Abstract
High-risk human papillomaviruses (HPVs) are the causative agents of cervical cancer. Here, we report that HPV16 E6E7 promotes cervical cancer cell proliferation by activating the pentose phosphate pathway (PPP). We found that HPV16 E6 activates the PPP primarily by increasing glucose-6-phosphate dehydrogenase (G6PD) enzyme activity. Mechanistically, HPV16 E6 promoted G6PD dimer formation by inhibiting its lactylation. Importantly, we suggest that G6PD K45 was lactylated during G6PD-mediated antioxidant stress. In primary human keratinocytes and an HPV-negative cervical cancer C33A cells line ectopically expressing HPV16 E6, the transduction of G6PD K45A (unable to be lactylated) increased GSH and NADPH levels and, correspondingly, decreasing ROS levels. Conversely, the re-expression of G6PD K45T (mimicking constitutive lactylation) in HPV16-positive SiHa cells line inhibited cell proliferation. In vivo, the inhibition of G6PD enzyme activity with 6-aminonicotinamide (6-An) or the re-expression of G6PD K45T inhibited tumor proliferation. In conclusion, we have revealed a novel mechanism of HPV oncoprotein-mediated malignant transformation. These findings might provide effective strategies for treating cervical and HPV-associated cancers.
Keywords: High-risk human papillomaviruses, Pentose phosphate pathway, Lactylation, glucose-6-phosphate dehydrogenase
1. Introduction
HPV16 is the predominant HPV type detected in cervical cancer (∼60%) [1] and the persistent expression of its E6 and E7 viral oncogenes or those of other high risk HPV types is a driving factor of cervical cancer [2]. These oncoproteins bind and destabilize multiple cellular targets, including the tumor suppressors p53 and pRB, leading to the dysregulation of multiple signaling pathways and immortalization [3]. However, the details are still lacking in the mechanism of carcinogenesis associated with HPV infection.
Metabolic reprogramming is a hallmark of cancer [4]. Tumor cells prefer to generate energy via glycolysis, even in the presence of oxygen (Warburg effect) [5]. The pentose phosphate pathway (PPP) is commonly overactivated in tumor cells. Glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme in the PPP, mediates the oxidative PPP to produce NADPH and maintain intracellular redox homeostasis [6]. The nonoxidative PPP provides a substrate for nucleotide synthesis [7]. Strong evidence suggests that an increased dependence on the PPP provides NADPH and metabolic intermediates for cancer cell proliferation and tumor development [8,9]. However, whether HPV activates the PPP to promote the HPV carcinogenesis remains unclear.
Lactylation is a novel posttranslational modification by the addition of lactyl groups to lysine (K) residues [10]. Histone lactylation modifications play important roles in regulating macrophage polarization and in inducing the expression of pluripotency genes during the reprogramming of Senescent cells into pluripotent stem cells [10,11]. Non-histone lactylation modifications stimulate HMGB1 release from macrophages [12]. Notably, global lactylation mapping analysis, identified via mass spectrometry, showed that several enzymes involved in the PPP, glycolysis, and tricarboxylic acid cycle are modified by lactylation [13]. However, whether G6PD can be modified by lactylation and the site of G6PD lactylation are unknown.
In the present study, we identified HPV16 E6 as a key regulator of PPP activation, particularly the oxidative PPP, to increase NADPH and GSH levels in response to oxidative stress. Mechanistically, HPV16 E6 inhibited G6PD K45 lactylation to increase G6PD enzyme activity. We further demonstrated that the HPV16 E6-mediated activation of G6PD is critical for its role in promoting cancer cell growth, both in vitro and in vivo.
2. Results
2.1. HPV16 E6E7 rewires the PPP
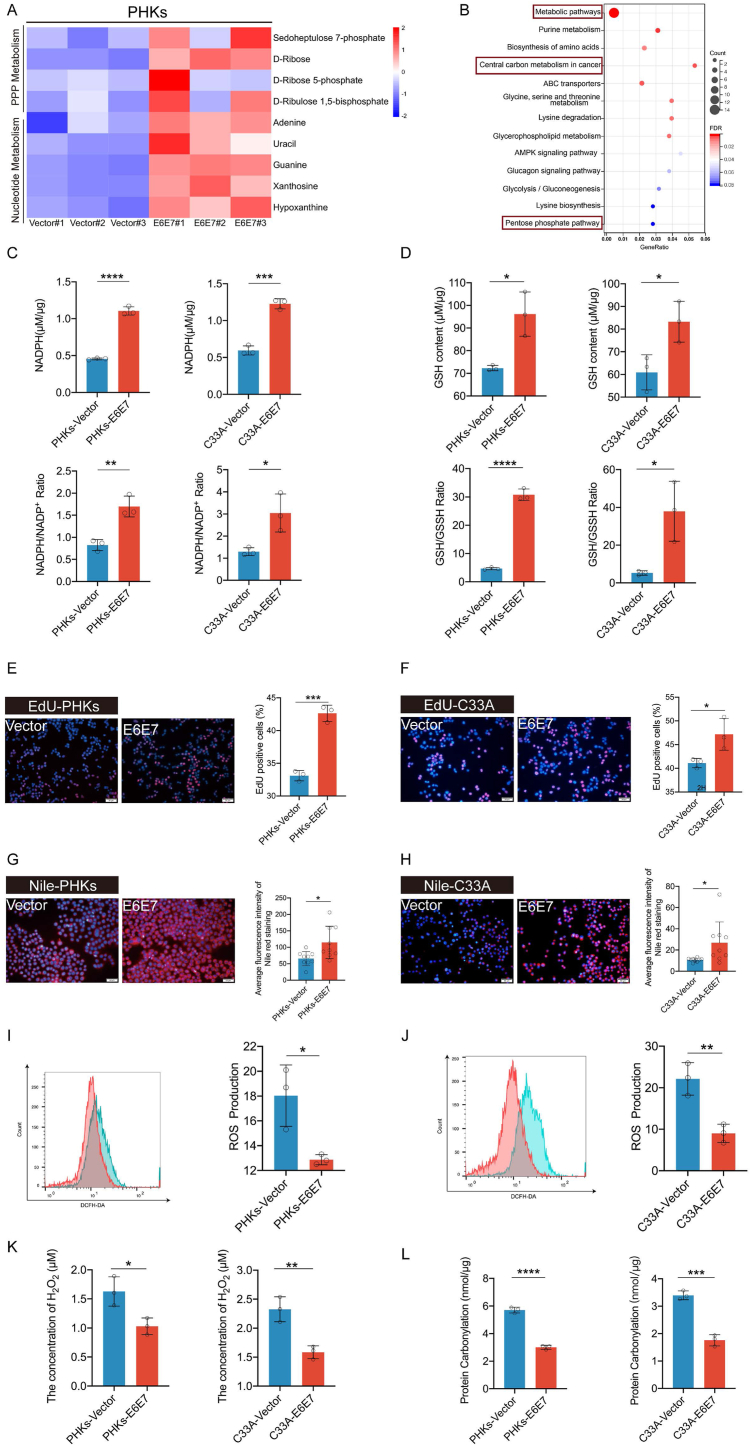
It has been long established that HPV16 E6E7 promotes cell proliferation [[14], [15], [16]]. As expected, in our current study, ectopic stable expression of HPV16 E6E7 from a recombinant lentivirus promoted cell proliferation in C33A cells (an HPV-negative cervical carcinoma cell line) and in primary human keratinocytes cells (PHKs) (Supplementary Figs. 1A–C), reduced apoptosis (Supplementary Fig. 1D) and maintained the growth and alleviated senescence of immortalized MEF cells (Supplementary Figs. 1E–F). To screen for metabolic pathways that are altered by E6E7 to promote cell proliferation, PHKs expressing E6E7 or the vector were used to identify differences in metabolites by using liquid chromatography-mass spectrometry (LC-MS). We observed an increase in the expression of substrates involved in PPP metabolism and nucleotide metabolism (Fig. 1A). Furthermore, KEGG analysis demonstrated enriched metabolites involved in metabolic pathways, central carbon metabolism in cancer, and the PPP (Fig. 1B).
Fig. 1.
HPV16 E6E7 enhances the PPP and biosynthesis in PHKs and C33A cells. Stable transduction of PHKs and C33A cells using a lentivirus expression vector or HPV16 E6E7 was performed. (A) Heat map of metabolites for which levels were upregulated in PHKs cells expressing HPV16 E6E7, compared to levels with the vector. (B) Differential metabolites were subjected to KEGG enrichment analysis. (C–D) Intracellular NADPH, NADPH/NADP+, GSH/GSSH, and GSH levels were detected in PHKs and C33A cells exogenously expressing the vector or HPV16 E6E7. (E–F) EdU was used to determine DNA synthesis in PHKs and C33A cells exogenously expressing the vector or HPV16 E6E7. ImageJ was used to analyze the proportion of EdU-positive cells. Representative images (left) and quantification results (right) are shown. (G–H) Detection of lipid synthesis via Nile red staining. Representative images (left) of average fluorescence quantification results (right) are shown. (I–J) DCFH-DA was used to examine ROS in the cytoplasm of C33A and PHKs cells. Representative images detected by performing flow cytometry are shown (left), and the corresponding histograms are shown on the right side of the image. Intracellular H2O2 is shown in (K). PHKs and C33A cells exogenously expressing the vector or HPV16 E6E7. A quantitative spectrophotometric assay involving the derivatization of protein carbonyl groups with 2,4-dinitrophenylhydrazine was used, as described in Section “Materials and Methods.“(L). Each dot represents an independent biological replicate in the plots. Data are presented as mean ± SD. Statistical significance was determined using unpaired two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with indicated groups. NS, not significant.
PPP provides precursors for nucleotide synthesis. Therefore, we investigated whether HPV reprogramed the PPP. We observed that NADPH, NADPH/NADP+, GSH/GSSH, and GSH levels and DNA synthesis were significantly increased in PHKs and C33A cells expressing HPV16 E6E7 (Fig. 1C–F). NADPH is required to scavenge reactive oxygen species (ROS) and is consumed during fatty acid synthesis [17]. Our results revealed that lipid synthesis levels were increased and ROS levels were reduced in cells expressing E6E7 (Fig. 1G–J). We also detect intracellular hydrogen peroxide (H2O2), intracellular H2O2 was significantly reduced in C33A and PHKs cells expressing HPV16 E6E7 compared to Vector (Fig. 1K). Protein carbonylation provide a comprehensive assessment of oxidative damage. It is a standard marker for oxidative stress [18]. Similarly, protein carbonylation showed lower levels in cells expressing HPV16 E6E7 (Fig. 1L). These findings show that HPVE16 E6E7 can reprogram the PPP.
2.2. HPV16 E6 reprograms the PPP
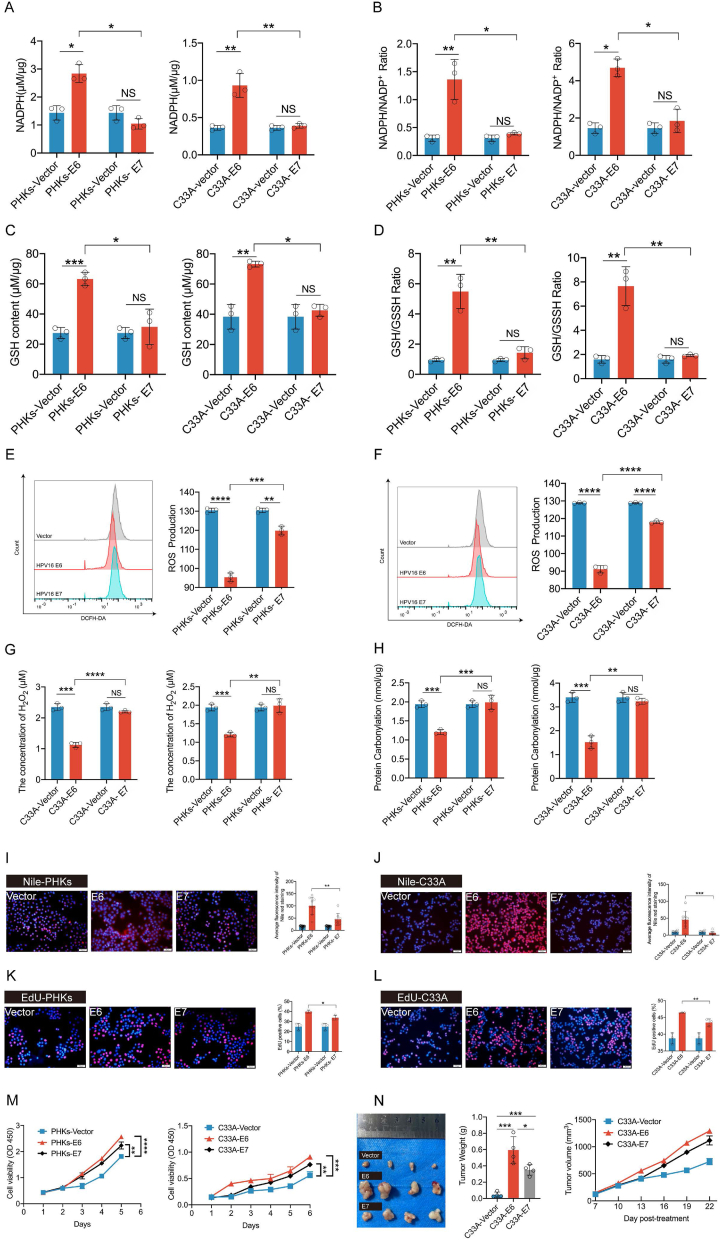
We further investigated this new role of the E6 and E7 oncoproteins in HPV infection. The stable transduction of PHKs and C33A cells using a lentivirus vector expressing HPV16 E6 and HPV16 E7 was achieved. These cells were then used to detect PPP-related metabolites. We found that NADPH, NADPH/NADP+, GSH/GSSH, and GSH levels were significantly higher in C33A and PHKs expressing E6 relative to those expressing E7 and that the corresponding ROS levels were significantly decreased (Fig. 2A–F). We also detect intracellular H2O2, intracellular H2O2 was significantly reduced in C33A and PHKs cells expressing HPV16 E6 compared to HPV16 E7 (Fig. 2G). Similarly, protein carbonylation showed lower levels in cells expressing HPV16 E6 (Fig. 2H). Moreover, both lipid synthesis Fig. 2E-F and DNA synthesis (Fig. 2I-L) were also significantly increased in HPV16 E6-expressing C33A and PHKs compared to HPV16 E7-expressing cells. We also observed that HPV16 E6-expressing PHKs and C33A cells exhibited higher proliferative capabilities than HPV16 E7-expressing cells (Fig. 2M). To collaborate the results in vitro, we conducted experiment in vivo. Consistently, in a xenograft experiment in which nude mice were injected with control C33A cells or those expressing HPV16 E6 or HPV16 E7, the growth rate and size of tumors derived from HPV16 E6-expressing cells were significantly increased compared to those in tumors derived from HPV16 E7-or vector-only cells (Fig. 2N). Thus, these results indicate that HPV16 E6 activates PPP, leading to cell proliferation and tumor growth.
Fig. 2.
HPV16 E6 primarily enhances the PPP and biosynthesis in PHKs and C33A cells. PHKs and C33A cells were stably transduced with a lentiviruses expression vector, HPV16 E6, or HPV16 E7. C33A/PHKs-HPV16 E6, C33A/PHKs -HPV16 E7, and control cells harboring an empty vector were tested for intracellular NADPH (A), NADPH/NADP+ (B), GSH (C) GSH/GSSH (D), ROS (E–F), H2O2 (G) and protein carbonylation (H). (I–J) Detection of lipid synthesis by performing Nile red staining. Representative images (left) of average fluorescence quantification results (right) are shown. (K–L) EdU was used to determine DNA synthesis in PHKs and C33A cells exogenously expressing HPV16 E6, HPV16 E7, or the vector. ImageJ was used to analyze the proportion of EdU-positive cells. Representative images (left) and quantification results (right) are shown. (M) Cell proliferation rates were determined by performing CCK8 assays. (N) Tumor masses in xenograft nude mice injected with C33A-HPV16 E6 (1 × 106) and C33A-HPV16 E7(5 × 106) cells compared to those in mice injected with the control vector cells. Each dot represents an independent biological replicate in the plots. Data are presented as mean ± SD. Statistical significance was determined using unpaired two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with indicated groups. NS, not significant.
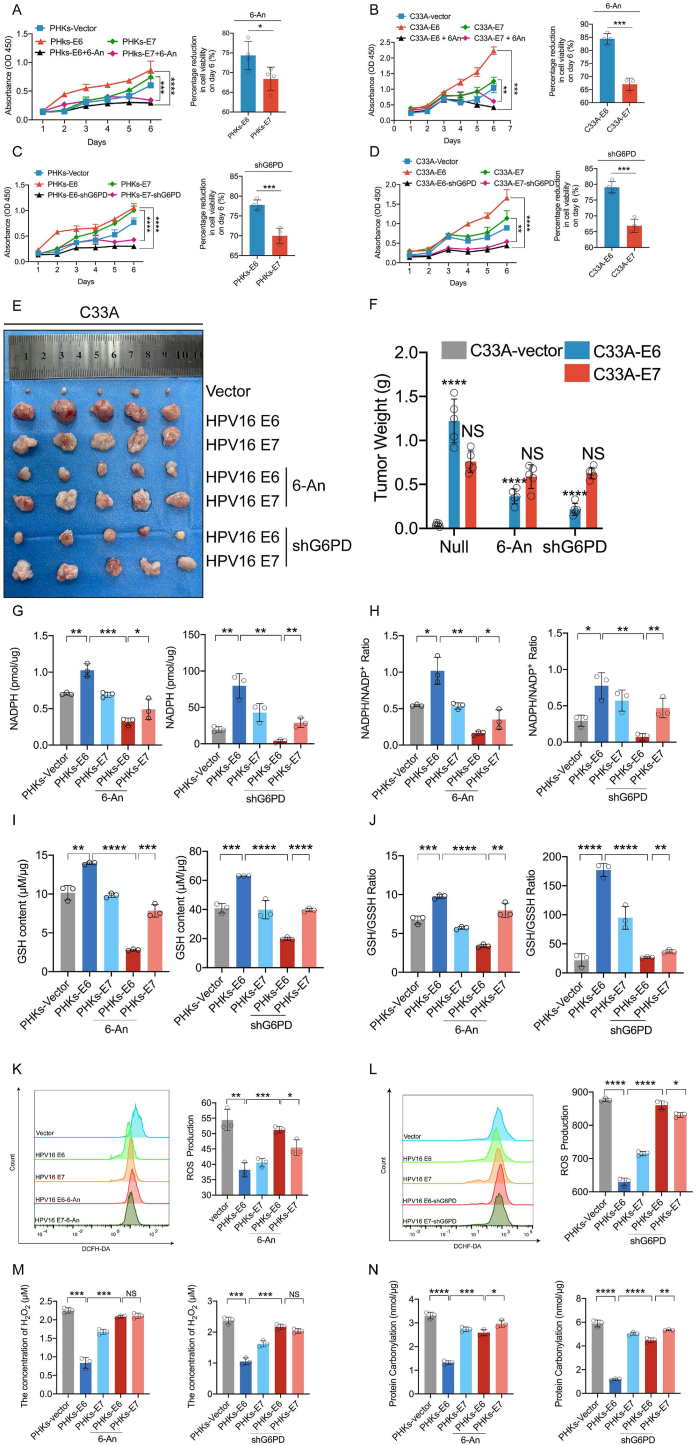
To substantiate our conclusion that E6 promotes cell proliferation via PPP activation, 6-An, a PPP inhibitor, was administered to C33A and PHKs expressing E6 or E7. The half-maximal inhibitory concentration (IC50) of 6-An to PHKs and C33A was 81.06 μM and 26.78 μM, respectively (Supplementary Figs. 2A and B). 6-An significantly reduced the proliferation of PHKs cells expressing E6 compared to E7 on day six (Fig. 3A-B). Consistently, knockdown of G6PD significantly inhibited E6-induced proliferation compared to inhibition of E7-induced proliferation (Fig. 3C, D). To collaborate these observations in vivo. In a xenograft experiment in which nude mice were injected with control C33A cells or those expressing HPV16 E6 or HPV16 E7, the size of tumors derived from HPV16 E6-expressing cells were smaller after treatment with 6-An or G6PD KD compared to tumors derived from cells expressing HPV16 E7 (Fig. 3E, F).
Fig. 3.
HPV16 E6 promotes cell proliferation mediated by G6PD. PHKs and C33A cells were stably transduced with lentiviruses expressing the vector, HPV16 E6, or HPV16 E7. These cells were then treated with the G6PD inhibitor 6-An (81.06 μM or 26.78 μM) or infected with a lentivirus expressing shG6PD. (A–D) Cell proliferation rates were determined by performing a CCK8 assay (left). The percentage reduction in cell viability on day 6 is shown independently (right). (E) In total, C33A-Vector, C33A-HPV16E6, and C33A-HPV16E7 cells (1 × 106) were inoculated subcutaneously into the right flanks of 4- to 5-week-old female nude mice (n = 5 each). Images are shown of nude mouse xenograft tumors derived from C33A-HPV16 E6 and C33A-HPV16 E7 cells treated with 4 mg/kg/3d 6-An. Tumor sizes were measured every 5 days for 3 weeks. Intracellular NADPH (G), NADPH/NADP+ (H), GSH (I), GSH/GSSH (J) ROS (K–L) H2O2 (M), and protein carbonylation levels (N) were tested. Each dot represents an independent biological replicate in the plots. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with indicated groups. Statistical significance was determined using unpaired two-tailed t-test. NS, not significant.
In addition, PHKs cells expressing HPV16 E6 showed significantly decreased NADPH, NADPH/NADP+, GSH/GSSH, and GSH levels after 6-An treatment compared to those in HPV16 E7-expressing cells (Fig. 3G–J). In contrast to the change in NADPH levels, the 6-An-mediated or G6PD KD-mediated increase in ROS levels in cells expressing E6 was more significant than those in cells expressing E7 (Fig. 3K-L). We also detect intracellular H2O2, intracellular H2O2 was significantly reduced in PHKs cells expressing HPV16 E6 compared to HPV16 E7 after 6-An treatment or G6PD-KD (Fig. 3M). Similarly, protein carbonylation showed lower levels in cells expressing HPV16 E6 after 6-An treatment or G6PD-KD (Fig. 3N). Collectively, these data demonstrate that HPV16 E6 promotes cell proliferation by reprogramming the PPP.
2.3. HPV16 E6 activates G6PD
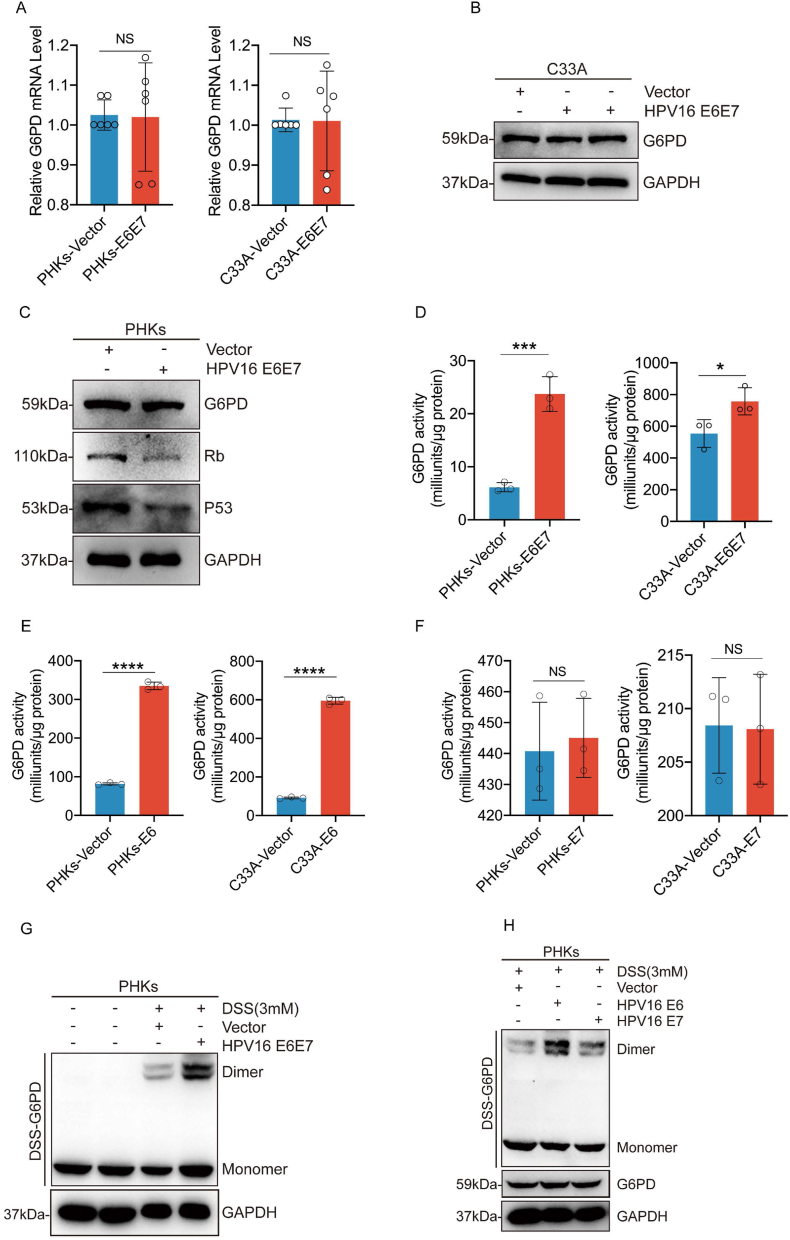
To determine the mechanism underlying HPV-mediated reprogramming of the PPP, we next investigated whether HPV16 E6E7 affects the expression of G6PD. We found that the enzymatic activity of G6PD was significantly increased in C33A and PHKs cells expressing HPV16 E6E7, but neither G6PD protein nor mRNA expression was affected (Fig. 4A–D). We also found that HPV16 E6, but not E7, significantly increased G6PD enzyme activity (Fig. 4E-F). G6PD exists as an inactivated monomer and enzymatically active dimer (17). Disuccinimidyl suberate (DSS) crosslinking analysis revealed that the ectopic expression of HPV16 E6E7 or HPV16 E6 resulted in increased levels of G6PD dimers. The HPV 16 E7 had only a minor positive effect (Fig. 4G-H). Thus, HPV16 E6 promotes the formation of active G6PD dimers.
Fig. 4.
HPV16 E6 increases G6PD enzyme activity by promoting the formation of G6PD dimers. PHKs and C33A cells were stably transduced with lentiviruses expressing the vector, HPV16 E6, HPV16 E7, or HPV16 E6E7. (A) mRNA levels of G6PD in the transduced cells were determined via qPCR. (B–C) Immunoblots were used to detect G6PD, Rb, and p53 levels in cell lysates. GAPDH served as a loading control. (D–F) G6PD enzyme activity was examined in these cells. (G–I) Cells were harvested and crosslinked using DSS (3 mM), followed by western blotting with an anti-G6PD antibody. Data are presented as mean ± SD. Each dot represents an independent biological replicate in the plots. *P < 0.05, ***P < 0.001, ****P < 0.0001 compared with indicated groups. Statistical significance was determined using unpaired two-tailed t-test. NS, not significant.
2.4. HPV16 E6 promotes G6PD dimer formation by inhibiting G6PD lactylation
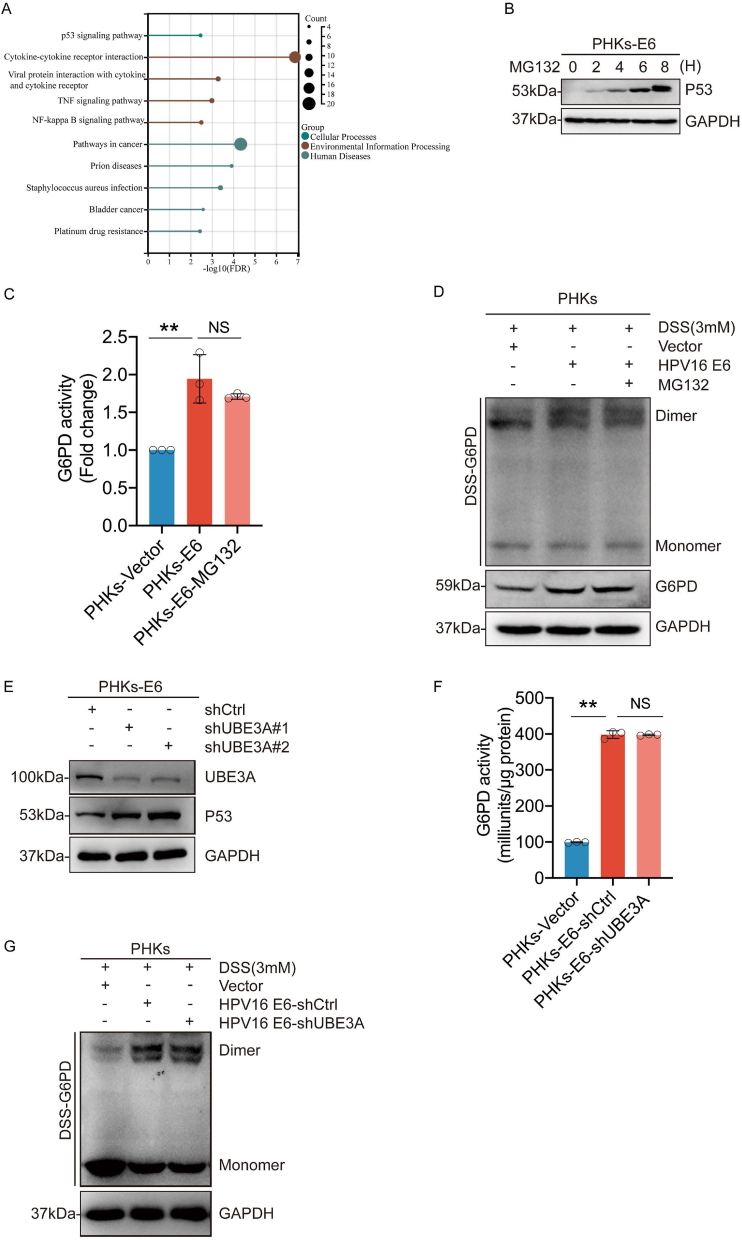
We then investigated how E6 promoted G6PD dimer formation. Published data from the GEO database were analyzed, and we found significant changes in the p53 signaling pathway (Fig. 5A). Previous reports have demonstrated that the oncoprotein HPV16 E6 promotes p53 degradation and that p53 binds directly to G6PD to prevent the formation of an active dimer [19]. Therefore, we explored whether HPV regulates G6PD enzymatic activity in a p53-dependent manner. MG132 was used to incubate PHKs cells expressing HPV16 E6 or vectors to inhibit the degradation of p53 mediated by E6 (Fig. 5B). We found that MG132 did not abolish the E6-induced increase in enzymatic activity or G6PD dimerization (Fig. 5C-D). In addition, we constructed PHKs cell lines in which UBE3A, an E3 ubiquitin ligase, was knocked down. Consistent with previous reports [20], the knockdown UBE3A significantly increased p53 levels. However, the knockdown of UBE3A did not inhibit the enzymatic activity of G6PD and the formation of dimer (Fig. 5E–G). These results suggest that HPV16 E6 regulates G6PD enzyme activity in a p53 independent manner.
Fig. 5.
HPV16 E6 regulates G6PD enzyme activity independent of p53. (A) KEGG enrichment analysis of the differential genes screened based on GEO (GSE58841). (B) PHKs-E6 cells were treated with 5 μM MG132 for the indicated duration. The whole-cell extracts (WCEs) were then collected for immunoblotting to detect p53 levels in the cells. (C) G6PD enzyme activity was assayed after treating PHKs-E6 cells with MG132 (5 μM) for 6 h. (D) PHKs-E6 cells were treated with 5 μM of MG132 for 6 h. Cells were harvested and crosslinked using DSS (3 mM), followed by western blotting with an anti-G6PD antibody. (E) PHKs-E6 cells were stably transduced with lentiviruses expressing shUBE3A. WCEs were analyzed via immunoblotting for UBE3A and p53. (F) PHKs-E6-shUBE3A cells were employed for the detection of G6PD enzyme activity. (G) Crosslinking with DSS followed by immunoblotting to detect dimeric and monomeric G6PD. Each dot represents an independent biological replicate in the plots. Data are presented as mean ± SD. **P < 0.01 compared with indicated groups. Statistical significance was determined using unpaired two-tailed t-test. NS, not significant. HPV, human papilloma virus; G6PD, glucose-6-phosphate dehydrogenase; DSS, disuccinimidyl suberate.
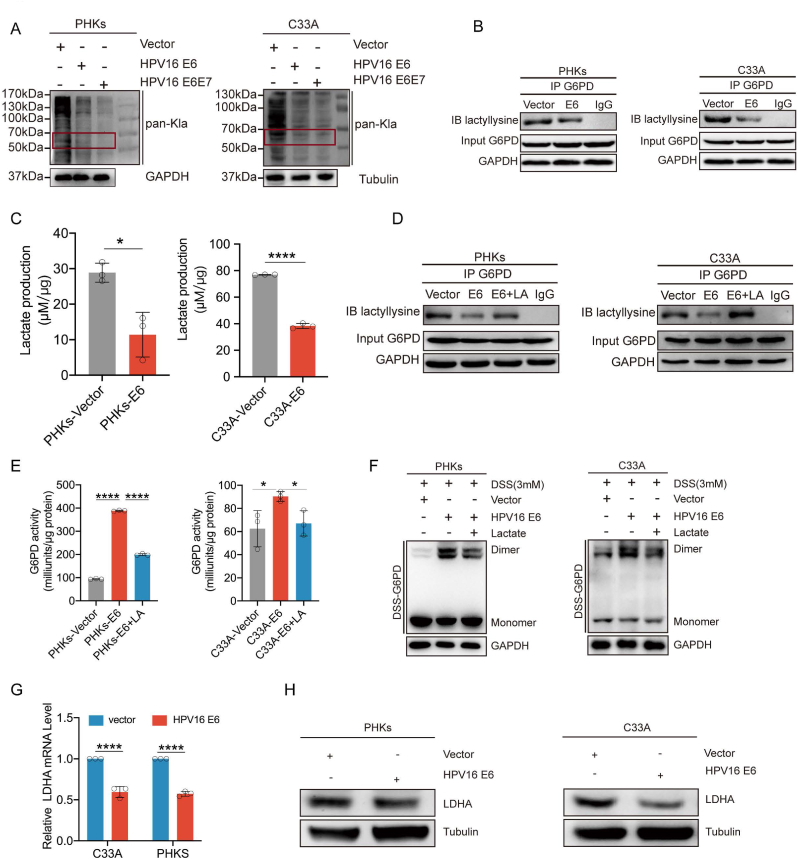
Recent studies have shown that lactylation also acts as a post-translational modification of lysine residues, and directly regulates enzyme activity [12,13]. Therefore, we investigated whether the enzymatic activity of G6PD was regulated by lactylation. First, C33A and PHKs cells expressing E6, E6E7, or the vector were used to determine the levels of global intracellular lactylation. We observed significantly reduced levels in cells expressing either E6 or E6E7 oncoproteins (Fig. 6A). The pan-lactylation antibody immunoprecipited significantly lower amounts of G6PD from C33A and PHKs cells ectopically expressing HPV16 E6 than from cells expressing E7 or vector-only without any changes in total G6PD (Fig. 6B). Consistently, the intracellular lactate levels were significantly lower in C33A and PHKs cells expressing E6 (Fig. 6C). Sodium lactate (LA) supplementation markedly increased the G6PD immunocomplex detected with the pan-lactylation antibody (Fig. 6D). To substantiate that the HPV16 E6-mediated inhibition of G6PD lactylation affects its enzymatic activity, we measured G6PD enzymatic activity after culturing E6-expressing cells in medium with adding sodium lactate. Our data showed that sodium lactate addition significantly diminished the enzymatic activity of G6PD (Fig. 6E). The DSS crosslinking assay also showed that lactate treatment reduced G6PD dimer formation (Fig. 6F). These results suggest that lactylation modifications inhibit the enzymatic activity of G6PD by reducing dimer formation.
Fig. 6.
HPV16 E6 inhibits G6PD lactylation modifications. (A) Pan-lactyation levels were detected in C33A and PHKs expressing HPV16 E6, HPV16 E7, and the vector via western blotting. (B) Immunoblotting for lactylation in the anti-G6PD immunoprecipitates. The immunoprecipitates were isolated from the C33A and PHKs cells overexpressing the vector and HPV16 E6. (C) Intracellular lactate levels were examined in C33A and PHKs cells overexpressing the vector and HPV16 E6. (D) Immunoblotting for lactylation in the anti-G6PD immunoprecipitates. The immunoprecipitates were isolated from C33A and PHKs cells overexpressing the vector and HPV16 E6 upon NaLa treatment (25 mM) for 24 h. (E) G6PD enzyme activity was assayed in C33A and PHKs cells overexpressing the vector and HPV16 E6 upon NaLa treatment (25 mM) for 24 h. (F) Cells were harvested and crosslinked using DSS (3 mM), followed by western blotting with an anti-G6PD antibody. (G–H) LDHA levels were detected in C33A cells expressing HPV16 E6, HPV16 E7, and the vector. mRNA levels of LDHA in the transduced cells were determined via qPCR. Each dot represents an independent biological replicate in the plots. Data are presented as mean ± SD. *P < 0.05, ****P < 0.0001 compared with indicated groups.
Studies have shown that intracellular lactate can be used as a substrate for lactylation. Therefore, we examined the expression of lactate dehydrogenase A (LDHA). qPCR (Fig. 6G) and immunoblots (Fig. 6H) showed that LDHA expression was significantly reduced in C33A cells and PHKs expressing E6 in comparison to that in cells with control vector. Taken together, these results demonstrate that HPV16 E6 inhibits G6PD lactylation modification by decreasing lactate production, which in turn increases dimer formation to enhance G6PD enzyme activity.
2.5. K45 is a key site for G6PD lactylation regulated by the HPV16 E6 oncoprotein
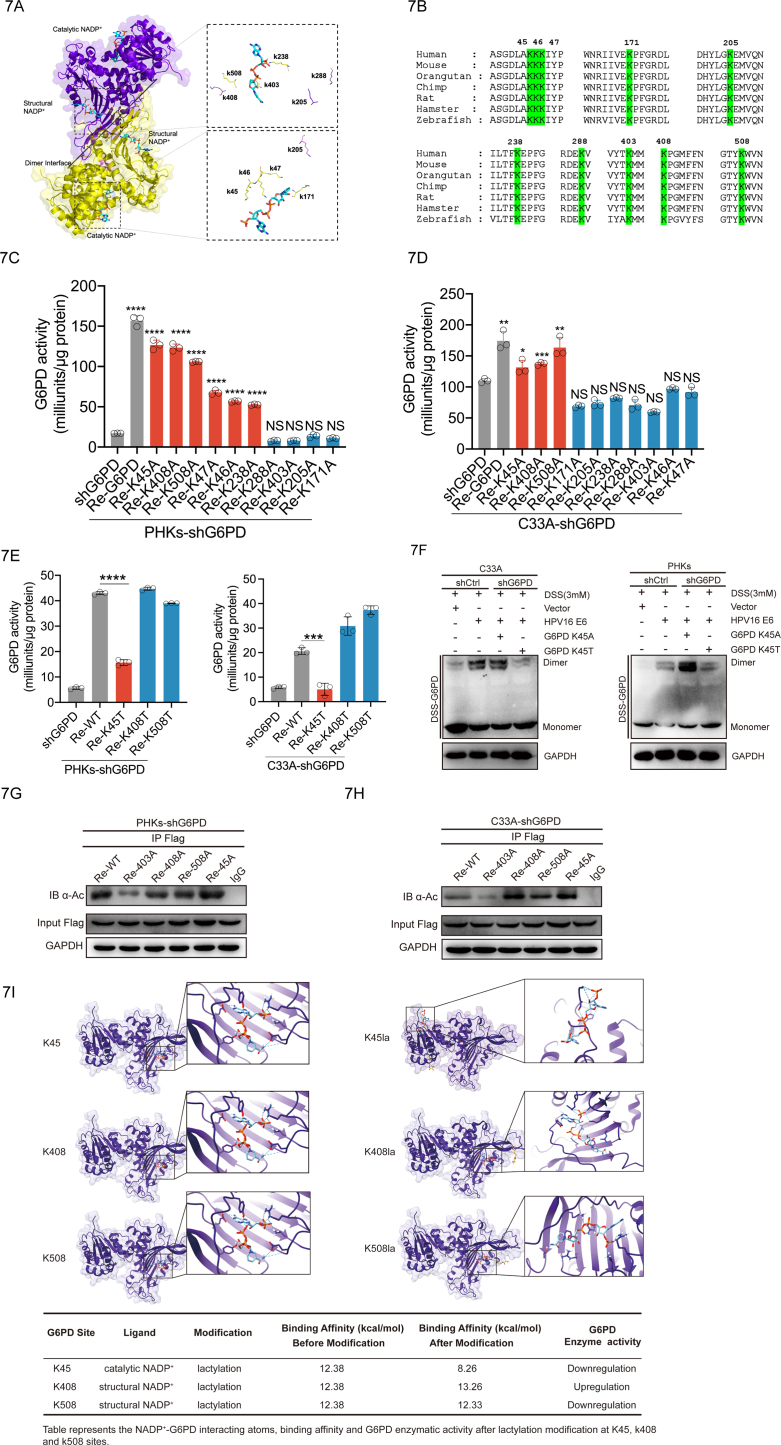
To identify the sites of G6PD lactylation, we analyzed the 3D protein structure of G6PD. X-ray structure of G6PD (PDB: 2BH9) [21,22] complexed with structural and catalytic NADP+ was achieved from the PDB database (https://www.pdbus.org/). Each subunit of the human G6PD has two NADP+-binding sites, a catalytic NADP+ coenzyme-binding domain and a structural NADP+-binding domain [23]. Knockdown of NADP+ inhibits the formation of G6PD dimer and reduces its enzymatic activity [24,25]. These studies suggest that NADP+ plays an important role in the regulation of G6PD dimerization and enzyme activity. Therefore, we selected 10 lysines near the NADP+ binding region as potential modification sites (Fig. 7A). The G6PD protein is highly conserved [26,27]; therefore, we speculated that the important regulatory sites targeted by lactylation might also be conserved. Sequence alignments from diverse species revealed that the same ten lysine residues (K45, K46, K47, K171, K205, K238, K288, K403, K408, and K508) were conserved (Fig. 7B). To determine which residues are targeted for lactylation, we mutated each of the 10 putative lysine residues to alanine (A) (Supplementary Fig. 2C) and assayed the activities of these mutations individually. We observed that ectopic expression of G6PD WT or K45A, K408A, K508A mutations significantly recovered G6PD activity in PHKs and C33A cells in which the endogenous G6PD was knocked down (Fig. 7C-D). The lactylation modification motif is structurally similar to threonine; therefore, the G6PD K45T, K408T, and K508T mutations were used to mimic sustained lactylation (Supplementary Fig. 2D). Only re-expression of the G6PD K45T mutation failed to restore the enzyme activity (Fig. 7E). To clarify further the function of K45 lactylation, intracellular GSH, NADPH, and ROS levels were examined in PHKs and C33A cells. We found that the NADPH, NADPH/NADP, GSH, and GSH/GSSG levels were significantly higher in cells re-expressing K45A than in cells re-expressing K45T in PHKs and C33A cells with endogenous G6PD knockdown (Supplementary Figs. S3A–D). Consistent with the NADPH and GSH changes, the re-expression of K45T correspondingly elevated ROS, H2O2 and protein carbonylation levels compared to those with K45A (Supplementary Figs. S3E–H). DSS crosslinking experiments further demonstrated that the K45T mutation displayed an impaired ability to form dimers, as compared to the K45A variant (Fig. 7F). Acetylation modifications also occur in lysine. K403 was reported to be a major G6PD acetylation site [28]. To further clarify that lysine 45 is a primary lactylation site, lysine at site 403, 408, 508 and 45 were mutated to alanine. C33A and PHKs cells re-expressing G6PD mutations were immunoprecipitated with anti-G6PD antibody and probed for acetylation (Kac) levels by Western blot. We found that the G6PD K45 mutation does not alter the level of G6PD acetylation (Fig. 7G-H). To further clarify that K45 is a potential lactylation modification site rather than an acetylation. We analyzed the ability of Acetyl-CoA or L-lactate to bind G6PD K45 with online molecular docking tools. We found that the docking score of L-lactate (208.85) to G6PD K45 was significantly higher than that of Acetyl-CoA (175.97) (Supplementary Fig. 4C). This indicates that G6PD K45 is more prone to lactylation modification rather than acetylation modification. Molecular docking was also used to simulate the binding conformation of G6PD and its coenzyme (catalytic NADP+ and structural NADP+) with or without K45la, K408la and K508la. After the Lys45 side chain modification, the catalytic NADP+ formed hydrogen bond with the modified Lys45, leading to an obvious binding site shift and a reduction in the binding affinity from 12.38 kcal/mol to 8.26 kcal/mol. In contrast, the binding affinity of K408la and K508la showed an increase or a slight decrease, respectively. Therefore, K45la distorted the original binding domain between catalytic NADP+ and G6PD, hence dimishing the G6PD activity (Fig. 7I). Taken together, these results demonstrate that HPV16 E6 inhibits the lactylation of G6PD K45, thereby enhancing its enzymatic activity by facilitating the formation of G6PD dimers.
Fig. 7.
The G6PD K45 lactylation modification reduces its enzymatic activity. (A) Schematic diagram of the G6PD structure (PDB: 2BH9). Each G6PD monomer consists of a catalytic NADP+ and a structural NADP+. Dual G6PD monomers are stacked into a dimer. (B) Species conservation analysis of potential l0 lactylation modification sequence sites for G6PD. (C–E) Re-expression of mutation in PHKs and C33A cells with the knockdown G6PD was used to detect G6PD enzyme activity. (F) Crosslinking with DSS, followed by immunoblotting to detect dimeric and monomeric G6PD. (G–H) Acetylation levels were blotted with a pan-anti-acetyllysine antibody (a-Ac). (I) 3D structure of ligand-receptor interactions shown in the left panel. The right panel shows the 2D representation of the interaction with ligands and the receptors in the binding pocket. Each dot represents an independent biological replicate in the plots. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with indicated groups. Statistical significance was determined using unpaired two-tailed t-test. NS, not significant.
2.6. G6PD activity is vital for HPV-regulated tumor proliferation in vitro and vivo
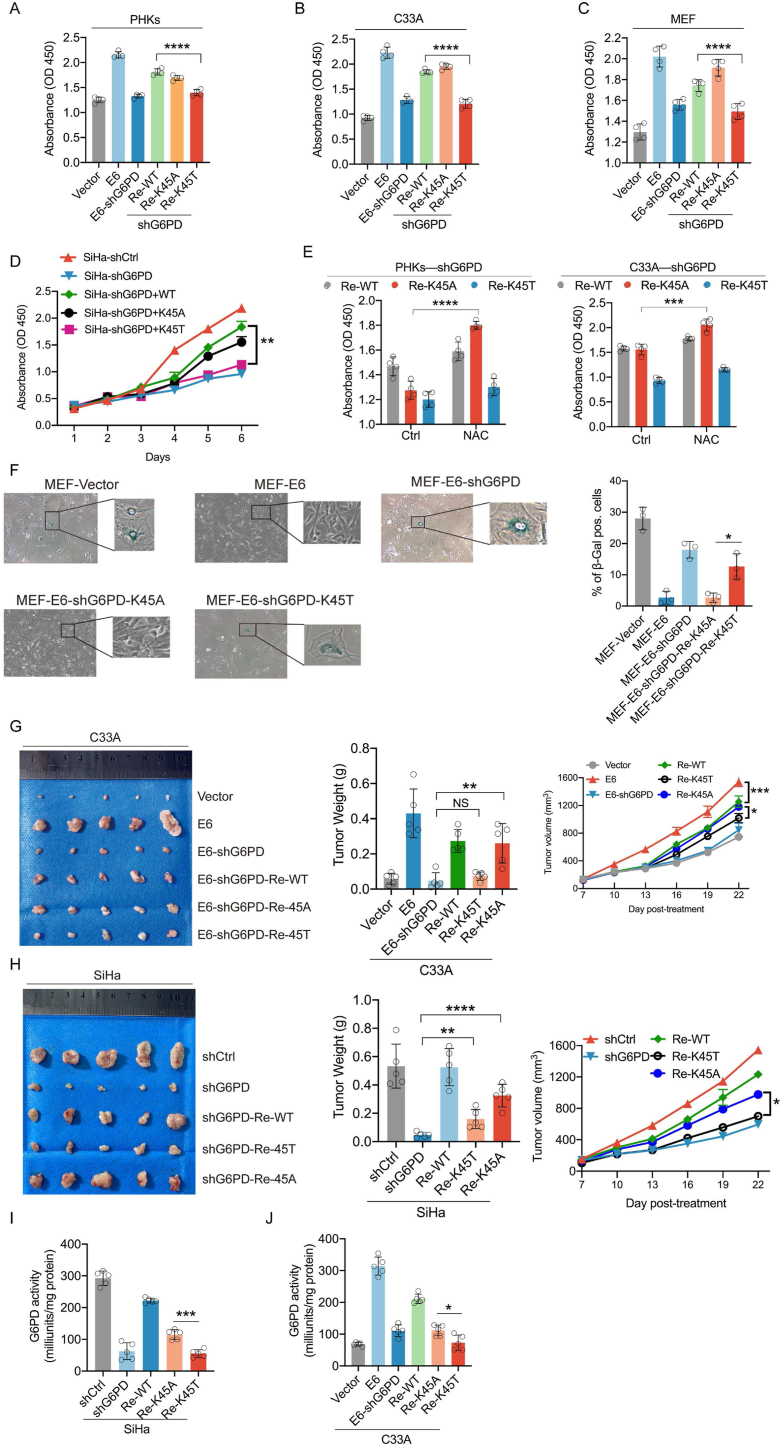
To provide insight to the role of the HPV16 E6–G6PD regulatory axis in HPV carcinogenesis, we investigated the effects of G6PD K45A and K45T mutations on cell growth in vitro. In HPV-16 E6-expressing PHKs, C33A cells, and mouse embryonic fibroblasts (MEFs), in which the endogenous G6PD was knocked down, ectopic expression of the WT G6PD or the K45A mutation, but not the K45T mutation, significantly enhanced cell growth (Fig. 8A–C). The same observation was made with HPV16+ cervical cancer SiHa cells with endogenous G6PD KD (Fig. 8D). G6PD is a critical enzyme for cellular PPP metabolism which produces the essential intracellular reductant NADPH to neutralize cellular ROS. Thus, the medium was supplemented with the ROS scavenger N-acetyl-l-cysteine (NAC) to investigate whether G6PD-mediated oxidative PPP metabolism was involved in the regulation of cell proliferation. Supplementing the medium with NAC further promoted the growth of cells expressing K45A compared to that in cells expressing K45T (Fig. 8E). We also clarified the role of the HPV16 E6–G6PD axis in senescence. β-Galactosidase staining showed that the re-expression of G6PD K45A, but not K45T, significantly reduced MEF senescence (Fig. 8F). Further in vivo xenograft experiments revealed that the stable over-expression of WT G6PD or the K45A mutation, but not K45T, in endogenous G6PD-knockdown SiHa and C33A cells, promoted tumor growth (Fig. 8G-H). Finally, G6PD enzyme activity was detected in C33A and SiHa cell-bearing xenograft tumors. We found that re-expression of the K45A mutation resulted in significantly higher G6PD enzymatic activity than that in turmors with K45T (Fig. 8I-J). In conclusion, these findings suggest that decreased lactylation modification of G6PD to increased enzyme activity, is important for HPV16 E6-mediated tumor cell growth in vitro and in vivo.
Fig. 8.
Elevated levels of G6PD K45 lactylation inhibit cell proliferation in vivo and in vitro (A–C) PHKs, C33A cells, and MEFs were transfected with the indicated plasmids. Cell proliferation was analyzed via cell viability assays. (D) SiHa (HPV16 positive) cells stably expressing shCtrl or shG6PD were further infected with lentiviruses expressing WT G6PD or its mutation, as indicated. (E) G6PD-knockdown cells or those cells rescued by WT G6PD or the K45T or K45A mutation were treated with NAC (2 mM), and cell proliferation was analyzed 5 days after treatment. (F) β-Galactosidase staining was used to detect the level of senescence in MEF cells. Right panel: analysis of β-galactosidase-positive cells. Left panel: representative images. (G–H) Tumors were weighed after mice were euthanized at the endpoint. (I–J) C33A and SiHa cell xenograft tumors were used to determine G6PD enzyme activity. Each dot represents an independent biological replicate in the plots. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with indicated groups. Statistical significance was determined using unpaired two-tailed t-test. NS, not significant. G6PD, glucose-6-phosphate dehydrogenase; NAC, N-acetyl-l-cysteine.
3. Discussion
Dysregulated energy metabolism is widely observed in the rapid proliferation of tumor cells [29], We previously reported that HPV activates the hexosamine biosynthetic pathway to increase the level of O-GlcNAc, promoting the transforming activities of viral oncogenes [14]. Other groups have also reported that HPV enhances aerobic glycolysis to increase ATP levels and promote cervical cancer progression [30]. But whether HPV16 E6E7 regulates energy metabolism through the PPP remains unclear. Furthermore, in addition to requiring energy, a large amount of substrates are required for macromolecule synthesis [31]. In the present study, we identified a significant increase in PPP-related metabolites using LC-MS in cells ectopically expressing HPV16 E6E7, with a corresponding significant increase in levels of the metabolite NADPH and DNA synthesis. These results demonstrate that activation of the PPP by HPV16 E6E7 not only alleviates intracellular oxidative stress and maintains intracellular redox homeostasis but also provides substrates for tumor cell proliferation.
Lactylation is a novel post-translational modification, and previous studies have primarily focused on histone lactylation. Histone lactylation modification promotes macrophage polarization from an M1 to M2 type [10] and regulates expression of the downstream repair-related genes Arg1 and KLF4 [32]. It also promotes the macrophage expression of profibrotic genes [33]. In oncology studies, histone lactylation has been found to promote the expression of the m6A modification-related proteins METTL3 and YTHDF2 to accelerate tumor progression [34,35]. In addition, non-histone lactylation modifications, including HMGB1 [12] and Snail1 [36] lactylation, have been determined to play important roles in promoting inflammatory responses and cardiac fibrosis, respectively.
In a recent proteome-wide lactylation study, numerous non-histone proteins were identified as having lactylation modification sites. KEGG enrichment analysis showed that lactylation-modified proteins were mainly enriched in glycolysis, the TCA, and the PPP [13]. Consistent with these data, we demonstrated that the PPP rate-limiting enzyme G6PD is modified by lactylation, which inhibits G6PD enzyme activity. We identified G6PD K45 as a potential lactylation modification site. Although sustained lactylation was simulated by mutating lysine to threonine, threonine did not exactly match the structure of the lactate group. Molecular docking was used to further analyze the change in binding affinity of coenzyme NADP+ to G6PD protein after lactylation modification at different sites. Only G6PD K45 lactylation modification was predicted to reduce the binding affinity significantly. Thus, we suggest that K45 lactylation modification reduces G6PD enzyme activity.
The rapid proliferation of tumor cells generates large amounts of ROS. High ROS levels are counteracted by increased antioxidant defense mechanisms [37]. Among the key mechanisms to counter oxidative stress is the diversion of the intermediate products of glycolysis to the oxidative PPP [38]. G6PD, the first NADPH-producing enzyme in the PPP, is considered the predominant contributor to the NADPH pool [39]. G6PD overexpression in multiple cancers is associated with a poor disease prognosis [7]. Chang et al. reported that HPV16 E6 increases G6PD expression [40]. However, we found that the expression of G6PD mRNA and protein was not significantly altered in C33A cells and PHKs expressing HPV16 E6 (Fig. 4A–C). G6PD expression was also found to be higher in human cervical squamous cell carcinoma tissue sections than in the normal cervical epithelium in our study (data not presented), possibly a result of a combination of multiple oncogenes. G6PD K45 has been consistently reported as a binding site that regulates G6PD enzyme activity (38, 39). In the present study, we demonstrated the lactylation modification of K45 inhibited the enzymatic activity of G6P, whereas HPV E6 prevent this modification and facilitate the formation of the active G6PD dimer.
In summary, we uncovered a mechanism underlying HPV carcinogenesis in the form of E6-stimulated G6PD enzyme activity, leading to elevated PPP activity. We further identified K45 as a potential site for the lactylation of G6PD. We believe that increased G6PD lactylation in HPV-infected cells or the inhibition of G6PD enzymatic activity with 6-An could be instrumental in reducing the oncogenic effects of HPV. The results of targeting G6PD enzymatic activity in mouse tumor models, described in this study, support this hypothesis and could ultimately prove to be of therapeutic value.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 82020108024) and the Science and Technology Department of Jilin Province (No. 20230101138JC). Science and Technology Department of Jilin Province (No. 20210402005 GH) and the National Key Research and Development Program of China (2023YFE0109800).
CRediT authorship contribution statement
Qingfei Meng: Writing – original draft. Yanghe Zhang: Investigation. Huihui Sun: Resources. Xiangzhe Yang: Data curation. Shiming Hao: Conceptualization. Bin Liu: Conceptualization, Validation. Honglan Zhou: Funding acquisition. Yishu Wang: Investigation. Zhi-Xiang Xu: Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103108.
Contributor Information
Honglan Zhou, Email: hlzhou@jlu.edu.cn.
Yishu Wang, Email: wangys@jlu.edu.cn.
Zhi-Xiang Xu, Email: zhixiangxu@jlu.edu.cn.
Materials and Methods
Plasmids. HPV16 E6, E7, E6E7, GV505 (G6PD), shRNA-Ctrl, shRNA-G6PD, and shRNA-UBE3A constructs were purchased from GeneChem (Shanghai, China).
Cell culture, reagents, and materials. C33A, SiHa, and MEF were maintained in Dulbecco Modified Eagle Medium (DMEM) with 10% FBS (Dalian Meilun Biotechnology Co., Ltd) at 37 °C in 5% CO2. Cell were frozen with serum-free cell cryopreservation (New Cell & Molecular Biotech, C40100, Suzhou, China).
PHKs were isolated from normal cervical epithelial obtained from iCell (HUM-iCell-f016). PHKs were cultured in EpiLife Medium (Thermo Fisher, MEPI500CA) with the addition of EpiLife Defined Growth Supplement (EDGS) (Thermo Fisher, S0125). The Coating Matrix (Thermo Fisher, R011K) was used to enhance the attachment, growth, and population doubling potential of human keratinocytes. For all the experiments, PHKs cultured between the third and fifth passages were used.
MG-132 (HY-13259), 6-Aminonicotinamide (HY–W010342), Puromycin (HY–K1057), Kanamycin (HY-16566), Hygromycin B (HY–K1051) Protein A/G Magnetic Beads (HY–K0202), N-Acetyl-l-cysteine ethyl ester (HY-134495), Recombinant Trypsin Solution (HY–129047B) were purchased from MCE. Mycoplasma removal reagents were purchased from umibio (Umibio, Cat. No: UR51002, China). DSS was purchased from Thermo Fisher Scientific (A39267). Water bath disinfecting solution was purchased from VivaCell (C3480-010, VivaCell, Shanghai, China). Sodium L-lactate was purchased from Sigma-Aldrich (SKU: 71718). SDS-PAGE gel preparation kit (SW108-01) was purchased from Seven/Abcells (Beijing, China).Cell culture dishes/plates (60 mm and 100 mm) and 20-mm glass-bottom dishes were obtained from NEST Biotechnology Co. Ltd (Wuxi, China). Cell culture dishes/plates (35 mm) was obtained from SAINING. 6-well plates purchased from BDBIO (H803-6, HangZhou China).
Animal experiments. All animal experiments were approved by the Animal Research Ethics Committee of Jilin University. Female BALB/c nude mice were purchased from Beijing HFK Bio-Technology Company (Beijing, China) and randomly assigned to experimental groups. For xenograft experiments, C33A or SiHa cell lines were infected with the indicated viruses expressing the proteins of interest or shRNAs. Equal numbers of established stable cells (1 × 106) were injected subcutaneously into nude mice. Starting at 10 or 12 days after injection, tumor volumes were measured every 2 or 3 days using a caliper and calculated using the following equation: volume = width × depth × length × 0.52.
Lentivirus production and stable knockdown/overexpression cell lines construction. To construct cell lines with stable knockdown (endogenous G6PD and UBE3A)/overexpression (HPV16 E6, E7,and E6E7), lentivirues were generated. Briefly, lentiviruses were produced by co-transfecting a lentiviral vector harboring shRNA/HPV16 E6, E7, E6E7, psPAX2 packaging plasmid, and pMD2.G envelope plasmid (Addgene) were co-transfected into HEK293T cells using Transfection Reagent (GeneCopoeia) according to the manufacturer's instructions. Fresh medium was changed 24 h later and lentivirus-containing supernatant medium was collected 48 h after transfection. To construct stable knockdown cells, target cells were infected with harvested lentivirus-containing supernatant for 24 h and subsequently subjected to selection with 2 mg/mL puromycin. Knockdown efficiency was confirmed by western blotting.
Immunoprecipitation. Immunoprecipitation was performed as described previously [14]. Briefly, about 500 μg of total cellular proteins were incubated with G6PD antibody overnight at 4 °C followed by adding 30 μL of Protein A/G Magnetic Beads (MCE HY-K0202). The rotary mixer (MX-RL-E, DLAB Scientific Co., Ltd, China) was placed in a 4-degree refrigerator for adequate mixing. The precipitates were washed seven times with lysis buffer and boiled in sodium dodecyl sulfate sample buffer. The supernatant was subjected to immunoblotting with the appropriate antibodies.
Western blot. cells were lysed with RIPA lysis buffer containing a cocktail of protease and phosphatase inhibitors (Solarbio). The protein concentration of lysates was measured by BCA (SW101-02, SEVEN, Beijing, China) assays and was adjusted to the same final concentration. After heat denaturation, equal amounts of protein in the lysates were separated by SDS-PAGE (and then transferred to a polyvinylidene fluoride (PVDF, Millipore) membrane. The membranes were blocked with 5% non-fat milk in tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 h at room temperature followed by incubation with the indicated primary antibodies at 4 °C overnight on a horizontal shaker (SK-D1810-S, DLAB Scientific Co., Ltd, China). After three washes with TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. The immunoreactive proteins were then detected using an enhanced chemiluminescent (ECL) substrate and visualized with enhanced chemiluminescence (ECL) system (Tanon). The following antibodies were used: p53 (cst # 2527S), Rb (cst # 9309S), LDHA (abcam # ab52488), UBE3A (cst # 7526S), Pan-L-lactyl-lysine (PTM BioLabs # PTM-1401RM), Alpha Tubulin (66031-1-Ig), GAPDH (proteintech # 60004-1-Ig), and G6PD (abcam # ab210702).
Quantitation of mRNA expression via real-time quantitative RT-PCR (qRT-PCR). Cells were washed with PBS (A19711, Chuanqiu Biotechnology Co. Ltd, China.) and suspended in 1 ml of AG RNAex Pro Reagent (AG21101, BIOTECHNOLOGY(HUNAN)CO., LTD,ChangSha,China) and then stored at −80 °C. Total RNA was purified from thawed samples using standard techniques, and cDNA was synthesized using the Hifair® Ⅲ 1st Strand cDNA Synthesis SuperMix Kit (Cat No. 11137ES60, Yeasen, Shanghai, China), according to the manufacturer's instructions. Real-time quantitative RT-PCR (qRT-PCR) was performed using a 7300 Real-Time PCR Detection System (Applied Biosystems). Sequences of primers used for real-time PCR. The sequences of primers used are listed. LDHA Forward: ATGGCAACTCTAAAGGATCA. Reverse: GCAACTTGCAGTTCGGGC. G6PD Forward: AAGAACGTGAAGCTCCCTGA. Reverse: AATATAGGGGATGGGCTTGG. GAPDH Forward: TGAAGGTCGGAGTCAACGGATT. Reverse: CTTCTCCATGGTGGTGAAGAC.
Cell viability assay. Two thousand cells were seeded in each well of a 96-well plate and allowed to adhere overnight. Cell viability was determined 6 days after treatment with or without 6-An using the CCK8 (KGA9310-1000, Keygen BioTECH) assay. The plates were read using a Synergy H1 microplate reader (BioTek Instruments) at a wavelength of 450 nm.
EdU incorporation assay. EdU incorporation assays were performed using a Cell-Light EdU Apollo 488 In Vitro Imaging Kit (Beyotime Company, Shanghai, China) according to the manufacturer's instructions. Images were acquired using an Olympus DP70 microscope (Olympus), and EdU-positive cells were counted.
Immunofluorescence. Cells cultured on coverslips were pre-seeded 1 day before immunofluorescence analysis, with a final confluence of 70–80%. The cells were fixed in 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 5 min, blocked with 5% bovine serum albumin, and incubated with the indicated antibodies, followed by Texas Red-conjugated anti-rabbit IgG and fluorescein isothiocyanate-conjugated anti-mouse IgG. The cells were mounted with DAPI-containing medium (Helixgen), and images were acquired using a microscope (Olympus).
Metabolic measurements. The extracellular lactate was measured using CheKine™ Micro Lactate Assay Kit (Abbkine) according to the manufacturer's instructions. Values were normalized to protein concentrations. Intracellular NADPH was measured using cell lysates with an NADPH assay kit (Beyotime) according to the manufacturer's instructions. The intracellular GSH and GSH/GSSH ratios were measured using cell lysates with a GSH assay kit (Beyotime) according to the manufacturer's instructions and normalized to the protein concentration. G6PD enzyme activity was determined by using a Solarbio kit (BC0265) according to the manufacturer's instruction.
Crosslinking assay. DSS crosslinking was used to detect the G6PD dimers. Briefly, cells were trypsinized and counted. Equal numbers of cells were collected for all experiments. Cells were washed with cold PBS, followed by suspension in conjugation buffer (20 mM HEPES, pH 8.0). The DSS solution prepared in DMSO was added to the cell suspension at a final concentration of 2 mM. After incubation at 37 °C for 30 min, the samples were boiled and used for Western blot assays.
Senescence β-galactosidase staining assay. The Senescence β-Galactosidase Staining Kit (Beyotime, China) was used to stain senescent cells. According to the manufacturer's instructions, the cells were fixed for 15 min, stained for 1 day, and analyzed. Senescent cells, identified based on blue staining, were captured using light microscopy.
Measurement of intracellular ROS. The amount of intracellular ROS was measured by detecting dichlorodihydrofluorescein, the cleavage product of carboxy-H2DCFDA (Invitrogen), following the manufacturer's instructions. Briefly, 2 × 105 cells were seeded in 6-well plates. Twenty-four hours later, cells were rinsed with PBS and loaded with 5 μM carboxy-H2DCFDA for 30 min at 37 °C. The cells were harvested, resuspended in PBS, and analyzed using FACS (BD Biosciences, USA; excitation and emission at 490 and 530 nm, respectively).
Measurement of intracellular H2O2. The concentration of H2O2 was measured by a micro H2O2 assay kit (BC3590, Solarbio, Beijing, China). Approximately 5x10^6 of cell samples were homogenised and then mixed in 1000 μL lysate. The supernatant was blended and homogenised with the same amount of H2O2 detection reagent after centrifuging for 5 min at 4 °C at 12,000g. The standard curve was used to calculate the accumulation of H2O2 and the absorbance of homogenate was detected at 415 nm.
Protein carbonylation measurements. Cells were grown in 100 mm culture dishes, trypsinized, pelleted, and dis-solved in 0.1% Triton-X 100 in PBS. Equal amounts of cellular protein from treated and untreated samples, as determined by a BCA protein assay, were then used to determine protein carbonyl levels. Each sample was divided into two and mixed with either 10 mM 2,4-dinitrophenylhydrazine in 2.5 M HCl or 2.5 M HCl alone and allowed to incubate in the dark for 1 h with vortex mixing every 15min. Trichloroacetic acid [20% (w/v)] was then added to samples to a final concentration of 10% (w/v), left on ice for 10 min, and then centrifuged for 5 min at 4 °C using a table top centrifuge to collect protein precipitates. Supernatant was discarded and a second wash with 10% (w/v) trichloroacetic acid was performed. Samples were then washed three times in ethanol-ethyl acetate (1:1; v/v) to remove free DNPH after which precipitates were dissolved in 6 M guanidine hydrochloride and left for 10 min with intermittent vortexing. Absorbance at 365 nm was then obtained using a (Tecan Infinite 200) spectropho-tometer. The difference in values from derivatized (DNPH) and control (only HCl) subsamples were then determined. The molar extinction coefficient of dinitrophenylhydrazine (ε of 22,000 M−1) was then used to calculate carbonyl values (nmol/μg protein). Determine the concentration of the carbonyl content by inserting the Corrected Absorbance into the following equation: CC (nmol/well) = [(A 365 nm)/22 mM−1cm−1 x 0.2893 cm] x (100 μL).
Liquid chromatography-mass spectrometry (LC-MS) analysis of cell metabolites. Approximately 5 × 106 cells were washed twice with cold PBS, and polar metabolites were immediately extracted with ice-cold 80% methanol. Samples were subjected to freeze–thaw cycles or sonication to extract the metabolites. The supernatants were collected and dried. The powder containing metabolites was dissolved in 80% methanol to run LC-MS. For the kinetic LC-MS analyses, a Shimadzu Nexera × 2 UHPLC combined with a Sciex 5600 Triple Time of Flight-Mass Spectrometry (TOFMS) was used, which was controlled by Sciex Analyst 1.7.1 instrument acquiring software. A Supelco Ascentis Express HILIC Acquity UPLC BEH Amide (150 cm × 2.1 mm, 1.7 μm) column was used with mobile phase (A) consisting of 5 mM ammonium formate and 0.05% formic acid; mobile phase (B) consisting of 90% acetonitrile (ACN) and 10% water. Gradient program: mobile phase (A) was held at 15% for 0 min and then increased to 30% in 7 min; then to 60% in 6 min and held for 1 min before returning initial condition. The column was held at 40 °C and 5 μl of sample was injected into the LC-MS with a flow rate of 0.2 ml/min. Automatic calibrations of TOFMS were achieved with average mass accuracy of <2 ppm. Data Processing Software included Sciex PeakView 2.2, MasterView 1.1, and MultiQuant 3.0.2.
Molecular Docking. Auto-Dock 4.0 was used to dock small molecules (substrates) into their enzyme structures with or without lysine lactylation (Kla) modification, respectively. The original G6PD enzyme structure (2BH9) was achieved from the PDB database (https://www.pdbus.org/). The enzyme structure with Kla was generated by modifying the lysine side chain at lysine 45, 408, and 508, respectively, using the ChimeraX-1.2.5 software. The structure of small molecules(catalytic NADP+ and structural NADP+) was isolated from 2BH9. In this study, the binding of NADP+ (catalytic or structural) and G6PD was simulated. Before the docking simulation, G6PD was placed into the substrate binding site of enzyme as the start point of docking. The conformation with the lowest binding energy of G6PD was considered as the enzyme-bound conformation. By comparing the binding energy and conformation of the enzymes with or without Kla modification, we infer the likely effect of the modification on the enzymatic activity.
Statistical analysis. A two-tailed Student's t-test was used to generate p values in studies in which statistical analyses were performed, except a two-way ANOVA was used for cell proliferation assay and tumor growth analysis. p values less than or equal to 0.05 were considered significant. Data with error bars represent mean ± SD. Statistical analysis and graphical presentation was performed using Prism 8.0 (GraphPad).
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supplementary Fig 1 (A) PHKs, MEF and C33A cells were stably transduced with lentiviruses expressing vector or HPV16 E7. (B–C) Cell proliferation rates were determined by CCK8 assay. (D) PI and Annexin-V staining was used to examine apoptosis in cytoplasm of C33A cells. Representative images detected by flow cytometry were shown (left). And the corresponding histogram was shown on the right side of the image. (E) Morphology of MEF-Vector and MEF-E6E7. (F) Cell area and Ratio length to width were used to assess the level of cellular senescence. (G) β-Galactosidase Staining was used to detect the level of senescence in MEF cells. Each dot represents an independent biological replicate in the plots. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with indicated groups. Statistical significance was determined using unpaired two-tailed t-test. NS, not significant.
Supplementary Fig. 2 (A-B) IC50 of 6-An against C33A and PHKs cells by CCK8 assay. (C) Overlap-PCR was used for the construction of mutation plasmids. Sanger sequencing was used to verify whether lysine was mutated to alanine (GCG). (D) Sanger sequencing was used to verify whether lysine was mutated to threonine (ACG) (right).
Supplementary Fig. 3 G6PD-knockdown cells and those cells rescued by the K45A or K45T mutation were subjected to determinations of intracellular NADPH (A), GSH (B), NADP/NADPH (C), GSH/GSSH (D), ROS (E-F) H2O2 (G) protein carbonylation levels (H). Each dot represents an independent biological replicate in the plots. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with indicated groups. NS, not significant. Statistical significance was determined using unpaired two-tailed t-test.
Supplementary Fig 4 (A) Illustration of Kla structure (left) and L-Threonine (right). (B) (①, ②) Overview structure, detailed 3D protein-ligand interaction analysis of NADP+ bound to G6PD. (③, ④) Overview structure, detailed 3D protein-ligand interaction analysis of NADP+ (K45T) bound to G6PD.(C) Online molecular docking tools (https://cadd.labshare.cn/cb-dock2/index.php) were used to analyze dock scores for acetyl coenzyme A, lactate and G6PD separately.
References
- 1.Schiffman M., et al. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Prim. 2016;2 doi: 10.1038/nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y., et al. Nuclear lactate dehydrogenase A senses ROS to produce α-hydroxybutyrate for HPV-induced cervical tumor growth. Nat. Commun. 2018;9(1):4429. doi: 10.1038/s41467-018-06841-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tommasino M. The human papillomavirus family and its role in carcinogenesis. Semin. Cancer Biol. 2014;26:13–21. doi: 10.1016/j.semcancer.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 6.Meng Q., et al. Recent findings in the regulation of G6PD and its role in diseases. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.932154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patra K.C., Hay N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014;39(8):347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma X., et al. Polo-like kinase 1 coordinates biosynthesis during cell cycle progression by directly activating pentose phosphate pathway. Nat. Commun. 2017;8(1):1506. doi: 10.1038/s41467-017-01647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng Q., et al. Lactylation stabilizes DCBLD1 activating the pentose phosphate pathway to promote cervical cancer progression. J. Exp. Clin. Cancer Res. : CR. 2024;43(1):36. doi: 10.1186/s13046-024-02943-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D., et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L., et al. Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat. Metab. 2020;2(9):882–892. doi: 10.1038/s42255-020-0267-9. [DOI] [PubMed] [Google Scholar]
- 12.Yang K., et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 2022;29(1):133–146. doi: 10.1038/s41418-021-00841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan N., et al. Cyclic immonium ion of lactyllysine reveals widespread lactylation in the human proteome. Nat. Methods. 2022;19(7):854–864. doi: 10.1038/s41592-022-01523-1. [DOI] [PubMed] [Google Scholar]
- 14.Zeng Q., et al. O-linked GlcNAcylation elevated by HPV E6 mediates viral oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 2016;113(33):9333–9338. doi: 10.1073/pnas.1606801113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Q., et al. LKB1 inhibits HPV-associated cancer progression by targeting cellular metabolism. Oncogene. 2017;36(9):1245–1255. doi: 10.1038/onc.2016.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamid N.A., Brown C., Gaston K. The regulation of cell proliferation by the papillomavirus early proteins. Cell. Mol. Life Sci. : CMLS. 2009;66(10):1700–1717. doi: 10.1007/s00018-009-8631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia A.A., Koperniku A., Ferreira J.C.B., Mochly-Rosen D. Treatment strategies for glucose-6-phosphate dehydrogenase deficiency: past and future perspectives. Trends Pharmacol. Sci. 2021;42(10):829–844. doi: 10.1016/j.tips.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butterfield D.A., Dalle-Donne I. Redox proteomics: from protein modifications to cellular dysfunction and disease. Mass Spectrom. Rev. 2014;33(1):1–6. doi: 10.1002/mas.21404. [DOI] [PubMed] [Google Scholar]
- 19.Jiang P., et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 2011;13(3):310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Zapien D., et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529(7587) doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotaka M., et al. Structural studies of glucose-6-phosphate and NADP+ binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005;61(Pt 5):495–504. doi: 10.1107/S0907444905002350. [DOI] [PubMed] [Google Scholar]
- 22.Koperniku A., Garcia A.A., Mochly-Rosen D. Boosting the discovery of small molecule inhibitors of glucose-6-phosphate dehydrogenase for the treatment of cancer, infectious diseases, and inflammation. J. Med. Chem. 2022;65(6):4403–4423. doi: 10.1021/acs.jmedchem.1c01577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horikoshi N., et al. Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency. Proc. Natl. Acad. Sci. U. S. A. 2021;118(4) doi: 10.1073/pnas.2022790118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Au S.W., et al. Solution of the structure of tetrameric human glucose 6-phosphate dehydrogenase by molecular replacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55(Pt 4):826–834. doi: 10.1107/s0907444999000827. [DOI] [PubMed] [Google Scholar]
- 25.Cancedda R., Ogunmola G., Luzzatto L. Genetic variants of human erythrocyte glucose-6-phosphate dehydrogenase. Discrete conformational states stabilized by NADP + and NADPH. Eur. J. Biochem. 1973;34(1):199–204. doi: 10.1111/j.1432-1033.1973.tb02746.x. [DOI] [PubMed] [Google Scholar]
- 26.Notaro R., Afolayan A., Luzzatto L. Human mutations in glucose 6-phosphate dehydrogenase reflect evolutionary history. Faseb. J. 2000;14(3):485–494. doi: 10.1096/fasebj.14.3.485. [DOI] [PubMed] [Google Scholar]
- 27.Kletzien R.F., Harris P.K., Foellmi L.A. Glucose-6-phosphate dehydrogenase: a "housekeeping" enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. Faseb. J. 1994;8(2):174–181. doi: 10.1096/fasebj.8.2.8119488. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y.P., et al. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. EMBO J. 2014;33(12):1304–1320. doi: 10.1002/embj.201387224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 30.Hu C., et al. HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m6A-MYC expression. Int. J. Biol. Sci. 2022;18(2):507–521. doi: 10.7150/ijbs.67770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu K., et al. HIF-1α regulates cellular metabolism, and Imatinib resistance by targeting phosphogluconate dehydrogenase in gastrointestinal stromal tumors. Cell Death Dis. 2020;11(7):586. doi: 10.1038/s41419-020-02768-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irizarry-Caro R.A., et al. TLR signaling adapter BCAP regulates inflammatory to reparatory macrophage transition by promoting histone lactylation. Proc. Natl. Acad. Sci. U. S. A. 2020;117(48):30628–30638. doi: 10.1073/pnas.2009778117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui H., et al. Lung myofibroblasts promote macrophage profibrotic activity through lactate-induced histone lactylation. Am. J. Respir. Cell Mol. Biol. 2021;64(1):115–125. doi: 10.1165/rcmb.2020-0360OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J., et al. Histone lactylation drives oncogenesis by facilitating m(6)A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021;22(1):85. doi: 10.1186/s13059-021-02308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong J., et al. Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell. 2022;82(9):1660–1677. doi: 10.1016/j.molcel.2022.02.033. [DOI] [PubMed] [Google Scholar]
- 36.Fan M., et al. Lactate promotes endothelial-to-mesenchymal transition via Snail1 lactylation after myocardial infarction. Sci. Adv. 2023;9(5):eadc9465. doi: 10.1126/sciadv.adc9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorrini C., Harris I.S., Mak T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013;12(12):931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 38.Cheung E.C., Vousden K.H. The role of ROS in tumour development and progression. Nat. Rev. Cancer. 2022;22(5):280–297. doi: 10.1038/s41568-021-00435-0. [DOI] [PubMed] [Google Scholar]
- 39.Berneburg I., Rahlfs S., Becker K., Fritz-Wolf K. Crystal structure of Leishmania donovani glucose 6-phosphate dehydrogenase reveals a unique N-terminal domain. Commun. Biol. 2022;5(1) doi: 10.1038/s42003-022-04307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang Y.F., et al. HPV16 E6 promotes the progression of HPV infection-associated cervical cancer by upregulating glucose-6-phosphate dehydrogenase expression. Front. Oncol. 2021:11. doi: 10.3389/fonc.2021.718781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.