Abstract
Background
Chronic inflammation persists in some people living with human immunodeficiency virus (HIV) during antiretroviral therapy and is associated with premature aging. The glycoprotein 120 (gp120) subunit of HIV-1 envelope sheds and can be detected in plasma, showing immunomodulatory properties even in the absence of detectable viremia. We evaluated whether plasma soluble gp120 (sgp120) and a family of gp120-specific anti–cluster A antibodies, linked to CD4 depletion in vitro, contribute to chronic inflammation, immune dysfunction, and subclinical cardiovascular disease in participants of the Canadian HIV and Aging Cohort Study with undetectable viremia.
Methods
Cross-sectional assessment of sgp120 and anti–cluster A antibodies was performed in 386 individuals from the cohort. Their association with proinflammatory cytokines and subclinical coronary artery disease was assessed using linear regression models.
Results
High levels of sgp120 and anti–cluster A antibodies were inversely correlated with CD4+ T cell count and CD4/CD8 ratio. The presence of sgp120 was associated with increased levels of interleukin 6. In participants with detectable atherosclerotic plaque and detectable sgp120, anti–cluster A antibodies and their combination with sgp120 levels correlated positively with the total volume of atherosclerotic plaques.
Conclusions
This study showed that sgp120 may act as a pan toxin causing immune dysfunction and sustained inflammation in a subset of people living with HIV, contributing to the development of premature comorbid conditions.
Keywords: HIV-1, soluble gp120, anti–cluster A antibodies, chronic inflammation, cardiovascular diseases
Soluble glycoprotein 120 (sgp120) is detected in the plasma of people living with HIV-1 with undetectable viremia. The presence of sgp120 and anti–cluster A antibodies is associated with correlates of immune dysfunction, chronic inflammation, and subclinical cardiovascular disease.
Graphical Abstract
Graphical Abstract.
While antiretroviral therapy (ART) efficiently inhibits viral replication, people living with human immunodeficiency virus (HIV; PLWH) have a 15-year gap in comorbidity-free years [1]. The underlying causes for this gap are numerous and include the presence of residual immune dysfunction and chronic antigenic stimulation by HIV, which contributes to a state of sustained inflammation [2]. Persistent immune dysfunction has also been associated with the immunological nonresponse that many PLWH experience [3–7]. These individuals, despite efficient viral control, have failure to restore circulating CD4+ T cells and have persistent immune activation, possibly leading to increased comorbid conditions.
The HIV-1 envelope (Env) glycoprotein, in particular the glycoprotein (gp) 120 domain of the gp120/gp41 heterodimer, besides its role in viral entry, has been shown to exhibit a variety of immunoregulatory activities that could contribute to HIV-1 pathogenesis and immune dysfunction. Owing to its noncovalent interaction with gp41, gp120 can be spontaneously released from the surface of virions and infected cells [8–10]. Consequently, variable amounts of soluble gp120 (sgp120) can be detected in plasma and tissues from PLWH, even during ART [7, 11, 12]. In several studies, sgp120 has been associated with HIV-1–induced immune dysfunction [4–7]. It has also been reported to exert proinflammatory activities, as binding of gp120 to CD4 on the surface of monocytes, macrophages, T cells and dendritic cells has been found to induce the production of cytokines, including interleukin 6 (IL-6), interleukin 10 and IL-1β, interferon α and γ, and tumor necrosis factor (TNF) α) [4, 13–15].
Notably, gp120 shed from productively infected cells has been shown to interact with CD4 present on uninfected bystander CD4+ T cells [15, 16]. This interaction leads to exposure of CD4-induced Env epitopes and sensitization of uninfected bystander CD4+ T cells to antibody-dependent cellular cytotoxicity (ADCC) mediated by HIV-positive plasma [16, 17]. Within HIV-positive plasma, antibodies targeting conserved CD4-induced gp120 cluster A epitopes have been shown to elicit potent ADCC activity against sgp120-coated cells [15, 18]. Antibodies recognizing the gp120 inner-domain cluster A region have been found to be responsible for most of the ADCC activity exhibited by chronically HIV-1–infected individuals, provided that the epitopes recognized by them are exposed [19].
Together, these observations suggest that sgp120 is associated with chronic inflammation, causing persistent antigen stimulation even in the context of prolonged ART and undetectable viremia in plasma, and a potential contributor to an increased risk of comorbid conditions. In the current study, we investigated whether the presence of sgp120 and ADCC-mediating antibodies, specifically anti–cluster A antibodies, were associated with correlates of immune dysfunction, chronic inflammation, and subclinical cardiovascular disease (CVD). To this end, we optimized an enzyme-linked immunosorbent assay (ELISA)–based assay to measure sgp120 in plasma samples from 386 PLWH with long-term ART treatment and undetectable viremia from the Canadian HIV and Aging Cohort Study (CHACS) [20].
METHODS
Ethics Statement
This research project was reviewed and approved by the Centre de Recherche du CHUM (CRCHUM) Research Ethics Board (Ethics Committee approval nos. 22.173, 11.063, and 11.062). Written informed consent was obtained from all study participants, and the research adhered to the standards indicated by the Declaration of Helsinki.
Study Design
We designed a cross-sectional study, nested within the CHACS. The specific objectives of the study were to quantify sgp120 in plasma of participants and study the relationship of sgp120 and anti–cluster A antibodies to correlates of immune dysfunction, inflammation, and subclinical CVD. Longitudinal samples from 9 sgp120-positive participants were also analyzed.
Study Population
The CHACS protocol has been described elsewhere [20]. Briefly, it is a prospective cohort study and biobank, ongoing since 2012 in 10 clinical sites across Canada. Inclusion criteria are to be ≥40 years old or to have lived with HIV for ≥15 years and have a life expectancy of >1 year at enrollment. Participants with no known CVD and a 10-year Framingham risk score of overt CVD ranging from 5% to 20%, no allergy to contrast medium, and no renal failure, were invited to participate in the cardiovascular imaging substudy, in which they underwent computed tomography coronary angiography (CCTA). Participants were selected for the present study if they were recruited into the Montréal and Québec CHACS study sites and had undetectable viral loads. For all participants, data on sociodemographic characteristics, HIV disease history, and traditional cardiovascular risk factors are available through the CHACS study database.
CCTA Protocol
CCTA was performed as described elsewhere [21–23]. A detailed description is provided in the Supplementary Materials.
ELISA Protocol
The sandwich ELISA was described elsewhere, with modifications [7, 15]. A detailed description is provided in the Supplementary Materials.
Protein Production and Purification
Production and purification of monomeric soluble HIV-1YU2 gp120 was described elsewhere [15, 24]. A detailed description is provided in the Supplementary Materials.
Multiplex Measurements of Soluble Inflammatory Markers
Customized Human Magnetic Luminex Assays (R&D Systems) were used to measure soluble inflammatory markers in plasma samples (see Supplementary Table 4 for complete analyte list). A detailed description is provided in the Supplementary Materials.
Measurement of HIV DNA
HIV DNA was measured as described elsewhere [25]. A detailed description is provided in the Supplementary Materials.
Statistical Analysis
Appropriate descriptive statistics were used for normally and nonnormally distributed variables, and variables were log-transformed, as needed. A detailed description of the statistical analysis performed is provided in the Supplementary Materials.
RESULTS
Plasma from 386 participants was available for measurements of sgp120 and anti–cluster A antibodies, 157 of whom also had soluble inflammatory markers measured using a multiplex platform. Of the 386, 145 took part in the cardiovascular imaging substudies. The baseline characteristics of all participants and those in each subgroup are summarized in Table 1. There were no differences in terms of sociodemographic characteristics, immune parameters, or traditional cardiovascular risk factors between the total study group and the 2 subgroups.
Table 1.
Characteristics of Participants in the Canadian HIV and Aging Cohort Study With Undetectable Human Immunodeficiency Virus Viremia, and participants to subgroups
| Characteristic | Study Participants, No. (%)a | P Valueb | Multiplex Substudy Participants, No. (%)a (n = 157) |
P Valueb | |
|---|---|---|---|---|---|
| Total Study Population (n = 386) |
Cardiovascular Imaging Substudy (n = 145) |
||||
| Age, mean (SD), years | 55.8 (7.6) | 56.6 (6.5) | .14 | 56.6 (6.6) | .07 |
| Male sex | 347 (90.0) | 134 (92.4) | .23 | 144 (91.7) | .39 |
| Ethnicity | |||||
| White | 317 (82.1) | 123 (84.8) | .39 | 131 (83.4) | .29 |
| Black | 32 (8.3) | 12 (8.2) | 15 (9.6) | ||
| Other | 37 (9.6) | 10 (6.9) | 11 (7.0) | ||
| Duration of HIV infection, median (IQR), years | 18.3 (12–25) | 18.3 (12–25) | .27 | 19.0 (14–25) | .52 |
| Nadir CD4+ cell count, median (IQR), cells/mL | 210 (120–310) | 200 (90–300) | .30 | 195 (100–300) | .11 |
| Nadir CD4+ cell count <200/mL | 155 (49.1) | 67 (52.8) | 74 (54.4%) | ||
| Absolute CD4+ cell count, mean (SD), cells/mL | 615 (250) | 620 (255) | .27 | 617 (251) | .50 |
| Absolute CD8+ cell count, mean (SD), cells | 763 (376) | 796 (382) | .10 | 809 (408) | .07 |
| CD4/CD8 ratio, mean (SD) | 0.96 (0.53) | 0.96 (0.47) | .92 | 0.93 (0.49) | .35 |
| Duration of ART, mean (SD), years | 13.9 (6.7) | 14.1 (6.8) | .58 | 14.1 (6.9) | .58 |
| Smoking status | |||||
| Never smoker | 128 (33.5) | 42 (29.4) | .22 | 48 (31.0) | .44 |
| Past or present smoker | 254 (66.5) | 101 (70.6) | 107 (69.0) | ||
| Intravenous drug use | 43 (11.2) | 14 (9.6) | .51 | 15 (9.5) | .42 |
| Alcohol abuse | 31 (8.0) | 11 (7.6) | .84 | 11 (7.0) | .57 |
| High blood pressure | 113 (29.7) | 45 (31.5) | .56 | 51 (32.7) | .31 |
| Diabetes mellitus | 37 (9.6) | 15 (10.4) | .71 | 18 (11.5) | .30 |
| Framingham risk score, median (IQR) | 9 (5–16) | 9 (6–13) | .72 | 9 (6–13) | .58 |
| Log anti–cluster A antibodies, mean (SD), RU | 2.4 (1.4) | 2.2 (1.2) | .22 | 2.3 (1.2) | .71 |
| sgp120 | |||||
| Undetectable | 279 (72.3) | 80 (55.2) | <.001 | 83 (52.9) | <.001 |
| Low | 68 (17.6) | 40 (27.6) | 48 (30.6) | ||
| High | 39 (10.1) | 25 (17.2) | 26 (16.56) | ||
Abbreviations: ART, antiretroviral therapy; gp120, glycoprotein 120; HIV, human immunodeficiency virus; IQR, interquartile range; RU, relative units; SD, standard deviation.
aData represent no. (%) of participants unless otherwise specified.
b P values were obtained using Fisher exact test for categorical variables and k-sample equality of median test for continuous variables, and they represent comparisons between subcohort participants and nonparticipants.
Measuring sgp120 in Plasma From PLWH With Undetectable Viremia
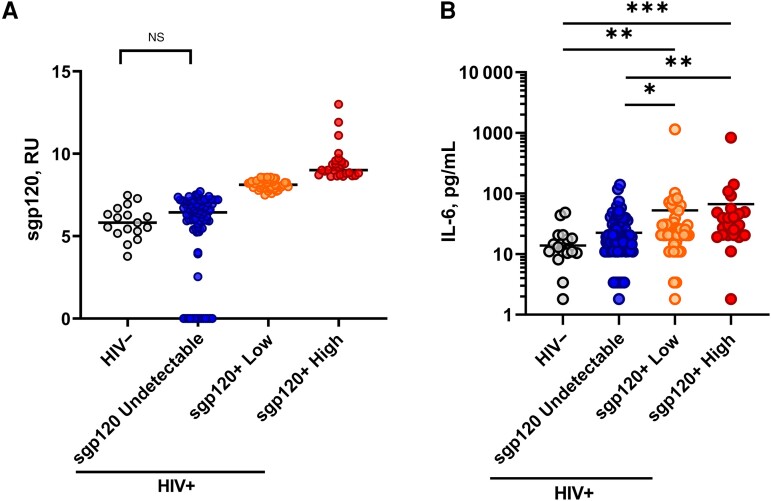
Based on a previous iteration [7], we optimized an ELISA to measure sgp120 in plasma (Supplementary Figure 1A). In this assay, we use a gp120 inner-domain–specific antibody (C11) that targets the highly conserved N-termini and 8-stranded β-sandwich structure of gp120 formed at the late stage of HIV-1 entry [26]. This epitope is therefore buried on the trimeric Env present on virions or infected cells but exposed on sgp120 [16, 26, 27]. The novelty of this assay is the use of the broadly neutralizing CD4 binding site (CD4BS) N6 antibody as a readout, which does not compete for C11 binding. This antibody was reported to target up to 98% of global HIV-1 isolates [28]. Using our assay, a strong linearity (r = 0.9836; P < .001) over a 500-fold range was observed between the signal obtained and the quantity of purified recombinant monomeric soluble HIV-1YU2 gp120 used (Supplementary Figure 1B). This assay enabled us to measure sgp120 in plasma samples from PLWH with undetectable viral loads compared with uninfected individuals (Figure 1A). We further stratified PLWH into 3 subgroups based on the positivity threshold established with the uninfected plasma samples: (1) undetectable sgp120, (2) low levels of sgp120, and (3) high levels of sgp120 (Figure 1A). Of the 386 plasma samples analyzed, 72.3% (n = 279) had undetectable sgp120, 17.6% (n = 68) had low levels, and 10.1% (n = 39) had high levels (Table 1).
Figure 1.
Detection of soluble glycoprotein 120 (sgp120) in people living with human immunodeficiency virus (HIV) with undetectable viremia is associated with inflammation. Representative stratification is shown for 157 people living with HIV, based on levels of sgp120 (A) and interleukin 6 (IL-6) (B). Statistical analysis was performed using Mann-Whitney U tests. *P < .05 ; **P < .01 ; ***P < .001 . Abbreviations: HIV−, HIV negative; HIV+, HIV positive; NS, not significant; RU, relative units.
Association of Anti–Cluster A Antibody Levels With Correlates of Immune Dysfunction
We have previously reported that the release of sgp120 from infected cells sensitizes uninfected bystander CD4+ T cells to ADCC mediated by HIV-positive plasma in vitro, but whether this happens in PLWH remains unclear [15–17]. We therefore measured the levels of anti–cluster A antibodies using an engineered stabilized gp120 inner-domain protein (ID2) exposing only the cluster A region [29, 30]. Figure 2 and Supplementary Table 1 present the associations between anti–cluster A antibodies and absolute CD4+ cell counts or the CD4/CD8 ratio (Supplementary Figure 1). In the total study population, after adjustment for potential confounders (including age, sex, ethnicity, smoking status, nadir CD4+ cell count, duration of ART, and levels of anti-CD4BS antibodies), each 1-log2 increase in anti-cluster A antibody levels was associated with a mean predicted decrease in CD4+ cell count of −15.3 ×106/mL (95% confidence interval [CI], −26.7 ×106/mL to −3.8 ×106/mL; P = .009) (Figure 2A).
Figure 2.
Anti–cluster A antibodies are inversely correlated with CD4+ T-cell counts in people living with human immunodeficiency virus (PLWH) presenting with high levels of soluble glycoprotein 120 (sgp120). Correlations between CD4+ T-cell counts and anti–cluster A antibody levels are shown for 386 PLWH, stratified by sgp120 levels. Correlations are shown for the total study population (A) and for PLWH with undetectable sgp120 (B), low levels of sgp120 (sgp120low) (C), or high levels of sgp120 (sgp120high) (D). Levels of anti–cluster A antibodies were log2 transformed. Univariable and multivariable linear regressions were performed, with the beta parameters representing the mean predicted change in absolute CD4+ cell counts for each 1-log2 increase in titers of anti–cluster A antibodies. Multivariable models are adjusted for age, sex, ethnicity, smoking status, duration of antiretroviral therapy, nadir CD4+ cell counts, and levels of anti–CD4 binding site antibodies. Abbreviations: CI, confidence interval; RU, relative units.
On stratification of the 386 PLWH by sgp120 levels, we observed that the mean predicted change in CD4+ cell counts varies by stratum of sgp120. For those with sgp120 below detection levels, each increase of 1-log2 in anti–cluster A antibodies is associated with a mean predicted decrease in absolute CD4+ cell count of −18.1 ×106/mL (P = .008) (Figure 2B), while the mean predicted decline for participants with high levels of sgp120 is −42.0 ×106/mL (P = .04) (Figure 2D). However, the P value for the interaction was 0.11 (Supplementary Table 1). A similar dynamic is observed with the CD4/CD8 ratio (presented in Supplementary Figure 2 and Supplementary Table 1); within the total study population, each 1-log2 increase in the levels of anti–cluster A antibodies is associated with a mean projected decrease in the CD4/CD8 ratio of −0.06 (95% CI, −0.08 to −0.03; P < .001) (Supplementary Figure 2A).
The magnitude of this association was more pronounced in the subgroup with high levels of sgp120, in which the mean predicted decline in CD4/CD8 ratio was −0.13 (95% CI, −0.22 to −0.04; P = .004) (Supplementary Figure 2D). However, this difference between the groups was not statistically significant (the P value for interaction between sgp120 levels and anti–cluster A antibodies was .21) (Supplementary Table 1). Of note, no association was observed between anti–cluster A antibodies and CD4+ T-cell count or CD4/CD8 ratio in individuals presenting with low levels of sgp120 (Figure 2C, Supplementary Figure 2C, and Supplementary Table 1). We also measured antibodies targeting the CD4BS, using the gp120 resurfaced stabilized core 3, a previously described probe exposing the CD4BS [31]. We found that only 66 of 386 individuals (17%) had detectable levels of CD4BS antibodies; from which 44 had undetectable sgp120 levels, 13 had low sgp120 levels, and 9 had high sgp120 levels. No associations between CD4BS antibody levels and CD4+ T-cell counts were observed.
Association Between sgp120 and Proinflammatory Markers
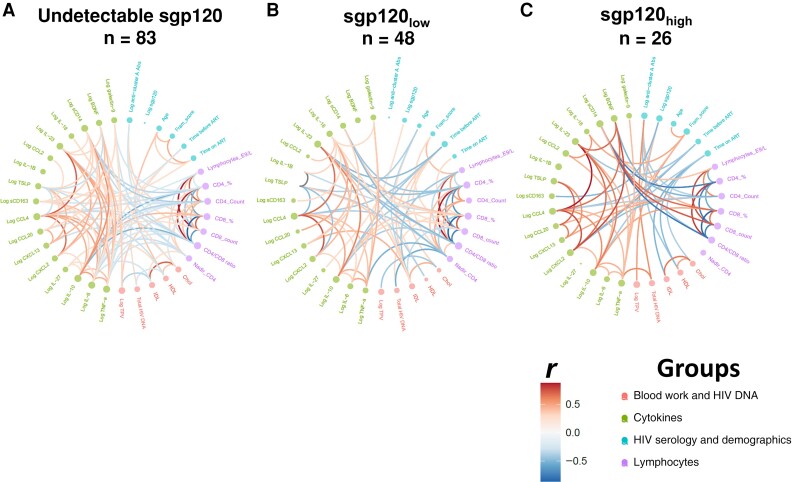
The HIV-1 gp120 is a pleiotropic molecule beyond its key role in viral entry [4, 7, 14, 15]. The presence of sgp120 was associated with increased levels of proinflammatory markers in the plasma of early and acute PLWH [7]. To test whether this observation could be extended to PLWH receiving long-term ART treatment, we performed multiplex measurements of various soluble markers associated with chronic inflammation (Supplementary Table 4) in a subset of 157 PLWH. Our results show that plasma levels of IL-6 are significantly higher in people with low or high levels of sgp120 than in the sgp120-undetectable group and uninfected controls (Figure 1B). Interestingly, we observed a positive correlation between sgp120 and TNF-α levels in the group with high sgp120 levels (Supplementary Table 3; r = 0.410, P = .04). Figure 3 shows the correlations between all inflammatory biomarkers measured by multiplex platform, as a function of detectable levels of sgp120. Edge bundling correlation plots show the effect of sgp120 on the network of associations between laboratory and clinical markers. While the undetectable group has weak associations among the parameters measured (Figure 3A), the presence of low (Figure 3B) or high (Figure 3C) levels of sgp120 intensifies the network of associations. Notably, the inverse association between soluble CD14 with lymphocyte counts and CD4/CD8 ratios is strengthened in the presence of sgp120. These results suggest that sgp120 acts as an effect modifier, modulating the associations among the different biomarkers analyzed in this study.
Figure 3.
Soluble glycoprotein 120 (sgp120) acts as an “effect modifier” of the associations between clinical and laboratory markers. Its presence modifies the network of associations between clinical and laboratory parameters in people living with human immunodeficiency virus (HIV) with undetectable viremia on stratification by undetectable sgp120 (A), low levels of sgp120 (sgp120low) (B), or high levels of sgp120 (sgp120high) (C). Spearman rank correlations were computed and graphed as edge bundling correlation plots, using program R v. 4.1.2. Statistical tests were 2 sided, and differences were considered significant at P < .05. Edges are only shown if P < .05, and nodes are sized according to the connecting edges’ r values. Nodes are color coded according to the grouping of variables. Abbreviations: Abs, antibodies; ART, antiretroviral-therapy; BDNF, brain-derived neurotrophic factor; Chol, cholesterol; Fram score, Framingham risk score; HDL, high-density lipoproteins; IL-1β, interleukin 1β; LDL, low-density lipoproteins; sCD14, soluble CD14; sCD163, soluble CD163; TNF, tumor necrosis factor; TPV, total atherosclerotic plaque volume; TSLP, thymic stromal lymphopoietin.
Association of sgp120 and Anti-Cluster A Antibodies with the Size of Coronary Artery Plaque in Participants With Subclinical CVD and Detectable sgp120
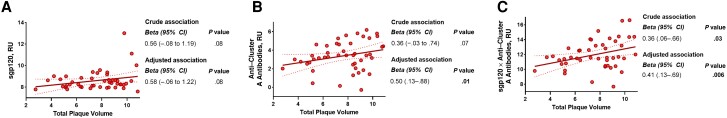
In the subgroup of 145 participants with available cardiovascular imaging, 97 (67%) presented with ≥1 coronary artery plaque measurable on CCTA, defining subclinical CVD. Levels of sgp120 (adjusted odds ratio, 1.04 [95% CI, 0.96–1.12]), anti–cluster A antibodies (1.01 [0.82–1.24), or a score defined by multiplying the levels of sgp120 and anti–cluster A antibodies (1.03 [0.96–1.1]) were not associated with the presence or absence of CVD. However, among the 46 individuals with detectable subclinical CVD and detectable sgp120, we found an association between the size of the coronary artery plaques and anti–cluster A antibodies (P = .01), as well as of the multiplicative score of anti–cluster A antibodies with levels of sgp120 (P = .006) (Figure 4 and Supplementary Table 2), after adjusting for potential confounders (age, sex, hypertension, diabetes mellitus, lipid levels, smoking status, alcohol use disorder, intravenous drug use, and duration of ART). We repeated those analyses using the low attenuation fraction of coronary artery plaque volume (a marker of high-risk plaque) as opposed to the total volume of coronary artery plaque, and the associations were unchanged (Supplementary Table 2).
Figure 4.
The combination of soluble glycoprotein 120 (sgp120) levels and anti–cluster A antibodies correlates positively with subclinical cardiovascular disease. Associations are displayed between the size of coronary artery plaque volume and sgp120 levels (A), anti–cluster A antibodies (B), and the multiplicative combination of both (C) in people living with human immunodeficiency virus who are positive for sgp120 and have detectable subclinical cardiovascular disease. Values for sgp120, anti–cluster A antibodies, and total plaque volume (mm3) were log2 transformed. Univariable and multivariable linear regressions were performed, with the beta parameters representing the mean predicted change in log total plaque volume for each 1-log2 increase in the exposure. Multivariable models are adjusted for age, sex, smoking status, low- or high-density lipoproteins, diabetes mellitus, hypertension, alcohol use disorder, intravenous drug use, and duration of antiretroviral therapy. Abbreviations: CI, confidence interval; RU, relative units.
Longitudinal Analysis of sgp120
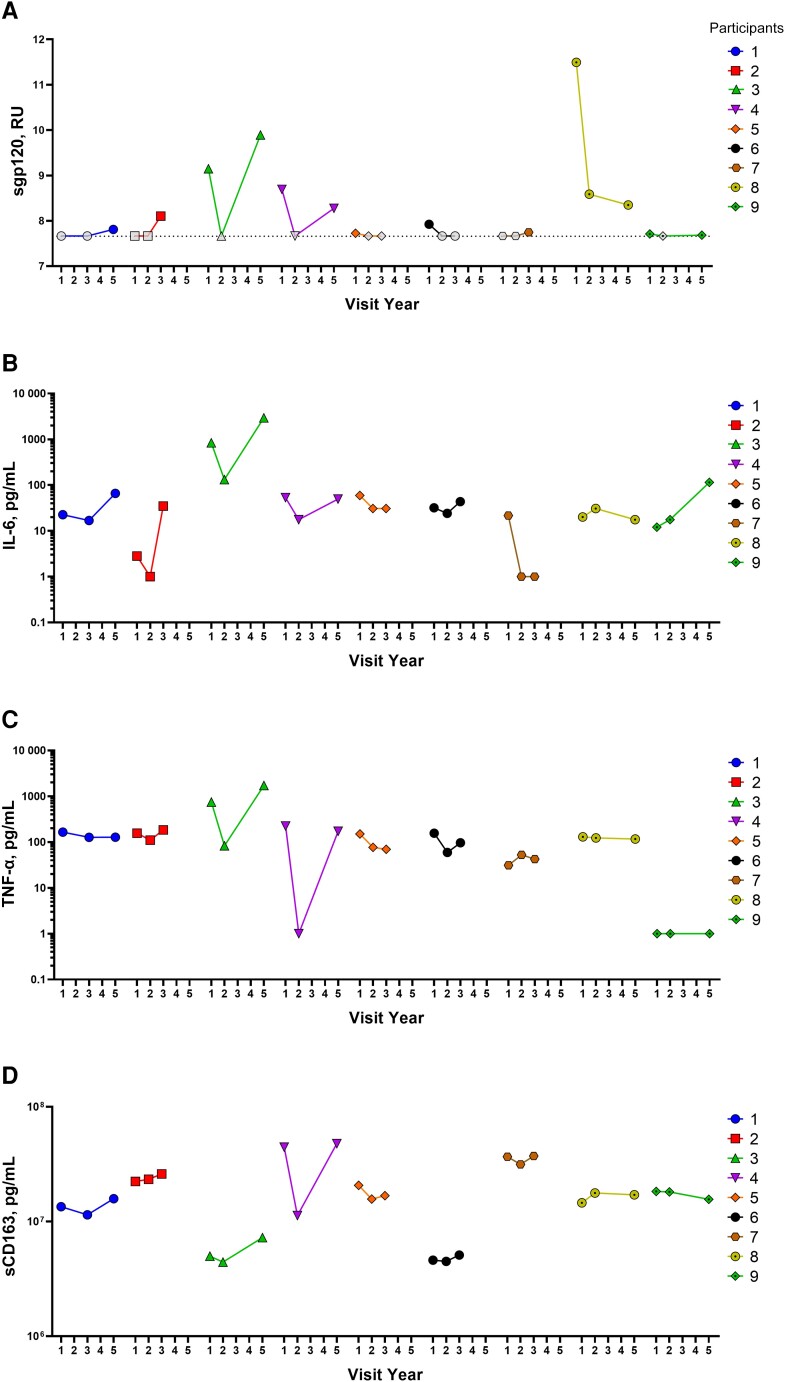
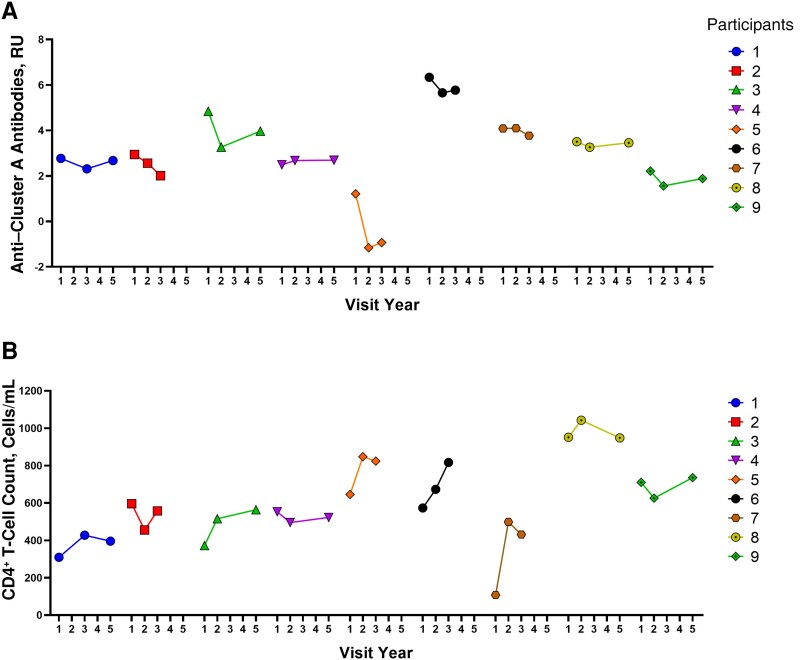
We analyzed plasma samples from 9 sgp120-positive PLWH in the CHACS cohort with no viremia. These individuals had 3 sample visits ranging from 1 to 5 years after the baseline assessment. We measured sgp120, anti–cluster A antibodies, and 3 inflammatory markers (IL-6, TNF-α, and soluble CD163 [sCD163]). We found that sgp120 levels are dynamic (Figure 5A). Interestingly, IL-6, TNF-α and sCD163 followed a similar pattern to that of sgp120 in most participants (Figure 5B–5D). While this does not prove causality, it suggests an association among these markers and sgp120. Further supporting our hypothesis that anti–cluster A antibodies are negatively associated with CD4+ T-cell counts, we observed opposite trends between anti–cluster A Abs and CD4+ T-cell counts in 6 of these 9 participants (Figure 6A and 6B).
Figure 5.
Longitudinal analysis of soluble glycoprotein 120 (sgp120) and some inflammatory markers. Levels of sgp120 (A), interleukin 6 (IL-6) (B), tumor necrosis factor (TNF) α (C), and soluble CD163 (sCD163) (D) were measured over time for 9 participants with detectable levels of sgp120. Dashed line in A represents seropositivity threshold established in plasma samples from uninfected participants. Abbreviation: RU, relative units.
Figure 6.
Longitudinal analysis of anti–cluster A antibodies and CD4+ T-cell counts. Levels of anti–cluster A antibodies (A) and CD4+ T-cell counts (B) were measured over time for 9 participants with detectable levels of soluble glycoprotein 120 (sgp120). Abbreviation: RU, relative units.
DISCUSSION
In this cross-sectional study, nested within a large observational cohort, we explored whether sgp120 could be associated with chronic inflammation and CVD in PLWH receiving long-term ART treatment. We demonstrated that sgp120 was detectable in the plasma of 107 of 386 participants (28%) with undetectable viremia and that these participants had more pronounced immune dysfunction associated with the presence of anti–cluster A antibodies, as well as distinct soluble inflammatory marker profiles. Longitudinal samples enabled us to evaluate sgp120 levels over time. We found that sgp120 levels are modulated over time and overall are positively associated with IL-6, TNF-α, and sCD163 but negatively associated with CD4+ T-cell counts. In addition, for those with detectable coronary atherosclerosis, anti–cluster A antibodies and their combination with sgp120 were associated with a significant increase in the size of coronary atherosclerotic plaques. Taken together, these results suggest that gp120 might represent a targetable pathway to decrease immune dysfunction, chronic inflammation, and CVD risk in PLWH.
We optimized an ELISA to detect sgp120 using C11, a monoclonal antibody recognizing a highly conserved gp120 region that is buried at the Env trimer interface and gets exposed on dissociation of the gp120 from gp41 [26], thus recognizing sgp120. We used the N6 broadly neutralizing antibody known to recognize 98% of global HIV-1 strains [28] as a readout. The use of a detection antibody targeting the CD4BS enables the detection of sgp120 which has the potential of binding the CD4 receptor whose interaction has been associated with immune activation and inflammation in vitro [4, 14, 15]. While sgp120 alone was not significantly associated with most clinical parameters measured, stratification of 386 PLWH based on sgp120 levels unveiled a striking inverse correlation between the levels of anti–cluster A antibodies with CD4+ cell count and CD4/CD8 ratio. These associations remained after adjustment for potential confounders (including age, sex, ethnicity, smoking status, nadir CD4, duration of ART, and levels of anti-CD4BS antibodies). This in vivo association is consistent with previously well-established in vitro evidence showing that binding of sgp120 to uninfected bystander CD4+ T cells leads to the death of these cells via ADCC on recognition by HIV-positive plasma [15–17].
We also observed a significant increase in IL-6 in individuals presenting detectable levels of sgp120. On one hand, plasma levels of IL-6 have been reported to be independently associated with frailty [32] as well as serious non-AIDS conditions or death among PLWH with undetectable viremia [33]. On the other hand, in vitro studies demonstrated that sgp120 binding to the CD4 receptor or CCR5/CXCR4 coreceptors on immune cells induces the liberation of inflammatory markers [4, 14, 15]. Together, these data suggest that sgp120 could contribute to the development of comorbid conditions by inducing proinflammatory cytokines in vivo. While we cannot establish a direct causal relationship between plasma sgp120 and the variation in inflammatory markers or the CD4+ T-cell count, our longitudinal samples strongly suggest an association between sgp120 and these markers over time.
Anti–cluster A antibodies, present in all PLWH [34, 35], and sgp120 were shown to eliminate uninfected bystander CD4+ T cells by ADCC in vitro. Here we extend these results by showing an association with CD4+ T-cell counts in a subset of PLWH (those presenting high levels of sgp120). Furthermore, the combination of these 2 parameters was associated with the size of atherosclerotic plaques, which defines subclinical CVD [20, 23], and this association remained after adjustment for potential confounding factors (age, sex, hypertension, diabetes mellitus, lipid levels, smoking status, alcohol use disorder, intravenous drug use, and duration of ART). This suggests that the levels of sgp120 and anti–cluster A antibodies could contribute to the development of CVD, which could be linked to depletion of uninfected bystander CD4+ cells, as it was shown that a low CD4/CD8 ratio was predictive of the presence of CVD in PLWH receiving ART [36, 37].
Notably, we propose that sgp120 acts as an “effect modifier,” potentially driving immune dysfunction, chronic inflammation, and the development of premature comorbid conditions. Thus, small molecules targeting gp120 could potentially “detoxify” its immunomodulatory effect and alleviate the development of premature comorbid conditions. One such molecule is the recently Food and Drug Administration–approved small-molecule gp120 inhibitor fostemsavir, a prodrug of temsavir (TMR), a novel attachment inhibitor that can prevent Env-CD4 interaction (BMS-663068, GSK3684934, Rukobia) and is currently used in combination with other ART for adults with multidrug-resistant HIV-1 [38, 39]. Intriguingly, many PLWH receiving fostemsavir exhibit clinical benefits that go beyond its capacity to decrease viral loads below detection levels [38, 39]. Our group has recently shown that TMR reduces the release of gp120 and that shed gp120 produced from TMR-treated cells has decreased capacity to interact with uninfected bystander CD4+ T cells, protecting these cells from ADCC responses and sgp120-mediated cytokine burst in human PBMCs [15]. Whether the use of TMR in treated PLWH presenting high levels of sgp120 could help alleviate sgp120-mediated chronic inflammation and immune dysfunction is yet to be shown.
The source of circulating sgp120 in long-term ART-treated PLWH with undetectable viral loads is not well understood. Although ART suppresses viral replication, it does not prevent proviral gene expression from the viral reservoir [40]. As such, a fraction of cells harboring intact or defective proviruses that persist during ART have been found to be transcriptionally competent, leading to the production of viral proteins and activation of the immune system [41, 42]. Interestingly, defective proviruses able to produce env transcripts have been found in a subset of PLWH receiving ART [43], raising the possibility that translation-competent proviruses could contribute to the production and release of sgp120, even during ART.
Accordingly, we did observe a significant association between the levels of HIV DNA and the combination of sgp120 and anti–cluster A antibodies (Figure 3 and Supplementary Table 3), although sgp120 alone was not significantly associated. Whether this is due to the limited number of individuals with detectable sgp120 in our cohort remains to be determined. Alternatively, the lack of association between sgp120 alone and the reservoir might be due to the fact that measures of HIV reservoir were performed in the blood and not in the tissues, which may be a preferential site for persistence of the active reservoir [44]. Finally, it has been demonstrated that the size of the viral reservoir in PLWH receiving ART was associated with subclinical CVD [25, 45]; whether this association is linked to the persistence of sgp120 in those individuals remains to be determined. Future studies are needed to elucidate the mechanism by which sgp120 persists over time and whether it may represent a potential therapeutic target to reduce the risk of early-onset comorbid conditions for PLWH.
Limitations of our study include its cross-sectional design, the underrepresentation of women, the reliance on subgroups in whom data was available for CVD assessment, and limited power for analysis of the sgp120 strata. While reverse causality is unlikely (CVD could not cause the presence of sgp120), a causal relationship cannot be inferred from this design, and replication of findings in independent populations is needed.
In conclusion, in this longitudinal and cross-sectional study, nested within a large cohort study of adults aging with HIV infection with undetectable viral load, we demonstrate that sgp120 is detectable in about one-third of participants, and that along with levels of anti–cluster A antibodies, it modulates immune and inflammation profiles. This may represent a novel treatment target for PLWH, to address the risk of inflammation-related comorbid conditions.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Mehdi Benlarbi, Centre de Recherche du CHUM, Montréal, Québec, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Québec, Canada.
Jonathan Richard, Centre de Recherche du CHUM, Montréal, Québec, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Québec, Canada.
Catherine Bourassa, Centre de Recherche du CHUM, Montréal, Québec, Canada.
William D Tolbert, Infectious Disease Division, Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Carl Chartrand-Lefebvre, Department of Radiology, Radiation Oncology and Nuclear Medicine, Université de Montréal, Montreal, Québec, Canada.
Gabrielle Gendron-Lepage, Centre de Recherche du CHUM, Montréal, Québec, Canada.
Mohamed Sylla, Centre de Recherche du CHUM, Montréal, Québec, Canada.
Mohamed El-Far, Centre de Recherche du CHUM, Montréal, Québec, Canada.
Marc Messier-Peet, Centre de Recherche du CHUM, Montréal, Québec, Canada.
Camille Guertin, Centre de Recherche du CHUM, Montréal, Québec, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Québec, Canada.
Isabelle Turcotte, Centre de Recherche du CHUM, Montréal, Québec, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Québec, Canada.
Rémi Fromentin, Centre de Recherche du CHUM, Montréal, Québec, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Québec, Canada.
Myriam Maude Verly, Centre de Recherche du CHUM, Montréal, Québec, Canada.
Jérémie Prévost, Centre de Recherche du CHUM, Montréal, Québec, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Québec, Canada.
Andrew Clark, ViiV Healthcare, Global Medical Affairs, Middlesex, United Kingdom.
Walther Mothes, Department of Microbial Pathogenesis, Yale University School of Medicine, New Haven, Connecticut, USA.
Daniel E Kaufmann, Centre de Recherche du CHUM, Montréal, Québec, Canada; Division of Infectious Diseases, Department of Medicine, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland.
Frank Maldarelli, HIV Dynamics and Replication Program, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Nicolas Chomont, Centre de Recherche du CHUM, Montréal, Québec, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Québec, Canada.
Philippe Bégin, Section of Allergy, Immunology and Rheumatology, Department of Pediatrics, CHU Sainte-Justine, Montréal, Québec, Canada; Department of Medicine, Faculty of Medecine, Centre Hospitalier de l’Université de Montréal, Montréal, Québec, Canada.
Cécile Tremblay, Centre de Recherche du CHUM, Montréal, Québec, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Québec, Canada.
Jean-Guy Baril, Clinique de Médecine Urbaine du Quartier Latin, Montréal, Québec, Canada; Département de Médecine Familiale, Université de Montréal, Montréal, Québec, Canada.
Benoit Trottier, Clinique de Médecine Urbaine du Quartier Latin, Montréal, Québec, Canada; Département de Médecine Familiale, Université de Montréal, Montréal, Québec, Canada.
Sylvie Trottier, Département de microbiologie-infectiologie et d'immunologie, Centre de recherche du centre hospitalier universitaire de Québec, Université Laval, Québec, Canada.
Ralf Duerr, Vaccine Center, NYU Grossman School of Medicine, NewYork, New York, USA; Department of Medicine, NYU Grossman School of Medicine, NewYork, New York, USA; Department of Microbiology, NYU Grossman School of Medicine, NewYork, New York, USA.
Marzena Pazgier, Infectious Disease Division, Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Madeleine Durand, Centre de Recherche du CHUM, Montréal, Québec, Canada; Department of Medicine, Faculty of Medecine, Centre Hospitalier de l’Université de Montréal, Montréal, Québec, Canada.
Andrés Finzi, Centre de Recherche du CHUM, Montréal, Québec, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Québec, Canada.
Notes
Acknowledgments. The authors thank the Centre de Recherche du CHUM (CRCHUM) biosafety level 3 platform. The graphical abstract and Supplementary Figure 1 were prepared using illustrations from BioRender. The following reagents were obtained through the National Institutes of Health HIV Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease: N6 monoclonal antibody heavy and light chains expression vectors (contributed by Drs Jinghe Huang and Mark Connors) and resurfaced stabilized core 3 protein, recombinant from HEK293 cells, ARP-12042 (contributed by Drs Zhi-Yong Yang, Peter Kwong, Gary Nabel, and John Mascola).
Author contributions. Conceptualization: M. B., J. R., W. M., M. D., and A. F. Methodology: M. B., J. R., and A. F. Investigation, M. B., J. R., C. B., C. C. L., G. G. L., M. E. F., M. M. P., C. G., I. T., R. F., M. M. V., and J. P. Resources: M. B., J. R., C. B., W. D. T., G. G. L., M. S., M. E. F., N. C., M. P., M. D., and A. F. Formal analysis: M. B. and M. D. Visualization: M. B., R. D., M. D., and A. F. Supervision: M. B., D. E. K., F. M., N. C., P. B., R. D., M. P., M. D., and A. F. Funding acquisition: M. D. and A. F. Writing—original draft: M. B., M. D., and A. F. Writing—review and editing: M. B., J. R., C. B., W. D. T., C. C. L., G. G. L., M. E. F., M. M. P., C. G., I. T., R. F., M. M. V., J. P., A. C., D. E. K., F. M., N. C., P. B., C. T., J. G. B., B. T., S. T., R. D., M. P., M. D., and A. F.
Disclaimer. The views expressed in this manuscript are those of the authors and do not reflect the official policy or position of the Uniformed Services University, the US Army, the Department of Defense, or the US government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability. Data and reagents are available on request.
Financial support. This work was supported by the National Institutes of Health (grants R01 AI148379 and R01 AI150322 to A. F., R01 AI129769 to M. P. and A. F., R01AI176531 to A. F., and N. C. and R01 AI116274 to M. P.); the Fonds de Recherche Quebec Santé AIDS and Infectious Diseases Network (grant to J. R., M. D., and A. F.); the Canadian Institutes of Health Research (foundation grant 352417 and research team grant 422148 to A. F., research team grant on HIV and health living [HAL398643 CIHR-IRSC:0635001811] to the Canadian HIV and Aging Cohort Study, and fellowship to M. B.); the Canada Foundation for Innovation (grant 41027 to D. E. K., N. C., and A. F.); the Canadian Institutes of Health Research HIV Clinical Trial Network (in-kind support [grant CTN-272] and pilot study CTNPT 052 to the Canadian HIV and Aging Cohort Study); the Enterprise for Research and Advocacy to Stop and Eradicate HIV (ERASE; grant UM1AI164562); the Delaney AIDS Research Enterprise to Cure HIV (DARE; grant UM1AI164560); and the Fonds de Recherche du Québec- Santé (clinician-researcher salary award to M. D.). A. F. is the recipient of a Canada Research Chair on Retroviral Entry (grant RCHS0235 950–232424). C. T. is the Pfizer/Université de Montreal Chair on HIV Translational Research. Funding to pay the Open Access publication charges for this article was provided by CIHR and NIH.
References
- 1. Marcus JL, Leyden WA, Alexeeff SE, et al. Comparison of overall and comorbidity-free life expectancy between insured adults with and without HIV infection, 2000–2016. JAMA Netw Open 2020; 3:e207954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodés B, Cadiñanos J, Esteban-Cantos A, Rodríguez-Centeno J, Arribas JR. Ageing with HIV: challenges and biomarkers. eBioMedicine 2022; 77:103896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lederman MM, Funderburg NT, Sekaly RP, Klatt NR, Hunt PW. Residual immune dysregulation syndrome in treated HIV infection. Adv Immunol 2013; 119:51–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chirmule N, Pahwa S. Envelope glycoproteins of human immunodeficiency virus type 1: profound influences on immune functions. Microbiol Rev 1996; 60:386–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Del Corno M, Donninelli G, Varano B, Da Sacco L, Masotti A, Gessani S. HIV-1 gp120 activates the STAT3/interleukin-6 axis in primary human monocyte-derived dendritic cells. J Virol 2014; 88:11045–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nazli A, Kafka JK, Ferreira VH, et al. HIV-1 gp120 induces TLR2- and TLR4-mediated innate immune activation in human female genital epithelium. J Immunol 2013; 191:4246–58. [DOI] [PubMed] [Google Scholar]
- 7. Rychert J, Strick D, Bazner S, Robinson J, Rosenberg E. Detection of HIV gp120 in plasma during early HIV infection is associated with increased proinflammatory and immunoregulatory cytokines. AIDS Res Hum Retroviruses 2010; 26:1139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol 1991; 65:2119–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X, Mahony E, Holm GH, Kassa A, Sodroski J. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology 2003; 313:117–25. [DOI] [PubMed] [Google Scholar]
- 10. Finzi A, Xiang SH, Pacheco B, et al. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell 2010; 37:656–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Santosuosso M, Righi E, Lindstrom V, Leblanc PR, Poznansky MC. HIV-1 envelope protein gp120 is present at high concentrations in secondary lymphoid organs of individuals with chronic HIV-1 infection. J Infect Dis 2009; 200:1050–3. [DOI] [PubMed] [Google Scholar]
- 12. Oh SK, Cruikshank WW, Raina J, et al. Identification of HIV-1 envelope glycoprotein in the serum of AIDS and ARC patients. J Acquir Immune Defic Syndr 1992; 5:251–6. [PubMed] [Google Scholar]
- 13. Herbein G, Khan KA. Is HIV infection a TNF receptor signalling-driven disease? Trends Immunol 2008; 29:61–7. [DOI] [PubMed] [Google Scholar]
- 14. Levast B, Barblu L, Coutu M, et al. HIV-1 gp120 envelope glycoprotein determinants for cytokine burst in human monocytes. PLoS One 2017; 12:e0174550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Richard J, Prévost J, Bourassa C, et al. Temsavir blocks the immunomodulatory activities of HIV-1 soluble gp120. Cell Chem Biol 2023; 30:540–52.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Richard J, Veillette M, Ding S, et al. Small CD4 mimetics prevent HIV-1 uninfected bystander CD4+ T cell killing mediated by antibody-dependent cell-mediated cytotoxicity. EBioMedicine 2016; 3:122–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richard J, Prevost J, Baxter AE, et al. Uninfected bystander cells impact the measurement of HIV-specific antibody-dependent cellular cytotoxicity responses. mBio 2018; 9:e00358-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bonsignori M, Pollara J, Moody MA, et al. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol 2012; 86:11521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding S, Veillette M, Coutu M, et al. A highly conserved residue of the HIV-1 gp120 inner domain is important for antibody-dependent cellular cytotoxicity responses mediated by anti-cluster A antibodies. J Virol 2016; 90:2127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Durand M, Chartrand-Lefebvre C, Baril JG, et al. The Canadian HIV and Aging Cohort Study—determinants of increased risk of cardio-vascular diseases in HIV-infected individuals: rationale and study protocol. BMC Infect Dis 2017; 17:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park JK, Kim JY, Kwon HM, et al. Multidetector computed tomography for the evaluation of coronary artery disease; the diagnostic accuracy in calcified coronary arteries, comparing with IVUS imaging. Yonsei Med J 2014; 55:599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Z, Boldeanu I, Nepveu S, et al. In vivo coronary artery plaque assessment with computed tomography angiography: is there an impact of iterative reconstruction on plaque volume and attenuation metrics? Acta Radiol 2017; 58:660–9. [DOI] [PubMed] [Google Scholar]
- 23. Boldeanu I, Sadouni M, Mansour S, et al. Prevalence and characterization of subclinical coronary atherosclerotic plaque with CT among individuals with HIV: results from the Canadian HIV and Aging Cohort study. Radiology 2021; 299:571–80. [DOI] [PubMed] [Google Scholar]
- 24. Coutu M, Finzi A. HIV-1 gp120 dimers decrease the overall affinity of gp120 preparations for CD4-induced ligands. J Virol Methods 2015; 215–216:37–44. [DOI] [PubMed] [Google Scholar]
- 25. Turcotte I, El-Far M, Sadouni M, et al. Association between the development of subclinical cardiovascular disease and human immunodeficiency virus (HIV) reservoir markers in people with HIV on suppressive antiretroviral therapy. Clin Infect Dis 2023; 76:1318–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tolbert WD, Gohain N, Alsahafi N, et al. Targeting the late stage of HIV-1 entry for antibody-dependent cellular cytotoxicity: structural basis for env epitopes in the C11 region. Structure 2017; 25:1719–31.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Veillette M, Desormeaux A, Medjahed H, et al. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol 2014; 88:2633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang J, Kang BH, Ishida E, et al. Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 2016; 45:1108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tolbert WD, Gohain N, Veillette M, et al. Paring down HIV env: design and crystal structure of a stabilized inner domain of HIV-1 gp120 displaying a major ADCC target of the A32 region. Structure 2016; 24:697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sherburn R, Tolbert WD, Gottumukkala S, et al. Incorporating the cluster A and V1V2 targets into a minimal structural unit of the HIV-1 envelope to elicit a cross-clade response with potent Fc-effector functions. Vaccines (Basel) 2021; 9:975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu X, Yang ZY, Li Y, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010; 329:856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Falutz J. Frailty in people living with HIV. Curr HIV/AIDS Rep 2020; 17:226–36. [DOI] [PubMed] [Google Scholar]
- 33. Grund B, Baker JV, Deeks SG, et al. Relevance of interleukin-6 and D-dimer for serious non-AIDS morbidity and death among HIV-positive adults on suppressive antiretroviral therapy. PLoS One 2016; 11:e0155100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Veillette M, Coutu M, Richard J, et al. The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-infected individuals. J Virol 2015; 89:545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Decker JM, Bibollet-Ruche F, Wei X, et al. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 2005; 201:1407–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Castilho JL, Shepherd BE, Koethe J, et al. CD4+/CD8+ ratio, Age, and risk of serious noncommunicable diseases in HIV-infected adults on antiretroviral therapy. AIDS 2016; 30:899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fernández Soto J, Romero-Jiménez MJ, Alarcón García JC, Bonet Estruch E, Sánchez Ramos JL, Castaño López M. Predictors of subclinical atherosclerosis in HIV. BMC Infect Dis 2023; 23:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lataillade M, Lalezari JP, Kozal M, et al. Safety and efficacy of the HIV-1 attachment inhibitor prodrug fostemsavir in heavily treatment-experienced individuals: week 96 results of the phase 3 BRIGHTE study. Lancet HIV 2020; 7:e740–e51. [DOI] [PubMed] [Google Scholar]
- 39. Kozal M, Aberg J, Pialoux G, et al. Fostemsavir in adults with multidrug-resistant HIV-1 infection. N Engl J Med 2020; 382:1232–43. [DOI] [PubMed] [Google Scholar]
- 40. Cohn LB, Chomont N, Deeks SG. The biology of the HIV-1 latent reservoir and implications for cure strategies. Cell Host Microbe 2020; 27:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Imamichi H, Smith M, Adelsberger JW, et al. Defective HIV-1 proviruses produce viral proteins. Proc Natl Acad Sci U S A 2020; 117:3704–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pollack RA, Jones RB, Pertea M, et al. Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe 2017; 21:494–506.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuniholm J, Armstrong E, Bernabe B, et al. Intragenic proviral elements support transcription of defective HIV-1 proviruses. PLoS Pathog 2021; 17:e1009982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Busman-Sahay K, Starke CE, Nekorchuk MD, Estes JD. Eliminating HIV reservoirs for a cure: the issue is in the tissue. Curr Opin HIV AIDS 2021; 16:200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McLaughlin MM, Ma Y, Scherzer R, et al. Association of viral persistence and atherosclerosis in adults with treated HIV infection. JAMA Netw Open 2020; 3:e2018099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.