Abstract
Background
Hereditary angioedema (HAE) is a rare autosomal dominant genetic disease characterised by acute episodes of non-pruritic skin and submucosal swelling caused by increase in vascular permeability.
Objective
Here we present the first complex analysis of the National HAE Slovakian cohort with the detection of 12 previously un-published genetic variants in SERPING1 gene.
Methods
In patients diagnosed with hereditary angioedema caused by deficiency or dysfunction of C1 inhibitor (C1–INH-HAE) based on clinical manifestation and complement measurements, SERPING1 gene was tested by DNA sequencing (Sanger sequencing/massive parallel sequencing) and/or multiplex ligation-dependent probe amplification for detection of large rearrangements.
Results
The Slovakian national cohort consisted of 132 living patients with confirmed HAE. We identified 51 index cases (32 families, 19 sporadic patients/112 adults, 20 children). One hundred seventeen patients had HAE caused by deficiency of C1 inhibitor (C1–INH-HAE-1) and 15 patients had HAE caused by dysfunction of C1 inhibitor (C1–INH-HAE-2). The prevalence of HAE in Slovakia has recently been calculated to 1:41 280 which is higher than average calculated prevalence. The estimated incidence was 1:1360 000. Molecular-genetic testing of the SERPING1 gene found 22 unique causal variants in 26 index cases, including 12 previously undescribed and unreported.
Conclusion
The first complex report about epidemiology and genetics of the Slovakian national HAE cohort expands the knowledge of the C1–INH-HAE genetics. Twelve novel causal variants were present in the half of the index cases. A higher percentage of inframe variants comparing to other studies was observed. Heterozygous deletion of exon 3 found in a large C1–INH-HAE-1 family probably causes the dysregulation of the splicing isoforms balance and leads to the decrease of full-length C1–INH level.
Keywords: Genetic testing, Angioedemas, Hereditary/epidemiology, Hereditary/genetics, Slovakia, Complement C1 inhibitor protein
Significance statement
SERPING1 gene is prone to mutation-inducing activity and origin of new variants that cause C1–INH-HAE. Our study focuses on the epidemiology and genetics of C1–INH-HAE in Slovakia. We report novel variants which can be included in clinical and research databases and used for evaluation of genotype-phenotype correlation of patients, especially in inframe variants. Exon 3 deletion in large hereditary angiodema (HAE) family with severe course of disease can be used for studying the splicing isoforms balance importance for full-length protein level.
Introduction
Hereditary angioedema (HAE, OMIM 106100) is a rare autosomal dominant genetic disease characterised by acute episodes of non-pruritic skin and submucosal swelling caused by the transient increase in vascular permeability. HAE can be caused by deficiency or dysfunction of C1 inhibitor (C1–INH-HAE-1 and C1–INH-HAE-2, respectively) due to genetic defects in SERPING1 gene.1 The prevalence of C1–INH-HAE varies from 1:50 000 to 1:100 000.2
C1 inhibitor (C1–INH), protease inhibitor, belongs to serpins and is responsible for the inhibition of the complement system.3 In the state of C1–INH dysfunction, the kallikrein-kinin system is overactivated and produces large amounts of bradykinin, which by binding to bradykinin B2 receptors increases the vascular permeability.4 C1–INH is encoded by SERPING1, and defects in this gene may lead to misfolded or truncated C1–INH, or to mRNA degradation by nonsense-mediated decay (NMD) preventing forming protein at all. Haploinsufficiency is a common feature in C1–INH-HAE, and, additionally, a variant product inactivates the wild-type allele in a dominant-negative manner.5 According to Drouet et al, 809 pathogenic or likely pathogenic variants were identified in SERPING1 gene that affect 1494 families.5 5.6% of causal variants originate de novo. Missense variants account for 32.2% of all variants and small deletions/duplications/insertions with subsequent frameshift for 36.2%.5 C1–INH-HAE-2 occurs due to missense variants in exon 8 of the SERPING1 gene which affect the reactive loop (active site of the molecule) and reduces the inhibitory effect of C1–INH on target proteins.1 In the study of Czech National HAE cohort (neighbouring country of Slovakia), missense variants carried 35.3% of probands, splicing variants 22.4%, frameshift variants 18.8%, gross deletions 16.5%, and nonsense variants 4.7%. Causative or probably causative variants were detected in 206 out of 207 (56 unique pathogenic or likely pathogenic sequence variants were found).6 In the study of Hungarian HAE patients (neighbouring country of Slovakia), missense variants were found in 30.1%, large deletions or duplications in 20.6%, frameshift variants in 19.1%, nonsense variants in 17.6%, and splicing variants in 11.8% of the index cases.7
According to the actual valid diagnostic criteria, the genetic confirmation in SERPING1 is not necessary for the final estimation of C1–INH-HAE diagnosis. However, the genetic analysis could significantly contribute to the diagnosis confirmation, especially in unclear or conflicting clinical and laboratory results.8
HAE has been found to be caused by different mechanisms due to deficiency in other genes—factor XII (F12),9 angiopoietin-1 (ANGPT1),10 plasminogen (PLG),11 kininogen (KNG1),12 myoferlin (MYOF),13 and heparan sulfate-glucosamine 3-O-sulfotransferase 6 (HS3ST6)14—in small amounts of patients (13.2%),5 with normal concentration and function of C1–INH (HAE with normal C1–INH).
Methods
Patients were recruited from a national survey of HAE in Slovak Republic with the diagnosis C1–INH-HAE. Most of the patients are followed by National Centre for Hereditary Angioedema in University Teaching Hospital in Martin (Slovakia) and were referred after national online survey by general practitioners, immunologists, dermatologists, or individuals who directly contacted the centre for evaluation, treatment, and genetic testing. Diagnosis of C1–INH-HAE was established according to international consensus guidelines, based on clinical symptoms and serum levels of functional and antigenic C1–INH.8 Patients were diagnosed as C1–INH-HAE-1 when functional and antigenic C1–INH were ≤50% of normal values. Patients were classified as C1–INH-HAE-2 when functional C1–INH was ≤50% and antigenic C1–INH was >50% of normal values. Informed written consent to mutational analysis from all patients was archived.
Genomic DNA for genetic testing was extracted from EDTA-anticoagulated whole blood. Sequencing of the coding region (exons 2–8) and exon-intron boundaries of SERPING1 (NM_000062.3) was performed using standard Sanger sequencing protocols or massive parallel sequencing (MPS) with a minimum of 20x depth of coverage, Clinical Exome Solution Kit, Sophia Genetics (Illumina NextSeq 550, San Diego, California, USA) with in silico Copy Number Variation analysis. Data were analyzed using Sophia DDM analysis software. Sanger sequencing analysis was used for confirmation of identified variants after MPS. Primer sequences and polymerase chain reaction (PCR) conditions are available on request. Multiplex ligand-dependent probe amplification (MLPA) was performed after the negative result of sequencing using SALSA MLPA P243 SERPING1 Kit (MRC Holland, The Netherlands) in order to search for large deletions or duplications. When the disease-causing variant of the family was identified, DNAs from family members were investigated by direct sequencing of the region (exon) carrying the variant or by MLPA as relevant. Identified variants nomenclature follows Human Genomic Variation Society (HGVS) recommendations.15 Coding DNA nucleotide numbering and protein sequence numbering were compared to GenBank reference sequence NM_000062.3 and NP_000053.2. Interpretation of identified variants was based on the criteria established by the American College of Medical Genetics and Genomics (ACMG)16 using Varsome database,17 InterVar tool18 and Uniprot database.19
Results
The Slovakian national C1–INH-HAE cohort consisted of 132 living patients (56 males and 76 females) with confirmed HAE. We identified 51 index cases (32 families and 19 sporadic patients/112 adults and 20 children). Among these, 117 patients were clinically, and laboratory confirmed to have C1–INH-HAE-1, while 15 patients had C1–INH-HAE-2. Deceased patients (n = 21) were excluded from the study, with 2 of them experiencing complications of HAE leading to death (laryngeal oedema). Fifteen patients died due to asphyxia before the clinical diagnosis of HAE. None of the patients died during the follow-up by National Centre for Hereditary Angioedema in University Teaching Hospital in Martin.
Molecular-genetic testing of the SERPING1 gene found 22 unique causal variants in 26 index cases, including 12 previously undescribed variants. Novel causal variants were present in 50% of index cases (n = 13) from our cohort and represent 54.6% of identified variants. In 44 patients, molecular-genetic testing was not performed due to their low compliance with the care and clinical management. We identified causal variants in 72 patients from 88 patients tested (81.8%). Patients with negative family history represented 30.1% of index cases. All variants were detected in a heterozygous state.
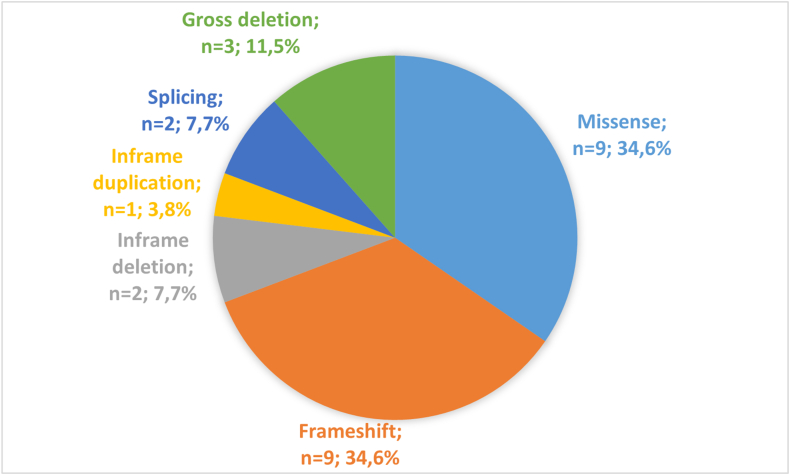
Both frameshift and missense variants were the most common in index cases (both n = 9, 34.6%), followed by gross deletions (n = 3, 11.5%) (Fig. 1). All frameshift variants led to incorporation of termination codon. We did not find any typical nonsense variants. From the other point of view, small deletions/duplications were present in 46.1% of cases.
Fig. 1.
Distribution of causal variants in SERPING1 gene according to the variant type. Frameshift and missense variants were the most common in index cases, followed by gross deletions. All frameshift variants led to incorporation of termination codon. We didn't find any typical nonsense variants
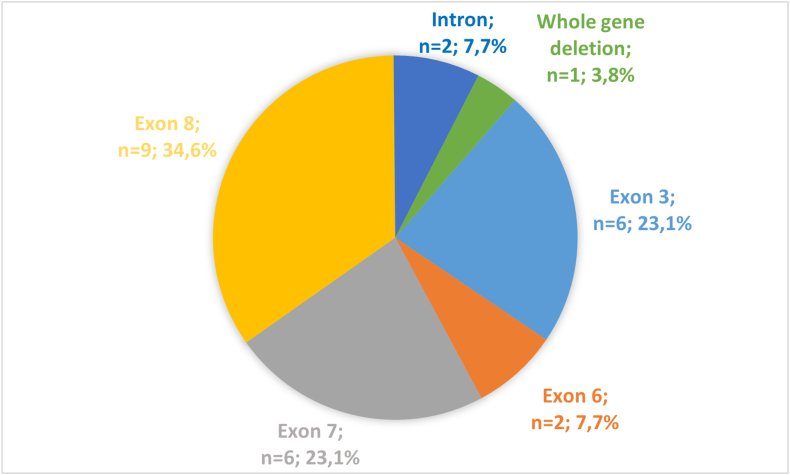
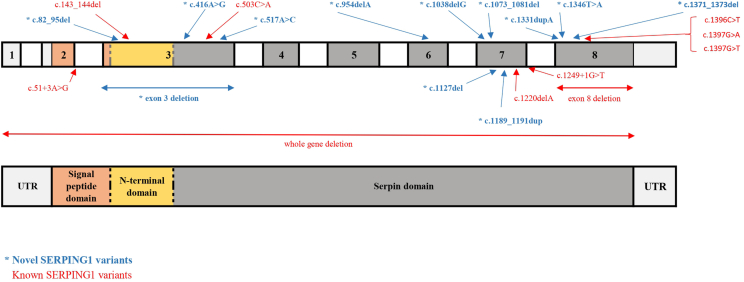
Exon 8 was affected in one-third of all cases (n = 9, 34.6%). Exons 3 and 7 were the second most affected (n = 6, 23.1%) (Fig. 2). The localization of the variants identified in Slovakian HAE patients on SERPING1 gene is shown in Fig. 3.
Fig. 2.
Distribution of causal variants of SERPING1 gene according to affected exons. Exon 8 was affected in one-third of all cases. Exons 3 and 7 were the second most affected, followed by exon 6 and intronic variants
Fig. 3.
Localization of variants identified in Slovakian HAE patients on SERPING1 gene. The upper part of the figure shows exons (colourful boxes) and introns (white boxes) of the SERPING1 gene with marked variants identified in our cohort; the lower part represents equivalent domains of the C1 inhibitor protein; UTR – untranslated region
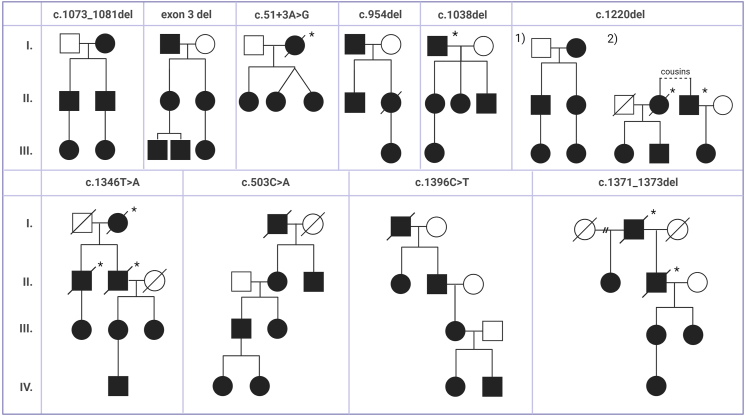
Table 1 presents the identified variants in SERPING1 gene within the Slovakian HAE cohort and Fig. 4 displays pedigrees of large HAE families with specific variant in SERPING1 gene. Three variants were associated with C1–INH-HAE-2 (c.1396C > T, c.1397G > A, c.1397G > T). We identified 10 variants (in 13 index cases) that are already known in literature (c.143_144del, c.503C > A, c.1220del, c.1396C > T, c.1397G > A, c.1397G > T, c.51+3A > G, c.1249+1G > T, exon 8 deletion, whole gene deletion).
Table 1.
Causal variants identified in the SERPING1 gene in Slovakian HAE cohort.
| Index cases | Exon | cDNA numbering (NM_000062.3) | Effect on protein | Variant type | ACMG evaluation | CADD | References | Related patients | Type of HAE |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | c.82_95del | p.Ser28GlyfsTer25 | Frameshift | pathogenic | not published | C1–INH-HAE-1 | ||
| 1 | 3 | c.143_144del | p.Thr48SerfsTer9 | Frameshift | pathogenic | 20, 21, 22, 23, 24 | 2 (father - without genetic confirmation, daughter) | C1–INH-HAE-1 | |
| 1 | 3 | c.416A > G | p.Glu139Gly | Missense | likely pathogenic | 17.6 | not published | C1–INH-HAE-1 | |
| 1 | 3 | c.503C > A | p.Ala168Asp | Missense | likely pathogenic | 22.6 | 7,25, 26, 27 | 7 (Fig. 4) | C1–INH-HAE-1 |
| 1 | 3 | c.517A > C | p.Ser173Arg | Missense | likely pathogenic | 25.8 | not published | 3 (father, son, daughter) | C1–INH-HAE-1 |
| 1 | 3 | Exon 3 deletion | Gross deletion | pathogenic | not published | 6 (Fig. 4) | C1–INH-HAE-1 | ||
| 2 | 6 | c.954del | p.Val319LeufsTer2 | Frameshift | pathogenic | not published | 4 (Fig. 4),2 (mother, daughter) | C1–INH-HAE-1 | |
| 1 | 7 | c.1038del | p.Gln346HisfsTer8 | Frameshift | pathogenic | not published | 4 (Fig. 4) | C1–INH-HAE-1 | |
| 1 | 7 | c.1073_1081del | p.Leu358_Pro360del | Inframe deletion | likely pathogenic | not published | 5 (Fig. 4) | C1–INH-HAE-1 | |
| 1 | 7 | c.1127del | p.Pro376LeufsTer21 | Frameshift | pathogenic | not published | C1–INH-HAE-1 | ||
| 1 | 7 | c.1189_1191dup | p.Thr397dup | Inframe duplication | likely pathogenic | not published | 3 (mother, son, daughter) | C1–INH-HAE-1 | |
| 2 | 7 | c.1220del | p.Gln407ArgfsTer24 | Frameshift | pathogenic | 28 | 5 (Fig. 4), 3 (Fig. 4) |
C1–INH-HAE-1 | |
| 1 | 8 | c.1331dup | p.Thr445AspfsTer28 | Frameshift | pathogenic | not published | C1–INH-HAE-1 | ||
| 1 | 8 | c.1346T > A | p.Leu449Gln | Missense | likely pathogenic | 27.6 | not published | 4 (Fig. 4) | C1–INH-HAE-1 |
| 1 | 8 | c.1371_1373del | p.Ala459del | Inframe deletion | pathogenic | not published ∗ | 4 (Fig. 4) | C1–INH-HAE-1 | |
| 1 | 8 | c.1396C > T | p.Arg466Cys | Missense | pathogenic | 25.4 | 7,26,28, 29, 30, 31, 32,36, 37, 38, 39 | 6 (Fig. 4) | C1–INH-HAE-2 |
| 3 | 8 | c.1397G > A | p.Arg466His | Missense | pathogenic | 24.0 | 20,23,26,29, 30, 31,39, 40, 41 | 2 (father, son) | C1–INH-HAE-2 |
| 1 | 8 | c.1397G > T | p.Arg466Leu | Missense | pathogenic | 23.8 | 4,24,33, 34, 35, 36, 37,40,42 | 3 (father, 2 daughters) | C1–INH-HAE-2 |
| 1 | 8 | Exon 8 deletion | Gross deletion | pathogenic | 28,41 | 2 (father, son) | C1–INH-HAE-1 | ||
| 1 | 1–8 | Exon 1–8 (whole gene) deletion | Gross deletion | pathogenic | 43 | C1–INH-HAE-1 | |||
| 1 | intronic | c.51+3A > G | Splicing | pathogenic | 21,38,41,44, 45, 46 | 3 (Fig. 4) | C1–INH-HAE-1 | ||
| 1 | intronic | c.1249+1G > T | Splicing | pathogenic | 20,38,47 | C1–INH-HAE-1 |
Index case represents the source patient, in whom the origin of the causal variant is observed (sporadic patient or first documented and genetically confirmed patient in the family). The cells in Effect on protein column is empty in case of splicing variants and gross deletions because of the variant effect nature. The resources are indicated in the Reference column. Related patients column contains the number of affected members in family with description of relations or reference to Fig. 4 where pedigrees are shown. Without genetic confirmation note indicates HAE association based on only typical clinical manifestation. The cell in this column is empty by the variants with the occurrence in sporadic patients. Missense variants were evaluated by in silico tool Combined Annotation Dependent Depletion (CADD). Other variant types were not evaluated by CADD, thus there are empty cells in the CADD column. Variant c.1371_1373del was not published in literature but was presented on European Society for Immunodeficiencies 2014 oral presentations – marked with asterisk (∗). Abbreviations: ACMG - American College of Medical Genetics and Genomics, CADD - Combined Annotation Dependent Depletion, C1–INH-HAE-1 - Hereditary angioedema caused by deficiency of C1 inhibitor, C1–INH-HAE-2 - Hereditary angioedema caused by dysfunction of C1 inhibitor, HAE – Hereditary angioedema
Fig. 4.
Pedigrees of large HAE families with specific causal variant in SERPING1 gene
HAE-affected family members carrying the causal variant are shown in black and healthy individuals are depicted by a blank symbol. Deceased family members are shown with a sloping line through the symbol. Divorce is shown as horizontal line connecting 2 symbols with 2 diagonal hash marks. Individuals with only clinical symptoms indicating HAE who were not genetically tested are marked by asterisk (∗). Variant c.1220del was found in 2 large families which are divided and marked as 1) and 2).
Whole gene deletion was associated with a severe course of disease. The patients had early onset of symptoms (from 1 to 5 years of age), developed from eight to 27 attacks per year with angioedemas of larynx.
On the other hand, missense variants c.1397G > A and c.1397G > T caused a milder course of disease (from 1 to 2 attacks per year) without laryngeal oedemas and the onset of symptoms between 3 to 21 years of age.
We report novel gross deletion (exon 3 deletion) in a large family with C1–INH-HAE-1 which is evaluated as a pathogenic according to ACMG criteria (PVS1, PM2, PP1, PP4). Patients in this family had early onset of symptoms (from 2 to 8 years of age), severe course of disease (10–25 attacks per year) with angioedemas of extremities, face, larynx, and abdominal symptoms.
Five of the novel variants introduce premature stop codons by creating frameshifts due to multiple nucleotide deletion (c.82_95del), single nucleotide deletions (c.954del, c.1038del, c.1127del c.1127del), or a single nucleotide duplication (c.1331dup). These variants are predicted to lead to degradation by nonsense mediated mRNA decay (NMD) leaving no transcripts for protein production.
Two novel variants are classified as inframe deletions (c.1371_1373del, c.1073_1081del) and 1 novel variant as inframe duplication (c.1189_1191dup). Co-segregation with disease in multiple affected family members were observed in all 3 variants. These variants are evaluated as likely pathogenic according to ACMG criteria (PM1, PM2, PM4, PP1, PP4).
Inframe duplication (c.1189_1191dup) caused onset of symptoms in early adulthood (from 17 to 20 years of age) and severe course of disease (20–30 attacks per year) with abdominal, orofacial, and laryngeal oedemas.
We report 3 novel missense variants. Novel missense variant c.1346T > A (p.Leu449Gln) was found in a large family with clinical diagnosis of C1–INH-HAE-1 (Fig. 4). Patients had an onset of symptoms between 2 to 26 years of age; male patient had only 1 attack per year, female patients from 20 to 51 attacks per year (the highest number in the whole cohort). Two other pathogenic missense variants at this nucleotide position were already reported: c.1346T > C22,26 and c.1346T > G.22,26,41,48 The amino acid (p.Leu449) is a part of the serpin domain (Fig. 3) and forms beta strand (breach region of the protein) assuming structural damage of the SERPING1 protein by amino acid change due to these missense variants. Leucine which has hydrophobic side chain is changed to glutamine with polar uncharged side chain due to c.1346T > A. Variant is evaluated as a likely pathogenic according to ACMG criteria (PM1, PM2, PP1, PP3, PP4). We evaluate this variant as causal considering segregation analysis in affected family, ACMG criteria, the presence of 2 missense pathogenic variants at the same nucleotide position, and amino acid change that probably affect protein function.
Novel missense variant c.416A > G was found in sporadic female patient with the diagnosis of C1–INH-HAE-1. The patient has been symptomatic from 32 years of age (her current age is 44 years). She experienced abdominal symptoms (abdominal pain, diarrhoea) with the frequency of 2 attacks per year and showed low (nearly zero) C1–INH concentration and its function. It codes amino acid with polar uncharged side chain p.Glu139. This amino acid is a part of the serpin domain (Fig. 3) and forms one of the helical structures. Variant is evaluated as a likely pathogenic according to ACMG criteria (PM1, PM2, PM6, PP4). We suppose the change to neutral and compact amino acid glycine affects the protein structure. We evaluate variant c.416A > G as causal considering ACMG criteria and amino acid change that probably affect protein function due to defect forming of the helical structure.
Novel missense variant c.517A > C was found in a family with the clinical diagnosis of C1–INH-HAE-1 (Fig. 4). It codes the amino acid p.Ser173Arg which is a part of the serpin domain (Fig. 3) and forms the helical structure. LOVD and Clinvar databases contain variant c.518G > A, which is located in the same codon as c.517A > C, causes protein change p.Ser173Asn and is not considered to affect protein function (VUS). Serin and asparagine both have polar uncharged side chain. On the other side, arginine is amino acid with positive electrically charged side chain. Variant is evaluated as a likely pathogenic according to ACMG criteria (PM1, PM2, PP1, PP3, PP4). We evaluate variant c.517A > C as causal considering segregation analysis in affected family and amino acid change that probably affect protein function due to defect forming of the helical structure.
We identified 2 variants (c.1397G > A, c.5C > T) in 1 patient with C1–INH-HAE-1. Variant c.5C > T, pAla2Val39,49 is classified as a variant of uncertain significance according the ACMG criteria (high allele frequency in European [non-Finnish] population 0.00127 and South Asian population 0.00358; ClinVar interpretation: 3x likely benign, 1x uncertain significance). We also identified a pathogenic variant c.1397G > A that fully explained the clinical manifestation. Although segregation analysis of c.5C > T in the family was not possible, we suppose this variant having no impact on clinical and laboratory manifestation in our patient (variant was not integrated in Table 1).
Discussion
The Slovakian national cohort consisted of 132 living patients with confirmed HAE. We identified 51 index cases (32 families and 19 sporadic patients). One hundred seventeen patients had clinically and laboratory confirmed C1–INH-HAE-1 and 15 patients C1–INH-HAE-2. The prevalence of C1–INH-HAE in the Slovak Republic according to this study is currently 1:41 280 and the incidence 1:1 360 000 (total population: 5 449 270 according to the 2021 population census). The prevalence is higher in comparison to data from other European countries: Sweden – 1:66 000,50 Italy – 1:65 000,51 Denmark – 1:70 900,20 Greece −1:90 000,52 Spain – 1:91 700,53 the Czech Republic – 1:52 307,6 and higher than average calculated prevalence 1:50 000.2
Two patients from our cohort died due to complications of HAE (laryngeal oedema). Both patients had low compliance with care and clinical management. In consulting family history with the patients from our cohort, we found 15 family members experiencing asphyxia leading to death before the clinical diagnosis of HAE. These patients had clinical manifestations of HAE (tissue swelling, oedema). We consider these events as complications of HAE (laryngeal oedema).
Variation type distribution in our cohort is different compared to neighbouring countries.6,7 We found a higher proportion of frameshift and inframe variants. Proportion of missense variants and gross deletions is similar to distribution according to the LOVD database.5 We found a lower proportion of splicing variants (7.7%). We assume that examination with RNA-based approach could help with identifying a causal splicing variant in a part of molecular-undiagnosed cases (16 HAE patients) from our cohort.
A large HAE family with exon 3 deletion is another interesting case to discuss. Exon 3 is involved in the formation of all 3 SERPING1 domains (Fig. 3). SERPING1 is a naturally alternatively spliced gene. Exon 3 skipping in hepatic cells and monocytes of healthy humans was already reported in the literature with presence in approximately one-third of all transcripts.45,54 The proportion of full-length and exon 3 skipped splicing isoforms has probably impact on the overall C1–INH level.54 We suppose that this variant identified in our cohort causes the dysregulation of the splicing isoforms balance which leads to the decrease of full-length protein level and the development of severe course of the disease.
Variants that cause premature introduction of a stop codon or NMD and gross deletions are assumed pathogenic and causal for developing HAE. In our cohort, these variants were present in 53.8% of cases. Determination of causality in missense variants is more problematic. The position of the variant is an important criterion. Modification of the peptide sequence within the serpin domain has a great impact on C1–INH dysfunction.5 Interestingly, 3 out of 4 missense variants causing C1–INH-HAE-1 detected in our cohort were novel. All our novel missense variants are rated as likely pathogenic according to ACMG criteria considering family history and segregation analysis.
A similar situation of problematic causality determination is characteristic for inframe variants. Our cohort has higher percentage of inframe variants (11,5%) comparing to other studies.5, 6, 7 Interpretation is challenging due to limitations of in-silico prediction tools and evaluation of impact on protein structure. All our novel inframe variants are rated as likely pathogenic according to ACMG criteria considering family history and segregation analysis.
Conclusions
Mutational heterogeneity of SERPING1 gene with high proportion of de novo variants was observed in many countries as well as in the Slovak Republic. Twenty-two unique causal variants including 12 previously undescribed expands the knowledge of the C1–INH-HAE genetics. Novel variants were present in the half of the index cases. A higher percentage of inframe variants comparing to other studies was observed. Three out of 4 missense variants causing C1–INH-HAE-1 detected in our cohort were novel. We report as first the heterozygous deletion of exon 3 in a large C1–INH-HAE-1 family with severe disease course which probably causes the dysregulation of the splicing isoforms balance and leads to the decrease of full-length C1–INH level. The identification of 12 previously unreported variants in SERPING1 gene could contribute to the current genetic databases and enlarge the understanding of the genetic background of C1–INH-HAE and help in the diagnostic approach in the patients with suspected HAE.
Abbreviations
ACMG, American College of Medical Genetics and Genomics; ANGPT1, angiopoietin-1 gene; CADD, Combined Annotation Dependent Depletion; C1–INH, C1 inhibitor; C1–INH-HAE-1, Hereditary angioedema caused by deficiency of C1 inhibitor; C1–INH-HAE-2, Hereditary angioedema caused by dysfunction of C1 inhibitor; EDTA, ethylenediaminetetraacetic acid; F12, factor XII gene; HAE, Hereditary angioedema; HGVS, Human Genomic Variation Society; HS3ST6, heparan sulfate-glucosamine 3-O-sulfotransferase 6 gene; KNG, kininogen gene; LOVD, Leiden open variation database; MLPA, multiplex ligand-dependent probe amplification; MPS, massive parallel sequencing; MYOF, myoferlin gene; NMD, nonsense-mediated decay; PCR, polymerase chain reaction; PLG, plasminogen gene; PM, moderate pathogenic criterion; PP, supporting pathogenic criterion; PVS, very strong pathogenic criterion; VUS, variant of uncertain significance
Acknowledgements
We are grateful to all general practitioners, dermatologists that have reported patients with susception on Hereditary angioedema; immunologists that have helped with data acquisition; laboratory technicians that have prepared and analyzed samples from patients.
Funding
This publication has been produced with the support of the Integrated Infrastructure Operational Program for the project: Systemic Public Research Infrastructure - Biobank for Cancer and Rare Diseases, ITMS: 313011AFG5, co-financed by the European Regional Development Fund.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author, MJ, upon reasonable request.
Authors’ contributions
AM, KH, MH, MJ designed study. MJ, TF, HG, LD, LVS, RLB were responsible for data acquisition. AM, MO, PB, KV, MJ performed statistical data analysis. AM, KV, MJ wrote the manuscript. KH, MH, PB edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Ethics statement
This is an observational study. Therefore, no ethical approval was required. Informed written consent to mutational analysis from all patients was archived. All participants gave their informed written consent for molecular genetic analysis of their samples. In addition, they provided written consent to collect and analyze their data.
Author’s consent
All authors declare that there is no conflict of interest regarding the publication of this paper. I, corresponding author on behalf of all contributing authors, hereby declare that the information given in this disclosure is true and complete to the best of my knowledge and belief.
Declaration of competing interest
AM has received honoraria for lectures (Takeda). KH has received consulting fees (Takeda). MH has received honoraria for lectures (ALK, Pleuran, Stallergenes, Takeda). TF has received speaker honoraria (Takeda). MJ has received consulting fees (Takeda, Glenmark, Zentiva, Pharming); honoraria for lectures, presentations (Takeda, Zentiva, Pharming, CSL Behring); support for attending meetings and/or travel (Takeda, Zentiva); served as principal investigator in clinical trials (Pharming, Takeda, BioCryst, Kalvista) and honoraria for participation on Advisory Boards (CSL Behring, Takeda, BioCryst, Kalvista, Pharming). HG, LD, LSV, RLB, MO, PB, KV have no conflict of interests to declare.
Footnotes
Full list of author information is available at the end of the article
Contributor Information
Karolina Vorcakova, Email: karolina.vorcakova@uniba.sk.
Milos Jesenak, Email: milos.jesenak@uniba.sk.
References
- 1.Busse P.J., Christiansen S.C. Hereditary angioedema. N Engl J Med. 2020;382:1136–1148. doi: 10.1056/NEJMra1808012. [DOI] [PubMed] [Google Scholar]
- 2.Lumry W.R., Settipane R.A. Hereditary angioedema: epidemiology and burden of disease. Allergy Asthma Proc. 2020;41(Suppl 1):S08–S13. doi: 10.2500/aap.2020.41.200050. [DOI] [PubMed] [Google Scholar]
- 3.Law R.H., Zhang Q., McGowan S., et al. An overview of the serpin superfamily. Genome Biol. 2006;7(5):216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X., Lei S., Xu Y., Liu S., Zhi Y. Mutation update of SERPING1 related to hereditary angioedema in the Chinese population. Hereditas. 2022;159(1):28. doi: 10.1186/s41065-022-00242-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drouet C.H., López-Lera A., Ghannam A., et al. SERPING1 variants and C1-INH biological function: a close relationship with C1-INH-HAE. Front Allergy. 2022;3:835503. doi: 10.3389/falgy.2022.835503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grombirikova H., Bily V., Soucek P., et al. Systematic approach revealed SERPING1 splicing-affecting variants to be highly represented in the Czech national HAE cohort. J Clin Immunol. 2023;43(8):1974–1991. doi: 10.1007/s10875-023-01565-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szabó E., Csuka D., Andrási N., Varga L., Farkas H., Szilágyi Á. Overview of SERPING1 variations identified in Hungarian patients with Hereditary angioedema. Front Allergy. 2022;3:836465. doi: 10.3389/falgy.2022.836465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maurer M., Magerl M., Betschel S., et al. The international WAO/EAACI guideline for the management of hereditary angioedema-The 2021 revision and update. Allergy. 2022;77(7):1961–1990. doi: 10.1111/all.15214. [DOI] [PubMed] [Google Scholar]
- 9.Dewald G., Bork K. Missense mutations in the coagulation factor XII (Hageman factor) gene in hereditary angioedema with normal C1 inhibitor. Biochem Biophys Res Commun. 2006;343(4):1286–1289. doi: 10.1016/j.bbrc.2006.03.092. [DOI] [PubMed] [Google Scholar]
- 10.Bafunno V., Firinu D., D'Apolito M., et al. Mutation of the angiopoietin-1 gene (ANGPT1) associates with a new type of hereditary angioedema. J Allergy Clin Immunol. 2018;141(3):1009–1017. doi: 10.1016/j.jaci.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Bork K., Wulf K., Steinmuller-Magin L., et al. Hereditary angioedema with a mutation in the plasminogen gene. Allergy. 2018;73(2):442–450. doi: 10.1111/all.13270. [DOI] [PubMed] [Google Scholar]
- 12.Bork K., Wulf K., Rossmann H., et al. Hereditary angioedema cosegregating with a novel kininogen 1 gene mutation changing the N-terminal cleavage site of bradykinin. Allergy. 2019;74(12):2479–2481. doi: 10.1111/all.13869. [DOI] [PubMed] [Google Scholar]
- 13.Ariano A., D'Apolito M., Bova M., et al. A myoferlin gain-of-function variant associates with a new type of hereditary angioedema. Allergy. 2020;75(11):2989–2992. doi: 10.1111/all.14454. [DOI] [PubMed] [Google Scholar]
- 14.Bork K., Wulf K., Mohl B.S., et al. Novel hereditary angioedema linked with a heparan sulfate 3-O-sulfotransferase 6 gene mutation. J Allergy Clin Immunol. 2021;148(4):1041–1048. doi: 10.1016/j.jaci.2021.01.011. [DOI] [PubMed] [Google Scholar]
- 15.den Dunnen J.T., Dalgleish R., Maglott D.R., et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37(6):564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 16.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and Genomics and the association for molecular pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopanos C., Tsiolkas V., Kouris A., et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35(11):1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q., Wang K. InterVar: clinical interpretation of genetic variants by ACMG-AMP 2015 guideline. Am J Hum Genet. 2017;100:267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.UniProt Consortium UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023;51:D523–D531. doi: 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bygum A., Fagerberg C.R., Ponard D., Monnier N., Lunardi J., Drouet C. Mutational spectrum and phenotypes in Danish families with hereditary angioedema because of C1 inhibitor deficiency. Allergy. 2011;66(1):76–84. doi: 10.1111/j.1398-9995.2010.02456.x. [DOI] [PubMed] [Google Scholar]
- 21.Mendoza-Alvarez A., Tosco-Herrera E., Muñoz-Barrera A., et al. A catalog of the genetic causes of hereditary angioedema in the Canary Islands (Spain) Front Immunol. 2022;13:997148. doi: 10.3389/fimmu.2022.997148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pappalardo E., Caccia S., Suffritti C., Tordai A., Zingale L.C., Cicardi M. Mutation screening of C1 inhibitor gene in 108 unrelated families with Hereditary angioedema: functional and structural correlates. Mol Immunol. 2008;45(13):3536–3544. doi: 10.1016/j.molimm.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Veronez C.L., Aabom A., Martin R.P., et al. Genetic variation of kallikrein-kinin system and related genes in patients with Hereditary angioedema. Front Med. 2019;6:28. doi: 10.3389/fmed.2019.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y.Y., Zhi Y.X., Yin J., et al. Mutational spectrum and geno-phenotype correlation in Chinese families with hereditary angioedema. Allergy. 2012;67(11):1430–1436. doi: 10.1111/all.12024. [DOI] [PubMed] [Google Scholar]
- 25.Gábos G., Moldovan D., Dobru D., et al. Mutational spectrum and genotype-phenotype relationships in a cohort of Romanian hereditary angioedema patients caused by C1 inhibitor deficiency. Rev Rom Med Lab. 2019;27(3):255–267. [Google Scholar]
- 26.Loules G., Zamanakou M., Parsopoulou F., et al. Targeted next-generation sequencing for the molecular diagnosis of hereditary angioedema due to C1-inhibitor deficiency. Gene. 2018;667:76–82. doi: 10.1016/j.gene.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 27.Suffritti C., Zanichelli A., Maggioni L., Bonanni E., Cugno M., Cicardi M. High-molecular-weight kininogen cleavage correlates with disease states in the bradykinin-mediated angioedema due to Hereditary C1-inhibitor deficiency. Clin Exp Allergy. 2014;44(12):1503–1514. doi: 10.1111/cea.12293. [DOI] [PubMed] [Google Scholar]
- 28.Gösswein T., Kocot A., Emmert G., et al. Mutational spectrum of the C1INH (SERPING1) gene in patients with hereditary angioedema. Cytogenet Genome Res. 2008;121(3-4):181–188. doi: 10.1159/000138883. [DOI] [PubMed] [Google Scholar]
- 29.Rijavec M., Korošec P., Šilar M., Zidarn M., Miljković J., Košnik M. Hereditary angioedema nationwide study in Slovenia reveals four novel mutations in SERPING1 gene. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maia L.S.M., Moreno A.S., Ferriani M.P.L., et al. Genotype-phenotype correlations in Brazilian patients with hereditary angioedema due to C1 inhibitor deficiency. Allergy. 2019;74(5):1013–1016. doi: 10.1111/all.13699. [DOI] [PubMed] [Google Scholar]
- 31.Veronez C.L., Mendes A.R., Leite C.S., Gomes C.P., Grumach A.S., Pesquero J.B. The panorama of Primary angioedema in the Brazilian population. J Allergy Clin Immunol Pract. 2021;9(6):2293–2304. doi: 10.1016/j.jaip.2020.11.039. e5. [DOI] [PubMed] [Google Scholar]
- 32.Kanepa A., Nartisa I., Rots D., Gailite L., Farkas H., Kurjane N. National survey on clinical and genetic characteristics of patients with hereditary angioedema in Latvia. Allergy Asthma Clin Immunol. 2023;19(1):28. doi: 10.1186/s13223-023-00783-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bissler J.J., Aulak K.S., Donaldson V.H., et al. Molecular defects in Hereditary angioneurotic edema. Proc Assoc Am Physicians. 1997;109(2):164–173. [PubMed] [Google Scholar]
- 34.Blanch A., Roche O., López-Granados E., Fontán G., López-Trascasa M. Detection of C1 inhibitor (SERPING1/C1NH) mutations in exon 8 in patients with Hereditary angioedema: evidence for 10 novel mutations. Hum Mutat. 2002;20(5):405–406. doi: 10.1002/humu.9073. [DOI] [PubMed] [Google Scholar]
- 35.Frangi D., Aulak K.S., Cicardi M., Harrison R.A., Davis A.E., 3rd A dysfunctional C1 inhibitor protein with a new reactive center mutation (Arg-444-->Leu) FEBS Lett. 1992;301(1):34–36. doi: 10.1016/0014-5793(92)80204-t. [DOI] [PubMed] [Google Scholar]
- 36.Hashimura C., Kiyohara C., Fukushi J.I., et al. Clinical and genetic features of Hereditary angioedema with and without C1-inhibitor (C1-INH) deficiency in Japan. Allergy. 2021;76(11):3529–3534. doi: 10.1111/all.15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pappalardo E., Cicardi M., Duponchel C., et al. Frequent de novo mutations and exon deletions in the C1 inhibitor gene of patients with angioedema. J Allergy Clin Immunol. 2000;106(6):1147–1154. doi: 10.1067/mai.2000.110471. [DOI] [PubMed] [Google Scholar]
- 38.Ponard D., Gaboriaud C., Charignon D., et al. SERPING1 mutation update: mutation spectrum and C1 inhibitor phenotypes. Hum Mutat. 2020;41(1):38–57. doi: 10.1002/humu.23917. [DOI] [PubMed] [Google Scholar]
- 39.Guryanova I., Suffritti C., Parolin D., et al. Hereditary angioedema due to C1 inhibitor deficiency in Belarus: epidemiology, access to diagnosis and seven novel mutations in SERPING1 gene. Clin Mol Allergy. 2021;19(1):3. doi: 10.1186/s12948-021-00141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuraw B.L., Herschbach J. Detection of C1 inhibitor mutations in patients with hereditary angioedema. J Allergy Clin Immunol. 2000;105(3):541–546. doi: 10.1067/mai.2000.104780. [DOI] [PubMed] [Google Scholar]
- 41.Förster T.M., Magerl M., Maurer M., et al. HAE patient self-sampling for biomarker establishment. Orphanet J Rare Dis. 2021;16(1):399. doi: 10.1186/s13023-021-02021-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto T., Horiuchi T., Miyahara H., et al. Hereditary angioedema in Japan: genetic analysis of 13 unrelated cases. Am J Med Sci. 2012;343(3):210–214. doi: 10.1097/MAJ.0b013e31822bdb65. [DOI] [PubMed] [Google Scholar]
- 43.Duponchel C., Di Rocco C., Cicardi M., Tosi M. Rapid detection by fluorescent multiplex PCR of exon deletions and duplications in the C1 inhibitor gene of Hereditary angioedema patients. Hum Mutat. 2001;17(1):61–70. doi: 10.1002/1098-1004(2001)17:1<61::AID-HUMU7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 44.Bafunno V., Bova M., Loffredo S., et al. Mutational spectrum of the C1 inhibitor gene in a cohort of Italian patients with Hereditary angioedema: description of nine novel mutations. Ann Hum Genet. 2014;78(2):73–82. doi: 10.1111/ahg.12052. [DOI] [PubMed] [Google Scholar]
- 45.Duponchel C., Djenouhat K., Frémeaux-Bacchi V., Monnier N., Drouet C., Tosi M. Functional analysis of splicing mutations and of an exon 2 polymorphic variant of SERPING1/C1NH. Hum Mutat. 2006;27(3):295–296. doi: 10.1002/humu.9414. [DOI] [PubMed] [Google Scholar]
- 46.Roche O., Blanch A., Duponchel C., Fontán G., Tosi M., López-Trascasa M. Hereditary angioedema: the mutation spectrum of SERPING1/C1NH in a large Spanish cohort. Hum Mutat. 2005;26(2):135–144. doi: 10.1002/humu.20197. [DOI] [PubMed] [Google Scholar]
- 47.Siddique Z., McPhaden A.R., Lappin D.F., Whaley K. An RNA splice site mutation in the C1-inhibitor gene causes type I hereditary angio-oedema. Hum Genet. 1991;88(2):231–232. doi: 10.1007/BF00206079. [DOI] [PubMed] [Google Scholar]
- 48.Speletas M., Szilagyi A., Psarros F., et al. Hereditary angioedema: molecular and clinical differences among European populations. J Allergy Clin Immunol. 2015;135(2):570–573. doi: 10.1016/j.jaci.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Rasmussen E.R., Aanæs K., Jakobsen M.A., Bygum A. Acquired complement C1 esterase inhibitor deficiency in a patient with a rare SERPING1 variant with unknown significance. BMJ Case Rep. 2019;12(9) doi: 10.1136/bcr-2019-231122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nordenfelt P., Dawson S., Wahlgren C.F., Lindfors A., Mallbris L., Bjorkander J. Quantifying the burden of disease and perceived health state in patients with hereditary angioedema in Sweden. Allergy Asthma Proc. 2014;35:185–190. doi: 10.2500/aap.2014.35.3738. [DOI] [PubMed] [Google Scholar]
- 51.Zanichelli A., Arcoleo F., Barca M.P., et al. A nationwide survey of hereditary angioedema due to C1 inhibitor deficiency in Italy. Orphanet J Rare Dis. 2015;10:11. doi: 10.1186/s13023-015-0233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Psarros F., Koutsostathis N., Farmaki E., Speletas M.G., Germenis A.E. Hereditary angioedema in Greece: the first results of the Greek hereditary angioedema registry. Int Arch Allergy Immunol. 2014;164:326–332. doi: 10.1159/000366276. [DOI] [PubMed] [Google Scholar]
- 53.Roche O., Blanch A., Caballero T., Sastre N., Callejo D., Lopez-Trascasa M. Hereditary angioedema due to C1 inhibitor deficiency: patient registry and approach to the prevalence in Spain. Ann Allergy Asthma Immunol. 2005;94:498–503. doi: 10.1016/S1081-1206(10)61121-0. [DOI] [PubMed] [Google Scholar]
- 54.Grymová T., Grodecká L., Souček P., Freiberger T. SERPING1 exon 3 splicing variants using alternative acceptor splice sites. Mol Immunol. 2019;107:91–96. doi: 10.1016/j.molimm.2019.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, MJ, upon reasonable request.