Abstract
Background
HER2DX, a multianalyte genomic test, has been clinically validated to predict breast cancer recurrence risk (relapse risk score), the probability of achieving pathological complete response post-neoadjuvant therapy (pCR likelihood score), and individual ERBB2 messenger RNA (mRNA) expression levels in patients with early-stage human epidermal growth factor receptor 2 (HER2)-positive breast cancer. This study delves into the comprehensive analysis of HER2DX’s analytical performance.
Materials and methods
Precision and reproducibility of HER2DX risk, pCR, and ERBB2 mRNA scores were assessed within and between laboratories using formalin-fixed paraffin-embedded (FFPE) tumor tissues and purified RNA. Robustness was appraised by analyzing the impact of tumor cell content and protocol variations including different instruments, reagent lots, and different RNA extraction kits. Variability was evaluated across intratumor biopsies and genomic platforms [RNA sequencing (RNAseq) versus nCounter], and according to protocol variations.
Results
Precision analysis of 10 FFPE tumor samples yielded a maximal standard error of 0.94 across HER2DX scores (1-99 scale). High reproducibility of HER2DX scores across 29 FFPE tumors and 20 RNAs between laboratories was evident (correlation coefficients >0.98). The probability of identifying score differences >5 units was ≤5.2%. No significant variability emerged based on platform instruments, reagent lots, RNA extraction kits, or TagSet thaw/freeze cycles. Moreover, HER2DX displayed robustness at low tumor cell content (10%). Intratumor variability across 212 biopsies (106 tumors) was <4.0%. Concordance between HER2DX scores from 30 RNAs on RNAseq and nCounter platforms exceeded 90.0% (Cohen’s κ coefficients >0.80).
Conclusions
The HER2DX assay is highly reproducible and robust for the quantification of recurrence risk, pCR likelihood, and ERBB2 mRNA expression in early-stage HER2-positive breast cancer.
Key words: HER2DX, breast cancer, gene expression, RNA, analytical validation
Highlights
-
•
HER2DX assay is highly reproducible and robust intra- and inter-laboratories.
-
•
HER2DX proved robust performance for different protocol variations including different platforms.
-
•
HER2DX has proven technical reliability in predicting outcomes for early-stage HER2-positive breast cancer.
Introduction
Women diagnosed with early-stage human epidermal growth factor receptor 2 (HER2)-positive breast cancer confront an array of therapeutic options.1 Yet, conventional diagnostic approaches rooted in anatomical–pathological tumor evaluation yield limited insights into prognosticating treatment responses.2 It is evident that some individuals receive inadequate treatment, while others endure unnecessary interventions. The emergence of molecular genomic techniques, including the molecular characterization of tumor tissue,3,4 is revolutionizing the landscape of clinical oncology diagnostics, ushering in improved patient outcomes through a rationale-driven approach to tailoring oncological interventions.
Embarking on this trajectory, the HER2DX genomic assay emerged in 2022, driven by the combination of 27 genes underlying 4 biological signatures, and clinical attributes.5, 6, 7, 8, 9, 10, 11 The test delivers two scores trained to predict both long-term prognosis (i.e. relapse risk score) and the likelihood of pathological complete response (i.e. pCR likelihood score) in early HER2-positive breast cancer following trastuzumab-based therapy. In addition, it also provides a third score showing the individual ERBB2 messenger RNA (mRNA) levels. By merging biological insights with clinical factors, including tumor size and nodal status, HER2DX enhances treatment decision making. Recent validations affirm its clinical utility across diverse contexts,5,8,9,11 including trials like APT and ATEMPT, identifying patients with high-risk disease within clinically low-risk scenarios.9,11 The HER2DX pCR likelihood score was validated in multiple clinical studies demonstrating its ability to inform neoadjuvant therapy strategies in the context of both European and US standard of care.5, 6, 7, 8,10 Notably, the 2022 SEOM-GEICAM-SOLTI guidelines endorsed its clinical utility in specific cases,12 and the St. Gallen International Consensus Conference for the Primary Therapy of Individuals with Early Breast Cancer 2023 identified HER2DX as a practice-changing finding.13
The pursuit of individualized patient management demands the establishment of rigorous performance criteria for clinical laboratory tests.3,4 Recognizing this crucial need, our investigation focused on an in-depth evaluation of the analytical performance of the HER2DX assay. By scrutinizing its precision, consistency, and reliability, we aimed to assess its suitability for routine diagnostics on patients undergoing HER2-positive breast cancer treatment care.
Materials and methods
In adherence to rigorous quality standards, our analytical validation strategy was designed to meet the stringent criteria outlined in the ISO15189 standard.
Tissue and RNA requirements
The HER2DX assay procedure utilizes formalin-fixed paraffin-embedded (FFPE) tissue samples, each previously diagnosed with viable invasive HER2-positive breast carcinoma. Pathological assessment by a qualified pathologist classifies the tumor as invasive carcinoma, encompassing various histological subtypes (ductal, lobular, mixed, or no special type). Identification and delineation (circling of tumor area) of the viable invasive breast carcinoma region are accomplished through a thorough review of hematoxylin–eosin (H&E)-stained slides, followed by meticulous circling of the designated area. To ensure sufficient material for the HER2DX test, each H&E-stained (4 μm) and consecutive unstained (10 μm) section requires a tumor surface area of ≥4 mm2 and tumor cellularity of ≥10%. A single 10-μm section is used for cases where the tumor surface area measured ≥100 mm2, and three sections for tumor surfaces ranging from 4 to 99 mm2. Subsequent steps involve targeted macrodissection removal of the tissue outside the area delineated in H&E-stained slides. The extraction of RNA from the remaining tumor tissue is scraped off the slide and transferred to an Eppendorf tube. Total RNA extraction is carried out by using the High Pure FFPET RNA Isolation Kit (Roche, Indianapolis, IN, USA) following manufacturer’s instructions. RNA is eluted into a 30-μl volume, accompanied by stringent checks to ensure compliance with specified concentration criteria (≥12.5 ng/μl) and purity standards [optical density (OD) 260/280 nm of 1.7-2.5].
The HER2DX assay
The HER2DX assay encompasses the quantification of mRNA levels of 27 target genes and 5 normalization genes with constitutive expression (GAPD, PUM1, ACTB, RPLP0, and PSMC4).5 These 27 genes constitute four different gene signatures, tracking immune infiltration (i.e. CD27, CD79A, HLA-C, IGJ, IGKC, IGL, IGLV3-25, IL2RG, CXCL8, LAX1, NTN3, PIM2, POU2AF1, and TNFRSF17), tumor cell proliferation (i.e. EXO1, ASPM, NEK2, and KIF23), luminal differentiation (i.e. BCL2, DNAJC12, AGR3, AFF3, and ESR1), and expression of the HER2 amplicon (i.e. ERBB2, GRB7, STARD3, and TCAP).
Isolated RNA is subjected to purity and concentration requirements described previously. If quality thresholds are not met, the pre-analytical steps are repeated on the remaining tumor tissue. The RNA samples undergo a hybridization process (devoid of reverse transcription or amplification). This procedure captures the essence of the measured genes and assay controls through capture and reporter probes. The multiplexed hybridizations unfold within a single tube, over a duration of 15-21 h at 65°C, employing 250 ng RNA. Integral to the assay, positive and negative controls are included to ensure compliance with global quality standards. Additionally, two reference RNA samples are simultaneously tested as part of each HER2DX test procedure.
Post-hybridization, the Food and Drug Administration-510K-cleared nCounter Analysis System (NanoString Technologies, Seattle, WA, USA) processes the target–probe complexes.14 Evaluation of the linearity of positive control target titration and the non-specific background attributed to negative control probes within each assay serves as the yardstick for performance assessment. As the assay is tailored for execution within local molecular pathology laboratories, an embedded software enforces automatic application of quality thresholds to the data collection process. A minimum threshold for the expression of normalized genes must be met in test sample data and the two reference control samples, ensuring gene expression values are of sufficient magnitude for precise algorithmic outcomes. Therefore, quality control metrics for the HER2DX assay have been established.
HER2DX scores
The HER2DX algorithm is an independent software developed and validated by Reveal Genomics. It is executed on the outputs from the nCounter Analysis System (NanoString Technologies, Seattle, WA, USA) to generate three scores (i.e. relapse risk score, pCR likelihood score, and ERBB2 mRNA score). The HER2DX relapse risk score is based on three gene expression-based signatures (immune, tumor cell proliferation, and tumor cell luminal differentiation), together with tumor stage (T1 versus T2 versus T3-4) and nodal stage (N0 versus N1 versus N2-3). The HER2DX pCR likelihood score is based on four gene expression-based signatures (immune, tumor cell proliferation, HER2 amplicon, and tumor cell luminal differentiation), together with tumor stage (T1 versus T2 versus T3-4) and nodal stage (N0 versus N1-3). The HER2DX ERBB2 mRNA score is based on the individual expression of the ERBB2 gene. Each score is scaled from 1 to 99, and pre-specified cut-offs (i.e. low versus high for HERDX relapse risk score, and low, medium, and high for HER2DX pCR likelihood score and ERBB2 mRNA score) were established.5
Precision evaluation within one laboratory
Ten different FFPE tumor samples from patients with newly diagnosed and untreated HER2-positive breast cancer (Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2024.102903) were used to determine the reproducibility of the HER2DX scores within the central laboratory (Genomics Core BM Lab at Hospital Clinic of Barcelona, herein referred to as central lab) where HER2DX is currently carried out for routine diagnostics. They were selected to represent the whole percentile spectrum for the relapse risk score percentile (two cases with percentiles between 10 and 20; two cases between 80 and 90; six cases between 32 and 77). RNA was purified twice and run in triplicate to determine gene expression and the HER2DX scores.
Precision evaluation across different laboratories
Thirty FFPE tumor samples and 20 RNAs from newly diagnosed HER2-positive breast cancer before any therapy were selected to evaluate the reproducibility between the central laboratory and the development laboratory (Translational Genomics and Targeted Therapies in Solid Tumors Lab, IDIBAPS/Hospital Clinic of Barcelona, herein referred to as development lab). The FFPE tumor tissues and the RNAs were from different patients and were selected to represent the whole spectrum of risk and pCR likelihood scores. Among the 30 FFPE tumor samples selected, 1 did not contain enough tumor tissue for the inter-laboratory comparison. A set of 30 RNAs already analyzed in both laboratories were run at the Genomics Lab at Vall d’Hebron University Hospital (VHIO), and 10 of them plus 20 other FFPE tumor samples at Center for Advanced Molecular Diagnostics (CAMD, Brigham and Women’s Hospital, Boston, MA, USA). Ten of these RNAs were also analyzed at Eremid Genomic Services Lab (Kannapolis, NC, USA). Criteria for selecting samples in these experiments are detailed in the Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2024.102903.
Robustness evaluation
To evaluate robustness, the HER2DX assay was carried out under different protocol modifications. Firstly, to assess the impact of adjacent non-tumor tissue, we carried out a spiking experiment in four FFPE samples by diluting the corresponding tumor RNA with different amounts of RNA extracted from stromal areas of the same FFPE block section. Secondly, the assay was carried out starting from different RNA concentrations (100, 250, and 500 ng) in four different samples. Thirdly, it was run by analyzing the same RNAs on two different nCounter instruments at the same laboratory (central lab). Fourthly, the assay was run with two different TagSet lots, one of which was defrosted twice. Fifthly, RNA from 24 samples was extracted with an additional RNA extraction kit (ReliaPrep™ FFPE Total RNA Miniprep System RNA Isolation Kit, Promega). The acceptance criteria were the same as those for the standard kit, requiring RNA concentrations of ≥12.5 ng/μl and an OD 260/280 nm ratio between 1.7 and 2.5.
Intratumor variability evaluation
HER2DX assay was conducted on extracted RNA at Oregon Health and Science University (Portland, OR, USA) from pretreatment FFPE biopsies from two distinct geographical areas within tumors (i.e. two different core biopsies) of patients enrolled in the 14-409 phase II study (NCT02326974).14 This study assessed the impact of HER2 heterogeneity on treatment response in stage II-III HER2-positive breast cancer patients treated with neoadjuvant trastuzumab emtansine and pertuzumab.
Variability evaluation across genomic platforms
A curated set of 30 RNA samples, which had previously undergone the HER2DX assay using the nCounter, were selected for RNA sequencing (RNAseq). The selection criteria were based on sensuring a diverse spectrum of HER2DX scores (Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2024.102903).
RNA sequencing procedures
For RNAseq analysis (utilizing the Illumina Exome Panel) (Illumina, Inc., San Diego, CA, USA), we used the Illumina RNA Prep protocol coupled with enrichment and Unique Dual Index (UDI) adapters during library preparation. The sequencing process (2 × 150 paired-end) was carried out on an Illumina NovaSeq sequencer, achieving a robust coverage of 0.5-1×. Library preparation and sequencing was carried out at the Cancer Genomics Laboratory at Vall d’Hebron Institute of Oncology (Barcelona, Spain).
Statistical analysis
Intra-run and inter-run reproducibility was measured by means of the standard error (SE) between replicates and the maximal range of difference (points of percentile). Cohen’s κ coefficient was used to assess the concordance between HER2DX groups intra- and inter-laboratories. The Pearson correlation and Bland–Altman plots were used to assess the agreement between measurements obtained at different laboratories. Student’s t-test was used to compare HER2DX scores under protocol variations. The variability of different conditions on HER2DX scores was limited to 10%. A simulation analysis (n = 1 × 106) based on the variability observed across the score distributions in the reproducibility experiments was carried out to model a potential real-world scenario and evaluate the diagnostic performance of the test. The accuracy, sensitivity, specificity, positive predictive value, and negative predictive value were calculated using simulations. The level of significance was set at P < 0.05 (two-sided). The test must demonstrate a sensitivity and specificity of at least 90% as acceptance criteria. All statistical analyses were conducted using R version 4.0.5 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Precision of HER2DX
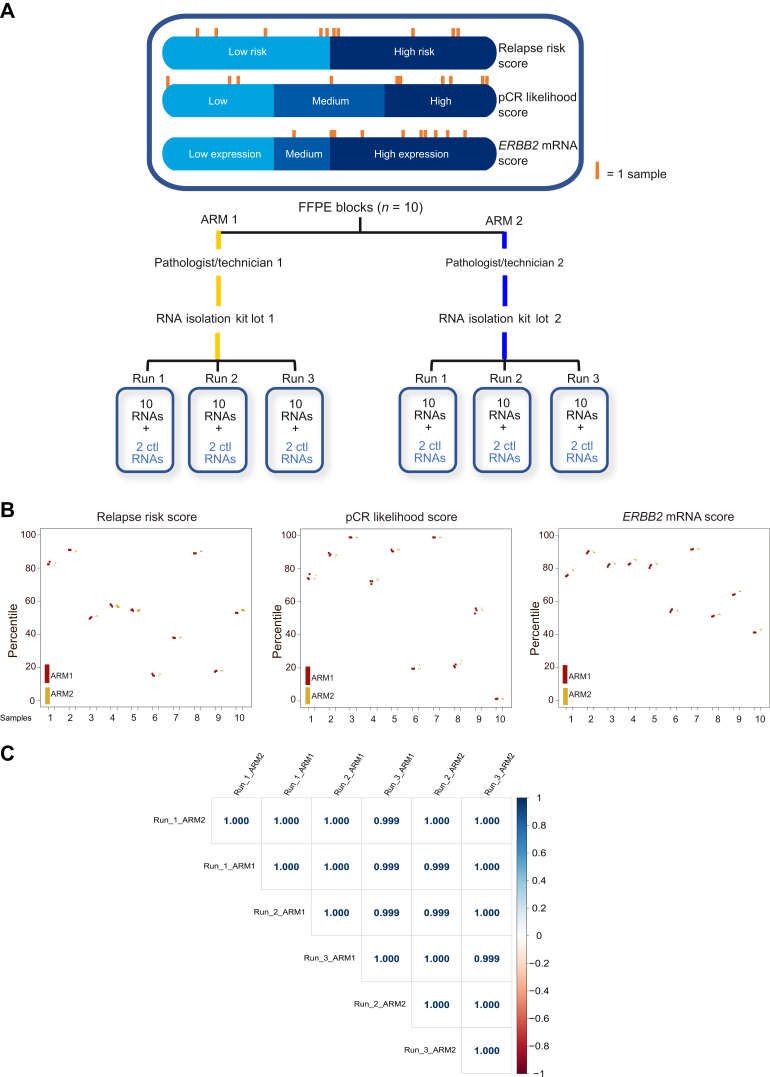
We first evaluated the precision of the HER2DX test by comparing its performance intra-laboratory, by comparing two different pathologist/technician teams and different RNA extraction kit lots (Figure 1A). Overall, the result revealed a maximal SE of 0.94 across the HER2DX scores (scale from 1 to 99) and an overall concordance of 99.4% (Cohen’s κ coefficient = 0.92) (Figure 1B). The gene expression Pearson correlation coefficient between runs was >0.99 (Figure 1C). This experiment demonstrated the consistent and reliable nature of the assay’s measurements within the same sample type and laboratory.
Figure 1.
Precision of HER2DX intra-laboratory. (A) Schematic outline of experimental design; (B) scatter plot revealing both intra- and inter-run variability across the three distinct HER2DX scores (i.e. relapse risk, pCR likelihood, and ERBB2 mRNA); (C) correlation matrix comparing runs from arm 1 and 2 with respect to the normalized gene expression of the 27 genes analyzed in the test. Correlation coefficients are shown.
Ctl, control; mRNA, messenger RNA; pCR, pathological complete response.
Precision and reproducibility of HER2DX scores across laboratories
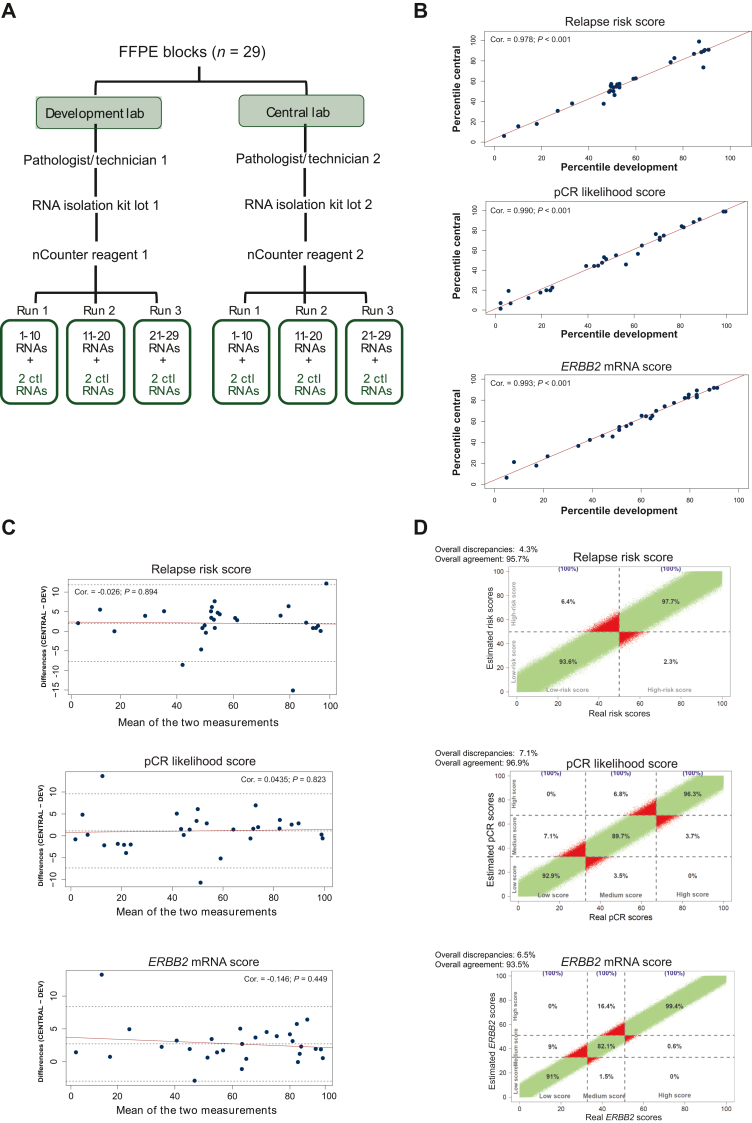
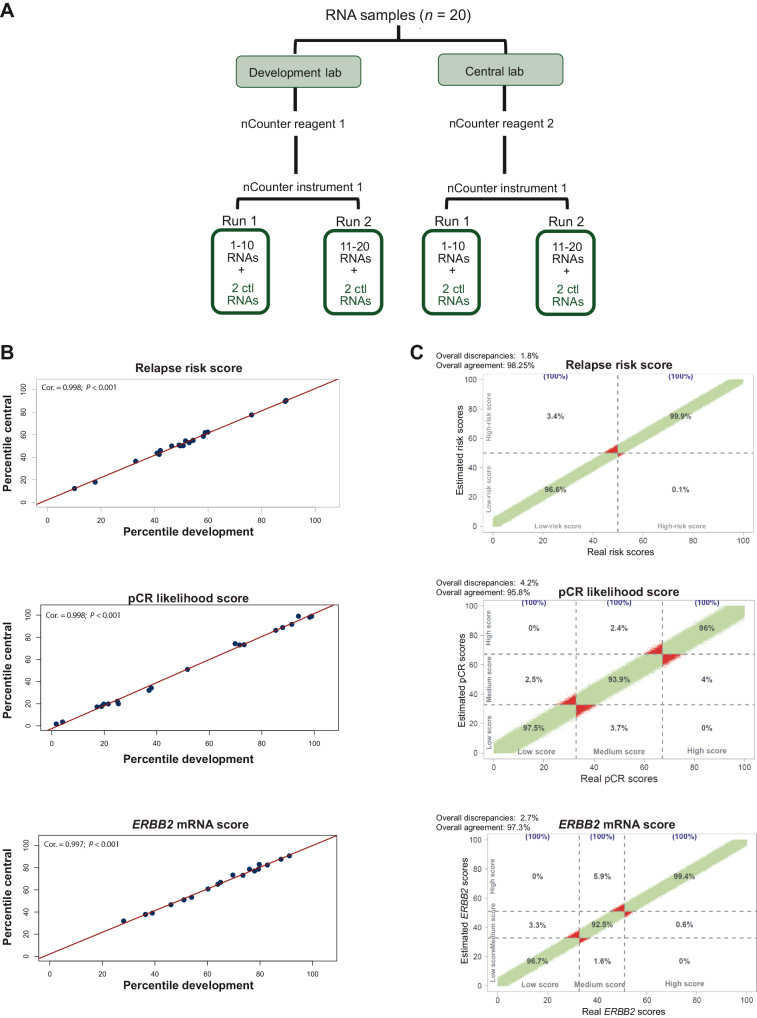
We extended our investigation to compare HER2DX results between two different laboratories, the central lab and the development lab. To accomplish this, 29 distinct FFPE tumor samples and 20 purified RNA samples were analyzed by both laboratories (Figures 2A and 3A). Both experiments (starting from FFPE and RNA) showcased a Pearson correlation coefficient >0.97 in all HER2DX scores (Figures 2B and 3B). A maximal SE of 5 units across the HER2DX scores (scale from 1 to 99) was observed (Figure 2B). The Bland–Altman plots showed no anomalous distributions of differences over the range of values (Figure 2C). The probability of identifying score differences >5 units between different laboratories was ≤5.2%.
Figure 2.
Testing the accuracy and reproducibility of HER2DX between laboratories from FFPE. (A) Overview of the experiment design when initiating from FFPE blocks; (B) correlation plots illustrating the concordance among the three scores (i.e. relapse risk, pCR likelihood, and ERBB2 mRNA) obtained in two distinct laboratories, both starting from FFPE blocks; (C) Bland–Altman plots including Pearson correlation coefficients offering a comprehensive analysis of the discrepancies between the three HER2DX scores obtained in both laboratories starting from FFPE blocks; (D) correlation plot between the three scores in the simulation data with 1 × 106 estimated scores.
Ctl, control; FFPE, formalin-fixed paraffin-embedded; mRNA, messenger RNA; pCR, pathological complete response.
Figure 3.
Testing the accuracy and reproducibility of HER2DX between laboratories from RNA. (A) Overview of the experiment design when initiating from RNAs; (B) correlation plots illustrating the concordance among the three scores (i.e. relapse risk, pCR likelihood, and ERBB2 mRNA) obtained in two distinct laboratories, both starting from FFPE blocks from RNAs; (C) correlation plot between the three scores in the simulation data with 1×106 estimated scores.
Cor., correlation; Ctl, control; FFPE, formalin-fixed paraffin-embedded; mRNA, messenger RNA; pCR, pathological complete response.
Using the variability observed across the three scores’ distribution in the 29 samples, a simulation was carried out to estimate the percentage of concordance in a potential real-world scenario. Overall, discrepancies in the group classification were observed in 4.3%, 7.1%, and 6.5% of the cases for the relapse risk, pCR likelihood, and ERRB2 mRNA scores, respectively (Figure 2D; Supplementary Tables S1-S3, available at https://doi.org/10.1016/j.esmoop.2024.102903). The diagnostic performance of the test based on this simulation is shown in Table 1.
Table 1.
Diagnostic values at reproducibility-simulation analysis
| Diagnostic values | Relapse risk score | pCR likelihood score | ERBB2 mRNA score |
|---|---|---|---|
| Sensitivity (%) | 97.7 | 92.9 | 91.0 |
| Specificity (%) | 93.6 | 98.2 | 99.6 |
| PPV (%) | 93.9 | 96.2 | 99.1 |
| NPV (%) | 97.6 | 96.6 | 95.8 |
| Accuracy (%) | 95.7 | 96.5 | 96.8 |
mRNA, messenger RNA; NPV, negative predictive value; pCR, pathological complete response; PPV, positive predictive value.
We further extended the HER2DX analysis by including an additional laboratory and carrying out a three-laboratory comparison by using 30 RNA samples. When contrasting HER2DX results between the three laboratories, no significant differences were found in any of the scores (analysis of variance tests, P ≥ 0.05) (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2024.102903).
To further test the performance of the HER2DX assay as a decentralized molecular test, a focused study was conducted involving two US-based laboratories (i.e. CAMD and Eremid), using 10 of the 30 original RNA samples. In comparison to the central laboratory, the Pearson correlation was >0.93, the mean difference <|3|, and the standard deviation (SD) <9.5 for all the three scores across both laboratories (Supplementary Figures S2 and S3, available at https://doi.org/10.1016/j.esmoop.2024.102903). These collective results underscore the consistency and minimal variability of the HER2DX assay when carried out in different laboratories.
An additional study was conducted involving re-analysis of 20 FFPE HER2-positive breast tumors across two laboratories (i.e. CAMD and development) (Supplementary Figure S4, available at https://doi.org/10.1016/j.esmoop.2024.102903). The results of the test revealed a correlation coefficient >0.96 in all HER2DX scores. The SDs (percentile units) were 6.3, 7.9, and 5.4 for risk, pCR, and ERBB2 mRNA scores, respectively (Supplementary Figure S4, available at https://doi.org/10.1016/j.esmoop.2024.102903). This additional assessment further substantiates the robustness and reliability of the HER2DX assay across varied settings and conditions.
Robustness in the context of low tumor content or protocol modifications
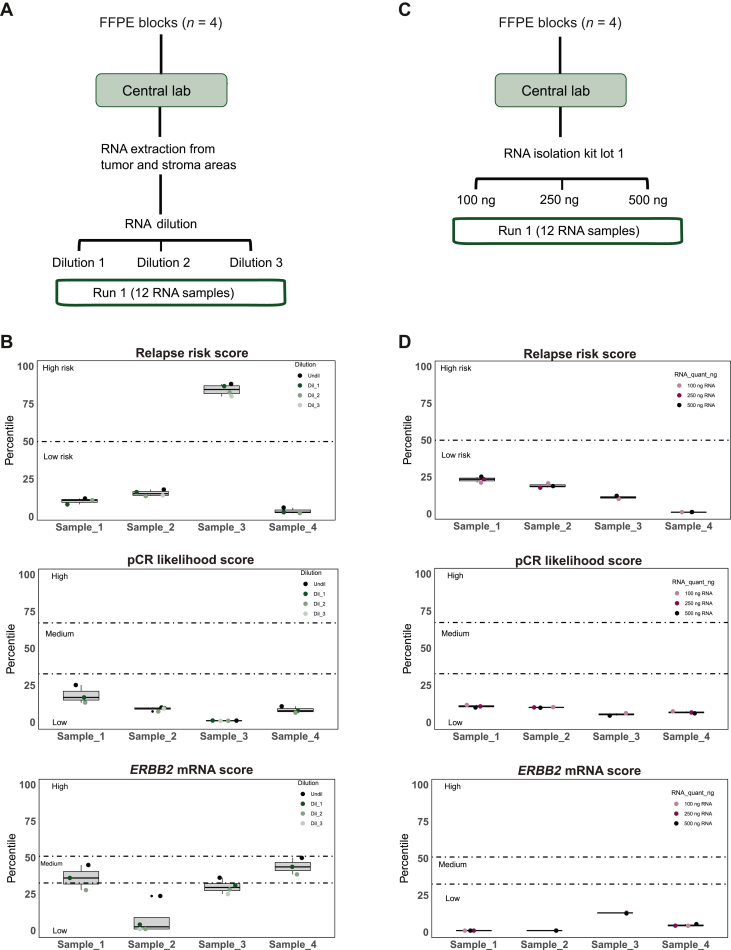
Robustness testing involved assessing potential interference from normal tissue. RNA from four FFPE tumor samples was spiked in non-tumor RNA (from stroma) from each corresponding sample (Figure 4A). Two or three dilutions were carried out summing a total of 14 samples with different % of tumor RNA content, which were run and analyzed (Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2024.102903). A 100% concordance was observed between relapse risk score and pCR likelihood score groups in all dilutions from all samples (Figure 4B). As expected, ERBB2 mRNA score decreased progressively with dilutions. The average difference in percentiles between the undiluted and the maximal diluted samples was 15.6 (range 11.2-22.6). The classifying group changed in two of the four samples in one of the dilutions (Figure 4B). HER2DX exhibited commendable performance even when analyzing samples with low tumor content, as low as 10%. We also evaluated HER2DX results in four samples with lower (100 ng) and higher (500 ng) RNA quantity than the standard (250 ng) (Figure 4C). No significant differences were observed (Figure 4D).
Figure 4.
Robustness of HER2DX: evaluation of non-tumor component and different starting RNA quantities. (A) Overview of the spiking experiment designed; (B) boxplot representing the percentiles for the three HER2DX scores (i.e. relapse risk score, pCR likelihood score, and ERBB2 score) after diluting tumor-derived RNA in stroma-derived RNA; (C) overview of the experiment design to evaluate the effect of starting from different RNA quantities; (D) boxplot representing the percentiles for the three HER2DX scores (relapse risk score, pCR likelihood score, and ERBB2 score) after starting from different quantities of RNA.
Ctl, control; FFPE, formalin-fixed paraffin-embedded; mRNA, messenger RNA; pCR, pathological complete response.
Furthermore, we examined HER2DX in multiple replicates of a single RNA analyzed in two different nCounter instruments within the central lab, with 20 and 24 replicates on instruments 1 and 2, respectively. HER2DX test was also evaluated by processing 10 RNA samples with two different TagSet lots and with two TagSet defrost cycles in 4 samples. No significant differences were observed when any of the protocol modifications was introduced (Supplementary Figure S5, available at https://doi.org/10.1016/j.esmoop.2024.102903). Furthermore, we investigated the influence of using different RNA extraction protocols by analyzing 24 samples extracted by two different kits. All samples exhibited 100% concordance for HER2DX risk and ERBB2 mRNA score groups, regardless of the protocol. The concordance for the pCR likelihood score was 91.7%, and correlation coefficients exceeded 0.99 for all three HER2DX scores (Supplementary Figure S6, available at https://doi.org/10.1016/j.esmoop.2024.102903).
Intratumor variability
We first explored intratumor variability by analyzing 208 different biopsies obtained from 104 unique tumors at the same time. The observed variability remained <4.0%, emphasizing the assay’s ability to yield consistent results even when applied to multiple regions within the same tumor. Concordance rates of the three HER2DX scores (relapse risk, pCR likelihood, and ERBB2 mRNA) between different biopsy sites were 91.3%, 82.7%, and 87.5%, respectively. No significant differential expression of HER2DX scores was observed across paired core biopsies [P = 0.148 (relapse risk score), P = 0.690 (pCR likelihood score), and P = 0.751 (ERBB2 mRNA score)]. The average score differences between biopsy cores (scale from 1– to 99) were 0.74 (relapse risk score), 0.30 (pCR likelihood score), and −0.24 (ERBB2 mRNA score). Correlation coefficients across paired core biopsies were 0.97 (relapse risk score), 0.91 (pCR likelihood score), and 0.92 (ERBB2 mRNA score) (Supplementary Figure S7, available at https://doi.org/10.1016/j.esmoop.2024.102903).
Subsequently, we delved into the within-tumor variations of HER2DX within patients who possessed paired tumor samples collected at distinct time points. To elaborate, seven individuals underwent an initial diagnostic core biopsy, followed by primary surgery with no intervening systemic treatment. HER2DX analysis was conducted on both the core biopsy and the surgical tumor specimens. The average time span between the core biopsy and the surgical specimens was 50.6 days (range 35-64 days). The collective findings unveiled a maximal SE of 6.1 across the HER2DX scores (measured on a scale of 1-99), coupled with an overall agreement rate of 86%. Notably, no correlation between the HER2DX scores and the duration between tissue acquisitions was observed (Supplementary Figure S8, available at https://doi.org/10.1016/j.esmoop.2024.102903).
Variability across genomic platforms
A pivotal aspect of our investigation involved evaluating the concordance of HER2DX scores obtained from two different genomic platforms. To this end, we leveraged the data from 30 RNA samples that had undergone both the HER2DX assay and RNAseq using the nCounter and Illumina platforms, respectively. Concordance between the two platforms in HER2DX relapse risk, pCR likelihood, and ERBB2 mRNA score groups was 96.7% (Cohen’s κ coefficient = 0.93), 96.7% (0.95), and 96.7% (0.94), respectively (Supplementary Figure S9, available at https://doi.org/10.1016/j.esmoop.2024.102903). Correlation of gene expression between platforms was high (mean correlation coefficient = 0.89, SD = 1.16). Only two genes (ASPM and NTN3) had a correlation coefficient <0.75. Coefficients of variation (CV) for each gene among the 30 samples within each platform were calculated and compared between platforms. Two genes (NTN3 and TCAP) had a CV difference >0.3 (Supplementary Figure S9, available at https://doi.org/10.1016/j.esmoop.2024.102903).
Discussion
The emergence of personalized patient care has underscored the importance of stringent performance criteria for clinical laboratory tests. In this context, the HER2DX genomic assay presents a significant advancement by providing accurate predictions of breast cancer recurrence risk, probability of achieving pCR after post-neoadjuvant therapy, and individual ERBB2 expression levels in early-stage HER2-positive breast cancer.5 This study aimed to comprehensively assess the analytical performance of HER2DX, focusing on precision, reproducibility, variability, and robustness.
Precision and reproducibility are fundamental attributes of any diagnostic assay, and our findings demonstrate the high degree of consistency and reliability of the HER2DX scores. The minimal SE observed across HER2DX scores within FFPE tumor samples indicates a high assay precision, and the strong correlation coefficients obtained between laboratories further affirm its reproducibility. Simulation analyses also underscored the accuracy of the scores, reinforcing the robustness of the assay’s measurements.
Intratumor variability analysis revealed that HER2DX maintains stability even within distinct geographical areas of tumors, with variations remaining <4.0%. Although the overall differences in scores between paired sites were low, our analysis indicates that borderline cases—those within 10 points of the classification cut-off—are particularly prone to reclassification upon re-sampling. This emphasizes the necessity for clinicians to consider the potential variability in HER2DX pCR scores when making treatment decisions based on borderline HER2DX pCR scores. Moreover, the assessment of within-patient variability, involving paired tumor samples collected at different time points, emphasized the consistent performance of HER2DX scores, irrespective of the duration between tissue acquisitions. The lack of correlation between HER2DX scores and acquisition interval further attests to the reliability of the assay.
A pivotal aspect of our investigation involved evaluating HER2DX performance across different genomic platforms. The high concordance and agreement rates observed between HER2DX scores obtained from nCounter and RNAseq platforms signify the assay’s consistent predictive capability across different technologies. Correlation coefficients for gene expression and CV further support the congruence of results between platforms, with only a few genes displaying minor discrepancies.
The robustness of HER2DX, particularly in the context of low tumor content, is noteworthy. The assay’s ability to maintain accuracy even when analyzing samples with as little as 10% tumor content highlights its utility in scenarios where limited tissue availability may be a concern. Of note, ERBB2 score is directly related to the % of tumor content. Additionally, our investigation addressed potential sources of variability, including instrument differences, reagent lots, and protocol modifications. The lack of significant variability observed across these factors underscores the stability and reliability of HER2DX measurements under diverse conditions.
Certain limitations should be acknowledged. The findings’ generalizability might be influenced by the specific subset of FFPE tumor samples and purified RNA analyzed, potentially affecting broader patient populations. External factors, such as sample handling and laboratory techniques, could introduce variability despite implemented quality controls. While high concordance was observed between genomic platforms, platform-specific characteristics should be considered during implementation. To date, the assay is offered using the nCounter platform, and future work will allow implementation of the assay on RNAseq-based platforms.
In conclusion, this comprehensive analysis reaffirms the analytical performance of the HER2DX genomic assay, enhancing its credibility as a valuable tool for guiding treatment decisions in early-stage HER2-positive breast cancer. The assay’s precision, reproducibility, low variability across laboratories and platforms, and robustness under different experimental conditions collectively underscore its potential to augment tailored treatment strategies. As we continue to advance toward more individualized approaches in clinical oncology, the HER2DX assay holds promise as a reliable and informative tool to improve patient outcomes.
Acknowledgments
Funding
This study was funded by Reveal Genomics S.L. (Barcelona, Spain). Their support extended beyond financial assistance, encompassing active engagement in multiple facets of the research process, such as study design, data collection, analysis, interpretation, and report composition. The study’s data were made fully accessible to all authors, collectively sharing the ultimate responsibility for the decision to present these findings for publication. FBM received funding from Fundación Científica Asociación Española Contra el Cáncer (Ayudas Investigador AECC 2021) [INVES21943BRAS]. AP received funding from Fundación CRIS contra el cáncer [PR_EX_2021-14], Agència de Gestió d'Ajuts Universitaris i de Recerca [2021 SGR 01156], Fundación Fero [BECA ONCOXXI21], Instituto de Salud Carlos III [grant number PI22/01017], Asociación Cáncer de Mama Metastásico IV Premios M. Chiara Giorgetti, Breast Cancer Research Foundation [BCRF-23-198], and RESCUER, funded by European Union’s Horizon 2020 Research and Innovation Programme [847912].
Disclosure
MMA is an employee at Reveal Genomics; GV has received a speaker’s fee from MSD, Pfizer, GSK, and Pierre Fabrer, has held an advisory role with AstraZeneca, and received consultant fees from Reveal Genomics; FBM has a HER2DX patent application EP21383165; PG is an employee at Reveal Genomics; OMS has declared travel expenses and consulting fees from Roche and Reveal, and speaker fees from Eisai, Daiichi, and Novartis; KP serves on the Scientific Advisory Board of Novartis, Ideaya Biosciences, and Scorpion Therapeutics, holds equity options in Scorpion Therapeutics and Ideaya Biosciences, and receives sponsored research funding from Novartis through DFCI; and has patents on S100A7 antibody and BET inhibitor resistance held by DFCI; JM is an employee at Reveal Genomics; WB is an employee at Reveal Genomics; CMP is an equity stockholder and consultant of BioClassifier LLC, and for Reveal Genomics, is also listed as an inventor on patent applications for the Breast PAM50 assay; PVG is one of the stockholders of Reveal Genomics; AP reports advisory and consulting fees from Roche, Pfizer, Novartis, Amgen, BMS, Puma, Oncolytics Biotech, MSD, Guardant Health, Peptomyc, and Lilly, lecture fees from Roche, Pfizer, Novartis, Amgen, BMS, and Daiichi Sankyo, institutional financial interests from Novartis, Roche, and Pfizer, stockholder and consultant of Reveal Genomics, SL; AP is also listed as an inventor on patent applications for the HER2DX assay; JSP is an equity stockholder and consultant for Reveal Genomics and is also listed as an inventor on patent applications for the Breast PAM50 assay; LP is listed as an inventor on HER2DX patent PCT/EP2021/070788 and is an employee at Reveal Genomics. All other authors have declared no conflicts of interest.
Supplementary data
References
- 1.Swain S.M., Shastry M., Hamilton E. Targeting HER2-positive breast cancer: advances and future directions. Nat Rev Drug Discov. 2023;22:101–126. doi: 10.1038/s41573-022-00579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schettini F., Prat A. Dissecting the biological heterogeneity of HER2-positive breast cancer. Breast. 2021;59:339–350. doi: 10.1016/j.breast.2021.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cronin M., Sangli C., Liu M.L., et al. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor–positive breast cancer. Clin Chem. 2007;53:1084–1091. doi: 10.1373/clinchem.2006.076497. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen T., Wallden B., Schaper C., et al. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer. 2014;14:177. doi: 10.1186/1471-2407-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prat A., Guarneri V., Pascual T., et al. Development and validation of the new HER2DX assay for predicting pathological response and survival outcome in early-stage HER2-positive breast cancer. EBioMedicine. 2022;75 doi: 10.1016/j.ebiom.2021.103801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waks A.G., Ogayo E.R., Paré L., et al. Assessment of the HER2DX assay in patients with ERBB2-positive breast cancer treated with neoadjuvant paclitaxel, trastuzumab, and pertuzumab. JAMA Oncol. 2023;9:835–840. doi: 10.1001/jamaoncol.2023.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bueno-Muiño C., Echavarría I., López-Tarruella S., et al. Assessment of a genomic assay in patients with ERBB2-positive breast cancer following neoadjuvant trastuzumab-based chemotherapy with or without pertuzumab. JAMA Oncol. 2023;9:841–846. doi: 10.1001/jamaoncol.2023.0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villacampa G., Tung N.M., Pernas S., et al. Association of HER2DX with pathological complete response and survival outcomes in HER2-positive breast cancer. Ann Oncol. 2023;34:783–795. doi: 10.1016/j.annonc.2023.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tolaney S.M., Tarantino P., Graham N., et al. Adjuvant paclitaxel and trastuzumab for node-negative, HER2-positive breast cancer: final 10-year analysis of the open-label, single-arm, phase 2 APT trial. Lancet Oncol. 2023;24:273–285. doi: 10.1016/S1470-2045(23)00051-7. [DOI] [PubMed] [Google Scholar]
- 10.Guarneri V., Brasó-Maristany F., Dieci M.V., et al. HER2DX genomic test in HER2-positive/hormone receptor-positive breast cancer treated with neoadjuvant trastuzumab and pertuzumab: a correlative analysis from the PerELISA trial. EBioMedicine. 2022;85 doi: 10.1016/j.ebiom.2022.104320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarantino P., Villacampa G., Graham N.T., et al. 6P - Combined analysis of the HER2DX genomic tool in adjuvant APT and ATEMPT trials. Ann Oncol. 2023;8 [Google Scholar]
- 12.Garcia-Saenz J.A., Blancas I., Echavarria I., et al. SEOM–GEICAM–SOLTI clinical guidelines in advanced breast cancer (2022) Clin Transl Oncol. 2023;25:2665. doi: 10.1007/s12094-023-03203-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curigliano G., Burstein H.J., Gnant M., et al. Understanding breast cancer complexity to improve patient outcomes: the St. Gallen International Consensus Conference for the Primary Therapy of Individuals with Early Breast Cancer 2023. Ann Oncol. 2023;34:970–986. doi: 10.1016/j.annonc.2023.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Geiss G.K., Bumgarner R.E., Birditt B., et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.