Abstract
Protein kinase C (PKC) isozymes are maintained in a ‘ready-to-go’ but ‘safe’ autoinhibited conformation until second messenger binding unleashes an autoinhibitory pseudosubstrate to allow substrate phosphorylation. But to gain this ‘ready-to-go’ conformation, PKC must be processed by a series of complex priming phosphorylations, the mechanism of which was enigmatic until now. Recent findings snapped the pieces of the phosphorylation puzzle into place to unveil a process that involves a newly-described motif (TOR interaction motif, TIM), a well-described kinase (mTORC2), and an oft-used mechanism (autophosphorylation) to prime PKC to signal. This review highlights new insights that have been elucidated for how phosphorylation controls PKC and discusses them in the context of common mechanisms for AGC kinase regulation by phosphorylation and autophosphorylation.
Keywords: autophosphorylation, mTORC2, PKC, phosphorylation
Protein kinases as signaling switches
Reversible protein phosphorylation, discovered almost 70 years ago by Fischer and Krebs [1], is the universal language of cell signaling, with the majority of proteins regulated at multiple sites by the opposing functions of protein kinases and protein phosphatases [2, 3]. Protein kinases are highly regulated enzymes, whose spatial and temporal activity is directed through various ON-OFF switch mechanisms to ensure kinases are activated in the proper cellular context [4-6]. Whereas kinases catalyze the transfer of phosphate groups to their substrates, phosphorylation is also the primary means by which kinases themselves are regulated and achieve catalytic competence. Phosphorylation dictates kinase function by affecting structure, localization, stability, and affinity for other proteins or second-messengers that direct spatiotemporal activity [6, 7]. The protein kinase C (PKC; Box 1) family of Ser/Thr kinases serves as a paradigm for how phosphorylation by both upstream kinases and autophosphorylation coordinately control kinase function. This review discusses the role of two upstream kinases, mechanistic target of rapamycin complex 2, (mTORC2; Box 2) and the phosphoinositide-dependent kinase 1 (PDK1), in converting newly-synthesized PKC into an autophosphorylation-competent species that modifies its own C-tail, the final step in producing the stable, signaling-competent enzyme that accumulates in cells. This review also highlights the role of autophosphorylation in regulating kinase function.
Box 1. Architecture of Protein Kinase C family members.
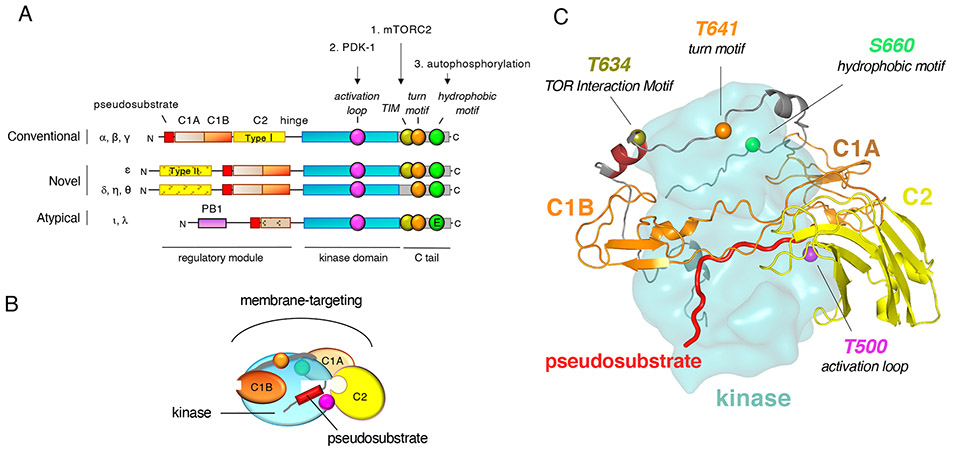
PKC isozymes are encoded by 9 genes that are grouped into three subclasses, conventional (α, β, γ), novel (δ, ε, η, θ), and atypical (ζ/λ, ι) (Figure IA) [8]. They share a common architecture of an N-terminal regulatory moiety that constrains the catalytic activity of a C-terminal kinase domain. All PKC isozymes have an autoinhibitory pseudosubstrate segment (red) that reversibly binds the kinase domain (cyan). In the absence of appropriate second messengers, it occupies the substrate-binding cavity to inhibit the kinase and is released following engagement of regulatory modules to appropriate second messengers or scaffolds, thus permitting substrate phosphorylation and downstream signaling. However, all regulatory domains participate in a network of interactions to maintain autoinhibition (Figure IB and IC) [56, 57]. The C1A domain (light orange) directly follows the pseudosubstrate in all PKC isozymes and functions with the pseudosubstrate to mediate autoinhibition [58]. The C1B domain (dark orange) serves as a diacylglycerol sensor in conventional and novel PKC isozymes, but not in atypical PKC isozymes because their C1 domain (striped) precludes ligand binding. The C2 domain (yellow) in conventional PKC isozymes is a Ca2+-sensor which targets these PKC isozymes to plasma membrane via a recognition motif for the plasma membrane-localized lipid phosphatidylinositol-4,5-bisphosphate (PIP2) [59]. Novel PKC isozymes also have a C2 domain, but it is a topological variant (Type II) allowing it to occupy the same predicted position in the tertiary structure of PKC despite preceding the pseudosubstrate in the primary structure [57]. The novel C2 domain lacks key Asp residues that coordinate Ca2+ and thus is not a Ca2+ sensor. The PB1 domain (purple) in atypical PKC isozymes, which respond to neither Ca2+ nor diacylglycerol, mediates binding to protein scaffolds, interactions that promote release of the pseudosubstrate to allow localized signaling [60, 61]. The kinase domain of PKC (cyan) is regulated by four constitutive post-translational phosphorylations that ‘process’ PKC into a mature, autoinhibited, and stable species. mTORC2 phosphorylates the newly-identified TOR interaction motif (TIM) (olive; Thr 634 in PKCβII) and turn motif (orange; Thr 641 in PKCβII) in the C-tail of certain PKCs (α, β, γ, ε, ι, and ζ), PDK1 catalyzes the phosphorylation on the activation loop (magenta; Thr 500 in PKCβII) in the kinase domain, and PKC autophosphorylates at the hydrophobic motif (green; Ser 600 in PKCβII). These phosphorylations are necessary for PKC to adopt the autoinhibited conformation, and aberrant PKC that cannot become autoinhibited is degraded by a quality control mechanism [28].
Box 2. Architecture of mTORC2.
mTOR is a large (289 kDa) multidomain Ser/Thr kinase that assembles into two distinct multi-subunit signaling complexes in the cell, mTORC1 and mTORC2 [62]. Whereas mTORC1 plays key roles in energy sensing and metabolism, the less well understood mTORC2 regulates AGC kinases and survival signaling [63]. mTORC2 shares core components mLST8 and the mTOR kinase with mTORC1; however, mTORC2 contains the adaptor Rictor instead of mTORC1 component Raptor, and additionally harbors the unique component Sin1 [64] (Figure IIA). mTORC1 activity is allosterically inhibited by Rapamycin and its derivatives, however the Sin1-Rictor interaction occludes the Rapamycin binding site on mTOR-FKBP12, rendering mTORC2 insensitive to acute rapamycin treatment [65]. Originally characterized as a modulator of hydrophobic motif phosphorylation on Akt [66], mTORC2 also regulates the phosphorylation and activity of other AGC kinases including SGK, PKN, and six of the nine PKC family members [23] (see main text). The hydrophobic motif of S6K, in contrast, is regulated by mTORC1. mTORC2 has been shown to directly phosphorylate two conserved sites, the TOR-Interaction Motif (TIM, PKCβII Thr634) and the turn motif (PKCβII Thr641), on the C-tail of these AGC kinases [9, 27, 67, 68]. Recruitment of its AGC kinase substrates is achieved by binding of the Sin1 CRIM domain to the kinase C-tail [9, 69, 70] (Figure IIB). For PKC and Akt, phosphorylation of the TIM and turn motif Thr facilitates phosphorylation of the activation loop by PDK1, which then triggers autophosphorylation of the hydrophobic motif to fully activate the kinase. Thus, one role of mTORC2 is to indirectly regulate hydrophobic motif phosphorylation by priming AGC kinases for activation.
PKC isozymes are primed to rapidly respond to signals that produce diacylglycerol and, for some family members Ca2+, to ‘message’ their engagement on cell surface receptors [8]. These Ser/Thr kinases are maintained in a ‘ready-to-go’ but autoinhibited conformation until second messenger binding releases an autoinhibitory pseudosubstrate segment from the kinase active site to allow substrate binding and phosphorylation (Figure 1, right panel). In order to gain this ‘ready-to-go’ conformation, PKC must be processed by a series of priming phosphorylations (Figure 1 left panel). Without these phosphorylations, PKC is in an open ‘unsafe’ conformation and is degraded [9]. Similarly, if the mature form is locked in an open and active conformation, it is dephosphorylated and degraded, a process referred to as downregulation. Thus, phosphorylation is critical to the stability and function of PKC. Deregulation of the inputs that control the activity or steady state levels of PKC result in pathophysiologies (Box 3).
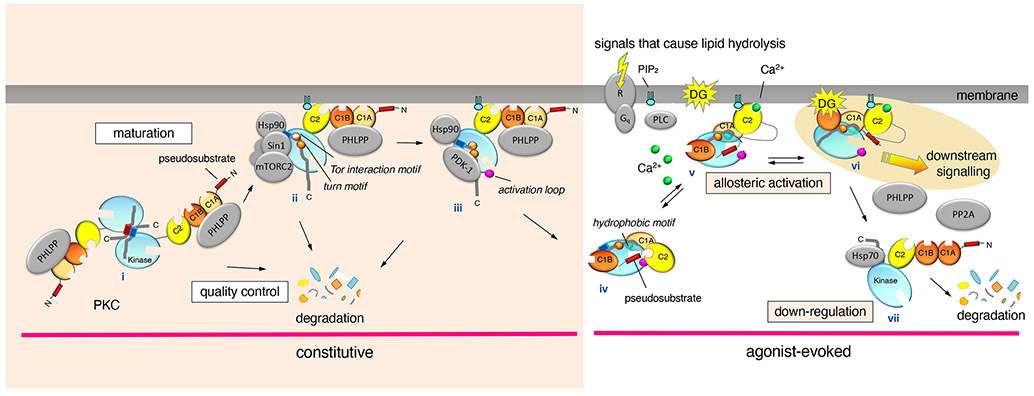
Figure 1. Lifecycle of Protein Kinase C.
PKC signaling involves constitutive priming phosphorylations and agonist-evoked activity. PKC is matured by phosphorylation following synthesis, conferring catalytic competence and protein stability. PKC is synthesized as a symmetrical homodimer (i). PKC is recruited to the kinase complex mTORC2 by Sin1 and the protein chaperone Hsp90 (ii). mTORC2 phosphorylation of the TIM and turn motif sites dissociates the dimer and exposes the C-tail for docking by PDK1 (iii). PDK1 phosphorylates the activation loop of PKC, which triggers intramolecular hydrophobic motif autophosphorylation. This phosphorylation confers full catalytic competence to PKC and simultaneously induces conformational changes that result in autoinhibition via the pseudosubstrate. PHLPP phosphatases survey the conformation of newly-synthesized PKC, dephosphorylating the hydrophobic motif of PKC enzymes which do not adopt the autoinhibited conformation. The fully phosphorylated, autoinhibited, and stable PKC is monomeric in the cytosol (iv). PKC is allosterically activated upon the generation of Ca2+, which recruits PKC to the plasma membrane via the C2 domain (v). C1B domain binding to diacylglycerol expels the pseudosubstrate to initiate downstream signaling. The active PKC conformation (vi) is sensitive to dephosphorylation by PHLPP and PP2A phosphatases and the dephosphorylated species is down-regulated by Hsp70-mediated proteasomal degradation (vii).
Box 3. PKC Dysregulation in Disease.
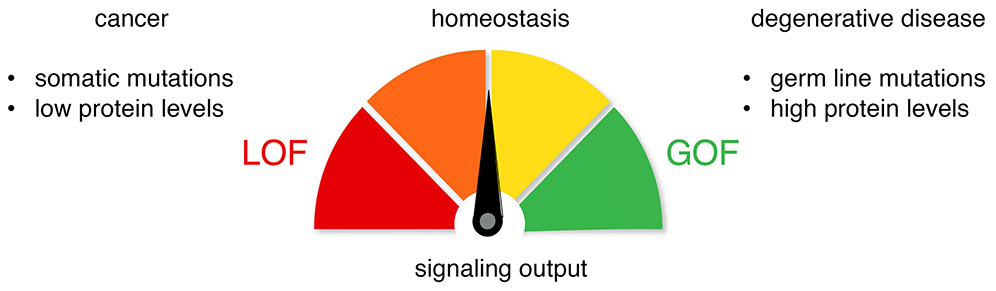
The signaling output of PKC in cells is exquisitely controlled by diverse mechanisms to maintain cellular homeostasis (Figure II). In addition to acute activation by second messengers, the steady-state levels of PKC are finely tuned. Mechanisms that promote phosphorylation (discussed in the main text) increase the steady-state levels of PKC whereas ones that promote the dephosphorylation reduce the steady-state levels of PKC by triggering its down-regulation. Indeed, an analysis of >5,000 tumor cell lines revealed a 1:1 correlation between phosphorylation of the hydrophobic motif of PKCα and PKCβII and total PKCα and PKCβII levels, underscoring the critical importance of hydrophobic motif phosphorylation in the stability of PKC [28]. Low PKC protein is associated with worsened survival in a number of cancers, including pancreatic, colon, and lung cancer [71]. Additionally, cancer-associated somatic mutations in PKC are generally loss-of-function (LOF). These findings have recently reframed PKC as a tumor suppressor. In contrast, gain-of-function (GOF) is associated with neurodegenerative disease. Notably, activity-enhancing germline mutations in PKCα are associated with Alzheimer’s Disease and ones that enhance PKCγ activity are associate with Spinocerebellar Ataxia Type 14 [72].
Activation loop phosphorylation
A nearly universal requirement for eukaryotic protein kinases is the structuring of a labile loop connecting the N- and C-lobes of the kinases, termed the activation loop (also T-loop). This segment near the entrance to the active site exists in a dynamic equilibrium of a DFG-IN conformation, in which a hydrophobic “regulatory spine” found in all kinases is aligned to poise the kinase for catalysis, and a DFG-OUT conformation in which the spine is not assembled and the kinase is inactive [4, 10]. Although kinases such as Aurora A can adopt the DFG-IN conformation by binding protein partners [11], alignment of the regulatory spine is most commonly achieved by phosphorylation, which either alone or in combination with other stabilizing factors [12-14], favors the DFG-IN conformation [4, 10]. Thus, activation loop phosphorylation is intrinsically linked to kinase activation and represents a central control step to achieve the active state. Illustrating the importance of structuring the activation segment for catalysis, protein pseudokinases, which possess a protein kinase fold but lack catalytic activity, are frequently rendered inactive as a result of mutations in prominent catalytic motifs such as the activation loop [15, 16]. Additionally, the activation loop is the most frequently mutated region in cancer kinomes [17]; modifications in its phosphorylation sites may mimic phosphorylation to hyperactivate oncoprotein kinases or prevent phosphorylation to inactivate tumor-suppressor kinases [18].
PDK1 – activation loop kinase
PDK1 is the “activation loop kinase” for PKC and many AGC kinases including protein kinase A (PKA), Akt, S6K, RSK, and SGK [19]. This single gene product, which is present in limiting quantities in cells (approximately 10 nM in HeLa cells [20]) is responsible for the phosphorylation of a disproportionate number of substrates (just the PKC isozymes alone account for on the order of 100 nM of substrate [20]). While the phosphorylation of all the client kinases occurs on the activation loop to promote alignment of residues for catalysis, different kinases take advantage of this step in various ways. In the case of PKC and PKA, phosphorylation by PDK1 occurs shortly after biosynthesis and is part of a conversion to a species that is poised to signal as soon as the appropriate second messengers are present [8]. But for kinases such as Akt, the PDK1-catalyzed phosphorylation is agonist-induced and transient [19].
Gateway to the activation loop
Although activation loop phosphorylation is necessary for PKC activity, it is not sufficient. PKC isozymes and many other AGC kinases including Akt and S6K require phosphorylation of a site termed the hydrophobic motif, a conserved feature in the C-terminal tail of nearly all AGC kinases [21]. Phosphorylation of this site is generally required for full kinase activation, however some AGC kinases, such as PKA, share the flanking hydrophobic sequence, but lack the phospho-acceptor Ser or Thr, and thus do not require hydrophobic motif phosphorylation for activity [4]. The identity of the direct hydrophobic motif kinase was long contentious, with evidence initially supporting an autophosphorylation mechanism and later revealing a requirement for the mTOR kinase [22, 23]. The resolution of this mechanism, described for PKC family kinases, linked the function of mTOR with both activation loop phosphorylation and hydrophobic motif autophosphorylation, providing an elegant example of how synergistic regulation of phosphorylation sites improves the fidelity of kinase maturation and activation [9].
mTORC2 – gatekeeper of the activation loop phosphorylation
The second mTOR-containing complex, mTORC2 (Box 2), was first characterized as a requirement for Akt phosphorylation and activity, as was later shown to be the case for several AGC kinases including PKC [23-25]. mTORC2 has been proposed to phosphorylate multiple sites on the C-tail including the turn motif, hydrophobic motif, and extreme C-tail sites on Akt [26]; garnering the moniker of “hydrophobic motif kinase”. Strong biochemical evidence for a direct mTORC2 phosphorylation, however, exists only for co-translational phosphorylation of the Akt turn motif [27]. Phosphorylation of the turn motif on Akt is not required for activity or its phosphorylation at the activation loop or hydrophobic motif [27], suggesting that mTORC2 regulates Akt by a distinct mechanism. For PKC, the turn motif site is also dispensable for its activity, while the hydrophobic motif is required for in vitro and cellular activity [28]. Curiously, PKC dephosphorylated exclusively at the activation loop and hydrophobic motif, but retaining the turn motif, is capable of re-autophosphorylation at the hydrophobic motif, whereas fully dephosphorylated PKC requires PDK1 phosphorylation of the activation loop in order to re-autophosphorylate and become active towards substrates [22, 29]. Therefore, the role of both mTORC2 and turn motif phosphorylation in PKC activation remained unclear for the 15 years following the discovery that mTORC2 promotes the maturation of PKC [23].
TOR interaction motif
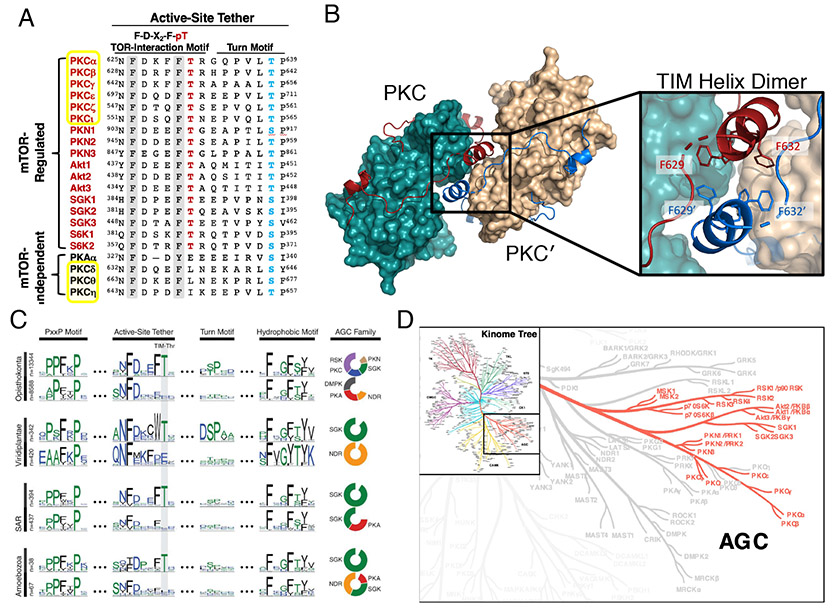
The identification of an invariant phosphorylation motif in the C-tail of every known mTORC2 client kinase, but not other AGC kinases, provided a breakthrough in solving the puzzle of how mTORC2 regulates PKC [9] (Figure 2A). This motif F-X3-F-pT is present in all mTORC2-regulated PKC isozymes, but not mTORC2-independent PKC isozymes. Indeed, AGC kinases can be divided into the ones that have the motif (such as most, but not all, PKC isozymes and Akt) and the ones that do not (such as PKA) (Figure 2B). The motif, termed the TOR Interaction Motif (TIM) is the most conserved phosphorylation element of the C-tail of AGC kinases, and is evolutionarily conserved to amoeba (Figure 2C). The motif is present in the yeast PKN1, from which the PKC family evolved [30], but was later lost in mTORC2-independent PKCs raising the question as to whether a new function and new regulatory mechanism may have evolved for these isozymes. The newly-identified phosphorylation site is positioned seven amino acids before the known turn motif site, on a segment of the kinase referred to as the Active Site Tether [21]. The solvent-exposed positioning of this Thr, unlike the kinase domain-facing turn motif Thr, likely accounts for why phosphate occupancy of the TIM site in mature PKC is low and its importance had not been previously appreciated.
Figure 2. Conservation of the TOR-Interaction Motif.
A) Protein sequence alignment showing the TOR-interaction motif and turn motif phosphorylation sites. mTOR-regulated AGC kinases exclusively conserve the TIM Thr. The TIM comprises an F-X3-F-pT motif in the C-terminal tail preceding the turn motif. Three PKC isozymes (δ1, η, θ) do not conserve a phospho-acceptor at the TIM, and these are not regulated by mTORC2. PKC isozymes are boxed in yellow. B) AGC branch of the human kinome tree showing the kinases conserving the TIM Thr (red). C) Evolutionary conservation of the TIM Thr in the AGC C-terminal tail in five major taxonomical groups. Sequence logos show major features, stratified by kinases that conserve (top) or do not conserve (bottom) the TIM Thr in the active-site tether for each group. Adapted from [9]. Reprinted with permission from AAAS.
Unveiling the functional relevance of the newly-identified TIM involved a key biochemical finding: overexpression of PDK1 completely bypassed the requirement for mTORC2 in allowing the hydrophobic motif phosphorylation of not only PKC, but also Akt [9]. Whereas PKC and Akt have no phosphate at the activation loop, turn motif, or hydrophobic motif in cells lacking functional mTORC2, overexpression of PDK1 promotes phosphorylation of the activation loop (the direct PDK1 site) and the hydrophobic motif (autophosphorylation). PDK1 does not, however, restore phosphorylation of the turn motif, consistent with this site being a bona fide mTORC2 site [27]. The PDK1-promoted phosphorylation of the hydrophobic motif of both PKC and Akt requires the intrinsic catalytic activity of each client kinase, validating previous studies identifying autophosphorylation as the mechanism of hydrophobic motif phosphorylation [22, 31, 32]. These studies revealed that mTORC2 facilitates the phosphorylation of the activation loop by PDK1, an event that promotes catalysis to allow autophosphorylation of the hydrophobic motif.
The final piece of the puzzle involved a structural insight: a crystal structure of the cleaved kinase domain of PKC revealed a previously ignored head-to-head dimer mediated by the newly-identified TIM [9, 33] present on the NFD helix of the C-tail (Figure 3). This dimer, not observed in other structures of the isolated PKC kinase domain, may have been fortuitously observed because the kinase domain was uniquely generated from full-length (and thus normally matured) PKCβII by limited proteolysis at the hinge (Figure I). Although phosphorylated (mature) PKC is a monomer [34], this structure of the isolated kinase introduced the idea that newly-synthesized PKC could be a dimer, mediated by the TIM helix and the substrate-binding CRIM domain of mTORC2 (Box 2). Whether this dimer is stabilized by protein scaffolds, potentially assisting in recruiting mTORC2, remains to be elucidated. The observation of this dimer interface led to the model that phosphorylation of the TIM and adjacent turn motif by mTORC2 would break apart the dimer, exposing the C-tail for PDK1 binding, and thus initiating the chain of priming phosphorylations. Indeed, biochemical studies revealed that mutation of the TIM and turn motif sites to Ala promoted dimerization of PKC and prevented processing by phosphorylation [9]. Furthermore, stapled peptides that competed with the dimer interface promoted the phosphorylation of PKC. Thus, the exact role of mTORC2 in the maturation of PKC was unveiled: TIM phosphorylation by mTORC2 dissociates an inactive PKC homodimer following synthesis, allowing PDK1 to phosphorylate the activation loop and triggering intramolecular autophosphorylation of the hydrophobic motif [9] (Figure 1, left panel). This mTORC2-catalyzed phosphorylation of the TIM is the first and rate-limiting step that allows PKC to become catalytically competent, stable, and poised to respond to second-messengers (Figure 1 right panel). Taken together, mTORC2 acts as the gatekeeper for PDK1 to phosphorylate the activation loop and prepare PKC to adopt its primed state.
Figure 3. The TOR-Interaction Motif Coordinates PKC Dimerization.
X-Ray structure (PDB ID: 2I0E; [33]) of the symmetrical PKCβII homodimer showing the PKC kinase domain (teal) and C-tail (red) with dimer partner (PKC’) kinase (tan) and C-tail (blue). The TIM helix forms a hydrophobic interface through conserved Phe resides; inset indicates F629-X2-F631-F632-T of the F-X3-F-pT. Phosphorylation of the TIM Thr may dissociate the dimer by perturbing this interaction. Adapted from [9]. Reprinted with permission from AAAS.
Figure I. Architecture of Protein Kinase C.
A) Domain composition of PKC isozymes showing: autoinhibitory pseudosubstrate (red), diacylglycerol- and phorbol ester-binding (solid orange) or non-ligand binding (stippled orange) C1 domains, Ca2+-sensing (solid yellow) or non-Ca2+-sensing (yellow stippled) C2 domain, kinase domain (cyan), and C-terminal tail (C-tail; grey). Also shown are the conserved phosphorylation sites: activation loop (fuchsia) by PDK1, the TOR-interaction motif (TIM; olive) and turn motif (orange) by mTORC2, and the hydrophobic motif (green) by cis-autophosphorylation. Atypical PKCs are not phosphorylated at their hydrophobic motif and instead have a Glu (E) at the phospho-acceptor position. Three novel PKCs (δ, η, θ) do not have the TIM Thr phosphorylation site and are not regulated by mTORC2. B) Cartoon depiction of mature PKC in the autoinhibited conformation. In the absence of second messengers, PKC is cytosolically localized and the pseudosubstrate restrains PKC activity. C) Molecular modeling of full-length, autoinhibited PKC βII, showing general packing architecture proposed for all PKC isozymes based on structures of individual domains [57]. The pseudosubstrate (red) is held in place by the C2 domain (yellow) and the C1 domains (orange) pack against the kinase domain, with the C-tail (grey) wrapping around the top of the upper N-lobe. Phosphorylation sites are shown as spheres: activation loop (fuchsia) and C-tail (grey) sites at TOR interaction motif (TIM; olive), turn motif (orange), and hydrophobic motif (green).
Determining the precise order, function, and consequence of individual phosphorylations was perhaps best suited for PKC, whose phosphorylations are ordered and constitutive; however, other AGC kinases with inducible phosphorylations dependent on multiple inputs may be more difficult to decipher. For example, the structural implications of TIM phosphorylation on Akt are not as well understood beyond the shared function of recruiting PDK1 to phosphorylate the activation loop to facilitate hydrophobic motif autophosphorylation. Given that the other mTORC2 phosphorylation site, the turn motif, is phosphorylated co-translationally [27], and that the phosphorylations of the activation loop and hydrophobic motif of Akt are agonist-evoked [19], there is important biology to be worked out surrounding the timing, localization, and structural requirements of these events. A general role of mTORC2 in facilitating the PDK1 step is supported by a recent study that showed that a Lys to Cys mutation in the PIF pocket (see Hydrophobic motif section) of yeast Gad8, an orthologue of human Akt and SGK1, also bypasses the mTORC2 requirement for activity of this AGC kinase [35]. This mutation likely unlatches the C-tail to render it more accessible for PDK1 binding. Additionally, other AGC kinases such as S6 kinases depend instead upon mTORC1 for phosphorylation of their hydrophobic motifs due to a C-terminus that blocks mTORC2 binding and an N-terminal TOR signaling motif that recruits mTORC1 [36]. Of note, a recent study revealed that specific mTOR complex-dependence (1 vs 2) influences the upstream signals that regulate the activation of distinct AGC kinases. Thus, the precise function of TIM phosphorylation in other AGC kinases that depend upon mTOR will be of great interest in future studies.
Autophosphorylation – how kinases self-activate
Activation of a growing number of protein kinases is recognized to involve autophosphorylation. The process of autophosphorylation presents an apparent paradox, as an inactive kinase must phosphorylate itself to become active [37]. This model invokes a “prone-to-autophosphorylate” conformation in which an inactive kinase is allosterically stabilized via dimerization or chaperone proteins to overcome the energy barrier for autophosphorylation, resulting in full activation [38]. The phenomenon of autophosphorylation is an evolutionarily ancient signaling mechanism: the His two-component system found in bacteria involves the autophosphorylation of a histidine kinase, which then transfers the phosphate group to a substrate effector to elicit a cellular response [39]. A recent analysis quantifying the extent of autophosphorylation in the eukaryotic protein kinome found that approximately 45% of the kinome is capable of autophosphorylation at the activation loop [38]. However, the extent and function of autophosphorylation at sites outside the activation loop is less extensively characterized.
Prior studies have established several mechanisms of kinase autophosphorylation, which can be described by three main criteria: 1) cis or trans autophosphorylation, referring to whether the reaction occurs intramolecularly or intermolecularly, respectively, 2) dimerization-dependent or -independent autophosphorylation, and 3) symmetric or asymmetric dimerization, indicating whether one or both molecules in a dimer becomes phosphorylated [38]. For example, a prominent mechanism of kinase activation involves symmetrical trans-autophosphorylation, as is the case with Chk2, where swapping of activation loop segments between dimerized kinases was determined structurally [40]. In contrast, EGF receptor family kinases engage in ligand-induced dimerization and asymmetrical trans-autophosphorylation, in which a head-to-head dimer containing an “activator” kinase phosphorylates and activates a “receiver” kinase by phosphorylating multiple Tyr residues on its C-tail [41]. Dimerization can also induce kinase autophosphorylation in cis; that is, dimerization stabilizes a prone-to-autophosphorylate state that facilitates an intramolecular autophosphorylation reaction. Allosteric activation of kinases in this manner is the basis for pseudokinases to participate in autophosphorylation reactions despite possessing no inherent catalytic activity towards substrates [42]. Kinase autophosphorylation can also be promoted allosterically by non-kinase proteins, as is the case for TPX2 activation of Aurora A trans-autophosphorylation at the mitotic spindle [43]. Finally, kinases may cis-autophosphorylate as a monomer, exemplified by the Hsp90-dependent autophosphorylation of GSK3β [44].
Which of these autophosphorylation mechanisms does PKC utilize to phosphorylate its own hydrophobic motif? It was previously determined that in vitro dephosphorylated PKC could re-autophosphorylate at the hydrophobic motif in a concentration-independent manner, indicative of dimerization-independent cis-autophosphorylation [31]. However, this mechanism was deduced from mature PKC, which had already undergone processing by mTORC2 and PDK1, and was subsequently dephosphorylated at the hydrophobic motif. The recent finding that newly-synthesized PKC is a symmetrical PKC homodimer, dissociated upon mTORC2 phosphorylation of the TIM, adds a new twist to the tale: PKC displays a unique autophosphorylation mechanism in which dimerization inhibits, rather than facilitates autophosphorylation. Dissociation of the dimer by mTORC2 exposes the PDK1 binding site in the C-tail, allowing phosphorylation of the activation loop, which triggers dimerization-independent cis-autophosphorylation. PKC hydrophobic motif (but not turn) phosphorylation is additionally dependent on Hsp90 binding of a PXXP motif in the C-terminal tail, suggesting that once the dimer is dissociated, Hsp90 protects PKC from down-regulation until it is fully phosphorylated [45].
If the hydrophobic motif is a conserved autophosphorylation site amongst AGC kinases, one might question how such a highly conserved motif can be phosphorylated by many AGC kinases with diverse substrate sequence preferences. Indeed, the PKC hydrophobic motif lacks the positively charged Arg or Lys residues preceding the phosphorylation site at the P-4 through P-2 positions in the PKC substrate consensus sequence [46]. Neither does the Akt hydrophobic possess the highly selected Arg at the P-3 position of its substrates [47]. However, sequence motifs are only one determinant by which kinases select their substrates, secondary to prior recruitment of the substrate and proper docking in an orientation amenable to phospho-transfer [48]. It is quite common that kinase autophosphorylation sites differ substantially from their sequence preferences [38]. Underscoring the degenerate nature of autophosphorylation sites, the Ser/Thr kinase HIPK2 autophosphorylates in cis at adjacent Tyr and Ser residues in its activation loop [49]. Thus, the strict sequences preferences of Ser/Thr vs. Tyr kinases can be broken in the case of cis-autophosphorylation. One explanation of this is that the prone-to-autophosphorylate state displays a different consensus motif than the fully activated form. This is certainly the case for DYRK kinases, which cis-autophosphorylate at the activation loop as a translational intermediate form in a one-time initiation event [50]. This is reminiscent of the nascent PKC protein that briefly exists as a dimer prior to an mTORC2 maturation event that yields a primed monomeric species. Another explanation, especially in the case of cis-autophosphorylation, is that the high effective concentration of the autophosphorylation site due to proximity on the same polypeptide increases the probability of a one-time phosphorylation event occurring if the reaction is sterically feasible. Thus, it may be that the ability to autophosphorylate is largely sequence-independent. In support of this, PKC autophosphorylates so rapidly during maturation that a partially phosphorylated species containing turn motif and activation loop phosphorylation, but not hydrophobic motif phosphorylation, cannot be isolated biochemically [51]. Instead, the first and rate-limiting step of PKC processing is mTORC2 phosphorylation of the TIM which dissociates the inactive dimer. Elucidating the mechanisms by which kinases autophosphorylate will contribute to the identification of new sites and functions, as well as the development of strategies to target these various kinase states and activation mechanisms.
Hydrophobic Motif
During the maturation of newly-synthesized PKC, phosphorylation of the hydrophobic motif performs two important functions by 1) rendering the kinase catalytically competent, and 2) stabilizing PKC in the autoinhibited state. Thus, while hydrophobic motif phosphorylation confers activity, it simultaneously induces conformational changes that produce an autoinhibited state that is inactive but protected from degradation [28]. This process ensures that all catalytically competent PKCs are autoinhibited and stabilized, but also that all PKCs which do not become autoinhibited are degraded. In this way, mature PKC signals only in the presence of second-messengers.
Hydrophobic motif phosphorylation activates AGC kinases by binding to a conserved PDK1-interacting-fragment (PIF) pocket on the N-lobe of the kinases, which works synergistically with the activation loop phosphorylation to produce an active conformation [52]. The role of the C-tail in activating AGC kinases is elegantly illustrated by the master activation loop kinase, PDK1, which is an AGC kinase lacking the portion of the C-tail that contains the TIM, turn, and hydrophobic motif sites [19]. To become activated and phosphorylate the activation loop, PDK1 must dock onto the hydrophobic motif sequence of its substrate kinase via its own PIF pocket. Thus, the unique C-tail of AGC kinases may have co-evolved with the kinase domain residues on the interaction surface, such that it could recruit the upstream activation loop kinase by the same mechanism that promotes its own activation. In the case of PKC, hydrophobic motif phosphorylation also induces conformational changes outside of the kinase domain. When the phosphorylated hydrophobic motif binds the PIF pocket [19], PKC rapidly autoinhibits, with the ligand-binding regulatory domains packing against the kinase domain and the pseudosubstrate segment occluding the active site. This autoinhibited or “closed” conformation protects PKC from degradation by masking proteolytically labile regions in the linker region between the catalytic and regulatory domains [53]. Autoinhibition furthermore masks the C-tail sites from phosphatases to protect the enzyme from dephosphorylation and subsequent down-regulation [22]. While the TIM phosphorylation initially dissociates the dimer during PKC processing, the adjacent turn motif (also phosphorylated by mTORC2) stabilizes conformational changes conferred by activation loop phosphorylation. Indeed, the activation loop phosphorylation of PKC is dispensable once the kinase is fully phosphorylated and matured. If only the hydrophobic motif is dephosphorylated, PKC retains activity and can independently re-autophosphorylate [31, 54]. However, if all sites are dephosphorylated, PKC cannot re-autophosphorylate and requires PDK1 to re-phosphorylate the activation loop before it can re-autophosphorylate at the hydrophobic motif and regain activity [22, 29, 54].
The autoinhibited conformation relies upon both autophosphorylation of the hydrophobic motif and the autoinhibitory pseudosubstrate. Mutation of the hydrophobic motif, residues in the kinase domain that disrupt catalysis, or the positively-charged residues in the pseudosubstrate result in an “open” and unstable PKC conformation [28]. While mutation of the pseudosubstrate itself does not impair hydrophobic motif autophosphorylation, impaired autoinhibition renders PKC sensitive to quality control degradation mediated by the PH domain leucine repeat protein phosphatase PHLPP [28]. PHLPP detects the open conformation of PKC and dephosphorylates the hydrophobic motif, priming PKC for further dephosphorylation and proteasomal degradation [28, 55]. Whether transient activation of PKC results in dephosphorylation of the hydrophobic motif followed by rapid re-autophosphorylation (before re-autoinhibition) remains to be elucidated, but prolonged activation results in dephosphorylation and degradation.
Taken together, the PKC C-tail acts as a multifunctional scaffold that not only activates the kinase when phosphorylated, but also recruits processing adaptors such as mTORC2, Hsp90, and PDK1, as well as recruiting quality control degradation machinery such as PHLPP. The PKC dimer also relies upon the C-tail, with the dimerization interface comprising the TIM helix in partner kinases, masking the C-tail from PDK1. Thus, PKC first needs to be phosphorylated by mTORC2 at the TIM to relieve dimerization and allow recruitment of PDK1 which phosphorylates the activation loop to promote autophosphorylation of the hydrophobic motif. Following the priming phosphorylation cascade, aberrantly active PKC that does not become autoinhibited presents an exposed C-tail, which can be dephosphorylated by PHLPP and targeted for degradation.
Concluding remarks
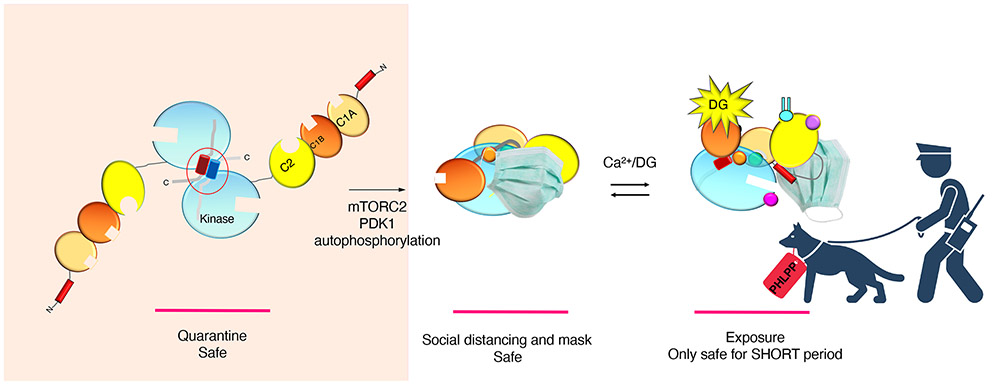
Why does PKC undergo such complex regulation by phosphorylation? One possibility is the newly-synthesized enzyme is kept in a safe ‘quarantine’ zone by forming a homodimer mediated by the TIM segment (Figure 4). This would protect the unphosphorylated enzyme from degradation until PDK1 is available to initiate activation loop phosphorylation, in turn allowing the stability-inducing hydrophobic motif phosphorylation. Thus, binding of mTORC2 would essentially release PKC from the safe quarantine zone by promoting the chain of phosphorylation events that ultimately result in a socially-distanced monomer that adopts the autoinhibited (masked) conformation and is now stable. Transient activation does not threaten the enzyme, but prolonged activation as occurs with phorbol esters results in dephosphorylation of the hydrophobic motif by PHLPP, preventing the pseudosubstrate from engaging in the active site (mask cannot go back on), and resulting in the ultimate degradation of PKC. Thus, mTORC2 serves to release newly-synthesized PKC from quarantine. Whether mTORC2 generally functions to release its client kinases from a stabilizing dimer following their biosynthesis is an open question (see Outstanding Questions).
OUTSTANDING QUESTIONS.
Is head-to-head dimerization a general mechanism to protect nascent kinases from degradation until appropriate phosphorylation/conformational changes take place?
Does mTORC2-catalyzed phosphorylation of the TIM serve as a general mechanism to facilitate activation loop phosphorylation?
What is the role of the TIM on kinases whose phosphorylation is agonist-evoked, such as Akt?
Is disruption of the TIM dimer interface a potential therapeutic strategy to promote the processing of PKC and increase its steady-state levels?
Is there an as yet-unidentified kinase, and potentially unidentified phosphorylation site, that promotes the processing of PKC isozymes that are insensitive to mTORC2 and lacking the TIM?
Figure 4. Model of PKC Release from Quarantine.
Newly-synthesized PKC exists an unphosphorylated homodimer that is transiently protected from degradation (quarantine safe). Upon phosphorylation by mTORC2, PDK1, and autophosphorylation, PKC emerges from quarantine as a monomer that is now protected from degradation by its pseudosubstrate “mask,” which retains PKC in the phosphatase-resistant autoinhibited conformation. Upon activation by second messengers, the pseudosubstrate “mask” is transiently removed to allow PKC phosphorylation of substrates in short bursts of activity. PHLPP polices for chronically-active PKC (as would occur in the presence of phorbol esters, or as a result of mutations that impair autoinhibition) and removes the hydrophobic motif phosphate, ultimately resulting in the degradation of the unmasked PKC.
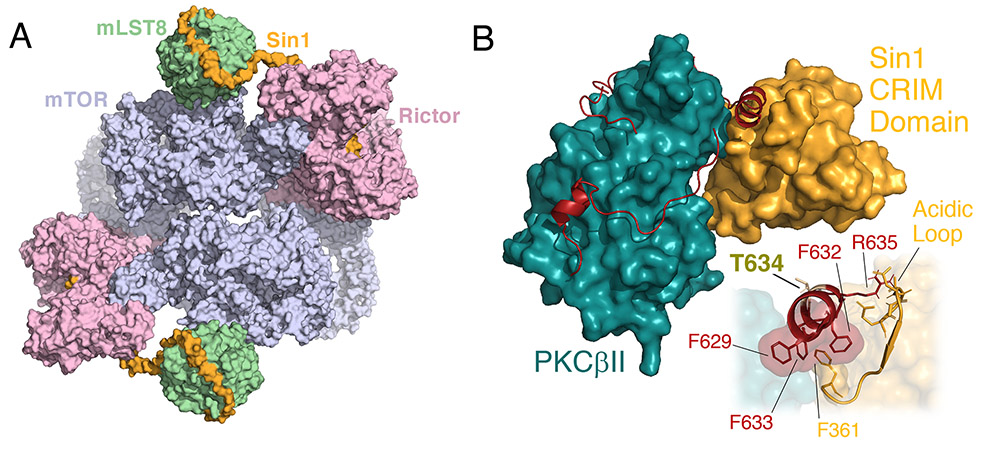
Figure II. Architecture of mTORC2.
A) Cryo-EM structure (PDB ID: 6ZWM) of the mTORC2 homodimer showing the mTOR kinase (lavender), Rictor (pink), mLST8 (green), and Sin1 (gold). Note: the Sin1 CRIM domain, which recruits mTORC2 to its substrates, is not resolved [75]. B) Docking of the Sin1 CRIM domain (gold; nuclear magnetic resonance structure, PDB ID: 2RVK) to the PKCβII catalytic domain (teal with C-tail in red; X-ray structure, PDB ID: 2I0E). Inset: Interactions of the CRIM domain acidic loop (gold) with the PKCβII TIM helix (red) are shown. The TOR-Interaction Motif (TIM) phosphorylation site (Thr634) is indicated as well as key residues in the binding interface.
Figure III. Dysregulation of PKC Signaling in Disease.
Deregulation of PKC results in pathophysiologies, with gain-of-function (GOF) generally associated with degenerative diseases and loss-of-function (LOF) with cancer.
Acknowledgements:
We thank Alexandr Kornev, Susan Taylor, and members of the Newton lab for helpful discussions. This work was supported by NIH R35 GM122523.
Abbreviations:
- GOF
gain-of-function
- LOF
loss-of-function
- mTORC2
mechanistic target of rapamycin complex 2
- PDK1
phosphoinositide-dependent kinase 1
- PHLPP
PH domain leucine rich repeat protein phosphatase
- PKC
protein kinase C
Glossary
- Activation loop
Flexible segment near the entrance to the active site that regulates activity, often in a phosphorylation-dependent manner. The activation loop begins with a conserved DFG motif and extends 20-30 residues to a conserved APE motif. Phosphorylation of this segment is critical for orientation of the DFG Asp that coordinates Mg2+ to facilitate ATP binding, as well as proper alignment of a separate HRD motif in which the Asp acts a catalytic base in the phosphorylation reaction.
- Down-regulation
Reduction in steady-state levels of a protein that can result from decreased transcription, destabilizing mutations, or altered post-translational modification leading to increased protein degradation. With reference to PKC, down-regulation signifies the loss of protein following prolonged treatment with activators such as phorbol esters.
- Gain-of-function (GOF) mutation
A mutation that results in increased function of a protein or the gain of a new molecular activity. These mutations are generally missense mutations that eliminate mechanisms of inhibition resulting in constitutive activation of the protein.
- Germline mutation
Inherited mutation that occurs in germ cells and is present in a majority of cells in the human body.
- Hydrophobic motif
Conserved phosphorylation site on the C-tail of AGC kinases such as PKC and Akt that is flanked by hydrophobic residues.
- Loss-of-function (LOF) mutation
A mutation that results in decreased protein expression or compromised protein function. LOF mutations can be missense mutations that result in a change in a single amino acid but are more commonly indels (insertions or deletions) or nonsense mutations that introduce a stop codon early in the gene.
- PDK1
An AGC family member composed of an N-terminal kinase domain and C-terminal PH domain that phosphorylates the activation loop of other members of this family, such as PKA, PKC and Akt, to structure the active site for catalysis.
- Somatic mutation
A mutation acquired in a cell from damage to DNA that is then passed on through cell division.
- TOR Interaction motif (TIM)
Newly-identified sequence (F-X3-F-pT) preceding the turn motif that is present on all known AGC kinases regulated by mTOR, where the Thr is directly phosphorylated by mTOR.
- Turn motif
Conserved phosphorylation site on the C-tail of AGC kinases such as PKC and Akt.
- Oncoprotein
The protein product of an oncogene, a gene that harbors frequent gain-of-function mutations or has increased expression in cancer leading to acquisition of phenotypes associated with cancer, such as increased proliferation or survival.
- Tumor suppressor
A gene that harbors frequent loss-of-function mutations or deletions in cancer; it generally acts to suppress proliferation and growth or promote cell death under certain physiological conditions.
Footnotes
None of the authors have any competing interests.
Gan et al. reported that PRR5L degradation promotes a mTORC2-dependent increase in the hydrophobic motif phosphorylation of PKCδ [73], but this may arise from the reported PRR5L suppression of the activity of the hydrophobic motif phosphatase PHLPP [74]; PKCδ hydrophobic motif is unaffected by Sin1 or rictor knockdown in MEFs [67]
References:
- 1.Fischer EH and Krebs EG (1955) Conversion Of Phosphorylase b to Phosphorylase a In Muscle Extracts. J. Biol. Chem 216, 121–132. [PubMed] [Google Scholar]
- 2.Hunter T (1995) Protein Kinases and Phosphatases: The Yin and Yang of Protein Phosphorylation and Signaling. Cell 80, 225–236. [DOI] [PubMed] [Google Scholar]
- 3.Humphrey SJ et al. (2015) Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation. Trends Endocrinol Metab 26 (12), 676–687. [DOI] [PubMed] [Google Scholar]
- 4.Taylor SS et al. (2012) Evolution of the eukaryotic protein kinases as dynamic molecular switches. Philos Trans R Soc Lond B Biol Sci 367 (1602), 2517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor SS et al. (2021) From structure to the dynamic regulation of a molecular switch: A journey over 3 decades. J Biol Chem 296, 100746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson LN et al. (1996) Active and Inactive Protein Kinases: Structural Basis for Regulation. Cell 85, 149–158. [DOI] [PubMed] [Google Scholar]
- 7.Pearlman SM et al. (2011) A mechanism for the evolution of phosphorylation sites. Cell 147 (4), 934–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton AC (2018) Protein kinase C: perfectly balanced. Crit Rev Biochem Mol Biol 53 (2), 208–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baffi TR et al. (2021) mTORC2 controls the activity of PKC and Akt by phosphorylating a conserved TOR interaction motif. Sci Signal 14 (678). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornev AP et al. (2006) Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A 103 (47), 17783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cyphers S et al. (2017) A water-mediated allosteric network governs activation of Aurora kinase A. Nat Chem Biol 13 (4), 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruff EF et al. (2018) A dynamic mechanism for allosteric activation of Aurora kinase A by activation loop phosphorylation. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilburt JAH et al. (2017) Dynamic Equilibrium of the Aurora A Kinase Activation Loop Revealed by Single-Molecule Spectroscopy. Angew Chem Int Ed Engl 56 (38), 11409–11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majumdar A et al. (2021) Allostery governs Cdk2 activation and differential recognition of CDK inhibitors. Nat Chem Biol 17 (4), 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon A et al. (2019) Tracing the origin and evolution of pseudokinases across the tree of life. Sci Signal 12 (578). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrestha S et al. (2020) Cataloguing the dead: breathing new life into pseudokinase research. Febs j 287 (19), 4150–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang LC et al. (2018) Integrative annotation and knowledge discovery of kinase post-translational modifications and cancer-associated mutations through federated protein ontologies and resources. Sci Rep 8 (1), 6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudson AM et al. (2018) Truncation- and motif-based pan-cancer analysis reveals tumor-suppressing kinases. Sci Signal 11 (526). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mora A et al. (2004) PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol 15 (2), 161–70. [DOI] [PubMed] [Google Scholar]
- 20.Hein MY et al. (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163 (3), 712–23. [DOI] [PubMed] [Google Scholar]
- 21.Kannan N et al. (2007) The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci U S A 104 (4), 1272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dutil EM et al. (1994) In vivo regulation of protein kinase C by trans-phosphorylation followed by autophosphorylation. J Biol Chem 269 (47), 29359–29362. [PubMed] [Google Scholar]
- 23.Guertin DA et al. (2006) Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 11 (6), 859–71. [DOI] [PubMed] [Google Scholar]
- 24.Frias MA et al. (2006) mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol 16 (18), 1865–70. [DOI] [PubMed] [Google Scholar]
- 25.Jacinto E et al. (2006) SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127 (1), 125–37. [DOI] [PubMed] [Google Scholar]
- 26.Liu P et al. (2014) Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature 508 (7497), 541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh WJ et al. (2010) mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. Embo J 29 (23), 3939–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baffi TR et al. (2019) Protein Kinase C Quality Control by Phosphatase PHLPP1 Unveils Loss-of-Function Mechanism in Cancer. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutil EM et al. (1998) Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1) [In Process Citation]. Curr Biol 8 (25), 1366–1375. [DOI] [PubMed] [Google Scholar]
- 30.Mellor H and Parker PJ (1998) The extended protein kinase C superfamily. Biochem J 332 (Pt 2), 281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behn-Krappa A and Newton AC (1999) The hydrophobic phosphorylation motif of conventional protein kinase C is regulated by autophosphorylation. Curr Biol 9 (14), 728–737. [DOI] [PubMed] [Google Scholar]
- 32.Toker A and Newton AC (2000) Akt/protein kinase B is regulated by autophosphorylation at the hypothetical PDK-2 site. J Biol Chem 275 (12), 8271–8274. [DOI] [PubMed] [Google Scholar]
- 33.Grodsky N et al. (2006) Structure of the catalytic domain of human protein kinase C beta II complexed with a bisindolylmaleimide inhibitor. Biochemistry 45 (47), 13970–81. [DOI] [PubMed] [Google Scholar]
- 34.Ziemba BP et al. (2014) Single-molecule studies reveal a hidden key step in the activation mechanism of membrane-bound protein kinase C-alpha. Biochemistry 53 (10), 1697–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pataki E et al. (2020) TOR Complex 2- independent mutations in the regulatory PIF pocket of Gad8AKT1/SGK1 define separate branches of the stress response mechanisms in fission yeast. PLoS Genet 16 (11), e1009196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali SM and Sabatini DM (2005) Structure of S6 kinase 1 determines whether raptor-mTOR or rictor-mTOR phosphorylates its hydrophobic motif site. J Biol Chem 280 (20), 19445–8. [DOI] [PubMed] [Google Scholar]
- 37.Lochhead PA (2009) Protein kinase activation loop autophosphorylation in cis: overcoming a Catch-22 situation. Sci Signal 2 (54), pe4. [DOI] [PubMed] [Google Scholar]
- 38.Beenstock J et al. (2016) How Do Protein Kinases Take a Selfie (Autophosphorylate)? Trends Biochem Sci 41 (11), 938–953. [DOI] [PubMed] [Google Scholar]
- 39.Mitrophanov AY and Groisman EA (2008) Signal integration in bacterial two-component regulatory systems. Genes Dev 22 (19), 2601–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai Z et al. (2009) Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol Cell 35 (6), 818–29. [DOI] [PubMed] [Google Scholar]
- 41.Lemmon MA et al. (2014) The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb Perspect Biol 6 (4), a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byrne DP et al. (2017) Pseudokinases: update on their functions and evaluation as new drug targets. Future Med Chem 9 (2), 245–265. [DOI] [PubMed] [Google Scholar]
- 43.Eyers PA et al. (2003) A novel mechanism for activation of the protein kinase Aurora A. Curr Biol 13 (8), 691–7. [DOI] [PubMed] [Google Scholar]
- 44.Lochhead PA et al. (2006) A chaperone-dependent GSK3beta transitional intermediate mediates activation-loop autophosphorylation. Mol Cell 24 (4), 627–33. [DOI] [PubMed] [Google Scholar]
- 45.Gould CM et al. (2009) The chaperones Hsp90 and Cdc37 mediate the maturation and stabilization of protein kinase C through a conserved PXXP motif in the C-terminal tail. J Biol Chem 284 (8), 4921–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barber KW et al. (2018) Kinase Substrate Profiling Using a Proteome-wide Serine-Oriented Human Peptide Library. Biochemistry 57 (31), 4717–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hutti JE et al. (2004) A rapid method for determining protein kinase phosphorylation specificity. Nat Methods 1 (1), 27–9. [DOI] [PubMed] [Google Scholar]
- 48.Miller CJ and Turk BE (2018) Homing in: Mechanisms of Substrate Targeting by Protein Kinases. Trends Biochem Sci 43 (5), 380–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saul VV et al. (2013) HIPK2 kinase activity depends on cis-autophosphorylation of its activation loop. J Mol Cell Biol 5 (1), 27–38. [DOI] [PubMed] [Google Scholar]
- 50.Lochhead PA et al. (2005) Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 121 (6), 925–36. [DOI] [PubMed] [Google Scholar]
- 51.Sonnenburg ED et al. (2001) The phosphoinositide-dependent kinase, PDK-1, phosphorylates conventional protein kinase C isozymes by a mechanism that is independent of phosphoinositide 3-kinase. J Biol Chem 276 (48), 45289–97. [DOI] [PubMed] [Google Scholar]
- 52.Biondi RM et al. (2000) Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. Embo J 19 (5), 979–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keranen LM and Newton AC (1997) Ca2+ differentially regulates conventional protein kinase Cs' membrane interaction and activation. J Biol Chem 272 (41), 25959–25967. [DOI] [PubMed] [Google Scholar]
- 54.Keranen LM et al. (1995) Protein kinase C is regulated in vivo by three functionally distinct phosphorylations. Curr Biol 5 (12), 1394–1403. [DOI] [PubMed] [Google Scholar]
- 55.Van AN et al. (2021) Protein kinase C fusion proteins are paradoxically loss-of-function in cancer. J Biol Chem, 100445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leonard TA et al. (2011) Crystal structure and allosteric activation of protein kinase C betaII. Cell 144 (1), 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones AC et al. (2020) Hypothesis: Unifying model of domain architecture for conventional and novel protein kinase C isozymes. IUBMB Life. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sommese RF et al. (2017) The Role of Regulatory Domains in Maintaining Autoinhibition in the Multidomain Kinase PKCalpha. J Biol Chem 292 (7), 2873–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Evans JH et al. (2006) Specific translocation of protein kinase Calpha to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Mol Biol Cell 17 (1), 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tobias IS and Newton AC (2016) Protein scaffolds control localized protein kinase C zeta Activity. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Graybill C et al. (2012) Partitioning-defective protein 6 (Par-6) activates atypical protein kinase C (aPKC) by pseudosubstrate displacement. J Biol Chem 287 (25), 21003–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu GY and Sabatini DM (2020) mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21 (4), 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu W and Hall MN (2020) Regulation of mTORC2 Signaling. Genes (Basel) 11 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Q et al. (2006) Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev 20 (20), 2820–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacinto E et al. (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6 (11), 1122–8. [DOI] [PubMed] [Google Scholar]
- 66.Sarbassov DD et al. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307 (5712), 1098–101. [DOI] [PubMed] [Google Scholar]
- 67.Ikenoue T et al. (2008) Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. Embo J 27 (14), 1919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Facchinetti V et al. (2008) The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. Embo J 27 (14), 1932–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cameron AJ et al. (2011) mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem J 439 (2), 287–97. [DOI] [PubMed] [Google Scholar]
- 70.Tatebe H et al. (2017) Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tovell H and Newton AC (2021) PHLPPing the balance: restoration of protein kinase C in cancer. Biochem J 478 (2), 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lorden G and Newton AC (2021) Conventional protein kinase C in the brain: repurposing cancer drugs for neurodegenerative treatment? Neuronal Signaling 5 (4), NS20210036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gan X et al. (2012) PRR5L degradation promotes mTORC2-mediated PKC-delta phosphorylation and cell migration downstream of Galpha12. Nat Cell Biol 14 (7), 686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Charbonnier LM et al. (2019) Functional reprogramming of regulatory T cells in the absence of Foxp3. Nat Immunol 20 (9), 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scaiola A et al. (2020) The 3.2-A resolution structure of human mTORC2. Sci Adv 6 (45). [DOI] [PMC free article] [PubMed] [Google Scholar]