Abstract
Vaccinia virus has two forms of infectious virions: the intracellular mature virus and the extracellular enveloped virus (EEV). EEV is critical for cell-to-cell and long-range spread of the virus. The B5R open reading frame (ORF) encodes a membrane protein that is essential for EEV formation. Deletion of the B5R ORF results in a dramatic reduction of EEV, and as a consequence, the virus produces small plaques in vitro and is highly attenuated in vivo. The extracellular portion of B5R is composed mainly of four domains that are similar to the short consensus repeats (SCRs) present in complement regulatory proteins. To determine the contribution of these putative SCR domains to EEV formation, we constructed recombinant vaccinia viruses that replaced the wild-type B5R gene with a mutated gene encoding a B5R protein lacking the SCRs. The resulting recombinant viruses produced large plaques, indicating efficient cell-to-cell spread in vitro, and gradient centrifugation of supernatants from infected cells confirmed that EEV was formed. In contrast, phalloidin staining of infected cells showed that the virus lacking the SCR domains was deficient in the induction of thick actin bundles. Thus, the highly conserved SCR domains present in the extracellular portion of the B5R protein are dispensable for EEV formation. This indicates that the B5R protein is a key viral protein with multiple functions in the process of virus envelopment and release. In addition, given the similarity of the extracellular domain to complement control proteins, the B5R protein may be involved in viral evasion from host immune responses.
Vaccinia virus has two forms of infectious virions: intracellular mature virus (IMV) and enveloped forms of virus that bear an additional membrane containing specific viral proteins. The enveloped virus is formed by the wrapping of IMV by trans-Golgi network vesicles containing viral envelope proteins (24, 34, 38). The enwrapped virion moves to the plasma membrane where it exits the cell and either remains associated with the outer plasma membrane as cell-associated enveloped virus (CEV) (3) or is released from the cell as extracellular enveloped virus (EEV). Formation of enveloped virus is critical for cell-to-cell and distant spread of virus in vitro and in vivo (31).
Six proteins encoded by vaccinia virus genes have been identified as constituents of this outer virus envelope including B5R (11, 20, 26), F13L (2, 17, 35, 36), A36R (30), A34R (5, 9, 27), A33R (33), and A56R (6, 37). Deletion of the B5R, F13L, and A36R genes result in decreased EEV formation, and these mutants form small plaques in tissue culture and are highly attenuated in vivo (2, 12, 30, 35, 42). In contrast, deletion of the EEV-specific protein encoded by the A34R gene results in increased liberation of EEV (27, 43). Enhanced EEV release is also seen with a point mutation in the sequence of A34R found in certain vaccinia virus strains (5). This increase in EEV is the result of a diminished retention of CEV at the cell surface.
The B5R open reading frame (ORF) encodes a 42-kDa glycosylated type I membrane protein that is found both on the membranes of infected cells and on EEV but not on IMV (11, 20, 26). The protein is highly conserved among multiple strains of vaccinia virus as well as other poxviruses including variola virus (12). The B5R protein ectodomain sequence is similar to the sequences of a group of proteins involved in the regulation of complement activation (11, 13, 41) and contains four conserved short consensus repeat (SCR) units found in eukaryotic complement control proteins (25). Interestingly, the B5R ORF is one of two vaccinia virus genes that encode proteins with similarity to the superfamily of complement control proteins. The other is a secreted 35-kDa protein encoded by the C3L ORF that has been shown to regulate complement activation (19, 22, 23, 28, 29). The family of complement control proteins is involved in the regulation of complement activation at early steps in a cascade of events that can ultimately lead to the lysis of a foreign organism or an infected cell. The B5R protein may protect infected cells and EEV from the host’s complement-mediated attack.
Because of the critical role the B5R protein plays in EEV morphogenesis and a possible, yet undetermined, role in counteracting the host’s complement defense, we have investigated the contribution of the extracellular portion of the protein to EEV formation. We report here our findings on mutant vaccinia viruses lacking the SCR domains. We studied the effect of the SCR deletion in two parental strains of vaccinia virus: a strain that releases high levels of EEV (IHD-J) and a strain that releases significantly less EEV (WR).
MATERIALS AND METHODS
Cells.
BSC-1, CV-1, and RK13 cells were maintained in minimum essential medium (MEM) with Earle’s salts (LTI) containing 10% fetal bovine serum (FBS), and STO cells were maintained in Dulbecco’s modified Eagle medium (LTI, Gaithersburg, Md.) containing 10% FBS. Cell lines were obtained from the American Type Culture Collection. Vaccinia virus infections of cells were carried out in media containing 2.5% FBS and incubated at 37°C in a 5% CO2 atmosphere.
Antibodies.
The polyclonal rabbit antibody C′-B5R was raised against a peptide corresponding to a region in the ectodomain of the B5R protein (Fig. 1) and has been previously described (20). The rat monoclonal antibody to the EEV-specific protein, P37, was a generous gift from G. Hiller (17).
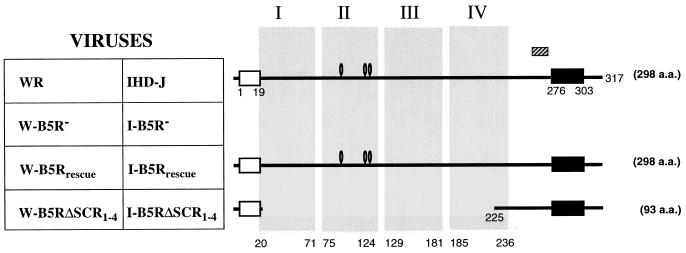
FIG. 1.
Schematic of the B5R proteins produced by WR- and IHDJ-based recombinant vaccinia viruses. Viruses have the wild-type B5R ORF (WR and IHD-J), B5R ORF deleted (W-B5R− and I-B5R−), wild-type B5R ORF reinserted into the deletion mutant (W-B5Rrescue and I-B5Rrescue), or an in-frame deletion of all four SCRs (W-B5RΔSCR1–4 and I-B5RΔSCR1–4). The schematic of the B5R proteins produced by these viruses shows the signal peptide (white box), transmembrane domain (black box), four putative SCR domains (shaded regions I to IV), potential N-linked glycosylation sites (0), and position of the 15-aa peptide used to generate antibody C′-B5R (hatched box). The numbers correspond to the residues of the translated protein. The predicted number of amino acids in the mature protein is indicated at the right.
Construction and in-frame deletion of the SCR domains of the B5R protein.
Construction of an in-frame mutation deleting the four putative SCRs of the B5R protein was accomplished by a series of restriction enzyme digestions of a previously described starting plasmid (pSI-80) that contains the B5R ORF and flanking vaccinia virus (strain WR) sequences (42). Initially, an NspI site at nucleotide position 37 in the vector portion of pSI-80 was eliminated by removal of a 477-bp KpnI/AatII region, followed by blunting with T4 DNA polymerase and ligation, resulting in plasmid pSI-112e. The in-frame deletion of the region encoding the four SCRs was achieved by digestion of pSI-112e with NspI, isolation of the 2,481- and 867-bp fragments, and ligation to form plasmid pSI-113b. Sequencing of the plasmid confirmed the orientation and the proper in-frame nature of the deletion of 612 bp comprising the four putative SCRs. Figure 1 schematically depicts the protein resulting from such an in-frame deletion.
Isolation and plaque purification of recombinant viruses.
The previously constructed B5R deletion mutants, W-B5R− and I-B5R− (42), derived from virus strains WR and IHD-J, respectively, were used as parental viruses for making the new recombinant viruses. The B5R deletion viruses have 710 bp of the B5R ORF deleted and replaced with guanine phosphoribosyltransferase (gpt) and β-galactosidase (β-gal) cassettes driven by vaccinia virus promoters. These viruses express no B5R protein and make small blue plaques with the addition of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and can grow in the presence of mycophenolic acid (42). To create the SCR-deleted viruses or to rescue the parental viruses with the wild-type gene, we utilized W-B5R− and I-B5R− viruses as the parental virus and introduced either the SCR-deleted ORF (pSI-113b) or the wild-type ORF (pSI-80) by homologous recombination following standard protocols (10). Recombinant viruses were then isolated by reverse gpt selection (18). This high-efficiency selection procedure takes advantage of the fact that the parental viruses (W-B5R− and I-B5R−) contain the gpt gene replacing the B5R ORF. Thus, the antimetabolite 6-thioguanine will inhibit the growth of parental (gpt+) virus, but not the growth of the new recombinant viruses in which the gpt gene is replaced by the B5R gene. Briefly, confluent monolayers of CV-1 cells were infected with parental virus (W-B5R− or I-B5R−) at a multiplicity of infection (MOI) of 0.05 PFU/cell. After 2 h, the infected cells were transfected with calcium phosphate-precipitated plasmid DNA. After 48 h, the infected monolayer was harvested and virus was released by successive freeze-thawing and sonication. Progeny virus was then subjected to three rounds of plaque purifications using STO cells. After an initial 2-h infection period, cells were overlaid with a 1:1 mixture of 2% low melting point (LMP) agarose (LTI) and 2× MEM (LTI) containing 5% FBS and 0.2 mM 6-thioguanine. After 48 to 72 h, individual plaques stained with neutral red were picked. After three successive rounds of plaque purifications, virus stocks were grown in BSC-1 cells. Rescue of the B5R deletion viruses with wild-type B5R ORF (pSI-80) resulted in a WR-based virus (vSI-21) which is hereafter referred to as W-B5Rrescue and the corresponding IHDJ-based virus (vSI-25) is called I-B5Rrescue. The WR-based virus containing deletions of all four SCRs (vSI-22) is referred to as W-B5RΔSCR1–4, and the corresponding IHDJ-based virus (vSI-26) is called I-B5RΔSCR1–4 (Fig. 1).
Southern blot.
Viral DNA was isolated from infected cells and digested with SnaBI (Promega) following standard protocols (10). The digested DNA was separated on 1.2% agarose gels, transferred to Immobilon-S membrane (Millipore), hybridized with a biotinylated B5R probe (a 905-bp fragment isolated from an EcoRV digestion of pSI-80), and visualized by nonradioactive means, using a NEBlot Phototope Kit (New England BioLabs).
Western blot.
Western blots were carried out using the polyclonal rabbit serum C′-B5R as previously described (20). For B5R expression in infected-cell lysates, individual wells of a 24-well plate containing BSC-1 cells were infected, incubated for 48 h, and lysed in 200 μl of lysis buffer (0.5% Triton X-100–20 mM Tris [pH 7.0] in phosphate-buffered saline [PBS]). For detecting B5R in EEV, the media from T150 flasks containing infected RK13 cells were removed at 48 h postinfection, clarified by low-speed centrifugation, and then spun in a Beckman SW41 rotor at 14,000 rpm for 1 h at 4°C. The virus was then repelleted through a 36% sucrose cushion in an SW41 rotor at 15,000 rpm for 80 min. Pellets were suspended in 200 μl of lysis buffer. Samples (14 μl) were mixed with Laemmli loading buffer and 2-mercaptoethanol, boiled, electrophoresed through 0.1% sodium dodecyl sulfate (SDS)–13% polyacrylamide gels, and transferred onto nitrocellulose (Schleicher & Schuell). The blot was probed with the antibody C′-B5R (1:3,000) followed by goat anti-rabbit immunoglobulin G conjugated to the enzyme horseradish peroxidase (Boehringer Mannheim) at a dilution of 1:10,000. Visualization of bands was accomplished by nonradioactive means, using Renaissance Chemiluminescence Reagent (DuPont NEN).
Plaque phenotype.
The plaque phenotype was examined on BSC-1 cells under both agarose and liquid overlays. A monolayer of cells was infected at a low MOI and then overlaid with a mixture of media and agarose, and 24 h later, representative individual plaques were photographed under phase contrast at a ×100 magnification. Monolayers contained in a six-well plate were infected and kept under liquid overlay for 48 h and then photographed after crystal violet staining.
PCR amplification of A34R ORF.
Viral DNA was subjected to PCR amplification (25 cycles, with 1 cycle consisting of 2 min at 94°C, 2 min at 55°C, and 1 min at 72°C) using A34R-specific oligonucleotides (olSI-73 [5′-CGGGATCCATGAAATCGCTTAATAG-3′] and olSI-74 [5′-GGAATTCCTCACTTGTAGAATTTTTTAACAC-3′]). The resulting 523-bp PCR product was separated on a low-melting-point gel, and the isolated product was digested with MseI as previously described (27). The resulting fragments were separated on a 4% Nusieve gel, and bands were visualized after ethidium bromide staining.
Analysis of IMV and EEV.
BSC-1 monolayers were infected at a high MOI to achieve one-step growth conditions as previously described (42). At various time points, supernatants were collected and the cells were harvested and lysed by freeze-thawing and sonication. Samples were titered on BSC-1 cells.
To compare the physical quantities of EEV released from cells infected with the recombinant viruses, virions were metabolically labeled during infection. Individual wells of a six-well plate containing monolayers of RK13 cells were infected at a MOI of 10. Six hours after infection, the inoculum was removed and replaced with 0.95 ml of methionine- and cysteine-free media (ICN) containing 100 μCi of Tran35S-Label (ICN) and supplemented with 0.05 ml of complete medium. At 24 h after infection, 0.8 ml of complete medium with 2.5% FBS was added and the incubation was continued for another 24 h. At 48 h postinfection, the cell supernatants were clarified by low-speed centrifugation and placed directly over a step gradient containing cesium chloride (CsCl) in 10 mM Tris (pH 9) at a density of 1.30 g/ml (2 ml), 1.25 g/ml (3 ml) and 1.20 g/ml (4 ml) as previously described (32). The loaded gradients were spun in an SW-41 rotor at 30,000 rpm for 2 h at 20°C. Fractions from the gradient were collected from the bottom of the tube, and an aliquot from each fraction was mixed with 3 ml of EcoLume (ICN) and counted in a Beckman liquid scintillation spectrometer. The densities of selected fractions were determined by measuring the refractive index.
Electron microscopy.
Monolayers of RK13 cells were infected with each WR-based virus and 16 h later fixed in 2.5% glutaraldehyde for 2 h at room temperature and then harvested by gentle scrapping. The cell pellet was postfixed in 1% osmium tetroxide and prestained overnight in saturated uranyl acetate and then embedded in Epon. Ultrathin sections were prepared from the embedded samples, collected on grids, and stained with lead citrate and examined with an electron microscope.
Immunofluorescence and phalloidin staining.
CV-1 cells were grown to 70% confluency on coverslips and were then either mock-infected or infected at a MOI of 5 PFU/cell. After infection, the virus inoculum was replaced with MEM–2% FBS, and the culture was incubated at 37°C for 7 h. Cells were fixed with 4% paraformaldehyde for 10 min at room temperature, washed with PBS, and made permeable with 0.1% Triton X-100 for 15 min at room temperature. After washing, cells were incubated for 5 min in PBS–0.1 M glycine. They were then incubated with a monoclonal antibody that detects the EEV-specific protein P37 (17). The hybridoma supernatant was diluted 1:50 in PBS containing 20% FBS and incubated with the cells for 30 min at room temperature. Staining was detected with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-rat antibody (Dako) used at a 1:50 dilution. To visualize filamentous actin, tetramethyl rhodamine isocyanate (TRITC)-phalloidin (Sigma) in PBS-FBS was added to the cells at a final concentration of 0.33 ng/ml and incubated for 30 min at room temperature. Cells were washed in PBS and mounted with Fluorsave (Calbiochem).
RESULTS
Construction of recombinant vaccinia viruses lacking the four putative SCR domains.
Because the B5R protein is important in the formation of EEV and thus critical for cell-to-cell and long-range spread of vaccinia virus, we sought to map domains of the B5R protein that are involved in the envelopment process. Since the B5R protein has a large, highly conserved ectodomain, we focused on this region. We constructed a transfer plasmid that replaced the wild-type B5R ORF with a mutated one lacking the four putative SCR units that comprise the majority of the extracellular domain of the B5R protein (Fig. 1). To analyze the effect this mutation would have on the resulting recombinant virus, we selected two distinct parental strains of vaccinia virus that differ widely in the amounts of EEV released. Strain IHD-J releases high levels of EEV and forms comet-shaped plaques in tissue culture, whereas strain WR releases significantly less EEV and forms large round plaques in tissue culture (1, 31). The mutated B5R ORF with an in-frame deletion of 204 amino acids (aa) was introduced into previously constructed B5R deletion viruses, W-B5R− and I-B5R− (42), and recombinant viruses W-B5RΔSCR1–4 and I-B5RΔSCR1–4 were isolated (Fig. 1). As a control, the wild-type B5R ORF was also reintroduced back into B5R deletion viruses W-B5R− and I-B5R−, resulting in W-B5Rrescue and I-B5Rrescue, respectively (Fig. 1).
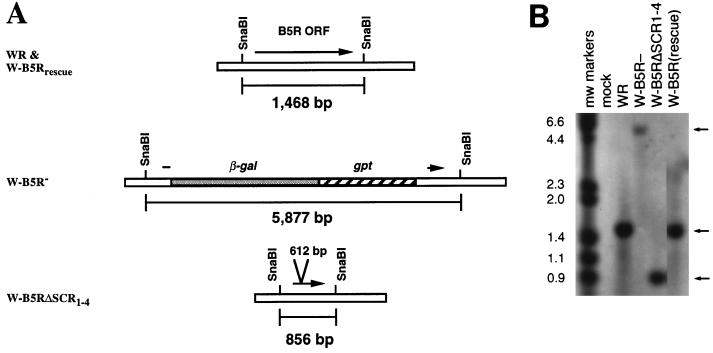
Southern blots of viral DNA isolated from infected cells confirmed that each mutation was properly introduced into the recombinant viruses (Fig. 2). Both the wild-type (WR) and revertant virus (W-B5Rrescue) gave the expected 1.5-kb SnaBI fragment, while the deletion mutant virus (W-B5R−) gave a 5.9-kb fragment due to the replacement of the B5R ORF with the gpt and lacZ genes. Recombinant virus W-B5RΔSCR1–4 produced the expected 0.85-kb fragment resulting from the deletion of 612 bp in the B5R gene. Southern blot data for IHDJ-derived viruses I-B5Rrescue and I-B5RΔSCR1–4 gave similar results (data not shown).
FIG. 2.
Southern blot analysis of recombinant viruses containing mutations in the B5R ORF. (A) Schematic diagram of B5R region of vaccinia virus genomic DNA. The positions of the SnaBI sites and the expected sizes of the corresponding fragments are indicated. The arrows represent the coding region of the B5R gene. (B) Southern blot of DNA digested with SnaBI from mock-infected cells (Mock), cells infected with the wild-type strain of vaccinia virus (WR), the B5R deletion virus (W-B5R−), the recombinant virus with all four SCRs deleted (W-B5RΔSCR1–4), and the B5R revertant virus (W-B5Rrescue). Blots were probed with a 905-bp fragment that overlaps a region of the B5R ORF present in all recombinants. DNA molecular sizes (in kilobase pairs) are indicated. Arrows indicate the positions of the SnaBI digestion products.
Expression of the mutated B5R protein.
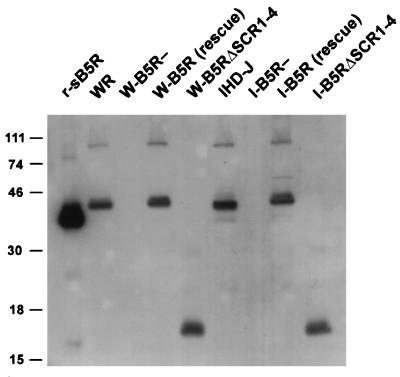
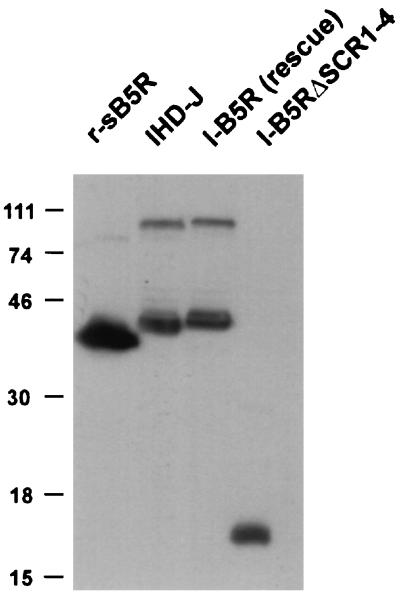
The mutated form of the protein is expected to produce a protein of 93 aa. The expression of this protein was demonstrated by Western blots of infected-cell lysates (Fig. 3). As anticipated, the revertant virus (W-B5Rrescue) produced a 42-kDa protein similar to that of the wild-type virus (WR), confirming that the B5R deletion had been corrected. W-B5RΔSCR1–4 produced a polypeptide of ∼16 kDa because the in-frame deletion results in a protein 204 aa smaller. The higher-molecular-weight bands present in the wild-type and recombinant proteins likely represent oligomeric forms of the proteins (11, 20). Similar results are shown for IHDJ-based viruses, I-B5Rrescue and I-B5RΔSCR1–4 (Fig. 3). Since the rescue viruses utilized the full-length B5R ORF derived from strain WR, the slight difference in the mobility of the B5R protein expressed from I-B5Rrescue compared to that from IHD-J is likely due to the known sequence differences between the B5R encoded by IHD-J and WR (12).
FIG. 3.
Western blot analysis of cell lysates from cells infected with wild-type and recombinant viruses. SDS-PAGE was performed on lysates from BSC-1 cells infected with the wild-type strains of vaccinia virus (WR and IHD-J), their respective B5R deletion viruses (W-B5R− and I-B5R−), the B5R revertant viruses (W-B5Rrescue and I-B5Rrescue), and the recombinant viruses with all four SCRs deleted (W-B5RΔSCR1–4 and I-B5RΔSCR1–4), and proteins were transferred to nitrocellulose and probed with antibody C′-B5R. r-sB5R refers to a recombinant soluble form of the B5R protein that has been previously described (20). An autoradiogram is shown. Numbers on the left refer to the molecular masses (in kilodaltons) of color protein markers (LTI).
Plaque phenotypes of recombinant viruses.
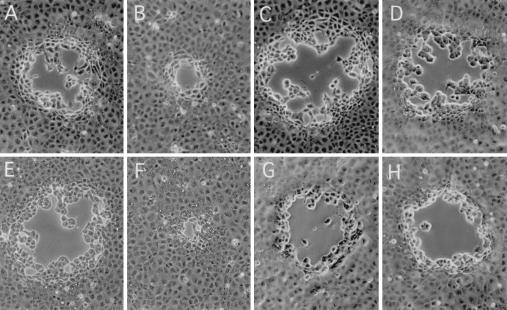
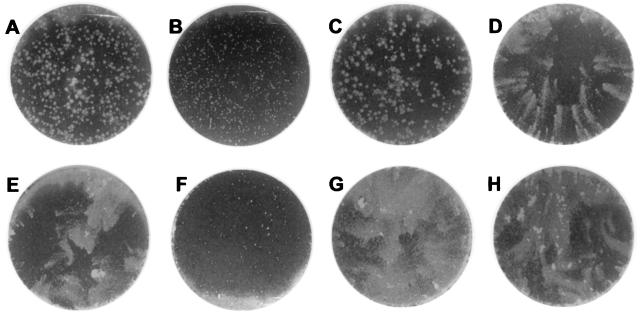
Plaquing of vaccinia virus under an agarose overlay prevents diffusion of EEV from the primary site of infection and can provide an indication of the efficiency of cell-to-cell spread. Figure 4 shows representative plaques from WR-based viruses (Fig. 4A to D) and IHDJ-based viruses (Fig. 4E to H). Wild-type viruses formed large plaques (Fig. 4A and E) as a consequence of efficient cell-to-cell spread, while the B5R deletion viruses formed small plaques (Fig. 4B and F). As expected, the rescued viruses, W-B5Rrescue and I-B5Rrescue, which had the complete wild-type B5R ORF reinserted into the parental deletion mutants, resulted in viruses that form large plaques (Fig. 4C and G). The recombinant viruses lacking the SCR domains, W-B5RΔSCR1–4 and I-B5RΔSCR1–4, also produced large plaques (Fig. 4D and H), indicating that a mutated B5R protein lacking the extracellular SCRs could support large plaque phenotype and efficient cell-to-cell spread. Because formation of enveloped virus is crucial for the efficient cell-to-cell spread of virus, these findings provided an initial indication that EEV was likely being formed in the absence of the four SCRs of the B5R protein.
FIG. 4.
Plaque morphology of the wild-type and mutant viruses under solid overlay. BSC-1 cell monolayers were infected with the panel of viruses and then overlaid with a 1:1 mixture of 2× media and 2% LMP agarose. At 24 h, representative plaques were photographed by phase-contrast microscopy (magnification, ×100). Viruses are WR (A), W-B5R− (B), W-B5Rrescue (C), W-B5RΔSCR1–4 (D), IHD-J (E), I-B5R− (F), I-B5Rrescue (G), and I-B5RΔSCR1–4 (H). Note the tiny plaques in the B5R deletion viruses (B and F) and the return of large plaques with the rescued virus (C and G) and the viruses lacking the SCR domains (D and H).
The amount of EEV released by infected cells into the culture medium can be qualitatively monitored by a standard plaque assay performed under liquid overlay. In this assay, viruses that release large amounts of EEV give rise to plaques with a characteristic comet shape. The comet tail is believed to form by small secondary plaques derived from EEV released by the primary infected cell. Vaccinia virus strain IHD-J produces large amounts of EEV and forms comet-shaped virus plaques (Fig. 5E). On the other hand, vaccinia virus strain WR produces large, round plaques (Fig. 5A) due to the majority of enveloped virus remaining as CEV (3). Figure 5 shows monolayers infected with WR-based viruses (Fig. 5A to D) and IHDJ-based viruses (Fig. 5E to H) under liquid overlay. Both W-B5R− and I-B5R− showed small plaques (Fig. 5B and F). As expected, W-B5Rrescue and I-B5Rrescue restored the round and comet plaque-forming phenotypes of their respective parental viruses (Fig. 5C and G). I-B5RΔSCR1–4 formed plaques with comets similar to those of wild-type IHD-J (Fig. 5H). Surprisingly, W-B5RΔSCR1–4, produced comet-like plaques (Fig. 5D), rather than the large round plaques characteristic of the wild-type WR strain.
FIG. 5.
Plaque formation by viruses under liquid overlay. BSC-1 cell monolayers were infected with the panel of viruses and incubated under liquid overlay. At 48 h postinfection, monolayers were stained with 0.1% crystal violet in 20% ethanol and photographed. Viruses are WR (A), W-B5R− (B), W-B5Rrescue (C), W-B5RΔSCR1–4 (D) IHD-J (E), I-B5R− (F), I-B5Rrescue (G), and I-B5RΔSCR1–4 (H). Comet-shaped plaques were formed by the IHDJ-based viruses (E to H) and by W-B5RΔSCR1–4, but not by WR or W-B5Rrescue.
To ensure that this comet-like phenotype of W-B5RΔSCR1–4 was not caused by a random mutation elsewhere in the genome of this plaque-purified virus, we carried out a completely independent infection-transfection procedure. This also resulted in progeny recombinant viruses with the identical comet-like phenotype under liquid overlay. We next determined whether alterations in the A34R gene might be responsible for this phenotype. Previous studies have mapped the difference between the round-plaque phenotype of strain WR and comet-forming strain IHD-J to a single amino acid change in the A34R gene that encodes an EEV membrane protein (5). We screened our panel of viruses to determine if any changes occurred at this site. The substitution of a lysine for a glutamic acid at codon 151 (K151E) in the A34R protein is due to a base change that results in the loss of a restriction site (MseI) present in the WR A34R ORF (27). Screening of PCR-amplified A34R ORFs by MseI digestion revealed that all the WR-based viruses showed the presence of the MseI restriction site, while all of the IHDJ-based viruses lacked the MseI site (data not shown). Therefore, the comet-like phenotype produced by W-B5RΔSCR1–4 could not be attributed to a K151E mutation in the A34R gene. Thus, the unexpected phenotype produced by W-B5RΔSCR1–4 is most likely due to the mutated B5R gene. This may indicate an unanticipated contribution of the SCR domains of the B5R protein in virus transmission.
Formation of EEV by the recombinant virus lacking the B5R SCR domains.
We compared the amounts of virus found in the media of infected cells following one-step growth conditions. The media from cells infected with W-B5Rrescue contained amounts of infectious virus similar to those of WR. Consistent with the comet-like phenotype, W-B5RΔSCR1–4 had ∼10 times more infectious virus in the supernatant than media from WR- and W-B5Rrescue-infected cells (data not shown). This suggests enhanced release of EEV from the WR-based virus lacking the SCRs. The media from cells infected with I-B5Rrescue and I-B5RΔSCR1–4 had amounts of virus similar to that of media from IHD-J, indicating that the SCRs were not required for infectious EEV formation (data not shown).
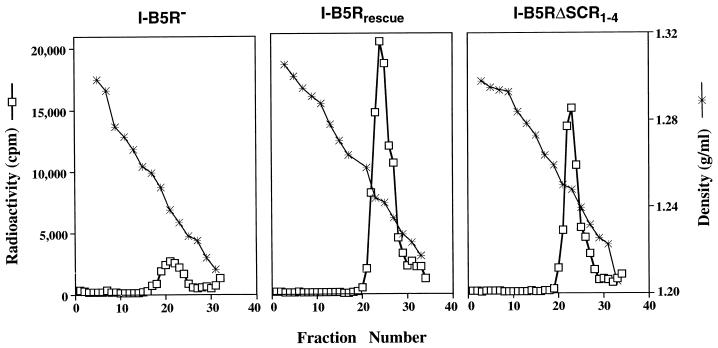
To verify that the virus found in the media from cells infected with the recombinant virus expressing a B5R protein lacking the four SCR domains was in fact EEV, we carried out analysis of metabolically labeled virus by sedimentation in CsCl gradients. Because of the additional membrane on EEV, its buoyant density differs from IMV, which allows them to be distinguished by gradient centrifugation (32). Figure 6 depicts the results of CsCl gradients of media from cells infected with the B5R deletion mutant (I-B5R−), the B5R revertant virus (I-B5Rrescue), and the recombinant virus with all four SCRs deleted (I-B5RΔSCR1–4). Fractions containing the highest radioactive counts were at the buoyant density that corresponds to EEV. These data clearly indicated that EEV formation was rescued in revertant virus (I-B5Rrescue) as well as in the recombinant virus with all four SCRs deleted (I-B5RΔSCR1–4). Thus, despite the high degree of conservation of the SCR region, the SCRs in the extracellular domain of the B5R protein are not required for EEV formation.
FIG. 6.
Equilibrium density centrifugation of extracellular virus. RK13 cells were infected with I-B5R−, I-B5Rrescue, and I-B5RΔSCR1–4 labeled with [35S]methionine and [35S]cysteine, and supernatant media was subjected to centrifugation in CsCl gradients. Fractions were collected from the bottom of tubes, and the levels of radioactivity of aliquots were counted in a liquid scintillation spectrometer. The densities of selected fractions were determined. Peak counts shown correspond to the characteristic density of EEV.
Incorporation of the mutated B5R protein into EEV particles.
To determine if the mutated B5R protein was incorporated into EEV, we isolated EEV released into the media from RK13 cells infected with IHD-J, I-B5Rrescue, or I-B5RΔSCR1–4. Similar amounts of B5R protein can be seen on Western blots containing similar quantities of infectious virus (Fig. 7). To test if the infectivity of each sample represented similar amounts of virions, the blot was stripped and reprobed with an antibody to an alternative EEV protein. Western blotting with antibody that recognizes the F13L gene product (P37) revealed a P37 band of equal intensity in each lane (data not shown). This indicates that the recombinant viruses and wild-type virus incorporate similar amounts of B5R protein into released EEV.
FIG. 7.
Western blot analysis of B5R protein incorporation into EEV. SDS-PAGE was performed on extracellular virus isolated from the media of RK13 cells infected with the wild-type virus (IHD-J), the B5R-revertant virus (I-B5Rrescue), and the recombinant virus with all four SCRs deleted (I-B5RΔSCR1–4), and proteins were transferred to nitrocellulose and probed with antibody C′-B5R. r-sB5R refers to a recombinant soluble form of the B5R protein that has been previously described (20). An autoradiogram is shown. Numbers on the left refer to the molecular masses (in kilodaltons) of color protein markers (LTI).
Demonstration of normal wrapping.
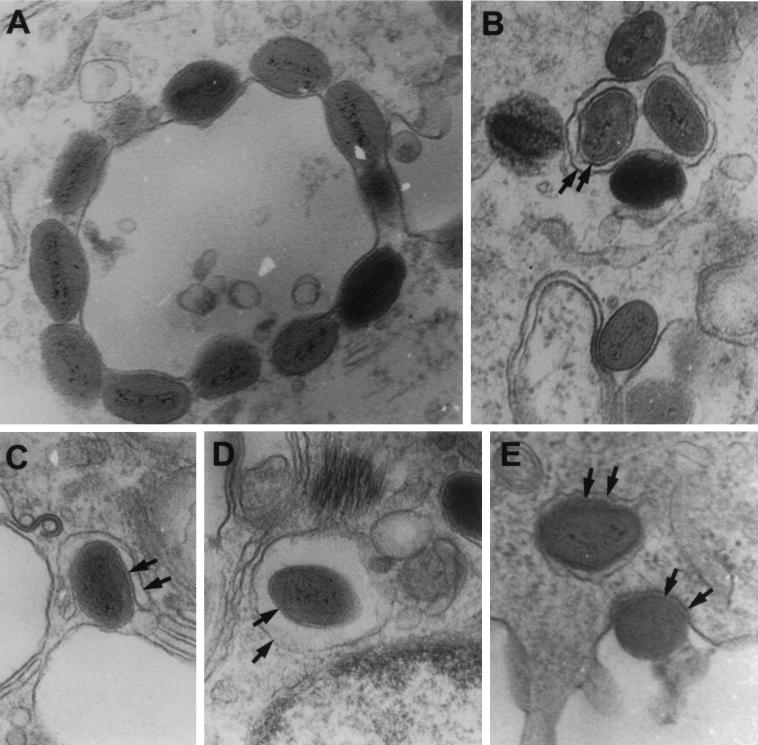
We then examined RK13 cells infected with W-B5RΔSCR1–4 by electron microscopy. We found all the stages of virus envelopment and release (Fig. 8). This process relies on the interaction of IMV particles with intracellular cisternae derived from the trans-Golgi network (Fig. 8A) that wrap the virions (Fig. 8C). Once the enwrapped virion (Fig. 8B and D) moves to the cell periphery, the external membrane fuses with the plasma membrane, releasing the enveloped virus to the extracellular space (Fig. 8E). These results indicate that despite the absence of the SCR domains in W-B5RΔSCR1–4, virus undergoes normal envelopment and exit.
FIG. 8.
Electron microscopy of cells infected with W-B5RΔSCR1–4. Thin sections of RK13 cells infected with W-B5RΔSCR1–4 were examined by electron microscopy. (A) IMV associating with a vesicle; (B) intracellular enveloped virus within a vesicle; (C) single IMV undergoing wrapping; (D) intracellular enveloped virus within a vesicle; (E) intracellular enveloped virus moving to the plasma membrane and an enveloped virus exiting from the cell. Arrows point to distinct membranes.
Cellular localization of a viral envelope protein.
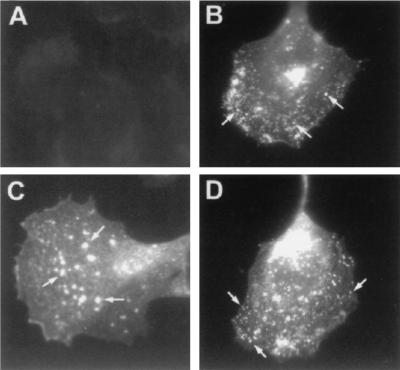
We next wished to study the distribution of enveloped virions in infected cells. Because of the large deletion in the ectodomain, the mutated B5R protein lacking the SCRs is not recognized by any of the B5R monoclonal antibodies we tested and the polyclonal rabbit antibody to B5R proved unsuitable for immunofluorescence staining (data not shown). We therefore used a monoclonal antibody recognizing the EEV-specific product of the F13L gene (P37) (17) to examine the immunofluorescence staining patterns of the panel of viruses. P37 staining of cells infected with the wild-type virus showed typical Golgi staining in a perinuclear position, as well as a more dispersed punctate pattern that presumably corresponds to intracellular enveloped virions (Fig. 9B). This is in contrast to W-B5R− in which the dispersed punctate staining was scarce and larger fluorescent dots were seen (Fig. 9C). These forms may correspond to the large vacuoles that have been previously described in cells infected with the B5R deletion virus (42). Cells infected with W-B5RΔSCR1–4 showed a P37 staining pattern similar to that of cells infected with wild-type virus, with typical Golgi staining, as well as smaller stained points dispersed in the cytoplasm and close to the periphery of the cell (Fig. 9D). This indicates that normal IMV wrapping and migration of intracellular enveloped virions was occurring in the presence of the mutated form of the protein.
FIG. 9.
Immunofluorescence staining with anti-P37 antibody. CV-1 cells were either mock infected (A) or infected with WR (B), W-B5R− (C), or W-B5RΔSCR1–4 (D). At 7 h postinfection, cells were fixed, made permeable, washed, and then incubated with a monoclonal antibody to P37, the major protein present in EEV. To visualize the primary antibody, rabbit anti-rat antibody coupled to FITC was used. Representative cells were photographed. Arrows indicate small punctate staining of WR and W-B5RΔSCR1–4 and large fluorescent dots in the B5R deletion virus (C).
Induction of actin tails by vaccinia virus.
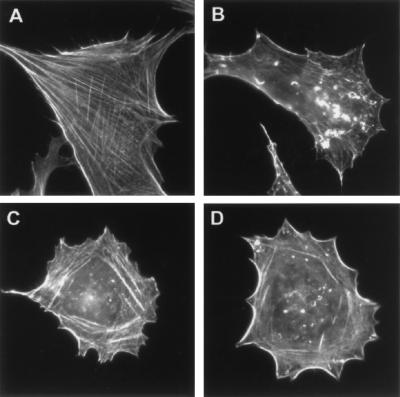
Since vaccinia virus infection induces the appearance of thick actin bundles (Fig. 10B) that are believed to propel virus out of the cell, a process that is dependent on the envelope wrapping process (7, 8, 15, 16, 40, 43), we analyzed the actin patterns induced by our panel of viruses. The cellular actin network can be visualized by staining with rhodamine-phalloidin (Fig. 10A). Similar to the observations made with a mutant virus defective in the wrapping process (3), cells infected with W-B5R− were devoid of recognizable actin bundles (Fig. 10C). Cells infected with W-B5RΔSCR1–4 were also severely impaired in thick actin bundle formation (Fig. 10D). This indicates that when the B5R protein lacking SCR domains is expressed, there is a defect in the induction of actin tails, despite the formation and release of significant amounts of extracellular virus. This finding is similar to a recent description of a mutant virus that undergoes wrapping and can exit the cell in the absence of actin bundles (43).
FIG. 10.
Phalloidin staining of infected cells. CV-1 cells were either mock infected (A) or infected with WR (B), W-B5R− (C), or W-B5RΔSCR1–4 (D). At 7 h postinfection, cells were fixed, made permeable, washed, and then incubated with TRITC-phalloidin and representative cells were photographed. Note the absence of thick actin bundles in cells infected with W-B5R− and W-B5RΔSCR1–4.
DISCUSSION
The vaccinia virus B5R protein is highly conserved and essential for EEV formation (12, 26, 42). To address the function of the putative SCR domains that comprise the majority of the extracellular portion of the B5R-protein, we generated recombinant vaccinia viruses (W-B5RΔSCR1–4 and I-B5RΔSCR1–4) in which the SCR domains were deleted. The virus expressed a smaller version of the protein because 80% of the extracellular portion of the protein was removed. The mutated protein retained a signal peptide, an intact transmembrane and cytosolic domains, as well as a 51-aa section of ectodomain just before the transmembrane region. In contrast to the B5R-deleted virus (12, 26, 42), the recombinant virus lacking the SCR domains underwent normal envelopment, produced significant amounts of extracellular virus, and was transmitted efficiently between cells. Thus, the SCR domains of B5R protein are not required for either formation or infectivity of enveloped virus, which strongly supports the concept that the carboxy-terminal portion of the glycoprotein is sufficient to mediate the steps required for EEV formation.
Different strains of vaccinia virus liberate different amounts of virus from infected cells. This feature can be monitored by a simple plaque assay, since high EEV producers, like strain IHD-J, give rise to comet-shaped plaques, whereas low EEV producers, like strain WR, give round plaques (1, 31). The difference in EEV production can be largely explained by different degrees of retention of enveloped virions at the cell surface (3). The differences between strain WR and IHD-J were previously mapped to a single amino acid change in the extracellular portion of the A34R gene, and the existence of at least a second gene influencing CEV release was predicted (5). In this study, the WR-based mutant virus lacking the four SCR domains unexpectedly produced comet-like plaques and liberated increased amounts of virus into the culture medium. It is unlikely that this was due to a random mutation elsewhere in the genome because several independently isolated plaques gave the identical phenotype. It is interesting that the mutations in the A34R and B5R genes that produce comet-forming phenotypes in strain WR are in the external portion of glycoproteins located on the EEV surface. The phenotype of W-B5RΔSCR1–4 suggests that the SCR domains might bind to the cell surface and thereby modulate the release of CEV from the cell surface. A second unexpected feature of W-B5RΔSCR1–4 was a decrease in virus-induced actin bundles that propel virus particles, which are thought to be involved in transmission of virus from cell to cell (8). The appearance of the bundles is dependent on virus wrapping, since they are not induced in conditions where wrapping is severely impaired, like deletion of F13L gene (3) or treatment of vaccinia virus-infected cells with a drug that inhibits wrapping events (7, 14). However, W-B5RΔSCR1–4 provides a second example in which the exit of EEV particles is not dependent on actin bundles. Recently, a virus with the A34R gene deleted was reported that did not induce actin bundle formation yet produced increased amounts of virus in the medium (43). The deletion of the SCRs in the B5R gene results in a similar behavior, with increased EEV production and a decrease in actin bundle formation. It is likely that a complex of several proteins in the virus envelope is required for the induction of the actin tails and that mutations in the luminal domains influence protein-protein interactions between these proteins.
The in vivo function(s) of the B5R protein’s SCR domains, a region conserved among not only vaccinia virus strains but orthopoxviruses in general, remains unknown (11, 12, 26, 41). Given its similarity to other complement regulatory proteins (11, 13, 26, 41), it is conceivable that the B5R SCR domains may be involved in the control of the complement-mediated host immune responses. Since the B5R protein is expressed on both the membranes of infected cells and on the outer membrane of EEV (11, 20, 26), it is well positioned as a viral defense molecule. Because vaccinia viruses lacking B5R fail to make EEV, the severe in vivo attenuation of B5R deletion viruses is likely due largely to the absence of EEV formation (12, 39, 42), which is central for viral dissemination in vivo. Our data, therefore, support the hypothesis that B5R has developed as a multifunctional protein, with an important role in the formation of EEV and, potentially, in regulation of host complement activation.
Recently Katz et. al. (21) constructed a recombinant vaccinia virus expressing a chimeric protein that fused the ectodomain of the HIV envelope protein to the transmembrane and cytoplasmic domains of the B5R protein. This fusion protein was detected on the membranes of infected cells and was incorporated into the outer envelope of EEV, indicating that the transmembrane and/or cytoplasmic domains of B5R contain an EEV targeting signal. However, in that construct, the chimeric human immunodeficiency virus-B5R protein was expressed along with the wild-type B5R protein and represented only ∼6% of the total amount of B5R protein expressed (21). Therefore, that study could not provide information about the B5R domains responsible for EEV formation. In our construct, the mutated version of the B5R protein lacking the SCR domains was introduced in place of the natural B5R locus and was expressed at a level similar to that of the wild-type B5R protein. Our results demonstrate that a small portion of B5R, including the cytoplasmic tail, transmembrane domain, and 51 residues in the juxta-membrane extracellular region, is sufficient for incorporation into EEV and sustains significant amounts of EEV formation.
The findings that the carboxy-terminal portion of the B5R contains the EEV targeting signal and also provides the requirements for EEV formation make it likely that expression of chimeric B5R-based proteins in the absence of wild-type B5R protein will be successful. This approach may provide an easy and efficient way to anchor foreign antigens to the virion surface that could be exploited for vaccine development. This design could offer several advantages over coexpression of the chimeric and wild-type B5R versions (21). First, high levels of the chimera can be expressed and targeted to the virus envelope in the absence of competing wild-type protein. Second, since plaque formation can be used as a selection criteria (4), the rescue of the large plaque phenotype should facilitate the isolation of recombinant viruses by reintroduction of the chimeric version in B5R deletion viruses. Finally, the deletion of the SCR repeats may be advantageous for a vaccine vector. Since attenuation of the virus may be desirable, viral genes that potentially counteract the host’s immune responses are good candidates for such manipulation.
In conclusion, this work provides evidence that the majority of the extracellular domain is required for induction of actin bundles, while it is dispensable for normal envelopment and egress of enveloped virus. Additionally, we provide data suggestive of a role of the protein in retention of enveloped virions at the cell surface. This work may have important implications related to vaccine development and suggests the possibility of targeting EEV to specific cells or organs by expression of proteins fused to the carboxy terminus of B5R. Furthermore, the recombinant viruses lacking the putative SCR domains of the B5R protein might also provide insight into the role of the SCR domains in pathogenesis, viral dissemination in vivo, and its potential role in the evasion from host immune responses.
ACKNOWLEDGMENTS
We thank Jeannie Chu for excellent technical assistance; the members of the Collman and Friedman laboratories for helpful discussions; and Ron Collman, Thandavarayan Nagashunmugam, John Lubinski, Lester Goldstein, and Harvey Friedman for critical review of the manuscript.
This study was supported by NIH grant AI-01324 to S.N.I. and Ministerio de Educación Cultura grant PB95-0237 to R.B.
REFERENCES
- 1.Appleyard G, Hapel A J, Boulter E A. An antigenic difference between intracellular and extracellular rabbitpox virus. J Gen Virol. 1971;13:9–17. doi: 10.1099/0022-1317-13-1-9. [DOI] [PubMed] [Google Scholar]
- 2.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco R, Moss B. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene. 1995;158:157–162. doi: 10.1016/0378-1119(95)00149-z. [DOI] [PubMed] [Google Scholar]
- 5.Blasco R, Sisler J R, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J Virol. 1993;67:3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown C K, Turner P C, Moyer R W. Molecular characterization of the vaccinia virus hemagglutinin gene. J Virol. 1991;65:3598–3606. doi: 10.1128/jvi.65.7.3598-3606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 8.Cudmore S, Reckmann I, Griffiths G, Way M. Vaccinia virus: a model system for actin-membrane interactions. J Cell Sci. 1996;109:1739–1747. doi: 10.1242/jcs.109.7.1739. [DOI] [PubMed] [Google Scholar]
- 9.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earl P L, Cooper N, Moss B. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates & Wiley Interscience; 1991. pp. 16.16.1–16.16.7. [Google Scholar]
- 11.Engelstad M, Howard S T, Smith G L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 12.Engelstad M, Smith G L. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 13.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. , 517–563. [DOI] [PubMed] [Google Scholar]
- 14.Hiller G, Eibl H, Weber K. Characterization of intracellular and extracellular vaccinia virus variants: N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine interferes with cytoplasmic virus dissemination and release. J Virol. 1981;39:903–913. doi: 10.1128/jvi.39.3.903-913.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiller G, Jungwirth C, Weber K. Fluorescence microscopical analysis of the life cycle of vaccinia virus in chick embryo fibroblasts. Virus-cytoskeleton interactions. Exp Cell Res. 1981;132:81–87. doi: 10.1016/0014-4827(81)90085-9. [DOI] [PubMed] [Google Scholar]
- 16.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology. 1979;98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 17.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isaacs S N, Kotwal G J, Moss B. Reverse guanine phosphoribosyltransferase selection of recombinant vaccinia viruses. Virology. 1990;178:626–630. doi: 10.1016/0042-6822(90)90367-z. [DOI] [PubMed] [Google Scholar]
- 19.Isaacs S N, Kotwal G J, Moss B. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc Natl Acad Sci USA. 1992;89:628–632. doi: 10.1073/pnas.89.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isaacs S N, Wolffe E J, Payne L G, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz E, Wolffe E J, Moss B. The cytoplasmic and transmembrane domains of the vaccinia virus B5R protein target a chimeric human immunodeficiency virus type 1 glycoprotein to the outer envelope of nascent vaccinia virions. J Virol. 1997;71:3178–3187. doi: 10.1128/jvi.71.4.3178-3187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotwal G J, Isaacs S N, McKenzie R, Frank M M, Moss B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science. 1990;250:827–830. doi: 10.1126/science.2237434. [DOI] [PubMed] [Google Scholar]
- 23.Kotwal G J, Moss B. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature. 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- 24.Krijnse-Locker J, Schleich S, Rodriguez D, Goud B, Snijder E J, Griffiths G. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J Biol Chem. 1996;271:14950–14958. doi: 10.1074/jbc.271.25.14950. [DOI] [PubMed] [Google Scholar]
- 25.Krych M, Atkinson J P, Holers V M. Complement receptors. Curr Opin Immunol. 1992;4:8–13. doi: 10.1016/0952-7915(92)90116-v. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Pomares L, Stern R J, Moyer R W. The ps/hr gene (B5R open reading frame homolog) of rabbitpox virus controls pock color, is a component of the extracellular enveloped virus, and is secreted into the medium. J Virol. 1993;67:5450–5462. doi: 10.1128/jvi.67.9.5450-5462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McIntosh A A, Smith G L. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J Virol. 1996;70:272–281. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenzie R, Kotwal G J, Moss B, Hammer C H, Frank M M. Regulation of complement activity by vaccinia virus complement-control protein. J Infect Dis. 1992;166:1245–1250. doi: 10.1093/infdis/166.6.1245. [DOI] [PubMed] [Google Scholar]
- 29.Miller C G, Shchelkunov S N, Kotwal G J. The cowpox virus-encoded homolog of the vaccinia virus complement control protein is an inflammation modulatory protein. Virology. 1997;229:126–133. doi: 10.1006/viro.1996.8396. [DOI] [PubMed] [Google Scholar]
- 30.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a M(r) 43-50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 31.Payne L G. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J Gen Virol. 1980;50:89–100. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 32.Payne L G, Norrby E. Presence of hemagglutinin in the envelope of extracellular vaccinia virus particles. J Gen Virol. 1976;32:63–72. doi: 10.1099/0022-1317-32-1-63. [DOI] [PubMed] [Google Scholar]
- 33.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmutz C, Payne L G, Gubser J, Wittek R. A mutation in the gene encoding the vaccinia virus 37,000-Mr protein confers resistance to an inhibitor of virus envelopment and release. J Virol. 1991;65:3435–3442. doi: 10.1128/jvi.65.7.3435-3442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmutz C, Rindisbacher L, Galmiche M C, Wittek R. Biochemical analysis of the major vaccinia virus envelope antigen. Virology. 1995;213:19–27. doi: 10.1006/viro.1995.1542. [DOI] [PubMed] [Google Scholar]
- 37.Shida H. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology. 1986;150:451–462. doi: 10.1016/0042-6822(86)90309-0. [DOI] [PubMed] [Google Scholar]
- 38.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van′t Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stern R J, Thompson J P, Moyer R W. Attenuation of B5R mutants of rabbitpox virus in vivo is related to impaired growth and not an enhanced host inflammatory response. Virology. 1997;233:118–129. doi: 10.1006/viro.1997.8556. [DOI] [PubMed] [Google Scholar]
- 40.Stokes G V. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J Virol. 1976;18:636–643. doi: 10.1128/jvi.18.2.636-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi-Nishimaki F, Funahashi S, Miki K, Hashizume S, Sugimoto M. Regulation of plaque size and host range by a vaccinia virus gene related to complement system proteins. Virology. 1991;181:158–164. doi: 10.1016/0042-6822(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 42.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolffe E J, Katz E, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]