Abstract
Objective.
Glutaminase (GLS) isoenzymes GLS1 and GLS2 catalyze the first step of glutaminolysis. GLS1 is requisite for Th17 cell differentiation, and its inhibition suppresses autoimmune disease in animals, but the function of GLS2 is not known. The aim of this study was to investigate the role of GLS2 in CD4+ T cell function and systemic lupus erythematosus (SLE) pathogenesis.
Methods.
We measured reactive oxygen species (ROS) levels, lipid peroxidation, and mitochondrial mass and polarization by flow cytometry, interleukin-2 (IL-2) production by a dual luciferase assay, and CpG DNA methylation of Il2 by a real-time polymerase chain reaction system. The impact of the overexpression of wild-type GLS1, wild-type GLS2, or mutated GLS2 at the PDZ domain–binding motif in CD4+ T cells was examined. Furthermore, GLS2 expression in CD4+ T cells from lupus-prone mice and patients with SLE was analyzed by Western blotting.
Results.
GLS2, but not GLS1, reduced ROS levels and lipid peroxidation and restored mitochondrial function in T cells. GLS2 promoted IL-2 production through the demethylation of the Il2 promoter. Mutation of the PDZ domain–binding motif abated the ability of GLS2 to regulate IL-2 and ROS levels. In lupus-prone mice and patients with SLE, the expression of GLS2 was decreased in CD4+ T cells. Finally, GLS2 overexpression corrected ROS levels and restored IL-2 production by CD4+ T cells from lupus-prone mice and SLE patients.
Conclusion.
Our findings suggest that GLS2 has a crucial role in IL-2 production by CD4+ T cells by supporting antioxidant defense, and they offer a new approach to correcting IL-2 production by T cells in SLE.
INTRODUCTION
Glutaminase (GLS) isoenzymes GLS1 and GLS2 are the first enzymes in the glutaminolysis pathway. Although both GLS1 and GLS2 convert glutamine into glutamate, these enzymes affect cell biology in distinct ways. Previous studies of various cancer cell lines have shown that the up-regulation of GLS1 promotes tumorigenesis, while the expression of GLS2 is linked to quiescent or differentiated cell states (1). Recently, we and other investigators have observed that GLS1 is essential for Th17 cell differentiation and that pharmacologic inhibition or genetic deletion of GLS1 ameliorated disease progression in lupus-prone MRL/lpr mice (2). GLS2, though, has been shown to decrease reactive oxygen species (ROS) levels through the glutathione (GSH)–dependent antioxidant system in various cancer cell lines (1). This observation is of interest because ROS have been shown to affect T cell signaling and impact T cell fate in healthy subjects and patients with systemic lupus erythematosus (SLE) (3). However, the impact of GLS2 on T cell function in SLE has not been addressed.
SLE is an autoimmune disease in which the immune system attacks its own tissues, causing widespread inflammation and tissue damage that is linked to significant morbidity and mortality (4). Recent studies have shown that insufficient production of interleukin-2 (IL-2) by effector CD4+ T cells contributes to the pathogenesis of SLE (4) and that treatment with low-dose IL-2 offers clinical benefit for a wide range of autoimmune diseases, including SLE (5). However, because IL-2 has a narrow therapeutic range and a short half-life, discovery of additional molecular pathways that would enable restoration of IL-2 production is desirable.
Here we report that GLS2, but not GLS1, reduces ROS levels and promotes the ability of CD4+ T cells to produce IL-2 by demethylating Il2. At the translational level, we demonstrate that GLS2 protein expression is decreased in CD4+ T cells from lupus-prone mice and patients with SLE and that overexpression of GLS2 reduces ROS levels and restores IL-2 production.
MATERIALS AND METHODS
Study participants.
Patients who fulfilled the American College of Rheumatology criteria for the diagnosis of SLE (6) and age-, sex-, and ethnicity-matched healthy individuals were enrolled after they provided informed consent. The Beth Israel Deaconess Medical Center (BIDMC) Institutional Review Board approved the study protocol (2006-P-0298). Demographic and clinical data and sample collection methods are described in Supplementary Table 1 and the Supplementary Methods, respectively, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.42112.
Mice.
C57BL/6J mice, MRL/MpJ-Faslpr/J (MRL/lpr) mice, and MRL/MpJ mice were purchased from The Jackson Laboratory. For in vitro experiments, C57BL/6J mice were euthanized at 6–8 weeks of age. MRL/lpr and MRL/MpJ mice were euthanized at 16 weeks of age. All mice were maintained in a specific pathogen–free animal facility (BIDMC). Experiments were approved by the Institutional Animal Care and Use Committee of BIDMC.
In vitro T cell stimulation.
Mouse or human naive CD4+ T cells and mouse or human CD4+ T cells were purified using specific T cell isolation kits (the mouse/human naive CD4+ T Cell Isolation Kit II and the mouse/human CD4+ T Cell Isolation Kit, respectively; Miltenyi Biotec). Purified mouse naive CD4+ T cells and mouse CD4+ T cells in RPMI 1640 medium with 10% fetal bovine serum, penicillin/streptomycin, and 2-mercaptoethanol were stimulated at 37°C with plate-bound goat anti-hamster antibodies crosslinking anti-CD3 (0.25 μg/ml; BioLegend) and anti-CD28 (0.5 μg/ml; BioXcell). Purified human naive CD4+ T cells and human CD4+ T cells in RPMI 1640 medium with 10% fetal bovine serum plus penicillin/streptomycin were stimulated with plate-bound anti-CD3 antibodies (1 μg/ml; OKT-3; BioXCell) and anti-CD28 antibodies (1 μg/ml; CD28.2; BioLegend) at 37°C.
Enzyme-linked immunosorbent assay (ELISA).
The Max Deluxe Set Mouse IL-2 ELISA platform (BioLegend) was used to detect IL-2 in culture supernatants. All procedures were performed according to the manufacturer’s instructions. Each assay was performed in duplicate independently.
Western blotting.
Cell lysates were separated on NuPAGE 4–12% Bis-Tris gel (ThermoFisher Scientific), and proteins were transferred to a nitrocellulose membrane. The following antibodies were used for protein detection on the ECL Western blot system (Cytiva): anti-GLS2 (Abcam), anti–β-actin (Sigma-Aldrich), horseradish peroxidase (HRP)–conjugated rabbit anti-goat IgG (R&D Systems), and HRP-conjugated goat anti-mouse IgG (Abcam). Bands on blots corresponding to proteins of interest were analyzed by ImageJ software (National Institutes of Health).
Flow cytometry.
The following antibodies were used for flow cytometry: CD90.2 (clone 53-2.1; BioLegend) and CD4 (clone GK1.5; ThermoFisher Scientific) for murine lymphocytes and CD3 (clone SK7; BioLegend) and CD4 (clone SK3; ThermoFisher Scientific) for human lymphocytes. Staining was performed using the Zombie UV Fixable Viability Kit (BioLegend) to eliminate dead cells. After cell surface markers were stained, CellRox Green Reagent (ThermoFisher Scientific) was used to detect ROS, Liperfluo (Dojindo) was used to detect lipid peroxidation, and staining with MitoTracker Deep Red and MitoTracker Green (Life Technologies) was performed to determine mitochondrial mass and polarization, respectively. In all transfection experiments, DsRed+CD4+ T cells were analyzed. The mean fluorescence intensity of DsRed expression in each experiment is shown in Supplementary Figure 1, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.42112. All procedures were performed according to manufacturers’ instructions. Detailed methods for cell staining are described in the Supplementary Methods.
Transfection of GLS2- and GLS2-overexpression vectors.
Mouse Gls1 and Gls2, human GLS1, or human GLS2 sequences were subcloned into the pIRES2-DsRed-Express vector by GenScript. All constructs were verified by DNA sequencing. The plasmids were transfected by means of the Amaxa mouse/human T cell Nucleofector kit, using the X-001 program on day 1 (for murine cells) and the T-023 program on day 2 (for human cells) of culture (Lonza).
Luciferase assay.
Murine Il2 and human IL2 promoter luciferase reporter constructs (pGL3_mIl2_vector and pGL3_hIL2_vector, respectively) were purchased from GenScript. The luciferase reporter plasmids were transfected as mentioned in the previous subsection. Each reporter experiment included 200 ng of Renilla luciferase construct as an internal control. Luciferase activity was quantified using a dual luciferase assay (Promega) on day 3 of culture, according to the manufacturer’s instructions.
Methylated CpG (methyl-CpG) DNA immunoprecipitation.
The methyl-CpG DNA immunoprecipitation assay (Zymo Research) was performed according to the manufacturer’s instructions. The detailed methods for obtaining methylated DNA are described in the Supplementary Methods, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.42112. Methylated DNA was subjected to polymerase chain reaction (PCR) analysis on an ABI OneStepPlus real-time PCR system. Sequences of PCR primers are listed in Supplementary Table 2.
Site-directed mutagenesis.
Site-directed mutagenesis for substituting the PDZ domain–binding motif in the mouse GLS2-overexpression vector with the corresponding sequence in the mouse Gls1 gene was performed using the Q5 site-directed mutagenesis kit (New England Biolabs) with the following primers: 5′-TTGCTATAAGGATCCGCCCCTCTCCCT-3′ and 5′-CCCGTCGAGATTCTCTTTGGACAGGGTCTCAGC-3′.
Statistical analysis.
Comparisons between 2 different groups were conducted using Student’s unpaired 2-tailed t-tests or, if the groups were related, Student’s paired t-tests. Comparisons between >2 groups were conducted using one-way analysis of variance with Tukey’s post hoc test for multiple comparisons. Statistical analyses were performed with GraphPad Prism 7.0 software (GraphPad Software). P values less than 0.05 were considered statistically significant.
RESULTS
Epigenetic influence of GLS2 on ROS levels and IL-2 production in murine CD4+ T cells.
GLS2 has been shown to decrease ROS levels in several cancer cell lines (1). To assess whether GLS2 reduces ROS levels in CD4+ T cells, naive CD4+ T cells from wild-type B6 mice were stimulated with anti-CD3 and anti-CD28 antibodies and transfected with either empty, GLS1-overexpression, or GLS2-overexpression plasmid vectors. As shown in Figure 1A, overexpression of GLS2, but not of GLS1, significantly reduced intracellular ROS levels. High levels of ROS cause lipid peroxidation (7) and eventually mitochondrial dysfunction (8). As expected, GLS2 overexpression decreased lipid peroxidation in CD4+ T cells (Figure 1B). A combination of MitoTracker Deep Red and MitoTracker Green staining revealed that GLS2 overexpression in CD4+ T cells was associated with a decreased percentage of CD4+ T cells with depolarized (i.e., dysfunctional) mitochondria (Figure 1C).
Figure 1.
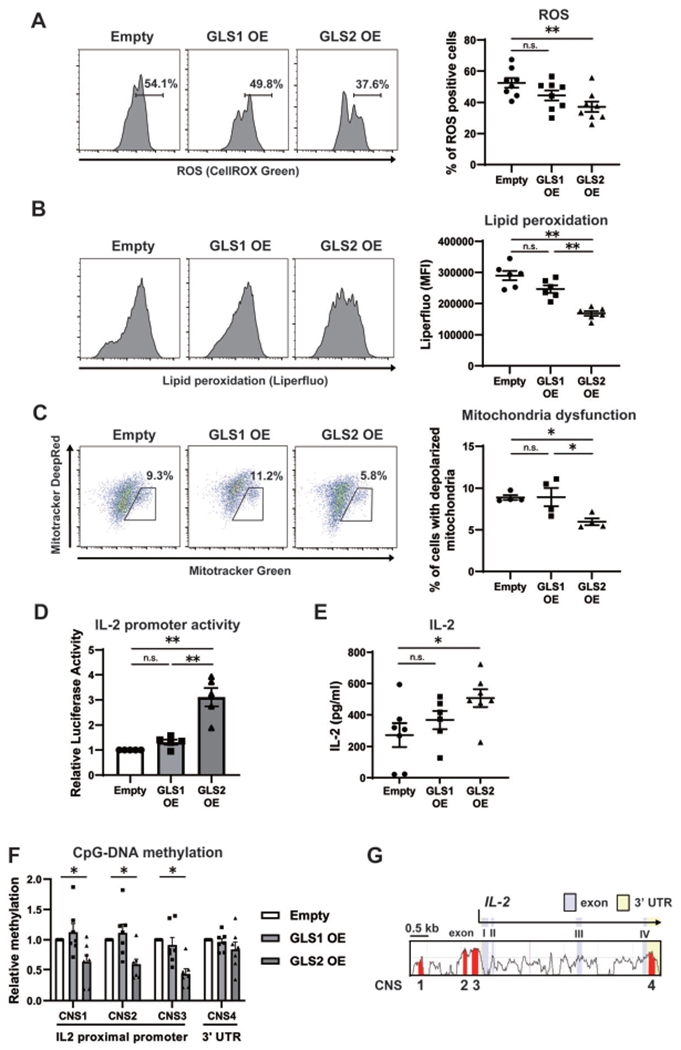
Epigenetic influence of glutaminase 2 (GLS2) on reactive oxygen species (ROS) levels and interleukin-2 (IL-2) production in murine CD4+ T cells. A-F, Naive CD4+ T cells from wild-type B6 mice were cultured with anti-CD3/anti-CD28 and transfected with empty vector (Empty), GLS1-overexpression plasmid (GLS1 OE), or GLS2-overexpression plasmid (GLS2 OE) on day 1. On day 3, CellRox Green Reagent was used to detect ROS (n = 8 samples per group) (A), Liperfluo was used to detect lipid peroxidation (n = 6 per group) (B), staining with MitoTracker Deep Red and Green was performed to assess mitochondrial polarization and mass, respectively (n = 4 per group) (C), a dual luciferase assay was used to assess IL-2 promoter activity (n = 5 per group) (D), an enzyme-linked immunosorbent assay was used to measure the IL-2 concentration in culture supernatants (n = 7 per group) (E), and an EZ DNA Methylation Kit was used to assess CpG DNA methylation (n = 7 per group) (F). In all transfection experiments, DsRed+CD4+ T cells were analyzed. Bars show the mean ± SEM; symbols represent individual samples. * = P < 0.05; ** = P < 0.01, by one-way analysis of variance with Tukey’s post hoc test for multiple comparisons. MFI = mean fluorescence intensity; NS = not significant. G, Alignment of the murine and human genes encoding IL-2 shows conserved noncoding sequences (CNS) (red) that were determined as regions of interest for further analysis of CpG DNA methylation. CNS1–CNS3 are located in the proximal promoter, which spans 2 kb, and CNS4 is located in the 3′-untranslated region (3′-UTR).
In a previous report, investigators showed that treatment with thiol N-acetylcysteine (NAC), a ROS scavenger and a precursor of GSH, reduced ROS levels and restored IL-2 production in T cells (9). Elsewhere, researchers found that thiol NAC suppressed SLE parameters when administered to patients with this disease (10). Overexpression of GLS2, but not of GLS1, significantly increased IL-2 promoter activity in CD4+ T cells and IL-2 concentrations in culture supernatants (Figures 1D and E). Because IL-2 expression is controlled by CpG DNA methylation of the Il2 promoter (11), we assessed CpG DNA methylation in Il2. We focused on 4 types of conserved noncoding sequence (CNS1–CNS4), which were identified (11) on the basis of the degree of sequence conservation and the presence of reported regulatory regions (Figure 1F). CNS1–CNS3 are located within the proximal promoter of Il2, whereas CNS4 localizes within the highly conserved 3′-untranslated regions. Overexpression of GLS2, but not of GLS1, resulted in significantly lower levels of CpG DNA methylation in CNS1, CNS2, and CNS3 but not in CNS4 (Figure 1G). Collectively, we have demonstrated that overexpression of GLS2, but not of GLS1, decreases ROS levels, lipid peroxidation, and mitochondrial dysfunction and results in increased IL-2 production by CD4+ T cells through demethylation of the Il2 promoter.
PDZ domain–binding motif and GLS2-mediated modulation of ROS levels and IL-2 production.
To understand how these differences between GLS1 and GLS2 occur, we focused on the structures of GLS1 and GLS2. The major amino acid sequences of GLS1 and GLS2 align closely, with most of the differences occurring in their C-termini and N-termini. In contrast to the C-terminus of GLS1, the C-terminus of GLS2 defines a PDZ domain–binding motif, which can bind to PDZ domain–containing proteins (12) (Figure 2A). Accordingly, we hypothesized that regulation of IL-2 and ROS by GLS2 depends on protein–protein interactions involving the PDZ domain. To investigate this, we generated a GLS2-overexpression vector in which the PDZ domain–binding motif was substituted with the corresponding sequence from GLS1 (Figure 2A). Intriguingly, mutation of the PDZ domain–binding motif abated the reduction in ROS levels (Figure 2B), lipid peroxidation (Figure 2C), and mitochondrial dysfunction (Figure 2D) and restored Il2 promoter activity (Figure 2E). Together, our results indicate that the PDZ domain–binding motif at the C-terminus mediates the effect of GLS2 on cellular ROS levels, mitochondrial functions, and, ultimately, IL-2 production.
Figure 2.
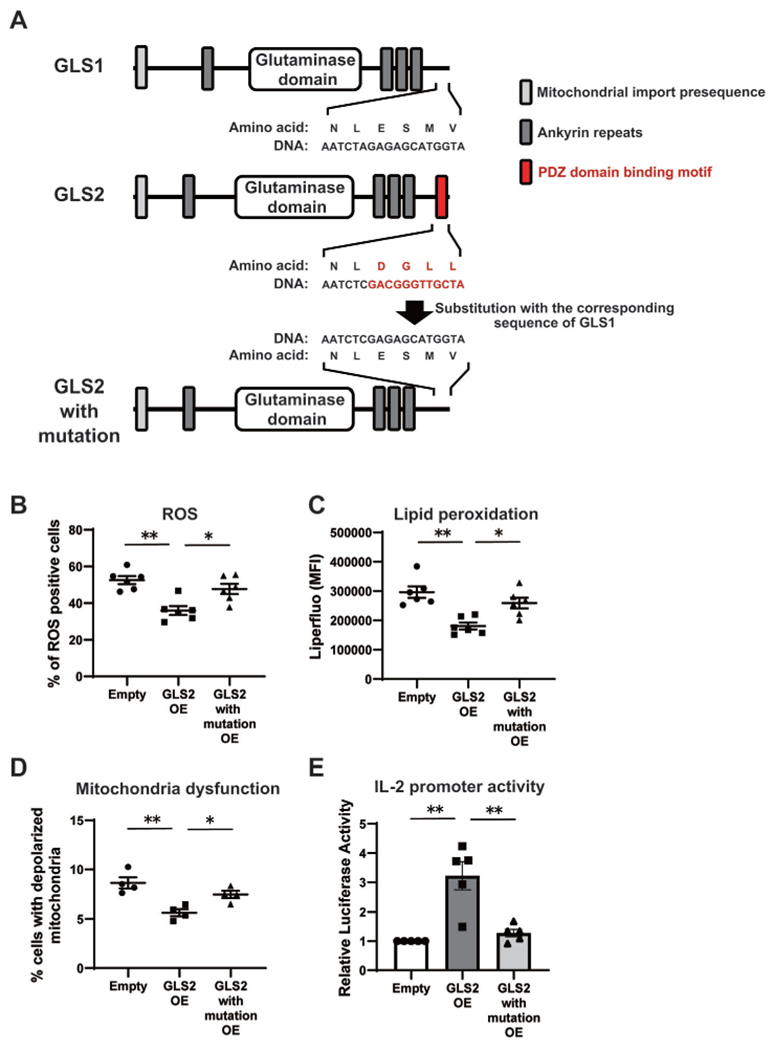
Role of the PDZ domain–binding motif in GLS2-mediated regulation of IL-2 and ROS levels in mice. A, Structures of GLS1 (isoform KGA), GLS2 (isoform GAB), and mutated GLS2, in which the PDZ domain–binding motif at the C-terminus (red) was substituted with that of GLS1, are shown. B–E, Naive CD4+ T cells from wild-type B6 mice were cultured with anti-CD3/anti-CD28 and transfected with empty vector, GLS2 OE, or mutated GLS2–overexpression plasmid (GLS2 with mutation OE) on day 1. On day 3, assessment of ROS (n = 6 samples per group) (B), lipid peroxidation (n = 6 per group) (C), mitochondrial polarization (n = 4 per group) (D), and Il2 promoter activity (n = 5 per group) (E) was performed. In all transfection experiments, DsRed+ CD4+ T cells were analyzed. Bars show the mean ± SEM; symbols represent individual samples. * = P < 0.05; ** = P < 0.01, by one-way analysis of variance with Tukey’s post hoc test for multiple comparisons. See Figure 1 for other definitions.
Effect of GLS2 on ROS levels and IL-2 production in CD4+ T cells from lupus-prone mice and patients with SLE.
Insufficient IL-2 production and higher cytoplasmic ROS levels in CD4+ T cells are key factors in the development of SLE (3,4). As shown in Figures 3A and B, GLS2 protein expression was decreased in CD4+ T cells isolated from MRL/lpr mice and patients with SLE, compared with expression in those from control MRL/MpJ mice and healthy individuals, respectively. Furthermore, there was no statistically significant difference in the expression of GLS2 in murine and human CD4+ T cells with or without CD3 and CD28 antibody stimulation (Supplementary Figure 2, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.42112), suggesting that the decreased expression of GLS2 in lupus T cells is not due to an increased frequency of activated T cells. Next, we transfected the GLS1- and GLS2-overexpression vectors into CD4+ T cells from MRL/lpr mice and patients with SLE. As expected, overexpression of GLS2, but not of GLS1, reduced ROS levels and restored IL-2 promoter activity in CD4+ T cells from MRL/lpr mice (Figures 3C and D) and patients with SLE (Figures 3E and F). These findings demonstrate that GLS2 corrects aberrant ROS levels and restores normal IL-2 production in SLE CD4+ T cells.
Figure 3.
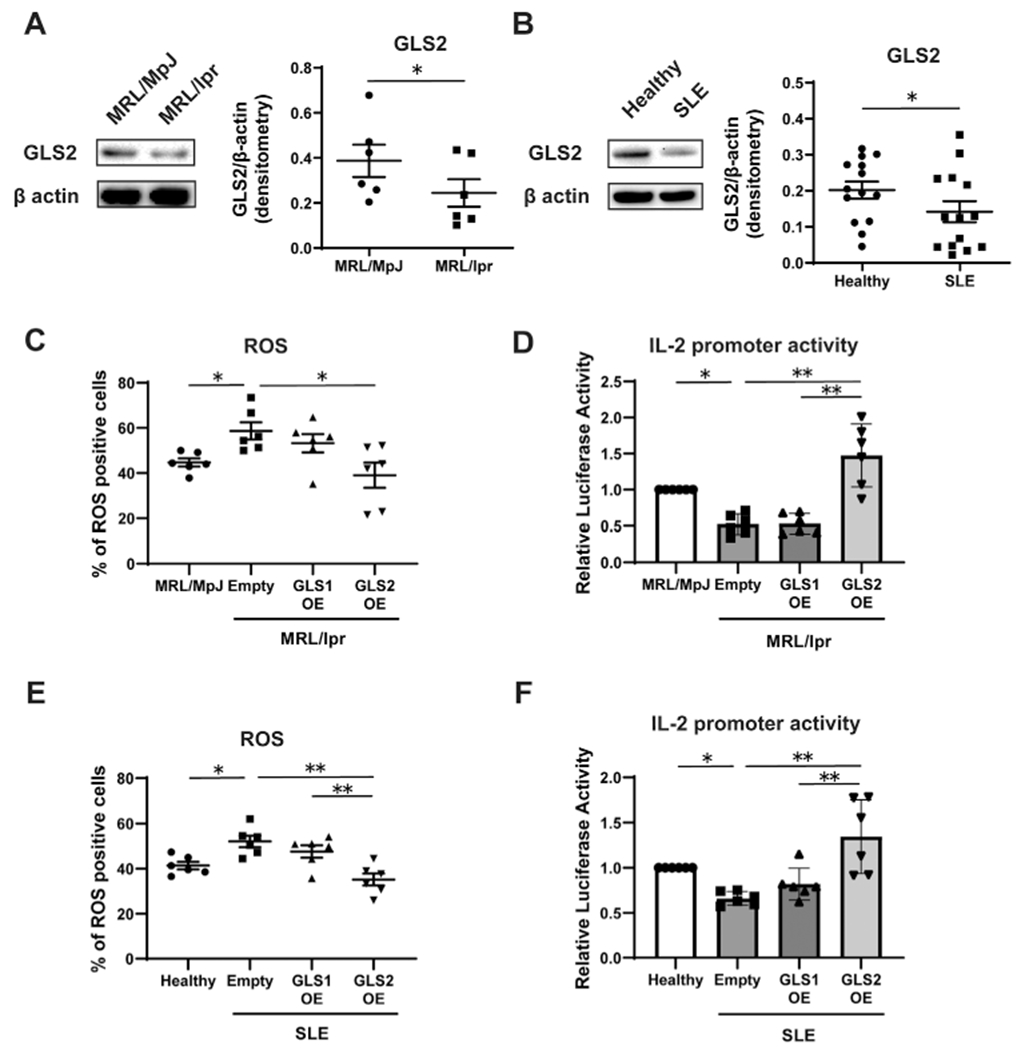
Effect of GLS2 on IL-2 production and ROS levels in CD4+ T cells from lupus-prone mice and patients with systemic lupus erythematosus (SLE). A and B, GLS2 and actin expression in purified CD4+T cells from 16-week-old MRL/MpJ-Faslpr/J (MRL/lpr) and MRL.MpJ (control) mice (n = 6 samples per group) (A) and healthy control participants and patients with SLE (n = 14 per group) (B) were analyzed by Western blotting. C–F, Naive CD4+ T cells from 16-week-old MRL.lpr mice, MRL.MpJ mice, healthy control participants, and patients with SLE were cultured with anti-CD3/anti-CD28. Cells from MRL.MpJ mice and healthy controls were transfected with empty vector, and cells from MRL.lpr mice (C and D) and patients with SLE (E and F) were transfected with empty vector, GLS1 OE, or GLS2 OE. On day 3, assessment of ROS (n = 6 per group) and Il2 and IL2 promoter activity (n = 6 per group) was performed. In all transfection experiments, DsRed+CD4+ T cells were analyzed. Bars show the mean ± SEM; symbols represent individual samples. * = P < 0.05; ** = P < 0.01, by Student’s paired t-test and one-way analysis of variance with Tukey’s post hoc test for multiple comparisons. See Figure 1 for other definitions.
DISCUSSION
In the current study, we demonstrated that GLS2 decreases ROS levels and restores IL-2 production through demethylation of the Il2 promoter and that the mechanism of this activity occurs at the PDZ domain–binding motif at the C-terminus of GLS2. We also observed that GLS2 corrects abnormal ROS levels and IL-2 production in CD4+ T cells from both lupus-prone MRL/lpr mice and patients with SLE.
Impaired IL-2 production and related signaling is considered a central defect in SLE because it is linked to a series of cellular abnormalities, and its restoration is expected to provide clinical benefit (3). Epigenetic events, including DNA methylation, contribute to the lineage commitment and function of helper T cells. Indeed, DNA demethylation in the IL2 promoter is a requisite for IL2 transcription initiation (13). Furthermore, we previously reported that CpG DNA methylation of the IL2 promoter is significantly increased in T cells from patients with active SLE (11).
High levels of cellular ROS have been linked to the development of a variety of diseases, including cancer and SLE (3). While moderate levels of ROS are necessary to promote T cell signaling, high levels of ROS can be detrimental to T cell survival. In fact, disruption in the GSH pathway, which causes excessive ROS levels, has been shown to reduce IL-2 production by T cells (14). In addition, cellular ROS themselves can induce specific hypermethylation by up-regulating DNA methyltransferases (DNMTs) (15). In previous studies, researchers observed that antioxidant treatment with thiol NAC reversed DNMT3a expression and DNA hypermethylation in cardiac tissues (16) and that forced expression of DNMT3a resulted in a significant increase in CpG DNA methylation of the IL2 promoter in primary human T cells (11). Because we showed that GLS2 is associated with a greater reduction in ROS levels, it is reasonable to hypothesize that GLS2 influences methylation of the IL2 promoter by regulating antioxidant capacity and the expression of DNMTs. Although further analysis is needed to confirm that ROS directly regulate DNA methylation in the IL2 promoter, we propose that GLS2 induces IL-2 production epigenetically by supporting antioxidant defense.
GLS1 and GLS2 exhibit a high degree of similarity in their amino acid sequences, particularly in the GLS enzymatic domain. However, studies of cancer cell lines have shown that GLS1 promotes tumor growth, while GLS2 suppresses tumor progression. Yet, it is unclear why GLS1 and GLS2 have contrasting roles. The main difference between GLS1 and GLS2 is the PDZ domain–binding motif at the C-terminus expressed by GLS2. We have found that the functional difference between the 2 isoforms is due to the presence of the PDZ domain–binding motif in the GLS2 isoenzyme, because its absence deprives GLS2 of its ability to suppress ROS levels and increase IL-2 production. Along this line, GLS2, but not GLS1, binds to small GTPase Rac1 and inhibits Rac1 activity, which in turn inhibits migration and metastasis of cancer cells through its C-terminus motif (17). Although both GLS isoenzymes generate glutamate, which is one of the precursors of the antioxidant GSH, we found that GLS2 is more effective than GLS1 in reducing ROS levels. This difference is linked to the presence of the PDZ domain–binding motif in the C-terminus of GLS2, which binds to still-unknown proteins that may affect ROS levels and GSH production. Furthermore, since PDZ domains often help tether receptors to cell membranes, GLS2 may alter cellular functions by interaction with tethering-related proteins.
In summary, we have presented a novel role for GLS2 in CD4+ T cell function. We showed that GLS2, through its PDZ domain–binding motif, reduces ROS levels, lipid peroxidation, and mitochondrial dysfunction and promotes IL-2 production through the demethylation of the Il2 promoter in CD4+ T cells. At a translational level, we demonstrated that GLS2 protein expression is down-regulated in CD4+ T cells from both lupus-prone MRL/lpr mice and patients with SLE and that GLS2 overexpression reduces ROS levels and restores IL-2 production. We propose that the GLS2-initated pathway represents a new therapeutic target in the treatment of SLE. It is important to confirm these results through further studies in vivo.
Supplementary Material
Acknowledgments
Dr. Hisada’s work was supported by the Uehara Memorial Foundation. Dr. Yoshida’s work was supported by the 2019 Gilead Sciences Research Scholars Program in Rheumatology. Dr. Scherlinger’s work was supported by the Société Française de Rhumatologie, Arthur-Sachs & Monahan fellowships, and the Philippe Foundation. Dr. Tsokos’ work was supported by the NIH (grant R37-AI-49954).
Footnotes
Author disclosures are available at https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fart.42112&file=art42112-sup-0001-Disclosureform.pdf.
REFERENCES
- 1.Wang Z, Liu F, Fan N, Zhou C, Li D, Macvicar T, et al. Targeting glutaminolysis: new perspectives to understand cancer development and novel strategies for potential target therapies [review]. Front Oncol 2020;10:589508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kono M, Yoshida N, Maeda K, Suárez-Fueyo A, Kyttaris VC, Tsokos GC. Glutaminase 1 inhibition reduces glycolysis and ameliorates lupus-like disease in MRL/lpr mice and experimental autoimmune encephalomyelitis. Arthritis Rheumatol 2019;71:1869–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perl A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus [review]. Nat Rev Rheumatol 2013;9:674–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsokos GC. Autoimmunity and organ damage in systemic lupus erythematosus. Nat Immunol 2020;21:605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharabi A, Tsokos MG, Ding Y, Malek TR, Klatzmann D, Tsokos GC. Regulatory T cells in the treatment of disease. Nat Rev Drug Discov 2018;17:823–44. [DOI] [PubMed] [Google Scholar]
- 6.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1982;25:1271–7. [DOI] [PubMed] [Google Scholar]
- 7.Su LJ, Zhang JH, Gomez H, Murugan R, Hong X, Xu D, et al. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid Med Cell Longev 2019;2019:5080843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu CC, Tseng LM, Lee HC. Role of mitochondrial dysfunction in cancer progression [review]. Exp Biol Med (Maywood) 2016;241:1281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadzic T, Li L, Cheng N, Walsh SA, Spitz DR, Knudson CM. The role of low molecular weight thiols in T lymphocyte proliferation and IL-2 secretion. J Immunol 2005;175:7965–72. [DOI] [PubMed] [Google Scholar]
- 10.Lai ZW, Hanczko R, Bonilla E, Caza TN, Clair B, Bartos A, et al. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: a randomized, double-blind, placebo-controlled trial. Arthritis Rheum 2012;64:2937–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedrich CM, Crispin JC, Rauen T, Ioannidis C, Apostolidis SA, Lo MS, et al. cAMP response element modulator α controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc Natl Acad Sci U S A 2012;109:16606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marquez J, Mates JM, Campos-Sandoval JA. Glutaminases. Adv Neurobiol 2016;13:133–71. [DOI] [PubMed] [Google Scholar]
- 13.Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J 2006;25:1081–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yarosz EL, Chang CH. The role of reactive oxygen species in regulating T cell-mediated immunity and disease. Immune Netw 2018;18:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kietzmann T, Petry A, Shvetsova A, Gerhold JM, Gorlach A. The epigenetic landscape related to reactive oxygen species formation in the cardiovascular system. Br J Pharmacol 2017;174:1533–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Gong L, Zhang P, Li Y, Liu B, Zhang L, et al. Epigenetic down-regulation of Sirt 1 via DNA methylation and oxidative stress signaling contributes to the gestational diabetes mellitus-induced fetal programming of heart ischemia-sensitive phenotype in late life. Int J Biol Sci 2019;15:1240–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Liu J, Zhao Y, Yue X, Zhu Y, Wang X, et al. Glutaminase 2 is a novel negative regulator of small GTPase Rac1 and mediates p53 function in suppressing metastasis. Elife 2016;5:e10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


