Abstract
Human cytomegalovirus (HCMV) gene expression is highly cell and tissue specific. Cell factor-mediated regulatory interactions are involved in regulating the restricted expression of the HCMV major immediate-early (IE) gene (J. F. Baskar, P. P. Smith, G. Nilaver, R. A. Jupp, S. Hoffmann, N. J. Peffer, D. J. Tenney, A. M. Colberg-Poley, P. Ghazal, and J. A. Nelson, 70:3207–3213, 1996). To gain an understanding of HCMV early gene activation, we studied the effect of each of the three major IE proteins, IE72, IE86, and IE55, on the HCMV DNA polymerase gene (pol; UL54) promoter. In transient-expression assays, the IE86 protein alone was able to transactivate the pol promoter, but IE72 and IE55 were not, in permissive U373MG cells. However, we were unable to detect IE86-mediated transactivation in nonpermissive HeLa or C33-A cells. Using electrophoretic mobility shift assays (EMSAs), we found that expression of the IE86 protein in U373MG cells resulted in specific binding of a DNA complex to an inverted-repeat element, IR1, of the pol promoter. Antibody supershifting and EMSA-Western blotting experiments further showed that IE86 and the cellular transcription factor Sp1 were components of the IR1 DNA-binding complex. Furthermore, we found that binding of DNA by Sp1 was dramatically increased in the presence of IE86. Interestingly, this IE86-induced DNA-binding activity of Sp1 was inhibited by a repressor activity presented in HeLa cells. In summary, our study suggests that a viral regulatory protein can modulate the DNA binding activity of a cellular transcription factor, resulting in cell-specific transactivation of viral genes.
Human cytomegalovirus (HCMV) is a ubiquitous member of the herpesvirus family and a medically important pathogen that typically causes asymptomatic infections in healthy individuals and severe complicating infections in utero as well as in immunocompromised and immunosuppressed patients (12, 21, 34, 36, 41, 46). HCMV infection of permissive cells in culture leads to an ordered sequential expression of viral genes which are divided into three kinetic classes: immediate-early (IE), early, and late (7, 8, 35, 58). The IE genes are expressed upon entry of the viral genome into the nucleus of an infected cell, and their expression requires specific cis-acting elements in the viral promoters as well as cellular and viral trans-acting factors. The IE genes have been extensively studied, and their expression is thought to be essential for viral replication (22, 28, 48–51, 53–55, 60, 61). The most important of the IE gene products are the 72-kDa IE1 (IE72; ppUL123) and the 86-kDa IE2 (IE86; ppUL122a) polypeptides that originate from the major IE gene region of HCMV and are transcribed under the control of a complex enhancer-promoter (3, 16, 40, 51, 56).
Early genes are expressed in a complex manner. Three subclasses of early genes have been defined (45, 48). Transcription of early genes is weak in the absence of the IE proteins. However, upon synthesis of IE viral proteins, some early genes are strongly activated (5, 33, 45, 59, 62). It has been demonstrated that IE86 alone strongly activates the UL4 and UL112-113 early genes in transiently transfected cells (5, 27, 42) while the IE72 protein can act synergistically with IE86 to activate these genes (5, 25, 45, 47, 62). The IE86 protein has been shown to bind to the cis-acting negative regulatory element on the UL4 promoter and negate its repressive effect (18). Transactivation of the UL112 gene is mediated through an interaction between IE86 and the cellular transcription factor CREB (1, 29, 30). An expression construct encoding the major IE proteins, IE72, IE86, and IE55, was shown to transactivate the pol promoter in the fully HCMV-permissive cells known as human foreskin fibroblasts (HFF) (24). This activation was mediated by a cis element, IR1, present in the pol promoter (24). However, no IE86 binding sequences had been identified in this promoter. Recently, it was shown that cellular transcription factors, such as ATF and Sp1, are involved in transactivation of the pol promoter (26, 32). HCMV infection upregulates the expression of genes encoding the cellular transcription factors NF-κB and Sp1 in human embryonic lung (HEL) fibroblasts (63, 64). Overall, these data suggest that the mechanism by which IE86 activates transcription of HCMV early genes seems to involve protein-DNA and multiple protein-protein interactions, i.e., interactions with upstream bound cellular transcription factors and with the basal transcription complex (14, 31, 43). Since the HCMV gene expression cascade is cell type restricted, it is possible that modulation of transcription by HCMV regulatory proteins and/or cell-specific transcription factors may determine the permissiveness of this gene expression cascade.
We were interested in understanding HCMV temporal gene regulation, especially the mechanism by which IE proteins mediate early gene activation, and in identifying cellular transcription factors involved in the DNA binding activity of IR1. Toward this goal, we studied the effect of each of the three major IE proteins on UL54 (pol) early gene promoter activity. Our results indicate that only IE86 alone is able to transactivate the UL54 (pol) promoter in permissive U373MG cells. Expression of IE86 resulted in the formation of a specific DNA complex on a cis element, IR1, of the pol promoter (24). To elucidate the IR1-bound proteins, we found that the IE86 protein and the cellular transcription factor Sp1 were components of the IR1-DNA complex. In addition, we found that binding of DNA by Sp1 was dramatically increased in the presence of IE86 in U373MG cells. However, this enhanced DNA binding by Sp1 was present only in the permissive U373MG cell line, not in nonpermissive HeLa cells. In contrast, this IE86-mediated binding of DNA by Sp1 was inhibited by a repressor activity present in HeLa cells. In summary, these data demonstrate that the cellular transcription factor Sp1 is involved in the DNA-binding activity of IR1 of the pol promoter. Furthermore, we demonstrate that a repressor activity present in HeLa cells somehow inhibits this IE86-induced IR1 DNA binding.
MATERIALS AND METHODS
Plasmid construction.
The UL54 (pol) promoter sequence from positions −425 to +15 and the UL112 promoter sequence from positions −352 to +37 were amplified by PCR with cosmid pCM1058 (a gift from Peter Ghazal) as a template. The sequences of the oligonucleotide primers used for amplification of the UL54 promoter were 5′-CCCAAGCTTGGGGGAATTCAACTCGTACAAGCAG-3′ (forward primer) and 5′-CCCAAGCTTGGGTCAGACGACGGTGGTCCC-3′ (reverse primer). These primers introduced HindIII restriction sites at the 5′ and 3′ ends of the UL54 (pol) promoter fragment, allowing insertion of this fragment into the pGL2-basic luciferase reporter plasmid (38) (Promega). The sequences of the oligonucleotide primers used for the amplification of the UL112 promoter were 5′-CGGGGTACCCCGCACAGAGGTAACAAC-3′ (forward primer) and 5′-GAAGATCTTCGGCGGTGGAGCGAGTGC-3′ (reverse primer). These primers introduced a KpnI and a BglII restriction site at the 5′ and 3′ ends of the UL112 promoter fragment, respectively, allowing directional insertion into the pGL2-basic luciferase reporter plasmid (38) (Promega). The PCR fidelity of the UL54 (pol) and UL112 promoter sequences was confirmed by sequencing. Expression vectors for each of the HCMV IE proteins—RSVIE72, RSVIE86, RSVIE55 (all gifts from Peter Ghazal), and pSVH, which expresses all three IE proteins from the major IE gene region (kindly provided by Richard M. Stenberg)—have been described previously (9, 24).
Transfection and infection assays.
U373MG, HFF, HeLa, and C33-A cells were cotransfected with the reporter construct and the indicated effector by use of the Profection mammalian transfection system (Promega). Forty hours posttransfection, cells were harvested and assayed for luciferase activity as described by the manufacturer (Analytical Luminescence Laboratory).
Establishment of stable U373MG and HeLa cell lines expressing IE86.
The RSVIE86 and pSV2Neo (Clontech Laboratories, Inc.) selection plasmids were cotransfected into U373MG and HeLa cells by the calcium phosphate method. Transfectants were selected in medium containing 0.6 mg of G418 per ml on the third day after transfection. G418-resistant clones were expanded, and 3 × 104 cells were seeded in triplicate in a 96-well plate. Cells were harvested and assayed for IE86 by Western blot analysis. Clones showing expression of IE86 protein were amplified and used for further studies.
Western blot analysis.
For each sample, 25 μg of total protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and transferred to a Hybond-ECL nitrocellulose membrane (Amersham). A monoclonal antibody specific for the HCMV IE proteins (MAB 810; Chemicon) was used. Proteins bound by primary antibodies were detected with an alkaline phosphatase-conjugated secondary antibody in accordance with the manufacturer’s protocol (Amersham).
Preparation of nuclear extracts.
U373MG, U373-IE86, HeLa, and HeLa-IE86 nuclear extracts were prepared in accordance with the protocol described by Dignam and coworkers (11).
EMSAs.
The sequences of the IR1 probes were 5′-GTTACAGGCTCCGCCTTC (forward) and 5′-GGAAGGCGGAGCCTGTA (reverse). The sequences of the IRmut probes were 5′-GTTACAGATATCGCCTTC (forward) and 5′-GGAAGGCGATATCTGTA (reverse) (underlined nucleotides are mutated). The sequences of the CRS probes were 5′-CGTTTAGTGAACCGTCAGAT (forward) and 5′-TCTGACGGTTCACTAAACG (reverse). The sequences of the CRSmut probes were 5′-GCGGCGGTGAACCGTCAGAT (forward) and 5′-TCTGACGGTTCACCGCCGC (reverse). The sequences of the EAIE2 probes were 5′-TAGCGTTGCGATTTGCAGTCCGCTCC (forward) and 5′-GGAGCGGACTGCAAATCGCAACGCT (reverse). The sequences of the EAIE2mut probes were 5′-TAGCGTTGTAACCCATAGTCCGCTCC (forward) and 5′-GGAGCGGACTATGGGTTACAACGCT (reverse). Sp1 and CREB consensus oligonucleotides were purchased from Promega. The probes and competitors were produced by annealing the appropriate oligonucleotides as described by Kerry and coworkers (24). For use as probes, the IR1 oligonucleotide was end labeled with [α-32P]dATP and the Sp1 oligonucleotide was labeled with [γ-32P]ATP. Five-microgram quantities of nuclear extracts were incubated with 1 μg of poly(dI-dC) · poly(dI-dC) and 10,000 cpm of labeled oligonucleotide for 30 min at room temperature in binding buffer (75 mM NaCl, 15 mM Tris [pH 7.5], 1.5 mM EDTA, 1.5 mM dithiothreitol, 7.5% glycerol, 0.3% Nonidet P-40, 20 μg of bovine serum albumin per ml). A 4% polyacrylamide gel was prerun in standard 0.25× Tris-borate-EDTA at 150 V for 1.5 h. Sample reactions were then subjected to PAGE. The gels were dried and subjected to autoradiography. In competition and antibody supershift experiments, a 50-fold excess of unlabeled oligonucleotides or 1 μg of monoclonal antibody specific for IE proteins (MAB 810, as described above) and polyclonal antibodies specific for NF-κB p65, NF-κB p50, Sp1, ATF, and CREB (Santa Cruz Biotechnology) were used, respectively. For electrophoretic mobility shift assays (EMSAs) and Western blotting experiments, after PAGE electrophoresis, the sample was transferred to DEAE and nitrocellulose membranes in regular Western Tris-glycine buffer. Recombinant IE86 protein (kindly provided by Peter Ghazal) was used as a control.
RESULTS
IE86 protein transactivates the pol promoter in permissive U373MG cells.
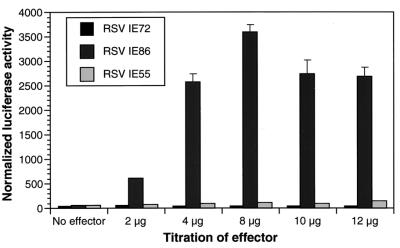
To gain an understanding of the mechanism by which IE86 transactivates the HCMV pol promoter, we first determined whether the −425/+15 pol (24) promoter construct responded to IE proteins in the U373MG cells. Indeed, we found that cotransfection with pSVH, a plasmid expressing the genes encoding three IE proteins, IE72, IE86, and IE55, from the endogenous genomic fragment under the control of their own major IE promoter (9), resulted in up to a 160-fold activation of the pol promoter, as measured by expression of the luciferase reporter (data not shown). This result is consistent with those reported for HFF (9, 51, 52). Thus, we were assured that the pol-luciferase reporter construct carried all regulatory elements previously shown to mediate the response to the IE proteins expressed from the pSVH expression vector (9, 48, 52). However, it remained unclear which of the three IE proteins play a major role in promoter activity. Therefore, we transfected expression constructs encoding the IE72, IE86, and IE55 cDNA sequences under the control of the heterologous Rous sarcoma virus (RSV) promoter. We found that the IE86 expression vector alone was capable of activating the pol promoter significantly (40- to 55-fold) (Fig. 1). Neither the IE72 nor the IE55 expression vector yielded significant activation (Fig. 1). Western blot analysis showed that pSVH expressed about 3- to 4-fold more IE86 protein than the RSVIE86 vector (data not shown), thus suggesting that the difference between pSVH (160-fold) and RSVIE86 (40- to 55-fold) activation of the pol promoter is likely due to higher levels of the IE86 protein. We conclude that among the IE gene products, the IE86 protein plays a major role in the transactivation of the pol promoter in permissive U373MG cells.
FIG. 1.
The IE86 protein is essential for the activation of the HCMV DNA polymerase (pol) gene promoter. U373MG cells were cotransfected with the pol-luciferase reporter and increasing amounts of the IE gene expression vectors RSVIE86, RSVIE72, and RSVIE55 and a β-galactosidase control vector. Luciferase activity was normalized to β-galactosidase activity. The data represent the results of three independent experiments.
Cell type-specific transactivation of the pol promoter by IE86.
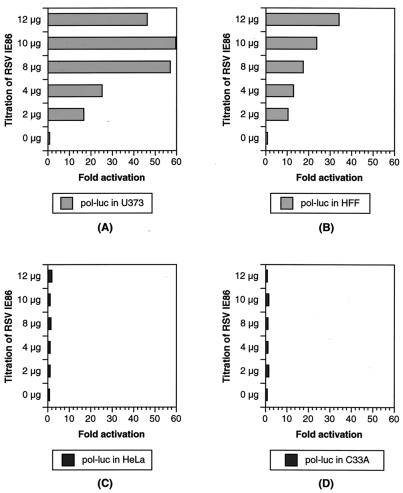
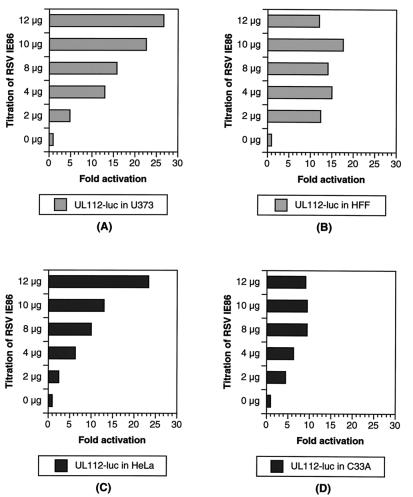
HCMV has a very restrictive species and cell tropism. The molecular events allowing the virus to productively infect some cell types but not others has not been investigated extensively. It has been shown that the HCMV gene products can act individually or in combination to regulate the expression of viral and cellular promoters in a manner that is dependent on the promoter, the host cell, and the IE gene products present (6). We were interested in determining if early gene promoters, in particular the pol promoter, were expressed in a cell type-specific fashion. Toward this goal, we analyzed the response of two early gene promoters, pol and UL112, to IE86 expression in permissive and nonpermissive cells. In permissive glioblastoma U373MG and HFF, the pol promoter was transactivated by IE86 60- and 35-fold, respectively (Fig. 2A and B) while the UL112 promoter was transactivated by IE86 28- and 18-fold, respectively (Fig. 3A and B).
FIG. 2.
Transactivation of the pol promoter by IE86 in the permissive U373MG and primary HFF cells. The pol-luciferase (pol-luc) and β-galactosidase reporters were cotransfected with increasing amounts of the RSVIE86 expression vector in permissive U373MG (A) and HFF (B) cells and in nonpermissive HeLa (C) and C33-A (D) cells as indicated. Luciferase activities were normalized to β-galactosidase activity. Units are fold activation. The data represent the results of three independent experiments.
FIG. 3.
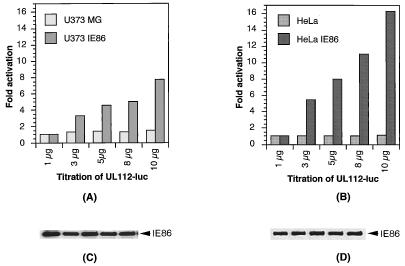
Transactivation of the UL112 promoter by IE86. The UL112-luciferase (UL112-luc) and β-galactosidase reporters were cotransfected with increasing amounts of the RSVIE86 expression vector in permissive U373MG (A) and HFF (B) cells and in nonpermissive HeLa (C) and C33-A (D) cells as indicated. Luciferase activities were normalized to β-galactosidase activity. Units are fold activation. The data represent the results of three independent experiments.
Interestingly, we were unable to detect IE86-mediated transactivation of the pol promoter in nonpermissive HeLa or C33-A epithelial cells (Fig. 2C and D). In contrast, the UL112 reporter was efficiently transactivated by cotransfection with the IE86-expressing vector in both HeLa and C33-A cells (Fig. 3C and D). The data shown are normalized for β-galactosidase levels expressed from a cotransfected control plasmid. Therefore, the lack of luciferase expression from the pol-luciferase reporter is not simply due to inefficient transfection. More importantly, the UL112-luciferase reporter was still activated by IE86 in those cells, which is consistent with the findings of Colberg-Poley and coworkers (6). pSVH, which coexpresses IE1 (IE72) and IE2 (IE86 and IE55), activates the pol promoter in U373MG cells (160-fold). In contrast, pSVH transactivates the pol promoter in HeLa cells by seven- to ninefold (data not shown). The 20-fold difference between the transactivation activities in permissive U373MG and nonpermissive HeLa cells may be a means of identifying the events required for IE86-mediated induction of pol promoter transcription.
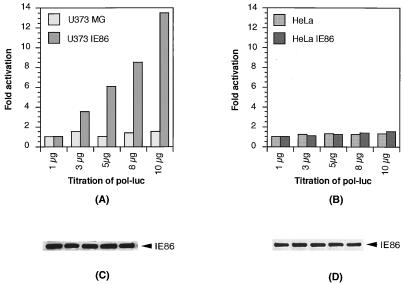
To ascertain that the difference between the responses of the pol promoter to IE86 in glial and in epithelial cells was not due to differences in expression levels, we tested the same reporter plasmids in cell lines stably expressing the IE86 protein. Several U373MG and HeLa cell clones constitutively expressing similar amounts of IE86 were transfected with increasing amounts of the pol-luciferase and UL112-luciferase reporters, respectively. The data representing three different stable clones showed that the luciferase gene driven by the pol promoter was activated in U373-IE86 cells but not in HeLa-IE86 cells (Fig. 4A and B) while the luciferase gene driven by UL112 promoter showed significant activation in both U373-IE86 and HeLa-IE86 cells (Fig. 5A and B). Western blot analysis showed that equal amounts of IE86 protein were expressed from the reporter-transfected stable cells (Fig. 4C and D; Fig. 5C and D). Therefore, we concluded that IE86 may transactivate the pol promoter in a cell type-specific manner.
FIG. 4.
Activation of the pol promoter in the U373MG stable cell line expressing IE86. The U373MG parental line and U373-IE86 stable cell line (A) or the HeLa parental line and HeLa-IE86 stable cell line (B) were transfected with increasing amounts of the pol-luciferase (pol-luc) reporter. Cells were harvested, and luciferase activity was assayed. The levels of expression of IE86 protein in the stable cell lines (C and D) were determined by Western blotting with a monoclonal antibody specific for the HCMV IE proteins.
FIG. 5.
Activation of the UL112 promoter in U373-IE86 and HeLa-IE86 stable cell lines. The U373MG parental cell line and U373-IE86 stable cell line (A) or the HeLa parental line and HeLa-IE86 stable cell line (B) were transfected with increasing amounts of the UL112-luciferase (UL112-luc) reporter. Cells were harvested, and luciferase activity was assayed. The levels of expression of IE86 protein in the stable cell lines (C and D) were determined by Western blotting with a monoclonal antibody specific for the HCMV IE proteins.
Identification of a cell-specific DNA-binding activity specific for the inverted-repeat element IR1.
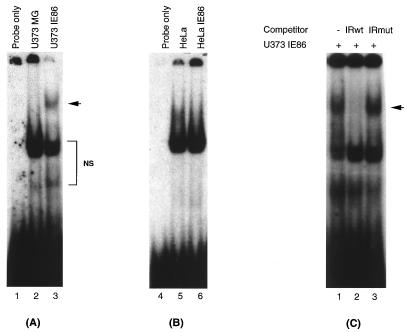
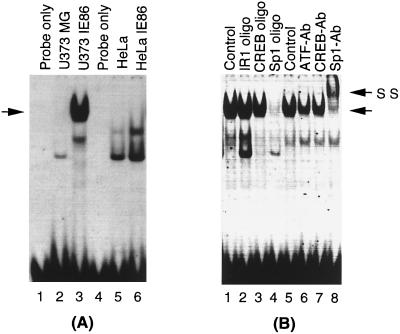
An inverted-repeat element, IR1, has been shown to mediate activation of the pol promoter by the HCMV IE proteins expressed from the pSVH vector. The cellular factors present in uninfected HFF cells bind to this IR1 element, and its binding increases with time upon viral infection (24). To determine whether IR1 binding was also present in U373MG cells and whether the IE86 protein was present in the IR1 DNA-binding complex, we conducted EMSAs with radioactively labeled IR1 oligonucleotides (24) and nuclear extracts from parental as well as IE86-expressing U373MG and HeLa cells. Interestingly, we found that a specific protein-DNA complex was present in the three different IE86-expressing U373MG cell clones but not in extracts from the parental cells (Fig. 6A). In contrast, HeLa cells did not show a unique complex in the presence of IE86 protein (Fig. 6B). The absence of the IE86-specific complex in HeLa cells is not due to a lack or lower level of IE86 protein since, as shown in Fig. 4C and D and 5C and D, the protein is expressed at similar levels in U373MG cells.
FIG. 6.
Identification of an IR1-specific DNA-binding complex in U373MG cells expressing IE86. Five-microgram quantities of nuclear extract from U373MG and U373-IE86 (A) or HeLa and HeLa-IE86 (B) cell lines were incubated with 32P-labeled IR1 oligonucleotide. IR1 binding was analyzed by EMSA. (C) U373MG-IE86 nuclear extract was incubated with labeled IR1 oligonucleotide and a 50-fold excess of either unlabeled wild-type (IRwt) or mutant (IRmut) IR1 oligonucleotides. The arrows indicate the specific complex. NS, nonspecific complexes.
To address whether the protein complex seen in IE86-expressing U373MG cell extracts was specific for the IR1 element, we conducted a competition experiment. As indicated in Fig. 6C, the wild-type IR1 oligonucleotide (at a 50-fold excess) was able to block the formation of the DNA-protein complex in the IE86-expressing U373MG nuclear extracts, but the same amount of this oligonucleotide carrying a mutation in the IR1 sequence was not. Therefore, we conclude that the unique complex is specific for the IR1 element. The data indicated that there was no specific IR1 binding in U373MG cells. However, expression of the IE86 protein in U373MG cells induced an IR1-specific DNA binding activity on the pol promoter.
IE86 protein is present in the IR1 DNA-binding complex.
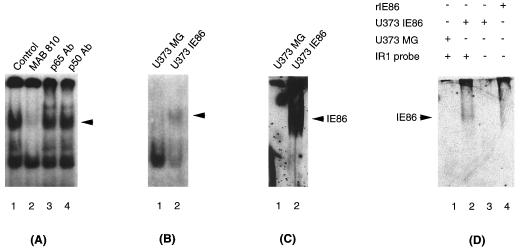
Since expression of IE86 protein in U373MG cells leads to IR1-specific DNA binding, we investigated whether the IE86 protein was present in the IR1 complex. To address this question, we performed antibody supershift experiments using an IE86-specific monoclonal antibody. As shown in Fig. 7A, addition of a monoclonal antibody that recognizes IE86 (MAb 810) disrupted the IR1-specific complex. In contrast, two other antibodies specific for two subunits of the cellular transcription factor NF-κB (p65Ab and p50Ab) had no effect on the IR1 complex. Titration of the IE86 antibody showed that the effect was specific for the IR1 complex (data not shown). To verify that IE86 is present in the IR1 DNA-binding complex, we conducted EMSA-Western blot analysis. As shown in Fig. 7B, the shifted IR1 probe was efficiently transferred onto a DEAE membrane (lane 2). The proteins present in the IR1 complex were transferred to a nitrocellulose membrane and then analyzed by Western blotting with the IE86-specific antibody (Fig. 7C and D, lanes 2). However, this antibody did not show a band in lanes containing U373-IE86 cell lysate or bacteria if the IR1 probe was not added (Fig. 7D, lanes 3 and 4), suggesting that the band recognized by the IE86-specific antibody represents the IE86 protein present in the IR1 DNA-protein complex but not free IE86 protein. Therefore, we concluded that IE86 is part of the IR1 DNA-binding complex.
FIG. 7.
IE86 antibody recognizes the IR1-DNA complex-bound IE86 protein. (A) U373-IE86 nuclear extract was incubated with labeled IR1 oligonucleotide and either a monoclonal antibody specific for the HCMV IE proteins (MAb 810) or polyclonal antibodies specific for NF-κB p65 and p50 subunits (p65 Ab and p50 Ab, respectively). U373-IE86 with the IR1 probe was used as a control. (B to D) EMSA was done as a control (see panel A, lane 1), and then the gel was blotted onto DEAE (B) and nitrocellulose (C and D) membranes. The nitrocellulose membranes were subsequently probed with a monoclonal antibody specific for IE86 (see Materials and Methods). U373-IE86 extracts and recombinant IE86 protein (rIE86) alone were used as controls in EMSA-Western blot analysis (D). Arrows indicate the IR1 DNA-binding complex (A and B) and IE86 protein present in the IR1 DNA-binding complex (C and D).
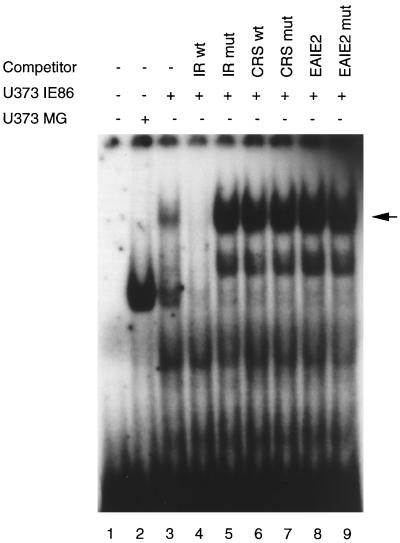
There remained the question of whether IE86 binds to the IR1 element directly or indirectly. To address the possibility of direct binding, we performed an EMSA using two well-characterized IE86-binding sequences, CRS and EAIE2, as competitor oligonucleotides. It has been shown that the IE86 protein efficiently binds to these two sites in in vitro biochemical assays. If the IE86 DNA-binding domain associates directly with the IR1 element, one would expect to see competition when excess amounts of the CRS or EAIE2 oligonucleotides are used. As shown in Fig. 8, neither wild-type nor mutant CRS or EAIE2 oligonucleotides could compete with the IR1 complex (lanes 6 to 9). However, the wild-type IR1, but not the mutant oligonucleotide, efficiently competed with the DNA-binding complex. The inability of the CRS and EAIE2 oligonucleotides to outcompete the IR1 DNA-binding complex could be due to their having lower binding affinities. However, the lack of a decrease in the IR1 DNA-binding complex suggests that the DNA-binding domain of the IE86 protein may not be required for its participation in the IR1 DNA-binding complex. It is thus possible that association of IE86 with the IR1 DNA response element occurs through a cellular factor.
FIG. 8.
Neither CRS nor EAIE2 IE86 cis elements can compete with the IR1-DNA complex. Nuclear extracts isolated from the U373-IE86 stable cell line were incubated with a radiolabeled IR1 oligonucleotide and a 50-fold excess of unlabeled CRS or EAIE2 competitor oligonucleotides as indicated. The arrow indicates the IR1 complex. wt, wild type; mut, mutant.
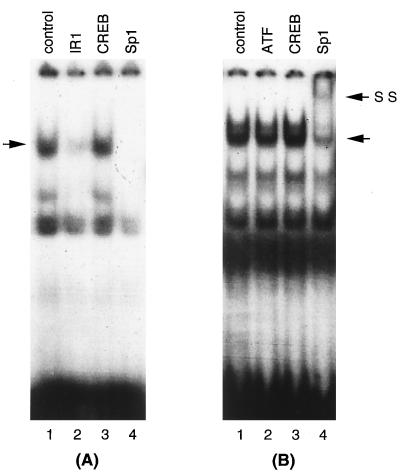
Cellular transcription factor Sp1 binds to the IR1 DNA element.
The results presented above suggest that the IE86 protein may associate with a cellular factor(s) to form a complex on the IR1 DNA element. We then proceeded to determine which cellular factor(s) is present in the complex. By computer analysis of the pol promoter sequence, we found that the IR1 element has similarity to the Sp1 cellular transcription factor binding site. Therefore, we conducted a competition experiment using the Sp1 consensus oligonucleotide and an antibody specific for Sp1 to elucidate whether the cellular transcription factor was involved in IE86-mediated IR1 complex formation. Oligonucleotides representing binding sites for CREB and ATF and antibodies specific for these transcription factors were used to broaden the survey. As indicated in Fig. 9, both IR1 and Sp1 consensus oligonucleotides (Fig. 9A, lanes 2 and 4, respectively) efficiently competed with the IR1 complex and Sp1 antibody supershifted the IR1 complex (Fig. 9B, lane 4). However, the CREB consensus oligonucleotide (Fig. 9A, lane 3) could not compete with the IR1 complex. The ATF and CREB antibodies did not supershift or disrupt the IR1 DNA-binding complex (Fig. 9B, lanes 2 and 3). These results indicate that the transcription factor Sp1 is in the IE86-mediated IR1 DNA-binding complex. Therefore, we conclude that Sp1 and IE86 are components of the IR1 DNA-binding complex.
FIG. 9.
The cellular transcription factor Sp1 binds to the IR1 element. U373-IE86 nuclear extracts were incubated with a radiolabeled IR1 oligonucleotide and a 50-fold excess of unlabeled IR1, CREB, or Sp1 competitor oligonucleotides (panel A, lanes 2 to 4, respectively) or 1 μg of polyclonal antibodies specific for ATF, CREB, or Sp1 (panel B, lanes 2 to 4, respectively). U373-IE86 nuclear extract alone was used as a control (lanes 1). The unlabeled arrows indicate specific complexes. SS, supershifted complex.
A repressor activity present in HeLa cells inhibits IE86-mediated binding of DNA by Sp1.
Sp1 was initially identified as a HeLa cell-derived factor (4, 23). Therefore, the following question remained: why was there no significant IE86-mediated transactivation of the pol promoter in HeLa cells? To address this question, we compared the DNA binding activity of Sp1 in U373-IE86 and HeLa-IE86 cells, using an Sp1 consensus oligonucleotide. The parental U373MG and HeLa cells were used as controls. As indicated in Fig. 10A, strong DNA binding was detected in U373-IE86 nuclear extracts (lane 3). The Sp1 consensus oligonucleotide specifically competed with this DNA-binding activity (Fig. 10B, lane 4), and this activity was supershifted by Sp1 antibody (Fig. 10B, lane 8). However, no DNA binding activity was detected for Sp1 in parental U373MG nuclear extracts (Fig. 10A, lane 2), indicating that the DNA binding activity of Sp1 was induced in the presence of IE86 protein. To address whether this IE86-modulated binding of DNA by Sp1 resulted from upregulation of Sp1 protein expression or enhancement of its DNA binding ability, we performed a Western blot analysis to measure Sp1 protein levels. However, no significant differences between the Sp1 protein levels of U373-IE86 and parental U373MG, HeLa, and HeLa-IE86 cells were found (data not shown), suggesting that the enhanced DNA binding activity of Sp1 in U373-IE86 cells was not due to increased protein levels. Surprisingly, we were unable to detect Sp1 binding in either HeLa-IE86 or parental HeLa cells (Fig. 10A, lane 5 and 6, and data not shown), suggesting that a factor(s) present in HeLa cells competed with Sp1 for its DNA binding.
FIG. 10.
IE86 protein enhances the DNA binding activity of Sp1. (A) U373MG (lane 2), U373-IE86 (lane 3), HeLa (lane 5), and HeLa-IE86 (lane 6) cell nuclear extracts were incubated with a radiolabeled Sp1 consensus oligonucleotide. (B) U373-IE86 nuclear extracts were incubated with the same Sp1 probe and a 50-fold excess of an unlabeled oligonucleotide (oligo) or polyclonal antibodies (Ab) as indicated. The unlabeled arrows indicate the Sp1 DNA-binding complex. SS, supershifted complex.
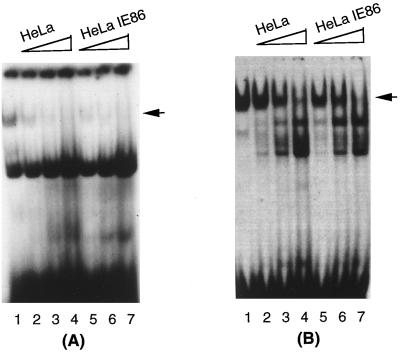
The Sp3 protein has been shown to compete with the Sp1 protein for the same DNA binding site (15). However, we were unable to detect Sp3 activity in the DNA-protein complexes formed in both HeLa and HeLa-IE86 extracts (data not shown). Another possibility is that a repressor activity present in HeLa cells inhibits the Sp1 binding activity. This would probably explain why the IE86 protein was unable to transactivate the pol promoter in HeLa cells. In this case, one would expect to see competition between Sp1 and this repressor for DNA binding. Therefore, we performed a competition experiment by individually titrating HeLa and HeLa-IE86 nuclear extracts to analyze whether they inhibited the IE86-mediated DNA binding activity of Sp1 in U373-IE86 nuclear extracts. As shown in Fig. 11A, the IE86-mediated Sp1-DNA binding to the IR1 probe was gradually inhibited by increasing amounts of either HeLa (compare lane 1 to lanes 2 to 4) or HeLa-IE86 (compare lane 1 to lanes 5 to 7) nuclear extract. A similar experiment performed with an Sp1 probe yielded an identical result (Fig. 11B). However, neither parental U373 nor U373-IE86 nuclear extract could inhibit the formation of IE86-induced Sp1 DNA-binding complex (data not shown). Overall, these data indicate that the repressor activity present in HeLa cells inhibits the IE86-mediated DNA binding activity of Sp1.
FIG. 11.
The factor(s) present in HeLa cells inhibits IE86-mediated DNA-binding activity of Sp1. U373-IE86 nuclear extract (2.5 μg) was incubated with radiolabeled IR1 (A) or Sp1 (B) consensus oligonucleotides, respectively. Increasing amounts of HeLa (lanes 2 to 4) or HeLa-IE86 (lanes 5 to 7) nuclear extract were added as indicated. U373-IE86 nuclear extract plus probe was used as a control (Lanes 1). The arrows indicate Sp1 DNA-binding complex.
DISCUSSION
This study demonstrates that IE86 is one of the essential regulatory proteins involved in the transactivation of the HCMV pol promoter in permissive U373MG cells. Comparison of HCMV-permissive U373MG and HFF cells with HCMV-nonpermissive HeLa and C33-A cells showed that the activation of the pol promoter by IE86 was restricted to the permissive cells. EMSA analysis suggested that IE86-mediated DNA binding of the cellular transcription factor Sp1 may be involved in transactivation of the pol promoter. However, a repressor activity present in HeLa cells prevented this Sp1-DNA binding, which may explain the inhibition of IE86-mediated transactivation of the pol promoter in HeLa cells.
The molecular mechanism of cell permissiveness in HCMV infection remains unknown. The gene expression cascade of HCMV initiates with the expression of the major IE gene. It has been shown that transcription from the major IE gene promoter (MIEP) is different in permissive and nonpermissive cells (39). Recently, Baskar and coworkers (2) provided evidence that the MIEP is regulated in a tissue-specific manner. The majority of tissue and cell types which display MIEP activity parallel tissues naturally infected by HCMV in the human host. Interestingly, many HCMV-infected human cells observed in vivo as well as infected nonpermissive rodent cell culture systems display abundant synthesis of the IE1 and IE2 regulatory proteins, but apparently without concomitant expression of other viral gene products. These observations suggest that sequential gene expression is blocked in an IE protein-independent way. This raises the question of why IE proteins are unable to activate early and late gene expression and suggests a potential mechanism involved in cell permissiveness of HCMV infection.
It is possible that early gene activation mediated by IE or other viral proteins (25) may require a cell- or tissue-specific factor(s) or the functional modification of a cellular transcription factor(s). It has been demonstrated that the pattern of HCMV infection in cultured epithelial and monocyte/macrophage cells is strikingly different from that in cultured fibroblasts (10, 13, 37). The HCMV genome can exist within those cells for prolonged periods with little or no viral gene expression. When the cells reach a certain stage of differentiation, the IE, early, and late genes are expressed sequentially (57). It has been speculated that accessory regulatory proteins may be required for triggering of full lytic-cycle progression in certain cell types as well as for reactivation of latent infections (6). These results suggest that cell differentiation and stimulation may induce cell- or tissue-specific factors which are required for sequential viral gene activation. Here we have provided evidence that the HCMV regulatory protein IE86 is able to modulate binding of DNA by the cellular transcription factor Sp1. This functional change in DNA binding activity may be induced by an interaction between Sp1 and the IE86 protein or by a posttranslational modification of Sp1 induced by IE86. Recently, Yurochko and colleagues (64) demonstrated in an in vitro assay using HEL fibroblasts that IE86 enhances Sp1-DNA binding. These data suggest that the viral regulatory protein may modulate the cellular machinery which mediates early gene activation in both moderately permissive U373MG cells and fully permissive HEL fibroblasts. Interestingly, they also found that viral infection upregulates Sp1 gene expression. In our study, we were unable to detect an increase in expression of the Sp1 protein. This difference suggests that IE86 itself does not induce expression of the Sp1 protein. Nevertheless, the viral infection event and/or viral regulatory proteins could modify the function of cellular transcription factors, thereby stimulating the sequential gene expression events associated with HCMV productive infection.
Alternatively, early gene promoters may be inhibited by a cell- or tissue-specific repressor(s). Huang and Stinski (18) recently demonstrated that a cellular repressor protein is present in abundance in nonpermissive HeLa cells. Binding of the cellular repressor protein to the upstream cis-acting negative regulatory element correlates with repression of transcription from the HCMV early UL4 promoter. Here we have provided further evidence that an activity present in HeLa cells significantly decreases IE86-mediated binding of DNA by Sp1. This repressor activity may compete with the IE86-mediated Sp1-IR1 binding activity directly or indirectly. It has been shown that Sp1 is heavily modified posttranslationally (19, 20). However, the mechanism involved in control of Sp1 activity is not completely understood. Nevertheless, our data suggest that a cell-specific repressor activity may be involved in the inhibition of HCMV early gene activation by IE86 in nonpermissive HeLa cells. This effect appears to be specific for the IR1 element of the pol promoter, since IE86 transactivates the pol promoter equally well in permissive U373MG and HFF cells. It is possible that this mechanism has a general role in cell permissiveness to HCMV infection.
Overall, early gene activation depends on the appropriate interaction between IE gene products and cellular transcription factors. However, the effects of IE proteins on early gene activation may be inhibited in certain cell types. This event would block the viral gene expression cascade that is necessary for productive infection. Since expression of early genes is required for viral DNA replication (17, 44), it is likely that their activation by IE proteins and other viral proteins (25) is also essential for cell permissivity. On the other hand, permissive cells may have a positive transcriptional regulatory factor(s) which is required for IE86-mediated transactivation. This factor(s) may negate the repressive effect of putative repressor factors on the HCMV gene expression cascade.
On the surface, the recent report by Luu and Flores (32) may appear to contradict our results. However, close examination suggests otherwise. Luu and Flores showed that there was a 7- to 10-fold activation of the pol promoter by the RL45 expression vector in HeLa cells. Indeed, we detected a similar activation of the pol promoter by the analogous IE gene expression vector pSVH in HeLa cells. In contrast, we have consistently found that pSVH transactivates the pol promoter in U373MG cells 160-fold. Therefore, we suggest that the finding of Luu and Flores that the association of Sp1 with viral IE proteins may play an important role in regulation of the pol gene transcription does support our data.
In summary, our results showed that IE86, in association with other viral as well as cellular proteins, may play a critical role in the activation of the HCMV pol promoter in permissive U373MG cells. This synergistic activation may require the functional modulation of the cellular transcription factor Sp1. Interestingly, we found that an activity present in nonpermissive HeLa cells inhibits IE86-mediated formation of the IE86-Sp1-IR1 DNA-binding complex. The cell-specific transactivation by IE86 suggests that early gene activation may also play a role in determining cell permissiveness. Identification of the factors responsible for cell-specific activation or inhibition of the HCMV genes should yield insight into the molecular mechanism of host cell restriction characteristic of the life cycle of HCMV.
ACKNOWLEDGMENTS
We thank Peter Ghazal for the RSV IE gene expression constructs, pCM1058, and recombinant IE86 protein and Richard M. Stenberg for plasmid pSVH. We also thank Bernd Stein, Robert Kovelman, and John Westwick for helpful comments and critical reviews of the manuscript.
REFERENCES
- 1.Arlt H, Lang D, Gebert S, Stamminger T. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J Virol. 1994;68:4117–4125. doi: 10.1128/jvi.68.7.4117-4125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baskar J F, Smith P P, Nilaver G, Jupp R A, Hoffmann S, Peffer N J, Tenney D J, Colberg-Poley A M, Ghazal P, Nelson J A. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J Virol. 1996;70:3207–3214. doi: 10.1128/jvi.70.5.3207-3214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 4.Briggs M R, Kadonaga J T, Bell S P, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 5.Chang C-P, Malone C L, Stinski M F. A human cytomegalovirus early gene has three inducible promoters that are regulated differentially at various times after infection. J Virol. 1989;63:281–290. doi: 10.1128/jvi.63.1.281-290.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colberg-Poley A M, Santomenna L D, Harlow P P, Benfield P A, Tenney D J. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMarchi J M. Human cytomegalovirus DNA: restriction enzyme cleavage and map location for immediate early, early and late RNAs. Virology. 1981;114:23–28. doi: 10.1016/0042-6822(81)90249-x. [DOI] [PubMed] [Google Scholar]
- 8.DeMarchi J M, Schmidt C A, Kaplan A S. Pattern of transcription of human cytomegalovirus in permissively infected cells. J Virol. 1980;35:277–286. doi: 10.1128/jvi.35.2.277-286.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depto A S, Stenberg R M. Regulated expression of the human cytomegalovirus pp65 gene: octamer sequence in the promoter is required for activation by viral gene products. J Virol. 1989;63:1232–1238. doi: 10.1128/jvi.63.3.1232-1238.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Detrick B, Rhame J, Wang Y, Nagineni C N, Hooks J J. Cytomegalovirus replication in human retinal pigment epithelial cells: altered expression of viral early proteins. Invest Ophthalmol Vis Sci. 1996;37:814–825. [PubMed] [Google Scholar]
- 11.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiala M, Payne J E, Berne T V, et al. Epidemiology of cytomegalovirus infection after transplantation and immunosuppression. J Infect Dis. 1975;132:421–433. doi: 10.1093/infdis/132.4.421. [DOI] [PubMed] [Google Scholar]
- 13.Fish K N, Britt W, Nelson J A. A novel mechanism for persistence of human cytomegalovirus in macrophage. J Virol. 1996;70:1855–1862. doi: 10.1128/jvi.70.3.1855-1862.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furnari B A, Poma E, Kowalik T F, Huong S-M, Huang E-S. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J Virol. 1993;67:4981–4991. doi: 10.1128/jvi.67.8.4981-4991.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang E S, Kowalik T F. The pathogenicity of human cytomegalovirus. In: Becker Y, Darai G, Huang E-S, editors. Molecular aspects of human cytomegalovirus diseases. Berlin, Germany: Springer-Verlag; 1993. pp. 3–45. [Google Scholar]
- 18.Huang L, Stinski M F. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J Virol. 1995;69:7612–7621. doi: 10.1128/jvi.69.12.7612-7621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson S P, MacDonald J J, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 20.Jackson S P, Tjian R. O-Glycosylation of eukaryotic transcription factors: implication for mechanism of transcriptional regulation. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson M A, Mills J. Serious cytomegalovirus disease in the acquired immunodeficiency syndrome (AIDS). Clinical findings, diagnosis, and treatment. Ann Intern Med. 1988;108:585–594. doi: 10.7326/0003-4819-108-4-585. [DOI] [PubMed] [Google Scholar]
- 22.Jahn G, Knust E, Schmolla H, Sarre T, Nelson J A, McDougall J K, Fleckenstein B. Predominant immediate-early transcripts of human cytomegalovirus AD 169. J Virol. 1984;49:363–370. doi: 10.1128/jvi.49.2.363-370.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadonaga J T, Carner K R, Masiarz F R, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 24.Kerry J A, Priddy M A, Stenberg R M. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J Virol. 1994;68:4167–4176. doi: 10.1128/jvi.68.7.4167-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerry J A, Priddy M A, Jervey T Y, Kohler C P, Staley T L, Vanson C D, Jones T R, Iskenderian A C, Anders D G, Stenberg R M. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J Virol. 1996;70:373–382. doi: 10.1128/jvi.70.1.373-382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerry J A, Priddy M A, Staley T L, Jones T R, Stenberg R M. The role of ATF in regulating the human cytomegalovirus DNA polymerase (UL54) promoter during viral infection. J Virol. 1997;71:2120–2126. doi: 10.1128/jvi.71.3.2120-2126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klucher K M, Sommer M, Kadonaga J T, Spector D H. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol Cell Biol. 1993;13:1238–1250. doi: 10.1128/mcb.13.2.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouzarides T, Bankier A T, Satchwell S C, Preddy E, Barrell B G. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. [DOI] [PubMed] [Google Scholar]
- 29.Lang D, Stamminger T. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol. 1993;67:323–331. doi: 10.1128/jvi.67.1.323-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukac D M, Manuppello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luu P, Flores O. Binding of SP1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter. J Virol. 1997;71:6683–6691. doi: 10.1128/jvi.71.9.6683-6691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marker S C, Howard R J, Simmons R L, et al. Cytomegalovirus infection: a quantitative prospective study of 320 consecutive renal transplants. Surgery. 1981;89:660–671. [PubMed] [Google Scholar]
- 35.McDonough S H, Spector D H. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology. 1983;125:31–46. doi: 10.1016/0042-6822(83)90061-2. [DOI] [PubMed] [Google Scholar]
- 36.Meyer J D, Spencer H C, Jr, Watts J C, et al. Cytomegalovirus pneumonia after human marrow transplantation. Ann Intern Med. 1975;82:181–188. doi: 10.7326/0003-4819-82-2-181. [DOI] [PubMed] [Google Scholar]
- 37.Minton E J, Tysoe C, Sinclair J H, Sissons J G P. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreira J L, Wirth M, Fitzek M, Hauser H. Evaluation of reporter genes in mammalian cell lines. Methods Mol Cell Biol. 1992;3:23–29. [Google Scholar]
- 39.Nelson J A, Reynolds-Kohler C, Smith B A. Negative and positive regulation by a short segment in the 5′-flanking region of the human cytomegalovirus major immediately-early gene. Mol Cell Biol. 1987;7:4125–4129. doi: 10.1128/mcb.7.11.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pizzorno M C, Mullen M-A, Chang Y-N, Hayward G S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubin R H, Cosimi A B, Tolkoff-Rubin N E, Russell P S, Hirsch M S. Infectious disease syndromes attributable to cytomegalovirus and their significance among renal transplant recipients. Transplantation. 1977;24:458–464. doi: 10.1097/00007890-197712000-00010. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz R, Sommer M H, Scully A, Spector D H. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J Virol. 1994;68:5613–5622. doi: 10.1128/jvi.68.9.5613-5622.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spaete R R, Mocarski E S. Regulation of cytomegalovirus gene expression: α and β promoters are trans activated by viral functions in permissive human fibroblasts. J Virol. 1985;56:135–143. doi: 10.1128/jvi.56.1.135-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spector D H, Klucher K M, Rabert D K, Wright D A. Human cytomegalovirus early gene expression. Curr Top Microbiol Immunol. 1990;154:21–45. doi: 10.1007/978-3-642-74980-3_2. [DOI] [PubMed] [Google Scholar]
- 46.Stango S, Pass R F, Cloud G, Britt W J, Henderson R E, Waltson P D, Veren D A, Page F, Alford C A. Primary cytomegalovirus infection in pregnancy. Incidence, transmission to fetus and clinical outcome. JAMA. 1986;256:1904–1908. [PubMed] [Google Scholar]
- 47.Staprans S I, Rabert D K, Spector D H. Identification of sequence requirements and trans-acting functions necessary for regulated expression of a human cytomegalovirus early gene. J Virol. 1988;62:3463–3473. doi: 10.1128/jvi.62.9.3463-3473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stenberg R M. Immediate-early genes of human cytomegalovirus: organization and function. In: Becker Y, Darai G, Huang E-S, editors. Molecular aspects of human cytomegalovirus diseases. Berlin, Germany: Springer-Verlag KG; 1993. pp. 330–359. [Google Scholar]
- 49.Stenberg R M, Thomsen D R, Stinski M F. Structural analysis of the major immediate early gene of human cytomegalovirus. J Virol. 1984;49:190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stenberg R M, Depto A S, Fortney J, Nelson J A. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J Virol. 1989;63:2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stinski M F, Thomsen D R, Stenberg R M, Goldstein L C. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1983;46:1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tenney D J, Colberg-Poley A M. Expression of the human cytomegalovirus UL36-38 immediate early region during permissive infection. Virology. 1991;182:199–210. doi: 10.1016/0042-6822(91)90663-v. [DOI] [PubMed] [Google Scholar]
- 55.Tenney D J, Colberg-Poley A M. Human cytomegalovirus UL36-38 and US3 immediate-early genes: temporally regulated expression of nuclear, cytoplasmic, and polysome-associated transcripts during infection. J Virol. 1991;65:6724–6734. doi: 10.1128/jvi.65.12.6724-6734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomsen D R, Stenberg R M, Goins W F, Stinski M F. Promoter-regulatory region of the major immediate early gene of human cytomegalovirus. Proc Natl Acad Sci USA. 1984;81:659–663. doi: 10.1073/pnas.81.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsutsui Y, Nogami-Satake T. Differential expression of the major immediate early gene of human cytomegalovirus. J Gen Virol. 1990;71:115–124. doi: 10.1099/0022-1317-71-1-115. [DOI] [PubMed] [Google Scholar]
- 58.Wathen M W, Stinski M F. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol. 1982;41:462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wathen M W, Thomsen D R, Stinski M F. Temporal regulation of human cytomegalovirus transcription at immediate early and early times after infection. J Virol. 1981;38:446–459. doi: 10.1128/jvi.38.2.446-459.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weston K. An enhancer element in the short unique region of human cytomegalovirus regulates the production of a group of abundant immediate early transcripts. Virology. 1988;162:406–416. doi: 10.1016/0042-6822(88)90481-3. [DOI] [PubMed] [Google Scholar]
- 61.Wilkinson G W G, Akrigg A, Greenaway P J. Transcription of the immediate early genes of human cytomegalovirus strain AD169. Virus Res. 1984;1:101–116. doi: 10.1016/0168-1702(84)90067-4. [DOI] [PubMed] [Google Scholar]
- 62.Wu, J., and M. S. Barbosa. Unpublished data.
- 63.Yurochko A D, Kowalik T F, Huong S-M, Huang E-S. Human cytomegalovirus upregulates NF-κB activity by transactivating the NF-κB p105/p50 and p65 promoters. J Virol. 1995;69:5391–5400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yurochko A D, Mayo M W, Poma E E, Baldwin A S, Jr, Huang E-S. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-κB promoters. J Virol. 1997;71:4638–4648. doi: 10.1128/jvi.71.6.4638-4648.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]