Summary
Congenital heart defects are the most prevalent human birth defects, and their incidence is exacerbated by maternal health conditions, such as diabetes during the first trimester (pregestational diabetes). Our understanding of the pathology of these disorders is hindered by a lack of human models and the inaccessibility of embryonic tissue. Using an advanced human heart organoid system, we simulated embryonic heart development under pregestational diabetes–like conditions. These organoids developed pathophysiological features observed in mouse and human studies before, including ROS-mediated stress and cardiomyocyte hypertrophy. scRNA-seq revealed cardiac cell-type-specific dysfunction affecting epicardial and cardiomyocyte populations and alterations in the endoplasmic reticulum and very-long-chain fatty acid lipid metabolism. Imaging and lipidomics confirmed these findings and showed that dyslipidemia was linked to fatty acid desaturase 2 mRNA decay dependent on IRE1-RIDD signaling. Targeting IRE1 or restoring lipid levels partially reversed the effects of pregestational diabetes, offering potential preventive and therapeutic strategies in humans.
Keywords: heart organoid, congenital heart defects, pregestational diabetes, pluripotent stem cell, heart development, very-long-chain fatty acid, omega-3 fatty acid
Graphical abstract

Highlights
-
•
Heart organoids to investigate pregestational diabetes-congenital heart defects
-
•
scRNA-seq revealed significant transcriptomic alterations in cardiac cell types
-
•
ER stress and disrupted lipid metabolism were significant features of PGD-CHD
-
•
Strategies reducing ER stress/restoring normal lipid balance were cardioprotective
Kostina et al. investigate how exposure to diabetes during early embryonic development affects the human heart in an innovative hPSC-derived heart organoid model. The authors discover that exposure to diabetes triggers ER stress and VLCFA dyslipidemia in the embryonic heart. Restoration of ER homeostasis and/or VLCFA levels using small molecules provided a significant degree of cardioprotection, suggesting new therapeutic alternatives.
Introduction
Congenital heart disease (CHD) constitutes the most common type of congenital defect in humans. Pregestational diabetes (PGD) mellitus, defined as diabetes of the mother before and during the first trimester of pregnancy, regardless of whether it is type 1 diabetes (T1D) or T2D, is one of the most outstanding nongenetic factors contributing to CHD and is present in a significant, growing population of diabetic female patients of reproductive age (Oyen et al., 2016). Newborns from mothers with PGD have an increased risk of CHD (up to 12%) versus ∼1% in the normal population (a 12-fold increase) (Basu and Garg, 2018). PGD is hard to manage clinically due to the sensitivity of the developing embryo to glucose oscillations and constitutes a critical health problem for the mother and the fetus. Preventive and therapeutic interventions are urgently needed to tackle this health problem.
Significant efforts have been devoted to understanding the molecular pathology of PGD-induced CHD using animal models, leading to the identification of increased reactive oxygen species (ROS) production, abnormal lipid metabolism, epigenetic reprogramming, and mitochondrial stress (Lehtoranta et al., 2017; Wu et al., 2016). The molecular links and the overall contribution of these abnormalities to the different phenotypes observed in PGD-induced CHD remain elusive, and despite these efforts, no advances have been translated to the clinical setting. A core issue is that it remains unclear to what extent rodent models recapitulate abnormalities present in human PGD-induced CHD, given critical species differences in heart size, cardiac physiology, electrophysiology, and bioenergetics (Hamlin and Altschuld, 2011). Furthermore, rodent models and many in vitro cell models rely on overtly aggressive diabetic conditions (∼25–50 mM blood glucose in diabetic mice versus ∼11–33 mM in human diabetic patients), leading to exaggerated cytotoxic phenotypes not relevant to the human clinical condition. The clinical practice largely precludes studies of PGD-induced CHD in humans or severely limits them; thus, direct studies of human embryos are not viable. The significantly limited access to human tissues for research of early-stage disease and mechanisms of PGD-induced CHD results in an overreliance on animal models.
Novel stem cell–based technologies have enabled the creation of engineered, highly complex, human organ–like three-dimensional tissues in vitro, with properties that recapitulate the physiological setting to a significant extent (Kretzschmar and Clevers, 2016). These organoids are particularly useful for studying unapproachable disease states in humans or states for which animal models are not well suited (Bredenoord et al., 2017; Lewis-Israeli et al., 2021a). Organoids can also be powerful tools to verify in human models what we have learned in animal studies, complementing them (Kostina et al., 2023). Although organoids have been used to model a wide range of human tissues and conditions, their application to cardiovascular studies has been sorely lacking until very recently (Hofbauer et al., 2021; Rossi et al., 2021). We recently established an advanced heart organoid model that recapitulates human heart development during the first trimester reliably or accurately, including critical steps such as chamber formation, vascularization, cardiac tissue organization, and relevant cardiac cell types (Lewis-Israeli et al., 2021b; Volmert et al., 2023).
In this report, we describe the application of our heart organoid model to the study of PGD-induced CHD. We found that heart organoids could be used to model critical aspects of CHD and recapitulated hallmarks of PGD-induced CHD found in mice and humans.
Results
Human heart organoids faithfully recapitulate pathological hallmarks of PGD-induced congenital heart disease
To model PGD conditions in the embryonic heart, we developed a high glucose and high insulin medium, with concentrations of these molecules similar to those previously reported in patients (Guemes et al., 2016). Human heart organoids (hHOs) were exposed to normoglycemic medium (NHOs) or hyperglycemic medium (PGDHOs) from their assembly at day 0 of differentiation and maintained in these conditions throughout in vitro development until assayed for a series of biochemical and cellular hallmarks of PGD-induced CHD described earlier (Lehtoranta et al., 2013). To determine the presence of cardiomyocyte (CM) hypertrophy, a well-described effect of embryonic diabetic cardiomyopathy (Al-Biltagi et al., 2021), we dissociated day 14 NHOs and PGDHOs into single-cell suspensions and performed immunofluorescence staining in individual CMs for TNNT2. A significant enlargement of CMs was observed in PGDHOs compared to NHOs (Figures 1A and 1B), resulting in a 27% ± 9.5% increase in CM size. To identify the total cell number for NHOs and PGDHOs, we used the PicoGreen assay. The mean cell number for NHOs was 180,198 ± 51,846 cells per organoid, whereas the mean for PGDHOs was 366,216 ± 132,720 cells per organoid. Thus, PGDHOs demonstrated significantly increased cellular content compared to NHOs, suggesting CM hypertrophy as well as hyperplasia (data not shown). Mitochondrial staining using the molecular probe Mitotracker revealed abnormal mitochondrial morphology and mitochondrial swelling in PGDHOs compared to NHOs (Figures 1C and 1D), suggesting mitochondrial dysfunction due to diabetic conditions and pointing potential metabolic phenotypes. It has been previously described in murine PGD models that PGD induces oxidative stress in the developing embryo (Wu et al., 2016). Analysis of the production of ROS in NHOs and PGDHOs revealed an increase in cellular ROS in PGDHOs compared with controls (Figures 1E and 1F). We also characterized the dynamic expression of key developmental transcription factors (HAND1, HAND2, NKX2-5, TBX5, and GATA4) (Figures 1G–1K) in NHOs and PGDHOs between days 0 and 14. NHOs displayed a clear transition in the expression of the first heart field marker HAND1, which was highly expressed between days 2 and 8, and the expression of the second heart field marker HAND2, which was highly expressed from day 8 onward. Both heart field markers were downregulated in PGDHOs compared to NHOs at critical time points. NKX2-5 and TBX5 both showed overexpression in the PGDHOs, which was not present in their control counterparts, the first showing an early overexpression on day 6 and the latter showing an overexpression on days 12 and 14. GATA4 also presented abnormal expression over time (Figure 1K). These data were consistent with previous observations of developmental transcription factor dysregulation in PGD conditions (Lehtoranta et al., 2017). We also investigated the expression of glucose transporters SLC2A1 and SLC2A4 (Figures 1L and 1M). NHOs demonstrate a clear transition between the glucose transporter SLC2A1 (highly expressed in the fetal heart) and the glucose transporter SLC2A4 (highly expressed in the adult heart) between days 6 and 8. However, PGDHOs show downregulation of SLC2A1 in the early days of differentiation, hinting at possible dysfunction in glycolysis and glucose transport (Figure 1L). Normal hHOs typically exhibit a well-developed primitive vascular network around the myocardial tissue during differentiation (Lewis-Israeli et al., 2021b). NHOs and PGDHOs showed drastic differences in the formation of this vascular network. The endothelial cell (EC) marker PECAM1 revealed a sophisticated plexus of vascular endothelial tissue covering large portions of the control NHOs (Figure 1N). Vascularization in PGDHOs appeared less organized than in NHOs, showing fewer regions of PECAM1 staining and a lack of an interconnected network (Figure 1N). To compare functional properties of control and diabetic hHOs, calcium transient analysis by live organoid imaging was conducted. The beating frequency of the control organoids was ∼120 bpm compared to ∼60 bpm in diabetic organoids (Figures 1O and 1P). Fetal heart rate can range from 110 to >160 bpm, with slower fetal heart rate associated with poor pregnancy outcomes (Hornberger, 2006). PGDHOs also showed a higher frequency of arrhythmic events when compared to NHOs (Figure 1O). Overall, these data demonstrate substantial morphological and molecular changes specific to PGDHOs that are consistent with observations of fetal tissue in animal and human models.
Figure 1.
Human heart organoids recapitulate phenotypic hallmarks of PGD in the developing human heart
(A) Immunofluorescence images of CMs from NHOs and PGDHOs stained with the CM marker TNNT2 (red) and the nuclear marker DAPI (blue); scale bar: 10 μm.
(B) Quantification of CM area from NHOs and PGDHOs; n = 4 organoids, n = 145 CMs for NHOs and n = 193 CMs for PGDHOs; nested t test.
(C) Immunofluorescence of mitochondria using MitoTracker showing mitochondrial swelling in PGDHOs; arrowheads indicate swollen mitochondria; scale bar: 10 μm.
(D) Quantification of mitochondrial swelling in NHOs and PGDHOs; n = 4; nested t test.
(E) Live ROS imaging using CellROX Green; scale bar: 10 μm.
(F) Quantification of ROS content in NHOs and PGDHOs; n = 3; nested t test.
(G–M) Time course qRT-PCR gene expression analysis of key developmental transcription factors (G–K) and glucose transporters (L and M) from days 0 to 14 of differentiation; n = 9 (3 biological replicates of 3 pooled organoids).
(O) Calcium transient live imaging using Fluo-4 from NHOs (left) and PGDHOs (right).
(P) Quantification of beats per minute in NHOs and PGDHOs; n = 7; value = mean ± SD, unpaired t tests.
Single-cell RNA sequencing (scRNA-seq) reveals cardiac cell-type-specific responses to PGD in heart organoids
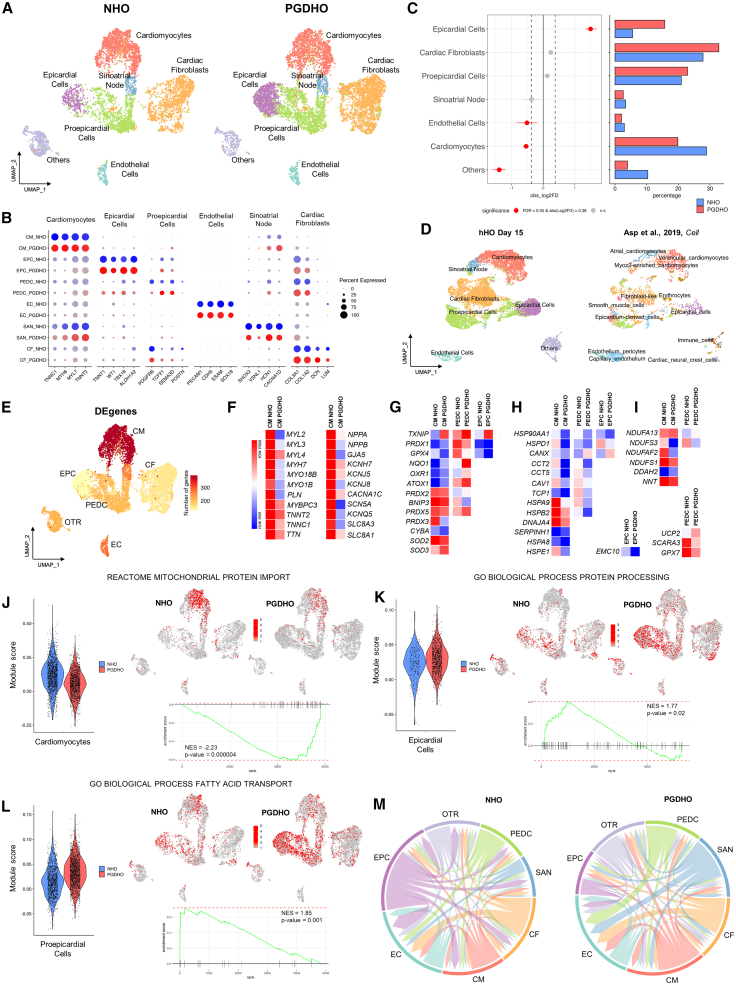
To explore the effects of PGD on cardiac lineage specification within heart organoids, we performed scRNA-seq on NHOs and PGDHOs at day 15 of differentiation. Computational analysis identified a total of 7 separate cell clusters representing distinct cardiac cell populations in both NHOs and PGDHOs. These clusters included CMs, sinoatrial node-related cells, epicardial cells (EPCs), proepicardial cells (PEDCs), ECs, cardiac fibroblasts (CFs), and a mixed population of mesenchymal cardiac progenitors, which were still poorly committed (labeled as others [OTR]) (Figure 2A). A summary of key markers related to identified cell populations can be found in Figure 2B. Although Seurat cluster analysis yielded one cluster of CMs, when mapping atrial (MYH6, MYL7, GJA5, NPPA, NR2F2) and ventricular (MYH7, MYL3, TNNC1, HSPB7, IRX3) specific markers, we could identify the presence of two CM populations, suggesting early stages of atrial and ventricular CM specification in hHOs (Figure S1A). All of the cell clusters were present in NHOs and PGDHOs. The main effects of PGD were a significant decrease in the number of CMs (29% in NHOs versus 20% in PGDHOs) and a very large increase in EPCs (5% in NHOs versus 16% in PGDHOs) (Figure 2C), hinting at dysregulation of CM and epicardial differentiation. The number of ECs was slightly decreased in PGDHOs (3% in NHOs versus 2% in PGDHOs) (Figure 2C), corresponding to the poor vascular network revealed by PECAM1 staining (Figure 1K). To examine the extent of similarity between hHO and the human developing heart, we integrated and compared transcriptomic profiles of hHO with previously published human fetal heart transcriptomic datasets (Asp et al., 2019). We discovered significant overlap in cell populations among the two datasets (Figure 2D) for CMs, EPCs, PEDCs (called epicardium-derived and smooth muscle cells in the human heart dataset), CFs (called fibroblast-like cells in the human heart dataset), and ECs. The lack of several cell types present in the human embryonic heart dataset but not in hHOs may be because day 15 hHOs represent earlier stages of cardiac development (6.5 postconception weeks [PCW] or ∼45 days postconception for human embryonic hearts versus ∼25 days postconception for organoids). This assumption is supported by the observation that cells in the PEDC and CM clusters in hHO are still not divided into distinct proepicardial-derived and atrial/ventricular populations as seen in 6.5 PCW human hearts. Differential expression analysis demonstrated that the CM cluster harbored the majority of differentially expressed genes (DEGs), indicating the high sensitivity of the CM population to diabetic conditions (Figure 2E). A summary of the most significant differentially regulated genes in CMs identified downregulation of sarcomeric and cytoskeletal gene expression in PGDHOs, particularly in genes frequently associated with familial cardiomyopathies and hypertrophy (e.g., MYH7, TNNT2, MYBPC3, MYL2, MYL3) (Maron et al., 2012) (Figure 2F, left; Table S1). Genes encoding natriuretic peptides NPPA and NPPB, highly expressed during heart development and implicated in cardiac dysfunction (Man et al., 2018), were also significantly downregulated in PGD CMs. Furthermore, alterations in expression profile of key CM ion channels were present in PGDHOs (Figure 2F, right; Table S1). In accordance with the revealed increase in cellular ROS in PGDHOs, DEG analysis identified dysregulation in the expression of ROS-associated molecular markers in CM, EPC, and PEDC clusters (Figure 2G; Table S2). In particular, all of the noted cell clusters were characterized by the increased expression of TXNIP, known to be implicated in mitochondrial death signaling and increasing damage from oxidative stress in the context of T2D and myocardial ischemia (Lane et al., 2013). The expression levels of one of the main lipid antioxidant mediators, GPX4, were significantly lower in CMs, EPCs, and PEDCs of PGDHOs. GPX4 deficiency has been shown to contribute to myocardial metabolic dysfunction (Baseler et al., 2013). PEDCs of PGDHOs demonstrated downregulation of GPX7-endoplasmic reticulum (ER)-localized protein involved in cellular response to oxidative stress (Figure 2H; Table S2). Antioxidant enzymes PRDX2, PRDX3, and PRDX5 were decreased in the CM cluster and increased in PEDCs, suggesting that the CM population is less protected from oxidative stress. Genes implicated in proper protein folding and ER function were also significantly downregulated, predominantly in CMs in PGDHOs, whereas EMC10, required for ER protein synthesis, had decreased expression in EPCs (Figure 2H; Table S2). Finally, dysregulated expression in genes encoding NADH dehydrogenases and other mitochondria-localized genes (NNT, DDAH2, UCP2) were identified for CMs and PEDCs (Figure 2I; Table S2), further implying metabolic dysfunction in PGDHOs.
Figure 2.
scRNA-seq reveals cardiac cell-type-specific responses to PGD in human heart organoids
(A) Uniform manifold approximation and projection (UMAP) showing the integrated cell map, which consists of 7 clusters. Clusters were annotated based on known marker genes. Cells are colored by clusters. A total of 5,951 cells from 4 pooled dissociated NHO organoids and 7,463 cells from 4 pooled dissociated PGDHO organoids were sequenced.
(B) Dot plot depicting representative marker genes across cell clusters. Dot size is proportional to percentage of cells in the cluster expressing specific genes. Color indicates samples NHOs (blue) and PGDHOs (red). Color intensity indicates average expression.
(C) Left: Relative differences in cell proportion for each cluster between NHOs and PGDHOs. Clusters colored red have a false discovery rate <0.05 and mean |log2 fold enrichment| > 0.38 compared to NHOs (permutation test; n = 10,000). Right: Bar plot displaying the cell-type abundance for NHOs and PGDHOs. Color bars indicate samples NHOs (blue) and PGDHOs (red).
(D) UMAP projection of integrated heart organoid dataset (left) and 6.5 PCW (or ∼45 days postconception) human embryonic heart dataset (right). The color corresponds to the cell annotation identified for organoids (left UMAP) or taken from the respective publication (right UMAP).
(E) UMAP displaying the number of DEGs in each cell type.
(F–I) Heatmaps of DEGs in CM, EPC, and PEDC clusters. Data displayed as log2 and quantile normalized counts based on pseudobulk differential expression analysis.
(J–L) Pathway enrichment analysis (fast gene set enrichment analysis–multilevel defining representation of DEGs in pathways from database MsigDB) in PGDHOs versus NHOs in CM (J), EPC (K) and PEDC (L) clusters. Violin plots depicting cells per sample by gene set module score. Feature plots displaying enrichment of pathway gene sets in cell populations. Enrichment plots of pathway-related gene sets overrepresented in NHOs and PGSHOs. NES, enrichment score normalized to mean enrichment of random samples of the same size (value represents the enrichment score after normalization).
(M) Summary chord diagrams of cell-type interactions; strong decrease in the number of interactions from EPCs toward all other cell types.
To better understand the molecular differences between heart organoids grown in healthy and PGD conditions, and to determine key biological processes affected by PGD, we performed multidatabase pathway analysis for DEGs for the main identified clusters. As illustrated in Figures 2J, S1B, and S1C, pathway enrichment analysis for the CM cluster revealed a decrease associated with PGD conditions in mitochondrial protein import, mitochondrial matrix, and oxidative phosphorylation. Fatty acid (FA) metabolism, protein folding, and chaperone complex processes were also strongly downregulated in PGD CMs (Figures S1D–S1F). These data again point to developmental defects in CMs under PGD conditions related to mitochondria and ER function, and FA-associated metabolic processes. The cardiac conduction system development process was also reduced in PGDHO CMs (Figure S1G). The role of Myc in mitochondrial biogenesis and regulating energy metabolism by directly binding the promotors of key factors involved in glucose metabolism has been demonstrated for adult myocardium (Ahuja et al., 2010). Pathway analysis revealed the downregulation of a subgroup of genes regulated by Myc in PGDHO CMs (Figure S1H). Upregulated DEGs of CM between NHOs and PGDHOs were mainly related to SMAD protein signaling transduction (Figure S1I). Pathway enrichment analysis for EPCs identified upregulation of DEGs related to protein processing, extracellular matrix (ECM) receptor interaction, and sulfur compound transport (Figures 2K, S2A, and S2B). Remarkably, sulfur is an essential part of vital antioxidant enzymes preventing deleterious effects of ROS (Mukwevho et al., 2014). PEDCs were characterized by an increase in processes of FA transport (Figure 2L), and regulation of insulin-like growth factor (IGF) transport uptake and by IGF binding protein (Figure S2C). Interestingly, when all of the clusters were considered together, the top disturbed processes upregulated in PGDHOs were key developmental pathways such as transforming growth factor β (TGF-β) signaling, regulation of bone morphogenetic protein (BMP) signaling, and cellular response to retinoic acid (Figures S2D–S2F). A list of all of the top processes altered in hHOs under diabetic conditions is presented in Figure S3A.
Next, we applied LIANA and MultiNicheNet computational frameworks to identify potential interactions between cell types in NHOs and PGDHOs and differentially expressed ligand-receptor pairs. We found that EPCs in PGDHOs presented significantly reduced signaling activity to other cell types compared to EPCs in NHOs (Figure 2M). Together with the finding of an increased percentage of EPC in PGDHOs, this observation supports the idea that EPC dysfunction may be a critical contributor to PGD-induced CHD. Furthermore, we found that most of the predicted interactions between EPC and CFs were downregulated and related to ECM, including collagen-integrin ligand-receptor pairs (Figure S3B, left). CFs play essential roles in myocardial function, primarily synthesis and remodeling of the ECM, suggesting that improper EPC–CF interactions associated with diabetic conditions may negatively affect heart development. Diabetic-related changes in the expression of ligand-receptor pairs in EPCs and CMs were predominantly associated with the WNT pathway (EPC WNT5A-CM FZD1), cell communication with ECM (EPC COL8A1/COL1A1-CM DDR1), and cardiac hypertrophy–linked genes (Xiang et al., 2018) (EPC LGI2-CM ADAM23). As for CMs, vascular endothelial growth factor A, which triggers the EC vascularization signaling axis and its downstream effectors, showed a decrease in PGDHOs, potentially contributing to poor vasculature (Figure S3B, right). Ligand-receptor pairs and downstream signals related to BMP and TGF-β networks were upregulated in CMs correlating with pathway enrichment analysis results. Moreover, the ligand-receptor pair LRIG1-ERBB4 and its downstream response were downregulated in CMs. ERBB4 is a crucial receptor in early heart development involved in CM differentiation and ventricular trabeculation (Odiete et al., 2012) (Figure S4B, right).
Overall, our scRNA-seq analysis revealed critical differences in transcriptomic profiles of normoglycemic and diabetic organoids affecting early heart development, particularly CM, EPC, and PEDC populations, and pointed to the potential dysregulation of redox and mitochondrial homeostasis, ER, and FA metabolism as important contributors in the development of PGD-induced CHD.
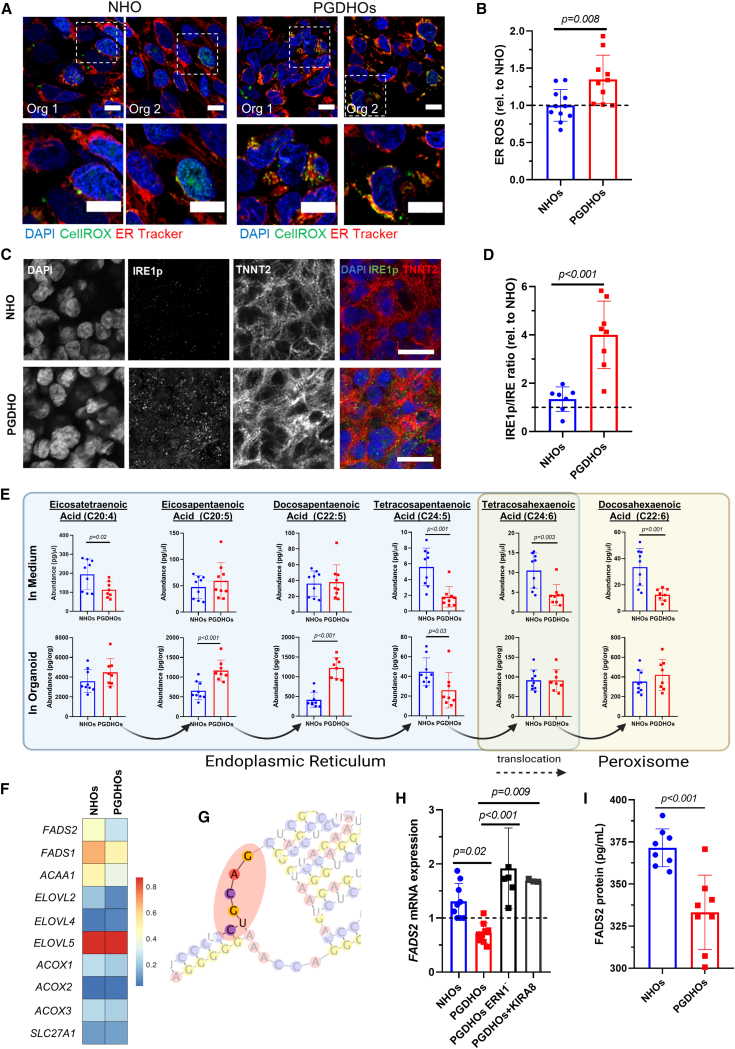
PGD triggers ER stress and very-long-chain FA (VLCFA) dyslipidemia in human heart organoids
Previous work has shown a strong association between ER stress and cardiomyopathies (Wu et al., 2016). Our data suggested that ER stress may also be a factor in PGD-induced CHD. We decided to explore the potential connection between ROS production and ER stress in CMs as a result of diabetic conditions. hHOs were co-stained with CellROX Green and ER Tracker Red, a highly selective dye for the ER, and imaged under normoglycemic and PGD conditions. PGDHOs exhibited increased cellular ROS predominantly accumulated in the ER (Figures 3A and 3B), suggesting potential ER dysfunction. Interestingly, ER stress has not been described in animal models of PGD and may be a human-specific phenotype of interest for therapeutic and preventive interventions. Inositol-requiring enzyme 1 (IRE1, gene name ERN1) is the main evolutionarily conserved sensor for ER stress. IRE1 phosphorylation within the ER membrane induces the unfolded protein response (UPR) through at least two pathways, one of them involving the splicing of XBP1 (X-box binding protein) mRNA and activation of downstream transcription effectors. The second pathway is called regulated IRE1-dependent decay (RIDD) and targets mRNAs for degradation in an XBP1-independent manner. Confocal imaging for activation of IRE1 demonstrated an increase in the ratio of the phosphorylated isoform (IRE1p) when compared to its unphosphorylated counterpart, confirming that PGDHOs have higher ER stress than NHOs (Figures 3C, 3D, and S4A). Another molecular phenotype identified in our scRNA-seq analysis was lipid metabolism. Liquid chromatography-mass spectrometry (LC-MS) lipidomic profiling revealed significant differences between PGDHOs and NHOs affecting VLCFA trafficking and synthesis (Figure 3E). Organoids were grown in fully defined medium, so no exogenous sources of VLCFAs or other lipids were available aside from essential FAs present in the medium. All of the other FAs present are therefore synthesized by the organoids to be used intracellularly or to be secreted into the extracellular space. The intracellular levels of two important omega-3 FAs, eicosapentaenoic acid (EPA) and docosapentaenoic acid (DPA), were significantly higher in PGDHOs (Figure 3E), whereas the levels of docosahexaenoic acid (DHA; another important omega-3 FA synthesized from EPA and DPA), were significantly lower in the medium for PGDHOs (Figure 3E). These data showed that the PGD conditions resulted in lipid dysregulation in VLCFAs, particularly in omega-3 FAs, which have been previously shown to be instrumental during development of the heart (Maron et al., 2012). The mechanistic implication of these changes in cardiac physiology remains unclear, since omega-3 FAs can be used as building blocks in cell membranes, signaling lipids, and antioxidants (Wasserman et al., 2020).
Figure 3.
PGD conditions trigger ER stress and VLCFA lipid imbalance in human heart organoids
(A) Immunofluorescence images of day 14 NHOs and PGDHOs showing colocalization of ROS (CellROX, green) and ER marker (ER Tracker, red); nuclear marker DAPI (blue); scale bar: 10 μm.
(B) Quantification ROS localized in the ER; n = 10 organoids per condition; value = mean ± SD, unpaired t tests.
(C) Immunofluorescence images of day 14 NHOs and PGDHOs stained for phosphorylated IRE1 (IRE1p, green), CM marker TNNT2 (red), and nuclear marker DAPI; n = 6; scale bar: 20 μm.
(D) Quantification of the ratio of phosphorylated IRE1 to unphosphorylated IRE1 compared to NHOs, measured by immunofluorescence image analysis; n = 7 for NHOs, n = 8 for PGDHOs; value = mean ± SD, unpaired t tests.
(E) LC-MS lipidomic analysis for VLCFA concentrations from day 15 organoids and their corresponding medium; n = 9 organoids per condition, value = mean ± SD, unpaired t test, ∗p < 0.05. Arrows indicate the direction of biosynthetic pathway of VLCFA, their transformation through elongation, and desaturation.
(F) Heatmap representing expression level of key enzymes involved in LCFA and VLCFA biosynthesis.
(G) Predicted secondary FADS2 mRNA structure.
(H) qRT-PCR gene expression analysis of FADS2; n = 6 and n = 3 biological replicates of 3 pooled organoids for ERN1 knockdown and KIRA8 respectively; value = mean ± SD.
(I) FADS2 protein measurement by ELISA; n = 8 organoids per condition; value = mean ± SD.
Next, we attempted to find a link between ER stress and disrupted VLCFA metabolism. The ER plays a significant role in the synthesis of lipid precursors, such as LCFAs necessary for the synthesis of more complex lipids, and we hypothesized that ER stress may interfere with this balance. We started by looking at IRE1-induced activation of the UPR, which can lead to apoptosis or reestablishment of protein homeostasis. To determine whether IRE1 activation was causing apoptosis, we used a transgenic induced pluripotent stem cell (iPSC) line expressing Flip-GFP, a nonfluorescence engineered GFP variant that turns fluorescent upon caspase activation and apoptosis. Flip-GFP organoids cultured either under control or diabetic conditions exhibited no fluorescence changes, indicating that there is no significant PGD-induced apoptosis (Figure S4B). Doxorubicin-treated NHOs and PGDHOs were used as a positive control for apoptosis (Figure S4B). Furthermore, analysis of UPR-specific genes, either induced by IRE1 or some other ER stress sensor (ATF6, PERK) demonstrated the absence of UPR pathway activation (Figure S4C). IRE1 signaling through XBP1 splicing also remained unchanged between NHOs and PGDHOs (Figure S4D). We then decided to investigate whether the IRE1-RIDD pathway was activated due to ER stress, and whether that was the process affecting lipid biosynthesis. RIDD has been reported to preferentially induce the degradation of ER-localized mRNA (Hollien et al., 2009), and most steps of endogenous FA biosynthesis take place in the smooth ER. We performed gene expression analysis for 10 ER-localized lipid biosynthesis enzymes in NHOs and PGDHOs and found increased expression of ELOVL5 (ELOVL FA elongase 5) and decreased expression of ELOVL2 and ACAA1 (3-ketoacyl-coenzyme A thiolase), and desaturases FADS1 (fatty acid desaturase 1) and FADS2 (also known as delta-6 desaturase [D6D]) in PGD conditions (Figure 3F; Table S3). (D6D) encoded by the FADS2 gene catalyzes the key initial rate-limiting step of VLCFA biosynthesis before exporting FAs to the peroxisome (tetracosapentaenoic acid to tetracosahexaenoic acid) and is a main determinant of VLCFA levels. Alterations in D6D activity alter FA profiles and are associated with metabolic and inflammatory diseases (Arshad et al., 2019). LC-MS analysis revealed significant differences in this metabolic step under the control of D6D both in organoids and medium (Figure 3E). Interestingly, the FADS2 mRNA possesses four RIDD consensus sequences for degradation in its 3′ and 5′ UTR regions (5′-CUGCAG-3′ motif located in the loop portion of a hairpin structure) (Figure S4E), and at least one of them is in a clear hairpin loop (Figure 3G). To confirm that FADS2 mRNA downregulation is IRE1-RIDD dependent, we analyzed FADS2 expression in a knockdown ERN1 (IRE1) iPSC line and in the presence of IRE1 monoselective inhibitor KIRA8. ERN1 knockdown PGDHOs exhibited restored levels of FADS2 expression comparable with NHOs and confirmed the dependence of FADS2 expression on IRE1-RIDD activation (Figure 3H). The same effect was found when IRE1 activity was inhibited using the specific inhibitor KIRA8 (Figure 3H) in the absence of XBP1 splicing (Figure S4D). D6D protein abundance was measured by ELISA and was found to be significantly lower in PGDHOs compared to NHOs, confirming the pathologic phenotype (Figure 3I). The expression of FADS1, ACAA1, and ELOVL2 in heart organoids with ERN1 knockdown was comparable with levels in PGDHOs, suggesting that these enzymes are not regulated by IRE1-RIDD and confirming the importance of FADS2 in this setting (Figures S4F–S4H). Taken together, these results show that PGD causes ROS-induced ER stress and IRE-RIDD activation leading to D6D deficiency in diabetic heart organoids. This in turn affects VLCFA biosynthesis and causes a crucial VLCFA imbalance during early cardiac development.
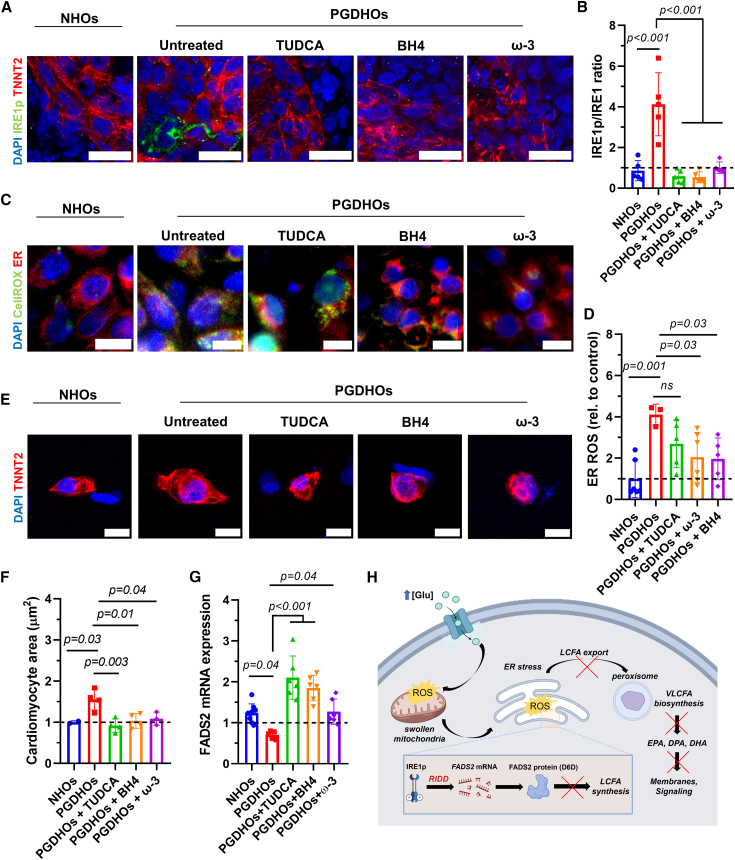
Reducing ER stress mitigates deleterious effects of PGD in human heart organoids
To ameliorate the phenotypic effects of PGD in the developing heart and the resulting ROS overproduction and ER stress, we tested several therapeutic compounds based on our mechanistic findings involving IRE1 and omega-3 FAs. These included the chemical chaperone tauroursodeoxycholic acid (TUDCA), known to reduce oxidative and ER stress, a combination of omega-3 FAs (DHA, DPA, EPA) designed to provide antioxidant resilience and relieve lipid imbalance, and sapropterin (BH4), an orally active synthetic form of tetrahydrobiopterin, which is a critical cofactor of endothelial NO synthase. Endothelial NO synthase has been described as a potential mitigating agent for PGD-induced CHD in murine models (Engineer et al., 2018). Treatment of PGDHOs with TUDCA, BH4, and omega-3 FAs caused a significant reduction in phosphorylated IRE1 in PGDHO organoids (Figure 4A). The ratio of IRE1p to IRE1 was significantly lower in all of the treated organoids as well, suggesting a reduction in IRE1 signaling activation (Figure 4B). Co-staining of organoids with CellROX Green and ER Tracker revealed reduced stress due to ROS both in the ER and cytosol for BH4 and omega-3 FAs treatments, but not for TUDCA (Figures 4C and 4D). Immunofluorescence imaging of individual CMs from dissociated organoids revealed that the treatment of PGDHOs with TUDCA, BH4, and omega-3 FAs resulted in a significant reduction in CM hypertrophy, suggesting an amelioration in the hypertrophic response (Figures 4E and 4F). Notably, treating PGDHOs with TUDCA, BH4, or omega-3 FAs affected mRNA levels of FADS2 (Figure 4G), likely due to the reduction in IRE1 signaling. Figure 4H shows a general mechanistic schematic summarizing our findings in the context of PGD-induced CHD.
Figure 4.
Strategies to reduce ER stress and lipid imbalance mitigate the deleterious effects of PGD in human heart organoids
(A) Immunofluorescence images of day 14 NHOs and PGDHOs treated with TUDCA, BH4, or omega-3 FAs, stained with phosphorylated IRE1 (IRE1p, green), CM marker TNNT2 (red), and nuclear marker DAPI (blue); n = 6; scale bar: 25 μm.
(B) Quantification of the ratio of phosphorylated IRE1 to unphosphorylated IRE1, measured by immunofluorescence image analysis; n = 6; value = mean ± SD, 1-way ANOVA.
(C) Immunofluorescence images of ROS (CellROX, green) and ER marked (ER Tracker, red) in day 14 NHOs and PGDHOs; n = 7; scale bar: 10 μm.
(D) Quantification of ROS content compared to NHOs; n = 6; value = mean ± SD, 1-way ANOVA.
(E) Immunofluorescence images of CMs dissociated from day 14 NHOs, PGDHOs; n = 4; scale bar: 10 μm.
(F) Quantification of CM area compared to NHOs; n = 4; value = mean ± SD, 1-way ANOVA.
(G) qRT-PCR gene expression analysis of FADS2 in NHOs and PGDHOs; n = 6 biological replicates of 3 pooled organoids; value = mean ± SD.
(H) Schematic diagram summarizing the mechanism of PGD-induced CHD.
Overall, the therapeutic agents tested were able to decrease all of the tested hallmarks of PGD-induced CHD, confirming our mechanistic hypothesis and validating the findings in mouse models and BH4. This is a potentially important finding for clinical translation because it suggests that dietary supplementation (e.g., omega-3 FAs) of diabetic mothers in the first trimester of pregnancy may be able to prevent or treat the early stages of PGD-induced CHD with no known drawbacks.
Discussion
We used a novel and powerful heart organoid technology based on recent work (Lewis-Israeli et al., 2021b) to the study of critical mechanisms of PGD-induced CHD in humans, and report, for the first time, that ER stress and VLCFA imbalance are critical factors contributing to these congenital disorders. Maternal diabetes is one of the most common causes of newborn CHD, yet the ability to study the etiology of these disorders in humans is greatly limited due to the inaccessibility to human fetal hearts at crucial stages of development. hHOs provide a necessary platform on which to investigate the underlying mechanisms between the association of PGD and CHD in humans without the drawbacks of animal models.
To model the effects of PGD, hHO culture conditions were modified to accurately reflect reported physiological levels of glucose and insulin in normal and diabetic mothers. The larger size of diabetic hHOs suggested signs of cardiac hypertrophy, a first hallmark of maternal PGD (Gandhi et al., 1995) on the heart of the embryo, which was confirmed by studying CM size. Furthermore, differences were noted in regard to ROS production and mitochondria of PGDHOs, revealing increased oxidative stress and mitochondrial swelling, which are also hallmarks of diabetic embryonic cardiomyopathies (Hornberger, 2006). Mouse models of PGD have shown dysregulation of transcription factors key to heart development (Kumar et al., 2007). In line with this, time course qPCR analysis revealed disparities in the regulation of GATA4, NKX2-5, and TBX5 at different time points throughout development, highlighting the importance of fine transcription control during early development. PGDHOs also presented arrhythmias and reduction in beat frequency, a phenotype that has not been described in diabetic embryonic cardiomyopathy but has been observed in neonatal rats from diabetic mothers (Alam et al., 2022).
Single-cell transcriptomic analysis on PGDHOs and NHOs revealed a reduction in CM numbers, a significant expansion of epicardial tissue, and the absence of a well-developed vasculature at early developmental stages. Furthermore, important candidate genes and signaling pathways were identified for the first time in this context in EPCs, PEDCs, and CMs, providing new hypotheses for future work and therapeutic interventions. A robust decrease in cell-to-cell communication from EPCs toward the rest of the heart was revealed, suggesting the epicardium plays a substantial role in signal transductions at this stage. PEDCs are a transitory embryonic cell population, being a source of epicardium, CFs, and smooth muscle cells (Niderla-BieliNska et al., 2019). Genes involved in FA metabolism and protein processing were downregulated in the PEDC cluster, likely underlying the further improper specification of epicardium. Numerous ligand–receptor interactions were observed to be changed in diabetic organoids, implying that the hyperglycemic environment disturbs cell-to-cell communications during development of the heart. In the case of CMs, major changes involved sarcomeric proteins, but also other proteins involved in protein folding, oxidative stress response, and mitochondrial metabolism.
Glycolysis constitutes a significant source of energy and intermediate metabolites in the embryonic heart, but FA oxidation is also important for mitochondrial growth and cardiac maturation as the heart develops (Zhong et al., 2016). Our model confirmed that heart organoids exposed to diabetic conditions exhibited increased accumulation of ROS, but also showed that a significant portion of ROS was localized to the ER and could be impairing its function. Numerous studies have shown that elevated ROS produces ER protein oxidation (Cui et al., 2022), leading to a condition known as ER stress. The ER plays an important role in lipid metabolism because a significant part of lipid synthesis occurs in the smooth ER. Out of the three sensor systems for ER stress that can activate the UPR, only IRE1 has been shown to be strongly associated with lipid metabolism (Volmer et al., 2013). The lipid metabolic profiles of PGDHOs revealed a clear dysregulation of VLCFA synthesis particularly affecting omega-3 polyunsaturated FAs (PUFA). We and others have shown that VLCFAs are critical during development of the heart for structural and signaling reasons (Wasserman et al., 2020), and the involvement of VLCFAs and ER stress has not been described before in PGD-CHD. The endogenous synthesis of these PUFAs mostly takes place in the ER, and along with the observed ROS accumulation in the ER, suggests a major ER-induced dyslipidemia in PGDHOs. We could pinpoint this dysregulation to the targeted degradation of FADS2, a key lipid biosynthesis enzyme present in the ER, by the RIDD pathway, and thus for the first time, we demonstrated a direct role of IRE1-RIDD in lipid metabolism dysregulation in PGD-induced CHD. Interestingly, IRE1-RIDD contribution has been described for several other cardiac pathologies (Binder et al., 2019).
Our findings show that targeting the IRE1 pathway, or downstream VLFCA dysregulation, could be significant for therapeutic targeting. VLCFA lipid imbalance is treatable by means of dietary supplementation (pregnant diabetic mothers and their newborns present better outcomes when supplemented with DHA). Alternatives to treat ER stress by chemically targeting IRE1 also exist and could be translated to the clinic. In an attempt to remedy the effects of ER stress, we tested several potentially therapeutic compounds on PGDHOs, including BH4, a recently identified molecule that can reduce the incidence of PGD-induced CHD in mice (Engineer et al., 2018), a mixture of omega-3 FAs, and TUDCA (an IRE1 modulator). Both BH4 and omega-3 PUFAs ameliorated diabetic phenotypes, whereas TUDCA (an IRE1 modulator) ameliorated CM hypertrophy. All of the compounds also restored FADS2 levels. These data suggest that although BH4 and omega-3 FAs help ameliorate the effects of PGD on the developing heart via a reduction in ROS, the therapeutic role of TUDCA may be more closely involved in transcription factor regulation. Lastly, the data suggest that the increased levels of phosphorylated IRE1 may be a key player in the mechanism behind PGD-associated CHD.
In summary, we established the value of a heart organoid model of PGD and CHD, and our findings unveiled a novel ROS-induced ER stress mechanism underlying critical VLCFA balance in PGD-CHD. These findings represent a new route for developing future preventive and therapeutic strategies, thus helping to reduce the incidence of CHD across the population and provide proof-of-concept of the utility of organoid models for human disease modeling and drug development.
Experimental procedures
Resource availability
Data are available in the main text or in the supplemental information. All unique reagents generated for this study are available from the lead contact without restriction.
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author Aitor Aguirre: aaguirre@msu.edu.
Materials availability
No new reagents were generated for this report.
Data and code availability
RNA-seq datasets have been deposited at the NCBI GEO repository under accession code GSE201343. No new code was generated in this report.
Stem cell culture
An in-house human iPSC line was used in this study (iPSC-L1). For apoptosis experiments, a transgenic iPSC-L1 line expressing Flip-GFP (Zhang et al., 2019) was used. Human PSCs (hPSCs) were cultured in Essential 8 Flex medium containing 1% penicillin-streptomycin (Gibco) on 6-well plates coated with growth factor–reduced Matrigel (Corning) in an incubator at 37°C, 5% CO2, until 60%–80% confluency was reached, at which point cells were split into new wells using ReLeSR passaging reagent (Stem Cell Technologies). Accutase (Innovative Cell Technologies) was used to dissociate iPSCs for spheroid formation. hPSCs were then seeded at 10,000 cells per well in round-bottom ultra-low attachment 96-well plates (Costar) on day −2 at a volume of 100 μL per well. The plates were then centrifuged at 100 × g for 3 min and placed in an incubator at 37°C, 5% CO2. After 24 h (day −1), 50 μL of media was carefully removed from each well, and 200 μL of fresh Essential 8 Flex medium was added for a final volume of 250 μL per well. The plates were returned to the incubator for an additional 24 h.
Heart organoid differentiation and PGD modeling
PGD was modeled as previously described (Lewis-Israeli et al., 2022). Briefly, diabetic conditions were simulated by using basal RPMI media with 11.1 mM glucose and 1.14 nM insulin and compared with control media containing 3.5 mM glucose and 170 pmol insulin, henceforth referred to as diabetic and healthy culture media, respectively. The differentiation of the healthy or diabetic organoids was conducted as previously described (Lewis-Israeli et al., 2021b). Briefly, on day 0 of differentiation, 166 μL of media was removed from each well and 166 μL of diabetic or healthy culture media without any insulin containing CHIR99021 (Selleck) was added at a final concentration of 4 μM per well along with BMP4 at 0.36 p.m. and activin A at 0.08 pmol, for 24 h. On day 1, 166 μL of media was replaced with fresh culture media without insulin. On day 2, 166 μL of media was replaced with fresh culture media minus insulin containing Wnt-C59 (Selleck), for a final concentration of 2 μM. On day 4, 166 μL of media was removed and replaced with fresh culture media without insulin. On day 6, 166 μL of media was replaced with fresh culture media with respective diabetic and healthy insulin in the culture media. On day 7, a second 2-μM CHIR99021 exposure was conducted for 1 h in fresh culture media. Subsequently, media was changed every 48 h until organoids were ready for analysis. For IRE1 inhibition organoids were treated with 1 μM monoselective inhibitor KIRA8 (Selleck) for 5 days. Organoids were analyzed on day 14 unless otherwise indicated.
Immunofluorescence and confocal microscopy
hHOs were fixed in 4% paraformaldehyde solution. Fixation was followed by washes in PBS-glycine (20 mM) and incubation in blocking/permeabilization solution containing 10% donkey normal serum, 0.5% Triton X-100, 0.5% BSA in PBS on a thermal mixer at minimum speed at 4°C overnight. hHOs were then incubated with primary antibodies in antibody solution (1% donkey normal serum, 0.5% Triton X-100, 0.5% BSA in PBS) on a thermal mixer at 4°C for 24 h. Primary antibody exposure was followed by incubation with secondary antibodies in antibody solution for 24 h in the dark. Stained hHOs were mounted on glass microscope slides using Vectashield Vibrance Antifade Mounting Medium (Vector Laboratories). Samples were imaged using confocal laser scanning microscopy (Nikon Instruments A1 Confocal Laser Microscope). The images were analyzed using Fiji (https://imagej.net/Fiji).
Statistics and reproducibility
All of the analyses were performed using Excel and GraphPad. Statistical significance was evaluated with unpaired Student’s t test or one-way ANOVA, as appropriate. All data are presented as mean ± SD and represent a minimum of 3 independent experiments 3 technical replicates per experiment unless otherwise stated.
Acknowledgments
We thank the MSU Collaborative Mass Spectrometry Core, Genomics Core, Advanced Imaging Core, and the IQ Microscopy Core. Work in Dr. Aguirre’s laboratory was supported by the NIH under award numbers K01HL135464 and R01HL151505, by a Pilot and Feasibility Grant from the Michigan Diabetes Research Center, University of Michigan (NIH grant P30-DK020572), by the American Heart Association under award number 19IPLOI34660342, and by the Spectrum-MSU Alliance Foundation. Work in the Chan laboratory was supported by NSF CBET 1802992. We also wish to thank members of the Chan laboratory Kevin Y. Chen and Sardar M. Murtaza for helping to identify RIDD targets, and all of the members of the Aguirre laboratory for their valuable comments and advice.
Author contributions
A. Kostina, Y.R.L.-I., M.A., and A.A. designed the experiments and conceptualized the work. A. Kostina and Y.R.L.-I., performed all of the experiments and data analyses. M.A., M.A.G., H.L., and A.H.W. performed the cell and organoid culture and qPCR. A. Kiselev performed the scRNA-seq computational analysis. B.D.V. performed the live calcium recordings. T.L. performed LC-MS and data analyses. A.R.H. performed the qPCR and lentivirus experiments. A. Kostina, Y.R.L.-I., I.O., S.P., C.C., and A.A. wrote the manuscript and provided valuable suggestions and advice. A.A. supervised all of the work.
Declaration of interests
The authors declare no competing interests.
Published: February 8, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2024.01.003.
Supplemental information
References
- Ahuja P., Zhao P., Angelis E., Ruan H., Korge P., Olson A., Wang Y., Jin E.S., Jeffrey F.M., Portman M., Maclellan W.R. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J. Clin. Invest. 2010;120:1494–1505. doi: 10.1172/JCI38331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Biltagi M., El Razaky O., El Amrousy D. Cardiac changes in infants of diabetic mothers. World J. Diabetes. 2021;12:1233–1247. doi: 10.4239/wjd.v12.i8.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M.J., Uppulapu S.K., Tiwari V., Varghese B., Mohammed S.A., Adela R., Arava S.K., Banerjee S.K. Pregestational diabetes alters cardiac structure and function of neonatal rats through developmental plasticity. Front. Cardiovasc. Med. 2022;9 doi: 10.3389/fcvm.2022.919293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad Z., Rezapour-Firouzi S., Ebrahimifar M., Mosavi Jarrahi A., Mohammadian M. Association of delta-6-desaturase expression with aggressiveness of cancer, diabetes mellitus, and multiple sclerosis: a narrative review. Asian Pac. J. Cancer Prev. APJCP. 2019;20:1005–1018. doi: 10.31557/APJCP.2019.20.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp M., Giacomello S., Larsson L., Wu C., Fürth D., Qian X., Wärdell E., Custodio J., Reimegård J., Salmén F., et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell. 2019;179:1647–1660.e19. doi: 10.1016/j.cell.2019.11.025. [DOI] [PubMed] [Google Scholar]
- Baseler W.A., Dabkowski E.R., Jagannathan R., Thapa D., Nichols C.E., Shepherd D.L., Croston T.L., Powell M., Razunguzwa T.T., Lewis S.E., et al. Reversal of mitochondrial proteomic loss in Type 1 diabetic heart with overexpression of phospholipid hydroperoxide glutathione peroxidase. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013;304:R553–R565. doi: 10.1152/ajpregu.00249.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu M., Garg V. Maternal hyperglycemia and fetal cardiac development: Clinical impact and underlying mechanisms. Birth Defects Res. 2018;110:1504–1516. doi: 10.1002/bdr2.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder P., Wang S., Radu M., Zin M., Collins L., Khan S., Li Y., Sekeres K., Humphreys N., Swanton E., et al. Pak2 as a novel therapeutic target for cardioprotective endoplasmic reticulum stress response. Circ. Res. 2019;124:696–711. doi: 10.1161/CIRCRESAHA.118.312829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenoord A.L., Clevers H., Knoblich J.A. Human tissues in a dish: the research and ethical implications of organoid technology. Science. 2017;355 doi: 10.1126/science.aaf9414. [DOI] [PubMed] [Google Scholar]
- Cui X., Zhang Y., Lu Y., Xiang M. ROS and endoplasmic reticulum stress in pulmonary disease. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.879204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engineer A., Saiyin T., Lu X., Kucey A.S., Urquhart B.L., Drysdale T.A., Norozi K., Feng Q. Sapropterin Treatment Prevents Congenital Heart Defects Induced by Pregestational Diabetes Mellitus in Mice. J. Am. Heart Assoc. 2018;7 doi: 10.1161/JAHA.118.009624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi J.A., Zhang X.Y., Maidman J.E. Fetal cardiac hypertrophy and cardiac function in diabetic pregnancies. Am. J. Obstet. Gynecol. 1995;173:1132–1136. doi: 10.1016/0002-9378(95)91339-4. [DOI] [PubMed] [Google Scholar]
- Güemes M., Rahman S.A., Hussain K. What is a normal blood glucose? Arch. Dis. Child. 2016;101:569–574. doi: 10.1136/archdischild-2015-308336. [DOI] [PubMed] [Google Scholar]
- Hamlin R.L., Altschuld R.A. Am Heart Assoc; 2011. Extrapolation from Mouse to Man. [DOI] [PubMed] [Google Scholar]
- Hofbauer P., Jahnel S.M., Papai N., Giesshammer M., Deyett A., Schmidt C., Penc M., Tavernini K., Grdseloff N., Meledeth C., et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021;184:3299–3317.e22. doi: 10.1016/j.cell.2021.04.034. [DOI] [PubMed] [Google Scholar]
- Hollien J., Lin J.H., Li H., Stevens N., Walter P., Weissman J.S. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberger L.K. BMJ Publishing Group Ltd; 2006. Maternal Diabetes and the Fetal Heart. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostina A., Volmert B., Aguirre A. Human heart organoids: current applications and future perspectives. Eur. Heart J. 2023 doi: 10.1093/eurheartj/ehad841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K., Clevers H. Organoids: modeling development and the stem cell niche in a dish. Dev. Cell. 2016;38:590–600. doi: 10.1016/j.devcel.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Kumar S.D., Dheen S.T., Tay S.S.W. Maternal diabetes induces congenital heart defects in mice by altering the expression of genes involved in cardiovascular development. Cardiovasc. Diabetol. 2007;6:34. doi: 10.1186/1475-2840-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T., Flam B., Lockey R., Kolliputi N. TXNIP shuttling: missing link between oxidative stress and inflammasome activation. Front. Physiol. 2013;4:50. doi: 10.3389/fphys.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtoranta L., Koskinen A., Vuolteenaho O., Laine J., Kytö V., Soukka H., Ekholm E., Räsänen J. Gestational hyperglycemia reprograms cardiac gene expression in rat offspring. Pediatr. Res. 2017;82:356–361. doi: 10.1038/pr.2017.42. [DOI] [PubMed] [Google Scholar]
- Lehtoranta L., Vuolteenaho O., Laine V.J., Koskinen A., Soukka H., Kytö V., Määttä J., Haapsamo M., Ekholm E., Räsänen J. Maternal hyperglycemia leads to fetal cardiac hyperplasia and dysfunction in a rat model. Am. J. Physiol. Endocrinol. Metab. 2013;305:E611–E619. doi: 10.1152/ajpendo.00043.2013. [DOI] [PubMed] [Google Scholar]
- Lewis-Israeli Y.R., Abdelhamid M., Olomu I., Aguirre A. Modeling the Effects of Maternal Diabetes on the Developing Human Heart Using Pluripotent Stem Cell-Derived Heart Organoids. Curr. Protoc. 2022;2:e461. doi: 10.1002/cpz1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Israeli Y.R., Wasserman A.H., Aguirre A. Heart organoids and engineered heart tissues: Novel tools for modeling human cardiac biology and disease. Biomolecules. 2021;11:1277. doi: 10.3390/biom11091277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Israeli Y.R., Wasserman A.H., Gabalski M.A., Volmert B.D., Ming Y., Ball K.A., Yang W., Zou J., Ni G., Pajares N., et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021;12:5142. doi: 10.1038/s41467-021-25329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man J., Barnett P., Christoffels V.M. Structure and function of the Nppa-Nppb cluster locus during heart development and disease. Cell. Mol. Life Sci. 2018;75:1435–1444. doi: 10.1007/s00018-017-2737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron B.J., Maron M.S., Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J. Am. Coll. Cardiol. 2012;60:705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- Mukwevho E., Ferreira Z., Ayeleso A. Potential role of sulfur-containing antioxidant systems in highly oxidative environments. Molecules. 2014;19:19376–19389. doi: 10.3390/molecules191219376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niderla-BieliŃska J., Jankowska-Steifer E., Flaht-Zabost A., Gula G., Czarnowska E., Ratajska A. Proepicardium: Current Understanding of its Structure, Induction, and Fate. Anat. Rec. 2019;302:893–903. doi: 10.1002/ar.24028. [DOI] [PubMed] [Google Scholar]
- Odiete O., Hill M.F., Sawyer D.B. Neuregulin in cardiovascular development and disease. Circ. Res. 2012;111:1376–1385. doi: 10.1161/CIRCRESAHA.112.267286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øyen N., Diaz L.J., Leirgul E., Boyd H.A., Priest J., Mathiesen E.R., Quertermous T., Wohlfahrt J., Melbye M. Prepregnancy Diabetes and Offspring Risk of Congenital Heart Disease: A Nationwide Cohort Study. Circulation. 2016;133:2243–2253. doi: 10.1161/CIRCULATIONAHA.115.017465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G., Broguiere N., Miyamoto M., Boni A., Guiet R., Girgin M., Kelly R.G., Kwon C., Lutolf M.P. Capturing cardiogenesis in gastruloids. Cell Stem Cell. 2021;28:230–240.e6. doi: 10.1016/j.stem.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer R., van der Ploeg K., Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. USA. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmert B., Kiselev A., Juhong A., Wang F., Riggs A., Kostina A., O'Hern C., Muniyandi P., Wasserman A., Huang A., et al. A patterned human primitive heart organoid model generated by pluripotent stem cell self-organization. Nat. Commun. 2023;14:8245. doi: 10.1038/s41467-023-43999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman A.H., Venkatesan M., Aguirre A. Bioactive Lipid Signaling in Cardiovascular Disease, Development, and Regeneration. Cells. 2020;9:1391. doi: 10.3390/cells9061391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Reece E.A., Zhong J., Dong D., Shen W.-B., Harman C.R., Yang P. Type 2 diabetes mellitus induces congenital heart defects in murine embryos by increasing oxidative stress, endoplasmic reticulum stress, and apoptosis. Am. J. Obstet. Gynecol. 2016;215:366.e1–366.e10. doi: 10.1016/j.ajog.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M., Luo H., Wu J., Ren L., Ding X., Wu C., Chen J., Chen S., Zhang H., Yu L., et al. ADAM23 in Cardiomyocyte Inhibits Cardiac Hypertrophy by Targeting FAK - AKT Signaling. J. Am. Heart Assoc. 2018;7 doi: 10.1161/JAHA.118.008604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Schepis A., Huang H., Yang J., Ma W., Torra J., Zhang S.-Q., Yang L., Wu H., Nonell S., et al. Designing a green fluorogenic protease reporter by flipping a beta strand of GFP for imaging apoptosis in animals. J. Am. Chem. Soc. 2019;141:4526–4530. doi: 10.1021/jacs.8b13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Xu C., Gabbay-Benziv R., Lin X., Yang P. Superoxide dismutase 2 overexpression alleviates maternal diabetes-induced neural tube defects, restores mitochondrial function and suppresses cellular stress in diabetic embryopathy. Free Radic. Biol. Med. 2016;96:234–244. doi: 10.1016/j.freeradbiomed.2016.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets have been deposited at the NCBI GEO repository under accession code GSE201343. No new code was generated in this report.