Abstract
Sequences present at the genomic termini of herpesviruses become linked during lytic-phase replication and provide the substrate for cleavage and packaging of unit length viral genomes. We have previously shown that homologs of the consensus herpesvirus cleavage-packaging signals, pac1 and pac2, are located at the left and right genomic termini of human herpesvirus 6 (HHV-6), respectively. Immediately adjacent to these elements are two distinct arrays of human telomeric repeat sequences (TRS). We now show that the unique sequence element formed at the junction of HHV-6B genome concatemers (pac2-pac1) is necessary and sufficient for virally mediated cleavage of plasmid DNAs containing the HHV-6B lytic-phase origin of DNA replication (oriLyt). The concatemeric junction sequence also allowed for the packaging of these plasmid molecules into intracellular nucleocapsids as well as mature, infectious viral particles. In addition, this element significantly enhanced the replication efficiency of oriLyt-containing plasmids in virally infected cells. Experiments revealed that the concatemeric junction sequence possesses an unusual, S1 nuclease-sensitive conformation (anisomorphic DNA), which might play a role in this apparent enhancement of DNA replication—although additional studies will be required to test this hypothesis. Finally, we also analyzed whether the presence of flanking viral TRS had any effect on the functional activity of the minimal concatemeric junction (pac2-pac1). These experiments revealed that the TRS motifs, either alone or in combination, had no effect on the efficiency of virally mediated DNA replication or DNA cleavage. Taken together, these data show that the cleavage and packaging of HHV-6 DNA are mediated by cis-acting consensus sequences similar to those found in other herpesviruses, and that these sequences also influence the efficiency of HHV-6 DNA replication. Since the adjacent TRS do not influence either viral cleavage and packaging or viral DNA replication, their function remains uncertain.
Human herpesvirus 6 (HHV-6) is a ubiquitous T-lymphotropic betaherpesvirus which has been etiologically linked to acute febrile illnesses in young children, including exanthem subitum (35). The virus has also been linked to cases of central nervous system disease (notably encephalitis [19]), as well as to serious complications in immunosuppressed individuals, such as pneumonitis (5) and bone marrow failure (10).
The HHV-6 genome is a double-stranded DNA (dsDNA) molecule of approximately 160 kbp and is composed of a single long unique sequence (U) flanked by identical direct repeats (DRL and DRR), with the arrangement DRL-U-DRR (12). This genome structure is similar to that of human herpesvirus 7 (HHV-7) (24) but distinct from those of all other human herpesviruses. The genetic homology between HHV-6 and HHV-7 also extends to the sequences which are located at the genomic termini. Both viruses contain homologs of the consensus herpesvirus cleavage-packaging motifs, pac1 and pac2, at their left and right termini, respectively (29). During viral DNA replication these sequences are brought into close proximity at the junction between concatemeric viral genomes to form a unique junctional element (pac2-pac1) which is absent in the unit length virus genome (29). Based on the positional and sequence similarity of this element to structures found in other human herpesviruses (6), this junctional sequence has been proposed to be the substrate for cleavage and packaging of unit length viral genomes from the concatemeric product that is generated during viral DNA replication.
In the intact HHV-6B genome, the pac2 and pac1 elements are located immediately adjacent to (TTAGGG)n motifs, identical to the human telomeric repeat sequence (TRS) (29). pac2 is located immediately 3′ to a tandem array of 15 to 60 perfect direct repeats of the telomeric hexamer at the right genome terminus of HHV-6 (this repeat arrangement is referred to below as the simple TRS [S-TRS] motif). In contrast, pac1 is located immediately 5′ to a tandem, directly repeated array of both perfect and degenerate copies of the telomeric hexamer (this repeat arrangement is referred to below as the complex TRS [C-TRS] motif). Since TRS elements are also conserved in HHV-6A and HHV-7, as well as in Marek’s disease virus (MDV), a T-cell-tropic avian alphaherpesvirus (15), it seems reasonable to conclude that these motifs may possess important functional or structural properties.
In the present study, we derived plasmids containing the HHV-6B lytic-phase origin of DNA replication (oriLyt), together with a minimal junctional sequence (DRR-DRL) that was previously cloned from concatemeric viral DNA (this DNA fragment contains the pac2-pac1 motif but lacks the flanking TRS arrays). We then tested the ability of these plasmid constructs to undergo site-specific cleavage and packaging in HHV-6B infected cells. These experiments showed that the HHV-6B DRR-DRL junction is necessary for virally mediated cleavage of replicated plasmid DNAs, as well as for the packaging of such DNAs into intracellular nucleocapsids and mature, infectious virus particles. These data suggest that the cleavage and packaging of HHV-6B occur via a conventional herpesvirus mechanism. Somewhat less expected was the observation that the concatemeric junction element enhanced the replication efficiency of oriLyt-containing plasmids in virally infected cells (by approximately four- to sixfold). Further analysis of the junctional DNA motif revealed that it possesses an unwound S1 nuclease-sensitive conformation (anisomorphic DNA [34])—a structure that has previously been shown to be susceptible to cleavage by a conformation-specific cellular endonuclease (33). It is possible that DNA cleavage at the concatemeric junction leads to the generation of dsDNA breaks that could enhance the putative recombination-dependent phase of herpesvirus DNA replication (18). Future studies will be needed to address this issue.
Finally, since pac2 and pac1 are flanked by adjacent TRS motifs in the HHV-6 genome, we tested whether these TRS arrays had any effect on cleavage, packaging, or replication of plasmids containing HHV-6B oriLyt and pac2-pac1. Neither a single TRS motif (S-TRS or C-TRS) nor the combination of both such elements (S-TRS plus C-TRS) had any demonstrable effect on the efficiency of cleavage, packaging, or replication of plasmid DNAs in HHV-6B-infected cells. Thus, the function of the TRS motifs in HHV-6 remains uncertain.
MATERIALS AND METHODS
Cells and viruses.
J-Jahn cells (human T cells; a gift of C. Hall) were propagated in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM glutamine, and antibiotics (penicillin and streptomycin [50 U/ml and 50 μg/ml, respectively]). The HHV-6B strain R1 (a gift of C. Hall) was used (9).
Plasmid construction.
pΔ2 contains a functional HHV-6B oriLyt DNA fragment of 1.65 kb and has been described previously (9). pΔ2C was derived by insertion of a concatemeric HHV-6 junctional sequence into the EcoRI-to-PstI sites of pΔ2 (plasmid pC61 was used as the source of the concatemeric junction, and this element corresponds to a pac2-pac1 junction spanning roughly 170 bp of viral sequence that was originally derived by PCR [29]). pO contains the same HHV-6B oriLyt fragment, which was excised as a ClaI fragment from pΔ2 and inserted into the AccI site of pGEM-T. pCO was derived from pC62 by the insertion of this same oriLyt fragment into the AccI site (plasmid pC62 contains a concatemeric insert identical to that of plasmid pC61, but oriented in the opposite direction in pGEM-T [29]). The organization of plasmids pO, pCO, pΔ2, and pΔ2C is shown in Fig. 1.
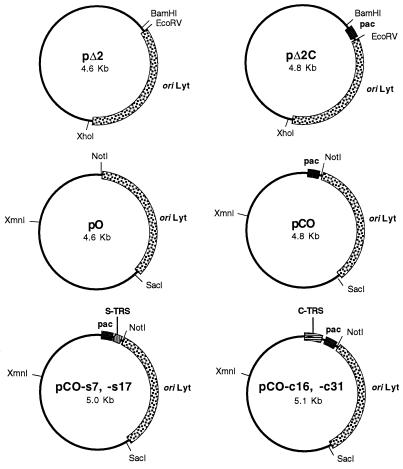
FIG. 1.
Plasmid clones used in this study. Plasmids pO, pCO, and derivatives of pCO were generated in pGEM-T vector, while plasmids pΔ2 and pΔ2C were generated in pKS. Key sites and features of the plasmids are marked; these include (i) oriLyt (a 1.6-kb DNA fragment corresponding to a functional lytic-phase origin of DNA replication from HHV-6B [9]), (ii) pac (a 0.2-kb DNA fragment corresponding to the DRR-DRL junction from HHV-6B, which contains the putative pac1 and pac2 signals; see text for details [29]), (iii) S-TRS (a tandem array of TRSs identical to the S-TRS array found at the right genome terminus of HHV-6 [29]), and (iv) C-TRS (an interspersed array of degenerate TRSs identical to the C-TRS array found at the left genome terminus of HHV-6 [29]). Note that the spacing of the S-TRS and C-TRS motifs relative to the putative pac1 and pac2 signals is identical to that found in the viral genome. Plasmid nomenclature is as follows: all plasmids containing the concatemeric junction element (pac) have the letter “C” in their designations; plasmids containing the S-TRS motif have the suffix -s7 or -s17 (indicating 7 or 17 copies of the TRS motif, respectively); plasmids containing the C-TRS motif have the suffix -c16 or -c31 (indicating 16 or 31 copies of the TRS motif, respectively).
Plasmids containing C-TRS and/or S-TRS arrays were derived from plasmid pC62 by using synthetic double-stranded oligonucleotides which were self-ligated prior to insertion into pC62. To create the S-TRS array, oligonucleotides were inserted into the EcoRI site in pC62, which is located immediately 5′ to the pac2 motif in this plasmid. The oligonucleotides inserted at this site were H6PC7, AATTT(GGGTTA)10G, and H6PC8, AATTC(TAACCC)10A. (The underlined nucleotides represent the EcoRI-compatible sticky end that is created upon the annealing of these oligonucleotides to one another.) It should also be noted that misalignment of the oligonucleotides can occur, leading to plasmids containing less than the expected number of GGGTTA repeat motifs (e.g., plasmid pC62-s7, which contains only seven copies of this motif). In addition, insertion of H6PC7-H6PC8 into the EcoRI site of pC62 results in re-creation of the EcoRI site at the 5′ end of the insert and loss of the site at the 3′ end. This facilitates the sequential insertion of multiple copies of this oligonucleotide into pC62, as was performed to produce plasmid pC62-s17, which contains 17 copies of the GGGTTA motif. Finally, plasmids pCO-s7 and -s17 were produced by insertion of HHV-6 oriLyt into pC62-s7 and -17 (this was achieved by precisely the same method used to generate pCO from pC62; see Fig. 1).
Derivation of plasmids containing the complex TRS motif was achieved by a similar, but slightly more complex, strategy. First, the unique AvrII site in plasmid pC62 (which is located immediately 3′ to the pac1 motif) was converted into a BstXI site by ligation of a dsDNA fragment into the AvrII site. This dsDNA fragment was derived from the self-complementary oligonucleotides BST-A (CTAGGGACCACGGGTTTGG) and BST-B (CTAGCCAAACCCGTGGTCC). (The underlined nucleotides represent the AvrII-compatible sticky end that is created upon the annealing of these oligonucleotides to one another.) The resulting plasmid was then digested with SacI and NsiI, blunt ended with T4 DNA polymerase, and religated, to remove the BstXI site present in the polylinker of pC62, thereby generating plasmid pC62/Bst. The single remaining BstXI site was then used for insertion of a dsDNA fragment corresponding to sequences from the C-TRS motif of HHV-6. The following self-complementary oligonucleotides were annealed and inserted: H6PC10, GACCTA(GGGTTA)3GACCTA(GGGTTA)3GGGCCCTAGGGT, and H6PC9, TAGGGCCC(TAACCC)3TAGGTC(TAACCC)TAGGTCACCC. (The underlined nucleotides represent the BstXI-compatible sticky end that is created upon the annealing of these oligonucleotides to one another.) Since BstXI generates asymmetric cohesive DNA ends, self-ligation of annealed and phosphorylated H6PC10 and H6PC9 results in the generation of tandem head-to-tail DNA repeats, as noted previously by Takeshita and colleagues (28). By using this approach, we were able to clone either two or four head-to-tail copies of the H6PC10-H6PC9 DNA fragment into plasmid pC62/Bst, thereby generating plasmids pC62-c16 and pC62-c31, respectively. Plasmids pCO-c16 and -s31 were then produced by insertion of HHV-6 oriLyt into pC62-c16 and -31 (as described for pCO). Finally, plasmid pCO-c16/s17 was constructed by digestion of pCO-s17 with NotI (which cuts 3′ to the S-TRS array) and NarI (which cuts between pac2 and pac1), followed by gel isolation of the small DNA fragment generated (this fragment spans the S-TRS motif and pac2). This fragment was then substituted for the corresponding segment of plasmid pCO-c31, thereby generating a construct which contained both the S-TRS and C-TRS motifs, arranged in the same orientation as that in which they are found in the HHV-6 genome (i.e., S-TRS, pac2-pac1, C-TRS).
Analysis of virally mediated DNA cleavage.
Briefly, 107 HHV-6B-infected J-Jahn cells (maintained in RPMI 1640 supplemented by 10% fetal bovine serum, 50 μg of penicillin/ml, 50 μg of streptomycin/ml, and 2 mM glutamine) were electroporated with 1 pmol of CsCl gradient-banded plasmid DNA (300 V; 960-μF pulse in RPMI 1640 with 10 mM dextrose and 0.1 mM dithiothreitol). Cells were harvested 96 h posttransfection, and extrachromosomal DNA was prepared by the Hirt procedure (14). The Hirt DNA was then digested with DpnI in order to eliminate any remaining input DNA that had not undergone replication in the J-Jahn cells and was then linearized prior to Southern blotting with a radiolabeled vector DNA probe.
Preparation of DNA from intracellular viral nucleocapsids.
HHV-6B-infected cells were transfected with test plasmids, and 96 h thereafter, the cells were pelleted, washed in Tris-buffered saline (25 mM Tris-HCl [pH 7.4], 137 mM NaCl, 20 mM KCl), repelleted, and resuspended in Tris-potassium-magnesium (TPM) buffer (10 mM Tris-HCl [pH 7.4], 10 mM KCl, 30 mM MgCl2). Cells were subjected to one cycle of freezing and thawing on dry ice-ethanol and were sonicated three times for 30 s. DNase I (Boehringer-Mannheim Biochemicals) was then added to these extracts to a final concentration of 75 μg/ml, and a 2-h incubation was performed at 37°C to allow for complete digestion of all nonencapsidated DNAs. After this step, viral nucleocapsids were disrupted by the addition of 0.5 volume of lysis buffer (1.8% sodium dodecyl sulfate, 30 mM EDTA, 30 mM Tris [pH 7.4], 300 μg of proteinase K/ml) and incubation at 37°C overnight, prior to conventional RNase digestion, phenol extraction, and precipitation of encapsidated DNAs.
Preparation of DNA from extracellular virus particles.
In some experiments, HHV-6B-infected J-Jahn cells were transfected with test plasmids, and extracellular virions were then isolated at the 96-h time point. Virus particles were prepared by high-speed centrifugation of cell culture supernatants, as previously described (29).
PFGE of nucleocapsid DNA.
HHV-6B-infected J-Jahn cells were transfected with test plasmids and collected at the 96-h time point. Cells (1.5 × 107) were then washed thoroughly, resuspended in 350 μl of TPM buffer, and frozen and thawed three times in a dry-ice–ethanol bath prior to sonication (three times for 30 s each) and mixing with an equal volume of 2% high-strength, analytical-grade agarose (Bio-Rad) in phosphate-buffered saline at 55°C. Agarose-embedded cells were then cast into three equal blocks in a Bio-Rad mold and lysed by incubation in pulsed-field gel electrophoresis (PFGE) lysis buffer (1% laurylsarcosine, 0.4 M EDTA [pH 9.0], and 1 mg of proteinase K/ml) for 24 h at 37°C, with one buffer change. Blocks were then rinsed five times, for 15 min each, in 1× Tris-EDTA at 50°C and were stored in Tris-EDTA at 4°C prior to use. For PFGE analysis, one-quarter of each block was sealed into each well of a 1% agarose gel made in 0.5× Tris-borate-EDTA, and electrophoresis was performed in a contour-clamped homogeneous electric field (CHEF) apparatus (Bio-Rad) with run conditions of 6 V/cm (approximately 200 V) for 20 h at 14°C, by using a straight pulse time of 8 s throughout the course of the run. Lambda-ladder and CHEF DNA size standards (both from Bio-Rad) were used as molecular-weight markers.
PCR amplification of plasmid sequences in extrachromosomal DNA.
Selected samples of DpnI-digested extrachromosomal DNA were subjected to PCR analysis for detection of replicated plasmid DNAs. PCR was performed under standard reaction conditions (a 55°C annealing temperature, Taq DNA polymerase, and 25 amplification cycles) with the primers AMP1 (AATGCTTAATCAGTGAGGCA) and AMP2 (TTACATCGAACTGGATCTCA). These primers generate an amplimer of approximately 730 bp, deriving from the bacterial gene for ampicillin resistance (note also that the primers span a total of six DpnI restriction sites, thereby ensuring that DpnI digestion of plasmid DNA will result in elimination of unreplicated plasmid targets).
Analysis of the left terminus of nucleocapsid DNA and of virion DNA.
Purified DNA from viral nucleocapsids, or from extracellular virions, was also used as source material for studies aimed at mapping precisely the sites of virally mediated cleavage of replicated plasmid DNA molecules (replicated pΔ2C was used as source material). Briefly, total virion or nucleocapsid DNA (including both wild-type viral genomes and plasmid concatemers) was blunt ended with T4 DNA polymerase (as described previously [29]) and then digested with NotI. NotI cleaves within the polylinker of pΔ2C, close to the anticipated site of cleavage at the pac2-pac1 junction, but it cleaves the wild-type viral genome only very infrequently, generating products of >20 kb. The cleaved virion or nucleocapsid DNA was then ligated into SmaI- and NotI-digested pSK vector, with the expectation that this would result in a strong selective bias for the insertion of the short plasmid-derived DNA fragments. Finally, the ligated DNA was used as a template for PCR amplification of left terminal DNA fragments, by using standard PCR amplification conditions (Taq DNA polymerase, a 55°C annealing temperature, and 35 amplification cycles). Primers used for PCR amplification of terminal DNA fragments were a plasmid-specific primer (the reverse universal primer [Stratagene]) and TER3, a primer specific for the HHV-6B left terminus (29).
S1 nuclease cleavage assays.
One or two micrograms of plasmid was digested with 1.5 U of S1 nuclease at 37°C for 30 min in a 50-μl volume. After ethanol precipitation, half of the S1-cut DNA was linearized at a unique site by digestion with an appropriate restriction enzyme, and the digests were resolved on a 1% agarose gel and stained with ethidium bromide. To investigate the role of the supercoiling energy in assuming unusual DNA structures, plasmids were also linearized by restriction digestion prior to S1 nuclease digestion. Sites of S1 nuclease cleavage were mapped approximately by agarose gel electrophoresis of the plasmid DNA fragments that were generated. Fine mapping of the cleavage sites was achieved by cloning the digested DNA species, followed by sequence analysis of randomly selected clones.
RESULTS
A 170-bp DNA fragment from HHV-6B (the DRR-DRL junction) contains the cis-acting signals necessary for cleavage and packaging.
We previously cloned DNA fragments corresponding to the DRR-DRL junction of HHV-6 genome concatemers from virally infected cells. The minimal, approximately 170-bp DNA element so derived was found to contain sequences homologous to the consensus pac2 and pac1 motifs that are present at the genomic termini of other herpesviruses, arranged in a unique configuration (pac2-pac1) that is not present within the unit length HHV-6 genome (29). These findings suggested that the pac2-pac1 motif (DRR-DRL junction) of HHV-6 might function to direct cleavage and packaging of viral genome units from the concatemeric product of viral DNA replication. To experimentally test this prediction, we cloned the 170-bp DRR-DRL junction fragment into plasmid vectors that contained a functional HHV-6B origin of lytic replication (oriLyt) (9).
Test plasmids containing HHV-6B oriLyt alone (pO and pΔ2) or in combination with the DRR-DRL junction element (pCO and pΔ2C) were generated in two different vector backbones (plasmids pO and pCO were generated in pGEM-T vector, while plasmids pΔ2 and pΔ2C were generated in pKS vector [Fig. 1]). These plasmids were then transfected into HHV-6B-infected J-Jahn T cells. Ninety-six hours later, extrachromosomal DNA was recovered, as well as DNA from intracellular viral nucleocapsids and (in some cases) DNA from mature extracellular viral particles. All DNAs were subjected to restriction by DpnI endonuclease, so as to eliminate unreplicated input molecules, and the DNAs were then linearized with appropriate restriction enzymes prior to Southern blot analysis using a plasmid-specific DNA probe. Both sets of plasmids gave essentially identical results in these assays. For simplicity, only the results of the experiments with plasmids pΔ2 and pΔ2C are shown in Fig. 2.
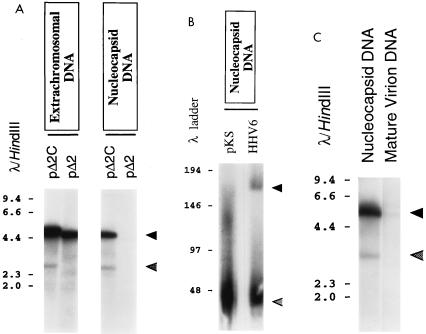
FIG. 2.
Analysis of virally mediated cleavage and packaging of test plasmids. (A) Analysis of the effect of the HHV-6B DRR-DRL genome junction on virally mediated cleavage and packaging of plasmid DNAs. Plasmids pΔ2 and pΔ2C were introduced into HHV-6B-infected J-Jahn T cells. Ninety-six hours later, the cells were collected and split into two equal fractions. Extrachromosomal DNAs were prepared from the first fraction, while nucleocapsids were prepared from the second fraction. These DNAs were then digested with DpnI and XhoI and subjected to Southern blot analysis with a radiolabeled pKS probe. Shown is a photograph of the resulting autoradiogram. Numbers correspond to the sizes of λ HindIII DNA fragments (in kilobases). Cleavage of replicated (DpnI-resistant) plasmid DNA with XhoI should give rise to unit length plasmid monomers (approximately 4.6 and 4.8 kbp for pΔ2 and pΔ2C, respectively) (solid arrowhead) and to two terminal fragments of roughly 3.0 kb and 1.8 kb, reflecting specific, virally mediated cleavage of packaged plasmid concatemers. Of these terminal fragments, only the 3-kb species should be detected by the plasmid probe used (lower, shaded arrowhead). (B) Analysis of the size of nucleocapsid DNA generated by plasmid pΔ2C. pΔ2C was introduced into HHV-6B-infected J-Jahn T cells, and 96 h later, the cells were collected and nucleocapsid DNA was isolated. Nondigested nucleocapsid DNA was then subjected to PFGE and to Southern blot analysis with either a probe specific for pΔ2C (pKS) or a probe specific for wild-type HHV-6 DNA (HHV6) (this DNA probe corresponds to the viral U94 open reading frame, or rep gene homolog). Shown is a photograph of the resulting autoradiogram. Numbers correspond to the sizes of concatemeric or unit length λ DNA molecules (in kilobases) that were used as molecular-weight markers. Arrowheads indicate the positions of the major packaged forms of pΔ2C DNA (approximately 43 kb; pKS probe) and wild-type viral DNA (approximately 160 kb; HHV6 probe). (C) Analysis of the effect of the HHV-6B DRR-DRL genome junction on packaging of plasmid DNAs into mature virus particles. Plasmid pΔ2C was introduced into HHV-6B-infected J-Jahn T cells. Ninety-six hours later, intracellular viral nucleocapsids and cell-free viral particles were prepared. DNA isolated from these particles was then digested with DpnI and XhoI and was subjected to Southern blot analysis as described for panel A. Shown is a photograph of the resulting autoradiogram. Numbers correspond to the sizes of λ HindIII DNA fragments (in kilobases). Upper (solid) arrowhead, replicated plasmid monomers; lower (shaded) arrowhead, terminal (cleaved) fragments.
In the extrachromosomal DNA fraction, replication of both plasmids was detected, as evidenced by the generation of DpnI-resistant plasmid monomers (Fig. 2A). This was expected, since both pΔ2 and pΔ2C contain HHV-6B oriLyt. In addition, a novel DpnI-resistant DNA fragment somewhat shorter than the linearized plasmid monomer was detected in extrachromosomal DNA from cells that had been transfected with plasmid pΔ2C (Fig. 2A). The size of this DNA fragment is consistent with specific virally mediated endonucleolytic cleavage at, or very close to, the pac2-pac1 junction.
To test whether the DRR-DRL junction element was sufficient for the packaging of replicated plasmid DNAs, we next examined DNA from intracellular viral nucleocapsids prepared from virally infected cells that had been transfected with pΔ2 and pΔ2C. As can be seen in Fig. 2A, only plasmid pΔ2C was successfully packaged into viral nucleocapsids. Densitometric analysis of the autoradiogram shown in Fig. 2A revealed that the ratio of the plasmid monomer to the shorter plasmid fragment was approximately 10 to 1. Since pΔ2C is roughly 5 kb, and since the data in Fig. 2A indicate the presence of 1 terminal plasmid fragment for every 10 internal plasmid molecules, the average length of the plasmid concatemers contained within the intracellular nucleocapsids can be estimated at only about 50 kb (note that the DNA probe used in Fig. 2A detects only one of the two expected terminal DNA fragments). This result suggests that most of the concatemeric plasmid DNA contained within intracellular nucleocapsids is considerably shorter than genome unit length (approximately 160 kb).
To confirm this prediction, intracellular viral nucleocapsids were harvested from virally infected cells that had been transfected with pΔ2C. Undigested nucleocapsid DNA was then subjected to PFGE and Southern blotting; the results are presented in Fig. 2B. In this experiment, two different radiolabeled DNA probes were used, one of which was specific for plasmid sequences (pKS) and one of which was specific for wild-type HHV-6 genomes (sequences recognized by this probe are absent from pΔ2C). As can be seen in Fig. 2B, the plasmid concatemers that were packaged into intracellular nucleocapsids varied considerably in size, spanning a range from roughly 40 to 160 kb. However, the predominant size of the packaged plasmid concatemers was approximately 43 kb, which represents nine tandem copies of pΔ2C (detected by the pKS probe) (Fig. 2B). In contrast, encapsidated wild-type viral genomes (detected by the HHV-6 probe) included a much higher proportion of full-length molecules (approximately 160 kb), although shorter, presumably defective molecules in the range 40 to 60 kb were also abundant.
Intracellular nucleocapsids containing plasmid concatemers of less than genome unit length are unlikely to develop into mature virus particles. We therefore compared the DNA content of nucleocapsids present in virally infected cells transfected with pΔ2C to the DNA content of extracellular virus particles released from the same cells. The results of this experiment are presented in Fig. 2C, which shows that the amount of plasmid DNA present in mature extracellular viral particles was dramatically less than that found within intracellular nucleocapsids. Thus, it is apparent that only a small fraction of the nucleocapsids containing plasmid DNA molecules ever develop into mature enveloped virions.
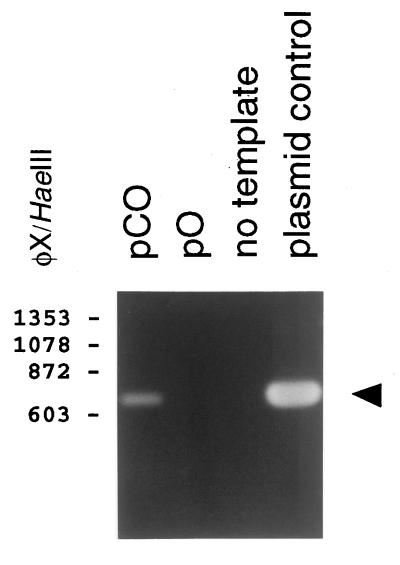
To verify that extracellular virions which contain replicated and packaged plasmid molecules are indeed infectious, we performed a serial passage experiment. Plasmids pCO and pO were transfected into HHV-6B-infected J-Jahn T cells. Ninety-six hours later, cell culture supernatants were collected, and these were then used to infect phytohemagglutinin (PHA)- and interleukin 2 (IL-2)-stimulated human peripheral blood mononuclear cells (PBMC). Seven days later, when a virally induced cytopathic effect (CPE) was observed in these cultures, cell culture supernatants were again collected and used to infect PBMC. Seven days thereafter (at peak CPE), extrachromosomal DNA was isolated from the cells and subjected to DpnI digestion and PCR amplification by using primers specific for the ampicillin resistance gene of pGEM-T (the vector used to construct pO and pCO). The results of this experiment are illustrated in Fig. 3. The data show that pCO, but not pO, was successfully transferred to the PBMC. Since the DRR-DRL junction element is present only in pCO, this result indicates that this sequence element is required for the packaging of replicated plasmid DNA into mature and infectious virus particles (i.e., particles capable of transferring the packaged plasmid DNA to new host cells).
FIG. 3.
Analysis of passage of amplicon-containing virus stocks. Plasmids pO and pCO were introduced into HHV-6B-infected J-Jahn T cells. Ninety-six hours later, cell-free virus particles were collected and used to infect PHA- and IL-2-stimulated PBMC. Seven days later, at the peak of the virally induced CPE, cell culture supernatants were again used to infect new PBMC. Finally, after another 7 days (again, at the peak of virally induced CPE), extrachromosomal DNA was prepared from these cells and digested with DpnI. The DNA was then subjected to PCR amplification using primers AMP1 and AMP2 (see Materials and Methods), and the PCR products were subjected to agarose gel electrophoresis. Shown is a photograph of the ethidium bromide-stained gel. Numbers correspond to the sizes of φX174/HaeIII DNA fragments (in base pairs). A PCR product of 730 bp, indicating the presence of plasmid DNA, was detected in cells that had been infected with supernatants from the pCO transfectants but not from the pO transfectants (arrowhead). A positive control (PCR amplification of plasmid DNA, in “plasmid control” lane) and a negative control (“no template” lane) are also shown.
Although plasmid pCO was able to function as a defective viral genome, or amplicon, in the presence of wild-type helper virus, the efficiency at which this plasmid was replicated and transferred to new host cells was very low. Serial passage of amplicon-containing virus stocks resulted in a level of plasmid DNA transfer that could be detected only by PCR-based methods and not (for example) by Southern blot analysis (data not shown). Similar results were also obtained with a number of other plasmid constructs analogous to pCO, including pΔ2C and derivatives of pΔ2C bearing a marker gene cassette (data not shown). In an attempt to understand the basis for this observation, we molecularly cloned and analyzed the terminal DNA sequences from intracellular viral nucleocapsids and from extracellular virus particles which contained replicated and packaged concatemers of plasmid pΔ2C.
Eleven terminal sequences from nucleocapsid DNA were cloned and sequenced, as well as nine sequences from extracellular virions (Fig. 4). Three of the 11 nucleocapsid-derived fragments (27%) and 4 of the 9 virion-derived clones (44%) were essentially identical (plus or minus 1 nucleotide) to previously obtained terminal sequences from the R1 isolate of HHV-6B (this isolate was the source of the DRR-DRL junction fragment incorporated into pΔ2C, pCO, and all derivatives of these plasmids; this sequence is shown as clone C62 at the top of Fig. 4). A majority of the nucleocapsid-derived clones (8 of 11, or 73%) contained substantial (≥6-nucleotide) changes relative to the bona fide viral terminus, whereas only 2 virion-derived clones (22%) were similarly divergent. Particularly noteworthy are three nucleocapsid-derived clones (E4, E13, and E14) which contained long (>30-nucleotide) stretches of nonviral DNA at their termini (these sequences, which are underlined in Fig. 4, were nonidentical to sequences of HHV-6 DNA, of plasmid DNA, or of any other DNA in the GenBank database). Two of these clones (E13 and E14) also lack a substantial part of the pac1 motif.
FIG. 4.
Analysis of left terminal sequences from packaged amplicons. Plasmid pΔ2C was transfected in HHV-6B-infected J-Jahn cells. Ninety-six hours later, viral nucleocapsids or extracellular virions were purified and used as a source of DNA for PCR amplification and cloning of terminal DNA fragments from packaged plasmid DNA concatemers. The sequences of 11 randomly selected clones derived from viral nucleocapsids (between E1 and E18) are shown, together with 9 clones from extracellular virions (between U1 and U11). These sequences are compared to that of the left genome terminus of the R1 isolate of HHV-6B (HHV-6B) and to that of the concatemeric junction fragment contained in plasmid pΔ2C (C62) (29). Dots, gaps introduced for purposes of alignment; uppercase letters, viral sequences extending beyond the left genome terminus; underlined sequences (in E4, E13, and E14), regions that are not homologous either to HHV-6 DNA or to plasmid (pΔ2C) DNA (or to any other DNA in GenBank).
The DRR-DRL junction element enhances replication efficiency of HHV-6B oriLyt-bearing plasmids in virally infected cells.
In examining the results of our experiments on virally mediated cleavage and packaging of plasmid DNA molecules, we noticed that the replication efficiency of plasmids containing the DRR-DRL junction element seemed to be enhanced relative to that of constructs which contained only HHV-6B oriLyt. To examine this further, we performed a number of additional experiments. The results of one such experiment are shown in Fig. 5.
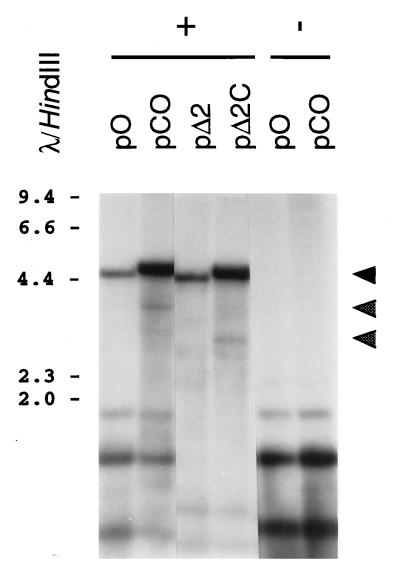
FIG. 5.
Analysis of virally mediated replication of test plasmids containing the HHV-6B DRR-DRL concatemeric junction. Plasmids were introduced into naive (minus sign) or HHV-6B-infected (plus sign) J-Jahn cells. Ninety-six hours later, cells were collected and extrachromosomal DNA was harvested. This DNA was digested with DpnI plus either XhoI (for pΔ2C and pΔ2C) or XmnI (for pO and pCO) and was subjected to Southern blot analysis with a radiolabeled pKS probe. Shown is a photograph of the resulting autoradiogram. Numbers correspond to the sizes of λ HindIII DNA fragments (in kilobases). Cleavage of replicated (DpnI-resistant) plasmid DNA with XhoI or XmnI should give rise to unit length plasmid monomers (approximately 4.6 to 4.8 kb) (top arrowhead) and to two terminal fragments of roughly 3.0 and 1.8 kbp (pΔ2C) or 3.8 and 1.0 kbp (pCO), reflecting specific, virally mediated cleavage of replicated plasmid concatemers. Of these terminal fragments, only the larger molecules should be detected by the plasmid probe used (middle and bottom arrowheads).
HHV-6B-infected J-Jahn cells were transfected with two pairs of test plasmids (pCO and pO, and pΔ2 and pΔ2C; see Fig. 1). Analysis of extrachromosomal DNA prepared from these transfectants revealed higher levels of DpnI-resistant (i.e., replicated) plasmid DNA in pCO and pΔ2C than in their counterparts, pO and pΔ2 (note the intensity of the upper DNA fragment in Fig. 5, which corresponds to the replicated and linearized plasmid DNA monomer). Densitometric analysis of the autoradiogram presented in Fig. 5 confirmed this observation and showed that the plasmids which contained the DRR-DRL junction element were replicated at approximately four- to sixfold higher efficiency than plasmids which contained oriLyt alone (note that replication efficiencies were normalized for total input DNA, which is represented by the lower, DpnI-sensitive bands in Fig. 5).
The DRR-DRL junction element possesses an unusual, S1 nuclease-sensitive conformation (anisomorphic DNA).
In seeking to understand how the DRR-DRL junction fragment might enhance virally mediated replication of plasmid DNA, we wondered whether this DNA sequence might assume an unusual conformation, as previously reported for the a sequences of herpes simplex virus type 1 (HSV-1) (34). Anisomorphic DNA has been shown to be a target for a cellular endonuclease (33), and unusual DNA structures have also been found to be required for the function of a cis-acting component of the Epstein-Barr virus (EBV) oriLyt element (22).
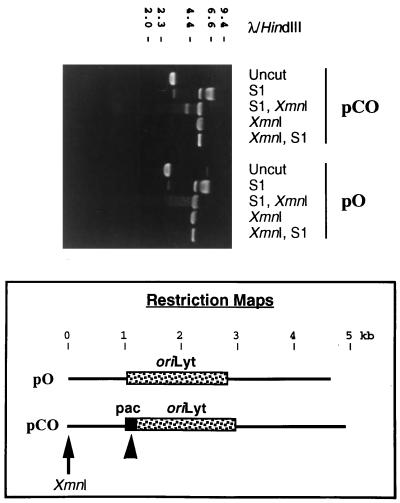
We therefore analyzed the conformation of the DRR-DRL junction element by determining its sensitivity to cleavage by S1 nuclease (34). Typical results are shown in Fig. 6. As expected, XmnI digestion of either plasmid resulted in linearization of the supercoiled plasmid DNA (Fig. 6; compare uncut lanes with XmnI lanes), while S1 nuclease treatment of the plasmid DNA resulted in its cleavage (Fig. 6, S1 lanes). In order to map the site(s) of S1-mediated cleavage, sequential digestions were performed with XmnI, which linearizes the plasmids at a known location, and with S1. Initial digestion of the plasmid DNAs with S1 nuclease, followed by digestion with XmnI (Fig. 6, lanes marked “S1, XmnI”), resulted in the production of linearized plasmid DNA (upper band) together with either a smear of smaller molecules (for pO), which was indicative of nonspecific cutting by S1, or the appearance of two additional DNA fragments (for pCO), which were the result of specific S1 nuclease cleavage (note that the smaller of these two cleavage products is not apparent in Fig. 6, although its size can be inferred from the size of the larger molecule). The restriction maps at the bottom of Fig. 6 show the estimated location of the specific S1 nuclease cleavage site within pCO, which mapped very approximately to the DRR-DRL junction. Furthermore, specific S1 nuclease cleavage of plasmid pCO was dependent upon superhelical tension, since no specific cleavage was observed if the DNA was first restricted with XmnI and then exposed to S1 (Fig. 6, lanes marked “XmnI, S1”).
FIG. 6.
The DRR-DRL concatemeric junction is hypersensitive to cleavage by S1 nuclease. Plasmids pO and pCO were subjected to agarose gel electrophoresis after one of the following experimental treatments: incubation in buffer alone (uncut), digestion with S1 nuclease (S1), digestion with S1 nuclease followed by XmnI (S1, XmnI), digestion with XmnI (XmnI), or digestion with XmnI followed by S1 nuclease (XmnI, S1). Shown is a photograph of the ethidium bromide-stained gel. Numbers correspond to the sizes of λ HindIII DNA fragments (in kilobases). Schematic representations of the two plasmids are shown below the gel. Arrow, unique XmnI site; arrowhead, position of the S1 nuclease-hypersensitive site within pCO; pac, DRR-DRL junction.
The precise site(s) of S1-mediated cleavage within pCO was fine mapped by molecular cloning and sequencing of the smaller product of S1- and XmnI-mediated cleavage. The results are presented in Fig. 7, which shows that the major site of S1-mediated cleavage was located within the pac2 sequence motif. Additional sites of cleavage mapped to pac1 and to the intervening region between these sequences. Thus, the DRR-DRL junction, and the pac2 motif in particular, does indeed correspond to a region of DNA with an unusual, S1-hypersensitive structure (previously defined as anisomorphic DNA [34]).
FIG. 7.
Sequence analysis of S1 nuclease products derived from the DRR-DRL genome junction. The small DNA fragment produced upon digestion of pCO with S1 and XmnI was molecularly cloned, and a total of seven such clones were then sequenced, in order to map precisely the site of S1 nuclease cleavage. The sequence of pC62 (used to create pCO) is shown, together with the positions of consensus DNA motifs (pac1 and pac2). Arrows indicate the sites of S1 nuclease-mediated cleavage for the clones that were sequenced (the number 3 means that 3 clones had an identical cleavage site).
The TRS motifs have no additional effect on viral DNA replication, DNA cleavage, or DNA packaging.
In wild-type HHV-6 genome concatemers, the junction of the two adjacent viral genomes is immediately flanked by arrays of TRSs found close to the two viral termini. Therefore, we decided to test whether these TRS arrays might influence either the replication, cleavage, or packaging of plasmids containing HHV-6B oriLyt and the DRR-DRL junction. To do this, a number of derivatives of plasmid pCO were constructed, including plasmids containing the S-TRS array that is located adjacent to pac2 (pCO-s7 and -s17), clones containing the C-TRS array found next to pac1 (pCO-c16 and -c31), and a clone containing both of these elements (pCO-c16/s17). These plasmids are shown schematically in Fig. 1, while the intermediates involved in their construction and the key sequences of the final clones are illustrated in Fig. 8.
FIG. 8.
Sequences of TRS-containing plasmids. (A) Sequences of plasmids which contain synthetic versions of the S-TRS array, found at the right genome end of HHV-6. The top two sequences correspond to the standard HHV-6 DRR-DRL junction. Plasmid pC62/pCO lacks TRS DNA, and plasmids pCO-s7 and pCO-s17 are derivatives of pCO that contain synthetic versions of the S-TRS motif. (B) Sequences of plasmids which contain synthetic versions of the C-TRS array, found at the left genome end of HHV-6. As in panel A, the upper sequences correspond to the standard viral DRR-DRL junction and the parental plasmid pC62, as well as its derivative pC62/Bst (the latter two have no TRS DNA; see text for details). Plasmids pCO-c16 and pCO-c31 were derived from pC62/Bst by insertion of synthetic versions of the C-TRS motif. The positions of previously identified direct repeat (DR) motifs in the HHV-6 C-TRS array are indicated above the HHV-6B sequence (DR2, DR3), and the dashes represent gaps introduced for purposes of sequence alignment (29). (C) Sequences of plasmids which contain synthetic versions of both the S-TRS and C-TRS arrays, found adjacent to the genome termini of HHV-6. As in panel A, the upper sequences correspond to the viral DRR-DRL junction and the parental plasmid pC62. Plasmid pCO-c16/s17 was derived from pCO-s17 and pCO-c16. In all panels the following conventions apply: uppercase letters, sequences which are identical to those found in viral DNA; lowercase letters, mismatched sequences; underlined sequences, restriction enzyme cleavage sites discussed in the text (AvrII, CCTAGG; BstXI, CCAXXXXXXTGG; EcoRI, GAATTC); [pac2→←pac1], core viral DRR-DRL junction.
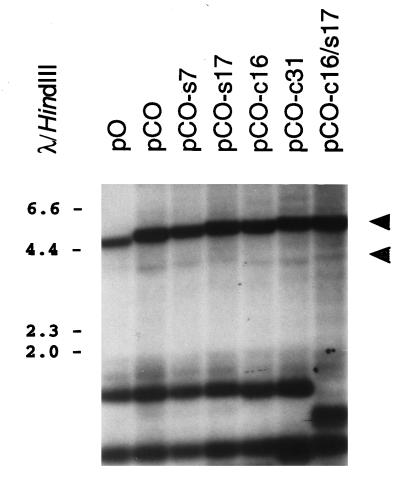
These plasmids were transfected into HHV-6B-infected J-Jahn cells, and extrachromosomal DNA was collected 4 days later for Southern blot analysis. The results, which are presented in Fig. 9, show that the addition of TRS motifs to plasmid pCO had no effect on the efficiency of plasmid replication in virally infected cells (compare the intensities of the upper, DpnI-resistant plasmid monomer bands in the various lanes). Likewise, there was no detectable difference in the efficiency of virally mediated endonucleolytic cleavage of the replicated plasmid DNA molecules (Fig. 9; compare the intensities of the slightly shorter, DpnI-resistant DNA fragments that are the products of the combination of virally mediated and restriction-mediated cleavage events). Densitometric analysis of the autoradiogram shown in Fig. 9, and of similar autoradiograms from additional experiments, confirmed that there was no difference in either DNA replication or DNA cleavage of derivatives of plasmid pCO which contained the TRS motifs, either alone or in combination (data not shown). Finally, serial passage of HHV-6B stocks generated from J-Jahn cells transfected with plasmid pCO-c16/s17 was also performed, by the same methods that were applied to analysis of amplicon stocks containing pCO (Fig. 3). In these experiments, pCO-c16/s17 was not found to be transferred to new host cells with any greater efficiency than plasmid pCO (data not shown). Thus, the TRS motifs appeared to have no significant effect on plasmid replication, cleavage, or packaging in HHV-6B-infected cells.
FIG. 9.
Analysis of virally mediated replication and cleavage of test plasmids containing the synthetic TRS arrays. Plasmids containing the HHV-6B oriLyt and DRR-DRL junction alone (pCO) or in combination with either the S-TRS (pCO-s7 and -s17) or C-TRS (pCO-c16 and -c31) array or both (pCO-c16/s17) were introduced into HHV-6B-infected J-Jahn cells. Ninety-six hours later, cells were collected and extrachromosomal DNA was harvested. This DNA was digested with DpnI plus XmnI and was subjected to Southern blot analysis with a radiolabeled pKS probe. Shown is a photograph of the resulting autoradiogram. Numbers correspond to the sizes of λ HindIII DNA fragments (in kilobases). Cleavage of replicated (DpnI-resistant) plasmid DNA with XmnI should give rise to unit length plasmid monomers (approximately 4.8 to 5.2 kb) (upper arrowhead) and to two terminal fragments of roughly 3.8 and 1.0 kb, reflecting specific cleavage of replicated plasmid concatemers. Of these terminal fragments, only the larger molecules were detected by the plasmid probe used (lower arrowhead).
DISCUSSION
Previous analyses of the genomic termini of HHV-6 had suggested that the substrate for cleavage and packaging of replicated viral DNA concatemers was probably formed by the unique juxtaposition of pac2 and pac1 consensus motifs located at the junction (DRR-DRL) between adjacent viral genomes. We have now experimentally tested this hypothesis. The data show that the HHV-6B DRR-DRL junction element is required in cis for specific, virally mediated cleavage of replicated plasmid DNA concatemers and for their incorporation into intracellular nucleocapsids as well as mature, extracellular virus particles (Fig. 2A and C and Fig. 3). This observation suggests that the cleavage and packaging of HHV-6 DNA proceeds via a conventional herpesvirus mechanism and that, by analogy with other herpesviruses, the cis-acting sequence required for cleavage-packaging is most probably formed by the pac2-pac1 motifs, acting in concert.
While the HHV-6B DRR-DRL junction element is sufficient for cleavage and packaging, the data (Fig. 2A and B) show that most of the plasmid concatemers that are found within intracellular nucleocapsids are much shorter than genome length. The packaging of sub-genome length plasmid concatemers into defective intracellular nucleocapsids has previously been reported for HSV-1 amplicons by Vlazny and colleagues (32). However, HSV-1 capsids appear to preferentially package plasmid concatemers of unit length, or near-unit length. For example, PFGE analysis of nondigested total intracellular DNA from HSV-1-infected cells that were transfected with HSV-1 amplicon constructs revealed a family of fragments migrating around 150 kb (146 to 164 kb), but no smaller bands (1). In contrast, the present data suggest that HHV-6B capsids efficiently package DNA molecules that are very much shorter than the full-length viral genome (Fig. 2B). As noted below, this may reflect a fundamental difference in the cleavage mechanisms that are used by these two viruses.
In any event, capsids containing DNA molecules shorter than the wild-type viral genome would not be likely to mature into infectious, extracellular virions (32). Consistent with this prediction, we found that the level of plasmid DNA which was incorporated into intracellular HHV-6B nucleocapsids greatly exceeded the amount of such DNA found in extracellular virus particles (Fig. 2C; compare the intensities of the plasmid DNA fragments detected in HHV-6B nucleocapsids and in extracellular virions; note that these two lanes represent roughly equivalent numbers of cells which were processed to yield either viral nucleocapsids or extracellular viral particles). Our data also suggest that only a fraction of the plasmid concatemers contained within extracellular virions are in fact capable of being transferred to new host cells. For example, we were unable to convincingly demonstrate the transfer of plasmid (amplicon) DNA to new host cells by standard Southern blot assay methods and could show such transfer only through the use of PCR amplification (Fig. 3). This may reflect, at least in part, the fact that the extracellular-particle-to-infectivity ratio for HHV-6B has been estimated at roughly 103:1 to 104:1 (26). Thus, of the relatively small amount of plasmid DNA found in extracellular virus particles (Fig. 2C), only 0.1 to 0.01% is presumably contained within infectious virions.
The HHV-6B-based amplicon system which we have constructed, using the viral oriLyt and DRR-DRL junction element, appears to be markedly less efficient in terms of gene transfer than comparable constructs based on HSV-1 DNA (27). Furthermore, the inefficiency of gene transfer by our test plasmids could not be increased through the addition of the flanking TRS arrays to the DRR-DRL junction (data not shown). Thus, there appear to be fundamental differences in the cleavage-packaging and/or replication mechanisms of HHV-6B and HSV-1 which presumably account for the striking inefficiency of HHV-6B-derived amplicon vectors. One previously reported difference between these viruses is their relative particle/infectivity ratios. Permissive cell cultures infected with both viruses yield similar total numbers of extracellular virus particles (108 to 109/ml), but the infectious yield of HHV-6B particles is between 1 and 3 log units lower than that for HSV-1 (26). In this regard, HHV-6B is rather more similar to another betaherpesvirus, human cytomegalovirus, which has been reported to have a particle/infectivity ratio of approximately 103:1 (2). The reason(s) for this difference remains unclear.
It is also intriguing that virally mediated cleavage of HHV-6B amplicon concatemers (plasmid pΔ2C) was found to be somewhat imprecise. A considerable degree of heterogeneity was detected among terminal clones derived from plasmid concatemers that were packaged into extracellular virions or intracellular nucleocapsids (Fig. 4). The nucleocapsid-derived sequences were especially divergent. Five of these termini contained quite large deletions (6 to 8 nucleotides) compared to the left genome end of HHV-6B DNA, and another three clones had acquired between 30 and 50 nucleotides of exogenous sequence, of neither viral nor plasmid origin, through an as-yet-unknown mechanism. The significance of these exogenous sequences is uncertain, but their acquisition could conceivably reflect the involvement of a recombination-dependent mechanism in the generation of the viral genomic termini—as has been proposed for Epstein-Barr virus (36). It is also interesting that two of these unusual clones (E13 and E14) (Fig. 4) are missing part of the pac1 motif.
Overall, our findings suggest that the cleavage machinery of HHV-6B operates with at least a moderate degree of flexibility and that it is capable of cleaving replicated DNA molecules at several positions along the DRR-DRL junction (at least when it operates on plasmid concatemers). Conceivably, this “sloppiness” might be due to the absence of viral TRS motifs from the DRR-DRL junction element present in the test plasmid, pΔ2C. However, analysis of terminal DNA fragments present in packaged plasmid concatemers generated from pCO-c16/s17 (which contains both of the viral TRS arrays) also showed evidence of imprecise cleavage at the DRR-DRL junction (data not shown). Thus, the TRS motifs do not appear to significantly enhance the specificity of virally mediated DNA cleavage at the DRR-DRL junction.
In addition to forming the site for virally mediated cleavage and packaging of plasmid concatemers, the DRR-DRL junction also acts in cis to enhance the replication efficiency of plasmids containing HHV-6 oriLyt (Fig. 5). It is possible that this apparent enhancement of plasmid replication is in fact a consequence of an increase in the stabilization of the products of DNA replication, due to their insertion into capsids. Alternatively, this result may reflect a true increase in the rate at which such plasmids are replicated by HHV-6B. At present this issue cannot be resolved, since packaging-defective mutants of HHV-6B, which could replicate plasmid DNAs without encapsidating them, do not exist. We therefore focused our attention on the possibility that the DRR-DRL junction might directly influence the efficiency with which HHV-6B replicates plasmid DNA molecules.
Herpesvirus DNA replication has recently been proposed to proceed via a biphasic mechanism that includes a late, recombination-dependent mode of replication analogous to that utilized by coliphage T4 (16, 21, 25). In fact, the terminal sequences of many herpesviruses have been shown to be involved in recombination processes. Most notably, the a sequence of HSV-1 (which includes the pac2 and pac1 motifs) is a preferred site for recombination during viral DNA replication (4), possibly because it contains sequence elements that are prone to breakage (31). Our observation that the core cleavage-packaging substrate (pac2-pac1) of HHV-6B forms an anisomorphic DNA conformation (Fig. 6 and 7) is therefore intriguing, since DNA structures of this type are known to be prone to cleavage by cellular endonucleases (33)—a process that could generate dsDNA breaks capable of strand invasion and recombination. Alternatively, unusual DNA structures such as anisomorphic DNA may contribute directly to replicator activity, as is the case in several other origins of DNA replication (13, 22). Further studies will be required to address the relationship, if any, between anisomorphic structures and DNA replication.
The HHV-6 TRS motifs appear to have no additional effect on the replication efficiency of plasmids containing HHV-6B DRR-DRL and oriLyt (Fig. 9), nor do they have any demonstrable effect on the efficiency or specificity of virally mediated DNA cleavage (Fig. 9 and data not shown). Thus, the functional significance of these sequences remains to be discovered. Possible functions for these motifs include a role in the regulation of viral gene expression (23) or in the stabilization of extrachromosomal HHV-6 episomes during viral latency, since telomeric sequences have been shown to stabilize episomal yeast artificial chromosomes in Saccharomyces cerevisiae (17). However, experiments designed to test whether HHV-6 TRSs could promote the episomal maintenance of plasmid DNAs in human cells failed to reveal any effect of these elements on plasmid maintenance or stability (3). A third potential function for the viral TRS motifs might be the promotion of site-specific integration of viral DNA into the telomers of host cell chromosomes, via homologous recombination, as suggested by Torelli and colleagues (30). Until recently, herpesvirus genome integration has been viewed as a genetic “accident” without biologic significance. However, studies by Hammerschmidt and colleagues have shown that (i) MDV genome integration is a common event in vitro (7) and (ii) chromosomally integrated MDV genomes can reactivate to yield infectious progeny (8). Unfortunately, it will not be possible to test whether the HHV-6 TRS motifs play any role in viral latency or integration until an in vitro model for latency is established.
In summary, the present studies have resulted in the identification of a functional, cis-acting substrate for HHV-6-mediated DNA cleavage and packaging. This sequence includes the pac2 and pac1 motifs previously identified in other herpesviruses but does not include the telomeric repeat motifs found in the a sequences of HHV-6 (29). The data indicate that the HHV-6 cleavage machinery is somewhat imprecise, at least when cutting plasmid substrates, and that the efficiency with which first-generation HHV-6 amplicon vectors are replicated in the presence of standard helper virus is unexpectedly low (far lower, for example, than comparable vectors based on HSV-1). While the mechanistic basis for this observation is uncertain, it may be instructive to consider that highly defective viral genomes occur naturally during passage of HSV-1 stocks at high multiplicities of infection (11) but that they have not been described either for HHV-6 or for other betaherpesviruses such as human cytomegalovirus (20).
ACKNOWLEDGMENTS
We thank Sandra Weller and Howard Federoff for sharing laboratory protocols; Joel Baines for helpful discussions; Caroline Hall for providing J-Jahn cells and the R-1 strain of HHV-6; Chin-To Fong, Steve Pollack, and Ernest Smith for assistance with PFGE experiments; and George Kampo and Jack Maniloff for preparation of oligonucleotides.
This work was supported by National Institutes of Health grants to S.D. (RO1 AI34231 and KO4 AI01240).
REFERENCES
- 1.Bataille D, Epstein A L. Equimolar generation of the four possible arrangements of adjacent L components in herpes simplex virus type 1 replicative intermediates. J Virol. 1997;71:7736–7743. doi: 10.1128/jvi.71.10.7736-7743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benyesh-Melnick M, Probstmeyer F, McCombs R, Brunschwig J P, Vonka V. Correlation between infectivity and physical virus particles in human cytomegalovirus. J Bacteriol. 1966;92:1555–1561. doi: 10.1128/jb.92.5.1555-1561.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulboaca, G. H., H. Deng, S. Dewhurst, and M. P. Calos. Telomeric sequences from human herpesvirus 6 do not mediate nuclear retention of episomal DNA in human cells. Submitted for publication. [DOI] [PubMed]
- 4.Chou J, Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985;41:803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- 5.Cone R W, Hackman R C, Huang M L, Bowden R A, Meyers J D, Metcalf M, Zeh J, Ashley R, Corey L. Human herpesvirus 6 in lung tissue from patients with pneumonitis after bone marrow transplantation. N Engl J Med. 1993;329:156–161. doi: 10.1056/NEJM199307153290302. [DOI] [PubMed] [Google Scholar]
- 6.Deiss L P, Chou J, Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986;59:605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delecluse H J, Hammerschmidt W. Status of Marek’s disease virus in established lymphoma cell lines: herpesvirus integration is common. J Virol. 1993;67:82–92. doi: 10.1128/jvi.67.1.82-92.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delecluse H J, Schuller S, Hammerschmidt W. Latent Marek’s disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 1993;12:3277–3286. doi: 10.1002/j.1460-2075.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewhurst S, Dollard S C, Pellett P E, Dambaugh T R. Identification of a lytic-phase origin of DNA replication in human herpesvirus 6B strain Z29. J Virol. 1993;67:7680–7683. doi: 10.1128/jvi.67.12.7680-7683.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drobyski W R, Dunne W M, Burd E M, Knox K K, Ash R C, Horowitz M M, Flomenberg N, Carrigan D R. Human herpesvirus-6 (HHV-6) infection in allogeneic bone marrow transplant recipients: evidence of a marrow-suppressive role for HHV-6 in vivo. J Infect Dis. 1993;167:735–739. doi: 10.1093/infdis/167.3.735. [DOI] [PubMed] [Google Scholar]
- 11.Frenkel N, Locker H, Vlazny D A. Studies of defective herpes simplex viruses. Ann N Y Acad Sci. 1980;354:347–370. doi: 10.1111/j.1749-6632.1980.tb27977.x. [DOI] [PubMed] [Google Scholar]
- 12.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 13.Hiasa H, Tanaka K, Sakai H, Yoshida K, Honda Y, Komano T, Godson G N. Distinct functional contributions of three potential secondary structures in the phage G4 origin of complementary DNA strand synthesis. Gene. 1989;84:17–22. doi: 10.1016/0378-1119(89)90134-0. [DOI] [PubMed] [Google Scholar]
- 14.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 15.Kishi M, Harada H, Takahashi M, Tanaka A, Hayashi M, Nonoyama M, Josephs S F, Buchbinder A, Schachter F, Ablashi D V, Wong-Staal F, Salahuddin S Z, Gallo R C. A repeat sequence, GGGTTA, is shared by DNA of human herpesvirus 6 and Marek’s disease virus. J Virol. 1988;62:4824–4827. doi: 10.1128/jvi.62.12.4824-4827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreuzer K N, Saunders M, Weislo L J, Kreuzer H W. Recombination-dependent DNA replication stimulated by double-strand breaks in bacteriophage T4. J Bacteriol. 1995;177:6844–6853. doi: 10.1128/jb.177.23.6844-6853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longtine M S, Enomoto S, Finstad S L, Berman J. Yeast telomere repeat sequence (TRS) improves circular plasmid segregation, and TRS plasmid segregation involves the RAP1 gene product. Mol Cell Biol. 1992;12:1997–2009. doi: 10.1128/mcb.12.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullers J A, Lakeman F D, Whitley R J. Human herpesvirus 6 is associated with focal encephalitis. Clin Infect Dis. 1995;21:571–576. doi: 10.1093/clinids/21.3.571. [DOI] [PubMed] [Google Scholar]
- 20.Ogura T, Tanaka J, Kamiya S, Sato H, Ogura H, Hatano M. Human cytomegalovirus persistent infection in a human central nervous system cell line: production of a variant virus with different growth characteristics. J Gen Virol. 1986;67:2605–2616. doi: 10.1099/0022-1317-67-12-2605. [DOI] [PubMed] [Google Scholar]
- 21.Pfuller R, Hammerschmidt W. Plasmid-like replicative intermediates of the Epstein-Barr virus lytic origin of DNA replication. J Virol. 1996;70:3423–3431. doi: 10.1128/jvi.70.6.3423-3431.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Portes-Sentis S, Sergeant A, Gruffat H. A particular DNA structure is required for the function of a cis-acting component of the Epstein-Barr virus OriLyt origin of replication. Nucleic Acids Res. 1997;25:1347–1354. doi: 10.1093/nar/25.7.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarisky R T, Weber P C. Role of anisomorphic DNA conformations in the negative regulation of a herpes simplex virus type 1 promoter. Virology. 1994;204:569–579. doi: 10.1006/viro.1994.1571. [DOI] [PubMed] [Google Scholar]
- 24.Secchiero P, Nicholas J, Deng H, Xiaopeng T, van Loon N, Ruvolo V R, Berneman Z N, Reitz M S, Jr, Dewhurst S. Identification of human telomeric repeat motifs at the genome termini of human herpesvirus 7: structural analysis and heterogeneity. J Virol. 1995;69:8041–8045. doi: 10.1128/jvi.69.12.8041-8045.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Severini A, Scraba D G, Tyrrell D L. Branched structures in the intracellular DNA of herpes simplex virus type 1. J Virol. 1996;70:3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiraki K, Mukai T, Okuno T, Yamanishi K, Takahashi M. Physicochemical characterization of human herpesvirus 6 infectivity. J Gen Virol. 1991;72:169–172. doi: 10.1099/0022-1317-72-1-169. [DOI] [PubMed] [Google Scholar]
- 27.Spaete R R, Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982;30:295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- 28.Takeshita S, Tezuka K, Takahashi M, Honkawa H, Matsuo A, Matsuishi T, Hashimoto-Gotoh T. Tandem gene amplification in vitro for rapid and efficient expression in animal cells. Gene. 1988;71:9–18. doi: 10.1016/0378-1119(88)90072-8. [DOI] [PubMed] [Google Scholar]
- 29.Thomson B J, Dewhurst S, Gray D. Structure and heterogeneity of the a sequences of human herpesvirus 6 strain variants U1102 and Z29 and identification of human telomeric repeat sequences at the genomic termini. J Virol. 1994;68:3007–3014. doi: 10.1128/jvi.68.5.3007-3014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torelli G, Barozzi P, Marasca R, Cocconcelli P, Merelli E, Ceccherini-Nelli L, Ferrari S, Luppi M. Targeted integration of human herpesvirus 6 in the p arm of chromosome 17 of human peripheral blood mononuclear cells in vivo. J Med Virol. 1995;46:178–188. doi: 10.1002/jmv.1890460303. [DOI] [PubMed] [Google Scholar]
- 31.Umene K. Herpes simplex virus type 1 variant a sequence generated by recombination and breakage of the a sequence in defined regions, including the one involved in recombination. J Virol. 1993;67:5685–5691. doi: 10.1128/jvi.67.9.5685-5691.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vlazny D A, Kwong A, Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci USA. 1982;79:1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wohlrab F, Chatterjee S, Wells R D. The herpes simplex virus 1 segment inversion site is specifically cleaved by a virus-induced nuclear endonuclease. Proc Natl Acad Sci USA. 1991;88:6432–6436. doi: 10.1073/pnas.88.15.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wohlrab F, McLean M J, Wells R D. The segment inversion site of herpes simplex virus type 1 adopts a novel DNA structure. J Biol Chem. 1987;262:6407–6416. [PubMed] [Google Scholar]
- 35.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;i:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 36.Zimmermann J, Hammerschmidt W. Structure and role of the terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J Virol. 1995;69:3147–3155. doi: 10.1128/jvi.69.5.3147-3155.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]