Abstract
We have identified a region of the herpes simplex virus major DNA-binding protein (ICP8) which is involved in cooperative binding to single-stranded DNA. This has been accomplished by analysis of ICP8 which was covalently modified by reaction with the extrinsic fluorophore fluorescein-5-maleimide (FM). Reaction conditions which result in the incorporation of 1 mol of FM per mol of ICP8 have been established. The binding properties of the modified protein were analyzed by polyacrylamide gel shift analysis with model oligonucleotides. This analysis indicates that while intrinsic binding is similar to that observed with unmodified protein, the cooperative binding of the modified protein to single-stranded DNA is significantly altered. Helix-destabilizing assays, whose results are a reflection of cooperative binding, also indicate that this property of ICP8 is decreased upon modification with FM. Mapping of the site of modification by cyanogen bromide cleavage and peptide sequencing has shown that the major site of modification is cysteine 254. This position in the primary structure of ICP8 is distinct from the regions previously shown to be involved in the interaction of this protein with single-stranded DNA.
The herpes simplex virus type 1 (HSV-1) UL29 gene product, commonly designated ICP8, is the major single-strand-DNA-binding protein (SSB) present in infected cells. ICP8 is one of seven HSV-1 gene products required for origin-dependent replication of viral DNA (7, 58). Functional ICP8 is absolutely required for viral growth, and conditional lethal mutants in ICP8 exhibit a DNA-negative phenotype as well as abnormalities in the production of viral proteins (19, 57). The protein is involved in the formation of prereplication complexes in infected cells (5, 11, 12, 17). ICP8 stimulates the activity of the viral DNA polymerase and the helicase activity of the HSV-1 origin-binding protein UL9 (1, 3, 8, 23, 41, 43, 47). ICP8 also optimizes the helicase and primase activities of the heterotrimeric HSV helicase-primase (10) in a species-specific manner, most likely through an interaction with the UL8 subunit of that complex (9, 53). Genetic studies have implicated a role for ICP8 in gene regulation in HSV-infected cells (18, 20–22, 57), and evidence that ICP8 promotes homologous pairing and strand transfer, suggesting a role in recombination, has been presented (2, 4, 13, 14).
The ability of ICP8 to interact with nucleic acids has been the subject of extensive study since its initial identification. Work from many laboratories has shown that ICP8 binds preferentially and cooperatively to single-stranded DNA (2, 13, 42, 45–49). This interaction is reminiscent of, although not identical to, that observed with the T4 gene 32 protein and the Escherichia coli single-strand-DNA-binding protein, based on results of agarose gel electrophoresis and electron microscopy. Nitrocellulose filter competition assays indicate that ICP8 is capable of binding to single-stranded polyribonucleotide homopolymers to an extent similar to that observed with single-stranded DNA. ICP8 also binds to duplex DNA and to short single-stranded oligonucleotides (34, 48). These interactions, however, are considerably weaker than those observed with single-stranded DNA. The presence of ICP8 destabilizes poly(dA)-poly(dT) duplexes (55), and it has been shown that ICP8 is capable of displacing short oligonucleotides annealed to long single strands of DNA (2). Finally, ICP8 has been shown to aid in the annealing of separated DNA strands under the proper conditions (13). These last two properties have been attributed to the cooperative nature of ICP8 binding.
The work described here was carried out in an effort to determine if the extrinsic, cysteine-specific fluorophore fluorescein-5-maleimide (FM) (Fig. 1) could be used as a reporter to identify and map domains of ICP8 that are involved in, or are conformationally altered by, protein-protein and/or protein-DNA interactions. FM modification of cysteines has previously been used to probe the structure of E. coli DNA polymerase III and the movement of E. coli ribosomal proteins within the 30S subunit (24, 25, 39). Further precedence for this type of analysis has been demonstrated by work involving the integrase protein of bacteriophage lambda (27, 44), myosin (26), the ATPase of E. coli (6), chymotrypsin (56), and the trp repressor (15). These studies all involved the modification of amino acid residues by chemical reagents (including maleimide derivatives) and/or fluorescent probes to gain insight into protein structure-function relationships.
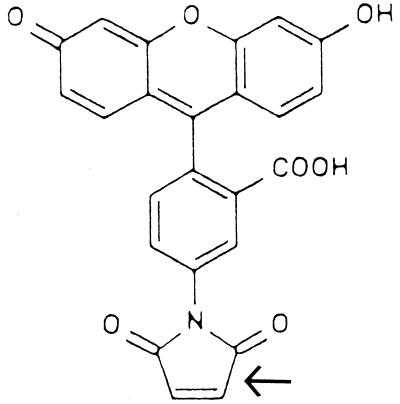
FIG. 1.
Structure of FM. The reactive maleimide moiety is indicated by the arrow.
In the course of the work presented here, the position of the cysteine side chain most reactive with FM was mapped and the interaction of the modified ICP8 was analyzed by polyacrylamide gel shift and helix-destabilizing assays. These experiments resulted in the identification of a residue within a region of ICP8 which is involved in the cooperative interaction of the protein with single-stranded DNA.
MATERIALS AND METHODS
Chemicals and reagents.
[α-32P]dATP and [3H]dTTP were purchased from New England Nuclear Laboratories. dCTP, dGTP, dATP, and dTTP were purchased from Sigma. The 17-base oligomer, 5′ GTTTTCCCAGTCACGAC 3′, was purchased from GIBCO/BRL. Deoxythymidine oligonucleotides were prepared by Michael Flora (Biomedical Instrumentation Center, Uniformed Services University of the Health Sciences, Bethesda, Md.). All enzymes were purchased from GIBCO/BRL. FM and anti-FM antibody were purchased from Molecular Probes Inc. Heparin-Sepharose, Sephadex G-50, and Seakem agarose were purchased from Pharmacia. Lambda bacteriophage double-stranded DNA, TPCK [l-1-chloro-3-(4-tosylamido)-4-phenyl-2-butanone N-tosyl-l-phenylalanine chloromethyl ketone], TLCK [l-1-chloro-3-(4-tosylamido)-7-amino-2-heptanone hydrochloride N-α-tosyl-l-lysine chloromethyl ketone], and bacteriophage M13mp19 single-stranded DNA were purchased from Boehringer Mannheim. Single-strand DNA agarose was purchased from GIBCO/BRL.
Cells and viruses.
Spodoptera frugiperda 21 cells were purchased from Clontech Laboratories. High Five insect cells were a gift from Edward Niles, State University of New York at Buffalo. Recombinant baculovirus expressing the HSV-1 UL29 gene under the control of the polyhedrin promoter was a gift from Mark Challberg.
ICP8 isolation.
ICP8 was isolated from S. frugiperda 21 or High Five insect cells infected with a recombinant baculovirus containing the HSV UL29 gene under the control of the baculovirus polyhedrin promoter. Insect cells were grown in monolayer culture to approximately 50% confluency in 10 25- by 150-mm dishes and infected with recombinant baculovirus at a multiplicity of infection of ∼10 PFU/cell. The flasks were incubated at 27°C for 1 h, the medium was removed, and 15 ml of fresh medium was added. Protein production was monitored by resolution of the proteins present in infected cell lysates by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Cells were harvested at peak protein production, usually 4 days postinfection, by centrifugation at 150 × g for 10 min.
The cells were lysed on ice by the addition of buffer containing 1.7 M KCl, 50 mM Tris (pH 7.5), 5 mM EDTA, 0.5 mM dithiothreitol (DTT), 0.2% Nonidet P-40, 20 μg of phenylmethylsulfonyl fluoride per ml, 2.5 μg of leupeptin per ml, 0.1 mM TLCK, and 0.1 mM TPCK. The resulting viscous solution was centrifuged at 75,000 × g for 6 h at 4°C. The supernatant was collected and centrifuged at 40,000 × g overnight. The second supernatant was collected and dialyzed against a buffer containing 150 mM KCl, 50 mM Tris (pH 7.5), 5 mM EDTA, 0.5 mM DTT, and 10% glycerol (buffer A) plus the protease inhibitors at the previous concentrations. The dialysate was centrifuged at 1,000 × g for 10 min. The supernatant was applied to a single-stranded-DNA agarose column (2.5 by 30 cm), washed with the dialysis buffer, and eluted with dialysis buffer containing 1 M KCl. Fractions (approximately 4 ml) were collected and analyzed by SDS-PAGE.
The fractions containing ICP8 were pooled and dialyzed overnight against buffer A. This dialysate was then applied to a heparin-Sepharose column (1 by 10 cm), and 1-ml fractions of the flowthrough were collected. Under these buffer conditions, ICP8 does not bind to heparin-Sepharose whereas contaminants are bound, in contrast to binding under buffer conditions with lower concentrations of salt and the presence of Nonidet P-40. Fractions were once again analyzed by SDS-PAGE and Western blot analysis with an anti-ICP8 antibody. ICP8-containing fractions were pooled and dialyzed against storage buffer (150 mM KCl, 10 mM Tris [pH 7.5], 1 mM EDTA, 0.1 mM DTT, 50% glycerol) and stored at −20°C. Approximately 70 mg of ICP8 was routinely purified from approximately 5 × 107 cells. The concentration of ICP8 was quantified by UV absorbance at 205 nm by the procedure of Scopes (51).
Anti-FM affinity chromatography.
An anti-FM affinity column was generated by coupling anti-FM antibody to cyanogen bromide (CNBr)-activated Sepharose 4B (Pharmacia) as per the manufacturer’s instructions. The resulting slurry was poured into a Bio-Rad Econocolumn with a final bed volume of 1 ml. The column was equilibrated with 10 mM CAPS (3-[cyclohexylamino]-1-propanesulfonic acid) buffer, pH 9.0. ICP8-FM was dialyzed in CAPS buffer overnight. Fifty micrograms of ICP8-FM was applied to the column, and 0.5-ml fractions were collected. Each fraction was monitored for protein by 280-nm-absorption readings on a Beckman DU 640 spectrophotometer. The amount of ICP8-FM in each fraction was calculated by using an extinction coefficient of 8.14 × 104 M−1 cm−1 for ICP8 (59).
39S monoclonal antibody.
The ICP8-specific 39S monoclonal antibody was prepared from the supernatant of cultures of the 39S mouse monoclonal cell line (52) obtained from the American Type Culture Collection (Rockville, Md.).
Modification of ICP8 with FM.
ICP8 in storage buffer was dialyzed against two changes of 2 M KCl in buffer A followed by dialysis against two 1-liter changes of 150 mM KCl in buffer A. The first dialysis was performed to eliminate ICP8 aggregation, which tends to occur at high concentrations of protein. Covalent modification of ICP8 with FM was carried out at ratios of 0.1 μmol of ICP8 to 0.5, 1.1, or 2.2 μmol of FM at 10°C for 50, 500, and 1,000 s. Reactions were quenched with a fivefold excess of β-mercaptoethanol, and ICP8-FM was separated from free FM by a 1-ml Sephadex G-50 spin column. Calculations of the moles of fluorophore bound per mole of ICP8 were determined by spectrophotometric readings by using the extinction coefficient for FM of 80,000 M−1 cm−1 at 490 nm. The concentration of ICP8 was quantified by UV absorbance at 280 nm. These readings were corrected to the absorbance of the protein at 205 nm to allow determination of the concentration of protein at this wavelength.
Cleavage and analysis of ICP8.
ICP8 was proteolytically cleaved with protease V8 or chemically cleaved with cyanogen bromide. A ratio of 100 μg of ICP8 to 1 μg of protease V8 was used. The reaction was allowed to proceed for 2 h at 37°C. Peptides were resolved onto an SDS–10% polyacrylamide gel and stained with Coomassie brilliant blue. CNBr cleavage of ICP8 was done at a ratio of 1 mol of ICP8 to 50 mol of CNBr at 25°C for 16 to 20 h. Approximately 100 μg of cleaved ICP8 was resolved on a Tricine-SDS-polyacrylamide gel. Tricine-SDS-polyacrylamide gels were prepared and run by the method of Schagger and von Jagow (50). Samples for N-terminal sequencing were generated by the transfer of peptides from the Tricine gel to polyvinylidene fluoride (PVDF) paper in 10 mM CAPS buffer, pH 11.0. Peptides containing the FM moiety were visualized by illumination with long-wavelength UV light and marked accordingly. The PVDF membrane was stained with Coomassie brilliant blue, and the relevant bands were excised for N-terminal amino acid sequencing.
Poly(dT) oligonucleotides.
Poly(dT) oligonucleotides 10, 25, and 50 bases in length were dissolved in 500 μl of distilled water and centrifuged at 14,000 × g for 10 min. The supernatant was withdrawn from any insoluble residue, extracted three times with 300 μl of water-saturated n-butanol, and evaporated to dryness. The oligomers were dissolved in 100 μl of a 0.5 M ammonium acetate buffer (pH 6.5), and 1 ml of 100% ethanol was added. The samples were placed at −80°C for 30 min, centrifuged at 14,000 × g for 1 min at 4°C, washed twice with 80% ethanol, and dried. The pellets were dissolved in water, and the concentrations of deoxythymidine oligonucleotides were determined spectrophotometrically with an extinction coefficient of 8.1 × 103 M−1 (nucleotide) cm−1 at 260 nm (31). Oligonucleotide (100 ng) was labeled at the 5′ terminus with [γ-32P]ATP (6,000 Ci/mmol) with T4 polynucleotide kinase. Unincorporated nucleotide was removed by spin column chromatography with Sephadex G-50. Labeled oligonucleotides were checked by gel electrophoresis to ensure uniformity of size and lack of contamination of the labeled product.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed with the poly(dT) oligonucleotides. Assay mixtures consisted of 10 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 150 mM KCl, 10% glycerol, and 10 mM β-mercaptoethanol in a total volume of 25 μl. The amounts of oligo(dT) used in the appropriate assay were 167 fmol of oligo(dT)10, 66 fmol of oligo(dT)25, and 33 fmol of oligo(dT)50. Various amounts of ICP8 or ICP8-FM were used as indicated in the Results. Reaction mixtures were incubated for 10 to 15 min on ice. Three microliters of sample dye (25% glycerol, 20 mM EDTA, 0.25% bromophenol blue [pH 7.0]) was added, and samples were immediately applied to a Tris-borate-EDTA (TBE)–6% polyacrylamide gel. The gel was run at 6 V/cm until the samples completely entered the gel, at which time the voltage was decreased to 3.4 V/cm and the gel was electrophoresed for approximately 12 to 18 h at 4°C. The gels were dried and exposed to X-ray film for autoradiography. The bands were excised from the dried gel and counted in a Wallac scintillation counter. The fraction of bound oligomer was determined by dividing the counts obtained from a single band by the total number of counts obtained per lane. Ninety to 100% of the input labeled oligonucleotide was accounted for in each lane.
Calculation of binding parameters.
The calculations of the intrinsic binding constant and cooperativity parameter were done by using the models of Monini and colleagues (38). The reaction of an oligonucleotide carrying two binding sites, a and b, is described by the fraction of DNA molecules carrying no bound protein (f0), one bound protein (f1), or two bound proteins (f2). These fractions can be expressed as a function of the association constants, ka and kb, the cooperativity parameter, ω, and the concentration of free protein. The number of molecules with one bound protein reaches a maximum and then declines as the concentration of protein is increased and the second binding site becomes occupied. At the point of maximal f1, the derivative of f1, [df1/dP], where P is total protein added, is 0. The value of f1 at this point can be used to determine the cooperativity parameter with ω equal to ([1/f1max] − 1)2, assuming ka is equal to kb. A cooperativity parameter equal to 1 represents no cooperativity, a value greater than 1 represents positive cooperativity, and a value below 1 represents negative cooperativity.
Helix-destabilizing experiments.
The generation of substrates for the helix-destabilizing assay involved the annealing of single-stranded M13mp19 DNA with a 17-base primer, 5′ GTTTTCCCAGTCACGAC 3′, which is complementary to residues 924 to 940 of the circular bacteriophage DNA. The primer was labeled at the 5′ terminus with [γ-32P]ATP (6,000 Ci/mmol) by using T4 polynucleotide kinase. Hybrids were formed by annealing 45 pmol of labeled primer with 4.5 pmol of M13mp19 single-stranded DNA in buffer containing 10 mM Tris-HCl (pH 7.5) and 5 mM MgCl2. The reaction mixture was heated to 90°C and allowed to slowly cool to 30°C. EDTA was added to a final concentration of 10 mM, and the excess oligonucleotide was removed by gel filtration through a 10-ml Bio-Gel A15m column equilibrated with 10 mM Tris-HCl (pH 7.5)–1 mM EDTA.
Helix-destablizing experiments were performed by the method of Boehmer and Lehman (2). Reactions were performed at 37°C for 15 min in buffer containing 20 mM Tris-HCl (pH 7.5), 5 mM DTT, 10% glycerol, 0.1 mg of bovine serum albumin, 640 ng of primed M13mp19 circular single-stranded DNA, and various amounts of ICP8 or ICP8-FM in a total reaction volume of 10 μl. Reactions were terminated by the addition of 1.5 μl of a solution containing 90 mM EDTA (pH 8.0), 12% SDS, 30% glycerol, 0.25% bromophenol blue, and 0.25% xylene cyanol buffer. The products were resolved by electrophoresis on a 10% polyacrylamide TBE gel and visualized by autoradiography. The amounts of free and annealed oligonucleotide were quantified by counting the radioactive bands excised from the gel.
RESULTS
Reaction of ICP8 with FM.
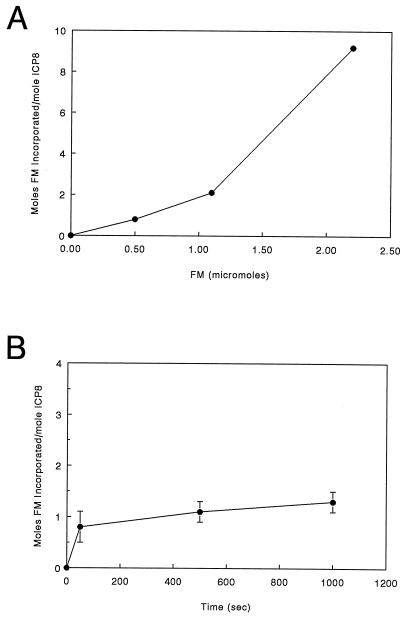
FM is a bulky charged molecule with an extensive conjugated ring system, which gives rise to its fluorescent properties (Fig. 1). The conditions under which reaction of ICP8 with FM were carried out result in exclusive covalent modification of cysteine residues (25, 38). Thus, we attempted to define conditions under which one or a few of the 23 cysteines present in ICP8 were modified. Such an approach would allow essentially site-specific modification of the native protein. Once such conditions were established, the modified protein would subsequently be characterized, and if its properties were found to be significantly different from those of the unmodified ICP8, the locations of the modified residues would be mapped. Experiments on the incorporation of FM into ICP8 indicated that the amount of fluorophore incorporated per mole of ICP8 was both time and concentration dependent. The results of these studies are presented in Fig. 2. They clearly show that conditions which result in the incorporation of approximately 1 mol of FM per mol of ICP8 at relatively low concentrations of FM have been established. In contrast, at higher ratios of FM to ICP8, signficant numbers of cysteine residues are modified. Extension of these labeling experiments with both longer times and increased concentrations of FM indicate that a maximum of 9 to 11 mol of FM can be incorporated per mol of ICP8 under these solution and temperature conditions (unpublished observations).
FIG. 2.
Reaction of ICP8 with FM. (A) Reaction of ICP8 in the presence of increasing concentrations of FM. All reactions were carried out with 0.1 μmol of ICP8 for 50 s at room temperature. (B) Time course of FM incorporation with 0.5 μmol of FM and 0.1 μmol of ICP8.
In an effort to determine if the approximately 1 mol of FM incorporated per mol of ICP8 represented a true average per molecule or if only a subpopulation was heavily labeled, we adsorbed the FM-labeled ICP8 to an anti-FM affinity column. Fifty micrograms of labeled protein was adsorbed to the column, which was then extensively washed. Essentially all of the modified protein appeared to bind to the column, since no protein was contained within the washes of the column after ICP8-FM application. One-half milliliter of a 0.2 M glycine–0.1 M HCl (pH 2.8) solution was then applied to the column, followed by 10 ml of 10 mM CAPS (pH 9.0) buffer (50), and 0.5-ml fractions were collected ICP8-FM (47 μg) was eluted from the column with glycine-HCl. Since approximately 94% of the ICP8-FM was retained on the anti-FM column and required glycine-HCl for elution, the assumption of an average value of approximately 1 mol of FM incorporated per mol of ICP8 is valid. Prior to the application of ICP8-FM, 50 μg of unmodified ICP8 was applied to the anti-FM affinity column under the same conditions as used in a control experiment. Less than 5% of the ICP8 was retained on the column.
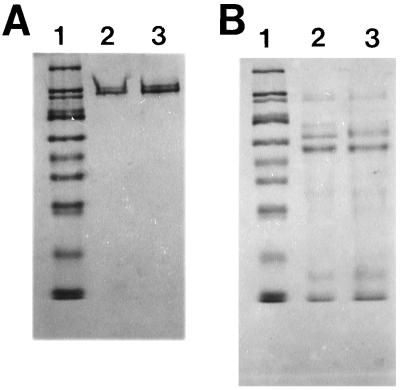
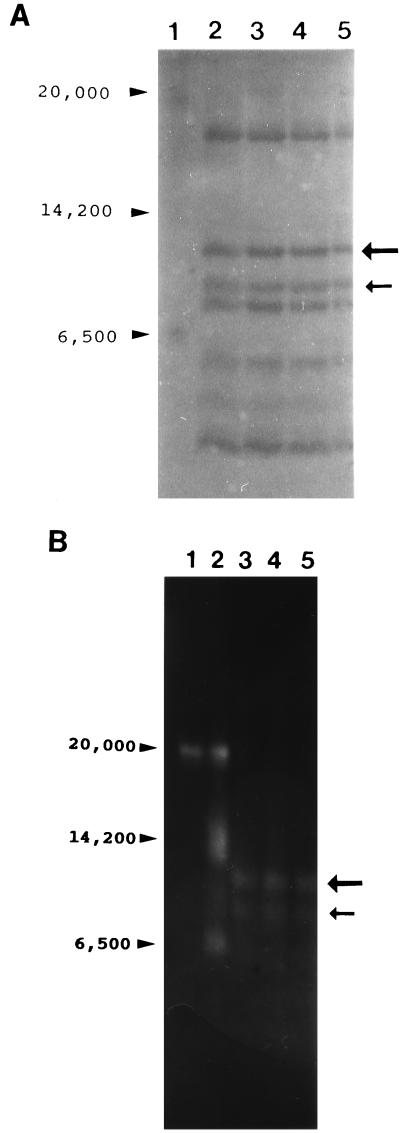
SDS-PAGE of the modified protein on 10% polyacrylamide gels showed that no degradation occurred as a result of the labeling procedure (Fig. 3A). It should be noted that both the unmodified and the modified ICP8 ran as a doublet in these gels. Observation of ICP8 doublets in SDS-polyacrylamide gels have been previously reported and have been attributed to the presence of two conformational forms of the protein, most likely reflecting the presence or absence of one or more disulfide bonds in the two species (30, 33). The similar migration patterns shown here indicate that modification with 1 mol of FM did not affect the protein at this level. In order to determine if the incorporation of 1 mol of FM caused a significant change in the conformation of ICP8, V8 protease digestions were carried out. As can be seen in Fig. 3B, while some differences are present, the overall patterns of digestion for modified and unmodified protein are very similar. The possibility of a major alteration in conformation was also addressed with the 39S anti-ICP8 monoclonal antibody (52). Immunoreactivity of this antibody with ICP8 has been shown to be sensitive to conformational changes in ICP8 temperature-sensitive mutants (57). No difference was observed between ICP8-FM and ICP8 based on reactivity with the 39S antibody (data not shown). Thus, by the criteria used, the incorporation of 1 mol of FM per mol of ICP8 does not significantly change the structure of the protein.
FIG. 3.
SDS-PAGE analysis of ICP8 modified with approximately 1 mol of FM/mol of protein. Lane 1 in both panels contains the following molecular weight markers: 205,000; 116,000; 97,000; 84,000; 66,000; 55,000; 45,000; 36,000; 29,000; and 24,000. (A) Lane 2, intact ICP8; lane 3, ICP8 FM. Note that both proteins show exactly the same pattern. The doublet, occasionally seen with ICP8, has been attributed to the presence of a relatively reduction-resistant disulfide linkage which FM modification would not disrupt. (B) V8 protease digests of ICP8 (lane 2) and ICP8-FM (lane 3). All ICP8 lanes in panels A and B contain 10 μg of protein.
EMSA.
We next wished to determine if the single-stranded-DNA binding activity of ICP8 was altered due to modification of the protein with 1 mol of FM. EMSAs were employed for this analysis, since both the intrinsic DNA dissociation constant and the cooperativity parameter of the interaction can be determined by this technique. An oligonucleotide providing a single ICP8 binding site is required for the calculation of the intrinsic dissociation constant, and the cooperativity parameter can be estimated with oligonucleotides containing two ICP8 binding sites.
Previous determinations of the site size of ICP8 binding to single-stranded DNA have ranged from 10 to 12 nucleotides to 40 nucleotides/protein. The most recent estimate of the ICP8 binding site size is 17 to 18 nucleotides based on a careful electron microscopic study using linear DNA fragments of defined lengths. Due to the circular nature of the observed complexes, however, it was surmised that the protein-DNA complexes had assumed a higher-order structure (4). These differences clearly depend in part on the experimental procedure used to determine this parameter (4, 40, 41, 45), since significant assay-dependent variation in the apparent binding site sizes of the T4 gene 32 protein, E. coli SSB, and the adenovirus SSB has also been reported (31, 32, 36). Assuming the lower-limit site size was correct, oligo(dT)10 was used in EMSAs as a potential single binding site substrate for ICP8. ICP8 (6 to 40 pmol) was titrated against a fixed amount (167 fmol) of oligo(dT)10, and the complexes were resolved by electrophoresis on a TBE–6% acrylamide gel. ICP8 did not bind to the oligo(dT)10 under the conditions of this assay, an assessment based on lack of any shifted bands at all of the ICP8 concentrations used (data not shown). When oligomers 17 nucleotides in length were used, the shifts seen were not reproducible and the stability of the oligonucleotide-ICP8 complexes as judged by nitrocellulose filter binding assays was very weak (12a).
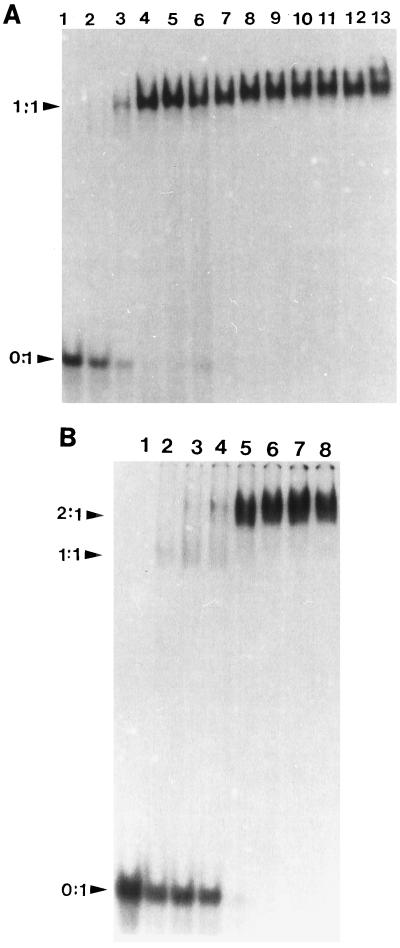
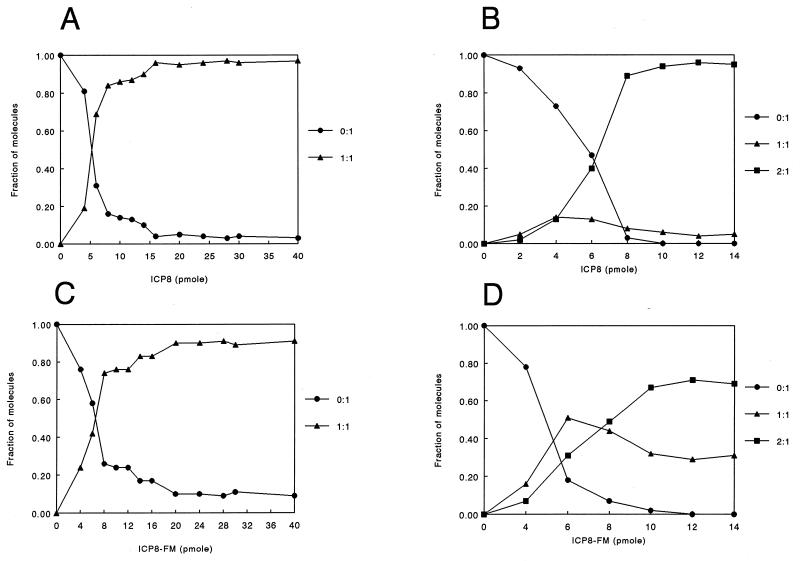
The ability of ICP8 to bind oligo(dT)25 was then examined, and the results from EMSAs and a plot of the fraction(s) of the molecular species detected are shown in Fig. 4A and 5A, respectively. In this case the observed shift was stable and reproducible and two species were seen in the presence of both ICP8 and ICP8-FM binding to the oligo(dT)25 (free oligonucleotide and bound oligonucleotide) (Fig. 5A and C). These results indicate that the oligo(dT)25 contained a single binding site for both proteins and that the additional length of the oligomer may be required for stable complex formation. The binding of ICP8 and ICP8-FM to oligo(dT)50, which presumably contains two sites, was investigated next. Two shifted bands, which correspond to one protein binding (1:1) and two proteins binding (2:1), were observed (Fig. 4B). Longer exposure of the autoradiograms did not reveal any additional bands, further suggesting that only two sites exist on this model binding substrate. Plots of the fractions of molecular species detected in the gel as a function of increasing concentration of ICP8 or ICP8-FM protein are shown in Fig. 5B and D. With ICP8, the lower of the two shifted bands, which corresponds to a 1:1 protein-to-oligonucleotide ratio, was present at relatively low levels and diminished as the amount of ICP8 increased. In contrast, two shifted bands (i.e., two species of bound oligonucleotide) were present at significant levels throughout the titration with ICP8-FM.
FIG. 4.
Typical EMSA data for ICP8 with oligo(dT)25 (A) and oligo(dT)50 (B). (A) Lanes 1 to 13 contain 66 fmol of labeled oligo(dT)25 and 0, 4, 6, 10, 12, 14, 16, 20, 24, 28, 30, and 40 pmol of ICP8, respectively. (B) Lanes 1 to 8 contain 33 fmol of oligo(dT)50 and 0, 2, 4, 6, 8, 10, 12, and 14 pmol of ICP8, respectively.
FIG. 5.
Quantification of gel shift data for ICP8 (A and B) and ICP8-FM (C and D). (A and C) Results with oligo(dT)25; (B and D) results with oligo(dT)50. Note the much higher level of the intermediate band with ICP8-FM than that with ICP8.
Calculation of the binding parameters of ICP8 and ICP8-FM for oligo(dT)25 and oligo(dT)50.
The dissociation constant of ICP8 and ICP8-FM was determined for the interaction of each protein with oligo(dT)25. The concentration of free protein at half saturation corresponds to the dissociation constant as described by Monini et al. (38). The dissociation constants were 2.1 × 10−7 M for ICP8 and 2.6 × 10−7 M for ICP8-FM. These values were calculated by averaging the Kd values obtained from triplicate experiments. These data suggest that the intrinsic DNA binding of ICP8-FM (interaction of the protein with DNA containing a single site) is not affected by the presence of the FM moiety.
Since oligo(dT)50 apparently contains two binding sites, estimates for the cooperativity parameters for ICP8 and ICP8-FM could be calculated from the EMSA data as described in Materials and Methods, by determining the values of f1max. The estimated cooperativity parameter determined for ICP8 was 38 with an f1max of 0.14% ± 0.06% taken from an average of triplicate experiments. This value indicates modest positive cooperativity in ICP8-ICP8 interactions with this model oligonucleotide. The estimated cooperativity parameter determined for ICP8-FM and the same oligonucleotide was 0.9 with an f1max of 0.51% ± 0.2% taken from an average of triplicate experiments. The value of this parameter suggests no cooperativity occuring between ICP8-FM molecules. The calculated macroscopic dissociation constant (intrinsic constant × cooperativity factor) for ICP8 binding to oligo(dT)50 was 2.1 × 10−7 M/38 = 5.6 × 10−9 M. The dissociation constant for ICP8-FM binding to an oligo(dT)50 was 2.6 × 10−7 M/0.9 = 2.9 × 10−7 M. Thus, these results indicate that there is an approximately 50-fold difference in the macroscopic dissociation constants of ICP8 and ICP8-FM with oligo(dT)50 as a model binding substrate.
Helix-destabilizing experiments.
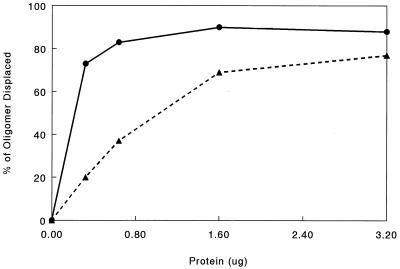
It has been suggested that the cooperative binding of ICP8 may be important in its helix-destabilizing activity (2, 4). Since the above-described data suggest that the cooperative binding of ICP8 is affected by FM modification, helix-destabilizing experiments comparing ICP8 and ICP8-FM should show a difference in the abilities of these protein species to displace an oligonucleotide annealed to M13 single-stranded DNA. The results from these experiments are shown in Fig. 6. The unmodified ICP8 removes the oligonucleotide in an essentially all-or-none fashion, whereas the ICP8-FM curve shows a distinct dose-dependent linear phase. Examination of the two curves suggests that ICP8 is at least fourfold more efficient than ICP8-FM at displacement of the oligonucleotide at low levels of protein. At higher concentrations of protein, the difference in oligonucleotide displacements by the two proteins is not as dramatic but is still significant. ICP8 displaced 90% of the oligonucleotide, whereas ICP8-FM displaced 75% of the oligonucleotide at the highest concentrations of protein used.
FIG. 6.
Results from a helix-destabilizing assay comparing the activities of ICP8 (circles) and ICP8-FM (triangles). The amounts of displaced versus amounts of annealed oligonucleotide were determined by scintillation counting of radioactive bands identified by autoradiography.
Mapping of the modified cysteine residue.
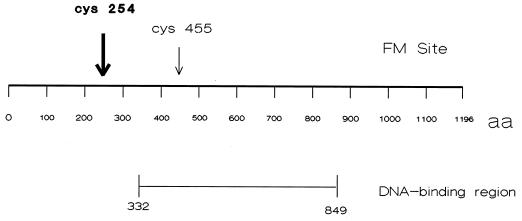
The above-described results show that modification of ICP8 with 1 mol of FM per mol of protein clearly affects cooperative binding of the protein. Therefore, mapping of the modified cysteine should identify a portion of the protein involved in this important property. ICP8-FM containing approximately 1 mol of incorporated FM was chemically cleaved with cyanogen bromide, the peptide fragments were separated on Tricine-SDS-polyacrylamide gels (Fig. 7A), and FM-containing fragments were visualized by illumination with long-wavelength UV light (Fig. 7B). Two peptides with approximate molecular weights of 12,000 and 10,000 were identified by this procedure. Densitometric scans of photographic negatives of the UV-illuminated gels revealed that approximately 86% of the fluorescence signal was located in the 12,000-kDa peptide compared to that in the 10,000-kDa peptide. These results were obtained from two separate experiments in which the scans were conducted in triplicate. Both peptides were excised, and the sequences of their N termini were determined (BIC/USUHS). The N-terminal amino acid sequence obtained from the 12,000-kDa peptide was PDFSRVIAEPFNANH. This sequence maps toward the N-terminus of ICP8. The predicted CNBr-derived peptide containing this sequence encompasses amino acid residues 204 through 318 and has a molecular weight of 12,430. This region of ICP8 contains one cysteine residue at position 254. The N-terminal sequence obtained from the 10,000-kDa peptide was VFSTNSALHLTEVDDAGPA. The predicted CNBr-derived peptide containing this sequence encompasses amino acids 375 through 465 in the primary structure of ICP8. The total molecular weight of this fragment is 10,327. There is one cysteine residue within this region at position 455. The positions of the major (amino acid 254; 86%) and minor (amino acid 455; 14%) labeling sites in the primary structure of ICP8 are indicated in Fig. 8.
FIG. 7.
(A) Replicate lanes (2 to 5) of a gel separating CNBr cleavage products of ICP8. The cleaved ICP8-FM was resolved on a Tricine-SDS-polyacrylamide gel and transferred to a PVDF membrane. The fluorescent bands were marked after UV illumination, and the membrane was stained with Coomassie brilliant blue. The large and small arrows indicate the positions of the bands showing major (86%) and minor (14%) fluorescence signals under UV light, respectively. Lane 1, molecular weight standards as indicated. (B) Visualization of FM-labeled ICP8 fragments in a Tricine-SDS-polyacrylamide gel. The gel was visualized by illumination from above with long-wavelength UV light and photographed. Lane 1, high-molecular-weight fluorescent standards (Sigma); lane 2, low-molecular-weight fluorescent standards; lanes 3 to 5, peptides obtained by CNBr cleavage of ICP8. Arrows indicate the positions of labeled peptides.
FIG. 8.
Schematic diagram showing the location of the major site of labeling of ICP8 by FM in relation to the DNA-binding region identified in previous studies. aa, amino acids.
DISCUSSION
Modification of ICP8 with FM has resulted in the generation of a protein whose single-stranded DNA-binding properties differ significantly from those of the unmodified protein based on results from polyacrylamide gel shift assays and helix-destabilizing assays. Mapping of the modified cysteine residues showed that the primary FM modification site was located on cysteine 254. The secondary site of modification is located on cysteine 455. Since an extrinsic fluorophore will modify residues that are within the same or similar environments, cysteines 254 and 455 are probably within similar environments in ICP8 and may be relatively close to each other in the tertiary structure of the protein.
In order to assess the formal possibility that the alteration in binding properties we observed was due to the 14% modification at Cys 455 rather than to the 86% modification at Cys 254, we used a probability approach (54) to determine the distribution of ICP8(254) and ICP8(455) bound to the oligo(dT)50 mer at saturation. The reasoning is as follows. If p is the mole fraction of ICP8(254) and q is the mole fraction of ICP8(455), then the probability of two ICP8(254) molecules binding to the same oligonucleotide is p × p, the probability of an ICP8(254) molecule and an ICP8 (455) molecule binding to the same oligonucleotide is p × q, and the probability of two ICP8(455) molecules binding to the same oligonucleotide is q × q. At saturation, these probabilities are normalized to unity. With values of 0.86 for p and 0.14 for q, this calculation predicts that at saturation, 84.1% of the DNA molecules would have two ICP8(254) molecules bound, 13.7% would have ICP8(254) and ICP8(455) bound, and 2.22% would have two ICP8(455) molecules bound. In the worst case, where binding of a single ICP8(455) molecule results in a loss of cooperativity and no shift to the fully bound state, one would see a minimum of 84% of the oligonucleotide fully shifted. The data in Fig. 5D show that this is not the case. No more than 65% of the oligo(dT)50 is fully shifted. Moreover, as the amount of modified ICP8 is increased in the titration, the ICP8(254) should out-compete the ICP8(455) and result in an increase in the amount of fully shifted oligonucleotide. Again, no such trend is evident in the data presented in Fig. 5D.
Thus, we believe that since ∼86% of the FM moiety is bound to cysteine 254 in ICP8, the results obtained in our experiments are primarily due to the modification at this residue and that the effects of this modification alter, but do not eliminate, binding. As shown in Fig. 8, the cysteine modified at position 254 lies outside the centrally located DNA-binding domain which has been identified by other studies (16, 19, 35, 55) and well outside a minimal DNA-binding domain (residues 563 to 636) recently identified in our laboratory (unpublished data). Thus, at least two regions of the protein are involved in different aspects of the interaction of ICP8 with single-stranded DNA, with the central region most likely being involved in intrinsic binding and the region around residue 254 being involved in cooperative binding.
The dissociation constants calculated from experiments monitoring the binding of a single protein to the oligo(dT)25 (2.1 × 10−7 M for ICP8 and 2.6 × 10−7 M for ICP8-FM) indicate that the modification of ICP8 by FM does not interfere with the intrinsic binding of the protein to single-stranded DNA. The difference seen between ICP8 and ICP8-FM by EMSA with a single-stranded DNA substrate 50 nucleotides in length may therefore be attributed to differences in levels of cooperativity. From calculations with data obtained with oligo(dT)50, the estimated cooperativity parameters were 38 for ICP8 and 0.9 for ICP8-FM. The estimated cooperativity parameter for ICP8 of 38 is low compared to that of T4 gene 32 protein (ω = ∼2,000) (31) and E. coli SSB (ω = ∼400− ∞, depending upon the salt concentration) (36). However, the ICP8 cooperativity value is similar to that obtained with the adenovirus DNA-binding protein (ω = 20 to 30) (32), human replication protein A (ω = ∼15) (28, 29), and replication protein A isolated from Drosophila melanogaster (ω = 10 to 300) (37). Thus, modest cooperativity may be a hallmark of single-strand-DNA-binding proteins in eukaryotic systems.
It should also be noted that the calculation represents the most conservative estimate of the cooperativity parameter. Since ICP8 binds to single-stranded DNA nonspecifically, the binding of the first molecule of ICP8 may occlude some potential binding sites for the second molecule of ICP8, resulting in negative cooperativity, and may therefore lead to an underestimate of the cooperativity parameter (29). However, until more information concerning this aspect of the mechanism of ICP8 binding is available, the currently estimated cooperativity parameters for ICP8 and ICP8-FM represent an almost 50-fold minimum difference in value, clearly indicating that the incorporation of FM has a major effect on the cooperativity of ICP8 binding.
The difference in levels of cooperativity seen between ICP8 and ICP8-FM may be due to a number of possibilities, including charge repulsion between ICP8 molecules, competition of the charged group for salt bridges, and prevention of a conformational change necessary for ICP8-ICP8 interactions. FM contains a carboxyl group that under the conditions used possesses a negative charge. The presence of this negative charge may cause charge-charge repulsions between ICP8 molecules, resulting in either no cooperative binding or negative cooperative binding interactions. Either the FM moiety itself or its negative charge present on cysteine 254 of ICP8 may prevent the formation of salt bridges or possibly disrupt hydrogen bond formation necessary for an interaction between ICP8 molecules. Finally, the bulky nature of the FM group may interfere with the ability of ICP8 to undergo a conformational change required for interaction between ICP8 molecules along the DNA strand.
Helix-destabilizing assays showed less efficient strand displacement by ICP8-FM than that by ICP8. These data confirm and expand results from other laboratories which suggested that cooperative binding is important in ICP8-promoted strand displacement (2, 4). However, cooperativity may not be the only parameter involved, since ICP8-FM can also displace the oligonucleotide at sufficiently high concentrations. This latter observation may be due either to the fact that ICP8-FM can bind with limited cooperativity to single-stranded DNA longer than 50 nucleotides or to the fact that the helix-destablizing activity of ICP8 involves a combination of the intrinsic DNA-binding and cooperative binding of the protein. In a similar fashion, we have found that ICP8-FM can stimulate the level of incorporation of nucleotides by the HSV DNA polymerase to a level similar to that observed with unmodified ICP8 in the context of a singly primed circular DNA template. However, the sizes of the products obtained in the presence of ICP8-FM are smaller than those obtained in the presence of unmodified ICP8 (unpublished observations). Thus, cooperativity may be required for optimization of only one of these polymerase-associated activities.
In summary, the work presented here has identified a region of ICP8 required for cooperative DNA binding which is distinct from the DNA-binding regions identified in other studies. This work has for the first time separated intrinsic DNA binding from cooperative DNA binding in ICP8, has allowed quantification of the binding parameters, and has provided a tool for the analysis of the role cooperativity may play in such functions as replication, recombination, and repair of HSV DNA as well as for the analysis of the role ICP8 plays in the expression of the viral genome. In all these cases, positioning and/or opening of DNA strands could be a key and limiting step. Thus, FM-modified ICP8 may be used as a reagent in in vitro assays specific for these processes. This work has also provided an obvious target for site-specific mutagenesis in which the role of cooperativity may ultimately be assessed in situ.
ACKNOWLEDGMENT
This work was supported by grant AI22468 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Boehmer P E, Lehman I R. Physical interaction between the herpes simplex virus 1 origin-binding protein and single-stranded DNA-binding protein ICP8. Proc Natl Acad Sci USA. 1993;90:8444–8448. doi: 10.1073/pnas.90.18.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boehmer P E, Lehman I R. Herpes simplex virus type 1 ICP8: helix-destabilizing properties. J Virol. 1993;67:711–715. doi: 10.1128/jvi.67.2.711-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehmer P E, Dodson M S, Lehman I R. The herpes simplex virus type-1 origin binding protein DNA helicase activity. J Biol Chem. 1993;268:1220–1225. [PubMed] [Google Scholar]
- 4.Bortner C, Hernandez T R, Lehman I R, Griffith J. Herpes simplex virus 1 single-strand DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J Mol Biol. 1993;231:241–250. doi: 10.1006/jmbi.1993.1279. [DOI] [PubMed] [Google Scholar]
- 5.Bush M, Yager D R, Gao M, Weisshart K, Marcy A I, Coen D M, Knipe D M. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J Virol. 1991;65:1082–1089. doi: 10.1128/jvi.65.3.1082-1089.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capaldi R A, Aggeler R, Turina P, Wilkens S. Coupling between catalytic sites and the proton channel in F1F0-type ATPases. Trends Biochem Sci. 1994;19:284–288. doi: 10.1016/0968-0004(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 7.Challberg M D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crute J J, Lehman I R. Herpes simplex 1 DNA polymerase: identification of an intrinsic 5′ to 3′ exonuclease with ribonuclease H activity. J Biol Chem. 1989;264:19266–19270. [PubMed] [Google Scholar]
- 9.Crute J J, Lehman I R. Herpes simplex virus helicase-primase. J Biol Chem. 1991;266:4484–4488. [PubMed] [Google Scholar]
- 10.Crute J J, Tsurumi T, Zhu L, Weller S K, Olivo P O, Challberg M D, Mocarski E S, Lehman I R. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc Natl Acad Sci USA. 1989;86:2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.deBruyn Kops A, Knipe D M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 12.deBruyn Kops A, Knipe D M. Preexisting nuclear architecture defines the intranuclear location of herpesvirus DNA replication structures. J Virol. 1994;68:3512–3526. doi: 10.1128/jvi.68.6.3512-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Dudas, K. C., J. Wang, and W. T. Ruyechan. Unpublished observations.
- 13.Dutch R E, Lehman I R. Renaturation of complementary DNA strands by herpes simplex virus type 1 ICP8. J Virol. 1993;67:6945–6949. doi: 10.1128/jvi.67.12.6945-6949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutch R E, Bianchi V, Lehman I R. Herpes simplex virus type I DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J Virol. 1995;69:3084–3089. doi: 10.1128/jvi.69.5.3084-3089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernando T, Royer C. Role of protein-protein interactions in the regulation of transcription by trp repressor investigated by fluorescence spectroscopy. Biochemistry. 1992;31:3429–3441. doi: 10.1021/bi00128a018. [DOI] [PubMed] [Google Scholar]
- 16.Gao M, Bouchey J, Curtin K, Knipe D M. Genetic identification of a portion of the herpes simplex virus ICP8 protein required for DNA-binding. Virology. 1988;163:319–329. doi: 10.1016/0042-6822(88)90272-3. [DOI] [PubMed] [Google Scholar]
- 17.Gao M, Knipe D M. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA-binding protein. J Virol. 1989;63:5258–5267. doi: 10.1128/jvi.63.12.5258-5267.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao M, Knipe D M. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J Virol. 1991;65:2666–2675. doi: 10.1128/jvi.65.5.2666-2675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao M, Knipe D M. Intragenic complementation of herpes simplex virus ICP8 DNA-binding protein mutants. J Virol. 1993;67:876–885. doi: 10.1128/jvi.67.2.876-885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godowski P J, Knipe D M. Mutations in the major DNA-binding protein gene of herpes simplex virus type 1 result in increased levels of viral gene expression. J Virol. 1983;47:478–486. doi: 10.1128/jvi.47.3.478-486.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godowski P J, Knipe D M. Identification of a herpes simplex virus function that represses late gene expression from parental viral genomes. J Virol. 1985;55:357–365. doi: 10.1128/jvi.55.2.357-365.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godowski P J, Knipe D M. Transcriptional control of herpesvirus gene expression: gene functions required for positive and negative regulation. Proc Natl Acad Sci USA. 1986;83:256–260. doi: 10.1073/pnas.83.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottlieb J, Marcy A I, Coen D M, Challberg M D. The herpes simplex virus UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol. 1990;64:5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griep M A, McHenry C S. The dimer of the beta subunit of Escherichia coli DNA polymerase III holoenzyme is dissociated into monomers upon binding of magnesium (II) Biochemistry. 1988;27:5210–5215. doi: 10.1021/bi00414a040. [DOI] [PubMed] [Google Scholar]
- 25.Griep M A, McHenry C S. Dissociation of the DNA polymerase beta 2 subunits is accompanied by a conformational change at distal cysteines 333. J Biol Chem. 1990;265:20356–20363. [PubMed] [Google Scholar]
- 26.Haugland R P. Myosin structure. Proximity measurements by fluorescence energy transfer. J Supramol Struct. 1975;3:338–347. doi: 10.1002/jss.400030405. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi Y, Nash H A. Nicking-closing activity associated with bacteriophage λ int gene product. Proc Natl Acad Sci USA. 1979;76:3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim C, Snyder R O, Wold M S. Binding properties of replication protein A from human and yeast cells. Mol Cell Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim C, Wold M S. Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 30.Knipe D M, Quinlan M P, Spang A E. Characterization of two conformational forms of the major DNA-binding protein encoded by herpes simplex virus 1. J Virol. 1982;44:736–741. doi: 10.1128/jvi.44.2.736-741.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowalczykowski S C, Eggleston A K. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 32.Kuil M E, van Amerongen H, van der Vliet P C, van Grondelle R. Complex formation between the adenovirus DNA-binding protein and single-stranded poly(rA): cooperativity and salt dependence. Biochemistry. 1989;28:9795–9800. doi: 10.1021/bi00451a038. [DOI] [PubMed] [Google Scholar]
- 33.Lee C K, Knipe D M. Thermolabile in vivo DNA binding activity associated with a protein encoded by mutants of herpes simplex type 1. J Virol. 1983;46:909–919. doi: 10.1128/jvi.46.3.909-919.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C K, Knipe D M. An immunoassay for the study of DNA-binding activities of herpes simplex virus protein ICP8. J Virol. 1985;54:731–738. doi: 10.1128/jvi.54.3.731-738.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leinbach S S, Heath L S. A carboxy-terminal peptide of the DNA-binding protein ICP8 of herpes simplex virus contains a single-stranded DNA-binding site. Virology. 1988;166:10–16. doi: 10.1016/0042-6822(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 36.Lohman T M, Overman L B, Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J Mol Biol. 1986;187:603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 37.Mitsis P G, Kowalczykowski S C, Lehman I R. A single-stranded DNA binding protein from Drosophila melanogaster: characterization of the heterotrimeric protein and its interaction with single-stranded DNA. Biochemistry. 1993;32:5257–5266. doi: 10.1021/bi00070a038. [DOI] [PubMed] [Google Scholar]
- 38.Monini P, Grossman S R, Pepinsky B, Androphy E J, Laimins L A. Cooperative binding of the E2 protein of bovine papillomavirus to adjacent E2-responsive sequences. J Virol. 1991;65:2124–2130. doi: 10.1128/jvi.65.4.2124-2130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odom O W, Dabbs E R, Dionne C, Muller M, Hardesty B. The distance between S1, S21, and the 3′ end of 16S RNA in 30S ribosomal subunits. The effect of polyuridylic acid and 50S subunits on these distances. Eur J Biochem. 1984;142:261–267. doi: 10.1111/j.1432-1033.1984.tb08280.x. [DOI] [PubMed] [Google Scholar]
- 40.O’Donnell M E, Elias P, Lehman I R. Processive replication of single-stranded DNA templates by the herpes simplex virus-induced DNA polymerase. J Biol Chem. 1987;62:4252–4259. [PubMed] [Google Scholar]
- 41.O’Donnell M E, Elias P, Funnell B E, Lehman I R. Interaction between the DNA polymerase and single-stranded DNA-binding protein (infected cell protein 8) of herpes simplex virus 1. J Biol Chem. 1987;262:4260–4266. [PubMed] [Google Scholar]
- 42.Powell K L, Littler E, Purifoy D J M. Nonstructural proteins of herpes simplex virus II. Major virus-specific DNA-binding protein. J Virol. 1981;39:894–902. doi: 10.1128/jvi.39.3.894-902.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabkin S D, Hanlon B. Herpes simplex virus DNA synthesis at a preformed replication fork in vitro. J Virol. 1990;64:4957–4967. doi: 10.1128/jvi.64.10.4957-4967.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross W, Landy A. Bacteriophage λ int protein recognizes two classes of sequence in the phage att site: characterization of arm-type sites. Proc Natl Acad Sci USA. 1982;79:7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruyechan W T. The major herpes simplex virus DNA-binding protein holds single-stranded DNA in an extended configuration. J Virol. 1983;46:661–666. doi: 10.1128/jvi.46.2.661-666.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruyechan W T. N-Ethylmaleimide inhibition of the DNA-binding activity of the herpes simplex virus type I major DNA-binding protein. J Virol. 1988;62:810–817. doi: 10.1128/jvi.62.3.810-817.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruyechan W T, Weir A C. Interaction with nucleic acids and stimulation of the viral DNA polymerase by the herpes simplex virus type 1 major DNA-binding protein. J Virol. 1984;52:727–733. doi: 10.1128/jvi.52.3.727-733.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruyechan W T, Chytil A, Fisher C M. In vitro characterization of a thermolabile herpes simplex virus DNA-binding protein. J Virol. 1986;59:31–36. doi: 10.1128/jvi.59.1.31-36.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruyechan W T, Olson J W. Surface lysine and tyrosine residues are required for interaction of the major herpes simplex virus type 1 DNA-binding protein with single-stranded DNA. J Virol. 1992;66:6273–6279. doi: 10.1128/jvi.66.11.6273-6279.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1-100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 51.Scopes R K. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974;59:277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- 52.Showalter S D, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanguy, Le Gac N, Villani G, Hoffman J-S, Boehmer P E. The UL8 subunit of the herpes simplex virus type-1 DNA helicase-primase optimizes utilization of DNA templates covered by the homologous single-strand DNA-binding protein ICP8. J Biol Chem. 1996;271:21645–21651. doi: 10.1074/jbc.271.35.21645. [DOI] [PubMed] [Google Scholar]
- 54.Von Hippel P H. On the molecular basis of the specificity of interaction of transcriptional proteins with genome DNA. In: Goldberger R F, editor. Biological regulation and development. Vol. 1. New York, N.Y: Plenum; 1979. pp. 279–347. [Google Scholar]
- 55.Wang Y, Hall J. Characterization of a major DNA-binding domain in the herpes simplex virus type 1 DNA-binding protein (ICP8) J Virol. 1990;64:2082–2089. doi: 10.1128/jvi.64.5.2082-2089.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber L D, Tulinsky A, Johnson D, El-Bayoumi M A. Expression of functionality of α-chymotrypsin. The structure of a fluorescent probe-α-chymotrypsin complex and the nature of its pH dependence. Biochemistry. 1979;18:1297–1303. doi: 10.1021/bi00574a028. [DOI] [PubMed] [Google Scholar]
- 57.Weller S K, Lee K J, Sabourin D J, Schaffer P A. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983;45:354–366. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu C A, Nelson N J, McGeoch D J, Challberg M D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xue, M., and W. T. Ruyechan. Unpublished data.