H3K36 methylation promotes balanced transcriptional output from chromosome 4 but is dispensable for dosage compensation.
Abstract
In Drosophila, two chromosomes require special mechanisms to balance their transcriptional output to the rest of the genome. These are the male-specific lethal complex targeting the male X chromosome and Painting of fourth targeting chromosome 4. Here, we explore the role of histone H3 methylated at lysine-36 (H3K36) and the associated methyltransferases—Set2, NSD, and Ash1—in these two chromosome-specific systems. We show that the loss of Set2 impairs the MSL complex–mediated dosage compensation; however, the effect is not recapitulated by H3K36 replacement and indicates an alternative target of Set2. Unexpectedly, balanced transcriptional output from the fourth chromosome requires intact H3K36 and depends on the additive functions of NSD and Ash1. We conclude that H3K36 methylation and the associated methyltransferases are important factors to balance transcriptional output of the male X chromosome and the fourth chromosome. Furthermore, our study highlights the pleiotropic effects of these enzymes.
INTRODUCTION
The evolution of sex chromosomes, for example, the X and Y chromosome pairs, often leads to sex-linked differences in gene dose. In Drosophila, the Y chromosome has few genes but is retained because of its specific roles in male fertility (1). This leads to a difference in X chromosome gene doses in males and females. Although some genes located on the X chromosome are expressed in a sex-specific manner, most require equal expression in both sexes (2, 3). Therefore, the transcriptional output from the genes on the single X chromosome in males has to match that of the genes from the two X chromosomes in females. In both sexes, the output from the X chromosome has to be balanced with the transcriptional output from the two sets of autosomal chromosomes (2–5). In Drosophila, the gene dosage problem is solved such that the transcriptional output from genes on the male X chromosome is increased by a factor of approximately 2 (4). This increased transcriptional output is partly mediated by chromosome-specific targeting and stimulation of the X-linked genes by the male-specific lethal (MSL) complex. The MSL complex consists of at least five proteins, originally identified through their MSL phenotype when lost, MSL1, MSL2, MSL3, MLE (the product of the maleless gene), and MOF (the product of the males absent on the first gene), and two long noncoding RNAs named RNA on the X (roX1 and roX2) (4, 6). The MSL complex binds the male X chromosome in two steps. First, it binds to roughly 250 specific sites, denoted chromatin entry sites, MSL recognition element, high-affinity sites (HASs), or pioneering sites on the X (Pion-X), largely overlapping but initially isolated and classified using different criteria (7–10). Next, the MSL complex spreads from these HASs to neighboring transcriptionally active genes. This spreading requires the presence of at least one of the two roX RNAs (11). The resulting X chromosome specific binding leads to histone 4 lysine 16 acetylation (H4K16ac), mediated by the acetyltransferase MOF (12, 13).
Importantly, at least one other chromosome has been exposed to a similar evolutionary process as the current X chromosome. The fourth chromosome of Drosophila melanogaster was ancestrally an X chromosome that has reverted to an autosome (14, 15). This sex chromosome reversal is likely to explain that the fourth chromosome is the subject of chromosome-specific targeting and regulatory mechanisms mediated by the protein named Painting of fourth (POF) (16–19). We have previously hypothesized that POF and its stimulatory function became trapped on the fourth chromosome when the latter reverted to being an autosome (16). The stimulatory effect of POF is comparable in level to the compensation mediated by the MSL complex on the X chromosome (8, 18, 20–22). The hypothesized origin of POF as a dosage compensation system is also supported by the fact that the POF-mediated compensation of the fourth chromosome is essential for the survival of flies with a monosomy of chromosome 4 (18). The fourth chromosome has several unique characteristics. It is the smallest chromosome in the Drosophila genome; it is replicated late; and it is enriched in heterochromatin protein 1 (HP1), the histone-lysine N-methyltransferase eggless (also known as SETDB1 homolog), and methylated H3K9 (23, 24).
Accurate targeting and precise stimulatory effect of these chromosome-specific systems require coordinated contributions of several factors. In addition to aforementioned DNA sequence motifs, these may include confinement to a specific nuclear compartment (25), long-range chromatin contacts (26), and the interaction between the chromodomain of MSL3 and histone H3 trimethylated at lysine-36 (H3K36me3). The latter was hypothesized to help the spreading of the MSL complex from the HASs to the neighboring transcriptionally active genes (27, 28). Consistent with this model, the loss of Set2 methyltransferase in third instar larvae and the drastic reduction of H3K36me3 are accompanied by partial loss of MSL2 and MSL3 binding to dosage-compensated genes (28), and mutations in the MSL3 chromodomain correlate with impaired spreading of the MSL complex from the HASs (27, 28). In contrast, structural studies suggest that the chromodomain of MSL3 has higher affinity to the more abundant histone H4 mono- and dimethylated at lysine-20 (H4K20) (29, 30). The rival hypothesis argues that the MSL3 chromodomain interacts with methylated H4K20 to present H4 tails for acetylation at lysine-16 by MOF (30). Thus, the extent and mechanisms by which H3K36 methylation and/or associated methyltransferases contribute to balanced transcriptional output from the male X and the fourth chromosomes remain open questions.
Three evolutionarily conserved Drosophila proteins—Absent, small, or homeotic discs 1 (Ash1), SET domain–containing 2 (Set2), and Nuclear receptor binding SET domain containing protein (NSD)—are believed to methylate H3K36. In vitro, Ash1 can add one or two methyl groups to H3K36 (31), and this activity is further enhanced by NuA4 complex subunit EAF3 homolog (also known as MRG15) and Chromatin assembly factor 1 p55 subunit (CAF1 p55) proteins with which Ash1 forms a complex (32–35). Flies lacking ash1 function show an approximated twofold reduction of bulk H3K36me1 but no detectable loss of overall H3K36me2 or H3K36me3 (33, 36). These ash1 mutants display multiple homeotic transformations and die at the larval stage due to erroneous repression of developmental genes by Polycomb group mechanisms (36–38). The Set2 protein can methylate H3K36 in vitro using mono- and dimethylated H3K36 as a substrate (39). Whether it can also add methyl groups to unmethylated H3K36 is not entirely clear, although it has been demonstrated that the human ortholog (SETD2) is able to do so in reconstituted reactions (40). Consistently, the Drosophila Set2 mutants have 10-fold lower levels of H3K36me3 overall and at specific genes (28, 36) but display no major changes in the bulk H3K36me2 and H3K36me1 (36). Together, these observations suggest that Set2 produces most of the Drosophila H3K36me3 and may contribute to cellular pools of H3K36me1 and H3K36me2. The loss of Set2 function is lethal in early pupa stage (28, 36), but the corresponding mutant shows no signs of excessive Polycomb repression (36), suggesting that Set2 and Ash1 affect Drosophila development in distinct ways. Less is known about the biochemical properties of the fly NSD. It has three closely related orthologs in mammals (NSD1, NSD2, and NSD3) whose methyltransferase activity in vitro has been extensively studied. These studies concur that NSD proteins can mono- and dimethylate H3K36 (41–43). However, methylation of other substrates is also suggested. A systematic screen indicates that these include K168 of histones H1.5 and H1.2, K169 of histone H1.3, and K44 of histone H4, as well as K1033 of the chromatin remodeller ATRX and K189 of the U3 small nucleolar RNA-associated protein 11 (43). NSD proteins are essential for proper mammalian development, and haploinsufficiencies in the NSD1 and NSD2 genes were linked to Sotos and Wolf-Hirschhorn genetic syndromes, respectively (44). Whether methylation of H3K36 or any of the substrates above are relevant for the developmental functions of the NSD proteins remains an open question. Unlike its mammalian counterparts, Drosophila NSD loss-of-function mutants are viable, fertile, and show no obvious morphological defects (36). Although early experiments in cultured cells suggested that NSD knockdown reduces bulk levels of H3K36me2 and H3K36me3 (39), the NSD loss-of-function mutants display no obvious changes in the overall levels of H3K36me1, H3K36me2, and H3K36me3 (36).
Whether H3K36 is the only (or even the major) physiological substrate is a question that appears equally relevant for Ash1 and Set2. One might expect that animals in which H3K36 is replaced with an amino acid that cannot be methylated will display defects similar to those seen in mutants that lack the corresponding methyltransferase. Mounting experimental evidence suggests the contrary. Thus, complete substitution of the zygotic H3 with a variant in which K36 is replaced with arginine (R) does not cause excessive repression of homeotic genes seen in ash1 mutants (36). Likewise, flies with replication-coupled histone H3.2 replaced with the H3K36R variant display no cryptic transcription initiation or altered splice site choice as reported for yeast Set2 or human SETD2 mutants (45). To investigate the role of H3K36 and the associated histone methyltransferases in chromosome-specific gene regulation, we evaluated relative levels of methylated H3K36 on individual chromosome arms and compared those to transcriptional imbalance caused by the loss of Set2, NSD, and Ash1 or the replacement of the histone H3.2 with a variant in which lysine-36 is substituted to arginine.
Confirming previous reports, we found that loss of Set2 impairs MSL complex–mediated dosage compensation. However, the effect is not recapitulated in H3K36R mutants and suggests an alternative target for Set2. This implies that the model in which Set2-mediated H3K36me3 helps the MSL complex to bind active genes may need to be revised. Unexpectedly, our results indicate that the balanced transcriptional output from the fourth chromosome depends on the additive contribution from Ash1 and NSD and requires intact H3K36. We conclude that H3K36 methylation and the associated methyltransferases are important factors for balanced transcriptional output of the male X chromosome and the fourth chromosome. Our study also emphasizes the importance of pleiotropic effects of these enzymes.
RESULTS
No evidence of enriched H3K36me3 on the male X chromosome
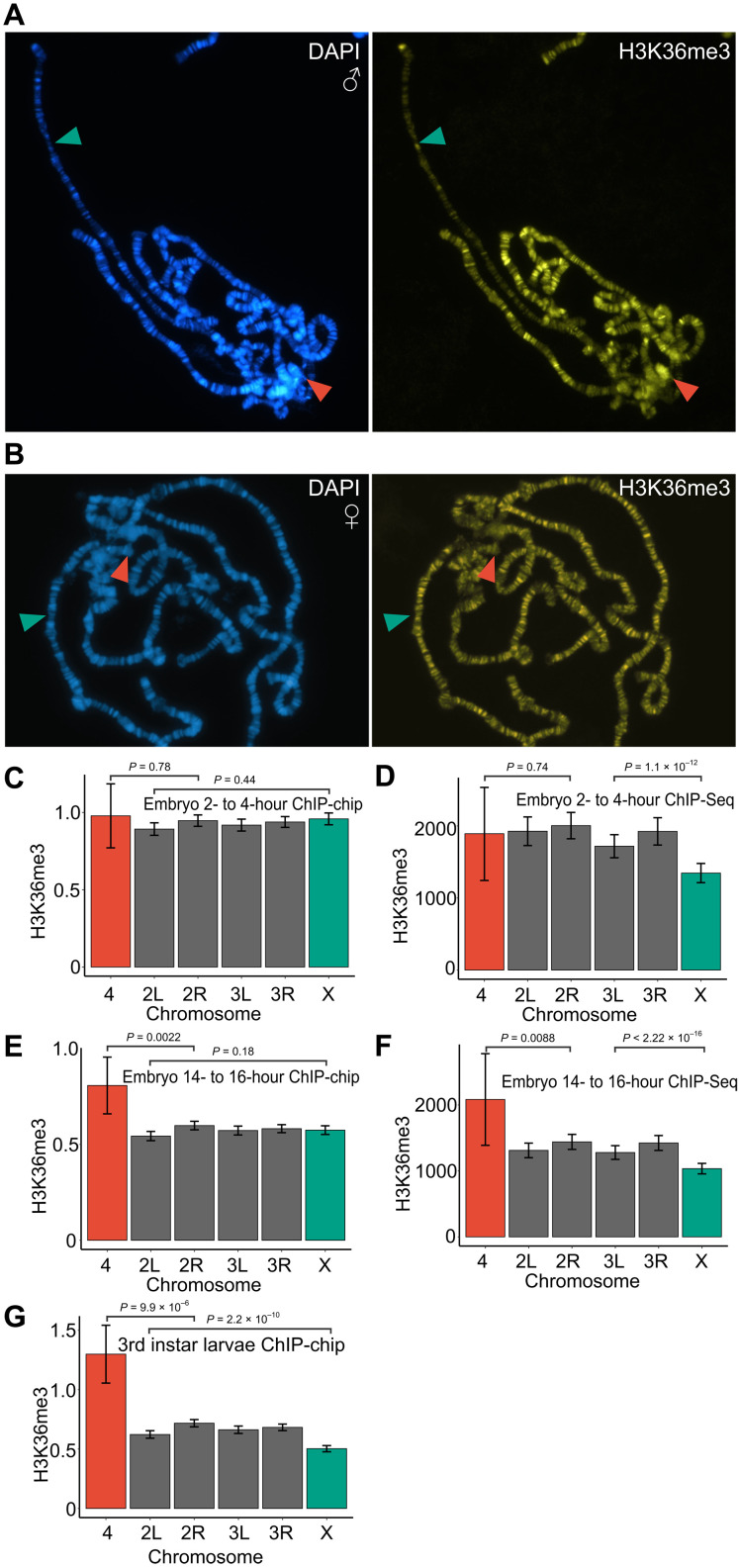
Drosophila chromosome-specific regulatory systems are accompanied by chromosome-specific enrichment of specific histone modifications. Thus, the male X chromosome is enriched in H4K16ac (46), and the fourth chromosome is enriched in H3K9me2/me3 (47). If the interaction between MSL3 and trimethylated H3K36 is important for dosage compensation, one may expect that the male X chromosome is enriched in H3K36me3. To test this conjecture, we stained polytene chromosomes from male third instar larvae with antibodies against mono-, di-, and trimethylated H3K36. Unexpectedly, H3K36me3 appeared less abundant on the male X chromosome (Fig. 1A and fig. S1A) compared to autosomes. The weaker anti-H3K36me3 immunostaining of the X chromosome was not observed in females (Fig. 1B). In males, the number of chromatids of the polytene X chromosome is half that of the autosomes. This difference may account for some of the reduced staining. Nevertheless, the decrease in H3K36me3 signal is more pronounced than that for H3K36me1 or H3K36me2 (fig. S1, A and B). This argues that the male polytene X chromosome has a lower level of H3K36me3 compared to the autosomes. Staining with antibodies against monomethylated and dimethylated H3K36 (H3K36me1 and H3K36me2) produced broad banding patterns uniformly distributed throughout chromosome arms (fig. S1B). However, the banding pattern of H3K36me2 showed little overlap with that of H3K36me1 (fig. S1, B and C). A noticeable difference was the conspicuously strong H3K36me2 staining of the pericentromeric region and the fourth chromosome (fig. S1, A and B). Increased staining of the fourth chromosome was also evident for H3K36me3 (Fig. 1A and fig. S1A). The apparent lower level of H3K36me3 on the male X chromosome is at odds with the model in which Set2-mediated H3K36me3 helps the MSL complex to bind active genes (27, 28).
Fig. 1. H3K36 trimethylation is enriched on the fourth chromosome and reduced on the X chromosome.
(A) Immunostaining of a male third instar larvae polytene chromosome shows an accumulation of H3K36me3 (yellow) on chromosome 4 (red arrowhead) and a reduction on the X chromosome (green arrowhead) as compared to autosomal signals. DAPI staining of DNA in blue indicates banding pattern. (B) Immunostaining of female third instar larvae polytene chromosome detects no differences between any chromosome arms. Average exon H3K36me3 enrichment scores per chromosome in 2- to 4-hour mixed-sex embryos (C and D), 14- to 16-hour mixed-sex embryos (E and F), and mixed-sex third instar larvae (G). Enrichment scores for the ChIP-chip experiments (C, E, and G) are the ChIP over input log2 ratio of the top 50% of the exon regions per gene. For ChIP-seq (D and F), the enrichment scores are reads adjusted for difference in position between ChIP and input. In ChIP-seq, only peaks in exons were used. Error bars (C to G) indicate the 95% confidence intervals. The statistical significances were determined by unpaired two-sample Wilcoxon tests comparing the X chromosome and chromosome 4 to the most similar autosome arms.
The interaction between MSL3 and trimethylated H3K36 may be critical at earlier stages of development, and an increased presence of the latter on the male X chromosome may not be apparent in salivary glands. To evaluate this possibility, we turned to published genome-wide mapping data from embryos and third instar D. melanogaster larvae of mixed sex (48, 49). Since H3K36me3 is predominantly enriched within the coding regions of genes (50), we calculated relative enrichment values of H3K36me3 within exons for each gene. Analyses of H3K36me3 at embryonic stages returned mixed results. Chromatin immunoprecipitation (ChIP)–chip mapping in early embryos (2 to 4 hours) indicate that the amount of H3K36me3 is uniformly the same for all chromosomes (Fig. 1C). In contrast, the ChIP-seq data show significantly reduced H3K36me3 ChIP signal on the X chromosome (Fig. 1D). In a late embryonic stage (14 to 16 hours), the fourth chromosome shows significantly stronger immunoprecipitation with H3K36me3 antibodies in both ChIP assays (Fig. 1, E and F), while only the ChIP-seq data show a reduced ChIP signal on the X chromosome (Fig. 1F). Confirming the observations from polytene chromosome staining, analysis of immunoprecipitations with chromatin from third instar larvae showed that H3K36me3 ChIP signals within gene bodies were significantly higher on the fourth chromosome and lower on the male X chromosome as compared to the autosomes (Fig. 1G).
While the reason for the discrepancy between ChIP-chip and ChIP-seq mapping remains unclear, it could be caused by the different ratio of males and females in the mixed-sex samples. In any case, neither of the methods detect the increased abundance of H3K36me3 on the X chromosome.
Set2 is required for balanced transcription of X-linked genes bound by the MSL complex
The level of H3K36me3 within genes correlates with their transcriptional activity (fig. S2), and Set2 was reported to help in maintaining the transcriptional balance of the male X chromosome. Yet, the level of H3K36me3 on this chromosome is, at best, the same, if not lower than it is on autosomes. Puzzled by this paradox, we decided to analyze transcriptional outputs in dissected brains from third instar male larvae mutants of the three known H3K36 specific methyltransferases: Set2, NSD, and Ash1. The genes encoding the three methyltransferases show similar relative mRNA abundance throughout development. The mRNA amounts peak in early embryos, reflecting maternal contribution, and reach the lowest level at early larval stages (fig. S3). In flies, Set2 is responsible for most of the H3K36me3, while the potential division of labor and redundancy in H3K36 methylation mediated by NSD and Ash1 is not fully understood. As reported previously (51), we found that NSD is enriched in pericentromeric heterochromatin and on the fourth chromosome (fig. S4), with a staining pattern similar to that of HP1 and H3K36me2 (fig. S1, A and B) (23, 51, 52).
In Set21 mutants, the MSL complex was reported to correctly target the high-affinity binding sites on the male X chromosome but reduce the binding to the adjacent transcriptionally active genes (28). The Set21 allele corresponds to a deletion of the N-terminal half of its open reading frame, including the catalytic SET domain (28), and leads to approximately a 10-fold reduction in global H3K36me3 (28, 36) and a nearly complete loss of H3K36me3 staining on polytene chromosomes (fig. S5). Homozygous Set21, but not ash122/ash19011 or NSDds46 (see the following section), displays a significant relative decrease in transcriptional output from the male X chromosome, suggesting an impaired dosage compensation (Fig. 2A).
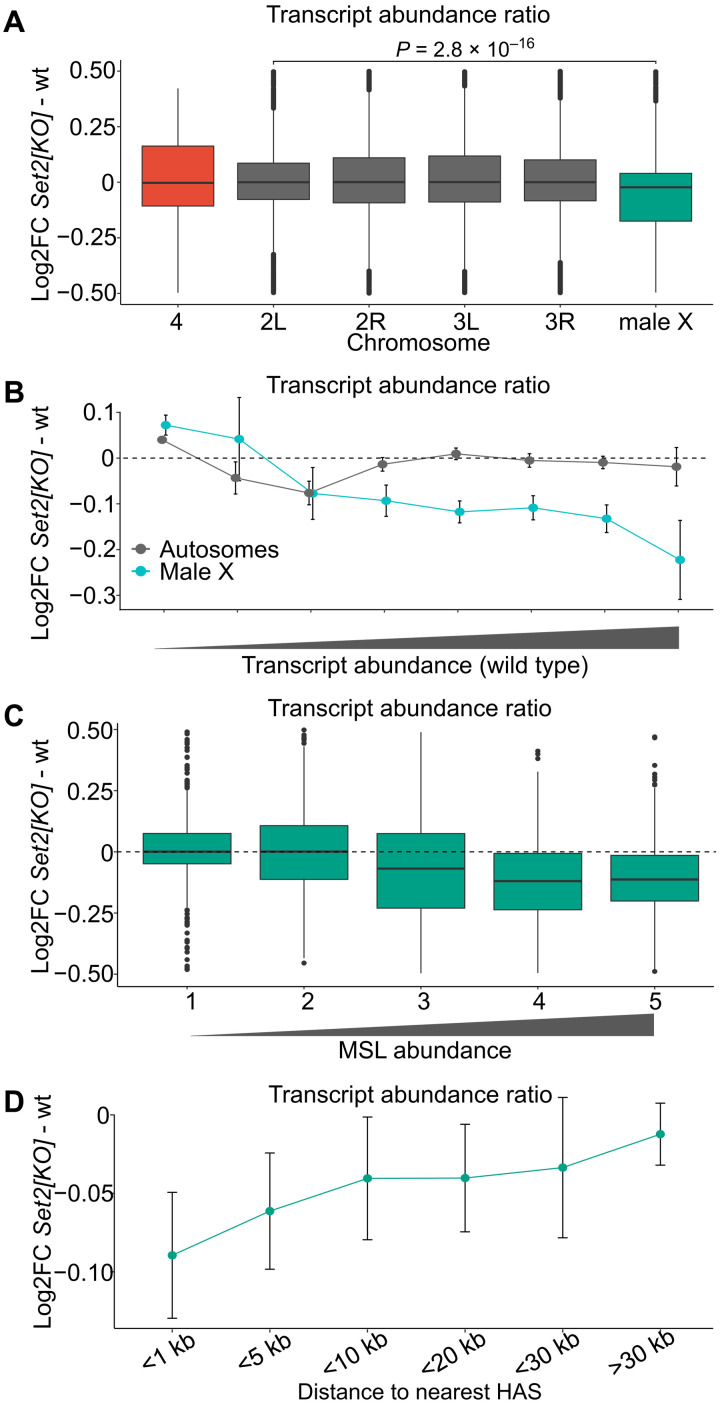
Fig. 2. Set2 is required for balanced transcription output from the single male X chromosome.
(A) Box plots of chromosome-specific transcript abundance ratios (log2 fold change) between Set21 and wild type for chromosome 4 (red), X chromosome (green), and the autosome arms (gray). The abundance of transcripts from X chromosome of the Set21 mutant is significantly reduced as compared to the autosomes. The likelihood that observed differences are due to chance was evaluated by unpaired two-sample Wilcoxon tests comparing values for the X chromosome and chromosome arm2L (the most similar autosome arm). (B) Transcript abundance in Set21 versus wild type plotted as fold change (log2). Genes were binned according to Flybase RNA-seq expression level intervals. The X chromosome genes with higher transcriptional output in the wild type are significantly more affected compared to those on autosomes. (C) Box plots of average transcript abundances of the X-chromosomal genes in Set21 versus wild type. The genes are grouped in equally sized bins based on their MSL1-binding strength; 1 (lowest) to 5 (highest). Genes with higher MSL1 levels show greater fold change compared to genes with no or low amounts of MSL1 bound. (D) Average log2 fold change for X-chromosomal genes binned by distance to HASs. In (B) and (D), the error bars indicate the 95% confidence intervals.
To evaluate the link, we divided genes according to their wild-type transcript levels and found that highly transcribed genes are those most affected by the Set2 loss (Fig. 2B). Highly transcribed genes require high levels of MSL complex binding (53). We therefore asked whether the reduced transcriptional output observed in Set21 correlates with binding levels of the MSL complex. To address this question, all X chromosome genes were divided into five bins based on their immunoprecipitation with antibodies against the MSL1 subunit of the MSL complex (54, 55). Thus, bin 1 included unbound and weakly bound genes, while bin 5 included genes highly enriched in MSL proteins. As illustrated in Fig. 2C, genes robustly enriched by MSL (bins 3 to 5) show reduced transcript abundance upon Set2 loss. To further substantiate this result, we grouped the genes by distance to the nearest HAS since it has been shown that genes more distal to HAS are less sensitive to loss of a functional MSL complex (8, 53). As expected, we observed that genes close to a HAS are, on average, more affected by the loss of Set2 compared to genes further away. Genes at distances of more than 30 kb from a HAS are essentially unaffected by the loss of Set2 (Fig. 2D). This is similar to what is seen after ablation of the roX1 roX2 noncoding RNAs (53). Together, our observations argue that the imbalanced transcriptional output from the Set21 male X chromosome is due to an impaired MSL complex–mediated dosage compensation.
H3K36R mutants show no imbalance of transcriptional output from the male X chromosome
It is tempting to speculate that the loss of Set2 leads to lower H3K36me3, which, in turn, impairs the binding of MSL3 (27, 28). If that is the case, we expect to see a similar transcriptional imbalance in the nonmodifiable H3K36R histone replacement mutant, ΔHisC; 12xH3K36R. These mutant flies carry the deletion of the histone gene cluster ΔHisC combined with a transgenic construct carrying 12 copies of the 5-kb histone repeat unit in which H3.2K36 is mutated to arginine (12xH3K36R) (56, 57).
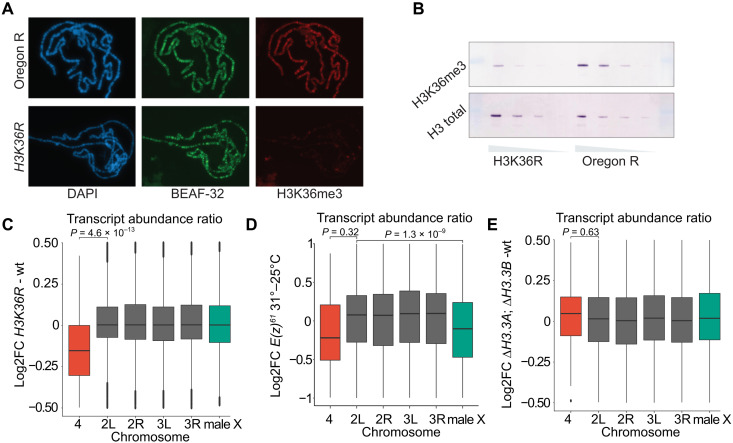
Polytene chromosomes of the ΔHisC; 12xH3K36R flies display a marked decrease of immunostaining with antibodies against H3K36me3 (Fig. 3A). This is in accordance to what has previously been shown (57) and is similar to that seen in Set21 mutants (fig. S5). Western blot analysis showed that H3K36me3 methylation is, as expected, strongly reduced in ΔHisC; 12xH3K36R flies (Fig. 3B). Notably, some H3K36me3 signal is still detected. This may correspond to the trimethylated H3.3 variant histone still present in ΔHisC; 12xH3K36R flies, but we cannot exclude the possibility that some of it may be caused by antibody cross-reactivity. Despite the fivefold reduction in the overall H3K36me3, we observed no X chromosome–specific transcriptional imbalance in ΔHisC; 12xH3K36R animals (Fig. 3C). This suggests that the imbalance in transcription of genes on male X chromosome after Set2 knockout is not linked to a concomitant loss of H3K36 methylation.
Fig. 3. Set2 contributes to increased transcriptional output from the male X chromosome independently of H3K36 methylation.
(A) Polytene chromosome staining of H3K36me3 in wild type (top row) and ΔHisC; 12xH3K36R histone replacement larvae (bottom row). Staining of boundary element-associated factor of 32kD (BEAF-32) serves as an internal control. (B) Western blot analyses of twofold dilutions of total protein lysates from brains and imaginal discs of third instar larvae show substantial decrease of H3K36me3 (top row) in ΔHisC; 12xH3K36R compared to wild type. Western blot with antibodies against total histone H3 serves as loading control. (C) Box plots of log2 fold change of transcript abundance per chromosome for ΔHisC; 12xH3K36R versus ΔHisC; 12xH3K36K (wt). Note that chromosome 4 shows a significant reduction in transcript abundance compared to the other chromosome arms. Here and below, unpaired two-sample Wilcoxon tests were used to determine the likelihood that observed differences are explained by chance. (D) Box plots of log2 fold change of transcript abundance per chromosome for the E(z)61 cell line at 31°C [strongly reduced E(z) function] versus 25°C. At restrictive temperature, the abundance of transcripts from X chromosome is significantly reduced compared to the major autosome arms. (E) Box plots of log2 fold change of transcript abundance per chromosome for ΔH3.3B; ΔH3.3A versus wild-type flies indicate no significant differences.
The interpretation of histone replacement experiments may, however, be less straightforward if it is the unmethylated H3K36 that is required for the process, which Set2 aims to prevent. Thus, recent structural studies suggest that unmodified H3K36 is required for unimpeded methylation of lysine-27 of histone H3 (H3K27) by the polycomb-repressive complex 2 (PRC2) (58, 59). PRC2 is required not only for epigenetic repression of many developmental genes (60) but also to suppress spurious transcription of inactive genes and intergenic regions (61). Conceivably, reduced activity of PRC2 in the ΔHisC; 12xH3K36R mutant may lead to an increased transcription of the X chromosome genes, which, in turn, may mask the effect of the impaired H3K36 methylation. To evaluate this possibility, we took advantage of the RNA sequencing (RNA-seq) data from (61) where the PRC2 function was disrupted by a temperature shift of a cell line homozygous for the temperature-sensitive E(z)61 allele (61, 62). We calculated transcript abundance ratios for all genes divided by chromosomes and found the transcription of genes on the X chromosome relative to autosomes to be reduced, not increased (Fig. 3D). Whether this relative reduction is a consequence of a more pronounced global derepression of low and nonexpressed genes on autosomes (61) remains an open question, which we have not attempted to investigate any further. Regardless, it counters the idea that imbalanced transcriptional output from the X chromosome of ΔHisC; 12xH3K36R males is masked by the concomitant reduction of PRC2 activity.
We next considered the possibility that methylation of variant histone H3 (H3.3) is critical for balanced transcriptional output from the male X chromosome and the regulatory mechanism mediated by Set2 methylation. It has previously been shown that the replication-independent histone variant H3.3 is enriched on the male X chromosome (63). We therefore analyzed gene transcription in brains from male third instar larvae with both H3.3 genes deleted (ΔH3.3B; ΔH3.3A). Despite the reported enrichment of H3.3 on the male X chromosome, we observed no reduction in X chromosome transcript abundance as compared to autosomes in the ΔH3.3B; ΔH3.3A double mutant (Fig. 3E). Together, the lack of an X chromosome–specific effect in ΔHisC; 12xH3K36R and in ΔH3.3B; ΔH3.3A mutants argues that Set2 contributes to increased expression of dosage-compensated genes independently of H3K36 methylation.
Ash1 and NSD together maintain balanced transcriptional output from the fourth chromosome
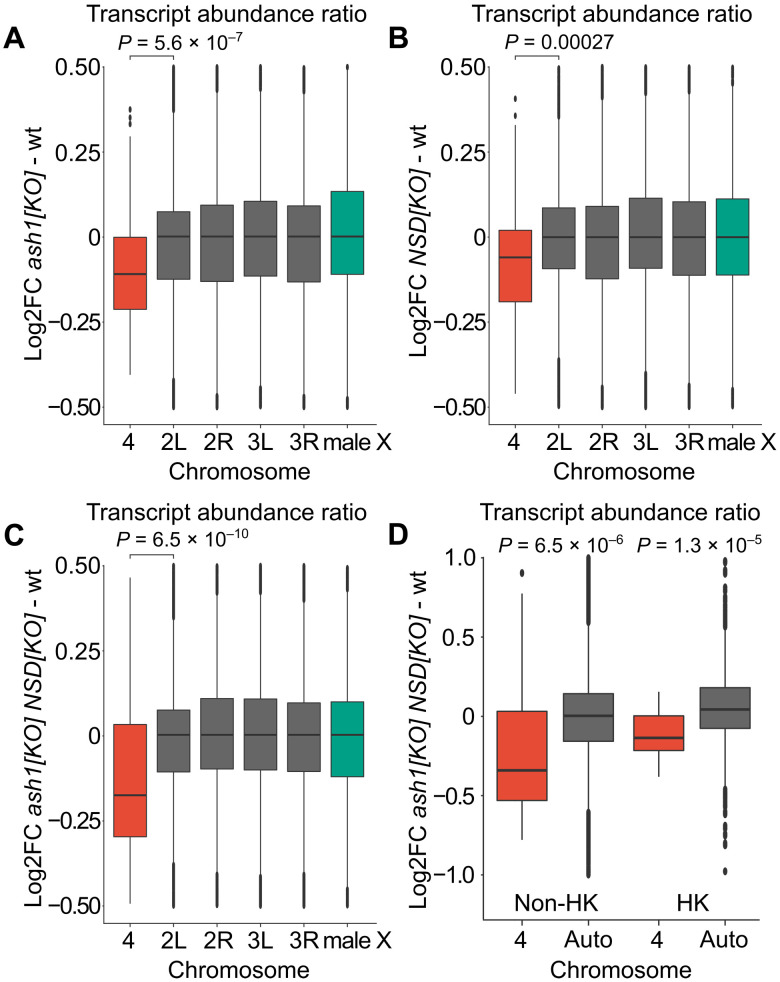
Immunostaining of polytene chromosomes and ChIP indicate that the fourth chromosome is enriched in both H3K36me2 (fig. S1, A and B) and H3K36me3 (Fig. 1 and fig. S1A). Despite this, Set2 knockout caused no imbalance in transcription of fourth chromosome genes compared to those on autosomes (Fig. 2A). In contrast, the ash122/ash19011 and the NSDds46 mutants displayed a small but significant imbalance in the transcriptional output from the fourth chromosome (Fig. 4, A and B). The effect was markedly more pronounced in flies that lacked both ash1 and NSD functions (ash122 NSDds46/ash19011 NSD ds46) (Fig. 4C). By dividing the genes into housekeeping and non-housekeeping classes, we found that the bulk of the change corresponded to imbalanced transcriptional output from the non-housekeeping genes (Fig. 4D). The magnitude of the drop in the relative transcript abundance of the non-housekeeping genes and the stronger effect on this class of genes resembled those observed in Pof mutants, the key component of the regulatory mechanism specific to chromosome 4 (20, 64). We therefore asked whether the reduced transcript output is accompanied by a lost binding of POF to the fourth chromosome. Polytene chromosome staining showed no detectable difference of POF binding to the fourth chromosome in ash1 NSD double mutant (fig. S6). To test whether the observed reduction in ash122 NSDds46/ash19011 NSD ds46 is a consequence of impaired methylation of H3K36, we included ΔHisC; 12xH3K36R for comparison. The transcriptional output from chromosome 4 dropped in the H3K36R histone replacement mutant, ΔHisC; 12xH3K36R, by a similar extent to that in the ash1 NSD double mutant, likewise without affecting the POF binding (Figs. 3C and 4C and fig. S6). Together, these observations argue that methylation of H3K36 by both Ash1 and NSD is required to balance transcriptional output from the fourth chromosome.
Fig. 4. Ash1 and NSD show additive effects and are required for balanced transcription output from genes on chromosome 4.
Box plots of chromosome-specific transcript abundance ratios (log2 fold change) compared to wild type for chromosome 4 (red), autosomes (gray), and the X chromosome (green). (A) In ash122/ash19011, the transcript abundance from chromosome 4 is reduced by 8.0% compared to the main autosome arms. (B) In NSDds46, the transcript abundance from chromosome 4 is reduced by 5.1% compared to the main autosome arms. (C) In ash122 NSDds46/ash19011 NSD ds46, transcript abundance is reduced by 14.9% compared to the main autosome arms. (D) Box plots showing chromosome 4 transcript abundance change in ash122 NSDds46/ash19011 NSD ds46 compared to wild type. The genes are grouped as non-housekeeping (left) and housekeeping genes (right). In ash122 NSDds46/ash19011 NSD ds46, the expression output is reduced by 21.2% in non-housekeeping genes, which is significantly greater than the 11.7% reduction observed in housekeeping genes. The statistical significances were determined by unpaired two-sample Wilcoxon tests. In (A) to (C), chromosome 4 is compared to chromosome arm 2L, and in (D), chromosome 4 is compared to all genes located on chromosomes 2 and 3.
DISCUSSION
Two main conclusions can be drawn from the observations presented here. First, transcriptome profiling of Set2-deficient animals indicates that this methyltransferase is required for the balanced transcriptional output from the single male X chromosome. However, the comparison with mutants where the major histone H3 isoform is replaced with a variant that carries arginine instead of lysine-36 provides evidence that the process does not involve methylation of H3K36. Second, the balanced transcriptional output from the fourth chromosome requires intact H3K36 and depends on the additive functions of NSD and Ash1.
The latter conclusion was unexpected. We and others have previously shown that Set2 is responsible for bulk of H3K36me3 (28, 36). Consistently, we see an almost complete loss of H3K36me3 signal on polytene chromosomes in Set21 mutants. In contrast, the loss of Set2 causes no major changes in the bulk H3K36me2 and H3K36me1 (36). We therefore speculate that Ash1 and NSD stimulate transcription of the fourth chromosome genes by dimethylation of H3K36. On polytene chromosomes, H3K36me2 appears enriched in pericentric heterochromatin. The regions enriched in H3K36me2 coincide with those stained with antibodies against H3K9me3 and HP1 (23, 52). We have previously shown that HP1 and POF act in concert to balance transcriptional output from the fourth chromosome (18, 64, 65). The reduced transcriptional output from the fourth chromosome upon Ash1 and NSD loss resembles that in the Pof mutants (20, 64). However, neither ablation of Ash1 and NSD nor H3K36R replacement affects POF binding, which argues that the stimulatory effect of H3K36 methylation happens downstream of POF recruitment to the fourth chromosome. Conceivably, di-methylated H3K36 counteracts methylation of H3K9 and/or HP1 binding and repression. Future studies are warranted to test these hypotheses.
The fourth chromosome is enriched in repetitive DNA and is therefore the Drosophila chromosome most similar in genetic composition to the human genome. In contrast to Drosophila, human NSD proteins are essential for proper development, and haploinsufficiencies in NSD1 and NSD2 have been linked to Sotos and Wolf-Hirschhorn genetic syndromes, respectively (44). It is tempting to speculate that the stimulatory effect of NSD proteins may be mechanistically similar but quantitatively more important for mammalian genes embedded in the repeat-rich genome. The Drosophila fourth chromosome displays a high and unusual tolerance to dosage differences and misexpression (18, 20, 65–67). Therefore, it is expected that the decreased transcriptional output from the fourth chromosome of the NSD mutants has no severe physiological consequences.
Paradoxically, our observations indicate that H3K36me3, at least in third instar larvae stage, is less abundant on the male X chromosome, while it is enriched on chromosome 4, yet it is not directly involved in either chromosome-specific regulatory system, suggesting that the differences represent a consequence rather than a cause of these chromosomes-specific functions. How could we explain the observed differences in H3K36me3 levels? Although still a matter of discussion and debate (4), it has been proposed that transcription elongation efficiency is higher on the male X chromosome compared to autosomes (68, 69). Likewise, we have previously shown that there is a significant reduction of the transcription elongation efficiency (elongation density index) on the fourth chromosome compared to the other autosomes (67). Considering that Set2 travels with RNA polymerase II, we speculate that the faster elongation speeds on the dosage-compensated X chromosome leave less time for Set2 to add three methyl groups to H3K36, resulting in the overall lower H3K36me3 levels. Conversely, the slower elongation on the fourth chromosome leads to more extensive H3K36 trimethylation.
Our observations indicate that the loss of Set2 impairs the MSL complex–mediated dosage compensation, while the H3.2K36R replacement does not. We hypothesize that Set2 exerts its functions by methylation of (as yet unknown) nonhistone targets similar to what has recently been suggested for Ash1 (36). How this contributes to dosage compensation at the mechanistic level remains to be found. Nevertheless, our findings imply that models that involve the binding of MSL complex to trimethylated H3K36 need to be revised.
MATERIALS AND METHODS
Fly strains and genetic crosses
All fly strains were cultivated and crossed at 25°C in vials containing potato mash yeast agar. For the RNA preparations, brains from male third instar larvae were collected. Oregon R flies were used as wild type in all experiments except in the histone preplacement comparison as described below. To generate the ash1 mutant larvae, ash122 flies (w/+; ash122, P[w+mW.hs=FRT(whs) 2A]/TM3, Ser e P[w+mC ActGFP]) (36) were crossed with w1118; Df(3L)Exel9011/TM3, Ser e P[w+mC ActGFP], hereafter ash19011 (36), and the ash122/ash19011 trans-heterozygote first instar larvae were isolated, based on the lack of a GFP signal under fluorescent stereomicroscopy, and grown to the third instar larvae at 25°C. The CRISPR-Cas9–generated NSD loss-of-function allele (NSDds46) has previously been described (36), and the ash1 NSD double mutant larvae (ash122 NSDds46/ash19011 NSD ds46) were generated as described previously (36). To obtain Set2 mutant larvae, we used the Set21 loss-of-function allele (68). Progeny from y1 w67c23 Set21/FM7c, P[GAL4-Kr.C]DC1 P[UAS-GFP.S65T]DC5 were screened under a fluorescent stereomicroscope, and non-GFP larvae (homozygous and hemizygous for Set21) were transferred to separate vials. To generate ΔHisC; 12xH3K36R and ΔHisC; 12xH3K36K larvae, the fly strain w; UAS-2xYFP ΔHisC/CyO, P[ftz-lacZ] was crossed with either y w; elav-Gal4, ΔHisC/CyO; VK33[H3K36Rx12]/TM6B,Tb (to supplement the progeny with the transgenic mutant Histone cluster where Lys36 residue of His3.2 was replaced by Arg) or y w; elav-Gal4, ΔHisC/CyO; VK33[H3K36Kx12]/TM6B,Tb (to supplement the progeny with the transgenic wild-type Histone cluster) (57). The larvae with the correct genotype were then selected on the basis of the presence of a GFP signal [hence homozygous for deletion of endogenous Histone cluster (ΔHisC)] and an absence of the Tb marker (hence presence of the transgenic Histone cluster). The ΔH3.3B; ΔH3.3A mutant larvae (70) were isolated on the basis of the lack of a GFP signal under fluorescent stereomicroscopy in the progeny from the cross w, ΔHis3.3B, hsp-Flp; Df(2L)His3.3A/CyO, P[w+mC ActGFP]JMR1 × w, ΔHis3.3B, hsp-Flp/Y; Df(2L)His3.3A/CyO, P[w+mC ActGFP]JMR1.
Immunostaining of polytene chromosomes
Immunostaining of polytene chromosomes was done essentially as described previously (67, 71). The antibodies used in the protocol are listed in table S1. Preparations were analyzed using a Zeiss Axiophot microscope (Plan Apochromat 40×/0.95 objective) equipped with a KAPPA DX20C CCD camera and with a Zeiss Apotome Microscope [Plan Apochromat 63×/1.40 oil DIC M27 objective; filter set: 63HE for red channel, 38HE for green channel, and 49 for 4′,6-diamidino-2-phenylindole (DAPI)] equipped with AxioCam MR R3 camera. For comparisons of targeting between different genotypes, the protocol was run in parallel, and nuclei with clear cytology were chosen on the basis of DAPI staining and photographed. At least 20 nuclei per slide were used in these comparisons and at least five slides per genotype. Images were processed with ZenPro software (v2.3, Zeiss).
Western blot
Total tissue extracts were prepared from hand-dissected brains and imaginal discs of third instar larvae. For analysis of the H3K36 methylation, protein extracts were loaded on a 15% SDS–polyacrylamide gel electrophoresis gel and blotted to a polyvinylidene difluoride membrane for 60 min at 200 mA. Primary and secondary antibodies were diluted in 1× PBS, 1% BSA, and 0.05% Tween-20. The antibodies used in the protocol are listed in table S1.
Library preparation
For library preparations, we chose brains from third instar larvae to test a diploid tissue, to reduce potential maternal effects, and since some of the used genotypes die in late larvae/pupae stage. For each sample, five male third instar larvae brains were homogenized, and RNA was purified using a Directzol microprep kit (Zymo Research) and using the standard protocol with on-column DNase treatment. RNA quality was determined with a Fragment Analyzer 5200 (Agilent Technologies Inc.) using the DNF-471 Standard Sensitivity RNA reagent kit. Total RNA libraries were synthesized using the Ovation RNA-seq system for Drosophila (NuGEN). Standard protocol procedure with integrated DNase treatment was used to make the sequencing libraries. Fragmentation was performed using a Covaris E220 Focused ultrasonicator with the recommended settings for 200–base pair (bp) target length. The library quality was evaluated on a Fragment Analyzer using the DNF-920 DNA reagent kit.
Sequencing and data analyses
In the first round, a total of five samples per genotype were sequenced (Oregon R, Set21, ash122/ash19011, NSDds46, ash122 NSDds46/ash19011 NSD ds46, ΔHisC; 12xH3K36R and ΔHisC; 12xH3K36K) on Illumina HiSeq2500 (Illumina Cambridge Ltd.) by Science for Life Laboratory, Stockholm, and 125-bp paired-end reads were obtained. In the second round, seven samples of ΔH3.3B; ΔH3.3A and an additional six Oregon R samples were sequenced at Science for Life Laboratory, Stockholm using Illumina NovaSeq SP (Illumina Cambridge Ltd.) with a paired-end read length of 150 bp. Reads from both rounds were mapped to the D. melanogaster genome version BDGP6 using STAR v2.5.3 (72) with default settings. Read counts were obtained with featureCounts v1.5.1 with default settings (73). Gene abundances were calculated using StringTie v1.3.3 (74). Replicate 3 of Set21 and replicate 5 of ΔHisC; 12xH3K36K did not pass the quality control in RSeQC (75) and were therefore excluded in the downstream analysis. DESeq2 v1.18 (76) was used for differential expression analysis using default parameters and normal log fold change shrinkage.
Distance to HASs and MSL gene enrichments
Genomic locations for 188 previously published HASs, compiled in (55), originating from (7, 8) were used in the analysis. The distance to the closest HAS was calculated for each gene on the X chromosome. The gene enrichment data for the MSL complex component MSL1 were from (55). The binding data were subdivided into five equally sized bins based on the average MSL1 enrichment, bin 1 comprising the genes with the lowest MSL1 enrichments, up to bin 5, which comprised genes with the highest MSL1 enrichments.
Classification of genes as housekeeping or non-housekeeping
Genes with expression levels greater than 6 in all 12 FlyAtlas-specified tissue types (77) are defined as housekeeping genes, and genes with expression levels greater than 6 in 11 or fewer tissue types are here defined as non-housekeeping genes or differentially expressed genes.
Wild-type gene expression and developmental RNA profiles
The average transcript per million (TPM) score for wild-type samples (Oregon R) was calculated for all genes and subsequently binned according to Flybase RNA-seq convention (78). The bins are 0 to 0 TPM, 1 to 3 TPM, 4 to 10 TPM, 11 to 25 TPM, 26 to 50 TPM, 51 to 100 TPM, 101 to 1000 TPM, and >1000 TPM for unexpressed, very low expression, low expression, moderate expression, moderately high expression, high expression, very high expression, and extremely high expression, respectively. The developmental RNA expression profiles of Set2, ash1, and NSD were compiled from the modENCODE development RNA-seq data (79).
Datasets and calculated gene enrichments
Raw and processed sequencing data generated were deposited to NCBI GEO under accession GSE166934. ChIP-chip datasets were obtained from GSE23457, GSE23458, and GSE32793. ChIP-seq datasets were obtained from GSE127177 (48) and, in processed form, provided by T. Schauer and P. Becker. RNA-seq data from the temperature-sensitive E(z)61 cell line were obtained from GSE61307 (61). The E(z)61 cell line was classified as male based on the expression of roX1, roX2, msl2, and traF. Enrichment scores for the ChIP-chip experiments is a log2 ratio of ChIP over input. A custom script was used to calculate the top 50% of the total exon region for each gene. For ChIP-seq, the Homer suite v4.11 (80) was used to obtain H3K36me3 peaks over input. The peak score used for downstream analysis is defined as position-adjusted reads from initial peak region. Only peaks originating in exons were used when calculating the chromosomal average.
Bioinformatics and plotting of the figures
All calculations after the compilation of the raw counts table were performed using R (81), and plots were generated using the ggplot2 R package (82).
Acknowledgments
We thank T. Schauer and P. Becker for sharing H3K36me3 ChIP-seq data; D. Schübeler for the NSD antibody; and M. Kuroda, G. Matera, and K. Basler for fly strains. We also thank the Science for Life Laboratory, Stockholm, Sweden; the National Genomics Infrastructure (NGI), Stockholm, Sweden; and UPPMAX, Uppsala, Sweden, for assistance with the RNA-seq and for providing the computational infrastructure. Funding: This work was supported by grants from the Swedish Research Council (2016-03306 to J.L. and 2017-03918 to Y.B.S.), Knut and Alice Wallenberg Stiftelse (2014.0018 to J.L. and Y.B.S.), Nils Erik Holmstens Forskningsstiftelse (to Y.B.S.), and Swedish Cancer Foundation (CAN 2017/342 to J.L.). Author contributions: J.L. and H.L. conceived the project. H.L. performed the RNA-seq experiments and all bioinformatics. A.G. and J.L. performed the immunostainings, and A.G. performed the Western blots. H.L. and E.D. performed the fly genetics. H.L., A.G., E.D., Y.B.S., and J.L. together analyzed the data. H.L., Y.B.S., and J.L. wrote the manuscript with contributions from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials and in the Gene Expression Omnibus database (GSE166934).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Table S1
REFERENCES AND NOTES
- 1.Bachtrog D., Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14, 113–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mank J. E., Sex chromosome dosage compensation: Definitely not for everyone. Trends Genet. 29, 677–683 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Stenberg P., Larsson J., Buffering and the evolution of chromosome-wide gene regulation. Chromosoma 120, 213–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuroda M. I., Hilfiker A., Lucchesi J. C., Dosage compensation in Drosophila—A model for the coordinate regulation of transcription. Genetics 204, 435–450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliver B., Sex, dose, and equality. PLoS Biol. 5, e340 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samata M., Akhtar A., Dosage compensation of the X chromosome: A complex epigenetic assignment involving chromatin regulators and long noncoding RNAs. Annu. Rev. Biochem. 87, 323–350 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Alekseyenko A. A., Peng S., Larschan E., Gorchakov A. A., Lee O. K., Kharchenko P., McGrath S. D., Wang C. I., Mardis E. R., Park P. J., Kuroda M. I., A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134, 599–609 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straub T., Grimaud C., Gilfillan G. D., Mitterweger A., Becker P. B., The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLOS Genet. 4, e1000302 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villa R., Schauer T., Smialowski P., Straub T., Becker P. B., PionX sites mark the X chromosome for dosage compensation. Nature 537, 244–248 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Kelley R. L., Meller V. H., Gordadze P. R., Roman G., Davis R. L., Kuroda M. I., Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98, 513–522 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Figueiredo M. L., Kim M., Philip P., Allgardsson A., Stenberg P., Larsson J., Non-coding roX RNAs prevent the binding of the MSL-complex to heterochromatic regions. PLOS Genet. 10, e1004865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith E. R., Pannuti A., Gu W. G., Steurnagel A., Cook R. G., Allis C. D., Lucchesi J. C., The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20, 312–318 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhtar A., Becker P. B., Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367–375 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Vicoso B., Bachtrog D., Numerous transitions of sex chromosomes in Diptera. PLOS Biol. 13, e1002078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vicoso B., Bachtrog D., Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 499, 332–335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim M., Ekhteraei-Tousi S., Lewerentz J., Larsson J., The X-linked 1.688 satellite in Drosophila melanogaster promotes specific targeting by Painting of fourth. Genetics 208, 623–632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson J., Chen J. D., Rasheva V., Rasmuson-Lestander A., Pirrotta V., Painting of fourth, a chromosome-specific protein in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 98, 6273–6278 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson A. M., Stenberg P., Bernhardsson C., Larsson J., Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J. 26, 2307–2316 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsson J., Svensson M. J., Stenberg P., Mäkitalo M., Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc. Natl. Acad. Sci. U.S.A. 101, 9728–9733 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenberg P., Lundberg L. E., Johansson A. M., Ryden P., Svensson M. J., Larsson J., Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLOS Genet. 5, e1000465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng X., Meller V. H., Molecularly severe roX1 mutations contribute to dosage compensation in Drosophila. Genesis 47, 49–54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Malone J. H., Powell S. K., Periwal V., Spana E., Macalpine D. M., Oliver B., Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8, e1000320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figueiredo M. L., Philip P., Stenberg P., Larsson J., HP1a recruitment to promoters is independent of H3K9 methylation in Drosophila melanogaster. PLOS Genet. 8, e1003061 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riddle N. C., Minoda A., Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Tolstorukov M. Y., Gorchakov A. A., Jaffe J. D., Kennedy C., Linder-Basso D., Peach S. E., Shanower G., Zheng H., Kuroda M. I., Pirrotta V., Park P. J., Elgin S. C., Karpen G. H., Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21, 147–163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valsecchi C. I. K., Basilicata M. F., Georgiev P., Gaub A., Seyfferth J., Kulkarni T., Panhale A., Semplicio G., Manjunath V., Holz H., Dasmeh P., Akhtar A., RNA nucleation by MSL2 induces selective X chromosome compartmentalization. Nature 589, 137–142 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Schauer T., Ghavi-Helm Y., Sexton T., Albig C., Regnard C., Cavalli G., Furlong E. E., Becker P. B., Chromosome topology guides the Drosophila dosage compensation complex for target gene activation. EMBO Rep. 18, 1854–1868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sural T. H., Peng S., Li B., Workman J. L., Park P. J., Kuroda M. I., The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat. Struct. Mol. Biol. 15, 1318–1325 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larschan E., Alekseyenko A. A., Gortchakov A. A., Peng S., Li B., Yang P., Workman J. L., Park P. J., Kuroda M. I., MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol. Cell 28, 121–133 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Kim D., Blus B. J., Chandra V., Huang P., Rastinejad F., Khorasanizadeh S., Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat. Struct. Mol. Biol. 17, 1027–1029 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore S. A., Ferhatoglu Y., Jia Y., Al-Jiab R. A., Scott M. J., Structural and biochemical studies on the chromo-barrel domain of male specific lethal 3 (MSL3) reveal a binding preference for mono- or dimethyllysine 20 on histone H4. J. Biol. Chem. 285, 40879–40890 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka Y., Katagiri Z., Kawahashi K., Kioussis D., Kitajima S., Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene 397, 161–168 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Lee Y., Yoon E., Cho S., Schmähling S., Müller J., Song J. J., Structural basis of MRG15-mediated activation of the ASH1L histone methyltransferase by releasing an autoinhibitory loop. Structure 27, 846–852.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Schmähling S., Meiler A., Lee Y., Mohammed A., Finkl K., Tauscher K., Israel L., Wirth M., Philippou-Massier J., Blum H., Habermann B., Imhof A., Song J.-J., Müller J., Regulation and function of H3K36 di-methylation by the trithorax-group protein complex AMC. Development 145, dev163808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang C., Yang F., Zhang Z., Zhang J., Cai G., Li L., Zheng Y., Chen S., Xi R., Zhu B., Mrg15 stimulates Ash1 H3K36 methyltransferase activity and facilitates Ash1 Trithorax group protein function in Drosophila. Nat. Commun. 8, 1649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou P., Huang C., Liu C.-P., Yang N., Yu T., Yin Y., Zhu B., Xu R.-M., Structural insights into stimulation of Ash1L’s H3K36 methyltransferase activity through Mrg15 binding. Structure 27, 837–845.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Dorafshan E., Kahn T. G., Glotov A., Savitsky M., Walther M., Reuter G., Schwartz Y. B., Ash1 counteracts Polycomb repression independent of histone H3 lysine 36 methylation. EMBO Rep. 20, e46762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tripoulas N. A., Hersperger E., La Jeunesse D., Shearn A., Molecular genetic analysis of the Drosophila melanogaster gene absent, small or homeotic discs1 (ash1). Genetics 137, 1027–1038 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klymenko T., Muller J., The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 5, 373–377 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell O., Wirbelauer C., Hild M., Scharf A. N., Schwaiger M., MacAlpine D. M., Zilbermann F., van Leeuwen F., Bell S. P., Imhof A., Garza D., Peters A. H., Schübeler D., Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 26, 4974–4984 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan W., Xie J., Long C., Erdjument-Bromage H., Ding X., Zheng Y., Tempst P., Chen S., Zhu B., Reinberg D., Heterogeneous nuclear ribonucleoprotein L is a subunit of human KMT3a/Set2 complex required for H3 Lys-36 trimethylation activity in vivo. J. Biol. Chem. 284, 15701–15707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rayasam G. V., Wendling O., Angrand P. O., Mark M., Niederreither K., Song L., Lerouge T., Hager G. L., Chambon P., Losson R., NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 22, 3153–3163 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Trojer P., Xu C.-F., Cheung P., Kuo A., Drury W. J. III, Qiao Q., Neubert T. A., Xu R.-M., Gozani O., Reinberg D., The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J. Biol. Chem. 284, 34283–34295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kudithipudi S., Lungu C., Rathert P., Happel N., Jeltsch A., Substrate specificity analysis and novel substrates of the protein lysine methyltransferase NSD1. Chem. Biol. 21, 226–237 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Kurotaki N., Imaizumi K., Harada N., Masuno M., Kondoh T., Nagai T., Ohashi H., Naritomi K., Tsukahara M., Makita Y., Sugimoto T., Sonoda T., Hasegawa T., Chinen Y., Tomita Ha H. A., Kinoshita A., Mizuguchi T., Yoshiura Ki K., Ohta T., Kishino T., Fukushima Y., Niikawa N., Matsumoto N., Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 30, 365–366 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Meers M. P., Henriques T., Lavender C. A., McKay D. J., Strahl B. D., Duronio R. J., Adelman K., Matera A. G., Histone gene replacement reveals a post-transcriptional role for H3K36 in maintaining metazoan transcriptome fidelity. eLife 6, e23249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner B. M., Birley A. J., Lavender J., Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69, 375–384 (1992). [DOI] [PubMed] [Google Scholar]
- 47.Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V., Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 111, 185–196 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Prayitno K., Schauer T., Regnard C., Becker P. B., Progressive dosage compensation during Drosophila embryogenesis is reflected by gene arrangement. EMBO Rep. 20, e48138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.modENCODE Consortium, Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M. L., Landolin J. M., Bristow C. A., Ma L., Lin M. F., Washietl S., Arshinoff B. I., Ay F., Meyer P. E., Robine N., Washington N. L., Di Stefano L., Berezikov E., Brown C. D., Candeias R., Carlson J. W., Carr A., Jungreis I., Marbach D., Sealfon R., Tolstorukov M. Y., Will S., Alekseyenko A. A., Artieri C., Booth B. W., Brooks A. N., Dai Q., Davis C. A., Duff M. O., Feng X., Gorchakov A. A., Gu T., Henikoff J. G., Kapranov P., Li R., MacAlpine H. K., Malone J., Minoda A., Nordman J., Okamura K., Perry M., Powell S. K., Riddle N. C., Sakai A., Samsonova A., Sandler J. E., Schwartz Y. B., Sher N., Spokony R., Sturgill D., van Baren M., Wan K. H., Yang L., Yu C., Feingold E., Good P., Guyer M., Lowdon R., Ahmad K., Andrews J., Berger B., Brenner S. E., Brent M. R., Cherbas L., Elgin S. C., Gingeras T. R., Grossman R., Hoskins R. A., Kaufman T. C., Kent W., Kuroda M. I., Orr-Weaver T., Perrimon N., Pirrotta V., Posakony J. W., Ren B., Russell S., Cherbas P., Graveley B. R., Lewis S., Micklem G., Oliver B., Park P. J., Celniker S. E., Henikoff S., Karpen G. H., Lai E. C., MacAlpine D. M., Stein L. D., White K. P., Kellis M., Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz S., Meshorer E., Ast G., Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 16, 990–995 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Alekseyenko A. A., Gorchakov A. A., Zee B. M., Fuchs S. M., Kharchenko P. V., Kuroda M. I., Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev. 28, 1445–1460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.James T. C., Eissenberg J. C., Craig C., Dietrich V., Hobson A., Elgin S. C., Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50, 170–180 (1989). [PubMed] [Google Scholar]
- 53.Kim M., Faucillion M. L., Larsson J., RNA-on-X 1 and 2 in Drosophila melanogaster fulfill separate functions in dosage compensation. PLOS Genet. 14, e1007842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kind J., Vaquerizas J. M., Gebhardt P., Gentzel M., Luscombe N. M., Bertone P., Akhtar A., Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 133, 813–828 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Philip P., Stenberg P., Male X-linked genes in Drosophila melanogaster are compensated independently of the male-specific lethal complex. Epigenetics Chromatin 6, 35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gunesdogan U., Jackle H., Herzig A., A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep. 11, 772–776 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKay D. J., Klusza S., Penke T. J., Meers M. P., Curry K. P., McDaniel S. L., Malek P. Y., Cooper S. W., Tatomer D. C., Lieb J. D., Strahl B. D., Duronio R. J., Matera A. G., Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell 32, 373–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jani K. S., Jain S. U., Ge E. J., Diehl K. L., Lundgren S. M., Muller M. M., Lewis P. W., Muir T. W., Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 116, 8295–8300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finogenova K., Bonnet J., Poepsel S., Schafer I. B., Finkl K., Schmid K., Litz C., Strauss M., Benda C., Muller J., Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3. eLife 9, e61964 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deevy O., Bracken A. P., PRC2 functions in development and congenital disorders. Development 146, dev181354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee H. G., Kahn T. G., Simcox A., Schwartz Y. B., Pirrotta V., Genome-wide activities of Polycomb complexes control pervasive transcription. Genome Res. 25, 1170–1181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones R. S., Gelbart W. M., Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 126, 185–199 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mito Y., Henikoff J. G., Henikoff S., Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37, 1090–1097 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Lundberg L. E., Stenberg P., Larsson J., HP1a, Su(var)3-9, SETDB1 and POF stimulate or repress gene expression depending on genomic position, gene length and expression pattern in Drosophila melanogaster. Nucleic Acids Res. 41, 4481–4494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johansson A. M., Stenberg P., Pettersson F., Larsson J., POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLOS Genet. 3, e209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.B. Hochman, The fourth chromosome of Drosophila melanogaster, in The Genetics and Biology of Drosophila, M. Ashburner, E. Novitski, Eds. (Academic Press, 1976), vol. 1B, pp. 903–928. [Google Scholar]
- 67.Johansson A. M., Stenberg P., Allgardsson A., Larsson J., POF regulates the expression of genes on the fourth chromosome in Drosophila melanogaster by binding to nascent RNA. Mol. Cell. Biol. 32, 2121–2134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larschan E., Bishop E. P., Kharchenko P. V., Core L. J., Lis J. T., Park P. J., Kuroda M. I., X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471, 115–118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prabhakaran M., Kelley R. L., Mutations in the transcription elongation factor SPT5 disrupt a reporter for dosage compensation in Drosophila. PLOS Genet. 8, e1003073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hodl M., Basler K., Transcription in the absence of histone H3.3. Curr. Biol. 19, 1221–1226 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Lundberg L. E., Kim M., Johansson A.-M., Faucillion M.-L., Josupeit R., Larsson J., Targeting of Painting of fourth to roX1 and roX2 proximal sites suggests evolutionary links between dosage compensation and the regulation of the fourth chromosome in Drosophila melanogaster. G3 3, 1325–1334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L., StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L., Wang S., Li W., RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chintapalli V. R., Wang J., Dow J. A., Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Thurmond J., Goodman J. L., Strelets V. B., Attrill H., Gramates L. S., Marygold S. J., Matthews B. B., Millburn G., Antonazzo G., Trovisco V., Kaufman T. C., Calvi B. R.; FlyBase Consortium , FlyBase 2.0: The next generation. Nucleic Acids Res. 47, D759–D765 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W., Brown J. B., Cherbas L., Davis C. A., Dobin A., Li R., Lin W., Malone J. H., Mattiuzzo N. R., Miller D., Sturgill D., Tuch B. B., Zaleski C., Zhang D., Blanchette M., Dudoit S., Eads B., Green R. E., Hammonds A., Jiang L., Kapranov P., Langton L., Perrimon N., Sandler J. E., Wan K. H., Willingham A., Zhang Y., Zou Y., Andrews J., Bickel P. J., Brenner S. E., Brent M. R., Cherbas P., Gingeras T. R., Hoskins R. A., Kaufman T. C., Oliver B., Celniker S. E., The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.R_Core_Team, (R Foundation for Statistical Computing, Vienna, Austria, 2016).
- 82.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, New York, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Table S1