Abstract
Learning how multicellular organs are developed from single cells to different cell types is a fundamental problem in biology. With the high-throughput scRNA-seq technology, computational methods have been developed to reveal the temporal dynamics of single cells from transcriptomic data, from phenomena on cell trajectories to the underlying mechanism that formed the trajectory. There are several distinct families of computational methods including Trajectory Inference (TI), Lineage Tracing (LT), and Gene Regulatory Network (GRN) Inference which are involved in such studies. This review summarizes these computational approaches which use scRNA-seq data to study cell differentiation and cell fate specification as well as the advantages and limitations of different methods. We further discuss how GRNs can potentially affect cell fate decisions and trajectory structures.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12551-023-01090-5.
Keywords: Single-cell RNA sequencing, Trajectory inference, Lineage tracing, Gene regulatory network inference
Introduction
Single-cell multi-omics measurements have opened up tremendous opportunities to study the temporal dynamics of cells. Sequencing experiments, such as scRNA-seq, provide a “snapshot" of cells’ transcriptomic profiles. These snapshots can potentially contain cells at different developmental stages, allowing us to characterize the entire cellular dynamic process. Computational algorithms have been developed to uncover biological artifacts of cells, such as cell type, state transitions, and lineages, as well as of genes, such as differential expressions and gene regulatory networks (GRNs). The unprecedented diversity and resolution of single-cell multi-omics analysis have revolutionized modern computational biology, and the field continues to evolve with new technologies and algorithms.
Various downstream analyses have been widely studied, centered around scRNA-seq technology. With the assumption that the sequenced samples contain cells from all different developmental stages, scRNA-seq data allows us to infer the so-called developmental trajectory of the cells, which ideally records the changes in cell states. This trajectory represents how cells’ overall transcriptomic profiles shift throughout cell differentiation, without uncovering the details of the lineage of each cell (Wagner and Klein 2020), or the interactions between specific genes. Specific regulatory interactions between genes can be inferred from the data as well to create a GRN, where each node represents a gene and the edges between nodes represent gene regulations. Meanwhile, the cells’ lineage history can be recorded using sequencing-based lineage tracing technologies that generate barcodes with unique marks denoting cells’ clonal information.
This review focuses on three major aspects of studying temporal dynamics from single-cell multi-omics sequencing technologies: Trajectory Inference (TI), Lineage Tracing (LT), and Gene Regulatory Network (GRN) Inference. TI and LT methods aim to learn cell temporal changes along either a “pseudotime” or real time, whereas GRN methods aim to learn the underlying mechanisms that govern the observed gene expression profiles and their dynamics. Both TI and GRN inference can be performed with only scRNA-seq datasets, while the reconstruction of the cell lineage is usually performed based on lineage barcodes. We discuss the assumptions and biological interpretations of each inference task, various computational approaches to solve the problem, and their advantages and limitations.A summary of these three computational tasks is in Table S1. Moreover, we specially discuss efforts to jointly model multiple aspects of temporal dynamics, which are often solved independently for the state-of-the-art. We aim to provide an overview of the commonalities and differences between different approaches and to highlight future directions for the integration of the three analyses and the comprehensive understanding of cells’ temporal dynamics.
Learning cell developmental trajectories using TI methods
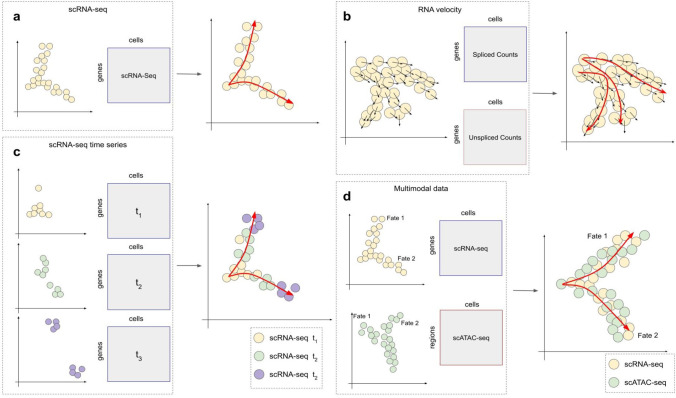
Trajectory inference (TI) methods are commonly used to learn the cell differentiation or developmental trajectories from scRNA-seq data (Fig. 1a). TI methods aim to find the trajectory backbone representing the major cell states and the dynamic paths between the states, and then “sort” the cells onto the backbone structure into a temporal ordering, and each cell is assigned a pseudotime. The trajectories represent how different cell states are connected. The inferred cell trajectories from the TI methods can vary in topology: linear, tree-shaped, and more complex ones such as cyclic and disconnected graphs. Numerous TI methods have been developed, with most methods focusing on inferring a specific type of topology. However, more recent methods aim to infer different types of topology using a single model without restricting the inferred trajectory’s topology. These methods tend to use more than one scRNA-seq dataset or incorporate other modalities besides transcriptomes (Hao et al. 2021; Lange et al. 2022; Welch et al. 2017; Zhang et al. 2022b; Zhang and Zhang 2021).
Fig. 1.
Inferring developmental trajectories from single-cell omics data. a Trajectory inference using a single batch of scRNA-seq data. b Trajectory inference using RNA velocity information. With the input of both single-cell spliced and unspliced RNA counts, RNA velocity can be calculated and can be used to infer the developmental trajectory. c Trajectory inference using scRNA-seq time series data. Given cell-by-gene matrices measured at different time points, the developmental trajectory of cells covering more developmental stages is inferred. d Trajectory inference using single-cell multimodal data such as scRNA-seq and scATAC-seq data. With multiple cell-by-feature (gene expression, chromatin accessibility, protein abundance, etc.) matrices, a joint developmental trajectory that combines the different modalities is inferred
The majority of trajectory inference methods are not applied directly to the input cell by the observed gene count matrix. Dimensionality reduction methods, such as principle component analysis (PCA) and independent component analysis (ICA), are applied to reduce the number of dimensions before learning the trajectory of cells. On the reduced dimensions of the dataset, some methods aim to infer an accurate pseudotime ordering of cells, which can, in turn, be translated to a linear trajectory (Saelens et al. 2019), such as scShaper (Smolander et al. 2021), SCORPIUS (Cannoodt et al. 2016) and MATCHER (Welch et al. 2017). Some other methods, including Slingshot (Street et al. 2018) and PAGA-Tree (Wolf et al. 2019), are able to infer tree-shaped trajectories where specific cell states can develop into multiple possible states. However, such methods do not take into account cyclic trajectories (cell cycle) and instead connect all cell states under a single tree graph. Some methods sought to resolve cell cycles from the single cell transcriptomic data such as reCAT (Liu et al. 2017). A few methods are able to detect most of the trajectory types, i.e. PAGA (Wolf et al. 2019) and StemID (Grün et al. 2016), but their performance can be inconsistent for different topology structures. Comprehensive comparisons of these methods (Saelens et al. 2019) indicate that there is no universal superior method for trajectory inference on all scRNA-seq datasets, due to the diversity and complexity of the underlying developmental trajectory. However, they provide a detailed method selection guide called dynguidelines to help select the best methods for different types of topologies. For example, PAGA and slingshot are two of the best-performing methods for tree topologies while SCORPIUS is better for lineage topologies. The authors also claimed that new methods should focus on improving the unbiased inference of trees, cyclic graphs, and disconnected topologies, which would allow the TI methods to perform well on experimental datasets with unknown developmental trajectories.
Recent developments in trajectory inference methods have expanded beyond the traditional deterministic modeling of the developmental trajectory. Probabilistic methods such as Palantir (Setty et al. 2019) and CSHMM (Lin and Bar-Joseph 2019) introduce uncertainty about the pseudotime of cells or their belonging to certain branches of the trajectory. In these models, each cell’s pseudotime can be drawn from a random variable, and different probabilistic models, such as the hidden Markov model, are used to represent the developmental trajectory of the cells. Meanwhile, some methods aim to use data types in addition to the mRNA counts in scRNA-seq data to build cell trajectories (Fig. 1b-d). First, with the abundance of unspliced and spliced mRNAs, the change in mRNA abundance, termed RNA velocity, can be inferred (Bergen et al. 2020; Manno et al. 2018). RNA velocity of all genes in a cell provides insights into the future state of the cell. Methods like CellRank (Lange et al. 2022) and CellPath (Zhang and Zhang 2021) utilize RNA velocity information to infer the developmental trajectories (Fig. 1b). CellRank is able to infer the developmental trajectory as well as cell fate specification probabilities. On the other hand, CellPath is able to infer multiple disconnected trajectories. A common advantage of methods that use RNA velocity is that they can learn the direction of the trajectories, compared to methods using only the spliced mRNA counts. However, RNA velocity inference methods also suffer from the high technical noise in scRNA-seq data (especially in unspliced counts) and model violations (Bergen et al. 2021). Recent methods, such as UniTVelo (Gao et al. 2022) and CellDancer (Li et al. 2023a), were developed with more realistic model assumptions, aiming to infer cell-specific, or temporal-regulated RNA velocity. Another type of approach to learning the directions of trajectories is to use time-series scRNA-seq data (Fig. 1c). By sequencing cells at different time points, multiple cell-by-gene matrices can be obtained to cover states of different developmental stages. Tempora (Tran and Bader 2020) and CSHMM (Lin and Bar-Joseph 2019) are methods developed for time-series data.
Single-cell multi-omics data, such as jointly profiled transcriptome and epigenome data, or jointly profiled transcriptome and proteome data, have been used to infer shared cell trajectories across modalities (Fig. 1d). Methods such as MATCHER (Welch et al. 2017) attempt to use manifold alignment to integrate different modalities, while other methods such as scDART (Zhang et al. 2022b) and Seurat v4 (Hao et al. 2021), learn a common embedding of the data and other TI methods can be applied to obtain the developmental trajectory. MultiVelo (Li et al. 2022) uses paired scRNA-seq and scATAC-seq data to infer RNA velocity as well as the temporal relationships between chromatin states changes and transcription kinetics. These methods have the potential to build a more accurate and comprehensive developmental trajectory by utilizing multimodal information.
Inferring the underlying developmental trajectory from a scRNA-seq dataset remains a challenging task, and successful trajectory inference relies on several assumptions: (1) The biological differentiation process must be dynamic, with gradual changes in gene expression during cell differentiation; (2) the dataset must contain enough cells with sufficient sampling depth to capture all transient states along the developmental trajectory. Furthermore, many trajectory methods require prior information, such as starting cells or clusters, to determine the directionality of the trajectory. With the increase of methods that use multi-modal data to perform TI, benchmarking studies are needed to compare the performance of such methods using different types of information with methods that use only mRNA counts. It is also important to explore the connection between cells’ pseudotime and other dynamic processes, such as cell divisions and gene regulatory programs, as these areas of research continue to develop.
Tracing lineage barcodes with scRNA-seq using CRISPR/Cas9 gene editing
Different from trajectory inference methods, which use the assumption of pseudotime, and require the datasets to contain cells at all developmental stages, lineage tracing techniques directly record cells’ true temporal orderings: the cell division histories. While traditional lineage tracing technologies can only measure a limited number of cells with low resolution, recent high-throughput single-cell sequencing technologies can jointly profile the transcriptomes and lineage information of cells at the same time. CRISPR/Cas9 is one well-known system for inducing guided genetic mutations on the target genome. By designing a target sequence and guide RNAs that bind the target sequence while attaching the Cas9 protein, the Cas9 protein can cut the target sequence at specific sites (Fig. 2 Step A). Then, the cell’s own DNA repair machinery will add or delete pieces of genetic material and these insertions or deletions are termed CRISPR/Cas9-induced mutations. The lineage tracing starts with injecting the target sequences, Cas9 proteins, and guide RNAs into the root cell, and the CRISPR/Cas9-induced mutations will occur during cell divisions. The induced genetic mutations can be passed down and accumulate during generations of cell divisions (Fig. 2 Step B). The mutated target sequence, or so-called lineage barcodes, can be sequenced together with the transcriptome using scRNA-seq technology. This framework allows us to obtain paired gene expression and clonal information on a single-cell level. It is worth mentioning some recent work on single-cell lineage tracing using endogenous mutations such as mitochondrial mutation variants (Lareau et al. 2020; Miller et al. 2022; Xu et al. 2019). However, such endogenous mutations are relatively noisy and uncontrollable. In comparison to the inferred lineages using CRISPR/Cas9 genetic barcodes, the inferred lineages from endogenous mutations tend to have much fewer internal nodes and worse resolution. Therefore, in this review, we will mainly focus on lineage tracing techniques based on CRISPR/Cas9-induced mutations.
Fig. 2.
Lineage reconstruction from CRISPR/Cas9 induced barcodes. Step A The lineage tracing system uses Cas9 proteins to generate double-stranded breaks that result in heritable insertions or deletions (mutations) after repair. Indels are induced at specific target sites of the barcode. Step B At the root, an unedited barcode, together with the Cas9 proteins and guide RNAs, is injected into the starting cell. Throughout generations of cell divisions, the Cas9 protein can bind to the designed barcode and induce mutations that are inherited and accumulated. Step C With the scRNA-seq experiment, the mutated barcodes of the present-time cells (leaf cells on the lineage tree) are sequenced. Step D Inferring the hidden lineage tree topology given the mutated barcodes of the leaf cells using computational methods
Works have been done to engineer the target sequence and other experimental setups to perform lineage tracing on various systems using CRISPR/Cas9-based lineage barcodes. scGESTALT (Raj et al. 2018) (single-cell Genome Editing of Synthetic Target Arrays for Lineage Tracing) uses multiple contiguous CRISPR/Cas9 targeting arrays to record the lineage of zebra fish and its brain development. scarTrace (Alemany et al. 2018), targets transgenic tandem fluorescent proteins and traces the lineages of different systems of zebrafish, while at the same time evaluating the efficiency of barcode generation using fluorescence intensity. CRISPR/Cas9 lineage tracing is also applied to other species such as mice (embryo (Chan et al. 2019), pancreatic cancer (Simeonov et al. 2021), etc.). With the joint profiles of lineage barcodes and gene expressions, a comprehensive cell fate map can be established by overlapping cell types onto the cell lineage tree.
The CRISPR/Cas9 lineage tracing system can potentially trace the lineage and transcriptomes of millions of single cells. Theoretically, the number of target sites and the number of mutations for each target site provide sufficient diversity to uniquely label every cell division in a lineage tree of thousands of leaves. However, in practice, perfectly tracing every cell division on the cell lineage tree is extremely challenging, due to the dynamic and uneven speed of cell divisions. Even under the assumption of a constant cell division rate, tuning the mutation rate of the CRISPR/Cas9 lineage tracing system to optimally generates a unique mutation pattern for every cell division is not realistic with current protocols. Other drawbacks of the CRISPR/Cas9 lineage tracing system (Salvador-Martínez et al. 2019), including excision dropouts (target deletion when two or more Cas9 proteins bind to neighboring target sites), biased distribution of mutated states, the limited capture efficiency of the sequencing experiment (which also causes dropout in the barcode data), etc., further add to the difficulties of inferring the correct lineage tree from the barcodes.
The lineage barcode of a cell can be represented computationally as a character vector of length equal to the number of target sites as designed by the CRISPR/Cas9 lineage recorder. Each character represents a state of the target site, which can be a mutated state, an unmutated state, or a dropout state. Therefore, the lineage barcode data can be represented as a cell-by-character matrix (Fig. 2 Step C). The objective is to infer the correct lineage tree that generates the barcode data observed at the leaf cells (Fig. 2 Step D). Different computational algorithms have been developed to infer the correct lineage tree from the CRISPR/Cas9-induced barcode data. Recently, a DREAM challenge was held to gather the community effort to compare the state-of-the-art lineage tree inference methods (Gong et al. 2021). DCLEAR (Gong et al. 2022) is a distance-based method that first calculates the pairwise distance between cells and then reconstructs the cell lineage using bottom-up (agglomerative) algorithms such as Neighbor-Joining (NJ) (Saitou and Nei 1987) or FastME (Lefort et al. 2015). Cassiopeia (Jones et al. 2020) is a parsimony-based method that aims at minimizing the number of mutations occurred on the reconstructed lineage tree. These methods were tested on both experimental and simulated datasets in the DREAM challenge and achieved the best performance benchmarked using Robinson-Foulds distance(Robinson and Foulds 1981) and Triplet distance, as described in Gong et al. (2021). More recently, integrated methods that combine lineage barcode and gene expression data are emerging, aiming to further improve the accuracy of cell lineage reconstruction. LinTIMaT (Zafar et al. 2020) develops a combined likelihood function and uses a local search framework to search for the tree with the maximum likelihood. LinRace (Pan et al. 2023) is another integrated method that first builds the lineage backbone using lineage barcode data and then refines subtrees using gene expression data and a likelihood-based local search program.
Using simulation tools and limited real datasets, we can compare how the integrated methods perform with the state-of-the-art barcode-based methods (Pan et al. 2022). These results showed the hypothetical optimal mutation rate to generate the barcode data and achieve the highest reconstruction accuracy. Even under ideal settings of mutation rate, barcode length, and other factors, the lineage reconstruction methods are still far from fully reconstructing the true lineages, mainly due to the large search space of possible tree structures with thousands of leaves (single cells). Meanwhile, although current methods can not reconstruct trees with high accuracy, the reconstructed trees are able to reflect the distributions of cell states under various subtrees.
Applying the CRISPR/Cas9-based lineage tracing systems to multiple species and resolving the gene expression distribution on the lineage tree led to the observation that in the reconstructed lineage tree, although a proportion of cells with the same cell type located in the same subtree, some cells of the same cell type are located in different subtrees, and the same subtree can have multiple cell types (Chan et al. 2019; Raj et al. 2018). This phenomenon, the partial consistency between transcriptome similarity and lineage similarity, is also observed in lineage-resolved species such as C. elegans (Tintori et al. 2016). Such inconsistency can be explained by asymmetric divisions of multipotent cells that develop into two daughter cells with different cell fates. Besides asymmetric divisions, varying differentiation speeds can also lead to diverse cell type distributions on the lineage tree.
The theoretical relationships between the pseudotime from TI methods and the lineage, however, are rarely discussed (Wagner and Klein 2020). A fully resolved lineage tree of C. elegans (Packer et al. 2019) demonstrates that the lineage and the transcriptomic trajectory can diverge and then converge at different developmental stages. Some methods have attempted to computationally model how cell state changes on the lineage tree, and utilize this model for various computational tasks, such as LinRace (Pan et al. 2023) (reconstructing cell division tree), CoSpar (Wang et al. 2022c) (learning cell transition map and predicting cell fate) and PhyloVelo (Wang et al. 2022a) (learning the phylogenetic velocity field which shows cell state trajectories). Moreover, LineageOT (Forrow and Schiebinger 2021) provides a unified framework of lineage and trajectory using optimal transport methods, but it is difficult to validate the model biologically. Overall, there is still much to uncover regarding different scenarios of coupling between cell divisions and transcriptomic trajectories.
Understanding temporal dynamics at molecular levels using GRNs
The developmental trajectories inferred from scRNA-seq or single-cell multi-omics data provide an understanding on cell state transitions, and the lineage trees inferred from lineage tracing barcodes show clonal relationships between cells from various cell states. Neither of these two explains the underlying molecular determinants of these dynamics, that is, what are the factors that regulate cell state transitions and determine the cell trajectories. It is commonly understood that the gene regulatory networks (GRNs) which represent relationships between genes are key to interpreting biological processes at molecular levels, and GRNs play a crucial role in forming cell states and cell trajectories (Guillemin and Stumpf 2020; Moris et al. 2016). It is also considered that these networks have dynamic interactions which regulate gene expression of different cell types in different developmental stages in a spatiotemporal manner (Cvekl and Zhang 2017; Kim et al. 2012), where GRNs can also vary depending on the spatial location of cells.
However, it is very difficult and time-consuming to experimentally measure GRNs, therefore, computational methods have been developed to infer GRNs from gene expression data, assuming that the regulatory dynamics can be observed through the changes in the gene expressions. With single-cell sequencing data quantifying the expression level of every gene in every cell, ideally, GRNs can be reconstructed on a whole-genome level. However, due to the complexity of the computational problem, all GRN inference methods start with a gene filtering step, to consider only a subset of genes of interest, or to remove genes that are hardly or lowly expressed in most of the cells. Then with the input of the filtered cell by gene matrix, GRN methods return a graph of directed or undirected connections between transcription factors and genes. GRN inference methods can be summarized into a few major categories (Nguyen et al. 2020): Boolean models (Hamey et al. 2017; Lim et al. 2016; Woodhouse et al. 2018) that model gene regulations as logical operations. These methods tend to use certain thresholds to binarize the gene expression levels, therefore require fewer parameters and can potentially avoid overfitting; Differential equation-based methods (Matsumoto et al. 2017; Ocone et al. 2015) that describe a gene’s expression as a function of other genes, and usually utilize pseudotime inferred by TI methods or time-series data, to characterize the causal relationships between genes; Correlation-based methods (Aibar et al. 2017; Chan et al. 2018; Liu et al. 2016) that calculates pairwise correlation metrics for the genes and build edges based on the rankings of each pair; and correlation ensembles with pseudotime (Deshpande et al. 2022; Gao et al. 2017; Specht and Li 2016; Xu et al. 2022) that calculates correlation scores in small windows of the pseudotime to take into account the temporal changes of the GRNs, and then combine the correlation matrices using ensemble strategies. More recently, methods have been developed to infer different GRNs for different cell types (Wang et al. 2022b; Zhang et al. 2023) or single cells (Zhang et al. 2022a).
Due to the lack of well-established and commonly acknowledged ground truth of GRNs, simulators were developed to generate simulated single-cell gene expression data with ground truth GRNs (Cannoodt et al. 2021; Dibaeinia and Sinha 2020; Li et al. 2022). The development of simulators enabled supervised learning methods to infer GRNs with models trained with simulated data (Shrivastava et al. 2022). Previous benchmarking works (Pratapa et al. 2020) used simulated datasets in addition to real datasets to compare the GRN inference methods in terms of accuracy, stability, and consistency for different runs. The benchmarking results suggest that for different kinds of datasets, the best-performing algorithms can be different, while the best overall performing methods are PIDC, GENIE3 and GRNBoost2 at the moment. However, even the top-performing methods have low accuracy, indicating that GRN inference is a challenging problem.
With the availability of single-cell multi-omics data, researchers are able to consider additional layers of the regulatory mechanisms besides transcription factors, such as epigenomics, translation, cell–cell interactions, and so on. There have been methods designed to infer GRNs from single-cell multi-omics data, especially scRNA-seq and scATAC-seq data. CellOracle (Kamimoto et al. 2023) infers a base network based on regulatory candidate genes by scanning for TF binding motifs within the regulatory DNA sequences (promoter/enhancers) of open chromatin sites, and then use the transcriptome profiles to further infer the actual GRN. Other methods such as scMEGA (Li et al. 2023b), FigR (Kartha et al. 2022), and Pando (Fleck et al. 2022) apply a similar strategy that first identifies candidate regulatory regions and then infers the relations between target genes, TF expression, and binding-site accessibility. In Velorama (Singh et al. 2022), RNA velocity is used instead of cell pseudotime to help improve GRN inference, especially for complex developmental trajectories. With the growing abundance of single-cell multimodal data, it is expected that more and more multimodal GRN inference methods will be developed in the near future.
GRN inference has been considered a separate computational task from trajectory inference and lineage tree reconstructions. Although some GRN inference methods use pseudotime information from TI methods as input, it is rarely studied how GRNs can affect trajectories. We discuss this aspect in the next section.
Outlook: towards a comprehensive understanding of cells’ temporal dynamics
In this section, we discuss how GRNs explain the observed gene expression data and potentially determine cell trajectories. The consistencies between gene expressions and the GRNs in specific systems have been discussed in existing work. For example, Laslo et al. (Laslo et al. 2008) reviewed how GRNs control the differentiation of myeloid and lymphoid cell fates within the immune system, which involves both intrinsic interactions between genes in the GRN as well as extrinsic signal inputs. More recently, Larsen et al. (Larsen et al. 2018) showed that there are moderate connections between E. coli static GRN and the gene expression profiles. In a recent study on Corynebacterium glutamicum (Parise et al. 2021), it is discussed that the GRN alone cannot fully explain the gene expression data. It is possible that such inconsistencies are caused by the inaccuracy of computationally inferred GRNs or even the GRNs obtained from databases. Existing experimental and theoretical research on the role of GRNs in determine cell trajectories are limited to specific TFs or very small GRN structures. Furthermore, it is likely that there exist other intrinsic and extrinsic factors that determine cell fates and the developmental trajectories together with GRNs. Therefore, further research is needed to investigate how GRNs control gene expression profiles of cells in a temporal process.
Along the developmental trajectory of cells, understanding how GRN controls the cells to take on specific branches of the trajectory, or cell fates, is of great interest. Existing efforts on studying how GRNs determine cell fates tend to only focus on one or a few key regulators, and try to understand their specific regulatory programs. For example, Nakajima et al. (Nakajima 2011) studied the differentiation and reprogramming of hematopoietic cells and identified the regulations of lineage-specific transcription factors such as C/EBP , PU.1, and GATA-1. Another study (Wang et al. 2014) that aims to dissect a regulator’s influence on rod and bipolar cell fate decisions in the vertebrate retina involves a detailed examination of key regulators’ enhancers and upstream transcription factors. These studies require the system of interest to be relatively easy to manipulate and observe the changes of regulator expressions and cell fates. The GRNs built from these works, despite involving only a few genes, can help determine the driving regulatory programs that influence the cells’ fate specifications.
Theoretically, it is possible to identify certain GRN structures and associated parameters that give rise to single-cell gene expression data with a specific trajectory topology (linear, tree, cyclic, etc.). In dyngen (Cannoodt et al. 2021), the authors defined “module networks" to model the regulatory cascades and feedback loops that lead to progressive changes in expression and cell fate decisions. These module networks tend to have only a few nodes and edges and are mapped to different kinds of trajectory topology. The actual GRN is generated from the module network by generating key transcription factors and adding target genes. With the several GRN-to-trajectory mappings provided, dyngen provides initial ideas on how the GRNs can control gene expression. In dyngen’s simulation framework, the key idea is to model the gene expression and developmental trajectory together with the GRN. In order to do that, the connections between target genes and upstream regulators are incorporated in calculating the expression changes for individual genes. Using the formula of the expected value of the total expression per unit time of a gene given the GRN, the expected developmental trajectory can be generated. For theoretical verification, we illustrate the ODE (ordinary differential equation) solutions of two GRNs, which are discussed in dyngen, that lead to the divergence (Fig. 3a) and convergence (Fig. 3b) of a bifurcating trajectory (Fig. 3). With a simple structure of two key genes down-regulating each other, we are able to generate two mutually exclusive cell fates while incorporating upstream or downstream genes that cause divergence or convergence. Although a full GRN contains a large number of genes and interactions, it is considered that at bifurcating points, the number of regulators that dominate cell fates is often very small. This is consistent with previous biological studies (Greulich et al. 2020).
Fig. 3.
Theoretical analysis of connections between GRN, developmental trajectory, and gene expression. In the GRNs, each node represents a gene (gene names are A, B, C) and the edges denote up-regulation (arrow) and down-regulation (block). For each gene, we use a total expression DE (differential equation) to model the changes of the gene expression, while also considering the regulation effects of promoters based on the GRN. We use a two-phase framework: an initial warm-up Burn Phase that only part of the network is active; a Transcription Phase that all network components are active. From the DE solution of the equations, discrete states can be defined. a A GRN that generates a bifurcation trajectory. Three cell states are defined from the DE solution: where only gene A is highly expressed; where gene A, B are highly expressed and where gene A, C are highly expressed. b A GRN that generates a bifurcation convergence trajectory. Three cell states are defined from the DE solution: where only gene A is highly expressed; where gene B is highly expressed and where gene C is highly expressed
While the toy GRN examples shown in Fig. 3 can theoretically lead to certain trajectory structures using differential equation models, when the network gets larger and more complex, it remains challenging to validate the consistency between the GRN and the observed cell trajectory. Indeed, GRNs may not be the only factor that determines gene expression and cell trajectories. Cell–cell interactions (CCIs) are considered to play a role in cell fate decisions (Greenwald and Rubin 1992; Kirouac et al. 2009). There exist methods that consider GRNs and cell–cell communications together to interpret cell fate decisions or cell–cell variations (Rommelfanger and MacLean 2021; Smith and Grima 2018) at the scale of small networks. For example, Rommelfanger and MacLean used a multiscale method to model GRNs using ODEs and the intercellular signal by Poisson processes (Rommelfanger and MacLean 2021). Future research is needed to expand these studies to larger GRN and CCI networks. Although it is challenging to study the effect the GRNs on cell trajectories for complex GRNs, it can be beneficial for current computational tasks to consider the relationship between the two tasks, GRN inference and TI. For example, some GRN inference methods have used the pseudotime from the TI methods, but the topology of the trajectories is not considered in GRN inference methods. Selecting the GRNs that can generate the observed trajectory topology can potentially yield more accurate GRNs.
Conclusions
Indeed, the integration of different models to jointly model single-cell temporal dynamics on a genome scale has the potential to greatly enhance our understanding of the underlying biological processes. As the technology continues to advance, we expect that more comprehensive models that can simultaneously account for gene expression changes, cell lineage, and gene regulatory networks will be developed. This will require not only more sophisticated computational algorithms but also a deeper biological understanding and knowledge of the systems being studied. Ultimately, we believe that these integrative models will provide powerful tools for developmental biologists to uncover the key events in cell differentiation and cell fate specifications, and shed light on the fundamental mechanisms that underlie development and disease.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
X.P. and X.Z. conceived the study and discussed the structure of the review. X.P. drafted the manuscript. X.P. and X.Z. refined the manuscript and approved the final manuscript.
Funding
This work was supported by the US National Science Foundation DBI-2019771, DBI-2145736, and National Institutes of Health grant R35GM143070.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, van den Oord J, Atak ZK, Wouters J, Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemany A, Florescu M, Baron CS, Peterson-Maduro J, van Oudenaarden A. Whole-organism clone trac- ing using single-cell sequencing. Nature. 2018;556(7699):108–112. doi: 10.1038/nature25969,10.1038/nature25969. [DOI] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to tran- sient cell states through dynamical modeling. Nat Biotechnol. 2020;38(12):1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- Bergen V, Soldatov RA, Kharchenko PV, Theis FJ (2021) RNA velocity—current challenges and future perspectives. Mol Syst Biol 17(8). 10.15252/msb.202110282 [DOI] [PMC free article] [PubMed]
- Cannoodt R, Saelens W, Deconinck L, Saeys Y (2021) Spearheading future omics analyses using dyngen, a multi- modal simulator of single cells. Nat Commun 12(1). 10.1038/s41467-021-24152-2 [DOI] [PMC free article] [PubMed]
- Cannoodt R, Saelens W, Sichien D, Tavernier S, Janssens S, Guilliams M, Lambrecht B, Preter KD, Saeys Y (2016) SCORPIUS improves trajectory inference and identifies novel modules in dendritic cell development. 10.1101/079509
- Chan MM, Smith ZD, Grosswendt S, Kretzmer H, Norman TM, Adamson B, Jost M, Quinn JJ, Yang D, Jones MG, Khodaverdian A, Yosef N, Meissner A, Weissman JS. Molecular recording of mammalian embryogenesis. Nature. 2019;570(7759):77–82. doi: 10.1038/s41586-019-1184-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TE, Pallaseni AV, Babtie AC, McEwen KR, Stumpf MP (2018) Empirical bayes meets information theoretical network reconstruction from single cell data. 10.1101/264853
- Cvekl A, Zhang X. Signaling and gene regulatory networks in mammalian lens development. Trends Genet. 2017;33(10):677–702. doi: 10.1016/j.tig.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A, Chu LF, Stewart R, Gitter A. Network inference with granger causality ensembles on single- cell transcriptomics. Cell Rep. 2022;38(6):110333. doi: 10.1016/j.celrep.2022.110333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibaeinia P, Sinha S. SERGIO: a single-cell expression simulator guided by gene regulatory networks. Cell Syst. 2020;11(3):252–271.e11. doi: 10.1016/j.cels.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck JS, Jansen SMJ, Wollny D, Zenk F, Seimiya M, Jain A, Okamoto R, Santel M, He Z, Camp JG, Treutlein B. Inferring and perturbing cell fate regulomes in human brain organoids. Nature. 2022 doi: 10.1038/s41586-022-05279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrow A, Schiebinger G (2021) LineageOT is a unified framework for lineage tracing and trajectory inference. Nat Commun 12(1). 10.1038/s41467-021-25133-1 [DOI] [PMC free article] [PubMed]
- Gao M, Qiao C, Huang Y (2022) UniTVelo: temporally unified RNA velocity reinforces single-cell trajectory in- ference. Nat Commun 13(1). 10.1038/s41467-022-34188-7 [DOI] [PMC free article] [PubMed]
- Gao NP, Ud-Dean SMM, Gandrillon O, Gunawan R. SINCERITIES: inferring gene regulatory networks from time-stamped single cell transcriptional expression profiles. Bioinformatics. 2017;34(2):258–266. doi: 10.1093/bioinformatics/btx575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, Granados AA, Hu J, Jones MG, Raz O, Salvador-Martínez I, Zhang H, Chow KHK, Kwak IY, Retkute R, Prusokas A, Prusokas A, Khodaverdian A, Zhang R, Rao S, Wang R, Rennert P, Saipradeep VG, Sivadasan N, Rao A, Joseph T, Srinivasan R, Peng J, Han L, Shang X, Garry DJ, Yu T, Chung V, Mason M, Liu Z, Guan Y, Yosef N, Shendure J, Telford MJ, Shapiro E, Elowitz MB, Meyer P. Benchmarked approaches for reconstruction of in vitro cell lineages and in silico models of c. elegans and m. musculus developmental trees. Cell Syst. 2021;12(8):810–826.e4. doi: 10.1016/j.cels.2021.05.008. [DOI] [PubMed] [Google Scholar]
- Gong W, Kim HJ, Garry DJ, Kwak IY (2022) Single cell lineage reconstruction using distance-based algorithms and the r package, DCLEAR. BMC Bioinformatics 23(1). 10.1186/s12859-022-04633-x [DOI] [PMC free article] [PubMed]
- Greenwald I, Rubin GM. Making a difference: the role of cell-cell interactions in establishing separate identities for equivalent cells. Cell. 1992;68(2):271–281. doi: 10.1016/0092-8674(92)90470-w. [DOI] [PubMed] [Google Scholar]
- Greulich P, Smith R, MacArthur BD (2020) The physics of cell fate. In: Phenotypic Switching. Elsevier, p 189–206. 10.1016/b978-0-12-817996-3.00003-7
- Grün D, Muraro MJ, Boisset JC, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H, de Koning EJ, van Oudenaarden A. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell. 2016;19(2):266–277. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin A, Stumpf MPH. Noise and the molecular processes underlying cell fate decision-making. Phys Biol. 2020;18(1):011002. doi: 10.1088/1478-3975/abc9d1. [DOI] [PubMed] [Google Scholar]
- Hamey FK, Nestorowa S, Kinston SJ, Kent DG, Wilson NK, Göttgens B. Reconstructing blood stem cell regulatory network models from single-cell molecular profiles. Proc Natl Acad Sci. 2017;114(23):5822–5829. doi: 10.1073/pnas.1610609114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MG, Khodaverdian A, Quinn JJ, Chan MM, Hussmann JA, Wang R, Xu C, Weissman JS, Yosef N (2020) Inference of single-cell phylogenies from lineage tracing data using cassiopeia. Genome Biol 21(1). 10.1186/s13059-020-02000-8 [DOI] [PMC free article] [PubMed]
- Kamimoto K, Stringa B, Hoffmann CM, Jindal K, Solnica-Krezel L, Morris SA. Dissecting cell identity via network inference and in silico gene perturbation. Nature. 2023;614(7949):742–751. doi: 10.1038/s41586-022-05688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartha VK, Duarte FM, Hu Y, Ma S, Chew JG, Lareau CA, Earl A, Burkett ZD, Kohlway AS, Lebofsky R, Buenrostro JD. Functional inference of gene regulation using single-cell multi-omics. Cell Genom. 2022;2(9):100166. doi: 10.1016/j.xgen.2022.100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Kim JR, Kim D, Lander AD, Cho KH (2012) Spatiotemporal network motif reveals the biological traits of developmental gene regulatory networks in drosophila melanogaster. BMC Syst Biol 6(1). 10.1186/1752-0509-6-31 [DOI] [PMC free article] [PubMed]
- Kirouac DC, Madlambayan GJ, Yu M, Sykes EA, Ito C, Zandstra PW. Cell–cell interaction net- works regulate blood stem and progenitor cell fate. Mol Syst Biol. 2009;5(1):293. doi: 10.1038/msb.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M, Bergen V, Klein M, Setty M, Reuter B, Bakhti M, Lickert H, Ansari M, Schniering J, Schiller HB, Pe’er D, Theis FJ. Cell Rank for directed single-cell fate mapping. Nat Methods. 2022;19(2):159–170. doi: 10.1038/s41592-021-01346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau CA, Ludwig LS, Muus C, Gohil SH, Zhao T, Chiang Z, Pelka K, Verboon JM, Luo W, Christian E, Rosebrock D, Getz G, Boland GM, Chen F, Buenrostro JD, Hacohen N, Wu CJ, Aryee MJ, Regev A, Sankaran VG. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat Biotechnol. 2020;39(4):451–461. doi: 10.1038/s41587-020-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen SJ, Röttger R, Schmidt HHHW, Baumbach J. E. coli gene regulatory networks are inconsistent with gene expression data. Nucleic Acids Res. 2018;47(1):85–92. doi: 10.1093/nar/gky1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslo P, Pongubala JM, Lancki DW, Singh H. Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Semin Immunol. 2008;20(4):228–235. doi: 10.1016/j.smim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Lefort V, Desper R, Gascuel O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program: Table 1. Mol Biol Evol. 2015;32(10):2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Virgilio MC, Collins KL, Welch JD. Multi-omic single-cell velocity models epigenome–transcriptome interactions and improves cell fate prediction. Nat Biotechnol. 2022;41(3):387–398. doi: 10.1038/s41587-022-01476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang Z, Squires M, Chen X, Zhang X (2022) scMultiSim: simulation of multi-modality single cell data guided by cell-cell interactions and gene regulatory networks. 10.1101/2022.10.15.512320
- Li S, Zhang P, Chen W, Ye L, Brannan KW, Le NT, ichi Abe J, Cooke JP, Wang G (2023a) A relay velocity model infers cell-dependent RNA velocity. Nat Biotechnol. 10.1038/s41587-023-01728-5 [DOI] [PMC free article] [PubMed]
- Li Z, Nagai JS, Kuppe C, Kramann R, Costa IG (2023b) scMEGA: single-cell multi-omic enhancer-based gene reg- ulatory network inference. Bioinformatics Adv 3(1). 10.1093/bioadv/vbad003 [DOI] [PMC free article] [PubMed]
- Lim CY, Wang H, Woodhouse S, Piterman N, Wernisch L, Fisher J, Göttgens B (2016) BTR: training asynchronous boolean models using single-cell expression data. BMC Bioinformatics 17(1). 10.1186/s12859-016-1235-y [DOI] [PMC free article] [PubMed]
- Lin C, Bar-Joseph Z. Continuous-state HMMs for modeling time-series single-cell RNA-seq data. Bioinformatics. 2019;35(22):4707–4715. doi: 10.1093/bioinformatics/btz296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li P, Zhu M, Wang X, Lu J, Yu T. Nonlinear network reconstruction from gene ex- pression data using marginal dependencies measured by DCOL. PLOS ONE. 2016;11(7):e0158247. doi: 10.1371/journal.pone.0158247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lou H, Xie K, Wang H, Chen N, Aparicio OM, Zhang MQ, Jiang R, Chen T (2017) Reconstructing cell cycle pseudo time-series via single-cell transcriptome data. Nat Commun 8(1). 10.1038/s41467-017-00039-z [DOI] [PMC free article] [PubMed]
- Manno GL, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, Fan J, Borm LE, Liu Z, van Bruggen D, Guo J, He X, Barker R, Sundström E, Castelo-Branco G, Cramer P, Adameyko I, Linnarsson S, Kharchenko PV. RNA velocity of single cells. Nature. 2018;560(7719):494–498. doi: 10.1038/s41586-018-0414-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Kiryu H, Furusawa C, Ko MSH, Ko SBH, Gouda N, Hayashi T, Nikaido I. SCODE:an efficient regulatory network inference algorithm from single-cell RNA-seq during differentiation. Bioinformatics. 2017;33(15):2314–2321. doi: 10.1093/bioinformatics/btx194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TE, Lareau CA, Verga JA, DePasquale EAK, Liu V, Ssozi D, Sandor K, Yin Y, Ludwig LS, Farran CAE, Morgan DM, Satpathy AT, Griffin GK, Lane AA, Love JC, Bernstein BE, Sankaran VG, van Galen P. Mitochondrial variant enrichment from high-throughput single-cell RNA sequencing re- solves clonal populations. Nat Biotechnol. 2022;40(7):1030–1034. doi: 10.1038/s41587-022-01210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris N, Pina C, Arias AM. Transition states and cell fate decisions in epigenetic landscapes. Nat Rev Genet. 2016;17(11):693–703. doi: 10.1038/nrg.2016.98. [DOI] [PubMed] [Google Scholar]
- Nakajima H. Role of transcription factors in differentiation and reprogramming of hematopoietic cells. Keio J Med. 2011;60(2):47–55. doi: 10.2302/kjm.60.47. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Tran D, Tran B, Pehlivan B, Nguyen T (2020) A comprehensive survey of regulatory network inference methods using single cell RNA sequencing data. Brief Bioinform 22(3). 10.1093/bib/bbaa190 [DOI] [PMC free article] [PubMed]
- Ocone A, Haghverdi L, Mueller NS, Theis FJ. Reconstructing gene regulatory dynamics from high-dimensional single-cell snapshot data. Bioinformatics. 2015;31(12):i89–i96. doi: 10.1093/bioinformatics/btv257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer JS, Zhu Q, Huynh C, Sivaramakrishnan P, Preston E, Dueck H, Stefanik D, Tan K, Trapnell C, Kim J, Waterston RH, Murray JI (2019) A lineage-resolved molecular atlas of c. elegans embryogenesis at single-cell resolution. Science 365(6459). 10.1126/science.aax1971 [DOI] [PMC free article] [PubMed]
- Pan X, Li H, Putta P, Zhang X (2023) LinRace: single cell lineage reconstruction using paired lineage barcode and gene expression data. 10.1101/2023.04.12.536601 [DOI] [PMC free article] [PubMed]
- Pan X, Li H, Zhang X. TedSim: temporal dynamics simulation of single-cell RNA sequencing data and cell division history. Nucleic Acids Res. 2022;50(8):4272–4288. doi: 10.1093/nar/gkac235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parise D, Parise MTD, Kataka E, Kato RB, List M, Tauch A, de CarvalhoAzevedo VA, Baumbach J. On the consistency between gene expression and the gene regulatory network of corynebacterium glutamicum. Netw Syst Med. 2021;4(1):51–59. doi: 10.1089/nsm.2020.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratapa A, Jalihal AP, Law JN, Bharadwaj A, Murali TM. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat Methods. 2020;17(2):147–154. doi: 10.1038/s41592-019-0690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Gagnon JA, Schier AF. Large-scale reconstruction of cell lineages using single-cell readout of transcriptomes and CRISPR–cas9 barcodes by scGESTALT. Nat Protoc. 2018;13(11):2685–2713. doi: 10.1038/s41596-018-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Foulds L. Comparison of phylogenetic trees. Math Biosci. 1981;53(1–2):131–147. doi: 10.1016/0025-5564(81)90043-2. [DOI] [Google Scholar]
- Rommelfanger MK, MacLean AL (2021) A single-cell resolved cell-cell communication model explains lineage commitment in hematopoiesis. Development 148(24):dev199779. 10.1242/dev.199779 [DOI] [PMC free article] [PubMed]
- Saelens W, Cannoodt R, Todorov H, Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol. 2019;37(5):547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salvador-Martínez I, Grillo M, Averof M, Telford MJ (2019) Is it possible to reconstruct an accurate cell lineage using CRISPR recorders? eLife 8. 10.7554/elife.40292 [DOI] [PMC free article] [PubMed]
- Setty M, Kiseliovas V, Levine J, Gayoso A, Mazutis L, Pe’er D. Characterization of cell fate probabilities in single-cell data with palantir. Nat Biotechnol. 2019;37(4):451–460. doi: 10.1038/s41587-019-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava H, Zhang X, Song L, Aluru S. GRNUlar: a deep learning framework for recovering Single-Cell gene regulatory networks. J Comput Biol. 2022;29(1):27–44. doi: 10.1089/cmb.2021.0437. [DOI] [PubMed] [Google Scholar]
- Simeonov KP, Byrns CN, Clark ML, Norgard RJ, Martin B, Stanger BZ, Shendure J, McKenna A, Lengner CJ. Single-cell lineage tracing of metastatic cancer reveals selection of hybrid EMT states. Cancer Cell. 2021;39(8):1150–1162.e9. doi: 10.1016/j.ccell.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Wu AP, Mudide A, Berger B (2022) Unraveling causal gene regulation from the RNA velocity graph using velorama. .10.1101/2022.10.18.512766
- Smith S, Grima R. Single-cell variability in multicellular organisms. Nat Commun. 2018;9(1):345. doi: 10.1038/s41467-017-02710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolander J, Junttila S, Venäläinen MS, Elo LL. scShaper: an ensemble method for fast and accurate linear trajectory inference from single-cell RNA-seq data. Bioinformatics. 2021;38(5):1328–1335. doi: 10.1093/bioinformatics/btab831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht AT, Li J (2016) LEAP: constructing gene co-expression networks for single-cell RNA-sequencing data using pseudotime ordering. Bioinformatics: btw729. 10.1093/bioinformatics/btw729 [DOI] [PMC free article] [PubMed]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S (2018) Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19(1). 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed]
- Tintori SC, Nishimura EO, Golden P, Lieb JD, Goldstein B. A transcriptional lineage of the early c. elegans embryo. Dev Cell. 2016;38(4):430–444. doi: 10.1016/j.devcel.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TN, Bader GD. Tempora: cell trajectory inference using time-series single-cell RNA sequencing data. PLOS Comput Biol. 2020;16(9):e1008205. doi: 10.1371/journal.pcbi.1008205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Klein AM. Lineage tracing meets single-cell omics: opportunities and challenges. Nat Rev Genet. 2020;21(7):410–427. doi: 10.1038/s41576-020-0223-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sengel C, Emerson MM, Cepko CL. A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev Cell. 2014;30(5):513–527. doi: 10.1016/j.devcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Hou L, Lu Z, Wang X, Zi Z, Zhai W, He X, Curtis C, Zhou D, Hu Z (2022a) Cell division history encodes directional information of fate transitions. .10.1101/2022.10.06.511094
- Wang L., Trasanidis N., Wu T., Dong G., Hu M., Bauer D.E., Pinello L. Dictys: dynamic gene regulatory network dissects developmental continuum with single-cell multi-omics. bioRxiv. 2022 doi: 10.1101/2022.09.14.508036. [DOI] [PubMed] [Google Scholar]
- Wang SW, Herriges MJ, Hurley K, Kotton DN, Klein AM. CoSpar identifies early cell fate biases from single-cell transcriptomic and lineage information. Nat Biotechnol. 2022;40(7):1066–1074. doi: 10.1038/s41587-022-01209-1. [DOI] [PubMed] [Google Scholar]
- Welch JD, Hartemink AJ, Prins JF (2017) MATCHER: manifold alignment reveals correspondence between single cell transcriptome and epigenome dynamics. Genome Biol 18(1). 10.1186/s13059-017-1269-0 [DOI] [PMC free article] [PubMed]
- Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, Theis FJ (2019) PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol 20(1). 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed]
- Woodhouse S, Piterman N, Wintersteiger CM, Göttgens B, Fisher J (2018) SCNS: a graphical tool for recon- structing executable regulatory networks from single-cell genomic data. BMC Syst Biol 12(1). 10.1186/s12918-018-0581-y [DOI] [PMC free article] [PubMed]
- Xu J, Nuno K, Litzenburger UM, Qi Y, Corces MR, Majeti R, Chang HY (2019) Single-cell lineage tracing by endogenous mutations enriched in transposase accessible mitochondrial DNA. eLife 8. 10.7554/elife.45105 [DOI] [PMC free article] [PubMed]
- Xu Q, Li G, Osorio D, Zhong Y, Yang Y, Lin YT, Zhang X, Cai JJ. scInTime: a computational method leveraging single-cell trajectory and gene regulatory networks to identify master regulators of cellular differentiation. Genes. 2022;13(2):371. doi: 10.3390/genes13020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafar H, Lin C, Bar-Joseph Z (2020) Single-cell lineage tracing by integrating CRISPR-cas9 mutations with transcriptomic data. Nat Commun 11(1). 10.1038/s41467-020-16821-5 [DOI] [PMC free article] [PubMed]
- Zhang Z, Zhang X. Inference of high-resolution trajectories in single-cell RNA-seq data by using RNA velocity. Cell Rep Methods. 2021;1(6):100095. doi: 10.1016/j.crmeth.2021.100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Han J., Song L., Zhang X. Inferring cell-specific gene regulatory networks from single cell gene expression data. bioRxiv. 2022 doi: 10.1101/2022.03.03.482887. [DOI] [Google Scholar]
- Zhang Z, Yang C, Zhang X (2022b) scDART: integrating unmatched scRNA-seq and scATAC-seq data and learning cross-modality relationship simultaneously. Genome Biol 23(1). 10.1186/s13059-022-02706-x [DOI] [PMC free article] [PubMed]
- Zhang S, Pyne S, Pietrzak S, Halberg S, McCalla SG, Siahpirani AF, Sridharan R, Roy S. Inference of cell type-specific gene regulatory networks on cell lineages from single cell omic datasets. Nat Commun. 2023;14(1):3064. doi: 10.1038/s41467-023-38637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.