosystems). All results were analyzed using the 2−△△CT method60 and normalized to the ribosomal protein L32 mRNA (gene-specific primer sequences can be found in Table S5).61–63
Statistical analysis
The results of the biological assay are presented as mean ± S.D. or S.E.M. Mice were assigned randomly to groups. A two-tailed Student’s t-test was used to evaluate the differences between two groups; a one-way ANOVA with Tukey’s range test was used to evaluate the differences among the three groups. A two-tailed Student t-test was used to compare uric acid-induced HepG2 cells to a control group. Results were considered significant at *p ≤ .05, **p ≤ .01, and ***p ≤ .001. Statistical analyses were performed using Prism 9, SPSS version 16.0, and Microsoft Excel 2017.
Supplementary Material
Acknowledgments
We thank the Ministry of Science and Technology (MOST), Republic of China (ROC), for supporting this study (MOST 106-2113-M-002-013-MY2; MOST 109-2636-M-002-005; MOST 112-2113-M-002-018-MY2). We thank mass spectrometry technical research services of NTU Consortia of Key Technologies and Center for Emerging Materials and Advanced Devices for metabolomics technology supply and data analysis. We thank Biotools Co., Ltd. for microbiota sequencing and analysis. We thank ez-Omics Co., Ltd. for visualization of the RNA-seq data.
Funding Statement
The work was supported by the Ministry of Science and Technology, Taiwan [MOST 109-2636-M-002-005]; Ministry of Science and Technology, Taiwan [MOST 106-2113-M-002-013-MY2].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
C.C.H. and Q.L. conceived the project, analyzed the data, and wrote the draft manuscript with input from all coauthors. R.A.C. wrote the manuscript as well as analyzed and visualized the data. Q.L. performed SPF C57BL/6Narl mouse experiments, fecal and microbial metabolite extraction, purification, and LC-MS analysis. H.B.Z., W.J.C., and Y.H.H performed gut microbial culture. H.Y.C. evaluated the inhibitory activity of metabolites on lipid accumulation in HepG2 cells. H.L.C. performed the C57BL/6Narl germ-free mouse experiments. L.K.S. performed the isotope tracking experiments, PCR analysis, and biosynthetic pathway analysis. L.H.L. extracted the protein and mRNA of the mouse liver and analyzed the RNA-seq data. S.M.T. visualization of the RNA-seq data. M.S.W., W.K.W., and P.Y.L. performed bacterial genome sequencing, genome assembly, and annotation. H.Y.W. and L.H.L. performed quantitative proteomic analysis.
Data and materials availability
The 16S rRNA gene amplicon sequencing, BCRC12585 genome sequencing, and RNA-seq data used in this study have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) under the BioProject ID PRJNA656418.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2300847
References
- 1.Bolte LA, Vich Vila A, Imhann F, Collij V, Gacesa R, Peters V, Wijmenga C, Kurilshikov A, Campmans-Kuijpers MJE, Fu J, et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut. 2021;70(7):1287–17. doi: 10.1136/gutjnl-2020-322670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan Y, Pedersen O.. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 3.Zmora N, Suez J, Elinav E.. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16(1):35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
- 4.Boutari C, Mantzoros CS. A 2022 update on the epidemiology of obesity and a call to action: as its twin COVID-19 pandemic appears to be receding, the obesity and dysmetabolism pandemic continues to rage on. Metabolism. 2022;133:155217. doi: 10.1016/j.metabol.2022.155217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiwari A, Balasundaram P. Public Health Considerations Regarding Obesity. StatPearls. Treasure Island (FL): StatPearls Publishing [PubMed] [Google Scholar]
- 6.Shapiro H, Suez J, Elinav E. Personalized microbiome-based approaches to metabolic syndrome management and prevention. J Diabetes. 2017;9(3):226–36. doi: 10.1111/1753-0407.12501. [DOI] [PubMed] [Google Scholar]
- 7.Tang WHW, Li DY, Hazen SL. Dietary metabolism, the gut microbiome, and heart failure. Nat Rev Cardiol 2019;16:137–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165(1):111–24. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koh A, Molinaro A, Ståhlman M, Khan MT, Schmidt C, Mannerås-Holm L, Wu H, Carreras A, Jeong H, Olofsson LE, et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell. 2018;175(4):947–61.e17. doi: 10.1016/j.cell.2018.09.055. [DOI] [PubMed] [Google Scholar]
- 10.Molinaro A, Nemet I, Bel Lassen P, Chakaroun R, Nielsen T, Aron-Wisnewsky J, Bergh P-O, Li L, Henricsson M, Køber L, et al. Microbially produced imidazole propionate is associated with heart failure and mortality. JACC Heart Fail. 2023;11(7):810–21. doi: 10.1016/j.jchf.2023.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosme P, Rodriguez AB, Espino J, Garrido M. Plant phenolics: bioavailability as a key determinant of their potential health-promoting applications. Antioxid (Basel). 2020;9(12):1263. doi: 10.3390/antiox9121263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar Singh A, Cabral C, Kumar R, Ganguly R, Kumar Rana H, Gupta A, Rosaria Lauro M, Carbone C, Reis F, Pandey AK, et al. Beneficial effects of dietary polyphenols on gut microbiota and strategies to improve delivery efficiency. Nutrients. 2019;11(9):11. doi: 10.3390/nu11092216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anhe FF, Pilon G, Roy D, Desjardins Y, Levy E, Marette A. Triggering Akkermansia with dietary polyphenols: A new weapon to combat the metabolic syndrome? Gut Microbes 2016; 7:146-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan MLY, Co VA, El-Nezami H. Dietary polyphenol impact on gut health and microbiota. Crit Rev Food Sci Nutr. 2021;61(4):690–711. doi: 10.1080/10408398.2020.1744512. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Daza MC, Pulido-Mateos EC, Lupien-Meilleur J, Guyonnet D, Desjardins Y, Roy D. Polyphenol-mediated gut microbiota modulation: toward prebiotics and further. Front Nutr. 2021;8:689456. doi: 10.3389/fnut.2021.689456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Qi Y, Zheng H. Dietary polyphenol, gut microbiota, and health benefits. Antioxid (Basel). 2022;11(6):11. doi: 10.3390/antiox11061212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerry RG, Patra JK, Gouda S, Park Y, Shin HS, Das G. Benefaction of probiotics for human health: a review. J Food Drug Anal. 2018;26(3):927–39. doi: 10.1016/j.jfda.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roager H M, Licht T R. (2018). Microbial tryptophan catabolites in health and disease. Nat Commun, 9(1), 3294 10.1038/s41467-018-05470-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cani PD, Knauf C. A newly identified protein from Akkermansia muciniphila stimulates GLP-1 secretion. Cell Metab. 2021;33(6):1073–5. doi: 10.1016/j.cmet.2021.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Markowiak-Kopeć P, Śliżewska K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients. 2020;12(4):12. doi: 10.3390/nu12041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donadio S, Monciardini P, Alduina R, Mazza P, Chiocchini C, Cavaletti L, Sosio M, Puglia AM. Microbial technologies for the discovery of novel bioactive metabolites. J Biotechnol. 2002;99(3):187–198. doi: 10.1016/S0168-1656(02)00209-2. [DOI] [PubMed] [Google Scholar]
- 22.Ma Y, Liu X, Wang J. Small molecules in the big picture of gut microbiome-host cross-talk. EBioMedicine. 2022;81:104085. doi: 10.1016/j.ebiom.2022.104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fobofou SA, Savidge T. Microbial metabolites: cause or consequence in gastrointestinal disease? Am J Physiol Gastrointest Liver Physiol. 2022;322(6):G535–G52. doi: 10.1152/ajpgi.00008.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdel-Razek AS, El-Naggar ME, Allam A, Morsy OM, Othman SI. Microbial natural products in drug discovery. Processes. 2020;8(4). doi: 10.3390/pr8040470. [DOI] [Google Scholar]
- 25.Markowiak P, Śliżewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients. 2017;9(9):1021. doi: 10.3390/nu9091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohland CL, Macnaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol. 2010;298(6):G807–19. doi: 10.1152/ajpgi.00243.2009. [DOI] [PubMed] [Google Scholar]
- 27.Rinschen MM, Ivanisevic J, Giera M, Siuzdak G. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol. 2019;20(6):353–67. doi: 10.1038/s41580-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyu Q, Kuo TH, Sun C, Chen K, Hsu CC, Li X. Comprehensive structural characterization of phenolics in litchi pulp using tandem mass spectral molecular networking. Food Chem. 2019;282:9–17. doi: 10.1016/j.foodchem.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Ibrahim SR, Mohamed GA. Litchi chinensis: medicinal uses, phytochemistry, and pharmacology. J Ethnopharmacol. 2015;174:492–513. doi: 10.1016/j.jep.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 30.Chukwuma CI, Izu GO, Chukwuma MS, Samson MS, Makhafola TJ, Erukainure OL. A review on the medicinal potential, toxicology, and phytochemistry of litchi fruit peel and seed. J Food Biochem. 2021;45(12):e13997. doi: 10.1111/jfbc.13997. [DOI] [PubMed] [Google Scholar]
- 31.Schellekens H, Torres-Fuentes C, van de Wouw M, Long-Smith CM, Mitchell A, Strain C, Berding K, Bastiaanssen TFS, Rea K, Golubeva AV, et al. Bifidobacterium longum counters the effects of obesity: partial successful translation from rodent to human. EBioMedicine. 2021;63:103176. doi: 10.1016/j.ebiom.2020.103176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maldonado-Gomez MX, Martinez I, Bottacini F, O’Callaghan A, Ventura M, van Sinderen D, Hillmann B, Vangay P, Knights D, Hutkins R, et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host & Microbe. 2016;20(4):515–26. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Anderson RC, Cookson AL, McNabb WC, Park Z, McCann MJ, Kelly WJ, Roy NC. Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol. 2010;10(1):316. doi: 10.1186/1471-2180-10-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazloom K, Siddiqi I, Covasa M. Probiotics: how effective are they in the fight against obesity? Nutrients. 2019;11(2):11. doi: 10.3390/nu11020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Shea EF, Cotter PD, Stanton C, Ross RP, Hill C. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int J Food Microbiol. 2012;152(3):189–205. doi: 10.1016/j.ijfoodmicro.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 36.Yoon SJ, Yu JS, Min BH, Gupta H, Won SM, Park HJ, Han SH, Kim B-Y, Kim KH, Kim BK, et al. Bifidobacterium-derived short-chain fatty acids and indole compounds attenuate nonalcoholic fatty liver disease by modulating gut-liver axis. Front Microbiol. 2023;14:1129904. doi: 10.3389/fmicb.2023.1129904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hevia H, Varela-Rey M, Corrales FJ, Berasain C, Martinez-Chantar ML, Latasa MU, Lu SC, Mato JM, García-Trevijano ER, Avila MA, et al. 5′-methylthioadenosine modulates the inflammatory response to endotoxin in mice and in rat hepatocytes. Hepatology. 2004;39(4):1088–1098. doi: 10.1002/hep.20154. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Wang F, Liang B, Su Y, Sun S, Xia S, Shao J, Zhang Z, Hong M, Zhang F, et al. Methionine metabolism in chronic liver diseases: an update on molecular mechanism and therapeutic implication. Signal Transduct Target Ther. 2020;5(1):280. doi: 10.1038/s41392-020-00349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Wang Y, Wu P. 5’-methylthioadenosine and cancer: old molecules, new understanding. J Cancer. 2019;10(4):927–36. doi: 10.7150/jca.27160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreu-Pérez P, Hernandez-Losa J, Moliné T, Gil R, Grueso J, Pujol A, Cortés J, Avila MA, Recio JA. Methylthioadenosine (MTA) inhibits melanoma cell proliferation and in vivo tumor growth. BMC Cancer. 2010;10(1):265. doi: 10.1186/1471-2407-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cirulli ET, Guo L, Leon Swisher C, Shah N, Huang L, Napier LA, Kirkness EF, Spector TD, Caskey CT, Thorens B, et al. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab. 2019;29(2):488–500.e2. doi: 10.1016/j.cmet.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perng W, Hector EC, Song PXK, Tellez Rojo MM, Raskind S, Kachman M, Cantoral A, Burant CF, Peterson KE. Metabolomic determinants of metabolic risk in Mexican adolescents. Obesity (Silver Spring). 2017;25(9):1594–1602. doi: 10.1002/oby.21926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simile MM, Banni S, Angioni E, Carta G, De Miglio MR, Muroni MR, Calvisi DF, Carru A, Pascale RM, Feo F, et al. 5′-methylthioadenosine administration prevents lipid peroxidation and fibrogenesis induced in rat liver by carbon-tetrachloride intoxication. J Hepatol. 2001;34(3):386–394. doi: 10.1016/S0168-8278(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 44.Azzu V, Vacca M, Kamzolas I, Hall Z, Leslie J, Carobbio S, Virtue S, Davies SE, Lukasik A, Dale M, et al. Suppression of insulin-induced gene 1 (INSIG1) function promotes hepatic lipid remodelling and restrains NASH progression. Mol Metab. 2021;48:101210. doi: 10.1016/j.molmet.2021.101210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong XY, Tang SQ. Insulin-induced gene: a new regulator in lipid metabolism. Peptides. 2010;31(11):2145–50. doi: 10.1016/j.peptides.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 46.Krapivner S, Popov S, Chernogubova E, Hellenius ML, Fisher RM, Hamsten A, van’t Hooft FM. Insulin-induced gene 2 involvement in human adipocyte metabolism and body weight regulation. J Clin Endocrinol Metab. 2008;93(5):1995–2001. doi: 10.1210/jc.2007-1850. [DOI] [PubMed] [Google Scholar]
- 47.Zadravec D, Brolinson A, Fisher RM, Carneheim C, Csikasz RI, Bertrand-Michel J, Borén J, Guillou H, Rudling M, Jacobsson A, et al. Ablation of the very-long-chain fatty acid elongase ELOVL3 in mice leads to constrained lipid storage and resistance to diet-induced obesity. FASEB J. 2010;24(11):4366–4377. doi: 10.1096/fj.09-152298. [DOI] [PubMed] [Google Scholar]
- 48.Ali II, D’Souza C, Singh J, Adeghate E. Adropin’s role in energy homeostasis and metabolic disorders. Int J Mol Sci. 2022;23(15):23. doi: 10.3390/ijms23158318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huynh FK, Neumann UH, Wang Y, Rodrigues B, Kieffer TJ, Covey SD. A role for hepatic leptin signaling in lipid metabolism via altered very low density lipoprotein composition and liver lipase activity in mice. Hepatology. 2013;57(2):543–54. doi: 10.1002/hep.26043. [DOI] [PubMed] [Google Scholar]
- 50.Avila MA, Garcia-Trevijano ER, Lu SC, Corrales FJ, Mato JM. Methylthioadenosine. Int J Biochem Cell Biol. 2004;36(11):2125–2130. doi: 10.1016/j.biocel.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 51.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108(15):6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1(3):1112–6. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 53.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol. 2016;34(8):828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Catalkaya G, Venema K, Lucini L, Rocchetti G, Delmas D, Daglia M, De Filippis A, Xiao H, Quiles JL, Xiao J, et al. Interaction of dietary polyphenols and gut microbiota: microbial metabolism of polyphenols, influence on the gut microbiota, and implications on host health. Food Front. 2020;1(2):109–33. doi: 10.1002/fft2.25. [DOI] [Google Scholar]
- 55.Mithul Aravind S, Wichienchot S, Tsao R, Ramakrishnan S, Chakkaravarthi S. Role of dietary polyphenols on gut microbiota, their metabolites and health benefits. Food Res Int. 2021;142:142. doi: 10.1016/j.foodres.2021.110189. [DOI] [PubMed] [Google Scholar]
- 56.Correa TAF, Rogero MM, Hassimotto NMA, Lajolo FM. The two-way polyphenols-microbiota interactions and their effects on obesity and related metabolic diseases. Front Nutr. 2019;6:188. doi: 10.3389/fnut.2019.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. 2004;70(1):167–173. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97(6):1166–77. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- 59.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Petersen C, Bell R, Klag KA, Lee SH, Soto R, Ghazaryan A, Buhrke K, Ekiz HA, Ost KS, Boudina S, et al. T cell–mediated regulation of the microbiota protects against obesity. Sci. 2019;365(6451). doi: 10.1126/science.aat9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–35. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 63.Nothias L, et al. Bioactivity-Based Molecular Networking for the Discovery of Drug Leads in Natural Product Bioassay-Guided Fractionation. J Nat Prod. 2018;81(4):758–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


