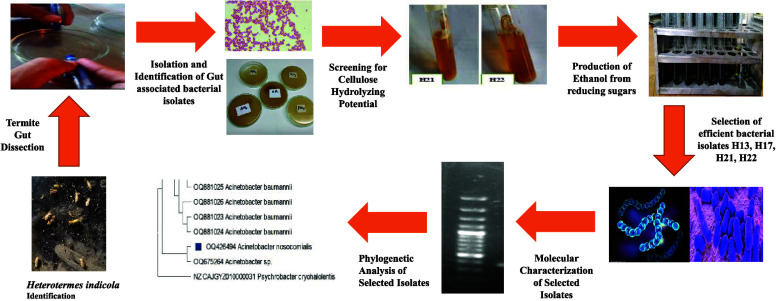
Abstract
Cellulose is the basic component of lignocellulosic biomass (LCB) making it a suitable substrate for bioethanol fermentation. Cellulolytic and ethanologenic bacteria possess cellulases that convert cellulose to glucose, which in turn yields ethanol subsequently. Heterotermes indicola is a subterranean termite that causes destructive damage by consuming wooden structures of infrastructure, LCB products, etc. Prospectively, the study envisioned the screening of cellulolytic and ethanologenic bacteria from the termite gut. Twenty six bacterial strains (H1–H26) based on varied colonial morphologies were isolated. Bacterial cellulolytic activity was tested biochemically. Marked gas production in the form of bubbles (0.1–4 cm) in Durham tubes was observed in H3, H7, H13, H15, H17, H21, and H22. Sugar degradation of all isolates was indicated by pink to maroon color development with the tetrazolium salt. Hallow zones (0.42–11 mm) by Congo red staining was exhibited by all strains except H2, H7, H8, and H19. Among the 26 bacterial isolates, 12 strains were identified as efficient cellulolytic bacteria. CMCase activity and ethanol titer of all isolates varied from 1.30 ± 0.03 (H13) to 1.83 ± 0.01 (H21) umol/mL/min and 2.36 ± 0.01 (H25) to 7.00 ± 0.01 (H21) g/L, respectively. Likewise, isolate H21 exhibited an ethanol yield of 0.40 ± 0.10 g/g with 78.38 ± 2.05% fermentation efficiency. Molecular characterization of four strains, Staphylococcus sp. H13, Acinetobacter baumanni H17, Acinetobacter sp. H21, and Acinetobacter nosocomialis H22, were based on the maximum cellulolytic index and the ethanol yield. H. indicola harbor promising and novel bacteria with a natural cellulolytic tendency for efficient bioconversion of LCB to value-added products. Hence, the selected cellulolytic bacteria can become an excellent addition for use in enzyme purification, composting, and production of biofuel at large.
1. Introduction
Lignocellulosic biomass (LCB) is considered a renewable and inexpensive resource in the world with energy content ($3–4/GJ).1−3 Pakistan, being an agricultural country, is one of those countries producing abundant lignocellulosic waste. In Pakistan, agricultural waste, especially fruit and vegetable waste, wheat husks, rice husks, cotton sticks, and sugar cane residues, is abundant.4 Annual massive accumulation of lignocellulosic waste in the form of fruit, vegetable, and agro crops has led to the spoilage of the valuable biomass, which can be processed in a wide range of value-added products, and deterioration of the environment and hence necessitates the requirement to look for new avenues for applicable utilization. The current estimates for lignocellulosic waste, wood, and wood-based residues are 20,494, 25,271, and 1,121 million tons, respectively.5 According to The Pakistan Business Council,6 Pakistan’s overall estimated biomass potential is 50,000 GW h/year or up to 36% of the country’s entire energy scenario.
Naturally, the cellulosic substrate is decomposed by the action of a mixture of hydrolytic enzymes (known as cellulases). The cellulase is a complex enzyme comprising endogenous (endoglucanase) and exogenous (cellobiohydrolase) that work synergistically in cellulose-degrading microorganisms.7,8 Cellulases of cellulolytic bacteria are involved in the hydrolysis of cellulose, producing sugar derivatives by breaking β-1,4-glycosidic linkage.9 Globally, cellulases are used due to their remarkable industrial applications, viz, biorefinery, paper, textile, feed, and agriculture industries, over decades. The worldwide demand for industrial cellulases reaches up to eight percent.10,11 A wide range of microbes, bacteria, and fungi have the ability to degrade the cellulose and cell wall components of plants. Among them, fungi is the main producer of cellulase.12,13
Cellulolytic enzyme systems are present naturally in many organisms and plants to degrade living as well as rotten plant materials. These organisms may serve as impending candidates for the production of biofuel from lignocellulosin substrate.7,14,15 Currently, attempts are made to screen substantial diversity of the environment and organisms to possess enzymes with amended characteristics of cellulolysis, a process of reliable determination and assessment of cellulase activity. A variety of aerobic, anaerobic, and facultative anaerobic microbes have the potential to breakdown cellulosic biomass.16,17 The metabolic, physiological, and efficient enzyme machinery of the microbe contributes for considerable carbon flow in the biosphere.7,15,18 As for animals, LCB is a poor nutritional food source and remains undigested due to lack of cellulolytic machinery and metabolic pathway. Termites play a havoc, being highly destructive to wooden structures and other cellulosic material, causing an estimated amount of $3 billion loss annually worldwide.19,20 They possess an ability to digest cellulose efficiently.21−23 Due to their lignocellulosic biomass recycling potential, they play a vital role in tropical, subtropical, and Mediterranean ecosystems.24,25 In the detritus food chain, they are the main decomposers due to their own cellulolytic system that is enhanced by the presence of cellulose-degrading microbes in their gut.26−28
Heterotermes indicola Wasmann, a subterranean termite, belongs to Rhinotermitidae, order Isoptera,29 and is termed as the lower termite that feeds only on wood. It is found in tropical to subtropical and warm temperate habitats in Pakistan, Sri Lanka, and India.30 Globally, it is known as the destructive lower termite that cause massive timber damage in agriculture and urban areas.31,32H. indicola has a highly organized nesting system with diffused colonies and earthen tunnels/galleries to hit timber. Moreover, it has the ability to spread up to 100 m or more by constructing satellite nests underground in woody material and rafters.33 This insect can also build hanging satellite tubes in search of moisture and food.34 The highly wood-damaging behavior of termites pointed to the gut’s powerful innate cellulolytic system. It may contribute to the fact that the gut of the lower termite has synergistic bacterial, fungal, and protozoan species, whereas higher termites possess only few bacterial species with no protozoans.35 The hind gut of H. indicola is larger than the midgut and contains a diverse range of bacteria and protozoans (symbiont). The microbial communities present in the hind gut of termite are linked to the digestion of wood (nutrient-deficient food source). Symbionts supplement this nutrient-deficient diet by synthesizing other necessary nutrients and stimulate reactions involved in the breakdown of all three major components of wood (cellulose, hemicellulose, and lignin phenolics).36,37 Symbiotic cellulase systems produce complex cellulolytic enzymes. Thus, it exhibits strong hydrolytic activity (40–88%) against the carboxymethyl cellulose (CMC) substrate in comparison with endogenous cellulase (40–85%) of total gut activity.38
It is evident that the termite gut is a complex microhabitat having distinct biotic and abiotic features that offer ecological niches to a diverse range of microbiota.39 The termite gut is the unique microhabitat for the micobiota and cellulolytic flagellates.40−42 As a result, scientists from all over the world are now particularly interested in the diversity and role of the microbial community in the termite gut.42−44 The microbiota present in the gut of lower termites belong to archaea, eukarya, and bacteria. Methanogen represented the intestinal group of archaea, whereas the protozoans, yeast, and fungi belong to eukarya. The gut bacteria may be Gram-positive, viz, bacteroides, firmicutes, acinetobacter, and spirochaetes. Gut bacteria along with symbiotic protists degrade aromatic compounds, hemicellulose, cellulose, and fixed nitrogen,45 while higher termites broke down only cellulose by relying on their own enzymes.46 The cellulolytic bacterial groups of the gut isolated from Coptotermes curvignathus were Bacillus cereus, Chryseobacterium kwangyangense, Acinetobacter sp., Enterobacter cloacae, and E. aerogenes.47−49 Ali et al.37 isolated five symbiotic bacterial strains Paenibacillus lactis, Lysinibacillus macrolides, Stenotrophomonas maltophilia, Lysinibacillus fusiformis, and Bacillus cereus from the gut of lower termite Psammotermes hypostoma. A cellulolytic Bacillus licheniformis HI-08 with 400 U/mL cellulase activity was reported from the wood-feeding lower termite Heterotermes indicola.50 The CMCase activity of 3.36 and 0.75 U/mL was recorded by Bacillus subtilis G4 and B. subtilis AS3 isolated from the termite gut.51,52 Javaheri-Kermani and Asoodeh53 reported a novel bifunctional (endo and exo glucanases) β-1,4-glucanase producing Bacillus sp. CF96 from lower termite Anacanthotermes sp. The purified β-1,4-glucanase has the ability to hydrolyze both CMC and avicel.54
The literature suggested that the cellulolytic bacterial isolates of termite gut possess great potential for breaking down LCB. There is a great chance to enhance the lignocellulose pretreatment by employing these effective bacterial cultures. Especially, in the canvas of biomass-to-bioenergy valorization, the search for efficient cellulases from the microbial systems is always in high demand. In this prospect, there is a need for exploring the cellylolytic potential of these enzymes for LCB hydrolysis and subsequent solvent production (e.g., acetone, butanol, ethanol, etc.).27,28,55,56 Hence, termites may be a desirable source of novel cellulolytic microorganisms and cellulases to be applied for the industrial conversion of biomass to biofuel. Termites are efficient wood decomposers57 that harbor diverse range of symbiotic cellulolytic microbiota. Termites opt different mechanisms for decomposition of wood such as direct ingestion and digestion, substrate modification (such as fragmentation and tunneling), and interactions with bacteria, fungi, and other saproxylic community members.58,59 Furthermore, these insects may have an impact on nitrogen dynamics in decaying woods by fixing nitrogen and releasing nutrients.60
The objectives of the current study are to screen and characterize molecularly the bacterial isolates from subterranean termite Heterotermes indicola for evaluation of their cellulolytic and ethanologenic potential. Microorganisms are exploited for bioconversion of natural renewable biomass to meet the demands of the future for energy and chemical precursors. However, termites possess excellent intestinal polysaccharide-degrading symbiosis (wide variety of bacteria and protozoans) over approximately 150 million years. Within the termite gut, ecosystem lignocellulose (cellulose and hemicellulose) is efficiently degraded while lignin contents are weakly attacked. The development of cellulose hydrolysis and biomass bioconversion processes/techniques, as explored in this study, may benefit from understanding of the principles of cellulose degradation in the naturally occurring polymer-degrading ecosystem from termite. The gut of termite may be a probable source to screen novel microbiota to be used for a wide range of industrial applications, viz, biofuel production and food industry.61
2. Results
2.1. Termite Collection
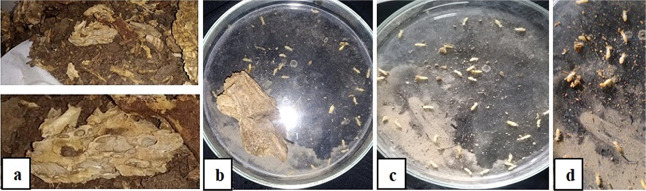
The termites collected were identified as Heterotermes indicola on the basis of large longer than broad yellowish brown rectangular heads and well developed slender mandibles that were slightly curved near the tip and were crossed while closing both. Labrum is tongue shaped with a needle-like tip (Figure 1).
Figure 1.
Heterotermes indicola. Termite collection (a), isolation of the termite (b, c), and identification of the soldier insect (d) (The photographs are original, unpublished and the effort of the first author).
2.2. Sampling of Termite for Bacterial Screening
Twenty six bacterial strains were identified from the termite’s gut and given codes H1–H26 based on the colonial morphology. The prefix H stands for Heterotermes indicola. All isolated bacteria were cultured on 2% cellulose-supplemented medium to get pure cultures and were preserved in the form of glycerol stock for further study.
2.3. Morphological Characterization of Isolated Bacteria
Colonial characteristics including colony size, color, elevation, margin, texture, optical features, pigmentation, and consistency of bacterial isolates on the cellulose-enriched medium were recorded in Table 1.
Table 1. Colonial Characteristics of Bacterial Isolates Following Streaking on Cellulose-Supplemented Agar Medium.
| sr. no | isolated bacteria | size | color of colony | margin | surface texture | elevation | optical feature | pigmentation | consistency |
|---|---|---|---|---|---|---|---|---|---|
| 1 | H1 | 0.5 mm | creamy white (pinkish) | entire | smooth | flat | opaque | no | brittle |
| 2 | H2 | 2 mm | off-white | undulate | smooth | flat | opaque | yes (light pink) | butyrous |
| 3 | H3 | 3 mm | creamy white | entire | granules | convex | opaque | no | butyrous |
| 4 | H4 | 1 mm | white | undulate | granules | flat | transparent | no | butyrous |
| 5 | H5 | 2 mm | off-white | undulate | smooth | concave | opaque | yes | viscous |
| 6 | H6 | 2 mm | creamy white | entire | smooth | dome | opaque | no | butyrous |
| 7 | H7 | 1.5 mm | creamy white | entire | smooth | dome | opaque | Yes (brownish) | viscous |
| 8 | H8 | pinpoint | off-white | undulate | granules | raised | opaque | no | butyrous |
| 9 | H9 | 3 mm | white | entire | smooth | flat | opaque | no | butyrous |
| 10 | H10 | pinpoint | White | undulate | granule | flat | transparent | no | butyrous |
| 11 | H11 | pinpoint | off-white | spreading | granule | irregular | opaque | no | butyrous |
| 12 | H12 | 2 mm | creamy white | entire | smooth | dome | opaque | no | butyrous |
| 13 | H13 | 2 mm | off-white | undulate | granule | concave | opaque | no | butyrous |
| 14 | H14 | 1 mm | white | entire | smooth | flat | opaque | no | butyrous |
| 15 | H15 | 3 mm | white | spreading | granule | flat | opaque | no | butyrous |
| 16 | H16 | 1 mm | white | undulate | granule | flat | transparent | no | butyrous |
| 17 | H17 | 3 mm | white | spreading | granule | flat | opaque | no | viscous |
| 18 | H18 | 3 mm | creamy white | undulate | smooth | flat | opaque | no | butyrous |
| 19 | H19 | 2 mm | white | entire | granule | flat | transparent | no | butyrous |
| 20 | H20 | 1 mm | white | spreading | granule | dome | opaque | no | butyrous |
| 21 | H21 | pinpoint | off-white | entire | smooth | flat | opaque | no | butyrous |
| 22 | H22 | 1 mm | off-white | undulate | granule | dome | opaque | no | butyrous |
| 23 | H23 | pinpoint | white | entire | smooth | flat | opaque | yes | viscous |
| 24 | H24 | pinpoint | white | undulate | granule | dome | transparent | no | butyrous |
| 25 | H25 | 2 mm | white | spreading | granule | irregular | opaque | no | viscous |
| 26 | H26 | 2 mm | off-white | entire | smooth | dome | opaque | no | butyrous |
The colonies vary from pinpoint to 2.5 cm as H8, H10, H11, H21, H23, and H24 were pinpoint, while H3, H9, H15, H17, and H18 showed 2.5 cm colony diameter. The color of the colonies were creamy white (H1, H3, H6, H7, H12, and H18), off-white (H2, H5, H8, H11, H13, H21, H22, and H26), and white for the remaining isolates. The margins of all colonies were entire, except H2, H4, H8, H10, H13, H16, H18, H22, and H24 (undulate) and H11, H15, H17, H20, and H25 (spreading). The textures of H1, H2, H5, H6, H7, H9, H12, H14, H18, H21, H23, and H26 were smooth, whereas other isolates were granular. The elevations observed were convex (H3), concave (H5 and H13), raised (H8), irregular (H11 and H25), dome-like (H6, H7, H12, H20, H22, H24, and H26), and flat for rest of the bacterial isolates. All isolates were opaque, except H4, H10, H16, H19, and H24 that were transparent. All bacterial isolates produced nonpigmented colonies except pigmented colonies of H2, H5, H7, and H23. Consistency of all isolates were butyreous except H5,H7, H17, H23, and H25 that were viscous.
2.4. Microscopic Examination of Bacterial Isolates
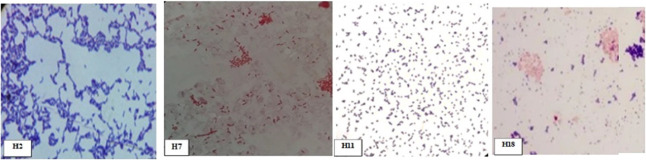
Microscopic cell characteristics of bacterial strains were studied by culturing on a cellulose-enriched medium. Cellular characteristics include cell shape, cell size, and cell type as shown in Table 2 and Figure 2. All bacterial strains were rod-shaped including long, short rods (with pointed or oval ends) except H1, H5, and H12 (diplococcic), H9, H13, H23, and H26 (coccus), and H7, H16, and H20 (spirila). The size ranged from 2 × 0.5 to 5.6 × 3 μm. The observed maximum size, i.e., 5.6 × 3 μm, was in H24, while a minimum of 2 × 0.5 μm was in H12. The size of spiral bacteria varied from 3 × 1 μm with one to two twists. All bacterial strains were Gram-positive, whereas H1, H3, H6, H7, H10, H13, H17, H21, H22, H23, and H24 were Gram-negative bacteria.
Table 2. Cellular Characteristics of Bacterial Isolates on Cellulose-Supplemented Medium.
| sr. no. | isolated bacteria | gram stain reaction | length (μm) | diameter (μm) | shape |
|---|---|---|---|---|---|
| 1 | H1 | gram-negative | 4.2 | 2 | diplococci |
| 2 | H2 | gram-positive | 5 | 2 0.7 | rods with pointed ends |
| 3 | H3 | gram-negative | 2 | 1.5 | short rod, chain |
| 4 | H4 | gram-positive | 5 | 2.5 | long rod, chain |
| 5 | H5 | gram-positive | 2 | 1 | diplococcic |
| 6 | H6 | gram-negative | 2.3 | 1.5 | single rod |
| 7 | H7 | gram-negative | 3 | 1.5 | spiral rod |
| 8 | H8 | gram-positive | 2 | 1 | single rod |
| 9 | H9 | gram-positive | 3 | 2 | coccus |
| 10 | H10 | gram-negative | 2 | 1.5 | single rod |
| 11 | H11 | gram-positive | 4 | 2.3 | cluster coccus |
| 12 | H12 | gram-positive | 2 | 0.5 | diplococci |
| 13 | H13 | gram-negative | 2.2 | 0.56 | coccus |
| 14 | H14 | gram-positive | 3 | 1.49 | cluster coccus |
| 15 | H15 | gram-positive | 4 | 2 | rods with oval ends |
| 16 | H16 | gram-positive | 2 | 1 | spiral rods |
| 17 | H17 | gram-negative | 5 | 2 | long rods, chain |
| 18 | H18 | gram-positive | 5 | 2.9 | rods with pointed ends |
| 19 | H19 | gram-positive | 5 | 1.89 | long rods |
| 20 | H20 | gram-positive | 3 | 1 | spiral rods |
| 21 | H21 | gram-negative | 2 | 0.58 | slightly curved rod |
| 22 | H22 | gram-negative | 2.5 | 0.5 | single rod |
| 23 | H23 | gram-negative | 3 | 2 | coccus |
| 24 | H24 | gram-negative | 5.6 | 3 | long rods, chain |
| 25 | H25 | gram-positive | 4 | 2 | short rods, chain |
| 26 | H26 | gram-positive | 3 | 1 | coccus |
Figure 2.
Gram reaction presented by different bacterial isolates as Gram-positive rods (H2, H18), Gram-negative rods (H7), and Gram-positive cocci (H11)(Photographs are original, unpublished and taken by the first author).
2.5. Analysis of Cellulose-Hydrolyzing Potential for Bacterial Isolates
In the present study, qualitative biochemical tests were performed to detect the utilization of cellulose by bacterial strains. These tests include gas production, color development, and clear zone formation via Durham tubes, tetrazolium chloride (TTC) indicator, and Congo red stain. Table 3 and Figure 2 interpreted the degradation of cellulose by different indicators. Bubbles of variable size 0.1–4 cm in Durham tubes were produced by H3, H7, H14, H15, H17, H21, H22, and H24 within 2–8 days. All bacteria produced a maroon color with TTC to depict the speedy degradation of cellulose except H23. Four bacterial isolates showed negative response, while 22 bacteria produced a clear zone as positive response by Congo red staining. The highest as well as lowest cellulolytic indexes were recorded as 5 (H21) and 0.67 (H9, H15).
Table 3. Biochemical Evaluation of Cellulolysis by Various Bacterial Isolatesa,b,c,d,e.
| sr. no. | bacterial isolates | bubble formation in Durham tubes | color development with TTC | clear zones diameter by Congo red staining (mm) | diameter of bacterial colonies | cellulolytic index |
|---|---|---|---|---|---|---|
| 1 | H1 | – | +++ (maroon) | 1.5 | 0.5 | 2.0 |
| 2 | H2 | – | +++ (maroon) | 2.0 | 0 | |
| 3 | H3 | ++ (1 cm, day 3) | +++ (maroon) | 7.0 | 3.0 | 1.33 |
| 4 | H4 | – | +++ (maroon) | 4.0 | 1.0 | 3.0 |
| 5 | H5 | – | +++ (maroon) | 4.0 | 2.0 | 1.0 |
| 6 | H6 | + (0.1 cm, day 8) | +++ (maroon) | 4.0 | 2.0 | 1.0 |
| 7 | H7 | – | +++ (maroon) | 1.5 | 0.0 | |
| 8 | H8 | – | +++ (maroon) | 0.1 | 0.0 | |
| 9 | H9 | – | +++ (maroon) | 5.0 | 3.0 | 0.67 |
| 10 | H10 | – | +++ (maroon) | 0.5 | 0.2 | 1.5 |
| 11 | H11 | – | +++ (maroon) | 0.42 | 0.2 | 1.1 |
| 12 | H12 | – | +++ (maroon) | 4.0 | 2.0 | 1.0 |
| 13 | H13 | +++ (1.5 cm, day 3) | +++ (maroon) | 4.0 | 2.0 | 1.0 |
| 14 | H14 | +++ (1.6 cm, day 2) | +++ (maroon) | 3.0 | 1.0 | 2.0 |
| 15 | H15 | + (0.1 cm, day 5) | +++ (maroon) | 5.0 | 3.0 | 0.67 |
| 16 | H16 | – | +++ (maroon) | 3.0 | 1.0 | 2.0 |
| 17 | H17 | + (0.2 cm, day 6) | +++ (maroon) | 11.0 | 3.0 | 2.67 |
| 18 | H18 | – | +++ (maroon) | 7.0 | 3.0 | 1.33 |
| 19 | H19 | – | +++ (maroon) | 2.0 | 0.0 | |
| 20 | H20 | – | +++ (maroon) | 5.0 | 1.0 | 4.0 |
| 21 | H21 | +++ (4 cm, day 2) | +++ (maroon) | 1.2 | 0.2 | 5.0 |
| 22 | H22 | +++ (3 cm, day 2) | +++ (maroon) | 5.0 | 1.0 | 4.0 |
| 23 | H23 | + (0.1 cm, day 2) | light pink, + + | 0.62 | 0.2 | 2.1 |
| 24 | H24 | ++ (1 cm, day 3) | +++ (maroon) | 0.82 | 0.2 | 3.1 |
| 25 | H25 | + (0.2 cm, day 4) | +++ (maroon) | 4.0 | 2.0 | 1.0 |
| 26 | H26 | + (0.1 cm, day 5) | +++ (maroon) | 6.0 | 2.0 | 2.0 |
Cellulolytic index = (clear zone diameter–bacterial colonies diameters)/ bacterial colonies diameter.
+++ Positive, strong response.
++ Positive, intermediate response.
+ Positive, weak response
– Negative response.
Figure 3.
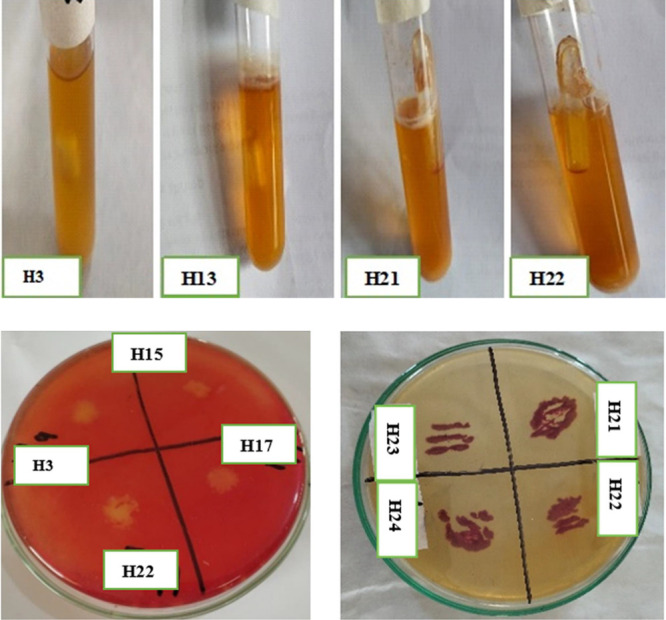
Bubble formation as observed in Durham tubes (upper row), clear zones by Congo red staining, and maroon color development with TTC (lower row) by different cellulolytic bacterial isolates (The photographs are original, unpublished and taken by the first author).
On the basis of qualitative analysis (color development by TTC indicator, gas production in Durham tubes, and hallow formation by Congo Red dye), 12 bacterial isolates (highlighted in Table 3) were selected for further study, i.e., cellulolytic and ethanologenic titer. Cellulolytic activity as depicted by conversion of cellulose to glucose is recorded in Table 4. The selected 12 bacterial isolates showed a tendency to release extracellular enzymes for cellulose hydrolysis. Bacterial isolates depicted the activity (μmol/min/mL) in the range of 1.30 ± 0.03 (H13) to 1.83 ± 0.01 (H21). Bacterial isolates H17 and H21 showed the maximum potential. The strains H17, H21, and H22 were selected for identification on the maximum cellulolytic potential.
Table 4. Estimation of Cellulolytic Potential of Different Bacterial Isolates to Hydrolyze Cellulosea.
| bacterial Isolates | enzyme activity (μmol/min/mL) |
|---|---|
| H3 | 1.65 ± 0.01B |
| H6 | 1.41 ± 0.01D |
| H13 | 1.30 ± 0.03E |
| H14 | 1.52 ± 0.01C |
| H15 | 1.67 ± 0.02B |
| H17 | 1.79 ± 0.024 |
| H21 | 1.83 ± 0.01A |
| H22 | 1.62 ± 0.02C |
| H23 | 1.44 ± 0.01D |
| H24 | 1.56 ± 0.01C |
| H25 | 1.54 ± 0.02C |
| H26 | 1.55 ± 0.01C |
Data embodied means ± SEM. Significance is recorded by different letters at p ≤ 0.05 by single-factor ANOVA.
2.6. Production of Ethanol from Cellulose as Substrate
Data related to ethanol titer by selected isolates are recorded in Table 5. Maximum ethanol was produced on different days by different isolates. Highest ethanol (g/L), i.e., 7.21 ± 0.01, 6.54 ± 0.01, 7.00 ± 0.01, and 5.64 ± 0.01, was generated by H13, H17, H21, and H22, respectively, on the eighth day of incubation followed by a decrease in content. In other strains, the maximum values were observed between days 7 and 8, and that was the indication of ethanol tolerance by bacterial isolates. Table 6 presents the calculated ethanol yield and fermentation efficiency (FE) in 2% CMC-supplemented fermentation medium. H21 showed a remarkable 0.40 g/g yield with 78.38% FE. Bacterial isolates H13, H17, and H22 exhibited 0.37–0.38 g/g yield with 71–73% FE. On the basis of the excellent ethanol yield, four isolates H13, H17, H21, and H22 were selected for molecular identification.
Table 5. Ethanol Titer (g/L) from Cellulose by Different Bacterial Isolatesa.
| days | H3 | H6 | H13 | H14 | H15 | H17 | H21 | H22 | H23 | H24 | H25 | H26 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.11 ± 0.02C | 2.04 ± 0.1H | 1.57 ± 0.021 | 1.20 ± 0.01H | 1.67 ± 0.01H | 1.97 ± 0.01H | 2.64 ± 0.01H | 1.73 ± 0.01J | 0.67 ± 0.03I | 0.71 ± 0.03J | 0.89 ± 002J | 2.18 ± 0.01I |
| 2 | 1.51 ± 0.03BC | 2.09 ± 0.01H | 2.33 ± 0.01H | 1.22 ± 0.01H | 1.70 ± 0.01H | 3.16 ± 0.01F | 3.18 ± 0.02G | 3.03 ± 0.01I | 1.70 ± 0.04F | 0.95 ± 0.01I | 1.48 ± 0.03H | 2.24 ± 0.01H |
| 3 | 2.86 ± 0.05AB | 2.54 ± 0.01G | 2.45 ± 0.02G | 1.99 ± 0.01F | 2.44 ± 0.02G | 3.42 ± 0.02E | 3.24 ± 0.01G | 3.40 ± 0.01H | 1.72 ± 0.01E | 1.04 ± 0.02H | 1.68 ± 0.03G | 2.63 ± 0.02F |
| 4 | 2.10 ± 0.02ABC | 4.64 ± 0.01D | 4.62 ± 0.03E | 3.30 ± 0.03D | 3.53 ± 0.01D | 4.11 ± 0.01C | 3.67 ± 0.02F | 3.91 ± 0.01F | 1.96 ± 0.03D | 3.12 ± 0.02E | 2.22 ± 0.02E | 2.67 ± 0.03F |
| 5 | 2.32 ± 0.02ABC | 5.08 ± 0.02C | 5.23 ± 0.02D | 4.00 ± 0.03C | 3.57 ± 0.04D | 4.17 ± 0.01C | 5.30 ± 0.02C | 4.39 ± 0.01D | 1.97 ± 0.03D | 3.23 ± 0.05D | 2.63 ± 0.03C | 3.18 ± 0.02D |
| 6 | 2.34 ± 0.04ABC | 5.20 ± 0.03B | 5.78 ± 0.01C | 4.02 ± 0.02C | 3.90 ± 0.03C | 4.29 ± 0.03B | 5.34 ± 0.02C | 5.00 ± 0.01C | 3.12 ± 0.02C | 3.37 ± 0.03C | 2.87 ± 0.02B | 3.52 ± 0.01C |
| 7 | 2.55 ± 0.01ABC | 5.54 ± 0.02A | 6.22 ± 0.03B | 4.84 ± 0.03A | 4.22 ± 0.01B | 4.36 ± 0.01B | 5.87 ± 0.07B | 5.42 ± 0.01B | 4.22 ± 0.02B | 3.68 ± 0.02B | 3.36 ± 0.03A | 5.63 ± 0.02A |
| 8 | 3.18 ± 0.01A | 5.52 ± 0.02A | 5.91 ± 0.03A | 4.59 ± 0.01B | 4.44 ± 0.01A | 6.54 ± 0.01A | 7.00 ± 0.01A | 5.64 ± 0.02A | 4.92 ± 0.02A | 4.57 ± 0.02A | 2.36 ± 0.01D | 3.62 ± 0.03B |
| 9 | 1.71 ± 0.03ABC | 3.64 ± 0.01F | 3.77 ± 0.01F | 2.67 ± 0.02E | 3.30 ± 0.02E | 3.58 ± 0.03D | 4.77 ± 0.02D | 4.09 ± 0.01E | 1.55 ± 0.04G | 1.73 ± 0.02F | 2.03 ± 0.01F | 2.98 ± 0.02E |
| 10 | 1.70 ± 0.04ABC | 4.36 ± 0.02E | 2.44 ± 0.01G | 1.76 ± 0.01G | 2.60 ± 0.04F | 2.17 ± 0.01G | 4.06 ± 0.03E | 3.59 ± 0.01G | 1.28 ± 0.02H | 1.07 ± 0.01G | 1.26 ± 0.02I | 2.51 ± 0.02G |
Values represent means ± SEM. Significance is recorded by different letters at p ≤ 0.05 by single-factor ANOVA.
Table 6. Calculated Ethanol Yield (g/g) and FE (%) from CMC by Different Bacterial Isolatesa,b,c.
| H3 |
H6 |
H13 |
H14 |
H15 |
H17 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| days | Yi | FE | Yi | FE | Yi | FE | Yi | FE | Yi | FE | Yi | FE |
| 1 | 0.21 ± 0.12 | 40.83 ± 0.21 | 0.16 ± 0.03 | 30.75 ± 0.19 | 0.15 ± 0.11 | 28.56 ± 0.01 | 0.17 ± 0.21 | 34.10 ± 0.19 | 0.21 ± 0.02 | 40.98 ± 2.90 | 0.25 ± 0.01 | 49.65 ± 1.45 |
| 2 | 0.25 ± 0.02 | 48.94 ± 0.17 | 0.15 ± 0.02 | 29.50 ± 0.14 | 0.21 ± 0.04 | 41.42 ± 0.01 | 0.16 ± 0.12 | 31.81 ± 0.20 | 0.19 ± 0.03 | 37.84 ± 2.02 | 0.29 ± 0.04 | 57.16 ± 1.25 |
| 3 | 0.21 ± 0.08 | 40.70 ± 0.23 | 0.17 ± 0.07 | 33.43 ± 0.19 | 0.16 ± 0.03 | 32.24 ± 0.07 | 0.23 ± 0.07 | 45.80 ± 1.27 | 0.23 ± 0.06 | 44.22 ± 1.05 | 0.30 ± 0.02 | 58.06 ± 3.04 |
| 4 | 0.24 ± 0.02 | 46.12 ± 0.02 | 0.29 ± 0.40 | 56.90 ± 0.21 | 0.31 ± 0.30 | 60.03 ± 0.06 | 0.30 ± 0.40 | 58.40 ± 1.55 | 0.27 ± 0.02 | 53.28 ± 3.87 | 0.34 ± 0.30 | 67.27 ± 1.77 |
| 5 | 0.27 ± 0.40 | 53.45 ± 0.08 | 0.31 ± 0.14 | 60.15 ± 0.05 | 0.31 ± 0.02 | 60.72 ± 0.05 | 0.33 ± 0.22 | 65.47 ± 3.92 | 0.27 ± 0.06 | 52.91 ± 2.44 | 0.33 ± 0.12 | 65.46 ± 1.71 |
| 6 | 0.28 ± 0.01 | 54.23 ± 0.10 | 0.31 ± 0.01 | 60.33 ± 0.18 | 0.34 ± 0.01 | 66.32 ± 0.02 | 0.33 ± 0.01 | 64.03 ± 1.10 | 0.28 ± 0.20 | 55.37 ± 2.85 | 0.32 ± 0.02 | 63.25 ± 3.15 |
| 7 | 0.27 ± 0.21 | 52.30 ± 0.15 | 0.32 ± 0.02 | 63.52 ± 0.41 | 0.37 ± 0.14 | 71.70 ± 0.05 | 0.33 ± 0.21 | 65.18 ± 1.62 | 0.29 ± 0.19 | 57.22 ± 1.78 | 0.30 ± 0.01 | 59.70 ± 1.50 |
| 8 | 0.29 ± 0.21 | 57.63 ± 0.26 | 0.31 ± 0.23 | 61.25 ± 0.11 | 0.33 ± 0.31 | 64.74 ± 0.32 | 0.32 ± 0.11 | 61.98 ± 1.58 | 0.29 ± 0.11 | 57.69 ± 1.88 | 0.38 ± 0.32 | 73.53 ± 0.15 |
| 9 | 0.13 ± 0.03 | 26.22 ± 0.05 | 0.20 ± 0.02 | 39.87 ± 0.17 | 0.21 ± 0.32 | 41.30 ± 0.59 | 0.17 ± 0.01 | 33.78 ± 1.34 | 0.21 ± 0.01 | 40.80 ± 2.10 | 0.21 ± 0.21 | 40.62 ± 1.13 |
| 10 | 0.12 ± 0.41 | 24.08 ± 0.11 | 0.24 ± 0.31 | 47.79 ± 0.19 | 0.14 ± 0.19 | 26.91 ± 0.12 | 0.11 ± 0.31 | 21.58 ± 1.34 | 0.16 ± 0.21 | 32.35 ± 2.29 | 0.13 ± 0.15 | 24.51 ± 0.62 |
| H21 |
H22 |
H23 |
H24 |
H25 |
H26 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| days | Yi | FE | Yi | FE | Yi | FE | Yi | FE | Yi | FE | Yi | FE |
| 1 | 0.24 ± 0.01 | 46.22 ± 1.31 | 0.16 ± 0.01 | 31.58 ± 1.51 | 0.46 ± 0.13 | 89.37 ± 0.04 | 0.12 ± 0.01 | 23.60 ± 2.45 | 0.20 ± 0.02 | 39.57 ± 3.31 | 0.28 ± 0.01 | 54.80 ± 1.58 |
| 2 | 0.25 ± 0.07 | 48.52 ± 1.43 | 0.27 ± 0.09 | 53.52 ± 0.89 | 0.42 ± 0.15 | 82.30 ± 0.03 | 0.14 ± 0.01 | 26.96 ± 0.04 | 0.29 ± 0.01 | 56.68 ± 2.17 | 0.27 ± 0.01 | 52.16 ± 1.95 |
| 3 | 0.25 ± 0.02 | 48.91 ± 1.14 | 0.28 ± 0.05 | 55.14 ± 2.77 | 0.31 ± 0.05 | 60.33 ± 0.18 | 0.14 ± 0.02 | 26.55 ± 0.12 | 0.31 ± 0.07 | 61.69 ± 0.21 | 0.29 ± 0.06 | 56.54 ± 0.90 |
| 4 | 0.27 ± 0.01 | 53.66 ± 1.37 | 0.30 ± 0.10 | 59.57 ± 2.40 | 0.23 ± 0.20 | 44.69 ± 0.05 | 0.39 ± 0.02 | 75.62 ± 4.28 | 0.32 ± 0.09 | 63.09 ± 0.57 | 0.27 ± 0.7 | 52.88 ± 1.45 |
| 5 | 0.38 ± 0.32 | 74.28 ± 1.80 | 0.32 ± 0.11 | 63.43 ± 2.24 | 0.16 ± 0.23 | 31.40 ± 0.52 | 0.36 ± 0.09 | 70.45 ± 0.03 | 0.33 ± 0.14 | 65.28 ± 2.46 | 0.29 ± 0.11 | 57.20 ± 1.44 |
| 6 | 0.37 ± 0.10 | 72.06 ± 2.60 | 0.36 ± 0.31 | 70.08 ± 2.30 | 0.24 ± 0.11 | 47.02 ± 1.05 | 0.37 ± 0.22 | 73.26 ± 2.68 | 0.33 ± 0.02 | 63.95 ± 3.08 | 0.30 ± 0.21 | 58.24 ± 0.12 |
| 7 | 0.39 ± 0.11 | 77.30 ± 1.62 | 0.37 ± 0.12 | 71.86 ± 3.10 | 0.30 ± 0.21 | 59.53 ± .95 | 0.38 ± 0.25 | 75.01 ± 0.18 | 0.35 ± 0.4 | 68.48 ± 0.09 | 0.36 ± 0.21 | 70.72 ± 2.53 |
| 8 | 0.40 ± 0.10 | 78.38 ± 2.05 | 0.37 ± 0.21 | 72.56 ± 1.70 | 0.35 ± 0.51 | 68.32 ± 1.18 | 0.39 ± 0.22 | 76.98 ± 0.21 | 0.23 ± 0.33 | 45.46 ± 0.20 | 0.26 ± 0.32 | 50.66 ± 0.61 |
| 9 | 0.31 ± 0.31 | 61.49 ± 1.74 | 0.27 ± 0.31 | 52.73 ± 2.13 | 0.10 ± 0.22 | 20.40 ± 0.33 | 0.14 ± 0.22 | 27.12 ± 0.17 | 0.19 ± 0.01 | 36.52 ± 2.36 | 0.20 ± 0.11 | 39.22 ± 1.31 |
| 10 | 0.26 ± 0.32 | 50.07 ± 0.06 | 0.24 ± 0.32 | 46.90 ± 1.73 | 0.08 ± 0.13 | 16.60 ± 0.30 | 0.08 ± 0.09 | 15.87 ± 0.21 | 0.12 ± 0.2 | 22.69 ± 2.89 | 0.19±0.22 | 37.83 ± 0.78 |
Values represent means ± SEM. Significance is recorded by different letters at p ≤ 0.05 by single-factor ANOVA.
Yi, ethanol yield.
FE, fermentation efficiency.
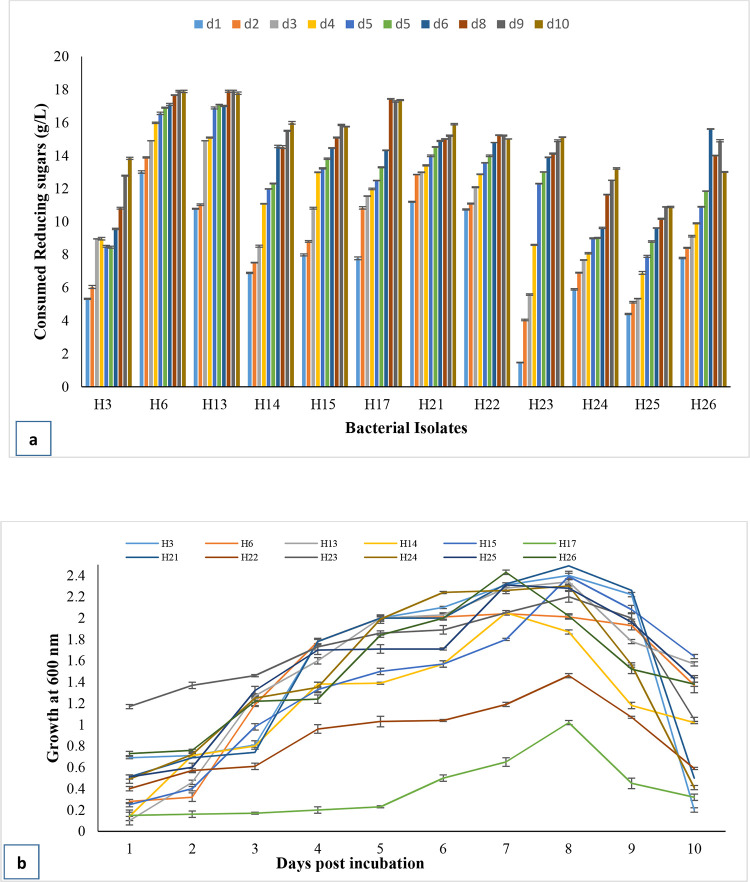
In the fermentation medium, carbon source was supplied in the form of CMC (2%) and yeast extract (0.65%). Figure 4 displays the correlation of reducing sugars in the fermentation medium and ethanol contents. It is hypothesized that ethanologenesis in the fermentation medium tends to cause a decrease in the remaining reducing sugars. The ethanol contents and the remaining reducing sugars in the medium are inversely proportional to each other. More or less inclination in reducing sugars with varied increase in ethanol contents was exhibited by different bacterial isolates. The increasing trend of ethanol contents from day 2 in H6, H13, H21, and H22 and less reducing sugars in the medium were detected, whereas, more reducing sugars with a slow increase in contents in the medium by H14, H23, H25, and H26 were recorded.
Figure 4.
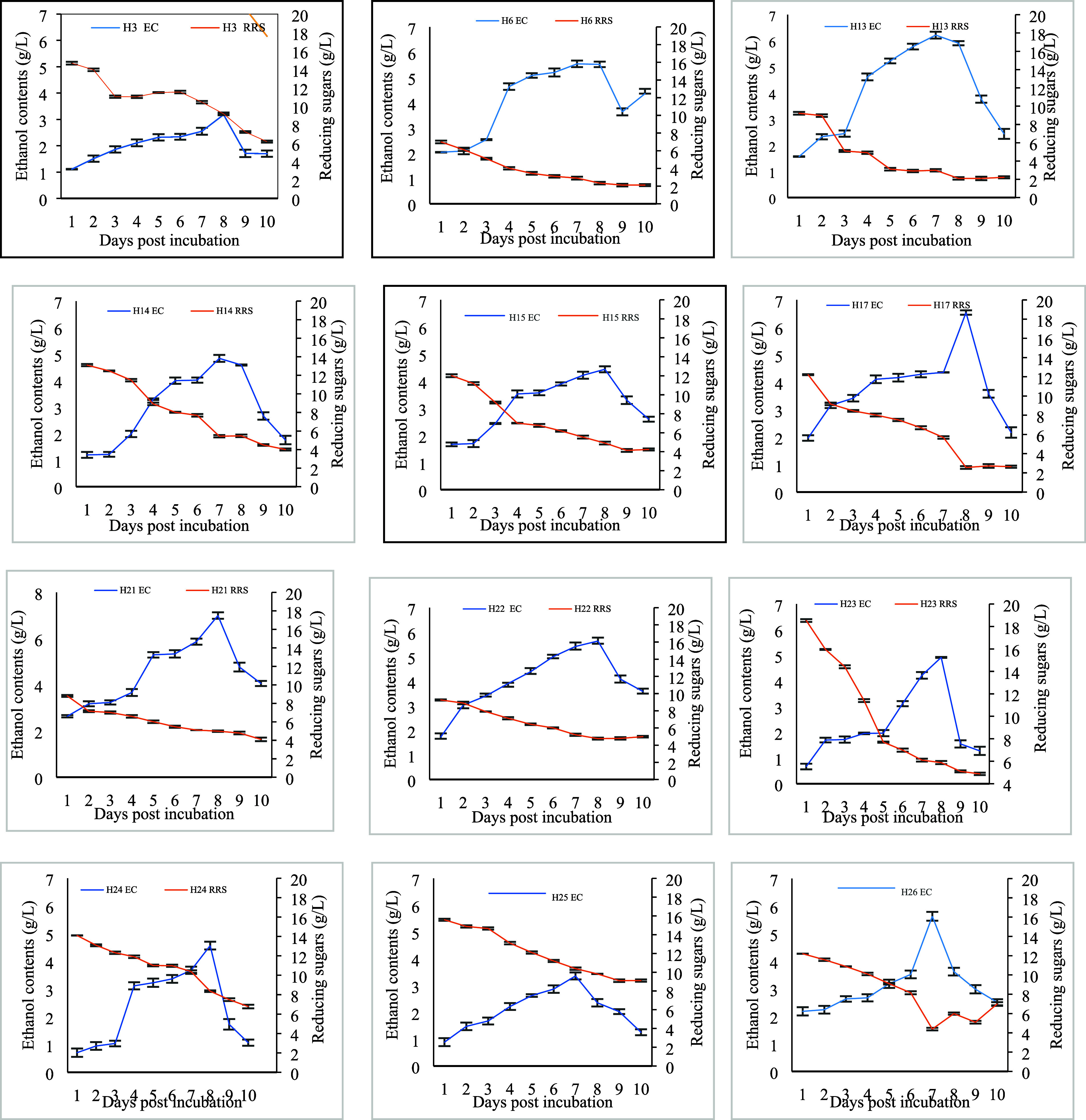
Ethanol and remaining reducing sugar contents in the fermentation medium supplemented with 2% cellulose by bacterial isolates. Bars denote average ± standard error mean.
Figure 5a reveals that consumption of reducing sugars tends to be increased as the experiment continued for 10 days. It is assumed that consumed reducing sugars may be used for ethanologenesis and bacterial biomass production. Growth tendencies are presented in Figure 5b. All bacterial isolates have a long lag phase up to day 3 except H6, H13, H15, H25, and H26 and a short decline phase, i.e., days 9 and 10. The log phase varied between days 3 and 8. Bacterial growth measurement exposed the fact that the highest ethanol contents were observed in the log phase with actively dividing cells by all bacterial isolates with a good end product of ethyl alcohol.
Figure 5.
Consumed reducing sugar contents (a) and growth (b) of selected bacterial isolates in a fermentation medium.
2.7. Molecular Characterization of Selected Bacterial Isolates
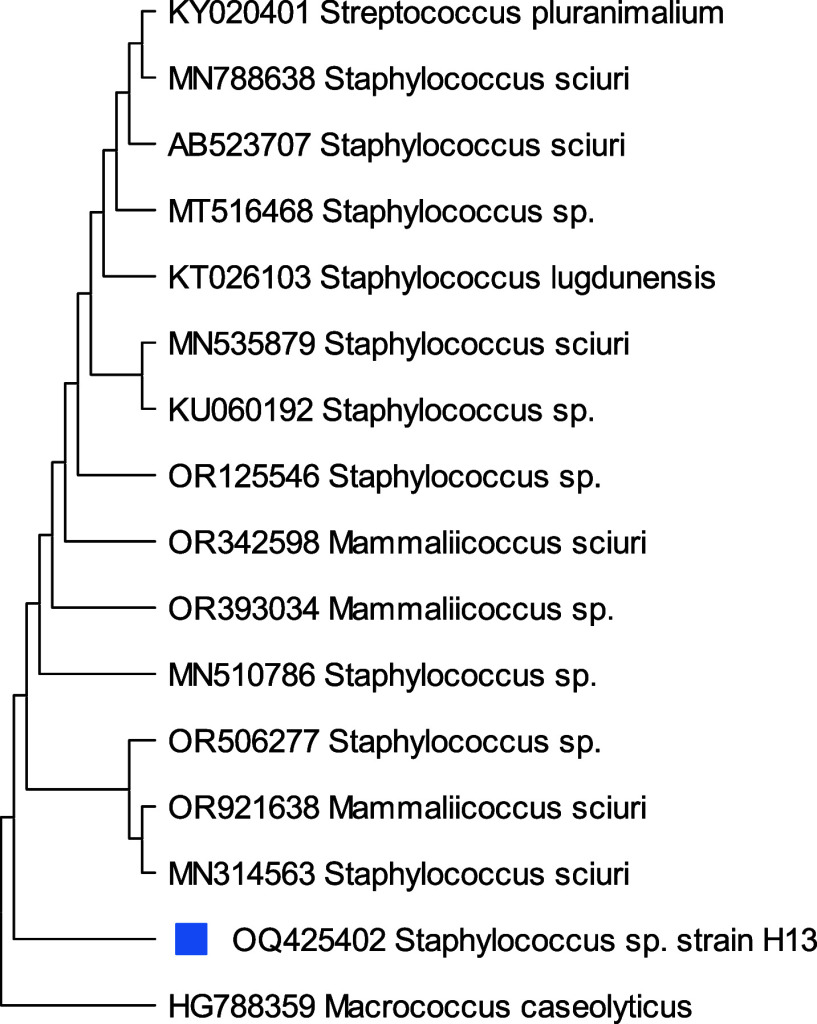
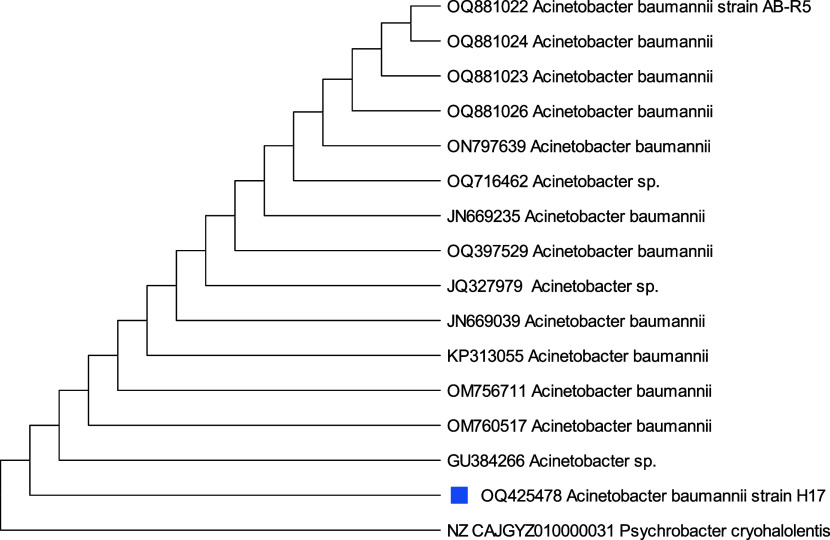
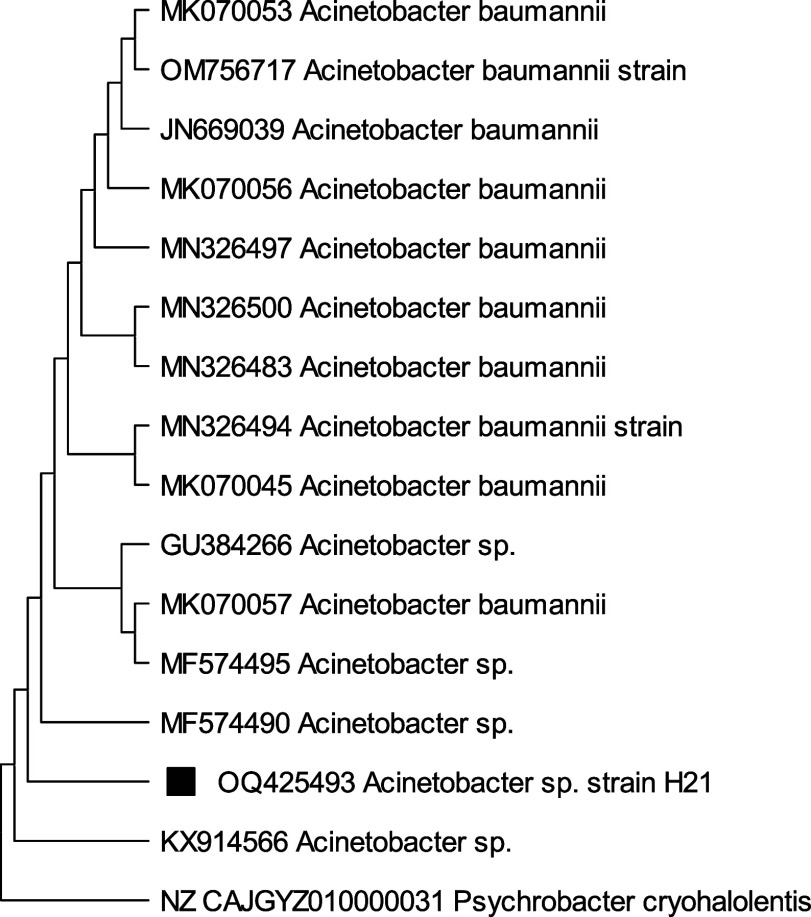
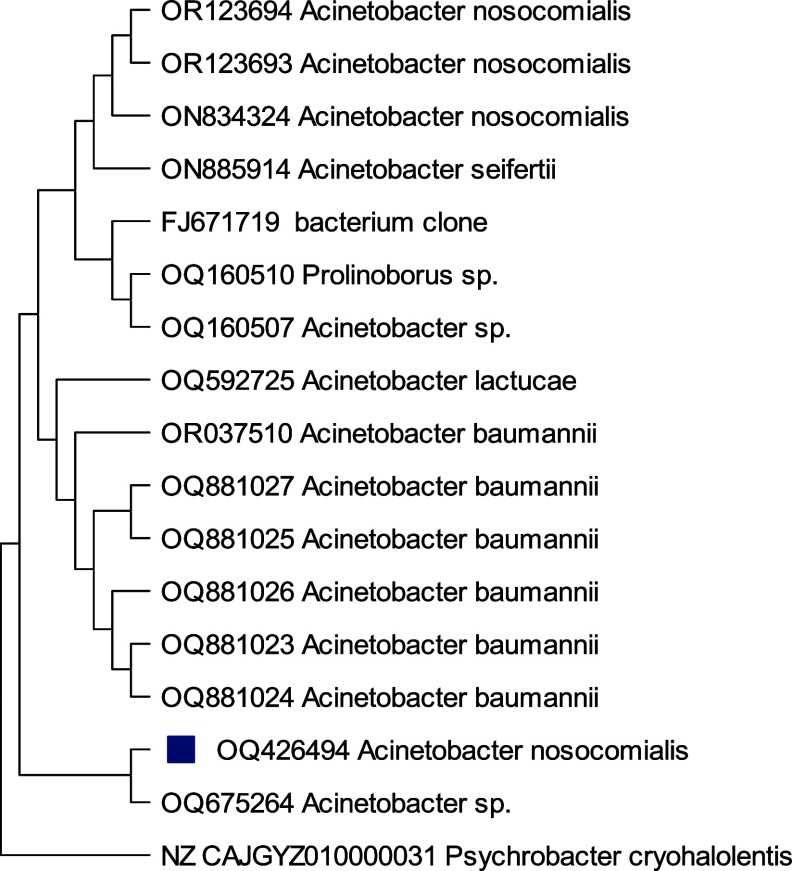
A phylogenetic study on isolates H13, H17, H21, and H22 based on 16S rRNA gene sequences specified that the similarity and the closest relative strains to selected isolates were Staphylococcus sp.H13(99.28%), Acinetobacter baumanni H17 (99.17%), Acinetobacter sp. H21 (98.01%), and Acinetobacter nosocomialis H22 (97.32%) with 98% similarity correspondingly. From phylogenetic trees, Figure 6 depicts that Staphylococcus sp., H13 differed from its closest clades. However, Staphylococcus sp. Strain H13 showed the highest similarity/identity with Staphylococcus sp. strain L11 with 28 bootstraps. Acinetobacter baumanni H17 showed the closest resemblance with Acinetocabter baumanni OIFC 189 clone with 57 bootstraps (Figure 7). Figures 8 and 9 indicated that the selected species Acinetocabter Sp. H21 and Acinetocabter nosocomialis H22 showed a close relation with Acinetocabter baumanni and Acinetocabter baumanni AbCTX5, respectively, with boot straps 41 and 33.
Figure 6.
Phylogenetic tree of Staphylococcus sp. H13 constructed by the neighbor-joining method with the bootstrap test showing replicate percentage and associated taxa. The tree with the highest log likelihood (−3137.86) is shown. Maximum Composite Likelihood tool computed evolutionary distances with 31 nucleotide sequence analysis by deletion of ambiguous pair wise positions. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 0.1031)]. Final data set has 1535 positions.
Figure 7.
Phylogenetic tree of Acinetobacter baumanni H17 by the neighbor-joining method with the bootstrap test showing replicate percentage and associated taxa. The tree with the highest log likelihood (−3696.54) is shown. Maximum Composite Likelihood tool computed evolutionary distances with 31 nucleotide sequence analysis by deletion of ambiguous pair wise positions. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 0.2439)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 41.21% sites). Final data set has 1560 positions.
Figure 8.
Phylogenetic tree of Acinetobacter sp. H21 by the neighbor-joining method with the bootstrap test showing replicate percentage and associated taxa. The tree with the highest log likelihood (−4277.18) is shown. Maximum Composite Likelihood tool computed evolutionary distances with 31 nucleotide sequence analysis by deletion of ambiguous pair wise positions. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 0.0916)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 24.25% sites). Final data set has 1501 positions.
Figure 9.
Phylogenetic tree of Acinetobacter nosocomialis H22 by the neighbor-joining method with the bootstrap test showing replicate percentage and associated taxa. The tree with the highest log likelihood (−3730.65) is shown. Maximum Composite Likelihood tool computed evolutionary distances with 31 nucleotide sequence analysis by deletion of ambiguous pair wise positions. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 0.5376)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 37.37% sites). Final data set has 1504 positions.
3. Discussion
The current study deals with the screening, identification, and characterization of cellulolytic and ethanologenic bacteria from termites. The termite collected from Jinnah Hospital and Botanical garden, Punjab University, Lahore was identified as Heterotermes indicola. The identification was made on the basis of behavior to make large number of tunnels as well as traveling long distances in tunnels and morphological features, viz, size and shape of the head, mandible, labrum, position of antennal segments, and tooth of workers and soldiers by following the web based keys.62−65Heteroterme indicola (Wasmann), Odontotermes obesus (Rambur), Microtermes obesi (Holmgren), and Coptotermes heimi (Wasmann) are reported as the most common and damaging species for wood and wooden infrastructures in Pakistan. Heterotermes indicola in Pakistani regions has been found to cause massive damage to wooden structures in houses. On the basis of massive wooden damage, it is ranked as the most destructive domestic pest in Lahore.66,67 The purpose of study is the isolation, characterization, and exploitation of termite harboring bacteria for cellulolysis and ethanologenesis. The gut of termites possesses large diversity of cellulolytic bacteria involved in digestion and degradation of lignolellulosic substrate and sugars.23,25,68−70 Hence, the study supported the idea of the presence of cellulose-degrading bacteria in the digestive tract of termites. Twenty six bacterial isolates were selected on CMC-supplemented medium from the samples of both sites. Few bacterial isolates were cultured in the laboratory. The gut of termite possesses diversified microbiota. The reason for less species of cultured bacteria may be the absence of real gut environment and natural nutrition.25,71,72 As the study involves the isolation of cellulolytic bacteria from the gut of termite, using microbiological pure culturing, it is not necessary to maintain/provide the gut environment to the isolate, though food in the form of cellulose was provided. The study was successfully conducted, aiming to generate knowledge and creativity based on the potential benefits from the natural gut microbiota from the termite gut. In the natural gut environment, these microbes have a symbiotic relationship. However, further research is needed to optimize the cellulolytic potential of the isolated bacteria and to be used as a consortium to improve the enzymatic hydrolysis of LCB at a commercial scale. Among bacteria isolated from H. indicola, the 17 isolates belong to bacilli and the remaining were cocci. Ali et al.37 isolated bacilli in subterranean termite Psammotermes hypostoma. In Macrotermes michaelseni, large frequencies of bacilli49 and cocci18 were observed. The diversity of bacteria in termites may differ due to variation in the food type, soil composition, and geographical regions.73
Bacterial isolates were isolated on CMC as the sole carbon source because of the only soluble form of cellulose.74−76 The current study deals with the estimation of the ethanologenic and cellulolytic potentials of bacteria. Hence, different biochemical tests were performed to assess the fermentative and cellulolytic abilities of carboxy methyl cellulose. Cellulolysis is the breakdown or digestion of cellulose microfibril into oligosaccharides and monosaccharids.77 Bacterial cellulases supported the cellulolysis of a complex substrate to digest them into monomers.78 Carbon dioxide production by fermentation of monomeric sugars in Durham tubes was assessed in the form of bubbles. Durham tube testing is not in common practice because slow fermenting bacteria cannot be detected efficiently.79 The sugar hydrolysis was also detected by two indicators, viz., TTC and Congo red stain. All isolates showed positive color development with TTC. Sugar hydrolysis by bacteria can be detected employing TTC which appeared as bright colored (light pink to dark maroon) formazones.80−82 In living cells, TTC is converted from colorless to colored triphenyl formazon because it is a good electron acceptor.83,84
Congo red staining appeared as a sensitive and rapid test for screening of cellulose-degrading bacteria.85 Except for four, all bacterial isolates formed varied zones of clearance around their colonies. The enzymes diffused into the medium by aging/lysis of bacterial cells to interact with the dye which in turn reduced the color of the dye.86 Congo red stain is a sulfonatedazo dye, and bacterial plasma membrane is impermeable to the dye. The high cellulolytic activity resulted in high zones of clearance and low retention of the dye.87−89 Hallow formation surrounding the bacterial colonies by Congo red is helpful to assess the cellulolytic potential and, in turn, cellulolytic index. For the computation of index, the diameter of hallows was divided by the bacterial colony diameter. Bacterial isolates showed varied cellulolytic index with the highest value 5.0 (H21). The hallow around the colonies after Congo red staining indicates production of extracellular cellulases by them.90 Cellulolytic bacteria produce cellulase enzymes that will hydrolyze the cellulose present in the medium. Cellulose hydrolysis is evident by hallow formation because of the reaction between Congo red and 1,4-glycosidic present in the cellulose polymer.91 The principle of Congo red staining is that the dye diffuses into the agar medium. The higher solubility of the enzymes will result in the form of larger hallow/clear zones (Jo et al.).92 . The variation in the cellulolytic index value can be attributed to the isolates’ capacity to hydrolyze the cellulose present in the medium through the release of endo-β-1,4-glucanase (CMCase). CMCase is an enzyme generated by the cellulolytic bacteria that breaks down the β-1,4 glycoside bond in the CMC medium.93,94 The isolates possessing cellulolytic index higher than 1.5 or 2.0 may be considered as an efficient cellulase producer.77 These findings corroborated with the cellulolytic index reported by Kakkar et al.69 in gut bacteria from Odontotermes parvidens, Raheli et al.25 in Macrotermes michaelseni, and in different termites.77
The bacterial strains isolated for the current study had efficient cellulase producers to convert cellulosic microfibrils into oligosaccharides and monosaccharides. The isolates H21 and H17 showed the highest activity as 1.83 ± 0.01, 1.79 ± 0.024 μmol/min/mL respectively. The results were corroborated with findings of Ali et al.37 who recorded the CMCase activity from 0.22 to 2.28 U/mL by Paenibacillus lactis, Lysinibacillus macrolides, Stenotrophomonas maltophilia, Lysinibacillus fusiformis, and Bacillus cereus isolated from lower termite Psammotermes hypostoma. On the contrary, Bacillus species B1, B2 and Brevibacillus sp. Br3 isolated from higher termite Bulbitermes sp. showed high 138.77 U/g endoglucanase, 32.16 U/g exoglcanases and 104.96 U/g xylanase activities under solid state fermentation, respectively.95,96 Similarly cellulolytic activity was observed as 0.9 μmol/mL/min CMCase by Paenibacillus sp.(97) 2.40 IU/mL CMCase as well as 1.43 FPU/mL by Phanerochaete chrysosporium,98 1.07 FPU/mL in alkaline rice straw99 and 1.9 FPU/mL by T. reesei in steam-treated wheat straw.100 The varied findings such as 0.71 FPU/mL was reported in powdered rice straw by C. thermocellum,101 10.5FPU/mL by A. cellulolyticus(102) and 154.58 U/gds with Trichoderma reesei in sugar cane bagasse.103Aspergillus niger released 0.1813 IU/mL cellulases,104 25.6 U/mL by Streptomyces,105 and 16.2 IU/g with Trichoderma reesei.106 Iqbal et al.107 investigated the maximum CMCase potential (480 ± 4.22 μM/mL/min) after seventh day of incubation at specific condition. All of these findings from the literature differed from the investigations of the present study.
The present study attempted to screen ethanologenic bacterial strains on 2% CMC-supplemented medium. Bacterial isolates H13, H17, H21, and H22 produced maximum ethanol g/L as 7.21 ± 0.03, 6.54 ± 0.01, 7.00 ± 0.01, and 5.64 ± 0.02 on day 8. Similar results, i.e., 10.8 g/L ethanol contents were recorded at a maximum concentration by Streptomyces sp. identified from Microcerotermes sp. The results of ethanol contents corroborated with the findings of 8.3 g/L employing B. subtilis in potato wastes,108 while the ethanol assay 15.73 ± 0.44, 14.22 ± 0.15, and 17.73 ± 0.25 g/L recorded by Chandel et al.109 via enzymes prepared from P. stipitis, A. oryzae MTCC 1846, and S. cerevisiae VS3 correspondingly differed from the findings of the current study. Ethanol yields (g/g) 0.37, 0.38, 0.40, and 0.37 with the percent FE of 71.70, 73.53, 78.38, 72.56 were calculated for H13, H17, H21, and H22 respectively. The findings of Rudolf et al.110 and Abedinifar et al.111 agreed with the results who reported an ethanol yield of 0.30 g/g from S. cerevisiae on sugar cane, 0.36–0.43 g/g by Mucor indicus, as well as 0.37–0.45 g/g via S. cerevisae in enzymatically treated rice straw. Zymomonas mobiliz generated 60.5 g/L ethanol content with 0.30 g/g yield using solid carob pods and wheat bran mixture under submerged fermentation.112,113 These findings varied from the current values in terms of ethanol titer. In the simultaneous saccharification and fermentation of potato peels by S. cerevisiae, an ethanol yield of 0.32 g/g was obtained by Chohan et al.114 In another report, Z. mobiliz fermented the potato peels for the highest ethanol yield after day 5, which was higher than S. cerevisiae after day 7. The selected bacterial isolates produced maximum ethanol on day 7–8 using CMC. The findings are comparable with the data obtained by Mazaheri and Pirouzi.115 The isolated microbes may tolerate the ethanol contents and ferment the sugars.
The present study endeavored to figure out the consumption of cellulose in the fermentation medium before and after the experiment by the reducing sugar analysis. During the fermentation study, the reducing sugars were utilized to produce ethanol contents. Both factors were inversely proportional to each other. Furthermore, an increasing trend in sugar consumption was noticed up to the termination of the experiment even though the ethanol contents decreased after day 8. Moreover, the maximum ethanol contents were detected in the exponential growth phase in all bacterial isolates. Bacterial cells are the competent source for production of cellulases in a liquid medium. The cellulolytic product, i.e., glucose, serves as the sole carbon source for bacterial biomass and fermentation, which in turn reduces the sugar yield. Similarly, the ethanol production rate is going to be decreased by the lowering sugar level.116 Contrary to this, high glucose concentrations may inhibit enzyme activity and growth of bacteria.117 The more effectual uptake and consumption of sugars by microbes are the main factors for efficient ethanol production.118Saccharomyces cerevisiae uses the Embden-Meyerhof-Parnas (EMP) glycolytic pathway, whereas Zymommonas mobiliz utilizes the Entner-Doudoroff (ED) pathway for ethanol yield.119Zymomonas mobiliz produces biomass by employing the ED pathway than the EMP pathway, utilizing Escherichia coli and S. cerevisiae. Subsequently, more carbon source will be available for fermentation with 2.5-fold greater ethanol productivity in Z. mobiliz than S. cerevisiae.120 Likewise, Clostridia harbors lignocellulosic enzymes that are secreted naturally to hydrolyze polymeric sugars to fermentable monomeric sugars, both hexoses and pentoses.121,122
Four selected competent bacterial strains were characterized molecularly as Staphylococcus sp. H13, Acinetobacter baumanni H17, Acinetobacter sp. H21, and Acinetobacter nosocomialis H22 with 98% similarity. Several researchers had reported different bacterial species from gut of termite capable of lignocellulose degradation such as Alcaligenes faecalis HI-1,123 Cellulomonas,124,125 Acinetobacter, Bacillus cereus, and Enterobacter aerogenes.48
From the past decade, several studies for screening of cellulolytic bacteria from termites indicated that bacteria have competence to hydrolyze crystalline cellulose completely.126,127 The bacteria grow rapidly than yeast and fungi and hence can be used widely for cellulase production and can be optimized efficiently for certain cultural conditions.128 The study was a stepping stone in establishing the large-scale process development of bioenergy production. As with every passing day, energy sources of the world are diminishing at alarmingly high speed, there is a great need to explore more efficient, sustainable, and renewable alternates.129,130 Cellulose and hemicellulose present in the LCB, in the prospect, can contribute as the prime source substrates for bioenergy generation. The systematic conversion of cellulose (via the employment of cellulolytic bacteria) into fermentable sugar is considered as the most critical step in the biofuel production.131 Once this step is optimized in as a cost-competitive way, it may lead to the establishment of eco-viable process development.132 Hence, these selected bacterial isolates may be proved as efficient candidates for cellulosic biomass transformation into bioethanol commercially and can be employed in plant waste degradation to yield fermentable sugars.
The cellulases obtained from microbes can be used in large scale for bioremediation of cellulosic wastes (Dixit et al.),133 for large-scale cellulosic substrate conversions into value-added products, e.g., biofuels (Ahmad et al.),129 biofertilizers (Yu et al.),134 animal feed (Azizi-Shotorkhoft et al.),135 pulp and paper (Karthika et al.),136 textile (Bussler et al.).137 As the study deals with the employment of native cellulolytic bacterial isolates, metabolic engineering can further enhance the cellulolytic and ethanologenic abilities of the microbes. In this regard, the detailed study on understanding of key metabolic pathways leads to desirable outcomes.
4. Conclusions
The present study supported the novel approach to search and screen competent Heterotermes indicola’s gut-associated bacteria possessing novel and efficient cellulolytic enzymes. The data revealed the competency of Acinetobacter sp. H21 screened from H. indicola’ gut with a cellulolytic index of 5.0 and CMCase activity as 1.83 ± 0.01umol/mL/min. Ethanol titer with Acinetobacter sp. H21 is observed as 7.00 ± 0.01 g/L having an ethanol yield of 0.40 g/g and 78.38% FE.
The gut of higher termites proved as a natural and potential source for screening of novel cellulolytic bacteria and enzymes. The screened cellulolytic bacteria possess the ability for biotransformation of cellulose into glucose and can be employed for biofuel production, enzyme purification, and composting.
5. Materials and Methods
5.1. Sampling of Termite for Bacterial Isolation
In the current study, H. indicola workers were collected from the highly infested, standing trees of Poplus euramericana using forceps and a chisel from two sampling areas, i.e., Botanical garden, University of Punjab (31° 520′ N, 74° 358′ E), and Jinnah hospital, (31° 495′ N, 74° 286′ E) Lahore, Pakistan on 13th July, 2021. At both locations, termites were found in wood, forming tunnels in soil leading to wood (Figure 1).
Random samples of Heterotermes indicola termites were taken from the decomposing logs present in both the study areas. A clean plastic container containing a minimum of several dozen termites, along with a sample of wood and soil, was brought back to the laboratory and kept at 26 °C and total darkness. The termite samples were identified morphologically based on the size and shape of heads and mandibles. The termites feed on the bark and soft parts around the base and the stems of trees. Twenty five termites were isolated and cleaned by disinfecting surfaces with 70% ethyl alcohol and stored at 4 °C by keeping in sterilized plastic bags. Within 48 h, termites were dissected for sampling of gut to be proceeded for bacterial isolation.
The gut of 24 termites were dissected using sterile dissecting tools (with 70% alcohol for 30 s) in a sterile air flow safety cabinet (laminar flow). The gut of the dissected termites was separated. The suspension was made in triplicates by crushing the 8 guts with a sterile glass rod in 2 mL of sterilized phosphate buffer saline (0.9% PBS) in a sterile glass vial. Phosphate buffer saline was prepared by mixing (g/L) Na2HPO4, 1.44, KCl, 0.2, NaCl, 8, KH2PO4, 0.24 by adjusting pH at 7.4. The suspension (0.1 mL) was plated on CMC-enriched media at 37 ± 1 °C for 1 day. Spread plate method was adopted for bacterial strain isolation from the gut contents. The composition (%) of CMC-supplemented medium was cellulose 2 g, peptone 1.5 g, yeast extract 1 g, MgSO4 0.05 g, (NH4)2SO4 0.1 g, KH2PO4 0.1 g, and CaCl2 0.1 g with agar 2 g.138 It could be assumed that the medium having CMC, yeast extract, and peptone served as the main carbon source for efficient ethanologenic fermentation. Bacterial isolates were selected based on the morphological characteristics of colonies. Streak plate method was adopted for the pure culturing of isolated strains.
5.2. Biochemical Evaluation for Sugar Degradation
The FE and sugar degrading ability were assessed by various biochemical testings. The isolated bacteria formed gas in Durham tubes, clear zones after Congo red staining, and maroon color by TTC. The medium for Durham tubes testing composed of (%) yeast extract 1, peptone 1.5, (NH4)2SO4 0.1, KH2PO4 0.1, MgSO4 0.05, and cellulose 2.138 The medium was dispensed in test tubes (10 mL) and Durham tubes (2 mL). The filled Durham tubes were placed inversely in a test tube containing liquid medium followed by inoculation of bacterial isolates. The fermentation medium dispensed test tubes were incubated at 37 ± 1 °C in static position for 10 days. The gas was detected in the form of bubbles in Durham tubes, and the length of the bubble was recorded.139
Congo red staining is established as a qualitative technique for the detection of sugar degradation during fermentation.18 Cellulose-supplemented medium was prepared by mixing yeast extract 1 g, MgSO4 0.05 g, cellulose 2 g, peptone 1.5 g, (NH4)2SO4 0.1 g, KH2PO4 0.1 g, and CaCl2 0.1 g with agar agar 2 g following the protocol laid by Zhang et al.138 All selected bacterial isolates were streaked on the cellulose-supplemented medium with autoclaved tooth picks for incubation for 16 hours at 37 ± 1 °C. One percent aqueous Congo red stain was flooded on streaked Petri plates with bacterial growth followed by reincubation for half an hour for 30 min at 37 ± 1 °C. The destaining of the bacterial culture was done by 1% aqueous NaCl solution. The sodium chloride-flooded bacterial cultures were incubated again for 30 min at 37 ± 1 °C for the removal of extra and unbound stain. For fine results, the destaining step was repeated three to four times. The sugar polymer degradation by bacterial cultures was evident in the form of hallow Petri plates.
The cellulose and TTC-enriched medium were used to evaluate the growth and color development of the bacterial isolates.138 Cellulose-supplemented medium (yeast extract 1 g, MgSO4 0.05 g, cellulose 2 g, peptone 1.5 g, (NH4)2SO4 0.1 g, KH2PO4 0.1 g, CaCl2 0.1 g, and agar 2 g) was mixed with TTC solution (10 mL of 0.5% aqueous TTC) after autoclaving. The inoculated Petri plates were kept for incubation overnight at 37 ± 1 °C. The development of pink to maroon color of colonies was interpreted as the positive test.
5.3. Screening of Bacterial Isolates Based on Morphology
Different cellulolytic bacteria were screened for identification on the basis of colonial and cellular features on 2% cellulose-supplemented medium.140 The observed colonial features for bacteria were color, size, elevation, texture, margin, optical feature, pigmentation, and consistency. For cellular characteristics, Gram’s reaction was performed. Sizes of stained bacterial cells were measured via micrometry using ocular and stage micrometers.
5.4. Fermentation Experiments
The composition of synthetic media with little modification was adopted by following the protocols of Chaudhary et al.130 The selected bacterial isolates were revived for the fermentation experiment in 2% CMC-enriched liquid medium. The bacterial cultures were incubated at 37 ± 1 °C for 24 h and kept on shaking. Synthetic medium contained (g/L) CMC 20, ZnCl2 0.00042, (NH4)2SO4 2.6, MgSO4.7H2O 0.8, KH2PO4 2.72, yeast extract 6.5, sodium citrate 6, CaCl2 0.3 and citric acid 1.5. One percent inoculum was used to start the fermentation. Fermentation was carried out in narrow-necked and screw-capped glass bottles. The fermentation of the inoculated synthetic medium continued statically at 37 ± 1 °C for 10 days. Estimation of reducing and ethanol contents were performed by drawing samples daily. Spectrophotometric measurement of bacterial growth was performed at 600 nm. Ethanol yield (Yi) and fermentation FE were computed by the following expressions;
5.5. Cellulolytic Activity
The basal medium for crude enzyme preparation include (g/L) magnesium sulfate 0.01, disodium hydrogen phosphate 0.7, sodium citrate 0.05, potassium dihydrogen phosphate 0.2, yeast extract 0.1, and CMC 2 and pH 7.0 according to protocols established by Abu-Gharbiya et al.141 The inoculated basal medium was agitated with 200 rpm at 37 ± 1 °C for 72 h. The supernatant after 15 min of centrifugation (1000 rpm) served as the bacterial crude enzyme. For the cellulolytic assay, the substrate buffer was prepared by mixing 2% CMC in acetate buffer (pH 5.0, 0.2 M). The ingredients of 0.2 M acetate buffer were (g/L) sodium acetate trihydrate 54.43, glacial acetic acid 12 mL. For the assay, 1.0 mL of substrate buffer was mixed with 0.5 mL of crude enzyme. The enzyme mixture was warmed for 30 min at 50 °C. Reducing sugars were measured by addition of 3 mL of DNS reagent followed by boiling in a water bath (5 min). The color change was measured spectrophotometrically at 640 nm. The color change in the DNS reagent indicated the conversion of cellulose into monomeric sugars.142 Cellulolytic potential was calculated by the following expression.
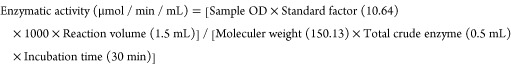 |
5.6. Molecular and Phylogenetic Characterization of Selected Bacterial Isolates
Four efficient bacterial isolates H13, H17, H21 and H22 were selected for 16s rRNA based molecular characterization on the basis of maximum CMCase potential, ethanol yield, and FE. The bacterial DNA from the colony was extracted with 5 mM NaOH and heated at 95 °C and 1 M Tris HCl followed by centrifugation. PCR master mixture (Thermo Fischer) and bacterial colony lysate (2 ul) were used for 16 S rRNA amplification. 35 polymerase chain reactions proceeded at 98 °C denaturation (10 s), 53 °C annealing (30 s) and 72 °C (1 min) extension temperatures. LabGenetix, Lahore, Pakistan facilitated the sequencing and BLAST homology querying of the amplified gene from bacterial isolates following the GenBank database (http://www.ncbi.nlm.nih.gov/blast). The sequenced genes were submitted to the NCBI database under GenBank accession numbers of OQ425402H13, OQ425478H17, OQ425493H21, and OQ426494H22. Based on the phylogenetic tree construction, the main branch with a bootstrap value of 100% was formed, namely, the genus branch cluster with the respective isolates.
The evolutionary history was inferred by using the Maximum Likelihood method, the Jukes-Cantor model,143 and the Kimura 2-parameter model.144 The percentage of trees in which the associated taxa clustered together is shown next to the branches. The initial tree(s) for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Jukes-Cantor model and Composite Likelihood (MCL) approach and then selecting the topology with the superior log likelihood value. Evolutionary analyses were conducted in MEGA11 software ver, 10 Biodesign Institute, Tempe, USA.145 The accuracy and authenticity of the phylogenetic tree was examined with higher bootstraps. To check the reliability of the phylogenetic tree branches, bootstrap analysis is considered as best tool which interpreted the best use of model data sets.146
5.7. Statistical Evaluation of Data
The experimental data was recorded by means with standard error of means. One-way analysis of variance (Duncan Multiple Range, Minitab Software, LLC, USA,Ver 17.1.1.) was used as a statistical tool for data evaluation.
Acknowledgments
The research work is a part of PhD thesis of the first author, S.A. The work was facilitated by the University of the Punjab and the University of Education, Lahore, Pakistan under supervision of A.A. and A.C. Z.H. and R.A.R. assisted the molecular study, while G.A., S.Q., S.A.A., and M.J.A. guided the manuscript preparation. This project was supported by Researchers Supporting Project number (RSP2024R5), King Saud University, Riyadh, Saudi Arabia.
The authors declare no competing financial interest.
References
- Lynd L. R.; Weimer P. J.; Van Zyl W. H.; Pretorius I. S. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. 2002, 66 (3), 506–577. 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. H. P. Reviving the carbohydrate economy via multi-product lignocellulose biorefineries. J. Ind. Microbiol. Biotechnol. 2008, 35 (5), 367–375. 10.1007/s10295-007-0293-6. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Wang Y. H.; Chu J.; Luo L. Z.; Zhuang Y. P.; Zhang S. L. Optimization of cellulase mixture for efficient hydrolysis of steam exploded corn stover by statistically designed experiments. Bioresour. Technol. 2009, 100, 819–825. 10.1016/j.biortech.2008.06.068. [DOI] [PubMed] [Google Scholar]
- Aziz N.; Sharif A.; Raza A.; Rong K. Revisiting the role of forestry, agriculture, and renewable energy in testing environment Kuznets curve in Pakistan: evidence from Quantile ARDL approach. Environ. Sci. Pollut. Res. 2020, 27, 10115–10128. 10.1007/s11356-020-07798-1. [DOI] [PubMed] [Google Scholar]
- Anwar Z.; Gulfraz M.; Irshad M. Agro-industrial lignocellulosic biomass a key to unlock the future bio-energy: a brief review. J. Radiat. Res. and Appl. Sci. 2014, 7, 163–173. 10.1016/j.jrras.2014.02.003. [DOI] [Google Scholar]
- The Pakistan Business Council . The State of Pakistan’s Agriculture 2023. An online report of the company under section 42 of the companies’ ordinance, 1984, 2023.
- Dashtban M.; Maki M.; Leung K. T.; Mao C.; Qin W. Cellulase activities in biomass conversion: measurement methods and comparison. Crit. Rev. Biotechnol. 2010, 30 (4), 302–309. 10.3109/07388551.2010.490938. [DOI] [PubMed] [Google Scholar]
- Obeng E. M.; Adam S. N. N.; Budiman C.; Ongkudon C. M.; Maas R.; Jose J. Lignocellulases: a review of emerging and developing enzymes, systems, and practices. Bioresour. Bioprocess. 2017, 4 (1), 16. 10.1186/s40643-017-0146-8. [DOI] [Google Scholar]
- Chantarasiri A. Diversity of cellulolytic bacteria isolated from a freshwater wetland reserve in Thailand and their cellulolytic activity. Appl. Ecol. Environ. Res. 2020, 18 (4), 5965–5983. 10.15666/aeer/1804_59655983. [DOI] [Google Scholar]
- Shweta A. Cellulases of bacterial origin and their applications: A review. Int. J. Sci. Res. 2014, 3 (10), 1652–1655. [Google Scholar]
- Menendez E.; Garcia-Fraile P.; Rivas R. Biotechnological applications of bacterial cellulases. AIMS Bioeng. 2015, 2 (3), 163–182. 10.3934/bioeng.2015.3.163. [DOI] [Google Scholar]
- McDonald J. E.; Rooks D. J.; McCarthy A. J.. Methods for the isolation of cellulose-degrading microorganisms. InMethods in enzymology; Academic Press, 2012; Vol. 510, pp. 349–374. [DOI] [PubMed] [Google Scholar]
- Esa F.; Tasirin S. M.; Abd Rahman N. Overview of bacterial cellulose production and application. Agric. Agric. Sci. Procedia 2014, 2, 113–119. 10.1016/j.aaspro.2014.11.017. [DOI] [Google Scholar]
- Lopez-Casado G.; Urbanowicz B. R.; Damasceno C. M.; Rose J. K. Plant glycosyl hydrolases and biofuels: a natural marriage. Curr. Opin. Plant Biol. 2008, 11 (3), 329–337. 10.1016/j.pbi.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Kudo T. Termite-microbe symbiotic system and its efficient degradation of lignocellulose. Biosci. Biotechnol. Biochem. 2009, 73 (12), 2561–2567. 10.1271/bbb.90304. [DOI] [PubMed] [Google Scholar]
- Chaudhary K.; Tauro P. Selective induction of β-glucosidase in Trichoderma reesei by xylan. Eur. J. Appl. Microbiol. 1982, 15, 185–187. 10.1007/BF00511246. [DOI] [Google Scholar]
- Mingardon F.; Bagert J. D.; Maisonnier C.; Trudeau D. L.; Arnold F. H. Comparison of family 9 cellulases from mesophilic and thermophilic bacteria. AEM. 2011, 77 (4), 1436–1442. 10.1128/AEM.01802-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. H. P.; Himmel M. E.; Mielenz J. R. Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 2006, 24 (5), 452–481. 10.1016/j.biotechadv.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Su N. Y.; Scheffrahn R. H. Economically important termites in the United States and their control. Sociobiology 1990, 17, 77–94. [Google Scholar]
- Aihetasham A.; Iqbal S. Feeding preferences of Microcerotermes championi (Snyder) for different wooden blocks dried at different temperatures under forced and choice feeding conditions in laboratory and field. Pakistan J. Zool. 2012, 44 (4), 1137–1144. [Google Scholar]
- Ohkuma M. Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl. Microbial. Biotechnol. 2003, 61, 1–9. 10.1007/s00253-002-1189-z. [DOI] [PubMed] [Google Scholar]
- Varghese L. M.; Agrawal S.; Sharma D.; Mandhan R. P.; Mahajan R. Cost-effective screening and isolation of xylano-cellulolytic positive microbes from termite gut and termitarium. 3 Biotech 2017, 7, 108. 10.1007/s13205-017-0733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonero-Pacheco J.; Aguilar J.; Raya M. C.; Trapero A.; Gaju-Ricart M.; Agustí-Brisach C. Diversity of cellulolytic microorganisms associated with the subterranean termite Reticulitermes grassei. J. Fungi 2023, 9 (3), 294. 10.3390/jof9030294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouquet P.; Traoré S.; Choosai C.; Hartmann C.; Bignell D. Influence of termites on ecosystem functioning. Ecosystem services provided by termites. Eur. J. Soil Biol. 2011, 47 (4), 215–222. 10.1016/j.ejsobi.2011.05.005. [DOI] [Google Scholar]
- Raheli N. M.; Lexa G. M.; George O. Cellulolytic activity of bacteria from the gut of termites (Macrotermes michaelseni) from Eldoret and Kakamega. Afr. J. Microbiol. Res. 2022, 16 (2), 82–87. 10.5897/AJMR2021.9573. [DOI] [Google Scholar]
- Watanabe H.; Noda H.; Tokuda G.; Lo N. A. cellulase gene of termite origin. Nature 1998, 394 (6691), 330–331. 10.1038/28527. [DOI] [PubMed] [Google Scholar]
- Sharma D.; Joshi B.; Bhatt M. R.; Joshi J.; Malla R.; Bhattarai T.; Sreerama L. Isolation of cellulolytic organisms from the gut contents of termites native to Nepal and their utility in saccharification and fermentation of lignocellulosic biomass. J. Biomass Biofuel 2015, 2, 11–20. 10.11159/jbb.2015.002. [DOI] [Google Scholar]
- Ferbiyanto A.; Rusmana I.; Raffiudin R. Characterization and identification of cellulolytic bacteria from gut of worker Macrotermes gilvus. Hayati J. Biosci. 2015, 22 (4), 197–200. 10.1016/j.hjb.2015.07.001. [DOI] [Google Scholar]
- Matsui T.; Tokuda G.; Shinzato N. Termites as functional gene resources. Recent Pat. Biotechnol. 2009, 3 (1), 10–18. 10.2174/187220809787172687. [DOI] [PubMed] [Google Scholar]
- Mani M. S., Ed. Ecology and biogeography in India, Vol. 23; Springer Science and Business Media, 2012. [Google Scholar]
- Manzoor F.; Mir N. Survey of termite infested houses, indigenous building materials and construction techniques in Pakistan. Pakistan J. Zool. 2010, 42 (6), 693–696. [Google Scholar]
- National Science Foundation . An annotated checklist of termites (Isoptera) from Sri Lanka; National Science Foundation, 2012, Retrieved 15 February 2017.
- Salihah Z.; Sattar A.; Farid A.; Shakoori A. R.. Termites of Pakistan and their control: Zoological Society of Pakistan; University of the Punjab: Lahore, Pakistan, 2012. [Google Scholar]
- Sen-Sarma P. K.Studies on wood destroying termites in relation to natural termite resistance of timber. Final Technical report 1968-73, 1975. [Google Scholar]
- Breznak J. A.; Brune A. Role of microorganisms in the digestion of lignocellulose by termites. Annu. Rev. Entomol. 1994, 39 (1), 453–487. 10.1146/annurev.en.39.010194.002321. [DOI] [Google Scholar]
- Peterson B. F.; Scharf M. E. Lower termite associations with microbes: synergy, protection, and interplay. Front. Microbiol. 2016, 7, 422. 10.3389/fmicb.2016.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali H. R.; Hemeda N. F.; Abdelaliem Y. F. Symbiotic cellulolytic bacteria from the gut of the subterranean termite Psammotermes hypostoma Desneux and their role in cellulose digestion. AMB Express 2019, 9, 111. 10.1186/s13568-019-0830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda G.; Lo N.; Watanabe H. Marked variations in patterns of cellulase activity against crystalline-vs. carboxymethyl-cellulose in the digestive systems of diverse, wood-feeding termites. Physiol. Entomol. 2005, 30, 372–380. 10.1111/j.1365-3032.2005.00473.x. [DOI] [Google Scholar]
- Scharf M. Termites as Targets and Models for Biotechnology. Annu. Rev. Entomol. 2015, 60, 77–102. 10.1146/annurev-ento-010814-020902. [DOI] [PubMed] [Google Scholar]
- Bourguignon T.; Lo N.; Dietrich C.; Šobotník J.; Sidek S.; Roisin Y.; Brune A.; Evans T. A. Rampant Host Switching Shaped the Termite Gut Microbiome. Curr. Biol. 2018, 28, 649–654. 10.1016/j.cub.2018.01.035. [DOI] [PubMed] [Google Scholar]
- Michaud C.; Hervé V.; Dupont S.; Dubreuil G.; Bézier A. M.; Meunier J.; Dedeine F. Efficient but occasionally imperfect vertical transmission of gut mutualistic protists in a wood-feeding termite. Mol. Ecol. 2020, 29 (2), 308–324. 10.1111/mec.15322. [DOI] [PubMed] [Google Scholar]
- Hervé V.; Liu P.; Dietrich C.; Sillam-Dussès D.; Stiblik P.; Šobotník J.; Brune A. Phylogenomic Analysis of 589 Metagenome-Assembled Genomes Encompassing All Major Prokaryotic Lineages from the Gut of Higher Termites. PeerJ. 2020, 8, e8614 10.7717/peerj.8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora J.; Kinjo Y.; Šobotník J.; Buček A.; Clitheroe C.; Stiblik P.; Roisin Y.; Žifčáková L.; Park Y. C.; Kim K. Y.; et al. The Functional Evolution of Termite Gut Microbiota. Microbiome 2022, 10, 78. 10.1186/s40168-022-01258-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do T. H.; Dao T. K.; Nguyen H. D.; Truong N. H. Understanding the Role of Free-Living Bacteria in the Gut of the Lower Termite Coptotermes gestroi Based on Metagenomic DNA Analysis. Insects 2023, 14 (11), 832. 10.3390/insects14110832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig H. Bacillus species in the intestine of termites and other soil invertebrates. J. Appl. Microbiol. 2006, 101 (3), 620–627. 10.1111/j.1365-2672.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- Brune A.; Dietrich C. The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu. Rev. Microbiol. 2015, 69, 145–166. 10.1146/annurev-micro-092412-155715. [DOI] [PubMed] [Google Scholar]
- Ramin M.; Alimon A. R.; Panandam J. M.; Sijam K.; Javanmard A.; Abdullah N. Digestion of rice straw and oil palm fronds by microflora from rumen and termite bacteria, in vitro. PJBS. 2008, 11 (4), 583–588. 10.3923/pjbs.2008.583.588. [DOI] [PubMed] [Google Scholar]
- Ramin M.; Alimon A. R.; Abdullah N. Identification of cellulolytic bacteria isolated from the termite Coptotermes curvignathus (Holmgren). J. rapid methods autom. Micribiol. 2009, 17 (1), 103–116. 10.1111/j.1745-4581.2009.00160.x. [DOI] [Google Scholar]
- Manjula A.; Pushpanathan M.; Sathyavathi S.; Gunasekaran P.; Rajendhran J. Comparative analysis of microbial diversity in termite gut and termite nest using ion sequencing. Curr. Microbial. 2016, 72, 267–275. 10.1007/s00284-015-0947-y. [DOI] [PubMed] [Google Scholar]
- Afzal M.; Qureshi M. Z.; Khan S. Production, purification and optimization of cellulase by Bacillus licheniformis HI-08 isolated from the hindgut of wood-feeding termite. Int. J. Agric. Biol. 2019, 21, 125–134. 10.17957/IJAB/15.0872. [DOI] [Google Scholar]
- Deka D.; Das S. P.; Sahoo N.; Das D.; Jawed M.; Goyal D.; Goyal A. Enhanced cellulase production from Bacillus subtilis by optimizing physical parameters for bioethanol production. Int. Sch. Res. Not. 2013, 2013, 1. 10.5402/2013/965310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. T. Isolation, screening and the influence of cultivation factors on cellulase of bacteria isolated from termites gut. Vietnam J. Sci. Technol. 2016, 54, 89. 10.15625/2525-2518/54/4A/11982. [DOI] [Google Scholar]
- Javaheri-Kermani M.; Asoodeh A. A novel beta-1, 4 glucanase produced by symbiotic Bacillus sp. CF96 isolated from termite (Anacanthotermes). Int. J. Biol. Macromol. 2019, 131, 752–759. 10.1016/j.ijbiomac.2019.03.124. [DOI] [PubMed] [Google Scholar]
- Ko K. C.; Han Y.; Choi J. H.; Kim G. J.; Lee S. G.; Song J. J. A novel bifunctional endo-/exo-type cellulase from an anaerobic ruminal bacterium. Appl. Microbiol. Biotechnol. 2011, 89, 1453–1462. 10.1007/s00253-010-2949-9. [DOI] [PubMed] [Google Scholar]
- Sreena C. P.; Sebastian D.. Cost effective cellulase production by Bacillus subtilis MUS1 using lignocellulosic biomass residues. In Conference: Biodiversity and Evaluation: Perspectives and Paradigm Shifts, 2015; pp. 268–270.
- Sreena C. P.; Vimal K. P.; Sebastian D. Production of cellulases and xylanase from Bacillus subtilis MU S1 isolated from protected areas of Munnar wildlife division. J. Microbiol. Biotechnol. Food Sci. 2016, 5 (6), 500. 10.15414/jmbfs.2016.5.6.500-504. [DOI] [Google Scholar]
- Griffiths H. M.; Ashton L. A.; Evans T. A.; Parr C. L.; Eggleton P. Termites can decompose more than half of deadwood in tropical rainforest. Curr. Biol. 2019, 29 (4), R118–R119. 10.1016/j.cub.2019.01.012. [DOI] [PubMed] [Google Scholar]
- Ulyshen M. D.; Müller J.; Seibold S. Bark coverage and insects influence wood decomposition: Direct and indirect effects. Applied Soil Ecology 2016, 105, 25–30. 10.1016/j.apsoil.2016.03.017. [DOI] [Google Scholar]
- Andringa J. I.; Zuo J.; Berg M. P.; Klein R.; Van’t Veer J.; De Geus R.; De Beaumont M.; Goudzwaard L.; van Hal J.; Broekman R.; Van Logtestijn R. S. Combining tree species and decay stages to increase invertebrate diversity in dead wood. Forest Ecol. Manag. 2019, 441, 80–88. 10.1016/j.foreco.2019.03.029. [DOI] [Google Scholar]
- Ulyshen M. D. Insect-mediated nitrogen dynamics in decomposing wood. Ecol. Entomol. 2015, 40, 97–112. 10.1111/een.12176. [DOI] [Google Scholar]
- Tokuda G.; Lo N.; Watanabe H.; Arakawa G.; Matsumoto T.; Noda H. Major alteration of the expression site of endogenous cellulases in members of an apical termite lineage. Mol. Ecol. 2004, 13 (10), 3219–3228. 10.1111/j.1365-294X.2004.02276.x. [DOI] [PubMed] [Google Scholar]
- Thakur M. L.Revision of the termite genus Odontotermes Holmgren (Isoptera: Termitidae: Macrotermitinae) from India. Zootaxa 1981, 2280. [Google Scholar]
- Roonwal M. L.; Chhotani O. B.. Isoptera (termites); Zoological Survey of India: Kolkata; 1989; Vol I, pp. 1–672. [Google Scholar]
- Chhotani O. B.The Fauna of India and the Adjacent Countries: Isoptera (Termites), V. 2 (Family Termitidae); Zoological Survey of India, 1997. [Google Scholar]
- Mahapatro G. K.; Kumar S. First report of a commonly prevalent termite Heterotermes indicola from Delhi region. Indian J. Agric. Sci. 2013, 83, 459–462. [Google Scholar]
- Manzoor F.; Sayyed A. H.; Rafique T.; Malik S. A. Toxicity and repellency of different insecticides against Heterotermes indicola (Isoptera: Rhinotermitidae). J. Anim. Plant Sci. 2012, 22 (1), 65–71. [Google Scholar]
- Rasib K. Z.; Hidayat W.; Aihetasham A. Feeding preferences and control of a Pakistani termite Odontotermes obesus (Rambur)(Isoptera, Rhinotermitidae). Annu. Res. Rev. Biol. 2017, 18, 1–13. 10.9734/ARRB/2017/36225. [DOI] [Google Scholar]
- Gupta P.; Samant K.; Sahu A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. Microbiol. 2012, 2012, 578925 10.1155/2012/578925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkar N.; Sanjeev K. G.; Baljeet S. S. Studies on cellulolytic activity and structure of symbiotic bacterial community in Odontotermes parvidens guts. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4 (10), 310–315. [Google Scholar]
- Oktiarni D.; Nofyan E.; Kasmiarti G. Isolation and identification cellulolytic bacteria from termite gut obtained from Indralaya peatland area. IOP Conf. Ser.: Earth Environ. Scii 2010, 926 (1), 012024 10.1088/1755-1315/926/1/012024. [DOI] [Google Scholar]
- Ntabo R.; Boga H. I.; Muigai A.; Mwirichia R.. Isolation and characterization of bacteria isolates from soil feeding termites and soil from Juja and Kakamega forest in Kenya; University of Embu, 2010. [Google Scholar]
- Kavitha D.; Vijayarani K.; Kumanan K. 16S rRNA typing of cellulolytic bacteria from the termite Odontotermes formosanus. Int. J. Vet. Anim. Sci. Res. 2014, 10 (5), 359–368. 10.5555/20153099865. [DOI] [Google Scholar]
- Ayitso A. S.; Onyango D. M. Isolation and identification by morphological and biochemical methods of antibiotic producing microorganisms from the gut of Macrotermes michaelseni in Maseno Kenya. J. Appl. Biol. Biotechnol. 2016, 4 (01), 27–33. 10.7324/JABB.2016.40105. [DOI] [Google Scholar]
- Sazci A.; Erenler K.; Radford A. Detection of cellulolytic fungi by using Congo red as an indicator: a comparative study with the dinitrosalicyclic acid reagent method. Appl. Environ. Microbiol. 1986, 61 (6), 559–562. 10.1111/j.1365-2672.1986.tb01729.x. [DOI] [Google Scholar]
- Sissons C. H.; Sharrock K. R.; Daniel R. M.; Morgan H. W. Isolation of cellulolytic anaerobic extreme thermophiles from New Zealand thermal sites. Appl. Environ. Microbiol. 1987, 53 (4), 832–838. 10.1128/aem.53.4.832-838.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romsaiyud A.; Songkasiri W.; Nopharatana A.; Chaiprasert P. Combination effect of pH and acetate on enzymatic cellulose hydrolysis. J. Environ. Sci. 2009, 21 (7), 965–970. 10.1016/S1001-0742(08)62369-4. [DOI] [PubMed] [Google Scholar]
- Hidayat M. R. Isolation and Identification of Cellulolytic Bacteria Symbiont from Various Termites on Different Nest Type in Bukit Baka Bukit Raya National Park, West Kalimantan, Indonesia. Walailak J. Sci. Technol. 2021, 18 (14), 12708–12712. 10.48048/wjst.2021.12708. [DOI] [Google Scholar]
- Peristiwati; Natamihardja Y. S.; Herlini H. Isolation and identification of cellulolytic bacteria from termites gut (Cryptotermes sp.). J. Phys.: Conf. Ser. 2018, 1013, 012173 10.1088/1742-6596/1013/1/012173. [DOI] [Google Scholar]
- Scheffers W. A. Alcoholic fermentation. Stud. Mycol. 1987, 30, 321–332. [Google Scholar]
- Thom S. M.; Horobin R. W.; Seidler E.; Barer M. R. Factors affecting the selection and use of tetrazolium salts as cytochemical indicators of microbial viability and activity. J. Appl. Microbiol. 1993, 74 (4), 433–443. 10.1111/j.1365-2672.1993.tb05151.x. [DOI] [PubMed] [Google Scholar]
- Caviedes L.; Delgado J.; Gilman R. H. Tetrazolium microplate assay as a rapid and inexpensive colorimetric method for determination of antibiotic susceptibility of Mycobacterium tuberculosis. Journal of Clinical Microbiology 2002, 40 (5), 1873–1874. 10.1128/JCM.40.5.1873-1874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abate G.; Aseffa A.; Selassie A.; Goshu S.; Fekade B.; WoldeMeskal D.; Miörner H. Direct colorimetric assay for rapid detection of rifampin-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 2004, 42 (2), 871–873. 10.1128/JCM.42.2.871-873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshana R. N.; Tadesse G.; Abate G.; Miörner H. Use of 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide for rapid detection of rifampin-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 1998, 36 (5), 1214–1219. 10.1128/JCM.36.5.1214-1219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.; Kong D. H.; Yun S. H.; Lee K. R.; Lee K. P.; Franzblau S. G.; Lee E. Y.; Chang C. L. Evaluation of a modified antimycobacterial susceptibility test using Middlebrook 7H10 agar containing 2, 3-diphenyl-5-thienyl-(2)-tetrazolium chloride. J. Microbiol. Methods. 2006, 66 (3), 548–551. 10.1016/j.mimet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Lu W. J.; Wang H. T.; Nie Y. F.; Wang Z. C.; Huang D. Y.; Qiu X. Y.; Chen J. C. Effect of inoculating flower stalks and vegetable waste with ligno-cellulolytic microorganisms on the composting process. J. Environ. Sci. Health Part B 2004, 39 (5–6), 871–887. 10.1081/LESB-200030896. [DOI] [PubMed] [Google Scholar]
- Jalandoni-Buan A. C.; Decena-Soliven A. L. A.; Cao E. P.; Barraquio V. L.; Barraquio W. L. Characterization and identification of Congo red decolorizing bacteria from monocultures and consortia. Philipp. J. Sci. 2010, 139 (1), 71–78. [Google Scholar]
- Stolz A. Basic and applied aspects in the microbial degradation of azo dyes. Applied microbiology and biotechnology 2001, 56, 69–80. 10.1007/s002530100686. [DOI] [PubMed] [Google Scholar]
- Fujimoto N.; Kosaka T.; Nakao T.; Yamada M. Bearing a high cellulose-degrading activity, which was isolated as a heat-resistant and micro-aerophilic microorganism from bovine rumen. Open Biotechnol. J. 2011, 5, 7–13. 10.2174/1874070701105010007. [DOI] [Google Scholar]
- Florencio C.; Couri S.; Farinas C. S. Correlation between agar plate screening and solid-state fermentation for the prediction of cellulase production by Trichoderma strains. Enzyme Res. 2012, 2012, 793708 10.1155/2012/793708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini A.; Aggarwal N. K.; Yadav A. Isolation and screening of cellulose hydrolyzing bacteria from different ecological niches. Bioeng. Biosci. 2017, 5 (1), 7–13. 10.13189/bb.2017.050102. [DOI] [Google Scholar]
- Zverlova V. V.; Holl W.; Schwarz H. Enzymes For Digestion Of Cellulose And Other Polysaccharides. In The Gut Of Longhorn Beetle Larvae, Rhagium Inquisitor L. (Col., Cerambycidae). Int. Biodeterior. Biodegad. 2003, 51, 175–179. 10.1016/S0964-8305(02)00139-7. [DOI] [Google Scholar]
- Jo W. S.; Park H. N.; Cho D. H.; Yoo Y. B.; Park S. C. Optimal Media Conditions for the Detection of Extracellular Cellulase Activity in Ganoderma neojaponicum. Micobiology 2011, 39 (2), 129–132. 10.4489/MYCO.2011.39.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe W. M. L. I.; Madusanka D. A. T.; Manage P. M. Isolation and Identification of Cellulase Producing and Sugar Fermenting Bacteria for Second-Generation Bioethanol Production. Int. J. Renew. Energy Develop. 2021, 10 (4), 699–711. 10.14710/ijred.2021.35527. [DOI] [Google Scholar]
- Ramadhani S. I.; Ardyati T.; Sjofjan O. Screening of Cellulolytic Bacteria from Sugarcane Waste (Bagasse) and Optimization of Cellulase Activity as Animal Feed: Screening of Cellulolytic Bacteria from Sugarcane Waste (Bagasse). J. Tropical Life Sci. 2023, 13 (3), 607–614. [Google Scholar]
- Kamsani N.; Salleh M. M.; Yahya A. Production of lignocellulolytic enzymes by microorganisms isolated from Bulbitermes sp. termite gut in solid-state fermentation. Waste Biomass Valor. 2016, 7, 357–371. 10.1007/s12649-015-9453-5. [DOI] [Google Scholar]
- Danso B.; Ali S. S.; Xie R.; Sun J. Valorisation of wheat straw and bioethanol production by a novel xylanase-and cellulase-producing Streptomyces strain isolated from the wood-feeding termite. Microcerotermes species. Fuel 2022, 310, 122333 10.1016/j.fuel.2021.122333. [DOI] [Google Scholar]
- Islam F.; Roy N. Screening, purification and characterization of cellulase from cellulase producing bacteria in molasses. BMC Res. Notes 2018, 11 (1), 445. 10.1186/s13104-018-3558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. H.; Ali S.; Fakhru’l-Razi A.; Alam Z. Use of fungi for the bioconversion of rice straw into cellulase enzyme. J. Environ. Sci. Health Part B 2007, 42 (4), 381–386. 10.1080/03601230701312647. [DOI] [PubMed] [Google Scholar]
- Sun W. C.; Cheng C. H.; Lee W. C. Protein expression and enzymatic activity of cellulases produced by Trichoderma reesei Rut C-30 on rice straw. Process Biochem. 2008, 43 (10), 1083–1087. 10.1016/j.procbio.2008.05.015. [DOI] [Google Scholar]
- Doppelbauer R.; Esterbauer H.; Steiner W.; Lafferty R. M.; Steinmüller H. The use of lignocellulosic wastes for production of cellulase by Trichoderma reesei. Appl. Microbiol. Biotechnol. 1987, 26, 485–494. 10.1007/BF00253537. [DOI] [Google Scholar]
- Xu C.; Qin Y.; Li Y.; Ji Y.; Huang J.; Song H.; Xu J. Factors influencing cellulosome activity in consolidated bioprocessing of cellulosic ethanol. Bioresour. Technol. 2010, 101 (24), 9560–9569. 10.1016/j.biortech.2010.07.065. [DOI] [PubMed] [Google Scholar]
- Prasetyo J.; Sumita S.; Okuda N.; Park E. Y. Response of cellulase activity in pH-controlled cultures of the filamentous fungus Acremonium cellulolyticus. Appl. Biochem. Biotechnol. 2010, 162, 52–61. 10.1007/s12010-009-8826-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhania R. R.; Sukumaran R. K.; Pillai A.; Prema P.; Szakacs G.; Pandey A.. Solid-state fermentation of lignocellulosic substrates for cellulase production by Trichoderma reesei NRRL 11460; CSIR, 2006.
- Acharya P. B.; Acharya D. K.; Modi H. A.. Optimization for cellulase production by Aspergillus niger using saw dust as substrate. Afr. J. Biotechnol. 2008, 7, (22), . [Google Scholar]
- El-Sersy N. A.; Abd-Elnaby H.; Abou-Elela G. M.; Ibrahim H. A.; El-Toukhy N. M. Optimization, economization and characterization of cellulase produced by marine Streptomyces ruber. Afr. J. Biotechnol. 2010, 9 (38), 6355–6364. [Google Scholar]
- Asquieri E. R.; Park Y. K. Production and characterization of extracellular cellulases from a thermostable Aspergillus sp. Rev. Microbiol. 1992, 23 (3), 183–188. [Google Scholar]
- Iqbal H. M.; Asgher M.; Ahmed I.; Hussain S. Media optimization for hyper-production of carboxymethyl cellulase using proximally analyzed agroindustrial residue with Trichoderma harzianum under SSF. Development 2010, 25, 37. [Google Scholar]
- Ali S. S.; Nugent B.; Mullins E.; Doohan F. M. Fungal-mediated consolidated bioprocessing: the potential of Fusarium oxysporum for the lignocellulosic ethanol industry. AMB Express 2016, 6, 13. 10.1186/s13568-016-0185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel A. K.; Chandrasekhar G.; Lakshmi Narasu M.; Venkateswar Rao L. Simultaneous saccharification and fermentation (SSF) of aqueous ammonia pretreated Saccharum spontaneum (wild sugarcane) for second generation ethanol production. Sugar Technol. 2010, 12, 125–132. 10.1007/s12355-010-0025-5. [DOI] [Google Scholar]
- Rudolf A.; Alkasrawi M.; Zacchi G.; Liden G. A comparison between batch and fed-batch simultaneous saccharification and fermentation of steam pretreated spruce. Enzyme Microb. Technol. 2005, 37 (2), 195–204. 10.1016/j.enzmictec.2005.02.013. [DOI] [Google Scholar]
- Abedinifar S.; Karimi K.; Khanahmadi M.; Taherzadeh M. J. Ethanol production by Mucor indicus and Rhizopus oryzae from rice straw by separate hydrolysis and fermentation. Biomass Bioenergy 2009, 33 (5), 828–833. 10.1016/j.biombioe.2009.01.003. [DOI] [Google Scholar]
- Mazaheri D.; Shojaosadati S. A.; Mousavi S. M.; Hejazi P.; Saharkhiz S. Bioethanol production from carob pods by solid-state fermentation with Zymomonas mobilis. Appl. Energy 2012, 99, 372–378. 10.1016/j.apenergy.2012.05.045. [DOI] [Google Scholar]
- Su R.; Ma Y.; Qi W.; Zhang M.; Wang F.; Du R.; Yang J.; Zhang M.; He Z. Ethanol production from high-solid SSCF of alkalinepretreated corncob using recombinant Zymomonas mobilis CP4. Bio Energy Res. 2013, 6, 292–299. 10.1007/s12155-012-9256-5. [DOI] [Google Scholar]
- Chohan N. A.; Aruwajoye G. S.; Sewsynker-Sukai Y.; Gueguim Kana E. B. Valorisation of potato peel wastes for bioethanol production using simultaneous saccharification and fermentation: Process optimization and kinetic assessment. Renew. Energy 2020, 146, 1031–1040. 10.1016/j.renene.2019.07.042. [DOI] [Google Scholar]
- Mazaheri D.; Pirouzi A. Valorization of Zymomonas mobilis for bioethanol production from potato peel: fermentation process optimization. Biomass Conversion and Biorefinery 2022, 12 (8), 3389–3398. 10.1007/s13399-020-00834-7. [DOI] [Google Scholar]
- Sofer S. S.; Zaborsky O. R.. Biomass conversion processes for energy and fuels; Springer Science and Business Media, 2012. [Google Scholar]
- Epps H. M.; Gale E. F. The influence of the presence of glucose during growth on the enzymic activities of Escherichia coli: comparison of the effect with that produced by fermentation acids. Biochem. J. 1942, 36 (7–9), 619. 10.1042/bj0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang A.; Lee T. S. Converting sugars to biofuels: ethanol and beyond. Bioeng. 2015, 2 (4), 184–203. 10.3390/bioengineering2040184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger G. A. Carbohydrate metabolism in Zymomonas mobilis: a catabolic highway with some scenic routes. FEMS Microbiol. Lett. 1996, 145 (3), 301–307. 10.1111/j.1574-6968.1996.tb08593.x. [DOI] [Google Scholar]
- Weber C.; Farwick A.; Benisch F.; Brat D.; Dietz H.; Subtil T.; Boles E. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl. Microbiol. Biotechnol. 2010, 87, 1303–1315. 10.1007/s00253-010-2707-z. [DOI] [PubMed] [Google Scholar]
- Mitchell W. J.; Albasheri K. A.; Yazdanian M. Factors affecting utilization of carbohydrates by clostridia. FEMS Microbiol. Rev. 1995, 17 (3), 317–329. 10.1111/j.1574-6976.1995.tb00215.x. [DOI] [Google Scholar]
- Lütke-Eversloh T.; Bahl H. Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr. Opin. Biotechnol. 2011, 22 (5), 634–647. 10.1016/j.copbio.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Iqtedar M.; Riaz S.; Mirza S. S.; Aftab M.; Kaleem A.; Abdullah R.; Aeihtesham A.; Aslam F.; Wasim M. Bioethanol production from urban cellulosic waste employing Alcaligenes faecalis HI-1 isolated from gut of termite Heterotermes indicola. Biosci. J. 2021, 37, e37023 10.14393/BJ-v37n0a2021-53598. [DOI] [Google Scholar]
- Wenzel M.; Schönig I.; Berchtold M.; Kämpfer P.; König H. Aerobic and facultatively anaerobic cellulolytic bacteria from the gut of the termite Zootermopsis angusticollis. J. Appl. Microbiol. 2002, 92 (1), 32–40. 10.1046/j.1365-2672.2002.01502.x. [DOI] [PubMed] [Google Scholar]
- Bakalidou A.; Kämpfer P.; Berchtold M.; Kuhnigk T.; Wenzel M.; König H. Cellulosimicrobium variabile sp. nov., a cellulolytic bacterium from the hindgut of the termite Mastotermes darwiniensis. Int. J. Syst. Evol. Microbiol. 2002, 52 (4), 1185–1192. 10.1099/00207713-52-4-1185. [DOI] [PubMed] [Google Scholar]
- Saha S.; Roy R. N.; Sen S. K.; Ray A. K. Characterization of cellulase-producing bacteria from the digestive tract of tilapia, Oreochromis mossambica (Peters) and grass carp, Ctenopharyngodon idella (Valenciennes). Aquac. Res. 2006, 37 (4), 380–388. 10.1111/j.1365-2109.2006.01442.x. [DOI] [Google Scholar]
- Doi R. H. Cellulases of mesophilic microorganisms: cellulosome and noncellulosome producers. Ann. N.Y. Acad. Sci. 2008, 1125 (1), 267–279. 10.1196/annals.1419.002. [DOI] [PubMed] [Google Scholar]
- Nakamura K.; Kitamura K.. Cellulases of Cellulomonas uda. In Methods in Enzymology; Academic Press, 1988; Vol. 160; pp 211–216. [Google Scholar]
- Ahmad Q. A.; Manzoor M.; Chaudhary A.; Yang S. T.; Shim H.; Qazi J. I. Bench-scale fermentation for second generation ethanol and hydrogen production by Clostridium thermocellum DSMZ. 1313 from sugarcane bagasse. Environ. Prog. Sustainable Energy 2021, 40, e13516 10.1002/ep.13516. [DOI] [Google Scholar]
- Chaudhary A.; Hussain A.; Shehzadi A.; Manzoor M.; Shahbaz M.; Deepanraj B. Production of ethanol from xylan by indigenous xylanolytic and ethanologenic bacteria isolated from fruit wastes Sustain. Energy Technol. Assess. 2023, 57, 103216 10.1016/j.seta.2023.103216. [DOI] [Google Scholar]
- Saleem A.; Hussain A.; Chaudhary A.; Ahmad Q. U. A.; Iqtedar M.; Javid A.; Akram A. M. Acid hydrolysis optimization of pomegranate peels waste using response surface methodology for ethanol production. Biomass Convers. Bioref. 2020, 12, 1513–1524. 10.1007/s13399-020-01117-x. [DOI] [Google Scholar]
- Chaudhary A.; Hussain A.; Ahmad Q. U. A.; Manzoor M.; Tahira S. A.; Karita S. Statistical optimization of alkaline treatment of pomegranate peel waste for bioethanol production. Biomass Conv. Boiref. 2022, 10.1007/s13399-022-02345-z. [DOI] [Google Scholar]
- Dixit M.; Gupta G. K.; Usmani Z.; Sharma M.; Shukla P. Enhanced bioremediation of pulp effluents through improved enzymatic treatment strategies: A greener approach. Renew. Sustainable Energy Rev. 2021, 152, 111664 10.1016/j.rser.2021.111664. [DOI] [Google Scholar]
- Yu Y. Y.; Li S. M.; Qiu J. P.; Li J. G.; Luo Y. M.; Guo J. H. Combination of agricultural waste compost and biofertilizer improves yield and enhances the sustainability of a pepper field. Journal of Plant Nutrition and Soil Science 2019, 182 (4), 560–569. 10.1002/jpln.201800223. [DOI] [Google Scholar]
- Azizi-Shotorkhoft A.; Mohammadabadi T.; Motamedi H.; Chaji M.; Fazaeli H. Isolation and identification of termite gut symbiotic bacteria with lignocellulose-degrading potential, and their effects on the nutritive value for ruminants of some by-products. Animal Feed Science and Technology 2016, 221, 234–242. 10.1016/j.anifeedsci.2016.04.016. [DOI] [Google Scholar]
- Karthika A.; Seenivasagan R.; Kasimani R.; Babalola O. O.; Vasanthy M. Cellulolytic bacteria isolation, screening and optimization of enzyme production from vermicompost of paper cup waste. Waste Management 2020, 116, 58–65. 10.1016/j.wasman.2020.06.036. [DOI] [PubMed] [Google Scholar]
- Bussler L.; Jacomini D.; Corrêa J. M.; Kadowaki M. K.; Maller A.; Simão R. D. C. G. Recombinant cellulase of Caulobacter crescentus: potential applications for biofuels and textile industries. Cellulose 2021, 28, 2813–2832. 10.1007/s10570-021-03700-5. [DOI] [Google Scholar]
- Zhang L.; Zhao H.; Gan M.; Jin Y.; Gao X.; Chen Q.; Guan J.; Wang Z. Application of simultaneous saccharification and fermentation (SSF) from viscosity reducing of raw sweet potato for bioethanol production at laboratory, pilot and industrial scales. Bioresour. Technol. 2011, 102 (6), 4573–4579. 10.1016/j.biortech.2010.12.115. [DOI] [PubMed] [Google Scholar]
- Meyer S. A.; Yarrow D. Validation of the names of three Candida species. Mycotaxon 1998, 66, 170–179. [Google Scholar]
- Benson H. J.; Benson H.; Brown A. E.; Brown A.. Benson’s microbiological applications, Laboratory manual in general microbiology, 9th Ed.; McGraw-Hill Higher Education, 1984. [Google Scholar]
- Abu-Gharbia M. A.; El-Sawy N. M.; Nasr A. M.; Zedan L. A. Isolation, optimization and characterization of cellulases and hemicellulases from Bacillus cereus LAZ 518 isolated from cow dung using corn cobs as lignocellulosic waste. J. Pharm. Appl. Chem. 2018, 4 (2), 1–13. 10.18576/JPAC/040201. [DOI] [Google Scholar]
- Miller G. L.; Blum R.; Glennon W. E.; Burton A. L. Measurement of carboxymethylcellulase activity. Anal. Biochem. 1960, 1 (2), 127–132. 10.1016/0003-2697(60)90004-X. [DOI] [Google Scholar]
- Jukes T. H.; Cantor C. R.. Evolution of protein molecules. In Mammalian Protein Metabolism, Munro H. N., Ed.; Academic Press: New York, 1969, pp. 21–132. [Google Scholar]
- Tamura K.; Stecher G.; Kumar S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022. 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evolution 1980, 16, 111–120. 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Stecher G.; Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33 (7), 1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]