Abstract
This study addressed the cytotoxic potential of four compounds: monocarbonyl curcuminoid, ethyl (2E)-2-benzylidene-3-oxobutanoate 1, 1,2-dimethoxy-12-methyl-13H- [1,3] benzodioxolo[5,6-c] phenanthridine 2, 3,5-dibenzyloxybenzyl bromide 3, and (E)-4-(4-chlorobenzylidene)-1-(4-nitrophenyl)hexan-3-one 4. In vitro cytotoxic assays were carried out in HL-60 and BJ cells using the MTT assay along with analysis of apoptosis with the annexin V detection kit. Additional network pharmacology and docking analyses were carried out. In the in vitro assays, compounds 2 and 4 displayed significant antiproliferative effects in HL-60 cells, exhibiting IC50 values of 5.02 and 9.50 μM, respectively. Compound 1 showed no activity, and compound 3 displayed toxicity in BJ cells. In addition, both compounds 2 and 4 induced apoptosis in HL-60 cells. Network pharmacology and docking analyses indicated that compounds 2 and 4 had synergistic effects targeting the CASP3 and PARP1 proteins. Notably, these proteins play pivotal roles in cancer-related pathways. Thus, by modulating these proteins, monocarbonyl curcuminoid has the potential to influence various cancer-related pathways. In summary, our novel findings provide valuable insights into the potential of these compounds to serve as novel anticancer therapeutic agents, warranting further mechanistic studies and clinical exploration.
1. Introduction
The second most common cause of death in the US is cancer, representing as well one of the largest global health risks.1 Between the ages of 0 and 74, the overall risk of cancer is 20.2%, with men having a higher risk (22.4%) than women (18.2%). In 2018, 18 million new cancer cases were diagnosed worldwide, the most prevalent being prostate, lung, and breast cancers (1.28 million, 2.09 million, and 2.09 million, respectively).2 Using current treatment strategies, roughly 33% of patients can be treated with localized treatment methods such as radiation or surgery. However, early micrometastasis is a limiting characteristic in the remaining cases, highlighting the need for novel chemotherapy administered systemically for effective cancer treatment. Furthermore, if the tumor is revealed at an advanced stage, chemotherapy alone is effective in less than 10% of patients.3
Multiple diseases, including Hodgkin’s multiple myeloma, leukemia, Hodgkin’s lymphoma, and non-Hodgkin lymphoma, are classified as blood malignancies.4 A malignant neoplasm known as leukemia is distinguishable by a widespread, accelerated, and uncontrolled proliferation of leukocytes.5 Leukemia was the 15th most often diagnosed cancer and the 11th most common cause of cancer mortality worldwide. According to GLOBOCAN, in 2018, leukemia was associated with 309,006 cancer-related fatalities and 437,033 new cases. Men experience a greater global burden of leukemia than women.6 In Pakistan, acute leukemia is more common than chronic leukemia and affects males more frequently than females. AML is the most typical type of childhood cancer, and children under the age of 15 are more prone to it.7
Based on the most recent cancer statistics, 8.2 million people worldwide experience permanent metastases from drug-resistant cancers. Since cancer-promoting cells (cancer stem cells) must be specifically eliminated, current treatments face great hurdles.8 Furthermore, chemotherapy is associated with multiple side effects, such as nausea, hair loss, and appetite loss. With traditional chemotherapy, disease recurrence is common. Thus, there has been considerable emphasis on developing novel efficient medications employing natural substances and botanicals.
The most prominent polyphenolic component is curcumin (diferuloylmethane), which has been isolated from the rhizomes of Curcuma longa by Vogel and Pelletier as early as 1815. Although curcumin has anticancer properties, its poor stability has been a major obstacle in therapeutic uses. Several curcumin analogues have been synthesized, among which bis-demethoxy curcumin and diacetyl curcumin were extensively studied and found to improve the antiproliferative capacity and metabolic stability of human cancer cells.9
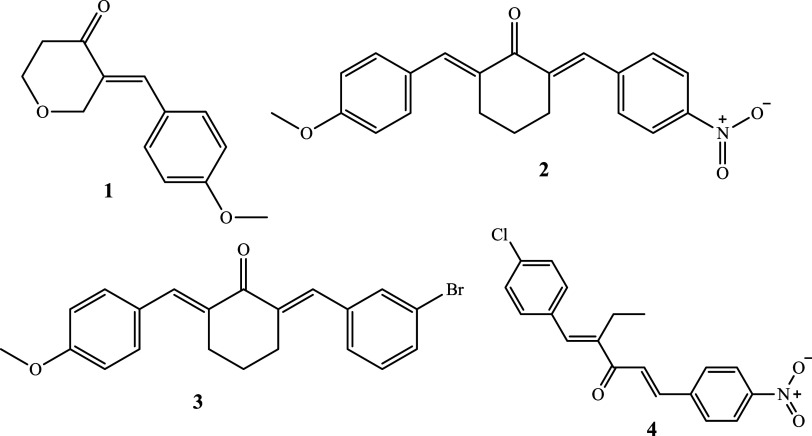
By interfering with the P-glycoprotein (P-gp)-mediated efflux pathway, several novel curcumin compounds have been shown to prevent the spread of breast cancer stem cells.10 These derivatives have a potential synergistic anticancer effect. When compared with other curcumin derivatives, these compounds possess greater affinity for binding to P-gp, in turn, inhibiting the growth of breast cancer stem cells.10 Accordingly, the present study addressed the anticancer potential of selected monocarbonyl curcuminoid 1–4 (Figure 1) in the HL 60 cell lines and BJ cells, addressing cytotoxicity and apoptosis as end points. In addition, network pharmacology approaches were also pursued in the study.
Figure 1.
Structures of test compounds.
2. Materials and Methods
2.1. Chemicals Used
Ethyl (2E)-2-benzylidene-3-oxobutanoate 1, 1,2-dimethoxy-12-methyl-13H- [1,3] benzodioxolo[5,6-c] phenanthridine 2, 3,5-dibenzyloxybenzyl bromide 3, and (E)-4-(4-chlorobenzylidene)-1-(4-nitrophenyl)hexan-3-one 4 were the four compounds used, and doxorubicin was used as a standard in this assay. Other chemical reagents we used include dimethyl sulfoxide (DMSO) HPLC grade (Sigma-Aldrich, Germany; Cat# 34869–2.5L), MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) (Invitrogen, Thermo Fisher Scientific; Cat# M6494), RPMI 1640 medium (Gibco; Cat# 11875093), fetal bovine serum (FBS) (Gibco, U.K.; 10500–064), Pen strep (Gibco; Cat# 15070–063), annexin-V (Invitrogen, Thermo Fisher Scientific; Cat# A13199), ethanol, and propidium iodide (Invitrogen, Thermo Fisher Scientific; Cat# P1304MP).
2.2. Cell Lines and Treatments
The HL-60 human promyelocytic leukemia cell line (ATCC CCL240TM) was obtained from the ATCC. RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% penicillin and streptomycin antibiotics were used to culture the HL-60 cells at 37 °C in a humid environment at 5% CO2. Only the exponential phase of cell growth was used for all experiments.11
2.3. MTT Cytotoxic Assay for HL-60 and BJ Cell Lines
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) tetrazolium test is a widely used standard method for evaluating the metabolic activity of cell cultures.12 The potential of cells to cleave the tetrazolium salt MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, into an insoluble blue MTT formazan product by the mitochondrial enzyme succinate dehydrogenase.13 The assay was used herein to measure cell viability.14 8 × 103 HL-60 cells were plated in each well of 96-well plates. The tested compounds were dissolved in ethanol and DMSO, and successive dilutions of the compounds were made. After 24 h, the cells were treated with the compounds at concentrations between 1.875 and 30.0 μM. MTT (3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide) solution (201 of 0.5 mg/mL) was added to each well after 24 h of incubation, with the incubation continuing for additional 4 h at 37 °C. Next, the cells were centrifuged for 5 min at 1200 rpm. The formazan crystals were dissolved in 100 μL of DMSO (spectrophotometric grade) in the dark for 5 min, and the absorbance (OD) of each well at 550 nm was read with a microplate reader. Nonlinear regression was used to estimate the proportion of cell survival. Cell survival was calculated using percent absorbance versus control (cells not treated with chemicals). By graphing percent survival vs concentrations, IC50 values for various compounds were determined. Doxorubicin was selected as the standard anticancer medication.11 The following is the formula that was used to calculate the cell viability.
2.4. Annexin V Assay
The apoptotic effects of compound 2 and compound 4 on HL-60 cells were investigated using the annexin V detection kit (BioLegend) in accordance with the manufacturer’s instructions. The human promyelocytic leukemia cell line was plated in 6-well plates at a density of 1.2 × 105 cells/well. The cells were exposed to the active substances (2 and 4) at an IC50 concentration for 24 h at 37 °C and 5% CO2. Next, the cells were centrifuged at 200 rmp and washed twice with PBS buffer. The pellets were resuspended and stained with propidium iodide and annexin V in annexin binding buffer (ABB). After double staining, the cells were incubated in the dark for 30 min and analyzed in a flow cytometry analyzer (BD FACSCelesta).15
2.5. Network Pharmacology Analysis
2.5.1. Prediction of Druglikeness Potential of Monocarbonyl Curcuminoids-Related Analogues
The canonical simplified molecular input line entry system (SMILES) notations for a diverse selection of chemical compounds were obtained from PubChem, a well-established chemical database. Subsequently, the SwissADME software tool was used to predict the druglikeness potential of these compounds. This evaluation encompassed key molecular properties, including molecular weight (MolWt), oral bioavailability (OB), the number of hydrogen bond acceptors (Num. H-bond acceptors), and the number of hydrogen bond donors (Num. H-bond donors).
To assess druglikeness, the compounds were screened based on their oral bioavailability (OB) values, with those exceeding 0.30 being considered. Additionally, stringent criteria were applied to ensure that the number of hydrogen bond acceptors did not surpass 10 and the number of hydrogen bond donors remained within the limit of 5. Furthermore, a criterion was established for molecular weight (MolWt), wherein compounds with molecular weights below 500 g/mol were selected. This methodical evaluation aimed to systematically identify and prioritize compounds with the potential for favorable druglike properties, laying the groundwork for subsequent analytical and investigatory endeavors in this study.
2.5.2. Target Prediction
The efficacy of monocarbonyl curcuminoid-related analogues was determined by analyzing interactions retrieved from two distinct platforms: STITCH16 and Swiss Target Prediction17 databases, with a focus on ″Homo sapiens″ species. For the STITCH database, targets with a combined score greater than or equal to 0.7 were selected for further analysis. Swiss Target Prediction, on the other hand, utilized the SMILES notations of selected compounds to identify potential targets based on a reverse pharmacophore matching approach. In this case, only target proteins with a probability value greater than or equal to 0.7 were considered as potential targets. while the cancer-related data was predicted with the GenCard database.
2.5.3. Compound–Target Network Construction
A Venn plot was constructed to identify overlapping genes between compounds and cancer. These overlapping genes were then regarded as potential targets of monocarbonyl curcuminoid-related analogues, signifying potential biomarkers for intervening in the pathophysiology of cancer. Cytoscape version 3.818 was used to construct a compound–target network. In this network, nodes represent compounds and their associated proteins, while solid black lines represent interactions between target proteins and compounds. Subsequently, network analysis assessed the degree of connectivity among compounds within the compound–target network.
2.6. Functional Annotation of Overlapped Genes
Once overlapping genes between treatment groups were identified, Gene Ontology (GO) and pathway enrichment analyses were conducted to unveil their underlying molecular functions (MFs), biological processes (BPs), cellular components (CCs), and key signaling pathways. The Database for Annotation, Visualization, and Integrated Discovery (DAVID)19 was used for the functional annotation of overlapping targets. The DAVID database generated several GO terms and KEGG pathways, with a stringent threshold of p-values less than 0.05 applied to screen for statistically significant KEGG pathways and GO terms. To visualize these findings, the ggplot2 package in R was used to present the top 20 GO terms and top 20 KEGG pathways based on their count and p-values <0.05.
2.6.1. Protein–Protein Interaction (PPI) Network Construction
Synergistic interactions within network pharmacology and protein–protein interaction (PPI) networks reveal critical insights for innovative therapeutic strategies. In network pharmacology, synergy is explored through the intricate mapping of drug–target interactions within biological networks. This mapping allows for the identification of potential synergistic drug pairs or combinations that target multiple nodes within these networks, potentially leading to more effective treatments with reduced side effects. PPI networks offer a window into the dynamic interactions between proteins within a cell, which is crucial for identifying how different drugs can influence these interactions in a potentially synergistic manner. For example, a study by Hu et al.20 demonstrated how targeting specific protein interactions within a PPI network could amplify the efficacy of drug combinations in oncology. In the current study, once common genes were identified, they were input into the STRING database to generate a PPI network.21 The functional interactions among overlapping targets were assessed based on a combined score threshold of 0.4. The resultant PPI network was then subjected to Cytoscape version 3.818 to identify hub genes. Hub genes are highly connected nodes with multiple interactions with other proteins, crucial for network integrity and stability. The degree method available in CytoHubba was used for selecting hub genes.
2.6.2. Compound–Target–Disease Network Construction
To explore the mode of action of the compounds against cancer, active ingredients–target protein and target protein–disease networks were constructed using Cytoscape version 3.8.18 These networks were merged to create a comprehensive compound–target–disease network. Nodes in this integrated network represent disease-related pathways, compounds, and proteins, while solid lines represent interactions between these nodes. This network sheds light on the synergistic effects of compounds when used in treating cancer.
2.6.3. Molecular Docking Analysis
To validate the accuracy of network pharmacology predictions, molecular docking analysis was performed to confirm interactions between key targets and compounds. This analysis aids in identifying potential drug combinations with synergistic effects on disease treatment. Autodock Vina 1.1.2 in PyRx 0.822 was used for docking analysis of the predicted X-ray crystal structure of hub proteins against active ingredients. The SDF formats of compounds were obtained from PubChem data and underwent energy minimization using OpenBabel, available in PyRX, to achieve stable conformations. Energy minimization consisted of 2000 steps, halting at an energy difference of <0.01 kcal/mol for a stable conformation. Energy-minimized ligands were converted to pdbqt format for docking. Binding pockets of target proteins were identified using the online CASTp tool.23 The target docking approach in PyRx 0.822 was used to calculate binding energies between ligand molecules and target proteins. Autodock Vina employed an empirical scoring function to determine protein–compound binding affinity, calculated by aggregating contributions from various individual terms. The docked complex with the lowest root-mean-square deviation (RMSD) was considered optimal, and binding energies below −5.00 kcal/mol were considered as strong binding, with values below −7.00 kcal/mol indicating very strong affinity. Finally, visualization of docked complexes was executed using Discovery Studio,24 PyMOL,25 and ChimeraX programs.26
3. Results
3.1. Effect of Monocarbonyl Curcuminoids (1–4) on HL-60 Cells
The anticancer effects of selected monocarbonyl curcuminoids 1–4 at various concentrations in the HL-60 cancer cell line are shown in Table 1. Doxorubicin was used as in a standard anticancer medication against the HL-60 cell line. The test compounds showed concentration-dependent cytotoxicity with maximal toxicity at 30 μM. Compounds 2, 3, and 4 demonstrated 94.65, 99.21, and 97.59% of the effect at 30 μM, respectively. However, compound 1 had no activity in the HL-60 cell line.
Table 1. Percent Inhibition by Monocarbonyl Curcuminoids (1–4) at Various Concentrations against HL-60 Cancer Cell Line.
| s. no | concentrations (μM) | percentage Inhibition ± SD | IC50 (μM) |
|---|---|---|---|
| compound 1 | 30 | inactive | |
| 15 | |||
| 7.5 | |||
| 3.5 | |||
| 1.875 | |||
| compound 2 | 30 | 94.65 ± 1.49 | 5.02 |
| 15 | 70.80 ± 3.18 | ||
| 7.5 | 66.14 ± 2.58 | ||
| 3.5 | 16.62 ± 4.57 | ||
| 1.875 | 2.36 ± 0.66 | ||
| compound 3 | 30 | 99.21 ± 5.66 | 8.42 |
| 15 | 77.30 ± 3.60 | ||
| 7.5 | 67.40 ± 3.70 | ||
| 3.5 | 17.90 ± 1.90 | ||
| 1.875 | 4.50 ± 0.75 | ||
| compound 4 | 30 | 97.59 ± 2.14 | 9.50 |
| 15 | 94.84 ± 3.52 | ||
| 7.5 | 13.48 ± 2.41 | ||
| 3.5 | 4.12 ± 2.46 | ||
| 1.875 | 4.12 ± 2.46 |
In the HL-60 cell line, IC50 values of 5.2, 8.42, and 9.50 μM were observed for compounds 2, 3, and 4, respectively. Doxorubicin, as a standard treatment, had an IC50 value of 0.17 μM.
The percent inhibition caused by compounds 2, 3, and 4 against the BJ cell line at 20 μM for 24 h is shown in Table 2. Compounds 2 and 4 were not toxic toward the BJ cell line, while compound 3 showed toxicity with 56% inhibition at 20 μM in the MTT assay.
Table 2. Percent Inhibition by 20 μM of Monocarbonyl Curcuminoids (1–3) against the BJ Cell Line.
| compounds | percent inhibition in the BJ cell line (20 μM) |
|---|---|
| compound 2 | 9.7% |
| compound 3 | 56% |
| compound 4 | 23.75% |
3.2. Apoptotic Effect of Compounds 2 and 4 on HL-60 and BJ Cell Lines
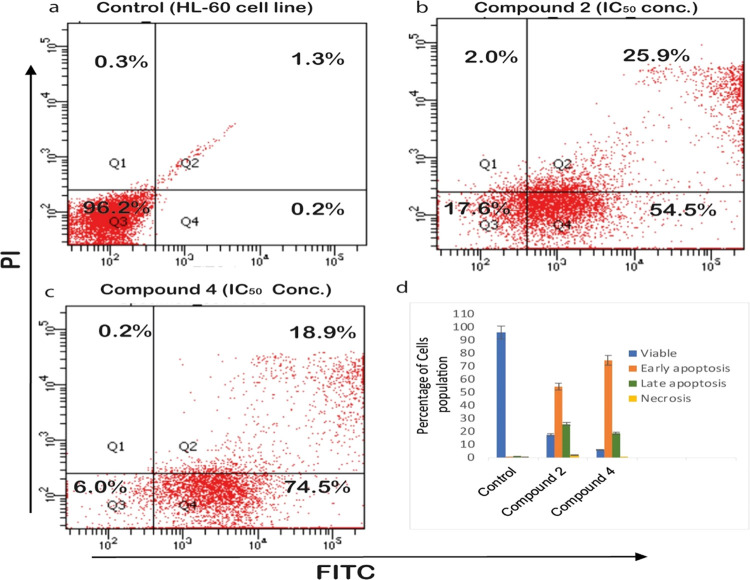
Compounds 2 and 4 were tested for their ability to cause apoptosis in the HL-60 cell line, using annexin V-FITC/PI assays to assess cell death mechanisms. The proportion of HL-60 cells in early and late apoptosis was determined after a 24 h treatment. Cells were divided into four quadrants (Q1, Q2, Q3, and Q4), as shown in Figure 2. The quadrants, Q1, Q2, Q3, and Q4, display cells in necrotic, late apoptotic, viable, and early apoptotic cells population, respectively. Compound 2 (Figure 2b) showed 54.5% early and 25.95% late apoptotic population of cells, while compound 4 exhibited 74.5% early and 18.9% late apoptotic cells in the HL-60 cell population. These results revealed that both compounds 2 and 4 induced apoptosis in the promyelocytic leukemic cell line (HL-60) after 24 h treatment.
Figure 2.
(a) Untreated control HL-60 cells and treated compound 2 (b) and compound 4 (c) using annexin V-FITC/PI double staining via flow cytometry.
3.3. Network Pharmacology Analysis
3.3.1. Predicting the Druglike Potential of Active Compounds
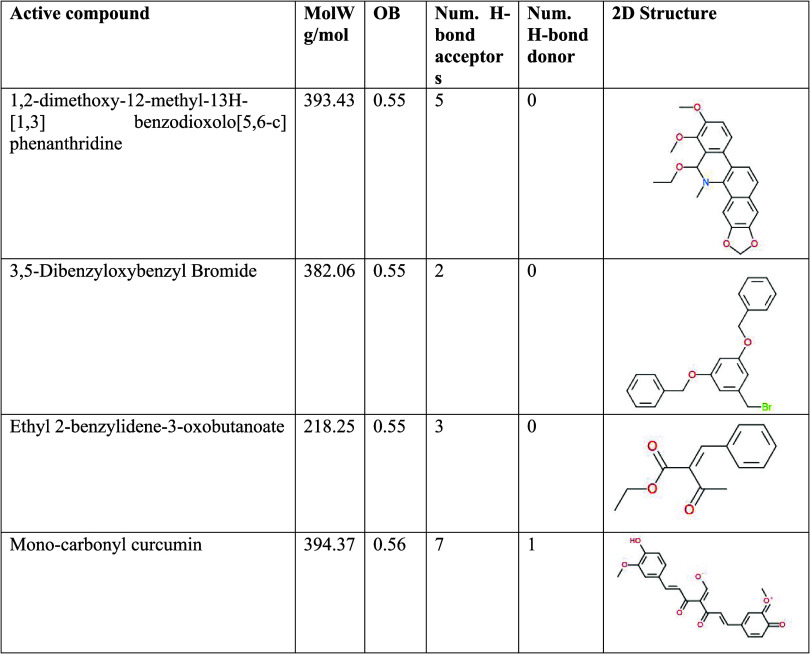
1,2-dimethoxy-12-methyl-13H-[1,3]benzodioxolo[5,6-c]phenanthridine, 3,5-dibenzyloxybenzyl bromide, ethyl 2-benzylidene-3-oxobutanoate, and monocarbonyl curcumin are the specific active compounds considered in this analysis. Their molecular weights range between 218.25 and 394.37 g/mol. Oral bioavailability (OB) values indicate the compound’s potential to be administered orally as a drug, with values between 0.55 and 0.56. The number of hydrogen bond acceptors and donors provides insights into the compounds’ capacity to form crucial interactions in biological systems, with values varying from 2 to 7 and 0 to 1, respectively. These properties collectively offer valuable insights into the druglikeness potential of these monocarbonyl curcuminoids-related analogues, laying the foundation for further investigations into their therapeutic properties and potential as drug candidates.
Table 3 provides a comprehensive overview of the physiochemical properties of monocarbonyl curcuminoids-related analogues, focusing on their MolW, OB, Num. H-bond acceptors, the Num. H-bond donor, and their respective 2D chemical structures. These properties were evaluated for a set of active compounds known to be associated with monocarbonyl curcuminoids.
Table 3. Physiochemical Properties of Monocarbonyl Curcuminoids-Related Analogues.
3.3.2. Target Prediction
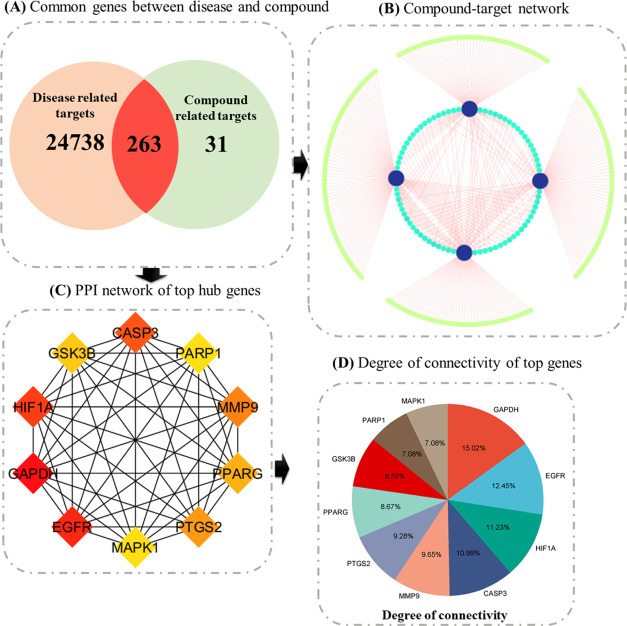
A comprehensive analysis was carried out with the retrieval of 25,001 disease-related target predictions from the GenCard data set. This was followed by Venn diagram analysis to identify shared genes between compound-related and disease-related targets. Subsequently, 294 potential target genes associated with four key compounds, sourced from the Swiss Target Prediction and STITCH databases, were identified. Of note, the Venn diagram revealed 263 potential anticancer genes specifically associated with curcumin. These promising genes were earmarked for further in-depth exploration.
Next, the compounds and their corresponding target genes were integrated into the Cytoscape platform to enhance network visualization. This approach enabled the identification of compounds occupying pivotal positions within the network and shed light on their potential synergistic effects. Hence, the compound–target network provided a holistic view of how these predicted targets could collaboratively contribute to the anticancer properties of these compounds.
3.3.3. Functional Annotation of Overlapped Targets
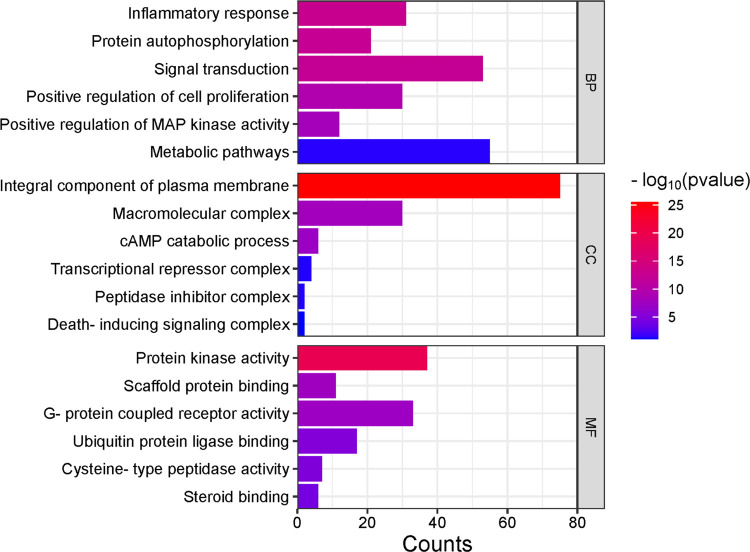
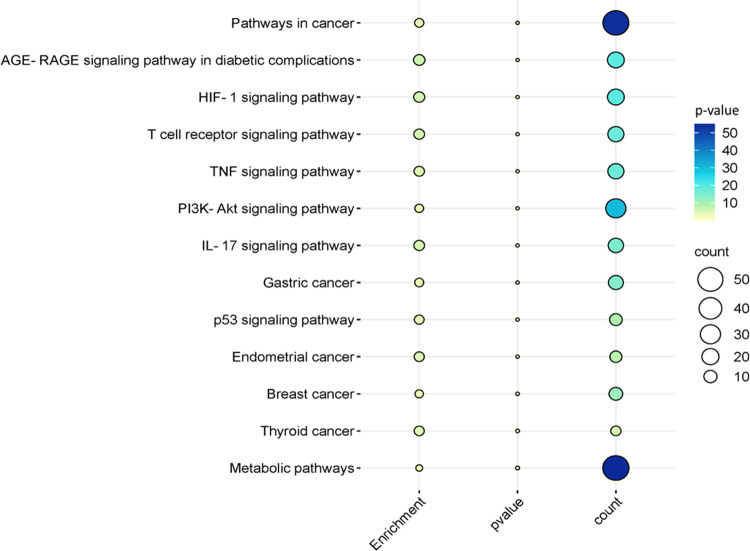
The exploration of the identified genes’ biological characteristics extended to a comprehensive analysis using Gene Ontology (GO) and KEGG pathway enrichment assessments via the DAVID resources (Figure 3). This analysis provided valuable insights into the potential roles and functions of these genes within various biological contexts. The KEGG pathway analysis unveiled a diverse spectrum of pathways in which these genes are actively involved. Notably, these pathways encompass both disease-related processes and fundamental cellular mechanisms. Among the disease-related pathways, diabetic cardiomyopathy stood out, indicating a potential link between the identified genes and this cardiac condition. Moreover, several cancer-related pathways were prominently identified, including colorectal and bladder cancer pathways, highlighting the potential significance of these genes in cancer biology. In addition to disease-associated pathways, the analysis illuminated the engagement of these genes in essential cellular processes. Metabolic pathways emerged as a central theme, emphasizing the genes’ involvement in core metabolic activities critical for cell function. Signaling pathways, such as HIF-1 and MAPK pathways, known for their pivotal roles in cellular responses to various stimuli, also featured prominently. The immune system’s involvement was evident through the IL-17 signaling pathway, suggesting a potential link between the identified genes and immune responses. Furthermore, pathways related to cellular regulation, such as apoptosis and autophagy, identified genes that control cell survival and homeostasis. The intricate network of molecular interactions, as exemplified by the Ras signaling pathway and microRNAs’ involvement in cancer, highlighted the complexity of these genes’ functions in cancer development and progression. Overall, this extensive analysis revealed a multifaceted landscape of the identified genes’ potential roles, encompassing not only disease-related processes but also fundamental cellular mechanisms. These findings provide a solid foundation for further investigations and experimental validations to uncover the precise functions and therapeutic implications of these genes in various biological contexts. The bubble map of top signification terms and pathways is shown in Figure 4.
Figure 3.
Bar plot representing a comprehensive visualization of the Gene Ontology (GO) analysis results, categorizing enriched terms into biological processes (BPs), cellular components (CCs), and molecular functions (MFs). Color intensity reflects the log-transformed values, indicating the importance of specific terms within each category.
Figure 4.
Bubble plot illustrating significant KEGG pathways. Each bubble corresponds to a pathway, and the color of each bubble represents the p-value of that pathway. Larger and more colorful bubbles indicate the number of genes (counts) involved in specific pathways, signifying their increased significance.
3.3.4. Identification of Hub Genes
The Protein–protein Interaction (PPI) network, composed of 259 nodes and 2635 edges, was meticulously constructed using the STRING database, reflecting the intricate interplay among the identified targets. Within this network, nodes represent proteins, while edges denote interactions between them. To discern the central players in this complex interaction landscape, the top 10 nodes were singled out based on their degree of connectivity. The degree, which refers to the number of interactions a node has with other nodes, serves as a critical measure of a node’s centrality within the network. These distinguished hub genes, listed alongside their respective degrees of connectivity within parentheses, include GAPDH (123), EGFR (102), HIF1A (92), CASP3 (90), MMP9 (79), PTGS2 (76), PPARG (71), GSK3B (70), PARP1 (58), and MAPK1 (58) (Figure 5).
Figure 5.
Integrated analysis of target genes and compound interactions. (A) Venn plot: This panel displays a Venn plot showcasing the common genes shared between compound-related and disease-related targets, providing insights into potential overlaps. (B) Compound–Target Network: In this subfigure, nodes represent compounds and their associated proteins. Node size reflects the degree of connectivity, emphasizing key interactions within the network. (C) Top 10 Hub genes: This panel highlights the top 10 genes ranked based on the degree algorithm, underlining their significance in the network. (D) Hub gene degrees: Shown as a bar plot, this subfigure represents the degree of connectivity for each hub gene, providing a visual perspective on their importance within the network.
The substantial degree of connectivity associated with these hub genes underscores their prominence in the PPI network and suggests their indispensable roles in mediating interactions between various proteins. Notably, most of the investigated compounds were found to selectively target these hub genes, further emphasizing their significance as primary molecular targets for the anticancer effects of the compounds under scrutiny. These hub genes represent key nodes within the intricate protein interaction network, potentially serving as linchpins in the mechanisms through which the compounds exert their anticancer properties. Consequently, these findings offer valuable insights into the molecular basis of the compounds’ anticancer effects and pave the way for more targeted therapeutic strategies in cancer treatment.
3.3.5. Compound–Target–Pathway Network
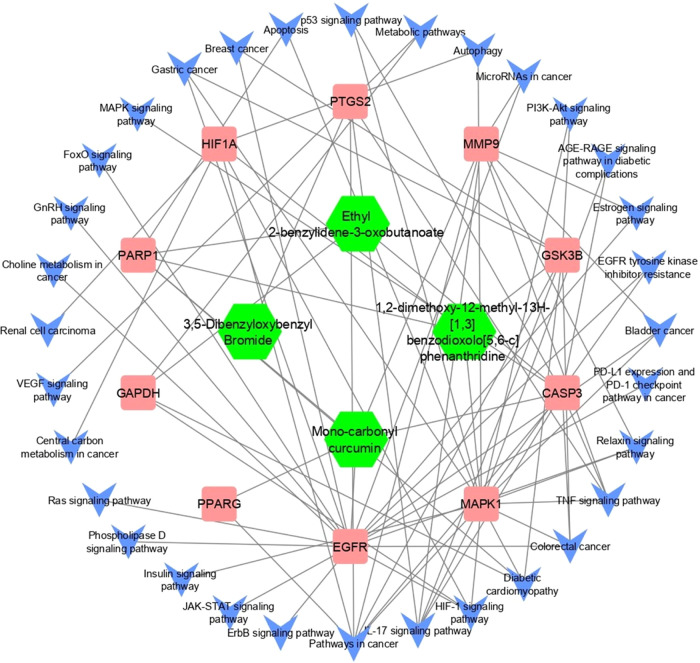
The construction of a holistic “compound–target–disease” network aimed to provide a more insightful grasp of the intricate mechanisms underlying the compound’s impact on cancer (Figure 6). In this network, pathways unrelated to cancer were systematically excluded, focusing solely on those directly associated with the disease. This comprehensive analysis integrated data from the compound–target network, protein–protein interaction (PPI) network, and the compound–target–disease network. Among the array of proteins, CASP3 and PARP1 emerged as top-ranking candidates, exhibiting both a heightened degree of connectivity within the PPI network and a significant convergence of compounds targeting these hub genes. Moreover, the GO analysis unveiled potential regulatory functions of these genes, particularly in cancer cell proliferation through processes like identical protein binding and positive regulation of apoptotic processes. Similarly, the KEGG pathway analysis highlighted the enrichment of CASP3 and PARP1 in pathways closely linked to carcinogenesis, metabolic pathways, and pathways integral to cancer progression, further emphasizing their pivotal roles in the complex network of cancer-related molecular interactions.
Figure 6.
Compound–target–disease network. In this schematic representation, hub proteins are symbolized by square shapes, targeted pathways are indicated by arrows, and active compounds are represented by hexagonal shapes. The connections and interactions among these elements reveal the intricate network of relationships underlying the impact of compounds on cancer.
3.3.6. Molecular Docking Analysis
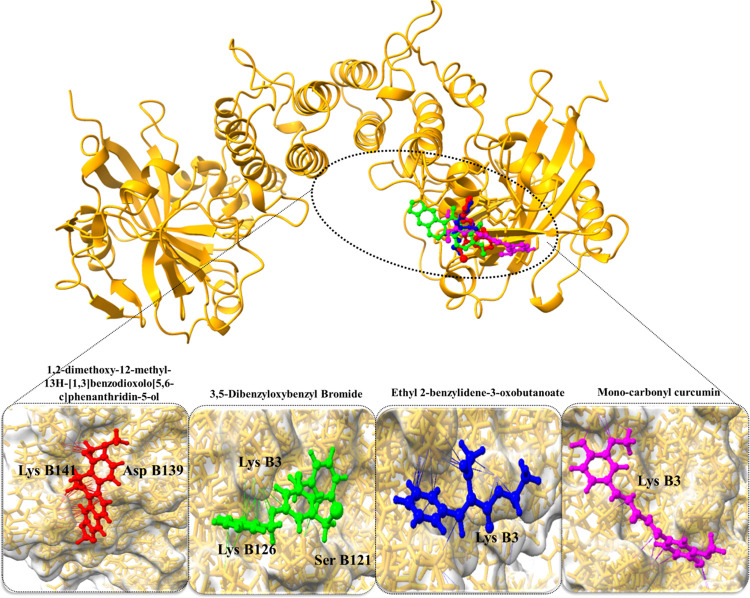
The molecular docking results provide valuable insights into the interactions between selected active compounds and two key hub proteins, PARP1 and CASP3, shedding light on their potential therapeutic effects in the context of cancer (Table 4). In the case of PARP1, the compound 1,2-dimethoxy-12-methyl-13H-[1,3]benzodioxolo[5,6-c]phenanthridin-5-ol displayed a substantial binding affinity of −7.38 kcal/mol. Notably, this compound formed crucial hydrogen bond interactions with Asp B139 and Lys B141 residues of PARP1, indicating strong binding capacity. Similarly, monocarbonyl curcumin exhibited a noteworthy binding affinity of −6.73 kcal/mol with PARP1, forming hydrogen bond interactions with Lys B126, Lys B3, and Ser B121 residues. These interactions suggest its potential to modulate the function of PARP1 effectively. Additionally, 3,5-dibenzyloxybenzyl bromide and ethyl 2-benzylidene-3-oxobutanoate displayed binding affinities of −6.43 kcal/mol and −5.89 kcal/mol, respectively, with specific interactions with the Lys B3 residue of PARP1 (Figure 7).
Table 4. Binding Affinity and RMSD of Monocarbonyl Curcuminoids-Related Analogues with Target Proteins.
| compound name | PubChem IDs | binding affinity (kcal/mol) | RMSD | H-bond interactions |
|---|---|---|---|---|
| PRAP1 (1uk0) | ||||
| 1,2-dimethoxy-12-methyl-13H-[1,3]benzodioxolo[5,6-c]phenanthridin-5-ol | 14818664 | –7.38 | 1.3 | Asp B139, Lys B141 |
| monocarbonyl curcumin | 969516 | –6.73 | 2.7 | Lys B126, Lys B3, Ser B121 |
| 3,5-dibenzyloxybenzyl bromide | 2761019 | –6.43 | 2.2 | Lys B3 |
| ethyl 2-benzylidene-3-oxobutanoate | 5376216 | –5.89 | 1.6 | Lys B3 |
| CASP3 (6zfl) | ||||
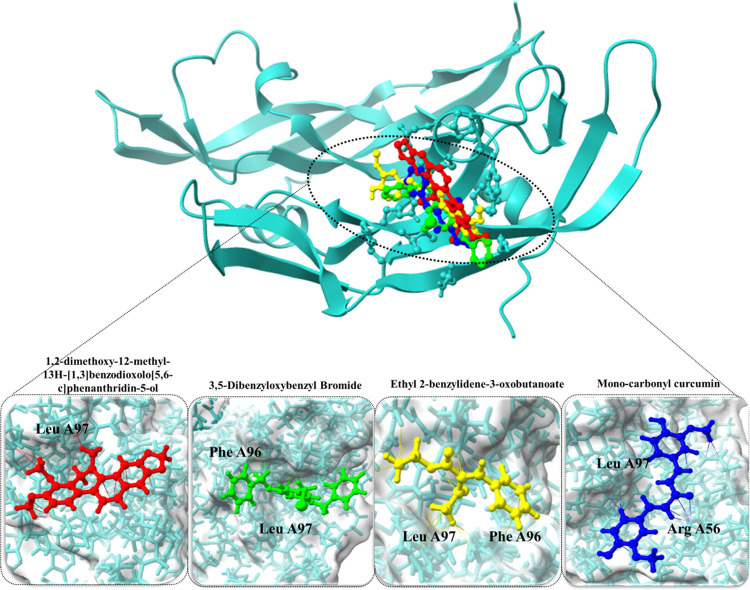
| monocarbonyl curcumin | 969516 | –8.56 | 1.9 | Arg A56, Leu A97 |
| 3,5-dibenzyloxybenzyl bromide | 2761019 | –8.32 | 1.2 | Phe A96, Leu A97 |
| 1,2-dimethoxy-12-methyl-13H-[1,3]benzodioxolo[5,6-c]phenanthridin-5-ol | 14818664 | –7.92 | 2.1 | Leu A97 |
| ethyl 2-benzylidene-3-oxobutanoate | 5376216 | –6.48 | 1.6 | Phe A96, Leu A97 |
Figure 7.
Docked complexes of PARP1 protein along with their strongest binding compounds.
For CASP3, monocarbonyl curcumin exhibited a remarkably strong binding affinity of −8.56 kcal/mol, indicating its potential as a modulator of CASP3 activity. This compound formed hydrogen bond interactions with Arg A56 and Leu A97 residues, underlining its precise binding to the protein’s active site. Similarly, 3,5-dibenzyloxybenzyl bromide demonstrated a robust binding affinity of −8.32 kcal/mol, with interactions involving Phe A96 and Leu A97 residues of CASP3. Additionally, 1,2-dimethoxy-12-methyl-13H-[1,3]benzodioxolo[5,6-c]phenanthridin-5-ol displayed a binding affinity of −7.92 kcal/mol, with interactions centered around the Leu A97 residue. Lastly, ethyl 2-benzylidene-3-oxobutanoate exhibited a binding affinity of −6.48 kcal/mol, forming hydrogen bond interactions with Phe A96 and Leu A97 residues of CASP3 (Figure 8).
Figure 8.
Docked complexes of CASP3 protein along with their strongest binding compounds.
These comprehensive docking results underscore the high binding affinities and specific interactions between the active compounds and key cancer-related proteins, PARP1 and CASP3. Such interactions suggest their potential as therapeutic agents capable of modulating the functions of these proteins within cancer-related pathways. Further experimental validation is essential to confirm and harness their therapeutic efficacy effectively.
4. Discussion
Using the MTT assay and apoptotic analyses, the current study established the anticancer effects of selected monocarbonyl curcuminoids in HL-60 and BJ cells. In addition, network pharmacology and docking analysis indicated synergistic effects, targeting both the CASP3 and PARP1 proteins by monocarbonyl curcuminoid.
The most prevalent technique for determining a substance’s ability to inhibit cell proliferation in cultured cells is the MTT tetrazolium assay.27 The test methodology relies on the enzymatic reduction of the light-colored tetrazolium salt to its spectrophotometrically quantifiable deep violet-blue formazan.12 Using the MTT cytotoxicity assay, we first identified the anticancer activity of the monocarbonyl curcuminoid derivatives and investigated their underlying anticancer activity in the HL-60 cell line.
Apoptosis, which is a process of planned cell death, can be brought on by a variety of factors, including cellular stress, DNA damage, and immune surveillance. Apoptosis is facilitated by several signaling pathways (known as intrinsic and extrinsic) that are activated by a variety of circumstances. Using annexin V-FITC/PI tests to analyze cell cycle arrest and cell death pathways, the apoptotic impact of compounds 2 and 4 on the HL-60 cell line was observed.15 Apoptosis testing showed that compounds 2 and 4 were effective against the HL-60 cell line after 24 h treatment. In contrast, compound 3 was harmful to the BJ cell line. During the 24 h treatment, no notable necrosis was found.
Network pharmacology analysis showed a wealth of information regarding the potential therapeutic properties of specific active compounds, shedding light on their druglike potential, target predictions, functional annotations of overlapped targets, identification of hub genes, and their role in the context of a compound–target–pathway network. Network pharmacology has the potential to uncover synergistic effects by analyzing how multiple drugs or compounds interact within biological networks.28−30 Such insights into synergistic interactions can lead to the development of more effective combination therapies, particularly in the context of complex diseases like cancer, where multiple molecular pathways are involved. The assessment of the druglike potential of the active compounds is a critical starting point. The molecular weights of these compounds fall within the range of 218.25 to 394.37 g/mol, indicating their compatibility with drug development. Moreover, their OB values of 0.55 to 0.56 suggest that they have the potential to be administered orally as drugs, which is a highly desirable characteristic for ease of patient use. Additionally, the number of hydrogen bond acceptors and donors provides insights into their ability to form crucial interactions within biological systems, further emphasizing their suitability as drug candidates. These properties collectively lay the foundation for further investigations into their therapeutic potential.
Target prediction analysis is a crucial step in understanding how these compounds exert their anticancer effects. The identification of 294 potential target genes associated with the four monocarbonyl curcuminoid compounds, including 263 potential anticancer genes, offers exciting avenues for further exploration. These genes represent potential molecular targets for the compounds, suggesting their roles in mediating the compounds’ anticancer properties.
The Cytoscape platform to integrate compound–target information significantly can enhance the understanding of interaction networks and potential synergistic effects. This visualization method not only positions the compounds within the network but also offers an overarching view of their predicted targets and their collaborative roles in anticancer activity, leading to the identification of key network components relevant to cancer therapy. Additionally, KEGG pathway analysis is crucial in elucidating the complex biological functions of genes and their roles in diverse physiological and pathological processes. Notably, the MAPK and PI3K-Akt signaling pathways, integral to cell growth and survival, have been extensively studied for their roles in cancer progression and have emerged as promising targets for therapeutic interventions.31,32 Yet, the cell cycle and apoptotic pathways are also crucial for maintaining tissue homeostasis, and their disruption can contribute to tumorigenesis.33,34 Furthermore, the Wnt, Notch, and HIF-1 signaling pathways have attracted attention given their roles in development, tissue regeneration, and cancer, offering potential avenues for targeted therapies.35−37 Overall, KEGG pathway analysis provides a robust foundation for understanding the molecular basis of diseases, including cancer, and informs the development of novel therapeutic strategies.
The identification of hub genes within the PPI network is a key highlight of this study. These hub genes, including GAPDH, EGFR, HIF1A, CASP3, MMP9, PTGS2, PPARG, GSK3B, PARP1, and MAPK1, exhibit significant degrees of connectivity within the network, emphasizing their central roles in mediating interactions among various proteins. Importantly, most of the investigated compounds selectively target these hub genes, underscoring their significance as primary molecular targets for the compounds’ anticancer effects. These hub genes serve as pivotal nodes within the intricate protein interaction network, potentially acting as linchpins in the mechanisms through which the compounds exert their anticancer properties.
The construction of a compound–target–pathway network offers valuable insights into the mechanisms through which specific compounds impact cancer. By mapping these compounds onto pathways directly associated with cancer, a more nuanced understanding of their potential therapeutic effects is achieved. Notably, CASP3 and PARP1 were identified as top-ranking candidates within this network. Their significant connectivity in the PPI network and the convergence of compounds targeting these hub genes highlight their potential roles in influencing cancer progression. Furthermore, the GO analysis was instrumental in elucidating the involvement of these genes in critical processes such as cancer cell proliferation and apoptosis. This is complemented by KEGG pathway analysis, which further delineates their roles in carcinogenesis and metabolic pathways, offering a broader perspective on the biological implications of targeting these genes. Moreover, the molecular docking analysis, focusing on the interactions between active compounds and key cancer-related proteins, specifically PARP1 and CASP3, provides essential insights. The observed high binding affinities and specific interactions underscore the potential efficacy of these compounds as therapeutic agents, particularly in modulating the functions of PARP1 and CASP3 within cancer-related pathways. These integrated analyses, encompassing druglike potential, target predictions, functional annotations, and molecular docking, collectively provide a robust framework that not only elucidates the anticancer properties of the investigated compounds but also merits further experimental exploration.
5. Conclusions
In this study, the evaluation of various curcuminoid analogues, 1–4, revealed that 1,2-dimethoxy-12-methyl-13H-[1,3]benzodioxolo[5,6-c]phenanthridine 2 and (E)-4-(4-chlorobenzylidene)-1-(4-nitrophenyl)hexan-3-one compound 4 demonstrated significant cytotoxic effects on the HL-60 cell line. These findings, derived from the MTT cytotoxicity assay and further validated by apoptosis analyses, highlight the potential of these compounds for cancer therapy. To deepen the understanding of these effects, molecular docking and network pharmacology were employed. This approach not only corroborated the in vitro results but also provided detailed insights into the molecular interactions of these compounds. Specifically, the molecular docking studies emphasized the strong binding affinities of compounds 2 and 4 with key target proteins. Furthermore, the integration of network pharmacology approaches with molecular docking analyses offered a comprehensive view on the therapeutic potential of these curcuminoid analogues. This combined methodology revealed that compounds 2 and 4, as monocarbonyl curcuminoids, effectively target critical proteins such as CASP3 and PARP1. Additionally, these compounds exhibit a synergistic effect, enhancing their anticancer properties when used in combination. These insights open promising avenues for further exploration. The potential of compounds 2 and 4, particularly when combined, to effectively target key proteins involved in cancer pathogenesis, underscores their significance as candidates for novel cancer therapies. This novel study contributes to the growing body of knowledge on cancer treatment and paves the way for future explorations into targeted, synergistic cancer therapeutics.
Acknowledgments
The authors extend their appreciation to Researchers Supporting Project number (RSPD2024R812), King Saud University, Riyadh, Saudi Arabia.
The authors declare no competing financial interest.
References
- Siegel R. L.; Miller K. D.; Fuchs H. E.; Jemal A. Cancer statistics, 2022. Ca-Cancer J. Clin. 2022, 72 (1), 7–33. 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- Mattiuzzi C.; Lippi G. Current cancer epidemiology. J. Epidemiol. Global Health 2019, 9 (4), 217. 10.2991/jegh.k.191008.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu E.; Sartorelli A.. Cancer Chemotherapy. In Lange’s Basic and Clinical Pharmacology; McGrawHill, 2018; pp 948–976. [Google Scholar]
- Shahrabi S.; Behzad M. M.; Jaseb K.; Saki N. Thrombocytopenia in leukemia: Pathogenesis and prognosis. Histol. Histopathol. 2018, 33 (9), 895–908. 10.14670/HH-11-976. [DOI] [PubMed] [Google Scholar]
- Mukherjee U., Pediatric Leukemia-An Overview.
- Bispo J. A. B.; Pinheiro P. S.; Kobetz E. K. Epidemiology and etiology of leukemia and lymphoma. Cold Spring Harbor Perspect. Med. 2020, 10 (6), a034819 10.1101/cshperspect.a034819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S.; Shah K. A.; Hussain H.; Haq A. U.; Ullah A.; Khan A.; Rahman N. U. Prevalence of Acute and Chronic Forms of Leukemia in Various Regions of Khyber Pakhtunkhwa, Pakistan: Needs Much More to be done!. Bangladesh J. Med. Sci. 2019, 18 (2), 222–227. 10.3329/bjms.v18i2.40689. [DOI] [Google Scholar]
- Koroth J.; Nirgude S.; Tiwari S.; Gopalakrishnan V.; Mahadeva R.; Kumar S.; Karki S. S.; Choudhary B. Investigation of anti-cancer and migrastatic properties of novel curcumin derivatives on breast and ovarian cancer cell lines. BMC Complementary Altern. Med. 2019, 19 (1), 273 10.1186/s12906-019-2685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile V.; Ferrari E.; Lazzari S.; Belluti S.; Pignedoli F.; Imbriano C. Curcumin derivatives: molecular basis of their anti-cancer activity. Biochem. Pharmacol. 2009, 78 (10), 1305–1315. 10.1016/j.bcp.2009.06.105. [DOI] [PubMed] [Google Scholar]
- Mbese Z.; Khwaza V.; Aderibigbe B. A. Curcumin and its derivatives as potential therapeutic agents in prostate, colon and breast cancers. Molecules 2019, 24 (23), 4386. 10.3390/molecules24234386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksuzoglu E.; Ertan-Bolelli T.; Can H.; Tarhan M.; Ozturk K.; Yildiz I. Antitumor activities on HL-60 human leukemia cell line, molecular docking, and quantum-chemical calculations of some sulfonamide-benzoxazoles. Artif. Cells, Nanomed., Biotechnol. 2017, 45 (7), 1388–1396. 10.1080/21691401.2016.1241796. [DOI] [PubMed] [Google Scholar]
- Grela E.; Kozłowska J.; Grabowiecka A. Current methodology of MTT assay in bacteria–A review. Acta Histochem. 2018, 120 (4), 303–311. 10.1016/j.acthis.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Vajrabhaya L.-o.; Korsuwannawong S. Cytotoxicity evaluation of a Thai herb using tetrazolium (MTT) and sulforhodamine B (SRB) assays. J. Anal. Sci. Technol. 2018, 9 (1), 15 10.1186/s40543-018-0146-0. [DOI] [Google Scholar]
- Ogbole O. O.; Segun P. A.; Adeniji A. J. In vitro cytotoxic activity of medicinal plants from Nigeria ethnomedicine on Rhabdomyosarcoma cancer cell line and HPLC analysis of active extracts. BMC Complementary Altern. Med. 2017, 17 (1), 494 10.1186/s12906-017-2005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan İ.; Koçak P.; Yıldırım M.; Ünsal N.; Yılmaz H.; Telci D.; Şahin F. Garlic (Allium sativum)-derived SEVs inhibit cancer cell proliferation and induce caspase mediated apoptosis. Sci. Rep. 2021, 11 (1), 14773 10.1038/s41598-021-93876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller D.; Grosdidier A.; Wirth M.; Daina A.; Michielin O.; Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42 (W1), W32–W38. 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M.; von Mering C.; Campillos M.; Jensen L. J.; Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008, 36 (Database issue), D684–D688. 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13 (11), 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G.; Sherman B. T.; Hosack D. A.; Yang J.; Gao W.; Lane H. C.; Lempicki R. A.. DAVID: database for annotation, visualization, and integrated discovery Genome Biol., 20034 (9), R60. [PubMed] [Google Scholar]
- Hu H.; Wang H.; Yang X.; Li Z.; Zhan W.; Zhu H.; Zhang T. Network pharmacology analysis reveals potential targets and mechanisms of proton pump inhibitors in breast cancer with diabetes. Sci. Rep. 2023, 13 (1), 7623 10.1038/s41598-023-34524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C.; Jensen L. J.; Snel B.; Hooper S. D.; Krupp M.; Foglierini M.; Jouffre N.; Huynen M. A.; Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33 (Database issue), D433–D437. 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S.; Olson A. J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Tian W.; Chen C.; Lei X.; Zhao J.; Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46 (W1), W363–W367. 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studio D.Discovery studio Accelrys [2.1] 2008.
- Yuan S.; Chan H. S.; Hu Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2017, 7 (2), e1298 10.1002/wcms.1298. [DOI] [Google Scholar]
- Goddard T. D.; Huang C. C.; Meng E. C.; Pettersen E. F.; Couch G. S.; Morris J. H.; Ferrin T. E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27 (1), 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai Y.; Pathak R.; Kumari N.; Sah D. K.; Pandey S.; Kalra N.; Soni R.; Dwarakanath B.; Bhatt A. N. Mitochondrial biogenesis and metabolic hyperactivation limits the application of MTT assay in the estimation of radiation induced growth inhibition. Sci. Rep. 2018, 8 (1), 1531 10.1038/s41598-018-19930-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor F.; Asif M.; Ashfaq U. A.; Qasim M.; Tahir ul Qamar M. Machine learning for synergistic network pharmacology: a comprehensive overview. Briefings Bioinf. 2023, bbad120 10.1093/bib/bbad120. [DOI] [PubMed] [Google Scholar]
- Noor F.; Tahir ul Qamar M.; Ashfaq U. A.; Albutti A.; Alwashmi A. S.; Aljasir M. A. Network pharmacology approach for medicinal plants: review and assessment. Pharmaceuticals 2022, 15 (5), 572. 10.3390/ph15050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor F.; Rehman A.; Ashfaq U. A.; Saleem M. H.; Okla M. K.; Al-Hashimi A.; AbdElgawad H.; Aslam S. Integrating network pharmacology and molecular docking approaches to decipher the multi-target pharmacological mechanism of Abrus precatorius L. acting on diabetes. Pharmaceuticals 2022, 15 (4), 414. 10.3390/ph15040414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R. Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol. Res. 2012, 66 (2), 105–143. 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Thorpe L. M.; Yuzugullu H.; Zhao J. J. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15 (1), 7–24. 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M.; Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 2009, 9 (3), 153–166. 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A.; Dixit V. M. Death receptors: signaling and modulation. Science 1998, 281 (5381), 1305–1308. 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- Clevers H.; Nusse R. Wnt/β-catenin signaling and disease. Cell 2012, 149 (6), 1192–1205. 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Aster J. C.; Blacklow S. C. Targeting the Notch pathway: twists and turns on the road to rational therapeutics. J. Clin. Oncol. 2012, 30 (19), 2418–2420. 10.1200/JCO.2012.42.0992. [DOI] [PubMed] [Google Scholar]
- Semenza G. L. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 2010, 20 (1), 51–56. 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]