Abstract
Marek’s disease virus (MDV) induces the rapid development of overwhelming T-cell lymphomas in chickens. One of its candidate oncogenes, meq (MDV Eco Q) which encodes a bZIP protein, has been biochemically characterized as a transcription factor. Interestingly, MEQ proteins are expressed not only in the nucleoplasm but also in the coiled bodies and the nucleolus. Its novel subcellular localization suggests that MEQ may be involved in other functions beyond its transcriptional potential. In this report we show that MEQ proteins are expressed ubiquitously and abundantly in MDV tumor cell lines. Overexpression of MEQ results in transformation of a rodent fibroblast cell line, Rat-2. The criteria of transformation are based on morphological transfiguration, anchorage-independent growth, and serum-independent growth. Furthermore, MEQ is able to distend the transforming capacity of MEQ-transformed Rat-2 cells through inhibition of apoptosis. Specifically, MEQ can efficiently protect Rat-2 cells from cell death induced by multiple modes including tumor necrosis factor alpha, C2-ceramide, UV irradiation, and serum deprivation. Its antiapoptotic function requires new protein synthesis, as treatment with a protein synthesis inhibitor, cycloheximide, partially reversed MEQ’s antiapoptotic effect. Coincidentally, transcriptional induction of bcl-2 and suppression of bax are also observed in MEQ-transformed Rat-2 cells. Taken together, our results suggest that MEQ antagonizes apoptosis through regulation of its downstream target genes involved in apoptotic and/or antiapoptotic pathways.
Marek’s disease virus (MDV) is one of the most potent oncogenic viruses, and it induces the rapid onset of T-cell lymphomas and a demyelinating disease (for reviews, see references 7, 20, and 38). Recent studies have revealed several candidate viral genes involved in the oncogenic process (for a review, see reference 24). Among these potential oncogenes, meq (MDV Eco Q) is most consistently expressed in all tumor and transformed cell lines (19, 37, 47). Furthermore, Xie et al. (52) recently used an antisense strategy to show that MEQ is required for the maintenance of the transformed state of an MDV tumor cell line, MSB1. MEQ has been biochemically characterized as a transcription factor (39). Its N-terminal basic region-leucine zipper (bZIP) domain has homology with the Jun/Fos family of transcription factors. The C-terminal transactivation domain is rich in proline residues with unique two and one-half repeats (19). There are two potential DNA response elements, namely, MERE1 (GAGTGATGA[C/G]TCATC) and MERE2 (PuACACACPy) (40), to which MEQ can bind and thereupon regulate both viral and host gene expression. Interestingly, MEQ localizes not only to the nucleus but also to the nucleolus and coiled bodies (27). Its novel subnuclear localization implies certain uncharacterized functions beyond its transcriptional potential. There are two clusters of arginine and lysine residues in the basic region of MEQ, namely, basic region 1 (BR1) and BR2. BR2 has been mapped to be the major nuclear localization signal and the sole nucleolar localization signal, while BR1 provides an auxiliary signal for nuclear entry.
Despite the revelation of the biochemical properties of MEQ, whether MEQ transforms cells and how it would exert this action remain undefined. The present report begins to address these questions. As there is no efficient chicken in vitro T-cell transformation system available, we followed the approaches successfully applied to other herpesviruses, such as Epstein-Barr virus (EBV). In the EBV-transformed cell lines, nine proteins and two small nuclear RNAs are expressed during latency. Among those latent proteins at least five of them, namely, EBNA-LP, -2, -3A, and -3C and LMP1, have been implicated in cell immortalization and/or transformation (10, 14, 21, 28, 48). Some of them were initially identified as oncogenes based on their ability to transform rodent fibroblast cell lines (50). The transformed cell lines were useful in further defining the mechanisms of action of the oncogenes.
In this report, we demonstrate that MEQ proteins are expressed ubiquitously and abundantly in MDV tumor cell lines. Overexpression of MEQ alone leads to transformation of Rat-2 cells, based on morphological characteristics, anchorage-independent growth, and serum-independent growth. Furthermore, MEQ protects the transformed cells from apoptosis induced by a variety of modes including tumor necrosis factor alpha, (TNF-α), C2-ceramide, UV irradiation, and serum withdrawal. Consistent with the antiapoptotic effect are the transcriptional induction of bcl-2 and suppression of bax, presumably induced by MEQ in those cells.
MATERIALS AND METHODS
Cells.
Rat-2 cells were maintained in Dulbecco modified Eagle medium (DMEM [high glucose]) supplemented with 10% calf serum. Chicken embryo fibroblasts (CEF) and duck embryo fibroblasts (DEF) were maintained in medium 199-DMEM (1:1) supplemented with 2% chicken serum and 5% calf serum. RP1 (31), RP4 (32), RP19 (34), MSB-1 (1), RP9 (35), RP13 (33), and CU cell lines (6, 42) were maintained as previously described.
Antibodies.
MEQ antisera (27) were used at a 1:200 dilution for immunofluorescence staining and at a 1:4,000 dilution for Western blotting, and antibromodeoxyuridine (BrdU) monoclonal antibody (MAb) (Amersham) was used undiluted.
Western blotting.
Total cell lysates were prepared in lysis buffer at 10 × 106 to 25 × 106 cells/ml. Cell lysates approximately equivalent to 0.5 × 106 to 1 × 106 cells were loaded in each lane for sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE). The gels were then transferred to Immobilon polyvinylidene difluoride membranes (Millipore). The membranes were then blocked with 5% bovine serum albumin (BSA)–phosphate-buffered saline containing 0.1% Tween 20 (PBST). The Western blotting was performed as previously described (19). Briefly, the blots were incubated with primary antibodies in PBST for 1 h, washed three times, and followed by incubation with alkaline phosphatase-conjugated secondary antibodies for 1 h, washed three times, and then exposed to substrates (BCIP [5-bromo-4-chloro-3-indolylphosphate] and Nitro Blue Tetrazolium). The Tropix Western-Light chemiluminescent detection system was employed to analyze Bcl-2 and Bax blots. The procedures were performed as described by the manufacturer.
Transforming assays.
Anchorage-independent growth was measured by soft agar colony assay to evaluate transforming potential. Briefly, this assay was performed in six-well plates with a base of 2 ml of medium containing 4% fetal bovine serum with 0.5% Noble agar (Difco). Cells were seeded in 2 ml of medium containing 4% fetal bovine serum with 0.35% agar at 104 cells/ml and layered onto the base. The number of colonies was scored under a microscope after 3 to 4 weeks.
Apoptotic assays.
Cells were treated with different apoptosis-promoting vehicles, including serum withdrawal (3 days), mouse TNF-α (mTNF-α) (1 ng/ml) plus serum withdrawal (24 h), C2-ceramide (10 μM) plus serum withdrawal (12 h), UV irradiation (50 J/m2, 24 h), and cycloheximide (CHX) (24 h). Apoptosis was evaluated by two different assays. First, a terminal deoxynucleotidyl transferase (TdT) assay was performed with the ApopTag in situ apoptosis detection kit (Oncor) according to the manufacturer’s specifications. Second, DAPI (4′,6-diamidino-2-phenylindole) staining was performed to examine the chromosomal pattern as described by Liu et al. (27).
BrdU incorporation assay.
DNA synthesis activity can be monitored by incorporation of BrdU. Briefly, cells were grown on coverslips inside the six-well plates. Serum deprivation was imposed for 3 days before BrdU (Amersham) was added to the media for 12 h. Cells were fixed with 1% formaldehyde in phosphate-buffered saline (PBS) for 20 min, washed with PBS and treated with 1 N HCl for 10 min, washed, blocked with 3% BSA–PBS for 1 h, and stained with anti-BrdU MAb (Amersham) for 1 h at 37°C followed by fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin Gs for 1 h at room temperature.
Indirect immunofluorescence.
Immunofluorescence staining was performed as previously described (27) with some modifications. Briefly, cells were seeded at 5 × 105/well in six-well plates with coverslips inside. Media were aspirated, and the cells were washed with PBS twice before the cells were fixed with 3.7% formaldehyde–PBS for 20 min. After another PBS wash, the cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min followed by blocking with 3% BSA–PBST for 1 h. Cells were then incubated with primary antibodies for 1 h. After two washes with PBST, the secondary antibodies conjugated with fluorescein isothyocyanate or Texas red (Vector Labs) were applied for another hour, and the cells were examined under a fluorescence microscope (40× objective; Nikon).
Reverse transcriptase-polymerase chain reaction (RT-PCR).
Total cell RNA was isolated from cells with TRIzol reagent (Life Technologies). Reverse transcription was then achieved with a Boehringer Mannheim reverse transcription kit. Primers for meq (5′ sequence, GCCATGGCTCAGGAGCCAGAGCCG; 3′ sequence, GGGAATTCTATATAACTAGGGGAGAA), bcl-2 (5′ sequence, AACCATGGCGCACGCTGGGAGA; 3′ sequence, GAATTCACTTGTGGCCCAGATA), bax (5′ sequence, CCTCGAGCCATGGACGGGTCCGGGGAGCAGCTTGGGAGC; 3′ sequence, CAGATCTCAGCCCATCTT), and the gene for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (5′ sequence, CGGAGTCAACGGATTTGGTCGTAT; 3′ sequence, AGCCTTCTCCATGGTGGTGAAGAC) were used in subsequent PCR to detect the transcriptional level of these genes. Only 16 cycles were programmed for amplification to avoid overamplification of the transcripts in lower abundance.
RESULTS
MEQ proteins are abundantly expressed in MDV cell lines.
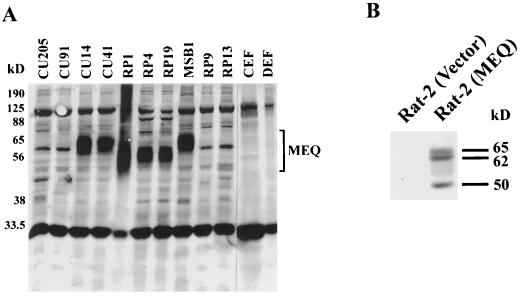
Although meq transcripts have been detected persistently in the MDV tumor samples and in the cell lines, MEQ’s expression at the protein level has not been addressed due to the lack of avid antibodies. Recently, we generated rabbit antisera against bacterially expressed MEQ (first 168 amino acids) proteins (27). The results of Western blotting are consistent with the expression data based on Northern blots published previously (19). Briefly, MEQ is highly expressed in all MDV tumor cell lines, such as RP1, RP4, RP19, and MSB1, as well as MDV-infected T-cell lines CU14 and CU41. However, MEQ is not detectable in normal CEF, DEF, or other cell lines derived from reticuloendotheliosis virus or avian leukosis virus transformation, including CU205, CU91, RP9, and RP13 (Fig. 1A). MEQ protein migrates in denaturing gel as a broad band whose molecular mass ranges between 50 and 75 kDa, and the mean size varies among different cell lines, presumably due to posttranslational modifications and a disorderly proline-rich structure. Recent findings of alternatively spliced MEQ products (26, 37) and the existence of rearranged genome in some cell lines (26) should also contribute to the variations in MEQ sizes.
FIG. 1.
Constitutive expression of MEQ proteins in the MDV tumor cell lines and MEQ-transformed Rat-2 cells. Total cell lysates were prepared from MDV cell lines (RP1, RP4, RP19, and MSB1), MDV-infected T-cell lines (CU14 and CU41), reticuloendotheliosis virus cell lines (CU91, CU205, and RP13), avian leukosis virus cell line (RP9), and normal avian embryo fibroblasts (CEF and DEF) (A) as well as from MEQ-transformed and vector-infected Rat-2 cells (B). Cell lysates equivalent to approximately 0.5 × 106 to 1 × 106 cells per lane were analyzed by SDS–10% PAGE. The gels were transferred to Immobilon polyvinylidene difluoride membranes and blotted with MEQ antisera (1:4,000 dilution). The Western blots were subsequently detected with the conventional alkaline phosphatase method.
MEQ is capable of transforming a rodent fibroblast cell line Rat-2 when overexpressed.
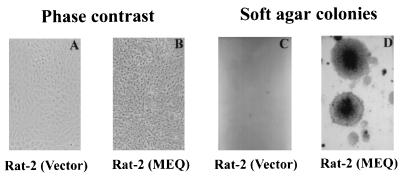
As described before, several lines of evidence suggest that MEQ may play a role in oncogenesis. First, MEQ’s structure is similar to those of the Jun/Fos oncoproteins and expression of MEQ is heightened in T-cell lymphomas but not in chronically infected cells (19, 47). Second, recent observations have demonstrated that MEQ antisense transcripts reversed the transformed phenotype of MSB1 (52). However, due to the lack of an efficient chicken T-cell transformation system, it is presently not possible to directly demonstrate the oncogenicity of MEQ in its natural target cells. We therefore utilized Rat-2 fibroblast transformation assays. For this experiment we first introduced meq into the murine retroviral vector pBabe-puro (30) and transfected the resulting construct, pBabe-MEQ, into a packaging cell line, Ψ2. The viral supernatants were then collected and used to infect Rat-2 cells. After selection with puromycin, the positive clones were pooled to avoid clonal variations that might complicate the interpretation of the results. Several passages later, the MEQ-infected Rat-2 cells became morphologically transformed; they are round to deformed and nonrefractile (Fig. 2B), as opposed to being spindle-shaped and refractile, like the untransformed vector-infected Rat-2 cells (Fig. 2A). In addition, they were devoid of contact inhibition, and numerous heaped-up foci could be observed. Moreover, they became resistant to trypsinization, presumably due to altered expressions of extracellular matrix proteins, and produced huge colonies in soft agar (Fig. 2D). Our indirect immunofluorescence staining (27) and Western blotting (Fig. 1B) showed that MEQ is expressed in these cells at a moderate to high level. Interestingly, three discrete bands were detected by MEQ antisera in pBabe-MEQ-transformed cells as opposed to the broad diffuse band found in all MDV cell lines as shown in Fig. 1A. The less complicated pattern is in part attributed to the inability of MEQ cDNA to undergo alternative splicing. Taken together, these results provide the first direct evidence that unspliced meq behaves like an oncogene and can transform Rat-2 cells, when overexpressed.
FIG. 2.
Transformation of Rat-2 cells by MEQ. Rat-2 cells were infected with supernatants containing viruses derived from either pBabe-puro vector or pBabe-MEQ construct. The transforming potential of MEQ was evaluated by morphology (A and B) and soft agar colony assay (C and D). Magnification, ×55.
MEQ promotes serum-independent growth of MEQ-transformed Rat-2 cells.
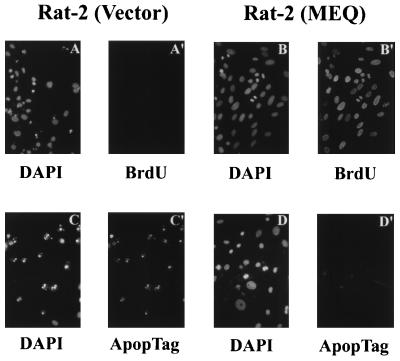
In addition to the morphological transformation described above, MEQ-transformed Rat-2 cells continued to proliferate in the absence of serum (Fig. 3). This observation was validated with further experiments using BrdU incorporation (to measure DNA synthesis) and DAPI staining (to analyze chromosomal structure). As shown in Fig. 4C′, after serum withdrawal for 3 days, more than 75% of MEQ-transformed Rat-2 cells showed signs of BrdU incorporation and showed mitotic figures as revealed by DAPI staining (Fig. 4B). In contrast, the vector-infected Rat-2 cells underwent either growth arrest (with very little BrdU incorporation) (Fig. 4A′) or apoptosis (Fig. 4A). Apoptosis was confirmed by TdT assay with the ApopTag in situ apoptosis detection kit (Fig. 4C, C′, D and D′). These findings suggest that MEQ-transformed Rat-2 cells either can synthesize their own growth factors through an autocrine loop or may bypass the need of growth factor stimulation through constitutive activation of mitogenic pathways downstream of growth factor receptors. Similar scenarios are often found in the cases for transcription factor-derived oncogenes such as Jun and Fos (2, 11).
FIG. 3.
MEQ-induced serum-independent growth in MEQ-transformed Rat-2 cells. A total of 2 × 105 MEQ-transformed and the same number of vector-infected Rat-2 cells were cultured in the absence of serum for up to 4 days. The cell number was counted at 24 h intervals, in duplicate.
FIG. 4.
MEQ-mediated BrdU incorporation and inhibition of apoptosis in serum-starved MEQ-transformed Rat-2 cells. BrdU was added to the media of MEQ-transformed and vector-infected Rat-2 cells for 12 h after cells were serum starved for 3 days. Cells were then fixed and stained with anti-BrdU MAb (A′ and B′) and counterstained with DAPI (A and B). Meanwhile, a TdT assay was used to evaluate apoptosis. Briefly, MEQ-transformed and vector-infected Rat-2 cells were also serum starved for 3 days, fixed and stained with ApopTag (C′ and D′) and counterstained with DAPI (C and D). Magnification, ×216.
MEQ displays antiapoptotic potential.
Oncogenic herpesviruses generally encode separate gene products to induce host cell immortalization and/or transformation as well as to block apoptosis (see Discussion). In some cases, however, both properties can be derived from the same viral gene products, such as LMP1 (16, 21).
The above-described experiments implicate MEQ in the abrogation of serum withdrawal-induced apoptosis of Rat-2 cells. We then asked whether MEQ is antiapoptotic. Rat-2 cells and their MEQ-transformed counterparts were treated with a number of reagents in addition to being subjected to serum withdrawal. The hallmarks of apoptosis include (i) the formation of distinct ladders of nucleosomal DNA fragments (180 to 200 bp), which can be analyzed by DNA fragmentation or TdT assays, (ii) chromosomal condensation and/or nuclear membrane breakdown, and (iii) formation of apoptotic bodies, which can be evaluated by DAPI staining. The antiapoptotic effects of MEQ were measured as described above.
Serum withdrawal.
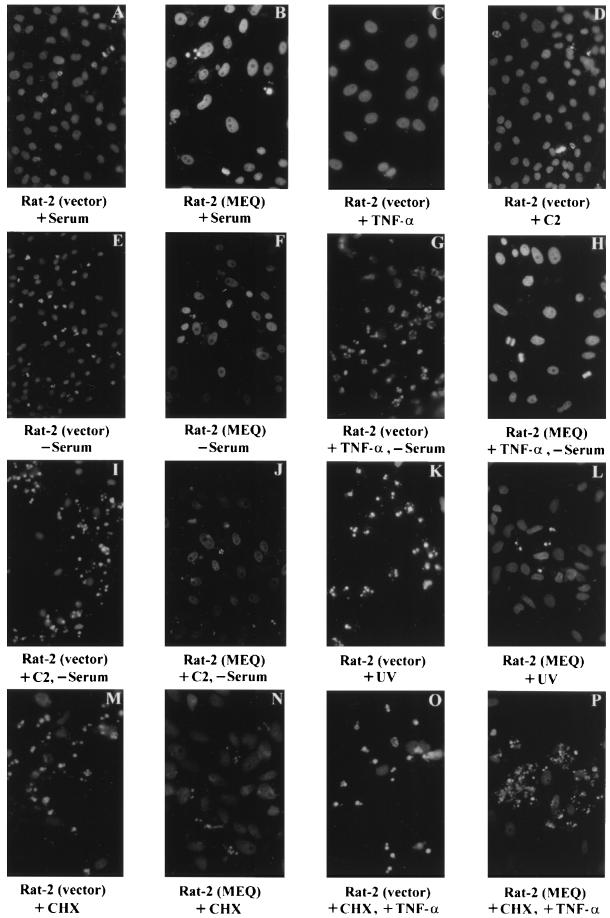
As shown in Fig. 5A and B, in the presence of serum, 0 to 10% of cells are apoptotic in both vector-infected and MEQ-transformed Rat-2 cells. Upon serum withdrawal for 3 days, 60 to 70% of the vector-infected Rat-2 cells became apoptotic (Fig. 5E), whereas only 5 to 10% of MEQ-transformed Rat-2 cells underwent apoptosis (Fig. 5F).
FIG. 5.
Protection of Rat-2 cells from apoptosis by MEQ. MEQ-transformed and vector-infected Rat-2 cells were treated with a variety of apoptosis-inducing vehicles including serum withdrawal, TNF-α, C2-ceramide (C2), UV irradiation, and CHX. Apoptosis was assessed with DAPI staining. Magnification, ×344.
TNF-α treatment.
Treatment with mTNF-α (1 ng/ml) under normal serum condition did not induce apoptosis in Rat-2 (vector) cells (Fig. 5C). However, treatment with mTNF-α in the absence of serum rapidly induced apoptosis in vector-infected Rat-2 cells within 18 to 24 h, which represents a significant shortening of the period required for serum withdrawal to induce apoptosis. A total of 60 to 70% of vector-infected Rat-2 cells became apoptotic (Fig. 5G), compared to only 5 to 10% of MEQ-transformed Rat-2 cells (Fig. 5H).
C2-ceramide treatment.
Similarly, when a downstream mediator of the TNF-α pathway, C2-ceramide (10 μM), was administered in the presence of serum, no apoptosis was induced in vector-infected Rat-2 cells. Meanwhile, the apoptotic effect of serum withdrawal could also be accelerated by C2-ceramide and seemed to be much stronger than that of TNF-α. Apoptosis was observed only 12 h after C2-ceramide treatment in addition to serum withdrawal. Between 70 and 85% of vector-infected Rat-2 cells appeared apoptotic (Fig. 5I), compared to only 0 to 5% of MEQ-transformed Rat-2 cells (Fig. 5J).
UV irradiation.
Likewise, when cells were subjected to UV irradiation (50 J/m2), 80 to 90% of vector-infected Rat-2 cells underwent apoptosis 24 h later (Fig. 5K); conversely, only 10 to 20% of MEQ-transformed Rat-2 cells were found to be apoptotic (Fig. 5L).
CHX treatment.
Interestingly, the blocking of TNF-α-induced apoptosis by MEQ apparently requires protein synthesis, as the addition of the protein synthesis inhibitor CHX reduces this block. MEQ did not appear to efficiently block apoptosis induced by CHX (5 μg/ml, 24 h) (Fig. 5N), especially in the presence of TNF-α (Fig. 5P). Under these conditions, there seemed to be more apoptotic cells in MEQ-transformed Rat-2 cells (Fig. 5P) than in vector-infected Rat-2 cells (Fig. 5O). This, however, is due to the fact that most of vector-infected Rat-2 cells treated with CHX and TNF-α underwent apoptosis and were already detached from the petri dish within 12 h.
In summary (Fig. 6), our data strongly suggest that MEQ is capable of antagonizing apoptosis mediated through TNF-α pathways (TNF-α and C2-ceramide treatments), UV irradiation, and serum withdrawal. This antiapoptotic process requires new protein synthesis and is consistent with MEQ being a transcription factor that induces the expression of genes involved in apoptosis, a notion that is supported by experiments described in the following section.
FIG. 6.
Statistical analysis of MEQ-mediated inhibition of apoptosis. The percent apoptotic cells was calculated based on the results shown in Fig. 5. C2, C2-ceramide.
MEQ up-regulates bcl-2 expression and down-regulates bax expression.
Since MEQ is a transcription factor (39) whose subcellular localization is primarily in the nucleus/nucleolus (27), we postulate that the antiapoptotic effect is likely mediated through regulation of genes involved in apoptosis and/or cell survival at the transcriptional level. Among these, bcl-2 and bax are two candidate genes that are shown to play triggering roles in cell survival and apoptosis, respectively.
As an initial step to explore the possible MEQ-regulated target genes that antagonize apoptosis, we performed Western blotting on vector-infected Rat-2 cells and MEQ-transformed Rat-2 cells to examine the expression levels of Bcl-2 and Bax. As shown in Fig. 7A (lane 3), vector-infected Rat-2 cells express a very low level of Bcl-2 but a high level of Bax. In contrast, MEQ-transformed Rat-2 cells in the presence of serum (Fig. 7A, lane 1) expressed significantly higher levels of Bcl-2, while Bax expression was completely turned off. When MEQ-transformed Rat-2 cells were treated with C2-ceramide in the absence of serum, the level of Bcl-2 expression was reduced but was still significantly higher than that of vector-infected Rat-2 cells (Fig. 7A, lane 2). Bax, on the other hand, remains down-regulated (Fig. 7A, lane 2). We further performed an RT-PCR experiment to determine whether the expression of Bcl-2 and Bax is regulated at the transcriptional or translational level. As shown in Fig. 7B, the transcription of bcl-2 is clearly enhanced, whereas the transcription of bax is completely down modulated in MEQ-transformed Rat-2 cells. We were unable to recover enough Rat-2 cells treated with C2-ceramide in the absence of serum to perform either Western blotting or RT-PCR, since most of the cells became apoptotic and detached from the plate. These findings suggest that MEQ directly or indirectly regulates the expression of bcl-2 and bax genes at the transcriptional level, thereby contributing to its antiapoptotic effects.
FIG. 7.
Induction of bcl-2 and suppression of bax expressions in MEQ-transformed Rat-2 cells. (A) Total cell lysates were prepared from MEQ-transformed Rat-2 cells grown in the presence of serum (lane 1) or after treatment with C2-ceramide in the absence of serum (lane 2) and vector-infected Rat-2 cells (in the presence of serum) (lane 3) and analyzed by SDS-PAGE. The Western blots were stained with rabbit anti-Bcl-2 and -Bax antisera and then detected with a Tropix Western-Light chemiluminescent kit. (B) RT-PCR was performed on total RNA extracted from MEQ-transformed Rat-2 cells grown in the presence of serum (lane 1) or after treatment with C2-ceramide in the absence of serum (lane 2). RT-PCR was similarly carried out with vector-infected Rat-2 cells grown in the presence of serum (lane 3). GAPDH gene expression was used as an internal control for the quality and quantity of the RT-PCR products.
DISCUSSION
In this study we analyzed the transforming potential of MEQ, a putative oncoprotein of MDV. We show that the meq gene effectively transforms established Rat-2 fibroblasts but only marginally transforms primary embryo fibroblasts (data not shown). Thus, unlike most retroviral oncogenes, but similar to herpesviral oncogenes, MEQ alone is not capable of transforming primary cells and may require additional cooperating oncogenes to display its full transforming activity (10, 14, 21, 28, 48). Nevertheless, our data suggest that MEQ is potentially oncogenic. In our study, injection of chickens with replication-defective virus carrying MEQ yielded a low incidence (5%) of sarcomas, which eventually metastasized to internal organs such as the liver, spleen, and lungs (unpublished results), consistent with its being a weak oncogene.
Oncogenesis is a complex process involving multiple steps, and individual oncogenes act on different steps: some override the G1/S restriction and thus activate cell cycle progression, some affect cell-cell communication, and others prevent cell death (4, 23). In this report we show that MEQ is able to induce morphological changes and anchorage-independent growth and to protect transformed cells from apoptosis. MEQ is a transcription factor in the family of the Jun/Fos oncoproteins. MEQ dimerizes with Jun with great affinity and enhances its transactivation (39). We presume that some of the growth stimulation functions may be attributed to MEQ’s ability to activate the oncoprotein Jun or other bZIP proteins. This hypothesis, however, has yet to be experimentally demonstrated. A striking observation from this study is that Rat-2 cells transformed by MEQ are resistant to apoptosis induced by a variety of stimuli including additions of C2-ceramide or TNF-α, UV irradiation, and serum (growth factor) starvation. Some of these apoptotic regimens utilize common signaling pathways leading to apoptosis. For instance, it has been shown that the sphingolipid ceramide is a downstream effector of TNF receptor (TNF-R) (15). Administration of exogenous C2-ceramide thus effectively activates the apoptotic pathway, bypassing the activation of TNF-R. However, in our study, we found that neither TNF-α nor C2-ceramide alone could induce apoptosis in Rat-2 cells. Efficient induction of apoptosis by these agents occurred only in the absence of serum, which contains growth factors, or in the presence of the protein synthesis inhibitor CHX.
How does MEQ perturb the apoptotic pathway? Analysis of Bcl-2 expression in MEQ-transformed Rat-2 cells reveals significant elevation at both protein and RNA levels. Conversely, the levels of Bax protein and RNA are reduced. It is well documented that overexpression of Bcl-2 in many cell types can antagonize apoptosis induced by serum deprivation, UV irradiation, TNF-α, and C2-ceramide treatments (13, 49). It is thought that Bcl-2 may prevent apoptosis through interaction with the upstream activators (CED-4) of ICE/caspases (9, 44, 51) or by blocking the release of cytochrome c from mitochondria (22, 53). However, the exact mechanisms remain to be elucidated. Bax, on the other hand, is a key effector molecule in executing the apoptotic process (36). In our studies, the induction of bcl-2 and the suppression of bax at the transcriptional level appear to correlate with the inhibition of apoptosis in MEQ-transformed Rat-2 cells. It is not clear whether this modulation is through direct binding of MEQ to the promoter of these genes or through other factors activated by MEQ. We note that MEQ binding sites (MERE1 and -2) are found in the promoter of the human bcl-2 gene. Whether the same motifs are present in the promoters of rat bcl-2 and bax genes remains to be established. It is possible that other Bcl-2- and Bax-like molecules are regulated by MEQ as well.
While an antiapoptotic property of a viral gene product may impact its transforming potential, most herpesviruses, oncogenic or not, have evolved a number of ways to prevent infected cells from premature cell death during viral replication and/or latency. Several mechanisms have been utilized by herpesviruses to dodge apoptosis of the host cells (for a review, see reference 46). First, some herpesviruses encode Bcl-2 homologs, such as EBV BHRF-1 (17), herpesvirus saimiri ORF16 (43), and human herpesvirus 8 KSbcl-2 (8). These molecules presumably function to antagonize apoptosis in a manner similar to that of Bcl-2. Second, some herpesviruses encode p53-binding proteins, such as EBV EBNA-LP (45) and BZLF-1/ZEBRA (54). p53 is known to trigger apoptosis at least in part by transactivating the expression of Bax and down-modulating the expression of Bcl-2 (29). Whether these herpesviral p53-binding proteins can inhibit apoptosis through p53 sequestration remains to be elucidated. It is noteworthy that MEQ has been found to interact with p53 in vitro (5). The up-regulation of Bcl-2 and down-regulation of Bax observed in MEQ-transformed Rat-2 cells might thus be mediated by p53 inhibition, although there is very little p53 expressed in Rat-2 and MEQ-transformed Rat-2 cells. Third, some herpesviruses encode death effector domain-containing molecules, such as equine herpesvirus 2 E8 protein, that interfere with signaling transduction of Fas-TNF-R (3, 18) pathways. Last, some herpesviruses encode transcription factors to block apoptosis, such as EBV LMP1 (16, 41), herpes simplex virus type 1 ICP4 (25), and cytomegalovirus IE1 and IE2 (55). LMP1 has been known to induce the expression of Bcl-2 in B cells (16) and A20, a zinc finger protein that can inhibit apoptosis, in epithelial cells (12). The target genes involved in ICP4-, IE1-, and IE2-mediated protection against apoptosis have not been identified yet. Our data add MEQ to the growing list of antiapoptotic herpesviral gene products. While the focus of this report is on the transforming potential, MEQ may function to prolong the cell life span during MDV replication as well.
In summary, this report provides the first investigation of the biological properties of MEQ, and MDV gene product implicated in oncogenic and latent processes of MDV. MEQ has been well delineated as a transcription factor with the characteristics of nuclear localization, dimerization, transactivation and/or repression, and DNA-binding activity. Here, it is shown that MEQ is mitogenic and antiapoptotic. The former is reflected by its ability to increase growth rate, induce BrdU incorporation, and elicit serum-independent growth. The latter is manifested by its potential to protect cells from apoptosis induced by serum starvation and by treatments with a number of apoptosis-inducing reagents. Our studies provide a framework to understand the mechanisms of MEQ as an effector in MDV oncogenesis.
ACKNOWLEDGMENTS
We thank K. A. Schat for CU cell lines and J. P. Morgenstern for the pBabe vector. We also thank K. Everiss and A. W. Grasso for critical reading of the manuscript.
This work was supported by grants from the USDA (93-37204-9340 to L.F.L. and H.-J.K.), the NCI (CA46613 to H.-J.K.), and the Council for Tobacco Research (4034 to H.-J.K.). J.-L.L. is a recipient of a USDA fellowship.
REFERENCES
- 1.Akiyama Y, Kato S. Two cell lines from lymphomas of Marek’s disease. Biken J. 1974;17:105–116. [PubMed] [Google Scholar]
- 2.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 3.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G-H, Senkenvich T G, Alnemri E S, Moss B, Lenardo M J, Tomaselli K J, Cohen J I. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop J M. Molecular themes in oncogenesis. Cell. 1991;64:235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- 5.Brunovskis, P. 1997. Personal communication.
- 6.Calnek B W, Shek W R, Schat K A. Spontaneous and induced herpesvirus genome expression in Marek’s disease tumor cell lines. Infect Immun. 1981;34:483–491. doi: 10.1128/iai.34.2.483-491.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calnek B W. Marek’s disease—a model for herpesvirus oncology. Crit Rev Microbiol. 1985;12:293–320. doi: 10.3109/10408418509104432. [DOI] [PubMed] [Google Scholar]
- 8.Cheng E H, Nickolas J, Bellows D S, Hayward G S, Guo H G, Reitz M S, Hardwick J M. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinnaiyan A M, O’Rourke K, Lane B R, Dixit V M. Interactions of CED-4 with CED-3 and CED-9: a molecular framework for cell death. Science. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- 10.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of B lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran T, Vogt P K. Dangerous liaisons: Fos and Jun, oncogenic transcription factors. In: McNight S L, Yamamota K, editors. Transcription regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 797–836. [Google Scholar]
- 12.Fries K L, Miller W E, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale A J, Smith C A, Sutherland L C, Stoneman V E A, Longthorne V L, Culhane A C, Williams G T. Apoptosis: molecular regulation of cell death. Eur J Biochem. 1996;236:1–26. doi: 10.1111/j.1432-1033.1996.00001.x. [DOI] [PubMed] [Google Scholar]
- 14.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature (London) 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 15.Hannun Y A, Obeid L M. Ceramide: an intracellular signal for apoptosis. Trends Biochem Sci. 1995;20:73–77. doi: 10.1016/s0968-0004(00)88961-6. [DOI] [PubMed] [Google Scholar]
- 16.Henderson S, Rowe M, Gregory C, Croom C D, Wang F, Longnecker R, Kieff E, Rickinson A. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 17.Henderson S, Huen D, Rowe M, Dawson C, Johnson G, Rickinson A. Epstein-Barr virus-encoded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc Natl Acad Sci USA. 1993;90:8479–8483. doi: 10.1073/pnas.90.18.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu S, Vincenz C, Buller M, Dixit V M. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J Biol Chem. 1997;272:9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 19.Jones D, Lee L, Liu J-L, Kung H-J, Tillotson J K. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc Natl Acad Sci USA. 1992;89:4042–4046. doi: 10.1073/pnas.89.9.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato S, Hirai K. Marek’s disease virus. Adv Virus Res. 1985;30:225–277. doi: 10.1016/s0065-3527(08)60452-2. [DOI] [PubMed] [Google Scholar]
- 21.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 23.Kung H-J, Liu J-L. Retroviral oncogenesis. In: Nathanson N, et al., editors. Viral pathogenesis. Philadelphia, Pa: Lippincott-Raven Publishers; 1997. pp. 235–266. [Google Scholar]
- 24.Kung H-J, Tanaka A, Nonoyama M. Two gene families of Marek’s disease virus with a potential role in tumor induction in chicken. Int J Oncol. 1995;6:997–1002. doi: 10.3892/ijo.6.5.997. [DOI] [PubMed] [Google Scholar]
- 25.Leopardi R, Roizman B. The herpes simplex virus major regulatory protein ICP4 blocks apoptosis induced by the virus or by hyperthermia. Proc Natl Acad Sci USA. 1996;93:9538–9587. doi: 10.1073/pnas.93.18.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, D. Unpublished results.
- 27.Liu J-L, Lee L F, Ye Y, Qian Z, Kung H-J. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J Virol. 1997;71:3188–3196. doi: 10.1128/jvi.71.4.3188-3196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyashita T, Krajewski S, Kraikjeski M, Wang H G, Lin H K, Liebermann D A, Hoffman B, Reed J C. Tumor-suppressor p53 is a regulator of bcl-2 and bax gene-expression in vitro and in vivo. Oncogene. 1994;9:1799–1805. [PubMed] [Google Scholar]
- 30.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nazerian K, Stephens E A, Sharma J M, Lee L F, Gailitis M, Witter R L. A nonproducer T lymphoblastoid cell line from Marek’s disease transplantable tumor (JMV) Avian Dis. 1977;21:69–76. [PubMed] [Google Scholar]
- 32.Nazerian K, Payne W, Lee L F, Witter R L. Abstracts of the 78th Annual Meeting of the American Society for Microbiology 1978. Washington, D.C: American Society for Microbiology; 1978. Marek’s disease virus (MDV) transformed lymphoblastoid cell line in a syngeneic system, abstr. S373; p. 274. [Google Scholar]
- 33.Nazerian K, Witter R L, Crittenden L B, Noori-Dalloii M R, Kung H-J. An IgM-producing B lymphoblastoid cell line established from lymphomas induced by a non-defective reticuloendotheliosis virus. J Gen Virol. 1982;58:351–360. doi: 10.1099/0022-1317-58-2-351. [DOI] [PubMed] [Google Scholar]
- 34.Nazerian K, Elmubarak A K, Sharama J M. Establishment of B-lymphoblastoid cell lines from Marek’s disease virus-induced tumors in turkeys. Int J Cancer. 1982;29:63–68. doi: 10.1002/ijc.2910290111. [DOI] [PubMed] [Google Scholar]
- 35.Okazaki W, Witter R L, Romero C, Nazerian K, Sharama J M, Fadly A, Ewert D. Induction of lymphoid leukosis transplantable tumors and the establishment of lymphoblastoid cell lines. Avian Pathol. 1980;9:311–329. doi: 10.1080/03079458008418416. [DOI] [PubMed] [Google Scholar]
- 36.Oltvai Z N, Milliman C L, Korsmeyer S J. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 37.Peng Q, Zeng M, Bhuiyan Z A, Ubakata E, Tanaka A, Nonoyama M, Shirazi Y. Isolation and characterization of Marek’s disease virus (MDV) cDNAs mapping to the Bam HI-I2, BamHI-Q2, and BamHI-L fragments of the MDV genome form lymphoblastoid cells transformed and persistently infected with MDV. Virology. 1995;213:590–599. doi: 10.1006/viro.1995.0031. [DOI] [PubMed] [Google Scholar]
- 38.Powell P C. Marek’s disease virus in the chicken. Adv Viral Oncol. 1985;5:103–127. [Google Scholar]
- 39.Qian Z, Brunovskis P, Rauscher III F, Lee L, Kung H-J. Transactivation activity of Meq, a Marek’s disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J Virol. 1995;69:4037–4044. doi: 10.1128/jvi.69.7.4037-4044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian Z, Brunovskis P, Lee L, Vogt P K, Kung H-J. Novel DNA binding specificities of a putative herpesvirus bZIP oncoprotein. J Virol. 1996;70:7161–7170. doi: 10.1128/jvi.70.10.7161-7170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowe M, Peng-Pilon M, Huen D S, Hardy R, Croom-Carter D, Lundgren E, Rickinson A B. Upregulation of bcl-2 by the Epstein-Barr virus latent membrane protein LMP1: a B-cell-specific response that is delayed relative to NF-κB activation and to induction of cell surface markers. J Virol. 1994;68:5602–5612. doi: 10.1128/jvi.68.9.5602-5612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schat K A, Pratt W D, Morgan R, Weinstock D, Calnek B W. Stable transfection of reticuloendotheliosis virus-transformed lymphoblastoid cell lines. Avian Dis. 1992;36:432–439. [PubMed] [Google Scholar]
- 43.Smith C A. A novel viral homologue of Bcl-2 and Ced-9. Trends Cell Biol. 1995;5:344. doi: 10.1016/s0962-8924(00)89061-3. [DOI] [PubMed] [Google Scholar]
- 44.Spector M S, Desnoyers S, Hoeppner D J, Hengartner M O. Interaction between the C. elegans cell-death regulators CED-9 and CED-4. Nature (London) 1997;385:653–656. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- 45.Szekely L, Selivanova G, Magnusson K P, Klein G, Wiman K G. EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc Natl Acad Sci USA. 1993;90:5455–5459. doi: 10.1073/pnas.90.12.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tillotson J K, Kung H-J, Lee L F. The 3rd International Symposium on Marek’s Disease. Osaka, Japan: Japanese Association on Marek’s Disease; 1988. Accumulation of viral transcripts coding for a DNA binding protein in Marek’s disease tumor cells; pp. 128–134. [Google Scholar]
- 48.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaux D L, Strasser A. The molecular biology of apoptosis. Proc Natl Acad Sci USA. 1996;93:2239–2244. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 51.Wu D, Wallen H D, Nunez G. Interaction and regulation of subcellular localization of CED-4 by CED-9. Science. 1997;275:1126–1129. doi: 10.1126/science.275.5303.1126. [DOI] [PubMed] [Google Scholar]
- 52.Xie Q, Anderson A S, Morgan R W. Marek’s disease virus (MDV) ICP4, pp38, and meq genes are involved in the maintenance of transformation of MDCC-MSB1 MDV-transformed lymphoblastoid cells. J Virol. 1996;70:1125–1131. doi: 10.1128/jvi.70.2.1125-1131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T-I, Jones D P, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Q, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]