Summary
Mutations in the AAA+ ATPase p97 cause multisystem proteinopathy 1, which includes amyotrophic lateral sclerosis; however, the pathogenic mechanisms that contribute to motor neuron loss remain obscure. Here, we use two induced pluripotent stem cell models differentiated into spinal motor neurons to investigate how p97 mutations perturb the motor neuron proteome. Using quantitative proteomics, we find that motor neurons harboring the p97 R155H mutation have deficits in the selective autophagy of lysosomes (lysophagy). p97 R155H motor neurons are unable to clear damaged lysosomes and have reduced viability. Lysosomes in mutant motor neurons have increased pH compared with wild-type cells. The clearance of damaged lysosomes involves UBXD1-p97 interaction, which is disrupted in mutant motor neurons. Finally, inhibition of the ATPase activity of p97 using the inhibitor CB-5083 rescues lysophagy defects in mutant motor neurons. These results add to the evidence that endo-lysosomal dysfunction is a key aspect of disease pathogenesis in p97-related disorders.
Keywords: p97, ALS, lysosome, lysophagy, autophagy, proteomics, galectin, mitochondria
Highlights
-
•
Proteomics of p97 R155H motor neurons finds deficits in lysosome quality control
-
•
p97 R155H motor neurons are unable to clear damaged lysosomes through lysophagy
-
•
Increased lysosomal pH in p97 mutant motor neurons
-
•
p97 catalytic inhibitor rescues lysosome defects in mutant motor neurons
In this article Raman and colleagues show that the p97 R155H mutation that causes ALS disrupts the selective autophagy of lysosomes (lysophagy). Using distinct iPSC cell models differentiated into lower motor neurons and quantitative proteomics, they find that p97 mutant motor neurons are unable to clear damaged lysosomes. They further show that inhibition of p97 catalytic activity using small molecule inhibitors rescues lysophagy defects.
Introduction
p97 (also known as valosin containing protein, VCP) is an evolutionarily conserved type II AAA+ ATPase with important roles in ubiquitin-dependent protein quality control. p97 is ubiquitously expressed and forms a homohexamer wherein each monomer is composed of two ATPase domains (D1 and D2) as well as an N-terminal regulatory domain (NTD) (DeLaBarre and Brunger, 2003). ATP hydrolysis by p97 has been demonstrated to enable substrate unfolding by threading the ubiquitylated substrate through the central pore of the hexamer (Cooney et al., 2019; Twomey et al., 2019). This “unfoldase” activity is suited for the extraction of substrates from membranes and multi-protein complexes prior to proteasomal degradation (Ye et al., 2017). Key to p97 function and specificity is its ability to form distinct complexes with a host of adaptor proteins. Over 40 adaptors have been identified that enable p97 recruitment to ubiquitylated substrates (Buchberger et al., 2015). We and others have shown that p97-adaptor complexes are important for many cellular processes including regulation of organelle contact (Ganji et al., 2023), cell cycle (Kochenova et al., 2022), DNA damage response (Franz et al., 2016), and autophagy (van den Boom and Meyer, 2018) (see Ahlstedt et al., 2022; van den Boom and Meyer, 2018 for review). Thus, loss of p97 impacts the degradation of a large cohort of the ubiquitin-modified proteome.
In addition to proteasomal degradation, p97 plays an important role in the endo-lysosomal system and autophagy (Krick et al., 2010; Ramanathan and Ye, 2012) with recent reports suggesting a role in lysophagy, the autophagic process of turning over damaged lysosomes (Arhzaouy et al., 2019; Kravić et al., 2022; Papadopoulos et al., 2017). p97 in conjunction with a trimeric adaptor complex composed of the adaptors UBX domain protein 1 (UBXD1), YOD1 deubiquitinase (YOD1), and phospholipase A2 activating protein (PLAA), collectively known as the endo-lysosomal damage repair (ELDR) complex (Papadopoulos et al., 2017), mediates removal of ubiquitylated intermediates on the lysosomal membrane enabling lysophagy (Kravić et al., 2022).
Mutations in p97 cause a heterogeneous disease known as multisystem proteinopathy 1 (MSP-1), which includes Paget’s disease of the bone, inclusion body myopathy, frontotemporal dementia, and amyotrophic lateral sclerosis (ALS) (Johnson et al., 2010; Watts et al., 2004). Tissue from patients with p97 mutations contain TDP-43 and ubiquitin-positive inclusions linking p97-related disease with sporadic forms of these diseases (Neumann et al., 2007). Certain disease-related p97 mutations have increased ATPase activity in vitro (Manno et al., 2010); however, whether this translates to a toxic gain-of-function or a dominant negative effect remains debated (Ahlstedt et al., 2022; van den Boom and Meyer, 2018; Franz et al., 2016). The majority of mutations occur in the NTD and N-terminal-D1 linker region, which is important for binding to some classes of adaptors (Pfeffer et al., 2022). Indeed, these mutations alter adaptor binding, with some adaptors having increased association (e.g., UFD1-NPL4 [Blythe et al., 2019]) and others with decreased association (e.g., UBXD1 [Ritz et al., 2011]). Thus, for some p97 mutations, there may be loss of targeting to some substrates leading to their accumulation or precocious degradation of others due to increased targeting; however, this has not been exhaustively tested in relevant cell types.
The cellular processes affected by mutant p97 that lead to disease are still intensely debated. Models in immortalized cell types such as U2OS and NSC-34 have found defects in lysophagy leading to an accumulation of damaged lysosomes and increased cell death (Ferrari et al., 2022; Papadopoulos et al., 2017). Drosophila and rodent models of p97 disease have shown defects in protein degradation, autophagy, and mitochondrial function (Gonzalez and Wang, 2020; Kim et al., 2013; Nalbandian et al., 2012). Induced pluripotent stem cell (iPSC)-derived motor neuron models of p97 disease have heterogeneous phenotypes as well. Initial studies utilizing iPSC-derived motor neurons from a patient with the R155H mutation found increased levels of TDP-43, ubiquitin, and autophagy-related proteins (Dec et al., 2014). Another study found TDP-43 mislocalization, endoplasmic reticulum (ER) stress induction, and mitochondrial dysfunction in motor neurons from patients carrying the R155C and R191Q mutations (Hall et al., 2017). A recent study used patient-derived iPSCs with R155H mutation and matched wild-type corrected lines and found no changes in ER stress or TDP-43 mislocalization; however, aberrant cell-cycle protein levels were determined to be contributors to decreased viability (Wang et al., 2022).
To begin to understand how p97 mutations impact the cellular proteome, we performed quantitative proteomics on two distinct p97 R155H iPSC-derived motor neuron cell lines. The first iPSC line was derived from a patient harboring the R155H mutation that was corrected to the wild-type allele using CRISPR-Cas9 editing. The second iPSC line was created by the iPSC neurodegenerative disease initiative (iNDI), wherein the R155H mutation was introduced into the endogenous p97 genomic locus of KOLF2.1 cells to create both heterozygous and homozygous lines (Pantazis et al., 2022; Ramos et al., 2021). These two cell-based models allow us to interrogate how the cellular proteome is impacted by p97 mutation alone (KOLF2.1 lines) as well as how patient-specific genetic backgrounds modify the p97 R155H phenotype. From proteomic studies, we found that each genetic background had over 200 differentially expressed proteins, but only a few proteins were common between the patient and KOLF2.1 cell lines. For example, KOLF2.1 R155H cells had a pronounced defect in mitochondrial homeostasis; surprisingly, the patient-derived cells did not have this phenotype. An unbiased clustering analysis identified autophagy and lysosomal proteins as the most significantly altered in both the mutant lines. Of the few commonly altered proteins, we identified multiple components of lysosome repair machinery.
We find that while wild-type motor neurons were able to effectively clear damaged lysosomes following agent L-leucyl leucine-O-methyl ester (LLOME) treatment, p97 mutant neurons were not. Interestingly, while LLOME treatment stimulated the association of wild-type p97 with UBXD1, this complex was less effectively formed by p97 R155H, suggesting defective ELDR complex formation. Because mutant p97 has increased ATPase activity, previous studies have utilized p97 inhibitors to reverse disease phenotypes (Harley et al., 2021; Wang et al., 2022; Zhang et al., 2017). We co-treated motor neurons with LLOME and CB-5083, a highly specific competitive p97 inhibitor, and found that inhibition of mutant p97 rescued lysophagy defects and prevented LLOME-induced cell death. Overall, our studies suggest that p97 R155H mutation negatively impacts lysosome quality control in human motor neurons.
Results
Differentiation of two isogenic iPSC p97 R155H models into functional motor neurons
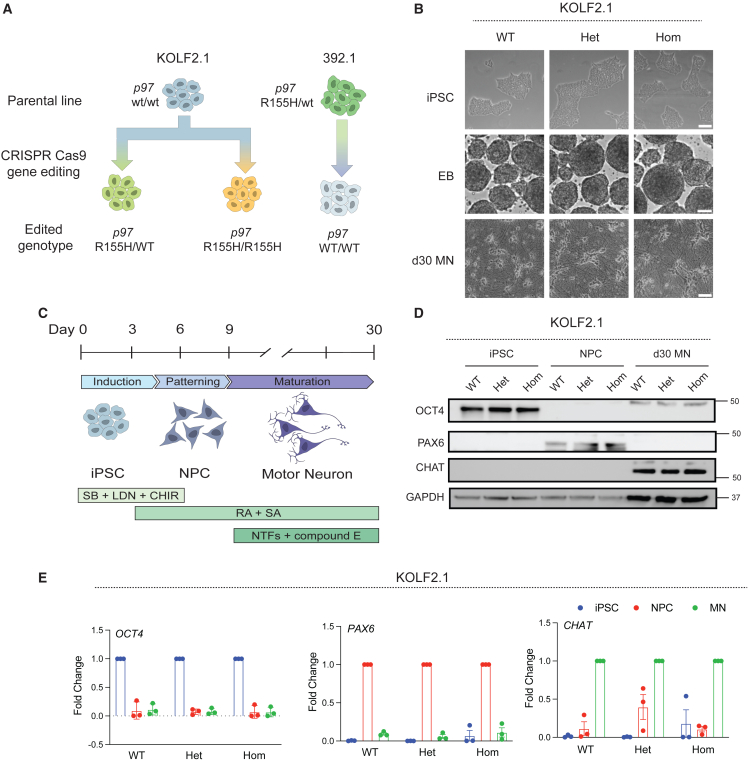
We used two orthogonal models to investigate how the p97 R155H mutation impacted the motor neuron proteome. The first was a wild-type KOLF2.1 iPSC line that was CRISPR edited to introduce the R155H mutation into the endogenous p97 locus that was developed by iNDI (Pantazis et al., 2022) (Figure 1A). We used two clonal lines (heterozygous and homozygous mutants) for our studies (referred to as KOLF2.1 wild-type, heterozygous, and homozygous). The second was the 392.1 iPSC line from a 48-year-old male diagnosed with ALS, dementia, and Paget’s disease, harboring the p97 R155H mutation. The R155H mutation was reverted to wild-type by CRISPR/Cas9 editing (referred to as 392.1 wild-type and R155H) (Figure 1A). Genotypes for both cell lines were confirmed by DNA sequencing (Figures S1A and S2A). A normal karyotype was confirmed for parental lines (Figures S1B and S2B). Wild-type and mutant iPSCs were indistinguishable from each other in colony morphology, the ability to form embryoid bodies, and the expression of pluripotency markers (Figures 1B, S1C, S2C, and S2D). We established a spinal motor neuron differentiation protocol using previous studies (Du et al., 2015; Qu et al., 2014; Shimojo et al., 2015). iPSCs were induced to neural ectoderm fate using dual-SMAD inhibition and a WNT pathway agonist (Figure 1C). After 3 days, retinoic acid (RA) and smoothened agonist (SAG, an activator of the SHH pathway) were added to create NES+, SOX2+, PAX6+ neural precursor cells (NPCs) (Figures 1D, 1E, S1D, S2E, and S2F). NPCs were further patterned using RA and SAG to a ventral, caudal fate for 3 days to create motor neuron progenitors (Figure 1C). Progenitors were replated onto poly L-ornithine (PLO), laminin, and fibronectin-coated plates and cultured in motor neuron maturation media containing neurotrophic factors (neurotrophin-3 [NT-3], brain-derived neurotrophic factor [BDNF], and glial-derived neurotrophic factor [GDNF]), RA, SAG, and the gamma-secretase inhibitor, compound E to activate the notch pathway. Twenty-one days of maturation produced >90% class III β-tubulin (TUJ1), neurofilament heavy chain (SMI32), choline acetyltransferase (CHAT)-positive motor neurons (Figures 1D, 1E, S1E, and S2G).
Figure 1.
Characterization of IPSC-derived motor neurons
(A) Two isogenic iPSC models were generated using gene editing to introduce the R155H mutation into the p97 endogenous locus (KOLF2.1) and correct the R155H mutation (392.1) to wild type.
(B) Phase contrast images of iPSCs (top row), embryoid bodies (middle row), and day 30 motor neurons (bottom row). Scale bars, 50 μm (top and bottom rows) and 100 μm (middle row).
(C) Schematic of the motor neuron differentiation protocol.
(D) Immunoblot of iPSCs, NPCs, and day 30 motor neurons. The faster migration band in PAX6 immunoblots likely represents another isoform.
(E) qPCR of wild-type, heterozygous, and homozygous KOLF2.1 iPSCs, NPCs, and motor neurons and markers for each stage. CHIR, CHIR99021; EB, embryoid body; LDN, LDN193189; MN, motor neuron; NPC, neural precursor cell; NTFs, neurotrophic factors; RA, retinoic acid; SA, smoothened agonist; SB, SB431542. See also Figures S1 and S2.
Characterization of cellular phenotypes in p97 R155H motor neurons
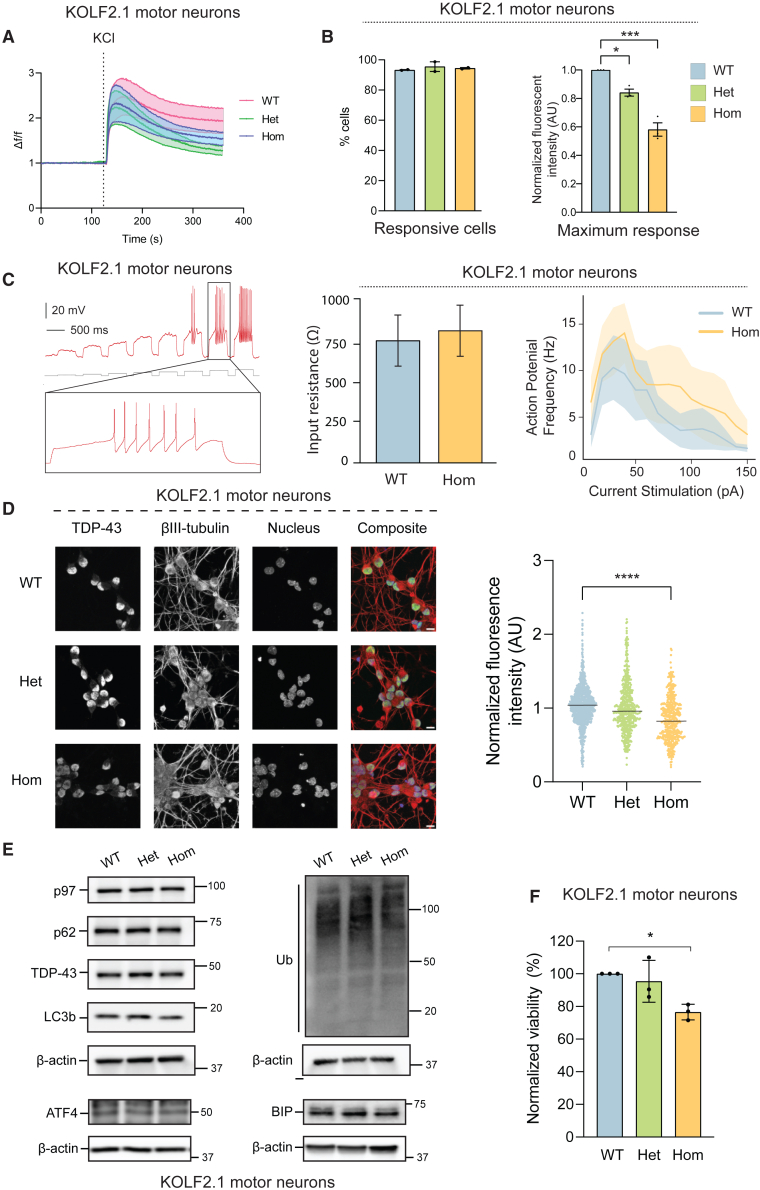
We next characterized cellular phenotypes attributed to mutant p97. We confirmed that mature KOLF2.1 motor neurons were functional using Fluo-4 a.m. live-cell calcium imaging, which showed >90% of cells responded to 80 mM KCl depolarization (Figures 2A and 2B). Interestingly, KOLF2.1 heterozygous and homozygous motor neurons had lower maximal calcium transients in response to KCl, suggesting altered calcium dynamics (Figure 2B, right). We next performed whole-cell patch clamp in wild-type and homozygous KOLF2.1 motor neurons at day 20. Both genotypes had equivalent input resistance (a measure of neuronal polarization) and had spontaneous action potentials (APs) and trains of action potentials in response to depolarizing current (Figure 2C, left and middle); however, homozygous motor neurons trended toward increased AP frequency though this did not reach statistical significance (Figure 2C right). These data suggest that our differentiation process produces functional motor neurons and that p97 mutation impacts their electrical activity as reported by others (Hall et al., 2017; Wang et al., 2022).
Figure 2.
p97 R155H motor neurons recapitulate ALS phenotypes
(A) Representative average Fluo-4 fluorescence values from KOLF2.1 motor neurons after 80 mM KCl stimulation (dotted line). The shaded area represents the 95% confidence interval. N = >50 cells per condition.
(B) Percent of KOLF2.1 motor neurons responsive to KCl stimulation (left). Maximal normalized Fluo-4 fluorescence in motor neurons. N > 150 cells, three independent experiments per condition.
(C) Sample trace from a wild-type (WT) KOLF2.1 motor neuron in response to a series of depolarizing current steps via whole-cell patch clamp (left). Input resistance from WT and homozygous motor neurons (middle). Input-output curves of motor neurons in WT and homozygous motor neurons at day 20 (right).
(D) Representative images of TDP-43 immunofluorescence in KOLF2.1 motor neurons (left panels). Quantification of nuclear TDP-43 fluorescence (right). N = >5,000 cells, four independent experiments. Data points represent individual cells; lines represent means. Scale bar, 10 μm.
(E) Representative immunoblots of TDP-43, autophagy markers (p62, LC3b), ER stress markers (ATF4, BIP), and total ubiquitylated proteins (Ub) in KOLF2.1 motor neurons. N = 3 independent experiments.
(F) Normalized viability of KOLF2.1 motor neurons at day 30. All data expressed as means ± SEM unless otherwise indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. One-way ANOVA with Dunnett’s multiple comparison test (B, D, F). See also Figure S3.
TAR DNA binding protein 43 (TDP-43) is a nuclear RNA binding protein that regulates the splicing of hundreds of genes (Tollervey et al., 2011) and its mislocalization to the cytosol and aggregation is prominent feature in ALS and MSP-1 (Neumann et al., 2007). We investigated TDP-43 localization in our iPSC lines using confocal microscopy and found that both mutant 392.1 and KOLF2.1 motor neurons had depleted levels of TDP-43 in the nucleus compared with wild-type (Figures 2D and S3A). However, total TDP-43 levels were equivalent between lines suggesting that nuclear depletion was not due to loss of protein (Figures 2E and S3B).
A key cellular function of p97 is ER-associated degradation (ERAD), which is the retrotranslocation of misfolded, proteins from the ER for proteasomal degradation. Whether p97 mutation negatively impacts ERAD is debated (Hall et al., 2017; Wang et al., 2022). In our models, we found no change in several markers of ER stress including the transcription factor ATF4 or the ER chaperone BIP (Figures 2E and S3B). Furthermore, we found no significant change in total ubiquitin conjugates or autophagy markers p62 or LC3B (Figures 2E and S3B). We measured cell viability and found that KOLF2.1 and 392.1 p97 R155H motor neurons had decreased survival relative to wild type in agreement with previous studies (Hall et al., 2017; Wang et al., 2022) (Figures 2F and S3C).
Unbiased quantitative proteomics reveals distinct proteome alterations between KOLF2.1 and patient cell lines
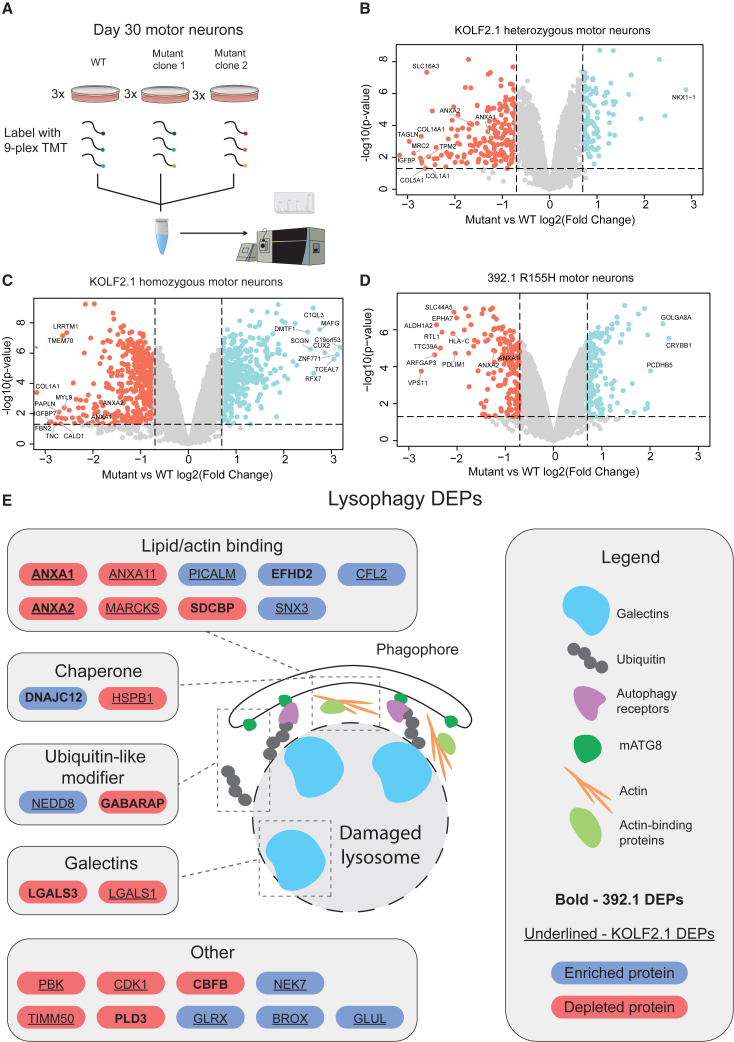
To interrogate changes in motor neurons due to p97 mutations at the proteome level, we employed unbiased quantitative proteomics using tandem mass tags (TMTs) (Thompson et al., 2003). Peptides from day 30 motor neurons from wild-type and p97 R155H (KOLF2.1 or 392.1) were labeled with TMTs and analyzed by liquid chromatography and mass spectrometry (LC/MS-MS) (Figure 3A and Table S1) (McAlister et al., 2014; Ting et al., 2011). More than 200 proteins were significantly altered (Log2 fold change mutant: wild type > 0.7) between mutants and wild-type in both cell lines suggesting broad changes at the protein level despite relatively mild phenotypic alterations (Figures 3B–3D). Gene ontology (GO) analysis of differentially expressed proteins (DEPs) found different pathways altered in KOLF2.1 and 392.1 mutant motor neurons despite the same R155H mutation (Figures S4A–S4C). KOLF2.1 homozygous motor neurons showed significant depletion of mitochondrial proteins (Figure S4B) that are part of complexes regulating import of proteins into mitochondria, and subunits of electron transport chain complexes I-IV was observed (Figure S4D). These results were further validated by immunoblot (Figure S4E). Notably, KOLF2.1 mutant motor neurons had significantly decreased mitochondrial membrane potential as measured using MitoTracker red (Figures S4F and S4G). No significant changes were observed in mitochondrial morphology (Figure S4H). Interestingly, these mitochondrial changes were not seen in the 392.1 mutant motor neurons (Figure S4C, and data not shown). Therefore, we sought to identify altered pathways that were shared between KOLF2.1 and 392.1 motor neurons. We utilized weighted gene correlation network analysis (WGCNA) adapted for proteomic studies to segment proteins into modules with similar expression profiles in an unbiased manner (Wu et al., 2020). This methodology uses a dynamic branch-cutting algorithm to separate clusters identified via a topological overlap matrix into distinct modules (Figure S5A and Table S2). Inspection of module eigenprotein expression revealed agreement between KOLF2.1 and 392.1 wild-type lines, but a divergence in the mutant neurons (Figure S5B). To identify modules that represented proteins changing in a similar manner between cell lines, modules were ranked by a composite score based on concordance between genetic backgrounds and average log2 fold change (Figures S5D–S5F and Table S2). The member proteins of the top three ranked modules were used for GO analysis to identify enriched terms. Multiple GO terms related to autophagy and lysosomal homeostasis were enriched (Figures S5D and S5E middle panels).
Figure 3.
Quantitative proteomics identifies autophagy defects in p97 R155H motor neurons
(A) Schematic of the experimental setup for the KOLF2.1 cells. Three replicates each of wild type (WT) and two clones of heterozygous or homozygous p97 R155H motor neurons were used in two 9-plex TMT experiments. Three replicates of WT and p97 R155H 392.1 motor neurons were used for a 6-plex TMT experiment.
(B–D) Volcano plots for the indicated cell line. Cutoff values of 0.7 log2 fold change and 0.05 p value were used to determine differentially expressed proteins (DEPs).
(E) Lysophagy-related proteins that were significantly altered in R155H motor neurons. Red and blue backgrounds indicate depleted and enriched proteins, respectively. See also Figures S4 and S5.
The identification of several pathways relating to lysosomal homeostasis was particularly interesting as p97 plays a critical role in the turnover of lysosomes and depletion of p97 leads to the persistence of damaged lysosomes (Papadopoulos et al., 2017). We compared our proteomics dataset to two published studies that identified proteins enriched on damaged lysosomes (Eapen et al., 2021) or were ubiquitylated following lysosomal damage (Kravić et al., 2022). We found over 20 DEPs that were differentially modulated in KOLF2.1 and/or 392.1 motor neurons suggesting that p97 R155H hinders the ability of motor neurons to respond to damaged lysosomes (Figure 3E).
Damaged lysosomes in motor neurons preferentially recruit LGALS8
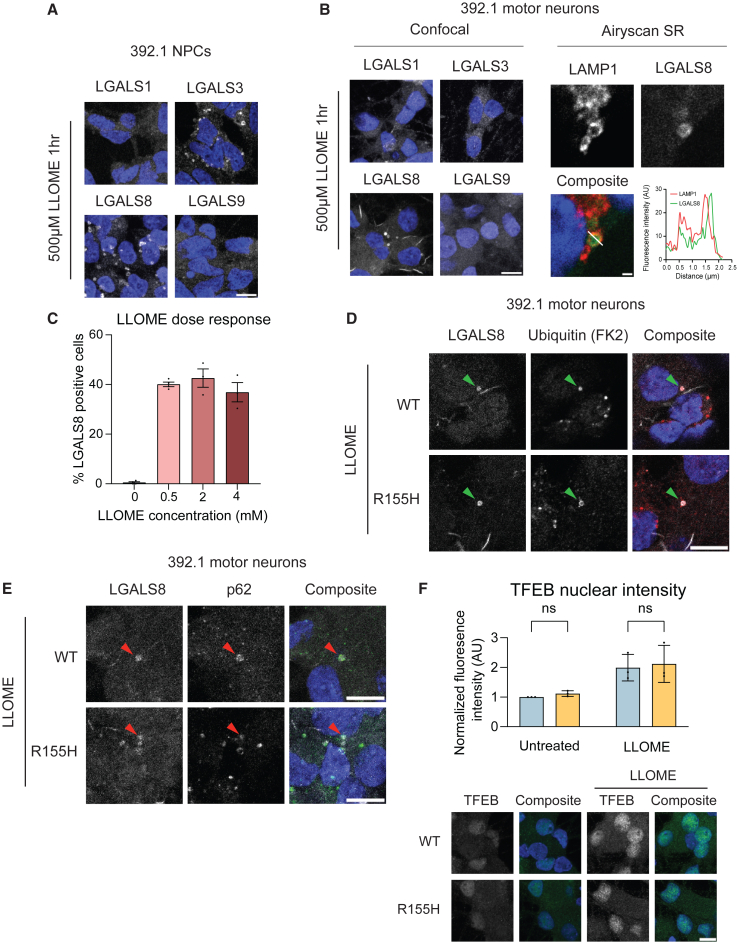
The alterations in autophagy and endocytosis pathways prompted us to investigate lysosomal turnover in p97 mutant motor neurons. L-leucine leucyl-O-methyl ester (LLOME) is a lysomotropic agent that permeabilizes lysosomal membranes to initiate galectin 3 (LGALS3) and autophagy component (p62 and LC3B) recruitment for lysosomal turnover (Eapen et al., 2021; Jia et al., 2020). Galectin recruitment to lysosomes is recognized as a sensitive measure of lysosomal damage (Aits et al., 2015); however, to our knowledge, lysophagy and markers of damaged lysosomes have not been evaluated in human spinal motor neurons. Indeed, other galectins such as LGALS1, LGALS8, and LGALS9 have also been shown to associate with lysosomes following LLOME treatment (Du Rietz et al., 2020). We evaluated the recruitment of LGALS 1, 3, 8, and 9 following LLOME treatment in both NPCs and d30 motor neurons. Surprisingly, we found that while NPCs recruited both LGALS3 and 8 to damaged lysosomes (Figure 4A), motor neurons selectively recruited LGALS8 (Figure 4B). This is in agreement with single-cell expression data of spinal cord cell types (Russ et al., 2021) that indicated that LGALS8 has highest expression in motor neurons compared with other galectins. We note that in addition to punctate localization, LGALS8 immunostaining also showed a non-punctate linear pattern both in untreated and treated conditions (Figure 4B). To ensure that LGALS8 puncta were damaged lysosomes, we performed Airyscan super resolution microscopy on motor neurons stained for the lysosomal protein LAMP1 and LGALS8 after LLOME treatment. All LGALS8 puncta colocalized with LAMP1 while the linear “dashes” did not (Figure 4B right panels). We found that 500 μM LLOME was sufficient to induce LGALS8 puncta formation in approximately 40% of cells and this did not increase with increasing dose (Figure 4C). LGALS8 puncta also colocalized with ubiquitin and p62, demonstrating that LGALS8 faithfully represents damaged lysosomes (Figures 4D and 4E). Non-punctate LGALS8 structures were not quantified in our subsequent studies. Previous studies have shown that the transcription factor TFEB translocates from lysosomes to the nucleus to induce lysosome biogenesis upon LLOME treatment (Fujita et al., 2013). We observed equivalent levels of increased nuclear TFEB in both wild-type and mutant motor neurons following LLOME treatment (Figure 4F).
Figure 4.
Motor neurons preferentially recruit LGALS8 to ubiquitylated lysosomes
(A) Representative images of galectin immunofluorescence in NPCs following 1 h of 500-μM LLOME treatment. N = 2 independent experiments. Scale bar, 10 μm.
(B) Representative images of galectin immunofluorescence in motor neurons following 1 h of 500-μM LLOME (left). Airyscan images of motor neurons following LLOME treatment co-stained with LAMP1 and LGALS8 show colocalization (right). N = 2 independent experiments. Scale bars, 10 μm (left) and 1 μm (right).
(C) Percent cells with LGALS8 puncta after 1 h of LLOME at indicated doses. N = 3 independent experiments.
(D and E) Representative images of LLOME-treated motor neurons co-stained for LGALS8 and ubiquitin (D) or p62 (E) demonstrating colocalization. N = 3 independent experiments. Scale bar, 10 μm.
(F) Representative images of TFEB staining. Quantification of nuclear TFEB fluorescence intensity. Scale bar, 10 μm. N = 3 independent experiments. All data expressed as means ± SEM. ns = nonsignificant. One-way ANOVA with Dunnett’s multiple comparison test (C, F). See also Figure S6.
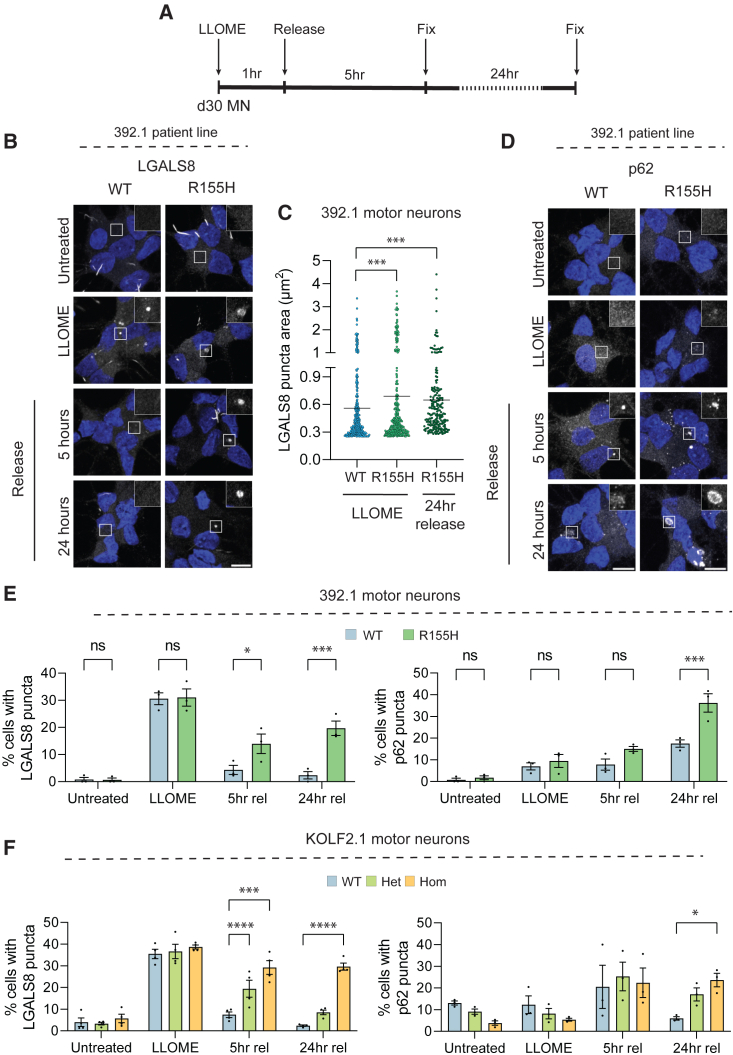
p97 R155H disrupts lysophagy in motor neurons
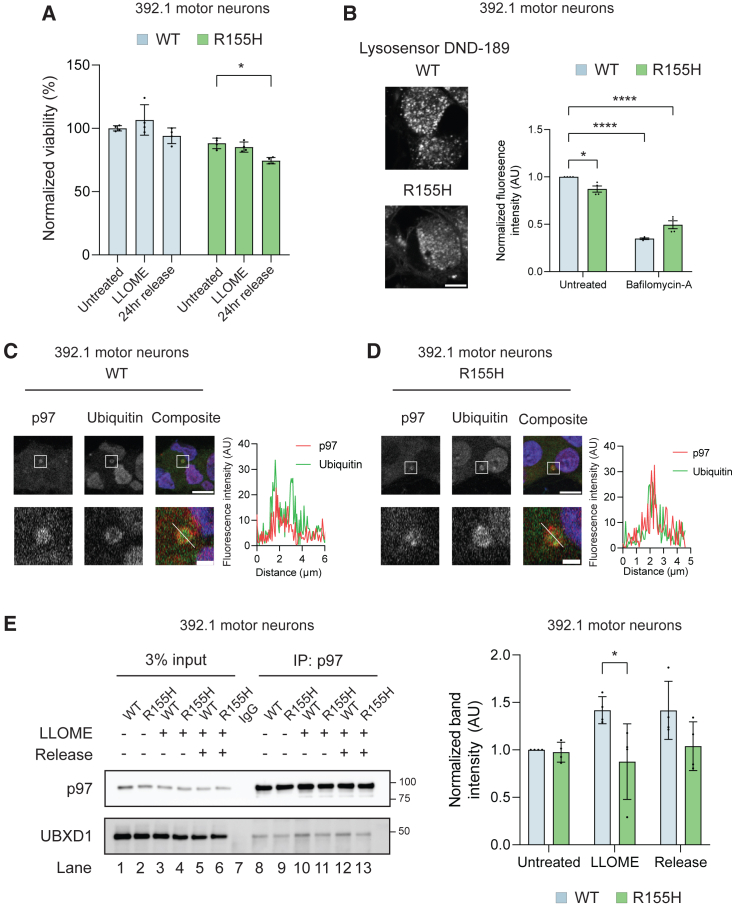
We utilized LGALS8 puncta formation as a measure of lysosomal damage in the following studies. We asked whether mutant motor neurons had defects in lysophagy. Motor neurons were treated with LLOME and then allowed to recover for 5 or 24 h and lysosome repair was monitored by LGALS8 recruitment (Figure 5A). Wild-type and mutant motor neurons had an equal number of LGALS8 puncta following treatment; however, after 5 h of recovery, wild-type neurons had completely cleared damaged lysosomes while the mutants had LGALS8 puncta that persisted 24 h after treatment (Figures 5B, 5E left, and 5F left). Notably, LGALS8-positive puncta were larger in mutant motor neurons during treatment and after release (Figure 5C). p62/SQSTM1 is a known autophagy receptor that is recruited to damaged lysosomes via interaction with ubiquitylated cargo (Fujita et al., 2013). Immunofluorescence for p62 demonstrated equal recruitment 5 h after treatment between wild-type and mutant; however, mutant motor neurons had significantly increased p62 puncta 24 h after treatment (Figures 5D, 5E right, and 5F right). Failure to clear damaged lysosomes can lead to cell death (Boya and Kroemer, 2008). We assessed viability following LLOME treatment and found that mutant motor neurons had increased cell death following release while wild-type neurons had no change (Figures 6A and S6A). We next asked whether disrupted levels of proteins involved in lysosome repair may also impact lysosome function in resting cells. We interrogated lysosomal pH using the pH-sensitive dye LysoSensor DND-189, which accumulates in acidic compartments and fluoresces at low pH. Following subthreshold LLOME or bafilomycin-A1 (inhibitor of lysosomal vacuolar H+ ATPase) treatment, we found that the LysoSensor fluorescence was significantly decreased demonstrating compromised pH (Figures 6B and S6B). Notably, mutant motor neurons had decreased fluorescence at basal levels compared with wild-type (Figures 6B and S6B).
Figure 5.
p97 R155H motor neurons are unable to clear damaged lysosomes and accumulate p62 following LLOME treatment
(A) Schematic of experimental design. Day 30 motor neurons were fixed either before LLOME treatment, directly after treatment, or 5 and 24 h after release.
(B) Representative images of 392.1 motor neurons stained for LGALS8 before, during, and after LLOME treatment. Scale bar, 10 μm.
(C) LGALS8 puncta size in wild-type and R155H 392.1 motor neurons following LLOME treatment and release. Data expressed as individual LGALS8 puncta; lines represent means.
(D) Representative images of 392.1 motor neurons stained for p62. Scale bar, 10 μm.
(E) Quantification of (B) (left) and (C) (right) in 392.1 motor neurons. N = 3 independent experiments.
(F) Quantification of LGALS8 puncta (left) and p62 puncta (right) in KOLF2.1 motor neurons. N = 3 independent experiments. D30 MN – day 30 motor neurons. 5h rel – 5-h release, 24h rel – 24-h release. Data expressed as means ± SEM unless otherwise indicated. ns = nonsignificant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. One-way ANOVA with Sidak’s multiple comparison test (B). Two-way ANOVA with Sidak’s (E) or Dunnett’s (F) multiple comparison test. See also Figure S6.
Figure 6.
p97 R155H sensitizes motor neurons to lysosomal damage, decreases lysosomal acidity, and disrupts UBXD1 recruitment
(A) Normalized viability in 392.1 motor neurons in untreated, LLOME treated, or after release. N = 4 independent experiments.
(B) Representative images of LysoSensor DND-189 live-cell imaging in 392.1 motor neurons (left). Quantification of DND-189 fluorescence in untreated and bafilomycin-A treated cells. N = 4 independent experiments. Scale bar, 5 μm.
(C and D) Representative images of wild-type (C) and p97 R155H (D) 392.1 motor neurons co-stained with p97 and ubiquitin showing colocalization (left). Line graph of the fluorescence intensity of p97 and ubiquitin as indicated in the bottom right image panel. N = 3 independent experiments. Scale bars, 10 μm (upper panels) and 1 μm (lower panels).
(E) Immunoblot of endogenous p97 immunoprecipitation in 392.1 motor neurons before, during, and after LLOME treatment (left). Release condition represents 5 h of recovery. Quantification of UBXD1 band intensities normalized to immunoprecipitated p97 (left). Data expressed as normalized band intensities ±SD. N = 4 independent experiments. All data expressed as means ± SEM unless otherwise indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Two-way ANOVA with Dunnett’s (A) or Sidak’s (B, E) multiple comparison test. See also Figure S6.
Ubiquitylation of lysosomal proteins is a key step in lysophagy and is required for lysosomal clearance (Chauhan et al., 2016). To determine if damaged lysosomes in mutant neurons were ubiquitylated, we co-stained for LGALS8 and ubiquitin. We found that LGALS8 puncta colocalized with ubiquitin in both wild-type and mutant motor neurons (Figures S6C and S6D). We next asked if mutant p97 was recruited to damaged lysosomes. However, both wild-type and mutant p97 were equally recruited to ubiquitylated lysosomes (Figures 6C and 6D). Mutations in p97 have been reported to alter the association with the adaptor UBXD1, a component of the ELDR complex (Ritz et al., 2011). We immunoprecipitated endogenous p97 from wild-type and mutant motor neurons before, during, and after LLOME treatment. At basal levels, both wild-type and mutant p97 associated with UBXD1; however, during LLOME treatment, wild-type p97 increased association with UBXD1 while mutant p97 did not (Figure 6E, compare lanes 10 with 11 and 12 with 13 and S6E, compare lane 13 with 14 and 15). Our studies suggest that p97 R155H motor neurons have perturbed lysosomal function and are unable to restore lysosomal homeostasis.
Inhibition of p97 rescues lysophagy defects in mutant motor neurons
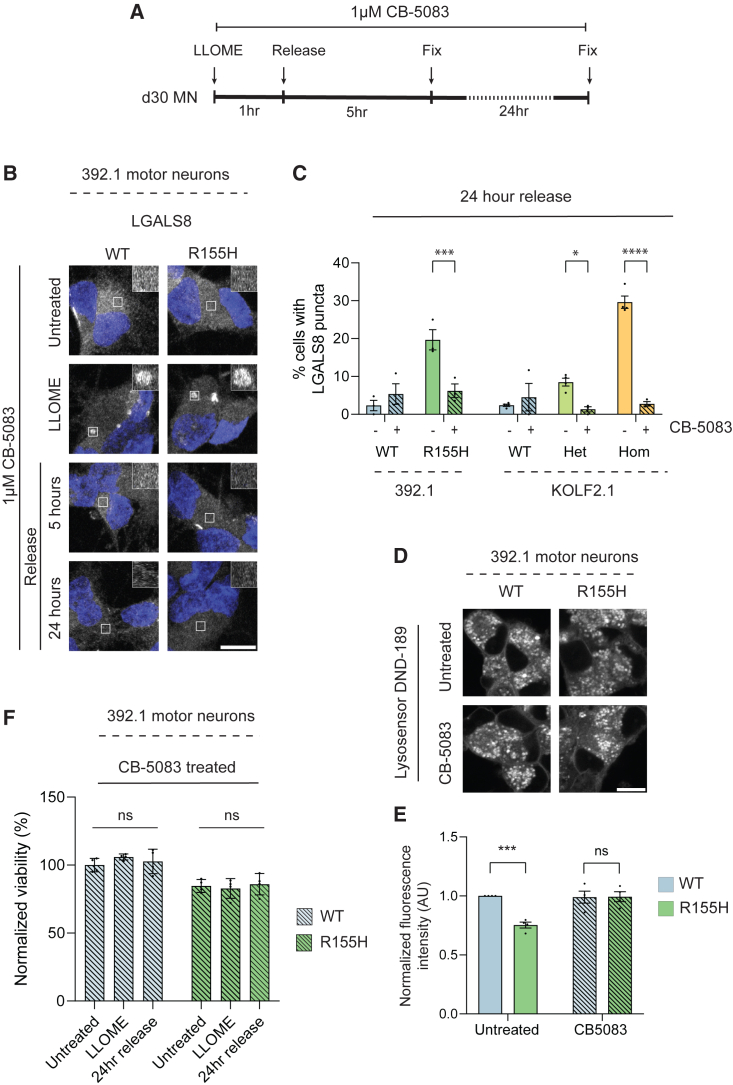
p97 R155H has increased ATPase activity in vitro, and multiple studies have used p97 inhibitors to rescue disease phenotypes including in iPSC-derived motor neurons (Harley et al., 2021; Wang et al., 2022). However, it is unknown if p97 inhibition rescues defective lysophagy. We attempted to prevent the persistence of damaged lysosomes in mutant motor neurons using a low dose of CB-5083, a highly specific p97 ATP-competitive inhibitor (Zhou et al., 2015) (Figure 7A). p97 inhibition by itself did not cause LGALS8 or p62 puncta formation in CB-5083-treated cells (Figures 7B, S7A, and S7E). Notably, CB-5083 treatment caused mutant neurons to accelerate the clearance of damaged lysosomes in a manner that was comparable to wild-type neurons (Figures 7B, 7C, and S7A). Indeed, 24 h following LLOME treatment, mutant neurons were indistinguishable from wild-type (Figure 7C). LGALS8 puncta in mutant motor neurons were larger than those in wild-type during CB-5083 treatment, similar to the pattern seen in non-CB-5083-treated neurons (Figure S7B). Previous studies have shown that p97 inhibition can stall lysophagy, which we did not observe at the low 1-μM dose. We asked whether higher doses of CB-5083 were also able to rescue lysophagy defects; however, at 2-μM and 4-μM CB-5083, we found that CB-5083 was unable to rescue the phenotype and LGALS8 puncta persisted in both wild-type and mutant motor neurons up to 24 h (Figure S7C). Doses higher than 4 μM caused significant cell death (data not shown).
Figure 7.
p97 inhibition rescues persistence of damaged lysosomes
(A) Schematic of experimental design.
(B) Representative images LGALS8 puncta in 392.1 motor neurons in untreated, LLOME treated, or after release. All conditions contain CB-5083. Scale bar, 10 μm.
(C) Quantification of LGALS8 puncta in 392.1 and KOLF2.1 motor neurons 24 h after LLOME treatment with and without CB-5083 co-treatment. N = 3 independent experiments.
(D) Representative images of LysoSensor DND-189 live-cell imaging in untreated and CB-5083-treated 392.1 motor neurons.
(E) Quantification of DND-189 fluorescence in untreated (solid bars) and CB-5083 treated (shaded bars) motor neurons. N = 4 independent experiments. Scale bar, 10 μm.
(F) Quantification of 392.1 motor neuron viability before, during, and after LLOME and CB-5083 co-treatment. N = 4 independent experiments. All data expressed as means ± SEM unless otherwise indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Two-way ANOVA with Sidak’s (C, D) or Dunnett’s (F) multiple comparison test. See also Figure S7.
While wild-type neurons treated with CB-5083 were able to clear LGALS8 puncta at a similar rate to untreated neurons, they showed persistent and elevated levels of p62 puncta 24 h after LLOME (Figures S7D–S7F). Indeed, increased p62 puncta were observed upon LLOME-CB-5053 co-treatment compared with LLOME alone (Figure S7F). This may be a consequence of general perturbations in autophagy due to p97 inhibition. We next asked if p97 inhibition could rescue lysosomal pH in mutant motor neurons. CB-5083 treatment restored basal lysosomal pH in mutant motor neurons to that of wild-type neurons (Figures 7D, 7E, and S7G). Furthermore, CB-5083 treatment prevented cell death in p97 mutant motor neurons during release from LLOME (Figures 7F and S7H). Thus, we conclude that p97 inhibition partially rescues defective lysophagy.
Discussion
p97 mutations cause MSP-1 in humans, which is a heterogeneous disease involving bone, muscle, spinal cord, and brain. Disease onset and symptom progression is highly variable even between members of the same family, possibly due to unknown risk alleles that modulate disease pathogenesis (Al-Obeidi et al., 2018). Due to this heterogeneity, inferring pathways altered by p97 mutations has been challenging. Previous iPSC-derived motor neuron models of p97 disease have been discordant, with some indicating ER stress and mitochondrial dysfunction (Hall et al., 2017) while others attribute disease to aberrant cell-cycle protein expression (Wang et al., 2022). Similar to previous studies, we found that p97 R155H altered different pathways in different genetic backgrounds. While the KOLF2.1 cell line with p97 R155H displayed mitochondrial deficits (Figures S4B and S4D), the 392.1 patient cell line had no significant alterations to the mitochondrial proteome (Figure S4C).
Unbiased clustering of proteins via WGCNA found that proteins relating to autophagy and endocytosis were altered (Figure 3H). Autophagy has been intensely investigated in ALS as multiple ALS-linked mutations are found in autophagy-related proteins (Chua et al., 2022). One of these proteins, p62 (SQSTM1), causes multisystem proteinopathy 2 (MSP-2), suggesting that autophagy defects are a common feature in MSPs. Mutations in p97 and inhibition of p97 disrupt autophagy in cells (Gonzalez et al., 2014; Hill et al., 2021) and animals (Ching and Weihl, 2013; Ju et al., 2009); however, only recently has this disruption been linked to defects in lysophagy (Ferrari et al., 2022; Papadopoulos et al., 2017). Multiple components of lysosome repair machinery were common between genetic backgrounds, including annexins (ANXA1, 2, and 11), which were depleted in mutant motor neurons (Figure 3E). Annexins are calcium-dependent phospholipid binding proteins whose depletion delayed lysosome repair independent of ESCRT machinery (Rescher and Gerke, 2004; Yim et al., 2022). Mutations in ANXA11 cause ALS (Smith et al., 2017) and a yet unnamed subtype of MSP (Leoni et al., 2021). ANXA11 contains a low complexity domain that associates with RNA and tethers RNA granules to lysosomes for transport, which is thought to be disrupted by disease-linked mutations (Liao et al., 2019). While ANXA2 does not have a low complexity domain, it is known to bind mRNA (Filipenko et al., 2004) but whether it plays a similar role to ANXA11 remains to be determined.
Here, we show that motor neurons specifically recruit LGALS8 to damaged lysosomes after LLOME treatment while other galectins including LGALS3 remain diffuse (Figure 4B). LGALS8 has a unique role in regulating mTOR inactivation and has been suggested to promote lysophagy over membrane repair compared with other galectins (Jia et al., 2018). Our finding that motor neurons preferentially recruit LGALS8 may indicate that lysophagy dominates membrane repair when faced with lysosomal damage; however, further study of lysosome repair and turnover in motor neurons is required. Galectin accumulation has been noted in skeletal muscle tissue from patients with p97 mutations (Papadopoulos et al., 2017) and spinal cord tissue from patients with sporadic ALS (Kato et al., 2001). Indeed, changes in galectin levels in cerebrospinal fluid and blood have been proposed as potential biomarkers for ALS disease progression (Ashraf and Baeesa, 2018; Zhou et al., 2010).
The cellular response to lysosome damage is increasingly recognized as an important homeostatic pathway that, when disrupted, may lead to disease (Yang and Tan, 2023; Zoncu and Perera, 2022). Proper lysosome turnover requires a multi-step process that includes p97-mediated extraction of ubiquitylated substrates (Kravić et al., 2022). We found that in motor neurons, mutant p97 prevents timely clearance of damaged LGALS8-positive lysosomes (Figure 5). While mutant p97 was recruited to ubiquitylated lysosomes, it did not increase in association with UBXD1 and presumably the ELDR complex (Figures 6 and S6). Without proper ELDR complex assembly, damaged lysosomes are unable to be turned over, leading to persistent damaged lysosomes (Papadopoulos et al., 2017).
In vitro studies of p97 mutations have shown increased ATPase activity leading to the hypothesis that these mutations are gain-of-function (Manno et al., 2010). In vivo studies have challenged this notion, as knocking out neuronal p97 in mice recapitulates MSP-1 phenotypes (Wani et al., 2021). p97 inhibitors have been used to attempt to rescue mutant p97-mediated dysfunction with success (Harley et al., 2021; Wang et al., 2022; Zhang et al., 2017). We found that treatment with low doses of CB-5083 allowed mutant p97 motor neurons to clear LGALS8 puncta at a similar rate as wild-type motor neurons (Figure 7). Interestingly, previous studies have found that p97 inhibition has both decreased LGALS3 puncta clearance (Magnaghi et al., 2013; Papadopoulos et al., 2017) and increased autophagic flux (Anderson et al., 2015). We found no lysophagy defect in CB-5083-treated wild-type motor neurons (Figures 7B and 7C). However, at higher doses lysophagy was equally inhibited in both wild-type and mutant motor neurons. Neurons may be able to better tolerate p97 inhibition than dividing cells such as HeLa or U2OS, which have been primarily used in previous studies. In wild-type and mutant neurons, p97 inhibition increased p62 accumulation (Figure S7). The divergence of LGALS8 and p62 puncta here suggests there are additional processes at play that require autophagic clearance. p62 is utilized for ubiquitin-mediated autophagy, which includes stress granule clearance and mitophagy: two processes that require p97 (Buchan et al., 2013; Tanaka et al., 2010; Zaffagnini and Martens, 2016). The additional rescue of lysosomal pH and viability suggests that p97 inhibition may afford therapeutic benefit (Figure 7 and S7).
While we focus here on the R155H mutation, as 90% of individuals with MSP-1 have this mutation, it is likely that other similar mutations in the N-D1 linker region that perturb UBXD1 interaction also impact lysophagy. Further studies with distinct mutations are required to validate this. Our findings add to the increasing evidence that lysosomal homeostasis, particularly lysophagy, are critical components to neuronal health and disruptions in this process lead to disease.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Malavika Raman (Malavika.raman@tufts.edu).
Material availability
KOLF2.1 cell lines are available through iNDI. Please contact Conrad Weihl for 391.1 patient iPSCs. Please contact the corresponding author for all other reagent requests.
Data and code availability
Proteomic data are available via ProteomeXchange with identifier PXD048185. Any other data are available from the corresponding author upon request.
Induced pluripotent stem cell culture
The KOLF2.1 iPSCs and mutants were acquired from Jackson Laboratories as a part of the iNDI project (Pantazis et al., 2022); 392.1 iPSCs and the CRISPR-corrected line were received from the Weihl laboratory at WUSTL. The patient is a 48-year-old man with a family history of dementia, weakness, and Paget’s disease of the bone. He has history of Paget’s disease of the bone beginning 1 year prior and presents with subacute onset of slowly progressive right lower extremity and left upper extremity weakness. His EMG/NCS demonstrated active denervation in three body regions including his thoracic paraspinous muscles consistent with a diagnosis of ALS. He developed difficulty with breathing at the age of 52 and currently uses non-invasive ventilation at night. He has no symptoms of dementia currently. He has a heterozygous R155H mutation in p97. iPSCs were maintained in mTeSR (05825 STEMCELL) plus media on Matrigel (354277 Corning)-coated plates for no more than 10 passages at 37°C in humidified incubators. iPSCs were passaged using ReLeSR (05873 STEMCELL) once they reached ∼80% confluency; 10 μM Rho kinase inhibitor Y-27632 (ROCK inhibitor) was added for 24 h after each split. Cell morphology was routinely monitored for abnormalities. Karyotyping for each cell line was performed by the WiCell Research Institute.
Neural ectoderm induction and NPC maintenance
We used a modified version of previously published motor neuron protocols (Du et al., 2015; Qu et al., 2014; Shimojo et al., 2015). Once iPSCs reached 80% confluency, the media was replaced with freshly made induction media (Table S1). Cells were kept in induction media with daily media changes for 3 days. Induction media was replaced with induction-patterning media for an additional 3 days with daily media changes to induce PAX6+ NPCs. At this stage, NPCs were passaged by incubating with 0.05% trypsin-EDTA (25300054 Thermo) for 4–8 min at 37°C. NPCs were collected using DMEM (SH30243.01 Cytiva) with 10% FBS (26140079 Gibco) and 1% P/S (PSL01 Caisson) to inactivate trypsin and centrifuged at 300 × g for 5 min. The media was aspirated, and cells were resuspended in fresh expansion media supplemented with 10 μM ROCK inhibitor and plated at 1:4 or 1.5 × 105 cells/cm2 on Matrigel-coated plates. After 24 h, the media was replaced with fresh expansion media without ROCK inhibitor. NPCs were banked at this stage in complete expansion media with 10% DMSO in liquid nitrogen storage. NPCs were thawed, centrifuged at 300 × g for 5 min, and plated in expansion media supplemented with 10 μM ROCK inhibitor. Twenty-four hours later the media was replaced with fresh expansion media without ROCK inhibitor. NPCs were passaged for no more than five passages or 14 days. Further passaging increases spontaneous differentiation and cultures become more heterogeneous.
Motor neuron differentiation and maturation
NPCs were plated at 1.5 × 105/cm2 in 12-well plates and allowed to grow for 3 days. After 3 days, expansion media was replaced with patterning media for 3 days with daily media changes. After 3 days, the cells were split as described previously, and resuspended in maturation media supplemented with 10 μM ROCK inhibitor. Cells were plated on triple-coated PLO/laminin/fibronectin plates at 4 × 10 4/cm2 for biochemistry applications or 1 × 10 4/cm2 on acid-washed #1.5 glass coverslips (200121 Azer). Half media change was performed the following day with maturation media supplemented with 10 μM ROCK inhibitor. Half the media was replaced 2 days later with maturation media supplemented with 40 mM BrdU to inhibit proliferating cells. Half media changes were performed every 2–3 days for an additional 19 days. Media composition can be found in the supplementary methods.
Live-cell imaging
Starting on day 9 of differentiation, progenitors were plated on 4-chambered live-cell imaging dishes (D35C4-20-1-N Cellvis) that were triple coated (PLO/laminin/fibronectin). Cells were maintained as previously described. Cells were treated as previously described. Cells were loaded with LysoSensor DND-189 (L7535 Molecular Probes) or Fluo-4 (F14201 Invitrogen) at 1 μM for 30 min or MitoTracker red (M22425 Cell Signaling) at 100 nM for 45 min at 37°C. Cells were then washed once with PBS and the media was replaced with imaging media (complete 3M without phenol red). Cells loaded with Fluo-4 were incubated at room temperature for 20 min before imaging. Samples were imaged immediately in a temperature-controlled chamber attached to a Zeiss LSM800 Airyscan confocal microscope. Images were taken at ×20 (Fluo-4) or ×63 (LysoSensor and MitoTracker) magnification. Image analysis was performed in ImageJ using custom macro scripts.
Statistics and reproducibility
For all experiments, n ≥ 3 or more biological replicates for each condition examined except immunofluorescence characterization experiments in Figures 4, S1, and S2 (n = 2). Fold changes, SEM, SD, and statistical analyses were performed using GraphPad Prism version 9.4.1 for Windows (GraphPad Software). Statistical tests and n values are mentioned in the figure legends.
Acknowledgments
We thank members of the Raman lab for critical reading of the manuscript. This work is supported by NIH grants GM127557 and NS123631 and Research Grant 1070479 from the Muscular Dystrophy Association to M.R. M.A.J. is an IRACDA scholar funded by GM133314. This work was funded in part by NIH grant GM67945 (S.P.G.) and GM132129 (J.A.P.), AA026256, NS105628, NS102937, MH128235, and MH122379 (J.M.) and AG031867 and AR073317 (C.C.W.).
Author contributions
J.K. and M.R. conceived the studies. J.K. performed all studies and data analysis, M.J. performed qRT-PCR and assisted in validation studies. C.P. and J.M. performed electrophysiology and analysis, J.P. and S.P.G. assisted with proteomic studies. C.C.W. generated the patient iPSC line. J.K. and M.R. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 8, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2024.01.002.
Supplemental information
References
- Ahlstedt B.A., Ganji R., Raman M. The functional importance of VCP to maintaining cellular protein homeostasis. Biochem. Soc. Trans. 2022;50:1457–1469. doi: 10.1042/BST20220648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aits S., Kricker J., Liu B., Ellegaard A.-M., Hämälistö S., Tvingsholm S., Corcelle-Termeau E., Høgh S., Farkas T., Holm Jonassen A., et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy. 2015;11:1408–1424. doi: 10.1080/15548627.2015.1063871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Obeidi E., Al-Tahan S., Surampalli A., Goyal N., Wang A.K., Hermann A., Omizo M., Smith C., Mozaffar T., Kimonis V. Genotype-phenotype study in patients with VCP valosin-containing protein mutations associated with multisystem proteinopathy. Clin. Genet. 2018;93:119–125. doi: 10.1111/cge.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.J., Le Moigne R., Djakovic S., Kumar B., Rice J., Wong S., Wang J., Yao B., Valle E., Kiss von Soly S., et al. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell. 2015;28:653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhzaouy K., Papadopoulos C., Schulze N., Pittman S.K., Meyer H., Weihl C.C. VCP maintains lysosomal homeostasis and TFEB activity in differentiated skeletal muscle. Autophagy. 2019;15:1082–1099. doi: 10.1080/15548627.2019.1569933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf G.M., Baeesa S.S. Investigation of Gal-3 Expression Pattern in Serum and Cerebrospinal Fluid of Patients Suffering From Neurodegenerative Disorders. Front. Neurosci. 2018;12:430. doi: 10.3389/fnins.2018.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe E.E., Gates S.N., Deshaies R.J., Martin A. Multisystem Proteinopathy Mutations in VCP/p97 Increase NPLOC4·UFD1L Binding and Substrate Processing. Structure. 2019;27:1820–1829.e4. doi: 10.1016/j.str.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boom J., Meyer H. VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol. Cell. 2018;69:182–194. doi: 10.1016/j.molcel.2017.10.028. [DOI] [PubMed] [Google Scholar]
- Boya P., Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- Buchan J.R., Kolaitis R.-M., Taylor J.P., Parker R. Eukaryotic Stress Granules Are Cleared by Autophagy and Cdc48/VCP Function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A., Schindelin H., Hänzelmann P. Control of p97 function by cofactor binding. FEBS Lett. 2015;589:2578–2589. doi: 10.1016/j.febslet.2015.08.028. [DOI] [PubMed] [Google Scholar]
- Chauhan S., Kumar S., Jain A., Ponpuak M., Mudd M.H., Kimura T., Choi S.W., Peters R., Mandell M., Bruun J.-A., et al. TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev. Cell. 2016;39:13–27. doi: 10.1016/j.devcel.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching J.K., Weihl C.C. Rapamycin-induced autophagy aggravates pathology and weakness in a mouse model of VCP-associated myopathy. Autophagy. 2013;9:799–800. doi: 10.4161/auto.23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua J.P., De Calbiac H., Kabashi E., Barmada S.J. Autophagy and ALS: mechanistic insights and therapeutic implications. Autophagy. 2022;18:254–282. doi: 10.1080/15548627.2021.1926656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney I., Han H., Stewart M.G., Carson R.H., Hansen D.T., Iwasa J.H., Price J.C., Hill C.P., Shen P.S. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science. 2019;365:502–505. doi: 10.1126/science.aax0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dec E., Ferguson D., Nalbandian A., Gargus M., Katheria V., Rana P., Ibrahim A., Hatch M., Lan M., Llewellyn K.J., et al. Disease-specific Induced Pluripotent Stem Cell Modeling: Insights into the Pathophysiology of Valosin Containing Protein (VCP) Disease. J. Stem Cell Res. Ther. 2014;4:1000168. [Google Scholar]
- DeLaBarre B., Brunger A.T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- Du Z.-W., Chen H., Liu H., Lu J., Qian K., Huang C.-L., Zhong X., Fan F., Zhang S.-C. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 2015;6:6626. doi: 10.1038/ncomms7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Rietz H., Hedlund H., Wilhelmson S., Nordenfelt P., Wittrup A. Imaging small molecule-induced endosomal escape of siRNA. Nat. Commun. 2020;11:1809. doi: 10.1038/s41467-020-15300-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen V.V., Swarup S., Hoyer M.J., Paulo J.A., Harper J.W. Quantitative proteomics reveals the selectivity of ubiquitin-binding autophagy receptors in the turnover of damaged lysosomes by lysophagy. Elife. 2021;10:e72328. doi: 10.7554/eLife.72328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari V., Cristofani R., Cicardi M.E., Tedesco B., Crippa V., Chierichetti M., Casarotto E., Cozzi M., Mina F., Galbiati M., et al. Pathogenic variants of Valosin-containing protein induce lysosomal damage and transcriptional activation of autophagy regulators in neuronal cells. Neuropathol. Appl. Neurobiol. 2022;48:e12818. doi: 10.1111/nan.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipenko N.R., MacLeod T.J., Yoon C.-S., Waisman D.M. Annexin A2 Is a Novel RNA-binding Protein. J. Biol. Chem. 2004;279:8723–8731. doi: 10.1074/jbc.M311951200. [DOI] [PubMed] [Google Scholar]
- Franz A., Ackermann L., Hoppe T. Ring of Change: CDC48/p97 Drives Protein Dynamics at Chromatin. Front. Genet. 2016;7:73. doi: 10.3389/fgene.2016.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Morita E., Itoh T., Tanaka A., Nakaoka M., Osada Y., Umemoto T., Saitoh T., Nakatogawa H., Kobayashi S., et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A.E., Wang X. Drosophila VCP/p97 Mediates Dynein-Dependent Retrograde Mitochondrial Motility in Axons. Front. Cell Dev. Biol. 2020;8:256. doi: 10.3389/fcell.2020.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji R., Paulo J.A., Xi Y., Kline I., Zhu J., Clemen C.S., Weihl C.C., Purdy J.G., Gygi S.P., Raman M. The P97-UBXD8 Complex Modulates ER-Mitochondria Contact Sites by Modulating Membrane Lipid Saturation and Composition. Nature Communications. 2023;14:638. doi: 10.1038/s41467-023-36298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M.A., Feely S.M., Speziani F., Strickland A.V., Danzi M., Bacon C., Lee Y., Chou T.-F., Blanton S.H., Weihl C.C., et al. A novel mutation in VCP causes Charcot–Marie–Tooth Type 2 disease. Brain. 2014;137:2897–2902. doi: 10.1093/brain/awu224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C.E., Yao Z., Choi M., Tyzack G.E., Serio A., Luisier R., Harley J., Preza E., Arber C., Crisp S.J., et al. Progressive Motor Neuron Pathology and the Role of Astrocytes in a Human Stem Cell Model of VCP-Related ALS. Cell Rep. 2017;19:1739–1749. doi: 10.1016/j.celrep.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley J., Hagemann C., Serio A., Patani R. TDP-43 and FUS mislocalization in VCP mutant motor neurons is reversed by pharmacological inhibition of the VCP D2 ATPase domain. Brain Commun. 2021;3:fcab166. doi: 10.1093/braincomms/fcab166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S.M., Wrobel L., Ashkenazi A., Fernandez-Estevez M., Tan K., Bürli R.W., Rubinsztein D.C. VCP/p97 regulates Beclin-1-dependent autophagy initiation. Nat. Chem. Biol. 2021;17:448–455. doi: 10.1038/s41589-020-00726-x. [DOI] [PubMed] [Google Scholar]
- Jia J., Abudu Y.P., Claude-Taupin A., Gu Y., Kumar S., Choi S.W., Peters R., Mudd M.H., Allers L., Salemi M., et al. Galectins Control mTOR in Response to Endomembrane Damage. Mol. Cell. 2018;70:120–135.e8. doi: 10.1016/j.molcel.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J., Claude-Taupin A., Gu Y., Choi S.W., Peters R., Bissa B., Mudd M.H., Allers L., Pallikkuth S., Lidke K.A., et al. Galectin-3 Coordinates a Cellular System for Lysosomal Repair and Removal. Dev. Cell. 2020;52:69–87.e8. doi: 10.1016/j.devcel.2019.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J., et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju J.-S., Fuentealba R.A., Miller S.E., Jackson E., Piwnica-Worms D., Baloh R.H., Weihl C.C. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J. Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Kurita K., Seino T., Kadoya T., Horie H., Wada M., Kawanami T., Daimon M., Hirano A. Galectin-1 Is a Component of Neurofilamentous Lesions in Sporadic and Familial Amyotrophic Lateral Sclerosis. Biochem. Biophys. Res. Commun. 2001;282:166–172. doi: 10.1006/bbrc.2001.4556. [DOI] [PubMed] [Google Scholar]
- Kim N.C., Tresse E., Kolaitis R.-M., Molliex A., Thomas R.E., Alami N.H., Wang B., Joshi A., Smith R.B., Ritson G.P., et al. VCP Is Essential for Mitochondrial Quality Control by PINK1/Parkin and this Function Is Impaired by VCP Mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenova O.V., Mukkavalli S., Raman M., Walter J.C. Cooperative assembly of p97 complexes involved in replication termination. Nat. Commun. 2022;13:6591. doi: 10.1038/s41467-022-34210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravić B., Bionda T., Siebert A., Gahlot P., Levantovsky S., Behrends C., Meyer H. Ubiquitin profiling of lysophagy identifies actin stabilizer CNN2 as a target of VCP/p97 and uncovers a link to HSPB1. Mol. Cell. 2022;82:2633–2649.e7. doi: 10.1016/j.molcel.2022.06.012. [DOI] [PubMed] [Google Scholar]
- Krick R., Bremer S., Welter E., Schlotterhose P., Muehe Y., Eskelinen E.-L., Thumm M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J. Cell Biol. 2010;190:965–973. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni T.B., González-Salazar C., Rezende T.J.R., Hernández A.L.C., Mattos A.H.B., Coimbra Neto A.R., da Graça F.F., Gonçalves J.P.N., Martinez A.R.M., Taniguti L., et al. A Novel Multisystem Proteinopathy Caused by a Missense ANXA11 Variant. Ann. Neurol. 2021;90:239–252. doi: 10.1002/ana.26136. [DOI] [PubMed] [Google Scholar]
- Liao Y.-C., Fernandopulle M.S., Wang G., Choi H., Hao L., Drerup C.M., Patel R., Qamar S., Nixon-Abell J., Shen Y., et al. RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Cell. 2019;179:147–164.e20. doi: 10.1016/j.cell.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi P., D’Alessio R., Valsasina B., Avanzi N., Rizzi S., Asa D., Gasparri F., Cozzi L., Cucchi U., Orrenius C., et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- Manno A., Noguchi M., Fukushi J., Motohashi Y., Kakizuka A. Enhanced ATPase activities as a primary defect of mutant valosin-containing proteins that cause inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Gene Cell. 2010;15:911–922. doi: 10.1111/j.1365-2443.2010.01428.x. [DOI] [PubMed] [Google Scholar]
- McAlister G.C., Nusinow D.P., Jedrychowski M.P., Wühr M., Huttlin E.L., Erickson B.K., Rad R., Haas W., Gygi S.P. MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Anal. Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbandian A., Llewellyn K.J., Kitazawa M., Yin H.Z., Badadani M., Khanlou N., Edwards R., Nguyen C., Mukherjee J., Mozaffar T., et al. The Homozygote VCPR155H/R155H Mouse Model Exhibits Accelerated Human VCP-Associated Disease Pathology. PLoS One. 2012;7:e46308. doi: 10.1371/journal.pone.0046308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Mackenzie I.R., Cairns N.J., Boyer P.J., Markesbery W.R., Smith C.D., Taylor J.P., Kretzschmar H.A., Kimonis V.E., Forman M.S. TDP-43 in the Ubiquitin Pathology of Frontotemporal Dementia With VCP Gene Mutations. J. Neuropathol. Exp. Neurol. 2007;66:152–157. doi: 10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- Pantazis C.B., Yang A., Lara E., McDonough J.A., Blauwendraat C., Peng L., Oguro H., Kanaujiya J., Zou J., Sebesta D., et al. A reference human induced pluripotent stem cell line for large-scale collaborative studies. Cell Stem Cell. 2022;29:1685–1702.e22. doi: 10.1016/j.stem.2022.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos C., Kirchner P., Bug M., Grum D., Koerver L., Schulze N., Poehler R., Dressler A., Fengler S., Arhzaouy K., et al. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2017;36:135–150. doi: 10.15252/embj.201695148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer G., Lee G., Pontifex C.S., Fanganiello R.D., Peck A., Weihl C.C., Kimonis V. Multisystem Proteinopathy Due to VCP Mutations: A Review of Clinical Heterogeneity and Genetic Diagnosis. Genes. 2022;13:963. doi: 10.3390/genes13060963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q., Li D., Louis K.R., Li X., Yang H., Sun Q., Crandall S.R., Tsang S., Zhou J., Cox C.L., et al. High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat. Commun. 2014;5:3449. doi: 10.1038/ncomms4449. [DOI] [PubMed] [Google Scholar]
- Ramanathan H.N., Ye Y. The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res. 2012;22:346–359. doi: 10.1038/cr.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos D.M., Skarnes W.C., Singleton A.B., Cookson M.R., Ward M.E. Tackling neurodegenerative diseases with genomic engineering: A new stem cell initiative from the NIH. Neuron. 2021;109:1080–1083. doi: 10.1016/j.neuron.2021.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescher U., Gerke V. Annexins – unique membrane binding proteins with diverse functions. J. Cell Sci. 2004;117:2631–2639. doi: 10.1242/jcs.01245. [DOI] [PubMed] [Google Scholar]
- Ritz D., Vuk M., Kirchner P., Bug M., Schütz S., Hayer A., Bremer S., Lusk C., Baloh R.H., Lee H., et al. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ D.E., Cross R.B.P., Li L., Koch S.C., Matson K.J.E., Yadav A., Alkaslasi M.R., Lee D.I., Le Pichon C.E., Menon V., Levine A.J. A harmonized atlas of mouse spinal cord cell types and their spatial organization. Nat. Commun. 2021;12:5722. doi: 10.1038/s41467-021-25125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo D., Onodera K., Doi-Torii Y., Ishihara Y., Hattori C., Miwa Y., Tanaka S., Okada R., Ohyama M., Shoji M., et al. Rapid, efficient, and simple motor neuron differentiation from human pluripotent stem cells. Mol. Brain. 2015;8:79. doi: 10.1186/s13041-015-0172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B.N., Topp S.D., Fallini C., Shibata H., Chen H.-J., Troakes C., King A., Ticozzi N., Kenna K.P., Soragia-Gkazi A., et al. Mutations in the vesicular trafficking protein annexin A11 are associated with amyotrophic lateral sclerosis. Sci. Transl. Med. 2017;9:eaad9157. doi: 10.1126/scitranslmed.aad9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.-F., Karbowski M., Youle R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A.K.A., Hamon C. Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS. Anal. Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Ting L., Rad R., Gygi S.P., Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A.L., Župunski V., et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E.C., Ji Z., Wales T.E., Bodnar N.O., Ficarro S.B., Marto J.A., Engen J.R., Rapoport T.A. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science. 2019;365:eaax1033. doi: 10.1126/science.aax1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Li S., Wang T.Y., Lopez G.A., Antoshechkin I., Chou T.F. P97/VCP ATPase inhibitors can rescue p97 mutation-linked motor neuron degeneration. Brain Commun. 2022;4:fcac176. doi: 10.1093/braincomms/fcac176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani A., Zhu J., Ulrich J.D., Eteleeb A., Sauerbeck A.D., Reitz S.J., Arhzaouy K., Ikenaga C., Yuede C.M., Pittman S.K., et al. Neuronal VCP loss of function recapitulates FTLD-TDP pathology. Cell Rep. 2021;36:109399. doi: 10.1016/j.celrep.2021.109399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts G.D.J., Wymer J., Kovach M.J., Mehta S.G., Mumm S., Darvish D., Pestronk A., Whyte M.P., Kimonis V.E. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Wu J.X., Pascovici D., Wu Y., Walker A.K., Mirzaei M. Workflow for Rapidly Extracting Biological Insights from Complex, Multicondition Proteomics Experiments with WGCNA and PloGO2. J. Proteome Res. 2020;19:2898–2906. doi: 10.1021/acs.jproteome.0c00198. [DOI] [PubMed] [Google Scholar]
- Yang H., Tan J.X. Lysosomal quality control: molecular mechanisms and therapeutic implications. Trends Cell Biol. 2023;33:749–764. doi: 10.1016/j.tcb.2023.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Tang W.K., Zhang T., Xia D. A Mighty “Protein Extractor” of the Cell: Structure and Function of the p97/CDC48 ATPase. Front. Mol. Biosci. 2017;4:39. doi: 10.3389/fmolb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim W.W.-Y., Yamamoto H., Mizushima N. Annexins A1 and A2 are recruited to larger lysosomal injuries independently of ESCRTs to promote repair. FEBS Lett. 2022;596:991–1003. doi: 10.1002/1873-3468.14329. [DOI] [PubMed] [Google Scholar]
- Zaffagnini G., Martens S. Mechanisms of Selective Autophagy. J. Mol. Biol. 2016;428:1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Mishra P., Hay B.A., Chan D., Guo M. Valosin-containing protein (VCP/p97) inhibitors relieve Mitofusin-dependent mitochondrial defects due to VCP disease mutants. Elife. 2017;6:e17834. doi: 10.7554/eLife.17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-J., Wang J., Yao B., Wong S., Djakovic S., Kumar B., Rice J., Valle E., Soriano F., Menon M.-K., et al. Discovery of a First-in-Class, Potent, Selective, and Orally Bioavailable Inhibitor of the p97 AAA ATPase (CB-5083) J. Med. Chem. 2015;58:9480–9497. doi: 10.1021/acs.jmedchem.5b01346. [DOI] [PubMed] [Google Scholar]
- Zhou J.-Y., Afjehi-Sadat L., Asress S., Duong D.M., Cudkowicz M., Glass J.D., Peng J. Galectin-3 is a candidate biomarker for ALS: Discovery by a proteomics approach. J. Proteome Res. 2010;9:5133–5141. doi: 10.1021/pr100409r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Perera R.M. Built to last: lysosome remodeling and repair in health and disease. Trends Cell Biol. 2022;32:597–610. doi: 10.1016/j.tcb.2021.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic data are available via ProteomeXchange with identifier PXD048185. Any other data are available from the corresponding author upon request.