Abstract
Analysis of disease induction by simian immunodeficiency viruses (SIV) in macaques was initially hampered by a lack of molecularly defined pathogenic strains. The first molecularly cloned SIV strains inoculated into macaques, SIVmacBK28 and SIVmacBK44 (hereafter designated BK28 and BK44, respectively), were cases in point, since they failed to induce disease within 1 year postinoculation in any inoculated animal. Here we report the natural history of infection with BK28 and BK44 in inoculated rhesus macaques and efforts to increase the pathogenicity of BK28 through genetic manipulation and in vivo passage. BK44 infection resulted in no disease in four animals infected for more than 7 years, whereas BK28 induced disease in less than half of animals monitored for up to 7 years. Elongation of the BK28 transmembrane protein (TM) coding sequence truncated by prior passage in human cells marginally increased pathogenicity, with two of four animals dying in the third year and one dying in the seventh year of infection. Modification of the BK28 long terminal repeat to include four consensus nuclear factor SP1 and two consensus NF-κB binding sites enhanced early virus replication without augmenting pathogenicity. In contrast, in vivo passage of BK28 from the first animal to die from immunodeficiency disease (1.5 years after infection) resulted in a consistently pathogenic strain and a 50% survival time of about 1.3 years, thus corresponding to one of the most pathogenic SIV strains identified to date. To determine whether the diverse viral quasispecies that evolved during in vivo passage was required for pathogenicity or whether a more virulent virus variant had evolved, we generated a molecular clone composed of the 3′ half of the viral genome derived from the in vivo-passaged virus (H824) fused with the 5′ half of the BK28 genome. Kinetics of disease induction with this cloned virus (BK28/H824) were similar to those with the in vivo-passaged virus, with four of five animals surviving less than 1.7 years. Thus, evolution of variants with enhanced pathogenicity can account for the increased pathogenicity of this SIV strain. The genetic changes responsible for this virulent transformation included at most 59 point mutations and 3 length-change mutations. The critical mutations were likely to have been multiple and dispersed, including elongation of the TM and Nef coding sequences; changes in RNA splice donor and acceptor sites, TATA box sites, and Sp1 sites; multiple changes in the V2 region of SU, including a consensus neutralization epitope; and five new N-linked glycosylation sites in SU.
The simian immunodeficiency viruses (SIV) are a diverse collection of primate lentiviruses related to human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2, respectively), with SIVmac (from macaques) and SIVsm (from sooty mangabeys) together with HIV-2 comprising one of the five major primate lentivirus lineages (24). The infection of macaques with SIVmac has been widely used as a nonhuman model for investigating lentivirus pathogenesis, transmission, tissue tropism, viral sequence variation, antiviral therapy, and prophylaxis. Most molecularly cloned SIV have been obtained following isolation in tissue cultures; several SIVmac and SIVsm clones have been reported to express various degrees of attenuated pathogenicity compared to the virus isolates from which they were derived (3, 17, 26, 31, 38), such as the first SIV strain isolated, SIVmac251, which resulted in a 50% 1-year mortality rate in macaques (6, 7, 31). The molecular bases for such observed attenuation of SIV clones remain largely unknown.
After short-term evaluation of the initial molecularly cloned SIVmac strains failed to produce disease (20, 38), we sought to create pathogenic molecular clones of SIVmac to assist in the development of this AIDS model system. In one approach, two putative virulence determinants were investigated by the deliberate introduction of specific mutations. Initially, the prematurely truncated transmembrane protein (TM) gene of the clone SIVmacBK28 (29) was extended to encode full-length gp41 (21), similar to that found in HIV-1. We and others (5, 21, 28) showed that truncation and extension of the SIV TM open reading frame (ORF) represented a reversible adaptation for efficient replication in certain human cell lines and macaques, respectively, but that extension of the TM ORF did not result in acute disease induction (21, 28). Next, based upon the observation that HIV-1 clones and the acutely lethal SIVsmmPBj14 contain duplicated NF-κB sites and larger numbers of near-consensus SP1-like recognition sites (11) compared to SIVmacBK28 (29), we introduced reiterated transcription factor binding sites in an effort to augment the pathogenicity of SIVmacBK28. We now know that nef gene expression is critical for disease induction (27) and that changes in the nef gene that cause it to mimic the nef gene found in SIVsmmPBj14 (11) greatly enhance the virulence characteristics of the SIVmac239 clone (13).
Experimental in vivo passage has often been found to increase the virulence of laboratory-adapted virus isolates (16). Thus, our second approach for obtaining a pathogenic SIVmac clone was to augment the virulence of an existing clone by in vivo passage and then to isolate virus fragments bearing naturally selected mutations. The second approach was successful, and by construction of a virus chimera bearing a 3′-half genome from a macaque with immunodeficiency disease, a pathogenic molecular clone was obtained.
MATERIALS AND METHODS
Viruses and cell lines.
For SIVmacBK28 and SIVmacBK44, infectious stocks were prepared by transfection of the molecular clones λBK28 and λBK44, derived from isolate SIVmac251 (29), into HuT78 cells. For SIVmacBK28-TM41, infectious stocks were prepared as previously described (21) by transfection of the molecular clone pBK28-TM41 into HuT78 cells. For SIVmacF965, bone marrow (obtained at necropsy) from macaque F965 was cocultivated with human peripheral blood mononuclear cells (PBMC) for 14 days, followed by passage of cell-free virus on human PBMC for an additional 14 days. It should be noted that this inoculum was prepared in 1988, prior to the publication of studies demonstrating the selective pressures on the SIV TM coding sequence in vitro by human cells. Work by ourselves (21) and others (5, 28) revealed that rhesus PBMC provided a less selective environment for SIVmac propagation than did human cells, the latter of which were shown to select for genetic alterations, including truncation of the TM gene. However, since the inoculum was shown to be immediately pathogenic in vivo, we did not need to recreate the inoculum by growth on macaque cells. For SIVmacBK28/H824 and SIVmacBK28-2kB-4SP, infectious stocks were prepared by liposome-mediated transfection of plasmid DNA from pSIVmacBK28/H824 or linear concatameric pBK.1-5′half (see below) ligated with pBK28-3′-2kB-4SP into CEMx174 cells with Lipofectin reagent (GIBCO/BRL, Gaithersburg, Md.), according to the manufacturer’s instructions, and were passaged for 25 or 27 days, respectively. Cultures were monitored at 1- to 2-day intervals for indications of virus production by visual evaluation of syncytium formation and by a reverse transcriptase assay (46). In all cases, virus-containing supernatants were passed through 0.22-μm-pore-size filters, mixed with equal volumes of fetal calf serum, and then stored in liquid nitrogen. The amount of inoculum used for each animal is shown in Fig. 1.
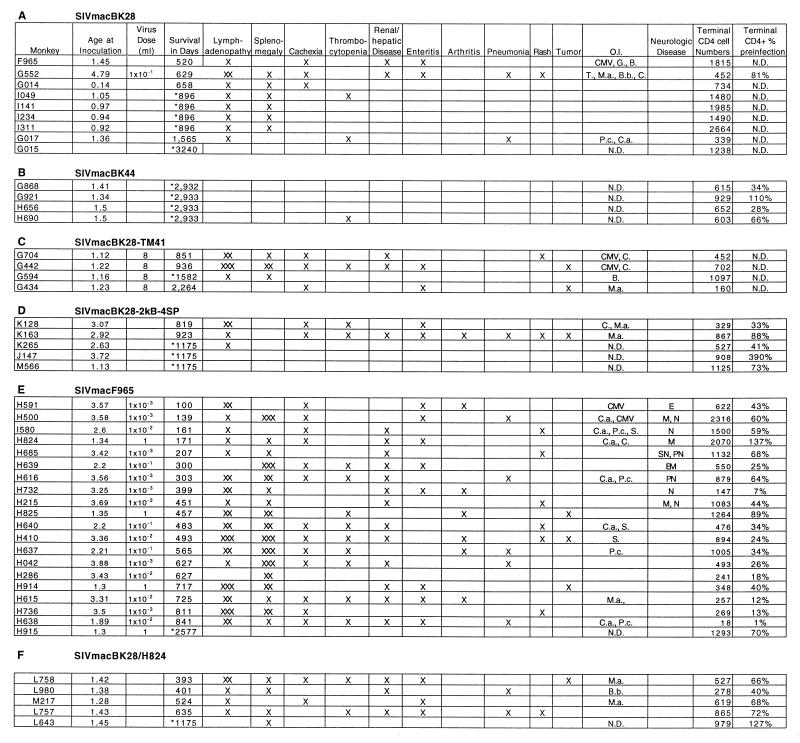
FIG. 1.
Pathology in macaques infected with molecularly cloned SIV strains. Age at inoculation is in years. An asterisk in the survival column identifies macaques that were sacrificed while clinically asymptomatic. Presentation of pathologic symptoms is indicated by ×. Diagnoses were made either during disease or at autopsy. Severities of lymphadenopathy and splenomegaly were graded by distribution and size as mild (×), moderate (××), and severe (×××). Thrombocytopenia was defined as platelet counts below 150,000/ml. Opportunistic infections (O.I.) associated with the following pathogens were denoted as follows: CMV, cytomegalovirus; G., Giardia; B., Balantidium; T., Trichomonas; M.a., Mycobacterium avium-intracellulare; B.b., Bordetella bronchiseptica; C., Cryptosporidium; P.c., Pneumocystis carinii; C.a., Candida albicans; S., Staphylococcus. Signs of neurological involvement were identified as neuritis (N), peripheral neuropathy (PN), spinal neuropathy (SN), meningitis (M), encephalitis (E), and encephalomalacia (EM). Terminal levels of CD4+ cells/mm3 of blood were determined by flow cytometric evaluation of samples stained with antibody FI-OKT4 in the clinical laboratory and were expressed as both absolute cell numbers (Terminal CD4 cell Numbers) and percentages of preinfection levels. N.D., not determined.
The HuT78 cell line was obtained from the American Type Culture Collection (Rockville, Md.), and the CEMx174 cell line was obtained from the AIDS Research and Reference Reagent Program (Rockville, Md.). Both cell lines were maintained in RPMI medium supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM l-glutamine.
Monkeys and in vivo inoculation experiments.
Indian rhesus macaques (Macaca mulatta) used in these studies were housed at the Tulane Regional Primate Research Center (TRPRC), Covington, La., and the Biomedical Primate Research Centre (BPRC), Rijswijk, The Netherlands. Following intravenous inoculation (dose indicated in Fig. 1), animals were assayed for infection by nested PCR amplification of PBMC DNA with primers specific for the viral long terminal repeat (LTR) (34a), by measurement of SIV gp140-specific antibodies in serum (42a), and by measurement of p27Gag levels in serum with the SIV Core Antigen Assay (Coulter Immunology, Hialeah, Fla.) (level of sensitivity, 0.03 ng/ml). Alternatively, animals were assayed for infection on the basis of antigenic cross-reactivity with HIV-1 by use of the Abbott HIV-1 Immunoassay (Abbott Laboratories, Abbott Park, Ill.) with incubation times modified for greater sensitivity (0.18 to 0.25 ng/ml) (9). Monkeys were observed daily by animal care staff, who noted any abnormal behavior. Full physical examinations were given by a veterinarian weekly for the first 4 weeks and monthly thereafter. To assess clinical status, weight, lymph node and spleen sizes, body temperature, general physical condition, and signs of opportunistic infections were monitored. Complete blood counts and enumeration of lymphocyte subsets were done periodically, and serum chemistries, fecal examinations, and radiography were done when necessary for clinical diagnosis. Animals which became moribund were humanely sacrificed.
Construction of viruses with additional NF-κB and SP1 binding sites in the 3′ LTR.
Site-directed mutagenesis was performed as previously described (19) to produce three synthetic LTRs, one in which a second NF-κB site was introduced, another in which the original SP1-like motifs from clone SIVmacBK28 were replaced with four consensus SP1 sites, and a third which contained both of these changes (Fig. 2).
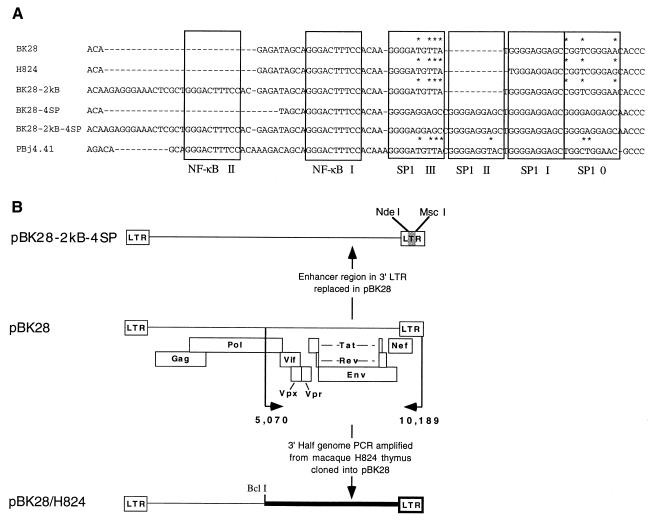
FIG. 2.
Recombinant LTR enhancer constructs. (A) Sequence alignments of SIVmacBK28 LTR U3 enhancer-promoter constructs and the acutely pathogenic SIVsmmPBj4.41. Gaps introduced to maintain alignment are indicated by dashes. Boxes enclose regions corresponding to transcription factor binding site consensus sequences. The consensus sequence (G/T)GGG(C/A)GG(G/A)(G/A)(C/T) was used to delineate potential binding sites for SP1, and the consensus sequence GGGACTTTCC was used to delineate potential binding sites for NF-κB. Bases which do not match the consensus sequence are indicated above the sequence with an asterisk. (B) Schematic representation of some molecularly cloned viruses used in this study. For the construction of chimera pBK28-2kB-4SP, the enhancer element region bounded by restriction sites NdeI and MscI was replaced with a mutagenized fragment described in Materials and Methods. For pBK28/H824, the 3′-half-genome fragments were PCR amplified from thymus DNA from SIVmacF965-infected macaque H824 and ligated into pUC19 to create 3′-half-genome clone pH824-3′. The 3′ BclI/EcoRI fragment was transferred into 5′-half-genome clone pBK.1-5′ to create the full-length proviral chimera.
Mutagenesis was performed by adding 15 ng of plasmid pTG637 (a derivative of pBK28 with only ∼1.5 kb of flanking cellular DNA, kindly provided by Marie-Paule Kieny) to 100 μl of a PCR reaction cocktail containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.25 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 100 nM each relevant primer (Fig. 3), and 2.5 U of AmpliTaq polymerase (Perkin Elmer/Roche Molecular Systems). Reaction mixtures were overlaid with 100 μl of mineral oil and subjected to 15 cycles of amplification (94°C for 0.75 min, 60°C for 1 min, and 72°C for 1 min). Mutant LTR fragments bearing two copies of the NF-κB binding sequence GGGACTTTCC were created with primers PENF-kB2, PENF-kB7, PENF-kB1, and PENF-kB8. Similarly, LTR fragments carrying four tandem repeats of the SP1 site GGGGAGGAGC were created with PENF-kB9 and PENF-kB10 and with PENF-kB7 and PENF-kB8.
FIG. 3.
Oligonucleotide primers for PCR amplification and mutagenesis.
The mutant LTR fragments were ligated into the EcoRI and SalI sites of pBluescript KS+ (Stratagene, La Jolla, Calif.), transformed into JM109 cells, and cultured at 37°C. Clones bearing only the desired mutations were selected after DNA sequencing. An LTR clone carrying two NF-κB sites (pBK28-3′LTR-2kB) was used as the template for an additional round of mutagenesis to combine the NF-κB and SP1 changes together in a single LTR by use of the SP1 mutagenic primers PENF-kB9 and PENF-kB10. The resulting product, containing two NF-κB sites and four SP1 sites, was cloned and sequenced as described above.
Mutant LTRs containing two NF-κB, four SP1, or both mutations were transferred into a pBluescript KS+ clone carrying bases 9,171 to 10,249 of SIVmacBK28 to create mutant 3′-end clones. Each of the mutant 3′-end clones was extended to span the entire 3′ half of the viral genome by insertion of the XbaI/BamHI fragment (bases 4,076 to 9,171) from pBK28.
A clone containing the 5′ half of the SIVmacBK28 viral genome (pBK.1-5′half) was prepared by removal of the 2,350-bp ClaI fragment (bases 8,059 to 10,405) from pBK.1 (a full-length subclone of pBK28 with only 150 bp of cellular flanking sequences, kindly provided by G. Viglianti). The 5′-half-genome plasmid and each of the 3′-half-genome plasmids, pBK.1-3′half, pBK28-3′-2kB, pBK28-3′-4SP, and pBK28-3′-2kB-4SP, were transformed into the dam host GM48. Infectious proviral DNA was assembled by digesting the 5′- and 3′-half-genome plasmids at the unique BclI site (base 5,113) and then ligating them together to create linear concatamers as previously described (11, 43). One month following transfection, the integrity of the 3′ LTR sequence was confirmed in vivo by DNA sequencing (data not shown).
PCR amplification of 5-kb proviral half genomes.
Genomic DNA was prepared from cryopreserved thymus tissue from macaque H824 by standard proteinase K digestion and phenol-chloroform extraction (49). To avoid preferential amplification of proviruses bearing large internal deletions, the genomic target DNA was diluted prior to amplification (14). One hundred nanograms of thymus DNA was used as a template in 100-μl PCR reaction mixtures containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 1% dimethyl sulfoxide, 100 nM (each) primers SD3 and SD2 (Fig. 1), and 2.5 U of AmpliTaq polymerase. Reaction mixtures were overlaid with 100 μl of mineral oil and subjected to 35 cycles of amplification (94°C for 0.75 min, 55°C for 1 min, and 72°C for 4 min) followed by 72°C for 10 min (first round) in a Thermal Cycler (Perkin-Elmer, Foster City, Calif.). Two microliters from each reaction mixture was transferred to fresh reaction tubes containing PCR reaction mixtures plus 100 nM (each) primers SD3 and SD9 (Fig. 3) and then reamplified for an additional 35 cycles under the same conditions (second round). As controls for contamination, reactions containing normal human DNA extracted in parallel with the H824 sample and reagents only were performed simultaneously. Positive reactions were identified by agarose gel electrophoresis with ethidium bromide staining.
Construction of chimeric provirus pBK28/H824.
3′-half-genome PCR products, amplified as described above from postmortem thymus DNA from macaque H824, were digested with EcoRI (at sites encoded by the PCR primers SD9 and SD12), ligated into the EcoRI site of pUC19, and then electroporated into 40 μl of J5 cells as previously described (1) (kindly provided by V. Hirsch, Georgetown University). Electroporation was performed at 400 Ω, 2.5 kV, and 25 μF in 0.2-cm electroporation cuvettes with a Gene Pulser and a Pulse Controller (Bio-Rad, Hercules, Calif.). Electroporated cells were incubated at 37°C for 1 h with Luria-Bertani medium and then spread onto Luria-Bertani agar plates containing 100 μg of carbenicillin per ml. Bacterial incubation on solid and in liquid media was performed at room temperature rather than at 37°C to enhance the stability of plasmids containing large lentivirus inserts (30; data not shown). The 3′-half-genome plasmid pH824-3′ was electroporated into the dam bacterial strain GM48.
The 5′-half-genome subclone pBK.1-5′ was derived from pBK.1 by replacement of the 3′ 5.3-kb BclI/EcoRI fragment (bases 5,113 to 10,404) with a BclI/EcoRI adapter made by annealing two partially complementary oligonucleotides, PE BclI-Eco-L1, and PE BclI-Eco-L2 (Fig. 3), which combined to create a 16-bp double-stranded DNA fragment with 5′ overhangs that are complementary to the BclI and EcoRI sites, respectively. Full-length chimeric provirus pBK28/H234-3′ was constructed by ligation of the 3′-half-genome fragments excised from pH824-3′ plasmids 1 to 5 (SIVmacBK28 nucleotide positions 5,113 to 10,189) into the BclI and EcoRI sites of plasmid pBK.1-5′.
DNA sequencing.
The complete nucleotide sequence of clone pBK28 was determined by deletion clone generation in phage M13 with the Bal 31 method and 35S-dideoxynucleotide sequencing. The sequence of the H824 3′-half genome was determined by a shotgun strategy of subcloning into an M13 vector with subsequent sequencing by use of Taq polymerase and fluorescent dideoxynucleotides, and the products were resolved with an Applied Biosystems 370A DNA sequencer. The nucleotide sequences of shorter segments of env were determined as part of another study (14a).
Nucleotide sequence accession numbers.
The pBK28 sequence has been deposited in GenBank under accession numbers including M15897. The H824 3′-half-genome sequence has been deposited in GenBank under accession no. U86638.
RESULTS
Infection of macaques with molecularly cloned SIVmacBK28 and SIVmacBK44: first in vivo passage.
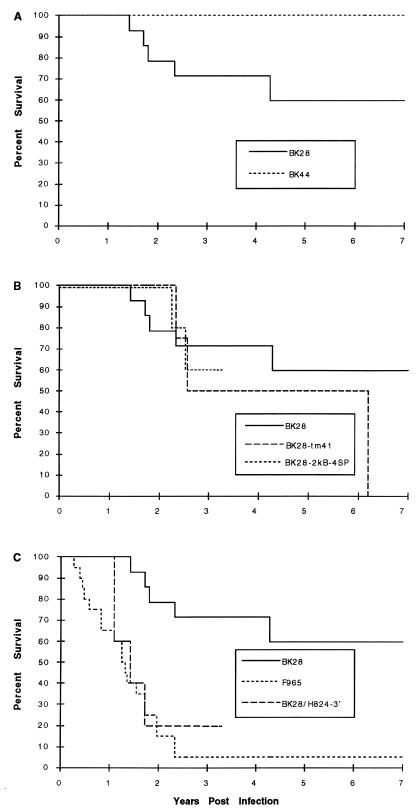
Over the course of several studies to evaluate the natural history of infection and virus stock titration, a total of 24 rhesus macaques at the TRPRC and the BPRC were infected with the molecularly cloned virus SIVmacBK28. Fourteen animals were sacrificed for tissue collection in the absence of overt manifestations of AIDS-like disease between 15 and 896 days postinfection (p.i.). Among the remaining 10 macaques, the median survival exceeded 7 years (range, 520 to >3,200 days) (Fig. 4A) and included the deaths of 5 macaques with AIDS-like symptoms after 520 to 1,565 days (4 of which are described in Fig. 1). Four additional macaques at the TRPRC were infected with SIVmacBK44, a clone derived from the same culture as SIVmacBK28 (29). None of these macaques died during the 7- year follow-up (Fig. 4A), and none developed p27Gag antigenemia within 2 weeks p.i. (Fig. 1 and Table 1).
FIG. 4.
Macaque survival after SIV infection. Graphs are Kaplan-Meier depictions of survival of SIV-infected macaques over time.
TABLE 1.
Frequency of acute antigenemia in SIV-infected macaquesa
| SIVmac inoculum | No. of monkeys that were:
|
% Positive monkeys | Mean p27 concn (ng/ml) | |
|---|---|---|---|---|
| p27 positive | p27 negative | |||
| BK28 | 4 | 12 | 25 | <0.03 |
| BK28-TM41 | 0 | 4 | 0 | <0.03 |
| BK44 | 0 | 4 | 0 | <0.03 |
| BK28-2kB-4Sp | 4 | 1 | 80 | 0.08 |
| F965 | 20 | 0 | 100 | 2.54 |
| BK28/H824 | 5 | 0 | 100 | 0.61 |
Antigenemia was determined by measurement of free p27Gag in serum or plasma from rhesus monkeys between 10 and 16 days p.i. by an antigen capture enzyme-linked immunosorbent assay. p27 was assayed with the SIV Core Antigen Assay; values greater than 0.03 ng/ml were considered positive.
Infection with SIVmacBK28 was generally characterized by asymptomatic infection and long-term survival with, on average, increasing levels of CD4+ cells (Fig. 5). Four of 16 animals tested developed viral p27Gag antigenemia 2 weeks p.i. (Table 1). The most common clinical findings in diseased macaques were mild lymphadenopathy and splenomegaly, with cachexia, enteritis, and thrombocytopenia being less frequently observed (Fig. 1). Opportunistic pathogens were detected in three macaques (Fig. 1). Hence, the molecular clones SIVmacBK28 and SIVmacBK44, derived following adaptation of the virus to growth in the human T-cell line HuT78, appeared to be less pathogenic than the SIVmac251 isolate from which they were derived.
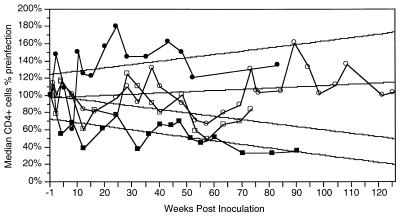
FIG. 5.
Median CD4+ cell levels in rhesus monkeys infected with four different SIV clones. Absolute numbers of circulating CD4+ cells were measured in rhesus monkeys infected with SIVmacBK28 (•), SIVmacF965 (▪), SIVmacBK28-2kB-4SP (○), or SIVmacBK28/H824 (□) and expressed relative to preinfection levels. Median values were calculated from all of the individual animal measurements (3 to 20 values for each point). Correlation coefficients (standard errors of the mean) were 0.30 (0.0033), 0.27 (0.001), 0.46 (0.0019), and 0.62 (0.0013) for animals infected with SIVmacBK28, SIVmacBK28-2kB-4SP, SIVmacBK28/H824, and SIVmac965, respectively.
Genetic manipulation of SIVmacBK28 in attempts to enhance pathogenicity. (i) Removal of the stop codon in the TM.
The portion of the env gene ORF encoding the TM is truncated as a result of adaptation to growth in some human T-cell lines (5, 21, 28). This ORF was extended by removal of the premature stop codon (21), and four macaques at the TRPRC were infected with this virus (SIVmacBK28-TM41). Median survival was reduced to 2.5 years p.i. (Fig. 4B); hence, a small increase in pathogenicity in vivo could have resulted from this change.
(ii) Effect of transcription factor site reiteration on pathogenicity.
The acutely lethal SIV pathogen, SIVsmmPBj14, was previously found to contain a second NF-κB binding site not found in other SIV strains (11). A further comparison to the SIVmacBK28 LTR sequence also revealed that it had fewer and less consensus-like nuclear factor SP1 binding sites (11, 29) (Fig. 2). Since similar changes had been known or suspected to have an impact on retrovirus pathogenicity (8, 10, 52), we sought to investigate the effect of NF-κB and SP1 binding site reiteration on the pathogenicity of SIVmacBK28. A series of mutant viruses containing different combinations of these sites in the 3′ LTR were generated. With site-directed mutagenesis, three variant LTRs were constructed, the first with two NF-κB sites and wild-type SIVmacBK28 SP1 sites, the second with four consensus SP1 sites and a single wild-type NF-κB site, and the third with both two NF-κB sites and four consensus SP1 sites; the placement of each site was chosen to match that found in the SIVsmmPBj14 sequence (Fig. 2). The wild-type and mutant LTRs were placed into the context of the full-length viral genome and transfected into cells to produce stocks of infectious virus. All four constructs displayed similar in vitro phenotypes characterized by syncytium induction and replication to high reverse transcriptase levels in HuT78 cells (data not shown).
Mutant virus derived from the construct containing the maximum number of NF-κB and SP1 sites (SIVmacBK28-2kB-4SP) was used to infect five rhesus macaques at the TRPRC to evaluate its pathogenicity in vivo. Four of the five were antigenemic on day 13 p.i. (Table 1), although none experienced acute onset of clinical disease. All were PCR positive for proviral DNA from PBMC at 285 days (data not shown). Two animals (K128 and K163) died with AIDS-like symptoms after 819 and 923 days, respectively (Fig. 1 and 4B). Throughout 125 weeks of hematological follow-up, levels of circulating CD4+ cells (and CD4+CD29+ lymphocytes; data not shown) in SIVmacBK28-2kB-4SP-infected macaques remained stable on average although depressed relative to levels in macaques infected with wild-type SIVmacBK28 (P < 0.05) (Fig. 5). However, the two rhesus macaques that died (K128 and K163) experienced greater than 80% declines in CD4+ lymphocyte levels, while CD4+ cell populations in the remaining macaques fluctuated near or above preinfection levels (data not shown). The induction of acute antigenemia in 80% of the animals infected with SIVmacBK28-2kB-4SP contrasted with the results for those infected with the parental virus SIVmacBK28 (25% of the animals; P < 0.025) (Table 1). Thus, replacement of the wild-type enhancer with a sequence containing two NF-κB and four consensus SP1 binding sites appeared to have enhanced early virus replication without substantially augmenting pathogenicity.
Second in vivo passage of SIVmacBK28: infection with SIVmacF965.
The deaths of three macaques in less than 2 years after infection with SIVmacBK28 contrasted with the long-term asymptomatic survival more typical of infection with this virus and suggested that virulent viruses or virus mixtures may have developed within these relatively short-lived animals. To test this hypothesis, virus was isolated from the bone marrow of the macaque with the shortest survival (520 days p.i., animal F965), expanded in vitro through two 14-day passages in human PBMC, and inoculated into a second group of macaques.
Twenty macaques at the TRPRC were inoculated with isolate SIVmacF965. In contrast to the results for animals inoculated with the parental molecularly cloned virus, death with AIDS-like symptoms occurred in 95% of infected macaques within 100 to 841 days (median, 457 days), with 85% (17 of 20) dying by 2 years p.i. and only one surviving more than 2.3 years (Fig. 1 and 4C). Thus, overall median survival was reduced about threefold from approximately 1,500 days with SIVmacBK28 infection to 470 days (one-tailed t test, assuming unequal variance; P < 0.005). As also found previously (40), survival was independent of infectious dose over a 1,000-fold range (Fig. 1). Furthermore, all SIVmacF965-infected macaques were positive for viral p27Gag antigenemia during acute infection (Table 1). Among eight animals examined in detail, antigenemia persisted throughout infection in the most rapidly declining animals, whereas clearance of antigen or clearance followed by a return of antigen was seen in the remaining macaques examined (data not shown). Production of anti-SIV antibodies was undetectable by Western blotting throughout infection in the macaques with the shortest survival, while broad antibody responses against several SIV proteins (p17Gag, p27Gag, gp35/41Env, and gp120Env) were detected in the macaques with longer survival (data not shown).
The majority of SIVmacF965-infected macaques with clinical disease displayed marked declines in CD4+ cell levels (and CD4+CD29+ cell levels; data not shown) relative to animals infected with SIVmacBK28 and SIVmacBK28-2kB-4SP (P < 0.05) (Fig. 5). Common symptoms were moderate to severe lymphadenopathy, splenomegaly, cachexia, enteritis, renal and hepatic disease, arthritis, and rashes (Fig. 1). Thrombocytopenia was present in 9 of 15 macaques that survived more than 300 days but was not detected in those with shorter survival. Opportunistic pathogens were detected in eight macaques, and lymphomas were found in three (Fig. 1).
Infection with chimeric virus BK28/H824.
We next evaluated the hypothesis that the enhanced pathogenicity of the SIVmacF965 strain could be embodied within individual virus variants rather than requiring a pool of equally virulent but antigenically diverse variants for rapid disease induction. Chimeric proviruses were generated by combining the 5′ half of SIVmacBK28 (nucleotide positions 1 to 5,113) with four distinct 3′-half genomes (nucleotide positions 5,113 to 10,189) obtained by direct PCR amplification of thymus DNA from SIVmacF965-infected macaque H824 (Fig. 2). This macaque was chronologically the first to die following infection with SIVmacF965, at 173 days p.i. Each chimera was transfected into CEMx174 cells, but only one (hereafter referred to as BK28/H824) gave rise to infectious virus, as measured by reverse transcriptase activity, syncytium induction, and cell-free infectivity (data not shown). BK28/H824 displayed a phenotype in cultures distinct from that of SIVmacBK28. While both replicated to high reverse transcriptase levels and produced large multinucleated syncytia in CEMx174 cells, BK28/H824 did not replicate in HuT78 cells, even after deliberate truncation of the TM from 41 to 32 kDa (data not shown).
Five rhesus macaques at the TRPRC were inoculated with the molecularly cloned chimeric virus BK28/H824. All five became antigenemic, with measurable serum p27Gag levels on day 13 p.i. but at a mean level lower than that found in SIVmacF965-infected animals (Table 1). Death with AIDS-like symptoms occurred in four of these infected macaques within 393 to 635 days (median, 462 days), with only animal L643 surviving more than 2 years (Fig. 1 and 4C). Symptoms were similar to those detected following infection with the ancestral SIVmacF965 isolate (Fig. 1).
Coincident with acute antigenemia were marked declines in circulating CD4+ cell levels in all infected animals beginning at 2 weeks p.i. (Fig. 5) as well as in CD4+CD29+ cell levels (data not shown). During the first 2 years of hematological follow-up, four of the five macaques experienced large declines (greater than 40 to 80%) in circulating CD4+ and CD4+CD29+ cell levels (Fig. 5; see Fig. 1 for data from individual animals). On average, CD4+ and CD4+CD29+ cell levels in BK28/H824-infected macaques were depressed relative to levels observed following SIVmacBK28 and SIVmacBK28-2kB-4SP infections (P < 0.05), although less so than following infection with SIVmacF965 (Fig. 5).
Nucleotide changes in the H824 3′-half genome associated with disease induction.
Fifty-nine point mutations over 5,111 bases (1.15% divergence), 12- and 1-base deletions, and a 3-base insertion distinguished the 3′-half genomes of H824 and BK28 (Fig. 6). A summary of these changes is shown in Table 2. Several changes that might conceivably contribute to the increased virulence of the BK28/H824 chimera were found. Mutations were noted in the consensus splice donor for tat and rev and in the acceptor for tat, rev, and nef, the latter of which also extended the TM coding sequence to encode a putative gp41 versus the gp32 encoded by BK28.
FIG. 6.
Deduced protein and LTR sequences from clone H824 compared to those of parental clone BK28. Sequences that differ from the BK28 sequence are indicated for each sequence element, whereas identical sequences are indicated by dots and gaps are indicated by dashes. ΔPol and ΔLTR indicate that the N-terminal sequence of Pol and the 3′ end of the LTR were not replaced in the BK28/H824 chimera. Predicted N-glycosylation sites that differ between the two sequences are underlined. A summary of the changes is shown in Tables 2 and 3.
TABLE 2.
Genetic changes distinguishing the 3′-half genomes of BK28 and H824a
| Segment | Length (nt/aa) | No. (%) of:
|
Comments regarding changes in H824 relative to BK28 | |
|---|---|---|---|---|
| nt changes | aa changes | |||
| Δpol | 314/104 | 2 (0.6) | 1 (1.0) | |
| vif | 642/214 | 5 (0.8) | 3 (1.4) | One new N-linked glycosylation site |
| vpx | 336/112 | 3 (0.9) | 0 (0.0) | |
| vpr | 291/97 | 1 (0.3) | 1 (0.5) | One N-linked glycosylation site lost |
| tat1 | 296/99 | 3 (1.0) | 3 (3.0) | Mutation in consensus splice donor led to amino acid change |
| rev1 | 70/23 | 2 (2.9) | 1 (4.2) | Mutation in consensus splice donor led to amino acid change |
| gp120 | 1,539/513 | 25b (1.6) | 20 (3.9) | Five new N-linked glycosylation sites; neutralization site in V2 altered |
| tat2 | 97/32 | 1 (1.0) | 1 (3.1) | Mutation in consensus splice donor led to reversion of stop codon and elongation of TM |
| rev2 | 254/85 | 2 (0.8) | 1 (1.2) | Mutation in consensus splice donor led to reversion of stop codon and elongation of TM |
| gp32/41 | Stop codon reverted to encode gp41 | |||
| nef | 741/247 | 9c (1.2) | 25b (10.1) | Repair of a frameshift mutation, resulting in replacement of 15 amino acids and extension of the protein by 16 amino acids |
| ΔLTR | 747 | 9 (1.2) | One base change in Sp1 site brings it closer to consensus; another Sp1 site has a 1-base deletion; TATAA consensus changed to CATAA | |
Δ, analysis of only a portion of the gene or LTR segment; nt, nucleotide; aa, amino acid.
In addition to the point mutations tallied, one four-codon deletion and a one-codon insertion were also detected.
Relative to the SIV database consensus, a deletion was present in BK28 but was repaired in H824.
As a result of a single base deletion, BK28 encoded a truncated Nef protein with a nonconsensus string of 15 amino acids following the frameshift at the C-terminal end. This mutation was exquisitely repaired in vivo by the insertion of the consensus (T) nucleotide in the H824 sequence. In the LTR, the TATA box consensus sequence was changed from TATAA to CATAA, and two mutations were noted in Sp1 sites, an A→G mutation in position 10 of SP1 0, a change that similarly agrees with the consensus, and a deletion of a G between Sp1 III and Sp1 III, which reduces the spacing between these sites (Fig. 2A).
Env glycosylation site changes.
Mutations resulting in the loss of one and the gain of five new sites for potential N-linked glycosylation occurred within the region from V1 to V5 of the H824 gp120 coding sequence. Given the involvement of N-linked glycosylation in virus replication and host interactions (2, 15, 18), conservation of these sites was evaluated in the other characterized SIV clones as well as BK28-infected macaques to investigate a possible association with SIV pathogenicity. We used DNA sequence analysis (14a) and allele-specific amplification (39) (Table 3). The site at position 114 was quantitatively lost by 267 days following infection in animal F965 (Table 3), as a result of an S→N change that simultaneously created the new site at position 116. The same mutation occurred during viral growth within two of the three other animals evaluated (G015 and I049 but not 2BS). Animal 2BS was the only other animal of this group to succumb to an AIDS-like illness; hence, a shift from position 114 to position 116 was not closely linked to disease induction in this study.
TABLE 3.
N-linked glycosylation sites in Env and proviral DNA load in SIV-infected macaquesa
| SIV or animal | Days p.i. | Proviral load/105 cells | Survival (days) | Amino acid position
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 114 | 116 | 173 | 256 | 417 | 481 | ||||
| SIV | |||||||||
| smmPBj4.41 | + | − | + | + | + | + | |||
| smmH4 | + | − | + | + | − | + | |||
| smmB670 | + | − | + | + | + | + | |||
| mneC18 | + | − | + | + | + | + | |||
| mneC18 in vivo | + | +/− | + | + | 1+ | + | |||
| mac239 | + | − | + | − | − | + | |||
| mac239 in vivo | + | − | + | − | +/− | + | |||
| mac142 | + | − | + | + | − | − | |||
| mac1A11 | + | − | + | − | − | + | |||
| macBK28 | + | − | − | − | − | − | |||
| Animal | |||||||||
| 2BS (BK28) | 123 | 8 | 855 | 100 | 0 | 0 | 0 | 0 | 0 |
| 2BS | 686 | 111 | 100 | 0 | 100 | 0 | 100 | 100 | |
| G015 (BK28) | 724 | 1 | 3,238b | 63 | 37 | 0 | 0 | 37 | 88 |
| I049 (BK28) | 897 | 130 | 897b | 69 | 31 | 100 | 100 | 100 | 100 |
| F965 (BK28) | 124 | 24 | 520 | 70 | 30 | 78 | 0 | 0 | 78 |
| F965 | 267 | 240 | 0 | 100 | 100 | 0 | 100 | 100 | |
| F965 (con) | − | + | + | + | + | + | |||
| H591 (F965) | 100 | 68 | 100 | 0 | 100 | 100 | 82 | 100 | 100 |
| H500 (F965) | 140 | 65 | 140 | 33 | 67 | 100 | 100 | 100 | 100 |
| I580 (F965) | 161 | 42 | 161 | 31 | 69 | 100 | 100 | 100 | 100 |
| H824 (F965) | 171 | 82 | 171 | 0 | 100 | 100 | 100 | 100 | 100 |
| H824 (clone) | − | + | + | + | + | + | |||
| H042 (F965) | 158 | 32 | 627 | 0 | 100 | 100 | 100 | 100 | 100 |
| H042 | 627 | 130 | 0 | 100 | 100 | 100 | 100 | 100 | |
| H914 (F965) | 717 | 12 | 717 | 90 | 10 | 100 | 89 | 100 | 100 |
| H615 (F965) | 158 | 87 | 725 | 27 | 73 | 100 | 100 | 100 | 100 |
| H615 | 725 | 166 | 33 | 67 | 100 | 100 | 83 | 100 | |
| H915 (F965) | 45 | 6 | 2,576b | 0 | 100 | 100 | 100 | 100 | 100 |
| H915 | 875 | 5 | 0 | 100 | 100 | 100 | 100 | 100 | |
The presence of potential sites for N-linked glycosylation, NX(S/T), at six positions in the SU envelope protein was analyzed for cloned viruses and for viruses following in vivo passage. For animals inoculated with SIVmacBK28 or SIVmacF965, the inoculum is indicated in parentheses. The presence or absence of N-linked glycosylation sites is indicated by + and −, respectively (determined by allele-specific amplification), or by the percentage of molecules sequenced that encoded the sites. Acquisition of a site in a minority of animals following in vivo replication is indicated by +/−. The site at amino acid 417 in SIVmneC18 was shifted two residues C terminal by in vivo mutation (1+). Amino acid positions were numbered according to the SIVmacBK28 sequence (the prototype SIVmac251 sequence in GenBank). Cloned virus sequences for SIVmacBK28 and SIVmacF965 were obtained from this study and from the Los Alamos HIV Sequence Database (37).
Sacrificed while asymptomatic (not because of disease).
The N-linked glycosylation site at position 173 was found in all of the other SIV strains evaluated and in 100% of the viral genomes evaluated in animals F965 and 2BS, at 267 and 686 days after infection, respectively, and in animal I049, when sacrificed while asymptomatic 896 days p.i. This site was not found in animal G015 at 724 days p.i.; this animal was later sacrificed when asymptomatic more than 8 years after infection. A significant correlation was found between the presence of this site and the proviral DNA load in the PBMC of infected macaques. The site was found in five of six macaques with high virus loads (50 to 240 proviruses/105 cells, including 2BS, I049, and F965) but in only one of seven macaques with low provirus loads (0.6 to 10 proviruses/105 cells, including G015) (chi-square test, P < 0.025).
Similar patterns of acquisition were found at N-linked glycosylation sites at positions 417 (100% conversion in 2BS, I049, and F965 but 37% in G015) and 481 (100% in 2BS, I049, and F965 and 88% in G015), although the larger set of animals for which provirus load was known was not evaluated for the presence of these sites. The site at 417 was also acquired by virus within a subset of macaques infected with the pathogenic molecular clone SIVmac239, the parental virus of which also lacked the glycosylation sites at positions 256 and 417 (Table 3) (4). The pattern of all six sites appearing in SIVmacF965 was generally maintained through in vivo passage of this strain (Table 3).
DISCUSSION
The initial objective of this study was to obtain a pathogenic molecular clone of SIVmac to be used for the investigation of SIV pathogenesis. Initial animal inoculations with molecular clones SIVmacBK28 and SIVmacBK44 resulted in long-term asymptomatic survival and no disease at all induced by SIVmacBK44, even after follow-up for up to 7 years. SIVmacBK44 thus appears to embody a truly minimally pathogenic phenotype. Three approaches were used to enhance the pathogenicity of the slightly more pathogenic SIVmacBK28: (i) extension of the env TM coding sequence truncated by passage in a human T-cell line (HuT78), (ii) deliberate introduction of specific mutations designed to augment the LTR enhancer, and (iii) in vivo passage followed by cloning and testing of virus fragments bearing naturally selected mutations.
Extension of the TM may have resulted in some enhancement of pathogenicity. The gene reverts rapidly to an extended form in vivo (21, 28), indicating that the truncated form of the protein results in attenuated virus replication in vivo. Thus, the slightly enhanced pathogenicity of the virus may have been due to a more fulminant replication early in infection, although the latter was not reflected in the detectable production of p27Gag, since none of the animals infected with SIVmacBK28-TM41 became antigenemic.
The introduction of a second NF-κB binding site and replacement of the three sequences resembling SP1 binding sites with four consensus SP1 binding sites in the SIVmacBK28 LTR enhancer simulated the LTR structure of the acute pathogen SIVsmmPBj14 (11). This change increased virus replication early after infection, reflected in the detection of antigenemia, but only marginally enhanced viral pathogenicity in vivo. These results are in agreement with those of other studies indicating that the acute pathogenicity of SIVsmmPBj14 was not due to changes in the LTR alone (41) but rather was due primarily to point mutation changes within the nef gene (13).
Passage of SIVmacBK28 in vivo resulted in the development of a consistent SIV pathogen derived from a human PBMC coculture of bone marrow cells from the first animal to die from this inoculum, 17 months p.i. (isolate SIVmacF965). Passage of the SIVmacF965 virus resulted in 85% mortality within 2 years in a second group of macaques. Unclear at this point was whether viral genetic and antigenic diversity was required for disease induction, as would be predicted by the “Lilliputian” hypothesis or “antigenic threshold” theory for AIDS pathogenesis (35, 42), or whether outgrowth of “virulent variants,” as has been found for immunodeficiency-inducing feline leukemia virus variants (23, 35, 36, 44), was responsible for the increased pathogenicity of the strain. The latter hypothesis was supported by the next finding that, like the uncloned progenitor virus SIVmacF965, a BK28/H824 virus chimera in which the 3′-half genome was derived from an animal infected with the SIVmacF965 strain resulted in the death of 80% of infected macaques with AIDS-like symptoms within 2 years. Thus, the BK28/H824 chimera embodied the essential pathogenic potential of the uncloned SIVmacF965 isolate and localized the major pathogenic determinants to the 3′ half of the viral genome.
The appearance of putative N-linked glycosylation sites in SIV Env suggests that they may contribute to the pathogenic phenotype of H824. However, the prevalence of the N-linked glycosylation sites at positions 116, 173, 256, 417, and 481 in other SIV strains, including the minimally pathogenic strain SIVmac1A11 (34), and the maintenance of these sites upon passage of the SIVmacF965 strain in vivo are also consistent with the hypothesis that they may be natural features of SIV Env. It is possible that SIVmacBK28 lost some of these N-linked glycosylation sites during prolonged passage in a transformed human cell line prior to its molecular cloning (29) and that those sites were later recovered as the virus readapted to replication in vivo. Thus, such changes may be necessary for increased virus replication but not sufficient for enhanced pathogenicity.
One of the changes that occurred during the evolution of the H824 pathogen was also deliberately introduced in parallel experiments designed to increase the pathogenicity of BK28 (i.e., extension of the TM coding sequence in SIVmacBK28-TM41). Some changes also occurred in the enhancer region in vivo, distinct from but within a region modified in a second deliberate construct, SIVmacBK28-2kB-4SP. Whereas neither deliberately introduced change was sufficient to confer the full pathogenic phenotype, at least the TM extension was likely to have contributed, since viruses with only this change were slightly more pathogenic than parental virus BK28. It is likely, therefore, that H824 corresponds to a relatively highly evolved pathogen, with multiple mutations contributing to its more pathogenic phenotype. Similarly, when the roles of individual mutations in the phenotype of the feline leukemia virus-feline AIDS pathogen were dissected, a series of both essential and virulence-enhancing mutations were identified (12).
As in the current study, in vivo passage of primate lentiviruses, especially serial passage, has been found to yield virus isolates with properties distinct from those of the parental viruses. The acutely pathogenic SIVsmmPBj14 isolate was derived following a single in vivo passage of the minimally pathogenic SIVsmm9 (17), and molecular clones of that isolate have been shown to recapitulate the rapidly fatal phenotype (11). Increased neurovirulence was observed following serial transfer of SIVmac239 from bone marrow to brain in macaques (51), and serial intravenous passage of microglia-associated virus following infection of rhesus macaques with uncloned SIVmac251 yielded a virus stock with neuroinvasive and neurovirulent properties (53). Increased pathogenicity was also observed during serial passage of virus from blood to blood following infection of a rhesus macaque with the minimally pathogenic clone SHIV-89.6 (carrying env from a T-cell and macrophage-tropic HIV-1 clone) (47). Similarly, increased virulence was detected following each serial transfer of the minimally pathogenic clone SHIV-HXBc2 from bone marrow to bone marrow in pigtailed macaques (25). However, while in vivo passage has been used on several occasions to obtain more highly pathogenic SIV or simian-human immunodeficiency virus (SHIV) isolates, in few cases have pathogenic molecular clones resulted.
Studies of HIV-1-infected individuals reveal that viruses that appear to be more virulent in vitro (by virtue of their broadened host range and cytopathogenicity associated with fast replication and syncytium formation) are found in about half of individuals progressing to AIDS but not in long-term asymptomatic individuals (50). The fact that syncytium-inducing variants are not found in all individuals progressing to AIDS raises the concern that they arise only following the failure of the immune system rather than being a cause of the failure. Furthermore, the relentless, massive turnover of virus that occurs in HIV-1-infected individuals (22, 45, 54) emphasizes the fact that minor changes in the growth properties of the virus could have a major impact on the delicate balance of virus and the available target cell reservoir (48); indeed, the diversity of the virus population present within an infected individual can be enormous. However, direct analysis of viral quasispecies in HIV-1-infected individuals reveals a correlation between outgrowth of a relatively uniform virus population in quickly progressing individuals and more rapid and greater diversification of the virus population in slowly progressing or nonprogressing individuals (32, 33, 55). The data from the present study provide a precedent that the formation of a diversified lentivirus population, while one of the more insidious properties thought to be crucial to escape from the immune system and fulminant persistence in the host, is not a prerequisite for the development of AIDS.
ACKNOWLEDGMENT
This work was supported by grants from the U.S. Public Health Service.
REFERENCES
- 1.Barry P, Pratt-Lowe E, Luciw P A. Electroporation of T-cell and macrophage cell lines. Methods in electroporation. Richmond, Calif: Bio-Rad Laboratories; 1988. [Google Scholar]
- 2.Botarelli P, Houlden B A, Haigwood N L, Servis C, Montagna D, Abrignani S. N-glycosylation of HIV-gp120 may constrain recognition by T lymphocytes. J Immunol. 1991;147:3128–3132. [PubMed] [Google Scholar]
- 3.Burns D P W. Ph.D. thesis. Cambridge, Mass: Harvard University; 1992. [Google Scholar]
- 4.Burns D P W, Desrosiers R C. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 1991;65:1843–1854. doi: 10.1128/jvi.65.4.1843-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakrabarti L, Emerman M, Tiollais P, Sonigo P. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J Virol. 1989;63:4395–4403. doi: 10.1128/jvi.63.10.4395-4403.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 7.Daniel M D, Letvin N L, Sehgal P K, Hunsmann G, Schmidt D K, King N W, Desrosiers R C. Long-term persistent infection of macaque monkeys with the simian immunodeficiency virus. J Gen Virol. 1987;68:3183–3189. doi: 10.1099/0022-1317-68-12-3183. [DOI] [PubMed] [Google Scholar]
- 8.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, Lawson J S, Cunningham A, Dwyer D, Dowton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 9.Delaporte E, Peeters M, Simon F, Dupont A, Schrijvers D, Kerouedan D, Josse R, Merlin M, Trebucq A, Collet M, Bedjabaga L, Honore C, Larouzé B, Brun-Vézinet F. Interpretation of antibodies reacting solely with human retroviral core proteins in western equatorial Africa. AIDS. 1989;3:179–182. doi: 10.1097/00002030-198903000-00010. [DOI] [PubMed] [Google Scholar]
- 10.Delassus S, Meyerhans A, Cheynier R, Wain-Hobson S. Absence of selection of HIV-1 variants in vivo based on transcription/transactivation during progression to AIDS. Virology. 1992;188:811–818. doi: 10.1016/0042-6822(92)90536-x. [DOI] [PubMed] [Google Scholar]
- 11.Dewhurst S, Embretson J E, Anderson D C, Mullins J I, Fultz P N. Sequence analysis and acute pathogenicity of molecularly cloned SIV(SMM-PBj14) Nature. 1990;345:636–640. doi: 10.1038/345636a0. [DOI] [PubMed] [Google Scholar]
- 12.Donahue P R, Quackenbush S L, Gallo M V, deNoronha C M C, Overbaugh J, Hoover E A, Mullins J I. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-F AIDS. J Virol. 1991;65:4461–4469. doi: 10.1128/jvi.65.8.4461-4469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Z, Lang S M, Sasseville V G, Lackner A A, Ilyinskii P O, Daniel M D, Jung J U, Desrosiers R C. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell. 1995;82:665–674. doi: 10.1016/0092-8674(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 14.Edmonson P F, Mullins J I. Efficient amplification of half-genome sized fragments of human immunodeficiency virus from infected tissue samples. Nucleic Acids Res. 1992;20:4933. doi: 10.1093/nar/20.18.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Edmonson, P. F., and J. I. Mullins. Unpublished data.
- 15.Fenouillet E, Jones I, Powell B, Schmitt D, Kieny M P, Gluckman J C. Functional role of the glycan cluster of the human immunodeficiency virus type 1 transmembrane glycoprotein (gp41) ectodomain. J Virol. 1993;67:150–160. doi: 10.1128/jvi.67.1.150-160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Fields’ virology. 2nd ed. Vol. 2. New York, N.Y: Raven Press; 1990. [Google Scholar]
- 17.Fultz P N, McClure H M, Anderson D C, Switzer W M. Identification and biologic characterization of an acutely lethal variant of simian immunodeficiency virus from sooty mangabeys (SIV/SMM) AIDS Res Hum Retroviruses. 1989;5:397–409. doi: 10.1089/aid.1989.5.397. [DOI] [PubMed] [Google Scholar]
- 18.Haggerty S, Dempsey M, Bukrinsky M I, Gou L, Stevenson M. Posttranslational modifications within the HIV-1 envelope glycoprotein which restrict assembly and CD4-dependent infection. AIDS Res Hum Retroviruses. 1991;7:501–510. doi: 10.1089/aid.1991.7.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch V M. Ph.D. thesis. Cambridge, Mass: Harvard University; 1987. [Google Scholar]
- 21.Hirsch V M, Edmonson P, Murphey-Corb M, Arbielle B, Johnson P R, Mullins J I. SIV adaption to human cells. Nature. 1989;341:572–574. doi: 10.1038/341573a0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 22.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 23.Hoover E A, Mullins J I, Quackenbush S L, Gasper P W. Experimental transmission and pathogenesis of immunodeficiency syndrome in cats. Blood. 1987;70:1880–1892. [PubMed] [Google Scholar]
- 24.Jin M J, Rogers J, Phillips-Conroy J E, Allan J S, Desrosiers R C, Shaw G M, Sharp P M, Hahn B H. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J Virol. 1994;68:8454–8460. doi: 10.1128/jvi.68.12.8454-8460.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joag S V, Li Z, Foresman L, Stephens E B, Zhao L J, Adany I, Pinson D M, McClure H M, Narayan O. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pigtailed macaques. J Virol. 1996;70:3189–3197. doi: 10.1128/jvi.70.5.3189-3197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R C. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 27.Kestler H W I, Ringler D J, Mori K, Panicali D P, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for the development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 28.Kodama T, Wooley D P, Naidu Y M, Kestler H W, Daniel M D, Desrosiers R C. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornfeld H, Riedel N, Viglianti G A, Hirsch V, Mullins J I. Cloning of HTLV-4 and its relation to simian and human immunodeficiency viruses. Nature. 1987;326:610–613. doi: 10.1038/326610a0. [DOI] [PubMed] [Google Scholar]
- 30.Kusumi K, Conway B, Cunningham S, Berson A, Evans C, Iversen A K N, Colvin D, Gallo M V, Coutre S, Shpaer E G, Faulkner D V, deRonde A, Volkman S, Williams C, Hirsch M S, Mullins J I. Human immunodeficiency virus type 1 envelope gene structure and diversity in vivo and after cocultivation in vitro. J Virol. 1992;66:875–885. doi: 10.1128/jvi.66.2.875-885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letvin N L, Daniel M D, Sehgal P K, Desrosiers R C, Hunt R D, Waldron L M, MacKey J J, Schmidt D K, Chalifoux L V, King N W. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science. 1985;230:71–73. doi: 10.1126/science.2412295. [DOI] [PubMed] [Google Scholar]
- 32.Liu S-L, Schacker T, Musey L, McElrath M J, Corey L, Mullins J I. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukashov V V, Kuiken C L, Goudsmit J. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J Virol. 1995;69:6911–6916. doi: 10.1128/jvi.69.11.6911-6916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marthas M L, Sujipto S, Higgins J, Lohman B, Torten J, Luciw P A, Marx P A, Pederson N C. Immunization with a live, attenuated simian immunodeficiency virus (SIV) prevents early disease but not infection in rhesus macaques challenged with pathogenic SIV. J Virol. 1990;64:3694–3700. doi: 10.1128/jvi.64.8.3694-3700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Martin-Amedee A, Lacour N, Martin L N, Clements J E, Bohm R B, Jr, Davison B, Harrison R, Murphey-Corb M. Genotypic analysis of infant macaques infected transplacentally and orally. J Med Primatol. 1996;25:225–235. doi: 10.1111/j.1600-0684.1996.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 35.Mullins J I. Molecular aspects of the FeLV and SIV AIDS models. In: Girard M, Valette L, editors. Retroviruses of human AIDS and related animal diseases. 3. Pasteur Vaccins. 1988. pp. 43–49. , Paris, France. [Google Scholar]
- 36.Mullins J I, Chen C S, Hoover E A. Disease-specific and tissue-specific production of unintegrated feline leukaemia virus variant DNA in feline AIDS. Nature. 1986;319:333–336. doi: 10.1038/319333a0. [DOI] [PubMed] [Google Scholar]
- 37.Myers G, Korber B, Hahn B H, Jeang K-T, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N. Human retroviruses and AIDS 1995. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 38.Naidu Y M, Kestler III H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K S. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton C R, Graham A, Heptinstall L E, Powell S J, Summers C, Kalsheker N, Smith J C, Markham A F. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niphius H, Holterman L, Haaft P T, Dubbes R, Koornstra W, Heeney J L. The rate of progression to AIDS is independent of the dose of the primary inoculum of SIV in rhesus monkeys. J Med Primatol. 1995;4:196–197. [Google Scholar]
- 41.Novembre F J, Johnson P R, Lewis M G, Anderson D C, Klumpp S, McClure H M, Hirsch V M. Multiple viral determinants contribute to pathogenicity of the acutely lethal simian immunodeficiency virus SIVsmmPBj variant. J Virol. 1993;67:2466–2474. doi: 10.1128/jvi.67.5.2466-2474.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nowak M A, Anderson R M, McLean A R, Wolfs T F, Goudsmit J, May R M. Antigenic diversity thresholds and the development of AIDS. Science. 1991;254:963–969. doi: 10.1126/science.1683006. [DOI] [PubMed] [Google Scholar]
- 42a.Ohkawa S, Wilson L A, LaRosa G, Javaherian K, Martin L N, Murphey-Corb M. Immune responses induced by prototype vaccines for AIDS in monkeys. AIDS Res Hum Retroviruses. 1994;10:27–37. doi: 10.1089/aid.1994.10.27. [DOI] [PubMed] [Google Scholar]
- 43.Overbaugh J, Dewhurst S, Mullins J I. Proviral DNA cloning. In: Aldovini A, Walker B D, editors. Techniques in HIV research. New York, N.Y: Stockton Press; 1990. pp. 131–145. [Google Scholar]
- 44.Overbaugh J, Donahue P R, Quackenbush S L, Hoover E A, Mullins J I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988;239:906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- 45.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 46.Popovic M, Sarngadharan M G, Read E, Gallo R C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 47.Reimann K A, Li J T, Voss G, Lekutis C, Tenner-Racz K, Racz P, Lin W, Montefiori D C, Lee-Parritz D E, Lu Y, Collman R G, Sodroski J, Letvin N L. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J Virol. 1996;70:3198–3206. doi: 10.1128/jvi.70.5.3198-3206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodrigo, A. G., and J. I. Mullins. A numerical model for HIV-related CD4+ T-lymphocyte decline and pathogenesis. Submitted for publication.
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma D P, Zink M C, Anderson M, Adams R, Clements J E, Joag S V, Narayan O. Derivation of neurotropic simian immunodeficiency virus from exclusively lymphocytotropic parental virus: pathogenesis of infection in macaques. J Virol. 1992;66:3550–3556. doi: 10.1128/jvi.66.6.3550-3556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sodroski J, Patarca R, Lenz J, Trus M, Crowther R, Perkins D, Gallo R C, Wong-Staal F, Josephs S, Gelmann E P, Haseltine W A. Long terminal repeat regions of murine leukemia virus and human T-cell leukemia virus as potential leukemogenic and pathogenic determinants. In: Gallo R C, Essex M E, Gross L, editors. Human T-cell leukemia/lymphoma virus. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 149–155. [Google Scholar]
- 53.Watry D, Lane T E, Streb M, Fox H S. Transfer of neuropathogenic simian immunodeficiency virus with naturally infected microglia. Am J Pathol. 1995;146:914–923. [PMC free article] [PubMed] [Google Scholar]
- 54.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics of HIV-1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 55.Wolinsky S M, Korber B T M, Neumann A U, Daniels M, Kuntsman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J T, Koup R A. Adaptive evolution of HIV-1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]