Abstract
The use of retroviral vectors for gene transfer into animals has been severely hampered by the lack of provirus transcription in the early embryo and embryonic stem (ES) cells. This primary block in provirus expression is maintained in differentiated cells by a cis-acting mechanism that is not well characterized. Retroviral vectors based on the murine embryonal stem cell virus (MESV), which overcome the transcriptional block in ES cells, were constructed to investigate this secondary mechanism. These vectors transferred G418 resistance to ES cells with the same efficiency as to fibroblasts, but overall transcript levels were greatly reduced. A mosaic but stable expression pattern was observed when single cells from G418-resistant clones were replated in G418 or assayed for expression of LacZ or interleukin-3. The expression levels in independent clones were variable and correlated inversely with methylation. However, a second, more pronounced, block to transcription was found upon differentiation induction. Differentiation of the infected ES cells to cells permissive for retroviral expression resulted in repression and complete extinction of provirus expression. Extinction was not accompanied by increased levels of methylation. Provirus expression is thus regulated by two independent cis-acting mechanisms: (i) partial repression in the undifferentiated state, accompanied by increased methylation but compatible with long-term, low expression of retroviral genes, and (ii) total repression and extinction during early stages of differentiation, apparently independent of changes in methylation. These results indicate a time window early during the transition from an undifferentiated to a differentiated stage in which provirus expression is silenced. The mechanisms are presently unknown, but elucidation of these events will have an important impact on vector development for targeting stem cells and for gene therapy.
Due to their high efficiency of transfer into a wide range of cell types, their precise integration into the host genome, and their stable expression compared to transfected DNA, retroviral vectors have proven invaluable in studies aimed at understanding gene function and control in normal development and oncogenesis. In these studies, retroviruses have been implemented in three important ways: (i) as vectors to introduce and express different genes into several cell types (13, 34, 58), (ii) as markers to trace differentiation lineages (10, 33, 60), and (iii) as insertional mutagens to mutate and tag genes associated with specific phenotypes (38, 55, 64). The extended use of retroviruses in these types of studies has been significantly hampered by a block to permissive infection in totipotent embryonic carcinoma (EC) and embryonic stem (ES) cells and in the early embryo (18, 30, 51).
Two blocks to permissive provirus expression in ES cells and their differentiated derivatives have been described. The first block is at the level of transcription; this is apparent immediately after infection and is attributable to trans-acting factors (22, 29, 36, 61, 65). In contrast, the second restriction is poorly characterized, occurs at an unknown time point, and acts in cis to maintain the initial block in transcription. The second block was identified in early studies which showed that differentiation induction or cell fusion relieved the first block to virus infection, as demonstrated by de novo infection, but did not permit expression of provirus that had integrated prior to differentiation (4, 19, 21, 43).
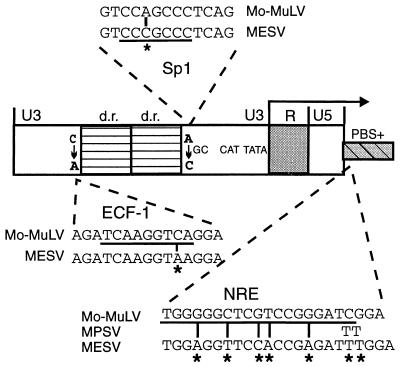
Through the analysis of retroviral mutants, we have developed a retrovirus that overcomes the block to retroviral transcription in ES cells (17, 22, 26, 56) and thus have been able to define some of the mechanisms responsible for the primary block to expression. This virus, the murine embryonal stem cell virus (MESV), differs from the prototype Moloney murine leukemia virus (Mo-MuLV), whose expression is restricted in EC and ES cells, in three important ways (Fig. 1): (i) the presence of a high-affinity binding site for the Sp1 transcription factor (23, 49); (ii) the disruption of the binding site for the embryonic long terminal repeat (LTR)-binding protein, also known as EC cell factor I, which acts as a transcriptional repressor (1, 67, 68); and (iii) most importantly, the elimination of a negative regulatory element (NRE) coincident with the proline tRNA primer-binding site (PBS) of Mo-MuLV (5, 22, 36, 70). The NRE-binding factor (factor A) is a potent repressor of LTR-mediated transcription in ES cells (9, 47). Importantly, activating mutations in the LTR are not sufficient to overcome the repressor activity of the NRE (12, 23); thus, only retroviruses such as MESV with mutations both in the LTR and the NRE are transcriptionally active in ES cells.
FIG. 1.
Schematic diagram of the mutations in MESV LTR and downstream sequences that allow efficient expression in ES cells. Within the LTR sequences, MESV is identical to MPSV, except for the loss of the second direct repeat (d.r.) in MESV. Only the critical point mutations between Mo-MuLV and MESV are shown.
Although the mechanisms which govern the first block in expression of retroviral vectors in ES cells are thus well understood, very little is known regarding the cis-acting mechanism which maintains the block in expression in a permissive cell environment. The importance of methylation in reinforcing the expression block has long been discussed, due to a high correlation between degree of methylation and expression of the provirus (9, 24, 32). However, although several studies have shown that abolition of methylation by treatment with the methyltransferase inhibitor 5-azacytidine reactivates provirus expression in a permissive state, these studies are not conclusive, as the actual extent of virus activation could not be measured due to virus spread (19, 31, 63). Another uncertainty has been the time point at which this second maintenance block occurs. The ability to productively infect early differentiated cultures of EC and ES cells defined a relatively early stage (<7 to 10 days postdifferentiation) where both blocks are relieved (4, 56). Because methylation occurs at approximately 8 days postinfection in the undifferentiated stage, that is, in a delayed response to the first block, it has been suggested that this event is coincident with the second block (19, 43). Alternatively, one cannot exclude the possibility that the second block does not occur in the undifferentiated state but occurs only after differentiation has been induced. Along the same line, it is possible that independent blocks may occur at both time points. Importantly, without retroviral vectors that are permissive for transcription in undifferentiated ES cells, neither the importance of methylation nor the events leading to extinction of provirus expression in the differentiated stage can be studied.
In the study presented here, we developed MESV-based retroviral vectors that are able to efficiently transfer and express genes in undifferentiated ES cells. By using various marker genes, expression levels in individual cells before and after differentiation were monitored. Experiments were designed to test (i) whether LTR-mediated transcription in ES cells is subjected to extinction by secondary events, (ii) the role of methylation in provirus repression, and (iii) the influence of differentiation induction both in vitro and in vivo on provirus expression.
(Part of this work was completed as part of the doctoral thesis of A.S. from the Fachbereich Chemie, Universität Hamburg.)
MATERIALS AND METHODS
Cell lines and virus preparation and infection.
Three different ES cell lines derived from mouse strain 129 were used in the experiments described here: CCE (51), W9.5 (C. Stewart, Frederick, Md.), and R1 (40). Cell lines were normally maintained on embryonic fibroblasts as a feeder. G418-resistant feeders were derived from transgenic mice expressing the Tn5 neomycin resistance (Neor) gene. To facilitate experimental procedures, cells were removed from the feeder and the medium was supplemented with 1,000 U of leukemia inhibitory factor (LIF) per ml to maintain the undifferentiated phenotype. All ES cell lines were maintained in Dulbecco modified Eagle medium with 15% fetal calf serum (FCS), 1 mM glutamine, 1 mM sodium pyruvate, 1% nonessential amino acids, and 150 μM α-monothioglycerol.
The packaging cell lines GP+E86 and GP-envAm12 (39) were used to generate infectious virus particles containing the MESV-based vectors. Recombinant DNA constructs were transfected into GP+E86 cell lines by electroporation (6), and 24-h supernatants were used to infect the amphotropic producer GP+envAm12 cells. The presence of intact provirus was ascertained by Southern blot analysis.
Construction of MESV-based retroviral vectors.
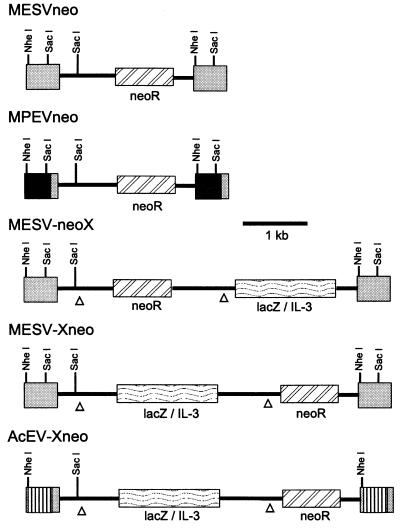
The MESV plasmid p5Gneo (22) was used as the basis for all vectors in this study. MESVneo (p5O-neo; R228) is a minimal MESV vector in which all sequences from the NarI site in gag to the ClaI site in env of MESV were deleted, leaving basically only LTR and packaging sequences from MESV. The entire wild-type Tn5 coding region of the Neor gene, in which the AUG has been altered for higher expression in eukaryotes, was inserted at the NarI-ClaI junction site. MPEVneo (p5O-Mneo; R229) is identical to MESVneo except that the 3′ LTR was replaced with that of myeloproliferative sarcoma virus (MPSV) (66) at the NheI site. One round of replication results in an MESV vector with MPSV sequences in the U3 region of the LTR. Due to our previous success in using splicing vectors for expressing two genes, thus eliminating the problem of promoter interference, the 3′ splicing signals of MPSV (34) were inserted into the NarI-ClaI junction of MESVneo, in which a polylinker had been inserted. The Neor gene was inserted either downstream or upstream of these signals to generate MESV-Xneo (p5O-Xneo) and MESV-neoX (p5O-neoX), respectively, which express the Neor gene from a spliced or full-length message, respectively. The murine interleukin-3 (IL-3) or the lacZ cDNA was introduced into p5O-Xneo or p5O-neoX. As splicing is relatively inefficient, the second gene is expressed at significantly reduced levels; however, these levels are sufficient for obtaining biological activity and are sometimes better suited to evaluate the normal function of a gene. To replace the β-actin promoter-enhancer with that of the MESV LTR, a 277-bp XhoI-HinfI fragment of pP1-CAT (37) containing sequences +1 to −277 of the β-actin gene (50) was inserted in the Sau3A-BssHII site of the 3′ LTR of MESV-Xneo.
Virus titration and monitoring of expression levels.
Supernatants containing pseudotyped retroviral vectors were harvested from amphotropic packaging cell lines, and titers were determined on either ES or NIH 3T3 cells plated in 24-well plates. As ES cells show spreading, titers were determined by end point dilution of virus (fivefold serial dilutions). All infections were performed in triplicate. At 24 h after infection, G418 (400 μg/ml) selection was applied, and G418-resistant colonies were counted at 10 to 14 days postinfection and expressed as Neor CFU.
Expression levels in infected cells were determined by IL-3 activity in supernatants of cells with vectors carrying both the IL-3 and Neor cDNAs. After G418 selection, mass cultures or clonal cell lines were seeded at a density of 5 × 104/ml. A 24-h cell-free supernatant was collected from confluent cultures and tested for biological activity by titration (serial threefold dilutions) on the IL-3-dependent 32D target cells (200 cells/well) in Terasaki plates. Cell numbers were determined at various time points. One unit was empirically set as the amount required for half-maximal stimulation of the target cells. This assay system has a high sensitivity and allows detection of small amounts of activity (0.1 U/ml). In differentiation assays of ES cells, cell counts were determined immediately after the supernatant was harvested for IL-3 assays, and thus IL3 activity is expressed as units per milliliter per 106 producing cells.
Virus expression in individual cells was determined by staining for β-galactosidase activity in vectors expressing both the Escherichia coli lacZ gene and Neor. Cells plated at different concentrations (102 to 104 cells/plate) were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (6). Cells were incubated in the staining solution for 14 to 20 h before enumeration of the fraction of LacZ-positive cells. With the use of a light microscope, a minimum of 200 colonies were scored for all cell lines tested.
In vitro and in vivo differentiation of ES cells.
In vitro differentiation was induced by removal of feeder in the absence of LIF and plating on gelatinized plates in Dulbecco modified Eagle medium supplemented with 20% FCS, 1 mM glutamine, 1 mM sodium pyruvate, 1% nonessential amino acids, and 450 μM α-monothioglycerol. Medium changes were performed every day.
The tetraploid aggregation technique (41) was used to obtain ES cell-derived hematopoietic cells. Briefly, primary embryos from CD1 outbred mice were collected at day 1.5 postcoitus as two-cell-stage embryos and electrofused (nonelectrolyte conditions, 93 V, 30 μs, 0.6 to 0.8 V AC, CF 100 pulser [BLS, Budapest, Hungary]). On the following day, two tetraploid embryos were aggregated with 10 to 15 vector-carrying R1 ES cells by the sandwich technique and cultured overnight. Day 3.5 blastocysts were transferred to day 2.5 pseudopregnant foster mothers. At day 15.5 of gestation, the foster mothers were sacrificed and the fetuses were analyzed. Cell suspensions from fetal livers were prepared and split into appropriate aliquots for glucose phosphate isomerase (GPI) analysis, DNA and RNA extraction, and cell culture. A GPI assay was performed as described elsewhere (41) to confirm ES origin. R1 ES cells are of GPI genotype AA, whereas CD1 embryos were obtained from matings of GPI AB females with GPI BB males, producing either GPI AB or BB genotypes. Freeze-thawed cell lysates from all specimens were prepared in sample buffer and loaded on a cellulose acetate isoelectric focusing gel. Controls were wild-type GPI AB or BB CD1 embryos.
Culture of fetal liver cells.
Aliquots of fetal liver cell suspensions were used to initiate mass cultures (Iscove modified Eagle medium, 10% FCS plus 2 mM glutamine, with or without 1% BPV conditioned medium, as a source of murine IL-3) or single-cell assays. Colony assays of fetal liver cells were performed by the original procedure of Iscove and Sieber (28). Briefly, cells were seeded at 1.25 × 106 cells/ml/dish in 0.8% Methocel (Fluka) in Iscove modified Eagle medium supplemented with 10% FCS, 2 mM glutamine, iron-saturated transferrin (0.23 mg/ml), and 10−4 M α-monothioglycerol. BPV conditioned medium was used as a source of murine IL-3 (10 U/ml; 1 U was standardized as half-maximum stimulation of 104 32D cells). Human recombinant erythropoietin (Boehringer Mannheim) was used at a final concentration of 5 U/ml/dish.
RESULTS
MESV-based vectors have a high efficiency of transfer into ES cells.
A series of recombinant retroviral vectors based on the previously described MESV (22) were constructed and are depicted in Fig. 2. These vectors incorporated all three regions (described above) that are necessary to overcome the retroviral transcription block in ES cells. In some constructs, the MESV LTR was replaced with that of MPSV. As the MESV LTR originated from an MPSV mutant with high expression levels in PCC4 EC cells (17, 26), it was of interest to determine if the loss of one direct repeat also improved expression in ES cells. The bacterial lacZ gene, encoding β-galactosidase (β-Gal), and the murine IL-3 cDNA, encoding the growth factor IL-3, were chosen as secondary markers. They offer the advantages of detecting expression in individual cells and providing a very sensitive assay for measuring expression, respectively.
FIG. 2.
Schematic diagram of MESV-based vectors used in experiments. Depicted are the integrated proviruses after one round of replication. In MPEV and AcEV, the U3 region of the LTR is derived from MPSV (dark gray) or the actin promoter (striped). X denotes any cDNA, in this case that for either IL-3 or lacZ. Triangles mark positions of splicing signals. Details are presented in Materials and Methods.
In initial experiments, the relative efficiency of Neor transfer into ES cells compared to fibroblasts was determined with the new MESV vectors. As shown in Table 1, the ratio of Neor CFU per milliliter in infected ES cells to that in NIH 3T3 cells was greater than 1 when either MESVneo (MESV LTR) or MPEVneo (MPSV LTR) was used. This is in striking contrast to the case for a similar construct, MPSVneo, also driven by the MPSV LTR but with an MPSV PBS (34), which showed a greater than 4 orders of magnitude reduction in efficiency of transfer to ES cells. These results underline the importance of the NRE downstream of the 5′ LTR in regulating LTR-mediated transcription efficiency in ES cells. No significant difference between the MESV and MPSV LTRs was observed.
TABLE 1.
MESV and MPEV vectors transfer Neor as efficiently to ES cells as to fibroblasts
| Vector | LTR | PBS | cDNA insert
|
Neor CFU/mla
|
Neor ratio (ES cells/fibroblasts) | Avg Neor ratio | ||
|---|---|---|---|---|---|---|---|---|
| 5′ | 3′ | Fibroblastsb | ES cellsc | |||||
| MPSV-neoR control | MPSV | MPSV | Neor | 1.9 × 103 | 1 | 5.3 × 10−4 | 4.8 × 10−4 | |
| 2.3 × 103 | 1 | 4.3 × 10−4 | ||||||
| MPEV-neoR | MPSV | MESV | Neor | 4.0 × 103 | 4.5 × 103 | 1.12 | 1.05 | |
| 1.5 × 103 | 2.4 × 103 | 1.60 | ||||||
| 3.8 × 103 | 1.6 × 102 | 0.42 | ||||||
| MESV-neoR | MESV | MESV | Neor | 1.2 × 104 | 1.3 × 104 | 1.10 | 1.43 | |
| 4.2 × 106 | 5.5 × 106 | 1.30 | ||||||
| 1.9 × 103 | 3.7 × 103 | 1.90 | ||||||
| MPEV-IL3neoR | MPSV | MESV | IL-3 | Neor | 2.2 × 103 | 3.0 × 102 | 0.14 | 0.19 |
| 1.2 × 103 | 2.0 × 102 | 0.18 | ||||||
| 7.1 × 102 | 1.3 × 102 | 0.13 | ||||||
| 7.3 × 102 | 9.2 × 101 | 0.13 | ||||||
| 2.9 × 102 | 9.6 × 101 | 0.33 | ||||||
| MPEV-lacZneoR | MPSV | MESV | lacZ | Neor | 1.5 × 104 | 2.3 × 103 | 0.17 | 0.20 |
| 1.5 × 103 | 3.1 × 102 | 0.20 | ||||||
| 3.1 × 102 | 1.5 × 103 | 0.20 | ||||||
| 2.5 × 103 | 1.0 × 104 | 0.24 | ||||||
| AcEV-lacZneoR | β-Actin | MESV | lacZ | Neor | 1.6 × 103 | 1.7 × 102 | 0.11 | 0.66 |
| 6.2 × 103 | 7.5 × 103 | 1.20 | ||||||
Virus titers, as measured by transfer of Neor to the indicated cells, were determined as described in Materials and Methods.
NIH 3T3 fibroblasts.
CCE ES cells.
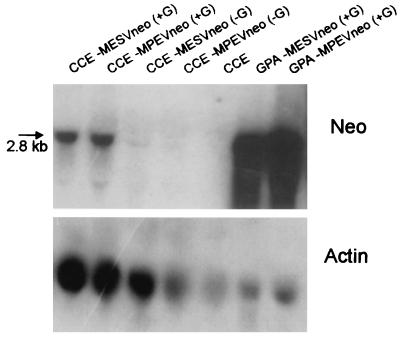
A fivefold reduction in transfer efficiency to ES cells was seen with retroviral constructs with the Neor gene expressed from a spliced mRNA (Table 1). Similar results were obtained whether the IL-3 or the lacZ gene was incorporated into the vector. Viral titers on fibroblasts of such constructs did not vary significantly from those with vectors in which the Neor gene was expressed from a genomic-length mRNA. These results suggest that the absolute levels of vector expression may vary between the two cell lines. As Neor transfer efficiency does not reflect absolute expression levels but rather reflects a nonlinear threshold, we used the MESV-IL3neoR vector to quantitate expression levels in fibroblasts versus ES cells. IL-3 expression levels in transduced fibroblasts and R1 ES cells were measured after G418 selection. Levels of biologically active IL-3 from ES cells expressing the high-expression vector MESV-IL3neoR were approximately 50-fold lower than those from fibroblasts transduced with the same vector (an average of 11 U/ml for six independent ES clones [range, 1.6 to 50 U/ml] versus 500 U/ml for fibroblasts). These results were consistent with an observed difference in RNA transcript levels of MESVneo in ES cells versus fibroblasts (Fig. 3) and thus are not caused by differences in translation efficiencies or protein stability.
FIG. 3.
MESV and MPSV expression levels are approximately 50-fold higher in fibroblasts than in ES cells. RNA was extracted from both infected and noninfected CCE ES cells and from infected GP-envAm12 fibroblasts (GPA). Total RNA (15 μg) was size separated by gel electrophoresis, transferred to a nylon membrane, and hybridized with either a Neor or a β-actin probe. Cells were infected with either MESVneo or MPEVneo, as indicated, with a multiplicity of infection of 2. Parallel cultures were either selected with G418 (+G) or not (−G).
In conclusion, both MESVneo and MPEVneo vectors can transduce G418 resistance to ES cells with the same efficiency as to fibroblasts. However, by using vectors carrying the IL-3 cDNA, it could be shown that the absolute expression levels of these vectors are 50-fold lower in ES cells than in fibroblasts.
MESV-based constructs in undifferentiated ES cells exhibit a stable but mosaic expression pattern that is integration site dependent.
The difference in expression levels of the integrated provirus in ES versus NIH 3T3 cells could be due to an overall reduced rate of transcription, as proposed to occur with a graded mode of enhancer action (35), or could be due to a mosaic expression pattern generated by a binary mode of enhancer action (69). In the latter mode, the stability of expression would depend on the ability of the integrated provirus to create and maintain a domain permissive for transcription. To determine which mechanism was responsible for the low levels of expression in ES cells, NIH 3T3 fibroblasts and W9.5 ES cells were infected with MESV-lacZneoR. Neor clones were isolated, checked for the integrity of the construct by Southern blot analysis (data not shown), and seeded onto Neor feeders at various concentrations (102 to 104 cells/plate). Colonies were stained for LacZ expression. Whereas microscopic inspection revealed a homogenous and 100% staining pattern in infected fibroblast cultures of six independent colonies, a heterogeneous pattern of β-Gal expression was observed in W9.5 cells. The percentage of β-Gal-positive colonies for six independent clones ranged from 5 to 80% (see Table 3; undifferentiated controls). Within a positive clone, the staining of individual cells ranged from 10 to 100% and the staining intensity varied from weakly positive to strong, roughly correlating with the incidence of positive cells (data not shown). Significantly, the distribution pattern of β-Gal-positive colonies for any individual clone, as well as their staining intensity, was stable in up to 10 independent experiments. Similar results were obtained in MESV-lacZneoR-infected CCE ES cells. Notably, the frequencies of extinction were comparable in both assays (Table 2). Thus, transcription repression occurred in a stochastic, discontinuous fashion. As each clone represents a unique single integration site, the provirus expression pattern must also be dependent on the integration site. Thus, whether measured by β-Gal expression or G418 resistance, a high frequency of extinction of LTR-mediated transcription was observed in infected ES cells.
TABLE 3.
Extinction of MPEV transcription during differentiation as assessed by LacZ expression
| ES cell line | Clone | % of LacZ+ colonies in un- differentiated controlsa | % of LacZ+ colonies in differentiated cultures relative to that in undif- ferentiated culturesb on day:
|
|||
|---|---|---|---|---|---|---|
| 3–4 | 5 | 6 | 7–8 | |||
| W9.5 | 0 | 0 | 0 | 0 | 0 | |
| W9.5-MPEVlacZneoR | 2 | 71 ± 14 | 97 ± 9 | 84 ± 29 | 64 ± 13 | 75 ± 27 |
| 4 | 47 ± 13 | 79 ± 53 | 66 ± 12 | 43 ± 12 | 41 ± 9 | |
| 5 | 22 ± 11 | 10 ± 5 | 7 ± 2 | 0 | 0 | |
| 1 | 18 ± 4 | 31 ± 16 | 45 ± 24 | 0 | 0 | |
| 3 | 15 ± 6 | 101 ± 59 | 44 ± 34 | 19 ± 7 | 48 ± 26 | |
| 6 | 8 ± 3 | 38 ± 27 | 0 | 0 | 0 | |
Cells were plated at different cell concentrations and analyzed for LacZ expression. The mean values (± standard deviations) from 9 or 10 independent experiments are presented.
The percentages of LacZ-positive cells at different time points in a differentiating culture were compared with those in parallel undifferentiated cultures. Mean values (± standard deviations) from three to five independent experiments are shown.
TABLE 2.
MPEV exhibits a mosaic expression pattern in ES cells
| ES cell line and clone | Cloning efficiency (%) (% LacZ+ colonies)a
|
Neor colonies/ total colonies | LacZ+ colonies/ total colonies
|
||
|---|---|---|---|---|---|
| Without G418 | With G418 | Without G418 | With G418 | ||
| CCE | 33.0 (0) | 0.0 (0) | NAb | NA | NA |
| CCE-MPEVlacZneoR | |||||
| 6 | 18.2 (5.2) | 6.5 (2.2) | 0.36 | 0.35 | 0.36 |
| 9 | 27.3 (2.4) | 5.1 (0.6) | 0.19 | 0.09 | 0.12 |
| 13 | 30.6 (8.5) | 14.6 (3.9) | 0.48 | 0.28 | 0.27 |
Cells were plated on inactivated feeder cells in either the absence or presence of G418. Results are averages from two independent experiments.
NA, not applicable.
In addition to determination of the number of β-Gal-positive Neor colonies, colonies plated without selection were also scored for β-Gal expression to determine if transcription repression was reversible. If irreversible, a culture held under G418 selection would constantly lose a subset of cells in which transcription was completely repressed. Cells held without selection would thus maintain a constantly increasing, nonexpressing population. Significantly, the percentages of colonies were similar whether determined from Neor colonies or unselected colonies (Table 2). These results suggest that total extinction has not occurred. To rule out that this result was not due to differences in the assay systems or to the time frame analyzed, parallel cultures of four independent clones of MPEV-lacZneoR were maintained with or without G418 selection over a period of 3 weeks. At 2- to 4-day intervals, cell aliquots were taken and cloning efficiency in the absence or presence of G418 was determined. Although cultures maintained without G418 gave rise to approximately 80% fewer and smaller colonies than parallel cultures held with G418, no change in the ratio of Neor colonies between the two cultures was observed with time for any of the four clones tested. These results show that MESV vector expression is low and is subject to stochastic repression but is stable. The low absolute expression levels, measured by both RNA and protein levels, in infected ES cultures compared to fibroblasts is thus probably in part due to a relatively high repression frequency. The variability of expression levels between clones indicates the importance of the proviral integration site in determining the frequency of extinction.
Expression levels correlate with methylation of the proviral genome.
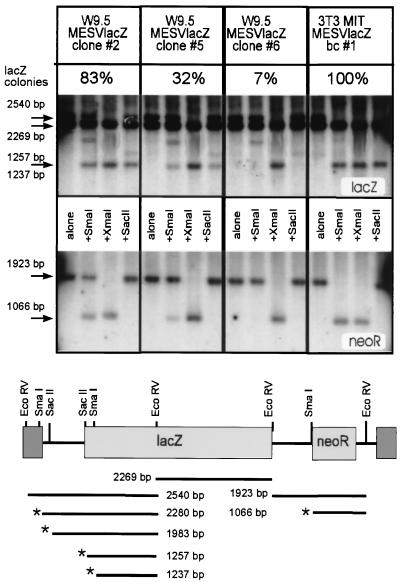
A high level of methylation has been correlated with inactivated provirus in EC and ES cells (19, 43, 63). To determine if MESV expression levels could be linked to methylation levels, we examined the methylation pattern of the provirus in infected ES cells. For this purpose, genomic DNA was isolated from three different clones of MESV-lacZneoR-infected W9.5 ES cells with either high (clone 2), intermediate (clone 5), or low (clone 6) LacZ activity. Parallel cultures were stained for β-Gal to ascertain the functional activity of the construct within the experiment. DNAs were subjected to double digestion with the restriction enzyme EcoRV and either the methylation-sensitive SacII or SmaI-enzyme. The methylation-insensitive SmaI isochizomer XmaI was used as a control. In combination with probes specific for the Neor and lacZ genes, this restriction analysis allows the detection of methylated sites in the provirus (Fig. 4). DNA of infected NIH 3T3 fibroblasts, displaying 100% LacZ activity in staining assays, was fully digested by the methylation-sensitive SmaI enzyme, resolving the 2.5-kb EcoRV band detectable with a lacZ probe to a 1.2-kb fragment (the second 2.2-kb EcoRV band does not contain a SmaI site) (Fig. 4, upper gels) or resolving the 1.9-kb Neor-specific EcoRV fragment to a 1.1-kb band (Fig. 4, lower gels). In contrast, DNAs from all three infected W9.5 ES clones showed partial to almost complete resistance to SmaI digestion, as evidenced by the persistence of the 2.5-kb band after SmaI digestion. DNA extracted from clone 2, with 83% β-Gal-positive colonies, showed only partial resistance to SmaI digestion, whereas those from clones 5 and 6, with 32 and 7% β-Gal-positive colonies, respectively, exhibited almost complete SmaI resistance (i.e., the degree of methylation was even more pronounced). All methylation sites tested throughout the retroviral genome were affected. Similar results were obtained with SacII and hybridization with the lacZ probe (Fig. 4). In the three clones examined, the degree of methylation and the levels of LacZ activity were inversely correlated. Even though no causal relationship can be inferred, it can still be concluded that the low rates of LTR-mediated transcription are accompanied by increased methylation.
FIG. 4.
MESV proviral DNA is highly methylated in ES but not fibroblast cultures. Genomic DNA was digested with EcoRV alone or together with SmaI, XmaI (control), or SacII and size separated by gel electrophoresis. After transfer to a nylon membrane, the DNA was hybridized with a lacZ probe and subsequently with a Neor probe, as indicated. The schematic diagram shows the fragments expected if digestion with either SmaI or SacII is not blocked or is only partially blocked by methylation. The percentage of LacZ-positive colonies determined at the time of DNA isolation for each clone is shown. bp, base pairs.
In vitro differentiation results in extinction of MESV vector expression.
ES cell differentiation is known to be accompanied by vast changes in gene expression due to establishment of tissue-specific gene expression patterns. It has previously been shown that despite these changes, Mo-MuLV provirus blocked by the cis-acting, secondary block in ES or EC cells remains transcriptionally inert after cell differentiation into a virus-permissive stage. To test if the MESV-based vectors are able to escape this irreversible block to extinction, infected ES cells were subjected to in vitro differentiation.
In a simple in vitro assay, six clones with different basal MESV LacZ expression levels were seeded at different cell densities (102 to 104 cells/plate) under either differentiating (gelatinized tissue culture plates without feeder or LIF) or, as a control, nondifferentiating (with feeders plus LIF) conditions. Parallel cultures were stained for β-Gal at 24-h intervals from day 3 to 8. As summarized in Table 3, provirus expression was repressed in all clones analyzed. Indeed, in half of the clones the retroviral vector underwent complete silencing during differentiation. Even in clones in which β-Gal activity was detected, the levels were reduced by up to 60% of that in the undifferentiated controls. With only one exception (clone 3), expression was completely lost in all clones with 25% or fewer β-Gal-positive cells. Thus, not only did differentiation induction fail to increase virus expression, in most cases complete extinction occurred.
To rule out the possibility that the lack of β-Gal detection was due to metabolic limitations, we applied a similar differentiation protocol to ES cells transduced with a MESV-IL3neoR construct. In parallel cultures, R1-MESV-IL3neoR ES cells were exposed to differentiation- and non-differentiation-inducing conditions. Conditioned media for confluent cultures were tested for IL-3 activity on 32D indicator cells and corrected for cell counts (units per milliliter per 106 cells). The results are shown in Table 4. Consistent with results obtained from lacZneoR ES cells, expression of the provirus decreased after in vitro differentiation in all three independent clones analyzed. IL-3 levels were decreased by 60 to 95% compared to those in undifferentiated controls, although in contrast to the lacZ analysis, activity was detected in all clones. This may reflect the highly sensitive method of detection.
TABLE 4.
Extinction of MPEV transcription during differentiation as assessed by IL-3 expression
| ES cell line | Clone | IL-3 activity (U/ml/106 cells) in undifferentiat- ed controlsa | % of IL-3 activity in differentiated cultures relative to that in undif- ferentiated culturesa on day:
|
|||
|---|---|---|---|---|---|---|
| 4 | 5 | 8 | 12 | |||
| R1 | 0 | 0 | 0 | 0 | 0 | |
| R1-MESVIL3neoR | 10 | 11.6 ± 4.0 | 68 ± 17 | 52 ± 10 | 42 ± 7 | 6 ± 2 |
| 6 | 1.9 ± 0.5 | 66 ± 9 | 47 ± 7 | 62 ± 20 | 38 ± 14 | |
| 4 | 2.9 ± 1.3 | 75 ± 21 | 44 ± 4 | 49 ± 23 | 19 ± 4 | |
IL-3 activity in conditioned medium from differentiated and undifferentiated cultures was measured by using 32D indicator cells. Results are mean values (± standard deviations) from two independent experiments.
Importantly, no upregulation of expression was observed in either the W9.5 or R1 ES cells expressing the lacZ or IL-3 vector, respectively, in a total of 15 independent integration sites analyzed. This is consistent with a cis-mediated block that prevents the upregulation of expression, even if the appropriate transcription factors are expressed or if the chromosome environment has become more permissive for expression during differentiation. The expression block observed in the differentiated cells probably occurred during early stages of differentiation induction, since expression in undifferentiated clones remained stable.
Silencing of MESV vectors during in vitro differentiation is not correlated with the degree of methylation.
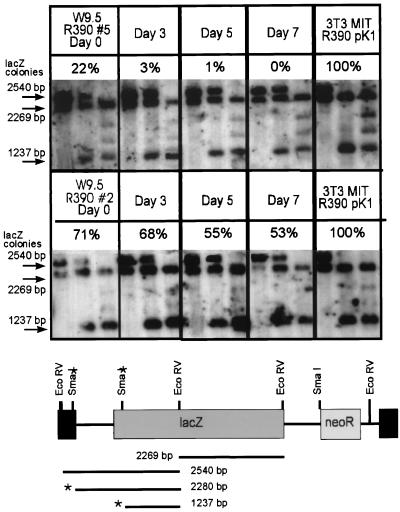
To determine if the methylation pattern of the proviral DNA had altered during differentiation, DNA was prepared from parallel cultures in a time course experiment. DNA digests of either clone 2, in which LacZ expression levels were basically maintained throughout differentiation (75% of that for the undifferentiated control), or clone 5, in which LacZ expression is completely silenced during differentiation, were prepared as described above and probed with a lacZ fragment. No obvious change in methylation was detected in either clone (Fig. 5), although in both cases an approximately 25 to 50% reduction in the total number of clones expressing LacZ was observed. This suggests that de novo methylation occurs after transcriptional extinction and is not a causative factor, in agreement with earlier studies (19, 43).
FIG. 5.
Methylation patterns of provirus in ES cell clones are not altered during differentiation. Genomic DNA was isolated from differentiating cultures at days 0, 3, 5, and 7 and digested with either EcoRV alone (first lane of each group) or together with SmaI (second lane of each group) or, as a control, XmaI (third lane of each group). After transfer to a nylon membrane, the DNA was hybridized with a lacZ probe as indicated. The schematic diagram shows the expected fragments if digestion with SmaI is not blocked or is only partially blocked by methylation. The percentage of LacZ-positive colonies determined at the time of DNA isolation for each clone is shown.
Extinction of MESV expression after in vivo differentiation.
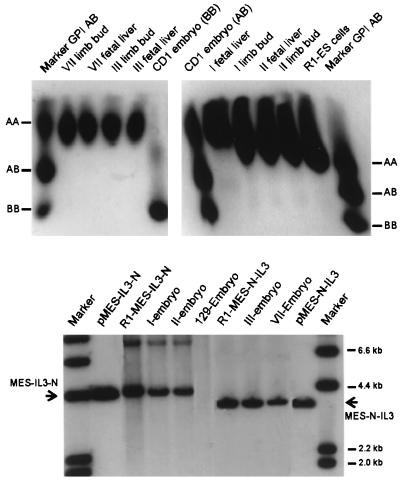
Although the results obtained from in vitro differentiation assays suggested that the integrated provirus was partially or fully blocked in expression as a result of differentiation induction, the inability to direct differentiation in this system makes it hard to assess if the differentiation state is truly permissive for retrovirus expression. We therefore took advantage of the fact that ES cells have totipotent differentiation capacity in vivo and can contribute to chimera formation. The tetraploid aggregation technique of Nagy et al. (42) allows development of 100% of ES cell derived fetal liver hematopoietic cells, an optimal environment for expression of MESV constructs (7, 14). R1 ES clones, carrying either the high- or low-expression MESV-IL3 vector, were aggregated to primary morulae with a tetraploid set of chromosomes by electrofusion of two cell-stage embryos. As expected, embryos developing from these aggregates were of completely diploid ES cell origin and contained a single intact provirus (Fig. 6).
FIG. 6.
Embryos derived by the tetraploid aggregation technique are completely of R1 ES cell origin and contain an intact provirus copy. Embryo origin was confirmed by using a GPI assay (see Materials and Methods). CD1 embryos used for aggregation express either AB or BB isoforms, due to mating with preselected GPI BB males, whereas R1 ES cells express exclusively the A isoform. All embryos used for these studies (I, II, III, and VII) expressed the A isoform. Protein extracted from either limb bud or fetal liver was analyzed. The phenotypes of CD1 embryos (BB or AB) are also shown. The presence of an intact provirus in the ES-derived embryos was confirmed by Southern blot analysis. Genomic DNA was isolated and restricted with NheI, which cuts once in each LTR. As controls, both plasmid DNA (pMES-IL3-N or pMES-N-IL3) and the original transduced R1 ES cell lines were used. DNA immobilized on a nylon membrane after gel electrophoresis was hybridized with a Neor-specific probe.
Embryos derived from either R1-MESV-IL3neoR (high expression) or R1-MESV-neoRIL3 (low expression) ES cells were sacrificed at day 15.5 of gestation and monitored for provirus expression. Previously, we have shown that low levels (<0.5 U) of endogenously synthesized IL-3 are sufficient to support growth of hematopoietic cells (34) and thus that monitoring of factor independent growth of hematopoietic cells is a sensitive assay to detect retrovirus-mediated expression of IL-3 in the transgenic animals. Thus, fetal liver cells were either cultivated as mass cultures or plated in hematopoietic colony assays for functional analysis of IL-3 expression. Fetal liver cells from four embryos from two independent transgenic lines were seeded at high densities (1 × 106 to 5 × 106 cells/ml) into medium with or without IL-3 as an exogenous growth factor. Whereas all control cultures in the presence of exogenous IL-3 proliferated over an observation period of 6 weeks until termination of the experiment, mass cultures initiated without IL-3, and thus dependent on auto- or paracrine IL-3 production, died within a week. Results from mass cultures were confirmed by plating fetal liver cells of three tetraploid aggregation embryos from two independent lines in hematopoietic colony assays either in the absence or presence of IL-3 and erythropoietin. Neither factor-independent colonies nor changes in colony distribution could be observed in vector-carrying fetal liver cells (Table 5). Since hematopoietic cells produce the trans-acting factors required for high-level MESV expression (7), the lack of retrovirus expression must be due to a cis-acting mechanism, thus extending our in vitro results.
TABLE 5.
MESV vectors are not expressed in ES cell-derived hemopoietic fetal liver cells permissive for infection
| Fetal liver cells | Vector | Growth factor(s) | No. of coloniesa
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Σ | G | M | GM | Mixed | BFUe | Other | |||
| Controls (129 mice) | None | None | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IL-3 | 27 | 4 | 8 | 10 | 1.3 | 0.3 | 2.8 | ||
| IL-3 + Epod | 23 | 3 | 8 | 7 | 3.5 | 0.3 | 1.3 | ||
| 129-CD1 Ib | MESV-IL3neoR | None | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IL-3 | 36 | 8 | 4.5 | 22 | 0.5 | 0 | 1 | ||
| IL-3 + Epo | 42 | 6.5 | 7 | 23 | 3.5 | 0 | 1.5 | ||
| 129-CD1 III | MESV-neoRIL3 | None | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IL-3 | 33 | 11 | 5 | 17 | 0 | 0 | 0 | ||
| IL-3 + Epo | 35 | 8 | 4 | 19 | 2 | 0.5 | 1.5 | ||
| 129-CD1 VII | MESV-neoRIL3 | None | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IL-3 | 26 | 8 | 1 | 14 | 3 | 0 | 0 | ||
| IL-3 + Epo | NTc | NT | NT | NT | NT | NT | NT | ||
Cell suspensions of day 15.5 fetal liver cells were prepared and seeded at 1.25 × 105 cells/ml/dish. G, granulocyte CFU; M, macrophage CFU; GM, granulocyte/macrophage CFU; BFUe, blast-forming unit, erythroid.
129-CD1 tetraploid aggregation chimeras are of 100% ES cell origin as determined by GPI analysis.
NT, not tested.
Epo, erythropoietin.
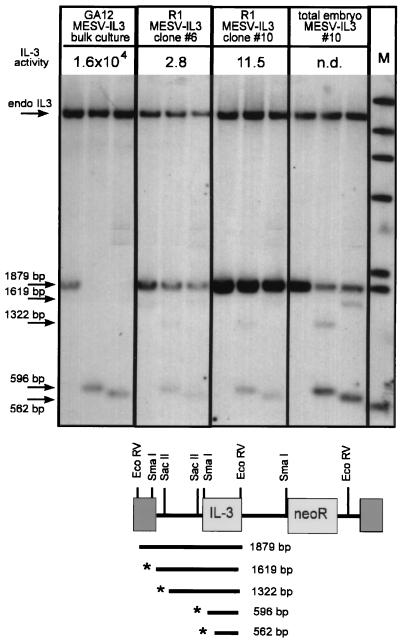
Global demethylation occurs in the morula during embryogenesis, followed by de novo methylation in the pregastrula stage. It was therefore of interest to see if the methylation pattern of the provirus was altered after in vivo differentiation. DNA was prepared from either total embryos or fetal liver cells. Analysis of DNA from the undifferentiated MESV-IL3neoR ES cells used for the chimeras showed a high degree of methylation, as indicated by the quite high levels of undigested DNA remaining after digestion with either the methylation-sensitive SmaI or SacII enzyme (Fig. 7). In contrast, the extent of proviral DNA methylation in the total embryo was somewhat reduced. Comparing the intensities of the 1,619- and 562-bp SmaI fragments with that of the undigested 1,879-bp band, the fraction of unmethylated DNA in the embryo is approximately 10-fold higher than in the original undifferentiated ES cell clone 10 used for aggregation. Although the overall degree of methylation was modulated during in vivo differentiation, the MESV vector did not regain activity in a normally permissive differentiation stage.
FIG. 7.
The methylation pattern of the MESV-IL3neoR provirus is altered after in vivo differentiation and does not correlate with changes in expression. DNA was prepared as described in the legend to Fig. 4. DNA was digested with EcoRV either alone (lanes 1, 4, 7, and 10), with SacII (lanes 2, 5, 8, and 11), or with SmaI (lanes 3, 6, 9, and 12). DNA was hybridized with IL-3. The EcoRV band corresponding to the endogenous (endo) IL-3 gene is indicated; it does not contain internal sites for either SmaI or SacII. IL-3 expression levels are indicated. Lane M, molecular size marker. n.d., not determined.
Exchange of the MESV promoter-enhancer with that of the β-actin housekeeping gene does not increase the transcription frequency or inhibit extinction of the provirus.
Two levels of repression have been characterized in the above-described studies: one acting in the undifferentiated cells, resulting in a low mosaic pattern of expression, and a second that occurs during differentiation and leads to a shut-down of transcription. Although separate events, they may both be influenced by the viral enhancer sequences. Replacement of the viral enhancer with that of a cellular gene may protect the provirus from repression by flanking chromatin. We thus proposed that if the MESV enhancer region was replaced with that of a housekeeping gene, expression would be not only initiated but also maintained. We chose the strong β-actin enhancer-promoter due to the high number of potential binding sites for the Sp1 transcription factor, which is expressed at high levels in ES cells. The strategy used to introduce the β-actin promoter-enhancer in the U3 region of the 3′ LTR is described in Materials and Methods. The 3′ LTR of MESV-lacZneoR was exchanged with the actin LTR to create AcEV-lacZneoR (Fig. 2).
Titers of pseudotyped AcEV-lacZneoR vectors on both fibroblasts and ES cells were measured. Similar to MESV vectors, the vector was able to transfer G418 resistance to both cell lines at the same frequency (Table 1). Replating and staining of five independent Neor colonies gave results similar to those described above: LacZ expression was observed in only 30 to 78% of the cells of the replated colonies (Table 6). A correlation between expression and methylation similar to that in cells transduced with MESV vectors was observed (data not shown). Finally, in vitro differentiation resulted in an average 75% decrease in expression frequencies compared to that for nondifferentiated controls (Table 6). In conclusion, exchanging the promoter-enhancer region of MESV with that of the β-actin gene did not either increase expression levels in undifferentiated cells or inhibit extinction in differentiating cells.
TABLE 6.
Replacement of the retroviral enhancer by the β-actin promoter does not increase expression levels or alleviate extinction during differentiationa
| Retroviral vector | No. of clones tested | % of LacZ+ colonies in:
|
Ratiod | |
|---|---|---|---|---|
| Undifferentiated culturesb | Differentiated culturesc | |||
| None | 1 | 0 | 0 | |
| MPEV-lacZneoR | 6 | 70 ± 21 | 18 ± 17 | 0.25 |
| AcEV-lacZneoR | 8 | 60 ± 22 | 16 ± 19 | 0.27 |
CCE ES cells were infected with the indicated vector, and independent clones selected by growth in G418 were isolated and tested for LacZ staining before and after differentiation induction.
Mean values (± standard deviations) obtained at day 0 before differentiation induction in two or three independent experiments.
Mean values (± standard deviations) obtained at either day 5 or 7 after differentiation induction in two or three independent experiments.
Ratio of LacZ+ colonies in differentiated cultures to those in undifferentiated cultures.
DISCUSSION
Early studies of retroviral infection of embryonal cells revealed two levels at which retrovirus expression is blocked, a trans-acting repression that could be relieved by cell differentiation and a cis-acting repression that prevented provirus expression in a retrovirus-permissive background (4, 21, 43, 56). In contrast to Mo-MuLV-based retroviral vectors, MESV is permissive for LTR-mediated expression in embryonic cells and thus circumvents the first block to expression. The goal of this work was to determine if vectors based on MESV could be used to examine the second block to provirus transcription, which results in the irreversible extinction of transcription by a cis-acting mechanism. Although the existence of this secondary block to retroviral expression was described over 10 years ago, it is still poorly understood (4, 19, 43). Increasing interest in extinction of retroviral transcription has developed due to the observation that similar mechanisms are also found in other cell types, including hematopoietic stem cells and primary fibroblasts and myoblasts (11, 27, 33, 44, 54).
Our results identify two mechanisms by which provirus expression is repressed. One occurs in the undifferentiated cell, is associated with methylation, and is compatible with stable, long-term expression. The second occurs during early stages of differentiation, is not associated with increased methylation, and leads to complete extinction of provirus expression. Significantly, the repression observed in the undifferentiated state did not result in extinction of provirus expression. Cell populations maintained without selective pressure continued to maintain the same proportion of cells expressing the provirus, suggesting a stochastic and reversible repression. This is in contrast to provirus expression after differentiation induction, where a striking decline in transcription frequency was observed. This was also observed in ES-derived hematopoietic cells, which are normally a highly permissive environment for MESV expression. The latter mechanism of repression is thus most likely responsible for the irreversible extinction of provirus expression in differentiated ES cells and the developing embryo. Extinction must occur during early stages of differentiation, as previous studies have shown that EC and ES cells are permissive for expression after 7 to 10 days of differentiation (4, 56). Thus, the developmental window in which extinction is most active may be quite small. This is in agreement with retroviral infection studies of developing embryos (53). It is likely that the extinction process is part of a protective mechanism of the embryo. The role of methylation remains unclear, but it appears to correlate with transcription repression but not with complete extinction.
Although provirus extinction after in vivo differentiation of Mo-MuLV-infected embryos is well documented (30), retroviral vectors expressing a transgene from an internal promoter were not silenced in mice generated by infection of preimplantation embryos or via ES cell chimeras (59, 62). Expression of the transgene via either β-globin or a thymidine kinase promoter was readily observed in each of three independent transgenic lines investigated for each construct. In the study presented here, over 18 independent integration sites were investigated after either in vitro or de novo differentiation, and in all cases MESV expression was either not detectable or reduced by more than 100-fold compared to that normally observed in fibroblasts or other permissive cells (e.g., hematopoietic cells). Repression was not specific to the viral promoter-enhancer, as complete exchange of the MESV promoter-enhancer domain with that of an endogenous cellular gene (the β-actin gene) did not alter the overall extinction frequency.
As the extent of repression for both events is dependent on the integration site, the surrounding chromatin structure of the integrated provirus must be a key determinant in the repression process. Inactivation of a gene in various numbers of cells upon integration near inactivating chromatin has been described for Drosophila (25), yeast (2), and mice (52) and has been termed position effect variegation (PEV). Although the initiating events (e.g., telomeric positioning or X chromosome inactivation) in these various phenomena may be different, they may have a mechanism in common with the silencing described here that is increased during differentiation. Silencing in PEV is facilitated by a large number of protein factors, many of which are known to be components of the chromatin. It has been postulated that transcriptional promoters and enhancers act to suppress PEV by antagonizing repression by flanking chromatin (3, 16, 45, 46, 69). Significantly, several of the transcription factors that bind to the MESV or Mo-MuLV enhancer domain are not present or are present at only low levels in ES cells, thus preventing enhancer function (8, 20, 61). We therefore postulate that MESV provirus lacks the appropriate enhancer elements necessary to antagonize the repressive activity of chromatin in ES cells.
In conclusion, we have defined two stages at which provirus transcription is repressed. Significantly, repression that occurs during early stages of differentiation is the most predominant and leads to extinction of expression. Experiments are being carried out to test whether incorporation of enhancer elements known to open or remodel chromatin structure within the MESV LTR can overcome this cis-acting repression of expression in early embryonic cells (15, 48, 57).
ACKNOWLEDGMENTS
C.L. thanks Alan Bernstein, Andras Nagy, and members of their laboratory at the Samuel Lumenfeld Research Center at Mt. Sinai Hospital in Toronto for sharing the aggregation technology and providing a stimulating work atmosphere. We thank Manuel Grez for providing the p5Gneo vector and Colin Stewart and Andras Nagy for providing ES cell lines. We are also indebted to Alexandra Mittel and Marion Nissen for expert technical assistance.
This work was supported by a research grant (Sto 225/2) and a Habilitation Stipend (La 279/A) from the Deutsche Forschungsgemeinschaft. A.S. was supported by the Boehringer-Ingelheim-Fonds. The Heinrich-Pette-Institut is financially supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit.
REFERENCES
- 1.Akgün E, Ziegler M, Grez M. Determinants of retrovirus gene expression in embryonal carcinoma cells. J Virol. 1991;65:382–388. doi: 10.1128/jvi.65.1.382-388.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allshire R C, Javerzat J-P, Redhead N J, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio O M, Gottschling D E. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 4.Asche W, Colletta G, Warnecke G, Nobis P, Pennie S, King R M, Ostertag W. Lack of retrovirus gene expression in somatic cell hybrids of Friend cells and teratocarcinoma cells with a teratocarcinoma phenotype. Mol Cell Biol. 1984;4:923–930. doi: 10.1128/mcb.4.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barklis E, Mulligan R C, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–399. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- 6.Baum C, Forster P, Hegewisch-Becker S, Harbers K. An optimized electroporation protocol applicable to a wide range of cell lines. BioTechniques. 1994;17:1058–1062. [PubMed] [Google Scholar]
- 7.Baum C, Hegewisch-Becker S, Eckert H-G, Stocking C, Ostertag W. Novel retroviral vectors for efficient gene expression of the multi-drug resistance (mdr-1) gene in early hemopoietic cells. J Virol. 1995;69:7541–7547. doi: 10.1128/jvi.69.12.7541-7547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baum C, Itoh K, Meyer J, Laker C, Ito Y, Ostertag W. The potent enhancer activity of the polycythemic strain of spleen focus-forming virus in hematopoietic cells is governed by a binding site for Sp1 in the upstream control region and by a unique enhancer core motif, creating an exclusive target for PEBP/Cbf. J Virol. 1997;71:6323–6331. doi: 10.1128/jvi.71.9.6323-6331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berwin B, Barklis E. Retrovirus-mediated insertion of expressed and non-expressed genes at identical chromosomal locations. Nucleic Acids Res. 1993;21:2399–2407. doi: 10.1093/nar/21.10.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cepko C L, Ryder E F, Austin C P, Walsh C, Fekette M D. Lineage analysis using retroviral vectors. Methods Enzymol. 1993;225:933–960. doi: 10.1016/0076-6879(93)25059-b. [DOI] [PubMed] [Google Scholar]
- 11.Challita P-M, Kohn D B. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc Natl Acad Sci USA. 1994;92:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Challita P-M, Skelton D, El-Khoueiry A, Yu X-J, Weinberg K, Kohn D B. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dzierzak E A, Papayannopoulou T, Mulligan R. Lineage-specific expression of a human β-globin gene in murine bone marrow transplant recipients reconstituted with retrovirus-transduced stem cells. Nature. 1988;331:35–41. doi: 10.1038/331035a0. [DOI] [PubMed] [Google Scholar]
- 14.Eckert H-G, Stockschläder M, Just U, Hegewisch-Becker S, Grez M, Uhde A, Zander A, Ostertag W, Baum C. High-dose multidrug resistance in primary human hematopoietic progenitor cells transduced with optimized retroviral vectors. Blood. 1996;88:3407–3415. [PubMed] [Google Scholar]
- 15.Ellis J, Tan-Un K C, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenfeld G. Chromatin as an essential part of the transcription mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 17.Franz T, Hilberg F, Seliger B, Stocking C, Ostertag W. Retroviral mutants efficiently expressed in embryonal carcinoma cells. Proc Natl Acad Sci USA. 1986;83:3292–3296. doi: 10.1073/pnas.83.10.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautsch J W. Embryonal carcinoma stem cells lack a function required for virus replication. Nature. 1980;285:110–112. doi: 10.1038/285110a0. [DOI] [PubMed] [Google Scholar]
- 19.Gautsch J W, Wilson M C. Delayed de novo methylation in teratocarcinoma suggests additional tissue-specific mechanisms for controlling gene expression. Nature. 1983;301:32–37. doi: 10.1038/301032a0. [DOI] [PubMed] [Google Scholar]
- 20.Golemis E A, Speck N A, Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990;64:534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greiser-Wilke I, Ostertag W, Goldgarb P, Lang A, Furasawa M, Conscience J F. Inducibility of spleen focus forming virus by BUdR is controlled by the differentiated state of the cell. Proc Natl Acad Sci USA. 1981;78:2995–2999. doi: 10.1073/pnas.78.5.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grez M, Akgün E, Hilberg F, Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc Natl Acad Sci USA. 1990;87:9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grez M, Zörnig M, Nowock J, Ziegler M. A single point mutation activates the Moloney murine leukemia virus long terminal repeat in embryonal stem cells. J Virol. 1991;65:4691–4698. doi: 10.1128/jvi.65.9.4691-4698.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harbers K, Schnieke A, Stuhlmann H, Jähner D, Jaenisch R. DNA methylation and gene expression: endogenous retroviral genomes become infectious after molecular cloning. Proc Natl Acad Sci USA. 1981;78:7609–7613. doi: 10.1073/pnas.78.12.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henikoff S. Position effect and related phenomena. Curr Opin Genet Dev. 1992;2:907–912. doi: 10.1016/s0959-437x(05)80114-5. [DOI] [PubMed] [Google Scholar]
- 26.Hilberg F, Stocking C, Ostertag W, Grez M. Functional analysis of a retroviral host-range mutant: altered long terminal repeat sequences allow expression in embryonal carcinoma cells. Proc Natl Acad Sci USA. 1987;84:5232–5236. doi: 10.1073/pnas.84.15.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoeben R C, Migchielsen A A J, van der Jagt R C M, van Ormondt H, van der Eb A J. Inactivation of the Moloney murine leukemia virus long terminal repeat in murine fibroblast cell lines is associated with methylation and dependent on its chromosomal position. J Virol. 1991;65:904–912. doi: 10.1128/jvi.65.2.904-912.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iscove N N, Sieber F. Erythroid progenitors in mouse bone marrow detected by macroscopic colony formation in culture. Exp Hematol. 1975;3:32–43. [PubMed] [Google Scholar]
- 29.Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci USA. 1976;73:1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaenisch R, Fan H, Croker B. Infection of preimplantation mouse embryos and of newborn mice with leukemia virus: tissue distribution of viral DNA and RNA and leukemogenesis in the adult animal. Proc Natl Acad Sci USA. 1975;72:4008–4012. doi: 10.1073/pnas.72.10.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jähner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 32.Jähner D, Stuhlmann H, Stewart C, Harbers K, Löhler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 33.Keller G, Snodgrass R. Life span of multipotential hematopoietic stem cells in vivo. J Exp Med. 1990;171:1407–1418. doi: 10.1084/jem.171.5.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laker C, Stocking C, Bergholz U, Hess N, DeLamarter J, Ostertag W. Autocrine stimulation after transfer of the granulocyte/macrophage colony stimulating factor gene and autonomous growth are distinct but interdependent steps in the oncogenic pathway. Proc Natl Acad Sci USA. 1987;84:8458–8462. doi: 10.1073/pnas.84.23.8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewin B. Chromatin and gene expression: constant questions but changing answers. Cell. 1994;79:397–406. doi: 10.1016/0092-8674(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 36.Loh T P, Sievert L L, Scott R W. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonal carcinoma cells. Mol Cell Biol. 1990;10:4045–4057. doi: 10.1128/mcb.10.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohse P, Arnold H H. The down-regulation of the chicken cytoplasmic β actin during myogenic differentiation does not require the gene promoter but involves the 3′ end of the gene. Nucleic Acids Res. 1988;16:2787–2803. doi: 10.1093/nar/16.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohuizen M v, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in Eμ-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 39.Markowitz D, Goff S, Bank A. Construction and use of a safe and efficient amphotropic packaging cell line. Virology. 1988;164:400–406. [PubMed] [Google Scholar]
- 40.Nagy A, Gocza E, Mentez-Diaz E, Prideaux R, Ivanyi E, Markkula M, Rosant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 41.Nagy A, Rossant J. Production of completely ES cell-derived fetuses. In: Joyner A L, editor. Gene targeting. Oxford, United Kingdom: IRL Press; 1993. pp. 147–179. [Google Scholar]
- 42.Nagy A, Rossant J, Nagy R, Abranow-Newerly W, Roder J C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in the suppression of Moloney murine leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- 44.Palmer T D, Rosman G J, Osborne W R, Miller A D. Genetically modified skin fibroblasts persist long after transplantation but gradually inactivate introduced genes. Proc Natl Acad Sci USA. 1991;88:1330–1334. doi: 10.1073/pnas.88.4.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paranjape S M, Kamakaka R T, Kodonaga J T. Role of chromatin structure in the regulation of gene expression. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 46.Paro R. Propagating memory of transcriptional states. Trends Genet. 1995;118:295–297. doi: 10.1016/s0168-9525(00)89081-2. [DOI] [PubMed] [Google Scholar]
- 47.Petersen R, Kempler G, Barklis E. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol Cell Biol. 1991;11:1214–1221. doi: 10.1128/mcb.11.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phi-Van L, von Kries J P, Ostertag W, Strätling W H. The chicken lysozyme 5′ matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol. 1990;10:2302–2307. doi: 10.1128/mcb.10.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prince V E, Rigby P W J. Derivatives of Moloney murine sarcoma virus capable of being transcribed in embryonal carcinoma stem cells have gained a functional Sp1 binding site. J Virol. 1991;65:1803–1811. doi: 10.1128/jvi.65.4.1803-1811.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quischke W W, Lin Z-Y, DePonti-Zilli L, Paterson B M. The β actin promoter: high levels of transcription depend upon a CCAAT binding factor. J Biol Chem. 1989;264:9539–9546. [PubMed] [Google Scholar]
- 51.Robertson E, Bradley A, Kuehn M, Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vectors. Nature. 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 52.Robertson G, Garrick D, Wu W, Kearns M, Martin D. Position-dependent variegation of globin transgene expression in mice. Proc Natl Acad Sci USA. 1995;92:5371–5375. doi: 10.1073/pnas.92.12.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savatier P, Morgenstern J, Beddington R S P. Permissiveness to murine leukemia virus expression during implantation and early postimplantation mouse development. Development. 1990;109:655–665. doi: 10.1242/dev.109.3.655. [DOI] [PubMed] [Google Scholar]
- 54.Scharfmann R, Axelrod J H, Verma I M. Long-term in vivo expression of retrovirus-mediated gene transfer in mouse fibroblast implants. Proc Natl Acad Sci USA. 1991;88:4626–4630. doi: 10.1073/pnas.88.11.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schnieke A, Harbers K, Jaenisch R. Embryonic lethal mutation in mice induced by retroviral insertion into the α-1(I) collagen gene. Nature. 1983;304:315–320. doi: 10.1038/304315a0. [DOI] [PubMed] [Google Scholar]
- 56.Seliger B, Kollek R, Stocking C, Franz T, Ostertag W. Viral transfer, transcription, and rescue of a selectable myeloproliferative sarcoma virus in embryonal cell lines: expression of the mos oncogene. Mol Cell Biol. 1986;6:286–293. doi: 10.1128/mcb.6.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kodonaga J T, Jones K A. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 58.Shimotono K, Temin H. Formation of infectious progeny after insertion of herpes Tk gene into DNA of an avian retrovirus. Cell. 1981;46:19–29. doi: 10.1016/0092-8674(81)90034-9. [DOI] [PubMed] [Google Scholar]
- 59.Soriano P, Cone R D, Mulligan R C, Jaenisch R. Tissue-specific and ectopic expression of genes introduced into transgenic mice by retroviruses. Science. 1986;234:1409–1413. doi: 10.1126/science.3024318. [DOI] [PubMed] [Google Scholar]
- 60.Soriano P, Jaenisch R. Retroviruses as probes for mammalian development: allocation of cells to the somatic and germ cell lineages. Cell. 1986;46:18–29. doi: 10.1016/0092-8674(86)90856-1. [DOI] [PubMed] [Google Scholar]
- 61.Speck N A, Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeats of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987;7:435–439. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart C L, Schuetze S, Vanek M, Wagner E F. Expression of retroviral vectors in transgenic mice obtained by embryo infection. EMBO J. 1987;6:383–388. doi: 10.1002/j.1460-2075.1987.tb04766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stewart C L, Stuhlmann H, Jahner D, Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci USA. 1982;79:4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stocking C, Bergholz U, Friel J, Klingler K, Wagener T, Starke C, Kitamura T, Miyajima A, Ostertag W. Distinct classes of factor-independent mutants can be isolated after retroviral mutagenesis of a human myeloid stem cell line. Growth Factors. 1993;8:197–209. doi: 10.3109/08977199309011023. [DOI] [PubMed] [Google Scholar]
- 65.Stocking C, Grez M, Ostertag W. Regulation of retrovirus infection and expression in embryonic and hematopoietic stem cells. In: Doerfler W, Boehm P, editors. Virus strategies. Heidelberg, Germany: VCH Verlag; 1993. pp. 433–455. [Google Scholar]
- 66.Stocking C, Kollek R, Bergholz U, Ostertag W. Long terminal repeat sequences impart hemopoietic transformation properties to the myeloproliferative sarcoma virus. Proc Natl Acad Sci USA. 1985;82:4098–4102. doi: 10.1073/pnas.82.17.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsukiyama T, Niwa O, Yokoro K. Analysis of the binding proteins and activity of the long terminal repeat of Moloney murine leukemia virus during differentiation of mouse embryonal carcinoma cells. J Virol. 1991;65:2979–2986. doi: 10.1128/jvi.65.6.2979-2986.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsukiyama T, Ueda H, Hirose S, Niwa O. Embryonal long terminal repeat-binding protein is a murine homolog of FTZ-F1, a member of the steroid receptor superfamily. Mol Cell Biol. 1992;12:1286–1291. doi: 10.1128/mcb.12.3.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I K. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 70.Weiher H, Barklis E, Ostertag W, Jaenisch R. Two distinct sequence elements mediate retroviral gene expression in embryonal carcinoma cells. J Virol. 1987;61:2742–2746. doi: 10.1128/jvi.61.9.2742-2746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]