Abstract
Salmonella enterica is distributed worldwide and is a common cause of bacterial food poisoning in humans and a serious public health problem. Although duck meat consumption has recently increased in Korea, studies on the epidemiological relationship between S. enterica contamination in duck farms are scarce. Salmonella enterica serovar Albany isolates recovered from duck farms were analyzed using two typing methods — IR Biotyper® (IRBT) and multilocus variable-number tandem repeat analysis (MLVA). The clustering results were compared with the epidemiological survey findings and the antimicrobial resistance profiles. From April 2019 to October 2020, 20 individual feces per farm from 5–6-week-old ducks were collected repeatedly from 105 duck farms. Salmonella spp. isolated from duck feces were identified using PCR and multilocus sequence typing to investigate the prevalence and distribution of the Salmonella serovars. The prevalence of S. enterica was 19%, and S. enterica Albany was the predominantly recovered isolate. The S. enterica Albany isolates underwent antimicrobial susceptibility testing to determine the minimum inhibitory concentration. MLVA and IRBT methods established relatedness and diversity among the S. enterica Albany isolates. Multidrug-resistant S. enterica Albany was distributed in all the farms. Antimicrobial resistance profiles reflected the duck farm characteristics and isolates recovered from the same farm showed an identical profile. Isolates repeatedly recovered from the same farm also showed identical IRBT clusters and MLVA groups. These findings suggest that the isolates remained on the duck farm and re-infected new duck flocks. Thus, proper cleaning and disinfection is required before the farms are repopulated.
Keywords: Salmonella enterica Albany, Duck, Typing method, Epidemiological survey, Antimicrobial resistance
1. Introduction
Salmonella. enterica Albany represents a risk to human and animal health, considering its identification in ducks [1,2]. A previous study reported a high Salmonella contamination in duck meat [3]. Another study showed that S. enterica Albany contamination of poultry carcasses caused public health problems [1]. However, few studies have investigated the epidemiology of S. enterica Albany in ducks.
Strain typing is defined as the epidemiological evaluation of isolates recovered from farms. It can be applied to confirm the characteristics of isolates [4,5]. Strain typing can be performed using multilocus variable number tandem repeat analysis (MLVA) or Fourier transform infrared (FTIR) spectroscopy. Bacterial genomes contain polymorphic tandem repeats at an identical locus (for example, variable-number tandem repeats (VNTRs)), which differ between strains [6]. MLVA takes advantage of these repeats to distinguish strains and can be easily performed by conventional PCR with identical primers [7]. VNTR analysis is useful for assessing genetic relatedness between strains and can provide accurate pathogen typing and phylogenetic analysis [8]. The discovery of VNTR loci has enabled genotyping via MLVA of multiple Salmonella serovars [6,9] and is considered a highly discriminatory method [6].
FTIR spectroscopy analyzes molecular vibrations caused by infrared light absorption using the principle that different chemical structures vibrate at different frequencies [5]. Microorganisms can be characterized according to their strain-specific absorbance patterns in the infrared spectrum [10]. FTIR spectroscopy has recently become an alternative to bacterial typing due to its ease of use and high discriminatory power in recognizing clonal relationships among bacterial isolates [11]. The IR Biotyper® system (IRBT, Bruker Daltonics GmbH & Co. KG, Bremen, Germany), based on FTIR technology, is a device that enables easy and fast strain typing [12]. For example, following species-level identification using the MALDI Biotyper® (Bruker Daltonics GmbH & Co. Billerica, MA, USA), the strains can be typed and grouped by measuring their specific molecular vibration fingerprints [10]. IRBT technology can be applied to the bacterial surface in a non-destructive manner, facilitating high-resolution analysis to evaluate the similarities between different isolates at the strain level [12].
Antimicrobial susceptibility testing can confirm the current prevalence rate and help identify the characteristics of resistant S. Albany strains in duck farms. β-lactams, aminoglycosides, and fluoroquinolones are often used to treat systemic bacterial infections on Korean poultry farms [13,14]. However, continuous antibiotic therapy increases public health problems by fostering antibiotic resistance and increasing the prevalence of multidrug-resistant Salmonella [13]. Investigating the prevalence rate and resistance profile of S. enterica Albany with antimicrobial resistance is needed to control multidrug-resistant Salmonella.
This study was aimed to (1) confirm the distribution of Salmonella serovars, the prevalence of S. enterica Albany, and the resistance profiles of isolates recovered from ducks; (2) evaluate strain-level typing using IRBT and MLVA for S. enterica Albany isolates; (3) compare the discrimination and concordance of IRBT and MLVA; and (4) compare the clustering results of S. enterica Albany generated via IRBT and MLVA with the epidemiological relationship and antimicrobial resistance profiles between farms to facilitate the development of prevention and control strategies for Salmonella infection in duck farms.
Our study provides a platform for follow-up studies on food safety and public health issues caused by S. enterica Albany recovered from duck farms.
2. Materials and methods
2.1. Sample collection
Between April 2019 and October 2020, 20 individual feces from 5–6-week-old ducks per farm were collected from 105 duck farms. The freshly deposited feces were placed in a 50 mL conical tube. A mixed pool of 20 individual feces was applied as one sample, and feces from farms were collected repeatedly each time the ducks were repopulated. Considering the characteristics of duck farming, the ducks were first raised communally in one duck house and then moved to other duck houses at an average age of 10.2 days at each farm. Thus, fecal sampling was uniformly based on the first duck house. Feces were collected at each time point and transported directly to the laboratory, where they were processed separately.
2.2. Isolation and identification of Salmonella spp.
The 20 individual feces collected from each farm at each time point were pooled, and 5 g of the pooled sample was added to 45 mL of sterilized PBS. The suspension was mixed well, and 1 mL of it was added to 9 mL of buffered peptone water (BPW). This mixture was incubated at 37 °C for 24 h [15]. A 0.1 mL aliquot of the cultured BPW was added to 10 mL of Rappaport–Vassiliadis broth, incubated at 42 °C for 24 h, and then inoculated onto xylose–lysine–deoxycholate agar, which was incubated at 37 °C for 24 h. The suspect Salmonella colonies (red with a black center) were sub-cultured from a single colony onto 5% sheep blood agar plates. After incubation at 37 °C for 24 h, the cultures were examined in the MALDI Biotyper® (Bruker Daltonics GmbH & Co. Billerica, MA, USA). Isolates identified as Salmonella were serotyped using PCR as previously described [16,17] (Table 1). The one-step multiplex PCR used can identify major chicken S. enterica subsp. enterica serovars – Albany, Enteritidis, Gallinarum, Gallinarum biotype Gallinarum, Gallinarum biotype Pullorum, Heidelberg, Kentucky, and S. enterica groups 1 and 2 [17]. Isolates assigned to serovars S. enterica groups 1 and 2 via PCR were then examined using a multilocus sequence typing (MLST) method [16] to recognize serovars such as Dublin and Montevideo. The quality control of sequence-based MLST was performed for each run using the pGEM control DNA in the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). MLST was performed using analysis tools available from the MLST website (https://enterobase.warwick.ac.uk) [18]. Identification of the identical serovars among suspected Salmonella colonies was considered as one Salmonella spp. Isolates identified as S. enterica Albany were sub-cultured on tryptic soy agar for MLVA, IRBT, and antimicrobial susceptibility tests.
Table 1.
Oligonucleotide sequences of primers used for PCR-based serotyping.
| Primer | Sequence (5′–3′) | Amplicon size (bp) | Reference | |
|---|---|---|---|---|
| bcfC-F | GGGTGGGCGGAAAACTATTTC | 993 | [17] | |
| bcfC-R | CGGCACGGCGGAATAGAGCAC | |||
| heli-F | ACAGCCCGCTGTTTAATGGTG | 782 | ||
| heli-R | CGCGTAATCGAGTAGTTGCC | |||
| steB-F | TGTCGACTGGGACCCGCCCGCCCGC | 636 | ||
| steB-R | CCATCTTGTAGCGCACCAT | |||
| rhs-F | TCGTTTACGGCATTACACAAGTA | 402 | ||
| rhs-R | CAAACCCAGAGCCAATCTTATCT | |||
| sdf-F | TGTGTTTTATCTGATGCAAGAG | 293 | ||
| sdf-R | CGTTCTTCTGGTACTTCAGATGAC | |||
| gly-F | TTCCAATTGAAACGAGTGCGG | 170 | ||
| gly-R | ACTAACCGCTTGGGTTGTTGCTGT | |||
| fliB-F | GTCTGCTAACAGCACTAACTC | 551 | ||
| fliB-R | CCTGAGTTTTTGTTACTTCTACC | |||
| aroC-F | CCTGGCACCTCGCGCTATAC | 826 | [16] | |
| aroC-R | CCACACACGGATCGTGGCG | |||
| dnaN-F | ATGAAATTTACCGTTGAACGTGA | 833 | ||
| dnaN-R | AATTTCTCATTCGAGAGGATTGC | |||
| hemD-F | ATGAGTATTCTGATCACCCG | 666 | ||
| hemD-R | ATCAGCGACCTTAATATCTTGCCA | |||
| hisD-F | GAAACGTTCCATTCCGCGCAGAC | 894 | ||
| hisD-R | CTGAACGGTCATCCGTTTCTG | |||
| thrA-F | GTCACGGTGATCGATCCGGT | 852 | ||
| thrA-R | CACGATATTGATATTAGCCCG | |||
| sucA-F | AGCACCGAAGAGAAACGCTG | 643 | ||
| sucA-R | GGTTGTTGATAACGATACGTAC | |||
| purE-F | ATGTCTTCCCGCAATAATCC | 510 | ||
| purE-R | TCATAGCGTCCCCCGCGGAT | |||
2.3. Antimicrobial susceptibility testing
The minimum inhibitory concentration (MIC) was determined with the KRNV5F Sensititre panel (TREK Diagnostic Systems) following the manufacturer's instructions. The antibiotics used in the test were amoxicillin/clavulanic acid (AUG2, 2/1–32/16 μg/mL), ampicillin (AMP, 2–64 μg/mL), cefoxitin (FOX, 1–32 μg/mL), ceftazidime (TAZ, 1–16 μg/mL), ceftiofur (XNL, 0.5–8 μg/mL), cefepime (FEP, 0.25–16 μg/mL), meropenem (MERO, 0.25–4 μg/mL), trimethoprim/sulfamethoxazole (SXT, 0.12/2.38–4/76 μg/mL), sulfisoxazole (FIS, 16–256 μg/mL), chloramphenicol (CHL, 2–64 μg/mL), ciprofloxacin (CIP, 0.12–16 μg/mL), nalidixic acid (NAL, 2–128 μg/mL), streptomycin (STR, 16–128 μg/mL), gentamicin (GEN, 1–64 μg/mL), tetracycline (TET, 2–128 μg/mL), and colistin (COL, 2–16 μg/mL). The breakpoints for antibiotics used in the MIC test were obtained from the Clinical Laboratory Standards Institute (2019) [19] and the National Antimicrobial Resistance Monitoring System [20] (Table 2). Multiple drug resistance (MDR) was defined as resistance to three or more antimicrobial classes.
Table 2.
Breakpoints of antimicrobials used for MIC testing of Salmonella spp.
| Antimicrobial agents | Range tested (μg/mL) | Breakpointsa (μg/mL) |
|---|---|---|
| Amoxicillin/clavulanic acid | 2/1–32/16 | ≥32/16 |
| Ampicillin | 2–64 | ≥32 |
| Cefoxitin | 1–32 | ≥32 |
| Ceftazidime | 1–16 | ≥16 |
| Ceftiofur | 0.5–8 | ≥8 |
| Cefepime | 0.25–16 | ≥16 |
| Meropenem | 0.25–4 | ≥4 |
| Trimethoprim/sulfamethoxazole | 0.12/2.38–4/76 | ≥4/76 |
| Sulfisoxazole | 16–256 | ≥512 |
| Chloramphenicol | 2–64 | ≥32 |
| Ciprofloxacin | 0.12–16 | ≥1 |
| Nalidixic acid | 2–128 | ≥32 |
| Streptomycin | 16–128 | ≥32 |
| Gentamicin | 1–16 | ≥16 |
| Tetracycline | 2–128 | ≥16 |
| Colistin | 2–16 | ≥4 |
2.4. Strain typing via MLVA
Loci for the MLVA were selected by uploading the WGS of ATCC 51960 strain (GenBank accession number CP019177.1) in FASTA format to a tandem repeat finder program (https://tandem.bu.edu/trf/basic_submit). The potential discriminatory ability of the selected loci was evaluated by comparing their sequences with the genomic sequences of S. enterica Albany strains deposited in the NCBI database. For the four loci determined as VNTR markers, primer pairs were designed for a product size of 100–600 bp using the Primer Express 3.0 software (Applied Biosystems). The forward primers were labeled with a 5′ fluorescent reporter dye (FAM, NED, VIC, and PET) for analysis in a genetic analysis instrument (Table 3).
Table 3.
Primers for VNTR markers of Salmonella enterica Albany.
| VNTR markers | Primer sequences (5′–3′) | Repeat sequence | Product size (bp)b | Fragment size (bp)c | DId | Locationb |
|---|---|---|---|---|---|---|
| STTR7 | FAM-CGCGCAGCCGTTCTCACT | 39 bpa | 546 | 507–585 (4–6) | 0.1538 | 2,979,895–2980,440 |
| TGTTCCAGCGCAAAGGTATCTA | ||||||
| SATR1 | NED-GGATGTTCTGGCGGACATGG | CACGAC | 114 | 114–126 (5–7) | 0.5 | 871,156–871,269 |
| CGCCTTCCGGATGTATGTGA | ||||||
| SATR2 | VIC-AAAATCCCCGTAAATCCCGCT | TGCCTG | 158 | 158–170 (7–9) | 0.2821 | 1,155,894–1156,051 |
| AGGTGCAAAAGTGGCCTCA | ||||||
| SATR3 | PET-CCTCCTGCTGGAAAAATCGC | ATCAATCCGT | 129 | 119–139 (3–5) | 0.2821 | 4,021,834–4021,962 |
| TGGCGATGCAATGCGTCTTA |
Model of repeats: CAGCAGCCGCAACAGCCGGTAGCGCCGCAACCGCAGTAT (39 bp).
Product size and location in ATCC 51960 (GenBank accession number CP019177.1).
Observed fragment size (repeat copy no.).
Simpson's diversity index, each index was calculated using the results from the 13 unrelated Salmonella Albany isolates.
PCR was performed using a reaction mixture containing Maxime™ PCR PreMix (iNtRON Biotech), 5 μL of primer mix, and 1–5 ng of DNA template. The final volume (20 μL) was adjusted using RNase-free water. PCR amplification conditions were as follows: 95 °C for 10 min; 40 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s; and 72 °C for 10 min. Next, 10 μL of diluted size standard (20 μL size standard mixed in 1 mL HI-DI formamide) was added to the final PCR products (3–5 ng/μL). Following denaturation at 95 °C for 5 min and rapid cooling to 4 °C for 1 min, the samples were injected into a capillary electrophoresis system (Applied Biosystems 3500 XL Genetic Analyzer, Life Technology) to measure the fragment sizes. We included amplified control DNA (CEPH 1347-02 from Applied Biosystems) during every series of capillary runs. The data were analyzed using GeneMapper® (Life Technologies) to determine the size.
2.5. Strain typing via IRBT analysis
The S. enterica Albany isolates were incubated on TSA at 37 °C for 24 h. A 1 μL loopful of culture was inoculated into a 1.5 mL microtube containing 50 μL of 70% ethanol and mixed uniformly. After vortexing the mixture for 1 min, 50 μL of sterile water was added, and the resulting 100 μL solution was vortexed for 1 min. After loading three spots of 15 μL each of the well-mixed sample solutions on the target plate, the plate was dried in a 37 °C incubator for 10–30 min. The dried target plate was inserted into the IRBT spectrometer (Bruker Daltonics GmbH & Co. KG, Bremen, Germany), and data were acquired using IRBT software (version 4.0) with the default settings. The quality control was performed for each run using the Infrared Test Standards (IRTS 1 and IRTS 2) from the IR Biotyper. Spectra of the isolates were acquired using OPUS 7.5 software (Bruker Daltonics GmbH & Co. KG, Bremen, Germany). The spectra that met the default quality criteria [0.4 < absorption <2, signal/noise >40, signal/water >20, fringes ( × 10−6) < 100] were accepted. All data were analyzed using IRBT software by constructing the dendrograms and 2D scatter plots on average spectra. Dendrograms were constructed using Euclidian distances and the average linkage clustering method. 2D scatter plots for similarity analysis were determined by principal component analysis (PCA). Cutoff values were automatically proposed by the software.
3. Results
3.1. Prevalence and distribution of Salmonella serovars on duck farms
A total of 745 samples from 105 duck farms were examined: a mixed pool of 20 individual feces was applied as one sample; the number of sampling events varied in the range of 1–16 because samples were repeatedly collected whenever new duck flocks were placed in the same farm. The positivity rate for Salmonella was 19%, and 141 Salmonella spp. were isolated from 63 farms. Salmonella spp. was not detected in 42 of the 105 duck farms and was detected more than once in 63 farms (Fig. 1A).
Fig. 1.
Prevalence and distribution of Salmonella serovars on duck farms. Positive rates of Salmonella spp. ranged from 0 to 100% in duck farms (A). A total of 11 serovars were detected, of which S. enterica Albany was the most common. The list includes farms where the same serotype was repeatedly recovered from existing and new duck flocks (asterisks) (B).
The isolates were assigned to 11 serovars (Fig. 1B). S. enterica Albany (66.7%; 94 isolates) was the most common serovar, followed by S. Enteritidis (12.1%; 17 isolates), S. London (4.3%; six isolates), S. Hadar (3.5%; five isolates), S. Typhimurium (3.5%; five isolates), S. Give (2.8%; four isolates), S. Indiana (2.8%; four isolates), S. Mbandaka (1.4%; two isolates), S. Zanzibar (1.4%; two isolates), S. Montevideo (0.7%; one isolate), and S. Virchow (0.7%; one isolate).
3.2. Antimicrobial susceptibility testing
A total of 94 S. enterica Albany isolates were tested for antimicrobial susceptibility. All 94 (100%) were resistant to FIS, 93 (98.9%) were resistant to NAL, 92 (97.9%) were resistant to SXT, 34 (36.2%) were resistant to AMP, 34 (36.2%) were resistant to CIP (36.2%), 30 (32.0%) were resistant to CHL, 28 were resistant to TET (29.8%), and 5 were resistant to STR (5.3%). No isolates were resistant to AUG2, FOX, TAZ, XNL, FEP, MERO, GEN, and COL (Table 4).
Table 4.
Minimum Inhibitory Concentration (MIC) (μg/mL) patterns for Salmonella enterica Albany isolated from duck feces.
Eleven antimicrobial resistance profiles were observed in the isolates (Table 5). All 94 isolates were resistant to two or more antimicrobial classes, and 37 were classified as MDR. FIS-SXT-NAL (42/94, 44.7%) was the most prevalent antimicrobial resistance profile, followed by FIS-SXT-NAL-CIP (15/94, 16.0%).
Table 5.
Antimicrobial resistance profiles of Salmonella enterica Albany isolated from duck feces.
| Antimicrobial agent (antimicrobial subclass) |
No. of resistant CLSI subclasses |
No. of resistant isolates |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Profiles | FIS (SAs) | SXT (SAs) | NAL (Qs) | CIP (Qs) | AMP (AP) | STR (AMGs) | CHL (PHs) | TET (TETs) | Total 94 | |
| A1 | R | R | R | S | S | S | S | S | 2 | 42 |
| A2 | R | R | R | R | S | S | S | S | 2 | 15 |
| A3 | R | R | R | S | R | S | S | S | 3 | 5 |
| A4 | R | R | R | S | S | R | S | S | 3 | 2 |
| A5 | R | S | S | S | S | R | R | R | 4 | 1 |
| A6 | R | R | R | R | R | S | R | S | 4 | 2 |
| A7 | R | R | R | S | R | S | R | R | 5 | 9 |
| A8 | R | S | R | R | R | S | R | R | 5 | 1 |
| A9 | R | R | R | R | R | S | R | R | 5 | 15 |
| A10 | R | R | R | S | R | R | R | R | 6 | 1 |
| A11 | R | R | R | R | R | R | R | R | 6 | 1 |
FIS, sulfisoxazole; SAs, sulfonamides; SXT, trimethoprim/sulfamethoxazole; NAL, nalidixic acid; Qs, quinolones; CIP, ciprofloxacin; AMP, ampicillin; AP, aminopenicillin; STR, streptomycin; AMGs, aminoglycosides; CHL, chloramphenicol; PHs, phenicols; TET(s), tetracycline(s); S, susceptible; R, resistance.
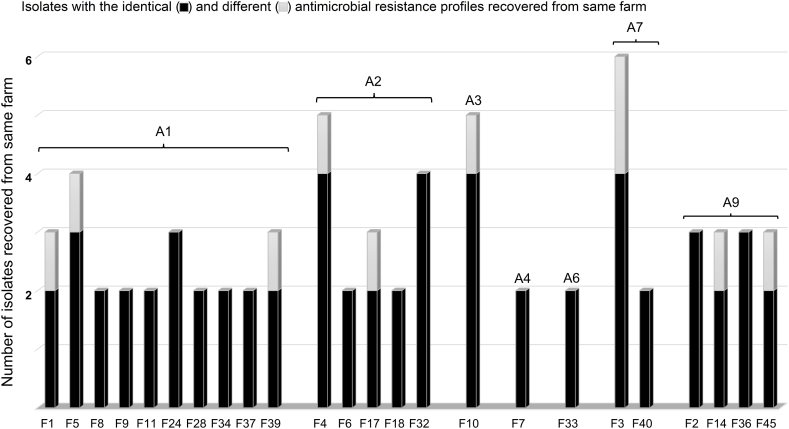
The antimicrobial resistance profiles of S. enterica Albany isolates repeatedly recovered from the same farm were similar (Table 6). Twenty-four farms where two or more S. enterica Albany isolates were detected had identical antibacterial resistance profiles (Fig. 2). These results confirm that S. enterica Albany isolates with identical antimicrobial resistance profiles were repeatedly recovered even when ducks raised on the farm were slaughtered, and a new flock of ducks was introduced to the same farm.
Table 6.
Description of Salmonella enterica Albany isolates used in the present study.
| Farms | Locations | Companies | Isolates | Collection dates | MLVA groups | IRBT cluster groups | Antimicrobial resistance profiles* |
|---|---|---|---|---|---|---|---|
| F1 | Jincheon | C1 | CB2 | 2019.04.18 | M1 | Cluster 6 | A7 |
| CB39 | 2019.08.26 | M1 | Cluster 4 | A1 | |||
| CB73 | 2019.11.01 | M1 | Cluster 4 | A1 | |||
| F2 | Eumseong | C2 | CB4 | 2019.04.23 | M1 | Cluster 5 | A9 |
| CB42 | 2019.08.26 | M1 | Cluster 5 | A9 | |||
| CB70 | 2019.10.21 | M1 | Cluster 5 | A9 | |||
| F3 | Jincheon | C2 | CB6 | 2019.04.24 | M3 | Cluster 9 | A7 |
| CB40 | 2019.08.26 | M3 | Cluster 9 | A11 | |||
| CB84 | 2019.12.20 | M3 | Cluster 9 | A7 | |||
| CB92 | 2020.02.27 | M3 | Cluster 10 | A7 | |||
| CB146 | 2020.08.14 | M3 | Cluster 10 | A7 | |||
| CB153 | 2020.09.08 | M3 | Cluster 14 | A10 | |||
| F4 | Jincheon | C1 | CB7 | 2019.04.29 | M1 | Cluster 4 | A1 |
| CB41 | 2019.08.26 | M1 | Cluster 4 | A2 | |||
| CB98 | 2020.03.25 | M1 | Cluster 8 | A2 | |||
| CB114 | 2020.06.08 | M1 | Cluster 8 | A2 | |||
| CB141 | 2020.08.06 | M1 | Cluster 8 | A2 | |||
| F5 | Cheongju | C1 | CB8 | 2019.05.20 | M1 | Cluster 4 | A1 |
| CB 37 | 2019.08.19 | M1 | Cluster 4 | A1 | |||
| CB48 | 2019.09.03 | M1 | Cluster 4 | A2 | |||
| CB136 | 2020.07.30 | M1 | Cluster 4 | A1 | |||
| F6 | Jincheon | C1 | CB9 | 2019.05.21 | M1 | Cluster 3 | A2 |
| CB140 | 2020.08.03 | M1 | Cluster 3 | A2 | |||
| F7 | Eumseong | C2 | CB10 | 2019.05.22 | M1 | Cluster 4 | A4 |
| CB76 | 2019.11.14 | M1 | Cluster 4 | A4 | |||
| F8 | Jincheon | C1 | CB21 | 2019.07.25 | M1 | Cluster 4 | A1 |
| CB54 | 2019.09.20 | M1 | Cluster 4 | A1 | |||
| F9 | Jincheon | C1 | CB19 | 2019.07.18 | M1 | Cluster 4 | A1 |
| CB93 | 2020.02.27 | M1 | Cluster 4 | A1 | |||
| F10 | Eumseong | C2 | CB20 | 2019.07.24 | M1 | Cluster 4 | A1 |
| CB52 | 2019.09.16 | M1 | Cluster 4 | A3 | |||
| CB81 | 2019.12.02 | M1 | Cluster 4 | A3 | |||
| CB112 | 2020.05.18 | M1 | Cluster 7 | A3 | |||
| CB132 | 2020.07.17 | M1 | Cluster 7 | A3 | |||
| F11 | Jincheon | C1 | CB7 | 2019.04.29 | M1 | Cluster 4 | A1 |
| CB22 | 2019.07.25 | M1 | Cluster 4 | A1 | |||
| F12 | Eumseong | C1 | CB25 | 2019.07.26 | M1 | Cluster 4 | A1 |
| F13 | Jincheon | C1 | CB28 | 2019.08.02 | M1 | Cluster 4 | A1 |
| F14 | Eumseong | C2 | CB11 | 2019.05.23 | M1 | Cluster 4 | A8 |
| CB29 | 2019.08.02 | M1 | Cluster 4 | A9 | |||
| CB127 | 2020.07.06 | M1 | Cluster 4 | A9 | |||
| F15 | Cheongju | C1 | CB30 | 2019.08.05 | M3 | Cluster 10 | A9 |
| F16 | Cheongju | C1 | CB31 | 2019.08.05 | M1 | Cluster 4 | A3 |
| CB36 | 2019.09.08 | M1 | Cluster 4 | A1 | |||
| F17 | Cheongju | C1 | CB63 | 2019.09.30. | M1 | Cluster 4 | A2 |
| CB69 | 2019.10.18 | M1 | Cluster 4 | A2 | |||
| CB148 | 2020.08.19 | M1 | Cluster 4 | A1 | |||
| F18 | Eumseong | C2 | CB33 | 2019.08.08 | M3 | Cluster 9 | A2 |
| CB94 | 2020.03.12 | M3 | Cluster 9 | A2 | |||
| F19 | Eumseong | C3 | CB38 | 2019.08.20 | M1 | Cluster 4 | A1 |
| F20 | Chungju | C1 | CB43 | 2019.08.26 | M1 | Cluster 4 | A1 |
| F21 | Jincheon | C4 | CB49 | 2019.09.05 | M1 | Cluster 7 | A9 |
| F22 | Jincheon | C1 | CB51 | 2019.09.16 | M1 | Cluster 4 | A1 |
| F23 | Cheongju | C1 | CB53 | 2019.09.16 | M1 | Cluster 4 | A1 |
| F24 | Eumseong | C1 | CB64 | 2019.10.04 | M1 | Cluster 1 | A1 |
| CB106 | 2020.04.16 | M1 | Cluster 1 | A1 | |||
| CB149 | 2020.08.24 | M1 | Cluster 1 | A1 | |||
| F25 | Jincheon | C3 | CB65 | 2019.10.08 | M2 | Cluster 2 | A7 |
| CB91 | 2020.02.20 | M1 | Cluster 16 | A1 | |||
| F26 | Jincheon | C1 | CB66 | 2019.10.08 | M1 | Cluster 4 | A1 |
| F27 | Eumseong | C6 | CB68 | 2019.10.18 | M1 | Cluster 7 | A1 |
| F28 | Cheongju | C1 | CB71 | 2019.10.25 | M1 | Cluster 4 | A1 |
| CB85 | 2019.12.27 | M1 | Cluster 4 | A1 | |||
| F29 | Eumseong | C2 | CB72 | 2019.10.31 | M5 | Cluster 9 | A9 |
| F30 | Goesan | C1 | CB26 | 2019.07.30 | M1 | Cluster 4 | A1 |
| F31 | Goesan | C1 | CB79 | 2019.11.25 | M1 | Cluster 13 | A1 |
| F32 | Eumseong | C1 | CB82 | 2019.12.04 | M1 | Cluster 4 | A2 |
| CB90 | 2020.01.30 | M1 | Cluster 4 | A2 | |||
| CB99 | 2020.03.27 | M1 | Cluster 4 | A2 | |||
| CB137 | 2020.07.31 | M1 | Cluster 4 | A2 | |||
| F33 | Jincheon | C1 | CB101 | 2020.04.02 | M1 | Cluster 4 | A6 |
| CB117 | 2020.06.17 | M1 | Cluster 4 | A6 | |||
| F34 | Eumseong | C1 | CB102 | 2020.04.06 | M1 | Cluster 4 | A1 |
| CB116 | 2020.06.16 | M1 | Cluster 4 | A1 | |||
| F35 | Cheongju | C3 | CB104 | 2020.04.13 | M1 | Cluster 4 | A9 |
| F36 | Cheongju | C2 | CB105 | 2020.04.13 | M1 | Cluster 7 | A9 |
| CB122 | 2020.06.25 | M1 | Cluster 7 | A9 | |||
| CB147 | 2020.08.18 | M1 | Cluster 7 | A9 | |||
| F37 | Jincheon | C1 | CB107 | 2020.04.17 | M1 | Cluster 4 | A1 |
| CB120 | 2020.06.25 | M1 | Cluster 4 | A1 | |||
| F38 | Jincheon | C2 | CB108 | 2020.04.20 | M1 | Cluster 4 | A1 |
| F39 | Eumseong | C2 | CB110 | 2020.04.23 | M1 | Cluster 4 | A1 |
| CB124 | 2020.07.02 | M1 | Cluster 12 | A1 | |||
| CB151 | 2020.08.27 | M1 | Cluster 11 | A4 | |||
| F40 | Eumseong | C2 | CB111 | 2020.04.23 | M4 | Cluster 9 | A7 |
| CB123 | 2020.07.02 | M4 | Cluster 9 | A7 | |||
| F41 | Jincheon | C2 | CB126 | 2020.07.06 | M3 | Cluster 10 | A7 |
| F42 | Jincheon | C1 | CB133 | 2020.07.20 | M1 | Cluster 4 | A1 |
| F43 | Cheongju | C1 | CB135 | 2020.07.30 | M1 | Cluster 7 | A1 |
| F44 | Jincheon | C5 | CB138 | 2020.07.31 | M4 | Cluster 9 | A9 |
| F45 | Eumseong | C2 | CB95 | 2020.03.17 | M1 | Cluster 15 | A5 |
| CB139 | 2020.07.31 | M3 | Cluster 7 | A9 | |||
| CB143 | 2020.08.10 | M3 | Cluster 7 | A9 | |||
| F46 | Eumseong | C2 | CB142 | 2020.08.10 | M1 | Cluster 4 | A1 |
A1, FIS-SXT-NAL; A2, FIS-SXT-NAL-CIP; A3, FIS-SXT-NAL-AMP; A4, FIS-SXT-NAL-STR; A5, FIS-STR-CHL-TET; A6, FIS-SXT-NAL-CIP-AMP-CHL; A7, FIS-SXT-NAL-AMP-CHL-TET; A8, FIS-NAL-CIP-AMP-CHL-TET; A9, FIS-SXT-NAL-CIP-AMP-CHL-TET; A10, FIS-SXT-NAL-AMP-STR-CHL-TET; A11, FIS-SXT-NAL-CIP-AMP-STR-CHL-TET.
Fig. 2.
Isolates of Salmonella enterica Albany repeatedly recovered from the same farm indicated identical antimicrobial resistance profiles (A1, FIS-SXT-NAL; A2, FIS-SXT-NAL-CIP; A3, FIS-SXT-NAL-AMP; A4, FIS-SXT-NAL-STR; A6, FIS-SXT-NAL-CIP-AMP-CHL; A7, FIS-SXT-NAL-AMP-CHL-TET; A9, FIS-SXT-NAL-CIP-AMP-CHL-TET). A5, A8, A10, and A11 antimicrobial resistance profiles had one isolate each and were not represented. Two or more isolates from 24 farms had identical antimicrobial resistance profiles.
3.3. Strain typing of S. enterica Albany via MLVA
MLVA typing assigned the 94 S. enterica Albany isolates to five groups. Most isolates (77 recovered from 39 duck farms) were included in the M1 group, followed by 12 isolates recovered from five duck farms in the M3 group. Three isolates recovered from two duck farms formed the M4 group, while the M2 and M5 groups had one isolate each (Table 7). Of the 26 farms where two or more isolates were recovered, 24 farms yielded one MLVA group in the same farm (Table 6). These results confirmed that isolates of an identical MLVA group were repeatedly recovered from different duck flocks at different times on the same farm.
Table 7.
MLVA groups of Salmonella enterica Albany isolated from duck feces.
| VNTR markers | No. of isolates | No. of farms | ||||
|---|---|---|---|---|---|---|
| Groups | STTR7 | SATR1 | SATR2 | SATR3 | Total (94) | Total (48a) |
| M1 | 5 | 6 | 7 | 4 | 77 | 39 |
| M2 | 5 | 6 | 8 | 4 | 1 | 1 |
| M3 | 5 | 6 | 6 | 4 | 12 | 5 |
| M4 | 6 | 6 | 6 | 4 | 3 | 2 |
| M5 | 5 | 7 | 6 | 5 | 1 | 1 |
Isolates of farms F25 and F45 were included in two MLVA groups and counted in duplicate.
3.4. Strain typing of S. enterica Albany via IRBT analysis
The 94 S. enterica Albany isolates were classified into 16 clusters via IRBT analysis (Fig. 3). Of the 94 isolates, 52 belonged to Cluster 4, forming the largest group; 10 isolates belonged to Cluster 7; nine isolates belonged to Cluster 9; four isolates belonged to Cluster 10; and two isolates belonged to Cluster 3. Clusters 1, 5, and 8 each included three isolates, and eight clusters (Clusters 2, 6, and 11–16) included only one isolate each. S. enterica Albany recovered from existing and new flocks of ducks on the same farm exhibited identical IRBT clusters. Of the 26 farms where two or more isolates were recovered, 19 farms yielded only one IRBT cluster, while 5 farms (F1, F3, F4, F10, F45) yielded isolates in more than two IRBT clusters, although some isolates did belong to the same IRBT cluster.
Fig. 3.
Dendrogram clustered using IRBT for 94 Salmonella enterica Albany isolates. The vertical line represents the cutoff value (0.126), and all isolates were classified into 16 clusters via IRBT. For ease of reading, clusters composed of more than two isolates are shaded in orange; two isolates are shaded in yellow; single isolates are shaded in green.
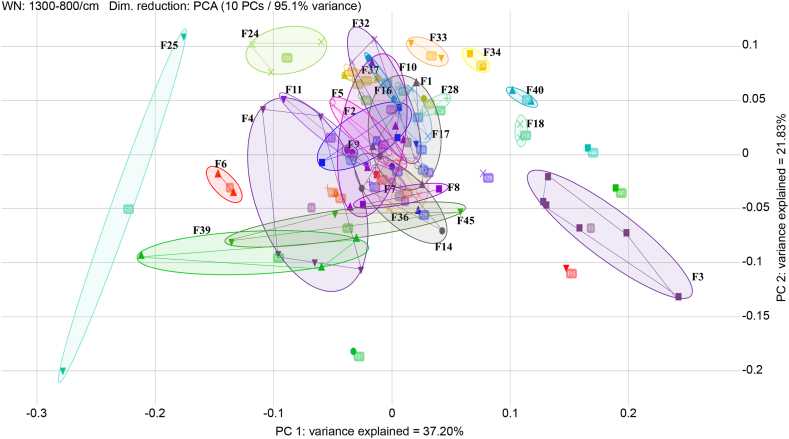
The result of IRBT typing is displayed in 2D scatter plot format. Isolates recovered from the same farm appear as point sets located close together and delimited by ellipses (Fig. 4). These results showed that S. enterica Albany isolates included in an identical cluster were repeatedly recovered from a farm even when a new flock was introduced after the existing flock was slaughtered.
Fig. 4.
Assessment of 2D scatter plot for 94 isolates of PCA using IRBT. The target was the farms. Isolates delimited using ellipses show the same farm, and points within the ellipse were located close together. Some farms overlap, but non-overlapping farms were distinguished in the spectra.
3.5. IRBT strain typing compared with MLVA
Table 6 shows a comparison of IRBT spectroscopic typing with MLVA results. There were 16 cluster groups in IRBT and five in MLVA. All isolates in Clusters 1, 3–6, 8, 11–13, 15, and 16 of IRBT corresponded to the M1 group, indicating that IRBT had a higher discriminatory power than MLVA for these isolates. The clustering results of IRBT and MLVA were consistent for Clusters 2 and 10 and M2 and M3. However, IRBT Clusters 7 and 9 corresponded to M1, M3, M4, and M5, indicating that MLVA had a higher discriminatory power than IRBT for these isolates.
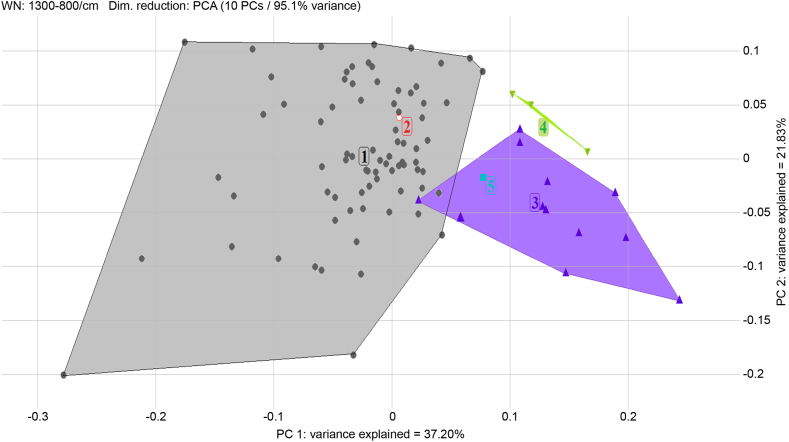
Of the 94 isolates from various farms, locations, and companies, 52 (55%) were included in Cluster 4 and M1 group, forming one large group (Fig. 3). Isolates recovered repeatedly from the same farm showed identical MLVA and IRBT typing results; for example, all isolates recovered from farm F2 were included in the M1 group and Cluster 5. Using both typing methods, isolates recovered from the same farm could be distinguished in more detail. Isolates recovered from farm F4 were divided into two isolates included in the M1 group/Cluster 4 and three other isolates included in the M1 group/Cluster 8 (Table 6). A 2D scatter plot of the IRBT typing demonstrated that three large MLVA groups (M1, M3, and M4) could be defined (Fig. 5).
Fig. 5.
Assessment of 2D scatter plot for 94 isolates of PCA using IRBT. The target was the MLVA groups. Isolates delimited using lines show the identical MLVA groups (M1, M3, and M4). These groups did not overlap, and a distinction was made in the spectra.
3.6. Strain typing and antimicrobial susceptibility
IRBT and MLVA typing results of the 94 isolates were statistically correlated with antimicrobial resistance profiles (Fisher's Exact Test, p < 0.05) but did not consistently correspond with each other (Fig. 6A and B). However, isolates repeatedly recovered from the same farm showed the same MLVA results, IRBT findings, and antimicrobial resistance profiles (Table 6). Among the 26 farms that yielded multiple S. enterica Albany isolates, 20 yielded isolates with identical MLVA results, IRBT findings, and antimicrobial resistance profiles (Fig. 6C).
Fig. 6.
Comparison of strain typing results and antimicrobial resistance profiles. The typing results of MLVA (A) and IRBT (B) for the 94 isolates did not consistently correspond with the antimicrobial resistance profiles. The isolates recovered from the same duck farms showed identical MLVA results, IRBT findings, and antimicrobial resistance profiles. M2 and M5 groups had one isolate each and were not represented (C).
4. Discussion
In this study, we examined 141 isolates of Salmonella spp. obtained from fecal samples of 5–6-week-old ducks from 105 duck farms between April 2019 and October 2020. S. enterica Albany isolates, the most recovered serovar, were tested for antimicrobial susceptibility to elucidate drug resistance profiles. The S. enterica Albany isolates were typed using IRBT and MLVA.
We isolated Salmonella spp. from 141 (19%) of 745 duck fecal samples, similar to the prevalence reported by a previous study [21]. In addition to the serovars reported in the previous study, we detected S. enterica Albany, S. Montevideo, S. Virchow, and S. Zanzibar, with S. enterica Albany as the most common serovar. The prevalence of S. enterica Albany has recently increased dramatically in poultry farms in Korea [4], but it had not been identified in chickens and ducks in Korea before 2016 [21]. S. enterica Albany has been commonly isolated from poultry and other livestock in Southeast Asia and Western countries, and it has recently been reported as an important serovar affecting humans [22,23]. Therefore, we investigated the characteristics of S. enterica Albany using phenotypic, genotypic, and spectroscopic techniques to develop prevention and control strategies for Salmonella infection.
All S. enterica Albany isolates recovered from ducks in this study were resistant to at least three of the antimicrobial agents tested. We detected resistance to FIS, NAL, SXT, AMP, CIP, CHL, TET, and STR in various patterns. In previous studies, S. enterica Albany isolated from poultry in Korea [4], Malaysia [24], and Cambodia [25] and from pigs in Taiwan [22] showed antimicrobial resistance, similar to the findings of our study. Our S. enterica Albany isolates had high resistance (close to 100%) to FIS, NAL, and SXT antibiotics and a low resistance of 5.3% STR. Approximately 30% of isolates were resistant to AMP, CHL, and TET, which was lower than approximately 90% of the isolates in previous studies [4,22,24,25]. These trends suggest that the amount of antibiotics used in duck production is lower than that used in chicken production in Korea [21]. Furthermore, 36.2% of isolates were resistant to CIP, which is higher than the approximately 6% of isolates found in previous studies in chickens [4,25]. S. enterica Albany isolated from ducks showed higher resistance to CIP than those in previous studies, possibly due to the overuse and abuse of fluoroquinolones for disease treatment in ducks [26]. Increased rates of decreased susceptibility to quinolones in salmonella have been reported with the prolonged use of excessive doses in poultry [4]. CIP is commonly used to treat non-typhoidal Salmonella infections (Bangera et al., 2019; [4,27], and poultry may have played a role in antibiotic-resistant Salmonella spp. infections in humans [28].
We found that 39.4% (37/94) of S. enterica Albany isolates were MDR strains. In previous studies, a majority of S. enterica Albany from both ducks [1] and broiler chickens [4] in Korea were MDR. The indiscriminate use of antibiotics has increased the number of MDR Salmonella strains and the antibiotic resistance rate [29]. In addition, the existence of MDR Salmonella strains suggests that they may pose a public health problem [30].
ACSuTN (ampicillin, chloramphenicol, sulfisoxazole, tetracycline, and nalicixic acid)-resistant S. enterica Albany accounted for 67.6% (25/37) of the MDR isolates. Resistance to these antimicrobials may be due to their long history of use in poultry farms to treat infections [4]. ACSSuT (ampicillin, chloramphenicol, streptomycin, sulfisoxazole, and tetracycline)-resistant S. Typhimurium was first reported in the United Kingdom in 1984 [31], with many ACSSuT-resistant Salmonella isolates reported subsequently [[32], [33], [34]]. ACSSuT Salmonella isolates with co-resistance to clinically important antibiotics (fluoroquinolone, cephalosporins, and colistin) complicate treatment and pose a real threat to global public health [4,30,33]. Two of our S. enterica Albany isolates showed an ACSSuT resistance profile, with one also showing co-resistance to ciprofloxacin.
Strain typing S. enterica Albany using MLVA and IRBT classified the isolates into five groups and 16 clusters, respectively. Based on the 2D scatter plot assessment of PCA (Fig. 5), IRBT could differentiate MLVA genotypic variation related to strain type due to the three separate data sets (M1, M3, and M4). High discriminating power for the M1, M3, and M4 groups of S. enterica Albany isolates was identified, suggesting that IRBT could be used to classify MLVA genotypes. This difference in discrimination power seems to be due to the typing principles – genotyping for MLVA and molecular structure analysis for IRBT. MLVA takes advantage of VNTRs to distinguish strains and can be easily performed at low cost using conventional PCR using identical primers [7]. Therefore, it is a popular method for subtyping for public health surveillance and outbreak investigations of Salmonella using DNA-based techniques [35]. On the other hand, IRBT analyzes molecular structures such as carbohydrate composition [36]. IRBT has recently become an alternate method for bacterial typing due to its ease of use and high discriminatory power in recognizing clonal relationships among bacterial isolates [11]. This is an automated system that is simple, quick, and reliable [37]. IRBT was used for transmission route analyses and outbreak investigations of strains [36]. A combination of these two typing methods with different principles could investigate the epidemiological relationship of isolates in more detail.
Strain typing using IRBT and MLVA showed that the S. enterica Albany isolates had high genetic and molecular structure homology despite being recovered from various farms with different locations and companies. S. enterica Albany isolates had 82% (77/94) similarity, as determined by MLVA, and 55% (52/94) similarity, as determined by IRBT. This suggests clonal dissemination of S. enterica Albany across duck farms. These results are consistent with the high genetic homology of S. enterica Albany isolated from domestic poultry farms analyzed via PFGE [4].
Moreover, we found that isolates repeatedly recovered from the same farm showed identical IRBT clusters or MLVA groups. Repeated acquisition of an identical strain may represent a risk of persistent infection due to the continued presence of the strain in the farm environment. This suggests that cleaning and disinfection before repopulation were performed improperly or inefficiently, resulting in Salmonella remaining in the environment and causing ongoing infection. Duck farms require proper sanitation after all ducks are removed to reduce Salmonella contamination of the environment.
Isolates repeatedly recovered from the same farm exhibited identical MLVA and IRBT clusters (Fig. 3). Moreover, isolates from the same farm could be distinguished in more detail using both typing methods (IRBT and MLVA). The two typing methods using different principles showed different results, and selecting one as a universal and ideally applicable typing method was impossible. Isolates could be distinguished more extensively to investigate the epidemiological relationship using both methods.
The typing results of IRBT and MLVA did not consistently correspond with the antimicrobial resistance profiles. However, isolates repeatedly recovered from duck farms did show identical MLVA results, IRBT findings, and antimicrobial resistance profiles (Fig. 6C). We noticed antimicrobial resistance diversity among isolates with identical typing results recovered from different duck farms. Since antibiotic consumption and use and resistance are related, changes in consumption composition may alter resistance patterns. In our study, different duck farms may have used different antibiotic treatment programs, causing different antimicrobial resistance profiles in S. enterica Albany isolates with the same strain typing.
The typing results of genotypic (MLVA) and spectroscopic (IRBT) techniques and the phenotypic results of antimicrobial resistance profiles were consistent for the same farm, which suggests that even if the existing duck flock was slaughtered and a new flock was stocked, re-infection with S. enterica Albany remaining on the farm could still occur.
5. Conclusions
The prevalence of S. enterica was 19%, and S. enterica Albany was predominantly distributed in duck farms in Korea. Of these 39.4% of S. enterica Albany isolates were classified as MDR. S. enterica Albany isolates repeatedly recovered from the same duck farm had identical antimicrobial resistance profiles, IRBT clusters, and MLVA groups. Therefore, re-infection with S. enterica Albany remaining in the environment is plausible, even if a new duck flock is introduced after slaughtering all ducks on the same farm. Farms require proper cleaning and disinfection before new duck flocks are repopulated. In this study, the epidemiological investigation of S. enterica Albany contamination was performed only in duck farms in Korea. Additional epidemiological analysis of duck carcasses in slaughterhouses is needed to investigate the potential of their transmission to humans via the food chain.
Funding
This research did not receive any specific grants from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics approval
The present study did not require ethical approval as we collected feces only from normal functioning farms and did not alter their routine.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Mina Han: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Munhui Chae: Methodology. Sangkab Lee: Investigation. Kyongok No: Conceptualization. Seongtae Han: Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Institute of Chungbuk Provincial Veterinary Service and Research for providing the environment and resources for the research and Jungeun Bae of Bruker Korea Co., Ltd. for technical support.
References
- 1.Silva-Hidalgo G., López-Moreno H.S., Ortiz-Navarrete V.F., Juárez-Barranco F., López-Valenzuela M. Fecal excretion of Salmonella Albany, its isolation in the diet and health repercussion on an ocelot (Leopardus pardalis) in captivity. Vet. Mex. 2012;43:59–69. [Google Scholar]
- 2.Kim T.S., Kim G.S., Son J.S., Lai V.D., Mo I.P., Jang H. Prevalence, biosecurity factor, and antimicrobial susceptibility analysis of Salmonella species isolated from commercial duck farms in Korea. Poultry Sci. 2021;100 doi: 10.1016/j.psj.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Little C.L., Richardson J.F., Owen R.J., de Pinna E., Threlfall E.J. Prevalence, characterisation and antimicrobial resistance of Campylobacter and Salmonella in raw poultrymeat in the UK, 2003–2005. Int. J. Environ. Health Res. 2008;18:403–414. doi: 10.1080/09603120802100220. [DOI] [PubMed] [Google Scholar]
- 4.Wei B., Shang K., Cha S.Y., Zhang J.F., Jang H.K., Kang M. Clonal dissemination of Salmonella enterica serovar Albany with concurrent resistance to ampicillin, chloramphenicol, streptomycin, Sulfisoxazole, tetracycline, and nalidixic acid in broiler chicken in Korea. Poultry Sci. 2021;100 doi: 10.1016/j.psj.2021.101141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X., Zhu L., Wang X., Li J., Tang B. Evaluation of IR biotyper for Lactiplantibacillus plantarum typing and its application potential in probiotic preliminary screening. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.823120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalai W., Martinez I., Bikandi J., Messadi L., Khazri I., Souissi N., Saidani M., Slim A.F., Boutiba-Ben Boubaker I., Garaizar J. Antimicrobial susceptibility and MLVA analysis of S. Typhimurium strains isolated from human and poultry samples in Tunisia. The Journal of Infection in Developing Countries. 2018;12:313–320. doi: 10.3855/jidc.10089. [DOI] [PubMed] [Google Scholar]
- 7.Van Belkum A. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA) FEMS Immunol. Med. Microbiol. 2007;49:22–27. doi: 10.1111/j.1574-695X.2006.00173.x. [DOI] [PubMed] [Google Scholar]
- 8.Lindstedt B.A. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis. 2005;26:2567–2582. doi: 10.1002/elps.200500096. [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Diao B., Cui Z., Yan M., Kan B. Genotyping of Salmonella Typhi using 8-loci multi locus VNTR analysis. Gut Pathog. 2016;8:14. doi: 10.1186/s13099-016-0094-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinkelacker A.G., Vogt S., Oberhettinger P., Mauder N., Rau J., Kostrzewa M., Rossen J.W.A., Autenrieth I.B., Peter S., Liese J. Typing and species identification of clinical Klebsiella isolates by Fourier transform infrared spectroscopy and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2018;56 doi: 10.1128/jcm.00843-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramzan M., Raza A., un Nisa Z., Ghulam Musharraf S. Recent studies on advance spectroscopic techniques for the identification of microorganisms: a review. Arab. J. Chem. 2023;3 doi: 10.1016/j.arabjc.2022.104521. [DOI] [Google Scholar]
- 12.Cordovana M., Mauder N., Join-Lambert O., Gravey F., LeHello S., Auzou M., Pitti M., Zoppi S., Buhl M., Steinmann J., Frickmann H., Dekker D., Funashima Y., Nagasawa Z., Soki J., Orosz L., Veloo A.C., Justesen U.S., Holt H.M., Liberatore A., Ambretti S., Pongolini S., Soliani L., Wille A., Rojak S., Hagen R.M., May J., Pranada A.B., Kostrzewa M. Machine learning-based typing of Salmonella enterica O-serogroups by the Fourier-transform infrared (FTIR) spectroscopy-based IR biotyper system. J. Microbiol. Methods. 2022;201 doi: 10.1016/j.mimet.2022.106564. [DOI] [PubMed] [Google Scholar]
- 13.Kang M.S., Kim A., Jung B.Y., Her M., Jeong W., Cho Y.M., Oh J.Y., Lee Y.J., Kwon J.H., Kwon Y.K. Characterization of antimicrobial resistance of recent Salmonella enterica serovar Gallinarum isolates from chickens in South Korea. Avian Pathol. 2010;39:201–205. doi: 10.1080/03079451003767261. [DOI] [PubMed] [Google Scholar]
- 14.Ministry of Agriculture, Food and Rural Affairs 2021년도 국가 항생제 사용 및 내성 모니터링 -동물, 축산물- (2021 Monitoring of national antimicrobial use and resistance -animal, retail meats-) 2021. https://www.mfds.go.kr/brd/m_231/view.do?seq=33053 2022 Aug 9.
- 15.Yue H., Zhang B., Zhu X., Zhang H., Tang C. Comparison of culture methods for isolation of salmonella in yak fecal samples. Indian J. Microbiol. 2014;54:223–226. doi: 10.1007/s12088-013-0423-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun Y.S., Chae S.J., Na H.Y., Chung G.T., Yoo C.K., Lee D.Y. Modified method of multilocus sequence typing (MLST) for serotyping in Salmonella species. J. Bacteriol. Virol. 2015;45:314–318. doi: 10.4167/jbv.2015.45.4.314. [DOI] [Google Scholar]
- 17.Zhu C., Yue M., Rankin S., Weill F.X., Frey J., Schifferli D.M. One-step identification of five prominent chicken Salmonella serovars and biotypes. J. Clin. Microbiol. 2015;53:3881–3883. doi: 10.1128/jcm.01976-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Z., Alikhan N.F., Mohamed K., Fan Y., Achtman M. The EnteroBase user's guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute . 29th ed. CLSI Document m100; 2019. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 20.Centers for Disease Control and Prevention. NARMS . vol. 2014. Centers for Disease Control and Prevention; Atlanta, GA: 2014. (Human Isolates Surveillance Report, Final Report). [Google Scholar]
- 21.Kim H., Lee J., Jang Y., Chang B., Kim A., Choe N. Prevalence and antimicrobial resistance of Salmonella spp. and Escherichia coli isolated from ducks in Korea. Korean Journal of Veterinary Research. 2016;56:91–95. doi: 10.14405/kjvr.2016.56.2.91. [DOI] [Google Scholar]
- 22.Kuo H.C., Lauderdale T.L., Lo D.Y., Chen C.L., Chen P.C., Liang S.Y., Kuo J.C., Liao Y.S., Liao C.H., Tsao C.S., Chiou C.S. An association of genotypes and antimicrobial resistance patterns among Salmonella isolates from pigs and humans in Taiwan. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuzihara T.O., Fernandes S.A., Franco B.D. Prevalence and dissemination of Salmonella serotypes along the slaughtering process in Brazilian small poultry slaughterhouses. J. Food Protect. 2000;63:1749–1753. doi: 10.4315/0362-028X-63.12.1749. [DOI] [PubMed] [Google Scholar]
- 24.Chuah L.O., Shamila Syuhada A.K., Mohamad Suhaimi I., Farah Hanim T., Rusul G. Genetic relatedness, antimicrobial resistance and biofilm formation of Salmonella isolated from naturally contaminated poultry and their processing environment in northern Malaysia. Food Res. Int. 2018;105:743–751. doi: 10.1016/j.foodres.2017.11.066. [DOI] [PubMed] [Google Scholar]
- 25.Vuthy Y., Lay K.S., Seiha H., Kerleguer A., Aidara-Kane A. Antibiotic susceptibility and molecular characterization of resistance genes among Escherichia coli and among Salmonella subsp. in chicken food chains. Asian Pac. J. Trop. Biomed. 2017;7:670–674. doi: 10.1016/j.apjtb.2017.07.002. [DOI] [Google Scholar]
- 26.Zhu D., Zheng M., Xu J., Wang M., Jia R., Chen S., Liu M., Zhao X., Yang Q., Wu Y., Zhang S., Huang J., Liu Y., Zhang L., Yu Y., Pan L., Chen X., Cheng A. Prevalence of fluoroquinolone resistance and mutations in the gyr A, par C and par E genes of Riemerella anatipestifer isolated from ducks in China. BMC Microbiol. 2019;19:1–8. doi: 10.1186/s12866-019-1659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bangera S.R., Umakanth S., Chowdhury G., Saha R.N., Mukhopadhyay A.K., Ballal M. Poultry. A receptacle for non-typhoidal Salmonellae and antimicrobial resistance. Iranian Jornal of Microbiology. 2019;11:31–38. [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann S., Batz M.B., Morris J.G. Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J. Food Protect. 2012;75:1292–1302. doi: 10.4315/0362-028X.JFP-11-417. [DOI] [PubMed] [Google Scholar]
- 29.Ma F., Xu S., Tang Z., Li Z., Zhang L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosafety and Health. 2021;3:32–38. doi: 10.1016/j.bsheal.2020.09.004. [DOI] [Google Scholar]
- 30.Angulo F.J., Nargund V.N., Chiller T.C. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J Vet Med B Infect Dis Vet Public Health. 2004;51:374–379. doi: 10.1111/j.1439-0450.2004.00789.x. [DOI] [PubMed] [Google Scholar]
- 31.Threlfall E.J., Rowe B., Ward L.R. A comparison of multiple drug resistance in salmonellas from humans and food animals in England and Wales, 1981 and 1990. Epidemiol. Infect. 1993;111:189–197. doi: 10.1017/S0950268800056892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y., Li M., Xu X., Fu Y., Xiong Z., Zhang L., Qu X., Zhang H., Wei Y., Zhan Z., Chen Z., Bai J., Liao M., Zhang J. High-levels of resistance to quinolone and cephalosporin antibiotics in MDR-ACSSuT Salmonella enterica serovar Enteritidis mainly isolated from patients and foods in Shanghai, China. Int. J. Food Microbiol. 2018;286:190–196. doi: 10.1016/j.ijfoodmicro.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 33.Dong N., Li Y., Zhao J., Ma H., Wang J., Liang B., Du X., Wu F., Xia S., Yang X., Liu H., Yang C., Qiu S., Song H., Jia L., Li Y., Sun Y. The phenotypic and molecular characteristics of antimicrobial resistance of Salmonella enterica subsp. enterica serovar Typhimurium in Henan Province, China. BMC Infect. Dis. 2020;20:511. doi: 10.1186/s12879-020-05203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Threlfall E.J., Frost J.A., Ward L.R., Rowe B. Increasing spectrum of resistance in multiresistant Salmonella typhimurium. Lancet. 1996;347:1053–1054. doi: 10.1016/S0140-6736(96)90199-3. [DOI] [PubMed] [Google Scholar]
- 35.Tang S., Orsi R.H., Luo H., Ge C., Zhang G., Baker R.C., Stevenson A., Wiedmann M. Assessment and comparison of molecular subtyping and characterization methods for Salmonella. Front. Microbiol. 2019;10:1591. doi: 10.3389/fmicb.2019.01591. https://doi:10.3389/fmicb.2019.01591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johler S., Stephan R., Althaus D., Ehling-Schulz M., Grunert T. High-resolution subtyping of Staphylococcus aureus strains by means of Fourier-transform infrared spectroscopy. Syst. Appl. Microbiol. 2016;39:189–194. doi: 10.1016/j.syapm.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Quintelas C., Ferreira E.C., Lopes J.A., Sousa C. An Overview of the Evolution of infrared spectroscopy applied to bacterial typing. Biotechnol. J. 2018;13 doi: 10.1002/biot.201700449. https://doi:10.1002/biot.201700449 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.