Abstract
Cell migration is a critical contributor to metastasis. Cytokine production and its role in cancer cell migration have been traditionally associated with immune cells. We find that the histone methyltransferase Mixed-Lineage Leukemia 1 (MLL1) controls 3D cell migration via cytokines, IL-6, IL-8, and TGF-β1, secreted by the cancer cells themselves. MLL1, with its scaffold protein Menin, controls actin filament assembly via the IL-6/8/pSTAT3/Arp3 axis and myosin contractility via the TGF-β1/Gli2/ROCK1/2/pMLC2 axis, which together regulate dynamic protrusion generation and 3D cell migration. MLL1 also regulates cell proliferation via mitosis-based and cell cycle–related pathways. Mice bearing orthotopic MLL1-depleted tumors exhibit decreased lung metastatic burden and longer survival. MLL1 depletion leads to lower metastatic burden even when controlling for the difference in primary tumor growth rates. Combining MLL1-Menin inhibitor with paclitaxel abrogates tumor growth and metastasis, including preexistent metastasis. These results establish MLL1 as a potent regulator of cell migration and highlight the potential of targeting MLL1 in patients with metastatic disease.
MLL1 regulates cytokine-driven cell migration and metastasis.
INTRODUCTION
Cell migration, particularly in three-dimensional (3D) environments, is a critical contributor to the metastatic spread of cancer cells from a primary tumor to distant sites (1). Despite metastatic disease causing the vast majority of cancer-related deaths, targeting metastasis remains challenging clinically (2). Cancer cell migration can be triggered and maintained by elevated levels of cytokines, thus playing a vital role in cancer metastasis (3–22). In solid tumors, these cytokines are presumed to be secreted by immune cells in the tumor microenvironment, although recent studies have shown that cancer cells themselves can secrete these cytokines (3, 4, 16, 19, 23–27). While the signaling cascades that regulate cell migration have been studied extensively, the epigenetic regulation of cell migration is still poorly understood (28). Despite identifying some epigenetic regulators of cancer cell migration, there is currently no known epigenetic regulator of cytokine-driven cell migration (fig. S1A). This is clinically important as inhibiting epigenetic drivers of cancer cell migration present potential targets and treatment venues targeting metastasis specifically. An epigenetic factor that is strongly correlated with cell migration–related genes in The Cancer Genome Atlas (TCGA) database is mixed-lineage leukemia 1 (MLL1 or MLL, gene name KMT2A). MLL1 is a histone methyltransferase, which, along with its scaffold protein Menin (29), is integral to methylation at the histone 3 lysine 4 (H3K4) site, leading to activation of transcription (30). The MLL1-Menin interaction has been shown to drive expression of several tumor-promoting genes (31). Notably, misregulation of H3K4 methylation has been associated with aberrant gene expression in breast cancer (32).
Here, we show that the MLL1-Menin interaction is essential for the migration of cancer cells and that the disruption of this interaction impairs cell migration and metastasis. In migration through 3D in vitro systems (hereby referred to as 3D cell migration), MLL1 depletion decreases migration by reducing protrusion generation via down-regulation of the cytokines, interleukins IL-6 and IL-8, and downstream proteins that activate actin filament assembly via an IL-6/8/signal transducers and activators of transcription 3 (STAT3)/actin-related protein 2/3 (Arp2/3) pathway. Pharmacological inhibitors permit finely tuning the inhibition of MLL1-Menin interaction and reveal a nonlinear mechanism of regulation of cell migration. A deep inhibition of the MLL1-Menin interaction also leads to reduced myosin II–based contractility via a transforming growth factor–β1 (TGF-β1)/GLI2/ROCK1/2 pathway, which is not present at lower drug dosage. These studies reveal that MLL1 plays a two-pronged role in cell migration by controlling both actin filament assembly and myosin-based contractility. Our in vivo studies support these mechanisms of cell migration and reveal that mice bearing orthotopic breast MLL1-depleted tumors grow substantially more slowly and induce lower metastatic burden than control tumors, leading to a longer survival. Reduced lung metastatic burden was observed even after accounting for the difference in the growth rate of primary tumors. In a syngeneic orthotopic mouse triple negative breast cancer (TNBC) model, concurrent administration of the MLL1-Menin inhibitor and paclitaxel led to a near-arrest of tumor growth and the extremely low metastatic burden. Last, the combination of MLL1-Menin inhibitor plus paclitaxel was also effective in reducing preexistent metastatic burden in tail vein model of syngeneic TNBC metastasis.
Overall, our studies show that MLL1 is an epigenetic regulator of 3D cancer cell migration, which exerts its effects by controlling the production of the key cytokines IL-6, IL-8, and TGF-β1. Our in vivo studies demonstrate a potential combination anticancer therapy that can reduce metastatic burden via two separate molecular mechanisms.
RESULTS
TCGA analysis suggests that MLL1 is an epigenetic regulator of cell migration
TNBC represents 15 to 20% of all breast cancer cases (33) and is an aggressive subtype with a poor prognosis (34) and a high incidence of metastases (35). Patients with TNBC relapse more frequently than other breast cancer subtypes (35), and despite administration of optimal chemotherapy regimens, only 30% of patients with metastatic TNBC show >5-year survival (36), highlighting the inadequacy of current standard-of-care treatments. In addition, targeting metastasis is a formidable challenge, and there is a dearth of antimetastatic therapies in the clinic (37). Thus, TNBC was chosen as the key focus for this study due to their propensity for metastatic spread. To identify potential epigenetic regulators of cell migration in TNBC, we calculated correlation coefficients of cell migration–related genes (ROCK1, ROCK2, LIMK1, RAC1, RHOA, CDC42, MYLK, WASF3, ACTR2, and ACTR3) from the Pan-Cancer Atlas (PanCanAtlas; TCGA) basal breast cancer dataset for 715 genes that encode for epigenetic factors (38). These epigenetic genes were then ranked on the basis of the summation of correlation coefficients (Fig. 1A), and the top 10 epigenetic factors were examined as potential candidates (fig. S1B). Only KMT2A (MLL1) was druggable and was the subject of cancer clinical trials, albeit these trials are in acute myeloid leukemia and are neither in solid tumors nor focused on metastatic prevention (fig. S1B). In addition, MLL1 was ranked higher than previously reported epigenetic factors of cell migration in fig. S1A (fig. S1C). Together, these findings indicated that MLL1 was a potential regulator of motility in TNBC cells and had high translational potential owing to ongoing clinical trials.
Fig. 1. MLL1-Menin interaction regulates cancer cell migration, proliferation, and tumor progression.
(A) Epigenetic factors in the TCGA basal breast cancer (BRCA) were queried for correlation with cell migration genes. MLL1 was among the best correlated epigenetic factors. (B) Cells embedded in 3D collagen gels were tracked overnight using live-cell phase-contrast microscopy. Resulting videos were analyzed to obtain cell trajectories, velocity, and diffusivity using the anisotropic persistent random walk model (APRW) cell migration model. (C) shMLL1 cells exhibited shorter trajectories than scrambled nontargeting shRNA control and WT cells. (D) shMLL1 cells had lower velocity, compared to control cells. Each dot in the box-and-whisker plot corresponds to one cell. The limits of the box represent the 75th and 25th percentiles, and the center line represents the median. The whiskers stretch from the 1st to the 99th percentile. (E) Pharmacological inhibition of MLL1-Menin interaction inhibits cell migration. (F) MLL1 depletion decreases cell proliferation. (G) shMLL1 tumors grew slower compared to scrambled control tumors. (H) Mice bearing shMLL1 tumors survive longer than control tumors [median survival of 89 days versus 64 days, respectively, and hazard ratio (HR) of 0.20]. (I) H&E staining of lungs show lighter staining for mice bearing shMLL1tumors, indicative of lower metastatic burden compared to scrambled control tumors. (J) Quantification of total metastatic burden per lung showed decreased metastatic burden in mice bearing shMLL1 tumors. (K) Histogram of lesion size of representative lungs shows that decreased metastatic burden in shMLL1 lungs was due to reduced number of lesions and decreased lesion size. (L) RNA sequencing (RNA-seq) of MLL1-Menin–inhibited cells revealed down-regulation of cell migration and metastasis-related pathways including the IL-6–JAK–STAT3 and TGF-β signaling pathways. ES, enrichment score. Data in this figure were generated with MDA-MB-231 cells in vivo [(G) to (K)] or embedded in 3D collagen gels [(B) to (F) and (L)] except (A) (TCGA). (B) and (K) were created with BioRender. **P < 0.01, ***P < 0.001, and ****P < 0.0001. NS, not significant.
In breast tumors, MLL1 (KMT2A) is expressed at the RNA level (fig. S1D), and 92% of the patients show medium-to-high MLL1 expression in breast cancer tissues (fig. S1E). MLL1 (KMT2A) had little overall prognostic value in overall breast cancer survival (fig. S1F) or in late-stage breast cancers, i.e., cancers that have invaded and spread to nearby lymph nodes or distant organs. However, MLL1 (KMT2A) was prognostic in earlier-stage breast cancer, i.e., cancers that have yet to spread from the original tumor site. Key motility genes ROCK1 and ROCK2 showed similar prognostic trends (fig. S1G), as did breast cancer metastasis suppressor BRMS1 (fig. S1H; although earlier stages show better prognosis with higher BRMS1 expression as BRMS1 expression suppresses metastasis). Gene set enrichment analysis (GSEA) showed that MLL1 expression was positively correlated with breast cancer metastasis and prognosis gene sets (fig. S2, A and B). Last, the expression of MLL1 in two metastatic TNBC cell lines (MDA-MB-231 and SUM-159) was compared against nontumorigenic breast epithelial cells (MCF-10A). The metastatic TNBC cells expressed higher levels of MLL1 than normal breast epithelial cells (fig. S2C). In sum, MLL1 is prognostic in breast cancers that have yet to metastasize, and its expression is higher in breast carcinoma cells compared to noncancer breast epithelial cells.
MLL1-Menin interaction regulates 3D cell migration and tumor progression
MLL1 was depleted in human TNBC (MDA-MB-231) cells via short hairpin RNA (shRNA) (fig. S2, D and E). Cells transduced with nontargeting scrambled control shRNA (scr. cont.), MLL1-depleted (shMLL1) cells, and wild-type (WT) cells were embedded in 3D collagen matrices, which mimic the 3D collagen–rich environment of the stromal matrix (39). Embedded cells were subjected to overnight live-cell tracking (Fig. 1B), and migration-related parameters were computed using a custom code (40). Live-cell microscopy permits the tracking of individual cells, fully dissociating any influence of proliferation on migration. Scr. cont. cells and WT cells were highly motile, as evident by longer trajectories (Fig. 1C), whereas all three MLL1 knockdowns were substantially less motile, indicating that MLL1 knockdown impaired cell migration. These observations were reflected when extracting cell velocities from cell trajectories (Fig. 1D): Control cells moved twice as fast as shMLL1 cells. The persistence time of migration (a measure of how long a cell takes to change direction), persistence speed, and cell diffusivity (an intrinsic measure of motility that is independent of elapsed time) (41) displayed similar trends (fig. S2, F to H). In addition, Menin was depleted separately using shRNA (fig. S2I): Menin-depleted cells displayed a loss in migration and decreased velocity similar to those displayed by MLL1-depleted cells (fig. S2J).
MLL1 was chosen over other epigenetic candidates as there are drugs against it in clinical trials (fig. S1B). Inhibition of the MLL1-Menin interaction (also referred to interchangeably as MLL1-Menin inhibition or MLL1 inhibition) was achieved using the drug MI-3454 (42), which led to decrease in cell motility similar to that observed with shMLL1 cells (Fig. 1E). Another MLL1-Menin inhibitor, MI-2-2 (43), also reduced cell migration, as evidenced by reduced velocity (fig. S2, K and L), diffusivity (fig. S2M), and shortened cell trajectories (fig. S2N). These observations were validated in another 3D in vitro system: tumor spheroids (fig. S2O). These double-layered spheroids consist of a core of cancer cells in Matrigel surrounded by an outer collagen layer (44). This system allows for the analysis of (i) invasion through a basement membrane–like material (Matrigel) and (ii) migration through collagen I (in yellow). MLL1-Menin inhibition reduced the dissemination of cells from the inner core into the outer collagen-rich corona (invasive front traced in orange). These results were confirmed in four other TNBC cell lines: BT-549, SUM-149, SUM-159 (all human TNBC), and 4T1 (mouse TNBC) (fig. S2P), as well as in HT-1080 fibrosarcoma cells (fig. S3A). Thus, MLL1-Menin interaction controls cell migration in a wide panel of cell lines. Henceforth, unless stated otherwise, we will use MDA-MB-231 cells embedded in 3D collagen gels. Any other cell lines/models used will be specified in the results and figure captions. Key details of each figure panel is summarized in table S1.
MLL1 is a component of the complex of proteins associated with Set1 (COMPASS) complex that incorporates other transcription factors such as WDR5 (45). Disruption of MLL1-WDR5 interaction reduced cell velocity (fig. S3B) to levels observed with MLL1-Menin inhibition or MLL1 depletion. MLL1 has been implicated as a driver in a subset of leukemias, where MLL1-fusion proteins drive leukemogenesis (46). In MLL1-fusion leukemias, the H3K79-specific methyltransferase Dot1L, which is not part of COMPASS, is necessary for progression and maintenance in MLL1-fusion leukemias (47). Dot1L inhibitors such as EPZ-5676 have shown efficacy in MLL1-rearranged acute myeloid leukemia and are being investigated in a phase 1b/2 clinical trial (NCT03724084). Treatment of breast cancer cells with Dot1L inhibitor at multiple doses did not alter cell migration (fig. S3C). In addition, depletion or inhibition of MLL1 in TNBC cells did not appreciably decrease the expression of Hox genes reported to be downstream of MLL1 in MLL-fusion leukemias (HOXA7, HOXA9, HOXA10, HOXA11, MEIS1, and PBX3) (48, 49), whose down-regulation has been shown to be a hallmark of MLL1-Menin inhibition in MLL-fusion leukemias (fig. S3, D to H). Hence, the underlying biology of MLL1-Menin–based cell migration is different from that of leukemogenesis of MLL1-fusion–driven leukemias. MLL1-Menin–based regulation of cell migration was also independent of 3D cell density (20 to 250 cells/mm3; fig. S3I) and collagen density in 3D gels (collagen, 1 to 3 mg/ml; fig. S3J). In sum, MLL1’s interaction with COMPASS members WDR5 and Menin regulates 3D cell migration. The underlying mechanism of MLL1-based cell migration is distinct from MLL1’s role in MLL1-fusion leukemias.
In addition to regulating cell migration, MLL1-Menin interaction regulates proliferation. MLL1 depletion decreased proliferation by 60% (Fig. 1F). These in vitro observations were validated in orthotopic TNBC mouse models to assess the role of MLL1-Menin–mediated migration and proliferation on tumor progression (tumor growth and metastasis). Further validation is shown below in syngeneic mouse models. In vivo modeling demonstrated that MLL1-depleted tumors grew more slowly (Fig. 1G) and mice bearing shMLL1 tumors exhibited increased survival (Fig. 1H and fig. S3L). Mice euthanized at a set time point (fig. S3M) showed decreased lung metastatic nodules in shMLL1 tumor–bearing mice, evident in darker hematoxylin and eosin (H&E) staining in lungs (Fig. 1I). Quantification of total lung metastatic burden showed an eightfold decrease in metastatic burden for mice bearing shMLL1 tumors compared to mice bearing control tumors (Fig. 1J). The size distribution of metastatic lesions was also determined for lungs shown in Fig. 1I. Control-bearing lungs not only had many more metastatic lesions (Fig. 1K, bottom), but these lesions were much larger than shMLL1-bearing lungs (Fig. 1K; indicated by a thick histogram tail for control mice). Thus, MLL1 regulates cell migration and proliferation in vitro, as well as tumor growth and metastasis in vivo. These studies were purely mechanistic, with an emphasis on validation of the role of MLL1 in tumor progression. The translation of these findings to preclinical in vivo models (see “MLL1-Menin inhibitors combined with paclitaxel block cell migration and proliferation” section) will demonstrate abrogation of tumor growth and metastasis when combining MLL1 inhibition with standard-of-care treatment.
Last, we have done an exhaustive assessment of any potential off-target effects. Disruption of the MLL1-Menin interaction was confirmed using the other MLL1-Menin inhibitors, MI-503 (fig. S4, A and B) (48), and VTP50469 (fig. S4, C and D) (49), in addition to MI-3454 (fig. S4E) and MI-2-2, for a total of four different MLL1-Menin inhibitors. More potent inhibitors (MI-3454, VTP50469, and MI-503) permit disruption of the MLL1-Menin interaction at a lower dose, minimizing potential off-target effects. To test for potential off-target effects of MI-2-2, we used a weak inhibitor of MLL1-Menin interaction, MI-nc, as a negative control. MI-nc has a similar structure to MI-2-2 but binds much more weakly to MLL1-Menin interaction and does not inhibit the MLL1-Menin interaction (50). MI-nc treatment did not affect cell migration (fig. S4F), suggesting that inhibition of cell migration by the MLL1-Menin inhibitors was due to the disruption of their interaction rather than off-target effects. In noncancerous MCF10A breast epithelial cells (fig. S2C), MLL1 inhibition had a markedly diminished impact on cell migration (fig. S4G) and proliferation (fig. S4, H and I) compared to TNBC cells, hinting at a potentially weaker impact of MLL1-Menin inhibition on noncancerous cells compared to tumor cells. Reduced proliferation in TNBC cells following MLL1-Menin inhibition was not accompanied by an increase in apoptosis (fig. S4J), indicating that decreased cell numbers and reduced cell motility were not due to decreased cell viability. Inhibition of cell migration was also readily and fully reversible upon withdrawal of drug treatment. Cells were pretreated with MLL1 inhibitor for 2 days before being seeded in 3D gels and subjected to further MLL1 inhibition (posttreatment) (fig. S4K). Pretreated cells without any posttreatment showed a full recovery in cell velocity within 2 to 3 days and had the same motility as untreated cells (fig. S4L). Both pretreated and nonpretreated cells receiving posttreatment showed similarly low motility. Similar results were also observed in HT-1080 fibrosarcoma cells. MLL1-Menin inhibition also did not further reduce the motility of shMLL1 cells (fig. S4M), implying that MLL1-Menin inhibitors had no effect when MLL1 was absent in cells. Thus, it is highly unlikely that the observed results were due to off-target effects. Decreased cell migration via pharmacological inhibition was due to the disruption of MLL1-Menin interaction rather than off-target effects, and administration of MLL1-Menin inhibitors was associated with low toxicity (indicated by low apoptosis rates). Further discussion pertaining to the assessment of off-target effects can be found in Methods. In sum, MLL1-Menin interaction regulates the 3D migration of TNBC cells in vitro and regulates metastatic burden in vivo.
MLL1-Menin interaction regulates IL-6/8 production and cell protrusion generation
Transcriptomic analysis showed that MLL1-Menin inhibition led to changes in key migration-related gene sets (Fig. 1L). Major cytokine-based cell migration–related pathways such as IL-6/Janus kinase (JAK)/STAT3 signaling, TGF-β signaling, and tumor necrosis factor–α (TNF-α) signaling via nuclear factor κB (NF-κB) were down-regulated following lower dose of MLL1 inhibitor (10 μM MI-2-2). Higher dose (30 μM) led to generally a much deeper repression of these pathways, as well as of other metastasis-related pathways such as epithelial-mesenchymal transition, apical junction transition, hypoxia, and Wnt β-catenin signaling pathways.
To determine whether MLL1-menin interaction regulated cell migration via the secretion of soluble factors such as cytokines, we collected conditioned medium (CM) from scrambled control cells embedded in 3D collagen gels (Fig. 2A). This CM was then added to shMLL1 cells in collagen gels, followed by overnight live-cell tracking. Addition of CM to shMLL1 cells fully rescued the motility lost by MLL1 knockdown (trajectories in Fig. 2B and cell velocities in Fig. 2C). CM did not further increase the motility of scrambled control cells. CM from shMLL1 cells (with higher cell density to account for the slower proliferation of shMLL1 cells and ensure similar cell numbers to scrambled control cells) did not increase velocity of scrambled control or shMLL1 cells compared to fresh medium (fig. S4N). Cells in 3D collagen gels display increased migration for increasing cell density (i.e., for increased number of cells per unit volume) (3). Cells at low density (LD; 10 cells/mm3) move less compared to the standard cell density used in this manuscript [100 cells/mm3; high density (HD)], but the motility of LD cells can be increased by the addition of CM from HD cells. To further confirm that MLL1-mediated cell motility was mediated via the secretion of soluble factors, we incubated LD cells with CM from scrambled control or shMLL1 cells at HD (fig. S4O). CM was collected after 3 days, allowing ample time for the secretion of cytokines critical for cell motility. The addition of CM from scrambled control cells to LD cells increased their motility (red) to levels comparable to cells at HD (black), evident in both the trajectories (fig. S4P) and cell velocities (fig. S4Q). However, CM from shMLL1 cells had no impact on the motility of cells at LD (green), indicating that CM from shMLL1 cells lacks secreted factors key for cell migration. In sum, MLL1-Menin interaction regulates cell migration via the secretion of soluble factors.
Fig. 2. MLL1-Menin interaction regulates cell motility and protrusion generation via IL-6,8/pSTAT3 signaling.
(A) CM was collected from scrambled control cells and added to shMLL1 or scrambled control cells. CM addition fully rescued cell migration of shMLL1 cells as seen in (B) trajectories and (C) velocities. (D) MLL1-Menin inhibition leads to down-regulation of key genes in the IL-6/JAK/STAT3 signaling pathway. MLL1 i, MLL1 inhibitor. (E) shMLL1 cells showed down-regulation of IL-6/STAT3–regulated actin assembly pathway. (F) MLL1 inhibition reduces IL-6 secretion. (G) MLL1 inhibited cells show decreased protrusion generation, which can be rescued by supplementation of IL-6/8. (H) A temporal heatmap of protrusion-related parameters shows reduced cumulative number of protrusions and the maximum length of protrusion generated per cell following MLL1 inhibition. Each row is one cell, and each block is one condition. (I) Cell motility was rescued by supplementing cells with IL-6/8 on top of MLL1 inhibition (10 μM MI-2-2). Both IL-6 and IL-8 are necessary and sufficient to rescue migration despite continuing inhibition of the MLL1-Menin interaction, evident in (J) velocity and (K) trajectories. (L) pSTAT3, not STAT3, levels show loss and rescue following MLL1 inhibition and IL-6/8 supplementation, respectively. (M) Quantification of IL-6/8 rescues Western blots, and bands from same blot are connected by a line. (N and O) Arp2 and Arp3 show the same trend as pSTAT3 following MLL1 inhibition and IL-6/8 supplementation. (P) STAT3 knockdown reduces cell migration. (Q) Motility rescued in MLL1-inhibited cells via IL-6/8 supplementation was lost by inhibiting STAT3. (R) Schematic illustration of MLL1-Menin–based regulation of cell migration. MLL1-Menin interaction controls the production of IL-6/8, which leads to pSTAT3. pSTAT3 drives actin filament assembly via Arp2/3. All data in this figure were generated with MDA-MB-231 cells embedded in 3D collagen gels. (A) and (L) were created with BioRender. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. NS, not significant.
MLL1-inhibited cells showed down-regulation of key members involved in the IL-6/STAT3–actin filament assembly pathway, including IL-6, STAT3, and ACTR2 (Fig. 2D and fig. S5A). The same IL-6–based actin assembly pathway was also down-regulated by other MLL1-Menin inhibitors, VTP50469 (fig. S5B) and MI-3454 (fig. S5C), as well as in all three shMLL1 clones (Fig. 2E). MLL1-Menin inhibition reduced the secretion of IL-6 (Fig. 2F) in a range of TNBC cells (MDA-MB-231, BT-549, SUM-149, and SUM-159) (fig. S5D). Secretion of a wider panel of cytokines was tested using a multiplex cytokine assay. A multiplexed antibody barcode microarray chip was prepared against IL-6 and IL-8, which has also been shown to be essential for 3D cell migration (3). IL-6 production was decreased by MLL1-Menin inhibition and by MLL1 depletion (fig. S5E). In contrast to IL-6, levels of IL-8 were only marginally reduced, mostly in MLL1-depleted cells (fig. S5F). Higher dosage of MLL1-Menin inhibitors somewhat reduced IL-8 production but were not as effective as shMLL1 cells that were a deep (>93%) MLL1 knockdown.
Cells produce dendritic protrusions to move effectively inside 3D collagen matrices, and, hence, cell migration is tightly connected to cell morphology (51): More protrusions correlate with more effective 3D cell migration (52). Cells embedded in 3D collagen gels produced large and dynamic protrusions, which were abrogated by MLL1-Menin inhibition (Fig. 2G). MLL1-Menin–inhibited cells supplemented with IL-6/8 displayed large protrusions, indicating that IL-6/8 supplementation rescued generation of cellular protrusions. A machine learning algorithm was used to further analyze cell and protrusion morphology from phase-contrast images recorded during overnight live-cell tracking. MLL1-Menin inhibition reduced the number of cells that generated protrusions and the number of cells that generated multiple protrusions (fig. S5, G and H). Decreased cell migration, measured as reduced mean squared displacements (MSDs), was accompanied by a decrease in the number and maximum length of protrusions generated by cells (Fig. 2H). Thus, MLL1-Menin interaction regulates cell migration via cytokine (IL-6 and IL-8) secretion and protrusion generation.
MLL1-Menin interaction regulates actin filament assembly via an IL-6/8/pSTAT3/Arp3 axis
Exogenous IL-6 and/or IL-8 were supplemented to rescue cell motility following impairment of cell migration by MLL1-Menin inhibition (Fig. 2I). Rescue wells were supplemented with recombinant human IL-6 and/or IL-8 at reported concentrations of these cytokines in 3D collagen gels (3). Supplementation of IL-6 increased cell velocity but did not fully rescue velocity to that observed with untreated cells. IL-8 also increased cell velocity; however, this increase was not statistically significant. MLL1-inhibited cells supplemented with IL-6 and IL-8 (referred to as IL-6/8, maroon; Fig. 2J) displayed increased cell motility compared to treated cells (red) and had velocities comparable to untreated control (black). Hence, cell motility lost by MLL1-Menin inhibition can be fully rescued by supplementation of IL-6 and IL-8. Addition of IL-6/8 to untreated cells (purple) did not further increase their velocity (fig. S5I), indicating that IL-6/8–mediated cell migration reaches a plateau, just as with CM. As a positive control for cytokine supplementation, addition of IL-6/8 to LD cells increased their motility (fig. S5J). These changes in cell velocity were reflected in cell trajectories (Fig. 2K), with supplementation of IL-6/8 leading to longer cell trajectories despite continuing MLL1 inhibition. Cell motility lost by MI-503 could also be similarly rescued by the supplementation of IL-6/8 (fig. S5K). Thus, IL-6/8 lies downstream of MLL1-Menin interaction and mediates MLL1-based cell migration.
Levels of phosphorylated STAT3 (pSTAT3), a key transcription factor downstream of IL-6 and IL-8 (53), were halved by MLL1-Menin inhibition (Fig. 2L, left lane versus middle lane). Supplementation of IL-6/8 restored pSTAT3 level (Fig. 2L, right lane; quantified in Fig. 2M) back to that observed in untreated cells. In contrast, total STAT3 expression was minimally changed, suggesting that MLL1 inhibition affects STAT3 phosphorylation rather than indirectly affecting phosphorylation via STAT3 expression. In addition to pSTAT3, Arp2 and Arp3, which are involved in actin filament assembly (54), were also down-regulated with MLL1-Menin inhibition (Fig. 2N, left lane versus middle lane) and rescued by supplementation of IL-6/8 (Fig. 2N, right lane; quantified in Fig. 2O). Arp2/3 is essential for protrusion generation (3), and decreased Arp2/3 is consistent with fewer and smaller protrusions generated by cells following MLL1-Menin inhibition (Fig. 2, G and H, and fig. S5, G and H). Supplementation of IL-6/8 to MLL1-inhibited cells rescued protrusion generation (Fig. 2G), and CM from scrambled control cells fully rescued protrusion generation of shMLL1 cells (fig. S5, L and M). To verify that STAT3 was indeed responsible for cell migration, we depleted STAT3 via shRNA (fig. S5N) and tracked the migration of shSTAT3 cells. STAT3 depletion reduced cell velocity to levels observed with MLL1 inhibition (Fig. 2P), showing that STAT3 plays a key role in cell migration. The same pathway involving JAK2, STAT3, WASF3, and ARP2/3 was also responsible for reduced cell migration in BT549 TNBC cells following MLL1-Menin inhibition (fig. S5O). To further confirm that increased STAT3 was responsible for increased cell motility downstream of IL-6/8 supplementation, we sought to determine whether rescued motility (Fig. 2J) could be lost by inhibiting STAT3 on top of cytokine supplementation (Fig. 2Q). Rescued motility in cells treated with MLL1 inhibitor + IL-6/8 (maroon) was lost by STAT3 inhibition on top of the rescue (brown, MLL1 inhibitor + IL-6/8 + STAT3 inhibitor), leading to similar cell motility as either MLL1 inhibition (red) or STAT3 inhibition (gray) alone. Thus, STAT3 lies downstream of IL-6 and regulates IL-6–mediated cell migration.
Together, our results establish that IL-6 and IL-8 lie downstream of MLL1-Menin interaction and STAT3 lies downstream of these cytokines (Fig. 2R). Inhibition of the MLL1-Menin interaction inhibits the production of IL-6 and IL-8, leading to lower pSTAT3, Arp2/3, and protrusion generation. These results suggest that MLL1-Menin interaction controls 3D cell motility by regulating STAT3/Arp2/3-based actin filament assembly and associated protrusion generation via IL-6/8 production.
MLL1-Menin interaction regulates TGF-β1–mediated myosin II contractility
In addition to providing a translational potential, pharmacological inhibition also permits for a finer modulation of MLL1-Menin interaction than what is possible with a knockdown. For example, increasing the concentration of MLL1-Menin inhibitors allowed for a gradual “titration” of cell migration (figs. S2L and S4, B, D, and E). Given that the MLL1/IL-6/8/STAT3/Arp2/3–based actin filament assembly is perturbed by MLL1 inhibitors at lower dosages, we next sought to determine whether additional migration-based pathways were also controlled by the same MLL1-Menin interaction (as seen in Fig. 1L). Cells treated with a higher (30 μM) dose of MI-2-2 displayed the same extent of migration inhibition; however, supplementation of IL-6/8 did not rescue cell motility (Fig. 3, A and B, dark green versus light green). Supplementation of IL-6/8 at three times the regular concentration (3× IL-6/8) also failed to rescue motility (fig. S5P). Hence, a “deeper” inhibition of the MLL1-Menin interaction affects cell migration by pathways other than IL-6/STAT3/Arp2/3–based actin filament assembly.
Fig. 3. MLL1-Menin–based regulation of TGF-β1–mediated cell migration is mechanistically nonlinear.
(A) In contrast to 10 μM MI-2-2 treatment, supplementation of IL-6/8 did not rescue cell migration in 30 μM MI-2-2–treated cells, shown by cell velocities and (B) trajectories. MLL1 i, MLL1 inhibitor. (C) pMLC was down-regulated at a deep MLL1-Menin inhibition (30 μM MI-2-2), but not at lower doses (10 μM MI-2-2). ROCK inhibitor (Y-27632)–treated cells were the positive control. (D) Quantification of nonlinear response of myosin contractility to MLL1 inhibition in (C). (E) The newest MLL1-Menin inhibitors (MI-3454 and VTP50469) also reduced pMLC2. (F) Heatmap of gene expression values for the genes involved in Hallmark cell migration (in purple) or proliferation (in green) gene sets shows two distinct gene expression patterns corresponding to low (mode-1) and high (mode-2) MLL1-Menin inhibitor dosage. TGF-β family members (labeled in black) were down-regulated with deep MLL1-Menin inhibition. (G) ChIP-Atlas (an online ChIP-seq database)–based analysis reveals that MLL1, Menin, and WDR5 bind to the promoter region of TGFB1. COMPASS members also bound to NFKB1 and RELA promoter sequences. (H) MLL1 knockdown/depletion reduces expression of genes central to TGF-β signaling and myosin contractility. (I) Multiplex cytokine analysis showed that TGF-β1 levels were unaffected with low-dose MLL1 inhibition but were reduced with high dosage. N.D., not detected. (J) Immunofluorescence microscopy of MLL1-inhibited cells showed that MLL1-Menin inhibition reduces cell size, increases cell roundedness, and disrupts the actin cytoskeleton (red). Mode-2 MLL1 inhibition (30 μM MI-2-2) reduces pMLC2 (green), while mode-1 (10 μM MI-2-2)–treated cells still show pMLC2. Immunofluorescence quantification affirms these observations. Data in this figure were generated with MDA-MB-231 cells embedded in collagen gels except (G) (ChIP-Atlas) and (K) (MDA-MB-231 cells in 2D). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. NS, not significant.
Cell migration requires both actin filament assembly (regulated here by the IL-6/8/STAT3/Arp2/3 axis) and actomyosin contractility (55). Actomyosin-based contractility is regulated via phosphorylation of myosin II (55), and levels of mono (S19) and di (T18 S19) phospho–myosin light chain II (pMLC2) were selectively reduced at a deep MLL1-Menin inhibition (Fig. 3C and fig. S5Q). pMLC2 (S19) levels showed a marked reduction upon increasing inhibition of the MLL1-Menin inhibition (quantified in Fig. 3D and fig. S5R) and were lower than that observed with Y-27632, a ROCK1/2 inhibitor. ROCK1 and ROCK2, major regulators of pMLC2, were also down-regulated (fig. S5R). These results were also validated with other MLL1-menin inhibitors, MI-503 (fig. S5S), VTP50469, and MI-3454 (Fig. 3E and quantified in fig. S5T). In addition, MLL1-depleted cells also exhibited reduced myosin contractility, indicated by reduced pMLC2 levels (fig. S5U). Together, these results indicate that, in addition to actin filament assembly, MLL1-Menin interaction controls myosin-based contractility.
Hierarchical clustering on a heatmap of migration- and proliferation-related genes identified two distinct sets of gene expression (Fig. 3F), one corresponding to low doses of MLL1-Menin inhibitors (10 μM MI-2-2, as well as 0.3 and 1 μM MI-503, hereby referred to as mode-1 of MLL1-Menin inhibition) and another corresponding to high doses (30 μM MI-2-2 and 3 μM MI-503, hereby referred to as mode-2 of MLL1-Menin inhibition), which forms a distinct pattern of gene expression corresponding to a deep inhibition of the MLL1-Menin interaction. Notably, several genes associated with TGF-β signaling (SMAD3, SMAD5, SMAD7, SKIL, and KLF10) and TGF-β target genes (ID1, ID2, and ID3) were differentially down-regulated in mode-2 but were unchanged in mode-1. To assess whether TGF-β–based signaling was a potential mediator of MLL1-based myosin contractility, we examined whether MLL1 (KMT2A) directly bound to the promoter of TGFB1 using ChIP-Atlas, a chromatin immunoprecipitation sequencing (ChIP-seq) database (56). Regions flanking the transcription start site (TSS), specifically ±2 kb of the TSS, were also checked for Menin (MEN1) and the COMPASS member WDR5 (that controls cell motility via its interaction with MLL1; fig. S3B). KMT2A bound directly to the TGFB1 promoter sequence (Fig. 3G) along with MEN1 and WDR5, indicating that COMPASS (and hence MLL1) directly binds to the TGFB1 promoter to potentially activate transcription. KMT2A and WDR5 also bound to the TGFB2 promoter (albeit less strongly; fig. S5V), while MEN1 did not show binding to this region. In addition, COMPASS members also bound to NFKB1 and RELA promoter sequences. Along with down-regulation of NFKB1 (in Figs. 2, D and E, and 3F), this hints at MLL1-based regulation of NF-κB signaling, which, in turn, controls IL-6/8 production (57). MLL1 depletion reduces levels of both these proteins, NF-κB1 (p50 and p105) and RELA (fig. S6, A and B). Consistent with NF-κB being upstream of IL-6, NF-κB inhibition in the absence of IL-6/8 supplementation did decrease motility (fig. S6C). However, NF-κB inhibition had no effect on cell motility in the presence of IL-6/8 supplementation.
At the transcript level, expression of both TGFB1 and TGFB2 were down-regulated following MLL1 depletion (Fig. 3H) or inhibition (fig. S6, D and E). However, only TGFB1 showed a nonlinear response with MLL1-Menin inhibition, mirroring the trend observed with myosin contractility. Downstream TGF-β family members were also down-regulated by MLL1 inhibition. GSEA enrichment plots show down-regulation of key cell migration–related pathways for mode-2 MLL1 inhibition cells, including TGF-β signaling (fig. S6F). In addition, levels of TGF-β receptors, TGFBR1 and TGFBR2, were unaffected by MLL1-menin inhibition (fig. S6G), indicating that the down-regulation of TGF-β signaling was potentially due to down-regulation of secreted factors. Hence, TGF-β1 (gene product of TGFB1) was hypothesized to be the likely regulator of pMLC2, which lays downstream of MLL1-Menin interaction. Genes related to myosin contractility such as RHOA, ROCK2, and MYLK were also down-regulated with MLL1 depletion (Fig. 3H) or MLL1-Menin inhibition (fig. S6H). Myosin light chain kinase (MYLK), essential for maintaining the phosphorylation of MLC2 (58), was down-regulated selectively at mode-2 but not mode-1 MLL1 inhibition, consistent with reduced pMLC2 levels observed exclusively with mode-2 inhibition in Fig. 3C. Similar results were also seen with the MLL1-Menin inhibitors VTP50469 (fig. S6I) and MI-3454 (fig. S6J), as well as in BT549 TNBC cells (fig. S6K). Secretion of TGF-β1, measured via a multiplex cytokine assay, was decreased by mode-2 MLL1-Menin inhibition but was unchanged with mode-1 inhibition (Fig. 3I). shMLL1 cells, which were hypothesized to phenocopy the mode-2, or deep, MLL1 inhibition, did not exhibit TGF-β1 secretion. In contrast, TGF-β2 levels were largely unchanged throughout (fig. S6L). None of the other migration- or proliferation-related cytokines tested by our multiplex assay showed a change with MLL1 depletion or inhibition (fig. S6M). MLL1-Menin inhibition also affected cell morphology in addition to cell migration machinery. Untreated cells were elongated, had high levels of pMLC2 (green, pointed to by arrowheads), and featured prominent actin fibers (red, Fig. 3J, left). Mode-2 MLL1 inhibition led to more rounded cells with virtually no pMLC2 staining (Fig. 3J, right). Mode-1 cells looked morphologically in between untreated and mode-2 cells but had higher levels of pMLC2 (Fig. 3J, middle). Only mode-2 inhibition showing a significant decrease in pMLC2 signal (Fig. 3J, bar plot).
Together, these results show that a deeper MLL1-Menin inhibition reduces myosin contractility, and its effect on 3D cell migration cannot be rescued solely by IL-6/8 supplementation. Concurrently, secretion of another cytokine, TGF-β1, and the expression of TGF-β family members were down-regulated at higher dosages of MLL1-Menin inhibitor.
TGF-β1 and IL-6/8 are necessary and sufficient for MLL1-Menin–based cell migration
To assess whether the reduction in motility in mode-2 was caused by loss of TGF-β1, we added exogenous TGF-β1 to MLL1-Menin–inhibited cells. Supplementation of either IL-6/8 (dark green) or TGF-β1 (brown) by itself did not change motility (velocities in Fig. 4A and trajectories in Fig. 4B). However, concurrent supplementation of both IL-6/8 and TGF-β1 (blue) restored cell motility to that observed with untreated control (black). This is consistent with the fact that both actin filament assembly via IL-6/8 and myosin contractility via TGF-β1 are required for cell motility. shMLL1 cells, which show reduced secretion of IL-6 (fig. S5E), IL-8 (fig. S5F), and TGF-β1 (Fig. 3I), exhibited a full rescue of motility after supplementation of IL-6/8 + TGF-β1 (Fig. 4C). As with mode-2 inhibition, supplementation of either IL-6/8 or TGF-β1 alone did not fully restore motility of shMLL1 cells. Thus, at mode-2 MLL1-Menin inhibition, TGF-β1 production is also decreased along with IL-6/8, and supplementation of these cytokines can fully rescue 3D cell migration.
Fig. 4. MLL1-Menin interaction regulates myosin-based contractility via a Gli2/ROCK1/2/pMLC2 axis.
(A and B) Supplementation of TGF-β1 and IL-6/8 is necessary and sufficient to fully rescued cell migration in mode-2 MLL1-inhibited cells and (C) shMLL1 cells. MLL1 i, MLL1 inhibitor. (D) IL-6/8 + TGF-β1 rescued protrusion generation in MLL1-inhibited cells. (E and F) pMLC2 was rescued with supplementation of TGF-β1 (either by itself or concurrently with IL-6/8), indicating restoration of myosin contractility. NES, normalized enrichment score. (G and H) pSTAT3 was rescued by supplementation of IL-6/8 (either by itself or concurrently with TGF-β1). (I) GLI2 is the most down-regulated gene following MLL1-Menin inhibition. (J) Gli2, ROCK1, and ROCK2 levels were rescued by replenishment of TGF-β1, indicating that these lie downstream of both the MLL1-Menin interaction and TGF-β1. (K) shMLL1 cells expressed low levels of Gli2, pSTAT3, and Arp3; all of which were rescued by supplementation of TGF-β1 + IL-6/8. (L and M) Inhibition of Gli2, but not STAT3, reduced pMLC2, indicating that myosin contractility was regulated by TGF-β1/Gli2 signaling, rather than IL-6/8/STAT3 signaling. (N) Gli2 inhibition reduced levels of ROCK1/2, necessary for myosin contractility, indicating that TGF-β1– and Gli2-regulated myosin contractility was mediated by ROCK1/2. (O) Gli2 knockdown reduces cell migration. (P) Motility rescued by TGF-β1 + IL-6/8 supplementation after MLL inhibition (blue) is lost by inhibiting either Gli2 (downstream of TGF-β1) or STAT3 (downstream of IL-6/8). Concurrent inhibition of Gli2 and STAT3 led to the lowest cell motility. (Q) Expression of key genes implicated in MLL1-Menin–regulated cell migration is positively correlated with MLL1 expression in TCGA. (R) Deep (mode-2) MLL1-Menin inhibition disrupts motility in a two-pronged manner. MLL1-Menin interaction controls the production of IL-6/8, which regulates motility via STAT3/Arp2/3–mediated protrusion generation. In addition, MLL1-Menin interaction also regulates the production of TGF-β1, which controls myosin contractility via a Gli2/ROCK1/2/pMLC2 axis. Data in this figure were generated with MDA-MB-231 cells embedded in 3D collagen gels except (Q) (TCGA). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. NS, not significant.
The rescue of cell migration via cytokine supplementation was accompanied by an increase in actin filament assembly and myosin contractility. Protrusion analysis showed that MLL1-Menin inhibition reduced the number of protrusions and maximum protrusion length (Fig. 4D). Supplementation of IL-6/8 + TGF-β1 rescued these parameters, with rescued cells generating similar sizes and number of protrusions as untreated control. Along with protrusion generation (actin filament assembly), myosin contractility was also restored in rescued cells (Fig. 4E and quantified in Fig. 4F). Supplementation of TGF-β1 (by itself or along with IL-6/8) increased pMLC2 levels. Supplementation with IL-6/8 did not fully rescue pMLC2, demonstrating that pMLC2 lost by MLL1-Menin inhibition can be regained by supplementing cells with TGF-β1. In contrast, IL-6/8 supplementation, but not TGF-β1, restored pSTAT3 levels (Fig. 4G and quantified in Fig. 4H), consistent with the restoration of protrusion generation.
Thus, MLL1-Menin interaction controls 3D cell migration via the secretion of cytokines IL-6, IL-8, and TGF-β1. Supplementation of exogenous IL-6, IL-8, and TGF-β1 rescued actin filament assembly and myosin contractility, respectively, thus restoring cell motility.
TGF-β1–based myosin contractility is mediated by a Gli2/ROCK1/2/pMLC2 axis
Polymerase chain reaction (PCR)–based assessment of several genes following mode-2 MLL1-Menin inhibition revealed GLI2 to be the most down-regulated gene (Fig. 4I, circled in red). Genes associated with TGF-β signaling and its target genes (TGFB1, ID1, SMAD3, and SMAD7) as well as genes involved in myosin contractility (RHOA, ROCK2, and MYLK) were also down-regulated. GLI2 was also the most differentially expressed gene between mode-2 versus mode-1 MLL1-Menin inhibition (fig. S7A). GLI2 has been reported to lie downstream of both TGF-β and Hedgehog signaling (59). Since MLL1-Menin inhibition does not induce an appreciable change in the genes involved in Hedgehog signaling (fig. S7, B and C), we reasoned that Gli2 mediates the TGF-β1–based regulation of myosin contractility.
Gli2 expression was down-regulated by MLL1-Menin inhibition (Fig. 4J and quantified in fig. S7D). Supplementation of IL-6/8 did not restore Gli2 expression, but supplementation of TGF-β1 (by itself or along with IL-6/8) restored Gli2 levels, indicating that Gli2 lies downstream of TGF-β1. shMLL1 cells expressed low levels of Gli2, pSTAT3, and Arp3, all of which were increased following IL-6/8 + TGF-β1 supplementation (Fig. 4K and quantified in fig. S7E). Gli2 inhibitor markedly reduced pMLC2 levels (Fig. 4L and quantified in Fig. 4M), indicating that Gli2 is a potent regulator of myosin contractility. STAT3 inhibition did not change pMLC2 levels appreciably, denoting little cross-talk between STAT3-mediated actin filament assembly and myosin contractility. Along with Gli2, expression of ROCK1 and ROCK2 was also reduced by MLL1-Menin inhibition and restored with TGF-β1 supplementation (Fig. 4J and quantified in fig. S7D). ROCK1 and ROCK2 are serine/threonine kinases that are responsible for phosphorylating myosin light chain and are essential for maintaining pMLC2 levels and myosin contractility (60). Gli2 inhibition reduced levels of both ROCK1 and ROCK2 (Fig. 4N and quantified in fig. S7F), indicating that Gli2-mediated myosin contractility was mediated by ROCK1/2. In contrast, STAT3 inhibition had little effect on ROCK1/2, consistent with STAT3 being a major regulator of actin filament assembly, but not myosin contractility. Thus, TGF-β1–mediated myosin contractility is regulated by the Gli2/ROCK1/2/pMLC2 axis. To verify that Gli2 is essential for cell migration, we depleted Gli2 via shRNA (fig. S7G). shGli2 cells were nonmotile and exhibited velocities similar to MLL1-Menin–inhibited cells (Fig. 4O). Gli2 inhibitor was used to verify that motility rescued by cytokine supplementation could be lost by downstream inhibition (Fig. 4P). Mode-2–inhibited cells showed reduced motility (green versus black), which was rescued after supplementation with IL-6/8 + TGF-β1 (blue). Inhibition of STAT3 (brown) or Gli2 (gray) on top of the MLL1 inhibition and cytokine supplementation reduced cell motility again, indicating that both STAT3 and Gli2 play a vital role in cell migration downstream of the cytokines. Maximum inhibition of cell migration was obtained by concurrent STAT3 and Gli2 inhibition (orange), which reduced the distribution of cell velocities to that observed with MLL1 inhibition (green).
To further substantiate our elucidated mechanism of MLL1-based cell migration, we examined basal breast cancer RNA sequencing (RNA-seq) data from PanCanAtlas (TCGA) to identify genes and gene sets that were correlated with MLL1 (KMT2A) expression (Fig. 4Q and fig. S7H). Top genes correlating with MLL1 expression included ROCK1 (0.9 log fold change in gene per log fold increase in MLL1 expression) and ROCK2 (0.7 log fold change) (Fig. 4Q). Other genes that were implicated in MLL1-Menin–mediated cell migration above and found to positively correlate with KMT2A expression included JAK2, STAT3, GLI2, and MYLK (although GLI2 and MYLK were just under the significance threshold). Epithelial-mesenchymal transition genes ZEB1 and ZEB2 were also significantly and positively correlated with MLL1 expression. In addition, focal adhesion, extracellular matrix (ECM) receptor interaction, myosin phosphorylation, and epithelial-mesenchymal transition gene sets were all significantly positively correlated with MLL1 expression in TCGA basal breast cancer dataset (fig. S7, H and I).
Thus, our results establish that TGF-β1 lies downstream of MLL1-Menin interaction and is depleted exclusively in mode-2 MLL1-menin inhibition (such as in shMLL1 cells, or by 30 μM MI-2-2 and 3 μM MI-503). Gli2 lies downstream of TGF-β1, while STAT3 lies downstream of IL-6/8 (Fig. 4R). Deep inhibition of the MLL1-Menin interaction inhibits the secretion of both IL-6/8 and TGF-β1, leading to lower pSTAT3 and Gli2 levels. Lower pSTAT3 levels lead to lesser actin filament assembly via reduced nucleator Arp2/3, while lower Gli2 levels correspond to decreased myosin contractility via ROCK1/2 and pMLC2. Therefore, MLL1-Menin interaction controls 3D cell motility by regulating both actin assembly via the IL-6/STAT3/Arp3 axis and myosin contractility via the TGF-β1/Gli2/ROCK1/2/pMLC2 axis.
MLL1-Menin interaction controls 3D cell proliferation
In addition to controlling cell migration, MLL1 also regulates proliferation. shMLL1 cells (Fig. 1F) or cells treated with MLL1-Menin inhibitors [MI-2-2 and MI-503 (Fig. 5A), VTP50469 (fig. S8A), and MI-3454 (fig. S8B)] showed reduced cell numbers. Unlike inhibition of cell migration (which was dose independent between mode-1 to mode-2), quantification of cell numbers in 3D collagen matrices showed that MLL1-Menin inhibition reduces cell proliferation in a dose-dependent manner (Fig. 5A). Mode-2 inhibition led to a near growth arrest, while mode-1 had minimal impact (fig. S8, C and D). Proliferation affected by MLL1-Menin inhibition was also readily reversible. Cells were pretreated with MLL1 inhibitor for 2 days before being seeded and subjected to further drug treatment. Pretreated cells without further treatment showed increased cell proliferation 2 days after drug washout (fig. S8E). In addition to TNBC cells, these results were also validated in fibrosarcoma cells (fig. S8F), and this change in proliferation was not accompanied by a change in cell cycle distribution (fig. S8G). Another TNBC cell line (BT-549) showed decreased cell numbers with minimal change in apoptosis levels (fig. S8, H and I), and a largely unchanged cell cycle distribution (fig. S8J). Thus, the MLL1-Menin interaction also controls cell proliferation.
Fig. 5. MLL1-Menin regulates cell proliferation via a multitude of proliferation- and cell cycle–related pathways.
(A) MLL1-Menin inhibition with MI-2-2 or MI-503 results in a dose-dependent growth suppression. (B) Gene-gene correlation analysis on the 3926 Hallmark gene set genes that were expressed revealed three distinct modules (proliferation, inflammation, and migration) and a mixed module. Key cell cycle and proliferation genes were substantially enriched in modules 1 and 3. RNA-seq analysis of (C) shMLL1 cells versus scrambled control cells or (D) MLL1-Menin–inhibited cells shows down-regulation of several key proliferation- and cell cycle–related pathways and up-regulation of antiproliferative pathways. (E) GSEA enrichment plots of cell cycle–related pathways for mode-2 MLL1-inhibited cells (30 μM MI-2-2 treatment). (F) Volcano plot of mode-2 MLL1 inhibition (30 μM MI-2-2) versus untreated (DMSO) control shows nearly 4000 genes that are affected by MLL1-Menin inhibition (log2 fold change > 1 and P < 0.05, plotted in blue). These genes include those that have been implicated in our MLL1-Menin–based regulation of cell migration and proliferation (validation, in black). In addition, a subset of these genes consists of proliferation-related genes that further extend the scope of MLL1-Menin interaction in regulation of proliferation (discovery, in yellow). MLL1 i, MLL1 inhibitor. (G) PCA analysis on all conditions shows that lower doses of MLL1-Menin inhibitors (mode-1) cluster closer to the untreated control than to higher drug dosages. Each oval encompasses all technical replicates for that condition. All data in this figure were generated with MDA-MB-231 cells embedded in 3D collagen gels. *P < 0.05, **P < 0.01, and ***P < 0.001. NS, not significant.
To assess the impact of MLL1-Menin interaction on proliferation, we performed correlation analysis on our RNA-seq data for the 3926 genes that are a part of Hallmark gene sets [Molecular Signatures Database (MSigDB) collection) and that were expressed in our samples. Proliferation genes formed the largest module (Fig. 5B), consisting mainly of mitosis and cell cycle–based gene sets. Gene sets pertaining to inflammation and cell migration formed the second module, while the third module was a mixture of all remaining gene sets including oncogene-based proliferation gene sets. Enrichment analysis on all proliferation-related pathways for shMLL1 and MLL1-Menin–inhibited cells emphasized the down-regulation of key cell cycle–based pathways including E2F targets (affecting G1/S transition), G2/M checkpoint, and mitotic spindle assembly (Fig. 5, C and D, GSEA enrichment plots in Fig. 5E and fig. S8K). Key proliferation pathways such as Myc targets, phosphatidylinositol 3-kinase (PI3K)–Akt–mammalian target of rapamycin (mTOR) signaling, and (genes up-regulated by) KRAS signaling were also diminished by MLL1 inhibition. In addition, antiproliferative pathways such as p53 pathway and DNA repair, which induce proliferative arrest, were up-regulated. MLL1-Menin inhibition reduced transcript levels of key markers of cellular proliferation, KI67 and PCNA (fig. S8, L and M) and affected a wide variety of cell cycle regulators at the transcript level, including cyclins and cyclin-dependent kinases (CDKs) (fig. S9A). CCND1 and CDK6, regulators of G1/S transition (61), as well as CCNB1 and CCNB2, major regulators of G2/M transition (61), were down-regulated by MLL1-Menin inhibition. Down-regulation of pathways and genes involved in both G1/S and G2/M transitions is consistent with decreased proliferation accompanied with minimal change in cell cycle distribution following MLL1-Menin inhibition. Members of the mitogen-activated protein kinase (MAPK) family (MAPK1, MAPK10, and MAPK12), which mediate cell proliferation (62), were also down-regulated with MLL1-Menin inhibition (fig. S9B). MAPK1 was also found to be positively correlated with MLL1 in our TCGA analysis (Fig. 4Q) along with G2/M checkpoint gene set (fig. S7, H and I). The same cell cycle factors were affected by MLL1 depletion in shMLL1 cells (fig. S9C) and by the MLL1-Menin inhibitors VTP50469 (fig. S9D) and MI-3454 (fig. S9E). Thus, MLL1-Menin interaction controls key proliferation-related genes and pathways, including markers of proliferation, cell cycle genes, and members of the MAPK family.
Transcriptomic analysis revealed that 3978 genes were significantly down-regulated (1994) or up-regulated (1984) in mode-2 MLL1 inhibition. These genes corroborated the abovementioned mechanism of MLL1-based cell migration (validation set, black points, labeled on the left side) and revealed further proliferation genes and transcription factors that were regulated by the MLL1-Menin interaction (discovery set, yellow points, labeled on the right side). Genes previously implicated in MLL1-menin–based cell migration including IL6, JAK2, STAT1, STAT2, NFKB1, GLI2, and ROCK2 were all down-regulated by MLL1-Menin inhibition (Fig. 5F). In addition, other genes shown to play a role in cell migration (HIF1A, MMP16, and MMP19), proliferation (KRAS, EGFR, SRC, PIK3CA, PIK3CB, CTNNB1, MDM2, MAP3K1, and MAP3K4), cell cycle and mitosis (CCNA2, CCNE2, CCNI2, BUB1, and PLK1), transcription and epigenetic factors (TEAD1, EZH2, MYC, and SETD2), and other genes necessary for tumor progression (CD274 and VEGFA) were also down-regulated by MLL1-Menin inhibition. Analysis of MLL1-Menin inhibition by MI-503 showed the same targets and underlying mechanism (fig. S9F). Principal components analysis (PCA) analysis showed that mode-1 MLL1-Menin inhibition (10 μM MI-2-2 and 0.3 μM MI-503) clustered closer to untreated control than to mode-2 MLL1-Menin inhibition (30 μM MI-2-2 and 3 μM MI-503) (Fig. 5G). PC1 accounted for the “depth” of MLL1-Menin inhibition (76% variance), while PC2 accounted for the differences between the two drugs, MI-2-2 and MI-503 (5% variance). Any cross-talk between cell migration– and proliferation-based pathways was determined by assessing if a rescue in cell migration affected proliferation or cell cycle distribution. CM from scrambled control cells did not rescue the proliferation defect of shMLL1 cells (fig. S9G), and neither did the supplementation of cytokines to MLL1-Menin–inhibited cells (fig. S9, H to J). This indicates that supplementation of cytokines/conditioned media can rescue cell motility, but not proliferation. Thus, the regulation of cell migration and proliferation by the MLL1-Menin interaction occurs via independent pathways. Hence, the MLL1-Menin interaction controls TNBC cell proliferation via a multitude of cell cycle–related and proliferation-based pathways. Further, this regulation of cell proliferation is uncoupled from its regulation of cell migration.
MLL1 regulates lung metastatic burden in vivo
Mice bearing MLL1-deficient tumors exhibited longer survival and lower metastatic burden (Fig. 1, G to K). However, MLL1-deficient tumors grew more slowly, and this reduced tumor growth rate could obscure the contribution of MLL1-regulated cell migration on metastatic burden. To account for this, we euthanized mice bearing shMLL1, scrambled control, and WT tumors when the tumors reached a set threshold size (1400 mm3) (Fig. 6, A to C). shMLL1 tumors grew for ~50% longer time than scrambled control or WT tumors (62, 41, and 43 days after tumor establishment, respectively). Lungs from mice bearing scrambled control and WT tumors showed more numerous and larger metastatic foci (evident as darker staining) than lungs from shMLL1 tumor–bearing mice (Fig. 6D and fig. S10A). Quantification of total lung metastatic burden showed three times lower lung metastatic burden for shMLL1 tumor–bearing mice compared to mice bearing scrambled control tumors (Fig. 6E). These metastatic lesions were not only fewer but also much smaller compared to WT or scrambled control cells (Fig. 6F). Fewer lesions (Fig. 6F, inset) may be due to lower cell migration or decreased survivability of cells in circulation. Given that shMLL1 mice were euthanized ~3 weeks after control mice, metastasized cells that have reached the lungs would have a longer time to proliferate and colonize. Hence, smaller metastases may not be due to lower proliferation of shMLL1 cells at the metastatic site. Immunohistochemistry (IHC) on scrambled control and shMLL1 tumors verified that the mechanism responsible for reduced tumor growth and metastasis in vivo is the same as that delineated in Figs. 2 to 5 (fig. S10C). Scrambled control tumors showed much stronger staining for pSTAT3 (which controls actin assembly and protrusion generation; Fig. 2), pMLC2 (which controls myosin contractility; Figs. 3 and 4), and Ki67 (marker of proliferation) than shMLL1 tumors. Scrambled control tumors also displayed an increased number of pMLC2-high cells at the tumor invasive front (here, the edge of the primary tumors; fig. S11A) compared to tumor bulk. This enrichment was notably reduced in MLL1-depleted tumors, with shMLL1 tumors showing very few pMLC2-high cells at the periphery (quantified in fig. S11B). Thus, MLL1 depletion inhibits metastasis independent of its effect on proliferation at the primary tumor site. Further, the underlying molecular mechanism in vivo is the same as that identified in our 3D in vitro systems.
Fig. 6. MLL1-depleted cancer cells exhibit decreased lung metastases in vivo.
(A) Orthotopic breast tumors were established in nonobese diabetic–severe combined immunodeficient gamma (NSG) mice by injecting cells into mammary fat pad. Mice were euthanized upon reaching a set threshold size (1400 mm3). shMLL1 mice were euthanized ~3 weeks after control mice. (B) WT and scrambled control tumors grew faster than shMLL1 tumors. (C) This difference in growth rate is also illustrated in tumor sizes at day 40. (D) shMLL1 tumor–bearing mice showed reduced lung metastatic burden despite being euthanized later and at the same primary tumor size as scrambled control and WT tumors. (E) Quantification of total metastatic burden per lung shows a threefold reduction in shMLL1 lungs compared to scrambled control. (F) Histogram of metastatic lesions of representative lungs shows that the reduced metastatic burden in shMLL1 lungs was due to fewer metastatic lesions that were also smaller. (G) Extravasation and metastatic outgrowth were assessed using a tail vein metastasis model. Mice were euthanized 6 weeks after injection. (H) Mice injected with control (scrambled control or WT) cells showed numerous and large metastatic lesions, while shMLL1 lungs showed barely any lesions. (I) Quantification of total metastatic burden per lung shows more than a 20-fold decrease for shMLL1 lungs compared to scrambled control. (J) Histogram of metastatic lesions for the representative lungs shows a markedly reduced lesion size distribution for shMLL1 lungs compared to scrambled control or WT. (K) Representation of the role of MLL1 in the metastatic cascade. Three different in vivo studies have been used to elucidate the contribution of MLL1-Menin interaction in driving primary tumor growth (left), cancer cell migration and invasion (center), as well as extravasation and metastatic outgrowth (right). All data in this figure were generated with MDA-MB-231 cells in NSG mice. (A), (G), and (K) were created with BioRender. *P < 0.05, **P < 0.01, and ****P < 0.0001. NS, not significant.
Next, the colonization and metastatic outgrowth of MLL1-depleted cells was assessed using a tail vein metastasis model (Fig. 6G). Lungs from mice injected with either scrambled control or WT cells displayed extensive metastasis with large colonies (Fig. 6H and fig. S11C; euthanized 6 weeks after injection). Lungs from mice injected with shMLL1 cells were mostly clear and looked similar to saline-injected lungs. Quantification of lung metastatic burden showed a 22-fold decrease for mice bearing MLL1-depleted tumors compared to scrambled control or WT control (Fig. 6I). Lungs in these mice showed markedly fewer and smaller metastatic colonies (Fig. 6J), indicating that MLL1 depletion plays a role in extravasation and metastatic outgrowth of lesions. Decreased metastatic burden with shMLL1-injected lungs was backed up by weaker staining for Ki67 via IHC for shMLL1 lungs (fig. S11E), indicating reduced proliferation among surviving shMLL1 lung metastatic colonies. Thus, MLL1 depletion reduces metastatic outgrowth of cancer cells and may also have an impact on extravasation of cancer cells.
Hence, MLL1 regulates metastasis of TNBC cells in multiple models of in vivo metastasis, and MLL1 depletion impairs multiple steps in the metastatic cascade (Fig. 6K). MLL1 regulates cell migration, a critical requirement for invasion and metastatic dissemination (1); extravasation and metastatic outgrowth; and proliferation, which leads to primary tumor growth and, hence, the number of cells invading into circulation.
MLL1-Menin inhibitors combined with paclitaxel block cell migration and proliferation
As new drugs in clinical trials are often evaluated in combination with existing standard of care, we assessed whether MLL1-Menin inhibitors could be used in conjunction with current standard of care in a potential neoadjuvant or adjuvant setting. Paclitaxel is a chemotherapeutic that is routinely administered to patients with TNBC, including in the metastatic setting (63). However, paclitaxel suffers from toxicity-based limitations that restrict its dosage, leading to poor efficacy in clinic. Doxorubicin, an anthracycline, is another chemotherapeutic routinely administered in TNBC (64). The impact of MLL1 depletion on diminishing tumor growth and metastasis in vivo hinted at the efficacy of MLL1-Menin inhibitors in a preclinical setting. The antiproliferative effect of mode-2 MLL1-Menin inhibition was additive with both paclitaxel (fig. S12A) and doxorubicin (fig. S12B). Mode-1 inhibition had a much smaller impact on cell numbers compared to either chemotherapeutic (fig. S12, C and D). Combination treatment of MLL1 inhibitor plus paclitaxel was also effective in decreasing cell migration (fig. S12E). The combination of MLL1 depletion in tumors and paclitaxel treatment (fig. S12F) led to a nearly flat growth curve (fig. S12G) and the smallest tumors (fig. S12, H and I). Lung sections of shMLL1 tumor–bearing mice treated with paclitaxel showed the lowest metastatic burden, with virtually no darker staining visible in the lung (fig. S13, A to C). This decrease was attributable to a reduction in both size and number of metastatic lesions (fig. S13D). Administration of paclitaxel to scrambled control tumors led to marginally lower metastatic burden than vehicle administration, which was not statistically significant. Treated mice also did not exhibit any toxicity associated with paclitaxel treatment (fig. S13E).
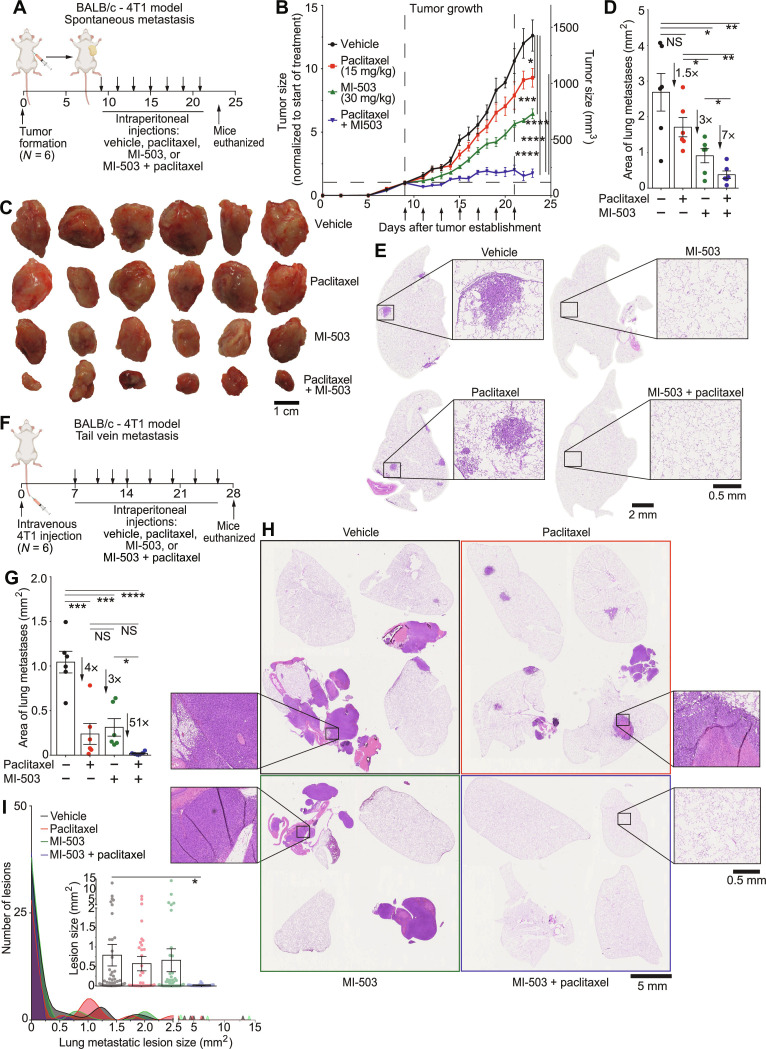
As cytokines play a critical role in immune responses, we assessed the combination of MLL1-Menin inhibition with paclitaxel in an immunocompetent model. BALB/c mice bearing syngeneic orthotopic 4T1 tumors were subjected to MI-503 + paclitaxel treatment, single-drug controls, or vehicle (Fig. 7A). MI-503 was the MLL1-Menin inhibitor used as MI-2-2 is not suitable for in vivo administration due to poor metabolic stability (48). Both single-drug controls slowed down tumor growth and the combination of MI-503 and paclitaxel nearly arrested tumor growth (Fig. 7B). The superior efficacy of MI-503 + paclitaxel was also evident in the final tumor sizes (Fig. 7C) and tumor weights (fig. S13G). Paclitaxel treatment did not reduce metastatic burden, but MLL1-Menin inhibition (either alone or in conjunction with paclitaxel) led to a sharp decrease in lung metastasis (fig. S13H), with combination-treated lungs exhibiting virtually no metastatic lesions (evident in purple staining in fig. S13I and metastatic burden quantified in Fig. 7D). High-resolution sample lungs (Fig. 7E) showed that vehicle-treated mice had large metastatic lesions (but fewer in number compared to MDA-MB-231 tumors) that was eliminated by MLL1-Menin inhibition (quantification and lesion size distribution in fig. S13J). IHC on vehicle- versus MI-503–treated tumors showed reduced levels of pSTAT3, pMLC2, and Ki67 (fig. S14A), consistent with the delineated in vitro mechanism for MLL1-Menin–regulated cell migration (via both pSTAT3-based actin assembly and pMLC2-based myosin contractility) and cell proliferation. Notably, the administration of MI-503 by itself or concurrently with paclitaxel did not lead to toxicity, as evident by a steady mouse body weight (fig. S14B).
Fig. 7. Concurrent paclitaxel and MLL1-Menin inhibition arrests tumor growth and abrogates metastasis.
(A) BALB/c mice bearing syngeneic orthotopic 4T1 tumors were injected with a MLL1-Menin inhibitor (MI-503, 30 mg/kg), paclitaxel (15 mg/kg), a combination of the two drugs, or a vehicle intraperitoneally every alternate day for seven times. (B and C) The combination of MI-503 and paclitaxel was effective in reducing tumor growth, with the growth of combination-treated tumors being essentially arrested. This efficacy of combination treatment was reflected in final tumor sizes. (D) Quantification of metastatic burden per lung showed that MI-503 treatment reduced metastatic burden, while administration of paclitaxel alone did not produce a significant decrease. Combination treatment (MI-503 + paclitaxel) led to a sevenfold decrease in metastatic burden compared to vehicle control. (E) Sample lungs from each condition show large but limited metastases in vehicle- and paclitaxel-treated mice. MI-503 treated mice—alone or in conjunction with paclitaxel—showed no major metastatic foci in their lungs. (F) BALB/c mice were injected with 4T1 cells intravenously via a tail vein injection to form lung metastases. One week after injection, they were treated with MI-503 (30 mg/kg), paclitaxel (15 mg/kg), a combination of the two drugs, or a vehicle intraperitoneally every 3 days for eight times. Mice were euthanized 4 weeks after metastasis formation. (G) Quantification of total metastatic lung burden per mouse showed that combination treatment virtually eliminated metastatic burden. (H) Sample lungs show that paclitaxel or MLL1-Menin inhibition led to decreased metastatic burden compared to vehicle. However, the clearest lungs were obtained in combination-treated mice. (I) Histogram of metastatic lesions for representative lungs in (H) demonstrates a reduction in both size and number of lesions with paclitaxel treatment and/or MLL1 depletion. All data in this figure were generated with 4T1 cells in BALB/c mice. (A) and (F) were created with BioRender. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. NS, not significant.
Last, to assess the potential for MLL1-Menin inhibition in more advanced stages of cancer with already established metastasis, we used a tail vein model (similar to that in Fig. 6, G to J) to form metastasis, which were then subjected to a similar combination treatment as above. Lung metastatic colonies were established in BALB/c mice via intravenous (tail vein) injection of 4T1 cells (Fig. 7F). Mice administered with vehicle showed extensive metastases upon excision of lungs, which was marginally reduced by single-drug administration (paclitaxel or MI-503; fig. S14D). Lungs excised from mice receiving combination treatment were virtually metastasis-free. Quantification of lung metastatic burden reflected these observations (Fig. 7G and full panel of lungs in fig. S15A). Single-drug treatment reduced metastatic burden by three- to fourfold. In contrast, the combination of MLL1-Menin inhibitor (MI-503) and paclitaxel led to synergistic (>50 fold) decrease in lung metastatic burden. High-resolution scans of lungs (Fig. 7H) showed that vehicle- or single drug–treated lungs had large metastatic lesions (longer histogram tails in Fig. 7I and inset bar plot), which was absent in MI-503 + paclitaxel–treated lungs. In addition to smaller lesions, combination-treated mice also exhibited fewer metastatic colonies (Fig. 7I, histogram). As was observed with the first MI-503 + paclitaxel treatment (Fig. 7, A to E and fig. S14B), neither treatment conditions led to toxicity or a loss of mouse body weight (fig. S15B).
In sum, our in vitro and in vivo results provide a preclinical rationale for the use of MLL1 inhibitors as antimetastatic agents, and it is compatible for administration in conjunction with the current TNBC chemotherapeutic, paclitaxel.
DISCUSSION
Biology of 3D cytokine-driven cell migration
Cytokines produced by cancer cells can drive their migration, but the epigenetic regulation of cytokine production and cell migration are relatively understudied areas of cancer cell biology. Our work identifies MLL1 and Menin as epigenetic regulators of cytokine-driven cell migration and metastasis. While MLL1 has been extensively studied in the context of MLL1-fusion–driven leukemias, their biology is profoundly different from the proposed mechanism of cell migration in breast cancer cells (fig. S3, C to H) (65). The metastatic cascade for liquid tumors is also different from that of solid tumors, particularly in the early stages, where migration plays an important role (66). Cell velocity has been used as the primary indicator of motility in this manuscript due to it being straightforward. Other motility-related parameters help better understand how perturbation of the MLL1-Menin interaction modulates cell migration (fig. S2, F to H). MLL1 knockdown decreases the extent of directed cell migration and increases “randomness,” indicated by the decrease in persistence time and diffusivity (41). MLL1 inhibition/depletion also primarily affects faster moving cells rather than the bulk average cells, which narrows the spread of cell velocities observed.
MLL1-Menin interaction mediates cell motility in part via IL-6/pSTAT3/Arp3–based protrusion generation in cancer cells. IL-6 has been linked to breast cancer progression and metastasis, including in TNBC (67). Serum levels of IL-6 have been investigated as a potential prognostic marker in breast cancer, and, until the current work, the upstream epigenetic regulators of this cytokine were unknown. The identification of these upstream proteins could provide additional avenues of therapy for cancers in which IL-6 plays a key role in progression.
In addition to establishing MLL1 as an important regulator of the actomyosin machinery, we also identify MLL1 as a regulator of myosin contractility. A deeper (mode-2) inhibition of the MLL1-Menin interaction leads to repression of additional genes, including genes associated with TGF-β signaling and myosin contractility. Gli2, which we identify as a regulator of actomyosin activity, belongs to the Gli family of transcription factors that have been reported to be up-regulated and drive tumor progression in a variety of cancers including basal cell carcinomas, medulloblastomas, and gliomas (68). Gli2 promotes the establishment of bone metastasis in breast cancer via parathyroid hormone–related protein (PTHrP)–mediated osteolysis (69). However, Gli2 has not been reported to be essential for cancer cell migration or play a role in other steps of the metastatic cascade in breast cancer. Our work connects Gli2 to the well-established ROCK/pMLC2 axis in myosin-based cell contractility. Our work also highlights the potential of anti-Gli2 therapies in blocking metastasis of breast cancer. Gli2 inhibitors would not only block the spread of cancer cells from the breast but also hamper the osteolysis-induced proliferation of cancer cells that colonize the bone.
Potential clinical implications
Despite the fact that metastasis causes the vast majority of cancer-related deaths, there are currently no U.S. Food and Drug Administration–approved therapies in clinic that target metastasis directly (70, 71). Rather, they target metastasis indirectly as a by-product of primary tumor shrinkage or attempt to shrink preexisting metastatic lesions. Epigenetic drugs have garnered interest as anticancer drugs, and many are currently in clinical trials (72). Validation of MLL1-Menin interaction as a regulator of tumor metastasis in our preclinical models could add an additional therapeutic target in TNBC, the subtype of breast cancer with the most limited treatment options. TNBC was chosen as the key focus for this study due to its relatively high metastatic rate. However, as shown, these observations could also be extended to other types of cancer. As this study investigates cell migration, a key step in metastatic dissemination, patients who are at the highest risk for metastatic spread (stage II and early stage III) were the targeted demographic of this study. MLL1 is expressed in breast cancers, and higher levels of MLL1 could be indicative of invasiveness and poor prognosis if the cancer has not spread. However, the lack of MLL1 expression being a clear prognostic indicator hints at a more multifaceted role for MLL1 in tumor progression than what is illustrated here.
MLL1 inhibitors are currently in phase 1/2 clinical trials in relapsed or refractory acute myeloid leukemia (NCT04067336, NCT04065399, and NCT04811560), an indication of their capability to be used in clinical settings. As MLL1 inhibitors are already in clinical trials, repurposing these drugs for breast cancer is an attractive proposition as it saves time and is cost-effective. Most clinical regimens are administered as combinations of drugs and/or antibodies, with combination therapies being better than monotherapies (73, 74). MLL1-Menin inhibitors could be incorporated into neoadjuvant chemotherapy regimens (including those containing taxanes and anthracyclines), preventing the establishment of metastatic lesions concurrently—and independently—of primary tumor management. Once undergone breast surgery, MLL1-Menin inhibitors could be administered as a part of an adjuvant chemotherapy regimen such as sequential anthracycline-taxane regimens (75). Along with management of residual disease, MLL1-Menin inhibitors could also target the outgrowth of metastatic lesions. Our data suggest that MLL1-Menin inhibitors could be used in conjunction with standard-of-care chemotherapeutics to treat established metastasis (Fig. 7, F to I). In addition to the abovementioned timing of administration, prophylactic administration of MLL1 inhibitors could also prevent against eventual development of metastasis caused by a small fraction of remaining cancer cells that are usually undetectable by current diagnostic methods. Last, the mouse models used in this manuscript primarily metastasizes to the lung, leading to assessment of lung metastatic burden as the primary indicator of metastasis. However, since cell migration is one of the first steps in the metastatic cascade (76), MLL1 could potentially play a vital role in metastasis to the lymph nodes and other organs as well.
METHODS
Cell culture
MDA-MB-231 and BT549 were purchased from American Type Culture Collection (ATCC) and cultured with Dulbecco’s modified Eagle’s medium (DMEM) [Corning; with glucose (4.5 g/liter), l-glutamine, and sodium pyruvate] supplemented with 10% fetal bovine serum (ATCC) and 1% penicillin-streptomycin (Gibco). 4T1 cells were purchased from ATCC and cultured with RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (ATCC), 1% penicillin-streptomycin (Gibco), 1% Hepes (Gibco), 1% glucose (Gibco), and 1% sodium pyruvate (Sigma-Aldrich). HT-1080 cells were purchased from ATCC and cultured with DMEM supplemented with 10% fetal bovine serum and 0.1% gentamycin (VWR). SUM149 (SUM149PT) and SUM159 (SUM159PT) cells (BioIVT) were cultured with DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. MCF10A cells were purchased from ATCC and cultured with MEGM Mammary Epithelial Cell Growth Medium BulletKit (Lonza/Clonetics) supplemented with not supplemented with cholera toxin (100 ng/ml; Millipore) and no GA-1000. These cells were cultured at 37°C and 5% CO2 for passage numbers less than 25 (15 for HT-1080) and tested for mycoplasma contamination every 6 months.
MLL1, MEN1, STAT3, and GLI2 depletion by shRNA
MLL1 was depleted using two separate shRNA constructs (MilliporeSigma):
1) Gene: KMT2A; TRC number: TRCN0000234741; target sequence: GATTATGACCCTCCAATTAAA (KD1 and KD2).
2) Gene: KMT2A; TRC number: TRCN0000005954; target sequence: GCACTGTTAAACATTCCACTT (KD3).
MEN1 was depleted using the shRNA construct (MilliporeSigma): gene: MEN1; TRC number: TRCN0000040141; target sequence: GCTGCGATTCTACGACGGCAT. STAT3 was depleted using the shRNA construct (MilliporeSigma): gene: STAT3; TRC number: TRCN0000329887; target sequence: GTTCCTGAACATGATGACCTA. GLI2 was depleted using the shRNA construct (MilliporeSigma): gene: GLI2; TRC number: TRCN0000033331; target sequence: GCACAATCTACGAAGAATCAA.
The bacterial glycerol stocks were grown overnight in sterile LB broth (Sigma-Aldrich) with ampicillin (100 μg/ml; Sigma-Aldrich). Plasmid was extracted from the bacteria using a midi prep kit (MACHEREY-NAGEL) per the manufacturer’s instructions. The shRNA construct/plasmid, along with pMD2.G [pMD.G vesicular stomatitis virus glycoprotein (VSV-G)] and pCMVDR8.91 (encoding Gag, Pol, Tat, and Rev), was cotransfected using Lipofectamine 3000. 293 T cells were transfected with 10 μg of shRNA, 5 μg of pCMVDR8.91, and 1 μg of pMD.G VSV-G. The viral harvest was collected 48 hours after transfection and filtered through a 0.45-μm filter. MDA-MB-231 cells were incubated for 30 min with DMEM supplemented with polybrene at a final concentration of 8 μg/ml. After incubation, the medium was replaced with fresh DMEM-polybrene, and 1 ml of filtered lentivirus was added (two separate replicates with one shMLL1 construct each). Cells were incubated with the lentivirus for 24 hours. Cells were selected with puromycin (2 μg/ml) 72 hours after transduction. Knockdowns were verified via Western blotting.
3D collagen gels and overnight live-cell tracking
Cells were trypsinized, counted, and resuspended in a 1:1 mixture of fresh medium and reconstitution buffer [2.2% (w/v) NaHCO3 (Sigma-Aldrich) and 4.8% (w/v) Hepes (Acros) in deionized water] at precalculated cell concentrations. High-concentration rat tail collagen I (Corning) was added and was neutralized immediately with 1 N of NaOH. The solution was mixed thoroughly but gently (to avoid inducing air bubbles) and plated into wells preheated at 37°C. Collagen gels were allowed to polymerize in the incubator for 1 hour, following which fresh medium was added on top of the gels. The final cell density was 100 cells/mm3, and the collagen concentration in the gel was 2 mg/ml, unless specified otherwise. For LD, cells were embedded at 10 cells/mm3.
Cell migration was assessed by overnight live-cell tracking. As proliferation can influence “apparent” cell migration in vitro, traditional cell migration assays (including transwell and wound healing) preclude a direct ability to dissociate the contributions of proliferation from measured invasion. Our live-cell microscopy platform (40, 41) enables us to track the migration of individual cells, without the potential confounding effect of cell proliferation. In addition, we have used 3D techniques for more accurate representations of the microenvironment compared to flat culture dishes. Cells were maintained at 37°C and 5% CO2 using a live-cell box (Pathology Devices Inc.) and imaged using a Cascade 1K charge-coupled device camera (Roper Scientific) mounted on a Nikon Eclipse TE200-E. Phase-contrast images were recorded with a 10× objective every 10 min for a time frame of 20 hours. Cells were tracked using Metamorph imaging software (Molecular Devices). A minimum of 100 cells were tracked (60 cells for LD) per condition. Migration parameters such as diffusivity, persistence time, and persistence speed were computed from measured cell trajectories using custom MATLAB codes (40). Cell velocity was calculated to obtain a more practical measure of motility and was calculated as follows
where ΔMSD is the absolute value of the difference in MSD between a time interval of T. To avoid errors introduced because of a non–steady-state system, a timescale was chosen that was longer than the persistence time but shorter than the total experiment time. T was set as half of the time length of experiment. For a total of N time points imaged, N/2 velocities were obtained (one each for the velocity between time points ti and tN/2+I; I ranging from 1 to N/2) that were then averaged to attain the reported velocities.
Generation of double-layered tumor spheroids
Tumor spheroids were prepared by immersing a core of cells in Matrigel into collagen gel (2 mg/ml) as described in (44). Briefly, 50,000 cells were suspended in 1 μl of Matrigel (Corning), pipetted into mineral oil (Sigma-Aldrich) to form a sphere, and incubated at 37°C for 1 hour to polymerize the Matrigel. An outer layer of collagen was added by dipping the Matrigel cores in cold liquid collagen mixture (described in previous section) and repeating the abovementioned steps again. This architecture of a Matrigel core and an external collagen I matrix better mimics physiological conditions experienced by cancer cells and models two critical steps in metastasis, i.e., the invasion of cancer cells through the basement membrane, followed by migration in the collagen-rich stromal matrix. These double-layered spheroids were added to 96 wells with 100 μl of fresh medium. Scrambled control and shMLL1 spheroids were imaged at ×4 magnification using phase-contrast microscopy.
Drug treatments and cytokine supplementations
MI-2-2 was purchased from Millipore; MI-503, VTP50469, S3I-201, Y-27632, paclitaxel, and doxorubicin were purchased from Selleckchem; MI-3454 was purchased from MedChemExpress; OICR-9429, EPZ-5676, MI-nc, and GANT-61 were purchased from Cayman Chemical; and blebbistatin was purchased from Sigma-Aldrich. These drugs were dissolved in dimethyl sulfoxide (DMSO) (except doxorubicin in water) and used at the concentrations shown in table S4. Untreated (control) cells were treated with an amount of DMSO corresponding to the highest concentration of drug that the cells were treated with (and, hence, the greatest volume of DMSO).
IL-6, IL-8, and TGF-β1 were purchased from R&D Systems and reconstituted according to the manufacturer’s instructions. IL-6 and IL-8 were supplemented at 375 and 150 pg/ml, respectively. These correspond to the levels that these cytokines are expressed when embedded in 3D collagen gels at 100 cells/mm3 (3). IL-8 levels were only marginally reduced with MLL1-Menin inhibition, but it was also supplemented to avoid being a bottleneck following IL-6 supplementation. For the 3× IL-6/8 rescue experiments, IL-6 and IL-8 were supplemented at 1125 and 450 pg/ml, respectively. TGF-β1 was supplemented at 10 ng/ml. The concentration reported in literature for cells on 2D matrices is 5 ng/ml (77). However, TGF-β1 has been reported to adsorb strongly to collagen matrices with over 6 ng/ml (of the initial 10 ng/ml) getting adsorbed (78). Hence, 10 ng/ml was supplemented to have ~4 ng/ml available to cells.
Once cells were embedded in 3D collagen gels, cells were allowed to polarize and acclimate to the gel for 12 hours. After this time, medium was exchanged for fresh medium containing the appropriate drugs and/or cytokines at the concentrations indicated above. Cells were incubated with the drugs for ~18 hours before being imaged overnight with live-cell tracking. Total drug treatment/cytokine supplementation time for RNA or protein level analysis was 2 days unless stated otherwise.
Assessing any potential off-target effects of drug treatment
The usage drugs as a primary method to elucidate the mechanism of MLL1-Menin–mediated regulation of cell migration and proliferation necessitate the confirmation that observed effects and delineated mechanisms are not partly or entirely due to off-target effects of the drug. The various steps taken to ensure the fidelity and robustness of these mechanisms include the following:
1) Choosing drug dosages that are consistent with that reported in literature. Drug dosages for the key drugs (MLL1-Menin inhibitors MI-2-2, MI-503, MI-3454, and VTP50469) were chosen in line with those reported in top journals (~10 to 20 references each). The top reference for choosing the dosage of each drug is presented in the previous section.
2) Using four-well characterized and highly cited MLL1-Menin inhibitors. We have used MI-2-2, MI-503, VTP50469, and MI-3454 (Fig. 1E and fig. S4, A, C, and E, respectively), which span three generations, are synthesized by different groups, and are highly cited. All four MLL1-Menin inhibitors lead to identical cellular responses in migration, proliferation, apoptosis, and gene expression,
3) Using MI-nc as a negative control (fig. S4F). MI-nc has a similar structure to MI-2-2 but binds much more weakly to MLL1-Menin and does not inhibit MLL1-Menin interaction. MI-nc treatment at concentrations identical to MI-2-2 had no effect on migration.
4) Validation of key proteins with knockdowns. MLL1 (fig. S2E), Menin (fig. S2I), and its key downstream effectors STAT3 (fig. S5N) and GLI2 (fig. S7G) were depleted using shRNA, and these cells were subjected to migration- and mechanism-related studies.
5) Testing the effect of MLL1-Menin inhibition on a normal noncancerous breast epithelial cell line, MCF10A (fig. S4, G to I). These cells were less susceptible to inhibition of proliferation and migration following MLL1-Menin inhibition.
6) Testing the impact of MLL1-Menin inhibitors on shMLL1 cells. MLL1-Menin inhibition did not further reduce the motility of shMLL1 cells (fig. S4M), indicating that MLL1-Menin inhibitors had no effect when MLL1 was not present in cells.
7) Confirming full reversibility of pharmacological inhibition of MLL1 on proliferation and migration (figs. S4, K and L, and S8, E and F). Withdrawal of drugs lead to the recommencement of cell migration and proliferation within 2 days, the same period of time needed for these drugs to repress migration and proliferation.
8) The absence of drug-induced apoptosis (fig. S4J). Reduced cell proliferation by MLL1-Menin was not accompanied by an increase in cellular apoptosis.
These data in addition to the extensive catalog of literature (see point 1) makes it highly unlikely that the observed effects are due to off-target effects.
In addition, note that drug availability is much more limited in 3D collagen matrices compared to a regular 2D system. All the reference articles used a guide to choosing the optimal concentration of drugs have been performed in 2D systems in which the concentration of drugs apparent to the cells is the same as the concentration of drugs at which it was added. However, for greater physiological relevance, we embed our cells in a 3D collagen I gel. Collagen is reported to be hydrophobic, leading to adsorption of hydrophobic molecules in the gel including hydrophobic drugs and proteins (see determination of TGF-β1 concentrations for supplementation in previous section). A similar effect would be in play regarding the drugs used in this manuscript, especially MLL1 inhibitors (which is a highly hydrophobic and insoluble in water), wherein the apparent concentration of the drugs felt by the cells would be lower than the reported concentration (it would be proportional to the amount of drugs added—amount adsorbed by gels).
Quantitative reverse transcription polymerase chain reaction
Cells were obtained by digesting the 3D collagen gels using collagenase. Gels were chopped into smaller pieces to increase their surface area and incubated with 1:1 ratio of collagen to type I collagenase (Gibco) dissolved in phosphate-buffered saline (PBS) at 2 mg/ml. After 1 hour, the solution was strained through a 40-μm strainer to remove any undigested collagen.
RNA was extracted using TRIzol (Thermo Fisher Scientific) and ethanol (Thermo Fisher Scientific) using a Direct-zol Mini Prep kit (Zymo). RNA concentration was measured using a NanoDrop spectrophotometer, and 1 μg of RNA was reverse-transcribed using iScript cDNA Synthesis Kit (Bio-Rad). Primers sequences were obtained from Harvard PrimerBank (sequences are listed in table S5) and synthesized by Integrated DNA Technologies. SYBR Green Master Mix (Bio-Rad) was used for fluorescence, and the reaction mixture was amplified (40 cycles) using the CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Gene expression was analyzed using Bio-Rad CFX Manager v3.1.
Immunofluorescence
Immunofluorescence was carried out as described in (79). Briefly, cells were seeded in six-well plates at a density of 10,000 cells/cm2. After drug treatment, cells were fixed using 4% paraformaldehyde (Thermo Fisher Scientific; diluted from 16%) for 10 min, followed by three PBS (Corning) washes for 5 min each. Cells were then permeabilized with 0.5% Triton X-100 (Thermo Fisher Scientific) for 10 min, followed by three PBS washes. Blocking was performed for 30 min using 5% bovine serum albumin (BSA) (Sigma-Aldrich), followed by overnight incubation with primary antibody [pMLC2 (Ser19) #3671, Cell Signaling Technology; 1:100] at 4°C. The next day, cells were washed three times with PBS and incubated with the secondary antibody (goat anti-rabbit Alexa Fluor 488, Thermo Fisher Scientific; 1:1000) and Alexa Fluor 568 Phalloidin (Thermo Fisher Scientific; for visualizing actin, 1:100) for 2 hours at room temperature. The solution was aspirated, and the cells were washed three times with PBS. DNA was stained using Hoechst 33342 (final concentration, 10 μg/ml; Thermo Fisher Scientific) for 10 min, followed by three PBS washes. The stained cells were visualized with a Nikon A1 confocal microscope using a Plan Fluor 100× oil objective. Images were analyzed using ImageJ (National Institutes of Health).
Western blotting
For assessment at the protein level, gels were digested, and lysates were obtained from the cell pellet using radioimmunoprecipitation assay buffer (Cell Signaling Technology) supplemented with 1 mM phenylmethylsulfonyl fluoride (Roche), protease inhibitor cocktail (Sigma-Aldrich), phosphatase inhibitor cocktail 3 (Sigma-Aldrich), and 4% SDS (Quality Biological; diluted from 20%). The lysate was sonicated (five short pulses at 30% power using Branson Sonifier 250), centrifuged at 10,000 rcf, and heated at 100°C for 5 min. Protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific), and 30 μg of protein was loaded per well/condition (unless stated otherwise). Lysates were loaded onto a 4 to 15% Mini-PROTEAN gel (Bio-Rad) and transferred onto a polyvinylidene difluoride membranes using a Trans-Blot Turbo transfer system (Bio-Rad). Membranes were blocked with 5% BSA (Sigma-Aldrich) in 0.1% Tween–tris-buffered saline (Bio-Rad) and incubated with primary antibodies. Primary antibodies were purchased from Cell Signaling Technology [β-actin–horseradish peroxidase (HRP), #5125; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), #5174; MLL1 C-terminal antigen, #14197; Menin, #6891; pStat3 (Tyr705), #9145; Stat3, #30835; ARP2, #5614; ARP3, #4738; pMLC2 (Ser19), #3671; pMLC2 (Thr18/Ser19), #3674; MLC2, #3672; tri-methyl-histone H3 (Lys4), #9751; ROCK1, #4035; ROCK2, #8236; NF-κB p65, #8242; and NF-κB1 p105/p50, #3035] or Novus Bio (Gli-2, #NB600) and used at the manufacturer’s recommended concentration for Western blotting. After overnight incubation in primary antibodies, membranes were washed with Tween–tris-buffered saline and incubated in secondary antibodies (anti-rabbit immunoglobulin G, HRP-linked antibody, #7074; or anti-mouse immunoglobulin G, HRP-linked antibody, #7076; both Cell Signaling Technology) for 1 hour. Bound antibodies were detected with ECL Western blotting substrate (Genesee) and imaged using a ChemiDoc imaging system (Bio-Rad). For total proteins corresponding to their phosphorylated forms, blots were stripped using Restore Western blot Stripping Buffer (Thermo Fisher Scientific) per the manufacturer’s recommendation, reblocked, and reprobed using the appropriate primary antibodies. Blot images were analyzed in ImageJ.
Enzyme-linked immunosorbent assay
IL-6 secretion was measured using a human IL-6 quantikine enzyme-linked immunosorbent assay kit (R&D Systems, #D6050) per the manufacturer’s instructions.
Protrusion analysis
Cell body and protrusion segmentation method
The method to extract morphological measures from the phase-contrast time-lapse videos consists of three steps: (i) cell body and protrusion segmentation, (ii) cell tracking, and (iii) merge of the tracks and the segmentations to calculate the morphological measures for each cell along the time dimension.
Cell body and cell protrusion segmentation
To segment each of the frames in the phase-contrast microscopy videos, we trained a U-Net–shaped convolutional neural network (80) with manually annotated videos. To build the training set, we extracted 25 short videos of 15 continuous temporal points from the MDA-MB-31 dataset. Then, we annotated those videos with a label of 1 for the cell body and a label of 2 for the protrusions (fig. S15, C to E). To fit the input size of the network, we cropped patches from each of the annotated frames where there was a cell either focused or nonfocused. Overall, we obtained a set with 5159 image patches from which 4642 and 516 were used as training and validation sets.
Our U-Net has four levels in the encoding path. Each level is composed of two 2D convolutional layers and a 2D max-pooling. As described in (80), the convolutional layers on each of the four levels have a size of 32-64-128-256 convolutional filters, respectively. These are then followed by a bottleneck with two convolutional layers of 512 convolutional filters. The decoding path is analog to the encoding path with 2D upsamplings instead of max-poolings. Each level of the encoding path has a skip connection to the corresponding level in the decoding path. All the convolutional layers have a kernel of size of 3 × 3 and Rectified Linear Units (ReLU) as activation function. The last convolutional layer of the U-Net is followed by a convolutional layer with a kernel of size of 1 × 1, one convolutional filter, and the sigmoid activation function. In our case, the input to the U-Net was a series of the temporal frames (t − 1, t, t + 1) to output the binary mask of the cell body and protrusions at the time point t. Therefore, the size of the images entering the U-Net was 320 pixels × 320 pixels × 3 pixels, and the output was 320 pixels × 320 pixels × 2 pixels. The training was conducted over a pretrained network. The pretrained weights were obtained from processing phase-contrast microscopy images of fibroblasts. We fine-tuned our network for 200 epochs, with a batch size of 10 and a decreasing learning rate starting from 5 × 10−6. The categorical cross-entropy was chosen as the loss function. At the end, we obtained a constant loss value of 4.1166 × 10−4 and an accuracy of 0.9880 for the training set and a loss value of 0.007 and an accuracy of 0.9878 for the validation set. We used the trained network to process the MDA-MB-231 image datasets. Each channel of the U-Net’s output was binarized with the threshold of 0.5. Then, we ensured that all the pixels segmented as cell bodies were not segmented as protrusions and that all protrusions corresponded to a cell body. Those cells with a size smaller than 127 μm2 and all the protrusions with a size smaller than 6 μm2 were discarded. These values were estimated from the manual annotations.
To get the temporal morphodynamics of the cells, cells were tracked using MetaMorph software (Molecular Devices, Sunnyvale, CA, USA). The obtained tracks were merged with the previous masks by filtering out all those segmentations that did not have a track. We revised all the final masks and tracks to correct for those spurious segmentations and reduce the noise in the results.
We computed the following morphodynamics metrics for each cell and time point t in the video: MSDs (in micrometers), number of protrusions, and the length of the longest protrusion. MSD was computed as the average of the squared displacements made in a given time window (i)
where x(t) represents the centroid of the cell body in Cartesian coordinates at the time point t. The MSD value is expected to increase with i when the cells express a persistent behavior or travel long distances. For each cell, we calculated the number of protrusions using the mask of the protrusions assigned to that cell. The number of protrusions [P(t)] was calculated as the number of connected components in the protrusions mask. The cumulative number of protrusions at each time point was calculated as
The length of each protrusion—each connected component—was given by the length of its skeleton. Then, the maximum length of each protrusion at each time point [L(t)] was calculated. Last, the cumulative maximum protrusion length is given as
As cells enter and exit the plane of focus, their segmentation can be missing for certain time points. In those cases, we averaged each variable considering their value in the former and the latter time points.
Multiplex cytokine assay
To perform profiling of secreted cytokines in the CM, multiplexed antibody barcode microarray chip was prepared as described previously (81). Briefly, a polydimethylsiloxane (PDMS; Momentive, USA) template was combined onto poly-l-lysine–coated glass slide, and capture antibodies (table S6) were flowed into guiding channels on the PDMS template for overnight at room temperature. After detaching the PDMS template, 3% BSA (MilliporeSigma, USA) in PBS solution was introduced onto surface of capture antibody-coated glass slide, and then glass slides were kept in −20°C fridge.
For running assay, the glass slide was overlaid with PDMS slab containing 24 sample-loading chambers, which were prepared by biopsy punch (3 mm in diameter; Ted Pella, Sweden). Twenty microliters of conditioned medium was added into the loading chambers, followed by incubation for 12 hours at 4°C. A cocktail of biotinylated detection antibodies (table S6) was introduced for 45 min at room temperature after detaching PDMS slab and rinsing three times with 3% BSA. Then, allophycocyanin (5 μg/ml)–labeled streptavidin (eBioscience, USA) was added. After 20 min of incubation at room temperature, the glass slide was again rinsed three times with 3% BSA, followed by blocking with 3% BSA for 30 min at room temperature. The glass slide was dried using nitrogen gas after dipping into 100% Dulbecco’s PBS, 50% Dulbecco’s PBS, and deionized water sequentially.
Scanning fluorescence images were obtained by using Genepix 4200A scanner (Molecular Devices, USA) with fluorescein isothiocyanate (488 nm) and allophycocyanin (635 nm) channels, and Genepix Pro software (Molecular Device, USA) was used to analyze and export fluorescence intensities.
Cell number measurement and proliferation assay
Cell numbers were counted by digesting the 3D collagen gels (described in previous section) and counting the number of cells using Trypan Blue (Thermo Fisher Scientific) stain. Cell proliferation (for kinetics) was determined using an MTT assay (Thermo Fisher Scientific) as described in (82). A total of 5000 cells were plated per well in 96-well plates. Once the cells had attached to the bottom, the existing medium was exchanged for fresh medium containing drugs. At set time points (0, 1, 2, and 3 days), medium was aspirated, and cells were incubated with a 100:11 mix of fresh medium and MTT solution (5 mg/ml) in PBS. After 3 hours of incubation, medium was aspirated, DMSO was added to solubilize the crystals, and absorbance was read at 550 nm. Apoptosis was measured using a propidium iodide kit (Abcam) per the manufacturer’s instructions.
RNA-seq: Sample preparation, sequencing, and analysis
RNA isolation, library preparation, and sequencing were performed by Quick Biology. Total RNA was isolated and deoxyribonuclease I–treated using QIAGEN RNeasy Plus Mini Kit (QIAGEN, Valencia, CA). The RNA integrity was checked by Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA), and samples with clean ribosomal RNA peaks [RNA integrity number (RIN) > 6] were used for library preparation. RNA-seq library was prepared according to KAPA mRNA HyperPrep kit with 200– to 300–base pair insert size (Roche, Wilmington, MA) using 250 ng of total RNAs as input. Final library quality and quantity were analyzed by Agilent Bioanalyzer 2100 and Life Technologies Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA). Paired-end reads (150 base pairs) were sequenced on Illumina HiSeq 4000 sequencer (Illumina Inc., San Diego, CA).
Transcript level counts were quantified from reads with salmon version 1.1.0 using human genome build h38. Quantification files generated by salmon were loaded into R/Bioconductor software. Data analysis was performed using with custom routines and standard packages. We perform paired differential expression analysis that retain cell line pairing using a multifactor design using the tximport (v 1.14.2) and DESeq2 (v 1.26.0) packages. The only protein coding genes were used. Genes with false discovery rate–adjusted P values below 0.05 and log fold change greater than 1 were called statistically significant. Pathway analysis was performed by the msigdbr (v 7.1.1) and fgsea (v 1.12.0) packages using hallmark pathways from the MSigDB version 7.0 (83). Volcano plot were created using the ggplot2 package (v 3.3.2), and heatmaps were created using the pheatmap R package (v 1.0.12). Venn diagrams were created by ggVennDiagram (v 0.3). Identification of significant Hallmark gene sets following RNA-seq–based gene clustering was done by EnrichR (https://maayanlab.cloud/Enrichr/). Relevant gene sets were sorted on the basis of their adjusted P value.
TCGA analysis
RNA-seq V2 data for basal breast cancer tumors from the PanCanAtlas (84) initiative of TCGA were used for analyses. Gene set statistics were computed with hallmark and curated gene sets from MSigDB and MLL1 expression using a one-sided Wilcoxon gene set test in limma (v 3.52.4). Gene sets with false discovery rate–adjusted P values below 0.05 were considered significantly enriched.
ChIP-Atlas analysis
ChIP-Atlas (56) was used to acquire ChIP-seq data and viewed in Integrative Genomics Viewer (ver. 2.10.0.06). ChIP-Atlas peak browser was used with the following parameters for hg38 data: antigen class as transcription factors and others, cell type class as all cell types, threshold for significance as 50, and antigen as KMT2A (MLL1), MEN1 (Menin), or WDR5. Cell type class was set as “all cell types” to ensure sufficient KMT2A/MEN1/WDR5 studies with breast samples alone (<5 each). Binding to a region of 2 kb flanking the TSS of each gene was considered as relevant. All cell types were chosen—as opposed to breast cancer cell lines only—because of the dearth of datasets that performed ChIP-seq for MLL1/KMT2A on breast cancer cell lines.
In vivo mouse modeling
All in vivo studies were performed in accordance with protocols approved by the Johns Hopkins University Animal Care and Use Committee (ACUC).
MLL1 knockdown tumors
Five- to 7-week-old female nonobese diabetic–severe combined immunodeficient gamma (NSG) mice were purchased from Johns Hopkins Medical Institution and housed under a 12-hour dark/light cycle at 25°C. One million MDA-MB-231 cells were suspended in a 1:1 mixture of PBS and Matrigel at 107 cells/ml. A total of 100 μl of cell suspension was injected into the second nipple of the left mammary fat pad to form orthotopic breast tumors. Tumor volume (measured with calipers) and body weight were monitored every 2 to 3 days. For survival study, mice were euthanized when they reached the maximum tumor size permitted in ACUC protocol.
Set tumor size threshold end-point studies
For tumor size control study, NSG mice were euthanized when their primary tumors reached 1400 mm3. Scrambled control and WT tumors grew faster and at an identical pace, and these mice were euthanized on days 41 and 43, respectively. shMLL1 tumors grew slower, and shMLL1 tumor–bearing mice were euthanized 62 days after tumor establishment. The lungs were excised, formalin-fixed, paraffin-embedded, sectioned, and stained with H&E. Lungs were imaged by the Johns Hopkins Oncology Tissue Services core. Lesion size and number were quantified using ImageJ (National Institutes of Health).
Tail vein injection of to assess extravasation and colonization
MDA-MB-231 shMLL1, scrambled control, or WT cells by were injected via the tail vein (500,000 cells per injection) in NSG mice (85). A total of 100 μl of cell suspension in PBS was injected into the tail vein of a mouse using a 30-gauge syringe. Saline with no cells was the negative control. Mice were euthanized 6 weeks after tumor establishment, and the lungs were excised, fixed, H&E-stained, and imaged.
MLL1 knockdown plus paclitaxel treatment
shMLL1 or scrambled control tumors were established in NSG mice as described above. Four weeks after tumor establishment, mice bearing both tumor types were randomly assigned to paclitaxel or vehicle treatment. Paclitaxel was administered at 25 mg/kg (86) by solubilizing in a vehicle [50% saline, 25% PEG-400 (Sigma-Aldrich), and 25% paclitaxel in DMSO]. Mice were injected intraperitoneally five times every 3 days. All mice were euthanized 4 days after the last injection, and their tumors were excised and weighed. The lungs were excised, formalin-fixed, H&E-stained, and fixed.
MI-503 plus paclitaxel treatment in syngeneic spontaneous metastasis model
Six-week-old female BALB/c mice were purchased from the Jackson laboratories and housed under a 12-hour dark/light cycle at 25°C. Mice were allowed to acclimate to the facility for a week before establishment of orthotopic breast tumors via the injection of 4T1 mouse TNBC cells (7000 cells/100 μl of injection, suspended in a 1:1 mixture of PBS: Matrigel at 7 × 104 cells/ml) as described above. Tumor volume (measured with calipers) and body weight were monitored every day. Nine days after tumor establishment, the mice were randomized and assigned to one of the treatment groups: vehicle (formulated as described above), paclitaxel (15 mg/kg), MLL1-Menin inhibitor MI-503 (30 mg/kg), or a combination of paclitaxel and MI-503. Half of the previously reported maximum tolerated doses (MTDs) were chosen for paclitaxel and MI-503 to avoid toxicity in combination-treated mice. Mice were injected intraperitoneally seven times every alternate day. Mice were euthanized 23 days after tumor establishment, when the tumors in the control (vehicle) group reached the maximum permitted tumor size of 1500 mm3. Tumor and organs were harvested from the mice as described above and subjected to the same analysis.
MI-503 plus paclitaxel treatment in syngeneic tail vein metastasis model
The efficacy of MLL1-Menin inhibition on established metastasis was evaluated using a tail vein forced metastasis model. This model was preferred to other metastasis models (such as spontaneous metastasis from a primary tumor followed by resection of tumor) as 4T1-BALB/c model metastasizes poorly (forms far fewer metastases compared to other metastatic cell lines, as observed in the previous section) and has rapid primary tumor growth (necessitating early resection, often within 3 weeks). Hence, a resection model might not have sufficient number of metastatic colonies to treat.
Six-week-old female BALB/c mice were purchased from the Jackson laboratories and housed as described above. After a week, all mice were injected with 4T1 mouse TNBC cells via the tail vein (150,000 cells/150 μl PBS) (85) using a 30-gauge syringe. Mouse body weight was monitored every other day. Seven days after tail vein injection of cells, the mice were randomized and assigned to one of the treatment groups: vehicle, paclitaxel (15 mg/kg), MLL1-Menin inhibitor MI-503 (30 mg/kg), or a combination of paclitaxel and MI-503. Mice were injected intraperitoneally eight times and euthanized 28 days after initial tail vein injection. Lungs were harvested from the mice as described above and subjected to the same analysis. Some lung lobes could be perfused with agarose due to extensive metastases, which prevented the flow of agarose solution into the lungs.
Immunohistochemistry
IHC was performed by Oncology Tissue Services Core (The Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins).
Ki67
Immunolabeling for Ki67 was performed on formalin-fixed, paraffin-4embedded sections on a Ventana Discovery Ultra autostainer (Roche Diagnostics). Briefly, following dewaxing and rehydration on board, epitope retrieval was performed using Ventana Ultra CC1 buffer (catalog no. 6414575001, Roche Diagnostics) at 96°C for 48 min. Primary antibody, anti-Ki67 (1:200 dilution; catalog no. Ab16667, lot number GR3185488-1, Abcam), was applied at 36oC for 60 min. Primary antibodies were detected using an anti-rabbit hydroquinone (HQ) detection system (catalog nos. 7017936001 and 7017812001, Roche Diagnostics), followed by ChromoMap DAB IHC detection kit (catalog no. 5266645001, Roche Diagnostics), counterstaining with Mayer’s hematoxylin, dehydration, and mounting.
pSTAT3
Immunostaining was performed at the Oncology Tissue Services Core of Johns Hopkins University School of Medicine. Immunolabeling for pSTAT3 was performed on formalin-fixed, paraffin-embedded sections on a Ventana Discovery Ultra autostainer (Roche Diagnostics). Briefly, following dewaxing and rehydration on board, epitope retrieval was performed using Ventana Ultra CC1 buffer (catalog no. 6414575001, Roche Diagnostics) at 96°C for 64 min. Primary antibody, anti-pSTAT3 (1:100 dilution; catalog no. 9145L, lot number 22, Cell Signaling Technology), was applied at 36°C for 60 min. Primary antibodies were detected using an anti-rabbit HQ detection system (catalog nos. 7017936001 and 7017812001, Roche Diagnostics), followed by ChromoMap DAB IHC detection kit (catalog no. 5266645001, Roche Diagnostics), counterstaining with Mayer’s hematoxylin, dehydration, and mounting.
pMLC2
Immunostaining was performed at the Oncology Tissue Services Core of Johns Hopkins University School of Medicine. Immunolabeling for pMLC2 was performed on formalin-fixed, paraffin-embedded sections on a Ventana Discovery Ultra autostainer (Roche Diagnostics). Briefly, following dewaxing and rehydration on board, epitope retrieval was performed using Ventana Ultra CC1 buffer (catalog no. 6414575001, Roche Diagnostics) at 96°C for 48 min. Primary antibody, pMLC2 (1:50 dilution; catalog no. 3671, lot number***, Cell Signaling Technology), was applied at 36°C for 60 min. Primary antibodies were detected using an anti-rabbit HQ detection system (catalog nos. 7017936001 and 7017812001, Roche Diagnostics), followed by ChromoMap DAB IHC detection kit (catalog no. 5266645001, Roche Diagnostics), counterstaining with Mayer’s hematoxylin, dehydration, and mounting.
Quantification of pMLC2 at tumor periphery and bulk
Quantification of pMLC2 IHC signal on tumor sections was performed using ImageJ. pMLC2 intensity ratio between the tumor edge and adjacent bulk was calculated as
Statistics
All in vitro experiments were repeated three times (n = 3) as biological repeats. Measurement for each replicate was performed on distinct samples, and representative experiments are shown in figures. Each data point is represented by an individual dot where possible. Barplots represent means ± SEM. For box-and-whisker plots, the upper limit of the box represents the 75th percentile, lower limit represents the 25th percentile, and the center line represents the median. The top and bottom whiskers stretch to the 99th and 1st percentiles, respectively. Experiments were analyzed using one-way analysis of variance (ANOVA) [wherever number of conditions ≥ 3, following output parameters in figure captions—F(DFn, DFd) and P value]. After confirming significance by ANOVA, significance between select key conditions were also analyzed by two-tailed t test (results depicted in figures, output parameters in figure captions). P < 0.05 was considered significant (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; NS, not significant). Paired t test was used where suitable (e.g., in rescues like Fig. 2M). All figure plotting and statistical calculations were performed using GraphPad Prism.
All migration and proliferation experiments were performed in technical replicates of 4 (N = 4) per condition. For migration, a minimum of 80 cells were tracked (40 cells for LD) per condition. The exact number of cells per condition is noted in the Supplementary Materials. Spheroid experiments were performed with a minimum of N = 6 per condition and n = 3. Quantitative reverse transcription PCR was performed with N ≥ 4 per gene.
All in vivo experimental conditions were performed with a minimum of five mice bearing one tumor each (N = 5). This N value provides a 73% power to detect a difference in mean tumor size between control and treated/shMLL1 groups. A priori power analysis was performed using t tests in G*Power 3.1.9.2 software using the following parameters: alpha as 0.05, power of 0.65, allocation ratio (N2/N1) as 1, effect size (ρ) as large, and analysis with one-tailed t test. For in vivo drug treatments, mice were randomized before initiation of treatment. For survival study, significance, hazard ratios, and 95% confidence intervals were calculated via log-rank (Mantel-Cox) test.
Acknowledgments
We are grateful to H. Easwaran, I. Godet, V. Rebecca, and P.-H. Wu for reading and editing the manuscript.
Funding: This work was supported by grants from the National Cancer Institute (U54CA143868 and U54CA268083), the National Institute on Aging (U01AG060903) and National Institutes of Health (U54AR081774) to D.W.; E.G.-d.-M. and A.M.-B. are grateful to Ministerio de Ciencia, Innovacion y Universidades, Agencia Estatal de Investigacion, under grants TEC2015-73064-EXP and PID2019-109820RB-I00, MINECO/FEDER, EU, cofinanced by European Regional Development Fund (ERDF), “A way of making Europe” to A.M.-B.; BBVA Foundation under a 2017 Leonardo Grant for Researchers and Cultural Creators to A.M.-B. They also acknowledge the support of NVIDIA Corporation with the donation of the Titan X (Pascal) GPU used for this research. E.G.-d.-M. acknowledges the support of the Gulbenkian Foundation (Fundação Calouste Gulbenkian) and the European Molecular Biology Organization (EMBO) Postdoctoral Fellowship (EMBO ALTF 174-2022).
Author contributions: Writing—original draft: P.R.N. and D.W. Conceptualization: P.R.N. and D.W. Investigation: P.R.N. and L.D. Writing—review and editing: P.R.N., D.K., R.F., A.M.-B., E.J.F., and D.W. Resources: R.F. and A.M.-B. Funding acquisition: R.F., A.M.-B., and D.W. Validation: P.R.N., L.D., and D.W. Data curation: E.G.-d.-M. Supervision: R.F., A.M.-B., and D.W. Formal analysis: P.R.N., L.D., E.G.-d.-M., A.M.-B., and E.J.F. Software: P.R.N., L.D., E.G.-d.-M., A.M.-B., and E.J.F. Project administration: P.R.N., R.F., and D.W. Visualization: P.R.N., L.D., E.G.-d.-M., R.F., E.J.F., and D.W.
Competing interests: One of the authors, R.F., is a shareholder in AtlasXomics. E.J.F. is on the Scientific Advisory Board for Resistance Bio and received consulting fees from Mestag Therapeutics and Merck. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq dataset generated in this study is publicly available online in the GEO database under accession number GSE180227.
Correction (19 April 2024): Shortly after publication, the authors alerted the Editorial Office to an omitted funding source. The funding source, National Institutes of Health grant U54AR081774, awarded to D.W., has been added to the Acknowledgments. The HTML and PDF versions of the paper have been corrected.
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
Tables S1 to S6
References
REFERENCES AND NOTES
- 1.Yamaguchi H., Wyckoff J., Condeelis J., Cell migration in tumors. Curr. Opin. Cell Biol. 17, 559–564 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Ganesh K., Massagué J., Targeting metastatic cancer. Nat. Med. 27, 34–44 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jayatilaka H., Tyle P., Chen J. J., Kwak M., Ju J., Kim H. J., Lee J. S. H., Wu P.-H., Gilkes D. M., Fan R., Wirtz D., Synergistic IL-6 and IL-8 paracrine signalling pathway infers a strategy to inhibit tumour cell migration. Nat. Commun. 8, 15584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgouli M., Herraiz C., Crosas-Molist E., Fanshawe B., Maiques O., Perdrix A., Pandya P., Rodriguez-Hernandez I., Ilieva K. M., Cantelli G., Karagiannis P., Mele S., Lam H., Josephs D. H., Matias-Guiu X., Marti R. M., Nestle F. O., Orgaz J. L., Malanchi I., Fruhwirth G. O., Karagiannis S. N., Sanz-Moreno V., Regional activation of myosin II in cancer cells drives tumor progression via a secretory cross-talk with the immune microenvironment. Cell 176, 757–774.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh S., Singh U. P., Grizzle W. E., Lillard J. W., CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab. Invest. 84, 1666–1676 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Wu Y., Zhou B. P., TNF-α/NF-κB/Snail pathway in cancer cell migration and invasion. Br. J. Cancer 102, 639–644 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell A. S., Albo D., Kimsey T. F., White S. L., Wang T. N., Macrophage inflammatory protein-3α promotes pancreatic cancer cell invasion. J. Surg. Res. 123, 96–101 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Chen J., Yao Y., Gong C., Yu F., Su S., Chen J., Liu B., Deng H., Wang F., Lin L., Yao H., Su F., Anderson K. S., Liu Q., Ewen M. E., Yao X., Song E., CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell 19, 541–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tardáguila M., Mira E., García-Cabezas M. A., Feijoo A. M., Quintela-Fandino M., Azcoitia I., Lira S. A., Mañes S., CX3CL1 promotes breast cancer via transactivation of the EGF pathway. Cancer Res. 73, 4461–4473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muscella A., Vetrugno C., Marsigliante S., CCL20 promotes migration and invasiveness of human cancerous breast epithelial cells in primary culture. Mol. Carcinog. 56, 2461–2473 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Wu J., Zhang P., CCL15/CCR1 axis is involved in hepatocellular carcinoma cells migration and invasion. Tumour Biol. 37, 4501–4507 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Maolake A., Izumi K., Shigehara K., Natsagdorj A., Iwamoto H., Kadomoto S., Takezawa Y., Machioka K., Narimoto K., Namiki M., Lin W. J., Wufuer G., Mizokami A., Tumor-associated macrophages promote prostate cancer migration through activation of the CCL22-CCR4 axis. Oncotarget 8, 9739–9751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y., Cheon S., Jung M. K., Song S. B., Kim D., Kim H. J., Park H., Bang S. I., Cho D., Interleukin-18 enhances breast cancer cell migration via down-regulation of claudin-12 and induction of the p38 MAPK pathway. Biochem. Biophys. Res. Commun. 459, 379–386 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Zhang X., Chen L., Dang W.-Q., Cao M.-F., Xiao J.-F., Lv S.-Q., Jiang W.-J., Yao X.-H., Lu H.-M., Miao J.-Y., Wang Y., Yu S.-C., Ping Y.-F., Liu X.-D., Cui Y.-H., Zhang X., Bian X.-W., CCL8 secreted by tumor-associated macrophages promotes invasion and stemness of glioblastoma cells via ERK1/2 signaling. Lab. Invest. 100, 619–629 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Razidlo G. L., Burton K. M., McNiven M. A., Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42. J. Biol. Chem. 293, 11143–11153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naldini A., Filippi I., Miglietta D., Moschetta M., Giavazzi R., Carraro F., Interleukin-1β regulates the migratory potential of MDAMB231 breast cancer cells through the hypoxia-inducible factor-1α. Eur. J. Cancer 46, 3400–3408 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Hedrick E., Safe S., Transforming growth factor β/NR4A1-inducible breast cancer cell migration and epithelial-to-mesenchymal transition is p38α (mitogen-activated protein kinase 14) dependent. Mol. Cell. Biol. 37, e00306–e00317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAndrews K. M., McGrail D. J., Ravikumar N., Dawson M. R., Mesenchymal stem cells induce directional migration of invasive breast cancer cells through TGF-β. Sci. Rep. 5, 16941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bin Fang W., Jokar I., Zou A., Lambert D., Dendukuri P., Cheng N., CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein- and p42/44 mitogen-activated protein kinase (MAPK)-dependent mechanisms. J. Biol. Chem. 287, 36593–36608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv W., Chen N., Lin Y., Ma H., Ruan Y., Li Z., Li X., Pan X., Tian X., Macrophage migration inhibitory factor promotes breast cancer metastasis via activation of HMGB1/TLR4/NF kappa B axis. Cancer Lett. 375, 245–255 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Huang C. Y., Fong Y. C., Lee C. Y., Chen M. Y., Tsai H. C., Hsu H. C., Tang C. H., CCL5 increases lung cancer migration via PI3K, Akt and NF-κB pathways. Biochem. Pharmacol. 77, 794–803 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Zipin-Roitman A., Meshel T., Sagi-Assif O., Shalmon B., Avivi C., Pfeffer R. M., Witz I. P., Ben-Baruch A., CXCL10 promotes invasion-related properties in human colorectal carcinoma cells. Cancer Res. 67, 3396–3405 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Eichbaum C., Meyer A.-S., Wang N., Bischofs E., Steinborn A., Bruckner T., Brodt P., Sohn C., Eichbaum M. H. R., Breast cancer cell-derived cytokines, macrophages and cell adhesion: Implications for metastasis. Anticancer Res. 31, 3219–3227 (2011). [PubMed] [Google Scholar]
- 24.Velasco-Velázquez M., Xolalpa W., Pestell R. G., The potential to target CCL5/CCR5 in breast cancer. Expert Opin. Ther. Targets 18, 1265–1275 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Brand S., Dambacher J., Beigel F., Olszak T., Diebold J., Otte J. M., Göke B., Eichhorst S. T., CXCR4 and CXCL12 are inversely expressed in colorectal cancer cells and modulate cancer cell migration, invasion and MMP-9 activation. Exp. Cell Res. 310, 117–130 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Boimel P. J., Smirnova T., Zhou Z. N., Wyckoff J., Park H., Coniglio S. J., Qian B.-Z., Stanley E. R., Cox D., Pollard J. W., Muller W. J., Condeelis J., Segall J. E., Contribution of CXCL12 secretion to invasion of breast cancer cells. Breast Cancer Res. 14, R23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bachmeier B. E., Mohrenz I. V., Mirisola V., Schleicher E., Romeo F., Höhneke C., Jochum M., Nerlich A. G., Pfeffer U., Curcumin downregulates the inflammatory cytokines CXCL1 and -2 in breast cancer cells via NFκB. Carcinogenesis 29, 779–789 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Chatterjee A., Rodger E. J., Eccles M. R., Epigenetic drivers of tumourigenesis and cancer metastasis. Cancer Biol. Ther. 51, 149–159 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Dou Y., Milne T. A., Ruthenburg A. J., Lee S., Lee J. W., Verdine G. L., Allis C. D., Roeder R. G., Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13, 713–719 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Shilatifard A., Glass C. K., Rosenfeld M. G., Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20, 341–348 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matkar S., Thiel A., Hua X., Menin: A scaffold protein that controls gene expression and cell signaling. Trends Biochem. Sci. 38, 394–402 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L., Jin M., Jeong K. W., Histone H3K4 methyltransferases as targets for drug-resistant cancers. Biology (Basel) 10, 581 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao H., He G., Yan S., Chen C., Song L., Rosol T. J., Deng X., Triple-negative breast cancer: Is there a treatment on the horizon? Oncotarget 8, 1913–1924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan H. G., Malmgren J. A., Impact of triple negative phenotype on breast cancer prognosis. Breast J. 14, 456–463 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Tseng L. M., Hsu N. C., Chen S. C., Lu Y. S., Lin C. H., Chang D. Y., Li H., Lin Y. C., Chang H. K., Chao T. C., Ouyang F., Hou M. F., Distant metastasis in triple-negative breast cancer. Neoplasma 60, 290–294 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Bianchini G., Balko J. M., Mayer I. A., Sanders M. E., Gianni L., Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 13, 674–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steeg P. S., Targeting metastasis. Nat. Rev. Cancer 16, 201–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medvedeva Y. A., Lennartsson A., Ehsani R., Kulakovskiy I. V., Vorontsov I. E., Panahandeh P., Khimulya G., Kasukawa T.; FANTOM Consortium, Drabløs F., EpiFactors: A comprehensive database of human epigenetic factors and complexes. Database 2015, bav067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nissen N. I., Karsdal M., Willumsen N., Collagens and cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J. Exp. Clin. Cancer Res. 38, 115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu P.-H., Giri A., Wirtz D., Statistical analysis of cell migration in 3D using the anisotropic persistent random walk model. Nat. Protoc. 10, 517–527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu P.-H., Giri A., Sun S. X., Wirtz D., Three-dimensional cell migration does not follow a random walk. Proc. Natl. Acad. Sci. U.S.A. 111, 3949–3954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klossowski S., Miao H., Kempinska K., Wu T., Purohit T., Kim E., Linhares B. M., Chen D., Jih G., Perkey E., Huang H., He M., Wen B., Wang Y., Yu K., Lee S. C.-W., Danet-Desnoyers G., Trotman W., Kandarpa M., Cotton A., Abdel-Wahab O., Lei H., Dou Y., Guzman M., Peterson L., Gruber T., Choi S., Sun D., Ren P., Li L.-S., Liu Y., Burrows F., Maillard I., Cierpicki T., Grembecka J., Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J. Clin. Invest. 130, 981–997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi A., Murai M. J., He S., Lund G., Hartley T., Purohit T., Reddy G., Chruszcz M., Grembecka J., Cierpicki T., Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia. Blood 120, 4461–4469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M.-H. Lee, Y. S. Rahmanto, G. Russo, P.-H. Wu, D. Gilkes, A. Kiemen, T. Miyamoto, Y. Yu, M. Habibi, I.-M. Shih, T.-L. Wang, D. Wirtz, Multi-compartment tumor organoids. bioRxiv 2020.11.03.367334 [Preprint] (2020). 10.1101/2020.11.03.367334. [DOI]
- 45.Shilatifard A., The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao R. C., Dou Y., Hijacked in cancer: The KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15, 334–346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokoyama A., Molecular mechanisms of MLL-associated leukemia. Int. J. Hematol. 101, 352–361 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Borkin D., He S., Miao H., Kempinska K., Pollock J., Chase J., Purohit T., Malik B., Zhao T., Wang J., Wen B., Zong H., Jones M., Danet-Desnoyers G., Guzman M. L., Talpaz M., Bixby D. L., Sun D., Hess J. L., Muntean A. G., Maillard I., Cierpicki T., Grembecka J., Pharmacologic inhibition of the menin-MLL interaction blocks progression of MLL leukemia In Vivo. Cancer Cell 27, 589–602 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krivtsov A. V., Evans K., Gadrey J. Y., Eschle B. K., Hatton C., Uckelmann H. J., Ross K. N., Perner F., Olsen S. N., Pritchard T., McDermott L., Jones C. D., Jing D., Braytee A., Chacon D., Earley E., McKeever B. M., Claremon D., Gifford A. J., Lee H. J., Teicher B. A., Pimanda J. E., Beck D., Perry J. A., Smith M. A., McGeehan G. M., Lock R. B., Armstrong S. A., A Menin-MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell 36, 660–673.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grembecka J., He S., Shi A., Purohit T., Muntean A. G., Sorenson R. J., Showalter H. D., Murai M. J., Belcher A. M., Hartley T., Hess J. L., Cierpicki T., Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 8, 277–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fraley S. I., Feng Y., Krishnamurthy R., Kim D.-H., Celedon A., Longmore G. D., Wirtz D., A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 12, 598–604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gagliardi P. A., Puliafito A., di Blasio L., Chianale F., Somale D., Seano G., Bussolino F., Primo L., Real-time monitoring of cell protrusion dynamics by impedance responses. Sci. Rep. 5, 10206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson D. E., O’Keefe R. A., Grandis J. R., Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 234–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campellone K. G., Welch M. D., A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agarwal P., Zaidel-Bar R., Diverse roles of non-muscle myosin II contractility in 3D cell migration. Essays Biochem. 63, 497–508 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., Kawaji H., Nakaki R., Sese J., Meno C., ChIP-Atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 19, e46255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsusaka T., Fujikawa K., Nishio Y., Mukaida N., Matsushima K., Kishimoto T., Akira S., Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl. Acad. Sci. U.S.A. 90, 10193–10197 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khuon S., Liang L., Dettman R. W., Sporn P. H. S., Wysolmerski R. B., Chew T. L., Myosin light chain kinase mediates transcellular intravasation of breast cancer cells through the underlying endothelial cells: A three-dimensional FRET study. J. Cell Sci. 123, 431–440 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Javelaud D., Alexaki V. I., Dennler S., Mohammad K. S., Guise T. A., Mauviel A., TGF-β/SMAD/GLI2 signaling axis in cancer progression and metastasis. Cancer Res. 71, 5606–5610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jerrell R. J., Parekh A., Matrix rigidity differentially regulates invadopodia activity through ROCK1 and ROCK2. Biomaterials 84, 119–129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pietenpol J. A., Stewart Z. A., Cell cycle checkpoint signaling: Cell cycle arrest versus apoptosis. Toxicology 181-182, 475–481 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Wei Z., Liu H. T., MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Mustacchi G., de Laurentiis M., The role of taxanes in triple-negative breast cancer: Literature review. Drug Des. Devel. Ther. 9, 4303–4318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Isakoff S. J., Triple-negative breast cancer: Role of specific chemotherapy agents. Cancer J. 16, 53–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meyer C., Kowarz E., Hofmann J., Renneville A., Zuna J., Trka J., Abdelali R. B., Macintyre E., De Braekeleer E., De Braekeleer M., Delabesse E., de Oliveira M. P., Cavé H., Clappier E., van Dongen J. J. M., Balgobind B. V., van den Heuvel-Eibrink M. M., Beverloo H. B., Panzer-Grümayer R., Teigler-Schlegel A., Harbott J., Kjeldsen E., Schnittger S., Koehl U., Gruhn B., Heidenreich O., Chan L. C., Yip S. F., Krzywinski M., Eckert C., Möricke A., Schrappe M., Alonso C. N., Schäfer B. W., Krauter J., Lee D. A., Stadt U. Z., Te Kronnie G., Sutton R., Izraeli S., Trakhtenbrot L., Nigro L. L., Tsaur G., Fechina L., Szczepanski T., Strehl S., Ilencikova D., Molkentin M., Burmeister T., Dingermann T., Klingebiel T., Marschalek R., New insights to the MLL recombinome of acute leukemias. Leukemia 23, 1490–1499 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Trendowski M., The inherent metastasis of leukaemia and its exploitation by sonodynamic therapy. Crit. Rev. Oncol. Hematol. 94, 149–163 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Hartman Z. C., Poage G. M., den Hollander P., Tsimelzon A., Hill J., Panupinthu N., Zhang Y., Mazumdar A., Hilsenbeck S. G., Mills G. B., Brown P. H., Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 73, 3470–3480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altaba A. R. I., Sánchez P., Dahmane N., Gli and hedgehog in cancer: Tumours, embryos and stem cells. Nat. Rev. Cancer 2, 361–372 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Sterling J. A., Oyajobi B. O., Grubbs B., Padalecki S. S., Munoz S. A., Gupta A., Story B., Zhao M., Mundy G. R., The hedgehog signaling molecule Gli2 induces parathyroid hormone-related peptide expression and osteolysis in metastatic human breast cancer cells. Cancer Res. 66, 7548–7553 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Gandalovičová A., Rosel D., Fernandes M., Veselý P., Heneberg P., Čermák V., Petruželka L., Kumar S., Sanz-Moreno V., Brábek J., Migrastatics-anti-metastatic and anti-invasion drugs: Promises and challenges. Trends Cancer 3, 391–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson R. L., Balasas T., Callaghan J., Coombes R. C., Evans J., Hall J. A., Kinrade S., Jones D., Jones P. S., Jones R., Marshall J. F., Panico M. B., Shaw J. A., Steeg P. S., Sullivan M., Tong W., Westwell A. D., Ritchie J. W. A., Cancer Research UK and Cancer Therapeutics CRC Australia Metastasis Working Group , A framework for the development of effective anti-metastatic agents. Nat. Rev. Clin. Oncol. 16, 185–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morel D., Jeffery D., Aspeslagh S., Almouzni G., Postel-Vinay S., Combining epigenetic drugs with other therapies for solid tumours — Past lessons and future promise. Nat. Rev. Clin. Oncol. 17, 91–107 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Mokhtari R. B., Homayouni T. S., Baluch N., Morgatskaya E., Kumar S., Das B., Yeger H., Combination therapy in combating cancer. Oncotarget 8, 38022–38043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nair P. R., Delivering combination chemotherapies and targeting oncogenic pathways via polymeric drug delivery systems. Polymers (Basel) 11, 630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stover D. G., Winer E. P., Tailoring adjuvant chemotherapy regimens for patients with triple negative breast cancer. Breast 24, S132–S135 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Clark A. G., Vignjevic D. M., Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 36, 13–22 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Chen Y., Huang S., Wu B., Fang J., Zhu M., Sun L., Zhang L., Zhang Y., Sun M., Guo L., Wang S., Transforming growth factor-β1 promotes breast cancer metastasis by downregulating miR-196a-3p expression. Oncotarget 8, 49110–49122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hempel U., Hintze V., Möller S., Schnabelrauch M., Scharnweber D., Dieter P., Artificial extracellular matrices composed of collagen i and sulfated hyaluronan with adsorbed transforming growth factor β1 promote collagen synthesis of human mesenchymal stromal cells. Acta Biomater. 8, 659–666 (2012). [DOI] [PubMed] [Google Scholar]
- 79.Nair P. R., Alvey C., Jin X., Irianto J., Ivanovska I., Discher D. E., Filomicelles deliver a chemo-differentiation combination of paclitaxel and retinoic acid that durably represses carcinomas in liver to prolong survival. Bioconjug. Chem. 29, 914–927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Falk T., Mai D., Bensch R., Çiçek Ö., Abdulkadir A., Marrakchi Y., Böhm A., Deubner J., Jäckel Z., Seiwald K., Dovzhenko A., Tietz O., Dal Bosco C., Walsh S., Saltukoglu D., Tay T. L., Prinz M., Palme K., Simons M., Diester I., Brox T., Ronneberger O., U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Lu Y., Chen J. J., Mu L., Xue Q., Wu Y., Wu P. H., Li J., Vortmeyer A. O., Miller-Jensen K., Wirtz D., Fan R., High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal. Chem. 85, 2548–2556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nair P. R., Karthick S., Spinler K. R., Vakili M. R., Lavasanifar A., Discher D. E., Filomicelles from aromatic diblock copolymers increase paclitaxel-induced tumor cell death and aneuploidy compared with aliphatic copolymers. Nanomedicine 11, 1551–1569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P., The molecular signatures database hallmark gene set collection. Cell Systems 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cancer Genome Atlas Research Network, Weinstein J. N., Collisson E. A., Mills G. B., Shaw K. R. M., Ozenberger B. A., Ellrott K., Shmulevich I., Sander C., Stuart J. M., The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang S., Zhang J. J., Huang X.-Y., Mouse models for tumor metastasis. Methods Mol. Biol. 928, 221–228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trail P. A., Willner D., Bianchi A. B., Henderson A. J., Smith M. D. T., Girit E., Lasch S., Hellström I., Hellström K. E., Enhanced antitumor activity of paclitaxel in combination with the anticarcinoma immunoconjugate BR96-doxorubicin. Clin. Cancer Res. 5, 3632–3638 (1999). [PubMed] [Google Scholar]
- 87.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grebien F., Vedadi M., Getlik M., Giambruno R., Grover A., Avellino R., Skucha A., Vittori S., Kuznetsova E., Smil D., Barsyte-Lovejoy D., Li F., Poda G., Schapira M., Wu H., Dong A., Senisterra G., Stukalov A., Huber K. V. M., Schönegger A., Marcellus R., Bilban M., Bock C., Brown P. J., Zuber J., Bennett K. L., al-awar R., Delwel R., Nerlov C., Arrowsmith C. H., Superti-Furga G., Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat. Chem. Biol. 11, 571–578 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Daigle S. R., Olhava E. J., Therkelsen C. A., Basavapathruni A., Jin L., Boriack-Sjodin P. A., Allain C. J., Klaus C. R., Raimondi A., Scott M. P., Waters N. J., Chesworth R., Moyer M. P., Copeland R. A., Richon V. M., Pollock R. M., Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 122, 1017–1025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Valencia A. M. J., Wu P.-H., Yogurtcu O. N., Rao P., DiGiacomo J., Godet I., He L., Lee M.-H., Gilkes D., Sun S. X., Wirtz D., Collective cancer cell invasion induced by coordinated contractile stresses. Oncotarget 6, 43438–43451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lauth M., Bergström Å., Shimokawa T., Toftgård R., Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc. Natl. Acad. Sci. U.S.A. 104, 8455–8460 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoshioka K., Foletta V., Bernard O., Itoh K., A role for LIM kinase in cancer invasion. Proc. Natl. Acad. Sci. U.S.A. 100, 7247–7252 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu W., Ma B., Ye H., Wang T., Wang X., Yang J., Wei Y., Zhu J., Chen L., Millepachine, a potential topoisomerase II inhibitor induces apoptosis via activation of NF-κB pathway in ovarian cancer. Oncotarget 7, 52281–52293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith L., Watson M. B., O’Kane S. L., Drew P. J., Lind M. J., Cawkwell L., The analysis of doxorubicin resistance in human breast cancer cells using antibody microarrays. Mol. Cancer Ther. 5, 2115–2120 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15
Tables S1 to S6
References